- Department of Neurology, The First Hospital of Jilin University, Changchun, China
Background: SCN1A and SCN2A genes have been reported to be associated with the efficacy of single and combined antiepileptic therapy, but the results remain contradictory. Previous meta-analyses on this topic mainly focused on the SCN1A rs3812718 polymorphism. However, meta-analyses focused on SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, or SCN2A rs2304016 polymorphisms are scarce or non-existent.
Objective: We aimed to conduct a meta-analysis to determine the effects of SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 polymorphisms on resistance to antiepileptic drugs (AEDs).
Methods: We searched the PubMed, Embase, Cochrane Library, WANFANG, and CNKI databases up to June 2020 to collect studies on the association of SCN1A and SCN2A polymorphisms with reactivity to AEDs. We calculated the pooled odds ratios (ORs) under the allelic, homozygous, heterozygous, dominant, and recessive genetic models to identify the association between the four single-nucleotide polymorphisms (SNPs) and resistance to AEDs.
Results: Our meta-analysis included 19 eligible studies. The results showed that the SCN1A rs2298771 polymorphism was related to AED resistance in the allelic, homozygous, and recessive genetic models (G vs. A: OR = 1.20, 95% CI: 1.012–1.424; GG vs. AA: OR = 1.567, 95% CI: 1.147–2.142; GG vs. AA + AG: OR = 1.408, 95% CI: 1.053–1.882). The homozygous model remained significant after Bonferroni correction (P < 0.0125). Further subgroup analyses demonstrated the significance of the correlation in the dominant model in Caucasians (South Asians) after Bonferroni correction (GG + GA vs. AA: OR = 1.620, 95% CI: 1.165–2.252). However, no association between SCN1A rs2298771 polymorphism and resistance to AEDs was found in Asians or Caucasians (non-South Asians). For SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 polymorphisms, the correlations with responsiveness to AEDs were not significant in the overall population nor in any subgroup after conducting the Bonferroni correction. The results for SCN1A rs2298771, SCN1A rs10188577, and SCN2A rs2304016 polymorphisms were stable and reliable according to sensitivity analysis and Begg and Egger tests. However, the results for SCN2A rs17183814 polymorphism have to be treated cautiously owing to the significant publication bias revealed by Begg and Egger tests.
Conclusions: The present meta-analysis indicated that SCN1A rs2298771 polymorphism significantly affects resistance to AEDs in the overall population and Caucasians (South Asians). There were no significant correlations between SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 polymorphisms and resistance to AEDs.
Introduction
Epilepsy is one of the most common chronic brain diseases, affecting over 70 million people worldwide (1). Antiepileptic drug (AED) therapy is the first-line treatment for patients with epilepsy (PWE); however, one-third of the individuals show drug resistance (2). The underlying mechanism of AED resistance has not been completely elucidated, but genetic factors may be important for the interpersonal differences seen in drug efficacy (3). The current relevant studies mainly focus on polymorphisms of genes that encode metabolic enzymes, transporters, and drug target molecules (4). The voltage-gated sodium (Nav) channel is an important target of AEDs. In the brain, Nav channels consist of a 260-kDa α subunit and four β subunits (β1–β4) of 33–36 kDa; the α subunit is the functional subunit and plays an important role in the generation and propagation of neuronal action potentials (5, 6).
As key genes in the encoding of Nav channels, SCN1A and SCN2A are associated with the efficacy, dosage, and toxicity of multiple AEDs (3). SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 are common single-nucleotide polymorphisms (SNPs) of these genes, and their association with AED resistance has been widely explored. However, the results are still contradictory. For example, Feng et al. (7) found that SCN1A rs10188577 polymorphism was related to valproic acid (VPA) resistance in Chinese children with generalized epilepsy (P = 0.035). However, another study showed that the association between SCN1A rs10188577 polymorphism and response to sodium channel blockers (SCB; including phenytoin, carbamazepine, oxcarbazepine, lamotrigin, topiramate, and valproic acid) was only marginally significant in Caucasians with epilepsy (P = 0.049) (8). Moreover, several other studies did not support this correlation (5, 9–11). A similar scenario can be found with SCN1A rs2298771, SCN2A rs17183814, and SCN2A rs2304016 polymorphisms (5, 7, 9–26). In 2013, a meta-analysis performed by Haerian et al. did not find a correlation between SCN1A rs2298771 and SCN2A rs17183814 polymorphisms and AEDs resistance (10). Another meta-analysis showed that the A allele of the SCN1A rs2298771, and the AA genotype in particular, significantly affected the responsiveness to SCB-AEDs (27).
The aforementioned findings are inconsistent. Furthermore, to date, there has been no meta-analysis on the association of SCN1A rs10188577 and SCN2A rs2304016 polymorphisms with responsiveness to AEDs. Therefore, we conducted a meta-analysis to clarify the association between the aforementioned four gene polymorphisms and AED efficacy.
Methods
This meta-analysis adheres to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
Search Strategy
PubMed, Embase, Cochrane Library, WANFANG, and CNKI databases were searched by two reviewers (M.L. and R.Z.) independently for eligible studies published until June 2020. We used the following search terms: (sodium channel 1.1 OR SCN1A OR sodium channel neuronal type I alpha subunit OR SCN2A OR sodium channel neuronal type 2 alpha subunit) AND (epilepsy OR seizure) AND [anti-epileptic drug(s) OR antiepileptic drug(s) OR anticonvulsant drugs OR drug-resistant OR resistance OR resistant OR refractory OR response OR drug responsiveness].
Selection Criteria
The included studies met the following criteria: (1) Design: well-designed cohort or case–control study; (2) Participants: the case group includes individuals who were diagnosed with epilepsy and who showed AED resistance, and the control group includes PWE sensitive to AEDs; (3) Outcome: the original paper describes detailed genotype data including at least one of the target SNPs (SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, or SCN2A rs2304016); (4) the diagnosis of epilepsy is in conformity with the International League against Epilepsy (ILAE) guidelines; (5) the genotype distributions of the control group satisfy the Hardy–Weinberg equilibrium (HWE; P > 0.01); (6) If studies had duplicate or overlapping data, we chose the largest study. Studies with the following characteristics were excluded: (1) case reports, editorials, comments, reviews, abstracts, or meta-analyses; (2) the control group is composed of healthy people rather than of PWE sensitive to AEDs; (3) articles without complete information.
Data Extraction and Quality Assessment
Two reviewers independently extracted the data from the studies that met the aforementioned criteria and resolved the discrepancies in data extraction through discussion. Data extraction was as follows: first author, publication year, country, ethnicity, age, gender, types of epilepsy and AED therapy, definition of drug resistance and drug responsiveness, number of cases and controls, genotype and allele frequency, HWE principle, and the Newcastle–Ottawa Scale (NOS) score (28). We used NOS to assess the quality of each included study and defined studies with an NOS score ≥6 as high-quality research.
Statistical Analysis
We used Stata 12.0 software to perform data analysis. Pooled odds ratios (ORs) and 95% CIs were used to assess the association of SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 polymorphisms with responsiveness to AEDs. The combined ORs were determined for the allelic, homozygous, heterozygous, dominant, and recessive genetic models. The statistical significance of the combined ORs was tested using the Z-test; P < 0.05 was deemed significant. We assessed heterogeneity among the studies using the Cochran Q test and I2 test. I2 >50% or P < 0.05 was regarded as significant heterogeneity. When heterogeneity was significant, the random-effects model was applied to pool the results; otherwise, the fixed-effects model was applied. Subgroup analyses were performed to assess the correlations in different ethnicities [Asians, Caucasians (South Asians), and Caucasians (non-South Asians)]. To evaluate the stability of our results, we deleted one study at a time sequentially and calculated the combined ORs of the remaining studies. In addition, we evaluated publication bias by drawing a funnel plot, and further confirmed it using Begg or Egger tests; P < 0.05 was defined as significant publication bias. Finally, the Bonferroni method was applied for multiple comparisons. We evaluated four SNPs in this meta-analysis; thus, P < 0.0125 after Bonferroni correction was considered significant (29, 30).
Results
Study Inclusion and Characteristics
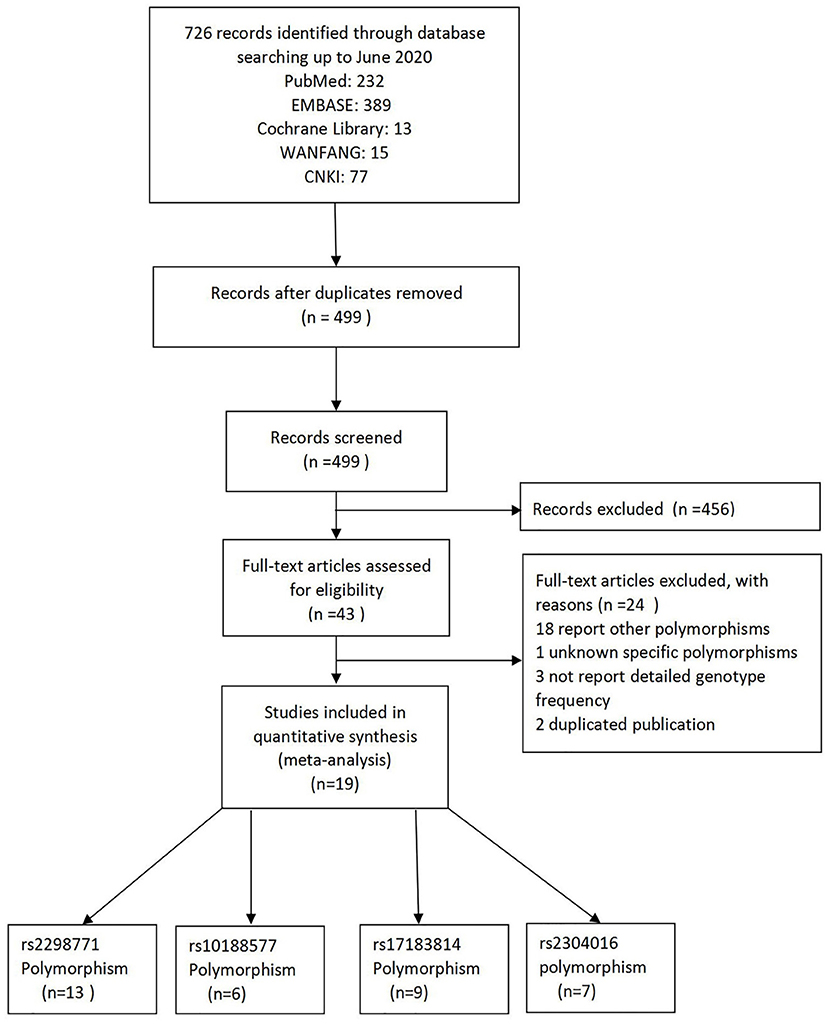
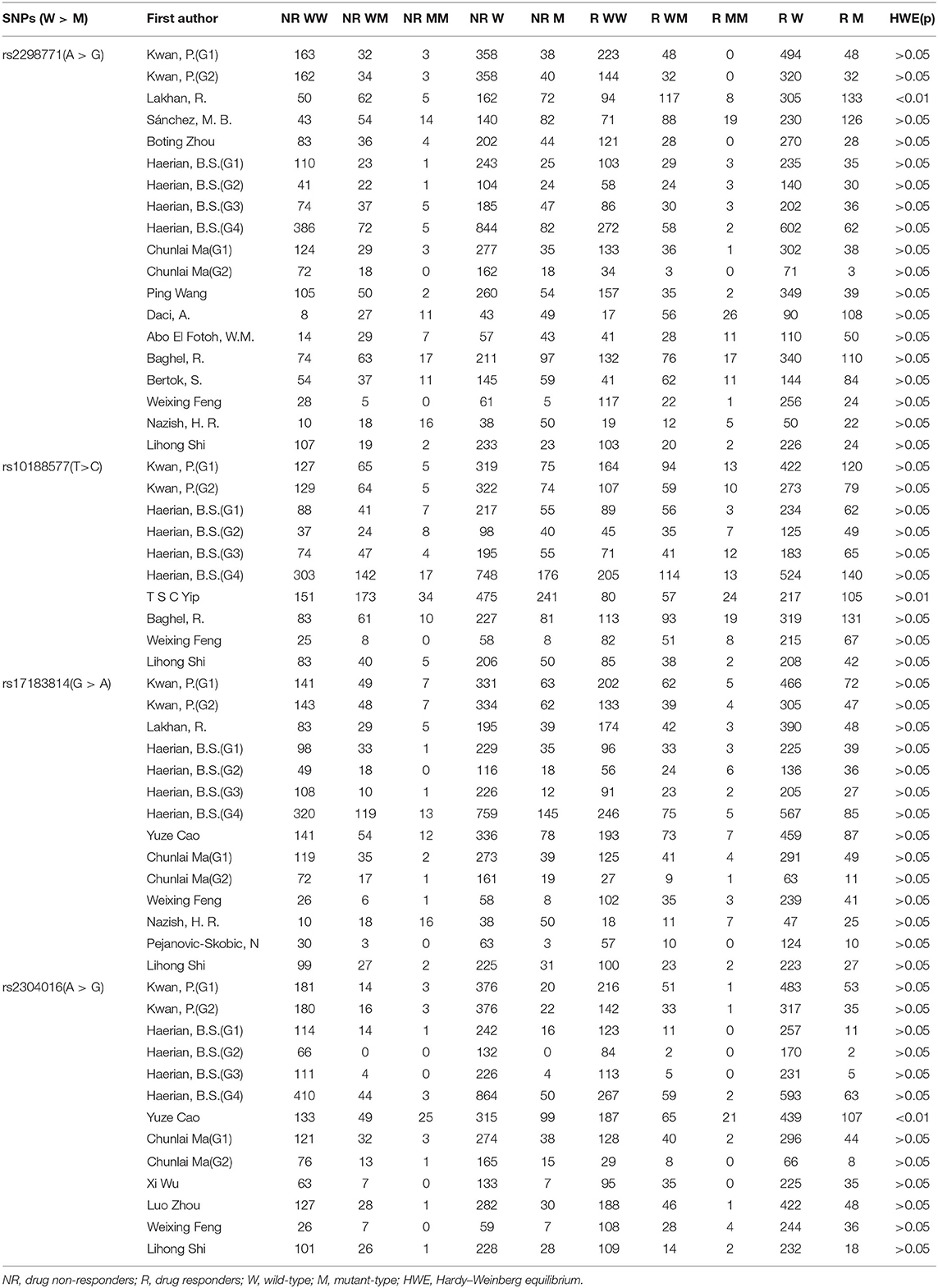
As a result of our retrieval strategy, a total of 726 records were identified. After eliminating 227 duplicate studies, we screened the titles and abstracts of the remaining 499 articles. A total of 456 studies were excluded based on the analysis of titles and abstracts. Then, full-text appraisal was performed for the remaining 43 articles, and 24 articles were excluded for different reasons. Of these, 18 articles reported other polymorphisms, 1 article did not explain specific polymorphisms, 3 articles did not provide detailed genotype frequency, and 2 articles had the same research data. Finally, 19 studies were included in our meta-analysis (5, 7–24). The screening process is shown in Figure 1. In addition to the one study that did not conform to the HWE principle, a total of 13 studies (18 groups) analyzed the SCN1A rs2298771 polymorphism; this represented 2,368 drug-resistant and 2,665 drug-responsive PWE. SCN1A rs10188577 was studied in 6 articles (10 groups), involving 1,860 drug-resistant and 1,790 drug-responsive PWE. Nine studies (14 groups) analyzed the SCN2A rs17183814 polymorphism, representing 1,973 drug-resistant and 2,172 drug-responsive PWE. Excluding one study that did not agree with the HWE principle, 7 studies (12 groups) assessed the SCN2A rs2304016 polymorphism; they consisted of 1,797 drug-resistant and 1,947 drug-responsive PWE. According to the Newcastle–Ottawa scale, the quality score of each included study was ≥6, indicating good quality overall. Table 1 displays the main characteristics and quality assessment results of the eligible studies. Table 2 shows SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 genotypes and allele distributions among drug non-responders and responders with epilepsy.
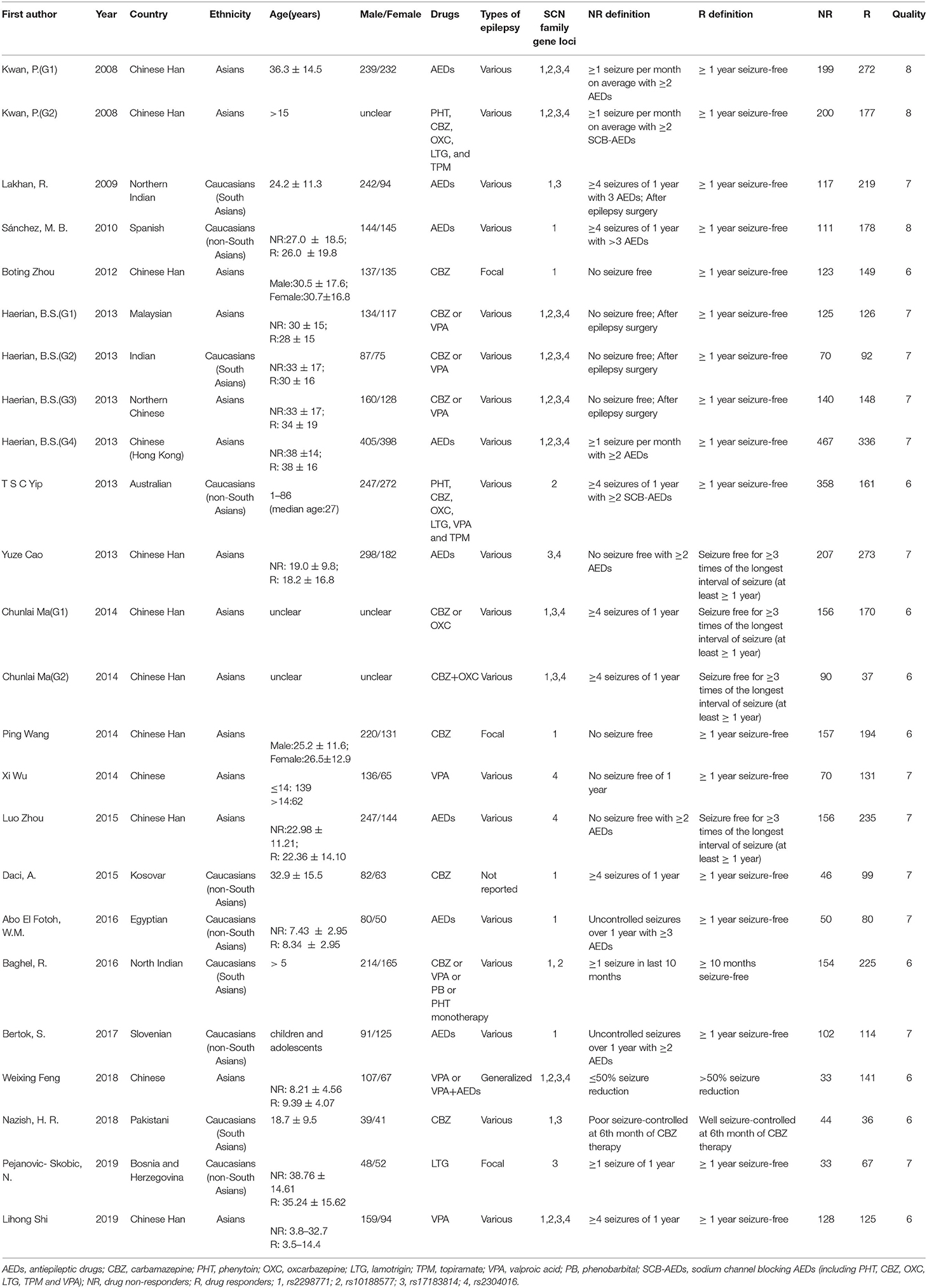
Table 1. Characteristics of studies on SCN1A and SCN2A polymorphisms and resistance to AEDs included in the meta-analysis.
Association Between the SCN1A rs2298771 Polymorphism and Resistance to AEDs
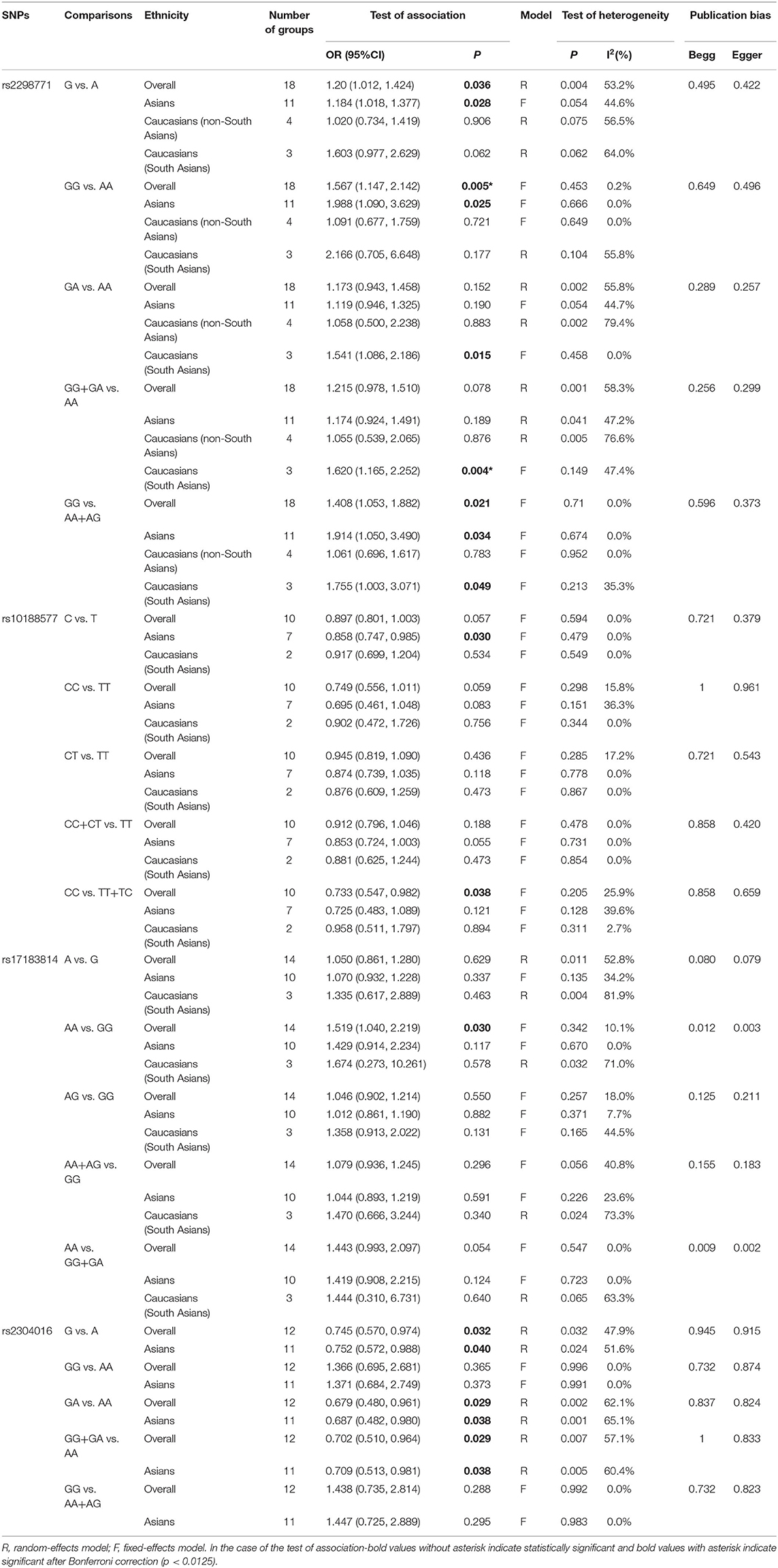
Comprehensive analysis showed a significant correlation between the SCN1A rs2298771 polymorphism and resistance to AEDs under the allelic, homozygous, and recessive genetic models (G vs. A: OR = 1.20, 95% CI: 1.012–1.424, P = 0.036, I2 = 53.2%; GG vs. AA: OR = 1.567, 95% CI: 1.147–2.142, P = 0.005, I2 = 0.2%; GG vs. AA + AG: OR = 1.408, 95% CI: 1.053–1.882, P = 0.021, I2 = 0.0%). Further subgroup analyses reported similar results among the Asians (G vs. A: OR = 1.184, 95% CI: 1.018–1.377, P = 0.028, I2 = 44.6%; GG vs. AA: OR = 1.988, 95% CI: 1.090–3.629, P = 0.025, I2 = 0.0%; GG vs. AA + AG: OR = 1.914, 95% CI: 1.050–3.490, P = 0.034, I2 = 0.0%). In addition, the correlation was also significant in Caucasians (South Asians) in the heterozygous, dominant, and recessive models (GA vs. AA: OR=1.541, 95% CI: 1.086–2.186, P = 0.015, I2 = 0.0%; GG + GA vs. AA: OR = 1.620, 95% CI: 1.165–2.252, P = 0.004, I2 = 47.4%; GG vs. AA + AG: OR = 1.755, 95% CI: 1.003–3.071, P = 0.049, I2 = 35.3%). However, no significant associations were found in Caucasians (non-South Asians). After Bonferroni correction, the associations between the SCN1A rs2298771 polymorphism and resistance to AEDs only remained significant in the overall population under the homozygous model and in Caucasians (South Asians) under the dominant model. Table 3 and Figure 2A display the results of the SCN1A rs2298771 polymorphism. The sensitivity analysis showed stable results (Figure 3A). Begg and Egger tests were performed to evaluate publication bias and indicated its absence (Table 3 and Figure 4A).
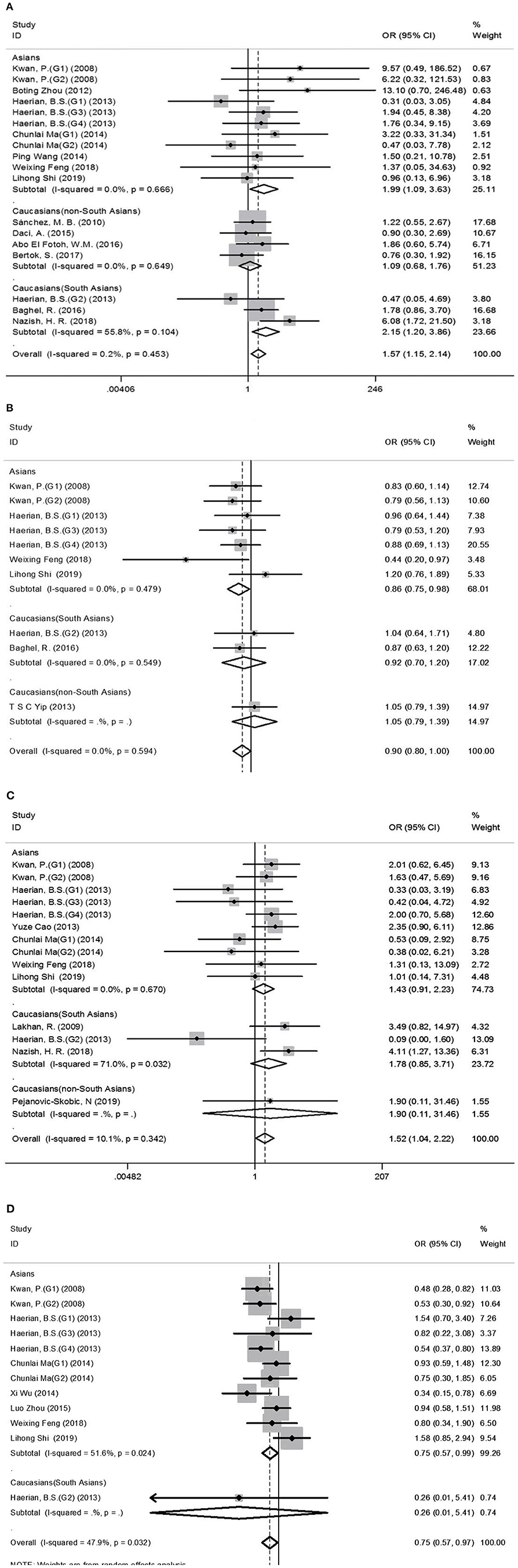
Figure 2. Forest plots for the association between the SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 polymorphisms and resistance to antiepileptic drugs. (A), SCN1A rs2298771 polymorphism (GG vs. AA). (B) SCN1Ars10188577 polymorphism (C vs. T). (C), SCN2A rs17183814 polymorphism (AA vs. GG). (D) SCN2A rs2304016 polymorphism (G vs. A). The sizes of the squares reflect the study's weight and horizontal lines represent 95% CI; the center of diamonds reflects the overall odds ratio (OR), and the horizontal span of the diamond represents the 95% CI of the OR.
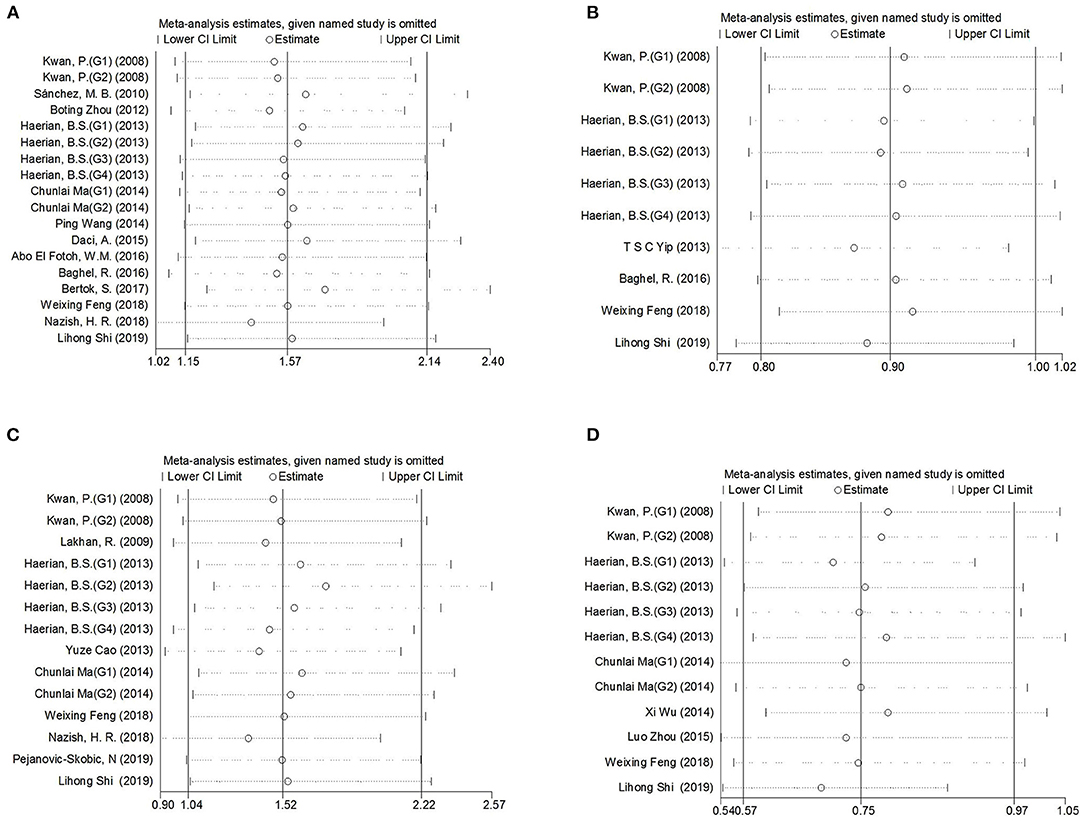
Figure 3. Sensitivity analysis indicated the stability of the results. (A) SCN1A rs2298771 polymorphism (GG vs. AA). (B) SCN1A rs10188577 polymorphism (C vs. T). (C), SCN2A rs17183814 polymorphism (AA vs. GG). (D), SCN2A rs2304016 polymorphism (G vs. A).
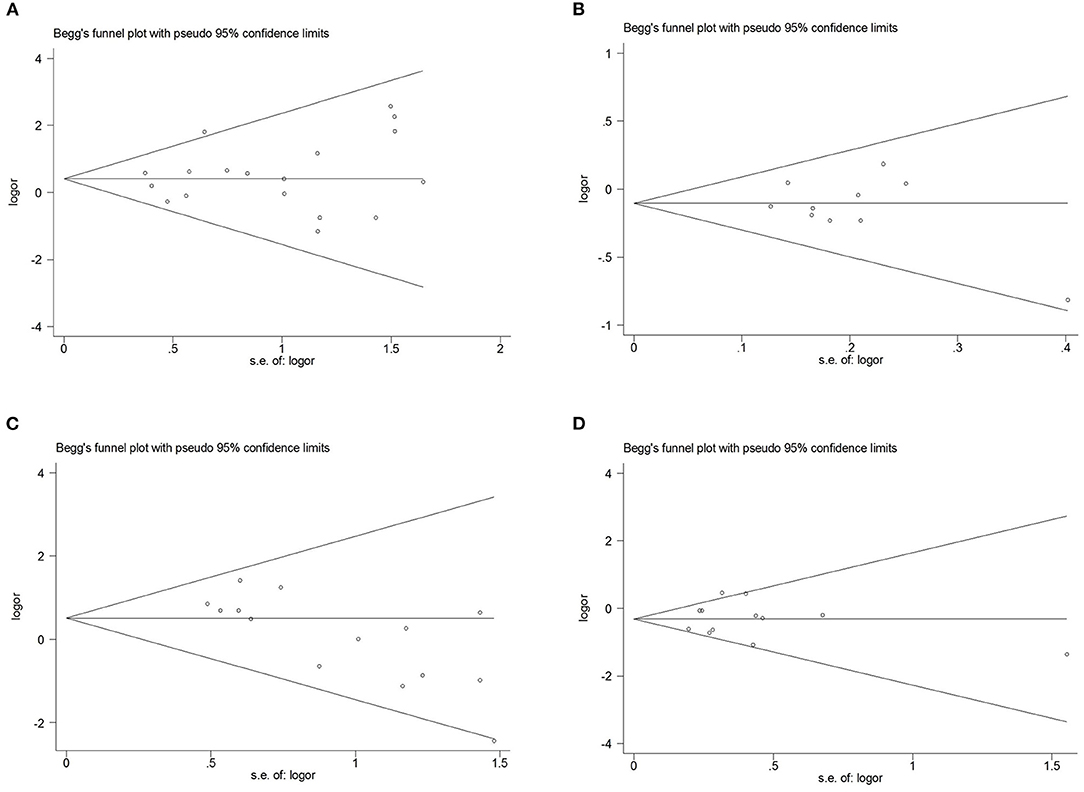
Figure 4. Begg's funnel plot for publication bias test. (A), SCN1A rs2298771 polymorphism (GG vs. AA). (B), SCN1A rs10188577 polymorphism (C vs. T). (C), SCN2A rs17183814 polymorphism (AA vs. GG). (D) SCN2A rs2304016 polymorphism (G vs. A).
Association Between the SCN1A rs10188577 Polymorphism and Resistance to AEDs
In the recessive genetic model, the CC genotype of the SCN1A rs10188577 was significantly associated with responsiveness to AEDs (OR = 0.733, 95% CI: 0.547–0.982, P = 0.038, I2 = 25.9%). Subgroup analyses indicated that the C allele was associated with sensitivity to AEDs in Asians with epilepsy in the allele model (OR = 0.858, 95% CI: 0.747–0.985, P = 0.030, I2 = 0.0%). However, after Bonferroni correction, there were no significant correlations between the SCN1A rs10188577 polymorphism and sensitivity to AEDs. These results are shown in Table 3 and Figure 2B. The results were stable and credible, as proved by the sensitivity analysis (Figure 3B). We did not detect any publication bias (Table 3 and Figure 4B).
Association Between the SCN2A rs17183814 Polymorphism and Resistance to AEDs
In the homozygous genetic model of the SCN2A rs17183814 polymorphism, the pooled OR value was 1.519 (95% CI: 1.040–2.219, P = 0.030) under the fixed-effects model. However, this correlation was not significant after Bonferroni correction. No association between the SCN2A rs17183814 polymorphism and resistance to AEDs was observed in the other four models or in further subgroup analyses. We found no significant heterogeneity (I2=10.1%; Table 3 and Figure 2C). The sensitivity analysis indicated stable results (Figure 3C). However, publication bias was observed according to Begg and Egger tests (Table 3 and Figure 4C). Therefore, these results should be applied cautiously.
Association Between the SCN2A rs2304016 Polymorphism and Resistance to AEDs
There was a significant correlation between the SCN2A rs2304016 polymorphism and AEDs response in the allelic, heterozygous, and dominant genetic models (G vs. A: OR = 0.745, 95% CI: 0.570–0.974, P = 0.032, I2 = 47.9%; GA vs. AA: OR = 0.679, 95% CI: 0.480–0.961, P = 0.029, I2 = 62.1%; GG + GA vs. AA: OR = 0.702, 95% CI: 0.510–0.964, P = 0.029, I2 = 57.1%). The subgroup analysis in Asians revealed similar results (G vs. A: OR = 0.752, 95% CI: 0.572–0.988, P = 0.040, I2 = 51.6%; GA vs. AA: OR = 0.687, 95% CI: 0.482–0.980, P = 0.038, I2 = 65.1%; GG + GA vs. AA: OR = 0.709, 95% CI: 0.513–0.981, P = 0.038, I2 = 60.4%; Table 3 and Figure 2D). After Bonferroni correction, the aforementioned correlation was not significant. The sensitivity analysis indicated stable results (Figure 3D). There was no significant publication bias (Table 3 and Figure 4D).
Discussion
The results of our meta-analysis showed that the G allele and the GG genotype of SCN1A rs2298771 increase the risk of resistance to AEDs for the overall population with epilepsy and for Asians with epilepsy. In addition, the AA genotype of SCN1A rs2298771 implies sensitivity to AEDs for Caucasians (South Asians). Regarding SCN1A rs10188577, the CC genotype for the overall population and the C allele for Asians reduce the risk of resistance to AEDs. The A allele and the AA genotype of SCN2A rs2304016 were more common in the overall population and Asians with drug-resistant epilepsy. The aforementioned results were stable and reliable after sensitivity analysis and Begg and Egger tests. Moreover, we found that the SCN2A rs17183814 polymorphism was related to resistance to AEDs under the homozygous genetic model, but there was significant publication bias, as shown by Begg and Egger tests; thus, this result was unreliable. Unfortunately, after Bonferroni correction, we found that only the AA genotype of SCN1A rs2298771 was associated with responsiveness to AEDs in all people and in Caucasians (South Asians), but the correlations between the other three SNPs and resistance to AEDs did not remain significant.
Epilepsy is known to be an ion channel disease. Nav channels are the main targets of many first-line AEDs (31–34). SCN1A and SCN2A genetic variants may change the response to AEDs (25). SCN1A rs2298771 (c.3184A>G/p.Thr1067Ala), located in the SCN1A exon region, is an A-to-G variant, which causes the substitution of alanine for threonine (27). Similarly, SCN2A rs17183814 (c.56G>A/p.Arg19Lys), situated in the SCN2A exon region, is a G-to-A SNP, which generates a conversion of arginine to lysine (12). These two SNPs may affect the structural and functional characteristics of Nav channels and further influence the therapeutic effects of AEDs (27). SCN1A rs10188577 (T>C) and SCN2A rs2304016 (IVS7-32A>G) are common SNPs of the intron region and are considered to be significant in the regulation of SCN1A and SCN2A expression, respectively (35).
The relationship between SCN1A and SCN2A polymorphisms and resistance to AEDs had been extensively studied, but the conclusions were inconsistent. Kwan et al. (9) found that the SCN1A rs2298771 polymorphism did not influence the response to AEDs. This finding was confirmed by later studies (5, 7, 10–15). However, some studies found that the SCN1A rs2298771 polymorphism was related to AED resistance (16–20). For example, E.F.W. Abo et al. reported that the AG genotype and the G allele frequency of SCN1A rs2298771 were significantly higher in AED-resistant patients than in drug responders (18). The SCN1A rs10188577 polymorphism was associated with AED resistance in studies performed by Feng et al. (7) and Yip et al. (8). However, several other studies did not support the aforementioned findings (5, 9–11). Lakhan et al. (12) found that A variant of SCN2A rs17183814 was more common in patients who were resistant to multiple AEDs. The study conducted by Nazish et al. (20) demonstrated that the GA and AA genotypes of rs17183814 were related to carbamazepine resistance. However, some studies did not find a similar relationship (5, 7, 9, 10, 14, 21, 24). Kwan et al. (9) and Li et al. (26) reported that the A allele and the AA genotype of SCN2A rs2304016 were risk factors for AED resistance. In contrast, Shi et al. (5) found that the rs2304016 G allele increased the risk of VPA resistance. Moreover, some studies have shown that the rs2304016 polymorphism does not affect the risk of AED resistance (7, 10, 14, 21–23, 25).
Recently, new case–control studies and cohort studies have been published. Hence, we performed an updated meta-analysis of the relationship between SCN1A rs2298771 and SCN2A rs17183814 polymorphisms and AED resistance. Furthermore, the study quantitatively evaluated the significance of SCN1A rs10188577 and SCN2A rs2304016 polymorphisms in AED resistance for the first time. The meta-analysis conducted by Haerian et al. did not find an association of SCN1A rs2298771 and SCN2A rs17183814 polymorphisms with response to AEDs (10). Our study, similar to this meta-analysis, did not limit the types of AEDs; however, we incorporated new studies (5, 7, 9, 11, 14–21, 24). Therefore, the conclusions of these two meta-analyses were not identical. Bao et al. (27) also performed a meta-analysis. However, they excluded studies that were not limited to SCB-AEDs; this did not happen in our study. Fortunately, their conclusions were similar to ours in that the AA genotype of SCN1A rs2298771 predisposed to responsiveness to AEDs. Thus, we speculate that the SCN1A rs2298771 polymorphism is related to resistance to multiple AEDs and not merely to SCB-AEDs.
There were certain limitations in our meta-analysis that should be considered while interpreting the results. First, the definition of treatment outcomes was different across the included studies, resulting in heterogeneity in grouping standards. Second, multivariate analysis to adjust for confounding factors, such as age, sex, type/duration/severity of epilepsy, and AEDs type, was not performed in the present meta-analysis because of the limited information offered by the included studies. Third, the Chinese were predominant among the Asians in the included studies, which constitutes a lack of ethnic diversity. Finally, we could not assess gene-to-gene and gene-to-environment interactions.
Conclusion
In conclusion, our meta-analysis demonstrated that SCN1A rs2298771 polymorphism was significantly associated with resistance to AEDs in overall population and Caucasians (South Asians). However, we could not identify significant correlations between SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 polymorphisms and resistance to AEDs according to current evidence. Considering the limitations of the present study, further large-scale and multi-ethnic studies on this topic are needed to verify our findings.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
ML, RZ, and WL conceived, designed, and drafted the article. ML, RZ, and YL reviewed the literatures and extracted the data. QZ and YL analyzed and interpreted data and edited pictures. All authors contributed to the critical revision of the article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate the authors of the primary studies included in our research.
References
1. Loscher W, Potschka H, Sisodiya SM, Vezzani A. Drug resistance in epilepsy: clinical impact, potential mechanisms, and new innovative treatment options. Pharmacol Rev. (2020) 72:606–38. doi: 10.1124/pr.120.019539
2. Thijs RD, Surges R, O'Brien TJ, Sander JW. Epilepsy in adults. Lancet. (2019) 393:689–701. doi: 10.1016/S0140-6736(18)32596-0
3. Lu Y, Su Q, Li M, Dayimu A, Dai X, Wang Z, et al. Association of SCN1A, SCN2A, and UGT2B7 polymorphisms with responsiveness to valproic acid in the treatment of epilepsy. Biomed Res Int. (2020) 2020:8096235. doi: 10.1155/2020/8096235
4. Markovic I, Pejanovic-Skobic N, Bozina N, Susak SI, Sporis D, Basic S. The lack of influence of IVS5-91 G>A polymorphism of the SCN1A gene on efficacy of lamotrigine in patients with focal epilepsy. Neurol Res. (2019) 41:930–5. doi: 10.1080/01616412.2019.1635321
5. Shi L, Zhu M, Li H, Wen Z, Chen X, Luo J, et al. SCN1A and SCN2A polymorphisms are associated with response to valproic acid in Chinese epilepsy patients. Eur J Clin Pharmacol. (2019) 75:655–63. doi: 10.1007/s00228-019-02633-0
6. Catterall WA, Kalume F, Oakley JC. NaV1.1 channels and epilepsy. J Physiol. (2010) 588(Pt 11):1849–59. doi: 10.1113/jphysiol.2010.187484
7. Feng W, Mei S, Zhu L, Yu Y, Yang W, Gao B, et al. Effects of UGT2B7, SCN1A and CYP3A4 on the therapeutic response of sodium valproate treatment in children with generalized seizures. Seizure. (2018) 58:96–100. doi: 10.1016/j.seizure.2018.04.006
8. Yip TS, O'Doherty C, Tan NC, Dibbens LM, Suppiah V. SCN1A variations and response to multiple antiepileptic drugs. Pharmacogenomics J. (2014) 14:385–9. doi: 10.1038/tpj.2013.43
9. Kwan P, Poon WS, Ng HK, Kang DE, Wong V, Ng PW, et al. Multidrug resistance in epilepsy and polymorphisms in the voltage-gated sodium channel genes SCN1A, SCN2A, and SCN3A: correlation among phenotype, genotype, and mRNA expression. Pharmacogenet Genomics. (2008) 18:989–98. doi: 10.1097/FPC.0b013e3283117d67
10. Haerian BS, Baum L, Kwan P, Tan HJ, Raymond AA, Mohamed Z. SCN1A, SCN2A and SCN3A gene polymorphisms and responsiveness to antiepileptic drugs: a multicenter cohort study and meta-analysis. Pharmacogenomics. (2013) 14:1153–66. doi: 10.2217/pgs.13.104
11. Baghel R, Grover S, Kaur H, Jajodia A, Rawat C, Srivastava A, et al. Evaluating the role of genetic variants on first-line antiepileptic drug response in North India: significance of SCN1A and GABRA1 gene variants in phenytoin monotherapy and its serum drug levels. CNS Neurosci Ther. (2016) 22:740–57. doi: 10.1111/cns.12570
12. Lakhan R, Kumari R, Misra UK, Kalita J, Pradhan S, Mittal B. Differential role of sodium channels SCN1A and SCN2A gene polymorphisms with epilepsy and multiple drug resistance in the north Indian population. Br J Clin Pharmacol. (2009) 68:214–20. doi: 10.1111/j.1365-2125.2009.03437.x
13. Sanchez MB, Herranz JL, Leno C, Arteaga R, Oterino A, Valdizan EM, et al. Genetic factors associated with drug-resistance of epilepsy: relevance of stratification by patient age and aetiology of epilepsy. Seizure. (2010) 19:93–101. doi: 10.1016/j.seizure.2009.12.004
14. Ma CL, Wu XY, Zheng J, Wu ZY, Hong Z, Zhong MK. Association of SCN1A, SCN2A and ABCC2 gene polymorphisms with the response to antiepileptic drugs in Chinese Han patients with epilepsy. Pharmacogenomics. (2014) 15:1323–36. doi: 10.2217/pgs.14.89
15. Daci A, Beretta G, Vllasaliu D, Shala A, Govori V, Norata GD, et al. Polymorphic variants of SCN1A and EPHX1 influence plasma carbamazepine concentration, metabolism and pharmacoresistance in a population of Kosovar Albanian epileptic patients. PLoS ONE. (2015) 10:e142408. doi: 10.1371/journal.pone.0142408
16. Boting Zhou. A study in the gene polymorphisms of SCN1A and GABA_ A receptor subunit and the antiepileptic efficacy and safety of carbamazepine (dissertation). Central South University, Changsha, China (2012).
17. Wang P, Zhou Q, Sheng Y, Tang B, Liu Z, Zhou B. Association between two functional SNPs of SCN1A gene and efficacy of carbamazepine monotherapy for focal seizures in Chinese Han epileptic patients. J Cent South Univ. (2014) 39:433–41. doi: 10.3969/j.issn.1672-7347.2014.05.001
18. Abo EFW, Abd ENS, Habib MS, ALrefai AA, Kasemy ZA. The potential implication of SCN1A and CYP3A5 genetic variants on antiepileptic drug resistance among Egyptian epileptic children. Seizure. (2016) 41:75–80. doi: 10.1016/j.seizure.2016.07.005
19. Bertok S, Dolzan V, Goricar K, Podkrajsek KT, Battelino T, Rener-Primec Z. The association of SCN1A p.Thr1067Ala polymorphism with epilepsy risk and the response to antiepileptic drugs in Slovenian children and adolescents with epilepsy. Seizure. (2017) 51:9–13. doi: 10.1016/j.seizure.2017.07.007
20. Nazish HR, Ali N, Ullah S. The possible effect of SCN1A and SCN2A genetic variants on carbamazepine response among Khyber Pakhtunkhwa epileptic patients, Pakistan. Ther Clin Risk Manag. (2018) 14:2305–13. doi: 10.2147/TCRM.S180827
21. Yuze Cao. Association of ABCC2, ABCB1, SCN1A, SCN2A, and GABRA1 polymorphisms with drug-resistant epilepsy in Chinese Han population (master's thesis). Central South University, Changsha, China (2013).
22. Xi Wu. Association of gene polymorphisms related to metabolism, transport and response pathway with the efficacy of sodium valproate in epilepsy (master's thesis). Central South University, Changsha, China (2014).
23. Zhou L, Cao Y, Long H, Long L, Xu L, Liu Z, et al. ABCB1, ABCC2, SCN1A, SCN2A, GABRA1 gene polymorphisms and drug resistant epilepsy in the Chinese Han population. Pharmazie. (2015) 70:416–20. doi: 10.1691/ph.2015.4849
24. Pejanovic-Skobic N, Markovic I, Bozina N, Basic S. Lack of association of SCN2A rs17183814 polymorphism with the efficacy of lamotrigine monotherapy in patients with focal epilepsy from Herzegovina area, Bosnia and Herzegovina. Epilepsy Res. (2019) 158:106221. doi: 10.1016/j.eplepsyres.2019.106221
25. Al-Eitan LN, Al-Dalalah IM, Aljamal HA. Effects of GRM4, SCN2A and SCN3B polymorphisms on antiepileptic drugs responsiveness and epilepsy susceptibility. Saudi Pharm J. (2019) 27:731–7. doi: 10.1016/j.jsps.2019.04.009
26. Li X, Zhang J, Wu X, Yan H, Zhang Y, He RH, et al. Polymorphisms of ABAT, SCN2A and ALDH5A1 may affect valproic acid responses in the treatment of epilepsy in Chinese. Pharmacogenomics. (2016) 17:2007–14. doi: 10.2217/pgs-2016-0093
27. Bao Y, Liu X, Xiao Z. Association between two SCN1A polymorphisms and resistance to sodium channel blocking AEDs: a meta-analysis. Neurol Sci. (2018) 39:1065–72. doi: 10.1007/s10072-018-3308-3
28. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z
29. Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. (2001) 125:279–84. doi: 10.1016/s0166-4328(01)00297-2
30. Goeman JJ, Solari A. Multiple hypothesis testing in genomics. Stat Med. (2014) 33:1946–78. doi: 10.1002/sim.6082
31. Alekov A, Rahman MM, Mitrovic N, Lehmann-Horn F, Lerche H. A sodium channel mutation causing epilepsy in man exhibits subtle defects in fast inactivation and activation in vitro. J Physiol. (2000) 529(Pt 3):533–9. doi: 10.1111/j.1469-7793.2000.00533.x
32. Vilin YY, Ruben PC. Slow inactivation in voltage-gated sodium channels: molecular substrates and contributions to channelopathies. Cell Biochem Biophys. (2001) 35:171–90. doi: 10.1385/CBB:35:2:171
33. Rogawski MA, Loscher W. The neurobiology of antiepileptic drugs. Nat Rev Neurosci. (2004) 5:553–64. doi: 10.1038/nrn1430
34. Xie X, Dale TJ, John VH, Cater HL, Peakman TC, Clare JJ. Electrophysiological and pharmacological properties of the human brain type IIA Na+ channel expressed in a stable mammalian cell line. Pflugers Arch. (2001) 441:425–33. doi: 10.1007/s004240000448
Keywords: antiepileptic drugs, responsiveness, resistance, polymorphisms, SCN1A, SCN2A
Citation: Li M, Zhong R, Lu Y, Zhao Q, Li G and Lin W (2021) Association Between SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 Polymorphisms and Responsiveness to Antiepileptic Drugs: A Meta-Analysis. Front. Neurol. 11:591828. doi: 10.3389/fneur.2020.591828
Received: 05 August 2020; Accepted: 27 November 2020;
Published: 14 January 2021.
Edited by:
Sherifa Ahmed Hamed, Assiut University Hospitals, EgyptReviewed by:
Rodrigo Secolin, Campinas State University, BrazilRam Lakhan, University of Maryland, Baltimore, United States
Copyright © 2021 Li, Zhong, Lu, Zhao, Li and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weihong Lin, lm065923@outlook.com
 Mengmeng Li
Mengmeng Li Weihong Lin
Weihong Lin