
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Neurosci. , 02 June 2023
Sec. Cellular Neurophysiology
Volume 17 - 2023 | https://doi.org/10.3389/fncel.2023.1166480
The communication between neurons and, in some cases, between neurons and non-neuronal cells, through neurotransmission plays a crucial role in various physiological and pathological processes. Despite its importance, the neuromodulatory transmission in most tissues and organs remains poorly understood due to the limitations of current tools for direct measurement of neuromodulatory transmitters. In order to study the functional roles of neuromodulatory transmitters in animal behaviors and brain disorders, new fluorescent sensors based on bacterial periplasmic binding proteins (PBPs) and G-protein coupled receptors have been developed, but their results have not been compared to or multiplexed with traditional methods such as electrophysiological recordings. In this study, a multiplexed method was developed to measure acetylcholine (ACh), norepinephrine (NE), and serotonin (5-HT) in cultured rat hippocampal slices using simultaneous whole-cell patch clamp recordings and genetically encoded fluorescence sensor imaging. The strengths and weaknesses of each technique were compared, and the results showed that both techniques did not interfere with each other. In general, genetically encoded sensors GRABNE and GRAB5HT1.0 showed better stability compared to electrophysiological recordings in detecting NE and 5-HT, while electrophysiological recordings had faster temporal kinetics in reporting ACh. Moreover, genetically encoded sensors mainly report the presynaptic neurotransmitter release while electrophysiological recordings provide more information of the activation of downstream receptors. In sum, this study demonstrates the use of combined techniques to measure neurotransmitter dynamics and highlights the potential for future multianalyte monitoring.
Neurons and glial cells are the key components of the human brain. As the basic working units of the brain, neurons communicate with one another through synapses, with most inter-neuronal communication taking place at chemical synapses (Majhi et al., 2019). An action potential arriving at the axon terminal causes the cell membrane to depolarize and opens voltage-gated calcium channels, initiating a signaling cascade that leads to the release of neurotransmitters into the synaptic cleft (Luo, 2020). These released neurotransmitters then diffuse across the synaptic cleft and bind to receptors on the postsynaptic membrane, causing the postsynaptic membrane to either depolarize or hyperpolarize, which allows for the transfer of information between neurons or the alteration of the target neuron’s cellular properties (Sudhof, 2008, 2013; Kaeser and Regehr, 2014; Luo, 2020). The proper regulation of neurotransmission is critical for the normal functioning of the central nervous system. Conversely, dysregulation of neurotransmission has been implicated in a variety of neurological and psychiatric conditions, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), depression, autism, schizophrenia, and addiction (Francis et al., 1999; Sarter et al., 2007; Ballatori et al., 2009). However, the synaptic properties of neurotransmission are not yet fully understood primarily due to the limitations of available tools for quantitative analysis of different neurotransmitters with high spatial and temporal resolution.
Many experimental methods, including molecular biology, electrophysiology, and imaging, are used to investigate changes in neurotransmission. The patch clamp technique is an essential electrophysiological tool that provides valuable insight into the functional activity of neurons and neurotransmitter dynamics by measuring electrical potentials and currents across the cell membrane, allowing scientists to decode intercellular and intracellular information (Reyes, 2019). Whole-cell electrophysiology has served as the gold standard to delineate synaptic properties of glutamatergic and GABAergic transmission because of its ability to functionally dissect synaptic transmission at molecular, cellular and network levels with remarkable sensitivity and high spatiotemporal localization (Neher, 2015; Jackman and Regehr, 2017). However, electrophysiology recordings acquire information from discrete electrode and only a limited number of electrodes could be applied simultaneously due to constrained space of the electrodes. For instance, although octuple patch-clamp recording has been developed to dissect neural circuits, they are technically challenging and are largely restricted to in vitro brain slice studies (Jiang et al., 2013; Wang et al., 2015), which hinders its wide use. Even with multiple-electrode arrays, researchers could only record at most hundreds of neurons at a time. Additionally, the technique may not be suitable for cells with minimal or no neurotransmitter-induced responses (Dani and Bertrand, 2007; Nadim and Bucher, 2014). However, a deep understand of neurotransmission in brains requires the researchers to capture as much information (ideally all brain cells) as possible simultaneously. To overcome these limitations, researchers have developed new tools to monitor neurotransmitters with high sensitivity, specificity, and high spatial and temporal resolution. Over the last decade, two major groups of genetically encoded neurotransmitter sensors have been developed, including G-protein-coupled receptor (GPCR) and bacterial periplasmic binding protein (PBP) sensors for glutamate (Marvin et al., 2013), GABA (Marvin et al., 2019), dopamine (Patriarchi et al., 2018; Sun et al., 2018, 2020), acetylcholine (Jing et al., 2018; Borden et al., 2020), norepinephrine (Feng et al., 2019), and serotonin (Unger et al., 2020; Wan et al., 2021). These sensors have been shown to effectively and specifically detect endogenous neurotransmitters in neuronal and non-neuronal tissue preparations in vitro, ex vivo, and in vivo (Zhu et al., 2020; Chen et al., 2021; Dong et al., 2022; Swanson et al., 2022).
The aim of this study was to compare the advantages and disadvantages of classical electrophysiology methods and newly developed genetically encoded neurotransmitter sensors. We chose acetylcholine (ACh), norepinephrine (NE), and serotonin (5-HT) in current study because they all belong to neuromodulatory transmitter systems and share a number of common features (Gu, 2002). Beyond the central nervous system, neuromodulatory transmitters are found and participate in many other tissue and organ functions (Berger et al., 2009; Dantzer, 2018). And deregulation of peripheral and non-neuronal cholinergic/adrenergic/serotoninergic signals is implicated in many non-neuronal pathological conditions, including cancer, diabetes, and cardiovascular diseases (Berger et al., 2009; Kruse et al., 2014; Herring et al., 2019; Sarrouilhe and Mesnil, 2019; Ye et al., 2021). We showed that both whole-cell patch clamp recordings and genetically encoded sensors (iAChSnFR, GRABNE, and GRAB5HT1.0) can be used in tandem in rat hippocampal slices without any interference. This comparison highlights the strengths and limitations of both approaches, providing researchers with guidance on the most appropriate tools for their specific needs and scientific goals.
Wild type male/female SD rats were used in this study. All rats were housed in a temperature-controlled room with a 12/12 h light/dark cycle, with humidity controlled as 55%, provided with food and water ad libitum. All procedures for animal surgery and maintenance were performed using protocols that were approved by the Animal Care and Use Committees at Shanghai Jiao Tong University School of Medicine (SYXK(Hu)2018-0027).
Organotypic hippocampal cultured slices were prepared from postnatal 5–7 days old male/female SD rats following our previous studies (Jing et al., 2018; Borden et al., 2020). Briefly, the hippocampi were dissected out in ice-cold HEPES-buffered Hanks’ solution (pH 7.35) under sterile conditions, sectioned into 400 μm slices on a tissue chopper, and explanted onto a Millicell-CM membrane (0.4 μm pore size; Millipore, Billerica, MA, USA). The membranes were then placed in 750 μl of MEM culture medium, containing (in mM): HEPES 30, heat-inactivated horse serum 20%, glutamine 1.4, D-glucose 16.25, NaHCO3 5, CaCl2 1, MgSO4 2, insulin 1 mg/ml, ascorbic acid 0.012% at pH 7.28 and osmolarity 320. Cultured slices were maintained at 35°C in a humidified incubator (ambient air enriched with 5% CO2).
Genetically encoded ACh, NE, and 5-HT fluorescent sensors were gifts from Dr. Julius Zhu at University of Virginia and then sub-cloned into Sindbis construct, and viral particles were produced following previous studies (Jing et al., 2018). In brief, fluorescent sensors were sub-cloned into Sindbis viral vector pSinREP5 with Xbal and Sphl restriction digestion. Expression of genetically encoded fluorescent sensors was performed as previously reported (Jing et al., 2018; Zhang et al., 2018, 2022). A glass pipette was used to penetrate the cultured hippocampal slices to deliver ∼50 nl of Sindbis solution by pressure injection to CA1, CA3, or DG cell layer to infect only a few CA1, CA3, or DG neurons (i.e., 1–5 neurons per slice/brain) as indicated. Experiments were performed 18 ± 4 h after Sindbis viral infection.
For the Sindbis virus-infected cultured rat hippocampal slices, wide-field epifluorescence imaging was performed using Hamamatsu ORCA FLASH4.0 camera (Hamamatsu Photonics, Japan), and sensor-expressing cells in cultured brain slices are excited by a 460-nm ultra-high power low-noise LED (Prizmatix, Givat-Shmuel, Israel). The patch recording pipettes (4-7 MΩ) were filled with intracellular solution 120 mM potassium gluconate, 4 mM KCl, 10 mM HEPES, 4 mM MgATP, 0.3 mM Na3GTP, and 10 mM sodium phosphocreatine (pH 7.25) for voltage-clamp recordings. Bath solution (29 ± 1.5°C) contained (in mM): NaCl 119, KCl 2.5, CaCl2 4, MgCl2 4, NaHCO3 26, NaH2PO4 1, and glucose 11, at pH 7.4 and gassed with 5% CO2/95% O2. Dual whole-cell recordings were obtained from two nearby infected and non-infected hippocampal CA1/CA3 pyramidal neurons with two Axopatch-200B patch clamp amplifiers (Molecular Devices, Sunnyvale, CA, USA) under visual guidance using fluorescence and transmitted light illumination. Sensor-expressing neurons were patch-clamp recorded and loaded with 5 μM Alexa Fluor 594 (Life Technologies) for ∼10 min to verify that imaged cells and patched cells were the same. Agonists, including ACh, NE, and 5-HT (Tocris Bioscience, Bristol, UK), were puff-applied with a glass pipette (1–1.5 μm in tip diameter) positioned ∼50 μm above the imaged neurons using 10 ms 30-kPa pressure pulses. The frame rate of the FLASH4.0 camera was set to 10 Hz and the sampling rate of electrophysiological recording was set to 10 KHz. To synchronize sensor image capture and whole-cell patch clamp recordings with puff experiments, the camera was set to external trigger mode and triggered by a custom-written IGOR Pro 6 program (WaveMetrics, Lake Oswego, OR, USA).
All results are reported as mean ± SEM. Animals or cells were randomly assigned into control or experimental groups and investigators were blinded to experiment treatments in cultured slices. Statistical significance of the means (p < 0.05; two sides) was determined using Student’s t-test or one-way ANOVA with Dunnett’s post-hoc test analysis.
Whole-cell patch clamp has proven ideal for studying the collective properties of neurons and neuronal networks in brain slices maintained in vitro, whereas recently developed genetically encoded sensors enabled us to directly visualize neurotransmitter/neuromodulator release with high temporal and spatial resolution. To directly compare the performance of the whole-cell patch clamp method and genetically encoded sensors in reporting neuromodulatory neurotransmitter release, we established a simultaneous electrophysiological recording and fluorescence imaging system. First, we expressed genetically encoded sensors in CA1/CA3 pyramidal neurons in cultured rat hippocampal slices with the Sindbis viral expression system that permitted a more rapid (∼18 h) and robust expression of the sensors (Jing et al., 2018). Next, we directly delivered specific neurotransmitters at the indicated concentration with an air puff system to evoke fluorescence and electrophysiological responses. The fluorescence responses captured by an epifluorescence microscope and whole-cell patch clamp recording showed that a brief 10 ms puff application of neurotransmitters at optimized concentrations could evoke both fluorescence response and membrane currents in the sensor expressing neurons (Figures 1B, 2B, 3B), whereas puff application of the control bath solution artificial cerebrospinal fluid (ACSF) induced no responses in the same neurons (Figures 2C, 4C). From here on, we applied this system to directly compare the properties of sensors and electrophysiological recordings in detecting ACh, NE, and 5-HT.
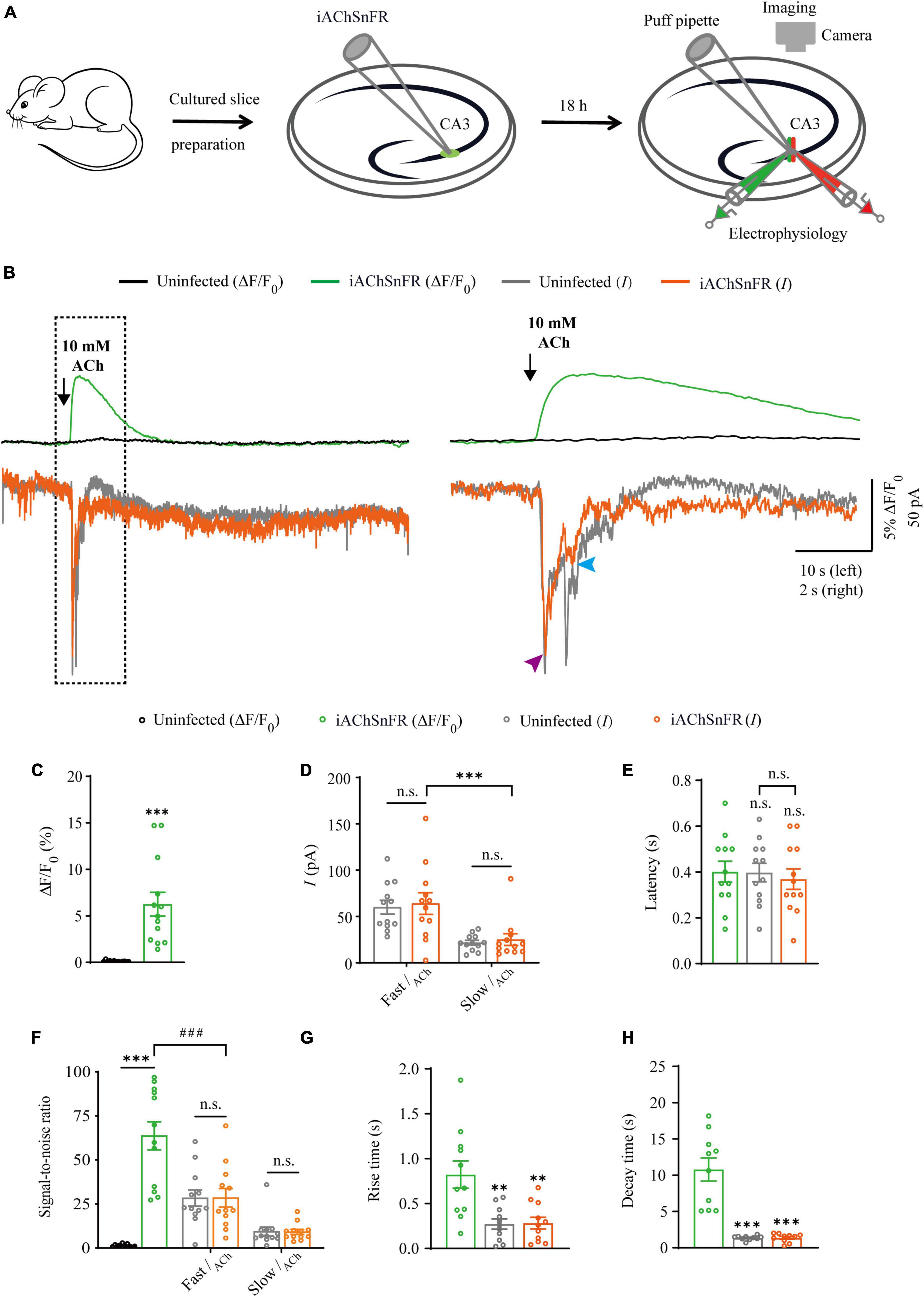
Figure 1. Simultaneous measurement of acetylcholine using iAChSnFR fluorescent sensor and electrophysiological recording in CA3 neurons. (A) Schematic drawing of the design of simultaneous sensor imaging and whole-cell patch clamp recording experiments in cultured rat hippocampal slice preparation. (B) Left, simultaneous fluorescence (upper panel) and currents (lower panel) of iAChSnFR-expressing and control non-expressing CA3 neurons to a brief puff application of ACh. Right, the responses in the left rectangle box are shown again on an expanded time scale. (C) Values for fluorescence responses of iAChSnFR-expressing CA3 neurons compared to non-expressing CA3 neurons (iAChSnFR: 6.25 ± 1.28%; Ctrl: 0.11 ± 0.02%; p < 0.0010; n = 13). (D) Values for the amplitudes of fast cholinergic currents (iAChSnFR: 63.92 ± 11.71 pA; Ctrl: 60.06 ± 7.46 pA; p = 0.79; n = 12) and slow cholinergic currents (iAChSnFR: 25.08 ± 6.36 pA; Ctrl: 21.73 ± 2.51 pA; p = 0.67; n = 12; Fast current vs. Slow current: p = 0.0010) in iAChSnFR-expressing CA3 neurons compared to non-expressing CA3 neurons. (E) Values for the latencies of fast cholinergic currents in non-expressing (Ctrl: 0.40 ± 0.04 s; p = 1.00; n = 12) and iAChSnFR-expressing (iAChSnFR: 0.37 ± 0.04 s; p = 0.82; n = 12; Ctrl current vs. iAChSnFR current: p = 0.85) CA3 neurons compared to those of fluorescence responses of iAChSnFR-expressing CA3 neurons (iAChSnFR: 0.40 ± 0.05 s; n = 12). (F) Values for the signal-to-noise ratio (SNR) of cholinergic fluorescence responses of iAChSnFR-expressing CA3 neurons compared to non-expressing CA3 neurons (iAChSnFR: 63.67 ± 7.94; Ctrl: 1.34 ± 0.23; p < 0.0010; n = 12), and that of fast (iAChSnFR: 28.47 ± 5.29; Ctrl: 28.36 ± 4.58; p = 0.99; n = 12) and slow (iAChSnFR: 9.13 ± 1.41; Ctrl: 9.37 ± 2.54; p = 0.94; n = 12) cholinergic currents of iAChSnFR-expressing CA3 neurons compared to non-expressing CA3 neurons. Values for the signal-to-noise ratio (SNR) of fast cholinergic currents in iAChSnFR-expressing CA3 neurons (iAChSnFR: 28.47 ± 5.29; p < 0.0010; n = 12) compared to those of fluorescence responses of iAChSnFR-expressing CA3 neurons (iAChSnFR: 63.67 ± 7.94; n = 12). (G) Values for rise time of the cholinergic currents in non-expressing (Ctrl: 0.27 ± 0.06 s; p = 0.0052; n = 11) and iAChSnFR-expressing (iAChSnFR: 0.28 ± 0.07 s; p = 0.0036; n = 11) CA3 neurons compared to those of fluorescence responses of iAChSnFR-expressing CA3 neurons (iAChSnFR: 0.82 ± 0.15 s; n = 11). (H) Values for decay time of the cholinergic currents in non-expressing (Ctrl: 1.36 ± 0.10 s; p < 0.0010; n = 10) and iAChSnFR-expressing (iAChSnFR: 1.37 ± 0.19 s; p < 0.0010; n = 10) CA3 neurons compared to those of fluorescence responses of iAChSnFR-expressing CA3 neurons (iAChSnFR: 10.79 ± 1.59 s; n = 10). Data are shown as mean ± SEM. ***p < 0.001, **p < 0.01, two-tailed Student’s unpaired t-test in panels (C,D,G,H), ###p < 0.001, one-way ANOVA with Dunnett’s post-hoc test in panels (E,F).
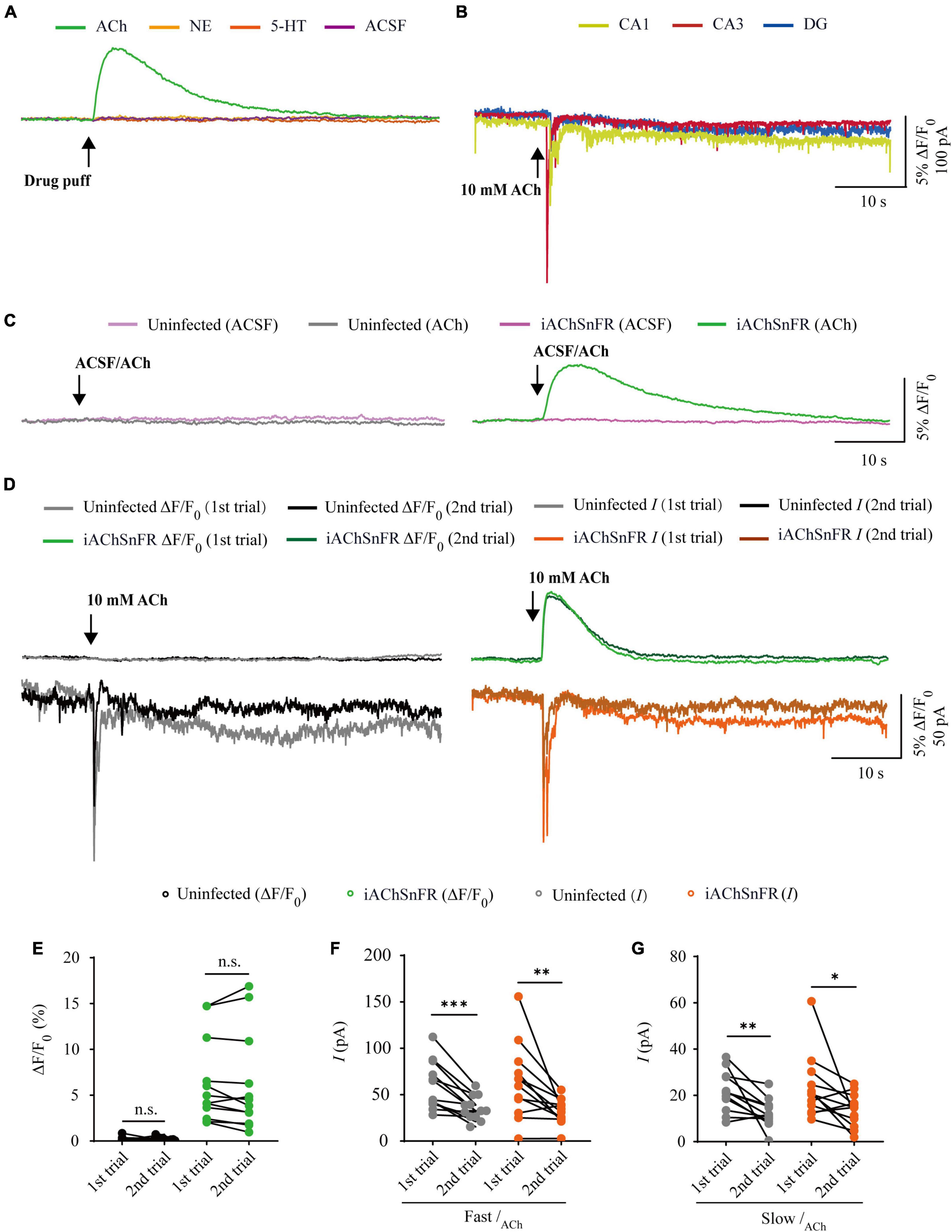
Figure 2. Comparison of iAChSnFR fluorescence and electrophysiological responses to two consecutive acetylcholine application. (A) Fluorescence responses of iAChSnFR-expressing cells to a brief drug puff (10 ms) application of 10 mM ACh, 200 μM norepinephrine (NE) and 100 μM serotonin (5-HT), respectively. (B) Electrophysiological responses of CA1, CA3, and DG neurons to a brief (10 ms) puff application of 10 mM ACh. (C) Fluorescence responses of control non-expressing (left) and iAChSnFR-expressing (right) CA3 neurons to a brief puff (10 ms) application of ACSF and 10 mM acetylcholine (ACh). (D) Fluorescence (upper panel) and electrophysiological responses (lower panel) of non-expressing (left) and iAChSnFR-expressing (right) CA3 neurons to two consecutive puff applications of ACh. (E) Values for the two consecutive fluorescence responses of non-expressing (first: 0.18 ± 0.07%; second: 0.19 ± 0.06%; p = 0.71; n = 12) and iAChSnFR-expressing (first: 6.39 ± 1.34%; second: 6.15 ± 1.56%; p = 0.38; n = 12) CA3 neurons. (F) Values for the two consecutive fast cholinergic currents in non-expressing (first: 60.06 ± 7.46 pA; second: 35.26 ± 3.74 pA; p < 0.0010; n = 12) and iAChSnFR-expressing (first: 63.92 ± 11.71 pA; second: 32.29 ± 4.02 pA; p = 0.0094; n = 12) CA3 neurons. (G) Values for the two consecutive slow cholinergic currents of non-expressing (first: 21.73 ± 2.51 pA; second: 12.48 ± 1.76 pA; p = 0.0063; n = 12) and iAChSnFR-expressing (first: 23.42 ± 4.00 pA; second: 13.67 ± 2.06 pA; p = 0.012; n = 12) CA3 neurons. Data are shown as mean ± SEM. ***p < 0.001, **p < 0.01, *p < 0.05, two-tailed Student’s paired t-test.
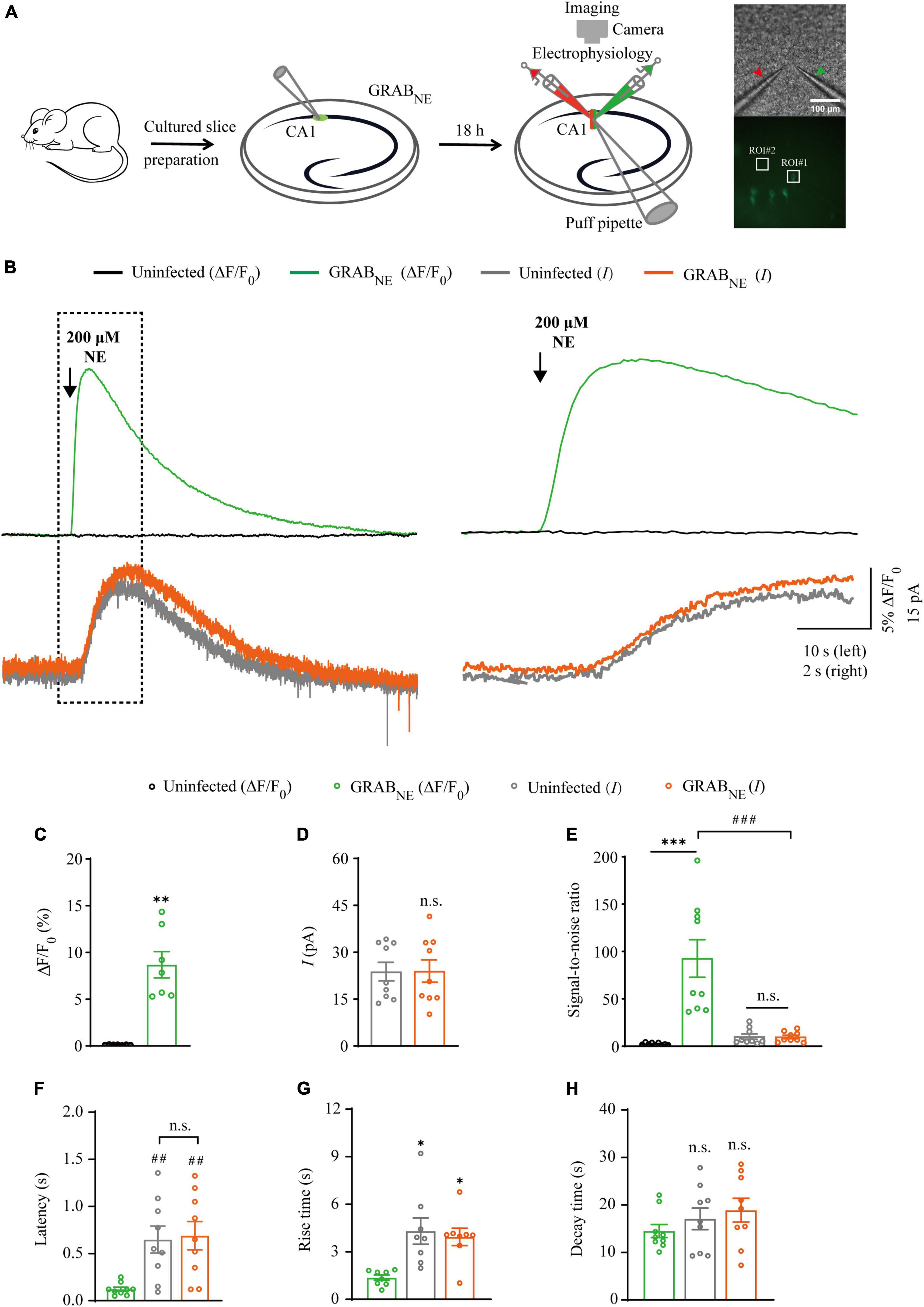
Figure 3. Simultaneous measurement of norepinephrine using GRABNE fluorescent sensor and electrophysiological recording in CA1 neurons. (A) Schematic drawing of the design of simultaneous imaging and electrophysiological recording experiments in cultured rat hippocampal slices. The red arrow indicates the patch electrode of control cell and green arrow indicates that of sensor-expressing cell. The white squares indicating regions of interest (ROIs). (B) Left, simultaneous fluorescence (upper panel) and electrophysiological (lower panel) responses of control non-expressing and GRABNE-expressing CA1 neurons to a brief puff application of NE. Right, the responses in the left rectangle box are shown again on an expanded time scale. (C) Values for the fluorescence responses of GRABNE-expressing CA1 neurons compared to non-expressing CA1 neurons (GRABNE: 8.68 ± 1.41%; Ctrl: 0.12 ± 0.01%; p < 0.0010; n = 7). (D) Values for the amplitudes of adrenergic currents in GRABNE expressing CA1 neurons compared to non-expressing CA1 neurons (GRABNE: 23.80 ± 2.95 pA; Ctrl: 23.99 ± 3.59 pA; p > 0.99; n = 9). (E) Values for the signal-to-noise ratio (SNR) of fluorescence responses of GRABNE-expressing CA1 neurons compared to non-expressing CA1 neurons (GRABNE: 92.61 ± 19.87; Ctrl: 2.43 ± 0.39; p < 0.0010; n = 9), and that of adrenergic currents of GRABNE-expressing CA1 neurons compared to non-expressing CA1 neurons (GRABNE: 9.78 ± 1.80; Ctrl: 10.15 ± 2.80; p = 0.67; n = 9). Values for the SNR of adrenergic currents in GRABNE-expressing CA1 neurons (9.78 ± 1.80; p < 0.0010; n = 9) compared to those of the fluorescence responses of GRABNE-expressing CA1 neurons (92.61 ± 19.87; n = 9). (F) Values for the latencies of adrenergic currents in non-expressing (Ctrl: 0.65 ± 0.14 s; p = 0.0091; n = 9) and GRABNE-expressing (GRABNE: 0.69 ± 0.15 s; p = 0.0051; n = 9; Ctrl vs. GRABNE current: p = 0.96; n = 9) CA1 neurons compared to those of fluorescence responses of GRABNE-expressing CA1 neurons (GRABNE: 0.12 ± 0.02 s; n = 9). (G) Values for rise time of the adrenergic currents in non-expressing (Ctrl: 4.31 ± 0.82 s; p = 0.014; n = 8) and GRABNE-expressing (GRABNE: 3.94 ± 0.55 s; p = 0.016; n = 8) CA1 neurons compared to those of fluorescence responses of GRABNE-expressing CA1 neurons (GRABNE: 1.37 ± 0.17 s; n = 8). (H) Values for decay time of the adrenergic currents in non-expressing (Ctrl: 17.08 ± 2.27 s; p = 0.21; n = 9) and GRABNE-expressing (GRABNE: 18.90 ± 2.50 s; p = 0.086; n = 9) neurons compared to those of fluorescence responses of GRABNE-expressing CA1 neurons (GRABNE: 14.50 ± 1.38 s; n = 9). Data are shown as mean ± SEM. ***p < 0.001, **p < 0.01, *p < 0.05, two-tailed Student’s unpaired t-test in panels (C,D,G,H), ###p < 0.001, ##p < 0.01, one-way ANOVA with Dunnett’s post-hoc test in panels (E,F).

Figure 4. Comparison of GRABNE fluorescence and electrophysiological responses to two consecutive norepinephrine application. (A) Fluorescence responses of GRABNE-expressing cells to a brief drug puff (10 ms) application of 200 μM NE, 10 mM ACh, 100 μM 5-HT and ACSF, respectively. (B) Electrophysiological responses of CA1, CA3, and DG neurons to a brief (10 ms) puff application of 200 μM NE. (C) Fluorescence responses of control non-expressing (left) and GRABNE-expressing (right) CA1 neurons to a brief puff (10 ms) application of ACSF and 200 μM NE. (D) Fluorescence (upper panel) and electrophysiological (lower panel) responses of non-expressing (left) and GRABNE-expressing (right) CA1 neurons to two consecutive puff (10 ms) of 200 μM NE. (E) Values for the two consecutive fluorescence responses of non-expressing (first: 0.13 ± 0.02%; second: 0.11 ± 0.01%; p = 0.10; n = 9) and GRABNE-expressing (first: 10.45 ± 2.75%; second: 10.41 ± 2.43%; p = 0.65; n = 9) CA1 neurons. (F) Values for the two consecutive adrenergic currents in non-expressing (first: 23.80 ± 2.95 pA; second: 13.63 ± 1.48 pA; p = 0.0039; n = 9) and GRABNE-expressing (first: 25.10 ± 3.21 pA; second: 13.75 ± 1.59 pA; p = 0.0078; n = 9) CA1 neurons. Data are shown as mean ± SEM. **p < 0.01, two-tailed Student’s paired t-test.
There are two types of genetically encoded fluorescent sensors for ACh, including GACh2.0/GACh3.0 (Jing et al., 2018, 2020) and iAChSnFR (Borden et al., 2020). GACh2.0 and GACh3.0 are constructed based on human muscarinic acetylcholine receptor 3 (hM3R), with circularly permutated GFP (cpGFP) inserted into the third intracellular loop (ICL3) of hM3R; whereas iAChSnFR is constructed using bacterial periplasmic binding protein (PBP) as the ACh recognition moiety and cpGFP as the fluorescent reporter module (Jing et al., 2018; Borden et al., 2020). Previously, it was reported that iAChSnFR showed higher sensitivity and faster on/off kinetics (Borden et al., 2020). Therefore, we chose iAChSnFR as the genetically encoded ACh sensor for the subsequent experiments. To directly compare iAChSnFR sensor with traditional electrophysiological recordings, we simultaneously made whole-cell recording and fluorescence imaging from pairs of neighboring iAChSnFR-expressing and control non-infected CA3 pyramidal neurons (Figure 1A), which had a robust current response to cholinergic stimulation in cultured hippocampal slices (Figure 2B), which is consistent with what has reported before (Grybko et al., 2010). A 10 ms ACh puff evoked a brief, large inward current (Figure 1B, pink arrow) followed by a prolonged, small inward current (Figure 1B, blue arrow) in both iAChSnFR-expressing and non-expressing neurons, presumably representing the activation of endogenous nicotinic and muscarinic receptors, respectively (Figures 1B, D). A concurrent fluorescence signal was observed only in iAChSnFR-expressing neurons but not in the control non-expressing CA3 neurons (Figures 1B, C, 2C). The latencies of the fast cholinergic currents and the fluorescence responses were the same in iAChSnFR-expressing neurons (Figures 1B, E), indicating that iAChSnFR could detect ACh as fast as endogenous cholinergic receptors. Furthermore, the signal-to-noise ratio (SNR) was calculated as the peak response dividing the standard error of baseline fluorescence/current. The SNR of the iAChSnFR fluorescence responses (∼60) seemed to be larger than that of the fast nicotinic-like cholinergic currents (∼28), and slow muscarinic-like cholinergic currents (∼9) (Figures 1B, F). To further compare the temporal responses, we compared rise time (10–90%) to the maximal responses and decay time (time from maximum to 50%) and found that iAChSnFR showed both much slower rise and decay time when compared to cholinergic currents (Figures 1G, H). This slow kinetic feature of iAChSnFR sensor may limit its use for accurately capturing high-frequency induced ACh release. Moreover, there was no difference in the amplitude, latency, SNR, rise, or decay time of the cholinergic currents in iAChSnFR-expressing and control non-expressing neurons (Figures 1D–H), further confirming that the expression of iAChSnFR did not affect the intrinsic properties of CA3 neurons. We further checked the specificity of iAChSnFR and found that iAChSnFR only showed fluorescent response to puffed ACh, but not NE, 5-HT, or control ACSF (Figure 2A). Moreover, we only detected fluorescent response in iAChSnFR-expressing neurons, but not nearby control uninfected neurons to puffed ACh (Figure 2C), confirming the good specificity of iAChSnFR. To further check the stability of iAChSnFR sensor and whole-cell recording in detecting ACh, we compared iAChSnFR fluorescence and electrophysiological responses to two consecutive ACh applications with an 8-min time interval. The second ACh puff evoked the same fluorescence responses, but smaller cholinergic currents (reduced by ∼40%) in iAChSnFR-expressing neurons compared to the first puff (Figures 2D–G), due presumably to the desensitization and endocytosis of endogenous receptors (Dani and Bertrand, 2007). These results indicated that iAChSnFR had better performance in monitoring presynaptic ACh signals repeatedly over long periods, while whole cell recordings could show the cell state of downstream receptor desensitization.
There is only one type of genetic fluorescent sensor for NE, including GRABNE1m [with a half maximal effective concentration (EC50) of 930 nM and a maximum ΔF/F0 of approximately 230% in response to a saturating concentration of NE] and GRABNE1h (with EC50 of ∼83 nM and a maximum ΔF/F0 of approximately 130%) (Feng et al., 2019), both of which are constructed based on the human α2 adrenergic G-protein-coupled receptor (α2AR). We chose GRABNE1m (from here on, GRABNE in brief) as the genetically encoded NE sensor for the subsequent experiments since it exhibited better performance with respect to ΔF/F0 and brightness (Feng et al., 2019). To evaluate the performance of GRABNE sensor and electrophysiological recording in detecting NE signal, we made simultaneous dual whole-cell recordings and fluorescence imaging in pairs of neighboring GRABNE-expressing and non-expressing CA1 pyramidal neurons (Figure 3A), which had larger current response (Figure 4B) to adrenergic stimulation compared to other regions in cultured hippocampal slices (Murchison et al., 2011). A 10 ms puff of 200 μM NE evoked a large outward current in both GRABNE-expressing and non-expressing neurons, presumably representing the activation of G-protein-coupled inwardly rectifying potassium channels (Figures 3B, D). A concurrent fluorescence signal was observed only in GRABNE-expressing neurons, but not in control non-expressing CA1 neurons (Figures 3B, C). The SNR of the GRABNE fluorescence responses (∼90) was larger than that of the adrenergic currents (∼10) (Figure 3E). The latencies of the adrenergic currents were longer than those of the fluorescence responses in GRABNE-expressing neurons (Figure 3F), indicating that GRABNE could detect NE signals faster than endogenous adrenergic receptors. To further compare the temporal responses, we compared rise time and decay time and found that GRABNE showed both much faster rise but comparable decay time to adrenergic currents of metabotropic receptors (Figures 3G, H), suggesting a relatively faster response time and better kinetics of GRABNE over whole-cell patch clamp in reporting NE signals. Moreover, there was no difference in the amplitude, latency, SNR, rise time or decay time of adrenergic currents in GRABNE-expressing and the neighboring non-expressing control neurons (Figures 3D–H), indicating that the expression of GRABNE had no obvious effect on the physiological properties of CA1 neurons. We further checked the specificity of GRABNE and found that this sensor only showed fluorescent response to puffed NE, but not ACh, 5-HT, or control ACSF (Figure 4A). Moreover, we only detected fluorescent response in GRABNE-expressing neurons to puffed NE, but not nearby control uninfected neurons to NE/ACSF (Figure 4C), confirming the good specificity of GRABNE. To further check the stability of both methods, we compared fluorescence and electrophysiological responses to two consecutive NE applications. Similarly, the second NE puff evoked almost the same fluorescence response, but smaller adrenergic currents (reduced by ∼45%) in GRABNE-expressing neurons compared to the first puff (Figures 4D–F), due presumably to the desensitization of endogenous receptors (Benarroch, 2009), suggesting the suitability of GRABNE in monitoring NE signals repeatedly over long periods.
There are two types of genetic fluorescent sensors for 5-HT, including GRAB5HT1.0 and iSeroSnFR. GRAB5HT1.0 is constructed based on the serotonin 2C receptor; whereas iSeroSnFR is generated using PBP as serotonin recognition moiety (Unger et al., 2020; Wan et al., 2021). Previously we directly compared GRAB5HT1.0 with iSeroSnFR in cultured rat hippocampal slices/mouse dorsal raphe neurons and found that GRAB5HT1.0 showed significantly larger fluorescence changes (ΔF/F0) compared to iSeroSnFR (Zhang et al., 2022). Therefore, we chose GRAB5HT1.0 as the genetically encoded serotonin sensor in our subsequent experiments. To compare GRAB5HT1.0 sensor imaging with patch-clamp recordings for their detection ability for 5-HT, we simultaneously made dual whole-cell recordings and fluorescence imaging from pairs of neighboring GRAB5HT1.0-expressing and non-expressing CA1 pyramidal neurons (Figure 5A), which had both inward and outward currents (Figure 6B), probably due to high expression of ionotropic and metabotropic receptors, such as 5-HT(3), 5-HT(1A), 5-HT(4), and 5-HT(7) subtypes (Bickmeyer et al., 2002; Eydipour et al., 2017). A 10 ms 5-HT (100 μM) puff evoked a brief, small inward current followed by a prolonged outward current in both GRAB5HT1.0-expressing and non-expressing neurons, presumably representing the activation of endogenous ionic and G-protein coupled 5-HT receptors, respectively (Figures 5B, D). A concurrent fluorescence signal was observed only in GRAB5HT1.0-expressing neurons but not in control non-expressing CA1 neurons (Figures 5B, C). The SNR of the GRAB5HT1.0 fluorescence responses (∼65) seemed to be larger than that of the fast (∼13) and slow serotoninergic currents (∼12) (Figure 5E). The latencies of the fast serotoninergic currents were longer than those of the fluorescence responses (Figure 5F), indicating that GRAB5HT1.0 detected 5-HT signals faster than endogenous serotoninergic receptors. To further compare the temporal responses, we compared rise time and decay time and found that GRAB5HT1.0 showed both much slower rise but not decay time when compared to adrenergic currents of ionotropic and metabotropic receptors (Figures 5G, H), suggesting a little slower kinetics of GRAB5HT1.0 over whole-cell patch clamp in reporting 5-HT signals. There was no significant difference in the amplitude, latency, SNR, rise time or decay time of serotoninergic currents in GRAB5HT1.0-expressing and non-expressing neurons (Figures 5D–H), further confirming that the expression of fluorescence sensor GRAB5HT1.0 did not change the intrinsic properties of the CA1 neurons. We further checked the specificity of GRAB5HT1.0 and found that this sensor only showed fluorescent response to puffed 5-HT, but not ACh, NE, or control ACSF (Figure 6A). Moreover, we only detected fluorescent response in GRAB5HT1.0-expressing neurons to puffed 5-HT, but not nearby control uninfected neurons to 5-HT/ACSF (Figure 6C), confirming the good specificity of GRAB5HT1.0. Similar with what we observed with iAChSnFR and GRABNE, the second 5-HT puff evoked the same fluorescence responses but smaller currents (reduced by ∼42%) in GRAB5HT1.0-expressing neurons compared to the first puff (Figures 6D–G), due presumably to the desensitization of endogenous receptors (Berger et al., 2009), indicative of the suitability of GRAB5HT1.0 in monitoring puffed 5-HT or presynaptic 5-HT release repeatedly over long periods.
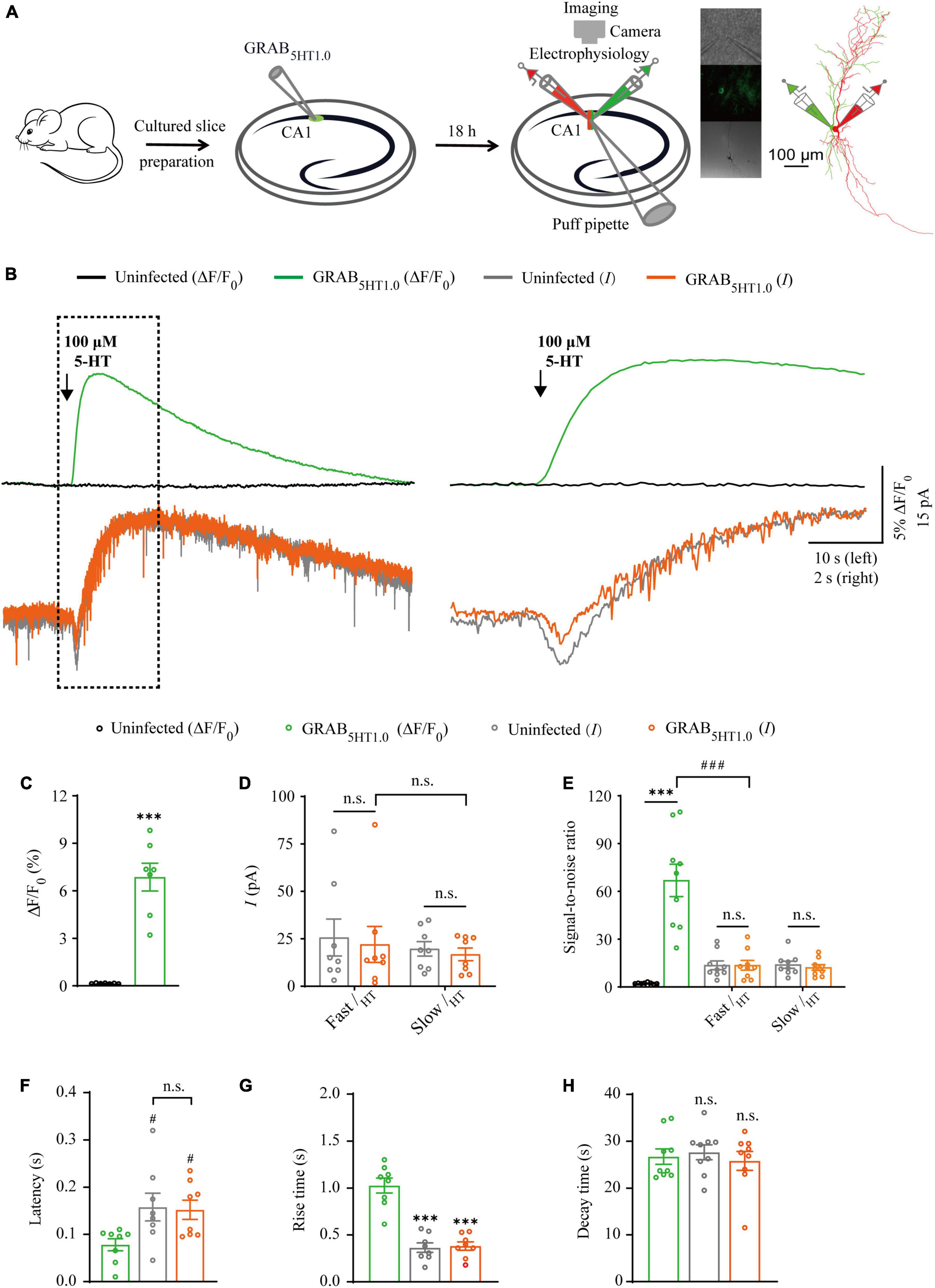
Figure 5. Simultaneous measurement of serotonin using GRAB5HT1.0 fluorescent sensor and electrophysiological recording in CA1 neurons. (A) Schematic drawing outlining the design of simultaneous sensor imaging and electrophysiological recording experiments in cultured rat hippocampal slices. (B) Left, simultaneous fluorescence (upper panel) and electrophysiological (lower panel) responses of neighboring paired non-expressing and GRAB5HT1.0-expressing CA1 neurons to a brief puff application of 5-HT. Right, the responses in the left rectangle box are shown again in an expanded time scale. (C) Values for the fluorescence responses of GRAB5HT1.0-expressing CA1 neurons compared to non-expressing CA1 neurons (GRAB5HT1.0: 6.86 ± 0.88%; Ctrl: 0.15 ± 0.01%; p < 0.0010; n = 7). (D) Values for the amplitudes of fast (GRAB5HT1.0: 22.02 ± 9.45 pA; Ctrl: 25.66 ± 9.75 pA; p = 0.96; n = 8) and slow serotoninergic currents (GRAB5HT1.0: 16.80 ± 3.32 pA; Ctrl: 19.69 ± 3.83 pA; p = 0.57; n = 8; Fast current vs. Slow current: p > 0.99) in GRAB5HT1.0-expressing CA1 neurons compared to non-expressing CA1 neurons. (E) Values for the signal-to-noise ratio (SNR) of serotoninergic fluorescence responses of GRAB5HT1.0-expressing CA1 neurons compared to non-expressing CA1 neurons (GRAB5HT1.0: 66.85 ± 10.14; Ctrl: 2.16 ± 0.22; p < 0.0010; n = 9), and that of fast (GRAB5HT1.0: 13.58 ± 3.04; Ctrl: 13.60 ± 2.72; p = 1.00; n = 9) and slow (GRAB5HT1.0: 12.17 ± 1.81; Ctrl: 13.94 ± 2.23; p = 0.56; n = 9) serotoninergic currents of GRAB5HT1.0-expressing CA1 neurons compared to non-expressing CA1 neurons. Values for the SNR of fast serotoninergic currents in GRAB5HT1.0-expressing CA1 neurons (current: 13.58 ± 3.04; p < 0.0010; n = 9) compared to those of fluorescence responses of GRAB5HT1.0-expressing CA1 neurons (fluorescence: 66.85 ± 10.14; n = 9). (F) Values for latencies of fast serotoninergic currents in non-expressing (Ctrl: 0.16 ± 0.03 s; p = 0.032; n = 8) and GRAB5HT1.0-expressing (GRAB5HT1.0: 0.15 ± 0.02 s; p = 0.047; n = 8; Ctrl current vs. GRAB5HT1.0 current: p = 0.98) CA1 neurons compared to those of fluorescence responses of GRAB5HT1.0-expressing CA1 neurons (GRAB5HT1.0: 0.08 ± 0.01 s, n = 8). (G) Values for rise time of the serotoninergic currents in non-expressing (Ctrl: 0.36 ± 0.05 s; p < 0.0010; n = 8) and GRAB5HT1.0-expressing (GRAB5HT1.0: 0.38 ± 0.04 s; p < 0.0010; n = 8) CA1 neurons compared to those of fluorescence responses of iAChSnFR-expressing CA1 neurons (GRAB5HT1.0: 1.00 ± 0.08 s; n = 8). (H) Values for decay time of the serotoninergic currents in non-expressing (Ctrl: 27.65 ± 1.60 s; p = 0.57; n = 9) and GRABNE-expressing (GRAB5HT1.0: 25.82 ± 2.02 s; p = 0.91; n = 9) CA1 neurons compared to those of fluorescence responses of GRABNE-expressing CA1 neurons (GRAB5HT1.0: 26.73 ± 1.65 s; n = 9). Data are shown as mean ± SEM. ***p < 0.001, two-tailed Student’s unpaired t-test in panels (C,D,G,H), ###p < 0.001, #p < 0.05, one-way ANOVA with Dunnett’s post-hoc test in panels (E,F).
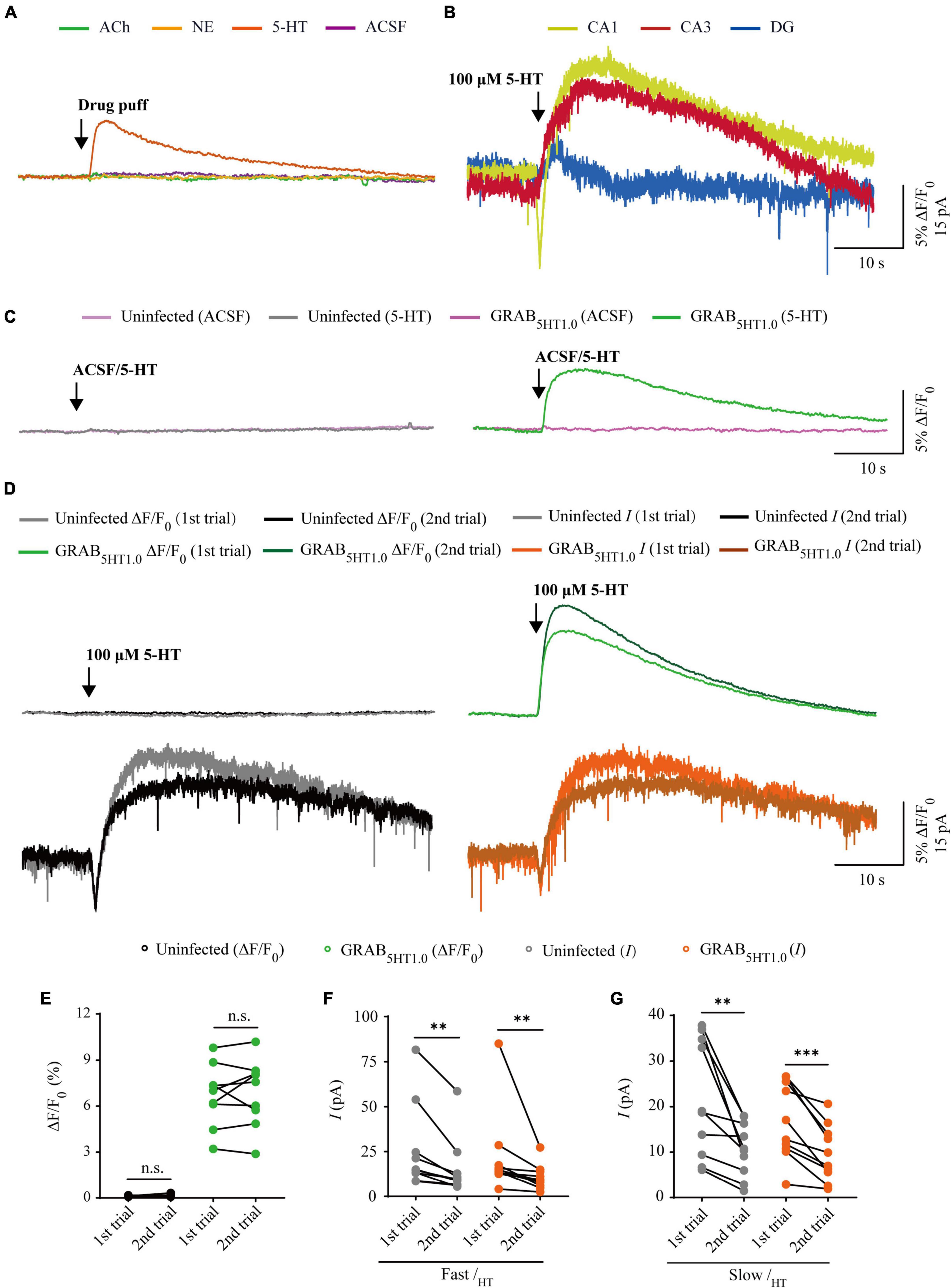
Figure 6. Comparison of GRAB5HT1.0 fluorescence and electrophysiological responses to two consecutive serotonin application. (A) Fluorescence responses of GRAB5HT1.0-expressing cells to a brief drug puff (10 ms) application of 100 μM 5-HT, 10 mM ACh, 200 μM NE, and ACSF, respectively. (B) Electrophysiological responses of CA1, CA3, and DG neurons to a brief (10 ms) puff application of 100 μM 5-HT. (C) Fluorescence responses of control non-expressing (left) and GRAB5HT1.0-expressing (right) CA1 neurons to a brief puff (10 ms) application of ACSF and 100 μM 5-HT. (D) Fluorescence (upper panel) and electrophysiological (lower panel) responses of non-expressing (left) and GRAB5HT1.0-expressing (right) CA1 neurons to two consecutive brief puff of 5-HT. (E) Values for the two consecutive fluorescence responses of control non-expressing CA1 neurons (first: 0.13 ± 0.01%; second: 0.14 ± 0.03%; p = 0.51; n = 9) and GRAB5HT1.0-expressing CA1 neurons (first: 6.70 ± 0.68%; second: 6.87 ± 0.73%; p = 0.65; n = 9). (F) Values for the two consecutive fast inward serotoninergic currents in control non-expressing (first: 25.47 ± 7.52 pA; second: 15.30 ± 5.12 pA; p = 0.0020; n = 10) and GRAB5HT1.0-expressing (first: 21.88 ± 7.23 pA; second: 10.63 ± 2.22 pA; p = 0.0020; n = 10) CA1 neurons. (G) Values for the two consecutive slow outward serotoninergic currents of control non-expressing (first: 21.63 ± 4.07 pA; second: 10.61 ± 1.88 pA; p = 0.0062; n = 10) and GRAB5HT1.0-expressing (first: 16.76 ± 2.64 pA; second: 9.73 ± 1.95 pA; p < 0.0010; n = 10) CA1 neurons. Data are shown as mean ± SEM. ***p < 0.001, **p < 0.01, two-tailed Student’s paired t-test.
In this study, we compared the performance of whole-cell patch clamp recordings and three genetically encoded fluorescence sensors, namely, iAChSnFR (Borden et al., 2020), GRABNE (Feng et al., 2019), and GRAB5HT1.0 (Wan et al., 2021), in detecting ACh, NE, and 5-HT in cultured rat hippocampal slices. We conducted simultaneous electrophysiology recordings and fluorescence sensor imaging in hippocampal slices because they maintained the general synaptic organization and connections of the brain, which were ideal for repeated imaging and electrophysiology experiments (Jing et al., 2018; Borden et al., 2020). We tested the sensors in different regions of the hippocampus, including CA1, CA3, and DG, and found differences in cholinergic, adrenergic, and serotoninergic currents (Figures 2B, 4B, 6B), but not fluorescence differences among these regions. As a result, we chose CA3 neurons for ACh detection and CA1 neurons for NE/5-HT detection. We also tried to puff different concentrations of ACh/NE/5-HT and found that genetically encoded sensors, but not whole-cell recordings, responded to very low concentrations of neurotransmitters (Feng et al., 2019; Borden et al., 2020; Wan et al., 2021; Zhang et al., 2022). We optimized the concentrations of ACh, NE, and 5-HT to 10 mM, 200 μM, and 100 μM, respectively, at which concentrations both fluorescence and electrophysiology responses could be detected. Moreover, we also found all the neurotransmitter sensors had very good specificity. For instance, iAChSnFR only showed fluorescent responses to puffed ACh, but not NE, 5-HT, and ACSF (Figure 2A), and this also applied to GRABNE and GRAB5HT1.0 (Figures 4A, 6A).
A 10 ms drug puff evoked robust fluorescence changes and inward/outward current in sensor-expressing neurons, representing the detection of neurotransmitters and the downstream activation of endogenous neurotransmitter receptors. Furthermore, we compared the responses of the fluorescence sensors and electrophysiological recording to two consecutive drug applications. The second ACh/NE/5-HT puff evoked the same fluorescence responses, but smaller currents in iAChSnFR/GRABNE/GRAB5HT1.0-expressing neurons compared to the first puff, probably due to the desensitization of endogenous receptors. However, the genetically encoded sensors did not trigger downstream receptor endocytosis signaling pathways, probably as a result of the mutations brought in the process of sensor constructions, making them ideal for monitoring puffed ACh/NE/5-HT or presynaptic neurotransmitter release over a long period repeatedly. Thus, these sensors permitted stable real-time measurements of the dynamic presynaptic release of the neurotransmitters. For example, iAChSnFR was used to visualize short-term depression of ACh vesicle release in mouse MEC neurons and 10 s was calculated as the time to relieve cholinergic presynaptic depression (Borden et al., 2020). Furthermore, combined with super-resolution and deconvolution microscopy, the genetically encoded sensors enabled the investigation of the fundamental properties of neurotransmission, such as the transmitter diffusion range and found that acetylcholine and monoamines employed spatially restricted transmission mode and diffused at individual release sites with a spread length constant of ∼0.75 μm (Zhu et al., 2020). When combined with high-resolution imaging and analysis algorithms, the genetically encoded sensors will unveil more properties of neurotransmitters such as the number of release sites, release pool size, release probability, quantal size, and refilling rate, that are crucial for comprehending the underlying mechanisms of behaviors and neurological diseases (Zhu et al., 2020; Chen et al., 2021; Lin et al., 2021; Zheng et al., 2022).
On the other hand, the genetically encoded neurotransmitter sensors have limitations and drawbacks that need to be considered when using them in future research studies. The kinetics of the newly developed genetically encoded neurotransmitter sensors is only rapid enough to report slow neurotransmitter releases generated by low- or moderate- but not high-frequency physiological stimulation. For example, both GACh2.0 and iAChSnFR failed to accurately report neurotransmitter release under high-frequency stimulation (>1 Hz) due to their long decay time (Jing et al., 2018; Borden et al., 2020). Moreover, the performance of the GPCR-based sensors can also be affected by agonists or antagonists that normally bind to their corresponding endogenous GPCRs, and this may limit the application of these sensors in certain pharmacological studies such as drug screening (Wu et al., 2022). In addition, the majority of PBP-based sensors, such as iSeroSnFR, iGABASnFR, and iATPSnFR (Lobas et al., 2019; Marvin et al., 2019; Unger et al., 2020), have relatively poor affinity for neurotransmitters because of their low sequence homogeneity to endogenous neurotransmitter receptors (Marvin et al., 2013). In addition, the ACh sensor iAChSnFR could detect both ACh and its precursor choline, limiting its application in high specificity requiring scenarios (Borden et al., 2020). Furthermore, the ectopic expression of genetically encoded sensors might interfere with endogenous signals since they are engineered neurotransmitter receptors and may differ from the endogenous receptor expression in some brain regions (Tubio et al., 2010; Wu et al., 2022). In this way, the overexpression of the genetically encoded sensors might not be appropriate for analyzing the spatiotemporal organization of neurotransmitter release (Liu et al., 2021). Finally, despite their cell specificity, remarkable sensitivity and excellent spatiotemporal resolution, the genetically encoded sensors could only detect the neurotransmitters released to the extracellular space but not the downstream signals that reflect the valid response from the target cells triggered by the binding of neurotransmitters, which may not reflect the real state of cells and cause overstatement of the importance of neurotransmitters in some behavior and diseases.
The classical patch clamp recording has remarkable sensitivity and temporal resolution and is a powerful tool to study the transmission properties of glutamatergic and gamma-aminobutyric acidergic transmitters (Jackman and Regehr, 2017; Chen et al., 2021). This is no doubt that patch-clamp recording is still the most direct and effective way of studying electrical signals in the brain (Reyes, 2019). However, this electrophysiology application is unsuitable for cells with minimal or no neuromodulator-induced electrophysiological responses. In this scenario, G-protein-activated potassium channels (GIRK2) were virally overexpressed in different brain regions of interest to provide a rapid electrophysiological readout of neuromodulator receptor activation (Marcott et al., 2014; Mamaligas and Ford, 2016). Despite its high informativity, the application of electrophysiology has strict requirement for experimental conditions, involves a complex experimental procedure, and is labor intensive but with low screening throughput (Sakmann, 2013; Wu et al., 2022). Moreover, both sensors and electrophysiological recordings only measure relative changes in ACh/NE/5-HT levels and do not provide concentration profiles. Fast-scan cyclic voltammetry (FSCV) is an electrochemical method commonly used to measure neuromodulator dynamics both in vitro and in vivo (Puthongkham and Venton, 2020; Venton and Cao, 2020). FSCV measures currents from direct oxidation of electroactive molecules and these currents are converted to concentrations using calibration values. Thus, genetically encoded sensors and whole-cell patch clamp recording could be combined with FSCV to study neurotransmitters dynamics in the future to understand their functional significance under both physiological and pathological conditions.
In conclusion, the combination of different techniques for the study of neurotransmitters offers a valuable opportunity to gain new insights into the complex mechanisms of synaptic communication in the central nervous system. The classical patch clamp recording and genetically encoded sensors each have their own unique advantages and limitations, and it is important to carefully consider these when selecting the best tool for a particular study. By combining these techniques, researchers can obtain a more comprehensive and in-depth understanding of neurotransmitter dynamics in both physiological and pathological conditions, and further advance our knowledge of the underlying mechanisms of synaptic transmission.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the Animal Care and Use Committees at Shanghai Jiao Tong University School of Medicine and South China University of Technology.
PZ, YZ, and AC conceived and supervised the project. KZ performed the experiments related to sensor cloning and virus preparation. YH performed the simultaneous patch-clamp and sensor imaging experiments in slices. KZ, YH, PZ, YZ, and AC wrote the manuscript with input and help from other members from YZ and AC Labs. All authors contributed to data interpretation and data analysis.
This work was supported by the Program of National Natural Science Foundation of China (82102184), Shanghai high-level local university construction project (PT21011), Shanghai 2023 “Science and Technology Innovation Action Plan” Natural Science Foundation Project (23ZR1436800), Alzheimer’s Association Research Fellowship (AARF-19-619387), and the Junior Thousand Talents Program of China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ballatori, N., Krance, S. M., Notenboom, S., Shi, S., Tieu, K., and Hammond, C. L. (2009). Glutathione dysregulation and the etiology and progression of human diseases. Biol. Chem. 390, 191–214. doi: 10.1515/BC.2009.033
Benarroch, E. E. (2009). The locus ceruleus norepinephrine system: Functional organization and potential clinical significance. Neurology 73, 1699–1704. doi: 10.1212/WNL.0b013e3181c2937c
Berger, M., Gray, J. A., and Roth, B. L. (2009). The expanded biology of serotonin. Annu. Rev. Med. 60, 355–366. doi: 10.1146/annurev.med.60.042307.110802
Bickmeyer, U., Heine, M., Manzke, T., and Richter, D. W. (2002). Differential modulation of I(h) by 5-HT receptors in mouse CA1 hippocampal neurons. Eur. J. Neurosci. 16, 209–218. doi: 10.1046/j.1460-9568.2002.02072.x
Borden, P., Zhang, P., Shivange, A., Marvin, J., Cichon, J., and Dan, C. (2020). A fast genetically encoded fluorescent sensor for faithful in vivo acetylcholine detection in mice, fish, worms and flies. bioRxiv [Preprint]. doi: 10.1101/2020.02.07.939504
Chen, J., Cho, K. E., Skwarzynska, D., Clancy, S., Conley, N. J., Clinton, S. M., et al. (2021). The property-based practical applications and solutions of genetically encoded acetylcholine and monoamine sensors. J. Neurosci. 41, 2318–2328. doi: 10.1523/JNEUROSCI.1062-19.2020
Dani, J. A., and Bertrand, D. (2007). Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu. Rev. Pharmacol. Toxicol. 47, 699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214
Dantzer, R. (2018). Neuroimmune interactions: From the brain to the immune system and vice versa. Physiol. Rev. 98, 477–504. doi: 10.1152/physrev.00039.2016
Dong, C., Zheng, Y., Long-Iyer, K., Wright, E. C., Li, Y., and Tian, L. (2022). Fluorescence imaging of neural activity, neurochemical dynamics, and drug-specific receptor conformation with genetically encoded sensors. Annu. Rev. Neurosci. 45, 273–294. doi: 10.1146/annurev-neuro-110520-031137
Eydipour, Z., Vaezi, G., Nasehi, M., Haeri-Rouhani, S. A., and Zarrindast, M. R. (2017). Different Role of CA1 5HT3 serotonin receptors on memory acquisition deficit induced by total (TSD) and REM Sleep Deprivation (RSD). Arch. Iran Med. 20, 581–588.
Feng, J., Zhang, C., Lischinsky, J. E., Jing, M., Zhou, J., Wang, H., et al. (2019). A genetically encoded fluorescent sensor for rapid and specific in vivo detection of norepinephrine. Neuron 102, 745–761.e8. doi: 10.1016/j.neuron.2019.02.037
Francis, P. T., Palmer, A. M., Snape, M., and Wilcock, G. K. (1999). The cholinergic hypothesis of Alzheimer’s disease: A review of progress. J. Neurol. Neurosurg. Psychiatry 66, 137–147. doi: 10.1136/jnnp.66.2.137
Grybko, M., Sharma, G., and Vijayaraghavan, S. (2010). Functional distribution of nicotinic receptors in CA3 region of the hippocampus. J. Mol. Neurosci. 40, 114–120. doi: 10.1007/s12031-009-9266-8
Gu, Q. (2002). Neuromodulatory transmitter systems in the cortex and their role in cortical plasticity. Neuroscience 111, 815–835. doi: 10.1016/s0306-4522(02)00026-x
Herring, N., Kalla, M., and Paterson, D. J. (2019). The autonomic nervous system and cardiac arrhythmias: Current concepts and emerging therapies. Nat. Rev. Cardiol. 16, 707–726. doi: 10.1038/s41569-019-0221-2
Jackman, S. L., and Regehr, W. G. (2017). The mechanisms and functions of synaptic facilitation. Neuron 94, 447–464. doi: 10.1016/j.neuron.2017.02.047
Jiang, X., Wang, G., Lee, A. J., Stornetta, R. L., and Zhu, J. J. (2013). The organization of two new cortical interneuronal circuits. Nat. Neurosci. 16, 210–218. doi: 10.1038/nn.3305
Jing, M., Li, Y., Zeng, J., Huang, P., Skirzewski, M., Kljakic, O., et al. (2020). An optimized acetylcholine sensor for monitoring in vivo cholinergic activity. Nat. Methods 17, 1139–1146. doi: 10.1038/s41592-020-0953-2
Jing, M., Zhang, P., Wang, G., Feng, J., Mesik, L., Zeng, J., et al. (2018). A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat. Biotechnol. 36, 726–737. doi: 10.1038/nbt.4184
Kaeser, P. S., and Regehr, W. G. (2014). Molecular mechanisms for synchronous, asynchronous, and spontaneous neurotransmitter release. Annu. Rev. Physiol. 76, 333–363. doi: 10.1146/annurev-physiol-021113-170338
Kruse, A. C., Kobilka, B. K., Gautam, D., Sexton, P. M., Christopoulos, A., and Wess, J. (2014). Muscarinic acetylcholine receptors: Novel opportunities for drug development. Nat. Rev. Drug Discov. 13, 549–560. doi: 10.1038/nrd4295
Lin, L., Gupta, S., Zheng, W. S., Si, K., and Zhu, J. J. (2021). Genetically encoded sensors enable micro- and nano-scopic decoding of transmission in healthy and diseased brains. Mol. Psychiatry 26, 443–455. doi: 10.1038/s41380-020-00960-8
Liu, C., Goel, P., and Kaeser, P. S. (2021). Spatial and temporal scales of dopamine transmission. Nat. Rev. Neurosci. 22, 345–358. doi: 10.1038/s41583-021-00455-7
Lobas, M. A., Tao, R., Nagai, J., Kronschlager, M. T., Borden, P. M., Marvin, J. S., et al. (2019). A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nat. Commun. 10:711. doi: 10.1038/s41467-019-08441-5
Majhi, S., Bera, B. K., Ghosh, D., and Perc, M. (2019). Chimera states in neuronal networks: A review. Phys. Life Rev. 28, 100–121. doi: 10.1016/j.plrev.2018.09.003
Mamaligas, A. A., and Ford, C. P. (2016). Spontaneous synaptic activation of muscarinic receptors by striatal cholinergic neuron firing. Neuron 91, 574–586. doi: 10.1016/j.neuron.2016.06.021
Marcott, P. F., Mamaligas, A. A., and Ford, C. P. (2014). Phasic dopamine release drives rapid activation of striatal D2-receptors. Neuron 84, 164–176. doi: 10.1016/j.neuron.2014.08.058
Marvin, J. S., Borghuis, B. G., Tian, L., Cichon, J., Harnett, M. T., Akerboom, J., et al. (2013). An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10, 162–170. doi: 10.1038/nmeth.2333
Marvin, J. S., Shimoda, Y., Magloire, V., Leite, M., Kawashima, T., Jensen, T. P., et al. (2019). A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods 16, 763–770. doi: 10.1038/s41592-019-0471-2
Murchison, C. F., Schutsky, K., Jin, S. H., and Thomas, S. A. (2011). Norepinephrine and ss(1)-adrenergic signaling facilitate activation of hippocampal CA1 pyramidal neurons during contextual memory retrieval. Neuroscience 181, 109–116. doi: 10.1016/j.neuroscience.2011.02.049
Nadim, F., and Bucher, D. (2014). Neuromodulation of neurons and synapses. Curr. Opin. Neurobiol. 29, 48–56. doi: 10.1016/j.conb.2014.05.003
Neher, E. (2015). Merits and limitations of vesicle pool models in view of heterogeneous populations of synaptic vesicles. Neuron 87, 1131–1142. doi: 10.1016/j.neuron.2015.08.038
Patriarchi, T., Cho, J. R., Merten, K., Howe, M. W., Marley, A., Xiong, W. H., et al. (2018). Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360:eaat4422. doi: 10.1126/science.aat4422
Puthongkham, P., and Venton, B. J. (2020). Recent advances in fast-scan cyclic voltammetry. Analyst 145, 1087–1102. doi: 10.1039/c9an01925a
Reyes, A. D. (2019). A breakthrough method that became vital to neuroscience. Nature 575, 38–39. doi: 10.1038/d41586-019-02836-6
Sarrouilhe, D., and Mesnil, M. (2019). Serotonin and human cancer: A critical view. Biochimie 161, 46–50. doi: 10.1016/j.biochi.2018.06.016
Sarter, M., Bruno, J. P., and Parikh, V. (2007). Abnormal neurotransmitter release underlying behavioral and cognitive disorders: Toward concepts of dynamic and function-specific dysregulation. Neuropsychopharmacology 32, 1452–1461. doi: 10.1038/sj.npp.1301285
Sudhof, T. C. (2008). Neurotransmitter release. Handb. Exp. Pharmacol. 184, 1–21. doi: 10.1007/978-3-540-74805-2_1
Sudhof, T. C. (2013). Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690. doi: 10.1016/j.neuron.2013.10.022
Sun, F., Zeng, J., Jing, M., Zhou, J., Feng, J., Owen, S. F., et al. (2018). A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell 174, 481–496.e19. doi: 10.1016/j.cell.2018.06.042
Sun, F., Zhou, J., Dai, B., Qian, T., Zeng, J., Li, X., et al. (2020). Next-generation GRAB sensors for monitoring dopaminergic activity in vivo. Nat. Methods 17, 1156–1166. doi: 10.1038/s41592-020-00981-9
Swanson, J. L., Chin, P. S., Romero, J. M., Srivastava, S., Ortiz-Guzman, J., Hunt, P. J., et al. (2022). Advancements in the quest to map, monitor, and manipulate neural circuitry. Front. Neural Circuits 16:886302. doi: 10.3389/fncir.2022.886302
Tubio, M. R., Fernandez, N., Fitzsimons, C. P., Copsel, S., Santiago, S., Shayo, C., et al. (2010). Expression of a G protein-coupled receptor (GPCR) leads to attenuation of signaling by other GPCRs: Experimental evidence for a spontaneous GPCR constitutive inactive form. J. Biol. Chem. 285, 14990–14998. doi: 10.1074/jbc.M109.099689
Unger, E. K., Keller, J. P., Altermatt, M., Liang, R., Matsui, A., Dong, C., et al. (2020). Directed evolution of a selective and sensitive serotonin sensor via machine learning. Cell 183, 1986–2002.e26. doi: 10.1016/j.cell.2020.11.040
Venton, B. J., and Cao, Q. (2020). Fundamentals of fast-scan cyclic voltammetry for dopamine detection. Analyst 145, 1158–1168. doi: 10.1039/c9an01586h
Wan, J., Peng, W., Li, X., Qian, T., Song, K., Zeng, J., et al. (2021). A genetically encoded sensor for measuring serotonin dynamics. Nat. Neurosci. 24, 746–752. doi: 10.1038/s41593-021-00823-7
Wang, G., Wyskiel, D. R., Yang, W., Wang, Y., Milbern, L. C., Lalanne, T., et al. (2015). An optogenetics- and imaging-assisted simultaneous multiple patch-clamp recording system for decoding complex neural circuits. Nat. Protoc. 10, 397–412. doi: 10.1038/nprot.2015.019
Wu, Z., Lin, D., and Li, Y. (2022). Pushing the frontiers: Tools for monitoring neurotransmitters and neuromodulators. Nat. Rev. Neurosci. 23, 257–274. doi: 10.1038/s41583-022-00577-6
Ye, D., Xu, H., Tang, Q., Xia, H., Zhang, C., and Bi, F. (2021). The role of 5-HT metabolism in cancer. Biochim. Biophys. Acta Rev. Cancer 1876:188618. doi: 10.1016/j.bbcan.2021.188618
Zhang, K., Wang, J. G., Wang, J. Y., Luo, J., Fan, L., Cheng, A. B., et al. (2022). Comparison of two genetically encoded fluorescent sensors to detect 5-HT in mouse brain. Progr. Modern Biomed. 22, 2601–2606. doi: 10.13241/j.cnki.pmb.2022.14.001
Zhang, L., Zhang, P., Wang, G., Zhang, H., Zhang, Y., Yu, Y., et al. (2018). Ras and Rap signal bidirectional synaptic plasticity via distinct subcellular microdomains. Neuron 98, 783–800.e4. doi: 10.1016/j.neuron.2018.03.049
Zheng, W. S., Zhang, Y., Zhu, R. E., Zhang, P., Gupta, S., Huang, L., et al. (2022). GESIAP: A versatile genetically encoded sensor-based image analysis program. bioRxiv [Preprint]. doi: 10.1101/2022.10.05.511006
Keywords: genetically encoded sensors, whole-cell patch clamp recording, acetylcholine (ACh), norepinephrine (NE), serotonin
Citation: Zhang K, Han Y, Zhang P, Zheng Y and Cheng A (2023) Comparison of fluorescence biosensors and whole-cell patch clamp recording in detecting ACh, NE, and 5-HT. Front. Cell. Neurosci. 17:1166480. doi: 10.3389/fncel.2023.1166480
Received: 15 February 2023; Accepted: 15 May 2023;
Published: 02 June 2023.
Edited by:
Haitao Wang, Southern Medical University, ChinaReviewed by:
Lichao Yang, Xiamen University, ChinaCopyright © 2023 Zhang, Han, Zhang, Zheng and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aobing Cheng, ZXljaGVuZ2FvYmluZ0BzY3V0LmVkdS5jbg==; Yuqiong Zheng, emhlbmd5dXFpb25nQHJlbmppLmNvbQ==; Peng Zhang, YmVuemhwQHNoc211LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.