- 1Key Laboratory of Zoonosis Research, Ministry of Education, College of Basic Medical Science, Jilin University, Changchun, China
- 2Department of Anesthesiology, China-Japan Union Hospital of Jilin University, Changchun, China
- 3Center of Cardiovascular Disease, The First Hospital of Jilin University, Changchun, China
Alzheimer’s disease (AD) is the most common neurodegenerative disease. In recent years, multiple pathway analyses of AD genome-wide association studies (GWAS) have been conducted, and provided strong support for immune pathways in AD. Rheumatoid arthritis (RA) is a chronic autoimmune disease. It is reported that antirheumatic drugs had protective effect on dementia in RA patients. However, observational studies have reported a controversial inverse relationship between AD and RA. In addition, Mendelian randomization studies have also been performed to evaluate the association of RA with AD. However, these studies reported inconsistent association of RA with AD. Until now, it is still unclear that AD is a causally associated with RA. Here, we performed a Mendelian randomization study to investigate the causal association of AD with RA. We analyzed the large-scale AD GWAS dataset (74,046 individuals) and RA GWAS dataset (58,284 individuals) from the European descent. However, we did not identify any significant association of AD with RA using inverse-variance weighted meta-analysis (IVW), weighted median regression and MR-Egger regression.
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disease in the elderly (Hu et al., 2017; Liu et al., 2017a, 2018). Until now, it is still largely unknown about the exact AD genes (Jiang et al., 2017). In recent years, multiple large-scale genome-wide association studies (GWAS) have been performed, and successfully identified common AD genes including CR1, BIN1, CLU, PICALM, MS4A4/MS4A6E, CD2AP, CD33, EPHA1, ABCA7, SORL1, HLA-DRB5/DRB1, PTK2B, SLC24A4-0RING3, DSG2, INPP5D, MEF2C, NME8, ZCWPW1, CELF1, FERMT2, and CASS4 (Harold et al., 2009; Hollingworth et al., 2011; Naj et al., 2011; Lambert et al., 2013; Liu et al., 2014b; Li et al., 2016; Jun et al., 2017; Sims et al., 2017). Importantly, some of these genes have been successfully validated (Liu et al., 2012, 2013b,c,d, 2014a,b,c, 2015, 2017a,b; Lambert et al., 2013; Bao et al., 2015; Chen et al., 2015; Li et al., 2015; Shen et al., 2015; Xiang et al., 2015; Zhang et al., 2015; Li et al., 2016; Liu and Jiang, 2016; Zhang et al., 2016; Jiang et al., 2017; Jun et al., 2017; Sims et al., 2017). In addition, multiple pathway analyses of AD GWAS have been conducted, and provided strong support for immune pathways in AD (Hong et al., 2010; Jones et al., 2010; Lambert et al., 2010; Liu et al., 2012). Yokoyama et al. (2016) performed a genetic association study to evaluate the genetic overlap between AD and seven immune-mediated diseases including Crohn’s disease, ulcerative colitis, rheumatoid arthritis (RA), type 1 diabetes, celiac disease, and psoriasis (Jiang et al., 2016; Yokoyama et al., 2016). They identified eight genetic variants associated with both AD and immune-mediated diseases (Jiang et al., 2016; Yokoyama et al., 2016). However, epidemiological studies have reported a controversial inverse relationship between AD and RA (Ferraccioli et al., 2012; Kao et al., 2016; Ungprasert et al., 2016).
Mendelian randomization could determine the causal inferences, and has been used to evaluate the association between RA and AD (Policicchio et al., 2017; Bae and Lee, 2018). Policicchio et al. (2017) selected 62 RA SNPs (P < 5.00E-08, a genome-wide significance level) as instrumental variables, and identified no evidence of a causal association between RA and AD. Bae and Lee (2018) selected 80 RA SNPs as instrumental variables. They selected three methods including IVW, weighted median, and MR-Egger (Bae and Lee, 2018). Both the IVW (beta = −0.039, P = 0.021) and weighted median (beta = −0.078, P = 0.001) indicated significant association of RA with AD (Bae and Lee, 2018). In summary, both studies evaluated the causal association of RA with AD, and reported inconsistent findings (Policicchio et al., 2017; Bae and Lee, 2018). Importantly, both studies did not evaluate the causal association of AD with RA. Until now, it is still unclear whether AD is a causally associated with RA. Here, we performed a Mendelian randomization study to investigate the causal association of AD with RA.
Materials and Methods
AD GWAS Dataset
The instrumental variables are AD variants at a genome-wide significance level P < 5.00E-08 identified by previous GWAS. The AD GWAS dataset is from the International Genomics of Alzheimer’s Project (IGAP) (Lambert et al., 2013). In stage 1, the IGAP analyzed a total of 17,008 AD cases and 37,154 controls of European descent (The European Alzheimer’s disease Initiative – EADI, the Alzheimer Disease Genetics Consortium – ADGC, The Cohorts for Heart and Aging Research in Genomic Epidemiology consortium – CHARGE, The Genetic and Environmental Risk in AD consortium – GERAD) (Lambert et al., 2013). In stage 2, IGAP analyzed additional independent 8,572 AD cases and 11,312 controls (Lambert et al., 2013). Here, we aimed to selected the independent AD variants at a genome-wide significance level P < 5.00E-08 in this AD dataset (Lambert et al., 2013).
RA GWAS Dataset
The RA GWAS dataset is from a previous RA GWAS meta-analysis in a total of >100,000 subjects of European and Asian ancestries (29,880 RA cases and 73,758 controls) (Okada et al., 2014). The summary statistics of RA GWAS meta-analysis included trans-ethnic RA GWAS meta-analysis (19,234 RA cases and 61,565 controls), European RA GWAS meta-analysis (14,361 RA cases and 43,923 controls), and Asian RA GWAS meta-analysis (4,873 RA cases and 17,642 controls) (Okada et al., 2014). Here, we selected the European RA GWAS meta-analysis, as the AD GWAS dataset was also from European samples.
Mendelian Randomization Analysis
Here, we selected three Mendelian randomization analysis methods including inverse-variance weighted meta-analysis (IVW), weighted median regression and MR-Egger regression, as did in recent studies (Bae and Lee, 2018; Jiang et al., 2018). In addition, we selected the MR-Egger intercept test to assess the instrumental variable assumptions, and provide a statistical test for the presence of potential pleiotropy (Bae and Lee, 2018; Jiang et al., 2018). The odds ratio (OR) as well as 95% confidence interval (CI) of RA correspond to the genetically determined increase in AD. Meanwhile, we performed a sensitivity analysis using a leave-one-out permutation. All analyses were conducted using the R package “MendelianRandomization” (Yavorska and Burgess, 2017). The significance level for significant association of AD with RA was P < 0.05.
Results
Association of AD Variants With RA
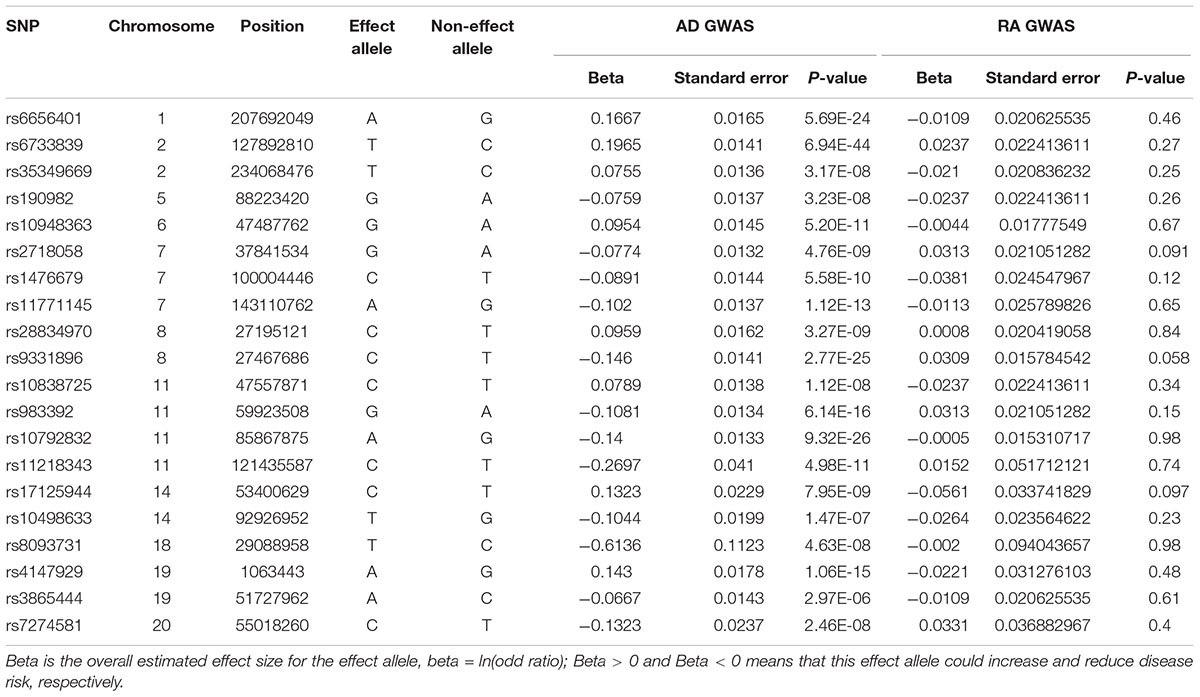
The meta-analysis of stage 1 and stage 2 in IGAP identified 21 independent AD variants at the genome-wide significance level P < 5.00E-08. Of the 21 AD risk variants, we extracted the summary statistics of 20 variants in RA GWAS. Only one variant rs10745742 and its proxy variants with r2 > = 0.8 in HaploReg v4.1 in 1000 Genomes Project (CEU) (Ward and Kellis, 2012), were not available in RA GWAS dataset. Hence, our analysis will focus on these 20 variants. Here, we provided more detailed information about these 20 variants in Table 1.
Association of AD With RA
In brief, we did not identify any significant association of AD with RA including the IVW (OR = 0.95, 95% CI: 0.88–1.03, P = 0.451), weighted median (OR = 0.96, 95% CI: 0.85–1.07, P = 0.217), and MR-Egger (OR = 0.98, 95% CI: 0.78–1.22, P = 0.827). In addition, MR-Egger intercept test did not show significant pleiotropy (MR-Egger intercept β = −0.003; P = 0.804). Hence, the estimates from these methods were consistent in terms of direction and magnitude. The leave-one-out permutation analysis showed that the direction and precision of the genetic estimates between AD and RA remained largely unchanged. Supplementary Figure S1 shows the individual causal estimates from each of the 20 genetic variants using different methods.
Discussion
Observational studies have evaluated the association between AD and RA. However, these studies reported inconsistent findings. Chou et al. (2016) conducted a nested case-control study by analyzing more than 8.5 million commercially insured adults. They found that AD was more prevalent among RA patients compared with those without RA (Chou et al., 2016). RA population had an increased AD risk (Chou et al., 2016). Kao et al. (2016) performed a case-control study to evaluate the relationship between prior RA and AD using 2271 patients with AD as cases and 6813 patients without AD as controls. They found an inverse association between prior RA and AD (Kao et al., 2016). Ungprasert et al. (2016) conducted a systematic review and meta-analysis of three cohort studies and two cross-sectional studies. They identified significant increased risk of dementia in RA cases (Ungprasert et al., 2016). Policicchio et al. (2017) a systematic review and meta-analysis of eight case-control and two population-based studies. They found that RA was associated with lower AD incidence (Policicchio et al., 2017).
Genetic association studies also have evaluated the association between AD and immune pathways. These findings are consistent. Several pathway analyses of AD GWAS dataset have identified some immune pathways in AD, including Natural killer cell mediated cytotoxicity (hsa04650), Antigen processing and presentation (hsa04612), retinoic acid-inducible gene-I (RIG-I)-like receptor signaling (hsa04622), asthma (hsa05310), hematopoietic cell lineage (hsa04640), graft-versus-host disease (hsa05332), allograft rejection (hsa05330), autoimmune thyroid disease (hsa05320), and type I diabetes mellitus (hsa04940) (Hong et al., 2010; Jones et al., 2010; Lambert et al., 2010; Liu et al., 2012). Yokoyama et al. (2016) found genetic overlap between AD and immune-mediated diseases (Jiang et al., 2016).
Until now, two Mendelian randomization studies have also been performed to evaluate the association between RA and AD (Policicchio et al., 2017; Bae and Lee, 2018). However, these two studies reported inconsistent findings. Policicchio et al. (2017) reported no evidence of a causal association between RA and AD. Bae and Lee (2018) identified a significant causal association of RA with AD. Judge et al. (2017) found that antirheumatic drugs had protective effect on dementia in RA patients. The classical disease-modifying antirheumatic drug (cDMARDs) users, especially the methotrexate users, had a reduced dementia risk (Judge et al., 2017).
In summary, AD and RA are the most common neurodegenerative disease and a chronic autoimmune disease, respectively (Liu et al., 2013a). Until now, observational studies, genetic association studies, and Mendelian randomization studies have reported inconsistent association between AD and RA. Here, we conducted a Mendelian randomization study to investigate the causal association of AD with RA. We did not identify any significant association of AD with RA.
Our Mendelian randomization study may have several strengths. First, we selected large-scale AD GWAS dataset (74,046 individuals) and RA GWAS dataset (58,284 individuals) from the European descent. This could reduce the influence of the population stratification. Second, the instruments consisted of 20 independent AD genetic variants, which could reduce the influence on of linkage disequilibrium. Third, we selected three Mendelian randomization methods, as did in recent studies.
Author Contributions
QC and BL designed the study, collected the samples, and clinic information. QC, BL, ZX, LZ, and FL analyzed the data and wrote the paper.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81320108025 and 81472662).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CW and handling Editor declared their shared affiliation at the time of the review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2018.00627/full#supplementary-material
FIGURE S1 | Causal estimates from single genetic variant using different Mendelian randomization analysis methods.
References
Bae, S. C., and Lee, Y. H. (2018). Causal association between rheumatoid arthritis and a decreased risk of Alzheimer’s disease: a Mendelian randomization study. Z. Rheumatol. doi: 10.1007/s00393-018-0504-8 [Epub ahead of print].
Bao, X., Liu, G., Jiang, Y., Jiang, Q., Liao, M., Feng, R., et al. (2015). Cell adhesion molecule pathway genes are regulated by cis-regulatory SNPs and show significantly altered expression in Alzheimer’s disease brains. Neurobiol. Aging 36, 2904.e1–2904.e7. doi: 10.1016/j.neurobiolaging.2015.06.006
Chen, H., Wu, G., Jiang, Y., Feng, R., Liao, M., Zhang, L., et al. (2015). Analyzing 54,936 samples supports the association between CD2AP rs9349407 polymorphism and Alzheimer’s disease susceptibility. Mol. Neurobiol. 52, 1–7. doi: 10.1007/s12035-014-8834-2
Chou, R. C., Kane, M., Ghimire, S., Gautam, S., and Gui, J. (2016). Treatment for rheumatoid arthritis and risk of Alzheimer’s disease: a nested case-control analysis. CNS Drugs 30, 1111–1120. doi: 10.1007/s40263-016-0374-z
Ferraccioli, G., Carbonella, A., Gremese, E., and Alivernini, S. (2012). Rheumatoid arthritis and Alzheimer’s disease: genetic and epigenetic links in inflammatory regulation. Discov. Med. 14, 379–388.
Harold, D., Abraham, R., Hollingworth, P., Sims, R., Gerrish, A., Hamshere, M. L., et al. (2009). Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41, 1088–1093. doi: 10.1038/ng.440
Hollingworth, P., Harold, D., Sims, R., Gerrish, A., Lambert, J. C., Carrasquillo, M. M., et al. (2011). Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 43, 429–435. doi: 10.1038/ng.803
Hong, M. G., Alexeyenko, A., Lambert, J. C., Amouyel, P., and Prince, J. A. (2010). Genome-wide pathway analysis implicates intracellular transmembrane protein transport in Alzheimer disease. J. Hum. Genet. 55, 707–709. doi: 10.1038/jhg.2010.92
Hu, Y., Cheng, L., Zhang, Y., Bai, W., Zhou, W., Wang, T., et al. (2017). Rs4878104 contributes to Alzheimer’s disease risk and regulates DAPK1 gene expression. Neurol. Sci. 38, 1255–1262. doi: 10.1007/s10072-017-2959-9
Jiang, Q., Hu, Y., Jin, S., and Liu, G. (2018). Genetically increased serum calcium levels reduce Alzheimer’s disease risk. bioRxiv [Preprint]. doi: 10.1101/255059
Jiang, Q., Hu, Y., and Liu, G. (2016). Association of Alzheimer disease susceptibility variants and gene expression in the human brain. JAMA Neurol. 73:1255. doi: 10.1001/jamaneurol.2016.2796
Jiang, Q., Jin, S., Jiang, Y., Liao, M., Feng, R., Zhang, L., et al. (2017). Alzheimer’s disease variants with the genome-wide significance are significantly enriched in immune pathways and active in immune cells. Mol. Neurobiol. 54, 594–600. doi: 10.1007/s12035-015-9670-8
Jones, L., Holmans, P. A., Hamshere, M. L., Harold, D., Moskvina, V., Ivanov, D., et al. (2010). Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PLoS One 5:e13950. doi: 10.1371/journal.pone.0013950
Judge, A., Garriga, C., Arden, N. K., Lovestone, S., Prieto-Alhambra, D., Cooper, C., et al. (2017). Protective effect of antirheumatic drugs on dementia in rheumatoid arthritis patients. Alzheimers Dement. 3, 612–621. doi: 10.1016/j.trci.2017.10.002
Jun, G. R., Chung, J., Mez, J., Barber, R., Beecham, G. W., Bennett, D. A., et al. (2017). Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 13, 727–738. doi: 10.1016/j.jalz.2016.12.012
Kao, L. T., Kang, J. H., Lin, H. C., Huang, C. C., Lee, H. C., and Chung, S. D. (2016). Rheumatoid arthritis was negatively associated with Alzheimer’s disease: a population-based case-control study. PLoS One 11:e0168106. doi: 10.1371/journal.pone.0168106
Lambert, J. C., Grenier-Boley, B., Chouraki, V., Heath, S., Zelenika, D., Fievet, N., et al. (2010). Implication of the immune system in Alzheimer’s disease: evidence from genome-wide pathway analysis. J. Alzheimers Dis. 20, 1107–1118. doi: 10.3233/JAD-2010-100018
Lambert, J. C., Ibrahim-Verbaas, C. A., Harold, D., Naj, A. C., Sims, R., Bellenguez, C., et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458. doi: 10.1038/ng.2802
Li, X., Shen, N., Zhang, S., Liu, J., Jiang, Q., Liao, M., et al. (2015). CD33 rs3865444 polymorphism contributes to Alzheimer’s disease susceptibility in Chinese, European, and North American populations. Mol. Neurobiol. 52, 414–421. doi: 10.1007/s12035-014-8880-9
Li, Y., Song, D., Jiang, Y., Wang, J., Feng, R., Zhang, L., et al. (2016). CR1 rs3818361 polymorphism contributes to Alzheimer’s disease susceptibility in Chinese population. Mol. Neurobiol. 53, 4054–4059. doi: 10.1007/s12035-015-9343-7
Liu, G., Bao, X., Jiang, Y., Liao, M., Jiang, Q., Feng, R., et al. (2015). Identifying the association between Alzheimer’s disease and Parkinson’s disease using genome-wide association studies and protein-protein interaction network. Mol. Neurobiol. 52, 1629–1636. doi: 10.1007/s12035-014-8946-8
Liu, G., and Jiang, Q. (2016). Alzheimer’s disease CD33 rs3865444 variant does not contribute to cognitive performance. Proc. Natl. Acad. Sci. U.S.A. 113, E1589–E1590. doi: 10.1073/pnas.1600852113
Liu, G., Jiang, Y., Chen, X., Zhang, R., Ma, G., Feng, R., et al. (2013a). Measles contributes to rheumatoid arthritis: evidence from pathway and network analyses of genome-wide association studies. PLoS One 8:e75951. doi: 10.1371/journal.pone.0075951
Liu, G., Jiang, Y., Wang, P., Feng, R., Jiang, N., Chen, X., et al. (2012). Cell adhesion molecules contribute to Alzheimer’s disease: multiple pathway analyses of two genome-wide association studies. J. Neurochem. 120, 190–198. doi: 10.1111/j.1471-4159.2011.07547.x
Liu, G., Li, F., Zhang, S., Jiang, Y., Ma, G., Shang, H., et al. (2014a). Analyzing large-scale samples confirms the association between the ABCA7 rs3764650 polymorphism and Alzheimer’s disease susceptibility. Mol. Neurobiol. 50, 757–764. doi: 10.1007/s12035-014-8670-4
Liu, G., Wang, H., Liu, J., Li, J., Li, H., Ma, G., et al. (2014b). The CLU gene rs11136000 variant is significantly associated with Alzheimer’s disease in Caucasian and Asian populations. Neuromolecular Med. 16, 52–60. doi: 10.1007/s12017-013-8250-1
Liu, G., Xu, Y., Jiang, Y., Zhang, L., Feng, R., and Jiang, Q. (2017a). PICALM rs3851179 variant confers susceptibility to Alzheimer’s disease in Chinese population. Mol. Neurobiol. 54, 3131–3136. doi: 10.1007/s12035-016-9886-2
Liu, G., Yao, L., Liu, J., Jiang, Y., Ma, G., Chen, Z., et al. (2014c). Cardiovascular disease contributes to Alzheimer’s disease: evidence from large-scale genome-wide association studies. Neurobiol. Aging 35, 786–792. doi: 10.1016/j.neurobiolaging.2013.10.084
Liu, G., Zhang, F., Jiang, Y., Hu, Y., Gong, Z., Liu, S., et al. (2017b). Integrating genome-wide association studies and gene expression data highlights dysregulated multiple sclerosis risk pathways. Mult. Scler. 23, 205–212. doi: 10.1177/1352458516649038
Liu, G., Zhang, L., Feng, R., Liao, M., Jiang, Y., Chen, Z., et al. (2013b). Lack of association between PICALM rs3851179 polymorphism and Alzheimer’s disease in Chinese population and APOEepsilon4-negative subgroup. Neurobiol. Aging 34, 1310.e9–1310.e10. doi: 10.1016/j.neurobiolaging.2012.08.015
Liu, G., Zhang, S., Cai, Z., Li, Y., Cui, L., Ma, G., et al. (2013c). BIN1 gene rs744373 polymorphism contributes to Alzheimer’s disease in East Asian population. Neurosci. Lett. 544, 47–51. doi: 10.1016/j.neulet.2013.02.075
Liu, G., Zhang, S., Cai, Z., Ma, G., Zhang, L., Jiang, Y., et al. (2013d). PICALM gene rs3851179 polymorphism contributes to Alzheimer’s disease in an Asian population. Neuromolecular Med. 15, 384–388.
Liu, G., Zhang, Y., Wang, L., Xu, J., Chen, X., Bao, Y., et al. (2018). Alzheimer’s disease rs11767557 variant regulates EPHA1 gene expression specifically in human whole blood. J. Alzheimers Dis. 61, 1077–1088. doi: 10.3233/JAD-170468
Naj, A. C., Jun, G., Beecham, G. W., Wang, L. S., Vardarajan, B. N., Buros, J., et al. (2011). Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43, 436–441. doi: 10.1038/ng.801
Okada, Y., Wu, D., Trynka, G., Raj, T., Terao, C., Ikari, K., et al. (2014). Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381. doi: 10.1038/nature12873
Policicchio, S., Ahmad, A. N., Powell, J. F., and Proitsi, P. (2017). Rheumatoid arthritis and risk for Alzheimer’s disease: a systematic review and meta-analysis and a Mendelian Randomization study. Sci. Rep. 7:12861. doi: 10.1038/s41598-017-13168-8
Shen, N., Chen, B., Jiang, Y., Feng, R., Liao, M., Zhang, L., et al. (2015). An updated analysis with 85,939 samples confirms the association between CR1 rs6656401 polymorphism and Alzheimer’s disease. Mol. Neurobiol. 51, 1017–1023. doi: 10.1007/s12035-014-8761-2
Sims, R., Van Der Lee, S. J., Naj, A. C., Bellenguez, C., Badarinarayan, N., Jakobsdottir, J., et al. (2017). Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49, 1373–1384. doi: 10.1038/ng.3916
Ungprasert, P., Wijarnpreecha, K., and Thongprayoon, C. (2016). Rheumatoid arthritis and the risk of dementia: a systematic review and meta-analysis. Neurol. India 64, 56–61. doi: 10.4103/0028-3886.173623
Ward, L. D., and Kellis, M. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934. doi: 10.1093/nar/gkr917
Xiang, Z., Xu, M., Liao, M., Jiang, Y., Jiang, Q., Feng, R., et al. (2015). Integrating genome-wide association study and brain expression data highlights cell adhesion molecules and purine metabolism in Alzheimer’s disease. Mol. Neurobiol. 52, 514–521. doi: 10.1007/s12035-014-8884-5
Yavorska, O. O., and Burgess, S. (2017). MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 46, 1734–1739. doi: 10.1093/ije/dyx034
Yokoyama, J. S., Wang, Y., Schork, A. J., Thompson, W. K., Karch, C. M., Cruchaga, C., et al. (2016). Association between genetic traits for immune-mediated diseases and Alzheimer disease. JAMA Neurol. 73, 691–697. doi: 10.1001/jamaneurol.2016.0150
Zhang, S., Li, X., Ma, G., Jiang, Y., Liao, M., Feng, R., et al. (2016). CLU rs9331888 polymorphism contributes to Alzheimer’s disease susceptibility in Caucasian but not East Asian populations. Mol. Neurobiol. 53, 1446–1451. doi: 10.1007/s12035-015-9098-1
Keywords: Alzheimer’s disease, rheumatoid arthritis, genome-wide association study, Mendelian randomization, autoimmune disease
Citation: Cai Q, Xin Z, Zuo L, Li F and Liu B (2018) Alzheimer’s Disease and Rheumatoid Arthritis: A Mendelian Randomization Study. Front. Neurosci. 12:627. doi: 10.3389/fnins.2018.00627
Received: 12 July 2018; Accepted: 21 August 2018;
Published: 12 September 2018.
Edited by:
Yan Huang, Harvard Medical School, United StatesCopyright © 2018 Cai, Xin, Zuo, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Liu, YmluX2xpdTAxQDEyNi5jb20=
 Qixuan Cai1
Qixuan Cai1 Bin Liu
Bin Liu