- 1Sechenov Institute of Evolutionary Physiology and Biochemistry of the Russian Academy of Sciences, Saint Petersburg, Russia
- 2Department of Anatomy and Neurobiology, University of Maryland School of Medicine, Baltimore, MD, United States
- 3Institute of Translational Biomedicine, Saint Petersburg State University, Saint Petersburg, Russia
- 4School of Medicine, Universidad Central del Caribe, Bayamón, PR, United States
- 5Nevsky Center of Scientific Collaboration, Saint Petersburg, Russia
- 6Böblingen Dental, Böblingen, Germany
A product of the immediate early gene Arc (Activity-regulated cytoskeleton-associated protein or Arc protein) of retroviral ancestry resides in the genome of all tetrapods for millions of years and is expressed endogenously in neurons. It is a well-known protein, very important for synaptic plasticity and memory consolidation. Activity-dependent Arc expression concentrated in glutamatergic synapses affects the long-time synaptic strength of those excitatory synapses. Because it modulates excitatory-inhibitory balance in a neuronal network, the Arc gene itself was found to be related to the pathogenesis of epilepsy. General Arc knockout rodent models develop a susceptibility to epileptic seizures. Because of activity dependence, synaptic Arc protein synthesis also is affected by seizures. Interestingly, it was found that Arc protein in synapses of active neurons self-assemble in capsids of retrovirus-like particles, which can transfer genetic information between neurons, at least across neuronal synaptic boutons. Released Arc particles can be accumulated in astrocytes after seizures. It is still not known how capsid assembling and transmission timescale is affected by seizures. This scientific field is relatively novel and is experiencing swift transformation as it grapples with difficult concepts in light of evolving experimental findings. We summarize the emergent literature on the subject and also discuss the specific rodent models for studying Arc effects in epilepsy. We summarized both to clarify the possible role of Arc-related pseudo-viral particles in epileptic disorders, which may be helpful to researchers interested in this growing area of investigation.
1. Introduction
It is known that a significant part (about 8%) of the cell genome in mammals is represented by endogenous retroviruses (ERVs), thus inheriting the ancient germ-line cell infections by retroviruses and the transmission of their genome to the descendants (1–3). While most ERVs are not replication-competent due to broken or tightly controlled viral genes, weakened control can activate ancient retroviral genes and their coded proteins in cancers and senescent cells (4, 5). Besides that, some also retain activity and are important biochemical players in everyday life. For example, ancient viral envelope proteins in the outer cellular layer of the placenta help fusion of the trophoblast cells and became a crucial element in allowing normal pregnancy (6, 7) others determine the fusion of muscle cells and muscle sexual dimorphism (8, 9). Unfortunately, many ERV-related proteins are also implicated in different neurological diseases (10, 11). However, there is not much information regarding their possible role in the pathogenesis of epilepsy.
Actively regulated cytoskeletal-associated (ARC) protein, a protein encoded by the ARC- gene, was characterized in the end of the last century (12, 13). ARC–protein is localized at the synaptic contacts of neurons, and its synthesis depends on the NMDA receptor. This protein plays a critical role in the molecular processes associated with learning and memory and may also serve as a marker of plastic changes in the brain (14, 15).
It is known that the epileptic activity of neurons depends on excitatory-inhibitory synaptic balance (16, 17). Some proteins are well-known regulators of excitatory gain: Pyk2 and Src kinase proteins are the part of NMDA receptor complex (18). PYk2/Src are direct effectors, triggered in active neurons, directly modulating hippocampal excitatory synapses (19). On the other hand, it was determined also that synaptic modulation depends on Arc protein (Activity-regulated cytoskeleton-associated protein, product of endogenous retroviral Arc gene) with recognized activity-dependent expression in neuronal synapses and its expression level is regulated by N-methyl-D-aspartate (NMDA) receptor activation (12). Recently, it was found that Arc protein forms full retrovirus-like capsid in synapses, and may transfer short RNAs from one cell to a post synaptic cell in this way later affecting synaptic morphology (20–22). Interestingly, Arc protein expression changes drastically during epileptic seizures (21, 23).
The study of Arc protein and its synaptic effects is a novel rapidly changing field, but this knowledge is of utmost importance to the understanding of epilepsy because it determines the balance of excitatory and inhibitory synaptic inputs to seizure-prone neurons and the stability of a neuronal network at whole. Here we summarize the emergent literature on this last subject, representing in bullet points Arc's known effects and properties, and then briefly reviewing Arc involvement in epilepsy and relevant experiments in rodent models.
1.1. Arc known properties
Mammalian Activity-Regulated Cytoskeleton-Associated protein (Arc, and its variants, products of immediate early Arc gene) is known for its exquisite importance for synapse maturation, synaptic plasticity, learning, and memory, while the dysregulation of Arc expression can have vast consequences for normal brain function, triggering aberrant wiring of neuronal circuits (24, 25). Arc/Arg3.1 mediates activity-dependent elimination of redundant climbing fiber to Purkinje cell synapses in the developing cerebellum (26). In glutamate neurons, following activation of the NMDA receptor, Arc mRNA became significantly upregulated in the nucleus before being transported to the dendrites for translation (12, 27). At postsynaptic sites Arc mRNA localizes in the PSD-95-NMDAR complexes and binds to inactive CaMKIIβ (not bound to calmodulin) (28, 29). Activation of calcium entry via ionotropic glutamate receptors, especially the NMDA subtype, during normal synaptic transmission and in seizures results in calmodulin-dependent dissociation of Arc mRNA from CaMKIIβ and PKA-dependent induction of Arc protein expression (29, 30). Later it happens that spines on dendrites, where Arc was produced, change their morphology increasing the spine density and proportion of thin spines, together with the endocytosis of AMPA receptors leading to decreased synaptic efficacy (21). Arc hyperexpression facilitates not only AMPA endocytosis but also downregulates transcription of the GluA1 subunit of the AMPA receptor which favors synaptic downscaling (31, 32). In this way, Arc participates in synaptic long-term potentiation and the consolidation of long-term memory (33). Interestingly, Arc transcription exhibited distinct temporal kinetics depending on the activation of excitatory inputs that convey functionally distinct information (34).
1.2. The mechanism of Arc protein function
Upon hyperexpression, in local dendritic compartments, Arc protein assembles into retrovirus-like capsids packing predominantly Arc mRNA. These capsids leave neurons wrapped in extracellular vesicles and can transmit mRNA to nearby cells (20, 22). Curiously, activation of metabotropic glutamate receptors (mGluR) facilitates Arc mRNA translation in capsid “infected” neurons (20, 22). Arc capsid-mediated transfection was not yet observed in the mammalian brain, but in mice, Arc expression in DRG neurons results in capsid formation modulating skin vasodilation (35). Arc mRNA transfer between cells can probably explain why after hyperactivity Arc accumulates not only in neurons but also in astrocytes, while it is originated from nearby neurons (27, 36, 37). Mammalian Arc protein lacks zinc fingers but has positively charged motifs binding polyanionic mRNA (38–40). This probably allows nonspecific packing of neuronal host mRNAs other than Arc mRNAs, because half of the RNAs encapsidated by retroviruses are host-derived RNAs (41). Extracellular vesicles are supposed to participate in the spread of different neurodegenerative pathologies over time in an activity-dependent manner via synaptic connections (42, 43). Generally, it looks probable that Arc capsids may mediate activity-dependent intercellular paracrine transfer of genetic information, which may alter neighboring cell response to network activity.
1.3. Arc protein forms virus-like particles and their history
Recently it became clear that Arc protein is not only some important regulatory synaptic protein but turns out to be repurposed retrotransposon protein that mediates intercellular mRNA transfer (22, 44, 45). This mechanism involves formation (mainly by glutamatergic neurons) and expression in their synapses of pseudo-viral, retrovirus-like particles (of about 60 nm diameter) made by Arc protein multimers, which encapsulates specific neuronal mRNA and then are trafficked across synaptic boutons (20, 46). The median part of the Arc protein sequence has similarity to modern retroviruses (for example HIV) Group-specific antigen (Gag). Like HIV Gag, Arc forms capsomeres (Figure 1) which self-assemble into capsids of about 30 nM in diameter, while the multimerization of Arc is mediated by its N-terminal helical coil motif (48–50). Arc protein ensembles form multiple capsomeres with symmetric pentameric structures (Figure 1), resembling some viral ion channels (48). Arc variants are found in both birds and humans, but not fish. It is hypothesized that Arc was inserted into the ancestral genome of all tetrapods (amphibians, reptiles, birds, mammals) around 350–400 million years ago. In humans, Arc is found in the greatest amount in brain structures associated with memory, which may be associated with synaptic plasticity and consolidation of memories (44, 51).
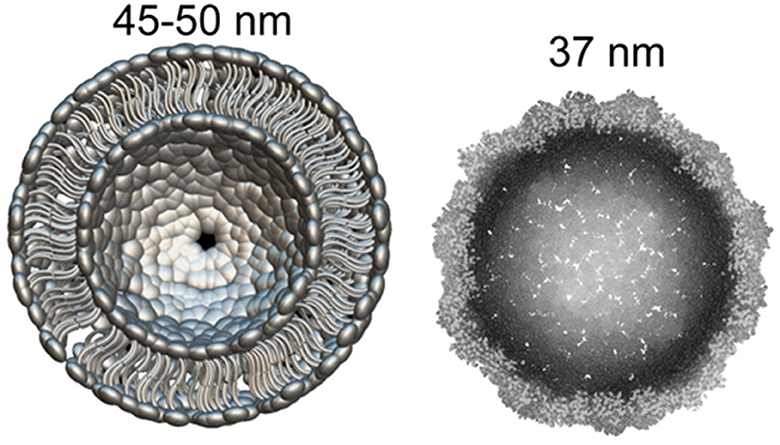
Figure 1. What can be found in synapse: the diameter of glutamatergic synaptic vesicle (Left) is approximately 45–50 nM [model drawn according to cryo-EM data by Du et al. (47)]. dArc1 capsid (Right)—a viral-like particle made by polymerization of 240 Arc-protein formed capsomeres (47, 48).
Of course, virus-like transport of mRNA can be employed not only in long-term memory, and Arc can be presented better as a multifunctional signaling hub (52). For example, it was shown that Arc can transport mRNAs from mutated genes related to schizophrenia (53). Arc may also play a role in the immunity and activity-dependent β-amyloid generation (30, 54).
1.4. Evolution of Arc proteins
Evolutionary analysis shows that Arc is derived from a lineage of Ty3/gypsy retrotransposons, which are also ancestors to retroviruses, that have been repurposed during the evolution to mediate intercellular communication in the nervous system (20, 22). Thus, animals before tetrapods most probably use Ty3/gypsy variants for memory consolidation. On the other hand, the tetrapod Arc protein structure is similar to the one which has been found in Schizophora (true) flies (Dipteros), thus it may have been transferred to a common ancestor of these insects independently (22, 48). At least two Arc homologs (dArc1 and dArc2) are found in flies, which arose by genomic duplication of an ancestral dArc gene but were not detected in any other dipteran (e.g., mosquitoes) or protostome species (20, 22). In Drosophila Arc1 protein forms capsid-like structures that bind mRNA in neurons and are loaded into extracellular vesicles. These vesicles pass from motor neurons to muscles. The disruption of the transfer, in turn, blocks synaptic plasticity (20). Thus, the transsynaptic mechanism of mRNA transport involving retrovirus-like Arc- capsids and extracellular vesicles can be considered proven (55).
While Arc genes originated independently, they still share significant homology in the retroviral Gag domain, and thus the ability to form capsids. Interestingly, the protein participating in memory consolidation in fish retains some immunoreactivity to Arc (56). This domestication of proteins using transposable elements is a well-known phenomenon known as lateral gene transfer that can enrich the recipient and provide a mechanism for evolutionary flexibility (57, 58). About one-half of the mammalian genome consists of DNA with viral or transposon origin and about 8% belong to ancient retroviruses (2, 59). According to the Gene Expression Omnibus (GEO) database for gene expression profiling, the Arc gene was identified as a candidate gene involved in the pathogenesis of various neurological diseases, including epilepsy and a number of others, such as depression.
2. Possible involvement of Arc in the mechanisms of neuropsychic disorders
2.1. Human brain pathologies associated with ARC-protein
Actively regulated cytoskeletal-associated (ARC) protein, encoded by the ARC gene, was characterized at the end of the last century (12, 13). This protein is localized in the synaptic contacts of neurons, and its synthesis is dependent on the NMDA receptor's activity. This protein plays a critical role in the molecular processes associated with learning and memory and may also serve as a marker of plastic changes in the brain (15).
Impaired Arc protein synthesis is associated with various brain pathologies, including memory disorders, Alzheimer's disease, autism spectrum disorders, schizophrenia, and epilepsy (60–62). Arc cellular pathways have emerged as key regulators of synaptic plasticity, and are becoming known for being central players in genetic risk for many neural disorders (20).
Thus, in a study on a large sample of patients, it was shown that both rare mutations and epigenetic regulation of ARC contribute to the pathogenesis of schizophrenia, at least in some patients (20, 63). It has also been shown that small mutations affecting one or more nucleotides are extremely widespread among glutamatergic postsynaptic proteins, including proteins associated with ARC and NMDAR, regulated by synaptic activity. At the same time, genes affected by mutations in schizophrenia overlap with genes affected by mutations in autism spectrum disorders and some other brain pathologies (64).
Patients with epilepsy have an increased likelihood of experiencing psychotic symptoms, many of which are similar to those of depression. However, many psychiatric comorbidities, not just the symptoms of depression, are more common in patients with epilepsy (65). It remains an open question whether depression is a risk factor for the development of epilepsy.
Interesting results were obtained by measuring the level of Arc/Arg3.1 in the blood plasma of children with a diagnosed autism spectrum disorder (ASDs) (66). The average level of Arc/Arg3.1 protein in blood plasma in autism was significantly higher than in the control (healthy) group. However, no significant association was found between plasma Arc/Arg3.1 protein levels and measures of autism severity (66). This suggests that Arc/Arg3.1 can be used as an early biomarker for diagnosing autism (66).
Autism and Angelman syndrome (AS) share many common characteristics, although Angelman syndrome is not usually included in ASD (67, 68). Angelman Syndrome is a neurological disorder caused by a mutation of the E3 ubiquitin ligase UBE3A, a gene whose mutation is associated with autism spectrum disorders. In childhood, seizures are observed in approximately 80–90% of patients with AS (69). Arc is one of the target proteins of the UBE3A gene. Since Arc is involved in learning and memory, its expression directly affects the manifestation of AS, including the epileptic seizures, typical of this syndrome. But the function of UBE3A during nervous system development and how UBE3A mutations give rise to cognitive impairment in individuals with AS and ASDs remains unclear. Nevertheless, we know that experience-driven neuronal activity induces UBE3A transcription. UBE3A then regulates excitatory synapse development by controlling the degradation of Arc (67). Disruption of UBE3A function in neurons leads to an increase in Arc expression and a concomitant decrease in the number of AMPA receptors at excitatory synapses. This deregulation of AMPA receptor expression at synapses may contribute to the brain pathologies that occur in AS and possibly other ASDs (67). Most researchers agree that the genesis of epileptic seizures in AS has a complex genesis (17). It is known that in patients with AS, there is a global decrease in the volume of a number of subcortical structures and an increase in the volume of gray matter. The degree of the abnormality correlates with the severity of seizures, suggesting that the occurrence of seizures may be directly related to morphological changes in the brain (17).
Fragile X Syndrome (FXS) is one of the most common inherited forms of developmental delay (70). Epilepsy is reported in up to 20% of individuals with fragile X syndrome (71). It was shown that the FMR1 gene, which produces the FMRP protein is responsible for FXS. One of the biological manifestations of FXS is elevated levels of metabotropic glutamate receptor (mGluR)-dependent long-term depression (LTD), (mGluR-LTD) a type of synaptic plasticity which is characterized by a reduction in the synaptic response at the excitatory synapses and overexpression of several proteins including Arc (72, 73). This abnormal overexpression of Arc leads to increased endocytosis of AMPARs and increased mGluR-LTD. In addition, altered dendritic spine morphology was observed not only in animal models of FXS but also in humans with this disorder (70, 74).
Epilepsy does not have a clearly identifiable cause in about half of patients. Also, this disease can be associated with various factors, including various genetic abnormalities. Some types of epilepsy are linked to certain genes, but generally, genetic factors can make a person more sensitive to the mechanisms that cause seizures. In addition, epilepsy has a wide comorbidity with other brain pathologies. Thus, we most likely cannot speak about the direct responsibility of the genetic pathologies of Arc for the formation of one form or another of epilepsy in humans, but Arc can certainly be associated with epilepsy.
2.2. Expression of Arc protein in neurons activated by epileptic seizures
Arc depletion may affect memory loss in the post-ictal state. It is known that memory problems experienced by people with epilepsy are characterized by difficulty in retrieving episodes or events that happened before a seizure and even general semantic information (43). Genetic interference of activity-dependent Arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and blocks the consolidation of long-term memory (33, 75). Can it be associated with the dysregulation of Arc protein expression during a seizure? It can be assumed that new methods of in vivo molecular imaging can help answer this question to some extent (76, 77).
Arc is an activity-dependent immediate early gene, its mRNA is translocated only to dendritic spines of active neurons where it is translated to Arc protein, which then multimerizes in capsids forming viral-like particles implanted with mRNA. Then in the form of viral-like particles, Arc-mRNA is loaded into extracellular vesicles and trafficked to postsynaptic boutons, participating in regulating dendritic spine morphology and the receptor content of glutamatergic synapses controlling synaptic plasticity changes and memory (20, 21, 31, 47, 78, 79). Notably Arc expression is augmented in synapses of recently activated neurons of the epileptic seizure focal zone but rapidly declines, probably due to dysregulation during aberrant neuronal overexcitation (80). For example, temporal lobe epilepsy originates from the mesolimbic network and provokes damage to the hippocampus including the dentate gyrus, which is very vulnerable to status epilepticus (81–83).
It is generally accepted that hippocampal sclerosis provokes the hippocampus to generate seizures resulting from the loss of interneurons and pyramids accompany the formation of recurrent synaptic circuits by dentate gyrus cells (DGC) (84, 85). Arc upregulation in DGC between seizures accompanies increased spine density which enhances excitatory input from the entorhinal cortex, which precedes the formation of recurrent mossy fiber synapses (80, 86). Conversely, hippocampal regions not affected by seizures preserve normal Arc expression (80).
Many authors studied the association between Arc expression and epileptogenesis. It is known that spine loss and other dendritic abnormalities occur in epilepsy which to some extent may be also associated with Arc (87). Of course, the Arc gene affects a whole galaxy of genes, and therefore the phenotypic consequences of its mutations are very diverse (21, 37, 88). Also, some genes and their products are necessary for Arc functioning and transport, and the disruption of these genes lead also to changes in neural net excitation (74).
2.3. Genetic studies of Arc expression during epileptogenesis
As part of the analysis of the role of Arc in epileptogenesis on clinical material, we analyzed the transcriptomic data from the National Center of Biotechnology Information (NCBI) public repository Gene Expression Omnibus (GEO) to evaluate the ARC expression in brain areas that are damaged in epilepsy (89). The RNA sequencing data were searched in the GEO Browser for the terms “epilepsy”, “epileptic”, and “seizure”. Afterward, we searched the GEO database for the terms “kainate”, “electroconvulsive”, and “pentylenetetrazol” to receive the data for the most common models of epilepsy. Each GEO dataset included in the analysis should meet the following criteria: (1) expression data in raw counts, fragments per kilobase per million mapped fragments (FPKM), or transcripts per million (TPM); (2) a clear explanation of sample origin (the datasets which comprise samples described like a “brain sample,” when the brain structure was not specified, were excluded); (3) at least three samples per study group; (4) the expression patterns were studied in brain structures, data for other tissues were excluded.
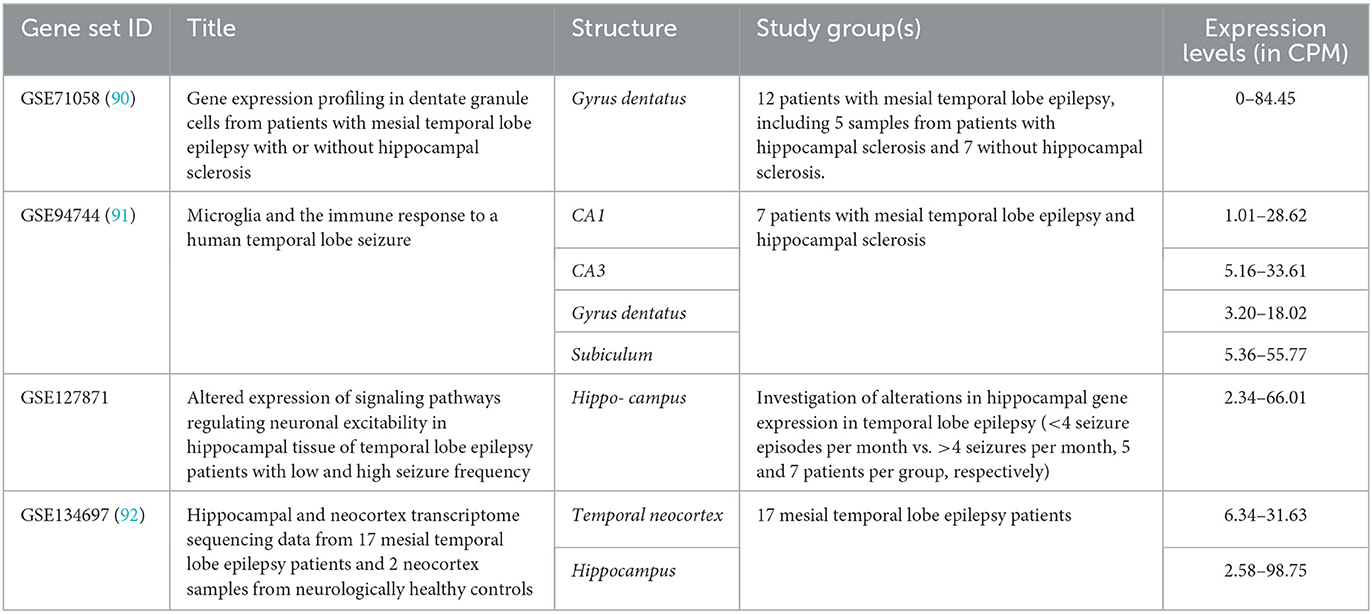
Four relevant datasets for the ARC expression pattern in human brain parts were mined (Table 1). In all datasets, only patients with temporal lobe epilepsy were included, and hippocampal structures were studied, except 17 neocortical temporal lobe samples in GSE134697. Further, no healthy or non-epileptic controls were included in the studies in the appropriate number (n ≥ 3) that did not allow for comparison of damaged tissues with normal tissues from healthy subjects.
No differences in ARC expression levels in mesial temporal lobe epilepsy patients with or without concomitant hippocampal sclerosis were identified in GSE71058 by an edgeR likelihood ratio test (Padj > 0.05) (93). Likewise, there were no significant differences in ARC expression levels between patients with low and high seizure frequencies in GSE127871. All data were count per million (CPM) normalized by edgeR package CPM function and demonstrated the congruent ARC expression levels (93). Applying the Expression Atlas recommendations, the expression levels may be interpreted as low expression (i.e. CPM between 0.5 and 10) or medium (CPM between 10 and 1,000) in all studied structures (94).
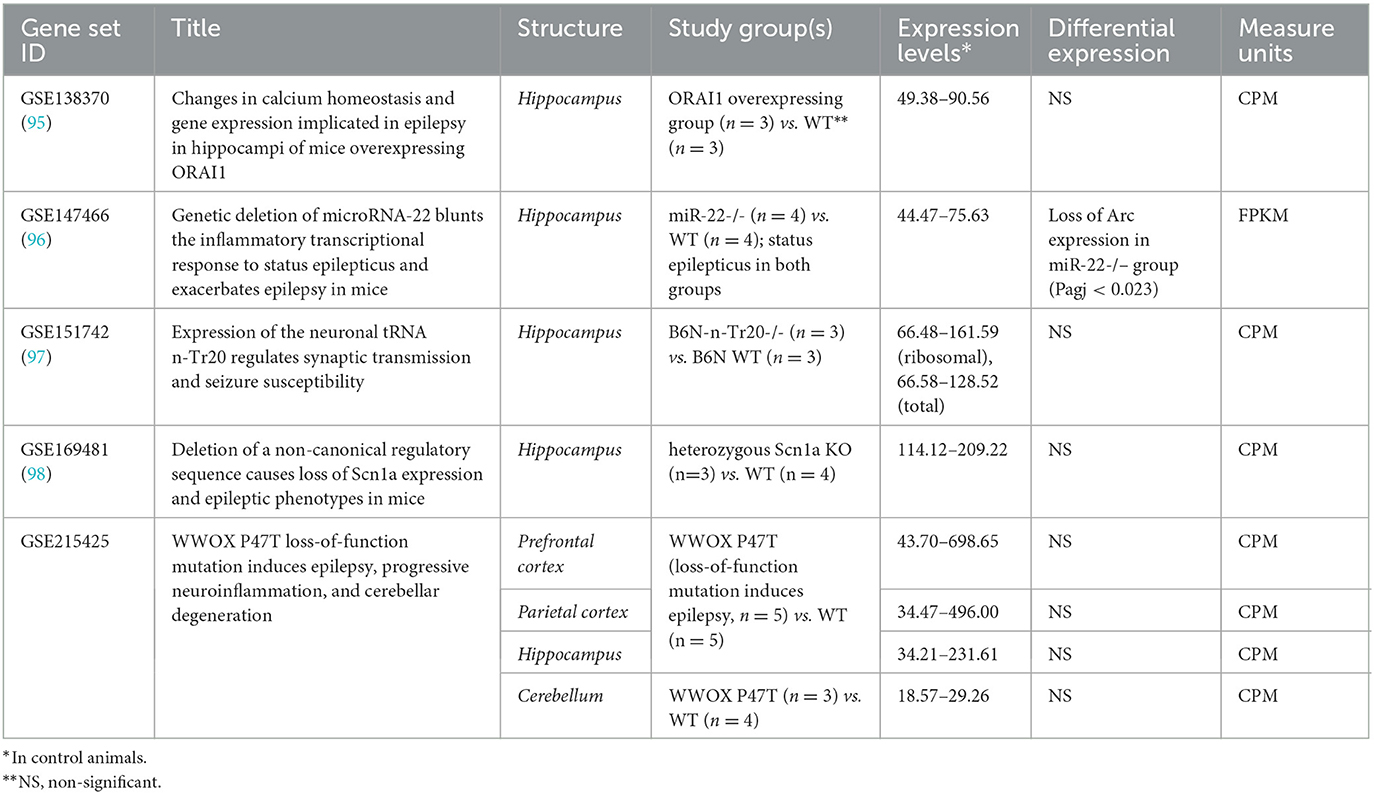
In mice, transcriptome modifications were studied in the hippocampus, neocortical structures, and cerebellum in genetic models of epilepsy (Table 2). No differences in Arc expression levels between control and study groups were identified in any study when likelihood ratio test (for raw counts) or empirical Bayes statistics eBayes function in limma package (for log 2 FPKM-normalized data) were applied (99). The only exception is the dramatic loss of Arc in the hippocampal transcriptome of mice lacking miR-22 during the epileptogenesis after kainate injection compared to the identically treated wild-type animals (Padj < 0.05). miR-22 loss results in an exacerbated epilepsy phenotype in kainate-induced epilepsy and is associated with the reduction of an inflammatory response at the transcriptional level (96).
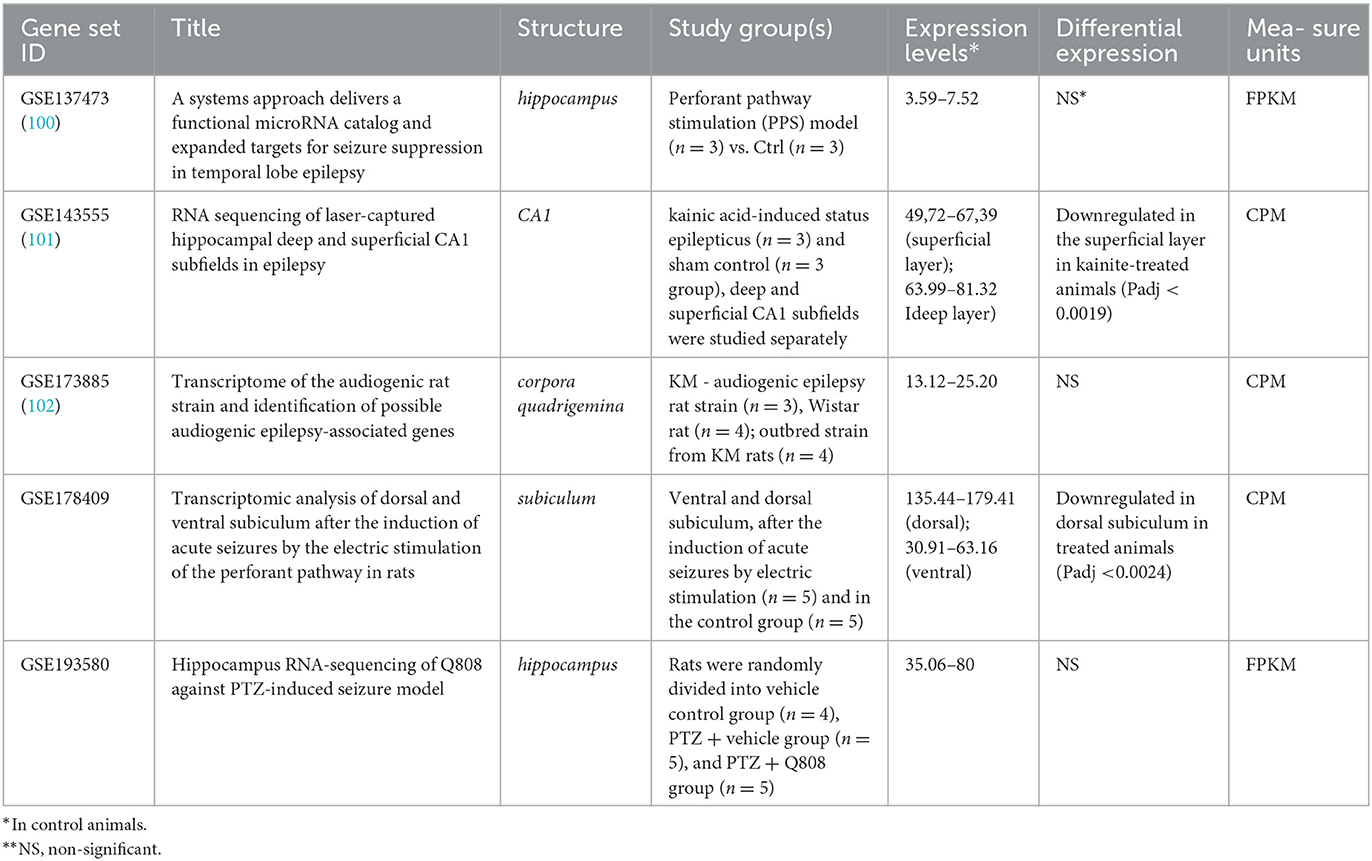
In contrast, the significant downregulation of Arc expression was identified in CA1 superficial layer in response to kainate treatment (Padj < 0.0019) in rats (Table 3). As well, Arc expression was downregulated in the dorsal subiculum (Padj < 0.0024) in response to the induction of acute seizures in rats by electric stimulation. No differences in Arc expression were identified between control and epileptic model rats when the whole hippocampus or corpora quadrigemina were studied. So, the demonstrated discrepancy between results revealed in mouse and rat models may be caused by expression examination in the whole hippocampus instead of the separated parts of this complex structure.
Genetic studies of Arc expression during epileptogenesis in humans and animals are still rare to provide non-contradictory conclusions on Arc's role in seizure-related pathologies. According to available datasets for the Arc expression, only in mice and rats, but not in humans, the downregulation of Arc expression was revealed in response to seizures. Further accumulation of more detailed data on Arc expression in specific regions of the brain is likely to allow more specific conclusions to be drawn in the future.
3. Animal epilepsy model
3.1. Arc gene expression in rodent epilepsy models
Practically all rodent epilepsy seizure induction models affect Arc expression. In rodent models, Arc gene expression can be stimulated by practically any method which induces seizure activity in the brain, at least in the hippocampus and cortex, but Arc expression is time-dependent and the stimulation later turns to depression (80, 103, 104).
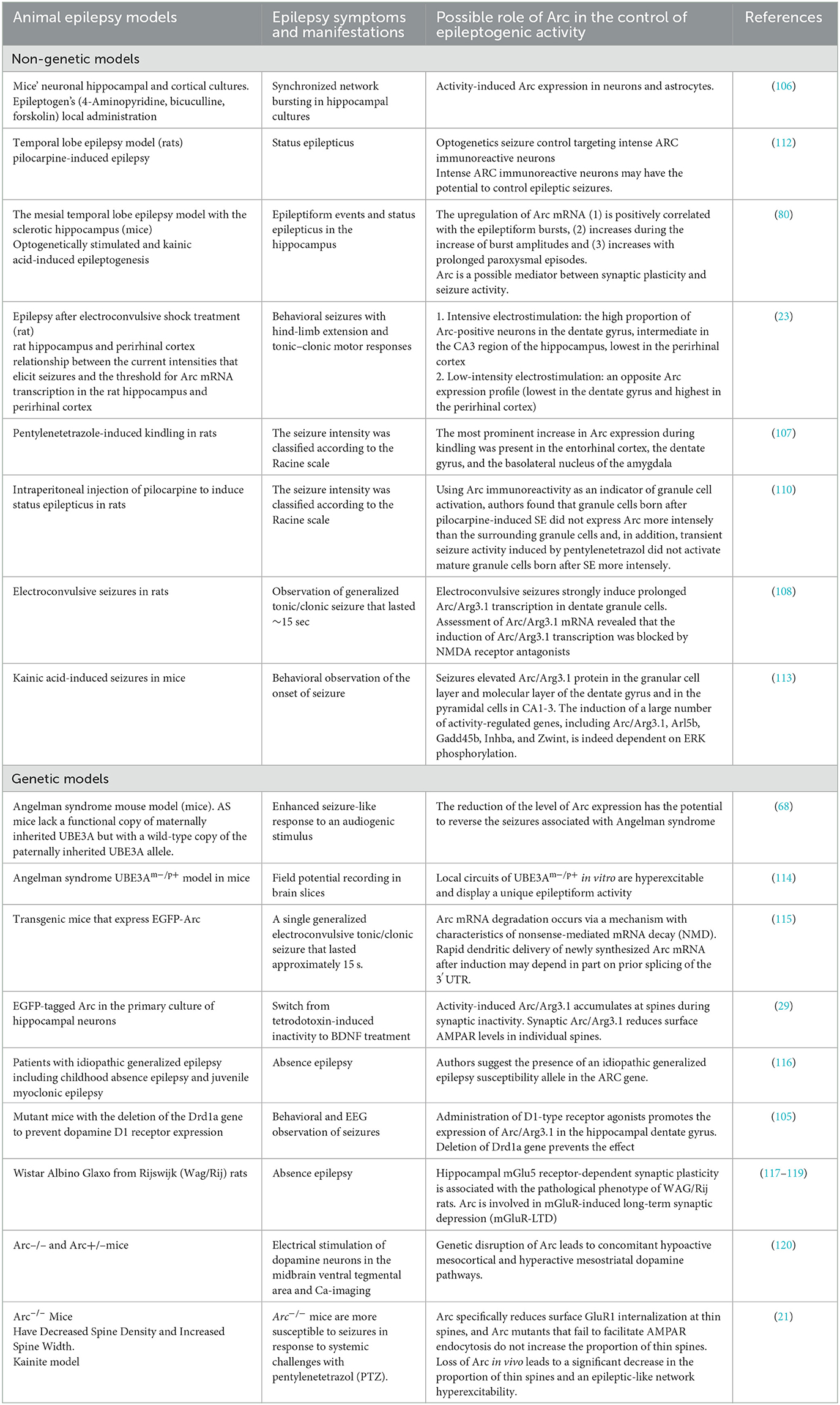
Initial hyperproduction of Arc can be seen in epilepsy models with kainate, D1-receptor agonists, 4-Aminopyridine (4AP), Bicuculline (Bic), and Forskolin, pilocarpine, pentylenetetrazole, kindling, activation of mGluR, electroconvulsive stimulation (Also Arc mRNA and proteins are rapidly induced in the striatum after acute cocaine administration (80, 105–111). Details on epileptic models inducing Arc expression are presented in Table 4.
Arc expression demonstrates complex behavior in seizure generation. The threshold of stimulation intensity (depolarization threshold) required for induction of Arc expression varies between brain regions (23). Particularly in the case of most intensive electroconvulsive stimulation, the proportion of Arc-positive neurons following seizures was highest in the dentate gyrus, intermediate in the CA3 region of the hippocampus, and lowest in the perirhinal cortex (23). In contrast, low-intensity seizure-inducing electrostimulation caused an opposite Arc expression profile (lowest in the dentate gyrus and highest in the perirhinal cortex), which indicates for Arc expression may be serving as a transcriptional threshold mechanism in CNS (23). Accordingly, Arc mRNA upregulation is positively correlated to seizure burst ratio, burst amplitudes, and length of paroxysmal episodes (80).
3.2. Rodent models with Arc knock-out
Peebles et al. studied spine morphology and the general stability of the glutamatergic neuronal network and found both dependent on Arc expression, employing a mouse model (21). They have confirmed that Arc expression leads to an increase in spine density, but generally decreases synaptic efficacy by reducing surface GluR1. Authors have shown that regulated synaptic strength in neuronal networks determined by Arc is very important for network stability. Studying kainite-elicited seizure activity in WT and Arc–/– mice, authors have shown that Arc–/– mice are more susceptible to kainite-elicited seizures and neuronal changes associated with epilepsy. Also, Arc–/– mice had aberrant spontaneous cortical network discharge activity, highly associated with epilepsy (21). On the other side, prenatal or perinatal deletion of Arc/Arg3.1 alters cortical network activity without excessive disruption of the balance of excitation and inhibition in the brain (88). Furthermore, Arc knockouts elevate AMPA receptor level expression in some brain regions including the nucleus accumbens which reduces the symptoms of epilepsy-associated pathologies (109). Pilocarpine-induced temporal lobe epilepsy causes a time-dependent decrease in Arc expression in hippocampal tissue (121).
Epilepsy is prevalent and often medically intractable in Angelman syndrome (AS). There are different models of AS. AS mouse models associated with UBE3A gene-deficient function (UBE3Am-/p+) shows reduced excitatory neurotransmission but a lower seizure threshold. Genetically decreased Arc expression additionally reduces abnormal EEGs and seizures in mice with Angelman syndrome associated with UBE3A gene-deficient function, because both Arc and UBE3A regulate surface expression of AMPA receptors. In another AS model, the so-called fragile X syndrome (FXS) mouse model, on the contrary, increased Arc is responsible for seizure phenotype (68, 114).
Comorbidity of schizophrenia and epilepsy are relatively common in clinical practice and animal models. Two different models of schizophrenia with seizures were developed, conditional KO (late-cKO) mice, in which Arc/Arg3.1 was deleted during late postnatal development, to investigate the causal relationship between Arc/Arg3.1 deletion and schizophrenia-linked neurophysiological and behavioral phenotypes. Nevertheless, in an animal study genetic deletion of Arc/Arg3.1 per se did not cause schizophrenia-like behavior, and a significantly higher dosage of kainic acid was required to elicit epileptic seizures in the KO mice (88). This completely contradicts the results obtained in another model of Arc/Arg3.1 knockout mice by Managò et al. showing genetic mutation disrupting Arc produced a hyperactive phenotype and amphetamine supersensitivity consistent with rodents' correlates of schizophrenia-like symptoms (120). The authors also describe important differences in the dopaminergic system in KO and wild-type mice. Most likely, this inconsistency can be explained by the different strategies used to create the two knockout mouse models.
4. Discussion and conclusion
The literature sources cited unanimously emphasize the paramount significance of Arc protein in regulating synaptic strength [reviewed by Zhang and Bramham (122)], which in turn determines the stability of the neuronal network, thus affecting epileptic seizure susceptibility (Figure 2) (21, 122). While the established mechanism of Arc-mediated influence relies on presynaptic NMDA-receptor mediated ionic currents in the glutamatergic pathway, it is also the primary mechanism that exerts direct cross-synaptic influence over postsynaptic spines morphology and their AMPARs (21). These spines are typically found at the postsynaptic site of most excitatory synapses in the mammalian brain and may be present on excitatory as well as inhibitory neurons (123). This implies that changes in Arc-protein-mediated activity can potentially affect the excitatory-inhibitory balance of the neuronal network, either reducing seizure threshold and promoting stability or inducing lability (88, 114).
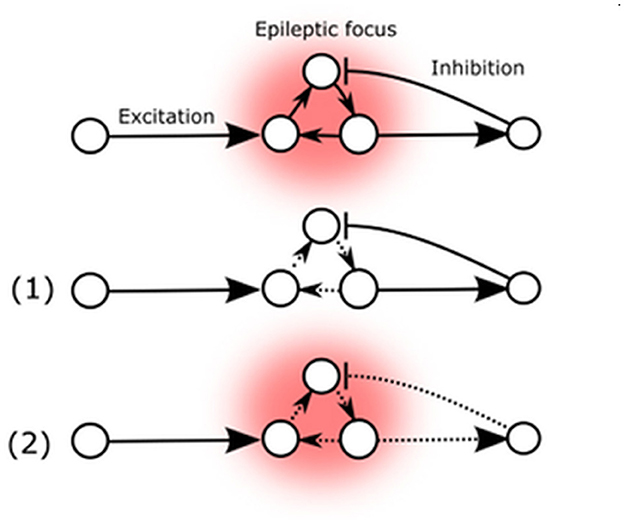
Figure 2. Arc mediated changes of seizure susceptibility. (1) Elevated Arc expression in epileptic focus weakens external AMPA-mediated excitatory influence. (2) Arc expression also decreases excitatory feedback to inhibitory neurons.
These two possibilities are represented by the potential Arc-mediated changes (Figure 2). The primary known effect of the Arc protein is to decrease the strength of excitatory synapses, which could potentially reduce the destabilizing flow of excitatory signals to the existing epileptic focus or decrease auto-excitation by reducing internal excitatory connections (1). Conversely, Arc-mediated changes could also decrease excitatory feedback to inhibitory neurons, thereby reducing their inhibitory influence on the existing epileptic focus (2).
Arc knockouts will remove such effects: the literature reviewed and knockout experiments consistently show this two-sided effect on seizure susceptibility, if some existent epileptiform activity is present. While Arc definitely is activated in neurons participating in epileptic discharges, there is no evidence of the direct induction of epilepsy by Arc-protein mediated mechanisms.
These allow us to conclude that Arc activity-regulated expression directly participates in seizure susceptibility. Also, epileptic activation of Arc-mediated changes during the seizure may affect memory consolidation and many other important neuronal functions in epilepsy.
Author contributions
Conceptualization: DS, MI, AV, ANV, and VT. Writing: MI, DS, AI, AV, ANV, and VT. Review and editing and figures preparation: DS, AI, and MI. Editing: JA, AI, ES, LR, and PS. Supervision and project administration: MI, VT, and AV. Funding acquisition: MI, AV, and ANV. All authors have read and agreed to the published version of the manuscript.
Funding
This work was sponsored by the National Institute of General Medical Science (NIGMS) of the National Institute of Health (NIH) under awards U54GM133807 and SC3GM143983 which provided funding to JA and MI and NIH NIGMS RCMI award number G12MD007583 granted to the UCC. AV and ANV were supported by the project ID: 94030300 of the St. Petersburg State University, St. Petersburg, Russia. DS was supported by IEPHB Research program 075-0152-22-00.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
References
1. Löwer R. The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol. (1999) 7:350–6. doi: 10.1016/S0966-842X(99)01565-6
2. Mustafin RN, Khusnutdinova EK. Involvement of transposable elements in neurogenesis. Vavilovskii Zhurnal Genet Selektsii. (2020) 24:209–18. doi: 10.18699/VJ20.613
3. Smit TK, Bos P, Peenze I, Jiang X, Estes MK, Steele AD. Seroepidemiological study of genogroup I and II calicivirus infections in South and southern Africa. J Med Virol. (1999) 59:227–31. doi: 10.1002/(SICI)1096-9071(199910)59:2<227::AID-JMV17>3.0.CO;2-8
4. Gonzalez-Cao M, Iduma P, Karachaliou N, Santarpia M, Blanco J, Rosell R. Human endogenous retroviruses and cancer. Cancer Biol Med. (2016) 13:483–8. doi: 10.20892/j.issn.2095-3941.2016.0080
5. Liu X, Liu Z, Wu Z, Ren J, Fan Y, Sun L, et al. Resurrection of endogenous retroviruses during aging reinforces senescence. Cell. (2023) 186:287–304.e26. doi: 10.1016/j.cell.2022.12.017
6. Denner J. Expression function of endogenous retroviruses in the placenta. APMIS. (2016) 124:31–43. doi: 10.1111/apm.12474
7. Frank JA, Singh M, Cullen HB, Kirou RA, Benkaddour-Boumzaouad M, Cortes JL, et al. Evolution antiviral activity of a human protein of retroviral origin. Science. (2022) 378:422–8. doi: 10.1126/science.abq7871
8. Redelsperger F, Raddi N, Bacquin A, Vernochet C, Mariot V, Gache V, et al. Genetic evidence that captured retroviral envelope syncytins contribute to myoblast fusion and muscle sexual dimorphism in mice. PLoS Genet. (2016) 12:1006289. doi: 10.1371/journal.pgen.1006289
9. Frese S, Ruebner M, Suhr F, Konou TM, Tappe KA, Toigo M, et al. Long-term endurance exercise in humans stimulates cell fusion of myoblasts along with fusogenic endogenous retroviral genes in vivo. PLoS ONE. (2015) 10:0132099. doi: 10.1371/journal.pone.0132099
10. Küry P, Nath A, Créange A, Dolei A, Marche P, Gold J, et al. Human endogenous retroviruses in neurological diseases. Trends Mol Med. (2018) 24:379–94. doi: 10.1016/j.molmed.2018.02.007
11. Römer C. Viruses and endogenous retroviruses as roots for neuroinflammation and neurodegenerative diseases. Front Neurosci. (2021) 15:175. doi: 10.3389/fnins.2021.648629
12. Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, et al. Arc a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. (1995) 14:433–45. doi: 10.1016/0896-6273(95)90299-6
13. Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl, D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci. (1995) 92:5734–5738. doi: 10.1073/pnas.92.12.5734
14. Diering Remembering GH and forgetting in sleep: Selective synaptic plasticity during sleep driven by scaling factors Homer1a and Arc. Neurobiol Stress. (2022) 22:100512. doi: 10.1016/j.ynstr.2022.100512
15. Savino R, Polito AN, Marsala G, Ventriglio A, Di Salvatore M, De Stefano MI, et al. Agomelatine: a potential multitarget compound for neurodevelopmental disorders. Brain Sci. (2023) 13:734. doi: 10.3390/brainsci13050734
16. Casillas-Espinosa PM, Powell KL, O'Brien TJ. Regulators of synaptic transmission: roles in the pathogenesis and treatment of epilepsy. Epilepsia. (2012) 53:41–58. doi: 10.1111/epi.12034
17. Du X, Wei L, Yang B, Long S, Wang J, Sun A, et al. Cortical and subcortical morphological alteration in Angelman syndrome. J Neurodev Disord. (2023) 15:7. doi: 10.1186/s11689-022-09469-3
18. Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SGN. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. (2000) 3:661–9. doi: 10.1038/76615
19. Giralt A, Brito V, Chevy Q, Simonnet C, Otsu Y, Cifuentes-DÍaz C, et al. Pyk2 modulates hippocampal excitatory synapses and contributes to cognitive deficits in a Huntington's disease model. Nat Commun. (2017) 8:15592. doi: 10.1038/ncomms15592
20. Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson, T. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell. (2018) 172:262. doi: 10.1016/j.cell.2017.12.022
21. Peebles CL, Yoo J, Thwin MT, Palop JJ, Noebels JL, Finkbeiner S. Arc regulates spine morphology and maintains network stability in vivo. Proc Natl Acad Sci. (2010) 107:18173–8. doi: 10.1073/pnas.1006546107
22. Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, et al. The neuronal gene arc encodes a repurposed retrotransposon gag protein that mediates intercellular RNA transfer. Cell. (2018) 172:275–88.e18. doi: 10.1016/j.cell.2017.12.024
23. Chawla MK, Gray DT, Nguyen C, Dhaliwal H, Zempare M, Okuno H, et al. Seizure-induced arc mRNA expression thresholds in rat hippocampus and perirhinal cortex. Front Syst Neurosci. (2018) 12:53. doi: 10.3389/fnsys.2018.00053
24. Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. (2011) 14:279–84. doi: 10.1038/nn.2708
25. Korb E, Finkbeiner S. Arc in synaptic plasticity: from gene to behavior. Trends Neurosci. (2011) 34:591–8. doi: 10.1016/j.tins.2011.08.007
26. Mikuni T, Uesaka N, Okuno H, Hirai H, Deisseroth K, Bito H, et al. Arc/Arg31 is a postsynaptic mediator of activity-dependent synapse elimination in the developing cerebellum. Neuron. (2013) 78:1024–35. doi: 10.1016/j.neuron.2013.04.036
27. Rodriguez JJ, Davies HA, Silva AT, De Souza IEJ, Peddie CJ, Colyer FM, et al. Long-term potentiation in the rat dentate gyrus is associated with enhanced Arc/Arg31 protein expression in spines, dendrites and glia. Eur J Neurosci. (2005) 21:2384–96. doi: 10.1111/j.1460-9568.2005.04068.x
28. Raju CS, Fukuda N, López-Iglesias C, Göritz C, Visa N, Percipalle P. In neurons P, activity-dependent association of dendritically transported mRNA transcripts with the transacting factor CBF-A is mediated by A2RE/RTS elements. Mol Biol Cell. (2011) 22:1864. doi: 10.1091/mbc.e10-11-0904
29. Okuno H, Akashi K, Ishii Y, Yagishita-Kyo N, Suzuki K, Nonaka M, et al. Inverse synaptic tagging of inactive synapses via dynamic interaction of Arc/Arg31 with CaMKIIβ. Cell. (2012) 149:886–98. doi: 10.1016/j.cell.2012.02.062
30. Wu J, Petralia RS, Kurushima H, Patel H, Jung MY, Volk L, et al. Arc/Arg31 regulates an endosomal pathway essential for activity-dependent β-amyloid generation. Cell. (2011) 147:615–28. doi: 10.1016/j.cell.2011.09.036
31. Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, et al. Arc/Arg31 interacts with the endocytic machinery to regulate AMPA receptor trafficking . Neuron. (2006) 52:445–59. doi: 10.1016/j.neuron.2006.08.033
32. Korb E, Wilkinson CL, Delgado RN, Lovero KL. Finkbeiner, S. Arc in the nucleus regulates PML-dependent GluA1 transcription and homeostatic plasticity. Nat Neurosci. (2013) 16:874–83. doi: 10.1038/nn.3429
33. Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF. Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. (2000) 20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000
34. Lituma PJ, Singer RH, Das S, Castillo PE. Real-time imaging of Arc/Arg3.1 transcription ex vivo reveals input-specific immediate early gene dynamics. Proc Natl Acad Sci. (2022) 119:e2123373119. doi: 10.1073/pnas.2123373119
35. de la Peña JB, Barragan-Iglesias P, Lou TF, Kunder N, Loerch S, Shukla T, et al. Intercellular arc signaling regulates vasodilation. J Neurosci. (2021) 41:7712–26. doi: 10.1523/JNEUROSCI.0440-21.2021
36. Bloomer WAC, VanDongen HMA, VanDongen, AMJ. Arc/Arg31 translation is controlled by convergent N-methyl-D-aspartate and Gs-coupled receptor signaling pathways. J Biol Chem. (2008) 283:582–92. doi: 10.1074/jbc.M702451200
37. Leung HW, Foo G, VanDonge A. Arc regulates transcription of genes for plasticity, excitability and Alzheimer's disease. Biomedicines. (2022) 10:1–44. doi: 10.3390/biomedicines10081946
38. Lingappa JR, Reed JC, Tanaka M, Chutiraka K, Robinson BA. How HIV-1 gag assembles in cells: Putting together pieces of the puzzle. Virus Res. (2014) 193:89–107. doi: 10.1016/j.virusres.2014.07.001
39. Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. (1999) 73:8527–40. doi: 10.1128/JVI.73.10.8527-8540.1999
40. Muriaux D, Mirro J, Nagashima K, Harvin D, Rein A. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J Virol. (2002) 76:11405–13. doi: 10.1128/JVI.76.22.11405-11413.2002
41. Eckwahl MJ, Telesnitsky A, Wolin SL. Host RNA packaging by retroviruses: a newly synthesized story. MBio. (2016). 7:15. doi: 10.1128/mBio.02025-15
42. Coleman BM. Hill AF. Extracellular vesicles–Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol. (2015) 40:89–96. doi: 10.1016/j.semcdb.2015.02.007
43. Thompson PJ. Corcoran R. Everyday memory failures in people with epilepsy. Epilepsia. (1992) 33:S18–20.
44. Day C, Shepherd JD. Arc: building a bridge from viruses to memory. Biochem. (2015) 469:e1–e3. doi: 10.1042/BJ20150487
45. Hantak MP, Einstein J, Kearns RB. Shepherd JD. Intercellular communication in the nervous system goes viral. Trends Neurosci. (2021) 44:248–59. doi: 10.1016/j.tins.2020.12.003
46. Shepherd, J.D. Arc—an endogenous neuronal retrovirus? Semin Cell Dev Biol. (2018) 77:73–8. doi: 10.1016/j.semcdb.2017.09.029
47. Du J, Vegh V, Reutens DC. Small changes in synaptic gain lead to seizure-like activity in neuronal network at criticality. Sci Rep. (2019) 9:9. doi: 10.1038/s41598-018-37646-9
48. Erlendsson S, Morado DR, Cullen HB, Feschotte C, Shepherd JD. Briggs JAG. Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins. Nat Neurosci. (2020) 23:172–5. doi: 10.1038/s41593-019-0569-y
49. Budnik V, Thomson T. Structure of an Arc-ane virus-like capsid. Nat Neurosci. (2020) 23:153–4. doi: 10.1038/s41593-019-0580-3
50. Eriksen MS, Nikolaienko O, Hallin EI, Grødem S, Bustad HJ, Flydal MI, et al. Arc self-association and formation of virus-like capsids are mediated by an N-terminal helical coil motif. FEBS. (2021) 288:2930–55. doi: 10.1111/febs.15618
51. Jenks KR, Kim T, Pastuzyn ED, Okuno H, Taibi AV, Bito H, et al. Arc restores juvenile plasticity in adult mouse visual cortex. Proc Natl Acad Sci. (2017) 114:9182–7. doi: 10.1073/pnas.1700866114
52. Nikolaienko O, Patil S, Eriksen MS. Bramham CR. Arc protein: a flexible hub for synaptic plasticity and cognition. Semin Cell Dev Biol. (2018) 77:33–42. doi: 10.1016/j.semcdb.2017.09.006
53. Zhang W, Wu J, Ward MD, Yang S, Chuang YA, Xiao M, et al. Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron. (2015) 86:490–500. doi: 10.1016/j.neuron.2015.03.030
54. Ufer F, Vargas P, Engler JB, Tintelnot J, Schattling B, Winkler H, et al. Arc/Arg3.1 governs inflammatory dendritic cell migration from the skin and thereby controls T cell activation. Sci Immunol. (2016) 1:aaf8665. doi: 10.1126/sciimmunol.aaf8665
55. Marsh APL, Edwards TJ, Galea C, Cooper HM, Engle EC, Jamuar SS, et al. DCC mutation update: Congenital mirror movements, isolated agenesis of the corpus callosum, and developmental split brain syndrome. Hum Mutat. (2018) 39:23–39. doi: 10.1002/humu.23361
56. Mans RA, Hinton KD, Rumer AE, Zellner KA, Blankenship EA. Expression of an arc-immunoreactive protein in the adult zebrafish brain increases in response to a novel environment. Sch Contrib from Membsh Others Artic. (2018) 76:2018. Available online at: https://digitalcommons.gaacademy.org/gjs/vol76/iss2/2/
57. Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. (2015) 16:472–82. doi: 10.1038/nrg3962
58. Emamalipour M, Seidi K, Zununi Vahed S, Jahanban-Esfahlan A, Jaymand M, Majdi H, et al. Horizontal gene transfer: from evolutionary flexibility to disease progression. Front Cell Dev Biol. (2020) 8:229. doi: 10.3389/fcell.2020.00229
59. Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. (1999) 9:657–63. doi: 10.1016/S0959-437X(99)00031-3
60. Potasiewicz A, Faron-Gorecka A, Popik P, Nikiforuk A. Repeated treatment with alpha 7 nicotinic acetylcholine receptor ligands enhances cognitive processes and stimulates Erk1/2 and Arc genes in rats. Behav Brain Res. (2021) 409:113338. doi: 10.1016/j.bbr.2021.113338
61. Boldridge M, Shimabukuro J, Nakamatsu K, Won C, Jansen C, Turner H, et al. Characterization of the C-terminal tail of the Arc protein. PLoS ONE. (2020) 15:239870. doi: 10.1371/journal.pone.0239870
62. Wang YY, Hsu SH, Tsai HY, Cheng FY. Cheng MC. Transcriptomic proteomic analysis of CRISPR/Cas9-mediated ARC-knockout HEK293 cells. Int J Mol Sci. (2022) 23:4498. doi: 10.3390/ijms23094498
63. Chuang YA, Hu TM, Chen CH, Hsu SH, Tsai HY, Cheng MC. Rare mutations and hypermethylation of the ARC gene associated with schizophrenia. Schizophr Res. (2016) 176:106–13. doi: 10.1016/j.schres.2016.07.019
64. Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. (2014) 506:179. doi: 10.1038/nature12929
65. Rodríguez C, Areces D, García T, Cueli M, Gonzalez-Castro P. Neurodevelopmental disorders: An innovative perspective via the response to intervention model. World J psychiatry. (2021) 11:1017–26. doi: 10.5498/wjp.v11.i11.1017
66. Alhowikan AM. activity-regulated cytoskeleton-associated protein dysfunction may contribute to memory disorder and earlier detection of autism spectrum disorders. Med Princ Pract. (2016) 25:350–4. doi: 10.1159/000445351
67. Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, et al. The angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. (2010) 140:704–16. doi: 10.1016/j.cell.2010.01.026
68. Mandel-Brehm C, Salogiannis J, Dhamne SC, Rotenberg A, Greenberg ME. Seizure-like activity in a juvenile Angelman syndrome mouse model is attenuated by reducing Arc expression. Proc Natl Acad Sci. (2015) 112:5129–34. doi: 10.1073/pnas.1504809112
69. Samanta D. Pharmacotherapeutic management of seizures in patients with Angleman syndrome. Expert Opin Pharmacother. (2022) 23:1511–22. doi: 10.1080/14656566.2022.2105141
70. Crawford DC, Acuña JM. Sherman SL. fMR1 and the fragile X syndrome: human genome epidemiology review. Genet Med. (2001) 3:359. doi: 10.1097/00125817-200109000-00006
71. Berry-Kravis E. Epilepsy in fragile X syndrome. Dev Med Child Neurol. (2002) 44:724–8. doi: 10.1111/j.1469-8749.2002.tb00277.x
72. Westmark CJ. Fragile X APP: a decade in review, a vision for the future. Mol Neurobiol. (2018) 56:3904–21. doi: 10.1007/s12035-018-1344-x
73. Wilkerson JR, Albanesi JP, Huber KM. Roles for Arc in metabotropic glutamate receptor-dependent LTD and synapse elimination: implications in health and disease. Semin Cell Dev Biol. (2018) 77:51–62. doi: 10.1016/j.semcdb.2017.09.035
74. Fernández E, Collins MO, Frank RAW, Zhu F, Kopanitsa MV, Nithianantharajah J, et al. Arc requires PSD95 for assembly into postsynaptic complexes involved with neural dysfunction and intelligence. Cell Rep. (2017) 21:679–91. doi: 10.1016/j.celrep.2017.09.045
75. Guzowski JF. Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus. (2002) 12:86–104. doi: 10.1002/hipo.10010
76. Borovkova M, Sieryi O, Lopushenko I, Kartashkina N, Pahnke J, Bykov A, et al. Screening of Alzheimer's disease with multiwavelength stokes polarimetry in a mouse model. IEEE Trans Med Imaging. (2022) 41:977–82. doi: 10.1109/TMI.2021.3129700
77. Kalchenko V, Harmelin A, Israeli D, Kateb B, Meglinski I, Tang Q, et al. Transcranial dynamic fluorescence imaging for the study of the epileptic seizures. In: Functional Brain Mapping: Methods and Aims. Tsytsarev V, Zhong N, Yamamoto V, (eds). Singapore: Springer Nature Singapore Pte Ltd. (2020). p. 49–66. doi: 10.1007/978-981-15-6883-1_3
78. Dynes JL. Steward O. Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J Comp Neurol. (2007) 500:433–47. doi: 10.1002/cne.21189
79. Bye CM, McDona RJ. A specific role of hippocampal NMDA receptors and arc protein in rapid encoding of novel environmental representations and a more general long-term consolidation function. Front Behav Neurosci. (2019) 13:8. doi: 10.3389/fnbeh.2019.00008
80. Janz P, Hauser P, Heining K, Nestel S, Kirsch M, Egert U, et al. Position- time-dependent arc expression links neuronal activity to synaptic plasticity during epileptogenesis. Front Cell Neurosci. (2018) 12:244. doi: 10.3389/fncel.2018.00244
81. Engel JJ. Mesial temporal lobe epilepsy: what have we learned? Neuroscientist. (2001) 7:340–52. doi: 10.1177/107385840100700410
82. Walker C. Hippocampal sclerosis: causes and prevention. Semin Neurol. (2015) 35:193–200. doi: 10.1055/s-0035-1552618
83. Postnikova TY, Diespirov GP, Amakhin DV, Vylekzhanina EN, Soboleva EB, Zaitsev AV. Impairments of long-term synaptic plasticity in the hippocampus of young rats during the latent phase of the lithium-pilocarpine model of temporal lobe epilepsy. Int J Mol Sci. (2021) 22:13355. doi: 10.3390/ijms222413355
84. Malmgren K. Thom, Hippocampal sclerosis–origins M, and imaging. Epilepsia. (2012) 53 Suppl 4:19–33. doi: 10.1111/j.1528-1167.2012.03610.x
85. Thom, M. Review: Hippocampal sclerosis in epilepsy: a neuropathology review. Neuropathol Appl Neurobiol. (2014) 40:520–43. doi: 10.1111/nan.12150
86. Janz P, Savanthrapadian S, Häussler U, Kilias A, Nestel S, Kretz O, et al. Synaptic remodeling of entorhinal input contributes to an aberrant hippocampal network in temporal lobe epilepsy. Cereb Cortex. (2017) 27:2348–64. doi: 10.1093/cercor/bhw093
87. Swann JW. Hablitz, Cellular abnormalities JJ, and synaptic plasticity in seizure disorders of the immature nervous system. Ment Retard Dev Disabil Res Rev. (2000) 6:258–67. doi: 10.1002/1098-2779(2000)6:4<258::AID-MRDD5>3.0.CO;2-H
88. Gao X, Grendel J, Muhia M, Castro-Gomez S, Susens U, Isbrandt D, et al. Disturbed prefrontal cortex activity in the absence of schizophrenia-like behavioral dysfunction in Arc/Arg31 deficient mice. J Neurosci. (2019) 39:8149. doi: 10.1523/JNEUROSCI.0623-19.2019
89. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. (2013) 41:gks1193. doi: 10.1093/nar/gks1193
90. Griffin NG, Wang Y, Hulette CM, Halvorsen M, Cronin KD, Walley NM, et al. Differential gene expression in dentate granule cells in mesial temporal lobe epilepsy with and without hippocampal sclerosis. Epilepsia. (2016) 57:376–85. doi: 10.1111/epi.13305
91. Morin-Brureau M, Milior G, Royer J, Chali F, LeDuigou C, Savary E, et al. Microglial phenotypes in the human epileptic temporal lobe. Brain. (2018) 141:3343–60. doi: 10.1093/brain/awy276
92. Kjær C, Barzaghi G, Bak LK, Goetze JP, Yde CW, Woldbye D, et al. Transcriptome analysis in patients with temporal lobe epilepsy. Brain. (2019) 142:E55. doi: 10.1093/brain/awz265
93. Robinson MD, McCarthy DJ. Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
94. Papatheodorou I, Moreno P, Manning J, Fuentes AMP, George N, Fexova S, et al. Expression alas update: from tissues to single cells. Nucleic Acids Res. (2020) 48:D77–83. doi: 10.1093/nar/gkz947
95. Majewski L, Wojtas B, Maciag F, Kuznicki J. Changes in calcium homeostasis and gene expression implicated in epilepsy in hippocampi of mice overexpressing ORAI1. Int J Mol Sci. (2019). 20. doi: 10.3390/ijms20225539
96. Almeida Silva LF, Reschke CR, Nguyen NT, Langa E, Sanz-Rodriguez A, Gerbatin RR, et al. Genetic deletion of microRNA-22 blunts the inflammatory transcriptional response to status epilepticus and exacerbates epilepsy in mice. Mol Brain. (2020) 13:114. doi: 10.1186/s13041-020-00653-x
97. Kapur M, Ganguly A, Nagy G, Adamson SI, Chuang JH, Frankel WN, et al. Expression of the neuronal tRNA n-Tr20 regulates synaptic transmission and seizure susceptibility. Neuron. (2020) 108:193. doi: 10.1016/j.neuron.2020.07.023
98. Haigh JL, Adhikari A, Copping NA, Stradleigh T, Wade AA, Catta-Preta R, et al. Deletion of a non-canonical regulatory sequence causes loss of Scn1a expression and epileptic phenotypes in mice. Genome Med. (2021) 13:884. doi: 10.1186/s13073-021-00884-0
99. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W., Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47–e47. doi: 10.1093/nar/gkv007
100. Venø MT, Reschke CR, Morris G, Connolly NMC, Su J, Yan Y, et al. A systems approach delivers a functional microRNA catalog and expanded targets for seizure suppression in temporal lobe epilepsy. Proc Natl Acad Sci. (2020) 117:15977–88. doi: 10.1073/pnas.1919313117
101. Cid E, Marquez-Galera A, Valero M, Gal B, Medeiros DC, Navarron CM, et al. Sublayer- and cell-type-specific neurodegenerative transcriptional trajectories in hippocampal sclerosis. Cell Rep. (2021) 35:429560. doi: 10.1101/2021.02.03.429560
102. Chuvakova LN, Funikov SY, Rezvykh AP, Davletshin AI, Evgen'ev MB, Litvinova SA, et al. Transcriptome of the krushinsky-molodkina audiogenic rat strain and identification of possible audiogenic epilepsy-associated genes. Front Mol Neurosci. (2021) 14:738930. doi: 10.3389/fnmol.2021.738930
103. Park AY, Park YS, So D, Song IK, Choi JE, Kim HJ, et al. Activity-regulated cytoskeleton-associated protein (Arc/Arg3.1) is transiently expressed after heat shock stress and suppresses heat shock factor 1. Sci Rep. (2019) 9:1. doi: 10.1038/s41598-019-39292-1
104. Wall MJ, Collins DR, Chery SL, Allen ZD, Pastuzyn ED, George AJ, et al. The temporal dynamics of arc expression regulate cognitive flexibility. Neuron. (2018) 98:1124–32.e7. doi: 10.1016/j.neuron.2018.05.012
105. Gangarossa G, Benedetto Di, O'Sullivan M, Dunleavy M, Alcacer C, Bonito-Oliva A, et al. Convulsant doses of a dopamine D1 receptor agonist result in erk-dependent increases in Zif268 and Arc/Arg31 expression in mouse dentate gyrus. PLoS ONE. (2011) 6:e19415. doi: 10.1371/journal.pone.0019415
106. Jiang Y, VanDongen AMJ. Neuronal activity-dependent accumulation of arc in astrocytes. bioRxiv. (2020) 2020:11.10.376756. doi: 10.1101/2020.11.10.376756
107. Szyndler J, Maciejak P, Wislowska-Stanek A, Lehner M. Plaznik A. Changes in the Egr1 and Arc expression in brain structures of pentylenetetrazole-kindled rats. Pharmacol Rep. (2013) 65:368–78. doi: 10.1016/S1734-1140(13)71012-0
108. Chotiner JK, Nielson J, Farris S, Lewandowski G, Huang F, Banos K, et al. Assessment of the role of MAP kinase in mediating activity-dependent transcriptional activation of the immediate early gene Arc/Arg31 in the dentate gyrus in vivo. Learn Mem. (2010) 17:117–29. doi: 10.1101/lm.1585910
109. Penrod RD, Thomsen M, Taniguchi M, Guo Y, Cowan CW, Smith LN. The activity-regulated cytoskeleton-associated protein, Arc/Arg3.1, influences mouse cocaine self-administration. Pharmacol Biochem Behav. (2020) 188:172818. doi: 10.1016/j.pbb.2019.172818
110. Gao F, Song X, Zhu D, Wang X, Hao A, Victor Nadler J. Zhan, Dendritic morphology RZ, synaptic transmission, and activity of mature granule cells born following pilocarpine-induced status epilepticus in the rat. Front Cell Neurosci. (2015) 9:384. doi: 10.3389/fncel.2015.00384
111. Coenen AML. Van Luijtelaar EM. Genetic animal models for absence epilepsy: a review of the WAG/Rij strain of rats. Behav Genet. (2003) 33:635–55. doi: 10.1023/a:1026179013847
112. Borges FS, Gabrick EC, Protachevicz PR, Higa GSV, Lameu EL, Rodriguez PXR, et al. Intermittency properties in a temporal lobe epilepsy model. Epilepsy Behav. (2023) 139:109072. doi: 10.1016/j.yebeh.2022.109072
113. Blüthgen N, Van Bentum M, Merz B, Kuhl D, Hermey G. Profiling the MAPK/ERK dependent and independent activity regulated transcriptional programs in the murine hippocampus in vivo. Sci Reports. (2017) 7:1–14. doi: 10.1038/srep45101
114. Chung L, Bey AL, Towers AJ, Cao X, Kim IH, Jiang Y. Lovastatin suppresses hyperexcitability and seizure in Angelman syndrome model. Neurobiol Dis. (2018) 110:12–9. doi: 10.1016/j.nbd.2017.10.016
115. Steward O, Matsudaira Yee K, Farris S, Pirbhoy PS, Worley P, Okamura K, et al. Bito, Delayed degradation H, and impaired dendritic delivery of intron-lacking EGFP-Arc/Arg3.1 mRNA in EGFP-Arc transgenic mice. Front Mol Neurosci. (2018) 10:435. doi: 10.3389/fnmol.2017.00435
116. Haug K, Kremerskothen J, Hallmann K, Sander T, Dullinger J, Rau B, et al. Mutation screening of the chromosome 8q243-human activity-regulated cytoskeleton-associated gene (ARC) in idiopathic generalized epilepsy. Mol Cell Probes. (2000) 14:255–60. doi: 10.1006/mcpr.2000.0314
117. Di Cicco G, Marzano E, Iacovelli L, Celli R, van Luijtelaar G, Nicoletti F, et al. Group I metabotropic glutamate receptor-mediated long term depression is disrupted in the hippocampus of WAG/Rij rats modelling absence epilepsy. Neuropharmacology. (2021) 196:108686. doi: 10.1016/j.neuropharm.2021.108686
118. Mano T, Albanese A, Dodt HU, Erturk A, Gradinaru V, Treweek JB, et al. Whole-brain analysis of cells and circuits by tissue clearing and light-sheet microscopy. J Neurosci. (2018) 38:9330–7. doi: 10.1523/JNEUROSCI.1677-18.2018
119. Van Luijtelaar G, Hramov A, Sitnikova E. Koronovskii A. Spike-wave discharges in WAG/Rij rats are preceded by delta and theta precursor activity in cortex and thalamus. Clin Neurophysiol. (2011) 122:687–95. doi: 10.1016/j.clinph.2010.10.038
120. Managò F, Mereu M, Mastwal S, Mastrogiacomo R, Scheggia D, Emanuele M, et al. Genetic disruption of Arc/Arg31 in mice causes alterations in dopamine and neurobehavioral phenotypes related to schizophrenia. Cell Rep. (2016) 16:2116–28. doi: 10.1016/j.celrep.2016.07.044
121. Xing J, Han D, Xu D, Li X, Sun L. CREB protects against temporal lobe epilepsy associated with cognitive impairment by controlling oxidative neuronal damage. Neurodegener Dis. (2020) 19:225–7. doi: 10.1159/000507023
122. Zhang H, Bramham CR. Arc/Arg3.1 function in long-term synaptic plasticity: Emerging mechanisms and unresolved issues. Eur J Neurosci. (2020) 54:6696–712. doi: 10.1111/ejn.14958
Keywords: Arc/Arg3.1, epilepsy, learning, memory, retrovirus, seizures, capsid
Citation: Sibarov DA, Tsytsarev V, Volnova A, Vaganova AN, Alves J, Rojas L, Sanabria P, Ignashchenkova A, Savage ED and Inyushin M (2023) Arc protein, a remnant of ancient retrovirus, forms virus-like particles, which are abundantly generated by neurons during epileptic seizures, and affects epileptic susceptibility in rodent models. Front. Neurol. 14:1201104. doi: 10.3389/fneur.2023.1201104
Received: 06 April 2023; Accepted: 02 June 2023;
Published: 07 July 2023.
Edited by:
Evgenia Sitnikova, Institute of Higher Nervous Activity and Neurophysiology (RAS), RussiaReviewed by:
Ramachandran Prakasam, Washington University in St. Louis, United StatesIgor Meglinski, Aston University, United Kingdom
Copyright © 2023 Sibarov, Tsytsarev, Volnova, Vaganova, Alves, Rojas, Sanabria, Ignashchenkova, Savage and Inyushin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vassiliy Tsytsarev, dHN5dHNhcmV2QHVtYXJ5bGFuZC5lZHU=
 Dmitry A. Sibarov
Dmitry A. Sibarov Vassiliy Tsytsarev
Vassiliy Tsytsarev Anna Volnova
Anna Volnova Anastasia N. Vaganova
Anastasia N. Vaganova Janaina Alves4
Janaina Alves4 Alla Ignashchenkova
Alla Ignashchenkova Mikhail Inyushin
Mikhail Inyushin