- 1College of Food Science and Engineering, Qingdao Agricultural University, Qingdao, China
- 2Department of Neurology, Affiliated Hospital of Qingdao University, Qingdao, China
- 3Department of Anesthesiology, Affiliated Hospital to Qingdao University, Qingdao, China
Chronic neuropathic pain, a type of chronic and potentially disabling pain caused by a disease or injury of the somatosensory nervous system, spinal cord injury, or various chronic conditions, such as viral infections (e.g., post-herpetic neuralgia), autoimmune diseases, cancers, and metabolic disorders (e.g., diabetes mellitus), is one of the most intense types of chronic pain, which incurs a major socio-economic burden and is a serious public health issue, with an estimated prevalence of 7–10% in adults throughout the world. Presently, the available drug treatments (e.g., anticonvulsants acting at calcium channels, serotonin-noradrenaline reuptake inhibitors, tricyclic antidepressants, opioids, topical lidocaine, etc.) for chronic neuropathic pain patients are still rare and have disappointing efficacy, which makes it difficult to relieve the patients’ painful symptoms, and, at best, they only try to reduce the patients’ ability to tolerate pain. Long non-coding RNAs (lncRNAs), a type of transcript of more than 200 nucleotides with no protein-coding or limited capacity, were identified to be abnormally expressed in the spinal cord, dorsal root ganglion, hippocampus, and prefrontal cortex under chronic neuropathic pain conditions. Moreover, a rapidly growing body of data has clearly pointed out that nearly 40% of lncRNAs exist specifically in the nervous system. Hence, it was speculated that these dysregulated lncRNAs might participate in the occurrence, development, and progression of chronic neuropathic pain. In other words, if we deeply delve into the potential roles of lncRNAs in the pathogenesis of chronic neuropathic pain, this may open up new strategies and directions for the development of novel targeted drugs to cure this refractory disorder. In this article, we primarily review the status of chronic neuropathic pain and provide a general overview of lncRNAs, the detailed roles of lncRNAs in the nervous system and its related diseases, and the abnormal expression of lncRNAs and their potential clinical applications in chronic neuropathic pain. We hope that through the above description, readers can gain a better understanding of the emerging roles of lncRNAs in chronic neuropathic pain.
Introduction
Chronic Neuropathic Pain
An epidemiological survey has clearly revealed that nearly 19% of adult Europeans suffered from chronic pain of moderate to severe intensity, which not only seriously affected the physical and mental quality of their social and working lives but also represents a significant public health issue that can be costly to the healthcare system (Rapo-Pylkko et al., 2017). Among all the different types of chronic pain, neuropathic pain is relatively common, but difficult to treat and manage (Baron, 2006, 2009). Moreover, neuropathic pain occurs in approximately 7–10% of the general population; therefore, neuropathic pain remains one of the most serious public health problems (van Hecke et al., 2014). Traditionally, the International Association for the Study of Pain (IASP) proposed a clinical description for neuropathic pain, which is defined as “pain initiated or caused by a primary lesion or dysfunction in the nervous system” (Scholz et al., 2019). However, this definition is broad, covering over 100 conditions, and has been controversial in recent years (National Institute of Neurological Disorders Stroke rt PASSG, 2005; Meacham et al., 2017). The main reason is that many scholars believe that the meaning of the term “dysfunction” is still relatively vague (Baron, 2009). Furthermore, it does not distinguish well between neuropathic pain and nociceptive pain (Vranken, 2012). Recently, a new precise definition of neuropathic pain has been put forward to avoid the above shortcomings, and to further beneficially support clinical and research purposes (Lauria et al., 2012; Treede et al., 2019). The revised definition is that “pain arose as a direct consequence of a lesion or disease affecting the somatosensory system,” which definitely indicates that the peripheral or central somatosensory system must be involved (Scholz et al., 2019; Treede et al., 2019). According to the underlying etiology and the anatomical location of the specific lesion, clinical neuropathic pain is haply divided into four main types, namely, painful peripheral neuropathies, central pain syndromes, complex painful neuropathic disorders, and mixed-pain syndromes (Baron, 2009; Nijs et al., 2016). Painful peripheral neuropathies are caused by ischemic, traumatic, inflammatory, infectious, metabolic, or compressive damage to the peripheral nervous system (PNS), such as polyneuritis, phantom limb pain, diabetic peripheral neuralgia, post-herpetic neuralgia, etc. (Beran, 2015; Watson and Dyck, 2015). Central pain syndromes result from the dysfunction of or damage to the central nervous system (CNS) pain conduction pathway, mainly involving cerebrovascular disease, spinal cord trauma, etc. (Petzke, 2010). Complex painful neuropathic disorders are also called “complex regional pain syndromes (CRPS),” which may result in trauma and affect the limbs (Baron and Wasner, 2001; Borchers and Gershwin, 2014). In addition, mixed-pain syndromes are the combination of nociceptive and neuropathic pain (Baron, 2009; Picelli et al., 2016).
Generally, patients with neuropathic pain may show distinct negative and positive sensory symptoms or signs in the nerve distribution area (Baron, 2006). Firstly, the negative sensory symptoms predominately contain hypoesthesia, pallhypoesthesia, hypoalgesia, thermohypoesthesia, etc. (Baron, 2006; Shaygan et al., 2014). Secondly, the positive sensory symptoms primarily include paresthesia, paroxysmal pain, superficial pain, and evoked pain. This would stimulate induced hypersensitivity and pain, such as light-pressure, gentle static pressure, and sharp pinprick (Baron, 2006; Soler et al., 2017). However, in a clinical setting, patients often were diagnosed as hyperesthesias that are mainly referred to as paresthesia and hyperalgesia (Zimmermann, 2001). The two types of hyperesthesia need to be clearly distinguished. Paresthesia (also known as mechanodynamic painful paresthesia) is defined as the non-noxious stimuli (e.g., light touch, warm, cool), and normally would not cause painful sensation in the normal nerve areas, but it could cause strongly painful sensation in the damaged nerve areas (Baron, 2009; Arle et al., 2016). Nevertheless, hyperalgesia is defined as an increased pain sensitivity or painful summation to mildly noxious stimuli (Baron, 2009; Jensen and Finnerup, 2014). To date, pharmacotherapy is the main therapeutic strategy in neuropathic pain patients. However, these drugs and agents lack specificity and efficiency, and could reduce the pain to a more tolerable level (Teixeira, 2009; Helfert et al., 2015). Thus, it is urgently needed to further explore the potential molecular mechanisms of the neuropathic pain development, which may provide a rational solution for clinical treatment and novel drug discovery in neuropathic pain.
Long Non-coding RNAs (lncRNAs)
With the development of larger-scale sequencing techniques and bioinformatics methods, the Encyclopedia of DNA Elements (ENCODE) project, the most comprehensive effort yet for surveying transcription in human cells, has clearly uncovered that only approximately 2% of the whole human genome encodes functional protein-coding genes, while the remaining genome is actively transcribed into a diversity of RNAs that were defined as non-coding RNAs (ncRNAs) and were previously believed to be non-functional “dark matter” (Qu and Fang, 2013). However, an accumulating body of evidence in recent years reported that, similar to protein-coding genes, ncRNAs are also involved in a wide variety of biological processes, such as cell proliferation, cellular structure integrity, cellular development, differentiation, stem cell pluripotency, signaling transductions, genomic imprinting, the maintenance of genomic integrity, reprogramming, heat shock response, etc. (Nicolas, 2017; Sherstyuk et al., 2018). Therefore, it was no surprise that abnormal expression profiles of ncRNAs were implicated in the initiation, development, and progression processes of many human diseases, including cardiovascular syndromes, diabetes, autoimmune diseases, nervous system disorders, cancers, and so on (Huang et al., 2013). However, among all classifications of ncRNAs, lncRNAs, a heterogeneous group of transcripts that are greater than 200 nucleotides (nt) in length, account for more than 80% of ncRNAs (Jarroux et al., 2017). Because lncRNAs occupy the largest class of ncRNAs and represent the most prevalent and functionally diverse class of ncRNAs, they attract interest in the scientific community in the past decades (Roberts et al., 2014).
Since Brannan and his colleagues reported the first lncRNA H19 in 1990, thousands of lncRNAs have been identified in a large diversity of species, including mammalian animals, plants, yeast, prokaryotes, and even viruses, by a large number of scientists (Jarroux et al., 2017). It is reported that the lncRNA is bidirectional transcribed, depending on their proximity to the nearest protein-coding transcripts. It can be grouped into five main categories: sense lncRNAs (lncRNAs initiate inside or 5’ of a protein-coding gene, then are transcribed in the same direction as protein-coding genes, and ultimately overlap at least one protein-coding exon), antisense lncRNAs (lncRNAs initiate inside or 3’ of a protein-coding gene, then are transcribed in the opposite direction of protein-coding genes, and ultimately overlap at least one protein-coding exon), bidirectional lncRNAs (the expression of the lncRNA and its neighboring protein-coding genes on the opposite strand is initiated in close genomic proximity), intronic lncRNAs (the entire sequence of the lncRNA falls within the intron of a protein-coding gene), and intergenic lncRNAs (also termed large intervening ncRNAs or lincRNAs, in which the entire sequence of the lncRNA falls between two protein-coding genes as a distinct unit) (Figure 1A; Ma et al., 2013; St Laurent et al., 2015). In addition, an increasing number of studies have found that lncRNAs play versatile roles in many aspects of gene regulation through four functional mechanisms: signaling and acting as a decoy, guide, and scaffolding molecule (Figure 1B; Akhade et al., 2017; Delas and Hannon, 2017). First, as a signaling molecule, lncRNAs are usually transcribed in a specific time and space, and their transcripts can integrate developmental cues, interpret cell status, or respond to multiple stimuli and further regulate the expression of other genes. Therefore, lncRNAs are seen as an important marker in a biological event (Wang and Chang, 2011). Second, as a decoy molecule, lncRNA can block the binding of transcriptional repressors to their homologous target gene promoters, as well as protein molecules, chromatin modifications, etc., and thereby lncRNAs could ultimately inhibit their functions (Ma et al., 2012). Third, as a guide molecule, lncRNAs are able to directly bind to the protein molecules and then guide the protein complexes containing the above-described protein-bound molecules to be precisely localized on a specific target (Wang and Chang, 2011). On the other hand, lncRNA can also recruit more transcription-related and epigenetic-related regulatory factors widely and further guide their localizations (Goff and Rinn, 2015). Finally, as a scaffold molecule, lncRNAs have many different structural domains that can combine different effector molecules to achieve the assembly of macromolecules at the same time. In fact, lncRNA is a complex assembly center platform, which can recruit many related molecules together to further activate or inhibit the expression of other genes (Bonasio et al., 2010; Ma et al., 2012). It is extremely important for the transmission of many biological signals, intermolecular interactions, and the precise regulation of the specificity and dynamics of the signal itself (Wang and Chang, 2011; Moran et al., 2012). Hence, based on the multiple functions of lncRNAs, researchers have gradually shifted their focus of research over the last few decades from previous studies on protein-coding gene-induced diseases to studies on lncRNA-induced diseases (Moran et al., 2012; Delas and Hannon, 2017).
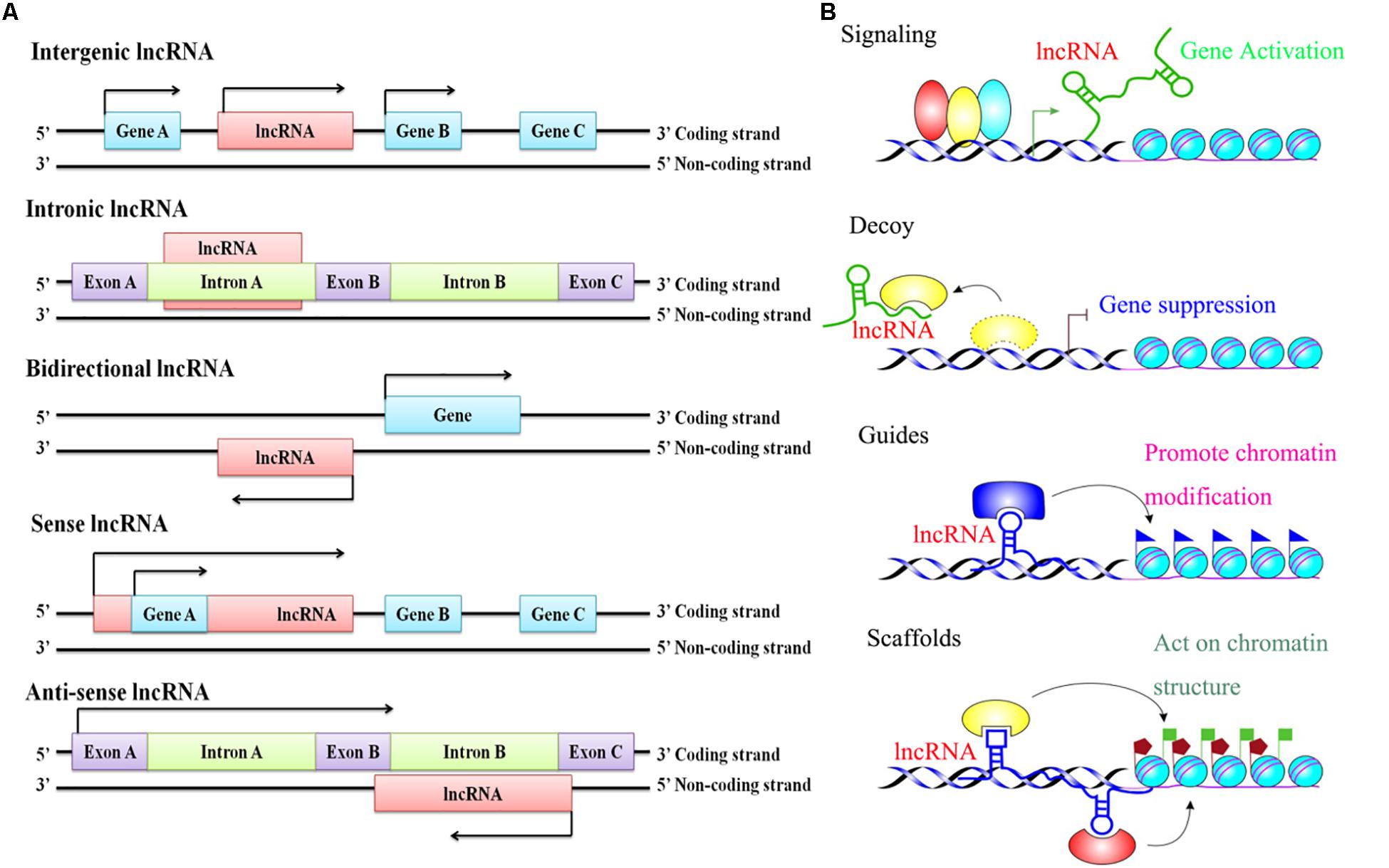
Figure 1. The classification and functional mechanisms of lncRNAs. (A) Classification of lncRNAs according to the relative proximity to the nearest protein-coding transcripts; (B) Functions of lncRNAs in regulating target genes.
The Roles of lncRNAs in the Nervous System and Its Related Diseases
The human nervous system is the most highly evolved and sophisticated biological system that plays a leading role in the regulation of physiological and functional activities in the body (Herculano-Houzel, 2009; Herculano-Houzel et al., 2014). Generally, the functions and physiological processes of different organs and systems in the human body are not carried out in isolation, but they are closely connected, interacting and cooperating under the direct or indirect control of the nervous system, which makes the human body become a complete and unified organism to achieve and maintain normal life activities when the human body encounters interoceptive and environmental stimuli (Herculano-Houzel, 2012; Kohl and Dulac, 2018). It is well-known that the human nervous system is mainly composed of nerve tissues that contain two basic types of cells, neurons and glial cells, and is divided into two major parts, the CNS and the PNS (Garner and Mayford, 2012; Zimmerman et al., 2017). The CNS, containing the brain and the spinal cord, is where we receive sensory information, generate thoughts and emotions, and store memories (Kaucka and Adameyko, 2014; Engelhardt et al., 2017). The perfect presentation of CNS and PNS functions predominately depends on the dynamic neural networks organized by neuronal and glial cells (Guarino et al., 2017). However, the development of these cells requires a precise spatiotemporal regulation of stem/progenitor cell proliferation and differentiation at various levels, such as epigenetic, transcriptional, or post-transcriptional processes, and then forms appropriate connections with each other (Qureshi and Mehler, 2010). In recent years, emerging data have reported that an increasing number of lncRNAs were found to play vital roles in mediating the developmental complexity of the nervous system, and these abundant lncRNAs are precisely, dynamically, and specifically expressed in the brain via temporal and spatial expression patterns (Qureshi and Mehler, 2013). In fact, it is not difficult to understand that the development of the nervous system is actually a complex neural network formed by the tight gene regulation of its constituent cells (Garner and Mayford, 2012). Nevertheless, based on the explanation of the functions of lncRNAs in the above, it can be known that the regulatory mechanisms of lncRNAs may participate in the entire process of nervous system development (Wu et al., 2013). Therefore, not surprisingly, the dysregulation of or mutations in lncRNA gene loci might directly affect nervous system development, function, maintenance, and differentiation and be intimately associated with the molecular pathophysiology of a broad array of the most devastating neurological diseases, such as glioma, neuropathic pain, autism spectrum disorder (ASD), Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), Asperger’s syndrome, schizophrenia, developmental delay, amyotrophic lateral sclerosis (ALS), depression, autism, and others (Riva et al., 2016; Wan et al., 2017).
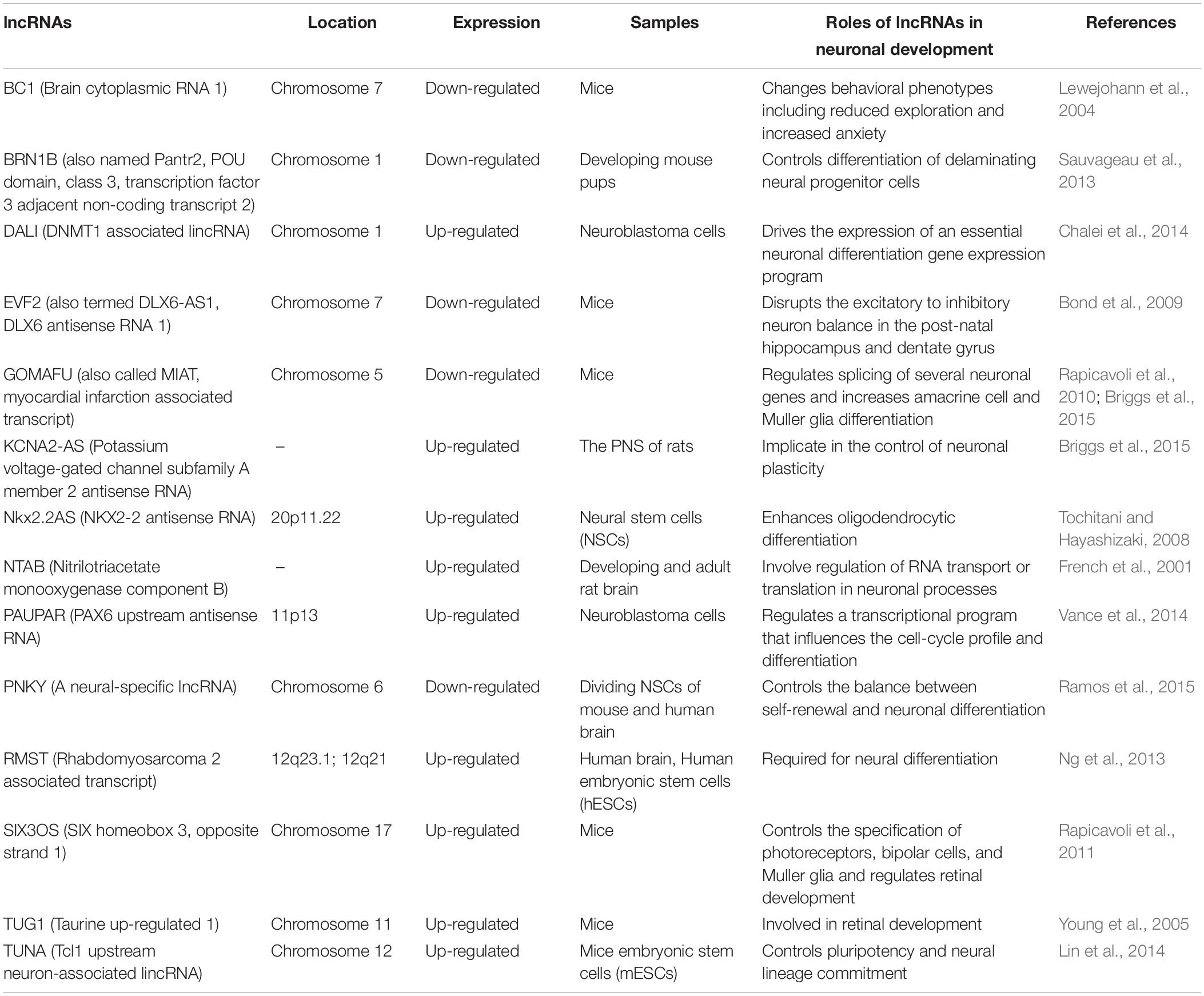
First, we explored the roles of lncRNA in neuronal development (Wu et al., 2013; Ramos et al., 2016). Several lines of evidence have demonstrated that the profiles of lncRNA expression present a dynamic change during the process of neuronal development (Rani et al., 2016). For example, Mercer et al. discovered that 1328 lncRNAs were identified in the mouse, whereas among these lncRNAs, 849 lncRNAs were located in the mouse brain with 623 lncRNAs exhibiting selective profiles for specific regions (e.g., olfactory bulb, hippocampus, cortex, or cerebellum), cell types (e.g., neurons, glia cells), and subcellular compartments (e.g., dendrite, axon, Nissl body, soma, etc.), which suggested that these lncRNAs might be involved in regulating specific processes through cell- type-, subcellular- compartment-, and developmental-stage-specific manners (Bussotti et al., 2016). Amaral et al. revealed that the lncRNA Sox2OT was expressed in regions of constitutive adult neurogenesis and mediated the expression of the Sox2 gene, which is a key transcription factor (TF) that is responsible for the neural induction and maintenance of neural stem and progenitor cells, indicating that the lncRNA Sox2OT might participate in neural cell fate decisions (Amaral et al., 2009). Other studies related to lncRNAs in neuronal development are shown in Table 1. Throughout the study of these lncRNAs in neurodevelopment, it is known that the methods of exploring the physiological functions of lncRNAs are basically the same. Researchers often use qRT-PCR to detect changes in the expression of lncRNAs in blood samples from patients with neurological diseases, in cell model samples, or in animal model samples. Moreover, a dual-luciferase reporter assay was applied to verify the target interaction between lncRNAs and their downstream target genes. In addition, CCK8, colony formation assay, and flow cytometry were utilized to assess the influences of lncRNA on neuronal development.
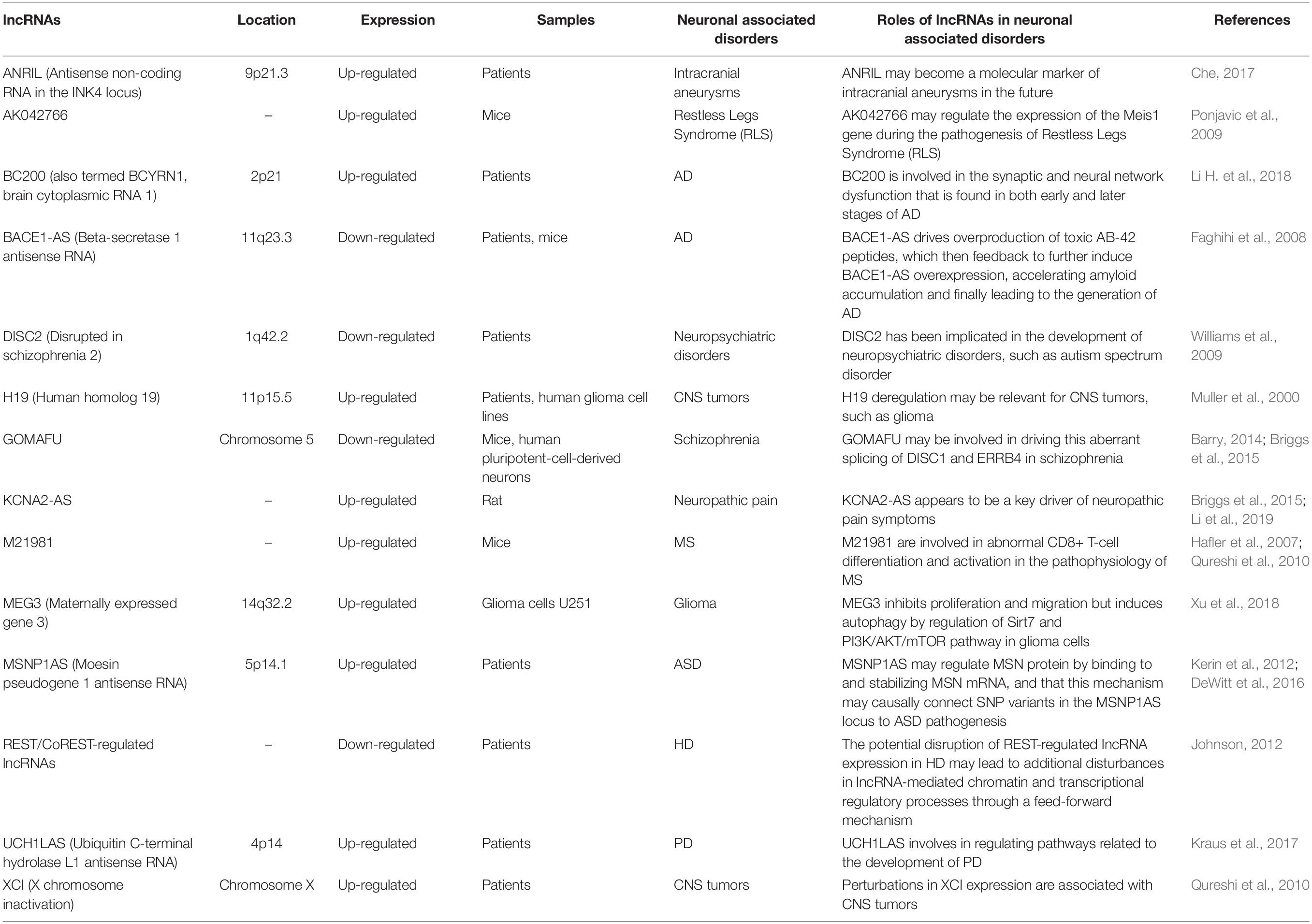
Second, we continue to investigate the effects of lncRNAs on neurological diseases (Riva et al., 2016). For instance, over-expressed lncRNA HOXA transcript antisense RNA, myeloid-specific 1 (HOTAIRM1), facilitates tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme (Li Q. et al., 2018). LncRNA small nucleolar RNA host gene 12 (SNHG12) accelerates angiogenesis following ischemic stroke via a miR-150/VEGF pathway, which further clarifies the mechanism of angiogenesis after ischemic stroke and provides a target for the treatment of this disease (Zhao M. et al., 2018). The imbalance of lncRNA AK127244 might have a particular influence on language development and behavior or mood in neuropsychiatric disorders (Duong et al., 2015). Nuclear factor-k-gene binding (NF-kB) interacting lncRNA (NKILA) promotes the endoplasmic reticulum stress/autophagy pathway and suppresses the NF-kB pathway after intracerebral hemorrhage (Jia et al., 2018). Other studies linked to lncRNAs in neuronal-associated disorders are detailed in Table 2.
The Abnormal Expression of lncRNAs and Their Potential Clinical Applications in Chronic Neuropathic Pain
Chronic neuropathic pain generally resulting from injury to or disease of the nervous system has always been very difficult in clinical treatment, mainly because almost all patients with chronic neuropathic pain are notoriously resistant to the actions of currently available analgesics, such as non-steroidal anti-inflammatory drugs and opioids, which could exert effective therapeutic roles in nociceptive pain (Jackson, 2006). Pharmacological (e.g., tricyclic antidepressants, anticonvulsants, selective serotonin norepinephrine reuptake inhibitors, and topical lidocaine), non-pharmacological (e.g., behavioral, cognitive, integrative, and physical approaches), and interventional (e.g., neuroablative and neuromodulation techniques) therapies are currently used in the treatment of chronic neuropathic pain. These treatments cannot completely control the symptoms of patients with chronic neuropathic pain, which may be associated with the diversity of pathophysiological situations, different pain etiologies, genetic predispositions, etc. (Gilron et al., 2015). Thus, chronic neuropathic pain remains a distressing and debilitating disease, there is still no panacea for the treatment of chronic neuropathic pain in the clinic, and it is imperative to further develop chronic neuropathic pain drugs (Jensen and Finnerup, 2007; Xu et al., 2016). In recent years, a large number of studies have revealed that many prominent and well-characterized mechanisms were observed in chronic neuropathic pain, including (1) ongoing ectopic activity in peripheral nerves, which resulted in excitation–inhibition imbalance; (2) the aberrant production and release of anti-inflammatory or proinflammatory cytokines at the site of injury, which triggers peripheral sensitization or central sensitization; (3) the impairment of endogenous inhibitory mechanisms of nociception sensitivity-induced microglial sensitivity after nerve injury, which enhanced the post-injury pain sensitivity; and (4) microglial activation after nerve injury, which contributed to the pathogenesis of pain hypersensitivity (Gilron et al., 2015). Therefore, in the full understanding of the pathogenesis of chronic neuropathic pain, we can develop corresponding new therapeutic drugs for chronic neuropathic pain patients according to their relevant mechanisms (Gilron et al., 2006; Vranken, 2012). Furthermore, most extensive evidence has definitely pointed out that there were many dysregulated lncRNAs expressed in damaged nerves, primary sensory dorsal root ganglion neurons, spinal cord dorsal roots, the prefrontal cortex, the post-synaptic dorsal horn, and even in higher-order neurons up to the cortical level after peripheral nerve injury; therefore, it was speculated that these dysregulated lncRNAs might be involved in the pathogenesis of chronic neuropathic pain (Li et al., 2019). Furthermore, this speculation was also demonstrated by a large number of researchers. For instance, Liu et al. constructed a mouse model of spared nerve injury-induced neuropathic pain and found, by microarray technology, that there were 22,213 abnormally expressed lncRNAs, which might be involved in the activities of cytokines (IL-17A and IL-17F) and chemokines (CCL-2, CCL-5, and CCL-7), in the spinal cord, suggesting that differentially expressed lncRNAs played a pivotal role in the progression of neuropathic pain (Liu Z. et al., 2017). Additionally, Jiang et al. identified 511 differentially expressed (>2-fold change) lncRNAs (366 up-regulated and 145 down-regulated) in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain, and these lncRNAs might be related to Toll-like receptor signaling, cytokine–cytokine receptor interaction, and peroxisome proliferator-activated receptor signaling pathways, which indicated that abnormally expressed lncRNAs might be implicated in the pathogenesis of chronic neuropathic pain (Jiang et al., 2015). With the deepening of lncRNA research, scientists are no longer limited to the study of lncRNA expression profiles of chronic neuropathic pain, and they have begun to specifically explore the biological function of these differentially expressed lncRNAs in chronic neuropathic pain (Wu et al., 2019). Thus, the amassing literature referring to lncRNAs in chronic neuropathic pain is presented in detail in Table 3 and Figure 2.
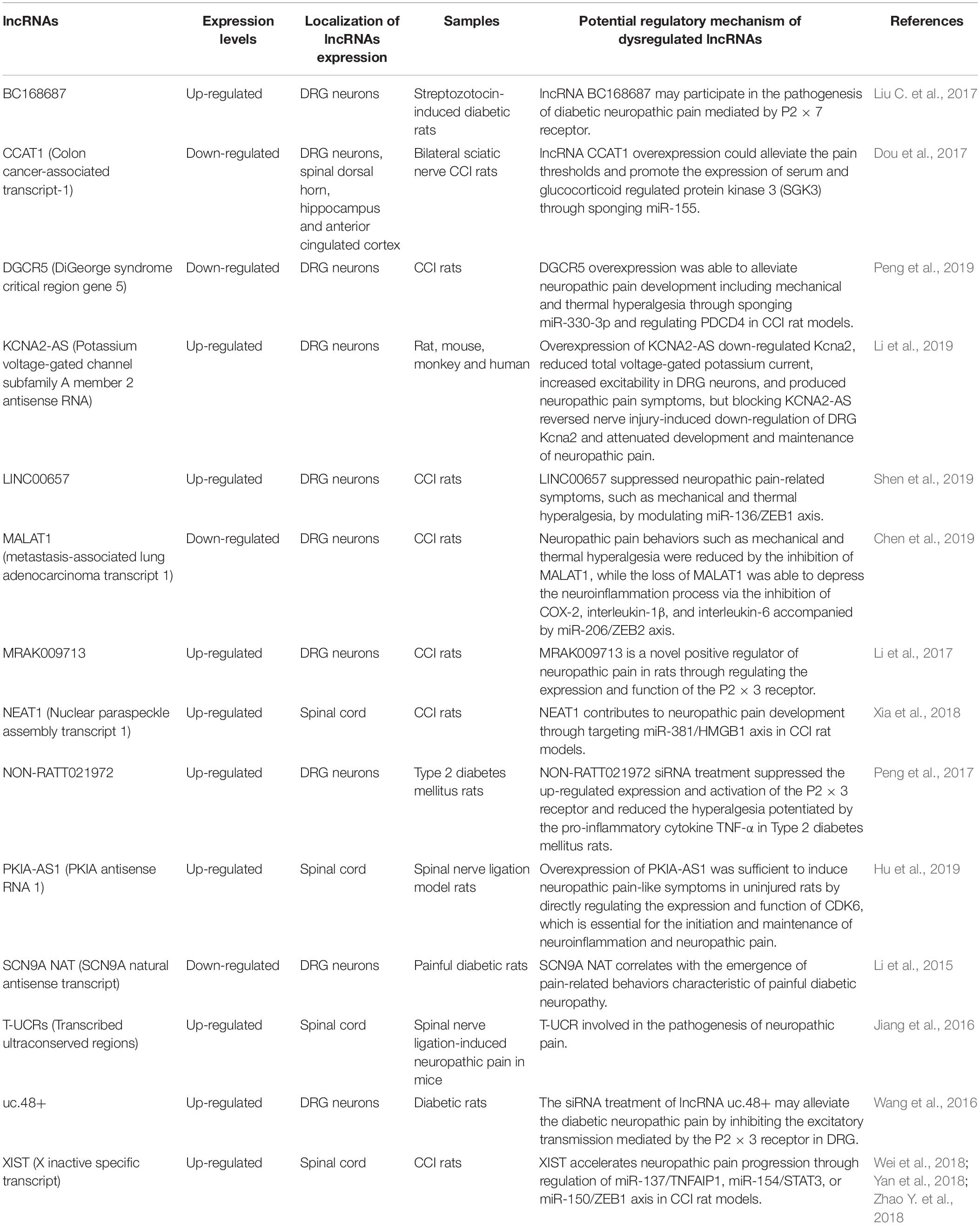
Table 3. Comprehensive analysis of aberrant lncRNA expression profiles in chronic neuropathic-related conditions.
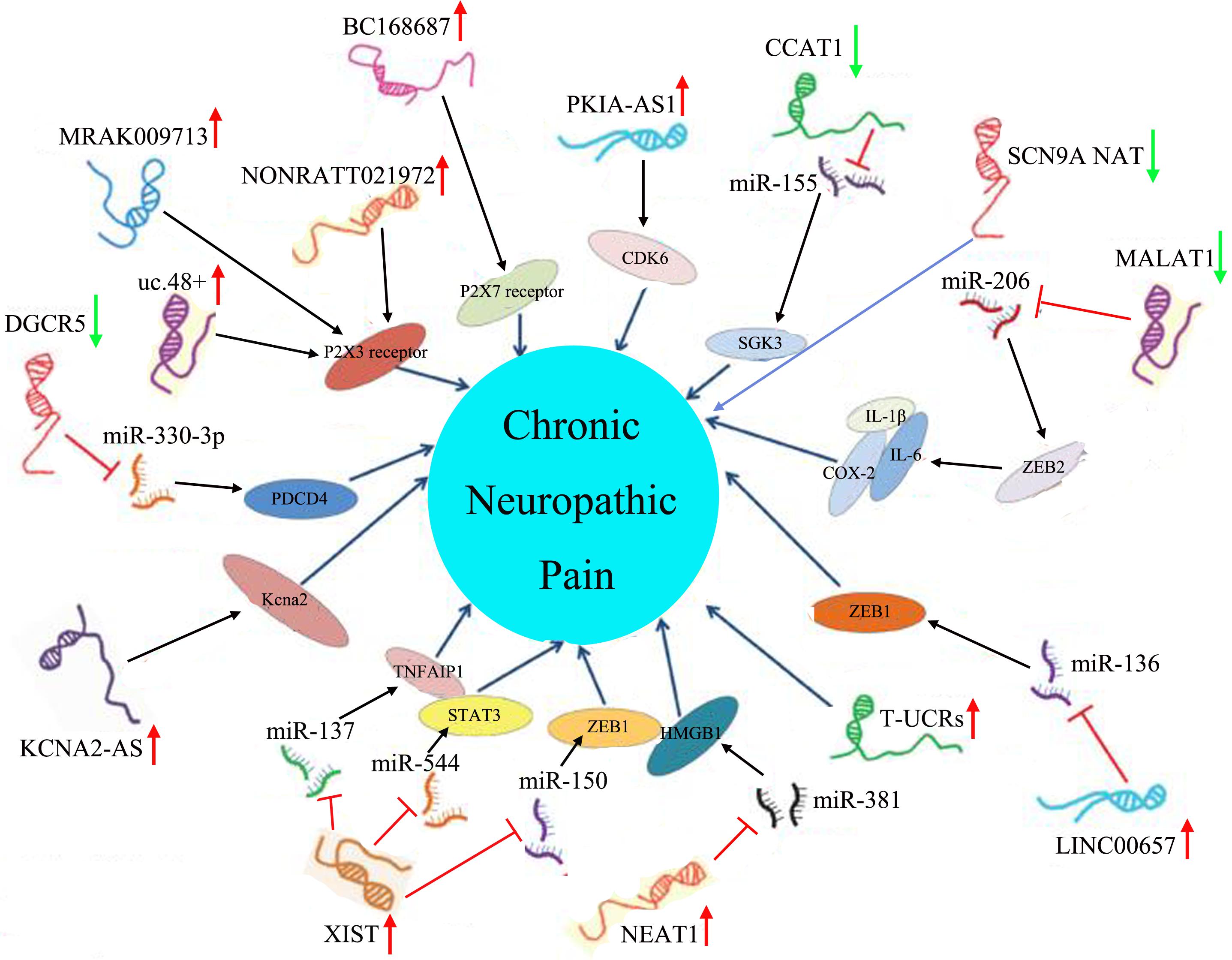
Figure 2. Aberrant lncRNA expression and their downstream molecular pathways in chronic neuropathic pain. The red arrow represented an up-regulated trend of lncRNAs, and the green arrow represented a down-regulated trend of lncRNAs.
It should be pointed out that all the current biological functions related to lncRNAs in chronic neuropathic pain are based on cellular or animal models, and research on the human body can be limited only to their expression levels, but not their biological functions (Kumar et al., 2018). The major reason is that chronic neuropathic pain is usually prone to irreversible damage in the human body, which is difficult for us to control (Gierthmuhlen and Baron, 2016). Hence, animal models of chronic neuropathic pain can be mainly grouped into four categories, including nerve injury models [such as spinal cord injury, excitotoxins, spinal hemi-section, thalamic syndrome, sciatic nerve chronic constriction injury (CCI), spinal nerve ligation, spared nerve injury, sciatic inflammatory neuritis, etc.], drug-induced chronic neuropathic pain models (such as anti-cancer-agent-induced neuropathy, vincristine-induced neuropathic pain, cisplatin-induced neuropathic pain, etc.), disease-induced neuropathy models (such as diabetes-induced neuropathy, the post-herpetic neuralgia model, human-immunodeficiency-virus-induced neuropathy, etc.), and miscellaneous models [such as chronic ethanol consumption/withdrawal-induced neuropathy, pyridoxine (vitamin B6)-induced neuropathy, inherited-induced neuropathies, etc.] (Kumar et al., 2018). Based on these models, whether the roles of lncRNAs in chronic neuropathic pain can be well reflected in the human body still needs to be further explored.
Conclusion and Future Directions
Chronic neuropathic pain is one of the most challenging and refractory neurological diseases in a great number of patients (Gierthmuhlen and Baron, 2016). In the clinic, chronic neuropathic pain patients are often characterized by symptoms of spasticity, muscle weakness, dysesthesia, spontaneous pain, allodynia, hyperalgesia, poor proprioception, and so on (Gilron et al., 2015). Currently, in response to these symptoms of chronic neuropathic pain, there are still no clinically effective drugs for their treatment (Gilron et al., 2006). Previous studies have precisely revealed that the available analgesics, such as non-steroidal anti-inflammatory drugs and opioids, which could effectively improve the clinical signs or symptoms of nociceptive pain, did not produce an adequate effect on chronic neuropathic pain patients (Vranken, 2012). Emerging lines of evidence have also uncovered that, despite a mass of pharmacological studies performed in chronic neuropathic pain, these newly developed treatment strategies, including pharmacological, non-pharmacological, and interventional methods, remain unable to adequately alleviate the patients’ pain or reduce the pain only to a range that the patients can tolerate (Jackson, 2006). Moreover, on the other hand, epidemiological surveys have reported that the prevalence of chronic neuropathic pain in the gross population is approximately 7–10% around the world (Zimmermann, 2001; van Hecke et al., 2014). Therefore, chronic neuropathic pain represents a worldwide challenging issue that is a socio-economic burden not only for patients and caregivers but also for the healthcare system (Baron, 2009). Currently, an increasing number of clinicians believe that patients with chronic neuropathic pain have not been treated well because the pathogenesis of the disease is not completely understood (Vranken, 2012). In-depth studies of the pathogenesis of chronic neuropathic pain may provide a solid theoretical basis for the development of new drugs or agents. lncRNAs, belonging to a family of RNAs characterized as transcripts more than 200 nucleotides in length, were discovered to participate in the cellular and molecular pathological processes of multiple nervous system diseases, including chronic neuropathic pain, by regulating the expression of neuronal-function-related genes (Qureshi et al., 2010; Wu et al., 2019). Hence, many researchers have turned their attention to the study of lncRNAs, with the thought that unveiling the underlying mechanism of how lncRNAs are involved in the occurrence, development and progression of chronic neuropathic pain may offer novel avenues to prevent and treat this refractory disease (Qureshi and Mehler, 2013).
In recent years, small-molecule targeted therapies using miRNAs or lncRNAs have been carried out in many disease areas. These molecules attracted so much attention due to their biology functions in regulating protein-coding genes at different levels, including epigenetic level, transcription level, post-transcriptional level, etc. In addition, their expressions not only have temporal and spatial specificity, but also cell and tissue specificity. Thus, these biological characteristics enable drug developers to find more accurate targets when developing new drugs or agents. Nonetheless, it is undeniable that there are still many challenges in the application of small molecules in the treatment of chronic neuropathic pain, for example, the transformation between experiment research and clinical application. Given that chronic neuropathic pain can cause irreversible damage to the human body, almost all models for chronic neuropathic pain research are based on cell or animal experiments. However, there are some differences between in vitro experiments and in vivo experiments, and these differences are unpredictable. Therefore, how the results obtained by small-molecule targeted therapy in in vitro experiments can be transformed and applied to human body is still a big problem.
Author Contributions
XJ and YZ proposed the concept, collected the literatures, and edited the manuscript. WW drafted the manuscript.
Funding
This study was supported by the National Key Research and Development Program under Grant No. 2016YFD0400105, the National Natural Science Foundation of China under Grant No. 81703228, the National Science Foundation of Shandong Province under Grant No. ZR2016CQ10, and the China Postdoctoral Science Foundation under Grant No. 2019M652320.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Akhade, V. S., Pal, D., and Kanduri, C. (2017). Long noncoding RNA: genome organization and mechanism of action. Adv. Exp. Med. Biol. 1008, 47–74. doi: 10.1007/978-981-10-5203-3_2
Amaral, P. P., Neyt, C., Wilkins, S. J., Askarian-Amiri, M. E., Sunkin, S. M., Perkins, A. C., et al. (2009). Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 15, 2013–2027. doi: 10.1261/rna.1705309
Arle, J. E., Mei, L., Carlson, K. W., and Shils, J. L. (2016). High-frequency stimulation of dorsal column axons: potential underlying mechanism of paresthesia-free neuropathic pain relief. Neuromodulation 19, 385–397. doi: 10.1111/ner.12436
Baron, R. (2006). Mechanisms of disease: neuropathic pain–a clinical perspective. Nat. Clin. Pract. Neurol. 2, 95–106. doi: 10.1038/ncpneuro0113
Baron, R. (2009). Neuropathic pain: a clinical perspective. Handb. Exp. Pharmacol. 194, 3–30. doi: 10.1007/978-3-540-79090-7_1
Baron, R., and Wasner, G. (2001). Complex regional pain syndromes. Curr. Pain Headache Rep. 5, 114–123.
Barry, G. (2014). Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatry 19, 410–416. doi: 10.1038/mp.2013.196
Bonasio, R., Tu, S., and Reinberg, D. (2010). Molecular signals of epigenetic states. Science 330, 612–616. doi: 10.1126/science.1191078
Bond, A. M., Vangompel, M. J., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027. doi: 10.1038/nn.2371
Borchers, A. T., and Gershwin, M. E. (2014). Complex regional pain syndrome: a comprehensive and critical review. Autoimmun. Rev. 13, 242–265. doi: 10.1016/j.autrev.2013.10.006
Briggs, J. A., Wolvetang, E. J., Mattick, J. S., Rinn, J. L., and Barry, G. (2015). Mechanisms of long non-coding rnas in mammalian nervous system development. Plast. Dis. Evol. Neuron 88, 861–877. doi: 10.1016/j.neuron.2015.09.045
Bussotti, G., Leonardi, T., Clark, M. B., Mercer, T. R., Crawford, J., Malquori, L., et al. (2016). Improved definition of the mouse transcriptome via targeted RNA sequencing. Genome Res. 26, 705–716. doi: 10.1101/gr.199760.115
Chalei, V., Sansom, S. N., Kong, L., Lee, S., Montiel, J. F., Vance, K. W., et al. (2014). The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 3:e04530.
Che, J. (2017). Molecular mechanisms of the intracranial aneurysms and their association with the long noncoding ribonucleic acid ANRIL-A review of literature. Neurol. India 65, 718–728.
Chen, Z. L., Liu, J. Y., Wang, F., and Jing, X. (2019). Suppression of MALAT1 ameliorates chronic constriction injury-induced neuropathic pain in rats via modulating miR-206 and ZEB2. J. Cell Physiol. [Epub ahead of print].
Delas, M. J., and Hannon, G. J. (2017). lncRNAs in development and disease: from functions to mechanisms. Open Biol. 7:170121. doi: 10.1098/rsob.170121
DeWitt, J. J., Hecht, P. M., Grepo, N., Wilkinson, B., Evgrafov, O. V., Morris, K. V., et al. (2016). Transcriptional gene silencing of the autism-associated long noncoding RNA MSNP1AS in human neural progenitor cells. Dev. Neurosci. 38, 375–383. doi: 10.1159/000453258
Dou, L., Lin, H., Wang, K., Zhu, G., Zou, X., Chang, E., et al. (2017). Long non-coding RNA CCAT1 modulates neuropathic pain progression through sponging miR-155. Oncotarget 8, 89949–89957.
Duong, L. T., Hoeffding, L. K., Petersen, K. B., Knudsen, C. D., Thygesen, J. H., Klitten, L. L., et al. (2015). Two rare deletions upstream of the NRXN1 gene (2p16.3) affecting the non-coding mRNA AK127244 segregate with diverse psychopathological phenotypes in a family. Eur. J. Med. Genet. 58, 650–653. doi: 10.1016/j.ejmg.2015.11.004
Engelhardt, B., Vajkoczy, P., and Weller, R. O. (2017). The movers and shapers in immune privilege of the CNS. Nat. Immunol. 18, 123–131. doi: 10.1038/ni.3666
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
French, P. J., Bliss, T. V., and O’Connor, V. (2001). Ntab, a novel non-coding RNA abundantly expressed in rat brain. Neuroscience 108, 207–215. doi: 10.1016/s0306-4522(01)00408-0
Garner, A., and Mayford, M. (2012). New approaches to neural circuits in behavior. Learn. Mem. 19, 385–390. doi: 10.1101/lm.025049.111
Gilron, I., Baron, R., and Jensen, T. (2015). Neuropathic pain: principles of diagnosis and treatment. Mayo Clin. Proc. 90, 532–545. doi: 10.1016/j.mayocp.2015.01.018
Gilron, I., Watson, C. P., Cahill, C. M., and Moulin, D. E. (2006). Neuropathic pain: a practical guide for the clinician. CMAJ 175, 265–275. doi: 10.1503/cmaj.060146
Goff, L. A., and Rinn, J. L. (2015). Linking RNA biology to lncRNAs. Genome Res. 25, 1456–1465. doi: 10.1101/gr.191122.115
Guarino, D., Nannipieri, M., Iervasi, G., Taddei, S., and Bruno, R. M. (2017). The Role of the Autonomic Nervous System in the Pathophysiology of Obesity. Front. Physiol. 8:665. doi: 10.3389/fphys.2017.00665
Hafler, D. A., Compston, A., Sawcer, S., Lander, E. S., Daly, M. J., et al. (2007). Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 357, 851–862. doi: 10.1056/nejmoa073493
Helfert, S. M., Reimer, M., Hoper, J., and Baron, R. (2015). Individualized pharmacological treatment of neuropathic pain. Clin. Pharmacol. Ther. 97, 135–142. doi: 10.1002/cpt.19
Herculano-Houzel, S. (2009). The human brain in numbers: a linearly scaled-up primate brain. Front. Hum. Neurosci. 3:31. doi: 10.3389/neuro.09.031.2009
Herculano-Houzel, S. (2012). Neuronal scaling rules for primate brains: the primate advantage. Prog. Brain Res. 195, 325–340. doi: 10.1016/b978-0-444-53860-4.00015-5
Herculano-Houzel, S., Avelino-de-Souza, K., Neves, K., Porfirio, J., Messeder, D., Mattos Feijo, L., et al. (2014). The elephant brain in numbers. Front. Neuroanat. 8:46. doi: 10.3389/fnana.2014.00046
Hu, J. Z., Rong, Z. J., Li, M., Li, P., Jiang, L. Y., Luo, Z. X., et al. (2019). Silencing of lncRNA PKIA-AS1 Attenuates Spinal Nerve Ligation-Induced Neuropathic Pain Through Epigenetic Downregulation of CDK6 Expression. Front. Cell Neurosci. 13:50. doi: 10.3389/fncel.2019.00050
Huang, Y., Wang, J. P., Yu, X. L., Wang, Z. V., Xu, T. S., and Cheng, X. C. (2013). Non-coding RNAs and diseases. Mol. Biol. 47, 531–543.
Jackson, K. C. II (2006). Pharmacotherapy for neuropathic pain. Pain Pract. 6, 27–33. doi: 10.1111/j.1533-2500.2006.00055.x
Jarroux, J., Morillon, A., and Pinskaya, M. (2017). History. Discovery, and classification of lncRNAs. Adv. Exp. Med. Biol. 1008, 1–46. doi: 10.1007/978-981-10-5203-3_1
Jensen, T. S., and Finnerup, N. B. (2007). Management of neuropathic pain. Curr. Opin. Support. Palliat. Care 1, 126–131.
Jensen, T. S., and Finnerup, N. B. (2014). Allodynia and hyperalgesia in neuropathic pain: clinical manifestations and mechanisms. Lancet Neurol. 13, 924–935. doi: 10.1016/s1474-4422(14)70102-4
Jia, J., Zhang, M., Li, Q., Zhou, Q., and Jiang, Y. (2018). Long noncoding ribonucleic acid NKILA induces the endoplasmic reticulum stress/autophagy pathway and inhibits the nuclear factor-k-gene binding pathway in rats after intracerebral hemorrhage. J. Cell Physiol. 233, 8839–8849. doi: 10.1002/jcp.26798
Jiang, B. C., Sun, W. X., He, L. N., Cao, D. L., Zhang, Z. J., and Gao, Y. J. (2015). Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Mol. Pain 11:43.
Jiang, B. C., Yang, T., He, L. N., Tao, Y. X., and Gao, Y. J. (2016). Altered T-UCRs expression profile in the spinal cord of mice with neuropathic pain. Transl. Perioper. Pain Med. 1, 1–10.
Johnson, R. (2012). Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol. Dis. 46, 245–254. doi: 10.1016/j.nbd.2011.12.006
Kaucka, M., and Adameyko, I. (2014). Non-canonical functions of the peripheral nerve. Exp. Cell Res. 321, 17–24. doi: 10.1016/j.yexcr.2013.10.004
Kerin, T., Ramanathan, A., Rivas, K., Grepo, N., Coetzee, G. A., and Campbell, D. B. (2012). A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci. Transl. Med. 4:128ra40. doi: 10.1126/scitranslmed.3003479
Kohl, J., and Dulac, C. (2018). Neural control of parental behaviors. Curr. Opin. Neurobiol. 49, 116–122. doi: 10.1016/j.conb.2018.02.002
Kraus, T. F. J., Haider, M., Spanner, J., Steinmaurer, M., Dietinger, V., and Kretzschmar, H. A. (2017). Altered long noncoding rna expression precedes the course of parkinson’s disease-a preliminary report. Mol. Neurobiol. 54, 2869–2877. doi: 10.1007/s12035-016-9854-x
Kumar, A., Kaur, H., and Singh, A. (2018). Neuropathic Pain models caused by damage to central or peripheral nervous system. Pharmacol. Rep. 70, 206–216. doi: 10.1016/j.pharep.2017.09.009
Lauria, G., Faber, C. G., Merkies, I. S., and Waxman, S. G. (2012). Diagnosis of neuropathic pain: challenges and possibilities. Expert Opin. Med. Diagn. 6, 89–93. doi: 10.1517/17530059.2012.661712
Lewejohann, L., Skryabin, B. V., Sachser, N., Prehn, C., Heiduschka, P., Thanos, S., et al. (2004). Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav. Brain Res. 154, 273–289. doi: 10.1016/j.bbr.2004.02.015
Li, G., Jiang, H., Zheng, C., Zhu, G., Xu, Y., Sheng, X., et al. (2017). Long noncoding RNA MRAK009713 is a novel regulator of neuropathic pain in rats. Pain 158, 2042–2052. doi: 10.1097/j.pain.0000000000001013
Li, H., Zheng, L., Jiang, A., Mo, Y., and Gong, Q. (2018). Identification of the biological affection of long noncoding RNA BC200 in Alzheimer’s disease. Neuroreport 29, 1061–1067. doi: 10.1097/wnr.0000000000001057
Li, Q., Dong, C., Cui, J., Wang, Y., and Hong, X. (2018). Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 37:265.
Li, Q. S., Cheng, P., Favis, R., Wickenden, A., Romano, G., and Wang, H. (2015). SCN9A variants may be implicated in neuropathic pain associated with diabetic peripheral neuropathy and pain severity. Clin. J. Pain 31, 976–982. doi: 10.1097/ajp.0000000000000205
Li, Z., Li, X., Chen, X., Li, S., Ho, I. H. T., Liu, X., et al. (2019). Emerging roles of long non-coding RNAs in neuropathic pain. Cell Prolif. 52:e12528. doi: 10.1111/cpr.12528
Lin, N., Chang, K. Y., Li, Z., Gates, K., Rana, Z. A., Dang, J., et al. (2014). An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1005–1019. doi: 10.1016/j.molcel.2014.01.021
Liu, C., Tao, J., Wu, H., Yang, Y., Chen, Q., Deng, Z., et al. (2017). Effects of LncRNA BC168687 siRNA on diabetic neuropathic pain mediated by P2X7 receptor on SGCs in DRG of rats. Biomed. Res. Int. 2017:7831251.
Liu, Z., Liang, Y., Wang, H., Lu, Z., Chen, J., Huang, Q., et al. (2017). LncRNA expression in the spinal cord modulated by minocycline in a mouse model of spared nerve injury. J. Pain Res. 10, 2503–2514. doi: 10.2147/jpr.s147055
Ma, H., Hao, Y., Dong, X., Gong, Q., Chen, J., Zhang, J., et al. (2012). Molecular mechanisms and function prediction of long noncoding RNA. ScientificWorldJournal. 2012:541786.
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 925–933.
Meacham, K., Shepherd, A., Mohapatra, D. P., and Haroutounian, S. (2017). Neuropathic pain: central vs. Peripheral mechanisms. Curr. Pain Headache Rep. 21:28.
Moran, V. A., Perera, R. J., and Khalil, A. M. (2012). Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 40, 6391–6400. doi: 10.1093/nar/gks296
Muller, S., Zirkel, D., Westphal, M., and Zumkeller, W. (2000). Genomic imprinting of IGF2 and H19 in human meningiomas. Eur. J. Cancer. 36, 651–655. doi: 10.1016/s0959-8049(99)00328-7
National Institute of Neurological Disorders Stroke rt PASSG, (2005). Recombinant tissue plasminogen activator for minor strokes: the national institute of neurological disorders and stroke rt-PA stroke study experience. Ann. Emerg. Med. 46, 243–252. doi: 10.1016/j.annemergmed.2005.02.013
Ng, S. Y., Bogu, G. K., Soh, B. S., and Stanton, L. W. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. doi: 10.1016/j.molcel.2013.07.017
Nicolas, F. E. (2017). Role of ncRNAs in development. Diagnosis and treatment of human cancer. Recent Pat Anticancer Drug Discov. 12, 128–135. doi: 10.2174/1574892812666170105113415
Nijs, J., Leysen, L., Adriaenssens, N., Aguilar Ferrandiz, M. E., Devoogdt, N., Tassenoy, A., et al. (2016). Pain following cancer treatment: guidelines for the clinical classification of predominant neuropathic, nociceptive and central sensitization pain. Acta Oncol. 55, 659–663. doi: 10.3109/0284186x.2016.1167958
Peng, C., Zhang, C., Su, Z., and Lin, D. (2019). DGCR5 attenuates neuropathic pain through sponging miR-330-3p and regulating PDCD4 in CCI rat models. J. Cell Physiol. 234, 7292–7300. doi: 10.1002/jcp.27487
Peng, H., Zou, L., Xie, J., Wu, H., Wu, B., Zhu, G., et al. (2017). lncRNA NONRATT021972 siRNA decreases diabetic neuropathic pain mediated by the P2X3 receptor in dorsal root ganglia. Mol. Neurobiol. 54, 511–523. doi: 10.1007/s12035-015-9632-1
Picelli, A., Buzzi, M. G., Cisari, C., Gandolfi, M., Porru, D., Bonadiman, S., et al. (2016). Headache, low back pain, other nociceptive and mixed pain conditions in neurorehabilitation. Evidence and recommendations from the Italian consensus conference on pain in neurorehabilitation. Eur. J. Phys. Rehabil. Med. 52, 867–880.
Ponjavic, J., Oliver, P. L., Lunter, G., and Ponting, C. P. (2009). Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5:e1000617. doi: 10.1371/journal.pgen.1000617
Qu, H., and Fang, X. (2013). A brief review on the human encyclopedia of DNA Elements (ENCODE) project. Genomics Proteomics Bioinform. 11, 135–141. doi: 10.1016/j.gpb.2013.05.001
Qureshi, I. A., Mattick, J. S., and Mehler, M. F. (2010). Long non-coding RNAs in nervous system function and disease. Brain Res. 1338, 20–35. doi: 10.1016/j.brainres.2010.03.110
Qureshi, I. A., and Mehler, M. F. (2010). Impact of nuclear organization and dynamics on epigenetic regulation in the central nervous system: implications for neurological disease states. Ann. N. Y. Acad. Sci. 1204(Suppl.), E20–E37.
Qureshi, I. A., and Mehler, M. F. (2013). Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics 10, 632–646. doi: 10.1007/s13311-013-0199-0
Ramos, A. D., Andersen, R. E., Liu, S. J., Nowakowski, T. J., Hong, S. J., Gertz, C., et al. (2015). The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447. doi: 10.1016/j.stem.2015.02.007
Ramos, A. D., Attenello, F. J., and Lim, D. A. (2016). Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci. Lett. 625, 70–79. doi: 10.1016/j.neulet.2015.12.025
Rani, N., Nowakowski, T. J., Zhou, H., Godshalk, S. E., Lisi, V., Kriegstein, A. R., et al. (2016). A Primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron 90, 1174–1188. doi: 10.1016/j.neuron.2016.05.005
Rapicavoli, N. A., Poth, E. M., and Blackshaw, S. (2010). The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 10:49. doi: 10.1186/1471-213X-10-49
Rapicavoli, N. A., Poth, E. M., Zhu, H., and Blackshaw, S. (2011). The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 6:32. doi: 10.1186/1749-8104-6-32
Rapo-Pylkko, S., Haanpaa, M., and Liira, H. (2017). A one-year follow-up study of chronic pain in community-dwelling older adults with and without neuropathic pain. BMC Geriatr. 17:152. doi: 10.1186/s12877-017-0537-x
Riva, P., Ratti, A., and Venturin, M. (2016). The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr. Alzheimer Res. 13, 1219–1231. doi: 10.2174/1567205013666160622112234
Roberts, T. C., Morris, K. V., and Weinberg, M. S. (2014). Perspectives on the mechanism of transcriptional regulation by long non-coding RNAs. Epigenetics 9, 13–20. doi: 10.4161/epi.26700
Sauvageau, M., Goff, L. A., Lodato, S., Bonev, B., Groff, A. F., Gerhardinger, C., et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2:e01749.
Scholz, J., Finnerup, N. B., Attal, N., Aziz, Q., Baron, R., Bennett, M. I., et al. (2019). The IASP classification of chronic pain for ICD-11: chronic neuropathic pain. Pain 160, 53–59. doi: 10.1097/j.pain.0000000000001365
Shaygan, M., Boger, A., and Kroner-Herwig, B. (2014). Neuropathic sensory symptoms: association with pain and psychological factors. Neuropsychiatr. Dis. Treat. 10, 897–906.
Shen, F., Zheng, H., Zhou, L., Li, W., Zhang, Y., and Xu, X. (2019). LINC00657 expedites neuropathic pain development by modulating miR-136/ZEB1 axis in a rat model. J. Cell Biochem. 120, 1000–1010. doi: 10.1002/jcb.27466
Sherstyuk, V. V., Medvedev, S. P., and Zakian, S. M. (2018). Noncoding RNAs in the regulation of pluripotency and reprogramming. Stem Cell Rev. 14, 58–70. doi: 10.1007/s12015-017-9782-9
Soler, M. D., Morina, D., Rodriguez, N., Sauri, J., Vidal, J., Navarro, A., et al. (2017). Sensory symptom profiles of patients with neuropathic pain after spinal cord injury. Clin. J. Pain 33, 827–834. doi: 10.1097/ajp.0000000000000467
St Laurent, G., Wahlestedt, C., and Kapranov, P. (2015). The Landscape of long noncoding RNA classification. Trends Genet. 31, 239–251. doi: 10.1016/j.tig.2015.03.007
Teixeira, M. J. (2009). Challenges in the treatment of neuropathic pain. Drugs Today 45(Suppl. C), 1–5.
Tochitani, S., and Hayashizaki, Y. (2008). Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem. Biophys. Res. Commun. 372, 691–696. doi: 10.1016/j.bbrc.2008.05.127
Treede, R. D., Rief, W., Barke, A., Aziz, Q., Bennett, M. I., Benoliel, R., et al. (2019). Chronic pain as a symptom or a disease: the IASP Classification of Chronic Pain for the International Classification of Diseases (ICD-11). Pain 160, 19–27.
van Hecke, O., Austin, S. K., Khan, R. A., Smith, B. H., and Torrance, N. (2014). Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain 155, 654–662. doi: 10.1016/j.pain.2013.11.013
Vance, K. W., Sansom, S. N., Lee, S., Chalei, V., Kong, L., Cooper, S. E., et al. (2014). The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 33, 296–311. doi: 10.1002/embj.201386225
Vranken, J. H. (2012). Elucidation of pathophysiology and treatment of neuropathic pain. Cent. Nerv. Syst. Agents Med. Chem. 12, 304–314. doi: 10.2174/187152412803760645
Wan, P., Su, W., and Zhuo, Y. (2017). The role of long noncoding RNAs in neurodegenerative diseases. Mol. Neurobiol. 54, 2012–2021. doi: 10.1007/s12035-016-9793-6
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. doi: 10.1016/j.molcel.2011.08.018
Wang, S., Xu, H., Zou, L., Xie, J., Wu, H., Wu, B., et al. (2016). LncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signal. 12, 139–148. doi: 10.1007/s11302-015-9488-x
Watson, J. C., and Dyck, P. J. (2015). Peripheral neuropathy: a practical approach to diagnosis and symptom management. Mayo Clin. Proc. 90, 940–951. doi: 10.1016/j.mayocp.2015.05.004
Wei, M., Li, L., Zhang, Y., Zhang, Z. J., Liu, H. L., and Bao, H. G. (2018). LncRNA X inactive specific transcript contributes to neuropathic pain development by sponging miR-154-5p via inducing toll-like receptor 5 in CCI rat models. J. Cell Biochem. [Epub ahead of print].
Williams, J. M., Beck, T. F., Pearson, D. M., Proud, M. B., Cheung, S. W., and Scott, D. A. (2009). A 1q42 deletion involving DISC1. DISC2, and TSNAX in an autism spectrum disorder. Am. J. Med. Genet. A 149A, 1758–1762. doi: 10.1002/ajmg.a.32941
Wu, P., Zuo, X., Deng, H., Liu, X., Liu, L., and Ji, A. (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 97, 69–80. doi: 10.1016/j.brainresbull.2013.06.001
Wu, S., Bono, J., and Tao, Y. X. (2019). Long noncoding RNA (lncRNA): a target in neuropathic pain. Expert Opin. Ther. Targets 23, 15–20. doi: 10.1080/14728222.2019.1550075
Xia, L. X., Ke, C., and Lu, J. M. (2018). NEAT1 contributes to neuropathic pain development through targeting miR-381/HMGB1 axis in CCI rat models. J. Cell Physiol. 233, 7103–7111. doi: 10.1002/jcp.26526
Xu, D. H., Chi, G. N., Zhao, C. H., and Li, D. Y. (2018). Long noncoding RNA MEG3 inhibits proliferation and migration but induces autophagy by regulation of Sirt7 and PI3K/AKT/mTOR pathway in glioma cells. J. Cell Biochem. [Epub ahead of print].
Xu, L., Zhang, Y., and Huang, Y. (2016). Advances in the treatment of neuropathic pain. Adv. Exp. Med. Biol. 904, 117–129.
Yan, X. T., Lu, J. M., Wang, Y., Cheng, X. L., He, X. H., Zheng, W. Z., et al. (2018). XIST accelerates neuropathic pain progression through regulation of miR-150 and ZEB1 in CCI rat models. J. Cell Physiol. 233, 6098– 6106.
Young, T. L., Matsuda, T., and Cepko, C. L. (2005). The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 15, 501–512. doi: 10.1016/j.cub.2005.02.027
Zhao, M., Wang, J., Xi, X., Tan, N., and Zhang, L. (2018). SNHG12 promotes angiogenesis following ischemic stroke via regulating miR-150/VEGF pathway. Neuroscience 390, 231–240. doi: 10.1016/j.neuroscience.2018.08.029
Zhao, Y., Li, S., Xia, N., Shi, Y., and Zhao, C. M. (2018). Effects of XIST/miR-137 axis on neuropathic pain by targeting TNFAIP1 in a rat model. J. Cell Physiol. 233, 4307–4316. doi: 10.1002/jcp.26254
Zimmerman, C. A., Leib, D. E., and Knight, Z. A. (2017). Neural circuits underlying thirst and fluid homeostasis. Nat. Rev. Neurosci. 18, 459–469. doi: 10.1038/nrn.2017.71
Keywords: chronic neuropathic pain, long non-coding RNAs (lncRNAs), neuronal associated disorders, DRG neurons, glioma cells
Citation: Wu W, Ji X and Zhao Y (2019) Emerging Roles of Long Non-coding RNAs in Chronic Neuropathic Pain. Front. Neurosci. 13:1097. doi: 10.3389/fnins.2019.01097
Received: 03 June 2019; Accepted: 30 September 2019;
Published: 18 October 2019.
Edited by:
Dinesh Upadhya, Manipal Academy of Higher Education, IndiaReviewed by:
Somasish Ghosh Dastidar, Duke University, United StatesGabriele Zanirati, Institute of the Brain of Rio Grande do Sul (InsCer), Brazil
Copyright © 2019 Wu, Ji and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Ji, eGlhb2p1bmppMTAxQDE2My5jb20=; Yang Zhao, emhhb3kxOTc5QDE2My5jb20=
 Wei Wu
Wei Wu Xiaojun Ji2*
Xiaojun Ji2*