Approaches to manipulating microRNAs in neurogenesis
- 1 Department of Cell and Developmental Biology, Weill Medical College of Cornell University, New York, NY, USA
- 2 Weill Cornell Graduate School of Medical Sciences, New York, NY, USA
The specification of motor neuron (MN) subtypes and columnar organization in developing spinal cord is controlled by multiple transcription factors. FoxP1 drives specification of lateral motor neuron (LMN) subtypes, and we demonstrated in our previous work that FoxP1 expression levels are regulated by the microRNA miR-9. Here we show that ectopic FoxP1 expression in the chick spinal cord can rescue Lhx3 and Hb9 expression in MNs altered by miR-9 over-expression, demonstrating that FoxP1 is a critical functional interaction partner for miR-9 in LMN development. Moreover, we have optimized a technique called a miRNA sponge in vitro, to permit easy discovery of the role of individual miRNA in vivo using a loss-of-function approach. We here show that narrow spacing between binding sites, inclusion of a coding gene, and optimizing the number of miRNA binding sites can significantly increase the blocking ability of a sponge. We go on to show that a miR-9 sponge reduces detectable miR-9 in the ventral horn, preventing miR-9 silencing of FoxP1 in vivo, and in turn modifies MN subtypes in the spinal cord. Our designs for optimized sponges provide a knockdown tool that is ready to be used to study the function of miRNA in vivo, and in particular for generating transgenic animal models.
Introduction
In developing spinal cords, motor neurons (MNs) are specified from progenitors under the control of cross-interactions of multiple transcription factors (Jessell, 2000; Goulding, 2009). MN subtypes are further organized into columns that project axons into distinct target muscles. For instance, medial motor column (MMC) neurons innervate axial muscles, and lateral motor columns (LMC) innervate limb muscles (Dasen et al., 2003, 2005; Shah et al., 2004; Wu et al., 2008; Jung et al., 2010). Previous studies have shown that proper expression levels of FoxP1 are critical for determining LMC or MMC MN fate (Dasen et al., 2008; Rousso et al., 2008). Furthermore, our own work indicates that microRNA (miRNA) miR-9 plays a role in optimizing FoxP1 expression levels and subsequently modifying MN subtypes (Otaegi et al., 2011).
MicroRNAs are a class of ~22 nucleotides (nt) endogenous non-coding RNAs (Lee et al., 1993; Wightman et al., 1993). Mature miRNAs bind to complementary sequences in the 3′-untranslated region (3′-UTR) of target messenger RNAs (mRNAs) and negatively regulate these targets by degrading the mRNA or repressing its translation (Carthew and Sontheimer, 2009; Kim et al., 2009). Emerging studies have shown that miRNAs have a broad impact on embryogenesis (Huang et al., 2011; Suh and Blelloch, 2011), tumor formation (Esquela-Kerscher and Slack, 2006; Zhang et al., 2007), and are implicated in numerous human diseases (Huang et al., 2011).
Examining the functions of specific miRNAs, particularly using genetic tools, has been challenging. The functions of several miRNAs have been demonstrated by generating miRNA knockout mice (van Rooij et al., 2007; Xiao et al., 2007; Ventura et al., 2008; Wang et al., 2008). However, many miRNAs are transcribed from multiple loci, or have families with similar sequences. To avoid functional compensation, one has to make double or triple knockout animals, which is costly and technically challenging. Furthermore, many miRNAs are transcribed or cleaved from intragenic regions, complicating efforts to target them for deletion. Alternative approaches are required to investigate miRNA functions, particularly in animal models.
A recent technological advance called the miRNA “sponge” has been shown to block miRNA activity in both cultured cells and animal models (Ebert et al., 2007; Gentner et al., 2008; Loya et al., 2009; Ebert and Sharp, 2010). A miRNA sponge is an exogenous transcript that contains multiple binding sites complementary to a mature miRNA. The miRNA sponge can bind to endogenous miRNA and block that miRNA’s silencing activity. Optimized sponge design should block the activity of a whole miRNA family that consists of identical or nearly identical isoforms of the mature miRNA.
To examine miR-9 function in MN subtype formation, we have developed optimized sponges to effectively block miR-9 silencing activity in vitro and in vivo. We show that a shorter spacing between binding sites and the inclusion of a coding gene in the transcript increase the efficacy of the sponge, as does an optimized number of binding sites. Moreover, we have found that miR-9 sponge can repress the function of exogenous or endogenous miR-9 in vivo, and can subsequently modify MN subtypes. Our studies provide a guideline for investigating miRNA functions in cellular and animal models using optimized miRNA sponge tools.
Materials and Methods
DNA Constructs
For luciferase experiments, Gga-miR-9-1, Gga-miR-9-1-Mut, and FoxP1 3′-UTR Luciferase constructs were used as described (Otaegi et al., 2011). Briefly, a 289 nt genomic sequence including the gga-miR-9-1 precursor was cloned into the pCAGIG expression vector (mature miR-9 sequence: 5′-TCTTTGGTTATCTAGCTGTATGA). The mutated version is identical except for three altered nucleotides in the seed sequences of mature miR-9 and three matching nucleotides in the opposite side of the precursor stem-loop (mutant miR-9 sequence: 5′-TCCTTAGGTATCTAGCTGTATGA). The FoxP1 3′-UTR construct contains ~1.8 kb of the chick FoxP1 3′-UTR including two central predicted miR-9 binding sites.
For sponge oligo design, binding sites were designed as the reverse complement of the full length mature miRNA sequence with one deletion and two mismatches to create a bulge as described in Gentner et al. (2008). Spacer regions of different size were designed using the siRNA Wizard scrambled design tool (http://www.sirnawizard.com/scrambled.php). Forward and reverse oligos were ordered with one or two binding sites and spacers of different lengths (Table 1). Oligos were annealed and ligated together in pBSIIKS+ (Stratagene) at SpeI/XbaI sites. Constructs were sub-cloned into XbaI to finally contain 6, 12, or 24 binding sites. These sponges were then cloned into pCDNA3.1+ (Invitrogen) or attached to iCre as a 3′-UTR and then cloned into pCDNA3.1+ for luciferase experiments. For in vivo electroporation experiments, all constructs were cloned into the pCAGIG expression vector.
Luciferase Assays
Neuro2a cells were transfected using Lipofectamine 2000 (Invitrogen) using the manufacturer’s protocol. Plasmids were quantified by UV spectrophotometry and used for transfection in a 3:1 ratio (miRNA:target luciferase constructs; Figure 2C); 9:3:1 ratio (sponge:miRNA:target luciferase constructs, Figures 2D and 3B,D); 1.5:3:1 ratio (sponge:miRNA:target luciferase constructs in Figure 3F). pGL4.13 firefly luciferase (Promega) was used for 3′-UTRs of targets. pGL4.73 renilla luciferase (Promega) was used as a transfection control. Luciferase was measured using the Dual-Luciferase Reporter Assay kit (Promega) using the manufacturer’s protocol and read on a Victor3 1420 multilabel counter (Perkin Elmer). Results are expressed as Firefly luciferase normalized to Renilla.
In ovo Electroporation
Electroporation was performed as previously described (Otaegi et al., 2011). Briefly, Hamburger and Hamilton (HH) stage 10–12 chick embryos were electroporated unilaterally (five 50-ms pulses at 25 V) with 1–3 μg/μl DNA or as indicated, using an ECM830 electro-squareporator (BTX). Embryos were analyzed 24 h later at HH20, or 48 h later at HH24.
Tissue Preparation and Immunohistochemistry
Spinal cord tissues from the chick embryos were fixed in 4% paraformaldehyde (PFA) in PBS overnight, incubated in 30% sucrose in PBS, embedded in OCT, and stored at −80°C until use. The spinal cords were sectioned (14–18 μm) using a cryostat.
For antigen recovery, sections were incubated in heated (95–100°C) antigen recovery solution (1 mM EDTA and 5 mM Tris, pH 8.0) for 20 min and cooled down for 20–30 min. Before applying antibodies, sections were blocked in 10% normal goat serum in PBS with 0.1% Tween 20 for 1 h. Sections were incubated with primary antibodies at 4°C overnight and visualized using goat anti-rabbit IgG Alexa Fluor 488, goat anti-mouse IgG Alexa Fluor 594, or anti-guinea pig IgG Alexa Fluor 594 (1:350; Invitrogen) for 1 h at room temperature. Images were captured using a Leica digital camera under a fluorescent microscope (Leica). Primary antibodies against the following antigens were used: Lhx3 (1:20, Developmental Studies Hybridoma Bank), FoxP1 (1:100; kindly provided by Dr. B. Novitch, University of California, Los Angeles, Los Angeles, CA, USA), green fluorescent protein (GFP; 1:500; Rockland), and HB9 (1:20; Developmental Studies Hybridoma Bank).
Motor Neuron Quantification
The total number of labeled motor neurons on electroporated and non-electroporated sides of the spinal cord was quantified from at least four sections (14–16 μm) showing strong eGFP expression collected from at least five electroporated embryos. Data is shown as the difference in total number of a specific MN subtype between the electroporated and non-electroporated side of the spinal cord. Statistical significance between experimental and control groups was analyzed by two-tailed t-test. Data were shown as mean ± SEM.
FoxP1 fluorescence intensity was measured in at least three sections from each of at least three embryos. At HH stages 20 and 24, FoxP1 is expressed only in the ventral horn throughout the cervical, thoracic, and lumbar regions of the spinal cord. Fluorescence intensity was measured using ImageJ (NIH) in identically sized regions encompassing all FoxP1+ neurons in the ventral horn and recorded as the intensity of the electroporated side divided by that of the non-electroporated side of each sample.
Results
FoxP1 is a Specific Target for MiR-9 in Motor Neuron Development
Our previous work showed that miR-9 regulates MN subtypes by targeting chick FoxP1 (cFoxP1) via two miR-9 binding sites in its 3′-UTR (Figure 1A; Otaegi et al., 2011). Over-expression of miR-9 significantly decreased numbers of FoxP1+ MNs but increased Lhx3+ and HB9+ MNs in chick spinal cords, electroporated at HH stages 10–11, and analyzed at stage 24 (Figures 1B,C,F,G). As previously reported, we observed a reduction in the number of FoxP1+ cells, and corresponding increases in the number of Lhx3+ and Hb9+ cells on the electroporated side of the spinal cord. If FoxP1 is the critical target of miR-9, then restoring the FoxP1 coding sequence (CDS) alone, without its miR-9-sensitive 3′-UTR in miR-9 over-expressing spinal cords should rescue altered MN subtypes. Co-electroporating miR-9 with a small amount of FoxP1 CDS was able to rescue miR-9’s effects on MN production, as detected by a reduced increase in the number of Lhx3+ and HB9+ MNs, compared to miR-9 alone (Figures 1D,F,G). We next electroporated FoxP1 CDS alone. Consistent with previous results (Dasen et al., 2008; Rousso et al., 2008), over-expression of FoxP1 CDS reduced Lhx3+ and HB9+ MNs (Figures 1E,H). Our results indicate that miR-9 and FoxP1 have opposite effects on MN production and FoxP1 CDS specifically rescues miR-9 effects on MN subtypes.
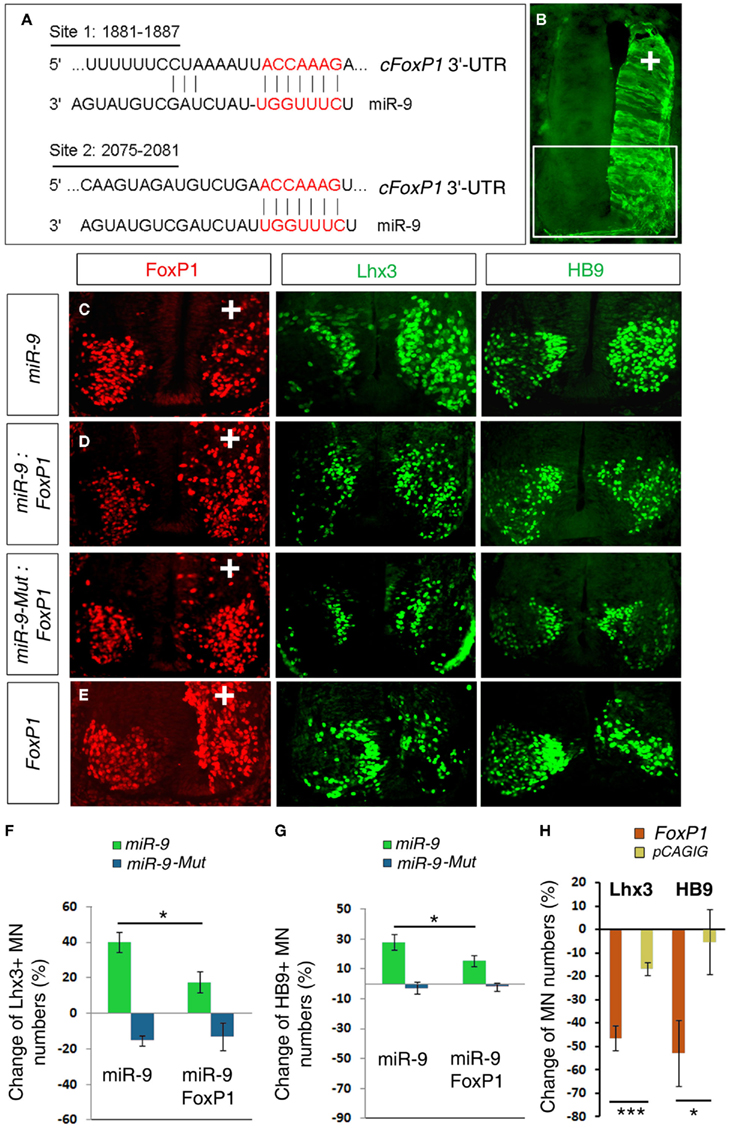
Figure 1. miR-9 regulates MN subtypes by a specific interaction with FoxP1 in chick spinal cords. (A) Schematic of miR-9 binding sites in the 3′-UTR of chick FoxP1 (cFoxP1). The 3′-UTR of cFoxP1 is 3877 nt with miR-9 binding sites at positions 1881 and 2075. (B) In ovo electroporation performed in chick spinal cords at HH stages 10–11 and analyzed at stage 24. The electroporated side can be visualized by a reporter eGFP expression. In all subsequent images the highlighted area in the spinal cord is shown and “+” indicates the electroporated side of the spinal cord. (C) Over-expression of miR-9 caused reduced numbers of FoxP1+ and increased numbers of Lhx3+ and HB9+ MNs. (D) Adding FoxP1 using co-electroporation of miR-9 and FoxP1 coding sequence (CDS) restored numbers of Lhx3+ and HB9+ MNs closer to control numbers. Co-electroporation of inactive mutated-miR-9 (miR-9-Mut) and FoxP1 caused similar phenotypes to expression of FoxP1 alone. (E) Over-expression of FoxP1 caused a significant reduction in Lhx3+ and HB9+ MNs. (F,G) Quantification of MN numbers affected by miR-9 or miR-9-Mut alone or co-expressed with FoxP1 CDS. n > 5, Lhx3, *p < 0.02; HB9, *p < 0.03. (H) Quantification of MN marker changes due to FoxP1 over-expression. n > 5, ***p < 0.002, *p < 0.05.
Blocking miR-9 Silencing Activities Using a miRNA Sponge
MicroRNA sponges, transcripts with complementary sequence to mature miRNAs, have been used as a loss-of-function approach to study miRNA function in vitro and in vivo (Ebert et al., 2007; Gentner et al., 2008; Loya et al., 2009; Ebert and Sharp, 2010). Previous work has shown that sponges containing bulged miRNA binding sites can block a miRNA from silencing its endogenous targets (Figure 2A).
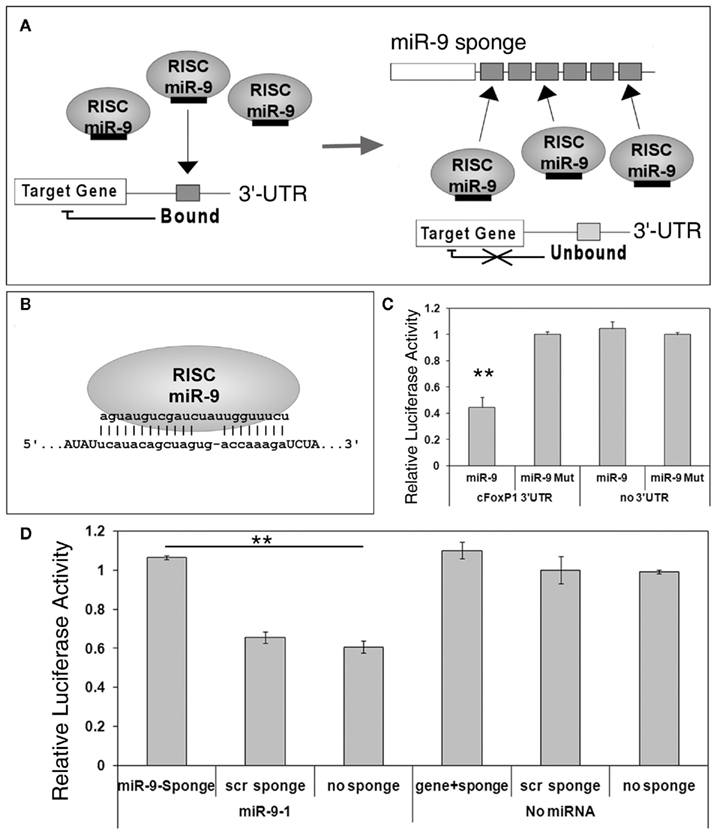
Figure 2. miR-9 sponges prevent miR-9 from silencing its targets. (A) A scheme of miRNA sponge function. Under normal circumstances, miRNA such as miR-9, loaded in RNA induced silencing complexes (RISCs), will bind to their endogenous targets and silence target genes. In the presence of sponge transcripts, however, miR-9 will bind to the exogenous binding sites and be unavailable for silencing endogenous targets. (B) A miR-9 sponge binding site, designed with an internal bulge, permits a specific interaction with endogenous miR-9. (C) FoxP1 3′-UTR luciferase activity was significantly reduced when co-expressed with miR-9, but not miR-9 mutation (miR-9-Mut). n = 3, **p < 0.002. (D) miR-9 sponges, but not sponges with scrambled binding sites (scr sponge), were able to relieve repression of miR-9 on FoxP1 3′-UTR, as detected by increased luciferase activity. n = 3, **p < 0.001.
To block miR-9 silencing activities in vitro and in vivo, we designed a complementary sequence called miR-9 sponge, which contains no other predicted miRNA binding sequences (Figure 2B; Table 1). We first tested miR-9 targeting effects on FoxP1 3′-UTR using luciferase assays. The 3′-UTR of cFoxP1 was cloned into a luciferase construct. The activity of luciferase was greatly reduced by miR-9, but not by a miR-9 construct with three deactivating mutations in the seed sequence (miR-9-Mut), when the luciferase construct contained the cFoxP1 3′-UTR (Figure 2C). Luciferase constructs containing no 3′-UTR were unaffected by miR-9. Our results indicate that miR-9 specifically targets FoxP1 3′-UTR.
Since miRNA sponges are able to block miRNA repression of a target gene, we next co-transfected miR-9 and miR-9 sponge in a luciferase assay. The miR-9 sponge transcript containing six closely spaced (6 nt spacer) binding sites was able to rescue the knockdown effect of miR-9 on FoxP1 3′-UTR, while a similarly designed sponge construct with scrambled binding sites had no effect (Figure 2D). These results indicate that miRNA sponges can be used to block miRNA silencing activities on target genes.
Optimization of miR-9 Sponge Design to Block miR-9 Silencing Activity
In order to achieve the most effective and repeatable knockdown of endogenous miRNA activities in vitro and in vivo using miRNA sponges, we optimized sponge design using miR-9 sponge as an example.
Existing sponges have used 6–8 nt separation space between miRNA binding sites. A recent study has shown that the RNA induced silencing complex (RISC) provides RNase protection for 46–62 nt in the 3′-UTR of target mRNAs (Chi et al., 2009). This suggests that using increased spacing between miRNA binding sites in sponge design might allow more RISCs to bind and thus increase miRNA knockdown effects. To test this, we generated miR-9 sponge constructs with 6, 29, and 42 nt spacers separating miR-9 binding sites (the spacers contain no miRNA binding sites). These spacers created binding sites of different lengths for the RISC: 28, 51, and 64 nt respectively (Figure 3A). Luciferase assays demonstrated that all three sponge constructs were able to interfere with miR-9 induced silencing activity (Figure 3B). There was no discernable blockade of miR-9 silencing from sponges with scrambled binding sites. Opposite to our prediction, using a shorter spacer between binding sites in miR-9 sponges generated a stronger blockade of miR-9 activities.
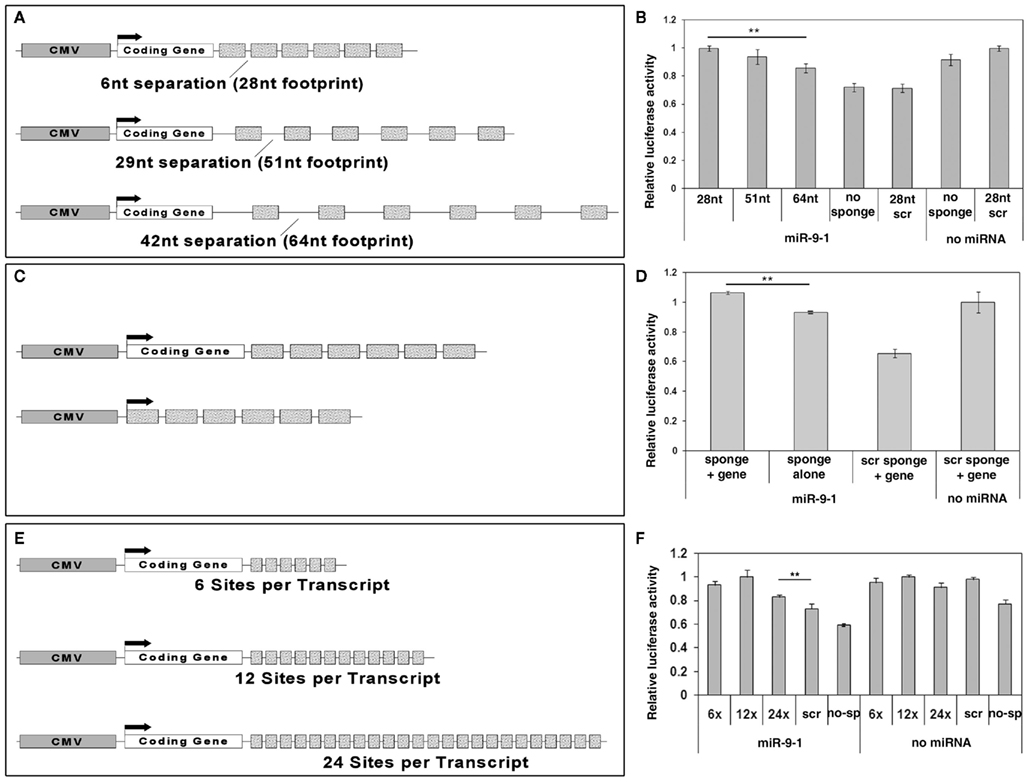
Figure 3. Optimization of miRNA sponge designs. (A) A scheme of miR-9 sponge constructs with different spacers (separation) in between binding sites. The CMV promoter was used to drive sponge expression. 6, 29, or 42 nucleotides (nt) spacers with a 22 nt binding site permit a 28, 51, or 64 nt region respectively for one RISC to bind. (B) All miR-9 sponges blocked miR-9 silencing activity on FoxP1 3′-UTR in luciferase assays. Shorter spacers provided stronger blockade. n > 6, **p < 0.005. (C) Schematics of miR-9 sponge constructs with or without a coding gene. (D) Sponge constructs with a coding gene showed significantly stronger blockade than those without. n = 3, **p < 0.001. (E) Schematics of miR-9 sponge constructs with 6, 12, or 24 binding sites. (F) All sponges showed significant rescue with the optimal number of binding sites being 12. n = 3, **p < 0.001.
Some sponge designs have put miRNA binding sites in the 3′-UTR of a coding gene (typically a reporter such as GFP), while others have simply generated a non-coding transcript. To test whether a coding gene is able to maximize sponge function, we generated miR-9 sponge constructs with and without a coding gene, here using improved Cre (iCre) as an example. Using the most effective 6 nt spacer sponges we tested relative luciferase activity using iCre-miR-9 sponge or miR-9 sponge alone (Figure 3C). miR-9 sponge alone was able to interfere with miR-9 silencing effects. But the presence of the coding gene iCre resulted in stronger rescue (Figure 3D).
Previous studies have shown blocking effects using different numbers of binding sites in miRNA sponges, typically ranging from 4 to 16 binding sites (Ebert and Sharp, 2010). To test whether the number of binding sites in a sponge may affect the sponge’s blocking ability, we generated constructs containing 6, 12, or 24 binding sites per sponge transcript (Figure 3E). miR-9 sponge with 12 binding sites showed higher rescue effect on miR-9 silencing activity than sponges with 6 binding sites. miR-9 sponge with 24 binding sites did not display the highest rescue effect (Figure 3F). Moreover, when a higher concentration of sponges was used, all three constructs were able to fully rescue miR-9 silencing activity (data not shown). These results suggest that the optimal number of binding sites is greater than 6, and somewhere around 12.
Taken together these results show that a miR-9 sponge optimized for the most efficient function in vitro contains more than 6 but fewer than 24 binding sites, short spacers between binding sites, and is expressed as the 3′-UTR of a coding gene.
miR-9 Sponge Blocks miR-9 Activities in MN Subtype Specification
To test whether miR-9 sponge functions in vivo, we examined the effect of miR-9 and miR-9 sponge on FoxP1 expression levels and MN subtype development in the chick spinal cord. A sponge containing six miR-9 binding sites with the shortest spacers or a similar scrambled control sponge were first co-electroporated with miR-9 in the chick neural tube at stage HH10–12 and analyzed at stage HH 24. Consistent with our previous results, over-expressing miR-9 with control sponges caused a significant reduction in the number of FoxP1+ MNs and subsequently a great increase of Lhx3+ and HB9+ MNs (Figures 4A,E). Interestingly, co-electroporation of miR-9 sponge and miR-9 was able to partially rescue the reduction of FoxP1+ MN numbers and resulted in milder changes of Lhx3+ and HB9+ MNs, suggesting that miR-9 sponge can block exogenous miR-9 silencing activities on the target gene FoxP1 (Figures 4B,E). miR-9 sponge and scrambled control sponge were next co-electroporated with miR-9-Mut construct in the spinal cord. Neither miR-9-Mut nor scrambled sponge displayed significant activity on FoxP1, Lhx3, or HB9 expression (Figures 4C,E). However, miR-9 sponge caused an increase in FoxP1 expression levels, as detected by FoxP1 fluorescent signal intensity on the electroporated side of the spinal cord compared to the non-electroporated side (Figure 4D). At the same time, slightly fewer Lhx3+ MNs were detected (Figure 4E). Furthermore, in situ hybridization for miR-9 showed that detectable miR-9 was reduced in the ventral horn on the electroporated side of the spinal cord in the presence of miR-9 sponge (Figure 4F). Together, these data suggest that the sponge may be leading to a reduced concentration of mature miR-9, permitting higher expression of FoxP1 and altering specification of Lhx3+ spinal MNs.
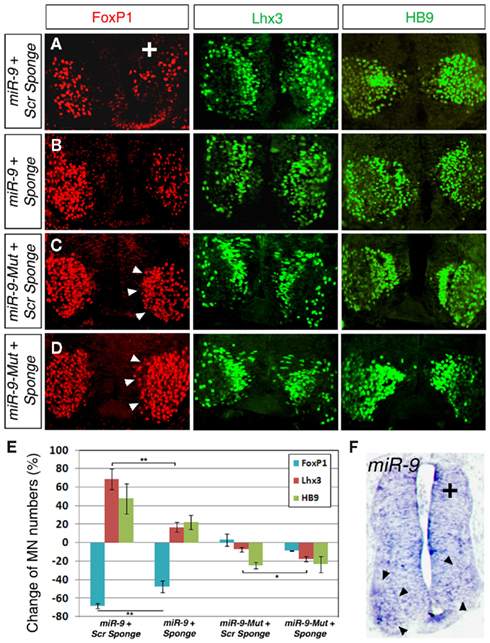
Figure 4. miR-9 sponges block miR-9 silencing activity in specifying MN subtypes in developing spinal cord. (A) Co-electroporation of a scrambled (scr) sponge with miR-9 caused a decrease in numbers of FoxP1+ MNs and corresponding increase in Lhx3+ and HB9+ MNs. (B) Co-electroporation of a miR-9 sponge with miR-9 partially rescued the silencing effects of miR-9, as detected by increased numbers of FoxP1+ MNs and decreased numbers of Lhx3+ MNs. Numbers of HB9+ MNs appeared to be less increased. (C) Co-electroporation of a scrambled sponge with mutated-miR-9 (miR-9-Mut) caused no changes in MN numbers or FoxP1 intensity (arrowheads). (D) Co-electroporation of a miR-9 sponge with miR-9-Mut was able to block endogenous miR-9 and caused increased FoxP1 expression levels (arrowheads). (E) Quantification of MN numbers affected by miR-9 sponge or scrambled sponges when co-electroporated with miR-9 or miR-9-Mut. n > 5, FoxP1, **p < 0.007; Lhx3 (with miR-9), **p < 0.006; Lhx3 (with miR-9-Mut), *p < 0.04. (F) Electroporation of miR-9 sponge alone. In situ hybridization for miR-9 detected miR-9 in the ventral horn (arrowheads) of the non-electroporated side, but was unable to detect miR-9 on the electroporated side.
To further examine this finding, chick embryos were electroporated with mir-9 sponge alone or scrambled sponge alone at low (0.5 μg/μl) and high (1.5 μg/μl) concentrations, evaluated at HH 20, a time when FoxP1 expression is present, but endogenous miR-9 is weak or absent; and at HH 24, when both are expressed (Otaegi et al., 2011). FoxP1 fluorescent intensity was quantified in a region containing all FoxP1+ MNs in the ventral horn on the electroporated versus non-electroporated side. FoxP1 intensity was not changed by sponges on the electroporated side at HH 20, when miR-9 is weak or absent (Figures 5A,C,I). At HH 24, a low concentration of miR-9 Sponge was unable to create an effect (Figures 5E,I), but at high concentration, FoxP1 intensity was significantly increased (Figures 5G,I). Scrambled sponge showed no effect in any condition (Figures 5B,D,F,H,I). This demonstrates that ectopic expression of miR-9 sponge blocks endogenous miR-9 activity in a concentration dependent manner, and releases its silencing of target gene FoxP1.
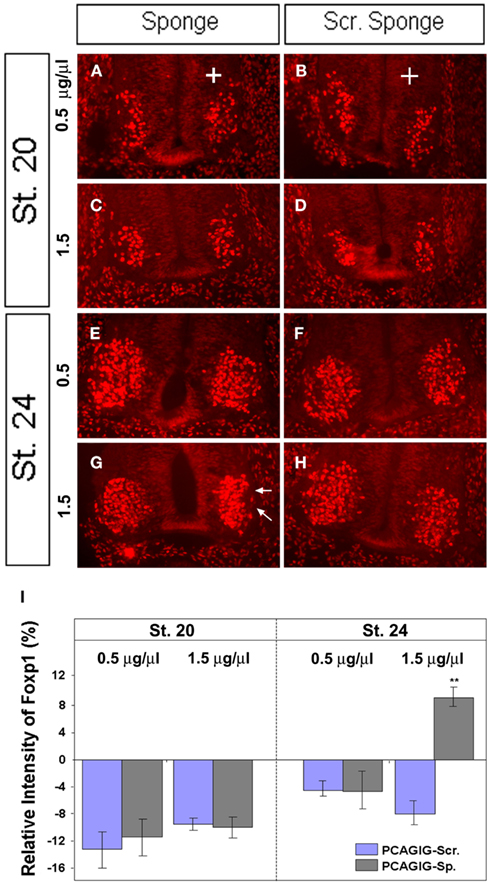
Figure 5. miR-9 sponges release repression by endogenous miR-9 in a concentration dependent manner. (A + B) Electroporation of miR-9 sponges or scrambled sponges at low concentration (0.5 μg/μl) showed no difference at HH 20. (C + D) Electroporation of miR-9 sponges or scrambled sponges at high concentration (1.5 μg/μl) showed no difference at HH 20. (E + F) Electroporation of miR-9 sponges or scrambled sponges at low concentration showed no difference at HH 24. (G + H) Electroporation of miR-9 or scrambled sponges at high concentration. miR-9 sponge showed a significant increase in FoxP1 signal intensity at HH 24 (arrows). (I) Quantification of pixel intensity on the electroporated side compared to the non-electroporated side of the same spinal cord section. n > 5, **p < 0.01.
Together, these results indicate that miR-9 sponge reduces miR-9 presence, effectively blocking its silencing activity in vivo and revealing the role of endogenous miR-9 in MN fate decisions (Figure 6).
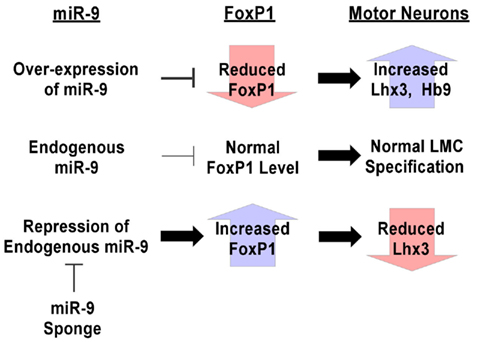
Figure 6. Summary of techniques for manipulation of miRNA expression levels in vivo. Over-expression of miR-9 silences endogenous FoxP1, causing increases in numbers of Lhx3+ and HB9+ MNs, demonstrating the ability of miR-9 to target those pathways. Conversely, miR-9 sponges are able to block miR-9 silencing of endogenous FoxP1, causing increased FoxP1 expression levels and a reduction in Lhx3+ MNs. These design principles can be used for generating tools to investigate the roles of miRNA in systems to which they can be introduced.
Discussion
Designing strategies to manipulate miRNA expression levels in vivo is critical to understanding their function. In this study we have optimized the design of miRNA sponges that can be used to block miRNA silencing activity in vitro and in vivo. We have used these sponges to block the function of miR-9 in the chick spinal cord, revealing miR-9’s role as a direct regulator of FoxP1 expression and MN development. Our results are consistent with previous reports showing that FoxP1 upregulation leads to altered specification of the LMC (Dasen et al., 2008; Rousso et al., 2008). Furthermore, our results provide a useful tool for further examination of miRNA function using loss-of-function approaches in animal models.
Accumulating evidence has demonstrated the importance of miRNAs in normal development and under disease conditions. Our previous work and this study have shown that the fate determination of MN subtypes in the spinal cord is controlled by both transcription factors and interactions between miRNAs and target genes (Otaegi et al., 2011). FoxP1 expression level is important for specifying LMC MNs. It appears that the proper protein output of FoxP1 is tuned by miR-9, which has overlapping expression with FoxP1 in LMC MNs. Over-expression of miR-9 silences FoxP1 translation and causes a reduction in LMC MNs (Otaegi et al., 2011). In this study, we have developed a miRNA sponge tool to block endogenous miR-9 activity. Blocking endogenous miR-9 allows stronger expression of FoxP1 and a mild reduction in Lhx3-expressing MNs. miR-9 expression mostly overlaps with FoxP1 expressing MNs in the LMC, so it is not unexpected that the LMC MN fate is not grossly altered by blockade of miR-9. Nevertheless, the subtle changes we see demonstrate that we are able to increase and decrease the activity of a specific miRNA in animal tissues to study its function in vivo (Figure 6).
Investigating the role of miRNA in development using a loss-of-function approach is a powerful tool. Modified oligos antisense to the mature miRNAs have been successfully transfected into cultured cells (Meister et al., 2004; Cheng, 2005). A similar technique using tiny locked nucleic acids (LNAs) antisense to the mature miRNA has shown some promise in animal models, but the oligos cannot reach some tissues, most notably the central nervous system (CNS; Obad et al., 2011). Cell permeable antisense oligos called antagomirs have also been used to study short term loss of miRNA functions in many animal tissues. However, like tiny LNAs, antagomirs will not reach cells in the brain due to the blood–brain barrier (Krützfeldt et al., 2005).
MicroRNA sponges offer a promising technique for gaining insight into miRNA functions in vitro and in vivo. Many model systems would readily express sponges, transcripts with sequence complementary to a specific miRNA. The sponges are able to reduce the amount of detectable miRNA in the cell, preventing the miRNA from regulating its putative targets. Due to the highly similar sequences of mature miRNAs from different precursors, a single miRNA sponge can allow blockade of a whole family of miRNAs. Moreover, because miRNA sponges can be easily cloned into plasmid DNA, sponges allow transfection in cells, electroporation of tissues, and generation of transgenic animals to block miRNA activity in vivo. For example, a miR-223 sponge was introduced into mouse hematopoietic stem cells using a viral transgene insertion (Gentner et al., 2008). Also, conditional transgenic flies were generated by ectopic expression of miR-8 sponges driven by a UAS/Gal4 system (Loya et al., 2009). Plasmid based sponges do have some of the delivery limitations of the oligo-based approaches. However, because they are not chemically modified, sponges can be expressed as transgenes, electroporated into tissues, or infected into cultured cells for sustained expression.
Early attempts at sponge technology have used a fragment of a target gene’s 3′-UTR, or a transcript containing a number of sites designed to be targeted by the miRNA (Ebert and Sharp, 2010). We have worked to improve on sponge designs to generate the strongest blockade of miRNA induced silencing using miR-9 as an example. We have hypothesized that due to the RISC footprint previously published in RNase protection assays (Chi et al., 2009), a larger spacer in between binding sites could provide more efficient rescue. Our results tell a different story, however, with shorter spacing producing stronger miRNA blockade. This could be due to changes in the outcome of miRNA binding. With high complementarity to the miRNA, it is likely that sponges get cleaved and degraded not long after being bound by one or a few miRNA. Perhaps with the binding sites so close together, there is some folding required to permit more RISCs to bind, which prevents or delays sponge transcript degradation. This could permit longer sequestration of a larger number of mature miRNAs.
Moreover, we have predicted that attaching sponge binding sites to the 3′-UTR of a coding gene would provide stronger miRNA blockade than just as a free-floating transcript. As we expected, this is the case. This result may be due to increased stability of the transcript, or an improved ability to be transported to the cytoplasm where most silencing takes place (Ohrt et al., 2008). Finally, we considered whether adding more binding sites per sponge transcript would allow stronger blockade. This also is the case up to a point. It appears that a medial number of binding sites, such as 12 copies, is sufficient to significantly block miRNA silencing activity in vitro. More binding sites do not seem to show stronger blockade. Perhaps as predicted in previous work, more binding sites per transcript might lead to faster sponge degradation and lower blocking ability (Ebert and Sharp, 2010). On the other hand, we have found that a construct with only a few binding sites can fully rescue miRNA silencing activity when it is expressed at a high concentration. Therefore, to achieve high blockade of miRNA activities, a high expression level of miRNA sponges is required in cells and in tissues.
All of the designs we show here are able to partially relieve miR-9 silencing in vitro, but some designs showed stronger blockade than others. Achieving the strongest possible blockade is critical for in vivo applications, and one of our designs is able to block endogenous miR-9 activity in vivo. miRNA’s seed sequence based targeting mechanism is fairly well conserved, so this result suggests that our design could work for most miRNAs in most systems. Once properly designed, a sponge can be constitutively expressed in specific tissues in animal models using electroporation, or a wealth of genetic tools, such as the Cre/loxP system in mice, to achieve functional blockade of specific miRNAs in a spatial and temporal manner opening up a new way to investigate miRNA functions in development and diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Sun laboratory for their valuable discussions and advice. This work was supported by the Ellison Medical Foundation (Tao Sun), an award from the Hirschl/Weill-Caulier Trust (Tao Sun), and an R01MH083680 grant from the NIH/NIMH (Tao Sun).
References
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655.
Cheng, A. M. (2005). Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 33, 1290–1297.
Chi, S. W., Zang, J. B., Mele, A., and Darnell, R. B. (2009). Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460, 479–486.
Dasen, J. S., De Camilli, A., Wang, B., Tucker, P. W., and Jessell, T. M. (2008). Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 134, 304–316.
Dasen, J. S., Liu, J. P., and Jessell, T. M. (2003). Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926–933.
Dasen, J. S., Tice, B. C., Brenner-Morton, S., and Jessell, T. M. (2005). A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123, 477–491.
Ebert, M. S., Neilson, J. R., and Sharp, P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726.
Ebert, M. S., and Sharp, P. A. (2010). MicroRNA sponges: progress and possibilities. RNA 16, 2043–2050.
Esquela-Kerscher, A., and Slack, F. J. (2006). Oncomirs – microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269.
Gentner, B., Schira, G., Giustacchini, A., Amendola, M., Brown, B. D., Ponzoni, M., and Naldini, L. (2008). Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods 6, 63–66.
Goulding, M. (2009). Circuits controlling vertebrate locomotion: moving in a new direction. Nat. Rev. Neurosci. 10, 507–518.
Huang, Y., Shen, X., Zou, Q., Wang, S., Tang, S., and Zhang, G. (2011). Biological functions of microRNAs: a review. J. Physiol. Biochem. 67, 129–139.
Jessell, T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29.
Jung, H., Lacombe, J., Mazzoni, E. O., Liem, K. F. Jr., Grinstein, J., Mahony, S., Mukhopadhyay, D., Gifford, D. K., Young, R. A., Anderson, K. V., Wichterle, H., and Dasen, J. S. (2010). Global control of motor neuron topography mediated by the repressive actions of a single Hox gene. Neuron 67, 781–796.
Kim, V. N., Han, J., and Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139.
Krützfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., and Stoffel, M. (2005). Silencing of microRNAs in vivo with ‘antagomirs.’ Nature 438, 685–689.
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854.
Loya, C. M., Lu, C. S., Van Vactor, D., and Fulga, T. A. (2009). Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat. Methods 6, 897–903.
Meister, G., Landthaler, M., Dorsett, Y., and Tuschl, T. (2004). Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10, 544–550.
Obad, S., Dos Santos, C. O., Petri, A., Heidenblad, M., Broom, O., Ruse, C., Fu, C., Lindow, M., Stenvang, J., Straarup, E. M., Hansen, H. F., Koch, T., Pappin, D., Hannon, G. J., and Kauppinen, S. (2011). Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 43, 371–378.
Ohrt, T., Mutze, J., Staroske, W., Weinmann, L., Hock, J., Crell, K., Meister, G., and Schwille, P. (2008). Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 36, 6439–6449.
Otaegi, G., Pollock, A., Hong, J., and Sun, T. (2011). MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J. Neurosci. 31, 809–818.
Rousso, D. L., Gaber, Z. B., Wellik, D., Morrisey, E. E., and Novitch, B. G. (2008). Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron 59, 226–240.
Shah, V., Drill, E., and Lance-Jones, C. (2004). Ectopic expression of Hoxd10 in thoracic spinal segments induces motoneurons with a lumbosacral molecular profile and axon projections to the limb. Dev. Dyn. 231, 43–56.
Suh, N., and Blelloch, R. (2011). Small RNAs in early mammalian development: from gametes to gastrulation. Development 138, 1653–1661.
van Rooij, E., Sutherland, L. B., Qi, X., Richardson, J. A., Hill, J., and Olson, E. N. (2007). Control of stress-dependent cardiac growth and gene expression by a MicroRNA. Science 316, 575–579.
Ventura, A., Young, A. G., Winslow, M. M., Lintault, L., Meissner, A., Erkeland, S. J., Newman, J., Bronson, R. T., Crowley, D., Stone, J. R., Jaenisch, R., Sharp, P. A., and Jacks, T. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17~92 family of miRNA clusters. Cell 132, 875–886.
Wang, S., Aurora, A. B., Johnson, B. A., Qi, X., Mcanally, J., Hill, J. A., Richardson, J. A., Bassel-Duby, R., and Olson, E. N. (2008). The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271.
Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862.
Wu, Y., Wang, G., Scott, S. A., and Capecchi, M. R. (2008). Hoxc10 and Hoxd10 regulate mouse columnar, divisional and motor pool identity of lumbar motoneurons. Development 135, 171–182.
Xiao, C., Calado, D. P., Galler, G., Thai, T.-H., Patterson, H. C., Wang, J., Rajewsky, N., Bender, T. P., and Rajewsky, K. (2007). MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131, 146–159.
Keywords: miRNAs, miR-9, FoxP1, motor neurons, spinal cord, miRNA sponges
Citation: Otaegi G, Pollock A and Sun T (2012) An optimized sponge for microRNA miR-9 affects spinal motor neuron development in vivo. Front. Neurosci. 5:146. doi: 10.3389/fnins.2011.00146
Received: 02 November 2011;
Paper pending published: 07 December 2011;
Accepted: 19 December 2011;
Published online: 05 January 2012.
Edited by:
Yanhong Shi, City of Hope, USAReviewed by:
Jenny Hsieh, University of Texas Southwestern Medical Center, USABenedikt Berninger, Ludwig-Maximilians-Universität München, Germany
Copyright: © 2012 Otaegi, Pollock and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Andrew Pollock and Tao Sun, Weill Cornell Graduate School of Medical Sciences, 1300 York Avenue, Box 60, New York, NY 10065, USA. e-mail: anp2016@med.cornell.edu; tas2009@med.cornell.edu
 Gaizka Otaegi1
Gaizka Otaegi1