
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci., 14 August 2020
Sec. Neurogenomics
Volume 14 - 2020 | https://doi.org/10.3389/fnins.2020.00818
 Jun Gao1,2†
Jun Gao1,2† Fuyi Xu1†
Fuyi Xu1† Athena Starlard-Davenport1
Athena Starlard-Davenport1 Diane B. Miller3
Diane B. Miller3 James P. O’Callaghan3
James P. O’Callaghan3 Byron C. Jones1
Byron C. Jones1 Lu Lu1*
Lu Lu1*Gulf War illness (GWI) is a chronic and multi-symptomatic disorder with persistent neuroimmune symptomatology. Chemokine receptor 6 (CCR6) has been shown to be involved in several inflammation disorders in humans. However, the causative relationship between CCR6 and neuroinflammation in GWI has not yet been investigated. By using RNA-seq data of prefrontal cortex (PFC) from 31 C57BL/6J X DBA/2J (BXD) recombinant inbred (RI) mouse strains and their parental strains under three chemical treatment groups – saline control (CTL), diisopropylfluorophosphate (DFP), and corticosterone combined with diisopropylfluorophosphate (CORT+DFP), we identified Ccr6 as a candidate gene underlying individual differences in susceptibility to GWI. The Ccr6 gene is cis-regulated and its expression is significantly correlated with CORT+DFP treatment. Its mean transcript abundance in PFC of BXD mice decreased 1.6-fold (p < 0.0001) in the CORT+DFP group. The response of Ccr6 to CORT+DFP is also significantly different (p < 0.0001) between the parental strains, suggesting Ccr6 is affected by both host genetic background and chemical treatments. Pearson product-moment correlation analysis revealed 1473 Ccr6-correlated genes (p < 0.05). Enrichment of these genes was seen in the immune, inflammation, cytokine, and neurological related categories. In addition, we also found five central nervous system-related phenotypes and fecal corticosterone concentration have significant correlation (p < 0.05) with expression of Ccr6 in the PFC. We further established a protein-protein interaction subnetwork for the Ccr6-correlated genes, which provides an insight on the interaction of G protein-coupled receptors, kallikrein-kinin system and neuroactive ligand-receptors. This analysis likely defines the heterogeneity and complexity of GWI. Therefore, our results suggest that Ccr6 is one of promising GWI biomarkers.
Gulf War illness (GWI) is the term used to describe a chronic and multi-symptomatic disorder affecting returning military veterans of the 1990–1991 Gulf War (Binns et al., 2014). The symptoms of GWI vary somewhat among individuals and typically include unexplained fatigue, chronic diarrhea musculoskeletal pain, headaches, cognitive dysfunction, rashes and respiratory problems, gastrointestinal, and dermatologic complaints (White et al., 2016; Maule et al., 2018). Although some views ascribed GWI to post-traumatic stress disorder (PTSD) or psychiatric condition to the consequence of wars, accumulated evidence shows that GWI is a neuroimmune disorder resulting from chemical exposures and the physiological stressors incurred in the war theater (O’Callaghan et al., 2015; White et al., 2016). Animal model behavioral data mirror GWI neurobehavioral deficits in terms of impaired memory and cognition, as well as increased anxiety and depressive-like mood (Abdullah et al., 2011; Parihar et al., 2013; Hattiangady et al., 2014; Zakirova et al., 2015; Carreras et al., 2018; Carpenter et al., 2020).
The neurotoxicant exposures encountered by GW military personnel during deployment, including carbamates, organophosphates (OPs), and other pesticides; OP nerve agents (sarin/cyclosarin); and pyridostigmine bromide (PB) (White et al., 2016; Maule et al., 2018). Accumulated neuroimaging studies have demonstrated abnormalities in the brains of veterans with GWI (Binns et al., 2014) including strong evidence for neuroinflammation (Alshelh et al., 2020). Brain pathology of reduced white and gray matter volumes also can be detected nearly two decades later in sarin and cyclosarin-exposed ill GW veterans (Chao et al., 2011). Studies revealed that brain chemistry is abnormal mainly in prefrontal cortex (PFC) and different subregions that mediate various characteristics of the chronic pain, such as sensory and affective dimensions, anxiety and depression (Apkarian et al., 2005). Changes in neurotransmitters, gene expression, glial cells, and neuroinflammation occur in the PFC during acute and chronic pain, which result in alterations to its structure, activity, and connectivity (Ong et al., 2019). Moreover, cortical regions involved in fatigue, pain, and hyperalgesia, also have been reported to be associated with diminished white matter integrity in GW veterans (GWV) (Rayhan et al., 2013). However, heterogeneous symptom presentation and lack of biomarkers in PFC that identify a distinct pathophysiological process in GWI still remain challenging.
Chronic inflammation is a component of the pathophysiology of GWI (Johnson et al., 2016). The sarin surrogate diisopropylfluorophosphate (DFP), an irreversible acetylcholinesterase (AChE) inhibitor, results in brain-wide neuroinflammation that is markedly enhanced in the mouse model by prior exposure to CORT (O’Callaghan et al., 2015; Locker et al., 2017; Koo et al., 2018). High circulating glucocorticoids exaggerates the neuroinflammatory response as measured by the expression of genes for multiple cytokines and chemokines (e.g., Tnf-α, Il6, Ccl2, Il-1β, Lif, and Osm) (O’Callaghan et al., 2015; Jones et al., 2020). Neuroinflammation disorder is induced by chemical exposure and has been linked to cytokine-induced ‘sickness’ behavior of GWI in veterans (Dantzer and Kelley, 2007; Dantzer et al., 2008; O’Callaghan et al., 2015); however, the underlying causes have not been fully elucidated.
Chemokine receptor 6 (CCR6) contributes to steady-state cell chemotaxis in supporting immunity and regulating immune homeostasis during inflammation (Ranasinghe and Eri, 2018). Genetic associations have been identified between CCR6 polymorphisms and immune system disorders in humans including rheumatoid arthritis (RA) and Crohn’s disease (Cheng et al., 2015; Julian et al., 2017). GWI is also characterized by gastrointestinal disorders such as inflammatory bowel disease (IBD) like Crohn’s disease (Ranasinghe and Eri, 2018; Seth et al., 2019). In addition, RA is reported to overlap with specific druggable components of GWI, and some immunosuppressants have been approved by the Food and Drug Administration (FDA) as the best available candidates for treating GWI symptoms (Craddock et al., 2015). However, the causative relation between CCR6 and GWI has not been reported yet.
The C57BL/6J X DBA/2J (BXD) recombinant inbred (RI) mouse strains, which are unique mosaic of alleles derived from the parental C57BL/6J (B6) and DBA/2J (D2) strains have been constructed as a high precision genetic reference population for systems genetics in unraveling the genetic architecture of polygenic traits (Ashbrook et al., 2019). The BXD family consists of more than 150 BXD fully inbred strains that segregate for ∼6 million genetic variants and thus can be used as an informative murine genetic reference panel. The application of the BXD strains provides a unique mouse model to investigate the role of Ccr6 in individual differences to GWI susceptibility.
In this study, we assessed the expression of Ccr6 in the PFC of the GWI BXD model with different chemical treatments. Furthermore, we sought to identify the eQTL for Ccr6, analyze correlated genes and potential pathways, and to construct a protein-protein interaction (PPI) subnetwork that may contribute to individual differences in GWI.
Four hundred-nine mice from 31 BXD strains and their parental strains (B6 and D2) were used in this study. The animals were randomly chosen at 2–4 months of age at testing and 2–3 animals per strain, sex and treatment group were used (Supplementary Data 1). All animals were housed in individually ventilated cage (IVC) system in the Animal Care Facility at the University of Tennessee Health Science Center (UTHSC, Memphis, TN, United States). The vivarium is a temperature (20 ± 2°C) and humidity (35%) controlled environment under a 12 h light/12 h dark cycle. The animals had free access to food and water throughout the experiment. Nine days before the euthanasia, every mouse was single caged, and received corresponding treatment after 2 days adaptation. The euthanasia was carried out in a separate procedure room. All animal procedures were carried out in accordance with the UTHSC guidelines on the humane treatment of experimental animals and with the explicit approval of the Institutional Animal Care and Use Committee (IACUC).
The experimental animals were divided into three treatment groups (Jones et al., 2020) as follows:
(1) Control group (CTL): These strains received plain tap water for fluid (Day 1–7). On the 8th day, the animals were injected with saline (0.9% NaCl) and euthanized by cervical dislocation 6 h after injection.
(2) Diisopropylfluorophosphate group (DFP): These strains received plain tap water for fluid (Day 1–7). On the 8th day, the animals were injected with 4 mg/kg DFP, i.p., 6 h after injection, the animals were euthanized by cervical dislocation followed by decapitation.
(3) Corticosterone + Diisopropylfluorophosphate group (CORT+DFP): These strains received tap water containing 20 mg% CORT dissolved in 0.6% (v/v) EtOH vehicle for 8 days. On the 8th day, the animals were injected with 4mg/kg DFP, i.p., 6 h after injection, the animals were euthanized by cervical dislocation followed by decapitation.
The chemicals DFP (Sigma, St. Louis, MO, United States), CORT (Steraloids, Inc., Newport, RI, United States) and other reagents were analytical grade.
The mice were euthanized by cervical dislocation followed by decapitated that is descripted in our previous publication (Jones et al., 2020) and the whole brain was immediately removed from the skull. The PFC was dissected with a 90° cut 1 mm from the posterior edge of olfactory bulb and another 90° cut 2 mm caudal from the first cut. The PFC was weighed and snap frozen in dry ice bath with isopentane and stored at −80°C until RNA extraction.
Total RNA was extracted from 20 mg frozen PFC tissue per sample using RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The concentration and purity of the RNA was measured using NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States). The RNA integrity (RIN) was assessed using Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, United States). 1 mg qualified RNA (per sample) with OD260/280 > 1.8, OD260/230 > 2.0, and RNA Integrity Number (RIN) >8.0 was used for library preparation and sequencing. The RNA-seq libraries were prepared using the NEBNext® Ultra RNA Library Prep Kit at Novogene Corporation Inc. Paired-end sequencing was performed on an Illumina Novaseq Platform (Illumina, San Diego, CA, United States) by reading 150 bases at each end of a fragment. Overall, each library generated an average of 40 million raw reads.
Raw reads, stored in fastq format, were filtered by removing the adaptor and low-quality reads for further analysis. To generate clean reads, allowing for reads containing over 50% bases with quality greater than 5 and less than 10% “N” bases to be included. The clean reads were then mapped onto the mouse reference genome (version: GRCm38) using the STAR aligner (v2.5.0a) (Dobin et al., 2013). FeatureCount (v0.6.1) (Liao et al., 2014) program was used to get the gene level reads count based on the gene model annotation file downloaded from the Ensembl genome browser1. Raw read count was normalized by DESeq2 R package (v1.22.2) (Love et al., 2014) and batch was added as a covariate for data normalization. Differential expression of Ccr6 was calculated between the three groups (DFP vs. CTL, CORT+DFP vs. CTL, and DFP vs. CORT+DFP) by unpaired t-test.
A two between-subjects variables (strain, treatment) design was used to assess the main effects and interaction on Ccr6 transcript abundance using ANOVA function in R software (R Core Team, 2013). The accepted level of significance for all tests was p < 0.05.
Broad sense heritability (h2) is a concept that summarizes how much of the variance in a quantitative trait is due to variation in genetic factors. It was calculated from the ANOVA results using the following formula (Hegmann and Possidente, 1981): 0.5 VA/(0.5 VA + VE), where VA is the additive genetic variance (variances of the strain means) and VE is the average environmental variance (variance within strains). The factor of 0.5 in this formula was applied to adjust for the 2-fold increase in the additive genetic variance among the inbred strains relative to outbred populations (Lu et al., 2018).
eQTL mapping is a regression analysis to determine the relationship between differences in a trait and differences in alleles at markers across the genome. The eQTL mapping of Ccr6 in three groups (CTL, DFP, and CORT+DFP) were conducted through the WebQTL module on GeneNetwork website2 according to the published methods (Mulligan et al., 2017; Williams and Williams, 2017). The input expression values of Ccr6 was normalized with TPM (transcripts per million) method (Wagner et al., 2012; Vera Alvarez et al., 2019) and log2 (TPM + 1) transformed. Simple interval mapping yielded a likelihood ratio statistic (LRS) score, providing us a quantitative measure of confidence of linkage between the observed phenotype and a genomic region. The genome-wide significance (p < 0.05) for each eQTL was determined with 1000 permutation tests.
Single nucleotide polymorphisms (SNPs) and insertion-deletions (InDels) in the Ccr6 gene and its surrounding up- and down-stream regions between the B6 and D2 were extracted from the Mouse Genome Project database3 (Keane et al., 2011; Yalcin et al., 2011).
To assess the relationship between the expression of Ccr6 and related traits across the BXD cohort, we queried the BXD archival phenotypes from the GeneNetwork and analyzed for Pearson product-moment correlation to the expression of Ccr6 in PFC. The top 500 Pearson product-moment correlations were filtered and p < 0.05 were considered significant.
In order to identify the Ccr6 correlated genes across the PFC transcriptomes in the treatment groups, we conducted Pearson product-moment correlations of the strain means between the expression of Ccr6 and the expression of all the other genes across the mouse genome to produce sets of genetically correlated genes on GeneNetwork. Genes significantly correlated with expression of Ccr6 (p < 0.05) were used for the gene set enrichment analysis, in which, Riken cDNA clones, intragenic sequences, and predicted genes were eliminated.
Gene set enrichment analysis was performed to investigate the gene ontology (GO, biological processes) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of the Ccr6 correlated genes. We submitted the gene set of each treatment group to the Webgestalt website4 (Liao et al., 2019) for analysis. The p-value generated from the test was automatically adjusted to account for multiple comparisons using the Benjamini and Hochberg correction (Benjamini and Hochberg, 1995). A minimum overlap of five genes and False Discovery Rate (FDR) < 0.05 was required to determine the genes significantly overrepresented in those categories.
PPI analysis of the Ccr6 correlated genes was based on the online STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (Szklarczyk et al., 2019), which contains known and predicted PPIs information by consolidating known and predicted protein-protein association data for a large number of organisms (Szklarczyk et al., 2016). In this study, we first constructed the PPI network by extracting the target gene lists from the database with required the highest score of confidence interaction of 0.9. Then Markov Cluster Algorithm (MCL) clustering was used for subnetwork construction, in which the inflation parameter used a default setting 3. The narrowed subnetwork genes were further used for GO and KEGG analysis to gain insight into the biological functions and pathways of Ccr6 correlated genes.
In this study, a total of 409 mice were used for PFC harvest and expression profiling across the treatments. Overall, the expression of Ccr6 in CORT+DFP group showed significant decrease in most of the BXD strains (Figure 1A). However, this effect was not consistent for DFP treatment, in which Ccr6 mRNA levels decreased in 18 strains (e.g., BXD29, BXD83, BXD65), but increased in the rest of 15 strains (e.g., BXD66, D2, BXD48). This finding further supports the assertion of O’Callaghan et al. (2015) that exposure to OPs plus high circulating glucocorticoids may be an essential condition for GWI. Next, we compared the expression of Ccr6 between the different treatment groups. Results showed that Ccr6 significantly decreased in the CORT+DFP group when compared with CTL group (Fold change = 1.60, p < 0.0001) and DFP group (Fold change = 1.42, p < 0.05) (Figure 1B), respectively.
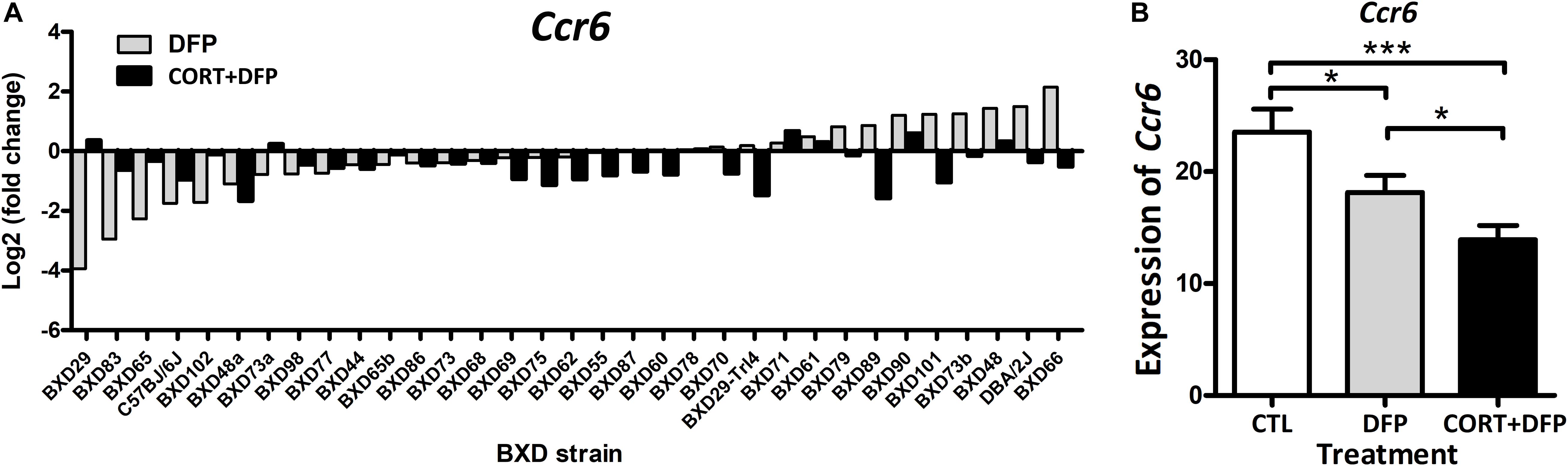
Figure 1. The expression of Ccr6 across the BXD strains. (A) The relative fold change of Ccr6 expression in the DFP and CORT+DFP groups compared to the CTL across the BXD RI strains. (B) Comparison of the expression (Mean ± SEM) of Ccr6 between the treatments (CTL, DFP, and CORT+DFP) by unpaired t-test. *p < 0.05, ***p < 0.0001. The Ccr6 expression is a normalized value by the DEseq2.
In order to determine the effects of strain and treatment on the expression of Ccr6 further, we conducted two way ANOVA which showed both factors have significant effects on the Ccr6 expression [Treatment: F(2, 310) = 22.20, p < 10E-10; Strain: F(32, 310) = 18.15, p < 3E-16]. The strain × treatment interaction was also significant [F(64, 310) = 2.40, p < 4.0E-7]. In addition, we calculated the heritability (h2) for each treatment group with h2 = 0.29 for CTL, 0.27 for DFP and 0.22 for CORT+DFP, suggesting both genetic and environmental factors contribute to the expression differences of Ccr6 among the BXD strains.
Ccr6 is located on chromosome 17 at 8.236 Mb of mice. Interval mapping indicated a genome-wide significant eQTL with a LRS of 53.5 in CTL (Figure 2A), 34.6 in DFP (Figure 2B), and 26 in CORT+DFP (Figure 2C) on chromosome 17 at 7.713 Mb. This locus is located 0.5 megabases (Mb) upstream of Ccr6, indicating that Ccr6 is cis-regulated in the PFC for all three groups (Figure 2). Next, we grouped the mice according to their genotype (B and D type) at the QTL peak position (rs48543649, Chr 17 at 8.199 Mb) which is near the physical position of Ccr6. Statistical analysis revealed that the mRNA levels of Ccr6 showed a significant difference (p < 0.0001) between B and D alleles in all three treatment groups (Figures 2D–F), with mice carrying the D allele evincing higher expression level of Ccr6.
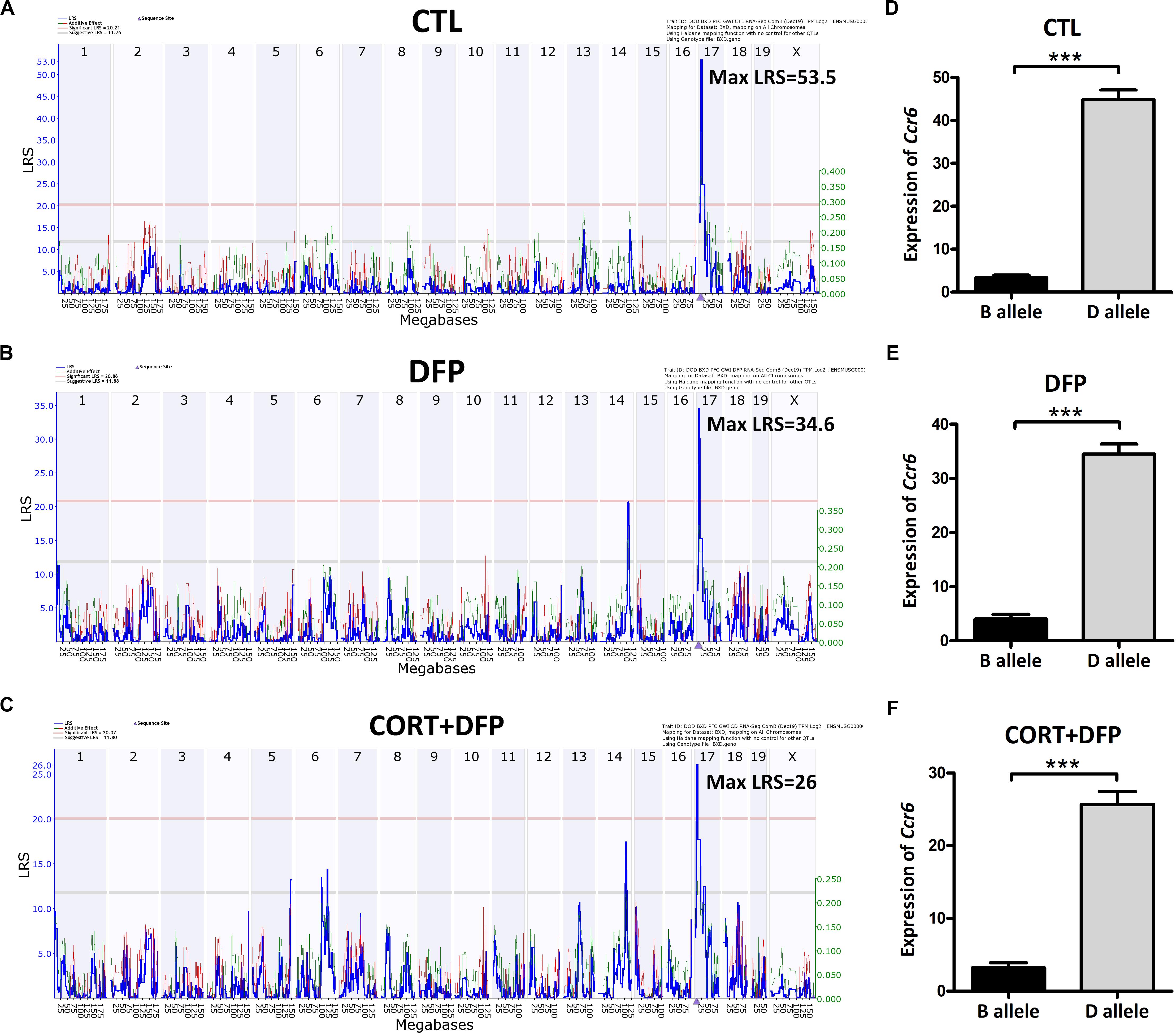
Figure 2. eQTLs mapping of Ccr6. eQTLs mapping demonstrates Ccr6 is cis regulated in the prefrontal cortex mRNA. (A) CTL (Max likelihood ratio statistic score (LRS) = 53.5), (B) DFP (Max LRS = 34.6), (C) CORT+DFP (Max LRS = 26.0). Chromosome number can be found across the top of the plot with megabases (Mb) on the x-axis. The y-axis contains the LRS. The location of Ccr6 is marked by an arrowhead found on the x-axis is at Chr 17 at 8.236 Mb. (D–F) The expression (Mean ± SEM) of Ccr6 is significantly different between B and D allele by unpaired t-test, ***p < 0.0001. The Ccr6 expression is a normalized value by the DEseq2.
Ccr6 is cis regulated, which means that sequence variants within or nearby Ccr6 likely affect its expression. Therefore, we explored nearby genetic variants using the database of Mouse genome project6. We identified 31 SNPs and 3 InDels (Table 1) between the parental strains B6 and D2, in which one is a synonymous variant (rs49056705), three are 5ˊ UTR variants (rs33886456, rs33640330, and rs33573638), and the rest of them are located within 5000 bp upstream of Ccr6. We also identified one trans-eQTL achieved statistical significance in DFP group, which located on Chr14 at 100–110 Mb. This eQTL interval contains 90 genes, of which, Kctd12 and Mycbp2 correlated with Ccr6 (p < 0.05), Slain1 and Ednrb harbor nonsynonymous mutations, suggesting they could be upstream candidate regulators.
To our knowledge, GWI is a chronic disease with significant neurological pathophysiology. In our mouse model, exposure to CORT+DFP treatment increased expression of proinflammatory cytokine genes, which is consistent with the neuroimmune basis of GWI (O’Callaghan et al., 2015). Additionally, our results show that CORT+DFP has the greatest effect on the expression of Ccr6. The question then becomes does CORT+DFP-related expression of Ccr6 associate with other central nervous system (CNS) phenotypes? After multiple testing correction (FDR < 0.05), we obtained a total of 117 CNS related phenotypes that were significantly correlated with the expression of Ccr6 in the CORT+DFP group (P < 0.05) (Supplementary Data 2). We listed five CNS-related phenotypes in BXD RI strains, including brain to body weight ratio (Figure 3A), novel open field behavior (Figure 3B), anxiety assay (Figure 3C), acoustic startle response (Figure 3D), learning and memory (Figure 3E), as well as fecal corticosterone concentration (Figure 3F) significantly correlated (p < 0.05) with the expression of Ccr6. These correlated phenotypes can be found on the GeneNetwork website with the access numbers of 17494, 11530, 12365, 13355, 20585, and 20113, respectively.
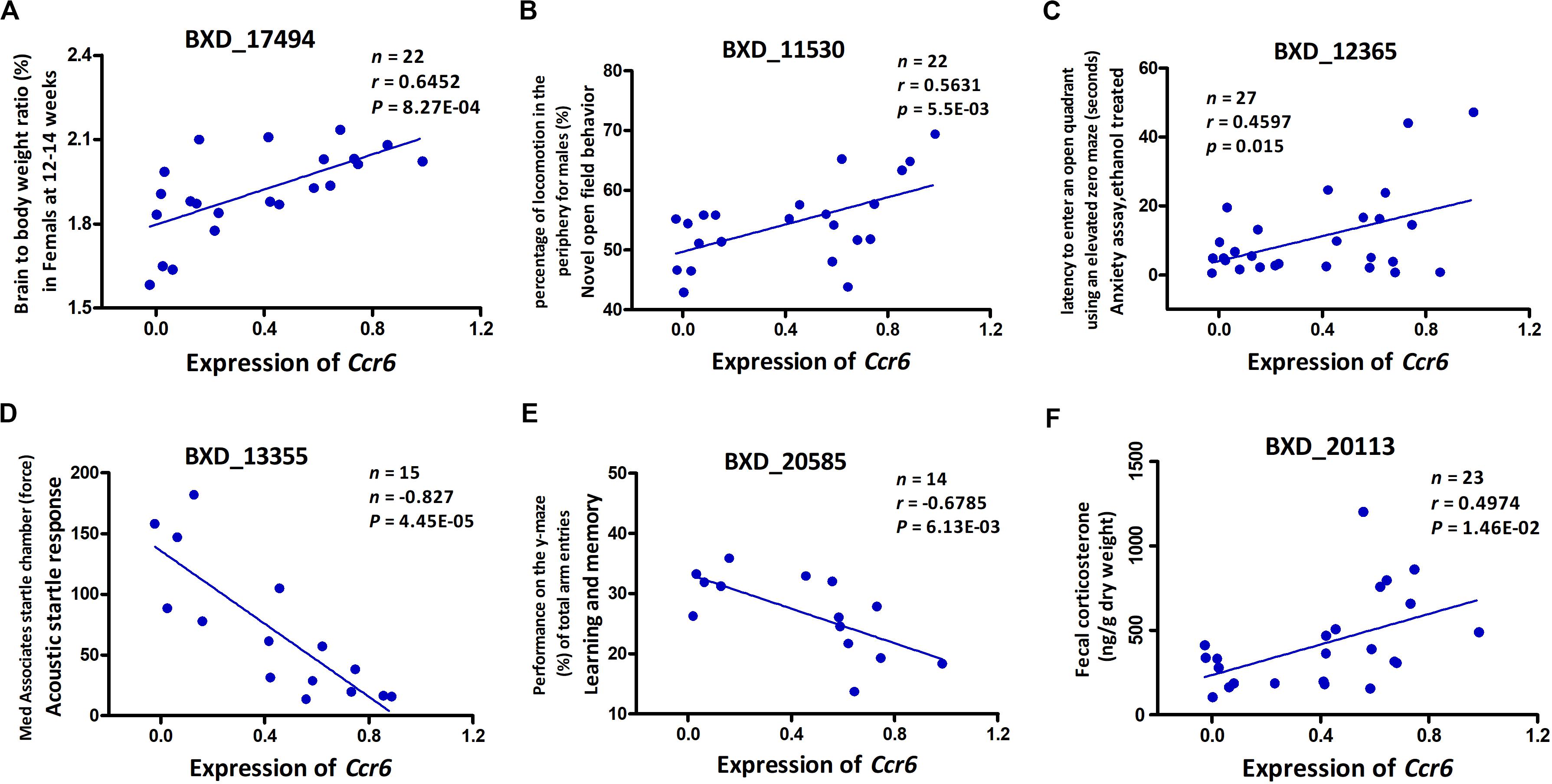
Figure 3. Phenotype Pearson correlation analysis with the expression of Ccr6. Five BXD central nervous system (CNS) related phenotypes including brain to body weight ratio (A), novel open field behavior (B), anxiety assay (C), acoustic startle response (D), and learning and memory (E) were significantly correlated with the expression of Ccr6 in PFC. The fecal CORT content was positively correlated with the expression of Ccr6 (F). GeneNetwork BXD phenotype identifiers (e.g., ID “BXD_17494”) are at the top of each plot. n, Number of strains, p, p-value.
To understand the biological processes and gene pathways of Ccr6 correlated genes, we performed correlation analysis and identified 1755, 7193, and 3996 genes that are significantly correlated (p < 0.05) with Ccr6 in the CTL, DFP, and CORT+DFP group, respectively. After removing Riken cDNA clones, intragenic sequences, predicted genes, 806 (CTL), 3983 (DFP), and 1473 (CORT+DFP) genes were separately submitted to Webgestalt web site7 for gene function enrichment analysis.
The gene set enrichment results (Supplementary Data 3) showed Ccr6 correlated genes were significantly enriched in immune and inflammation-related GO terms in the CTL group. For the DFP group, the enrichment results demonstrate a high degree of neurological association with the Ccr6 correlated genes. For the CORT+DFP group, we obtained a total of 47 significantly enriched GO terms (FDR < 0.05) and 23 KEGG pathways (FDR < 0.05). The top 20 GO and KEGG categories are listed in Figure 4. Of which, immune, inflammation, and cytokine terms were further highlighted.
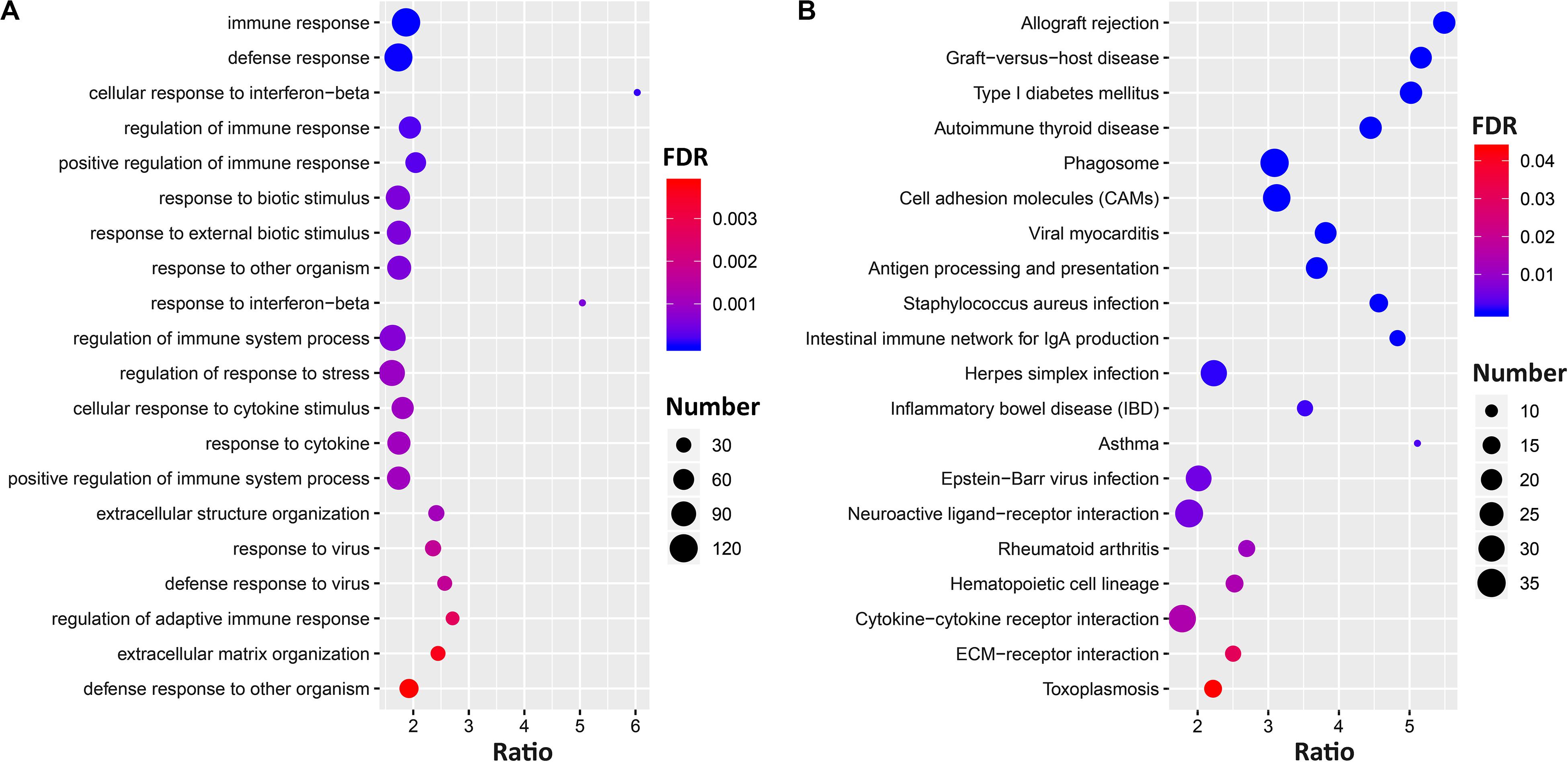
Figure 4. The gene set enrichment analysis of Ccr6 correlated genes. The bubble plot shows the top 20 GO (A) and KEGG pathways (B) of Ccr6 correlated genes in the CORT+DFP group.
To further dissect the potential interactions of the Ccr6 correlated genes in the CORT+DFP group, we uploaded the above 1473 genes correlated to Ccr6 into STRING5 to search for PPI. By performing MCL clustering, we identified a Ccr6 PPI subnetwork, which includes 38 genes (Figure 5). These genes are highly inter-connected (interaction score ≥ 0.9).
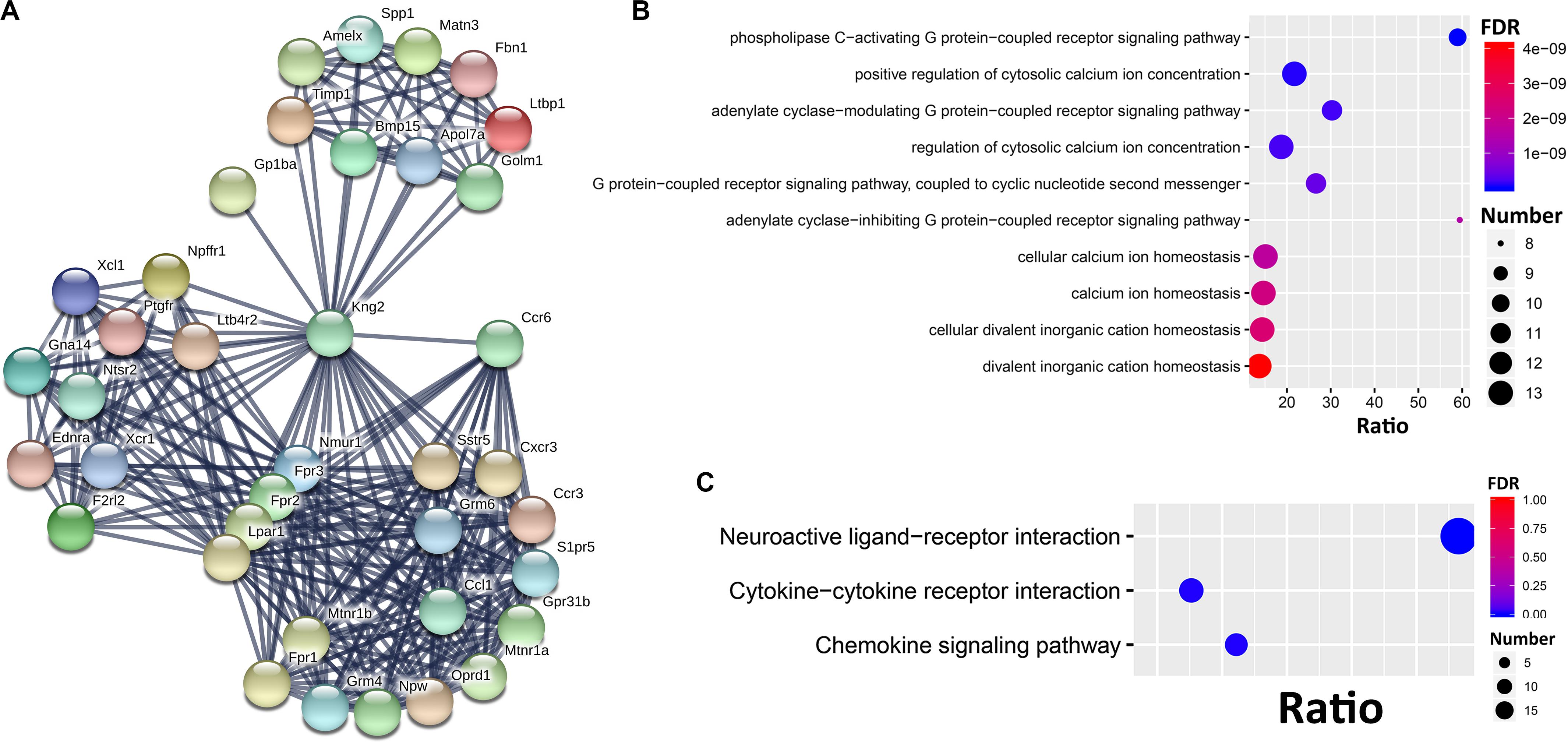
Figure 5. PPI subnetwork of Ccr6 correlated genes. (A) Ccr6 PPI subnetwork includes 38 genes with the interaction score ≥0.9. The nodes represent genes while edges represent PPIs between two genes. Bubble plot of the Top 10 GO (B) and 3 KEGG (C) enrichment categories for the 38 PPI subnetwork genes.
Next, we performed GO and KEGG enrichment analysis for these 38 genes. The top 10 significant GO terms (FDR < 0.05) showed that those genes are mainly involved in G protein-coupled receptor signaling pathways (Oprd1, Ednra, Nmur1, Ntsr2, Fpr1, F2rl2, Lpar1, Fpr3, Gna14, Fpr2, Grm4, Sstr5, Grm6, Ptgfr, Cxcr3, and Mtnr1a) and cytosolic calcium ion concentration (Xcl1, Ednra, Fpr1, Ccl1, Ccr3, Ccr6, F2rl2, Lpar1, Ptgfr, Fpr3, Kng2, and Fpr2) (Figure 5B). The top 10 KEGG pathways (Figure 5C) had 3 achieve significance (FDR < 0.05), including one neuroactive ligand-receptor interaction pathway (Ltb4r2, Oprd1, Npffr1, S1pr5, Ednra, Grm4, Nmur1, Sstr5, Ntsr2, Fpr1, F2rl2, Grm6, Lpar1, Ptgfr, Mtnr1b, Fpr3, Mtnr1a, and Fpr2), and two cytokine-related pathways (Xcl1, Ccl1, Ccr3, Ccr6, Bmp15, Cxcr3, and Xcr1).
Compared to nondeployed veterans, at least one fourth of the 697,000 U.S. veterans suffered from GWI when they returned from the theater of operations (Binns et al., 2014). These veterans also reported higher rates of amyotrophic lateral sclerosis (ALS) (Coffman et al., 2005; Horner et al., 2008), brain cancer (Barth et al., 2009), repeated seizures, neuralgia or neuritis, stroke (Kang et al., 2009), and migraine headaches (Unwin et al., 1999; Kang et al., 2000; Steele, 2000; Gray et al., 2002). Accumulating Studies clearly supports the links between adverse neurological outcomes and chemical exposures of GWV (Binns et al., 2014; White et al., 2016). Deployed GWV had significantly lower scores on tests of verbal memory, verbal learning, motor speed, and attention than nondeployed due to the pesticides and PB exposures (Toomey et al., 2009). Sarin/cyclosarin exposed GWV showed signs of reduced total gray and white matter volumes in the brain compared to unexposed controls and worse on a continuous performance test of attention (Chao et al., 2011).
Accurate diagnosis and treatment of GWI patients require an in-depth understanding of the cause of the disease. Although some of the individual differences in susceptibility to GWI may be explained by different exposures or different dose effects, much of it cannot be, and leaves genetics as a significant contributor to individual differences in susceptibility and response to the exposures. The BXD mouse strains put the investigator at great advantage for systems genetics analysis of complex traits such as GWI and those traits that have modest heritability (Williams et al., 2001). In this study, we used BXD strains to explore the etiologic agents and pathways that underlie the “sickness” behavior of GWI. Indeed overall, we have supported the notion that this disease is the result of genetic–environment interaction.
Our results indicate Ccr6 as one reasonable candidate gene that underlies individual differences in susceptibility to GWI. In addition, we also found five CNS-related phenotypes that show the wide-ranging effects of GWI. Furthermore, we identified 31 SNPs and 3 InDels that differ in response to CORT+DFP between B6 and D2 inbred strains. CCR6 may turn out to be a target of therapeutic approaches to GWI.
The results of gene set enrichment analysis highlighted the categories of Ccr6 correlated genes related to immune, inflammation, cytokine, and neurological aspects. Accumulating evidence suggest that Ccr6 plays a major role in driving T-helper differentiation in inflammatory diseases and maintaining leukocyte homeostasis (Ranasinghe and Eri, 2018). Ccr6 regulates the migration of inflammatory and regulatory T cells (Th17 and Treg), which play opposite roles in autoimmune diseases (Yamazaki et al., 2008). Although Ccr6-mediated Th17 migration to inflamed tissues may be important for driving CNS inflammation, Ccr6 expression is deemed to be more critical to Treg cells than to Th17 cells, because this subset suppresses inflammatory T cell proliferation and promotes disease resolution (Ranasinghe and Eri, 2018). The use of Ccr6–/– mice in experimental autoimmune encephalomyelitis (EAE) study, an animal model of brain inflammation for the study of human CNS diseases characterized by mononuclear cell infiltration and demyelination, showed delayed disease onset and more neurological damage and increased mortality compared to wild-type mice (Villares et al., 2009). Severe phenotype in Ccr6–/– EAE mice is linked to increased inflammatory activity in target tissues. This suggests that Ccr6 is necessary for Treg recruitment and initiates a feedback anti-inflammatory mechanism which compensatorily downregulates the CNS inflammatory activity (Yamazaki et al., 2008; Villares et al., 2009). Although the immune regulation mechanism of CCR6 has not been fully elucidated, the CCR6/CCL20 axis is an important chemokine receptor-ligand and may present a therapeutic target for the treatment of human disorders (Ranasinghe and Eri, 2018).
As we understand, GWI is a complex trait with underlying gene-environment and likely gene-gene interactions. Indeed, we identified a Ccr6 PPI subnetwork that includes 38 genes enriched in G protein-coupled receptor (GPCR) signaling pathways and cytosolic calcium ion concentration signaling pathway. Astrocytes and microglia are the most prominent target cells for inflammation in the CNS. Their responses upon activation include downregulation of ATP-induced Ca2+ signaling, G protein activities and release of pro-inflammatory cytokines (Hansson et al., 2018). Many druggable targets for treatment of common diseases involve GPCRs that mediate therapeutic effects of ∼34% of the marketed drugs (Hauser et al., 2018). Further understanding of genetic factors and regulation networks of Ccr6 is likely to advance drug treatment of GWI in the future.
Another gene worth noting is Kng2, that appears as a hub in the Ccr6 PPI subnetwork, implicating that the Kallikrein-kinin system (KKS) mediate the pathophysiological features of neurological disorders, including GWI. Despite a paucity of literature on Kng2, pharmacological research in mice and human genetic analyses suggest that the KKS may regulate anxiety (Nokkari et al., 2018; Rouhiainen et al., 2019). On the other hand, Ccr6 correlated genes in PFC were observed to be significantly enriched in neuroactive ligand-receptor interaction pathways, of which, three formylpeptide receptors (Fpr1, Fpr2, Fpr3) are critical mediators of myeloid cell trafficking in microbial infection, inflammation, and immune responses (Krepel and Wang, 2019). Fpr2 also proved to mediate anxiety as shown by effects reported for Fpr agonists (Zhao et al., 2016). Although the role of the Ccr6 PPI subnetwork genes and their contribution to the neuroinflammation of GWI are not fully elucidated, it provides a new insight to the complexity of GWI.
A suitable animal model of GWI should show evidence of the illness acutely and have it persist to model the entire 30-year course of the symptoms exhibited by ill veterans. The data in the present manuscript models the acute condition. We also have an extension of this model that represents the chronic “primed” inflammatory condition (manuscript in internal review). The chronic model is based on the paradigm of systemic challenge with lipopolysaccharide (LPS) (Kelly et al., 2018). It is important to subsequently investigate the changes of Ccr6 mRNA levels at later time points in the chronic model to verify the possibility that CCR6 can be a marker for GWI or similar ailment. Given the complexity and heterogeneity of GWI, we cannot exclude the existence of other regulatory elements. For example, genes such as Tnf-α, Il6, Il1β, and Spon1 may also be involved in the neuroinflammatory response of GWI (Jones et al., 2020).
In this study, we identified Ccr6 involvement in the neuroimmune response to CORT+DFP treatment in the BXD mouse model of GWI. Genetic factors and treatments both impact on the expression of Ccr6 in PFC, which may contribute to CORT+DFP neuroinflammation in BXD strains. In humans, CCR6-mediates the migration of inflammatory and regulatory T cells and regulates CNS inflammation, which indicates it may be a promising therapeutic target of GWI. Our study also suggests the polymorphisms of Ccr6 and synergy interaction of the related GPCRs, KKS system, and neuroactive ligand-receptor may contribute to the heterogeneity and complexity of GWI and related sickness behaviors.
The datasets generated for this study can be found in the online repositories. The names of the repository/repositories and accession number(s) can be found below: www.genenetwork.org, GN880 (http://gn1.genenetwork.org/webqtl / main. py ? FormID = sharinginfo &GN _AccessionId = 880 ), GN881 (http://gn1.genenetwork.org/webqtl/main.py?FormID= sharinginfo&GN_AccessionId=881), and GN882 (http://gn1. genenetwork . org /w ebqtl / main . py ? FormID = sharinginfo & GN _AccessionId=882).
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC).
LL, BJ, DM, and JO’C conceived the study and oversaw the execution of the experimental work. JG and FX performed data analysis and prepared the figures and tables. LL, JG, and FX wrote the manuscript. BJ, LL, JO’C, and AS-D edited the manuscript. All authors read and approved the final version of the manuscript.
This research was supported in part by the CDMRP Grant W81XWH-17-10472.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention. This work was supported by the Assistant Secretary of Defense for Health Affairs, through the Gulf War Illness Research Program. Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2020.00818/full#supplementary-material
DATA S1 | Sample information.
DATA S2 | Ccr6 correlated phenotypes in BXD mice.
DATA S3 | Enrichment results of Ccr6 correlated genes.
Abdullah, L., Crynen, G., Reed, J., Bishop, A., Phillips, J., Ferguson, S., et al. (2011). Proteomic CNS profile of delayed cognitive impairment in mice exposed to Gulf War agents. Neuromol. Med. 13, 275–288. doi: 10.1007/s12017-011-8160-z
Alshelh, Z., Albrecht, D. S., Bergan, C., Akeju, O., Clauw, D. J., Conboy, L., et al. (2020). In-vivo imaging of neuroinflammation in veterans with Gulf War illness. Brain Behav. Immun. 87, 498–507. doi: 10.1016/j.bbi.2020.01.020
Apkarian, A. V., Bushnell, M. C., Treede, R.-D., and Zubieta, J.-K. (2005). Human brain mechanisms of pain perception and regulation in health and disease. Eur. J. Pain 9, 463–484.
Ashbrook, D. G., Arends, D., Prins, P., Mulligan, M. K., Roy, S., Williams, E. G., et al. (2019). The expanded BXD family of mice: a cohort for experimental systems genetics and precision medicine. BioRxiv [Preprint] doi: 10.1101/672097
Barth, S. K., Kang, H. K., Bullman, T. A., and Wallin, M. T. (2009). Neurological mortality among US veterans of the Persian Gulf War: 13-year follow-up. Am. J. Ind. Med. 52, 663–670. doi: 10.1002/ajim.20718
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Binns, J. H., Bloom, F. E., Bunker, J. A., Crawford, F., Golomb, B. A., Graves, J. C., et al. (2014). Gulf War illness and the health of Gulf War veterans: Research update and recommendations, 2009-2013. Boston, MA: US Government Printing Office.
Carpenter, J., Gordon, H., Ludwig, H., Wagner, J., Harn, D., Norberg, T., et al. (2020). Neurochemical and neuroinflammatory perturbations in two Gulf War Illness models: modulation by the immunotherapeutic LNFPIII. Neurotoxicology 77, 40–50. doi: 10.1016/j.neuro.2019.12.012
Carreras, I., Aytan, N., Mellott, T., Choi, J.-K., Lehar, M., Crabtree, L., et al. (2018). Anxiety, neuroinflammation, cholinergic and GABAergic abnormalities are early markers of Gulf War illness in a mouse model of the disease. Brain Res. 1681, 34–43. doi: 10.1016/j.brainres.2017.12.030
Chao, L. L., Abadjian, L., Hlavin, J., Meyerhoff, D. J., and Weiner, M. W. (2011). Effects of low-level sarin and cyclosarin exposure and Gulf War Illness on brain structure and function: a study at 4 T. Neurotoxicology 32, 814–822. doi: 10.1016/j.neuro.2011.06.006
Cheng, P., Zhang, Y., Huang, H., Zhang, W., Yang, Q., Guo, F., et al. (2015). Association between CCR6 and rheumatoid arthritis: a meta-analysis. Int. J. Clin. Exp. Med. 8, 5388–5396.
Coffman, C. J., Horner, R. D., Grambow, S. C., and Lindquist, J. (2005). Estimating the occurrence of amyotrophic lateral sclerosis among Gulf War (1990–1991) veterans using capture-recapture methods. Neuroepidemiology 24, 141–150. doi: 10.1159/000083297
Craddock, T. J., Harvey, J. M., Nathanson, L., Barnes, Z. M., Klimas, N. G., Fletcher, M. A., et al. (2015). Using gene expression signatures to identify novel treatment strategies in gulf war illness. BMC Med. Genomics 8:36. doi: 10.1186/s12920-015-0111-3
Dantzer, R., and Kelley, K. W. (2007). Twenty years of research on cytokine-induced sickness behavior. Brain Behav. Immun. 21, 153–160. doi: 10.1016/j.bbi.2006.09.006
Dantzer, R., O’connor, J. C., Freund, G. G., Johnson, R. W., and Kelley, K. W. (2008). From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 9, 46–56. doi: 10.1038/nrn2297
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Gray, G. C., Reed, R. J., Kaiser, K. S., Smith, T. C., and Gastañaga, V. M. (2002). Self-reported symptoms and medical conditions among 11,868 Gulf War-era veterans: the Seabee Health Study. Am. J. Epidemiol. 155, 1033–1044. doi: 10.1093/aje/155.11.1033
Hansson, E., Björklund, U., Skiöldebrand, E., and Rönnbäck, L. (2018). Anti-inflammatory effects induced by pharmaceutical substances on inflammatory active brain astrocytes—promising treatment of neuroinflammation. J. Neuroinflammation 15:321.
Hattiangady, B., Mishra, V., Kodali, M., Shuai, B., Rao, X., and Shetty, A. K. (2014). Object location and object recognition memory impairments, motivation deficits and depression in a model of Gulf War illness. Front. Behav. Neurosci. 8:78. doi: 10.3389/fnbeh.2014.00078
Hauser, A. S., Chavali, S., Masuho, I., Jahn, L. J., Martemyanov, K. A., Gloriam, D. E., et al. (2018). Pharmacogenomics of GPCR drug targets. Cell 172, 41–54.e19. doi: 10.1016/j.cell.2017.11.033
Hegmann, J. P., and Possidente, B. (1981). Estimating genetic correlations from inbred strains. Behav. Genet. 11, 103–114. doi: 10.1007/bf01065621
Horner, R. D., Grambow, S. C., Coffman, C. J., Lindquist, J. H., Oddone, E. Z., Allen, K. D., et al. (2008). Amyotrophic lateral sclerosis among 1991 Gulf War veterans: evidence for a time-limited outbreak. Neuroepidemiology 31, 28–32. doi: 10.1159/000136648
Johnson, G. J., Slater, B. C., Leis, L. A., Rector, T. S., and Bach, R. R. (2016). Blood biomarkers of chronic inflammation in Gulf War Illness. PLoS One 11:e0157855. doi: 10.1371/journal.pone.0157855
Jones, B. C., Miller, D. B., Lu, L., Zhao, W., Ashbrook, D. G., Xu, F., et al. (2020). Modeling the genetic basis of individual differences in susceptibility to Gulf War Illness. Brain Sci. 10:143. doi: 10.3390/brainsci10030143
Julian, B., Gao, K., Harwood, B. N., Beinborn, M., and Kopin, A. S. (2017). Mutation-induced functional alterations of CCR6. J. Pharmacol. Exp. Ther. 360, 106–116. doi: 10.1124/jpet.116.237669
Kang, H. K., Li, B., Mahan, C. M., Eisen, S. A., and Engel, C. C. (2009). Health of US veterans of 1991 Gulf War: a follow-up survey in 10 years. J. Occup. Environ. Med. 51, 401–410. doi: 10.1097/jom.0b013e3181a2feeb
Kang, H. K., Mahan, C. M., Lee, K. Y., Magee, C. A., and Murphy, F. M. (2000). Illnesses among united states veterans of the gulf war:: a population-based survey of 30,000 veterans. J. Occup. Environ. Med. 42, 491–501. doi: 10.1097/00043764-200005000-00006
Keane, T. M., Goodstadt, L., Danecek, P., White, M. A., Wong, K., Yalcin, B., et al. (2011). Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294.
Kelly, K. A., Michalovicz, L. T., Miller, J. V., Castranova, V., Miller, D. B., and O’Callaghan, J. P. (2018). Prior exposure to corticosterone markedly enhances and prolongs the neuroinflammatory response to systemic challenge with LPS. PLoS One 13:e0190546. doi: 10.1371/journal.pone.0190546
Koo, B. B., Michalovicz, L. T., Calderazzo, S., Kelly, K. A., Sullivan, K., Killiany, R. J., et al. (2018). Corticosterone potentiates DFP-induced neuroinflammation and affects high-order diffusion imaging in a rat model of Gulf War Illness. Brain Behav. Immun. 67, 42–46. doi: 10.1016/j.bbi.2017.08.003
Krepel, S. A., and Wang, J. M. (2019). Chemotactic ligands that activate G-protein-coupled formylpeptide receptors. Int. J. Mol. Sci. 20:3426. doi: 10.3390/ijms20143426
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z., and Zhang, B. (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205.
Locker, A. R., Michalovicz, L. T., Kelly, K. A., Miller, J. V., Miller, D. B., and O’Callaghan, J. P. (2017). Corticosterone primes the neuroinflammatory response to Gulf War Illness-relevant organophosphates independently of acetylcholinesterase inhibition. J. Neurochem. 142, 444–455. doi: 10.1111/jnc.14071
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Lu, Y., Zhou, D., King, R., Zhu, S., Simpson, C. L., Jones, B. C., et al. (2018). The genetic dissection of Myo7a gene expression in the retinas of BXD mice. Mol. Vis. 24, 115–126.
Maule, A. L., Janulewicz, P. A., Sullivan, K. A., Krengel, M. H., Yee, M. K., Mcclean, M., et al. (2018). Meta-analysis of self-reported health symptoms in 1990–1991 Gulf War and Gulf War-era veterans. BMJ Open 8:e016086. doi: 10.1136/bmjopen-2017-016086
Mulligan, M. K., Mozhui, K., Prins, P., and Williams, R. W. (2017). “GeneNetwork: a toolbox for systems genetics,” in Systems Genetics, eds K. Schughart and R. Williams (Berlin: Springer), 75–120. doi: 10.1007/978-1-4939-6427-7_4
Nokkari, A., Abou-El-Hassan, H., Mechref, Y., Mondello, S., Kindy, M. S., Jaffa, A. A., et al. (2018). Implication of the Kallikrein-Kinin system in neurological disorders: quest for potential biomarkers and mechanisms. Prog. Neurobiol. 165, 26–50. doi: 10.1016/j.pneurobio.2018.01.003
O’Callaghan, J. P., Kelly, K. A., Locker, A. R., Miller, D. B., and Lasley, S. M. (2015). Corticosterone primes the neuroinflammatory response to DFP in mice: potential animal model of Gulf War Illness. J. Neurochem. 133, 708–721. doi: 10.1111/jnc.13088
Ong, W.-Y., Stohler, C. S., and Herr, D. R. (2019). Role of the prefrontal cortex in pain processing. Mol. Neurobiol. 56, 1137–1166. doi: 10.1007/s12035-018-1130-9
Parihar, V. K., Hattiangady, B., Shuai, B., and Shetty, A. K. (2013). Mood and memory deficits in a model of Gulf War illness are linked with reduced neurogenesis, partial neuron loss, and mild inflammation in the hippocampus. Neuropsychopharmacology 38, 2348–2362. doi: 10.1038/npp.2013.158
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ranasinghe, R., and Eri, R. (2018). Pleiotropic immune functions of chemokine receptor 6 in health and disease. Medicines 5:69. doi: 10.3390/medicines5030069
Rayhan, R. U., Stevens, B. W., Timbol, C. R., Adewuyi, O., Walitt, B., Vanmeter, J. W., et al. (2013). Increased brain white matter axial diffusivity associated with fatigue, pain and hyperalgesia in Gulf War illness. PLoS One 8:e58493. doi: 10.1371/journal.pone.0058493
Rouhiainen, A., Kulesskaya, N., Mennesson, M., Misiewicz, Z., Sipilä, T., Sokolowska, E., et al. (2019). The bradykinin system in stress and anxiety in humans and mice. Sci. Rep. 9:19437.
Seth, R. K., Maqsood, R., Mondal, A., Bose, D., Kimono, D., and Holland, L. A. (2019). Gut DNA virome diversity and its association with host bacteria regulate inflammatory phenotype and neuronal immunotoxicity in experimental Gulf War Illness. Viruses 11:968. doi: 10.3390/v11100968
Steele, L. (2000). Prevalence and patterns of Gulf War illness in Kansas veterans: association of symptoms with characteristics of person, place, and time of military service. Am. J. Epidemiol. 152, 992–1002. doi: 10.1093/aje/152.10.992
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., and Huerta-Cepas, J. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613.
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., and Simonovic, M. (2016). The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368.
Toomey, R., Alpern, R., Vasterling, J. J., Baker, D. G., Reda, D. J., and Lyons, M. J. (2009). Neuropsychological functioning of US Gulf War veterans 10 years after the war. J. Int. Neuropsychol. Soc. 15, 717–729. doi: 10.1017/s1355617709990294
Unwin, C., Blatchley, N., Coker, W., Ferry, S., Hotopf, M., and Hull, L. (1999). Health of UK servicemen who served in Persian Gulf War. Lancet 353, 169–178. doi: 10.1016/s0140-6736(98)11338-7
Vera Alvarez, R., Pongor, L. S., Mariño-Ramírez, L., and Landsman, D. (2019). TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics 35, 1960–1962. doi: 10.1093/bioinformatics/bty896
Villares, R., Cadenas, V., Lozano, M., Almonacid, L., Zaballos, A., Martínez-A, C., et al. (2009). CCR6 regulates EAE pathogenesis by controlling regulatory CD4+ T-cell recruitment to target tissues. Eur. J. Immunol. 39, 1671–1681. doi: 10.1002/eji.200839123
Wagner, G. P., Kin, K., and Lynch, V. J. (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285. doi: 10.1007/s12064-012-0162-3
White, R. F., Steele, L., O’Callaghan, J. P., Sullivan, K., Binns, J. H., and Golomb, B. A. (2016). Recent research on Gulf War illness and other health problems in veterans of the 1991 Gulf War: effects of toxicant exposures during deployment. Cortex 74, 449–475. doi: 10.1016/j.cortex.2015.08.022
Williams, R. W., Gu, J., Qi, S., and Lu, L. (2001). The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2:RESEARCH0046.
Williams, R. W., and Williams, E. G. (2017). “Resources for systems genetics,” in Systems Genetics, eds K. Schughart and R. Williams (Berlin: Springer), 3–29. doi: 10.1007/978-1-4939-6427-7_1
Yalcin, B., Wong, K., Agam, A., Goodson, M., Keane, T. M., and Gan, X. (2011). Sequence-based characterization of structural variation in the mouse genome. Nature 477, 326–329. doi: 10.1038/nature10432
Yamazaki, T., Yang, X. O., Chung, Y., Fukunaga, A., Nurieva, R., and Pappu, B. (2008). CCR6 regulates the migration of inflammatory and regulatory T cells. J. Immunol. 181, 8391–8401. doi: 10.4049/jimmunol.181.12.8391
Zakirova, Z., Tweed, M., Crynen, G., Reed, J., Abdullah, L., and Nissanka, N. (2015). Gulf War agent exposure causes impairment of long-term memory formation and neuropathological changes in a mouse model of Gulf War Illness. PLoS One 10:e0119579. doi: 10.1371/journal.pone.0119579
Zhao, H., Sonada, S., Yoshikawa, A., Ohinata, K., and Yoshikawa, M. (2016). Rubimetide, humanin, and MMK1 exert anxiolytic-like activities via the formyl peptide receptor 2 in mice followed by the successive activation of DP1, A2A, and GABAA receptors. Peptides 83, 16–20. doi: 10.1016/j.peptides.2016.07.001
Keywords: Ccr6, GWI, DFP, CORT, BXD strain, RNA-seq, neuroinflammation
Citation: Gao J, Xu F, Starlard-Davenport A, Miller DB, O’Callaghan JP, Jones BC and Lu L (2020) Exploring the Role of Chemokine Receptor 6 (Ccr6) in the BXD Mouse Model of Gulf War Illness. Front. Neurosci. 14:818. doi: 10.3389/fnins.2020.00818
Received: 02 June 2020; Accepted: 13 July 2020;
Published: 14 August 2020.
Edited by:
Igor Ponomarev, Texas Tech University Health Sciences Center, United StatesReviewed by:
Clarissa Carlin Parker, Middlebury College, United StatesCopyright © 2020 Gao, Xu, Starlard-Davenport, Miller, O’Callaghan, Jones and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Lu, bGx1QHV0aHNjLmVkdQ==; bHVsdUB1dGhzYy5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.