- 1Department of Human Genetics, McGill University, Montreal, QC, Canada
- 2Montreal Neurological Institute, McGill University, Montreal, QC, Canada
- 3Department of Neurology and Neurosurgery, McGill University, Montreal, QC, Canada
Restless legs syndrome (RLS) is a common sleep-related disorder for which the underlying biological pathways and genetic determinants are not well understood. The genetic factors so far identified explain less than 10% of the disease heritability. The first successful genome-wide association study (GWAS) of RLS was reported in 2007. This study identified multiple RLS associated risk variants including some within the non-coding regions of MEIS1. The MEIS1 GWAS signals are some of the strongest genetic associations reported for any common disease. MEIS1 belongs to the homeobox containing transcriptional regulatory network (HOX). Work in C. elegans showed a link between the MEIS1 ortholog and iron homeostasis, which is in line with the fact that central nervous system (CNS) iron insufficiency is thought to be a cause of RLS. Zebrafish and mice have been used to study the MEIS1 gene identifying an RLS-associated-SNP dependent enhancer activity from the highly conserved non-coding regions (HCNR) of MEIS1. Furthermore, this gene shows a lower expression of mRNA and protein in blood and thalamus of individuals with the MEIS1 RLS risk haplotype. Simulating this reduced MEIS1 expression in mouse models resulted in circadian hyperactivity, a phenotype compatible with RLS. While MEIS1 shows a strong association with RLS, the protein's function that is directly linked to an RLS biological pathway remains to be discovered. The links to iron and the enhancer activity of the HCNRs of MEIS1 suggest promising links to RLS pathways, however more in-depth studies on this gene's function are required. One important aspect of MEIS1's role in RLS is the fact that it encodes a homeobox containing transcription factor, which is essential during development. Future studies with more focus on the transcriptional regulatory role of MEIS1 may open novel venues for RLS research.
Introduction
Restless legs syndrome is a common neurological disorder. The prevalence of RLS cases based on the minimum diagnostic criteria of the international RLS study group (IRLSSG) was estimated between 3.9 and 14.3% of the adult population. It is more common in women than men and the prevalence increases with age in the European and North American populations (1). RLS is more prevalent in people with iron deficiency or kidney disease (2). Twin studies and a familial aggregation analysis estimated the heritability of RLS between 54.0 and 69.4%, thus there is a strong genetic element to the disease (3–5). RLS is a complex condition and environmental factors also contribute to its development.
The first attempts to identify RLS genetic risk factors used genome wide linkage (GWL) approaches in large multiplex families, most of which had an autosomal dominant mode of inheritance and one with autosomal recessive inheritance pattern. The GWL identified loci with large genomic regions, but no causative variant was identified by this approach and the results have often not been reproducible (5–12). Considering the complex nature of RLS, the genetic studies on RLS moved forward to association studies in search for common variants with low to moderate effect size.
Significance of MEIS1 in the RLS Genetics Studies
RLS is the first common sleep disorder for which genome wide association studies (GWAS) was performed and genetic risk loci identified. In 2007, the first genome-wide association study on RLS using 401 patients with familial RLS and 1,644 control individuals of German and French-Canadian origin identified common variants in three noncoding genomic regions (13). The strongest association signal found is a 32 kb linkage disequilibrium block in the intron 8 of MEIS1 gene (13). This association has been replicated in several follow up studies in both familial and sporadic RLS cases (odds ratio 1.92, 95% CI 1.85–1.99, p-value = 2.00E−280, from the latest report in 2017) (14–17). Furthermore, common genetic variants with low effect size were identified for RLS in 18 additional loci that each confer a small risk for the disease (17). MEIS1 is a homeobox transcription factor that belongs to the three amino acid loop extension (TALE) family of homeodomain proteins; it is known to have functions in hematopoiesis and vascular patterning (18, 19). This protein forms heterodimeric or heterotrimeric complexes with PBX or HOX proteins for higher DNA binding specificity and affinity (20). It also plays roles in neurodevelopment as well as the development of proximodistal limb axis, with high expression in dopaminergic neurons of substantia nigra and red nucleus (21–24).
Rare coding variants of MEIS1 were also proposed to contribute to be the cause of RLS. An Arg272-to-His (p.R272H) was found in one of 71 familial probands with RLS. However, a case-control genotyping study of this mutation across a North American cohort failed to validate this variant (25). In another study, an excess of rare null alleles specific to MEIS1 isoform 1 was observed in RLS cases compared to controls in a burden test on a German population (26). Lastly, in a study conducted by Xiong et al. the thirteen MEIS1 exons (and their respective splice junctions) were sequenced in 285 familial probands with a confirmed clinical diagnosis and no variants were identified (27). Hence coding variants are at most a very rare cause of RLS. This is not surprising as the gene is involved in many different developmental processes, so functional coding variants would likely have many additional manifestations, in addition to RLS.
Reduced MEIS1 Expression may Contribute to the Development of RLS
After the publication of the first GWAS on RLS, a subsequent study by Xiong et al. (27), used human lymphoblastoid cell lines (LCL) as well as two different brain regions (thalamus and pons) from RLS patients for an expression study. A q-RT-PCR followed by western blot analysis showed that the patients who harbor the MEIS1 risk haplotype (GG/GG, rs12469063–rs2300478) express lower levels of MEIS1 mRNA and protein in LCL and thalamus (Figure 1). The authors argued that lower MEIS1 expression in a subset of individuals can contribute to the development of RLS symptoms (27).
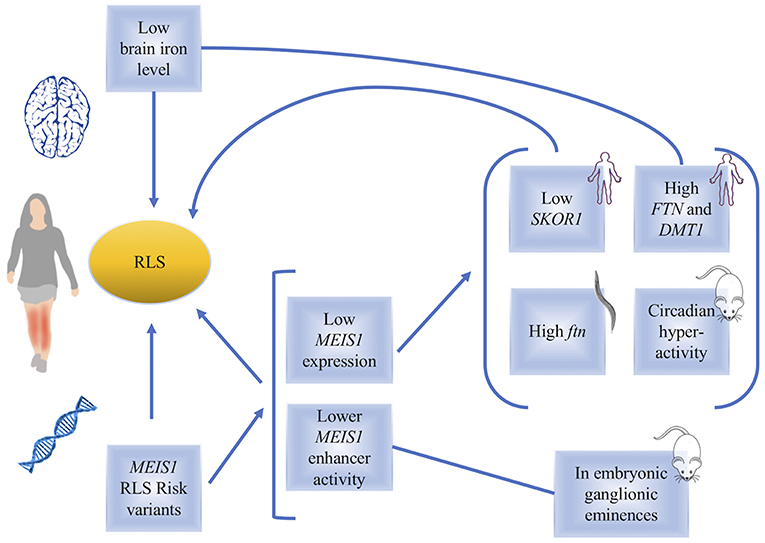
Figure 1. A diagram, summarizing the links between MEIS1 and restless legs syndrome. Low brain iron level or genetic variations in MEIS1 have been found in RLS patients. The consequences of low MEIS1 expression have been identified in human, C. elegans and mouse models (indicated in the figure).
Given the lower MEIS1 expression in a subgroup of patients, in vivo studies on mouse models with heterozygous Meis1 knockout were conducted (23, 28). Young male and female mice showed hyperactivity with no effect on anxiety-related behaviors, providing a potential animal model to study RLS (Figure 1). Furthermore, considering the age-related manifestation of RLS symptoms in human, effects of Meis1 haploinsufficiency was also studied in middle aged mice in a study by Salminen et al. (29). Meis1 haploinsufficiency was associated with a sex dependent increase in the activity more specific to the initial of the rest phase of animal, similar to the circadian rhythm of RLS symptoms observed in human patients. Effects on sensorimotor system in a sex dependent manner were also reported (29).
The Links Between MEIS1 and Iron Metabolism
Observations made using a Caenorhabditis elegans model showed that post developmental inactivation of Unc-62 (MEIS1 ortholog) makes it one of the 64 genes that increase the worm's life span. Unc-62 in the worms has 25% identity with MEIS1 and almost 80% identity with its homeodomain. Adding iron chelators in the culture media of the worms resulted in changes in the effect of Unc-62 inactivation on worms' lifespan, suggesting that Unc-62 is involved in iron metabolism. The link between Unc-62 and iron was also measured through its impact on the expression of ferritin. It was found that post developmental inactivation of Unc-62 using RNAi resulted in higher expression of ferritin, a protein that plays a key role in iron metabolism through its ability to store excess iron and to release it into a soluble and nontoxic form (30). Thalamus samples obtained from RLS patients also showed that individuals with the MEIS1 risk haplotype, who also have a reduced expression of MEIS1, show an increased expression of both ferritin (FTN) light and heavy chains. Furthermore, DMT1 gene expression was higher in these individuals. DMT1 is a proton-coupled metal transporter that carries iron from the extracellular domain to the cytoplasm (Figure 1). This finding also supports a link between MEIS1 and iron metabolism where DMT1 transports iron into the brain (31). More work remains to be done to clarify the exact role of MEIS1 in iron metabolism relevant to RLS (32). Recent studies suggest that DMT1 is expressed in endosomes of brain capillary endothelial cells denoting the blood-brain barrier (BBB) (33). The best well-established neurobiological abnormality in RLS is reduced brain iron, despite a normal peripheral iron level. The fact that some, but not all RLS patients, respond to intravenous (IV) iron provides an opportunity to interrogate the underlying pathways differing between responders versus non-responders. This led some studies to focus their attention toward the iron uptake at the blood-brain barrier (34). A preliminary analysis of MEIS1 expression showed elevated MEIS1 levels in the microvasculature isolated from RLS brain tissue by comparison to the tissues of control individuals. Moreover, a cell culture model of the BBB showed that treatment with an iron chelator increased the MEIS1 expression, while iron loading conversely decreased MEIS1 expression (unpublished data presented in a review article on the links between iron and RLS by Connor et al.; the small sample size in this report indicates that more investigations remain to be done to further validate this observation) (35, 36). These data suggest a novel role for MEIS1 in the BBB that warrants further examination. There are also observations revealing peripheral hypoxia to be associated with RLS symptoms (37). Hypoxia pathway is activated in a number of cell types of RLS patients; this activation can result from or be related to cellular iron deficiency (38). Another study used LCLs and showed MEIS1 down regulation by RNAi techniques resulted in an increase in transferrin-2 receptor and ferroportin and a decrease in hepcidin mRNA expression (39, 40). The authors suggest that MEIS1 might control cellular iron transfer to mitochondria and cellular export of iron (40). Putting together all these findings, the data suggest that decreased acquisition of iron by the brain cells is an RLS related pathophysiology, for which a possible role for MEIS1 can be accounted.
MEIS1 has an Allele Dependent Cis-regulatory Function in Telencephalon (a Study in Mice and Zebrafish)
A cluster of highly conserved non-coding regions (HCNRs) in the MEIS1 locus suggests the presence of cis-regulatory elements (23). Considering that most variants found by GWAS are located in the regulatory regions, Spieler et al. conducted a study to identify the cis-regulatory role of the common intronic variants in MEIS1 HCNRs (23). They studied an RLS associated variant (rs12469063), which is in the HCNR 617 of MEIS1, in transgenic mice and zebrafish using a reporter assay. They found that in mice, rs12469063 lies within a region of high interspecies conservation with neural enhancer activity and has an allele-specific functional impact. This study found that the risk allele of rs12469063 decreases the enhancer activity of this region in LGE and MGE (lateral and medial ganglionic eminences). The effect of rs12469063 on MEIS1 enhancer function in the LGE/MGE region suggests that RLS may involve the basal ganglia because these regions give rise to the basal ganglia [also discussed in a review by Salminen et al. (41)]. The enhancer activity of HCNR harboring the RLS associated rs12469063 happens in this mouse model during development, which suggests its predisposition to RLS occurs during embryonic development (Figure 1). Affinity chromatography showed that CREB1 has higher binding affinity to the RLS risk allele compared to the protective allele of rs12469063. A reporter screen in the zebrafish confirms the enhancer activity of HCNR observed in the mice and found two more transcriptionally active enhancers to this region. HCNR 617 harboring rs12469063 is the only RLS-SNP dependent enhancer region (23, 42).
MEIS1 Regulates SKOR1
Expression studies of the RLS associated loci of BTBD9, MAP2K5, and SKOR1 (previously called LBXCOR1) did not find changes in their levels of expression in lymphoblasts or two brain regions (pons and thalamus) of RLS patients (43). However, RLS patients with the MEIS1 risk haplotype were observed to have a reduced expression of SKOR1, in addition to a reduced expression of MEIS1 (Figure 1) (43). Follow up studies using siRNA targeting MEIS1, electromobility shift assays and luciferase reporter assays suggested the expression of SKOR1 to be under the regulatory control of MEIS1. This transcription factor action of MEIS1 is due to its direct binding on two distinct promoter regions of SKOR1 (43). Hence the dysregulation of MEIS1 might predispose to RLS both directly and indirectly, possibly throughout its regulatory role on other genes like SKOR1. A new SNP reported in this study is 8.7 kb upstream SKOR1 ATG start site, which acts as a regulatory SNP (rSNP). The risk allele in this locus reduces the binding affinity of MEIS1 to the SKOR1 promotor and results in reduced expression of SKOR1 (43). SKOR1 acts as the transcriptional corepressor of LBX1, a homeodomain transcription factor. Skor1 expression in the Mice embryonic CNS is present in a certain subset of post-mitotic neurons generated posterior to the midbrain-hindbrain border. Skor1 is selectively expressed in the dorsal horn interneurons of developing spinal cord in Mice, where Lbx1 is required for proper specification. It is suggested that SKOR1 probably mediates the sensory inputs of RLS, among others. Despite the importance of SKOR1 in RLS genetic, only little is known regarding the actual function of this gene in RLS underlying pathways. The current literature only suggests that MEIS1 dysregulation may causes SKOR1 dysregulation possibly leading to the sensory phenotypes of RLS (44).
MEIS1 and Other Sleep Related Disorders Like Insomnia, PLMS and RBD
Insomnia is characterized by problems in falling asleep or maintaining asleep. With a heritability estimate of 38 and 59% in men and women, respectively, genetic factors must play a crucial role in insomnia. Recent genetic studies of insomnia using cases from the UK biobank showed that MEIS1 has the strongest association signal, suggesting MEIS1 may be a shared genetic risk factor for RLS and insomnia (45–47). Some reports argue that the phenotype overlap could only drive some, and not all of the MEIS1's association with insomnia, thus suggesting that MEIS1 has a pleiotropic effect on RLS and insomnia (45). However, other reports suggest that the association of MEIS1 with insomnia only comes from the inclusion of RLS cases (48). Such inconsistencies might be due to the heterogeneous phenotypic definition of insomnia itself which can lead to the inclusion of a substantial number of RLS cases. Furthermore, GWAS on periodic leg movement during sleep (PLMS), which are present in approximately 80% of RLS cases, also shows association with MEIS1 (49–53). This pleiotropic effect can arise from MEIS1's wide expression pattern during the development (54). So far, no genetic links between RBD (REM sleep behavior disorder) and MEIS1 have been reported.
Conclusion
MEIS1 region is one of the several loci found to be associated with RLS genetic, which overall explain less than 10% of RLS heritability. The many roles for MEIS1 in development make the study of its role in RLS challenging. MEIS1 establishes motor neuron pool identity and their target-muscle connectivity (55), it also regulates the proximodistal limb axis development (21). This protein is highly expressed in dopaminergic neurons of the substantia nigra and red nucleus, though what it does in these cells remains unknown (22–24). Meanwhile, the biology of RLS is poorly understood, with the most consistent abnormality being altered iron homeostasis with brain iron deficiency (56–59). Data presented in this report suggest that the role of MEIS1 in RLS involves, among possibly other functions, altered iron homeostasis via altered transcriptional regulatory activity in RLS pathways. More in depth follow up studies on the function of MEIS1 with more focus on its regulatory role as a transcription factor might shed more light on the underlying pathways involved in RLS. To reach this goal, it will be essential to have access to RLS patients brain material carrying different genotypes of the RLS GWAS signals and to detailed clinical data to be used as covariates in the analyses. This combination would increase the likelihood of identifying elements that are critical to the onset and progression of RLS. This clinical data includes patient iron levels, their response to medication, age at onset, familial or sporadic RLS, presence or absence of PLMS, diagnosis by a physician and information about the patients' other health conditions. The next step could involve the use of model organisms to further validate and investigate RLS related mechanisms.
Author Contributions
FS contributed to the original concept of the article, did the literature search, wrote the original version of the manuscript and reviewed it as it progressed. PD and GR contributed to the original concept of the manuscript and reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Ohayon MM, O'Hara R, Vitiello MV. Epidemiology of restless legs syndrome: a synthesis of the literature. Sleep Med Rev. (2012) 16:283–95. doi: 10.1016/j.smrv.2011.05.002
2. Trenkwalder C, Allen R, Hogl B, Paulus W, Winkelmann J. Restless legs syndrome associated with major diseases: a systematic review and new concept. Neurology. (2016) 86:1336–43. doi: 10.1212/WNL.0000000000002542
3. Desai AV, Cherkas LF, Spector TD, Williams AJ. Genetic influences in self-reported symptoms of obstructive sleep apnoea and restless legs: a twin study. Twin Res. (2004) 7:589–95. doi: 10.1375/1369052042663841
4. Xiong L, Jang K, Montplaisir J, Levchenko A, Thibodeau P, Gaspar C, et al. Canadian restless legs syndrome twin study. Neurology. (2007) 68:1631–3. doi: 10.1212/01.wnl.0000261016.90374.fd
5. Chen S, Ondo WG, Rao S, Li L, Chen Q, Wang Q. Genomewide linkage scan identifies a novel susceptibility locus for restless legs syndrome on chromosome 9p. Am J Hum Genet. (2004) 74:876–85. doi: 10.1086/420772
6. Desautels A, Turecki G, Montplaisir J, Sequeira A, Verner A, Rouleau GA. Identification of a major susceptibility locus for restless legs syndrome on chromosome 12q. Am J Hum Genet. (2001) 69:1266–70. doi: 10.1086/324649
7. Bonati MT, Ferini-Strambi L, Aridon P, Oldani A, Zucconi M, Casari G. Autosomal dominant restless legs syndrome maps on chromosome 14q. Brain J Neurol. (2003) 126(Pt 6):1485–92. doi: 10.1093/Brain/Awg137
8. Levchenko A, Provost S, Montplaisir JY, Xiong L, St-Onge J, Thibodeau P, et al. A novel autosomal dominant restless legs syndrome locus maps to chromosome 20p13. Neurology. (2006) 67:900–1. doi: 10.1212/01.wnl.0000233991.20410.b6
9. Pichler I, Marroni F, Volpato CB, Gusella JF, Klein C, Casari G, et al. Linkage analysis identifies a novel locus for restless legs syndrome on chromosome 2q in a South Tyrolean population isolate. Am J Hum Genet. (2006) 79:716–23. doi: 10.1086/507875
10. Kemlink D, Plazzi G, Vetrugno R, Provini F, Polo O, Stiasny-Kolster K, et al. Suggestive evidence for linkage for restless legs syndrome on chromosome 19p13. Neurogenetics. (2008) 9:75–82. doi: 10.1007/s10048-007-0113-1
11. Levchenko A, Montplaisir JY, Asselin G, Provost S, Girard SL, Xiong L, et al. Autosomal-dominant locus for restless legs syndrome in French-Canadians on chromosome 16p12.1. Mov Disord Off J Mov Disord Soc. (2009) 24:40–50. doi: 10.1002/mds.22263
12. Jimenez-Jimenez FJ, Alonso-Navarro H, Garcia-Martin E, Agundez JAG. Genetics of restless legs syndrome: an update. Sleep Med Rev. (2018) 39:108–21. doi: 10.1016/j.smrv.2017.08.002
13. Winkelmann J, Schormair B, Lichtner P, Ripke S, Xiong L, Jalilzadeh S, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. (2007) 39:1000–6. doi: 10.1038/ng2099
14. Kemlink D, Polo O, Frauscher B, Gschliesser V, Hogl B, Poewe W, et al. Replication of restless legs syndrome loci in three European populations. J Med Genet. (2009) 46:315–8. doi: 10.1136/jmg.2008.062992
15. Winkelmann J, Czamara D, Schormair B, Knauf F, Schulte EC, Trenkwalder C, et al. Genome-wide association study identifies novel restless legs syndrome susceptibility loci on 2p14 and 16q12.1. PLoS Genet. (2011) 7:e1002171. doi: 10.1371/journal.pgen.1002171
16. Yang Q, Li L, Chen Q, Foldvary-Schaefer N, Ondo WG, Wang QK. Association studies of variants in MEIS1, BTBD9, and MAP2K5/SKOR1 with restless legs syndrome in a US population. Sleep Med. (2011) 12:800–4. doi: 10.1016/j.sleep.2011.06.006
17. Schormair B, Zhao C, Bell S, Tilch E, Salminen AV, Putz B, et al. Identification of novel risk loci for restless legs syndrome in genome-wide association studies in individuals of European ancestry: a meta-analysis. Lancet Neurol. (2017) 16:898–907. doi: 10.1016/S1474-4422(17)30327-7
18. Azcoitia V, Aracil M, Martinez AC, Torres M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev Biol. (2005) 280:307–20. doi: 10.1016/j.ydbio.2005.01.004
19. Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. (2004) 23:450–9. doi: 10.1038/sj.emboj.7600038
20. Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. (2006) 291:193–206. doi: 10.1016/J.Ydbio.2005.10.032
21. Mercader N, Leonardo E, Azpiazu N, Serrano A, Morata G, Martinez C, et al. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. (1999) 402:425–9. doi: 10.1038/46580
22. Barber BA, Liyanage VR, Zachariah RM, Olson CO, Bailey MA, Rastegar M. Dynamic expression of MEIS1 homeoprotein in E14.5 forebrain and differentiated forebrain-derived neural stem cells. Ann Anat. (2013) 195:431–40. doi: 10.1016/j.aanat.2013.04.005
23. Spieler D, Kaffe M, Knauf F, Bessa J, Tena JJ, Giesert F, et al. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. (2014) 24:592–603. doi: 10.1101/gr.166751.113
24. Mignot E. A step forward for restless legs syndrome. Nat Genet. (2007) 39:938–9. doi: 10.1038/ng0807-938
25. Vilarino-Guell C, Chai H, Keeling BH, Young JE, Rajput A, Lynch T, et al. MEIS1 p.R272H in familial restless legs syndrome. Neurology. (2009) 73:243–5. doi: 10.1212/WNL.0b013e3181ae7c79
26. Schulte EC, Kousi M, Tan PL, Tilch E, Knauf F, Lichtner P, et al. Targeted resequencing and systematic in vivo functional testing identifies rare variants in MEIS1 as significant contributors to restless legs syndrome. Am J Hum Genet. (2014) 95:85–95. doi: 10.1016/j.ajhg.2014.06.005
27. Xiong L, Catoire H, Dion P, Gaspar C, Lafreniere RG, Girard SL, et al. MEIS1 intronic risk haplotype associated with restless legs syndrome affects its mRNA and protein expression levels. Hum Mol Genet. (2009) 18:1065–74. doi: 10.1093/hmg/ddn443
28. Meneely S, Dinkins ML, Kassai M, Lyu S, Liu Y, Lin CT, et al. Differential dopamine D1 and D3 receptor modulation and expression in the spinal cord of two mouse models of restless legs syndrome. Front Behav Neurosci. (2018) 12:199. doi: 10.3389/fnbeh.2018.00199
29. Salminen AV, Garrett L, Schormair B, Rozman J, Giesert F, Niedermeier KM, et al. Meis1: effects on motor phenotypes and the sensorimotor system in mice. Dis Model Mech. (2017) 10:981–91. doi: 10.1242/dmm.030080
30. Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. (1996) 1275:161–203. doi: 10.1016/0005-2728(96)00022-9
31. Catoire H, Dion PA, Xiong L, Amari M, Gaudet R, Girard SL, et al. Restless legs syndrome-associated MEIS1 risk variant influences iron homeostasis. Ann Neurol. (2011) 70:170–5. doi: 10.1002/ana.22435
32. Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, et al. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. (1997) 388:482–8. doi: 10.1038/41343
33. Skjorringe T, Burkhart A, Johnsen KB, Moos T. Divalent metal transporter 1 (DMT1) in the brain: implications for a role in iron transport at the blood-brain barrier, and neuronal and glial pathology. Front Mol Neurosci. (2015) 8:19. doi: 10.3389/fnmol.2015.00019
34. Wade QW, Chiou B, Connor JR. Iron uptake at the blood-brain barrier is influenced by sex and genotype. Adv Pharmacol. (2019) 84:123–45. doi: 10.1016/bs.apha.2019.02.005
35. Connor JR, Patton SM, Oexle K, Allen RP. Iron and restless legs syndrome: treatment, genetics and pathophysiology. Sleep Med. (2017) 31:61–70. doi: 10.1016/j.sleep.2016.07.028
36. Gonzalez-Latapi P, Malkani R. Update on restless legs syndrome: from mechanisms to treatment. Curr Neurol Neurosci Rep. (2019) 19:54. doi: 10.1007/s11910-019-0965-4
37. Salminen AV, Rimpila V, Polo O. Peripheral hypoxia in restless legs syndrome (Willis-Ekbom disease). Neurology. (2014) 82:1856–61. doi: 10.1212/WNL.0000000000000454
38. Patton SM, Ponnuru P, Snyder AM, Podskalny GD, Connor JR. Hypoxia-inducible factor pathway activation in restless legs syndrome patients. Eur J Neurol. (2011) 18:1329–35. doi: 10.1111/j.1468-1331.2011.03397.x
39. N Silver RA, CJ Earley. MEIS1 as a potential mediator of the RLS-iron pathology. Mov Disord. (2010) 25:S513–4.
40. Guo S, Huang J, Jiang H, Han C, Li J, Xu X, et al. Restless legs syndrome: from pathophysiology to clinical diagnosis and management. Front Aging Neurosci. (2017) 9:171. doi: 10.3389/fnagi.2017.00171
41. Salminen AV, Lam DD, Winkelmann J. Role of MEIS1 in restless legs syndrome: from GWAS to functional studies in mice. Adv Pharmacol. (2019) 84:175–84. doi: 10.1016/bs.apha.2019.03.003
42. Allen RP, Donelson NC, Jones BC, Li Y, Manconi M, Rye DB, et al. Animal models of RLS phenotypes. Sleep Med. (2017) 31:23–8. doi: 10.1016/j.sleep.2016.08.002
43. Catoire H, Sarayloo F, Mourabit Amari K, Apuzzo S, Grant A, Rochefort D, et al. A direct interaction between two restless legs syndrome predisposing genes: MEIS1 and SKOR1. Sci Rep. (2018) 8:12173. doi: 10.1038/s41598-018-30665-6
44. Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y. Corl1, a novel neuronal lineage-specific transcriptional corepressor for the homeodomain transcription factor Lbx1. J Biol. Chem. (2005) 280:3645–55. doi: 10.1074/jbc.M411652200
45. Hammerschlag AR, Stringer S, de Leeuw CA, Sniekers S, Taskesen E, Watanabe K, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. (2017) 49:1584–92. doi: 10.1038/ng.3888
46. Lane JM, Jones SE, Dashti HS, Wood AR, Aragam KG, van Hees VT, et al. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet. (2019) 51:387–93. doi: 10.1038/s41588-019-0361-7
47. Lane JM, Liang J, Vlasac I, Anderson SG, Bechtold DA, Bowden J, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. (2017) 49:274–81. doi: 10.1038/ng.3749
48. El Gewely M, Welman M, Xiong L, Yin S, Catoire H, Rouleau G, et al. Reassessing GWAS findings for the shared genetic basis of insomnia and restless legs syndrome. Sleep. (2018) 41. doi: 10.1093/sleep/zsy164
49. Moore Ht, Winkelmann J, Lin L, Finn L, Peppard P, Mignot E. Periodic leg movements during sleep are associated with polymorphisms in BTBD9, TOX3/BC034767, MEIS1, MAP2K5/SKOR1, and PTPRD. Sleep. (2014) 37:1535–42. doi: 10.5665/sleep.4006
50. Stefansson H, Rye DB, Hicks A, Petursson H, Ingason A, Thorgeirsson TE, et al. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. (2007) 357:639–47. doi: 10.1056/NEJMoa072743
51. Winkelman JW, Blackwell T, Stone K, Ancoli-Israel S, Tranah GJ, Redline S, et al. Genetic associations of periodic limb movements of sleep in the elderly for the MrOS sleep study. Sleep Med. (2015) 16:1360–5. doi: 10.1016/j.sleep.2015.07.017
52. Haba-Rubio J, Marti-Soler H, Marques-Vidal P, Tobback N, Andries D, Preisig M, et al. Prevalence and determinants of periodic limb movements in the general population. Ann Neurol. (2016) 79:464–74. doi: 10.1002/ana.24593
53. Aurora RN, Kristo DA, Bista SR, Rowley JA, Zak RS, Casey KR, et al. The treatment of restless legs syndrome and periodic limb movement disorder in adults–an update for 2012: practice parameters with an evidence-based systematic review and meta-analyses: an American Academy of Sleep Medicine Clinical Practice Guideline. Sleep. (2012) 35:1039–62. doi: 10.5665/sleep.1988
54. Royo JL, Bessa J, Hidalgo C, Fernandez-Minan A, Tena JJ, Roncero Y, et al. Identification and analysis of conserved cis-regulatory regions of the MEIS1 gene. PLoS ONE. (2012) 7:e33617. doi: 10.1371/journal.pone.0033617
55. Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. (2005) 123:477–91. doi: 10.1016/j.cell.2005.09.009
56. Allen RP, Earley CJ. The role of iron in restless legs syndrome. Mov Disord Off J Mov Disord Soc. (2007) 22(Suppl 18):S440–8. doi: 10.1002/mds.21607
57. Ondo W, Romanyshyn J, Vuong KD, Lai D. Long-term treatment of restless legs syndrome with dopamine agonists. Arch Neurol. (2004) 61:1393–7. doi: 10.1001/archneur.61.9.1393
58. Godau J, Klose U, Di Santo A, Schweitzer K, Berg D. Multiregional brain iron deficiency in restless legs syndrome. Mov Disord Off J Mov Disord Soc. (2008) 23:1184–7. doi: 10.1002/mds.22070
Keywords: restless legs syndrome, MEIS1, iron, sleep disorders, neurogenetics
Citation: Sarayloo F, Dion PA and Rouleau GA (2019) MEIS1 and Restless Legs Syndrome: A Comprehensive Review. Front. Neurol. 10:935. doi: 10.3389/fneur.2019.00935
Received: 24 May 2019; Accepted: 12 August 2019;
Published: 28 August 2019.
Edited by:
Yue Huang, Beijing Tiantan Hospital, ChinaReviewed by:
Stefan Clemens, East Carolina University, United StatesHuifang Shang, Sichuan University, China
Copyright © 2019 Sarayloo, Dion and Rouleau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guy A. Rouleau, Z3V5LnJvdWxlYXVAbWNnaWxsLmNh
 Faezeh Sarayloo
Faezeh Sarayloo Patrick A. Dion
Patrick A. Dion Guy A. Rouleau
Guy A. Rouleau