- Blue Brain Project, Brain Mind Institute, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Ion channels are membrane proteins that selectively conduct ions across the cell membrane. The flux of ions through ion channels drives electrical and biochemical processes in cells and plays a critical role in shaping the electrical properties of neurons. During the past three decades, extensive research has been carried out to characterize the molecular, structural, and biophysical properties of ion channels. This research has begun to elucidate the role of ion channels in neuronal function and has subsequently led to the development of computational models of ion channel function. Although there have been substantial efforts to consolidate these findings into easily accessible and coherent online resources, a single comprehensive resource is still lacking. The success of these initiatives has been hindered by the sheer diversity of approaches and the variety in data formats. Here, we present “Channelpedia” (http://channelpedia.net), which is designed to store information related to ion channels and models and is characterized by an efficient information management framework. Composed of a combination of a database and a wiki-like discussion platform Channelpedia allows researchers to collaborate and synthesize ion channel information from literature. Equipped to automatically update references, Channelpedia integrates and highlights recent publications with relevant information in the database. It is web based, freely accessible and currently contains 187 annotated ion channels with 45 Hodgkin–Huxley models.
Introduction
The mammalian brain expresses approximately up to 500 ion channel genes and their variants (Ashburner et al., 2000). Ion channels are trans-membrane proteins that control the active and passive electrical behavior of a cell by selectively conducting ions across the cell membrane. The enigma of ion channel function and the differential influence of these ion channels on neuronal function has been the topic of much research over the past three decades. Researchers have been working extensively to address the genetic, proteomic, structural, biophysical, and functional properties of these ion channels and to build computational models that capture their biophysical and kinetic behavior. Hodgkin and Huxley (1952) developed a mathematical model of ionic conductance to demonstrate the role of ion channels in the electrical behavior of excitable nerve cells. Since then, the Hodgkin–Huxley model has been widely used to build ion channel models and to construct biologically realistic neuron models. Ion channel models and their integration into neuronal models have allowed for a better understanding of (a) the role of any particular ion channel in generating the different electrical behaviors of neurons, (b) the differential role of ion channels in different neurons, (c) the effect of neuromodulators on neuronal activity at the microcircuit and network levels.
The experimental and computational approaches employed to unravel the enigma of ion channels have produced a significant volume of scientific literature. Ion channel research alone produces approximately 800 publications every month (Figure 1A). Effective organization of this information requires constant maintenance, dedication and follow-up due to sheer volume of data produced, the continuously evolving data formats, and the inherent diversity of research methodology. Ion channel models are an example of a continuously evolving data format, since they go through a number of iterations to capture the complex kinetics of an ion channel by successively adding biophysical details. There have been several efforts made to document and archive ion channel information (Brammar, 1998; Hille, 2001; Beeman and Bower, 2004; Hines et al., 2004; Li and Gallin, 2004; Harmar et al., 2009). IUPHAR (Harmar et al., 2009) is currently the most comprehensive resource available for ion channels but lacks the computational models necessary for neuronal modeling. ModelDB (Hines et al., 2004), in contrast, contains some ion channel models but is not designed to manage these models effectively. Further, it does not contain ion channel related information nor cross-references to other ion channel resources like Rat Genome and Nucleotide databases. These resources have a rigid database structure, which makes them easily accessible and searchable but unsuitable for unstructured data as curators can only add information to predefined fields. Generally file upload and attachments are the sole means by which unstructured data can be supported in this type of information system. The rigidity of these resources makes them ideal core sources for data mining, but additional online tools are required to create meta-platforms that integrate multiple resources with unstructured information. Currently, Wikipedia (Contributors, 2004) allows for the systematic addition of novel information and also provides an ideal platform for unstructured data. Moreover, multiple contributors on Wikipedia can speed up the process of consolidating the data. A disadvantage of Wikipedia-like platforms, however, is that they do not handle structured data effectively. Thus, efforts to summarize the ion channel knowledge base have been inadequate due to the lack of a resource that supports both unstructured and structured data.
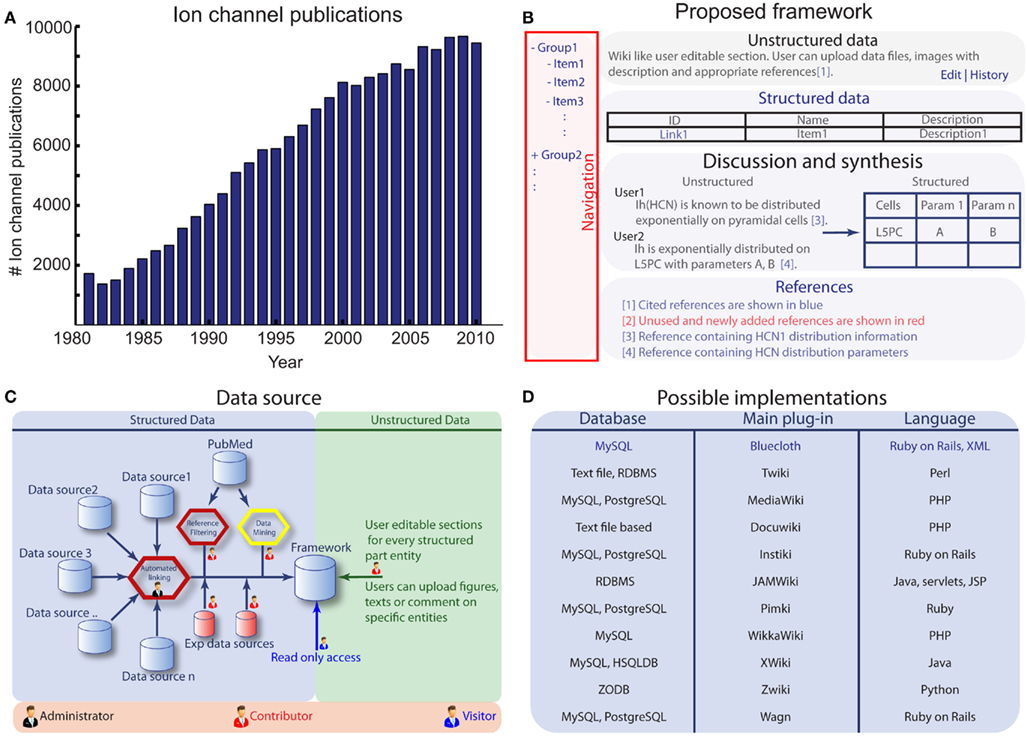
Figure 1. (A) Histogram of yearly ion channel publications, generated from 180,000 ion channel references present in the Channelpedia reference database. (B) The proposed framework for scientific information management. (C) The data source and possible user credentials. (D) The possible choices for current implementation.
We propose a novel approach to the management of ion channel related information through Channelpedia which is a freely accessible web application that combines the functionality of unstructured wiki-like data and yet has the advantages of a structured database. It provides a framework which enables the collective contribution of researchers to build a comprehensive resource for ion channel information. Additionally, it has a built-in referencing system that automatically filters new publications from PubMed and adds them to their respective categories, thus automating the acquisition and sorting of newly acquired information. It also notifies curators and researchers of newly published and relevant data, thus streamlining the organization of the ion channel knowledgebase.
Methods and Results
Overview
The information management framework underlying Channelpedia consists of five main modules: (Figure 1B). (1) Navigation, (2) Unstructured data, (3) Structured data, (4) Discussion and synthesis, (5) Reference management.
Easy navigation through data is an essential component of information management and requires logical grouping and hierarchical ordering. The current version of Channelpedia supports easy navigation to 187 ion channels in different categories and sub categories.
Data can be organized into two main categories: (1) Unstructured; data that are not quantifiable and have storage formats that are prone to change over time, (2) Structured; data that are quantifiable and have storage formats that rarely change. For example, the distribution profiles of ion channels on neurons are not fully constrained, generally being described in qualitative terms such as “Nav1.6 is known to be expressed in the axon initial segment of L2/3 pyramidal neurons” and are thus categorized under unstructured data. In contrast, data about gene ID and sequence of most of the ion channels would fall under structured data since their data format is fully constrained. Within Channelpedia, unstructured data are stored as editable text and images and structured data contains predefined fields for data with known formats and links to other online resources.
Data synthesis is a proposed conceptual process where unstructured data could be converted into structured data. Channelpedia provides an environment conducive to discussion among contributors which may allow for data synthesis. For example, a discussion on HCN distribution could conclude as an exponential distribution over apical dendrites and these parameters could be stored in structured data instead of descriptive language such as “HCN is known to be distributed exponentially on pyramidal cells” (Figure 1B).
To integrate existing information on ion channels, Channelpedia connects many different specialized databases and supports different user groups to manage the data (Figure 1C). The information source for all data is managed through the reference management module. This module also automatically finds relevant literature from PubMed by integrating an automated web crawler to download relevant article abstracts which are then added to their respective categories. Contributors are notified through this module whenever there are new publications to be curated.
Database, formatted text support, reference crawler, web interface, and a web server are the basic components required for the proposed framework underlying Channelpedia. There are many tools available to support these functionalities (Figure 1D), one of them being Ruby on Rails with MySQL database which is used for the implementation of Channelpedia.
Data Source and Supported User Roles
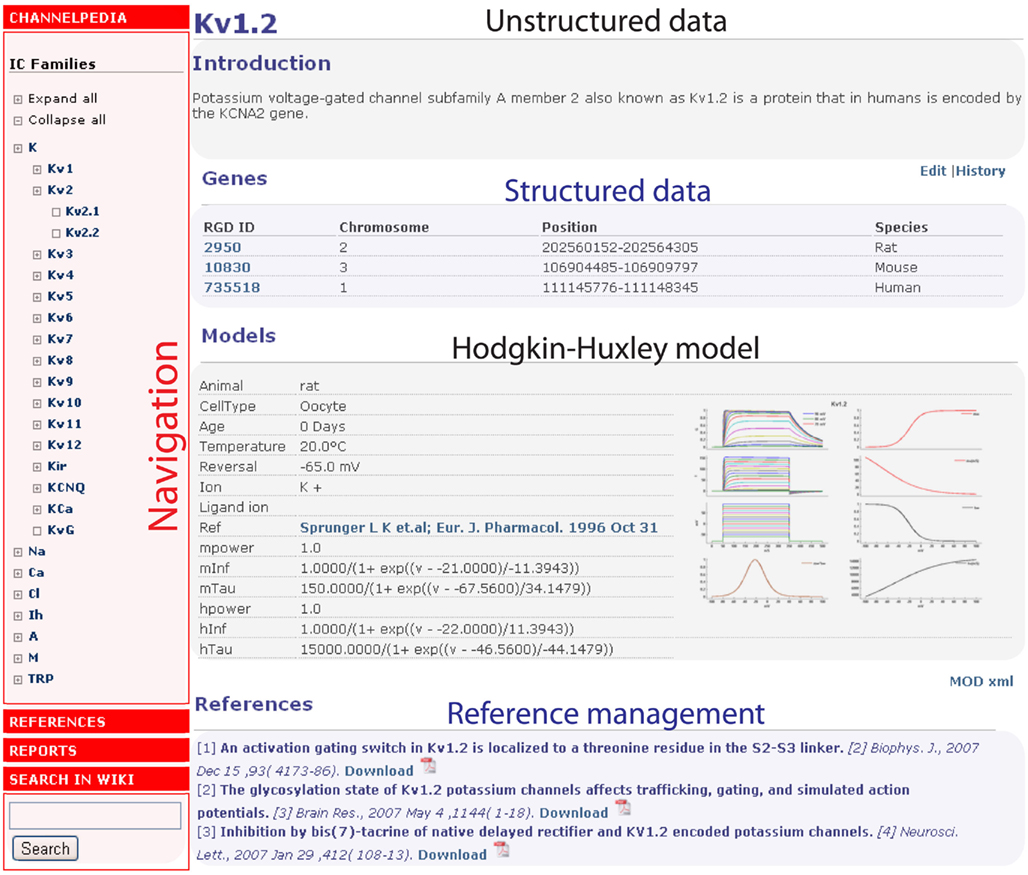
The current version of Channelpedia (Figure 2) contains the following sections to store unstructured and structured information: Introduction, Genes, Ontologies, Interactions, Structure, Expression, Distribution, Function, Kinetics, and Models. Structured data contains genetic information such as gene ID, symbol, name, synonyms, and descriptions from the Rat Genome Database (http://rgd.mcw.edu/; Twigger et al., 2007) which is then cross-referenced to other online resources using Ruby scripts (Matsumoto, 2011). Information related to genes and transcript sequences are obtained from Ensembl (Flicek et al., 2011); protein accession and peptide sequences are obtained from Uniprot (Jain et al., 2009); published interactions are obtained from the IntAct database (Aranda et al., 2010); complete GO annotations (process, Function, and Component) are obtained from the Gene Ontology database (Ashburner et al., 2000); and 3D structures of the channels were queried from the protein data bank (PDB; Berman et al., 2000).
The Channelpedia framework supports three different user credentials: (1) Visitor; can access all the data but are not allowed to make any changes, (2) Contributor; needs a framework account to update or add new information from literature or to upload experimental data, (3) Administrator; requires a framework account to add/delete/change user credentials and database structure.
Contributors and existing online resources are the two main sources of data. Unstructured data (Figure 3) is populated by contributors who can freely edit formatted text and upload images without violating copyright agreements. Structured data contains data from existing online resources and is managed by administrators using automated scripts. Experimental data from literature is also uploaded by contributors and stored as structured data.
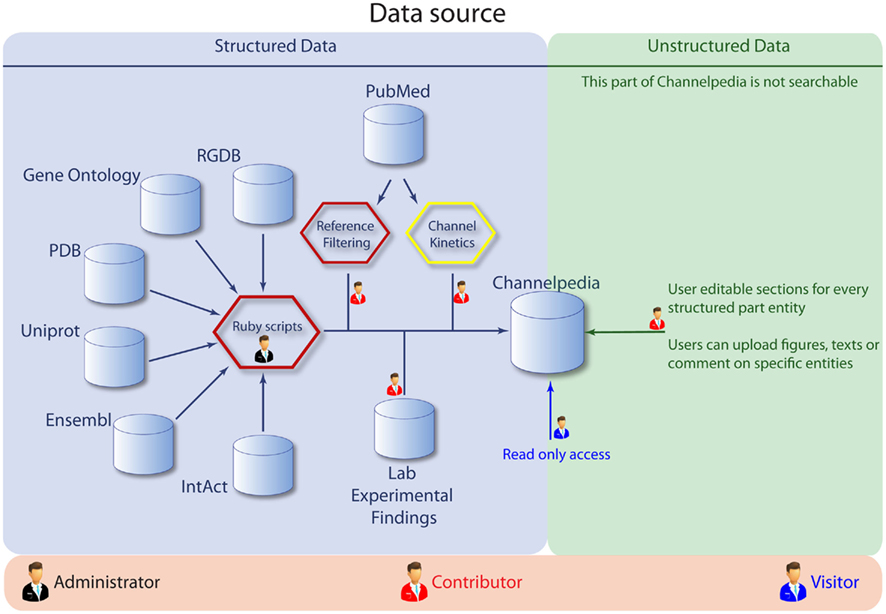
Figure 3. The data sources for Channelpedia, with a breakup of areas accessible to administrators, contributors, and visitors.
Models
The ion channel models were built with a Hodgkin and Huxley (1952) formulation. The net current flow Iion and the ionic conductance gion across the membrane for a given ion is described as

where m and h represent the proportion of open activation and inactivation gates respectively. P and Q are the numbers of independent gates required for the observed time-course of activation and inactivation. Vm is the membrane potential, and Eion denotes the Nernst potential for the ion. The variable 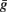 scales for maximum channel conductance. The general description in Eq. 1 can be applied for different ion species. Gating variables m and h are modeled as first-order kinetics
scales for maximum channel conductance. The general description in Eq. 1 can be applied for different ion species. Gating variables m and h are modeled as first-order kinetics
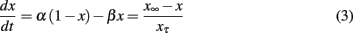
where x denotes either m or h in Eq. 2. α and β are rate constants for gate opening and closing, respectively, and both are functions of Vm. Note that the final expression in Eq. 3 presents the equivalent form with steady state activation/inactivation parameter (x∞) and time constant (xτ).
Published journal articles were manually searched for kinetic parameters (x∞, xτ). A custom Matlab tool was used to approximate the unavailable parameters and to recreate the voltage response for each channel. For more details refer to http://channelpedia.net. Apart from description of kinetics and experimental details like temperature, cell-type, and age of the animal, Channelpedia also contains a plot of the model response to a step voltage protocol (Figure 4). Authorized users can upload models to Channelpedia using a customized XML schema. Uploaded models are available in XML format, ChannelML (v1.8.1; Gleeson et al., 2010), or Neuron NMODL descriptions for all users (Hines and Carnevale, 2000, 2001).
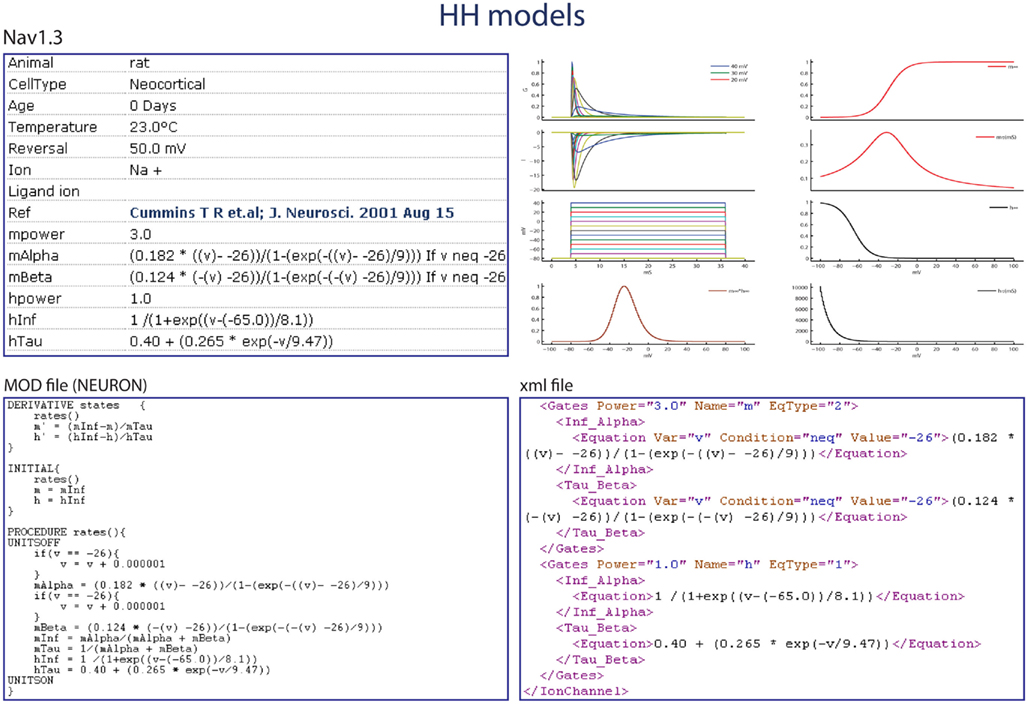
Figure 4. Example of the Nav1.3 ion channel kinetics model in Channelpedia, with the activation and inactivation kinetics and time constants, representation as a mod file in the NEURON simulation environment, and in a custom XML format.
Reference Management
References are handled in Channelpedia in a two step process. The first step includes a weekly automated download and keyword based classification of ion channel related abstracts from PubMed (Figure 5). Channelpedia contains ∼180,000 abstracts and they are available under the reference module of Channelpedia and categorized under different sections for each ion channel subtype. The second step requires contributors to manually identify the paper of interest from automatically categorized list and select them for respective ion channel page. These references are initially highlighted in red on the ion channel page and are marked up to blue when they are used (referenced) by contributors in the ion channel page (Figure 5). This feature enables verified integration of new information to ion channel page and allows contributors to identify unread publications for curation.
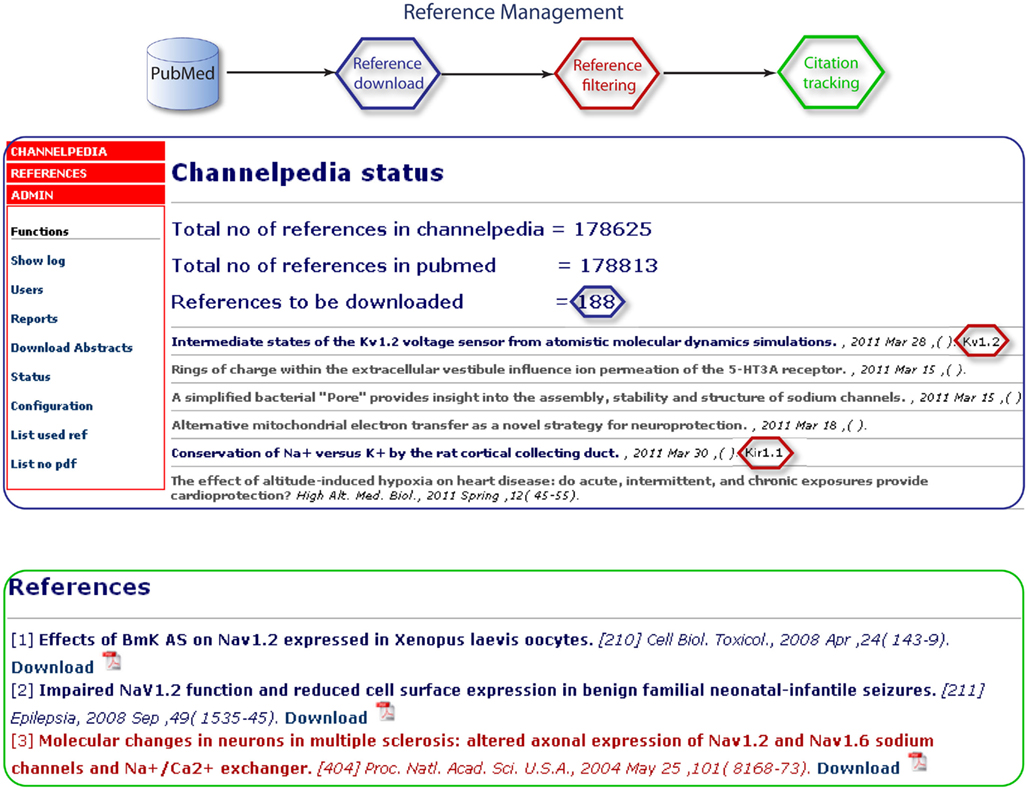
Figure 5. The reference management structure, showing the step-by-step process to download a reference from PubMed and its usage in Channelpedia.
Implementation
MySQL is used as a database backend and the Ruby on Rails framework implements the structure of the application. Channelpedia also uses several plugins or gems for other functionalities. The list of Ruby gems used are: Nokogiri for XML parsing, OpenURI as a wrapper for net/http, Hpricot for HTML parsing, Mechanize for web automation, bluecloth for wiki formatting, and Attachment_fu plugin for file uploads (Figure 6).
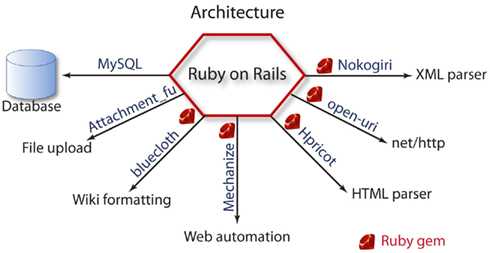
Figure 6. The overall architecture of Channelpedia, and tools used to manage the database, file upload, wiki formatting, web automation, HTML, and XML parsing.
Discussion and Future Directions
In this study we have proposed “Channelpedia” which utilizes an alternate approach to information management and facilitates the building of an ion channel knowledge base to accommodate both structured and unstructured data. The structured data reside in “queryable” database structures whereas unstructured data remain in wiki-like formatted text and image format. Although there is no automatic way to convert unstructured data into structured data, the framework is designed in such a way that by adding relevant database tables and tags to these tables, it will be possible to move the desired data into a structured format.
For example, Channelpedia currently only supports Hodgkin–Huxley models but by adding a database table “Markov Models” with appropriate entities, Channelpedia will be able to support Markov models in the structured database. Channelpedia is currently unable to store raw data, analysis code, analysis results, and hypotheses in a machine-readable format instead of descriptive language. Ideally, a provision should be made to do this, but significant technological and administrative advancement is required to tackle this problem.
Temperature coefficient factor (Q10) is used to capture the effect of temperature on gating kinetics. In most of the ion channel models Q10 factor is approximated to 2.3, whereas in reality it could vary between 2 and 30 (Dhaka et al., 2006). To keep such modifications and assumptions traceable we propose to add a separate model for each assumption.
Ion channel kinetics are often modified to achieve desired results in neuronal modeling. It becomes difficult to find the kinetics of the original model after several such modifications. For example, the Kv2.1 ion channel model used in (Johnson and Mcintyre, 2008) contains an activation parameter v1/2 = −22 mV, citing (Chan et al., 2007) with v1/2 = 17.5 mV, whereas, experimental data reports this value to be −18 mV (Baranauskas et al., 1999). Channelpedia with its Wiki-like functionality can provide an ideal platform to track such changes.
Data on ion channel interactions, 3D structure, expression in brain regions, cellular distribution, and function in neuronal activity are currently limited in Channelpedia. Although references are automatically curated, it would require a significant amount of manual effort to summarize published literature. We are actively adding more information from literature and expect more user contributions to make it a more reliable and comprehensive database for ion channels.
Currently Channelpedia supports very limited application programming interfaces (APIs) but future versions of Channelpedia will include support for ontology and generic querying to access any aspect of the data. Using ontologies along with APIs would be an ideal way to connect multiple Channelpedia-like, small, but specialized databases.
Channelpedia exemplifies the combined functionality of structured and unstructured data management merged with intelligent automated reference handling. A generic implementation of this tool, which users can customize to their specific needs, could make this approach generally useful to other domains of research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was performed within the framework of the Blue Brain Project and was supported by the respective EPFL project fund. We thank all the mentioned contributors for their help in the development of Channelpedia. We thank Shruti Muralidhar and Dr. Abdel Elhamdani for their comments on the manuscript, and Melissa Cochrane for her help in editing the article.
Author Contributions
Channelpedia was conceived and constructed as part of the Blue Brain Project’s general aim to build brain models with biological accuracy and its specific aim to model each type of neuron according to the specific ion channels expressed. A number of researchers have contributed to the construction of Channelpedia: for literature search (Rajnish Ranjan, Srikanth Ramaswamy, Geetanjali Saha, Mehgana Katiki, Albert Gidon, Etay Hay, Shaul Druckmann, and Michael Schartner); for prototype design and implementation (Rajnish Ranjan, Albert Gidon, James G. King, Etay Hay, Srikanth Ramaswamy, Felix Schürmann, Sean Hill, Shaul Druckmann, Adnan Abid, and Thomas McColgan); for ion channel models (Rajnish Ranjan, Albert Gideon, Etay Hay, Shaul Druckmann, and Mehgana Katiki); for cross referencing (Rajnish Ranjan and Georges Khazen); for data entry (Rajnish Ranjan, Georges Khazen, and Martin Telefont); for maintenance (Rajnish Ranjan and Luca Gambazzi); for the final implementation (Rajnish Ranjan, Luca Gambazzi, James G. King, and John Kenyon). Rajnish Ranjan, Srikanth Ramaswamy, Henry Markram, and Georges Khazen wrote the manuscript.
References
Aranda, B., Achuthan, P., Alam-Faruque, Y., Armean, I., Bridge, A., Derow, C., Feuermann, M., Ghanbarian, A. T., Kerrien, S., Khadake, J., Kerssemakers, J., Leroy, C., Menden, M., Michaut, M., Montecchi-Palazzi, L., Neuhauser, S. N., Orchard, S., Perreau, V., Roechert, B., Van Eijk, K., and Hermjakob, H. (2010). The IntAct molecular interaction database in 2010. Nucleic Acids Res. 38, D525–D531.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29.
Baranauskas, G., Tkatch, T., and Surmeier, D. J. (1999). Delayed rectifier currents in rat globus pallidus neurons are attributable to Kv2.1 and Kv3.1/3.2 K(+) channels. J. Neurosci. 19, 6394–6404.
Beeman, D., and Bower, J. M. (2004). Simulator-independent representation of ionic conductance models with channeldb. Neurocomputing 1085–1090.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242.
Brammar, E. C. C. A. W. J. (1998). Ion Channel Facts Book Voltage-Gated Channels. San Diego: Academic Press.
Chan, C. S., Guzman, J. N., Ilijic, E., Mercer, J. N., Rick, C., Tkatch, T., Meredith, G. E., and Surmeier, D. J. (2007). “Rejuvenation” protects neurons in mouse models of Parkinson’s disease. Nature 447, 1081–1086.
Contributors, W. (2004). “Ion Channel” Wikipedia, the Free Encyclopedia. Available at: http://en.wikipedia.org/wiki/Ion_channel [accessed February 10, 2011].
Dhaka, A., Viswanath, V., and Patapoutian, A. (2006). Trp ion channels and temperature sensation. Annu. Rev. Neurosci. 29, 135–161.
Flicek, P., Amode, M. R., Barrell, D., Beal, K., Brent, S., Chen, Y., Clapham, P., Coates, G., Fairley, S., Fitzgerald, S., Gordon, L., Hendrix, M., Hourlier, T., Johnson, N., Kahari, A., Keefe, D., Keenan, S., Kinsella, R., Kokocinski, F., Kulesha, E., Larsson, P., Longden, I., Mclaren, W., Overduin, B., Pritchard, B., Riat, H. S., Rios, D., Ritchie, G. R., Ruffier, M., Schuster, M., Sobral, D., Spudich, G., Tang, Y. A., Trevanion, S., Vandrovcova, J., Vilella, A. J., White, S., Wilder, S. P., Zadissa, A., Zamora, J., Aken, B. L., Birney, E., Cunningham, F., Dunham, I., Durbin, R., Fernandez-Suarez, X. M., Herrero, J., Hubbard, T. J., Parker, A., Proctor, G., Vogel, J., and Searle, S. M. (2011). Ensembl 2011. Nucleic Acids Res. 39, D800–D806.
Gleeson, P., Crook, S., Cannon, R. C., ‘Hines, M. L., Billings, G. O., Farinella, M., Morse, T. M., Davison, A. P., Ray, S., Bhalla, U. S., Barnes, S. R., Dimitrova, Y. D., and Silver, R. A. (2010). NeuroML: a language for describing data driven models of neurons and networks with a high degree of biological detail. PLoS Comput. Biol. 6, e1000815. doi:10.1371/journal.pcbi.1000815
Harmar, A. J., Hills, R. A., Rosser, E. M., Jones, M., Buneman, O. P., Dunbar, D. R., Greenhill, S. D., Hale, V. A., Sharman, J. L., Bonner, T. I., Catterall, W. A., Davenport, A. P., Delagrange, P., Dollery, C. T., Foord, S. M., Gutman, G. A., Laudet, V., Neubig, R. R., Ohlstein, E. H., Olsen, R. W., Peters, J., Pin, J. P., Ruffolo, R. R., Searls, D. B., Wright, M. W., and Spedding, M. (2009). IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 37, D680–D685.
Hines, M. L., and Carnevale, N. T. (2000). Expanding neuron’s repertoire of mechanisms with NMODL. Neural Comput. 12, 995–1007.
Hines, M. L., and Carnevale, N. T. (2001). NEURON: a tool for neuroscientists. Neuroscientist 7, 123–135.
Hines, M. L., Morse, T., Migliore, M., Carnevale, N. T., and Shepherd, G. M. (2004). ModelDB: a database to support computational neuroscience. J. Comput. Neurosci. 17, 7–11.
Hodgkin, A. L., and Huxley, A. F. (1952). A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. (Lond.) 117, 500–544.
Jain, E., Bairoch, A., Duvaud, S., Phan, I., Redaschi, N., Suzek, B. E., Martin, M. J., Mcgarvey, P., and Gasteiger, E. (2009). Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10, 136. doi:10.1186/1471-2105-10-136
Johnson, M. D., and Mcintyre, C. C. (2008). Quantifying the neural elements activated and inhibited by globus pallidus deep brain stimulation. J. Neurophysiol. 100, 2549–2563.
Li, B., and Gallin, W. J. (2004). VKCDB: voltage-gated potassium channel database. BMC Bioinformatics 5, 3. doi:10.1186/1471-2105-5-3
Matsumoto, Y. M. (2011). Ruby A Programmer’s Best Friend. Available at: http://www.ruby-lang.org/en/
Keywords: ion channel, kinetics, Hodgkin–Huxley model, database, information management, structured wiki
Citation: Ranjan R, Khazen G, Gambazzi L, Ramaswamy S, Hill SL, Schürmann F and Markram H (2011) Channelpedia: an integrative and interactive database for ion channels. Front. Neuroinform. 5:36. doi: 10.3389/fninf.2011.00036
Received: 29 April 2011;
Accepted: 12 December 2011;
Published online: 30 December 2011.
Edited by:
Andrew P. Davison, CNRS, FranceReviewed by:
Robert C. Cannon, Textensor Limited, UKNicholas T. Carnevale, Yale University School of Medicine, USA
Padraig Gleeson, University College London, UK
Copyright: © 2011 Ranjan, Khazen, Gambazzi, Ramaswamy, Hill, Schürmann and Markram. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Henry Markram, Blue Brain Project, Brain Mind Institute, Ecole Polytechnique Fédérale de Lausanne, AAB 110, Station 15, CH-1015 Lausanne, Switzerland. e-mail:aGVucnkubWFya3JhbUBlcGZsLmNo