- 1Laboratory of Immune-Biology, Department of Biomedical and Clinical Sciences, University of Milan, Milan, Italy
- 2Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, Milan, Italy
- 3Laboratory of Immunology, Department of Pathophysiology and Transplantation, University of Milan, Milan, Italy
- 4Department of Medical Biotechnology and Translational Medicine, Aldo Ravelli Center for Neurotechnology and Experimental Brain Therapeutics, University of Milan, Milan, Italy
- 5Department of Pathophysiology and Transplantation, “Dino Ferrari” Center, University of Milan, Milan, Italy
- 6Don C. Gnocchi Foundation, Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Foundation, Milan, Italy
Introduction: COVID-19 typically causes Q7 respiratory disorders, but a high proportion of patients also reports neurological and neuromuscular symptoms during and after SARSCoV-2 infection. Despite a number of studies documenting SARS-CoV-2 infection of various neuronal cell populations, the impact of SARS-CoV-2 exposure on motor neuronal cells specifically has not been investigated so far.
Methods: Thus, by using human iPSC-derived motor neurons (iPSC-MNs) we assessed: (i) the expression of SARS-CoV-2 main receptors; (ii) iPSC-MN infectability by SARS-CoV-2; and (iii) the effect of SARS-CoV-2 exposure on iPSC-MN transcriptome.
Results: Gene expression profiling and immunofluorescence (IF) analysis of the main host cell receptors recognized by SARS-CoV-2 revealed that all of them are expressed in iPSC-MNs, with CD147 and NRP1 being the most represented ones. By analyzing SARS-CoV-2 N1 and N2 gene expression over time, we observed that human iPSC-MNs were productively infected by SARS-CoV-2 in the absence of cytopathic effect. Supernatants collected from SARS-CoV-2-infected iPSC-MNs were able to re-infect VeroE6 cells. Image analyses of SARS-CoV-2 nucleocapsid proteins by IF confirmed iPSC-MN infectability. Furthermore, SARS-CoV-2 infection in iPSCMNs significantly altered the expression of genes (IL-6, ANG, S1PR1, BCL2, BAX, Casp8, HLA-A, ERAP1, CD147, MX1) associated with cell survival and metabolism, as well as antiviral and inflammatory response.
Discussion: These results suggest for the very first time that SARS-CoV-2 can productively infect human iPSC-derived MNs probably by binding CD147 and NRP1 receptors. Such information will be important to unveil the biological bases of neuromuscular disorders characterizing SARS-CoV-2 infection and the so called long-COVID symptoms.
Introduction
SARS-CoV-2 infection typically causes respiratory disorders, but a surprisingly high proportion of patients also reports neurological and neuromuscular complications during and after the acute phase of Corona Virus Disease 19 (COVID-19) (Koralnik and Tyler, 2020; Mao et al., 2020; Chou et al., 2021; Nalbandian et al., 2021). These include cerebrovascular disease, seizures, meningitis, encephalitis, loss of smell (anosmia), taste (ageusia) and myositis (Pezzini and Padovani, 2020; Klein et al., 2021). Moreover, even several months after recovery, almost 30% of the patients display prolonged symptoms including deficits of memory and attention, insomnia, anxiety, depression, a dysexecutive syndrome consisting of inattention, disorientation, and poor movement coordination (Jaywant et al., 2021; Méndez et al., 2021; Ferrucci et al., 2022), and also ataxia, muscle aches and joint pains (Davis et al., 2021; Huang et al., 2021; Tomasoni et al., 2021).
Besides the Central Nervous System (CNS), the documented symptoms also imply an alteration of the peripheral nervous system (PNS) following SARS-CoV-2 infection, which could trigger or worsen neurodegenerative disorders, as recently reported by Serrano-Castro et al. (2020). Supporting this hypothesis, peripheral nerve damage and a more generalized acute polyneuropathy, known as the Guillain-Barré syndrome (GBS) (Needham et al., 2020; Pezzini and Padovani, 2020; Abu-Rumeileh et al., 2021; Raahimi et al., 2021; Taguchi et al., 2022; Stępień and Pastuszak, 2023), have been reported in SARS-CoV-2-infected patients. In three COVID-19 patients, a possible association with a new diagnosis of myasthenia gravis was documented (Restivo et al., 2020; Muhammed et al., 2021). Furthermore, peripheral motor neuropathy has been described before the onset of the typical flu-like symptoms of COVID-19 (Caress et al., 2020; Zhao et al., 2020) and diagnostic criteria for acute polyradiculoneuropathy have been described as well (Alberti et al., 2020). Further supporting the detrimental effect of coronaviruses on PNS, both GBS and acute motor axonal neuropathy (AMAN) have been associated to SARS and MERS infections (Kim et al., 2017; Andalib et al., 2021).
The aforementioned sequelae contribute to defining a clinical picture commonly referred to as “Long COVID-19” or “Post COVID-19” (Carfì et al., 2020; Garrigues et al., 2020),1 which might also occur as a consequence of direct infection/exposure of neuronal cells to SARS-CoV-2 (Lyons-Weiler, 2020). Indeed, ex vivo, in vivo, and in vitro studies suggest that SARS-CoV-2 is able to infect different kinds of neuronal populations, with different degrees of success (Pellegrini et al., 2020; Ramani et al., 2020; Zhang et al., 2020; Song et al., 2021; Lopez et al., 2022; Lyoo et al., 2022; Stein et al., 2022; Kettunen et al., 2023). Our understanding of the mechanisms through which SARS-CoV-2 can enter the nervous system and infect nerve cells is still not completely clear. Studies from both post-mortem COVID-19 patients and animal models suggest that SARS-CoV-2 exploits different neuroinvasive strategies and accession routes (Nampoothiri et al., 2020; Zhou et al., 2020; Anand et al., 2021). These include: (i) hematogenous transport of infected immune cells via the circulatory system of the brain tissue; (ii) infection of the nasal olfactory epithelium to reach the brain by axonal transport along the olfactory nerve; (iii) retrograde virus spread from the lungs to the CNS through the vagus nerve; (iv) entry from the ocular epithelium; and (v) virus invasion through impairment of the blood-brain barrier (BBB).
The molecular mechanisms sustaining SARS-CoV-2 infection of nerve cells are yet to be defined as well. For instance, angiotensin converting enzyme 2 (ACE2), the major actor involved in multi-organ SARS-CoV-2 infection, shows low expression levels in the human brain (Hamming et al., 2004). Thus, despite the initial concept of ACE2 distribution as the major determinant of SARS-CoV-2 infectivity and spread, other factors have been called-on as responsible for SARS-CoV-2 neurovirulence. This is the case of Neuropilin 1 (NRP1) (Cantuti-Castelvetri et al., 2020; Daly et al., 2020), CD147 (BSG) (Chen et al., 2020), transmembrane serine protease 2 (TMPRSS2) (Qiao et al., 2020), and Furin (Coutard et al., 2020; Ou et al., 2020), which show higher and broader patterns of expression in neuronal cells compared to ACE2.2
Although public health measures and vaccination campaigns have significantly contributed to limiting COVID-19 spread and severity, SARS-CoV-2-induced neurological manifestations still represent a threat for long-term health, as their biological bases remain largely unknown. In particular, the documented neuromuscular dysfunctions related with COVID-19 suggest the occurrence of widespread alterations possibly affecting all the motor unit components. Nonetheless, the impact of SARS-CoV-2 exposure on motor neuronal cells specifically has not been investigated so far. Therefore, as summarized in Figure 1, by using an in vitro model of human motor neurons differentiated from induced pluripotent stem cells (iPSC-MNs), we aimed to assess, for the first time (i) the expression of SARS-CoV-2 main receptors; (ii) iPSC-MN infectability by SARS-CoV-2; and (iii) the effect of SARS-CoV-2 exposure on iPSC-MN transcriptome.
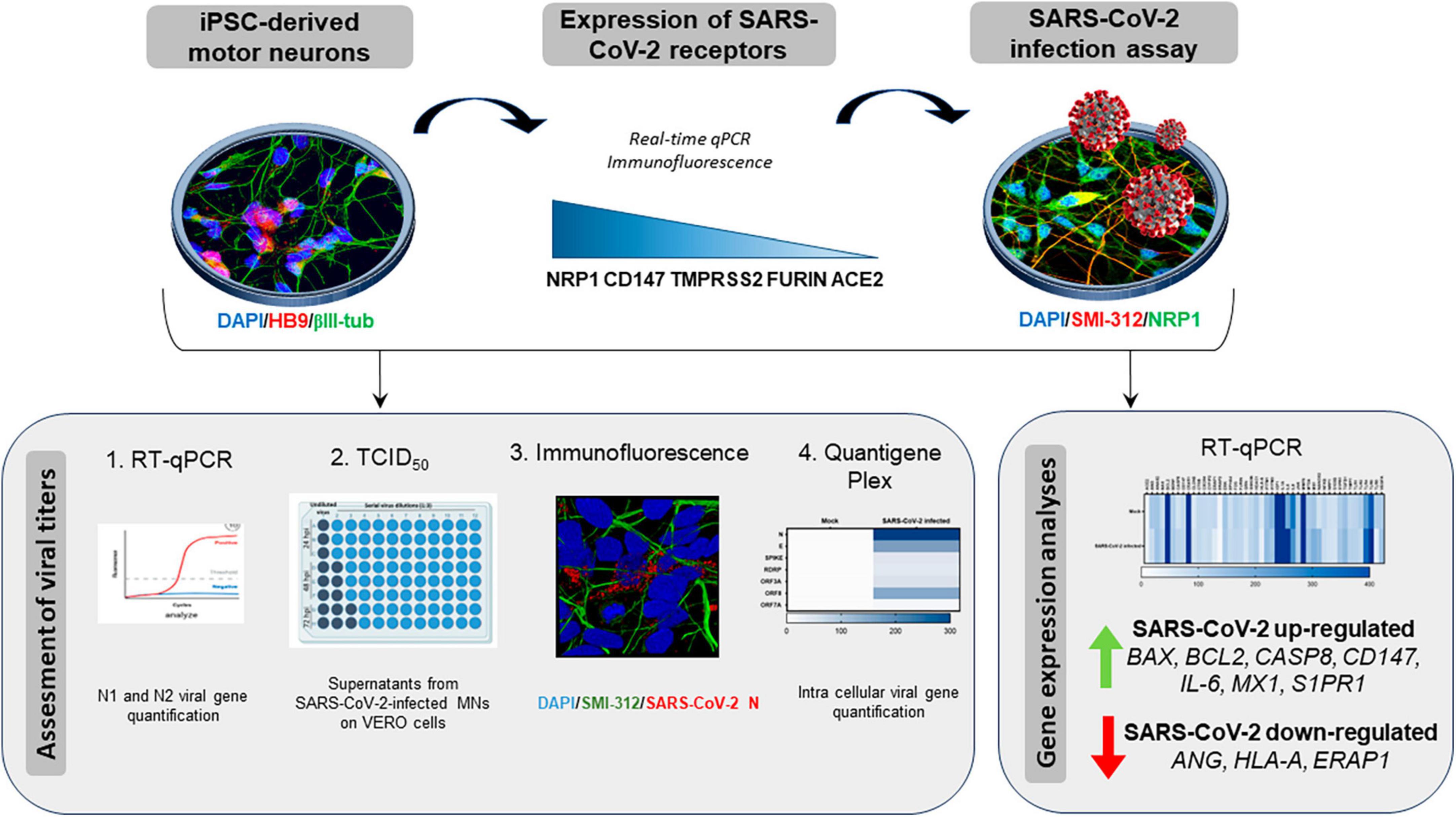
Figure 1. Graphical representation of the study workflow. SARS-CoV-2 host receptor gene expression was assessed on iPSC-derived motor neurons (iPSC-MNs) by RT-qPCR and immunofluorescence. iPSC-MN were then in vitro infected by SARS-CoV-2 and viral infection/replication was assessed by a multidisciplinary approach using RT-qPCR, TCID50, immunofluorescence and quantigene Plex techniques. The effect of viral infection on iPSC-MN homeostasis was determined by analyzing the alteration of their transcriptome.
Materials and methods
Cell lines and culture
VeroE6 cells (ATCC, VA, USA) and Human Lung Carcinoma Cells Expressing Human Angiotensin-Converting Enzyme 2 (A549-hACE2) (BEI Resources, Catalog No. NR-53821) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) (Euroclone, Milan, Italy) containing 4 mM L-glutamine, supplemented with 10% FBS, 100 μ/mL penicillin and 100 μg/mL streptomycin at 37°C and 5% CO2. Cells were regularly passaged and tested for the presence of mycoplasma contamination.
iPSC generation and MN differentiation
Induced Pluripotent Stem Cells (iPSC) were reprogrammed from 3 healthy donors (Supplementary Table 1) after obtaining informed consent and approval from local ethics committee (approval number 2022_03_15_12). For fibroblast/blood cell reprogramming the CytoTune®-iPSC 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific) was used as previously described (Bardelli et al., 2020). After picking and selecting about 6 clones per sample, one clone for each cell line was further grown in Essential 8 medium (Thermo Fisher Scientific) and fully characterized for the expression of stemness markers (TRA-1-60, OCT3/4, SOX2, NANOG) and for the capacity to spontaneously differentiate into the three germ layers lineages as assessed by the expression of alpha-feto protein (AFP) as endodermal, βIII tubulin as ectodermal and desmin as mesodermal markers and already shown in Bossolasco et al. (2018) and Gumina et al. (2019). Genome integrity of the iPSC clones was determined by Q-banding karyotype analysis (Bossolasco et al., 2018).
Induced Pluripotent Stem Cells were differentiated into motoneurons (MNs) as previously described (Bardelli et al., 2020). Briefly, iPSCs were grown in suspension for 21 days to obtain embryoid bodies (EBs) in HuES medium (DMEM/F12, 20% knock-out serum replacement, 2 mM L-glutamine, 10 μ/ml penicillin, 10 μg/ml streptomycin, 0.1 mM MEM NEAA, 110 μM β-mercaptoethanol) for the first 3 days and then in neural induction medium (DMEM/F12, 2 mM L-glutamine, 10 μ/ml penicillin, 10 μg/ml streptomycin, 0.1 mM MEM NEAA, 2 μg/ml heparin, 1% N2 supplement), supplemented with specific factors. EBs were dissociated with 0.05% trypsin and cells were plated on poly-D-lysine/laminin-coated (Thermo Fisher Scientific) coverslips and cultured in neural differentiation medium (Neurobasal medium, 2 mM L-glutamine, 10 μ/ml penicillin, 10 μg/ml streptomycin, 0.1 mM MEM NEAA, 1% N2 supplement, all from Thermo Fisher Scientific) with the addition of specific factors for 13 days to obtain iPSC-MNs.
In vitro SARS-CoV-2 infection assay
The European (EU–B.1) SARS-CoV-2 lineage was a kind gift of Dr. Davide Mileto, Clinical Microbiology, ASST Fatebenefratelli-Sacco, Milan, Italy. All the experiments with SARS-CoV-2 were performed in a BSL3 facility.
In order to generate a viral stock, SARS-CoV-2 was expanded in VeroE6 cells and infectious viral particles concentration was assessed by 50% tissue culture infectious dose (TCID50) assay, as elsewhere described (Fenizia et al., 2022).
Induced Pluripotent Stem Cells-MNs were in vitro Mock- or SARS-CoV-2-infected with 1 multiplicity of infection (MOI). After an overnight incubation, cells were thoroughly washed three times with pre-warmed PBS and replenished with the complete neural differentiation medium. Supernatants were collected at 6 (T0), 24 (T1), 48 (T2), and 72 (T3) hours post-infection (hpi) to monitor viral replication and to perform SARS-CoV-2 infection assays on VeroE6 cells for each iPSC-MN line (Supplementary Table 1 and Supplementary Figure 1).
At 48 hpi, iPSC-MNs were fixed for immunofluorescence (IF) analyses, while cells harvested at 72 hpi were lysed for RNA extraction and appropriately stored at −80°C for further processing, as specified below.
At 72 hpi, cell viability was assessed by Trypan Blue exclusion assay. Briefly, iPSC-MNs were incubated in Accutase (Thermo Fisher Scientific) for 5 min at 37°C. Then, an equal volume of fresh medium was added to the wells to stop the dissociation reaction and the cells were detached and centrifuged for 8 min at 1200 rpm. The supernatant was carefully discarded, and cells were resuspended in 1 mL of fresh medium. Ten μl of cell suspension were incubated with 10 μl of 0.4% Trypan Blue (Merck-Sigma, Milan, Italy) in 96-well plates. Ten μl of the mix were loaded on chamber slides and counted with the T20 Automated Cell Counter (Bio-Rad Laboratories, Hercules, CA, USA).
Viral replication assessment
For SARS-CoV-2 replication assessment, RNA was extracted from iPSC-MN supernatants using the Maxwell® RSC Instrument with Maxwell® RSC Viral Total Nucleic Acid Purification Kit (Promega, Fitchburg, WI, USA). Viral RNA was reverse transcribed in a single-step RT-qPCR (GoTaq 1-Step RT-qPCR; Promega) on a CFX96 instrument (Bio-Rad, Hercules, CA, USA) using primers specifically designed to target two regions of the nucleocapsid (N1 and N2) gene (Fenizia et al., 2020) (2019-nCoV CDC qPCR Probe Assay emergency kit; IDT, Coralville, IA, USA), together with primers for the human RNase P gene. Viral copy number quantification (viral copy number/μl) was assessed by creating a standard curve from the quantified 2019-nCoV_N positive Plasmid Control (IDT).
VeroE6 cells were in vitro infected with supernatants collected from SARS-CoV-2-infected iPSC-MNs at different time points (24, 48, 72 hpi) and infectious viral particles concentration was assessed by TCID50, as previously described. Briefly, VeroE6 cells were seeded at 2 × 104 cells per well in a 96-well plate and cultured with serial dilutions (1:3) of the iPSC-MN supernatants collected at different time points. After 72 hpi, VeroE6 cell supernatants were removed, cells fixed by 4% paraformaldehyde (PFA–Sigma-Aldrich, MO, USA) for 1 h at RT and then stained with 0.2% crystal violet solution (Sigma-Aldrich) to assess cell death and to calculate TCID50.
QuantiGene plex gene expression assay
SARS-CoV-2 infection was further assessed on 5 × 104 iPSC-MNs by QuantiGene Plex assay (Thermo Scientific), which uses signal amplification rather than target amplification for direct measurement of RNA transcripts directly from lysed cells. The following SARS-CoV-2 viral genes were analyzed: ORF7A, ORF3A, ORF8, RDRP, E, and N. Signal was detected using a Luminex instrument and results were calculated relative to GAPDH, and PPIB as housekeeping genes, and expressed as ΔCt.
Gene expression analyses
Total RNA was extracted from iPSC-MNs as previously described by Limanaqi et al. (2022). Gene expression analyses of the main SARS-CoV-2 receptors (ACE2, CD147, NRP1) and peptidases (FURIN, TMPRSS2) as well as N (nucleocapsid)1, N2, S (spike)1, S2 and E (envelope)1 SARS-CoV-2 sequences was performed by Real-time qPCR (CFX96 connect, Bio-Rad, Hercules, CA, USA) using SYBR Green PCR mix (Promega), according to the following thermal profile: initial denaturation (95°C, 15 min), and 40 cycles with denaturation (15 s at 95°C), annealing (1 min at 60°C) and extension (20 s at 72°C). A Ct value of 35 or higher was considered negative. Melting curves were also analyzed for amplicon characterization. Results for gene expression analyses were calculated by the 2–ΔΔCt equation and presented as the average of the relative expression units to an internal reference sample and normalized to the expression of the GAPDH housekeeping gene. Samples with GAPDH Ct values above 20 were excluded from the analysis. Already optimized primers were purchased (PrimePCR, Bio-Rad, Segrate, Italy). Gene expression analyses of the main SARS-CoV-2 receptors was also assessed on RNA extracted from A549-hACE2 cells, as positive control.
The expression of 46 genes related to inflammatory, apoptotic, and antiviral pathways were analyzed by a PCR array including a set of optimized Real-time PCR primers (Bio-Rad) for the targets reported in Supplementary Table 2. Gene expression analyses were performed in iPSC-MNs at 72 hpi in duplicates. Results were analyzed using the SABiosciences online software, expressed by the 2–ΔΔCt equation and presented as the average of the relative expression units to an internal reference sample and normalized to the expression of the GAPDH and ACTB housekeeping genes.
Immunofluorescence assays
Induced Pluripotent Stem Cells-MNs were seeded on coverslips in a 24-well plate, cultured until differentiation, and infected as specified above. At 48 hpi, cells were fixed in PBS containing 4% PFA at RT for 10 min, followed by permeabilization with 0,1% TritonX-100 in PBS for 10 min. Cells were treated with 1% BSA in PBS for blocking at RT for 1 h, and incubated at 4°C overnight with specific primary antibodies. The following primary antibodies were used: anti-beta III Tubulin (1:500, Abcam, Cambridge, UK), anti-SMI-312 (1:1000, Covance, Princetown, NJ, USA) and anti-ChAT (1:200, Chemicon) to assess iPSC-MN differentiation; anti-ACE2 (1:200, Prodotti Gianni), anti-CD147 (1:100, Thermo Fisher Scientific) anti-NRP1 (1:100, Thermo Fisher Scientific) and anti-N Nucleocapsid SARS-CoV-2 (1:1000, BEI Resources) to assess SARS-CoV-2 receptors and infection. Coverslips were then stained with secondary antibodies (Alexa Fluor 488 or 647, 1:500, Abcam) for 45 min at RT and mounted using a medium containing DAPI (Enzo Life Sciences, Milan, Italy). Confocal images were acquired on a TCS SP8 System equipped with a DMi8 inverted microscope and a HC PL APO 40 × /1.30 Oil CS2 (Leica Microsystems, Wetzlar, Germany) at a resolution of 1024 × 1024 pixels (single stack).
Statistical analyses
Overall, we performed 14 SARS-CoV-2 independent experiments by using iPSC-derived MNs from 3 healthy donors. The different analyses were assessed on such samples according to the scheme reported in Supplementary Table 1.
Statistical analyses were performed using GraphPad Prism 8. Results are expressed as mean ± SEM of the indicated n-values. The two-tailed Student’s t-test was used with a p-value threshold of 0.05.
Results
Expression of SARS-CoV-2 receptors in iPSC-derived MNs
To assess the potential susceptibility of motor neuronal cells to SARS-CoV-2 infection, we differentiated iPSC from 3 healthy donor individuals (1 male and 2 females, 37–49 years of age at biopsy collection; Supplementary Table 1) into motor neurons (iPSC-MNs) expressing neuronal (bIII-tubulin and SMI-312) and motoneuronal (ChAT, HB9) markers (Figure 2A). By Real-time qPCR we validated the gene expression of the main receptors used by the virus and observed that all SARS-CoV-2 receptors (ACE2, CD147, NRP1) and peptidases (TMPRSS2, FURIN) analyzed were expressed in iPSC-MNs, although with different degrees (Figure 2B). In particular, by assessing the expression of these receptors in A549-hACE2 cells, a cell line used as positive control, ACE2 and Furin gene expression was significantly lower in iPSC-MNs compared to A549-hACE2 cells (Figure 2B). NRP1 gene expression was instead comparable in iPSC-MNs and A549-hACE2 cells, while CD147 and TMPRSS2 gene expression was significantly higher in iPSC-MNs than in A549-hACE2 cells (Figure 2B). Gene expression data on SARS-CoV-2 human receptors were further confirmed by IF analysis for ACE2, CD147 and NRP1 markers on both iPSC-MNs (Figure 2C) and A549-hAC2 cells (Figure 2D). Indeed, as for the gene expression analysis, ACE2 fluorescence intensity was significantly lower in iPSC-MNs compared to A549-hACE2 cells (Supplementary Figure 1).
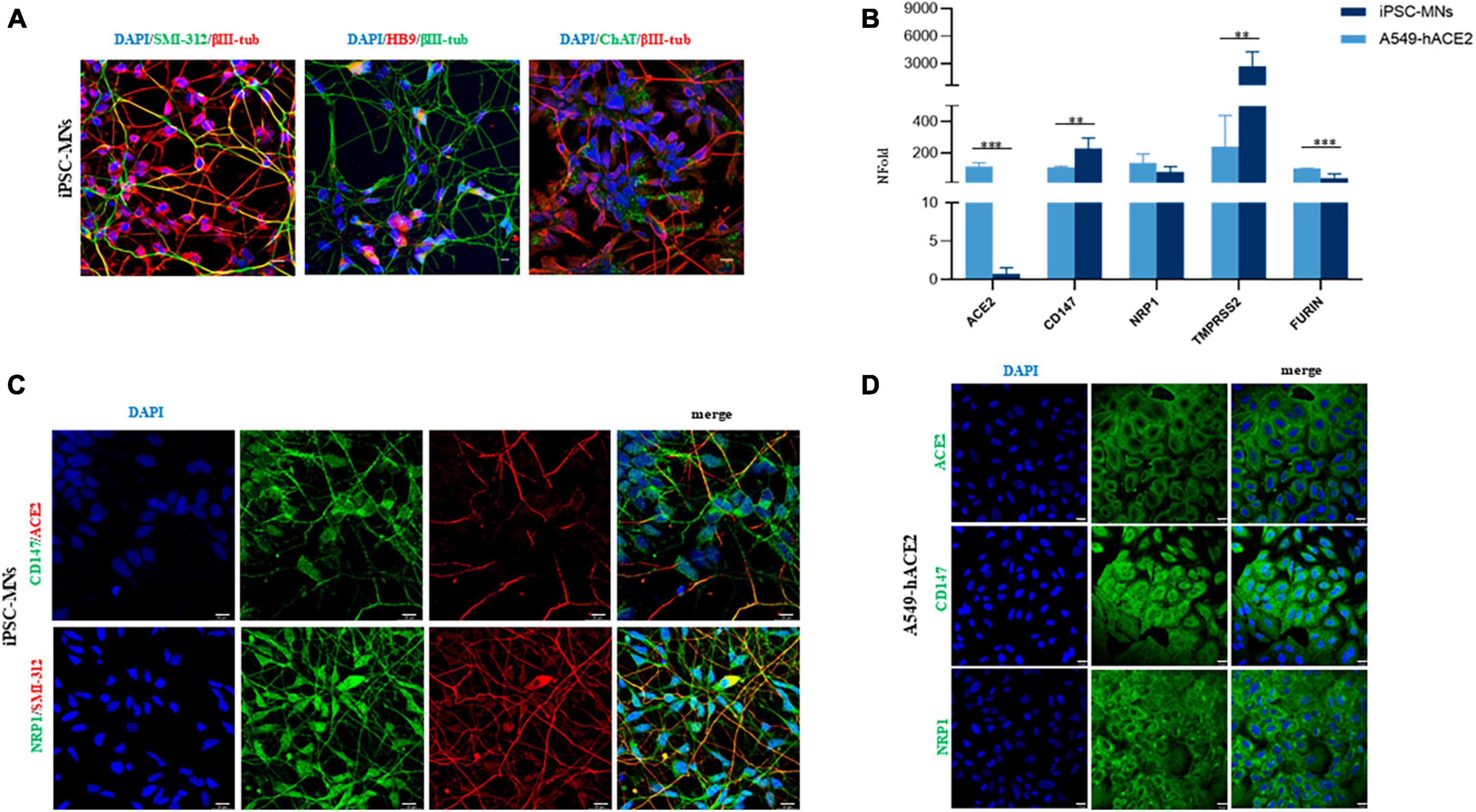
Figure 2. Expression of SARS-CoV-2 human receptors on iPSC-MNs and A549-hACE2 cells. (A) Representative images of differentiated iPSC-MNs obtained from 3 healthy control individuals. Expression of neuronal (βIII-Tubulin, red and green, and SMI-312, green) and motoneuronal (HB9, red and ChAT, green) markers is shown in merged images. Nuclei were stained with DAPI (blue). Bar, 10 μm. (B) Gene expression analyses of ACE2, CD147, NRP1, TMPRSS2, and FURIN in A549-hACE2 cells and iPSC- MNs by Real-time qPCR. Results are presented as mean ± SEM; n ≥ 4 for each cell line/iPSC-MN; the Student’s t-test was used with the p-value threshold of 0.05. Significance is indicated as follows: **p < 0.01; ***p < 0.001. Representative immunofluorescence images for CD147 and NRP1 markers (green) in iPSC-MNs and in A549-hACE2 cells are shown in panels (C,D), respectively. The expression of ACE2 is shown in red (C) and in green (D). Nuclei were stained with DAPI (blue). The neuronal marker SMI-312 (red) is shown only in (C). Bars correspond to 20 μm in both (C,D).
SARS-CoV-2 viral replication in iPSC-MNs
To assess whether iPSC-MNs are productively infected by SARS-CoV-2, different experimental approaches were employed. We first assessed that cell viability, measured by Trypan blue assay, was not significantly modified in the 3 different iPSC-MN lines by comparing mock- and SARS-CoV-2-infected cells which showed more than 90% viability (Supplementary Table 3), indicating a lack of cytopathic effect.
By analyzing N1 and N2 viral nucleocapsid gene expression by Real-time qPCR in supernatants from iPSC-MN cultures over a timeframe of 72 hpi, we observed that human iPSC-MNs were productively infected by SARS-CoV-2 in a time-dependent manner (Figure 3A), although viral replication was not accompanied by cytopathic effect as assessed by crystal violet assay (data not shown). Moreover, levels of viral replication were modest (at 72 hpi: mean viral copy number/μl ± SEM, N1 = 4464.2 ± 1281.7; N2 = 19139.4 ± 5157.0) compared with SARS-CoV-2-susceptible VeroE6 cells (at 72 hpi: mean viral copy number/μl ± SEM, N1 = 18.18e + 06 ± 3.2 e + 06; N2 = 80.93e + 06 ± 17.29 e + 06) (Supplementary Figure 2).
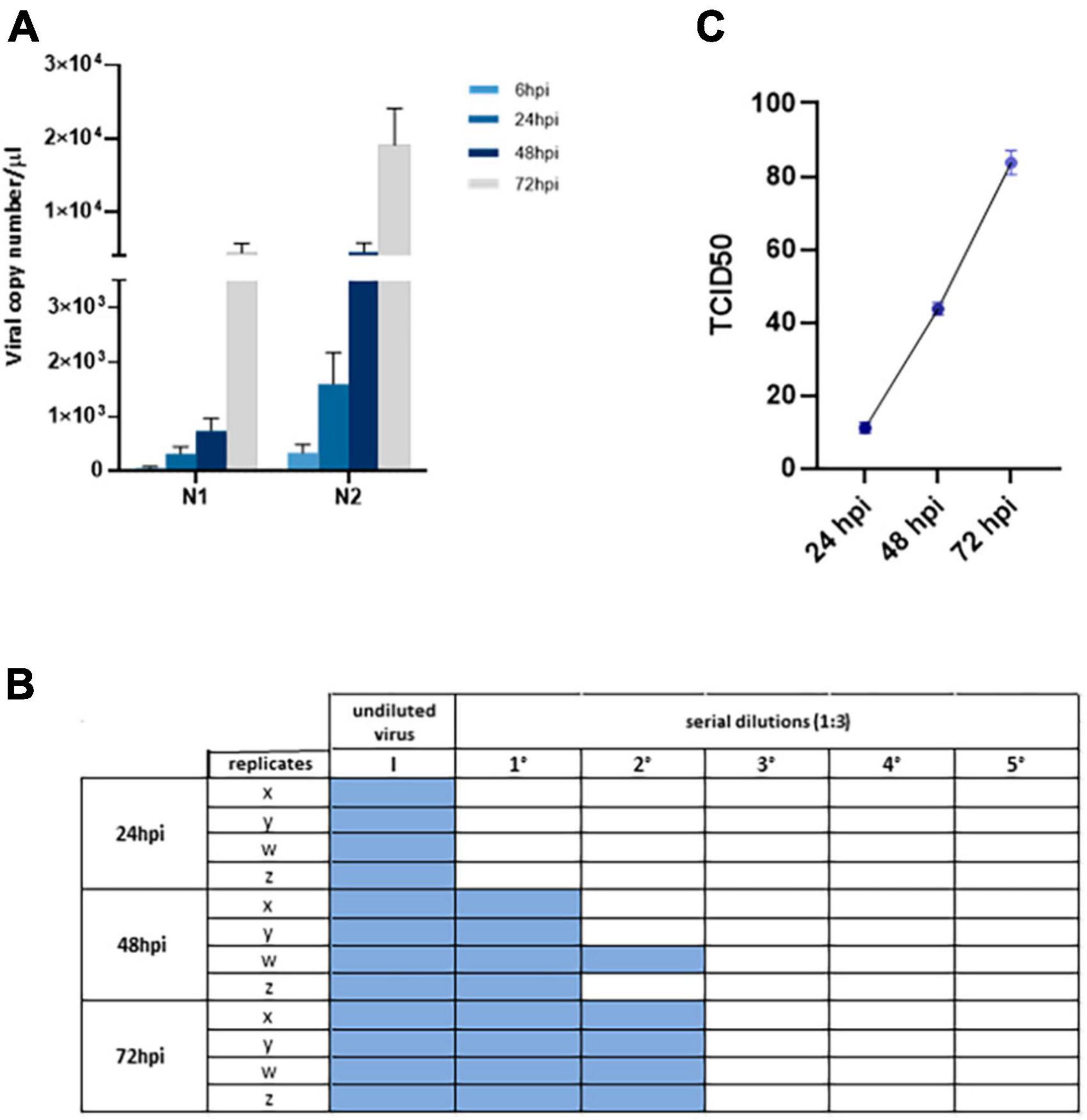
Figure 3. Assessment of viral replication in iPSC-MNs. (A) Upon in vitro challenge of iPSC-MNs with 1 MOI of SARS-CoV-2, the infection was monitored at 6, 24, 48, and 72 hpi. Results correspond to the absolute viral copy number/μl of the SARS-CoV-2 N1 and N2 target sequences from cell supernatants that were quantified through a single-step Real-time qPCR by referring to a standard curve for Ct values (IDT, Coralville, IA, USA). Results are presented as mean ± SEM from ≥4 independent replicates each on iPSC-MNs derived from the 3 enrolled healthy subjects (Supplementary Table 1). (B) TCID50 analyses on VeroE6 cell infectability after exposure to iPSC-MN-infected supernatants, specifically the undiluted virus (I) and five serial dilutions (1:3), at 24, 48, and 72 hpi. Viral infection was assessed by cytopathic effect on VeroE6 cells as represented by colored well. The plate is representative of a single experiment which was performed once in quadruplicate (x, y, z, w) for each of the three different iPSC-MN lines. (C) Titration of SARS-CoV-2 virus in VeroE6 supernatants at 24, 48, and 72 hpi from data shown in (B). Data are shown as TCID50. Results are presented as mean ± SEM from four independent replicates each on iPSC-MNs derived from the three enrolled healthy subjects.
To further verify the productive infectability of iPSC-MNs, we then exposed VeroE6 cells to supernatants collected from infected iPSC-MNs at different time points (24, 48, and 72 hpi). VeroE6 cell infection resulted in an evident cytopathic effect which, as expected, increased according to the supernatant collection period over time (Figure 3B), from 9.5 TCID50 at 24 hpi to 85.4 TCID50 at 72 hpi (Figure 3C), mirroring the results obtained by Real-time qPCR.
To further validate these results, the expression of some viral RNA targets was investigated in iPSC-MNs at intracellular level by three different methods: QuantiGene assay, Real-time qPCR and immunofluorescence (IF). As reported in the heatmap, the RNA of all the viral targets analyzed by QuantiGene (N, E, SPIKE, RDRP, ORF3A, ORF8, and ORF7A) were exclusively detected in SARS-CoV-2-infected cells, although at different levels (Figure 4A). The mRNA expression of N1, S1, S2, and E2 in infected iPSC-MNs was further confirmed by Real-time qPCR (Figure 4B).
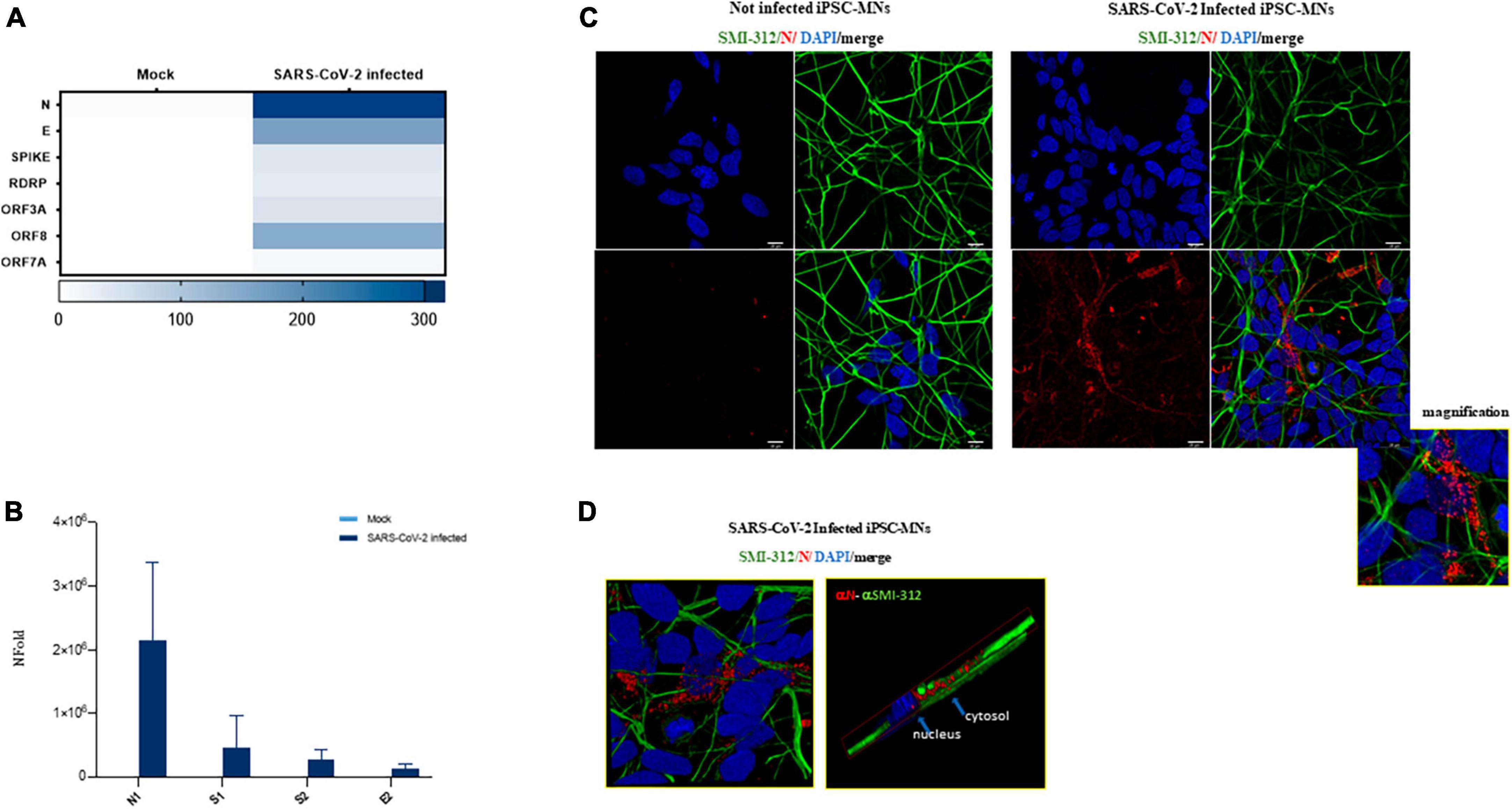
Figure 4. Assessment of viral replication in iPSC-MNs. (A) Expression of SARS-CoV-2 viral genes ORF7A, ORF3A, ORF8, RDRP, S, E, and N by QuantiGene Plex Gene expression technology in uninfected (Mock) and SARS-CoV-2-infected iPSC-MNs. Results shown on the heatmap correspond to the mean ± SEM from ≥1 independent replicates on each iPSC-MNs derived from 2 healthy control subjects (Supplementary Table 1). (B) Real-time qPCR expression analyses of N1, S1, S2, and E2 viral genes in uninfected (Mock) and SARS-CoV-2-infected iPSC-MNs. Results are presented as mean ± SEM from ≥4 independent replicates on iPSC-MNs derived from the 3 healthy control subjects (Supplementary Table 1). (C) Representative immunofluorescence images of N protein (red) and neuronal SMI-312 marker (green) in Mock- and SARS-CoV-2-infected iPSC-MNs at 48 hpi. Nuclei were stained with DAPI (blue). Bars correspond to 20 μm. (D) Representative magnified immunofluorescence images and 3D reconstruction for N protein and SMI-312 marker in SARS-CoV-2-infected iPSC-MNs at 48 hpi.
Finally, by IF assay, the nucleocapsid (N) protein was detected exclusively in SARS-CoV-2-infected iPSC-MNs (Figure 4C), mainly at perinuclear level in the soma and along the neurite extensions (Figure 4D), though the percentage of infected cells seems to be very low.
Effect of SARS-CoV-2 infection on iPSC-MN gene expression
In order to assess whether SARS-CoV-2 infection fosters changes in iPSC-MNs gene expression, we evaluated the expression profile of a set of genes involved in the antiviral and immune-related response. Overall, among the 46 analyzed targets by a custom array (Supplementary Table 2), we observed a widespread alteration of gene expression following SARS-CoV-2 exposure at 72 hpi, suggesting a deep alteration of cell homeostasis (Figure 5A). Notably, SARS-CoV-2-induced deregulation was significant for 10 genes involved in different intracellular pathways with both up-regulated (BAX, BCL2, CASP8, CD147, IL-6, MX1, S1PR1) and down-regulated (ANG, HLA-A, ERAP1) expression, as shown in Figure 5B. Notably, despite a slight rise in CASP8 expression, the BCL2/BAX ratio was significantly increased in SARS-CoV-2 infected iPSC-MNs insinuating different plausible speculations about the effect triggered by viral entry on the apoptotic pathway (Supplementary Figure 3).
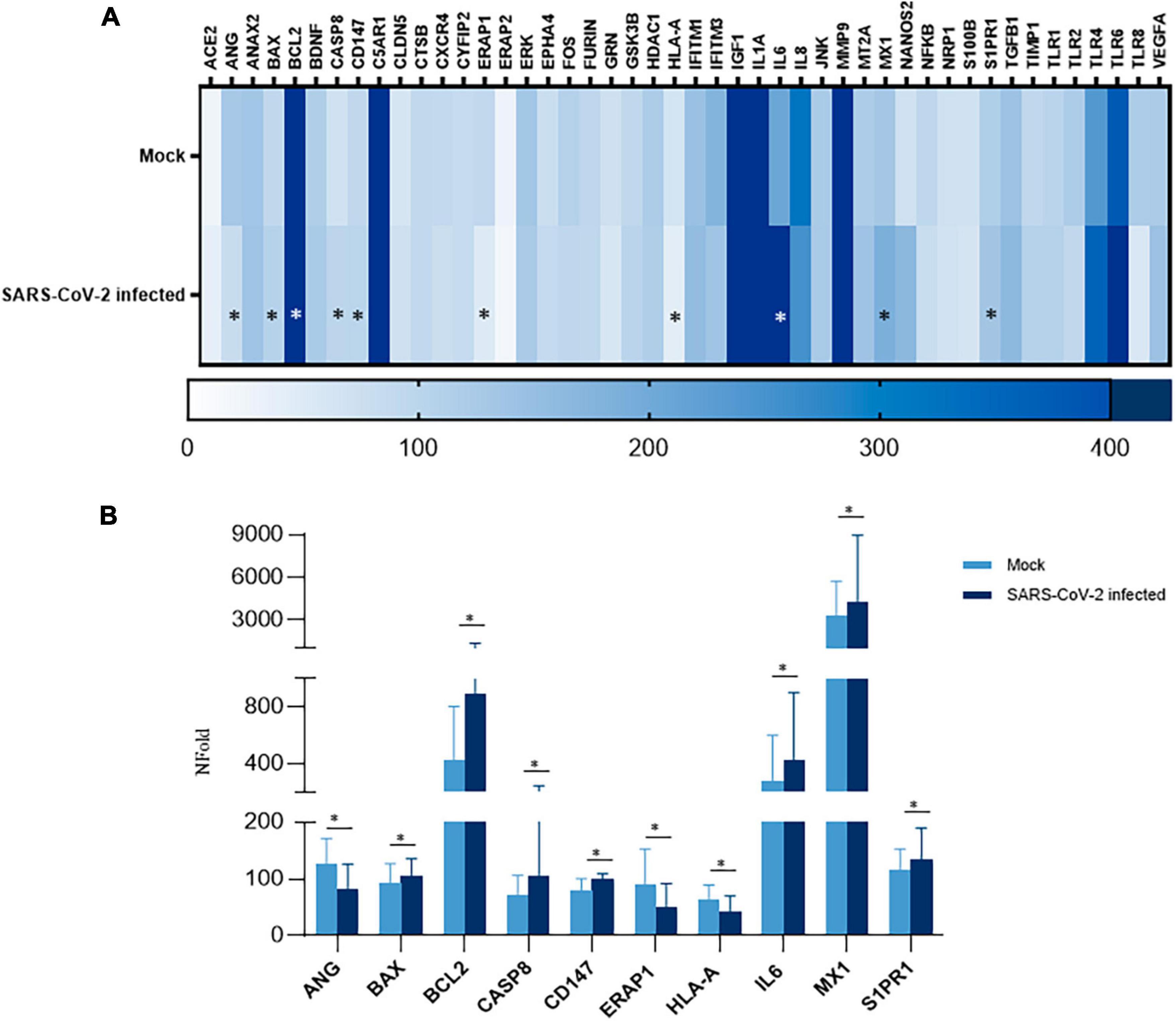
Figure 5. Gene expression analyses in SARS-CoV-2-infected iPSC-MNs. (A) Heatmap representation of Real-time qPCR expression data of 46 genes in uninfected (Mock) and SARS-CoV-2-infected iPSC-MNs at 72 hpi. (B). Expression analyses of the statistically significant genes in SARS-CoV-2-infected vs. uninfected (Mock) iPSC-MNs at 72 hpi are shown in (A). mRNA quantification was performed by Real-time qPCR and calculated by the 2–ΔΔCt equation. Results in (A,B) correspond to the mean ± SEM from ≥4 independent replicates on each iPSC-MN derived from the three enrolled healthy subjects (Supplementary Table 1); the Student’s t-test was used with the p-value threshold of 0.05. Significance is indicated as follows: *p < 0.05.
Discussion
Clinical observations from COVID-19 patients support evidence for the damages caused by SARS-CoV-2 infection on both central and peripheral components of the NS. Documented neurological abnormalities include postural tremor (13.8%), motor/sensory deficits (7.6%) (Pilotto et al., 2021), as well as persistent muscle pain (myalgia), muscle weakness (mild to severe), fatigue, exercise intolerance and arthralgia (Carfì et al., 2020; Pleguezuelos et al., 2021; Soares et al., 2022). These are more frequent in severe manifestations of the disease and can differ among individuals (Montalvan et al., 2020; Niazkar et al., 2020). Rarely, clinically defined cases of acute disseminated encephalomyelitis, Guillain-Barré syndrome, and acute necrotizing encephalopathy have also been reported in COVID-19 patients (Lou et al., 2021). Furthermore, COVID-19 has been shown to significantly affect amyotrophic lateral sclerosis (ALS) patients, causing a rapid neurological deterioration, accompanied by a marked decline in fine motor skills of hand and leg strength (Fu et al., 2022). The damage of motor neurons, in turn, leads to the deterioration of muscle function, resulting in physical weakness, muscle atrophy, and paralysis. In addition, as reported by Li et al. (2022), SARS-CoV-2 infection may stimulate the ALS-associated amyloid aggregation of host proteins, providing molecular evidence for the role of SARS-CoV-2 in triggering neurodegeneration and MNDs.
Despite a number of studies documenting SARS-CoV-2 infection of various neuronal cell populations (Pellegrini et al., 2020; Ramani et al., 2020; Zhang et al., 2020; Lopez et al., 2022; Stein et al., 2022), to date, neither MN susceptibility to SARS-CoV-2 infection nor the molecular consequences of viral exposure on their homeostasis have been investigated. Therefore, it is still debated if the documented neurological/neuromuscular manifestations are secondary to a direct MN viral invasion and/or a collateral injury driven by an uncontrolled innate immune response fostering a pro-inflammatory milieu (immune mediated cytokines release), which is a hallmark of SARS-CoV-2 infection and COVID-19 severity.
In this study, we demonstrated for the first time that human iPSC-MNs are permissive to SARS-CoV-2 entry and production of infectious viral particles which are released in the cell culture supernatant. In fact, by using different experimental approaches, we verified that the virus is able to infect and replicate within this neuronal cell model. However, the levels of viral replication and the percentage of infected cells are significantly lower compared to those of susceptible cells, such as VeroE6 ones, possibly justifying the absence of cytopathic effect in iPSC-MNs. However, one should consider that this cellular model lacks the immunological component which, following SARS-CoV-2 infection, may favor the onset of a pro-inflammatory environment that is advantageous for viral infection/replication, and subsequent neuronal damage. It is therefore possible that in a pro-inflammatory setting, (i.e., in patients with pre-existing neuromuscular conditions) the rate of viral entry and replication would be substantially higher.
By profiling the expression of the main host cell receptors exploited by SARS-CoV-2, we confirmed previous data that ACE2 is scarcely expressed in neurons (Piras et al., 2022), here extending the observation to motor neuronal cells. We therefore speculate that iPSC-MN infection mainly relies on CD147 and/or NRP1 binding, as their expression on iPSC-MNs is high. Further confirming this assumption, it has already been demonstrated that these proteins have higher and broader patterns of expression in the human brain (Qiao et al., 2020), advocating for their role as putative mediators of SARS-CoV-2 entry into human nerve cells. However, further analyses will be necessary to validate this hypothesis.
Interestingly, we also observed that SARS-CoV-2 infection promotes an alteration of iPSC-MNs transcriptome, involving different intracellular pathways. First, SARS-CoV-2 infection was associated with upregulation of MX1, a protective factor whose expression was reported to be switched-on in several viral infections including SARS-CoV-2 (Sironi et al., 2014; Haller et al., 2018; Bizzotto et al., 2020; Haller and Kochs, 2020). This suggests that SARS-CoV-2 prompts the activation of the antiviral response in iPSC-MNs.
Second, in SARS-CoV-2-infected iPSC-MNs we observed a conspicuous transcriptional upregulation of the pro-inflammatory cytokine IL-6. Remarkably, significantly elevated IL-6 levels have been reported in COVID-19 patients, which is associated with adverse clinical outcome (Coomes and Haghbayan, 2020). In the NS, IL-6 levels increase in case of injury and/or inflammation, which may have both beneficial and detrimental effects on nerve cell survival and healing, depending on the cell type and context (Kummer et al., 2021). In the frame of neurodegenerative or neuropathic disorders, IL-6 exacerbates neurodegeneration and cell death, while blockade of IL-6 signaling improves the locomotor function in mice with spinal cord injury (Tan et al., 2013). It is therefore plausible to assume that the neuromuscular complications of SARS-CoV-2 infection are due, at least in part, to an increased production of this inflammatory mediator, which is to some extent self-powered by infected MNs.
The expression of genes involved in the apoptotic pathway, such as BCL2, BAX, and CASP8, was upregulated as well. Similar results were recently documented by Li et al. (2020) in SARS-CoV-2 infected lung epithelial cells where an increase of Caspase 8 responsible for cell death and inflammation was observed. Nonetheless, we also observed that the ratio between the anti-apoptotic BCL2 and pro-apoptotic BAX genes was significantly increased in SARS-CoV-2 infected iPSC-MNs, suggesting that programmed cell death is somehow prevented following infection in these neuronal cells. Further analyses will be necessary to verify if this result reflects the virus’ ability to manipulate, delay, or inhibit the host defense response, that usually exploits the apoptotic process to disrupt virus multiplication and propagation, thereby preserving other cells and surrounding tissue.
We also reported a modulation in the expression of factors involved in the antigen processing and presentation pathway. In particular, a statistically significant reduction in the expression of both HLA-A (MHC-I) and ERAP1 was observed. This is not surprising, as modifications in HLA-I and ERAPs expressions are found in many viral diseases. The downregulation of these targets represents one of the immune evasion strategies most widely used by viruses to block antigen presentation as well as CD8 + and NK cell response (Jonjić et al., 2008; Chaudhry et al., 2020; Mancini and Vidal, 2020). SARS-CoV-2 is no exception, as it was documented to downregulate cell surface MHC-I as a strategy to escape immune responses in various peripheral cell models (Zhang et al., 2020; Yoo et al., 2021). The role of MHC-I expression/activation in diseased MNs remains contradictory, as both detrimental and protective effects have been documented (Nardo et al., 2016). Besides immunological mechanisms, MHC-I has been firmly implicated in neuronal plasticity, regulation of synaptic density and axonal regeneration in the CNS and PNS, both during development and in brain diseases. Therefore, the downregulation of MHC-I along with ERAP1 detected in our model of SARS-CoV-2-infected iPSC-MNs might represent: (i) an efficient antagonism of adaptive immune responses favoring successful viral replication, and/or also (ii) a consequence of virus-induced alterations of neuronal homeostasis. This calls for additional focused investigations on the role and functional significance of neuronal MHC-I expression in the frame of SARS-CoV-2 infection.
In line with the possible alterations of neuronal homeostasis associated with SARS-CoV-2 infection in iPSC-MNs, we also detected a consistent downregulation of ANG (Angiogenin) gene expression. ANG is expressed in neurons during neuro-ectodermal differentiation and it exerts both neurotrophic and neuroprotective functions in the light of its role in multiple steps of RNA metabolism or processing (Thiyagarajan et al., 2012). Again, ANG acts as a circulating protein induced during inflammation and exhibits microbicidal activity against bacterial and fungal systemic pathogens, thus contributing to systemic responses to infection (Hooper et al., 2003; Noschka et al., 2021). Our data suggest that SARS-CoV-2 infection in iPSC-MNs goes along with impaired ANG transcription, which might contribute to sustaining both virus-induced immune antagonism and dysregulation of neurotrophic responses.
Finally, in our model of SARS-CoV-2-infected iPSC-MNs, we detected an upregulation of CD147 and S1PR1 (Sphingosine 1-phosphate Receptor 1) genes. CD147 silencing was shown to reduce SARS-CoV-2 replication in epithelial lung cells (Fenizia et al., 2021). Remarkably, CD147 upregulation has been documented in neurons, axons, and capillaries of Alzheimer’s disease brain tissues (Nahalkova et al., 2010). These data configure CD147 as a potential target at the crossroad of SARS-CoV-2 infection and related neurological abnormalities, which remains to be confirmed in other neuronal models.
S1P is a bioactive sphingolipid with pleiotropic functions in many tissues, including the NS, where it regulates neurogenesis and inflammation through the S1P/S1PR axis (Meacci et al., 2020). For instance, accumulation of S1P in the brain promotes glial cell activation, TRL4 upregulation, and IL-6 secretion (Dusaban et al., 2017; O’Sullivan et al., 2018). In SARS-CoV-2 infection the study of the role of S1P at peripheral level has led to contradictory results (Marfia et al., 2021; Torretta et al., 2021). Remarkably, Fingolimod (FTY720), an approved drug for the treatment of multiple sclerosis (MS), which promotes the irreversible internalization and degradation of bound S1PR, has also been tested for COVID-19 treatment, in the light of its potential ability to inhibit the cytokine storm (Meacci et al., 2020). Thus, the observed S1PR1 upregulation might play an important role in SARS-CoV-2 infection in MNs, possibly by fostering pro-inflammatory cytokine release.
Overall, these data are in line with those profiled in other SARS-CoV-2 infected neuronal cell models (Kathuria et al., 2020; Gugliandolo et al., 2021; Samudyata et al., 2021; Valeri et al., 2021), suggesting an embedded mess-up of the transcriptional machinery affecting the cellular equilibrium. Notwithstanding, it appears rather difficult to pinpoint a shared and precise expression profile fostered by SARS-CoV-2 infection in this motor neuronal cell model.
The aforementioned lack of the immune system, a certain level of heterogeneity in cell differentiation, and the restricted number of enrolled subjects represent limiting factors in the interpretation of the results. Last but not least, the lack of an in vivo blood-CNS barrier represents a major inherent limitation of the present model. However, we believe that the present results contribute to adding to previous evidence that supports the ability of SARS-CoV-2 to affect different neuronal populations after entering the CNS parenchyma via direct or indirect BBB impairment (Pellegrini et al., 2020; Song et al., 2021; Lyoo et al., 2022; Stein et al., 2022; Kettunen et al., 2023). Indeed, our data are the first to document the susceptibility of iPSC-MNs to productive SARS-CoV-2 infection and the ensuing alteration of their homeostasis. Yet, the neuroinvasive routes of SARS-CoV-2 entry within the gray matter of the spinal cord specifically, remain to be investigated by employing in vivo models. Remarkably, during the revision of the present paper, a preprint was published on bioRxiv, documenting that SARS-CoV-2 can productively infect spinal cord neurons in mice models (Joyce et al., 2022). The study suggest that SARS-CoV-2 infection of spinal cord motor neurons might occur following virus spread from brainstem neurons. It also detected productive infection in sensory ganglionic neurons of the PNS (Joyce et al., 2022), suggesting the existence of an alternative, retrograde axonal pathway that fosters trans-synaptic viral spread from periphery to sensory ganglionic neurons, interneurons and motor neurons. Coupled with our results, these data suggest that SARS-CoV-2 is able to establish productive infection in unassessed nervous system sites, potentially causing sensory and neuromuscular symptoms associated with COVID-19 (Jacob et al., 2022). Further research is needed to understand how changes in motor neurons might contribute to the acute and long term neurologic sequelae. These findings could help uncover the biological basis of neuromuscular disorders and identify new therapeutic targets to counteract the neurological symptomatology.
Consent for publication
All authors give consent for the publication of the manuscript.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the Local Ethics Committee (approval number 2022_03_15_12). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
GC: Formal analysis, Investigation, Methodology, Writing – original draft. CC: Formal analysis, Investigation, Methodology, Writing – original draft. FL: Formal analysis, Methodology, Writing – original draft, Conceptualization, Data curation. SI: Methodology, Data curation, Writing – original draft. MG: Methodology, Writing – original draft, Formal analysis, Investigation. CV: Formal analysis, Investigation, Writing – original draft, Data curation. CM: Investigation, Writing – original draft, Methodology. SS: Investigation, Writing – original draft, Data curation. SZ: Data curation, Investigation, Writing – original draft, Methodology. DT: Writing – original draft, Conceptualization. VS: Funding acquisition, Writing – review and editing. MC: Funding acquisition, Writing – review and editing. AR: Writing – review and editing. MB: Writing – review and editing, Conceptualization, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Partially supported by grants from Fondazione Alessandro e Vincenzo Negroni Prati Morosini and Fondazione Romeo and Enrica Invernizzi, by EU funding within the Next Generation EU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT) and by the Italian Ministry of Health. SS was recipient of a fellowship from the Ph.D. program in Experimental Medicine of the University of Milan, Milan.
Acknowledgments
We acknowledge Dr. Patrizia Bossolasco for her expertise in iPSC generation. We also acknowledge the support of the APC central fund of the University of Milan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2023.1285836/full#supplementary-material
Footnotes
References
Abu-Rumeileh, S., Abdelhak, A., Foschi, M., Tumani, H., and Otto, M. (2021). Guillain–Barré syndrome spectrum associated with COVID-19: an up-to-date systematic review of 73 cases. J. Neurol. 268, 1133–1170. doi: 10.1007/s00415-020-10124-x
Alberti, P., Beretta, S., Piatti, M., Karantzoulis, A., Piatti, M., Santoro, P., et al. (2020). Guillain-Barré syndrome related to COVID-19 infection. Neurol. Neuroimmunol. Neuroinflamm. 7, e741. doi: 10.5005/jp-journals-10071-23723
Anand, H., Ende, V., Singh, G., Qureshi, I., Duong, T., and Mehler, M. (2021). Nervous system-systemic crosstalk in SARS-CoV-2/COVID-19: A unique dyshomeostasis syndrome. Front. Neurosci. 15:727060. doi: 10.3389/fnins.2021.727060
Andalib, S., Biller, J., Di Napoli, M., Moghimi, N., McCullough, L., Rubinos, C., et al. (2021). Peripheral nervous system manifestations associated with COVID-19. Curr. Neurol. Neurosci. Rep. 21:9.
Bardelli, D., Sassone, F., Colombrita, C., Volpe, C., Gumina, V., Peverelli, S., et al. (2020). Reprogramming fibroblasts and peripheral blood cells from a C9ORF72 patient: A proof-of-principle study. J. Cell Mol. Med. 24, 4051–4060. doi: 10.1111/jcmm.15048
Bizzotto, J., Sanchis, P., Abbate, M., Lage-Vickers, S., Lavignolle, R., Toro, A., et al. (2020). SARS-CoV-2 Infection Boosts MX1 Antiviral Effector in COVID-19 Patients. iScience 23:101585. doi: 10.1016/j.isci.2020.101585
Bossolasco, P., Sassone, F., Gumina, V., Peverelli, S., Garzo, M., and Silani, V. (2018). Motor neuron differentiation of iPSCs obtained from peripheral blood of a mutant TARDBP ALS patient. Stem Cell Res. 30, 61–68. doi: 10.1016/j.scr.2018.05.009
Cantuti-Castelvetri, L., Ojha, R., Pedro, L., Djannatian, M., Franz, J., Kuivanen, S., et al. (2020). Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860.
Caress, J., Castoro, R., Simmons, Z., Scelsa, S., Lewis, R., Ahlawat, A., et al. (2020). COVID-19–ASSOCIATED GUILLAIN-BARRÉ syndrome: The early pandemic experience. Muscle Nerve 62, 485–491.
Carfì, A., Bernabei, R., Landi, F., and Gemelli Against COVID-19 Post-Acute Care Study Group. (2020). Persistent symptoms in patients after acute COVID-19. JAMA 324, 603–605.
Chaudhry, M., Casalegno-Garduno, R., Sitnik, K., Kasmapour, B., Pulm, A., Brizic, I., et al. (2020). Cytomegalovirus inhibition of extrinsic apoptosis determines fitness and resistance to cytotoxic CD8 T cells. Proc. Natl. Acad. Sci. U. S. A. 117, 12961–12968.
Chen, T., Wu, D., Chen, H., Yan, W., Yang, D., Chen, G., et al. (2020). Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ 368, m1091.
Chou, S., Beghi, E., Helbok, R., Moro, E., Sampson, J., Altamirano, V., et al. (2021). Global Incidence of Neurological Manifestations Among Patients Hospitalized With COVID-19—A Report for the GCS-NeuroCOVID Consortium and the ENERGY Consortium. JAMA Netw. Open 4, e2112131. doi: 10.1001/jamanetworkopen.2021.12131
Coomes, E., and Haghbayan, H. (2020). Interleukin-6 in Covid-19: A systematic review and meta-analysis. Rev. Med. Virol. 30, 1–9.
Coutard, B., Valle, C., de Lamballerie, X., Canard, B., Seidah, N., and Decroly, E. (2020). The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 176:104742. doi: 10.1016/j.antiviral.2020.104742
Daly, J., Simonetti, B., Klein, K., Chen, K., Williamson, M., Antón-Plágaro, C., et al. (2020). Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370, 861–865.
Davis, H., Assaf, G., McCorkell, L., Wei, H., Low, R., Re’em, Y., et al. (2021). Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. eClinicalMedicine 38:101019. doi: 10.1016/j.eclinm.2021.101019
Dusaban, S., Chun, J., Rosen, H., Purcell, N., and Brown, J. (2017). Sphingosine 1-phosphate receptor 3 and RhoA signaling mediate inflammatory gene expression in astrocytes. J. Neuroinflammation 14:111.
Fenizia, C., Biasin, M., Cetin, I., Vergani, P., Mileto, D., Spinillo, A., et al. (2020). Analysis of SARS-CoV-2 vertical transmission during pregnancy. Nat. Commun. 11:5128.
Fenizia, C., Galbiati, S., Vanetti, C., Vago, R., Clerici, M., Tacchetti, C., et al. (2021). SARS-CoV-2 Entry: At the Crossroads of CD147 and ACE2. Cells 10:1434.
Fenizia, C., Galbiati, S., Vanetti, C., Vago, R., Clerici, M., Tacchetti, C., et al. (2022). Cyclosporine A Inhibits Viral Infection and Release as Well as Cytokine Production in Lung Cells by Three SARS-CoV-2 Variants. Microbiol. Spectr. 10, e0150421. doi: 10.1128/spectrum.01504-21
Ferrucci, R., Dini, M., Rosci, C., Capozza, A., Groppo, E., Reitano, M., et al. (2022). One-year cognitive follow-up of COVID-19 hospitalized patients. Euro. J. Neurol. 29, 2006–2014.
Fu, Y., Xu, H., and Liu, S. J. (2022). COVID-19 and neurodegenerative diseases. Eur. Rev. Med. Pharmacol. Sci. 26, 4535–4544.
Garrigues, E., Janvier, P., Kherabi, Y., Le Bot, A., Hamon, A., Gouze, H., et al. (2020). Post-discharge persistent symptoms and health-related quality of life after hospitalization for COVID-19. J. Infect. 81, e4–e6.
Gugliandolo, A., Chiricosta, L., Calcaterra, V., Biasin, M., Cappelletti, G., Carelli, S., et al. (2021). SARS-CoV-2 infected pediatric cerebral cortical neurons: Transcriptomic analysis and potential role of toll-like receptors in pathogenesis. IJMS 22:8059. doi: 10.3390/ijms22158059
Gumina, V., Colombrita, C., Fallini, C., Bossolasco, P., Maraschi, A., Landers, J., et al. (2019). TDP-43 and NOVA-1 RNA-binding proteins as competitive splicing regulators of the schizophrenia-associated TNIK gene. Biochim. Biophys. Acta Gene Regul. Mech. 1862:194413. doi: 10.1016/j.bbagrm.2019.194413
Haller, O., Arnheiter, H., Pavlovic, J., and Staeheli, P. (2018). The discovery of the antiviral resistance gene Mx: A story of great ideas, great failures, and some success. Annu. Rev. Virol. 5, 33–51.
Haller, O., and Kochs, G. (2020). Mx genes: host determinants controlling influenza virus infection and trans-species transmission. Hum. Genet. 139, 695–705. doi: 10.1007/s00439-019-02092-8
Hamming, I., Timens, W., Bulthuis, M., Lely, A., Navis, G., and van Goor, H. (2004). Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637. doi: 10.1002/path.1570
Hooper, L., Stappenbeck, T., Hong, C., and Gordon, J. (2003). Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4, 269–273. doi: 10.1038/ni888
Huang, C., Huang, L., Wang, Y., Li, X., Ren, L., Gu, X., et al. (2021). 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet 397, 220–232.
Jacob, S., Kapadia, R., Soule, T., Luo, H., Schellenberg, K., Douville, R., et al. (2022). Neuromuscular Complications of SARS-CoV-2 and Other Viral Infections. Front. Neurol. 13:914411. doi: 10.3389/fneur.2022.914411
Jaywant, A., Vanderlind, W., Alexopoulos, G., Fridman, C., Perlis, R., and Gunning, F. (2021). Frequency and profile of objective cognitive deficits in hospitalized patients recovering from COVID-19. Neuropsychopharmacology 46, 2235–2240. doi: 10.1038/s41386-021-00978-8
Jonjić, S., Babić, M., Polić, B., and Krmpotić, A. (2008). Immune evasion of natural killer cells by viruses. Curr. Opin. Immunol. 20, 30–38.
Joyce, J., Moore, G., Goswami, P., Harrell, T., Taylor, T., Hawks, S., et al. (2022). SARS-CoV-2 infects peripheral and central neurons before viremia, facilitated by neuropilin-1. bioRxiv [Preprint]. doi: 10.1101/2022.05.20.492834
Kathuria, A., Lopez-Lengowski, K., Watmuff, B., and Karmacharya, R. (2020). Comparative transcriptomic analysis of cerebral organoids and cortical neuron cultures derived from human induced pluripotent stem cells. Stem Cells Dev. 29, 1370–1381.
Kettunen, P., Lesnikova, A., Räsänen, N., Ojha, R., Palmunen, L., Laakso, M., et al. (2023). SARS-CoV-2 infection of human neurons is TMPRSS2 independent, requires endosomal cell entry, and can be blocked by inhibitors of host phosphoinositol-5 kinase. J. Virol. 97, e0014423. doi: 10.1128/jvi.00144-23
Kim, J., Heo, J., Kim, H., Song, S. H., Park, S., Park, T., et al. (2017). Neurological complications during treatment of middle east respiratory syndrome. J. Clin. Neurol. 13:227.
Klein, R., Soung, A., Sissoko, C., Nordvig, A., Canoll, P., Mariani, M., et al. (2021). COVID-19 induces neuroinflammation and loss of hippocampal neurogenesis [Internet]. Res. Sq. 3:1031824.
Koralnik, I., and Tyler, K. (2020). COVID-19: A global threat to the nervous system. Ann. Neurol. 88, 1–11. doi: 10.1002/ana.25807
Kummer, K., Zeidler, M., Kalpachidou, T., and Kress, M. (2021). Role of IL-6 in the regulation of neuronal development, survival and function. Cytokine 144:155582.
Li, S., Zhang, Y., Guan, Z., Li, H., Ye, M., Chen, X., et al. (2020). SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Sig. Transduct. Target Ther. 5:235.
Li, Y., Lu, S., Gu, J., Xia, W., Zhang, S., Zhang, S., et al. (2022). SARS-CoV-2 impairs the disassembly of stress granules and promotes ALS-associated amyloid aggregation. Protein Cell 13, 602–614. doi: 10.1007/s13238-022-00905-7
Limanaqi, F., Zecchini, S., Dino, B., Strizzi, S., Cappelletti, G., Utyro, O., et al. (2022). Dopamine reduces SARS-CoV-2 replication in vitro through downregulation of D2 receptors and upregulation of type-i interferons. Cells 11:1691. doi: 10.3390/cells11101691
Lopez, G., Tonello, C., Osipova, G., Carsana, L., Biasin, M., Cappelletti, G., et al. (2022). Olfactory bulb SARS-CoV-2 infection is not paralleled by the presence of virus in other central nervous system areas. Neuropathol. Appl. Neurobiol. 48:12752.
Lou, J., Movassaghi, M., Gordy, D., Olson, M., Zhang, T., Khurana, M., et al. (2021). Neuropathology of COVID-19 (neuro-COVID): clinicopathological update. Free Neuropathol. 2:2. doi: 10.17879/freeneuropathology-2021-2993
Lyons-Weiler, J. (2020). Pathogenic priming likely contributes to serious and critical illness and mortality in COVID-19 via autoimmunity. J. Transl. Autoimmunity 3:100051. doi: 10.1016/j.jtauto.2020.100051
Lyoo, K., Kim, H., Lee, B., Che, Y., Kim, S., Song, D., et al. (2022). Direct neuronal infection of SARS-CoV-2 reveals cellular and molecular pathology of chemosensory impairment of COVID-19 patients. Emerg. Microb. Infect. 11, 407–412. doi: 10.1080/22221751.2021.2024095
Mancini, M., and Vidal, S. (2020). Mechanisms of Natural Killer Cell Evasion Through Viral Adaptation. Annu. Rev. Immunol. 38, 511–539.
Mao, L., Jin, H., Wang, M., Hu, Y., Chen, S., He, Q., et al. (2020). Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 77:683.
Marfia, G., Navone, S., Guarnaccia, L., Campanella, R., Mondoni, M., Locatelli, M., et al. (2021). Decreased serum level of sphingosine-1-phosphate: a novel predictor of clinical severity in COVID-19. EMBO Mol. Med. 13, e13424. doi: 10.15252/emmm.202013424
Meacci, E., Garcia-Gil, M., and Pierucci, F. (2020). SARS-CoV-2 Infection: A role for S1P/S1P receptor signaling in the nervous system? Int. J. Mol. Sci. 21:6773. doi: 10.3390/ijms21186773
Méndez, R., Balanzá-Martínez, V., Luperdi, S., Estrada, I., Latorre, A., and González-Jiménez, P. (2021). Short-term neuropsychiatric outcomes and quality of life in COVID-19 survivors. J Intern. Med. 290, 621–631. doi: 10.1111/joim.13262
Montalvan, V., Lee, J., Bueso, T., De Toledo, J., and Rivas, K. (2020). Neurological manifestations of COVID-19 and other coronavirus infections: A systematic review. Clin. Neurol. Neurosurg. 194:105921.
Muhammed, L., Baheerathan, A., Cao, M., Leite, M., and Viegas, S. (2021). MuSK Antibody–Associated Myasthenia Gravis With SARS-CoV-2 Infection: A Case Report. Ann. Intern. Med. 174, 872–873. doi: 10.7326/L20-1298
Nahalkova, J., Volkmann, I., Aoki, M., Winblad, B., Bogdanovic, N., Tjernberg, L., et al. (2010). CD147, a γ-secretase associated protein is upregulated in Alzheimer’s disease brain and its cellular trafficking is affected by presenilin-2. Neurochem. Int. 56, 67–76. doi: 10.1016/j.neuint.2009.09.003
Nalbandian, A., Sehgal, K., Gupta, A., Madhavan, M., McGroder, C., Stevens, J., et al. (2021). Post-acute COVID-19 syndrome. Nat. Med. 27, 601–615.
Nampoothiri, S., Sauve, F., Ternier, G., Fernandois, D., Coelho, C., Imbernon, M., et al. (2020). The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis. bioRxiv [Preprint]. doi: 10.1101/2020.06.08.139329
Nardo, G., Trolese, M., and Bendotti, C. (2016). Major Histocompatibility Complex I Expression by Motor Neurons and Its Implication in Amyotrophic Lateral Sclerosis. Front. Neurol. 13:89. doi: 10.3389/fneur.2016.00089
Needham, E., Newcombe, V., Michell, A., Thornton, R., Grainger, A., Anwar, F., et al. (2020). Mononeuritis multiplex: an unexpectedly common feature of severe COVID-19. medRxiv [Preprint]. doi: 10.1101/2020.07.19.20149898
Niazkar, H., Zibaee, B., Nasimi, A., and Bahri, N. (2020). The neurological manifestations of COVID-19: a review article. Neurol. Sci. 41, 1667–1671.
Noschka, R., Gerbl, F., Löffler, F., Kubis, J., Rodríguez, A., Mayer, D., et al. (2021). Unbiased identification of angiogenin as an endogenous antimicrobial protein with activity against virulent mycobacterium tuberculosis. Front. Microbiol. 11:618278. doi: 10.3389/fmicb.2020.618278
O’Sullivan, S., O’Sullivan, C., Healy, L., Dev, K., and Sheridan, G. (2018). Sphingosine 1-phosphate receptors regulate TLR4-induced CXCL5 release from astrocytes and microglia. J. Neurochem. 144, 736–747. doi: 10.1111/jnc.14313
Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11:1620.
Pellegrini, L., Albecka, A., Mallery, D., Kellner, M., Paul, D., Carter, A., et al. (2020). SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell 27, 951–961. doi: 10.1016/j.stem.2020.10.001
Pezzini, A., and Padovani, A. (2020). Lifting the mask on neurological manifestations of COVID-19. Nat. Rev. Neurol. 16, 636–644.
Pilotto, A., Cristillo, V., Cotti Piccinelli, S., Zoppi, N., Bonzi, G., Sattin, D., et al. (2021). Long-term neurological manifestations of COVID-19: prevalence and predictive factors. Neurol. Sci. 42, 4903–4907.
Piras, M., Cau, F., Manchia, M., Paribello, P., Saba, L., Suri, J., et al. (2022). Strong ACE-2 expression in the choroidal vessels: do high choroid plexuses serve as a gateway for SARS-CoV-2 infection on the human brain? Eur. Rev. Med. Pharmacol. Sci. 26, 3025–3029. doi: 10.26355/eurrev_202204_28633
Pleguezuelos, E., Del Carmen, A., Llorensi, G., Carcole, J., Casarramona, P., Moreno, E., et al. (2021). Severe loss of mechanical efficiency in COVID-19 patients. J. Cach. Sarcop. Muscle 12, 1056–1063.
Qiao, J., Li, W., Bao, J., Peng, Q., Wen, D., Wang, J., et al. (2020). The expression of SARS-CoV-2 receptor ACE2 and CD147, and protease TMPRSS2 in human and mouse brain cells and mouse brain tissues. Biochem. Biophys. Res. Commun. 533, 867–871. doi: 10.1016/j.bbrc.2020.09.042
Raahimi, M., Kane, A., Moore, C., and Alareed, A. (2021). Late onset of Guillain-Barré syndrome following SARS-CoV-2 infection: part of ‘long COVID-19 syndrome’? BMJ Case Rep. 14, e240178.
Ramani, A., Müller, L., Ostermann, P., Gabriel, E., Abida-Islam, P., Müller-Schiffmann, A., et al. (2020). SARS -CoV-2 targets neurons of 3D human brain organoids. EMBO J. 39:6230.
Restivo, D., Centonze, D., Alesina, A., and Marchese-Ragona, R. (2020). Myasthenia Gravis Associated With SARS-CoV-2 Infection. Ann. Intern. Med. 173, 1027–1028.
Samudyata, S., Oliveira, A., Malwade, S., de Sousa, N., Goparaju, S., Orhan, F., et al. (2021). SARS-CoV-2 neurotropism and single cell responses in brain organoids containing innately developing microglia. bioRxiv [Preprint]. doi: 10.1101/2021.07.07.451463
Serrano-Castro, P., Estivill-Torrús, G., Cabezudo-García, P., Reyes-Bueno, J., Ciano Petersen, N., Aguilar-Castillo, M., et al. (2020). Impact of SARS-CoV-2 infection on neurodegenerative and neuropsychiatric diseases: A delayed pandemic? Neurología 35, 245–251. doi: 10.1016/j.nrl.2020.04.002
Sironi, M., Biasin, M., Cagliani, R., Gnudi, F., Saulle, I., Ibba, S., et al. (2014). Evolutionary Analysis Identifies an MX2 Haplotype Associated with Natural Resistance to HIV-1 Infection. Mol. Biol. Evol. 31, 2402–2414. doi: 10.1093/molbev/msu193
Soares, M., Eggelbusch, M., Naddaf, E., Gerrits, K., Schaaf, M., Borst, B., et al. (2022). Skeletal muscle alterations in patients with acute Covid-19 and post-acute sequelae of Covid-19. J. Cach. Sarcop. Muscle 13, 11–22.
Song, E., Zhang, C., Israelow, B., Lu-Culligan, A., Prado, A., Skriabine, S., et al. (2021). Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 218, e20202135.
Stein, S., Ramelli, S., Grazioli, A., Chung, J., Singh, M., Yinda, C., et al. (2022). SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 612, 758–763. doi: 10.1038/s41586-022-05542-y
Stępień, J., and Pastuszak, Ż (2023). Electroneurological changes in peripheral nerves in patients post-COVID. J. Neurophysiol. 129, 392–398. doi: 10.1152/jn.00396.2022
Taguchi, M., Bonner, K., and Memon, A. (2022). Late-Onset Guillain-Barré Syndrome and Right Facial Nerve Palsy after COVID-19 Infection. Case Rep. Neurol. 14, 12–18. doi: 10.1159/000521245
Tan, Y., Uchida, K., Nakajima, H., Guerrero, A., Watanabe, S., Hirai, T., et al. (2013). Blockade of Interleukin 6 Signaling Improves the Survival Rate of Transplanted Bone Marrow Stromal Cells and Increases Locomotor Function in Mice With Spinal Cord Injury. J. Neuropathol. Exp. Neurol. 72, 980–993. doi: 10.1097/NEN.0b013e3182a79de9
Thiyagarajan, N., Ferguson, R., Subramanian, V., and Acharya, K. (2012). Structural and molecular insights into the mechanism of action of human angiogenin-ALS variants in neurons. Nat. Commun. 3:1121. doi: 10.1038/ncomms2126
Tomasoni, D., Bai, F., Castoldi, R., Barbanotti, D., Falcinella, C., Mulè, G., et al. (2021). Anxiety and depression symptoms after virological clearance of COVID-19: A cross-sectional study in Milan, Italy. J. Med. Virol. 93, 1175–1179. doi: 10.1002/jmv.26459
Torretta, E., Garziano, M., Poliseno, M., Capitanio, D., Biasin, M., Santantonio, T., et al. (2021). Severity of COVID-19 patients predicted by serum sphingolipids signature. Int. J. Mol. Sci. 22:10198. doi: 10.3390/ijms221910198
Valeri, A., Chiricosta, L., Calcaterra, V., Biasin, M., Cappelletti, G., Carelli, S., et al. (2021). Transcriptomic analysis of HCN-2 cells suggests connection among oxidative stress, senescence, and neuron death after SARS-CoV-2 infection. Cells 10:2189. doi: 10.3390/cells10092189
Yoo, J., Sasaki, M., Cho, S., Kasuga, Y., Zhu, B., Ouda, R., et al. (2021). SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 12:6602. doi: 10.1038/s41467-021-26910-8
Zhang, B., Chu, H., Han, S., Shuai, H., Deng, J., Hu, Y., et al. (2020). SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 30, 928–931.
Zhao, H., Shen, D., Zhou, H., Liu, J., and Chen, S. (2020). Guillain-Barré syndrome associated with SARS-CoV-2 infection: causality or coincidence? Lancet Neurol. 19, 383–384.
Keywords: SARS-CoV-2 infection, iPSC-derived motor neurons, long-COVID, neuroinflammation, neuromuscular disorders, COVID-19
Citation: Cappelletti G, Colombrita C, Limanaqi F, Invernizzi S, Garziano M, Vanetti C, Moscheni C, Santangelo S, Zecchini S, Trabattoni D, Silani V, Clerici M, Ratti A and Biasin M (2023) Human motor neurons derived from induced pluripotent stem cells are susceptible to SARS-CoV-2 infection. Front. Cell. Neurosci. 17:1285836. doi: 10.3389/fncel.2023.1285836
Received: 30 August 2023; Accepted: 17 November 2023;
Published: 05 December 2023.
Edited by:
Sara Salinas, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Larisa Ryskalin, University of Pisa, ItalyLara Gibellini, University of Modena and Reggio Emilia, Italy
Copyright © 2023 Cappelletti, Colombrita, Limanaqi, Invernizzi, Garziano, Vanetti, Moscheni, Santangelo, Zecchini, Trabattoni, Silani, Clerici, Ratti and Biasin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mara Biasin, bWFyYS5iaWFzaW5AdW5pbWkuaXQ=
†These authors share senior authorship
 Gioia Cappelletti
Gioia Cappelletti Claudia Colombrita2
Claudia Colombrita2 Fiona Limanaqi
Fiona Limanaqi Sabrina Invernizzi
Sabrina Invernizzi Micaela Garziano
Micaela Garziano Claudia Vanetti
Claudia Vanetti Claudia Moscheni
Claudia Moscheni Daria Trabattoni
Daria Trabattoni Vincenzo Silani
Vincenzo Silani Mario Clerici
Mario Clerici Antonia Ratti
Antonia Ratti Mara Biasin
Mara Biasin