- Laboratory of Entomology, Wageningen University and Research, Wageningen, Netherlands
Sensory processes have often been argued to play a central role in the selection of ecological niches and in the formation of new species. Butterflies are among the best studied animal groups with regards to their evolutionary and behavioral ecology and thereby offer an attractive system to investigate the role of chemosensory genes in sympatric speciation. We focus on two Pieris butterflies with overlapping host-plant ranges: P. brassicae and P. rapae. Host-plant choice in lepidopterans is largely based on their olfactory and gustatory senses. Although the chemosensory responses of the two species have been well characterized at the behavioral and physiological levels, little is known about their chemoreceptor genes. Here, we compared the chemosensory genes of P. brassicae and P. rapae to investigate whether differences in these genes might have contributed to their evolutionary separation. We identified a total of 130 and 122 chemoreceptor genes in the P. brassicae genome and antennal transcriptome, respectively. Similarly, 133 and 124 chemoreceptors were identified in the P. rapae genome and antennal transcriptome. We found some chemoreceptors being differentially expressed in the antennal transcriptomes of the two species. The motifs and gene structures of chemoreceptors were compared between the two species. We show that paralogs share conserved motifs and orthologs have similar gene structures. Our study therefore found surprisingly few differences in the numbers, sequence identities and gene structures between the two species, indicating that the ecological differences between these two butterflies might be more related to a quantitative shift in the expression of orthologous genes than to the evolution of novel receptors as has been found in other insects. Our molecular data supplement the wealth of behavioral and ecological studies on these two species and will thereby help to better understand the role of chemoreceptor genes in the evolution of lepidopterans.
1. Introduction
Sympatric speciation has been of major interest in evolutionary biology as it allows to investigate selective pressures in the absence of geographical constraints (Fitzpatrick et al., 2008; Foote, 2018). Butterflies and moths have a long history as model species, both for ecology-driven speciation as well as for speciation through sexual selection (Kawahara et al., 2019; De Pasqual et al., 2021).
Changes in the chemosensory system have often been argued to be major drivers of speciation of plant-feeding insects as these heavily rely on chemical cues for selecting an optimal host-plant and for finding mates (Knolhoff and Heckel, 2014; Khallaf and Knaden, 2022). In the genus Drosophila, several cases of host specialization have occurred, among which the adaption of D. sechellia to the toxic noni fruit has been analyzed most thoroughly. This host switch from a broad range of decaying fruits to a single normally toxic plant was accompanied by a loss of one specific chemoreceptor, point mutations in another receptor as well as changes in the expression levels of several receptor genes (Auer et al., 2020). Similarly, the fly Scaptomyza flava, which is a close relative to Drosophila within the family Drosophilidae, has recently evolved an herbivorous life style. This switch from microbe-feeding to a plant-based diet was accompanied by a loss or pseudogenization of several olfactory genes related to the detection of microbial volatiles and a gene duplication event for another receptor involved in the detection of plant volatiles (Goldman-Huertas et al., 2015). In lepidopterans, chemoreceptor genes have been extensively analyzed in different moths’ species. The two closely related species Helicoverpa armigera and H. assulta, for example, have strongly diverged in their host-plant range. While H. armigera is a worldwide pest with over 200 known host-plants, H. assulta is restricted to plants of the Solanaceous family. Interestingly, these two species also differ in their set of expressed antennal chemoreceptors with several unique genes in both species (Zhang et al., 2015). Taken together, these examples across different insect orders indicate that both rapid gene loss, as well as gene duplication events accompanied by changes in chemoreceptor expression levels are common hallmarks of host-plant shifts and other ecological adaptions in insects.
Therefore, the comparison of chemoreceptors occurring in closely related species can reveal important information on the differences between these species with regards to their host-plant selection or sexual communication. Chemoreceptors in insects can be classified into three classes: odorant receptors (ORs), ionotropic receptors (IRs), and gustatory receptors (GRs). Although a large number of studies have identified many chemoreceptors in different species of Diptera, Lepidoptera, Hemiptera, and Hymenoptera, the specific function of these receptors often remains unknown (Howlett et al., 2012; Zhou et al., 2012; Engsontia et al., 2014; Koenig et al., 2015; Wang et al., 2015; Zhao et al., 2016, 2020).
Before the large-scale application of genome sequencing in insects, transcriptome analysis helped tremendously in identifying chemoreceptors in different insect species (Zhu J. Y. et al., 2018; Wan et al., 2019). Yet, transcriptomes on their own are usually not sufficient to reveal all chemoreceptors given that some chemoreceptors have a very low abundance. Hence, the combination of genome and transcriptome data greatly benefits the identification of chemoreceptors (Montagné et al., 2015).
The cabbage white butterflies, Pieris spp., are important model species with regards to the co-evolutionary arms-race with their host plants, their natural enemies and their adaptations to novel environments (Edger et al., 2015; Sikkink et al., 2017; Zhu F. et al., 2018; Okamura et al., 2019). Pieris brassicae and P. rapae are very closely related species sharing comparable appearances, host plants and geographical distributions (Feltwell, 1982; Ryan et al., 2019; EPPO, 2022). Both species have characterized chemosensory systems and are able to detect similar volatiles and tastants in their environment (van Loon et al., 1992a; Miles et al., 2005). Specifically, the two species use glucosinolates as cues during caterpillar feeding and ovipositing in female butterflies, leading to similar choice of host-plant species (Smallegange et al., 2007). Moreover, both species occur in highly similar habitat types with regards to climate and succession levels. Although the two Pieris species share many behavioral and physiological similarities, they also show some distinctions. For instance, P. brassicae butterflies deposit clustered eggs and the caterpillars feed gregariously on their host plant, while P. rapae butterflies lay their eggs individually and the caterpillars feed solitarily on their host plant (Davies and Gilbert, 1985). In addition, the two species have quite different anti-aphrodisiac pheromone components. P. brassicae employs brassica lactone (Yildizhan et al., 2009) as sex pheromone and benzyl cyanide (Andersson et al., 2003) as an anti-aphrodisiac pheromone, which repels males from mating with gravid females, while P. rapae uses ferrulactone (Yildizhan et al., 2009) as sex pheromone and, methyl salicylate and indole as anti-aphrodisiac pheromone components (Andersson et al., 2003). The similarities and differences between the two Pieris species make the comparison of chemoreceptor genes valuable to understand the evolution and divergence of chemoreception in butterfly species.
It is widely acknowledged that chemoreceptors for host-plant seeking, feeding and pheromone perception drive sympatric speciation across different insect orders (Smadja and Butlin, 2009; Khallaf and Knaden, 2022). To unravel the role of the chemosensory system in defining the ecological niches utilized by the two Pieris species, we identified chemoreceptors in the P. brassicae and P. rapae genomes by homolog searching with queries from other lepidopteran species and conducted a phylogenetic analysis with the identified chemoreceptor sequences of the two Pieris species and seven other lepidopterans. The genome-guided antennal transcriptomes of both Pieris species were assembled and the chemoreceptors were identified as well. Moreover, homolog comparisons of each of the three chemoreceptor families in the two Pieris species were performed by constructing phylogenetic trees, illustrating the gene structures and searching conserved motifs to find similarities and differences within the same gene family. The gene expression levels of chemoreceptors were also compared to potentially explain the ecological overlaps and differences between the two Pieris species.
2. Materials and methods
2.1. Insects and tissue collection
Insects were taken from a laboratory colony of the Laboratory of Entomology, Wageningen University and Research, Netherlands. Caterpillars were reared on Brussels sprouts plants, Brassica oleracea var. gemmifera cv. Cyrus, Brussels sprouts at 22 ± 2°C, RH 70 ± 10%, light 14 h: dark 10 h until pupation. After eclosion butterflies are transferred to another cage and fed with 5% sugar water at the same environmental condition. Brussels sprouts plants were provided to collect eggs.
Adult male and female butterflies were reared in the same cage for 3 days to ensure mating. Males and females were collected to dissect antennae for transcriptome analysis. Antennae from five male and five female butterflies were pooled for each biological replicate. Three biological replicates were collected for the antennal transcriptomes of P. brassicae and P. rapae.
2.2. RNA isolation
Antennal samples were ground in Eppendorf tubes in liquid nitrogen and isolated and purified with ZYMO Quick-RNA/Insect Kit (ZYMO, Irvine, CA, USA). After the manual grinding, 450 μL lysis buffer was added. The RNA samples were isolated and purified according to the manual, followed by dissolving the RNA samples in 20 μL RNase-free H2O through centrifugation. The final RNA samples were stored at −80°C. After that the RNA concentrations were determined using a DeNovix (Wilmington, NC, USA) spectrophotometer and RNA integrity was determined by denaturing gel electrophoresis. Paired-end sequencing (150 bp) was performed by Illumina NovaSeq6000 platform in the Novogene UK sequencing center (London).
2.3. Average nucleotide identity measurement
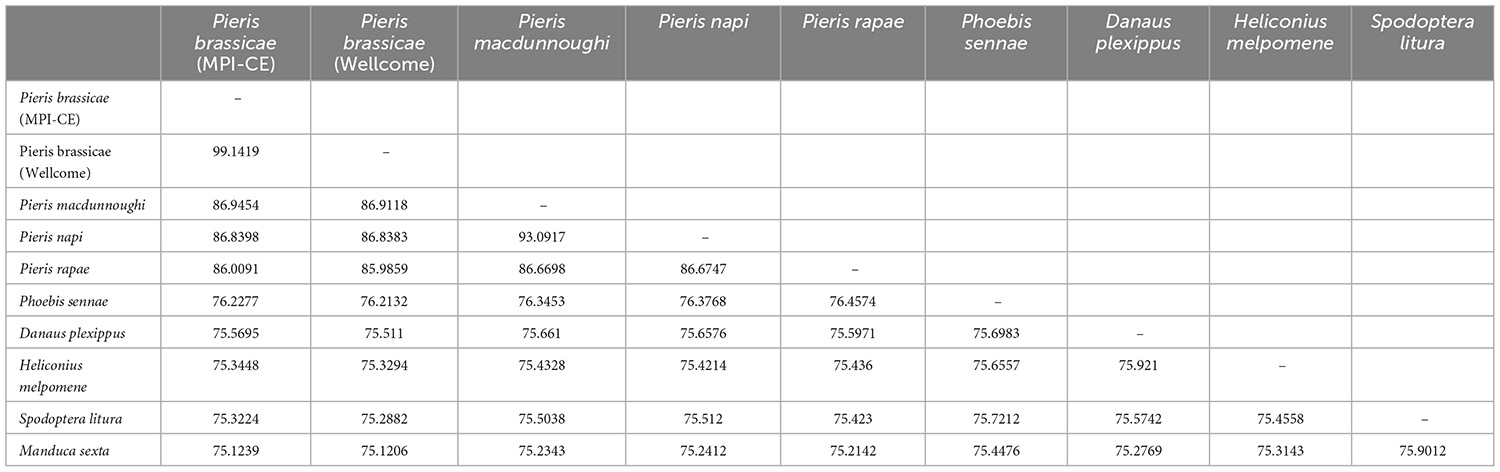
Genomes of four different Pieris species: P. brassicae (GCA_942653925, MPI Chemical Ecology and GCA_905147105.1, Wellcome Sanger Institute), P. rapae (GCA_001856805.1) (Shen et al., 2016), P. napi (GCA_905475465.1), P. macdunnoughi (GCA_905332375.1), as well as the genomes of the outgroup butterflies Heliconius melpomene (GCA_000313835.2), and Danaus plexippus (GCA_009731565.1), Manduca sexta (GCF_014839805.1), Spodoptera litura (GCF_002706865.1) and Phoebis sennae (GCA_001586405.1) were collected to evaluate genome identity. The average nucleotide identity (ANI) was measured using fastANI v1.33 (Jain et al., 2018) using default settings. The detailed information of genomes that were used in this study is shown in Supplementary material 1.
2.4. Transcriptome assembly
The transcriptome was assembled via a genome-guided assembly method. The raw data of different transcriptomes were filtered by Fastp v0.20.0 (Chen et al., 2018) to remove short and low-quality reads and adaptors, the filtered data was subsequently assessed by FastQC v0.11.9 (Andrews, 2010). The filtered data was mapped to the corresponding genomes of P. brassicae (GCA_942653925) and P. rapae (GCA_001856805.1) with Hisat2 v2.1.0 by reporting alignments tailored for transcript assemblers (Kim et al., 2015), followed by transforming and sorting the mapped reads by SAMtools v1.10 (Li et al., 2009), the genome-guided assembly and quantification was achieved by StringTie v2.1.3b using default parameters (Pertea et al., 2015) and non-guided assembly was achieved by Trinity v2.8.6 (Grabherr et al., 2011). Transcripts per million (TPM) was employed to measure the expression levels of chemoreceptor genes in the antennal transcriptomes. In order to compare the expression of chemoreceptors between P. brassicae and P. rapae, we normalized the expression values of all the chemoreceptors to tubulin and rsps20, based on previous studies (Shakeel et al., 2015; Cusumano et al., 2018; Yin et al., 2020; Yang et al., 2021).
2.5. Identification of chemoreceptors
The protein sequences of ORs were collected from eight lepidopteran species: Bombyx mori (Tanaka et al., 2009), M. sexta (Koenig et al., 2015), S. littoralis (Walker et al., 2019), S. exigua (Zhang et al., 2018), H. melpomene (Heliconius Genome, 2012), Plutella xylostella (Engsontia et al., 2014), D. plexippus (Zhan et al., 2011), and Helicoverpa armigera (Liu et al., 2014), as queries to do homolog searching against the genomes of P. brassicae and P. rapae. The protein sequences of GRs were collected from nine lepidopteran species, i.e., B. mori (Wanner and Robertson, 2008), M. sexta (Koenig et al., 2015), S. littoralis (Walker et al., 2019), S. exigua (Zhang et al., 2018), H. melpomene (Briscoe et al., 2013), P. xylostella (Engsontia et al., 2014), and D. plexippus (Zhan et al., 2011; Briscoe et al., 2013), H. armigera (Liu et al., 2014) and Cydia pomonella (Walker et al., 2016), as queries for homolog searching. The protein sequences of IRs were collected from a total of 62 lepidopteran species, originating from three different datasets: IR sequences of 29 lepidopteran species were taken from the study by Liu et al. (2018) and IRs of 32 lepidopteran species were collected from the study by Yin et al. (2021), in addition, we also included the sequences of S. littoralis (Walker et al., 2019) as queries to search against the genomes of P. brassicae and P. rapae to find the candidate orthologs. Another butterfly species Ph. sennae (GCA_001586405.1) was introduced as an outgroup of Pieris to analyze the gene duplication cases.
Exonerate v2.2.0 (Slater and Birney, 2005) was applied to search against the genomes of P. brassicae (GCA_942653925) and P. rapae (GCA_001856805.1) with the collected queries to identify chemoreceptors in the two species, using the protein2genome model. Three best matched results were outputted for each query. The search results were sorted by scores. The targets sharing over 100 amino acids in a row and locating closely on the same scaffold were considered as the same receptor gene. The sequences with the highest scores were extracted, and the redundant alignment results and sequence lengths less than 100 amino acids were removed. The output results were collected as queries to further search in both genome-guided and non-guided transcriptome assemblies. The transcriptome searching results were then filtered by removing redundant and short results which are shorter than 100 amino acids. The transcripts were aligned to determine if some of them are from the same genes when they share 50 amino acids in a row. If any, the overlapped sequences will be assembled. The output transcripts and those outputs having no transcriptome targets were collected to search in NCBI database by blastp to determine if the outputs are full-length sequences. If not, the first high-similarity targets will be collected to repeat the previous steps until no longer sequence can be acquired. The identified chemosensory genes were also filtered with pfam domains (7tm_6 for ORs, Lig_chan for IRs, and 7tm_7 for GRs) to ensure the identified sequences are chemoreceptors. Diamond v0.9.25 (Buchfink et al., 2015) was later employed to identify potential chemoreceptors in the antennal transcriptomes by searching against these assemblies with an e-value of 10–5 using the identified chemoreceptors from the genomes as queries.
The protein sequences of identified chemoreceptors were collected to search against the genomes of S. litura, Papilio glaucus (GCA_000931545.1), Ph. sennae (GCA_001586405.1), Vanessa tameamea (GCF_002938995.1), Delias pasithoe (GCA_010014985.1), and Calepholis nemesis (GCA_002245505.1) in addition to the P. brassicae and P. rapae genomes, to identify the potential gene duplication cases using Exonerate v2.2.0 (Slater and Birney, 2005). The searching was performed under the protein2genome model and only the best matched target for each query was reported.
2.6. Phylogenetic analysis
The protein sequences of ORs, IRs and GRs from the seven lepidopteran species B. mori, M. sexta, S. littoralis, S. exigua, H. melpomene, P. xylostella, and D. plexippus were collected for phylogenetic analysis. The collected chemoreceptor sequences together with those of P. brassicae and P. rapae were aligned by MAFFT v7.475 (Katoh and Standley, 2013). The alignment output was imported into FastTree v2.1.11 (Price et al., 2010) for phylogenetic tree construction under the default settings with the maximum likelihood method. Olfactory receptors were rooted to Orco genes, while IRs were rooted to the iGluR clade and GRs were rooted to the sugar receptor clade. The output of FastTree was visualized and presented by the iTOL online tool v6.5.8 (Letunic and Bork, 2019).
2.7. Illustration of gene structures and motifs
The protein sequences of the identified chemoreceptors from the genomes were collected and imported to The MEME Suite version 5.4.1 (Bailey et al., 2015). The settings of discovery mode, sequence alphabet and site distribution were set to default, the number of motifs that should be found was set to 10. The gene structure information of the chemoreceptors was obtained by searching against the corresponding genomes (GCA_942653925 and GCA_001856805.1) with Exonerate v2.2.0 (Slater and Birney, 2005). The output of motif searching, gene structure as well as phylogenetic tree of chemoreceptors of the two Pieris species were integrated using TBtools v1.09876 (Chen et al., 2020). Transmembrane domains of all Piers chemoreceptors that we identified were predicted by TOPCONS (Tsirigos et al., 2015) and TMHMM 2.0 (Möller et al., 2001).
3. Results
3.1. Average nucleotide identity among butterfly species
Average nucleotide identity was measured to evaluate the closeness of the phylogenetic relationship among butterflies and especially for P. brassicae and P. rapae. The P. brassicae genome of (GCA_942653925) has a very high identity with the latest released genome version of P. brassicae (GCA_905147105.1) by the Wellcome Sanger Institute of 99.14%, confirming the validity of both genome assemblies. P. brassicae shares relatively high similarities with the other three species belonging to the genus Pieris: 86.95, 86.84, and 86.01% with P. macdunnoughi, P. napi, and P. rapae, respectively, indicative of a very close relationship among the four Pieris species. The ANI decreases with increasing phylogenetic distance within the Lepidoptera (Table 1). For instance, the P. brassicae genome is less identical with D. plexippus (75.57%), H. melpomene (75.34%), S. litura (75.32%), and M. sexta (75.12%). Ph. sennae also belongs to the Pieridae and has a slightly higher ANI (76.23%) with the four Pieris species than those species belonging to other families.
3.2. Identification of ORs
All identified ORs (see Supplementary materials 2, 3) are named after D. plexippus because the ORs of P. brassicae and P. rapae share a relatively close relationship with D. plexippus according to phylogenetic analysis (Figure 1A). A total of 60 ORs were identified in both P. brassicae and P. rapae (Figure 1A) including the conserved odorant receptor coreceptor (Orco). The clustering of PbraOrco and PrapOrco with the Orco genes of other lepidopteran species was supported by high bootstrap values (Figure 1B). The reported pheromone receptors of moths such as B. mori, M. sexta, S. littoralis, S. exigua, and P. xylostella also cluster (Figure 1C), and show four HmelORs and one DpleOR as potential novel pheromone receptors. No pheromone receptors of P. brassicae and P. rapae were found in the pheromone receptor clade. Some gene duplication cases were found in the phylogenetic tree as well, such as the OR7/8/9 clade which is formed by the OR7, OR8, and OR9 of the two species with high bootstrap values.
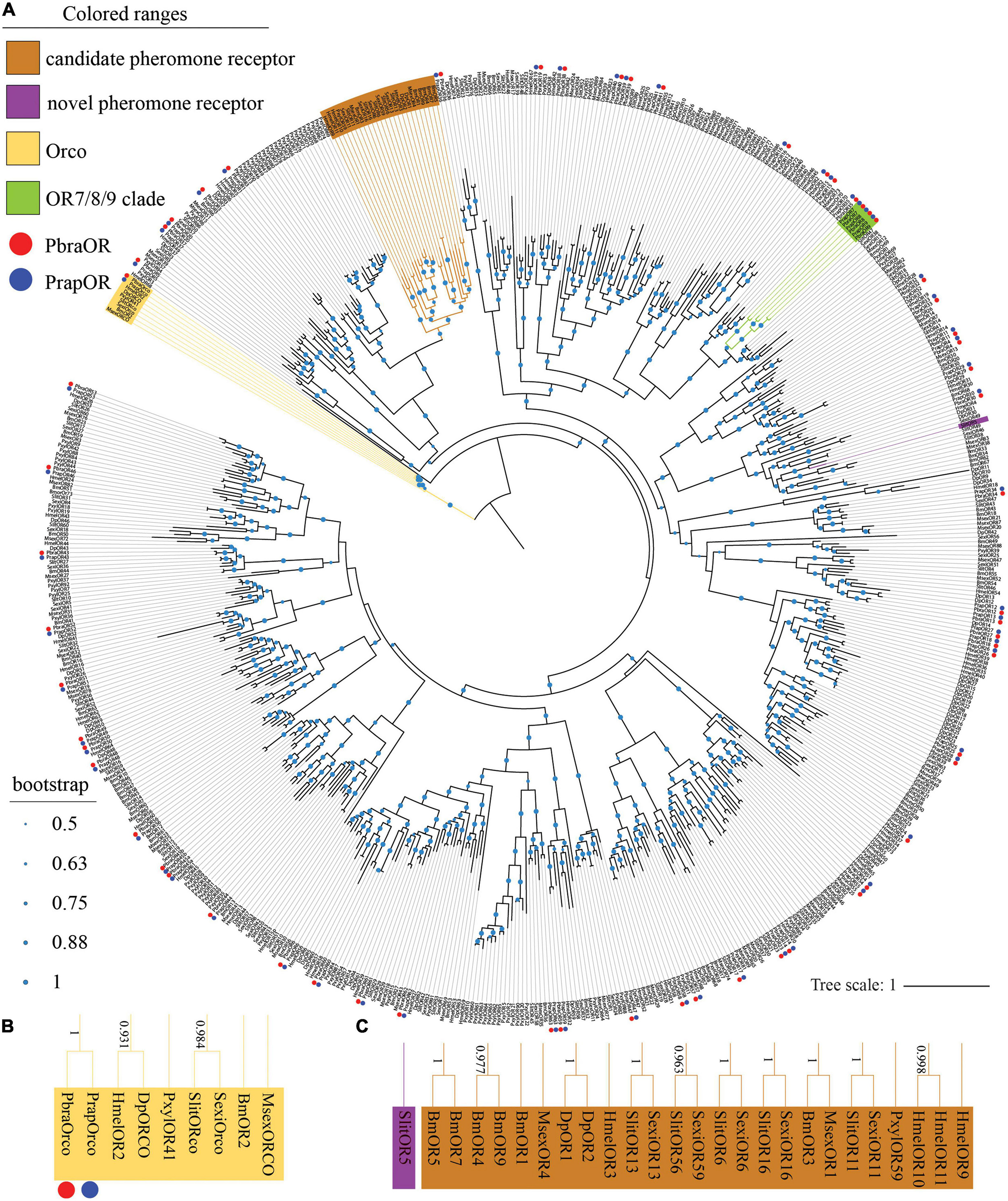
Figure 1. Phylogenetic tree of lepidopteran odorant receptor genes. (A) The phylogeny was rooted using the Orcos. PbraOR, Pieris brassicae OR; PrapOR, P. rapae OR; BmOR, Bombyx mori OR; MsexOR, Manduca sexta OR; DpOR, Danaus plexippus OR; PxylOR, Plutella xylostella OR; HmelOR, Heliconius melpomene OR; SexiOR, Spodoptera exigua; SlitOR, and S. littoralis. The candidate pheromone receptor clade is labeled in light brown, the novel pheromone receptor clade was labeled in purple, the Orco clade is labeled in yellow and the OR7/8/9 clade is labeled in green. The green dots in the middle of the branches of the phylogenetic tree indicate the bootstrap values. The scale bar represents the number of substitutions per site. The Orco clade and candidate pheromone receptor/novel pheromone receptor clade are magnified in panels (B,C), respectively, the bootstrap values higher than 0.5 are shown alongside the branches. PbraORs and PrapORs are labeled with red and blue dots, respectively.
3.3. Identification of IRs
Thirty-one PbraIRs and 34 PrapIRs (see Supplementary materials 2, 3) were identified from the P. brassicae and P. rapae genome, respectively. PbraIRs and PrapIRs were named after H. melpomene since HmelIRs are closer to most PbraIRs and PrapIRs in the phylogenetic tree compared to IRs from other species (Figure 2A). Sixteen out of the 31 PbraIRs and 17 out of the 34 PrapIRs were annotated in the antennal IRs clade (Figure 2A, yellow and green in the phylogenetic tree). Moreover, five PbraIRs (PbraIR8a, PbraIR25a, PbraIR25a, PbraIR76b, and PbraIR93a) and six PrapIRs (PrapIR8a, PrapIR25a1, PrapIR25a2, Prap76b1, PrapIR76b2, and PrapIR93a) were identified as ionotropic receptor co-receptors (Ircos) (Figure 2B). PrapIR76b1 and PrapIR76b2 share a very high similarity of more than 90%, their corresponding DNA sequences have a length of 2,072 bp on the same scaffold with the same direction, although there is only one IR76b gene found in P. brassicae genome. PrapIR25a1 and PrapIR25a2 share a lower identity of 76% with a 1,509 bp interval and, PbraIR25a1 and PbraIR25a2 share a 75% similarity with a 10,008 bp interval on the same scaffold with the same direction. Fifteen out of 31 PbraIRs and 17 out of 34 PrapIRs were annotated in divergent IRs clade in blue in the phylogenetic tree (Figure 2). The orthologs of PrapIR7d.4 and PrapIR100e were not found in the P. brassicae genome. Thirty-two out of 34 PrapIRs have been identified previously (Liu et al., 2018), the two newly identified genes are PrapIR4 and PrapIR25a2.
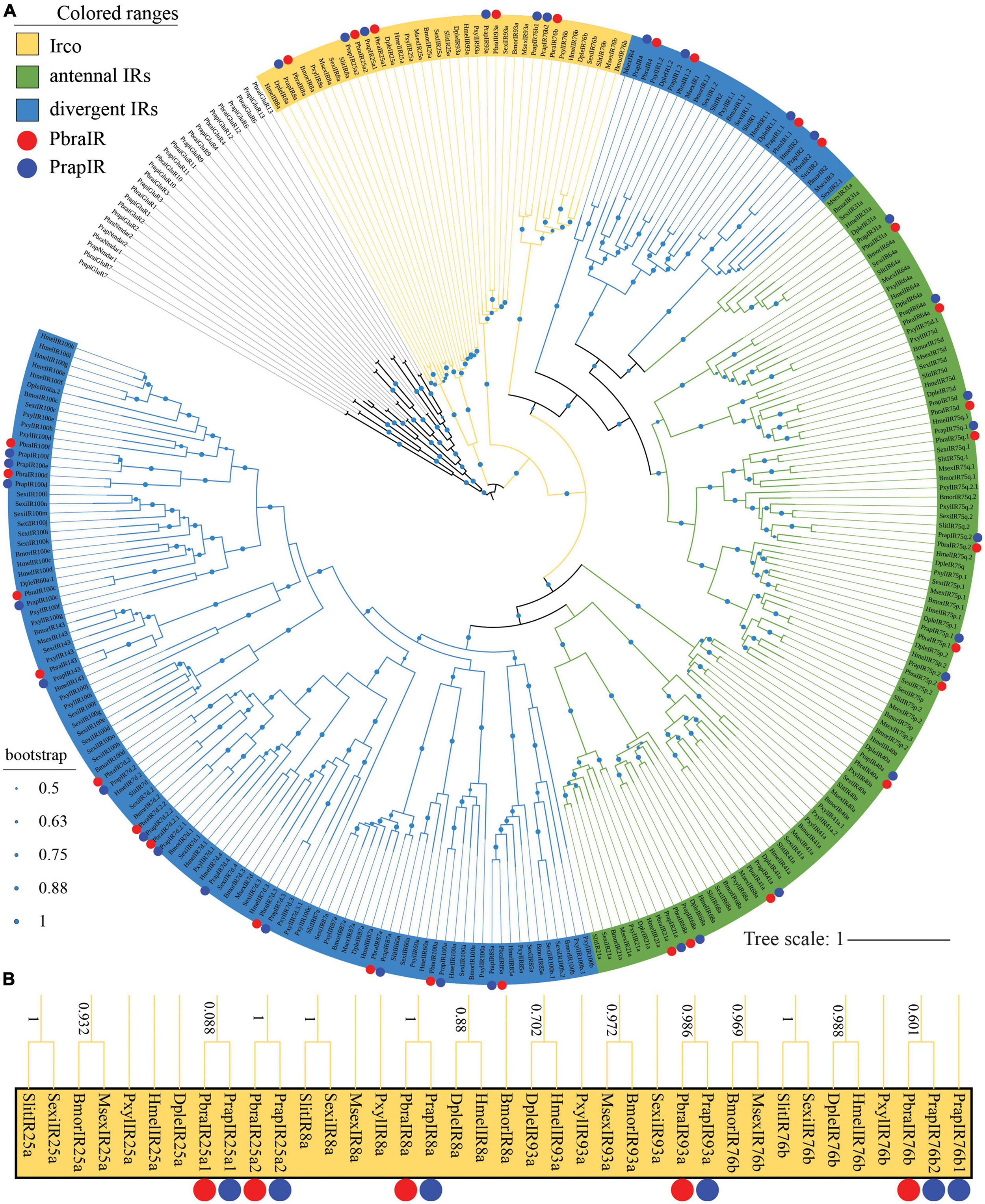
Figure 2. Phylogenetic tree of lepidopteran ionotropic receptor genes. (A) The phylogeny was rooted using iGluRs of P. brassicae and P. rapae. PbraIR, Pieris brassicae IR; PrapIR, P. rapae IR; BmorIR, Bombyx mori IR; MsexIR, Manduca sexta IR; DpleIR, Danaus plexippus IR; PxylIR, Plutella xylostella IR; HmelIR, Heliconius melpomene IR; SexiIR, Spodoptera exigua IR; SlitIR, and S. littoralis IR. The Irco clade is labeled in yellow, the antennal IR clade is labeled in green and the divergent IR clade is labeled in blue. The green dots in the middle of the branches of the phylogenetic tree indicate the bootstrap values that are higher than 0.5. The scale bar represents the number of substitutions per site. (B) Irco clade is magnified, and bootstrap values higher than 0.5 are shown alongside the branches. PbraIRs and PrapIRs are labeled with red and blue dots, respectively.
3.4. Identification of GRs
Thirty-nine GRs (see Supplementary materials 2, 3) were identified in both the P. brassicae and P. rapae genomes. All identified GRs were named after H. melpomene because most PbraGRs and PrapGRs clustered with HmelGRs (Figure 3A), just like the IRs presented above. Based on former publications (Wanner and Robertson, 2008; Yang et al., 2020), three PbraGRs and PrapGRs were annotated as CO2 receptors (Figure 3A, in yellow), three PbraGRs and PrapGRs were annotated as fructose/inositol receptors (Figure 3A, in blue) and nine PbraGRs and PrapGRs were annotated as sugar receptors (Figures 3A, B in pink) and a sinigrin receptor of P. rapae (Yang et al., 2021) was indicated by the blue arrow in Figure 3A.
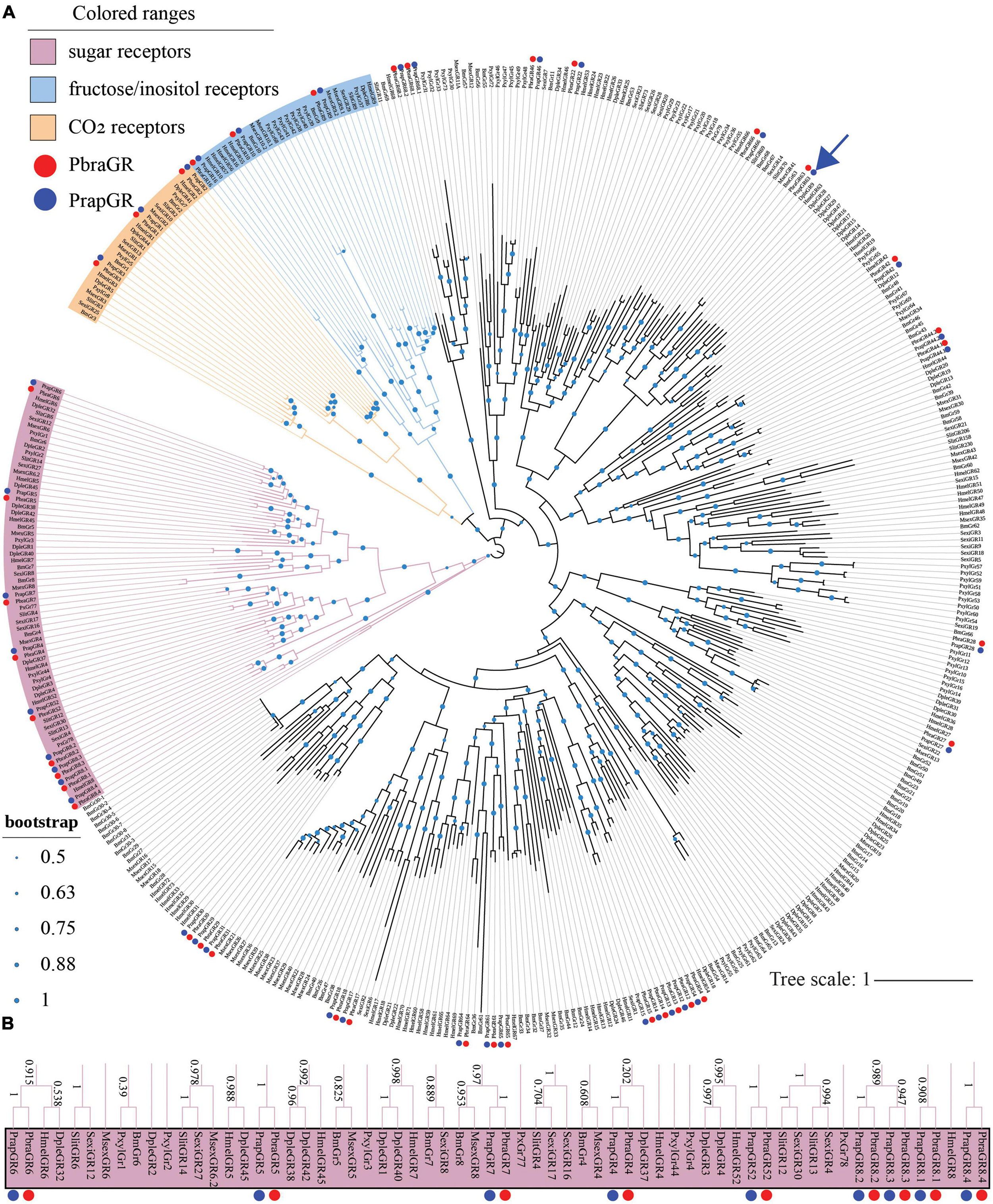
Figure 3. Phylogenetic tree of lepidopteran gustatory receptors. The phylogeny was rooted using sugar receptors. PbraGR, Pieris brassicae GR; PrapGR, P. rapae GR; BmGR, Bombyx mori GR; MsexGR, Manduca sexta GR; DpleIR, Danaus plexippus GR; PxylGR, Plutella xylostella GR; HmelGR, Heliconius melpomene GR; SexiGR, Spodoptera exigua GR; SlitGR, and S. littoralis GR. (A) The CO2 receptor clade is labeled in yellow; the sugar receptor clade is labeled in magenta; the fructose/inositol clade is labeled in blue; the deorphanized sinigrin receptor in P. rapae (Yang et al., 2021) is indicated by a blue arrow. The green dots in the middle of the branches of the phylogenetic tree indicate the bootstrap values higher than 0.5. The scale bar represents the number of substitutions per site. (B) The sugar receptor clade is magnified, and bootstrap values are shown alongside the branches.
3.5. Gene structure and conserved motif of chemoreceptors
Based on the chemoreceptor sequences identified from the genomes of the two Pieris species, we compared the full-length gene structures and analyzed the conserved protein motifs and the arrangements of motifs. We found that most chemoreceptors have a comparable gene structure with other orthologs and with the same number of introns but sometimes with different lengths. For example, PrapOR58 and PbraOR60 have much longer introns than their orthologs (Figure 4). Interestingly, we found that most of IRs in the divergent IRs clade annotated in Figure 3 have no intron at all, such as IR7d.3, IR100c, IR100d, IR100e, IR100f, IR87a, IR7d.2, IR7d.2.1, IR7d.2.2, and IR143; three IRs, i.e., IR85a, IR100a, and IR7d.4, have only one very short intron. Notably, the orthologs having no or only one short intron share highly similarly conserved motifs and arrangements. The other IRs in the divergent IRs clade, i.e., IR1.1, IR1.2, IR2, and IR4, have multiple introns similar to those in antennal IRs clade (Figure 5). However, we also found some exceptions in GRs that showed different number of introns, specifically PbraGR1 has 8 introns while PrapGR1 has 9 introns (Figure 6).
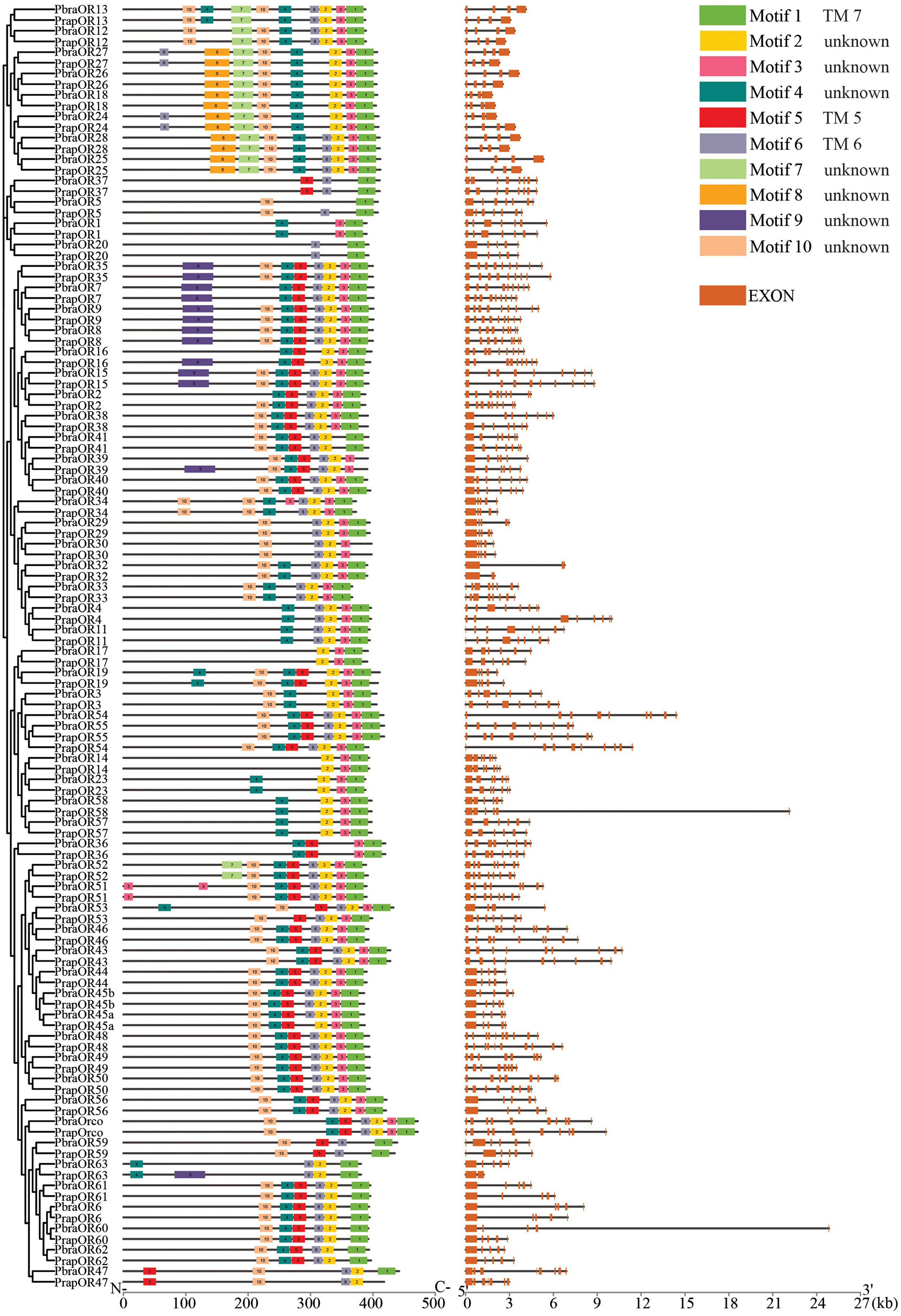
Figure 4. Conserved motifs and arrangements and gene structures of odorant receptors of Pieris brassicae and P. rapae. Scale bars under conserved motifs and gene structures represent the protein length from N- to C- terminus and the transcript length from the 5′ end to the 3′ end. The legend indicates the conserved motifs that are found in the chemoreceptors and the exons that are found in the transcript.
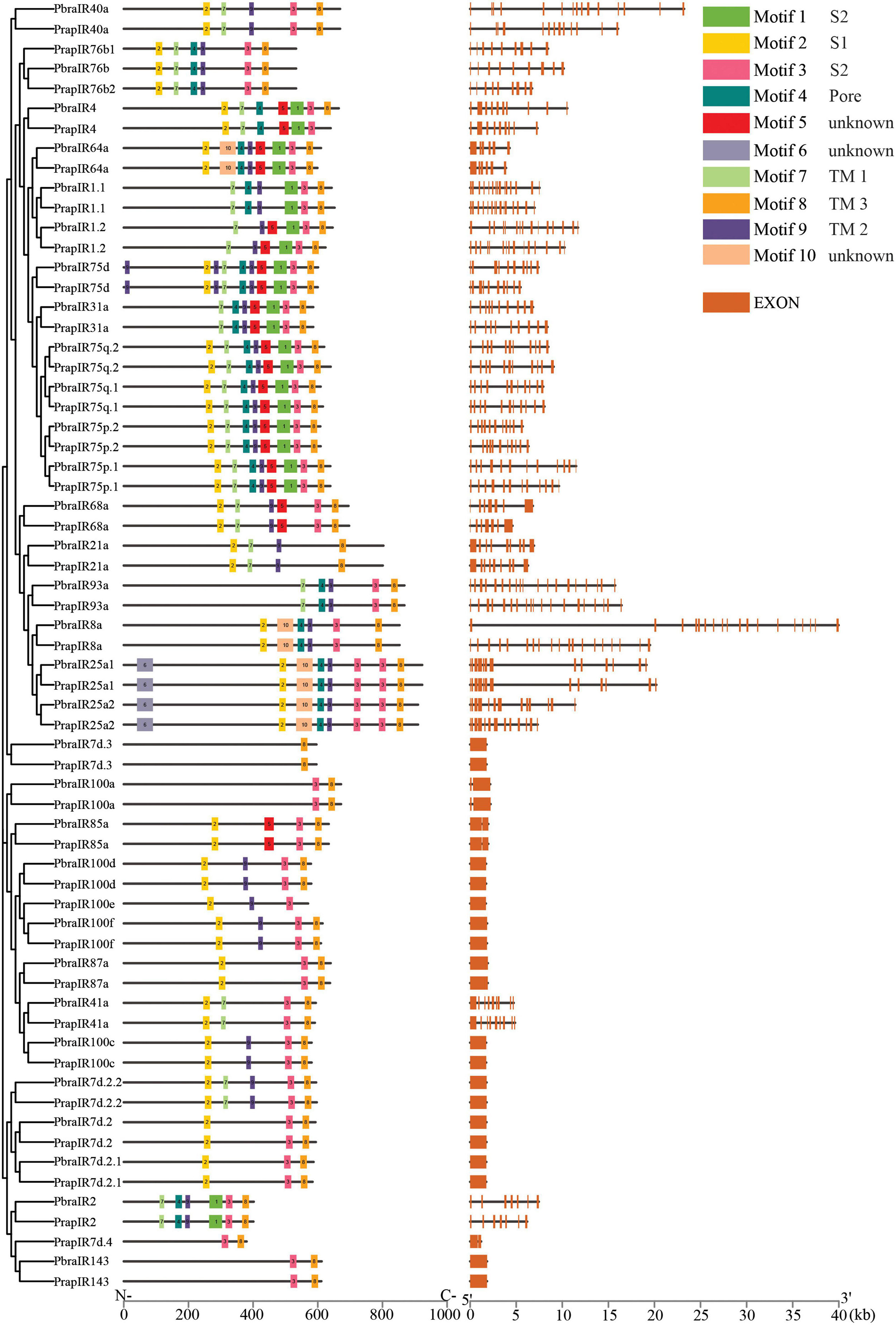
Figure 5. Conserved motifs and gene structures of ionotropic receptors of Pieris brassicae and P. rapae. Scale bars under conserved motifs and gene structures represent the number of amino acids and the number of nucleotides from the 5′ end to the 3′ end.
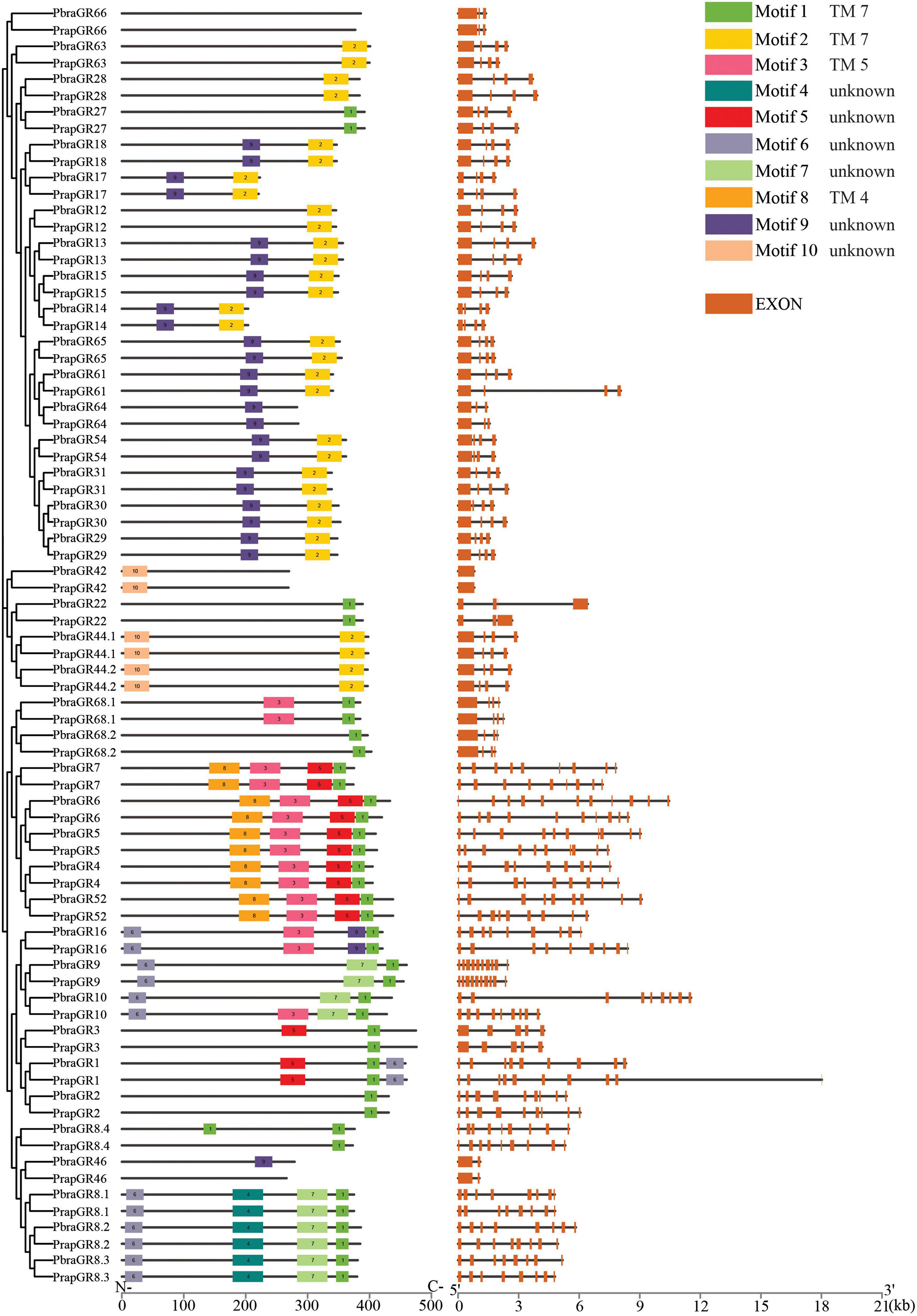
Figure 6. Conserved motifs and gene structures of gustatory receptors of Pieris brassicae and P. rapae. Scale bars under conserved motifs and gene structures represent the number of amino acids and the number of nucleotides from the 5′ end to the 3′ end.
Multiple motifs (sequences see Supplementary material 4) are found in the chemoreceptor protein sequences. Motifs 1, 2, 3, 4, and 5 are the most abundant conserved motifs and are also found in most ORs at similar position of the sequences in both species (Figure 4). According to our transmembrane domain prediction results (Supplementary material 5), we found that ORs exhibit six to seven transmembrane domains. Interestingly, the location of Motif 1 is highly consistent with that of the last transmembrane (TM) domain 7, location of Motif 6 overlaps with TM 6, and Motif 5 covers TM 5 in most ORs. The other conserved motifs are located in intracellular or extracellular loops.
In the IRs Motif 8 is present in all sequences, while Motif 2 is found in most IR sequences. In contrast, Motif 6 is only present in sequences of IR25a1/2 of both species (Figure 5), which is located in amino-terminal domain (ATD). According to the transmembrane predictions, IRs have three to four transmembrane domains. Here, locations of identified conserved motifs are highly consistent with known domains in IRs (Benton et al., 2009; Croset et al., 2010). Motifs 7, 9, and 8 are TM 1, TM 2, and TM 3, respectively. Motif 4 overlaps with the pore domain (P). In addition, Motif 2 was found to locate in ligand-binding domain (LBD) S1 and, Motif 1, and Motif 3 were found to locate in LBD S2.
Compared to ORs and IRs which have four to nine conserved motifs, GRs have fewer conserved motifs in their sequences; for instance, no conserved motif has been found in GR6 and over half of the GRs have only 1–2 conserved motifs (Figure 6). The most common motifs 1 and 2 are only found in about half of the GR sequences. Transmembrane domain prediction results showed that most GRs have six to eight transmembrane domains. Locations of Motif 1 and Motif 2 match well with TM 7 (or the last transmembrane domain) of GRs. Motif 3 and Motif 8 covers TM 5 and TM 4, respectively while the two conserved motifs did not strictly match with the transmembrane domains of GRs.
3.6. Gene duplication and microsynteny
Based on the identification and phylogenetic analysis of chemoreceptors, we found some gene duplication cases. Clade OR18/OR26/OR27, clade OR48/OR49/OR50, and clade OR7/OR8/OR9 are positioned closely on the same scaffold and share high similarities of protein sequences in both Pieris species. Similar cases can also be found in IRs and GRs. IR25a1/IR25a2 and IR100d/IR100f of both species and PrapIR76b1/PrapIR76b2 are placed closely on the same scaffold and show a very high sequence identity. This was also found for GR8.1/GR8.2/GR8.3. Interestingly, while most chemoreceptors mentioned above have the same number of exons, OR49/OR50 have eight exons and are different from OR48 which has ten exons.
In order to clearly elucidate the potential gene duplication and microsyntenic pattern of chemoreceptors in Pieris species, we selected OR7/OR8/OR9 (Figure 7A) and IR25a1/IR25a2 (Figure 7B) as targets to extract their relative positions and gene structures in P. brassicae, P. rapae, P. napi, and P. macdunnoughi. We found that the four selected Pieris species have these target genes, and these paralogs are closely located on the same scaffold in the same order and orthologs share the same number of exons and even share similar exon lengths. In contrast, the ortholog of OR7 is not present in the Pieris outgroup species Ph. sennae and no IR25a duplication was found in any of the selected non-Pieris lepidopteran species (S. litura, Pa. glaucus, V. tameamea, Ph. sennae, D. pasithoe, and C. nemesis).
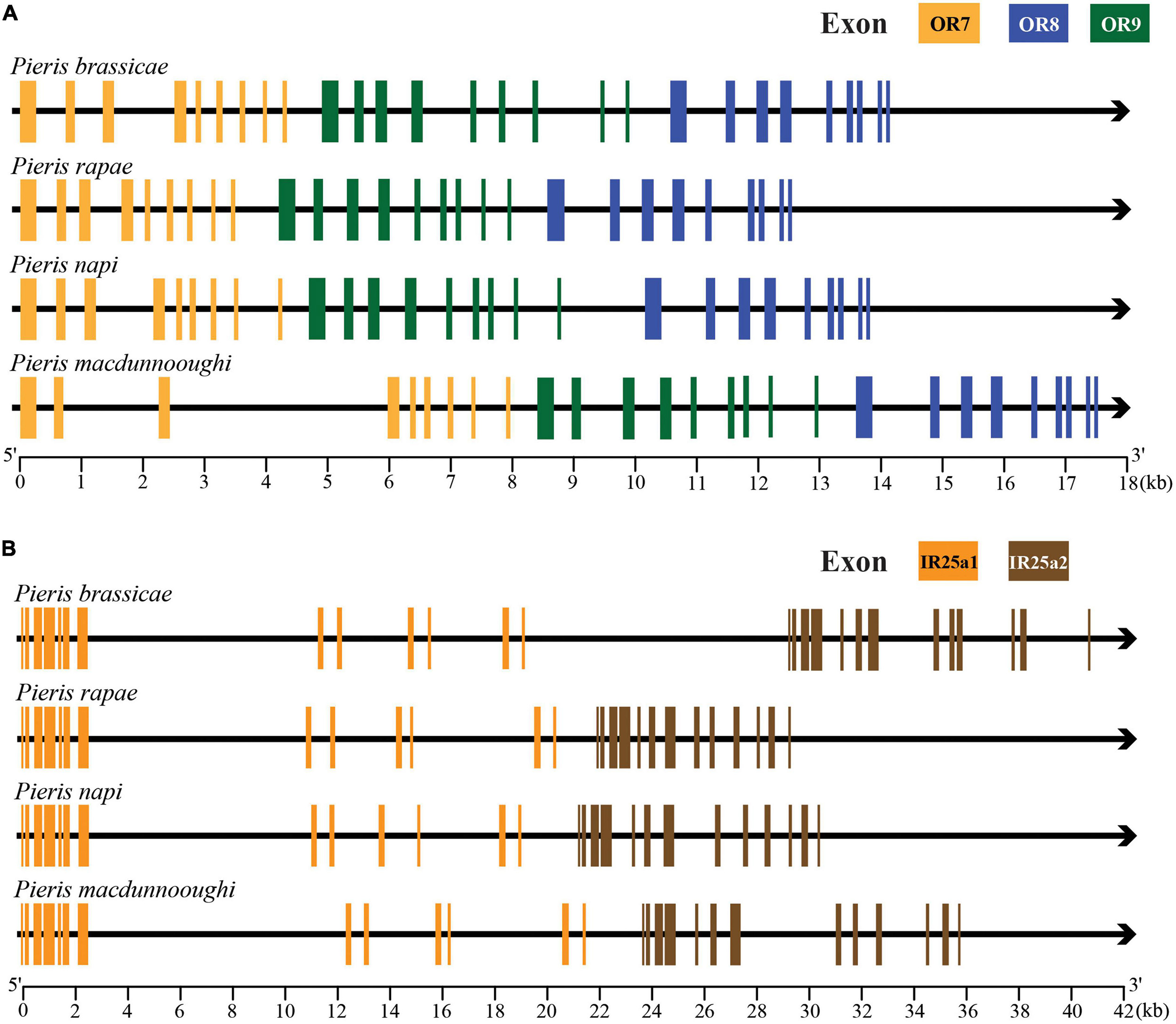
Figure 7. Microsyntenic patterns of OR7/8/9 and IR25a in Pieris. (A) Gene structures and positions of clade OR7/OR8/OR9. Exons of OR7, OR8, and OR9 are labeled in yellow, blue and green, respectively. (B) Gene structures and positions of clade IR25a1/IR25a2. The exons of the IR25a1 and IR25a2 are labeled in orange and brown, respectively. The black arrows indicate introns and directions from 5′ end to the 3′ end. The scales at the bottom of gene structures indicate the length of depicted genes in nucleotides.
3.7. Expression of chemoreceptors in adult transcriptome
We compared the chemoreceptor identification results of the de novo assembly and genome-guided assembly and found the results are identical, although de novo assembly did not always reveal the complete sequences. Sixty PbraORs and 60 PrapORs are identified in the adult antennal transcriptome of P. brassicae and P. rapae, respectively, Orco is the highest expressed gene among chemoreceptors in the transcriptomes. However, out of the 60 genes, the expression levels of 25 ORs were found to be significantly different between the transcriptomes of the two Pieris species. Twenty-two of the 25 differently expressed ORs showed higher expression levels in the P. rapae antennal transcriptome, OR27, OR47, and OR54 had higher expression levels in the P. brassicae antennal transcriptome (Figure 8).
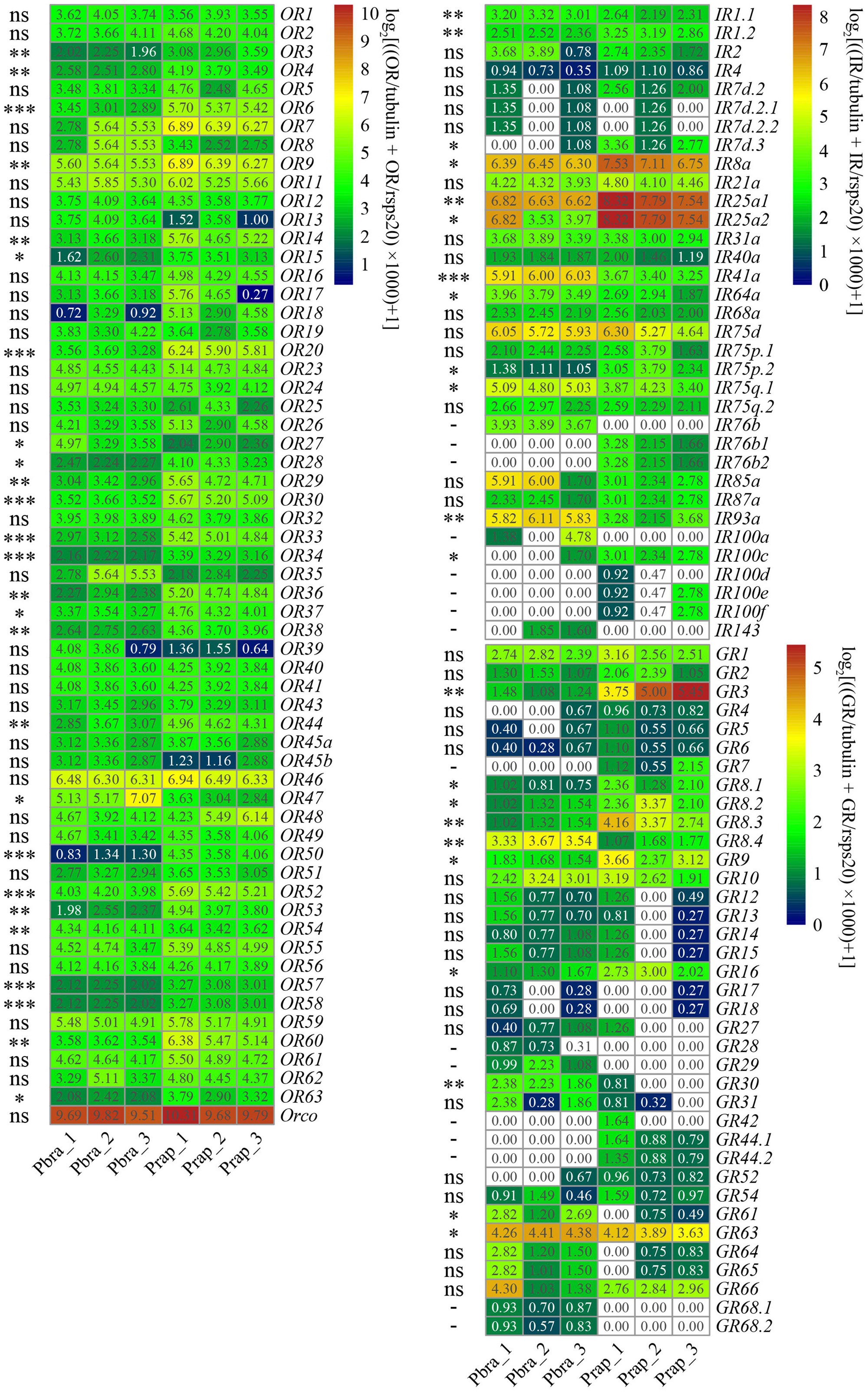
Figure 8. Heatmap of transcriptome of genes encoding chemoreceptors in antennae of Pieris brassicae and P. rapae. The TPM values were normalized by log2 [(TPM chemoreceptor/TPM tubulin + TPM chemoreceptor/TPM rsps20) × 1,000 + 1]. The scales of transformed normalized heatmap values of ORs, IRs, and GRs are 1–10, 0–8, and 0–5, respectively. The white cells indicate that the genes were not found in the transcriptome. values less than 2.0 are in white for ORs and values less than 1.0 are in white for IRs and GRs. Significant difference was detected by t-test with normalized TPM values and labeled as * (P < 0.05), ** (P < 0.01), *** (P < 0.001) or ns (no significance, P > 0.05), –(significant difference was not calculated because the genes were not found).
Twenty-nine and 31 IRs were identified from the P. brassicae and P. rapae adult antennal transcriptomes, respectively. Ircos PbraIR8a, PbraIR25a1, PbraIR93a and those IRs in the antennal IRs clade PbraIR41a, PbraIR75d, PbraIR75q.1, and PbraIR85a were highly expressed in the antenna of P. brassicae. Out of these, 12 IRs were found differently expressed between the two transcriptomes. Seven (IR1.2, IR7d.3, IR8a, IR25a1, IR25a2, IR75p.2, and IR100c) out of the 12 IRs were found to be higher expressed in the P. rapae antennal transcriptome, such as IR8a, IR25a1, and IR25a2 than P. brassicae and five of the 12 IRs (IR1.1, IR41a, IR64a, IR75q.1, and IR93a) had higher expression levels in the P. brassicae antennal transcriptome. In addition, IR76b1, IR76b2, and IR 100d/e/f were not found in the antennal transcriptome of P. brassicae; some of these genes (IR76b1, IR76b2, and IR100e) were only found in the P. rapae genome. IR76b, IR100a, and IR143 are exclusively found in the P. brassicae transcriptome (Figure 8).
In total, 33 GRs were identified from both the P. brassicae and the P. rapae adult antennal transcriptome, although several genes were only identified in one of the two species. Compared to ORs and IRs, GRs have comparatively lower expression in the adult antennal transcriptomes of both species according to normalized TPM values. The sinigrin receptor GR63 was highly expressed in both antennal transcriptomes of P. brassicae and P. rapae. Ten GRs were found differently expressed between the two transcriptomes. Six (GR3, GR8.1, GR8.2, GR8.3, GR9, and GR16) of these ten GRs showed higher expression in the P. rapae antennal transcriptome, the other four GRs (GR8.4, GR30, GR61, and GR63) showed higher expression in the P. brassicae antennal transcriptome. Notably, four PbraGRs (PbraGR7, PbraGR42, PbraGR44.1, and PbraGR44.2) and four PrapGRs (PrapGR28, PrapGR29, PrapGR68.1, and PrapGR68.2) were not detected in the corresponding antennal transcriptomes. These undetected GRs are displayed in white cells in Figure 8.
4. Discussion
In order to clarify the role that chemoreception might play in the ecological adaptation of P. brassicae and P. rapae, we compared the qualitative and quantitative differences in the receptor sets of these two species through genome and transcriptome analyses. In this study we measured the ANI among several butterfly and moth species and found that Pieris species share relatively high sequence identities in comparison to the other lepidopterans. P. brassicae and P. rapae are closely related species, having a similar geographical distribution and host-plant range, but differ in their oviposition strategy and their larval feeding behavior (Feltwell, 1982; Davies and Gilbert, 1985; Ryan et al., 2019). Here, we identified 60 PbraORs, 31 PbraIRs, and 39 PbraGRs from the P. brassicae genome and 60 PrapORs, 34 PrapIRs, and 39 PrapGRs from the P. rapae genome by searching against genomes with lepidopteran homologs. The gene structures and conserved motifs of chemoreceptors in the two Pieris species are identical to their orthologs in the other species. We then analyzed the adult transcriptome data based on the identified chemoreceptors in the genomes of the two Pieris species. We found that 60 ORs, 29 IRs, and 33 GRs were expressed in the P. brassicae antennae, while 60 ORs, 31 IRs, and 33 GRs were expressed in the antennae of P. rapae. The co-receptors such as Orco, IR8a, and IR25a were also found in the transcriptome with very high expression levels. For most chemoreceptor genes homologs were found in both antennal transcriptomes. However, the expression levels of some specific receptor genes differed between the transcriptomes of the two species.
The electrophysiological and behavioral responses to odorants and tastants of the two Pieris species have been extensively investigated over the last decades. These studies showed different sensitivities to amino acids and volatiles for P. brassicae and P. rapae. Specifically, histidine, phenylalanine, and tryptophan are the amino acids that elicit the strongest responses in the lateral sensilla of P. brassicae caterpillars while the three amino acids caused the weakest responses in the same sensillum of P. rapae (van Loon and van Eeuwijk, 1989). Similarly, electroantennogram (EAG) analysis showed that P. brassicae and P. rapae butterflies responded to the same tested chemicals, but with a different responsiveness: trans-hex-2-enal elicited the strongest response in P. brassicae whereas P. rapae showed similarly strong responses to trans-hex-2-enal, hexan-1-ol, and hexanal without significant difference among the three chemicals (van Loon et al., 1992b). However, the mechanism underlying the different responses to these compounds is still unclear. In our study we found significant differences in the expression of several gustatory and ORs and it is plausible that these differences are responsible for the observed contrasts in the chemosensory responses of the two species (Figure 8).
In recent years, identification of chemoreceptors has been performed extensively in a wide range of insect species and some receptors have been functionally characterized as well. The glucosinolates such as glucobrassicin and sinigrin strongly elicit egg-laying behavior in P. rapae and P. brassicae (Renwick and Radke, 1983; Traynier and Truscott, 1991; van Loon et al., 1992a). Sinigrin was recently found as an agonist of a gustatory receptor in P. rapae. The sinigrin receptor GR28 (named GR63 in this study) was characterized at a molecular level by expression in Xenopus oocytes and two-electrode voltage clamp recordings, showing that GR28/GR63 recognizes sinigrin specifically (Yang et al., 2021). Interestingly, we found that GR63 is one of the most highly expressed GR genes in both P. brassicae and P. rapae, with similar expression levels in both species, which corresponds with their similar host-plant choice (Du et al., 1995).
Functional characteristics of IRs seem to be much more divergent than ORs and GRs and even go beyond chemoreception (Wicher and Miazzi, 2021). For example, the Drosophila IR25a is involved in setting the circadian clock by detecting temperature through sensors on the antennae (Chen et al., 2015) and sensing moisture (Knecht et al., 2017). ORs could have originated from GRs in arthropods (Robertson et al., 2003; Robertson, 2019) while IRs are already expressed in the olfactory organs of protostomes, indicating that IRs could be as old as the protostomes and IR25a could be the oldest one (Croset et al., 2010). Interestingly we found that two IR25a paralogs are located on the same scaffold in close proximity in Pieris and we therefore designated the IR25a paralogs as a duplication event. This IR25a duplication only occurred in the four Pieris species (P. brassicae, P. rapae, P. napi, and P. macdunnoughi) which we tested here, but not in any of the other butterfly and moth species that we analyzed. Beyond Pieris, Irco duplications were also found in parasitoid wasp Microplitis mediator (Wang et al., 2015; Wang S. N. et al., 2016) and Aphidius gifuensis (Kang et al., 2017). Here we speculate that the second IR25a gene in Pieris evolved after the speciation from the last common Pieris ancestors, but before the divergence of Pieris. The physiological difference between the two IR25a genes is still unclear and needs to be clarified in the future, although we speculate that the differentiation of IR25a in the Pieris species would improve the efficiency of their interaction with both the biotic as well the abiotic environment considering the multiple functions of this receptor in insects (Ni et al., 2016; Shan et al., 2019).
The evolution of chemoreceptor families is very complicated and often related to specific ecological pressures (Hansson and Stensmyr, 2011). Similar to the duplication of IR25a, we also found cases in other chemoreceptor families, such as the clade OR7/OR8/OR9. These duplications were also found in other Pieris species, but not in other Pieridae species, which suggests that these selected genes also arose in the last common ancestor of the Pieris species. Gene duplication events in olfactory receptors are often related to specific adaptations such as a shift in diet preference (Goldman-Huertas et al., 2015) and it is conceivable that the fact that this specific duplication which can be found in all Pieris species, but not in the close relatives, might be related to some of the multiple adaptation events that took place in the co-evolution of the Pieridae and the Brassicales plants (Edger et al., 2015).
Similarly, to the role gene duplication plays for natural selection, sex pheromone perception has been argued to have strongly contributed to speciation in some lepidopterans, especially moths (Smadja and Butlin, 2009; Leary et al., 2012). Pheromone receptors (PRs) were found and investigated in many species of various insect orders already (Sakurai et al., 2004; Wicher et al., 2017; Wang et al., 2021). However, we did not find any potential PR in the Pieris species in this study, even though our phylogenetic analysis did detect all the previously identified pheromone receptors in the species we used for our comparison (Kaissling and Kasang, 1978; Chisholm et al., 1983; Tumlinson et al., 1989; Kalinová et al., 2001). Until now, the pheromone components of only a few butterfly species have been reported (Meinwald et al., 1969), among them P. brassicae and P. rapae. The large cabbage white, P. brassicae, uses brassica lactone as a sex pheromone and benzyl cyanide as an anti-aphrodisiac compound, while the small cabbage white, P. rapae, uses ferrulactone as a sex pheromone and methyl salicylate and indole as anti- aphrodisiacs (Andersson et al., 2003; Yildizhan et al., 2009). The identified pheromone components of the different butterfly species analyzed so far show a diversity of chemical structures and are mostly unrelated to known sex pheromone components of moths. In addition, no butterfly PR has been identified and functionally deorphanized although abundant PRs have been characterized in different moth species (Krieger et al., 2005; Mitsuno et al., 2008). It is therefore likely that the butterfly PRs are not phylogenetically closely related to the moths’ PRs, but might rather be an independent lineage that detect compounds with a different chemical structure (Bastin-Heline et al., 2019).
In addition to the molecular basis of pheromone perception, the ORs for the recognition of general odors such as plant volatiles and floral odorants in moths have been extensively deorphanized (Tanaka et al., 2009; Cao et al., 2016; de Fouchier et al., 2017; Liu et al., 2020); however, in butterflies, the other large group of lepidopterans, no ORs have been deorphanized so far. This knowledge gap makes it challenging to comprehensively assess the significance of chemoreception in butterflies and hinders a comparison between butterflies and moths, which would help to better understand the divergence between these two groups.
According to previous reports, ORs are highly divergent among insects, sharing low identities with their paralogs even in the same species (Clyne et al., 1999). However, we found that the paralogs among the two Pieris species share highly conserved motifs and a highly similar motif layout, indicating that chemoreceptors are relatively conserved between the two species. Compared to ORs, IRs are more conserved across insect species with specific domains such as ATD, ligand-binding domain (LBD), transmembrane domain (TMD) and pore (P), which was also confirmed by our motif searching results of the IRs. Similar to ORs, GRs were thought to be highly divergent (Clyne et al., 2000), although we found that GRs share similar motifs and motif layouts with their paralogs in the two Pieris species with only a few exceptions found in this study. This suggests that GRs are also rather conserved chemoreceptors between the two Pieris species from the perspective of their protein sequences (Robertson, 2019). These similarities in the gene structure and in the number of chemoreceptor genes in the two Pieris species contrasts to some extent with the rapid gene loss and duplication of chemoreceptor genes that has been found in some Drosophila species (Gardiner et al., 2008; Goldman-Huertas et al., 2015; Auer et al., 2020). However, in many of the well-studied examples the different fly species are not only ecological but also geographically separated (Khallaf and Knaden, 2022). This makes it difficult to directly compare the evolution of sensory genes in drosophilid flies with the two Pieris species, which are not only overlapping in large parts of their geographic range, but also with regards to their behavioral ecology. Interestingly, the differential expression of genes instead of the de novo gene gain could also lead to the divergence of populations within the same species (Sikkink et al., 2017).
The behavior of insects is greatly influenced by the expression of genes encoding for chemoreceptors. The silencing or knocking out of chemoreceptor genes can greatly change the physiology and behavior of insects across different orders (Trible et al., 2017; Yan et al., 2017; Fandino et al., 2019). The higher expression of genes encoding for GRs in female butterflies also facilitates finding an optimal oviposition site (Briscoe et al., 2013). In our study, most chemoreceptors had similar expression levels in the two Pieris species and we speculate that those genes that had similar expression levels, conserved motifs and gene structures, represent a conserved set of chemoreceptors, which are involved in the behavioral traits shared between both species while the differential expression of certain other chemoreceptor orthologs could be involved in more species specific adaptations (Teets et al., 2012; Birnbaum and Abbot, 2020). To fully identify all candidate chemoreceptors in the two Pieris butterfly species, we employed antennal transcriptomic data, which included both male and female antennae samples. Although, combing male and female tissues has provided us with greater certainty on the identification of individual genes, we were unable to compare differential expression of chemoreceptors between the sexes of the two species. Analyzing the sex-biased expression in butterfly chemoreceptors will be an interesting topic for future research, especially in comparison to similarly biased receptors in different moths’ species.
In this study, we identified and systemically compared the chemoreceptors of P. brassicae and P. rapae based on genomic data and antennal transcriptomes. By searching for conserved motifs of chemoreceptors and gene structures, we found that although all chemoreceptors are highly similar to their orthologs in other closely related species, several genes varied significantly in their expression levels. The comparison of the chemoreceptors between these two closely related species, therefore, highlights both the similarities and differences in the structure and expression of chemoreceptors and provides a molecular basis for any further investigation on the evolution of these two ecologically and agriculturally important butterflies.
Data availability statement
The datasets presented in this study can be found in NCBI online database. Accession numbers of sequences and BioProject can be found in this article/Supplementary material.
Author contributions
QW collected and analyzed the data, with the support of AH, and wrote the initial draft of the manuscript. All authors contributed to the final revision and involved in the initial planning and design of the study.
Funding
This project was funded by the NWO VENI grant (016.Veni.192.116) to AH.
Acknowledgments
We would especially like to thank Heiko Vogel and Yu Okamura at the Max Planck Institute for Chemical Ecology, for unreservedly providing the genomic data of P. brassicae. We very much appreciate the critical reviews and very helpful comments of Joop van Loon and Eveline Verhulst at the Laboratory of Entomology. Finally, we would also like to thank Hangwei Liu at Agricultural Genomics Institute at Shenzhen, CAAS and Longfei Chen at Zhejiang University for their professional suggestions on bioinformatical tools.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2023.1155405/full#supplementary-material
References
Andersson, J., Borg-Karlson, A. K., and Wiklund, C. (2003). Antiaphrodisiacs in pierid butterflies: A theme with variation! J. Chem. Ecol. 29, 1489–1499.
Andrews, S. (2010). FASTQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Auer, T. O., Khallaf, M. A., Silbering, A. F., Zappia, G., Ellis, K., Alvarez-Ocana, R., et al. (2020). Olfactory receptor and circuit evolution promote host specialization. Nature 579, 402–408. doi: 10.1038/s41586-020-2073-7
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bastin-Heline, L., De Fouchier, A., Cao, S., Koutroumpa, F., Caballero-Vidal, G., Robakiewicz, S., et al. (2019). A novel lineage of candidate pheromone receptors for sex communication in moths. eLife 8:e49826. doi: 10.7554/eLife.49826
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Birnbaum, S. S. L., and Abbot, P. (2020). Gene expression and diet breadth in plant-feeding insects: Summarizing trends. Trends Ecol. Evol. 35, 259–277. doi: 10.1016/j.tree.2019.10.014
Briscoe, A. D., Macias-Munoz, A., Kozak, K. M., Walters, J. R., Yuan, F., Jamie, G. A., et al. (2013). Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9:e1003620. doi: 10.1371/journal.pgen.1003620
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Cao, S., Liu, Y., Guo, M., and Wang, G. (2016). A conserved odorant receptor tuned to floral volatiles in three Heliothinae species. PLoS One 11:e0155029. doi: 10.1371/journal.pone.0155029
Chen, C., Buhl, E., Xu, M., Croset, V., Rees, J. S., Lilley, K. S., et al. (2015). Drosophila ionotropic receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520. doi: 10.1038/nature16148
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chisholm, M. D., Steck, W. F., Underhill, E. W., and Palaniswamy, P. (1983). Field trapping of diamondback moth Plutella xylostella using an improved four-component sex attractant blend. J. Chem. Ecol. 9, 113–118. doi: 10.1007/BF00987775
Clyne, P. J., Warr, C. G., and Carlson, J. R. (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. doi: 10.1126/science.287.5459.1830
Clyne, P. J., Warr, C. G., Freeman, M. R., Lessing, D., Kim, J., and Carlson, J. R. (1999). A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron 22, 327–338. doi: 10.1016/S0896-6273(00)81093-4
Croset, V., Rytz, R., Cummins, S. F., Budd, A., Brawand, D., Kaessmann, H., et al. (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6:e1001064. doi: 10.1371/journal.pgen.1001064
Cusumano, A., Zhu, F., Volkoff, A. N., Verbaarschot, P., Bloem, J., Vogel, H., et al. (2018). Parasitic wasp-associated symbiont affects plant-mediated species interactions between herbivores. Ecol. Lett. 21, 957–967. doi: 10.1111/ele.12952
Davies, C. R., and Gilbert, N. (1985). A comparative study of the egg-laying behavior and larval development of Pieris rapae L. and P. brassicae L. on the same host plants. Oecologia 67, 278–281. doi: 10.1007/BF00384299
de Fouchier, A., Walker, W. B. III, Montagne, N., Steiner, C., Binyameen, M., Schlyter, F., et al. (2017). Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat. Commun. 8:15709. doi: 10.1038/ncomms15709
De Pasqual, C., Groot, A. T., Mappes, J., and Burdfield-Steel, E. (2021). Evolutionary importance of intraspecific variation in sex pheromones. Trends Ecol. Evol. 36, 848–859. doi: 10.1016/j.tree.2021.05.005
Du, Y. J., Van Loon, J. J. A., and Renwick, J. A. A. (1995). Contact chemoreception of oviposition-stimulating glucosinolates and an oviposition-deterrent cardenolide in two subspecies of Pieris napi. Physiol. Entomol. 20, 164–174. doi: 10.1111/j.1365-3032.1995.tb00813.x
Edger, P. P., Heidel-Fischer, H. M., Bekaert, M., Rota, J., Glockner, G., Platts, A. E., et al. (2015). The butterfly plant arms-race escalated by gene and genome duplications. Proc. Natl. Acad. Sci. U.S.A. 112, 8362–8366. doi: 10.1073/pnas.1503926112
Engsontia, P., Sangket, U., Chotigeat, W., and Satasook, C. (2014). Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: Implications for their adaptation and speciation. J. Mol. Evol. 79, 21–39. doi: 10.1007/s00239-014-9633-0
Fandino, R. A., Haverkamp, A., Bisch-Knaden, S., Zhang, J., Bucks, S., Nguyen, T. A. T., et al. (2019). Mutagenesis of odorant coreceptor Orco fully disrupts foraging but not oviposition behaviors in the hawkmoth Manduca sexta. Proc. Natl. Acad. Sci. U.S.A. 116, 15677–15685. doi: 10.1073/pnas.1902089116
Feltwell, J. (1982). Large white butterfly: The biology, biochemistry and physiology of Pieris brassicae (Linnaeus). The Hague: Dr W. Junk Publishers. doi: 10.1007/978-94-009-8638-1_10
Fitzpatrick, B. M., Fordyce, J. A., and Gavrilets, S. (2008). What, if anything, is sympatric speciation? J. Evol. Biol. 21, 1452–1459. doi: 10.1111/j.1420-9101.2008.01611.x
Foote, A. D. (2018). Sympatric speciation in the genomic era. Trends Ecol. Evol. 33, 85–95. doi: 10.1016/j.tree.2017.11.003
Gardiner, A., Barker, D., Butlin, R. K., Jordan, W. C., and Ritchie, M. G. (2008). Drosophila chemoreceptor gene evolution: Selection, specialization and genome size. Mol. Ecol. 17, 1648–1657. doi: 10.1111/j.1365-294X.2008.03713.x
Goldman-Huertas, B., Mitchell, R. F., Lapoint, R. T., Faucher, C. P., Hildebrand, J. G., and Whiteman, N. K. (2015). Evolution of herbivory in Drosophilidae linked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet. Proc. Natl. Acad. Sci. U.S.A. 112, 3026–3031. doi: 10.1073/pnas.1424656112
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hansson, B. S., and Stensmyr, M. C. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi: 10.1016/j.neuron.2011.11.003
Heliconius Genome, C. (2012). Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98. doi: 10.1038/nature11041
Howlett, N., Dauber, K. L., Shukla, A., Morton, B., Glendinning, J. I., Brent, E., et al. (2012). Identification of chemosensory receptor genes in Manduca sexta and knockdown by RNA interference. BMC Genomics 13:211. doi: 10.1186/1471-2164-13-211
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Kaissling, K. E., and Kasang, G. (1978). A new pheromone of the silkworm moth Bombyx mori. Naturwissenschaften 65, 382–384. doi: 10.1007/BF00439702
Kalinová, B., Hoskovec, M., Liblikas, I., Unelius, C. R., and Hansson, B. S. (2001). Detection of sex pheromone components in Manduca sexta (L.). Chem. Senses 26, 1175–1186. doi: 10.1093/chemse/26.9.1175
Kang, Z. W., Tian, H. G., Liu, F. H., Liu, X., Jing, X. F., and Liu, T. X. (2017). Identification and expression analysis of chemosensory receptor genes in an aphid endoparasitoid Aphidius gifuensis. Sci. Rep. 7:3939. doi: 10.1038/s41598-017-03988-z
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kawahara, A. Y., Plotkin, D., Espeland, M., Meusemann, K., Toussaint, E. F. A., Donath, A., et al. (2019). Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc. Natl. Acad. Sci. U.S.A. 116, 22657–22663. doi: 10.1073/pnas.1907847116
Khallaf, M. A., and Knaden, M. (2022). Evolutionary neuroecology of olfactory-mediated sexual communication and host specialization in Drosophila – a review. Entomol. Exp. Appl. 170, 289–302. doi: 10.1111/eea.13143
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Knecht, Z. A., Silbering, A. F., Cruz, J., Yang, L., Croset, V., Benton, R., et al. (2017). Ionotropic receptor-dependent moist and dry cells control hygrosensation in Drosophila. eLife 6:e26654. doi: 10.7554/eLife.26654
Knolhoff, L. M., and Heckel, D. G. (2014). Behavioral assays for studies of host plant choice and adaptation in herbivorous insects. Annu. Rev. Entomol. 59, 263–278. doi: 10.1146/annurev-ento-011613-161945
Koenig, C., Hirsh, A., Bucks, S., Klinner, C., Vogel, H., Shukla, A., et al. (2015). A reference gene set for chemosensory receptor genes of Manduca sexta. Insect Biochem. Mol. Biol. 66, 51–63. doi: 10.1016/j.ibmb.2015.09.007
Krieger, J., Grosse-Wilde, E., Gohl, T., and Breer, H. (2005). Candidate pheromone receptors of the silkmoth Bombyx mori. Eur. J. Neurosci. 21, 2167–2176. doi: 10.1111/j.1460-9568.2005.04058.x
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E. Jr., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 109, 14081–14086. doi: 10.1073/pnas.1204661109
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, N. Y., Xu, W., Dong, S. L., Zhu, J. Y., Xu, Y. X., and Anderson, A. (2018). Genome-wide analysis of ionotropic receptor gene repertoire in Lepidoptera with an emphasis on its functions of Helicoverpa armigera. Insect Biochem. Mol. Biol. 99, 37–53. doi: 10.1016/j.ibmb.2018.05.005
Liu, N. Y., Xu, W., Papanicolaou, A., Dong, S. L., and Anderson, A. (2014). Identification and characterization of three chemosensory receptor families in the cotton bollworm Helicoverpa armigera. BMC Genomics 15:597. doi: 10.1186/1471-2164-15-597
Liu, X. L., Zhang, J., Yan, Q., Miao, C. L., Han, W. K., Hou, W., et al. (2020). The molecular basis of host selection in a crucifer-specialized moth. Curr. Biol. 30, 4476–4482.e5. doi: 10.1016/j.cub.2020.08.047
Meinwald, J., Meinwald, Y. C., and Mazzocchi, P. H. (1969). Sex pheromone of the queen butterfly: Chemistry. Science 164, 1174–1175. doi: 10.1126/science.164.3884.1174
Miles, C. I., Campo, M. L. D., and Renwick, J. A. A. (2005). Behavioral and chemosensory responses to a host recognition cue by larvae of Pieris rapae. J. Comp. Physiol. A 191, 147–155. doi: 10.1007/s00359-004-0580-x
Mitsuno, H., Sakurai, T., Murai, M., Yasuda, T., Kugimiya, S., Ozawa, R., et al. (2008). Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Möller, S., Croning, M. D., and Apweiler, R. (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17, 646–653. doi: 10.1093/bioinformatics/17.7.646
Montagné, N., De Fouchier, A., Newcomb, R. D., and Jacquin-Joly, E. (2015). Advances in the identification and characterization of olfactory receptors in insects. Prog. Mol. Biol. Transl. Sci. 130, 55–80. doi: 10.1016/bs.pmbts.2014.11.003
Ni, L., Klein, M., Svec, K. V., Budelli, G., Chang, E. C., Ferrer, A. J., et al. (2016). The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. eLife 5:e13254. doi: 10.7554/eLife.13254
Okamura, Y., Sato, A., Tsuzuki, N., Murakami, M., Heidel-Fischer, H., and Vogel, H. (2019). Molecular signatures of selection associated with host plant differences in Pieris butterflies. Mol. Ecol. 28, 4958–4970. doi: 10.1111/mec.15268
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Renwick, J. A. A., and Radke, C. D. (1983). Chemical recognition of host plants for oviposition by the cabbage butterfly, Pieris rapae (Lepidoptera: Pieridae). Environ. Entomol. 12, 446–450. doi: 10.1093/ee/12.2.446
Robertson, H. M. (2019). Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 64, 227–242. doi: 10.1146/annurev-ento-020117-043322
Robertson, H. M., Warr, C. G., and Carlson, J. R. (2003). Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 100, 14537–14542. doi: 10.1073/pnas.2335847100
Ryan, S. F., Lombaert, E., Espeset, A., Vila, R., Talavera, G., Dinca, V., et al. (2019). Global invasion history of the agricultural pest butterfly Pieris rapae revealed with genomics and citizen science. Proc. Natl. Acad. Sci. U.S.A. 116, 20015–20024. doi: 10.1073/pnas.1907492116
Sakurai, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., Tanoue, S., et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 101, 16653–16658. doi: 10.1073/pnas.0407596101
Shakeel, M., Zhu, X., Kang, T., Wan, H., and Li, J. (2015). Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J. Asia Pac. Entomol. 18, 123–130. doi: 10.1016/j.aspen.2015.01.001
Shan, S., Wang, S. N., Song, X., Khashaveh, A., Lu, Z. Y., Dhiloo, K. H., et al. (2019). Antennal ionotropic receptors IR64a1 and IR64a2 of the parasitoid wasp Microplitis mediator (Hymenoptera: Braconidate) collaboratively perceive habitat and host cues. Insect Biochem. Mol. Biol. 114:103204. doi: 10.1016/j.ibmb.2019.103204
Shen, J., Cong, Q., Kinch, L. N., Borek, D., Otwinowski, Z., and Grishin, N. V. (2016). Complete genome of Pieris rapae, a resilient alien, a cabbage pest, and a source of anti-cancer proteins. F1000Res. 5:2631. doi: 10.12688/f1000research.9765.1
Sikkink, K. L., Kobiela, M. E., and Snell-Rood, E. C. (2017). Genomic adaptation to agricultural environments: Cabbage white butterflies (Pieris rapae) as a case study. BMC Genomics 18:412. doi: 10.1186/s12864-017-3787-2
Slater, G. S., and Birney, E. (2005). Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6:31. doi: 10.1186/1471-2105-6-31
Smadja, C., and Butlin, R. K. (2009). On the scent of speciation: The chemosensory system and its role in premating isolation. Heredity 102, 77–97. doi: 10.1038/hdy.2008.55
Smallegange, R. C., van Loon, J. J. A., Blatt, S. E., Harvey, J. A., Agerbirk, N., and Dicke, M. (2007). Flower vs. leaf feeding by Pieris brassicae: Glucosinolate-rich flower tissues are preferred and sustain higher growth rate. J. Chem. Ecol. 33, 1831–1844. doi: 10.1007/s10886-007-9350-x
Tanaka, K., Uda, Y., Ono, Y., Nakagawa, T., Suwa, M., Yamaoka, R., et al. (2009). Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 19, 881–890. doi: 10.1016/j.cub.2009.04.035
Teets, N. M., Peyton, J. T., Colinet, H., Renault, D., Kelley, J. L., Kawarasaki, Y., et al. (2012). Gene expression changes governing extreme dehydration tolerance in an Antarctic insect. Proc. Natl. Acad. Sci. U.S.A. 109, 20744–20749. doi: 10.1073/pnas.1218661109
Traynier, R. M., and Truscott, R. J. (1991). Potent natural egg-laying stimulant for cabbage butterfly Pieris rapae. J. Chem. Ecol. 17, 1371–1380. doi: 10.1007/BF00983770
Trible, W., Olivos-Cisneros, L., Mckenzie, S. K., Saragosti, J., Chang, N. C., Matthews, B. J., et al. (2017). Orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell 170, 727–735.e10. doi: 10.1016/j.cell.2017.07.001
Tsirigos, K. D., Peters, C., Shu, N., Käll, L., and Elofsson, A. (2015). The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407. doi: 10.1093/nar/gkv485
Tumlinson, J. H., Brennan, M. M., Doolittle, R. E., Mitchell, E. R., Brabham, A., Mazomenos, B. E., et al. (1989). Identification of a pheromone blend attractive to Manduca sexta (L.) males in a wind tunnel. Arch. Insect Biochem. Physiol. 10, 255–271. doi: 10.1002/arch.940100402
van Loon, J. J. A., Blaakmeer, A., Griepink, F. C., van Beek, T. A., Schoonhoven, L. M., and de Groot, A. (1992a). Leaf surface compound from Brassica oleracea (Cruciferae) induces oviposition by Pieris brassicae (Lepidoptera: Pieridae). Chemoecology 3, 39–44. doi: 10.1007/BF01261455
van Loon, J. J. A., Frentz, W. H., and van Eeuwijk, F. A. (1992b). Electroantennogram responses to plant volatiles in two species of Pieris butterflies. Entomol. Exp. Appl. 62, 253–260. doi: 10.1111/j.1570-7458.1992.tb00665.x
van Loon, J. J. A., and van Eeuwijk, F. A. (1989). Chemoreception of amino acids in larvae of two species of Pieris. Physiol. Entomol. 14, 459–469. doi: 10.1111/j.1365-3032.1989.tb01115.x
Walker, W. B. III, Gonzalez, F., Garczynski, S. F., and Witzgall, P. (2016). The chemosensory receptors of codling moth Cydia pomonella-expression in larvae and adults. Sci. Rep. 6:23518. doi: 10.1038/srep23518
Walker, W. B. III, Roy, A., Anderson, P., Schlyter, F., Hansson, B. S., and Larsson, M. C. (2019). Transcriptome analysis of gene families involved in chemosensory function in Spodoptera littoralis (Lepidoptera: Noctuidae). BMC Genomics 20:428. doi: 10.1186/s12864-019-5815-x
Wan, F., Yin, C., Tang, R., Chen, M., Wu, Q., Huang, C., et al. (2019). A chromosome-level genome assembly of Cydia pomonella provides insights into chemical ecology and insecticide resistance. Nat. Commun. 10:4237. doi: 10.1038/s41467-019-12175-9
Wang, Q., Xiao, Y., An, X. K., Shan, S., Khashaveh, A., Gu, S. H., et al. (2021). Functional characterization of a candidate sex pheromone receptor AlinOR33 involved in the chemoreception of Adelphocoris lineolatus. J. Agric. Food Chem. 69, 6769–6778. doi: 10.1021/acs.jafc.1c01319
Wang, S. N., Peng, Y., Lu, Z. Y., Dhiloo, K. H., Gu, S. H., Li, R. J., et al. (2015). Identification and expression analysis of putative chemosensory receptor genes in Microplitis mediator by antennal transcriptome screening. Int. J. Biol. Sci. 11, 737–751. doi: 10.7150/ijbs.11786
Wang, S. N., Peng, Y., Lu, Z. Y., Dhiloo, K. H., Zheng, Y., Shan, S., et al. (2016). Cloning and expression profile of ionotropic receptors in the parasitoid wasp Microplitis mediator (Hymenoptera: Braconidae). J. Insect Physiol. 90, 27–35. doi: 10.1016/j.jinsphys.2016.05.002
Wang, Y., Chen, Q., Zhao, H., and Ren, B. (2016). Identification and comparison of candidate olfactory genes in the olfactory and non-olfactory organs of elm pest Ambrostoma quadriimpressum (Coleoptera: Chrysomelidae) based on transcriptome analysis. PLoS One 11:e0147144. doi: 10.1371/journal.pone.0147144
Wanner, K. W., and Robertson, H. M. (2008). The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17, 621–629. doi: 10.1111/j.1365-2583.2008.00836.x
Wicher, D., and Miazzi, F. (2021). Functional properties of insect olfactory receptors: Ionotropic receptors and odorant receptors. Cell Tissue Res. 383, 7–19. doi: 10.1007/s00441-020-03363-x
Wicher, D., Morinaga, S., Halty-Deleon, L., Funk, N., Hansson, B., Touhara, K., et al. (2017). Identification and characterization of the bombykal receptor in the hawkmoth Manduca sexta. J. Exp. Biol. 220, 1781–1786. doi: 10.1242/jeb.154260
Yan, H., Opachaloemphan, C., Mancini, G., Yang, H., Gallitto, M., Mlejnek, J., et al. (2017). An engineered Orco mutation produces aberrant social behavior and defective neural development in ants. Cell 170, 736–747.e9. doi: 10.1016/j.cell.2017.06.051
Yang, J., Guo, H., Jiang, N. J., Tang, R., Li, G. C., Huang, L. Q., et al. (2021). Identification of a gustatory receptor tuned to sinigrin in the cabbage butterfly Pieris rapae. PLoS Genet. 17:e1009527. doi: 10.1371/journal.pgen.1009527
Yang, K., Gong, X. L., Li, G. C., Huang, L. Q., Ning, C., and Wang, C. Z. (2020). A gustatory receptor tuned to the steroid plant hormone brassinolide in Plutella xylostella (Lepidoptera: Plutellidae). eLife 9:e64114. doi: 10.7554/eLife.64114
Yildizhan, S., van Loon, J., Sramkova, A., Ayasse, M., Arsene, C., Ten Broeke, C., et al. (2009). Aphrodisiac pheromones from the wings of the small cabbage white and large cabbage white butterflies, Pieris rapae and Pieris brassicae. Chembiochem 10, 1666–1677. doi: 10.1002/cbic.200900183
Yin, J., Sun, L., Zhang, Q., and Cao, C. (2020). Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene 749:144712. doi: 10.1016/j.gene.2020.144712
Yin, N. N., Nuo, S. M., Xiao, H. Y., Zhao, Y. J., Zhu, J. Y., and Liu, N. Y. (2021). The ionotropic receptor gene family in Lepidoptera and Trichoptera: Annotation, evolutionary and functional perspectives. Genomics 113, 601–612. doi: 10.1016/j.ygeno.2020.09.056
Zhan, S., Merlin, C., Boore, J. L., and Reppert, S. M. (2011). The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185. doi: 10.1016/j.cell.2011.09.052
Zhang, J., Wang, B., Dong, S., Cao, D., Dong, J., Walker, W. B., et al. (2015). Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PLoS One 10:e0117054. doi: 10.1371/journal.pone.0117054
Zhang, Y. N., Qian, J. L., Xu, J. W., Zhu, X. Y., Li, M. Y., Xu, X. X., et al. (2018). Identification of chemosensory genes based on the transcriptomic analysis of six different chemosensory organs in Spodoptera exigua. Front. Physiol. 9:432. doi: 10.3389/fphys.2018.00432
Zhao, H., Du, Y., Gao, P., Wang, S., Pan, J., and Jiang, Y. (2016). Antennal transcriptome and differential expression analysis of five chemosensory gene families from the Asian honeybee Apis cerana cerana. PLoS One 11:e0165374. doi: 10.1371/journal.pone.0165374
Zhao, Y., Cui, K., Li, H., Ding, J., Mu, W., and Zhou, C. (2020). Identification and expression analysis of chemosensory receptor genes in Bradysia odoriphaga (Diptera: Sciaridae). J. Econ. Entomol. 113, 435–450. doi: 10.1093/jee/toz286
Zhou, X., Slone, J. D., Rokas, A., Berger, S. L., Liebig, J., Ray, A., et al. (2012). Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 8:e1002930. doi: 10.1371/journal.pgen.1002930
Zhu, F., Cusumano, A., Bloem, J., Weldegergis, B. T., Villela, A., Fatouros, N. E., et al. (2018). Symbiotic polydnavirus and venom reveal parasitoid to its hyperparasitoids. Proc. Natl. Acad. Sci. U.S.A. 115, 5205–5210. doi: 10.1073/pnas.1717904115
Keywords: Pieris, sympatric speciation, sensory drive, chemoreceptors, genome and transcriptome, lepidoptera
Citation: Wang Q, Dicke M and Haverkamp A (2023) Sympatric Pieris butterfly species exhibit a high conservation of chemoreceptors. Front. Cell. Neurosci. 17:1155405. doi: 10.3389/fncel.2023.1155405
Received: 31 January 2023; Accepted: 17 April 2023;
Published: 11 May 2023.
Edited by:
Xin-Cheng Zhao, Henan Agricultural University, ChinaReviewed by:
Carolina Gomez-Diaz, University of Oviedo, SpainCamille Meslin, INRA Centre Versailles-Grignon, France
Copyright © 2023 Wang, Dicke and Haverkamp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexander Haverkamp, YWxleGFuZGVyLmhhdmVya2FtcEB3dXIubmw=
 Qi Wang
Qi Wang Marcel Dicke
Marcel Dicke Alexander Haverkamp
Alexander Haverkamp