- 1Department of Neurosurgery, Shanxi Provincial People's Hospital, The Affiliated People’s Hospital of Shanxi Medical University, Taiyuan, China
- 2Key Laboratory of Cellular Physiology at Shanxi Medical University, Ministry of Education, Taiyuan, Shanxi, China
Background and objectives: Observational studies have suggested that a multitude of pathological processes and biomolecules are involved in the initiation and development of epilepsy, and ULK3 is linked to the nervous system. However, it remains uncertain whether this association between ULK3 and epilepsy is causal and the direction of any causal relationship. This study employs a two-sample Mendelian randomization (MR) method to investigate the relationship between ULK3 and the risk of epilepsy.
Methods: We analyzed genome-wide association study (GWAS) summary statistics for ULK3 (sample size = 3,301), focal epilepsy (sample size = 39,348), and generalized epilepsy (sample size = 33,446). Bidirectional MR analyses were conducted to explore these relationships. We selected a set of single nucleotide polymorphisms (SNPs) with an association threshold of less than 1 × 10−5 as instrumental variables for further analysis. Various MR methods, including Inverse Variance Weighted, Weighted Median, MR-Egger Regression, Simple Model, Weighted Model, and Robust Adjustment Profile Score were used. Sensitivity analyses were performed to ensure the robustness of the results.
Results: Our MR analyses revealed a causal relationship where an increased level of ULK3 was associated with a decreased risk of focal epilepsy (odds ratio = 0.92, 95% confidence interval: 0.86–1.00, p = 0.041). No significant heterogeneity (Q = 7.85, p = 0.165) or horizontal pleiotropy (Egger regression intercept = 0.0191, p = 0.415) was detected. However, in the reverse analysis, we found no significant causal effect of focal epilepsy on ULK3 (p > 0.05). Furthermore, no significant causation was identified between ULK3 and generalized epilepsy (p > 0.05).
Conclusion: This study suggests a causal relationship between ULK3 and the risk of focal epilepsy from a genetic perspective. Nevertheless, further investigation is needed to understand the role of ULK3 in epilepsy fully.
1 Introduction
Epilepsy, one of the most prevalent and debilitating neurological disorders, is characterized by active, transient, reproducible, and often paroxysmal disruptions of the central nervous system (CNS) (1). It can be broadly classified into three groups based on genetic factors, underlying mechanisms, and clinical effects: focal epilepsy, generalized epilepsy, and unknown epilepsy, depending on the type of seizure. Focal epilepsy originates within networks limited to one hemisphere and in subcortical structures. It may be discretely localized or more widely distributed. In contrast, generalized epilepsy originates from some point within, involving bilateral cerebral cortex and subcortical structures, and rapidly spreads to bilaterally distributed networks (2). According to the Global Epilepsy Burden Report, approximately 130,000 individuals succumb to epilepsy-related complications annually, with half a million people worldwide diagnosed with epilepsy, resulting in 125,000 annual deaths (3). Understanding the causes and mechanisms underlying epilepsy is crucial to identifying novel opportunities for ameliorating the condition.
ULK3, a serine/threonine protein kinase located in the cytoplasm, plays a key role in regulating Sonic Hedgehog (SHH) signaling and autophagy (4). On the one hand, ULK3 can positively regulate the transmission of SHH signaling, as a regulator of the SHH signaling pathway that regulates various developmental processes, tissue homeostasis, and adult neurogenesis, in addition to processes such as protein autophosphorylation and regulation of the smoothened signaling pathway (5, 6). On the other hand, studies had demonstrated that ULK3 involved in autophagy as a positive regulator of GLI1 (GLI family zinc finger 1) (5). These findings highlight ULK3’s role in both autophagy and cell division processes. Autophagy dysfunction is associated with various human diseases, including cancer, heart disease, autoimmune disorders, and neurological conditions, such as epilepsy (7). In neurons, autophagy is dynamically regulated in different cellular compartments, including somatic cells, axons, and dendrites. Dysfunctional autophagy in epilepsy is primarily due to an imbalance between excitation and inhibition in the brain. Autophagy has been considered a potential target for neurological disease treatment (8). Consequently, we conducted this study to assess the causal relationship between ULK3 and epilepsy, with the aim of improving epilepsy treatment strategies.
Mendelian randomization (MR) is a method that utilizes genetic variants as instrumental variables (IVs) to determine whether a risk factor causally affects a health outcome (9). The Mendelian randomization is based on Mendel’s second law, the independent separation of genetic alleles when DNA is passed from parent to offspring during gametic formation (10). Mendelian randomization analysis can provide critical evidence for the potential causal effects of many modifiable exposures, including traditional epidemiological risk factors, lifestyle factors, and drug targets (11). It is designed to provide unbiased assessments of causality as much as possible (12). To address the gaps in our understanding of the relationship between ULK3 and epilepsy, we conducted a two-sample MR analysis to investigate their causal connection.
2 Methods
2.1 Data source
The genome-wide association study (GWAS) data for epilepsy, including generalized epilepsy (cases = 3,769) and focal epilepsy (cases = 9,671), was obtained from the 2018 International Alliance against Epilepsy (ILAE) Complex Epilepsy Alliance, consisting of 15,212 cases of epilepsy and 29,677 controls (13). The genetic variation data for ULK3 were sourced from the human plasma protein genome map,1 derived from a study of 3,301 healthy individuals, linking genetic variations to disease and drug databases (14).
2.2 Study design
We conducted a two-sample MR analysis using the GWAS data for epilepsy and the ULK3 data. To ensure the robustness of our results, we performed multiple sensitivity analyses. MR research necessitates meeting three core conditions: (1) IVs are closely related to ULK3; (2) IVs are independent of confounding factors; (3) IVs affect focal epilepsy and generalized epilepsy exclusively through ULK3 (Figure 1) (15).
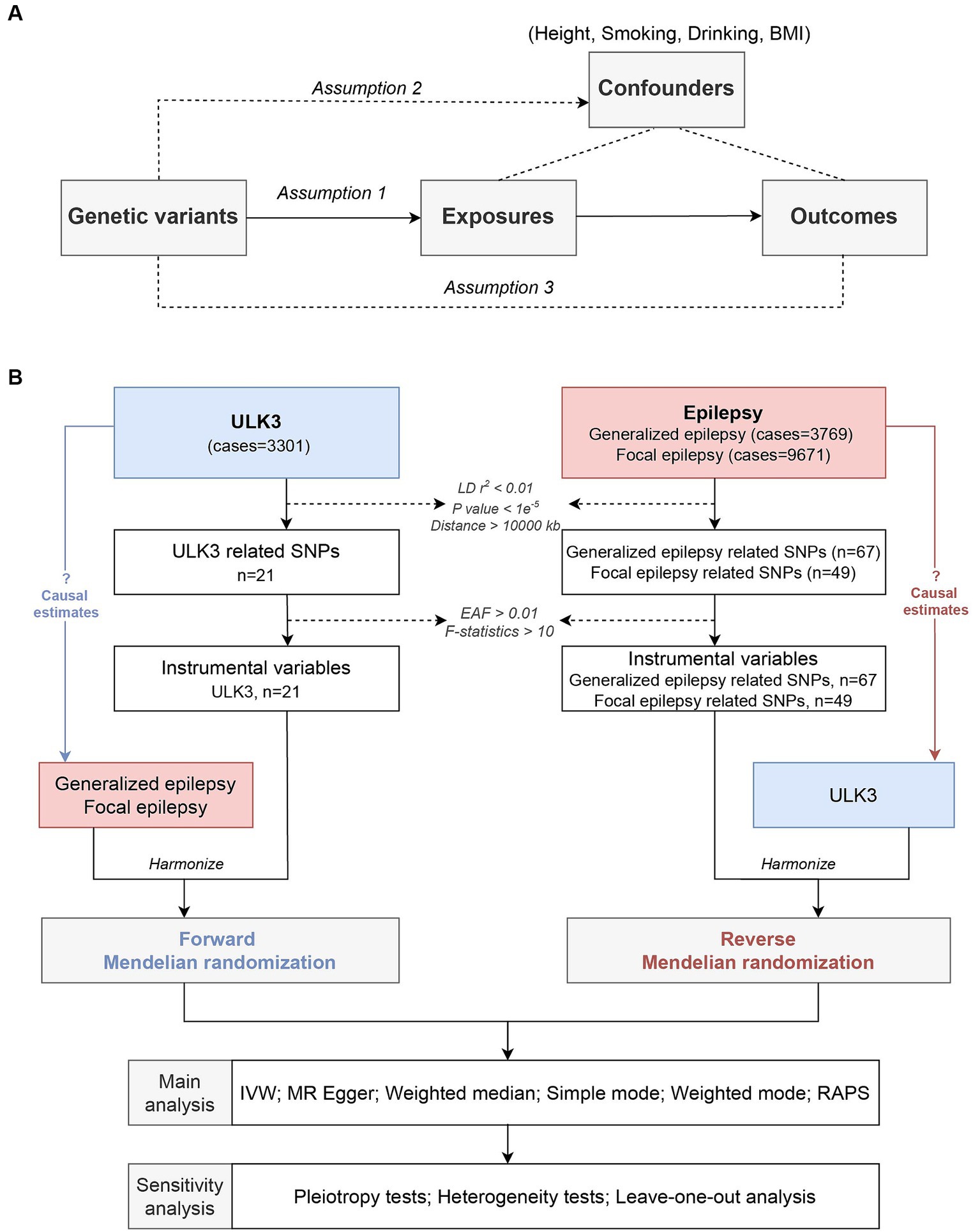
Figure 1. Study design. (A) Schematic diagram of the assumptions in Mendelian randomization (MR) model. (B) Flow diagram of this current MR framework.
2.3 Instrumental variable selection
We rigorously screened single nucleotide polymorphisms (SNPs), selecting those with a genome-wide significance threshold (p < 1e−5). To ensure the independence of IVs, we selected SNPs with an R2 of ≥0.01 within a 10,000 kb-sized window (16). We used PhenoScannerV2 to remove SNPs related to IVs (17), such as height (18), BMI (19), smoking (20), drinking (21), and other potential confounding factors. We calculated the F-statistic of IVs, selecting SNPs with F ≥ 10 and a minor allele frequency ≥ 0.01 to minimize the impact of weak IVs on bias (22). The remaining SNPs were used for MR analysis.
2.4 Statistical analysis
To assess the causal relationship between ULK3 and epilepsy, we performed MR analysis using the following methods: Inverse Variance Weighted (IVW): This method aggregates two or more random variables to minimize the sum variance and calculate the Wald ratio of the causal effect of each SNP (23). IVW results served as a primary basis, with other methods providing Supplementary Data Sheet 1 (24). MR-Egger Regression: This method fits regression models to test the existence of SNPs’ horizontal pleiotropy and quantifies SNPs with an intercept term. An intercept term of 0 indicates no horizontal pleiotropy (25). Weighted Median: This method provides a consistent and reliable causal estimate even when only half of the effective IVs are available (26). Simple Mode: Simple mode is not as powerful as IVW, but it provides robustness for pleiotropy. When the largest group of instruments with MR estimates is valid, Weighted Mode based causal estimation consistently estimates the causal effect mode (27). Robust Adjustment Profile Score (RAPS): This method is robust to system multiplicity and heterogeneity, reducing the influence of weak IVs and enhancing statistical effectiveness (28).
To ensure the robustness of our analysis, we conducted several sensitivity analyses: Cochran’s Q Statistics were used to evaluate heterogeneity among IVs (29). The intercept term of MR-Egger regression was calculated to assess horizontal pleiotropy (30). Leave-One-Out (LOO) Analysis was employed to eliminate SNPs one by one, evaluating whether the results of MR depended on a specific SNP (31).
The analysis was conducted using R (version 4.3.1) and the Two-sample MR analysis package (version 0.5.7). All datasets were analyzed in both forward and reverse directions, with a significance threshold of p-value < 0.05 applied.
3 Results
3.1 Selection of IVs
A total of 21 SNPs met the genome-wide significance threshold for IVs related to ULK3. After matching with the GWAS data for focal epilepsy and generalized epilepsy, six SNPs remained. None of these SNPs were associated with confounding factors in PhenoScanner, and all were employed in subsequent MR analysis. The F-statistic, with a value of 26.98, indicated the presence of strong instruments.
3.2 ULK3 and focal epilepsy
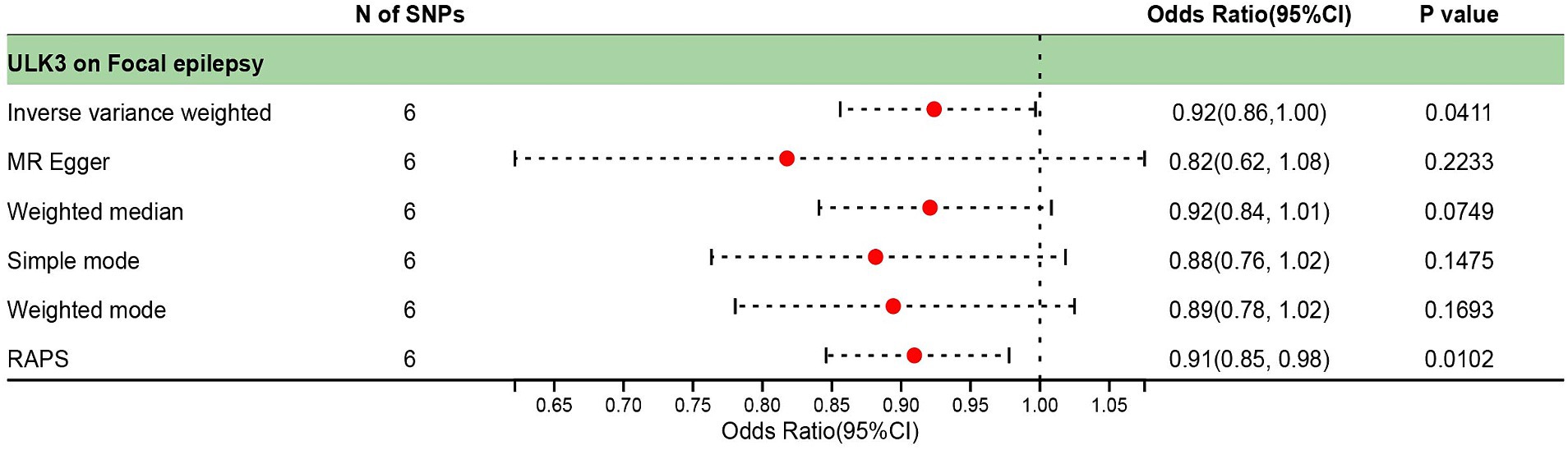
The F-statistic for individual SNPs ranged from 19.98 to 28.98, suggesting that the causal association was unlikely to be affected by weak instrumental variable bias (Supplementary Table 1). Using the IVW method, we found a significant association between ULK3 and an increased risk of focal epilepsy (odds ratio (OR) = 0.924, 95% confidence interval (95% CI): 0.622–0.856, p = 0.041). The MR-Egger, weighted median, and weighted model methods yielded more conservative estimates that did not reach statistical significance (Figure 2). No evidence of pleiotropy (intercept = 0.019, p = 0.414) or heterogeneity (p = 0.165) was observed. Scatterplots and funnel plots displayed a symmetrical distribution of points of causal effects, suggesting minimal susceptibility to potential bias. Additionally, the LOO analysis found no significant disproportionate effect for any SNP on the causal estimates (Figure 3; Table 1).
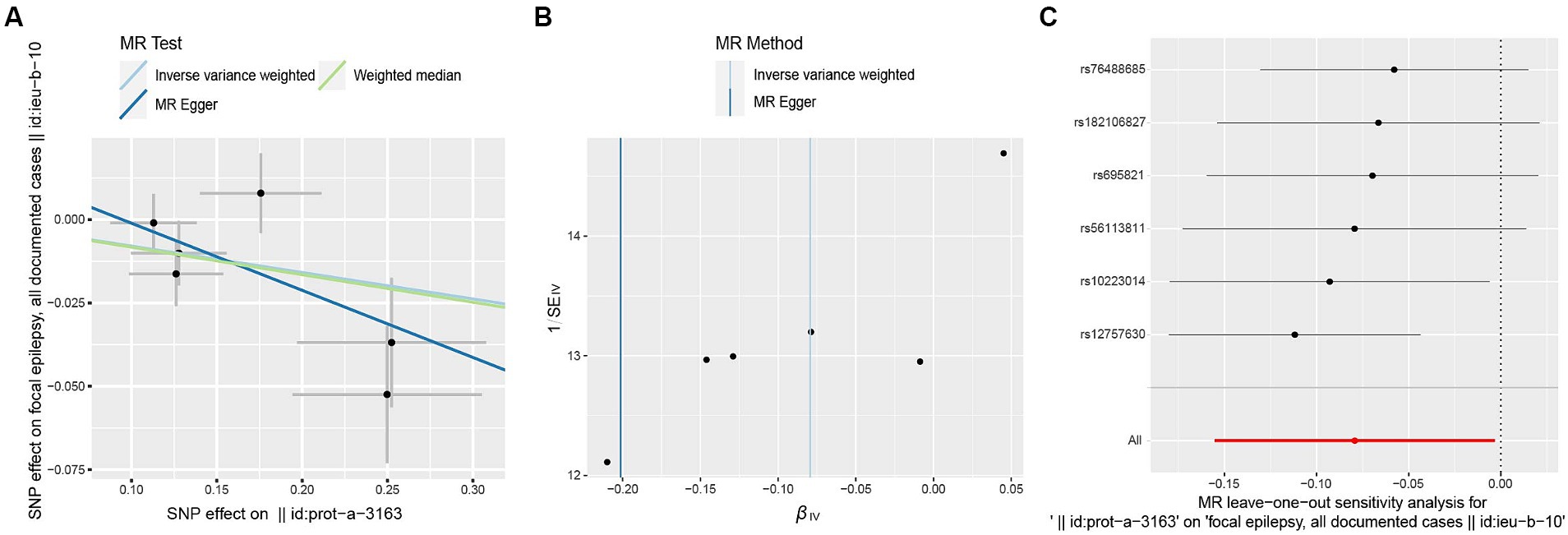
Figure 3. Scatter plot (A), funnel plot (B), leave-one-out analysis (C) of the suggestive causal effect of ULK3 on focal epilepsy.
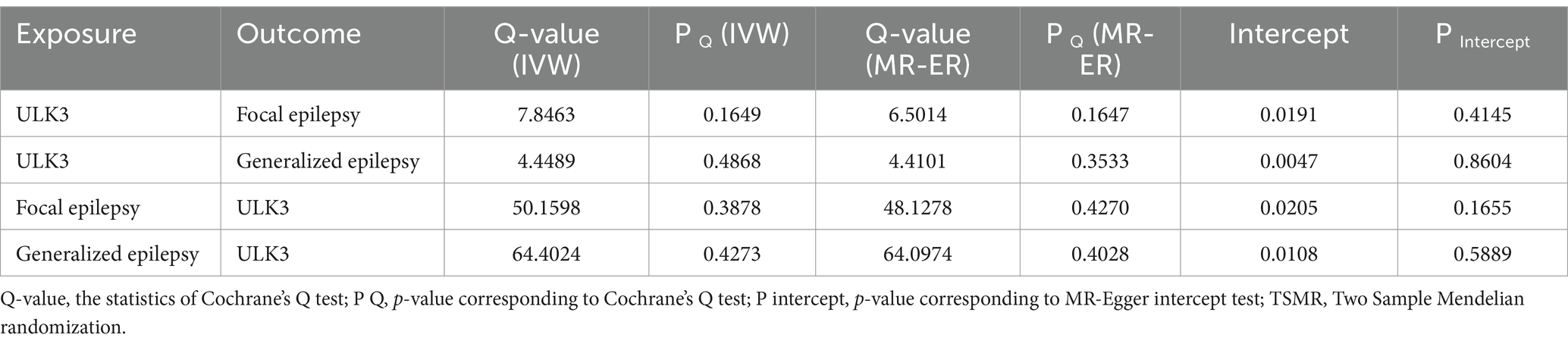
Table 1. Heterogeneity and pleiotropy tests for bidirectional TSMR analyses between ULK3 and epilepsy.
3.3 ULK3 and generalized epilepsy
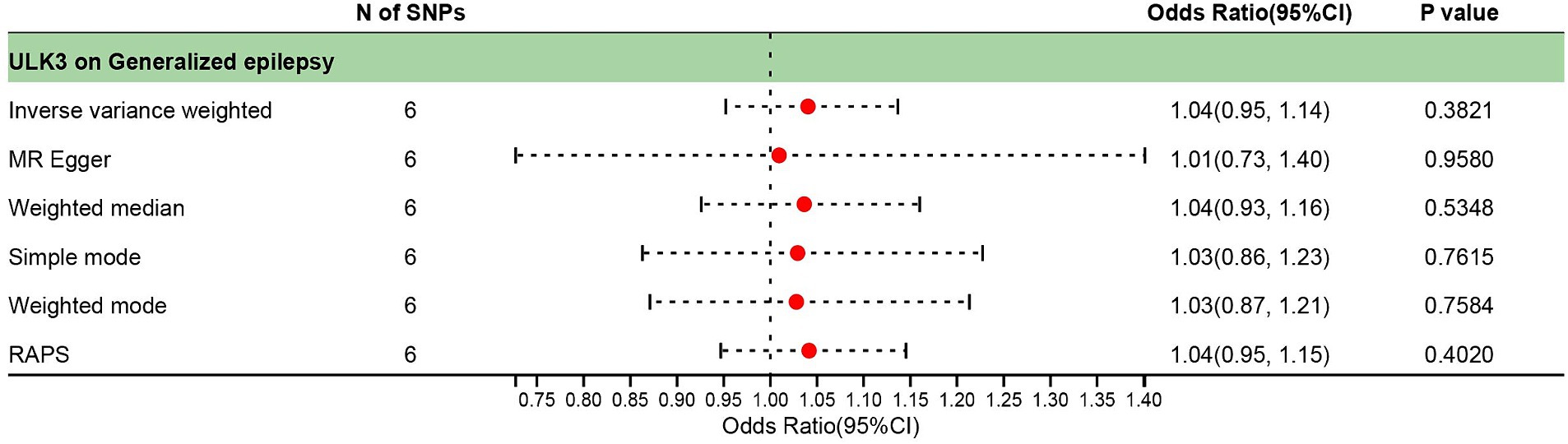
The F-statistic of individual SNPs ranged from 19.98 to 28.98 (Supplementary Table 2). Using IVW, MR-Egger, weighted median, and weighted model methods, no significant causal relationship was found between ULK3 and the risk of generalized epilepsy (OR = 1.04, 95% CI: 0.727–0.952, p = 0.382; Figure 4). No evidence of pleiotropy (intercept = 0.005, p = 0.860) or heterogeneity (p = 0.487) was detected (Figure 5; Table 1).
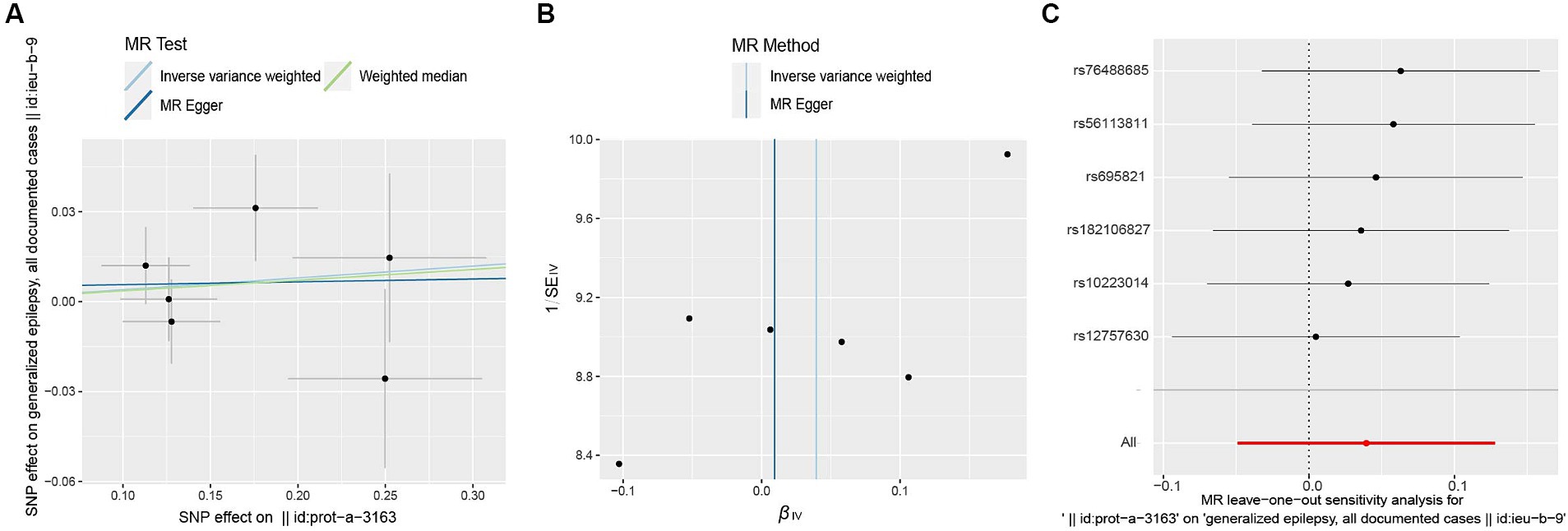
Figure 5. Scatter plot (A), funnel plot (B), leave-one-out analysis (C) of the suggestive causal effect of ULK3 on generalized epilepsy.
3.4 Reverse MR analysis
To investigate whether focal epilepsy and generalized epilepsy had any causal effect on ULK3, a reverse MR analysis was conducted. By identifying SNPs closely associated with epilepsy as genetic tools for exposure, the F-statistic for epilepsy was well above 10 (focal epilepsy: 33.55, generalized epilepsy: 71.76; Supplementary Tables 3, 4), indicating no evidence of weak instrument bias. Focal epilepsy (OR = 1.15, 95% CI: 0.471–0.963, p = 0.121) and generalized epilepsy (OR = 1.03, 95% CI: 0.510–0.933, p = 0.548) showed no significant causal effect on ULK3 (Figures 6, 7; Table 1).
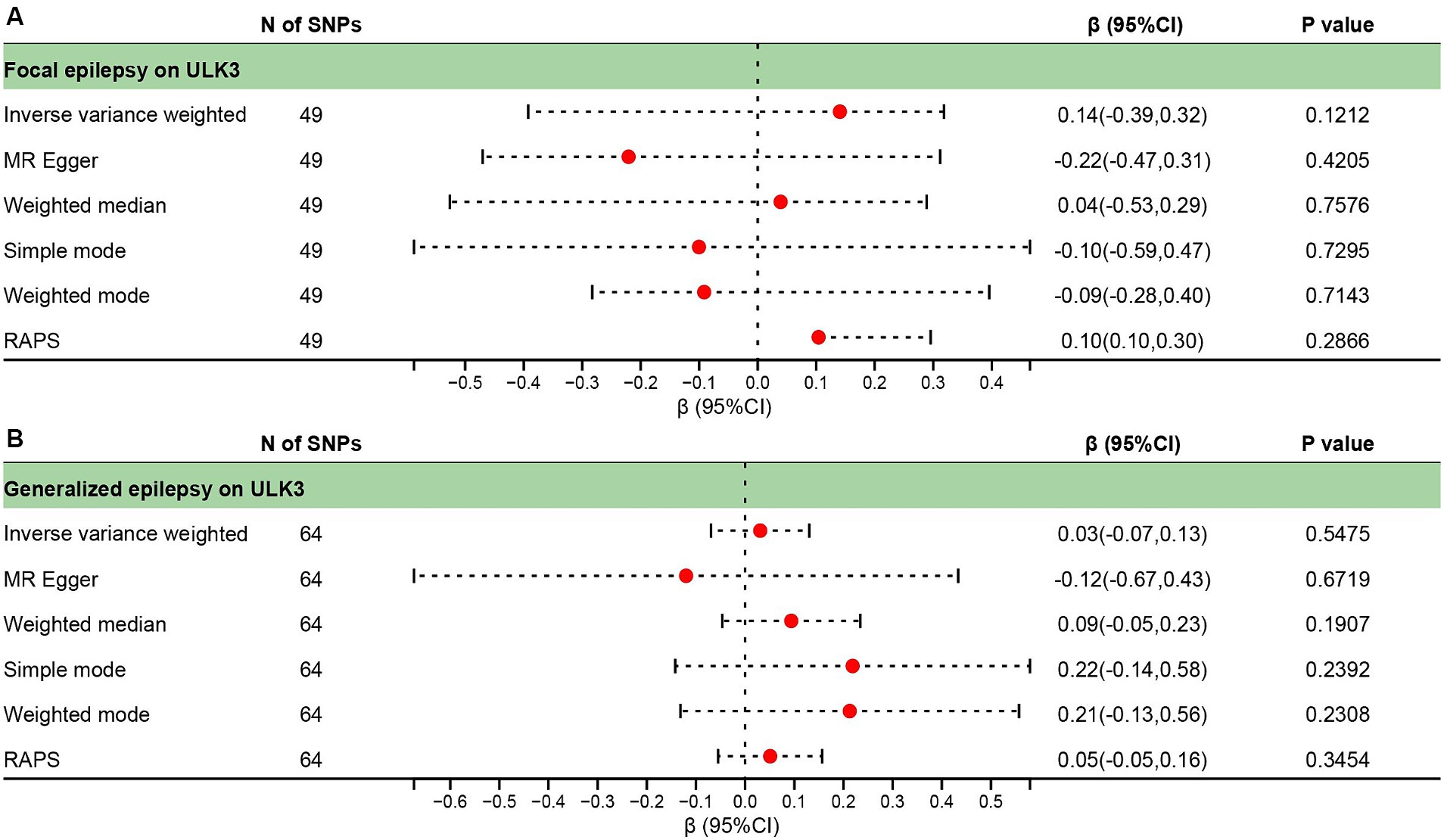
Figure 6. Forest plot of reverse causal effect between ULK3 and focal epilepsy (A), ULK3 and generalized epilepsy (B).
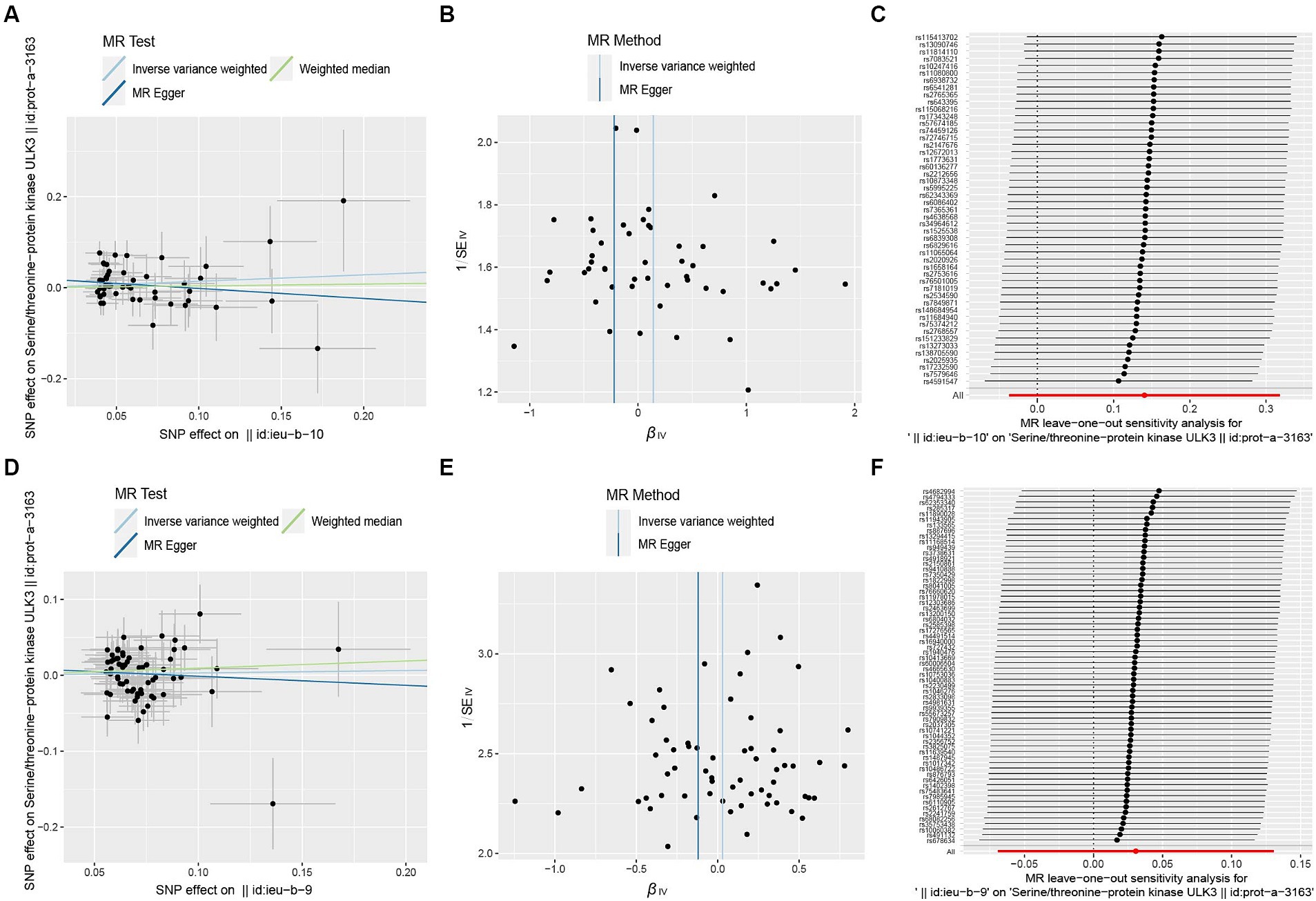
Figure 7. Scatter plot (A,D), funnel plot (B,E), leave-one-out analysis (C,F) of the suggestive causal effect of ULK3 on focal and generalized epilepsy, respectively.
4 Discussion
In this study, we leveraged subtype classification data from a substantial sample size within the 2018 International Union Against Epilepsy (ILAE) to enhance the reliability of our subtype-specific data. Using bidirectional MR analysis, we explored the association between epilepsy and ULK3. In the forward MR analysis, we observed that a higher serum level of ULK3 was associated with a protective effect against focal epilepsy. However, we found no causal relationship between ULK3 and generalized epilepsy.
Recently, the role of ULK3 in epilepsy development remains unclear. There is no significant relationship has not been reported previously. ULK3 positively regulates SHH signaling as a pathway regulator (32). The SHH signaling pathway is crucial for axon formation, proliferation, survival, and differentiation (33). SHH can inhibit the activity of glutamate transporters in neurons (6), potentially impacting the development of epileptic-like activity. Recent studies (5) have demonstrated that ULK3 is involved in autophagy as a positive regulator of autophagy. The elevation of autophagy-related proteins is associated with neuronal plasticity and epileptic behavior (34).
Epilepsy represents a multifaceted neurological disorder characterized by the involvement of multiple genes, proteins, and signaling pathways in its pathogenesis. Among these contributing factors, ULK3 is identified as potentially influential, particularly within specific types of epilepsy. It is important to recognize that distinct types of epilepsy may engage varying genetic, molecular, and cellular mechanisms. Prior studies have postulated that the differentiation between generalized epilepsy and focal epilepsy manifests as variances in the brain structure of affected individuals. In cases of generalized epilepsy, the epileptogenic network implicates bilateral thalamic cortical structures and exhibits widespread distribution throughout the brain (35), often linked to genetic factors. Conversely, focal epilepsy entails neural circuits within the cerebral hemispheres, typically the neocortex or limbic cortex, which could be incited by local injury, inflammation, tumors, or other brain abnormalities (36). Notably, focal seizures commonly manifest as a primary symptom in patients with glioma (37). Consequently, epilepsy emerges as a diverse condition influenced by a spectrum of genetic, environmental, and physiological factors. While elevated levels of ULK3 may correlate with reduced susceptibility to partial epilepsy in certain individuals, it is essential to acknowledge that other factors, such as additional genetic components and environmental influences, may exert a more substantial influence on the risk of generalized epilepsy, thus resulting the causative association of ULK3 inconclusive.
However, our study has several limitations. The study population consisted mainly of individuals of European ancestry, which may limit the generalizability of our findings to other populations. Additionally, the subtypes we studied, generalized epilepsy and focal epilepsy, had relatively small case numbers, necessitating analysis of larger sample sizes in future research to increase result confidence. In addition, the GWAS data in this study are based on results from a cross-sectional study, and longitudinal analysis of ULK3 and epilepsy is required to confirm our hypothesis. Besides, this study includes circulating levels of ULK3 protein, not protein levels in the brain or cerebrospinal fluid, which may be indirect for the assessment of epilepsy risk. Unmeasured and residual confounding factors that have not been accounted for may introduce bias into the overall outcome estimate.
5 Conclusion
This study marks the first exploration of a causal relationship between ULK3 and focal epilepsy. It contributes to our understanding of the pathological basis of epilepsy. Our findings provide valuable insights and impetus for further research.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the patients/participants or patients/participants' legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
BL: Writing – review & editing, Supervision, Formal analysis, Conceptualization. KF: Writing – original draft. XZ: Writing – review & editing, Data curation. YZ: Writing – review & editing, Data curation. SB: Writing – review & editing, Data curation. ZL: Writing – review & editing, Data curation. SX: Writing – review & editing, Data curation. ZS: Writing – review & editing, Data curation. HC: Writing – review & editing, Methodology. HZ: Writing – review & editing, Methodology. SZ: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Shanxi Province Key Research and Development Project (project no. 201803D31150).
Acknowledgments
The authors would like to thank the International Alliance against Epilepsy (ILAE) Complex Epilepsy Alliance for providing the GWAS summary statistics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2024.1376314/full#supplementary-material
Footnotes
References
1. Thijs, RD, Surges, R, O'Brien, TJ, and Sander, JW. Epilepsy in adults. Lancet. (2019) 393:689–701. doi: 10.1016/S0140-6736(18)32596-0
2. Berg, AT, Berkovic, SF, Brodie, MJ, Buchhalter, J, Cross, JH, van Emde, BW, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE commission on classification and terminology, 2005-2009. Epilepsia. (2010) 51:676–85. doi: 10.1111/j.1528-1167.2010.02522.x
3. Singh, G, and Sander, JW. The global burden of epilepsy report: implications for low-and middle-income countries. Epilepsy Behav. (2020) 105:106949. doi: 10.1016/j.yebeh.2020.106949
4. Piirsoo, A, Kasak, L, Kauts, ML, Loog, M, Tints, K, Uusen, P, et al. Protein kinase inhibitor SU6668 attenuates positive regulation of Gli proteins in cancer and multipotent progenitor cells. Biochim Biophys Acta. (2014) 1843:703–14. doi: 10.1016/j.bbamcr.2014.01.003
5. Maloverjan, A, Piirsoo, M, Michelson, P, Kogerman, P, and Osterlund, T. Identification of a novel serine/threonine kinase ULK3 as a positive regulator of hedgehog pathway. Exp Cell Res. (2010) 316:627–37. doi: 10.1016/j.yexcr.2009.10.018
6. Feng, S, Ma, S, Jia, C, Su, Y, Yang, S, Zhou, K, et al. Sonic hedgehog is a regulator of extracellular glutamate levels and epilepsy. EMBO Rep. (2016) 17:682–94. doi: 10.15252/embr.201541569
7. Yang, Y, and Klionsky, DJ. Autophagy and disease: unanswered questions. Cell Death Differ. (2020) 27:858–71. doi: 10.1038/s41418-019-0480-9
8. Bar-Yosef, T, Damri, O, and Agam, G. Dual role of autophagy in diseases of the central nervous system. Front Cell Neurosci. (2019) 13:196. doi: 10.3389/fncel.2019.00196
9. Bowden, J, and Holmes, MV. Meta-analysis and Mendelian randomization: a review. Res Synth Methods. (2019) 10:486–96. doi: 10.1002/jrsm.1346
10. Larsson, SC, Butterworth, AS, and Burgess, S. Mendelian randomization for cardiovascular diseases: principles and applications. Eur Heart J. (2023) 44:4913–24. doi: 10.1093/eurheartj/ehad736
11. Sekula, P, Del Greco, MF, Pattaro, C, and Köttgen, A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. (2016) 27:3253–65. doi: 10.1681/ASN.2016010098
12. Davey Smith, G, and Hemani, G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. (2014) 23:R89–98. doi: 10.1093/hmg/ddu328
13. Abou-Khalil, B, Auce, P, Avbersek, A, Bahlo, M, Balding, DJ, Bast, T, et al. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. (2018) 9:5269. doi: 10.1038/s41467-018-07524-z
14. Sun, BB, Maranville, JC, Peters, JE, Stacey, D, Staley, JR, Blackshaw, J, et al. Genomic atlas of the human plasma proteome. Nature. (2018) 558:73–9. doi: 10.1038/s41586-018-0175-2
15. Larsson, SC . Mendelian randomization as a tool for causal inference in human nutrition and metabolism. Curr Opin Lipidol. (2021) 32:1–8. doi: 10.1097/MOL.0000000000000721
16. Machiela, MJ, and Chanock, SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. (2015) 31:3555–7. doi: 10.1093/bioinformatics/btv402
17. Kamat, MA, Blackshaw, JA, Young, R, Surendran, P, Burgess, S, Danesh, J, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. (2019) 35:4851–3. doi: 10.1093/bioinformatics/btz469
18. McGowan, ME . Final adult height of patients with epilepsy. Dev Med Child Neurol. (1983) 25:591–4. doi: 10.1111/j.1469-8749.1983.tb13816.x
19. Zhou, K, Yang, H, Chen, R, Wang, W, and Qu, Z. Causal relationship among obesity and body fat distribution and epilepsy subtypes. Front Neurol. (2022) 13:984824. doi: 10.3389/fneur.2022.984824
20. Larsson, SC, and Burgess, S. Appraising the causal role of smoking in multiple diseases: a systematic review and meta-analysis of Mendelian randomization studies. EBioMedicine. (2022) 82:104154. doi: 10.1016/j.ebiom.2022.104154
21. Samokhvalov, AV, Irving, H, Mohapatra, S, and Rehm, J. Alcohol consumption, unprovoked seizures, and epilepsy: a systematic review and meta-analysis. Epilepsia. (2010) 51:1177–84. doi: 10.1111/j.1528-1167.2009.02426.x
22. Burgess, S, Small, DS, and Thompson, SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. (2017) 26:2333–55. doi: 10.1177/0962280215597579
23. Patsalos, PN, Stephenson, TJ, Krishna, S, Elyas, AA, Lascelles, PT, and Wiles, CM. Side-effects induced by carbamazepine-10,11-epoxide. Lancet. (1985) 326:1432. doi: 10.1016/s0140-6736(85)92602-9
24. Jia, Y, Guo, D, Zhang, K, Yang, P, Zang, Y, Sun, L, et al. Causal associations of serum matrix metalloproteinase-8 level with ischaemic stroke and ischaemic stroke subtypes: a Mendelian randomization study. Eur J Neurol. (2021) 28:2543–51. doi: 10.1111/ene.14878
25. Wang, YZ, and Shen, HB. Challenges and factors that influencing causal inference and interpretation, based on Mendelian randomization studies. Zhonghua Liu Xing Bing Xue Za Zhi. (2020) 41:1231–6. doi: 10.3760/cma.j.cn112338-20200521-00749
26. Bowden, J, Davey Smith, G, Haycock, PC, and Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
27. Ellingjord-Dale, M, Papadimitriou, N, Katsoulis, M, Yee, C, Dimou, N, Gill, D, et al. Coffee consumption and risk of breast cancer: a Mendelian randomization study. PLoS One. (2021) 16:e0236904. doi: 10.1371/journal.pone.0236904
28. Zhao, Q, Chen, Y, Wang, J, and Small, DS. Powerful three-sample genome-wide design and robust statistical inference in summary-data Mendelian randomization. Int J Epidemiol. (2019) 48:1478–92. doi: 10.1093/ije/dyz142
29. Burgess, S, Zuber, V, Gkatzionis, A, and Foley, CN. Modal-based estimation via heterogeneity-penalized weighting: model averaging for consistent and efficient estimation in Mendelian randomization when a plurality of candidate instruments are valid. Int J Epidemiol. (2018) 47:1242–54. doi: 10.1093/ije/dyy080
30. Hemani, G, Bowden, J, and Davey, SG. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. (2018) 27:R195–208. doi: 10.1093/hmg/ddy163
31. Hemani, G, Zheng, J, Elsworth, B, Wade, KH, Haberland, V, Baird, D, et al. The MR-base platform supports systematic causal inference across the human phenome. eLife. (2018) 7:e34408. doi: 10.7554/eLife.34408
32. Goruppi, S, Clocchiatti, A, Bottoni, G, Di Cicco, E, Ma, M, Tassone, B, et al. The ULK3 kinase is a determinant of keratinocyte self-renewal and tumorigenesis targeting the arginine methylome. Nat Commun. (2023) 14:887. doi: 10.1038/s41467-023-36410-6
33. Hooper, JE, and Scott, MP. Communicating with hedgehogs. Nat Rev Mol Cell Biol. (2005) 6:306–17. doi: 10.1038/nrm1622
34. Wu, Q, Zhang, M, Liu, X, Zhang, J, and Wang, H. CB2R orchestrates neuronal autophagy through regulation of the mTOR signaling pathway in the hippocampus of developing rats with status epilepticus. Int J Mol Med. (2020) 45:475–84. doi: 10.3892/ijmm.2019.4439
35. Sharma, AK, Reams, RY, Jordan, WH, Miller, MA, Thacker, HL, and Snyder, PW. Mesial temporal lobe epilepsy: pathogenesis, induced rodent models and lesions. Toxicol Pathol. (2007) 35:984–99. doi: 10.1080/01926230701748305
36. Zhu, H, Wang, W, and Li, Y. Molecular mechanism and regulation of autophagy and its potential role in epilepsy. Cells. (2022) 11:2621. doi: 10.3390/cells11172621
Keywords: ULK3, Mendelian randomization, epilepsy, causality, genetics
Citation: Liu B, Fan K, Zheng X, Zhang Y, Bai S, Liu Z, Xu S, Su Z, Cao H, Zhang H and Zhang S (2024) Genetic associations between ULK3 and epilepsy: a two-sample Mendelian randomization study. Front. Neurol. 15:1376314. doi: 10.3389/fneur.2024.1376314
Edited by:
Félix Javier Jiménez-Jiménez, Hospital Universitario del Sureste, SpainReviewed by:
Meizhen Sun, First Hospital of Shanxi Medical University, ChinaYoujie Zeng, Central South University, China
Copyright © 2024 Liu, Fan, Zheng, Zhang, Bai, Liu, Xu, Su, Cao, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baolai Liu, bGl1YmFvbGFpNkAxNjMuY29t
†These authors have contributed equally to this work
 Baolai Liu
Baolai Liu Keyi Fan2†
Keyi Fan2† Shengxiao Zhang
Shengxiao Zhang