- Department of Neurology, Second Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
Background: Parkinson's disease (PD) and dystonia are closely related in terms of pathophysiology and clinical manifestations, but their common genetic characteristics remain unclear. Some genome-wide association studies (GWASs) and replication studies have revealed correlations between single nucleotide polymorphisms (SNPs) of the ARSG, BDNF, NALCN, OR4X2, KIAA1715, and OR4B1 genes and dystonia. This study was conducted to assess the association between these genetic loci and PD in a population from Eastern China.
Methods: We genotyped the SNPs (rs11655081 of ARSG; rs6265 of BDNF; rs61973742, rs1338051, rs9518384, and rs9518385 of NALCN; rs67863238 of OR4X2; rs10930717 of KIAA1715; and rs35875350 of OR4B1) in a cohort of 474 patients with PD and 439 healthy controls from East China. To determine the genotypes of these SNPs, we used an Agena MassARRAY Typer 4.0. Odds ratios (ORs) and 95% CIs were computed to evaluate the correlations between these SNPs and the risk of PD.
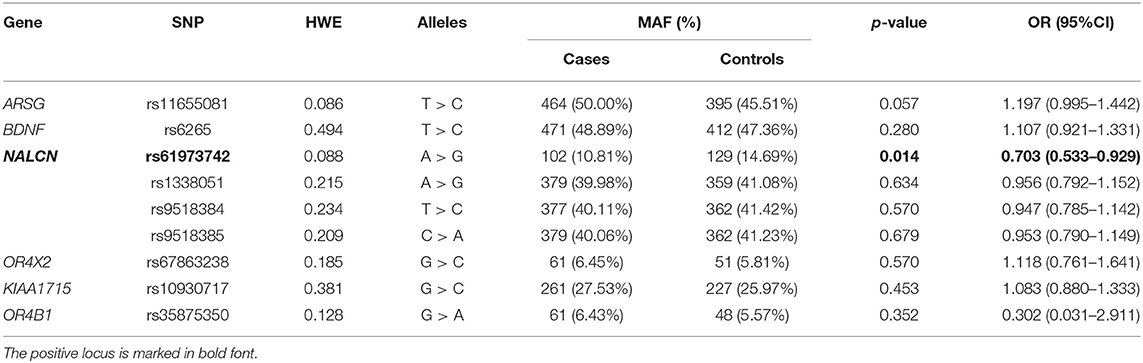
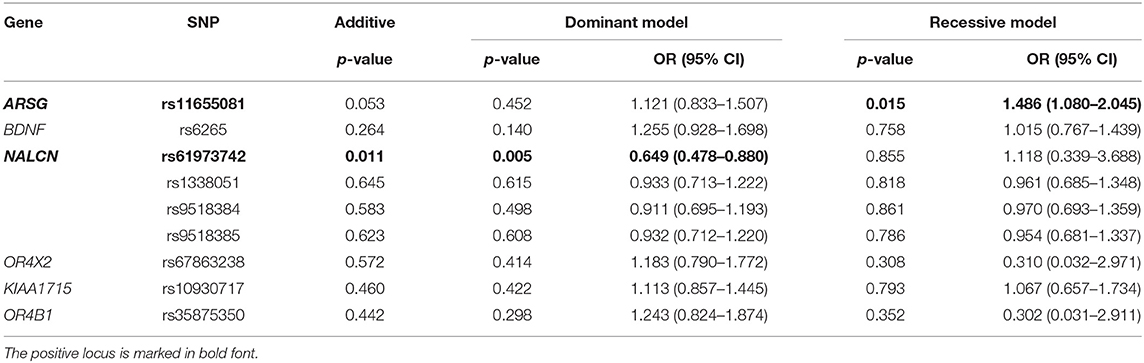
Results: There were significant differences in the genotype distribution (OR = 0.649, 95% CI = 0.478–0.880) and minor allele frequency (MAF) (OR = 0.703, 95% CI = 0.533–0.929) of SNP rs61973742 (NALCN) between patients with PD and healthy controls. A significant difference was detected in the genotype distribution of rs11655081 (ARSG) (OR = 1.486, 95% CI = 1.080–2.045).
Conclusion: Single nucleotide polymorphisms rs11655081 (ARSG) and rs61973742 (NALCN) may be associated with PD. The C allele of rs11655081 may increase the risk of PD, whereas the G allele of rs61973742 may be a protective factor.
Introduction
Parkinson's disease (PD) is the second most common neurodegenerative disease, affecting ~2–3% of the population aged ≥65 years old (1). The clinical characteristics of PD are described as parkinsonism (classical motor features, such as bradykinesia, muscular rigidity, rest tremor, and postural imbalance) and non-motor features, such as emotional disorders, sleep disorders, and autonomic dysfunction (2). The cause of PD is still not clear, but genetic and environmental factors have been shown to work together in its risk (3). Among these genetic factors, increasing SNP loci were found to be related to the onset of PD in genome-wide association studies (GWASs) (4–6). Dystonia is a neurological condition characterized by involuntary movements or postures due to persistent or intermittent muscle contractions (7). Dystonia can be found both in isolation and in combination with other movement disorders (e.g., dystonia with parkinsonism and myoclonus) (8). Dystonia may occur in idiopathic PD and is more frequently seen as a complication in the treatment of PD (9). The etiology of dystonia remains unknown. However, increasing evidence suggests that genetic variants may function in dystonia, and DYT genes are usually the cause of monogenic dystonia (7). Moreover, previous studies have indicated that dystonia and PD may share some common genetic risk factors. For example, parkinsonism is generally detected with mutations in DYT genes (such as DYT/PARK-TAF1, DYT5/PARK- GCH1, DYT/PARK-ATP1A3, and DYT/PARK-PRKRA) (10). On the other hand, patients with PD having mutations in PARK1, PARK6, PARK7, and PARK8 appear to often manifest dystonia (3).
Some SNP loci in genes, such as TOR1A and GCH1 may increase the genetic burden in dystonia (11, 12). Among these SNPs, BDNF rs6265 was identified to be probably related to many diseases, such as PD (13). Recent studies have revealed more SNPs that may be related to the onset of dystonia. Two GWASs have been used to identify gene alterations that may predispose to dystonia (14). In the GWAS executed by Lohmann et al., Arylsulfatase G (ARSG) was found to play a role in dystonia (15). According to the GWAS conducted by Mok et al., the most statistically significant variants in dystonia included NALCN (rs61973742, rs1338051, rs9518385, and rs9518384), OR4X2 (rs67863238), KIAA1715 (rs10930717), and OR4B1 (rs35875350) (16). Among these genes, NALCA has been found to be associated with PD (17), while others have hardly been studied in patients with PD. To explore the genetic associations between these SNPs and PD, we performed a case-control study.
Materials and Methods
Subjects
Our study included 474 patients with PD without dystonia from the Neurology Department at the Second Affiliated Hospital of Zhejiang University. Patients with PD caused by secondary causes of Parkinsonism were excluded. Patients with other neurodegenerative diseases, such as essential tremor (ET), multiple system atrophy (MSA), corticobasal degeneration (CBD), and Wilson's disease (WD) were excluded. A total of 439 healthy individuals were recruited as the healthy controls. Both the patients and healthy controls were Han people from Eastern China.
Ethical approval was obtained from the Medical Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine. Informed consent was obtained from all patients and healthy controls prior to blood withdrawal.
Genotyping
Peripheral blood samples were collected from each patient. A DNA rapid extraction kit (BioTeKe Corporation, China) was used to extract DNA. AssayDesigner 3.1 (San Diego, CA: Agena Corporation) was used to design the genotyping assays. The following nine loci were tested: rs11655081 of ARSG; rs6265 of BDNF; rs61973742, rs1338051, rs9518384, and rs9518385 of NALCN; rs67863238 of OR4X2; rs10930717 of KIAA1715, and rs35875350 of OR4B1. Agena MassARRAY Typer 4.0 (San Diego, CA: Agena Corporation) was used to determine the genotypes of these SNPs.
Statistical Analysis
The t-test was used to assess differences in age between patients with PD and controls. The chi-square test was used to detect the sex distribution and Hardy–Weinberg equilibrium (HWE) in healthy controls. Allelic, minor allele dominant, and recessive models were applied, and association analyses were performed using the chi-square test or Fisher's exact test. The odds ratio (OR) was used to assess the strength of polymorphisms and PD susceptibility. The Cochran-Armitage trend test (CATT) was used to calculate the additive model. All statistical analyses were performed using SPSS version 25.0 (Armonk, NY: IBM Corporation), and statistical significance was set at p < 0.05. The power was computed using the Quanto version 1.2.4.
Results
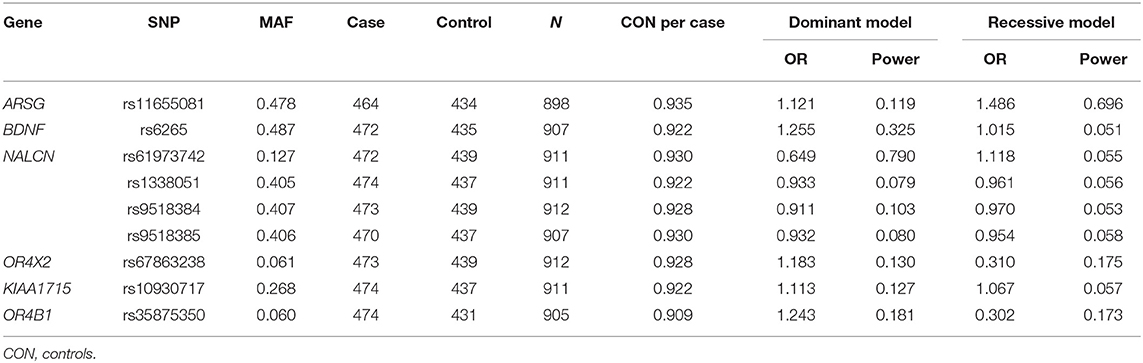
A total of 474 patients with PD and 439 healthy controls were included. There were no significant differences in sex or age distribution between patients and controls (Table 1). High genotyping quality was achieved in all genotyping reactions. No significant deviation from the HWE was detected in the genotype frequency distributions of controls (Table 2). There were significant differences in the genotypes and allele frequencies of SNP rs61973742 (NALCN) between patients with PD and controls, as shown in Tables 2, 3. Using allelic, additive, minor allele dominant, and recessive models, we found that the minor allele frequency (MAF) of rs61973742 in PD cases was 10.81%, which was lower than that in healthy controls (p = 0.014, OR = 0.703, 95% CI = 0.533–0.929), and a difference in genotype distribution was observed in the additive (p = 0.011) and dominant models (p = 0.005, OR = 0.649, 95% CI = 0.478–0.880). The results also demonstrated an increase in the frequency of the CC genotype (recessive model) for SNP ARSG rs11655081 in patients with PD compared with healthy controls (p = 0.015, OR = 1.486, 95% CI = 1.080–2.045). The power of the recessive model in rs11655081 reached nearly 70% and the power of the dominant model of rs6197374 was nearly 80% (Table 4). There were no significant differences in the allele frequencies or genotype distributions of the SNP rs6265 of BDNF; rs1338051, rs9518384, and rs9518385 of NALCN; rs67863238 of OR4X2; rs10930717 of KIAA1715; and rs35875350 of OR4B1 between patients and controls. The MAF in our population was consistent with 1,000 Genomes in PubMed (Supplementary Table 1).
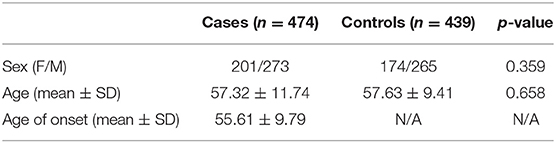
Table 1. Demographic characteristics of patients with Parkinson's disease (PD) and healthy controls.
Discussion
This study was conducted to assess the association of nine dystonia-related genetic loci in a Chinese population of patients with PD. We found that rs11655081 of ARSG and rs61973742 of NALCN were significantly correlated with PD. The C allele of rs11655081 may increase the susceptibility to PD, while the G allele of rs61973742 may decrease it.
The proteins encoded by ARSG hydrolyze sulfate esters; therefore, they participate in cell signaling, protein degradation, and hormone biosynthesis (18, 19). GWAS by Lohmann et al. suggested that rs11655081 of ARSG was related to the musician's dystonia and writer's cramp (15). A study in a Dutch patient group with musician's dystonia and writer's cramp found an accumulation of rare single nucleotide variants in the coding region of ARSG, which indicated that ARSG may function in the task-induced focal dystonia (20). A cohort containing 206 blepharospasm (BSP) patients and 206 controls from Greek was sequenced, and a subtle trend for the correlation of rs11655081 and BSP susceptibility was found (p = 0.088, OR = 0.64, 95% CI = 0.38–1.07) (21). No studies evaluating the relationship between rs11655081 and PD have been conducted.
According to a GWAS performed by Mok et al. in a British cohort with cervical dystonia (CD), the cluster of variants of NALCN was nearest to the genome-wide significance threshold. A replication case-control study in a Spanish population did not report any association between CD and NALCN rs61973742 (p = 0.1155, OR = 0.7098) (22). A study by Zhou et al. (23) in a Chinese population did not suggest that the SNP rs61973742 of NALCN plays a role in focal CD. However, this SNP showed a potential effect in the same direction as the previously reported GWAS. NALCN encodes a protein associated with sodium leak channels, and it is predominantly expressed in the nervous system and is involved in regulating the resting membrane potential and neuronal excitability. Previous studies found that the protein encoded by the ANO3 (DYT24) gene, which functions as calcium-activated chloride channels, can participate in the modulation of neuronal excitability, and its mutations have been identified in sporadic CD (24). Based on this, Mok et al. (16) assumed that NALCN may be a candidate gene for dystonia. Few animal studies found that akinesia and freezing were the main phenotypes of NALCN-deficient strains; therefore, they put forward a hypothesis that the NALCN may be involved in movement disorders clinically characterized by akinesia and freezing gait in humans, such as PD (25, 26). Moreover, previous studies suggested that NALCA may contribute to both disordered movement and psychiatric features in PD through gene-gene interactions (17). The specific function of our SNP loci in NALCN remains unknown.
The BDNF gene has been found to influence synaptic plasticity, and it has been indicated to play a role in a wide range of neurological diseases and injuries (27). An increasing number of studies have focused on the relationship between the risk of PD and BDNF SNPs, especially rs6265, although strong evidence is yet to be presented (28). Michałowska et al. (29) identified that the variation of rs6265 increased the risk of PD. However, Pal et al. (28) and Svetel et al. (30) found that the allele frequency and genotype distribution in patients with PD and healthy controls did not deviate significantly. Our results are consistent with these latter findings. To date, studies concerning the role of rs6265 in dystonia have yielded contradictory results (14, 31–33). Therefore, the role of BDNF in both PD and dystonia requires further exploration.
Our study aimed to examine the associations between SNPs in the selected genes and PD in an Eastern Chinese population. There were some limitations to our study. First, the clinical manifestation of PD is diverse; therefore, it would be better to set subtypes, such as early- or late-onset PD, and PD patients with or without dystonia, according to the potential function of the SNPs. In addition, new association studies have been performed. For example, SNPs in COL8A1, DENND1A, and GABBR2 were found to be associated with cervical dystonia in a multicenter GWAS (5). These loci have not been replicated in case-control studies. Given the racial heterogeneity and limited sample size, more replication studies are needed to assess whether these loci and variants are common genetic factors between PD and dystonia.
Conclusion
In this study, we found that rs11655081 and rs61973742 may be associated with PD. The C allele of rs11655081 may increase the risk, and the G allele of rs61973742 may be a protective factor. No significant differences were detected either in allele frequencies or genotype distributions in the SNP rs6265 of BDNF; rs1338051, rs9518384, and rs9518385 of NALCN; rs67863238 of OR4X2; rs10930717 of KIAA1715; and rs35875350 of OR4B1 between patients and controls.
Data Availability Statement
All data generated or used during the study are available from the corresponding author by request.
Ethics Statement
The studies involving human participants were reviewed and approved by Medical Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JT contributed to conception and design of the study. C-YJ and RZ organized the database. W-YY, TG, and RZ performed the statistical analysis. W-YY and S-SJ wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (Grant Numbers 81771216 and 81520108010) and the Key R&D Program of Zhejiang Province (Grant Number 2020C03020).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge all the participants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2021.711050/full#supplementary-material
References
1. Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, et al. Parkinson disease. Nat Rev Dis Primers. (2017) 3:17013. doi: 10.1038/nrdp.2017.13
2. Kalia LV, Lang AE. Parkinson's disease. Lancet. (2015) 386:896–912. doi: 10.1016/S0140-6736(14)61393-3
3. Shetty AS, Bhatia KP, Lang AE. Dystonia and Parkinson's disease: what is the relationship? Neurobiol Dis. (2019) 132:104462. doi: 10.1016/j.nbd.2019.05.001
4. Cui X, Xu C, Zhang L, Wang Y. Identification of Parkinson's disease-causing genes via omics data. Front Genet. (2021) 12:712164. doi: 10.3389/fgene.2021.712164
5. Sun YV Li C, Hui Q, Huang Y, Barbano R, Rodriguez R, et al. A multi-center genome-wide association study of cervical dystonia. Mov Disord. (2021) 36:2795–801. doi: 10.1002/mds.28732
6. Pihlstrøm L, Fan CC, Frei O, Tan M, Karunamuni RA, Blauwendraat C, et al. Genetic stratification of age-dependent parkinson's disease risk by polygenic hazard score. Mov Disord. (2021) 37:62–9. doi: 10.1002/mds.28808
7. Balint B, Mencacci NE, Valente EM, Pisani A, Rothwell J, Jankovic J, et al. Dystonia. Nat Rev Dis Primers. (2018) 4:25. doi: 10.1038/s41572-018-0023-6
8. Albanese A, Bhatia K, Bressman SB, Delong MR, Fahn S, Fung VS, et al. Phenomenology and classification of dystonia: a consensus update. Mov Disord. (2013) 28:863–73. doi: 10.1002/mds.25475
9. Tolosa E, Compta Y. Dystonia in Parkinson's disease. J Neurol. (2006) 253 Suppl 7:Vii7–13. doi: 10.1007/s00415-006-7003-6
10. Phukan J, Albanese A, Gasser T, Warner T. Primary dystonia and dystonia-plus syndromes: clinical characteristics, diagnosis, and pathogenesis. Lancet Neurol. (2011) 10:1074–85. doi: 10.1016/S1474-4422(11)70232-0
11. Giri S, Ghosh A, Roy S, Sankhla CS, Das SK, Ray K, et al. Association of TOR1A and GCH1 polymorphisms with isolated dystonia in India. J Mol Neurosci. (2020). doi: 10.1007/s12031-020-01653-1
12. Siokas V, Dardiotis E, Tsironi EE, Tsivgoulis G, Rikos D, Sokratous M, et al. The role of TOR1A polymorphisms in dystonia: a systematic review and meta-analysis. PLoS ONE. (2017) 12:e0169934. doi: 10.1371/journal.pone.0169934
13. Zintzaras E, Hadjigeorgiou GM. The role of G196A polymorphism in the brain-derived neurotrophic factor gene in the cause of Parkinson's disease: a meta-analysis. J Hum Genet. (2005) 50:560–6. doi: 10.1007/s10038-005-0295-z
14. Siokas V, Aloizou AM, Tsouris Z, Michalopoulou A, Mentis AA, Dardiotis E. Risk factor genes in patients with dystonia: a comprehensive review. Tremor Other Hyperkin Mov. (2018) 8:559. doi: 10.5334/tohm.437
15. Lohmann K, Schmidt A, Schillert A, Winkler S, Albanese A, Baas F, et al. Genome-wide association study in musician's dystonia: a risk variant at the arylsulfatase G locus? Mov Disord. (2014) 29:921–7. doi: 10.1002/mds.25791
16. Mok KY, Schneider SA, Trabzuni D, Stamelou M, Edwards M, Kasperaviciute D, et al. Genomewide association study in cervical dystonia demonstrates possible association with sodium leak channel. Mov Disord. (2014) 29:245–51. doi: 10.1002/mds.25732
17. Kasap M, Dwyer DS. Na(+) leak-current channel (NALCN) at the junction of motor and neuropsychiatric symptoms in Parkinson's disease. J Neural Transm. (2021) 128:749–62. doi: 10.1007/s00702-021-02348-6
18. Ferrante P, Messali S, Meroni G, Ballabio A. Molecular and biochemical characterisation of a novel sulphatase gene: arylsulfatase G (ARSG). Eur J Hum Genet. (2002) 10:813–8. doi: 10.1038/sj.ejhg.5200887
19. Sardiello M, Annunziata I, Roma G, Ballabio A. Sulfatases and sulfatase modifying factors: an exclusive and promiscuous relationship. Hum Mol Genet. (2005) 14:3203–17. doi: 10.1093/hmg/ddi351
20. Nibbeling E, Schaake S, Tijssen MA, Weissbach A, Groen JL, Altenmüller E, et al. Accumulation of rare variants in the arylsulfatase G (ARSG) gene in task-specific dystonia. J Neurol. (2015) 262:1340–3. doi: 10.1007/s00415-015-7718-3
21. Siokas V, Kardaras D, Aloizou AM, Asproudis I, Boboridis KG, Papageorgiou E, et al. Lack of association of the rs11655081 ARSG gene with blepharospasm. J Mol Neurosci. (2019) 67:472–6. doi: 10.1007/s12031-018-1255-3
22. Gomez-Garre P, Huertas-Fernandez I, Caceres-Redondo MT, Alonso-Canovas A, Bernal-Bernal I, Blanco-Ollero A, et al. Lack of validation of variants associated with cervical dystonia risk: a GWAS replication study. Mov Disord. (2014) 29:1825–8. doi: 10.1002/mds.26044
23. Zhou Q, Yang J, Cao B, Chen Y, Wei Q, Ou R, et al. Association analysis of NALCN polymorphisms rs1338041 and rs61973742 in a Chinese population with isolated cervical dystonia. Parkinson's Dis. (2016) 2016:9281790. doi: 10.1155/2016/9281790
24. Lohmann K, Klein C. Genetics of dystonia: what's known? What's new? What's next? Mov Disord. (2013) 28:899–905. doi: 10.1002/mds.25536
25. Kasap M, Bonnett K, Aamodt EJ, Dwyer DS. Akinesia and freezing caused by Na(+) leak-current channel (NALCN) deficiency corrected by pharmacological inhibition of K(+) channels and gap junctions. J Comp Neurol. (2017) 525:1109–21. doi: 10.1002/cne.24119
26. Bonnett K, Zweig R, Aamodt EJ, Dwyer DS. Food deprivation and nicotine correct akinesia and freezing in Na(+) -leak current channel (NALCN)-deficient strains of Caenorhabditis elegans. Genes Brain Behav. (2014) 13:633–42. doi: 10.1111/gbb.12153
27. Deng P, Anderson JD Yu AS, Annett G, Fink KD, Nolta JA. Engineered BDNF producing cells as a potential treatment for neurologic disease. Expert Opin Biol Ther. (2016) 16:1025–33. doi: 10.1080/14712598.2016.1183641
28. Pal P, Sadhukhan T, Chakraborty S, Sadhukhan S, Biswas A, Das SK, et al. Role of apolipoprotein E, cathepsin D, and Brain-derived neurotrophic factor in Parkinson's disease: a study from Eastern India. Neuromol Med. (2019) 21:287–94. doi: 10.1007/s12017-019-08548-4
29. Michałowska M, Chalimoniuk M, Jówko E, Przybylska I, Langfort J, Toczylowska B, et al. Gene polymorphisms and motor levodopa-induced complications in Parkinson's disease. Brain Behav. (2020) 10:e01537. doi: 10.1002/brb3.1537
30. Svetel M, Pekmezovic T, Markovic V, Novaković I, Dobričić V, Djuric G, et al. No association between brain-derived neurotrophic factor G196A polymorphism and clinical features of Parkinson's disease. Eur Neurol. (2013) 70:257–62. doi: 10.1159/000352033
31. Siokas V, Kardaras D, Aloizou AM, Asproudis I, Boboridis KG, Papageorgiou E, et al. BDNF rs6265 (Val66Met) Polymorphism as a risk factor for blepharospasm. Neuromol Med. (2019) 21:68–74. doi: 10.1007/s12017-018-8519-5
32. Sako W, Murakami N, Izumi Y, Kaji R. Val66Met polymorphism of brain-derived neurotrophic factor is associated with idiopathic dystonia. J Clin Neurosci. (2015) 22:575–7. doi: 10.1016/j.jocn.2014.08.014
Keywords: Parkinson's disease, dystonia, single nucleotide polymorphism, ARSG, NALCN
Citation: Yang W-Y, Jiang S-S, Pu J-L, Jin C-Y, Gao T, Zheng R, Tian J and Zhang B-R (2022) Association Between Dystonia-Related Genetic Loci and Parkinson's Disease in Eastern China. Front. Neurol. 12:711050. doi: 10.3389/fneur.2021.711050
Received: 17 May 2021; Accepted: 22 December 2021;
Published: 22 February 2022.
Edited by:
Steven Frucht, NYU Grossman School of Medicine, United StatesReviewed by:
Zhong Pei, Sun Yat-sen University, ChinaJean-Marc Burgunder, Clinic Siloah, Switzerland
Copyright © 2022 Yang, Jiang, Pu, Jin, Gao, Zheng, Tian and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Rong Zhang, YnJ6aGFuZyYjeDAwMDQwO3pqdS5lZHUuY24=; Jun Tian, anVudGlhbiYjeDAwMDQwO3pqdS5lZHUuY24=
†These authors have contributed equally to this work
 Wen-Yi Yang
Wen-Yi Yang Si-Si Jiang†
Si-Si Jiang† Jia-Li Pu
Jia-Li Pu Chong-Yao Jin
Chong-Yao Jin Ting Gao
Ting Gao Ran Zheng
Ran Zheng Jun Tian
Jun Tian Bao-Rong Zhang
Bao-Rong Zhang