
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurol. , 20 February 2025
Sec. Stroke
Volume 16 - 2025 | https://doi.org/10.3389/fneur.2025.1540307
 Qiong Yang1†
Qiong Yang1† Haixin Sun2,3†
Haixin Sun2,3† Xinran Ma1
Xinran Ma1 Lu Tang1
Lu Tang1 Xiaolu Liu1
Xiaolu Liu1 Xin Huang1
Xin Huang1 Xiao Huang1
Xiao Huang1 Yong Chen1
Yong Chen1 Danyang Tian1
Danyang Tian1 Xiangzhu Zeng4
Xiangzhu Zeng4 Nan Li5
Nan Li5 Wenzhi Wang2,3
Wenzhi Wang2,3 Dongsheng Fan1,6,7*
Dongsheng Fan1,6,7*Background: The island sign is a predictor of hematoma expansion and worse outcomes in patients of spontaneous primary intracerebral hemorrhage (ICH). The biological mechanism of the island sign remains unclear, but its presence might be influenced by the underlying vasculopathy related to Apolipoprotein E (APOE) genotypes. Therefore, we aimed to research the association between APOE genotypes and the island sign.
Methods: We enrolled patients with primary supratentorial ICH in a multicenter cohort in northern China with baseline noncontrast CT images performed within 14 days after symptoms onset and APOE genotype available. The island sign was rated on the CT images according to validated criteria. Univariable and multivariable analyses were used to identify the association between APOE genotypes and the island sign, stratified by the ICH location.
Results: Among 460 patients enrolled, 122 were lobar ICH. In all patients, after adjusting for age, sex, hypertension, and time to CT, the presence of the APOE ε4 allele (OR 2.020, 95% CI 1.064–3.834, p = 0.032) was associated with the island sign, whereas the presence of the APOE ε2 allele (OR 0.734, 95% CI 0.339–1.593, p = 0.435) was not. After stratifying by ICH location, multivariable analysis revealed that APOE ε4 (OR 3.510, 95% CI 1.393–8.846, p = 0.008), rather than ε2 (OR 0.621, 95% CI 0.203–1.901, p = 0.404), was associated with the island sign in lobar ICH patients. Neither the ε2 nor the ε4 allele was associated with the island sign among nonlobar ICH patients.
Conclusion: The APOE ε4 allele was associated with the island sign in lobar ICH patients. Our findings indicate that the presence of the island sign may be influenced by the underlying vasculopathy related to APOE ε4, which increases amyloid deposition in the cerebral vasculature.
Intracerebral hemorrhage (ICH) comprises 10–15% of all strokes worldwide. This severe form of stroke has an early-term mortality of approximately 30–40% (1). A meta-analysis revealed an overall incidence of ICH of 24.6 per 100,000 person-years (2, 3) and that Asian populations are twice as likely to experience ICH as white populations (2, 3). Given the growing aging population and the widespread use of anticoagulants, the ICH incidence is expected to remain substantial, despite ongoing public health efforts to improve hypertension management (1).
Hematoma expansion (HE) prevails in 20% of ICH patients and predicts worse outcomes (4). Preventing HE appears to be an appealing therapeutic strategy, but how to early identify high risk patients when they present with ICH remains challenging. Previous reports proposed imaging predictors for identifying hematomas that have the potential to expand, such as the spot sign observed in CT angiography and several noncontrast CT features including the island sign (5, 6). The island sign, characterized by multifocal small bleeding around the main hematoma, can reflect a hematoma with an extremely irregular shape (7–9). The exact mechanisms underlying the formation of the island sign remain unclear; one explanation is that as the main hematoma, which represents rupture and bleeding of a single blood vessel, expands, active bleeding from adjacent arterioles may cause island-like hematomas, forming the island sign (7). Another explanation is that the island sign may be caused by rupture of several arterioles leading to multifocal active bleeding (7).
The apolipoprotein E (APOE) gene is an important genetic risk factor for ICH (10, 11). Previous studies revealed that the presence of APOE ε2 and ε4 increases the risk of lobar ICH (12–14). Moreover, APOE ε2 is linked with larger ICH volumes (15), hematoma expansion (16) and the presence of CTA spot signs (17) in lobar ICH patients; APOE ε4 is associated with functional dependency and poor survival after ICH (10); and both ε2 and ε4 are associated with a greater risk of ICH recurrence (10).
The underlying mechanism by which APOE alleles influence ICH may be related to their effects on cerebral amyloid angiopathy (CAA). CAA, defined by the deposition of beta-amyloid proteins in the walls of small cortical and leptomeningeal vessels in the brain (18), is an important cause of lobar ICH in elders (19). The APOE ε4 allele is an established risk factor for CAA (18). The presence of APOE ε4 enhances the severity of amyloid deposition in the cerebral vasculature which may accelerate the vascular damage and cause vascular rupture, whereas APOE ε2 is predominantly related to the rupture and bleeding of these amyloid-laden vessels (20). Besides, previous studies (21–25) and meta-analyses (26) have shown that irregular borders are among the most common imaging features of CAA related ICH, though the feature has not been clearly defined.
Therefore, we conducted a prospective, multicenter study of ICH cohort to test the hypothesis that the APOE genotype is associated with the island sign in lobar ICH patients.
Data from a prospective multicenter cohort of acute primary ICH patients who were recruited from 19 hospitals across Beijing, Hebei, and Inner Mongolia in northern China between 2015 and 2019 were analyzed. The main inclusion criteria were as follows: (1) primary spontaneous supratentorial ICH and (2) available APOE genotype data. Patients were excluded if they had any of the following characteristics: (1) secondary ICH due to vascular malformation, tumor, trauma or hemorrhagic cerebral infarction, etc.; (2) no noncontrast CT scan performed within 14 days after the onset of symptoms or low-quality images; (3) head surgery performed before the baseline CT scan; or (4) an unknown exact time of onset (to the minute).
This cohort study was approved by the Ethics Committee of Peking University Third Hospital [(2014)-191-3] (Clinical Trial Registry on clinicaltrials.gov, NCT02361411). Informed consent in writing was obtained from all patients or their representatives.
Individual patient data, including age, sex, vascular risk factors, history of previous ICH, and medication history, were systematically and prospectively collected and documented by trained neurologists at the time of the index symptomatic ICH, based on medical records or information provided by patients or their relatives. The National Institutes of Health Stroke Scale (NIHSS) score was assessed upon admission. Medical records were reviewed to obtain the time to initial CT imaging.
CT images were examined by trained study personnel to identify the location and volume of the ICH and to assess for the presence of intraventricular hemorrhage (IVH). The ICH location was defined as supratentorial (lobar or nonlobar) or infratentorial (cerebellum or brainstem) based on the Cerebral Hemorrhage Anatomical RaTing Instrument (CHARTS) (27). The volume of the ICH was calculated from the baseline CT images with the ABC/2 method.
The definition of island sign was (1) the presence of three or more small, scattered hematomas separate from the main hematoma or (2) the presence of four or more small hematomas, some or all of which might be connected to the main hematoma (7). The presence of the island sign was assessed according to published criteria (7) by 2 experienced investigators (Figure 1). Discrepancies were resolved through consensus after the investigators reviewed all the scans. All researchers evaluating the imaging were blinded to both the clinical information and the patient’s APOE genotype.
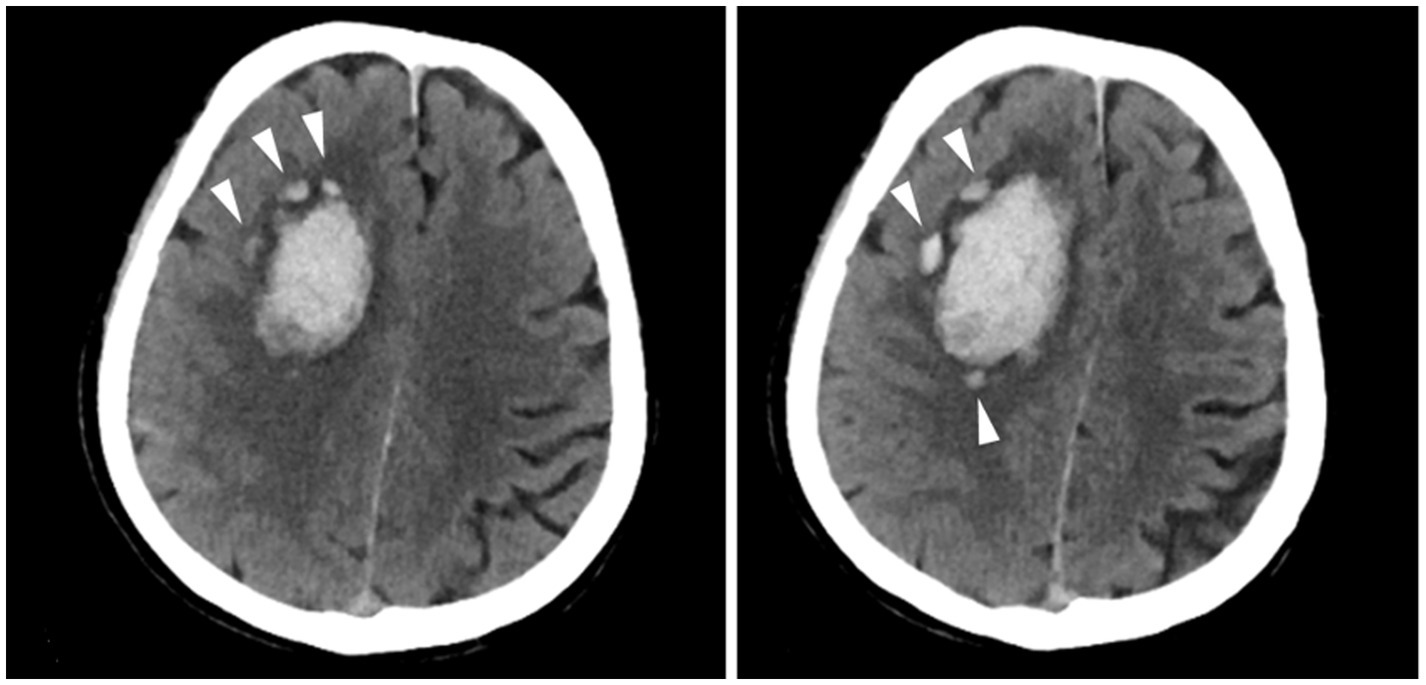
Figure 1. Examples of the island sign. A patient with the island sign (three or more small, scattered hematomas separate from the main hematoma).
With DNA extracted from whole blood samples donated by the patient at enrollment, APOE gene loci (rs7412 and rs429358) were tested and then, translated to APOE genotypes (ε2/ε2, ε3/ε2, ε2/ε4, ε3/ε3, ε3/ε4, and ε4/ε4). Participants carrying the ɛ2/ɛ2, ε3/ε2, and ɛ2/ɛ4 genotypes were defined as APOE-ɛ2 carriers, whereas those carrying the ɛ2/ɛ4, ɛ3/ɛ4, or ɛ4/ɛ4 genotype were defined as APOE-ɛ4 carriers. All laboratory staffs performing genotyping were blinded to the clinical data and CT images.
Categorical variables were presented as counts with percentages (%), while continuous variables were reported as medians with interquartile ranges (IQRs) due to the nonnormal distribution of the data.
Univariable and multivariable logistic regression analyses were conducted to assess the associations between the presence of APOE ε2 and/or ε4 alleles and the island sign. Multivariable model 1 included prespecified predictors, which included age, sex, hypertension, time to CT, and the presence of APOE ε2 or ε4 allele. Multivariable model 2 included the aforementioned prespecified predictors along with variables that had a p value <0.1 in the univariable analysis. We also performed these analyses after stratifying by location, i.e., lobar versus nonlobar ICH. Finally, we conducted subgroup analyses for patients whose time to CT was within 6 h of ICH onset. Statistical significance was defined for p < 0.05. All analyses were performed with SPSS (version 26.0).
A total of 460 patients were eligible for analysis, with a median age of 60 (51, 73) years, and 297 of them were male (64.6%). Among them, 122 patients had lobar ICH, and 338 patients had nonlobar ICH. Figure 2 shows the flow chart for patient inclusion. The included patients had less hypertension, less diabetes mellitus, lower percentage of moderate to severe alcohol consumption, and shorter time to CT than excluded patients (Supplementary Table S1).
The baseline characteristics of all patients stratified by ICH location were shown in Table 1. Patients in the lobar ICH group were older and had less hypertension, but had a greater ICH volume and a higher percentage of previously ICH, island sign and APOE ε4 carriers, compared to those in the nonlobar ICH group (Table 1). Specifically, the island sign was presented in 32 (26.2%) and 31 (9.2%) of patients in the lobar and nonlobar ICH groups, respectively, while 30 patients (24.6%) in the lobar group and 51 patients (15.1%) in the nonlobar group were APOE ε4 carriers.
In the univariable analysis, greater age (OR 1.026, 95% CI 1.006–1.046, p = 0.011), the presence of hypertension (OR 0.477, 95% CI 0.275–0.825, p = 0.008), greater ICH volume (OR 1.040, 95% CI 1.029–1.051, p < 0.001), the presence of IVH (OR 1.731, 95% CI 1.006–2.980, p = 0.048) and the presence of the APOE ε4 allele (OR 1.923, 95% CI 1.037–3.565, p = 0.038) were associated with the presence of the island sign (Table 2).
In the multivariable analysis, after adjusting for prespecified predictors such as age, sex, hypertension and time to CT, the presence of the APOE ε4 allele (OR 2.020, 95% CI 1.064–3.834, p = 0.032), but not the presence of the APOE ε2 allele (OR 0.734, 95% CI 0.339–1.593, p = 0.435), was associated with the island sign (Table 3). Moreover, after adjusting both for the prespecified predictors and variables with p < 0.1 in the univariable analysis, including age, sex, hypertension, ICH volume, IVH, and time to CT, the presence of APOE ε4 (OR 2.114, 95% CI 1.038–4.307, p = 0.039), but not the presence of APOE ε2 (OR 0.608, 95% CI 0.252–1.467, p = 0.268), was associated with the island sign (Table 3).
In the univariable analysis, a larger ICH volume (OR 1.042, 95% CI 1.022–1.061, p < 0.001) and the presence of the APOE ε4 allele (OR 3.597, 95% CI 1.487–8.699, p = 0.004) were associated with the presence of the island sign (Table 2).
In the multivariable analysis, after adjusting for the prespecified predictors such as age, sex, hypertension and time to CT, the presence of the APOE ε4 allele (OR 3.510, 95% CI 1.393–8.846, p = 0.008), but not the presence of the APOE ε2 allele (OR 0.621, 95% CI 0.203–1.901, p = 0.404), was associated with the island sign (Table 3). Moreover, after adjusting for the prespecified predictors and variables with p < 0.1 in the univariable analysis, including age, sex, hypertension, diabetes mellitus, previous ICH, ICH volume and time to CT, the presence of the APOE ε4 allele (OR 3.605, 95% CI 1.152–11.279, p = 0.028), but not that of the APOE ε2 allele (OR 0.320, 95% CI 0.070–1.463, p = 0.142), was associated with the island sign (Table 3).
According to the univariable analysis, greater ICH volume (OR 1.034, 95% CI 1.020–1.048, p < 0.001) and the presence of IVH (OR 2.344, 95% CI 1.113–4.936, p = 0.025) were associated with the island sign (Table 2).
In the multivariable analysis, after adjusting for prespecified predictors, including age, sex, hypertension and time to CT, the presence of neither APOE ε4 (OR 0.673, 95% CI 0.193–2.351, p = 0.535) nor APOE ε2 (OR 0.577, 95% CI 0.166–2.002, p = 0.386) was associated with the island sign (Table 3). Moreover, the presence of neither APOE ε4 (OR 1.003, 95% CI 0.275–3.665, p = 0.996) nor APOE ε2 (OR 0.638, 95% CI 0.174–2.338, p = 0.498) was associated with the island sign after adjusting for the prespecified predictors and variables with p < 0.1 in the univariable analysis, including age, sex, hypertension, ICH volume, IVH and time to CT (Table 3).
In the univariable analysis, the presence of neither APOE ε4 (OR 1.648, 95% CI 0.611–4.447, p = 0.324) nor APOE ε2 (OR 1.703, 95% CI 0.663–4.373, p = 0.269) was associated with the island sign in all ICH patients (Supplementary Table S2); similar results were obtained in the multivariable analysis (Supplementary Table S3). Subgroup analyses for lobar and nonlobar ICH patients also failed to uncover an association between the presence of either APOE ε4 or ε2 allele and the presence of the island sign (Supplementary Tables S2, S3).
Our findings demonstrated the association between APOE ε4 and the island sign on CT imaging in patients with lobar ICH rather than nonlobar ICH. However, no association between APOE ε2 and the island sign was observed in either lobar or nonlobar ICH patients.
Our study revealed for the first time that the APOE ε4 allele was associated with the island sign in lobar ICH patients. Though the pathophysiological mechanism remains unclear, the role of APOE in the island sign might be consistent with APOE in CAA and ICH. The histopathologic mechanisms of APOE ε4 and ε2 appear different in CAA. APOE ε4 allele increases the deposition of amyloid protein in the wall of small cortical and meningeal vessels, making it vulnerable to rupture, whereas APOE ε2 mainly causes blood vessels with amyloid deposition to rupture and bleed (20). Vascular damage caused by amyloid deposition, which is accelerated by APOE ε4, impacts a substantial portion of the leptomeningeal and cortical arterioles in patients with CAA (28). We hypothesized this damage makes them more susceptible to multifocal bleeding, leading to the formation of the island sign. In addition, the island sign may contribute to some of the ambiguous imaging features of irregular borders in CAA related ICH observed in previous studies (21–26).
Furthermore, we explored the associations between the island sign and APOE across different time windows including 14 days and 6 h. Previously, Li et al. proposed that the island sign mainly appears on images taken within 6 h, reflecting early (within 24 h) hematoma expansion (7). Our study showed that the association between ε4 and the island sign was significant within 14 days but not within 6 h of ICH onset, although the OR values were similar. On one hand, the avalanche effect caused by acute cerebral hemorrhage is an important prerequisite for the formation of the island sign, for which the first 6 h constitutes the peak window (7, 29). On the other hand, the post hoc analysis of the TICH-2 trial found that in patients with lobar CAA related ICH, the risk of hematoma expansion increased with time from symptom onset, indicating a longer time window of hematoma expansion, which was different from nonlobar ICH and lobar non-CAA related ICH (28). The mechanisms leading to prolonged hematoma expansion in CAA related ICH may be that the bleeding related to CAA originates from leptomeningeal vessels which form an effective collateral network. As a result, the vasoconstrictive response involved in hemostasis may be less effective (28). In addition, the lobar location of the hemorrhage provides more space and reduces the likelihood of tamponade, which can help stop the bleeding, compared to non-lobar locations (28). This also suggest that the time window for the presence of the island sign may exceed 6 h.
Our study did not find a link between APOE ε2 and the island sign, though previous reports have indicated that APOE ε2 was related to spot sign, as well as greater hematoma volume and hematoma expansion (15–17). Theoretically, APOE ε2 might be expected to be associated with the island sign within the first 6 h in lobar ICH, similar to its association with the spot sign (17). The proposed mechanism may involve APOE ε2 predisposing to additional vessel rupture, leading to hematoma expansion and the formation of the island sign. In the subgroup analysis of our study, we did observe an increased odds ratio of the association between APOE ε2 and the island sign in 6 h, but it was statistically insignificant which may be attributed to the limited number of patients imaged during that time frame. Further research is needed to clarify this potential relationship. In contrast, over the longer 14-day time window, we observed a significant association between the island sign and APOE ε4, rather than APOE ε2. This could be due to vascular amyloid changes induced by APOE ε4, which contribute to the formation of multiple small bleedings surrounding the main hematoma over a prolonged period (beyond 6 h), resulting in the island sign.
The strengths of our study included that the use of data from a multicenter prospective cohort and a thorough evaluation of the neuroimaging. This study added the knowledge of effect of APOE ε4 genotype on the island sign over a 14-day period in lobar ICH patients, providing insights into the biological mechanisms underlying the island sign. Given the role of APOE ε4 in CAA and its link to increased recurrence risk in ICH patients, the presence of the island sign may also predict a higher likelihood of ICH recurrence which needs to be clarified in future research. Furthermore, with bedside genotyping available, there would be room of optimization of acute management of ICH by the combination use of island sign and the APOE gene, in terms of risk stratification and an early bundle of care focused on blood pressure control (30), individualized anti-coagulation strategy for patients at high thromboembolic risk including atrial fibrillation, venous thromboembolism, etc. (1). Additionally, the island sign can be easily determined on CT scans, which could potentially improve the management strategies and prognostic prediction in the acute setting when APOE genotype test is not available.
(1) Our cohort did not have complete MRI data, preventing us from identifying a subgroup of patients with CAA based on the Boston criteria. (2) The limited number of patients with available baseline CT images within 6 h hindered a thorough investigation of the relationship between the APOE allele and the island sign. This was partly due to late arrivals at the hospital, referrals from other facilities, and the inability to secure initial CT images from those hospitals. Further research is necessary to more concretely establish the role of APOE to validate our findings.
In conclusion, the APOE ε4 allele is associated with the presence of the island sign in lobar ICH patients. Given the known effect of APOE ε4 on amyloid deposition in the cerebral vasculature, our findings indicate that APOE genotype-related vasculopathies may influence the presence of the island sign.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/omix/, OMIX008306.
The studies involving humans were approved by the Medical Scientific Research Ethics Committee of Peking University Third Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
QY: Methodology, Resources, Writing – original draft. HS: Data curation, Methodology, Resources, Writing – original draft. XM: Writing – review & editing. LT: Resources, Writing – review & editing, Methodology. XL: Resources, Writing – review & editing. XinH: Resources, Writing – review & editing. XiaH: Resources, Writing – review & editing. YC: Resources, Writing – review & editing. DT: Writing – review & editing, Resources. XZ: Methodology, Writing – review & editing. NL: Writing – review & editing, Data curation, Methodology. WW: Writing – review & editing. DF: Writing – review & editing, Conceptualization, Project administration, Supervision.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by National Natural Science Foundation of China (grant numbers 81901204) and Beijing Municipal Science and Technology Commission (grant numbers D141100000114005).
We are grateful to the patients and investigators in every center for their involvement.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2025.1540307/full#supplementary-material
1. Greenberg, SM, Ziai, WC, Cordonnier, C, Dowlatshahi, D, Francis, B, Goldstein, JN, et al. 2022 guideline for the management of patients with spontaneous intracerebral hemorrhage: a guideline from the American Heart Association/American Stroke Association. Stroke. (2022) 53:e282–361. doi: 10.1161/STR.0000000000000407
2. van Asch, CJ, Luitse, MJ, Rinkel, GJ, van der Tweel, I, Algra, A, and Klijn, CJ. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: a systematic review and meta-analysis. Lancet Neurol. (2010) 9:167–76. doi: 10.1016/S1474-4422(09)70340-0
3. Sheth, KN. Spontaneous intracerebral hemorrhage. New Engl J Med. (2022) 387:1589–96. doi: 10.1056/NEJMra2201449
4. Morotti, A, Boulouis, G, Dowlatshahi, D, Li, Q, Shamy, M, Al-Shahi Salman, R, et al. Intracerebral haemorrhage expansion: definitions, predictors, and prevention. Lancet Neurol. (2023) 22:159–71. doi: 10.1016/S1474-4422(22)00338-6
5. Huang, YW, Huang, HL, Li, ZP, and Yin, XS. Research advances in imaging markers for predicting hematoma expansion in intracerebral hemorrhage: a narrative review. Front Neurol. (2023) 14:1176390. doi: 10.3389/fneur.2023.1176390
6. Morotti, A, Arba, F, Boulouis, G, and Charidimou, A. Noncontrast ct markers of intracerebral hemorrhage expansion and poor outcome: a meta-analysis. Neurology. (2020) 95:632–43. doi: 10.1212/WNL.0000000000010660
7. Li, Q, Liu, QJ, Yang, WS, Wang, XC, Zhao, LB, Xiong, X, et al. Island sign: an imaging predictor for early hematoma expansion and poor outcome in patients with intracerebral hemorrhage. Stroke. (2017) 48:3019–25. doi: 10.1161/STROKEAHA.117.017985
8. Wei, Y, Zhu, G, Gao, Y, Chang, J, Zhang, H, Liu, N, et al. Island sign predicts hematoma expansion and poor outcome after intracerebral hemorrhage: a systematic review and meta-analysis. Front Neurol. (2020) 11:429. doi: 10.3389/fneur.2020.00429
9. Deng, L, Zhang, G, Wei, X, Yang, WS, Li, R, Shen, YQ, et al. Comparison of satellite sign and island sign in predicting hematoma growth and poor outcome in patients with primary intracerebral hemorrhage. World Neurosurg. (2019) 127:e818–25. doi: 10.1016/j.wneu.2019.03.273
10. Guo, H, You, M, Wu, J, Chen, A, Wan, Y, Gu, X, et al. Genetics of spontaneous intracerebral hemorrhage: risk and outcome. Front Neurosci. (2022) 16:874962. doi: 10.3389/fnins.2022.874962
11. Wahab, KW, Tiwari, HK, Ovbiagele, B, Sarfo, F, Akinyemi, R, Traylor, M, et al. Genetic risk of spontaneous intracerebral hemorrhage: systematic review and future directions. J Neurol Sci. (2019) 407:116526. doi: 10.1016/j.jns.2019.116526
12. Biffi, A, Sonni, A, Anderson, CD, Kissela, B, Jagiella, JM, Schmidt, H, et al. Variants at apoe influence risk of deep and lobar intracerebral hemorrhage. Ann Neurol. (2010) 68:934–43. doi: 10.1002/ana.22134
13. Marini, S, Crawford, K, Morotti, A, Lee, MJ, Pezzini, A, Moomaw, CJ, et al. Association of apolipoprotein e with intracerebral hemorrhage risk by race/ethnicity: a meta-analysis. JAMA Neurol. (2019) 76:480–91. doi: 10.1001/jamaneurol.2018.4519
14. Sawyer, RP, Sekar, P, Osborne, J, Kittner, SJ, Moomaw, CJ, Flaherty, ML, et al. Racial/ethnic variation of apoe alleles for lobar intracerebral hemorrhage. Neurology. (2018) 91:e410–20. doi: 10.1212/WNL.0000000000005908
15. Biffi, A, Anderson, CD, Jagiella, JM, Schmidt, H, Kissela, B, Hansen, BM, et al. Apoe genotype and extent of bleeding and outcome in lobar intracerebral haemorrhage: a genetic association study. Lancet Neurol. (2011) 10:702–9. doi: 10.1016/S1474-4422(11)70148-X
16. Brouwers, HB, Biffi, A, Ayres, AM, Schwab, K, Cortellini, L, Romero, JM, et al. Apolipoprotein e genotype predicts hematoma expansion in lobar intracerebral hemorrhage. Stroke. (2012) 43:1490–5. doi: 10.1161/STROKEAHA.111.643262
17. Brouwers, HB, Biffi, A, McNamara, KA, Ayres, AM, Valant, V, Schwab, K, et al. Apolipoprotein e genotype is associated with ct angiography spot sign in lobar intracerebral hemorrhage. Stroke. (2012) 43:2120–5. doi: 10.1161/STROKEAHA.112.659094
18. Koemans, EA, Chhatwal, JP, van Veluw, SJ, van Etten, ES, van Osch, MJP, van Walderveen, MAA, et al. Progression of cerebral amyloid angiopathy: a pathophysiological framework. Lancet Neurol. (2023) 22:632–42. doi: 10.1016/S1474-4422(23)00114-X
19. Grangeon, L, Roussel, M, Gillibert, A, Verdalle-Cazes, M, Dolores, M, Ozkul-Wermester, O, et al. Applicability of the Edinburgh ct criteria for lobar intracerebral hemorrhage associated with cerebral amyloid angiopathy. Clin Neuroradiol. (2023) 33:455–65. doi: 10.1007/s00062-022-01230-6
20. Carpenter, AM, Singh, IP, Gandhi, CD, and Prestigiacomo, CJ. Genetic risk factors for spontaneous intracerebral haemorrhage. Nat Rev Neurol. (2016) 12:40–9. doi: 10.1038/nrneurol.2015.226
21. Brown, RT, Coates, RK, and Gilbert, JJ. Radiographic-pathologic correlation in cerebral amyloid angiopathy. A review of 12 patients. J Can Assoc Radiol. (1985) 36:308–11.
22. Izumihara, A, Ishihara, T, Iwamoto, N, Yamashita, K, and Ito, H. Postoperative outcome of 37 patients with lobar intracerebral hemorrhage related to cerebral amyloid angiopathy. Stroke. (1999) 30:29–33. doi: 10.1161/01.STR.30.1.29
23. Minakawa, T, Takeuchi, S, Sasaki, O, Koizumi, T, Honad, Y, Fujii, Y, et al. Surgical experience with massive lobar haemorrhage caused by cerebral amyloid angiopathy. Acta Neurochir. (1995) 132:48–52. doi: 10.1007/BF01404847
24. Wagle, WA, Smith, TW, and Weiner, M. Intracerebral hemorrhage caused by cerebral amyloid angiopathy: radiographic-pathologic correlation. AJNR Am J Neuroradiol. (1984) 5:171–6.
25. Xu, HQ. Intracerebral hemorrhage related with cerebral amyloid angiopathy. Zhonghua Shen Jing Jing Shen Ke Za Zhi. (1990) 23:345, 384–7.
26. Samarasekera, N, Rodrigues, MA, Toh, PS, and Al-Shahi, R. Imaging features of intracerebral hemorrhage with cerebral amyloid angiopathy: systematic review and meta-analysis. PLoS One. (2017) 12:e0180923. doi: 10.1371/journal.pone.0180923
27. Charidimou, A, Schmitt, A, Wilson, D, Yakushiji, Y, Gregoire, SM, Fox, Z, et al. The cerebral haemorrhage anatomical rating instrument (charts): development and assessment of reliability. J Neurol Sci. (2017) 372:178–83. doi: 10.1016/j.jns.2016.11.021
28. Seiffge, DJ, Polymeris, AA, Law, ZK, Krishnan, K, Zietz, A, Thilemann, S, et al. Cerebral amyloid angiopathy and the risk of hematoma expansion. Ann Neurol. (2022) 92:921–30. doi: 10.1002/ana.26481
29. Schlunk, F, and Greenberg, SM. The pathophysiology of intracerebral hemorrhage formation and expansion. Transl Stroke Res. (2015) 6:257–63. doi: 10.1007/s12975-015-0410-1
Keywords: APOE genotype, island sign, intracerebral hemorrhage, genetics, cerebral amyloid angiopathy
Citation: Yang Q, Sun H, Ma X, Tang L, Liu X, Huang X, Huang X, Chen Y, Tian D, Zeng X, Li N, Wang W and Fan D (2025) Apolipoprotein E genotype is associated with island sign in lobar intracerebral hemorrhage. Front. Neurol. 16:1540307. doi: 10.3389/fneur.2025.1540307
Received: 05 December 2024; Accepted: 04 February 2025;
Published: 20 February 2025.
Edited by:
Minghuan Wang, Huazhong University of Science and Technology, ChinaReviewed by:
Qi Li, Second Affiliated Hospital of Anhui Medical University, ChinaCopyright © 2025 Yang, Sun, Ma, Tang, Liu, Huang, Huang, Chen, Tian, Zeng, Li, Wang and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsheng Fan, ZHNmYW4yMDEwQGFsaXl1bi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.