
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neurosci. , 17 February 2012
Sec. Neurogenesis
Volume 6 - 2012 | https://doi.org/10.3389/fnins.2012.00025
This article is part of the Research Topic microRNAs in neural stem cells and neurogenesis View all 10 articles
Adult neurogenesis generates functional neurons from neural stem cells present in specific brain regions. It is largely confined to two main regions: the subventricular zone of the lateral ventricle, and the subgranular zone of the dentate gyrus (DG), in the hippocampus. With age, the function of the hippocampus and particularly the DG is impaired. For instance, adult neurogenesis is decreased with aging, in both proliferating and differentiation of newborn cells, while in parallel an age-associated decline in cognitive performance is often seen. Surprisingly, the synaptogenic potential of adult-born neurons is only marginally influenced by aging. Therefore, although proliferation, differentiation, and synaptogenesis of adult-born new neurons in the DG are closely related to each other, they are differentially affected by aging. In this review we discuss the crucial roles of a novel class of recently discovered modulators of gene expression, the small non-coding RNAs, in the regulation of adult neurogenesis. Multiple small non-coding RNAs are differentially expressed in the hippocampus. In particular a subgroup of the small non-coding RNAs, the microRNAs, fine-tune the progression of adult neurogenesis. This makes small non-coding RNAs appealing candidates to orchestrate the functional alterations in adult neurogenesis and cognition associated with aging. Finally, we summarize observations that link changes in circulating levels of steroid hormones with alterations in adult neurogenesis, cognitive decline, and vulnerability to psychopathology in advanced age, and discuss a potential interplay between steroid hormone receptors and microRNAs in cognitive decline in aging individuals.
In humans, aging is being intensively studied, among other reasons because humans are reaching more advanced ages and the effects of a substantially larger aging population on the society have risen significantly. Aging theories traditionally associate a slow accumulation of loss of function and plasticity in cells and organs with aging. Therefore, factors that control the rates of cellular mitogenesis, differentiation, and cell death are considered important regulators of the aging process (Bowen and Atwood, 2004). Many physical changes take place in our bodies as we age, such as hair loss, endocrine changes, motor deficits, and sensory changes resulting in a reduced acuity of vision and impairment in hearing. However, the well-reported age-related decline in cognition and memory is arguably one of the aging symptoms that worries humans the most, possibly because memory is so central to our personal identity and relations (Shoemaker, 1959).
The human brain coordinates our cognitive abilities and in particular hippocampal and neocortical areas associated with memory and cognition are highly vulnerable to aging (Hof and Morrison, 2004; Small et al., 2011). Early studies have shown that aging results in a decline in hippocampal functions such as spatial navigation (Barnes, 1979; Gage et al., 1984; Markowska et al., 1989). Although the hippocampus has been traditionally evaluated as a single structure, it is now widely accepted that the hippocampus is a complex functional circuit, composed of molecular and functionally diverse regions (Gilbert et al., 2001; Wu et al., 2008; Brickman et al., 2010; Datson et al., 2010; Small et al., 2011). Studies in rodents and monkeys have shown selective regional differences in sensitivity to advancing age in the hippocampus (Small et al., 2004). Interestingly, the hippocampal dentate gyrus (DG) is particularly affected by aging (Small et al., 2002, 2004; Wu et al., 2008). In the following section we will review in more detail literature that link adult hippocampal neurogenesis (AHN) with cognitive functions that decline with age.
It is now clearly established that new neurons continue to be generated in the adult brain throughout life by neurogenesis from neural stem cells (NSCs). This phenomenon is largely confined to two main regions: the subventricular zone (SVZ) of the lateral ventricle, and the subgranular zone (SGZ) of the DG. In other adult cortical regions limited neurogenesis may occur, but only under specific conditions [(Acharya et al., 2008; Lucassen et al., 2010; Bruel-Jungerman et al., 2011) and references therein]. In the SGZ, NSCs give rise to transit amplifying neural progenitors that in turn differentiate into new immature neurons. These newly generated neurons migrate short distances into the granular cell layer of the DG and mature locally into dentate granule cells (GCs) within a period of 4–5 weeks (for reviews Zhao et al., 2008; Lucassen et al., 2010). During maturation, newly generated GCs become functionally integrated into pre-existing hippocampal circuits, receiving synaptic inputs mainly from the entorhinal cortex via the perforant path and extend their axons (mossy fibers) to establish synapses onto CA3 pyramidal cells. Both phenotypic maturation and functional integration are tightly regulated processes and are strongly dependent on cellular activity and connectivity with pre-existing networks (Zhao et al., 2006; Toni et al., 2008; Deng et al., 2010; Kelsch et al., 2010; Toni and Sultan, 2011).
The process of AHN consists of several steps: proliferation of progenitor cells; early selective elimination by apoptosis; fate decision and commitment to a neuronal phenotype; morphological and physiological maturation with the development of functional neuronal characteristics and a second selection by synaptic integration into pre-existing hippocampal circuits (Sierra et al., 2010; Encinas and Sierra, 2011). Moreover, AHN generates a whole range of neurogenic cell types that are differentially regulated and may play specific roles in the overall process (Figure 1A). During this slow maturation process, many of the newborn neurons are selected and more than 50% of the newborn GCs die within the first few weeks after birth (Cameron et al., 1993; Biebl et al., 2000; Kempermann et al., 2003; Hattiangady and Shetty, 2008). The rapid decline during early stages results from active elimination by apoptosis (Sun et al., 2004; Sierra et al., 2010). On the other hand, those immature neurons that develop synapses and are recruited into functionally active hippocampal circuits stand a better chance to survive (Kempermann et al., 1997; Gould et al., 1999). Thus, survival and death of newborn neurons in the DG are not only closely interconnected (Kuhn et al., 2005), the balance between these two processes is fine-tuned by neuronal activity and cognitive experience (Dupret et al., 2007). For more information about this topic, we refer the readers to some recent reviews (Deng et al., 2010; Kempermann, 2011; Ming and Song, 2011; Mongiat and Schinder, 2011).
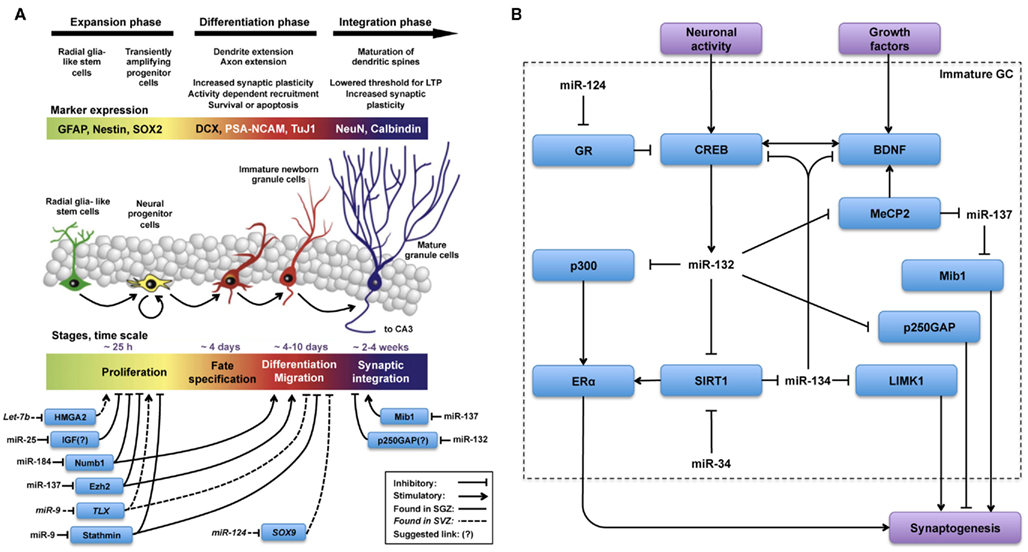
Figure 1. MicroRNas are key regulators in all phases of the adult neurogenesis cascade. Schematic illustration, adapted from (Lucassen et al., 2010), summarizing (A) miRs and targets involved in the regulation of different phases of adult neurogenesis and (B) miRs and targets hypothesized to be involved in the regulation of synaptogenesis during functional integration of adult-born new neurons. (A) The overall picture indicates that regulation by miR is less well-characterized in the integration phase as opposed to expansion and differentiation phases. (B) Regulation of synaptogenesis by miR-132 and miR-137 has been studied in AHN and in other contexts as well. From these observations, described in the text, we hypothesize that the regulatory network(s) depicted in (B) could be engaged in fine-tuning synaptogenesis during the functional integration phase of AHN.
Although the exact role of newborn neurons in the DG is still under debate, recent data support a functional role for adult-born neurons in learning and memory processes as reviewed in detail elsewhere (Aasebo et al., 2011; Aimone et al., 2011; Koehl and Abrous, 2011; Sahay et al., 2011). We here focus briefly on DG-dependent memory functions that may decrease with aging. The emerging consensus is that adult-born neurons in the DG play a crucial role in pattern separation, a memory mechanism that permits the differential representation of similar stimuli encoded by hippocampal circuits (Aimone et al., 2009; Sahay et al., 2011; Small et al., 2011). In humans, pattern separation can be assessed by a combination of functional magnetic resonance imaging and specific memory tasks (Bakker et al., 2008). Decreased performance in these tasks has been registered in aging subjects, positioning the DG as a key region in age-associated cognitive decline (Erickson and Barnes, 2003; Kempermann et al., 2003; Small et al., 2004; Toner et al., 2009). Importantly, impairment in tasks associated with pattern separation in humans may be an early indicator of DG dysfunction and possibly of early Alzheimer symptoms as altered performance in these tasks has been associated with changes in the activity of the entorhinal cortex, the main input to the DG (Albert, 1996), probably affecting newborn GCs survival (Stone et al., 2011).
A steep decline inz AHN associated with aging in the DG is well conserved in mammals and has been extensively reviewed elsewhere (Kempermann et al., 1997, 2003; Rao et al., 2006; Ben Abdallah et al., 2010; Encinas et al., 2011; Kempermann, 2011). The most dramatic changes in AHN associated with aging in fact take place already early in life, when the decrease in AHN rate is exponential and becomes stabilized in early adult life, remaining active for the rest of the lifespan in rodents. Interestingly, this decrease in AHN associated with aging appears to result from a decrease in proliferation and differentiation (Heine et al., 2004; Kuhn et al., 2005) and an increase in quiescence of NSCs (Hattiangady and Shetty, 2008).
Surprisingly, synaptogenesis in newborn GCs is less affected by aging than proliferation. Similar levels of synaptogenesis, as measured by dendritic spine densities in newborn GCs, are found both in old and young animals (Morgenstern et al., 2008; Toni et al., 2008; Toni and Sultan, 2011). As mentioned before and consistent with preserved synaptogenic potential in the aging DG, early-response gene expression and electrophysiological recordings have indicated that newborn GCs are more likely to respond to spatial processing than older GCs (Marrone et al., 2011). These observations suggest that even in the context of an otherwise declining function associated with aging, adult-born neurons remain functional and excitable and may therefore maintain their role in information processing. Although these are intriguing observations, future studies should have to address whether synapse formation or elimination still proceed at the same pace in the aging. Moreover, studies examining the expression of immediate early genes should be interpreted with care. Although immediate early genes are induced by activity, their expression does not provide a direct indication of information processing because it is unclear whether their activation is due to neuronal firing, synaptic plasticity, or subthreshold depolarizations (Guzowski, 2002; Loebrich and Nedivi, 2009; Schoenenberger et al., 2009). Overall, the concept that although in decreased numbers, newborn GCs are still efficiently integrated into hippocampal circuits in the aging DG is consistent with theories proposing that AHN could be a promising substrate to restore function in the aging DG (Yamashima et al., 2007; Marlatt and Lucassen, 2010). Moreover, it is in agreement with an earlier hypothesis that AHN may create a neurogenic reserve that buffers age-related cognitive decline (Kempermann, 2008).
In order to fully understand the tight selection process newborn GCs undergo, it is important to mention that newborn GCs exhibit a period of enhanced excitability and plasticity when they are between 2 weeks and 5 months old. Interestingly, this critical period of enhanced excitability and plasticity is associated with a period of intense synaptogenesis (Ramirez-Amaya et al., 2006; Tashiro et al., 2006; Zhao et al., 2006). During this period, newborn GCs can bypass apoptosis helped by NMDAR-mediated neuronal activity. They actively compete among themselves and possibly with their pre-existing mature counterparts for survival and connectivity to the network (Song et al., 2005; Tashiro et al., 2006; Faulkner et al., 2008; Toni et al., 2008). In this respect, studies aimed to characterize the maturation of synapses in adult-born neurons in the DG have found that they receive a diversity of inputs similar to mature granule neurons. Initially, immature neurons contact preferentially multiple synapse boutons. As new neurons maturate, spines form synapses preferentially with boutons devoid of other synaptic partners (Toni et al., 2007). These observations indicate that the connectivity of new GCs changes in time and suggest the existence of synaptic competition at the level of glutamatergic inputs into new neurons.
Although adult-born neurons ultimately blend into a phenotype that is functionally indistinguishable from older DG granule neurons generated during embryonic development and early postnatal life (Laplagne et al., 2006), adult-born neurons are intrinsically different form their pre-existing counterparts because they undergo a complex maturation process within the context of already functional hippocampal circuits. This multi-step maturation involves the transition across several phenotypic and physiologically dissimilar cell types within an otherwise mature hippocampus (Figure 1A). Particularly in terms of synaptogenesis, their first output synapses are formed with regulatory interneurons and as their axons develop, they form synapses with more diverse partners, including input pyramidal cells in the CA3. This shift in connectivity partners is unique for adult-born new GCs and may hallmark their physiology and functional integration within the DG (Ramirez-Amaya et al., 2006; Toni and Sultan, 2011). Moreover, as some of the transitional phenotypes show differential physiological properties, these intermediate cell types may have particular functions within the DG as well (Esposito et al., 2005). In particular, more immature phenotypes may modulate the activity of interneurons and neighboring granule neurons connected to the same interneurons. This transient state may have a potential effect on information coding by the DG (Michel et al., 2010).
In conclusion, proliferation, differentiation, survival, and synaptogenesis seem to be differentially affected by aging during the complex maturation and selection process through which newborn cells establish their final connectivity in the DG. In the next section we will review recent evidence suggesting that various small non-coding RNAs may be key players in the regulation of AHN.
The small non-coding RNAs are involved in a variety of gene expression regulatory mechanisms in the cell, such as alternative splicing, ribosomal RNA (rRNA) modifications, and repression of messenger RNA (mRNA) expression by RNA interference, a regulatory mechanism mediated by RNA–RNA interactions first observed in C. elegans (Lee et al., 1993; Fire et al., 1998).
Small non-coding RNAs can de classified into several major classes, i.e., small nucleolar RNAs (snoRNAs), endogenous small interfering RNAs (siRNAs), piwi-interacting RNAs (piRNAs), microRNas (miRs), transfer RNAs (tRNAs), rRNAs, spliceosomal RNAs, RNase P/MRP genes (Kim et al., 2009; Wright and Bruford, 2011). Other, less well-characterized small non-coding RNAs classes are the small modulatory RNAs (smRNAs), repeat-associated small interfering RNA (rasiRNA) that associate with piRNAs in protective mechanisms against transposable elements in the germ line (Saito et al., 2006) and the smallest members of the family, the 17–18 nucleotide long transcription initiation RNAs (tiRNAs) and small RNAs positioned at splice sites (spliRNAs), thought to be involved in the regulation of nucleosome positioning (Taft et al., 2009). With respect to the regulation of adult NSCs, the small double-stranded RNA (dsRNA) NRSE can trigger gene expression of neuron-specific genes through interaction with the NRSF/REST transcriptional machinery, resulting in the transition from NSCs into cells with a neuronal identity. The mechanism of action appears to be mediated through a dsRNA/protein interaction, rather than through RNA interference (Kuwabara et al., 2004).
Small nucleolar RNAs, are derived from protein coding and non-protein coding transcripts. They are involved in sequence-specific 2′-O-methylation (box C/D snoRNAs) or in the isomerization of specific uridines to pseudouridines (box H/ACA snoRNAs) in target RNAs (Henras et al., 2004). Some snoRNAs show tissue- and/or context-dependent expression, especially in the brain (Cavaille et al., 2000; Rogelj et al., 2003; Smalheiser et al., 2011) and may target other RNAs, including spliceosomal and tRNAs (Henras et al., 2004). Except for snoRNA MBII-52, involved in the alternative splicing of the serotonin receptor 2C (Cavaille et al., 2000; Filipowicz, 2000), very little is known about the function of snoRNAs in the brain (Cao et al., 2006). Although not directly linked to AHN, an increased expression level of snoRNA host-gene growth arrest specific 5 (GAS5) has been correlated to age-dependent spatial memory deficit in mice (Verbitsky et al., 2004). GAS5 is a gene with a complex structure, whose introns encode the snoRNAs SNORD44, 47, 74–81 involved in rRNA biosynthesis by 2′-O-methylating pre-rRNAs (Smith and Steitz, 1998). GAS5’s 5′ end harbors a terminal oligopyrimidine and its exons do not encode a protein (Smith and Steitz, 1998). Interestingly, Kino et al. (2010) have demonstrated that GAS5 is able to block the transcriptional activity of the glucocorticoid receptor (GR) and other steroid hormone receptors. In line with this crosstalk between GAS5 and steroid hormone receptors, psychogenic stressors and the subsequent release of stress hormone corticosterone, upregulate GAS5 levels in the hippocampus (Meier et al., 2010). Interestingly, treatment of cortical NSCs with ciliar neurotrophic factor (CNTF) induces NSCs to drift into the astrocytic lineage and strongly induces GAS5 expression (Sauvageot et al., 2005). Recently, we have observed that GAS5 is expressed in human hippocampal NSCs in culture (Schouten et al., unpublished data). Future studies will have to address the question whether GAS5 has a regulatory role in steroid hormone responsiveness in hippocampal NSCs.
Endogenous siRNAs are substrates of the ribonuclease Dicer and act through the RNA interference pathway, usually by perfect match with the target mRNA, resulting in mRNA degradation. They also seem to be involved in epigenetic regulation of target sequences by yet not well-characterized mechanisms (Lippman et al., 2004; Kim et al., 2006a).
Piwi-interacting RNAs use the RNA interference pathway as well, but they are not processed by Dicer and are involved in the silencing of transposons, primarily in the germline (Malone et al., 2009) and are also involved in epigenetic regulation events such as DNA methylation and histone modification (Yin and Lin, 2007). Interestingly, recent studies have reported the expression of a restricted group of piRNAs in the hippocampus, with at least one of this piRNAs (DQ541777) being expressed in the dendritic compartment of hippocampal neurons. Suppression of this piRNA by antisense oligonucleotides suggested a role in dendritic spine shape regulation (Lee et al., 2011).
MicroRNas are approximately 22 nucleotides long single stranded small non-coding RNAs. miRs are processed by Dicer, bind the RNA-induced silencing complex (RISC) and act through RNA interference by imperfect match recognition of target sites in the 3′UTRs of mRNAs, resulting in repression of target mRNA expression. Since their first discovery almost two decades ago, hundreds of miRs have been identified, in a wide range of organisms and are the best characterized members of the small non-coding RNA family. They play important roles in almost all biological processes studied, from development to cell death and metabolic control (Kim et al., 2009). Over 60% of all mammalian mRNAs seem to be under the control of miRs, adding an extra layer of control to the already complex regulatory mechanism of gene expression (Bartel, 2009; Friedman et al., 2009; Fabian et al., 2010). The miR seed region (5′ region nucleotides 2–8) usually binds to mRNA by almost perfect base-pairing, and the miR 3′ region binds mRNAs with less accurate base-pairing. Due to the limited size of miRs and their low mRNA binding specificity, miRs target several mRNAs and one mRNA can be targeted by multiple miRs (Lim et al., 2005; Selbach et al., 2008). In the canonical description of the miR pathway, target recognition by the miR leads to a decrease in the abundance of proteins encoded by the target. This has been explained by several mechanisms including posttranscriptional degradation of the target, translational repression, and deadenylation-dependent target decay through partially complementary miR target sites in mRNA untranslated regions (Maroney et al., 2006; Petersen et al., 2006; Wu et al., 2006). Challenging this canonical view of the miR pathway, recent observations suggest that in quiescent cells, a cellular state that may be relevant for aging, miRs induce upregulation of their targets by induction of protein translation, while in cycling/proliferating cells miRs inhibit translation (Vasudevan et al., 2008).
Simultaneously with the regulation that individual miRs exert on their specific targets, the miR pathway is regulated at different levels including miR biogenesis and decay (Krol et al., 2010b). The ribonucleases (RNases) III Drosha and Dicer as well as Argonaute 2 (Ago2), appear to be essential for miR biogenesis. In mammals, the presence of Dicer is essential for miR biogenesis, as Dicer-deficient mice die at the embryonic stage (Bernstein et al., 2001). Therefore, specific Dicer deletion and its consequential loss of miRs have been extensively used to characterize the global role of miRs in neurogenesis. Applying this experimental approach, studies have shown that miRs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early embryonic neurogenesis (De Pietri Tonelli et al., 2008). Using a similar approach in mature hippocampal neurons, more recent studies have demonstrated an essential role for miRs in learning and memory (Konopka et al., 2010). Accessory proteins of the miR pathway, such as the DiGeorge syndrome critical region gene 8 (DGCR8) protein, exportin-5 (Exp-5), TAR RNA binding protein (TRBP), and fragile X mental retardation protein (FMRP) are important in miR biogenesis as well and are affected in a variety of human pathologies (Perron and Provost, 2009). Interestingly, schizophrenia is associated with an increase in cortical miR biogenesis in the adult CNS. This induction of miR biogenesis is linked to an elevation in primary miR processing and corresponds with an increase in the microprocessor component DGCR8 (Beveridge et al., 2010).
In contrast to miR biogenesis, miR decay has received much less attention. This is probably because miRs are considered to be highly stable molecules. Nevertheless, several examples of regulation of miR turnover are known (Krol et al., 2010b). Interestingly, recent studies have shown that neurons actively degrade miRs upon synaptic stimulation (Krol et al., 2010a). In these studies, blocking glutamate receptors prevented the turnover of miR-124, -128, -134, and -138, while the addition of glutamate accelerated it. Notably, the behavior of miR-132 was opposite to that of the other miRs. Its degradation was induced by blocking glutamate receptors and not by the addition of glutamate. These findings suggest a difference in the mechanisms regulating the turnover of miR-132 as compared to other neuronal miRs such as miR-134 or -138.
The actions of several individual miRs on the proliferation, differentiation, and synaptogenesis stages of adult neurogenesis have been intensively studied (Figure 1A). For example, Szulwach et al. (2010) found that miR-137 targets Ezh2 mRNA, thereby promoting proliferation and inhibiting differentiation of NSCs in the SGZ. miR-137 also inhibits dendrite formation through inhibition of its target Mib1 in newborn immature neurons (Smrt et al., 2010). Based on these observations it would be possible to speculate that changes in miR-137 levels could be partially responsible for the age-dependent decrease in proliferation of NSCs in the SGZ. In this respect, it would be appealing to investigate whether levels of miR-137 and its targets Ezh2 and Mib1 change with aging in NSCs and immature neurons. On the other hand, assuming that one miR alone would be responsible for regulating NSC proliferation, differentiation, and integration may be an oversimplification. Indeed, in addition to miR-137, other miRs have been shown to have a regulatory function in the proliferation stage of AHN (Table 1). For example, miR let-7b reduces stem cell numbers and self-renewal in NSC of the SVZ through its target Hmga2 (Nishino et al., 2008). Supporting a possible role in aging, Nishino et al. demonstrated that changes in let-7b and Hmga2 expression during aging contribute to decline in NSC function. Moreover, miR-137 and let-7b converge on molecular pathways that involve TLX, a member of the nuclear receptor family central in the control of adult NSC renewal and fate determination. Let-7b regulates NSC proliferation and differentiation by targeting, among other mRNAs, TLX (Zhao et al., 2010). Interestingly, TLX represses the expression of mir-137 by recruiting the histone lysine-specific demethylase 1 (LSD1) to genomic regions of miR-137, providing a clear example of crosstalk between miRs and epigenetic regulatory mechanisms (Sun et al., 2011). A possible interaction between of miR-137 and let-7b in regulating NSC function is interesting for multiple reasons. One is that different miRs could agonize or antagonize on particular NSC functions (i.e., proliferation, differentiation, synaptogenesis), potentiating or counteracting their individual effects. Another reason is that the levels of some individual miRs could be altered by aging, while others would not. In this latter scenario, the potentiating or counteracting effects of two miRs on NSCs would dynamically change with aging. Of course, the same questions could be applied not only to miR-137 and let-7b but to other miRs as well. Therefore, we will discuss in greater detail miRs that regulate adult neurogenesis at different stages, their mRNA targets and the subsequent effects.
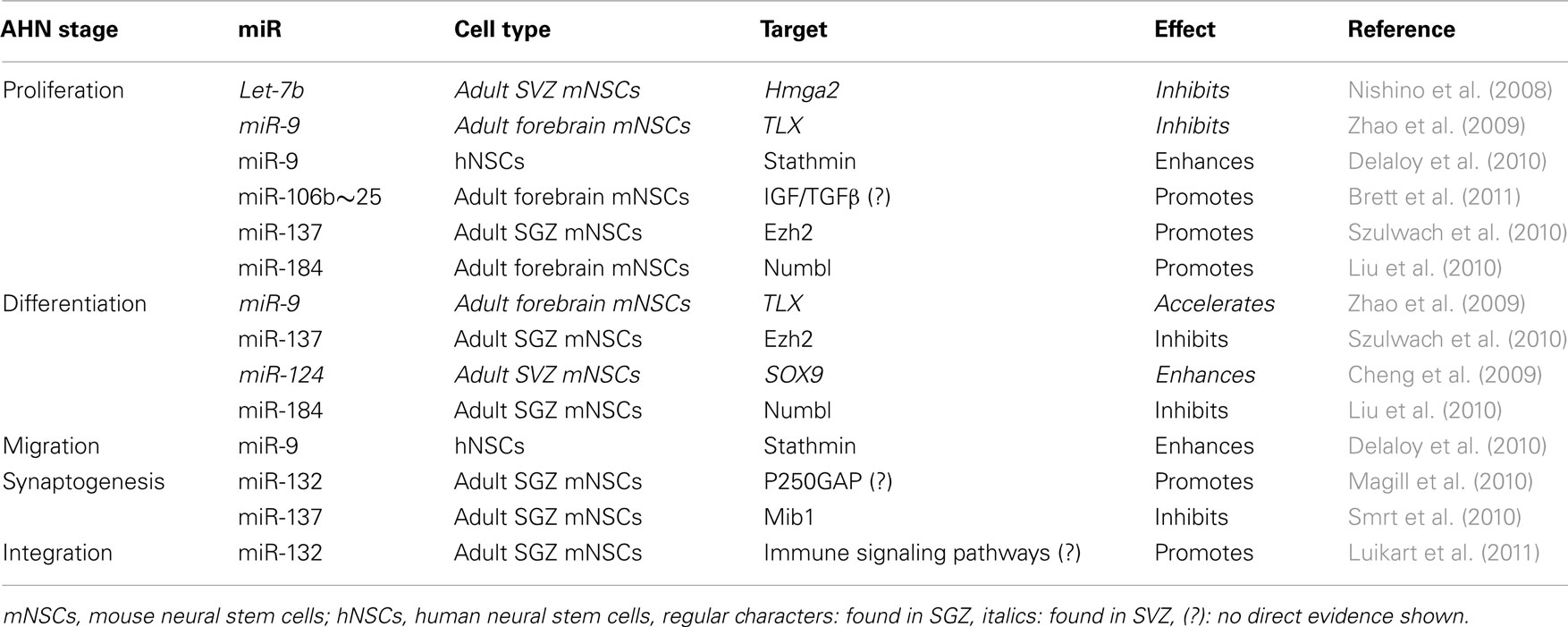
Table 1. All the stages of SGZ or SVZ neurogenesis are regulated by miRs, repressing their target mRNAs to be translated.
Another miR with important functions in NSCs is miR-184. The methyl-CpG binding protein 1 (MBD1) regulates gene expression via epigenetic mechanisms and miR-184 is directly repressed by MBD1 in NSCs, providing and interesting example of crosstalk between epigenetic regulation and miRs. Acting through inhibition of its target mRNA NumbI, high levels of miR-184 promoted proliferation but inhibited differentiation of NSCs. Therefore, MBD1, miR-184, and NumbI form a regulatory network that controls the balance between proliferation and differentiation of NSCs (Liu et al., 2010).
The miR cluster miR-106b∼25 and miR-25 in particular seems to be relevant in the regulation of NSC proliferation, since inhibition of miR-25 expression resulted in decreased NSC proliferation. Brett et al. (2011) proposed that miR-25 would regulate NSC proliferation through a number of potential targets involved in insulin/insulin-like growth factor-1 (IGF) signaling, a pathway implicated in aging. Unfortunately no direct experimental evidence of this regulation was provided in these studies. Still, the concept of miR-106b∼25-dependent regulation of insulin/IGF signaling in NSCs is attractive because a direct link between insulin/IGF signaling activation and subsequent increase in NSC proliferation has been demonstrated before (Groszer et al., 2001, 2006; Sinor and Lillien, 2004; Koltai et al., 2011; Rafalski and Brunet, 2011).
Two other well-characterized brain-specific miRs regulate NSCs functions. miR-124 overexpression in HeLa cells, resulted in an expression profile similar to that of brain tissue (Lim et al., 2005). Moreover, introducing miR-9 and miR-124 into human fibroblasts caused these cells to develop into functional neurons (Yoo et al., 2011). These two examples illustrate the potential of miR-9 and miR-124 to profoundly drive cells into a neuronal fate. Therefore, it seems reasonable that miR-9 and miR-124 fulfill a similar role in NSCs of the SGZ. Supporting this hypothesis, miR-9 and miR-124 where found to be abundantly expressed in the human hippocampus and were differentially expressed in fetal and normal aged hippocampus (Lukiw, 2007). miR-9 has been linked to enhanced proliferation and migration by regulation of its target Stathmin in embryonic derived NSCs (Delaloy et al., 2010). In the embryonic ventricular zone, miR-9 inhibits proliferation and enhances differentiation through regulation of TLX (Zhao et al., 2009). In the adult SVZ, miR-124, enhances differentiation of NSCs through regulation of its target the transcription factor sex determining region Y-box 9 (SOX9; Cheng et al., 2009).
As suggested by the examples discussed before, it is important to realize that single miRs could have opposite effects, depending on the presence of their specific targets in a particular cell type (Zhu et al., 2011). Therefore, in spite of the well-established roles of several miRs in NSCs of different ages and origins, the question still remains whether they have similar functions in any of the various cell types involved in AHN (Figure 1A). In summary, several miRs have been shown to tightly regulate many targets involved in NSC proliferation, differentiation, and maturation. In addition, multiple miRs and other small non-codingRNAs are differentially expressed in the aged hippocampus. This makes small non-coding RNAs appealing candidates to regulate various stages of AHN that may be involved in age-related decrease in cognitive functions.
After the literature overview presented in the previous section and summarized in Figure 1A, it becomes apparent that many miRs play key roles in the initial phases of AHN. Besides their regulatory roles in proliferation, differentiation and migration (Table 1) some miRs, including the previously discussed miR-137, have also been identified as regulators of synaptogenesis and neuronal integration of newborn immature neurons in the SGZ (Smrt et al., 2010). More specifically, by deleting the locus encoding miR-132 Magill et al. (2010) demonstrated a dramatic decrease in dendrite length, arborization, and spine density of the newborn immature neurons. Using lentiviral and retroviral reporters of miR-132 activity, Luikart et al. (2011) showed miR-132 is “at the right place and right time” to regulate the integration of newborn immature neurons. In addition to morphological changes, Luikart et al. showed that newly born GCs have impaired synaptic connectivity after miR-132 inactivation. As mentioned before, newborn neurons in the aging DG are as capable of synapse formation and functional integration as neurons born in a younger DG (Marrone et al., 2011). Therefore, if miR-132 would be a key factor in maintaining synaptogenic potential in newborn neurons during aging, its levels should be unaffected by aging in these cells. Indeed, recent studies have shown that miR-132 levels remain unchanged in fetal, aged, and even Alzheimer’s patient hippocampus (Lukiw, 2007).
Assuming that some of the miR-132’s regulatory capabilities and target networks are conserved between mature and newborn neurons, some of the observations on the function of miR-132 in synaptogenesis of mature neurons could be extrapolated to adult-born neurons in the DG. This assumption has the limitation discussed before for other miRs, i.e., the overall effect of a particular miR may depend on the specific repertoire of targets expressed in the cell type of interest. Yet, in mature neurons much more details are known about the cellular compartments, pathways, and networks in which miR-132 functions (Siegel et al., 2011; Figure 1B).
Overexpression of miR-132 in cultured hippocampal neurons revealed that miR-132 modulates short-term synaptic plasticity (Lambert et al., 2010), and overexpression in vivo triggers an increase in dendritic spine density and impaired novel object recognition memory (Hansen et al., 2010). Besides these relevant functions of miR-132 plays in regulating synaptic plasticity and memory, miR-132 expression is strongly regulated by neuronal activity (Magill et al., 2010; Luikart et al., 2011). Upon activation of cortical neurons, cAMP-response element binding protein (CREB) induces miR-132 expression through the CaMK-MEK/ERK–CREB pathway (Vo et al., 2005), a mechanism probably also present in hippocampal, olfactory bulb, and striatal neurons and neurons of the visual cortex (Nudelman et al., 2010; Mellios et al., 2011; Tognini et al., 2011). Aging in humans is strongly associated with changes in the circadian clock, resulting in strong sleep alterations in the elderly (Viola et al., 2011). In rodents, sleep deprivation strongly inhibits AHN through HPA axis-dependent and -independent mechanisms (Mirescu et al., 2006; Mueller et al., 2008). Again, miRs may provide a link between alterations in the circadian clock and human health disorders associated with aging. In particular, miR-219 and miR-132 modulate the circadian clock. From these two, only miR-132 is induced by light via a MAPK/CREB-dependent mechanism, and modulates clock-gene expression and attenuates the entraining effects of light on the circadian clock (Cheng et al., 2007). These findings have suggested that approaches to increase the robustness of the circadian clock by controlling miR expression may counteract the fragmentation of the sleep–wake cycle associated with aging (Hansen et al., 2011)
As all miRs, miR-132 targets multiple mRNAs including p250GAP (Vo et al., 2005; Wayman et al., 2008), MeCP2 (Klein et al., 2007), SIRT1 (Strum et al., 2009), p120RasGAP (Anand et al., 2010), and p300 (Lagos et al., 2010). miR-132 and its target p250GAP play a key role in activity dependent structural and functional plasticity in hippocampal neurons (Wayman et al., 2008). P250GAP is highly abundant in the postsynaptic density, where it interacts with multiple proteins involved in synaptic plasticity such as the tyrosine kinase Fyn (Taniguchi et al., 2003), β-catenin (Murase et al., 2002; Yu and Malenka, 2003), the NR2B subunit of the NMDA receptor and the PSD-95 scaffolding protein (Okabe et al., 2003). Other p250GAP partners within the Rho family, including RhoA, Rac1, and Cdc42 regulate actin cytoskeleton organization (Nakazawa et al., 2003; Impey et al., 2010). These findings suggest that miR-132 is a central regulator of synaptic plasticity, capable of linking synaptic activity with changes in synaptic structure by repressing p250GAP expression and thereby altering the composition of the synapse in an activity dependent manner.
Another target of miR-132 is MeCP2 (Klein et al., 2007), capable of binding methylated DNA and either repressing or activating transcription (Chahrour et al., 2008). Important in synaptic plasticity, mutations, and/or altered levels of MeCP2 have been linked to severe neurodevelopmental disorders such as Rett syndrome, Angelman’s syndrome, and autism (Gonzales and LaSalle, 2010). Blocking miR-132 in primary cortical neurons elevates MeCP2 expression and subsequently increases BDNF levels, while loss of MeCP2 reduces BDNF and miR-132 levels, indicating a feedback loop that involves miR-132 and regulates MeCP2 expression (Klein et al., 2007). Importantly, MeCP2 depletion in human and mouse brain causes an increase in expression of two neuronal gene transcriptional repressors REST (RE1 silencing transcription factor), and CoREST and is associated with a change in the histone modification profile to a more active conformation, suggesting that MeCP2 is a central regulator of epigenetic processes in the brain (Abuhatzira et al., 2007). This demonstrates another example of crosstalk between epigenetic regulation and miRs that could have a relevant role in AHN, since epigenetic regulation seems to play an important role in AHN and neuropsychiatric disorders (Hsieh and Eisch, 2010).
Highlighting the relevance of the crosstalk between classical epigenetic mechanisms and miRs, MeCP2 is a key regulator of miR-137 expression too (Szulwach et al., 2010; Wu et al., 2010). Therefore, miR-132 could regulate miR-137 expression through MeCP2, finally resulting in modulation of the miR-137 target Mib1. This miR-132–MeCP2–miR-137–Mib1 pathway could result in an inhibition of immature neuron maturation (Figure 1B), which would contradict the previously described pro-maturation effect of miR-132. This may not be completely unexpected, as exemplified by several seemingly paradoxical effects of miRs on NSC proliferation discussed before. Nevertheless, one simple explanation could be that miR-132 and miR-137 are not expressed simultaneously during maturation of NSCs. Therefore, in future studies the potential interactions between signaling pathways modulated by miR-132 and miR-137 would have to be carefully validated experimentally.
Pharmacological or genetic upregulation of the sirtuin (silent mating type information regulation 2 homolog) pathway, associated with anti-aging effects of calorie restriction, has shown promising results in laboratory models of aging (Bonda et al., 2011). The sirtuin SIRT1, is involved in NSC fate determination and SIRT1 is required for NSCs to adopt an astrocytic fate at the expense of the neuronal lineage under oxidative stress (Prozorovski et al., 2008). Interestingly, SIRT1 has been identified as a target of miR-132 (Strum et al., 2009). Moreover, SIRT1 limits the expression of another brain-specific miR, miR-134. SIRT1 deficiency results in miR-134 upregulation and concomitant, downregulation of CREB and BDNF, thereby impairing synaptic plasticity (Gao et al., 2010). Therefore, the miR-132-SIRT1 pathway could connect miR-132 to another regulator of dendritic spine development, miR-134 (Figure 1B). As miR-132 inhibits SIRT1 (Strum et al., 2009), it would relieve SIRT1-mediated repression of miR-134 resulting in increased levels of miR-134 and decreased CREB and BDNF expression and described by Gao et al. Alternatively, miR-134 negatively regulates Limk1, decreasing the size of dendritic spines (Schratt et al., 2006). The theoretical anti-synaptogenic effect that miR-132 could exert via both the miR-132-SIRT1-miR-134-Limk1 and miR-132-MeCP2-miR-137-Mib1 pathways, contrasts with the observations made with miR-132 overexpression in vivo, which triggers an increase in dendritic spine density (Hansen et al., 2010) and highlights a complex homeostatic balance network in which miR-132 may function by competing or interacting with other miRs to fine-tune expression of relevant targets during synaptogenesis (Figure 1B).
Underlining the complexity of synaptogenesis regulation by miRs other targets downstream of miR-134, including BDNF and CREB (Gao et al., 2010), complete a complex self-regulatory circle in the following theoretical pathway: miR-132–SIRT1–miR-134–CREB–miR-132. Simpler feedback regulatory loops controlling self-expression are well-characterized features of miR pathways in NSCs (Zhao et al., 2009; Eendebak et al., 2011). Therefore, further experiments would be needed to verify the proposed interaction between miR-132 and miR-134 through SIRT1 and CREB in NSCs. Notably, an increase in chromatin instability and DNA breaks correlates with aging in mammals. In response to DNA damage, SIRT1 relocates to DNA breaks to promote repair, resulting in transcriptional changes that parallel those observed in the aging mouse brain (Oberdoerffer et al., 2008). Although SIRT1 levels change with aging in various brain areas in mice (Lafontaine-Lacasse et al., 2010) the hippocampus was not analyzed in this study. Extrapolating from observations showing a preserved synaptogenic and functional activation potential of newborn GCs in the aging DG (Marrone et al., 2011) one would predict SIRT1 levels to be unaffected by aging in adult-born immature neurons of the SGZ. This would be in agreement with results discussed before showing that miR-132 expression seems to be unaffected by aging.
In summary, miRs are strategically placed within intrinsic regulatory networks that coordinate AHN. It is evident that levels of some miRs can change with aging, affecting the expression of their specific targets, while levels of other miRs seem to be unaffected. Thus, the next question is which could be the factors regulate expression of specific miRs during aging. In the following section we will discuss the possibility that steroid hormones could be involved in this regulation.
Healthy aging in humans is associated with a decline in plasma concentrations of several hormones and a gradual loss of function of endocrine systems (Lamberts et al., 1997). Traditionally, the development of physical frailty and a gradual loss in cognitive function that aging brings about, has been considered to be physiological and unavoidable. In recent years, however, it has become evident that it might not be necessary to accept the stereotype of aging as an unalterable process of decline and loss (Lamberts, 2002), particularly in terms of cognitive abilities, i.e., memory functions, that so profoundly mark our individual experiences and feelings of well-being in late life.
In humans, the adrenal glands synthesize and secrete large amounts the chemical precursors of sex steroid hormones (dehydroepiandrosterone and its sulfate) and neuroactive steroids. More than 30% of total androgens in elderly men and more than 90% of estrogen in postmenopausal women are derived from these precursors (Lamberts, 2002). In aging, a progressive and continuous decline in circulating levels of these precursors has been observed, while levels of the glucocorticoid stress hormone cortisol, synthesized primarily in the adrenal glands, show a parallel linear increase in some aging humans (Kalmijn et al., 1998; McEwen et al., 1999; Yen, 2001). Importantly, estrogens and glucocorticoids are strong regulators of the miR biogenesis pathway. Both hormones have been shown to control the expression of Dicer-1 and other key enzymes in miR synthesis in different experimental systems (Yamagata et al., 2009; Smith et al., 2010b). These observations suggest that steroid hormones may be crucial in favoring the expression of miR sets or “signatures” involved in the coordination of gene networks (Castellano et al., 2009; Eendebak et al., 2011). Although the effects of steroid hormones are strongly tissue and cell type specific, these observations suggest that steroid hormone regulation of miR biogenesis could be involved in the changes in miR expression associated with aging in the brain (Somel et al., 2010; Eda et al., 2011; Khanna et al., 2011; Wang et al., 2011b).
Low levels of circulating estrogens in post-menopause females have been linked to cognitive deficits (Smith et al., 2010a). In rats, estrogen replacement after ovariectomy increases LTP and dendritic spine density in hippocampal neurons, suggesting a key role of estrogen signaling in synaptic plasticity (Smith et al., 2010a). The estrogen receptor α (ERα) is a steroid hormone receptor that can be acetylated – and thereby activated – by p300, a target of miR-132 (Kim et al., 2006b). In addition, SIRT1 is found to promote ERα expression (Yao et al., 2010). Overall, these data indicate yet another potential pathway regulating synaptogenesis, in which miR-132 could be central.
Cortisol production by the adrenals influences memory and cognition during aging. Higher cortisol levels are associated with a poorer memory performance and a higher likelihood of memory decline, especially in women. These detrimental effects of cortisol seem to be directed at the hippocampus (McEwen et al., 1999; Li et al., 2006). In healthy elderly individuals, cortisol levels seemed to be associated with cognitive impairment (Kalmijn et al., 1998). Therefore, stress and resulting increases in glucocorticoid levels may have important consequences on the degree and speed of decline in memory and other cognitive abilities in the elderly (Lamberts, 2002). Although increasing levels of glucocorticoids are not always found in aged individuals, high levels of glucocorticoids are associated with synaptic loss in the hippocampus, hippocampal atrophy, and cognitive decline during aging in some individuals. These observations have led to the suggestion that glucocorticoids may contribute to, or accelerate aspects of aging (Nichols et al., 2001). Therefore, although stress and increased glucocorticoid levels may not contribute to aging in all individuals, they could decrease structural plasticity and the brain’s vulnerability to disease (Radley and Morrison, 2005; Korosi et al., 2011) resulting in a pro-aging activity in vulnerable individuals (Wolkowitz et al., 2009).
Despite the previously discussed inhibition of AHN by glucocorticoids in the DG, the relationship between plasma glucocorticoid levels, receptor expression and AHN is complex. Interestingly, studies from our lab have demonstrated that a brief treatment with the GR antagonist mifepristone rapidly reverses the deleterious effects of chronic stress on AHN (Oomen et al., 2007), strongly suggesting that the GR is involved in chronic glucocorticoid hormone suppression of AHN. The observations that GR expression, particularly in the DG, is increased in depressed elderly women and within this group correlates positively with age, suggests that GR activity could be linked to disease mechanisms during aging (Wang et al., 2011a). Gene profiling studies in chronically stressed animals have shown that CREB is central in the signaling pathways regulated by the GR in the DG (Datson et al., 2010).
As we have discussed before, CREB is part of a central pathway in the regulation of AHN (Merz et al., 2011) and this pathway crosstalks to several miRs involved in the regulation of NSC proliferation, differentiation, and synaptogenesis (Figure 1B), in particular the neuronal activity-induced, miR-132 (Nudelman et al., 2010). Notably, GR activation suppresses miR-132 expression and results in a decrease in BDNF and glutamate receptors (Kawashima et al., 2010). These observations suggest that high glucocorticoid levels observed in many aging individuals may result in a GR-dependent inhibition of miR-132 expression and reduced glutamate receptor expression in adult-born immature neurons of the DG. This hormone and miR mediated pathway could induce significant changes (e.g., reduced synaptogenic potential) in adult-born neurons in susceptible patients that are not observed in healthy aging individuals.
Moreover, glucocorticoids may influence not only cell birth and death, but also pathways that regulate GC differentiation and survival (Sousa and Almeida, 2002). These glucocorticoid-mediated pathways may involve some miRs with key roles in NSCs. For example, both glucocorticoid and mineralocorticoid receptors are targets of miR-124 (Cheng et al., 2009; Vreugdenhil et al., 2009; Sober et al., 2010). Interestingly, recent observations in human NSCs have shown that GR activation inhibits the expression of neuronal differentiation markers (Anacker et al., 2011). Therefore, in a likely scenario, the gradual increases in miR-124 that takes place during neuronal differentiation of adult NSCs (Cheng et al., 2009), may help to fine-tune GR expression to a physiological range that promotes a pro-neuronal phenotype.
In conclusion, we have reviewed literature that supports a role for AHN in DG-dependant cognitive functions related to memory, and have discussed how alterations in this process may be related to aging-associated cognitive decline. Furthermore, we have described how miRs could be placed among the factors that control the generation of adult-born neurons in the hippocampus. Moreover, we propose that miRs are strategically positioned within regulatory networks that fine-tune at different levels the proliferation, differentiation, survival, and synaptogenesis of adult-born neurons. Finally, we reviewed evidence suggesting that aging-associated changes in circulating levels of steroid hormones, in particular estrogens and glucocorticoids are associated with cognitive decline and proposed that these changes may impact on AHN through signaling networks that involve miRs. In the future, new experimental efforts will address whether this hypothesis holds true and if so, how could we use it to design new therapeutic interventions that may help us reach a successful healthy aging.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work is part of the research program VIDI H64.09.016 (to Carlos P. Fitzsimons), financed by the Netherlands Organization for Scientific Research. Paul J. Lucassen is supported by the HersenStichting Nederland, IPF, the EU (NEURAD), and ISAO. The authors want to thank Wendy Timmermans and Dr. S. A. Fratantoni for critically reading the manuscript.
Aasebo, I. E., Blankvoort, S., and Tashiro, A. (2011). Critical maturational period of new neurons in adult dentate gyrus for their involvement in memory formation. Eur. J. Neurosci. 33, 1094–1100.
Abuhatzira, L., Makedonski, K., Kaufman, Y., Razin, A., and Shemer, R. (2007). MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics 2, 214–222.
Acharya, M. M., Hattiangady, B., and Shetty, A. K. (2008). Progress in neuroprotective strategies for preventing epilepsy. Prog. Neurobiol. 84, 363–404.
Aimone, J. B., Deng, W., and Gage, F. H. (2011). Resolving new memories: a critical look at the dentate gyrus, adult neurogenesis, and pattern separation. Neuron 70, 589–596.
Aimone, J. B., Wiles, J., and Gage, F. H. (2009). Computational influence of adult neurogenesis on memory encoding. Neuron 61, 187–202.
Albert, M. S. (1996). Cognitive and neurobiologic markers of early Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 93, 13547–13551.
Anacker, C., Zunszain, P. A., Cattaneo, A., Carvalho, L. A., Garabedian, M. J., Thuret, S., Price, J., and Pariante, C. M. (2011). Antidepressants increase human hippocampal neurogenesis by activating the glucocorticoid receptor. Mol. Psychiatry 16, 738–750.
Anand, S., Majeti, B. K., Acevedo, L. M., Murphy, E. A., Mukthavaram, R., Scheppke, L., Huang, M., Shields, D. J., Lindquist, J. N., Lapinski, P. E., King, P. D., Weis, S. M., and Cheresh, D. A. (2010). MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 16, 909–914.
Bakker, A., Kirwan, C. B., Miller, M., and Stark, C. E. (2008). Pattern separation in the human hippocampal CA3 and dentate gyrus. Science 319, 1640–1642.
Barnes, C. A. (1979). Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat. J. Comp. Physiol. Psychol. 93, 74–104.
Ben Abdallah, N. M., Slomianka, L., Vyssotski, A. L., and Lipp, H. P. (2010). Early age-related changes in adult hippocampal neurogenesis in C57 mice. Neurobiol. Aging 31, 151–161.
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366.
Beveridge, N. J., Gardiner, E., Carroll, A. P., Tooney, P. A., and Cairns, M. J. (2010). Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry 15, 1176–1189.
Biebl, M., Cooper, C. M., Winkler, J., and Kuhn, H. G. (2000). Analysis of neurogenesis and programmed cell death reveals a self-renewing capacity in the adult rat brain. Neurosci. Lett. 291, 17–20.
Bonda, D. J., Lee, H. G., Camins, A., Pallas, M., Casadesus, G., Smith, M. A., and Zhu, X. (2011). The sirtuin pathway in ageing and Alzheimer disease: mechanistic and therapeutic considerations. Lancet Neurol. 10, 275–279.
Bowen, R. L., and Atwood, C. S. (2004). Living and dying for sex. A theory of aging based on the modulation of cell cycle signaling by reproductive hormones. Gerontology 50, 265–290.
Brett, J. O., Renault, V. M., Rafalski, V. A., Webb, A. E., and Brunet, A. (2011). The microRNA cluster miR-106b∼25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 3, 108–124.
Brickman, A. M., Stern, Y., and Small, S. A. (2010). Hippocampal subregions differentially associate with standardized memory tests. Hippocampus 21, 923–928.
Bruel-Jungerman, E., Lucassen, P. J., and Francis, F. (2011). Cholinergic influences on cortical development and adult neurogenesis. Behav. Brain Res. 221, 379–388.
Cameron, H. A., Woolley, C. S., Mcewen, B. S., and Gould, E. (1993). Differentiation of newly born neurons and glia in the dentate gyrus of the adult rat. Neuroscience 56, 337–344.
Cao, X., Yeo, G., Muotri, A. R., Kuwabara, T., and Gage, F. H. (2006). Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 29, 77–103.
Castellano, L., Giamas, G., Jacob, J., Coombes, R. C., Lucchesi, W., Thiruchelvam, P., Barton, G., Jiao, L. R., Wait, R., Waxman, J., Hannon, G. J., and Stebbing, J. (2009). The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc. Natl. Acad. Sci. U.S.A. 106, 15732–15737.
Cavaille, J., Buiting, K., Kiefmann, M., Lalande, M., Brannan, C. I., Horsthemke, B., Bachellerie, J. P., Brosius, J., and Huttenhofer, A. (2000). Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. U.S.A. 97, 14311–14316.
Chahrour, M., Jung, S. Y., Shaw, C., Zhou, X., Wong, S. T., Qin, J., and Zoghbi, H. Y. (2008). MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229.
Cheng, H. Y., Papp, J. W., Varlamova, O., Dziema, H., Russell, B., Curfman, J. P., Nakazawa, T., Shimizu, K., Okamura, H., Impey, S., and Obrietan, K. (2007). microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829.
Cheng, L. C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408.
Datson, N. A., Speksnijder, N., Mayer, J. L., Steenbergen, P. J., Korobko, O., Goeman, J., De Kloet, E. R., Joels, M., and Lucassen, P. J. (2010). The transcriptional response to chronic stress and glucocorticoid receptor blockade in the hippocampal dentate gyrus. Hippocampus 22, 359–371.
De Pietri Tonelli, D., Pulvers, J. N., Haffner, C., Murchison, E. P., Hannon, G. J., and Huttner, W. B. (2008). miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135, 3911–3921.
Delaloy, C., Liu, L., Lee, J. A., Su, H., Shen, F., Yang, G. Y., Young, W. L., Ivey, K. N., and Gao, F. B. (2010). MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335.
Deng, W., Aimone, J. B., and Gage, F. H. (2010). New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat. Rev. Neurosci. 11, 339–350.
Dupret, D., Fabre, A., Dobrossy, M. D., Panatier, A., Rodriguez, J. J., Lamarque, S., Lemaire, V., Oliet, S. H., Piazza, P. V., and Abrous, D. N. (2007). Spatial learning depends on both the addition and removal of new hippocampal neurons. PLoS Biol. 5, e214. doi:10.1371/journal.pbio.0050214
Eda, A., Takahashi, M., Fukushima, T., and Hohjoh, H. (2011). Alteration of microRNA expression in the process of mouse brain growth. Gene 485, 46–52.
Eendebak, R. J., Lucassen, P. J., and Fitzsimons, C. P. (2011). Nuclear receptors and microRNAs: who regulates the regulators in neural stem cells? FEBS Lett. 585, 717–722.
Encinas, J. M., Michurina, T. V., Peunova, N., Park, J. H., Tordo, J., Peterson, D. A., Fishell, G., Koulakov, A., and Enikolopov, G. (2011). Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 8, 566–579.
Encinas, J. M., and Sierra, A. (2011). Neural stem cell deforestation as the main force driving the age-related decline in adult hippocampal neurogenesis. Behav. Brain Res. 227, 433–439.
Erickson, C. A., and Barnes, C. A. (2003). The neurobiology of memory changes in normal aging. Exp. Gerontol. 38, 61–69.
Esposito, M. S., Piatti, V. C., Laplagne, D. A., Morgenstern, N. A., Ferrari, C. C., Pitossi, F. J., and Schinder, A. F. (2005). Neuronal differentiation in the adult hippocampus recapitulates embryonic development. J. Neurosci. 25, 10074–10086.
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379.
Faulkner, R. L., Jang, M. H., Liu, X. B., Duan, X., Sailor, K. A., Kim, J. Y., Ge, S., Jones, E. G., Ming, G. L., Song, H., and Cheng, H. J. (2008). Development of hippocampal mossy fiber synaptic outputs by new neurons in the adult brain. Proc. Natl. Acad. Sci. U.S.A. 105, 14157–14162.
Filipowicz, W. (2000). Imprinted expression of small nucleolar RNAs in brain: time for RNomics. Proc. Natl. Acad. Sci. U.S.A. 97, 14035–14037.
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811.
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105.
Gage, F. H., Kelly, P. A., and Bjorklund, A. (1984). Regional changes in brain glucose metabolism reflect cognitive impairments in aged rats. J. Neurosci. 4, 2856–2865.
Gao, J., Wang, W. Y., Mao, Y. W., Graff, J., Guan, J. S., Pan, L., Mak, G., Kim, D., Su, S. C., and Tsai, L. H. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109.
Gilbert, P. E., Kesner, R. P., and Lee, I. (2001). Dissociating hippocampal subregions: double dissociation between dentate gyrus and CA1. Hippocampus 11, 626–636.
Gonzales, M. L., and LaSalle, J. M. (2010). The role of MeCP2 in brain development and neurodevelopmental disorders. Curr. Psychiatry Rep. 12, 127–134.
Gould, E., Beylin, A., Tanapat, P., Reeves, A., and Shors, T. J. (1999). Learning enhances adult neurogenesis in the hippocampal formation. Nat. Neurosci. 2, 260–265.
Groszer, M., Erickson, R., Scripture-Adams, D. D., Dougherty, J. D., Le Belle, J., Zack, J. A., Geschwind, D. H., Liu, X., Kornblum, H. I., and Wu, H. (2006). PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proc. Natl. Acad. Sci. U.S.A. 103, 111–116.
Groszer, M., Erickson, R., Scripture-Adams, D. D., Lesche, R., Trumpp, A., Zack, J. A., Kornblum, H. I., Liu, X., and Wu, H. (2001). Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2186–2189.
Guzowski, J. F. (2002). Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus 12, 86–104.
Hansen, K. F., Sakamoto, K., and Obrietan, K. (2011). MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 3, 10.
Hansen, K. F., Sakamoto, K., Wayman, G. A., Impey, S., and Obrietan, K. (2010). Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 5, e15497. doi:10.1371/journal.pone.0015497
Hattiangady, B., and Shetty, A. K. (2008). Aging does not alter the number or phenotype of putative stem/progenitor cells in the neurogenic region of the hippocampus. Neurobiol. Aging 29, 129–147.
Heine, V. M., Maslam, S., Joels, M., and Lucassen, P. J. (2004). Prominent decline of newborn cell proliferation, differentiation, and apoptosis in the aging dentate gyrus, in absence of an age-related hypothalamus-pituitary-adrenal axis activation. Neurobiol. Aging 25, 361–375.
Henras, A. K., Dez, C., and Henry, Y. (2004). RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 14, 335–343.
Hof, P. R., and Morrison, J. H. (2004). The aging brain: morphomolecular senescence of cortical circuits. Trends Neurosci. 27, 607–613.
Hsieh, J., and Eisch, A. J. (2010). Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: unraveling the genome to understand the mind. Neurobiol. Dis. 39, 73–84.
Impey, S., Davare, M., Lasiek, A., Fortin, D., Ando, H., Varlamova, O., Obrietan, K., Soderling, T. R., Goodman, R. H., and Wayman, G. A. (2010). An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol. Cell. Neurosci. 43, 146–156.
Kalmijn, S., Launer, L. J., Stolk, R. P., De Jong, F. H., Pols, H. A., Hofman, A., Breteler, M. M., and Lamberts, S. W. (1998). A prospective study on cortisol, dehydroepiandrosterone sulfate, and cognitive function in the elderly. J. Clin. Endocrinol. Metab. 83, 3487–3492.
Kawashima, H., Numakawa, T., Kumamaru, E., Adachi, N., Mizuno, H., Ninomiya, M., Kunugi, H., and Hashido, K. (2010). Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience 165, 1301–1311.
Kelsch, W., Sim, S., and Lois, C. (2010). Watching synaptogenesis in the adult brain. Annu. Rev. Neurosci. 33, 131–149.
Kempermann, G. (2008). The neurogenic reserve hypothesis: what is adult hippocampal neurogenesis good for? Trends Neurosci. 31, 163–169.
Kempermann, G. (2011). Seven principles in the regulation of adult neurogenesis. Eur. J. Neurosci. 33, 1018–1024.
Kempermann, G., Gast, D., Kronenberg, G., Yamaguchi, M., and Gage, F. H. (2003). Early determination and long-term persistence of adult-generated new neurons in the hippocampus of mice. Development 130, 391–399.
Kempermann, G., Kuhn, H. G., and Gage, F. H. (1997). More hippocampal neurons in adult mice living in an enriched environment. Nature 386, 493–495.
Khanna, A., Muthusamy, S., Liang, R., Sarojini, H., and Wang, E. (2011). Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging (Albany NY) 3, 223–236.
Kim, D. H., Villeneuve, L. M., Morris, K. V., and Rossi, J. J. (2006a). Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 13, 793–797.
Kim, M. Y., Woo, E. M., Chong, Y. T., Homenko, D. R., and Kraus, W. L. (2006b). Acetylation of estrogen receptor alpha by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol. Endocrinol. 20, 1479–1493.
Kim, V. N., Han, J., and Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139.
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3, ra8.
Klein, M. E., Lioy, D. T., Ma, L., Impey, S., Mandel, G., and Goodman, R. H. (2007). Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 10, 1513–1514.
Koehl, M., and Abrous, D. N. (2011). A new chapter in the field of memory: adult hippocampal neurogenesis. Eur. J. Neurosci. 33, 1101–1114.
Koltai, E., Zhao, Z., Lacza, Z., Cselenyak, A., Vacz, G., Nyakas, C., Boldogh, I., Ichinoseki-Sekine, N., and Radak, Z. (2011). Combined exercise and insulin-like growth factor-1 supplementation induces neurogenesis in old rats, but do not attenuate age-associated DNA damage. Rejuvenation Res. 14, 585–596.
Konopka, W., Kiryk, A., Novak, M., Herwerth, M., Parkitna, J. R., Wawrzyniak, M., Kowarsch, A., Michaluk, P., Dzwonek, J., Arnsperger, T., Wilczynski, G., Merkenschlager, M., Theis, F. J., Kohr, G., Kaczmarek, L., and Schutz, G. (2010). MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30, 14835–14842.
Korosi, A., Naninck, E. F., Oomen, C. A., Schouten, M., Krugers, H., Fitzsimons, C., and Lucassen, P. J. (2011). Early-life stress mediated modulation of adult neurogenesis and behavior. Behav. Brain Res. 227, 400–409.
Krol, J., Busskamp, V., Markiewicz, I., Stadler, M. B., Ribi, S., Richter, J., Duebel, J., Bicker, S., Fehling, H. J., Schubeler, D., Oertner, T. G., Schratt, G., Bibel, M., Roska, B., and Filipowicz, W. (2010a). Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631.
Krol, J., Loedige, I., and Filipowicz, W. (2010b). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610.
Kuhn, H. G., Biebl, M., Wilhelm, D., Li, M., Friedlander, R. M., and Winkler, J. (2005). Increased generation of granule cells in adult Bcl-2-overexpressing mice: a role for cell death during continued hippocampal neurogenesis. Eur. J. Neurosci. 22, 1907–1915.
Kuwabara, T., Hsieh, J., Nakashima, K., Taira, K., and Gage, F. H. (2004). A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell 116, 779–793.
Lafontaine-Lacasse, M., Richard, D., and Picard, F. (2010). Effects of age and gender on Sirt 1 mRNA expressions in the hypothalamus of the mouse. Neurosci. Lett. 480, 1–3.
Lagos, D., Pollara, G., Henderson, S., Gratrix, F., Fabani, M., Milne, R. S., Gotch, F., and Boshoff, C. (2010). miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12, 513–519.
Lambert, T. J., Storm, D. R., and Sullivan, J. M. (2010). MicroRNA132 modulates short-term synaptic plasticity but not basal release probability in hippocampal neurons. PLoS ONE 5, e15182. doi:10.1371/journal.pone.0015182
Lamberts, S. W., Van Den Beld, A. W., and Van Der Lely, A. J. (1997). The endocrinology of aging. Science 278, 419–424.
Laplagne, D. A., Esposito, M. S., Piatti, V. C., Morgenstern, N. A., Zhao, C., Van Praag, H., Gage, F. H., and Schinder, A. F. (2006). Functional convergence of neurons generated in the developing and adult hippocampus. PLoS Biol. 4, e409. doi:10.1371/journal.pbio.0040409
Lee, E. J., Banerjee, S., Zhou, H., Jammalamadaka, A., Arcila, M., Manjunath, B. S., and Kosik, K. S. (2011). Identification of piRNAs in the central nervous system. RNA 17, 1090–1099.
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854.
Li, G., Cherrier, M. M., Tsuang, D. W., Petrie, E. C., Colasurdo, E. A., Craft, S., Schellenberg, G. D., Peskind, E. R., Raskind, M. A., and Wilkinson, C. W. (2006). Salivary cortisol and memory function in human aging. Neurobiol. Aging 27, 1705–1714.
Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., Bartel, D. P., Linsley, P. S., and Johnson, J. M. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773.
Lippman, Z., Gendrel, A. V., Black, M., Vaughn, M. W., Dedhia, N., Mccombie, W. R., Lavine, K., Mittal, V., May, B., Kasschau, K. D., Carrington, J. C., Doerge, R. W., Colot, V., and Martienssen, R. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476.
Liu, C., Teng, Z. Q., Santistevan, N. J., Szulwach, K. E., Guo, W., Jin, P., and Zhao, X. (2010). Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6, 433–444.
Loebrich, S., and Nedivi, E. (2009). The function of activity-regulated genes in the nervous system. Physiol. Rev. 89, 1079–1103.
Lucassen, P. J., Meerlo, P., Naylor, A. S., Van Dam, A. M., Dayer, A. G., Fuchs, E., Oomen, C. A., and Czeh, B. (2010). Regulation of adult neurogenesis by stress, sleep disruption, exercise and inflammation: implications for depression and antidepressant action. Eur. Neuropsychopharmacol. 20, 1–17.
Luikart, B. W., Bensen, A. L., Washburn, E. K., Perederiy, J. V., Su, K. G., Li, Y., Kernie, S. G., Parada, L. F., and Westbrook, G. L. (2011). miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS ONE 6, e19077. doi:10.1371/journal.pone.0019077
Lukiw, W. J. (2007). Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport 18, 297–300.
Magill, S. T., Cambronne, X. A., Luikart, B. W., Lioy, D. T., Leighton, B. H., Westbrook, G. L., Mandel, G., and Goodman, R. H. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107, 20382–20387.
Malone, C. D., Brennecke, J., Dus, M., Stark, A., Mccombie, W. R., Sachidanandam, R., and Hannon, G. J. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535.
Markowska, A. L., Stone, W. S., Ingram, D. K., Reynolds, J., Gold, P. E., Conti, L. H., Pontecorvo, M. J., Wenk, G. L., and Olton, D. S. (1989). Individual differences in aging: behavioral and neurobiological correlates. Neurobiol. Aging 10, 31–43.
Marlatt, M. W., and Lucassen, P. J. (2010). Neurogenesis and Alzheimer’s disease: biology and pathophysiology in mice and men. Curr. Alzheimer Res. 7, 113–125.
Maroney, P. A., Yu, Y., Fisher, J., and Nilsen, T. W. (2006). Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 13, 1102–1107.
Marrone, D. F., Ramirez-Amaya, V., and Barnes, C. A. (2011). Neurons generated in senescence maintain capacity for functional integration. Hippocampus. doi: 10.1002/hipo.20959. [Epub ahead of print].
McEwen, B. S., De Leon, M. J., Lupien, S. J., and Meaney, M. J. (1999). Corticosteroids, the aging brain and cognition. Trends Endocrinol. Metab. 10, 92–96.
Meier, I., Fellini, L., Jakovcevski, M., Schachner, M., and Morellini, F. (2010). Expression of the snoRNA host gene gas5 in the hippocampus is upregulated by age and psychogenic stress and correlates with reduced novelty-induced behavior in C57BL/6 mice. Hippocampus 20, 1027–1036.
Mellios, N., Sugihara, H., Castro, J., Banerjee, A., Le, C., Kumar, A., Crawford, B., Strathmann, J., Tropea, D., Levine, S. S., Edbauer, D., and Sur, M. (2011). miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat. Neurosci. 14, 1240–1242.
Merz, K., Herold, S., and Lie, D. C. (2011). CREB in adult neurogenesis – master and partner in the development of adult-born neurons? Eur. J. Neurosci. 33, 1078–1086.
Michel, B. F., Luciani, V., Geda, Y. E., Sambuchi, N., Paban, V., and Azorin, J. M. (2010). In Alzheimer’s disease, the clinical expression of behavioral and psychological signs and symptoms is early and specific of neuropathological stages. Encephale 36, 314–325.
Ming, G. L., and Song, H. (2011). Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702.
Mirescu, C., Peters, J. D., Noiman, L., and Gould, E. (2006). Sleep deprivation inhibits adult neurogenesis in the hippocampus by elevating glucocorticoids. Proc. Natl. Acad. Sci. U.S.A. 103, 19170–19175.
Mongiat, L. A., and Schinder, A. F. (2011). Adult neurogenesis and the plasticity of the dentate gyrus network. Eur. J. Neurosci. 33, 1055–1061.
Morgenstern, N. A., Lombardi, G., and Schinder, A. F. (2008). Newborn granule cells in the ageing dentate gyrus. J. Physiol. (Lond.) 586, 3751–3757.
Mueller, A. D., Pollock, M. S., Lieblich, S. E., Epp, J. R., Galea, L. A., and Mistlberger, R. E. (2008). Sleep deprivation can inhibit adult hippocampal neurogenesis independent of adrenal stress hormones. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1693–R1703.
Murase, S., Mosser, E., and Schuman, E. M. (2002). Depolarization drives beta-catenin into neuronal spines promoting changes in synaptic structure and function. Neuron 35, 91–105.
Nakazawa, T., Watabe, A. M., Tezuka, T., Yoshida, Y., Yokoyama, K., Umemori, H., Inoue, A., Okabe, S., Manabe, T., and Yamamoto, T. (2003). p250GAP, a novel brain-enriched GTPase-activating protein for Rho family GTPases, is involved in the N-methyl-d-aspartate receptor signaling. Mol. Biol. Cell 14, 2921–2934.
Nichols, N. R., Zieba, M., and Bye, N. (2001). Do glucocorticoids contribute to brain aging? Brain Res. Brain Res. Rev. 37, 273–286.
Nishino, J., Kim, I., Chada, K., and Morrison, S. J. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135, 227–239.
Nudelman, A. S., Dirocco, D. P., Lambert, T. J., Garelick, M. G., Le, J., Nathanson, N. M., and Storm, D. R. (2010). Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 20, 492–498.
Oberdoerffer, P., Michan, S., Mcvay, M., Mostoslavsky, R., Vann, J., Park, S. K., Hartlerode, A., Stegmuller, J., Hafner, A., Loerch, P., Wright, S. M., Mills, K. D., Bonni, A., Yankner, B. A., Scully, R., Prolla, T. A., Alt, F. W., and Sinclair, D. A. (2008). SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135, 907–918.
Okabe, T., Nakamura, T., Nishimura, Y. N., Kohu, K., Ohwada, S., Morishita, Y., and Akiyama, T. (2003). RICS, a novel GTPase-activating protein for Cdc42 and Rac1, is involved in the beta-catenin-N-cadherin and N-methyl-D-aspartate receptor signaling. J. Biol. Chem. 278, 9920–9927.
Oomen, C. A., Mayer, J. L., De Kloet, E. R., Joels, M., and Lucassen, P. J. (2007). Brief treatment with the glucocorticoid receptor antagonist mifepristone normalizes the reduction in neurogenesis after chronic stress. Eur. J. Neurosci. 26, 3395–3401.
Perron, M. P., and Provost, P. (2009). Protein components of the microRNA pathway and human diseases. Methods Mol. Biol. 487, 369–385.
Petersen, C. P., Bordeleau, M. E., Pelletier, J., and Sharp, P. A. (2006). Short RNAs repress translation after initiation in mammalian cells. Mol. Cell 21, 533–542.
Prozorovski, T., Schulze-Topphoff, U., Glumm, R., Baumgart, J., Schroter, F., Ninnemann, O., Siegert, E., Bendix, I., Brustle, O., Nitsch, R., Zipp, F., and Aktas, O. (2008). Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 10, 385–394.
Radley, J. J., and Morrison, J. H. (2005). Repeated stress and structural plasticity in the brain. Ageing Res. Rev. 4, 271–287.
Rafalski, V. A., and Brunet, A. (2011). Energy metabolism in adult neural stem cell fate. Prog. Neurobiol. 93, 182–203.
Ramirez-Amaya, V., Marrone, D. F., Gage, F. H., Worley, P. F., and Barnes, C. A. (2006). Integration of new neurons into functional neural networks. J. Neurosci. 26, 12237–12241.
Rao, M. S., Hattiangady, B., and Shetty, A. K. (2006). The window and mechanisms of major age-related decline in the production of new neurons within the dentate gyrus of the hippocampus. Aging Cell 5, 545–558.
Rogelj, B., Hartmann, C. E., Yeo, C. H., Hunt, S. P., and Giese, K. P. (2003). Contextual fear conditioning regulates the expression of brain-specific small nucleolar RNAs in hippocampus. Eur. J. Neurosci. 18, 3089–3096.
Sahay, A., Wilson, D. A., and Hen, R. (2011). Pattern separation: a common function for new neurons in hippocampus and olfactory bulb. Neuron 70, 582–588.
Saito, K., Nishida, K. M., Mori, T., Kawamura, Y., Miyoshi, K., Nagami, T., Siomi, H., and Siomi, M. C. (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222.
Sauvageot, C., Dahia, P. L., Lipan, O., Park, J. K., Chang, M. S., Alberta, J. A., and Stiles, C. D. (2005). Distinct temporal genetic signatures of neurogenic and gliogenic cues in cortical stem cell cultures. J. Neurobiol. 62, 121–133.
Schoenenberger, P., Gerosa, D., and Oertner, T. G. (2009). Temporal control of immediate early gene induction by light. PLoS ONE 4, e8185. doi:10.1371/journal.pone.0008185
Schratt, G. M., Tuebing, F., Nigh, E. A., Kane, C. G., Sabatini, M. E., Kiebler, M., and Greenberg, M. E. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289.
Selbach, M., Schwanhausser, B., Thierfelder, N., Fang, Z., Khanin, R., and Rajewsky, N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63.
Siegel, G., Saba, R., and Schratt, G. (2011). microRNAs in neurons: manifold regulatory roles at the synapse. Curr. Opin. Genet. Dev. 21, 491–497.
Sierra, A., Encinas, J. M., Deudero, J. J., Chancey, J. H., Enikolopov, G., Overstreet-Wadiche, L. S., Tsirka, S. E., and Maletic-Savatic, M. (2010). Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7, 483–495.
Sinor, A. D., and Lillien, L. (2004). Akt-1 expression level regulates CNS precursors. J. Neurosci. 24, 8531–8541.
Smalheiser, N. R., Lugli, G., Thimmapuram, J., Cook, E. H., and Larson, J. (2011). Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA 17, 166–181.
Small, S. A., Chawla, M. K., Buonocore, M., Rapp, P. R., and Barnes, C. A. (2004). Imaging correlates of brain function in monkeys and rats isolates a hippocampal subregion differentially vulnerable to aging. Proc. Natl. Acad. Sci. U.S.A. 101, 7181–7186.
Small, S. A., Schobel, S. A., Buxton, R. B., Witter, M. P., and Barnes, C. A. (2011). A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 12, 585–601.
Small, S. A., Tsai, W. Y., Delapaz, R., Mayeux, R., and Stern, Y. (2002). Imaging hippocampal function across the human life span: is memory decline normal or not? Ann. Neurol. 51, 290–295.
Smith, C. C., Vedder, L. C., Nelson, A. R., Bredemann, T. M., and Mcmahon, L. L. (2010a). Duration of estrogen deprivation, not chronological age, prevents estrogen’s ability to enhance hippocampal synaptic physiology. Proc. Natl. Acad. Sci. U.S.A. 107, 19543–19548.
Smith, L. K., Shah, R. R., and Cidlowski, J. A. (2010b). Glucocorticoids modulate microRNA expression and processing during lymphocyte apoptosis. J. Biol. Chem. 285, 36698–36708.
Smith, C. M., and Steitz, J. A. (1998). Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 18, 6897–6909.
Smrt, R. D., Szulwach, K. E., Pfeiffer, R. L., Li, X., Guo, W., Pathania, M., Teng, Z. Q., Luo, Y., Peng, J., Bordey, A., Jin, P., and Zhao, X. (2010). MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070.
Sober, S., Laan, M., and Annilo, T. (2010). MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochem. Biophys. Res. Commun. 391, 727–732.
Somel, M., Guo, S., Fu, N., Yan, Z., Hu, H. Y., Xu, Y., Yuan, Y., Ning, Z., Hu, Y., Menzel, C., Hu, H., Lachmann, M., Zeng, R., Chen, W., and Khaitovich, P. (2010). MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 20, 1207–1218.
Song, H., Kempermann, G., Overstreet Wadiche, L., Zhao, C., Schinder, A. F., and Bischofberger, J. (2005). New neurons in the adult mammalian brain: synaptogenesis and functional integration. J. Neurosci. 25, 10366–10368.
Sousa, N., and Almeida, O. F. (2002). Corticosteroids: sculptors of the hippocampal formation. Rev. Neurosci. 13, 59–84.
Stone, S. S., Teixeira, C. M., Devito, L. M., Zaslavsky, K., Josselyn, S. A., Lozano, A. M., and Frankland, P. W. (2011). Stimulation of entorhinal cortex promotes adult neurogenesis and facilitates spatial memory. J. Neurosci. 31, 13469–13484.
Strum, J. C., Johnson, J. H., Ward, J., Xie, H., Feild, J., Hester, A., Alford, A., and Waters, K. M. (2009). MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 23, 1876–1884.
Sun, G., Ye, P., Murai, K., Lang, M. F., Li, S., Zhang, H., Li, W., Fu, C., Yin, J., Wang, A., Ma, X., and Shi, Y. (2011). miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2, 529.
Sun, W., Winseck, A., Vinsant, S., Park, O. H., Kim, H., and Oppenheim, R. W. (2004). Programmed cell death of adult-generated hippocampal neurons is mediated by the proapoptotic gene Bax. J. Neurosci. 24, 11205–11213.
Szulwach, K. E., Li, X., Smrt, R. D., Li, Y., Luo, Y., Lin, L., Santistevan, N. J., Li, W., Zhao, X., and Jin, P. (2010). Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141.
Taft, R. J., Glazov, E. A., Cloonan, N., Simons, C., Stephen, S., Faulkner, G. J., Lassmann, T., Forrest, A. R., Grimmond, S. M., Schroder, K., Irvine, K., Arakawa, T., Nakamura, M., Kubosaki, A., Hayashida, K., Kawazu, C., Murata, M., Nishiyori, H., Fukuda, S., Kawai, J., Daub, C. O., Hume, D. A., Suzuki, H., Orlando, V., Carninci, P., Hayashizaki, Y., and Mattick, J. S. (2009). Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 41, 572–578.
Taniguchi, S., Liu, H., Nakazawa, T., Yokoyama, K., Tezuka, T., and Yamamoto, T. (2003). p250GAP, a neural RhoGAP protein, is associated with and phosphorylated by Fyn. Biochem. Biophys. Res. Commun. 306, 151–155.
Tashiro, A., Sandler, V. M., Toni, N., Zhao, C., and Gage, F. H. (2006). NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature 442, 929–933.
Tognini, P., Putignano, E., Coatti, A., and Pizzorusso, T. (2011). Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nat. Neurosci. 14, 1237–1239.
Toner, C. K., Pirogovsky, E., Kirwan, C. B., and Gilbert, P. E. (2009). Visual object pattern separation deficits in nondemented older adults. Learn. Mem. 16, 338–342.
Toni, N., Laplagne, D. A., Zhao, C., Lombardi, G., Ribak, C. E., Gage, F. H., and Schinder, A. F. (2008). Neurons born in the adult dentate gyrus form functional synapses with target cells. Nat. Neurosci. 11, 901–907.
Toni, N., and Sultan, S. (2011). Synapse formation on adult-born hippocampal neurons. Eur. J. Neurosci. 33, 1062–1068.
Toni, N., Teng, E. M., Bushong, E. A., Aimone, J. B., Zhao, C., Consiglio, A., Van Praag, H., Martone, M. E., Ellisman, M. H., and Gage, F. H. (2007). Synapse formation on neurons born in the adult hippocampus. Nat. Neurosci. 10, 727–734.
Vasudevan, S., Tong, Y., and Steitz, J. A. (2008). Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle 7, 1545–1549.
Verbitsky, M., Yonan, A. L., Malleret, G., Kandel, E. R., Gilliam, T. C., and Pavlidis, P. (2004). Altered hippocampal transcript profile accompanies an age-related spatial memory deficit in mice. Learn. Mem. 11, 253–260.
Viola, A. U., Chellappa, S. L., Archer, S. N., Pugin, F., Gotz, T., Dijk, D. J., and Cajochen, C. (2011). Interindividual differences in circadian rhythmicity and sleep homeostasis in older people: effect of a PER3 polymorphism. Neurobiol. Aging. doi: 10.1016/j.neurobiolaging.2011.10. [Epub ahead of print].
Vo, N., Klein, M. E., Varlamova, O., Keller, D. M., Yamamoto, T., Goodman, R. H., and Impey, S. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16426–16431.
Vreugdenhil, E., Verissimo, C. S., Mariman, R., Kamphorst, J. T., Barbosa, J. S., Zweers, T., Champagne, D. L., Schouten, T., Meijer, O. C., De Kloet, E. R., and Fitzsimons, C. P. (2009). MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology 150, 2220–2228.
Wang, Q., Joels, M., Swaab, D. F., and Lucassen, P. J. (2011a). Hippocampal GR expression is increased in elderly depressed females. Neuropharmacology 62, 527–533.
Wang, W. X., Huang, Q., Hu, Y., Stromberg, A. J., and Nelson, P. T. (2011b). Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 121, 193–205.
Wayman, G. A., Davare, M., Ando, H., Fortin, D., Varlamova, O., Cheng, H. Y., Marks, D., Obrietan, K., Soderling, T. R., Goodman, R. H., and Impey, S. (2008). An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. U.S.A. 105, 9093–9098.
Wolkowitz, O. M., Burke, H., Epel, E. S., and Reus, V. I. (2009). Glucocorticoids. Mood, memory, and mechanisms. Ann. N. Y. Acad. Sci. 1179, 19–40.
Wright, M. W., and Bruford, E. A. (2011). Naming ‘junk’: human non-protein coding RNA (ncRNA) gene nomenclature. Hum. Genomics 5, 90–98.
Wu, H., Tao, J., Chen, P. J., Shahab, A., Ge, W., Hart, R. P., Ruan, X., Ruan, Y., and Sun, Y. E. (2010). Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 107, 18161–18166.
Wu, L., Fan, J., and Belasco, J. G. (2006). MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U.S.A. 103, 4034–4039.
Wu, W., Brickman, A. M., Luchsinger, J., Ferrazzano, P., Pichiule, P., Yoshita, M., Brown, T., Decarli, C., Barnes, C. A., Mayeux, R., Vannucci, S. J., and Small, S. A. (2008). The brain in the age of old: the hippocampal formation is targeted differentially by diseases of late life. Ann. Neurol. 64, 698–706.
Yamagata, K., Fujiyama, S., Ito, S., Ueda, T., Murata, T., Naitou, M., Takeyama, K., Minami, Y., O’Malley, B. W., and Kato, S. (2009). Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol. Cell 36, 340–347.
Yamashima, T., Tonchev, A. B., and Yukie, M. (2007). Adult hippocampal neurogenesis in rodents and primates: endogenous, enhanced, and engrafted. Rev. Neurosci. 18, 67–82.
Yao, Y., Li, H., Gu, Y., Davidson, N. E., and Zhou, Q. (2010). Inhibition of SIRT1 deacetylase suppresses estrogen receptor signaling. Carcinogenesis 31, 382–387.
Yen, S. S. (2001). Dehydroepiandrosterone sulfate and longevity: new clues for an old friend. Proc. Natl. Acad. Sci. U.S.A. 98, 8167–8169.
Yin, H., and Lin, H. (2007). An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450, 304–308.
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., Lee-Messer, C., Dolmetsch, R. E., Tsien, R. W., and Crabtree, G. R. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231.
Yu, X., and Malenka, R. C. (2003). Beta-catenin is critical for dendritic morphogenesis. Nat. Neurosci. 6, 1169–1177.
Zhao, C., Deng, W., and Gage, F. H. (2008). Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660.
Zhao, C., Sun, G., Li, S., Lang, M. F., Yang, S., Li, W., and Shi, Y. (2010). MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 1876–1881.
Zhao, C., Sun, G., Li, S., and Shi, Y. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371.
Zhao, C., Teng, E. M., Summers, R. G. Jr., Ming, G. L., and Gage, F. H. (2006). Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J. Neurosci. 26, 3–11.
Keywords: microRNA, neural stem cells, neurodegeneration, steroid hormones, adult neurogenesis, hippocampus, cognitive decline
Citation: Schouten M, Buijink MR, Lucassen PJ and Fitzsimons CP (2012) New neurons in aging brains: molecular control by small non-coding RNAs. Front. Neurosci. 6:25. doi: 10.3389/fnins.2012.00025
Received: 13 October 2011;
Accepted: 30 January 2012;
Published online: 17 February 2012.
Edited by:
Yanhong Shi, City of Hope, USAReviewed by:
Alejandro F. Schinder, Leloir Institute, ArgentinaCopyright: © 2012 Schouten, Buijink, Lucassen and Fitzsimons. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Carlos P. Fitzsimons, Center for Neuroscience, Swammerdam Institute for Life Sciences, University of Amsterdam, Room C3-271, Science Park 904, 1098 XH Amsterdam, Netherlands. e-mail:Yy5wLmZpdHpzaW1vbnNAdXZhLm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.