
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neurosci. , 21 May 2012
Sec. Neurogenesis
Volume 6 - 2012 | https://doi.org/10.3389/fnins.2012.00071
This article is part of the Research Topic microRNAs in neural stem cells and neurogenesis View all 10 articles
MicroRNAs (miRNAs) are short non-coding RNAs that regulate gene expression at the post-transcriptional level by mediating mRNA degradation or translational inhibition. MiRNAs are implicated in many biological functions, including neurogenesis. It has been shown that miRNAs regulate multiple steps of neurogenesis, from neural stem cell proliferation to neuronal differentiation and maturation. MiRNAs execute their functions in a dynamic and context-dependent manner by targeting diverse downstream target genes, from transcriptional factors to epigenetic regulators. Identifying context-specific target genes is instrumental for understanding the roles that miRNAs play in neurogenesis. This review summarizes our current state of knowledge on the dynamic roles that miRNAs play in neural stem cells and neurogenesis.
The human genome is composed of ∼20,000–25,000 protein-coding genes. To adapt to evolutionary pressure, various mechanisms have been evolved to create more diversified protein expression by post-transcriptional regulation, such as alternative splicing, mRNA transport and local translation, and non-coding RNA-mediated mRNA degradation and translational repression. MicroRNAs (miRNAs) are 20- to 22-nucleotide RNA molecules belonging to the family of small non-coding RNAs. They regulate gene expression by binding to the 3′ untranslated region (3′ UTR) of mRNAs. The base-pairing of a miRNA with the 3′ UTR of its target mRNA can lead to either mRNA degradation or translational inhibition. In the human genome, more than 60% of the transcribed mRNAs are under the control of miRNA regulation (Friedman et al., 2009).
The miRNA genes are typically located either within a host gene or in the intergenic regions. They are transcribed by RNA polymerase II (Pol II) to pri-miRNAs, which are further processed by Drosha and DiGeorge syndrome critical region gene 8 (DGCR8) or partner of Drosha (Pasha) in the nucleus to generate pre-miRNAs. Following exportation into the cytoplasm, pre-miRNAs are cleaved by Dicer to form the mature 20- to 22-nucleotide miRNAs. The mature miRNA is then loaded into a ribonucleoprotein complex, called the RNA-induced silencing complex (RISC), which is composed of the human immunodeficiency virus transactivating response RNA-binding protein (TRBP), Argonaute 2 (Ago2), and Dicer, to bind to the target mRNA to modulate gene expression (Bartel, 2004; Kim, 2005; Bushati and Cohen, 2007; Figure 1).
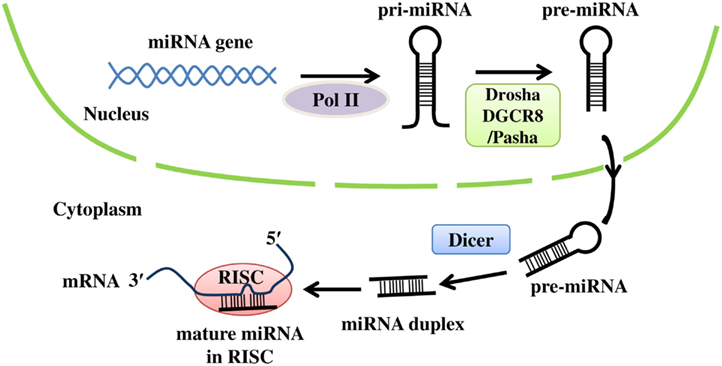
Figure 1. The biogenesis of miRNAs. Primary miRNA (pri-miRNA) is transcribed from the corresponding miRNA gene by RNA polymerase II (Pol II), and then processed by Drosha and DGCR8 (or Pasha) to form precursor miRNA (pre-miRNA). After pre-miRNA is exported into the cytoplasm, Dicer cleaves pre-miRNA to generate the mature miRNA duplex. Finally, the mature miRNA is loaded into RISC, where it binds to the 3′ UTR of target mRNA.
MicroRNAs have been found to participate in virtually all biological processes, including neurogenesis (Stefani and Slack, 2008; Liu and Zhao, 2009; Gao, 2010; Lau and Hudson, 2010; Li and Jin, 2010; Shi et al., 2010; Vreugdenhil and Berezikov, 2010). Neurogenesis is defined as the generation of mature and functional neurons from neural stem and progenitor cells, collectively referred to as NSCs in the following text. NSCs have the ability to maintain their undifferentiated state and to differentiate into neuronal and glial lineages (Gage, 2000). In the developing brain, NSCs are located in the ventricular zone of the lateral ventricles; in the adult brain, they are found in the subventricular zone (SVZ) of the lateral ventricles and subgranular zone (SGZ) of the hippocampal dentate gyrus. Neurogenesis is a multi-step process, including NSC proliferation, self-renewal, neuronal commitment, migration, maturation, and integration. MiRNAs are involved in multiple steps of neurogenesis process (Shi et al., 2008, 2010). This review will discuss recent progress on understanding the roles of miRNAs in cortical neurogenesis in embryonic and adult brains.
Many studies have used animals to model the effect of deficiency in the miRNA biogenesis pathway, which have revealed essential roles for miRNAs in neurogenesis. Ago2 is a RISC component that possesses endonuclease activity to cleave the miRNA target mRNA. Ago2-deficient mice exhibited defects in neural tube closure and mis-patterning of the forebrain (Liu et al., 2004). Dicer is another RISC component with RNase III enzymatic activity that cleaves the pre-miRNA to produce the mature miRNA. Loss of Dicer in zebrafish led to abnormal brain development and the phenotype could be rescued by a single miRNA, miR-430 (Giraldez et al., 2005). Conditional knockout of Dicer in mouse NSCs resulted in smaller cortex in the knockout mice (De Pietri Tonelli et al., 2008). Similar studies using the same knockout mice found that knockout of Dicer interfered NSC expansion and differentiation (Andersson et al., 2010; Kawase-Koga et al., 2010). Loss of Dicer expression in neurons also resulted in neuronal maturation defects, as revealed by reduced dendritic arborization and axonal path finding (Davis et al., 2008). In addition, microcephaly was observed in Dicer knockout mice as a result of apoptosis. Deletion of Dicer in dopaminergic neurons resulted in progressive loss of midbrain dopaminergic neurons and Parkinson-like symptoms in mice (Kim et al., 2007). In this study, miR-133b was found to specifically regulate dopaminergic neuronal maturation by targeting the transcription factor Pitx3. Another study deleted Dicer in dopaminoceptive neurons (Cuellar et al., 2008). These mice displayed many neurological disorders, including ataxia and clasping limbs, presumably due to reduced striatal medium spiny neurons. Together, these studies demonstrated important roles of the miRNA biogenesis pathway in neural development.
The brain enriched miRNA miR-9 has been extensively studied in neurogenesis. It is expressed in the brain as early as E10.5 and its expression is increased during embryonic brain development and decreased postnatally (Krichevsky et al., 2003, 2006; Smirnova et al., 2005; De Pietri Tonelli et al., 2008). At the onset of neurogenesis, miR-9 was found throughout the cortical wall with a higher expression in the ventricular zone (De Pietri Tonelli et al., 2008; Shibata et al., 2008). We showed that there is an inverse correlation between the expression of miR-9 and nuclear receptor TLX (or NR2E1) during NSC differentiation (Zhao et al., 2009). TLX has been shown to play an essential role in maintaining NSCs in the proliferative and self-renewable state (Shi et al., 2004; Liu et al., 2008; Zhang et al., 2008; Qu et al., 2010). miR-9 inhibits TLX expression by base-pairing to the 3′ UTR of TLX mRNA (Zhao et al., 2009). Overexpression of miR-9 dramatically decreased NSC proliferation and promoted both neuronal and astroglial differentiation. Conversely, miR-9 knockdown resulted in increased proliferation of NSCs. In utero electroporation of miR-9 led to precocious neuronal differentiation and outward cell migration (Zhao et al., 2009). The effect of miR-9 overexpression could be rescued by expression of a TLX construct lacking its 3′ UTR.
Consistent with the role of miR-9 in promoting neural differentiation in NSCs (Zhao et al., 2009), miR-9 also promotes neural differentiation of mouse embryonic stem cells by targeting sirtuin 1 (SIRT1; Saunders et al., 2010). The pro-differentiation role of miR-9 was further supported by the observation that miR-9 stimulates Cajal–Retzius cell differentiation by targeting Forkhead box protein G1 (Foxg1) expression (Shibata et al., 2008). Cajal–Retzius cells are reelin-producing neurons located in the embryonic marginal zone to set up the lamination of the cortical plate by limiting radial migration of newborn neurons (Meyer, 2010). Foxg1 is a transcription factor involved in telencephalic development and has been associated with Rett syndrome (Hebert and Fishell, 2008; Jacob et al., 2009). MiR-9 was shown to repress Foxg1 expression. Up-regulation of miR-9 significantly increased Cajal–Retzius cell differentiation, whereas down-regulation of miR-9 expression inhibited Cajal–Retzius differentiation.
Besides mammals, miR-9 also promotes neurogenesis in other organisms. In developing zebrafish, miR-9 could stimulate neurogenesis by down-regulating hairy-related 5 (her5), hairy-related 9 (her9), canopy1, fibroblast growth factor 8 (fgf8), and fibroblast growth factor receptor 1 (fgfr1) through their 3′ UTRs (Leucht et al., 2008). In Xenopus brains, miR-9 inhibited neural progenitor cell proliferation in the hindbrain by targeting Hairy 1, a member of the Hes (hairy and enhancer of split) family genes. Hairy 1 mediates this function of miR-9 by regulating the Wnt signaling (Bonev et al., 2011).
In addition to the main theme that miR-9 inhibits NSC proliferation and promotes neural differentiation, Delaloy et al. (2010) recently showed that miR-9 was expressed early on at the neurosphere stage upon neural induction of human embryonic stem cells. At this early stage, miR-9 participates in neural progenitor cell expansion by promoting progenitor proliferation. More recently, miR-9-2/3 knockout mice were generated by mutating two of the miR-9 genomic loci (miR-9-2 and miR-9-3; Shibata et al., 2011). In these mice, miR-9 was shown to regulate both neural progenitor proliferation and differentiation by regulating multiple downstream targets, including Foxg1, TLX, GS homeobox 2 (Gsh2), and Meis homeobox 2 (Meis2) in a cellular context-specific manner. Moreover, miR-9 suppressed neural progenitor apoptosis in the Xenopus forebrain by targeting Hairy 1. In this cellular context, Hairy 1 dictated miR-9 anti-apoptotic function by regulating the p53 pathway (Bonev et al., 2011).
Several mechanisms have been proposed to regulate miR-9 expression in NSCs and neurogenesis. We have demonstrated that TLX represses miR-9 expression by binding to the miR-9 genomic locus, in addition to inhibition of TLX expression by miR-9. TLX represses the transcription of miR-9 primary precursors, miR-9-1, and miR-9-2. Thus, miR-9 inhibits NSC proliferation and promotes neural differentiation by forming a feedback regulatory loop with TLX. In a recent study by Obernier et al. (2011) an inverse correlation was also observed between TLX and miR-9 expression in NSCs and transit amplifying progenitors (TAPs), supporting the idea that miR-9 targets TLX expression (Zhao et al., 2009). However, this study failed to reveal altered expression of miR-9 in TLX−/− neonatal subependymal zone of the lateral ventricles (Obernier et al., 2011), in contrast to the previous observation that miR-9 expression is increased in TLX−/− adult mouse brains (Zhao et al., 2009). This discrepancy may be explained by the differential expression levels of TLX in neonatal and adult brains. It has been shown that the expression of TLX peaks at E13.5 in embryonic brains, decreases to barely detectable levels at birth and increases again after birth with high levels of expression in adult brains (Monaghan et al., 1995; Li et al., 2008). The low expression of TLX in neonatal brains may explain the lack of change in miR-9 expression in TLX−/− neonatal subependymal zone.
Using a neuronal differentiation model in neuroblastoma cells, REST (RE1-silencing transcription factor), and cAMP response element-binding (CREB) were shown to coordinately regulate miR-9 expression by binding to one of the miR-9 genomic loci, miR-9-2 (Laneve et al., 2010). REST suppressed miR-9 expression by occupying the miR-9-2 promoter at the proliferative state and its removal during differentiation allowed CREB mediated activation of miR-9 expression. On the other hand, miR-9 targets REST expression through base-pairing to the 3′ UTR of REST (Packer et al., 2008), thus forming a feedback regulatory loop between miR-9 and REST.
Another mechanism for how miR-9 expression is regulated involves post-transcriptional regulation by fragile X mental retardation gene 1 (FXR1P; Xu et al., 2011). FXR1P is one of the three fragile X mental retardation proteins and its knockout leads to early death, while fragile X mental retardation protein (FMRP), or fragile X mental retardation gene 2 (FXR2P) knockout mice are viable with various degrees of defects in brain development (The Dutch-Belgian Fragile X Consortium et al., 1994; Gu et al., 2002; Mientjes et al., 2004). One key feature revealed by this study is that expression of FXR1P can increase the level of mature miR-9 by potentiating the processing of pre-miR-9 to mature miR-9. In contrast, knockdown of FXR1P dramatically decreased mature miR-9 level, but increased pre-miR-9 level. It is proposed that FXR1P regulates miRNA processing by direct binding to Dicer and modulating Dicer activity.
In summary, the regulation of miR-9 expression and function is a dynamic process. The function of miR-9 is cellular context-dependent, tightly coupled with the temporal and spatial expression patterns of both its downstream target genes and its upstream regulators (summarized in Figure 2). While miR-9 may facilitate progenitor expansion early on, its main role at later stages seems to promote neural differentiation both during the transition from embryonic stem cells to neural lineages and during fate determination from NSCs to differentiated neurons and glia. The regulatory loop between miR-9 and transcription factors, such as TLX, REST, and CREB, provides an efficient control for miR-9-mediated NSC fate determination and promotion of neuronal differentiation. The interaction of miRNAs and transcription factors in a feedback circuit may represent a general paradigm for neurogenesis.
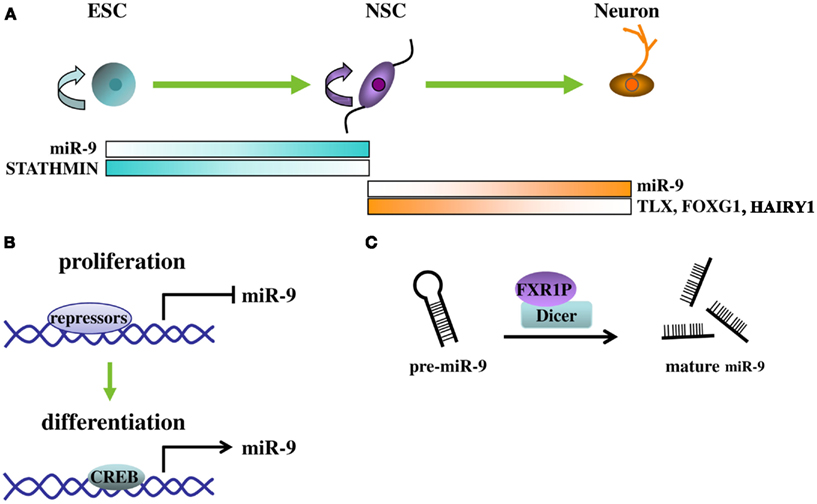
Figure 2. Regulation and functions of miR-9. (A) MiR-9 participates at different stages of neurogenesis from embryonic stem cells (ESC) to NSC and to neurons. At each stage, depending on the cellular context, different target genes are regulated by miR-9 and their expression levels are inversely correlated to that of miR-9. (B) Expression of mR-9 is controlled by transcriptional repressors and activators at the transcription level. During proliferative, transcriptional repressors, such as REST and TLX, bind to the promoter of miR-9 locus to repress its expression. During differentiating, transcriptional repressors dissociate from the miR-9 locus and leaving activators, such as CREB, to increase miR-9 expression. (C) At the miRNA processing level, binding of FXR1P with Dicer greatly increased mature miR-9 levels by presumably facilitating Dicer-mediated pre-miR-9 processing.
MiR-137 has been shown to inhibit proliferation and promote differentiation of NSCs derived from the SVZ of adult mouse brains (Silber et al., 2008). This observation is supported by a recent study that demonstrated a pro-differentiation role of miR-137 using NSCs from the ventricular zone of embryonic mouse brains (Sun et al., 2011). In the study by Sun et al., nuclear receptor TLX brings its transcriptional co-repressor LSD1, the lysine-specific histone demethylase, to the miR-137 promoter to suppress miR-137 expression. MiR-137 in turn inhibits the expression of LSD1 by base-pairing to the 3′ UTR of LSD1 mRNA. Increased expression of miR-137 led to reduced NSC proliferation and increased neural differentiation. The regulatory loop of TLX-miR-137-LSD1 ensures the coordinated expression of TLX/LSD1 and miR-137 during the transition of NSC proliferation and differentiation and provides a mechanism to control the dynamics between NSC proliferation and differentiation (Sun et al., 2011).
In another study, the methyl CpG binding protein 2 (MeCP2) cooperated with SRY-box containing gene 2 (Sox2) to bind to the promoter of miR-137 to repress the expression of miR-137 (Szulwach et al., 2010). Altered miR-137 expression in turn regulated adult hippocampal neurogenesis by targeting the enhancer of zeste homolog 2 (Ezh2), a histone methyltransferase that is important in maintaining the bivalent chromatin state of stem cells (Boyer et al., 2006). Thus, epigenetic regulation of miR-137 leads to chromatin state change through Ezh2, which in turn controls NSC proliferation and differentiation. In addition, miR-137 has been shown to reduce neuronal maturation by targeting an ubiquitin ligase, mindbomb homolog 1 (Mib1; Smrt et al., 2010). The diverse functions of miR-137 are reminiscent of the roles that miR-9 plays at different stages of neurogenesis.
MiR-184 was found to promote NSC proliferation but inhibit neuronal differentiation (Liu et al., 2010). Methyl CpG binding protein 1 (MBD1) regulates miR-184 expression in NSCs. Increased miR-184 expression was found in MBD1-deficient NSCs. Chromatin immunoprecipitation analysis revealed that MBD1 could directly bind to the miR-184 genomic loci. MBD1 functions by binding to the methylated DNA and resulting in the formation of silent chromatin, thus repressing gene expression (Wade, 2001; Bogdanovic and Veenstra, 2009). Numblike (Numbl) was identified as a target of miR-184 in this study (Liu et al., 2010). Overexpression of Numbl rescued the phenotypes caused by overexpression of miR-184 or knockout of MBD1 and resulted in a switch from NSC proliferation to neuronal differentiation, suggesting that Numbl is a functional target of miR-184 in adult hippocampal NSCs.
The lethal-7 (let-7) gene is one of the first two miRNAs discovered in Caenorhabditis elegans (C. elegans) and is highly conserved (Pasquinelli et al., 2000; Reinhart et al., 2000). Several mature miRNA sequences that differ only in a few nucleotides have been identified in mammals and are collectively referred to as let-7 family of miRNAs (Roush and Slack, 2008). Among them, let-7a, b, c, and e were shown to be expressed in the brain and up-regulated upon neuronal differentiation of mouse and human embryonic carcinoma cells (Sempere et al., 2004; Wulczyn et al., 2007).
Though expressed in various tissues, let-7 family members have been found to play significant roles in neurogenesis. The first such evidence is the interplay between let-7a and Lin-28 (Rybak et al., 2008), a pluripotent factor that is expressed in embryonic stem cells (Yu et al., 2007). When embryonic stem cells commit to the neural lineage, Lin-28 suppresses pre-let-7a expression through specific binding to pre-let-7a. Down-regulation of Lin-28 expression releases its repression of let-7a expression, allowing let-7a to induce neural differentiation. A recent study demonstrated that Musashi1 potentiates the inhibitory effect of Lin-28 on let-7 family miRNA production during early neural differentiation (Kawahara et al., 2011). However, another study argued that Lin-28 promoted neurogenesis independently of its role in regulating let-7 family miRNAs (Balzer et al., 2010). Lin-28 and let-7 family miRNAs may function during neural differentiation through both independent and concomitant actions.
Mechanisms of let-7 in controlling NSC differentiation were uncovered in several recent studies (Nishino et al., 2008; Schwamborn et al., 2009; Zhao et al., 2010). High mobility group AT-hook 2 (Hmga2), a member of the high mobility group A, exhibited high expression in NSCs, but declined its expression with age. In contrast, the expression of let-7 is more enriched in differentiated cells and let-7b was found to be highly up-regulated with age, among the let-7 family members. Overexpression of let-7b led to reduced self-renewal of NSCs and this reduction can be rescued by transduction of an Hmga2 expressing vector lacking its 3′ UTR. Let-7b has also been shown to promote neurogenesis by targeting the 3′ UTR of TLX and the cell cycle regulator, cyclin D1 (Zhao et al., 2010). Overexpression of let-7b in NSCs decreased proliferation and promoted both neuronal and astroglial differentiation. In vivo cell cycle analysis demonstrated that let-7b promoted cell cycle exit, thus paving the way for neural differentiation. Recently, it was shown that expression of let-7a can be activated by tripartite motif containing 32 (TRIM32), a homolog of Drosophila Brat/Mei-P26, that inhibits NSC proliferation (Schwamborn et al., 2009). TRIM32 inhibits NSC proliferation by mediating c-Myc degradation and promotes neuronal differentiation by inducing let-7a expression. Multiple let-7 miRNAs may show similar functions in promoting differentiation (Roush and Slack, 2008). However, the small differences in their seed regions may discriminate let-7 family members for their specific target genes, thus leading to distinctive biological consequences.
MiR-124 is also specifically expressed in the central nervous system (Lagos-Quintana et al., 2002; Deo et al., 2006). It was mostly expressed in post-mitotic neurons, although also found in neural precursors located in the ventricular zone, with higher expression when cells were more mature and farther away from the lateral ventricles (Maiorano and Mallamaci, 2009). MiR-124 expression increases during brain development (Krichevsky et al., 2003, 2006; Sempere et al., 2004; Smirnova et al., 2005). In mammalian neurons, miR-124 was found predominantly in the cellular fractions containing messenger ribonucleoprotein (mRNP) complex and polyribosome (Kim et al., 2004), where active translation takes place, supporting its involvement in regulating neuronal protein translation. Ectopic expression of miR-124 in HeLa cells was shown to suppress non-neuronal genes, while neuronal genes were largely stimulated, suggesting that miR-124 has a pro-neuronal role (Lim et al., 2005).
At least five mechanisms have been identified for miR-124-mediated promotion of neuronal differentiation (Figure 3A). The first one involves the transcriptional repressor REST/Small CTD Phosphatases 1 (SCP1) pathway (Visvanathan et al., 2007). SCP1 is a phosphatase that is restricted to non-neuronal tissues and recruited by REST to repress neuronal gene expression (Yeo et al., 2005). In chick embryos, SCP1 was highly expressed around the central canal of the spinal cord, but weakly expressed in the lateral neuronal zone (Visvanathan et al., 2007). Either overexpression of miR-124 or inactivation of SCP1 activity induced more neuronal differentiation. SCP1 is a direct target of miR-124, allowing binding of miR-124 to its 3′ UTR. The phenotype caused by miR-124 could be reversed by a SCP1 construct lacking its 3′ UTR. On the other hand, REST can repress miR-124 expression by binding to the repressor elements (RE1) at the miR-124 genomic loci (Conaco et al., 2006). In an independent study, miR-124 expression exerted no effect on neuronal differentiation in an earlier time window (Cao et al., 2007). Instead, miR-124 interrupted the basal lamina, a layer of extracellular matrix that borders the neural tubes, by targeting the basal lamina components, laminin gamma 1 (LAMC1) and integrin beta 1 (ITGB1; Cao et al., 2007).
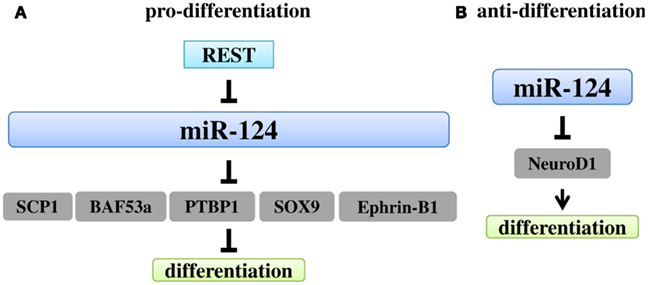
Figure 3. Regulation and functions of miR-124. (A). The major role of miR-124 in neurogenesis is to promote differentiation. REST can repress miR-124 expression. Through five mechanisms, miR-124 promotes differentiation, targeting SCP1, BAF53a, SOX9, PTBP1, and Ephrin-B1, respectively. (B) MiR-124 can also inhibit NSC differentiation by targeting NeuroD1, a pro-neuronal gene.
The second mechanism for miR-124-mediated neuronal differentiation is the repression of BAF complex 53 kDa subunit (BAF53a) by miR-124 to assist in the BAF53 subunit switch in ATP-dependent chromatin-remodeling (Yoo et al., 2009). During neuronal differentiation, neural progenitor specific BAF subunits (BAF53a and BAF45a) were replaced by the neuron-specific BAFs (BAF53b and BAF45b). MiR-124 binds to the 3′ UTR of BAF53a to reduce its expression, thus facilitating the replacement of BAF53 subunits. Overexpression of REST reactivated BAF53a expression in post-mitotic neurons, presumably by down-regulation of miR-124 expression (Yoo et al., 2009).
The third mechanism of miR-124 action was demonstrated in adult neurogenesis (Cheng et al., 2009). MiR-124 was shown to be up-regulated when transit amplifying cells became neuroblasts and maintained high expression in differentiated olfactory bulb neurons. Knockdown of miR-124 retained the proliferation of transit amplifying cells but could not recover the proliferative neuroblasts, suggesting that miR-124 plays an important role in the transition from proliferating to differentiating stage. Target identification revealed SRY-box containing gene 9 (Sox9) as a physiologically relevant target of miR-124 in neuronal differentiation. Sox9 was expressed in ependymal cells, SVZ astrocytes, transit amplifying cells, but not in neuroblasts. It has been shown to be an essential determinant in gliogenesis (Stolt et al., 2003). Knockdown of miR-124 expression increased Sox9 expression in neuroblasts. Overexpression of a 3′ UTR-deficient Sox9 construct prevented NSCs from neuronal differentiation. Together, these results suggest that miR-124 promotes neuronal fate determination by suppressing Sox9 expression.
The evidence for the fourth mechanism of miR-124 action comes from the observation that miR-124 regulates alternative splicing (Makeyev et al., 2007), which puts another layer of complexity in post-transcriptional control of gene expression. Polypyrimidine tract binding protein 1 (PTBP1) is a repressor of neuronal-specific splicing (Black, 2003). Down-regulation of PTBP1 by miR-124 caused up-regulation of polypyrimidine tract binding protein 2 (PTBP2), which in turn favored neuronal-specific splicing and neuronal differentiation (Makeyev et al., 2007).
The fifth mechanism concerning miR-124-mediated neuronal differentiation was proposed to involve the negative feedback regulation of ephrin-B1 and miR-124 (Arvanitis et al., 2010). Ephrin-B1 mRNA was found to be very unstable and deletion of its 3′ UTR strongly increased its stability. This observation prompted the authors to demonstrate that miR-124 controlled ephrin-B1 mRNA stability through its 3′ UTR. Meanwhile, activation of the ephrin-B1 signaling pathway can down-regulate miR-124 expression. MiR-124 induced neuronal differentiation and neurite growth is dependent on reduced level of ephrin-B1. Increased expression of miR-124 and neuronal differentiation was observed in ephrin-B1 knockout mice.
Recently miR-124 was found to regulate early neurogenesis in the optic vesicle and Xenopus forebrain by targeting the basic helix-loop-helix transcription factor, neurogenic differentiation 1 (NeuroD1; Liu et al., 2011). Both overexpression of miR-124 and down-regulation of NeuroD1 expression at an early developmental stage resulted in enhanced proliferation and decreased neurogenesis (Figure 3B). The effect on proliferation is likely a consequence of miR-124-mediated inhibition of differentiation by suppressing NeuroD1 expression, rather than a direct impact on fundamental aspects of stem cell proliferation.
In addition to its role in neuronal differentiation, miR-124 also regulates neuronal maturation. MiR-124 was shown to inhibit the formation of long-term facilitation (LTF) through down-regulating CREB expression (Rajasethupathy et al., 2009). It is also essential for the maturation and survival of hippocampal dentate gyrus neurons by targeting LIM/homeobox protein 2 (Lhx2; Sanuki et al., 2011).
With the development of induced lineage reprogramming technology, miR-124 was combined with two other transcription factors, myelin transcription factor 1-like (MYT1L) and POU class 3 homeobox 2 (POU3F2 or BRN2), to reprogram terminally differentiated dermal fibroblast to functional neurons (Ambasudhan et al., 2011). Similarly, functional neurons were generated by introducing miR-9*, miR-124, neurogenic differentiation 2 (NeuroD2), achaete–scute complex homolog 1 (ASCL1), and MYT1L in human fibroblast (Yoo et al., 2011). These studies further highlight the importance of miR-124 in neuronal differentiation.
MiR-125b was found to be up-regulated during neuronal differentiation of two human cell lines, neuroblastoma cell line SH-SY5Y and neural progenitor ReNcell VM cells (Le et al., 2009), suggesting that miR-125b may play a role in neuronal differentiation. Indeed, several validated target genes of miR-125b were found to repress neuronal gene expression, providing a mechanism for miR-125b-mediated promotion of neuronal differentiation.
MiR-125b has also been shown to play a role in neuronal maturation. Overexpression of miR-125b led to longer and thinner dendritic spines (Edbauer et al., 2010). The effect of miR-125b on neuronal maturation is dependent on the expression of FMRP. MiR-125b was found to be associated with FMRP and FMRP participates in miRNA processing pathways. Both miR-125b and FMRP are involved in the regulation of the N-Methyl-D-aspartic acid (NMDA) receptor subunit NR2A. The regulation of dendritic morphology by miR-125b and its interacting partner protein may serve as a general mechanism to regulate complex biological processes.
MiR-128 is one of the miRNAs that showed increased expression during brain development (Krichevsky et al., 2003; Smirnova et al., 2005). miR-128 is mainly expressed in mature neurons but not in astrocytes (Bruno et al., 2011). Overexpression of miR-128 could promote neuronal differentiation and increase dendritic length by regulating nonsense-mediated decay (NMD) activity. NMD degrades abnormally premature transcripts, characterized by stop codons at particular position in mRNA (Chan et al., 2007; Bhuvanagiri et al., 2010). Two players in NMD pathway, regulator of nonsense transcripts 1 (UPF1) and cancer susceptibility candidate 3 (CASC3 or MLN51), are the functional targets of miR-128. UPF1, a RNA helicase, is essential for remodeling the mRNA surveillance complex. MLN51, a core component of the exon-junction complex, promotes NMD activity. Knockdown of UPF1 and MLN51 by miR-128 dramatically inhibited NMD activity. Addition of UPF1 and MLN51 rescued miR-128-reduced NMD activity. These results demonstrated the importance of miR-128 in neuronal maturation.
A critical step for newborn neurons to integrate into the existing neural network is the development of dendritic synapses. MiR-132 was shown to enhance cortical dendritic development by down-regulating Rho GTPase activating protein 32 (ARHGAP32 or p250GAP; Vo et al., 2005; Wayman et al., 2008; Luikart et al., 2011). Overexpression of miR-132 in transgenic mice led to increased dendritic spine intensity and these mice displayed impaired novel object recognition memory (Hansen et al., 2010), whereas conditional knockout of miR-132 in hippocampal dentate gyrus led to reduced dendritic length, branching, and dendritic spine density of newborn dentate gyrus neurons (Magill et al., 2010). MiR-132 has also been shown to regulate dendritic spine width (Edbauer et al., 2010). MeCP2 has been identified to be an endogenous target of miR-132 in hippocampal neurons (Klein et al., 2007). Noticeably, Abnormal MeCP2 expression has been associated with Rett syndrome, an X-linked neurodevelopmental disorder (Guy et al., 2011; Samaco and Neul, 2011). Thus, dysregulation of miR-132 expression may contribute to Rett syndrome.
MiR-134 was identified as a brain-specific miRNA and is localized to neuronal dendrites (Schratt et al., 2006). It could decrease dendritic spine width of hippocampal neurons. LIM domain kinase 1 (Limk1) was identified to be a target of miR-134. Brain-derived neurotrophic factor (BDNF) could down-regulate miR-134 expression, thus increasing Limk1 expression to maintain normal dendritic spine volume. In addition to its role in dendritic spine morphogenesis, miR-134 also regulates neural progenitor cell proliferation and attenuate neuronal migration by targeting doublecortin (DCX) and chordin-like 1 (Chrd-1; Gaughwin et al., 2011).
Dendritic spine morphogenesis was also shown to be influenced by miR-138 through the depalmitoylation enzyme, acyl protein thioesterase 1 (APT1; Siegel et al., 2009). A functional screening in synaptosomes discovered that miR-138 was expressed in the synaptodendritic compartment. MiR-138 decreased dendritic spine volume in a dose-dependent manner by targeting APT1.
MicroRNAs play important roles at various stages of neurogenesis and exert different functions specific to the cell type throughout development. Therefore, when investigating miRNA functions, the expression of both miRNA and its target(s) must both be considered in a context-dependent manner (Table 1). Feedback regulatory circuit is often found in miRNA function and regulation (Figure 4). The regulatory loop can be interplay between miRNAs and transcription factors, epigenetic regulators, or other signaling molecules.
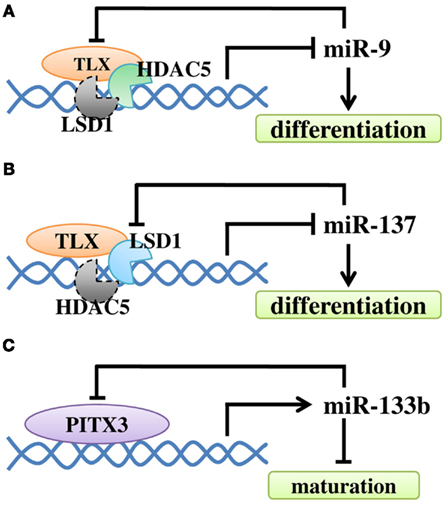
Figure 4. The regulatory loop between miRNAs and their regulators in NSC differentiation and neuronal maturation. (A) Negative feedback regulatory loop of miR-9 and TLX. TLX recruits HDAC5 to the miR-9 locus to inhibit miR-9 expression. MiR-9 can repress the level of TLX through its 3′ UTR. At the same time, LSD1 may be recruited. (B) TLX recruits LSD1 to the miR-137 locus to inhibit miR-137 expression. The expression of miR-137 can repress the level of LSD1. Similarly, HDAC5 may be recruited. (C) Transcription factor Pitx3 promotes miR-133b expression, while miR-133b can down-regulate Pitx3 level.
In the near future, more new miRNAs will be identified in neurogenesis due to the development of deep sequencing. One recent example is the discovery of miR-3099 and its role in neural differentiation (Ling et al., 2011). Thus, not only more new functions will be added to the existing miRNAs, but also new miRNAs will emerge to play important roles in neurogenesis in the future.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Grace Asuelime for comments. This work was supported by NIH NINDS R01 NS059546, RC1 NS068370, and California Institute for Regenerative Medicine TR2-01832.
Ambasudhan, R., Talantova, M., Coleman, R., Yuan, X., Zhu, S., Lipton, S. A., and Ding, S. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9, 113–118.
Andersson, T., Rahman, S., Sansom, S. N., Alsio, J. M., Kaneda, M., Smith, J., O’Carroll, D., Tarakhovsky, A., and Livesey, F. J. (2010). Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS ONE 5, e13453. doi:10.1371/journal.pone.0013453
Arvanitis, D. N., Jungas, T., Behar, A., and Davy, A. (2010). Ephrin-B1 reverse signaling controls a posttranscriptional feedback mechanism via miR-124. Mol. Cell. Biol. 30, 2508–2517.
Balzer, E., Heine, C., Jiang, Q., Lee, V. M., and Moss, E. G. (2010). LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development 137, 891–900.
Bhuvanagiri, M., Schlitter, A. M., Hentze, M. W., and Kulozik, A. E. (2010). NMD: RNA biology meets human genetic medicine. Biochem. J. 430, 365–377.
Black, D. L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336.
Bogdanovic, O., and Veenstra, G. (2009). DNA methylation and -CpG binding proteins: developmental requirements and function. Chromosoma 118, 549–565.
Bonev, B., Pisco, A., and Papalopulu, N. (2011). MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 20, 19–32.
Boyer, L. A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L. A., Lee, T. I., Levine, S. S., Wernig, M., Tajonar, A., Ray, M. K., Bell, G. W., Otte, A. P., Vidal, M., Gifford, D. K., Young, R. A., and Jaenisch, R. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353.
Bruno, I. G., Karam, R., Huang, L., Bhardwaj, A., Lou, C. H., Shum, E. Y., Song, H.-W., Corbett, M. A., Gifford, W. D., Gecz, J., Pfaff, S. L., and Wilkinson, M. f. (2011). Identification of a MicroRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 42, 500–510.
Cao, X., Pfaff, S. L., and Gage, F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536.
Chan, W.-K., Huang, L., Gudikote, J. P., Chang, Y.-F., Imam, J. S., Maclean, J. A., and Wilkinson, M. F. (2007). An alternative branch of the nonsense-mediated decay pathway. EMBO J. 26, 1820–1830.
Cheng, L.-C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408.
Conaco, C., Otto, S., Han, J. J., and Mandel, G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U.S.A. 103, 2422–2427.
Cuellar, T. L., Davis, T. H., Nelson, P. T., Loeb, G. B., Harfe, B. D., Ullian, E., and Mcmanus, M. T. (2008). Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 105, 5614–5619.
Davis, T. H., Cuellar, T. L., Koch, S. M., Barker, A. J., Harfe, B. D., Mcmanus, M. T., and Ullian, E. M. (2008). Conditional loss of dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28, 4322–4330.
De Pietri Tonelli, D., Pulvers, J. N., Haffner, C., Murchison, E. P., Hannon, G. J., and Huttner, W. B. (2008). miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135, 3911–3921.
Delaloy, C., Liu, L., Lee, J. A., Su, H., Shen, F., Yang, G. Y., Young, W. L., Ivey, K. N., and Gao, F. B. (2010). MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335.
Deo, M., Yu, J.-Y., Chung, K.-H., Tippens, M., and Turner, D. L. (2006). Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 235, 2538–2548.
Edbauer, D., Neilson, J. R., Foster, K. A., Wang, C.-F., Seeburg, D. P., Batterton, M. N., Tada, T., Dolan, B. M., Sharp, P. A., and Sheng, M. (2010). Regulation of synaptic structure and function by FMRP-associated MicroRNAs miR-125b and miR-132. Neuron 65, 373–384.
The Dutch-Belgian Fragile X ConsortiumBakker, C. E., Verheij, C., Willemsen, R., Van Der Helm, R., Oerlemans, F., Vermey, M., Bygrave, A., Hoogeveen, A., Oostra, B. A., Reyniers, E., De Boule, K., D’Hooge, R., Cras, P., Van Velzen, D., Nagels, G., Martin, J.-J., De Deyn, P. P., Darby, J. K., and Willems, P. J. (1994). Fmr1 knockout mice: a model to study fragile X mental retardation. Cell 78, 23–33.
Friedman, R. C., Farh, K. K.-H., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105.
Gao, F.-B. (2010). Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 5, 25.
Gaughwin, P., Ciesla, M., Yang, H., Lim, B., and Brundin, P. (2011). Stage-specific modulation of cortical neuronal development by mmu-miR-134. Cereb. Cortex 21, 1857–1869.
Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., Hammond, S. M., Bartel, D. P., and Schier, A. F. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838.
Gu, Y., Mcilwain, K. L., Weeber, E. J., Yamagata, T., Xu, B., Antalffy, B. A., Reyes, C., Yuva-Paylor, L., Armstrong, D., Zoghbi, H., Sweatt, J. D., Paylor, R., and Nelson, D. L. (2002). Impaired conditioned fear and enhanced long-term potentiation inFmr2 knock-out mice. J. Neurosci. 22, 2753–2763.
Guy, J., Cheval, H., Selfridge, J., and Bird, A. (2011). The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 27, null.
Hansen, K. F., Sakamoto, K., Wayman, G. A., Impey, S., and Obrietan, K. (2010). Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 5, e15497. doi:10.1371/journal.pone.0015497
Hebert, J. M., and Fishell, G. (2008). The genetics of early telencephalon patterning: some assembly required. Nat. Rev. Neurosci. 9, 678–685.
Jacob, F. D., Ramaswamy, V., Andersen, J., and Bolduc, F. V. (2009). Atypical Rett syndrome with selective FOXG1 deletion detected by comparative genomic hybridization: case report and review of literature. Eur. J. Hum. Genet. 17, 1577–1581.
Kawahara, H., Okada, Y., Imai, T., Iwanami, A., Mischel, P. S., and Okano, H. (2011). Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated Let-7 family MicroRNA biogenesis in early neural differentiation. J. Biol. Chem. 286, 16121–16130.
Kawase-Koga, Y., Low, R., Otaegi, G., Pollock, A., Deng, H., Eisenhaber, F., Maurer-Stroh, S., and Sun, T. (2010). RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J. Cell. Sci. 123, 586–594.
Kim, J., Inoue, K., Ishii, J., Vanti, W. B., Voronov, S. V., Murchison, E., Hannon, G., and Abeliovich, A. (2007). A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317, 1220–1224.
Kim, J., Krichevsky, A., Grad, Y., Hayes, G. D., Kosik, K. S., Church, G. M., and Ruvkun, G. (2004). Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 101, 360–365.
Kim, V. N. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385.
Klein, M. E., Lioy, D. T., Ma, L., Impey, S., Mandel, G., and Goodman, R. H. (2007). Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 10, 1513–1514.
Krichevsky, A. M., King, K. S., Donahue, C. P., Khrapko, K., and Kosik, K. S. (2003). A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9, 1274–1281.
Krichevsky, A. M., Sonntag, K. C., Isacson, O., and Kosik, K. S. (2006). Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 24, 857–864.
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739.
Laneve, P., Gioia, U., Andriotto, A., Moretti, F., Bozzoni, I., and Caffarelli, E. (2010). A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 38, 6895–6905.
Lau, P., and Hudson, L. D. (2010). MicroRNAs in neural cell differentiation. Brain Res. 1338, 14–19.
Le, M. T. N., Xie, H., Zhou, B., Chia, P. H., Rizk, P., Um, M., Udolph, G., Yang, H., Lim, B., and Lodish, H. F. (2009). MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol. Cell. Biol. 29, 5290–5305.
Leucht, C., Stigloher, C., Wizenmann, A., Klafke, R., Folchert, A., and Bally-Cuif, L. (2008). MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11, 641–648.
Li, W., Sun, G., Yang, S., Qu, Q., Nakashima, K., and Shi, Y. (2008). Nuclear receptor TLX regulates cell cycle progression in neural stem cells of the developing brain. Mol. Endocrinol. 22, 56–64.
Li, X., and Jin, P. (2010). Roles of small regulatory RNAs in determining neuronal identity. Nat. Rev. Neurosci. 11, 329–338.
Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., Bartel, D. P., Linsley, P. S., and Johnson, J. M. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773.
Ling, K.-H., Brautigan, P., Hahn, C., Daish, T., Rayner, J., Cheah, P.-S., Raison, J., Piltz, S., Mann, J., Mattiske, D., Thomas, P., Adelson, D., and Scott, H. (2011). Deep sequencing analysis of the developing mouse brain reveals a novel microRNA. BMC Genomics 12, 176. doi:10.1186/1471-2164-12-176
Liu, C., Teng, Z.-Q., Santistevan, N. J., Szulwach, K. E., Guo, W., Jin, P., and Zhao, X. (2010). Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6, 433–444.
Liu, C., and Zhao, X. (2009). MicroRNAs in adult and embryonic neurogenesis. Neuromolecular Med. 11, 141–152.
Liu, H. K., Belz, T., Bock, D., Takacs, A., Wu, H., Lichter, P., Chai, M., and Schutz, G. (2008). The nuclear receptor tailless is required for neurogenesis in the adult subventricular zone. Genes Dev. 22, 2473–2478.
Liu, J., Carmell, M. A., Rivas, F. V., Marsden, C. G., Thomson, J. M., Song, J.-J., Hammond, S. M., Joshua-Tor, L., and Hannon, G. J. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441.
Liu, K., Liu, Y., Mo, W., Qiu, R., Wang, X., Wu, J. Y., and He, R. (2011). MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 39, 2869–2879.
Luikart, B. W., Bensen, A. L., Washburn, E. K., Perederiy, J. V., Su, K. G., Li, Y., Kernie, S. G., Parada, L. F., and Westbrook, G. L. (2011). miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS ONE 6, e19077. doi:10.1371/journal.pone.0019077
Magill, S. T., Cambronne, X. A., Luikart, B. W., Lioy, D. T., Leighton, B. H., Westbrook, G. L., Mandel, G., and Goodman, R. H. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107, 20382–20387.
Maiorano, N., and Mallamaci, A. (2009). Promotion of embryonic cortico-cerebral neuronogenesis by miR-124. Neural Dev. 4, 40.
Makeyev, E. V., Zhang, J., Carrasco, M. A., and Maniatis, T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448.
Meyer, G. (2010). Building a human cortex: the evolutionary differentiation of Cajal-Retzius cells and the cortical hem. J. Anat. 217, 334–343.
Mientjes, E. J., Willemsen, R., Kirkpatrick, L. L., Nieuwenhuizen, I. M., Hoogeveen-Westerveld, M., Verweij, M., Reis, S., Bardoni, B., Hoogeveen, A. T., Oostra, B. A., and Nelson, D. L. (2004). Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum. Mol. Genet. 13, 1291–1302.
Monaghan, A. P., Grau, E., Bock, D., and Schutz, G. (1995). The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development 121, 839–853.
Nishino, J., Kim, I., Chada, K., and Morrison, S. J. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135, 227–239.
Obernier, K., Simeonova, I., Fila, T., Mandl, C., Holzl-Wenig, G., Monaghan-Nichols, P., and Ciccolini, F. (2011). Expression of Tlx in both stem cells and transit amplifying progenitors regulates stem cell activation and differentiation in the neonatal lateral subependymal zone. Stem cells 29, 1415–1426.
Packer, A. N., Xing, Y., Harper, S. Q., Jones, L., and Davidson, B. L. (2008). The Bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 28, 14341–14346.
Pasquinelli, A. E., Reinhart, B. J., Slack, F., Martindale, M. Q., Kuroda, M. I., Maller, B., Hayward, D. C., Ball, E. E., Degnan, B., Muller, P., Spring, J., Srinivasan, A., Fishman, M., Finnerty, J., Corbo, J., Levine, M., Leahy, P., Davidson, E., and Ruvkun, G. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89.
Qu, Q., Sun, G., Li, W., Yang, S., Ye, P., Zhao, C., Yu, R. T., Gage, F. H., Evans, R. M., and Shi, Y. (2010). Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat. Cell Biol. 12, 31–40.
Rajasethupathy, P., Fiumara, F., Sheridan, R., Betel, D., Puthanveettil, S. V., Russo, J. J., Sander, C., Tuschl, T., and Kandel, E. (2009). Characterization of small RNAs in aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63, 803–817.
Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R., and Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906.
Rybak, A., Fuchs, H., Smirnova, L., Brandt, C., Pohl, E. E., Nitsch, R., and Wulczyn, F. G. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10, 987–993.
Samaco, R. C., and Neul, J. L. (2011). Complexities of Rett syndrome and MeCP2. J. Neurosci. 31, 7951–7959.
Sanuki, R., Onishi, A., Koike, C., Muramatsu, R., Watanabe, S., Muranishi, Y., Irie, S., Uneo, S., Koyasu, T., Matsui, R., Cherasse, Y., Urade, Y., Watanabe, D., Kondo, M., Yamashita, T., and Furukawa, T. (2011). miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 14, 1125–1134.
Saunders, L. R., Sharma, A. D., Tawney, J., Nakagawa, M., Okita, K., Yamanaka, S., Willenbring, H., and Verdin, E. (2010). miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging 2, 415–431.
Schratt, G. M., Tuebing, F., Nigh, E. A., Kane, C. G., Sabatini, M. E., Kiebler, M., and Greenberg, M. E. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289.
Schwamborn, J. C., Berezikov, E., and Knoblich, J. A. (2009). The TRIM-NHL protein TRIM32 activates MicroRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925.
Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. (2004). Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5, R13.
Shi, Y., Chichung Lie, D., Taupin, P., Nakashima, K., Ray, J., Yu, R. T., Gage, F. H., and Evans, R. M. (2004). Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427, 78–83.
Shi, Y., Sun, G., Zhao, C., and Stewart, R. (2008). Neural stem cell self-renewal. Crit. Rev. Oncol. Hematol. 65, 43–53.
Shi, Y., Zhao, X., Hsieh, J., Wichterle, H., Impey, S., Banerjee, S., Neveu, P., and Kosik, K. S. (2010). MicroRNA regulation of neural stem cells and neurogenesis. J. Neurosci. 30, 14931–14936.
Shibata, M., Kurokawa, D., Nakao, H., Ohmura, T., and Aizawa, S. (2008). MicroRNA-9 modulates Cajal–Retzius cell differentiation by suppressing foxg1 expression in mouse medial pallium. J. Neurosci. 28, 10415–10421.
Shibata, M., Nakao, H., Kiyonari, H., Abe, T., and Aizawa, S. (2011). MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 31, 3407–3422.
Siegel, G., Obernosterer, G., Fiore, R., Oehmen, M., Bicker, S., Christensen, M., Khudayberdiev, S., Leuschner, P. F., Busch, C. J. L., Kane, C., Hubel, K., Dekker, F., Hedberg, C., Rengarajan, B., Drepper, C., Waldmann, H., Kauppinen, S., Greenberg, M. E., Draguhn, A., Rehmsmeier, M., Martinez, J., and Schratt, G. M. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716.
Silber, J., Lim, D., Petritsch, C., Persson, A., Maunakea, A., Yu, M., Vandenberg, S., Ginzinger, D., James, C. D., Costello, J., Bergers, G., Weiss, W., Alvarez-Buylla, A., and Hodgson, J. G. (2008). miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 6, 14. doi:10.1186/1741-7015-6-14
Smirnova, L., Grafe, A., Seiler, A., Schumacher, S., Nitsch, R., and Wulczyn, F. G. (2005). Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 21, 1469–1477.
Smrt, R. D., Szulwach, K. E., Pfeiffer, R. L., Li, X., Guo, W., Pathania, M., Teng, Z.-Q., Luo, Y., Peng, J., Bordey, A., Jin, P., and Zhao, X. (2010). MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind Bomb-1. Stem Cells 28, 1060–1070.
Stefani, G., and Slack, F. J. (2008). Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230.
Stolt, C. C., Lommes, P., Sock, E., Chaboissier, M.-C., Schedl, A., and Wegner, M. (2003). The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689.
Sun, G., Ye, P., Murai, K., Lang, M.-F., Li, S., Zhang, H., Li, W., Fu, C., Yin, J., Wang, A., Ma, X., and Shi, Y. (2011). miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2, 529.
Szulwach, K. E., Li, X., Smrt, R. D., Li, Y., Luo, Y., Lin, L., Santistevan, N. J., Li, W., Zhao, X., and Jin, P. (2010). Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141.
Visvanathan, J., Lee, S., Lee, B., Lee, J. W., and Lee, S. -K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749.
Vo, N., Klein, M. E., Varlamova, O., Keller, D. M., Yamamoto, T., Goodman, R. H., and Impey, S. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16426–16431.
Vreugdenhil, E., and Berezikov, E. (2010). Fine-tuning the brain: MicroRNAs. Front. Neuroendocrinol. 31, 128–133.
Wade, P. A. (2001). Methyl CpG-binding proteins and transcriptional repression. Bioessays 23, 1131–1137.
Wayman, G. A., Davare, M., Ando, H., Fortin, D., Varlamova, O., Cheng, H.-Y. M., Marks, D., Obrietan, K., Soderling, T. R., Goodman, R. H., and Impey, S. (2008). An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. U.S.A. 105, 9093–9098.
Wulczyn, F. G., Smirnova, L., Rybak, A., Brandt, C., Kwidzinski, E., Ninnemann, O., Strehle, M., Seiler, A., Schumacher, S., and Nitsch, R. (2007). Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 21, 415–426.
Xu, X.-L., Zong, R., Li, Z., Biswas, M. H. U., Fang, Z., Nelson, D. L., and Gao, F.-B. (2011). FXR1P but not FMRP regulates the levels of mammalian brain-specific microRNA-9 and microRNA-124. J. Neurosci. 31, 13705–13709.
Yeo, M., Lee, S. K., Lee, B., Ruiz, E. C., Pfaff, S. L., and Gill, G. N. (2005). Small CTD phosphatases function in silencing neuronal gene expression. Science 307, 596–600.
Yoo, A. S., Staahl, B. T., Chen, L., and Crabtree, G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646.
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., Lee-Messer, C., Dolmetsch, R. E., Tsien, R. W., and Crabtree, G. R. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231.
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir, G. A., Ruotti, V., Stewart, R., Slukvin, I. I., and Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920.
Zhang, C. L., Zou, Y., He, W., Gage, F. H., and Evans, R. M. (2008). A role for adult TLX-positive neural stem cells in learning and behaviour. Nature 451, 1004–1007.
Zhao, C., Sun, G., Li, S., Lang, M.-F., Yang, S., Li, W., and Shi, Y. (2010). MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 1876–1881.
Keywords: miRNA, neurogenesis, neural stem cell, proliferation, differentiation, maturation, gene regulation
Citation: Lang M-F and Shi Y (2012) Dynamic roles of microRNAs in neurogenesis. Front. Neurosci. 6:71. doi: 10.3389/fnins.2012.00071
Received: 23 January 2012; Paper pending published: 17 February 2012;
Accepted: 28 April 2012; Published online: 21 May 2012.
Edited by:
Xinyu Zhao, University of New Mexico, USAReviewed by:
Francesca Ciccolini, University of Heidelberg, GermanyCopyright: © 2012 Lang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Yanhong Shi, Department of Neurosciences, Beckman Research Institute of City of Hope, 1500 E Duarte Road, Duarte, CA 91010, USA. e-mail:eXNoaUBjb2gub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.