- 1Division of Neurology, Hospital for Sick Children, University of Toronto, Toronto, ON, Canada
- 2Department of Laboratory Medicine and Pathobiology, Temerty Faculty of Medicine, University of Toronto, Toronto, ON, Canada
- 3McMaster Children’s Hospital, McMaster University, Hamilton, ON, Canada
- 4Department of Pathology and Molecular Medicine, School of Medicine, Faculty of Health Sciences, Queen’s University, Kingston, ON, Canada
- 5Division of Cardiology, Hospital for Sick Children, University of Toronto, Toronto, ON, Canada
- 6Department of Pediatrics, Faculty of Medicine, University of Toronto, Toronto, ON, Canada
Introduction: Sudden Unexpected Death in Epilepsy (SUDEP) is the leading epilepsy-related cause of death, affecting approximately 1 per 1,000 individuals with epilepsy per year. Genetic variants that affect autonomic function, such as genes associated with cardiac arrhythmias, may predispose people with epilepsy to greater risk of both sudden cardiac death and SUDEP. Advances in next generation sequencing allow for the exploration of gene variants as potential biomarkers.
Methods: Genetic testing for the presence of cardiac arrhythmia and epilepsy gene variants was performed via genetic panels in 39 cases of SUDEP identified via autopsy by the Ontario Forensic Pathology Service. Variants were summarized by in-silico evidence for pathogenicity from 4 algorithms (SIFT, PolyPhen-2, PROVEAN, Mutation Taster) and allele frequencies in the general population (GnomAD). A maximum credible population allele frequency of 0.00004 was calculated based on epilepsy prevalence and SUDEP incidence to assess whether a variant was compatible with a pathogenic interpretation.
Results: Median age at the time of death was 33.3 years (range: 2, 60). Fifty-nine percent (n=23) were male. Gene panels detected 62 unique variants in 45 genes: 19 on the arrhythmia panel and 26 on the epilepsy panel. At least one variant was identified in 28 (72%) of decedents. Missense mutations comprised 57 (92%) of the observed variants. At least three in silico models predicted 12 (46%) cardiac arrhythmia panel missense variants and 20 (65%) epilepsy panel missense variants were pathogenic. Population allele frequencies were <0.00004 for 11 (42%) of the cardiac variants and 10 (32%) of the epilepsy variants. Together, these metrics identified 13 SUDEP variants of interest.
Discussion: Nearly three-quarters of decedents in this SUDEP cohort carried variants in comprehensive epilepsy or cardiac arrhythmia gene panels, with more than a third having variants in both panels. The proportion of decedents with cardiac variants aligns with recent studies of the disproportionate cardiac burden the epilepsy community faces compared to the general population and suggests a possible cardiac contribution to epilepsy mortality. These results identified 13 priority targets for future functional studies of these genes potential role in sudden death and demonstrates the necessity for further exploration of potential genetic contributions to SUDEP.
Introduction
Sudden death affects the epilepsy population at a rate 24 times the general population (1, 2), and sudden unexpected death in epilepsy (SUDEP) is the leading epilepsy-related cause of death, affecting approximately 1 per 1,000 individuals with epilepsy each year (3–5). SUDEP refers to deaths in people with epilepsy (PwE) without a known cause and is usually accompanied by an autopsy that identifies no anatomical or toxicological cause of death (6). SUDEP rates are higher in certain epilepsy groups, including those with poorly controlled seizures, epilepsy surgery candidates, and some genetic epilepsies, such as Dravet Syndrome (DS) (7, 8).
Understanding the pathophysiology of SUDEP is a top priority among people living with epilepsy, caregivers, and researchers. The United States National Institute of Neurological Disorders and Stroke (NINDS) released epilepsy research guidelines in 2007 which included the goals of identification of biomarkers for epileptogenesis for risk stratification, and identification of the mechanisms responsible for SUDEP (9). In 2022, the Ontario Brain Institute (OBI), in partnership with the Epilepsy Research Program (EpLink), published the top 10 unanswered questions in epilepsy according to patients, caregivers, and healthcare providers across Canada. The top priority related to the identification of genetic biomarkers to diagnose and treat epilepsy, and the fourth pertained to risk factors for SUDEP.1 The identification of biomarkers and risk factors for SUDEP is critical to providing appropriate care, as well as understanding how SUDEP risk can be mitigated.
Growing evidence implicates autonomic dysregulation (10, 11), especially cardiorespiratory dysfunction (12–14) in the mechanism of SUDEP, however, risk factors usually associated with these events have not been definitively linked to SUDEP. Despite this, several clinical risk factors for SUDEP have been identified, with the presence and frequency of bilateral tonic–clonic seizures (BTC) being the most robust (15–18). Other risk factors include nocturnal seizures, living alone or not sharing a bedroom, and failure to optimize treatment with anti-seizure medications (18–20). Despite best efforts to identify those at high risk of SUDEP, it is still not understood why many people at high risk will live for years with frequent BTC while others may die after just one or two seizures. The insufficiency of clinical risk factors emphasizes the need for the identification of different biomarkers to identify PwE at the greatest risk for SUDEP. Reliable biomarkers may inform future studies and effective SUDEP prevention strategies.
Cardiac arrhythmia genes are of particular interest as potential biomarkers given growing evidence for heart-brain interactions (21, 22) and similarities in the characteristics of SUDEP and sudden cardiac death (23, 24). Multiple genes have been identified that are associated with arrhythmogenic epilepsy; single gene variants that produce both epilepsy and cardiac arrhythmias (21, 25, 26). Many genes that produce epilepsy and cardiac phenotypes encode ion channels which are often expressed in both heart and brain tissue. Genetic epilepsies associated with channelopathies often have higher SUDEP rates (27), which suggests factors specific to channel disorders may have a role in SUDEP pathogenesis (28). Even when variants in cardiac genes are not associated with epilepsy or produce a cardiac phenotype in isolation, there is the possibility of polygenic risk whereby the presence of multiple gene variants in an individual confers an additive or multiplicative effect on SUDEP risk compared to one gene alone.
In humans, sudden cardiac death occurs 2.8 times more frequently in PwE compared to the general population (29), and a disproportionate number of PwE and individuals from SUDEP cohorts were found to have pathogenic variants in cardiac genes (30–34). Altered cardiac function has been observed in animal models of DS (35, 36) and SCN8A-related developmental and epileptic encephalography (DEE) (37) with cardiac arrhythmias often preceding SUDEP in these models. Likewise, seizures and epilepsy phenotypes have been observed in conjunction with sudden cardiac death in mouse models of Long QT Syndrome (38) and Catecholaminergic Polyventricular Tachycardia (39, 40).
Given the evidence of cardiac comorbidities among PwE and previous SUDEP cohorts, damaging cardiac gene variants are promising potential SUDEP biomarkers. We investigated the yield of cardiac- and epilepsy-related gene panels in a series of 39 SUDEP cases to explore whether those who die of SUDEP may be predisposed to sudden death by underlying genetic variants.
Methods
This study was approved by the Hospital for Sick Children Research Ethics Board.
Study cohort
The Ontario Forensic Pathology Service (OFPS) is tasked with investigation of all sudden or unexpected deaths in Ontario, Canada, which typically includes autopsy, toxicology investigation, and when appropriate genetic testing. This study retrospectively reviewed reports of SUDEP identified by the OFPS between January 2014 – June 2016. The OFPS shared the case descriptions of 83 decedents. Two neurologists with epilepsy subspecialty training and SUDEP research expertise reviewed the cases for SUDEP criteria and identified 39 cases with Definite or Definite Plus SUDEP classifications (6).
Data collected included case details, toxicology report, and genetic testing findings. Where available from the coroner’s warrant, a summary of the decedent’s medical history, medications, and circumstances surrounding the death were extracted from the autopsy reports.
Genetic analysis
DNA samples were extracted post-mortem from blood or frozen tissue. No samples from family members were provided. Next generation sequencing was performed by GeneDx and included the Comprehensive Arrhythmia Panel and the Comprehensive Epilepsy Panel including deletion/duplication analysis. At time of sequencing, the epilepsy and arrhythmia panels included 87 and 46 genes, respectively.
Complete coding regions and splice site junctions of the genes tested were enriched using a proprietary targeted capture system developed by GeneDx. Enriched targets were sequenced with paired-end reads on an Illumina platform. Bi-directional sequence reads were assembled and aligned to reference sequences based on NCBI RefSeq transcripts and human genome build GRCh37/UCSC hg19. After gene-specific filtering, data were analyzed to identify sequence variants and most deletions and duplications involving coding exons; however, technical limitations and inherent sequence properties effectively reduce this resolution for some genes. Alternative sequencing or copy number detection methods were used to analyze or confirm regions with inadequate sequence or copy number data by next generation sequencing.
Identified variants were classified according to American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guidelines (41), as well as by ClinVar2 interpretation, by pathogenicity predictions from four in-silico algorithms [SIFT3 (42), PROVEAN v1.1.34 (43), PolyPhen-25 (44), and MutationTaster6 (45)], and by population allele frequencies in the general population. Allele frequencies were calculated as the total group maximum filtering allele frequency in exomes and genomes reported by GnomAD v4.0.0.7 All data were retrieved January 10, 2024.
We used the frequency-based framework outlined by (46) to assess whether a variant was “too common” to be considered pathogenic; Assuming an epilepsy prevalence of 5/1000 (47) and a SUDEP incidence of 1/1000 per year (4, 5) we calculated a maximum credible population allele frequency of 0.00004. Variants that were predicted to be pathogenic by at least three in-silico algorithms and occurred below the population allele frequency threshold were classified as SUDEP variants of interest (VoI).
Statistical analyses
Continuous variables were compared between groups using Wilcoxon ranked sum test with continuity correction. Categorical variables were compared with Fisher’s exact test. All statistical analyses were performed using R (version 3.2.1.), and a p < 0.05 was used to indicate statistical significance. Percentages reported for clinical parameters are based on non-missing data.
Results
Cohort characteristics
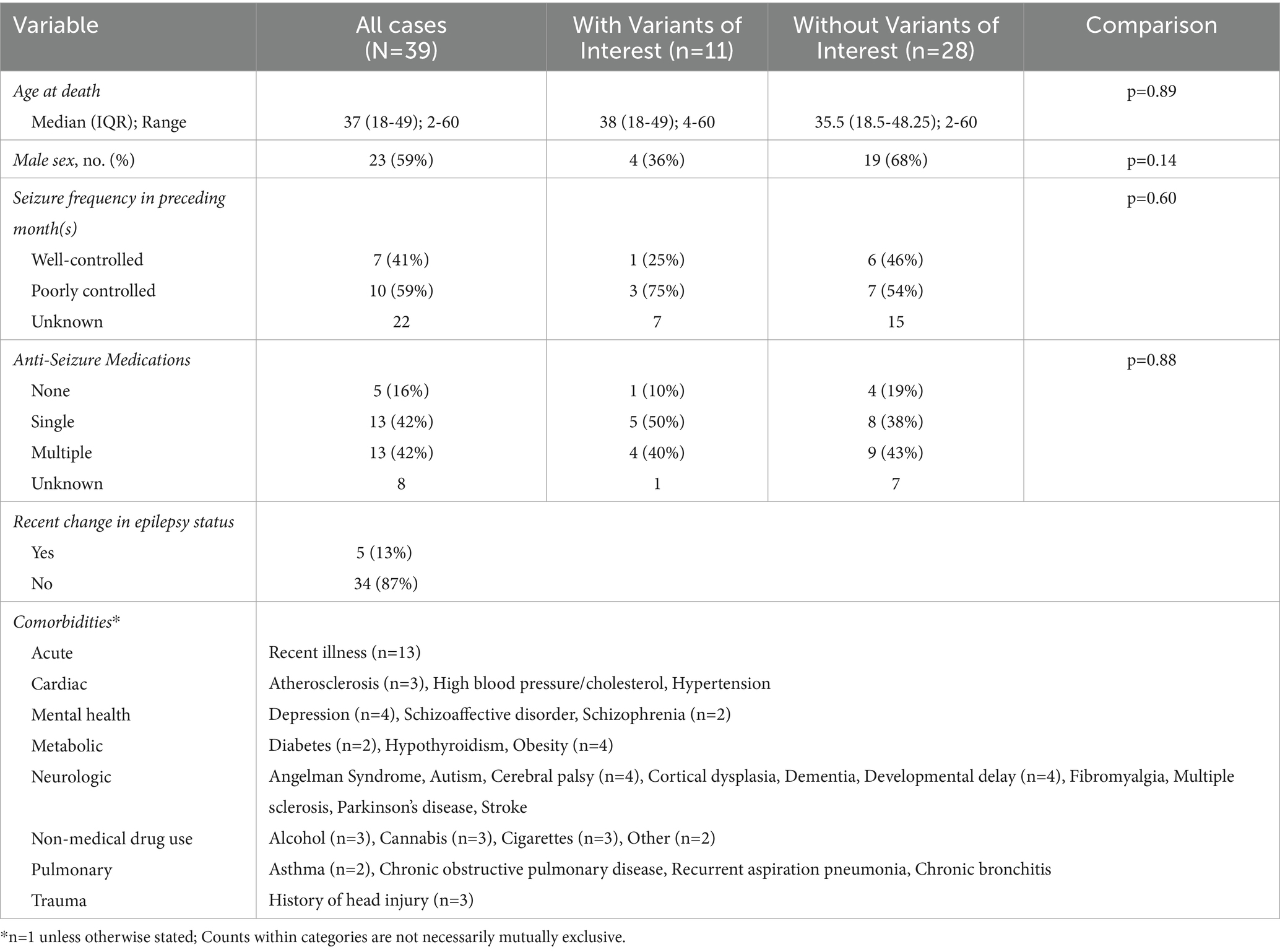
The SUDEP cohort consisted of 39 decedents. These SUDEP cases were included in a previous report on cause of death in PwE (48). All cases were classified as Definite or Definite Plus SUDEP. Median age at time of death was 33 years (IQR:18–49) and ranged from 2 to 60 years. Males accounted for 59% (n = 23). A qualitative summary of the cohort is presented in Table 1.
Seizure frequency data were available for 17 cases and reported as well-controlled (i.e., seizure free or infrequent) in 7 cases (41%) and poorly controlled (i.e., multiple per month) in 10 cases (59%). Data for anti-seizure medications (ASM) were available in 31 cases, with zero ASM reported actively taken in five cases (16%), a single ASM in 13 cases (42%), and two or more ASM in 13 cases (42%). The most frequently reported ASM were phenytoin (n = 8), valproate (n = 8), and carbamazepine (n = 5). Seizure status had recently changed for five (13%) cases, including one increase in seizure frequency and four who had weaned or changed their ASM. Toxicology and post-mortem reports indicated that three decedents were potentially non-adherent with ASM treatment.
Comorbidities were present in at least 29 cases (74%). The most common categories of comorbidities reported were neurologic (e.g., cerebral palsy), substance misuse, and acute (e.g., recent illness). Other comorbidities fell under cardiac, mental health, metabolic, pulmonary, and trauma categories (Table 1). Additional relevant medical history was limited; however, autopsy reports noted the following pertinent to cardiac history: one decedent experienced cardiac arrest after a seizure earlier in the year, and one decedent had a father who died of myocardial infarct. Lastly, one case experienced respiratory arrest after a seizure earlier in the year. All autopsy reports detailed unremarkable cardiac findings.
Sudden unexpected death in epilepsy characteristics
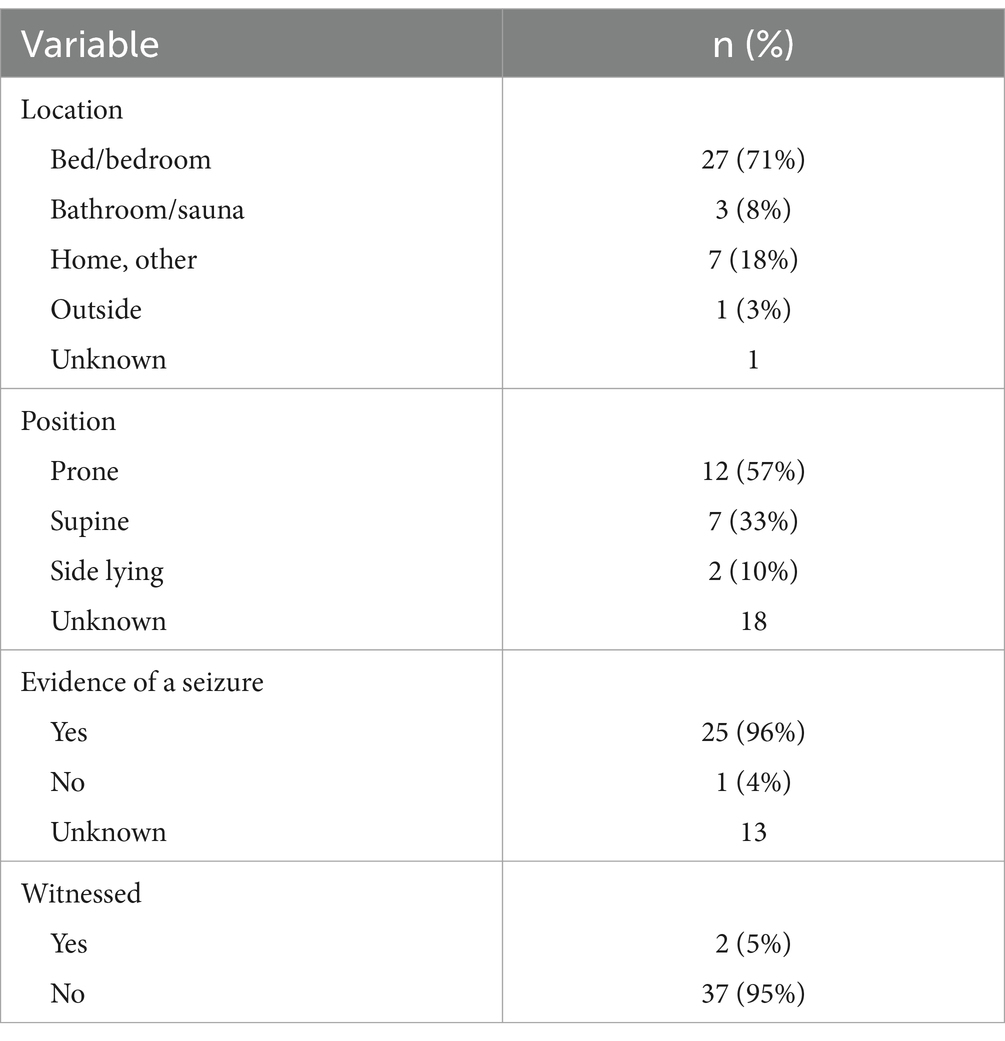
Circumstances of death are presented in Table 2. The location where the deceased was found was reported for 38 cases (97%). Most individuals (n = 27, 71%) were found in a bed – their own, a caregiver’s, or otherwise. Others were found in a bathroom (n = 3, 8%), elsewhere in their home (n = 7, 18%), or outside (n = 1, 3%). Body position was reported for 21 cases: 12 prone (57%), seven supine (33%), and two side-lying (10%). Evidence of a seizure immediately preceding SUDEP was reported for 25 cases. Examples of accepted evidence included a witnessed seizure, tongue bites, vomit, frothy fluid at the mouth, and found in an awkward position suggestive of recent convulsion. Only one case explicitly reported there was no evidence of a seizure; the remainder were unknown (n = 13). Only two deaths were witnessed, while 37 (95%) were unwitnessed.
Data on epilepsy diagnosis, etiology, and years lived with epilepsy were not available from autopsy reports. Limited data was available regarding seizure semiology and prescription drug use.
Gene panel findings
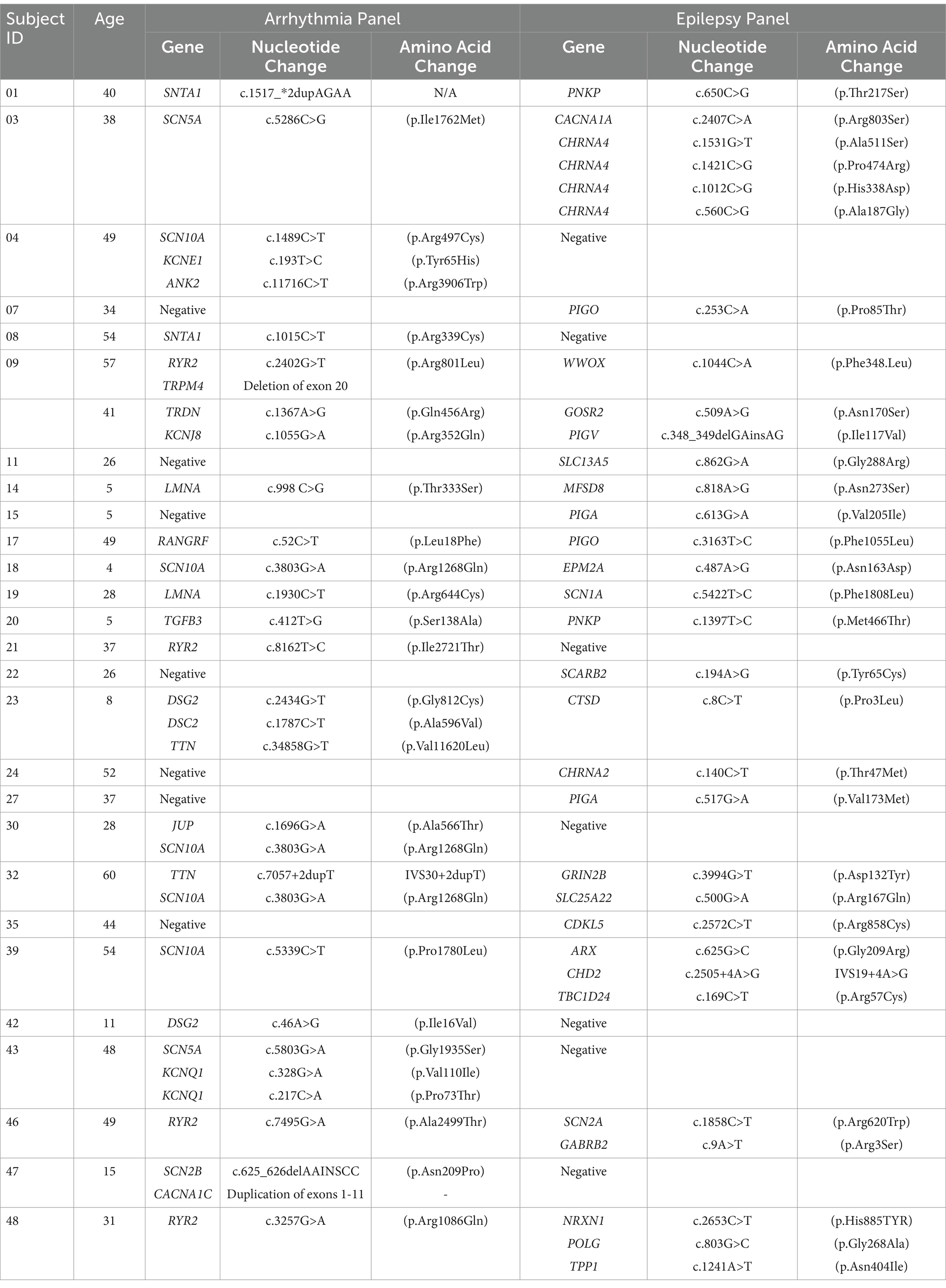
Genetic testing revealed 62 unique variants detected in 45 genes: 19 genes from the arrhythmia panel and 26 from the epilepsy panel. A full list of both panel’s genes tested is available in Supplemental Table S1. At least one variant was identified on either the epilepsy or cardiac panel in 28 (72%) decedents. Among those with at least one variant, 21 (54%) had variants in genes listed on the epilepsy panel and 21 had genes listed on the cardiac panel. A single variant was identified in 10 (36%) cases, while two or more variants were found in 18 (64%) cases. Among those with multiple variants, eight (21%) had multiple cardiac variants, six (15%) had multiple epilepsy variants, and 14 (36%) had at least one variant in both panels (Table 3). Missense variants comprised 57 (92%) of the observed variants. Other variant types included duplication of an exon (n = 1), deletion of an exon (n = 1), intronic variants (n = 2), and variants in untranslated regions (n = 1).
According to the ACMG/AMP guidelines 1 (2%) variant was classified as pathogenic – DSG2 c.2434G > T (p.Gly812Cys) – 4 (7%) as likely pathogenic, 5 (9%) as likely benign, and 47 (82%) as uncertain significance. Interpretations of the pathogenicity of the 57 missense variants by ClinVar were Benign or Likely Benign for five (9%), Variant of Uncertain Significance (VUS) for 27 (47%) and Conflicting Evidence for 25 (44%). Among the variants with Conflicting Evidence, 14 (56%) were predominantly Benign or Likely Benign. One likely pathogenic EPM2A variant associated with Lafora disease, and one likely pathogenic SCN1A variant associated with DS were identified on the epilepsy panel (Table 4).
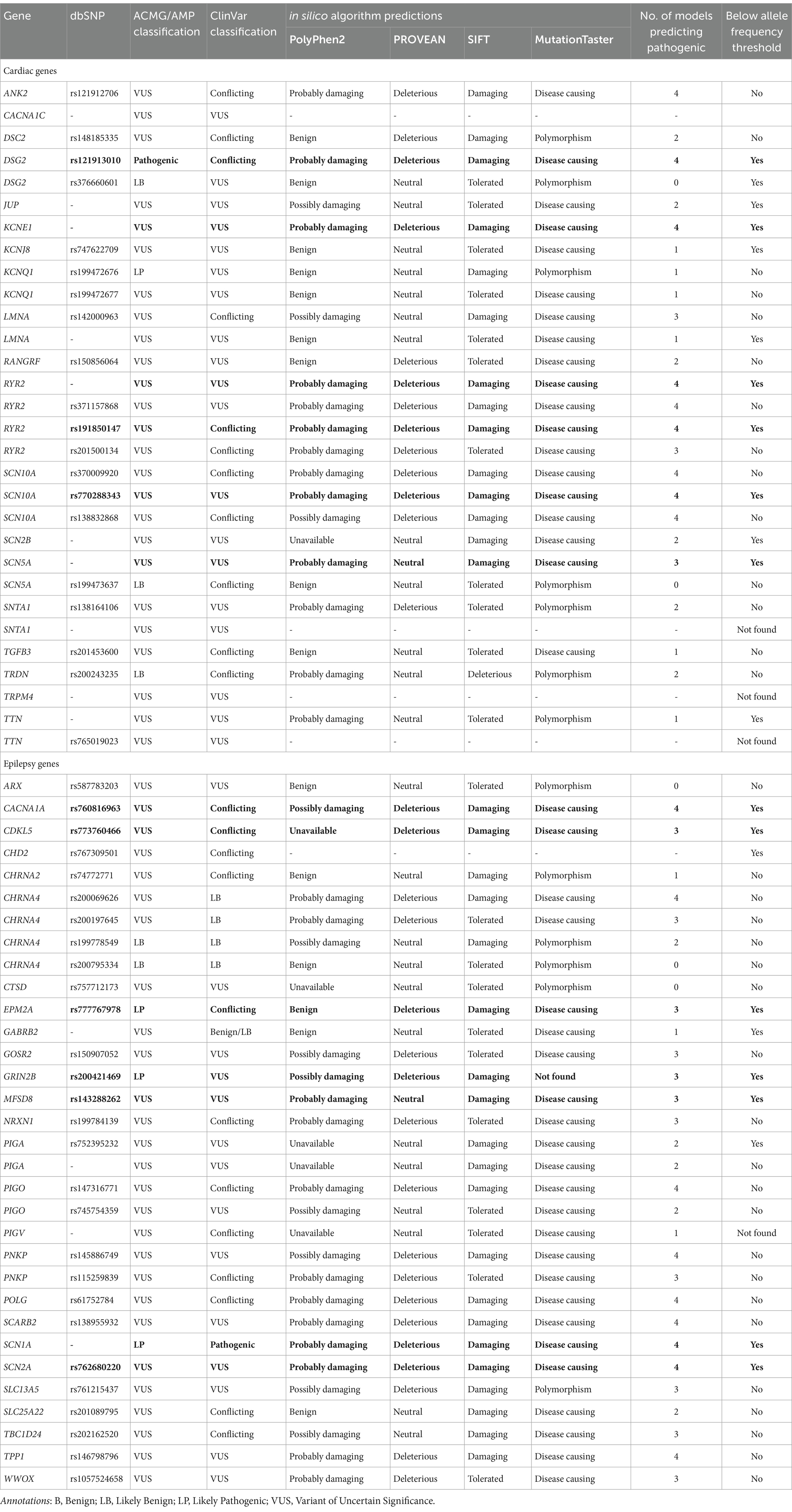
Table 4. Summary of genetic variants identified in the SUDEP cohort. Four in silico algorithms were applied, PolyPhen2, PROVEAN, SIFT, and MutationTaster. Genes are organized alphabetically by panel. Population allele frequency threshold was predetermined to be 0.00004. Variants of Interest (VoI) are bolded.
The functional impact of each missense variant was predicted by four in-silico algorithms: PROVEAN, SIFT, PolyPhen-2, and MutationTaster (Table 4). These tools were selected as they are commonly used to interpret missense variants in clinical settings (41). Analysis of the cardiac arrhythmia panel variants identified 12 of 26 (46%) predicted to be pathogenic by at least three models (i.e., Damaging, Probably damaging, Possibly damaging, Deleterious, or Disease causing). Analysis of the epilepsy panel variants identified 20 of 31 (65%) predicted to be pathogenic by at least three models.
A population allele frequency of any gene that was less than our calculated threshold (<0.00004), suggests they are less frequent than expected if the variants were benign. We identified 11 of 26 (42%) cardiac variants and 10 of 31 (32%) epilepsy variants with population allele frequencies below threshold. Of these variants, six cardiac variants and seven epilepsy variants were also predicted to be pathogenic by at least three in silico models. These 13 variants, identified across 11 individuals, were categorized as SUDEP variants of interest (VoI; Figure 1). Phenotypes commonly associated with each of the genes included among the 13 VoI are presented in Table 5; Protein expression profiles are provided in Supplemental Table S2.
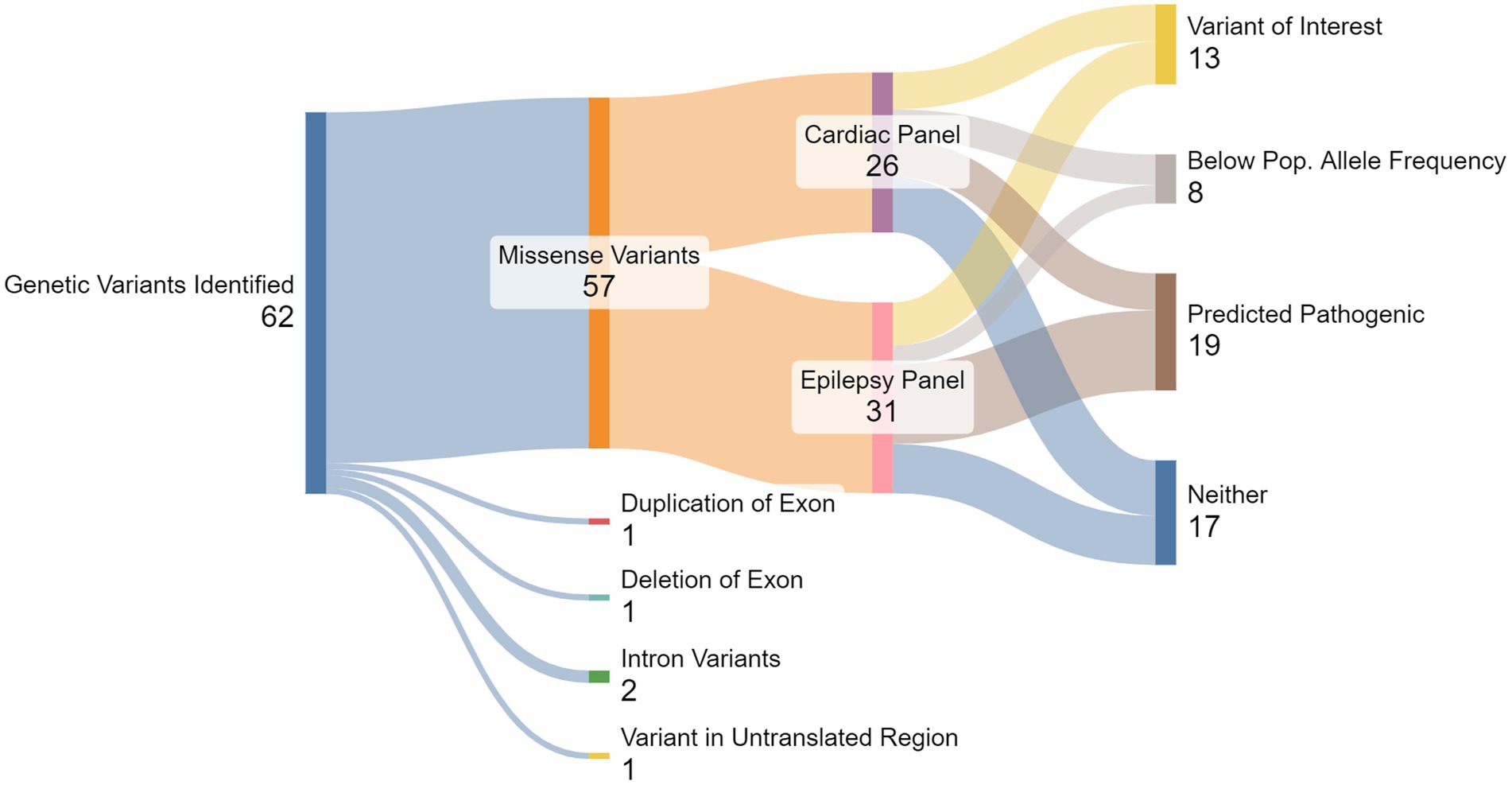
Figure 1. Summary of gene testing results. From the total 62 genetic variants detected, 57 were missense variants, comprised of 27 genes from the cardiac panel and 30 from the epilepsy panel. Multiple in silico algorithms predicted that a total of 32 variants were pathogenic, and a total of 22 variants were below the pre-determined population allele frequency threshold (<0.00004). Thirteen variants, seven from the cardiac panel and six from the epilepsy panel, were both predicted pathogenic and below population allele frequency threshold, identifying them as SUDEP Variants of Interest (VoI).
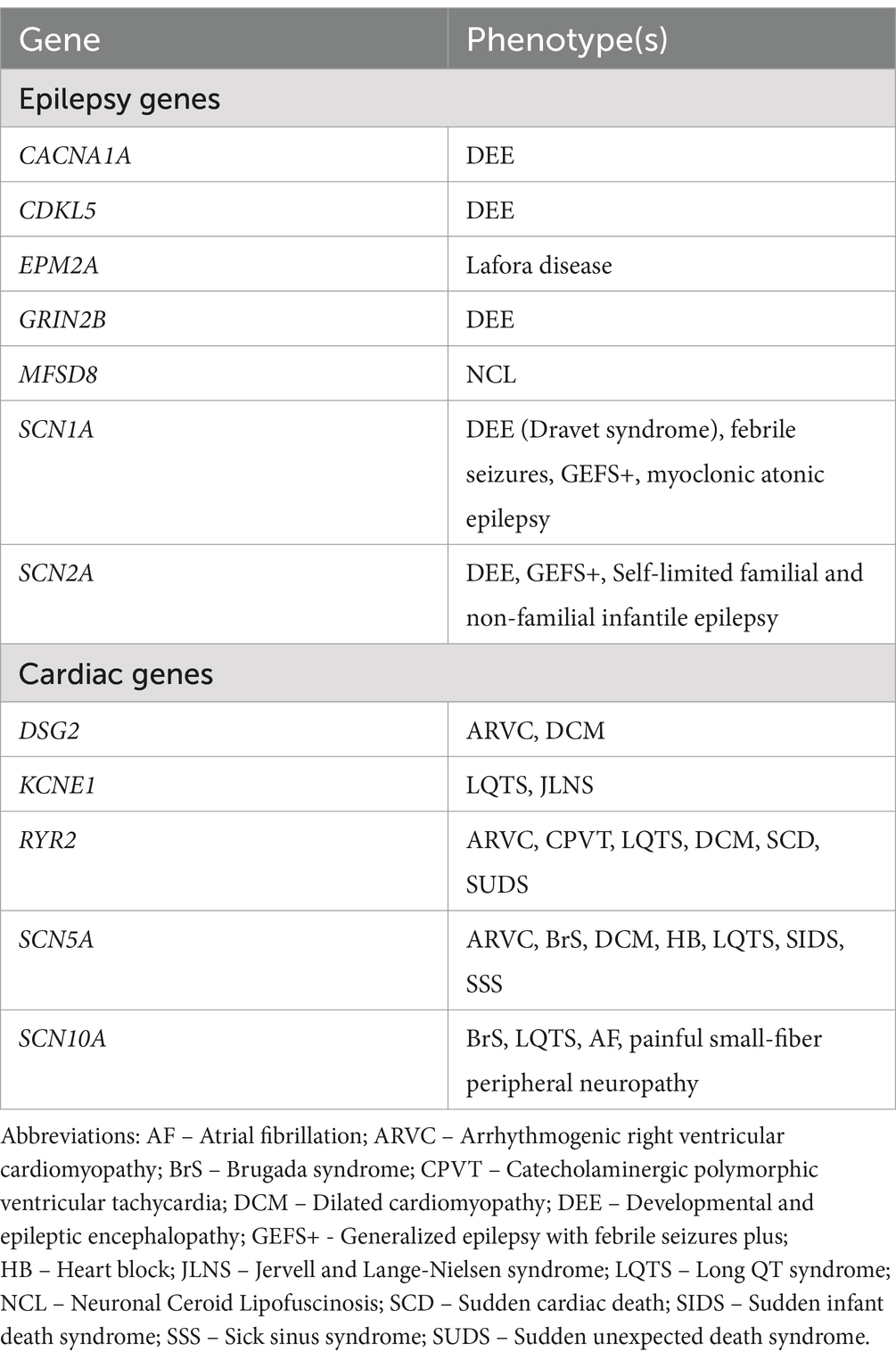
Table 5. Epilepsy and cardiac phenotypes commonly associated with the genes identified as Variants of Interest.
We compared demographic and clinical factors between decedents with SUDEP VoI to everyone else. Age, sex, seizure frequency, and number of ASM were not significantly different between those with VoI (n = 14) and those without (n = 25) (Age: W = 159, p = 0.89; Sex: Fisher’s exact test p = 0.14; Seizure frequency: Fisher’s exact test p = 0.60; ASM: Fisher’s exact test p = 0.88).
This cohort contained novel variants with no corresponding ClinVar entry. From the cardiac panel, two variants were identified – SCN10A c.5339C > T (p.Pro1780Leu), and SCN5A c.5286C > G, (p.Ile1762Met); Both were classified as SUDEP genes of interest. From the epilepsy panel one novel variant was identified – GRIN2B c.3994G > T (p.Asp132Tyr). The GRIN2B variant met our criteria for a SUDEP VoI.
An SCN10A variant c.3803G > A (p.Arg1268Gln) was the only identical variant documented in multiple SUDEP cases in our cohort (n = 3). Two additional SCN10A variants were identified in the cohort, making SCN10A the most frequent gene finding. Multiple variants were also reported in more than one decedent for RYR2 (n = 4), as well as DSG2, LMNA, PIGA, PIGO, PNKP, SCN5A, SNTA1, and TTN variants (n = 2). All other genes were observed in single cases.
Discussion
More than half of the decedents in this SUDEP cohort (21/39; 54%) had genetic variants in both epilepsy and cardiac gene panels. Over half of the missense variants identified on either panel were predicted to be pathogenic by three or more in silico algorithms, and over a third of the variants were compatible with pathogenicity based on population allele frequency (46). Together, these metrics identified 13 SUDEP Variants of Interest (VoI), 23% of all missense variants in this cohort. Our results contribute to a growing literature on the possible contribution of cardiac genetic variants to epilepsy mortality, and SUDEP, specifically.
The circumstances surrounding death in this cohort were typical of SUDEP characteristics; death was most frequently reported during the night, the deceased were found in bed, death was unwitnessed, and there was evidence of a preceding seizure. Interestingly, many of the typical features of sudden cardiac death are similar to those of SUDEP, including occurring during the night, the deceased is found in bed in a prone position, the death is unwitnessed, and post-mortem investigation fails to identify a cause of death (49–51).
It is well-established that epilepsy negatively affects heart health, independent of any present gene variants. Epidemiological studies have found increased rates of heart disease and sudden cardiac death in adults diagnosed with epilepsy compared to the general population (21). Post-mortem findings of PwE have identified myocardial structural changes such as fibrosis, myocyte vacuolization (52–54). These findings have led to the hypothesis that surges in catecholamines and hypoxemia due to repeated seizures lead to electrical and mechanical heart dysfunction (22, 55–58). In addition, while no single ASM, nor the presence of polytherapy or monotherapy have been reliably associated with SUDEP (59, 60), it should be noted that some ASM have been shown to modulate cardiac activity. For example, therapeutic doses of carbamazepine may induce bradyarrhythmia in some patients (61, 62) and both ASM use and sudden withdrawal of ASM have been shown to reduce heart-rate variability (63, 64) which has been associated with an increase in sudden cardiac death (65). These effects could be relevant in genetically susceptible individuals. Two of the most common ASMs reported in this cohort included phenytoin and carbamazepine, which in the setting of certain cardiac conditions (e.g., Brugada syndrome) should be avoided. Notably, at least three decedents in this cohort with either SCN5A or SCN10A variants reported using one of these two ASMs (cases: 03, 04, 32).
In part because of these heart-brain interactions, proving a cardiac genetic contribution to SUDEP is difficult. If such a relationship exists, a cardiac-related genetic contribution to SUDEP could take multiple forms: (1) a subclinical arrhythmogenic variant that increases the susceptibility to SUDEP (66); (2) a genetic variant that confers both a risk of epilepsy and a risk of sudden cardiac death (i.e., monogenic/arrhythmogenic epilepsy) (66); (3) the co-occurrence of different epilepsy-causing and arrhythmia-causing genes that independently increase the risk for sudden death (i.e., polygenic) (25). Polygenic risk for SUDEP may overlap with polygenic risk for sudden cardiac death and the proportion of SUDEP deaths that may be poly- or oligemic compared to monogenic is not known.
Sudden unexpected death in epilepsy variants of interest
At least three in silico algorithms classified 13 variants in the cardiac panel and 19 variants in the epilepsy panel as pathogenic, totaling 32 (56%). Based on population allele frequency, 12 variants in the cardiac panel and nine in the epilepsy panel were less than the threshold 0.00004, totaling 21 (37%). Together, these metrics identified a total of 13 (23%) of the 57 missense variants in this cohort as SUDEP VoI. The 13 VoI were identified in 11 decedents (two individuals had two: cases 03 and 46).
In both epilepsy and cardiac gene panels, ion channels were the most common protein types encoded by the in silico-predicted pathogenic genes, including sodium, calcium, potassium, and chloride channels. Other groups of proteins identified in this cohort included cell adhesion proteins & ion channel regulators, protein & solute transports, cell signaling molecules, cell replication & DNA repair proteins, and proteins regulating metabolism.
Among the 13 SUDEP VoI, nine of the genes encode ion channel subunits (five of the nine were from the cardiac panel), one encodes a cell signaling molecule (epilepsy panel), one encodes a cell adhesion protein (cardiac panel), one phosphatase (epilepsy panel), and one a protein/solute transport (epilepsy panel). Most of the genes with identified VoI are autosomal dominant inheritance, excepting MFSD8 (recessive), EPM2A (recessive), and CDKL5 (x-linked).
The exact mechanisms of disease are not known for some of the cardiac phenotypes associated with the genes in this panel. However, there are many ways that proteins linked to cardiac phenotypes could contribute to SUDEP. Ion channel dysfunction provides perhaps the most straightforward explanation of sudden death. Ion channels are critical to the formation and propagation of action potentials, both in the heart and brain. Dysregulation of these time- and voltage-dependent mechanisms could lead to heart failure directly or cardiorespiratory arrest via spreading depolarization in the autonomic nervous system – all proposed mechanisms of SUDEP (15, 67, 68). Proteins involved in cell adhesion (e.g., DSG2) or protein transport (e.g., MFSD8) could alter the structural architecture of the heart which could lead directly to a cardiac phenotype or could reduce resistance to mechanical stress which may be experienced during a seizure (69, 70). Regardless of the mechanism, a variant in an arrhythmogenic- or cardiomyopathy-related gene could predispose a person with epilepsy to sudden death by exacerbating the impacts of recurrent seizures on heart health or the effect of a single prolonged convulsive seizure.
Relation to previous work
A recent systematic review of the genetics of SUDEP concluded that the most prominent and frequent gene variants identified across studies were associated with ion channels – which may be co-expressed in the heart and brain and are causally related to some epilepsies (e.g., DS and SCN1A) and cardiac disorders (e.g., Brugada Syndrome and SCN5A) – and genes linked to cardiac arrhythmia phenotypes (25). However, most of the genes identified had VUS ratings by in silico pathogenicity models. Importantly, although protein dysfunction in these genes may lead to disease (channelopathies) associated with the heart or the brain, almost all these ion channels are expressed to some degree in both the heart and the brain (Supplemental Table S2), opening the possibility of a shared genetic susceptibility between epilepsy and cardiac arrhythmias.
Previous post-mortem investigations of SUDEP cohorts have reported many of the same genes identified in this SUDEP cohort. These include the epilepsy genes CACNA1A, CDKL5, CHRNA4, GRIN2B, NRXN1, PNKP, POLG, SCARB2, SCN1A, and SCN2A as well as the cardiac genes ANK2, KCNE1, KCNQ1, RYR2, SCN5A, and SCN10A (26, 30, 31, 33, 34, 71, 72). Many of these genes, from both panels, were reported across multiple studies, including the ion channel subunits for sodium (SCNs), potassium (KCNs), and calcium (CACNA1A, RYR2). These genes constitute a high priority for future investigations of genetic biomarkers for SUDEP.
Future work
More than half of our cohort had variants identified in the comprehensive cardiac arrhythmia panel, however, the role these genetic variants played in sudden death is unclear. Further work to determine the significance of the identified variants is needed. Ultimately, genetic risk factors could be combined with other clinical risk factors to develop individualized SUDEP risk prediction tools.
Concerted efforts are needed to investigate the contribution of cardiac genotypes to epilepsy mortality. Few of the genes identified in our cohort, as well as in the broader cardiac SUDEP literature, have had functional studies. Among the epilepsy panel findings in our cohort, we were only able to find functional studies for the two known disease-associated variants in the genes EPM2A (73) and SCN1A (74). Among the cardiac panel findings, we found functional studies for five variants in the genes ANK2 (75, 76), DSG2 (77), KCNQ1 (78), LMNA (79, 80), and SCN10A (81). Evaluation by in vivo and in vitro methods to assess potential pathogenic mechanisms are critical steps in validating the remainder of these gene variants as SUDEP biomarkers, particularly those found in multiple SUDEP cohorts.
Cardiac contributions to epilepsy mortality are well-recognized and beginning to receive appropriate research attention. However, SUDEP mechanisms are likely heterogeneous and only a subset is likely attributable to cardiac dysfunction. The MORTEMUS study, which systematically reviewed cardiorespiratory events captured in epilepsy monitoring units, showed that primary cardiac arrhythmia is rarely an isolated primary cause of SUDEP in a cohort of people admitted to epilepsy monitoring units for evaluation (12). Thus, genetic screening should be expanded to other potential mechanisms, such as respiratory disorders (33, 82, 83), the serotonergic system (67, 84, 85), and the immune system (86–88).
Limitations
This cohort was identified through a retrospective case series review. At the time of this study, GeneDx comprehensive epilepsy and cardiac panels included 87 and 46 genes, respectively. Currently, these panels include 144 and 58 genes, respectively. As is common in genetics case series, variants were assessed for pathogenicity through multiple in silico algorithms. Our study was the first to apply a population allele frequency threshold to filter variants of interest. Although the method used here was novel in the context of SUDEP genetics, the variants of interest we identified did not meet the definite criteria for pathogenicity based on the guidelines published by the ACMG/AMP (41) and there is not substantial evidence that these genes are causative of SUDEP. The majority of the gene variants identified in this cohort were classified as VUS. This classification pertains to their relationship to cardiac or epilepsy phenotypes, not SUDEP. However, they may still be relevant to SUDEP in some contexts. Particularly, the variants in the cardiac genes may be impactful in people with epilepsy, even if they produce subclinical phenotypes in otherwise healthy individuals. These variants may be worth further investigation; variants can be reclassified over time as more evidence becomes available.
Access to clinical data was limited, including epilepsy etiology, seizure semiology, and cardiac medical histories. Although autopsy reports did not identify a cause of death that would rule out SUDEP, we cannot be certain all relevant comorbidities were reported for all cases. Genetic testing of family members was not available, which could have provided clinically useful evidence for the relationship between cardiac arrhythmias and epilepsy. Based on our findings and the results of previous reports, genetic testing should be performed in first degree relatives after sudden death. SUDEP within a family with a common gene variant would be among the strongest evidence for a genetic influence on SUDEP. Moreover, it would allow for appropriate screening and management of relatives for those who genetic variants are identified. Few reports of multiple cases of SUDEP within a family have been published (89–91).
A persistent limitation across studies of SUDEP genetics is the lack of standardization in methods, making comparison difficult. First, populations and ascertainment methods differed, with a mix of prospective and retrospective sampling, with few reaching the level of population-based sampling. Second, different genes were assessed, and the method of genetic testing varied. Some studies used whole genome or whole exome sequencing, while others were limited to genetic panels provided by various services. Finally, the criteria for determining pathogenicity differed; most studies use some combination of in silico algorithms and some also compared to living epilepsy groups. Importantly, all studies examined at an outcome (SUDEP) within a pathogenic population (people with epilepsy), so it is possible that these variants may confer risk of epilepsy and be non-contributory to the SUD.
Conclusion
Targeted epilepsy and cardiac arrhythmia gene panels detected variants in 28 of 39 (72%) SUDEP cases. Over half of this cohort had epilepsy variants (54%) and over half had cardiac variants (54%), with more than a third having variants in both panels (36%). Nearly two-thirds of these variants were predicted to be pathogenic by at least three of four in silico algorithms, while one-third of the cardiac variants and one-fifth of the epilepsy variants had population allele frequencies below threshold. These results identified 13 priority targets for future functional studies of these genes potential role in sudden death.
The proportion of cases with cardiac variants aligns with recent studies of the disproportionate cardiac burden the epilepsy community faces compared to the general population and suggests a possible cardiac contribution to SUDEP. This data also supports the concept of a collaborative care approach between neurology and cardiology, as well as pathology and genetics disciplines. Standard cardiac screening such as 12-lead ECG or Holter monitor could be increasingly utilized in epilepsy care at little cost. Targeting these initiatives to individuals with channelopathies may be particularly effective, given their well-recognized role in severe epilepsies and cardiac arrhythmias.
This data demonstrates the necessity for further exploration of potential genetic contributions to SUDEP. Such endeavors will (1) allow for identification of high-risk individuals which can aid in enriched cohorts for clinical trials and (2) allow for design of targeted therapies to prevent SUDEP. Valuable information would be gained through protocols that identify genetic predispositions in individuals with epilepsy, and that additional value may be obtained from extending genetic screening to parents, siblings, and children after the sudden death of an individual with epilepsy. Epilepsy lags other disease fields, including cardiac arrhythmias, in incorporating genetic analyses as part of pathology analysis. Improvements in technology have made genetic testing fast and cost-effective, and uptake in the use of genetic analyses aligns with precision/individualized medicine goals adopted by many health care institutions. Widespread uptake of genetic screening in PwE could facilitate research efforts to identify SUDEP biomarkers.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving humans were approved by The Hospital for Sick Children Research Ethics Board. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from the Ontario Forensic Pathology Service (OFPS) which is tasked with investigation of all sudden or unexpected deaths in Ontario, Canada. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
AA: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation. AK: Writing – review & editing, Methodology, Conceptualization. AW: Writing – review & editing, Resources, Methodology, Investigation, Data curation. RW: Writing – review & editing, Visualization, Validation. KC: Writing – review & editing, Methodology, Data curation, Conceptualization. RH: Writing – review & editing. MP: Writing – review & editing, Resources, Methodology, Investigation, Data curation, Conceptualization. ED: Writing – review & editing, Supervision, Resources, Methodology, Funding acquisition, Data curation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Ontario Brain Institute and the Michael Bahen Chair in Epilepsy Research at the University of Toronto.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2024.1386730/full#supplementary-material
Footnotes
1. ^https://braininstitute.ca/epilepsy-psp
2. ^https://www.ncbi.nlm.nih.gov/clinvar/
4. ^https://www.jcvi.org/research/provean
5. ^http://genetics.bwh.harvard.edu/pph2/
References
1. Forsgren, L, Hauser, WA, Olafsson, E, Sander, JWA, Sillanpää, M, and Tomson, T. Mortality of epilepsy in developed countries: a review. Epilepsia. (2005) 46:18–27. doi: 10.1111/j.1528-1167.2005.00403.x
2. Ficker, DM, So, EL, Shen, WK, Annegers, JF, O’Brien, PC, Cascino, GD, et al. Population-based study of the incidence of sudden unexplained death in epilepsy. Neurology. (1998) 51:1270–4. doi: 10.1212/WNL.51.5.1270
3. Walczak, TS, Leppik, IE, D’Amelio, M, Rarick, J, So, E, Ahman, P, et al. Incidence and risk factors in sudden unexpected death in epilepsy: a prospective cohort study. Neurology. (2001) 56:519–25. doi: 10.1212/WNL.56.4.519
4. Sveinsson, O, Andersson, T, Carlsson, S, and Tomson, T. The incidence of SUDEP: a nationwide population-based cohort study. Neurology. (2017) 89:170–7. doi: 10.1212/WNL.0000000000004094
5. Keller, AE, Whitney, R, Li, SA, Pollanen, MS, and Donner, EJ. Incidence of sudden unexpected death in epilepsy in children is similar to adults. Neurology. (2018) 91:e107–11. doi: 10.1212/WNL.0000000000005762
6. Nashef, L, So, EL, Ryvlin, P, and Tomson, T. Unifying the definitions of sudden unexpected death in epilepsy. Epilepsia. (2012) 53:227–33. doi: 10.1111/j.1528-1167.2011.03358.x
7. Cooper, MS, McIntosh, A, Crompton, DE, McMahon, JM, Schneider, A, Farrell, K, et al. Mortality in Dravet syndrome. Epilepsy Res. (2016) 128:43–7. doi: 10.1016/j.eplepsyres.2016.10.006
8. Shmuely, S, Sisodiya, SM, Gunning, WB, Sander, JW, and Thijs, RD. Mortality in Dravet syndrome: a review. Epilepsy Behav. (2016) 64:69–74. doi: 10.1016/j.yebeh.2016.09.007
9. Kelley, MS, Jacobs, MP, and Lowenstein, DH. The NINDS epilepsy research benchmarks. Epilepsia. (2009) 50:579–82. doi: 10.1111/j.1528-1167.2008.01813.x
10. DeGiorgio, CM, Curtis, A, Hertling, D, and Moseley, BD. Sudden unexpected death in epilepsy: Risk factors, biomarkers, and prevention. Acta Neurol Scand. (2019) 139:220–30. doi: 10.1111/ane.13049
11. Szurhaj, W, Leclancher, A, Nica, A, Périn, B, Derambure, P, Convers, P, et al. Cardiac autonomic dysfunction and risk of sudden unexpected death in epilepsy. Neurology. (2021) 96:998. doi: 10.1212/WNL.0000000000011998
12. Ryvlin, P, Nashef, L, Lhatoo, SD, Bateman, LM, Bird, J, Bleasel, A, et al. Incidence and mechanisms of cardiorespiratory arrests in epilepsy monitoring units (MORTEMUS): a retrospective study. Lancet Neurol. (2013) 12:966–77. doi: 10.1016/S1474-4422(13)70214-X
13. Lhatoo, SD, Nei, M, Raghavan, M, Sperling, M, Zonjy, B, Lacuey, N, et al. Nonseizure SUDEP: sudden unexpected death in epilepsy without preceding epileptic seizures. Epilepsia. (2016) 57:1161–8. doi: 10.1111/epi.13419
14. Barot, N, and Nei, M. Autonomic aspects of sudden unexpected death in epilepsy (SUDEP). Clin Auton Res. (2019) 29:151–60. doi: 10.1007/s10286-018-0576-1
15. Nashef, L, Hindocha, N, and Makoff, A. Risk factors in sudden death in epilepsy (SUDEP): the quest for mechanisms. Epilepsia. (2007) 48:859–71. doi: 10.1111/j.1528-1167.2007.01082.x
16. Hughes, JR. A review of sudden unexpected death in epilepsy: prediction of patients at risk. Epilepsy Behav. (2009) 14:280–7. doi: 10.1016/j.yebeh.2008.12.004
17. Sveinsson, O, Andersson, T, Carlsson, S, and Tomson, T. Circumstances of SUDEP: a nationwide population-based case series. Epilepsia. (2018) 59:1074–82. doi: 10.1111/epi.14079
18. Sveinsson, O, Andersson, T, Mattsson, P, Carlsson, S, and Tomson, T. Clinical risk factors in SUDEP: a nationwide population-based case-control study. Neurology. (2020) 94:e419–29. doi: 10.1212/WNL.0000000000008741
19. Ryvlin, P, Cucherat, M, and Rheims, S. Risk of sudden unexpected death in epilepsy in patients given adjunctive antiepileptic treatment for refractory seizures: a meta-analysis of placebo-controlled randomised trials. Lancet Neurol. (2011) 10:961–8. doi: 10.1016/S1474-4422(11)70193-4
20. Harden, C, Tomson, T, Gloss, D, Buchhalter, J, Cross, JH, Donner, E, et al. Practice guideline summary: sudden unexpected death in epilepsy incidence rates and risk factors: report of the guideline development, dissemination, and implementation subcommittee of the American Academy of Neurology and the American Epilepsy Society. Epilepsy Curr. (2017) 17:180–7. doi: 10.5698/1535-7511.17.3.180
21. Costagliola, G, Orsini, A, Coll, M, Brugada, R, Parisi, P, and Striano, P. The brain-heart interaction in epilepsy: implications for diagnosis, therapy, and SUDEP prevention. Ann Clin Transl Neurol. (2021) 8:1557–68. doi: 10.1002/acn3.51382
22. Verrier, RL, Pang, TD, Nearing, BD, and Schachter, SC. The epileptic heart: concept and clinical evidence. Epilepsy Behav. (2020) 105:106946. doi: 10.1016/j.yebeh.2020.106946
23. Gould, L, Reid, C-A, Rodriguez, AJ, and Devinsky, O For SUDC Video Working Group. Video analyses of sudden unexplained deaths in toddlers. Neurology. (2024) 102:e208038. doi: 10.1212/WNL.0000000000208038
24. Devinsky, O, Friedman, D, Cheng, JY, Moffatt, E, Kim, A, and Tseng, ZH. Underestimation of sudden deaths among patients with seizures and epilepsy. Neurology. (2017) 89:886–92. doi: 10.1212/WNL.0000000000004292
25. Chahal, CAA, Salloum, MN, Alahdab, F, Gottwald, JA, Tester, DJ, Anwer, LA, et al. Systematic review of the genetics of sudden unexpected death in epilepsy: potential overlap with sudden cardiac death and arrhythmia-related genes. J Am Heart Assoc. (2020) 9:e012264. doi: 10.1161/JAHA.119.012264
26. Friedman, D, Kannan, K, Faustin, A, Shroff, S, Thomas, C, Heguy, A, et al. Cardiac arrhythmia and neuroexcitability gene variants in resected brain tissue from patients with sudden unexpected death in epilepsy (SUDEP). NPJ Genom Med. (2018) 3:9. doi: 10.1038/s41525-018-0048-5
27. Verducci, C, Friedman, D, Donner, E, and Devinsky, O. Genetic generalized and focal epilepsy prevalence in the north American SUDEP registry. Neurology. (2020) 94:e1757–63. doi: 10.1212/WNL.0000000000009295
28. Glasscock, E. Genomic biomarkers of SUDEP in brain and heart. Epilepsy Behav. (2014) 38:172–9. doi: 10.1016/j.yebeh.2013.09.019
29. Bardai, A, Blom, MT, Van Noord, C, Verhamme, KM, Sturkenboom, MCJM, and Tan, HL. Sudden cardiac death is associated both with epilepsy and with use of antiepileptic medications. Heart. (2015) 101:17–22. doi: 10.1136/heartjnl-2014-305664
30. Tu, E, Bagnall, RD, Duflou, J, and Semsarian, C. Post-mortem review and genetic analysis of sudden unexpected death in epilepsy (SUDEP) cases. Brain Pathol. (2011) 21:201–8. doi: 10.1111/j.1750-3639.2010.00438.x
31. Coll, M, Allegue, C, Partemi, S, Mates, J, Del Olmo, B, Campuzano, O, et al. Genetic investigation of sudden unexpected death in epilepsy cohort by panel target resequencing. Int J Legal Med. (2016) 130:331–9. doi: 10.1007/s00414-015-1269-0
32. Partemi, S, Vidal, MC, Striano, P, Campuzano, O, Allegue, C, Pezzella, M, et al. Genetic and forensic implications in epilepsy and cardiac arrhythmias: a case series. Int J Legal Med. (2015) 129:495–504. doi: 10.1007/s00414-014-1063-4
33. Bagnall, RD, Crompton, DE, Petrovski, S, Lam, L, Cutmore, C, Garry, SI, et al. Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann Neurol. (2016) 79:522–34. doi: 10.1002/ana.24596
34. Ge, Y, Ding, D, Zhu, G, Kwan, P, Wang, W, Hong, Z, et al. Genetic variants in incident SUDEP cases from a community-based prospective cohort with epilepsy. J Neurol Neurosurg Psychiatry. (2019) 91:126–31. doi: 10.1136/jnnp-2019-321983
35. Auerbach, DS, Jones, J, Clawson, BC, Offord, J, Lenk, GM, Ogiwara, I, et al. Altered cardiac electrophysiology and SUDEP in a model of Dravet syndrome. PLoS One. (2013) 8:e77843. doi: 10.1371/journal.pone.0077843
36. Kalume, F, Westenbroek, RE, Cheah, CS, Yu, FH, Oakley, JC, Scheuer, T, et al. Sudden unexpected death in a mouse model of Dravet syndrome. J Clin Invest. (2013) 123:1798–808. doi: 10.1172/JCI66220
37. Frasier, CR, Wagnon, JL, Bao, YO, McVeigh, LG, Lopez-Santiago, LF, Meisler, MH, et al. Cardiac arrhythmia in a mouse model of sodium channel SCN8A epileptic encephalopathy. Proc Natl Acad Sci USA. (2016) 113:12838–43. doi: 10.1073/pnas.1612746113
38. Goldman, AM, Glasscock, E, Yoo, J, Chen, TT, Klassen, TL, and Noebels, JL. Arrhythmia in heart and brain: KCNQ1 mutations link epilepsy and sudden unexplained. Sci Transl Med. (2009) 1:2ra6. doi: 10.1126/scitranslmed.3000289
39. Lehnart, SE, Mongillo, M, Bellinger, A, Lindegger, N, Chen, B-X, Hsueh, W, et al. Leaky ca 2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J Clin Invest. (2008) 118:346. doi: 10.1172/JCI35346
40. Aiba, I, Wehrens, XHT, and Noebels, JL. Leaky RyR2 channels unleash a brainstem spreading depolarization mechanism of sudden cardiac death. Proc Natl Acad Sci USA. (2016) 113:E4895–903. doi: 10.1073/pnas.1605216113
41. Richards, S, Aziz, N, Bale, S, Bick, D, Das, S, Gastier-Foster, J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. (2015) 17:405–24. doi: 10.1038/gim.2015.30
42. Ng, PC, and Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. (2001) 11:863–74. doi: 10.1101/gr.176601
43. Choi, Y, Sims, GE, Murphy, S, Miller, JR, and Chan, AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. (2012) 7:688. doi: 10.1371/journal.pone.0046688
44. Adzhubei, IA, Schmidt, S, Peshkin, L, Ramensky, VE, Gerasimova, A, Bork, P, et al. A method and server for predicting damaging missense mutations. Nat Methods. (2010) 7:248–9. doi: 10.1038/nmeth0410-248
45. Steinhuas, R, Proft, S, Schuelke, M, Cooper, DN, Schwarz, JM, and Seelow, D. MutationTaster2021. Nucleic Acids Res. (2021) 49:W446–51. doi: 10.1093/nar/gkab266
46. Whiffin, N, Minikel, E, Walsh, R, O’Donnell-Luria, AH, Karczewski, K, Ing, AY, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med. (2017) 19:1151–8. doi: 10.1038/gim.2017.26
47. Lhatoo, SD, Solomon, JK, McEvoy, AW, Kitchen, ND, Shorvon, SD, and Sander, JW. A prospective study of the requirement for and the provision of epilepsy surgery in the United Kingdom. Epilepsia. (2003) 44:673–6. doi: 10.1046/j.1528-1157.2003.46002.x
48. Keller, AE, Ho, J, Whitney, R, Li, SA, Williams, AS, Pollanen, MS, et al. Autopsy-reported cause of death in a population-based cohort of sudden unexpected death in epilepsy. Epilepsia. (2021) 62:472–80. doi: 10.1111/epi.16793
49. Ryvlin, P, and Cascino, GD. Sudden unexpected death in epilepsy patients is often misdiagnosed as sudden cardiac death. Neurol. Genet. (2017) 89:878–9. doi: 10.1212/WNL.0000000000004301
50. Dasheiff, RM. Sudden unexpected death in epilepsy: a series from an epilepsy surgery program and speculation on the relationship to sudden cardiac death. J Clin Neurophysiol. (1991) 8:216–22. doi: 10.1097/00004691-199104000-00010
51. Gami, AS, Olson, EJ, Shen, WK, Wright, RS, Ballman, KV, Hodge, DO, et al. Obstructive sleep apnea and the risk of sudden cardiac death: a longitudinal study of 10,701 adults. J Am Coll Cardiol. (2013) 62:610–6. doi: 10.1016/j.jacc.2013.04.080
52. Falconer, B, and Rajs, J. Post-mortem findings of cardiac lesions in epileptics: a preliminary report. Forensic Sci. (1976) 8:63–71. doi: 10.1016/0300-9432(76)90048-0
53. Leestma, JE, Walczak, T, Hughes, JR, Kalelkar, MB, and Teas, SS. A prospective study on sudden unexpected death in epilepsy. Ann Neurol. (1989) 26:195–203. doi: 10.1002/ana.410260203
54. Natelson, BH, Suarez, RV, Terrence, CF, and Turizo, R. Patients with epilepsy who die suddenly have cardiac disease. Arch Neurol. (1998) 55:857–60. doi: 10.1001/archneur.55.6.857
55. Blum, AS, Ives, JR, Goldberger, AL, Al-Aweel, IC, Krishnamurthy, KB, Drislane, FW, et al. Oxygen desaturations triggered by partial seizures: implications for cardiopulmonary instability in epilepsy. Epilepsia. (2000) 41:536–41. doi: 10.1111/j.1528-1157.2000.tb00206.x
56. Fineschi, V, Silver, MD, Karch, SB, Parolini, M, Turillazzi, E, Pomara, C, et al. Myocardial disarray: an architectural disorganization linked with adrenergic stress? Int J Cardiol. (2005) 99:277–82. doi: 10.1016/j.ijcard.2004.01.022
57. Moseley, B, Bateman, L, Millichap, JJ, Wirrell, E, and Panayiotopoulos, CP. Autonomic epileptic seizures, autonomic effects of seizures, and SUDEP. Epilepsy Behav. (2013) 26:375–85. doi: 10.1016/j.yebeh.2012.08.020
58. Nass, RD, Motloch, LJ, Paar, V, Lichtenauer, M, Baumann, J, Zur, B, et al. Blood markers of cardiac stress after generalized convulsive seizures. Epilepsia. (2019) 60:201–10. doi: 10.1111/epi.14637
59. Hesdorffer, DC, and Tomson, T. Sudden unexpected death in epilepsy: potential role of antiepileptic drugs. CNS Drugs. (2013) 27:113–9. doi: 10.1007/s40263-012-0006-1
60. Hesdorffer, DC, Tomson, T, Benn, E, Sander, JW, Nilsson, L, Langan, Y, et al. Do antiepileptic drugs or generalized tonic-clonic seizure frequency increase SUDEP risk? A combined analysis. Epilepsia. (2012) 53:249–52. doi: 10.1111/j.1528-1167.2011.03354.x
61. Kasarskis, EJ, Kuo, CS, Berger, R, and Nelson, KR. Carbamazepine-lnduced cardiac dysfunction: characterization of two distinct clinical syndromes. Arch Intern Med. (1992) 152:186–91. doi: 10.1001/archinte.1992.00400130184025
62. Kenneback, G, Bergfeldt, L, Tomson, T, Spina, E, and Edhag, O. Carbamazepine induced bradycardia - a problem in general or only in susceptible patients? A 24-h long-term electrocardiogram study. Epilepsy Res. (1992) 13:141–5. doi: 10.1016/0920-1211(92)90069-6
63. Kennebäck, G, Ericson, M, Tomson, T, and Bergfeldt, L. Changes in arrhythmia profile and heart rate variability during abrupt withdrawal of antiepileptic drugs. Seizure. (1997) 6:369–75. doi: 10.1016/S1059-1311(97)80036-2
64. Lotufo, PA, Valiengo, L, Bensenor, IM, and Brunoni, AR. A systematic review and meta-analysis of heart rate variability in epilepsy and antiepileptic drugs. Epilepsia. (2012) 53:272–82. doi: 10.1111/j.1528-1167.2011.03361.x
65. Bigger, JT, Fleiss, JL, Steinman, RC, Rolnitzky, LM, Kleiger, RE, and Rottman, JN. Frequency domain measures of heart period variability and mortality after myocardial infarction. Circulation. (1992) 85:164–71. doi: 10.1161/01.CIR.85.1.164
66. Bleakley, LE, Soh, MS, Bagnall, RD, Sadleir, LG, Gooley, S, Semsarian, C, et al. Are variants causing cardiac arrhythmia risk factors in sudden unexpected death in epilepsy? Front Neurol. (2020) 11:925. doi: 10.3389/fneur.2020.00925
67. Richerson, GB, and Buchanan, GF. The serotonin axis: shared mechanisms in seizures, depression, and SUDEP. Epilepsia. (2011) 52:28–38. doi: 10.1111/j.1528-1167.2010.02908.x
68. Aiba, I, and Noebels, JL. Spreading depolarization in the brainstem mediates sudden cardiorespiratory arrest in mouse SUDEP models. Sci Transl Med. (2015) 7:50. doi: 10.1126/scitranslmed.aaa4050
69. Kadam, SD, Sullivan, BJ, Goyal, A, Blue, ME, and Smith-Hicks, C. Rett syndrome and CDKL5 deficiency disorder: from bench to clinic. Int J Mol Sci. (2019) 20:5098. doi: 10.3390/ijms20205098
70. Crasto, S, My, I, and Di Pasquale, E. The broad spectrum of LMNA cardiac diseases: from molecular mechanisms to clinical phenotype. Front Physiol. (2020) 11:761. doi: 10.3389/fphys.2020.00761
71. Coll, M, Striano, P, Ferrer-Costa, C, Campuzano, O, Mates, J, del Olmo, B, et al. Targeted next-generation sequencing provides novel clues for associated epilepsy and cardiac conduction disorder/SUDEP. PLoS One. (2017) 12:e0189618. doi: 10.1371/journal.pone.0189618
72. Hata, Y, Yoshida, K, Kinoshita, K, and Nishida, N. Epilepsy-related sudden unexpected death: targeted molecular analysis of inherited heart disease genes using next-generation DNA sequencing. Brain Pathol. (2017) 27:292–304. doi: 10.1111/bpa.12390
73. García-Gimeno, MA, Viana, R, Rubio-Villena, C, Sánchez-Martín, P, Brewer, MK, Gentry, MS, et al.. Phenotypic characterization of a new EPM2A mutation (N163D). 2nd biennial international Lafora workshop; California: La Jolla (2016).
74. Vanoye, CG, Lossin, C, Rhodes, TH, and George, AL. Single-channel properties of human NaV1.1 and mechanism of channel dysfunction in SCN1A-associated epilepsy. J Gen Physiol. (2006) 127:1–14. doi: 10.1085/jgp.200509373
75. Mohler, PJ, Le Scouarnec, S, Denjoy, I, Lowe, JS, Guicheney, P, Caron, L, et al. Defining the cellular phenotype of “ankyrin-B syndrome” variants: human ANK2 variants associated with clinical phenotypes display a spectrum of activities in cardiomyocytes. Circulation. (2007) 115:432–41. doi: 10.1161/CIRCULATIONAHA.106.656512
76. Mohler, PJ, Splawski, I, Napolitano, C, Bottelli, G, Sharpe, L, Timothy, K, et al. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci USA. (2004) 101:9137–42. doi: 10.1073/pnas.0402546101
77. Schlipp, A, Schinner, C, Spindler, V, Vielmuth, F, Gehmlich, K, Syrris, P, et al. Desmoglein-2 interaction is crucial for cardiomyocyte cohesion and function. Cardiovasc Res. (2014) 104:245–57. doi: 10.1093/cvr/cvu206
78. Cordeiro, JM, Perez, GJ, Schmitt, N, Pfeiffer, R, Nesterenko, VV, Burashnikov, E, et al. Overlapping lqt1 and LQT2 phenotype in a patient with long QT syndrome associated with loss-of-function variations in KCNQ1 and KCNH2. Can J Physiol Pharmacol. (2010) 88:1181–90. doi: 10.1139/Y10-094
79. Capell, BC, and Collins, FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. (2006) 7:940–52. doi: 10.1038/nrg1906
80. Perrot, A, Hussein, S, Ruppert, V, Schmidt, HHJ, Wehnert, MS, Duong, NT, et al. Identification of mutational hot spots in LMNA encoding Lamin a/C in patients with familial dilated cardiomyopathy. Basic Res Cardiol. (2009) 104:90–9. doi: 10.1007/s00395-008-0748-6
81. Hu, D, Barajas-Martinez, H, Pfeiffer, R, Dezi, F, Pfeiffer, J, Buch, T, et al. Mutations in SCN10A are responsible for a large fraction of cases of brugada syndrome. J Am Coll Cardiol. (2014) 64:66–79. doi: 10.1016/j.jacc.2014.04.032
82. Shorvon, S, and Tomson, T. Sudden unexpected death in epilepsy. Lancet. (2011) 378:2028–38. doi: 10.1016/S0140-6736(11)60176-1
83. Hitiris, N, Suratman, S, Kelly, K, Stephen, LJ, Sills, GJ, and Brodie, MJ. Sudden unexpected death in epilepsy: a search for risk factors. Epilepsy Behav. (2007) 10:138–41. doi: 10.1016/j.yebeh.2006.11.010
84. Feng, HJ, and Faingold, CL. Abnormalities of serotonergic neurotransmission in animal models of SUDEP. Epilepsy Behav. (2017) 71:174–80. doi: 10.1016/j.yebeh.2015.06.008
85. Petrucci, AN, Joyal, KG, Purnell, BS, and Buchanan, GF. Serotonin and sudden unexpected death in epilepsy. Exp Neurol. (2020) 325:113145. doi: 10.1016/j.expneurol.2019.113145
86. Bhandare, AM, Kapoor, K, Farnham, MMJ, and Pilowsky, PM. Microglia PACAP and glutamate: friends or foes in seizure-induced autonomic dysfunction and SUDEP? Respir Physiol Neurobiol. (2016) 226:39–50. doi: 10.1016/j.resp.2016.01.003
88. Vezzani, A, Balosso, S, and Ravizza, T. Neuroinflammatory pathways as treatment targets and biomarkers in epilepsy. Nature reviews. Neurology. (2019) 15:459–72. doi: 10.1038/s41582-019-0217-x
89. Hindocha, N, Nashef, L, Elmslie, F, Birch, R, Zuberi, S, Al-Chalabi, A, et al. Two cases of sudden unexpected death in epilepsy in a GEFS+ family with an SCN1A mutation [1]. Epilepsia. (2008) 49:360–5. doi: 10.1111/j.1528-1167.2007.01439_2.x
90. FbA, N, Borlot, F, Cossette, P, Minassian, BA, and Andrade, DM. Two definite cases of sudden unexpected death in epilepsy in a family with a DEPDC5 mutation. Neurology. Genetics. (2015) 1:28. doi: 10.1212/NXG.0000000000000028
Keywords: SUDEP (sudden unexpected death in epilepsy), cardiac arrhythmias, biomarker, population allele frequencies, gene panel
Citation: Aschner A, Keller A, Williams A, Whitney R, Cunningham K, Hamilton RM, Pollanen M and Donner E (2024) Cardiac arrhythmia and epilepsy genetic variants in sudden unexpected death in epilepsy. Front. Neurol. 15:1386730. doi: 10.3389/fneur.2024.1386730
Edited by:
Christopher Michael DeGiorgio, University of California, Los Angeles, United StatesReviewed by:
Oscar Campuzano, University of Girona, SpainIrena Dolezalova, St. Anne’s University Hospital Brno, Czechia
Copyright © 2024 Aschner, Keller, Williams, Whitney, Cunningham, Hamilton, Pollanen and Donner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amir Aschner, YW1pci5hc2NobmVyQHNpY2traWRzLmNh; Elizabeth Donner, RWxpemFiZXRoLkRvbm5lckBTaWNra2lkcy5jYQ==
 Amir Aschner
Amir Aschner Anne Keller1
Anne Keller1 Robyn Whitney
Robyn Whitney Robert M. Hamilton
Robert M. Hamilton Elizabeth Donner
Elizabeth Donner