- 1Ministry of Education Key Laboratory of Intelligent Computing and Signal Processing, Information Materials and Intelligent Sensing Laboratory of Anhui Province, Anhui University, Hefei, China
- 2School of Artificial Intelligence, Anhui University, Hefei, China
- 3Institute of Artificial Intelligence, Hefei Comprehensive National Science Center, Hefei, China
- 4SEU-ALLEN Joint Center, Institute for Brain and Intelligence, Southeast University, Nanjing, China
Biomedical image registration refers to aligning corresponding anatomical structures among different images, which is critical to many tasks, such as brain atlas building, tumor growth monitoring, and image fusion-based medical diagnosis. However, high-throughput biomedical image registration remains challenging due to inherent variations in the intensity, texture, and anatomy resulting from different imaging modalities, different sample preparation methods, or different developmental stages of the imaged subject. Recently, Generative Adversarial Networks (GAN) have attracted increasing interest in both mono- and cross-modal biomedical image registrations due to their special ability to eliminate the modal variance and their adversarial training strategy. This paper provides a comprehensive survey of the GAN-based mono- and cross-modal biomedical image registration methods. According to the different implementation strategies, we organize the GAN-based mono- and cross-modal biomedical image registration methods into four categories: modality translation, symmetric learning, adversarial strategies, and joint training. The key concepts, the main contributions, and the advantages and disadvantages of the different strategies are summarized and discussed. Finally, we analyze the statistics of all the cited works from different points of view and reveal future trends for GAN-based biomedical image registration studies.
Introduction
The goal of biomedical image registration (BIR) is to estimate a linear or non-linear spatial transformation by geometrically aligning the corresponding anatomical structures between images. The images can be acquired across time, modalities, subjects, or species. By aligning the corresponding structures or mapping the images onto a canonical coordinate space, the registration allows quantitative comparison across the subjects imaged under different conditions. It enables the analysis of their distinct aspects in pathology or neurobiology in a coordinated manner (Oliveira and Tavares, 2014). In addition, image registration is also fundamental to image-guided intervention and radiotherapy.
In recent years, there has been a steady emergence of high-resolution and high-throughput biomedical imaging techniques (Li and Gong, 2012; Chen et al., 2021). Some commonly used macroscale imaging techniques include magnetic resonance imaging (MRI), computed tomography (CT), positron emission tomography (PET), and single photon emission computed tomography (SPECT) (Gering et al., 2001; Staring et al., 2009). However, mesoscale and microscale imaging techniques, such as serial two-photon tomography (STPT) (Ragan et al., 2012), fluorescence micro-optical sectioning tomography (FMOST) (Gong et al., 2016), volumetric imaging with synchronous on-the-fly scan and readout (VISOR) (Xu et al., 2021), and the electron microscope (EM) (Ruska, 1987), play pivotal roles in various neuroscience studies. The resulting exploration of the number, resolution, dimensionality, and modalities of biomedical images not only provides researchers with unprecedented opportunities to study tissue functions, diagnose diseases, etc. but also poses enormous challenges to image registration techniques.
A large number of image registration methods, ranging from the traditional iterative method to the one-shot end-to-end method (Klein S. et al., 2009; Qu et al., 2021), from the fully supervised strategy to the unsupervised strategy (Balakrishnan et al., 2019; He et al., 2021), have been developed to take full advantage of the rapidly accumulating biomedical images with different geometric and modalities. According to the different acquisition techniques of the images, these methods can also be classified into two main categories: mono-modal (intra-modal) registration and cross-modal (or inter-model) registration. Generally, the images of different modalities often vary substantially in their voxel intensity, image texture, and anatomical structures (e.g., due to uneven brain shrinkage resulting from different sample preparation methods). Therefore, cross-modal registration is even more challenging to achieve.
To the best of our knowledge, few studies focus on cross-modal medical image registration. Among the available reviews (Jiang S. et al., 2021), traditional feature-based cross-modal alignment methods have been reviewed in detail. Most of these traditional registration methods are based on iterative training, which is time consuming. The supervised alignment methods are limited by insufficient labels among the learning-based methods. However, unsupervised methods are proposed with various loss functions due to the absence of ground truth and supervision.
Additionally, these unsupervised methods do not perform as well as unimodal on cross-modal images due to too much variation between cross-modal appearances. Nevertheless, efforts are being directed toward removing the modal differences between cross-modalities. Among these various deep-learning-based methods, the Generative Adversarial Networks (GAN) (Goodfellow et al., 2014) have received increasing attention from researchers for their unique network structures and training strategies. In addition, the GAN-based methods have shown extraordinary potential in dealing with cross-modal registration. In particular, the conditional GAN (CGAN) can realize transformation between different styles of images, which provides a new solution to the difficult cross-modal registration method, which has been plagued by the different modality characteristics for a long time.
Since it was proposed, the GAN has been widely used for biomedical image analysis, such as classification, detection, and registration. Its outstanding performance in image synthesis, style translation (Kim et al., 2017; Jing et al., 2020), and the adversarial training strategy has attracted attention in many areas (Li et al., 2021). GAN has been applied to image registration tasks since 2018 (Yan et al., 2018). However, compared with other deep-learning-based registration methods, the GAN-based methods are still in their infancy, and their potential needs further exploration. To the best of our knowledge, there has not yet been a specific review on GAN in biomedical image registration. Therefore, we hereby try to provide an up-to-date and comprehensive review of existing GAN applications in biomedical image registration.
In the survey, we focus on both the GAN-based mono- and cross-modal biomedical image registrations but may highlight more on the contribution of the GAN-based cross-modal image registration. Cross-modal biomedical image registration is still facing many challenges compared with mature mono-modal biomedical image registration.
This review is structured as follows: Section Common GAN structures briefly introduces the basic theory of common GAN related to image registrations; Section Strategies of GAN based biomedical image registration provides a comprehensive analysis of four GAN-based registration strategies; in Section Statistics, we analyze the ratio distribution of some important characteristics of these studies, and in Section Future perspectives, we discuss some open issues and future research perspectives.
Common GAN structures
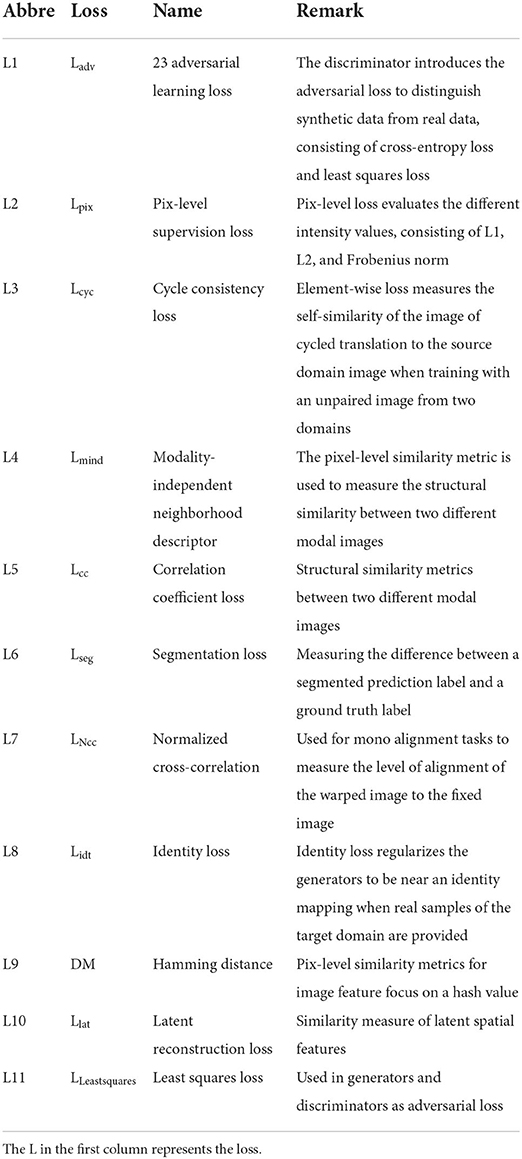
This section gives a brief introduction to the GAN structures used for image generation. The structures considered are often used directly or indirectly in the cross-modal biomedical image registration model. We summarize this literature, which considers GAN structures, in Tables 2–5. The section emphasizes the overall architecture, data flow, and objective function of GAN. The differences between the various methods are also presented.
Original GAN
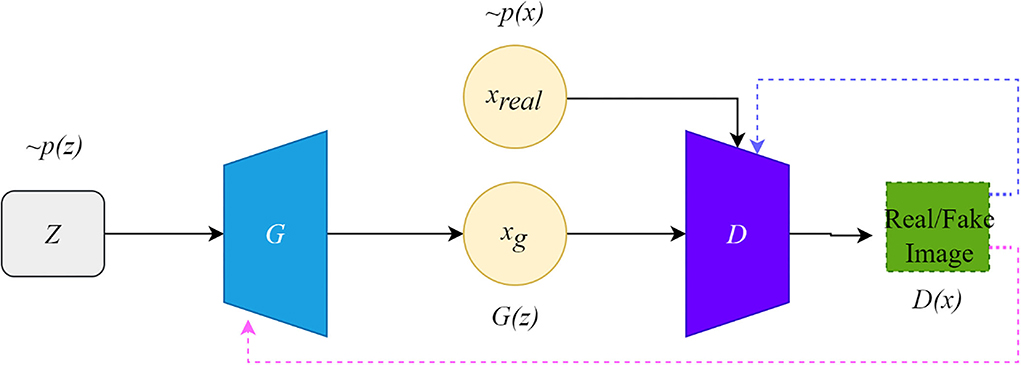
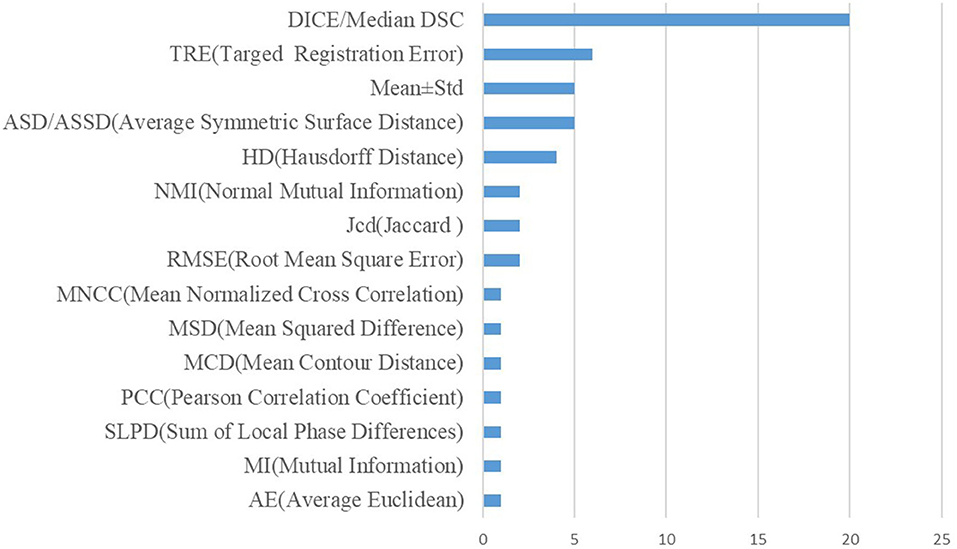
The framework of the original GAN is shown in Figure 1. The original GAN consists of two networks, the generator (G) and the discriminator (D), both of which are fully connected. The input to the generator is a random noise vector from the noise distribution ~p(z) (random noise is Gaussian noise or uniform noise). The generator can learn a mapping from the low-dimensional noise vector space to the high-dimensional data space. The input of the discriminator is the real data ~Pr(x) and the synthetic data ~Pg(x) by the generator. If the input to the discriminator is real data x, the purpose of the discriminator is to represent the probability that x comes from ~Pr(x) rather than ~Pg(x), and the discriminator should classify it as real data and return a value close to 1.
Conversely, if the input is synthetic data, the discriminator should classify it as false data and return a value close to 0. The false signal output from the discriminator is back propagated to the generator to update the network parameters. This framework is trained in an adversarial strategy corresponding to a two-player minimax game. The minimax GAN loss is equivalent to the game's rules, while the generator and the discriminator are equivalent to the two players. The goal of the generator is to minimize the loss by generating synthetic images that look as similar to the real images as possible to fool the discriminator.
In contrast, the discriminator maximizes the loss to maximize the probability of assigning the correct label to both the training examples and the samples from the generator. The training improves the performance of both the generator and the discriminator networks. Their loss functions can be formulated as follows:
where LD and LG are the loss functions of D and G, respectively, and D is the binary classifier; it is expected that the data distribution generated by G(z) is close to the real data when the model is optimized.
DCGAN
Compared with the original GAN, the deep convolutional generative adversarial networks (DCGAN) (Radford et al., 2015) add specific architectural constraints to GAN by replacing all the full-connected neural networks with CNN, which results in stable training. Figure 2A illustrates the structure of DCGAN, in which there are three important changes in the convolutional neural network (CNN) architecture. Firstly, the pooling layers in the discriminator and the generator are replaced by the stridden convolution and the fractionally strung convolutions, respectively, which allow the generator to learn the specific spatial upsampling from the input noise distribution to the output image. Secondly, batch normalization (Ioffe and Szegedy, 2015) is utilized to regulate poor initialization to prevent the generator from collapsing from all samples to a single point. Thirdly, the LeakyReLU (Maas et al., 2013) activation is adopted to replace the maxout activation in all layers of the discriminator, promoting the output of higher-resolution images.
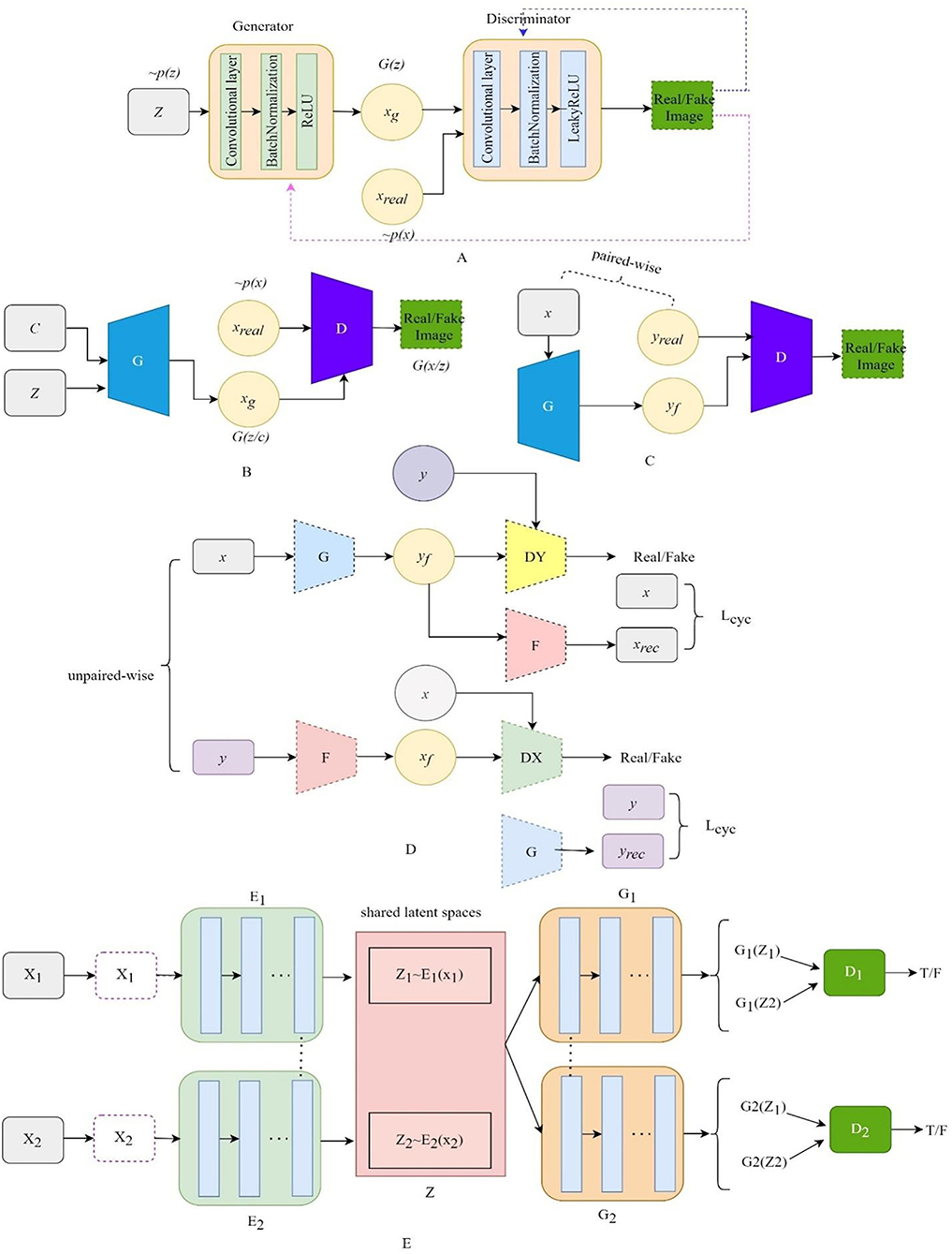
Figure 2. The architecture of the variant GANs. (A) The architecture of DCGAN, (B) the architecture of CGAN, (C) the architecture of Pix2Pix, (D) the architecture of CyclGAN, and (E) the architecture of UNIT.
CGAN
The structure of CGAN (Mirza and Osindero, 2014) is illustrated in Figure 2B. The CGAN performs the conditioning for the output mode by feeding the auxiliary information related to the desired properties y and noise vector z to the generator and the discriminator. The objective function of the CGAN can be formulated as follows:
where y is the auxiliary information, which could be a class label, an image, or even the data from different modes. For instance, Pix2Pix (Isola et al., 2017) translates the label image or edge image to an image with another style. InfoGAN (Chen et al., 2016) is regarded as a special CGAN whose condition is a conditional constraint on the random noise z for guiding the thickness, slope, and other features of the generated image.
Pix2Pix
Pix2Pix (Isola et al., 2017) is the first GAN framework for image-to-image translation, which can learn a mapping that transforms an image from one modality to another based on paired-wise images. The structure of Pix2Pix is depicted in Figure 2C. A paired-wise image means that the internal structures in the image pair are accurately aligned, while their texture, brightness, and other modality-related features differ. The objective loss combines CGAN with the L1 loss so that the generator is also asked to generate images as close as possible to the ground truth:
where x and y represent the images from the source and the target domain, respectively; the L1 loss is a pixel-level metric between the target domain image and the generated image to impose a constraint on G, which could recover the low-frequency part of the image; the adversarial loss could recovery the high-frequency part of the image, and λ is the adjustable parameter.
Cycle-GAN
CycleGAN (Zhu et al., 2017) contains two generators and two discriminators, which are self-bounded by an inverse loop to transform the image between the two domains. The structure of Cycle-GAN is illustrated in Figure 2D. One generator, G, translates the source domain image X to the target domain image Y. Another generator, F, learns the inverse mapping of G, which brings G(X) back to its original image X., i.e., x→ G(x)→ F[G(x)] ≈ x. Similarly, for y from the domain Y, F and G also satisfy the cycle-consistent, i.e., y→ F(y)→ G[F(y)] ≈ y. The cycle-consistent loss Lcycle measures the reconstructed image and the real image by pixel-level loss calculation to constrain the training of G and F, ensuring the consistency of its morphological structure in the transformation process. Two discriminators distinguish between the reconstructed image and the real image. The adversarial loss and the cycle-consistent loss are as follows:
The training procedure uses the least squares and replays buffer for training stability. UNET (Ronneberger et al., 2015) and PatchGAN (Li and Wand, 2016; Isola et al., 2017) are used to build the generator and the discriminator.
UNIT
UNIT (Liu et al., 2017) can also perform unpaired image-to-image transformation by combining two variational autoencoder generative adversarial networks (VAEGAN) (Xian et al., 2019), with each responsible for one modality but sharing the same latent space. The UNIT structure is illustrated in Figure 2E, which consists of six subnetworks: two domain image encoders, E1 and E2, two domain image generators, G1 and G2, and two adversarial domain discriminators, D1 and D2. The encoder–generator pair {E1, G1} constitutes a VAE for the X1 domain, called VAE1. For an input image x1∈X1, the VAE1 first maps x1 to a code in a latent space Z via the encoder E1 and then decodes a random-perturbed version of the code to reconstruct the input image via the generator G1. The image x2 in X2 can be translated to an image in X1 by applying G1(Z2). For real images sampled from the X1 domain, D1 should output true, whereas, for images generated by G1(Z2), it should output false. The cycle-consistency constraint exists in x1 = F2 − 1[F1 − 2(x1)], where F1 − 2 = G2[E1(X1)].
Strategies for GAN-based biomedical image registration
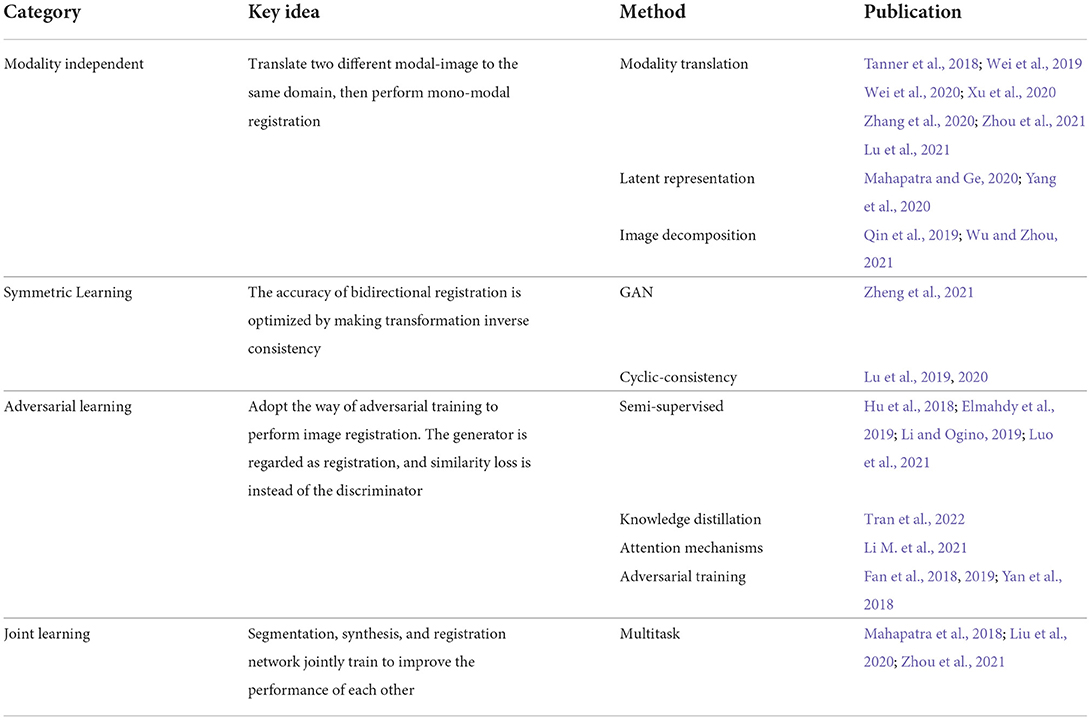
Cross-modal biomedical image registration using GAN has given rise to an increasing number of registration algorithms to solve the current problems mentioned in the introduction section. Based on the different strategies, the algorithms are divided into four categories: modality translation, symmetric learning, adversarial strategies, and joint training. A category overview of the biomedical image registration methods using GAN is shown in Table 1. In the table, we describe the key ideas of the four categories, respectively, and summarize the different implementation methods for each strategy. In the subsequent subsections, we review all the relevant works as classified in Table 1.
To present a comprehensive overview of all the relevant works on GANs in biomedical image registration, we searched science datasets, including Google Scholar, SpringerLink, and PubMed, for all relevant published articles from 2018 to 2021. The keywords included medical image registration/matching alignment, GAN (Generative Adversarial Networks), multimodal (cross-modality) medical image registration, GAN, adversarial medical image registration, segmentation, and registration. About 300 papers are indexed, including 36 papers that completely match our criteria. There are two requirements for our inclusion in the articles. The first is that the topic of the papers is image registration, and the second is that the method is of GAN based on and used to implement the registration strategy. To verify the comprehensiveness of the search, we also searched them separately in the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), the IEEE International Symposium on Biomedical Imaging (ISBI), and SPIE Medical Imaging to compare with the already searched papers. During the literature review process, we try to integrate all relevant papers to reach a reasonable conclusion; however, because this topic is still in its infancy, the number of published papers is minimal. Therefore, we are unable to conduct an experimental review on this topic because most of the searched articles have no open-source code, and the data are private.
Modality-Independent based strategy
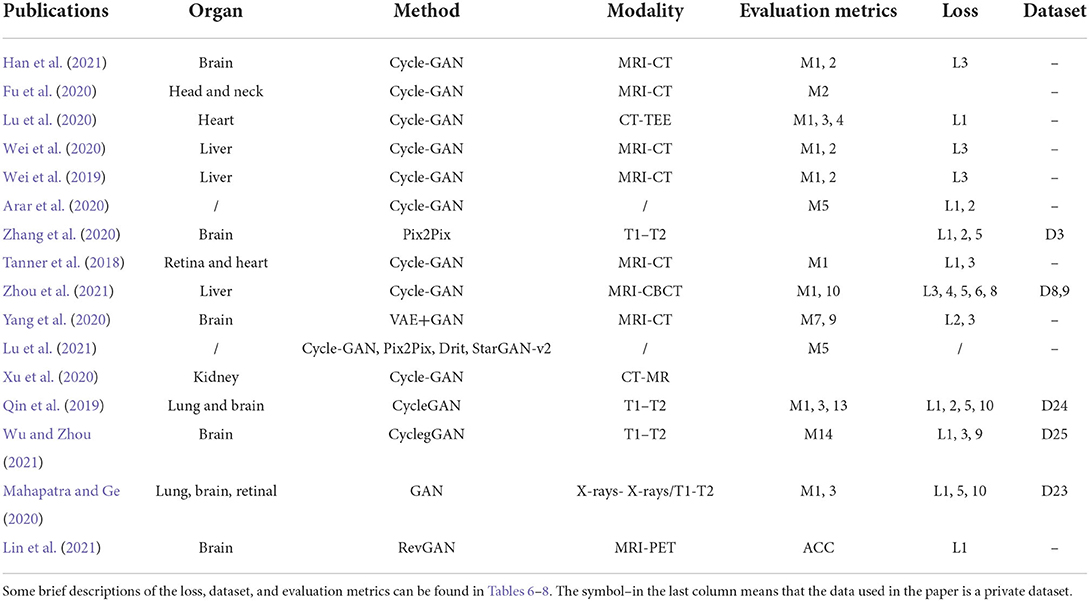
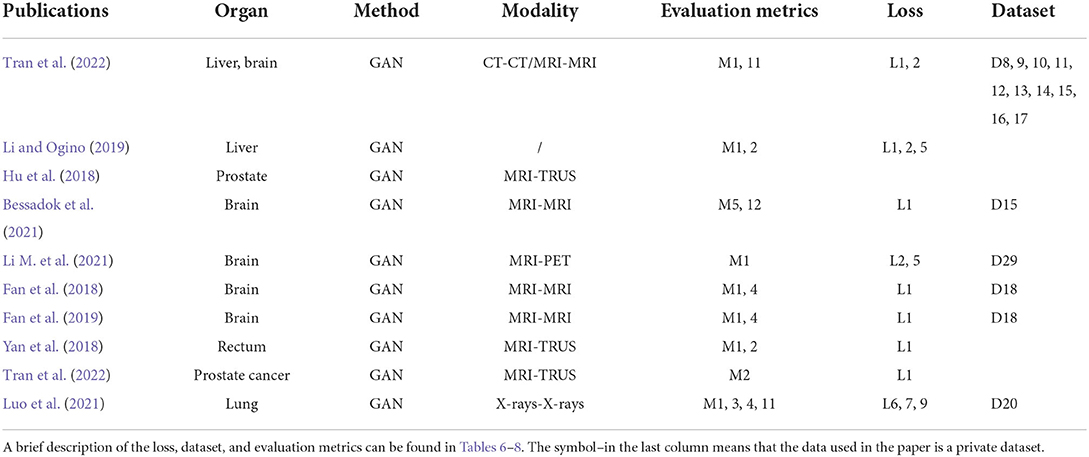
Biomedical image registration algorithms of modality-independent based strategies mainly focus on cross-modal images. The key idea of the strategy is to eliminate the variance between modalities so that cross-modality registration can be performed on modality-independent data. A modality-independent strategy can be implemented by translating cross-modality, image disentangling, and latent representation methods. This strategy can avoid the design of cross-modal similarity loss. It only uses robust mono-modal similarity loss to guide the optimization of the model. In Table 2, we provide an overview of the important elements of all the reviewed papers. Among these papers, 12 directly use Cycle-GAN as the baseline model, and seven are applied to the registration tasks of MRI-CT, with the organs covered by the brain, liver, retina, and heart.
Modality translation
To register MRI to CT, Tanner et al. (2018) make the first attempt at modality translation using Cycle-GAN and subsequently perform the mono-modality registration on two images in the same domain. The Patch-GAN uses N × N patches instead of a single value as the output of the discriminator for spatial corresponded-preservation. This pioneering work assessed the feasibility of this strategy. However, the mono-registration significantly relies on the quality of synthetic images. In a subsequent study, to constrain the geometric changes during modality translation, Wei et al. (2020) designed the mutual information (MI) loss to regularize the anatomy and between the classic mono-modal registration method ANTS (Avants et al., 2008, 2009) as the registration network. Xu et al. (2020) combined the deformation field from uni- and multimodal stream networks by dual stream fusion network for cross-modality registration. The uni-modal stream model preserves the anatomy during modality translation using the Cycle-GAN by combining several losses, including the modality independent neighborhood descriptor (MIND) (Heinrich et al., 2012), the correlation coefficient loss (CC), and the L2 loss. The basic structures of these methods are shown in Figure 3D. To further solve the uni-modal mismatch problem caused by the unrealistic soft-tissue details generated by the modality translation, the multimodal stream network is proposed on the UNET-based cross-modal network to learn the original information from both the fixed and moving images. The dual stream fusion network is responsible for fusing the deformation fields of the uni-modal and multimodal streams. The two registration streams' complementary functions preserve the edge details of images. However, the multimodal stream also learns some redundant features, which is not beneficial to the alignment. Unlike the aforementioned methods, Zhou et al. (2021) translate the CBCT and MRI to the CT modality by Cycle-GAN. The UNET-based segmentation network is trained to get the segmentation map of the synthetic CT image for guiding the robust point matching (RPM) registration. The systems combine the synthesis and segmentation networks to implement cross-modality image segmentation. Arar et al. (2020) assume that a spatial translation network (STN) (Jaderberg et al., 2015) registration network (R) and the CGAN-based translation net (T) are commutative, i.e., T°R=R°T. Based on this assumption, the optimized L1-reconstruction loss Lrecon(T, R) = ||ORT−Itarget||1+||OTR−Itarget||1, which encourages T to be geometrically preserved. Benefited from the anatomy-consistency constraints, the registration accuracy can be improved. However, the training of GAN may suffer from non-convergence, which may pose certain additional difficulties to the training of the registration network. Lu et al. (2021) assessed what role image translation plays in the cross-modal registration based on the performance of Cycle-GAN, which also shows the instability of this approach and the overdependence on the data.
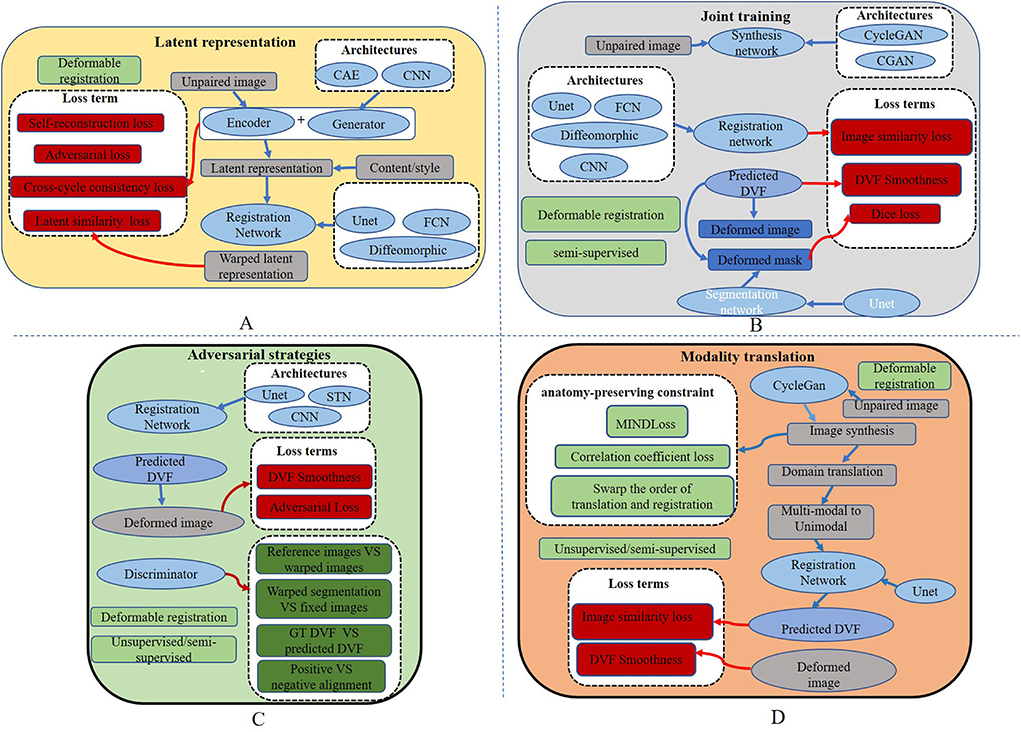
Figure 3. Overall structures of existing cross-modal image registration methods. (A) The overall structure of the latent representation method. (B) The overall structure of joint learning-based strategy. (C) The overall structure of adversarial learning-based strategy. (D) The basic structures of modality translation.
Image disentangling
Qin et al. (2019) try to learn a registration function in modal-independent latent shape space in an unsupervised manner. The proposed framework consists of three parts: a disentangling image network via unpaired modality translation, a deformable registration network in the disentangled latent space, and a GAN modal to learn a similarity metric in the image space implicitly. Several losses are used in the network to train the three parts, including the self-reconstruction loss, the latent reconstruction loss, the cross-cycle consistency, and the adversarial loss with similarity metrics defined in latent space. The work is capable of translating cross-modal images by image disentangling to obtain shape latent representation related to the image alignment. This method can alleviate unrealistic image generation from the Cycle-GAN-based approaches. However, the deformation field generated by latent shape representation introduces unsmooth edges. Wu and Zhou (2021) propose a fully unsupervised registration network through image disentangling. The proposed registration framework consists of two parts: one registration network aligns the image from x to y, and the other aligns the image from y to x. Each part consists of two subnetworks: an unsupervised deformable registration network and a disentangling representation network via unpaired image-to-image translation. Unlike Qin et al. (2019), the representation disentangling model aims to drive a deformable registration network for learning the mapping between the two modalities.
Latent representation
Mahapatra and Ge (2020) use a convolutional autoencoder (CAE) network to learn latent space representation for different modalities of images. The generator is fed into latent features from CAE to generate the deformation fields. The intensity and shape constraints are achieved by content loss, including the normal mutual information (NMI), the structural similarity index measure (SSIM) (Wang et al., 1987, 2004), and the visual graphics generator (VGG) (Simonyan and Zisserman, 2014) with L2 loss. The cycle consistency loss and the adversarial loss are used to constrain the deformation field consistency, which is calculated as follows:
where (G, F) represents the two generators, and represent IFlt and IRef as the real data of the discriminator, x and y represent the original images of the two modalities, and MSENorm is the MSE normalized to [0, 1]. Yang et al. (2020) transform image modality through a conditional auto-encoder generative adversarial network (CAE-GAN), which redesigns VAE (Kingma and Welling, 2013) and GAN to form the symmetric UNET. The registration network uses a traditional nonparametric deformable method based on local phase differences at multiple scales. The overall structure of the latent representation method is shown in Figure 3A.
Symmetric learning-based strategy
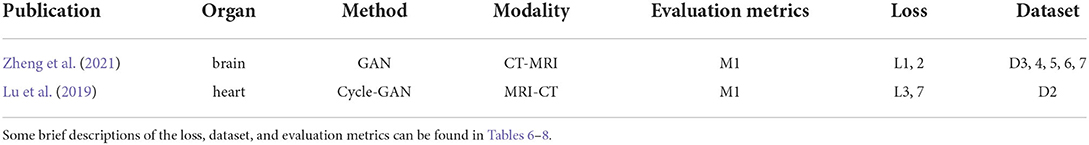
Table 3 lists two papers about using the symmetric learning-based GAN methods. Both of them perform image registration for CT-MRI. One is used on the brain, and another on the heart.
Cyclic-consistency
From the perspective of cyclic learning, symmetric learning can assist and supervise each other. The CIRNet (Lu et al., 2019) uses two cascaded networks with identical structures as the symmetric registration networks. The two cascaded networks share the weights. The L2 loss is used as Lcyc to enforce image A translating through two deformation fields ϕ1 and ϕ2, and A(ϕ1, ϕ2) = A. Lcyc is defined as follows:
where N represents the number of all the voxels, and Ω refers to all the voxels in the image. The total loss of the two registration networks is represented by Equations (13, 14), respectively:
where A(φ1) denotes a warped image produced by the module R1, A(φ1, φ2) denotes a warped image produced by the module R2, and Lreg(φ1) is the smooth regularizationLreg.
SymReg-GAN (Zheng et al., 2021) proposes a GAN-based symmetric registration to resolve the inverse-consistent translation between cross-modal images. A generator performs the modality translation, consisting of an affine-translation regressor and a non-linear-deformation regressor. The discriminator distinguishes between the translation estimation and the ground truth. The SymReg-GAN is trained by jointly utilizing labeled and unlabeled images. It encourages symmetry in registration by enforcing a condition that in the cycle composed of the transformation from one image to the other, its reverse transformation should bring the original image back. The total loss combines the symmetry loss, registration loss, and supervision loss into one. This method takes full advantage of both labeled and unlabeled data and resolves the limitation of iterative optimization by non-learning techniques. However, the spatial transformation and the modality transformation may not be the same, and even if the spatial transformation is symmetric, the transformation error may still be cyclic.
Adversarial learning-based strategy
Biomedical image registration algorithms of adversarial learning-based strategies utilize adversarial loss to drive the learning of registration networks such as GAN. Adversarial loss consists of two parts: the training aim of the generator is to generate an image that makes the discriminator consider it real, and the optimization objective of the discriminator is to distinguish between an image generated by the generator and a real image in the dataset as accurately as possible. Based on this strategy, several methods, as shown in Table 1, including semi-supervised, knowledge distillation, attention mechanisms, and adversarial training, are implemented to improve registration performance. The similarity loss is instead by learning a discriminator network. Although GAN can be trained unsupervised, paired training data may be more helpful for model convergence for the cross-modal registration modal. An overview of the crucial elements of all the reviewed papers is shown in Table 4. Five papers are for mono-modal image registration and four for cross-modal registration. The overall structure of this strategy is shown in Figure 3C.
Adversarial training
For image registration tasks, the effect of adversarial loss is to make the warped image closer to the target image (Yan et al., 2018). The role of the generator is to generate a deformation field, and the task of the discriminator is to identify the alignment image. For more stable loss, Wasserstein GAN (WGAN) is adopted. Since a discriminator can be considered a registration image quality assessor, the quality of a warped image can be improved with cross-modal similarity metrics. However, the training of GAN may suffer from non-convergence, which may pose additional difficulties in training the registration network. Compared to Fan et al. (2018, 2019) and Yan et al. (2018) select more reasonable reference data for training the discriminator for better model convergence.
Semi-supervised
Hu et al. (2018) use a biomechanical simulation deformation field to regularize the deformation fields formed by the alignment network. The generator is fed into simulated motion data to form a translation. The discriminator tries to distinguish the dense displacement field from ground truth deformation. Another similarity loss metric warps moving labels and fixed labels in a weakly supervised manner. Li and Ogino (2019) propose a general end-to-end registration network in which a CNN similar to UNET is trained to generate the deformation field. For better guiding, the anatomical shape alignment and masks of moving and fixed objects are also fed into the registration network. The input of the discriminator net is a positive and negative alignment pair, consisting of masks and images of the fixed and warped images for guiding a finer anatomy alignment. In addition, an encoder extracts anatomical shape differences as another registration loss. The studies by Elmahdy et al. (2019) and Luo et al. (2021) are similar to that of Li and Ogino (2019), in which the adversarial learning-based registration network joint segmentation and registration with segmentation label information as the input of the generator and the discriminator. The dice similarity coefficient (DSC) and the normalized cross-correlation (NCC) are added to the generator to avoid slow convergence and suboptimal registration.
Knowledge distillation
Knowledge distillation is a process of transferring knowledge from a cumbersome pre-trained model (i.e., the teacher network) to a light-weighted one (i.e., the student network). Tran et al. (2022) used knowledge distillation by adversarial learning to streamline the expensive and effective teacher registration model to a light-weighted student registration model. In their proposed method, the teacher network is the recursive cascaded network (RCN) (Zhao et al., 2019a) for extracting meaningful deformations, and the student network is a CNN-based registration network. When training the registration network, the teacher network and the student network are optimized by Ladv = Υlrec+(1−Υ)ldis, where lrec represents the reconstructed loss and the discriminator loss, and lrec and ldis are expressed as follows:
where denotes a warped image by the student network, CorrCoef[I1−I2] is the correlation between images I1 and I2, ∅s and ∅t denote the deformation of the teacher and the student networks, respectively, and denotes the joint deformation. Applying knowledge distillation by means of adversarial learning provides a new and efficient way to reduce computational costs and achieve competitive accuracy.
Attention mechanisms
To reduce feature loss of the upsampling process in a registration network, Li M. et al. (2021) proposed a GAN-based registration network combining UNET with dual attention mechanisms. The dual attention mechanisms consist of the channel attention mechanism and the location attention mechanism. Meanwhile, the residual structure is also introduced into the upsampling process for improving feature restoration.
Joint learning-based strategy
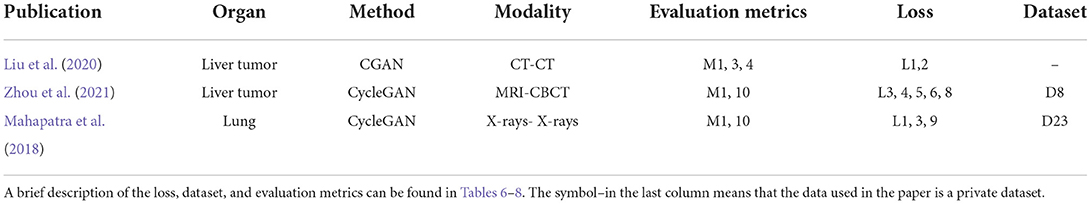
Joint learning of segmentation, registration, and synthesis networks can improve their performance for each other. An overview of the essential elements of all the reviewed papers is shown in Table 5. Two of which are for mono-model registration methods. The overall structure is shown in Figure 3B.
Multitask
Existing experiments show that a segmentation map can help registration by joint learning. However, in real registration tasks, segmentation labels may not be available. Liu et al. (2020) propose a joint system of segmentation, registration, and synthesis via multi-task learning. The objectives of the CGAN-based synthesis model and the registration model are optimized via a joint loss. The segmentation network is trained in a supervised manner. The segmentation module estimates the segmentation map for the moving, fixed, and synthesized images. During the training procedure, a dice loss is optimized between the segmentation maps of the warped moving image and the fixed image. The result proves that the segmentation task can improve registration accuracy. Zhou et al. (2021) take advantage of each other through the joint Cycle-GAN and UNET-based segmentation network to solve the missing label problem via Cycle-GAN's translating of the two modalities to the third one with a large number of available labels. Thus, the synthesis network improves the segmentation accuracy and further improves the accuracy of the RPM registration. Mahapatra et al. (2018) have trained the generation network to complete the alignment of the reference and moving images by combining the segmentation map that is used directly as the input to the generator, with no need to train an additional segmentation network. Segmentation and alignment are mutually driven. The ways of joining image registration task and image segmentation task may improve the accuracy by sharing the result of learning, which can expand the goal of the registration research.
Statistics
It is essential to conduct relevant analyses from a global perspective after a detailed study of each category of biomedical image registration strategies. In the past 4 years, more than half of the reviewed works have used the modality-independent-based strategy to solve cross-modal biomedical registration—the methods of Adversarial Learning Based Strategy account for 32%. From 2020 to 2021, the number of articles published on the modality-independent-based strategy was higher than others, peaking in 2020. However, there is a drop-down trend in 2021. As noted, no paper on the adversarial learning-based strategy was published in 2020. In the other years, the works on the adversarial learning-based strategy are published in a balanced proportion, with the detailed percentages shown in Figures 4, 5. In addition to analyzing the trends in the published number of papers and the popularity of the four strategy categories, we also analyzed the percentage distribution of the other characteristics, which is shown in Figure 6. A total of 75% of the works aim to solve the problem of the cross-modal domain of biomedical image registration, among which 46 and 42% adopt direct or indirect Cycle-GAN and GAN are part of the important structure of the registration framework. Cycle-GAN is utilized only for the cross-modal domain of biomedical image registration, whereas GAN is utilized for both cross-modal and uni-modal image registration. In the cross-modal bioimage registration, 33% of the works perform image registration between CT and MRI. The number of articles using MRI accounts for 97%. Regarding the region of interest (ROI), the brain and liver are the most studied sites. The brain is the top registration target in all works. The reason for the wide adoption of the brain consists of its clinical importance, availability in public datasets, and relative simplicity of registration.
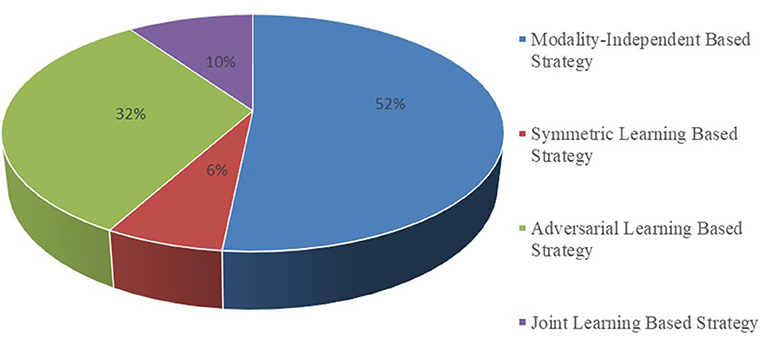
Figure 4. Proportional distribution pie chart of the number of publications on different implementation strategies.
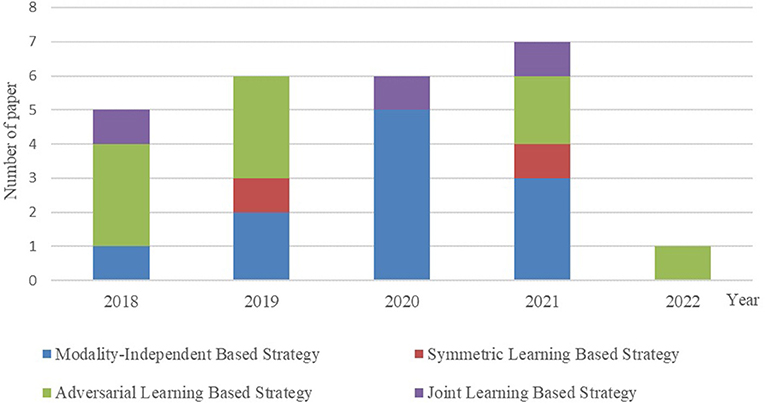
Figure 5. Contrast bar graph of the number of publications on the four strategies for the GAN-based biomedical image registration over the last 5 years from 2018–2022.
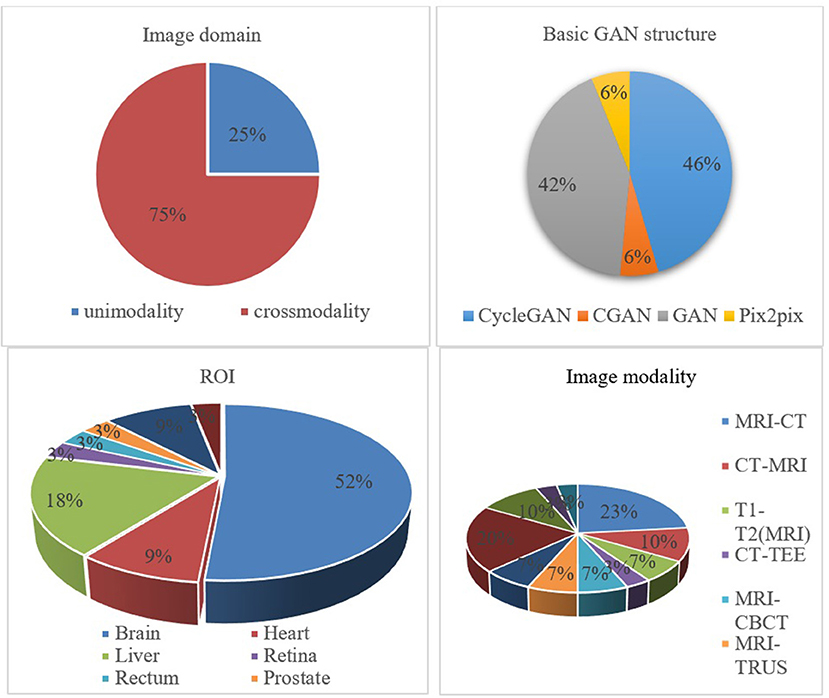
Figure 6. Percentage pie chart of various attributes of the GAN-based biomedical image registration methods.
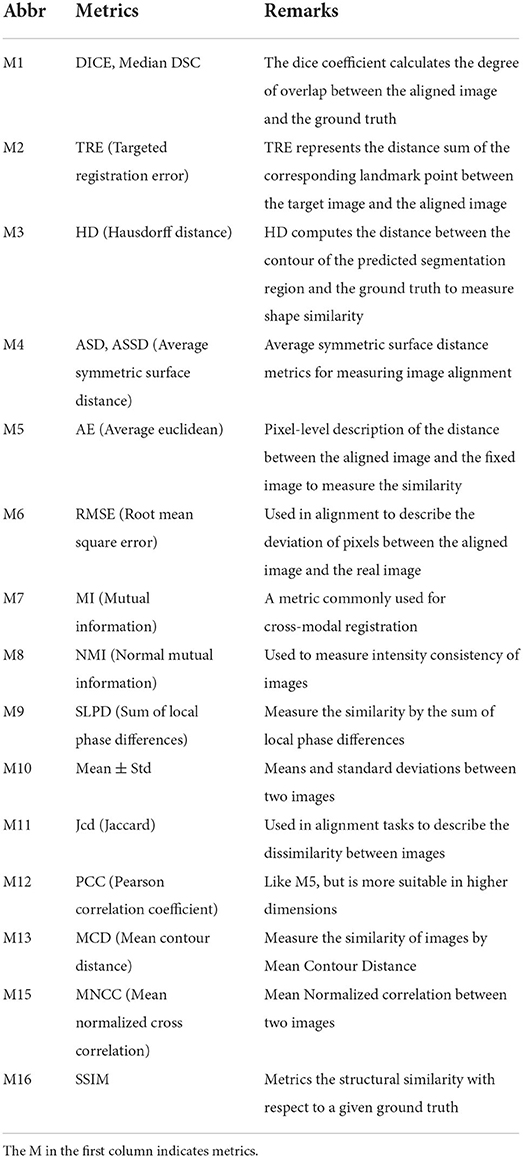
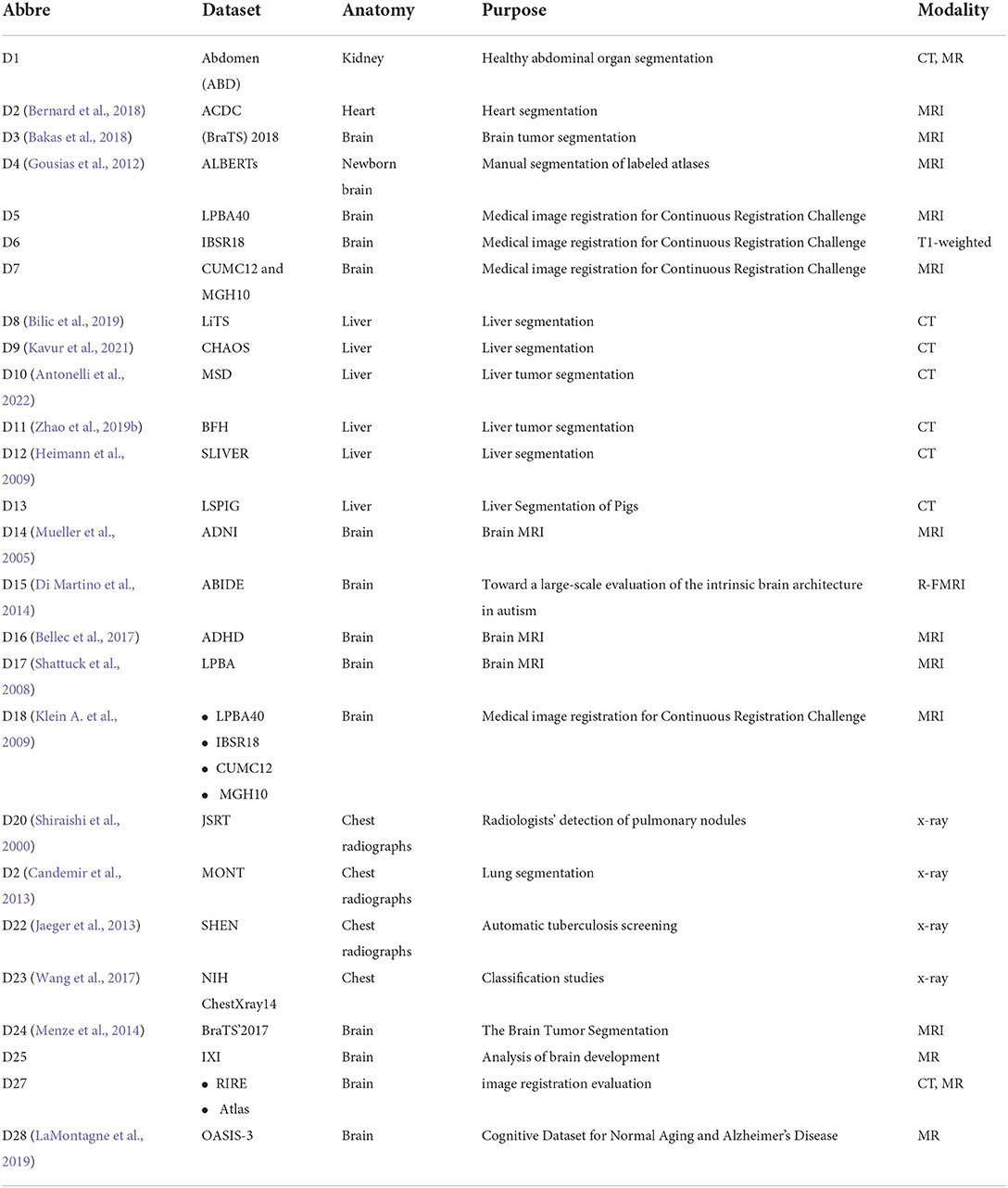
In addition, the metrics used in the cross-modal registration methods are shown in Figure 7. As seen in the figure, dice and TRE are the top two most frequently used metrics. The Dice coefficient calculates the degree of overlap between the aligned image and the ground truth, and the confusion matrix formula is as follows:
where “ref” refers to the reference image, and “w” represents a warped image. Obviously, when the two images overlap exactly, the Dice coefficient is 1. TRE represents the distance sum of the corresponding landmark between the target image and the aligned image and is expressed as follows:
where n is the number of landmarks, r is the reference image, w is the aligned image, i is the i-th corresponding point, and d indicates the Euclidean distance.
Future perspectives
Exploring in-between representatives of two modalities
Many existing modality-translation-based methods for cross-modality biomedical image registration rely on synthetic images to train the mono-modal registration network because it is difficult to develop cross-modality similarity measures. Although such a training scheme does not need to perform cross-modal similarity metrics to improve the image synthesis performance, it is still necessary to design various losses to constrain other feature changes. Additionally, is the synthetic modality useful for improving registration performance? As far as we know, this intensity information does not play a key role in improving image performance. Some shape features, such as edges and corners, are essential for image registration. An in-between representation was found (Lu et al., 2021), i.e., COMIR, which maps the modalities to their established “common ground.” An in-between representative with characteristics relevant to the accurate alignment would be good. In the future, more workers are expected to be carried out in this direction to find the in-between representatives.
Exploring quality assessment guided modal translation network
The image quality generated by the mode translation network directly affects the accuracy of the registration algorithm. Therefore, an important research direction is how to effectively and reasonably evaluate the quality of images generated by the GAN network. Additionally, an effective generated image quality evaluation method can be used to constrain the mode translation network's training process and improve the modal translation's effectiveness. There have recently been quality evaluation methods for images generated by GAN (Gu et al., 2020), but there is still a lack of quality evaluation methods for synthetic biomedical images.
Designing large-scale biomedical image generation GAN network
The size of images existing image generation networks can generate is minimal (Brock et al., 2019), but biomedical images are generally of high resolution, especially biological images used in neurological research. The training process of the existing GAN network is difficult to converge, especially with the increase in image size. The dimension of data space will dramatically increase. This challenge is difficult with current hardware levels and GAN-based image-synthesized methods. Therefore, designing an image synthesis network capable of synthesizing large-scale biomedical images is also a future direction.
Designing prior knowledge-guided registration methods
Traditional image registration models often use some standard landmarks like points and lines as guidance to optimize the model. Several recent studies have shown that a segmentation mask can be utilized in the discriminator (Luo et al., 2021) or generator (Mahapatra et al., 2018) for guiding the edge alignment. However, their works simply use only a segmentation mask as the edge space correspondence guidance. More space correspondence features are expected to be explored and verified.
Conclusions
This paper provides a comprehensive survey of cross-modal and mono-modal biomedical image registration approaches based on GAN. The commonly used GAN structures are summarized, followed by the analyses of the biomedical image registration studies of the modality-independent based strategy, the symmetric learning-based strategy, the adversarial learning-based strategy, and the joint learning-based strategy from different implementation methods and perspectives. In addition, we have conducted a statistical analysis of the existing literature in various aspects and have drawn the corresponding conclusions. Finally, we outline four interesting research directions for future studies.
Author contributions
TH, JW, and LQ contributed to the conception and design of this paper. TH completed the literature selection and the writing of the first draft. JW and ZJ improved the writing of the paper. LQ provided constructive comments and determined the final draft of the paper. All authors contributed to the drafting of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (61871411, 62271003, and 62201008), the Sci-Tech Innovation 2030 Agenda (2022ZD0205200 and 2022ZD0205204), the University Synergy Innovation Program of Anhui Province (GXXT-2021-001), and the Natural Science Foundation of the Education Department of Anhui Province (KJ2021A0017).
Acknowledgments
The authors acknowledge the high-performance computing platform of Anhui University for providing computing resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fninf.2022.933230/full#supplementary-material
References
Antonelli, M., Reinke, A., Bakas, S., Farahani, K., Kopp-Schneider, A., Landman, B. A., et al. (2022). The medical segmentation decathlon. Nat. Commun. 13, 4128. doi: 10.1038/s41467-022-30695-9
Arar, M., Ginger, Y., Danon, D., Bermano, A. H., and Cohen-Or, D. (2020). “Unsupervised multi-modal image registration via geometry preserving image-to-image translation,” in IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Seattle, WA), 13410–13419. doi: 10.1109/CVPR42600.2020.01342
Avants, B. B., Epstein, C. L., Grossman, M., and Gee, J. C. (2008). Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41. doi: 10.1016/j.media.2007.06.004
Avants, B. B., Tustison, N., and Song, G. (2009). Advanced normalization tools (ANTS). Insight J. 2, 1–35. doi: 10.54294/uvnhin
Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, M., Crimi, A., et al. (2018). Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge.
Balakrishnan, G., Zhao, A., Sabuncu, M. R., Guttag, J., and Dalca, A. V. (2019). VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38, 1788–1800. doi: 10.1109/TMI.2019.2897538
Bellec, P., Chu, C., Chouinard-Decorte, F., Benhajali, Y., Margulies, D. S., and Craddock, R. C. (2017). The neuro bureau ADHD-200 preprocessed repository. Neuroimage 144, 275–286. doi: 10.1016/j.neuroimage.2016.06.034
Bernard, O., Lalande, A., Zotti, C., Cervenansky, F., Yang, X., Heng, P.-A., et al. (2018). Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans. Med. Imaging 37, 2514–2525. doi: 10.1109/TMI.2018.2837502
Bessadok, A., Mahjoub, M. A., and Rekik, I. (2021). Brain graph synthesis by dual adversarial domain alignment and target graph prediction from a source graph. Med. Image Anal. 68, 101902–101902. doi: 10.1016/j.media.2020.101902
Bilic, P., Christ, P. F., Vorontsov, E., Chlebus, G., Chen, H., Dou, Q., et al. (2019). The Liver Tumor Segmentation Benchmark (Lits).
Brock, A., Donahue, J., and Simonyan, K. (2019). “Large scale GAN training for high fidelity natural image synthesis,” in International Conference on Learning Representations(ICLR) (New Orleans, LA).
Candemir, S., Jaeger, S., Palaniappan, K., Musco, J. P., Singh, R. K., Xue, Z., et al. (2013). Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 33, 577–590. doi: 10.1109/TMI.2013.2290491
Chen, W., Liu, M., Du, H., Radojević, M., Wang, Y., and Meijering, E. (2021). Deep-Learning-based automated neuron reconstruction from 3D microscopy images using synthetic training images. IEEE Trans. Med. Imaging 41, 1031–1042. doi: 10.1109/TMI.2021.3130934
Chen, X., Duan, Y., Houthooft, R., Schulman, J., Sutskever, I., and Abbeel, P. (2016). Infogan: interpretable representation learning by information maximizing generative adversarial nets. Adv. Neural Inf. Process. Syst. 29, 2180–2188.
Di Martino, A., Yan, C.-G., Li, Q., Denio, E., Castellanos, F. X., Alaerts, K., et al. (2014). The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 19, 659–667. doi: 10.1038/mp.2013.78
Elmahdy, M. S., Wolterink, J. M., Sokooti, H., Išgum, I., and Staring, M. (2019). “Adversarial optimization for joint registration and segmentation in prostate CT radiotherapy,” in Medical Image Computing and Computer-Assisted Intervention (MICCAI) (Shanghai), 366–374. doi: 10.1007/978-3-030-32226-7_41
Fan, J., Cao, X., Wang, Q., Yap, P. T., and Shen, D. (2019). Adversarial learning for mono- or multi-modal registration. Med. Image Anal. 58, 101545. doi: 10.1016/j.media.2019.101545
Fan, J., Cao, X., Xue, Z., Yap, P. T., and Shen, D. (2018). Adversarial similarity network for evaluating image alignment in deep learning based registration. Med. Image Comput. Comput. Assist. Interv. 11070, 739–746. doi: 10.1007/978-3-030-00928-1_83
Fu, Y., Lei, Y., Zhou, J., Wang, T., David, S. Y., Beitler, J. J., et al. (2020). Synthetic CT-aided MRI-CT image registration for head and neck radiotherapy. Int. Soc. Opt. Photon. 11317. doi: 10.1117/12.2549092
Gering, D. T., Nabavi, A., Kikinis, R., Hata, N., O'Donnell, L. J., Grimson, W. E. L., et al. (2001). An integrated visualization system for surgical planning and guidance using image fusion and an open MR. J. Mag. Reson. Imaging 13, 967–975. doi: 10.1002/jmri.1139
Gong, H., Xu, D., Yuan, J., Li, X., Guo, C., Peng, J., et al. (2016). High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 7, 12142. doi: 10.1038/ncomms12142
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2014). Generative adversarial nets. Adv. Neural Inf. Process. Syst. 27, 139–144. doi: 10.1145/3422622
Gousias, I. S., Edwards, A. D., Rutherford, M. A., Counsell, S. J., Hajnal, J. V., Rueckert, D., et al. (2012). Magnetic resonance imaging of the newborn brain: manual segmentation of labelled atlases in term-born and preterm infants. Neuroimage 62, 1499–1509. doi: 10.1016/j.neuroimage.2012.05.083
Gu, S., Bao, J., Chen, D., and Wen, F. (2020). “Giqa: generated image quality assessment,” in inj European Conference on Computer Vision, ECCV (Glasgow), 369–385. doi: 10.1007/978-3-030-58621-8_22
Han, R., Jones, C. K., Ketcha, M. D., Wu, P., Vagdargi, P., Uneri, A., et al. (2021). Deformable MR-CT image registration using an unsupervised end-to-end synthesis and registration network for endoscopic neurosurgery. Int. Soc. Opt. Photon. 11598. doi: 10.1117/12.2581567
He, Y., Li, T., Ge, R., Yang, J., Kong, Y., Zhu, J., et al. (2021). Few-shot learning for deformable medical image registration with perception-correspondence decoupling and reverse teaching. IEEE J. Biomed. Health Inform. 26, 1177–1187. doi: 10.1109/JBHI.2021.3095409
Heimann, T., Van Ginneken, B., Styner, M. A., Arzhaeva, Y., Aurich, V., Bauer, C., et al. (2009). Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans. Med. Imaging 28, 1251–1265. doi: 10.1109/TMI.2009.2013851
Heinrich, M. P., Jenkinson, M., Bhushan, M., Matin, T., Gleeson, F. V., Brady, M., et al. (2012). MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration. Med. Image Anal. 16, 1423–1435. doi: 10.1016/j.media.2012.05.008
Hu, Y., Gibson, E., Ghavami, N., Bonmati, E., Moore, C. M., Emberton, M., et al. (2018). “Adversarial deformation regularization for training image registration neural networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Granada), 774–782. doi: 10.1007/978-3-030-00928-1_87
Ioffe, S., and Szegedy, C. (2015). Batch normalization: accelerating deep network training by reducing internal covariate shift. PMLR. 27, 448–456.
Isola, P., Zhu, J.-Y., Zhou, T., and Efros, A. A. (2017). “Image-to-Image translation with conditional adversarial networks,” in IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Hawaii, HI), 1125–1134. doi: 10.1109/CVPR.2017.632
Jaderberg, M., Simonyan, K., and Zisserman, A. (2015). Spatial transformer networks. Adv. Neural Inform. Proc. Syst. 28, 607.
Jaeger, S., Karargyris, A., Candemir, S., Folio, L., Siegelman, J., Callaghan, F., et al. (2013). Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging 33, 233–245. doi: 10.1109/TMI.2013.2284099
Jiang, S., Wang, C., and Huang, C. (2021). “Image registration improved by generative adversarial networks,” in International Conference on Multimedia Modeling (Prague), 26–35. doi: 10.1007/978-3-030-67835-7_3
Jing, Y., Yang, Y., Feng, Z., Ye, J., Yu, Y., and Song, M. (2020). Neural style transfer: a review. IEEE Trans. Vis. Comput. Graph. 26, 3365–3385. doi: 10.1109/TVCG.2019.2921336
Kavur, A. E., Gezer, N. S., Bariş, M., Aslan, S., Conze, P.-H., Groza, V., et al. (2021). CHAOS challenge-combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 69, 101950. doi: 10.1016/j.media.2020.101950
Kim, T., Cha, M., Kim, H., Lee, J. K., and Kim, J. (2017). “Learning to discover cross-domain relations with generative adversarial networks,” in International Conference on Machine Learning (Sydney) 1857–1865.
Kingma, D. P., and Welling, M. (2013). Auto-encoding variational bayes. arXiv preprint. arXiv:1312.6114.
Klein, A., Andersson, J., Ardekani, B. A., Ashburner, J., Avants, B., Chiang, M.-C., et al. (2009). Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46, 786–802. doi: 10.1016/j.neuroimage.2008.12.037
Klein, S., Staring, M., Murphy, K., Viergever, M. A., and Pluim, J. P. W. (2009). Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29, 196–205. doi: 10.1109/TMI.2009.2035616
LaMontagne, P. J., Benzinger, T. L. S., Morris, J. C., Keefe, S., Hornbeck, R., Xiong, C., et al. (2019). OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer disease. Alzheimers Dementia. doi: 10.1101/2019.12.13.19014902
Li, A. A., and Gong, H. (2012). Progress on whole brain imaging methods at the level of optical microscopy. Prog. Biochem. Biophys. 39,498–504. doi: 10.3724/SP.J.1206.2012.00237
Li, C., and Wand, M. (2016). “Precomputed real-time texture synthesis with markovian generative adversarial networks,” in European Conference on Computer Vision (Amsterdam), 702–716. doi: 10.1007/978-3-319-46487-9_43
Li, M., Wang, Y., Zhang, F., Li, G., Hu, S., and Wu, L. (2021). “Deformable medical image registration based on unsupervised generative adversarial network integrating dual attention mechanisms,” in International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI) (Shenyang) 1–6. doi: 10.1109/CISP-BMEI53629.2021.9624229
Li, X., Jiang, Y., Rodriguez-Andina, J. J., Luo, H., Yin, S., and Kaynak, O. (2021). When medical images meet generative adversarial network: recent development and research opportunities. Discover Art. Intell. 1, 1–20. doi: 10.1007/s44163-021-00006-0
Li, Z., and Ogino, M. (2019). “Adversarial learning for deformable image registration: application to 3d ultrasound image fusion,” in Smart Ultrasound Imaging and Perinatal, Preterm and Paediatric Image Analysis (Cham: Springer), 56–64. doi: 10.1007/978-3-030-32875-7_7
Lin, W., Lin, W., Chen, G., Zhang, H., Gao, Q., Huang, Y., et al. (2021). Bidirectional mapping of brain MRI and PET with 3D reversible GAN for the diagnosis of Alzheimer's disease. Front. Neurosci. 15, 357. doi: 10.3389/fnins.2021.646013
Liu, F., Cai, J., Huo, Y., Cheng, C.-T., Raju, A., Jin, D., et al. (2020). “Jssr: a joint synthesis, segmentation, and registration system for 3d multi-modal image alignment of large-scale pathological ct scans,” in European Conference on Computer Vision (Glasgow) 257–274. doi: 10.1007/978-3-030-58601-0_16
Liu, M.-Y., Breuel, T., and Kautz, J. (2017). Unsupervised image-to-image translation networks. Adv. Neural Inf. Process. Syst. 30, 721–730. doi: 10.1007/978-3-319-70139-4
Lu, J., Öfverstedt, J., Lindblad, J., and Sladoje, N. (2021). Is Image-to-Image Translation the Panacea for Multimodal Image Registration? A Comparative Study.
Lu, Y., Li, B., Liu, N., Chen, J. W., Xiao, L., Gou, S., et al. (2020). CT-TEE image registration for surgical navigation of congenital heart disease based on a cycle adversarial network. Comput. Math. Methods Med. 2020, 4942121. doi: 10.1155/2020/4942121
Lu, Z., Yang, G., Hua, T., Hu, L., Kong, Y., Tang, L., et al. (2019). “Unsupervised three-dimensional image registration using a cycle convolutional neural network,” in IEEE International Conference on Image Processing (ICIP) (Taipei), 2174–2178. doi: 10.1109/ICIP.2019.8803163
Luo, Y., Cao, W., He, Z., Zou, W., and He, Z. (2021). Deformable adversarial registration network with multiple loss constraints. Comput. Med. Imaging Graph. 91, 101931. doi: 10.1016/j.compmedimag.2021.101931
Maas, A. L., Hannun, A. Y., and Ng, A. Y. (2013). “Rectifier nonlinearities improve neural network acoustic models,” in Proceeding ICML (Shenzhen).
Mahapatra, D., and Ge, Z. (2020). Training data independent image registration using generative adversarial networks and domain adaptation. Patt. Recognit. 100, 107109. doi: 10.1016/j.patcog.2019.107109
Mahapatra, D., Ge, Z., Sedai, S., and Chakravorty, R. (2018). “Joint registration and segmentation of Xray images using generative adversarial networks,” in Machine Learning in Medical Imaging (Granada), 73–80. doi: 10.1007/978-3-030-00919-9_9
Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., et al. (2014). The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34, 1993–2024. doi: 10.1109/TMI.2014.2377694
Mueller, S. G., Weiner, M. W., Thal, L. J., Petersen, R. C., Jack, C. R., Jagust, W., et al. (2005). Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's disease neuroimaging initiative (ADNI). Alzheimers Dementia 1, 55–66. doi: 10.1016/j.jalz.2005.06.003
Oliveira, F. P., and Tavares, J. M. (2014). Medical image registration: a review. Comput. Methods Biomech. Biomed. Engin. 17, 73–93. doi: 10.1080/10255842.2012.670855
Qin, C., Shi, B., Liao, R., Mansi, T., Rueckert, D., and Kamen, A. (2019). “Unsupervised deformable registration for multi-modal images via disentangled representations,” in International Conference on Information Processing in Medical Imaging (Hong Kong), 249–261. doi: 10.1007/978-3-030-20351-1_19
Qu, L., Li, Y., Xie, P., Liu, L., Wang, Y., et al. (2021). Cross-Modality Coherent Registration of Whole Mouse Brains. doi: 10.1038/s41592-021-01334-w
Radford, A., Metz, L., and Chintala, S. (2015). “Unsupervised representation learning with deep convolutional generative adversarial networks,” in International Conference on Learning Representations(ICLR) (San Diego, CA).
Ragan, T., Kadiri, L. R., Venkataraju, K. U., Bahlmann, K., Sutin, J., Taranda, J., et al. (2012). Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 9, 255–258. doi: 10.1038/nmeth.1854
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-Net: convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention (MICCAI) (Munich), 234–241. doi: 10.1007/978-3-319-24574-4_28
Ruska, E. (1987). The development of the electron microscope and of electron microscopy. Rev. Mod. Phys. 59, 627. doi: 10.1103/RevModPhys.59.627
Shattuck, D. W., Mirza, M., Adisetiyo, V., Hojatkashani, C., Salamon, G., Narr, K. L., et al. (2008). Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 39, 1064–1080. doi: 10.1016/j.neuroimage.2007.09.031
Shiraishi, J., Katsuragawa, S., Ikezoe, J., Matsumoto, T., Kobayashi, T., Komatsu, K.-I., et al. (2000). Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists' detection of pulmonary nodules. Am. J. Roentgenol. 174, 71–74. doi: 10.2214/ajr.174.1.1740071
Simonyan, K., and Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
Staring, M., van der Heide, U. A., Klein, S., Viergever, M. A., and Pluim, J. P. (2009). Registration of cervical MRI using multifeature mutual information. IEEE Trans. Med. Imaging 28, 1412–1421. doi: 10.1109/TMI.2009.2016560
Tanner, C., Ozdemir, F., Profanter, R., Vishnevsky, V., Konukoglu, E., and Goksel, O. (2018). Generative Adversarial Networks for MR-CT Deformable Image Registration.
Tran, M. Q., Do, T., Tran, H., Tjiputra, E., Tran, Q. D., and Nguyen, A. (2022). Light-weight deformable registration using adversarial learning with distilling knowledge. IEEE Trans. Med. Imaging 41, 1443–1453. doi: 10.1109/TMI.2022.3141013
Wang, L. T., Hoover, N. E., Porter, E. H., and Zasio, J. J. (1987). “SSIM: a software levelized compiled-code simulator,” in Proceedings of the 24th ACM/IEEE Design Automation Conference (Miami Beach, FL), 2-8. doi: 10.1145/37888.37889
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., and Summers, R. M. (2017). “Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Miami Beach, FL) 2097–2106. doi: 10.1109/CVPR.2017.369
Wang, Z., Bovik, A. C., Sheikh, H. R., and Simoncelli, E. P. (2004). “Image quality assessment: from error visibility to structural similarity,” in IEEE transactions on image processing. 13, 600–612.
Wei, D., Ahmad, S., Huo, J., Huang, P., Yap, P. T., Xue, Z., et al. (2020). SLIR: synthesis, localization, inpainting, and registration for image-guided thermal ablation of liver tumors. Med. Image Anal. 65, 101763. doi: 10.1016/j.media.2020.101763
Wei, D., Ahmad, S., Huo, J., Peng, W., Ge, Y., Xue, Z., et al. (2019). “Synthesis and inpainting-based MR-CT registration for image-guided thermal ablation of liver tumors,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Shanghai), 512–520. doi: 10.1007/978-3-030-32254-0_57
Wu, J., and Zhou, S. (2021). A disentangled representations based unsupervised deformable framework for cross-modality image registration. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 3531–3534. doi: 10.1109/EMBC46164.2021.9630778
Xian, Y., Sharma, S., Schiele, B., and Akata, Z. (2019). “f-vaegan-d2: a feature generating framework for any-shot learning,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Oxford), 10275–10284. doi: 10.1109/CVPR.2019.01052
Xu, F., Shen, Y., Ding, L., Yang, C.-Y., Tan, H., Wang, H., et al. (2021). High-throughput mapping of a whole rhesus monkey brain at micrometer resolution. Nat. Biotechnol. 39, 1521–1528. doi: 10.1038/s41587-021-00986-5
Xu, Z., Luo, J., Yan, J., Pulya, R., Li, X., Wells, W., et al. (2020). “Adversarial uni-and multi-modal stream networks for multimodal image registration,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 222–232. doi: 10.1007/978-3-030-59716-0_22
Yan, P., Xu, S., Rastinehad, A. R., and Wood, B. J. (2018). “Adversarial image registration with application for MR and TRUS image fusion,” in International Workshop on Machine Learning in Medical Imaging, 197–204. doi: 10.1007/978-3-030-00919-9_23
Yang, H., Qian, P., and Fan, C. (2020). An indirect multimodal image registration and completion method guided by image synthesis. Comput. Math. Methods Med. 2020, 2684851. doi: 10.1155/2020/2684851
Zhang, X., Jian, W., Chen, Y., and Yang, S. (2020). Deform-GAN: An Unsupervised Learning Model for Deformable Registration.
Zhao, S., Dong, Y., Chang, E. I., and Xu, Y. (2019a). “Recursive cascaded networks for unsupervised medical image registration,” in Proceedings of the IEEE/CVF International Conference on Computer Vision (Seoul) 10600–10610. doi: 10.1109/ICCV.2019.01070
Zhao, S., Lau, T., Luo, J., Eric, I., Chang, C., and Xu, Y. (2019b). Unsupervised 3D end-to-end medical image registration with volume tweening network. IEEE J. Biomed. Health Inform. 24, 1394–1404. doi: 10.1109/JBHI.2019.2951024
Zheng, Y., Sui, X., Jiang, Y., Che, T., Zhang, S., Yang, J., et al. (2021). SymReg-GAN: symmetric image registration with generative adversarial networks. IEEE Trans. Patt. Anal. Mach. Intell. 44, 5631–5646. doi: 10.1109/TPAMI.2021.3083543
Zhou, B., Augenfeld, Z., Chapiro, J., Zhou, S. K., Liu, C., and Duncan, J. S. (2021). Anatomy-guided multimodal registration by learning segmentation without ground truth: application to intraprocedural CBCT/MR liver segmentation and registration. Med. Image Anal. 71, 102041. doi: 10.1016/j.media.2021.102041
Keywords: cross-modal, biomedical image registration, Generative Adversarial Networks, image translation, adversarial training
Citation: Han T, Wu J, Luo W, Wang H, Jin Z and Qu L (2022) Review of Generative Adversarial Networks in mono- and cross-modal biomedical image registration. Front. Neuroinform. 16:933230. doi: 10.3389/fninf.2022.933230
Received: 30 April 2022; Accepted: 13 October 2022;
Published: 22 November 2022.
Edited by:
Zhi Zhou, Microsoft, United StatesReviewed by:
Fazhi He, Wuhan University, ChinaDong Huang, Fourth Military Medical University, China
Copyright © 2022 Han, Wu, Luo, Wang, Jin and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Qu, cXVsZWlAYWh1LmVkdS5jbg==
 Tingting Han
Tingting Han Jun Wu
Jun Wu Wenting Luo1
Wenting Luo1 Zhe Jin
Zhe Jin Lei Qu
Lei Qu