
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Neural Circuits, 11 September 2018
Volume 12 - 2018 | https://doi.org/10.3389/fncir.2018.00068
Cre-LoxP conditional knockout animals have become a prominent tool to understand gene function in discrete cell-types and neural circuits. However, this technology has significant limitations including off target cre-dependent recombination. The Rgs9cre strain has been used to generate a conditional knockout in striatal medium spiny neurons, but, as presented in the current study, off target recombination in the germline results in nonconditional deletion of LoxP alleles. Using a Rem2 conditional allele, germline deletion (GD) was observed in a sex dependent manner. When Cre and LoxP alleles were co-inherited from the female parent, 27 of 29 LoxP alleles were recombined, but when co-inherited from the male parent, 5 of 36 LoxP alleles were recombined. Rem2 expression measured by RT-qPCR confirmed nonconditional recombination in extrastriatal nuclei. Cre-LoxP is a powerful technique to modify genomic DNA (gDNA), however careful characterization of these mice is required to confirm control of conditional recombination.
Conditional knockouts using Cre-LoxP technology have been instrumental in dissecting the function of proteins in neurophysiology by allowing gene deletion or gene expression with temporal, tissue and cell-type specificity. This technology exploits the function of a bacteriophage P1 topoisomerase, Cre-recombinase (Cre), to recombine genomic DNA (gDNA) that is flanked by a pair of Cre recognition sequences call LoxP sites. In many experimental designs, mutant mice containing a floxed (i.e., flanked by LoxP) gene of interest are bred with mice expressing Cre driven by a tissue or cell-type specific promoter. These Cre-driver lines are generated by identifying a gene with advantageous expression patterns and inserting the Cre coding sequence into either the endogenous gene locus or a bacterial artificial chromosome (BAC) harboring the gene promoter of interest (the BAC is subsequently introduced into the host genome; Parkitna et al., 2009). Although Cre-LoxP conditional knockout mice have been transformative in neuroscience research, significant limitations exist (Schmidt-Supprian and Rajewsky, 2007; Harno et al., 2013). One such limitation is the potential for germline recombination of conditional alleles due to transient Cre expression in the germline (for a detailed description see, Song and Palmiter, 2018), a phenomenon documented with increasing frequency in the literature (Rempe et al., 2006; Zeller et al., 2008; Friedel et al., 2011; Tsai et al., 2012; Kobayashi and Hensch, 2013; Zhang et al., 2013; He et al., 2017; Song and Palmiter, 2018). The current report describes germline recombination in the Rgs9cre line (Dang et al., 2006), which has been widely used to manipulate medium spiny neurons in the striatum.
All animal studies were conducted in accordance to the National Institutes of Health’s Guidelines for Animal Care and Use and all experimental protocols were approved by the National Institute on Alcohol Abuse and Alcoholism Animal Use and Care Committee. Rem2 conditional knockout mice (B6.Cg-Rem2tm3551(T2A-mkate2)Arte; Liput et al., 2016), which contain LoxP sequences flanking exons 2 and 3 (Figure 1B), were bred with Rgs9cre mice, which have a targeted insertion of an IRES-Cre-polyA sequence on the 3′ end of the translated region of exon 19. Mice were bred using the mating scheme, Rem2LoxP,LoxPRgs9WT/WT × Rem2LoxP/WTRgs9Cre/WT (Figure 1A), and multiple breeding cages were generated so offspring inherited the Rgs9Cre allele from the male or female parent.
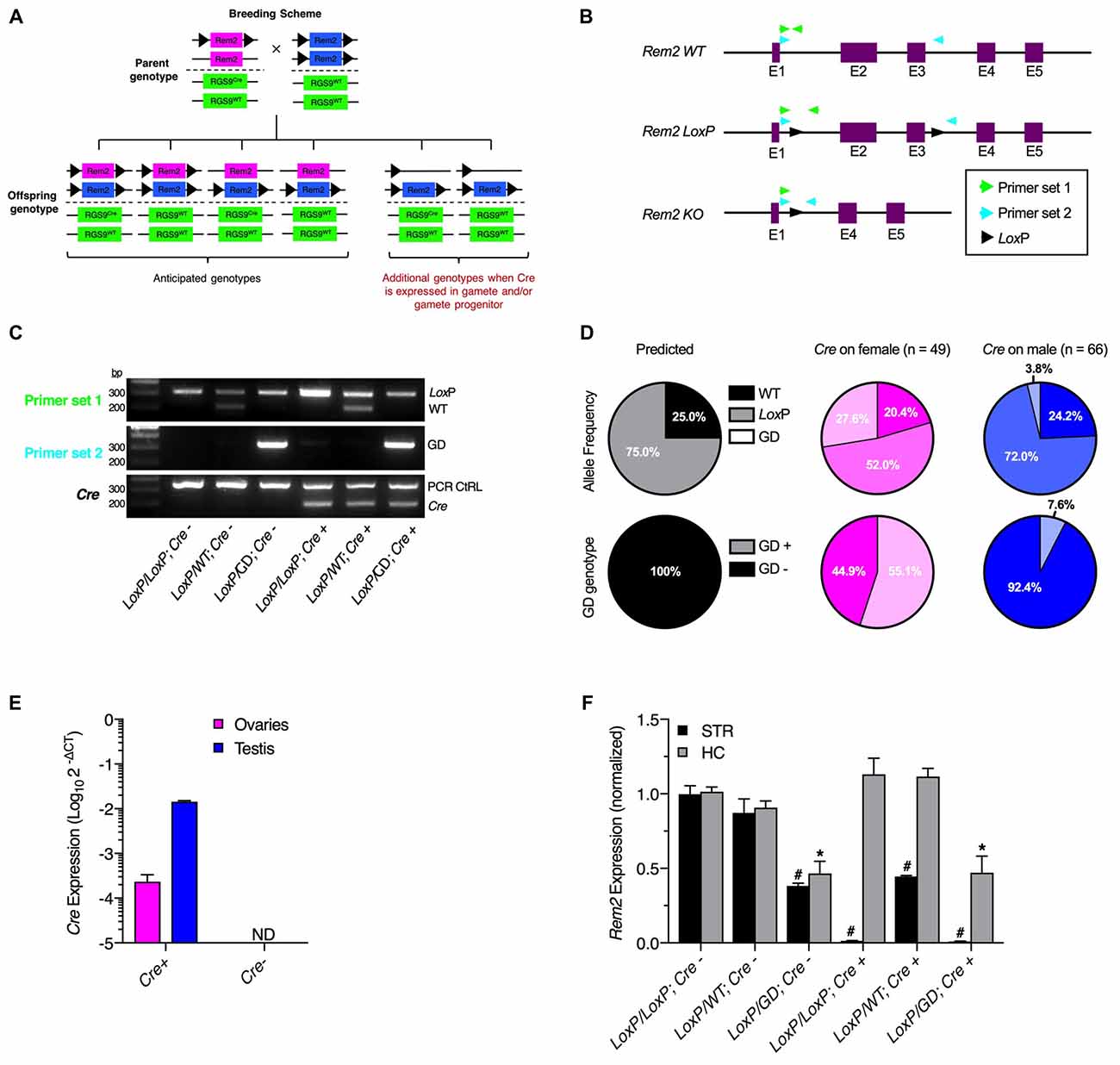
Figure 1. Germline recombination of Rem2 conditional alleles by Rgs9cre driver mice. (A) Summary of anticipated and observed genotypes. (B) Schematic of Rem2 gene showing the wildtype allele, conditional allele with LoxP sequences flanking exons two and three, and knockout allele following Cre-recombinase (Cre) dependent deletion. Two primer sets were used to distinguish the three alleles. Primer set one flanked the 5′ LoxP site, while primer set two flanked the entire floxed region. Note that the complementary DNA sequence for the reverse primer of Primer set 1 is excised following recombination, which results in failed polymerase chain reaction (PCR) amplification. (C) PCR results from genomic DNA (gDNA) purified from ear biopsies. PCR using primer set 1 produced two possible amplicons corresponding to the wildtype (221 bp) and LoxP conditional (327 bp) alleles. PCR with primer set 2 resulted in three possible amplicons corresponding to the wildtype (1,915 bp; not shown), LoxP conditional (2,065 bp; not shown), and Cre-dependent deleted (338 bp) alleles. The Cre PCR reaction contained primers within the Cre coding sequence (200 bp) and PCR control primers (350 bp). (D) Allele frequency and germline genotype frequency predicted for the cross, Rem2LoxP/LoxPRgs9WT/WT × Rem2LoxP/WTRgs9Cre/WT, and observed frequencies when the Cre gene was inherited from the female or male parent. (E) RT-qPCR from gonad cDNA showing Cre expression in Cre+ mice. (F) RT-qPCR from striatum and hippocampus cDNA showing nonconditional Rem2 knockout. #p < 0.05 compared to LoxP/LoxP; Cre-STR; *p < 0.05 compared to LoxP/LoxP; Cre-HC. GD, germline deletion. ND, not detected.
gDNA was isolated from ear biopsies using the DNeasy kit (Qiagen) and polymerase chain reaction (PCR) was performed using GoTaq DNA polymerase (Promega). For genotyping the Rem2 alleles, primers were designed to either flank the 5′ LoxP site (primer set 1; fwd: 5′-acaaacactcacgcatgcg-3′, rev: 5′-cctacattaggcctagtgagttgg-3′) or the entire floxed DNA sequence containing exons two and three (fwd: 5′-acaaacactcacgcatgcg-3′, rev: 5′-gctacttgctcaactaggagcc-3′, Figure 1B). The genotyping reaction for cre recombinase contained primers for the Cre coding sequence (fwd:5′-agcctgttttgcacgttcacc-3′, rev: 5′-ggtttcccgcagaacctgaa-3′) and PCR control primers (fwd: 5′-cctagcacccacccaaagagctg-3′, rev: 5′-ggtcctcactggcagcagctgca-3′).
For Rem2 mRNA quantification, striata and hippocampi were dissected and placed in Trizol regent for isolation of total RNA using the RNeasy Lipid Tissue kit (Qiagen). For Cre mRNA quantification, total RNA was purified from ovaries and testis according to the RNeasy Mini kit (Qiagen). After purification, 500 ng (brain tissues) or 1 μg (ovaries and testis) of RNA was reverse transcribed using the QuantiTect Reverse Transcription kit (Qiagen) and diluted to an estimated 5 ng/μL (cDNA was not diluted for Cre reactions). qPCR was performed in triplicate on a StepOnePlus thermocycler using 2 μL (5 μL for Cre reactions) cDNA template, Universal Master Mix II, and predesigned TaqMan Gene Expression Assays (all from ThermoFisher Scientific) for Rem2 (Mm00600529_m1, spanning exons three and four), Cre (Mr00635245_cn), and actb (4352933E). Amplification was quantified using the 2−ΔCT method with actb serving as the normalization control.
RT-qPCR data was plotted as mean ± SEM and analyzed by two-way ANOVA. For all tests α was set at 0.05.
To examine germline recombination, male and female mice homozygous for a Rem2 floxed allele (Liput et al., 2016) were bred with mice heterozygous for both the Rem2 floxed and Rgs9cre alleles (Rem2LoxP/LoxPRgs9WT/WT × Rem2LoxP/WTRgs9Cre/WT; Figure 1A). Genotypes were determined by PCR using gDNA from ear biopsies. For the Rem2 allele, two primer sets were designed to flank either the 5′ LoxP site (primer set 1) or the entire floxed region (primer set 2; Figure 1B). Using these primer sets and Cre primers, PCR product mass revealed six possible genotypes (Figure 1C) as opposed to the four genotypes predicted by Mendelian genetics (Figure 1A). Germline deleted (GD) alleles were detected in many animals by primer set 2, a genotype that cannot be identified by primer set 1 due to excision of the complementary DNA for the reverse primer. Importantly, this result shows that amplifying a single LoxP site is insufficient for genotype identification. Allele and genotype frequencies were calculated for offspring generated when the Rgs9cre allele was inherited from the female or male parent (Figure 1D). Frequency of the GD allele was 27.6% when Cre was inherited from the female parent, while 3.8% when Cre was inherited from the male parent. Thus, a GD genotype occurred in 55.1% of offspring generated from a Cre+ female parent and in 7.6% of offspring generated from a Cre+ male parent.
If recombined alleles detected in ear biopsies is due to germline recombination, then mRNA should be present in the gonads of Rgs9cre mice. To test this prediction, RT-qPCR was performed using gonadal cDNA from Rgs9cre mice (Figure 1E). Cre expression was observed in the gonads of female and male Cre+, but not Cre−, mice. Although germline recombination was rarely observed when Cre was inherited from the male parent, Cre mRNA was more abundant in the male gonads, suggesting that Cre is expressed in somatic cells of the testis.
To verify genotypes, RT-qPCR was used to quantify Rem2 mRNA expression in the striatum, where Cre is expressed in the Rgs9cre mouse line, and in the hippocampus, where Cre is not expressed in the Rgs9cre mouse line (Figure 1F; Rahman et al., 1999; Dang et al., 2006). Consistent with GD, mice with a LoxP/GD; Cre− genotype had approximately 50% Rem2 expression in the striatum and hippocampus compared to LoxP/LoxP; Cre− control mice. Additionally, mice with a LoxP/GD; Cre+ genotype did not express Rem2 in the striatum and had approximately 50% Rem2 expression in the hippocampus.
The current report demonstrates germline recombination of Rem2 conditional alleles when breeding with the Rgs9cre driver line, which occurred at higher frequency when the Cre allele was present on the female parent. As 27 of 29 conditional Rem2 alleles inherited from the Cre+ female breeder were GD, breeding in this manner is impractical for generating experimental animals. Although GD occurred with Cre+ male breeders, the frequency was much lower with 5 of 36 conditional alleles recombined, thus representing a more favorable breeding strategy. A Cre-dependent recombination event resulting in a GD genotype determination could conceivably occur at one of several developmental stages other than in the germline (i.e., in lineage specific stem cells or differentiated somatic cells present in the tissue biopsy). However, none of the 115 mice genotyped for the current study were genotyped GD homozygous, which indicates Cre expression and recombination occurred in the germline, prior to fertilization. Additionally, some mice had a GD allele, but were Cre negative, further indicating that recombination occurred during gametogenesis prior to generation of the final haploid gametes.
This report, in combination with others, demonstrate the need for careful characterization of Cre-driver lines. In many cases, Cre expression patterns are validated by crossing a Cre-driver mouse with a floxed reporter (i.e., LacZ or fluorescent protein) mouse and observing reporter expression in mice from the F1 generation (Madisen et al., 2010). This strategy fails to detect germline recombination because the Cre gene and reporter gene are inherited from separate parents, but germline recombination occurs in cases of co-inheritance of the two transgenes from one parent (Schmidt-Supprian and Rajewsky, 2007). In order to screen for germline recombination using a reporter strategy, mice from the F1 generation that have a Cre+/−; Reporter+/− genotype need to be bred with a wildtype mouse and reporter expression examined in the generated offspring (Song and Palmiter, 2018). Although, the utility of this strategy is questionable because Cre may have unequal sensitivity for various floxed conditional alleles (Schmidt-Supprian and Rajewsky, 2007).
As put forth by others (Schmidt-Supprian and Rajewsky, 2007; Kobayashi and Hensch, 2013; Song and Palmiter, 2018), several strategies can be employed to screen for germline recombination. First, genotyping requires the use of primers that flank the entire floxed locus of a conditional gene in combination with primers flanking a single LoxP site (Figure 1A for example). This genotyping strategy should be carried out for all conditional alleles due to the possibility of differential sensitivity to Cre-mediated recombination (Schmidt-Supprian and Rajewsky, 2007). Second, male and female breeders harboring the Cre gene should be examined to determine the frequency of germline recombination, which can be different between the male and female germline, as reported here for the RGS9cre driver line. Third, PCR from gDNA or RT-qPCR can be performed using an array of tissue types to verify the specificity of recombination (although this strategy alone will inevitably miss recombination in underrepresented cell-types due to non-germline off target Cre activity). Lastly, it is important to report detailed breeding schemes used for generation of experimental animals and methods for validating conditional knockouts so that the possibility of GD deletion can be assessed when interpreting the presented data (Song and Palmiter, 2018).
DL designed the experiments, collected and analyzed data and wrote the manuscript.
This work was supported by the National Institutes of Health, National Institute on Alcohol Abuse and Alcoholism, Division of Intramural Clinical and Biological Research (DICBR).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author would like to thank David Lovinger, Ph.D., Margret Davis, Ph.D. and Karina Abrahao, Ph.D. for valuable discussion on germline recombination.
Dang, M. T., Yokoi, F., Yin, H. H., Lovinger, D. M., Wang, Y., and Li, Y. (2006). Disrupted motor learning and long-term synaptic plasticity in mice lacking NMDAR1 in the striatum. Proc. Natl. Acad. Sci. U S A 103, 15254–15259. doi: 10.1073/pnas.0601758103
Friedel, R. H., Wurst, W., Wefers, B., and Kuhn, R. (2011). Generating conditional knockout mice. Methods Mol. Biol. 693, 205–231. doi: 10.1007/978-1-60761-974-1_12
Harno, E., Cottrell, E. C., and White, A. (2013). Metabolic pitfalls of CNS Cre-based technology. Cell Metab. 18, 21–28. doi: 10.1016/j.cmet.2013.05.019
He, Y., Sun, X., Wang, L., Mishina, Y., Guan, J.-L., and Liu, F. (2017). Male germline recombination of a conditional allele by the widely used Dermo1-cre (Twist2-cre) transgene. Genesis 55:e23048. doi: 10.1002/dvg.23048
Kobayashi, Y., and Hensch, T. K. (2013). Germline recombination by conditional gene targeting with Parvalbumin-Cre lines. Front. Neural Circuits 7:168. doi: 10.3389/fncir.2013.00168
Liput, D. J., Lu, V. B., Davis, M. I., Puhl, H. L., and Ikeda, S. R. (2016). Rem2, a member of the RGK family of small GTPases, is enriched in nuclei of the basal ganglia. Sci. Rep. 6:25137. doi: 10.1038/srep25137
Madisen, L., Zwingman, T. A., Sunkin, S. M., Oh, S. W., Zariwala, H. A., Gu, H., et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. doi: 10.1038/nn.2467
Parkitna, J. R., Engblom, D., and Schutz, G. (2009). Generation of Cre recombinase-expressing transgenic mice using bacterial artificial chromosomes. Methods Mol. Biol. 530, 325–342. doi: 10.1007/978-1-59745-471-1_17
Rahman, Z., Gold, S. J., Potenza, M. N., Cowan, C. W., Ni, Y. G., He, W., et al. (1999). Cloning and characterization of RGS9–2: a striatal-enriched alternatively spliced product of the RGS9 gene. J. Neurosci. 19, 2016–2026. doi: 10.1523/jneurosci.19-06-02016.1999
Rempe, D., Vangeison, G., Hamilton, J., Li, Y., Jepson, M., and Federoff, H. J. (2006). Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis 44, 44–49. doi: 10.1002/gene.20183
Schmidt-Supprian, M., and Rajewsky, K. (2007). Vagaries of conditional gene targeting. Nat. Immunol. 8, 665–668. doi: 10.1038/ni0707-665
Song, A. J., and Palmiter, R. D. (2018). Detecting and avoiding problems when using the Cre-lox system. Trends Genet. 34, 333–340. doi: 10.1016/j.tig.2017.12.008
Tsai, P. T., Hull, C., Chu, Y., Greene-Colozzi, E., Sadowski, A. R., Leech, J. M., et al. (2012). Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 488, 647–651. doi: 10.1038/nature11310
Zeller, A., Crestani, F., Camenisch, I., Iwasato, T., Itohara, S., Fritschy, J. M., et al. (2008). Cortical glutamatergic neurons mediate the motor sedative action of diazepam. Mol. Pharmacol. 73, 282–291. doi: 10.1124/mol.107.038828
Keywords: conditional knockout, Cre recombinase, germline, LoxP, RGS9, striatum, medium spiny neuron, basal ganglia
Citation: Liput DJ (2018) Cre-Recombinase Dependent Germline Deletion of a Conditional Allele in the Rgs9cre Mouse Line. Front. Neural Circuits 12:68. doi: 10.3389/fncir.2018.00068
Received: 01 May 2018; Accepted: 07 August 2018;
Published: 11 September 2018.
Edited by:
Fu-Ming Zhou, University of Tennessee Health Science Center, United StatesReviewed by:
Michael J. Schmeisser, Medizinische Fakultät, Universitätsklinikum Magdeburg, GermanyCopyright © 2018 Liput. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel J. Liput, bGlwdXRkakBtYWlsLm5paC5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.