
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Neurosci. , 06 April 2022
Sec. Cellular Neuropathology
Volume 16 - 2022 | https://doi.org/10.3389/fncel.2022.865005
This article is part of the Research Topic Unfolded Protein Response (UPR): An Impending Target for Multiple Neurological Disorders View all 10 articles
Background: Aberrant endoplasmic reticulum stress (ERS) plays an important role in multiple cardiovascular diseases. However, their implication in intracranial aneurysms (IAs) remains unclear. We designed this study to explore the general expression pattern and potential functions of ERS in IAs.
Methods: Five Gene Expression Omnibus (GEO) microarray datasets were used as the training cohorts, and 3 GEO RNA sequencing (RNA-seq) datasets were used as the validating cohorts. Differentially expressed genes (DEGs), functional enrichment, Lasso regression, logistic regression, ROC analysis, immune cell profiling, vascular smooth muscle cell (VSMC) phenotyping, weighted gene coexpression network analysis (WGCNA), and protein-protein interaction (PPI) analysis were applied to investigate the role of ERS in IA. Finally, we predicted the upstream transcription factor (TF)/miRNA and potential drugs targeting ERS.
Results: Significant DEGs were majorly associated with ERS, autophagy, and metabolism. Eight-gene ERS signature and IRE1 pathway were identified during the IA formation. WGCNA showed that ERS was highly associated with a VSMC synthesis phenotype. Next, ERS-VSMC-metabolism-autophagy PPI and ERS-TF-miRNA networks were constructed. Finally, we predicted 9 potential drugs targeting ERS in IAs.
Conclusion: ERS is involved in IA formation. Upstream and downstream regulatory networks for ERS were identified in IAs. Novel potential drugs targeting ERS were also proposed, which may delay IA formation and progress.
Intracranial aneurysm (IA) is a life-threatening, complicated, and multifactorial disease that forms owing to the interaction among hemodynamics, genetics, and environmental factors. Immune/inflammation infiltration, cell death, lipid metabolism, oxidative stress, proteolytic activity, and iron accumulation are major histopathological features of IAs (Frösen et al., 2012). The recruitment and infiltration of immune cells have been confirmed to be a key phase in IA formation and development (Hosaka and Hoh, 2014; Signorelli et al., 2018). Recent studies suggest that vascular smooth muscle cell (VSMC) phenotype transformation is crucial to vascular wall remodeling of IA (Starke et al., 2014). Dysregulated autophagy can alter the VSMC phenotype, impair arterial wall function, and contribute to IA formation. Metabolism is also closely associated with the degeneration of IA arterial wall (Frösen et al., 2013). Therefore, it is urgent to investigate the complete mechanisms behind IA formation.
Endoplasmic reticulum stress (ERS) is various physiological or molecular disturbances that unbalance the unfolded-protein-response-regulated endoplasmic reticulum homeostasis (Ren et al., 2021). As a fundamental organelle, the dysfunction of the endoplasmic reticulum can affect multiple biological processes. Relevant studies show that ERS participates in the formation and development of cardiovascular diseases (Ren et al., 2021). Increased ERS markers have been reported in aortic aneurysm walls (Clément et al., 2019). Furthermore, stress-induced ERS can promote VSMC apoptosis, endothelial dysfunction, inflammation infiltration, and ultimately induce aortic aneurysm formation (Jia et al., 2015, 2017). Identifying the associations between ERS and IA may provide a better understanding of IA etiology.
In our study, 5 Gene Expression Omnibus (GEO) microarray datasets were selected as training cohorts, while 3 GEO RNA sequencing (RNA-seq) datasets were selected as validating cohorts. The association between ERS and IA formation was first confirmed by functional enrichment of differential expression genes (DEGs). Afterward, we constructed an ERS signature gene set, identified classical ERS pathways, generated an ERS-VSMC-metabolism-autophagy regulated network, predicted upstream transcription factor (TF) and microRNA targets of ERS genes, and explored the relationship between ERS and single nucleotide polymorphisms (SNPs) in IA diseases. Finally, potential drugs targeting ERS were predicted to inhibit IA formation and development.
Eight public IA datasets were downloaded from the GEO1, including 5 microarray datasets (GSE75436, GSE54083, GSE26969, GSE13353, GSE15629) and 3 RNA-sequencing datasets (GSE158558, GSE122897 and GSE66240). The patients involved in the database have obtained ethical approval. The raw data were merged and normalized using the “limma” R package (Ritchie et al., 2015). Batch effects were eliminated using the Combat algorithm (Leek et al., 2012). Of the 181 samples enrolled in our study, microarray data (55IAs and 42 controls) were used as the training set, and RNA-seq data (53 IAs and 31 controls) were used as the validating sets.
Principal components analysis (PCA) was employed to visualize the disparity between IA and control groups using the “factoextra” R package. DEG screening was conducted using the “limma” package (P < 0.05 and log2-fold change > 1 or < −1). Furthermore, we analyzed DEG functions by Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA) analysis (P < 0.05).
Two ERS-related gene sets (GO RESPONSE TO ENDOPLASMIC RETICULUM STRESS and GO REGULATION OF RESPONSE TO ENDOPLASMIC RETICULUM STRESS) were downloaded from Molecular Signature Database (MSigDB) v7.0. The Lasso regression was performed to identify the ERS-related DEGs with the highest IA predictive values. The predictive ability was further evaluated by univariate logistic analysis. Next, using these genes, we quantified ERS expression levels of all samples by Gene Set Variation Analysis (GSVA) scores (Hänzelmann et al., 2013).
Three ERS-related signaling pathways (GOBP ATF6 MEDIATED UNFOLDED PROTEIN RESPONSE, GOBP IRE1 MEDIATED UNFOLDED PROTEIN RESPONSE, GOBP PERK MEDIATED UNFOLDED PROTEIN RESPONSE) were downloaded from MsigDB v7.0. The GSVA scores were performed to quantify the expression level of these pathways in all samples. Pearson correlation analysis was performed between ERS pathways and signature genes.
Immunocyte infiltration of arterial walls was estimated using the “xCell” R package, which uses gene expression profiles to predict enrichment of 64 immune and stromal cell types (Aran et al., 2017). The VSMC phenotype was identified by 7 feature genes [SDC1, RBP1, MMP14, CDH2, MGP, PDGFA, MYH9 (Nakahara et al., 1992; Shanahan et al., 1993; Orlandi et al., 2002; Lyon et al., 2010; Chaterji et al., 2014; Shao et al., 2020)] and quantified by GSVA scores.
Weighted gene coexpression network analysis (WGCNA) was performed using the “WGCNA” R package (Langfelder and Horvath, 2008). An optimal soft threshold β was set to attain a scale-free topology network. Next, we evaluated the correlation between “ERS” and other pathophysiological traits. “ERS,” “VSMC synthesis,” and “IA” traits had the same high-associated modules (P < 0.001 and r > 0.45), which were assumed to be the key modules involved in IA formation and progression. The gene function of key modules was analyzed using GO and KEGG enrichment.
Apart from ERS, the DEG functions also included metabolism and autophagy. To evaluate the association between ERS, metabolism, and autophagy, we downloaded 948 metabolism-related genes from the KEGG database2, and 232 autophagy-related genes from the HADb database3. GSVA scoring and Pearson correlation analysis were then performed. After identifying the correlation, the aforementioned genes, together with high ERS-VSMC-IA-associated module genes were then imported into the STRING database4. Protein-protein interaction (PPI) networks were further visualized by Cytoscape software (version 3.9.0). Furthermore, NetworkAnalyst5 (Zhou et al., 2019), a comprehensive network visual analytics platform for gene expression analysis, was applied to predict upstream TFs and miRNAs of ERS. Finally, based on ERS signature genes, we constructed ERS-TF-miRNA networks.
The 80 TFs and 142 nearby genes of regulatory regions which overlapped with IA-associated SNPs, were downloaded from Laarman’s study (Laarman et al., 2018). The integration analysis was used between TFs of the ERS signature and TFs of regulatory regions. The correlation analysis was performed between the ERS signature and genes in proximity to regulatory regions.
The Connectivity Map (CMAP) website6 was applied to explore small molecule drugs with the potential to inhibit IA formation and development. The drugs with negative Raw_cs and high fdr_q_nlog10 values were considered as potential therapeutic agents because they could suppress the expression of ERS signature genes.
All statistical analyses were conducted using the R software (version 4.0.2). The Wilcox test was applied to compare the difference of continuous variables between the two groups. P < 0.05 was considered statistically significant. Data were visualized using the R package “ggplot2.” Heatmaps were drawn using the “pheatmap” R package. Volcano plots were generated using the “ggrepel” R package.
The study was designed as indicated in the flow chart (Figure 1). We sought to explore the role of ERS in IA formation by comprehensive analysis based on microarray and RNA-sequencing datasets. In total, we collected 55 cases of IA and 42 cases of normal arteries as controls in microarray training cohorts, and 53 cases of IA and 31 cases of normal arteries as controls in RNA-seq validation cohorts (Figure 2A). For both training and validation cohorts, similar distributions of different samples were observed in normalized data after preprocessing (Figure 2B). PCA analysis showed that the IA group could be discriminated from the controls at the transcript level (Figure 2C). On filtering with the limma package, 1,628 up-regulated genes and 2,013 down-regulated genes were found in the training cohort. 590 up-regulated genes and 685 down-regulated genes were found in the validation cohort (Figure 2D). Heatmaps were used to visualize the expression of DEGs in all cases (Figure 2E).
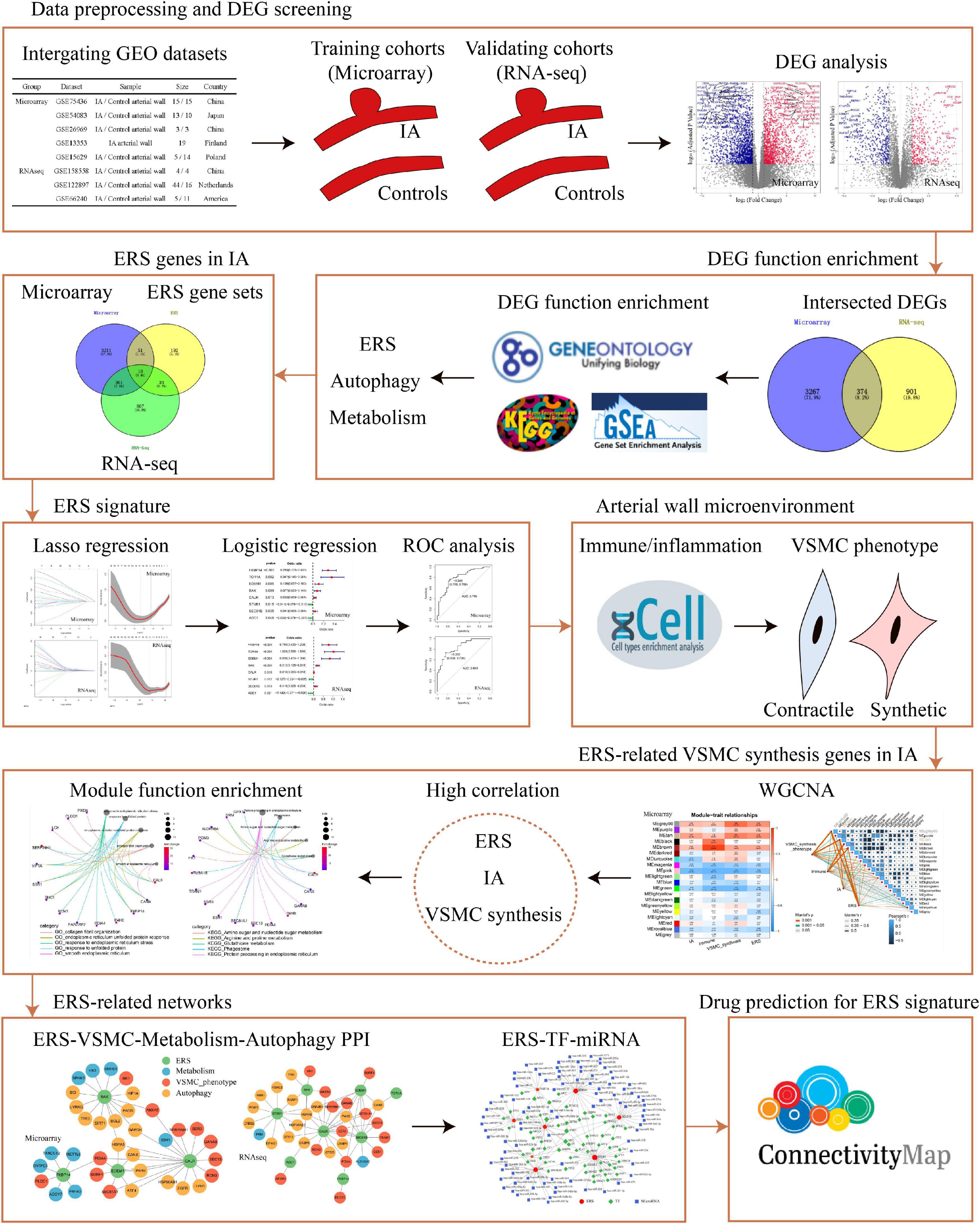
Figure 1. The flow chart of data analysis. DEG, differential expression gene; RNA-seq, RNA sequencing; ERS, endoplasmic reticulum stress; ROC, receiver operating characteristic; VSMC, vascular smooth muscle cell; PPI, protein–protein Interaction; TF, transcription factor.
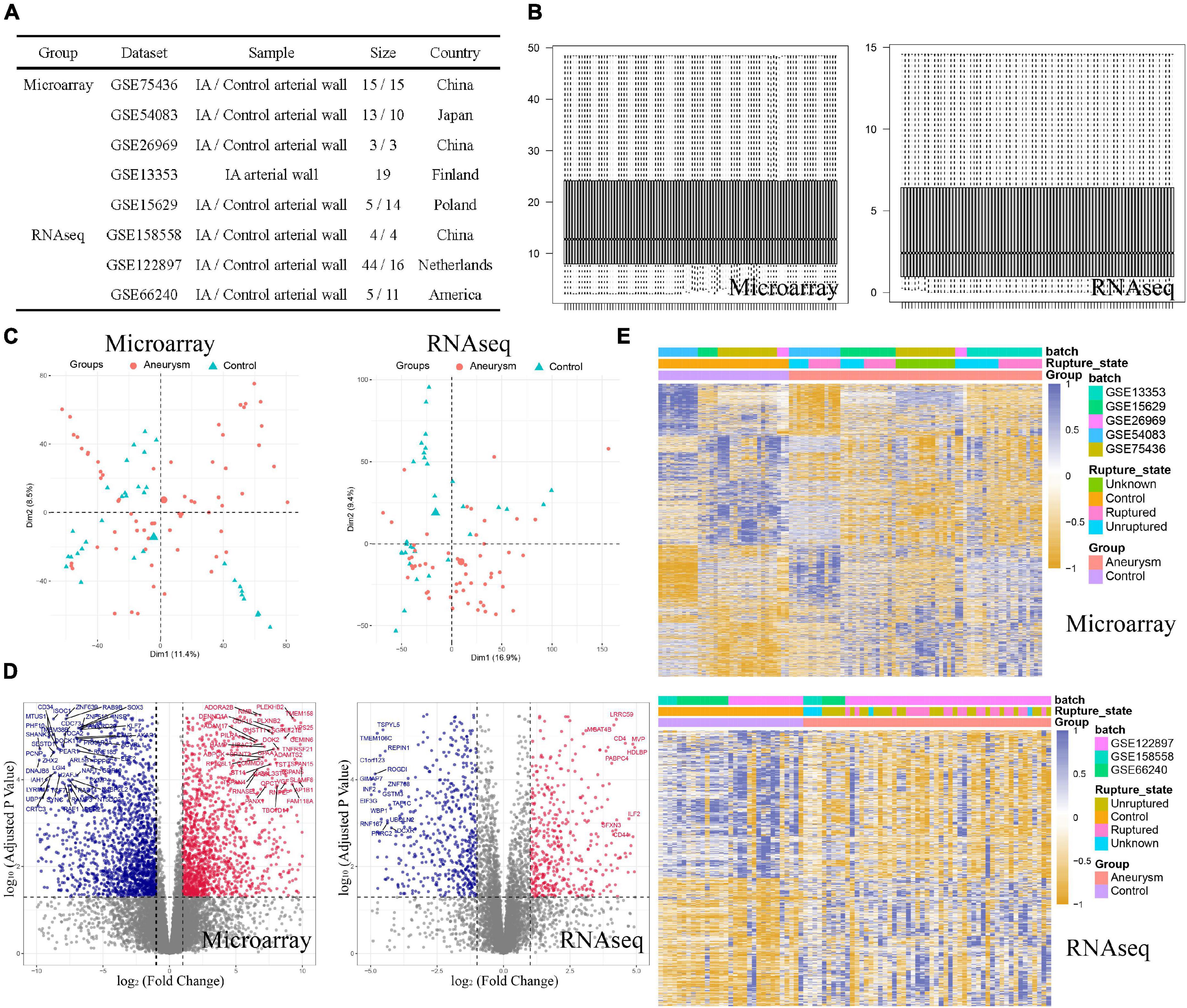
Figure 2. Data preprocessing and DEG screening in both training and validating cohorts. (A) Basic information of included dataset. (B) Boxplots of normalized data showed similar distributions of different samples. (C) PCA plots showed the IA group could be discriminated from the controls. (D) Volcano plots visualized the fold change and P-value of all genes between the two groups. Red plots were upregulated genes and blue plots were downregulated genes. (E) Heatmaps visualized the expression level of DEGs.
To explore disease progression in IA, we performed functional enrichment analysis for intersected DEGs between the training and validation cohorts. Among GO enrichment terms, the most overrepresented were ERS, response to unfolded protein, autophagosome, and similar pathways (Figure 3A). In the KEGG pathway analysis, DEGs were notably enriched in protein processing in ER, metabolism process, phagosome, antigen processing and presentation, and others (Figure 3B). In GSEA biological process results, endomembrane system and response to stimulus terms showed higher expression in the IA group (Figure 3C), whereas cellular macromolecule metabolic process was more frequent in the normal artery control group (Figure 3D). Overall, DEGs were functionally enriched in ERS, autophagy, and metabolism-related processes. Further correlation analysis showed that the expression of autophagy and metabolism was positively associated with ERS, separately (Supplementary Figures 1A,B).
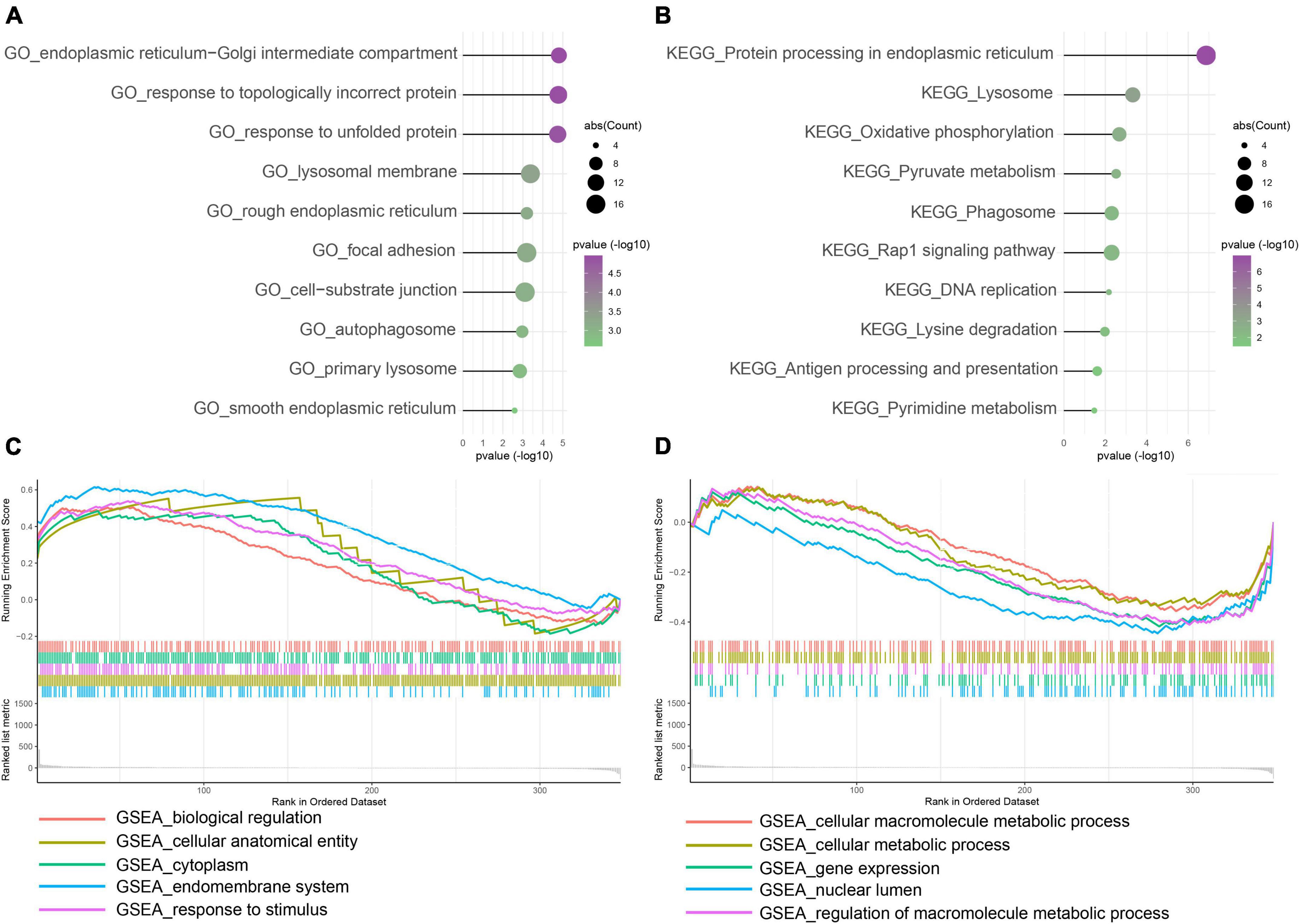
Figure 3. Function enrichment of intersected DEGs between the training and control group. (A) Gene Ontology (GO) enrichment analysis. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. (C,D) Gene Set Enrichment Analysis (GSEA). Enrichment showed that DEG function mainly focused on ERS, unfolded protein response (UPR), autophagy, immune/inflammation, and metabolism.
Considering the prominent role of ERS in DEG functional enrichment, we sought to determine diagnostic values of ERS in IA by constructing an ERS signature. Firstly, we selected 18 IA-related ERS genes by intersecting ERS gene sets and DEGs of training and validation cohorts. Next, these 18 genes were used for LASSO regression to select the most valuable predictive genes, and an 8-gene ERS signature was constructed (Figures 4A,B). FKBP14, TOR1A, EDEM1, BAX, CALR, SEC61B were upregulated, whereas STUB1 and ADD1 were downregulated in IAs (Supplementary Figure 3A). Univariate Logistic regression showed FKBP14, TOR1A, EDEM1, BAX, CALR, and SEC61B may promote IA formation, while STUB1 and ADD1 can prevent it (Figure 4C). ROC curve analysis showed that GSVA scores of the ERS signature could predict IA formation, with areas under the curve (AUC) of 0.799 and 0.845 in the training and validation cohorts, respectively (Figure 4D). Heatmaps were used to visualize ERS signature expression in all cases (Figure 4E).
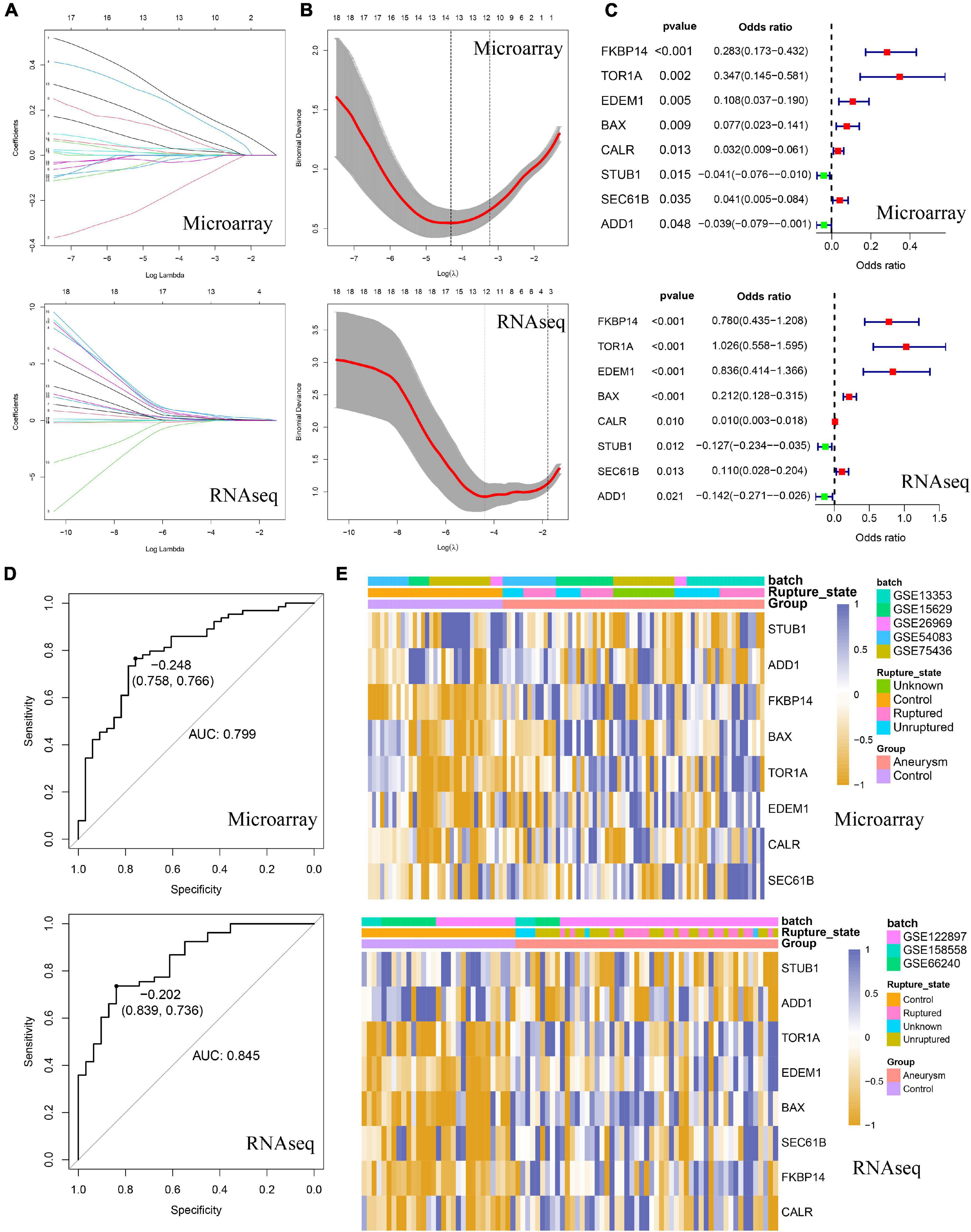
Figure 4. Identification of the ERS-related signature in both training and validating cohorts. (A) The coefficient profiles of the LASSO regression model. (B) Cross-validation for tuning parameter screening in the LASSO regression model. (C) Univariate Logistic regression identified 8 ERS genes’ odds ratios (ORs) and 95% confidence intervals (Cls) after LASSO regression filtration. (D) ROC curve analysis for Gene Set Variation Analysis (GSVA) scores of 8 ERS genes. (E) The heatmap visualized the expression level of 8 ERS genes.
There were 3 ERS-related classical signaling pathways, including the ATF6 pathway, IRE1 pathway, and Perk pathway. The expression of the IRE1 pathway was significantly higher in IAs than controls, whereas the ATF6 pathway and perk pathway did not show significant differences between the two groups (Figure 5A). IRE1 pathway showed high correlations to FKBP14, BAX, and SEC61B expression (Correlation coefficient > 0.3, Figure 5B).
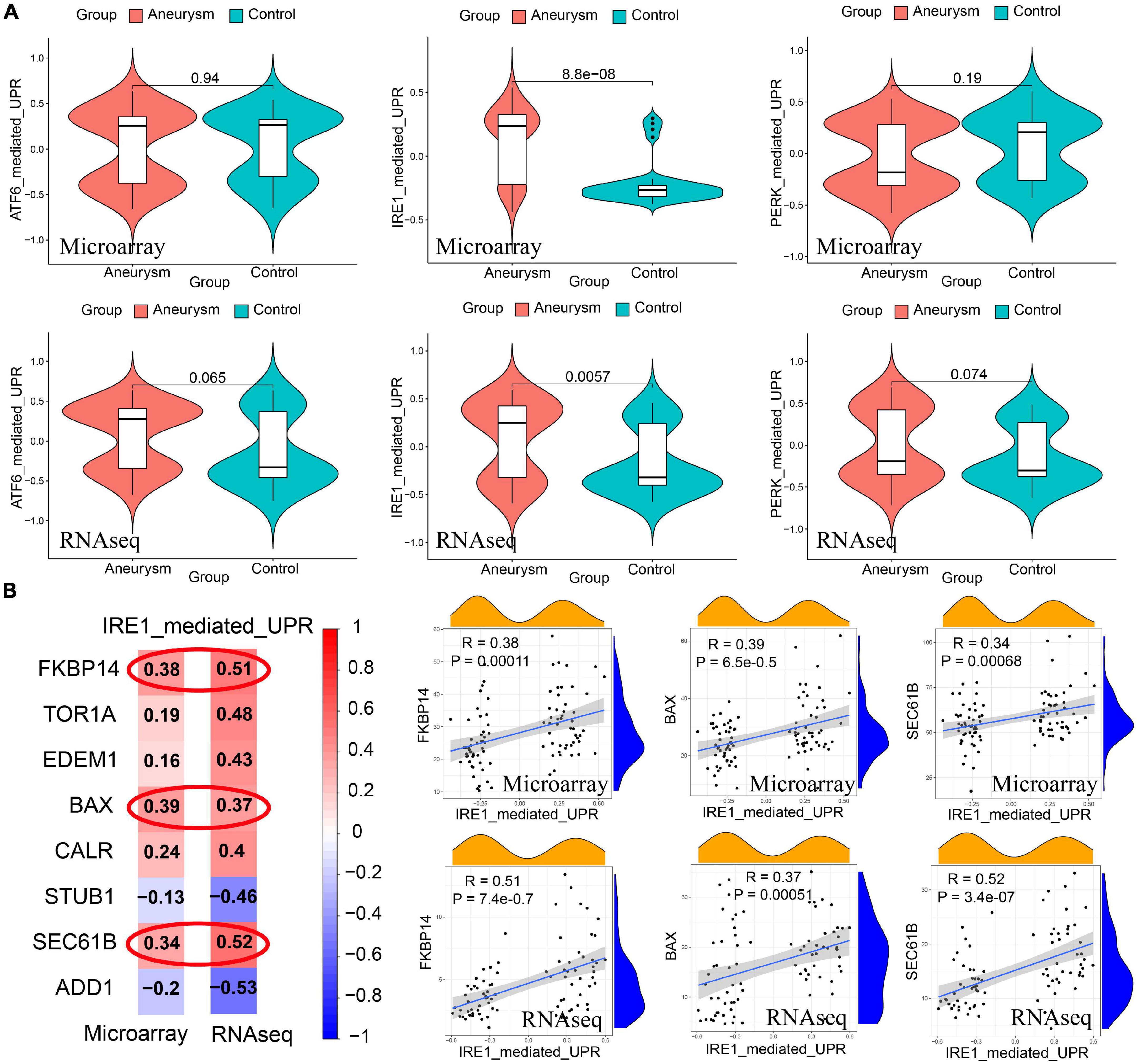
Figure 5. Identification of ERS-related signaling pathways in both training and validating cohorts. (A) GSVA scores of 3 pathway expression. IRE1 pathway had a higher expression level in IAs than the level in controls. The expression of the ATF6 pathway and PERK pathway did not show significant differences between the two groups. (B) Pearson correlation between IRE1 pathway and signature genes. The expression of FKBP14, BAX, and SEC61B had high correlations to IRE1 pathway expression (Correlation coefficient > 0.3 in both training and validating cohorts).
Since immune infiltration/inflammation and VSMC phenotype are tightly associated with IA formation and progression, we further investigated the arterial wall microenvironment. For both training and validation cohorts, Xcell immune profiling results showed more immune/inflammation-related cell types and higher immune scores in IA (Figures 6A,B). VSMC phenotype analysis revealed that IA cohorts expressed more VSMC-synthesis-phenotype-feature genes and higher synthesis-phenotype GSVA scores (Figures 6C,D).
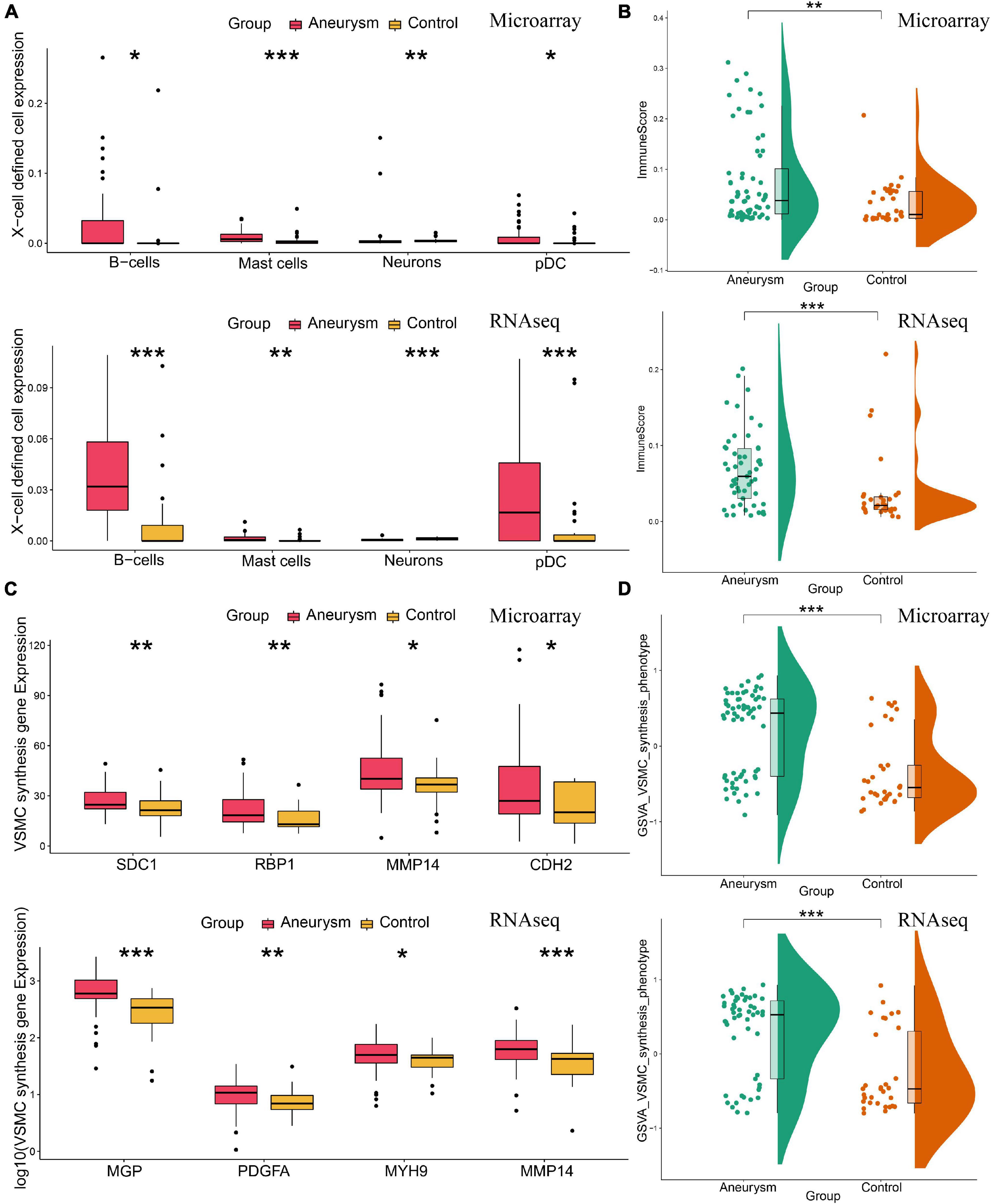
Figure 6. Annotation of the arterial wall microenvironment of immune/inflammation infiltrating and VSMC phenotype in both training and validating cohorts. (A) The expression level of X cell-defined immunocytes and neurons. (B) The total enrichment scores of the immune microenvironment. IA lesions had higher immune expression than control arteries. (C) The expression of feature genes of VSMC synthesis phenotype. (D) The GSVA scores of VSMC synthesis phenotype. IA lesions had more VSMC synthesis phenotype than control arteries. *P < 0.05; **P < 0.01; ***P < 0.001.
Considering ERS, immune/inflammation, and VSMC phenotype are all involved in IA formation, we sought to conduct WGCNA coexpression analysis to identify the relationship among those. Powers β = 5 or 4 were selected as the software threshold for scale-free network construction in training and validation cohorts, respectively (Figure 7A and Supplementary Figure 2A). In the training cohort, 20 modules were identified, and in the validation cohort, 13 modules were identified by clustering dendrogram (Figure 7B and Supplementary Figure 2B). IA, VSMC synthesis, and ERS had the same highest-correlated modules (MEgray60 and MEtan), indicating strong associations among these traits (Figures 7C,D). A similar result was also observed in the validation cohort (Supplementary Figures 2C,D). By intersecting the two most relevant modules in the training and validation group, we identified 85 ERS-related VSMC phenotype genes involved in IA formation. GO enrichment analysis showed these genes mainly focused on collagen fibril organization, smooth endoplasmic reticulum, and others (Figure 7E). KEGG pathway analysis showed metabolism, phagosome, and protein processing in the endoplasmic reticulum were more enriched among these genes (Figure 7F).
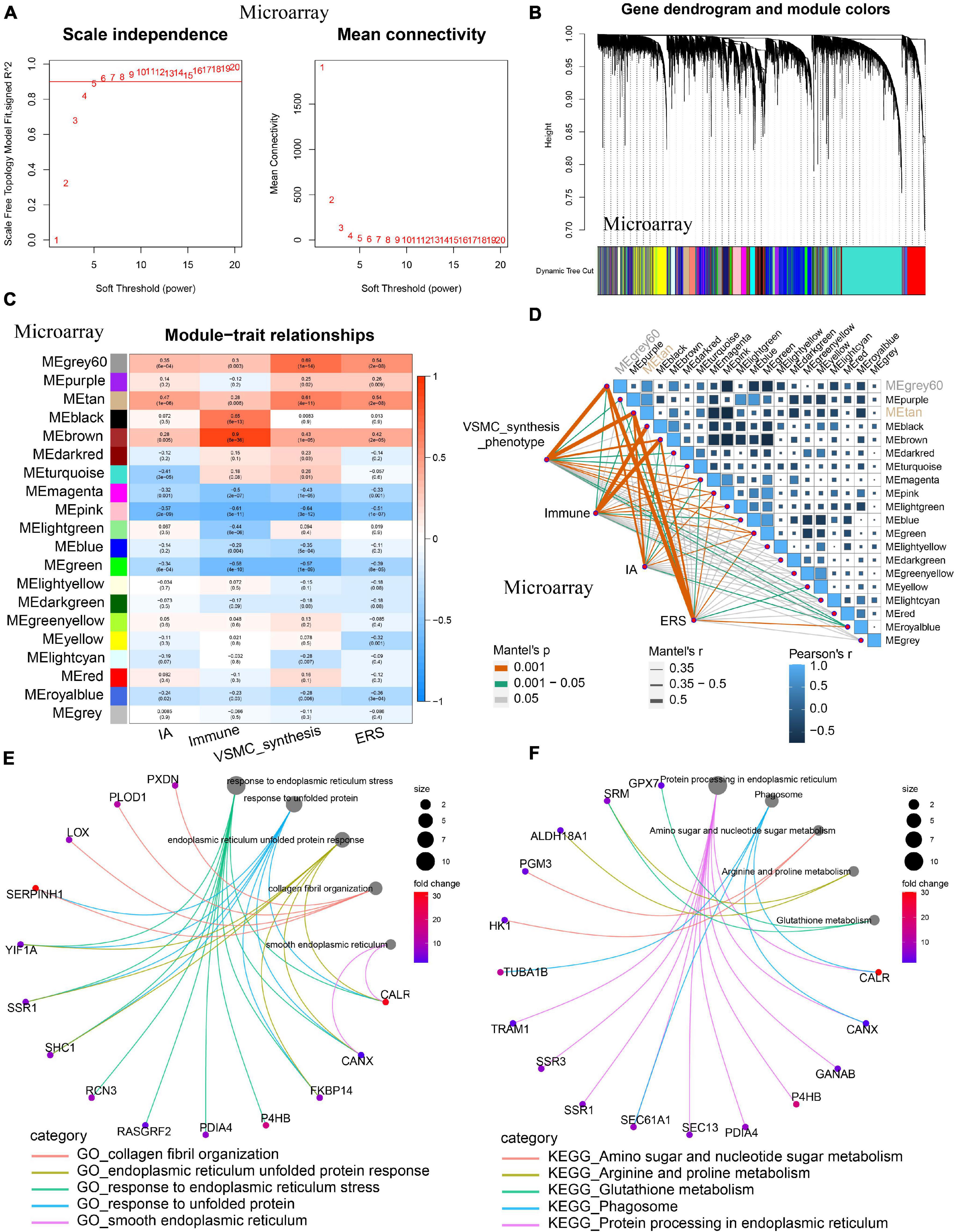
Figure 7. Co-expression analysis identifying ERS-related VSMC phenotype genes in the training cohort. (A) A scale-free network construction (power threshold β = 5). (B) Gene dendrogram generating gene modules. (C) and (D) Correlation analysis between modules and pathophysiological traits. IA occurrence, VSMC synthesis, and ERS had the same two highest correlation modules (MEgray60 and MEtan). (E) GO enrichment analysis of the intersection between the training cohort (MEgray60 and MEtan) and the validating cohort (MEbrown and MEpink). (F) KEGG enrichment analysis of the intersection between the training cohort (MEgray60 and MEtan) and (MEbrown and MEpink). Function enrichment showed that these modules mainly focused on smooth endoplasmic reticulum, collagen fibril organization, metabolism, and phagosome.
After identifying the correlation among ERS, VSMC phenotype, metabolism, and autophagy in IA formation, we constructed PPI networks among those pathophysiological traits. Within DEGs targeting the ERS signature, a total of 11 were involved in the VSMC synthesis phenotype, 9 were correlated to metabolism, and 15 were associated with autophagy in the training cohort (Figure 8A). The validating cohort also showed similar ERS-VSMC-metabolism-autophagy PPI networks (Figure 8B).
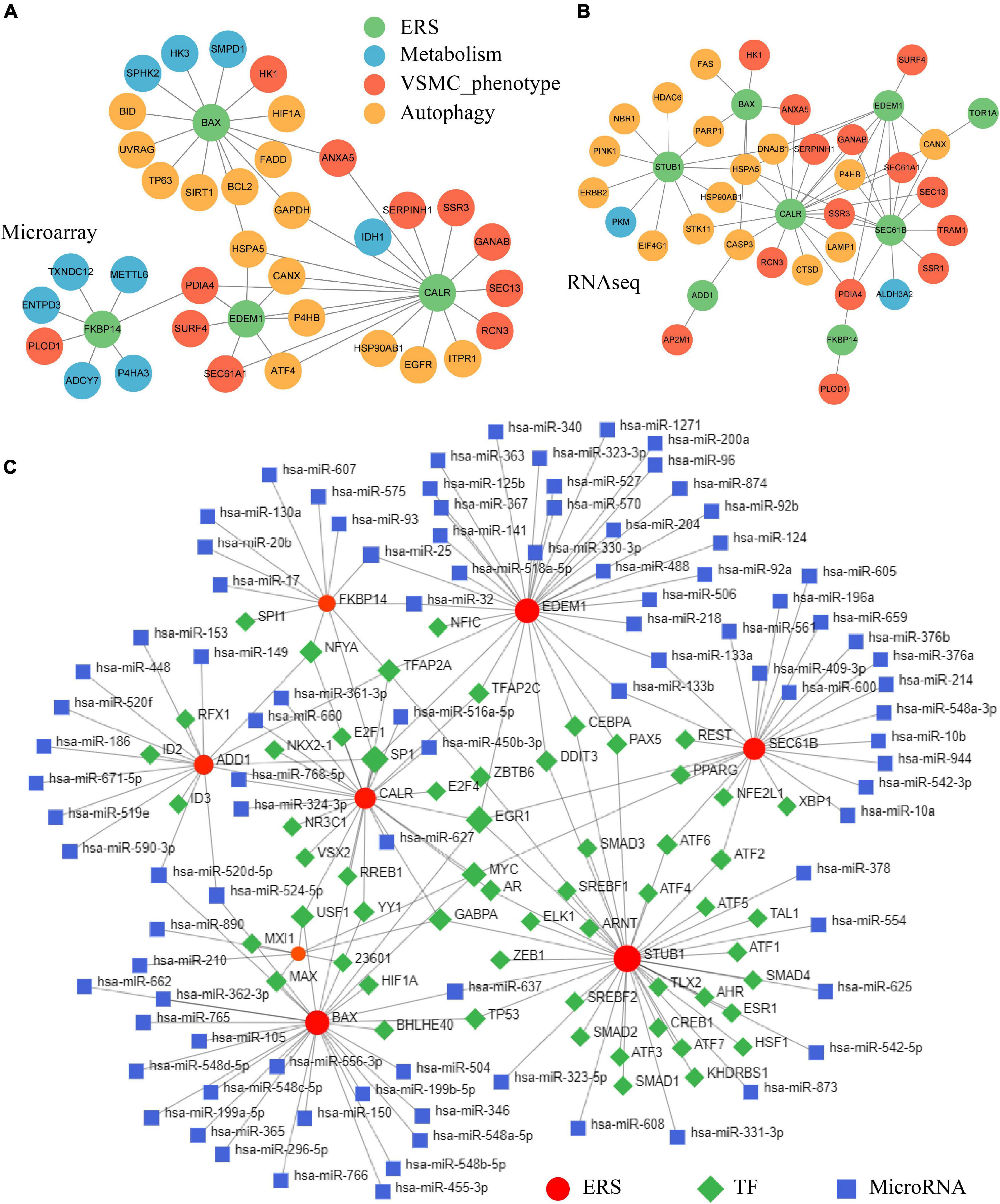
Figure 8. Constructing ERS-VSMC-metabolism-autophagy PPI and ERS-TF-miRNA networks. (A) PPI network among ERS, metabolism, VSMC phenotype, and autophagy-related genes in the training cohorts. (B) PPI network among ERS, metabolism, VSMC phenotype, and autophagy-related genes in the validating cohorts. (C) Prediction of upstream TFs and microRNAs for ERS-related genes.
The NetworkAnalyst online tool was used to predict ERS upstream TF and miRNA. Eight ERS signature genes had identified TFs. NFYA, TFAP2A, SP1, EGR1, MYC, GABPA, and USF1 were common TFs among at least 3 genes. ERS signature genes of EDEM1 and BAX had the most predicted miRNAs including hsa-miR-25, hsa-miR-32, hsa-miR-520d-5p, hsa-miR-524-5p, hsa-miR-637, hsa-miR-133b and hsa-miR-133a (Figure 8C).
We then investigated TFs and nearby genes of ERS-associated non-coding SNPs in IAs. Seventeen TFs were identified to co-regulate ERS and non-coding SNPs. Among these, MYC had the most ERS target genes and TF binding sites (Supplementary Table 1). Moreover, 6 nearby genes were found to differentially express (Supplementary Figure 3B). Correlation analysis showed the tight connectivity between 8 ERS signature genes and 6 nearby genes, in which KCTD15 had the most significant correlations with ERS (Supplementary Figure 3C).
To predict small molecule drugs with the potential to inhibit IA ERS, we uploaded the ERS signature into the CMAP online tool. We identified 9 drugs (thioperamide, tracazolate, cephaeline, GW-843682X, aminopurvalanol-a, geranylgeraniol, hydroflumethiazide, BRD-K76674262, everolimus) with the negative Raw_cs and the top fdr_q_nlog10 values, suggesting they could inhibit the expression of the ERS signature (Figure 9 and Supplementary Table 2).
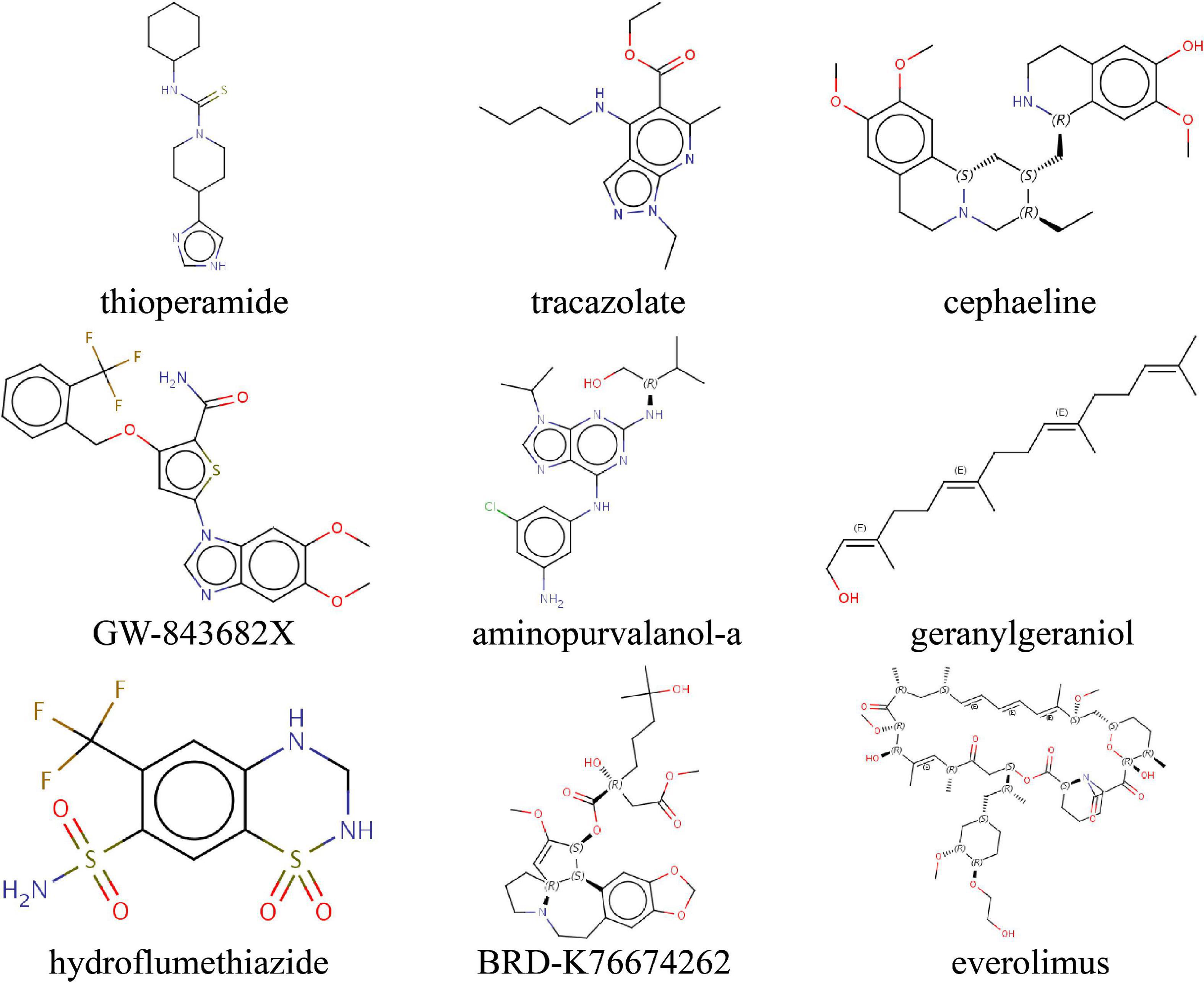
Figure 9. Identifying the molecular structure of 9 small component drugs targeting ERS-related gens in IAs by CMAP.
Endoplasmic reticulum stress is an imbalance of the endoplasmic reticulum homeostasis caused by an accumulation of unfolded or misfolded proteins. Multiple pathologies can induce ERS, including pressure overload, metabolic disorders, atherosclerosis, ischemia-reperfusion injury, endothelial dysfunction, and others. Long-term ERS promotes abnormal inflammation and apoptosis in the vascular wall, leading to disturbances in cardiovascular function (Ren et al., 2021). Previous studies have shown that excessive ERS is closely associated with various cardiovascular diseases, including heart failure, cardiomyopathy, hypertension, stroke, and the like (Ren et al., 2021).
In the present study, the role of ERS in IA formation was explored using bioinformatics analysis for the first time. We firstly identified ERS by functional enrichment of DEGs, constructed an ERS signature gene set. Afterward, we generated ERS-VSMC-metabolism-autophagy PPI networks and predicted ERS-related upstream TF and microRNAs. The relationship between ERS and non-coding SNPs was then explored. Finally, potential drugs targeting ERS were predicted to inhibit IA formation and development.
Recently, accumulating evidence demonstrates that ERS plays an important role in aneurysm formation and development. In our research, multiple types of DEG functional enrichment analyses showed ERS was related to IA pathogenesis. Similarly, Clément et al. (2019) found increased expression of ERS markers in VSMCs of dissected aortic aneurysms. Jia et al. (2015) proved that stress-induced ERS contributed to thoracic aortic aneurysm and dissection formation. They also reported that ERS-dependent microparticles promote endothelial dysfunction during the formation process of thoracic aortic aneurysm and dissection (Jia et al., 2017). In addition, several studies showed that ERS inhibition could attenuate the formation and development of abdominal aortic aneurysms (Li et al., 2017; Ni et al., 2018).
The ERS signature gene set was then constructed in IA, including FKBP14, TOR1A, EDEM1, BAX, CALR, STUB1, SEC61B, and ADD1. These genes have been confirmed to be related to multiple cardiovascular diseases. Among those, BAX, whose protein belongs to the BCL2 family, is an apoptosis activator. One study showed that overexpressed Bax regulated intimal hyperplasia of VSMCs in arteriosclerosis (Hayakawa et al., 1999). Another study showed that upregulated Bax was associated with the presence of cystic medial degeneration of the aorta (Ihling et al., 1999). Calreticulin (CALR) encoded by the CALR gene, is a highly conserved chaperone protein primarily expressed in the endoplasmic reticulum. Previous studies indicate that CALR can coordinate vascular function and heterocellular calcium signaling (Biwer et al., 2018). STUB1, encoding the protein of STIP1 Homology And U-Box Containing Protein 1, was down-regulated during the IA process in our studies. A prior study showed that the decreased STUB1 in VSMCs inhibited thrombosis in flow loops (Shashar et al., 2017). Adducin 1 (ADD1), belonging to the cytoskeletal protein family, was also expressed at lower levels in vascular walls of IA patients. A sequencing study suggested that ADD1 polymorphism significantly increased the susceptibility to ischemic and hemorrhagic strokes (Kalita et al., 2011).
To update, there are three classic signaling pathways in ERS, including ATF6 pathway, IRE1 pathway, and Perk pathway. They act as proximal sensors of unfolded protein response (UPR) (Wu and Kaufman, 2006). Our results showed that the IRE1 pathway was highly expressed in IA lesions and had strong correlations to the gene expression of BAX, FKBP14, and SEC61B. Previous research has demonstrated that proapoptotic BAX moduled UPR by direct interaction with IRE (Hetz et al., 2006). The overexpression of BAX inhibitor-1 could inhibit IRE and reversed hyperglycemia in diet-induced obesity mice (Bailly-Maitre et al., 2010). Whether these ERS genes could promote IA formation by the IRE1 pathway deserves further basic experimental study.
VSMC phenotype transformation, from contractility to synthesis, is involved in IA formation and development. Our results suggest a strong association between ERS and VSMC synthesis in IA pathogenesis. This relationship has already been confirmed in a previous study. Zhang et al. (2020) showed that the microgravity regulated ERS to induce VSMC phenotype transform. Zhao et al. (2020) identified that Matrine inhibited VSMC phenotype transformation via ERS-dependent Notch signaling. Chattopadhyay et al. (2021) found that UPR could drive cholesterol-induced VSMC phenotype transformation.
Considering metabolism and autophagy were also enriched in IAs, we constructed ERS-VSMC-metabolism-autophagy PPI networks. Body metabolism disorders have been discovered to involve the pathological processes of IA. Frösen et al. (2013) found that lipid accumulation and its oxidation in the IA wall, together with low plasma levels of acquired antibodies against oxidized lipids, were associated with IA wall degeneration and rupture. Semmler et al. (2008) demonstrated that polymorphisms of homocysteine metabolism were possible risk factors for IA formation. Besides, growing evidence showed that autophagy was also involved in IA formation, development, and rupture. Sun et al. (2017) proved that ruptured IA tissues had more expression of autophagy-related genes, including LC3, Atg5, and Atg14, followed by unruptured IA and control artery tissues. In vitro experiments showed that activated VSMC autophagy could enhance the VSMC proliferation and migration, and induce IA formation (Zhang et al., 2019). Furthermore, the relationships among ERS, metabolism, and autophagy have been demonstrated in other diseases. There are mutual regulations between ERS and metabolism. Fu et al. (2011) showed that aberrant lipid metabolism would cause ERS in obesity. Henkel et al. (2017) proved that ERS regulated hepatic bile acid metabolism in mice. As for autophagy, it is generally considered the last means to restore the homeostasis of the endoplasmic reticulum (Henkel et al., 2017). Together, we speculated ERS can influence metabolism/autophagy/VSMC phenotype and thus contribute to IA formation, which needs further basic research.
The dysregulation of upstream TF and microRNA for ERS also has a crucial impact on the formation and development of cardiovascular diseases. Our TF prediction showed that NFYA, TFAP2A, SP1, EGR1, MYC, GABPA, and USF1 were common TFs with at least 3 ERS genes in IAs. Among these, SP1, whose encoding protein is involved in cell differentiation and growth, has been confirmed to be associated with ERS and VSMC phenotype switching. Dauer et al. (2017) proved that inhibition of SP1 prevented endoplasmic reticulum homeostasis. Hu et al. (2021) found that SP1 regulated migration and phenotype switching of VSMCs through the MAPK pathway in aortic dissections. Tang et al. (2017) identified that microRNA-124 controlled VSMC phenotypic switching via SP1. EGR, belonging to the early growth response family, was found to be related to ERS and aneurysm formation. Previous studies showed that ERS can activate EGR1 transcription via the MAPK pathway (Shan et al., 2019). Other studies prove that EGR1 upregulation leads to aortic aneurysm formation and EGR1 downregulation can reverse this process (Lin et al., 2020; Shin et al., 2020). In addition, we predicted 91 upstream microRNAs for ERS. Seven microRNAs had 2 target ERS genes. Among these, hsa-miR-25, hsa-miR-133b, and hsa-miR-133a have been confirmed to independently predict aneurysm occurrence or prevent aneurysm development (Li et al., 2014; Plana et al., 2020; Akerman et al., 2021). Furthermore, upregulated hsa-miR-637 can aggravate ERS-induced apoptosis (Kong et al., 2020).
IA-associated SNPs were reported to be enriched in Cow regulatory regions (Laarman et al., 2018). The relationship between non-coding SNPs and ERS was investigated. Integration analysis showed that 17 TFs co-regulated ERS and regulatory regions in IAs. Among these, MYC, whose encoded a nuclear phosphoprotein, with a role in cycle progression, apoptosis, and cellular transformation, had the most ERS target genes and TF binding sites. Previous research had confirmed that MYC was involved in ERS. Dong et al. (2019) found that the IRE1 ERS sensor could activate natural killer cell immunity by MYC regulation. Jayasooriya et al. (2018) found that camptothecin enhanced MYC-mediated ERS and led to autophagy. In addition, Li et al. (2020) proved that the downregulating MYC-mediated ENC1 could prevent IA formation. Correlation analysis showed the tight connectivity between ERS signature genes and nearby genes of regulatory regions. KCTD15, a potassium channel encoding gene, had the most significant associations with ERS. Previous research found that potassium channels have the modification of gating properties under the ERS and were involved in the cerebral vasospasm after subarachnoid hemorrhage (Sobey and Faraci, 1998; Khodaee et al., 2014). The role of KCTD15 in IA formation deserves further research.
Previous studies reported that ERS was the potential therapeutic target for aneurysms. In this research, we predicted 9 small molecule drugs for IAs. These drugs have shown the potential to inhibit ERS progress. Cephaeline and BRD-K76674262, belonging to protein synthesis inhibitors, could inhibit tumor viability, migration, and proliferation (Silva et al., 2021). Han et al. (2013) proved ERS increased protein synthesis leading to cell death, and presented, limiting protein synthesis would be therapeutic for ERS-caused diseases. Everolimus, an mTOR inhibitor, is used in immunosuppressive treatment after organ transplantation and anticancer treatment for advanced renal cell cancers (Patel and Kobashigawa, 2006; Mariniello et al., 2012). Previous studies have found bidirectional crosstalk between ERS and mTOR (Appenzeller-Herzog and Hall, 2012). Persistent mTOR activation could induce ERS occurrence (Wang et al., 2016). Of note, everolimus has been shown capable of limiting aortic aneurysm dilatation in apolipoprotein E-deficient mouse (Moran et al., 2013). IA progress may be delayed by these compounds.
Our study had some limitations. One major limitation was the lack of basic experimental data to confirm and support our findings. Another limitation was the lack of IA-associated clinical data, like size, location, number, and others, to further explore the association between ERS and IA. Additionally, the predicted TF, miRNA, and drugs remain to be further explored to understand their real-world roles in IA formation and development.
Our results strongly suggest that ERS is involved in IA formation. Upstream and downstream regulatory networks for ERS were identified in IAs. Novel potential drugs targeting ERS were also proposed, which may delay IA formation and progress.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by open-source GEO database. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
BC, YX, and LZ designed and drafted the manuscript. BC, HZ, LY, XZ, and LZ organized figure legends and revised the article. BC conducted data analysis. All authors have read and approved the final manuscript.
This work was supported by the Nature Science Foundation of China (No. 81402249 to LZ), the Natural Science Foundation of Hunan Province (No. 2019JJ50963 to LZ), and Fundamental Research Funds for the Central Universities of Central South University (No. 160171016 to BC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge GEO databases for their platforms and contributors for uploading their meaningful datasets. We would like to express our very great appreciation to F. H. Chen for his valuable and constructive suggestions during the development of this research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2022.865005/full#supplementary-material
Supplementary Figure 1 | Correlation analysis among ERS, metabolism, and autophagy in both training and validating cohorts. Autophagy (A) and metabolism (B) were positively associated with ERS, separately.
Supplementary Figure 2 | Co-expression analysis identifying ERS-related VSMC phenotype genes in the validating cohort. (A) A scale-free network construction (power threshold β = 4). (B) Gene dendrogram generating gene modules. (C,D) Correlation analysis between modules and pathophysiological traits. IA occurrence, VSMC synthesis, and ERS had the same two highest correlation modules (MEbrown and MEpink).
Supplementary Figure 3 | Correlation analysis between ERS and nearby genes of regulatory regions in both training and validating cohorts. (A) The expression level of ERS signature genes. IA lesions had higher expression of FKBP14, TOR1A, EDEM1, BAX, CALR, SEC61B, and lower expression of STUB1 and ADD1. (B) Venn diagrams of differential expressed genes (DEGs) and nearby genes. Six nearby DEGs of regulatory regions were gained. (C) Correlation analysis between ERS signature genes and nearby genes. KCTD15 had the most significant associations with ERS.
Supplementary Table 1 | TF intersection between ERS signature genes and regulatory regions.
Supplementary Table 2 | Identified 9 small molecular drugs by CMAP.
ADD1, adducin 1; AUC, areas under the curve; CALR, calreticulin; CMAP, connectivity map; DEG, differential expression gene; ERS, endoplasmic reticulum stress; GEO, Gene Expression Omnibus; GO, Gene Ontology; GSEA, Gene Set Enrichment Analysis; GSVA, Gene Set Variation Analysis; IA, intracranial aneurysm; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCA, principal components analysis; PPI, protein–protein Interaction; RNA-seq, RNA sequencing; SNP, single nucleotide polymorphism; TF, transcription factor; UPR, unfolded protein response; VSMC, vascular smooth muscle cell; WGCNA, weighted gene coexpression network analysis.
Akerman, A. W., Collins, E. N., Peterson, A. R., Collins, L. B., Harrison, J. K., DeVaughn, A., et al. (2021). miR-133a replacement attenuates thoracic aortic aneurysm in mice. J. Am. Heart Assoc. 10:e019862. doi: 10.1161/jaha.120.019862
Appenzeller-Herzog, C., and Hall, M. N. (2012). Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 22, 274–282. doi: 10.1016/j.tcb.2012.02.006
Aran, D., Hu, Z., and Butte, A. J. (2017). xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18:220. doi: 10.1186/s13059-017-1349-1
Bailly-Maitre, B., Belgardt, B. F., Jordan, S. D., Coornaert, B., von Freyend, M. J., Kleinridders, A., et al. (2010). Hepatic Bax inhibitor-1 inhibits IRE1alpha and protects from obesity-associated insulin resistance and glucose intolerance. J. Biol. Chem. 285, 6198–6207. doi: 10.1074/jbc.M109.056648
Biwer, L. A., Good, M. E., Hong, K., Patel, R. K., Agrawal, N., Looft-Wilson, R., et al. (2018). Non-Endoplasmic reticulum-based calr (Calreticulin) can coordinate heterocellular calcium signaling and vascular function. Arterioscler. Thromb. Vasc. Biol. 38, 120–130. doi: 10.1161/atvbaha.117.309886
Chaterji, S., Lam, C. H., Ho, D. S., Proske, D. C., and Baker, A. B. (2014). Syndecan-1 regulates vascular smooth muscle cell phenotype. PLoS One 9:e89824. doi: 10.1371/journal.pone.0089824
Chattopadhyay, A., Kwartler, C. S., Kaw, K., Li, Y., Kaw, A., Chen, J., et al. (2021). Cholesterol-Induced phenotypic modulation of smooth muscle cells to macrophage/fibroblast-like cells is driven by an unfolded protein response. Arterioscler. Thromb. Vasc. Biol. 41, 302–316. doi: 10.1161/atvbaha.120.315164
Clément, M., Chappell, J., Raffort, J., Lareyre, F., Vandestienne, M., Taylor, A. L., et al. (2019). Vascular smooth muscle cell plasticity and autophagy in dissecting aortic aneurysms. Arterioscler. Thromb. Vasc. Biol. 39, 1149–1159. doi: 10.1161/atvbaha.118.311727
Dauer, P., Gupta, V. K., McGinn, O., Nomura, A., Sharma, N. S., Arora, N., et al. (2017). Inhibition of Sp1 prevents ER homeostasis and causes cell death by lysosomal membrane permeabilization in pancreatic cancer. Sci. Rep. 7:1564. doi: 10.1038/s41598-017-01696-2
Dong, H., Adams, N. M., Xu, Y., Cao, J., Allan, D. S. J., Carlyle, J. R., et al. (2019). The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc. Nat. Immunol. 20, 865–878. doi: 10.1038/s41590-019-0388-z
Frösen, J., Tulamo, R., Heikura, T., Sammalkorpi, S., Niemelä, M., Hernesniemi, J., et al. (2013). Lipid accumulation, lipid oxidation, and low plasma levels of acquired antibodies against oxidized lipids associate with degeneration and rupture of the intracranial aneurysm wall. Acta Neuropathol. Commun. 1:71. doi: 10.1186/2051-5960-1-71
Frösen, J., Tulamo, R., Paetau, A., Laaksamo, E., Korja, M., Laakso, A., et al. (2012). Saccular intracranial aneurysm: pathology and mechanisms. Acta Neuropathol. 123, 773–786. doi: 10.1007/s00401-011-0939-3
Fu, S., Yang, L., Li, P., Hofmann, O., Dicker, L., Hide, W., et al. (2011). Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473, 528–531. doi: 10.1038/nature09968
Han, J., Back, S. H., Hur, J., Lin, Y. H., Gildersleeve, R., Shan, J., et al. (2013). ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490. doi: 10.1038/ncb2738
Hänzelmann, S., Castelo, R., and Guinney, J. (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14:7. doi: 10.1186/1471-2105-14-7
Hayakawa, Y., Takemura, G., Misao, J., Kanoh, M., Ohno, M., Ohashi, H., et al. (1999). Apoptosis and overexpression of bax protein and bax mRNA in smooth muscle cells within intimal hyperplasia of human radial arteries : analysis with arteriovenous fistulas used for hemodialysis. Arterioscler. Thromb. Vasc. Biol. 19, 2066–2077. doi: 10.1161/01.atv.19.9.2066
Henkel, A. S., LeCuyer, B., Olivares, S., and Green, R. M. (2017). Endoplasmic reticulum stress regulates hepatic bile acid metabolism in mice. Cell Mol. Gastroenterol. Hepatol. 3, 261–271. doi: 10.1016/j.jcmgh.2016.11.006
Hetz, C., Bernasconi, P., Fisher, J., Lee, A. H., Bassik, M. C., Antonsson, B., et al. (2006). Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 312, 572–576. doi: 10.1126/science.1123480
Hosaka, K., and Hoh, B. L. (2014). Inflammation and cerebral aneurysms. Transl. Stroke Res. 5, 190–198. doi: 10.1007/s12975-013-0313-y
Hu, C., Huang, W., Xiong, N., and Liu, X. (2021). SP1-mediated transcriptional activation of PTTG1 regulates the migration and phenotypic switching of aortic vascular smooth muscle cells in aortic dissection through MAPK signaling. Arch. Biochem. Biophys. 711:109007. doi: 10.1016/j.abb.2021.109007
Ihling, C., Szombathy, T., Nampoothiri, K., Haendeler, J., Beyersdorf, F., Uhl, M., et al. (1999). Cystic medial degeneration of the aorta is associated with p53 accumulation, Bax upregulation, apoptotic cell death, and cell proliferation. Heart 82, 286–293. doi: 10.1136/hrt.82.3.286
Jayasooriya, R., Dilshara, M. G., Karunarathne, W., Molagoda, I. M. N., Choi, Y. H., and Kim, G. Y. (2018). Camptothecin enhances c-Myc-mediated endoplasmic reticulum stress and leads to autophagy by activating Ca(2+)-mediated AMPK. Food Chem. Toxicol. 121, 648–656. doi: 10.1016/j.fct.2018.09.057
Jia, L. X., Zhang, W. M., Li, T. T., Liu, Y., Piao, C. M., Ma, Y. C., et al. (2017). ER stress dependent microparticles derived from smooth muscle cells promote endothelial dysfunction during thoracic aortic aneurysm and dissection. Clin. Sci. (Lond) 131, 1287–1299. doi: 10.1042/cs20170252
Jia, L. X., Zhang, W. M., Zhang, H. J., Li, T. T., Wang, Y. L., Qin, Y. W., et al. (2015). Mechanical stretch-induced endoplasmic reticulum stress, apoptosis and inflammation contribute to thoracic aortic aneurysm and dissection. J. Pathol. 236, 373–383. doi: 10.1002/path.4534
Kalita, J., Somarajan, B. I., Kumar, B., Mittal, B., and Misra, U. K. (2011). A study of ACE and ADD1 polymorphism in ischemic and hemorrhagic stroke. Clin. Chim. Acta 412, 642–646. doi: 10.1016/j.cca.2010.12.022
Khodaee, N., Ghasemi, M., Saghiri, R., and Eliassi, A. (2014). Endoplasmic reticulum membrane potassium channel dysfunction in high fat diet induced stress in rat hepatocytes. Excli J. 13, 1075–1087.
Kong, Q., Zhang, Z., and Liang, Z. (2020). Upregulating miR-637 aggravates endoplasmic reticulum stress-induced apoptosis in gastric cancer cells by suppressing Calreticulin. Anim. Cells Syst. (Seoul) 24, 267–274. doi: 10.1080/19768354.2020.1816579
Laarman, M. D., Vermunt, M. W., Kleinloog, R., de Boer-Bergsma, J. J., Brain Bank, N., Rinkel, G. J. E., et al. (2018). Intracranial aneurysm-associated single-nucleotide polymorphisms alter regulatory DNA in the human circle of willis. Stroke 49, 447–453. doi: 10.1161/strokeaha.117.018557
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E., and Storey, J. D. (2012). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. doi: 10.1093/bioinformatics/bts034
Li, P., Zhang, Q., Wu, X., Yang, X., Zhang, Y., Li, Y., et al. (2014). Circulating microRNAs serve as novel biological markers for intracranial aneurysms. J. Am. Heart Assoc. 3:e000972. doi: 10.1161/jaha.114.000972
Li, X., Zhao, H., Liu, J., and Tong, J. (2020). Long Non-coding RNA MIAT knockdown prevents the formation of intracranial aneurysm by downregulating ENC1 via MYC. Front. Physiol. 11:572605. doi: 10.3389/fphys.2020.572605
Li, Y., Lu, G., Sun, D., Zuo, H., Wang, D. W., and Yan, J. (2017). Inhibition of endoplasmic reticulum stress signaling pathway: a new mechanism of statins to suppress the development of abdominal aortic aneurysm. PLoS One 12:e0174821. doi: 10.1371/journal.pone.0174821
Lin, H., You, B., Lin, X., Wang, X., Zhou, D., Chen, Z., et al. (2020). Silencing of long non-coding RNA Sox2ot inhibits oxidative stress and inflammation of vascular smooth muscle cells in abdominal aortic aneurysm via microRNA-145-mediated Egr1 inhibition. Aging (Albany NY) 12, 12684–12702. doi: 10.18632/aging.103077
Lyon, C. A., Koutsouki, E., Aguilera, C. M., Blaschuk, O. W., and George, S. J. (2010). Inhibition of N-cadherin retards smooth muscle cell migration and intimal thickening via induction of apoptosis. J. Vasc. Surg. 52, 1301–1309. doi: 10.1016/j.jvs.2010.05.096
Mariniello, B., Rosato, A., Zuccolotto, G., Rubin, B., Cicala, M. V., Finco, I., et al. (2012). Combination of sorafenib and everolimus impacts therapeutically on adrenocortical tumor models. Endocr. Relat. Cancer 19, 527–539. doi: 10.1530/erc-11-0337
Moran, C. S., Jose, R. J., Moxon, J. V., Roomberg, A., Norman, P. E., Rush, C., et al. (2013). Everolimus limits aortic aneurysm in the apolipoprotein E-deficient mouse by downregulating C-C chemokine receptor 2 positive monocytes. Arterioscler. Thromb. Vasc. Biol. 33, 814–821. doi: 10.1161/atvbaha.112.301006
Nakahara, K., Nishimura, H., Kuro-o, M., Takewaki, S., Iwase, M., Ohkubo, A., et al. (1992). Identification of three types of PDGF-A chain gene transcripts in rabbit vascular smooth muscle and their regulated expression during development and by angiotensin II. Biochem. Biophys. Res. Commun. 184, 811–818. doi: 10.1016/0006-291x(92)90662-5
Ni, X. Q., Lu, W. W., Zhang, J. S., Zhu, Q., Ren, J. L., Yu, Y. R., et al. (2018). Inhibition of endoplasmic reticulum stress by intermedin1-53 attenuates angiotensin II-induced abdominal aortic aneurysm in ApoE KO Mice. Endocrine 62, 90–106. doi: 10.1007/s12020-018-1657-6
Orlandi, A., Francesconi, A., Clément, S., Ropraz, P., Spagnoli, L. G., and Gabbiani, G. (2002). High levels of cellular retinol binding protein-1 expression in leiomyosarcoma: possible implications for diagnostic evaluation. Virchows Arch. 441, 31–40. doi: 10.1007/s00428-001-0576-7
Patel, J. K., and Kobashigawa, J. A. (2006). Everolimus: an immunosuppressive agent in transplantation. Expert Opin. Pharmacother. 7, 1347–1355. doi: 10.1517/14656566.7.10.1347
Plana, E., Gálvez, L., Medina, P., Navarro, S., Fornés-Ferrer, V., Panadero, J., et al. (2020). Identification of novel microRNA profiles dysregulated in plasma and tissue of abdominal aortic aneurysm patients. Int. J. Mol. Sci. 21:4600. doi: 10.3390/ijms21134600
Ren, J., Bi, Y., Sowers, J. R., Hetz, C., and Zhang, Y. (2021). Endoplasmic reticulum stress and unfolded protein response in cardiovascular diseases. Nat. Rev. Cardiol. 18, 499–521. doi: 10.1038/s41569-021-00511-w
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Semmler, A., Linnebank, M., Krex, D., Götz, A., Moskau, S., Ziegler, A., et al. (2008). Polymorphisms of homocysteine metabolism are associated with intracranial aneurysms. Cerebrovasc. Dis. 26, 425–429. doi: 10.1159/000155638
Shan, J., Dudenhausen, E., and Kilberg, M. S. (2019). Induction of early growth response gene 1 (EGR1) by endoplasmic reticulum stress is mediated by the extracellular regulated kinase (ERK) arm of the MAPK pathways. Biochim. Biophys. Acta Mol. Cell Res. 1866, 371–381. doi: 10.1016/j.bbamcr.2018.09.009
Shanahan, C. M., Weissberg, P. L., and Metcalfe, J. C. (1993). Isolation of gene markers of differentiated and proliferating vascular smooth muscle cells. Circ. Res. 73, 193–204. doi: 10.1161/01.res.73.1.193
Shao, Y., Li, G., Huang, S., Li, Z., Qiao, B., Chen, D., et al. (2020). Effects of extracellular matrix softening on vascular smooth muscle cell dysfunction. Cardiovasc. Toxicol. 20, 548–556. doi: 10.1007/s12012-020-09580-8
Shashar, M., Belghasem, M. E., Matsuura, S., Walker, J., Richards, S., Alousi, F., et al. (2017). Targeting STUB1-tissue factor axis normalizes hyperthrombotic uremic phenotype without increasing bleeding risk. Sci. Transl. Med. 9:eaam8475. doi: 10.1126/scitranslmed.aam8475
Shin, S. J., Hang, H. T., Thang, B. Q., Shimoda, T., Sakamoto, H., Osaka, M., et al. (2020). Role of PAR1-Egr1 in the initiation of thoracic aortic aneurysm in Fbln4-Deficient mice. Arterioscler. Thromb. Vasc. Biol. 40, 1905–1917. doi: 10.1161/atvbaha.120.314560
Signorelli, F., Sela, S., Gesualdo, L., Chevrel, S., Tollet, F., Pailler-Mattei, C., et al. (2018). Hemodynamic stress, inflammation, and intracranial aneurysm development and rupture: a systematic review. World Neurosurg. 115, 234–244. doi: 10.1016/j.wneu.2018.04.143
Silva, L. C., Borgato, G. B., Wagner, V. P., Martins, M. D., Rocha, G. Z., Lopes, M. A., et al. (2021). Cephaeline is an inductor of histone H3 acetylation and inhibitor of mucoepidermoid carcinoma cancer stem cells. J. Oral Pathol. Med. Online ahead of print. doi: 10.1111/jop.13252
Sobey, C. G., and Faraci, F. M. (1998). Subarachnoid haemorrhage: what happens to the cerebral arteries? Clin. Exp. Pharmacol. Physiol. 25, 867–876. doi: 10.1111/j.1440-1681.1998.tb02337.x
Starke, R. M., Chalouhi, N., Ding, D., Raper, D. M., McKisic, M. S., Owens, G. K., et al. (2014). Vascular smooth muscle cells in cerebral aneurysm pathogenesis. Transl. Stroke Res. 5, 338–346. doi: 10.1007/s12975-013-0290-1
Sun, L., Zhao, M., Zhang, J., Lv, M., Li, Y., Yang, X., et al. (2017). MiR-29b downregulation induces phenotypic modulation of vascular smooth muscle cells: implication for intracranial aneurysm formation and progression to rupture. Cell Physiol. Biochem. 41, 510–518. doi: 10.1159/000456887
Tang, Y., Yu, S., Liu, Y., Zhang, J., Han, L., and Xu, Z. (2017). MicroRNA-124 controls human vascular smooth muscle cell phenotypic switch via Sp1. Am. J. Physiol. Heart Circ. Physiol. 313, H641–H649. doi: 10.1152/ajpheart.00660.2016
Wang, J., Yang, X., and Zhang, J. (2016). Bridges between mitochondrial oxidative stress, ER stress and mTOR signaling in pancreatic β cells. Cell. Signal. 28, 1099–1104. doi: 10.1016/j.cellsig.2016.05.007
Wu, J., and Kaufman, R. J. (2006). From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ. 13, 374–384. doi: 10.1038/sj.cdd.4401840
Zhang, J., Jin, J., and Yang, W. (2019). [Autophagy regulates the function of vascular smooth muscle cells in the formation and rupture of intracranial aneurysms]. Zhejiang Da Xue Xue Bao Yi Xue Ban 48, 552–559.
Zhang, R., Jiang, M., Zhang, J., Qiu, Y., Li, D., Li, S., et al. (2020). Regulation of the cerebrovascular smooth muscle cell phenotype by mitochondrial oxidative injury and endoplasmic reticulum stress in simulated microgravity rats via the PERK-eIF2α-ATF4-CHOP pathway. Biochim Biophys. Acta Mol. Basis Dis. 1866:165799. doi: 10.1016/j.bbadis.2020.165799
Zhao, L., Cai, H., Tang, Z., Cui, Q., Liu, Z., and Lu, S. (2020). Matrine suppresses advanced glycation end products-induced human coronary smooth muscle cells phenotype conversion by regulating endoplasmic reticulum stress-dependent Notch signaling. Eur. J. Pharmacol. 882:173257. doi: 10.1016/j.ejphar.2020.173257
Keywords: intracranial aneurysm, endoplasmic reticulum stress, bioinformatics, drug prediction, unfolded protein response
Citation: Chen B, Zhou H, Zhou X, Yang L, Xiong Y and Zhang L (2022) Comprehensive Analysis of Endoplasmic Reticulum Stress in Intracranial Aneurysm. Front. Cell. Neurosci. 16:865005. doi: 10.3389/fncel.2022.865005
Received: 29 January 2022; Accepted: 07 March 2022;
Published: 06 April 2022.
Edited by:
Safikur Rahman, Babasaheb Bhimrao Ambedkar Bihar University, IndiaReviewed by:
Esmerina Tili, The Ohio State University, United StatesCopyright © 2022 Chen, Zhou, Zhou, Yang, Xiong and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyuan Xiong, eGlvbmd5dWFueXVhbl9wdWJAMTI2LmNvbQ==; Liyang Zhang, emhhbmdsaXlhbmdAY3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.