- 1Institute of Neuropathology, Faculty of Medicine, University of Freiburg, Freiburg, Germany
- 2Faculty of Biology, University of Freiburg, Freiburg, Germany
- 3Center for Basics in NeuroModulation (NeuroModulBasics), Faculty of Medicine, University of Freiburg, Freiburg, Germany
- 4CIBSS-Centre for Integrative Biological Signalling Studies, University of Freiburg, Freiburg, Germany
Microglia build the first line of defense in the central nervous system (CNS) and play central roles during development and homeostasis. Indeed, they serve a plethora of diverse functions in the CNS of which many are not yet fully described and more are still to be discovered. Research of the last decades unraveled an implication of microglia in nearly every neurodegenerative and neuroinflammatory disease, making it even more challenging to elucidate molecular mechanisms behind microglial functions and to modulate aberrant microglial behavior. To understand microglial functions and the underlying signaling machinery, many attempts were made to employ functional in vitro studies of microglia. However, the range of available cell culture models is wide and they come with different advantages and disadvantages for functional assays. Here we aim to provide a condensed summary of common microglia in vitro systems and discuss their potentials and shortcomings for functional studies in vitro.
Introduction
Research of the last century has unraveled the crucial role of microglia—the resident tissue macrophages of the central nervous system (CNS)—during development, immune defense, and homeostasis, but also in various neuropathologies including Alzheimer’s disease and multiple sclerosis (Prinz et al., 2019; Sierra et al., 2019). The study of neurodegenerative and neuroinflammatory diseases requires reliable in vitro systems to model microglia physiology and functionality and to facilitate a successful transfer to animal models and later clinical studies (Timmerman et al., 2018). However, to develop such in vitro surrogate systems for microglia has proven to be an extraordinary challenge many research groups around the world have attempted to solve. Some approaches rely on direct primary microglia isolation and purification from different developmental stages, others are based on oncogenically transformed cell lines, or co-cultures such as mixed glial-cultures, which are commonly used (Hansson, 1984; Blasi et al., 1990; Bohlen et al., 2019). Further on, during the last decade induced pluripotent stem cell (iPSC) based models have been developed, including the establishment of 3D CNS organoid cultures (Lancaster et al., 2013; Lancaster and Knoblich, 2014; Ormel et al., 2018), which aim to provide novel, elegant techniques to study brain tissue and also microglia in vitro. Although impressive technological advances have led to a deeper understanding of brain resident phagocytes on a single cell level (Hammond et al., 2019; Li et al., 2019; Masuda et al., 2019) there is not yet an ultimate cell culture model available to fulfill all research needs and to overcome shortcomings of maintaining microglia in vitro. In this review we want to summarize the available in vitro models for microglia which all have certain advantages and disadvantages and highlight which research questions the different models are best suited for.
Primary Cell Cultures
Cultivation of freshly isolated microglia were thought to represent the closest surrogate system to the in vivo conditions (Figure 1). The isolation of microglia from different developmental stages requires the dissociation of the CNS tissue into a single-cell suspension either via mechanical dissociation or enzymatic digestion (Haimon et al., 2018; Ocañas et al., 2022), followed by different purification methods such as fluorescence activated cell sorting (FACS; Hickman et al., 2013; Bennett et al., 2016; Pan and Wan, 2020) or magnetic activated cell sorting (MACS; Gordon et al., 2011; Nikodemova and Watters, 2012). These sorting—and dissociation—based cultures offer an easy approach to obtain a pure microglia culture without contamination by other CNS-resident cells. However, sorted cells often develop an altered metabolism and display artificial activation (Gosselin et al., 2017; Haimon et al., 2018; Llufrio et al., 2018; Mattei et al., 2020; Ocañas et al., 2022), increased motility, and altered phagocytosis speeds (Montilla et al., 2020). Even though they cannot model a fully accurate in vivo phenotype, they offer a surrogate for many in vitro microglia studies. FACS or MACS purification of microglia for cell cultures often results in a low cell number. Pan and Wan reported ~7 × 104 microglia via FACS and ~11.7 × 104 microglia via MACS sorting (Pan and Wan, 2020). Therefore, primary cultures from MACS or FACS-sorted microglia would require a high number of animals to allow performance of functional assays. Moreover, human brain tissue is very limited, thus functional in vitro studies with sorted human primary microglia are barely possible. Culture of sorted microglia from adult CNS tissue does not allow a long-term culture approach, resulting in a short time window for functional assays and screenings. A rather different approach to gain primary microglia is the use of a primary mixed glial culture (DuBois et al., 1985; Murphy et al., 1990; Chen et al., 2013; Figure 2). Here microglia grow on a confluent glial cell layer, and are mechanically shaken off to gain pure microglia in monocultures which can be used for functional assays. Mixed glial cultures are most often generated from neonatal or embryonic CNS tissue, hence the generated microglia have a rather activated and immature phenotype and do not serve as an ideal tool to mimic adult microglia. This further points to the importance for considering the developmental stage of microglia used for the culture system according to the research question. Nevertheless, neonatal microglia from mixed glial cultures are still widely used (Georgieva et al., 2018) offering advantages such as a high cell yield after a short cultivation period. Primary microglia cell culture systems are very liable to various factors including the choice of culture flask coating or the supplements in the media itself such as serum, growth factors, or added metabolites. The media used to culture microglia can be supplemented with various necessary growth factors and cytokines such as macrophage colony stimulating factor (M-CSF) and Interleukin-34 (IL-34; Bohlen et al., 2017), which are important for microglia differentiation (Ginhoux et al., 2010; Greter et al., 2012; Wang et al., 2012) and promote culture growth and survival (Bohlen et al., 2017), or transforming growth factor beta (TGF-β), which has been shown to be an essential cytokine for adult microglia homeostasis and maturation (Butovsky et al., 2014; Zöller et al., 2018). Additionally supplementation with lipids such as cholesterol have also proven to favor microglia survival in vitro (Bohlen et al., 2017). Beside media supplementation, various studies were able to demonstrate that different culture coatings such as polyethyleneimine (PEI; Sepulveda-Diaz et al., 2016) and poly-D-lysine (PDL; Lian et al., 2016) have proven to selectively promote microglia attachment and culture growth. PEI coating is a positively charged polymer promoting a high cell yield of pure neonatal microglia cells (Sepulveda-Diaz et al., 2016). In contrast poly-l-lysine (PLL) or PDL coating can be used to not only culture microglia but also neurons and astrocytes (Skaper et al., 2012). On the contrary, these different media conditions also show the impact and artificial modulation of media supplementation on the functionality of the cells such as phagocytic capacity and proliferation (Bohlen et al., 2017), making it difficult to find the right balance between optimal culture conditions and inducing an artificial phenotype. Further shortcomings are the downregulation of important signature markers on primary microglia already hours after culturing them in vitro (Bohlen et al., 2017; Gosselin et al., 2017). These markers include microglia signature genes such as transmembrane protein 119 (Tmem119) and purinergic receptor (P2ry12). These studies showed that sorted primary microglia undergo detrimental transcriptomic alterations in vitro, making it sometimes difficult to use primary microglial cultures for exploring their functions since they do not fully recapitulate the in vivo microglia phenotype (Bohlen et al., 2017; Gosselin et al., 2017). For example, Montilla et al. (2020) showed that MACS sorted microglia from rats (P10-P12) lack the ability to degrade and phagocytose myelin in vitro, probably due to the reduction of CD68, an important protein associated with phagocytic activity located on the lysosomal membrane. Primary cultures are extensively pushed forward by new approaches to get more organotypic culture conditions. With the available primary microglia monocultures, a lot of various assays are possible such as microglia activation studies and screening for small molecules (Song et al., 2016; Figuera-Losada et al., 2017; Telpoukhovskaia et al., 2020), which all use primary microglia from neonates (P1-P4). Various studies revealed differences between primary cultures and other surrogate systems in distinct functional assays. One study for example showed differences in activation between primary cultured microglia and the murine oncogenically transformed microglial cell line BV2, with a weaker inflammatory response of BV2 cells than primary microglia upon stimulation (Luan et al., 2022). Another advantage of primary microglia is the possibility to culture cells from genetically modified mice and transgenic disease models for neurodegenerative diseases such as Alzheimer’s disease or multiple sclerosis. Moreover, the use of primary cells allows the generation of cell cultures to compare and study microglial functions across various species, including rodents (Bohlen et al., 2017; Montilla et al., 2020), primates (Zuiderwijk-Sick et al., 2007), and humans (Rustenhoven et al., 2016; Tewari et al., 2021), but also primary cultures of zebrafish or leech microglia might be possible in the future. Further on, new and modified primary microglia culture models are still needed to optimize in vitro microglia studies and also to facilitate cross-species characterization.
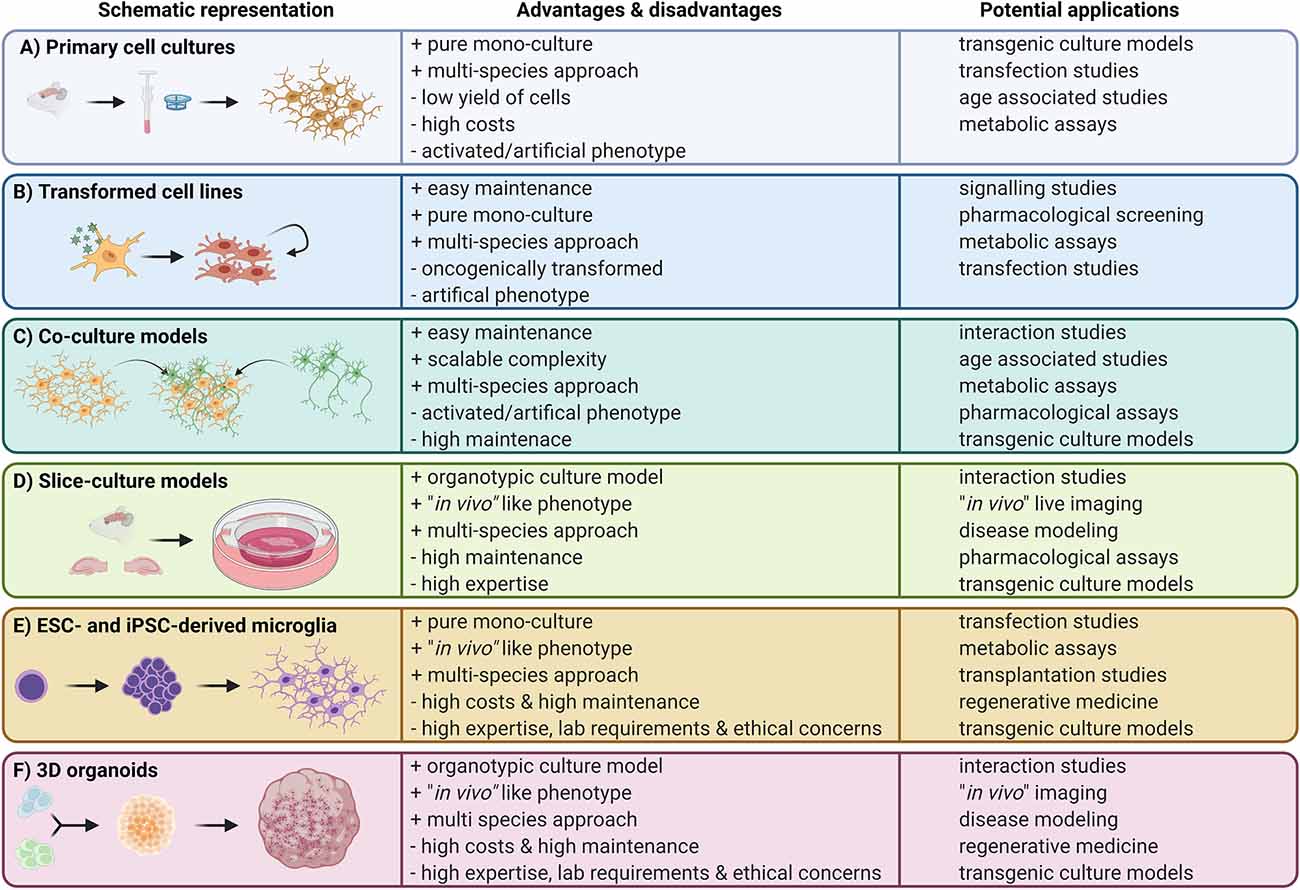
Figure 1. Overview of available microglia in vitro approaches. Left panels: schematic representation of the cell culture model; middle panels: advantages and disadvantages of the in vitro approach; right panels: potential applications.
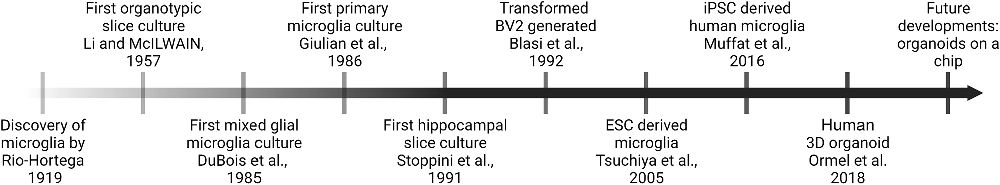
Figure 2. Timeline of selected milestones in the development of microglia in vitro models over the last decades.
Oncogenically Transformed Cell Lines
To avoid high costs, low cell numbers and time-consuming isolation and purification protocols of primary microglia cultures, immortalized cell lines were generated, such as the murine BV2 or N9 cells (Figure 1). These cell lines were generated via viral transduction of isolated embryonic microglia with different oncogenes such as v-raf/v-myc (Blasi et al., 1990; Figure 2). Oncogenically transformed cell lines proliferate rapidly with a short generation time and can be sub-cultured indefinitely, thus allowing easy culture maintenance and a high cell yield (Blasi et al., 1990). However, this is often at the expense of an artificial phenotype and an unnaturally high proliferation rate with no contact inhibition (Bocchini et al., 1992; Napoli et al., 2009). Already in the first publications describing the BV2 cell line and its usage for in vitro studies, the authors pointed out that these cells rather resemble activated microglia (Bocchini et al., 1992). Of course, transformed microglial cell lines provide certain similarities to their ex vivo counterparts such as expression of ionized calcium-binding adapter molecule 1 (Iba-1) and inflammatory cytokine secretion upon lipopolysaccharides (LPS) stimulation (Horvath et al., 2008), as well as reactivity to fibrillar Aβ (1–42; Kopec and Carroll, 1998). Furthermore they are easy to manipulate, and allow big in vitro screens due to their unlimited availability (Pluvinage et al., 2019). Unfortunately, the immortalized microglia cell lines also show alterations in biological processes such as loss of contact inhibition, differentially regulated metabolism, and distinct cytokine production dynamics (Horvath et al., 2008; Das et al., 2016; Melief et al., 2016). Additionally, the unnaturally high rate of proliferation always promotes undesired genetic mutations, which could potentially lead to an even more artificial phenotype. Despite all their disadvantages, the BV2 and N9 cell lines are widely used as a first model system to understand molecular mechanisms of microglial immune activation and the underlying signaling pathways, most likely due to the high cell yield and easy maintenance. For example, Duan et al. (2013) revealed an increased synthesis and secretion of pro nerve growth factor (proNGF) by BV2 and N9 cells induced by LPS stimulation via Western blot, a method requiring a high number of cells, which no other approach can offer so far. This revealed a potential target for therapeutic manipulation. BV2 and N9 cells were developed almost 30 years ago and there are more than 3,000 publications available up to date using these cell lines. Even though their high proliferation and activated phenotype do not completely resemble adult microglia or aged microglia, they are often used to study microglia functions during neurodegenerative diseases including Parkinson’s disease (Gu et al., 2018). Beside the murine BV2 and N9 cell lines, there are also cell lines for different species available, for example the immortalized (HAPI) cell line derived from neonatal rat brains (Cheepsunthorn et al., 2001) or the transformed human HMO6 cells generated from embryonic telencephalon tissue (Nagai et al., 2001). Nagai et al. (2001) could show that the generated HMO6 cell line showed similar expression profiles upon activation compared to primary human microglia, providing an easy to use model. Moreover, other studies highlight the ability to perform cross species experiments. By transplanting HMO6 cells in a rat middle cerebral artery occlusion model (MCAO), Narantuya and colleagues could demonstrate the importance of transplanted microglia in stroke animals, in showing reduced ischemic deficits and apoptotic cells in rats transplanted with HMO6 (Narantuya et al., 2010). Oncogenically transformed microglial cell lines bridge the gap of a low cell yield for microglial in vitro studies, but come with the risk of artificial activation and an altered behavior due to the oncogenic transformation and excessive proliferation rate.
Co-Culture Models
To address the lack of tissue specific cues and to understand direct cell-cell interactions, co-culture systems have been developed (Figure 1). Co-culture systems are commonly used to provide a more organotypic environment by mimicking direct interactions between cells inside their respective niche, for example microglia with astrocytes, oligodendrocytes, neurons or other CNS associated cells (Roqué and Costa, 2017; Goshi et al., 2020). Co-cultures are often used to model and understand interactions during different neuroinflammatory or neurodegenerative diseases. Due to a vast variety of co-culturing combinations, available protocols, and adjustments, this leads to an open-ended complexity of culture systems used, making it hard to compare and transfer results from one study to the next. Co-cultures offer a strong basis for interaction investigations ranging from synergic molecular interplays during homeostasis to pathological interactions during activation, inflammation, and disease. Without going into the details of the wide spectrum of studies, we would like to point out some studies using complex co-culture systems. As an example, Goshi et al. were able to use a tri-culture model of microglia, neurons and astrocytes to study the neuroinflammatory interplay after LPS stimulation, and identified classical hallmarks in response to LPS (Goshi et al., 2020). Other groups use oncogenically transformed cell lines for co-cultures. For example one study used BV2 cells and PC12 cells as a surrogate for a microglia and neuron co-culture to describe the function of neuron-derived exosomes driving microglia towards a more activated phenotype with an increased release of proinflammatory cytokines (Yin et al., 2020). Further on, more complex co-cultures are available, such as a model to study the interaction of primary murine microglia and T-cells in a biomimetic 3D extracellular matrix (ECM) culture system (Frühauf et al., 2021). In this study, Frühauf et al. (2021) were able to improve survival of T-cells when co-cultured with microglia and provide a basis to study T-cell activation by microglia. Co-culture systems are also often used to study the interaction of CNS-resident cells as well as immune cells in disease models such as brain tumors. Leite and colleagues used a co-culture model comprising human glioblastoma (GBM) cell lines and a human microglia cell line (CHME3) as a model to study their interaction (Leite et al., 2020). They were able to show a higher GBM proliferation and migration rate and further an increased protective effect of microglia to GBM cells pointing to a potential role of microglia promoting their survival and immune escape. Overall co-culture models have been used to study microglial interactions with other cells for years now and with scalable complexity and feasibility they will still be a useful alternative to more complex models in future studies.
Slice Culture Models
Taking a cell out of its homeostatic tissue niche and placing it into a plastic dish with an artificial environment entails many problems. Thus, the development of organotypic slice cultures (OSCs) from brain tissue in 1957 marked an important milestone for in vitro studies of microglia and other CNS cells leaving them in their CNS environment where interactions with neighboring cells can be observed and modulated (Li and McIlwain, 1957; Figure 2). In 1971, Okamoto and Quastel (1973) published a protocol employing cerebellar slices from guinea pigs, which was adapted to a variety of species and proved to be a valuable tool to study myelination, demyelination, and remyelination processes (Jarjour et al., 2012; Doussau et al., 2017). In 1991, Stoppini et al. (1991) developed a protocol for organotypic hippocampal slice cultures (OHSC) that has since then been widely used until this day (Figure 2). This protocol involves culturing hippocampal sections on a semi-permeable membrane creating an interface between air and culture medium. Similar to the concept of OHSCs, organotypic entorhinal slice cultures (OESCs) combine cortex and hippocampus and are ideal to study processes after neuronal axotomies (Del Turco and Deller, 2007). Microglia in OHSCs transition from a reactive and inflammatory to a largely homeostatic phenotype similar to in vivo conditions over the course of 3 weeks (Delbridge et al., 2020). OHSC microglia are of mesodermal origin, which together with the preserved cytoarchitecture and the possibility to perform long-term cultures, makes them an interesting in vitro alternative to in vivo experiments (Figure 1). The drawback of this model is that OSCs require the usage of immature neonatal tissue (P1-P4) as these tissues still have a higher proliferative capacity, which does not represent an adult or aging CNS or even age-associated diseases (Figure 1). Only a few studies described the successful culture of hippocampal slices from adult mice or rats and most of them only for a short period of time (Xiang et al., 2000; Humpel, 2015; Croft et al., 2019). Moreover, OSCs are usually performed with a high serum content (around 25%) which is far from the physiological situation in the brain, hampers reproducibility and renders pharmacologic small molecules tests difficult as these bind to albumin (Croft et al., 2019). Additionally, some reports have shown that this culture system is translatable to human settings allowing the survival of post-mortem or fetal OHSCs for several weeks (Lyman et al., 1991; Eugène et al., 2014). OHSCs harbor the advantage that they can be swiftly prepared from different transgenic lines and are relatively easy to maintain in culture, making them a suitable tool for many labs worldwide to study microglia function and especially their interaction with neighboring CNS cells.
ESC- and iPSC-Derived Microglia
Embryonic stem cells (ESCs) are pluripotent cells derived from the inner cell mass of the blastocyst, which can be maintained and passaged in vitro (Evans and Kaufman, 1981; Figure 1). Since then, numerous studies using ESCs as a tool to differentiate a plethora of cell types in vitro opened new avenues and approaches for biomedical studies (Wobus and Boheler, 1999; Cyranoski, 2018; Zakrzewski et al., 2019). Several protocols are nowadays available to differentiate ESCs into neural progenitors, neurons, astrocytes, oligodendrocytes, and also ES cell-derived microglia (ESdM; Tsuchiya et al., 2005; Napoli et al., 2009; Beutner et al., 2010). Tsuchiya et al. (2005) already observed the presence of ESdM next to neural cells derived from embryonic bodies in 2005 (Figure 2). To obtain monocultures of ESdM, ESCs are first expanded before their differentiation into embryoid bodies is induced by withdrawal of leukemia inhibitory factor (LIF). The differentiation of neural progenitors is achieved by changing the ES cell medium to ITSFn medium composed of DMEM/F12 supplemented with insulin, sodium selenite transferrin, and fibronectin. Many studies reported microglia-like cells in the neural or even neuronal differentiated cultures from ES cells as contaminating subpopulations (Tsuchiya et al., 2005; Napoli et al., 2009), other studies showed direct ESdM differentiation protocols (Muffat et al., 2016). All of these studies reported to obtain a high cell density of ESdM coupled with morphology and marker expression profile comparable to microglia in vivo, including the expression of macrophage markers such as macrophage-1 antigen (Mac-1) or Iba-1 and a characteristically low expression of the pan-hematopoietic marker CD45. Together with an analogous transcriptomic signature to primary microglia, these similarities seem to make them a great in vitro model to study microglia physiology (Figure 1; Beins et al., 2016). The drawback of ESdM is their expensive generation and the limited availability of transgenically modified ESdM (Figure 1). Moreover, similar to BV2 cells and primary neonatal microglia, ESdM also showed differences to their in vivo counterparts, for example the production of high levels of inducible nitric oxide synthase (iNOS) in response to interferon gamma (IFN-γ) stimulation which stands in contrast to observations made for microglia in the adult mouse brain (Napoli et al., 2009). Generation of microglia-like cells from ESCs is holding a huge potential for regenerative medicine and thus, the model was soon adapted to human ESCs. However there are ethical concerns regarding the use of human ESCs whereas they need to be directly generated from human embryos (Thomson et al., 1998; Cowan et al., 2004). This constituted a major roadblock for ESCs in clinical approaches making it very challenging to employ human ESC-derived microglia as a possible therapeutic approach and not only as an in vitro model for studying microglial function.
To circumvent the ethical concerns and in the attempt of generating patient-specific cell sources, it was a major breakthrough when the first induced pluripotent stem cells (iPSCs) were established. iPSC-derived microglia (IdM) can be obtained by inducing the formation of embryoid bodies from iPSCs and further using a specialized neuro-glial differentiation medium supplemented with M-CSF and IL-34. From this structure, round motile cells expressing microglia surface markers delaminate from the outer border and can be harvested (Muffat et al., 2016; Figure 2). In that way, IdM, at least to some degree, mirror the normal microglia development by passing through a myeloid cell intermediate (Abud et al., 2017). IdM recapitulate both phenotypic and functional characteristics of their in vivo counterparts like expression of P2ry12 and Tmem119 (Abud et al., 2017; Quarta et al., 2019). However gene clustering analyses have shown that IdM most closely resemble neonatal microglia rather than adult microglia hence raising the question whether they in fact constitute a model to study adult mature microglia and especially microglia during age-associated neurodegenerative diseases such as Alzheimer’s or Parkinson’s disease (Abud et al., 2017). Even though human IdM allow the unique opportunity to generate large cell numbers from specific patients, iPSC differentiation protocols require a large amount of expertise in cell culture and labor (2–8 weeks depending on species and protocol; Muffat et al., 2016). Nevertheless, iPSCs allow researchers to recapitulate and study microglia with the genetic background of a patient suffering from a specific neurological disorder like Parkinson’s disease and then compare these cells to healthy unaffected donor cells. This unique possibility is not feasible with freshly isolated primary cells from the patients, which are scarcely available and not applicable by using ESdM (Badanjak et al., 2021). Another advantage of the IdM protocols is that the cells can be maintained in serum free culture conditions similar to the endogenous CNS environment and are easier to reproduce, as serum is known to add strong batch effects to a protocol (Figure 1). This model is difficult to employ for the analysis of microglia from transgenic lines as the several weeklong protocol to obtain microglia-like cells from iPSCs needs to be repeated for every transgenic line used (Figure 1). In contrast to primary cells, new iPSCs would need to be transfected for each transgenic line.
3D Cerebral Organoids
Recent technical advances in 3D cell culture models have allowed researchers to study the interplay between microglia and other CNS inhabiting cells by using iPSC-derived brain organoids (Figure 1). These surrogate systems can offer the study of molecular interactions of complex neurologic diseases including autism spectrum disorders or microcephaly (Lancaster et al., 2013; Mariani et al., 2015). This revolutionary advancement in creating iPSC-derived brain organoids is reflected by an exponential increase in publications and studies employing this technique over the last 10 years (Pacitti et al., 2019). Cerebral organoids can be derived from either ESCs or iPSCs. Microglia-like cells innately developing in organoid cultures display a typical ramified morphology, which is much more similar to the in vivo morphology compared to other cell culture systems. Furthermore, the cells can be monitored to take over many of their usual homeostatic tasks in the brain such as synaptic pruning of neuronal networks or inflammatory cytokine secretion in response to LPS by transcribing high levels of Il1β and Il6 (Ormel et al., 2018; Figure 2). While similar, the described pro-inflammatory cytokine response to LPS challenge is significantly higher compared to adult microglia raising once more the question whether these microglia like cells in brain organoids do indeed resemble mature adult microglia or rather immature developing microglia being in a pre-activated state (Figure 1). Although hierarchical clustering revealed that organoid-derived microglia cluster closer to adult microglia than for example IdM, they start expressing signature markers like P2RY12 and TMEM119 only after more than 100 days making this protocol very long and laborious (Ormel et al., 2018). Nevertheless, organoid cultures represent a fascinating opportunity to study cell-cell interactions of different CNS cells during developmental stages or after genetic modifications (Figure 1). One major challenge of this new technology is to make it reproducible enough with sufficient consistency for standardized drug testing (Tachibana, 2018). Furthermore, not all CNS cells develop easily in cerebral organoids, for example endothelial cells and other cells comprising the neurovascular unit are difficult to introduce in this model (Cakir et al., 2019). The latest technical advancement of 3D cultures are organoids-on-chips which attempt to solve this problem by providing the needed architecture and structural organization of stem cells and differentiated cells on a microfabricated device adapted to dynamically accommodate the respective research question (Karzbrun et al., 2018; Park et al., 2019).
Conclusion and Outlook
Microglia cell culture approaches have been used for many decades, offering a wide tool box to study molecular mechanisms and cell-cell interactions in vitro. Even though researchers can choose from a wide range of models and numerous approaches to maintain rodent as well as human microglia in vitro, the available models are still far from offering an ideal surrogate system, perfectly mimicking microglia in vivo. Many obstacles, including artificial activation, low cell yield, loss of signature genes and alterations in activation profiles need to be overcome by development of new in vitro approaches and improvement of existing culture systems. Furthermore, researchers need to carefully evaluate, depending on their study type, which cell culture system is the most suitable one to answer their questions. Studies exploring age-associated diseases will struggle with cell culture models where microglia display a rather immature phenotype such as those derived from mixed glial cultures, ESCs, iPSCs brain organoids (Figure 1). Technical advances in high throughput approaches and the continuous development of new transgenic animal models to study microglia are supporting the field to employ in vivo studies and analysis of freshly isolated microglia. However, there is still a huge demand for microglia cell culture models, especially if it comes to large pharmaceutical screening studies or the understanding of complex signaling pathways. Furthermore, there is constant demand for cell culture models which allow us to efficiently use microglia from scarce specimens such as human tissue samples and moreover an ethical requirement to reduce and refine animal studies. Hence the field is in urgent need to develop new innovative in vitro or even in silico surrogate systems mimicking the in vivo counterparts as closely as possible to avoid artificial results. Improvement of cell culture platforms will further facilitate an easier and more reliable transfer of results from “the dish” into the living model organisms and finally from there into clinical studies. Therefore, it will be the task of future studies and ongoing research to develop and adapt in vitro approaches to study microglial function in vitro as closely as possible to the in vivo situation.
Author Contributions
PA and PP wrote the manuscript and designed the figures. KK conceptualized, wrote and edited this review. All authors contributed to the article and approved the submitted version.
Funding
KK was supported by a project grant of the Fritz Thyssen Foundation, by the German Research Foundation (DFG) with a project grant (Project ID 432207796), as well as project grants within the SFB/TRR167 (Project ID 259373024) and the CRC1479 (Project ID 441891347), and by the DFG under Germany’s Excellence Strategy (grant no. CIBSS—EXC-2189, Project ID 390939984).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank all members of the Kierdorf lab and especially the members of the “cell culture team” who are joining us on our way to develop new in vitro approaches for microglia. All figures were created with Biorender. We apologize to all colleagues in the microglia field and beyond whose work we could not cite here due to space limitations.
References
Abud, E. M., Ramirez, R. N., Martinez, E. S., Healy, L. M., Nguyen, C. H. H., Newman, S. A., et al. (2017). iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293.e9. doi: 10.1016/j.neuron.2017.03.042
Badanjak, K., Mulica, P., Smajic, S., Delcambre, S., Tranchevent, L.-C., Diederich, N., et al. (2021). iPSC-derived microglia as a model to study inflammation in idiopathic Parkinson’s disease. Front. Cell Dev. Biol. 9:740758. doi: 10.3389/fcell.2021.740758
Beins, E., Ulas, T., Ternes, S., Neumann, H., Schultze, J. L., and Zimmer, A. (2016). Characterization of inflammatory markers and transcriptome profiles of differentially activated embryonic stem cell-derived microglia. Glia 64, 1007–1020. doi: 10.1002/glia.22979
Bennett, M. L., Bennett, F. C., Liddelow, S. A., Ajami, B., Zamanian, J. L., Fernhoff, N. B., et al. (2016). New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. U S A 113, E1738–E1746. doi: 10.1073/pnas.1525528113
Beutner, C., Roy, K., Linnartz, B., Napoli, I., and Neumann, H. (2010). Generation of microglial cells from mouse embryonic stem cells. Nat. Protoc. 5, 1481–1494. doi: 10.1038/nprot.2010.90
Blasi, E., Barluzzi, R., Bocchini, V., Mazzolla, R., and Bistoni, F. (1990). Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J. Neuroimmunol. 27, 229–237. doi: 10.1016/0165-5728(90)90073-v
Bocchini, V., Mazzolla, R., Barluzzi, R., Blasi, E., Sick, P., and Kettenmann, H. (1992). An immortalized cell line expresses properties of activated microglial cells. J. Neurosci. Res. 31, 616–621. doi: 10.1002/jnr.490310405
Bohlen, C. J., Bennett, F. C., and Bennett, M. L. (2019). Isolation and culture of microglia. Curr. Protoc. Immun. 125:e70. doi: 10.1002/cpim.70
Bohlen, C. J., Bennett, F. C., Tucker, A. F., Collins, H. Y., Mulinyawe, S. B., and Barres, B. A. (2017). Diverse requirements for microglial survival, specification and function revealed by defined-medium cultures. Neuron 94, 759–773.e8. doi: 10.1016/j.neuron.2017.04.043
Butovsky, O., Jedrychowski, M. P., Moore, C. S., Cialic, R., Lanser, A. J., Gabriely, G., et al. (2014). Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143. doi: 10.1038/nn.3599
Cakir, B., Xiang, Y., Tanaka, Y., Kural, M. H., Parent, M., Kang, Y.-J., et al. (2019). Development of human brain organoids with functional vascular-like system. Nat. Methods 16, 1169–1175. doi: 10.1038/s41592-019-0586-5
Cheepsunthorn, P., Radov, L., Menzies, S., Reid, J., and Connor, J. R. (2001). Characterization of a novel brain-derived microglial cell line isolated from neonatal rat brain. Glia 35, 53–62. doi: 10.1002/glia.1070
Chen, S.-H., Oyarzabal, E. A., and Hong, J.-S. (2013). Preparation of rodent primary cultures for neuron-glia, mixed glia, enriched microglia and reconstituted cultures with microglia. Methods Mol. Biol. 1041, 231–240. doi: 10.1007/978-1-62703-520-0_21
Cowan, C. A., Klimanskaya, I., McMahon, J., Atienza, J., Witmyer, J., Zucker, J. P., et al. (2004). Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 350, 1353–1356. doi: 10.1056/NEJMsr040330
Croft, C. L., Futch, H. S., Moore, B. D., and Golde, T. E. (2019). Organotypic brain slice cultures to model neurodegenerative proteinopathies. Mol. Neurodegener. 14:45. doi: 10.1186/s13024-019-0346-0
Cyranoski, D. (2018). How human embryonic stem cells sparked a revolution. Nature 555, 428–430. doi: 10.1038/d41586-018-03268-4
Das, A., Kim, S. H., Arifuzzaman, S., Yoon, T., Chai, J. C., Lee, Y. S., et al. (2016). Transcriptome sequencing reveals that LPS-triggered transcriptional responses in established microglia BV2 cell lines are poorly representative of primary microglia. J. Neuroinflammation 13:182. doi: 10.1186/s12974-016-0644-1
Del Turco, D., and Deller, T. (2007). Organotypic entorhino-hippocampal slice cultures–a tool to study the molecular and cellular regulation of axonal regeneration and collateral sprouting in vitro. Methods Mol. Biol. 399, 55–66. doi: 10.1007/978-1-59745-504-6_5
Delbridge, A. R. D., Huh, D., Brickelmaier, M., Burns, J. C., Roberts, C., Challa, R., et al. (2020). Organotypic brain slice culture microglia exhibit molecular similarity to acutely-isolated adult microglia and provide a platform to study neuroinflammation. Front. Cell. Neurosci. 14:592005. doi: 10.3389/fncel.2020.592005
Doussau, F., Dupont, J.-L., Neel, D., Schneider, A., Poulain, B., and Bossu, J. L. (2017). Organotypic cultures of cerebellar slices as a model to investigate demyelinating disorders. Expert Opin. Drug Discov. 12, 1011–1022. doi: 10.1080/17460441.2017.1356285
Duan, L., Chen, B.-Y., Sun, X.-L., Luo, Z.-J., Rao, Z.-R., Wang, J.-J., et al. (2013). LPS-induced proNGF synthesis and release in the N9 and BV2 microglial cells: a new pathway underling microglial toxicity in neuroinflammation. PLoS One 8:e73768. doi: 10.1371/journal.pone.0073768
DuBois, J. H., Hammond-Tooke, G. D., and Cuzner, M. L. (1985). Expression of major histocompatibility complex antigens in neonate rat primary mixed glial cultures. J. Neuroimmunol. 9, 363–377. doi: 10.1016/s0165-5728(85)80036-9
Eugène, E., Cluzeaud, F., Cifuentes-Diaz, C., Fricker, D., Le Duigou, C., Clemenceau, S., et al. (2014). An organotypic brain slice preparation from adult patients with temporal lobe epilepsy. J. Neurosci. Methods 235, 234–244. doi: 10.1016/j.jneumeth.2014.07.009
Evans, M. J., and Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. doi: 10.1038/292154a0
Figuera-Losada, M., Thomas, A. G., Stathis, M., Stockwell, B. R., Rojas, C., and Slusher, B. S. (2017). Development of a primary microglia screening assay and its use to characterize inhibition of system xc^- by erastin and its analogs. Biochem. Biophys. Rep. 9, 266–272. doi: 10.1016/j.bbrep.2016.12.009
Frühauf, M., Zeitschel, U., Höfling, C., Ullm, F., Rabiger, F. V., Alber, G., et al. (2021). Construction of a 3D brain extracellular matrix model to study the interaction between microglia and T cells in co-culture. Eur. J. Neurosci. 53, 4034–4050. doi: 10.1111/ejn.14978
Georgieva, M., Leeson-Payne, A., Dumitrascuta, M., Rajnicek, A., Malcangio, M., and Huang, W. (2018). A refined rat primary neonatal microglial culture method that reduces time, cost and animal use. J. Neurosci. Methods 304, 92–102. doi: 10.1016/j.jneumeth.2018.04.017
Ginhoux, F., Greter, M., Leboeuf, M., Nandi, S., See, P., Gokhan, S., et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. doi: 10.1126/science.1194637
Gordon, R., Hogan, C. E., Neal, M. L., Anantharam, V., Kanthasamy, A. G., and Kanthasamy, A. (2011). A simple magnetic separation method for high-yield isolation of pure primary microglia. J. Neurosci. Methods 194, 287–296. doi: 10.1016/j.jneumeth.2010.11.001
Goshi, N., Morgan, R. K., Lein, P. J., and Seker, E. (2020). A primary neural cell culture model to study neuron, astrocyte and microglia interactions in neuroinflammation. J. Neuroinflammation 17:155. doi: 10.1186/s12974-020-01819-z
Gosselin, D., Skola, D., Coufal, N. G., Holtman, I. R., Schlachetzki, J. C. M., Sajti, E., et al. (2017). An environment-dependent transcriptional network specifies human microglia identity. Science 356:eaal3222. doi: 10.1126/science.aal3222
Greter, M., Lelios, I., Pelczar, P., Hoeffel, G., Price, J., Leboeuf, M., et al. (2012). Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060. doi: 10.1016/j.immuni.2012.11.001
Gu, C., Hu, Q., Wu, J., Mu, C., Ren, H., Liu, C.-F., et al. (2018). P7C3 inhibits LPS-induced microglial activation to protect dopaminergic neurons against inflammatory factor-induced cell death in vitro and in vivo. Front. Cell. Neurosci. 12:400. doi: 10.3389/fncel.2018.00400
Haimon, Z., Volaski, A., Orthgiess, J., Boura-Halfon, S., Varol, D., Shemer, A., et al. (2018). Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 19, 636–644. doi: 10.1038/s41590-018-0110-6
Hammond, T. R., Dufort, C., Dissing-Olesen, L., Giera, S., Young, A., Wysoker, A., et al. (2019). Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6. doi: 10.1016/j.immuni.2018.11.004
Hansson, E. (1984). Cellular composition of a cerebral hemisphere primary culture. Neurochem. Res. 9, 153–172. doi: 10.1007/BF00964164
Hickman, S. E., Kingery, N. D., Ohsumi, T. K., Borowsky, M. L., Wang, L., Means, T. K., et al. (2013). The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16, 1896–1905. doi: 10.1038/nn.3554
Horvath, R. J., Nutile-McMenemy, N., Alkaitis, M. S., and Deleo, J. A. (2008). Differential migration, LPS-induced cytokine, chemokine and NO expression in immortalized BV-2 and HAPI cell lines and primary microglial cultures. J. Neurochem. 107, 557–569. doi: 10.1111/j.1471-4159.2008.05633.x
Humpel, C. (2015). Organotypic vibrosections from whole brain adult Alzheimer mice (overexpressing amyloid-precursor-protein with the Swedish-Dutch-Iowa mutations) as a model to study clearance of beta-amyloid plaques. Front. Aging Neurosci. 7:47. doi: 10.3389/fnagi.2015.00047
Jarjour, A. A., Zhang, H., Bauer, N., Ffrench-Constant, C., and Williams, A. (2012). in vitro modeling of central nervous system myelination and remyelination. Glia 60, 1–12. doi: 10.1002/glia.21231
Karzbrun, E., Kshirsagar, A., Cohen, S. R., Hanna, J. H., and Reiner, O. (2018). Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 14, 515–522. doi: 10.1038/s41567-018-0046-7
Kopec, K. K., and Carroll, R. T. (1998). Alzheimer’s beta-amyloid peptide 1-42 induces a phagocytic response in murine microglia. J. Neurochem. 71, 2123–2131. doi: 10.1046/j.1471-4159.1998.71052123.x
Lancaster, M. A., and Knoblich, J. A. (2014). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340. doi: 10.1038/nprot.2014.158
Lancaster, M. A., Renner, M., Martin, C.-A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Leite, D. M., Zvar Baskovic, B., Civita, P., Neto, C., Gumbleton, M., and Pilkington, G. J. (2020). A human co-culture cell model incorporating microglia supports glioblastoma growth and migration and confers resistance to cytotoxics. FASEB J. 34, 1710–1727. doi: 10.1096/fj.201901858RR
Li, Q., Cheng, Z., Zhou, L., Darmanis, S., Neff, N. F., Okamoto, J., et al. (2019). Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10. doi: 10.1016/j.neuron.2018.12.006
Li, C. L., and McIlwain, H. (1957). Maintenance of resting membrane potentials in slices of mammalian cerebral cortex and other tissues in vitro. J. Physiol. 139, 178–190. doi: 10.1113/jphysiol.1957.sp005885
Lian, H., Roy, E., and Zheng, H. (2016). Protocol for primary microglial culture preparation. Bio. Protoc. 6:e1989. doi: 10.21769/BioProtoc.1989
Llufrio, E. M., Wang, L., Naser, F. J., and Patti, G. J. (2018). Sorting cells alters their redox state and cellular metabolome. Redox Biol. 16, 381–387. doi: 10.1016/j.redox.2018.03.004
Luan, W., Li, M., Wu, C., Shen, X., and Sun, Z. (2022). Proteomic dissimilarities of primary microglia and BV2 cells under stimuli. Eur. J. Neurosci. 55, 1709–1723. doi: 10.1111/ejn.15637
Lyman, W. D., Tricoche, M., Hatch, W. C., Kress, Y., Chiu, F. C., and Rashbaum, W. K. (1991). Human fetal central nervous system organotypic cultures. Brain Res. Dev. Brain Res. 60, 155–160. doi: 10.1016/0165-3806(91)90044-j
Mariani, J., Coppola, G., Zhang, P., Abyzov, A., Provini, L., Tomasini, L., et al. (2015). FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 162, 375–390. doi: 10.1016/j.cell.2015.06.034
Masuda, T., Sankowski, R., Staszewski, O., Böttcher, C., Amann, L., Sagar, , et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. doi: 10.1038/s41586-019-0924-x
Mattei, D., Ivanov, A., van Oostrum, M., Pantelyushin, S., Richetto, J., Mueller, F., et al. (2020). Enzymatic dissociation induces transcriptional and proteotype bias in brain cell populations. Int. J. Mol. Sci. 21:E7944. doi: 10.3390/ijms21217944
Melief, J., Sneeboer, M. A. M., Litjens, M., Ormel, P. R., Palmen, S. J. M. C., Huitinga, I., et al. (2016). Characterizing primary human microglia: a comparative study with myeloid subsets and culture models. Glia 64, 1857–1868. doi: 10.1002/glia.23023
Montilla, A., Zabala, A., Matute, C., and Domercq, M. (2020). Functional and metabolic characterization of microglia culture in a defined medium. Front. Cell. Neurosci. 14:22. doi: 10.3389/fncel.2020.00022
Muffat, J., Li, Y., Yuan, B., Mitalipova, M., Omer, A., Corcoran, S., et al. (2016). Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 22, 1358–1367. doi: 10.1038/nm.4189
Murphy, T. H., Schnaar, R. L., and Coyle, J. T. (1990). Immature cortical neurons are uniquely sensitive to glutamate toxicity by inhibition of cystine uptake. FASEB J. 4, 1624–1633.
Nagai, A., Nakagawa, E., Hatori, K., Choi, H. B., McLarnon, J. G., Lee, M. A., et al. (2001). Generation and characterization of immortalized human microglial cell lines: expression of cytokines and chemokines. Neurobiol. Dis. 8, 1057–1068. doi: 10.1006/nbdi.2001.0437
Napoli, I., Kierdorf, K., and Neumann, H. (2009). Microglial precursors derived from mouse embryonic stem cells. Glia 57, 1660–1671. doi: 10.1002/glia.20878
Narantuya, D., Nagai, A., Sheikh, A. M., Masuda, J., Kobayashi, S., Yamaguchi, S., et al. (2010). Human microglia transplanted in rat focal ischemia brain induce neuroprotection and behavioral improvement. PLoS One 5:e11746. doi: 10.1371/journal.pone.0011746
Nikodemova, M., and Watters, J. J. (2012). Efficient isolation of live microglia with preserved phenotypes from adult mouse brain. J. Neuroinflammation 9:147. doi: 10.1186/1742-2094-9-147
Ocañas, S. R., Pham, K. D., Blankenship, H. E., Machalinski, A. H., Chucair-Elliott, A. J., and Freeman, W. M. (2022). Minimizing the ex vivo confounds of cell-isolation techniques on transcriptomic and translatomic profiles of purified microglia. eNeuro 9:ENEURO.0348-21.2022. doi: 10.1523/ENEURO.0348-21.2022
Okamoto, K., and Quastel, J. H. (1973). Spontaneous action potentials in isolated guinea-pig cerebellar slices: effects of amino acids and conditions affecting sodium and water uptake. Proc. R. Soc. Lond. B Biol. Sci. 184, 83–90. doi: 10.1098/rspb.1973.0032
Ormel, P. R., Vieira de Sá, R., van Bodegraven, E. J., Karst, H., Harschnitz, O., Sneeboer, M. A. M., et al. (2018). Microglia innately develop within cerebral organoids. Nat. Commun. 9:4167. doi: 10.1038/s41467-018-06684-2
Pacitti, D., Privolizzi, R., and Bax, B. E. (2019). Organs to cells and cells to organoids: the evolution of in vitro central nervous system modelling. Front. Cell Neurosci. 13:129. doi: 10.3389/fncel.2019.00129
Pan, J., and Wan, J. (2020). Methodological comparison of FACS and MACS isolation of enriched microglia and astrocytes from mouse brain. J. Immunol. Methods 486:112834. doi: 10.1016/j.jim.2020.112834
Park, S. E., Georgescu, A., and Huh, D. (2019). Organoids-on-a-chip. Science 364, 960–965. doi: 10.1126/science.aaw7894
Pluvinage, J. V., Haney, M. S., Smith, B. A. H., Sun, J., Iram, T., Bonanno, L., et al. (2019). CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 568, 187–192. doi: 10.1038/s41586-019-1088-4
Prinz, M., Jung, S., and Priller, J. (2019). Microglia biology: one century of evolving concepts. Cell 179, 292–311. doi: 10.1016/j.cell.2019.08.053
Quarta, A., Le Blon, D., D’aes, T., Pieters, Z., Hamzei Taj, S., Miró-Mur, F., et al. (2019). Murine iPSC-derived microglia and macrophage cell culture models recapitulate distinct phenotypical and functional properties of classical and alternative neuro-immune polarisation. Brain Behav. Immun. 82, 406–421. doi: 10.1016/j.bbi.2019.09.009
Roqué, P. J., and Costa, L. G. (2017). Co-culture of neurons and microglia. Curr. Protoc. Toxicol. 74, 11.24.1–11.24.17. doi: 10.1002/cptx.32
Rustenhoven, J., Park, T. I.-H., Schweder, P., Scotter, J., Correia, J., Smith, A. M., et al. (2016). Isolation of highly enriched primary human microglia for functional studies. Sci. Rep. 6:19371. doi: 10.1038/srep19371
Sepulveda-Diaz, J. E., Ouidja, M. O., Socias, S. B., Hamadat, S., Guerreiro, S., Raisman-Vozari, R., et al. (2016). A simplified approach for efficient isolation of functional microglial cells: application for modeling neuroinflammatory responses in vitro. Glia 64, 1912–1924. doi: 10.1002/glia.23032
Sierra, A., Paolicelli, R. C., and Kettenmann, H. (2019). Cien años de microglía: milestones in a century of microglial research. Trends Neurosci. 42, 778–792. doi: 10.1016/j.tins.2019.09.004
Skaper, S. D., Argentini, C., and Barbierato, M. (2012). “Culture of neonatal rodent microglia, astrocytes and oligodendrocytes from cortex and spinal cord,” in Neurotrophic Factors: Methods and Protocols Methods in Molecular Biology, ed S. D. Skaper (Totowa, NJ: Humana Press), 67–77. doi: 10.1007/978-1-61779-536-7_7
Song, G. J., Nam, Y., Jo, M., Jung, M., Koo, J. Y., Cho, W., et al. (2016). A novel small-molecule agonist of PPAR-γ potentiates an anti-inflammatory M2 glial phenotype. Neuropharmacology 109, 159–169. doi: 10.1016/j.neuropharm.2016.06.009
Stoppini, L., Buchs, P. A., and Muller, D. (1991). A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods 37, 173–182. doi: 10.1016/0165-0270(91)90128-m
Tachibana, C. Y. (2018). Stem-cell culture moves to the third dimension. Nature 558, 329–331. doi: 10.1038/d41586-018-05380-x
Telpoukhovskaia, M. A., Liu, K., Sayed, F. A., Etchegaray, J. I., Xie, M., Zhan, L., et al. (2020). Discovery of small molecules that normalize the transcriptome and enhance cysteine cathepsin activity in progranulin-deficient microglia. Sci. Rep. 10:13688. doi: 10.1038/s41598-020-70534-9
Tewari, M., Khan, M., Verma, M., Coppens, J., Kemp, J. M., Bucholz, R., et al. (2021). Physiology of cultured human microglia maintained in a defined culture medium. Immunohorizons 5, 257–272. doi: 10.4049/immunohorizons.2000101
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. doi: 10.1126/science.282.5391.1145
Timmerman, R., Burm, S. M., and Bajramovic, J. J. (2018). An overview of in vitro methods to study microglia. Front. Cell. Neurosci. 12:242. doi: 10.3389/fncel.2018.00242
Tsuchiya, T., Park, K. C., Toyonaga, S., Yamada, S. M., Nakabayashi, H., Nakai, E., et al. (2005). Characterization of microglia induced from mouse embryonic stem cells and their migration into the brain parenchyma. J. Neuroimmunol. 160, 210–218. doi: 10.1016/j.jneuroim.2004.10.025
Wang, Y., Szretter, K. J., Vermi, W., Gilfillan, S., Rossini, C., Cella, M., et al. (2012). IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13, 753–760. doi: 10.1038/ni.2360
Wobus, A. M., and Boheler, K. R. (1999). Preface. Cells Tissues Organs 165, 129–130. doi: 10.1159/000016692
Xiang, Z., Hrabetova, S., Moskowitz, S. I., Casaccia-Bonnefil, P., Young, S. R., Nimmrich, V. C., et al. (2000). Long-term maintenance of mature hippocampal slices in vitro. J. Neurosci. Methods 98, 145–154. doi: 10.1016/s0165-0270(00)00197-7
Yin, Z., Han, Z., Hu, T., Zhang, S., Ge, X., Huang, S., et al. (2020). Neuron-derived exosomes with high miR-21-5p expression promoted polarization of M1 microglia in culture. Brain Behav. Immun. 83, 270–282. doi: 10.1016/j.bbi.2019.11.004
Zakrzewski, W., Dobrzyński, M., Szymonowicz, M., and Rybak, Z. (2019). Stem cells: past, present and future. Stem Cell Res. Ther. 10:68. doi: 10.1186/s13287-019-1165-5
Zöller, T., Schneider, A., Kleimeyer, C., Masuda, T., Potru, P. S., Pfeifer, D., et al. (2018). Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun. 9:4011. doi: 10.1038/s41467-018-06224-y
Keywords: microglia, in vitro models, cell culture systems, primary cell culture, transformed cell lines, organotypic cultures, CNS organoid, ipsc-derived microglia
Citation: Aktories P, Petry P and Kierdorf K (2022) Microglia in a Dish—Which Techniques Are on the Menu for Functional Studies? Front. Cell. Neurosci. 16:908315. doi: 10.3389/fncel.2022.908315
Received: 30 March 2022; Accepted: 11 May 2022;
Published: 03 June 2022.
Edited by:
Liviu-Gabriel Bodea, University of Queensland, AustraliaReviewed by:
Björn Spittau, Bielefeld University, GermanyYukari Shigemoto-mogami, National Institute of Health Sciences (NIHS), Japan
Copyright © 2022 Aktories, Petry and Kierdorf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katrin Kierdorf, a2F0cmluLmtpZXJkb3JmQHVuaWtsaW5pay1mcmVpYnVyZy5kZQ==
† These authors have contributed equally to this work
 Philipp Aktories1,2†
Philipp Aktories1,2† Philippe Petry
Philippe Petry Katrin Kierdorf
Katrin Kierdorf