- Galway Neuroscience Centre, School of Natural Sciences, National University of Ireland Galway, Galway, Ireland
The accumulation of iron within the brain occurs in many chronic disorders including Alzheimer’s and Parkinson’s disease and multiple sclerosis. Outside the CNS, a link between levels of iron and the unfolded protein response has already been established. To determine if such a relationship operates in within the brain, we used our ex vivo hippocampal slice-based model of iron accumulation. Ferrocene addition caused accumulation of iron within slices and loss of oligodendrocytes, an effect that was partially inhibited when ferrocene and ER stressor tunicamycin (Tm) were added together. An upward trend (not found to be statistically significant) in the expression of UPR transcripts in response to ferrocene was demonstrated using real-time PCR, while a significant upregulation of mRNA for B cell immunoglobulin-binding protein (BiP) and C/EBP homologous binding protein (CHOP) occurred following exposure to Tm. In silico analysis revealed consensus DNA-binding sequences for UPR-associated transcription factors within the promoter regions of eight iron-regulatory genes. In addition, dual-staining for CHOP and oligodendrocyte transcription factor 2 (OLIG2) or Ionized calcium binding adaptor molecule 1 (Iba1) showed nuclear expression of CHOP in some oligodendrocyte-lineage cells in response to Tm or Tm+ferrocene, but CHOP was rarely found in microglia. Co-expression of UPR-associated activated transcription factor 6 (ATF6) was detected in the nuclei of some oligodendrocyte-lineage cells exposed to Tm alone, or to Tm and ferrocene, but rarely in microglia. These data highlight the therapeutic potential of targeting UPR-associated proteins when developing novel treatments for chronic brain disorders that are affected by dysregulated iron.
Introduction
The unfolded protein response (UPR) is a ‘check and balance’ program activated upon disturbed homeostasis in the endoplasmic reticulum. Occasional but infrequent hints have documented an interplay between the UPR and iron metabolism (Ye and Connor, 2000; You et al., 2003; Oliveira et al., 2009; Vecchi et al., 2009). However, reports are few on the possible links between the UPR and iron within the brain (Liu and Connor, 2012; Rahman et al., 2017, 2018; Lumsden et al., 2018). A better understanding of this cross-talk might provide further clues regarding disease pathogenesis, given that aberrant iron metabolism and increased expression of markers of the UPR have been independently reported in several neurodegenerative diseases (Hetz and Mollereau, 2014), including multiple sclerosis (Stephenson et al., 2014), Alzheimer’s disease and Parkinson’s disease (Mercado et al., 2013; Ward et al., 2014). Researchers have exploited glial and neuronal mono- and dual-cultures in vitro, as well as ex vivo systems to investigate the regulation of iron within the brain [see (Healy et al., 2017) for comprehensive review of this topic]. As a foundation for future studies, the overall goal of this work was therefore to confirm and characterize the reciprocal relationship between UPR and iron homeostasis within our hippocampal slice-based model (Healy et al., 2016) of the accumulation of iron within the CNS.
Methods
Ethics Statement
The research described was approved by the Animal Care Research Ethics Committee Filing ID 12/FEB/04) and was carried out in accordance with European directive 2010/63/EU.
Hippocampal Slice Culture and Reagent Exposure
Neonatal hippocampal slices were maintained as previously described (Healy et al., 2016).
Five μg/ml tunicamycin (Tm, T7765, Sigma-Aldrich, Dublin, Ireland) exposure was used to activate a UPR response, while iron loading was induced by a 12-h treatment with 1 μM ferrocene, a membrane-permeable compound that enables uptake of iron by cells (F408, Sigma-Aldrich, Dublin, Ireland). Slices were harvested immediately after iron loading.
Analysis of RNA Transcripts
RNA was isolated from control and treated slice cultures using TRIreagent (T9424, Sigma-Aldrich, Dublin, Ireland) as per the manufacturer’s instructions. Following DNAse I treatment (18068-015, Thermofisher-Scientific, Dublin, Ireland) and cDNA synthesis using manufacturer protocol (1804014, Superscript II Reverse Transcriptase, Invitrogen, Dublin, Ireland), real-time PCR was accomplished using Applied Biosystems 4346906 MicroAmp®Fast Optical 96-Well Reaction Plate and Fast SYPR green master mix (applied Biosystems, 4385612) and the Step One real-time PCR System (Thermo-Fisher).
Unless otherwise indicated, oligonucleotide primer sequences (Sigma-Aldrich, Dublin, Ireland) used were designed using Primer3. Primer sequence details are summarized in the Supplementary Table 1. Results were quantified using the ΔΔCT method following standardization relative to β-actin as an internal control.
Tissue Viability
Tissue viability was measured as described in Healy et al. (2016), using a lactate dehydrogenase (LDH) and MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide] assay.
Immunohistochemistry and Imaging
Tissue was fixed and stained as previously described (Healy et al., 2016). Briefly, slices were fixed in 4% paraformaldehyde (PFA) for 30 min followed by three washes in PBS and then blocked and permeabilised for 1 h with 10% normal goat serum and 0.4% Triton X-100 in PBS. Subsequently, the slices were incubated with primary antibody (for single and dual labeling) in 2.5% NGS (or 2.5% horse serum) and 0.1% Triton X-100 in PBS for 48 h at 4°C and then washed three times for 15 min before incubation with the appropriate secondary antibody at 4°C overnight. Source information and dilutions used for all antibodies is available in the Supplementary Materials. Slice cultures were mounted in Vectashield containing diamino-2-phenylindole (DAPI) (H-1200, Vector Laboratories, Peterborough, United Kingdom) to allow visualization of nuclei. Negative “no primary antibody” controls were included. Slides were stored in the dark at 4°C until imaged. All samples were imaged on a laser scanning confocal microscope (Olympus Fluoview 1000) using a 40 × oil-immersion lens (numerical aperture of 1.3).
In silico Analysis of UPR Transcription Factor Binding Sites
The ALGGEN PROMO algorithm1 was used to determine if any UPR transcription factor consensus DNA binding sequences were present within the promoter regions of genes that encode iron homeostasis regulators. Only hits that had a dissimilarity index <15% and a random expectation <0.5 were included in this analysis.
Statistical Analysis
Data was analyzed using Prism 6 (GraphPad Software). All measurements were expressed as mean ± standard error of the mean (SEM) unless otherwise indicated. Normality was assessed using the Shapiro-Wilk test. Statistical analyses were carried out using Student’s t-test, Mann-Whitney test or an ANOVA with Dunnet post-test, as appropriate, and as indicated in figure legends. Differences were considered statistically significant if P<0.05.
Results
UPR Transcript Expression in Cultured Slices Exposed to Ferrocene Remains Unchanged
Real-time PCR analysis of changes in the transcripts for B-cell immunoglobulin binding protein (BiP), calreticulin (CRT), X-box binding protein 1 (XBP1), activated transcription factor 6 (ATF6), activated transcription factor and C/EBP homologous binding protein (CHOP) revealed an approximate 1.4–2.1-fold increase, which was not found to be statistically significant following three replicate experiments (Figure 1A).
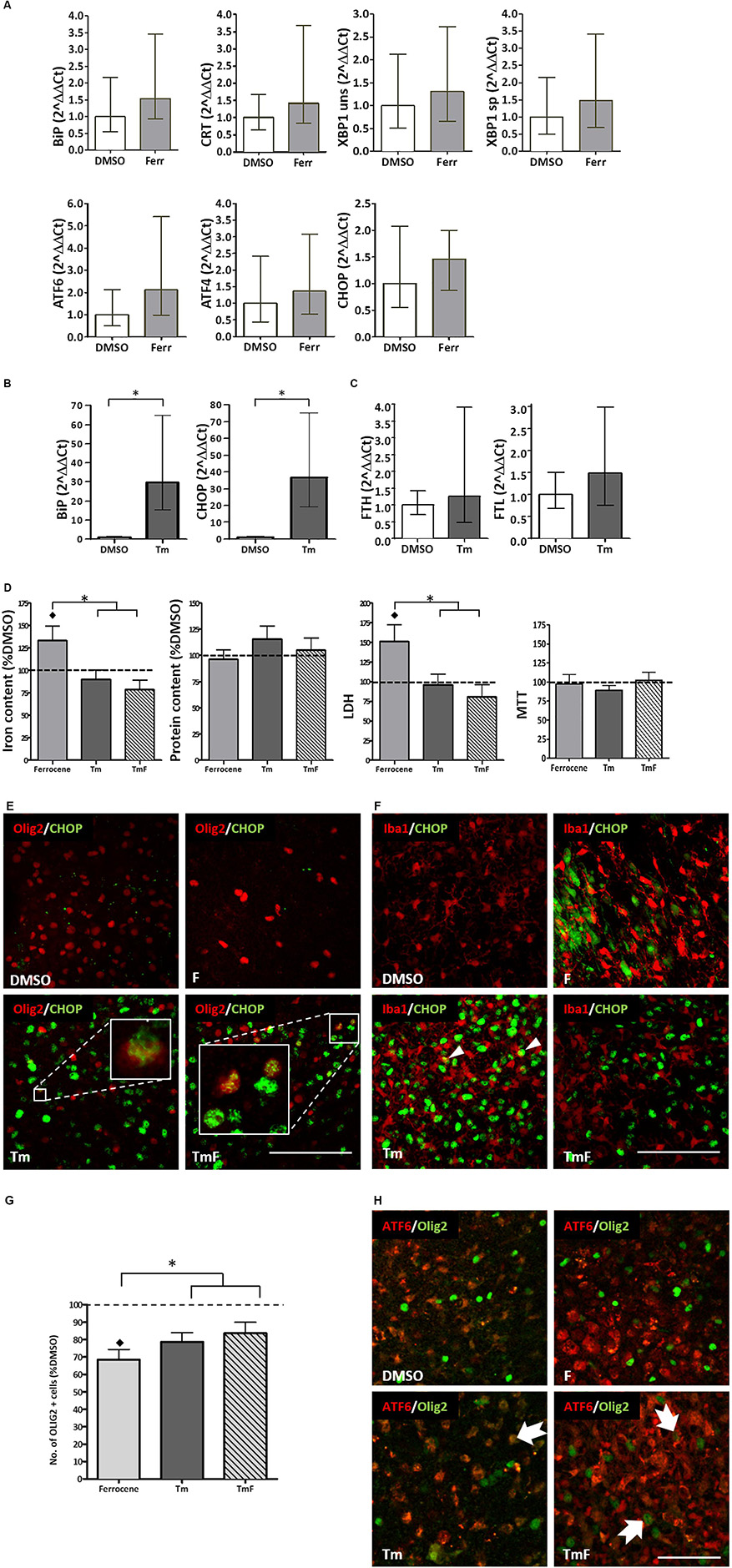
Figure 1. Cross-talk between regulators of the UPR and iron homeostasis. (A) Real-time PCR analysis of RNA samples derived from cultured slices treated for 12 h with 1 μM ferrocene. An upward trend in the expression of BiP, CRT, XBP1, ATF6, ATF4, and CHOP was detected (n = 3 experimental replicates). (B) Increased BiP and CHOP transcripts detected following 12 h treatment with 5 μg/ml Tm (n = 3). Data is expressed as mean ± 95% confidence interval following completion of three experimental repeats. ∗denotes p<0.05. (C) Upward trend in the expression of FTL and FTH transcripts detected following 12 h Tm treatment (5 μg/ml, n = 2 experimental repeats). (D) Effect of Tm exposure (5 μg/ml, 12 h) on iron and protein content and on tissue viability (n = 3 experimental replicates). Cultures were loaded with 1 μM ferrocene (F), 5 μg/ml Tm or both treatments concurrently (TmF). Data is expressed as percentage of DMSO control, which is indicated by the dashed line. Values are mean + SEM from three experiments. ♢ denotes a significant difference (p<0.05) when compared with DMSO and ∗denotes significant difference between F-treated slices and those treated with Tm or TMF (p<0.05). (E) CHOP expression (green) was detected within the nuclei of olig2-positive cells (red) in hippocampal slices treated with Tm or TmF (lower panels), but was not seen within the Olig2-positive population of control or F-treated slices (upper panels). Scale bar = 100 μm (F) When cultured slices were dual-labeled using antibodies to Iba1 (red) and CHOP (green, RH panels), CHOP was not found in DMSO-exposed slices but was seen in non-Iba1-positive slices treated with F or TmF and occasionally in Iba1-positive nuclei in slices treated with Tm alone (white arrowheads). Scale bar = 100 μm. (G) The significant loss of oligodendrocyte-lineage cells caused by exposure to F is partially reversed when slices are co-treated with Tm or TmF (n = 2 experimental replicates). ♢denotes a significant difference (p<0.05) when compared with DMSO; ∗denotes a significant difference between F-treated slices and those treated with Tm or TMF (p<0.05). (H) Cytoplasmic expression of ATF6 (red) detected in the non-olig2-expressing cells of DMSO- and F-treated hippocampal slices (top panels). Treatment with Tm or TmF led to nuclear expression of ATF6 within olig2-positive cells (green, arrows). Scale bar = 50 μm.
UPR Induction Does Not Increase Expression of Iron Storage Protein Transcripts
The ability of cultured hippocampal slices to mount a response to ER stress was confirmed when a significant increase in BiP and CHOP transcription (approximately 30-fold for both), was detected following a 12-h treatment with 5 μg/ml tunicamycin (Tm, Figure 1B). Tm inhibits N-glycosylation of proteins trafficked through the ER (Foufelle and Fromenty, 2016) hence causing a build-up of mis-folded proteins and ER stress. However, Tm was not found, after two experimental repeats, to cause a significant increase in the expression of ferritin light chain (FTL) or ferritin heavy chain (FHL) transcripts (Figure 1C).
UPR Activation Prevents Ferrocene-Induced Iron Accumulation and Glial Perturbations
In line with our previous findings (Healy et al., 2016), 1 μM ferrocene produced a 1.3-fold increase in the levels of iron in slice cultures when compared to vehicle (Figure 1D). Similarly, we confirmed that ferrocene-induced a significant 1.5-fold increase in LDH release, and, once again, did not report any iron-induced loss in metabolic activity as assessed by MTT assay (Figure 1D). Interestingly, concurrent exposure to 5 μg/ml Tm (TmF) resulted in amelioration in iron accumulation and toxicity (Figure 1D). TmF treatment led to a significant reduction in iron content and a significant decrease in LDH release, when compared with ferrocene alone, but not when compared with the DMSO control. In keeping with our data relating to FTL and FTH transcriptional expression, Tm exposure alone did not lead to a significant difference in iron content. Tm treatment alone also had no effect on tissue viability when compared to DMSO control conditions. There were no significant differences in metabolic activity as measured using an MTT assay, or in protein content between groups (Figure 1D).
UPR Transcription Factor Expression in Glia Following Tm and Iron Exposure
Classically, CHOP is upregulated in response to phosphorylation of PERK and is frequently exploited as a marker of activation of the UPR. Immunofluorescent staining demonstrated an absence of CHOP expression in control and ferrocene-treated slice cultures, but robust upregulation of CHOP in Tm-treated tissue. Dual immunofluorescent staining confirmed CHOP expression in Olig2-positive cells within Tm-treated slices, which appeared to increase when slices were co-exposed to Tm and Ferrocene (Figure 1E). Less frequently, CHOP was found to be present in microglia within cultured slices exposed to Ferrocene, Tm or both (Figure 1F). Similar to our prior finding of significantly reduced numbers of microglial endpoints, following slice treatment with F, we also found, following two experimental repeats, fewer end-points after F treatment, which was rescued when tissue was treated with TmF (data not shown). ATF6 is the second major regulator of the UPR whose protein expression we analyzed by immunofluorescence within cultured control and treated slices. While non-nuclear ATF6 was detected in control DMSO-treated cultures, an increase in nuclear ATF6 staining was observed following treatment with Tm, F, or a combination of both reagents (not shown).
In our previous study, we reported a significant loss of oligodendrocyte-lineage cells following iron loading (Healy et al., 2016). This result was confirmed here (Figure 1G). Intriguingly, slice exposure to Tm or TmF reversed this loss, to the extent that Olig2-lineage cells responded by increasing their number significantly (Figure 1G). Dual immunofluorescence was then used to determine if the localization of ATF6 expression within Olig2-positive cells was affected by the same conditions. Our images showed that while nuclear (active) ATF6 was not detectable within DMSO-treated Olig2-lineage cells, it was present in Tm-, F- or TmF-treated cultures (Figure 1H).
In silico Analysis of Putative Transcription Factor Binding Sites Within Promoter Regions of Iron-Regulatory Genes
Using the ALGGEN PROMO algorithm to predict consensus transcription factor (TF) DNA binding sequences, we identified a number of putative binding sites for UPR-associated transcription within the promoter regions of iron homeostasis genes (Table 1). None of the TFs were found to be commonly expressed on all the promoter regions examined. Activated transcription factor 3 (ATF3) and CCAAT-enhancer-binding protein alpha (C/EBP-α) consensus-binding sequences were found on six of the eight genes (Table 1). Consensus sequences for XBP1, ATF and ATF2 were found within two promoter regions; the CREB consensus sequence was found on three genes and C/EBP-β was not found on any. Therefore, there was not a consistent pattern of transcription factor hits across the different genes (Table 1).
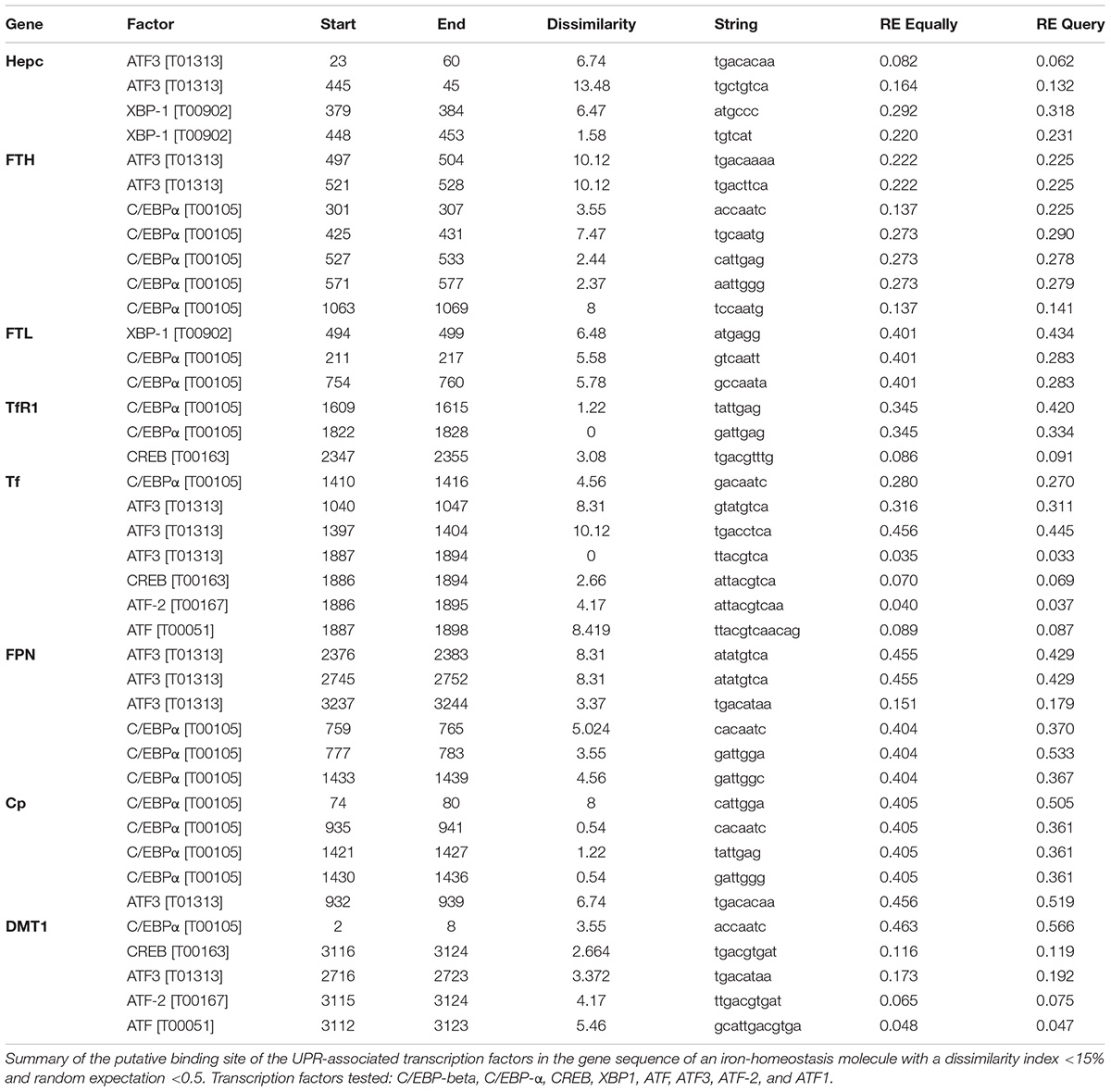
Table 1. Putative binding sites of UPR transcription factors in the promoters of genes encoding iron homeostasis proteins.
Discussion
In an attempt to elucidate whether or not cross-talk between iron-regulatory proteins and ER stress, we exploited our rat hippocampal slice-based model of iron accumulation. Following slice exposure to Tm, F or both, real-time PCR and fluorescent immunohistochemistry yielded the following primary findings: (1) Tm exposure does not affect the level of iron in hippocampal slices, but it triggers a UPR that appears to prevent iron overload and possibly ameliorates toxic effects of iron on oligodendrocytes and microglia; (2) iron overload causes an upward trend in the levels of transcripts encoding the major markers of the UPR, although this was not statistically significant; (3) the observation that Tm treatment led to no significant change in the expression of FTL or FTH transcripts was surprising, since the promoter regions of eight genes that encode iron-regulatory proteins, including FTL and FTH, contain putative binding sites for ER stress-associated transcription factors; (4) transcription factor ATF6 is activated following exposure of cultured hippocampal slices to Tm, F and TmF; (5) Activated ATF6 is detectable within the Tm- and TmF-treated oligodendrocycte population but rarely in similarly treated microglia.
While intriguing, it remains unclear how concurrent iron accumulation and UPR activation, demonstrated here, leads to the ameliorated accumulation of iron. Outside the brain, the main description of the intersection between the UPR and iron metabolism is the action of CHOP on hepcidin (via modulation of C/EBPα binding and/or through CREBH) in the liver and spleen (Oliveira et al., 2009; Vecchi et al., 2009) although ER stress associated chaperones calreticulin and BiP have also been implicated in iron UPR cross-talk [reviewed in (Oliveira et al., 2011)]. More recently, an increase in hepcidin expression leading to accumulated iron within the brain of rats subjected to sub-arachnoid hemorrhage, was found to be regulated by CHOP (Zhao et al., 2018).
Our observation that CHOP immunofluorescence appears to be increased in Olig-2 positive cells in slices that were co-exposed to Tm and Ferrocene compared with only Tm is consistent with these studies and it hints at the potential therapeutic benefit of UPR proteins induced by Tm to protect against iron-induced toxicity within oligodendrocyte lineage cells. However, further work is required to elucidate or reasonably speculate on such a mechanism.
Author Contributions
SH carried out the PCR. SH and JM completed the tissue staining. JM and UF conceived and oversaw research described. SH, JM, and UF wrote the manuscript.
Funding
SH was supported by a College of Science Fellowship from The National University of Ireland, Galway. SH and JM were supported by the Foundation Office of the National University of Ireland, Galway.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2018.00969/full#supplementary-material
Footnotes
References
Foufelle, F., and Fromenty, B. (2016). Role of endoplasmic reticulum stress in drug-induced toxicity. Pharmacol. Res. Perspect. 4:e00211. doi: 10.1002/prp2.211
Healy, S., McMahon, J., Owens, P., and FitzGerald, U. (2016). Significant glial alterations in response to iron loading in a novel organotypic hippocampal slice culture model. Sci. Rep. 6:36410. doi: 10.1038/srep36410
Healy, S., McMahon, J. M., and FitzGerald, U. (2017). Modelling iron mismanagement in neurodegenerative disease in vitro: paradigms, pitfalls, possibilities & practical considerations. Prog. Neurobiol. 158, 1–14. doi: 10.1016/j.pneurobio.2017.08.004
Hetz, C., and Mollereau, B. (2014). Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 15, 233–249. doi: 10.1038/nrn3689
Liu, Y., and Connor, J. R. (2012). Iron and ER stress in neurodegenerative disease. Biometals 25, 837–845. doi: 10.1007/s10534-012-9544-8
Lumsden, A. L., Rogers, J. T., Majd, S., Newman, M., Sutherland, G. T., Verdile, G., et al. (2018). Dysregulation of neuronal iron homeostasis as an alternative unifying effect of mutations causing familial Alzheimer’s disease. Front. Neurosci. 12:533. doi: 10.3389/fnins.2018.00533
Mercado, G., Valdes, P., and Hetz, C. (2013). An ERcentric view of Parkinson’s disease. Trends Mol. Med. 19, 165–175. doi: 10.1016/j.molmed.2012.12.005
Oliveira, S. J., de Sousa, M., and Pinto, J. P. (2011). ER stress and iron homeostasis: a new frontier for the UPR. Biochem. Res. Int. 2011:896474. doi: 10.1155/2011/896474
Oliveira, S. J., Pinto, J. P., Picarote, G., Costa, V. M., Carvalho, F., Rangel, M., et al. (2009). ER stress-inducible factor CHOP affects the expression of hepcidin by modulating C/EBPalpha activity. PLoS One 4:e6618. doi: 10.1371/journal.pone.0006618
Rahman, S., Archana, A., Jan, A. T., and Minakshi, R. (2018). Dissecting Endoplasmic Reticulum Unfolded Protein Response (UPR(ER)) in managing clandestine modus operandi of Alzheimer’s disease. Front. Aging Neurosci. 10:30. doi: 10.3389/fnagi.2018.00030
Rahman, S., Jan, A. T., Ayyagari, A., Kim, J., Kim, J., and Minakshi, R. (2017). Entanglement of UPR(ER) in aging driven Neurodegenerative diseases. Front. Aging Neurosci. 9:341. doi: 10.3389/fnagi.2017.00341
Stephenson, E., Nathoo, N., Mahjoub, Y., Dunn, J. F., and Yong, V. W. (2014). Iron in multiple sclerosis: roles in neurodegeneration and repair. Nat. Rev. Neurol. 10, 459–468. doi: 10.1038/nrneurol.2014.118
Vecchi, C., Montosi, G., Zhang, K., Lamberti, I., Duncan, S. A., Kaufman, R. J., et al. (2009). ER stress controls iron metabolism through induction of hepcidin. Science 325, 877–880. doi: 10.1126/science.1176639
Ward, R. J., Zucca, F. A., Duyn, J. H., Crichton, R. R., and Zecca, L. (2014). The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 13, 1045–1060. doi: 10.1016/S1474-4422(14)701176
Ye, Z., and Connor, J. R. (2000). Identification of iron responsive genes by screening cDNA libraries from suppression subtractive hybridization with antisense probes from three iron conditions. Nucleic Acids Res. 28, 1802–1807. doi: 10.1093/nar/28.8.1802
You, K. R., Liu, M. J., Han, X. J., Lee, Z. W., and Kim, D. G. (2003). Transcriptional regulation of the human transferrin gene by GADD153 in hepatoma cells. Hepatology 38, 745–755. doi: 10.1053/jhep.2003.50367
Keywords: iron, unfolded protein response, UPR, tunicamycin, ferrocene
Citation: Healy S, McMahon J and FitzGerald U (2018) UPR Induction Prevents Iron Accumulation and Oligodendrocyte Loss in ex vivo Cultured Hippocampal Slices. Front. Neurosci. 12:969. doi: 10.3389/fnins.2018.00969
Received: 13 September 2018; Accepted: 04 December 2018;
Published: 18 December 2018.
Edited by:
Isabella Zanella, Università degli Studi di Brescia, ItalyReviewed by:
Safikur Rahman, Yeungnam University, South KoreaRaffaella Gozzelino, Centro de Estudos de Doenças Crónicas (CEDOC), Portugal
Copyright © 2018 Healy, McMahon and FitzGerald. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Una FitzGerald, dW5hLmZpdHpnZXJhbGRAbnVpZ2Fsd2F5Lmll
 Sinead Healy
Sinead Healy Jill McMahon
Jill McMahon Una FitzGerald
Una FitzGerald