- 1 Department of Animal and Human Biology, University of Turin, Turin, Italy
- 2 Neuroscience Institute Cavalieri Ottolenghi, Orbassano, Italy
Current advances in imaging techniques have extended the possibility of visualizing small structures within large volumes of both fixed and live specimens without sectioning. These techniques have contributed valuable information to study neuronal plasticity in the adult brain. However, technical limits still hamper the use of these approaches to investigate neurogenic regions located far from the ventricular surface such as parenchymal neurogenic niches, or the scattered neuroblasts induced by brain lesions. Here, we present a method to combine confocal laser scanning microscopy (CLSM) and serial section reconstruction in order to reconstruct large volumes of brain tissue at cellular resolution. In this method a series of thick sections are imaged with CLSM and the resulting stacks of images are registered and 3D reconstructed. This approach is based on existing freeware software and can be performed on ordinary laboratory personal computers. By using this technique we have investigated the morphology and spatial organization of a group of doublecortin (DCX)+ neuroblasts located in the lateral striatum of the late post-natal guinea pig. The 3D study unraveled a complex network of long and poorly ramified cell processes, often fascicled and mostly oriented along the internal capsule fiber bundles. These data support CLSM serial section reconstruction as a reliable alternative to the whole mount approaches to analyze cyto-architectural features of adult germinative niches.
Introduction
Newly generated neuronal cells are constantly added to the mammalian olfactory bulb (OB) and dentate gyrus (DG) during adult life (Kriegstein and Alvarez-Buylla, 2009). Comparative analyses in different mammalian species either in physiologic or pathologic conditions, indicate that neurogenesis can also occur in other brain regions (Lindsey and Tropepe, 2006; Gould, 2007). Nonetheless, out of the OB and DG neurogenic niches, most of adult-generated neurons show short survival life span and elusive identity. In this context, a detailed description of newborn neurons morphology and neuroanatomical organization in both physiological and pathological conditions is a fundamental step to understand their identity and function.
The study of neuroanatomy has been classically carried out through histological sections cut with different orientations and thickness. Although stereological methods contribute much information on the intact 3D structures, the global architecture is mostly lost in sectioned samples. To retrieve information over this higher anatomical level, the structures of interest must be either entirely imaged or reconstructed from serially sectioned material. Recent advances in light microscopy and in molecular and genetic manipulations have greatly extended the possibility of imaging large volumes of both fixed and live neural tissue at cellular resolution, enabling the visualization of complex 3D objects such as neuronal or vascular networks (Mizrahi, 2007; Lu et al., 2009; Tsai et al., 2009; Wilt et al., 2009; Khairy and Keller, 2011). These imaging techniques represent a pivotal innovation for multiple neuroanatomical fields ranging from the definition of comprehensive maps of neuronal connections, the so called “connectome,” to the study of neuronal plasticity (Hofer et al., 2009; DeFelipe, 2010). Nonetheless, the relatively limited depth at which objects can be observed with conventional microscopy, and the difficulties of using immunohistochemistry in large tissue volumes, still hamper a general use of these techniques among brain regions and animal species. Given the periventricular location of the subventricular zone (SVZ) neurogenic niche, whole mount preparations have been used in mice to deal with several architectural aspects of this germinative region, ranging from the orientation of the chains of migrating neuroblasts to the spatial organization of progenitor cells (Doetsch and Alvarez-Buylla, 1996; Tavazoie et al., 2008; Mirzadeh et al., 2010). However, the whole mount preparation is less suitable to study neurogenic regions located far from the ventricular surface such as parenchymal neurogenic niches (Luzzati et al., 2006, 2007), or scattered neuroblasts induced by a brain lesion (Arvidsson et al., 2002; Parent et al., 2002). In these latter cases, reconstruction from serial sections represents a reliable alternative. In the classic version of this technique, supported by several commercial and freeware software, planar images are taken from an ordered series of sections and digitally registered. Individual structures can be then converted into virtual 3D objects through different segmentation methods (Fiala, 2005; Cardona et al., 2010). A major drawback of reconstructing from serial sections is that since each slice is represented by a single image, the z-axis resolution directly depends on the slice thickness. Accordingly, serial section reconstruction at cellular or sub-cellular resolution is usually performed by registering electron microscopy (EM) images (Fiala, 2005; Cardona et al., 2010). This technique provided much information on the composition of SVZ and DG adult germinative niches (Doetsch et al., 1997; Seri et al., 2004) and on the organization of chains of neuroblasts in the rabbit brain parenchyma (Luzzati et al., 2003). However, limiting the section thickness critically increases the number of sections to acquire and register, and thus it considerably lengthens the reconstruction of a given volume (Cardona et al., 2010). A possible implementation of this approach can be obtained matching the use of laser scanning confocal microscopy (CLSM) to serial section reconstruction, thus allowing the use of thick sections without losing resolution along the z-axis. To date, only a few attempts have been made to use CLSM stacks instead of planar images in serial section reconstructions but they were not employed to perform reconstructions at cellular resolution (Capek et al., 2009).
Here, we describe a new method in which serial section reconstruction is combined to CLSM in order to produce multi scale 3D reconstructions of large volumes of brain tissue at cellular resolution. The in silico part of the analysis is performed on standard laboratory personal computers (PC) and it is based on the use of freeware software including: serial section editors, volume integration of tiled optical sections, image analysis and object tracing in large confocal data sets. In this paper, we apply this method to characterize the morphology and the organization of a population of DCX+ neuroblasts located in the ventro-lateral caudate putamen complex of the guinea pig. The obtained results support the CLSM serial section reconstruction method as a useful tool to analyze neuroanatomical features of adult germinative niches.
Materials and Methods
Experiments were conducted in accordance with current European Union and Italian law, under authorization of the Italian Ministry of Health number 66/99-A. All experiments were designed to minimize the numbers of animals used and their discomfort.
Results and data-analyses here presented were obtained from two female albino Dunkin-Hartley guinea pig (Cavia porcellus) 50 days old that was purchased from a local breeder. The animal was deeply anesthetized with a ketamine/xylazine solution (100 and 33 mg/kg body weight, respectively) and transcardially perfused with ice-cold saline solution (0.9% NaCl), followed by a freshly prepared solution of 4% paraformaldehyde (PFA) plus 2% picric acid in 0.1 M sodium phosphate buffer, pH 7.4. Brains were then postfixed overnight, cryoprotected, frozen at −80°C, and cryostat 40 μm sectioned along a coronal plane in four series.
Free-floating sections were incubated for 48 h at 4°C in a solution of 0.01 M PBS, pH 7.4, containing 1% Triton X-100, normal horse serum and either anti-DCX antibody 1:500 (goat polyclonal sc-8066; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or myelin associated glycoprotein (MAG; mouse monoclonal, Chemicon international, Billerica, MA, USA). After rinsing in PBS solution, sections were incubated with an anti-goat or anti-mouse cyanine 3 (Cy3)-conjugated (1:800; Jackson ImmunoResearch, West Grove, PA, USA). Sections were then coverslipped with antifade mounting medium Dabco (Sigma) and analyzed with a laser scanning Olympus Optical (Milan, Italy) Fluoview confocal system (Olympus Optical). In one series of sections the anti-DCX antibodies were revealed with biotin–avidin system. In this case sections were rinsed in PBS, incubated with the anti-goat biotinylated secondary antibody for 1 h (1:250; Vector Laboratories, Burlingame, CA, USA), rinsed, and incubated in avidin–biotin complex (1:400; Vector Laboratories). The reaction product was visualized with 0.15 mg/ml 3,3-diaminobenzidine (DAB) in PBS with 0.01% H2O2. Sections were then serially mounted onto Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA, USA), air dried, dehydrated in graded alcohols, cleared in xylenes, and coverslipped using DPX mounting medium (Aldrich, Milwaukee, WI, USA).
Images (1024 × 1024) were taken with the following objectives (4×, NA 0.13; 10×, NA 0.3; 20×, NA 0.7; 100×, NA 1.3). The actual value of z in the voxel size of 20× and 100× CLSM reconstruction was calculated a posteriori by dividing the number of the optical planes for the total length along the z-axis. Considering that the z-step used for the acquisition of the 20× and 100× images were 2 and 0.7 μm respectively, we can calculate slice shrinkage of 66% along the z-axis.
Image analyses were conducted on a laptop DELL Vostro, Intel Core i5 M 520 2.40 GHz × 4, 5.87 GB RAM and running 64 bit version of Windows 7. General adjustments to color, contrast, and brightness were made with Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA, USA).
Results
In the brain of 2-month-old guinea pigs several doublecortin (DCX+) positive newborn neurons were identified along the central part of the external capsule (EC) and in surrounding regions of the lateral striatum (LS; Figure 1 and data not shown). These cells spanned about two millimeters along the antero-posterior axis, however most of them were found at the level of the decussation of the anterior commissure. At this level, the DCX+ cells occupied the entire dorso-ventral extent of the LS, while at more rostral and caudal positions they were restricted to the ventral EC.
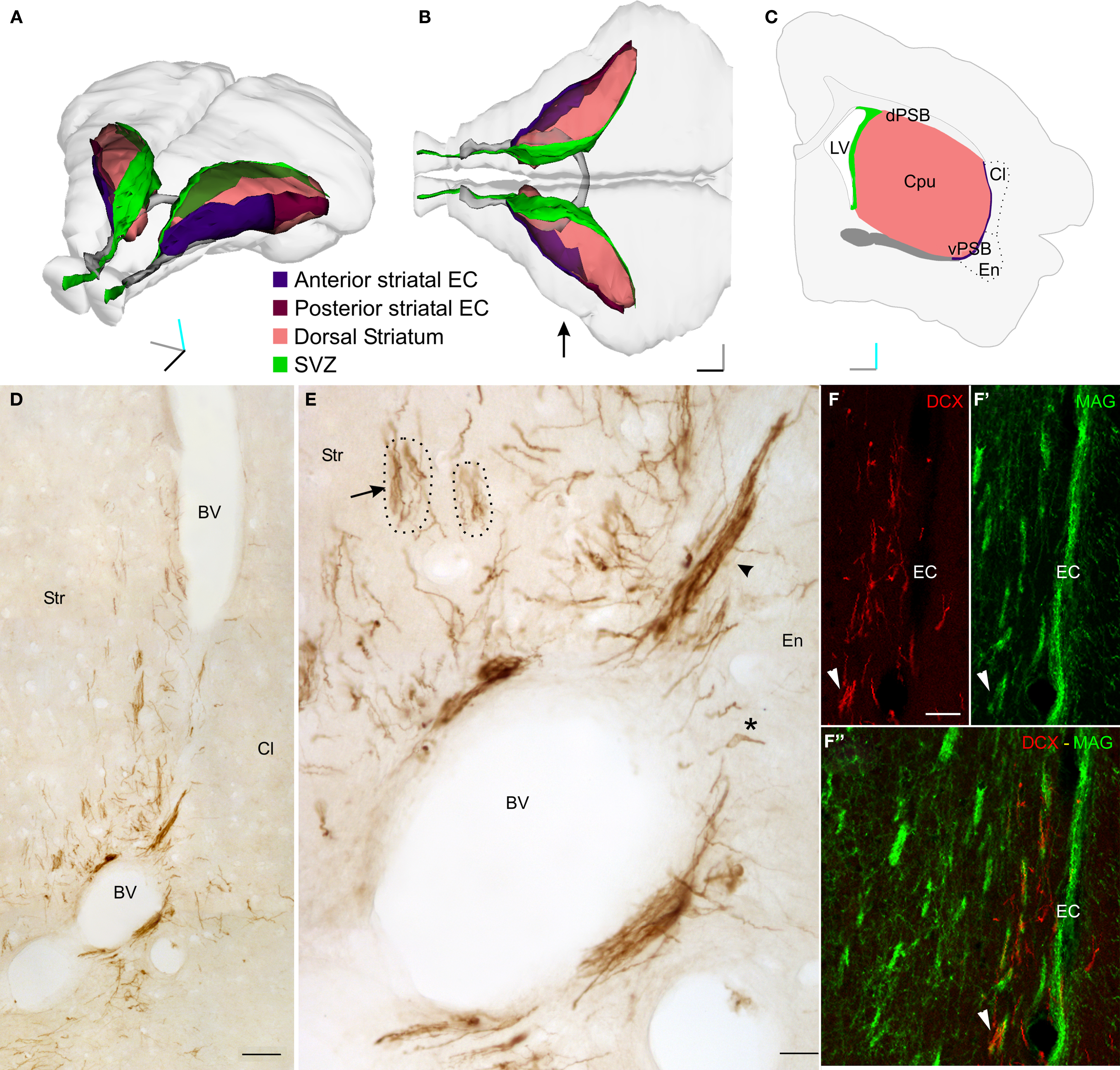
Figure 1. Dcx+ cells in the lateral striatum (LS). (A–B) Perspective (A) and top (B) views of a 3D reconstruction of the adult guinea pig brain showing the SVZ (green), the dorsal striatum (pink), the anterior commissure (gray), the striatal tract of the external capsule (anterior: violet; posterior: maroon). This reconstruction was produced with Reconstruct through coronal sections available from the University of Wisconsin and Michigan State Comparative Mammalian Brain Collections (www.brainmuseum.org). (C) Scheme of a single coronal section of the guinea pig brain, about 300 micrometer rostral to the decussation of the anterior commissure. (D,E) DCX staining in a coronal section of the LS. DCX positive cells can be organized either as individual (asterisk) or clustered (arrowheads) elements. In addition, several cell processes, often fascicled (arrow), can be observed. Dotted lines indicate the approximate limits of two internal capsule fiber bundles associated with fascicled DCX processes. (F–F“) Double labeling for MAG and DCX showing the close association between fascicles of DCX+ processes and internal capsule fiber bundles (arrowhead). Orientation Bars: Gray line: medial; cyan line: dorsal; dark line: rostral. BV: blood vessel; Cl: claustrum; dPSB: dorsal pallial subpallial boundary; vPSB: ventral pallial subpallial boundary; EC: external capsule; Str: Striatum; Scale bars: 80 μm in (D); 20 μm in (E); 50 μm in (F–F“).
A time course analysis with BrdU indicated that these cells were newly generated (data not shown).
In coronal sections containing the LS, the DCX immunostaining identified cells organized as cluster or individual elements (Figures 1D,E). Clustered elements were restricted to the ventral end of the EC, while individual elements were scattered along the EC and in the surrounding striatal parenchyma (Figures 1D,E). The cell processes of the individual elements were often fascicled and closely associated with the internal capsule fiber bundles (arrow in Figures 1E,F–F"). This neurogenic system, which will be described in detail in a separate study (Luzzati et al., unpublished results), offers a good opportunity to show how the method of CLSM serial section reconstruction we developed is suitable to investigate the distribution, organization, and morphological features of immunolabeled newborn neurons located deep within the brain parenchyma.
Choosing the Section Thickness and Cutting Plane
When planning a serial section reconstruction, firstly the section thickness must be chosen. The use of thick sections (60–100 μm) reduces the number of gaps in the reconstruction, as well as the number of sections to acquire and register. This leads to a more uniform and quickly performed reconstruction. At the same time, thick sections tend to deform after drying, especially near cavities like the lateral ventricles or large blood vessels. Furthermore, section thickness must not exceed the penetration capacity of the antibodies and must be consistent with the optical properties of the used lenses.
In our specific case we found 40 μm thickness as the best compromise to obtain limited deformations after drying, good antibody penetration and high signal to noise ratio of the acquired images. In addition, in order to increase antibody penetration, and to enable a better control of section uniformity, immunohistochemistry was performed on free-floating sections.
Another important point to be considered in serial section studies regards the selection of the cutting plane. In the case of the current reconstruction, the minimum number of sections containing all the DCX+ cells of the LS would be obtained by cutting parallel to the EC. However, by this way the fascicles of DCX+ processes would be nearly parallel to the slice surfaces, where small tissue loss and irregularities can occur. This increases the level of ambiguity in the identification of corresponding neurites in subsequent slices. Starting from these considerations, we chose to cut the brain along the coronal plane. This way, most of the DCX+ elements of the LS region were comprised within 25 coronal sections, covering a tract of 1 mm along the rostro-caudal axis.
Rough Reconstruction of DCX Staining in the LS by Conventional Serial Section Reconstruction (Voxel Size: 1.4 μm × 1.4 μm × 40 μm)
Before producing high-resolution CSLM serial sections reconstructions, it is usually convenient to produce a rough reconstruction in order to map the region of interest and to control the slices order. This can be done by conventional serial section reconstruction, in which each slice is represented by a single image spanning its entire thickness (Figures 2A,B). In our material, the use of 40 μm thick slices resulted in a relatively low resolution along the z-axis. This resolution was only suitable to reconstruct large objects, having cross-sectional profiles comparable to the section thickness, such as large blood vessels (Figure 3 gray and golden) or the DCX stained area as a whole (Figure 3 violet and red).
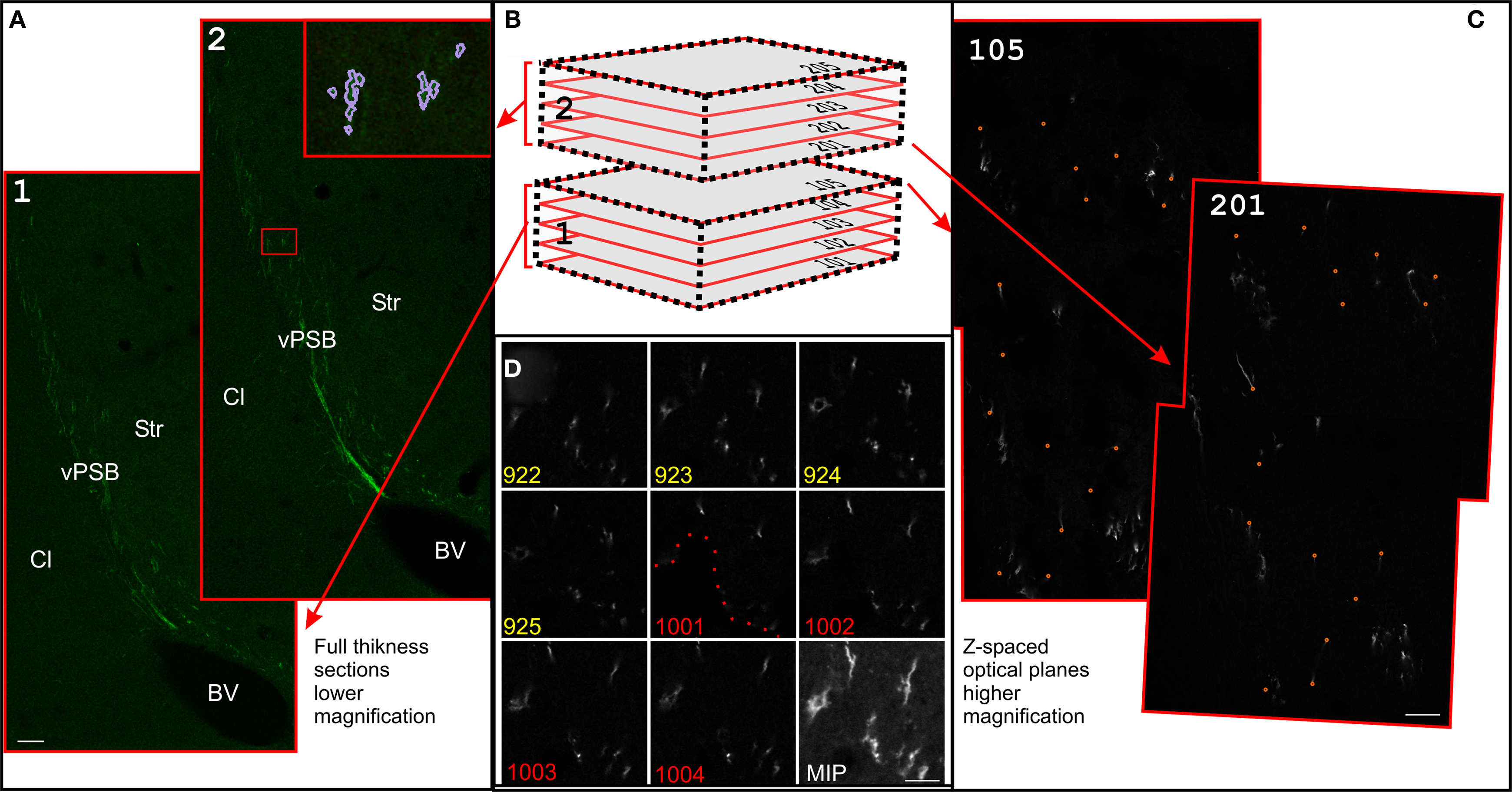
Figure 2. Serial section reconstructions of thick sections. (A) Full thickness projection of the DCX staining in two subsequent slices at low magnification (1.2). In the inset in 2, traces made in Reconstruct around the DCX staining are shown (violet). The original traces were thickened for clarity. Slices 1 and 2 are respectively slices 13 and 12 of the reconstruction shown in Figure 3. (B) Scheme of two subsequent thick slices. This material can be either entirely imaged (left) or re-sampled with CLSM in z-spaced thin optical planes (right). In the CLSM reconstruction these optical planes are numbered with a first digit indicating the slice number and subsequent digits indicating the optical plane number within each slice (101–205). (C) Superficial optical planes at the surface of subsequent slices at high magnification. Corresponding fiducial marks used for alignment are shown with small orange circles. Optical planes 105 and 201 are respectively optical plane 829 and 901 of the reconstruction shown in Figure 6. (D) Detail of the last four optical planes of slice number 9 (922–925), and first four optical planes of slice number 10 (1001–1004) of the 100× reconstruction shown in Figure 6. Dotted line show the border of a hollow at the surface of the section number 10. Scale Bars: 80 μm in (A); 7 μm in (C); 10 μm in (D).
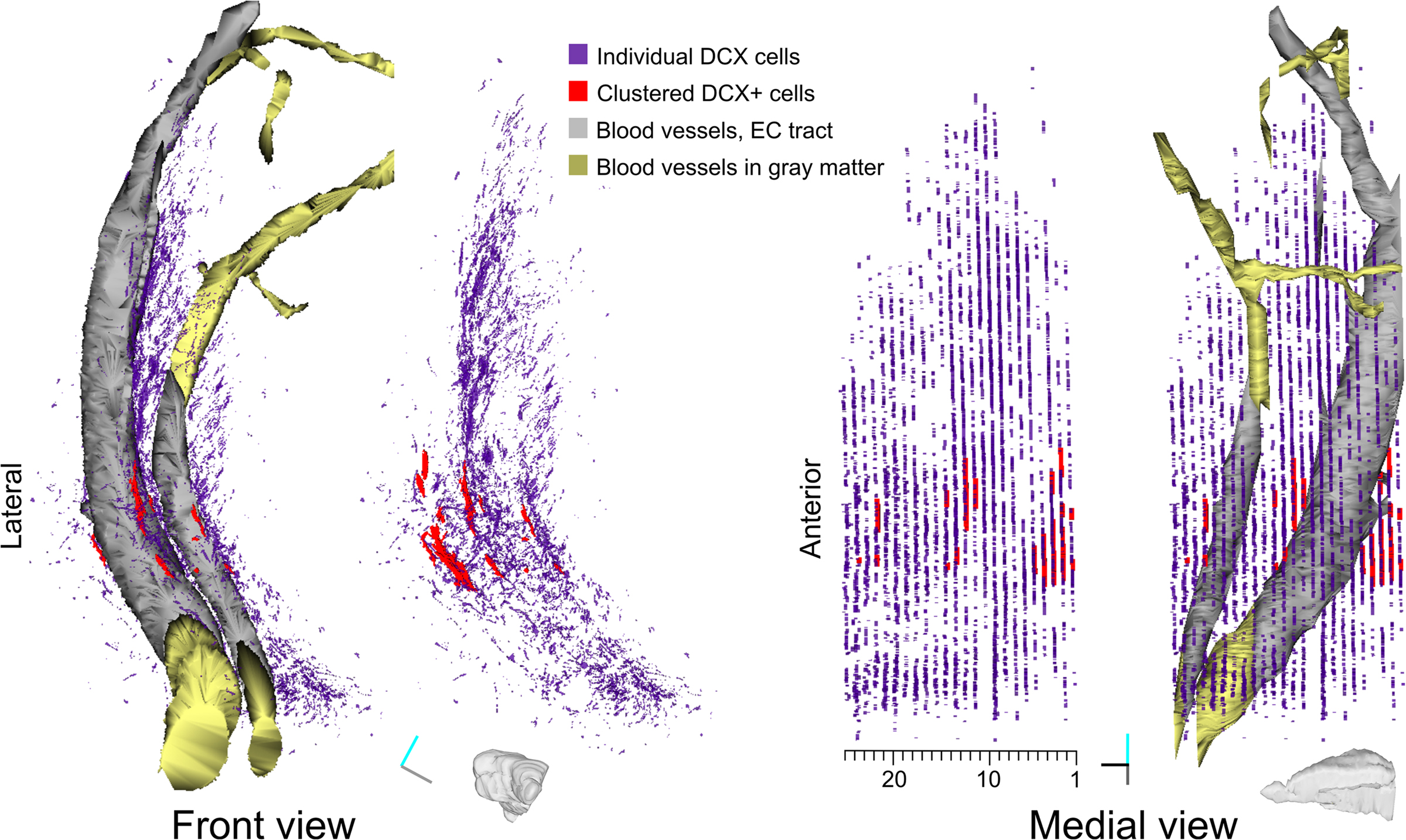
Figure 3. Reconstruction of DCX staining distribution in LS. Front and medial views of a 3D model of DCX staining distribution in the LS of guinea pig. For both views the model is shown either with or without blood vessels. The model is about 30° rotated medially, to give a more comprehensive visualization on the medial view. This model is the result of the tracing of individual DCX+ cells and processes (violet), clustered DCX+ cells (red), blood vessels passing through the EC (gray and gold). Tracts of the blood vessels running in the EC or in the gray matter are depicted as separate Boissonnat surfaces colored in gray and golden respectively. Both DCX+ staining and clustered DCX+ cells are represented as traces slabs. A scale indicating the number of individual sections in the reconstruction is shown under the medial view of the model. Orientation Bars: gray line: medial; cyan line: dorsal; dark line: rostral.
This reconstruction was quickly performed in Reconstruct1 (Fiala, 2005; Lu et al., 2009; see Table 1). This open source software was initially developed for EM serial section reconstruction, and subsequently implemented for light microscopy analyses. Reconstruct allows performing all the steps required for a serial section analysis: multi-fields montage (or stitching), section alignment (or registration), image segmentation and 3D representation as detailed below.
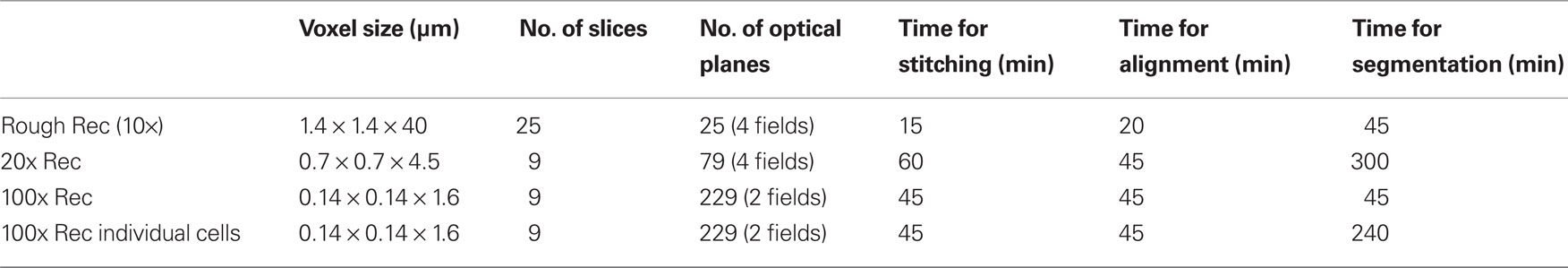
Table 1. Voxel size, number of slices, and optical planes, and approximate time required to perform stitching, alignment, and segmentation in each reconstruction described in this study.
Multi-field montage
Bi-dimensional images of the full section thickness were acquired on a CLSM with a 10× objective (pixel size 1.4 pixel/μm; Figure 2A). Since the size of the LS was larger than the microscope field of view, we collected multiple images per slice at each magnification. The montage (or stitching) of multiple fields in a single mosaic image was automatically performed using the 2D/3D stitching plugins bundled with Fiji, an image-processing package based on Image J2 (Preibisch et al., 2009). Alternatively, manual stitching can be performed directly in Reconstruct.
Section alignment
All stitched images were then imported in Reconstruct. In this software each image is represented as a domain that is part of a section. Each section has a specific number indicating its position into a series of sections. After importing, adjacent sections were randomly rotated and translated with respect to each other. Reconstruct offers several possibilities to perform section alignment, which include both rigid-body (translation, rotation) and non-linear transformations (slant, scaling, deformation, and bending). All these operations can be performed manually, or based on imposed fiducial marks.
Since each slice has its unique texture and is independently deformed by cutting, slide attachment and drying, during the alignment process there is a high risk of introducing biases by propagating deformations, shifts or distortions (Fiala, 2005; Capek et al., 2009). Here, due to the low number of slices, and to their good shape preservation, the alignment process was entirely performed with rigid-body transformations.
Image segmentation
Three-dimensional objects drawing requires the clustering of pixels into salient image regions, a process called image segmentation. The production of algorithms to automatically perform segmentation is constantly growing. However, one of the main limits of these algorithms lies in the difficulty to delineate an object without a priori knowledge of its possible form. In the next paragraphs we will briefly describe the use of some segmentation methods specifically designed for neuronal cells. For a more general discussion on segmentation the reader can refer to a recent review (Khairy and Keller, 2011).
For the rough 3D reconstruction of the LS, we applied a simple and flexible way to extract 3D object surfaces based on the manual or semi-automatic definition of the object contour in each section. In Reconstruct this segmentation method consists of drawing traces around the cross-section profiles of objects (inset in Figure 2A). The name applied to a specific trace defines the object to which the trace belongs. Traces can be drawn manually or semi-automatically with the wildfire “region growing” tool, which identifies the object boundaries through user-defined constraints including: hue, saturation, brightness, minimum area and distance from other traces. An implementation of the wildfire uses traces drawn in one section to automatically seed a new “region growing” process in subsequent sections (Lu et al., 2009). This function enables to rapidly follow a given object through multiple sections. For the LS reconstruction we used this automatic wildfire tool to trace the blood vessels passing through the EC and having at least a 25-μm diameter. Two blood vessels were recognized without ambiguity throughout sections, thus we could define each of them as individual objects. For visualization purpose, the blood vessel tracts running through the EC or in the gray matter were kept as separate objects (Figure 3 gray and golden respectively). By contrast, the relationships of the traces of DCX stained structures in adjacent slices could not be inferred; thus we collected all of them in two objects: (1) individual cells and processes, which were traced by using the conventional wildfire tool (Figure 3, violet), (2) clustered DCX+ cells, which were manually drawn (Figure 3, red).
3D representation
In Reconstruct traced objects can be 3D represented in multiple forms. Here, the blood vessels were represented as realistic 3D surfaces using the Boissonnat surface function (Figure 3). This function interpolates the object traces to create a 3D surface. This method is suitable to produce realistic representations of complex shapes, including branched structures. However, the Boissonnat surface was not appropriate for the 3D representation of the DCX staining, since the relationships between traces in adjacent sections were highly ambiguous. For this reason, the DCX staining was represented as traces slabs (Figure 3, violet and red). With this option traces are filled with color and extruded along the z-axis for a user-defined length. To more clearly discriminate between traces of clustered and individual DCX+ cells, the traces slabs of these objects were given different colors, transparency and extrusion thickness (Figure 3).
Overall, the rough reconstruction of the LS showed the clustered DCX+ cells were mostly restricted to the ventral EC, closely associated to two large caliber blood vessels, while the individual DCX+ cells and their processes were widely distributed in a strip of LS extending about 300 μm from the EC. Within the LS, a dense patch of DCX+ cells and processes stand out in the caudo-dorsal sector of the reconstructed volume (sections #4–12).
CLSM Serial Section Reconstruction of the Shape of DCX+ Cells
The main limit of the above described reconstruction concerns the low resolution along the z-axis (Figure 3). This results in a nearly complete loss of information about the 3D shape of the DCX-labeled cells. One possible way to circumvent this problem is the use of the CLSM. This implies the optical sectioning of a slice along the z-axis in series of images that can be processed to extract volume information.
Here, by using multiple available freeware software, we developed a procedure to use CLSM image stacks in serial section reconstructions (Figures 2B,C). Through this method we were able to obtain detailed 3D reconstructions of the DCX staining in the LS at cellular resolution.
Images acquisition and multi-fields montage
When planning a CLSM serial section reconstruction, firstly it is important to carefully evaluate the acquisition area and the voxel size, in order to use the minimum required data set. This will greatly improve data handling and interpretation.
To unravel the general 3D organization of the DCX staining in the LS, we selected, from the above described rough reconstruction, a sequence of nine sections in which a dense patch of DCX+ cells and processes, including numerous fiber fascicles, were found (Figure 3, sections #4–12).
A first set of images was acquired with a 20× objective along the entire dorso-ventral extent of the LS (4 fields/slice; voxel size 0.7 μm × 0.7 μm × 4.5 μm; Figure 4). Then, we acquired higher resolution images by using a 100× objective, to obtain a more detailed view of a selected group of DCX+ fascicles (2 field/slice; 229 optical planes; voxel size 0.14 μm × 0.14 μm × 1.6 μm; Figure 5). In both cases (20× and 100× reconstructions) acquisition of multiple fields-of-view and subsequent stitching of overlapping stacks was required. It is to note that although Reconstruct is able to import stacks, it is not suitable to stitch them. Mosaic montage of stacks can be performed with freeware tools, such as the 3D stitching plugins of Fiji (Preibisch et al., 2009), or VIAS 2.43 (Rodriguez et al., 2003). The 3D stitching plugins of Fiji operate with minimum user intervention and are also designed to work with tile scans. However, in our study, we preferred VIAS 2.4 since with this software overlapping stacks are montaged either manually or semi-automatically. This helps to maintain the superficial planes of the resulting mosaic stack as flat as possible. This is particularly important when large numbers of stacks are used. In addition, in the VIAS workspace only projections of the stacks on the x–y, y–z, and x–z planes are imported, thus reducing memory requirement and enabling a more fluid image manipulation. The output of VIAS is a sequence of mosaic optical planes defining a single volume (Figures 2D and 4A). If correctly numbered the sequences of optical sections derived from all the physical slices can be directly imported in Reconstruct for alignment. The numbering of slices must be consistent with the direction of the reconstruction so that the last optical plane of a slice is followed by the first optical plane of the next slice (Figures 2B,D).
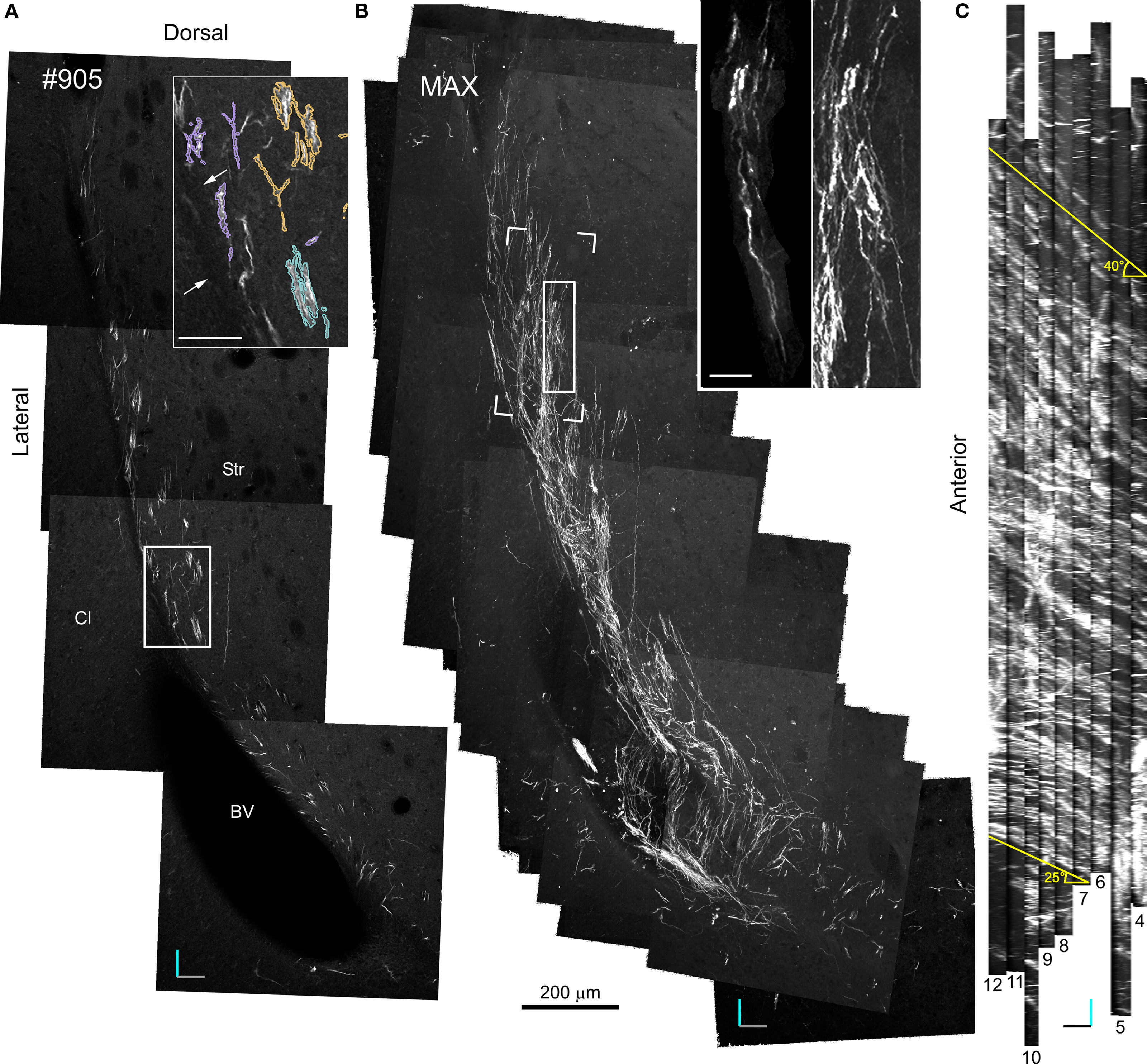
Figure 4. 20× Confocal laser scanning microscopy serial section reconstruction. (A) Image of the fifth optical plane of section #9 in the 20× CLSM serial section reconstruction. Note that this image is the result of four separate fields-of-view stitched together with Vias. In the inset, DCX+ processes belonging to distinct fascicles are outlined by differently colored traces. (B) Maximal intensity projections (MIP) along the z-axis of 79 optical planes taken from nine subsequent sections (#4–12). In the inset: on the right a magnified view of the area outlined by a continuous line in the main image, on the left a DCX+ cell in a fascicle (the pink fascicle of Figures 5 and 7) within the same area after a manual cut-off of the unwanted parts section by section. The open box indicates the position of the reconstruction shown in Figures 6 and 7. (C) MIP along the y-axis, note the numerous parallel fascicles of DCX+ processes. The slope of these fascicles on the rostro-caudal axis is indicated. Orientation Bars: gray line: medial; cyan line: dorsal; dark line: rostral; Scale bars: 50 μm in the inset in A; 25 μm in the inset in B.
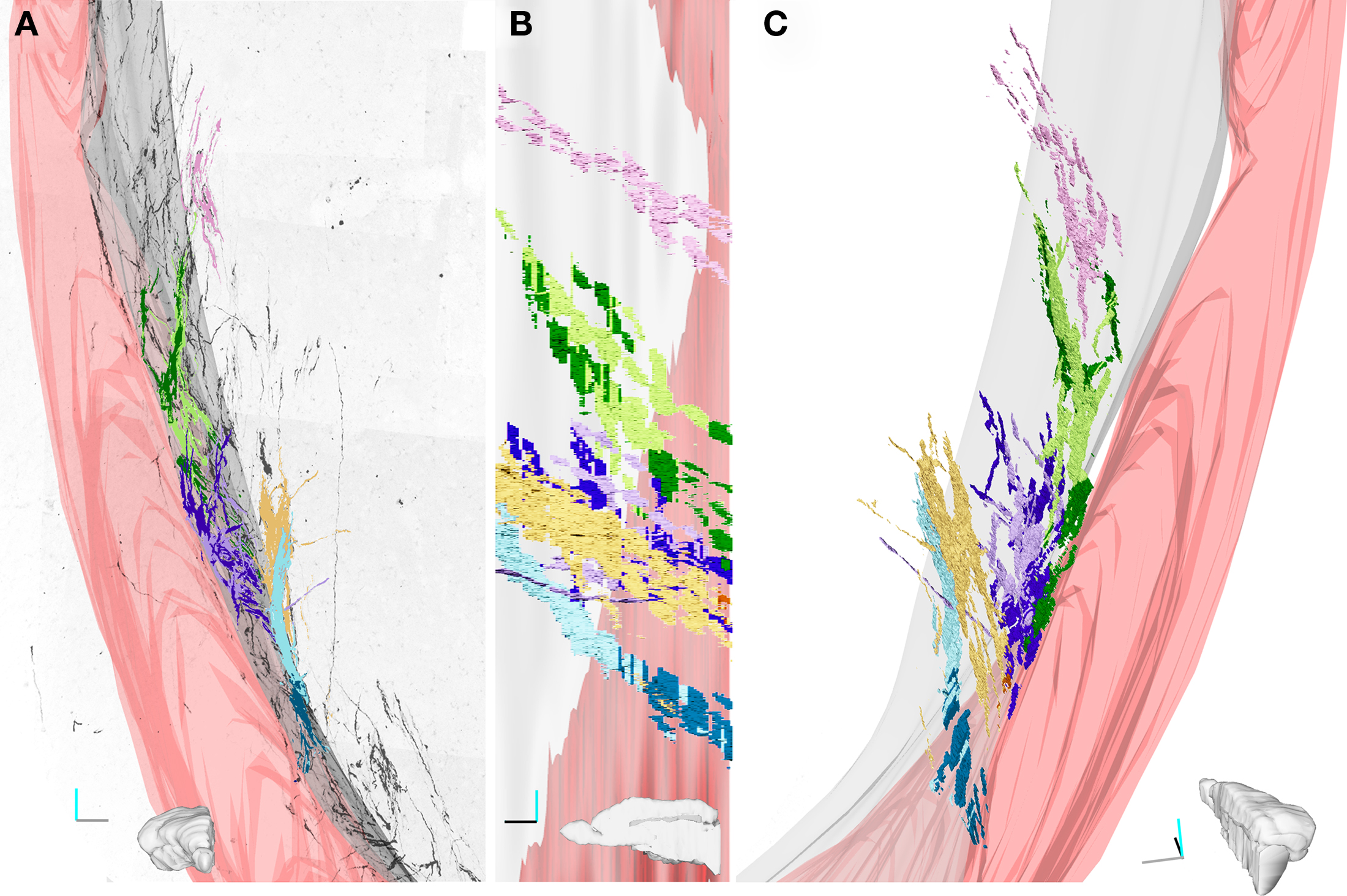
Figure 5. Three-dimensional reconstruction of four fascicles of DCX+ processes. (A–C). Four fascicles of DCX+ processes were traced in the stack shown in Figure 4. Each fascicles is represented by a different color and its tracts running within the EC are in darker colors. The 3D model was rendered in Blender (www.blender.org). In A the front view of the 3D model is superposed to an inverted image of the MIP of the stack along the z-axis. (B) medial view. (C) Caudal view slightly rotated medially. Only one fascicle (pink) runs exclusively in the striatal parenchyma, while the others run partly inside the EC. The Lilac and green fascicle extend always close to the EC, while the yellow and cyan contact the EC caudally and then turn medially within the striatal parenchyma. The medial surface of the EC is in gray; the surface of a big blood vessel running through the EC is in pink. Orientation Bars: gray line: medial; cyan line: dorsal; dark line: caudal.
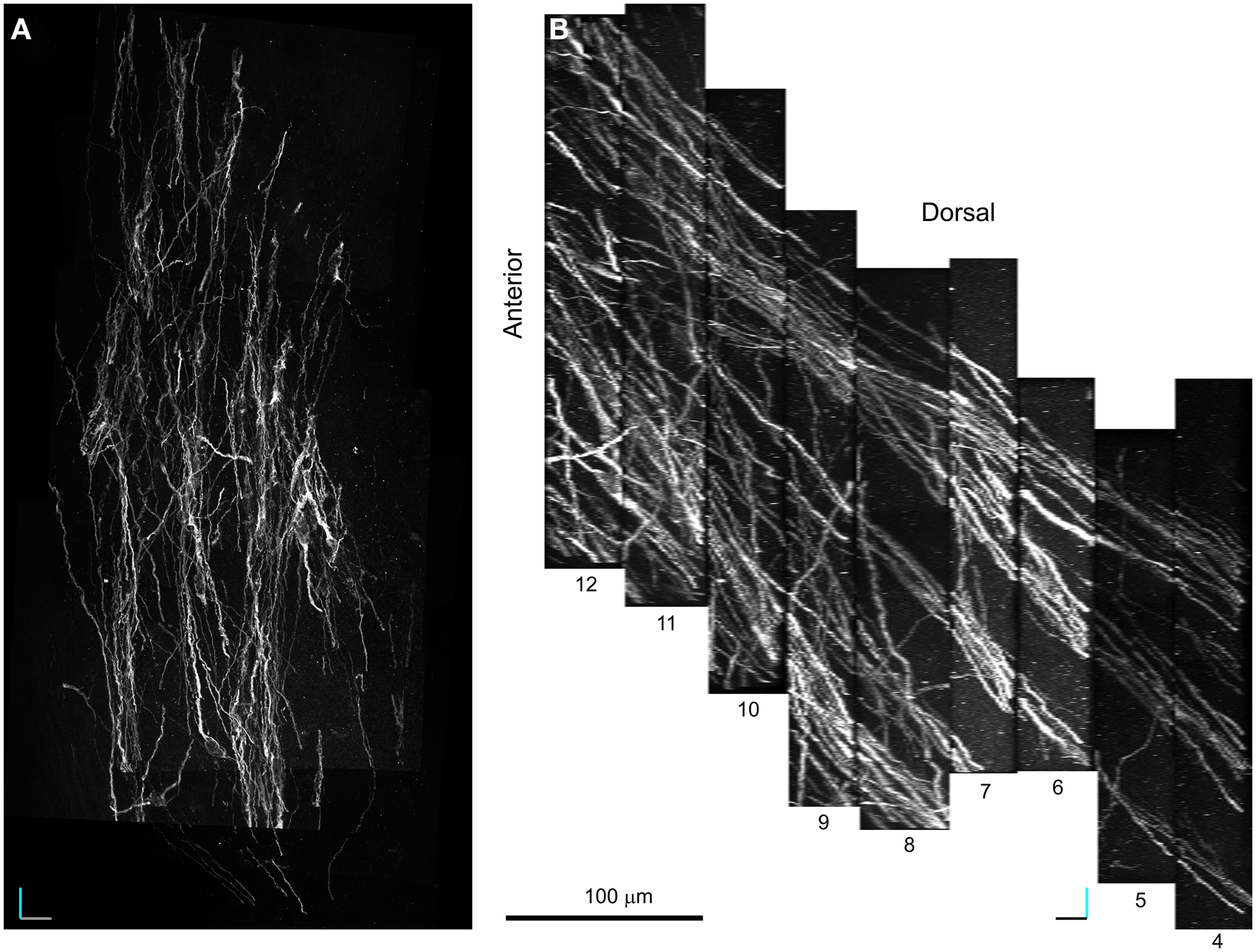
Figure 6. 100× Confocal laser scanning microscopy serial section reconstruction. (A–B) MIP along the z(A) and y (B) axis of a stack of 229 optical planes taken from nine subsequent optical slices (voxel size is 0.14 μm × 0.14 μm × 1.6 μm). This volume includes four fascicles of DCX+ processes one of which is the pink colored fascicle shown in Figure 5. The position of the reconstructed volume relative to the 20× reconstruction is indicated by the open box in Figure 4B. Note that fascicles of DCX+ processes extend parallel in the rostro-caudal direction within a network of less densely packed cell processes more randomly oriented. Orientation Bars: gray line: medial; cyan line: dorsal; dark line: caudal.
Alignment
Superficial optical planes of subsequent slices had very similar staining patterns, thus they were appropriate for alignment based on imposed fiducial marks. In this procedure, three or more couples of corresponding points are marked in two adjacent sections and used as reference to perform registration (Figure 2C). As previously mentioned, this procedure can involve rigid-body or non-linear transformations. In Reconstruct transformations applied to the superficial optical plane of a slice needs to be manually propagated to the others. This can be done through the command Propagate from the sub-menu Movement of the menu Section. It is to note that the slice surface is rarely planar and occasionally corresponding points must be searched in deeper optical planes, particularly at higher magnification (Figure 2D). Overall for both the 20× and 100× reconstructions, the alignment procedure was performed trough 8 rigid-body transformations, resulting in two series of 79 and 229 aligned optical planes respectively.
Segmentation and 3D Representation of 20× CLSM Serial Section Reconstruction: Spatial Organization of the Fascicles of DCX+ Cells (Voxel Size 0.7 μm × 0.7 μm × 4.5 μm)
Specific aim of the 20× CLSM reconstruction was to unravel the general 3D organization of the DCX staining in the LS, focusing on the fiber fascicles (Figures 4 and 5). The voxel size of this reconstruction (0.7 μm × 0.7 μm × 4.5 μm) was adequate to recognize DCX+ cell bodies and processes when isolated. By contrast, within fascicles DCX labeled structures were often too close to be resolved (Figure 4A). To visualize the entire data set as a single volume, the aligned Reconstruct series of images was exported and assembled in a 570-MB 8-bit stack (2104 × 3596 × 79 pixels) using Image J. As for any CLSM stacks, this data set can be visualized in several ways such as cross-sectional views, maximal intensity projections (MIP) or alpha bending. All these type of visualization are supported by freeware software packages such as the volume viewer of Image J or V3D Neuron4 (Peng et al., 2010). The MIP projection on the y, z plane of the entire 20× reconstruction stack showed that the DCX+ fascicles had a slope gradually increasing from 25° ventrally to 40° dorsally, in respect to the rostro-caudal axis (Figure 4C). This specific organization is closely reminding that of internal capsule fibers bundles (Figure A1 in Appendix). For more in depth observation of the 3D stack we used V3D Neuron. This latter software has a user-friendly interface that easily enables the rotation, zoom and volume cut of image stacks. In particular, volume cut is useful to analyze individual structures, since it reduces the visualized area allowing a better discrimination of small objects within a large volume (Figures 4B,C). Nonetheless, since volume-cut acts along a fixed plane, it can be inadequate for the analysis of highly convoluted and densely packed objects. In this case, it is possible to manually cut-off the unwanted parts of the image stack, section by section (compare right and left insets in Figure 4B). Alternatively, since this procedure can be relatively time consuming, it might be preferable to highlight the structures of interest through image segmentation. In the 20× reconstruction we used image segmentation to track the pathway of five fascicles of DCX+ cell processes. To this aim, the fascicles contour in each optical plane was traced with the wildfire tool of Reconstruct (inset in Figure 4A). The drawn traces were represented as differently colored traces slabs in the 3D environment (Figure 5). The parts of the DCX+ fascicles running within the EC were rendered in darker colors. We did not need to counter stain the tissue in order to identify the EC, since it appeared as a darker region in the background (arrows in Figure 4A). Four of the reconstructed DCX+ fascicles run partly inside the EC and partly in the striatal parenchyma. In particular, the two more ventral fascicles (yellow and cyan, Figure 5) contacted the EC caudally, then proceeding dorsally into the striatal parenchyma.
Although the slope and curvature of these fascicles are reminding those of internal capsule fibers bundles (Figure 5; Figure A1 in Appendix), these latter proceed in an opposite direction (rostral-to-ventral) from their EC origin. It is thus possible that these DCX+ fascicles entered within an internal capsule fiber bundle originating more rostrally and then followed it backward (ventral-to-rostral).
Overall, the 20× reconstruction indicates the 3D organization of the DCX+ fiber fascicles is closely reminding that of the internal capsule fiber bundles, with which they are closely associated.
Segmentation and 3D Representation of the 100× CLSM Serial Section Reconstruction: Organization of DCX+ Cells within and Among Fascicles. (Voxel Size 0.14 μm × 0.14 μm × 1.6 μm)
The purpose of the 100× CLSM reconstruction was to obtain a more detailed representation of the morphology of the DCX+ cells in the LS, and in parallel to understand the organization of cell processes within and among the DCX+ fascicles.
The acquired volume laid in the dorsal LS (Figure 4B) and included four fascicles of DCX+ processes, one of which was previously reconstructed at lower resolution (Figure 5, pink). To visualize the entire data set as a single volume, the aligned Reconstruct series of images was exported and assembled into a 991-MB 8-bit stack (1516 × 2995 × 229 pixels) using Image J. In the reconstructed volume four main parallel fascicles of DCX+ processes extended in the rostro-caudal direction within a network of less densely packed cell processes showing variable orientation (Figure 5). The resolution of this reconstruction (voxel size: 0.14 μm × 0.14 μm × 1.6 μm) enabled a detailed discrimination of both isolated and fascicled DCX+ cell processes. Nonetheless, the definition of clean-cut boundaries between individual DCX+ cells was not always possible since in regions of close proximity (that likely include cell contacts), the fluorescence of different processes tended to amalgamate.
For this reason, to reconstruct the path of the DCX+ processes, within and among fascicles, we traced the DCX+ processes of each fascicles as a unique network. To this aim we used Neuronstudio 0.9.925 (Rodriguez et al., 2003), a freeware software specifically designed to automatically trace neuronal elements. Starting from seeding points, the medial axis of a neuritic process is automatically skeletonized as a chain of nodes, and the diameter of each node is computed using the Rayburst sampling algorithm (Rodriguez et al., 2006). Using Neuronstudio we produced separate groups of tracings for each of the four fascicles that were comprised in the acquired volume. Often, from single seeding points, the growing algorithm trace spreads to multiple cells of the same fascicle and occasionally it extended also to other fascicles. In this latter case, bridging processes were manually interrupted at the first contacting point with the un-traced fascicles.
Digital reconstructions of the four fascicles were stored as . swc files (Cannon et al., 1998), imported in V3D neuron, and differently colored. As visible in Figures 7A–C, these fascicles run mostly independently, being interconnected only by a few DCX+ processes coursing in the mediolateral plane. Processes interconnecting multiple fascicles could be recognized because they were labeled in multiple colors.
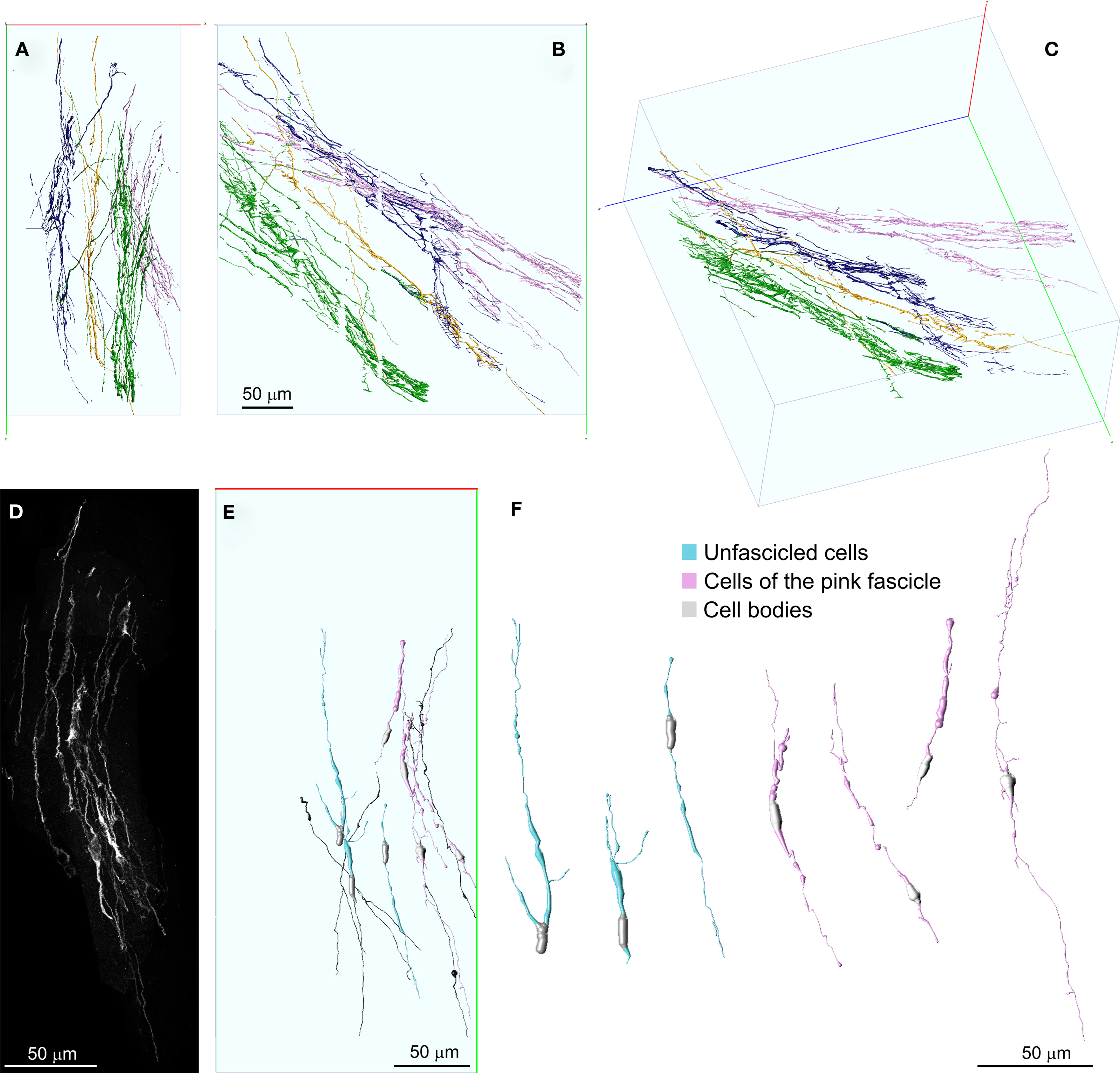
Figure 7. Detailed 3D representation of DCX+ cells and processes within and among fascicles. (A–C) Front (A), lateral (B), and perspective (C) views of DCX+ processes within and among four separate fascicles, represented in different colors. Fascicles were automatically segmented in Neuronstudio and rendered in V3D neuron. No manual editing has been performed in order to connect corresponding processes at the inter-slice gaps. The numerous small orthogonal branches in (B) are artifacts of the automatic tracing algorithm. (D) MIP of the DCX+ processes composing the pink colored fascicle after a manual cut-off of surrounding areas section by section. (E,F) 3D reconstructions of individual DCX+ cells isolated (cyan) or being part of the pink colored fascicle (pink). Some individual DCX+ process, whose cell body was not included in the reconstructed volume, are also shown (dark) in (E). These cells were reconstructed in Neuromantic and rendered in V3D neuron. Note these cells are mostly bipolar and show long and poorly ramified processes. Orientation Bars: gray line: medial; cyan line: dorsal; dark line: caudal.
Given that Neuronstudio works optimally only when the fluorescence completely fill the cell, and that DCX staining does not satisfy this pre-requisite, to entirely reconstruct individual DCX+ cells we switched to a manual tracing method. To this aim we used Neuromantic 1.7.56, a freeware software that is specifically designed for manual or semi-automatic tracing of neuronal cells. In this software neuronal cells are traced accordingly with the .swc file format, in which traces are made by cylindrical segments of specific length and diameter. Neuromantic has several advantages for manual tracing over Neuronstudio including: the selective visualization of traced neuronal segments close to the focal plane, the possibility to color segments differentially by depth and to automatically focus at specific segments. Due to the above mentioned resolution limits of our data set, only a few DCX+ cells could be entirely reconstructed. In Figure 7 the reconstruction of seven cells is illustrated. The cell bodies of these cells were recognized as cytoplasmic swellings with a superficial DCX staining. Four of the reconstructed DCX+ cells were part of the pink colored fascicle shown in Figure 4 (Figures 7E,F pink), while the others were not associated to any fiber fascicles (Figures 7E,F cyan). The reconstructed cells showed mostly a bipolar morphology, consistent with that of immature or migrating neuroblasts. Nonetheless, one cell showed two very long opposing processes, suggesting it was at a more differentiated stage (Figures 7E,F). It is to note that the reconstructed cells represent a biased sample of the entire population, since they were chosen among the more isolated DCX+ cells. Overall, the 100× reconstruction unveils that the fiber fascicles were made by long and poorly ramified processes, running mostly within the same fascicle, but occasionally interconnecting distinct fascicles.
Discussion
Newborn neuronal cells have been described in the mature striatum of different mammalian species, raising hopes to exploit this neurogenic activity for brain repair (Arvidsson et al., 2002; Bedard et al., 2002; Luzzati et al., 2006, 2007). However, much information is still lacking regarding the identity and function of newborn striatal cells (Liu et al., 2009; Kernie and Parent, 2010). Detailed neuroanatomical analyses of the newly formed elements regarding their spatial organization and relationships with specific circuits, can help to unravel the role of these cells both in physiological and pathological conditions.
In the present paper we describe a method to combine CLSM with serial section reconstruction, in order to obtain 3D reconstructions of large tissue volumes at cellular resolution. Through this method we provided the first description of the morphology and neuroanatomical organization of a population of newborn neuroblasts occurring in the mammalian striatum. Data obtained from this study indicate that newborn cells in LS of the guinea pig are mostly bipolar elements, showing poorly ramified processes that organize to form long fascicles. Interestingly, these fascicles follow the path of the fiber bundles of the internal capsule, suggesting newborn cells may play a role within this striatal compartment. Given that only very few DCX+ cells and no DCX+ bundle can be entirely included in a single 40 μm thick section, the use of classical anatomical approaches to describe this system would have required much more time (to find the right planes of sectioning, to look for the right section), and would have led to a less precise and complete picture.
Current advances in two-photon excitation microscopy allow the visualization of large tissue volumes at cellular resolution within the living brain (Sigler and Murphy, 2010). Although this technique represents a pivotal innovation for the study of neuroanatomy and neuronal plasticity, it has been little used in studies of adult neurogenesis (Mizrahi, 2007). A major limiting factor is that two-photon microscopy can image only within a few hundred micrometers of depth (Sigler and Murphy, 2010), confining the analyses to relatively superficial regions, such as the mice neocortex or the OB (Mizrahi, 2007). Ex vivo whole mount preparation of both fixed and living specimens is a possible approach to overcome depth limits. This approach has been successfully used to study the periventricular SVZ and it could be potentially extended to other brain regions (Tavazoie et al., 2008; Tsai et al., 2009; Lacar et al., 2010). Nonetheless, besides superficial regions such as the SVZ, the specimen thickness in whole mount preparations can be too large for the penetration capacity of most histological staining methods, in particular for immunofluorescence labeling. This can be a major limiting factor, especially in mammalian species in which genetically labeled models are not currently available. The DCX+ neuroblasts in the LS of the guinea pig represent a good example of a cell population that cannot be visualized through current whole mount approaches since these cells are scattered over a large area, and require immunohistochemistry to be labeled.
The serial section reconstruction represents a reliable alternative to visualize at cellular resolution large volumes of tissue, independently from depth and from the staining method. The main drawback of the classic version of this technique is that an inverse relationship exists between z-axis resolution and section thickness. Although much effort has been devoted to the automation of image acquisition and registration, serial section reconstruction at cellular resolution is a considerable time-consuming approach (Fiala, 2005; Cardona et al., 2010).
Here, we present an implementation of the serial section reconstruction method in which CLSM stacks of images are registered in order to use thick slices to produce reconstructions at cellular resolution. The basic idea behind this method is to fragment the region of interest in the minimum number of slices enabling their staining and high-resolution imaging. These slices are then optically sectioned with CLSM and virtually stitched along the z-axis using a serial section reconstruction editor. The reduction in the number of slices per volume consistently speed up image acquisition and registration. Moreover, the intrinsic registration of the optical planes within each slice and the need of less transformations per volume, increase also the reliability of the reconstruction. Indeed, this reduces the risks of introducing biases during slice registration by propagating deformations, shifts or distortions (Fiala, 2005; Capek et al., 2009). The good shape preservation of the thick slices further contribute to the precision of the reconstruction. Overall, the advantages of our implementation of serial section reconstruction are well demonstrated by the 100× reconstruction in which we obtained a series of 229 aligned optical sections by using only eight rigid-body transformations.
The main drawback of CLSM serial section reconstruction in respect to whole mount imaging is the occurrence of small gaps within the reconstructed volume. These gaps lie between slices and are the result of tissue loss during cutting and further tissue processing. The impact of these superficial artifacts has been extensively evaluated for stereological methods, and it has been shown to be strongly dependent on tissue quality, cutting angle, and embedding media (Baryshnikova et al., 2006). In the context of CLSM serial section reconstruction, slice surface artifacts cause a variable reduction of the actual resolving power at the inter-slice interface. Depending on dimensions, orientation and density of the structures of interest, inter-slice gaps differentially hamper the identification of corresponding structures between consecutive slices. In particular, tiny structures running parallel to the cutting plane are more likely to be affected by slice surface artifacts. In such cases, the identification of corresponding structures between subsequent slices will be strongly dependent on their density. In general since multiple factors influence the reconstruction of specific objects, the impact of slice surface artifacts should be determined case by case. In the present paper we were able to reconstruct a network of fascicled DCX+ processes that were relatively densely packed, taking advantage from their straight orientation and choosing a perpendicular cutting plane. This minimized the effects of surface artifacts.
It is to note that despite the periodic drop of the resolving power between slices, the resolution within slices obtained with CLSM serial section reconstruction can be higher than in whole mount imaging. Indeed, since the signal to noise ratio suffers from considerable degradation with depth, the resolution limit of light microscopes is higher for slice than for whole mount imaging (Wilt et al., 2009; Sigler and Murphy, 2010). Moreover, since our method is compatible with virtually any stack-based microscopy technique, it can be adapted for the recently developed super-resolution fluorescence microscopy that can resolve particles separated by few tens of nanometers (Schermelleh et al., 2010). Considering that these applications so far efficiently work only within a limited depth range, stack-based serial sections reconstruction might increase the volume that can be visualized with these methods (Schermelleh et al., 2010).
A direct demonstration of the resolution capacity of CLSM serial section reconstruction was not the specific aim of the present study. Accordingly, the resolution used here does not represent the maximum resolution of CLSM, but rather the minimal resolution required to reconstruct the network of DCX+ processes within and among fascicles. The reconstruction of individual elements within this network was often impaired by the presence of close contacts between their processes. Indeed, in contrast to the EM, closely apposed membranes cannot be resolved with CSLM without specific markers. For analyses at sub-cellular resolution, higher resolving power can be achieved even with ordinary CLSM in example through higher NA of the objective, closer optical planes, or the use of image deconvolution.
Confocal laser scanning microscopy serial section reconstruction is a modification of both whole mount imaging and classic serial section reconstruction and accordingly it takes advantage from diverse software developed for these two methods. Although here we used specific sets of software, we would like to emphasize that other applications can be used to perform the stitching, registration and segmentation of series of CLSM stacks. In example, a method of CLSM serial section reconstruction has been recently proposed by Capek et al. (2009) by using the unreleased software Rapid 3D. This method has been designed to reconstruct large volumes such as entire brains or embryos, and to date it has been used only for low magnification reconstructions. Nonetheless, it should be interesting to test Rapid 3D to produce reconstructions at cellular resolution. In addition, several useful stitching, registration and segmentation plugins are bundled with the free software package Fiji. Among them, TrakEM2 is a powerful Image J plugin that is closely related to Reconstruct7 (Cardona et al., 2010).
In conclusion, we showed that LSCM serial section reconstruction is a profitable effective way to analyze diverse architectural aspects of the adult brain. In particular, we propose it is suitable to investigate neuroanatomical features of adult neurogenic niches. Future development of this method may include imaging at higher resolution, within larger volumes and using multiple wavelengths.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Supports Contributed by Compagnia di San Paolo (NEUROTRANSPLANT 2004.2019; 2008.2192); Università di Torino 2007; PRIN 2007; Regione Piemonte – Ricerca Sanitaria Finalizzata (2009).
Footnotes
References
Arvidsson, A., Collin, T., Kirik, D., Kokaia, Z., and Lindvall, O. (2002). Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat. Med. 8, 963–970.
Baryshnikova, L. M., Von Bohlen Und Halbach, O., Kaplan, S., and Von Bartheld, C. S. (2006). Two distinct events, section compression and loss of particles (“lost caps”), contribute to z-axis distortion and bias in optical disector counting. Microsc. Res. Tech. 69, 738–756.
Bedard, A., Cossette, M., Levesque, M., and Parent, A. (2002). Proliferating cells can differentiate into neurons in the striatum of normal adult monkey. Neurosci. Lett. 328, 213–216.
Cannon, R. C., Turner, D. A., Pyapali, G. K., and Wheal, H. V. (1998). An on-line archive of reconstructed hippocampal neurons. J. Neurosci. Methods 84, 49–54.
Capek, M., Bruza, P., Janacek, J., Karen, P., Kubinova, L., and Vagnerova, R. (2009). Volume reconstruction of large tissue specimens from serial physical sections using confocal microscopy and correction of cutting deformations by elastic registration. Microsc. Res. Tech. 72, 110–119.
Cardona, A., Saalfeld, S., Preibisch, S., Schmid, B., Cheng, A., Pulokas, J., Tomancak, P., and Hartenstein, V. (2010). An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biol. 8, e1000502. doi: 10.1371/journal.pbio.1000502
DeFelipe, J. (2010). From the connectome to the synaptome: an epic love story. Science 330, 1198–1201.
Doetsch, F., and Alvarez-Buylla, A. (1996). Network of tangential pathways for neuronal migration in adult mammalian brain. Proc. Natl. Acad. Sci. U S A. 93, 14895–14900.
Doetsch, F., Garcia-Verdugo, J. M., and Alvarez-Buylla, A. (1997). Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J. Neurosci. 17, 5046–5061.
Fiala, J. C. (2005). Reconstruct: a free editor for serial section microscopy. J. Microsc. 218, 52–61.
Hofer, S. B., Mrsic-Flogel, T. D., Bonhoeffer, T., and Hübener, M. (2009). Experience leaves a lasting structural trace in cortical circuits. Nature 457, 313–317.
Kernie, S. G., and Parent, J. M. (2010). Forebrain neurogenesis after focal Ischemic and traumatic brain injury. Neurobiol. Dis. 37, 267–274.
Kriegstein, A., and Alvarez-Buylla, A. (2009). The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184.
Lacar, B., Young, S. Z., Platel, J. C., and Bordey, A. (2010). Imaging and recording subventricular zone progenitor cells in live tissue of postnatal mice. Front. Neurosci. 4:43.
Lindsey, B. W., and Tropepe, V. (2006). A comparative framework for understanding the biological principles of adult neurogenesis. Prog. Neurobiol. 80, 281–307.
Liu, F., You, Y., Li, X., Ma, T., Nie, Y., Wei, B., Li, T., Lin, H., and Yang, Z. (2009). Brain injury does not alter the intrinsic differentiation potential of adult neuroblasts. J. Neurosci. 29, 5075–5087.
Lu, J., Fiala, J. C., and Lichtman, J. W. (2009). Semi-automated reconstruction of neural processes from large numbers of fluorescence images. PLoS ONE 4, e5655. doi: 10.1371/journal.pone.0005655
Luzzati, F., De Marchis, S., Fasolo, A., and Peretto, P. (2006). Neurogenesis in the caudate nucleus of the adult rabbit. J. Neurosci. 26, 609–621.
Luzzati, F., De Marchis, S., Fasolo, A., and Peretto, P. (2007). Adult neurogenesis and local neuronal progenitors in the striatum. Neurodegener. Dis. 4, 322–327.
Luzzati, F., Peretto, P., Aimar, P., Ponti, G., Fasolo, A., and Bonfanti, L. (2003). Glia-independent chains of neuroblasts through the subcortical parenchyma of the adult rabbit brain. Proc. Natl. Acad. Sci. U.S.A. 100, 13036–13041.
Mirzadeh, Z., Doetsch, F., Sawamoto, K., Wichterle, H., and Alvarez-Buylla, A. (2010). The subventricular zone en-face: wholemount staining and ependymal flow. J. Vis. Exp. 39, pii: 1938.
Mizrahi, A. (2007). Dendritic development and plasticity of adult-born neurons in the mouse olfactory bulb. Nat. Neurosci. 10, 444–452.
Parent, J. M., Vexler, Z. S., Gong, C., Derugin, N., and Ferriero, D. M. (2002). Rat forebrain neurogenesis and striatal neuron replacement after focal stroke. Ann. Neurol. 52, 802–813.
Peng, H., Ruan, Z., Long, F., Simpson, J. H., and Myers, E. W. (2010). V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 28, 348–353.
Preibisch, S., Saalfeld, S., and Tomancak, P. (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465.
Rodriguez, A., Ehlenberger, D., Kelliher, K., Einstein, M., Henderson, S. C., Morrison, J. H., Hof, P. R., and Wearne, S. L. (2003). Automated reconstruction of three-dimensional neuronal morphology from laser scanning microscopy images. Methods 30, 94–105.
Rodriguez, A., Ehlenberger, D. B., Hof, P. R., and Wearne, S. L. (2006). Rayburst sampling, an algorithm for automated three-dimensional shape analysis from laser scanning microscopy images. Nat. Protoc. 1, 2152–2161.
Schermelleh, L., Heintzmann, R., and Leonhardt, H. (2010). A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190, 165–175.
Seri, B., Garcia-Verdugo, J. M., Collado-Morente, L., McEwen, B. S., and Alvarez-Buylla, A. (2004). Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J. Comp. Neurol. 478, 359–378.
Sigler, A., and Murphy, T. H. (2010). In vivo 2-photon imaging of fine structure in the rodent brain: before, during, and after stroke. Stroke 41, S117–S123.
Tavazoie, M., Van der Veken, L., Silva-Vargas, V., Louissaint, M., Colonna, L., Zaidi, B., Garcia-Verdugo, J. M., and Doetsch, F. (2008). A specialized vascular niche for adult neural stem cells. Cell Stem Cell 3, 279–288.
Tsai, P. S., Kaufhold, J. P., Blinder, P., Friedman, B., Drew, P. J., Karten, H. J., Lyden, P. D., and Kleinfeld, D. (2009). Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J. Neurosci. 29, 14553–14570.
Keywords: neurogenesis, striatum, whole mount, confocal laser scanning microscopy, serial section reconstruction
Citation: Luzzati F, Fasolo A and Peretto P (2011) Combining confocal laser scanning microscopy with serial section reconstruction in the study of adult neurogenesis. Front. Neurosci. 5:70. doi: 10.3389/fnins.2011.00070
Received: 23 February 2011;
Accepted: 03 May 2011;
Published online: 13 May 2011.
Edited by:
Silvia De Marchis, University of Turin, ItalyReviewed by:
Vincent Prevot, INSERM, FranceJoao R. L. Menezes, Universidade Federal do Rio de Janeiro, Brazil
Copyright: © 2011 Luzzati, Fasolo and Peretto. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Federico Luzzati, Neuroscience Institute Cavalieri Ottolenghi, Regione Gonzole, 10, 10043 Orbassano, Turin, Italy. e-mail: federico.luzzati@unito.it

 Aldo Fasolo1 and Paolo Peretto1,2
Aldo Fasolo1 and Paolo Peretto1,2