- Department of Cell Biology and Imaging, Institute of Zoology and Biomedical Research, Jagiellonian University, Cracow, Poland
Heme oxygenase (HO) has been shown to control various cellular processes in both mammals and Drosophila melanogaster. Here, we investigated how changes in HO levels in neurons and glial cells during development affect adult flies, by using the TARGET Drosophila system to manipulate the expression of the ho gene. The obtained data showed differences in adult survival, maximum lifespan, climbing, locomotor activity, and sleep, which depended on the level of HO (after ho up-regulation or downregulation), the timing of expression (chronic or at specific developmental stages), cell types (neurons or glia), sex (males or females), and age of flies. In addition to ho, the effects of changing the mRNA level of the Drosophila CNC factor gene (NRF2 homolog in mammals and master regulator of HO), were also examined to compare with those observed after changing ho expression. We showed that HO levels in neurons and glia must be maintained at an appropriate physiological level during development to ensure the well-being of adults. We also found that the downregulation of ho in either neurons or glia in the brain is compensated by ho expressed in the retina.
1. Introduction
Heme oxygenase (HO) maintains and modulates a broad spectrum of processes and among them, the most important ones are the degradation of intracellular heme and the subsequent generation of the products: carbon monoxide (CO), ferrous ion (Fe2+), and biliverdin (BV). Heme homeostasis is crucial for controlling cellular stress and processes modulated or dependent on it (Duvigneau et al., 2019).
In mammals, two active isoforms of HO have been identified: inducible HO-1 and constitutive HO-2, which differ in tissue distribution and function. The expression of HO-1 is detectable in most tissues, but it is exceptionally high in the spleen and liver. In contrast, HO-2 is abundant in the brain and testes and it is vital for their physiological functions (Ewing and Maines, 1997; Zakhary et al., 1997; Boehning and Snyder, 2003; Adachi et al., 2004; Alam et al., 2004; Mancuso, 2004; Andoh et al., 2006; Shibahara et al., 2007). The importance of HO-1 was demonstrated in the study using HO-1 knockout (HO-1-null) mice, which revealed its central role in development and iron homeostasis (Poss and Tonegawa, 1997a,b; Zhao et al., 2009; Kovtunovych et al., 2010). In humans, HO-1 deficiency results in early death, severe growth retardation, and multiple other disorders (Yachie et al., 1999; Kawashima et al., 2002; Radhakrishnan et al., 2011), confirming what has been found in animal studies.
In Drosophila melanogaster, the HO-1 homolog (dHO) is encoded only by a single gene (ho). HO in Drosophila has a unique structure and slower activity rate than mammalian and bacterial HOs (Zhang et al., 2004). It controls viability, development, iron accumulation, cell death (Cui et al., 2008), DNA damage signaling (Ida et al., 2013), protection against DNA damage caused by UV and white light (Damulewicz et al., 2017a,b), phototransduction, DNA repair, immune responses (Damulewicz et al., 2019), and circadian clock (Mandilaras and Missirlis, 2012; Klemz et al., 2017; Damulewicz et al., 2017a, 2019). The expression of ho is controlled by the circadian clock (Ceriani et al., 2002; Damulewicz et al., 2017a; Abaquita et al., 2021) and induced by many factors (Damulewicz et al., 2017b; Abaquita et al., 2021). Several processes, such as autophagy and apoptosis, have been reported to be associated and regulated by changes in the level of ho mRNA (Damulewicz et al., 2019; Abaquita et al., 2021, 2023). In all these studies Drosophila transgenic lines were used to induce higher or lower ho mRNA levels to identify HO biological functions. The genetic modifications employed in earlier studies were chronic, however, which means that changes in the ho expression were activated during embryonic development and maintained until adulthood. It has not been examined how modifications in ho expression at different developmental stages affect the life parameters of Drosophila adults.
Heme, the HO target for degradation, is a vital component for insect molting and metamorphosis. It is utilized as the prosthetic group of hemoproteins, including P450 enzymes involved in the synthesis of ecdysone and juvenile hormone (Feyereisen, 1999; Braz et al., 2001; Warren et al., 2002; Namiki et al., 2005; Rewitz et al., 2006; Chung et al., 2009; Iga and Kataoka, 2012; Sztal et al., 2012). Excessive heme degradation by sustained upregulation of HO in an anti-oxidative defense strategy may be detrimental to heme regulatory functions (Kimpara et al., 2000; Atamna et al., 2002; Zhu et al., 2002; Atamna, 2004; Sengupta et al., 2005; Smith et al., 2011). On the other hand, downregulation of ho can result in heme overload, which eventually promotes its abnormal binding and oxidative stress (Schmitt et al., 1993; Yoshida and Migita, 2000; Kikuchi et al., 2005; Kumar and Bandyopadhyay, 2005; Yang et al., 2008; Guenthner et al., 2009; Vallelian et al., 2015; Klemz et al., 2017; Chiabrando et al., 2018). Moreover, the dysfunction of heme metabolism may influence many molecular processes controlled by its activity-dependent end-products (Chiabrando et al., 2018; Duvigneau et al., 2019; Luu Hoang et al., 2021; Yang and Wang, 2022). Interestingly, the CNC factor, the Drosophila counterpart of the nuclear erythroid factor 2-related factor 2 (NRF2) that regulates HO-1, also plays an important role in the fly’s growth, development, and aging (Loboda et al., 2016).
In the nervous system, heme controls a variety of biological processes like energy production (Padmanaban et al., 1989; Sabová et al., 1993; Giraud et al., 1998; Mense and Zhang, 2006; Azuma et al., 2008; Kim et al., 2012), ion channels activity, gene expression, and miRNA processing (Smith et al., 2011). These processes are crucial for neuronal survival and differentiation (see Chiabrando et al., 2018). An imbalance in heme synthesis and degradation can mediate oxidative stress (Kagan et al., 2001; Klouche et al., 2004; Kumar and Bandyopadhyay, 2005; Schaer et al., 2013; Chiabrando et al., 2014; Wegiel et al., 2015), proteostasis failure (see Chiabrando et al., 2018), or mitochondrial decay and degeneration (Atamna, 2004). Increasing data show that both HO deficiency (see Mena et al., 2015) and overexpression (see Schipper et al., 1995, 2006; Song et al., 2007; Schipper et al., 2009; Ohnishi et al., 2010; Tronel et al., 2013; Barone et al., 2014; Chiang et al., 2018; Nitti et al., 2018; Waza et al., 2018; Schipper et al., 2019; Luu Hoang et al., 2021; Wu and Hsieh, 2022; Yang and Wang, 2022) are associated with neurodegenerative disorders. The brain is, therefore, at risk when HO is either at its high or low level. Moreover, there is no data about the effect of age-dependent (or stage-dependent) function of HO. In the present study we examined how manipulations of ho mRNA level at specific developmental stage affect adult flies. Knowing how HO is important for maintaining brain functions, we targeted separately neurons and glia, which play different functions in the brain and interact with each other.
We used the temporal and regional gene expression targeting (TARGET) system to manipulate gene expression spatially and temporally, and to do that a temperature-sensitive (ts) variant of the GAL80 protein was added to GAL4/UAS system. The GAL80ts molecule represses GAL4 at low temperatures, while at higher temperature GAL80ts is inactivated, which allows GAL4 to bind to UAS and to begin transcription of the target gene (McGuire et al., 2004). Here, the mRNA level of ho or cnc in the brain was modified chronically without GAL80ts or modified at specific developmental stages (either during larval, pupal, or adult life), using the tubulin promoter-GAL80ts transgene, and the longevity, fitness, and sleeping pattern of adult flies were examined.
The obtained results showed that changes in ho or cnc expression in the brain at different developmental stages adversely affect adult life compared to chronic expression. The influence of ho or cnc on the life expectancy, climbing performance, activity, and sleep pattern of Drosophila adults vary according to their expression level (upregulation or downregulation), cell types (neurons or glia), sex (males or females), and age of adult flies.
2. Materials and methods
2.1. Animals
The following Drosophila strains were used in the study: repo-Gal4 (pan-glial cell marker, BDRC no. 7415), elav-Gal4 (pan-neuronal cell marker), tubGal80ts; repo-Gal4, tubGal80ts; elav-Gal4, UAS-ho (Cui et al., 2008), UAS-hoRNAi (Cui et al., 2008), UAS-cnc, UAS-cncRNAi, UAS-GFP.Valium10 (BDRC no. 35786), and wild type (CS). The strains UAS-ho and UAS-hoRNAi were kindly provided by Dr. Taketani (Kyoto Institute of Technology, Japan). Constructs are inserted on the 3rd chromosome in w1118 background.
To study the effect of overexpression/silencing of ho or cnc at different developmental stages, flies crossed with strains containing tubGal80ts were kept at different temperature conditions: (i) larva-specific, at 29°C from embryo to pupariation and then at 18°C; (ii) pupa-specific, at 18°C from embryo to pupariation followed by 29°C during the pupal stage and at 18°C after eclosion; (iii) adult-specific, at 18°C from embryo until eclosion and then at 29°C. For chronic overexpression/silencing, flies without tubGal80ts were kept constantly at 25°C. Control flies, both parental strains backcrossed to wild-type flies, were kept in the same temperature conditions as the experimental flies.
2.2. Survival assay
After eclosion, flies were separated according to sex and kept in groups of 30 individuals in specific temperature conditions. Every day the number of dead flies was counted, and every 3 days, flies were transferred to the new vials with fresh food. The experiment continued until the death of the last fly in the vial.
2.3. Climbing assay (negative geotaxis test)
Males aged 7, 14, and 30 days-old were transferred into an empty vial without using CO2 to avoid effects of this type of anaesthesia on locomotion and climbing ability. After a short recovery, flies were gently tapped to the bottom of a vial, and those that climbed vertically beyond the 5 cm marked line in 15 s were counted. The experiment was carried out in a dark room with red light only, which is invisible to flies. Every trial with 30 flies was repeated three times. Every experimental group had three repetitions.
2.4. Locomotor activity and sleep
The locomotor activity was recorded using a Drosophila Activity Monitoring System (DAMS, Trikinetics). Males, 2 days-old, were anesthetized with CO2, placed on ice and then transferred into glass tubes (one fly per tube) sealed at both ends by food (agar, sugar, yeasts) and by a foam stopper. These tubes were placed in monitors (maximum 32 tubes per monitor) equipped with infrared light-emitting diodes and detectors that were connected to a computer. Whenever the fly passed the emitter/detector, the infrared beam was interrupted, transmitting a signal to the computer. To analyze the circadian period of locomotor activity, flies were maintained for 7 days under LD12:12 and next under constant darkness (DD) for 7 days. The locomotor activity was scored every 1 min. To study sleep, the activity of flies was analyzed on the second day of LD12:12 conditions. Sleep was measured as intervals of at least 5 min of inactivity.
Data were analyzed in Excel by using “Befly!” software (Department of Genetics, Leicester University). Lomb–Scargle normalized periodogram was used to determine rhythmic flies; flies with a period value lower than 10 (confidence level 0.05) were regarded as arrhythmic. Flies that did not survive until the end of each experiment were removed from analyses. Every experiment was repeated three times with at least 60 flies per group.
2.5. RNA extraction, cDNA synthesis, and qPCR
To compare ho or cnc expression between experimental groups, samples were collected at different developmental stages: the whole body of the 3rd instar larvae or pupae (5 individuals per sample, 3 repetitions), and heads of 5 days-old males and females (20 individuals per sample, 3 repetitions), separately. All samples were collected 1 h after the lights-on. We decided to examine gene expression in the whole body of larvae and pupae, and in heads of adults. In case of larvae we did not want to exclude motoneurons and glia located in the peripheral nervous system. There are about 700 neurons and 77 glial cells in each segment of the larval body (Bittern et al., 2021). Since we were not sure which cells in the central or peripheral nervous system were more affected in the experiments we decided to use whole body samples, although other tissues may have higher levels of both gene mRNA than neurons and glial cells. In case of pupae the nervous system is changed from larval to adult one and it is very difficult to dissect brains in early pupae. The experiments were carried out on wandering L3 larvae and in case of pupae in the pupal early stage when pupae changed color from white to brown (prepupae or P2 according to Bainbridge and Bownes, 1981). Total RNA was isolated using the Trizol method, purity was checked using NanoDrop and confirmed as acceptable with 260/280 and 260/230 ratios higher than 1.8. Total RNA (500 ng) was used for reverse transcription. Gene expression was measured using Kapa Sybr Green (KAPA Biosystems, Cape Town, South Africa) using specific primer sequences (ho: For 5′-ACCATTTGCCCGCCGGGATG-3′, Rev 5′-AGTGCGACGGCCAGCTTCCT-3′, gene accession no. CG14716; and cnc: For 5′-GAGGTGGAAATCGGAGATGA-3′, Rev 5′-CTGCTTGTAGAGCACCTCAGC-3′, gene accession no. CG43286) on StepOnePlus Real-Time PCR System. Rpl32 was used as a reference gene (For 5′-TATGCTAAGCTGTCGCACAAATG-3′, Rev 5′-AGCACGTGTATAAAAAGTGCCA-3′, gene accession no. CG7939). A standard curve was used to calculate gene expression levels. The number of target gene copies was normalized to the reference gene.
2.6. Statistical analysis
All data were examined for distribution normality, and statistical tests were chosen accordingly. The Kaplan–Meier curve was used to present data on the survival percentage of flies plotted against their age (in days). The survival (in %) in every phase of the aging process and the maximum lifespan between genotypes (i.e., experimental group versus control 1 and experimental group versus control 2) were analyzed using the log-rank test performed with the R/R Studio free statistical version 4.2.0.1 The non-parametric analysis of variance (ANOVA) was used to analyse climbing performance and qPCR results (Dunn’s test). For locomotor activity and sleep analysis, the parametric ANOVA was used (Dunnett’s test). Statistical analyses were performed with the GraphPad Prism 7.05 software. Detailed statistics is provided in Supplementary Tables S1, S5, S7–S9.
3. Results
3.1. Effects of changing ho or cnc mRNA level in the brain at different developmental stages on the longevity of adult flies
The role of HO or its upstream regulator CNC in the brain during development was first examined on the survival pattern and maximum lifespan of adult flies. For developmental studies, the TARGET system is commonly used, however, flies are cultured at high or low temperatures which may affect their longevity. Universally, adult flies go through three phases of aging under an optimal temperature condition: health span (4–30 days after eclosion or DAE), transition phase (31–60 DAE), and senescence phase (61–120 DAE) (Phom et al., 2014). Here, we used 25°C as the optimal temperature for culturing of flies with chronic GAL4 activation, 18°C to inhibit Gal4 system, and 29°C to inactivate Gal80ts and activate cell-specific gene expression. For transgenic groups maintained at 18°C or 29°C, we used the following descriptions to identify the DAE range for each phase: (i) health span for those DAE with no natural death, (ii) transition phase for those DAE with a slight decline in the mortality curve showing 10% death, and (iii) senescence phase for those DAE with a steady decline in mortality curve represented by the window between the end of the transition phase till maximum lifespan of the fly (Ayajuddin et al., 2022). The DAE range for each phase could change after exposure to lower or higher temperature conditions than the optimal one (Mołoń et al., 2020). Effects of disrupting the expression of ho or cnc in the brain during development on the life expectancy of adults were analysed in each phase. In general, the adult longevity of Drosophila changed when ho or cnc mRNA level was changed during development. Interestingly, we observed opposite effects between chronic modification and those targeted at specific developmental stages (Table 1; Figures 1, 2; Supplementary Table S1). Survival was mostly better in adults when ho was either overexpressed or silenced chronically in the brain. Whereas low or unaffected survival was mainly detected when ho overexpression or silencing was explicitly done in the larval, pupal, or adult stage. We also observed differences in the effects of changing ho expression level in neurons or glia during development on adult longevity.
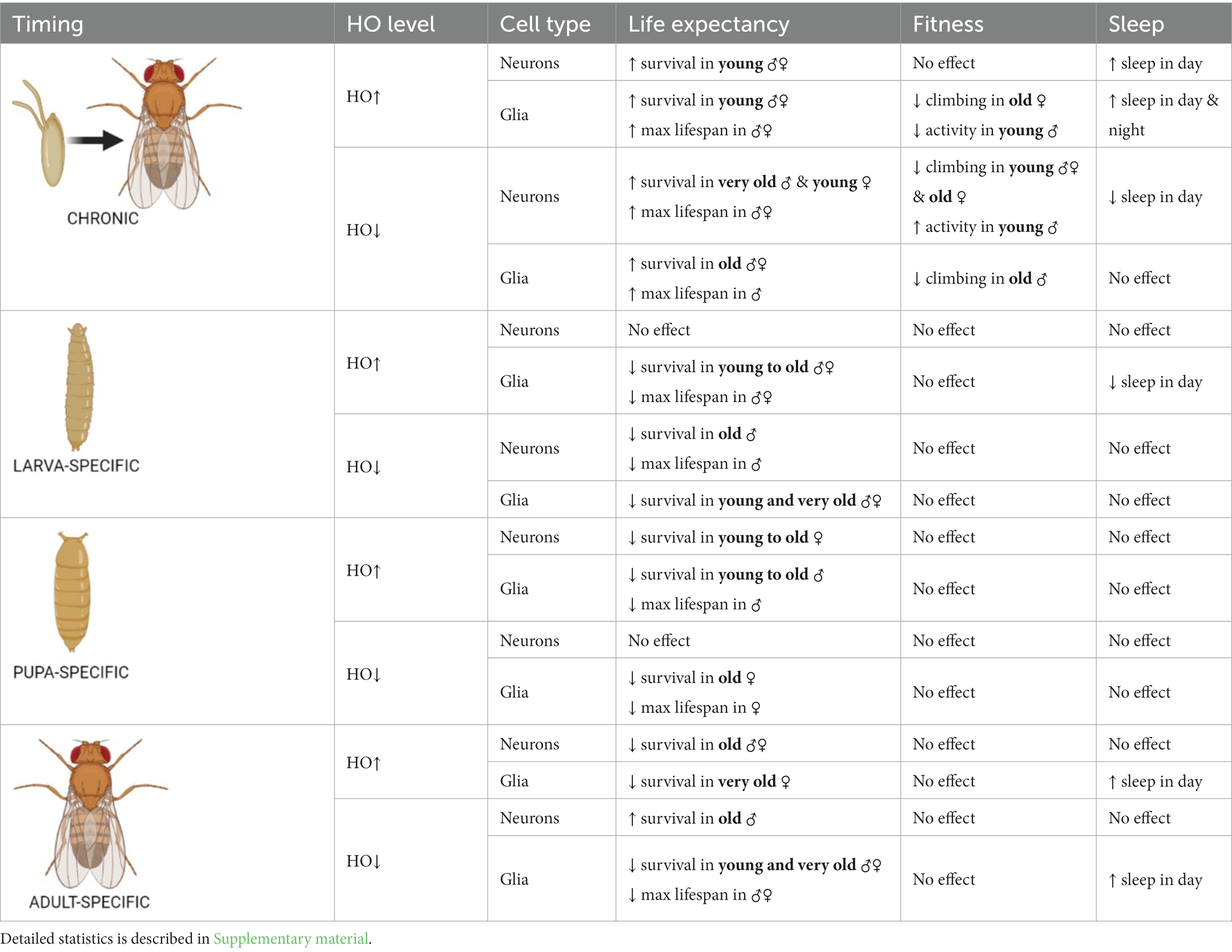
Table 1. Summary of the effects of ho mRNA level changes in neurons or glia at specific developmental stages on adult longevity, fitness and sleep.
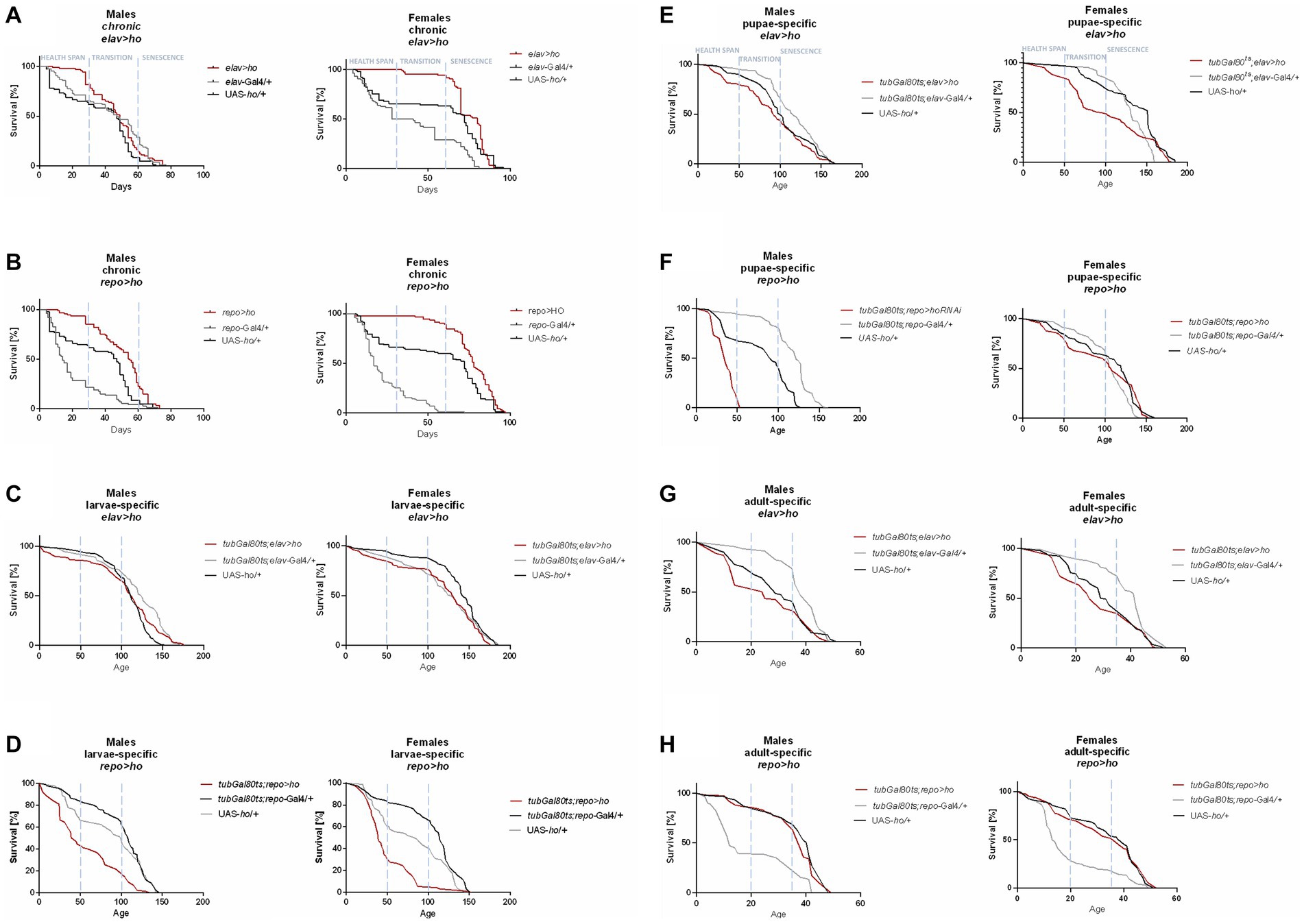
Figure 1. The survival curve of Drosophila adults is altered by ho overexpression in the brain (specifically in neurons or glia) at different developmental stages: chronic (A,B), larva-specific (C,D), pupa-specific (E,F), and adult-specific (G,H).
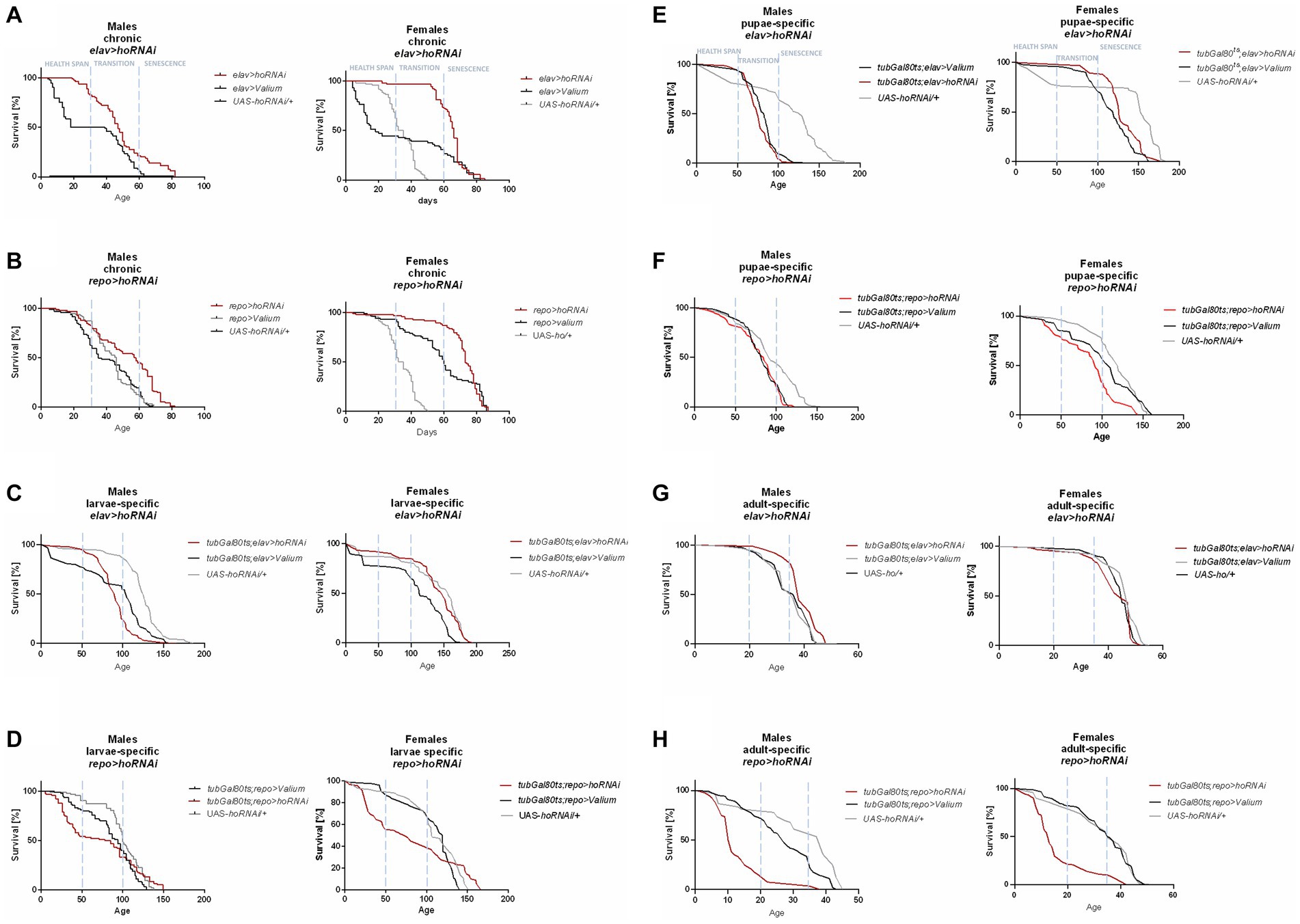
Figure 2. The survival curve of Drosophila adults is altered by ho silencing in the brain (specifically in neurons or glia) at different developmental stages: chronic (A,B), larva-specific (C,D), pupa-specific (E,F), and adult-specific (G,H).
Flies with chronic ho overexpression in neurons (elav > ho) had higher survival during the health span in both sexes, but over time, changes did not vary with the parental controls, except in females during senescence phase (Figure 1A). Similarly, chronic ho overexpression in glia (repo > ho) resulted in a higher percentage of survival during the health span stage for both male and female adults (Figure 1B). Upregulation of ho in neurons during the larval stage had the same effect (Figure 1C), while in glia, it resulted in a significantly lower chance of survival in the health span to the transition phase for both sexes which extended in males until senescence phase (Figure 1D). Pupa-specific ho overexpression in neurons resulted in a lower percentage of survival in the health span and the transition phase but only for females (Figure 1E), while in glia, a lower percentage of survival was observed in similar pattern but with male adults (Figure 1F). Activating ho overexpression in neurons during the adult stage resulted in lower survival during the health span in males only (Figure 1G), while in glia, adults did not have any changes in their survival pattern except for males in senescence phase (Figure 1H).
Chronic ho silencing either in neurons (elav > hoRNAi) or glia (repo > hoRNAi) produced similar survival patterns. Female adults survived better before senescence phase, while male adults had a higher percentage of survival during the senescence phase (Figures 2A,B). Larva-specific ho silencing in neurons did not give any significant differences in survival patterns, except in males with a lower percentage of survival during the transition phase (Figure 2C). On the contrary, larva-specific ho silencing in glia had lower survival during the health span and the senescence phase for both males and females (Figure 2D). The survival pattern of adults was not changed after downregulating ho expression in neurons during the pupal stage (Figure 2E), while in glia, only female adults showed a significantly lower chance of survival during the transition phase (Figure 2F). Silencing ho in neurons during the adult stage resulted in higher survival in the transition phase (only males) (Figure 2G). On the other hand, pan-glial ho silencing during the adult stage exhibited lower chances of survival during the health span and the senescence phase for both male and female adults (Figure 2H).
Similarly, flies with modified cnc expression levels in the brain were observed to have opposite effects between chronic expression and those at specific developmental stages (Table 2). The survival of adults was better when cnc was either overexpressed or silenced continuously in comparison to those modifications activated specifically in the larval, pupal, or adult stage. The survival of chronic elav > cnc flies was significantly higher in male adults during health span while female adults during the senescence phase (Figure 3A). In chronic repo > cnc flies, the higher percentage of survival was almost detectable in all phases except for females in senescence stage (Figure 3B). Upregulation of cnc in glia during the larval stage did not produce any significant differences in survival patterns (Figure 3D), while in neurons, only males had lower percentages of survival during the transition to the senescence phase (Figure 3C). Pupa-specific cnc overexpression in neurons did not produce any significant differences, except in females with a lower percentage of survival during the transition phase (Figure 3E). On the contrary, female adults with pupa-specific cnc overexpression in glia had a lower percentage of survival in all phases (Figure 3F). Activating cnc overexpression in neurons during the adult stage did not produce any significant differences in survival patterns, except males in the senescence phase (Figure 3G), while in glia, a significantly lower survival was observed only in males during the transition phase (Figure 3H).
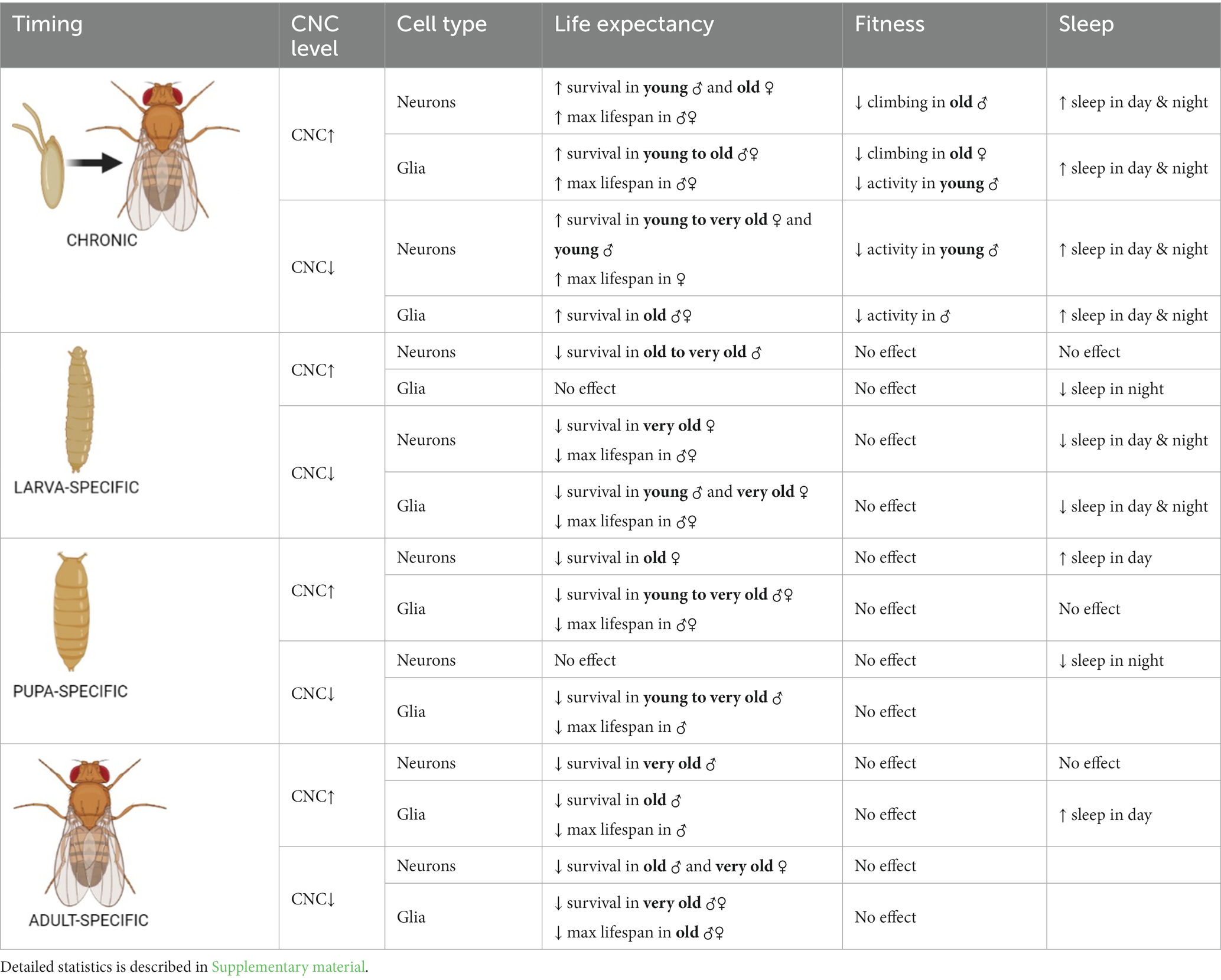
Table 2. Summary of the effects of cnc mRNA level changes in neurons or glia at specific developmental stages on adult longevity, fitness and sleep.
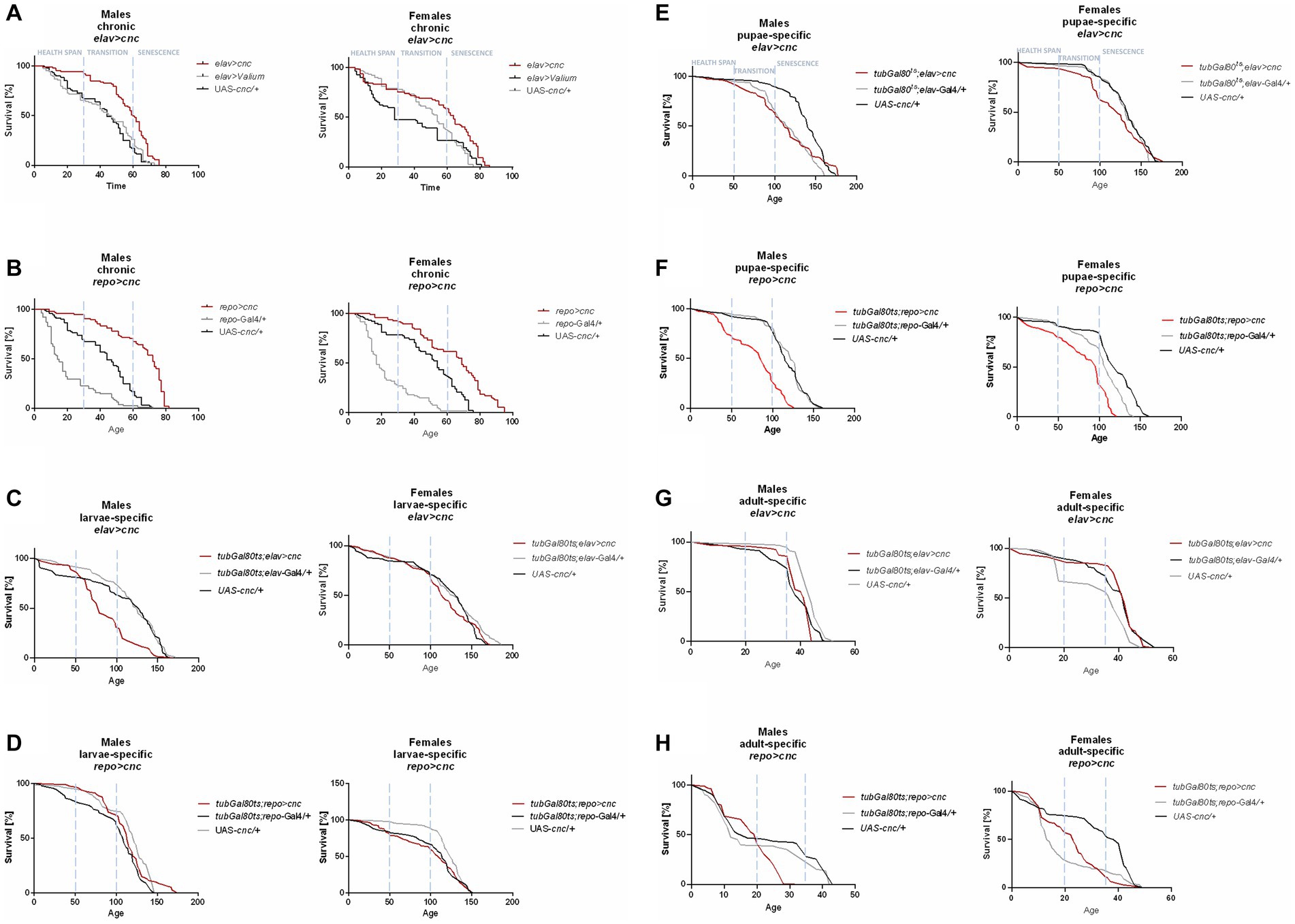
Figure 3. The survival curve of Drosophila adults is altered by cnc overexpression in the brain (specifically in neurons or glia) at different developmental stages: chronic (A,B), larva-specific (C,D), pupa-specific (E,F), and adult-specific (G,H).
For chronic elav > cncRNAi flies, a higher percentage of survival was observed in all phases for female adults and only in the health span for males (Figure 4A). In chronic repo > cncRNAi flies, a better percentage of survival was detectable in the transition to the senescence phase for females only (Figure 4B). Larva-specific cnc silencing in neurons obtained a lower percentage of survival in the senescence phase (only females) (Figure 4C), whereas in glia, male adults had a lower percentage of survival in the health span stage and females in the senescence phase as well (Figure 4D). After downregulating cnc expression in the glia during the pupal stage, only males showed significantly lower survival in the transition to the senescence phase (Figure 4F). In contrast, cnc silencing in neurons at the pupal stage did not affect survival at all (Figure 4E). A significantly lower likelihood of survival was found during the transition phase for males and the senescence phase for females after silencing cnc in neurons during the adult stage (Figure 4G). In comparison, adult-specific pan-glial cnc silencing generated a lower percentage of survival only during the senescence phase for both sexes (Figure 4H).
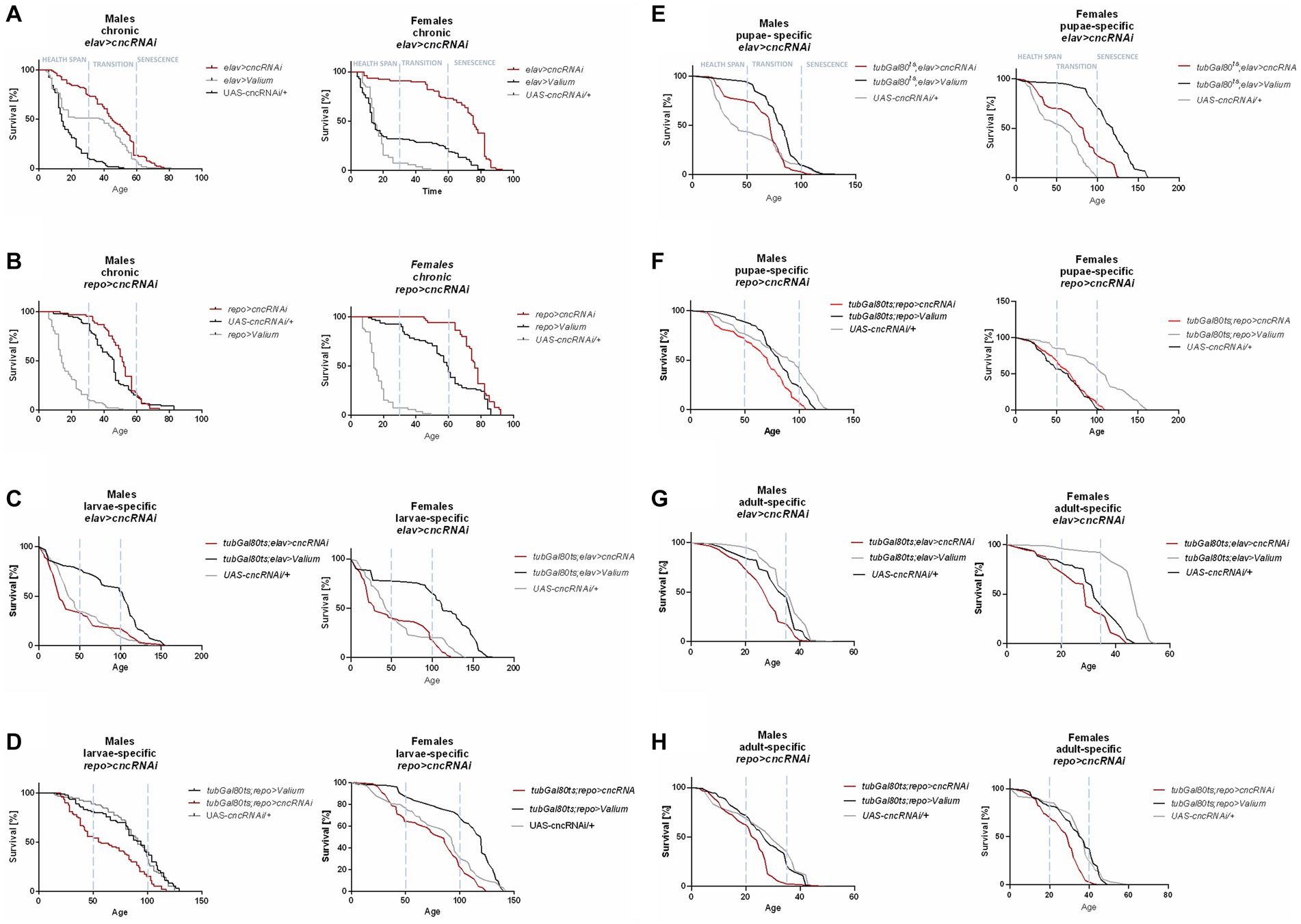
Figure 4. The survival curve of Drosophila adults is altered by cnc silencing in the brain (specifically in neurons or glia) at different developmental stages: chronic (A,B), larva-specific (C,D), pupa-specific (E,F), and adult-specific (G,H).
The maximum lifespan of adults in each experimental condition reflected differences when the expression level of ho or cnc was targeted chronically or at a specific developmental stage (Supplementary Table S1). Under 25°C, the maximum lifespan in chronic experiments was 73–97 DAE. For adult-specific experiments, when flies were kept at 29°C during adult life, aging was faster (Mołoń et al., 2020). It may explain why adults survived only 32–52 DAE. For larva- and pupa-specific experiments that were kept at 18°C during their adult life, longevity generally increased (Mołoń et al., 2020). In this case, adults survived maximum 194 DAE. The effects of modifying ho expression level were not adequately strong to cause changes in the maximum lifespan of adult flies, regardless of the timing (chronically or at specific developmental stages) when overexpression/silencing was done. Most changes in the maximum lifespan were detected only in flies with ho silencing in the glia. Silencing of ho in the glia chronically, in pupae or adults did not increase survival compared with parental controls, while larva-specific ho silencing in the glia extended lifespan of adult flies. In the case of the chronic modifications in cnc expression, we found that flies with cnc overexpression in either neurons or glia survived significantly longer than controls. For flies with pan-neuronal overexpression at specific developmental stages, the flies treated during the adult stage showed a significantly shorter maximum lifespan compared to the control groups. It had a similar pattern with adult-specific repo > cnc. Also, both larva- and pupa-specific overexpression of cnc in glia did not survive longer than their parental controls. The downregulation of cnc in neurons or glia mostly led to a shorter maximum lifespan, regardless of which developmental stage was treated, except for chronic elav > cncRNAi which had an extended maximum lifespan. Differences in maximum lifespan between male and female adults were also observed. In most cases, females had longer maximum lifespans compared to males.
3.2. Effects of changing ho or cnc expression in the brain at different developmental stages on the fitness of adult flies
Changing the ho or cnc mRNA level, specifically during the larval, pupal, or adult stage, did not cause any significant differences in the climbing performance of adult flies (Supplementary Figures S2–S4 and Supplementary Table S5). The effects of ho or cnc expression modifications on the climbing behavior of flies were visible only after chronic overexpression or silencing of either ho or cnc in the brain. Flies with sustained upregulation of ho expression, elav > ho, did not show any changes in their climbing ability (Figure 5A and Supplementary Table S4), while repo > ho had a significantly lower percentage of climbing at 30 DAE, which were observed only in female adults (Figure 5B). For elav > hoRNAi flies, climbing defects were observed in males at 7 DAE, whereas in females, the effect was stronger as their climbing performance at 7, 14, and 30 DAE was decreased (Figure 5C). In repo > hoRNAi flies, only males had a lower percentage of climbing at 30 DAE (Figure 5D). In flies with chronic cnc overexpression, sex-dependent effects were observed at 30 DAE. Only a few males of elav > cnc were able to climb up the 5 cm demarcation line (Figure 5E), while the same effect was observed in repo > cnc females (Figure 5F). Continuous cnc silencing in both neurons and glia did not bring any significant results regarding climbing performance (Figures 5G,H).
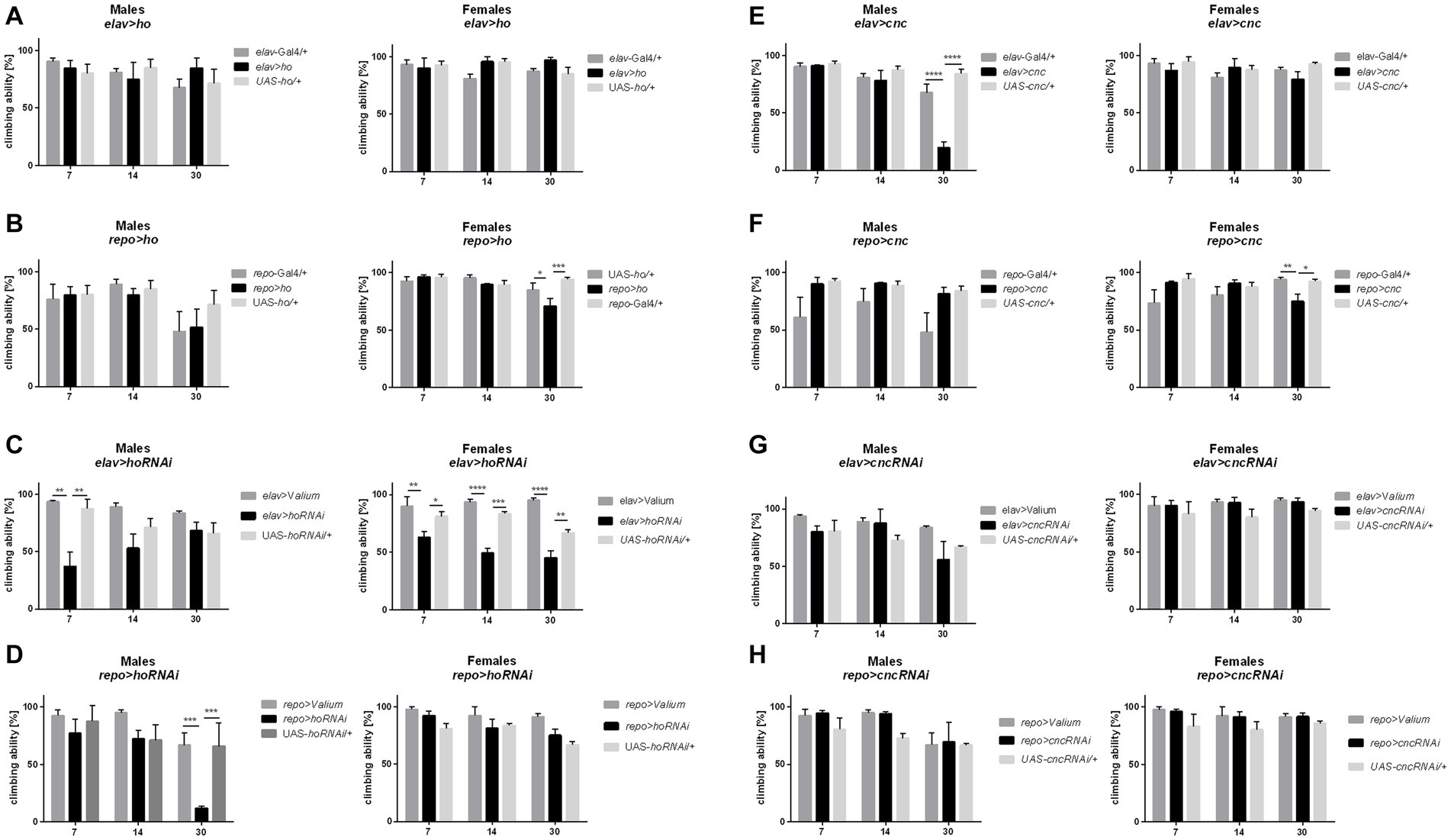
Figure 5. The climbing performance of Drosophila adults 7, 14, and 30 days after eclosion (DAE) was affected by chronic changes in ho (A–D) or cnc (E–H) expression in the brain, specifically in neurons or glia. Data are shown as means ± SD. Statistically significant differences between genotypes are indicated with asterisks (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
The locomotor activity of flies after changing ho or cnc mRNA level in the brain is summarized in Table 3 (Supplementary Figure S6 and Supplementary Table S7). Significant differences in the total activity were mostly observed in flies with chronic modifications of ho expression in the brain. Only elav > ho flies showed identical activity patterns to their parental controls, while repo > ho flies were generally less active. In the case of elav > hoRNAi flies, they were typically more active. Whereas the total activity of repo > hoRNAi flies was not significantly different from the control groups. Flies with modifications in cnc expression in the brain showed changes in their locomotor activity compared to ho since we found significant differences not only in chronic experiments but also in those at specific developmental stages. In chronically treated elav > cnc flies, no significant difference was observed in total activity, while repo > ho flies exhibited a reduction in their total activity. In flies with chronic cnc silencing either in neurons or glia, the total activity was lower when compared to parental controls. When changes in cnc expression level were introduced at different developmental stages of Drosophila, we found higher total activity in larva-specific cnc overexpression in glia, larva-specific cnc silencing in either neurons or glia and adult-specific cnc silencing in glia.
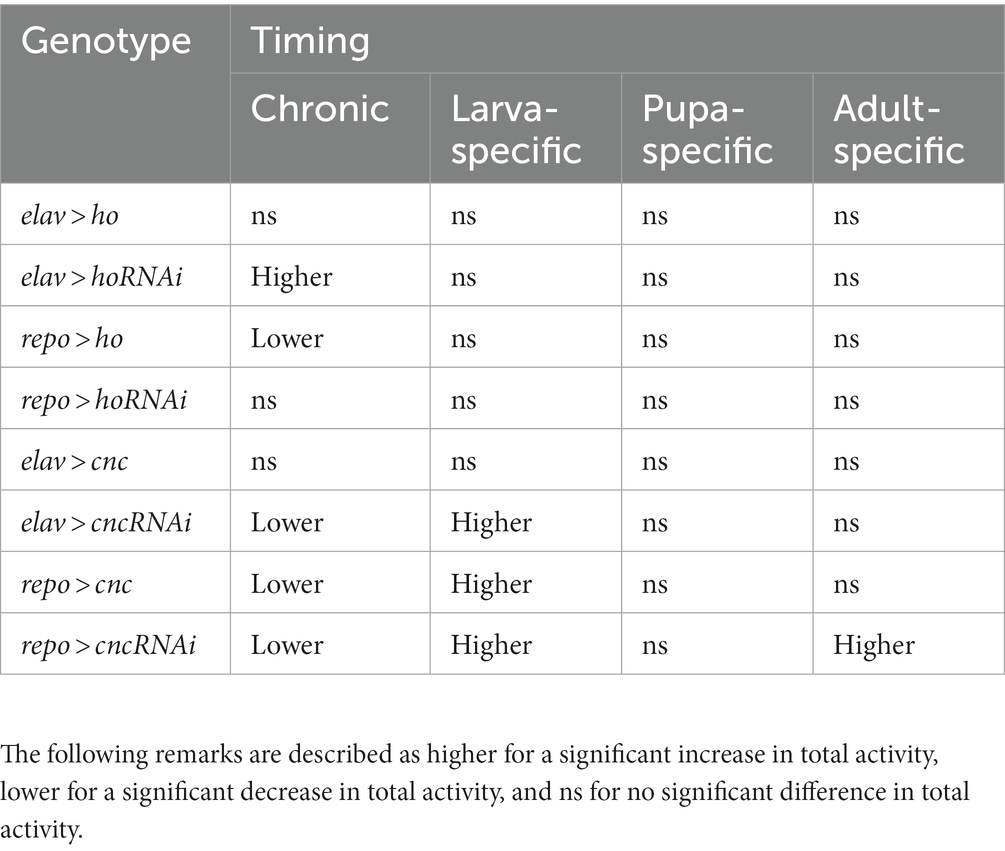
Table 3. Summary of the effects of chronic changes of ho or cnc mRNA level in neurons or glia on locomotor activity pattern in Drosophila adults.
3.3. Effects of changing ho or cnc expression in the brain at different developmental stages on sleep in adults
Sleep was also checked in adults with modifications of ho expression during their development. We found significant differences in sleep when ho expression was upregulated or downregulated in the brain at different developmental stages (Table 4; Supplementary Table S8).
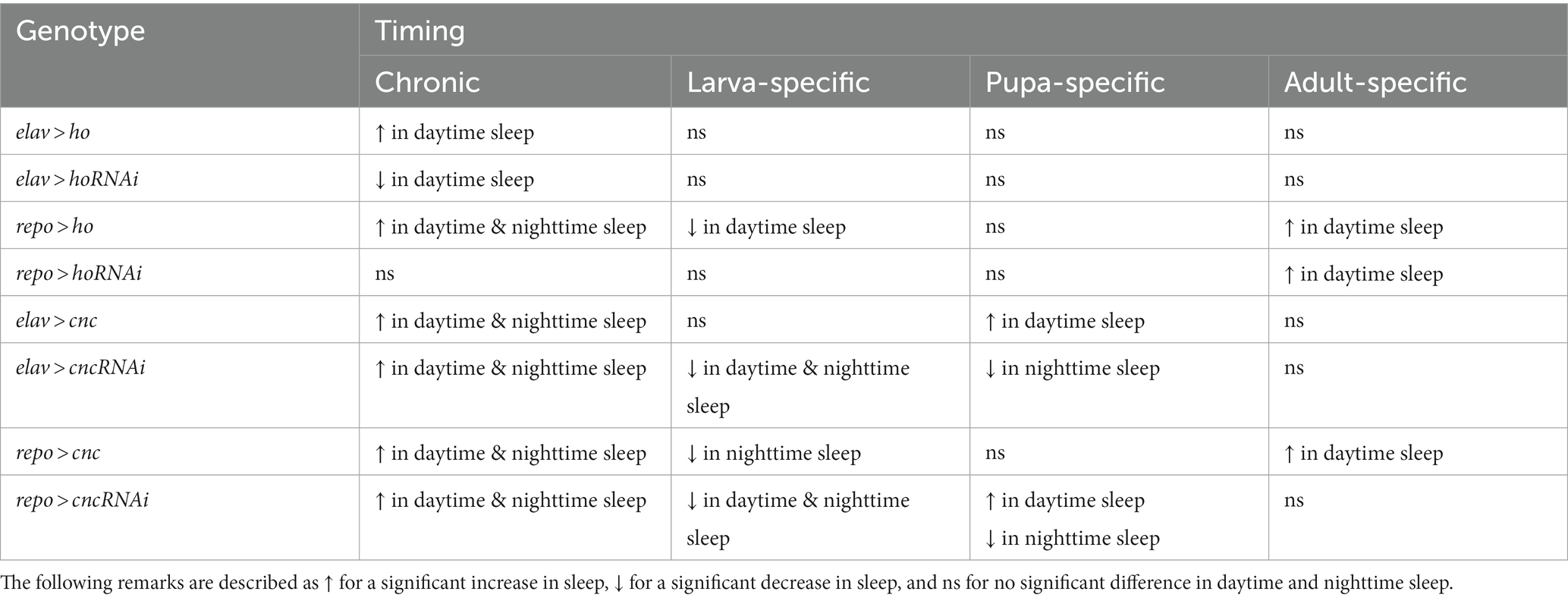
Table 4. Summary of the effects of changing ho mRNA level in neurons or glia at specific developmental stages on sleeping pattern (daytime and nighttime sleep) of Drosophila adults.
When ho was chronically overexpressed in the brain, changes in sleep patterns were significant compared to their parental controls. In elav > ho flies, daytime sleep was higher while nighttime sleep was unaffected (Figure 6A and Supplementary Table S8). On the other hand, daytime and nighttime sleep increased in repo > ho adults (Figure 6B). Pan-neuronal ho overexpression at specific developmental stages did not produce any significant results in sleep patterns. On the contrary, different responses were observed after overexpressing ho in the glia at specific developmental stages. Nighttime sleep was not affected by expression changes made during the larval, pupal, or adult stages. But then, opposite effects were observed regarding daytime sleep when pan-glial ho overexpression was induced at the larval or adult stage. Daytime sleep was reduced in flies with larva-specific ho overexpression in the glia, while it was increased in flies treated during the adult stage. In pupa-specific ho overexpression in glia, the daytime sleep was not changed at all. The effects of ho silencing in the brain on sleep in adults were rather weak (Figures 6C,D). We only found significant differences in daytime sleep in chronically treated elav > hoRNAi, which was shorter, and in adult-specific ho silencing in glia which had longer daytime sleep.
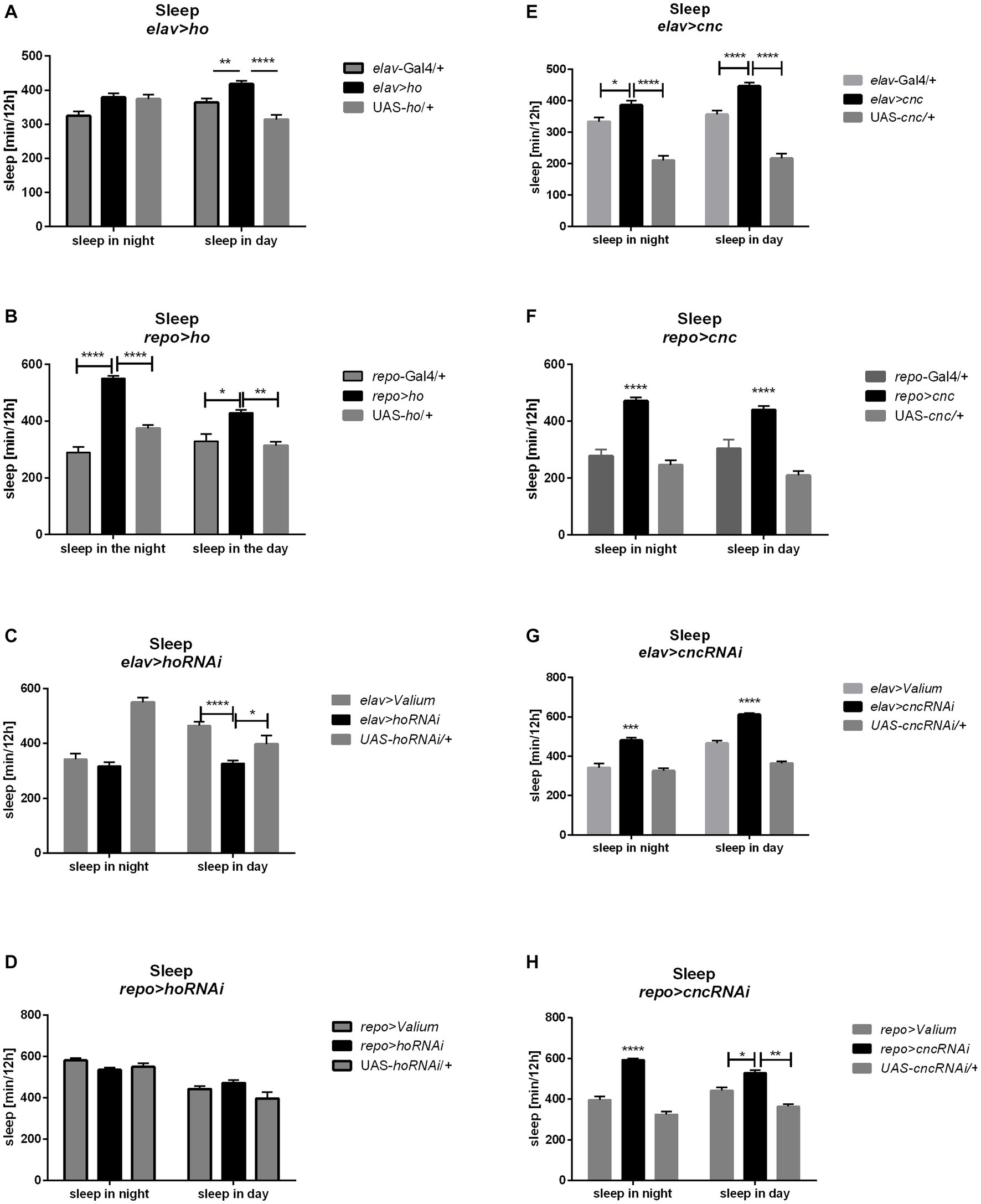
Figure 6. The sleep level of Drosophila adults is altered by chronic changes in ho (A–D) or cnc (E–H) mRNA level in the brain, specifically in neurons or glia. Data are expressed as means ± SE. Statistically significant differences between genotypes are indicated with asterisks (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
The chronic overexpression of cnc in neurons or glia brough similar results in daytime and nighttime sleep, which were longer in comparison to their respective parental controls (Figures 6E,F). On the other hand, continuous silencing of cnc in neurons or glia showed similar effects, increasing daytime and nighttime sleep (Figures 6G,H). Activating cnc overexpression at specific developmental stages generated different effects. The sleeping pattern was changed only when pan-neuronal overexpression of cnc was carried out at the pupal stage and daytime sleep was increased in this case. For pan-glial overexpression of cnc, we only found significant differences in sleep after overexpression at the larval stage (with decreased nighttime sleep), and at the adult stage (increased daytime sleep). On the other hand, continuous silencing of cnc in neurons or glia brough similar effects because daytime and nighttime sleep increased in both cases. It was the opposite after larvae-specific silencing of cnc in neurons or glia since both daytime and nighttime sleep was decreased. Pupae-specific downregulation of cnc in the brain reduced nighttime sleep, however, when directed to glia, it also increased daytime sleep. The partial suppression of cnc in either neurons or glia during the adult stage did not bring any significant changes in the sleeping behavior of adult flies.
3.4. The mRNA level of ho and cnc at different developmental stages
To have baseline data about the transcriptional regulation of ho and cnc during development in Drosophila, we examined ho and cnc mRNA levels at different life stages of wild-type flies. Both ho and cnc mRNA levels were similarly maintained at higher levels during the larval stage compared to the pupal and adult stages under optimal temperature conditions (Figure 7A and Supplementary Table S8). At 29°C, there were no significant changes in ho and cnc mRNA levels between CS larvae (whole body), pupae (whole body), and adults (head) (Figure 7B and Supplementary Table S9). At 18°C, both ho and cnc mRNA were significantly higher in adults (head) compared to larvae (whole body) and pupae (whole body) (Figure 7C and Supplementary Table S9).
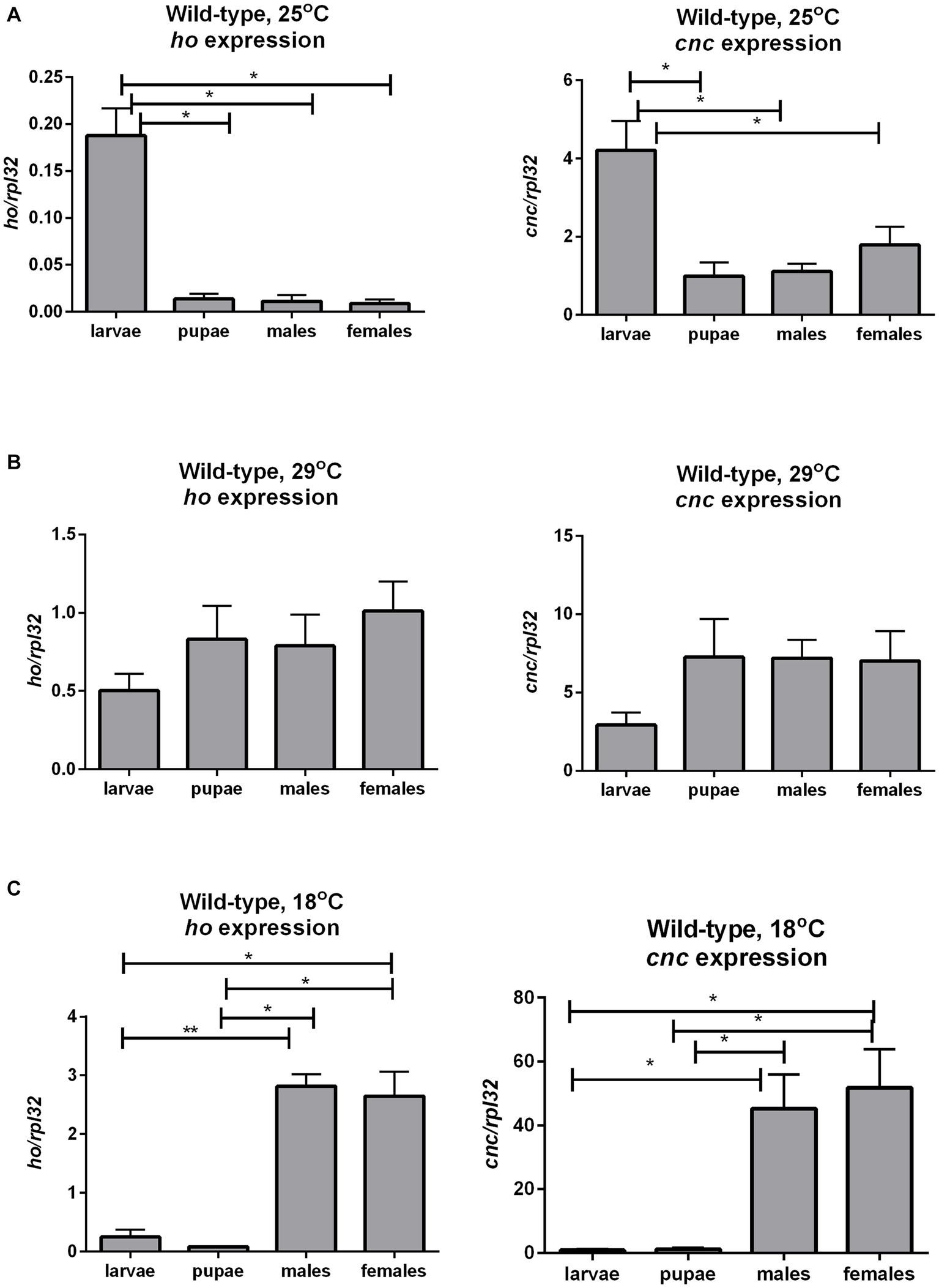
Figure 7. The mRNA level of ho and cnc in larvae, pupae, and in heads of adults (males and females) of wild-type (CS) flies changes with ambient temperature conditions: 25°C (A), 29°C (B), and 18°C (C). Data are expressed as means ± SD. Statistically significant differences between groups are indicated with asterisks (*p < 0.05 and **p < 0.01).
The mRNA level of ho was also examined in larvae (whole body), pupae (whole body), and adults (heads of males and females, separately) after modifying its expression chronically or at specific developmental stages. Surprisingly, we did not find any significant differences in ho mRNA levels in larvae, pupae, and heads of adults after changing ho expression, specifically during the larval, pupal, or adult stage (Supplementary Table S8). Nevertheless, we found a trend of ho mRNA increase in larvae, pupae, or adults in flies with ho overexpression and unaffected or lesser ho mRNA in flies with RNAi. However, the differences were not statistically significant. We only observed significant changes in ho mRNA levels in male and female adults (heads) after chronic expression. The continuous ho overexpression in both neurons and glia induced significantly higher ho mRNA levels in the heads of males and females (Figures 8A,B and Supplementary Table S9). On the contrary, we observed significantly high ho mRNA levels in the heads of female elav > hoRNAi and male repo > hoRNAi flies (Figures 8C,D and Supplementary Table S9). We suspected compensatory effects after partially suppressing ho in neurons or glia. We found that the high expression of ho originated from the retina (Figure 9 and Supplementary Table S9). In the brain, ho mRNA level did not change. It seems that the compensatory effect occurs when the compound eye is fully developed as we did not find any changes in ho mRNA level in larvae or pupae. However, it is worth mentioning that we manipulated ho expression only in specific cell types, neurons or glia, and this change can be difficult to detect in whole head samples.
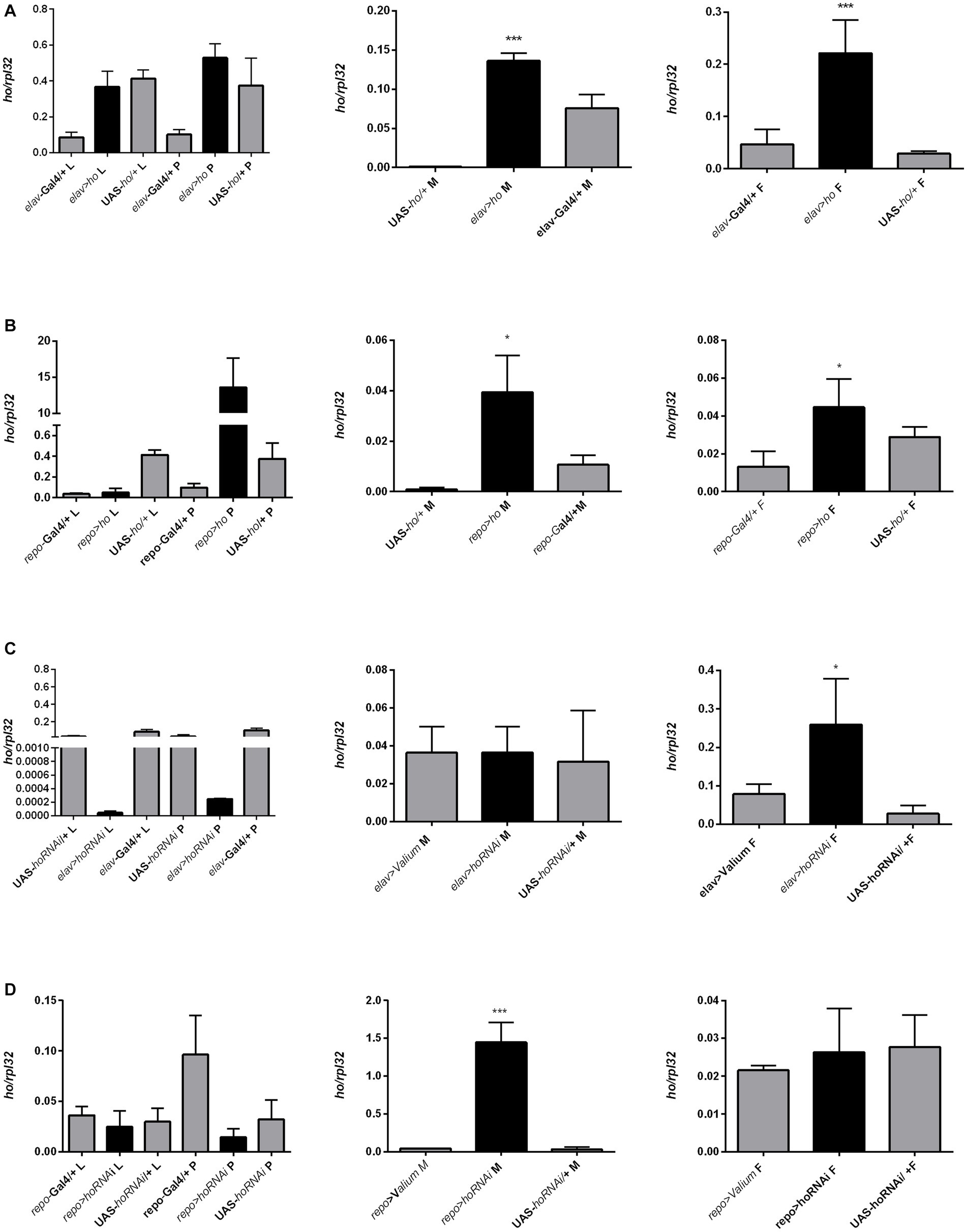
Figure 8. The ho expression pattern in larvae (L), pupae (P), and in heads of Drosophila adults (M for males, F for females) after chronic overexpression or silencing of ho in either neurons (A,C) or glia (B,D). Data are shown as means ± SE. Statistically significant differences between genotypes are indicated with asterisks (*p < 0.05 and ***p < 0.001).
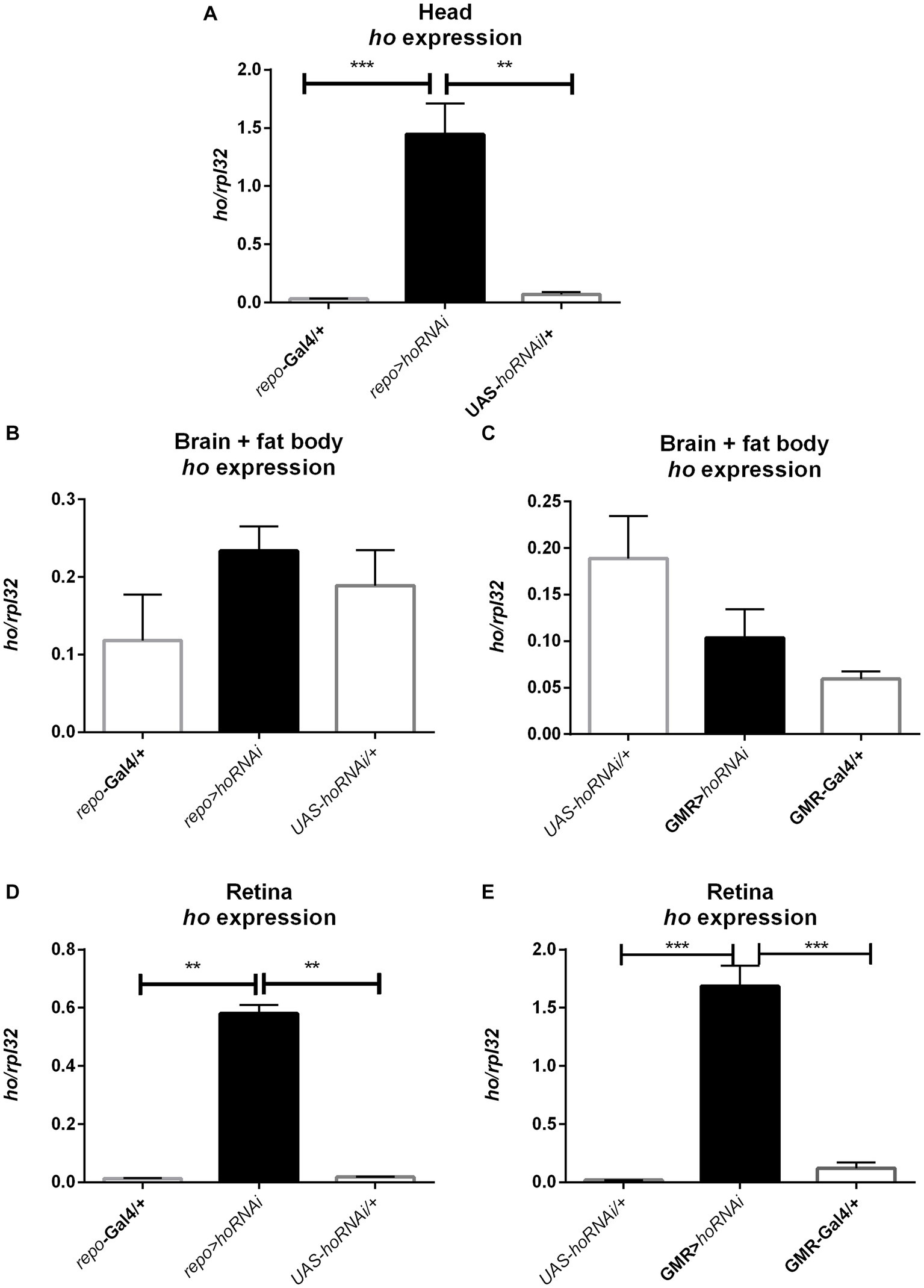
Figure 9. The increase of ho mRNA in the heads of adult flies with chronic ho silencing in glia (A) is not from the brain (B,C) but mostly from the retina (D,E). Data are presented as means ± SE. Statistically significant differences between genotypes are indicated with asterisks (**p < 0.01 and ***p < 0.001).
We also quantified cnc mRNA levels in larvae (whole body), pupae (whole body), and adults (head) after the chronic modification of cnc expression in the brain, however, significant differences between experimental and control groups were only detected in adults. Chronic overexpression of cnc in neurons elevated cnc mRNA levels in both males and females (Figure 10A and Supplementary Table S9). In glia, cnc overexpression was only detected in females; however, we observed a similar trend in males (Figure 10B and Supplementary Table S9). The reduction of cnc transcript level was detected only in males and females after chronic cnc silencing in glia (Figure 10C and Supplementary Table S8). We found no significant differences when cnc was silenced in neurons (Figure 10D and Supplementary Table S9). Nonetheless, cnc mRNA levels were similar to controls.
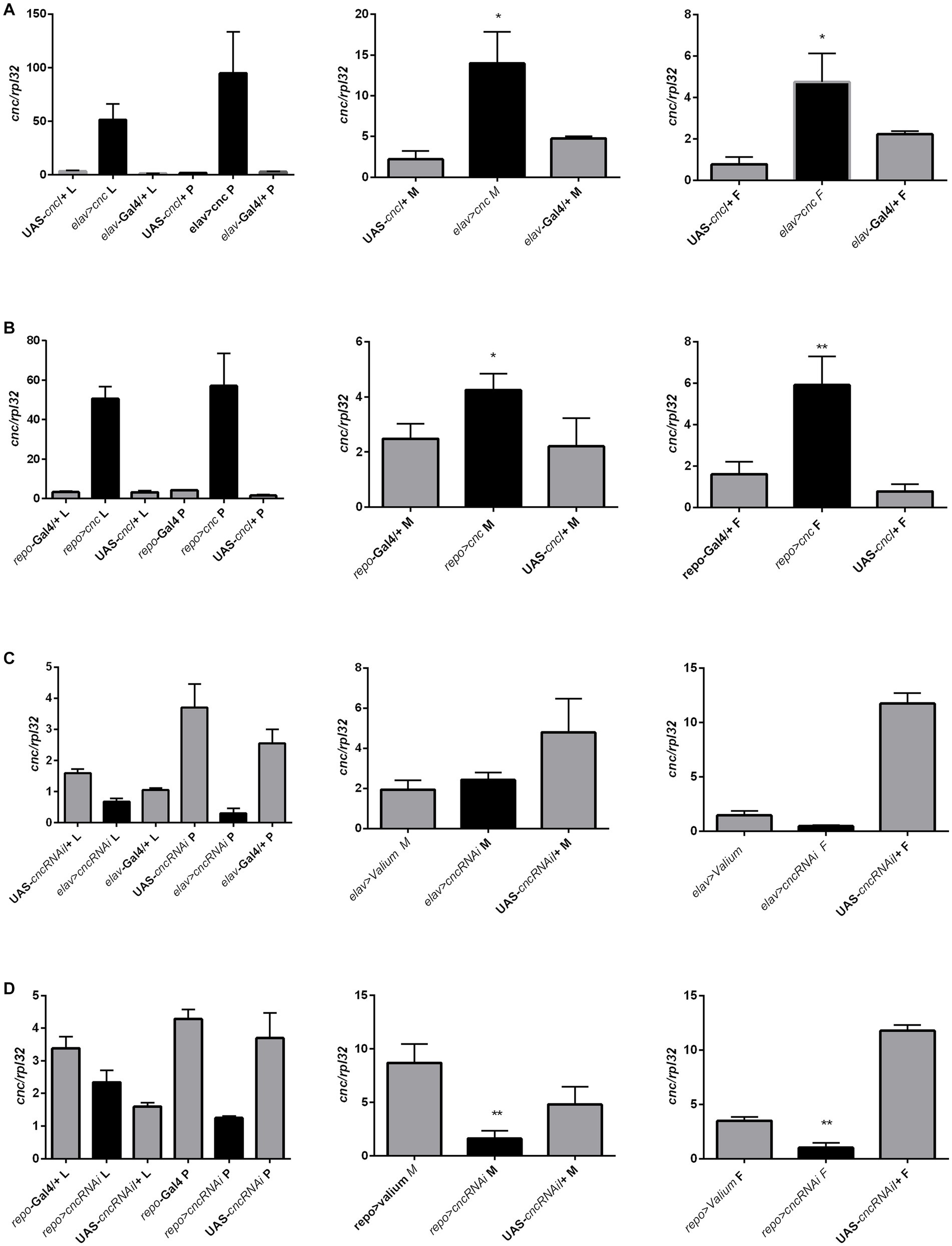
Figure 10. The cnc expression pattern in larvae (L), pupae (P), and heads of Drosophila adults (M for males, F for females) after chronic overexpression or silencing of ho in either neurons (A,C) or glia (B,D). Data are expressed as means ± SE. Statistically significant differences between genotypes are indicated with asterisks (*p < 0.05 and ***p < 0.001).
4. Discussion
In the present study, we showed that disruption of the normal physiological level of heme oxygenase (HO) during development affects the adult life of Drosophila melanogaster.
First we found that the expression of ho and cnc varies between developmental stages and depends on temperature. At 25°C it was the highest in larvae and lower in pupae and adults. At 18°C, however, the expression level was reversed with the highest level in adults and at 29°C the mRNA level was similar during all developmental stages. This indicates that temperature exposure during rearing can affect experiments involving genetic manipulations. In addition, the TARGET system, which is commonly used in fly research, also needs to be used with a great care, since stage-specific genetic modification requires temperature changes between 18°C and 29°C, which are not optimal for flies, and can affect physiological processes in the adult life. Moreover, the temperature of 29°C drives expression of thermo-responsive miRNA, which affects gene expression and transposition (Chen et al., 2015; Fast and Rosenkrantz, 2018).
The effect of HO on adult longevity (survival and maximum lifespan) was more pronounced and consistent than on fitness (climbing ability and locomotor activity) and sleep of adult flies. We found that chronic modifications of ho expression (either upregulated or downregulated) in the brain results in higher survival compared with controls. It is possible that continuous overexpression or silencing of ho through development to adulthood affects many crucial processes dependent on HO level (Zhang et al., 2004; Cui et al., 2008; Ida et al., 2013; Klemz et al., 2017; Damulewicz et al., 2017a,b, 2019; Abaquita et al., 2021, 2023), including the circadian clock (Mandilaras and Missirlis, 2012; Klemz et al., 2017; Damulewicz et al., 2017a, 2019). The clock disruption can alter circadian rhythms in many physiological and behavioral processes (Ceriani et al., 2002). It may also explain why fitness was significantly changed in flies with chronic ho overexpression or silencing in the brain compared with changes that were larva, pupa-, and adult-specific.
When changes in ho expression were induced at specific developmental stages, we observed an opposite effect on adult survival. The survival was mostly decreased compared to their respective controls. This indicates that HO in the brain is tightly regulated during development to adulthood since changing ho expression at different developmental stages can disrupt the longevity of adults. There are, however, exemption, because the survival did not change after increasing ho expression in neurons at the larval stage and decreasing it in neurons at the pupal stage. It is important to highlight these findings because, for instance, inducing ho expression in larvae does not affect longevity, fitness, and sleep of adults. This suggests that a high physiological level of HO is important for larvae. After checking the ho mRNA level at different developmental stages, we found that the ho gene is highly expressed in larvae under physiological conditions. It has also been reported that ho mRNA level is high in the third-instar larvae (L3, specifically at puff stage 1–9), using a high-throughput expression analysis (Brown et al., 2014) which may be associated with the physiological demand for the synthesis of hormones (i.e., juvenile hormone and ecdysteroids) regulating molting and metamorphosis (Massie et al., 1985; Llorens et al., 2015). The synthesis of both ecdysone and juvenile hormone requires functional cytochrome P450 which contains heme as a cofactor. The high expression of ho in larvae is probably related to its cytoprotective and antioxidative functions to prevent free heme deposition (Linzke et al., 2014). HO might have the same functions in pupae (Brown et al., 2014), however, partial suppression of ho is not critical according to our results. Active heme metabolism is essential during the larval stage as HO deficiency in L3 leads to the increase of heme and iron pools (Cui et al., 2008). The same authors also reported that 95% knockdown of the ho gene is lethal at the larval stage since a few viable L3 were observed and they did not survive metamorphosis.
All available data regarding the level of ho and cnc expression at specific stages of development are not consistent, however. According to the modENCODE Anatomy RNA-Seq, in L3 wandering larvae ho is at its high level in the digestive system and fat body and it is moderate in the nervous system, while FlyAtlas Anatomical Expression Data indicate extremely high ho mRNA level in the fat body, but not in the digestive system. We decided to use whole body of larvae and pupae to analyse the whole nervous system, including the peripheral one. Because of that mRNA levels of ho and cnc, which we measured, did not show statistically significant changes between experimental and control groups. However, we used qPCR to quantify mRNA, which provides precise data on gene expression. Because of that we could focus on one selected gene, since primers were designed specifically for the gene of interest and reaction specificity was checked with melt curve analysis. The obtained results showed a growing effect of silencing/overexpression of ho and cnc from larvae to pupae, and in adults statistically significant changes were detected. Moreover, our results confirmed the modENCODE Temporal Expression Data that in larvae the mRNA level of ho is higher than in pupae and adults, and almost the same in males and females.
Similarly, mammalian HO-1 (homolog of HO in Drosophila) is highly induced during embryonic development supporting the survival of the embryo (Ihara et al., 1998; Watanabe et al., 2004; Zhao et al., 2009; Zenclussen et al., 2015; Loboda et al., 2016). Deletion of HO-1 in mice leads to high prenatal lethality and surviving adults with HO-1 deficiency are smaller, they breed poorly and are less active. They also have several abnormalities leading to high mortality (Poss and Tonegawa, 1997a,b; Kovtunovych et al., 2010). In contrast, HO-1 overexpression improves embryonic development (Zenclussen et al., 2006, 2015). It has been suggested that dysfunction of HO can affect the release of its metabolite CO (Zenclussen et al., 2011, 2015) which acts against oxidative stress (Linzke et al., 2014) and probably maintains homeostasis of glucose (Klemz et al., 2017).
We also observed sex- and age-specific differences in the longevity and fitness of Drosophila adults. Regarding maximum lifespan, females had usually longer lifespans compared to males. It was previously shown that Drosophila females live longer, and their aging is delayed because of higher antioxidative enzyme expression in old age (Deepashree et al., 2017; Niveditha et al., 2017). In addition, there are sex-specific differences in stress response and stress adaptation (see Tower et al., 2020), including the regulation of the growth and cell signaling pathways, organ homeostasis and metabolism (Magwere et al., 2004; Millington and Rideout, 2018), and the HO system (Barañano and Snyder, 2001; Han E. et al., 2005; Han Y. et al., 2005; Ren et al., 2021). Another evidence is that the survival pattern of females is less affected than males, specifically when ho expression is reduced in neurons at the larval or adult stage and induced in glia at the pupal stage. One exemption was when ho was silenced in glia at the pupal stage, and this affected only females but not males. We also found that increasing ho expression in glia decreases climbing ability in 30 days-old females while decreasing ho expression adversely affects the climbing performance of 30 days-old males.
It seems that the influence of HO in the brain on the survival of aging flies depends on which cells in the nervous system are affected. In our chronic experiments, neuronal ho overexpression resulted in a higher percentage of survival of young flies while neuronal ho silencing promoted better survival of aging flies. Glial ho expression changes were critical for aging flies only. It means that HO in neurons and glial cells has distinct functions during development until adulthood which has already been reported in mammals (see Schipper et al., 1995, 2006; Song et al., 2007; Schipper et al., 2009; Ohnishi et al., 2010; Tronel et al., 2013; Barone et al., 2014; Mena et al., 2015; Chiang et al., 2018; Nitti et al., 2018; Waza et al., 2018; Schipper et al., 2019; Luu Hoang et al., 2021; Wu and Hsieh, 2022; Yang and Wang, 2022).
Sleep is very sensitive to changes in an organism’s homeostasis and is regulated by the circadian clock. Here, the sleep pattern was changed in adults after ho overexpression or silencing in the brain. In our chronic expression experiments, different sleep patterns were observed depending on the level of ho expression in neurons and glia. Overexpression of ho in the glia increased both daytime and nighttime sleep, which did not change when ho expression in the glia was downregulated. Flies with ho overexpression in neurons had longer daytime sleep compared to controls, but when ho was silenced, daytime sleep was reduced. When ho expression was changed at specific developmental stages, the sleep pattern was either unaffected or results were inconsistent.
The effects observed in adult flies after ho overexpression or silencing in either neurons or glia at specific developmental stages, were not always clear and consistent with those after chronic treatments. It might be due to high and low temperatures used to induce and suppress, respectively ho expression. We found differences in the relative abundance and expression pattern of ho mRNA after exposing immature and adult Drosophila to low or high temperatures than to the optimal one. Under heat stress aging and metabolism are faster, while in lower temperatures, both processes are slow down (Mołoń et al., 2020). For that reason, HO activity can also change under different temperature conditions.
In addition, the high-throughput expression analyses of the Fly Cell Atlas project (Li et al., 2022) revealed that basal ho expression in neurons is maintained at low to moderate levels. Dysregulation of heme metabolism may be stressful for neurons and glia. The appropriate level of HO in the brain is so important that there are compensatory mechanisms that help to maintain its expression. When ho was partially silenced in neurons or glia, we surprisingly observed overexpression of ho in heads. More detailed analysis showed that the retina produces high amount of HO that compensates the lack of this enzyme in other cells. We did not identify the cell type, which is responsible for this process, as silencing in both glia and photoreceptors showed increased ho expression in the retina. Additional evidence suggesting that this mechanism depends on the retina comes from developmental studies—we did not observe this compensatory effect in larvae, which do not have compound eyes. It started to progress in pupae and then it was strongly manifested in adults. This phenomenon is very interesting as in most cases genetic compensation is observed only in mutants but not after knockdowns and it occurs through the overexpression of different genes to avoid the negative effect of mutation (El-Brolosy and Stainier, 2017).
Interestingly, disrupting the cnc expression, a gene encoding one of the active isoforms of Drosophila CNC in the brain also affected adult flies, depending on the timing, expression level, cell types, sex, and age of adults. However, the obtained results were different than after manipulating ho expression. We expected that CNC-dependent genes like ho, should have similar effects as after changing cnc expression (Loboda et al., 2016). CNC is a multifunctional transcription factor that regulates growth and development, proteasome stability and detoxification (Loboda et al., 2016). Its function begins during oogenesis (Guichet et al., 2001). We found that cnc is highly expressed in larvae, but its transcript level is comparably lower in pupae and adults. The CNC transcription factors play an important role in aging (Loboda et al., 2016), which can explain the differences in adult life parameters between cnc and ho after modifying their expression in the brain.
In conclusion, adult life depends on adequate HO levels in the brain during development. Our findings indicate that the appropriate level of HO at the early stage of life can prevent health problems during aging. It seems to be similar in flies and mammals, because multiple cellular processes are affected by heme metabolism dysfunction, leading to differences in longevity, fitness, and sleep under various conditions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://doi.org/10.57903/UJ/UD75IP, UD75IP.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
BB and MD carried out all experiments. MD performed all data analyses and prepared figures. TA summarized the results and wrote the first draft of the manuscript. EP finalized the concept of the study, validated all results, interpreted the data, and together with MD, prepared the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Science Centre in Poland, grant OPUS (Funding Number: UMO/2018/29/B/NZ3/02591) granted to EP.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2023.1239101/full#supplementary-material
Footnotes
References
Abaquita, T. A. L., Damulewicz, M., Bhattacharya, D., and Pyza, E. (2021). Regulation of heme oxygenase and its cross-talks with apoptosis and autophagy under different conditions in Drosophila. Antioxidants 10:1716. doi: 10.3390/antiox10111716
Abaquita, T. A. L., Damulewicz, M., Tylko, G., and Pyza, E. (2023). The dual role of heme oxygenase in regulating apoptosis in the nervous system of Drosophila melanogaster. Front. Physiol. 14:1060175. doi: 10.3389/fphys.2023.1060175
Adachi, T., Ishikawa, K., Hida, W., Matsumoto, H., Masuda, T., Date, F., et al. (2004). Hypoxemia and blunted hypoxic ventilatory responses in mice lacking heme oxygenase-2. Biochem. Biophys. Res. Commun. 320, 514–522. doi: 10.1016/j.bbrc.2004.05.195
Alam, J., Igarashi, K., Immenschuh, S., Shibahara, S., and Tyrrell, R. M. (2004). Regulation of heme oxygenase-1 gene transcription: recent advances and highlights from the International Conference (Uppsala, 2003) on heme oxygenase. Antioxid. Redox Signal. 6, 924–933. doi: 10.1089/ars.2004.6.924
Andoh, Y., Mizutani, A., Ohashi, T., Kojo, S., Ishii, T., Adachi, Y., et al. (2006). The antioxidant role of a reagent, 2′,7′-dichlorodihydrofluorescin diacetate, detecting reactive-oxygen species and blocking the induction of heme oxygenase-1 and preventing cytotoxicity. J. Biochem. 140, 483–489. doi: 10.1093/jb/mvj187
Atamna, H. (2004). Heme, iron, and the mitochondrial decay of ageing. Ageing Res. Rev. 3, 303–318. doi: 10.1016/j.arr.2004.02.002
Atamna, H., Killilea, D. W., Killilea, A. N., and Ames, B. N. (2002). Heme deficiency may be a factor in the mitochondrial and neuronal decay of aging. Proc. Natl. Acad. Sci. U. S. A. 99, 14807–14812. doi: 10.1073/pnas.192585799
Ayajuddin, M., Phom, L., Koza, Z., Modi, P., Das, A., Chaurasia, R., et al. (2022). Adult health and transition stage-specific rotenone-mediated Drosophila model of Parkinson’s disease: impact on late-onset neurodegenerative disease models. Front. Mol. Neurosci. 15:896183. doi: 10.3389/fnmol.2022.896183
Azuma, M., Kabe, Y., Kuramori, C., Kondo, M., Yamaguchi, Y., and Handa, H. (2008). Adenine nucleotide translocator transports haem precursors into mitochondria. PLoS One 3:e3070. doi: 10.1371/journal.pone.0003070
Bainbridge, S. P., and Bownes, M. (1981). Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66, 57–80.
Barañano, D. E., and Snyder, S. H. (2001). Neural roles for heme oxygenase: contrasts to nitric oxide synthase. Proc. Natl. Acad. Sci. U. S. A. 98, 10996–11002. doi: 10.1073/pnas.191351298
Barone, E., Di Domenico, F., Mancuso, C., and Butterfield, D. A. (2014). The Janus face of the heme oxygenase/biliverdin reductase system in Alzheimer disease: it’s time for reconciliation. Neurobiol. Dis. 62, 144–159. doi: 10.1016/j.nbd.2013.09.018
Bittern, S., Pogodalla, N., Ohm, H., Brüser, L., Kottmeier, R., Schirmeier, S., et al. (2021). Neuron-glia interaction in the Drosophila nervous system. Dev. Neurobiol. 81, 438–452. doi: 10.1002/dneu.22737
Boehning, D., and Snyder, S. H. (2003). Novel neural modulators. Annu. Rev. Neurosci. 26, 105–131. doi: 10.1146/annurev.neuro.26.041002.131047
Braz, G. R. C., Abreu, L., Masuda, H., and Oliveira, P. L. (2001). Heme biosynthesis and oogenesis in the blood-sucking bug, Rhodnius prolixus. Insect Biochem. Mol. Biol. 31, 359–364. doi: 10.1016/S0965-1748(00)00129-6
Brown, J. B., Boley, N., Eisman, R., May, G. E., Stoiber, M. H., Duff, M. O., et al. (2014). Diversity and dynamics of the Drosophila transcriptome. Nature 512, 393–399. doi: 10.1038/nature12962
Ceriani, M. F., Hogenesch, J. B., Yanovsky, M., Panda, S., Straume, M., and Kay, S. A. (2002). Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22, 9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002
Chen, J., Nolte, V., and Schlotterer, C. (2015). Temperature-Related Reaction Norms of Gene Expression: Regulatory Architecture and Functional Implications. Molecular Biology and Evolution. 32, 2393–2402. doi: 10.1093/molbev/msv120
Chiabrando, D., Fiorito, V., Petrillo, S., and Tolosano, E. (2018). Unraveling the role of heme in neurodegeneration. Front. Neurosci. 12:712. doi: 10.3389/fnins.2018.00712
Chiabrando, D., Vinchi, F., Fiorito, V., Mercurio, S., and Tolosano, E. (2014). Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 5:61. doi: 10.3389/fphar.2014.00061
Chiang, S.-K., Chen, S.-E., and Chang, L.-C. (2018). A dual role of heme oxygenase-1 in cancer cells. Int. J. Mol. Sci. 20:39. doi: 10.3390/ijms20010039
Chung, H., Sztal, T., Pasricha, S., Sridhar, M., Batterham, P., and Daborn, P. J. (2009). Characterization of Drosophila melanogaster cytochrome P450 genes. Proc. Natl. Acad. Sci. U. S. A. 106, 5731–5736. doi: 10.1073/pnas.0812141106
Cui, L., Yoshioka, Y., Suyari, O., Kohno, Y., Zhang, X., Adachi, Y., et al. (2008). Relevant expression of Drosophila heme oxygenase is necessary for the normal development of insect tissues. Biochem. Biophys. Res. Commun. 377, 1156–1161. doi: 10.1016/j.bbrc.2008.10.133
Damulewicz, M., Loboda, A., Jozkowicz, A., Dulak, J., and Pyza, E. (2017a). Interactions between the circadian clock and heme oxygenase in the retina of Drosophila melanogaster. Mol. Neurobiol. 54, 4953–4962. doi: 10.1007/s12035-016-0026-9
Damulewicz, M., Loboda, A., Jozkowicz, A., Dulak, J., and Pyza, E. (2017b). Haeme oxygenase protects against UV light DNA damages in the retina in clock-dependent manner. Sci. Rep. 7:5197. doi: 10.1038/s41598-017-05418-6
Damulewicz, M., Świątek, M., Łoboda, A., Dulak, J., Bilska, B., Przewłocki, R., et al. (2019). Daily regulation of phototransduction, circadian clock, DNA repair, and immune gene expression by heme oxygenase in the retina of Drosophila. Genes 10:6. doi: 10.3390/genes10010006
Deepashree, S., Shivanandappa, T., and Ramesh, S. R. (2017). Life history traits of an extended longevity phenotype of Drosophila melanogaster. Curr. Aging Sci. 10, 224–238. doi: 10.2174/1874609810666170209111350
Duvigneau, J. C., Esterbauer, H., and Kozlov, A. V. (2019). Role of heme oxygenase as a modulator of heme-mediated pathways. Antioxidants 8:475. doi: 10.3390/antiox8100475
El-Brolosy, M. A., and Stainier, D. Y. R. (2017). Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 13:e1006780. doi: 10.1371/journal.pgen.1006780
Ewing, J. F., and Maines, M. D. (1997). Histochemical localization of heme oxygenase-2 protein and mRNA expression in rat brain. Brain Res. Protocol. 1, 165–174. doi: 10.1016/S1385-299X(96)00027-X
Fast, I., and Rosenkrantz, D. Temperature-dependent small RNA expression in Drosophila melanogaster. (2018). RNA Biology. 15, 308–313. doi: 10.1080/15476286.2018.1429881
Feyereisen, R. (1999). Insect P450 enzymes. Annu. Rev. Entomol. 44, 507–533. doi: 10.1146/annurev.ento.44.1.507
Giraud, S., Bonod-Bidaud, C., Wesolowski-Louvel, M., and Stepien, G. (1998). Expression of human ANT2 gene in highly proliferative cells: GRBOX, a new transcriptional element, is involved in the regulation of glycolytic ATP import into mitochondria. J. Mol. Biol. 281, 409–418. doi: 10.1006/jmbi.1998.1955
Guenthner, C. J., Bickar, D., and Harrington, M. E. (2009). Heme reversibly damps PERIOD2 rhythms in mouse suprachiasmatic nucleus explants. Neuroscience 164, 832–841. doi: 10.1016/j.neuroscience.2009.08.022
Guichet, A., Peri, F., and Roth, S. (2001). Stable anterior anchoring of the oocyte nucleus is required to establish dorsoventral polarity of the Drosophila egg. Dev. Biol. 237, 93–106. doi: 10.1006/dbio.2001.0354
Han, E., Platonov, A. G., Akhalaya, M. Y., Son, J. Y., An, J. Y., and Yun, Y. S. (2005). Tissue-specific changes in heme oxygenase activity and level of nonprotein thiols in C57Bl/6 mice after whole-body γ-irradiation. Bull. Exp. Biol. Med. 140, 341–344. doi: 10.1007/s10517-005-0485-x
Han, Y., Platonov, A., Akhalaia, M., Yun, Y.-S., and Song, J.-Y. (2005). Differential effect of γ-radiation-induced heme oxygenase-1 activity in female and male C57BL/6 mice. J. Korean Med. Sci. 20, 535–541. doi: 10.3346/jkms.2005.20.4.535
Ida, H., Suyari, O., Shimamura, M., Tien Tai, T., Yamaguchi, M., and Taketani, S. (2013). Genetic link between heme oxygenase and the signaling pathway of DNA damage in Drosophila melanogaster. Tohoku J. Exp. Med. 231, 117–125. doi: 10.1620/tjem.231.117
Iga, M., and Kataoka, H. (2012). Recent studies on insect hormone metabolic pathways mediated by cytochrome P450 enzymes. Biol. Pharm. Bull. 35, 838–843. doi: 10.1248/bpb.35.838
Ihara, N., Akagi, R., Ejiri, K., Kudo, T., Furuyama, K., and Fujita, H. (1998). Developmental changes of gene expression in heme metabolic enzymes in rat placenta. FEBS Lett. 439, 163–167. doi: 10.1016/S0014-5793(98)01324-6
Kagan, V. E., Kozlov, A. V., Tyurina, Y. Y., Shvedova, A. A., and Yalowich, J. C. (2001). Antioxidant mechanisms of nitric oxide against iron-catalyzed oxidative stress in cells. Antioxid. Redox Signal. 3, 189–202. doi: 10.1089/152308601300185160
Kawashima, A., Oda, Y., Yachie, A., Koizumi, S., and Nakanishi, I. (2002). Heme oxygenase-1 deficiency: the first autopsy case. Hum. Pathol. 33, 125–130. doi: 10.1053/hupa.2002.30217
Kikuchi, G., Yoshida, T., and Noguchi, M. (2005). Heme oxygenase and heme degradation. Biochem. Biophys. Res. Commun. 338, 558–567. doi: 10.1016/j.bbrc.2005.08.020
Kim, H. J., Khalimonchuk, O., Smith, P. M., and Winge, D. R. (2012). Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta 1823, 1604–1616. doi: 10.1016/j.bbamcr.2012.04.008
Kimpara, T., Takeda, A., Yamaguchi, T., Arai, H., Okita, N., Takase, S., et al. (2000). Increased bilirubins and their derivatives in cerebrospinal fluid in Alzheimer’s disease. Neurobiol. Aging 21, 551–554. doi: 10.1016/s0197-4580(00)00128-7
Klemz, R., Reischl, S., Wallach, T., Witte, N., Jürchott, K., Klemz, S., et al. (2017). Reciprocal regulation of carbon monoxide metabolism and the circadian clock. Nat. Struct. Mol. Biol. 24, 15–22. doi: 10.1038/nsmb.3331
Klouche, K., Morena, M., Canaud, B., Descomps, B., Beraud, J. J., and Cristol, J. P. (2004). Mechanism of in vitro heme-induced LDL oxidation: effects of antioxidants. Eur. J. Clin. Investig. 34, 619–625. doi: 10.1111/j.1365-2362.2004.01395.x
Kovtunovych, G., Eckhaus, M. A., Ghosh, M. C., Ollivierre-Wilson, H., and Rouault, T. A. (2010). Dysfunction of the heme recycling system in heme oxygenase 1-deficient mice: effects on macrophage viability and tissue iron distribution. Blood 116, 6054–6062. doi: 10.1182/blood-2010-03-272138
Kumar, S., and Bandyopadhyay, U. (2005). Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 157, 175–188. doi: 10.1016/j.toxlet.2005.03.004
Li, H., Janssen, J., Waegeneer, M., Kolluru, S. S., and Davie, K. (2022). Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 375:eabk2432. doi: 10.1126/science.abk2432
Linzke, N., Schumacher, A., Woidacki, K., Croy, B. A., and Zenclussen, A. C. (2014). Carbon monoxide promotes proliferation of uterine natural killer cells and remodeling of spiral arteries in pregnant hypertensive heme oxygenase-1 mutant mice. Hypertension 63, 580–588. doi: 10.1161/HYPERTENSIONAHA.113.02403
Llorens, J. V., Metzendorf, C., Missirlis, F., and Lind, M. I. (2015). Mitochondrial iron supply is required for the developmental pulse of ecdysone biosynthesis that initiates metamorphosis in Drosophila melanogaster. J. Biol. Inorg. Chem. 20, 1229–1238. doi: 10.1007/s00775-015-1302-2
Loboda, A., Damulewicz, M., Pyza, E., Jozkowicz, A., and Dulak, J. (2016). Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell. Mol. Life Sci. 73, 3221–3247. doi: 10.1007/s00018-016-2223-0
Luu Hoang, K. N., Anstee, J. E., and Arnold, J. N. (2021). The diverse roles of heme oxygenase-1 in tumor progression. Front. Immunol. 12:658315. doi: 10.3389/fimmu.2021.658315
Magwere, T., Chapman, T., and Partridge, L. (2004). Sex differences in the effect of dietary restriction on life span and mortality rates in female and male Drosophila melanogaster. J. Gerontol. A 59, B3–B9. doi: 10.1093/gerona/59.1.B3
Mancuso, C. (2004). Heme oxygenase and its products in the nervous system. Antioxid. Redox Signal. 6, 878–887. doi: 10.1089/ars.2004.6.878
Mandilaras, K., and Missirlis, F. (2012). Genes for iron metabolism influence circadian rhythms in Drosophila melanogaster†. Metallomics 4, 928–936. doi: 10.1039/c2mt20065a
Massie, H. R., Aiello, V. R., and Williams, T. R. (1985). Iron accumulation during development and ageing of Drosophila. Mech. Ageing Dev. 29, 215–220. doi: 10.1016/0047-6374(85)90020-X
McGuire, S. E., Roman, G., and Davis, R. L. (2004). Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 20, 384–391. doi: 10.1016/j.tig.2004.06.012
Mena, N. P., Urrutia, P. J., Lourido, F., Carrasco, C. M., and Núñez, M. T. (2015). Mitochondrial iron homeostasis and its dysfunctions in neurodegenerative disorders. Mitochondrion 21, 92–105. doi: 10.1016/j.mito.2015.02.001
Mense, S. M., and Zhang, L. (2006). Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 16, 681–692. doi: 10.1038/sj.cr.7310086
Millington, J. W., and Rideout, E. J. (2018). Sex differences in Drosophila development and physiology. Curr. Opin. Physiol. 6, 46–56. doi: 10.1016/j.cophys.2018.04.002
Mołoń, M., Dampc, J., Kula-Maximenko, M., Zebrowski, J., Mołoń, A., Dobler, R., et al. (2020). Effects of temperature on lifespan of Drosophila melanogaster from different genetic backgrounds: links between metabolic rate and longevity. Insects 11:470. doi: 10.3390/insects11080470
Namiki, T., Niwa, R., Sakudoh, T., Shirai, K., Takeuchi, H., and Kataoka, H. (2005). Cytochrome P450 CYP307A1/spook: a regulator for ecdysone synthesis in insects. Biochem. Biophys. Res. Commun. 337, 367–374. doi: 10.1016/j.bbrc.2005.09.043
Nitti, M., Piras, S., Brondolo, L., Marinari, U. M., Pronzato, M. A., and Furfaro, A. L. (2018). Heme oxygenase 1 in the nervous system: does it favor neuronal cell survival or induce neurodegeneration? Int. J. Mol. Sci. 19:2260. doi: 10.3390/ijms19082260
Niveditha, S., Deepashree, S., Ramesh, S. R., and Shivanandappa, T. (2017). Sex differences in oxidative stress resistance in relation to longevity in Drosophila melanogaster. J. Comp. Physiol. B 187, 899–909. doi: 10.1007/s00360-017-1061-1
Ohnishi, M., Katsuki, H., Unemura, K., Izumi, Y., Kume, T., Takada-Takatori, Y., et al. (2010). Heme oxygenase-1 contributes to pathology associated with thrombin-induced striatal and cortical injury in organotypic slice culture. Brain Res. 1347, 170–178. doi: 10.1016/j.brainres.2010.05.077
Padmanaban, G., Venkateswar, V., and Rangarajan, P. N. (1989). Haem as a multifunctional regulator. Trends Biochem. Sci. 14, 492–496. doi: 10.1016/0968-0004(89)90182-5
Phom, L., Achumi, B., Alone, D. P., Muralidhara,, and Yenisetti, S. C. (2014). Curcumin’s neuroprotective efficacy in Drosophila model of idiopathic Parkinson’s disease is phase specific: implication of its therapeutic effectiveness. Rejuvenation Res. 17, 481–489. doi: 10.1089/rej.2014.1591
Poss, K. D., and Tonegawa, S. (1997a). Heme oxygenase 1 is required for mammalian iron reutilization. Proc. Natl. Acad. Sci. U. S. A. 94, 10919–10924. doi: 10.1073/pnas.94.20.10919
Poss, K. D., and Tonegawa, S. (1997b). Reduced stress defense in heme oxygenase 1-deficient cells. Proc. Natl. Acad. Sci. U. S. A. 94, 10925–10930. doi: 10.1073/pnas.94.20.10925
Radhakrishnan, N., Yadav, S. P., Sachdeva, A., Wada, T., and Yachie, A. (2011). An interesting tetrad of asplenia, inflammation, hemolysis, and nephritis. Pediatr. Hematol. Oncol. 28, 723–726. doi: 10.3109/08880018.2011.613979
Ren, H., Han, R., Liu, X., Wang, L., Koehler, R. C., and Wang, J. (2021). Nrf2-BDNF-TrkB pathway contributes to cortical hemorrhage-induced depression, but not sex differences. J. Cereb. Blood Flow Metab. 41, 3288–3301. doi: 10.1177/0271678X211029060
Rewitz, K. F., Rybczynski, R., Warren, J. T., and Gilbert, L. I. (2006). Identification, characterization and developmental expression of Halloween genes encoding P450 enzymes mediating ecdysone biosynthesis in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 36, 188–199. doi: 10.1016/j.ibmb.2005.12.002
Sabová, L., Zeman, I., Supek, F., and Kolarov, J. (1993). Transcriptional control of AAC3 gene encoding mitochondrial ADP/ATP translocator in Saccharomyces cerevisiae by oxygen, heme and ROX1 factor. Eur. J. Biochem. 213, 547–553. doi: 10.1111/j.1432-1033.1993.tb17793.x
Schaer, D. J., Buehler, P. W., Alayash, A. I., Belcher, J. D., and Vercellotti, G. M. (2013). Hemolysis and free hemoglobin revisited: exploring hemoglobin and hemin scavengers as a novel class of therapeutic proteins. Blood 121, 1276–1284. doi: 10.1182/blood-2012-11-451229
Schipper, H. M., Bennett, D. A., Liberman, A., Bienias, J. L., Schneider, J. A., Kelly, J., et al. (2006). Glial heme oxygenase-1 expression in Alzheimer disease and mild cognitive impairment. Neurobiol. Aging 27, 252–261. doi: 10.1016/j.neurobiolaging.2005.01.016
Schipper, H. M., Cissé, S., and Stopa, E. G. (1995). Expression of heme oxygenase-1 in the senescent and Alzheimer-diseased brain. Ann. Neurol. 37, 758–768. doi: 10.1002/ana.410370609
Schipper, H. M., Song, W., Tavitian, A., and Cressatti, M. (2019). The sinister face of heme oxygenase-1 in brain aging and disease. Prog. Neurobiol. 172, 40–70. doi: 10.1016/j.pneurobio.2018.06.008
Schipper, H. M., Song, W., Zukor, H., Hascalovici, J. R., and Zeligman, D. (2009). Heme oxygenase-1 and neurodegeneration: expanding frontiers of engagement. J. Neurochem. 110, 469–485. doi: 10.1111/j.1471-4159.2009.06160.x
Schmitt, T. H., Frezzatti, W. A., and Schreier, S. (1993). Hemin-induced lipid membrane disorder and increased permeability: a molecular model for the mechanism of cell lysis. Arch. Biochem. Biophys. 307, 96–103. doi: 10.1006/abbi.1993.1566
Sengupta, A., Hon, T., and Zhang, L. (2005). Heme deficiency suppresses the expression of key neuronal genes and causes neuronal cell death. Brain Res. Mol. Brain Res. 137, 23–30. doi: 10.1016/j.molbrainres.2005.02.007
Shibahara, S., Han, F., Li, B., and Takeda, K. (2007). Hypoxia and heme oxygenases: oxygen sensing and regulation of expression. Antioxid. Redox Signal. 9, 2209–2226. doi: 10.1089/ars.2007.1784
Smith, A. G., Raven, E. L., and Chernova, T. (2011). The regulatory role of heme in neurons. Metallomics 3, 955–962. doi: 10.1039/c1mt00085c
Song, L., Song, W., and Schipper, H. M. (2007). Astroglia overexpressing heme oxygenase-1 predispose co-cultured PC12 cells to oxidative injury. J. Neurosci. Res. 85, 2186–2195. doi: 10.1002/jnr.21367
Sztal, T., Chung, H., Berger, S., Currie, P. D., Batterham, P., and Daborn, P. J. (2012). A cytochrome P450 conserved in insects is involved in cuticle formation. PLoS One 7:e36544. doi: 10.1371/journal.pone.0036544
Tower, J., Pomatto, L. C. D., and Davies, K. J. A. (2020). Sex differences in the response to oxidative and proteolytic stress. Redox Biol. 31:101488. doi: 10.1016/j.redox.2020.101488
Tronel, C., Rochefort, G. Y., Arlicot, N., Bodard, S., Chalon, S., and Antier, D. (2013). Oxidative stress is related to the deleterious effects of heme oxygenase-1 in an in vivo neuroinflammatory rat model. Oxidative Med. Cell. Longev. 2013:264935. doi: 10.1155/2013/264935
Vallelian, F., Deuel, J. W., Opitz, L., Schaer, C. A., Puglia, M., Lönn, M., et al. (2015). Proteasome inhibition and oxidative reactions disrupt cellular homeostasis during heme stress. Cell Death Differ. 22, 597–611. doi: 10.1038/cdd.2014.154
Warren, J. T., Petryk, A., Marqués, G., Jarcho, M., Parvy, J.-P., Dauphin-Villemant, C., et al. (2002). Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 99, 11043–11048. doi: 10.1073/pnas.162375799
Watanabe, S., Akagi, R., Mori, M., Tsuchiya, T., and Sassa, S. (2004). Marked developmental changes in heme oxygenase-1 (HO-1) expression in the mouse placenta: correlation between HO-1 expression and placental development. Placenta 25, 387–395. doi: 10.1016/j.placenta.2003.10.012
Waza, A. A., Hamid, Z., Ali, S., Bhat, S. A., and Bhat, M. A. (2018). A review on heme oxygenase-1 induction: is it a necessary evil. Inflamm. Res. 67, 579–588. doi: 10.1007/s00011-018-1151-x
Wegiel, B., Hauser, C. J., and Otterbein, L. E. (2015). Heme as a danger molecule in pathogen recognition. Free Radic. Biol. Med. 89, 651–661. doi: 10.1016/j.freeradbiomed.2015.08.020
Wu, Y.-H., and Hsieh, H.-L. (2022). Roles of heme oxygenase-1 in neuroinflammation and brain disorders. Antioxidants 11:923. doi: 10.3390/antiox11050923
Yachie, A., Niida, Y., Wada, T., Igarashi, N., Kaneda, H., Toma, T., et al. (1999). Oxidative stress causes enhanced endothelial cell injury in human heme oxygenase-1 deficiency. J. Clin. Invest. 103, 129–135. doi: 10.1172/JCI4165
Yang, J., Kim, K. D., Lucas, A., Drahos, K. E., Santos, C. S., Mury, S. P., et al. (2008). A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol. Cell. Biol. 28, 4697–4711. doi: 10.1128/MCB.00236-08
Yang, Q., and Wang, W. (2022). The nuclear translocation of heme oxygenase-1 in human diseases. Front. Cell Dev. Biol. 10:890186. doi: 10.3389/fcell.2022.890186
Yoshida, T., and Migita, C. T. (2000). Mechanism of heme degradation by heme oxygenase. J. Inorg. Biochem. 82, 33–41. doi: 10.1016/s0162-0134(00)00156-2
Zakhary, R., Poss, K. D., Jaffrey, S. R., Ferris, C. D., Tonegawa, S., and Snyder, S. H. (1997). Targeted gene deletion of heme oxygenase 2 reveals neural role for carbon monoxide. Proc. Natl. Acad. Sci. U. S. A. 94, 14848–14853. doi: 10.1073/pnas.94.26.14848
Zenclussen, M. L., Anegon, I., Bertoja, A. Z., Chauveau, C., Vogt, K., Gerlof, K., et al. (2006). Over-expression of heme oxygenase-1 by adenoviral gene transfer improves pregnancy outcome in a murine model of abortion. J. Reprod. Immunol. 69, 35–52. doi: 10.1016/j.jri.2005.10.001
Zenclussen, M. L., Casalis, P. A., El-Mousleh, T., Rebelo, S., Langwisch, S., Linzke, N., et al. (2011). Haem oxygenase-1 dictates intrauterine fetal survival in mice via carbon monoxide. J. Pathol. 225, 293–304. doi: 10.1002/path.2946
Zenclussen, M. L., Linzke, N., Schumacher, A., Fest, S., Meyer, N., Casalis, P. A., et al. (2015). Heme oxygenase-1 is critically involved in placentation, spiral artery remodeling, and blood pressure regulation during murine pregnancy. Front. Pharmacol. 5:291:291. doi: 10.3389/fphar.2014.00291
Zhang, X., Sato, M., Sasahara, M., Migita, C. T., and Yoshida, T. (2004). Unique features of recombinant heme oxygenase of Drosophila melanogaster compared with those of other heme oxygenases studied. Eur. J. Biochem. 271, 1713–1724. doi: 10.1111/j.1432-1033.2004.04077.x
Zhao, H., Wong, R. J., Kalish, F. S., Nayak, N. R., and Stevenson, D. K. (2009). Effect of heme oxygenase-1 deficiency on placental development. Placenta 30, 861–868. doi: 10.1016/j.placenta.2009.07.012
Keywords: neurons, glia, cap’n’collar, survival, behavior, sleep, aging
Citation: Bilska B, Damulewicz M, Abaquita TAL and Pyza E (2023) Changes in heme oxygenase level during development affect the adult life of Drosophila melanogaster. Front. Cell. Neurosci. 17:1239101. doi: 10.3389/fncel.2023.1239101
Edited by:
Weilin Liu, Fujian University of Traditional Chinese Medicine, ChinaReviewed by:
Christian Wegener, Julius Maximilian University of Würzburg, GermanyAnnekathrin Widmann, University of Göttingen, Germany
Copyright © 2023 Bilska, Damulewicz, Abaquita and Pyza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elzbieta Pyza, ZWx6YmlldGEucHl6YUB1ai5lZHUucGw=
†These authors have contributed equally to this work
 Bernadetta Bilska
Bernadetta Bilska Milena Damulewicz
Milena Damulewicz Terence Al L. Abaquita
Terence Al L. Abaquita Elzbieta Pyza
Elzbieta Pyza