Corrigendum: A Student's Guide to Neural Circuit Tracing
- 1Neurobiology of Vital Systems Node, Faculty of Medicine and Health Sciences, Macquarie University, Sydney, NSW, Australia
- 2The School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, United Kingdom
- 3CNRS, Hindbrain Integrative Neurobiology Laboratory, Neuroscience Paris-Saclay Institute (Neuro-PSI), Université Paris-Saclay, Gif-sur-Yvette, France
The mammalian nervous system is comprised of a seemingly infinitely complex network of specialized synaptic connections that coordinate the flow of information through it. The field of connectomics seeks to map the structure that underlies brain function at resolutions that range from the ultrastructural, which examines the organization of individual synapses that impinge upon a neuron, to the macroscopic, which examines gross connectivity between large brain regions. At the mesoscopic level, distant and local connections between neuronal populations are identified, providing insights into circuit-level architecture. Although neural tract tracing techniques have been available to experimental neuroscientists for many decades, considerable methodological advances have been made in the last 20 years due to synergies between the fields of molecular biology, virology, microscopy, computer science and genetics. As a consequence, investigators now enjoy an unprecedented toolbox of reagents that can be directed against selected subpopulations of neurons to identify their efferent and afferent connectomes. Unfortunately, the intersectional nature of this progress presents newcomers to the field with a daunting array of technologies that have emerged from disciplines they may not be familiar with. This review outlines the current state of mesoscale connectomic approaches, from data collection to analysis, written for the novice to this field. A brief history of neuroanatomy is followed by an assessment of the techniques used by contemporary neuroscientists to resolve mesoscale organization, such as conventional and viral tracers, and methods of selecting for sub-populations of neurons. We consider some weaknesses and bottlenecks of the most widely used approaches for the analysis and dissemination of tracing data and explore the trajectories that rapidly developing neuroanatomy technologies are likely to take.
Introduction
The relationship between structure and function is a central theme in the field of biology. In the same way that deciphering the crystal structure of DNA propelled research toward the mechanics of inheritance in the 20th century (Watson and Crick, 1953), it is widely believed that elucidation of the structural architecture of the brain will fundamentally alter neuroscience in the 21st (Rubinov and Bullmore, 2013; Lerner et al., 2016; Mikula, 2016). Here, we review the approaches used by contemporary neuroscientists to identify connectivity patterns between components of neural circuits, the trajectories that rapidly developing neuroanatomy technologies are likely to take, and some bottlenecks that may hinder this mission.
It is unknown at what point in history the brain was first recognized as the control center for the body. Although sometimes attributed to Hippocrates and Galen around 2,000 years ago, the association between traumatic brain injury and distinct functional deficits was clearly understood by Egyptian physicians 30 centuries before that (Stiefel et al., 2006), and archeological evidence indicates widespread neurosurgical practice in diverse cultures since time immemorial (Andrushko and Verano, 2008, reviewed by Kshettry et al., 2007; Moghaddam et al., 2015). What is clear is that the association between brain structure and function is a relatively recent realization: this consensus was only reached in the late 1800s after nearly 100 years of disagreement between those who, like influential French physiologist and child prodigy Marie-Jean-Pierre Flourens, believed that the brain, like the mind, was indivisible (Pearce, 2009), and those like Frantz Joseph Gall who proposed that the brain is composed of distinct functional compartments, and that the relative contribution of each is to some extent independent from the others [see Ferrier (1884) for contemporaneous review].1
At the same time that Broca and others were cementing the idea of cerebral localization at a macroscopic scale, the first histologists were using the microscope to discover the delicate structure of neural tissue and developing theories about the cellular basis of brain function. The “neuron theory” developed by Ramon y Cajal, Waldeyer-Hartz and others argued that a particular cell type, the neuron, was the functional unit of the nervous system, from which axons grew and relied upon for nutrition; that neurons were discontinuous and formed physical contacts at which communication occurred; and that information flowed across neurons in one direction, from the dendrite toward the axonal terminals (reviewed by Fishman, 1994; Llinas, 2003; Guillery, 2005; Lopez-Munoz et al., 2006). These central tenets are now universally accepted, with caveats (Guillery, 2005), but spawned an acrimonious and dogmatic battle between “reticularists” and “neuronists” that persisted from the late 1870s until the invention of the electron microscope and subsequent visualization of mammalian synapses in the 1950s.2
The Field of Connectomics
Since then, generations of neuroscientists have used progressively more selective labeling techniques and more powerful microscopes to reveal the patterns of synaptic connectivity that are thought to underlie the functional properties of neural circuits. This effort has given rise to the field of “connectomics,” a now standalone sub-discipline of neuroscience with the stated aim of understanding the “structural architecture of nervous system connectivity in all animals at all resolutions” (reviewed by Hagmann, 2005; Sporns et al., 2005; Swanson and Bota, 2010; Catani et al., 2013). Connectomics generates simplified circuit diagrams at macroscopic (brain-wide), mesoscopic (circuit level) or nanoscopic (synapse level) resolutions that classify neurons in terms of their connectivity to each other (Figure 1: discussed by Branco and Staras, 2009). However, these efforts have been hampered by both the technical complexity involved in accurately identifying synaptic connections and the sheer magnitude of the task: full connectomic reconstruction of the human brain would require the mapping of approximately 86 billion neurons (Azevedo et al., 2009; Herculano-Houzel, 2009) and the identification of the thousands of inputs and outputs that connect each one (Nimchinsky et al., 2002).
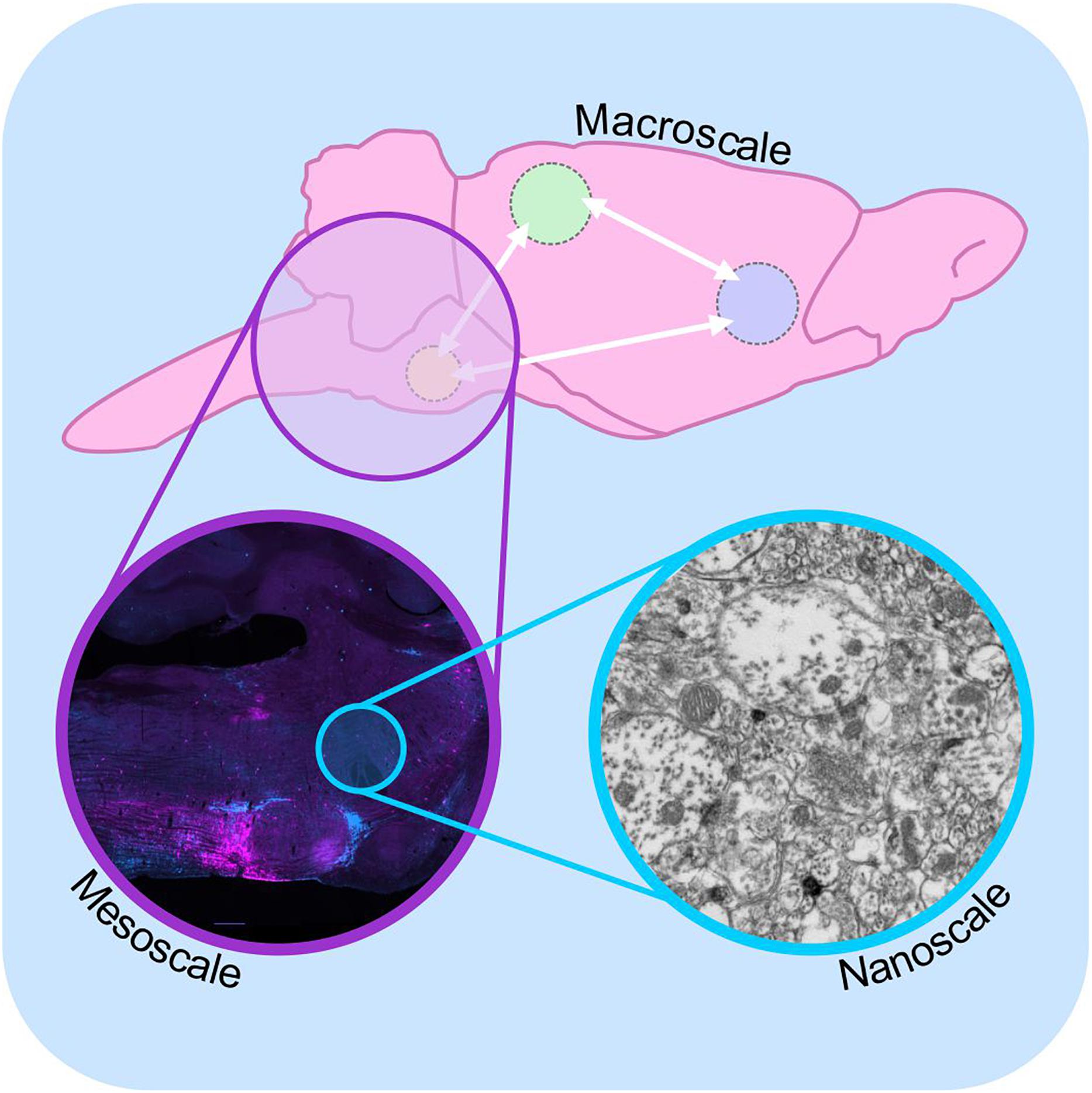
Figure 1. Macro-, meso-, and microscale connectomics. Schematic diagram illustrating connectome analysis at macroscale (top), mesoscale (purple inset), and nanoscale resolutions (blue inset). Macroscale approaches (top) examine communication between regions of the brain on a global level, using approaches such as diffusion-weighted magnetic resonance imaging. Mesoscale connectomics interrogates neural circuitry at the cellular level, employing light microscopy to map the distribution of synaptically linked neurons, in this case monosynaptic inputs to putative sympathetic premotor neurons (see Menuet et al., 2017, modified with permission). Nanoscale connectomics assesses individual synaptic contacts using electron microscopy. Nanoscale image shows electron micrograph of synapses and polysialyic acid immunoreactivity: see Bokiniec et al. (2017).
As a result of this complexity, the only entire connectomes thus far cataloged belong to tiny animals: the first (nearly) complete map of the entire nervous system of an individual species was made by White et al. (1986) from reconstructed serial electron micrographs of the roundworm Caenorhabditis elegans (later completed by Varshney et al., 2011), and more recently larval and adult Drosophila melanogaster (Ohyama et al., 2015; Zheng et al., 2018) and the larval sea-squirt Ciona intestinalis (Ryan et al., 2016). The “nanoscale” approach used to map these connectomes offers comprehensive knowledge of every neuronal connection, and is well-suited to components of relatively small local circuits such as in the retina, in which the inputs and outputs are understood (Briggman et al., 2011; Denk et al., 2012).
However, ultrastructural strategies are poorly suited to the interrogation of large or dispersed circuits because of the enormous investments of time and infrastructure required to acquire and handle the data (Lichtman and Denk, 2011; Wanner et al., 2015). These technical limitations seem unlikely to be resolved in the foreseeable future; despite recent innovations such as the development of serial block-face and Focused Ion Beam electron microscopy, which have reduced the acquisition time for a cubic millimeter of tissue from ∼18 to ∼1.5 years (Wanner et al., 2015; Xu et al., 2017), obstacles to the analysis and even the storage of high resolution microscopy data remain. For example, the raw dataset for a single human brain would require ∼175 exabytes of storage space, costing 2–8 billion Euros (Mikula, 2016). By comparison, the entire storage capacity of the planet in 2011 was ∼295 exabytes (Hilbert and Lopez, 2011).
These limitations have provided a stimulus for the development of genetically modified viral tracers that can be used to identify components of a given neuronal circuit without requiring direct visualization of synaptic contacts. The major advantages of this approach are its compatibility with light microscopy, greatly reducing the imaging, analysis and storage burden inherent to ultrastructural analysis, and its applicability to mapping networks that are dispersed throughout the brain.
Anterograde and Retrograde Tracers
Early tracing approaches involved the physical or electrical lesion of a region of interest, which rendered the degenerating axons differentially susceptible to impregnation with metallic silver (Hoff, 1932). Although crude, this approach was eventually refined to the point where terminal boutons could be resolved (Glees, 1946; Nauta, 1952; Fink and Heimer, 1967). Visualization of Wallerian degeneration was replaced by approaches that did not require destructive lesions, relying instead on the axonal transport of injected/applied materials from the site of injection to either the cell body (retrograde tracers) or axonal processes (anterograde tracers; Figure 2). The earliest of these tracers were radiolabeled amino acids such as tritiated leucine and proline that were injected into neural tissue then incorporated into polypeptides in the soma and transported to axons and terminal processes where they were identified by autoradiography (Grafstein, 1967; Cowan et al., 1972). These tracers were soon surpassed by materials that could be detected with conventional light microscopy via histological or immunohistochemical (IHC) processing, intrinsic fluorescence, or conjugation with a fluorophore or enzymatically active probe. This variety of tracer constitutes what are now referred to as “conventional tracers” (in contrast to viral tracers). The variety and utility of conventional tracing techniques are broad and have been extensively reviewed elsewhere (Köbbert et al., 2000; Vercelli et al., 2000; Lanciego and Wouterlood, 2011): here, we provide a simple overview of some of the most popular conventional tracers for comparison with the viral tracers discussed in upcoming sections.
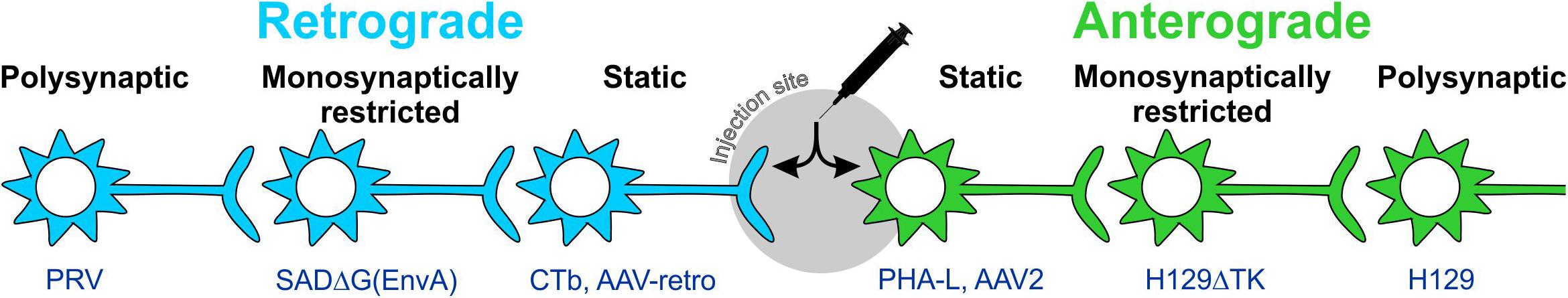
Figure 2. Anterograde and retrograde labeling with static and trans-synaptic tracers. Tracers are categorized as anterograde or retrograde based on their direction of travel within neurons. Anterograde tracers (green) are taken up by neuronal cell bodies at the injection site and travel down the axon to terminal processes. Retrograde tracers (blue) are taken up by terminals and travel back to the cell body. Static tracers remain within the first neurons they enter, while other tracers can spread trans-synaptically and may or may not be monosynaptically restricted. CTb: cholera toxin B subunit; H129ΔTK: tyrosine kinase-deleted H129 herpes; PHA-L: Phaseolus vulgaris-leucoagglutinin; PRV: pseudorabies virus; SADΔG(EnvA): glycoprotein-deleted EnvA-pseudotyped rabies.
Conventional (Mainly) Retrograde Tracers
The glycoprotein and enzyme horse radish peroxidase (HRP) was found in the early 1970s to be an effective retrograde tracer, having been taken up into neurons non-selectively by passive endocytosis (Kristensson and Olsson, 1971; LaVail and LaVail, 1972; Köbbert et al., 2000; Vercelli et al., 2000). HRP is visualized as it catalyzes, together with hydrogen peroxide, the oxidation of chromogenic substrates such as 3,3′-diaminobenzidine and tetramethylbenzidine. The resultant staining is limited to the cell soma and primary dendrites (Köbbert et al., 2000) and, on its own, HRP demonstrates a relatively low sensitivity due to inefficient uptake by neurons at the injection site. However, conjugation of HRP to the plant lectin wheat germ agglutinin (WGA), itself a neuronal tracer, significantly improved both uptake and transport within neurons (Staines et al., 1980; Köbbert et al., 2000). WGA binds to N-acetylglucosamine and the plasma membrane-bound sugar sialic acid, and is rapidly actively transported in both the anterograde and retrograde directions, providing more extensive (but not complete) labeling of the neuron compared to HRP (Schwab et al., 1978; Dumas et al., 1979; Levy et al., 2017). WGA-HRP conjugates are also capable of limited trans-synaptic travel (Goshgarian and Buttry, 2014; Sillitoe, 2016), introducing some ambiguity to the interpretation of data. WGA is now available in a fluorophore-conjugated preparation, eliminating the need for immunohistochemical processing for visualization.
Cholera toxin subunit B (CTb) was introduced as a retrograde tracer in 1977 (Stoeckel et al., 1977; Vercelli et al., 2000). Trojanowski et al. (1981, 1982) then sought to improve the sensitivity of HRP using CTb-HRP conjugates, and found that when the number and detail of labeled neurons was compared, CTb-HRP significantly out-performed free HRP. They attributed this to the GM1 ganglioside (sugar) mediated uptake of CTb in comparison to the non-specific endocytosis of HRP (Trojanowski et al., 1981; Trojanowski et al., 1982; Lanciego and Wouterlood, 2011). Unconjugated CTb can be detected via immunohistochemistry, however, the development of CTb-conjugated fluorophores (Conte et al., 2009a, b) or even magnetically opaque labels visible to MRI (Wu et al., 2011) has extended the scope of CTb. The signal strength of fluorescently conjugated CTb, its rapid transport (2–7 days), low toxicity, and ease of use also makes it suitable for identification of neurons for subsequent electrophysiological recordings in vitro (Korim et al., 2014; Bou Farah et al., 2016) or in vivo (Yamashita and Petersen, 2016), and has elevated fluorescently conjugated CTb variants as a go-to retrograde tracer for many researchers (Parker et al., 2015; Zhao et al., 2017), although it should be noted that, like most conventional tracers, CTb transport is to some extent bidirectional (Noseda et al., 2010). In our experience, CTb-conjugated fluorophores can suffer quenching when used in conjunction with in situ hybridization, and as the fluorophore prevents binding of anti-CTb antibodies, quenched signal cannot be boosted with anti-CTb IHC, so for dual labeling of mRNA and CTb the unconjugated form is preferable.
A number of inorganic tracers are also widely used. Hydroxystilbamidine (FluoroGoldTM) is a fluorescent inorganic compound directly visible with fluorescence microscopy. Following uptake into nerve terminals by fluid phase endocytosis, FluoroGoldTM is transported retrogradely to cell bodies in vesicles and accumulates in the cytoplasm, where it remains detectable for months (Köbbert et al., 2000; Lanciego and Wouterlood, 2011). The intense, bleach-resistant labeling achieved using FluoroGoldTM, and the availability of anti-FluoroGoldTM antibodies for further amplification, has resulted in it becoming the “gold-standard” tracer in rodents, against which newly developed tracers are compared (Lanciego and Wouterlood, 2011; Tervo et al., 2016).
Other noteworthy inorganic fluorescent tracers are Fast Blue, Diamidino Yellow, True Blue and the carbocyanines DiI and DiO (Bentivoglio et al., 1980; Bonhoeffer and Huf, 1980; Kuypers et al., 1980; Puigdellivol-Sanchez et al., 1998; Schofield, 2008; Yu et al., 2015). Although no longer widely used for tracing projections in the central nervous system, they (along with organic tracers such as CTb, WGA, etc.) remain popular choices for the identification of autonomic, motor or sensory innervation of peripheral targets, into which they can be injected in relatively large volumes (Furukawa et al., 2008; Zele et al., 2010; Yu et al., 2015; Pidsudko et al., 2019; Rytel et al., 2019). Interestingly, because they are not dependent on active transport, inorganic dyes may also be used for identification of projections in fixed tissue post mortem. Typically applied as crystals to the surface of formalin-fixed tissue blocks, highly lipophilic dyes such as DiI (red) and DiO (green) move evenly throughout cells in both anterograde and retrograde directions via the lipid portion of neuronal membranes, resulting in complete labeling of the soma and dendritic tree (Thanos et al., 1991, 1992; Köbbert et al., 2000; Boon et al., 2019; Trivino-Paredes et al., 2019). However, the lipophilic nature of carbocyanine dyes makes them difficult to use in conjunction with standard IHC protocols that use lipid-solubilizing detergents to facilitate antibody penetration (Elberger and Honig, 1990; Matsubayashi et al., 2008).
Fluorescent latex microspheres 30–90 nm in diameter (RetrobeadsTM) were recognized as retrograde tracers in the mid-1980s (Katz et al., 1984; Katz and Iarovici, 1990) and, like FluoroGoldTM, are highly resistant to fading and offer long-term labeling. Although somewhat difficult to use, based on their tendency to clump in the injection pipette, latex beads offer stable fluorescence with minimal diffusion from the injection site, exclusive retrograde travel with minimal entry to undamaged fibers of passage, and are non-toxic to neurons so can be used in long duration experiments (Köbbert et al., 2000; Vercelli et al., 2000; Lanciego and Wouterlood, 2011). Their distinctive bright punctate appearance makes them easy to distinguish, meaning they can be mixed with an anterograde tracer of the same color [e.g., to simultaneously identify retro- and anterograde projections from a single brain region (Turner et al., 2013)].
Conventional (Mainly) Anterograde Tracers
Phaseolus vulgaris-leucoagglutinin (PHA-L) is one of the earliest and most widely used anterograde tracers (Gerfen and Sawchenko, 1984; Köbbert et al., 2000) and, like WGA, PHA-L binds to membrane bound carbohydrates to gain cell entry (in this case N-acetyl D-glucosamine and mannose). PHA-L is detected using IHC, revealing detailed cell morphology, including axon terminal branches to the level of terminal boutons (Lanciego and Wouterlood, 2011; Wouterlood et al., 2014). PHA-L requires longer post-injection survival times to achieve transport compared other conventional tracers, typically 10 to 20 days (Vercelli et al., 2000).
Emerging in the mid-1980s, dextran-amines (DAs) were rapidly adopted and remain widely used as conventional axonal tracers (Gimlich and Braun, 1985; Glover et al., 1986; Brandt and Apkarian, 1992; Veenman et al., 1992; Wouterlood et al., 2014). DAs enter injured neurons at the injection site and spread evenly throughout the entire neuron via diffusion, resulting in a Golgilike level of staining detail (Glover et al., 1986; Fritzsch, 1993; Glover, 1995; Köbbert et al., 2000; Lanciego and Wouterlood, 2011; Wouterlood et al., 2014).
Despite the common perception that DAs are preferential anterograde tracers, many studies indicate bidirectional travel (Schmued et al., 1990; Fritzsch, 1993; Glover, 1995; Zhang et al., 2017), including the original description of their axonal transport by Glover et al. (1986). Their retrograde capabilities have been exploited both for conventional tracing (Sivertsen et al., 2014, 2016; Lunde et al., 2019) and for delivery of calcium-sensitive indicators for optical recording of neurons selected by axonal trajectory (O’Donovan et al., 1993; McPherson et al., 1997).
There is a perception that the molecular weight of DA-conjugates contributes to their directional selectivity, with smaller molecules exhibiting superior performance as a retrograde tracer (Reiner et al., 2000; Lanciego and Wouterlood, 2011). However, the influence, if any, of molecular weight on directional specificity is probably overstated, and may instead reflect differences in speed of transport, which is distinctly faster for smaller compounds (Fritzsch, 1993), combined with differences in volume of synaptic terminals compared to cell bodies (Glover, personal communication).
Like CTb, fluorophore-labeled dextran amine variants are now widely used instead of or in addition to biotinylated versions that require histological processing for visualization, and we and others have used tetramethylrhodamine-conjugated dextran for juxtacellular labeling during electrophysiological recordings (Noseda et al., 2010; Dempsey et al., 2015).
Limitations of Conventional Tracers
Despite their ongoing popularity, the major limitations of conventional tracers are worthy of consideration:
(1) Conventional tracers can be taken up by fibers of passage (Dado et al., 1990; Chen and Aston-Jones, 1995; Conte et al., 2009a), which can lead to incorrect identification of projections. [Notably, canine adenovirus (CAV) can also be taken up by fibers of passage (Schwarz et al., 2015)].
(2) The spread of many conventional tracers around the injection site results in intense and diffuse labeling that may reflect deposition in the extracellular matrix or take-up by neurons or glia. Such non-specific labeling makes it difficult to reliably identify labeled neurons within ∼1 mm of the injection site. Thus the historical use of conventional tracers has probably overemphasized the relative significance of distant inputs/outputs compared to those originating from local interneurons; contemporary connectomic studies indicate that long-distance projections are relatively rare compared to short-distance connections (Oh et al., 2014; Henriksen et al., 2016; van den Heuvel et al., 2016; Dempsey et al., 2017).
(3) Tracer uptake relies predominantly on sugars that are located on the glycocalyx of most, if not all neurons, or on common mechanisms such as endocytosis. Consequently, restricted uptake by functionally or neurochemically/genetically homogeneous neuronal populations is not possible.
(4) The direction of axonal transport is rarely exclusive, which complicates circuit analysis; the archetypal retrograde and anterograde tracers, CTb and BDA respectively, both label axons traveling in the “wrong” direction (Luppi et al., 1987; Schmued et al., 1990; Fritzsch, 1993; Glover, 1995; Angelucci et al., 1996; Noseda et al., 2010; Zhang et al., 2017).
Viral Tracers
Recombinant viral vectors that drive the expression of fluorescent “reporter” proteins in transduced neurons have been widely adopted by neuroscientists because of their directional specificity, the high (in most cases permanent) levels of reporter expression obtained, and the absence of transduction of fibers of passage (comprehensively reviewed by Callaway, 2005; Luo et al., 2008; Betley and Sternson, 2011; Nassi et al., 2015). Here, we will examine some widely used variants and consider the extent to which they address the limitations of conventional tracers.
Viral tracers may be divided into two distinct classes: static vectors that remain locked within the targeted cell population and essentially function like conventional tracers, which are usually replication-deficient, and vectors that spread through linked circuits via trans-synaptic travel, which are almost always replication-competent. Each class contains vectors that can be used to label selectively in an antero- or retrograde direction.
Static Viral Tracers
Recombinant vectors derived from human immunodeficiency virus (HIV), herpes simplex virus type 1 (HSV-1), human and canine adenoviridae (Ad), adeno-associated virus (AAV), semliki forest virus, sindbis virus (SIN), and rabies have all been developed as alternatives to conventional chemical tracers (Chamberlin et al., 1998; Furuta et al., 2001; Wickersham et al., 2007a; Junyent and Kremer, 2015; Lerner et al., 2016; Jia et al., 2017; Farmer et al., 2019). A detailed review of the biology of each is beyond the scope of the current article; the following resources provide useful overviews of the most commonly used replication deficient viral vectors (Ad: Akli et al., 1993, AAV: Kuo et al., 1995; Drouin and Agbandje-McKenna, 2013; Junyent and Kremer, 2015, HIV: Lundberg et al., 2008; Murlidharan et al., 2014, HSV-1: Neve, 2012). HIV, HSV, Ad, SIN, and AAV vectors differ in their maximum genetic payload, whether transgenes are integrated into the host genome, and the onset and duration of gene expression, but share common features: the wild-type virus is modified to remove genes required for viral replication and replaced with a genetic sequence that encodes a reporter protein under the control of a ubiquitous (e.g., cytomegalovirus, CaMKII), pan-neuronal (e.g., neuron-specific enolase, synapsin) or cell-type specific (e.g., PRSx8, GAD1) promoter or, if used in transgenic animals (reviewed by Wang et al., 2011), genetically restricted expression systems such as Cre-LoxP or FLP-FRT (Blomer et al., 1997; Hwang et al., 2001; Jakobsson et al., 2003; Schnutgen et al., 2003; Gofflot et al., 2011; Liu et al., 2013; Fenno et al., 2014; Luo et al., 2018). In contrast, glycoprotein-deleted rabies is a static retrograde vector that retains its capacity for replication, but has been modified so that it can no longer spread trans-synaptically (Larsen et al., 2007; Wickersham et al., 2007a). This has the advantage of self-amplification, resulting in very high levels of reporter expression, but results in cytotoxicity within a few weeks of infection and, as transcription is not promoter-dependent, cannot be targeted to particular cell types without further genomic alterations (discussed below). Furthermore, as the rabies genome is RNA based, strategies for selective recombination within subpopulations of infected neurons (e.g., Cre-LoxP) are not possible.
The mode of use of static viral tracers is similar to conventional tracers; vectors are injected in small volumes into the region or organ of interest under anesthesia, and experimental animals are allowed to recover with appropriate post-operative care. Following a period sufficient for transgene translation, the animal is euthanized and perfused with fixative, and the CNS removed for histological processing, which may include IHC amplification of reporter signal, followed by imaging. The major differences compared to conventional tracers lie in the duration of recovery (although reporter expression is visible within 12 h of HSV or rabies injections, protein transcription is typically optimal 10–20 days after transduction by AAV and lentiviral vectors), the volume of injectate, which is typically larger for conventional tracers (300–1000 nl vs. 20–100 nl for CNS injections, 1–5 μl for peripheral injections into target organs), and the level of biological containment required. When injected into the periphery, a further consideration is the age of the animal; in our experience peripheral injections of rabies and HSV-1 vectors do not result in detectable neuronal labeling when administered to mice older than post-natal day 7, perhaps reflecting the immaturity of the innate and adaptive immune responses in the early post-natal period.
Like conventional tracers, cell entry is mediated via interactions between glycans or proteins expressed on the cell membrane and components of the viral vector (Lykken et al., 2018). The tendency of a given vector to label in an anterograde or retrograde direction therefore reflects the cellular distribution of cognate receptors to its particular surface proteins; binding with receptors expressed on the soma results in anterograde labeling (e.g., AAV serotype 2, 5, 7, 8, and rh.10, human adenovirus), whereas binding with receptors preferentially expressed at the axon terminal results in retrograde labeling (e.g., HSV-1, rabies and canine adenovirus) (Frampton et al., 2005; Berges et al., 2007; Salinas et al., 2009; Penrod et al., 2015; Castle et al., 2016; Hirschberg et al., 2017; Sathiyamoorthy et al., 2017; Farmer et al., 2019). Note that viral titer may also play a role in the direction of travel; human adenovirus becomes a retrograde tracer at high titers as the recruited immune response limits their efficacy (Howorth et al., 2009). Perhaps unsurprisingly, variation of viral surface proteins by natural selection or human manipulation can alter their affinity for cellular binding partners, changing the tropism for different cell types (e.g., neuronal versus non-neuronal) or different neuronal compartments (e.g., pre-synaptic versus post-synaptic) (Kanaan et al., 2017; Lykken et al., 2018). The diverse capsid sequences found in the dozen or so naturally occurring primate AAVs that have been developed as vectors for gene transfer exhibit dramatically different tropisms for different cell types (reviewed by Drouin and Agbandje-McKenna, 2013; Castle et al., 2016) and hosts (Watakabe et al., 2015). When injected into the rodent central nervous system, most result in anterograde labeling, but the AAV1, 5 and 9 serotypes exhibit both anterograde and retrograde transport, with the degree of retrograde tropism varying according to the region studied and construct used (Rothermel et al., 2013; Castle et al., 2016). Altered directional tropism can also be conferred by pseudotyping viral vectors with chimeric envelope proteins from other viruses; this approach has been used with success to switch between anterograde and retrograde tropism in HIV-1 and vesicular stomatitis virus vectors, and to generate rabies variants that can gain cellular access at the soma instead of the axon terminal (Blomer et al., 1997; Wickersham et al., 2007b; Kato et al., 2011; Beier et al., 2011, 2013).
Many different viruses have been developed as potential vehicles for gene delivery, but in recent years AAV vectors have emerged as front-runners (Kanaan et al., 2017). Although the maximum size of the AAV payload is limited compared to lentiviral, adenoviral and HSV vectors, they are easy to work with, have low toxicity and immunogenicity, and result in rapid (∼14 days) and permanent transgene expression in post-mitotic cells such as neurons (reviewed by Chamberlin et al., 1998; Betley and Sternson, 2011; Sun and Schaffer, 2018). These properties have made AAV vectors attractive candidates in gene therapy for cancer, metabolic, and neurological diseases (Weinberg et al., 2013; O’Connor and Boulis, 2015; Santiago-Ortiz and Schaffer, 2016; Grimm and Buning, 2017), stimulating investment in AAV production and targeting technologies that are likely to further accelerate their development. AAV capsids have been manipulated to improve their transduction ability for both gene therapy and neuroanatomical applications (Tervo et al., 2016; Chan et al., 2017). Of particular relevance to the neuroscientist is the recent development of a synthetic AAV capsid that drives retrograde transduction with particularly high efficiency and selectivity (Tervo et al., 2016). This capsid, known as AAV-retro, results in transduction profiles that closely resemble conventional retrograde tracers such as FluoroGoldTM (Tervo et al., 2016), is compatible with small injection volumes, and is not associated with significant toxicity (Sun et al., 2019). These features, and the fact that it can be made in any viral production facility that produces conventional AAV vectors, indicate that the popularity of AAVs will continue to increase.
The AAV-retro variant, and other synthetic AAV capsids, were made using in vivo directed evolution, a process in which error-prone PCR is used to introduce mutations to the cap gene (Drouin and Agbandje-McKenna, 2013). Mutant viral particles are then injected into organoids or animal models and the tissue of interest is harvested, from which AAV variants are isolated and sequenced to identify mutations with the desired tropism (Kotterman and Schaffer, 2014; Sun and Schaffer, 2018). In producing AAV-retro, AAV variants with different cap mutations were injected into either the substantia nigra or the cerebellar cortex and harvested from the striatum or inferior olive, respectively, revealing candidate capsid sequences with high selectivity for retrograde transport (Tervo et al., 2016). Given the many dozens of unstudied capsid subtypes that lie within the 13 major groups of primate AAV (Gao et al., 2004), the many thousands of potential capsids that lie within putative ancestral primate AAV libraries (Santiago-Ortiz et al., 2015), and the perhaps millions of capsid variants yet to be isolated and characterized from non-primate AAVs (Smith et al., 2016), it seems likely that AAV capsids with useful tropisms will continue to be discovered and developed for neuroscience applications. This may even lead to development of capsid variants with selectivity for particular neuronal cell types, bypassing the need for recombinase-driving transgenic animals or cell-type specific promoters, the Achilles’ heel of AAV due to its small payload size (although the selectivity of synthetic AAV promoters is also rapidly improving: see Jüttner et al., 2019), with obvious benefits to both research and gene therapy applications.
Identification of Synaptic Contacts
Irrespective of whether anterograde labeling is achieved by conventional tracers or vector-mediated reporter expression, the identification of terminal appositions under light microscopy remains a challenge because of their small size (Burette et al., 2015). The development of vectors that selectively label presynaptic terminals (e.g., synaptophysin-GFP or synaptophysin-mRuby) makes disambiguation of terminals from axons and fibers easier (Li et al., 2010; Oh et al., 2014; Beier et al., 2015; Lerner et al., 2015), but the fact remains that <50% of “close appositions” constitute functional synapses (Pilowsky et al., 1992; Descarries and Mechawar, 2000; da Costa and Martin, 2011), and so their presence needs to be interpreted with caution. In recent years a number of elegant approaches have been developed to circumvent this limitation, the ultimate of which is GRASP (GFP Reconstitution Across Synaptic Partners), a dual vector system where each vector drives the expression of one half of a GFP-derived dimer that only becomes fluorescent when both halves bind. When presynaptic neurons are targeted with one and post-synaptic neurons with the other, the only place at which the two components can become physically close enough to bind is the synaptic cleft, and so GFP fluorescence denotes synaptic contact (Feinberg et al., 2008; Kim et al., 2011). The interested reader is directed to recent reviews that considers alternative methods for synaptic complementation (Wickersham and Feinberg, 2012; Luo et al., 2018).
Trans-Synaptic Circuit Tracing
Replication-deficient viral vectors that drive reporter expression are ideal anterograde tracers, because they can target genetically restricted cells in circumscribed brain regions and result in unambiguous labeling of the entire neuron and/or its synaptic terminals. However, for the reasons outlined above, such fine detail is not easy to extract across whole brains and, moreover, anterograde labeling experiments are generally conducted in an effort to determine the post-synaptic targets of a given population of neurons, not to map the distribution or number of synaptic contacts arising from them. For the purposes of identifying post-synaptic targets, a better solution would be a tracer that labels the entire post-synaptic cell, which can be unambiguously and rapidly detected using light microscopy under relatively low magnification instead of the high-resolution methods required to reliably detect individual synapses. One approach that can achieve this exploits the trans-synaptic trafficking of Cre recombinase that occurs when expressed at high levels by replication-deficient viral vectors. Zingg et al. (2017) found that AAV-mediated Cre recombinase expressed in presynaptic neurons could drive Cre-dependent transgene expression in post-synaptic neurons, and that the degree of post-synaptic transgene expression, which presumably reflects the degree of Cre recombinase trafficking, varies depending on AAV serotype. This provides a useful tool for identification and manipulation of post-synaptic neurons, although the approach is somewhat confounded by the partial retrograde tropism of the AAV1 and AAV9 serotypes (Rothermel et al., 2013; Castle et al., 2016), which coincidentally produced the highest Cre recombinase trafficking (Zingg et al., 2017), leading to ambiguity over whether tagged neurons represent post- or pre-synaptic network components.
Another strategy that avoids having to identify individual synapses is to infect target populations with a replication-competent neurotrophic virus and to trace circuit components via the trans-synaptic infection of viral progeny (comprehensively reviewed by Nassi et al., 2015). This approach, initially developed using wild-type viruses that exhibit selectively retrograde (rabies, pseudorabies) or anterograde tropism (HSV-1 strain H129) (Strack et al., 1989b; Zemanick et al., 1991; Card and Enquist, 1995; Ugolini, 1995; Aston-Jones and Card, 2000; Kelly and Strick, 2000), represents a significant advance over static tracers in the sense that labeling can be safely interpreted as indicating synaptic contact. However, tracing using intact replication-competent viruses suffers two major drawbacks: first, it is never clear whether connections are mono- or polysynaptic, because the replicating virus continues to propagate along the synaptic hierarchy. Second, wild-type replication-competent viruses cannot be used to selectively target groups of neurons embedded in the CNS; direct injection of these vectors gives rise to non-selective infection, and so their use is best restricted to circuits that begin (i.e., sensory) or end (i.e., motor) in the periphery.
Anterograde Trans-Synaptic Tracing via HSV-1 Strain H129
Investigators have attempted to sidestep the drawbacks listed above by modifying viral constructs to alter their tropisms (so that target populations of neurons can be selectively infected following central injection), by comparing the results of viral and conventional tracing studies (to gain insight into which projections are likely to represent monosynaptic connections), and by performing detailed time-course studies to map the likely hierarchy of neuronal connections. All of these approaches have been performed with H129, a strain of HSV-1 that was originally isolated from a patient who died from acute necrotizing encephalitis (Dix et al., 1983) and is notable for its predominantly anterograde direction of trans-synaptic travel (Archin and Atherton, 2002; Wojaczynski et al., 2015). H129 has been used for the mapping of neural circuits for over 20 years (Sun et al., 1996; Garner and LaVail, 1999; Labetoulle et al., 2000); it exhibits rapid replication and spread, with reporter-driving variants producing detectable fluorescence at the site of primary infection within 24 h of innoculation. In our hands, H129 produces detectable trans-synaptic labeling of visual pathways within 48 h of intraocular injection, and the progression of labeling suggests another synapse is “jumped” every 48 h thereafter, although signal strength is variable and benefits from IHC amplification.
As infection progresses, H129 causes cytopathic changes in cell morphology and, if left unchecked, eventual death of the animal, imposing ethical and practical constraints on the duration of experiments (Lo and Anderson, 2011; McGovern et al., 2012b, Wojaczynski et al., 2015). Inoculated rodents begin to show signs of neurological disease that may include hemiparesis, ataxia and drastic weight loss, which varies in its onset time, severity, and prevalence depending on the site of initial infection (Archin and Atherton, 2002; Rinaman and Schwartz, 2004; McGovern et al., 2012a). Interestingly, a significant proportion of rodents inoculated with H129 never develop any detectable infection: in our hands intraocular infection fails to “take” in almost half of cases, similar to the 36% failure rate reported by Lo and Anderson (2011), who also found a surprisingly high failure rate following direct brain injection (18%). The transient nature of the primary viral infection and host defense mechanisms may contribute to variability in uptake and differences in reported strength of labeling.
The specificity of H129 for tracing subpopulations of neurons has been improved by modification of the viral genome: Lo and Anderson (2011) produced a H129 strain dependent on Cre recombinase for replication and transcription of a red reporter. This was achieved by the insertion of a floxed STOP cassette upstream of the tdTomato and TK genes (the TK gene being necessary for replication). Infection of Cre-containing neurons therefore results in permanent removal of the STOP cassette, restoration of viral replication and transcription of the reporter gene, whereas initial infection is unable to take hold in neurons that do not synthesize Cre recombinase. Reporter expression therefore indicates that a neuron either contains Cre recombinase or lies downstream from a Cre-containing neuron, allowing the identification of post-synaptic circuit components. A similar strategy was devised by McGovern et al. (2015b), in which the H129 genome was modified to encode a floxed GFP lying upstream of a red reporter gene (H129-HCMV-loxP-EGFP-HCMVpA-loxP-tdTomato-SV40pA, Figure 3). In this case the GFP gene is expressed in naïve neurons but is excised by Cre, resulting in a switch in reporter color in Cre-synthesizing neurons and their (polysynaptic) downstream partners. These modifications allow H129 to be targeted to subpopulations of central neurons, allowing identification of connected circuits, but these variants are still compromised by ambiguity regarding the number of synapses that link labeled neurons to the originally targeted population and the systemic illness that results from unrestricted central infection. These limitations have been recently overcome by deletion of a gene required for viral replication, tyrosine kinase (TK), which arrests the spread of H129. When supplied by a helper vector in trans, TK transiently restores the ability of H129 to replicate and to spread into monosynaptically connected neurons. As these do not contain TK, the H129 becomes trapped and so viral reporter expression can be safely interpreted as denoting monosynaptic spread (Zeng et al., 2017; Li et al., 2018).
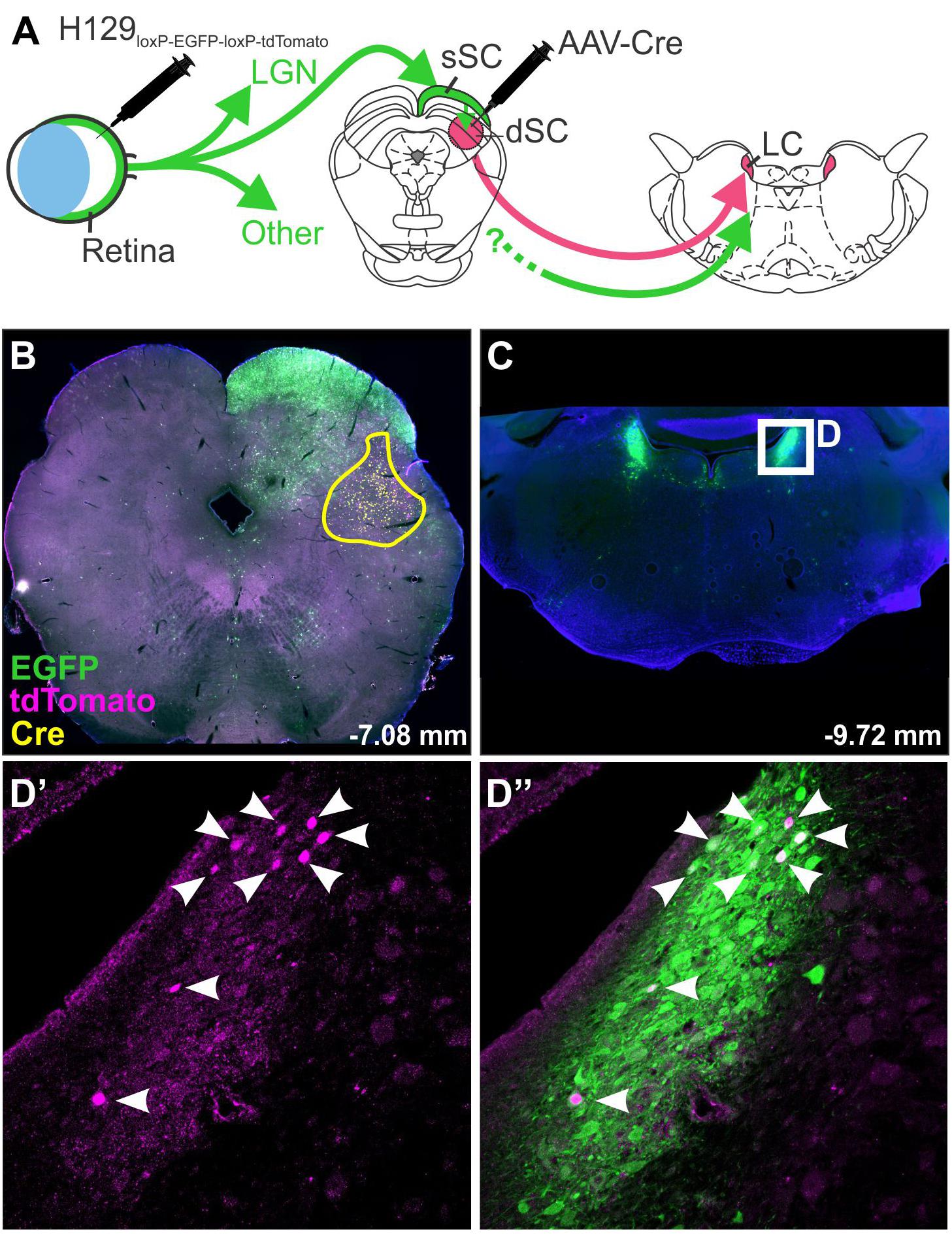
Figure 3. H129 tracing of a polysynaptic pathway. Demonstration of H129 infection of a polysynaptic pathway from the retina, through the deep superior colliculus (dSC), to the locus coeruleus (LC). (A) Intravitreal injections of H129-HCMV-loxP-EGFP-HCMVpA-loxP-tdTomato-SV40pA initially infected retinal ganglion cells, driving EGFP expression in retinorecipient nuclei and downstream neurons. When AAV-Cre was injected into the dSC prior to H129 infection (yellow boundary in B) the genomes of H129 virions passing through the dSC were cleaved, resulting in tdTomato expression in post-synaptic neurons. Red neurons in downstream regions, such as LC (Low power micrograph in C, magnified view in D’), can therefore be interpreted as being part of a pathway passing through the dSC. Co-expression of tdTomato and EGFP was common (see merged image, D”) and may indicate incomplete H129 cleavage or multiple pathways converging on the same region. LGN: lateral geniculate nucleus; sSC: superficial layer of the superior colliculus.
Retrograde Poly-Synaptic Viral Tracers
Rabies and pseudorabies virus (PRV, a member of the alphaherpes virus family) were developed as retrograde trans-synaptic tracers in the latter decades of the 20th century (Strack et al., 1989a; Ugolini, 1995). PRV, HSV-1 and other alphaherpes viridae rely on interactions between the viral envelope glycoprotein complex and cell surface receptors to gain intracellular access (reviewed by Frampton et al., 2005; Sathiyamoorthy et al., 2017); in neuronal tissue, nectin-1, a synaptic adhesion molecule that is predominantly expressed on the presynaptic membrane and colocalizes well with synaptophysin, is the primary mediator of cell entry and infection, without which little neuronal infection occurs (Geraghty et al., 1998; Campadelli-Fiume et al., 2000; Mizoguchi et al., 2002; Takai et al., 2008). In addition to the nectin-1- pathway, it is understood that direct cell-to-cell transfer can occur via non-synaptic mechanisms, and that direction of transport may be bi-directional, particularly in wild-type PRV strains, although this confound is reduced in the PRV-Bartha laboratory strain (Card et al., 1998, 1990, 1991, 1999; Pickard et al., 2002; Ugolini, 2011). As a result, PRV experiments are often limited to short incubations and low viral doses in an attempt to reduce the incidence of non-synaptic release of viral particles via cytotoxic cell death.
PRV spreads rapidly, causing observable transgene expression within 6 h in vitro, 1 day in vivo, and demonstrating trans-synaptic travel within 2 days in vivo (Card et al., 1990; Kobiler et al., 2010). The speed and directional selectivity of PRV has made it the historical tracer of choice for both peripheral and central tracing studies (Martin and Dolivo, 1983; Strack et al., 1989a; Jansen et al., 1995; Smith et al., 1998; DeFalco et al., 2001) although, due to its polysynaptic nature, PRV poses the same challenge as H129 – the differentiation of mono- and polysynaptic connections. As with H129, time-course analyses have sought to address this issue, as well as co-injection of PRV with conventional tracers to identify regions sequentially labeled along polysynaptic pathways (Card et al., 1990; Smith et al., 1998; Kim et al., 1999; Aston-Jones and Card, 2000; DeFalco et al., 2001; Card and Enquist, 2014). Replicating PRV viruses cause neuronal damage and ultimately death of the animal within 5 to 6 days following peripheral injections and 4 days following intracerebral injections (Ugolini et al., 1987; Card et al., 1990, 1992; Card and Enquist, 2014; Oyibo et al., 2014). A less virulent strain has been developed in which the only immediate early gene of PRV, IE80, has been deleted (“IE80-null PRV,” Oyibo et al., 2014). Neurons infected with IE80-null PRV have been reported to maintain physiological properties similar to non-infected neurons up to 6 months post infection, with injected mice surviving the same period of time, demonstrating a promising reduction in toxicity.
As with H129 (McGovern et al., 2015a), strains of PRV encoding different transgenes have been used in dual-tracing experiments in which the aim is to assess whether two populations of neurons receive input from a common, third region (Jansen et al., 1995; Banfield et al., 2003). For example, Jansen et al. (1995) used two strains of PRV-Bartha encoding different viral antigens, one encoding the viral glycoprotein gC, and the other encoding β-galactosidase. One strain was injected into the stellate ganglion, while the second was injected into the adrenal gland and IHC used to identify single- and double-labeled neurons. While dual tracing studies appear deceptively simple, a number of important factors must be taken into consideration when selecting the viral vectors to use. The core issue is “superinfection inhibition”; the limited capacity of a single neuron to be infected by more than one virus (Kobiler et al., 2010; Card and Enquist, 2014). Alterations to the viral genome, including the transgene expressed and the location of its insertion, influence the virulence and efficiency of expression, which may place a vector at a disadvantage compared to a “fitter” strain (Kobiler et al., 2010; Card and Enquist, 2014), and so isogenic strains of viruses should be used to minimize these differences (Smith et al., 2000; Banfield et al., 2003). The speed of travel and therefore time of arrival of a virus to the population of interest is also critical; Banfield et al. (2003) compared the dual-infection of rat dorsal root ganglion neurons in vitro by PRV152 and PRV614 introduced asynchronously. They found that when the vectors were added simultaneously, almost all neurons were double-labeled, but that a delay of 2 h between injections reduced double-labeling by ∼70%, and that a 4-h delay resulted in almost none. Furthermore, there is a limit to the number of copies of a transgene that a neuron can produce: Kobiler et al. (2010) injected three isogenic strains of PRV that expressed the “Brainbow” cassette and, based on the combinations of fluorophores expressed, estimated that no more than seven genomes could be expressed in each cell. Therefore, vectors should be chosen that are as closely matched as possible for genomic sequence, virulence, speed of transduction and transgene expression, and should arrive at the common region of interest at as close a timepoint as possible.
Retrograde Trans-Synaptic Tracing via Glycoprotein-Deleted Rabies
The evolution of the TK-deleted H129 virus parallels the development of glycoprotein-deleted rabies, the first virus to be developed as a genetically restricted monosynaptic tracer (reviewed by Luo et al., 2008, 2018; Callaway and Luo, 2015). It was developed from the Street Alabama, Dufferin B19 (SADB19) strain of rabies, a virus isolated from a symptomatic dog in the 1930s and maintained on cultured rodent tissue for decades thereafter, over which time it became rodent-specific, attenuated, and incapable of horizontal or vertical transmission (Vos et al., 1999; Beckert et al., 2009). These features make recombinant SADB19 not only a safer option as a research tool, but also led to its development as a live oral vaccine for wild foxes across central Europe, with over 50 million doses airdropped since the 1980s (Geue et al., 2008).
Genetically modified SADB19 has provided a new generation of tracing tools that avoid the shortcomings of classic replication-competent neurotrophic viruses. At the heart of this technology is the key role the rabies glycoprotein plays in the infection and exclusively retrograde trans-synaptic spread of rabies, which occurs through still-undefined mechanisms (Lentz et al., 1982; Mebatsion et al., 1996; Etessami et al., 2000; Schnell et al., 2010; Ugolini, 2010). Deletion of the glycoprotein gene renders the virus, so-called SADΔG, incapable of spreading from one neuron to another but does not affect its capacity for replication (Mebatsion et al., 1996; Etessami et al., 2000; Wickersham et al., 2007a), meaning that SADΔG variants becomes trapped inside infected neurons.
The key insight made by Wickersham et al. (2007b) is that SADΔG’s ability to trans-synaptically migrate can be transiently restored by expression of the rabies glycoprotein in infected neurons in trans (Marshel et al., 2010). For example, when motoneurons are induced to express rabies glycoprotein using an AAV vector, subsequent infection of those neurons by intramuscular injection of SADΔG results in retrograde infection of monosynaptic input neurons in the spinal cord and brainstem (Esposito et al., 2014).
This approach was further refined by restricting initial access of SADΔG to targeted populations of neurons by pseudotyping glycoprotein-deleted rabies with the envelope protein of a virus that is unable to infect naïve mammalian neurons, avian sarcoma and leukosis virus (EnvA). Infection by pseudotyped SADΔG, notated as SADΔG(EnvA), can be controlled by selective expression of its cognate receptor, TVA, on target neurons (Wickersham et al., 2007b; Marshel et al., 2010). The starting point for monosynaptic retrograde tracing strategies that use SADΔG(EnvA) can be defined by co-expression of the genes that encode TVA and the rabies glycoprotein in target neurons, often referred to as “seed” or “starter” neurons. Direct CNS injection of SADΔG(EnvA) selectively infects seed neurons at the injection site, wherein virions replicate, incorporate the glycoprotein, and spread to monosynaptically connected pre-synaptic (“input”) neurons. To distinguish input neurons from seed neurons, a reporter that differs in color from that contained in the SADΔG genome is by convention included within the TVA/glycoprotein cassette (Figure 4).
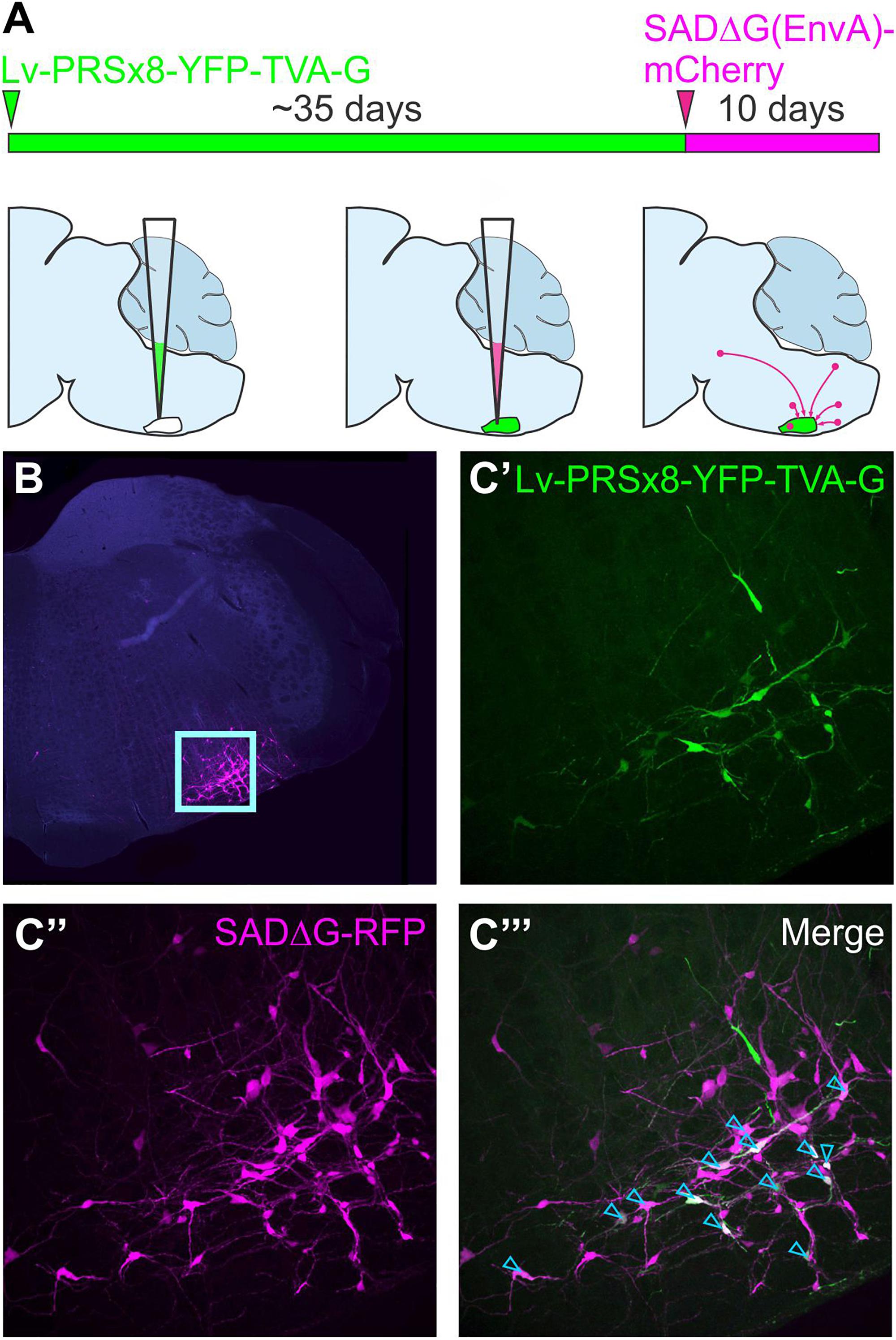
Figure 4. Monosynaptically restricted retrograde tracing using glycoprotein-deleted rabies. (A) Experimental strategy: the starting point is co-expression of genes that encode TVA and the rabies glycoprotein in target neurons, in this case delivered by a lentiviral vector that targets putative sympathetic premotor neurons in the ventrolateral medulla (Lv-PRSx8-YFP-TVA-G). Subsequent infection of TVA/G-expressing neurons by SADΔG(EnvA) microinjection leads to trans-synaptic infection of pre-synaptic (“input”) neurons. Panel (B) shows low power micrograph with infected neurons concentrated in the ventrolateral medulla (boxed region, enlarged in C) “Seed” neurons may be distinguished from “input” neurons by co-expression of the reporters contained in the lentiviral (C’) and rabies constructs (C”), which appear white in the merged image (C”’, denoted by blue arrowheads). Modified with permission from Menuet et al. (2017).
In theory, this approach could be used to identify monosynaptic inputs to any population in which it is possible to selectively drive TVA and the rabies glycoprotein. Investigators from diverse branches of neuroscience have used innovative approaches to limit seeding to their populations of interest, using transgenic mice (Takatoh et al., 2013), lentiviral, AAV or HSV-1 viral vectors (Brennand et al., 2011; Liu et al., 2013; Yonehara et al., 2013; Dempsey et al., 2017; Menuet et al., 2017), and even in vivo transfection of single functionally identified neurons (Rancz et al., 2011; Velez-Fort et al., 2014; Wertz et al., 2015).
SADΔG(EnvA) represents a current state-of-the-art technique for interrogation of circuit structure; its promise is reflected by its rapid adoption by diverse branches of neuroscience, high citation rates of studies that use it, and the continued investment in its refinement. However, it is not without its limitations: first, the proportion of monosynaptic input neurons labeled by SADΔG(EnvA) is low and subject to variability, with the ratio of input neurons per starter neuron ranging from less than 10 to several hundred (Rancz et al., 2011; Velez-Fort et al., 2014; Stornetta et al., 2016). Although some of this variability reflects actual differences in the numbers of afferent neurons that converge upon different cell types (Velez-Fort et al., 2014), as discussed at length by Callaway and Luo (2015), the strength of glycoprotein/TVA expression on the target neuron, the titer of SADΔG(EnvA) (and therefore initial number of replicating virions within starter cells), and the duration of the experiment also interact to determine labeling efficiency. Efforts to improve labeling efficiency have focussed on modification of the rabies glycoprotein, resulting in a codon-optimized chimera of the SADB19 and Pasteur G glycoproteins known as “oG” (Kim et al., 2016), or generation of glycoprotein-deleted variants of other rabies strains (Reardon et al., 2016), both of which have improved the efficiency of the original system.
A second caveat to the use of glycoprotein-deleted rabies variants lies in uncertainty regarding potential biases in its tropism. Although validation of trans-synaptic infection of excitatory and inhibitory neurons was included in the initial description of this system (Wickersham et al., 2007b), the potential for differential selectivity according to proximity or phenotype has been raised by a number of investigators (Callaway and Luo, 2015; Reardon et al., 2016). Recent work by Sun et al. (2019) has confirmed these concerns, showing differential labeling of neurons by AAV-retro, SADΔG and conventional tracers, as well as by polysynaptic strains of rabies and pseudorabies, which they speculate reflects variable expression of cognate receptors to viral proteins in different neuronal populations.
A third weakness of the SADΔG system is its neurotoxicity, which is low compared to H129 but nonetheless results in microglial infiltration, gene dysregulation, and cell death within a few weeks of infection (Wickersham et al., 2007a; Sun et al., 2019), and is a major barrier to its integration with functional tools. Again, the development of glycoprotein-deleted versions of other rabies strains has been reported to reduce toxicity (Reardon et al., 2016), and much has been made of a self-inactivating form that clears infected neurons of histotoxic elements shortly after infection (Ciabatti et al., 2017), although a recent early report by Wickersham et al. throws the validity of this approach into doubt (preprint by Matsuyama et al., 2019). Perhaps more promising is the development of so-called “double-deletion rabies” variants (SADΔGL), in which the “L” gene responsible for transcription and replication of the viral genome is deleted in addition to that of the glycoprotein, resulting in a virus with negligible gene transcription and toxicity but which can nonetheless drive expression of recombinase proteins at levels sufficient to drive site-specific recombination in transgenic animals (Chatterjee et al., 2018). Although the recent work by Chatterjee et al. (2018) supports the suitability of SADΔGL as an alternative to AAV-retro, the applicability of this system as a monosynaptically restricted tracer is still unknown.
Finally, in contrast to PRV and H129, the cellular mechanisms responsible for the transmission of rabies are yet to be comprehensively delineated, resulting in some skepticism regarding its supposed retrograde trans-synaptic exclusivity (eloquently summarized in a recent blog by Svoboda, 2019). The strongest evidence in favor of the exclusivity of this mechanism is broad coherence between the results of rabies tracing, ultrastructural, and electrophysiology studies (considered extensively by Ugolini, 2011), and in particular the absence of rabies virions from the extracellular space (which would indicate non-synaptic transmission). However, as pointed out by Svoboda, without a complete understanding of the rabies life cycle it remains difficult to account for discrepancies in connectivity schemes suggested by the results of viral tracing and electrophysiological mapping studies, and so some caution should be exercised in the interpretation of these data.
Imaging, Analysis and Presentation of Connectomic Data
The repurposing of viral vectors as instruments for scientific research has transformed the field of neuroanatomy in less than 20 years. The mass production of viral vectors by commercial “vector cores” has lowered economic barriers to their adoption by researchers, while the development of technologies that permit high-throughput manipulation of viral genomes and selection of useful traits, such as directed evolution and genetic barcoding, has made it easier than ever for vector biologists to prototype new viral tools. These factors, coupled with the scientific merit of the approach, will drive innovation in this space for the foreseeable future.
It is therefore surprising that the technologies used by researchers to capture, analyze and present anatomical data have changed so little over the same period: with few exceptions, researchers continue to present their data in terms of the numbers of labeled neurons that lie within each region of the brain, averaged across replicates and estimated by comparing images of histological sections to plates from a 2-D reference atlas. As we have argued before (Dempsey et al., 2017), expressing rich datasets in such crude terms precludes their independent analysis by other researchers, while potential future reclassification of brain taxonomy threatens the shelf-life of the data. Moreover, it is a laborious and error-prone process, because accuracy is contingent on cutting histological sections in perfect alignment with the reference atlas while maintaining correct ordering and orientation. This task sounds straightforward but is hard to achieve in reality – it took George Paxinos over 50 attempts to get a perfect example for the original Paxinos and Watson rat brain atlas (G. Paxinos, personal communication) – and small deviations from the orthogonal make correct alignment with the reference atlas impossible; for example, if a section through the widest part of the rat brain is cut at 5° mediolateral from true coronal alignment, the lateral border of the cortex on one side of the brain will lie 1.4 mm rostral to the other.
How should this problem be addressed? In our view, it will not be resolved by improving imaging technologies per se – if one’s objective is to simply map populations of labeled neurons, then improvements in optical resolution are unlikely to help much (although the lowered cost, improved speed and spectral sensitivity of modern microscopes present benefits of their own). Nor are recently developed tissue processing techniques that permit continuous imaging of thick volumes of brain likely to provide a simple solution (Chung et al., 2013; Renier et al., 2014; Tomer et al., 2014): although useful for imaging small volumes of brain, they are slow, labor- and resource-intensive, and inherently difficult to align to a reference atlas. Instead, a new generation of tools is required that can automate and standardize the analysis of tracing data by automatically registering histological tissue into 3-d volumetric brain atlases, extracting the locations of labeled neurons and expressing them in Cartesian co-ordinates as well as identifying the regions in which they reside. This development would simplify analysis, reduce variability, and permit the sharing and independent analysis of connectivity data. Although a number of investigators have highlighted the importance of this mission (Osten and Margrie, 2013; Renier et al., 2016; Furth et al., 2018; Puchades et al., 2019), to date the tools available are hard to use and far from intuitive.
Conclusion
Connectomic data will not, by itself, explain how brains work: connectivity is but one parameter in an overlapping spectrum of classifications used by neuroscientists to bracket neurons. However, when combined with electrophysiological properties, synaptic strength, neurotransmitter content, receptor expression and developmental lineage, connectivity data may provide clues to the processes that underlie the function of specific brain circuits and provide insights into the general rules that underlie brain circuit formation, growth and plasticity (Rockland, 2015).
Nevertheless, controversy about the utility of extensive connectomic data remains unresolved (Carandini, 2012; Denk et al., 2012; Rockland, 2015; Jonas and Kording, 2017) (interested readers are also encouraged to watch the debates between Anthony Movshon and Sebastian Seung hosted by Neuwrite at Columbia University in 20123 and between Anthony Movshon and Moritz Helmstaedter, held as part of the Cognitive Neuroscience Society annual meeting in 20164). Similar skepticism characterized the early years of the Human Genome Project: the scientific value of sequencing the whole human genome, the exploratory nature of the Project, the seemingly insurmountable technical challenges and high cost projections were a cause for concern for scientists, politicians and tax-payers alike (reviewed by Gannett, 2016). However, the completion of the Human Genome Project not only dramatically advanced sequencing techniques (e.g., personal genealogical genotyping is now commercially viable), but also accelerated the incorporation of genetic technologies to biology at large and spawned gene therapy (Lander, 2011). Whether the discoveries that emerge from the contemporary neuroscience equivalents of the Human Genome Project will lead to analogous translational technologies remains to be seen, but the connectomic movement has already transformed neuroscience.
Author Contributions
All authors contributed to the planning, discussion, and drafting of the manuscript.
Funding
This research was generously supported by the Hillcrest Foundation (IPAP2019/0933) and by the National Health and Medical Research Council of Australia (APP1127817). CS is a Macquarie University and University of Bristol Ph.D. scholar.
Conflict of Interest Statement
SL was a Ph.D. student at the time of writing. He is now an employee of the Olympus Australia. He has approved the submission of the manuscript but has not provided any material input since becoming an Olympus Australia employee.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
H129 virus was kindly shared by Stuart Mazzone and Alice McGovern, The University of Melbourne. We thank Professor Anthony Pickering for his helpful suggestions and comments.
Footnotes
- ^ Ironically, despite having already been discredited as a serious science by then, phrenology contributed to the general acceptance of this view by fostering the notion that individual variation in discrete brain functions are reflected by skull topography (summarized by Gross, 2009).
- ^ Opponents to the neuronist school have been characterized as subscribing to a chaotic and somewhat absurd view of the brain. Guillery (2005) makes a compelling case for reassessment of this historical debate, highlighting discoveries that were made after the description of the physical structure of the synapse (e.g., gap junctions, axo-axonic contacts) that fundamentally undermine some tenets of the neuron theory. Guillery argues that both models were ultimately wrong, and that a less dogmatic post-neuronist view is more appropriate.
- ^ https://youtu.be/fRHzkRqGf-g
- ^ https://youtu.be/uSbNRyY2QH0
References
Akli, S., Caillaud, C., Vigne, E., Stratford-Perricaudet, L. D., Poenaru, L., Perricaudet, M., et al. (1993). Transfer of a foreign gene into the brain using adenovirus vectors. Nat. Genet. 3, 224–228. doi: 10.1038/ng0393-224
Andrushko, V. A., and Verano, J. W. (2008). Prehistoric trepanation in the Cuzco region of Peru: a view into an ancient Andean practice. Am. J. Phys. Anthropol. 137, 4–13. doi: 10.1002/ajpa.20836
Angelucci, A., Clascá, F., and Sur, M. (1996). Anterograde axonal tracing with the subunit B of cholera toxin: a highly sensitive immunohistochemical protocol for revealing fine axonal morphology in adult and neonatal brains. J. Neurosci. Methods 65, 101–112. doi: 10.1016/0165-0270(95)00155-7
Archin, N. M., and Atherton, S. S. (2002). Rapid spread of a neurovirulent strain of HSV-1 through the CNS of BALB/c mice following anterior chamber inoculation. J. Neurovirol. 8, 122–135. doi: 10.1080/13550280290049570
Aston-Jones, G., and Card, J. P. (2000). Use of pseudorabies virus to delineate multisynaptic circuits in brain: opportunities and limitations. J. Neurosci. Methods 103, 51–61. doi: 10.1016/s0165-0270(00)00295-8
Azevedo, F. A., Carvalho, L. R., Grinberg, L. T., Farfel, J. M., Ferretti, R. E., Leite, R. E., et al. (2009). Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 513, 532–541. doi: 10.1002/cne.21974
Banfield, B. W., Kaufman, J. D., Randall, J. A., and Pickard, G. E. (2003). Development of pseudorabies virus strains expressing red fluorescent proteins: new tools for multisynaptic labeling applications. J. Virol. 77, 10106–10112. doi: 10.1128/jvi.77.18.10106-10112.2003
Beckert, A., Geue, L., Vos, A., Neubert, A., Freuling, C., and Muller, T. (2009). Genetic stability (in vivo) of the attenuated oral rabies virus vaccine SAD B19. Microbiol. Immunol. 53, 16–21. doi: 10.1111/j.1348-0421.2008.00085.x
Beier, K. T., Saunders, A., Oldenburg, I. A., Miyamichi, K., Akhtar, N., Luo, L., et al. (2011). Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc. Natl. Acad. Sci. U.S.A. 108, 15414–15419. doi: 10.1073/pnas.1110854108
Beier, K. T., Saunders, A. B., Oldenburg, I. A., Sabatini, B. L., and Cepko, C. L. (2013). Vesicular stomatitis virus with the rabies virus glycoprotein directs retrograde transsynaptic transport among neurons in vivo. Front. Neural Circ. 7:11. doi: 10.3389/fncir.2013.00011
Beier, K. T., Steinberg, E. E., DeLoach, K. E., Xie, S., Miyamichi, K., Schwarz, L., et al. (2015). Circuit architecture of VTA dopamine neurons revealed by systematic input-output mapping. Cell 162, 622–634. doi: 10.1016/j.cell.2015.07.015
Bentivoglio, M., Kuypers, H. G., Catsman-Berrevoets, C. E., Loewe, H., and Dann, O. (1980). Two new fluorescent retrograde neruonal tracers which are transported over long distances. Neurosci. Lett. 18, 25–30. doi: 10.1016/0304-3940(80)90208-6
Berges, B. K., Wolfe, J. H., and Fraser, N. W. (2007). Transduction of brain by herpes simplex virus vectors. Mol. Ther. 15, 20–29. doi: 10.1038/sj.mt.6300018
Betley, J. N., and Sternson, S. M. (2011). Adeno-associated viral vectors for mapping, monitoring, and manipulating neural circuits. Hum. Gene Ther. 22, 669–677. doi: 10.1089/hum.2010.204
Blomer, U., Naldini, L., Kafri, T., Trono, D., Verma, I. M., and Gage, F. H. (1997). Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J. Virol. 71, 6641–6649.
Bokiniec, P., Shahbazian, S., McDougall, S. J., Berning, B. A., Cheng, D., Llewellyn-Smith, I. J., et al. (2017). Polysialic acid regulates sympathetic outflow by facilitating information transfer within the nucleus of the solitary tract. J. Neurosci. 37, 6558–6574. doi: 10.1523/JNEUROSCI.0200-17.2017
Bonhoeffer, F., and Huf, J. (1980). Position-dependent propoerties of retinal axons and their growth-cones. J. Neurosci. Methods 45, 35–40.
Boon, J., Clarke, E., Kessaris, N., Goffinet, A., Molnar, Z., and Hoerder-Suabedissen, A. (2019). Long-range projections from sparse populations of GABAergic neurons in murine subplate. J. Comp. Neurol. 527, 1610–1620. doi: 10.1002/cne.24592
Bou Farah, L., Bowman, B. R., Bokiniec, P., Karim, S., Le, S., Goodchild, A. K., et al. (2016). Somatostatin in the rat rostral ventrolateral medulla: origins and mechanism of action. J. Comp. Neurol. 524, 323–342. doi: 10.1002/cne.23846
Branco, T., and Staras, K. (2009). The probability of neurotransmitter release: variability and feedback control at single synapses. Nat. Rev. Neurosci. 10, 373–383. doi: 10.1038/nrn2634
Brandt, H. M., and Apkarian, A. V. (1992). Biotin-dextran: a sensitive anterograde tracer for neuroanatomic studies in rat and monkey. J. Neurosci. Methods 45, 35–40. doi: 10.1016/0165-0270(92)90041-b
Brennand, K. J., Simone, A., Jou, J., Gelboin-Burkhart, C., Tran, N., Sangar, S., et al. (2011). Modelling schizophrenia using human induced pluripotent stem cells. Nature 473, 221–225. doi: 10.1038/nature09915
Briggman, K. L., Helmstaedter, M., and Denk, W. (2011). Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188. doi: 10.1038/nature09818
Burette, A., Collman, F., Micheva, K. D., Smith, S. J., and Weinberg, R. J. (2015). Knowing a synapse when you see one. Front. Neuroanat. 9:100. doi: 10.3389/fnana.2015.00100
Callaway, E. M. (2005). A molecular and genetic arsenal for systems neuroscience. Trends Neurosci. 28, 196–201. doi: 10.1016/j.tins.2005.01.007
Callaway, E. M., and Luo, L. (2015). monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J. Neurosci. 35, 8979–8985. doi: 10.1523/jneurosci.0409-15.2015
Campadelli-Fiume, G., Cocchi, F., Menotti, L., and Lopez, M. (2000). The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10, 305–309.
Carandini, M. (2012). From circuits to behavior: a bridge too far? Nat. Neurosci. 15, 507–509. doi: 10.1038/nn.3043
Card, J. P., and Enquist, L. W. (1995). Neurovirulence of pseudorabies virus. Crit. Rev. Neurobiol. 9, 137–162.
Card, J. P., and Enquist, L. W. (2014). Transneuronal circuit analysis with pseudorabies viruses. Curr. Protoc. Neurosci. 68, 1.5.1–1.5.39. doi: 10.1002/0471142301.ns0105s68
Card, J. P., Enquist, L. W., and Moore, R. Y. (1999). Neuroinvasiveness of pseudorabies virus injected intracerebrally is dependent on viral conventration and terminal field density. J. Comp. Neurol. 407, 438–452. doi: 10.1002/(sici)1096-9861(19990510)407:3<438::aid-cne11>3.0.co;2-2
Card, J. P., Levitt, P., and Enquist, L. W. (1998). Different patterns of neuronal infection after intracerebral injections of two strains of pseudorabies virus. J. Virol. 72, 4434–4441.
Card, J. P., Rinaman, L., Schwaber, J. S., Miselis, R. R., Whealy, M. E., Robbins, A. K., et al. (1990). Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J. Neurosci. 10, 1974–1994. doi: 10.1523/jneurosci.10-06-01974.1990
Card, J. P., Whealy, M. E., Robbins, A. K., and Enquist, L. W. (1992). Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J. Virol. 66, 3032–3041.
Card, J. P., Whealy, M. E., Robbins, A. K., Moore, J. P., and Enquist, L. W. (1991). Two ?-herpesvirus strains are transported differentially in the rodent visual system. Neuron 6, 957–969. doi: 10.1016/0896-6273(91)90236-s
Castle, M. J., Turunen, H. T., Vandenberghe, L. H., and Wolfe, J. H. (2016). Controlling AAV tropism in the nervous system with natural and engineered capsids. Methods Mol. Biol. 1382, 133–149. doi: 10.1007/978-1-4939-3271-9_10
Catani, M., Thiebaut de Schotten, M., Slater, D., and Dell’Acqua, F. (2013). Connectomic approaches before the connectome. Neuroimage 80, 2–13. doi: 10.1016/j.neuroimage.2013.05.109
Chamberlin, N. L., Du, B., de Lacalle, S., and Saper, C. B. (1998). Recombinant adeno-associated virus vector: use for transgene expression and anterograde tract tracing in the CNS. Brain Res. 793, 169–175. doi: 10.1016/s0006-8993(98)00169-3
Chan, K. Y., Jang, M. J., Yoo, B. B., Greenbaum, A., Ravi, N., Wu, W. L., et al. (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179. doi: 10.1038/nn.4593
Chatterjee, S., Sullivan, H. A., MacLennan, B. J., Xu, R., Hou, Y., Lavin, T. K., et al. (2018). Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons. Nat. Neurosci. 21, 638–646. doi: 10.1038/s41593-018-0091-7
Chen, S., and Aston-Jones, G. (1995). Evidence that cholera toxin B subunit (CTb) can be avidly taken up and transported by fibers of passage. Brain Res. 674, 107–111. doi: 10.1016/0006-8993(95)00020-q
Chung, K., Wallace, J., Kim, S. Y., Kalyanasundaram, S., Andalman, A. S., Davidson, T. J., et al. (2013). Structural and molecular interrogation of intact biological systems. Nature 497, 332–337. doi: 10.1038/nature12107
Ciabatti, E., Gonazález-Rueda, A., Mariotti, L., Morgese, F., and Tripodi, M. (2017). Life-long genetic and functional access to neural circuits using self-inactivating rabies virus. Cell 170, 382–392. doi: 10.1016/j.cell.2017.06.014
Conte, W. L., Kamishina, H., and Reep, R. L. (2009a). Multiple neuroanatomical tract-tracing using fluorescent Alexa Fluor conjugates of cholera toxin subunit B in rats. Nat. Protoc. 4, 1157–1166. doi: 10.1038/nprot.2009.93
Conte, W. L., Kamishina, H., and Reep, R. L. (2009b). The efficacy of the fluorescent conjugates of cholera toxin subunit B for multiple retrograde tract tracing in the central nervous system. Brain Struct. Funct. 213, 367–373. doi: 10.1007/s00429-009-0212-x
Cowan, W. M., Gottlieb, D. I., Hendrickson, A. E., Price, J. L., and Woolsey, T. A. (1972). The autoradiographic demonstration of axonal connections in the central nervous system. Brain Res. 37, 21–51. doi: 10.1016/0006-8993(72)90344-7
da Costa, N. M., and Martin, K. A. (2011). How thalamus connects to spiny stellate cells in the cat’s visual cortex. J. Neurosci. 31, 2925–2937. doi: 10.1523/JNEUROSCI.5961-10.2011
Dado, R. J., Burstein, R., Cliffer, K. D., and Giesler, G. J. Jr. (1990). Evidence that fluoro-gold can be transported avidly through fibers of passage. Brain Res. 533, 329–333. doi: 10.1016/0006-8993(90)91358-n
DeFalco, J., Tomishima, M., Liu, H., Zhao, C., Cai, X., Marth, J. D., et al. (2001). Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291, 2608–2613. doi: 10.1126/science.1056602
Dempsey, B., Le, S., Turner, A., Bokiniec, P., Ramadas, R., Bjaalie, J. G., et al. (2017). Mapping and analysis of the connectome of sympathetic premotor neurons in the rostral ventrolateral medulla of the rat using a volumetric brain atlas. Front. Neural Circ. 11:9. doi: 10.3389/fncir.2017.00009
Dempsey, B., Turner, A. J., Le, S., Sun, Q. J., Bou Farah, L., Allen, A. M., et al. (2015). Recording, labeling, and transfection of single neurons in deep brain structures. Physiol. Rep. 3:e12246. doi: 10.14814/phy2.12246
Denk, W., Briggman, K. L., and Helmstaedter, M. (2012). Structural neurobiology: missing link to a mechanistic understanding of neural computation. Nat. Rev. Neurosci. 13, 351–358. doi: 10.1038/nrn3169
Descarries, L., and Mechawar, N. (2000). Ultrastructural evidence for diffuse transmission by monoamine and acetylcholine neurons of the central nervous system. Prog. Brain Res. 125, 27–47. doi: 10.1016/s0079-6123(00)25005-x
Dix, R. D., McKendall, R. R., and Baringer, R. (1983). Comparitive neurovirulence of herpes simplex virus type 1 strains after peripheral or intracerebral inoculation of BALB/c mice. Infect. Immun. 40, 103–112.
Drouin, L. M., and Agbandje-McKenna, M. (2013). Adeno-associated virus structural biology as a tool in vector development. Future Virol. 8, 1183–1199. doi: 10.2217/fvl.13.112
Dumas, M., Schwab, M. E., and Thoenen, H. (1979). Retrograde axonal transport of specific macromolecules as a tool for characterizing nerve terminal membranes. J. Neurobiol. 10, 179–197. doi: 10.1002/neu.480100207
Elberger, A. J., and Honig, M. G. (1990). Double-labeling of tissue containing the carbocyanine dye DiI for immunocytochemistry. J. Histochem. Cytochem. 38, 735–739. doi: 10.1177/38.5.2110209
Esposito, M. S., Capelli, P., and Arber, S. (2014). Brainstem nucleus MdV mediates skilled forelimb motor tasks. Nature 508, 351–356. doi: 10.1038/nature13023
Etessami, R., Conzelmann, K. K., Fadai-Ghotbi, B., Natelson, B., Tsiang, H., and Ceccaldi, P. E. (2000). Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J. Gen. Virol. 81, 2147–2153. doi: 10.1099/0022-1317-81-9-2147
Farmer, D. G. S., Pracejus, N., Dempsey, B., Turner, A., Bokiniec, P., Paton, J. F. R., et al. (2019). On the presence and functional significance of sympathetic premotor neurons with collateralized spinal axons in the rat. J. Physiol. 597, 3407–3423. doi: 10.1113/JP277661
Feinberg, E. H., Vanhoven, M. K., Bendesky, A., Wang, G., Fetter, R. D., Shen, K., et al. (2008). GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. doi: 10.1016/j.neuron.2007.11.030
Fenno, L. E., Mattis, J., Ramakrishnan, C., Hyun, M., Lee, S. Y., He, M., et al. (2014). Targeting cells with single vectors using multiple-feature Boolean logic. Nat. Methods 11, 763–772. doi: 10.1038/nmeth.2996
Ferrier, D. (1884). Cerebral localisation. a review and forecast. being the marshall hall prize oration. Med. Chir. Trans. 67, 33–49. doi: 10.1177/095952878406700103
Fink, R. P., and Heimer, L. (1967). Two methods for selective silver impregnation of degenerating axons and their synaptic endings in the central nervous system. Brain Res. 4, 369–374. doi: 10.1016/0006-8993(67)90166-7
Fishman, R. A. (1994). Origins of neuroscience: a history of explorations into brain function.By Stanley Finger, New York, Oxford University Press, 1994, 462 pp, illustrated, $75.00. Ann. Neurol. 36:807. doi: 10.1002/ana.410360532
Frampton, A. R. Jr., Goins, W. F., Nakano, K., Burton, E. A., and Glorioso, J. C. (2005). HSV trafficking and development of gene therapy vectors with applications in the nervous system. Gene Ther. 12, 891–901. doi: 10.1038/sj.gt.3302545
Fritzsch, B. (1993). Fast axonal diffusion of 3000 molecular weight dextran amines. J. Neurosci. Methods 50, 95–103. doi: 10.1016/0165-0270(93)90060-5
Furth, D., Vaissiere, T., Tzortzi, O., Xuan, Y., Martin, A., Lazaridis, I., et al. (2018). An interactive framework for whole-brain maps at cellular resolution. Nat. Neurosci. 21, 139–149. doi: 10.1038/s41593-017-0027-7
Furukawa, H., Saito, A., Mol, W., Sekido, M., Sasaki, S., and Yamamoto, Y. (2008). Double innervation occurs in the facial mimetic muscles after facial-hypoglossal end-to-side neural repair: rat model for neural supercharge concept. J. Plast. Reconstr. Aesthet. Surg. 61, 257–264. doi: 10.1016/j.bjps.2007.01.075
Furuta, T., Tomioka, R., Taki, K., Nakamura, K., Tamamaki, N., and Kaneko, T. (2001). In vivo transduction of central neurons using recombinant Sindbis virus: golgi-like labeling of dendrites and axons with membrane-targeted fluorescent proteins. J. Histochem. Cytochem. 49, 1497–1508.
Gannett, L. (2016). “The human genome project,” in The Stanford Encyclopedia of Philosophy, ed. E. N. Zalta, (Stanford, CA: Metaphysics Research Lab, Stanford University).
Gao, G., Vandenberghe, L. H., Alvira, M. R., Lu, Y., Calcedo, R., Zhou, X., et al. (2004). Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78, 6381–6388. doi: 10.1128/jvi.78.12.6381-6388.2004
Garner, J. A., and LaVail, J. H. (1999). Differential anterograde transport of HSV type 1 viral strains in the murine optic pathway. J. Neurovirol. 5, 140–150. doi: 10.3109/13550289909021996
Geraghty, R. J., Krummenacher, C., Cohen, G. H., Eisenberg, R. J., and Spear, P. G. (1998). Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280, 1618–1620. doi: 10.1126/science.280.5369.1618
Gerfen, C. R., and Sawchenko, P. E. (1984). An anterograde neuroanatomical tracing method that shows the detailed morphology of neurons, their axons and terminals: immunohistochemical localization of an axonally transported plant lectin, Phaseolus vulgaris leucoagglutinin (PHA-L). Brain Res. 290, 219–238. doi: 10.1016/0006-8993(84)90940-5
Geue, L., Schares, S., Schnick, C., Kliemt, J., Beckert, A., Freuling, C., et al. (2008). Genetic characterisation of attenuated SAD rabies virus strains used for oral vaccination of wildlife. Vaccine 26, 3227–3235. doi: 10.1016/j.vaccine.2008.04.007
Gimlich, R. L., and Braun, J. (1985). Improved fluorescent compounds for tracing cell lineage. Dev. Biol. 109, 509–514. doi: 10.1016/0012-1606(85)90476-2
Glees, P. (1946). Terminal degeneration within the central nervous system as studied by a new silver method. J. Neuropathol. Exp. Neurol. 5, 54–59. doi: 10.1097/00005072-194601000-00005
Glover, J. C. (1995). Retrograde and anterograde axonal tracing with fluorescent dextrans in the embryonic nervous system. Neurosci. Protoc. 95, 1–13.
Glover, J. C., Petursdottir, G., and Jansen, J. K. S. (1986). Fluorescent dextran amines used as axonal tracers in the nervous system of chicken embryo. J. Neurosci. Methods 18, 243–254. doi: 10.1016/0165-0270(86)90011-7
Gofflot, F., Wendling, O., Chartoire, N., Birling, M. C., Warot, X., and Auwerx, J. (2011). Characterization and validation of cre-driver mouse lines. Curr. Protoc. Mouse Biol. 1, 1–15. doi: 10.1002/9780470942390.mo100103
Goshgarian, H. G., and Buttry, J. L. (2014). The pattern and extent of retrograde transsynaptic transport of WGA-Alexa 488 in the phrenic motor system is dependent upon the site of application. J. Neurosci. Methods 222, 156–164. doi: 10.1016/j.jneumeth.2013.11.003
Grafstein, B. (1967). Transport of protein by goldfish optic nerve fibers. Science 157, 196–198. doi: 10.1126/science.157.3785.196
Grimm, D., and Buning, H. (2017). Small but increasingly mighty: latest advances in AAV vector research, design, and evolution. Hum. Gene Ther. 28, 1075–1086. doi: 10.1089/hum.2017.172
Gross, C. G. (2009). “Phrenology,” in Encyclopedia of Neuroscience, ed. L. R. Squire, (Oxford: Academic Press), 701–705.
Guillery, R. W. (2005). Observations of synaptic structures: origins of the neuron doctrine and its current status. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1281–1307. doi: 10.1098/rstb.2003.1459
Hagmann, P. (2005). From Diffusion MRI to Brain Connectomics. Ph.D thesis, Ecole Polytechnique Fédérale de Lausanne, Lausanne.
Henriksen, S., Pang, R., and Wronkiewicz, M. (2016). A simple generative model of the mouse mesoscale connectome. eLife 5:e12366. doi: 10.7554/eLife.12366
Herculano-Houzel, S. (2009). The human brain in numbers: a linearly scaled-up primate brain. Front. Hum. Neurosci. 3:31. doi: 10.3389/neuro.09.031.2009
Hilbert, M., and Lopez, P. (2011). The world’s technological capacity to store, communicate, and compute information. Science 332, 60–65. doi: 10.1126/science.1200970
Hirschberg, S., Li, Y., Randall, A., Kremer, E. J., and Pickering, A. E. (2017). Functional dichotomy in spinal-vs prefrontal-projecting locus coeruleus modules splits descending noradrenergic analgesia from ascending aversion and anxiety in rats. eLife 6:e29808. doi: 10.7554/eLife.29808
Hoff, E. C. (1932). Central nerve terminals in the mammalian spinal cord and their examination by experimental degeneration. Proc. R. Soc. Lond. Ser. BContain. Pap. Biol. 111, 175–188. doi: 10.1098/rspb.1932.0049
Howorth, P. W., Thornton, S. R., O’Brien, V., Smith, W. D., Nikiforova, N., Teschemacher, A. G., et al. (2009). Retrograde viral vector-mediated inhibition of pontospinal noradrenergic neurons causes hyperalgesia in rats. J. Neurosci. 29, 12855–12864. doi: 10.1523/JNEUROSCI.1699-09.2009
Hwang, D. Y., Carlezon, W. A. Jr., Isacson, O., and Kim, K. S. (2001). A high-efficiency synthetic promoter that drives transgene expression selectively in noradrenergic neurons. Hum. Gene Ther. 12, 1731–1740. doi: 10.1089/104303401750476230
Jakobsson, J., Ericson, C., Jansson, M., Bjork, E., and Lundberg, C. (2003). Targeted transgene expression in rat brain using lentiviral vectors. J. Neurosci. Res. 73, 876–885. doi: 10.1002/jnr.10719
Jansen, A. S. P., Van Nguyen, X., Karpitskiy, X., Mettenleiter, T. C., and Loewy, A. D. (1995). Central command neurons of the sympathetic nervous system: basis of the fight-or-flight response. Science 270, 253–260.
Jia, F., Miao, H., Zhu, X., and Xu, F. (2017). Pseudo-typed semliki forest virus delivers EGFP into neurons. J. Neurovirol. 23, 205–215. doi: 10.1007/s13365-016-0486-8
Jonas, E., and Kording, K. P. (2017). Could a neuroscientist understand a microprocessor? PLoS Comput. Biol. 13:e1005268. doi: 10.1371/journal.pcbi.1005268
Junyent, F., and Kremer, E. J. (2015). CAV-2–why a canine virus is a neurobiologist’s best friend. Curr. Opin. Pharmacol. 24, 86–93. doi: 10.1016/j.coph.2015.08.004
Jüttner, J., Szabo, A., Gross-Scherf, B., Morikawa, R. K., Rompani, S. B., Hantz, P., et al. (2019). Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci. 22, 1345–1356. doi: 10.1038/s41593-019-0431-2
Kanaan, N. M., Sellnow, R. C., Boye, S. L., Coberly, B., Bennett, A., Agbandje-McKenna, M., et al. (2017). Rationally engineered AAV capsids improve transduction and volumetric spread in the CNS. Mol. Ther. Nucleic Acids 8, 184–197. doi: 10.1016/j.omtn.2017.06.011
Kato, S., Kobayashi, K., Inoue, K., Kuramochi, M., Okada, T., Yaginuma, H., et al. (2011). A lentiviral strategy for highly efficient retrograde gene transfer by pseudotyping with fusion envelope glycoprotein. Hum. Gene Ther. 22, 197–206. doi: 10.1089/hum.2009.179
Katz, L. C., Burkhalter, A., and Dreyer, W. J. (1984). Fluorescent latex microspheres as a retrograde neuronal marker for in vivo and in vitro studies of visual cortex. Nature 310, 498–500. doi: 10.1038/310498a0
Katz, L. C., and Iarovici, D. M. (1990). Green fluorescent latex microspheres: a new retrograde tracer. Neuroscience 34, 511–520. doi: 10.1016/0306-4522(90)90159-2
Kelly, R. M., and Strick, P. L. (2000). Rabies as a transneuronal tracer of circuits in the central nervous system. J. Neurosci. Methods 103, 63–71. doi: 10.1016/s0165-0270(00)00296-x
Kim, E. J., Jacobs, M. W., Ito-Cole, T., and Callaway, E. M. (2016). Improved monosynaptic neural circuit tracing using engineered rabies virus glycoproteins. Cell Rep. 15, 692–699. doi: 10.1016/j.celrep.2016.03.067
Kim, J., Zhao, T., Petralia, R. S., Yu, Y., Peng, H., Myers, E., et al. (2011). mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat. Methods 9, 96–102. doi: 10.1038/nmeth.1784
Kim, J. S., Moore, R. Y., Enquist, L. W., and Card, J. P. (1999). Circuit-specific co-infection of the neurons in the rat central nervous system with two pseudorabies virus recombinants. J. Virol. 75, 9521–9531.
Köbbert, C., Apps, R., Bechmann, I., Lanciego, J. L., Mey, J., and Thanos, S. (2000). Current concepts in neuroanatomical tracing. Prog. Neurobiol. 62, 327–351. doi: 10.1016/s0301-0082(00)00019-8
Kobiler, O., Lipman, Y., Therkelsen, K., Daubechies, I., and Enquist, L. W. (2010). Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat. Commun. 1:146. doi: 10.1038/ncomms1145
Korim, W. S., Bou Farah, L., McMullan, S., and Verberne, A. J. (2014). Orexinergic activation of medullary premotor neurons modulates the adrenal sympathoexcitation to hypothalamic glucoprivation. Diabetes Metab. Res. Rev. 63, 1895–1906. doi: 10.2337/db13-1073
Kotterman, M. A., and Schaffer, D. V. (2014). Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 15, 445–451. doi: 10.1038/nrg3742
Kristensson, K., and Olsson, Y. (1971). Retrograde axonal transport of protein. Brain Res. 29, 363–365. doi: 10.1016/0006-8993(71)90044-8
Kshettry, V. R., Mindea, S. A., and Batjer, H. H. (2007). The management of cranial injuries in antiquity and beyond. Neurosurg. Focus 23:E8.
Kuo, H., Ingram, D. K., Crystal, R. G., and Mastrangeli, A. (1995). Retrograde transfer of replication deficient recombinant adenovirus vector in the central nervous system for tracing studies. Brain Res. 705, 31–38. doi: 10.1016/0006-8993(95)01065-3
Kuypers, H. G., Bentivoglio, M., Castman-Berreveoets, C. E., and Bharos, A. T. (1980). Double retrograde neuronal labeling through divergent axon collaterals, using two fluorescent tracers with the same excitation wavelength which label different features of the cell. Exp. Brain Res. 40, 383–392.
Labetoulle, M., Kucera, P., Ugolini, G., Lafay, F., Frau, E., Offret, H., et al. (2000). Neuronal pathways for the propagation of herpes simplex virus type 1 from one retina to the other in a murine model. J. Gen. Virol. 81, 1201–1210. doi: 10.1099/0022-1317-81-5-1201
Lanciego, J. L., and Wouterlood, F. G. (2011). A half century of experimental neuroanatomical tracing. J. Chem. Neuroanat. 42, 157–183. doi: 10.1016/j.jchemneu.2011.07.001
Lander, E. S. (2011). Initial impact of the sequencing of the human genome. Nature 470, 187–197. doi: 10.1038/nature09792
Larsen, D. D., Wickersham, I. R., and Callaway, E. M. (2007). Retrograde tracing with recombinant rabies virus reveals correlations between projection targets and dendritic architecture in layer 5 of mouse barrel cortex. Front. Neural Circ. 1:5. doi: 10.3389/neuro.04.005.2007
LaVail, J. H., and LaVail, M. M. (1972). Retrograde axonal transport in the central nervous system. Science 176, 1416–1417. doi: 10.1126/science.176.4042.1416
Lentz, T. L., Burrage, T. G., Smith, A. L., Crick, J., and Tignor, G. H. (1982). Is the acetylcholine receptor a rabies virus receptor? Science 215, 182–184. doi: 10.1126/science.7053569
Lerner, T. N., Shilyansky, C., Davidson Thomas, J., Evans Kathryn, E., Beier Kevin, T., Zalocusky Kelly, A., et al. (2015). Intact-Brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell 162, 635–647. doi: 10.1016/j.cell.2015.07.014
Lerner, T. N., Ye, L., and Deisseroth, K. (2016). Communication in neural circuits: tools. Oppor. Chall. Cell 164, 1136–1150. doi: 10.1016/j.cell.2016.02.027
Levy, S. L., White, J. J., Lackey, E. P., Schwartz, L., and Sillitoe, R. V. (2017). WGA-Alexa conjugates for axonal tracing. Curr. Protoc. Neurosci. 79, 12821–212824. doi: 10.1002/cpns.28
Li, L., Tasic, B., Micheva, K. D., Ivanov, V. M., Spletter, M. L., Smith, S. J., et al. (2010). Visualizing the distribution of synapses from individual neurons in the mouse brain. PLoS One 5:e11503. doi: 10.1371/journal.pone.0011503
Li, X., Chen, W., Pan, K., Li, H., Pang, P., Guo, Y., et al. (2018). Serotonin receptor 2c-expressing cells in the ventral CA1 control attention via innervation of the Edinger-Westphal nucleus. Nat. Neurosci. 21, 1239–1250. doi: 10.1038/s41593-018-0207-0
Lichtman, J. W., and Denk, W. (2011). The big and the small: challenges of imaging the brain’s circuits. Science 334, 618–623. doi: 10.1126/science.1209168
Liu, Y.-J., Ehrengruber Markus, U., Negwer, M., Shao, H.-J., Cetin Ali, H., and Lyon David, C. (2013). Tracing inputs to inhibitory or excitatory neurons of mouse and cat visual cortex with a targeted rabies virus. Curr. Biol. 23, 1746–1755. doi: 10.1016/j.cub.2013.07.033
Llinas, R. R. (2003). The contribution of santiago ramon y cajal to functional neuroscience. Nat. Rev. Neurosci. 4, 77–80. doi: 10.1038/nrn1011
Lo, L., and Anderson, D. J. (2011). A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron 72, 938–950. doi: 10.1016/j.neuron.2011.12.002
Lopez-Munoz, F., Boya, J., and Alamo, C. (2006). Neuron theory, the cornerstone of neuroscience, on the centenary of the Nobel Prize award to Santiago Ramon y Cajal. Brain Res. Bull. 70, 391–405. doi: 10.1016/j.brainresbull.2006.07.010
Lundberg, C., Bjorklund, T., Carlsson, T., Jakobsson, J., Hantraye, P., Deglon, N., et al. (2008). Applications of lentiviral vectors for biology and gene therapy of neurological disorders. Curr. Gene Ther. 8, 461–473. doi: 10.2174/156652308786847996
Lunde, A., Okaty, B. W., Dymecki, S. M., and Glover, J. C. (2019). Molecular profiling defines evolutionarily conserved transcription factor signatures of major vestibulospinal neuron groups. eNeuro 6:e0475–0418.2019. doi: 10.1523/ENEURO.0475-18.2019
Luo, L., Callaway, E. M., and Svoboda, K. (2008). Genetic dissection of neural circuits. Neuron 57, 634–660. doi: 10.1016/j.neuron.2008.01.002
Luo, L., Callaway, E. M., and Svoboda, K. (2018). Genetic dissection of neural circuits: a decade of progress. Neuron 98, 256–281. doi: 10.1016/j.neuron.2018.03.040
Luppi, P. H., Sakai, K., Salvert, D., Fort, P., and Jouvet, M. (1987). Peptidergic hypothalamic afferents to the cat nucleus raphe pallidus as revealed by a double immunostaining technique using unconjugated cholera toxin as a retrograde tracer. Brain Res. 402, 339–345. doi: 10.1016/0006-8993(87)90041-2
Lykken, E. A., Shyng, C., Edwards, R. J., Rozenberg, A., and Gray, S. J. (2018). Recent progress and considerations for AAV gene therapies targeting the central nervous system. J. Neurodev. Disord. 10:16. doi: 10.1186/s11689-018-9234-0
Marshel, J. H., Mori, T., Nielsen, K. J., and Callaway, E. M. (2010). Targeting single neuronal networks for gene expression and cell labeling In Vivo. Neuron 67, 562–574. doi: 10.1016/j.neuron.2010.08.001
Martin, X., and Dolivo, M. (1983). Neuronal and transneuronal tracing in the trigeminal system of the rat using the herpes virus suis. Brain Res. 273, 253–276. doi: 10.1016/0006-8993(83)90850-8
Matsubayashi, Y., Iwai, L., and Kawasaki, H. (2008). Fluorescent double-labeling with carbocyanine neuronal tracing and immunohistochemistry using a cholesterol-specific detergent digitonin. J. Neurosci. Methods 174, 71–81. doi: 10.1016/j.jneumeth.2008.07.003
Matsuyama, M., Jin, L., Lavin, T. K., Sullivan, H. A., Hou, Y., Lea, N. E., et al. (2019). “Self-inactivating” Rabies Viruses are Just First-Generation, ΔG Rabies Viruses. Available at: https://www.biorxiv.org/content/10.1101/550640v1 (accessed July 24, 2019).
McGovern, A. E., Davis-Poynter, N., Farrell, M. J., and Mazzone, S. B. (2012a). Transneural tracing of airways-related sensory circuitry using herpes simplex virus 1, strain H129. Neuroscience 207, 148–166. doi: 10.1016/j.neuroscience.2012.01.029
McGovern, A. E., Davis-Poynter, N., Rokoczy, J., Phipps, A., Simmons, D. G., and Mazzone, S. B. (2012b). Anterograde neuronal circuit tracing using a genetically modified herpes simplex virus expressing EGFP. J. Neurosci. Methods 209, 158–167. doi: 10.1016/j.jneumeth.2012.05.035
McGovern, A. E., Davis-Poynter, N., Yang, S. K., Simmons, D. G., Farrell, M. J., and Mazzone, S. B. (2015a). Evidence for multiple sensory circuits in the brain arising from the respiratory system: an anterograde viral tract tracing study in rodents. Brain Struct. Funct. 220, 3683–3699. doi: 10.1007/s00429-014-0883-9
McGovern, A. E., Driessen, A. K., Simmons, D. G., Powell, J., Davis-Poynter, N., Farrell, M. J., et al. (2015b). Distinct brainstem and forebrain circuits receiving tracheal sensory neuron inputs revealed using a novel conditional anterograde transsynaptic viral tracing system. J. Neurosci. 35, 7041–7055. doi: 10.1523/JNEUROSCI.5128-14.2015
McPherson, D. R., McClellan, A. D., and O’Donovan, M. J. (1997). Optical imaging of neuronal activity in tissue labeled by retrograde transport of Calcium Green Dextran. Brain Res. Brain Res. Protoc. 1, 157–164. doi: 10.1016/S1385-299X(96)00024-4
Mebatsion, T., Konig, M., and Conzelmann, K. K. (1996). Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84, 941–951. doi: 10.1016/s0092-8674(00)81072-7
Menuet, C., Le, S., Dempsey, B., Connelly, A. A., Kamar, J. L., Jancovski, N., et al. (2017). Excessive respiratory modulation of blood pressure triggers hypertension. Cell Metab. 25, 1–10. doi: 10.1016/j.cmet.2017.01.019
Mikula, S. (2016). Progress towards mammalian whole-brain cellular connectomics. Front. Neuroanat. 10:62. doi: 10.3389/fnana.2016.00062
Mizoguchi, A., Nakanishi, H., Kimura, K., Matsubara, K., Ozaki-Kuroda, K., Katata, T., et al. (2002). Nectin: an adhesion molecule involved in formation of synapses. J. Cell Biol. 156, 555–565. doi: 10.1083/jcb.200103113
Moghaddam, N., Mailler-Burch, S., Kara, L., Kanz, F., Jackowski, C., and Lösch, S. (2015). Survival after trepanation—Early cranial surgery from Late Iron Age Switzerland. Int. J. Paleopathol. 11, 56–65. doi: 10.1016/j.ijpp.2015.08.002
Murlidharan, G., Samulski, R. J., and Asokan, A. (2014). Biology of adeno-associated viral vectors in the central nervous system. Front. Mol. Neurosci. 7:76. doi: 10.3389/fnmol.2014.00076
Nassi, J. J., Cepko, C. L., Born, R. T., and Beier, K. T. (2015). Neuroanatomy goes viral! Front. Neuroanat. 9:80. doi: 10.3389/fnana.2015.00080
Nauta, W. J. (1952). Selective silver impregnation of degenerating axons in the central nervous system. Stain Technol. 27, 175–179. doi: 10.3109/10520295209105080
Neve, R. L. (2012). Overview of gene delivery into cells using HSV-1-based vectors. Curr. Protoc. Neurosci. Chapter 4:Unit 4.12. doi: 10.1002/0471142301.ns0412s61
Nimchinsky, E. A., Sabatini, B. L., and Svoboda, K. (2002). Structure and function of dendritic spines. Annu. Rev. Physiol. 64, 313–353.
Noseda, R., Kainz, V., Jakubowski, M., Gooley, J. J., Saper, C. B., Digre, K., et al. (2010). A neural mechanism for exacerbation of headache by light. Nat. Neurosci. 13, 239–245. doi: 10.1038/nn.2475
O’Connor, D. M., and Boulis, N. M. (2015). Gene therapy for neurodegenerative diseases. Trends Mol. Med. 21, 504–512. doi: 10.1016/j.molmed.2015.06.001
O’Donovan, M. J., Ho, S., Sholomenko, G., and Yee, W. (1993). Real-time imaging of neurons retrogradely and anterogradely labelled with calciumsensitive dyes. J. Neurosci. Methods 46, 91–106. doi: 10.1016/0165-0270(93)90145-H
Oh, S. W., Harris, J. A., Ng, L., Winslow, B., Cain, N., Mihalas, S., et al. (2014). A mesoscale connectome of the mouse brain. Nature 508, 207–214. doi: 10.1038/nature13186
Ohyama, T., Schneider-Mizell, C. M., Fetter, R. D., Aleman, J. V., Franconville, R., Rivera-Alba, M., et al. (2015). A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639. doi: 10.1038/nature14297
Osten, P., and Margrie, T. W. (2013). Mapping brain circuitry with a light microscope. Nat. Methods 10, 515–523. doi: 10.1038/nmeth.2477
Oyibo, H. K., Znamenskiy, P., Oviedo, H. V., Enquist, L. W., and Zador, A. M. (2014). Long-term Cre-mediated retrograde tagging of neurons using a novel recombinant pseudorabies virus. Front. Neuroanat. 8:86. doi: 10.3389/fnana.2014.00086
Parker, L. M., Kumar, N. N., Lonergan, T., McMullan, S., and Goodchild, A. K. (2015). Distribution and neurochemical characterization of neurons in the rat ventrolateral medulla activated by glucoprivation. Brain Struct. Funct. 220, 117–134. doi: 10.1007/s00429-013-0642-3
Pearce, J. M. (2009). Marie-Jean-Pierre Flourens (1794-1867) and cortical localization. Eur. Neurol. 61, 311–314. doi: 10.1159/000206858
Penrod, R. D., Wells, A. M., Carlezon, W. A. Jr., and Cowan, C. W. (2015). Use of adeno-associated and herpes simplex viral vectors for In Vivo neuronal expression in mice. Curr. Protoc. Neurosci. 73, 4.37.1–4.37.31. doi: 10.1002/0471142301.ns0437s73
Pickard, G. E., Smeraski, C. A., Tomlinson, C. C., Banfield, B. W., Kaufman, J., Wilcox, C. L., et al. (2002). Intravitreal injection of the attenuated pseudorabies virus PRV-Bartha results in infection of the hamster suprachiasmatic nucleus only by retrograde transsynaptic transport via autonomic circuits. J. Neurosci. 22, 2701–2710. doi: 10.1523/jneurosci.22-07-02701.2002
Pidsudko, Z., Listowska, Z., Franke-Radowiecka, A., Klimczuk, M., Zalecki, M., and Kaleczyc, J. (2019). Distribution and chemical coding of urinary bladder apex-projecting neurons in aorticorenal and testicular ganglia of the male pig. Pol. J. Vet. Sci. 22, 427–430. doi: 10.24425/pjvs.2019.129227
Pilowsky, P., Llewellyn-Smith, I. J., Lipski, J., and Chalmers, J. (1992). Substance P immunoreactive boutons form synapses with feline sympathetic preganglionic neurons. J. Comp. Neurol. 320, 121–135. doi: 10.1002/cne.903200109
Puchades, M. A., Csucs, G., Ledergerber, D., Leergaard, T. B., and Bjaalie, J. G. (2019). Spatial registration of serial microscopic brain images to three-dimensional reference atlases with the QuickNII tool. PLoS One 14:e0216796. doi: 10.1371/journal.pone.0216796
Puigdellivol-Sanchez, A., Prats-Galino, A., Ruano-Gil, D., and Molander, C. (1998). Efficacy of the fluorescent dyes Fast Blue, Fluoro-Gold, and diamidino yellow for retrograde tracing to dorsal root ganglia after subcutaneous injection. J. Neurosci. Methods 86, 7–16. doi: 10.1016/s0165-0270(98)00137-x
Rancz, E. A., Franks, K. M., Schwarz, M. K., Pichler, B., Schaefer, A. T., and Margrie, T. W. (2011). Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nat. Neurosci. 14, 527–532. doi: 10.1038/nn.2765
Reardon, T. R., Murray, A. J., Turi, G. F., Wirblich, C., Croce, K. R., Schnell, M. J., et al. (2016). Rabies virus CVS-N2c(DeltaG) strain enhances retrograde synaptic transfer and neuronal viability. Neuron 89, 711–724. doi: 10.1016/j.neuron.2016.01.004
Reiner, A., Veenman, C. L., Medina, L., Jiao, Y., Del Mar, N., and Honig, M. G. (2000). Pathway tracing using biotinylated dextran amines. J. Neurosci. Methods 103, 23–37. doi: 10.1016/s0165-0270(00)00293-4
Renier, N., Adams, E. L., Kirst, C., Wu, Z., Azevedo, R., Kohl, J., et al. (2016). Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802. doi: 10.1016/j.cell.2016.05.007
Renier, N., Wu, Z., Simon, D. J., Yang, J., Ariel, P., and Tessier-Lavigne, M. (2014). iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. doi: 10.1016/j.cell.2014.10.010
Rinaman, L., and Schwartz, G. (2004). Anterograde transneuronal viral tracing of central viscerosensory pathways in rats. J. Neurosci. 24, 2782–2786. doi: 10.1523/jneurosci.5329-03.2004
Rothermel, M., Brunert, D., Zabawa, C., Díaz-Quesada, M., and Wachowiak, M. (2013). Transgene expression in target-defined neuron populations mediated by retrograde infection with adeno-associated viral vectors. J. Neurosci. 33, 15195–15206. doi: 10.1523/JNEUROSCI.1618-13.2013
Rubinov, M., and Bullmore, E. T. (2013). Fledgline pathoconnectomics of psychiatric disorders. Trends Cogn. Sci. 17, 641–647. doi: 10.1016/j.tics.2013.10.007
Ryan, K., Lu, Z., and Meinertzhagen, I. A. (2016). The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. eLife 5:e16962. doi: 10.7554/eLife.16962
Rytel, L., Snarska, A., Gonkowski, S., Wojtkiewicz, J., Szenci, O., and Sobiech, P. (2019). Identification of neuropeptide y in superior cervical ganglion neurons that project to the oesophagus - A combined immunohistochemical labelling and retrograde tracing study in pigs. Acta Vet. Hung. 67, 98–105. doi: 10.1556/004.2019.011
Salinas, S., Bilsland, L. G., Henaff, D., Weston, A. E., Keriel, A., Schiavo, G., et al. (2009). CAR-associated vesicular transport of an adenovirus in motor neuron axons. PLoS Pathog. 5:e1000442. doi: 10.1371/journal.ppat.1000442
Santiago-Ortiz, J., Ojala, D. S., Westesson, O., Weinstein, J. R., Wong, S. Y., Steinsapir, A., et al. (2015). AAV ancestral reconstruction library enables selection of broadly infectious viral variants. Gene Ther. 22, 934–946. doi: 10.1038/gt.2015.74
Santiago-Ortiz, J. L., and Schaffer, D. V. (2016). Adeno-associated virus (AAV) vectors in cancer gene therapy. J. Control. Release 240, 287–301. doi: 10.1016/j.jconrel.2016.01.001
Sathiyamoorthy, K., Chen, J., Longnecker, R., and Jardetzky, T. S. (2017). The COMPLEXity in herpesvirus entry. Curr. Opin. Virol. 24, 97–104. doi: 10.1016/j.coviro.2017.04.006
Schmued, L., Kyriakidis, K., and Heimer, L. (1990). In vivo anterograde and retrograde axonal transport of the fluorescent rhodamine-dextran-amine, Fluoro-Ruby, within the CNS. Brain Res. 526, 127–134. doi: 10.1016/0006-8993(90)90258-D
Schnell, M. J., McGettigan, J. P., Wirblich, C., and Papaneri, A. (2010). The cell biology of rabies virus: using stealth to reach the brain. Nat. Rev. Microbiol. 8, 51–61. doi: 10.1038/nrmicro2260
Schnutgen, F., Doerflinger, N., Calleja, C., Wendling, O., Chambon, P., and Ghyselinck, N. B. (2003). A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat. Biotechnol. 21, 562–565. doi: 10.1038/nbt811
Schofield, B. R. (2008). Retrograde axonal tracing with fluorescent markers. Curr. Protoc. Neurosci. Chapter 1:Unit1.17. doi: 10.1002/0471142301.ns0117s43
Schwab, M. E., Javoy-Agid, F., and Agid, Y. (1978). Labeled wheat germ agglutinin (WGA) as a new, highly sensitive retrograde tracer in the rat brain hippocampal system. Brain Res. 152, 145–150. doi: 10.1016/0006-8993(78)90140-3
Schwarz, L. A., Miyamichi, K., Gao, X. J., Beier, K. T., Weissbourd, B., DeLoach, K. E., et al. (2015). Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature 524, 88–92. doi: 10.1038/nature14600
Sillitoe, R. V. (2016). Mossy fibers terminate directly within purkinje cell zones during mouse development. Cerebellum 15, 14–17. doi: 10.1007/s12311-015-0712-6
Sivertsen, M. S., Glover, J. C., and Perreault, M. C. (2014). Organization of pontine reticulospinal inputs to motoneurons controlling axial and limb muscles in the neonatal mouse. J. Neurophysiol. 112, 1628–1643. doi: 10.1152/jn.00820.2013
Sivertsen, M. S., Perreault, M. C., and Glover, J. C. (2016). Pontine reticulospinal projections in the neonatal mouse: internal organization and axon trajectories. J. Comp. Neurol. 524, 1270–1291. doi: 10.1002/cne.23904
Smith, B. N., Banfield, B. W., Smeraski, C. A., Wilcox, C. L., Dudek, F. E., Enquist, L. W., et al. (2000). Pseudorabies virus expressing enhanved green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. U.S.A. 97, 9264–9269. doi: 10.1073/pnas.97.16.9264
Smith, J. E., Jansen, A. S., Gilbey, M. P., and Loewy, A. D. (1998). CNS cell groups projecting to sympathetic outflow of tail artery: neural circuits involved in heat loss in the rat. Brain Res. 786, 153–164. doi: 10.1016/s0006-8993(97)01437-6
Smith, R. H., Hallwirth, C. V., Westerman, M., Hetherington, N. A., Tseng, Y.-S., Cecchini, S., et al. (2016). Germline viral “fossils” guide in silico reconstruction of a mid-Cenozoic era marsupial adeno-associated virus. Sci. Rep. 6:28965. doi: 10.1038/srep28965
Sporns, O., Tononi, G., and Kotter, R. (2005). The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1:e42. doi: 10.1371/journal.pcbi.0010042
Staines, W. A., Kimura, H., Fibiger, H. C., and McGeer, E. G. (1980). Peroxidase-labeled lectin as a neuroanatomical tracer: evaluation in a CNS pathway. Brain Res. 197, 485–490. doi: 10.1016/0006-8993(80)91133-6
Stiefel, M., Shaner, A., and Schaefer, S. D. (2006). The edwin smith papyrus: the birth of analytical thinking in medicine and otolaryngology. Laryngoscope 116, 182–188. doi: 10.1097/01.mlg.0000191461.08542.a3
Stoeckel, K., Schwab, M., and Thoenen, H. (1977). Role of gangliosides in the uptake and retrograde axonal transport of cholera and tetanus toxin as compared to nerve growth factor and wheat germ agglutinin. Brain Res. 132, 273–285. doi: 10.1016/0006-8993(77)90421-8
Stornetta, R. L., Inglis, M. A., Viar, K. E., and Guyenet, P. G. (2016). Afferent and efferent connections of C1 cells with spinal cord or hypothalamic projections in mice. Brain Struct. Funct. 221, 4027–4044. doi: 10.1007/s00429-015-1143-3
Strack, A. M., Sawyer, W. B., Hughes, J. H., Platt, K. B., and Loewy, A. D. (1989a). A general pattern of CNS innervation of the sympathetic outflow demonstrated by transneuronal pseudorabies viral infections. Brain Res. 491, 156–162. doi: 10.1016/0006-8993(89)90098-x
Strack, A. M., Sawyer, W. B., Platt, K. B., and Loewy, A. D. (1989b). CNS cell groups regulating the sympathetic outflow to adrenal gland as revealed by transneuronal cell body labeling with pseudorabies virus. Brain Res. 491, 274–296. doi: 10.1016/0006-8993(89)90063-2
Sun, L., Tang, Y., Yan, K., Yu, J., Zou, Y., Xu, W., et al. (2019). Differences in neurotropism and neurotoxicity among retrograde viral tracers. Mol. Neurodegener. 14:8. doi: 10.1186/s13024-019-0308-6
Sun, N., Cassell, M. D., and Perlman, S. (1996). Anterograde transneuonal transport of herpes simplex virus type 1 strain H129 in the murine visual system. J. Virol. 70, 5405–5413.
Sun, S., and Schaffer, D. V. (2018). Engineered viral vectors for functional interrogation, deconvolution, and manipulation of neural circuits. Curr. Opin. Neurobiol. 50, 163–170. doi: 10.1016/j.conb.2017.12.011
Svoboda, K. (2019). “Using rabies virus for tracing neural connections: caveats and limitations exposed by studies of barrel cortex circuits,” in Spikesphotons, Wordpress.com. Available at: https://spikesphotons.blog/2019/05/13/using-rabies-virus-for-tracing-neural-connections-caveats-and-limitations-exposed-by-studies-of-barrel-cortex-circuits/ (accessed July 24, 2019).
Swanson, L. W., and Bota, M. (2010). Foundational model of structural connectivity in the nervous system with a schema for wiring diagrams, connectome, and basic plan architecture. Proc. Natl. Acad. Sci. U.S.A. 107, 20610–20617. doi: 10.1073/pnas.1015128107
Takai, Y., Miyoshi, J., Ikeda, W., and Ogita, H. (2008). Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 9, 603–615. doi: 10.1038/nrm2457
Takatoh, J., Nelson, A., Zhou, X., Bolton, M. M., Ehlers, M. D., Arenkiel, B. R., et al. (2013). New modules are added to vibrissal premotor circuitry with the emergence of exploratory whisking. Neuron 77, 346–360. doi: 10.1016/j.neuron.2012.11.010
Tervo, D. G., Hwang, B. Y., Viswanathan, S., Gaj, T., Lavzin, M., Ritola, K. D., et al. (2016). A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382. doi: 10.1016/j.neuron.2016.09.021
Thanos, S., Rohrbach, J. M., and Thiel, H. J. (1991). Postmortem preservation of ganglion cells in the human retina. A morphometric investigation with the carbocyanine dye DiI. Retina 11, 318–327. doi: 10.1097/00006982-199111030-00009
Thanos, S., Vanselow, J., and Mey, J. (1992). Ganglion cells in the juvenile chick retina and their ability to regenerate axons in vito. Exp. Eye Res. 54, 377–391. doi: 10.1016/0014-4835(92)90050-3
Tomer, R., Ye, L., Hsueh, B., and Deisseroth, K. (2014). Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697. doi: 10.1038/nprot.2014.123
Trivino-Paredes, J. S., Nahirney, P. C., Pinar, C., Grandes, P., and Christie, B. R. (2019). Acute slice preparation for electrophysiology increases spine numbers equivalently in the male and female juvenile hippocampus: a DiI labeling study. J. Neurophysiol. doi: 10.1152/jn.00332.2019 [Epub ahead of print].
Trojanowski, J. Q., Gonatas, J. O., and Gonatas, N. K. (1981). Conjugates of horseradish peroxidase (HRP) with cholera toxin and wheat germ agglutinin are superior to free HRP as orthogradely transported markers. Brain Res. 223, 381–385. doi: 10.1016/0006-8993(81)91151-3
Trojanowski, J. Q., Gonatas, J. O., and Gonatas, N. K. (1982). Horseradish peroxidase (HRP) conjugates of cholera toxin and lectins are more sensitive retrogradely transported markers than free HRP. Brain Res. 231, 33–50. doi: 10.1016/0006-8993(82)90005-1
Turner, A., Kumar, N., Farnham, M., Lung, M., Pilowsky, P., and McMullan, S. (2013). Rostroventrolateral medulla neurons with commissural projections provide input to sympathetic premotor neurons: anatomical and functional evidence. Eur. J. Neurosci. 38, 2504–2515. doi: 10.1111/ejn.12232
Ugolini, G. (1995). Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J. Comp. Neurol. 356, 457–480. doi: 10.1002/cne.903560312
Ugolini, G. (2010). Advances in viral transneuronal tracing. J. Neurosci. Methods 194, 2–20. doi: 10.1016/j.jneumeth.2009.12.001
Ugolini, G. (2011). Rabies virus as a transneuronal tracer of neuronal connections. Adv. Virus Res. 79, 165–202. doi: 10.1016/B978-0-12-387040-7.00010-X
Ugolini, G., Kuypers, H. G. J. M., and Simmons, A. (1987). Retrograde transneuronal transfer of Herpes simplex virus type 1 (HSV 1) from motoneurons. Brain Res. 422, 242–256. doi: 10.1016/0006-8993(87)90931-0
van den Heuvel, M. P., Bullmore, E. T., and Sporns, O. (2016). Comparative connectomics. Trends Cogn. Sci. 20, 345–361. doi: 10.1016/j.tics.2016.03.001
Varshney, L. R., Chen, B. L., Paniagua, E., Hall, D. H., and Chklovskii, D. B. (2011). Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput. Biol. 7:e1001066. doi: 10.1371/journal.pcbi.1001066
Veenman, C. L., Reiner, A., and Honig, M. G. (1992). Biotinylated dextran amine as an anterograde tracer for single- and double-labeling studies. J. Neurosci. Methods 41, 239–254. doi: 10.1016/0165-0270(92)90089-v
Velez-Fort, M., Rousseau, C. V., Niedworok, C. J., Wickersham, I. R., Rancz, E. A., Brown, A. P., et al. (2014). The stimulus selectivity and connectivity of layer six principal cells reveals cortical microcircuits underlying visual processing. Neuron 83, 1431–1443. doi: 10.1016/j.neuron.2014.08.001
Vercelli, A., Repici, M., Garbossa, D., and Grimaldi, A. (2000). Recent techniques for tracing pathways in the central nervous system of developing and adult mammals. Brain Res. Bull. 51, 11–28. doi: 10.1016/s0361-9230(99)00229-4
Vos, A., Neubert, A., Aylan, O., Schuster, P., Pommerening, E., Muller, T., et al. (1999). An update on safety studies of SAD B19 rabies virus vaccine in target and non-target species. Epidemiol. Infect. 123, 165–175. doi: 10.1017/s0950268899002666
Wang, Y., Yau, Y.-Y., Perkins-Balding, D., and Thomson, J. G. (2011). Recombinase technology: applications and possibilities. Plant Cell Rep. 30, 267–285. doi: 10.1007/s00299-010-0938-1
Wanner, A. A., Kirschmann, M. A., and Genoud, C. (2015). Challenges of microtome-based serial block-face scanning electron microscopy in neuroscience. J. Microsc. 259, 137–142. doi: 10.1111/jmi.12244
Watakabe, A., Ohtsuka, M., Kinoshita, M., Takaji, M., Isa, K., Mizukami, H., et al. (2015). Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neurosci. Res. 93, 144–157. doi: 10.1016/j.neures.2014.09.002
Watson, J. D., and Crick, F. H. (1953). Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738.
Weinberg, M. S., Samulski, R. J., and McCown, T. J. (2013). Adeno-associated virus (AAV) gene therapy for neurological disease. Neuropharmacology 69, 82–88. doi: 10.1016/j.neuropharm.2012.03.004
Wertz, A., Trenholm, S., Yonehara, K., Hillier, D., Raics, Z., Leinweber, M., et al. (2015). PRESYNAPTIC NETWORKS. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science 349, 70–74. doi: 10.1126/science.aab1687
White, J. G., Southgate, E., Thomson, J. N., and Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340.
Wickersham, I. R., and Feinberg, E. H. (2012). New technologies for imaging synaptic partners. Curr. Opin. Neurobiol. 22, 121–127. doi: 10.1016/j.conb.2011.12.001
Wickersham, I. R., Finke, S., Conzelmann, K. K., and Callaway, E. M. (2007a). Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 4, 47–49. doi: 10.1038/nmeth999
Wickersham, I. R., Lyon, D. C., Barnard, R. J., Mori, T., Finke, S., Conzelmann, K. K., et al. (2007b). Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647. doi: 10.1016/j.neuron.2007.01.033
Wojaczynski, G. J., Engel, E. A., Steren, K. E., Enquist, L. W., and Patrick Card, J. (2015). The neuroinvasive profiles of H129 (herpes simplex virus type 1) recombinants with putative anterograde-only transneuronal spread properties. Brain Struct. Funct. 220, 1395–1420. doi: 10.1007/s00429-014-0733-9
Wouterlood, F. G., Bloem, B., Mansvelder, H. D., Luchicchi, A., and Deisseroth, K. (2014). A fourth generation of neuroanatomical tracing techniques: exploiting the offspring of genetic engineering. J. Neurosci. Methods 235, 331–348. doi: 10.1016/j.jneumeth.2014.07.021
Wu, C. W., Vasalatiy, O., Liu, N., Wu, H., Cheal, S., Chen, D. Y., et al. (2011). Development of a MR-visible compound for tracing neuroanatomical connections in vivo. Neuron 70, 229–243. doi: 10.1016/j.neuron.2011.03.010
Xu, C. S., Hayworth, K. J., Lu, Z., Grob, P., Hassan, A. M., García-Cerdán, J. G., et al. (2017). Enhanced FIB-SEM systems for large-volume 3D imaging. eLife 6:e25916. doi: 10.7554/eLife.25916
Yamashita, T., and Petersen, C. (2016). Target-specific membrane potential dynamics of neocortical projection neurons during goal-directed behavior. eLife 5:e15798. doi: 10.7554/eLife.15798
Yonehara, K., Farrow, K., Ghanem, A., Hillier, D., Balint, K., Teixeira, M., et al. (2013). The first stage of cardinal direction selectivity is localized to the dendrites of retinal ganglion cells. Neuron 79, 1078–1085. doi: 10.1016/j.neuron.2013.08.005
Yu, Y.-L., Li, H.-Y., Zhang, P.-X., Yin, X.-F., Han, N., Kou, Y.-H., et al. (2015). Comparison of commonly used retrograde tracers in rat spinal motor neurons. Neural Regen. Res. 10, 1700–1705. doi: 10.4103/1673-5374.167772
Zele, T., Sketelj, J., and Bajrovic, F. F. (2010). Efficacy of fluorescent tracers in retrograde labeling of cutaneous afferent neurons in the rat. J. Neurosci. Methods 191, 208–214. doi: 10.1016/j.jneumeth.2010.06.021
Zemanick, M. C., Strick, P. L., and Dix, R. D. (1991). Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc. Natl. Acad. Sci. U.S.A. 88, 8048–8051. doi: 10.1073/pnas.88.18.8048
Zeng, W.-B., Jiang, H.-F., Gang, Y.-D., Song, Y.-G., Shen, Z.-Z., Yang, H., et al. (2017). Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Mol. Neurodegener. 12:38. doi: 10.1186/s13024-017-0179-7
Zhang, W. J., Xu, D. S., Cui, J. J., Jing, X. H., Xu, N. G., Liu, J. H., et al. (2017). Anterograde and retrograde tracing with high molecular weight biotinylated dextran amine through thalamocortical and corticothalamic pathways. Microsc. Res. Tech. 80, 260–266. doi: 10.1002/jemt.22797
Zhao, Z., Wang, L., Gao, W., Hu, F., Zhang, J., Ren, Y., et al. (2017). A central catecholaminergic circuit controls blood glucose levels during stress. Neuron 95, 138–152.e5. doi: 10.1016/j.neuron.2017.05.031
Zheng, Z., Lauritzen, J. S., Perlman, E., Robinson, C. G., Nichols, M., Milkie, D., et al. (2018). A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174, 730–743.e22. doi: 10.1016/j.cell.2018.06.019
Keywords: neuroanatomy, viral tracers, anterograde tracer, retrograde tracers, synaptic contacts, connectome analysis
Citation: Saleeba C, Dempsey B, Le S, Goodchild A and McMullan S (2019) A Student’s Guide to Neural Circuit Tracing. Front. Neurosci. 13:897. doi: 10.3389/fnins.2019.00897
Received: 31 May 2019; Accepted: 12 August 2019;
Published: 27 August 2019.
Edited by:
Vaughan G. Macefield, Baker Heart and Diabetes Institute, AustraliaReviewed by:
Patrice G. Guyenet, University of Virginia, United StatesEberhard Weihe, University of Marburg, Germany
Alice McGovern, The University of Melbourne, Australia
Copyright © 2019 Saleeba, Dempsey, Le, Goodchild and McMullan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simon McMullan, simon.mcmullan@mq.edu.au
†These authors have contributed equally to this work
 Christine Saleeba
Christine Saleeba Bowen Dempsey
Bowen Dempsey Sheng Le1
Sheng Le1 Ann Goodchild
Ann Goodchild Simon McMullan
Simon McMullan