- 1Department of Orthopedics, Tianjin Medical University General Hospital, Tianjin, China
- 2Department of Orthopedics, International Science and Technology Cooperation Base of Spinal Cord Injury, Tianjin Key Laboratory of Spine and Spinal Cord Injury, Tianjin Medical University General Hospital, Tianjin, China
- 3The Affiliated Hospital of Medical School, Ningbo University, Ningbo, China
N6-methyladenosine (m6A), an essential post-transcriptional modification in eukaryotes, is closely related to the development of pathological processes in neurological diseases. Notably, spinal cord injury (SCI) is a serious traumatic disease of the central nervous system, with a complex pathological mechanism which is still not completely understood. Recent studies have found that m6A modification levels are changed after SCI, and m6A-related regulators are involved in the changes of the local spinal cord microenvironment after injury. However, research on the role of m6A modification in SCI is still in the early stages. This review discusses the latest progress in the dynamic regulation of m6A modification, including methyltransferases (“writers”), demethylases (“erasers”) and m6A -binding proteins (“readers”). And then analyses the pathological mechanism relationship between m6A and the microenvironment after SCI. The biological processes involved included cell death, axon regeneration, and scar formation, which provides new insight for future research on the role of m6A modification in SCI and the clinical transformation of strategies for promoting recovery of spinal cord function.
Introduction
N6-methyladenosine (m6A) modification, a type of posttranscriptional modification, has been confirmed to be involved in the post-transcriptional regulation of gene (Roundtree et al., 2017a; Zhao et al., 2017). It was first discovered in mammals in the 1970s (Desrosiers et al., 1974). Notably, m6A is the most common reversible modification found in higher eukaryotic mRNAs (Desrosiers et al., 1974). The dynamic modification of m6A depends on the action of intracellular methylase and demethylase. The former includes methyltransferase-like (METTL) 3, METTL14, Wilms tumor 1-associating protein (WTAP), etc. And the latter includes Fat mass and obesity-associated protein (FTO) and human AlkB homolog 5 (ALKBH5). In addition, m6A-binding proteins also affect RNA metabolism, such as YT521-B homology domain protein family members (YTHDF1-3/YTHDC1-2), heterogeneous nuclear ribonucleoprotein C (HNRNPC) and insulin-like growth factor 2 mRNA-binding proteins 1/2/3 (IGF2BP1/2/3) (Dominissini et al., 2012; Wang et al., 2014; Liu et al., 2015; Huang et al., 2018; Zaccara and Jaffrey, 2020). Moreover, recent studies have demonstrated that m6A is closely related to biological processes of the nervous system, such as brain and cerebellum development, axonal and synaptic formation, gliogenesis, etc (Walters et al., 2017; Yoon et al., 2017; Ma et al., 2018; Xu et al., 2020; Zhao F. et al., 2021).
Spinal cord injury (SCI), a catastrophic condition resulting from a combination of factors, is associated with high rates of disability and fatality and always reduces patient quality of life and imposes a financial burden on families (National SCI Statistical Center [NSCISC], 2016; Ahuja et al., 2017; Tran et al., 2018). Notably, there are no established strategies for completely alleviating SCI and no ideal methods for completely restoring the function of the spinal cord (Venkatesh et al., 2019). Traumatic spinal cord injury is a common type of SCI in clinic (Ahuja et al., 2017). It has two progressive phases: primary injury and secondary injury (Tator, 1995; McDonald and Sadowsky, 2002). The former describes the damage inflicted by direct impact, and the severity of primary injury is proportional to the magnitude of the force applied and the location of the injury (McDonald and Sadowsky, 2002). Secondary injury occurs shortly after primary injury and is accompanied by a series of microenvironmental changes, such as localized hemorrhage and ischemia, inflammation, ionic and neural factor imbalance, glial scarring, and programmed cell death (PCD) (McDonald and Sadowsky, 2002; Fan et al., 2018). Therefore, reducing secondary injury and enhancing functional recovery are key for treating SCI. Fully elucidating the pathogenic mechanisms of SCI is especially critical. Recent studies have found that after SCI, the overall m6A level in the lesion site is increased, and the content of related regulatory factors, such as METTL3 and METTL14, are increased (Xing et al., 2021; Wang et al., 2021; Gao et al., 2022). Furthermore, it was discovered that the specific knockout of mettl14 helps functional recovery after SCI and reduces neuronal apoptosis (Wang et al., 2021; Gao et al., 2022). However, the function of m6A modification in SCI has yet to be fully elucidated. The pathological changes in nerve-related cells and repair processes after SCI may be related to RNA m6A modification, and determining how m6A modification influences these changes may provide insights into novel therapeutic strategies for SCI.
In this review, we summarize the current state of research on m6A modification and emphasize the regulatory mechanism of this type of modification in various pathological processes associated with dysfunction of the nervous system after injury and subsequent tissue repair after SCI to provide a theoretical basis for future research on SCI.
The regulatory mechanism of N6-methyladenosine modification
Since the discovery of m6A modification, researchers have continued to explore its mechanism and function. With the emergence of various sequencing technologies, such as m6A-seq, MeRIP-seq, m6A-CLIP, and m6A-sensitive HRM analysis, etc., it has been found that m6A modification is ubiquitous in coding and non-coding RNAs (Dominissini et al., 2012; Coker et al., 2019; Wang and Jia, 2020). The deposition of m6A on RNA affects mRNA metabolism, including mRNA nuclear export, splicing, translation, transcription, and degradation (Roundtree et al., 2017b; Huang et al., 2018; Liu J. et al., 2020; Cho et al., 2021; Mendel et al., 2021). Interestingly, numerous studies have confirmed that m6A modification sites are conserved in mRNA and that m6A preferentially binds to regions near stop codons or 3′ and 5′ untranslated regions (Meyer et al., 2012; Meyer et al., 2015). Notably, the conserved mRNA sequence to which m6A binds is generally “RRACH,” where R represents adenine or guanine and H can represent adenine, cytosine, or uracil (Harper et al., 1990). Moreover, successful methylation of the sixth N of adenylate is inextricably linked to m6A-regulating factors, including “writers,” “erasers,” and “readers” (Zaccara et al., 2019).
Writers
Intracellular RNA methylation often requires co-catalysis by various enzymes, which are named “writers” (Oerum et al., 2021). The methyltransferase complex, which consists of a heterodimeric core formed by METTL3-METTL14 and additional enzymes, such as WTAP (Liu et al., 2014; Ping et al., 2014), normally catalyzes m6A modification (Liu et al., 2014). METTL3, which has been widely studied since it was first discovered in 1997, is known to be the catalytic core of the methylase complex (Bokar et al., 1997; Oerum et al., 2021). Another enzyme, METTL14, plays a synergistic role with METTL3, as both are essential components of the methylase complex (Wang et al., 2016). Binding of METTL14 to RNA enhances the methylase activity of METTL3 and stabilizes the complex structure (Wang et al., 2016).
In addition to Mettl3/14, the role of other writers is also worth exploring. First, WTAP plays a regulatory role in the methylase complex, linking the complex to RNA, and deletion of WTAP results in in aberrant gene expression and alternative splicing (Ping et al., 2014). Recent research on the development and progression of ataxia and neuronal degeneration has revealed that WTAP expression is associated with disease progression and prognosis (Yang et al., 2022). WTAP-deficient mice not only had lower methylation levels in cerebellar Purkinje cells, but they also developed cerebellar atrophy and ataxia over time (Yang et al., 2022). Moreover, METTL16, another member of the METTL family, binds to U6 snRNA, ncRNAs, lncRNAs, and pre-mRNAs to catalyze methyl synthesis and is implicated in RNA splicing and translating (Pendleton et al., 2017; Warda et al., 2017; Satterwhite and Mansfield, 2022). Additionally, METTL16 can promote translation initiation by interacting with eukaryotic initiation factor 3a/b and rRNA in the cytoplasmic matrix, which is dependent on Mtase domain of METTL16 (Su et al., 2022). Furthermore, translation-related rRNAs can be methylated by another methylase, METTL5. METTL5 is essential for cell activity and differentiation potential and is required for effective translation (Ignatova et al., 2020). Mettl5 deficiency reduces overall translation rate, cell pluripotency, and differentiation potential in mouse embryonic stem cells (Ignatova et al., 2020). Additionally, cell translation and proliferation are related to ZCCHC4, a novel m6A writer that can interact with human 28S rRNA and mRNAs in vitro and in vivo (Ma et al., 2019). A study shows that ZCCHC4 knockout eliminates m6A modification in 28S rRNA, reduces global translation, and inhibits cell proliferation (Ma et al., 2019).
Erasers
Demethylases can remove methyl groups from nucleotides, and the discovery of m6A demethylases, generally known as “erasers,” reveals that the m6A modification of RNA may be reversed dynamically (Yu et al., 2018). FTO and ALKBH5, both of which are AlkB proteins, can effectively decreased m6A levels (Jia et al., 2011; Zheng et al., 2013). FTO was the first demethylase to be discovered (Jia et al., 2011). Guifang Jia identified the enzyme “FTO” as m6A demethylase in 2011 and established that m6A is the predominant FTO substrate in the nucleus in vivo and in vitro (Jia et al., 2011). In addition to fat metabolism, FTO has recently been shown to be involved in nervous system pathologies in different contexts (Fischer et al., 2009; Li et al., 2017; Walters et al., 2017; Zhuang et al., 2019).
AlkB homolog 5, another enzyme capable of reversing m6A modification, has also been implicated in posttranscriptional RNA regulation, including mRNA splicing, stability, export and RNA metabolism (Zheng et al., 2013; Covelo-Molares et al., 2021). Inactivation of ALKBH5 causes an increase in m6A levels on mRNAs, and studies have shown that ALKBH5 is essential for the progression of non-neoplastic and neoplastic diseases of the reproductive, immune, circulatory, and nervous systems (Zheng et al., 2013; Cheng et al., 2021; Dong et al., 2021).
Readers
Eukaryotes produce a variety of proteins that can bind to the m6A modification site and affect RNA translation, splicing, and disintegration and other biological processes (Shi et al., 2017). These proteins are referred to as “readers” and include, most notably, YTH domain family protein 1/2/3(YTHDF1/2/3), YTH domain containing 1/2(YTHDC1/2), HNRNPC, and IGF2BP1/2/3 (Dominissini et al., 2012; Wang et al., 2014; Liu et al., 2015; Huang et al., 2018; Zaccara and Jaffrey, 2020).
YTHDF2 interacts with the m6A modification site on RNA, increasing the likelihood of RNA degradation (Wang et al., 2014). YTHDF2 exerts its effect through several pathways. For instance, YTHDF2 accelerates RNA degradation by recruiting the CCR4/NOT complex (Du et al., 2016). It was also shown that YTHDF2 regulates m6A-mediated RNA decay through the YTHDF2-HRSP12-RNase P/MRP axis (Park et al., 2019). Additionally, after YTHDF1 binds to m6A-tagged mRNAs in the cytoplasm, it stimulates ribosome occupancy of its target mRNA and acts in concert with initiation factors to improve the efficiency of mRNA translation (Wang et al., 2015). YTHDF3, another m6A binder, has been found to have two functions (Shi et al., 2017). It can work with YTHDF1 and YTHDF2 to increase mRNA translation or speed up methylated mRNA degradation, respectively (Shi et al., 2017). Furthermore, YTHDC1, a particular nuclear ribonucleic acid-binding protein, promotes alternative splicing by attracting the RNA splicing factor SRSF3 and preventing SRSF10 from binding to mRNAs in the nucleus (Xiao et al., 2016). It also regulates mRNA export from the nucleus to the cytoplasm (Roundtree et al., 2017b). Another member of this family, YTHDC2, is capable of altering the translation efficiency and mRNA abundance of its targets (Hsu et al., 2017). In addition, HNRNPC is also a common nuclear protein that detects and binds to m6A-modified sequences in mRNAs and lncRNAs, affecting target RNA abundance and splicing (Liu et al., 2015). In contrast to YTHDF2, IGF2BP1/2/3 are novel m6A readers that can protect m6A-modified mRNAs from degradation (Huang et al., 2018). They help thousands of potential mRNA targets remain stable and undergo translation (Huang et al., 2018). Recently, a novel m6A “reader,” Prrrc2a, which is strongly associated with oligodendrocyte formation and axonal myelination, was identified by Wu R. et al. (2019). Their study found that Prrc2a can stabilize Oligo2 mRNA after binding to the m6A site (Wu R. et al., 2019). Additionally, when Prrc2a was removed, mice showed developmental abnormalities, such as enlarged lateral ventricles and significantly reduced myelin sheaths (Wu R. et al., 2019).
N6-methyladenosine modification after spinal cord injury
The nervous system is a multicellular network, and the close interactions among numerous nerve cells, such as neurons and glial cells, is essential for the coordination of its functions (Sousa et al., 2017). Direct damage to the spinal cord can disrupt the blood–spinal cord barrier and cause local blood supply insufficiency, directly resulting in cell death (Ahuja et al., 2017). Notably, the subsequent changes in the internal environment of the spinal cord broaden the scope of injury, and local structures undergo corresponding changes, including scar formation and axonal regeneration (Hara et al., 2017; Fan et al., 2018). M6A modifications are at higher levels in the nervous system (Meyer et al., 2012). Changes in M6A content and associated regulatory factors influence nervous system development and function. For instance, METTLl14 deficiency reduced m6A levels in mouse cerebral cortex and prolonged cortical neurogenesis (Yoon et al., 2017). A study has also demonstrated that the deletion of the methylase METTL3 results in ataxia, hypoplastic development of the mouse cerebellum, and an increase in the apoptosis of immature granulosa cells (Wang et al., 2018). Another study found that peripheral nerve damage raised the levels of FTO, G9a protein, and decreased Ehmt2 mRNA m6A methylation level, all of which contributed to the development of neuropathic pain. Additionally, it was shown that reducing FTO expression in the dorsal root ganglion can reduce neuropathic pain caused by injury (Li et al., 2020).
Recent studies have also reported that after SCI, the levels of m6A as well as writers, such as mettl3 and mettl14, in tissues rise dramatically and specific knockout of methylase can alleviate the severity of SCI (Wang et al., 2021; Xing et al., 2021; Gao et al., 2022). This indicates that dynamic m6A modification has a strong potential to regulate the injury mechanism after SCI and influencing functional recovery.
N6-methyladenosine modification and cell death after spinal cord injury
The structural and functional integrity of the spinal cord are the foundations for proper physiological activity (Ahuja et al., 2017). However, SCI is a multistep disorder usually accompanied by massive neuronal cell death, which is one of the reasons why SCI is difficult to treat (Anjum et al., 2020). In addition to the cell destruction induced by direct impact, secondary injury changes the internal environment and structure of the spinal cord and induces PCD of nerve cells (Fan et al., 2018; Shi et al., 2021). Therefore, preserving nerve cells and reducing or even eliminating cell death are critical for the treatment of SCI. To achieve better treatment outcomes, it is essential to explore the mechanism of PCD after SCI.
Programmed cell death is tightly linked to m6A modification (Wang et al., 2020; Lan et al., 2021; Shen et al., 2021; Liu et al., 2022). Apoptosis is a common form of PCD in the nervous system (Fricker et al., 2018). A study showed that knockout of mettl3 results in massive apoptosis of newborn cerebellar granule cells, resulting in dysplasia in the mouse cerebellum (Wang et al., 2018). Similarly, Mettl3 deficiency in the mouse hippocampus increases local apoptosis and alter the cell cycle (Zhao F. et al., 2021). In addition, after ischemic brain injury, overexpression of YTHDC1 reduces neuronal apoptosis (Zhang et al., 2020).
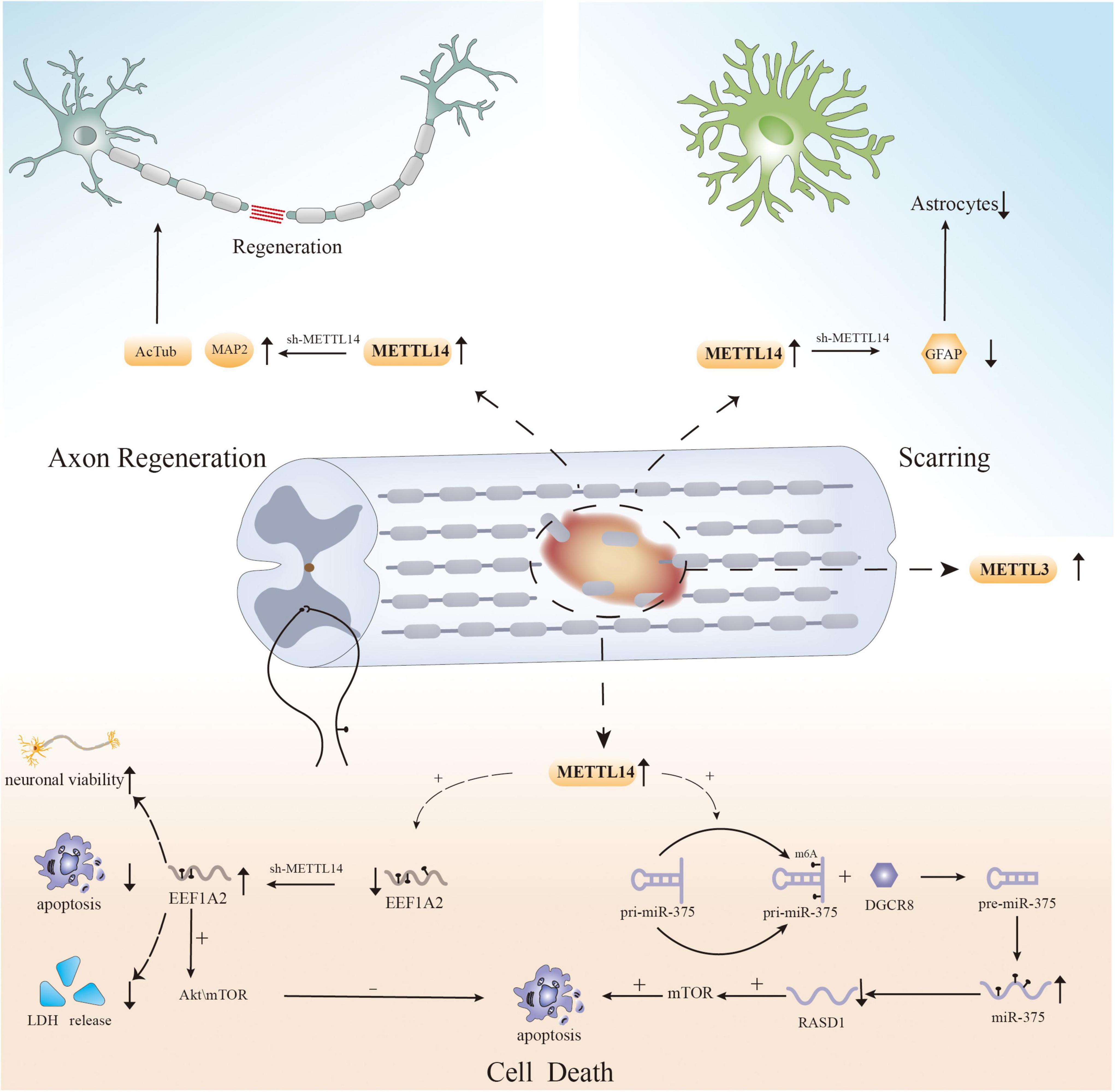
Recently, several studies have shown that methylation regulators can influence cell survival after SCI by regulating m6A levels (Figure 1). Haoyu Wang et al. verified that significant neuronal death and cell dysfunction occur at the site of injury in a rat spinal cord contusion model (Wang et al., 2021). Moreover, m6A levels were increased, and the expression of the “writer” mettl14 is increased (Wang et al., 2021). Surprisingly, inhibiting local Mettl14 expression lowers overall m6A levels and the severity of SCI in experimental animals while also promoting motor function recovery after injury (Wang et al., 2021). To explore the changes at the cellular level, the researchers performed HE staining and immunofluorescence (Wang et al., 2021). The results showed the presence of fewer reactive astrocytes in the injury area and more surviving neurons in the mettl14 knockout group compared to the control group (Wang et al., 2021). More importantly, further experiments also showed that overexpression of Mettl14 can induce apoptosis in vitro, as Mettl14 can promote the conversion of pri-miR-375 to miR-375, which is related to apoptosis and inhibits neural recovery (Wang et al., 2021). In addition, increased expression of METTL14 during SCI mediates the m6A modification of EEF1A2, which accelerates neuronal degeneration through the apoptotic pathway and impairs recovery after injury (Gao et al., 2022). EEF1A2 expression is reduced after SCI, while silencing of mettl14 increases EEF1A2 levels, decreases inflammatory cytokine production, and reduces neuronal degeneration in the spinal cord (Gao et al., 2022).
The above experiments show that the regulation of cell death after SCI, particularly neuronal apoptosis, is influenced by RNA m6A modification, providing a new direction for reducing cellular dysfunction and promoting functional recovery. However, apoptosis is not the only cause of cell loss after injury, and previous studies have shown that other forms of PCD, such as ferroptosis, autophagy, and necroptosis, also mediate cell death after SCI (Fan et al., 2016; Zhou et al., 2020; Feng et al., 2021; Shi et al., 2021). There have been multiple studies on the effect of m6A modification on PCD in different disorders (Yang et al., 2019; Lan et al., 2021; Shen et al., 2021); however, there has been no research on the relationship between m6A modification and other forms of PCD after SCI. Therefore, to properly elucidate the pathogenic mechanism of SCI, researchers must examine the role of m6A modification in other types of PCD after SCI.
N6-methyladenosine modification and axonal regeneration after spinal cord injury
Another cause of functional deficiency following SCI is the disruption of spinal nerve continuity (Ramer et al., 2014; Tran et al., 2018; Varadarajan et al., 2022). Unfortunately, compared to that of the peripheral nervous system, the axonal regeneration capacity of the central nervous system is extremely limited (Hutson and Di Giovanni, 2019; Avraham et al., 2021). Failure of regeneration results in permanent loss of neurological function. Although we have conducted in-depth research on the internal and external environment and regeneration mechanisms after axonal injury, complete axonal regeneration is difficult to achieve (Liu et al., 2011; Varadarajan et al., 2022). Studies have shown that changes related to gene expression can effectively regulate axonal regeneration, which involves physiological processes such as translation and transcription (Moore et al., 2009; Song et al., 2015; Mahar and Cavalli, 2018). Recent research on m6A modification also revealed that RNA modification can influence axonal regeneration, providing a solid theoretical basis for our ongoing research on axonal regeneration (Weng et al., 2018; Zhang et al., 2021; Qi et al., 2022).
In the nervous system, regeneration of neuronal axons is likewise affected by m6A modification. Following sciatic nerve damage, the levels of m6A-tagged transcripts associated with axonal regeneration are increased in mouse dorsal root neurons, facilitating axonal regeneration. Primary neurite length is considerably decreased in vitro when METTL14 is knocked out, as is the capacity to increase the axon length in vivo. In addition to that in the peripheral nervous system, Pten deletion-induced axonal regeneration in CNS neurons is considerably impeded following METTL14 loss. Furthermore, the YTHDF1 reader is required for injury-induced protein translation and axonal regeneration in neurons (Weng et al., 2018). Additionally, another study pointed out that FTO can reduce RNA m6A levels in axons and dynamically regulate local protein translation (Yu et al., 2018). After inhibition of intraneuronal axonal FTO expression by rhein, m6A levels are significantly decreased, and axonal elongation is inhibited (Yu et al., 2018). Interestingly, Mengru Zhuang’s team discovered that the m6A-binding protein YTHDF1 recognizes transcripts and regulates the translation of Robo3.1, which is modified by m6A, provides axonal pathfinding guiding signals, and affects the guidance of crossing axons of spinal cord commissure neurons (Zhuang et al., 2019). In addition, YTHDF1 and YTHDF2 are highly expressed in cerebellar granule cell axons in vitro and in vivo, and knock out of these proteins might enhance axonal development (Yu et al., 2021). To govern neuronal axonal development, YTHDF1 and YTHDF2 synergistically regulate Wnt5a signaling, which is involved in axonal guidance and can influence axonal development (Yu et al., 2021).
Recently, m6A modification was shown to have the potential to regulate axonal regeneration after SCI (Figure 1). In an experiment on SCI in zebrafish and mice, MeRIP-seq and RNA-seq analysis of injured tissue after SCI revealed that RNAs that showed obvious differences in m6A levels, such as hsp90ab1, taf1, igf2bp1, and tp53, were associated with axonal growth and neuronal development (Xing et al., 2021). Simultaneously, the expression of METTL3 was found to be upregulated in local tissues in mouse and zebrafish SCI models, as well as in neural stem cell and astrocyte SCI models (Xing et al., 2021). This is the first study on the role of RNA m6A modification in SCI, and the results suggest that dynamic changes in the methylation of associated genes have an effect on axonal regeneration (Xing et al., 2021). In addition, specific knockout of METTL14 can significantly increase the expression of AcTub and MAP2 after SCI, which are two markers associated with axons whose expression is decreased after SCI. These findings indicate that METTL14 is involved in the regulation of axons after SCI (Wang et al., 2021). And another study found that METTL14 catalyzes the m6A methylation of EEF1A2 mRNA (Gao et al., 2022). Knockdown of mettl14 can increase the level of EEF1A2, and the opposite occurs after mettl14 overexpression (Gao et al., 2022). Moreover, the reduction in EEF1A2 expression after SCI inhibits the Akt/mTOR pathway, which previous studies have shown to affect pathway regeneration (Zhao Y. et al., 2021; Gao et al., 2022). Therefore, m6A modification may have an effect on nerve recovery. The results of the abovementioned experiments suggest that m6A modification could be a potential strategy for affecting axonal regeneration after SCI.
N6-methyladenosine modification and scarring after spinal cord injury
One of the secondary characteristics of SCI is the aggregation of a considerable number of reactive astrocytes, which always results in localized scarring (Hara et al., 2017). Spatially, scars can be used to isolate damaged tissue and prevent damage from spreading further (Tran et al., 2018). In addition to exerting a protective effect, scars inhibit nerve regeneration, which is closely related to the recovery of spinal cord function (Silver and Miller, 2004). Recently, research has shown that scar formation after injury does not necessarily hinder axonal regeneration but may actually promote recovery (Anderson et al., 2016). Compared to that of astrocytes, the role of pericytes in scar formation has received less attention. Pericytes are also crucial for the scarring process (Göritz et al., 2011; Dias et al., 2018). Therefore, research on scar formation from the perspective of m6A modification could open up a new field of research related to SCI.
N6-methyladenosine modification can regulate the physiological functions of astrocytes (Huang et al., 2020; Teng et al., 2021). In a study on major depressive disorder, it was verified that circSTAG1 can bind to the demethylase ALKBH5 in the mouse hippocampus, decreasing ALKBH5 levels to alter the m6A level of FAAH mRNA and limit FAAH expression (Huang et al., 2020). Ultimately, astrocyte dysfunction and astrocyte loss are reduced (Huang et al., 2020). Additionally, METTL14 knockdown reduces m6A levels in the substantia nigra, decreases TH expression, and enhances microglial and astrocyte survival (Teng et al., 2021).
Recently, several studies have shown that changes in m6A modification affect the aggregation of astrocytes following SCI (Figure 1; Wanner et al., 2013). Lingyan Xing et al. found that the expression of METTL3 in astrocytes increases dramatically after SCI, possibly affecting the activation and proliferation of cells (Xing et al., 2021). Although more research is needed, the results indicate a new direction for the study of astrocytes after SCI. Moreover, another study reported that GFAP expression was decreased and the number of astrocytes produced at the injury site was reduced in an SCI model with selective deletion of Mettl14 compared to the control group (Wang et al., 2021). Surprisingly, in vitro, lack of Mettl14 was shown to reduce the apoptosis of C8-D1A murine astrocytes after simulation of SCI-induced apoptosis with H2O2 (Wang et al., 2021). This implies that m6A modification is linked to astrocyte survival after SCI, which can alter scar formation. However, since there are only few related studies, the relationship between m6A and astrocytes after SCI still needs to be further explored.
While astrocytes are involved in scarring postinjury, the role of pericytes in SCI cannot be ignored (Dias et al., 2018). Pericytes are involved in the establishment of the blood–brain barrier and the blood–spinal cord barrier, as well as the stability of the internal environment of the brain and spinal cord (Cheng et al., 2018; Sweeney et al., 2019). Previous studies have shown that pericytes are closely related to the formation of scars and the recovery of function after SCI (Dias et al., 2018; Hesp et al., 2018; Zhu et al., 2022). Some studies have confirmed that m6A modification in pericytes is involved in the occurrence and development of hypertension and diabetes (Wu Q. et al., 2019; Suo et al., 2022). For instance, Qingbin Wu et al. discovered that in pericytes, mRNAs undergo m6A modification in coding regions under hypertensive conditions. Subsequent GO and KEGG enrichment analyses revealed that the differentially expressed genes are linked to hypertension genes and pathways. This suggests that changes in m6A modification in pericytes play a role in the pathogenesis of vascular diseases such as hypertension (Wu Q. et al., 2019). Moreover, a recent study found that diabetes-induced pericyte dysfunction is associated with changes in RNA m6A levels, which are regulated by m6A-related enzymes and proteins (Suo et al., 2022). Selective METTL3 silencing can reduce YTHDF2-induced degradation of PKC, FAT4, and PDGFRA mRNA, reducing the occurrence of diabetes-induced vascular complications and pericyte dysfunction (Suo et al., 2022).
Future directions related to the role of N6-methyladenosine modification after spinal cord injury
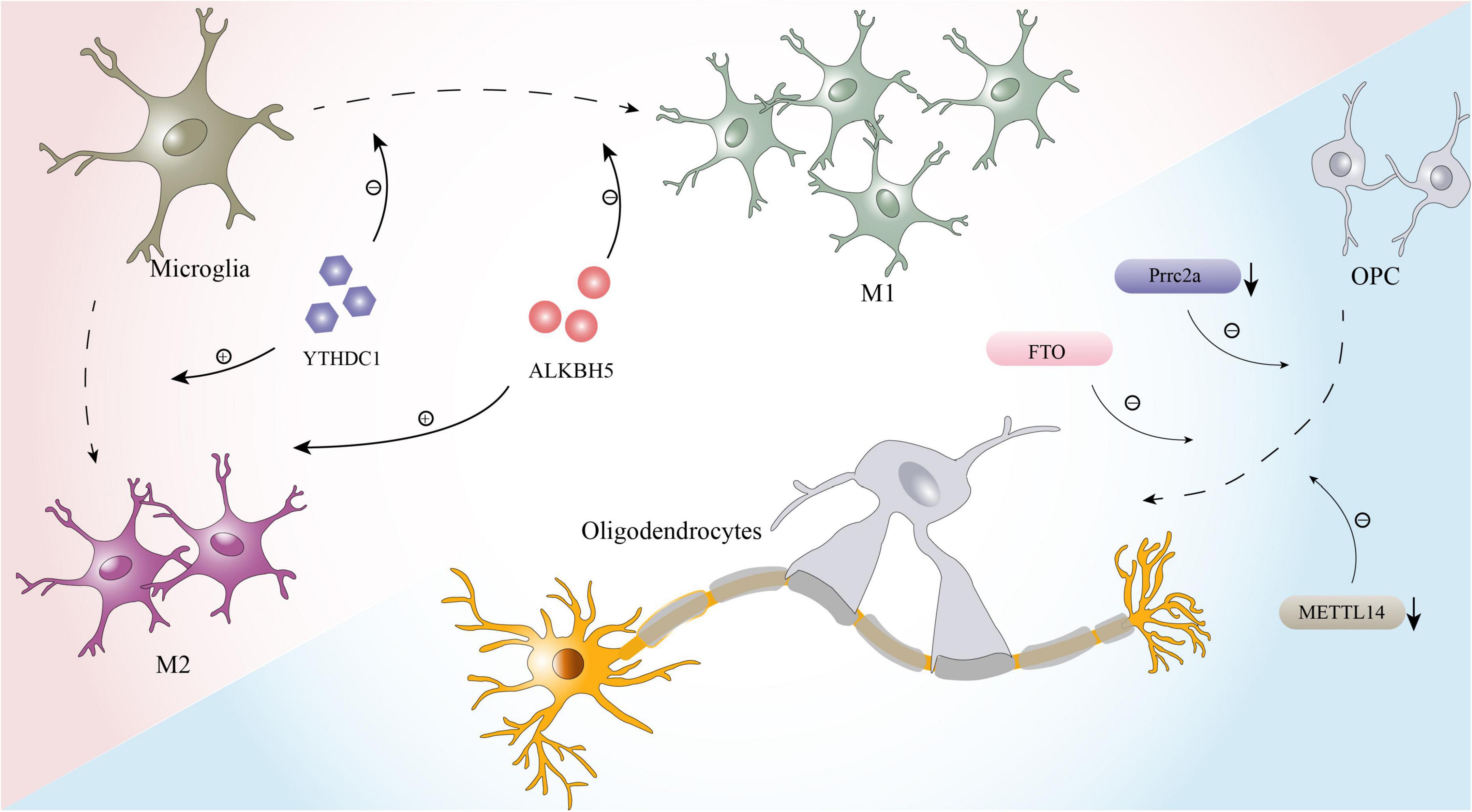
In addition to the pathological processes mentioned above, the effects of local inflammation and myelination dysfunction on prognosis after SCI should not be ignored (Plemel et al., 2014; Zrzavy et al., 2021), and m6A modification is also likely to be involved in these effects. Microglia, which are key factors affecting inflammation after SCI, have two polarization states, the proinflammatory M1 phenotype and the anti-inflammatory M2 phenotype (Lan et al., 2017). After injuries such as stroke, cerebral hemorrhage, SCI, M1 polarization of microglia is often induced (Fan et al., 2018; Liao et al., 2020; Sun et al., 2020). While M1 microglia play a defensive role, they also aggravate neuroinflammation and nerve cell damage, affecting the recovery of nervous system function (Fan et al., 2018). Therefore, reducing the polarization of M1 glial cells or driving their conversion to the anti-inflammatory M2 phenotype can aid nerve recovery and lessen secondary damage (Liu W. et al., 2020). According to recent studies, m6A modification plays a critical role in glial phagocytosis and polarization (Figure 2; Li et al., 2021; Zhou et al., 2021; Chen et al., 2022). A study on uveitis found that deletion of the m6A reader YTHDC1 enhances the M1 polarization of microglia and accelerates inflammation (Zhou et al., 2021). Furthermore, another bioinformatics study showed that m6A has a high potential to modulate the microglia-mediated inflammatory response. A large number of m6A-modified transcripts are among the genes that are differentially expressed between different subtypes of microglia (Li et al., 2021). Researchers have also observed that when microglia are active, m6A levels of the transcripts of many pro- and anti-inflammatory components are altered (Li et al., 2021).
Furthermore, oligodendrocytes, whose primary function is to form the myelin sheath of axons and contribute to the efficient and rapid transmission of information, are inextricably linked to myelin regeneration during the process of nerve repair after SCI (Bradl and Lassmann, 2010; Sankavaram et al., 2019). In recent years, it was proven that m6A modification plays a key role in the development and maturation of oligodendrocytes and maintains the normal function of oligodendrocytes (Figure 2; Wu R. et al., 2019; Xu et al., 2020). For example, Prrrc2a, a novel m6A “reader” identified by Wu R. et al. (2019) is strongly associated with oligodendrocyte formation and axonal myelination. When prrc2a is specifically knocked out, the proliferation and differentiation of OPCs are affected, and the number of mature oligodendrocytes is markedly reduced (Wu R. et al., 2019). Moreover, axons in the corpus callosum exhibit hypomyelination (Wu R. et al., 2019). Interestingly, Xu et al. (2020) performed RNA-seq and m6A-seq of OPCs and successfully induced the differentiation of OPCs from neonatal mice into oligodendrocytes. When METTL14 is inactivated by Cre-loxP, the number of mature oligodendrocytes in postnatal mice is significantly reduced, but the formation and proliferation of OPCs are not affected (Xu et al., 2020).
Conclusion
This review discusses in detail the current status of research on m6A modification and the relationship between m6A modification and pathophysiological processes after SCI, including cell death, axonal regeneration, and scarring. Although there has been research on the role of m6A modification in some neurological diseases, such as Alzheimer’s disease and stroke, research on the role of this posttranslational modification in SCI is still in its infancy. Research on this topic is limited to bioinformatics analysis of gene expression and differential expression at the tissue and cell levels, and studies on the specific mechanism of m6A modification after SCI are extremely rare. Simultaneously, the only m6A modification-regulating molecules that have been studied after SCI are “writers,” and more research on the impact of demethylases and binding proteins after SCI is needed. The importance of m6A modification in neurological diseases cannot be overstated. This dynamic modification could be a possible target for influencing the pathological process of SCI and promoting recovery of spinal cord function. Clearly, the role of m6A modification in SCI needs to be explored further.
Author contributions
DL, BF, and JL contributed the central idea and wrote the manuscript. JM and TS collected the related data. XZ and SF participated in key revisions of the manuscript and finalized the final version. All authors contributed to the revision of the manuscript and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (82102563) and the Tianjin Key Medical Discipline (Specialty) Construct Project (TJYXZDXK-027A).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahuja, C. S., Wilson, J. R., Nori, S., Kotter, M. R. N., Druschel, C., Curt, A., et al. (2017). Traumatic spinal cord injury. Nat. Rev. Dis. Primers 3:17018. doi: 10.1038/nrdp.2017.18
Anderson, M. A., Burda, J. E., Ren, Y., Ao, Y., O’Shea, T. M., Kawaguchi, R., et al. (2016). Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200. doi: 10.1038/nature17623
Anjum, A., Yazid, M. D., Fauzi Daud, M., Idris, J., Ng, A. M. H., Selvi Naicker, A., et al. (2020). Spinal cord injury: pathophysiology, multimolecular interactions, and underlying recovery mechanisms. Int. J. Mol. Sci. 21:7533. doi: 10.3390/ijms21207533
Avraham, O., Feng, R., Ewan, E. E., Rustenhoven, J., Zhao, G., and Cavalli, V. (2021). Profiling sensory neuron microenvironment after peripheral and central axon injury reveals key pathways for neural repair. ELife 10:e68457. doi: 10.7554/eLife.68457
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247.
Bradl, M., and Lassmann, H. (2010). Oligodendrocytes: biology and pathology. Acta Neuropathol. 119, 37–53.
Chen, T., Zhu, W., Wang, C., Dong, X., Yu, F., Su, Y., et al. (2022). ALKBH5-Mediated mA Modification of A20 Regulates Microglia Polarization in Diabetic Retinopathy. Front. Immunol. 13:813979. doi: 10.3389/fimmu.2022.813979
Cheng, J., Korte, N., Nortley, R., Sethi, H., Tang, Y., and Attwell, D. (2018). Targeting pericytes for therapeutic approaches to neurological disorders. Acta Neuropathol. 136, 507–523. doi: 10.1007/s00401-018-1893-0
Cheng, P., Han, H., Chen, F., Cheng, L., Ma, C., Huang, H., et al. (2021). Amelioration of acute myocardial infarction injury through targeted ferritin nanocages loaded with an ALKBH5 inhibitor. Acta Biomater 140, 481–491. doi: 10.1016/j.actbio.2021.11.041
Cho, S., Lee, G., Pickering, B. F., Jang, C., Park, J. H., He, L., et al. (2021). mTORC1 promotes cell growth via mA-dependent mRNA degradation. Mol. Cell 81, 2064–2075. doi: 10.1016/j.molcel.2021.03.010
Coker, H., Wei, G., and Brockdorff, N. (2019). m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta. Gene Regul. Mech. 1862, 310–318. doi: 10.1016/j.bbagrm.2018.12.002
Covelo-Molares, H., Obrdlik, A., Poštulková, I., Dohnálková, M., Gregorová, P., Ganji, R., et al. (2021). The comprehensive interactomes of human adenosine RNA methyltransferases and demethylases reveal distinct functional and regulatory features. Nucleic Acids Res. 49, 10895–10910. doi: 10.1093/nar/gkab900
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975.
Dias, D. O., Kim, H., Holl, D., Werne Solnestam, B., Lundeberg, J., Carlén, M., et al. (2018). Reducing pericyte-derived scarring promotes recovery after spinal cord injury. Cell 173, 153–165. doi: 10.1016/j.cell.2018.02.004
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Dong, F., Qin, X., Wang, B., Li, Q., Hu, J., Cheng, X., et al. (2021). ALKBH5 Facilitates Hypoxia-Induced Paraspeckle Assembly and IL8 Secretion to Generate an Immunosuppressive Tumor Microenvironment. Cancer Res. 81, 5876–5888. doi: 10.1158/0008-5472.Can-21-1456
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7:12626. doi: 10.1038/ncomms12626
Fan, B., Wei, Z., Yao, X., Shi, G., Cheng, X., Zhou, X., et al. (2018). Microenvironment imbalance of spinal cord injury. Cell Transplant. 27, 853–866. doi: 10.1177/0963689718755778
Fan, H., Zhang, K., Shan, L., Kuang, F., Chen, K., Zhu, K., et al. (2016). Reactive astrocytes undergo M1 microglia/macrohpages-induced necroptosis in spinal cord injury. Mol. Neurodegener. 11:14. doi: 10.1186/s13024-016-0081-8
Feng, Z., Min, L., Chen, H., Deng, W., Tan, M., Liu, H., et al. (2021). Iron overload in the motor cortex induces neuronal ferroptosis following spinal cord injury. Redox Biol. 43:101984. doi: 10.1016/j.redox.2021.101984
Fischer, J., Koch, L., Emmerling, C., Vierkotten, J., Peters, T., Brüning, J. C., et al. (2009). Inactivation of the Fto gene protects from obesity. Nature 458, 894–898.
Fricker, M., Tolkovsky, A. M., Borutaite, V., Coleman, M., and Brown, G. C. (2018). Neuronal Cell Death. Physiol. Rev. 98, 813–880.
Gao, G., Duan, Y., Chang, F., Zhang, T., Huang, X., and Yu, C. (2022). METTL14 promotes apoptosis of spinal cord neurons by inducing EEF1A2 m6A methylation in spinal cord injury. Cell Death Discov. 8:15. doi: 10.1038/s41420-021-00808-2
Göritz, C., Dias, D. O., Tomilin, N., Barbacid, M., Shupliakov, O., and Frisén, J. (2011). A pericyte origin of spinal cord scar tissue. Science 333, 238–242. doi: 10.1126/science.1203165
Hara, M., Kobayakawa, K., Ohkawa, Y., Kumamaru, H., Yokota, K., Saito, T., et al. (2017). Interaction of reactive astrocytes with type I collagen induces astrocytic scar formation through the integrin-N-cadherin pathway after spinal cord injury. Nat. Med. 23, 818–828. doi: 10.1038/nm.4354
Harper, J. E., Miceli, S. M., Roberts, R. J., and Manley, J. L. (1990). Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucl. Acids Res. 18, 5735–5741.
Hesp, Z. C., Yoseph, R. Y., Suzuki, R., Jukkola, P., Wilson, C., Nishiyama, A., et al. (2018). Proliferating NG2-Cell-dependent angiogenesis and scar formation alter axon growth and functional recovery after spinal cord injury in mice. J. Neurosci. 38, 1366–1382. doi: 10.1523/JNEUROSCI.3953-16.2017
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi: 10.1038/cr.2017.99
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295.
Huang, R., Zhang, Y., Bai, Y., Han, B., Ju, M., Chen, B., et al. (2020). N-Methyladenosine Modification of Fatty Acid Amide Hydrolase Messenger RNA in Circular RNA STAG1-Regulated Astrocyte Dysfunction and Depressive-like Behaviors. Biol. Psychiatry 88, 392–404. doi: 10.1016/j.biopsych.2020.02.018
Hutson, T. H., and Di Giovanni, S. (2019). The translational landscape in spinal cord injury: focus on neuroplasticity and regeneration. Nat. Rev. Neurol. 15, 732–745. doi: 10.1038/s41582-019-0280-3
Ignatova, V. V., Stolz, P., Kaiser, S., Gustafsson, T. H., Lastres, P. R., Sanz-Moreno, A., et al. (2020). The rRNA m(6)A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 34, 715–729. doi: 10.1101/gad.333369.119
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Lan, H., Liu, Y., Liu, J., Wang, X., Guan, Z., Du, J., et al. (2021). Tumor-Associated Macrophages Promote Oxaliplatin Resistance METTL3-Mediated mA of TRAF5 and Necroptosis in Colorectal Cancer. Mol. Pharm. 18, 1026–1037. doi: 10.1021/acs.molpharmaceut.0c00961
Lan, X., Han, X., Li, Q., Yang, Q.-W., and Wang, J. (2017). Modulators of microglial activation and polarization after intracerebral haemorrhage. Nat. Rev. Neurol. 13, 420–433.
Li, L., Zang, L., Zhang, F., Chen, J., Shen, H., Shu, L., et al. (2017). Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411.
Li, Q., Wen, S., Ye, W., Zhao, S., and Liu, X. (2021). The potential roles of mA modification in regulating the inflammatory response in microglia. J. Neuroinflamm. 18:149. doi: 10.1186/s12974-021-02205-z
Li, Y., Guo, X., Sun, L., Xiao, J., Su, S., Du, S., et al. (2020). N-Methyladenosine Demethylase FTO Contributes to Neuropathic Pain by Stabilizing G9a Expression in Primary Sensory Neurons. Adv. Sci. 7:1902402. doi: 10.1002/advs.201902402
Liao, S., Wu, J., Liu, R., Wang, S., Luo, J., Yang, Y., et al. (2020). A novel compound DBZ ameliorates neuroinflammation in LPS-stimulated microglia and ischemic stroke rats: Role of Akt(Ser473)/GSK3β(Ser9)-mediated Nrf2 activation. Redox Biol. 36:101644. doi: 10.1016/j.redox.2020.101644
Liu, J., Dou, X., Chen, C., Chen, C., Liu, C., Xu, M. M., et al. (2020). -methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586.
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, K., Tedeschi, A., Park, K. K., and He, Z. (2011). Neuronal intrinsic mechanisms of axon regeneration. Ann. Rev. Neurosci. 34, 131–152. doi: 10.1146/annurev-neuro-061010-113723
Liu, L., Li, H., Hu, D., Wang, Y., Shao, W., Zhong, J., et al. (2022). Insights into N6-methyladenosine and programmed cell death in cancer. Mol. Cancer 21:32.
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Liu, W., Rong, Y., Wang, J., Zhou, Z., Ge, X., Ji, C., et al. (2020). Exosome-shuttled miR-216a-5p from hypoxic preconditioned mesenchymal stem cells repair traumatic spinal cord injury by shifting microglial M1/M2 polarization. J. Neuroinflamm. 17:47. doi: 10.1186/s12974-020-1726-7
Ma, C., Chang, M., Lv, H., Zhang, Z.-W., Zhang, W., He, X., et al. (2018). RNA mA methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19:68.
Ma, H., Wang, X., Cai, J., Dai, Q., Natchiar, S. K., Lv, R., et al. (2019). N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88–94.
Mahar, M., and Cavalli, V. (2018). Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 19, 323–337.
Mendel, M., Delaney, K., Pandey, R. R., Chen, K.-M., Wenda, J. M., Vågbø, C. B., et al. (2021). Splice site mA methylation prevents binding of U2AF35 to inhibit RNA splicing. Cell 184, 3125–3142. doi: 10.1016/j.cell.2021.03.062
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Moore, D. L., Blackmore, M. G., Hu, Y., Kaestner, K. H., Bixby, J. L., Lemmon, V. P., et al. (2009). KLF family members regulate intrinsic axon regeneration ability. Science 326, 298–301. doi: 10.1126/science.1175737
National SCI Statistical Center [NSCISC] (2016). Spinal cord injury (SCI) 2016 facts and figures at a glance. J. Spinal Cord Med. 39, 493–494. doi: 10.1080/10790268.2016.1210925
Oerum, S., Meynier, V., Catala, M., and Tisné, C. (2021). A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucl. Acids Res. 49, 7239–7255. doi: 10.1093/nar/gkab378
Park, O. H., Ha, H., Lee, Y., Boo, S. H., Kwon, D. H., Song, H. K., et al. (2019). Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 74, 494–507.
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 169, 824–835. doi: 10.1016/j.cell.2017.05.003
Ping, X.-L., Sun, B.-F., Wang, L., Xiao, W., Yang, X., Wang, W.-J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Plemel, J. R., Keough, M. B., Duncan, G. J., Sparling, J. S., Yong, V. W., Stys, P. K., et al. (2014). Remyelination after spinal cord injury: is it a target for repair? Prog. Neurobiol. 117, 54–72.
Qi, Z., Wang, S., Li, J., Wen, Y., Cui, R., Zhang, K., et al. (2022). Protective role of mRNA demethylase FTO on axon guidance molecules of nigro-striatal projection system in manganese-induced parkinsonism. J. Hazard. Mater. 426:128099. doi: 10.1016/j.jhazmat.2021.128099
Ramer, L. M., Ramer, M. S., and Bradbury, E. J. (2014). Restoring function after spinal cord injury: towards clinical translation of experimental strategies. Lancet. Neurol. 13, 1241–1256. doi: 10.1016/S1474-4422(14)70144-9
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017a). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Roundtree, I. A., Luo, G.-Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017b). YTHDC1 mediates nuclear export of N-methyladenosine methylated mRNAs. ELife 6:e31311. doi: 10.7554/eLife.31311
Sankavaram, S. R., Hakim, R., Covacu, R., Frostell, A., Neumann, S., Svensson, M., et al. (2019). Adult neural progenitor cells transplanted into spinal cord injury differentiate into oligodendrocytes, enhance myelination, and contribute to recovery. Stem Cell Rep. 12, 950–966.
Satterwhite, E. R., and Mansfield, K. D. (2022). RNA methyltransferase METTL16: Targets and function. Wiley Interdiscip. Rev. RNA 13:e1681. doi: 10.1002/wrna.1681
Shen, M., Li, Y., Wang, Y., Shao, J., Zhang, F., Yin, G., et al. (2021). N-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 47:102151. doi: 10.1016/j.redox.2021.102151
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Shi, Z., Yuan, S., Shi, L., Li, J., Ning, G., Kong, X., et al. (2021). Programmed cell death in spinal cord injury pathogenesis and therapy. Cell Prolif. 54:e12992.
Silver, J., and Miller, J. H. (2004). Regeneration beyond the glial scar. Nat. Rev. Neurosci. 5, 146–156.
Song, Y., Sretavan, D., Salegio, E. A., Berg, J., Huang, X., Cheng, T., et al. (2015). Regulation of axon regeneration by the RNA repair and splicing pathway. Nat. Neurosci. 18, 817–825.
Sousa, A. M. M., Meyer, K. A., Santpere, G., Gulden, F. O., and Sestan, N. (2017). Evolution of the human nervous system function, structure, and development. Cell 170, 226–247.
Su, R., Dong, L., Li, Y., Gao, M., He, P. C., Liu, W., et al. (2022). METTL16 exerts an mA-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol. 24, 205–216. doi: 10.1038/s41556-021-00835-2
Sun, Z., Wu, K., Gu, L., Huang, L., Zhuge, Q., Yang, S., et al. (2020). IGF-1R stimulation alters microglial polarization via TLR4/NF-κB pathway after cerebral hemorrhage in mice. Brain Res. Bull. 164, 221–234. doi: 10.1016/j.brainresbull.2020.08.026
Suo, L., Liu, C., Zhang, Q.-Y., Yao, M.-D., Ma, Y., Yao, J., et al. (2022). METTL3-mediated -methyladenosine modification governs pericyte dysfunction during diabetes-induced retinal vascular complication. Theranostics 12, 277–289. doi: 10.7150/thno.63441
Sweeney, M. D., Zhao, Z., Montagne, A., Nelson, A. R., and Zlokovic, B. V. (2019). Blood-Brain barrier: from physiology to disease and back. Physiol. Rev. 99, 21–78. doi: 10.1152/physrev.00050.2017
Tator, C. H. (1995). Update on the pathophysiology and pathology of acute spinal cord injury. Brain Pathol. 5, 407–413.
Teng, Y., Liu, Z., Chen, X., Liu, Y., Geng, F., Le, W., et al. (2021). Conditional deficiency of m6A methyltransferase Mettl14 in substantia nigra alters dopaminergic neuron function. J. Cell. Mol. Med. 25, 8567–8572. doi: 10.1111/jcmm.16740
Tran, A. P., Warren, P. M., and Silver, J. (2018). The biology of regeneration failure and success after spinal cord injury. Physiol. Rev. 98, 881–917. doi: 10.1152/physrev.00017.2017
Varadarajan, S. G., Hunyara, J. L., Hamilton, N. R., Kolodkin, A. L., and Huberman, A. D. (2022). Central nervous system regeneration. Cell 185, 77–94. doi: 10.1016/j.cell.2021.10.029
Venkatesh, K., Ghosh, S. K., Mullick, M., Manivasagam, G., and Sen, D. (2019). Spinal cord injury: pathophysiology, treatment strategies, associated challenges, and future implications. Cell Tissue Res. 377, 125–151. doi: 10.1007/s00441-019-03039-1
Walters, B. J., Mercaldo, V., Gillon, C. J., Yip, M., Neve, R. L., Boyce, F. M., et al. (2017). The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology 42, 1502–1510. doi: 10.1038/npp.2017.31
Wang, C.-X., Cui, G.-S., Liu, X., Xu, K., Wang, M., Zhang, X.-X., et al. (2018). METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 16:e2004880. doi: 10.1371/journal.pbio.2004880
Wang, H., Yuan, J., Dang, X., Shi, Z., Ban, W., and Ma, D. (2021). Mettl14-mediated m6A modification modulates neuron apoptosis during the repair of spinal cord injury by regulating the transformation from pri-mir-375 to miR-375. Cell Biosci. 11:52. doi: 10.1186/s13578-020-00526-9
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. doi: 10.1038/nature18298
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, X., Wu, R., Liu, Y., Zhao, Y., Bi, Z., Yao, Y., et al. (2020). m 6 A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 16, 1221–1235. doi: 10.1080/15548627.2019.1659617
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wang, Y., and Jia, G. (2020). Detection methods of epitranscriptomic mark N6-methyladenosine. Essays Biochem. 64, 967–979.
Wanner, I. B., Anderson, M. A., Song, B., Levine, J., Fernandez, A., Gray-Thompson, Z., et al. (2013). Glial scar borders are formed by newly proliferated, elongated astrocytes that interact to corral inflammatory and fibrotic cells via STAT3-dependent mechanisms after spinal cord injury. J. Neurosci. 33, 12870–12886. doi: 10.1523/JNEUROSCI.2121-13.2013
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Höbartner, C., et al. (2017). Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. doi: 10.15252/embr.201744940
Weng, Y.-L., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018). Epitranscriptomic mA regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325. doi: 10.1016/j.neuron.2017.12.036
Wu, Q., Yuan, X., Han, R., Zhang, H., and Xiu, R. (2019). Epitranscriptomic mechanisms of N6-methyladenosine methylation regulating mammalian hypertension development by determined spontaneously hypertensive rats pericytes. Epigenomics 11, 1359–1370. doi: 10.2217/epi-2019-0148
Wu, R., Li, A., Sun, B., Sun, J.-G., Zhang, J., Zhang, T., et al. (2019). A novel mA reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29, 23–41. doi: 10.1038/s41422-018-0113-8
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 61, 507–519. doi: 10.1016/j.molcel.2016.01.012
Xing, L., Cai, Y., Yang, T., Yu, W., Gao, M., Chai, R., et al. (2021). Epitranscriptomic m6A regulation following spinal cord injury. J. Neurosci. Res. 99, 843–857. doi: 10.1002/jnr.24763
Xu, H., Dzhashiashvili, Y., Shah, A., Kunjamma, R. B., Weng, Y.-L., Elbaz, B., et al. (2020). mA mRNA methylation is essential for oligodendrocyte maturation and cns myelination. Neuron 105, 293–309. doi: 10.1016/j.neuron.2019.12.013
Yang, S., Wei, J., Cui, Y.-H., Park, G., Shah, P., Deng, Y., et al. (2019). mA mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 10:2782. doi: 10.1038/s41467-019-10669-0
Yang, Y., Huang, G., Jiang, X., Li, X., Sun, K., Shi, Y., et al. (2022). Loss of Wtap results in cerebellar ataxia and degeneration of Purkinje cells. J. Genet. Genomics *, doi: 10.1016/j.jgg.2022.03.001
Yoon, K.-J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal control of mammalian cortical neurogenesis by ma methylation. Cell 171, 877–889. doi: 10.1016/j.cell.2017.09.003
Yu, J., Chen, M., Huang, H., Zhu, J., Song, H., Zhu, J., et al. (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucl. Acids Res. 46, 1412–1423. doi: 10.1093/nar/gkx1182
Yu, J., She, Y., Yang, L., Zhuang, M., Han, P., Liu, J., et al. (2021). The m A Readers YTHDF1 and YTHDF2 Synergistically Control Cerebellar Parallel Fiber Growth by Regulating Local Translation of the Key Wnt5a Signaling Components in Axons. Adv. Sci. 8:e2101329. doi: 10.1002/advs.202101329
Zaccara, S., and Jaffrey, S. R. (2020). A Unified Model for the Function of YTHDF Proteins in Regulating mA-Modified mRNA. Cell 181, 1582–1595. doi: 10.1016/j.cell.2020.05.012
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol Cell Biol. 20, 608–624. doi: 10.1038/s41580-019-0168-5
Zhang, L., Hao, D., Ma, P., Ma, B., Qin, J., Tian, G., et al. (2021). Epitranscriptomic Analysis of m6A Methylome After Peripheral Nerve Injury. Front. Genet. 12:686000. doi: 10.3389/fgene.2021.686000
Zhang, Z., Wang, Q., Zhao, X., Shao, L., Liu, G., Zheng, X., et al. (2020). YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 11:977. doi: 10.1038/s41419-020-03186-2
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nature Reviews. Mol. Cell Biol. 18, 31–42. doi: 10.1038/nrm.2016.132
Zhao, F., Xu, Y., Gao, S., Qin, L., Austria, Q., Siedlak, S. L., et al. (2021). METTL3-dependent RNA mA dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol. Neurodegener. 16:70. doi: 10.1186/s13024-021-00484-x
Zhao, Y., Wang, Q., Xie, C., Cai, Y., Chen, X., Hou, Y., et al. (2021). Peptide ligands targeting FGF receptors promote recovery from dorsal root crush injury via AKT/mTOR signaling. Theranostics 11, 10125–10147. doi: 10.7150/thno.62525
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C.-M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zhou, H., Xu, Z., Liao, X., Tang, S., Li, N., and Hou, S. (2021). Low Expression of YTH Domain-Containing 1 Promotes Microglial M1 Polarization by Reducing the Stability of Sirtuin 1 mRNA. Front. Cell. Neurosci. 15:774305. doi: 10.3389/fncel.2021.774305
Zhou, K., Zheng, Z., Li, Y., Han, W., Zhang, J., Mao, Y., et al. (2020). TFE3, a potential therapeutic target for Spinal Cord Injury via augmenting autophagy flux and alleviating ER stress. Theranostics 10, 9280–9302. doi: 10.7150/thno.46566
Zhu, S., Chen, M., Ying, Y., Wu, Q., Huang, Z., Ni, W., et al. (2022). Versatile subtypes of pericytes and their roles in spinal cord injury repair, bone development and repair. Bone Res. 10:30. doi: 10.1038/s41413-022-00203-2
Zhuang, M., Li, X., Zhu, J., Zhang, J., Niu, F., Liang, F., et al. (2019). The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucl. Acids Res. 47, 4765–4777. doi: 10.1093/nar/gkz157
Keywords: epigenetics, N6-methyladenosine (m6A), post-transcriptional modification, nervous system, spinal cord injury (SCI)
Citation: Liu D, Fan B, Li J, Sun T, Ma J, Zhou X and Feng S (2022) N6-methyladenosine modification: A potential regulatory mechanism in spinal cord injury. Front. Cell. Neurosci. 16:989637. doi: 10.3389/fncel.2022.989637
Received: 08 July 2022; Accepted: 05 September 2022;
Published: 23 September 2022.
Edited by:
Fengquan Zhou, Zhejiang University, ChinaReviewed by:
Fang Y. E., Sun Yat-sen University, ChinaEsra Yalcin, Boston Children’s Hospital and Harvard Medical School, United States
Copyright © 2022 Liu, Fan, Li, Sun, Ma, Zhou and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianhu Zhou, emhvdXhpYW5odUBuYnUuZWR1LmNu; Shiqing Feng, c3FmZW5nQHRtdS5lZHUuY24=
†These authors have contributed equally to this work
‡These authors have contributed equally to this work and share first authorship
 Derong Liu
Derong Liu Baoyou Fan1,2‡
Baoyou Fan1,2‡ Xianhu Zhou
Xianhu Zhou Shiqing Feng
Shiqing Feng