
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neural Circuits , 22 June 2022
Volume 16 - 2022 | https://doi.org/10.3389/fncir.2022.702901
 Adeola Oyeyinka†
Adeola Oyeyinka† Mehul Kansal†
Mehul Kansal† Sean M. O’Sullivan
Sean M. O’Sullivan Claudia Gualtieri
Claudia Gualtieri Zachary M. Smith
Zachary M. Smith Fernando J. Vonhoff*
Fernando J. Vonhoff*Exposure to alcohol has multiple effects on nervous system function, and organisms have evolved mechanisms to optimally respond to the presence of ethanol. Sex differences in ethanol-induced behaviors have been observed in several organisms, ranging from humans to invertebrates. However, the molecular mechanisms underlying the dimorphic regulation of ethanol-induced behaviors remain incompletely understood. Here, we observed sex differences in ethanol sedation sensitivity in Drosophila Genome Reference Panel (DGRP) lines of Drosophila melanogaster compared to the absence of dimorphism in standard laboratory wildtype and control lines. However, in dose response experiments, we were able to unmask dimorphic responses for the control mutant line w1118 by lowering the testing ethanol concentration. Notably, feminization of the small population of Corazonin (Crz) neurons in males was sufficient to induce female-like sedation sensitivity. We also tested the role of the transcription factor apontic (apt) based on its known expression in Crz neurons and its regulation of sedation responses. Interestingly, loss of function apt mutations increased sedation times in both males and females as compared to controls. No significant difference between male and female apt mutants was observed, suggesting a possible role of apt in the regulation of dimorphic ethanol-induced responses. Thus, our results shed light into the mechanisms regulating sex-differences in ethanol-induced behaviors at the cellular and molecular level, suggesting that the genetic sex in a small neuronal population plays an important role in modulating sex differences in behavioral responses to ethanol.
Exposure to ethanol has significant effects on neuronal and brain function in humans and animals. Ethanol effects range from stimulating at acute and low concentrations, to depressant at chronic and high concentrations, involving the possible development of alcohol use disorders (AUDs) (Pohorecky, 1991). The literature surrounding the effects of ethanol in different organisms have shown that the molecular and neuronal basis underlying ethanol responses are evolutionary conserved, including in invertebrates such as Caenorhabditis elegans (Pandey et al., 2021), honey bees (Mustard et al., 2019), and Drosophila fruit flies (reviewed in Scholz and Mustard, 2013; Lathen et al., 2020). Fruit flies have been a useful animal model to study responses to ethanol, not only because they encounter ethanol in fermented food under natural conditions, but also because of the high level of conservation in behavioral responses to ethanol (hyperactivity, tolerance, and sedation) as well as in the molecular mechanisms identified in both mammals and flies (Grotewiel and Bettinger, 2015; Park et al., 2017).
Whereas early studies in flies focused on genes involved in ethanol metabolism such as alcohol dehydrogenase (ADH) (Cavener, 1979) and aldehyde dehydrogenase (ALDH) (Heinstra et al., 1989), recent studies have diversified their molecular candidates, including molecules involved in memory formation and the reward system, such as the neurotransmitters dopamine (Bainton et al., 2000; Scaplen et al., 2020) and octopamine (Scholz et al., 2000; Scholz, 2005), as well as the neuropeptides NPF (Shohat-Ophir et al., 2012) and Corazonin (Crz) (McClure and Heberlein, 2013; Sha et al., 2014; Varga et al., 2016). In fact, Crz neurons have been the focus of recent studies involving male flies and ethanol-dependent behaviors. This is in part because activation of Crz neurons in males causes ejaculation (Tayler et al., 2012; Zer-Krispil et al., 2018), which is described to be rewarding and to reduce motivation to consume ethanol (Zer-Krispil et al., 2018). By contrast, silencing of Crz neurons increases ethanol sedation times (McClure and Heberlein, 2013), making Crz-dependent signaling a key player in the male reward and stress mechanism (Khan et al., 2021; Zandawala et al., 2021).
Interestingly, although sex-dependent responses to ethanol have been observed in humans (Miller et al., 2009; Ceylan-Isik et al., 2010) and flies (Devineni and Heberlein, 2012), most studies include either females or males, and only a few include both. Clear dimorphic differences in ethanol-induced pharmacokinetics and behaviors have been previously reported in flies, with males showing increased hyperactivity and longer sedation times as compared to females (Devineni and Heberlein, 2012). However, the molecular mechanisms underlying the dimorphic regulation of ethanol responses remain poorly understood. At the molecular level, some of the observed sex differences in ethanol responses have been linked to the female-specific splicing factor Transformer (Tra), which regulates splicing of the neural sex determination gene fruitless (fru). fru encodes a male-specific group of putative transcription factors called FruM (Demir and Dickson, 2005; Kimura et al., 2005; Stockinger et al., 2005). Although sex determining genetic factors are required pan-neuronally, including the ∼1500 fru-expressing neurons to regulate dimorphic ethanol responses, it remains unclear whether genetic sex plays a role in a smaller subset of neurons to determine dimorphism in ethanol-induced behaviors. Furthermore, the Myb/SANT-containing transcription factor apontic (apt) is known to be required in Crz neurons for ethanol sedation sensitivity in males, but its role in ethanol-induced differences between males and females remains unexplored (McClure and Heberlein, 2013).
Here, we investigate the molecular and cellular mechanisms underlying sexually dimorphic responses in ethanol sedation sensitivity. We used an established assay to test sedation sensitivity by exposing male and female flies to a high ethanol vapor concentration (65%–73%) (Sandhu et al., 2015). We show that dimorphism in ethanol sedation sensitivity is observed in four different lines from the Drosophila Genome Reference Panel (DGRP), whereas classic wildtype (Canton S and Oregon R) and control (w1118) lines show no dimorphic differences. A dimorphic sedation response could be observed in w1118 flies by lowering the ethanol concentration as indicated by our dose response experiments. Additionally, sex-reversing experiments demonstrate that feminization of Crz neurons in males is sufficient to reduce sedation time to female-like levels. Lastly, we observe no sex differences in ethanol sedation in apt mutant flies, suggesting a potential role of apt-dependent transcription in dimorphic ethanol-induced behaviors.
Drosophila Genome Reference Panel lines: DGRP-774 (RRID:BDSC_25205), DGRP-73 (RRID:BDSC_28131), DGRP-304 (RRID:BDSC_25177), and DGRP-229 (RRID:BDSC_29653), as well as CRZ-GAL4 (RRID:BDSC_51976), and UAS-Tra (RRID:BDSC_4590) lines were obtained from the Bloomington Drosophila Stock Center. The apt mutant lines (Gellon et al., 1997; Starz-Gaiano et al., 2008) as well as Oregon R, Canton S, and w1118 (RRID:BDSC_3605) were obtained from the Starz-Gaiano Lab at UMBC. DGRP lines are fully sequenced inbred lines derived from a natural population from Raleigh, NC, United States (Mackay et al., 2012). w1118 is a strain of flies that have a specific mutation in the white gene, which can potentially exist in any genetic background. Flies were reared at 25°C in a 12/12 h light/dark incubator on standard plastic vials. Our food recipe included 9900 ml of water, 800 ml of molasses, 652 g of cornmeal, 130 g of yeast, 34 g of agar, 57.6 ml propionic acid, and 13.4 g methyl paraben. Males from fly lines carrying the two apt mutant alleles tested here, aptKG05830 and apt167 were crossed to a fly stock that carried the X chromosome from the DGRP-774 line and visible markers on the second chromosome (+DGRP774; CyO/Sp). F1 CyO males (+DGRP774/>; apt/CyO) were backcrossed to the maker line (+DGRP774; CyO/Sp), and subsequently, F2 males and females were selected against Sp to make stable fly lines (+DGRP774; aptKG05830/CyO and +DGRP774; apt167/CyO) that were maintained and expanded in the lab. We used resources provided by flybase.org, which serves as an essential database for our studies (Larkin et al., 2021).
Behavioral assays were performed following established sedation protocols (Sandhu et al., 2015), with minor modifications. Briefly, 10 2-day to 4-day-old non-virgin females and males of each genotype were subjected to brief (less than 5 min) carbon dioxide anesthesia no less than 24 h before behavioral testing. Flies were transferred into clean polystyrene tubes (22.5 mm × 90 mm) and plugged using two Bonded Dense Weave Cellulose Acetate closures. The top of the first plug lined up with a line 2 cm down from the top of each tube. Flies were allowed to acclimate to room temperature (23°C) for 30 min. One milliliter of the testing ethanol concentration was added to the plug. Then, a second plug was added as low as 1 cm down from the top of each tube, creating a 1 cm wide vaporization chamber (Chan et al., 2014). Tubes were gently tapped every 5 min and the loss-of-righting reflex (LORR) was quantified (number of flies unable to right themselves) (van der Linde and Lyons, 2011) for a maximum of 60 min. Sedation time 50 (ST50) values represent the time required for 50% of flies to show LORR and were calculated using linear extrapolation.
Graphs and statistical analysis were performed using GraphPad Prism 9.3.1. Normality was tested using the Shapiro–Wilk test. Statistical significance was established using the parametric one-way ANOVA and Šidák or Holm-Šídák’s corrections for multiple comparisons. Data is presented as scatter plots showing the observed individual values along with descriptive statistics (mean and SEM). In all graphs, statistical significance is shown by ****p < 0.0001, ***p < 0.001, **p < 0.01, and *p < 0.05.
Control and experimental flies were separated by sex, snap frozen in an ethanol dry ice slush and homogenized in DNA/RNA Shield (Zymo Research). Total RNA was extracted using Quick-RNA MiniPrep (Zymo Research) according to manufacturer’s instructions and stored at −20°C. Total RNA was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad) according to manufacturer’s instructions. A Bio-Rad CFX96 Real-time Detection System was used for amplification (Saadin and Starz-Gaiano, 2018; Le et al., 2021). The sequence for primers targeting apt were obtained from Wang et al. (2020) and were normalized to endogenous actin42A transcript levels with primer pair sequences from Ponton et al. (2011). Experiments were performed on three biological replicates, each in technical triplicates using groups of three 3–7 days old flies for each genotype. The data were presented as fold change as compared to DGRP-774 males. Primers used for apt were: Forward, GGGAACGCACCCAGAACTG, Reverse, GACCGGCATCCAATCTCTTGT. Primers for actin42A were: Forward, GCGTCGGTCAATTCAATCTT, Reverse, AAGCTGCAACCTCTTCGTCA. Statistical analysis was performed by one-way ANOVA followed by Dunnett’s multiple comparisons test using GraphPad Prism 9.3.1.
In order to examine whether we can observe dimorphic responses to ethanol in adult flies, we first tested a range of wildtype lines for their sedation response following previously established protocols using a LORR assay (Sandhu et al., 2015). Based on previous studies, we exposed flies to a concentration of 73% ethanol and observed sigmoidal sedation kinetics that indicated dimorphic behavior in four lines from the DGRP: DGRP-774, -73, -304, and -229 (Mackay et al., 2012) previously used in degeneration studies (Ederer et al., 2018; Gabrawy et al., 2019, 2022). A well-established method to measure sedation response or sensitivity is to quantify the time required for 50% of flies to sedate (ST50) (McClure and Heberlein, 2013; Sandhu et al., 2015). In all four DGRP lines, males sedated more slowly than females indicated by a higher ST50 value (Figure 1). For example, DGRP-774 males had an ST50 of 27.65 ± 0.76 min (N = 20 runs, 10 flies in each run; mean ± SEM; see section “Materials and Methods”) whereas females had an ST50 of 22.28 ± 0.60 min (N = 20 runs; p = 0.0003, ANOVA) (Figures 1A, 2D). The total male average ST50 from all DGRP male trials was 30.63 ± 5.08 min (Figure 2D, blue-shaded area), whereas the total female average ST50 was calculated as 24.16 ± 3.86 min (Figure 2D, red-shaded area).
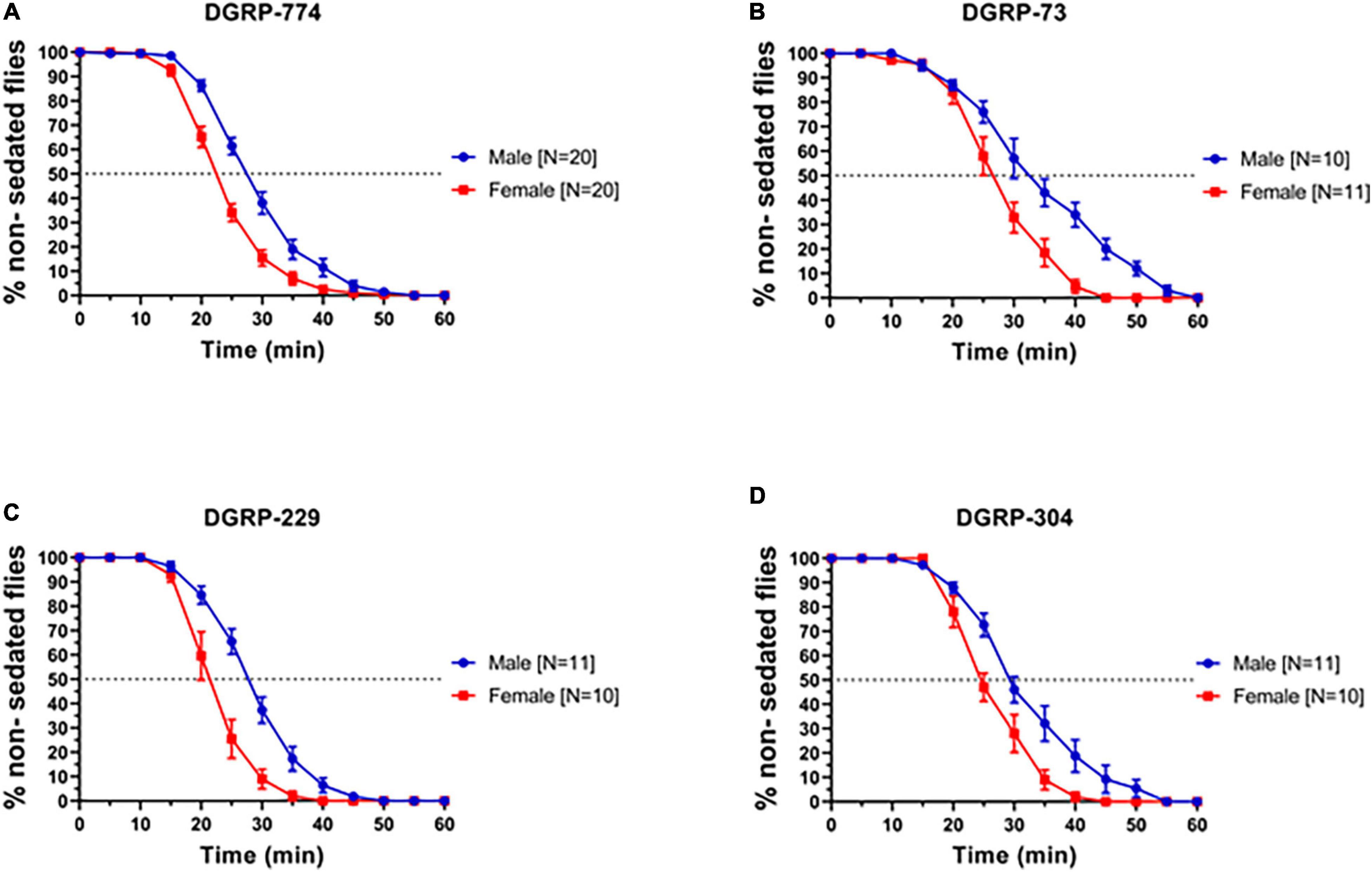
Figure 1. Drosophila Genome Reference Panel lines show dimorphic responses to ethanol sedation. Sedation curves of males and females exposed to 73% ethanol vapor for fly lines DGRP-774 (A), DGRP-73 (B), DGRP-229 (C), and DGRP-304 (D). Sample size (N) indicates the number of runs. Each run included a total of 10 flies per single run.
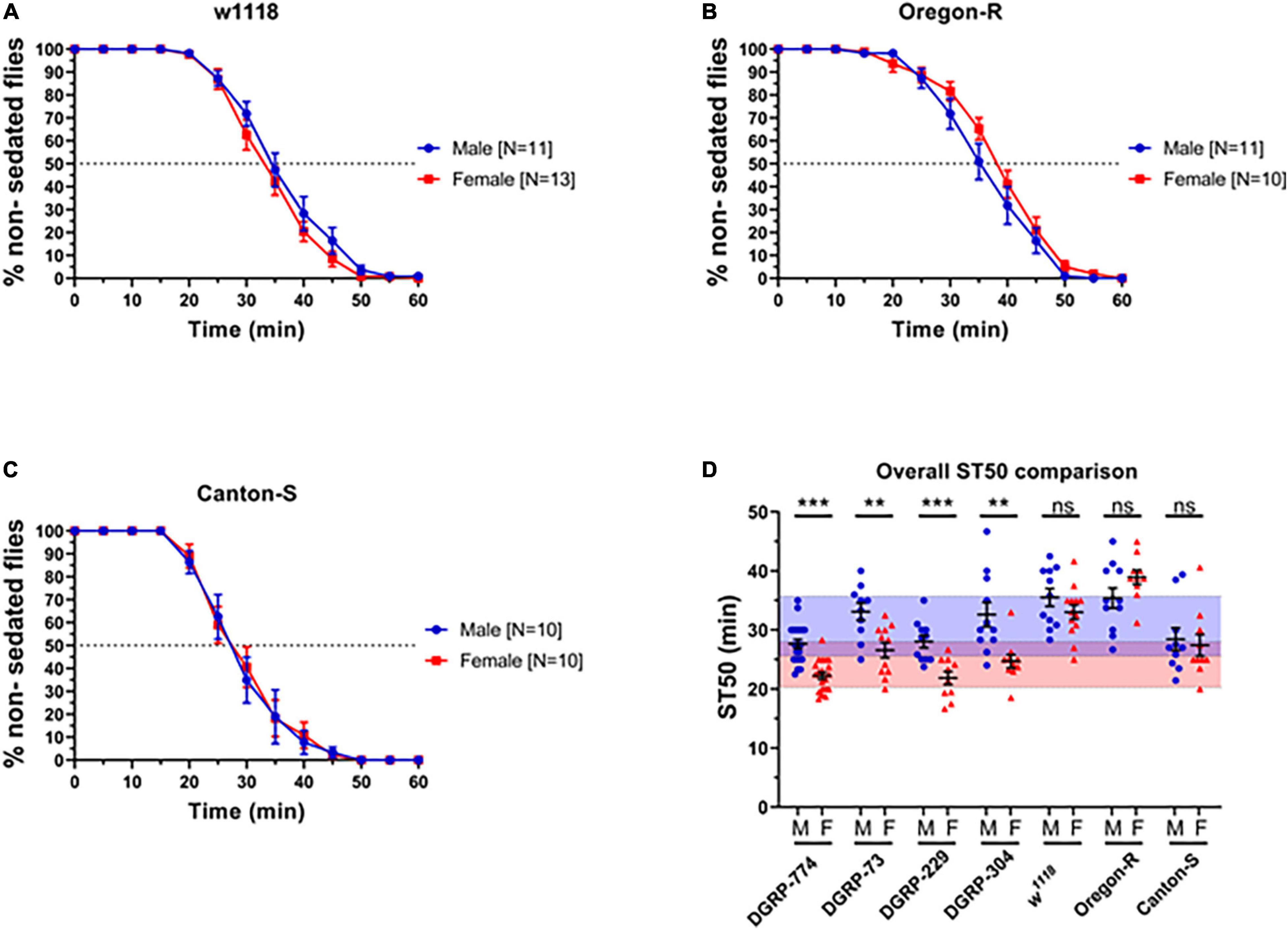
Figure 2. Classic control lines show no dimorphic responses to ethanol sedation when exposed to 73% ethanol vapor. Sedation curves of males and females for fly lines w1118 (A), Oregon R (B), and Canton-S (C). ST50 values of DGRP and control lines show that DGRP lines show dimorphic ethanol responses at 73%, whereas classic laboratory-adapted fly lines show no sex differences (D). Red-shaded area represents the total female average ST50 from all DGRP runs (24.16 ± 3.86 min). Blue-shaded area represents the total male average ST50 male (30.63 ± 5.08 min). Statistical significance was tested by one-way ANOVA with Šidák corrections and is shown by ***p < 0.001 and **p < 0.01. ns, not significant.
We next sought to determine the effect of genetic background by testing additional lines that have served as common control lines in fly research for much longer than the DGRP lines. These include the wildtype lines Canton-S and Oregon-R, as well as the white mutant w1118. Surprisingly, we did not observe dimorphic sedation rates using 73% ethanol in any of the classical control lines (Figure 2). For example, w1118 males had an ST50 of 35.51 ± 1.5 min (N = 11), while w1118 females had an ST50 of 33.01 ± 1.2 min (N = 13; p = 0.20) (Figure 2A). We hypothesized that the used ethanol concentration (73%) may be too high to dissect apart differences between male and female sedation responses. Therefore, we next performed a dose response assay on DGRP-774 and w1118 to test whether discernible effects of ethanol on dimorphism could be observed.
Indeed, we observed a dimorphic sedation response in w1118 at lower ethanol concentrations than DGRP-774 (Figure 3). While DGRP-774 showed significant differences between males and females in sedation rates at concentrations between 30% and 73% (Figure 3E), w1118 only showed significance at 50% and 65% (Figure 3F). The effect sizes with confidence interval as a function of ethanol percentage for ST50 values of DGRP-774 and w1118 flies were further analyzed to compare the strength of the observed dimorphic effects (Supplementary Figure 1). An ST50 was not determined for 30% as w1118 flies did not begin to sedate until ∼1 h after ethanol exposure. Thus, our data indicate that dimorphic differences can be masked at higher ethanol concentrations dependent on the genetic background.
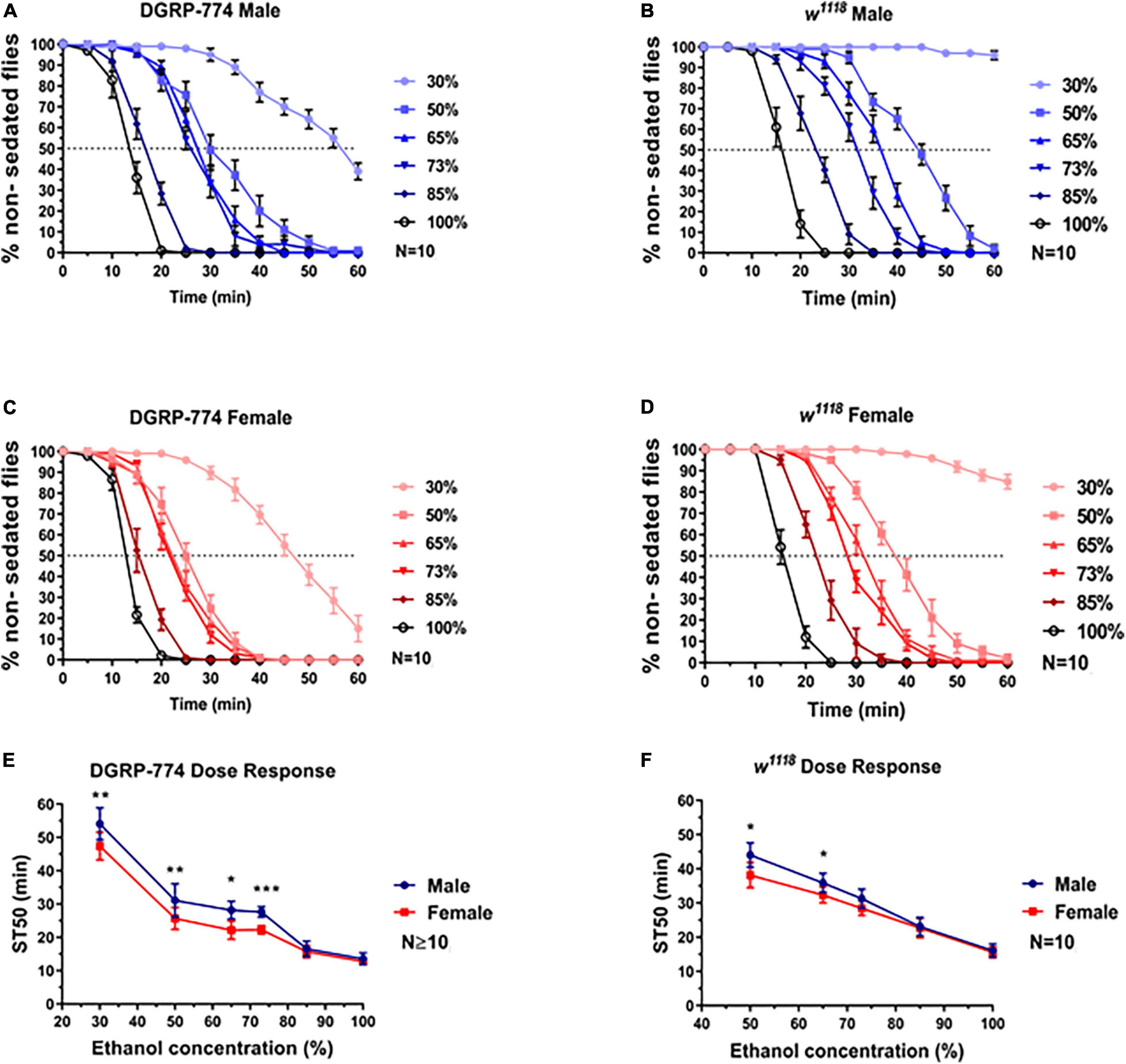
Figure 3. Dose required to observe sexual dimorphism varies by genetic background. While the DGRP-774 showed dimorphism at concentrations ≤73% (A,C,E), w1118 was not dimorphic until concentrations ≤65% (B,D,F). Statistical significance was tested by one-way ANOVA with Šidák corrections and is shown by ***p < 0.001, **p < 0.01, and *p < 0.05.
Next, we sought to further understand dimorphic responses to ethanol at the cellular and molecular level. Previous studies used sex-reversal experiments to elegantly demonstrate that masculinized females showed male-like responses to ethanol such as increased ST50 values as compared to control females (Devineni and Heberlein, 2012). Consistently, it was also shown that feminized males presented female-like responses. However, these experiments were based on the manipulation of the female-specific splicing factor Tra and Transformer 2 (Tra2) in all neurons or in a large population of neurons, such as in the cells expressing the neuronal sex determination gene fru. Therefore, we asked whether sex-reversing a small number of cells would be sufficient to regulate the observed sex-dependent responses in our ethanol sedation assay. Neurons expressing the neuropeptide Crz represent a good candidate to regulate dimorphic ethanol responses, as silencing of Crz neurons as well as Crz RNAi-knockdown have been previously shown to reduce ethanol sedation sensitivity (McClure and Heberlein, 2013). Adult brains contain 6-8 Crz neurons on each hemisphere (Nässel and Zandawala, 2020; Khan et al., 2021), representing a smaller set than the fru-expressing population.
In order to investigate whether sex-determining genetic factors function autonomously within Crz neurons to regulate ethanol sedation, we expressed the UAS-Tra construct via Crz-GAL4 to feminize Crz neurons. Males carrying both the UAS and the GAL4 constructs showed a significant reduction in sedation time compared to males from each control line (Figures 4A,C; p < 0.0001; ANOVA). Male heterozygous control animals with hybrid backgrounds showed higher ST50 values than females (UAS-Tra/+, 36.42 vs. 29.62 min, p = 0.0455; and Crz-GAL4/+, 39.09 vs. 34.35 min, p = 0.2223), whereas heterozygous males with feminized Crz neurons showed lower ST50 values than females from the same cross (UAS-Tra/crz-GAL4; 28.67 vs. 32.80 min, respectively). These results suggest that the sex determination pathway may function in a cell autonomous manner to modulate ethanol sedation sensitivity and that dimorphic ethanol responses can be regulated by the genetic sex of a small subpopulation of neurons.
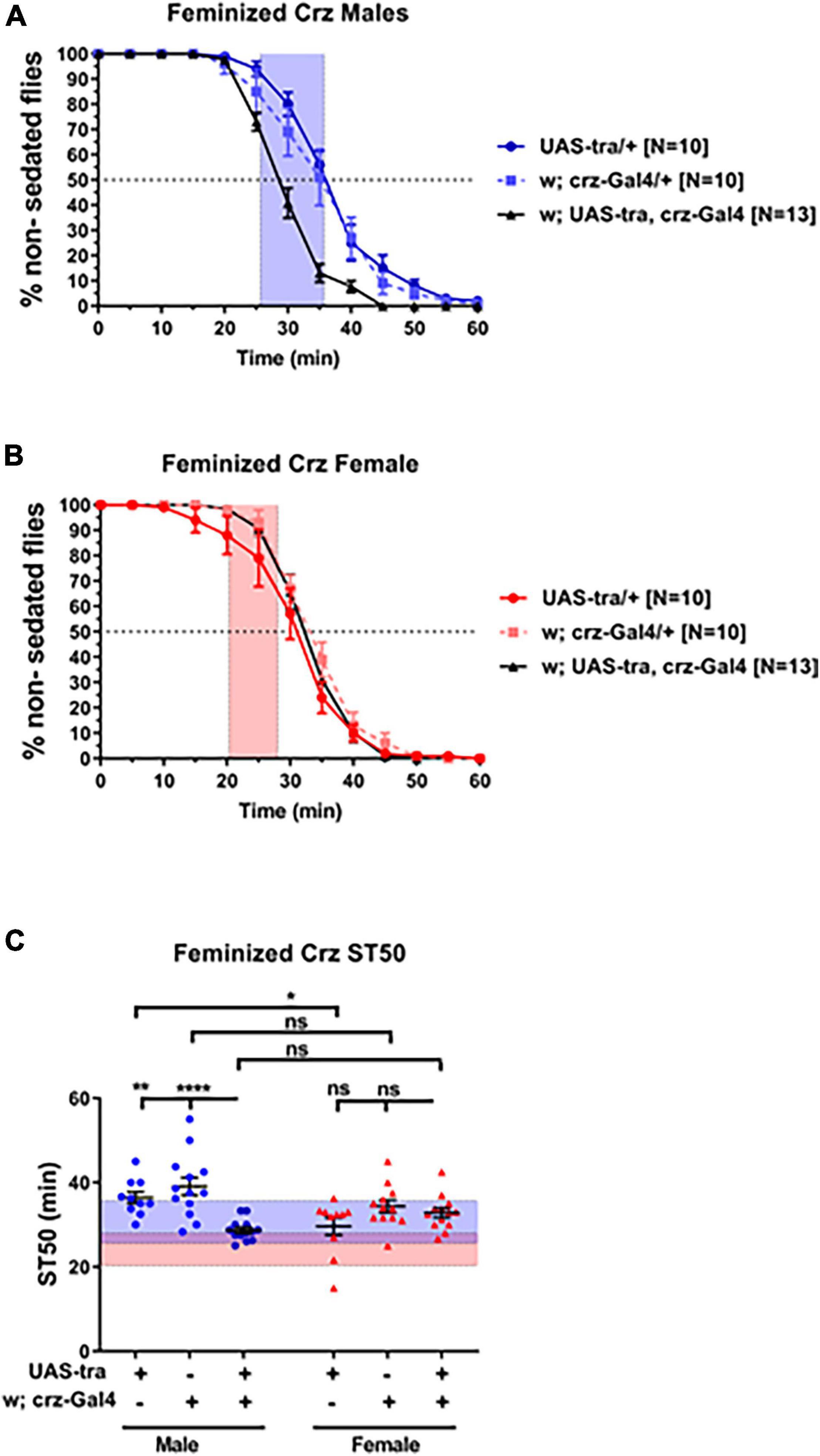
Figure 4. Feminization of Crz neurons reduces male sedation time. Expression of the feminizing factor UAS-Tra in Crz neurons using the Crz-GAL4 driver reduced sedation times in males (A) to female-like levels (B) when tested using 73% ethanol. Heterozygous animals were obtained from crosses between w1118 flies and either the GAL4 or the UAS line. For reference, red-shaded area in all panels represents the total female average ST50 at 73% from all DGRP runs (24.16 ± 3.86 min). Blue-shaded area represents the total male average ST50 male (30.63 ± 5.08 min). Statistical significance in ST50 values (C) was tested by one-way ANOVA with Šidák corrections and is shown by ****p < 0.0001, **p < 0.01, and *p < 0.05. ns, not significant.
We next sought to investigate the molecular mechanisms regulating dimorphic Crz function in ethanol sedation sensitivity. Although several genes have been identified to regulate ethanol sedation responses, few have been shown to function specifically in identified neurons. One such gene is the transcription factor apt, which has been shown to function in Crz neurons to promote sedation in males (McClure and Heberlein, 2013). However, whether apt function regulates ethanol sedation in females remains an unexplored question. Therefore, we tested the dimorphic effects of loss of apt on ethanol sedation sensitivity by using two apt loss-of-function alleles, which were crossed into the DGRP-774 genetic background (see section “Materials and Methods”). The hypomorphic allele aptKG05830 contains a P-element insertion 5′ to the start site of the apt-RA transcript (Bellen et al., 2004), whereas apt167 contains a point mutation in the DNA binding domain (Gellon et al., 1997) and is a lethal null allele. By crossing both lines, we obtained viable apt167/KG05830 heteroallelic mutants (Starz-Gaiano et al., 2008). Our results show that DGRP-774 control flies and aptKG05830/+ heterozygotes showed significant dimorphic responses, whereas no significant differences between male and female ST50 responses were observed in the other apt mutant alleles (Figure 5). Additional crosses were set to assess the effect of different genetic backgrounds on these apt flies, which included crosses to obtain heterozygous flies for the w1118 background or the CyO balancer on the second chromosome. We observed consistent trends in all cases, where aptKG05830/+ heterozygotes showed significant dimorphic responses, but no significant differences were observed in different apt167/+ heterozygotes (Supplementary Figure 2).
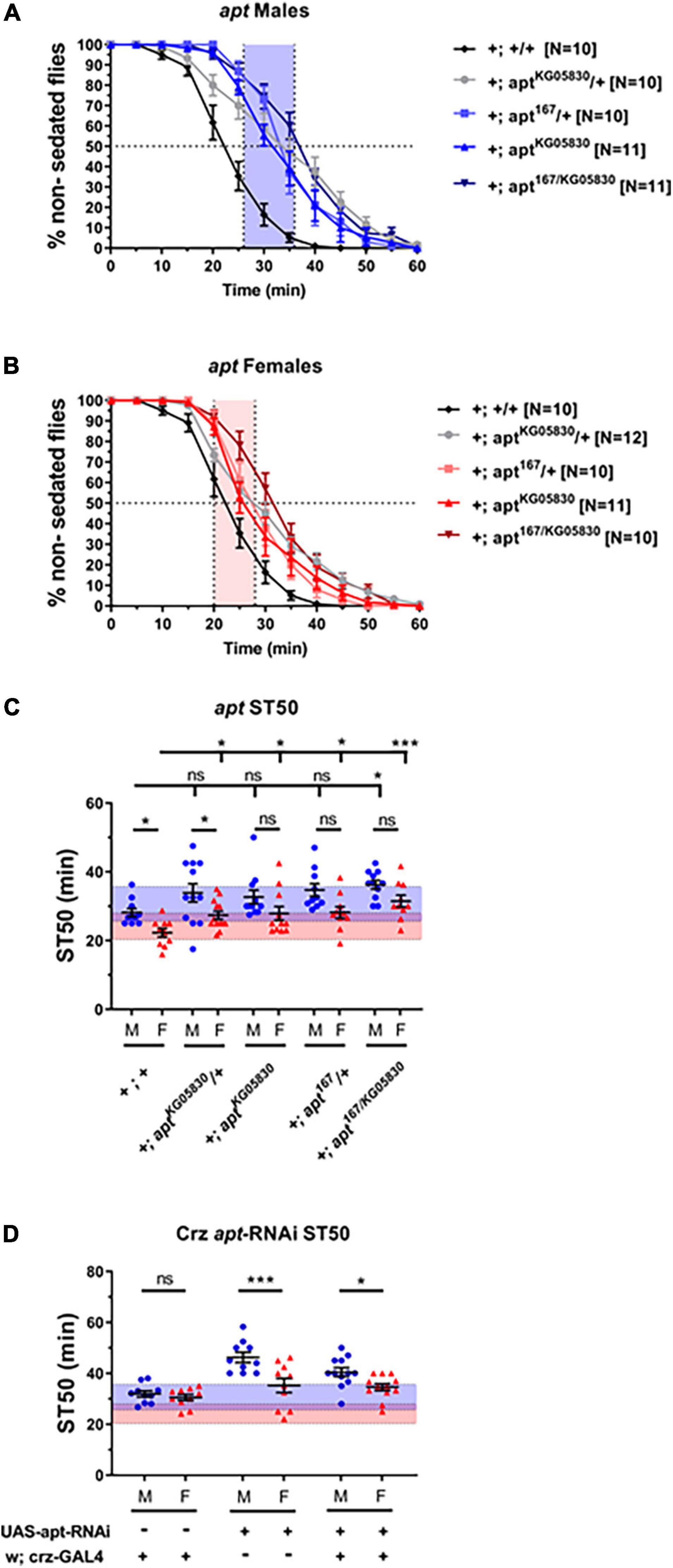
Figure 5. Flies bearing apt mutations do not show sexually dimorphic sedation responses. Flies bearing loss-of-function mutations in apt were tested using 65% ethanol show higher sedation times than DGRP-774 control flies in males (A) and females (B). Significant differences between male and female ST50 values are observed in DGRP-774 control flies and aptKG05830/+ heterozygotes, whereas no dimorphism was observed between apt167/KG05830 heteroallelic mutants, aptKG05830 homozygotes, and apt167/+ heterozygotes males and females (C). All apt mutants shown in panel (C) were crossed to generate stable lines that have the X chromosomal background of the DGRP-774 line (see section “Materials and Methods”). Heterozygous animals were obtained by crossing females from these stable apt lines with DGRP-774 males. Expression of an apt-RNAi construct via Crz-GAL4 did not disrupt sexually dimorphic sedation responses (D). For reference, the red-shaded and the blue-shaded area represents the female and male average ST50 at 73% as described in Figure 4. Statistical significance was tested by one-way ANOVA with Šidák corrections and is shown by ***p < 0.001 and *p < 0.05. ns, not significant.
Additionally, all apt mutant backgrounds showed slightly increased sedation times in males compared to DGRP-774 male controls, but a statistically significant difference was only observed in the apt167/KG05830 heteroallelic mutant males. By contrast, a significant difference in sedation times was observed for all apt mutant females tested when compared to DGRP-774 female controls (Figure 5C). These results suggest that apt functions in both males and females to promote dimorphic sedation responses, although an effect from the genetic background on the observed sedation responses in apt mutants cannot be fully ruled out. To better understand these results, we performed RT-qPCR analysis on apt transcript levels from whole animals in DGRP-774 controls and the different apt mutants. Our control data of apt expression was consistent with previously reported dimorphic differences in the Flyatlas2 dataset (Leader et al., 2018; Krause et al., 2021), showing a higher overall apt expression in males than in females (Supplementary Figure 2). Interestingly, the aptKG05830 allele led to a significant reduction of apt transcript levels in females, whereas it had a modest, more variable effect in decreasing apt levels in males. By contrast, male apt167/+ heterozygotes showed a significant decrease in apt levels, whereas female apt167/+ heterozygotes showed similar apt levels as female controls. These results suggest that the aptKG05830 allele affects females at the transcriptional level, while the effects from the apt167/+ allele may be at the functional level due to the mutated DNA binding domain.
Corazonin neurons have been previously described to express apt to regulate sedation responses in males (McClure and Heberlein, 2013). Therefore, we tested whether disruption of apt-dependent signaling exclusively in Crz neurons would be enough to phenocopy the disruption of dimorphic sedation responses we observed in apt mutants. Targeted apt-knockdown to Crz neurons did not affect the dimorphic differences in sedation responses as males expressing an apt-RNAi construct showed significantly higher ST50 values than females (40.45 vs. 34.61 min; p = 0.04, ANOVA; Figure 5D). These results suggest that apt-dependent signaling may be required in different parts of the nervous system beyond Crz neurons to regulate dimorphic sedation responses.
Our results indicate that sexually dimorphic responses to ethanol are genotype-dependent similar to previous studies testing several wildtype strains (Devineni and Heberlein, 2012) as well as DGRP lines when examined in a genome-wide association study (GWAS) aiming to identify candidate genes associated with natural variation in alcohol sensitivity (Morozova et al., 2015). Intriguingly, we observed no dimorphic differences in classic control lines when tested under the same conditions as the more recently isolated DGRP lines. Differences in dimorphic ethanol responses between laboratory-adapted fly populations have been reported in previous studies of the ADH locus (Oakeshott et al., 1984; Barbancho et al., 1987), but the precise underlying mechanisms remain unknown. Additionally, whereas some studies observed no dimorphic responses in w1118 flies (Devineni and Heberlein, 2012; Sandhu et al., 2015), we were able to observe dimorphism in this line after performing dose response experiments (Chan et al., 2014). However, the contribution of the genetic background to sedation responses was noticeable, particularly when fly lines carrying different genetic constructs in the w1118 background showed distinct effects on dimorphic sedation sensitivity. When tested with 73% ethanol, no dimorphism was observed in w1118 homozygotes and w; Crz-GAL4/+ heterozygotes, but it was observed in w; UAS-Tra/+ heterozygotes. Besides the overall genetic background, an additional explanation for this discrepancy could involve effects caused by differences in the insertion site as well as expression of the two genetic constructs. In addition to the presence or lack of GAL4 expression in these flies, both Crz-GAL4 and UAS-Tra constructs include a mini-white gene that leads to a variable level of eye pigmentation (Silicheva et al., 2010). A differential expression of mini-white may modulate various responses in w1118 flies as previously described for male-male courtship (Hing and Carlson, 1996), ethanol sensitivity (Chan et al., 2014), copulation success (Xiao et al., 2017), and neuronal degeneration (Ferreiro et al., 2017). Interestingly, whereas flies heterozygous for these constructs tend to show higher ST50 values than the calculated baseline from wildtype and w1118 homozygotes, males with both constructs (w; Crz-GAL4/UAS-Tra) to induce feminization of Crz neurons showed the opposite effect, namely a strong reduction of ethanol sensitivity. Therefore, although strong effects from the genetic background cannot be ruled out, the most parsimonious explanation for the decreased ST50 observed in Crz-GAL4/UAS-Tra males may involve the feminization of Crz neurons in these animals.
Given the effect that feminization of Crz neurons had on sedation times, future studies could examine whether masculinization of Crz neurons by UAS-tra2 RNAi expression (Devineni and Heberlein, 2012) would also be sufficient to delay sedation in females. Loss of Tra2 function causes females to produce male specific isoforms of Dsx and Fru thus masculinizing Crz neurons. Pan-neuronal masculinization produced weaker effects on sedation than feminization (Devineni and Heberlein, 2012), likely due to reduced RNAi penetrance or due to the presence of additional non-FruM neurons preventing complete neural masculinization. Additionally, future studies would focus on immunohistochemistry approaches or single-cell RNA-seq of masculinized and feminized Crz neurons, as evidence of Dsx or Fru expression in Crz neurons is still lacking. It is possible that a cascade of responsive genes downstream of Dsx or Fru would set up the differential responses in male and female brains, which ultimately control dimorphic ethanol-dependent behaviors. Due to the developmental role of these molecules, we expect Dsx and Fru to be expressed during larval or pupal stages at higher levels than during adulthood. Our results show how genetic sex regulates a specific subset of neurons in order to module alcohol-induced behaviors, thus providing insight into the molecular mechanisms underlying sexually dimorphic ethanol responses. Moreover, our results provide further evidence to support the observation that ethanol responses are strongly regulated by neuronal function, whereas physiological factors such as body weight have a weaker effect on ethanol-induced behaviors (data not shown; see also McClure et al., 2011).
From an evolutionary perspective, it seems that a variety of mechanisms to regulate the animals’ exposure to ethanol as well as ethanol-dependent behaviors have evolved in the nervous system in different animals. Most genes involved in behavioral responses to ethanol show high degrees of conservation among species. For example, ADH is expressed in most living organisms (Holmes, 1994), and most genes associated with ethanol-related behavior in flies have orthologs in C. elegans (Grotewiel and Bettinger, 2015; Scholz, 2019). However, the marine animal Trichoplax adhaerens, which lacks muscles and a nervous system, expresses ADH and ALDH, but not corazonin or apt, as revealed by genomic and transcriptomic analyses (Srivastava et al., 2008; Wong et al., 2019). This might suggest that corazonin and apt evolved at a later time during the establishment of more complex ethanol-dependent behaviors than just ethanol degradation. In contrast to mechanisms well conserved among species, the dopamine/ecdysteroid receptor (DopEcR), a G-protein coupled receptor for dopamine and the insect steroid hormone ecdysone, regulates ethanol responses in flies (Petruccelli et al., 2016; Aranda et al., 2017), but lacks mammalian orthologs, despite sharing more protein sequence similarity to vertebrate β-adrenergic receptors than to the Drosophila family of dopaminergic receptors (Petruccelli et al., 2020a). These results suggest that alternative mechanisms to ethanol exposure may exist, which go beyond ethanol processing and degradation. Such evolving mechanisms and molecules may be adopted by neuronal networks to facilitate appropriate behavioral responses in a changing environment and influence processes driven by natural selection. An intriguing question would address the evolutionary processes that led to the establishment of sexually dimorphic behaviors. Future studies using additional DGRP lines can be performed to map additional genes that contribute to dimorphism in the context of ethanol sedation sensitivity (Morozova et al., 2015; Engel et al., 2019; Petruccelli et al., 2020b). Furthermore, elucidating molecular and cellular mechanisms of sexual dimorphism may be relevant in understanding fundamental differences in not only brain function but also gender-dependent neurological diseases such as autism (Retico et al., 2016; Walsh et al., 2021) or Alzheimer’s disease (Podcasy and Epperson, 2016; Rahman et al., 2019) and their study in animal models, including rodents (Schäfer et al., 2007; Grabrucker et al., 2016) and flies (Bellosta and Soldano, 2019; Mariano et al., 2020; Jalali et al., 2021).
The aptKG05830 allele is located at the start site of the apt-RA transcript (Bellen et al., 2004), which was the only transcript previously described to be expressed in male brains (McClure and Heberlein, 2013). Recent tissue specific expression data in Flyatlas2 (Leader et al., 2018; Krause et al., 2021) confirms that apt-RA is the only transcript expressed in both male and female brains at comparable levels. Our RT-qPCR data shows differences in the way that each allele affects apt expression, showing the aptKG05830 allele a strong suppression of apt transcripts in females, whereas the apt167 allele showed a significantly decreased expression in males. These results suggest that apt expression may be regulated in a dimorphic manner and that apt mutations may have a different effect in males and females. Despite such sex-dependent differences in the regulation of apt, our results indicate that LOF mutations in apt affects both males and females, disrupting sexually dimorphic sedation responses. However, targeted apt-knockdown to Crz neurons in males and females did not disrupt dimorphism, suggesting that apt-dependent signaling is required in a wide range of populations of neurons that regulate dimorphic behaviors. Some potential candidates may include neurons involved in the reward system (e.g., NPF and octopamine) as well as Fru-expressing neurons as described above. Furthermore, future studies would contrast apt-dependent regulation of gene transcription in males and females during development and adulthood to potentially unravel different roles as previously described for Crz (McClure and Heberlein, 2013). Together these experiments will shed light into the molecular mechanisms underlying gene transcription within specific populations of neurons in a dimorphic manner to generate a proper behavioral response to alcohol exposure.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AO, MK, and SO’S were involved in performing sedation experiments. ZS, CG, and FV performed the molecular biology techniques and some sedation experiments. All authors were involved in collecting and analyzing data, contributed to the manuscript drafting and revising, and approved the submitted version.
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Support to AO was from the Meyerhoff Scholars Program and the LSAMP Program at UMBC, which is funded through an award from the National Science Foundation (Award #1002566). This research was partially funded by the UMBC- CNMS pre-professoriate fellowship to FV.
Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation (NSF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Prableen Chowdhary (Brewster Lab) and Sabeen Ikram (Green Lab) at UMBC for their guidance in the performance of the RT-qPCR experiments. We would also like to thank Noah Reger for his tremendous contributions to initial experimental design, data collection and analysis, as well as Michelle Starz-Gaiano and members of the Vonhoff Lab for helpful comments on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncir.2022.702901/full#supplementary-material
Supplementary Figure 1 | Estimation graphics of ST50 values from DGRP-774 (A) and w1118 (B) males and females. The mean difference for five ethanol concentrations (50, 65, 73, 85, and 100%) is shown in the Cumming estimation plots using the web application available at https://www.estimationstats.com (Ho et al., 2019). The raw data is plotted on the upper axes; each mean difference is plotted on the lower axes as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical error bars. The effect sizes and CIs are reported above as: effect size (CI width: lower bound; upper bound). A total of 5000 bootstrap samples were taken; the confidence interval is bias-corrected and accelerated. The p-value(s) reported are the likelihood(s) of observing the effect size(s) if the null hypothesis of zero difference is true. For each permutation p-value, 5000 reshuffles of the control and test labels were performed.
Supplementary Figure 2 | (A) Heterozygous apt mutant flies show similar trends in sedation responses between males and females in different genetic backgrounds. Animals heterozygous for the aptKG05830 allele and either for the w1118 background or the CyO balancer show sexually dimorphic sedation responses similar to aptKG05830/+ flies heterozygous for the DGRP-774 background as shown in Figure 5C. For the apt167 allele, the progeny from the same cross (between females from the apt167/CyO stable line and DGRP-774 males) was compared. Whereas no significant dimorphic responses were observed in apt167/+ heterozygotes, dimorphism was observed between female and male CyO/+ siblings. Statistical significance was tested by one-way ANOVA with Šidák corrections and is shown by ***p < 0.001, **p < 0.01, and *p < 0.05. (B) apt and actin42A transcript levels were measured by qPCR from replicates using 3–7 days old flies. Fold change expression is normalized to DGRP-774 males. Significance values are shown for comparisons in-sex to DGRP-774 flies and between sexes within each genotype.
Supplementary Table 1 | Details of the statistical analyses conducted in this study.
Aranda, G. P., Hinojos, S. J., Sabandal, P. R., Evans, P. D., and Han, K. A. (2017). Behavioral sensitization to the disinhibition effect of ethanol requires the dopamine/ecdysone receptor in Drosophila. Front. Syst. Neurosci. 11:56. doi: 10.3389/fnsys.2017.00056
Bainton, R. J., Tsai, L. T., Singh, C. M., Moore, M. S., Neckameyer, W. S., and Heberlein, U. (2000). Dopamine modulates acute responses to cocaine, nicotine and ethanol in Drosophila. Curr. Biol. 10, 187–194. doi: 10.1016/s0960-9822(00)00336-5
Barbancho, M., Sánchez-Cañete, F. J., Dorado, G., and Pineda, M. (1987). Relation between tolerance to ethanol and alcohol dehydrogenase (ADH) activity in Drosophila melanogaster: selection, genotype and sex effects. Heredity 58(Pt 3), 443–450. doi: 10.1038/hdy.1987.73
Bellen, H. J., Levis, R. W., Liao, G., He, Y., Carlson, J. W., Tsang, G., et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761–781. doi: 10.1534/genetics.104.026427
Bellosta, P., and Soldano, A. (2019). Dissecting the genetics of autism spectrum disorders: a Drosophila perspective. Front. Physiol. 10:987. doi: 10.3389/fphys.2019.00987
Cavener, D. (1979). Preference for ethanol in Drosophila melanogaster associated with the alcohol dehydrogenase polymorphism. Behav. Genet. 9, 359–365.
Ceylan-Isik, A. F., McBride, S. M., and Ren, J. (2010). Sex difference in alcoholism: who is at a greater risk for development of alcoholic complication? Life Sci. 87, 133–138. doi: 10.1016/j.lfs.2010.06.002
Chan, R. F., Lewellyn, L., DeLoyht, J. M., Sennett, K., Coffman, S., Hewitt, M., et al. (2014). Contrasting influences of Drosoiphila white/mini-white on ethanol sensitivity in two different behavioral assays. Alcohol. Clin. Exp. Res. 38, 1582–1593. doi: 10.1111/acer.12421
Demir, E., and Dickson, B. J. (2005). fruitless splicing specifies male courtship behavior in Drosophila. Cell 121, 785–794. doi: 10.1016/j.cell.2005.04.027
Devineni, A. V., and Heberlein, U. (2012). Acute ethanol responses in Drosophila are sexually dimorphic. Proc. Natl. Acad. Sci. U S A. 109, 21087–21092. doi: 10.1073/pnas.1218850110
Ederer, K. A., Jin, K., Bouslog, S., Wang, L., Gorman, G. S., Rowe, G. C., et al. (2018). Age- and genotype-specific effects of the angiotensin-converting enzyme inhibitor lisinopril on mitochondrial and metabolic parameters in Drosophila melanogaster. Int. J. Mol. Sci. 19:3351.
Engel, G. L., Taber, K., Vinton, E., and Crocker, A. J. (2019). Studying alcohol use disorder using Drosophila melanogaster in the era of ‘Big Data’. Behav. Brain Funct. 15:7.
Ferreiro, M. J., Pérez, C., Marchesano, M., Ruiz, S., Caputi, A., Aguilera, P., et al. (2017). Drosophila melanogaster white mutant w1118 undergo retinal degeneration. Front. Neurosci. 11:732. doi: 10.3389/fnins.2017.00732
Gabrawy, M. M., Campbell, S., Carbone, M. A., Morozova, T. V., Arya, G. H., Turlapati, L. B., et al. (2019). Lisinopril preserves physical resilience and extends life span in a genotype-specific manner in Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 74, 1844–1852. doi: 10.1093/gerona/glz152
Gabrawy, M. M., Khosravian, N., Morcos, G. S., Morozova, T. V., Jezek, M., Walston, J. D., et al. (2022). Genome-wide analysis in Drosophila reveals the genetic basis of variation in age-specific physical performance and response to ACE inhibition. Genes 13:143. doi: 10.3390/genes13010143
Gellon, G., Harding, K. W., McGinnis, N., Martin, M. M., and McGinnis, W. (1997). A genetic screen for modifiers of Deformed homeotic function identifies novel genes required for head development. Development 124, 3321–3331. doi: 10.1242/dev.124.17.3321
Grabrucker, S., Boeckers, T. M., and Grabrucker, A. M. (2016). Gender Dependent Evaluation of Autism like Behavior in Mice Exposed to Prenatal Zinc Deficiency. Front. Behav. Neurosci. 10:37. doi: 10.3389/fnbeh.2016.00037
Grotewiel, M., and Bettinger, J. C. (2015). Drosophila and Caenorhabditis elegans as Discovery Platforms for Genes Involved in Human Alcohol Use Disorder. Alcohol. Clin. Exp. Res. 39, 1292–1311. doi: 10.1111/acer.12785
Heinstra, P. W., Geer, B. W., Seykens, D., and Langevin, M. (1989). The metabolism of ethanol-derived acetaldehyde by alcohol dehydrogenase (EC 1.1.1.1) and aldehyde dehydrogenase (EC 1.2.1.3) in Drosophila melanogaster larvae. Biochem. J. 259, 791–797. doi: 10.1042/bj2590791
Hing, A. L., and Carlson, J. R. (1996). Male-male courtship behavior induced by ectopic expression of the Drosophila white gene: role of sensory function and age. J. Neurobiol. 30, 454–464. doi: 10.1002/(SICI)1097-4695(199608)30:4<454::AID-NEU2>3.0.CO;2-2
Ho, J., Tumkaya, T., Aryal, S., Choi, H., and Claridge-Chang, A. (2019). Moving beyond P values: data analysis with estimation graphics. Nat. Methods 16, 565–566. doi: 10.1038/s41592-019-0470-3
Holmes, R. S. (1994). Alcohol dehydrogenases: a family of isozymes with differential functions. Alcohol. Alcohol. Suppl. 2, 127–130.
Jalali, D., Guevarra, J. A., Martinez, L., Hung, L., and Vonhoff, F. J. (2021). Nutraceutical and probiotic approaches to examine molecular interactions of the amyloid precursor protein APP in Drosophila models of alzheimer’s disease. Int. J. Mol. Sci. 22:7022. doi: 10.3390/ijms22137022
Khan, Z., Tondravi, M., Oliver, R., and Vonhoff, F. J. (2021). Drosophila corazonin neurons as a hub for regulating growth, stress responses, ethanol-related behaviors, copulation persistence and sexually dimorphic reward pathways. J. Dev. Biol. 9:26. doi: 10.3390/jdb9030026
Kimura, K., Ote, M., Tazawa, T., and Yamamoto, D. (2005). Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature 438, 229–233. doi: 10.1038/nature04229
Krause, S. A., Overend, G., Dow, J. A. T., and Leader, D. P. (2021). FlyAtlas 2 in 2022: enhancements to the Drosophila melanogaster expression atlas. Nucleic Acids Res. 50, D1010–D1015. doi: 10.1093/nar/gkab971
Larkin, A., Marygold, S. J., Antonazzo, G., Attrill, H., Dos Santos, G., Garapati, P. V., et al. (2021). FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 49, D899–D907. doi: 10.1093/nar/gkaa1026
Lathen, D. R., Merrill, C. B., and Rothenfluh, A. (2020). Flying together: Drosophila as a tool to understand the genetics of human alcoholism. Int. J. Mol. Sci. 21:6649. doi: 10.3390/ijms21186649
Le, N., Hufford, T. M., Park, J. S., and Brewster, R. M. (2021). Differential expression and hypoxia-mediated regulation of the N-myc downstream regulated gene family. FASEB J. 35:e21961. doi: 10.1096/fj.202100443R
Leader, D. P., Krause, S. A., Pandit, A., Davies, S. A., and Dow, J. A. T. (2018). FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 46, D809–D815. doi: 10.1093/nar/gkx976
Mackay, T. F., Richards, S., Stone, E. A., Barbadilla, A., Ayroles, J. F., Zhu, D., et al. (2012). The Drosophila melanogaster genetic reference panel. Nature 482, 173–178.
Mariano, V., Achsel, T., Bagni, C., and Kanellopoulos, A. K. (2020). Modelling learning and memory in Drosophila to understand intellectual disabilities. Neuroscience 445, 12–30. doi: 10.1016/j.neuroscience.2020.07.034
McClure, K. D., and Heberlein, U. (2013). A small group of neurosecretory cells expressing the transcriptional regulator apontic and the neuropeptide corazonin mediate ethanol sedation in Drosophila. J. Neurosci. 33, 4044–4054.
McClure, K. D., French, R. L., and Heberlein, U. (2011). A Drosophila model for fetal alcohol syndrome disorders: role for the insulin pathway. Dis. Model. Mech. 4, 335–346. doi: 10.1242/dmm.006411
Miller, M. A., Weafer, J., and Fillmore, M. T. (2009). Gender differences in alcohol impairment of simulated driving performance and driving-related skills. Alcohol. Alcohol. 44, 586–593. doi: 10.1093/alcalc/agp051
Morozova, T. V., Huang, W., Pray, V. A., Whitham, T., Anholt, R. R., and Mackay, T. F. (2015). Polymorphisms in early neurodevelopmental genes affect natural variation in alcohol sensitivity in adult Drosophila. BMC Genomics 16:865. doi: 10.1186/s12864-015-2064-5
Mustard, J. A., Oquita, R., Garza, P., and Stoker, A. (2019). Honey bees (Apis mellifera) show a preference for the consumption of ethanol. Alcohol. Clin. Exp. Res. 43, 26–35. doi: 10.1111/acer.13908
Nässel, D. R., and Zandawala, M. (2020). Hormonal axes in Drosophila: regulation of hormone release and multiplicity of actions. Cell Tissue Res. 382, 233–266. doi: 10.1007/s00441-020-03264-z
Oakeshott, J. G., Gibson, J. B., and Wilson, S. R. (1984). Selective effects of the genetic background and ethanol on the alcohol dehydrogenase polymorphism in Drosophila melanogaster. Heredity 53(Pt 1), 51–67. doi: 10.1038/hdy.1984.62
Pandey, P., Singh, A., Kaur, H., Ghosh-Roy, A., and Babu, K. (2021). Increased dopaminergic neurotransmission results in ethanol dependent sedative behaviors in Caenorhabditis elegans. PLoS Genet. 17:e1009346. doi: 10.1371/journal.pgen.1009346
Park, A., Ghezzi, A., Wijesekera, T. P., and Atkinson, N. S. (2017). Genetics and genomics of alcohol responses in Drosophila. Neuropharmacology 122, 22–35.
Petruccelli, E., Brown, T., Waterman, A., Ledru, N., and Kaun, K. R. (2020b). Alcohol causes lasting differential transcription in Drosophila Mushroom Body Neurons. Genetics 215, 103–116. doi: 10.1534/genetics.120.303101
Petruccelli, E., Lark, A., Mrkvicka, J. A., and Kitamoto, T. (2020a). Significance of DopEcR, a G-protein coupled dopamine/ecdysteroid receptor, in physiological and behavioral response to stressors. J. Neurogenet. 34, 55–68. doi: 10.1080/01677063.2019.1710144
Petruccelli, E., Li, Q., Rao, Y., and Kitamoto, T. (2016). The unique dopamine/ecdysteroid receptor modulates ethanol-induced sedation in Drosophila. J. Neurosci. 36, 4647–4657. doi: 10.1523/JNEUROSCI.3774-15.2016
Podcasy, J. L., and Epperson, C. N. (2016). Considering sex and gender in Alzheimer disease and other dementias. Dialog. Clin. Neurosci. 18, 437–446. doi: 10.31887/DCNS.2016.18.4/cepperson
Pohorecky, L. A. (1991). Stress and alcohol interaction: an update of human research. Alcohol. Clin. Exp. Res. 15, 438–459.
Ponton, F., Chapuis, M. P., Pernice, M., Sword, G. A., and Simpson, S. J. (2011). Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57, 840–850. doi: 10.1016/j.jinsphys.2011.03.014
Rahman, A., Jackson, H., Hristov, H., Isaacson, R. S., Saif, N., Shetty, T., et al. (2019). Sex and gender driven modifiers of Alzheimer’s: the role for estrogenic control across age, race, medical, and lifestyle risks. Front. Aging Neurosci. 11:315. doi: 10.3389/fnagi.2019.00315
Retico, A., Giuliano, A., Tancredi, R., Cosenza, A., Apicella, F., Narzisi, A., et al. (2016). The effect of gender on the neuroanatomy of children with autism spectrum disorders: a support vector machine case-control study. Mol. Autism 7:5. doi: 10.1186/s13229-015-0067-3
Saadin, A., and Starz-Gaiano, M. (2018). Cytokine exocytosis and JAK/STAT activation in the Drosophila ovary requires the vesicle trafficking regulator α-Snap. J. Cell Sci. 131:jcs217638.
Sandhu, S., Kollah, A. P., Lewellyn, L., Chan, R. F., and Grotewiel, M. (2015). An inexpensive, scalable behavioral assay for measuring ethanol sedation sensitivity and rapid tolerance in Drosophila. J. Vis. Exp. 2015:52676. doi: 10.3791/52676
Scaplen, K. M., Talay, M., Nunez, K. M., Salamon, S., Waterman, A. G., Gang, S., et al. (2020). Circuits that encode and guide alcohol-associated preference. Elife 9:e48730. doi: 10.7554/eLife.48730
Schäfer, S., Wirths, O., Multhaup, G., and Bayer, T. A. (2007). Gender dependent APP processing in a transgenic mouse model of Alzheimer’s disease. J. Neural Transm. 114, 387–394. doi: 10.1007/s00702-006-0580-9
Scholz, H. (2005). Influence of the biogenic amine tyramine on ethanol-induced behaviors in Drosophila. J. Neurobiol. 63, 199–214. doi: 10.1002/neu.20127
Scholz, H. (2019). Unraveling the mechanisms of behaviors associated with AUDs using flies and worms. Alcohol. Clin. Exp. Res. 43, 2274–2284. doi: 10.1111/acer.14199
Scholz, H., and Mustard, J. A. (2013). Invertebrate models of alcoholism. Curr. Top. Behav. Neurosci. 13, 433–457. doi: 10.1007/7854_2011_128
Scholz, H., Ramond, J., Singh, C. M., and Heberlein, U. (2000). Functional ethanol tolerance in Drosophila. Neuron 28, 261–271.
Sha, K., Choi, S. H., Im, J., Lee, G. G., Loeffler, F., and Park, J. H. (2014). Regulation of ethanol-related behavior and ethanol metabolism by the Corazonin neurons and Corazonin receptor in Drosophila melanogaster. PLoS One 9:e87062. doi: 10.1371/journal.pone.0087062
Shohat-Ophir, G., Kaun, K. R., Azanchi, R., Mohammed, H., and Heberlein, U. (2012). Sexual deprivation increases ethanol intake in Drosophila. Science 335, 1351–1355. doi: 10.1126/science.1215932
Silicheva, M., Golovnin, A., Pomerantseva, E., Parshikov, A., Georgiev, P., and Maksimenko, O. (2010). Drosophila mini-white model system: new insights into positive position effects and the role of transcriptional terminators and gypsy insulator in transgene shielding. Nucleic Acids Res. 38, 39–47. doi: 10.1093/nar/gkp877
Srivastava, M., Begovic, E., Chapman, J., Putnam, N. H., Hellsten, U., Kawashima, T., et al. (2008). The Trichoplax genome and the nature of placozoans. Nature 454, 955–960. doi: 10.1038/nature07191
Starz-Gaiano, M., Melani, M., Wang, X., Meinhardt, H., and Montell, D. J. (2008). Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev. Cell 14, 726–738. doi: 10.1016/j.devcel.2008.03.005
Stockinger, P., Kvitsiani, D., Rotkopf, S., Tirián, L., and Dickson, B. J. (2005). Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807. doi: 10.1016/j.cell.2005.04.026
Tayler, T. D., Pacheco, D. A., Hergarden, A. C., Murthy, M., and Anderson, D. J. (2012). A neuropeptide circuit that coordinates sperm transfer and copulation duration in Drosophila. Proc. Natl. Acad. Sci. U S A. 109, 20697–20702.
van der Linde, K., and Lyons, L. C. (2011). Circadian modulation of acute alcohol sensitivity but not acute tolerance in Drosophila. Chronobiol. Int. 28, 397–406.
Varga, K., Nagy, P., Arsikin Csordás, K., Kovács, A. L., Hegedûs, K., and Juhász, G. (2016). Loss of Atg16 delays the alcohol-induced sedation response via regulation of Corazonin neuropeptide production in Drosophila. Sci. Rep. 6:34641. doi: 10.1038/srep34641
Walsh, M. J. M., Wallace, G. L., Gallegos, S. M., and Braden, B. B. (2021). Brain-based sex differences in autism spectrum disorder across the lifespan: a systematic review of structural MRI, fMRI, and DTI findings. Neuroimage Clin. 31:102719. doi: 10.1016/j.nicl.2021.102719
Wang, X. F., Liu, J. X., Ma, Z. Y., Shen, Y., Zhang, H. R., Zhou, Z. Z., et al. (2020). Evolutionarily conserved roles for apontic in induction and subsequent decline of cyclin E expression. iScience 23:101369. doi: 10.1016/j.isci.2020.101369
Wong, Y. Y., Le, P., Elkhatib, W., Piekut, T., and Senatore, A. (2019). Transcriptome profiling of Trichoplax adhaerens highlights its digestive epithelium and a rich set of genes for fast electrogenic and slow neuromodulatory cellular signaling. [Preprint].
Xiao, C., Qiu, S., and Robertson, R. M. (2017). The white gene controls copulation success in Drosophila melanogaster. Sci. Rep. 7:7712.
Zandawala, M., Nguyen, T., Balanyà Segura, M., Johard, H. A. D., Amcoff, M., Wegener, C., et al. (2021). A neuroendocrine pathway modulating osmotic stress in Drosophila. PLoS Genet. 17:e1009425. doi: 10.1371/journal.pgen.1009425
Keywords: invertebrate, neurobiology, neuroethology, sex-reversal, sexual-dimorphism, insects, neuromodulation
Citation: Oyeyinka A, Kansal M, O’Sullivan SM, Gualtieri C, Smith ZM and Vonhoff FJ (2022) Corazonin Neurons Contribute to Dimorphic Ethanol Sedation Sensitivity in Drosophila melanogaster. Front. Neural Circuits 16:702901. doi: 10.3389/fncir.2022.702901
Received: 30 April 2021; Accepted: 23 May 2022;
Published: 22 June 2022.
Edited by:
Wolfgang Stein, Illinois State University, United StatesReviewed by:
Adrian Rothenfluh, The University of Utah, United StatesCopyright © 2022 Oyeyinka, Kansal, O’Sullivan, Gualtieri, Smith and Vonhoff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernando J. Vonhoff, ZnZvbmhvZmZAdW1iYy5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.