
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neurol., 20 April 2015
Sec. Sleep Disorders
Volume 6 - 2015 | https://doi.org/10.3389/fneur.2015.00080
This article is part of the Research TopicCircadian lessons from owls and larks: The impact of circadian phenotype on health, well-being, and performanceView all 16 articles
There is evidence of a link between the circadian system and psychiatric diseases. Studies in humans and mammals suggest that environmental and/or genetic disruption of the circadian system leads to an increased liability to psychiatric disease. Disruption of clock genes and/or the clock network might be related to the etiology of these pathologies; also, some genes, known for their circadian clock functions, might be associated to mental illnesses through clock-independent pleiotropy. Here, we examine the features which we believe make Drosophila melanogaster a model apt to study the role of the circadian clock in psychiatric disease. Despite differences in the organization of the clock system, the molecular architecture of the Drosophila and mammalian circadian oscillators are comparable and many components are evolutionarily related. In addition, Drosophila has a rather complex nervous system, which shares much at the cell and neurobiological level with humans, i.e., a tripartite brain, the main neurotransmitter systems, and behavioral traits: circadian behavior, learning and memory, motivation, addiction, social behavior. There is evidence that the Drosophila brain shares some homologies with the vertebrate cerebellum, basal ganglia, and hypothalamus-pituitary-adrenal axis, the dysfunctions of which have been tied to mental illness. We discuss Drosophila in comparison to mammals with reference to the: organization of the brain and neurotransmitter systems; architecture of the circadian clock; clock-controlled behaviors. We sum up current knowledge on behavioral endophenotypes, which are amenable to modeling in flies, such as defects involving sleep, cognition, or social interactions, and discuss the relationship of the circadian system to these traits. Finally, we consider if Drosophila could be a valuable asset to understand the relationship between circadian clock malfunction and psychiatric disease.
Mental health diseases (i.e., depressive syndromes, bipolar disorders, and schizophrenia) make up about 20% of all illnesses and approximately one person in four is afflicted by one form or other of this kind of disease during their lifetime. It is widely accepted that these disorders are complex pathologies, influenced by the interplay between several genes and multiple environmental factors (1, 2). Different lines of evidence suggest a link between the endogenous circadian system and at least some forms of these diseases (3–5).
In mammals, the circadian system is physiologically composed by an hierarchical network of clocks in which time-keeping molecular and cellular processes are integrated at the organismic level. The central circadian clock maps to the suprachiasmatic nucleus (SCN) of the hypothalamus, harboring an autonomous oscillator which is responsible for the synchronization with the daily environmental variations, such as the light: dark (LD) cycle [reviewed in Ref. (6, 7)] (Figure 1A). The SCN sends direct and indirect signals to all the peripheral clocks located in the brain and body, coordinating their rhythm and phase, so that they oscillate in phase with the SCN itself, with each other, and with the environment. Furthermore, the organism’s circadian clock-controlled phenotypes, such as the sleep/wake cycle, body temperature, and metabolism, are synchronized with the 24 h environmental variations.
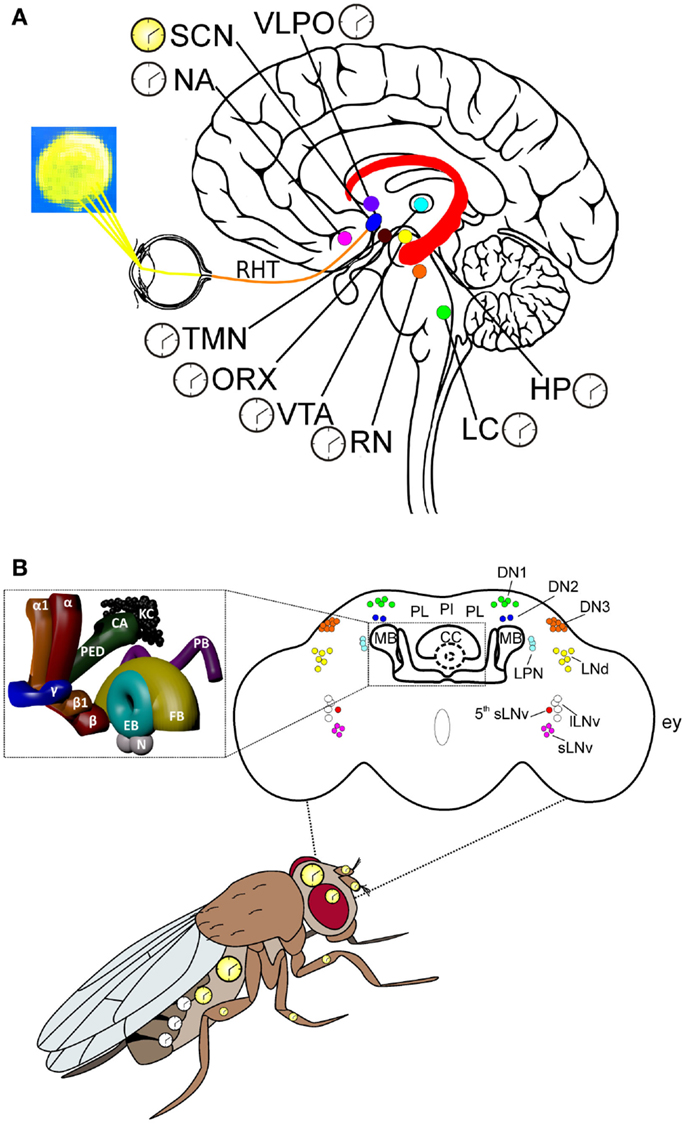
Figure 1. Key centers of the mammalian and Drosophila brains and the circadian system in Drosophila. (A) Some of the key areas of the human brain involved in the control of arousal/sleep, circadian rhythms, and cognitive processes related to motivation, emotions, and learning/memory. Hippocampus (HP): this relatively large structure is part of the limbic system and is involved in stabilizing information in the consolidation of short-term to long-term memory. The HP is also implicated in spatial navigation. Locus coeruleus (LC): norepinephrine from the LC mediates arousal, and primes the brain’s neurons to be activated by stimuli, and is involved with physiological responses to stress and panic. Nucleus accumbens (NA): this region of the brain is involved in the cognitive elaboration of associative learning, motivation, pleasure, and addiction/reward. Orexinergic neurons (ORX): these neurons produce the neurotransmitter orexin/hypocretin, which is involved in regulating arousal, wakefulness, and appetite. Suprachiasmatic nucleus (SCN): these neurons are situated directly above the optic chiasm and are the seat of the central clock (yellow), which controls circadian rhythms through the action of different peptides and neurotransmitters on many other regions of the brain, which contain subsidiary clocks (white). Raphe nuclei (RN): these serotoninergic nuclei are involved in a reciprocal feedback loop with the SCN to which they send information regarding levels of alertness; the SCN in turn sends connections to the RN; thus, influencing serotonin levels, which are involved in regulating sleep/wake states. Ventrotegmental area (VTA): the dopaminergic neurons of this area of the brain are involved in cognitive processes related to addiction/reward and motivation. Tuberomammillary nuclei (TMN): these histaminergic nuclei are involved in the control of arousal, learning, memory, sleep, and energy balance. Retinohypothalamic tract (RHT): the retinohypothalamic tract originates in the intrinsically photosensitive retinal ganglion cells, which contain the photopigment melanopsin. The RHT axons, through the optic nerve and the optic chiasm, project to the suprachiasmatic nuclei. Ventrolateral preoptic nucleus (VLPO): the VLPO is active during sleep and releases mainly GABA and galanin, which inhibit neurons that are involved in wakefulness and arousal (i.e., ORX, RN, LC, TMN). The latter groups of neurons are involved in a reciprocal feedback loop with the VLPO, thus contributing to the regulation of sleep. (B) The circadian system organization in Drosophila, with multiple oscillators located in the brain and body. The clocks in the brain, and in most of the peripheral tissues, are autonomous (yellow), while those located in the oenocytes resemble the mammalian subsidiary clocks (white), whose phase is controlled by the central brain clock; see text for details. Upper part: schematic representation of the adult fly brain, in which the relative positions of the circadian neurons, the mushroom bodies (MBs), the central complex (CC), the pars lateralis (PL), and the pars intercelebralis (PI) are reported [modified from Ref. (8, 9)]. The inset shows a 3D-reconstruction of the MBs and CC. KC: kenyon cells; CA: calyx; PED: pedunculus; α: α lobe; α1: α′ lobe; β: β lobe; β1: β′ lobe; γ: γ lobe; EB: ellipsoid body; FB: fan-shaped body; N: noduli; PB: protocerebral bridge; lLNv: large ventral lateral neurons; sLNv: small LNvs; 5th sLNv: the 5th PDF-negative sLNv; LPN: lateral posterior neuron; LNd: dorsal LNs; DN1: dorsal neurons group 1; DN2: DN group 2; DN3: DN group 3; ey: relative position of the compound eye respect to the brain.
In humans, clinical observations have shown that many psychiatric patients display abnormalities in circadian parameters such as cycles in body temperature, melatonin levels, blood pressure, cortisol secretion, and sleep/wake cycles (4, 10, 11). Several single nucleotide polymorphisms at the level of genes involved in the control of the circadian clock have been associated to different forms of psychiatric disorders (4, 12, 13). In addition, a post-mortem transcriptome analysis performed in individuals affected by depressive disorders showed that in different brain regions several circadian clock genes oscillate with a lower amplitude with respect to controls (14). On the other hand, in otherwise asymptomatic subjects, environmental conditions such as shift-work, chronic jet-lag, or “social” jet-lag, which are responsible for a misalignment between the environment and the internal circadian system, might represent risk factors for deterioration of mental health, cognition, and mood (15–17). Studies in mammalian models have shown that some genes such as mPeriod2, mClock, and mRev-erb α, which are fundamental elements of the molecular clockwork (see below), are also involved in the control of behaviors, which are considered psychiatric-like hallmarks in the modeling of mental illnesses in non-primate animals (18–22). Recent analyses have linked mCLOCK and mPER2 activities to the circadian regulation of dopamine synthesis (mCLOCK) and catabolism (mPER2) in brain regions considered fundamental in the control of psychiatric-like behaviors (18–21, 23), while REV-ERB α (22) has been demonstrated to be involved in the control of the hippocampal adult neurogenesis. Overall, these data have given rise to the hypothesis that an environmental and/or a genetically induced disruption of the circadian system might be associated with an increased susceptibility to psychiatric disease. This dysregulation of the circadian system might affect brain neuronal pathways and networks, i.e., through alterations in: monoamine neurotransmitter modulation, regulation of the hypothalamus-pituitary-adrenal axis (HPA), and neurogenesis, which are considered to be among the possible causes of these complex pathologies [reviewed in Ref. (24)]. However, the complexity of the circadian system organization in mammals, with the SCN coordinating all the other brain peripheral clocks, makes it difficult to determine whether the master clock exerts a direct influence on these neurological phenomena. In addition, some molecular components, known mainly for their involvement in circadian clock functions, might be associated to psychiatric illnesses through pleiotropic effects (i.e., additional clock-independent functions) (25).
Here, we speculate whether a relatively simple model organism, such as the fruit fly Drosophila melanogaster, might be informative in the study of the possible relationship between the circadian clock and human psychiatric disease. The Drosophila circadian clock is one of the best characterized at the molecular, physiological, and behavioral levels. Notwithstanding significant differences in the organization of the multiple-clock system at the organismic level, the overall molecular architecture of the Drosophila and mammalian circadian oscillators are comparable and many components are evolutionarily conserved. Recently, Drosophila has been proposed as a model organism for the study of psychiatric illnesses. Significantly, current research work is contributing to the definition of the details of the full set of a fly’s behavior, which includes motivation, social behavior, as well as some aspects of addiction, which are likely relevant features of neuropsychiatric disorders. In this review, we will describe the Drosophila characteristics regarding: (i) the organization of the brain and neurotransmitter systems, (ii) the architecture of the circadian clock, (iii) the clock-controlled behaviors, mainly in comparison with mammals. We will also sum up current knowledge relative to behavioral endophenotypes, which are amenable to modeling in flies, such as defects involving sleep, cognition, or social interactions, and discuss the relationship of the circadian system to these traits. Finally, we will speculate on the possible strategies based on the use of Drosophila as model organism to understand the relationship between circadian clock dysregulation and psychiatric disease.
From an evolutionary and phylogenetic standpoint, the invertebrate D. melanogaster is a member of the protostomes whereas vertebrates are deuterostomes; both of which are related by their belonging to the Bilateria (animals showing bilateral symmetry). The relevance of this distinction lies in the hypothesis, originally expressed by Anton Dohrn in 1875, that the vertebrate nervous system looks essentially like a dorsoventrally inverted version of the invertebrate nervous system. This idea has found solid support at the cellular and molecular levels (26). Furthermore, the dorsoventral and anteroposterior patterning of protostomes and deuterostomes show deep homology (i.e., phylogenetic conservation of genetic regulatory networks), so much so, that the organization of the central nervous system into forebrain, midbrain, and hindbrain is thought to have originated before the protostome–deuterostome split, which is estimated to have occurred between 600 and 800 million years ago [reviewed in Ref. (27)].
The rostral to caudal organization of the Drosophila brain1 can be schematized as follows:
This is the most anterior neuropil of the brain and contains many complex substructures (Figure 1B).
(1) The Mushroom bodies (MBs): studies in Drosophila and other insects, (i.e., cockroaches and honey bees), suggest that the MBs are involved in olfactory learning and memory (LM) (28) but probably also in place memory, associative memory, context dependent sensory filtering, as well as playing a role in motor control (29–33). Drosophila MBs are formed by a calyx-shaped neuropil, situated in a posterior–dorsal region of the protocerebrum. The calyx then continues anteriorly into a pedunculus, which then divides into a dorsal lobe (consisting in two subdivisions, called α and α′) and a medial lobe (consisting of three subdivisions, called β, β′, and γ lobes); the β subdivision corresponds to the α subdivision of the dorsal lobe and the β′ subdivision corresponds to the dorsal lobe’s α′ subdivision (Figure 1B). At the cellular level, the MB consists of about 2500 intrinsic neurons, called Kenyon cells, which originate from globuli cells situated above the calyx.
(2) The Central complex (CC): the CC is the most central and the only unpaired neuropil in the insect brain (Figure 1B). It receives multimodal inputs from most parts of the brain and has been proposed as a higher center for locomotor control, which regulates several aspects of walking and flying behavior and has also been suggested to act as a higher center for the integration of visual input as well as playing a role in spatial visual memory and place learning (34). Furthermore, there is evidence that dopaminergic neurons of the CC are involved in the control of arousal, wakefulness, and aggression [reviewed in Ref. (35)].The CC consists of four interconnected substructures: the protocerebral bridge (PB), the fan-shaped body (FB), the ellipsoid body (EB), and the noduli (N) (Figure 1B). The EB is an almost circular neuropil, lying anterior to the FB. The EB receives terminals of neurons, which originate in the protocerebrum, and it shares dendrites with parts of the FB, which lies posterior to the EB. The FB contains arborizations of terminals and dendrites linking the FB to lateral regions of the protocerebrum. The N are two nodular neuropils that receive connections from the PB neurons that also provide collaterals to the FB. The PB forms a handlebar-shaped commissural connection between the two dorsal lobes of the protocerebrum, and they also provide axons that project into the FB, from where they project further into the EB and the N.
(3) The pars intercerebralis (PI) and pars lateralis (PL) together with their projections to the corpora cardiaca, corpora allata, and prothoracic gland, constitute the Drosophila neuroendocrine system (36, 37). Most of the neurosecretory cells (NSC) are contained within the small groups of cells of the PI/PL, which are located in the dorsal medial region of the protocerebrum. The neurohemal secretory cells of the corpora cardiaca, corpora allata, and prothoracic gland form a ring-like structure partially surrounding the dorsal blood vessel. NSCs of the PI and PL secrete insulin-like peptides, FMRFamide-like peptides, pigment-dispersing hormone, corazonin, ovary ecdysteroidogenic hormone, and myomodulin (38). The corpora cardiaca/corpora allata produce juvenile hormone which, together with ecdysone, produced and released by the prothoracic gland, controls growth and molting. Recent studies demonstrated that the PI is involved in multiple behaviors and physiological phenomena, such as sleep (39, 40), locomotion (41, 42), circadian locomotor activity (43), and metabolism (44, 45), and it is considered a region functionally analogous to the mammalian hypothalamus (36).
This is the second division of the supraesophageal ganglion. The antennal lobes, which are glomerular neuropils receiving mainly olfactory receptor terminals, are part of the deutocerebrum. The deutocerebrum also receives mechanosensory input from the head surface as well as input from the optic lobes, the EB and FB of the central complex, and dendrites from ascending pathways.
This is the third segmental preoral ganglion, which lies ventrally on either side of the gut. The tritocerebrum, similarly to the deutocerebrum, also gives rise to descending neurons and receives connections from various regions of the more anterior neuropils.
Aside from the tripartite organization of the Drosophila brain, recent work has drawn attention to the notion that there may be functional homology between the protocerebral neuropils and corresponding structures in the vertebrate forebrain. In particular, Strausfeld and Hirth (46) discuss evidence suggesting a deep homology of the arthropod central complex and vertebrate basal ganglia. On the other hand, Wirmer et al. (37) review the evidence suggesting the functional homology of the insect NSC of the PI/PL-copora cardiaca system and the vertebrate hypothalamus–pituitary axis. While, Farris (30) presents arguments in favor of a structural and functional homology between the MBs and the vertebrate cerebellum. What these considerations imply is that, to all intents and purposes, the fly brain can be considered a “simplified” miniature version of the vertebrate brain (47, 48). As such, it can be expected that, probably at a very basic level, the neural circuitries governing context-dependent sensory integration, LM, action and directed behavior are probably conserved from Drosophila to vertebrates. In fact, there is strong evidence for a conservation of the principle neurotransmitters and the associated receptor/signaling systems, with particular regard to the dopaminergic (49), serotoninergic (50), GABAergic (51), and adrenergic systems (52). In the case of adrenergic signaling, it is, however, important to point out that, although at the behavioral level, there is good evidence for a functional conservation of the so called “fight or flight” response, motivation, and aggression (in vertebrates mainly modulated by the adrenergic system); in Drosophila, these behaviors are instead modulated by octopamine and tyramine. Nonetheless, these two neurotransmitters are structurally and functionally related to adrenaline and noradrenaline, respectively (52), since they are all products of the metabolic transformation of the same amino acid, tyrosine. In fact, tyramine is the decarboxylation product of tyrosine, and octopamine is the β-hydroxylation product of tyramine.
In Drosophila, as in mammals, circadian rhythms at the molecular and cellular levels are driven by interlocking autoregulatory transcriptional/translational feedback loops (TTLs). These have been recently reviewed [e.g., Ref. (53, 54)], and here we present a simplified model of the two major TTLs (Figures 2A,B). In D. melanogaster, the transcription factors dCLOCK (dCLK) and dCYCLE (dCYC) act as a heterodimer (dCLK/dCYC), promoting the transcription of the dperiod (dper) and dtimeless (dtim) genes (Figure 2B). In mammals, the orthologs mCLK (or mNPAS2 in the forebrain) and mBMAL1 exert the function of positive regulators activating the transcription of the three mammalian orthologs of dperiod (mPer1, mPer2, and mPer3) and the two mCryptochrome genes (mCry1 and mCry2) (Figure 2A). In mammals, the mCry genes replace dtim in the main TTL. Once translated, dPER and dTIM (mPERs and mCRYs in mammals) are targeted by different kinases and phosphatases, which mediate the timing of their nuclear translocation, stability, and action as negative feedback elements of dCLK/dCYC (or mCLK/mBMAL1 in mammals) regulatory activity. Among the kinases, it is worth underlining the roles played by dSHAGGY, homologous to mammalian glycogen synthase kinase-3 (mGSK3) (55), which is involved in the phosphorylation of dTIM and dPER (mPERs and mCRYs in mammals), and by dDOUBLETIME [dDBT, homologous to mammalian Casein Kinase 1 ε (CK1ε) (56)], which targets dPER (57). These factors are involved in the regulation of dPER and dTIM (mPERs and mCRYs in mammals) stability and nuclear entry and contribute to the fine-tuning of circadian rhythmicity (58, 59) (Figures 2A,B). dCLK/dCYC (mCLK/mBMAL1) are also the positive regulators of a second TTL, which (auto)controls the rhythmic expression of dClk in flies and mBmal1 in mammals. In Drosophila, this TTL is under negative control by dVRILLE (dVRI), which probably competes with the positive regulator dPDP1 to bind sequence elements in the promoter region of dClk (60, 61) (Figure 2B). In mammals, the second TTL is controlled by the nuclear hormone receptors mRORs and mREV-ERBs, which act as transcriptional repressors and activators of mBmal1, respectively (62, 63) (Figure 2A).
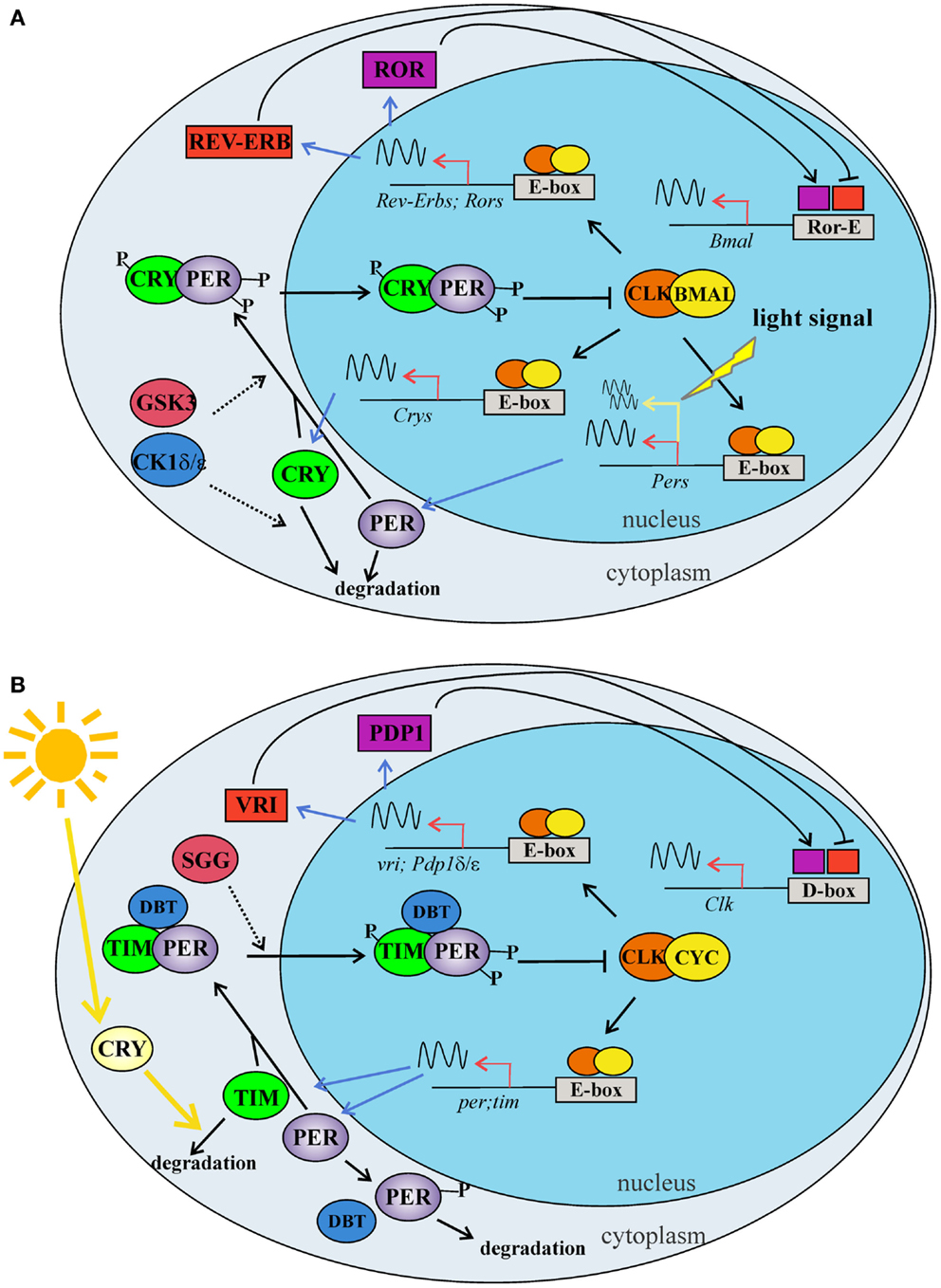
Figure 2. The two major TTLs of the circadian molecular clock in mammals (A) and Drosophila (B). (A) The first mammalian TTL includes BMAL1 and CLK, which act as heterodimer, binding the enhancer boxes (E-boxes) in the promoter of Per and Cry clock genes. PER and CRY proteins dimerize and enter into the nucleus, where inhibit the CLK -BMAL1 activity. A second loop modulates Bmal1 expression: CLK-BMAL1 dimers induce the transcription of Rev-erbα and Ror nuclear orphan receptor genes. REV-ERBs and RORs compete for the same element (Ror-E) in the Bmal1 promoter, controlling Bmal1 transcription. Phosphorylation mediated by CKs (δ/ε) and GSK3β modulate clock protein activities regulating protein–protein interactions, nuclear translocation, and degradation. Within the master clock, at the cell level, the light stimulus induces the transcription of the Per genes via a signal transduction cascade. (B) In the first TTL of Drosophila, CLK and CYC form a dimer, which binds the E-boxes in the promoter of per and tim clock genes. PER and TIM proteins interact in a complex, enter into the nucleus, and inhibit the CLK-CYC activity. A second TTL modulates Clk expression: CLK-CYC dimer induces the transcription of vri and Pdp1 δ/ε genes. VRI and PDP1 δ/ε compete for the same element (D-box) in the Clk promoter, controlling Clk transcription. Phosphorylation mediated by DBT and SGG modulate clock protein activities, regulating protein–protein interactions, nuclear translocation, and degradation. In the cell, light activates the internal photoreceptor CRY, which associates with TIM and mediates its degradation. BMAL: brain and muscle ARNT-Like 1; CKδ: casein kinase; CLOCK: circadian locomotor output cycles Kaput; CRY: cryptochrome; CYC: cycle; DBT: doubletime; GSK3β: glycogen synthase kinase 3 beta; PDP1: PAR domain protein 1; PER: period; REV-ERB: nuclear receptor subfamily 1, group D; ROR: RAR-related orphan receptor; TIM: timeless; VRI: vrille; SGG: Shaggy. Dashed arrows indicate phosphorylation, while sinusoidal lines indicate transcription activity.
In mammals, the light signal reaches the SCN via the retinohypothalamic tract (RHT) and synchronizes the master clock, promoting the transcription of the mPer1 and mPer2 genes, via the activation of a signal transduction cascade [reviewed in Ref. (10); Figures 1A and 2A]. On the contrary, in Drosophila, the ability to synchronize the clock with the 24 h environmental LD cycles is cell-autonomous and is mainly due to the light-mediated degradation of dTIM which, in turn, affects the stability of dPER. The majority of evidence concerning the molecular mechanism involved in light resetting suggests that this is mediated by the internal blue-light photoreceptor dCRY (64, 65) (Figure 2B). dCRY resets the molecular clock through a light dependent association with dTIM, which in turn activates the proteasome-mediated degradation of dTIM in a process involving the ubiquitin ligase dJETLAG (66, 67).
In Drosophila, the circadian system at the organismal level is composed by multiple oscillators located in the brain and peripheral tissues (Figure 1B). The brain master clock consists of ~150 neurons (of the ~250,000 Drosophila adult brain neurons), organized in bilateral pairs of clusters. In each brain hemisphere, these have been anatomically classified into 5 lateral neuron (LN) groups [including 4 large ventral lateral neurons (lLNvs), 4 small ventral lateral neurons (sLNvs), a 5th sLNv, 6 dorsal lateral neurons (LNds), and 3 lateral posterior neurons (LPNs)], and 3 clusters of dorsal neurons (DNs 1, 2, and 3) [reviewed in Ref. (68)] (Figure 1B). The anatomical subdivision does not correspond to a functional classification, and the single neurons within each cluster can show differences in protein/neuropeptide expression profiles, properties, and activities [reviewed in Ref. (68–70)]. For example, both the 4 lLNvs and the 4 sLNvs express the neuropeptide pigment dispersing factor (PDF). In the physiology of the Drosophila central circadian clock, PDF plays multiple roles: (i) as an output neurotransmitter, considered to be the equivalent of the mammalian circadian neuropeptide vasoactive intestinal peptide (71, 72); (ii) as a synchronizer of the oscillations of the clock neurons; (iii) playing a part in the signal transduction of light input into the circadian-neuron circuitry [reviewed in Ref. (68, 73)]. Some LNds express the long form of neuropeptide F (NPF), while sLNvs and one LNd produce the short form (sNPF). These neuropeptides are homologous to mammalian neuropeptide Y (NPY), which is involved in the control of both sleep and feeding in humans and rodents (74, 75). The role of the different neuronal clock clusters in regulating circadian activity has been investigated mainly by evaluating daily cycles of locomotor activity as a readout. In laboratory conditions (i.e., LD cycles with abrupt LD transitions), the locomotor activity of flies shows a bimodal profile, with one peak in the morning and a second peak in the evening. Several studies indicated that these two peaks are governed by specific morning-(M) and evening-(E) coupled clocks, which have been located to the 4 sLNvs (M), and in the 5th sLNv, the LNds, and the DNs (E), (76, 77). While initial studies suggested a dominant role of the M clock in the control of the endogenous rhythm (78), recent data indicate that the network of all circadian clock neurons contributes to the generation of behavioral rhythmicity, in both constant darkness (DD) and LD conditions (79, 80).
Recent work has begun to clarify how the master clock communicates with other brain regions to give rise to circadian rhythmic locomotor behavior (43). It has, in fact, been demonstrated that the time-of-day information generated by the circadian clock network of the sLNvs, and probably the LNds, is sent to the DN1s, which in turn contact specific neurons (identified by the driver Kurs58Gal4) of the PI (Figure 1B). The Kurs58 neurons probably consist of at least two different neuronal populations, which seem to have opposite roles in the control of the sleep/wake rhythm, and their relative contribution might vary during the 24 h (43). Interestingly, Cavanaugh and colleagues identified DH44, the Drosophila homolog of the mammalian stress hormone Corticotropin releasing factor, as a possible candidate signaling molecule important in the PI-modulation of locomotor activity rhythms. In addition, since DH44 receptors have been identified in a group of cells of the lateral protocerebrum, probably involved in stress-induced locomotor activity (81), the authors suggest a possible parallelism with mammals in which the stress glucocorticoids released from the HPA axis show a circadian production and act as synchronizing signals for the peripheral and possibly also for the central clocks (43, 82).
In Drosophila, in addition to the clock neurons, glial cells (in particular astrocytes) contribute to the daily control of locomotor activity rhythms (83, 84). In this activity, ebony, which encodes for an enzyme involved in dopamine and histamine recycling, seems to play a key role as an output gene (83). Moreover, glia might act by modulating PDF transport and/or release from vLN projections, indicating the importance of a glia-to-neuron communication in the control of behavioral rhythmicity (84). Interestingly, a role for the glial cells has been suggested also for circadian behavior in mammals (85).
In contrast to mammals, in which virtually every area of the brain possesses a functioning peripheral clock (Figure 1A), in Drosophila, brain regions outside the master clock network and glia apparently do not contain all the molecular elements necessary for a functional clock. However, a study performed in 2000 revealed that several independent dper-promoter- and dtim-promoter-Gal4 lines (tools commonly used in Drosophila circadian studies) showed activity in non-master clock brain neurons and structures. Among them, the PI, the EB, and the FB were labeled in the perGal4 strains, while neurons located dorsolaterally to the antennal lobe, near the tritocerebrum, and the subesophageal ganglion were marked in the timGal4 lines. Such neurons are normally not (or weakly) detected with anti-dPER or anti-dTIM antibodies (86). In 2008, Yoshii and colleagues demonstrated the presence of dCRY in about 12 neurons per brain hemisphere, which resemble the R4 EB neurons of the central complex (87). Moreover, recently, dPER and dCRY proteins were detected in two neurons located in the dorsal-anterior-lateral (DAL) protocerebrum and implicated in a form of long-term memory (88).
As in mammals, non-central-brain peripheral circadian clocks are present in multiple districts of the Drosophila body, such as the compound eyes, antennae (89, 90), gustatory neurons (91), prothoracic gland (92), Malpighian tubules (93), and oenocytes (94) (Figure 1B). In the simplified model of the mammalian circadian system, the SCN master clock controls the phase of all the peripheral clocks, with the exception of the semi-autonomous oscillator in the olfactory bulb (95). This organization is not completely transposable to Drosophila. Experimental evidence instead suggests that in Drosophila, some peripheral clocks, such as those of the antenna (which control circadian odor-sensitivity), those of the proboscis (controlling the gustatory physiology rhythms), and those of the Malpighian tubules (the renal organ of the fly), are autonomous systems which might oscillate in phase with the master clock by directly perceiving and responding to the same environmental stimuli (91, 96, 97). However, there are at least two exceptions: it has been demonstrated that the central clock controls both the non-autonomous clock in the prothoracic gland, which regulates the rhythm of pupal eclosion (92), and the phase of clock timing in the oenocytes (which are involved in the synthesis of sex pheromones secreted on the cuticle surface), via PDF signaling (98). Interestingly, the pheromones synthesized by the oenocytes are not only involved in mating (98) but, to a lesser extent, also in male aggressiveness (99).
In humans and vertebrate models, complex behaviors such as sleep, as well as LM, appear to be influenced in some way by the circadian system or genes, even if the mechanistic relationship is still unclear. Both sleep and LM are often impaired in psychiatric patients. In particular, the majority of patients with schizophrenia, bipolar, and depressive disorders refer sleep disturbances [insomnia or sleepiness; reviewed in Ref. (100, 101)]. Neuropsychiatric patients may also be characterized by impairments in the selection of positive or negative information from working memory and in their capability to access episodic memories (102). In addition, many psychopathologies may display impairments in interpersonal interactions (103). As illustrated in the following sections, several data suggest an involvement of the circadian clock or genes in the determination of these complex behaviors also in Drosophila.
Sleep is controlled by both circadian and homeostatic systems. In the mammalian brain, the core sleep circuit is formed by interconnected sleep and arousal centers, which include hyptothalamic GABAergic sleep-promoting and orexin-positive wake-promoting areas, which project to multiple brain regions [reviewed in Ref. (104, 105)] (Figure 1A). Recently, glial astrocytes have been implicated in the regulation of sleep homeostasis (106). In mammals, several lines of evidence indicate that the circadian clock controls the timing of the sleep-wake cycle. For example, in humans, mutations, which alter the phosphorylation site of PER2 or of the CKs, have a causal role in circadian-based sleep disorders such as the advanced and delayed sleep phase syndromes [reviewed in Ref. (24)]. In addition, studies on mammalian models suggest that mutations at the level of some circadian clock genes affect sleep homeostasis. For example, Clk mutant mice sleep on average 2 h less than wild-type individuals (107), while both Bmal1 knock-out (KO) and Cry1/Cry2 double KO mice show an increase in their total sleep time (108, 109). However, this is not a general rule for circadian clock genes since both single or double gene mutants for mPer1 and/or mPer2 do not show abnormalities in sleep homeostasis (110). These data seem to suggest a pleiotropic non-circadian effect of some of the circadian genes in the control of sleep homeostasis.
More than a decade ago, it was shown that D. melanogaster has a sleep-like condition [reviewed in Ref. (111)] associated to a decrement in both sensory responsiveness and brain activity (112, 113). Behaviorally, each bout of the fly sleep-like state is defined as a period of inactivity lasting ≥5 min, and day or night time sleep are calculated by summing up the total bouts of sleep time occurring in the light or dark period, respectively (114, 115). Using this behavioral parameter and the powerful transgenic toolbox available for this insect, it has been demonstrated that, as in humans, in fruit flies GABAergic neurotransmission and serotoninergic signaling promote sleep (116, 117), while dopaminergic neurons stimulate arousal and wakefulness (118, 119). Differently to humans, however, Drosophila does not seem to have an orexin-based wake-promoting neurotransmitter system (120), which might be substituted instead by PDF, together with octopamine (40, 121). Nonetheless, recent work has extended the parallelism between mammalian and Drosophila sleep physiology demonstrating, via both behavioral and electrophysiological recordings, that also in flies sleep intensity varies during the 24 h (122). In particular, deeper sleep phases occur mainly during the night and have been hypothesized as being associated to a synaptic downscaling process, similar to that proposed for mammals (122). It is noteworthy that the presence of similarities between mammalian and Drosophila sleep allowed the design of a strategy based on the analysis of the sleep-like state in genetically modified or pharmacologically treated flies, which then led to the identification of amylase as a possible biomarker of sleepiness in humans (123).
As in mammals, Drosophila sleep is subject to both circadian and homeostatic regulation (124, 125). It has also been shown that some clock genes, such as dcycle and dClk, might be involved in sleep homeostasis, while others such as dper appear not to have a role in the control of this phenomenon (124, 126, 127), again suggesting a non-circadian role for those circadian genes, which do affect sleep homeostasis. Brain areas involved in fly sleep regulation include the PI, which harbors both Dilp2-positive wake-promoting and EGFR ligand-expressing sleep-promoting neurons (39, 40), the sleep-promoting regions of the MBs (128, 129), and the FB of the central complex (130, 131) (Figure 1B). In addition, part of the circadian clock network is important in sleep regulation, with opposing effects. It has been demonstrated that 4 sLNvs promote sleep during the night, while the lLNv neurons have a role in wakefulness and arousal (132–136) (Figure 1B). In particular, the sleep promoting role of the sLNv neurons seems to be mediated by the neuropeptide sNPF, which likely acts with other neuromodulators in multiple brain areas involved in the sleep/wake state (136). The neuropeptide involved in the wake promoting role of the lLNvs is PDF, which probably has an effect on sLNvs and other brain regions expressing the PDF receptor, such as those involved in locomotion control (i.e., the EB) (132). In addition, lLNvs express the sNPF receptor, probably important for the sLNv–lLNv coordination (136). Moreover lLNvs produce the Rdl GABAA and GABAB-R2 receptors, which are a likely target of the GABAergic sleep-promoting neurons (132, 133, 137). Interestingly, hyperexcitation of the LNv leads to a more fragmented nocturnal sleep of flies (133). Finally, several lines of evidence indicate that the role of LNvs in sleep regulation and homeostasis is independent from their role as circadian clock neurons, since genetic manipulations of LNvs which alter sleep homeostasis do not modify their circadian functions, evaluated both as circadian locomotor activity or circadian expression of molecular clock components (133, 136, 137).
In healthy individuals, human cognitive processes are tightly connected to the sleep/wake cycle, and during the 24 h day they show a progressive deterioration associated to the increased amount of time spent awake (138). However, the decrement in cognitive performance is not linear with increasing sleep pressure, and several lines of evidence indicate the impact of circadian regulation in cognition, with circadian variations in the ability to learn as well as memory acquisition and retrieval (139–142).
In mammals, the hippocampus (HP) plays an important role in learning processes, formation of new memories, as well as in consolidation from short- to long-term memories (Figure 1A). Similarly to the majority of the brain regions in mammals, the HP expresses clock genes in a rhythmic manner (143). In addition, the HP possibly receives both direct and indirect circadian input from the SCN and other peripheral oscillators [reviewed in Ref. (141)]. Mice mutants for many circadian clock genes (such as mBmal1, mCry1, mCry2 and mNpas2) show impairments in different types of LM tests (144–146). Although such data do not exclude per se non-circadian pleiotropic effects of these mutations, nevertheless, the importance of a functional circadian system in hippocampal-dependent LM has been suggested from studies in humans which indicate that both shift-work and chronic jetlag cause cognitive impairments (16, 147). Similar indications have been obtained both following manipulation of the environmental LD cycle and in SCN-lesioned animal models [reviewed in Ref. (141)]. However, the importance of the SCN in these processes remains controversial (141).
Hippocampus-dependent LM has been associated to several biological processes and phenomena, which rhythmically occur in this structure. For example, the formation and persistence of long-term memory has been associated to the signal transduction pathway, involving the cyclic adenosine monophosphate (cAMP)/cAMP response element binding protein (mCREB) and mitogen activated protein kinase (mMAPK) (148–150). In the HP, cAMP/mCREB and mMAPK are rhythmically expressed (150, 151) and are also part of the molecular clock (152–155). For example, the phosphorylation of mBMAL1 by mMAPK inhibits the mBMAL1/mCLK-dependent transcription of the mPer and mCry genes (156). The mPer1 and mPer2 promoters contain cAMP responsive element (CRE) sites that bind mCREB in order to enhance their transcription (157). It has been demonstrated that the disruption of mMAPK oscillations interferes with the persistence of long-term memory (158), and both mPer1 and mPer2 KOs show defects in hippocampal-dependent learning tasks and reduced levels of phosphorylated mCREB have been demonstrated in mPer2 KOs [reviewed in Ref. (141)]. Other phenomena linked to LM processes, which show daily rhythmicities, are synaptic morphology (159) and adult neurogenesis (160). In particular, there are several indications concerning the circadian regulation of both phenomena. About 24 h rhythms in neuronal proliferation and changes in dendritic complexity and spine density have been observed in several rodent species (161–165). Circadian disruption caused by phase shifts negatively impacts both neurogenesis and synaptic complexity and can reduce the performance in hippocampal-dependent memory tasks in several species [reviewed in Ref. (141)]. Moreover, adult neurogenesis is impaired in mice harboring mutations in different circadian clock genes, such as mPer2 and mRev-erb α (166).
Drosophila melanogaster shows several types of LM, such as olfactory LM, courtship conditioning LM, spatial and visual LM [reviewed in Ref. (167, 168)]. Most of these memories are characterized by different phases: short (minutes), middle (hours), and long, which can persist for several days (169, 170). The possible involvement of the circadian clock in fly LM performance has been mainly evaluated using the olfactory and courtship conditioning paradigms. In the olfactory LM assay, flies learn to associate conditioned stimuli (CS i.e., an odorant stimulus) to an unconditioned stimulus (US i.e., an electrical shock) (171), while in the courtship conditioning paradigm attractive pheromones act as the CS, while aversive pheromones act as the US (172). The MBs are the key structures for olfactory LM, with the γ lobes mainly required for short-term memory (STM) formation, and α/β neurons involved in the establishment of long-term memories (LTMs) (28, 173) (Figure 1B). In addition to MBs, EB also appear to contribute to LTM (174) (Figure 1B). In the courtship conditioning paradigm, the antennal lobes (and perhaps also the optic lobes) seem to be particularly important in the formation of the first part of memory (up to 30 min), while the MBs have been implicated in the subsequent phases (from 30 min to several days) (175). As in mammals, the cAMP signaling pathway plays a central role in Drosophila memory and, while STM involves cAMP-dependent modifications of existing proteins [e.g., ion channel activities, either directly or indirectly via phosphorylation by PKA (170, 176)], LTM requires cAMP/CREB-mediated transcription (177).
Several lines of evidence suggest that the circadian clock impacts olfactory LM in flies. Under 12:12 LD cycles, Drosophila shows a 24 h rhythm in learning efficiency, which has been hypothesized to be associated with the rhythmic variation in abundance of the Drosophila mCREB homolog, dCREB2, in adult heads (178). Interestingly, dCREB2 appears to be under circadian control in several brain regions, including the MBs (179), and plays a crucial role in circadian rhythmicity, since mutations in dCREB2 alter circadian locomotor activity probably by modifying the transcriptional oscillations of the dper gene (180). In addition, it has been demonstrated that in wild-type flies, olfactory STM is under circadian control, with a peak of memory performance at the beginning of the night, both in LD and in DD conditions (181). The STM rhythmicity was lost in LL conditions and in per0 and tim0 mutant flies tested in DD. Since cryb mutants, which possess a non-functional peripheral oscillator in the antenna, maintain STM rhythmicity in DD, the authors suggest that the circadian STM modulation is due to the central oscillator, which might govern the availability of molecules involved in the memory-based signal transduction cascade (181). The circadian clock gene dper has also been implicated in Drosophila LTM, in both the olfactory learning and courtship conditioning paradigms (88, 182, 183). The role of dper in olfactory and courtship LTMs appeared to be independent from the circadian clock, since other clock mutants, such as tim0, Clk-Jrk, and cyc0, showed normal LTM (88, 182). Although the protocol differences in the two LM paradigms complicate the direct comparison of the results, it is interesting to note that dper is required outside the MBs in both cases. In particular, the per-dependent control of olfactory LTM maps at the level of the DAL neurons in the dorsal lateral protocerebrum (88), while for the courtship LTM the presence of dper is required at the level of the FB of the central complex (88, 183). The activity of these neuronal structures seems to be mainly important for the LTM recall, rather than LTM formation and storage (88, 183). In addition, both types of memory required dCREB2 activity, although the mechanistic link with dper has not yet been explored (88, 183).
A suite of social behaviors, including courtship, aggression, mating, the recognition of conspecifics, have been identified in flies [reviewed in Ref. (184)]. In Drosophila, several lines of evidence suggest a link between social behavior and the circadian clock. For example, behaviors such as courtship and mating show a circadian timing (185–188). In addition, an interaction between social behavior and the circadian clock has been demonstrated by Levine et al. (189), who showed that the phase of circadian locomotor activity is modulated by social context. In fact, the locomotor activity phase of wild-type flies was more dispersed in the presence of per0 (arrhythmic) mutant flies and advanced in the presence of perShort (perS) individuals, which are characterized by a circadian clock with a short periodicity (190). These effects were found to be dependent on many variables, such as the size and relative genetic composition of the group, the time of the day, and the ability to perceive odors, since anosmic flies were insensitive to the modification of the social context (189).
Following evaluation of the relationship between mating behavior and the circadian clock, it was shown that the production of sex pheromones in males, which occurs at the level of specific cuticular cells named oenocytes, is clock-regulated and controlled by a peripheral circadian clock within the oenocytes themselves (94). The oenocyte molecular clock appears to be less autonomous with respect to those of the other peripheral tissues, since the master clock is able to modulate the phase of oenocytes via PDF, which might act as a neuroendocrine signal (98). In flies, social context can act as an input signal to the clock (zeitgeber), influencing the expression of circadian clock genes at the level of the central nervous system as well as in the periphery (94, 98), thus modulating the circadian locomotor behavior (189), as well as other behavioral outputs such as mating and possibly other aspects of social behavior. For instance, it is interesting to note that oenocytes are also important in the regulation of male–male social interactions, since mutant males lacking oenocytes show low levels of aggression and high levels of male–male courtship compared to wild-type flies (99).
To our knowledge, the only studies conducted in Drosophila, which could be thought of as addressing the link between the circadian clock and neuropsychiatric diseases, were performed independently by two different research groups some years ago (191, 192). Both analyses evaluated the effects of the drugs, lithium and valproate (192), or lithium only (191), on Drosophila circadian locomotor activity.
Lithium and valproate are mood stabilizers that have been widely used for the treatment of bipolar disorder (193–195). To a more limited degree, valproate is also used in the treatment of schizophrenia (196). Although the mechanisms of action of lithium and valproate are still not completely understood, it is known that both types of drugs affect several biological phenomena which might be related to their therapeutic effects, including neurotransmitter release, monoamine metabolism, neuronal excitability, adult neurogenesis, as well as different circadian parameters (197–201). These effects are possibly the result of a direct inhibitory activity of both drugs on the G-proteins, myo-inositol monophosphatase (mIMP), and the mGSK-3α and mGSK-3β isoforms (197, 200).
Similarly to mammals, in Drosophila, adults chronic administration of lithium, within doses normally employed to treat human mood disorders, determines an increase in the periodicity of free-running circadian locomotor activity rhythms (191, 192). Analogous results were obtained following valproate treatments, although the effect was weaker with respect to that observed in the case of lithium; in particular, valproate was also more toxic to the flies (190). These reports further suggest that lithium possibly affects fly circadian periodicity by acting on dSHAGGY, the Drosophila ortholog of mGSK-3β (Figures 2A,B). As mentioned earlier, this kinase is part of the molecular clockwork and acts by phosphorylating dPER and dTIM, thereby regulating their daily nuclear translocation which contributes to the fine tuning of circadian rhythmicity (58, 59). Dokucu and colleagues showed that flies heterozygous for a null shaggy mutation as well as individuals overexpressing dshaggy, specifically in pdf-expressing neurons, showed a lengthening of their circadian periodicity after chronic exposure to lithium (190). It is worth underlining that heterozygote shaggy null mutants are characterized by longer circadian periodicity compared to controls, while overexpression of dshaggy in pdf-neurons leads to short-period circadian locomotor activity. Furthermore, Padiath and colleagues showed that lithium-treated flies were characterized by a reduced activity of the fly mGSK-3β homolog (189).
Even though the above studies evaluated a single behavioral trait, these data still suggest that Drosophila might represent a useful translational animal model to screen for candidate drugs suitable for the treatment of neuropsychiatric disorders. In addition, the similarity between the effects obtained on the circadian parameters and the molecular target/s in both mammals and flies supports the idea that Drosophila might be used in the evaluation of the link between the circadian clock and neuropsychiatric diseases.
The multifactorial nature of mental illnesses makes the study of neuropsychiatric diseases in animal models extremely difficult. One of the major problems arises from the lack of specific biomarkers for the different types of these disorders. In addition, psychiatric patients are characterized by several behavioral traits and it is difficult to have reliable tests, which mimic psychiatric-like behaviors in non-human organisms. On the other hand, specifically in the case of flies, it is practically feasible to subject experimental groups of animals to batteries of behavioral tests, the main characteristics of which are described below. The value of these assays does not lie so much in the information that each can provide singularly but, in keeping with the complex behavioral aspects typical of neuropsychiatric illnesses, in the information that the whole set can provide collectively.
Susceptibility of flies to seizures can be evaluated following hyperstimulation by mechanical shock (202). Following mechanical hyperstimulation, modifications of the original protocol allow flies to be assayed so that following the mechanical shock, flies are not only assayed for the time taken to recover from the temporary seizure, but they are also video-recorded in order to evaluate the time taken to climb to different heights of the vial in which they are contained (203). The assay performed in this way allows not only the determination of the post-hyperstimulation recovery of flies from seizure, but also the time taken to regain locomotor coordination.
Nociception can be determined using a heat-plate test paradigm [i.e., as described in Ref. (204)]. In this paradigm, avoidance of noxious heat is determined by placing groups of flies in a sealed experimental chamber, in the dark. The bottom of one end of the chamber is heated to 46°C using a computer controlled-thermoelectric (Peltier) element, and following a period of 4 min the distribution of flies within the chamber and the percentage of avoidance are estimated based on counting the number of flies that fail to avoid the noxious temperature, compared to the total number of flies in the chamber.
Recent advances in the field of machine vision and video-tracking techniques have led to the development of an open source software project called Ctrax (205). Continuous high definition filming and tracking of up to 20 flies moving simultaneously in a single arena for periods of up to 1 h is a realistic experimental condition. The tracking data are then subjected to analysis to provide (i) qualitative and quantitive data on social interactions occurring between pairs of flies during the whole tracking period; (ii) basic locomotor information from the single flies, such as distance traveled, speed of locomotion, turning angle, and time spent moving. From the latter, it is also possible to extrapolate information regarding cognitive aspects of locomotion, which relate to decision making processes active during the exploration of the arena by the individual flies.
Optokinetic response and attention can be determined by using a “maze” approach as described in Ref. (206). This apparently simple paradigm, in its basic form, provides a measure of the integrity of the neuronal circuitry underlying the perception and elaboration of visual information. Through manipulation of the experimental conditions, it is also possible to evaluate the integrity of attention-like processes.
Circadian analysis can be conducted as described in detail in (115, 205, 207). Whichever approach is adopted, the analysis of sleep patterns, based on the criteria originally defined by Shaw et al. (125), can be performed using the open source PySolo software package, as mentioned above (114, 125).
The determination of food preference in groups of flies can be established using the two-taste discrimination test (208). The test allows the evaluation of two kinds of output. (i) On the one hand it is possible to precisely quantify the preference between attractively tasting and unpleasantly flavored feeding solutions. (ii) The second possibility is to determine the tendency of flies under certain conditions (i.e., genetically, pharmacologically and/or environmentally determined) to prefer feeding solutions laced with known quantities of addictive compounds, such as ethanol, with respect to the standard feeding solution (209). The latter modality of conducting the test allows the evaluation of addiction/reward-seeking behavior of flies.
A rather recent approach consists in a learning paradigm, which the authors (210) called no-idleness learning. The response (or rather, lack of response) of an organism to unrelentingly uncontrollable noxious environmental variations is governed by a special kind of LM called learned uncontrollability. Under such conditions, an animal can learn that there is no patterned behavioral response which can be used to avoid the noxious effects of the essentially unpredictable environmental variations. In particular, in Drosophila, learned helplessness appears to consist in a cognitive element, which is the learned uncontrollability, and a motivational component, which consists in the decreased behavioral activity of the animal under the stressful conditions (209).
Drosophila has been recently proposed as a model organism to study neuropsychiatric disorders (211–213). The main reasons which justify this choice are that: (i) flies show a relatively simple brain, which shows interesting functional homologies with important regions of the mammalian brain; (ii) the fundamental neurobiological processes and neurotransmitter systems are conserved; (iii) flies are characterized by a set of complex behaviors such as sleep/wake cycles, LM and social interactions, which can be modulated by experience. Similar endophenotypes are often impaired in neuropsychiatric patients. Drosophila, therefore, represents a powerful tool for the comprehension of the molecular, cellular, and neural circuit mechanisms, which form the basis of such behaviors in both normal and impaired conditions (211).
In the study of the possible link between psychiatric diseases and the circadian clock, the use of Drosophila might provide an advantage for three main reasons: (i) the Drosophila circadian system organization is simpler compared to that of mammals; (ii) genetic mutations at the level of circadian clock genes, which cause modifications in the timing of the circadian clock, are available such as, for example, the two dper gene mutants, which show shortening (perS) or lengthening (perLong, perL) of the clock periodicity (190); (iii) Drosophila has homologs to most of the candidate genes associated with psychiatric diseases, as reported in case-control and genome-wide associations (GWA) studies (212, 214, 215). For these genes, Drosophila null or knock-down mutants are already available or easily obtainable (i.e., from the public fly mutant repositories). It is, for example, noteworthy to mention dDmca1D, the fly homolog of mCACNA1C (or mCaV1.2), a gene encoding for an L-Type voltage-gated calcium channel which has been found in several GWA analyses to be associated with both schizophrenia and bipolar disorders (216, 217). Interestingly, it was demonstrated that in mammals the expression of mCav1.2 is rhythmic and modulated by the circadian clock element mREV-ERBα. In addition, mCav1.2 appears to be involved in resetting of the circadian clock by light (218). In Drosophila, it is relatively easy to construct strains in which null mutations in psychiatric-disease-candidate genes are carried in a “sensitized” genetic background (i.e., having a slightly perturbed circadian clock, as in the case of the perS and perL mutants). The double or single mutants can then be tested in stress-inducing environments, which might be represented, for example, by extreme LD cycles or a condition of sexual deprivation (209). This approach would open the possibility of evaluating both Gene X Environment (G X E) and Gene X Gene X Environment (G X G X E) interactions, which probably constitute the basis of the multifactorial nature of psychiatric diseases.
In Drosophila, experimental designs exploring G X E and G X G X E interactions could be used to test the hypotheses which link the circadian clock to psychiatric disorders in humans. For example, it should be possible to evaluate the idea that an increased risk for psychiatric disease is associated with an environmentally and/or genetically induced misalignment between the central and the peripheral clocks. One of the possibilities to approach this problem in Drosophila could be to analyze the effects of such perturbations on parameters of social interaction and their underlying neuronal circuitries. In fact, as illustrated in the previous sections, the relationship between the master clock and the peripheral oenocyte clocks, which have been shown to play an important role in modulating fly social interactions, resembles the situation in mammals in which the autonomous central clock governs the phase of “slave” peripheral clocks. Moreover, the hypothesis of a direct influence of the master clock on psychiatric disease might be evaluated using the behavioral outputs of the learning and short-term memory phenotypes, which are modulated by the clock both in Drosophila and mammals. However, differently from mammals, the Drosophila brain regions involved in the control of these phenotypes apparently do not contain a running clock, but are directly or indirectly influenced by the master clock. Therefore, this represents a simplified system to evaluate G X E and G X G X E interactions. Finally, another possibility is to conduct the G X E and G X G X E analyses using the “omic” approach, i.e., by evaluating several relatively simple traits in parallel in a “behavioromic” type of approach. With the Drosophila powerful transgenic toolbox, it should then be possible to identify the specific neuronal circuitry governing the selected behaviors and to characterize the activity of such neuronal circuits during development and adulthood as well as in response to pharmacological treatments.
As has already transpired from this, as well as from other excellent reviews, it is clear that in Drosophila, given a behavioral paradigm it is straightforward to choose a genetic approach in order to define the molecular, cellular, and circuit mechanisms, as well as the pathogenesis of impairments caused by specific genetic, pharmacological, and/or environmental manipulations. In particular, Drosophila is a choice model organism in which to test the in vivo therapeutic potential of large numbers of chemicals as a first tier approach in the translation to mammalian models and humans. In this respect, the possibility of testing relevant behavioral end points in a high throughput manner is of particular importance. Currently, high throughput designs have been described for the evaluation of: (i) circadian patterns of activity/sleep (115); (ii) optomotor performance and attention (219); (iii) locomotor activity and social interactions following simultaneous tracking of multiple flies (205). In conclusion, we argue that the molecular/genetic, cellular, and neurobiological features of Drosophila make this a choice translational model in which to test the causal link between genetic and/or environmental manipulations leading to perturbations of the circadian system and defects in a suite of behavioral traits which, collectively, would address some of the key neurological features involved in neuropsychiatric diseases. Such investigations could prove of extreme value in pointing the way for more focused studies in model organisms evolutionarily closer to humans.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Matteo Simonetti for the graphical support. We would also like to acknowledge the use of the following open-source software packages: Zotero (http://www.zotero.org; a bibliographical database and citation manager), Blender (http://www.blender.org; a 3D design and rendering software), GIMP (http://www.gimp.org; an image manipulation program), and Inkscape (https://inkscape.org/en/; a vector graphics editor). No funding was obtained specifically for this review.
1. Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet (2012) 13:537–51. doi: 10.1038/nrg3240
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Lau JYF, Eley TC. The genetics of mood disorders. Annu Rev Clin Psychol (2010) 6:313–37. doi:10.1146/annurev.clinpsy.121208.131308
3. Karatsoreos IN. Links between circadian rhythms and psychiatric disease. Front Behav Neurosci (2014) 8:162. doi:10.3389/fnbeh.2014.00162
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Landgraf D, McCarthy MJ, Welsh DK. Circadian clock and stress interactions in the molecular biology of psychiatric disorders. Curr Psychiatry Rep (2014) 16:483. doi:10.1007/s11920-014-0483-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Gonzalez R. The relationship between bipolar disorder and biological rhythms. J Clin Psychiatry (2014) 75:e323–31. doi:10.4088/JCP.13r08507
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron (2012) 74:246–60. doi:10.1016/j.neuron.2012.04.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol (2010) 72:517–49. doi:10.1146/annurev-physiol-021909-135821
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Helfrich-Förster C. Neurobiology of the fruit fly’s circadian clock. Genes Brain Behav (2005) 4:65–76. doi:10.1111/j.1601-183X.2004.00092.x
9. Hermann C, Yoshii T, Dusik V, Helfrich-Förster C. Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J Comp Neurol (2012) 520:970–87. doi:10.1002/cne.22742
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Bunney WE, Bunney BG. Molecular clock genes in man and lower animals: possible implications for circadian abnormalities in depression. Neuropsychopharmacology (2000) 22:335–45. doi:10.1016/S0893-133X(99)00145-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Emens J, Lewy A, Kinzie JM, Arntz D, Rough J. Circadian misalignment in major depressive disorder. Psychiatry Res (2009) 168:259–61. doi:10.1016/j.psychres.2009.04.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Etain B, Milhiet V, Bellivier F, Leboyer M. Genetics of circadian rhythms and mood spectrum disorders. Eur Neuropsychopharmacol (2011) 21(Suppl 4):S676–82. doi:10.1016/j.euroneuro.2011.07.007
13. Partonen T. Clock gene variants in mood and anxiety disorders. J Neural Transm (2012) 119:1133–45. doi:10.1007/s00702-012-0810-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A (2013) 110:9950–5. doi:10.1073/pnas.1305814110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Driesen K, Jansen NWH, Kant I, Mohren DCL, van Amelsvoort LGPM. Depressed mood in the working population: associations with work schedules and working hours. Chronobiol Int (2010) 27:1062–79. doi:10.3109/07420528.2010.489877
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Cho K. Chronic “jet lag” produces temporal lobe atrophy and spatial cognitive deficits. Nat Neurosci (2001) 4:567–8. doi:10.1038/88384
17. Levandovski R, Dantas G, Fernandes LC, Caumo W, Torres I, Roenneberg T, et al. Depression scores associate with chronotype and social jetlag in a rural population. Chronobiol Int (2011) 28:771–8. doi:10.3109/07420528.2011.602445
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. McClung CA, Sidiropoulou K, Vitaterna M, Takahashi JS, White FJ, Cooper DC, et al. Regulation of dopaminergic transmission and cocaine reward by the clock gene. Proc Natl Acad Sci U S A (2005) 102:9377–81. doi:10.1073/pnas.0503584102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Roybal K, Theobold D, Graham A, DiNieri JA, Russo SJ, Krishnan V, et al. Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci U S A (2007) 104:6406–11. doi:10.1073/pnas.0609625104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Hampp G, Ripperger JA, Houben T, Schmutz I, Blex C, Perreau-Lenz S, et al. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol (2008) 18:678–83. doi:10.1016/j.cub.2008.04.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Mukherjee S, Coque L, Cao J-L, Kumar J, Chakravarty S, Asaithamby A, et al. Knockdown of clock in the ventral tegmental area through RNA interference results in a mixed state of mania and depression-like behavior. Biol Psychiatry (2010) 68:503–11. doi:10.1016/j.biopsych.2010.04.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Schnell A, Chappuis S, Schmutz I, Brai E, Ripperger JA, Schaad O, et al. The nuclear receptor REV-ERBα regulates Fabp7 and modulates adult hippocampal neurogenesis. PLoS One (2014) 9:e99883. doi:10.1371/journal.pone.0099883
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Sidor MM, Spencer SM, Dzirasa K, Parekh PK, Tye KM, Warden MR, et al. Daytime spikes in dopaminergic activity drive rapid mood-cycling in mice. Mol Psychiatry (2015) 5:1–14. doi:10.1038/mp.2014.167
24. Schnell A, Albrecht U, Sandrelli F. Rhythm and mood: relationships between the circadian clock and mood-related behavior. Behav Neurosci (2014) 128:326–43. doi:10.1037/a0035883
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Rosenwasser AM. Circadian clock genes: non-circadian roles in sleep, addiction, and psychiatric disorders? Neurosci Biobehav Rev (2010) 34:1249–55. doi:10.1016/j.neubiorev.2010.03.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Denes AS, Jékely G, Steinmetz PRH, Raible F, Snyman H, Prud’homme B, et al. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell (2007) 129:277–88. doi:10.1016/j.cell.2007.02.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. Farris SM. Evolutionary convergence of higher brain centers spanning the protostome-deuterostome boundary. Brain Behav Evol (2008) 72:106–22. doi:10.1159/000151471
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Zars T. Behavioral functions of the insect mushroom bodies. Curr Opin Neurobiol (2000) 10:790–5. doi:10.1016/S0959-4388(00)00147-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Farris SM. Structural, functional and developmental convergence of the insect mushroom bodies with higher brain centers of vertebrates. Brain Behav Evol (2008) 72:1–15. doi:10.1159/000139457
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Farris SM. Are mushroom bodies cerebellum-like structures? Arthropod Struct Dev (2011) 40:368–79. doi:10.1016/j.asd.2011.02.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Van Swinderen B. Fly memory: a mushroom body story in parts. Curr Biol (2009) 19:R855–7. doi:10.1016/j.cub.2009.07.064
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Mizunami M, Weibrecht JM, Strausfeld NJ. Mushroom bodies of the cockroach: their participation in place memory. J Comp Neurol (1998) 402:520–37. doi:10.1002/(SICI)1096-9861(19981228)402:4<520::AID-CNE6>3.3.CO;2-B
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
33. Strausfeld NJ, Hansen L, Li Y, Gomez RS, Ito K. Evolution, discovery, and interpretations of arthropod mushroom bodies. Learn Mem (1998) 5:11–37.
34. Ofstad TA, Zuker CS, Reiser MB. Visual place learning in Drosophila melanogaster. Nature (2011) 474:204–7. doi:10.1038/nature10131
35. Pfeiffer K, Homberg U. Organization and functional roles of the central complex in the insect brain. Annu Rev Entomol (2014) 59:165–84. doi:10.1146/annurev-ento-011613-162031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. De Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, et al. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol (2007) 302:309–23. doi:10.1016/j.ydbio.2006.09.035
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Wirmer A, Bradler S, Heinrich R. Homology of insect corpora allata and vertebrate adenohypophysis? Arthropod Struct Dev (2012) 41:409–17. doi:10.1016/j.asd.2012.04.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Hartenstein V. The neuroendocrine system of invertebrates: a developmental and evolutionary perspective. J Endocrinol (2006) 190:555–70. doi:10.1677/joe.1.06964
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci (2007) 10:1160–7. doi:10.1038/nn1957
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron (2010) 65:670–81. doi:10.1016/j.neuron.2010.01.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Gatti S, Ferveur JF, Martin JR. Genetic identification of neurons controlling a sexually dimorphic behaviour. Curr Biol (2000) 10:667–70. doi:10.1016/S0960-9822(00)00517-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Belgacem YH, Martin J-R. Neuroendocrine control of a sexually dimorphic behavior by a few neurons of the pars intercerebralis in Drosophila. Proc Natl Acad Sci U S A (2002) 99:15154–8. doi:10.1073/pnas.232244199
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Cavanaugh DJ, Geratowski JD, Wooltorton JRA, Spaethling JM, Hector CE, Zheng X, et al. Identification of a circadian output circuit for rest: activity rhythms in Drosophila. Cell (2014) 157:689–701. doi:10.1016/j.cell.2014.02.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
44. Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science (2002) 296:1118–20. doi:10.1126/science.1070058
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Broughton SJ, Piper MDW, Ikeya T, Bass TM, Jacobson J, Driege Y, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A (2005) 102:3105–10. doi:10.1073/pnas.0405775102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Strausfeld NJ, Hirth F. Deep homology of arthropod central complex and vertebrate basal ganglia. Science (2013) 340:157–61. doi:10.1126/science.1231828
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Miller SM, Ngo TT, van Swinderen B. Attentional switching in humans and flies: rivalry in large and miniature brains. Front Hum Neurosci (2011) 5:188. doi:10.3389/fnhum.2011.00188
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Van Swinderen B, Andretic R. Dopamine in Drosophila: setting arousal thresholds in a miniature brain. Proc Biol Sci (2011) 278:906–13. doi:10.1098/rspb.2010.2564
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Curran KP, Chalasani SH. Serotonin circuits and anxiety: what can invertebrates teach us? Invert Neurosci (2012) 12:81–92. doi:10.1007/s10158-012-0140-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Yamamoto S, Seto ES. Dopamine dynamics and signaling in Drosophila: an overview of genes, drugs and behavioral paradigms. Exp Anim (2014) 63:107–19. doi:10.1538/expanim.63.107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Enell L, Hamasaka Y, Kolodziejczyk A, Nässel DR. Gamma-aminobutyric acid (GABA) signaling components in Drosophila: immunocytochemical localization of GABA(B) receptors in relation to the GABA(A) receptor subunit RDL and a vesicular GABA transporter. J Comp Neurol (2007) 505:18–31. doi:10.1002/cne.21472
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Roeder T. Tyramine and octopamine: ruling behavior and metabolism. Annu Rev Entomol (2005) 50:447–77. doi:10.1146/annurev.ento.50.071803.130404
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Özkaya Ö, Rosato E. The circadian clock of the fly: a neurogenetics journey through time. Adv Genet (2012) 77:79–123. doi:10.1016/B978-0-12-387687-4.00004-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol (2013) 23:724–31. doi:10.1016/j.conb.2013.02.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Iitaka C, Miyazaki K, Akaike T, Ishida NA. Role for glycogen synthase kinase-3 in the mammalian circadian clock. J Biol Chem (2005) 280:29397–402. doi:10.1074/jbc.M503526200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, et al. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase iepsilon. Cell (1998) 94:97–107. doi:10.1016/S0092-8674(00)81225-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell (1998) 94:83–95. doi:10.1016/S0092-8674(00)81224-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell (2001) 105:769–79. doi:10.1016/S0092-8674(01)00383-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. A hierarchical phosphorylation cascade that regulates the timing of period nuclear entry reveals novel roles for proline-directed kinases and GSK-3beta/SGG in circadian clocks. J Neurosci (2010) 30:12664–75. doi:10.1523/JNEUROSCI.1586-10.2010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Cyran SA, Buchsbaum AM, Reddy KL, Lin M-C, Glossop NRJ, Hardin PE, et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell (2003) 112:329–41. doi:10.1016/S0092-8674(03)00074-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Glossop NRJ, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of clock in the Drosophila circadian oscillator. Neuron (2003) 37:249–61. doi:10.1016/S0896-6273(03)00002-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell (2002) 110:251–60. doi:10.1016/S0092-8674(02)00825-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron (2004) 43:527–37. doi:10.1016/j.neuron.2004.07.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell (1998) 95:669–79. doi:10.1016/S0092-8674(00)81637-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell (1998) 95:681–92. doi:10.1016/S0092-8674(00)81638-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Lin FJ, Song W, Meyer-Bernstein E, Naidoo N, Sehgal A. Photic signaling by cryptochrome in the Drosophila circadian system. Mol Cell Biol (2001) 21:7287–94. doi:10.1128/MCB.21.21.7287-7294.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science (2006) 312:1809–12. doi:10.1126/science.1124951
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Peschel N, Helfrich-Förster C. Setting the clock – by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett (2011) 585:1435–42. doi:10.1016/j.febslet.2011.02.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol (2010) 72:605–24. doi:10.1146/annurev-physiol-021909-135815
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Muraro NI, Pírez N, Ceriani MF. The circadian system: plasticity at many levels. Neuroscience (2013) 247:280–93. doi:10.1016/j.neuroscience.2013.05.036
71. Vosko AM, Schroeder A, Loh DH, Colwell CS. Vasoactive intestinal peptide and the mammalian circadian system. Gen Comp Endocrinol (2007) 152:165–75. doi:10.1016/j.ygcen.2007.04.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Talsma AD, Christov CP, Terriente-Felix A, Linneweber GA, Perea D, Wayland M, et al. Remote control of renal physiology by the intestinal neuropeptide pigment-dispersing factor in Drosophila. Proc Natl Acad Sci U S A (2012) 109:12177–82. doi:10.1073/pnas.1200247109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, Allada R. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol (2014) 12:e1001810. doi:10.1371/journal.pbio.1001810
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Vanden Broeck J. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides (2001) 22:241–54. doi:10.1016/S0196-9781(00)00376-4
75. Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides (2002) 23:773–80. doi:10.1016/S0196-9781(01)00647-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
76. Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature (2004) 431:869–73. doi:10.1038/nature02935
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature (2004) 431:862–8. doi:10.1038/nature02926
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell (1999) 99:791–802. doi:10.1016/S0092-8674(00)81676-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Dissel S, Hansen CN, Özkaya Ö, Hemsley M, Kyriacou CP, Rosato E. The logic of circadian organization in Drosophila. Curr Biol (2014) 24:2257–66. doi:10.1016/j.cub.2014.08.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science (2014) 343:1516–20. doi:10.1126/science.1251285
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Zhao Y, Bretz CA, Hawksworth SA, Hirsh J, Johnson EC. Corazonin neurons function in sexually dimorphic circuitry that shape behavioral responses to stress in Drosophila. PLoS One (2010) 5:e9141. doi:10.1371/journal.pone.0009141
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
82. Son GH, Chung S, Kim K. The adrenal peripheral clock: glucocorticoid and the circadian timing system. Front Neuroendocrinol (2011) 32:451–65. doi:10.1016/j.yfrne.2011.07.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron (2007) 55:435–47. doi:10.1016/j.neuron.2007.06.038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Ng FS, Tangredi MM, Jackson FR. Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr Biol (2011) 21:625–34. doi:10.1016/j.cub.2011.03.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Slat E, Freeman GM, Herzog ED. The clock in the brain: neurons, glia, and networks in daily rhythms. Handb Exp Pharmacol (2013) 217:105–23. doi:10.1007/978-3-642-25950-0_5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol (2000) 422:66–94. doi:10.1002/(SICI)1096-9861(20000619)422:1<66::AID-CNE5>3.0.CO;2-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol (2008) 508:952–66. doi:10.1002/cne.21702
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Chen C-C, Wu J-K, Lin H-W, Pai T-P, Fu T-F, Wu C-L, et al. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science (2012) 335:678–85. doi:10.1126/science.1212735
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature (1999) 400:375–8. doi:10.1038/22566
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
90. Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol (2004) 14:638–49. doi:10.1016/j.cub.2004.04.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. Chatterjee A, Tanoue S, Houl JH, Hardin PE. Regulation of gustatory physiology and appetitive behavior by the Drosophila circadian clock. Curr Biol (2010) 20:300–9. doi:10.1016/j.cub.2009.12.055
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Myers EM, Yu J, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr Biol (2003) 13:526–33. doi:10.1016/S0960-9822(03)00167-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Giebultowicz JM, Hege DM. Circadian clock in malpighian tubules. Nature (1997) 386:664. doi:10.1038/386664a0
94. Krupp JJ, Kent C, Billeter J-C, Azanchi R, So AK-C, Schonfeld JA, et al. Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster. Curr Biol (2008) 18:1373–83. doi:10.1016/j.cub.2008.07.089
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Granados-Fuentes D. The suprachiasmatic nucleus entrains, but does not sustain, circadian rhythmicity in the olfactory bulb. J Neurosci (2004) 24:615–9. doi:10.1523/JNEUROSCI.4002-03.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science (1997) 278:1632–5. doi:10.1126/science.278.5343.1632
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Ivanchenko M, Stanewsky R, Giebultowicz JM. Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J Biol Rhythms (2001) 16:205–15. doi:10.1177/074873001129001917
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. Krupp JJ, Billeter J-C, Wong A, Choi C, Nitabach MN, Levine JD. Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in Drosophila. Neuron (2013) 79:54–68. doi:10.1016/j.neuron.2013.05.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Wang L, Han X, Mehren J, Hiroi M, Billeter J-C, Miyamoto T, et al. Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat Neurosci (2011) 14:757–62. doi:10.1038/nn.2800
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Wulff K, Gatti S, Wettstein JG, Foster RG. Sleep and circadian rhythm disruption in psychiatric and neurodegenerative disease. Nat Rev Neurosci (2010) 11:589–99. doi:10.1038/nrn2868
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Jagannath A, Peirson SN, Foster RG. Sleep and circadian rhythm disruption in neuropsychiatric illness. Curr Opin Neurobiol (2013) 23:888–94. doi:10.1016/j.conb.2013.03.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Dere E, Pause BM, Pietrowsky R. Emotion and episodic memory in neuropsychiatric disorders. Behav Brain Res (2010) 215:162–71. doi:10.1016/j.bbr.2010.03.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders: DSM-5. 5th ed. Washington, DC: American Psychiatric Association (2013).
104. Rihel J, Schier AF. Sites of action of sleep and wake drugs: insights from model organisms. Curr Opin Neurobiol (2013) 23:831–40. doi:10.1016/j.conb.2013.04.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Alexandre C, Andermann ML, Scammell TE. Control of arousal by the orexin neurons. Curr Opin Neurobiol (2013) 23:752–9. doi:10.1016/j.conb.2013.04.008
106. Frank MG. Astroglial regulation of sleep homeostasis. Curr Opin Neurobiol (2013) 23:812–8. doi:10.1016/j.conb.2013.02.009
107. Naylor E, Bergmann BM, Krauski K, Zee PC, Takahashi JS, Vitaterna MH, et al. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci (2000) 20:8138–43.
108. Wisor JP, O’Hara BF, Terao A, Selby CP, Kilduff TS, Sancar A, et al. A role for cryptochromes in sleep regulation. BMC Neurosci (2002) 3:20. doi:10.1186/1471-2202-3-20
109. Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep (2005) 28:395–409.
110. Kopp C, Albrecht U, Zheng B, Tobler I. Homeostatic sleep regulation is preserved in mPer1 and mPer2 mutant mice. Eur J Neurosci (2002) 16:1099–106. doi:10.1046/j.1460-9568.2002.02156.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
111. Cirelli C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat Rev Neurosci (2009) 10:549–60. doi:10.1038/nrn2683
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
112. Nitz DA, van Swinderen B, Tononi G, Greenspan RJ. Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr Biol (2002) 12:1934–40. doi:10.1016/S0960-9822(02)01300-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
113. Van Swinderen B, Nitz DA, Greenspan RJ. Uncoupling of brain activity from movement defines arousal States in Drosophila. Curr Biol (2004) 14:81–7. doi:10.1016/S0960-9822(03)00987-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
114. Gilestro GF, Cirelli C. pySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics (2009) 25:1466–7. doi:10.1093/bioinformatics/btp237
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
115. Gilestro GF. Video tracking and analysis of sleep in Drosophila melanogaster. Nat Protoc (2012) 7:995–1007. doi:10.1038/nprot.2012.041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
116. Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci (2008) 11:354–9. doi:10.1038/nn2046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
117. Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol (2006) 16:1051–62. doi:10.1016/j.cub.2006.04.032
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
118. Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol (2005) 15:1165–75. doi:10.1016/j.cub.2005.05.025
119. Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci (2005) 25:7377–84. doi:10.1523/JNEUROSCI.2048-05.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
120. Sakurai T. The neural circuit of orexin (hypocretin): maintaining sleep and wakefulness. Nat Rev Neurosci (2007) 8:171–81. doi:10.1038/nrn2092
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
121. Crocker A, Sehgal A. Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J Neurosci (2008) 28:9377–85. doi:10.1523/JNEUROSCI.3072-08a.2008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
122. Van Alphen B, Yap MHW, Kirszenblat L, Kottler B, van Swinderen B. A dynamic deep sleep stage in Drosophila. J Neurosci (2013) 33:6917–27. doi:10.1523/JNEUROSCI.0061-13.2013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
123. Seugnet L, Boero J, Gottschalk L, Duntley SP, Shaw PJ. Identification of a biomarker for sleep drive in flies and humans. Proc Natl Acad Sci U S A (2006) 103:19913–8. doi:10.1073/pnas.0609463104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
124. Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, et al. Rest in Drosophila is a sleep-like state. Neuron (2000) 25:129–38. doi:10.1016/S0896-6273(00)80877-6
125. Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science (2000) 287:1834–7. doi:10.1126/science.287.5459.1834
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
126. Hendricks JC, Lu S, Kume K, Yin JCP, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms (2003) 18:12–25. doi:10.1177/0748730402239673
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
127. Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature (2002) 417:287–91. doi:10.1038/417287a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
128. Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature (2006) 441:757–60. doi:10.1038/nature04811
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
129. Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature (2006) 441:753–6. doi:10.1038/nature04739
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
130. Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol (2012) 22:2114–23. doi:10.1016/j.cub.2012.09.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
131. Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, et al. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci (2012) 15:1516–23. doi:10.1038/nn.3238
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
132. Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJL, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron (2008) 60:672–82. doi:10.1016/j.neuron.2008.10.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
133. Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou Y-T, Sharma VK, et al. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol (2008) 18:1537–45. doi:10.1016/j.cub.2008.08.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
134. Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABAA receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol (2009) 19:386–90. doi:10.1016/j.cub.2009.01.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
135. Shang Y, Haynes P, Pírez N, Harrington KI, Guo F, Pollack J, et al. Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci (2011) 14:889–95. doi:10.1038/nn.2860
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
136. Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, Griffith LC. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron (2013) 80:171–83. doi:10.1016/j.neuron.2013.07.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
137. Gmeiner F, Kolodziejczyk A, Yoshii T, Rieger D, Nassel DR, Helfrich-Forster C. GABAB receptors play an essential role in maintaining sleep during the second half of the night in Drosophila melanogaster. J Exp Biol (2013) 216:3837–43. doi:10.1242/jeb.085563
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
138. Diekelmann S. Sleep for cognitive enhancement. Front Syst Neurosci (2014) 8:46. doi:10.3389/fnsys.2014.00046
139. Wyatt JK, Ritz-De Cecco A, Czeisler CA, Dijk DJ. Circadian temperature and melatonin rhythms, sleep, and neurobehavioral function in humans living on a 20-h day. Am J Physiol (1999) 277:R1152–63.
140. Wright KP, Hull JT, Czeisler CA. Relationship between alertness, performance, and body temperature in humans. Am J Physiol Regul Integr Comp Physiol (2002) 283:R1370–7. doi:10.1152/ajpregu.00205.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
141. Smarr BL, Jennings KJ, Driscoll JR, Kriegsfeld LJ. A time to remember: the role of circadian clocks in learning and memory. Behav Neurosci (2014) 128:283–303. doi:10.1037/a0035963
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
142. Gaggioni G, Maquet P, Schmidt C, Dijk D-J, Vandewalle G. Neuroimaging, cognition, light and circadian rhythms. Front Syst Neurosci (2014) 8:126. doi:10.3389/fnsys.2014.00126
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
143. Jilg A, Lesny S, Peruzki N, Schwegler H, Selbach O, Dehghani F, et al. Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus (2009) 20(3):377–88. doi:10.1002/hipo.20637
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
144. Wardlaw SM, Phan TH, Saraf A, Chen X, Storm DR. Genetic disruption of the core circadian clock impairs hippocampus-dependent memory. Learn Mem (2014) 21:417–23. doi:10.1101/lm.035451.114
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
145. De Bundel D, Gangarossa G, Biever A, Bonnefont X, Valjent E. Cognitive dysfunction, elevated anxiety, and reduced cocaine response in circadian clock-deficient cryptochrome knockout mice. Front Behav Neurosci (2013) 7:152. doi:10.3389/fnbeh.2013.00152
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
146. Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, et al. Impaired cued and contextual memory in NPAS2-deficient mice. Science (2000) 288:2226–30. doi:10.1126/science.288.5474.2226
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
147. Marquie J-C, Tucker P, Folkard S, Gentil C, Ansiau D. Chronic effects of shift work on cognition: findings from the VISAT longitudinal study. Occup Environ Med (2014) 72:258–64. doi:10.1136/oemed-2013-101993
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
148. Athos J, Impey S, Pineda VV, Chen X, Storm DR. Hippocampal CRE-mediated gene expression is required for contextual memory formation. Nat Neurosci (2002) 5:1119–20. doi:10.1038/nn951
149. Kelleher RJ, Govindarajan A, Jung H-Y, Kang H, Tonegawa S. Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell (2004) 116:467–79. doi:10.1016/S0092-8674(04)00115-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
150. Kelly A, Laroche S, Davis S. Activation of mitogen-activated protein kinase/extracellular signal-regulated kinase in hippocampal circuitry is required for consolidation and reconsolidation of recognition memory. J Neurosci (2003) 23:5354–60.
151. Phan TH, Chan GC-K, Sindreu CB, Eckel-Mahan KL, Storm DR. The diurnal oscillation of MAP (mitogen-activated protein) kinase and adenylyl cyclase activities in the hippocampus depends on the suprachiasmatic nucleus. J Neurosci (2011) 31:10640–7. doi:10.1523/JNEUROSCI.6535-10.2011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
152. Antoun G, Cannon PB, Cheng H-YM. Regulation of MAPK/ERK signaling and photic entrainment of the suprachiasmatic nucleus circadian clock by Raf kinase inhibitor protein. J Neurosci (2012) 32:4867–77. doi:10.1523/JNEUROSCI.5650-11.2012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
153. Butcher GQ. Light stimulates MSK1 activation in the suprachiasmatic nucleus via a PACAP-ERK/MAP kinase-dependent mechanism. J Neurosci (2005) 25:5305–13. doi:10.1523/JNEUROSCI.4361-04.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
154. Dziema H, Oatis B, Butcher GQ, Yates R, Hoyt KR, Obrietan K. The ERK/MAP kinase pathway couples light to immediate-early gene expression in the suprachiasmatic nucleus. Eur J Neurosci (2003) 17:1617–27. doi:10.1046/j.1460-9568.2003.02592.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
155. Tischkau SA, Gallman EA, Buchanan GF, Gillette MU. Differential cAMP gating of glutamatergic signaling regulates long-term state changes in the suprachiasmatic circadian clock. J Neurosci (2000) 20:7830–7.
156. Sanada K, Okano T, Fukada Y. Mitogen-activated protein kinase phosphorylates and negatively regulates basic helix-loop-helix-PAS transcription factor BMAL1. J Biol Chem (2002) 277:267–71. doi:10.1074/jbc.M107850200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
157. Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A (2002) 99:7728–33. doi:10.1073/pnas.102075599
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
158. Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GC-K, Scheiner ZS, et al. Circadian oscillation of hippocampal MAPK activity and cAMP: implications for memory persistence. Nat Neurosci (2008) 11:1074–82. doi:10.1038/nn.2174
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
159. Caroni P, Donato F, Muller D. Structural plasticity upon learning: regulation and functions. Nat Rev Neurosci (2012) 13:478–90. doi:10.1038/nrn3258
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
160. Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat Rev Neurosci (2010) 11:339–50. doi:10.1038/nrn2822
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
161. Ikeno T, Weil ZM, Nelson RJ. Photoperiod affects the diurnal rhythm of hippocampal neuronal morphology of siberian hamsters. Chronobiol Int (2013) 30:1089–100. doi:10.3109/07420528.2013.800090
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
162. Perez-Cruz C, Simon M, Flügge G, Fuchs E, Czéh B. Diurnal rhythm and stress regulate dendritic architecture and spine density of pyramidal neurons in the rat infralimbic cortex. Behav Brain Res (2009) 205:406–13. doi:10.1016/j.bbr.2009.07.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
163. Gilhooley MJ, Pinnock SB, Herbert J. Rhythmic expression of per1 in the dentate gyrus is suppressed by corticosterone: implications for neurogenesis. Neurosci Lett (2011) 489:177–81. doi:10.1016/j.neulet.2010.12.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
164. Tamai S, Sanada K, Fukada Y. Time-of-day-dependent enhancement of adult neurogenesis in the hippocampus. PLoS One (2008) 3:e3835. doi:10.1371/journal.pone.0003835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
165. Smith JM, Hechtman A, Swann J. Fluctuations in cellular proliferation across the light/dark cycle in the subgranular zone of the dentate gyrus in the adult male Syrian hamster. Neurosci Lett (2010) 473:192–5. doi:10.1016/j.neulet.2010.02.039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
166. Borgs L, Beukelaers P, Vandenbosch R, Nguyen L, Moonen G, Maquet P, et al. Period 2 regulates neural stem/progenitor cell proliferation in the adult hippocampus. BMC Neurosci (2009) 10:30. doi:10.1186/1471-2202-10-30
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
167. Siwicki KK, Ladewski L. Associative learning and memory in Drosophila: beyond olfactory conditioning. Behav Processes (2003) 64:225–38. doi:10.1016/S0376-6357(03)00137-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
168. Vogt K, Schnaitmann C, Dylla KV, Knapek S, Aso Y, Rubin GM, et al. Shared mushroom body circuits underlie visual and olfactory memories in Drosophila. Elife (2014) 3:e02395. doi:10.7554/eLife.02395
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
169. Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell (1994) 79:35–47. doi:10.1016/0092-8674(94)90398-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
170. Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci (2003) 4:266–75. doi:10.1038/nrn1074
171. Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A (1985) 157:263–77. doi:10.1007/BF01350033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
172. Joiner MA, Griffith LC. Mapping of the anatomical circuit of CaM kinase-dependent courtship conditioning in Drosophila. Learn Mem (1999) 6:177–92.
173. Qin H, Cressy M, Li W, Coravos JS, Izzi SA, Dubnau J. Gamma neurons mediate dopaminergic input during aversive olfactory memory formation in Drosophila. Curr Biol (2012) 22:608–14. doi:10.1016/j.cub.2012.02.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
174. Wu C-L, Xia S, Fu T-F, Wang H, Chen Y-H, Leong D, et al. Specific requirement of NMDA receptors for long-term memory consolidation in Drosophila ellipsoid body. Nat Neurosci (2007) 10:1578–86. doi:10.1038/nn2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
175. McBride SM, Giuliani G, Choi C, Krause P, Correale D, Watson K, et al. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron (1999) 24:967–77. doi:10.1016/S0896-6273(00)81043-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
176. Gasque G, Labarca P, Delgado R, Darszon A. Bridging behavior and physiology: ion-channel perspective on mushroom body-dependent olfactory learning and memory in Drosophila. J Cell Physiol (2006) 209:1046–53. doi:10.1002/jcp.20764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
177. Xia S, Miyashita T, Fu T-F, Lin W-Y, Wu C-L, Pyzocha L, et al. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol (2005) 15:603–15. doi:10.1016/j.cub.2005.02.059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
178. Fropf R, Zhang J, Tanenhaus AK, Fropf WJ, Siefkes E, Yin JCP. Time of day influences memory formation and dCREB2 proteins in Drosophila. Front Syst Neurosci (2014) 8:43. doi:10.3389/fnsys.2014.00043
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
179. Tanenhaus AK, Zhang J, Yin JCP. In vivo circadian oscillation of dCREB2 and NF-κB activity in the Drosophila nervous system. PLoS One (2012) 7:e45130. doi:10.1371/journal.pone.0045130
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
180. Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron (1999) 22:777–87. doi:10.1016/S0896-6273(00)80736-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
181. Lyons LC, Roman G. Circadian modulation of short-term memory in Drosophila. Learn Mem (2008) 16:19–27. doi:10.1101/lm.1146009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
182. Sakai T, Tamura T, Kitamoto T, Kidokoro Y. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc Natl Acad Sci U S A (2004) 101:16058–63. doi:10.1073/pnas.0401472101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
183. Sakai T, Inami S, Sato S, Kitamoto T. Fan-shaped body neurons are involved in period-dependent regulation of long-term courtship memory in Drosophila. Learn Mem (2012) 19:571–4. doi:10.1101/lm.028092.112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
184. Sokolowski MB. Social interactions in “simple” model systems. Neuron (2010) 65:780–94. doi:10.1016/j.neuron.2010.03.007
185. Fujii S, Krishnan P, Hardin P, Amrein H. Nocturnal male sex drive in Drosophila. Curr Biol (2007) 17:244–51. doi:10.1016/j.cub.2006.11.049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
186. Tauber E, Roe H, Costa R, Hennessy JM, Kyriacou CP. Temporal mating isolation driven by a behavioral gene in Drosophila. Curr Biol (2003) 13:140–5. doi:10.1016/S0960-9822(03)00004-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
187. Hardeland R. Species differences in the diurnal rhythmicity of courtship behaviour within the melanogaster group of the genus Drosophila. Anim Behav (1972) 20:170–4. doi:10.1016/S0003-3472(72)80188-X
188. Sakai T, Ishida N. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc Natl Acad Sci U S A (2001) 98:9221–5. doi:10.1073/pnas.151443298
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
189. Levine JD, Funes P, Dowse HB, Hall JC. Resetting the circadian clock by social experience in Drosophila melanogaster. Science (2002) 298:2010–2. doi:10.1126/science.1076008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
190. Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A (1971) 68:2112–6. doi:10.1073/pnas.68.9.2112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
191. Padiath QS, Paranjpe D, Jain S, Sharma VK. Glycogen synthase kinase 3beta as a likely target for the action of lithium on circadian clocks. Chronobiol Int (2004) 21:43–55. doi:10.1081/CBI-120027981
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
192. Dokucu ME, Yu L, Taghert PH. Lithium- and valproate-induced alterations in circadian locomotor behavior in Drosophila. Neuropsychopharmacology (2005) 30:2216–24. doi:10.1038/sj.npp.1300764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
193. Bowden CL, Singh V. Valproate in bipolar disorder: 2000 onwards. Acta Psychiatr Scand Suppl (2005) 111 (Suppl 426):13–20. doi:10.1111/j.1600-0447.2005.00522.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
194. Tondo L, Baldessarini RJ. Reduced suicide risk during lithium maintenance treatment. J Clin Psychiatry (2000) 61(Suppl 9):97–104.
195. Cruceanu C, Alda M, Turecki G. Lithium: a key to the genetics of bipolar disorder. Genome Med (2009) 1:79. doi:10.1186/gm79
196. Schwarz C, Volz A, Li C, Leucht S. Valproate for schizophrenia. Cochrane Database Syst Rev (2008) 3:CD004028. doi:10.1002/14651858.CD004028.pub3
197. Oruch R, Elderbi MA, Khattab HA, Pryme IF, Lund A. Lithium: a review of pharmacology, clinical uses, and toxicity. Eur J Pharmacol (2014) 740:464–73. doi:10.1016/j.ejphar.2014.06.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
198. Chen G, Rajkowska G, Du F, Seraji-Bozorgzad N, Manji HK. Enhancement of hippocampal neurogenesis by lithium. J Neurochem (2000) 75:1729–34. doi:10.1046/j.1471-4159.2000.0751729.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
199. Kaladchibachi SA, Doble B, Anthopoulos N, Woodgett JR, Manoukian AS. Glycogen synthase kinase 3, circadian rhythms, and bipolar disorder: a molecular link in the therapeutic action of lithium. J Circadian Rhythms (2007) 5:3. doi:10.1186/1740-3391-5-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
200. Chiu C-T, Wang Z, Hunsberger JG, Chuang D-M. Therapeutic potential of mood stabilizers lithium and valproic acid: beyond bipolar disorder. Pharmacol Rev (2013) 65:105–42. doi:10.1124/pr.111.005512
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
201. Johansson A-S, Brask J, Owe-Larsson B, Hetta J, Lundkvist GBS. Valproic acid phase shifts the rhythmic expression of period2: luciferase. J Biol Rhythms (2011) 26:541–51. doi:10.1177/0748730411419775
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
202. Ganetzky B, Wu CF. Indirect suppression involving behavioral mutants with altered nerve excitability in Drosophila melanogaster. Genetics (1982) 100:597–614.
203. Ghezzi D, Arzuffi P, Zordan M, Da Re C, Lamperti C, Benna C, et al. Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat Genet (2011) 43:259–63. doi:10.1038/ng.761
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
204. Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, et al. A genome-wide Drosophila screen for heat nociception identifies α2δ3 as an evolutionarily conserved pain gene. Cell (2010) 143:628–38. doi:10.1016/j.cell.2010.09.047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
205. Branson K, Robie AA, Bender J, Perona P, Dickinson MH. High-throughput ethomics in large groups of Drosophila. Nat Methods (2009) 6:451–7. doi:10.1038/nmeth.1328
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
206. Evans O, Paulk AC, van Swinderen B. An automated paradigm for Drosophila visual psychophysics. PLoS One (2011) 6:e21619. doi:10.1371/journal.pone.0021619
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
207. Rosato E, Kyriacou CP. Analysis of locomotor activity rhythms in Drosophila. Nat Protoc (2006) 1:559–68. doi:10.1038/nprot.2006.79
208. Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A (2007) 104:8253–6. doi:10.1073/pnas.0702726104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
209. Shohat-Ophir G, Kaun KR, Azanchi R, Mohammed H, Heberlein U. Sexual deprivation increases ethanol intake in Drosophila. Science (2012) 335:1351–5. doi:10.1126/science.1215932
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
210. Yang Z, Bertolucci F, Wolf R, Heisenberg M. Flies cope with uncontrollable stress by learned helplessness. Curr Biol (2013) 23:799–803. doi:10.1016/j.cub.2013.03.054
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
211. O’Kane CJ. Drosophila as a model organism for the study of neuropsychiatric disorders. Curr Top Behav Neurosci (2011) 7:37–60. doi:10.1007/7854_2010_110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
212. Van der Voet M, Nijhof B, Oortveld MAW, Schenck A. Drosophila models of early onset cognitive disorders and their clinical applications. Neurosci Biobehav Rev (2014) 46(Pt 2):326–42. doi:10.1016/j.neubiorev.2014.01.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
213. Van Alphen B, van Swinderen B. Drosophila strategies to study psychiatric disorders. Brain Res Bull (2013) 92:1–11. doi:10.1016/j.brainresbull.2011.09.007
214. What is the PGC?. Psychiatric Genomics Consortium (2015). Available from: http://www.med.unc.edu/pgc/
215. Chang S-H, Gao L, Li Z, Zhang W-N, Du Y, Wang J. BDgene: a genetic database for bipolar disorder and its overlap with schizophrenia and major depressive disorder. Biol Psychiatry (2013) 74:727–33. doi:10.1016/j.biopsych.2013.04.016
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
216. Hamshere ML, Walters JTR, Smith R, Richards AL, Green E, Grozeva D, et al. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the schizophrenia PGC. Mol Psychiatry (2013) 18:708–12. doi:10.1038/mp.2012.67
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
217. Drago A, Giegling I, Schäfer M, Hartmann AM, Friedl M, Konte B, et al. AKAP13, CACNA1, GRIK4 and GRIA1 genetic variations may be associated with haloperidol efficacy during acute treatment. Eur Neuropsychopharmacol (2013) 23:887–94. doi:10.1016/j.euroneuro.2012.08.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
218. Schmutz I, Chavan R, Ripperger JA, Maywood ES, Langwieser N, Jurik A, et al. A specific role for the REV-ERBα-controlled L-type voltage-gated calcium channel CaV1.2 in resetting the circadian clock in the late night. J Biol Rhythms (2014) 29:288–98. doi:10.1177/0748730414540453
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
219. Van Swinderen B. The optomotor maze: a population assay for visual perception in Drosophila. Cold Spring Harb Protoc (2011) 2011:1337–9. doi:10.1101/pdb.prot066530
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: circadian clock, neuropsychiatric diseases, Drosophila melanogaster, gene X environment interactions, sleep, cognitive impairments, social interactions, behavioral traits
Citation: Zordan MA and Sandrelli F (2015) Circadian clock dysfunction and psychiatric disease: could fruit flies have a say? Front. Neurol. 6:80. doi: 10.3389/fneur.2015.00080
Received: 13 January 2015; Accepted: 24 March 2015;
Published online: 20 April 2015.
Edited by:
Roland Brandstaetter, University of Birmingham, UKReviewed by:
Monica M. C. Gonzalez, Instituto Ferrero de Neurologia y Sueno, ArgentinaCopyright: © 2015 Zordan and Sandrelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Federica Sandrelli, Department of Biology, University of Padova, via U. Bassi 58/B, Padova 35131, Italy e-mail:ZmVkZXJpY2Euc2FuZHJlbGxpQHVuaXBkLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.