- 1Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
- 2Department of Cell Research and Immunology, School of Molecular Cell Biology and Biotechnology, The George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel
- 3Department of Psychiatry and Biobehavioral Sciences, Semel Institute for Neuroscience and Human Behavior, University of California, Los Angeles, Los Angeles, CA, United States
- 4Department of Translational Molecular Medicine, John Wayne Cancer Institute at Providence Saint John’s Health Center, Santa Monica, CA, United States
Neural repair after stroke involves initiation of a cellular proliferative program in the form of angiogenesis, neurogenesis, and molecular growth signals in the surrounding tissue elements. This cellular environment constitutes a niche in which regeneration of new blood vessels and new neurons leads to partial tissue repair after stroke. Cancer metastasis has similar proliferative cellular events in the brain and other organs. Do cancer and CNS tissue repair share similar cellular processes? In this study, we identify a novel role of the regenerative neurovascular niche induced by stroke in promoting brain melanoma metastasis through enhancing cellular interactions with surrounding niche components. Repair-mediated neurovascular signaling induces metastatic cells to express genes crucial to metastasis. Mimicking stroke-like conditions in vitro displays an enhancement of metastatic migration potential and allows for the determination of cell-specific signals produced by the regenerative neurovascular niche. Comparative analysis of both in vitro and in vivo expression profiles reveals a major contribution of endothelial cells in mediating melanoma metastasis. These results point to a previously undiscovered role of the regenerative neurovascular niche in shaping the tumor microenvironment and brain metastatic landscape.
Introduction
In both health and disease, cellular interactions in the brain occur around a neurovascular niche of neurons, endothelial cells, astrocytes, and stromal cells that maintain blood–brain barrier function and neuronal signaling. Injury to this neurovascular niche during stroke elicits spontaneous but limited repair processes that drastically alter the local environment: creating a cellular niche for tissue repair. Immature neurons (neuroblasts) derived from neural stem cells in the subventricular zone migrate to injured brain regions and differentiate into neurons. These migrating neuroblasts tightly associate with the angiogenic vasculature, which secretes factors essential to post-stroke neurogenesis (Ohab and Carmichael, 2008; Lindvall and Kokaia, 2015). Reactive astrocytes and microglia interact extensively with neuroblasts during post-stroke neurogenesis and engage in signaling via cytokines and growth factors to mediate several parallel cellular functions after stroke, including increased angiogenesis (Ohab and Carmichael, 2008; Lindvall and Kokaia, 2015). Because these cellular elements collectively mediate repair after stroke through angiogenesis, neurogenesis, and tissue reorganization, this microenvironment has been termed the regenerative neurovascular niche (Brumm and Carmichael, 2012).
Brain metastasis presents analogous features with these regenerative neurovascular processes after stroke. In stroke tissue repair, neuroblast migration from a distant brain region into the regenerative neurovascular niche after stroke resembles the migration and localization of a metastatic cancer cell within the brain. Angiogenesis is an essential process in both brain metastasis and repair after stroke (Fukumura et al., 2010; Ergul et al., 2012). Reactive astrocytes interact with both neuroblasts and metastatic cells, and are essential in promoting neurogenesis and metastasis (Klein et al., 2015).
Compelling evidence suggests that similar signaling pathways regulate both stroke repair and brain metastasis (Yao Y. et al., 2016). Tumor suppressor phosphatase tensin homolog (PTEN) is commonly mutated in cancers, and PTEN loss activates oncogenes that reprogram the tumor microenvironment and lead to the formation of brain metastasis. PTEN inhibition after stroke promotes axon regrowth and neuronal repair (Bronisz et al., 2011; Song et al., 2012; Wikman et al., 2012; Zhang et al., 2015). VEGF and angiopoietin signaling mediate endothelial cell proliferation and migration in both cancer metastasis and neural repair after stroke (Hansen et al., 2008; Yang et al., 2010; Ziyad and Iruela-Arispe, 2011; Lange et al., 2016). Matrix metalloproteases (MMPs) also have dual roles in their proteolytic activity, promoting cancer migration and invasion and enhancing angiogenesis after stroke (Lutsenko et al., 2003; Ben-Baruch, 2008; Noel et al., 2008; Izraely et al., 2010; Gialeli et al., 2011; Weitzenfeld and Ben-Baruch, 2013). The TGF-β system regulates cancer cell proliferation in a context-dependent manner (Yang et al., 2008) and mediates tissue reorganization and recovery after stroke (Li et al., 2015). These studies suggest significant overlap in signaling mechanisms of cancer metastasis and tissue repair after stroke.
In short, while the processes of tissue regeneration and metastasis have essentially opposite functional outcomes, the niche components that drive them share similar cellular and molecular features. Both the metastatic tumor and regenerative neurovascular niches involve signaling among a principal cell type (the cancer cell or neuroblast), angiogenic vasculature, reactive astrocytes, and stromal cells. Based on the parallel biology between these two disease processes we asked: (a) could the regenerative neurovascular niche in stroke create a facilitative substrate for brain metastasis, (b) could the synergy between tissue repair processes and metastasis enhance metastatic cell-niche interactions, and (c) does the presence of repair-mediated neurovascular signaling alter the metastatic transcriptome to enhance metastasis?
Materials and Methods
Cell Culture
Human brain metastatic melanoma cells YDFR-CB3 were generated in the Witz laboratory as described earlier (Izraely et al., 2012; Klein-Goldberg et al., 2013). YDFR-CB3 cells were cultured in RPMI medium supplemented with 10% heat-inactivated fetal calf serum, 2 mmol/mL L-glutamine, 0.01M HEPES buffered saline, 100 units/mL penicillin, 0.1 mg/mL streptomycin, and 12.5 units/mL nystatin. The YDFR-CB3 cells were transduced with GFP virus (pCDH10-EF1-MCS-2A-copGFP) (Boiko et al., 2010; Quintana et al., 2012). To produce mCherry expressing cells, melanoma cells were transduced with a pQCXIN-mCherry plasmid and selected using 800 μg/mL G418 Sulfate (A.G. Scientific, Inc., San Diego, CA, United States). Immortalized human brain microvascular endothelial cells (hEC) were kindly provided by Dr. Clara Nahmias and Prof. Pierre-Olivier Couraud (Inserm, U1016, Institut Cochin, Paris, France; Cnrs, UMR8104, Paris, France; University Paris Descartes, Paris, France). hEC cells were cultured on collagen type I, rat tail (1:30; BD Biosciences, Bedford, MA, United States) in EBM-2 medium (Clonetics, Cambrex BioScience, Wokingham, United Kingdom) supplemented with 5% heat-inactivated FCS, 2 mmol/mL L-glutamine, 100 units/mL penicillin, 0.1 mg/mL streptomycin, 12.5 units/mL nystatin, 1 ng/mL basic FGF, 10 mM HEPES buffered saline and 1.4 μM hydrocortisone. Human astrocytes (HA; ScienCell Research Laboratories, Carlsbad, CA, United States) were cultured on 0.0015% poly-L-lysine (Sigma-Aldrich, St. Louis, MO, United States) in astrocyte growth medium (AM, ScienCell Research Laboratories) supplemented with 2% heat-inactivated FCS. Cells were routinely cultured in humidified air with 5% CO2 at 37°C.
Modeling Stroke and Metastasis
All animal procedures were carried out in accordance with the National Institutes of Health animal protection guidelines and protocols approved by University of California, Los Angeles Chancellor’s Animal Research Committee. NOD-scid gamma (NSG), a highly immunodeficient mouse model lacking T cells, B cells, and NK cells, was used for these studies (Jackson Laboratories). Inducing strokes in this immunodeficient mouse strain provides a mouse model that allows for the engraftment of human cancer cells without graft rejection and little to no involvement of innate or acquired immunity (Agliano et al., 2008; Chen Q. et al., 2009).
Ischemic strokes were induced in adult male NSG mice aged 2–4 months. Briefly, animals were anesthetized under 2.5–3% isoflurane and positioned on a stereotactic apparatus (Model 940, David Kopf Instruments). To expose the skull, a midline incision was made followed by a transverse incision above the zygomatic arch. A craniotomy was performed to expose the left middle cerebral artery (MCA). The proximal branch of the MCA was cauterized followed by bilateral occlusion of the jugular veins for 15 min. Body temperature was maintained at 37 ± 0.5°C throughout the procedure (RightTempTM, Kent Scientific, Torrington, CT, United States) as measured using a rectal probe. After cauterization, the skin was glued together to cover the skull and animals were returned to their home cages for recovery. Metastasis was modeled by injecting YDFR-CB3GFP+ cells (106 cells/50 μl of L15 medium) intracardially 7 days after stroke when the course of regeneration/tissue repair by means of angiogenesis and neurogenesis had reached peak activity (Ohab et al., 2006; Lamanna, 2012). On day 14 after stroke, animals were euthanized and the brains were processed for further experiments.
Sham surgeries were not performed. But surgical incision areas were examined for any tumor homing and no metastasis was observed in these regions.
Quantitative Analysis of Distribution of Metastasis
Two cohorts consisting of six to eight male NSG mice each were utilized to quantify the distribution of metastasis in different brain regions. One cohort of animals was subjected to stroke and metastasis and the other with metastasis alone. Brains were extracted, fixed in 4% paraformaldehyde and cryosectioned into 100 μm thick sagittal sections while frozen in OCT. Four sections per animal were used to quantify metastasis and the distribution of YDFR-CB3GFP+ cells in various brain regions. Sections were imaged, outlined, and GFP+ cells were manually counted using a stereoinvestigator (StereoInvestigator MBF Bioscience, Williston, VT, United States) on a Leica DMLB fluorescent microscope (Leica Microsystems, Wetzlar, Germany) with a 40× objective.
Immunohistochemistry and Confocal Imaging
Immunohistochemistry was performed on free-floating brain sections. Briefly, entire extracted brains were fixed in 4% paraformaldehyde for 24 h and then cryoprotected in 30% sucrose for 48 h. After cryosectioning, 50 μm thick sagittal sections were washed 3× with PBS (phosphate buffered saline) and blocked with 2% horse serum and 5% normal donkey serum in 0.3% Triton. The primary antibodies used are goat anti-DCX (Santa Cruz Biotechnology, CA); rat anti-PECAM (BD Pharminogen); rat anti-GFAP (Invitrogen, IL); Rb anti-albumin (Genetex, CA); mouse anti-MART-1 (Novus biologicals, CO); Rb anti-Tuc-4 (EMD Millipore, CA); mouse antiPSA-NCAM (Millipore, CA); Isolectin Dylight 649 (Vector labs, CA). Sections were incubated with primary antibodies overnight followed by 3× wash with PBS. Secondary antibodies were tested without primary antibodies for non-specific binding were negative (data not shown). Sections were stained with respective secondary antibodies, incubated for 1 h, and washed 3× with PBS. The stained sections were mounted on slides, dehydrated and coverslipped.
For the time course experiments assessing cellular relationships, GFAP, DCX, and lectin immunoreactive areas around the peri-infarct region were imaged. Images were captured of three fields at 100× for astrocytes and 40× for neuroblast and vasculature from three independent sagittal sections of each animal on a confocal microscope (Nikon C2). Z-stacked images of size 512 × 512 were captured. Scanning parameters were maintained constant across groups for consistency.
Human brain metastatic melanoma tissues excised from patients were used to assess neurovascular associations with the metastatic melanoma cells. Paraffin-embedded, slide-mounted human brain sections (5 μm thick) were first immersed in water at 85°C and washed three times, de-paraffinized and hydrated. Double antigen retrieval was performed first using Sodium citrate pH 6 followed by EDTA at pH 8 for 25 min each. As outlined above, immunohistochemistry was performed with MART-1, Tuc-4, and GFAP to identify melanoma cells, neuroblasts and astrocytes, respectively. The Tuc-4, DCX, and GFAP stains were optimized on human hippocampus sections prior to staining the human brain metastatic melanomas.
Microsphere Injections and Analysis
Blood–brain barrier leakage and melanoma extravasation were tested with an intracardiac injection of 100 μl of 0.2-μm diameter yellow–green polystyrene fluorescent microspheres (Molecular Probes) which are relatively smaller than metastatic melanoma cells in NSG mice 1 and 7 days after stroke. The microspheres were allowed to circulate for 8 min and the animals were euthanized and brains extracted for immunohistochemistry. 100 μm thick sections were immunostained for the vasculature using PECAM and imaged with Nikon C2 confocal microscope. Intravascular microsphere volume was analyzed (Volocity Improvision 5.5). On the 3D rendered image PECAM positive regions were first detected and selected then regions outside of the PECAM were cropped and the volume of GFP+ microspheres accumulated inside these PECAM vessels within the peri-infarct and corresponding contralateral regions was quantified. Schematics are shown in Figure 1J.
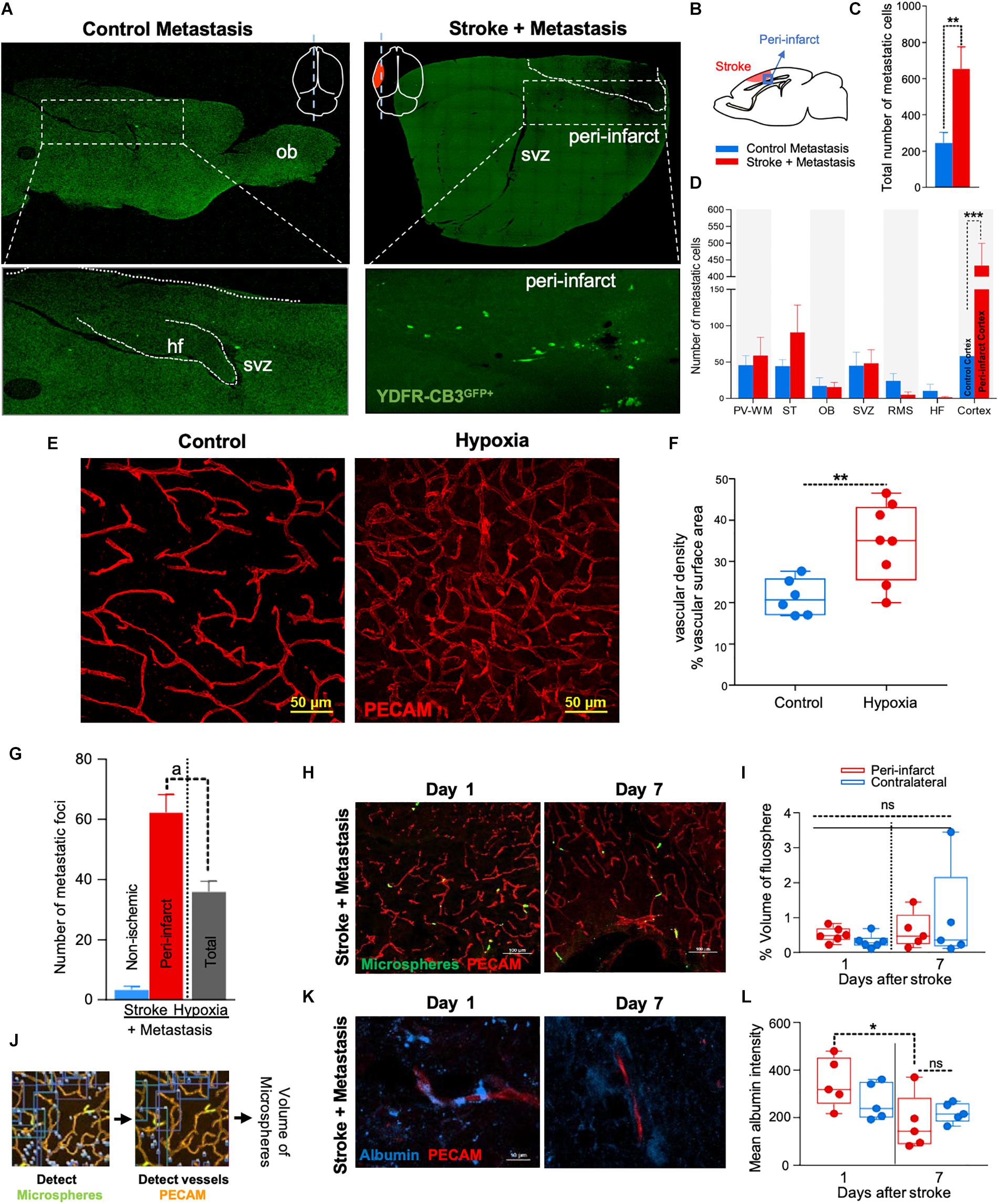
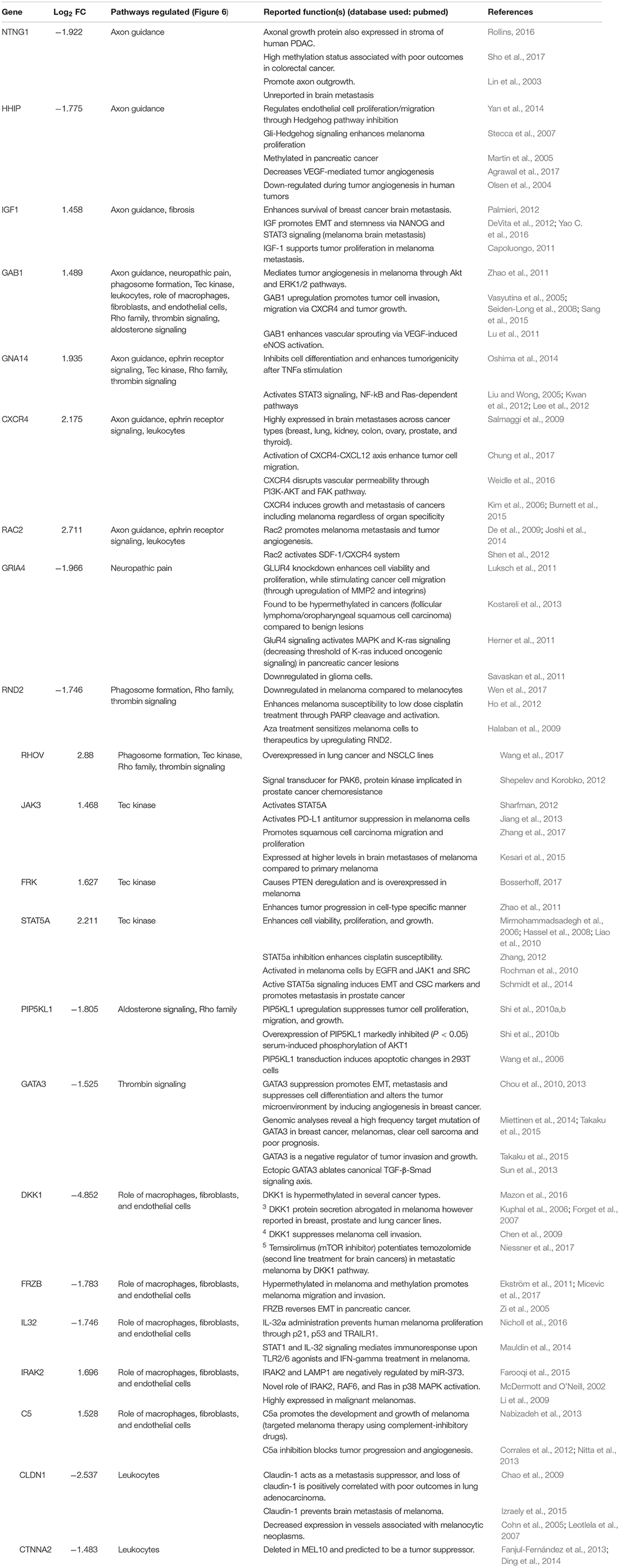
Figure 1. The regenerative neurovascular niche after stroke facilitates brain metastasis. (A) Representative brain images from control and stroke groups subjected to melanoma metastasis shown in the top panel. GFP+ cells identifies brain metastatic melanoma at high magnification in the bottom panel. (B) Schematics showing region from stroke brains and corresponding regions in control quantified. (C) Bar graphs of total number of metastatic cells in both control and stroke groups and (D) showing quantification and distribution of metastatic cells in different brain regions as mean ± SEM (n = 6–7, four sections per animal, ∗∗p = 0.0082, ∗∗∗p = 0.0012 Mann–Whitney, two-tailed t-test). PV-WM, periventricular white matter; ST, striatum; OB, olfactory bulb; SVZ, subventricular zone; RMS, rostral migratory stream; HF, hippocampus. (E) Representative images depicting vascular density in control and hypoxia + metastasis group. (F) Box and scatter plots with minimum and maximum percentage vascular density (surface area μm3) mean ± SEM (n = 6–8), (∗∗p = 0.0090 unpaired, two-tailed t-test) (G) Stroke increases. brain metastasis significantly more than hypoxia mediated angiogenesis. Bar graphs showing number of metastatic foci in stroke and hypoxic groups shown as mean ± SEM (n = 6–8, four sections/animal, ap = 0.003, Mann–Whitney, two-tailed t-test). (H) Representative images from days 1 and 7 after stroke showing localization of fluorescent microspheres in green and vasculature (PECAM) in red. (I) Box and scatter plots with minimum and maximum percentage volume of fluorescent microspheres localized to the peri-infarct and contralateral regions at days 1 and 7 after stroke, mean ± SEM (n = 5–6, one-way ANOVA, Holm-Sidak for multiple comparisons p = 0.46, ns, non-significant). (J) Schematics of the method of quantification of fluorescent microspheres volume localized around the peri-infarct regions. (K,L) Representative immunohistochemical images and intensity of endogenous albumin extravasated from days 1 and 7 after stroke and contralateral regions, mean ± SEM (n = 5, three regions/animal, ∗p = 0.0255, one-way ANOVA, Holm-Sidak for multiple comparisons).
In vivo Hypoxia Model
NOD-scid gamma mice were housed for 7 days in a hypoxic chamber (Coy Laboratory Products, Grass Lake, MI, United States) maintained with a continuous flow of 10% O2 mixed with N2 as background gas. Chamber O2 concentration was checked daily and CO2 was eliminated by refurbishing the hypoxic chamber with fresh O2. Humidity from respiration was controlled with silica desiccant. Animals were provided with food and water ad libitum throughout the experiment. After 7 days of hypoxia exposure, YDFR-CB3GFP+ cells (106 cells/50 μl of L15 medium) were intracardially injected. The animals were then housed in normoxia/ambient Oxygen levels until day 14. The animals were then euthanized and the brain extracted to analyze the metastatic response to hypoxia-mediated angiogenesis (Harik et al., 1996; Tarnawski et al., 2014).
Quantitative Analysis of Cellular Associations
Neurovascular associations with melanoma were assessed using DCX-RFP reporter mice. These mice were immunosuppressed with IP injections of Tacrolimus-FK-506 (Cell Signaling, Danvers, MA, United States) beginning with a daily intraperitoneal injection of Tacrolimus (3 mg/kg/day; dissolved in DMSO) 2 days prior to stroke and continuing until the mice were euthanized. The extracted whole brains were fixed, permeabilized, and cryosectioned into 50 μm thick sections. Cell-specific antibodies identified each of these cell types (neuroblasts, astrocytes, and vasculature) through immunohistochemistry.
The cellular association of brain metastatic melanoma with the cells in the regenerative neurovascular niche was assessed by measuring the distances between individual components (astrocytes, neuroblasts) and YDFR-CB3GFP+ cells using Imaris software version 8.3.1 (Bitplane®). Inflection point analysis and melanoma distance to vascular segments were measured using Volocity Improvision 6. Inflection point measures the number of irregularities within a curvature or change in direction of curvature (Bullitt et al., 2003, 2007). Inflection point ratios were obtained by counting the number of nodes on a vascular segment associated with a metastatic foci and normalizing it to a distant vasculature from the same animal. Vascular segments of the same linear length were compared for every observation for consistency.
In vitro Oxygen Glucose Deprivation (OGD) and Generation of Conditioned Medium
An oxygen-glucose deprivation (OGD) treatment of brain microenvironmental cells was used as an in vitro model for stroke (Yang et al., 2012). 5 × 105 human brain endothelial cells and astrocytes were seeded for 24 h. Then, cells were washed with PBSX1 and culture medium was replaced with 0.5% FCS supplemented DMEM medium lacking glucose. The cells were incubated in a hypoxia chamber (Hot box; Billups-Rothenberg Inc, Del Mar, CA, United States) with a gas mixture of: 1% O2, 5% CO2, and 94% N2 for 4 h in 37°C. Cells in control group were treated with 0.5% FCS supplemented DMEM medium with normal glucose levels. These cells were incubated in normoxia for 4 h in 37°C.
Following hypoxia or normoxia, both OGD and control culture media were replaced with a suitable 0.5% FCS supplemented medium (EBM2 for endothelial cells or AM for astrocytes) and incubated in normoxia for 20 h in 37°C, to simulate the reperfusion phase following stroke. Finally, conditioned media from control and OGD cultures was collected, centrifuged and filtered (0.45 μm; Whatman GmbH, Freiburg, Germany) on ice.
Viability Assay
To examine cell viability of astrocytes and BECs subjected to the OGD treatment, 5 × 103 cells were seeded overnight in coated 96 well-tissue plates. Cells were washed once with PBSX1 and exposed to OGD treatment or normoxia for 4 h with 100 μl relevant DMEM starvation medium. In these studies, AM is astrocyte medium (cat. no. 1801 ScienCell Research Laboratories) and EBM2 is Endothelial Cell Growth Basal Medium-2 (cat. no. 00190860 Lonza). After 4 h the medium was replaced with EBM-2 or AM containing 0.5% serum. Cell viability was determined after 44 h by using Cell Proliferation Kit (XTT, Biological industries, Kibbutz Beit Haemek, Israel), according to the manufacturer’s instructions. Absorbance at 450 nm (OD450) was determined for each well using an automated microplate reader (SpectraMax 190; Molecular Devices, Sunnyvale, CA, United States) and subtraction of non-specific readings (measured at 630 nm) was done automatically during the OD450 reading.
Adhesion Assay
Approximately 5 × 104 brain endothelial cells were cultured overnight to form a confluent monolayer, then washed with PBSX1 and subjected to OGD or normoxia for 4 h with 100 μl relevant DMEM starvation medium. After 4 h, the medium was replaced with 100 μl EBM-2 starvation medium containing 0.5% FCS (EC starvation medium) for 20 h.
At the end of the reperfusion phase to model what happens in the in vivo setting, brain endothelial cells, in vitro were gently washed twice with PBSX1. 1 × 105 m-Cherry labeled melanoma cells diluted in TBSX1 (Tris-buffered saline) containing 2 mM CaCl2 were added onto the endothelial monolayer and incubated for 30 min at 37°C to allow adhesion to occur. The total fluorescence signal of labeled cells added to the well before the removal of the non-adherent cells was measured by a fluorescent microplate reader (BioTek FL500, BioTek Instruments, Inc., Winooski, VT, United States) at wavelength of 490/530. Then, the wells were washed three times with PBSX1 to remove non-adherent cells and fluorescence of the adherent cells was measured at wavelength of 490/530. To obtain the percentage of adherent cells, the ratio between the OD of the adherent cells and that of the total cells plated was calculated and multiplied by 100. To determine VCAM-1 involvement, 5 μl/mL VCAM-1 blocking antibody (BD PharmingenTM, San Diego, CA, United States) was added to the m-Cherry labeled melanoma cells that were added onto the endothelial monolayer following the OGD treatment.
ECM Transmigration Assay
About 5 × 104 brain endothelial cells or astrocytes were cultured overnight. Cells were washed once with PBSX1 and subjected to OGD treatment or normoxia for 4 h with relevant DMEM starvation medium. After 4 h, the medium was replaced with EBM-2 starvation medium for endothelial cells or AM starvation medium containing 0.5% FCS for astrocytes for 20 h. Transwell inserts (6.5 mm diameter polycarbonate membrane with 8.0 μm pores, Corning Costar Corp., New York, NY, United States) were coated with 100 μg/mL collagen type I for 1 h at 37°C. After two washes with 200 μl PBSX1, 1 × 105 melanoma cells were added to the Transwell for 1.5 h at 37°C, and the cells were allowed to settle on the semipermeable membrane of the Transwell. The Transwells were placed in the well plates containing the OGD or control endothelial cells or astrocytes and melanoma cells were allowed to transmigrate through the ECM (Collagen) and membrane overnight toward the lower chamber.
At the end of the migration period, melanoma cells in the upper chamber that had not transmigrated were removed using cotton swabs, followed by a gentle wash of the bottom side of the Transwell inserts in PBSX1 and fixation in absolute methanol for 5 min. The Transwell inserts were washed in PBSX1 and stained with Diff-Quik® staining kit (Medion Diagnostics, Düdingen, Switzerland).
Transwells were photographed using Olympus IX53 inverted microscope fitted with an Olympus DP73 camera Olympus Inc., Hamburg, Germany), two fields per Transwell in duplicate Transwells. Cells per field were counted or analyzed using ImageJ software.
Cytokine Array
The supernatants from OGD and control astrocytes and endothelial cells were analyzed for the secretion of 102 cytokines, using the Proteome Profiler Human XL Cytokine Array Kit (R&D systems, Inc., Minneapolis, MA, United States), according to the manufacturer’s instructions. A minimum of a 20% difference in pixel density between OGD and control samples was accounted as a change in secretion level.
Tissue Processing and FACS
Male NSG mice aged 2–4 months were subjected to stroke followed by metastasis at day 7 as previously described. To eliminate any confounding factors and since estrogen is neuroprotective, male animals were used for this study. On day 14 after stroke, the animals were anesthetized with isoflurane, decapitated, and the peri-infarct areas were carefully dissected using a sterile lancet. Corresponding areas dissected from the non-stroke side served as a control. The dissected tissue was further cut into fine bits with a sterile blade, triturated with a pipette tip, and digested in 3 mL of collagenase (3 mg/mL, Roche Diagnostics) at 37°C for 25 min at 170 rpm. Hibernate buffer without calcium (Brainbits, Springfield, IL, United States) was used to maintain tissue viability and pH during the process of dissection and digestion. The digestate was triturated and washed twice at 3,000 rpm for 10 min with this hibernate medium. To enrich the human population and deplete potential mouse cells from the neural tissue, the pellet was re-suspended in low fluorescence hibernate media and the MACS- mouse cell depletion kit (Miltenyi Biotec, San Diego, CA, United States) was used as per manufacturer’s instructions. Brain metastatic melanoma cells were negatively selected through immunomagnetic separation as provided by the manufacturer’s protocol.
The enriched cell suspensions were stained with melanoma marker MCSP conjugated with PE (Human; MCSP-PE, 10 μL antibody/107 cells, Miltenyi Biotec, San Diego, CA, United States) by incubation at 4°C for 10 min. The cells were washed with 2 mL of Hibernate medium, re-suspended in low fluorescence hibernate medium, and maintained on ice during FACS isolation. Cell were co-stained with DAPI to sort for live cells. DAPI, PE, and GFP gates were set using positive and negative controls prior to sorting melanoma cells. Metastatic melanoma cells were collected via FACS (FACsARIA, Becton Dickinson, UCLA FACS Core) into low fluorescence medium.
RNA Isolation (Both in vivo and in vitro)
FACS-sorted cells were pooled from six to eight animals and the RNA was extracted using an RNA-Microprep kit (Zymo-Research) and eluted into 12 μL RNAse-free H2O. RNA-quality was verified on an Agilent Bioanalyzer using an Agilent picokit with relative concentrations that ranged from 170 to 1,733 pg/μL. Samples with RIN > 6 were sent for sequencing. For the extraction of RNA from in vitro melanoma cultures, YDFR-CB3 cells were treated with conditioned media of either OGD or normoxic conditions on hEC/h-Astro. Melanoma cells were then washed twice with PBSX1 and RNA was extracted using the NucleoSpin miRNA kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions.
RNA Sequencing and Transcriptome Analysis
RNA sequencing was done by the UCLA Neuroscience Genomics Core at UCLA. Briefly, for in vivo samples, library was prepared using Ovation RNA Ultra Low Input (500 pg) kit (Nugen manufacturer) and TruSeq Nano DNA library preparation kit (Illumina manufacturer). Amplified cDNA was purified and validated followed by library preparation. Preparation included 69 bp paired end reads. Samples were sequenced by Hi-Seq 2500 over 7 RapidRun lanes, corresponding to about 1.8 samples per lane (80 million reads per sample) for the in vivo datasets. In vitro samples were sequenced over 12 RapidRun lanes, corresponding to about 2.3 samples per lane (65 million reads per sample).
RNA sequencing performed on melanoma cells conditioned with either astrocyte/endothelial (OGD/control) conditioned media resulted in 60,906,978 to 25,890,188 with 40.18 to 75.37% of uniquely mapped reads. Samples with a minimum of 10 transcript counts were used and κ = 3 for in vitro batch correction was applied in edgeR package. Melanoma cells exposed to endothelial conditioned media with or without OGD resulted in 4717 significant DE (differentially expressed) genes (p < 0.05). Further filtering for log2 (fold change) ≥±0.5 resulted in 3,073 genes of which 712 were upregulated and 2,361 were downregulated. Melanoma cells exposed to astrocyte conditioned media with or without OGD resulted in 1,127 significant DE genes (p < 0.05). Further filtering for log2 (fold change) ≥ ±0.5 resulted in 362 genes of these, 252 were upregulated and 111 were downregulated.
Read Mapping, Assembly, and Expression Analysis
Short reads were aligned using STAR to the latest human reference genome (GRCh38). Fragment counts were determined by HTseq program using RefSeq gene and Ensembl transcript model (Conesa et al., 2016).
Differentially Expressed (DE) Gene Analysis Between in vivo and in vitro Datasets
Due to the large number of samples, unexpected batch effects were expected. Removal of unwanted variation (RUV) is effective to remove such batch effect. For in vitro datasets, raw counts were filtered (minimum 10 counts for at least eight samples), and then normalized by TMM followed by RUV correction with k = 3 (Risso et al., 2014). Differentially expressed genes were determined using edgeR. For in vivo samples, differential expression analysis was done without RUV correction (Robinson et al., 2010).
GSEA (Gene Set Enrichment Analysis)
Within this analysis, a hallmark gene set is defined by a collection that summarizes specific well-defined biological states, processes, or diseases with coherent expression changes in many molecules within a system (Subramanian et al., 2005; Liberzon et al., 2015). A unique Stroke-MET transcriptome was identified by subtracting metastatic melanoma gene expression in non-stroke vs. metastatic melanoma gene expression in the stroke side. The set of differentially regulated genes in the Stroke-MET transcriptome were probed for common regulatory elements, which might be controlling gene expression. These are termed upstream regulators, as they are predicted to regulate genes that are significantly induced or downregulated in the Stroke-MET transcriptome. These upstream regulators were tested for significance based on an enrichment score (Fisher’s exact test p-value overlap of predicted and observed regulated gene sets) and activation Z-scores (to test for their association with up or down-regulation of their associated genes) in the Stroke-MET transcriptome.
Network Generation and Properties
To explore the molecular interactions, transcription profiles that overlap between in vivo and in vitro datasets were analyzed. Overlapping molecular transcription profiles between in vivo and in vitro datasets with high focus numbers were analyzed for network properties. Network properties such as degree and betweenness centralities were assessed with CytoNCA (Tang et al., 2015).
SPIA (Single Primer Isothermal Amplification)
To further validate candidate gene expression profiles obtained from the RNAseq datasets, metastatic melanoma cells were FACS isolated as described above from independent experimental samples. The resultant RNA were in pg/μl range and this was amplified using the Nugen Ovation® Pico WTA System V2. RNA amplification was performed based on the protocol and instructions provided in the kit using the Ribo-SPIA technology.
qPCR Analysis of Molecules Associated With Both the Metastatic Microenvironment
The SPIA amplified RNA were used to test the gene expression fold change between the metastatic melanoma cells associated with the stroke and non-stroke sides of the brain. 1.25 ng/μl of cDNA was generated from each sample and used to detect expression using Roche Light Cycler 480 SYBR Green I Master Kit. RT-qPCRs were performed using custom primers and human-GAPDH as the house keeping gene.
CLARITY (Clear Lipid-Exchanged Acrylamide-Hybridized Rigid Imaging Compatible Tissue Hydrogel)
Brains exposed to both stroke and metastasis in DCX-RFP mice were extracted. Brains were left intact and were optically cleared using a CLARITY-PACT protocol modified for passive and quick clearing (Chung and Deisseroth, 2013; Tomer et al., 2014; Yang et al., 2014). Briefly, each DCX-RFP animals subjected to stroke and metastasis were perfused with 100 μl of vascular lectin (Vector Laboratories) prior to euthanization. The brains were then fixed with 4% paraformaldehyde and incubated at 40°C with 30 mL of cold hydrogel for 5 days. Prior to polymerization of the hydrogel, the tubes were vacuum sealed, purged with nitrogen gas and immediately sealed. The tubes were then incubated at 370°C for 4–5 h to allow for adequate polymerization of the hydrogel bound tissue. After polymerization, excess gel was carefully removed and the whole brain was cut into 2 mm thick sagittal sections using a brain mold. The brains were passively cleared with a solution of 200 mM boric acid, 8% SDS, pH 8.5 at 400°C. Clearing solution was refreshed every day until the sections were transparent. The tissue sections were rinsed with PBS and mounted in Focus Clear (CelExplorer Labs, Hsinchu, Taiwan) solution. The sections were imaged under a confocal microscope (Nikon C2) at 10× and the stacked images were constructed into video files.
Statistical Analysis
Data analysis was done with Microsoft Excel and GraphPad Prism version 7.00 (GraphPad Software, La Jolla, CA, United States). Statistical significance was determined by two-tailed unpaired Student’s t-test when comparing two groups, one-way ANOVA, or two-way ANOVA wherever appropriate for experiments consisting of more than two groups. Disease and time course interaction and effects were done followed with post hoc test with wither Holm-Sidak or Fisher’s least significant difference (LSD) and are specified on each figure legends. Significance values are presented as mean ± standard error of the mean (SEM) for the in vivo experiments and mean ± SD for the in vitro experiments.
Results
The Regenerative Neurovascular Niche Enhances Melanoma Brain Metastasis
NOD-scid gamma mice were subjected to distal middle cerebral artery occlusion (MCAo), a common, clinically relevant stroke model (Agliano et al., 2008; Chen B. et al., 2009), and injected with YDFR-CB3, a melanoma line selected for its ability to form brain macrometastases (Izraely et al., 2010). This is an ideal model to test the preferential localization of a metastatic tumor cell, since melanoma develops from the neural crest and commonly metastasizes to brain (Boiko et al., 2010; Izraely et al., 2012; Quintana et al., 2012; Marzese et al., 2015). YDFR-CB3GFP+ were administered via intracardiac injection 7 days post-stroke, when angiogenesis and neurogenesis peak and the regenerative neurovascular niche is most active (Morris et al., 2003; Ohab et al., 2006; Lamanna, 2012). The total number of metastatic cells in the brain was significantly greater in the stroke group compared to controls 7 days post-injection (p = 0.0082, control metastasis 245 ± 57, stroke + metastasis 653 ± 122) (Figures 1A–C and Supplementary Figure 1). The YDFR-CB3GFP+ cell distribution was quantified in the peri-infarct, where angiogenic vessels are predominantly found, and in other brain regions in which angiogenesis or tissue repair is absent (Cavaglia et al., 2001): periventricular white matter, striatum, hippocampus, and olfactory bulb. On average, 433 ± 67 YDFR-CB3GFP+ cells preferentially localized to the peri-infarct compared to other regions in stroke and 73 ± 23 cells were found in corresponding cortical regions in control metastasis (p = 0.0012, n = 6–7) (Figure 1D). This data indicates that metastatic melanoma preferentially localizes to the regenerative neurovascular niche.
Metastasis to the Regenerative Neurovascular Niche Occurs Independently of Hypoxia Mediated Angiogenesis
Within the regenerative neurovascular niche after stroke, there are several potential cellular signaling sources that might influence brain metastasis. Angiogenic endothelial cells may directly initiate melanoma cell localization and establish metastasis. Instead, melanoma cells may localize to the regenerative multicellular environment of endothelial cells, astrocytes and neuroblasts within the peri-infarct cortex, the combination of these cells constituting a niche in which cell–cell communication is the interaction of the cells (the niche) rather than that of a single cell (Carmichael, 2016). To test this, we used a model of global angiogenesis that does not produce the regenerative neurovascular niche or neurogenesis seen in stroke. Chronic hypobaric oxygen mediates angiogenesis by globally increasing cerebral vascular density (Figures 1E,F) (Harik et al., 1996; LaManna et al., 1998). If angiogenesis itself is inducing melanoma cell localization, this generalized angiogenic state would be expected to result in the preferential localization of vascular melanoma cells. A cohort of hypoxic animals subjected to 10% hypoxia for 7 days was injected with metastatic melanoma cells intracardially and the number of metastatic foci was assessed 7 days after injection. Each metastatic focus represents an incidence of metastasis and is defined by a cluster of melanoma cells that has localized and developed into a colony. The 62 ± 6 metastatic foci observed in the peri-infarct region was significantly higher than 36 ± 4 of the hypoxic cohort (p = 0.003, n = 6–8) (Figure 1G). This suggests that the regenerative response after stroke significantly enhances brain metastasis localization of melanoma compared to hypoxia-mediated angiogenesis alone.
Metastasis to the Regenerative Neurovascular Niche Occurs Independently of Vascular Hemodynamic Changes
Stroke alters blood flow dynamics and blood–brain barrier permeability in the peri-infarct tissue (Chen Q. et al., 2009) into which metastatic melanoma preferentially localize. To investigate whether this preferential localization of melanoma is simply due to the alterations in blood flow or vascular permeability changes induced by stroke, we injected two cohorts of animals with fluorescent microspheres (0.2 μm diameter) at 1 and 7 days after stroke. These microspheres lodge in the capillary bed and allow for tracking and quantification of differences in blood flow to different brain regions (Springer et al., 2000). The volume of microspheres accumulated in angiogenic vessels within the peri-infarct and corresponding contralateral regions was quantified. No significant difference was observed between microsphere accumulation in the ischemic and non-ischemic hemispheres at days 1 and 7 after stroke (Figures 1H–J). This suggests that blood flow to the peri-infarct is not significantly altered at these time periods after stroke and that blood flow changes are not the cause of the observed preferential melanoma cell metastasis to this region.
Vascular leakage due to blood–brain barrier breakdown leads to extravasation of plasma proteins (Abraham et al., 2002; Tenreiro et al., 2016). To determine if blood–brain barrier permeability changes could contribute to preferential localization of melanoma, we stained brain sections for endogenous albumin extravasated at days 1 and 7 after stroke (Abraham et al., 2002). The mean intensity of extravascular albumin was elevated at day 1, but significantly reduced at day 7 and was not different from the corresponding contralateral region at day 7 (p = 0.0255, n = 5) (Figures 1K,L). These findings support the idea that the localization of metastatic melanoma cells to the peri-infarct at day 7 are not due to vascular hemodynamic changes induced by stroke, and may be due to specific cellular or molecular features of these vessels within the angiogenic neurovascular niche.
Neuroblast-Associated Vessels Within the Regenerative Neurovascular Niche Mediate Brain Metastasis
Intercellular communication between cancer cells and host tissue is critical for metastatic localization and tumor progression (Termini et al., 2014). After stroke, neural stem cells in the subventricular zone divide and differentiate into immature neurons (termed neuroblasts), which migrate to brain tissue adjacent to the stroke site and closely associate with angiogenic blood vessels (Cook et al., 2017). We assessed the influence of neuroblasts on metastatic melanoma localization to angiogenic vessels within the regenerative neurovascular niche using genetic labeling: a DCX-RFP mouse, in which red fluorescent protein is expressed in neuroblasts and serves as an accurate reporter of post-stroke neurogenesis (Cook et al., 2017). In these studies, vessels are labeled with perfused lectin, and are rendered in white for visualization in the black photomicrograph background (Figures 2, 3).
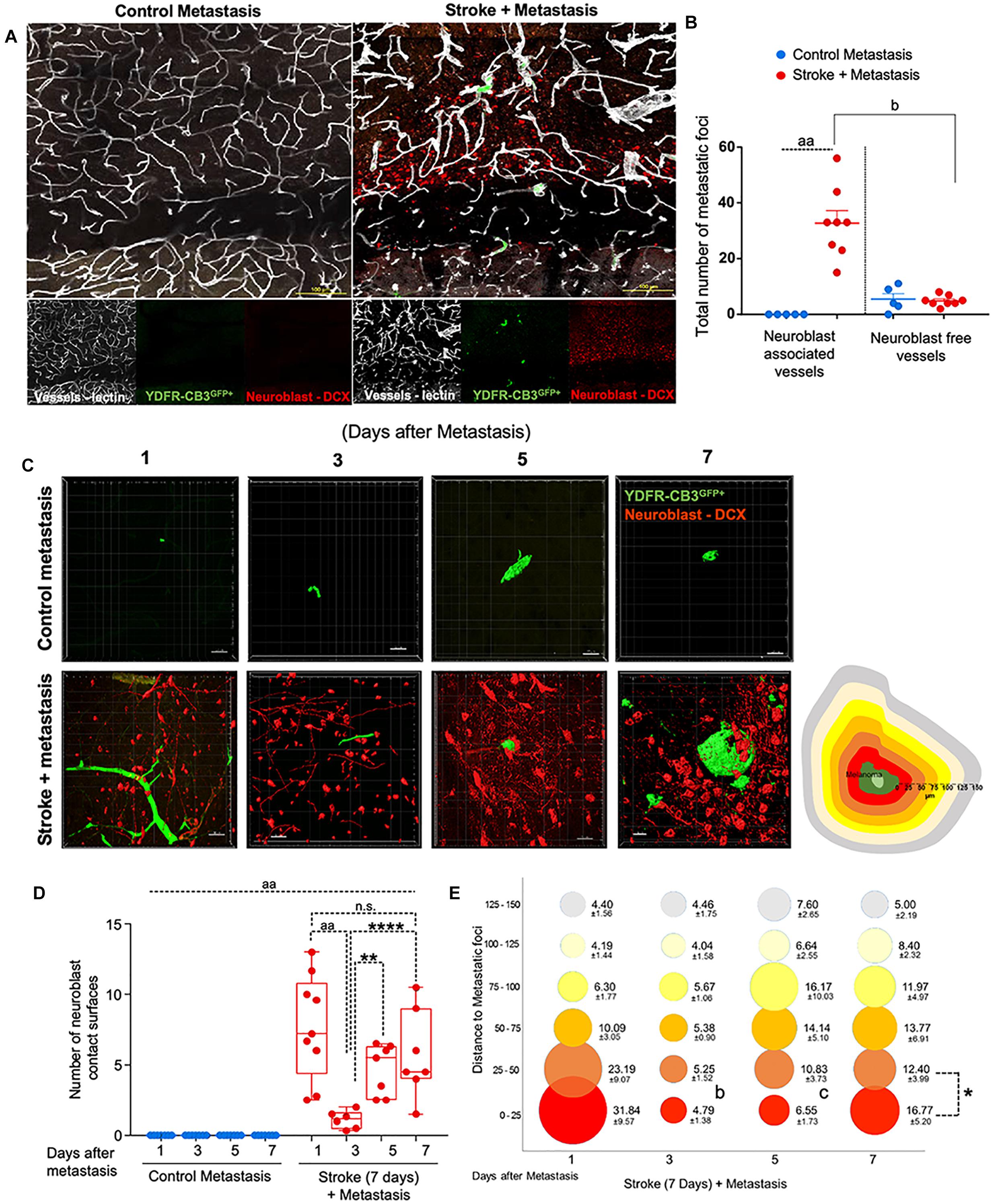
Figure 2. Repair augments direct cellular interactions between the metastatic cell and neurovascular niche. (A) Immunohistochemical analysis of metastatic melanoma cells shown as GFP+ green, lectin-labeled vasculature in white and DCX+ neuroblasts in red. (B) Quantitative analysis of metastatic cell interaction with vessels associated with or without neuroblasts shown as scatter plot with mean ± SEM (n = 5–8, four sections/animal, aap < 0.0001; bp = 0.0354, Kruskal–Wallis, one-way ANOVA, Dunn’s for multiple comparisons). (C) Representative brain images of metastatic foci (GFP+) and neuroblasts (RFP+) over time (left). Control groups did not associate with neuroblasts in the corresponding brain region. Schematic representation of distance bins (25 μm intervals) used to analyze neuroblast distribution around metastatic foci. Bin closest to melanoma is shown in bright red and increasing distances showing in gradients of yellow and the farthest depicted in gray (right). (D) Box and scatter plot showing number of neuroblast contact surfaces interacting with metastatic foci. Mean ± SEM (n = 6–9, ∗∗p = 0.0092, ∗∗∗∗p = 0.0006, aap < 0.0001; one-way ANOVA, Holm-Sidak post hoc test used for multiple comparisons). (E) Bubble plot depicting density and distribution of neuroblasts around metastatic foci at 25 μm distances Mean ± SEM (n = 5–8, bp = 0.033, cp = 0.033, ∗p < 0.0164; one-way ANOVA, Holm-Sidak post hoc test used for multiple comparisons). Bubble size represents number of neuroblasts and increasing distances are color coded as shown in schematic.
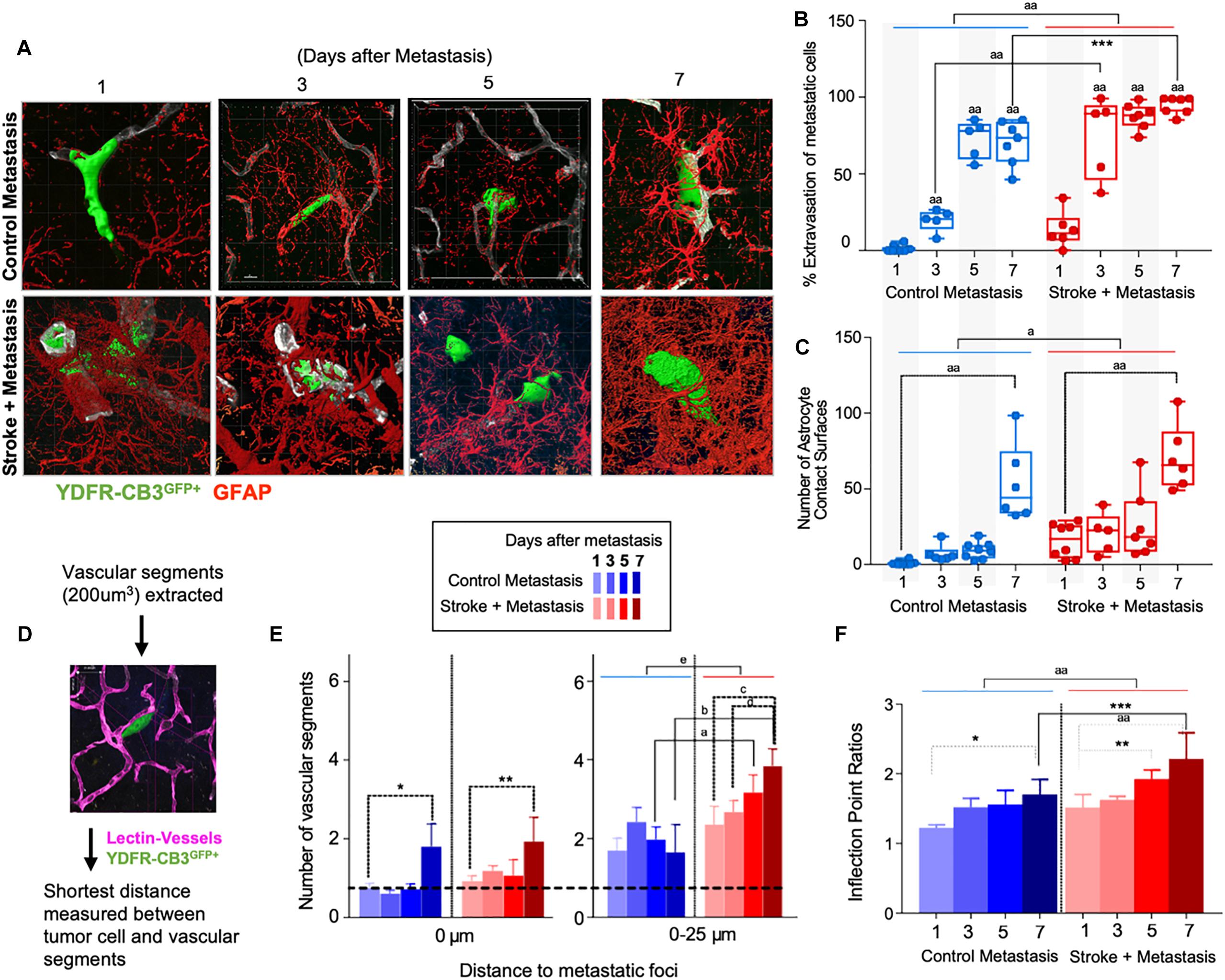
Figure 3. Repair enhances direct cellular interactions between the metastatic cell and astrocytes and increases neovascularization. (A) Representative images showing metastatic melanoma interaction with reactive astrocytes during melanoma extravasation. (B) Quantitative analysis of % volume of metastatic cell extravasated. Mean ± SEM [∗∗∗p = 0.0082, aap < 0.0001, two-way ANOVA, Holm-Sidak post hoc test used for multiple comparisons (n = 5–8)]. (C) Astrocyte coverage at corresponding time points around each metastatic foci. Mean ± SEM [aap < 0.0001, ap = 0.0006, two-way ANOVA, Holm-Sidak post hoc test used for multiple comparisons (n = 5–8)]. (D) Methodology of analysis of neovascularization and infection point analysis. (E) Changes in neovascularization response over time mean ± SEM, (n = 5–8), (∗p = 0.0424, ∗∗p = 0.0349, ap = 0.0454, bp = 0.0015, cp = 0.0163, dp = 0.0474, ep = 0.0009, two-way ANOVA, Fisher LSD). (F) Bar graphs showing temporal changes in inflection point ratios [∗p = 0.0032, ∗∗p = 0.0215; ∗∗∗p = 0.0012; aap < 0.0001; two-way ANOVA, Holm-Sidak post hoc test used for multiple comparisons, (n = 5–8)].
Within 1 day after metastasis, 33 ± 5 metastatic foci seeded in neuroblast-associated vessels in the peri-infarct of stroke animals (p = 0.0354, n = 5–8), while there was no such metastatic localization in the corresponding control cortex (p < 0.0001, n = 5–8) (Figures 2A,B). On average, five metastatic foci localized to vessels without neuroblasts in both control and stroke mice groups. This suggests that blood vessels that support post-stroke neurogenesis govern the preferential localization of metastatic melanoma in the regenerative neurovascular niche.
Tissue Repair Augments Direct Cellular Interactions Between Metastatic Cells and Neurovascular Niche Components
As metastatic melanoma cells localize to the regenerative neurovascular niche, they associate with distinct cellular niche elements over time, such as angiogenic vessels, astrocytes, and neuroblasts (Termini et al., 2014). These associations between metastatic cells and niche components at day 7 after metastasis are shown on CLARITY-processed tissue after stroke (Supplementary Video 1). To further determine cellular relationships between melanoma cells and this niche, we examined spatiotemporal interactions between the metastatic cells and neurovascular components at 1, 3, 5, and 7 days after metastasis in control and stroke cohorts. These time points reflect early events in metastasis that determine metastatic cell survival and extravasation.
Metastatic Melanoma Interact With Neuroblasts Within the Regenerative Neurovascular Niche
To examine direct interactions between metastatic cells and neuroblasts (the two principal cells of the metastatic and regenerative neurovascular niches) the number of neuroblasts-melanoma contact surfaces were quantified over time in the peri-infarct and corresponding control cortex at 1, 3, 5, and 7 days after metastasis. On average, 7.7 ± 1.2 contact surfaces from neuroblasts interacted with the metastatic foci at day 1. Following this, the number of interactions significantly reduced at day 3 to 1.1 ± 0.3 contact surfaces (p < 0.0001, n = 6–9). Further, at days 5 and 7, neuroblast contact surfaces steadily increased compared to day 3 (4.7 ± 0.7, p = 0.0092, and 5.7 ± 1.2, ∗∗∗∗p = 0.0006, respectively) in the stroke cohort. No interactions were found in the corresponding control regions due to the absence of neuroblasts in analyzed control regions (Figures 2C,D).
To determine the spatiotemporal distribution of neuroblasts around melanoma cells, we analyzed neuroblast counts at 25 μm distance intervals from the metastatic foci periphery (Figure 2E). In the stroke cohort, 32 ± 10 neuroblasts were found at 1 day after metastasis within a proximity of 25 μm. At day 3, 5 ± 1 neuroblasts surround metastatic foci in equal distributions at increasing distances. At days 5 and 7, more neuroblasts migrate closer to the metastatic foci, with 7 ± 2 neuroblasts surrounding metastatic cells at day 5 and 17 ± 5 neuroblasts at day 7 (p = 0.0164, n = 5–8).
This data suggests that neuroblasts around the metastatic foci associate with melanoma in two phases. At day 1, there is a vascular phase: melanoma cells form a large number of contacts with neuroblasts as they associate with neuroblast-associated vasculature in the peri-infarct (Figures 2A,B). After melanoma extravasation at day 3, melanoma separate from the neuroblasts in the niche, followed by a tissue phase: melanoma cell associations with neuroblasts increase until day 7 in the regenerative neurovascular niche (Figures 2C–E). These distinct phases in neuroblast-metastasis interaction are unique to the regenerative neurovascular niche. The corresponding cortical regions in the metastasis-only (no stroke) group had no metastatic foci associated with neuroblasts.
Tissue Repair After Stroke Enhances Association Between Extravasated Metastatic Cells and Surrounding Reactive Astrocytes
Since reactive astrocytes play a crucial role in the neurovascular niche and in brain metastasis (Klein et al., 2015), we assessed if the regenerative neurovascular niche enhances reactive astrocyte contact surfaces with the metastatic melanoma at 1, 3, 5, and 7 days after stroke. Extravasation of metastatic cells initiates after 3 days in controls and within 1 day in stroke group. A two-way ANOVA indicated that this accelerated time course of extravasation was temporally dependent upon the presence of regenerative neurovascular niche (p = 0.0008, variance 4.33%) (Figures 3A–C). Corresponding to the extravasation, astrocytic processes begin interacting with extravasated melanoma early at day 1 in the stroke group compared to day 3 in controls. Total astrocytic contacts increase from 7 ± 2 at 3 days to 9 ± 2 at 5 days to 53 ± 10 at 7 days in control metastasis (p < 0.0001, n = 5–8) while they increase from 20 ± 6 at day 3 to 26 ± 8 at day 5 to 70 ± 9 at day 7 in stroke+metastasis (p < 0.0001, n = 5–8). Astrocyte contact surfaces linearly increase as metastatic cells extravasate in both control and stroke (pooled slope: 7.706). The presence of a regenerative neurovascular niche significantly enhanced melanoma–astrocyte interactions independent of time (two-way AVOVA, p = 0.0006) (Figure 3).
Regenerative Vasculature Is More Susceptible to Metastatic Cell-Induced Remodeling
The progression of brain metastasis is dependent upon the hijacking or recruitment of adjacent vessels, a process known as vascular cooption (Qian et al., 2016). To examine the temporal increase in vessel recruitment with metastatic cells, the distance between individual vessel segments and metastatic cells were measured. After metastasis, direct vessel contact with metastatic cells significantly increased from day 1 to day 7 in both control and stroke cohorts with 0.75 ± 0.12 vessels in contact with metastatic melanoma at day 1 and 1.8 ± 0.58 at day 7 in controls (p = 0.0424, n = 5–8), and 0.93 ± 0.13 at day 1 and 1.93 ± 0.62 at day 7 in the stroke cohort (p = 0.0349, n = 5–8). While no difference in direct vessel associations was observed between cohorts, there was a significant increase in close proximity vessels in the stroke groups compared to controls. Within a periphery of 25 μm from the metastatic foci, vessel association significantly increased from day 1 to day 7 in the stroke groups compared to controls, with 1.65 ± 0.71 vessels at day 7 in control vs. 3.85 ± 0.43 at day 7 (p = 0.0015, n = 5–8) in stroke (Figures 3D,E). A two-way ANOVA indicated that the regenerative neurovascular niche significantly influenced vascular associations independent of temporal effects (p = 0.0009, variance 19.71%). This data suggests that vascular cooption is essential to metastasis as early as 7 days and that this vascular remodeling is significantly augmented in the regenerative neurovascular niche.
When metastatic cells’ co-opt or route along vasculature, they induce local irregularities in vessels (Bullitt et al., 2003, 2005). To assess these vessel irregularities, we utilized inflection points, which measure the number of irregularities or changes in direction within a curvature. We measured inflection points on vessels associated with metastatic cells and normalized over inflection points on vasculature not associated with metastatic cells. Metastasis alone significantly increased inflection points on associated vasculature at day 7 compared to day 1 (p = 0.0032, n = 5–7). However, after stroke, inflection point ratios progressively increased at day 7 compared to day 1 (p < 0.0001, n = 5–7) and were significantly higher compared to controls at day 7 (p = 0.0012, n = 5–7) (Figure 3F). A two-way ANOVA indicated that stroke significantly enhanced inflection points independent of time. This data suggests that metastasis enhances vascular irregularities as early as 7 days, and angiogenic vessels within the neurovascular niche not only preferentially localize melanoma cells but are then more susceptible to melanoma-induced vascular remodeling.
Human Brain Metastatic Melanoma Interact With Neuroblasts and Astrocytes
To identify the possible interactions of neuroblasts, astrocytes, and metastatic cells in human brain tumors, we immunostained representative human brain metastatic melanomas FFPE sections obtained from elective surgeries with biomarkers for neuroblasts (Tuc-4), astrocytes (GFAP) and melanoma (MART-1). Fifteen distinct patient samples of brain metastasis were immunostained and 4 of 15 tumors showed co-localization of metastatic with neuroblasts (Figure 4). The presence of interactions between human melanoma and surrounding neural cells implicated in stroke repair suggests that elements of the regenerative neurovascular niche play an important role in mediating melanoma metastasis.
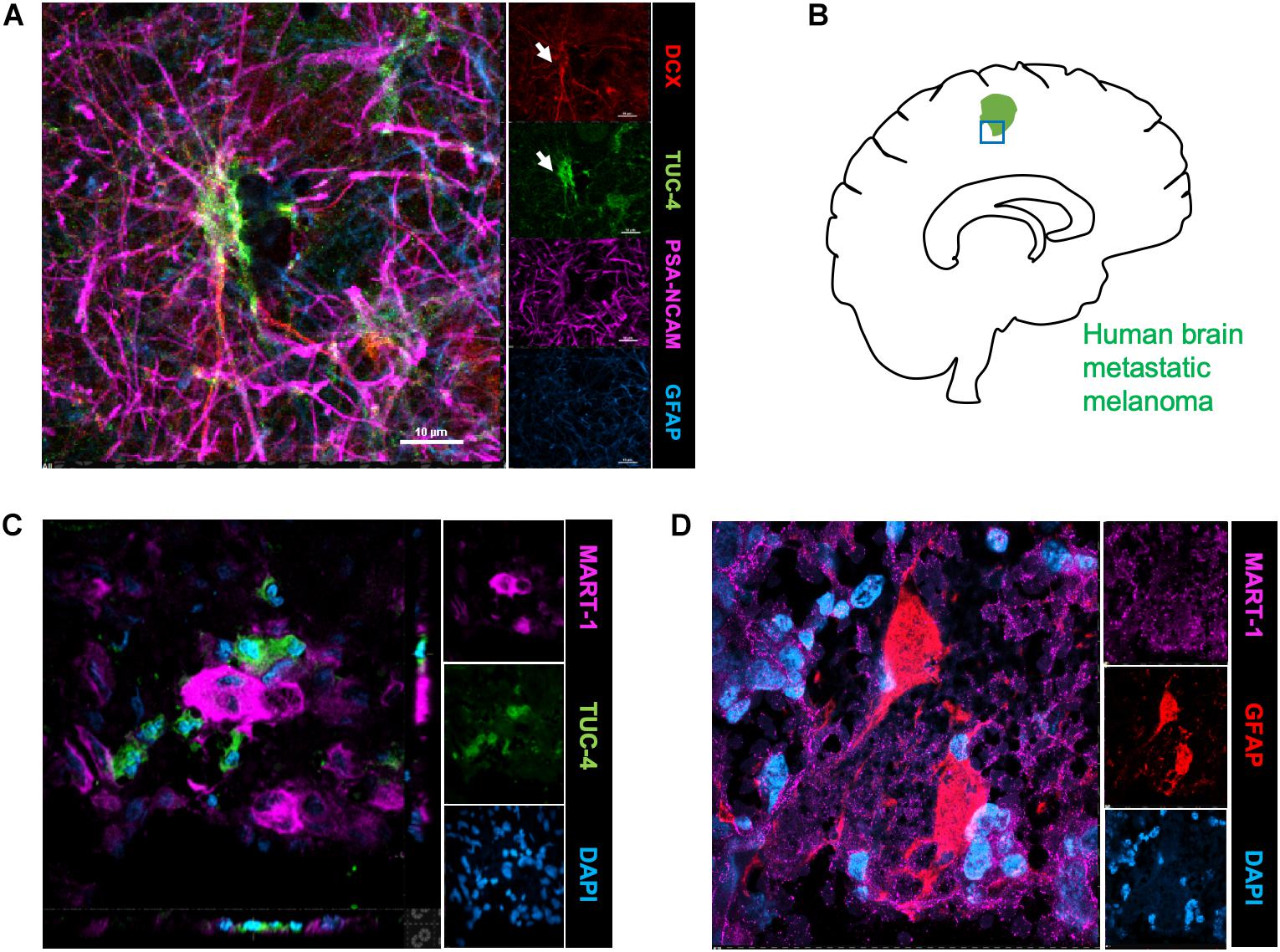
Figure 4. Human brain metastatic melanoma interact with neuroblasts and astrocytes. (A) Representative image depicting characterization of antibodies on human brain samples. Human hippocampus section showing staining for DCX, PSA-NCAM, GFAP, and Tuc-4. DCX positive neuroblasts colocalize with Tuc-4. (B) Schematics showing human brain tumor margin immunostained for neuroblasts and astrocyte association. (C) Immunohistochemical representative images of brain metastatic melanoma show novel interactions with neuroblasts excised from human patients (4/15 show close neuroblast–melanoma interactions). (D) Representative images of brain metastatic melanoma from human patients show interactions with astrocytes.
Isolation and Sequencing of Stroke Responsive Metastatic Melanoma Cells
FACS sort was performed on YDFR-CB3GFP+ cells isolated from both the peri-infarct region and the corresponding non-stroke side. We developed an efficient method to purify and enrich these metastatic cells using magnetic bead columns that negatively selected human cells in the elute and retained the mouse cells in columns bound to mouse cell specific antibodies conjugated to beads. This elute enriched for human cell population was gated for GFP and MCSP-PE channels in FACS. For each sample, metastatic cells isolated from two to three mice were pooled from the peri-infarct regions and six to eight animals were pooled from the contralateral side (n = 5–7) (Figure 5).
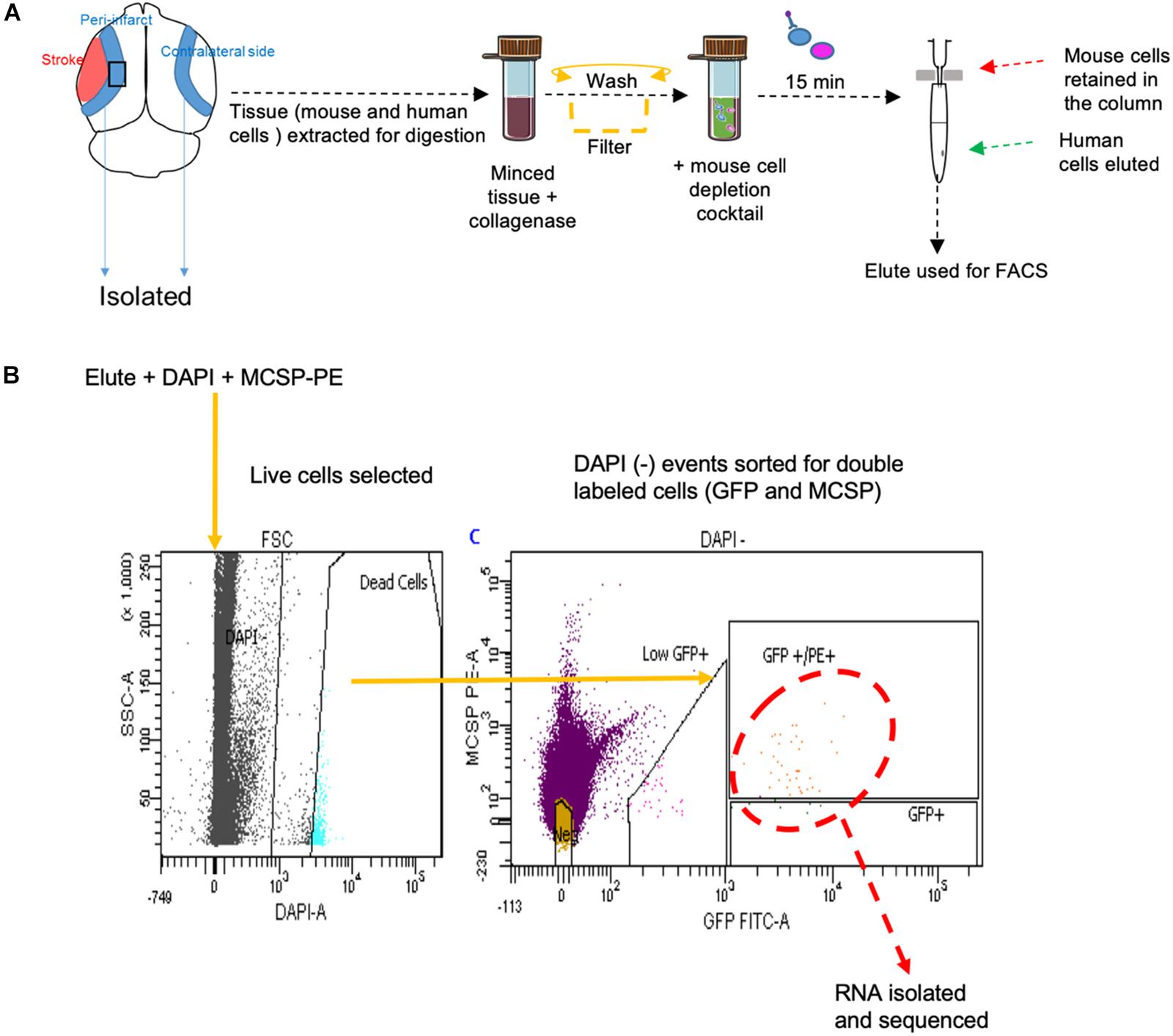
Figure 5. (A) Schematic representation of tissue processing and FACS sort methodology performed. (B) Gating areas used during FACS and the GFP and MCSP-PE double positive cells were isolated for RNA sequencing. MCSP, melanoma-associated chondroitin sulfate proteoglycan.
RNA sequencing performed on the metastatic melanoma cells isolated in vivo from the stroke and the non-stroke side of the brain resulted in 74–101.5 million reads per transcriptome. The percent of uniquely mapped reads ranged from 79.37 to 86.23%. Samples with a minimum of 10 transcript counts were used and batch correction was applied to remove unwanted variation with k = 2 (Risso et al., 2014). Differentially expressed genes between the metastatic cells isolated from the peri-infarct region (stroke side) and the corresponding non-stroke brain regions (contralateral hemisphere) were determined with the bioconductor edgeR package (Robinson et al., 2010).
Molecular Signaling Systems in Regeneration-Potentiated Brain Metastasis
To determine the unique gene expression changes that occur within metastatic melanoma in the presence of regenerative neurovascular signaling, we sequenced total RNA extracted from FACS-isolated, regeneration-potentiated metastatic melanoma cells in vivo from the peri-infarct cortex vs. metastatic melanoma cells not localized to the neurovascular niche after stroke. Cell-specific RNA-sequencing identified 4,083 differentially expressed genes between metastatic melanoma cells (stroke vs. non-stroke p ≤ 0.05). Filtering this significantly regulated gene list for log2 (fold change) ≥±0.5 resulted in 2,264 genes in regeneration-potentiated metastatic melanoma cells vs. brain metastases not in the stroke site. Because this transcriptome comes specifically from a subtraction of metastatic melanoma gene expression in non-stroke vs. the potentiated state of stroke, this is a unique transcriptome of melanoma specific to the regenerative neurovascular niche, termed the Stroke-MET transcriptome: 1,118 genes are upregulated and 1,146 are downregulated in metastatic melanoma within the regenerative neurovascular niche compared to melanoma brain metastases outside of this niche. The organization of differentially regulated genes in the Stroke-MET transcriptome was studied by association into top-scoring canonical molecular pathways (Figure 6). Canonical pathways strongly activated in the Stroke-MET transcriptome include axon guidance, ephrin receptor signaling, phagosome formation, and thrombin signaling. A volcano plot of the significant differentially expressed genes identifies those genes with the highest fold change and most significant expression in the Stroke-MET transcriptome (Figure 6). Twenty-two of these genes are reported to have extensive roles in determining metastasis as tabulated in Table 1.
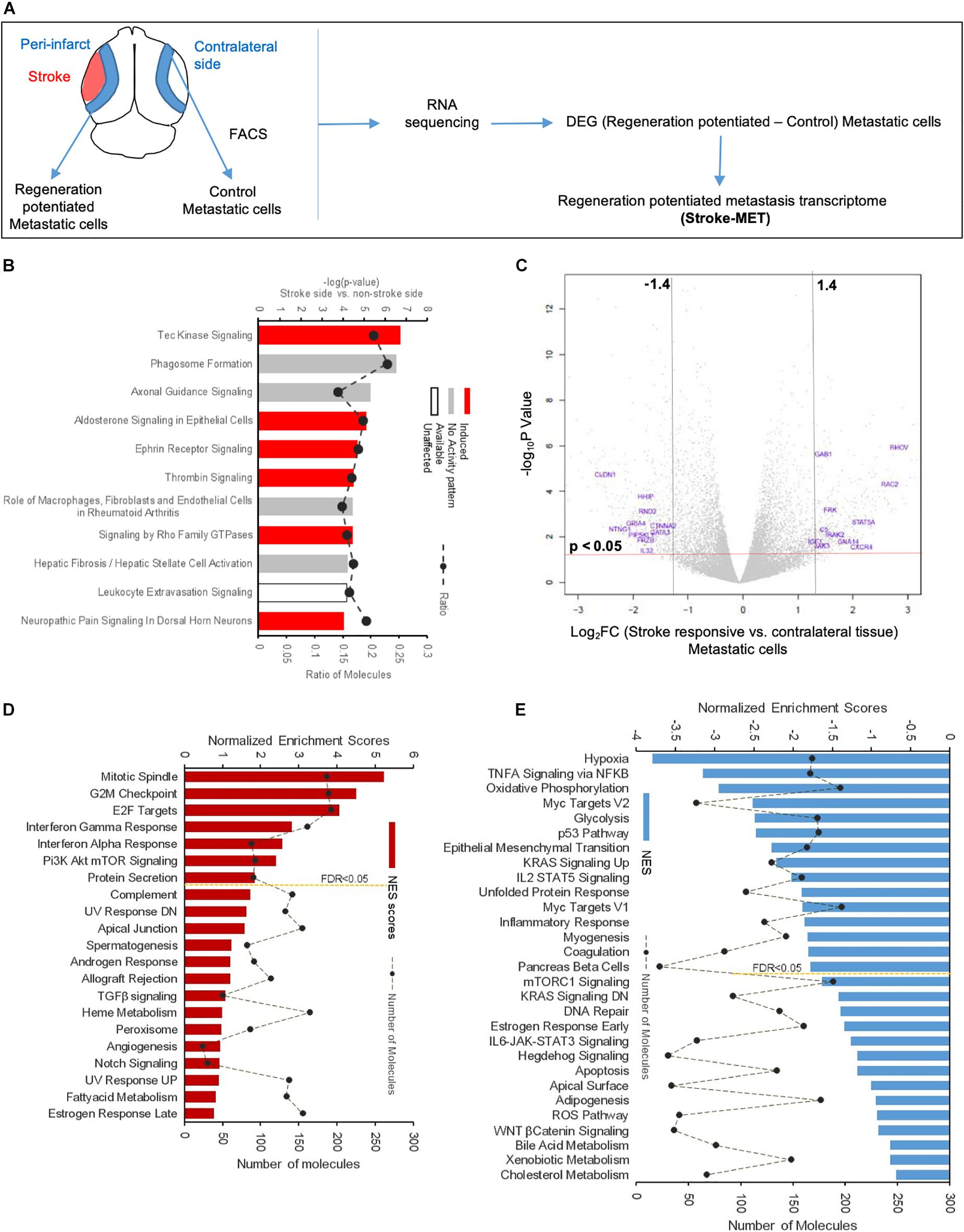
Figure 6. Molecular signaling systems in regeneration-potentiated brain metastasis. (A) Schematic representation of flow of transcription profiling and regenerative neurovascular niche components interacting with metastatic cells. Significant and differentially expressed genes between regeneration-potentiated and contralateral metastatic cells as determined by log2 fold change > 0.5 and p < 0.05 (Stroke-MET). (B) Top differentially regulated canonical pathways with largest negative logP shown in metastatic melanoma cells from the regenerative neurovascular niche compared to contralateral tissue. The dashed line represents the ratio of the number of molecules in the dataset over the number of molecules in the pathway. (C) Volcano plot showing p-values correlated with log2 fold change in metastatic cells isolated from regenerative neurovascular niche vs. contralateral side. Genes systematically identified from top pathways from (B), are marked in blue. (D,E) GSEA of hallmark genes up and downregulated in the Stroke-MET transcriptome. Estimated probability of FDR < 25% represent 3/4 chances of a gene set being valid.
We utilized Gene Set Enrichment Analysis (GSEA) to refine this dataset and identify genes with common biological functions based on curated datasets (Subramanian et al., 2005). Using directional p-values for differential expression in the Stroke-MET transcriptome, we identified 50 hallmark gene sets, of which 21 were upregulated and 29 were downregulated. At an FDR < 0.05, 7 upregulated and 15 downregulated gene sets were significantly enriched in Stroke-MET (Figures 6D,E). The gene-set enrichment analysis indicated an enriched up-regulation of PI3K-mTOR pathway (Sarris et al., 2012) and downregulation of p53 pathway (Oren, 2003) often associated with tumor progression in cancer. These analyses of gene classes and identification of specific genes uniquely expressed in stroke-melanoma metastasis vs. “general” brain metastasis identifies many that are associated with neuronal pathfinding, neuronal tropic and trophic interactions and neuronal cell motility, supporting a role of the metastatic cell in an analogous position in the regenerative neurovascular niche to the normal principle cell in this niche, the neuroblast.
Mimicking Stroke-Like Conditions With Oxygen-Glucose Deprivation (OGD) Facilitates Brain Metastasis
To model the influence of stroke on melanoma metastasis in vitro, we assessed the (a) metastatic properties and (b) molecular profile of melanoma cells in stroke-like conditions using a well-established in vitro model: OGD (Hillion et al., 2005). Metastatic cells were treated with conditioned media harvested from human brain endothelial cells (h-EC) or human astrocytes (h-Astro) subjected to either OGD or control conditions of normal oxygen and glucose levels.
Oxygen Glucose Deprivation (OGD) Enhances Chemoattraction of Melanoma Cells by Brain Cells
Oxygen Glucose Deprivation (OGD) Enhances Migration of Melanoma Cells
Since cell migration is an essential component of the metastatic cascade (van Zijl et al., 2011), we examined the directed migratory capacity of brain metastatic melanoma in response to cues from h-EC and h-Astro after OGD and control conditions. Melanoma cells were added upon a collagen-coated Transwell insert and allowed to migrate toward h-EC/h-Astro subjected to OGD or control conditions. While melanoma cells displayed significantly increased migration toward OGD-exposed h-EC compared to control conditions (p < 0.05), OGD-conditioned media from h-Astro did not significantly alter migration (Figure 7C). OGD exposed h-EC also displayed enhanced adhesion of metastatic melanoma cells, which was reduced by blocking VCAM-1 (Figure 8). Thus, metastatic characteristics in melanoma cells can be enhanced by recapitulating stroke-like conditions in vitro, and this in vitro results suggest that endothelial cells are key mediators of regeneration-potentiated metastasis.
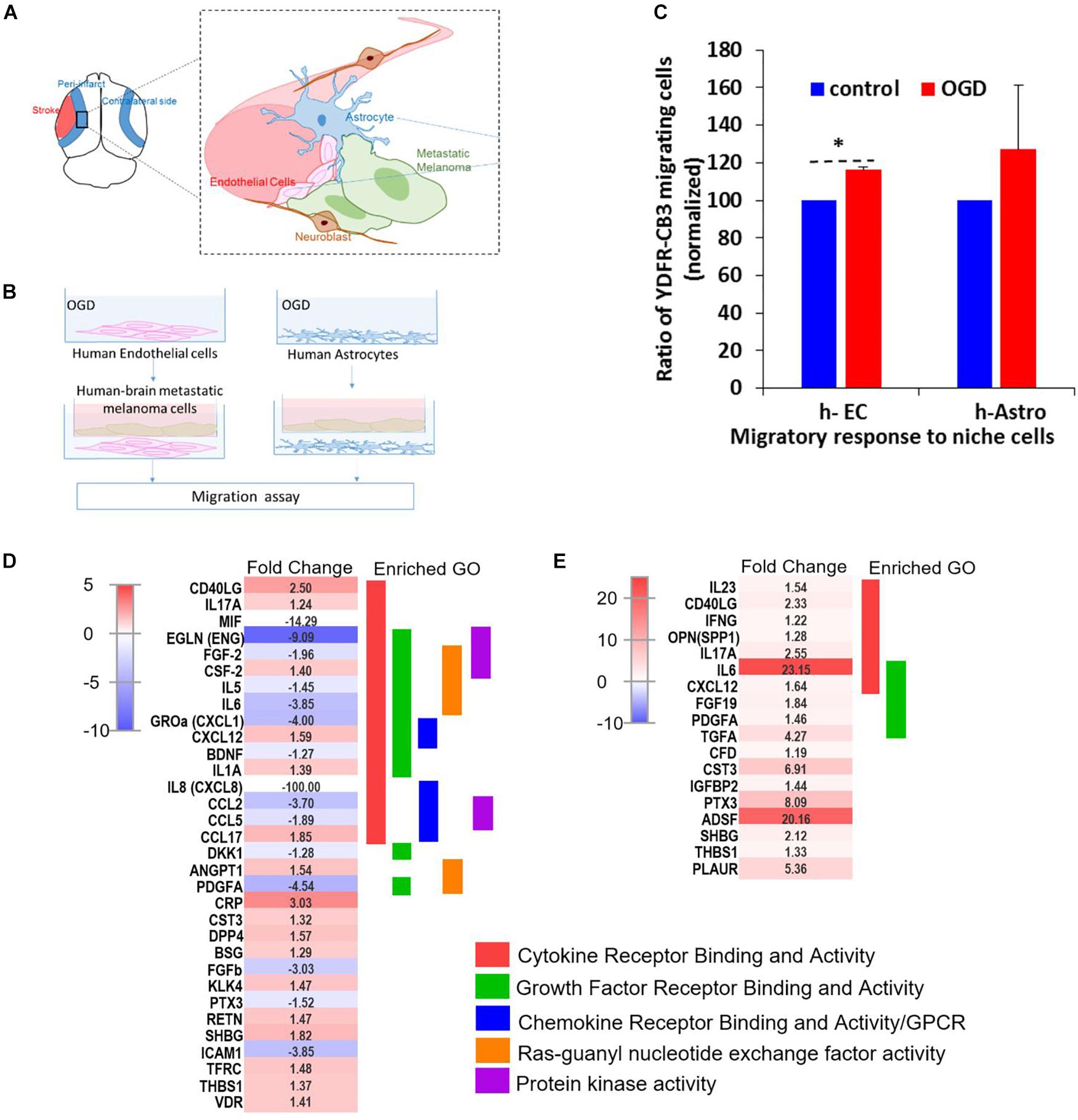
Figure 7. OGD alters chemokine/cytokine responses in the neurovascular niche and enhances migration of brain metastatic cells. (A) Schematics showing the interaction of human astrocytes (h-Astro) and human endothelial cells (h-EC) with metastatic melanoma cells. (B) Schematics of method of oxygen glucose deprivation (OGD) conditioned media exposed to metastatic melanoma cells. (C) Migratory response of metastatic melanoma cells exposed to OGD conditioned media from cells are shown as mean ± SD, ∗P < 0.05. (D,E) Heatmap of relative expression of secreted proteins profiled from the medium of h-Astro (E) and h-EC (D). Functionally grouped enriched pathways are shown in one color. Enrichment/depletion tests performed with ClueGO application on cytoscape, κ score ≥ 0.4.
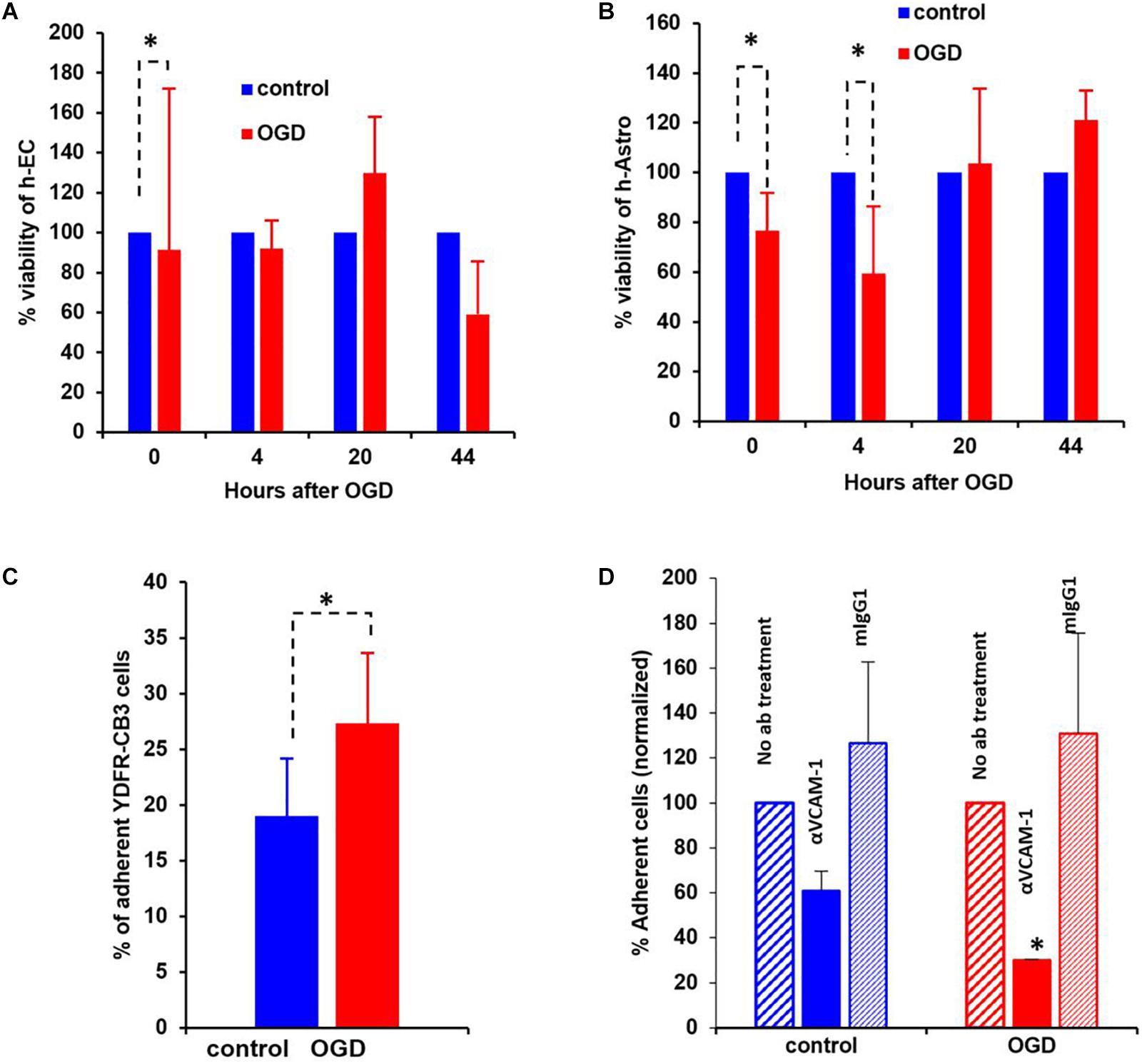
Figure 8. Oxygen-glucose deprivation (OGD) affects viability of human-endothelial cells (h-EC), astrocytes (h-Astro) and adhesion of metastatic melanoma cells. (A,B) Percentage viability of h-EC (A) and h-Astro (B) subjected to OGD for 4 h followed by a 4, 20, and 44 h reperfusion phase. Cell viability as measured by XTT assay. The bars represent the average % viability of OGD exposed cells, normalized to control cells subjected to normal oxygen and glucose levels, mean ± SD, ∗p < 0.05. The different time points indicate the hours in reperfusion after OGD/control conditions. (C) h-EC were subjected to OGD for 4 h followed by a 20 h reperfusion phase. m-Cherry expressing macro- and micro-metastatic melanoma cells were seeded on top of the h-EC monolayer and incubated for 30 min, to allow adhesion to occur. The fluorescence signal of labeled cells was measured before and after removal of non-adherent cells. The bars represent the % adherent cells in the wells. The graph represents an average of three independent experiments + SD. ∗P < 0.05. (D) m-Cherry expressing metastatic melanoma cells were seeded on top of the OGD or control h-EC monolayers and incubated for 30 min with 5 μg/mL VCAM-1 blocking Ab, an isotype control (mIgG1), or without IgG. The fluorescence signal of labeled cells was measured before and after removal of non-adherent cells. The bars represent % adherent melanoma cells seeded with anti-VCAM-1 blocking Ab or isotype control normalized to % adherent melanoma cells seeded without Ab. The graph represents an average of two independent experiments + SD. ∗p < 0.05.
OGD Alters Cytokine/Chemokine Responses in Astrocytes and Endothelial Cells in vitro
We next examined the secreted factors from OGD-exposed endothelial cells and astrocytes to determine signaling cues that play a role in the in vitro migration findings, and isolate the influence of individual cell types on metastatic cells. To identify these candidate factors, secreted and extracellular molecules from h-EC (OGD vs. control) and h-Astro (OGD vs. control) were measured using a cytokine array. Thirty-two cytokines (fold change < -1.2 or > 1.2) were differentially secreted from OGD vs. control h-EC and 19 were differentially secreted from OGD vs. control h-Astro (Figures 7D,E). To identify gene ontologies in these in vitro stroke-endothelial and stroke-astrocyte secretomes, enrichment depletion (simultaneous analysis of enriched: over-represented DE genes and depleted: under-represented genes between two states) (Rivals et al., 2007) was performed for both cellular processes and molecular functions. Both endothelial and astrocyte-derived OGD secretomes were enriched for cytokine and growth factor receptor activity while endothelial-derived secretomes displayed additional enrichment of chemokines, Ras-GTP, and protein kinase activity, further supporting the central role of endothelial cell in contributing to this pro-metastatic niche.
OGD Reduces Viability of Human Brain Endothelial Cells and Astrocytes
The viability of neurovascular niche cells was assessed under OGD conditions. Human brain h-EC and h-Astro were subjected to OGD or control (normoxic) conditions for 4 h followed by 4, 20, and 44 h of normoxia recapitulating reperfusion. Cell viability was then measured by XTT assay. The results showed that h-Astro were more sensitive to OGD than h-EC (Figures 8A,B). While the viability of the h-Astro was significantly reduced after 4 h of OGD conditions (t = 0, p < 0.05), a significant reduction in h-EC viability only occurred after 44 h (p < 0.05). When the normoxic phase was prolonged, “stroked” astrocytes showed a gradual recovery over time (20 and 44 h after start of the reperfusion phase). These findings mimic spontaneous recovery of neurovascular components observed after stroke.
OGD Enhances the Adhesive Capacity of h-EC to Metastatic Melanoma Cells
A crucial initial step in melanoma cell transmigration through the brain vasculature is the adhesion of cancer cells to the endothelium. To investigate whether OGD enhances the ability of h-EC to bind melanoma cells, we compared the adhesion of brain-metastasizing melanoma cells to OGD and control h-EC. The brain-metastasizing melanoma cells showed significantly greater adherence to OGD exposed h-EC than to control (Figures 8C,D). This result suggests that stroke-like conditions recapitulated by OGD facilitates the formation of brain metastasis by making h-EC more amenable to tumor cell adhesion. The expression of the endothelial cell adhesion molecules ICAM-1 and VCAM-1 was tested in control and OGD exposed h-EC. Flow cytometry analysis demonstrated a slight up-regulation in the expression of both molecules following OGD (data not shown). To further demonstrate the increased adherence of h-EC to metastatic melanoma cells, YDFR-CB3m-Cherry cells were co-cultured with h-EC and the above experiment was similarly performed in the presence of a VCAM-1 blocking antibody. Inhibiting VCAM-1 significantly decreased OGD (and not control) h-EC adherence to YDFR-CB3m-Cherry suggesting its importance in brain metastasis in the regenerative neurovascular niche (Figure 8D).
Transcription Landscapes From Regenerative Niche and Melanoma Brain Metastasis: in vivo and in vitro Overlaps
To delineate the influence of specific niche cells (endothelial/astrocytes) upon the metastatic transcriptome in vitro, we sequenced the total RNA from metastatic melanoma in conditioned media harvested from h-EC or h-Astro subjected to OGD or controls. The differentially expressed (DE) genes from OGDEC-MET/OGDAstro-MET were compared with the transcriptional in vivo profile of regeneration-potentiated melanoma (Stroke-MET) to determine overlap between the in vivo and in vitro studies.
We performed a rank–rank hypergeometric overlap (RRHO) that compares the ranked order and the log10-transformed t-test p-value of in vitro differentially expressed genes to in vivo differentially expressed genes in this melanoma cell line, and in stroke conditions. The heat maps show that top ranked genes in OGDEC-MET overlapped with those of Stroke-MET to a greater extent than OGDAstro-MET (Figures 9C,D). An assessment of overlapping canonical signaling systems reveals that the top highly up/down-regulated pathways all play important potential or predicted roles in governing melanoma metastasis: PPAR/RXRα, angiopoietin signaling, mTOR, Semaphorin signaling between the Stroke-MET and OGDEC-MET transcriptome, and ephrin receptor signaling in both Stroke-MET and OGDAstro-MET transcriptomes (Figures 9A,B). This analysis indicates that the in vitro melanoma profile, when exposed to stroke-conditioned endothelial cells, significantly overlaps with the in vivo melanoma gene expression profile in the stroke regenerative neurovascular niche, but not that of melanoma exposed to stroke-conditioned astrocytes.
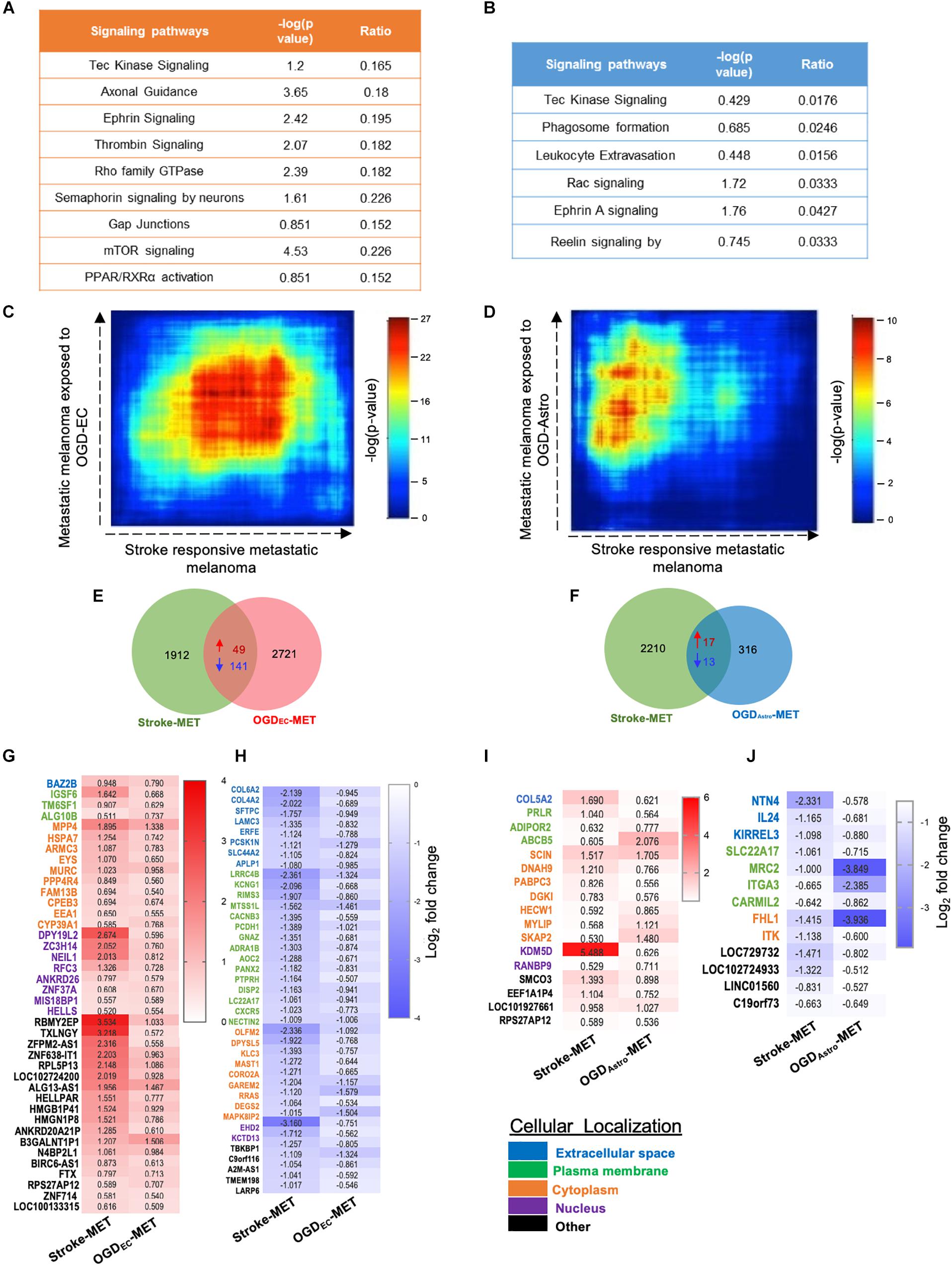
Figure 9. Overlapping transcription landscapes from regenerative NV niche after stroke and melanoma brain metastasis. (A,B) Common canonical signaling pathways between the in vivo Stroke-MET and in vitro OGD-ECMET/OGD-AstroMET transcriptome. (C,D) Rank–rank hypergeometric overlap (RRHO) analysis of common genes expression spread between Stroke-MET and OGD-ECMET/OGD-AstroMET transcriptome. RRHO analysis of common genes expression spread between Stroke-MET and OGD-AstroMET datasets. Red and blue pixels on the heatmap depict a high and low number of overlapping genes, respectively. Stroke-MET and OGDEC-MET display stronger hypergeometric overlap those with OGDAstro-MET transcriptome. (E,F) Overlapping up and downregulated genes between in vivo Stroke-MET and in vitro OGD-ECMET/OGD-AstroMET transcriptome are depicted in the Venn diagram. (G–J) Heatmap of log2 FC of overlapping genes between Stroke-MET and OGD-ECMET/OGD-AstroMET transcriptome. The up and down regulated overlapping genes are shown in red and blue a change in gradients of these color correspond to the level of log2 FC as shown on the scale bars. Genes classified based on the cellular location and the color coded as shown in the key at the bottom of the heatmaps : Blue: extracellular, green: plasma membrane, orange: cytoplasm, violet: nucleus, and black: other.
To move forward on this similarity, a comparison of genes similarly up and down regulated genes in in vivo regeneration-potentiated melanoma transcriptome (Stroke-MET) and in vitro OGDEC-MET transcriptome showed 343 overlapping significant genes (p < 0.05) of which 49 were upregulated and 141 were downregulated. Stroke-MET transcriptome compared to OGDAstro-MET transcriptome showed 45 significant genes, of which 17 were upregulated and 13 were downregulated (Figures 9E–J). This gene list is small and provides a tractable set of genes for mechanistic study in their effect on tumor metastasis and stroke neurovascular responses.
We previously showed that brain metastasizing melanoma express a unique set of genes distinct from that found in cutaneous melanoma (Izraely et al., 2012). Some of these genes are known as metastasis-mediating genes. Our objective was to examine whether the expression of this set of genes is altered in melanoma cells subjected to in vitro post-stroke reperfusion conditions, i.e., to supernatants of “stroked” astrocytes. We incubated “stroked” and control astrocytes in starvation medium for 20 h to allow secretion of post-stroke soluble factors into the medium, collected these supernatants, and added them into brain metastatic melanoma cultures. We observed an upregulation in the expression of ANGPTL4, COX2, and CYR61, and a slight downregulation in MMP1 expression. These results suggest that stroked astrocytes secrete factors that may induce the metastatic potential of melanoma cells (Figure 10).
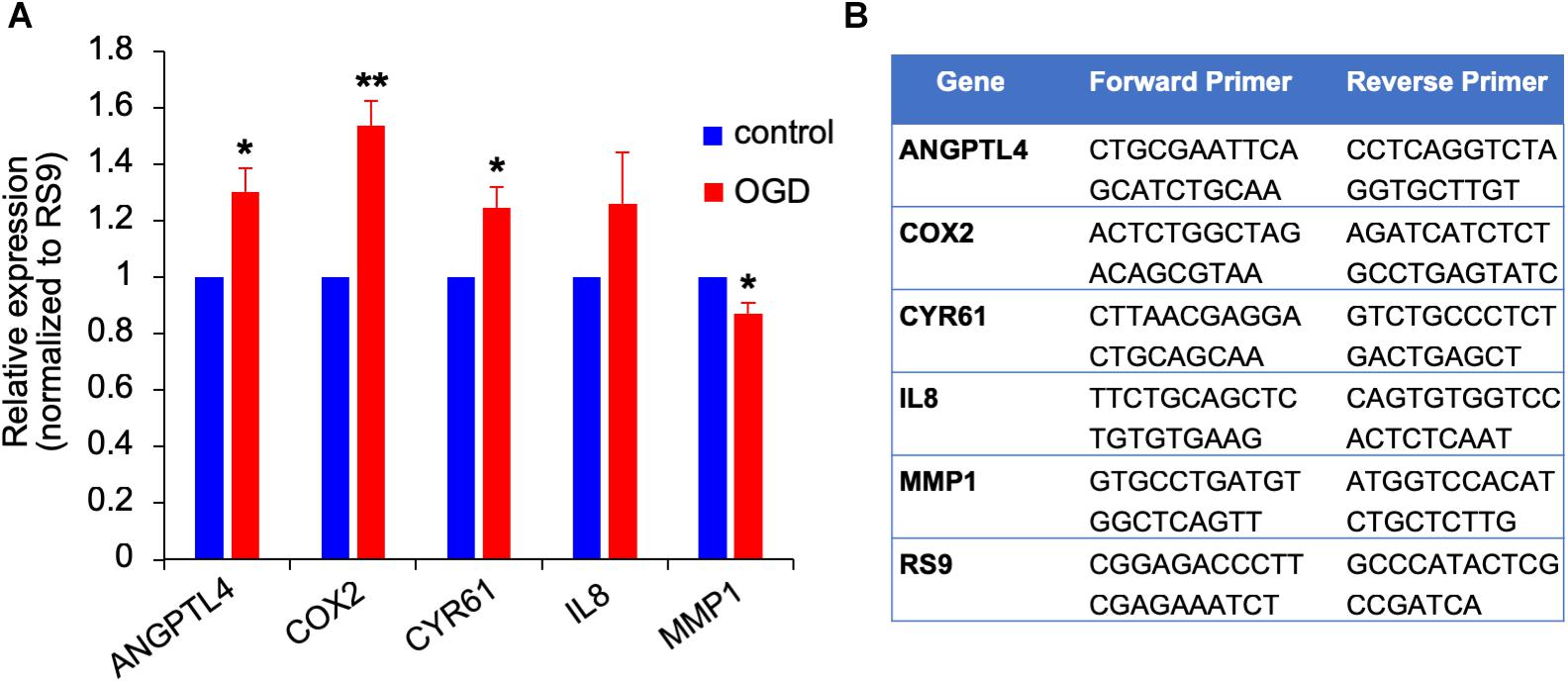
Figure 10. Gene expression changes in OGD exposed metastatic melanoma cells. (A) The effect of OGD exposed astrocyte secreted factors on melanoma gene expression. Astrocytes were subjected to OGD or control conditions for 4 h followed by a 20 h reperfusion phase. Metastatic melanoma cells were treated for 24 h with the OGD or control conditioned media collected from the reperfusion phase. Total RNA was extracted and gene expression was determined by RT-qPCR. mRNA expression levels were normalized to the expression of RS9. Bars represent gene expression in cells treated with conditioned medium of “stroked” astrocytes normalized to gene expression in cells treated with control astrocyte conditioned medium mean ± SD of three independent experiments, ∗p < 0.05, ∗∗p < 0.01. (B) Primer sequences used for the qPCR analysis shown in (A,B).
To identify putative molecular interactions that facilitate metastasis in stroke-like conditions, we profiled membrane and secreted proteins between endothelial cells/astrocytes, and metastatic melanoma in the regenerative neurovascular niche. An interaction map was generated of putative interactions between regeneration-potentiated melanoma cells and the two cells in the regenerative neurovascular niche that were determined to have a demonstrated in vitro effect on melanoma metastasis–astrocytes and endothelial cells. This approach used significantly regulated genes in in vivo regeneration potentiated melanoma that were membrane-bound or secreted, and had signaling partners on another cell type: astrocyte–melanoma and endothelial cell-melanoma exposed to OGD in vitro. A greater number of molecular interactions between endothelial cells and metastatic melanoma were identified compared to astrocytes. Interestingly PROM1 a surface marker of tumor initiating cancer cells (Raso et al., 2011) was found in both interactomes. IL-6 and CCL2 had the most number of signaling partners in both the interactomes. These putative molecular interactions can be used to explore the functional significance of target molecules, and characterize the dynamic interactions that occur between metastatic cells and the regenerative neurovascular niche to drive regeneration-potentiated metastasis (Figure 11).
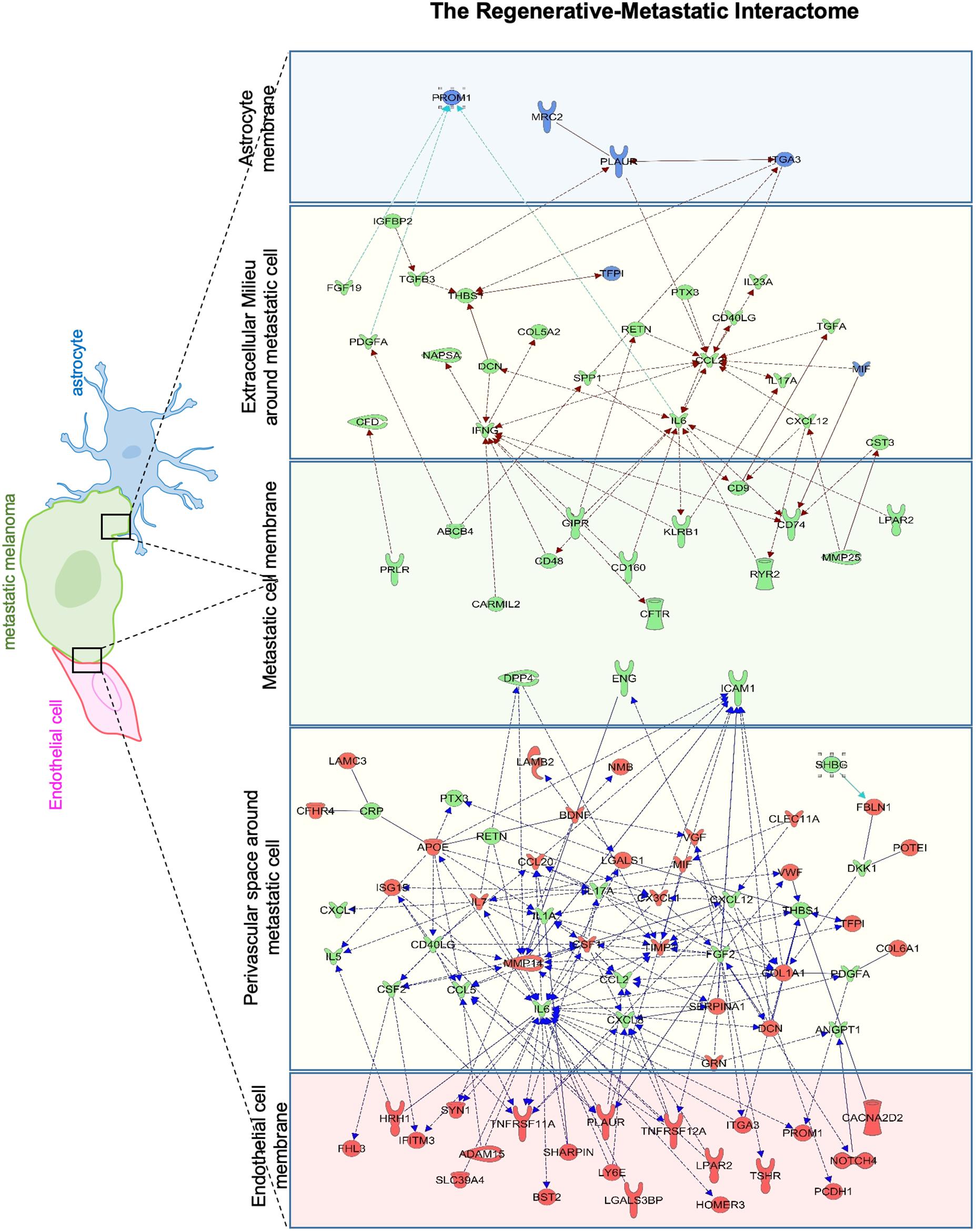
Figure 11. Astrocyte–metastatic cell and endothelial cell–metastatic cell interactome. Extracellular and plasma membrane bound proteins were chosen to determine putative gene interactions between metastatic melanoma cell with two cell types in the regenerative neurovascular niche–astrocyte/endothelial cells. Astrocyte–metastatic cell interactome display differentially expressed proteins in blue shows OGD exposed astrocyte secretome (blue panel) and their interactions with Genes in the metastatic melanoma cells from the Stroke-MET transcriptome colored in green (green panel). Endothelial cell–metastatic cell interactome differentially expressed proteins in red shows OGD exposed astrocyte secretome (red panel) and their interactions with genes in the metastatic melanoma cells from the Stroke-MET transcriptome colored in green (green panel). Yellow panels depicts molecules in the extracellular space. Molecules in yellow: secretome, red: OGD exposed endothelial cells, blue: OGD exposed astrocytes, green: stroke responsive melanoma Stroke-MET.
Network Analysis of Prognostic Gene Expression Profiles Identify Key Central Mediators
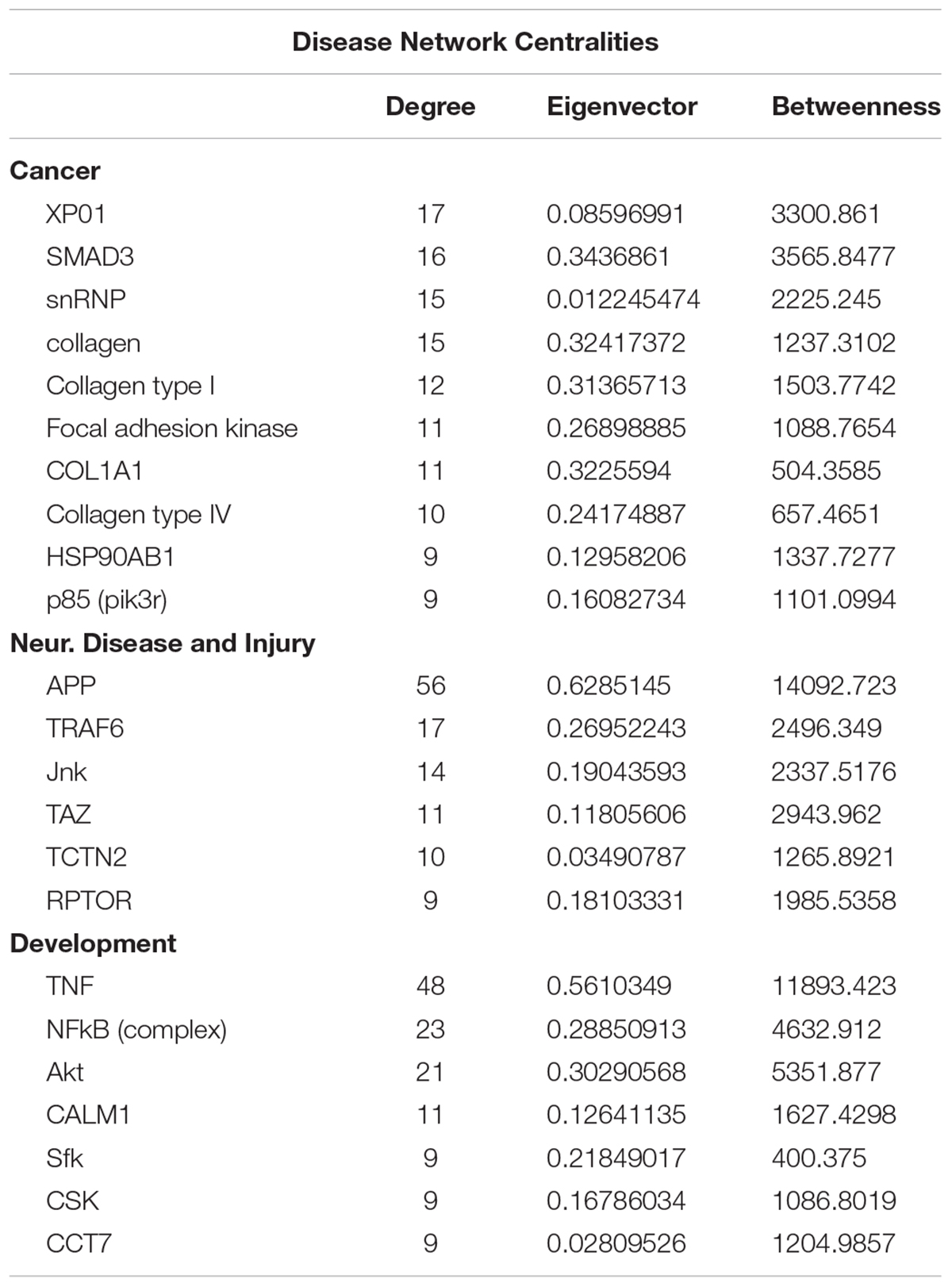
To gain insights into system-level properties of biologically important genes, the significantly regulated genes in regeneration-potentiated melanoma (Stroke-MET) were grouped into networks and assigned focus scores based on p-values. Using network analysis allows us to identify genes of a high functional and biological significance in the metastatic cells within the regenerative niche. Focus scores indicate the likelihood of genes within a network being found together, taking into account the size of the network based on relevance to disease or function (Long et al., 2004). The genes with specific connectivity belonging to disease classes from the Stroke-MET transcriptome with highest focus scores were used for centrality analysis (Cytoscape, CytoNCA). Centrality analysis identifies genes that are highly interconnected and influence a large number of other genes within a network. We identified these central genes from three classes of gene networks with coordinated functions and diseases including neurological disease and injury, development, and cancer (Tang et al., 2015). We determined the degree and betweenness centrality of the central genes, which are designated by large node size in the networks (Figures 12A–C). A gene’s degree and betweenness centrality represents its interconnectedness to other genes in the network, and its influence over adjacent genes, respectively (Ozgur et al., 2008). Centrality values are provided in Table 2. From these networks we identified APP, RPTOR, and TAZ (neurological injury network), extracellular matrix signatures such as collagen I, IV and FAK (cancer network) as central genes that are predicted master regulators specific to brain metastasis and potential targets for further experimental analysis.
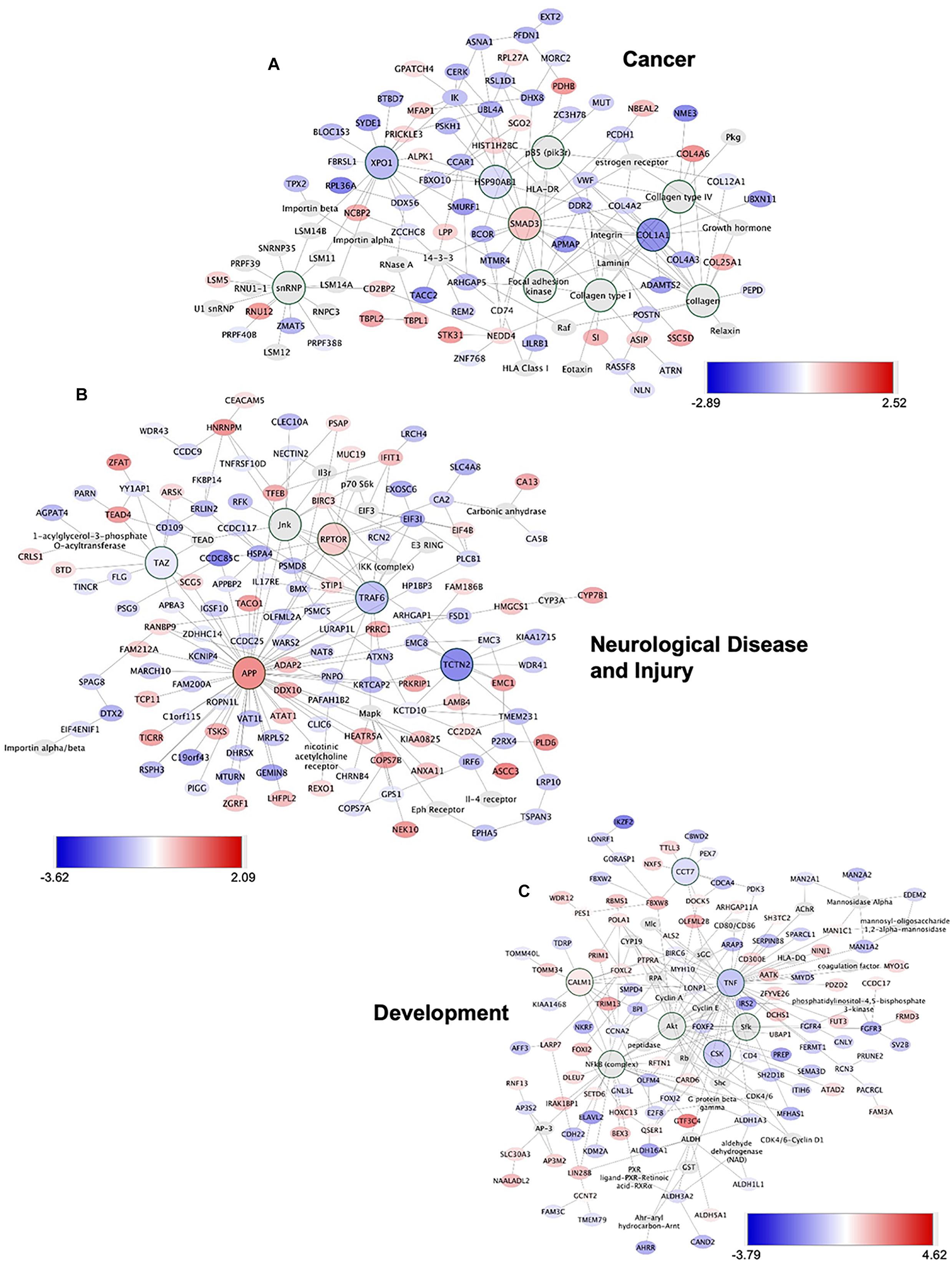
Figure 12. Gene network analysis of highly repetitive classes of coordinated diseases and functions from the Stroke-MET transcriptome. Network of gene interactions representing genes associated with “cancer” (A), “Neurological disease and injury” (B), “Development” (C). Red: upregulated genes, blue: downregulated genes, and larger nodes: central and highly connected genes. Nodes shown in gradients of red and blue are up- and down-regulated, respectively. Centrality values are provided in the Table 2. Central genes with higher connectivity with neighboring genes are shown as larger nodes in each category.
To further identify the unique molecular profile of metastatic melanoma cells within the regenerative neurovascular niche after stroke, we compared the differentially expressed genes of these cells in this niche to those in metastatic melanoma cells outside of this niche, in liver metastasis. To identify biological variation without interference from different isoform abundance, we determined differences at the gene-level that are identifiable based on a RefSeq model (Soneson et al., 2015). FPKM values above 4 were used as a cut off to identify unique molecules in brain metastasis and liver metastasis (Figures 13A,B). While gene-level overlap tests for overall changes in transcriptional output of a gene, individual transcripts may have different mechanisms or effects on compared conditions. To further to detect these changes in transcript isoforms, we used the Ensembl model at the transcript-level (Conesa et al., 2016). A core comparison analysis showed that top canonical pathways are composed of chemokines and cytokines in the brain compared to the liver metastasis (Figure 13C). Interestingly, genes and pathways involved in axon guidance and neuropathic pain are significantly enriched in the brain metastasis compared to the liver. GSEA analysis of hallmark gene sets show 4 upregulated and 15 downregulated pathways significantly enriched in the differentially expressed transcriptome of Stroke-MET vs. liver-metastasis (Figures 13D,E). Brain metastasis to the regenerative neurovascular niche exhibits a unique signaling signature involving repair pathways not observed in distant metastasis.
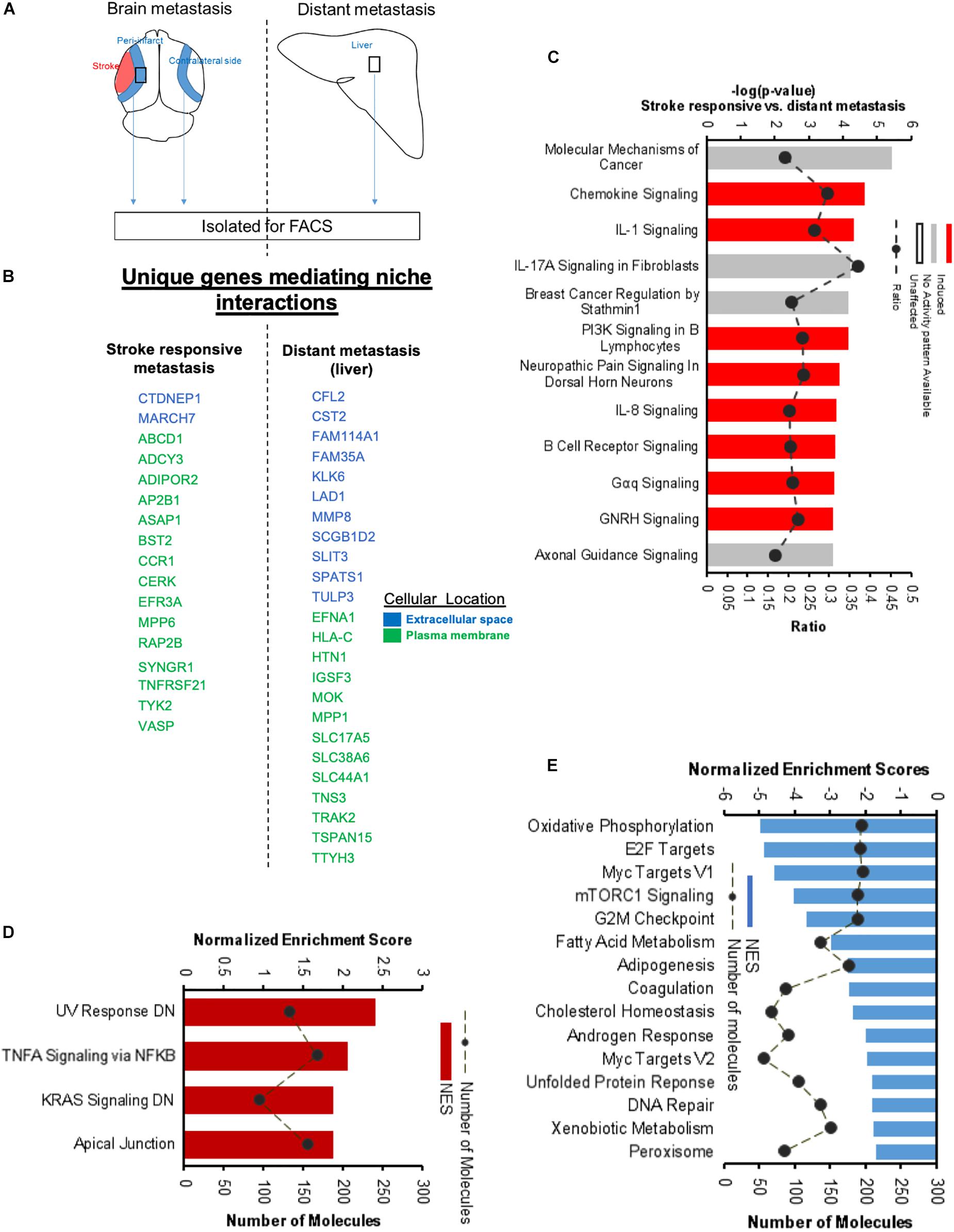
Figure 13. Unique molecular signaling systems in brain metastasis. (A) Schematic representation of brain and distant sites of metastasis compared. The significant and differentially expressed genes between Brain metastasis (stroke responsive metastatic cells) and distant metastasis (liver metastasis) were compared. (B) Unique extracellular and plasma membrane genes that interact with the surrounding niche in each of these two transcriptomes were identified at FPKM values > 4. (C) Top canonical pathways differentially regulated in brain metastatic melanoma over a distant metastatic site (liver) with the largest negative logP values are shown. The dashed line represents the ratio of the number of molecules in the dataset over the number of molecules in the pathway. (D,E) GSEA of hallmark genes up and downregulated in the differentially expressed genes of brain metastatic vs. liver metastasis.
Discussion
The dynamic cross-talk between cancer cells and neural niche elements provides insight into the influence of the tumor microenvironment on metastasis. Neuronal precursor cells, astrocytes, and endothelial cells have all been found to play a crucial role in creating a permissive niche that drives metastatic success (Psaila and Lyden, 2009; Venkatesh et al., 2015). In both neural injury and brain metastasis, the local microenvironment undergoes analogous cellular and molecular changes that influence cell proliferation, differentiation, and survival (Chen B. et al., 2009; Termini et al., 2014). Both tumor progression and regenerative tissue reorganization require angiogenic vasculature, co-localization of a progenitor or tumor cell, and dynamic cell–cell signaling (Ohab et al., 2006; Prakash and Carmichael, 2015; Carmichael, 2016). In this study, we provide compelling evidence that cues emanating from the regenerative neurovascular niche facilitate brain melanoma metastasis.
The present data indicate that cellular and molecular features of the regenerative neurovascular niche induce the preferential localization of melanoma to the peri-infarct over other brain regions independent of changes in cerebral blood flow, blood–brain barrier breakdown, and vascular permeability normally found after stroke. Metastatic melanoma localized specifically to regions on angiogenic blood vessels closely associated with neuroblasts. However, angiogenic vessels alone were not sufficient to produce the significant amount of brain metastasis seen in stroke. This differential localization of metastatic melanoma to blood vessels associated with migratory neuroblasts identifies a central cellular complex in the regenerative neurovascular niche that enhances melanoma metastasis. After the initial localization of metastatic cells to neuroblast-associated angiogenic vessels, there is a progressive increase in spatiotemporal associations between neuroblasts and melanoma. Interestingly, we confirmed that neuroblasts interact with metastatic melanoma cells in human brain melanoma metastases. Taken together, these findings point to a novel, potentially crucial role of neuroblasts in brain metastasis raising new questions as to how neuroblast–melanoma interactions facilitate brain metastasis on a cellular and molecular level.
Brain endothelial cells and astrocytes are key supportive elements involved in repair processes within the regenerative neurovascular niche after stroke that are vital for efficient metastasis (Ohab et al., 2006; Langley and Fidler, 2013). The regenerative neurovascular niche significantly enhances melanoma-vasculature interactions in vivo, providing a compelling role for endothelial cells in mediating this regeneration-potentiated metastasis. Metastatic melanoma cells within this specialized niche also modify phenotypic structure of the surrounding vasculature. Interactions with astrocytes occur after extravasation but the course of astrocytic interactions is unaltered in the presence of the regenerative neurovascular niche. Thus cross-talk between melanoma and the specialized vasculature that is associated with neuroblasts governs brain metastasis in the regenerative neurovascular niche.
To further tease out the influence of stroke repair on the metastatic transcriptome, we isolated regeneration-potentiated melanoma cells and compared these to brain metastatic melanoma cells outside of the regenerative neurovascular niche using RNAseq. We identify 22 differentially regulated genes in regeneration-potentiated metastatic cells that activate molecular programs strongly implicated within metastasis, including established oncogenes such as JAK3, STAT5A, RHOV, and RAC2.
To isolate molecular interactions between metastatic cells and individual cells in the regenerative neurovascular niche, we mimicked stroke-like conditions in vitro using OGD. Metastatic migration was significantly enhanced in response to OGD-exposed endothelial cells while the contribution from astrocytes was not significant, supporting our in vivo observations that endothelial cell interactions with melanoma are key to governing metastasis. We compared the transcriptomes of in vivo regeneration-potentiated melanoma cells with in vitro melanoma cells exposed to supernatant from OGD-exposed astrocytes and endothelial cells. In vivo regeneration-potentiated metastatic cells showed greater overlap in transcriptional profile with metastatic melanoma exposed to OGD-endothelial cell supernatants than cells exposed to OGD-astrocyte supernatant, suggesting that endothelial cells are the pivotal cell type in guiding regenerative-metastatic signaling. This parallels the central role of vasculature noted above in governing metastatic cell localization to the regenerative neurovascular niche.
We identified molecular interactomes between secreted and extracellular proteins from the endothelial cells and astrocytes to those on in vivo regeneration-potentiated melanoma cells. These include both established (IL23, IL17A) and novel (DPP4, CST3) candidates for signaling in metastasis. Molecular pathway analysis reveals central genes expressed by melanoma cells that might serve as hubs in organizing the pro-metastatic molecular response in regeneration-potentiated melanoma, such as APP, SMAD3, and RPTOR, supporting the hypothesis that stroke induces pro-metastatic alterations to the melanoma transcriptome.
Conclusion
These studies demonstrate that (a) the regenerative neurovascular niche enhances metastasis by facilitating melanoma interactions with regenerative neurovascular niche components, (b) regeneration-potentiated melanoma cells display a unique transcriptome with an enhancement of genes conducive to metastasis, (c) mimicking repair such as stroke-like conditions successfully enhances metastasis while allowing for the identification of cell-specific signaling molecules that play important roles in driving metastasis. These molecular targets may allow promotion of recovery after stroke and brain disease without affecting cancer-related mechanisms.
Author Contributions
RP, SI, DH, GC, IW, and SC conceptualized and designed the work. RP, SI, NT, RL, MR, OS-A, SB-M, TM, and SO collected the data. RP, SI, NT, RL, RK, DH, GC, IW, and SC analyzed and interpreted the data, and drafted and critically revised the manuscript. MR, OS-A, SB-M, TM, and SO analyzed the data. MM performed the experiments. RP, SI, NT, RL, RK, DH, GC, IW, SC, MR, OS-A, SB-M, TM, SO, and MM approved the final version of the manuscript to be published.
Funding
This study was funded by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2019.00297/full#supplementary-material
References
Abraham, C. S., Harada, N., Deli, M. A., and Niwa, M. (2002). Transient forebrain ischemia increases the blood-brain barrier permeability for albumin in stroke-prone spontaneously hypertensive rats. Cell. Mol. Neurobiol. 22, 455–462. doi: 10.1023/A:1021067822435
Agliano, A., Martin-Padura, I., Mancuso, P., Marighetti, P., Rabascio, C., Pruneri, G., et al. (2008). Human acute leukemia cells injected in NOD/LtSz-scid/IL-2Rgamma null mice generate a faster and more efficient disease compared to other NOD/scid-related strains. Int. J. Cancer 123, 2222–2227. doi: 10.1002/ijc.23772
Agrawal, V., Kim, D. Y., and Kwon, Y. G. (2017). Hhip regulates tumor-stroma-mediated upregulation of tumor angiogenesis. Exp. Mol. Med. 49:e289. doi: 10.1038/emm.2016.139
Ben-Baruch, A. (2008). Organ selectivity in metastasis: regulation by chemokines and their receptors. Clin. Exp. Metastasis 25, 345–356. doi: 10.1007/s10585-007-9097-3
Boiko, A. D., Razorenova, O. V., Swetter, S. M., Johnson, D. L., Ly, D. P., Butler, P. D., et al. (2010). Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature 466, 133–137. doi: 10.1038/nature09161
Bosserhoff, A. K. (2017). Melanoma Development: Molecular Biology, Genetics and Clinical Application. 2017. Cham: Springer International Publishing. doi: 10.1007/978-3-319-41319-8
Bronisz, A., Godlewski, J., Wallace, J. A., Merchant, A. S., Nowicki, M. O., Mathsyaraja, H., et al. (2011). Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat. Cell Biol. 14, 159–167. doi: 10.1038/ncb2396
Brumm, A. J., and Carmichael, S. T. (2012). Not just a rush of blood to the head. Nat. Med. 18, 1609–1610. doi: 10.1038/nm.2990
Bullitt, E., Aylward, S. R., Van Dyke, T., and Lin, W. (2007). Computer-assisted measurement of vessel shape from 3T magnetic resonance angiography of mouse brain. Methods 43, 29–34. doi: 10.1016/j.ymeth.2007.03.009
Bullitt, E., Gerig, G., Pizer, S. M., Lin, W., and Aylward, S. R. (2003). Measuring tortuosity of the intracerebral vasculature from MRA images. IEEE Trans. Med. Imaging 22, 1163–1171. doi: 10.1109/TMI.2003.816964
Bullitt, E., Zeng, D., Gerig, G., Aylward, S., Joshi, S., Smith, J. K., et al. (2005). Vessel tortuosity and brain tumor malignancy: a blinded study. Acad. Radiol. 12, 1232–1240. doi: 10.1016/j.acra.2005.05.027
Burnett, R. M., Craven, K. E., Krishnamurthy, P., Goswami, C. P., Badve, S., Crooks, P., et al. (2015). Organ-specific adaptive signaling pathway activation in metastatic breast cancer cells. Oncotarget 6, 12682–12696. doi: 10.18632/oncotarget.3707
Capoluongo, E. (2011). Insulin-like growth factor system and sporadic malignant melanoma. Am. J. Pathol. 178, 26–31. doi: 10.1016/j.ajpath.11.004
Carmichael, S. T. (2016). Emergent properties of neural repair: elemental biology to therapeutic concepts. Ann. Neurol. 79, 895–906. doi: 10.1002/ana.24653
Cavaglia, M., Dombrowski, S. M., Drazba, J., Vasanji, A., Bokesch, P. M., and Janigro, D. (2001). Regional variation in brain capillary density and vascular response to ischemia. Brain Res. 910, 81–93. doi: 10.1016/S0006-8993(01)02637-3
Chao, Y. C., Pan, S. H., Yang, S. C., Yu, S. L., Che, T. F., Lin, C. W., et al. (2009). Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am. J. Respir. Crit. Care Med. 179, 123–133. doi: 10.1164/rccm.200803-456OC
Chen, B., Friedman, B., Cheng, Q., Tsai, P., Schim, E., Kleinfeld, D., et al. (2009). Severe blood-brain barrier disruption and surrounding tissue injury. Stroke 40, e666–e674. doi: 10.1161/STROKEAHA.109.551341
Chen, J., Li, H., Chen, H., Hu, D., Xing, Q., Ren, G., and Luo, X. (2012). Dickkopf-1 inhibits the invasive activity of melanoma cells. Clin. Exp. Dermatol. 37, 404–410. doi: 10.1111/j.1365-2230.2011.04276.x
Chen, Q., Khoury, M., and Chen, J. (2009). Expression of human cytokines dramatically improves reconstitution of specific human-blood lineage cells in humanized mice. Proc. Natl. Acad. Sci. U.S.A. 106, 21783–21788. doi: 10.1073/pnas.0912274106
Chou, J., Lin, J. H., Brenot, A., Kim, J. W., Provot, S., and Werb, Z. (2013). GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell Biol. 15, 201–213. doi: 10.1038/ncb2672
Chou, J., Provot, S., and Werb, Z. (2010). GATA3 in development and cancer differentiation: cells GATA have it!. J. Cell Physiol. 222, 42–49. doi: 10.1002/jcp.21943
Chung, B., Esmaeili, A. A., Gopalakrishna-Pillai, S., Murad, J. P., Andersen, E. S., Kumar Reddy, N., et al. (2017). Human brain metastatic stroma attracts breast cancer cells via chemokines CXCL16 and CXCL12. NPJ Breast Cancer 3:6. doi: 10.1038/s41523-017-0008-8
Chung, K., and Deisseroth, K. (2013). CLARITY for mapping the nervous system. Nat. Methods 10, 508–513. doi: 10.1038/nmeth.2481
Cohn, M. L., Goncharuk, V. N., Diwan, A. H., Zhang, P. S., Shen, S. S., and Prieto, V. G. (2005). Loss of claudin-1 expression in tumor-associated vessels correlates with acquisition of metastatic phenotype in melanocytic neoplasms. J. Cutan. Pathol. 32, 533–536. doi: 10.1111/j.0303-6987.2005.00324.x
Conesa, A., Madrigal, P., Tarazona, S., Cervera, A., McPherson, A., Szczesniak, M. W., et al. (2016). A survey of best practices for RNA-seq data analysis. Genome Biol. 17:13. doi: 10.1186/s13059-016-0881-8
Cook, D. J., Nguyen, C., Chun, H. N., Llorente, L. I., Chiu, A. S., Machnicki, M., et al. (2017). Hydrogel-delivered brain-derived neurotrophic factor promotes tissue repair and recovery after stroke. J. Cereb. Blood Flow Metab. 37, 1030–1045. doi: 10.1177/0271678X16649964
Corrales, L., Ajona, D., Rafail, S., Lasarte, J. J., Riezu-Boj, J. I., Lambris, J. D., et al. (2012). Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J. Immunol. 189, 4674–4683. doi: 10.4049/jimmunol.1201654
De, P., Peng, Q., Traktuev, D. O., Li, W., Yoder, M. C., March, K. L., et al. (2009). Expression of RAC2 in endothelial cells is required for the postnatal neovascular response. Exp. Cell Res. 315, 248–263. doi: 10.1016/j.yexcr.2008.10.003
DeVita, V. T., Lawrence, T. S., and Rosenberg, S. A. (2012). Cancer: Principles & Practice of Oncology: Primer of the Molecular Biology of Cancer. Philadelphia, PA: Lippincott Williams & Wilkins.
Ding, L., Kim, M., Kanchi, K. L., Dees, N. D., Lu, C., Griffith, M., et al. (2014). Clonal architectures and driver mutations in metastatic melanomas. PLoS One 9:e111153. doi: 10.1371/journal.pone.0111153
Ekström, E. J., Sherwood, V., and Andersson, T. (2011). Methylation and loss of secreted frizzled-related protein 3 enhances melanoma cell migration and invasion. PLoS One 6:e18674. doi: 10.1371/journal.pone.0018674
Ergul, A., Alhusban, A., and Fagan, S. C. (2012). Angiogenesis: a harmonized target for recovery after stroke. Stroke 43, 2270–2274. doi: 10.1161/STROKEAHA.111.642710
Fanjul-Fernández, M., Quesada, V., Cabanillas, R., Cadiñanos, J., Fontanil, T., Obaya, A., et al. (2013). Cell-cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat. Commun. 4:2531. doi: 10.1038/ncomms3531
Farooqi, A. A., Tang, J. Y., Li, R. N., Ismail, M., Chang, Y. T., Shu, C. W., et al. (2015). Epigenetic mechanisms in cancer: push and pull between kneaded erasers and fate writers. Int. J. Nanomed. 10, 3183–3191. doi: 10.2147/IJN.S82527
Forget, M. A., Turcotte, S., Beauseigle, D., Godin-Ethier, J., Pelletier, S., Martin, J., et al. (2007). The Wnt pathway regulator DKK1 is preferentially expressed in hormone-resistant breast tumours and in some common cancer types. Br. J. Cancer 96, 646–653. doi: 10.1038/sj.bjc.6603579
Fukumura, D., Duda, D. G., Munn, L. L., and Jain, R. K. (2010). Tumor microvasculature and microenvironment: novel insights through intravital imaging in pre-clinical models. Microcirculation 17, 206–225. doi: 10.1111/j.1549-8719.2010.00029.x
Gialeli, C., Theocharis, A. D., and Karamanos, N. K. (2011). Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 278, 16–27. doi: 10.1111/j.1742-4658.2010.07919.x
Halaban, R., Krauthammer, M., Pelizzola, M., Cheng, E., Kovacs, D., Sznol, M., et al. (2009). Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: clinical implications. PLoS One 4:e4563. doi: 10.1371/journal.pone.0004563
Hansen, T. M., Moss, A. J., and Brindle, N. P. (2008). Vascular endothelial growth factor and angiopoietins in neurovascular regeneration and protection following stroke. Curr. Neurovasc. Res. 5, 236–245. doi: 10.2174/156720208786413433
Harik, N., Harik, S. I., Kuo, N. T., Sakai, K., Przybylski, R. J., and LaManna, J. C. (1996). Time-course and reversibility of the hypoxia-induced alterations in cerebral vascularity and cerebral capillary glucose transporter density. Brain Res. 737, 335–338. doi: 10.1016/0006-8993(96)00965-1
Hassel, J. C., Winnemöller, D., Schartl, M., and Wellbrock, C. (2008). STAT5 contributes to antiapoptosis in melanoma. Melanoma Res. 18, 378–385. doi: 10.1097/CMR.0b013e32830ce7d7
Herner, A., Sauliunaite, D., Michalski, C. W., Erkan, M., De, Oliveira T, Abiatari, I., et al. (2011). Glutamate increases pancreatic cancer cell invasion and migration via AMPA receptor activation and Kras-MAPK signaling. Int. J. Cancer 129, 2349–2359. doi: 10.1002/ijc.25898
Hillion, J. A., Takahashi, K., Maric, D., Ruetzler, C., Barker, J. L., and Hallenbeck, J. M. (2005). Development of an ischemic tolerance model in a PC12 cell line. J. Cereb. Blood Flow Metab. 25, 154–162. doi: 10.1038/sj.jcbfm.9600003
Ho, H., Aruri, J., Kapadia, R., Mehr, H., White, M. A., and Ganesan, A. K. (2012). RhoJ regulates melanoma chemoresistance by suppressing pathways that sense DNA damage. Cancer Res. 72, 5516–5528. doi: 10.1158/0008-5472.CAN-12-0775
Izraely, S., Klein, A., Meshel, T., Tsarfaty, G., Nahmias, C., Couraud, P. O., et al. (2012). The metastatic microenvironment: brain-residing melanoma metastasis and dormant micrometastasis. Int. J. Cancer 131, 1071–1082. doi: 10.1002/ijc.27324
Izraely, S., Klein, A., Sagi-Assif, O., Meshel, T., Tsarfaty, G., Hoon, D. S., et al. (2010). Chemokine-chemokine receptor axes in melanoma brain metastasis. Immunol. Lett. 130, 107–114. doi: 10.1016/j.imlet.2009.12.003
Izraely, S., Sagi-Assif, O., Klein, A., Meshel, T., Ben-Menachem, S., Zaritsky, A., et al. (2015). The metastatic microenvironment: claudin-1 suppresses the malignant phenotype of melanoma brain metastasis. Int. J. Cancer 136, 1296–1307. doi: 10.1002/ijc.29090
Jiang, X., Zhou, J., Giobbie-Hurder, A., Wargo, J., and Hodi, F. S. (2013). The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin. Cancer Res. 19, 598–609. doi: 10.1158/1078-0432.CCR-12-2731
Joshi, S., Singh, A. R., Zulcic, M., Bao, L., Messer, K., Ideker, T., et al. (2014). Rac2 controls tumor growth, metastasis and M1-M2 macrophage differentiation in vivo. PLoS One 9:e95893. doi: 10.1371/journal.pone.0095893
Kesari, S., Xiu, J., Hu, J., Michael, E. S., Piccioni, D., Reddy, SK. (2015). Available at: https://www.carislifesciences.com/wp-content/uploads/2015/06/Tumor-profiles-of-brain-metastases-from-NSCLC-breast-cancer-and-melanoma_ASCO-2015.pdf (accessed 28, 2017).
Kim, J., Mori, T., Chen, S. L., Amersi, F. F., Martinez, S. R., Kuo, C., et al. (2006). Chemokine receptor CXCR4 expression in patients with melanoma and colorectal cancer liver metastases and the association with disease outcome. Ann. Surg. 244, 113–120. doi: 10.1097/01.sla.0000217690.65909.9c
Klein, A., Schwartz, H., Meshel, T., Izraely, S., Bengaiev, R., Nahmias, C., et al. (2015). Astrocytes facilitate melanoma brain metastasis via secretion of IL-23. J. Pathol. 236, 116–127. doi: 10.1002/path.4509
Klein-Goldberg, A., Maman, S., and Witz, I. P. (2013). The role played by the microenvironment in site-specific metastasis. Cancer Lett. 352, 54–58. doi: 10.1016/j.canlet.2013.08.029
Kostareli, E., Holzinger, D., Bogatyrova, O., Hielscher, T., Wichmann, G., Keck, M., et al. (2013). HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J. Clin. Invest. 123, 2488–2501. doi: 10.1172/JCI67010
Kuphal, S., Lodermeyer, S., Bataille, F., Schuierer, M., Hoang, B. H., and Bosserhoff, A. K. (2006). Expression of Dickkopf genes is strongly reduced in malignant melanoma. Oncogene 25, 5027–5036. doi: 10.1038/sj.onc.1209508
Kwan, D. H., Yung, L. Y., Ye, R. D., and Wong, Y. H. (2012). Activation of Ras-dependent signaling pathways by G(14) -coupled receptors requires the adaptor protein TPR1. J. Cell Biochem. 113, 3486–3497. doi: 10.1002/jcb.24225
Lamanna, J. C. (2012). Angioplasticity and cerebrovascular remodeling. Adv. Exp. Med. Biol. 737, 13–17. doi: 10.1007/978-1-4614-1566-4_2
LaManna, J. C., Kuo, N. T., and Lust, W. D. (1998). Hypoxia-induced brain angiogenesis. Signals and consequences. Adv. Exp. Med. Biol. 454, 287–293. doi: 10.1007/978-1-4615-4863-8_34
Lange, C., Storkebaum, E., de Almodovar, C. R., Dewerchin, M., and Carmeliet, P. (2016). Vascular endothelial growth factor: a neurovascular target in neurological diseases. Nat. Rev. Neurol. 12, 439–454. doi: 10.1038/nrneurol.2016.88
Langley, R. R., and Fidler, I. J. (2013). The biology of brain metastasis. Clin. Chem. 59, 180–189. doi: 10.1373/clinchem.2012.193342
Lee, M. M., Chui, R. K., Tam, I. Y., Lau, A. H., and Wong, Y. H. (2012). CCR1-mediated STAT3 tyrosine phosphorylation and CXCL8 expression in THP-1 macrophage-like cells involve pertussis toxin-insensitive Gα(14/16) signaling and IL-6 release. J. Immunol. 189, 5266–5276. doi: 10.4049/jimmunol.1103359
Leotlela, P. D., Wade, M. S., Duray, P. H., Rhode, M. J., Brown, H. F., Rosenthal, D. T., et al. (2007). Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene 26, 3846–3856. doi: 10.1038/sj.onc.1210155
Li, L., Dragulev, B., Zigrino, P., Mauch, C., and Fox, J. W. (2009). The invasive potential of human melanoma cell lines correlates with their ability to alter fibroblast gene expression in vitro and the stromal microenvironment in vivo. Int. J. Cancer 125, 1796–1804. doi: 10.1002/ijc.24463
Li, S., Nie, E. H., Yin, Y., Benowitz, L. I., Tung, S., Vinters, H. V., et al. (2015). GDF10 is a signal for axonal sprouting and functional recovery after stroke. Nat. Neurosci. 18, 1737–1745. doi: 10.1038/nn.4146
Liao, Z., Lutz, J., and Nevalainen, M. T. (2010). Transcription factor Stat5a/b as a therapeutic target protein for prostate cancer. Int. J. Biochem. Cell Biol. 42, 186–192. doi: 10.1016/j.biocel.2009.11.001
Liberzon, A., Birger, C., Thorvaldsdottir, H., Ghandi, M., Mesirov, J. P., and Tamayo, P. (2015). The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. doi: 10.1016/j.cels.2015.12.004
Lin, J. C., Ho, W. H., Gurney, A., and Rosenthal, A. (2003). The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat. Neurosci. 6, 1270–1276. doi: 10.1038/nn1148
Lindvall, O., and Kokaia, Z. (2015). Neurogenesis following stroke affecting the adult brain. Cold Spring Harb. Perspect. Biol. 7:a019034. doi: 10.1101/cshperspect.a019034
Liu, A. M., and Wong, Y. H. (2005). Activation of nuclear factor κB by somatostatin type 2 receptor in pancreatic acinar AR42J cells involves Gα14and multiple signaling components. J. Biol. Chem. 280, 34617–34625. doi: 10.1074/jbc.m504264200
Long, F., Liu, H., Hahn, C., Sumazin, P., Zhang, M. Q., and Zilberstein, A. (2004). Genome-wide prediction and analysis of function-specific transcription factor binding sites. In Silico Biol. 4, 395–410.
Lu, Y., Xiong, Y., Huo, Y., Han, J., Yang, X., Zhang, R., et al. (2011). Grb-2-associated binder 1 (Gab1) regulates postnatal ischemic and VEGF-induced angiogenesis through the protein kinase A-endothelial NOS pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 2957–2962. doi: 10.1073/pnas.1009395108
Luksch, H., Uckermann, O., Stepulak, A., Hendruschk, S., Marzahn, J., Bastian, S., et al. (2011). Silencing of selected glutamate receptor subunits modulates cancer growth. Anticancer Res. 31, 3181–3192.
Lutsenko, S. V., Kiselev, S. M., and Severin, S. E. (2003). Molecular mechanisms of tumor angiogenesis. Biochemistry 68, 286–300.
Martin, S. T., Sato, N., Dhara, S., Chang, R., Hustinx, S. R., Abe, T., et al. (2005). Aberrant methylation of the Human Hedgehog interacting protein (HHIP) gene in pancreatic neoplasms. Cancer Biol. Ther. 4, 728–733. doi: 10.4161/cbt.4.7.1802
Marzese, D. M., Witz, I. P., Kelly, D. F., and Hoon, D. S. (2015). Epigenomic landscape of melanoma progression to brain metastasis: unexplored therapeutic alternatives. Epigenomics 7, 1303–1311. doi: 10.2217/epi.15.77
Mauldin, I. S., Craig, L. S., and Wang, E. (2014). TLR2/6 agonists and IFN-gamma treatment induces favorable immune cell recruiting signatures from melanoma associated with STAT1 and IL-32 signaling. J. ImmunoTher. Cancer 2(Suppl. 3):P225. doi: 10.1186/2051-1426-2-s3-p225
Mazon, M., Masi, D., and Carreau, M. (2016). Modulating dickkopf-1: a Strategy to Monitor or treat cancer? Cancers 8:E62. doi: 10.3390/cancers8070062
McDermott, E. P., and O’Neill, L. A. (2002). Ras participates in the activation of p38 MAPK by interleukin-1 by associating with IRAK, IRAK2, TRAF6, and TAK-1. J. Biol. Chem. 277, 7808–7815. doi: 10.1074/jbc.M108133200
Micevic, G., Theodosakis, N., and Bosenberg, M. (2017). Aberrant DNA methylation in melanoma: biomarker and therapeutic opportunities. Clin. Epigenet. 9:34. doi: 10.1186/s13148-017-0332-8
Miettinen, M., McCue, P. A., Sarlomo-Rikala, M., Rys, J., Czapiewski, P., Wazny, K., et al. (2014). GATA3: a multispecific but potentially useful marker in surgical pathology: a systematic analysis of 2500 epithelial and nonepithelial tumors. Am. J. Surg. Pathol. 38, 13–22. doi: 10.1097/PAS.0b013e3182a0218f
Mirmohammadsadegh, A., Hassan, M., Bardenheuer, W., Marini, A., Gustrau, A., Nambiar, S., et al. (2006). STAT5 phosphorylation in malignant melanoma is important for survival and is mediated through SRC and JAK1 kinases. J. Invest. Dermatol. 126, 2272–2280. doi: 10.1038/sj.jid.5700385
Morris, D. C., Yeich, T., Khalighi, M. M., Soltanian-Zadeh, H., Zhang, Z. G., and Chopp, M. (2003). Microvascular structure after embolic focal cerebral ischemia in the rat. Brain Res. 972, 31–37. doi: 10.1016/S0006-8993(03)02433-8
Nabizadeh, J., Akhir, M. D., Rolfe, B. E., et al. (2013). “Role of the Complement system in melanoma tumour growth,” in Proceedings of the 6th International Conference on Complement Therapeutics, (Kos).
Nicholl, M. B., Chen, X., Qin, C., Bai, Q., Zhu, Z., Davis, M. R., et al. (2016). IL-32α has differential effects on proliferation and apoptosis of human melanoma cell lines. J. Surg. Oncol. 113, 364–369. doi: 10.1002/jso.24142
Niessner, H., Kosnopfel, C., Sinnberg, T., Beck, D., Krieg, K., Wanke, I., et al. (2017). Combined activity of temozolomide and the mTOR inhibitor temsirolimus in metastatic melanoma involves DKK1. Exp. Dermatol. 26, 598–606. doi: 10.1111/exd.13372
Nitta, H., Wada, Y., Kawano, Y., Murakami, Y., Irie, A., and Taniguchi, K. (2013). Enhancement of human cancer cell motility and invasiveness by anaphylatoxin C5a via aberrantly expressed C5a receptor (CD88). Clin. Cancer Res. 19, 2004–2013. doi: 10.1158/1078-0432.CCR-12-1204
Noel, A., Jost, M., and Maquoi, E. (2008). Matrix metalloproteinases at cancer tumor-host interface. Semin. Cell Dev. Biol. 19, 52–60. doi: 10.1016/j.semcdb.2007.05.011
Ohab, J. J., and Carmichael, S. T. (2008). Poststroke neurogenesis: emerging principles of migration and localization of immature neurons. Neuroscientist 14, 369–380. doi: 10.1177/1073858407309545
Ohab, J. J., Fleming, S., Blesch, A., and Carmichael, S. T. (2006). A neurovascular niche for neurogenesis after stroke. J. Neurosci. 26, 13007–13016. doi: 10.1523/JNEUROSCI.4323-06.2006
Olsen, C. L., Hsu, P. P., Glienke, J., Rubanyi, G. M., and Brooks, A. R. (2004). Hedgehog-interacting protein is highly expressed in endothelial cells but down-regulated during angiogenesis and in several human tumors. BMC Cancer 4:43. doi: 10.1186/1471-2407-4-43
Oren, M. (2003). Decision making by p53: life, death and cancer. Cell Death Differ. 10, 431–442. doi: 10.1038/sj.cdd.4401183
Oshima, H., Ishikawa, T., Yoshida, G. J., Naoi, K., Maeda, Y., Naka, K., et al. (2014). TNF-α/TNFR1 signaling promotes gastric tumorigenesis through induction of Noxo1 and Gna14 in tumor cells. Oncogene 33, 3820–3829. doi: 10.1038/onc.2013.356
Ozgur, A., Vu, T., Erkan, G., and Radev, D. R. (2008). Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics 24, i277–i285. doi: 10.1093/bioinformatics/btn182
Palmieri, D. ed (2012). “An introduction to brain metastasis,”. Central Nervous System Metastasis, the Biological Basis and Clinical Considerations (Springer: Heidelberg) doi: 10.1007/978-94-007-5291-7
Prakash, R., and Carmichael, S. T. (2015). Blood-brain barrier breakdown and neovascularization processes after stroke and traumatic brain injury. Curr. Opin. Neurol. 28, 556–564. doi: 10.1097/WCO.0000000000000248
Psaila, B., and Lyden, D. (2009). The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer 9, 285–293. doi: 10.1038/nrc2621
Qian, C. N., Tan, M. H., Yang, J. P., and Cao, Y. (2016). Revisiting tumor angiogenesis: vessel co-option, vessel remodeling, and cancer cell-derived vasculature formation. Chin. J. Cancer 35, 10. doi: 10.1186/s40880-015-0070-2
Quintana, E., Piskounova, E., Shackleton, M., Weinberg, D., Eskiocak, U., Fullen, D. R., et al. (2012). Human melanoma metastasis in NSG mice correlates with clinical outcome in patients. Sci. Transl. Med. 4:159ra149. doi: 10.1126/scitranslmed.3004599
Raso, A., Mascelli, S., Biassoni, R., Nozza, P., Kool, M., Pistorio, A., et al. (2011). High levels of PROM1 (CD133) transcript are a potential predictor of poor prognosis in medulloblastoma. Neuro. Oncol. 13, 500–508. doi: 10.1093/neuonc/nor022
Risso, D., Ngai, J., Speed, T. P., and Dudoit, S. (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902. doi: 10.1038/nbt.2931
Rivals, I., Personnaz, L., Taing, L., and Potier, M. C. (2007). Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23, 401–407. doi: 10.1093/bioinformatics/btl633
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rochman, Y., Kashyap, M., Robinson, G. W., Sakamoto, K., Gomez-Rodriguez, J., Wagner, K. U., et al. (2010). Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7-induced signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 19455–19460. doi: 10.1073/pnas.1008271107
Rollins, D. (2016). Netrin G1 in Desmoplastic Fibroblasts Enhances Interactions with Pancreatic Cancer Cells (Order No. 10154210). Available at: https://search.proquest.com/docview/1836084588?accountid=14512 (accessed 28, 2017).
Salmaggi, A., Maderna, E., Calatozzolo, C., Gaviani, P., Canazza, A., Milanesi, I., et al. (2009). CXCL12, CXCR4 and CXCR7 expression in brain metastases. Cancer Biol. Ther. 8, 1608–1614. doi: 10.4161/cbt.8.17.9202
Sang, H., Li, T., Li, H., and Liu, J. (2015). Gab1 regulates proliferation and migration through the PI3K/Akt signaling pathway in intrahepatic cholangiocarcinoma. Tumour Biol. 36, 8367–8377. doi: 10.1007/s13277-015-3590-0
Sarris, E. G., Saif, M. W., and Syrigos, K. N. (2012). The biological role of PI3K pathway in lung cancer. Pharmaceuticals 5, 1236–1264. doi: 10.3390/ph5111236
Savaskan, N. E., Seufert, S., Hauke, J., Tränkle, C., Eyüpoglu, I. Y., and Hahnen, E. (2011). Dissection of mitogenic and neurodegenerative actions of cystine and glutamate in malignant gliomas. Oncogene 30, 43–53. doi: 10.1038/onc.2010.391
Schmidt, J. W., Wehde, B. L., Sakamoto, K., Triplett, A. A., Anderson, S. M., Tsichlis, P. N., et al. (2014). Stat5 regulates the phosphatidylinositol 3-kinase/Akt1 pathway during mammary gland development and tumorigenesis. Mol. Cell. Biol. 34, 1363–1377. doi: 10.1128/MCB.01220-13
Seiden-Long, I., Navab, R., Shih, W., Li, M., Chow, J., Zhu, C. Q., et al. (2008). Gab1 but not Grb2 mediates tumor progression in Met overexpressing colorectal cancer cells. Carcinogenesis 29, 647–655. doi: 10.1093/carcin/bgn009
Sharfman, W. H. (2012). “Immunotherapies for metastatic melanoma,” in Melanoma (emerging cancer therapeutics), eds W. H. Sharfman and J. Arbraham (New York, NY: Demos Medical Publishing).
Shen, L., Gao, Y., Qian, J., Wu, Y., Zhou, M., Sun, A., et al. (2012). The role of SDF-1α/Rac pathway in the regulation of endothelial progenitor cell polarity; homing and expression of Rac1, Rac2 during endothelial repair. Mol. Cell. Biochem. 365, 1–7. doi: 10.1007/s11010-011-1083-z
Shepelev, M. V., and Korobko, I. V. (2012). Pak6 protein kinase is a novel effector of an atypical Rho family GTPase Chp/RhoV. Biochemistry 77, 26–32. doi: 10.1134/S0006297912010038
Shi, L., Wang, K., Zhao, M., Yuan, X., and Huang, C. (2010a). Overexpression of PIP5KL1 suppresses the growth of human cervical cancer cells in vitro and in vivo. Cell Biol. Int. 34, 309–315. doi: 10.1042/CBI20090040
Shi, L., Zhao, M., Luo, Q., Ma, Y. M., Zhong, J. L., Yuan, X. H., et al. (2010b). Overexpression of PIP5KL1 suppresses cell proliferation and migration in human gastric cancer cells. Mol. Biol. Rep. 37, 2189–2198. doi: 10.1007/s11033-009-9701-5
Sho, S., Court, C. M., Winograd, P., Russell, M. M., and Tomlinson, J. S. (2017). A prognostic mutation panel for predicting cancer recurrence in stages II and III colorectal cancer. J. Surg. Oncol. 116, 996–1004. doi: 10.1002/jso.24781
Soneson, C., Love, M. I., and Robinson, M. D. (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 4:1521. doi: 10.12688/f1000research.7563.2
Song, Y., Ori-McKenney, K. M., Zheng, Y., Han, C., Jan, L. Y., and Jan, Y. N. (2012). Regeneration of drosophila sensory neuron axons and dendrites is regulated by the Akt pathway involving Pten and microRNA bantam. Genes Dev. 26, 1612–1625. doi: 10.1101/gad.193243.112
Springer, M. L., Ip, T. K., and Blau, H. M. (2000). Angiogenesis monitored by perfusion with a space-filling microbead suspension. Mol. Ther. 1, 82–87. doi: 10.1006/mthe.1999.0006
Stecca, B., Mas, C., Clement, V., Zbinden, M., Correa, R., Piguet, V., et al. (2007). Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc. Natl. Acad. Sci. U.S.A. 104, 5895–5900. doi: 10.1073/pnas.0700776104
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., and Gillette, M. A. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Sun, J., He, H., Pillai, S., Xiong, Y., Challa, S., Xu, L., et al. (2013). GATA3 transcription factor abrogates Smad4 transcription factor-mediated fascin overexpression, invadopodium formation, and breast cancer cell invasion. J. Biol. Chem. 288, 36971–36982. doi: 10.1074/jbc.M113.506535
Takaku, M., Grimm, S. A., and Wade, P. A. (2015). GATA3 in breast cancer: tumor suppressor or oncogene? Gene Expr. 16, 163–168. doi: 10.3727/105221615x14399878166113
Tang, Y., Li, M., Wang, J., Pan, Y., and Wu, F. X. (2015). CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 127, 67–72. doi: 10.1016/j.biosystems.2014.11.005
Tarnawski, A. S., Ahluwalia, A., and Jones, M. K. (2014). Angiogenesis in gastric mucosa: an important component of gastric erosion and ulcer healing and its impairment in aging. J Gastroenterol. Hepatol. 29(Suppl. 4), 112–123. doi: 10.1111/jgh.12734
Tenreiro, M. M., Ferreira, R., Bernardino, L., and Brito, M. A. (2016). Cellular response of the blood-brain barrier to injury: potential biomarkers and therapeutic targets for brain regeneration. Neurobiol. Dis. 91, 262–273. doi: 10.1016/j.nbd.2016.03.014
Termini, J., Neman, J., and Jandial, R. (2014). Role of the neural niche in brain metastatic cancer. Cancer Res. 74, 4011–4015. doi: 10.1158/0008-5472.CAN-14-1226
Tomer, R., Ye, L., Hsueh, B., and Deisseroth, K. (2014). Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697. doi: 10.1038/nprot.2014.123
van Zijl, F., Krupitza, G., and Mikulits, W. (2011). Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat. Res. 728, 23–34. doi: 10.1016/j.mrrev.2011.05.002
Vasyutina, E., Stebler, J., Brand-Saberi, B., Schulz, S., Raz, E., and Birchmeier, C. (2005). CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 19, 2187–2198. doi: 10.1101/gad.346205
Venkatesh, H. S., Johung, T. B., Caretti, V., Noll, A., Tang, Y., Nagaraja, S., et al. (2015). Neuronal activity promotes glioma growth through neuroligin-3 secretion. Cell 161, 803–816. doi: 10.1016/j.cell.2015.04.012
Wang, L., Gao, X., Gao, P., Deng, W., Yu, P., Ma, J., et al. (2006). Cell-based screening and validation of human novel genes associated with cell viability. J. Biomol. Screen. 11, 369–376. doi: 10.1177/1087057106286654
Wang, S., Fan, W., Wan, B., Tu, M., Jin, F., Liu, F., et al. (2017). Characterization of long noncoding RNA and messenger RNA signatures in melanoma tumorigenesis and metastasis. PLoS One 12:e0172498. doi: 10.1371/journal.pone.0172498
Weidle, U. H., Birzele, F., Kollmorgen, G., and Krüger, A. (2016). Molecular targets and pathways involved in liver metastasis of colorectal cancer. Clin. Exp. Metastasis 32, 623–635. doi: 10.1007/s10585-015-9732-3
Weitzenfeld, P., and Ben-Baruch, A. (2013). The chemokine system, and its CCR5 and CXCR4 receptors, as potential targets for personalized therapy in cancer. Cancer Lett. 352, 36–53. doi: 10.1016/j.canlet.2013.10.006
Wen, S. J., Zhang, W., Ni, N. N., Wu, Q., Wang, X. P., Lin, Y. K., et al. (2017). Expression of Rho GTPases family in melanoma cells and its influence on cytoskeleton and migration. Oncotarget 8, 30112–30122. doi: 10.18632/oncotarget.15618
Wikman, H., Lamszus, K., Detels, N., Uslar, L., Wrage, M., Benner, C., et al. (2012). Relevance of PTEN loss in brain metastasis formation in breast cancer patients. Breast Cancer Res. 14:R49. doi: 10.1186/bcr3150
Yan, G. N., Yang, L., Lv, Y. F., Shi, Y., Shen, L. L., Yao, X. H., et al. (2014). Endothelial cells promote stem-like phenotype of glioma cells through activating the hedgehog pathway. J. Pathol. 234, 11–22. doi: 10.1002/path.4349
Yang, B., Treweek, J. B., Kulkarni, R. P., Deverman, B. E., Chen, C. K., Lubeck, E., et al. (2014). Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958. doi: 10.1016/j.cell.2014.07.017
Yang, J. P., Liu, H. J., and Liu, X. F. (2010). VEGF promotes angiogenesis and functional recovery in stroke rats. J. Invest. Surg. 23, 149–155. doi: 10.3109/08941930903469482
Yang, L., Huang, J., Ren, X., Gorska, A. E., Chytil, A., Aakre, M., et al. (2008). Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 13, 23–35. doi: 10.1016/j.ccr.2007.12.004
Yang, L., Shah, K. K., and Abbruscato, T. J. (2012). An in vitro model of ischemic stroke. Methods Mol. Biol. 814, 451–466. doi: 10.1007/978-1-61779-452-0_30
Yao, Y., Ye, H., Qi, Z., Mo, L., Yue, Q., Baral, A., et al. (2016). B7-H4(B7x)-mediated cross-talk between glioma-initiating cells and macrophages via the IL6/JAK/STAT3 pathway lead to poor prognosis in glioma patients. Clin. Cancer Res. 22, 2778–2790. doi: 10.1158/1078-0432.CCR-15-0858
Yao, C., Su, L., Shan, J., Zhu, C., Liu, L., Liu, C., et al. (2016). IGF/STAT3/NANOG/Slug signaling axis simultaneously controls epithelial-mesenchymal transition and stemness maintenance in colorectal cancer. Stem Cells 34, 820–831. doi: 10.1002/stem.2320
Zhang, L., Zhang, S., Yao, J., Lowery, F. J., Zhang, Q., Huang, W. C., et al. (2015). Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104. doi: 10.1038/nature15376
Zhang, Y. (2012). STAT5a-targeting miRNA enhances chemosensitivity to cisplatin and 5-fluorouracil in human colorectal cancer cells. Mol. Med. Rep. 5, 1215–1219. doi: 10.3892/mmr.2012.801
Zhang, Z., Liu, F., Li, Z., Wang, D., Li, R., Sun, C., et al. (2017). Jak3 is involved in CCR7-dependent migration and invasion in metastatic squamous cell carcinoma of the head and neck. Oncol. Lett. 13, 3191–3197. doi: 10.3892/ol.2017.5861
Zhao, J., Wang, W., Ha, C. H., Kim, J. Y., Wong, C., Redmond, E. M., et al. (2011). Endothelial Grb2-associated binder 1 is crucial for postnatal angiogenesis. Arterioscler. Thromb. Vasc. Biol. 31, 1016–1023. doi: 10.1161/ATVBAHA.111.224493
Zi, X., Guo, Y., Simoneau, A. R., Hope, C., Xie, J., Holcombe, R. F., et al. (2005). Expression of Frzb/secreted frizzled-related protein 3, a secreted Wnt antagonist, in human androgen-independent prostate cancer PC-3 cells suppresses tumor growth and cellular invasiveness. Cancer Res. 65, 9762–9770. doi: 10.1158/0008-5472.CAN-05-0103
Keywords: angiogenesis, stroke, neuroblast, astrocytosis, gliosis
Citation: Prakash R, Izraely S, Thareja NS, Lee RH, Rappaport M, Kawaguchi R, Sagi-Assif O, Ben-Menachem S, Meshel T, Machnicki M, Ohe S, Hoon DS, Coppola G, Witz IP and Carmichael ST (2019) Regeneration Enhances Metastasis: A Novel Role for Neurovascular Signaling in Promoting Melanoma Brain Metastasis. Front. Neurosci. 13:297. doi: 10.3389/fnins.2019.00297
Received: 30 October 2018; Accepted: 15 March 2019;
Published: 09 April 2019.
Edited by:
Tibor Hortobágyi, University of Debrecen, HungaryReviewed by:
Monique Stins, Johns Hopkins University, United StatesRoger Chammas, University of São Paulo, Brazil
Ewa Czeslawa Izycka-Swieszewska, Medical University of Gdańsk, Poland
Copyright © 2019 Prakash, Izraely, Thareja, Lee, Rappaport, Kawaguchi, Sagi-Assif, Ben-Menachem, Meshel, Machnicki, Ohe, Hoon, Coppola, Witz and Carmichael. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: S. Thomas Carmichael, c2Nhcm1pY2hhZWxAbWVkbmV0LnVjbGEuZWR1
 Roshini Prakash
Roshini Prakash Sivan Izraely2
Sivan Izraely2 Nikita S. Thareja
Nikita S. Thareja Rex H. Lee
Rex H. Lee Riki Kawaguchi
Riki Kawaguchi Orit Sagi-Assif
Orit Sagi-Assif Shuichi Ohe
Shuichi Ohe Giovanni Coppola
Giovanni Coppola Isaac P. Witz
Isaac P. Witz S. Thomas Carmichael
S. Thomas Carmichael