
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neurol., 15 March 2017
Sec. Sleep Disorders
Volume 8 - 2017 | https://doi.org/10.3389/fneur.2017.00100
This article is part of the Research TopicIntrinsic ClocksView all 12 articles
Normal physiological functions require a robust biological timer called the circadian clock. When clocks are dysregulated, misaligned, or dampened, pathological consequences ensue, leading to chronic diseases and accelerated aging. An emerging research area is the development of clock-targeting compounds that may serve as drug candidates to correct dysregulated rhythms and hence mitigate disease symptoms and age-related decline. In this review, we first present a concise view of the circadian oscillator, physiological networks, and regulatory mechanisms of circadian amplitude. Given a close association of circadian amplitude dampening and disease progression, clock-enhancing small molecules (CEMs) are of particular interest as candidate chronotherapeutics. A recent proof-of-principle study illustrated that the natural polymethoxylated flavonoid nobiletin directly targets the circadian oscillator and elicits robust metabolic improvements in mice. We describe mood disorders and aging as potential therapeutic targets of CEMs. Future studies of CEMs will shed important insight into the regulation and disease relevance of circadian clocks.
The circadian clock is an intrinsic biological timing device operative in evolutionarily divergent species, ranging from microorganisms to human (1, 2). The clock drives daily oscillations of important molecular and physiological processes to anticipate and respond to the changing environment imposed by the rotation of the Earth. Consistent with its adaptive function, normal clock functions are required for organisms to survive and thrive. Coculture of cyanobacteria with varying period lengths demonstrated competitive growth advantage when inherent periodicity aligned with external light/dark rhythms (3), in accordance with findings from plant experiments (4). Likewise, circadian patterns of foraging and predator avoidance are well documented for animals in their natural habitats. For example, chipmunks whose central pacemaker, the hypothalamic suprachiasmatic nuclei (SCN), had been surgically removed suffered significantly higher mortality rate in the wild than those with fully functional clocks (5). The clock has also been postulated to protect early eukaryotes from irradiation during the day (6, 7). Despite the lack of acute lethality from genetic disruption of clock genes in laboratory animals, there exists a strong correlation, and in some cases causative relationship, between malfunctioning clocks and chronic diseases as well as aging (8, 9).
As we extend the list of clock-associated pathologies and probe for greater mechanistic understanding, the outstanding question remains whether and how to target the clock to combat disease and physiological decline (10–12). Except in the case of jet-lag, targeting the clock for health benefits will likely entail chronic intervention and gradual and systemic improvement of phenotypes and symptoms. Here, we highlight clock-associated metabolic disease, mood disorder, and aging as clock-associated processes characterized by dampened amplitude of circadian oscillation (13). Small-molecule enhancers of the circadian clock may strengthen the clock and clock-driven gene expression and physiology, retarding pathological deterioration. While this review will mainly focus on circadian amplitude enhancement, clock modulators capable of circadian phase and/or period modulation can show clinical utility in diseases states that are accompanied by circadian phase misalignment or abnormal periodicity (10, 14).
In the canonical mammalian clock, the molecular oscillator is the functional unit present in every cell of the body (15, 16). Comprised of interlocked feedback loops (Figure 1), molecular oscillators in individual tissues coordinate to govern highly tissue-specific expression programs of clock-controlled genes (CCGs). While 43% of genes have been shown to oscillate in at least one tissue in mice (17), indicating prevalent circadian gene regulation, the overlap of CCGs between tissues was found to be approximately 10% (18). At the system level, various tissue clocks are orchestrated by the SCN master pacemaker, a pair of neuron clusters bilaterally located in the anterior of the hypothalamus (19). The SCN displays tight coupling among its neurons (20) and functions to respond to photic signals to synchronize tissue and cellular clocks throughout the body via neural and hormonal signals.
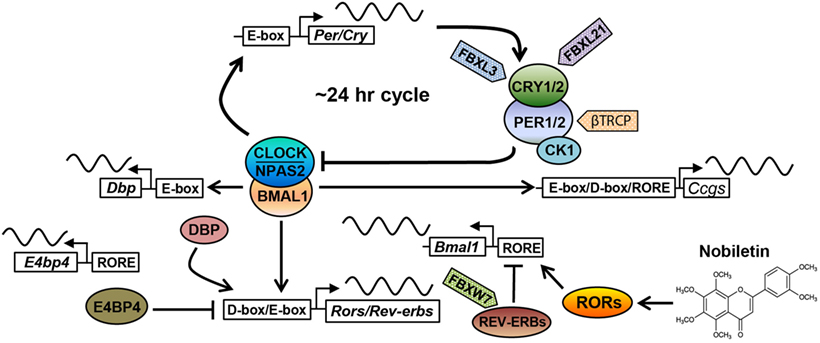
Figure 1. The core circadian oscillator and regulatory molecules. The circadian clock oscillator is comprised of a network of transcriptional–translational feedback loops including the core loop (BMAL1/CLOCK/NPAS2 and PERs/CRYs), the stabilization loop (BMAL1/CLOCK, REV-ERBs, and RORs), and the auxiliary loop (DBP, E4BP4, REV-ERBs, and RORs). Various protein regulators (F-box-containing E3 ligases are shown as examples) and small-molecule modulators (nobiletin is shown) have been identified to target core clock components, regulating circadian periodicity and amplitude. See the main text for details.
The molecular oscillator is composed of intersecting negative feedback loops to drive ~24-h gene expression rhythms (1). In the core loop, the positive arm consists of three bHLH-PAS transcription factors, including paralogous CLOCK/NPAS2 and their heterodimeric partner BMAL1 (Figure 1). CLOCK or NPAS2 each interacts with BMAL1 through the PAS and bHLH domains. After dimerization, CLOCK/BMAL1 and NPAS2/BMAL1 activate expression of Period (Per) and Cryptochrome (Cry) genes via E-box promoter elements. PER and CRY proteins themselves heterodimerize and translocate into the nucleus to inhibit transcriptional activities of CLOCK/BMAL1 and hence their own transcription. CRYs belong to the photosensing photolyase protein family that functions in DNA damage repair in bacteria and in circadian photic entrainment in flies (21). However, the mammalian CRY proteins appear to have lost the photosensing ability yet acquired function as the major transcriptional repressor in the circadian core loop. Crystal structure studies showed that CLOCK and BMAL1 interact via bHLH and two PAS domains in an asymmetrical fashion, characterized by a β-sheet/α-helix interaction involving respective BMAL1 and CLOCK PAS-B domains (22). On the β-sheet surface of CLOCK PAS-B, mutagenesis screen identified several residues whose mutations attenuated CRY inhibition of CLOCK/BMAL1 transactivation, suggesting CLOCK/CRY interactions. Several crystal structures of CRY proteins have been reported. The FAD-binding domain of CRY proteins appears to be a key nodal point recognized by both a CRY-stabilizing small molecule (23) or an CRY-degrading E3 ligase (24), and that PER binding to CRY precludes access for the E3 ligase FBXL3 (25), thus stabilizing CRY. Future structural studies of core clock complex formation on promoter DNA will advance our understanding of circadian oscillator function.
Several other feedback loops have been shown to stabilize and/or modulate the core feedback loop (Figure 1). In the principal stabilization loop, CLOCK/BMAL1 and NPAS2/BMAL1 activate highly cyclic expression of genes encoding the nuclear hormone receptors REV-ERBα/β (26). REV-ERBs and their antagonistic receptors RORα/β/γ compete for binding to shared consensus elements (RORE and RevDR2) on the promoter of Bmal1/Npas2 and other target genes throughout the genome to promote robust oscillatory gene expression (27–29). In another auxiliary loop (30), CLOCK/BMAL1 activates expression of genes encoding the PAR-bZip transcription factor DBP (D-box binding protein), which in turn drives Ror gene expression via their D-box promoter elements. In addition to Bmal1, REV-ERBs and RORs also govern the expression of the Nfil3 gene, which encodes a transcriptional suppressor (also known as E4BP4) that binds to the D-box to antagonize DBP transcriptional activity. Apart from these transcriptional feedback loops, other feedback mechanisms are also important, including a post-translational loop involving the NAD+-dependent sirtuin (SIRT) 1 deacetylase (31). CLOCK/BMAL1 activates the Nampt gene, which encodes the rate-limiting enzyme for NAD+ biosynthesis. The NAD+ level directly correlates with SIRT1 activity, which directly target core clock proteins including BMAL1 and PER2 (32, 33).
Degradation of core clock components has emerged as a key regulatory mode for circadian functions. Casein kinase 1 has been shown to phosphorylate PERs, thereby facilitating their proteasomal degradation by the F-box proteins β–TRCP1/2 (34). Likewise, the AMPK kinase phosphorylates CRYs to promote CRY degradation (35), mainly mediated by the F-box protein FBXL3 (36–38). FBXL21, a close homolog of FBXL3, was found to antagonize FBXL3 to decelerate CRY degradation in the nucleus, on the other hand, also accelerate CRY turnover in the cytoplasm (39, 40). Mice harboring hypomorphic mutations in Fbxl3 and Fbxl21 showed opposite effects on circadian period length, highlighting an important circadian function for ubiquitin-mediated proteasomal degradation. Autophagy is another major protein degradation mechanism, involving lysosomal degradation of protein cargo delivered via autophagosome (41). It was recently found that BMAL1 undergoes dual degradation by proteasome- and autophagosome-dependent pathways, and attenuation of both in ClockΔ19/+ heterozygous mice improves glucose homeostasis (42). Overall, the circadian clock system is regulated by an exceedingly complex array of molecular mechanisms encompassing all levels of gene expression, together ensuring temporal precision (~24 h) and oscillatory robustness (see below).
Amplitude denotes the robustness of circadian oscillation, measured by the difference between peak and trough of the circadian cycle. Whereas dampened circadian amplitude has been shown to closely correlate with chronic diseases and aging (10, 12, 43), the molecular and physiological mechanisms underlying circadian amplitude regulation are not well understood. Within the core oscillator, multiple lines of evidence indicated the importance of balancing positive vs. negative activities. For example, in mouse MEF cells, CLOCK/BMAL1 (positive factors) are in higher abundance than PER/CRY (the negative arm); as a result, overexpressing PER and CRY, but not CLOCK or BMAL1, strongly enhanced circadian amplitude (44). Such functional balance is further illustrated by the antagonistic transcriptional function of REV-ERBs and RORs in the secondary loop. Whereas ROR levels cycle only weakly, REV-ERB mRNA and protein levels are highly oscillatory. By directly competing for binding to promoter elements, they together govern a significant fraction of genome-wide circadian gene expression (29, 45). The clock is inherently a self-limiting, rhythmic machinery, namely, a limit cycle. Maintaining the “Yin–Yang” balance may lead to sustained oscillation, whereas brute force beyond a homeostatic range will dampen the overall amplitude of the following cycles. In other studies, CLOCK overexpression was found to enhance amplitude (46, 47), yet it remains unclear whether the primary mechanism involved is simply the greater level and activity of the positive transcription factor or an optimized functional balance.
More recent studies have provided insight into the functional complexity and dexterity of core clock components in amplitude regulation. In one study, REV-ERBα was found to be phosphorylated by cyclin-dependent kinase 1 (CDK1) at T275, a site not conserved in REV-ERBβ (48). Phosphorylated REV-ERBα was subsequently recognized by the F-box protein FBXW7 for proteasome degradation. Knockdown of CDK1 or FBXW7 reduced the amplitude of a circadian reporter in a dose-dependent manner, suggesting this REV-ERBα degradation pathway plays an important role in circadian amplitude. Another study described a “facilitated recruitment” mechanism where REV-ERBs are recruited to open chromatin following a rate-limiting step mediated by ROR/BMAL1 and transcription cofactors SRC-2/PBAF (49). It was posited that recruitment of the REV-ERB repressors by the activators ROR/BMAL1 ensures efficient and timely transcriptional shutdown, resulting in robust amplitude in target gene expression.
At intercellular and physiological/behavioral levels, oscillator coupling is of paramount importance to maintaining robust oscillation (50). The SCN rhythm is known to be exceptionally refractory to genetic perturbation compared with peripheral cells due to the tight coupling between SCN neurons (20). For example, several clock genes, including Per1 and Cry1, are required for sustained PER2:LUC reporter rhythms in dissociated fibroblast cells and SCN neurons. At the tissue level, whereas lung explants remained arrhythmic, SCN slices showed robust cycling of the PER2:LUC reporter. In accordance, Per1-null mice displayed clear rhythmic locomotor behavior, albeit with a short period length (51). These studies together indicate that intercellular synchronization between SCN neurons, likely involving vasoactive intestinal polypeptide (VIP) (48), strengthens system amplitude. Such coupling-induced rhythm stabilization can also be observed in peripheral cells, where single-cell reporter rhythms were less robust or stable compared with those in tissue slices (16, 52). Besides genetic perturbation, intercellular coupling can also confer protection against pharmacological disturbance and stochastic noise (53). Reciprocally, intercellular coupling can also facilitate noise-generated stochastic rhythm. While dispersed SCN neurons from Bmal1−/− mice showed no circadian rhythmicity, Bmal1−/− SCN slices displayed shorter and highly variable circadian rhythms (54). Such unstable rhythms were shown to be abolished by tetrodotoxin-induced uncoupling in the SCN slices, further indicating that intercellular coupling augments rhythmic stability and robustness.
More than half of top-selling drugs act on protein targets encoded by cyclically expressed genes (17), and xenobiotic metabolism is subjected to circadian regulation (55). These findings indicate a close circadian regulation of pharmacodynamics and pharmacokinetics (56–58). On the other hand, rather than aligning the timing of chronotherapy with intrinsic rhythms, a distinct strategy is to manipulate the clock or clock components to alleviate clock-regulated disease symptoms (10–12, 14). Behavioral or dietary manipulations have been shown to modulate circadian rhythms, such as light exposure (59–61), exercise (62) as well as feeding/fasting regimens (63). For example, a series of studies have shown that time-restricted feeding (TRF) can improve sleep and metabolic homeostasis and delay cardiac aging in Drosophila (13, 64) At the molecular level, TRF activates genes involved in circadian rhythms and mitochondrial electron transport chain complexes. Similarly, timed caloric restriction (CR) led to highly consolidated food intake, which enhanced the expression and amplitude of core clock genes and improved lipid homeostasis, eventually contributing to life span extension (63, 65). Finally, bright light and melatonin, both major circadian synchronizers that strengthen rhythms, have been shown to improve cognition and mood in the elderly (66). These studies exemplify the beneficial effects of enhancing the molecular and physiological rhythms on physiology and behavior.
Various chemical compounds capable of manipulating clocks have been discovered via either unbiased phenotypic screens or targeted approaches focusing on particular clock components (67–72). As described above, the clock is a self-limiting machine with a myriad of check-and-balance mechanisms governing its periodicity and robustness. Excessive functional manipulation, either stimulatory or inhibitory, of a specific clock protein may compromise the inherent balance within the clock, eventually diminishing or even abrogating the intended effects. Therefore, when searching for small molecules capable of enhancing circadian robustness, it is important to evaluate the sustained effects on reporter rhythms rather than assaying only the molecular function of individual clock components. Below, we describe our recent efforts to utilize phenotypic screening to identify chemical modifiers that enhance circadian amplitude.
In two separate screens using cell-based phenotypic assays, we reported a group of clock amplitude-enhancing small molecules dubbed CEMs. The first screen of 200,000, largely synthetic, compounds identified 4 CEMs that potentiated cellular and tissue reporter rhythms in both WT and ClockΔ19/+ heterozygous mutant backgrounds (73). In contrast to ClockΔ19/+ heterozygous cells that displayed attenuated but sustained circadian rhythms, ClockΔ19/Δ19 homozygous or Bmal1-null cells where the oscillators are essentially broken were refractory to CEM (14). CEM3, a benzimidazole compound, was uniquely able to further potentiate the robust reporter rhythms of the SCN pacemaker. In a second, smaller screen, a natural flavonoid compound called nobiletin (NOB) was identified as a novel CEM, along with its close analog tangeretin (74). NOB showed strong enhancing activities in circadian reporter cells, with an EC50 in the low micromolar range. NOB is a major polymethoxylated flavone found in citrus peels and exhibits a favorable pharmacokinetic profile devoid of significant toxicity (75). Previous studies have reported diverse biological activities against metabolic syndrome, oxidative stress, inflammation, and cancer (76–80); however, its molecular mechanism of action and direct protein targets were unknown.
A potential metabolic efficacy of NOB is intriguing and provides a focal point of connecting circadian manipulation and metabolic fitness. Previous research has established a regulatory role of the circadian clock in metabolic homeostasis (31). For example, the ClockΔ19/Δ19 mutant mice showed a broad array of metabolic dysfunctions, including blunted feeding rhythms, hyperphagia, exaggerated obesity risk under high-fat diet (HFD) feeding or at older ages, elevated blood glucose levels and hypoinsulinemia (81). Reciprocally, metabolism and/or nutrition also modulate our internal clocks (82, 83). For example, under ad libitum HFD feeding, mice showed a slight increase in the free-running period length (~23.8 h) compared with regular chow-fed animals (~23.6 h), and importantly a marked decrease in amplitude of circadian rhythms, including both clock gene oscillation in the periphery and feeding rhythms (82, 84). Both examples showed a correlation of circadian amplitude reduction and metabolic dysfunction, consistent with human studies where blunted insulin secretion rhythm associates with increased risk for diabetes (85).
We therefore examined the efficacy of NOB in two mouse metabolic disease models, namely the HFD-induced obese mice and db/db diabetic mice. Metabolic characterization illustrated that NOB effectively mitigated body weight gain without altering food intake, stimulated energy expenditure (EE) and circadian activity, enhanced glucose and insulin tolerance, and diminished lipid content in circulation and in liver (74). The alleviated liver steatosis phenotype was accompanied by restored oscillation of core clock components in mouse liver. In addition to energy homeostasis, NOB was also found to reduce serum ammonia levels in different diets and appeared to enhance urea cycle gene expression and function under HFD feeding (86). ClockΔ19/Δ19 homozygous mutant mice showed no or much diminished response to NOB, indicating clock requirement for NOB effects. Microarray analysis using mouse liver showed extensive remodeling of energy metabolic pathways including lipid metabolism and mitochondrial respiration. Together, these findings support the notion that clock enhancement by NOB contributes to metabolic improvement (87).
Importantly, NOB was found to directly activate ROR receptors via filter binding and functional studies including mammalian one-hybrid assays (74). This key finding highlights the role of RORs in circadian amplitude regulation and also sheds important insight on the functional complexity of NOB and ROR. First, despite the robust affinity of NOB–ROR interaction, the activation of ROR target genes, including core clock genes (e.g., Bmal1) and downstream output genes, was generally moderate (74). This observation is consistent with the limit cycle nature of the clock where the balance between positive and negative limbs is paramount to the overall amplitude. Second, a large number of ROR inverse agonists and REV-ERB agonists have been identified (71, 88). Despite opposite molecular functions relative to NOB as an ROR agonist, several of these compounds have been shown to improve energy metabolism in metabolic disease models (89, 90). This apparent paradox illustrates a potential functional dexterity of ROR (and also REV-ERB). It is possible that specific ligands, either agonists or antagonists, of ROR/REV-ERB can promote metabolic health, likely via distinct compound-specific mechanisms. A recent study (91) showed that three antagonists of RORγt employed divergent molecular mechanisms to affect its promoter binding and target gene expression and exhibited different degrees of mimicry with genetic RORγt disruption. These studies highlight the importance of in-depth mechanistic understanding of CEMs in circadian rhythms and downstream physiology.
Below we highlight two potential targets of CEMs, namely mood disorders and aging, where accumulating evidence indicates a strong correlation between pathophysiology and clock amplitude decline.
Mood disorders and circadian dysfunction are closely associated. Various manifestations of major mood disorders such as major depressive disorder, bipolar disorder, and seasonal affective disorder (BPD and SAD, respectively) exhibit diurnal rhythms, with the most severe symptoms typically occurring in the morning or around sunset (92, 93). In an early study comparing depressed, recovered, and healthy subjects, the depressed group exhibited blunted circadian rhythms, with a significant correlation to scores on depression severity (94). Recovered participants following 3 weeks of antidepressant treatment showed restored circadian amplitude, suggesting that depression is closely linked to circadian rhythmicity. In SAD patients suffering from depression during winter months with shorter daytime (95), circadian rhythms in feeding, sleep, body temperature, cortisol, and melatonin release, neurotransmitter (serotonin, norepinephrine, and dopamine) have been shown to be disturbed or dampened (96, 97). Another mood disorder is Sundowning syndrome, also referred to as “nocturnal delirium” (93). Sundowning syndrome is characterized by a worsening of behavior (i.e., aggression, restlessness, delirium, and agitation) in the late afternoon or early evening, particularly in the elderly population suffering from dementia. Clinical and preclinical data suggest that disturbances in sleep, environmental entrainment cues, and the SCN pacemaker all contribute to Sundowning syndrome (93). Specifically, sleep disruptions including impaired NREM sleep consolidation, sleep fragmentation, daytime sleeping, and reduced sleep efficiency are common among both the elderly and demented (98), and circadian amplitude disturbances manifested as sleep disruptions listed above can contribute to mood imbalance (99).
Mouse studies have begun to supply evidence for a possible causal relationship between clock function and mood. For example, behavioral assays using the ClockΔ19/Δ19 mice revealed manic-like behaviors similar to human bipolar mania (100), including hyperactivity, decreased sleep, hyperhedonia, and an increased preference for cocaine use. Disrupted circadian rhythms are also commonly found in human mania (94). More recently, the subcapsular cell hyperplasia associated with adrenal tissue remodeling was reported to enhance circadian amplitude of glucocorticoid rhythm, but not the total glucocorticoid levels (101). Interestingly, the enhanced stress hormone rhythm promotes anxiolytic function. It was postulated that the high-amplitude oscillation of the anxiogenic glucocorticoid, the descending phase in particular, endows a robust anxiolytic response to regulate mood balance.
Consistent with a close relationship between clock disruption and mood disorders, various treatment options are known to manipulate or enhance circadian and/or sleep cycles. Among the environmental therapies are bright light therapy, social rhythm therapy (SRT), and sleep deprivation. Bright light therapy is the treatment of choice for SAD and has also been applied to depression, bipolar disorder, and sleep–wake cycle disturbances (102). Bright light in the morning serves to advance the circadian phase to correct the phase delays commonly seen in SAD patients and may also function as a strong photic zeitgeber to improve daily rhythms. Likewise, SRT (103) entails social zeitgebers such as routine daily tasks to restore stability of biological rhythms in depression patients. Finally, a total sleep deprivation paradigm has also been developed to temporarily alleviate SAD symptoms. Its biological basis is not well understood, although it has been shown to impact neurotransmitter function and rapidly reset behavioral and circadian rhythms (104). Therefore, behavioral and environmental cues employed in these therapies reset and potentiate circadian rhythms, mainly at the behavioral levels, to counter the debilitating depressive tendency.
Various pharmacological agents have been used in mood disorders, including antidepressants, antimanic or mood-stabilizing drugs, and antipsychotics (Table 1). Lithium is a mood-stabilizing drug that has been used to treat bipolar disorder for more than 50 years. In addition to its mood-stabilizing effects, lithium has been reported to lengthen the free-running circadian period in mammals including hamsters and mice (105, 106). A potentially important target of lithium is GSK-3β (107), a kinase broadly acting in various signaling pathways. GSK-3β was previously shown to phosphorylate and stabilize REV-ERBα, and lithium treatment accelerated proteasomal degradation of REV-ERBα (108). More recently, lithium was found to activate Per2 gene expression and enhance the circadian reporter amplitude in both SCN and periphery (106). Another pharmacological treatment that affects the circadian system is valproic acid or valproate. Valproate is traditionally an anti-epileptic drug but has been repurposed as a mood-stabilizing drug. Valproate has been shown to alter circadian period (109) and acute valproate treatment of PER2:LUC bioluminescence experiments in skin fibroblasts yielded amplitude enhancement and induced phase-shifts, depending on the relative level of PER2:LUC protein expression (110). Previous mouse studies have also suggested antidepressive functions of NOB (111, 112) (Table 2). For example, NOB was found to improve mouse performance in forced swimming test and tail suspension tests, while pretreatments with drugs targeting monoaminergic systems disrupted the NOB effects (112). It will be interesting for future studies to investigate a role of circadian clocks in these NOB efficacies.
Gradual decline in metabolic, physiological, and behavioral functions with age leads to increasing risk of chronic disease and mortality (130). One physiological basis for such system-wide deterioration is age-related circadian attenuation (13, 43). Various clock-regulated physiological and behavioral processes are known to display reduced amplitude with age (43, 61, 131). For example, aging correlates with impaired rhythms in SCN firing rate, hormone secretion (e.g., cortisol and melatonin), and body temperature (132). Sleep fragmentation, characterized by multiple short periods of sleep episodes throughout the normal sleep phase and also sleep during the normal active phase, indicates amplitude dampening of the sleep/wake cycle and constitutes a well-documented characteristic of aging and various age-related diseases including Alzheimer’s disease (133). At the molecular level, there is also broad dysregulation of clock gene expression (61, 134, 135). Whereas peripheral clocks appear to suffer amplitude dampening (136, 137), the central clock neurons maintain robust molecular oscillation (135, 137). It is possible that cellular coupling and/or output pathways are compromised during aging, leading to systemic decline. In accordance, old age in both humans and mice is associated with delayed adaptation to phase shift cues (138, 139), suggesting that aging compromises circadian synchronization and weakens entraining response. Genetic studies have also provided evidence linking the clock and aging. The Bmal1-null mutant mice, exhibiting arrhythmic clock gene expression and defective clock-controlled physiological processes such as metabolism and activity (140, 141), suffered premature aging phenotypes such as sarcopenia, cataracts, and early mortality (142, 143). On the other hand, the αMUPA transgenic mice, as a long-living mouse model, displayed 24-h circadian periodicity regardless of age (144). These mice maintained robust behavioral and physiological rhythms, and core clock gene expression showed enhanced amplitude. Collectively, the evidence indicates that circadian robustness, involving both clock gene oscillation and systemic synchronization (145), may confer beneficial effects on life span and health span.
An established circadian output marker is melatonin (146), a sleep-regulating hormone in humans whose synthesis pathway is governed by the clock (147). Aging dampens the circadian peak (and amplitude) and daily total secretion of melatonin (148–150), contributing to lower sleep quality including decreased rapid eye movement, slow wave sleep, and increased stage 2 non-REM sleep in the elderly (151, 152).
Aging is associated with prevalent metabolic deterioration (130). For example, total EE declines during aging, as the elderly display diminished EE and gross energy intake (EI) compared with young adults (153). Such age-related energy imbalance, with EI > EE in the elderly and EI < EE in young adults, causes exaggerated body mass index during aging (154). Body temperature is a circadian output that shows a diurnal pattern with a dip during sleep (146, 155, 156). Thermogenesis plays a significant role in energy homeostasis, and age-related deterioration in energy homeostasis impairs circadian body temperature rhythm. For example, despite largely comparable basal body temperature, phase and amplitude of body temperature rhythm have been shown to significantly differ between the elderly and young- or middle-aged subjects (155, 157, 158). Liver and muscle play important roles in body temperature regulation, and attenuated skeletal muscle mass and mitochondrial function significantly contribute to dampened energy homeostasis and thermogenesis during aging (62, 157).
Caloric restriction universally prolongs life span (159). CR depletes white adipose tissue, especially the pro-inflammatory and diabetogenic visceral fat that accumulates over age (160). Timed CR leads to highly consolidated food intake within a few hours, enhancing the amplitude of circadian metabolic rhythms (63, 161) and core clock gene oscillation (65). CR involves several nutrient-sensing pathways including AMPK, AKT, and mTORC1, all of which have been reported to functionally interact with the clock (31, 42, 160, 161). In particular, the NAD+-dependent deacetylase SIRT proteins play important roles at the interface of energy homeostasis, clock, and aging (161, 162). Mammals express seven SIRT proteins (SIRT1–7), several of which have been implicated in circadian regulation of metabolism (32, 33, 163, 164). For example, SIRT1 directly deacetylates core clock components including BMAL1 and PER2, regulating their molecular function and CCG expression (32, 33). More recently, SIRT1 was found to interact with PGC-1α to control Clock and Bmal1 gene expression in the SCN, consequently regulating CLOCK/BMAL1 target genes (165). Various SIRT1-activating small molecules (e.g., resveratrol) have been shown to extend life span (166); resveratrol, in particular, has been shown to modulate physiological and behavioral rhythms and clock gene expression (167–169).
Circadian amplitude regulation and pharmacological modifiers are exciting research topics with promising translational potential. The list of CEMs will likely continue to grow, either from phenotypic screening, as in the case of NOB, or from targeted ligand development (14). On the other hand, pharmacological agents shown to target or mimic clock-enhancing pathways such as CR, TRF, and exercise are a rich venue for discovery of additional clock-targeting agents (63, 130, 161, 170). For example, a growing number of small molecules or drugs have been shown to extend life span and health span, including those deliberately designed to mimic CR and other manipulations (170, 171). Future studies should characterize their circadian clock effects and delineate molecular mechanisms.
Besides metabolic diseases, mood disorders, and aging, other chronic diseases such as neurodegenerative diseases (172, 173) have also been shown to correlate with dampened circadian amplitude or clock dysregulation and may represent new venues for studies of clock modifiers. In addition to antidepressive effects, several studies have shown neurological efficacies of NOB using transgenic disease models (Table 2). For example, 11-day oral administration of NOB resulted in an overall memory improvement in olfactory-bulbectomized (OBX) mice based on the step-through passive-avoidance task and the Y-maze test (111). OBX mice share clinical features with both human neurodegenerative diseases and major depression (174). The depression-like phenotype is thought to derive from pathological or compensatory mechanisms within the cortical–hippocampal–amygdala circuit, which typically involve deterioration of spine density and/or synaptic strength changes (175). Future studies are required to determine the specific role of circadian clocks and RORs in disease models.
Significant gaps of knowledge remain regarding circadian amplitude regulation, especially the mechanisms employed by CEMs. At the intracellular level, questions of particular interest include gene expression regulation, such as cofactor recruitment, epigenetic mechanisms, and chromosome dynamics (1). At the intercellular and system levels, other coupling molecules in addition to VIP and the communication between peripheral and central clocks are outstanding questions (50). It is conceivable that CEMs execute distinct mechanistic schemes to restore a robust overall output under disease or aging conditions. Exemplified by the complex and divergent ROR mechanisms when bound by distinct ligands (74, 90, 91), a detailed mechanistic understanding is important to fully exploit the therapeutic potential of individual CEMs.
In conclusion, circadian clocks safeguard physiological health, and dysregulated and dampened clocks can serve as therapeutic targets to mitigate disease symptoms. Exciting functional and mechanistic studies await to develop CEMs as novel preventive and therapeutic agents.
GG, S-HY, and ZC contributed to manuscript preparation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was in part supported by the Robert A. Welch Foundation (AU-1731) and NIH/NIA (R01AG045828) to ZC, and NIH/NIGMS (R01GM114424) to S-HY. GG is supported by R01AG045828-04S1.
1. Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet (2017) 18:164–79. doi:10.1038/nrg.2016.150
2. Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet (2005) 6:544–56. doi:10.1038/nrg1633
3. Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol (2004) 14:1481–6. doi:10.1016/j.cub.2004.08.023
4. Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science (2005) 309:630–3. doi:10.1126/science.1115581
5. DeCoursey PJ. Survival value of suprachiasmatic nuclei (SCN) in four wild sciurid rodents. Behav Neurosci (2014) 128:240–9. doi:10.1037/a0036696
6. Gehring W, Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J Mol Evol (2003) 57(Suppl 1):S286–9. doi:10.1007/s00239-003-0038-8
7. Chen Z, McKnight SL. A conserved DNA damage response pathway responsible for coupling the cell division cycle to the circadian and metabolic cycles. Cell Cycle (2007) 6:2906–12. doi:10.4161/cc.6.23.5041
8. Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet (2008) 9:764–75. doi:10.1038/nrg2430
9. Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science (2016) 354:994–9. doi:10.1126/science.aah4965
10. Schroeder AM, Colwell CS. How to fix a broken clock. Trends Pharmacol Sci (2013) 34:605–19. doi:10.1016/j.tips.2013.09.002
11. Wallach T, Kramer A. Chemical chronobiology: toward drugs manipulating time. FEBS Lett (2015) 589:1530–8. doi:10.1016/j.febslet.2015.04.059
12. Nohara K, Yoo SH, Chen ZJ. Manipulating the circadian and sleep cycles to protect against metabolic disease. Front Endocrinol (2015) 6:35. doi:10.3389/fendo.2015.00035
13. Manoogian EN, Panda S. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res Rev (2016) 1–9. doi:10.1016/j.arr.2016.12.006
14. Chen Z, Yoo SH, Takahashi JS. Small molecule modifiers of circadian clocks. Cell Mol Life Sci (2013) 70:2985–98. doi:10.1007/s00018-012-1207-y
15. Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell (1998) 93:929–37. doi:10.1016/S0092-8674(00)81199-X
16. Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A (2004) 101:5339–46. doi:10.1073/pnas.0308709101
17. Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A (2014) 111:16219–24. doi:10.1073/pnas.1408886111
18. Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, et al. Extensive and divergent circadian gene expression in liver and heart. Nature (2002) 417:78–83. doi:10.1038/nature744
19. Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH. Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb Perspect Biol (2017) 9:a027706. doi:10.1101/cshperspect.a027706
20. Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell (2007) 129:605–16. doi:10.1016/j.cell.2007.02.047
21. Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev (2003) 103:2203–37. doi:10.1021/cr0204348
22. Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, et al. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science (2012) 337:189–94. doi:10.1126/science.1222804
23. Nangle S, Xing W, Zheng N. Crystal structure of mammalian cryptochrome in complex with a small molecule competitor of its ubiquitin ligase. Cell Res (2013) 23:1417–9. doi:10.1038/cr.2013.136
24. Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, et al. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature (2013) 496:64–8. doi:10.1038/nature11964
25. Nangle SN, Rosensweig C, Koike N, Tei H, Takahashi JS, Green CB, et al. Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex. Elife (2014) 3:e03674. doi:10.7554/eLife.03674
26. Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol (2007) 3:630–9. doi:10.1038/nchembio.2007.37
27. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell (2002) 110:251–60. doi:10.1016/S0092-8674(02)00825-5
28. Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron (2004) 43:527–37. doi:10.1016/j.neuron.2004.07.018
29. Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, et al. Gene regulation. Discrete functions of nuclear receptor Rev-Erbalpha couple metabolism to the clock. Science (2015) 348:1488–92. doi:10.1126/science.aab3021
30. Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev (2001) 15:995–1006. doi:10.1101/gad.873501
32. Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell (2008) 134:317–28. doi:10.1016/j.cell.2008.06.050
33. Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell (2008) 134:329–40. doi:10.1016/j.cell.2008.07.002
34. Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol (2007) 8:139–48. doi:10.1038/nrm2106
35. Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science (2009) 326:437–40. doi:10.1126/science.1172156
36. Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science (2007) 316:900–4. doi:10.1126/science.1141194
37. Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science (2007) 316:897–900. doi:10.1126/science.1141138
38. Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell (2007) 129:1011–23. doi:10.1016/j.cell.2007.04.030
39. Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell (2013) 152:1091–105. doi:10.1016/j.cell.2013.01.055
40. Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell (2013) 152:1106–18. doi:10.1016/j.cell.2013.01.054
41. Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol (2010) 12:814–22. doi:10.1038/ncb0910-814
42. Jeong K, He B, Nohara K, Park N, Shin Y, Kim S, et al. Dual attenuation of proteasomal and autophagic BMAL1 degradation in Clock Delta19/+ mice contributes to improved glucose homeostasis. Sci Rep (2015) 5:12801. doi:10.1038/srep12801
43. Brown SA, Pagani L, Cajochen C, Eckert A. Systemic and cellular reflections on ageing and the circadian oscillator: a mini-review. Gerontology (2011) 57:427–34. doi:10.1159/000320673
44. Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem (2011) 286:7033–42. doi:10.1074/jbc.M110.207217
45. Takeda Y, Jothi R, Birault V, Jetten AM. RORgamma directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res (2012) 40:8519–35. doi:10.1093/nar/gks630
46. Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell (1997) 89:655–67. doi:10.1016/S0092-8674(00)80246-9
47. Allada R, Kadener S, Nandakumar N, Rosbash M. A recessive mutant of Drosophila Clock reveals a role in circadian rhythm amplitude. EMBO J (2003) 22:3367–75. doi:10.1093/emboj/cdg318
48. Zhao X, Hirota T, Han X, Cho H, Chong LW, Lamia K, et al. Circadian amplitude regulation via FBXW7-targeted REV-ERBalpha degradation. Cell (2016) 165:1644–57. doi:10.1016/j.cell.2016.05.012
49. Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, et al. Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol Cell (2015) 60:769–83. doi:10.1016/j.molcel.2015.10.024
50. Hogenesch JB, Herzog ED. Intracellular and intercellular processes determine robustness of the circadian clock. FEBS Lett (2011) 585:1427–34. doi:10.1016/j.febslet.2011.04.048
51. Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron (2001) 30:525–36. doi:10.1016/S0896-6273(01)00302-6
52. Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol (2004) 14:2289–95. doi:10.1016/j.cub.2004.11.057
53. van Ooijen G, Millar AJ. Non-transcriptional oscillators in circadian timekeeping. Trends Biochem Sci (2012) 37:484–92. doi:10.1016/j.tibs.2012.07.006
54. Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, et al. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol (2010) 8:e1000513. doi:10.1371/journal.pbio.1000513
55. Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab (2011) 13:125–37. doi:10.1016/j.cmet.2011.01.006
56. Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol (2007) 47:593–628. doi:10.1146/annurev.pharmtox.47.120505.105208
57. Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, et al. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A (2005) 102:3407–12. doi:10.1073/pnas.0409897102
58. Musiek ES, Fitzgerald GA. Molecular clocks in pharmacology. Handb Exp Pharmacol (2013) 217:243–60. doi:10.1007/978-3-642-25950-0_10
59. Belancio VP, Blask DE, Deininger P, Hill SM, Jazwinski SM. The aging clock and circadian control of metabolism and genome stability. Front Genet (2014) 5:455. doi:10.3389/fgene.2014.00455
60. Fonken LK, Nelson RJ. The effects of light at night on circadian clocks and metabolism. Endocr Rev (2014) 35:648–70. doi:10.1210/er.2013-1051
61. Banks G, Nolan PM, Peirson SN. Reciprocal interactions between circadian clocks and aging. Mamm Genome (2016) 27:332–40. doi:10.1007/s00335-016-9639-6
62. Schroder EA, Esser KA. Circadian rhythms, skeletal muscle molecular clocks, and exercise. Exerc Sport Sci Rev (2013) 41:224–9. doi:10.1097/JES.0b013e3182a58a70
63. Longo VD, Panda S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab (2016) 23:1048–59. doi:10.1016/j.cmet.2016.06.001
64. Gill S, Le HD, Melkani GC, Panda S. Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science (2015) 347:1265–9. doi:10.1126/science.1256682
65. Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, et al. Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab (2016) 23:143–54. doi:10.1016/j.cmet.2015.10.014
66. Riemersma-van der Lek RF, Swaab DF, Twisk J, Hol EM, Hoogendijk WJ, Van Someren EJ. Effect of bright light and melatonin on cognitive and noncognitive function in elderly residents of group care facilities: a randomized controlled trial. JAMA (2008) 299:2642–55. doi:10.1001/jama.299.22.2642
67. Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, et al. Identification of small molecule activators of cryptochrome. Science (2012) 337(6098):1094–7. doi:10.1126/science.1223710
68. Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A (2009) 106:15744–9. doi:10.1073/pnas.0908733106
69. Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature (2012) 485:62–8. doi:10.1038/nature11030
70. Antoch MP, Kondratov RV. Pharmacological modulators of the circadian clock as potential therapeutic drugs: focus on genotoxic/anticancer therapy. Handb Exp Pharmacol (2013) 217:289–309. doi:10.1007/978-3-642-25950-0_12
71. He B, Chen Z. Molecular targets for small-molecule modulators of circadian clocks. Curr Drug Metab (2016) 17:503–12. doi:10.2174/1389200217666160111124439
72. Trump RP, Bresciani S, Cooper AW, Tellam JP, Wojno J, Blaikley J, et al. Optimized chemical probes for REV-ERBalpha. J Med Chem (2013) 56:4729–37. doi:10.1021/jm400458q
73. Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A (2012) 109:101–6. doi:10.1073/pnas.1118034108
74. He B, Nohara K, Park N, Park YS, Guillory B, Zhao Z, et al. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab (2016) 23:610–21. doi:10.1016/j.cmet.2016.03.007
75. Evans M, Sharma P, Guthrie N. Bioavailability of citrus polymethoxylated flavones and their biological role in metabolic syndrome and hyperlipidemia. In: Noreddin A, editor. Readings in Advanced Pharmacokinetics – Theory, Methods and Applications. Rijeka, Croatia: InTech (2012). p. 1–19.
76. Lee YS, Cha BY, Choi SS, Choi BK, Yonezawa T, Teruya T, et al. Nobiletin improves obesity and insulin resistance in high-fat diet-induced obese mice. J Nutr Biochem (2013) 24:156–62. doi:10.1016/j.jnutbio.2012.03.014
77. Matsuzaki K, Miyazaki K, Sakai S, Yawo H, Nakata N, Moriguchi S, et al. Nobiletin, a citrus flavonoid with neurotrophic action, augments protein kinase A-mediated phosphorylation of the AMPA receptor subunit, GluR1, and the postsynaptic receptor response to glutamate in murine hippocampus. Eur J Pharmacol (2008) 578:194–200. doi:10.1016/j.ejphar.2007.09.028
78. Mulvihill EE, Assini JM, Lee JK, Allister EM, Sutherland BG, Koppes JB, et al. Nobiletin attenuates VLDL overproduction, dyslipidemia, and atherosclerosis in mice with diet-induced insulin resistance. Diabetes (2011) 60:1446–57. doi:10.2337/db10-0589
79. Walle T. Methoxylated flavones, a superior cancer chemopreventive flavonoid subclass? Semin Cancer Biol (2007) 17:354–62. doi:10.1016/j.semcancer.2007.05.002
80. Huang H, Li L, Shi W, Liu H, Yang J, Yuan X, et al. The multifunctional effects of nobiletin and its metabolites in vivo and in vitro. Evid Based Complement Alternat Med (2016) 2016:2918796. doi:10.1155/2016/2918796
81. Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science (2005) 308:1043–5. doi:10.1126/science.1108750
82. Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab (2007) 6:414–21. doi:10.1016/j.cmet.2007.09.006
83. Oosterman JE, Kalsbeek A, la Fleur SE, Belsham DD. Impact of nutrients on circadian rhythmicity. Am J Physiol Regul Integr Comp Physiol (2015) 308:R337–50. doi:10.1152/ajpregu.00322.2014
84. Pendergast JS, Branecky KL, Yang W, Ellacott KL, Niswender KD, Yamazaki S. High-fat diet acutely affects circadian organisation and eating behavior. Eur J Neurosci (2013) 37:1350–6. doi:10.1111/ejn.12133
85. Boden G, Chen X, Polansky M. Disruption of circadian insulin secretion is associated with reduced glucose uptake in first-degree relatives of patients with type 2 diabetes. Diabetes (1999) 48:2182–8. doi:10.2337/diabetes.48.11.2182
86. Nohara K, Shin Y, Park N, Jeong K, He B, Koike N, et al. Ammonia-lowering activities and carbamoyl phosphate synthetase 1 (Cps1) induction mechanism of a natural flavonoid. Nutr Metab (Lond) (2015) 12:23. doi:10.1186/s12986-015-0020-7
87. Bass J. Targeting time in metabolic therapeutics. Cell Metab (2016) 23:575–7. doi:10.1016/j.cmet.2016.03.011
88. Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov (2014) 13:197–216. doi:10.1038/nrd4100
89. Chang MR, He Y, Khan TM, Kuruvilla DS, Garcia-Ordonez R, Corzo CA, et al. Antiobesity effect of a small molecule repressor of RORgamma. Mol Pharmacol (2015) 88:48–56. doi:10.1124/mol.114.097485
90. Solt LA, Banerjee S, Campbell S, Kamenecka TM, Burris TP. ROR inverse agonist suppresses insulitis and prevents hyperglycemia in a mouse model of type 1 diabetes. Endocrinology (2015) 156:869–81. doi:10.1210/en.2014-1677
91. Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, et al. Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity (2014) 40:477–89. doi:10.1016/j.immuni.2014.04.004
92. McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther (2007) 114:222–32. doi:10.1016/j.pharmthera.2007.02.003
93. Bedrosian TA, Nelson RJ. Sundowning syndrome in aging and dementia: research in mouse models. Exp Neurol (2013) 243:67–73. doi:10.1016/j.expneurol.2012.05.005
94. Souetre E, Salvati E, Belugou JL, Pringuey D, Candito M, Krebs B, et al. Circadian rhythms in depression and recovery: evidence for blunted amplitude as the main chronobiological abnormality. Psychiatry Res (1989) 28:263–78. doi:10.1016/0165-1781(89)90207-2
95. Magnusson A, Boivin D. Seasonal affective disorder: an overview. Chronobiol Int (2003) 20:189–207. doi:10.1081/CBI-120019310
96. Partonen T, Lonnqvist J. Seasonal affective disorder. Lancet (1998) 352:1369–74. doi:10.1016/S0140-6736(98)01015-0
97. Lamont EW, Legault-Coutu D, Cermakian N, Boivin DB. The role of circadian clock genes in mental disorders. Dialogues Clin Neurosci (2007) 9:333–42.
98. Klaffke S, Staedt J. Sundowning and circadian rhythm disorders in dementia. Acta Neurol Belg (2006) 106:168–75.
99. Vandekerckhove M, Cluydts R. The emotional brain and sleep: an intimate relationship. Sleep Med Rev (2010) 14:219–26. doi:10.1016/j.smrv.2010.01.002
100. Roybal K, Theobold D, Graham A, DiNieri JA, Russo SJ, Krishnan V, et al. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci U S A (2007) 104:6406–11. doi:10.1073/pnas.0609625104
101. Ikeda Y, Kumagai H, Skach A, Sato M, Yanagisawa M. Modulation of circadian glucocorticoid oscillation via adrenal opioid-CXCR7 signaling alters emotional behavior. Cell (2013) 155:1323–36. doi:10.1016/j.cell.2013.10.052
102. Pail G, Huf W, Pjrek E, Winkler D, Willeit M, Praschak-Rieder N, et al. Bright-light therapy in the treatment of mood disorders. Neuropsychobiology (2011) 64:152–62. doi:10.1159/000328950
103. Frank E, Swartz HA, Kupfer DJ. Interpersonal and social rhythm therapy: managing the chaos of bipolar disorder. Biol Psychiatry (2000) 48:593–604. doi:10.1016/S0006-3223(00)00969-0
104. Dallaspezia S, Suzuki M, Benedetti F. Chronobiological therapy for mood disorders. Curr Psychiatry Rep (2015) 17:95. doi:10.1007/s11920-015-0633-6
105. LeSauter J, Silver R. Lithium lengthens the period of circadian rhythms in lesioned hamsters bearing SCN grafts. Biol Psychiatry (1993) 34:75–83. doi:10.1016/0006-3223(93)90259-G
106. Li J, Lu WQ, Beesley S, Loudon AS, Meng QJ. Lithium impacts on the amplitude and period of the molecular circadian clockwork. PLoS One (2012) 7:e33292. doi:10.1371/journal.pone.0033292
107. Gould TD, Manji HK. Glycogen synthase kinase-3: a putative molecular target for lithium mimetic drugs. Neuropsychopharmacology (2005) 30:1223–37. doi:10.1038/sj.npp.1300731
108. Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-Erbalpha is a critical lithium-sensitive component of the circadian clock. Science (2006) 311:1002–5. doi:10.1126/science.1121613
109. Landgraf D, Joiner WJ, McCarthy MJ, Kiessling S, Barandas R, Young JW, et al. The mood stabilizer valproic acid opposes the effects of dopamine on circadian rhythms. Neuropharmacology (2016) 107:262–70. doi:10.1016/j.neuropharm.2016.03.047
110. Johansson AS, Brask J, Owe-Larsson B, Hetta J, Lundkvist GB. Valproic acid phase shifts the rhythmic expression of Period2:Luciferase. J Biol Rhythms (2011) 26:541–51. doi:10.1177/0748730411419775
111. Nagase H, Omae N, Omori A, Nakagawasai O, Tadano T, Yokosuka A, et al. Nobiletin and its related flavonoids with CRE-dependent transcription-stimulating and neuritegenic activities. Biochem Biophys Res Commun (2005) 337:1330–6. doi:10.1016/j.bbrc.2005.10.001
112. Yi L-T, Xu H-L, Feng J, Zhan X, Zhou L-P, Cui C-C. Involvement of monoaminergic systems in the antidepressant-like effect of nobiletin. Physiol Behav (2011) 102:1–6. doi:10.1016/j.physbeh.2010.10.008
113. Moriya S, Tahara Y, Sasaki H, Hamaguchi Y, Kuriki D, Ishikawa R, et al. Effect of Quetiapine on Per1, Per2, and Bmal1 Clock gene expression in the mouse amygdala and hippocampus. J Pharm Sci (2014) 125:329–32. doi:10.1254/jphs.14071SC
114. Klemfuss H, Kripke DF. Antimanic drugs stabilize hamster circadian rhythms. Psychiatry Res (1995) 57:215–22. doi:10.1016/0165-1781(95)02687-R
115. Schaufler J, Ronovsky M, Savalli G, Cabatic M, Sarton SB, Singewald N, et al. Fluoxetine normalizes disrupted light-induced entrainment, fragmented ultradian rhythms and altered hippocampal clock gene expression in an animal model of high trait anxiety- and depression-related behavior. Ann Med (2016) 48:17–27. doi:10.3109/07853890.2015.1122216
116. Kiryanova V, Smith VM, Dyck RH, Antle MC. Circadian behavior of adult mice exposed to stress and fluoxetine during development. Psychopharmacology (2016) 234:793–804. doi:10.1007/s00213-016-4515-3
117. Possidente B, Lumia AR, McEldowney S, Rapp M. Fluoxetine shortens circadian period for wheel running activity in mice. Brain Res Bull (1992) 28:629–31. doi:10.1016/0361-9230(92)90114-D
118. Loo H, Hale A, D’Haenen H. Determination of the dose of agomelatine, a melatoninergic agonist and selective 5-HT(2C) antagonist, in the treatment of major depressive disorder: a placebo-controlled dose range study. Int Clin Psychopharmacol (2002) 17:239–47. doi:10.1097/00004850-200209000-00004
119. Kennedy SH, Emsley R. Placebo-controlled trial of agomelatine in the treatment of major depressive disorder. Eur Neuropsychopharmacol (2006) 16:93–100. doi:10.1016/j.euroneuro.2005.09.002
120. Olie JP, Kasper S. Efficacy of agomelatine, a MT1/MT2 receptor agonist with 5-HT2C antagonistic properties, in major depressive disorder. Int J Neuropsychopharmacol (2007) 10:661–73. doi:10.1017/S1461145707007766
121. Pitrosky B, Kirsch R, Malan A, Mocaer E, Pevet P. Organization of rat circadian rhythms during daily infusion of melatonin or S20098, a melatonin agonist. Am J Physiol (1999) 277:R812–28.
122. Leproult R, Van Onderbergen A, L’Hermite-Baleriaux M, Van Cauter E, Copinschi G. Phase-shifts of 24-h rhythms of hormonal release and body temperature following early evening administration of the melatonin agonist agomelatine in healthy older men. Clin Endocrinol (2005) 63:298–304. doi:10.1111/j.1365-2265.2005.02341.x
123. Courtet P, Olie E. Circadian dimension and severity of depression. Eur Neuropsychopharmacol (2012) 22:S476–81. doi:10.1016/j.euroneuro.2012.07.009
124. Roth T, Seiden D, Sainati S, Wang-Weigand S, Zhang J, Zee P. Effects of ramelteon on patient-reported sleep latency in older adults with chronic insomnia. Sleep Med (2006) 7:312–8. doi:10.1016/j.sleep.2006.01.003
125. Rajaratnam S, Polymeropoulos M, Fisher D, Roth T, Scott C, Birznieks G, et al. Melatonin agonist tasimelteon (VEC-162) for transient insomnia after sleep-time shift: two randomised controlled multicentre trials. Lancet (2009) 373:482–91. doi:10.1016/S0140-6736(08)61812-7
126. Onozuka H, Nakajima A, Matsuzaki K, Shin R-W, Ogino K, Saigusa D, et al. Nobiletin, a citrus flavonoid, improves memory impairment and ABeta pathology in a transgenic mouse model of Alzheimer’s disease. J Pharmacol Exp Ther (2008) 326:739–44. doi:10.1124/jpet.108.140293
127. Nakajima A, Aoyama Y, Shin E-J, Nam Y, Kim H-C, Nagai T, et al. Nobiletin, a citrus flavonoid, improves cognitive impairment and reduces soluble ABeta levels in a triple transgenic mouse model of Alzheimer’s disease. Behav Brain Res (2015) 289:69–77. doi:10.1016/j.bbr.2015.04.028
128. Nakajima A, Aoyama Y, Nguyen T-TL, Shin E-J, Kim H-C, Yamada S, et al. Nobiletin, a citrus flavonoid, ameliorates cognitive impairment, oxidative burden, and hyperphosphorylation of tau in senescence-accelerated mouse. Behav Brain Res (2013) 250:351–60. doi:10.1016/j.bbr.2013.05.025
129. Yabuki Y, Ohizumi Y, Yokosuka A, Mimaki Y, Fukunaga K. Nobiletin treatment improves motor and cognitive deficits seen in MPTP-induced Parkinson model mice. Neuroscience (2014) 259:126–41. doi:10.1016/j.neuroscience.2013.11.051
130. Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell (2013) 153:1194–217. doi:10.1016/j.cell.2013.05.039
131. Gibson EM, Williams WP III, Kriegsfeld LJ. Aging in the circadian system: considerations for health, disease prevention and longevity. Exp Gerontol (2009) 44:51–6. doi:10.1016/j.exger.2008.05.007
132. Hofman MA, Swaab DF. Living by the clock: the circadian pacemaker in older people. Ageing Res Rev (2006) 5:33–51. doi:10.1016/j.arr.2005.07.001
133. Ju YE, Lucey BP, Holtzman DM. Sleep and Alzheimer disease pathology – a bidirectional relationship. Nat Rev Neurol (2014) 10:115–9. doi:10.1038/nrneurol.2013.269
134. Kolker DE, Fukuyama H, Huang DS, Takahashi JS, Horton TH, Turek FW. Aging alters circadian and light-induced expression of clock genes in golden hamsters. J Biol Rhythms (2003) 18:159–69. doi:10.1177/0748730403251802
135. Wyse CA, Coogan AN. Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. Brain Res (2010) 1337:21–31. doi:10.1016/j.brainres.2010.03.113
136. Miyazaki M, Schroder E, Edelmann SE, Hughes ME, Kornacker K, Balke CW, et al. Age-associated disruption of molecular clock expression in skeletal muscle of the spontaneously hypertensive rat. PLoS One (2011) 6:e27168. doi:10.1371/journal.pone.0027168
137. Luo W, Chen WF, Yue Z, Chen D, Sowcik M, Sehgal A, et al. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell (2012) 11:428–38. doi:10.1111/j.1474-9726.2012.00800.x
138. Valentinuzzi VS, Scarbrough K, Takahashi JS, Turek FW. Effects of aging on the circadian rhythm of wheel-running activity in C57BL/6 mice. Am J Physiol (1997) 273:R1957–64.
139. Sellix MT, Evans JA, Leise TL, Castanon-Cervantes O, Hill DD, DeLisser P, et al. Aging differentially affects the re-entrainment response of central and peripheral circadian oscillators. J Neurosci (2012) 32:16193–202. doi:10.1523/JNEUROSCI.3559-12.2012
140. Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell (2000) 103:1009–17. doi:10.1016/S0092-8674(00)00205-1
141. Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature (2010) 466:627–31. doi:10.1038/nature09253
142. Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev (2006) 20:1868–73. doi:10.1101/gad.1432206
143. Kondratov RV, Vykhovanets O, Kondratova AA, Antoch MP. Antioxidant N-acetyl-l-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein BMAL1. Aging (Albany NY) (2009) 1:979–87. doi:10.18632/aging.100113
144. Gutman R, Genzer Y, Chapnik N, Miskin R, Froy O. Long-lived mice exhibit 24 h locomotor circadian rhythms at young and old age. Exp Gerontol (2011) 46:606–9. doi:10.1016/j.exger.2011.02.015
145. Ramkisoensing A, Meijer JH. Synchronization of biological clock neurons by light and peripheral feedback systems promotes circadian rhythms and health. Front Neurol (2015) 6:128. doi:10.3389/fneur.2015.00128
146. Hofstra WA, de Weerd AW. How to assess circadian rhythm in humans: a review of literature. Epilepsy Behav (2008) 13:438–44. doi:10.1016/j.yebeh.2008.06.002
147. Benloucif S, Burgess HJ, Klerman EB, Lewy AJ, Middleton B, Murphy PJ, et al. Measuring melatonin in humans. J Clin Sleep Med (2008) 4:66–9.
148. Sharma M, Palacios-Bois J, Schwartz G, Iskandar H, Thakur M, Quirion R, et al. Circadian rhythms of melatonin and cortisol in aging. Biol Psychiatry (1989) 25:305–19. doi:10.1016/0006-3223(89)90178-9
149. Waldhauser F, Weiszenbacher G, Tatzer E, Gisinger B, Waldhauser M, Schemper M, et al. Alterations in nocturnal serum melatonin levels in humans with growth and aging. J Clin Endocrinol Metab (1988) 66:648–52. doi:10.1210/jcem-66-3-648
150. Pierpaoli W, Regelson W. Pineal control of aging: effect of melatonin and pineal grafting on aging mice. Proc Natl Acad Sci U S A (1994) 91:787–91. doi:10.1073/pnas.91.2.787
151. Weitzman ED, Moline ML, Czeisler CA, Zimmerman JC. Chronobiology of aging: temperature, sleep-wake rhythms and entrainment. Neurobiol Aging (1982) 3:299–309. doi:10.1016/0197-4580(82)90018-5
152. Munch M, Knoblauch V, Blatter K, Schroder C, Schnitzler C, Krauchi K, et al. Age-related attenuation of the evening circadian arousal signal in humans. Neurobiol Aging (2005) 26:1307–19. doi:10.1016/j.neurobiolaging.2005.03.004
153. Black AE, Coward WA, Cole TJ, Prentice AM. Human energy expenditure in affluent societies: an analysis of 574 doubly-labelled water measurements. Eur J Clin Nutr (1996) 50:72–92.
154. Pannemans DL, Westerterp KR. Energy expenditure, physical activity and basal metabolic rate of elderly subjects. Br J Nutr (1995) 73:571–81. doi:10.1079/BJN19950059
155. Monk TH, Buysse DJ, Reynolds CF III, Kupfer DJ, Houck PR. Circadian temperature rhythms of older people. Exp Gerontol (1995) 30:455–74. doi:10.1016/0531-5565(95)00007-4
156. Duffy JF, Zeitzer JM, Rimmer DW, Klerman EB, Dijk DJ, Czeisler CA. Peak of circadian melatonin rhythm occurs later within the sleep of older subjects. Am J Physiol Endocrinol Metab (2002) 282:E297–303. doi:10.1152/ajpendo.00268.2001
157. Kenney WL, Munce TA. Invited review: aging and human temperature regulation. J Appl Physiol (1985) (2003) 95:2598–603. doi:10.1152/japplphysiol.00202.2003
158. Vitiello MV, Smallwood RG, Avery DH, Pascualy RA, Martin DC, Prinz PN. Circadian temperature rhythms in young adult and aged men. Neurobiol Aging (1986) 7:97–100. doi:10.1016/0197-4580(86)90146-6
159. Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet (2007) 8:835–44. doi:10.1038/nrg2188
161. Tevy MF, Giebultowicz J, Pincus Z, Mazzoccoli G, Vinciguerra M. Aging signaling pathways and circadian clock-dependent metabolic derangements. Trends Endocrinol Metab (2013) 24:229–37. doi:10.1016/j.tem.2012.12.002
162. Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol (2014) 24:464–71. doi:10.1016/j.tcb.2014.04.002
163. Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell (2014) 158:659–72. doi:10.1016/j.cell.2014.06.050
164. Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science (2013) 342:1243417. doi:10.1126/science.1243417
165. Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell (2013) 153:1448–60. doi:10.1016/j.cell.2013.05.027
166. Hubbard BP, Sinclair DA. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol Sci (2014) 35:146–54. doi:10.1016/j.tips.2013.12.004
167. Oike H, Kobori M. Resveratrol regulates circadian clock genes in Rat-1 fibroblast cells. Biosci Biotechnol Biochem (2008) 72:3038–40. doi:10.1271/bbb.80426
168. Pifferi F, Dal-Pan A, Menaker M, Aujard F. Resveratrol dietary supplementation shortens the free-running circadian period and decreases body temperature in a prosimian primate. J Biol Rhythms (2011) 26:271–5. doi:10.1177/0748730411401788
169. Pifferi F, Dal-Pan A, Languille S, Aujard F. Effects of resveratrol on daily rhythms of locomotor activity and body temperature in young and aged grey mouse lemurs. Oxid Med Cell Longev (2013) 2013:187301. doi:10.1155/2013/187301
170. Lopez-Otin C, Galluzzi L, Freije JM, Madeo F, Kroemer G. Metabolic control of longevity. Cell (2016) 166:802–21. doi:10.1016/j.cell.2016.07.031
171. Okada-Iwabu M, Yamauchi T, Iwabu M, Honma T, Hamagami K, Matsuda K, et al. A small-molecule AdipoR agonist for type 2 diabetes and short life in obesity. Nature (2013) 503:493–9. doi:10.1038/nature12656
172. Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci (2012) 13:325–35. doi:10.1038/nrn3208
173. Kudo T, Schroeder A, Loh DH, Kuljis D, Jordan MC, Roos KP, et al. Dysfunctions in circadian behavior and physiology in mouse models of Huntington’s disease. Exp Neurol (2011) 228:80–90. doi:10.1016/j.expneurol.2010.12.011
174. Czeh B, Fuchs E, Wiborg O, Simon M. Animal models of major depression and their clinical implications. Prog Neuropsychopharmacol Biol Psychiatry (2016) 64:293–310. doi:10.1016/j.pnpbp.2015.04.004
Keywords: circadian clock, small molecules, amplitude, metabolic disease, mood disorder, aging
Citation: Gloston GF, Yoo S-H and Chen Z (2017) Clock-Enhancing Small Molecules and Potential Applications in Chronic Diseases and Aging. Front. Neurol. 8:100. doi: 10.3389/fneur.2017.00100
Received: 21 January 2017; Accepted: 28 February 2017;
Published: 15 March 2017
Edited by:
Timo Partonen, National Institute for Health and Welfare, FinlandReviewed by:
Kyungjin Kim, Seoul National University, South KoreaCopyright: © 2017 Gloston, Yoo and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng (Jake) Chen, emhlbmcuY2hlbi4xQHV0aC50bWMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.