- 1Department of Neurodegenerative Diseases, Beckman Research Institute of City of Hope, Duarte, CA, United States
- 2Irell and Manella Graduate School of Biological Sciences, Beckman Research Institute of City of Hope, Duarte, CA, United States
Introduction
Tissue-resident macrophages (TRMs) are specialized myeloid cells that adapt to the local microenvironment and perform both core macrophage functions, such as phagocytosis and immune surveillance, as well as tissue-specific roles (Troutman et al., 2021). The identity of TRMs is established by a combination of their ontogeny (or lineage) and the surrounding tissue environment that provides distinct signaling cues to educate TRMs toward more specialized functions, such as synaptic pruning by microglia (Butovsky and Weiner, 2018; Prinz et al., 2019; Troutman et al., 2021; Paolicelli et al., 2022). Mechanistically, signal-induced transcription factor activity leads to tissue-specific chromatin remodeling and enhancer activation superimposed on core macrophage gene expression programs (Lavin et al., 2014; Troutman et al., 2021). However, human cell-based experimental systems to probe individual subtypes of TRMs, such as those of brain macrophages, as well as delineate the molecular mechanisms underlying TRM specialization are largely lacking. In this Opinion, we propose a platform of induced pluripotent stem cell (iPSC)-derived neuroimmune organoids to establish the diversity of human cell-based brain TRM models and study their roles in tissue homeostasis and disease.
Brain macrophage identity and diversity
The brain macrophage population is composed of parenchymal microglia and border-associated macrophages (BAMs), also known as central nervous system (CNS)-associated macrophages (CAMs), that together maintain homeostasis of the CNS and its surrounding tissues (Gosselin et al., 2017; Butovsky and Weiner, 2018; Kierdorf et al., 2019; Prinz et al., 2019; Troutman et al., 2021). Microglia develop from the yolk sac erythromyeloid progenitors during primitive hematopoiesis and populate the brain parenchyma early in development (Butovsky and Weiner, 2018; Ginhoux and Garel, 2018; Paolicelli et al., 2022). Although the microglial population is maintained by cell proliferation with no input from peripheral myeloid cells under homeostatic conditions, microglia display substantial spatial and temporal heterogeneity (Grabert et al., 2016; Butovsky and Weiner, 2018; Friedman et al., 2018; Hammond et al., 2019; Young et al., 2021; Paolicelli et al., 2022). For example, cerebellar microglia exhibit increased debris clearance as compared to forebrain microglia as well as depend on colony-stimulating factor 1 (CSF-1) signaling, whereas forebrain microglia survive CSF-1 depletion (Ayata et al., 2018; Kana et al., 2019). Moreover, lipid-droplet-accumulating microglia arise in aging, whereas disease-associated microglia (DAM) are characteristic of Alzheimer's disease (AD) (Deczkowska et al., 2018; McQuade and Blurton-Jones, 2019; Marschallinger et al., 2020; Silvin et al., 2022). BAMs comprise macrophages at the boundaries of the CNS, including meninges, vasculature, and the choroid plexus (Kierdorf et al., 2019; Mildenberger et al., 2022). Like microglia, BAMs originate from yolk sac erythromyeloid progenitors although the dynamics of BAM subtype specification and maintenance vary (Goldmann et al., 2016; Utz et al., 2020; Masuda et al., 2022). Whereas meningeal and choroid plexus macrophages are established prenatally, perivascular macrophages attain their full identity postnatally, when the Virchow-Robin space of their residence is established (Masuda et al., 2022). Moreover, while meningeal and perivascular macrophages are stably maintained into adulthood, choroid plexus macrophages are gradually replaced by peripheral bone marrow-derived cells (Goldmann et al., 2016; Prinz et al., 2019; Van Hove et al., 2019). BAMs also exhibit transcriptional heterogeneity, presumably indicating functional specialization to support their tissues of residence (Mrdjen et al., 2018; Li et al., 2019; Van Hove et al., 2019). The importance of tissue-specific signaling for maintaining distinct TRM populations in the brain is exemplified by microglial but not BAM dependence on tumor growth factor β (TGF-β) signaling (Thion and Garel, 2020; Utz et al., 2020; Brioschi et al., 2023). Importantly, TGF-β signaling promotes expression of the spalt-like transcription factor 1 (SALL1), a master regulator of microglia identity. SALL1 is not expressed in other macrophage populations except for Kolmer's epiplexus cells that populate the choroid plexus and might, in fact, be a subtype of microglia (Butovsky et al., 2014; Buttgereit et al., 2016; Van Hove et al., 2019; Troutman et al., 2021; Brioschi et al., 2023). Interestingly, culturing microglia ex vivo leads to downregulation of SALL1 expression and a substantial loss of microglia gene expression signatures (Gosselin et al., 2017).
Species-specific divergence of murine and human microglia
It is well-established that murine and human microglia exhibit considerable species-specific divergence, hindering therapeutic development targeting microglia (Smith and Dragunow, 2014; Galatro et al., 2017; Geirsdottir et al., 2019). For example, human microglia exhibit higher transcriptional heterogeneity than do murine microglia as well as distinct immune function- and aging-associated gene expression signatures (Galatro et al., 2017; Geirsdottir et al., 2019). Moreover, risk factor genes implicated in brain diseases, such as AD, that are associated with microglial functions are poorly conserved between rodents and humans (Mancuso et al., 2019; Wightman et al., 2021). A key microglial surface receptor triggering receptor expressed on myeloid cells 2 (TREM2) shares < 60% of amino acid identity between murine and human variants (Mancuso et al., 2019). Likewise, gene expression programs of mouse DAM and human AD microglia (HAM) implicated in AD progression share few similarities with each other (Srinivasan et al., 2020). Given these species-specific differences between murine and human myeloid cell biology, there is a great need to develop robust human cell-based models that could faithfully recapitulate human brain macrophage biology.
Human iPSC-derived macrophages and microglia
Since its inception, the iPSC technology has offered an almost unlimited access to in vitro models of human brain cells that are otherwise difficult to obtain from primary human brain tissue (Takahashi et al., 2007; Yu et al., 2007; Shi et al., 2017; Li et al., 2018; Li and Shi, 2020; Tong et al., 2021). Various protocols to differentiate macrophage- and microglia-like cells (iMGs) from iPSCs have been developed (Abud et al., 2017; Lee et al., 2018; McQuade et al., 2018; Pocock and Piers, 2018; Hasselmann and Blurton-Jones, 2020; Tang et al., 2022; Washer et al., 2022). Notably, iPSC differentiation into iMGs entails mesodermal specification and transition of the differentiating cells through an erythromyeloid-like progenitor, reminiscent of primitive hematopoiesis (Buchrieser et al., 2017; Lee et al., 2018). Embryonic-like origin of iMGs indicates their applicability to study microglial and BAM biology, especially given that peripheral macrophages arising via definitive hematopoiesis do not fully attain microglial identity when transplanted into the mouse brain and exhibit distinct gene expression profiles, such as high levels of apolipoprotein E (APOE) expression (Bennett et al., 2018). However, embryonic iMG ontogeny is insufficient to establish their brain macrophage identity. Indeed, iMGs cultured in isolation exhibit limited expression of key microglial markers SALL1 and the transmembrane protein 119 (TMEM119), indicating that such iMGs have not yet attained bona fide microglial identity (Hasselmann et al., 2019). To overcome this limitation, iMGs have been successfully transplanted into the rodent brain to derive chimeric mouse models (Hasselmann et al., 2019; Mancuso et al., 2019; Svoboda et al., 2019; Fattorelli et al., 2021). Transplanted iMGs exhibit complex morphology and increased expression of key microglial genes, including SALL1 and TMEM119, indicating the presence of correct intrinsic programs in iMGs that can be activated by the appropriate environment (Hasselmann et al., 2019). Although chimeric mouse models enable studying the interactions between human microglia and mouse brain cells, the implications of modeling human-to-mouse cell-to-cell interactions remain to be clarified. It is thus highly desired to develop human cell-based three-dimensional (3D) models of the brain tissue, so that iMGs could both mature to attain their full identity and establish interactions with other brain cell types that are of human origin.
Achieving iPSC-derived brain macrophage diversity using regionally-patterned neuroimmune brain organoids
Brain organoids (BOs) are iPSC-derived 3D self-organizing assemblies of brain cell types and are used to model human brain development and disease (Lancaster and Knoblich, 2014). Given that myeloid cells arise from the mesoderm, whereas BOs are derived from the neuroectodermal lineage, various strategies to obtain iMG-containing neuroimmune organoids have been developed (Abud et al., 2017; Ao et al., 2021; Popova et al., 2021; Xu et al., 2021; Cakir et al., 2022). A simple and well-controlled approach to introduce iMGs into BOs involves iMG differentiation from iPSCs followed by iMG seeding onto BOs and infiltration into the neural tissue, although other protocols have been developed as well (Abud et al., 2017; Xu et al., 2021; Cakir et al., 2022; Zhang et al., 2023). So far, neuroimmune organoids have primarily been established using BOs derived by unguided differentiation, which yields organoids resembling a mixture of multiple brain regions, as well as using cortical BOs (Abud et al., 2017; Ao et al., 2021; Xu et al., 2021; Cakir et al., 2022; Jin et al., 2022). Carefully crafted guided differentiation approaches can be used to obtain BOs that resemble distinct brain regions and associated tissues, such as the forebrain, cerebellum, retina, choroid plexus, and others (Lullo and Kriegstein, 2017; Pellegrini et al., 2020; Sridhar et al., 2020; Nayler et al., 2021; Lee et al., 2022). Introduction of iMGs into such regionally-patterned BOs would provide a distinct environment to educate iMGs, so that diverse brain macrophage populations may be established (Figure 1). For example, cortical, cerebellar, retinal, and spinal cord neuroimmune organoids may be used to educate iMGs toward different microglia subtypes, whereas choroid plexus neuroimmune organoids may promote the identity of choroid plexus macrophages. Moreover, 3D blood-brain barrier (BBB) spheroids and in vitro BBB models may be used to educate iMGs toward perivascular macrophage identity (Cho et al., 2017; Blanchard et al., 2020). Having established regionally-patterned neuroimmune organoids with their distinct iMG populations, in-depth analysis of iMG morphology, functionality, response to stimulation, and gene expression programs would reveal how tissue residency promotes iMG specialization and how it compares to that of in vivo brain macrophage specialization. Do iMGs exhibit increased phagocytic activity in cerebellar neuroimmune organoids as compared to iMGs in cortical neuroimmune organoids? Do iMGs resemble distinct choroid plexus macrophage states, including Kolmer's epiplexus cells, in choroid plexus neuroimmune organoids? Do iMGs regulate cerebrospinal fluid production in choroid plexus neuroimmune organoids? These and other questions could be addressed by using the diverse neuroimmune organoid platform to elucidate the homeostatic roles of brain macrophages.
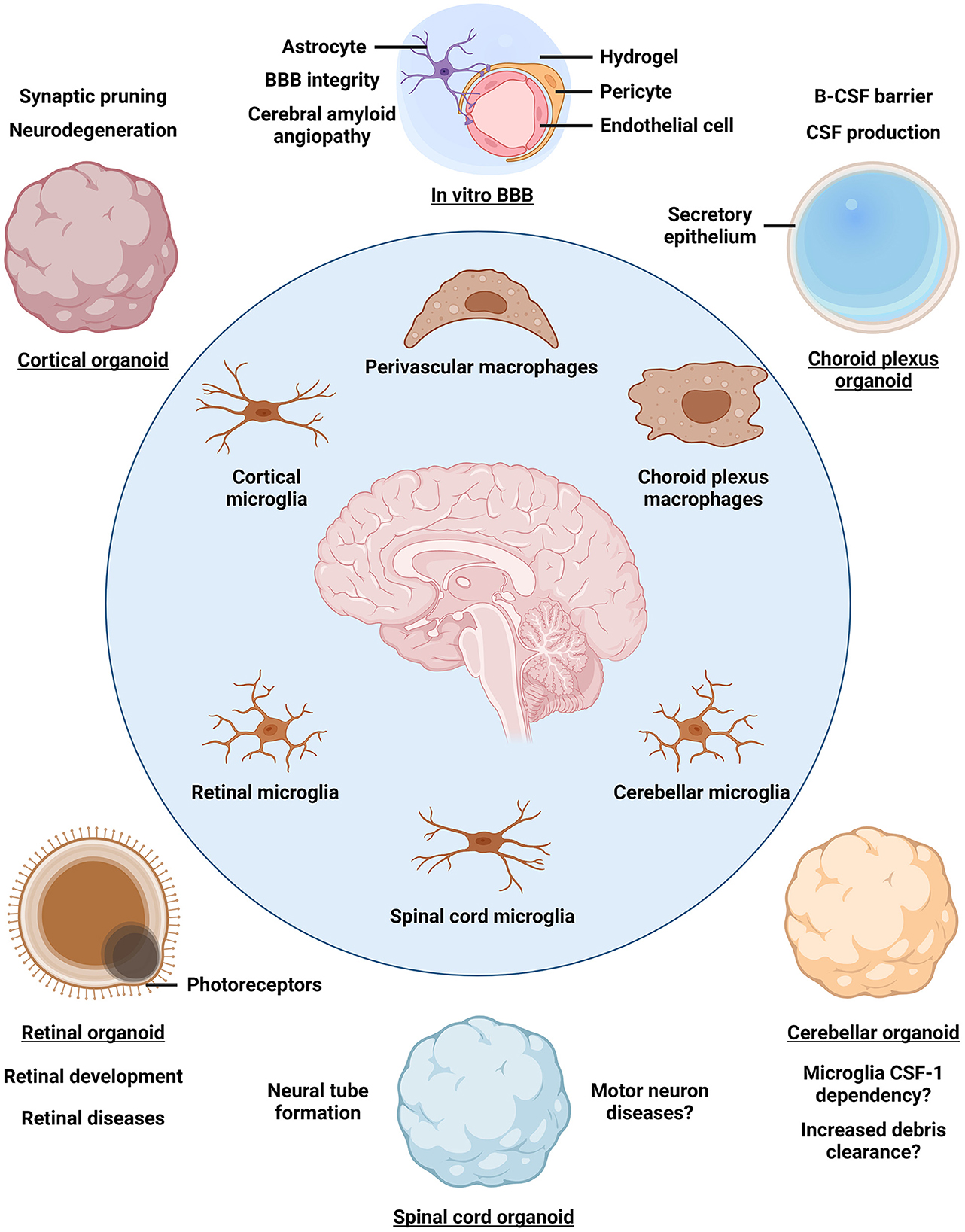
Figure 1. Organoid models for studying brain macrophage diversity. The identity of parenchymal microglia and border-associated macrophages is established by a combination of ontogeny and niche-specific environment that further educates brain macrophages to acquire tissue-relevant functionality. Incorporating induced pluripotent stem cell-derived macrophages into brain organoids patterned toward different brain regions may reveal niche-specific contributions to macrophage identity and function as well as enable disease modeling. BBB, blood-brain barrier; B-CSF barrier, blood-cerebrospinal fluid barrier; CSF, cerebrospinal fluid.
Modeling inflammation and brain diseases using neuroimmune organoids
In addition to studying the homeostatic roles of brain macrophages, neuroimmune organoids can be used to reveal macrophage phenotypes in the context of brain diseases (Figure 1). As the sole immune cells of the brain parenchyma, microglia mount an inflammatory response upon stimulation, which may be protective under normal conditions, but becomes excessive and detrimental in a diseased brain, causing widespread inflammation (Lyman et al., 2014). Indeed, neuroinflammation is considered a hallmark of AD and other brain diseases (Lyman et al., 2014; Kinney et al., 2018; Leng and Edison, 2021). Exposing neuroimmune organoids to bacterial-derived lipopolysaccharide, amyloid β, physical injury, or virus infection leads to iMG activation, indicating a common response to an inflammatory stimulus (Abud et al., 2017; Ao et al., 2021; Xu et al., 2021; Cakir et al., 2022). Regionally-patterned neuroimmune organoids may be derived from iPSCs of patients carrying disease mutations or genetic risk factors, such as APOE4, and tailored to define iMG roles in distinct processes associated with disease progression. For example, how do iMGs respond to vascular amyloid seeding in perivascular neuroimmune organoids, mimicking cerebral amyloid angiopathy characteristic of AD? Importantly, neuroimmune organoids may help clarify how activated microglia shift from playing protective roles at early stages of neurodegenerative diseases to exacerbating tissue damage by sustained pro-inflammatory cytokine secretion and gliosis as the disease progresses (Deczkowska et al., 2018; Kinney et al., 2018; Leng and Edison, 2021). Another common feature of neurodegenerative diseases is the breakdown of the BBB, leading to peripheral monocyte-derived macrophage infiltration into the brain (Fani Maleki and Rivest, 2019; Silvin et al., 2022). To model peripheral macrophage infiltration, neuroimmune organoids may be co-cultured with primary monocyte-derived macrophages isolated from the donors' blood. In this way, the interactions between brain-resident microglia and peripheral macrophages as well as their different roles in disease progression may be clarified. Indeed, a recent study indicates that the transcriptional cluster of DAM in neurodegeneration is, in fact, comprised of protective parenchymal microglia and detrimental inflammatory macrophages infiltrating into the brain (Silvin et al., 2022). Therefore, elucidating the contributions of different macrophage populations to disease progression may inform therapeutic development targeting neuroinflammation.
Discussion
Signaling from the tissue microenvironment is indispensable for establishing the TRM identity. Therefore, developing an array of iPSC-derived neuroimmune organoids resembling different brain regions and structures will provide a platform for studying brain macrophage diversity using iMGs. Moreover, neuroimmune organoids will enable modeling of human-specific cell-to-cell interactions between iMGs and other brain-resident cell types, so that novel molecular pathways that establish brain macrophage identity, specialization, and dysfunction in disease may be uncovered. We anticipate that the iPSC-derived iMG and organoid platforms will be widely applied to better define microglia and BAM states, perform high-throughput drug screening, and clarify disease-associated phenotypes. It will be important to compare any findings obtained from neuroimmune organoids to the transcriptomes of primary human brain macrophages to determine whether iMGs in neuroimmune organoids faithfully recapitulate human macrophage biology (Bian et al., 2020). Harnessing the human iPSC-based neuroimmune organoid technology will be especially beneficial for studying polygenic and sporadic diseases, such as AD, that cannot be easily recapitulated in animal models. In addition to the neuroimmune organoids discussed in this Opinion, further advances to the BO technology may enable modeling of meningeal structures and their resident dural and leptomeningeal macrophages (Kierdorf et al., 2019; Mildenberger et al., 2022), defining the roles of peripheral immune cells (Pasciuto et al., 2020; Zhang et al., 2022; Chen et al., 2023) and non-cellular factors (Chen et al., 2021; Liu et al., 2022) in microglia development and disease, and clarifying the transcriptomic and functional sexual dimorphism of microglia (Hanamsagar et al., 2017; Thion et al., 2018; Kelava et al., 2022). Finally, the same principles of using specialized neuroimmune organoids to establish a distinct iMG environment could also be applied to brain tumor organoids to better define the diversity and function of microglia and macrophages in the context of brain cancer (Hambardzumyan et al., 2016; Keane et al., 2021).
Author contributions
JC wrote the manuscript and prepared the figure. JC and YS edited and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Institute of Aging of the National Institutes of Health R01 AG056305, RF1 AG061794, R01 AG072291, and RF1 AG079307 to YS. JC is a predoctoral scholar in the Stem Cell Biology and Regenerative Medicine Research Training Program of the California Institute for Regenerative Medicine (CIRM).
Acknowledgments
The authors would like to thank Louise and Herbert Horvitz for their generosity and forethought and thank the Judy and Bernard Briskin Fund, the Christopher Family Endowed Innovation Fund, and the Sidell Kagan Foundation for their generous support. Figure 1 was created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abud, E. M., Ramirez, R. N., Martinez, E. S., Healy, L. M., Nguyen, C. H. H., Newman, S. A., et al. (2017). iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293. doi: 10.1016/j.neuron.2017.03.042
Ao, Z., Cai, H., Wu, Z., Song, S., Karahan, H., Kim, B., et al. (2021). Tubular human brain organoids to model microglia-mediated neuroinflammation. Lab Chip 21, 2751–2762. doi: 10.1039/D1LC00030F
Ayata, P., Badimon, A., Strasburger, H. J., Duff, M. K., Montgomery, S. E., Loh, Y., et al. (2018). Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 21, 1049–1060. doi: 10.1038/s41593-018-0192-3
Bennett, F. C., Bennett, M. L., Yaqoob, F., Mulinyawe, S. B., Grant, G. A., Gephart, M. H., et al. (2018). A combination of ontogeny and CNS environment establishes microglial identity. Neuron 98, 1170–1183. doi: 10.1016/j.neuron.2018.05.014
Bian, Z., Gong, Y., Huang, T., Lee, C. Z. W., Bian, L., Bai, Z., et al. (2020). Deciphering human macrophage development at single-cell resolution. Nature 582, 571–576. doi: 10.1038/s41586-020-2316-7
Blanchard, J. W., Bula, M., Davila-Velderrain, J., Akay, L. A., Zhu, L., Frank, A., et al. (2020). Reconstruction of the human blood-brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes. Nat. Med 26, 952–963. doi: 10.1038/s41591-020-0886-4
Brioschi, S., Belk, J. A., Peng, V., Molgora, M., Rodrigues, P. F., Nguyen, K. M., et al. (2023). A Cre-deleter specific for embryo-derived brain macrophages reveals distinct features of microglia and border macrophages. Immunity. 8, 28. doi: 10.1016/j.immuni.2023.01.028
Buchrieser, J., James, W., and Moore, M. D. (2017). Human induced pluripotent stem cell-derived macrophages share ontogeny with MYB-independent tissue-resident macrophages. Stem Cell Rep. 8, 334–345. doi: 10.1016/j.stemcr.2016.12.020
Butovsky, O., Jedrychowski, M. P., Moore, C. S., Cialic, R., Lanser, A. J., Gabriely, G., et al. (2014). Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143. doi: 10.1038/nn.3599
Butovsky, O., and Weiner, H. L. (2018). Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 19, 622–635. doi: 10.1038/s41583-018-0057-5
Buttgereit, A., Lelios, I., Yu, X., Vrohlings, M., Krakoski, N. R., Gautier, E. L., et al. (2016). Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 17, 1397–1406. doi: 10.1038/ni.3585
Cakir, B., Tanaka, Y., Ridvan Kiral, F., Xiang, Y., Dagliyan, O., Wang, J., et al. (2022). Expression of the transcription factor PU.1 induces the generation of microglia-like cells in human cortical organoids. Nat. Commun 13, 430. doi: 10.1038/s41467-022-28043-y
Chen, X., Firulyova, M., Manis, M., Herz, J., Smirnov, I., Aladyeva, E., et al. (2023). Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy. Nature 18, 1–10. doi: 10.1038/s41586-023-05788-0
Chen, X., Sun, G., Tian, E., Zhang, M., Davtyan, H., Beach, T. G., et al. (2021). Modeling sporadic Alzheimer's disease in human brain organoids under serum exposure. Adv. Sci. 8, e2101462. doi: 10.1002/advs.202101462
Cho, C.-F., Wolfe, J. M., Fadzen, C. M., Calligaris, D., Hornburg, K., Chiocca, E. A., et al. (2017). Blood-brain-barrier spheroids as an in vitro screening platform for brain-penetrating agents. Nat. Commun. 8, 15623. doi: 10.1038/ncomms15623
Deczkowska, A., Keren-Shaul, H., Weiner, A., Colonna, M., Schwartz, M., Amit, I., et al. (2018). Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell 173, 1073–1081. doi: 10.1016/j.cell.2018.05.003
Fani Maleki, A., and Rivest, S. (2019). Innate immune cells: monocytes, monocyte-derived macrophages and microglia as therapeutic targets for alzheimer's disease and multiple sclerosis. Front. Cell Neurosci. 13, 355. doi: 10.3389/fncel.2019.00355
Fattorelli, N., Martinez-Muriana, A., Wolfs, L., Geric, I., De Strooper, B., and Mancuso, R. (2021). Stem-cell-derived human microglia transplanted into mouse brain to study human disease. Nat. Protoc 16, 1013–1033. doi: 10.1038/s41596-020-00447-4
Friedman, B. A., Srinivasan, K., Ayalon, G., Meilandt, W. J., Lin, H., Huntley, M. A., et al. (2018). Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer's disease not evident in mouse models. Cell Rep. 22, 832–847. doi: 10.1016/j.celrep.2017.12.066
Galatro, T. F., Holtman, I. R., Lerario, A. M., Vainchtein, I. D., Brouwer, N., Sola, P. R., et al. (2017). Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 20, 1162–1171. doi: 10.1038/nn.4597
Geirsdottir, L., David, E., Keren-Shaul, H., Weiner, H, Cornelius Bohlen, S., Neuber, J., et al. (2019). Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell 179, 1609–1622. doi: 10.1016/j.cell.2019.11.010
Ginhoux, F., and Garel, S. (2018). The mysterious origins of microglia. Nat. Neurosci. 21, 897–899. doi: 10.1038/s41593-018-0176-3
Goldmann, T., Wieghofer, P., Jordão, M. J. C., Prutek, F., Hagemeyer, N., Frenzel, K., et al. (2016). Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805. doi: 10.1038/ni.3423
Gosselin, D., Skola, D., Coufal, N. G., Holtman, I. R., Schlachetzki, J. C. M., Sajti, E., et al. (2017). An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222. doi: 10.1126/science.aal3222
Grabert, K., Michoel, T., Karavolos, M. H., Clohisey, S., Baillie, J. K., Stevens, M. P., et al. (2016). Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 19, 504–516. doi: 10.1038/nn.4222
Hambardzumyan, D., Gutmann, D. H., and Kettenmann, H. (2016). The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 19, 20–27. doi: 10.1038/nn.4185
Hammond, T. R., Dufort, C., Dissing-Olesen, L., Giera, S., Young, A., Wysoker, A., et al. (2019). Single-Cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 50, 253–271. doi: 10.1016/j.immuni.2018.11.004
Hanamsagar, R., Alter, M. D., Block, C. S., Sullivan, H., Bolton, J. L., and Bilbo, S. D. (2017). Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. GLIA 65, 1504–1520. doi: 10.1002/glia.23176
Hasselmann, J., and Blurton-Jones, M. (2020). Human iPSC-derived microglia: a growing toolset to study the brain's innate immune cells. Glia 68, 721–739. doi: 10.1002/glia.23781
Hasselmann, J., Coburn, M. A., England, W., Velez, D. X. F., Shabestari, S. K., Tu, C. H., et al. (2019). Development of a chimeric model to study and manipulate human microglia in vivo. Neuron 103, 1016–1033. doi: 10.1016/j.neuron.2019.07.002
Jin, M., Xu, R., Wang, L., Alam, M. M., Ma, Z., Zhu, S., et al. (2022). Type-I-interferon signaling drives microglial dysfunction and senescence in human iPSC models of Down syndrome and Alzheimer's disease. Cell Stem Cell 29, 1135–1153. doi: 10.1016/j.stem.2022.06.007
Kana, V., Desland, F. A., Casanova-Acebes, M., Ayata, P., Badimon, A., Nabel, E., et al. (2019). CSF-1 controls cerebellar microglia and is required for motor function and social interaction. J. Exp Med. 216, 2265–2281. doi: 10.1084/jem.20182037
Keane, L., Cheray, M., Blomgren, K., and Joseph, B. (2021). Multifaceted microglia - key players in primary brain tumour heterogeneity. Nat. Rev. Neurol. 17, 243–259. doi: 10.1038/s41582-021-00463-2
Kelava, I., Chiaradia, I., Pellegrini, L., Kalinka, A. T., and Lancaster, M. A. (2022). Androgens increase excitatory neurogenic potential in human brain organoids. Nature 602, 112–116. doi: 10.1038/s41586-021-04330-4
Kierdorf, K., Masuda, T., Jordão, M. J. C., and Prinz, M. (2019). Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat. Rev. Neurosci. 20, 547–562. doi: 10.1038/s41583-019-0201-x
Kinney, J. W., Bemiller, S. M., Murtishaw, A. S., Leisgang, A. M., Salazar, A. M., and Lamb, B. T. (2018). Inflammation as a central mechanism in Alzheimer's disease. Transl. Res. Clin. Interv. 4, 575–590. doi: 10.1016/j.trci.2018.06.014
Lancaster, M. A., and Knoblich, J. A. (2014). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340. doi: 10.1038/nprot.2014.158
Lavin, Y., Winter, D., Blecher-Gonen, R., David, E., Keren-Shaul, H., Merad, M., et al. (2014). Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326. doi: 10.1016/j.cell.2014.11.018
Lee, C. Z. W., Kozaki, T., and Ginhoux, F. (2018). Studying tissue macrophages in vitro: are iPSC-derived cells the answer? Nat. Rev. Immunol. 18, 716–725. doi: 10.1038/s41577-018-0054-y
Lee, J.-H., Shin, H., Shaker, M. R., Kim, H. J., Park, S.-H., Kim, J. H., et al. (2022). Production of human spinal-cord organoids recapitulating neural-tube morphogenesis. Nat. Biomed Eng. 6, 435–448. doi: 10.1038/s41551-022-00868-4
Leng, F., and Edison, P. (2021). Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat. Rev. Neurol 17, 157–172. doi: 10.1038/s41582-020-00435-y
Li, L., Chao, J., and Shi, Y. (2018). Modeling neurological diseases using iPSC-derived neural cells: iPSC modeling of neurological diseases. Cell Tissue Res. 371, 143–151. doi: 10.1007/s00441-017-2713-x
Li, L., and Shi, Y. (2020). When glia meet induced pluripotent stem cells (iPSCs). Mol. Cell Neurosci. 109, 103565. doi: 10.1016/j.mcn.2020.103565
Li, Q., Cheng, Z., Zhou, L., Darmanis, S., Neff, N. F., Okamoto, J., et al. (2019). Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-Cell RNA sequencing. Neuron. 101, 207–223. doi: 10.1016/j.neuron.2018.12.006
Liu, C.-C., Zhao, J., Fu, Y., Inoue, Y., Ren, Y., Chen, Y., et al. (2022). Peripheral apoE4 enhances Alzheimer's pathology and impairs cognition by compromising cerebrovascular function. Nat. Neurosci. 25, 1020–1033. doi: 10.1038/s41593-022-01127-0
Lullo, D., and Kriegstein, E. (2017). The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci 18, 573–584. doi: 10.1038/nrn.2017.107
Lyman, M., Lloyd, D. G., Ji, X., Vizcaychipi, M. P., and Ma, D. (2014). Neuroinflammation: the role and consequences. Neurosci Res. 79, 1–12. doi: 10.1016/j.neures.2013.10.004
Mancuso, R., Van Den Daele, J., Fattorelli, N., Wolfs, L., Balusu, S., Burton, O., et al. (2019). Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat. Neurosci. 22, 2111–2116. doi: 10.1038/s41593-019-0525-x
Marschallinger, J., Iram, T., Zardeneta, M., Lee, S. E., Lehallier, B., Haney, M. S., et al. (2020). Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208. doi: 10.1038/s41593-019-0566-1
Masuda, T., Amann, L., Monaco, G., Sankowski, R., Staszewski, O., Krueger, M., et al. (2022). Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature 604, 740–748. doi: 10.1038/s41586-022-04596-2
McQuade, A., and Blurton-Jones, M. (2019). Microglia in Alzheimer's Disease: exploring how genetics and phenotype influence risk. J. Mol. Biol. 431, 1805–1817. doi: 10.1016/j.jmb.2019.01.045
McQuade, A., Coburn, M., Tu, C. H., Hasselmann, J., Davtyan, H., and Blurton-Jones, M. (2018). Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 13, 1–13. doi: 10.1186/s13024-018-0297-x
Mildenberger, W., Stifter, S. A., and Greter, M. (2022). Diversity and function of brain-associated macrophages. Curr. Opin. Immunol. 76, 102181. doi: 10.1016/j.coi.2022.102181
Mrdjen, D., Pavlovic, A., Hartmann, F. J., Schreiner, B., Utz, S. G., Leung, B. P., et al. (2018). High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395. doi: 10.1016/j.immuni.2018.01.011
Nayler, S., Agarwal, D., Curion, F., Bowden, R., and Becker, E. B. E. (2021). High-resolution transcriptional landscape of xeno-free human induced pluripotent stem cell-derived cerebellar organoids. Sci Rep 11, 1–17. doi: 10.1038/s41598-021-91846-4
Paolicelli, R. C., Sierra, A., Stevens, B., Tremblay, M. E., Aguzzi, A., et al. (2022). Microglia states and nomenclature: a field at its crossroads. Neuron 110, 3458–3483. doi: 10.1016/j.neuron.2022.10.020
Pasciuto, E., Burton, O. T., Roca, C. P., Lagou, V., Rajan, W. D., Theys, T., et al. (2020). Microglia Require CD4 T cells to complete the fetal-to-adult transition. Cell 182, 625–640. doi: 10.1016/j.cell.2020.06.026
Pellegrini, L., Bonfio, C., Chadwick, J., Begum, F., Skehel, M., and Lancaster, M. A. (2020). Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 369, eaaz5626. doi: 10.1126/science.aaz5626
Pocock, J. M., and Piers, T. M. (2018). Modelling microglial function with induced pluripotent stem cells: an update. Nat. Rev. Neurosci. 19, 445–452. doi: 10.1038/s41583-018-0030-3
Popova, G., Soliman, S. S., Kim, C. N., Keefe, M. G., Hennick, K. M., Jain, S., et al. (2021). Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell 28, 2153–2166. doi: 10.1016/j.stem.2021.08.015
Prinz, M., Jung, S., and Priller, J. (2019). Microglia biology: one century of evolving concepts. Cell 179, 292–311. doi: 10.1016/j.cell.2019.08.053
Shi, Y., Inoue, H., Wu, J. C., and Yamanaka, S. (2017). Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 16, 115–130. doi: 10.1038/nrd.2016.245
Silvin, A., Uderhardt, S., Piot, C., Mesquita, D., Yang, S., et al. (2022). Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity. 55, 1448–1465. doi: 10.1016/j.immuni.2022.07.004
Smith, A. M., and Dragunow, M. (2014). The human side of microglia. Trends Neurosci. 37, 125–135. doi: 10.1016/j.tins.2013.12.001
Sridhar, A., Hoshino, A., Finkbeiner, C. R., Chitsazan, A., Dai, L., Haugan, A. K., et al. (2020). Single-cell transcriptomic comparison of human fetal retina, hpsc-derived retinal organoids, and long-term retinal cultures. Cell Rep. 30, 1644–1659. doi: 10.1016/j.celrep.2020.01.007
Srinivasan, K., Friedman, B. A., Etxeberria, A., Huntley, M. A., van der Brug, M. P., Foreman, O., et al. (2020). Alzheimer's patient microglia exhibit enhanced aging and unique transcriptional activation. Cell Rep. 31, 107843. doi: 10.1016/j.celrep.2020.107843
Svoboda, D. S., Barrasa, M. I., Shu, J., Rietjens, R., Zhang, S., Mitalipova, M., et al. (2019). Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proc. Natl. Acad. Sci. U S A 116, 25293–25303. doi: 10.1073/pnas.1913541116
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. doi: 10.1016/j.cell.2007.11.019
Tang, Y. M., Pulimood, N. S., and Stifani, S. (2022). Comparing the characteristics of microglia preparations generated using different human ipsc-based differentiation methods to model neurodegenerative diseases. ASN Neuro. 14, 1–15. doi: 10.1177/17590914221145105
Thion, M. S., and Garel, S. (2020). Microglial ontogeny, diversity and neurodevelopmental functions. Curr. Opin. Genet. Dev. 65, 186–194. doi: 10.1016/j.gde.2020.06.013
Thion, M. S., Low, D., Silvin, A., Chen, J., Grisel, P., Schulte-Schrepping, J., et al. (2018). Microbiome influences prenatal and adult microglia in a sex-specific manner. Cell. 172, 500–516. doi: 10.1016/j.cell.2017.11.042
Tong, L. M., Fong, H., and Huang, Y. (2021). Human induced pluripotent stem cell–based modeling of Alzheimer's disease, a glial perspective. iPSCs Model. CNS. 10, 21–35. doi: 10.1016/B978-0-323-85764-2.00006-5
Troutman, T. D., Kofman, E., and Glass, C. K. (2021). Exploiting dynamic enhancer landscapes to decode macrophage and microglia phenotypes in health and disease. Mol. Cell 81, 3888–3903. doi: 10.1016/j.molcel.2021.08.004
Utz, S. G., See, P., Mildenberger, W., Thion, M. S., Silvin, A., Lutz, M., et al. (2020). Early fate defines microglia and non-parenchymal brain macrophage development. Cell. 181, 557–573. doi: 10.1016/j.cell.2020.03.021
Van Hove, H., Martens, L., Scheyltjens, I., De Vlaminck, K., Antunes, A. R. P., De Prijck, S., et al. (2019). A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22, 1021–1035. doi: 10.1038/s41593-019-0393-4
Washer, S. J., Perez-Alcantara, M., Chen, Y., Steer, J., James, W. S., Trynka, G., et al. (2022). Single-cell transcriptomics defines an improved, validated monoculture protocol for differentiation of human iPSC to microglia. Sci Rep. 12, 19454. doi: 10.1038/s41598-022-23477-2
Wightman, D. P., Jansen, I. E., Savage, J. E., Shadrin, A. A., Bahrami, S., Holland, D., et al. (2021). A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer's disease. Nat. Genet. 53, 1276–1282. doi: 10.1038/s41588-021-00921-z
Xu, R., Boreland, A. J., Li, X., Erickson, C., Jin, M., Atkins, C., et al. (2021). Developing human pluripotent stem cell-based cerebral organoids with a controllable microglia ratio for modeling brain development and pathology. Stem Cell Rep. 16, 1923–1937. doi: 10.1016/j.stemcr.2021.06.011
Young, A. M. H., Kumasaka, N., Calvert, F., Hammond, T. R., Knights, A., Panousis, N., et al. (2021). A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genet. 53, 861–868. doi: 10.1038/s41588-021-00875-2
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. doi: 10.1126/science.1151526
Zhang, W., Jiang, J., Xu, Z., Yan, H., Tang, B., Liu, C., et al. (2023). Microglia-containing human brain organoids for the study of brain development and pathology. Mol Psychiatry 28, 96–107. doi: 10.1038/s41380-022-01892-1
Keywords: microglia, iPSCs, brain organoids, macrophages, Alzheimer's disease, stem cells, brain diseases
Citation: Cerneckis J and Shi Y (2023) Modeling brain macrophage biology and neurodegenerative diseases using human iPSC-derived neuroimmune organoids. Front. Cell. Neurosci. 17:1198715. doi: 10.3389/fncel.2023.1198715
Received: 02 April 2023; Accepted: 09 May 2023;
Published: 05 June 2023.
Edited by:
Lotje D. de Witte, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Hiroyuki Konishi, Nagoya University, JapanCopyright © 2023 Cerneckis and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhong Shi, eXNoaUBjb2gub3Jn
 Jonas Cerneckis
Jonas Cerneckis Yanhong Shi
Yanhong Shi