- 1Department of Anatomy, Hebei Medical University, Shijiazhuang, China
- 2Neuroscience Research Center, Hebei Medical University, Shijiazhuang, China
- 3Hebei Key Laboratory of Neurodegenerative Disease Mechanism, Shijiazhuang, China
The importance of epitranscriptomics in regulating gene expression has received widespread attention. Recently, RNA methylation modifications, particularly N6-methyladenosine (m6A), have received marked attention. m6A, the most common and abundant type of eukaryotic methylation modification in RNAs, is a dynamic reversible modification that regulates nuclear splicing, stability, translation, and subcellular localization of RNAs. These processes are involved in the occurrence and development of many diseases. An increasing number of studies have focused on the role of m6A modification in Alzheimer’s disease, which is the most common neurodegenerative disease. This review focuses on the general features, mechanisms, and functions of m6A methylation modification and its role in Alzheimer’s disease.
Introduction
Epitranscriptomics, i.e., chemical modification used for RNA regulation, has recently emerged as a highly investigated subfield of neuroscience. In the early 1970s, RNA methylation modifications were discovered (Desrosiers et al., 1974). These include N6-methyladenosine (m6A), N1-methyladenosine (m1A), N6, 2-O-dimethyladenosine (m6Am), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (5hmC), and 7-methylguanine (m7G) (Chen et al., 2016). Among them, m6A is the most common and abundant type of eukaryotic methylation modification in RNAs, including mRNAs, long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), microRNAs (miRNAs), rRNAs, tRNAs, and small nuclear RNAs (snRNAs) (Yue et al., 2015; Du et al., 2018; Shi et al., 2019). The adenine in the RRACH sequence (R = adenine or guanine, and H = cytosine, adenine, or uracil) is usually the site of m6A modification (Figure 1; Deng et al., 2018). In mammals, m6A methylation modification is widely distributed in many tissues, particularly in the brain (Meyer and Jaffrey, 2014). The abundance of m6A and its emerging role as an important post-transcriptional regulator in the mammalian brain has gained wide attention in the field of neuroepigenetics (Chang et al., 2017).
Alzheimer’s disease (AD), the most frequently and commonly diagnosed dementia, is an increasing global health concern with a notable impact on human health (Guo et al., 2020). Despite significant advances in our understanding of AD pathogenesis and the definition of the disease since the first case reported by Alzheimer (1907), there are still no disease-modifying treatments (Pakavathkumar et al., 2017). Altered m6A-methylation has been considered associated with AD as many players in the m6A pathway have been implicated as critical factors in neuronal function (Shafik et al., 2021).
In this review, we will focus on the general features, functions, and adjustability of m6A methylation modifications and their role in AD.
RNA N6-Methyladenosine Methylation Modification Protein
The m6A modification of RNA has been proven to be reversible, as it is bidirectionally regulated by m6A methyltransferase and demethylase, which work with RNA m6A methylation-binding protein to regulate the fate of RNAs (Knuckles and Bühler, 2018; Widagdo and Anggono, 2018). These proteins have served as valuable tools for investigating the cellular and physiological roles of m6A methylation modification in the brain.
N6-Methyladenosine Methyltransferase
The m6A methyltransferases, also known as “Writers” in RNA m6A methylation modification, catalyze the transfer of a methyl group from S-adenosyl methionine (SAM) to adenine nucleotides of RNA substrates. Some “Writers,” including methyltransferases such as 3/14 (METTL3/14), Wilms’ tumor 1-associating protein (WTAP), KIAA1429, and RNA-binding motifs protein 15/15B (RBM15/15B), form the core components of m6A methyltransferase, which work together to catalyze methylation of RNA substrates (Liu et al., 2020). In addition, zinc finger CCCH-type containing 13 (ZC3H13) and E3 ubiquitin-protein ligase Hakai (HAKAI) are also components of this complex. Moreover, recent studies have found that methyltransferase-like 16 (METTL16) can catalyze the methylation of target RNAs alone, without relying on the above m6A methyltransferase complexes (Pendleton et al., 2017).
METTL3 (Figures 2, 3A) is the best-known m6A methyltransferase. It is identified as a SAM-binding component in the complex and has its own catalytic ability. Unlike METTL3, METTL14 (Figures 2, 3B) does not bind to the SAM domain performing its own m6A methyltransferase catalytic ability but plays a key role in substrate identification. Biochemical and structural studies have revealed that METTL3 and METTL14 form a heterodimer (Figure 3C; Wang et al., 2016) that has a higher methylation activity than METTL3 alone (Liu et al., 2014; Wang et al., 2017). WTAP (Figures 2, 3D) is a regulatory subunit of the complex that interacts with METTL3 and METTL14 and localizes the METTL3-METTL14 complex to the nucleus (Ping et al., 2014; Schwartz et al., 2014). KIAA1429 (Figures 2, 3E) can mediate preferential mRNA methylation in 3’UTR and near stop codon and is also known as vir-like m6A methyltransferase-associated (VIRMA), whose N-terminus has the ability to recruit the METTL3/METTL14/WTAP complex (Yue et al., 2018). RBM15/15B (Figures 2, 3F,G) was three- to four-fold higher at the RRACH sequence site than at the non-methylation site. Knockdown of RBM15/15B decreases m6A levels in cellular mRNA (Knuckles et al., 2018). ZC3H13 (Figures 2, 3H) and HAKAI (Figures 2, 3I) are also components of the methyltransferase complex. ZC3H13 anchors the complex to the nucleus (Wen et al., 2018), and HAKAI regulates m6A levels in Arabidopsis (Růžička et al., 2017). Additionally, METTL16 (Figures 1, 3J; Doxtader et al., 2018) is also known as RNA m6A methyltransferase, an enzyme that maintains SAM homeostasis (Shima et al., 2017; Warda et al., 2017; Aoyama et al., 2020). METTL16 does not form complexes with other m6A methyltransferases, and has a distinct set of targets for m6A modification, including the 3′-untranslated region (UTR) of MAT2A mRNA and U6 snRNA.
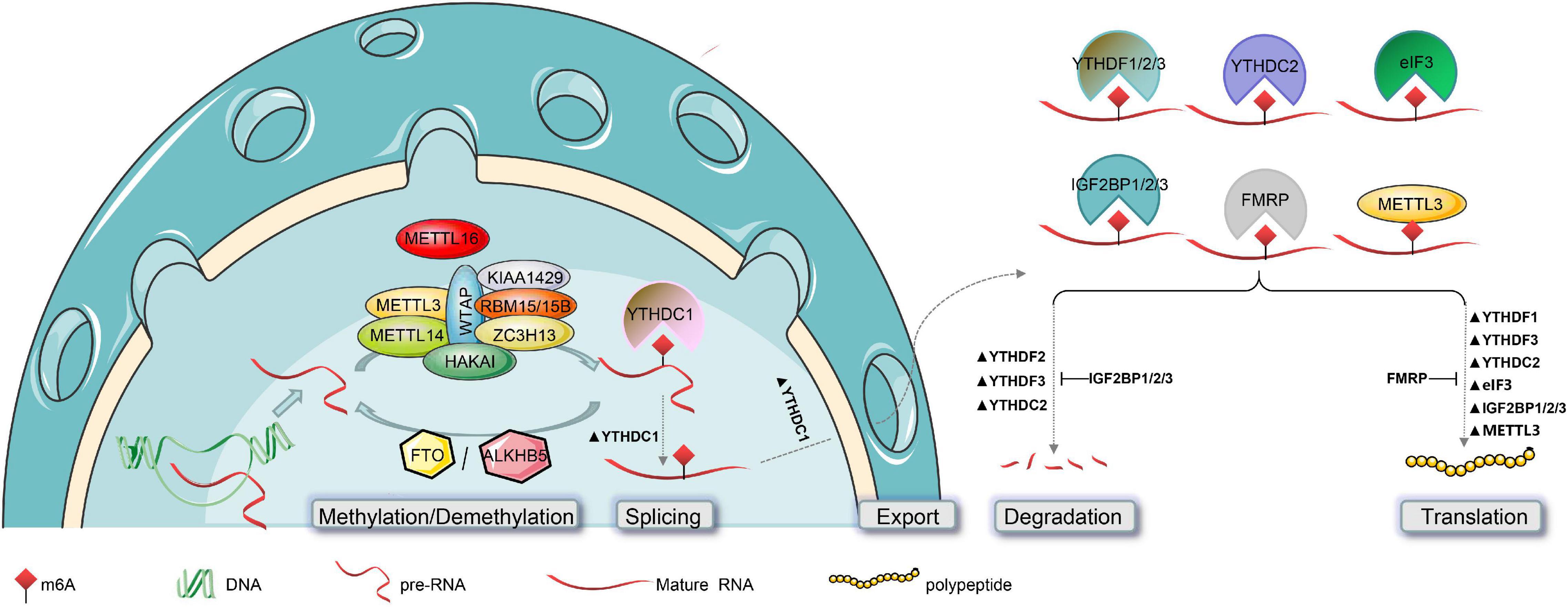
Figure 2. Post-transcriptional modification of RNA by m6A and its functions. The biogenesis of m6A in the mammalian nucleus is catalyzed by the methyltransferase complex (composed of METTL3, METTL14, WTAP, KIAA1429, RBM15/15B, ZC3H13, and HAKAI) or METTL16, and is reversed by m6A-demethylase (FTO or ALKBH5). The functional effects of m6A on RNAs can affect their nuclear splicing, nuclear export, stability, and translation efficiency. This is mediated by m6A “Readers,” such as YTHDC1 in the nucleus, and YTHDF1/2/3, YTHDC2, eIF3, IGF2BP1/2/3, and FMRP in the cytoplasm. Notably, in addition to catalyzing m6A methylation, METTL3 also directly promotes RNA translation.
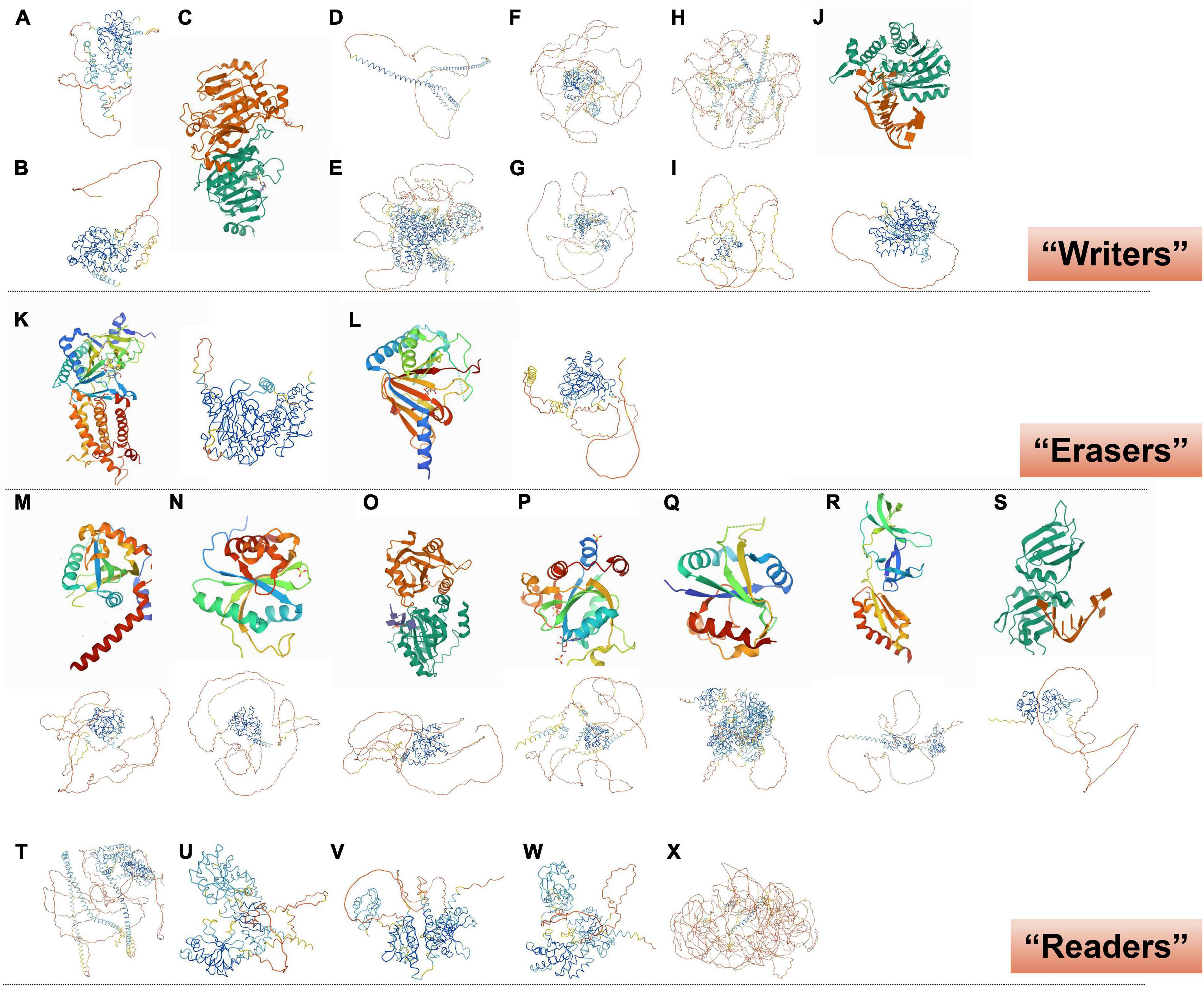
Figure 3. Crystal structure of RNA m6A methylation-modification enzymes. (A) METTL3 and (B) METTL14, predicted from “AlphaFold.” (C) SAM-bound METTL3-METTL14 complex (Wang et al., 2016). (D) WTAP, (E) KIAA1429, (F) RBM15, (G) RBM15B, (H) ZC3H13, and (I) HAKAI, predicted from “AlphaFold.” (J) Human METTL16 catalytic domain in complex with MAT2A 3’-UTR hairpin 6 (Upper; Doxtader et al., 2018); METTL16, predicted from “AlphaFold” (Lower). (K) FTO reveals the basis for its substrate specificity (Left; Han et al., 2010); FTO, predicted from “AlphaFold” (Right). (L) Human ALKBH5 in complex with citrate and acetate (Left; Feng et al., 2014); ALKBH5, predicted from “AlphaFold” (Right). (M) YTHDF1 YTH domain (Upper; Xu et al., 2015); YTHDF1, predicted from “AlphaFold” (Lower). (N) YTH-YTHDF2 in the free state (Upper; Li et al., 2014); YTHDF2, predicted from “AlphaFold” (Lower). (O) YTHDF3 YTH domain in complex with m6A RNA (Upper; Li et al., 2020); YTHDF3, predicted from “AlphaFold” (Lower). (P) YTHDC1 with m6A (Upper; Li Y. et al., 2021); YTHDC1, predicted from “AlphaFold” (Lower). (Q) Human YTHDC2 YTH domain (Upper; Ma et al., 2019); YTHDC2, predicted from “AlphaFold” (Lower). (R) N-terminal domain of FMRP (Upper; Myrick et al., 2015); FMRP, predicted from “AlphaFold” (Lower). (S) HNRNPA2B1 in complex with RNA (Upper; Wu et al., 2018); HNRNPA2B1, predicted from “AlphaFold” (Lower). (T) Eif3, (U) IGF2BP1, (V) IGF2BP2, (W) IGF2BP3, and (X) PRRC2A, predicted from “AlphaFold.”
N6-methyladenosine methyltransferases are mainly localized in the nucleus (Ping et al., 2014), which is consistent with the m6A-binding site found in nascent pre-mRNAs or pri-miRNAs (Ping et al., 2014; Ke et al., 2017; Slobodin et al., 2017). Interestingly, METTL3 catalyzes the methylation of mature mRNAs and has a non-methylating function in the cytoplasm (Lin et al., 2019; Liu et al., 2020). Lin et al. (2016) found that METTL3 was associated with ribosomes and promoted mRNA translation in the cytoplasm. Additionally, METTL16 functions independently of m6A methylation. In SAM-limiting conditions, METTL16 occupancy of a hairpin (hp1) in the MAT2A 3′-UTR induces splicing of the MAT2A-retained intron, which controls the production of SAM (Pendleton et al., 2017). Thus, while m6A methyltransferases generally affect RNA processing through m6A “Readers,” direct contributions of methyltransferases to RNA metabolism should not be overlooked (Pendleton et al., 2017; Doxtader et al., 2018).
N6-Methyladenosine Demethylase
The m6A demethylases, also known as “Erasers” in RNA m6A methylation modification, catalyze the demethylation of RNA substrates modified by m6A. In eukaryotes, fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5) were both found to be demethylases. These enzymes belong to the AlkB family of the dioxygenase superfamily and have a similar catalytic core, although they prefer different substrates and are expressed in different organs (Gerken et al., 2007; Zheng et al., 2013; Zou et al., 2016; Liu et al., 2020).
Fat mass and obesity (Figures 2, 3K; Han et al., 2010), also known as AlkB homolog 9 (ALKBH9), was the first discovered RNA m6A demethylase. FTO is mainly detected in the nucleus, similarly to the m6A methyltransferase (Jia et al., 2011). Its long stem-loop domain at the C-terminus enables substrate RNA demethylation (Jia et al., 2011; Bartosovic et al., 2017). However, Hess et al. (2013) only found 5,000 new m6A peaks in FTO-knockout mice, compared with 42,000 peaks in the control samples. These data indicated that FTO does not globally target all m6A-modified mRNAs, or which seems to imply that another m6A “Eraser,” ALKHB5, compensates for the lack of FTO. ALKBH5 (Figures 2, 3L) is similar to FTO and is also a Fe2+ and α-Ketoglutaric acid-dependent non-heme oxygenase (Zheng et al., 2013; Feng et al., 2014).
RNA N6-Methyladenosine Methylation-Binding Protein
The m6A binding proteins, also known as “Readers,” in RNA m6A methylation modification, specifically bind to the m6A methylation region, weakening the homologous binding to RNA reading proteins, and altering the secondary structure of RNA to alter protein–RNA interaction (Li et al., 2019). This function is widely implicated in the stability, translation, alternative splicing, and subcellular targeting of specific RNAs by recruiting or repelling some RNA-binding proteins (RBPs) or by altering the secondary structure of targeted RNAs (Adhikari et al., 2016; Wu et al., 2017; Widagdo and Anggono, 2018). Many “Readers,” including YTH domain-containing RNA-binding protein (YTP), fragile X mental retardation protein (FMRP), heterogeneous nuclear ribonucleoprotein (HNRNP), eukaryotic initiation factor 3 (eIF3), insulin-like growth factor 2 mRNA-binding protein (IGF2BP), and proline-rich coiled-coil 2A (PRRC2A), have already been identified.
YTH domain-containing RNA-binding protein includes YTH domain-containing family protein 1/2/3 (YTHDF1/2/3) (Figures 2, 3M–O; Li et al., 2014, 2020; Xu et al., 2015) and YTH domain-containing protein 1/2 (YTHDC1/2) (Figures 2, 3P,Q; Ma et al., 2019; Li Y. et al., 2021). Their YTH domains are capable of combining with the m6A RRACH sites to mediate RNA-specific binding, while their proline/glutamine/asparagine-enriched (P/Q/N-rich) domains regulate the subcellular localization of target RNA (Liao et al., 2018; Patil et al., 2018). YTHDF1/2/3 and YTHDC2 play specific roles in the cytoplasm, and YTHDC1 plays a role in the nucleus. It is generally considered that YTHDF1 enhances mRNA translation by promoting ribosome occupancy and interacting with initiation factors, YTHDF2 promotes mRNA degradation by localizing m6A-modified RNA to mRNA decay sites, and YTHDF3 enhances translation along with YTHDF1 and promotes degradation along with YTHDF2 in the cytoplasm (Wang et al., 2014; Li A. et al., 2017; Shi et al., 2017, 2018). However, Zaccara and Jaffrey (2020) showed that YTHDF1/2/3 co-regulated mRNA degradation rather than promoting mRNA translation in HeLa cells. Similar to YTHDF3, YTHDC2 in the cytoplasm accelerates degradation of the modified mRNA and enhances the translation of the corresponding protein by recognizing m6A (Hsu et al., 2017). Additionally, YTHDC1 regulates m6A-dependent mRNA splicing by recruiting splicing factors and mediates the nuclear export of m6A methylated mRNAs by interacting with the nuclear export adaptor protein SRSF3 (Xu et al., 2014; Xiao et al., 2016; Roundtree et al., 2017). Briefly, YTHDF1/2/3 and YTHDC2 promote the metabolism of m6A-modified mRNAs, but YTHDC1 regulates their splicing (Figure 2; Wang et al., 2014; Xiao et al., 2016; Hsu et al., 2017; Li A. et al., 2017; Shi et al., 2017, 2018; Zaccara and Jaffrey, 2020).
Fragile X mental retardation protein, an RBP (Figures 2, 3R; Myrick et al., 2015), negatively regulates the translation of mRNAs by interacting with m6A sites and then recruits RNA-induced silencing complexes and some miRNAs to arrest ribosomal elongation (Darnell et al., 2011; Suhl et al., 2014; Richter et al., 2015; Arguello et al., 2017; Chang et al., 2017).
Heterogeneous nuclear ribonucleoprotein is a group of RBPs that includes nearly 30 proteins, named A1 to U, which can interact with each other to form a complex. The most studied heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1) (Figure 3S; Wu et al., 2018) binds directly to a set of nuclear transcripts with m6A marks, and activates downstream variable shear events of partial genes (Alarcón et al., 2015; Geissler et al., 2016).
The eIF3 protein (Figures 2, 3T) facilitates the translation of mRNA by binding to the m6A sites of mRNA 5′-UTRs directly. In addition, IGF2BP, including IGF2BP1/2/3 (Figures 2, 3U–W), promotes mRNA stability and translation by recognizing the GG (m6A) C sequence (Huang et al., 2018). PRRC2A (Figure 3X) stabilizes mRNA expression by binding to the consensus GGACU motif in the coding sequence (CDS) region of mRNA in an m6A-dependent manner (Wu et al., 2019).
Adjustability of RNA N6-Methyladenosine Methylation
The dynamic nature of chemical modifications is an essential feature of functionality in the nervous system (Widagdo and Anggono, 2018). RNA m6A methylation, as the most abundant internal RNA modification, contributes markedly to this. Not surprisingly, RNA m6A methylation is precisely regulated.
Activity-Dependent Regulation of N6-Methyladenosine
Previous studies have demonstrated that cellular m6A levels are dynamically regulated in response to hypoxia, heat shock, and ultraviolet irradiation in cells (Dominissini et al., 2012; Meyer et al., 2015; Zhou et al., 2015; Zhang et al., 2016; Xiang et al., 2017; Lu et al., 2019). In the mammalian central nervous system, stimulus-dependent regulation of m6A has recently been shown to occur in response to behavioral training, cell microenvironment changes, and nerve injury (Widagdo et al., 2016; Engel et al., 2018; Shi et al., 2018; Weng et al., 2018; Zhang et al., 2018).
KCl is used to activate neurons by increasing membrane potentials in cells and opening voltage-gated calcium ion channels in cell membranes (Rosen et al., 1994; Lakk et al., 2017). m6A methylation of RNAs is upregulated following administration of KCl to primary neuronal cultures (Widagdo et al., 2016). Behavioral training is another way to stimulate neurons. Widagdo et al. (2016) discovered that the percentage of m6A-occupancies RNAs increased significantly in the medial prefrontal cortex after cued fear conditioning. A similar increase in the levels of m6A methylation was also observed in the dorsal hippocampus following contextual fear conditioning (Walters et al., 2017). Interestingly, activity-dependent m6A RNA modification has been found to occur in many immediate early genes and synaptic plasticity-related transcripts (Widagdo et al., 2016; Shi et al., 2018; Zhang et al., 2018). However, acute restraint-stress reduced global m6A levels in the mouse prefrontal cortex (Engel et al., 2018), which requires further investigation.
Tissue, Cellular/Subcellular, and Site-Specific Regulation of N6-Methyladenosine
More intriguingly, dynamic adjustability of m6A has been demonstrated in different brain regions, such as the prefrontal cortex, hippocampus, and amygdala (Widagdo et al., 2016; Walters et al., 2017; Engel et al., 2018). Shafik et al. (2021) revealed that this tissue specificity was most pronounced in the hypothalamus.
As different brain areas include different types of cells, it is reasonable to speculate that RNA m6A methylation may have cellular and subcellular specificity. Although the transcriptomic profiles of m6A in neuronal subpopulations have yet to be established, bioinformatics analysis showed RNA m6A methylation-enrichment in genes specific to neuronal subtypes (Chang et al., 2017), which implies a particular bias for m6A toward RNAs in neurons rather than in glial cells. Furthermore, the m6A-modified transcripts were widespread in a position distal from the neuronal cell body, indicating an interesting mode of m6A regulation outside of the nucleus. Indeed, some immunocytochemical assays and biochemical subcellular fractionation tests have revealed that some m6A “Writers” (METTL3, METTL14, METTL16), “Erasers” (FTO, ALKBH5), and “Readers” (YTHDF1/2/3) are present in the extra-somatic regions of neurons (Gershoni-Emek et al., 2016; Merkurjev et al., 2018; Yu et al., 2018; Nance et al., 2020). This localization of m6A “Writers” and “Erasers” in the extra-somatic regions may expedite the dynamic regulation efficiency of neurons (Widagdo and Anggono, 2018) in response to changes in the extracellular milieu, although most of the work may still be done in the soma. The localization of m6A in axons and its role in axonal growth has also been studied (Yu et al., 2018). An axonal elongation factor, Gap-43, was found to be an mRNA target of m6A. The local translation of Gap-43 was negatively modulated by m6A methylation and could be regulated by FTO in axons (Yu et al., 2018). In addition, the function of m6A has been observed in the synapses. In the pre-and postsynaptic compartments of neuron, most of the m6A target genes fell into the Gene Ontology functional terms “cell junction” and “synapse,” as well as surface receptor pathways, all of which maintain the functionality and integrity of synapses (Merkurjev et al., 2018).
Furthermore, the sites of m6A modification were found to be non-randomly distributed within genes (94.8%), where the proportions of CDS, UTRs, and introns were 50.9%, 41.9%, and 2.0%, respectively (Figure 4A; Meyer et al., 2012; Deng et al., 2018). Although the frequency of the RRACU sequence in last exons was the same as that in other exons (Figure 4B), m6A was found to be enriched within the last exons of a gene (Figures 4C,D; Ke et al., 2015).
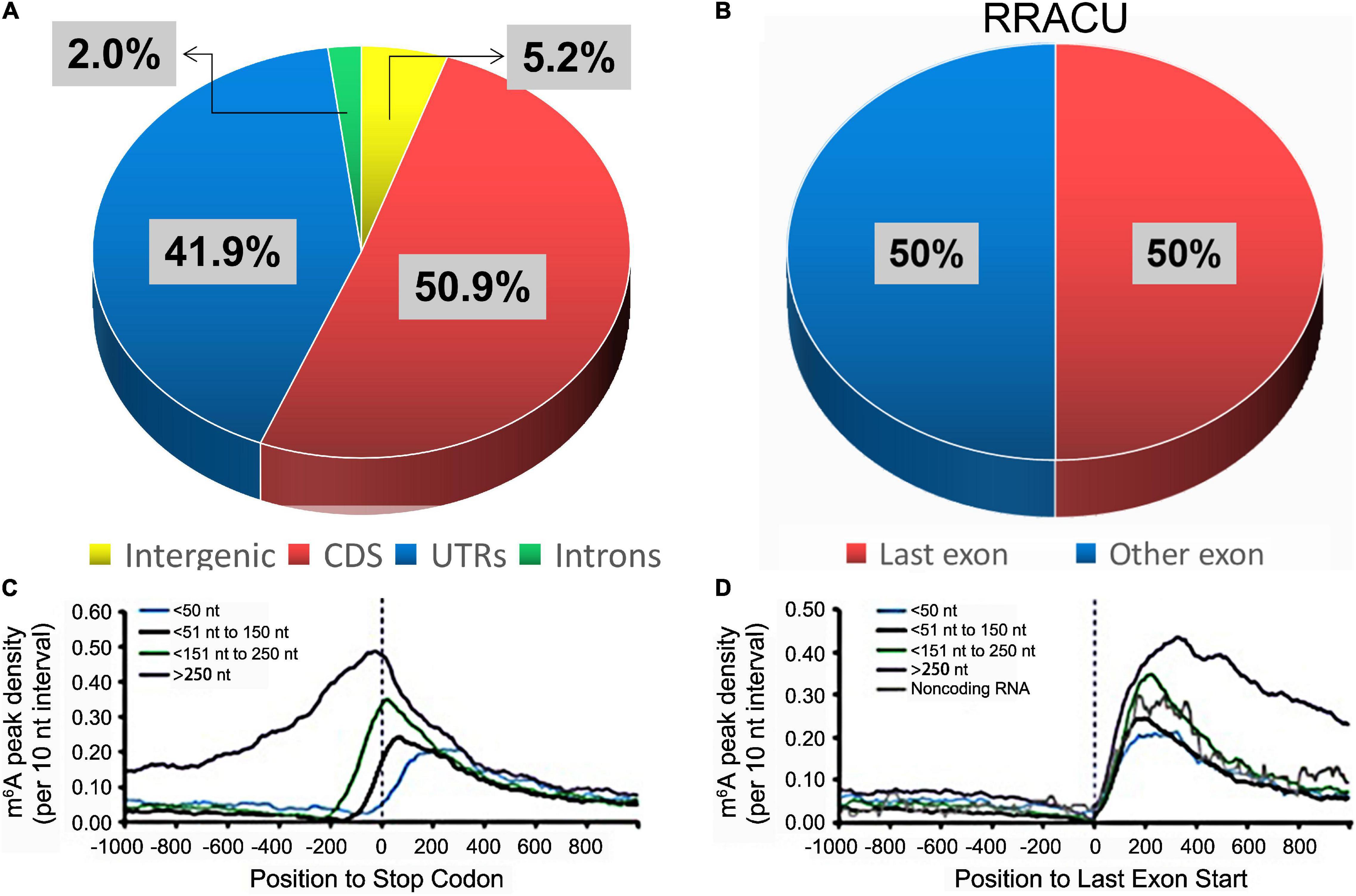
Figure 4. The distributions of m6A peaks. (A) The percentage of m6A peaks within distinct RNA sequence types. (B) The relative proportions of RRACU motifs in the last exon and other exons, with 100% representing all RRACU sequences in mRNA (Ke et al., 2015). (C,D) In the last exon, m6A density increased sharply (Ke et al., 2015). mRNAs were grouped according to their stop codon locations relative to the start of the last exon (D).
Developmental Stage-Specific Regulation of N6-Methyladenosine
Studies of m6A modifications have highlighted the need to maintain transcriptomic dynamics during neurodevelopment. The expression pattern of m6A indeed differs across various developmental phases (Meyer et al., 2012; Yoon et al., 2017). Shafik et al. (2021) also found that m6A exerted a critical function in both early and late brain development in a spatio-temporal fashion.
Prenatally, m6A modification is markedly increased throughout brain development (Meyer et al., 2012). Changing the m6A methylation modification by interfering with the expression levels of m6A “Writers” or “Erasers” leads to defects in brain development. The loss of m6A in embryonic neuronal progenitor cells by conditionally deleting METTL14 in embryonic mouse braiTablens resulted in delayed differentiation and prolonged cell cycle progression, extending cortical neurogenesis into the postnatal stages (Yoon et al., 2017). An abnormal increase of m6A by knocking out FTO caused cerebellar shrinkage and impaired spatial learning and memory (Li L. et al., 2017), which might be due to the increase in m6A promoting the decomposition of mRNAs encoding proteins with known functions in neuronal differentiation, stem cells, and the cell cycle (Yoon et al., 2017).
Postnatally, the function of m6A in neurogenesis was first uncovered in FTO-knockout mice (Li L. et al., 2017). Loss of FTO in FTO-knockout mice resulted in decreased proliferation and differentiation and reduced numbers of adult neural stem cells. In addition to the aforementioned central nervous system, loss of FTO in FTO-knockout mice also led to a shorter axonal length in mouse dorsal root ganglia neurons (Widagdo and Anggono, 2018). Taken together, these results suggest that balanced m6A modification plays a significant role in the establishment and development of the mammalian central and peripheral nervous systems.
MicroRNA Regulation of N6-Methyladenosine-Related Enzymes
MicroRNAs, which are approximately 22-nucleotide single-strand sequences, are a group of important post-transcriptional regulators in eukaryotes, which affect RNA m6A methylation by targeting m6A “Writers,” “Erasers,” and “Readers” in terms of both their functions and expression. It has been reported that manipulation of some miRNA sequences or their expression altered m6A methylation modification levels by regulating the binding of METTL3 to some mRNAs containing miRNA-binding sites (Chen et al., 2015). In addition, miR-33a, which targets the METTL3 3′-UTR, suppressed cell proliferation (Liu et al., 2020). miRNA-421-3p targeting the “Reader” YTHDF1 inhibited p65 mRNA translation to prevent inflammatory responses in cerebral ischemia/reperfusion injury (Zheng et al., 2020). miR-145 restrained the expression of YTHDF2 by targeting YTHDF2 mRNA, thus inhibiting cell proliferation. miR-744-5p targeting the “Reader” hnRNPC promoted ovarian cancer cell death (Chen et al., 2020).
RNA N6-Methyladenosine Methylation in Alzheimer’s Disease
Miao et al. (2020) observed that the frequency percentage of m6A in genes was positively correlated with the length and number of exons (Figures 5A,B) but negatively correlated with GC content and gene distance to the adjacent gene (Figures 5C,D), which implies that RNA m6A methylation is not random and disordered. Several recent reports have started to uncover the functional significance of m6A regulation in de novo RNA transcripts, including nuclear splicing, stability, translation, and subcellular localization, suggesting that m6A serves as a regulator to fine-tune many diseases precisely over time (Li et al., 2019). Indeed, m6A has been identified as a conserved epitranscriptomic modification in many neurodegenerative diseases, such as AD (Hess et al., 2013; Engel and Chen, 2018; Shafik et al., 2021).
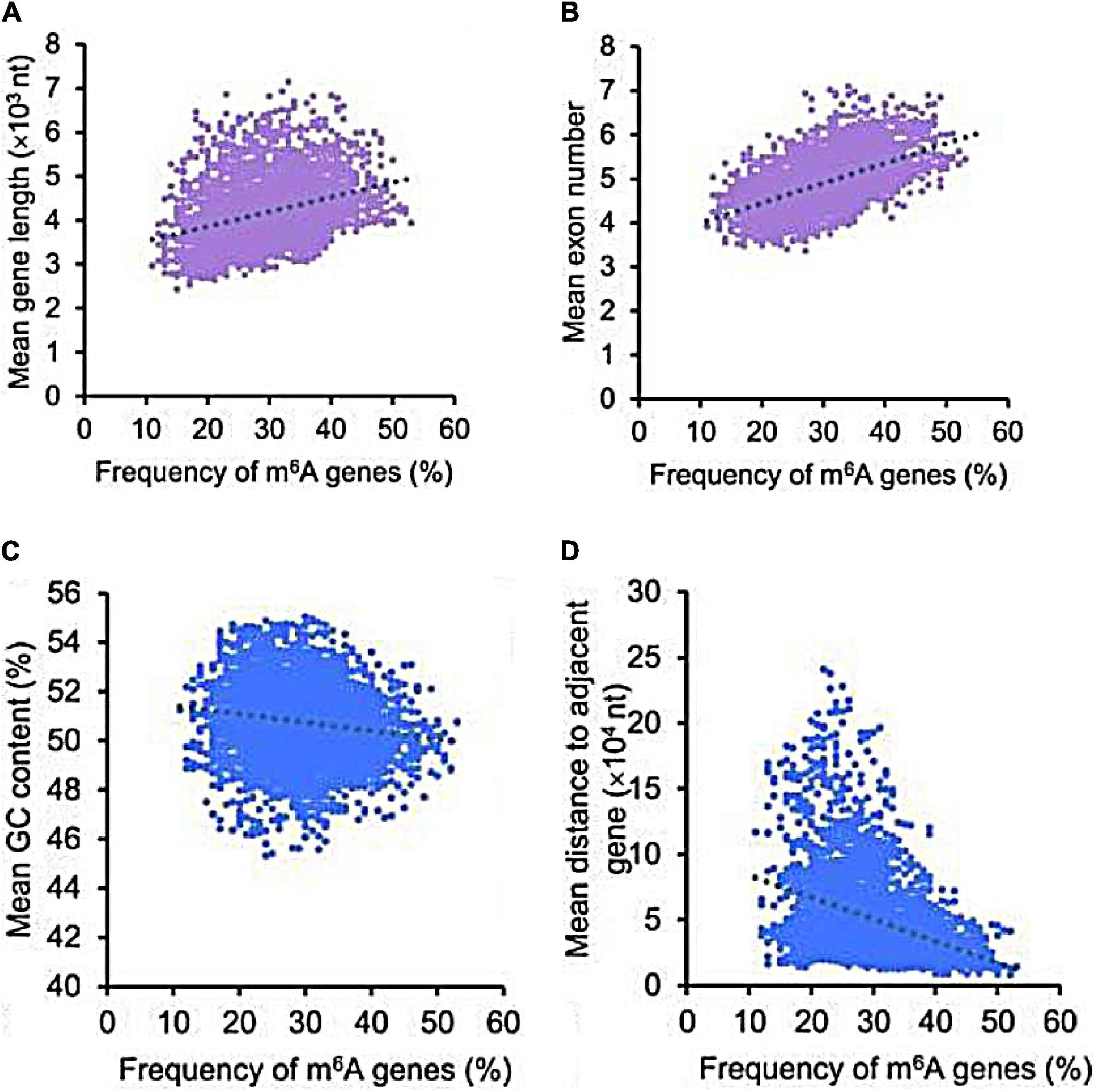
Figure 5. The correlation of m6A genes with multiple gene features. The frequency percentage of m6A genes was positively correlated with the length (A) and number of exons (B), but was negatively correlated with GC content (C) and gene distance to the adjacent gene (D) (Miao et al., 2020).
Just as m6A methylation, AD has tissue, cellular/subcellular, and site-specificity, associated with environment and neurodevelopment and regulated by miRNAs. The progressive degeneration of hippocampal neurons is the main feature of AD. Mutations in the APP, PS1, and PS2 genes are considered the main causes of familial AD (Popugaeva et al., 2018). Environmental stimulation, such as infections, trauma, and radiofrequency radiation (Dasdag et al., 2020), is thought to induce sporadic AD. Neurodevelopment-related signaling pathways, such as Notch/Wnt/Reelin intracellular signaling pathways, may represent a novel approach to the regulation of neurodegenerative processes in AD (Grilli et al., 2003). In addition, the above-mentioned miRNAs that regulate m6A are also considered to be involved in the pathogenesis of AD. Jaouen and Gascon (2016) described miR-33 function modulating ATP-binding cassette transporter A1 (ABCA1) and interfering with Aβ plaque formation through cholesterol metabolism regulation. Peña-Bautista et al. (2021) found hsa-miR-421 showed a positive correlation with some detected lipids (FA (16:0), FA (20:2), FA (18:2), FA (20:4), FA (20:3), FA (18:0), FA (14:0)) in AD plasma samples. Docosahexaenoic acids (DHA) are known to be beneficial in AD. miR-33a and miR-145 are regulated by DHA, and this regulation becomes disrupted in AD (Chiang, 2021).
RNA m6A methylation has been considered to be an important epigenetic marker associated with AD disturbances, including mitochondrial dysfunction, neuroinflammatory response, oxidative stress, neurotoxic substance deposition, and memory deficits. Here, we will introduce them to AD one by one in the following.
N6-Methyladenosine and Mitochondrial Dysfunction of Alzheimer’s Disease
The central nervous system requires approximately 20% of the body’s total basal oxygen consumption to support neuronal energy expenditure. Mitochondria are organelles that are responsible for energy production. Therefore, neurons are damaged by mitochondrial deficiency. Mitochondrial dysfunction was observed in the brains of AD patients, even before the appearance of neurofibrillary tangles and senile plaques (Lim et al., 2020).
Some m6A-related enzymes have been shown to affect mitochondrial function. The physiological role of METTL3 in mitochondria is under debate. Shi et al. (2021) considered that METTL3 preserved mitochondrial function in Down syndrome by reducing the expression of nuclear receptor-interacting protein 1 (NRIP1), a crucial gene in the regulation of the mitochondrial pathway. But Zhang et al. (2021) demonstrated that METTL3 and YTHDF2 cooperatively promoted mitochondrial dysfunction and inflammatory response during oxLDL-induced inflammation in monocytes. In addition, the demethylase FTO inhibitor MO-I-500 was found to ameliorate astrocyte mitochondrial dysfunction in streptozotocin-induced AD cell models (Cockova et al., 2021).
N6-Methyladenosine and Neuroinflammatory Response of Alzheimer’s Disease
An excessive neuroinflammatory response is harmful for the brain. Growing evidence shows that neuroinflammation is directly implicated in AD processes (Amor et al., 2014). Microglia are the main effectors in the neuroinflammatory process. Once overactivated, microglia may release proinflammatory cytokines and accelerate neurodegeneration.
Li Q. et al. (2021) identified a distinct m6A epitranscriptome in microglia. They found that m6A served as a novel and essential regulator of the anti-inflammatory and proinflammatory responses of microglia. An in silico analysis of immunoprecipitated methylated RNAs with microarrays also demonstrated that m6A methylation was increased in major inflammatory pathways (Chokkalla et al., 2019), indicating that RNA m6A methylation is closely related to neuroinflammation. Indeed, METTL3 was found to promote lipopolysaccharide (LPS)-induced neuroinflammation through the TRAF6/NF-κB pathway (Wen et al., 2020). LPS-induced neuroinflammation was considered might impair the efficient readout of neuronal genetic information and might contribute to a progressive disruption in the readout of genetic information in the AD brain (Zhao et al., 2017). The METTL3 knockdown was found to inhibit the inflammatory response by regulating the variable splicing of MyD88 (Feng et al., 2018).
N6-Methyladenosine and Oxidative Stress of Alzheimer’s Disease
Oxidative stress plays a crucial role in AD pathogenesis. The brain is more vulnerable to oxidative stress than other organs, and most of the components of neurons (proteins, lipids, and nucleic acids) can be oxidized in AD (Bonda et al., 2010).
It has been reported that m6A modification is affected by oxidative stress. The arsenic exposure hypothesis for AD provides a parsimonious testable hypothesis for the development and progression of this devastating disease to some degree (Gong and O’Bryant, 2010). Arsenite-induced oxidative stress possibly increases the levels of RNA m6A methylation by regulating m6A “Writers” or “Erasers,” particularly promoting METTL14 and WTAP expression (Zhao et al., 2019). Additionally, m6A modification is important in the regulation of oxidative stress. The m6A-binding protein YTHDF1/3 was found to promote stress granule formation (Fu and Zhuang, 2020). Although associations per se cannot prove cause-effect relationships, the development of pathological stress granules has been implicated in the onset and progression of AD (Ash et al., 2014). The m6A methyltransferase METTL3 was reported to attenuate oxidative stress and cell apoptosis in colistin-induced kidney injury by activating the antioxidant Keap1/Nrf2 pathway (Wang et al., 2019). The reduced neuronal m6A modification in the hippocampus caused by METTL3 knockdown led to extensive synaptic loss and neuronal death along with multiple AD-related cellular alterations, including oxidative stress and aberrant cell cycle events in vivo (Zhao et al., 2021).
N6-Methyladenosine and Pathologic Hallmark of Alzheimer’s Disease
Accumulation of insoluble neurotoxic aggregates, including extracellular amyloid (A)β plaques and intracellular tau neurofibrillary tangles, represents a major pathological hallmark of AD. Their accumulation leads to neuronal degeneration, synaptic dysfunction, and ultimately, dementia (Lane et al., 2018).
N6-Methyladenosine and Aβ Plaques
Folic acid, a water-soluble B vitamin, reduces the production of Aβ and slows the progression of AD by involving in generating S-adenosylmethionine (SAM), potentially enhancing the levels of RNA m6A methylation (Li et al., 2015; Li N. et al., 2021), implying that RNA m6A methylation may be involved in Aβ metabolism. Interestingly, Aβ treatment was found to have significantly reduced METTL3 and postsynaptic density-95 (PSD-95) expression in rat primary cortical neurons. On the contrary, METTL3 overexpression was found to rescue Aβ-induced synaptic PSD-95 loss in vitro. Importantly, METTL3 overexpression rescued synaptic damage and cognitive impairment in Aβ-induced AD mice. In addition, the demethylase FTO was reported to alleviate Aβ-induced cell degeneration via the PKA/CREB signaling pathway (Hu et al., 2020). In addition to targeting Aβ, m6A methylation also altered the expression levels of Aβ production-related proteins, such as Aβ precursor protein (APP) and the β-site APP-cleaving enzyme (BACE1) (Kolisnyk et al., 2017; Edens et al., 2019). The m6A “Reader” FMRP was found to regulate the local protein synthesis of neuronal synapses and change the nuclear output of m6A-dependent mRNA by regulating APP mRNA translation (Chang et al., 2017; Edens et al., 2019). The downregulation of the “Reader” HNRNPA2B1 was shown to promote abnormal splicing of BACE1 (Kolisnyk et al., 2017).
N6-Methyladenosine and Tau Neurofibrillary Tangles
In human postmortem AD samples, Huang et al. (2020) observed METTL3 accumulation in the insoluble fractions, which correlated positively with levels of insoluble tau neurofibrillary tangles. This was accompanied by an increased level and redistribution of METTL3 expression in the AD hippocampus, likely representing aberrant misfolding and/or aggregation of METTL3, perhaps similar to the frequent aggregation of misfolded proteins in AD (Huang et al., 2020). In the brain of an AD mouse model, upregulated FTO was also observed to activate the phosphorylation of tau and accelerate the pathological hallmarks of AD in an mTOR-dependent manner (Li et al., 2018). In the Drosophila AD model, which specifically expresses the human tau gene with the R406W mutation in the eye, found that the loss of m6A by loss of METTL3, METTL14 or YTHDF enhanced tau toxicity and had more severe locomotive defects (Shafik et al., 2021).
N6-Methyladenosine and Memory Disorder in Alzheimer’s Disease
Learning and memory impairments are the most important clinical symptoms of AD. Synaptic plasticity, that is, the adjustability of synaptic morphology and strength, is considered to be the basis of learning and memory. It is worth noting that short- and long-term memory requires multiple layers of regulation, from protein modifications at the synapse to RNA synthesis de novo in the nucleus (Cai et al., 2016; Holtmaat and Caroni, 2016; Karunakaran et al., 2016). The stability and translation of RNA transcripts have been shown to depend on m6A modification (Yue et al., 2015), which is strongly biased to neuronal genes and functions (Meyer et al., 2012; Schwartz et al., 2014). Thus, it is perhaps unsurprising that RNA m6A methylation modifications have been implicated in neural plasticity, thereby affecting learning and memory.
N6-Methyladenosine Methyltransferase and Memory Disorders
Increased m6A levels in adult neurons have been found to promote the transcriptome response to synaptic plasticity (Engel et al., 2018; Leighton et al., 2018). In contrast, reducing the m6A peaks in cellular mRNAs by knocking out some m6A methyltransferases, such as METTL3, METTL14, METTL16, WTAP, RBM15/15B, and HAKAI may result in memory disorders (Ping et al., 2014; Schwartz et al., 2014; Růžička et al., 2017; Knuckles et al., 2018; Zhang et al., 2018; Shafik et al., 2021). The most studied m6A “Writer,” METTL3, has been shown to have a direct effect on the regulation of hippocampal-dependent memory formation. The overexpression of METTL3 in the dorsal hippocampus of wildtype mice was found to enhance long-term memory consolidation significantly (Zhang et al., 2018; Table 1), whereas the knockout of METTL3 in the forebrain was found to inhibit memory consolidation, which could be restored by adequate training (Zhang et al., 2018; Zhao et al., 2021). METTL14-mediated RNA m6A modification is also critical for epitranscriptomic regulation of learning. The deletion of METTL14 was observed to reduce striatal m6A levels, increase neuronal excitability, and severely impair striatal-mediated learning-related behavior (Table 1; Koranda et al., 2018).
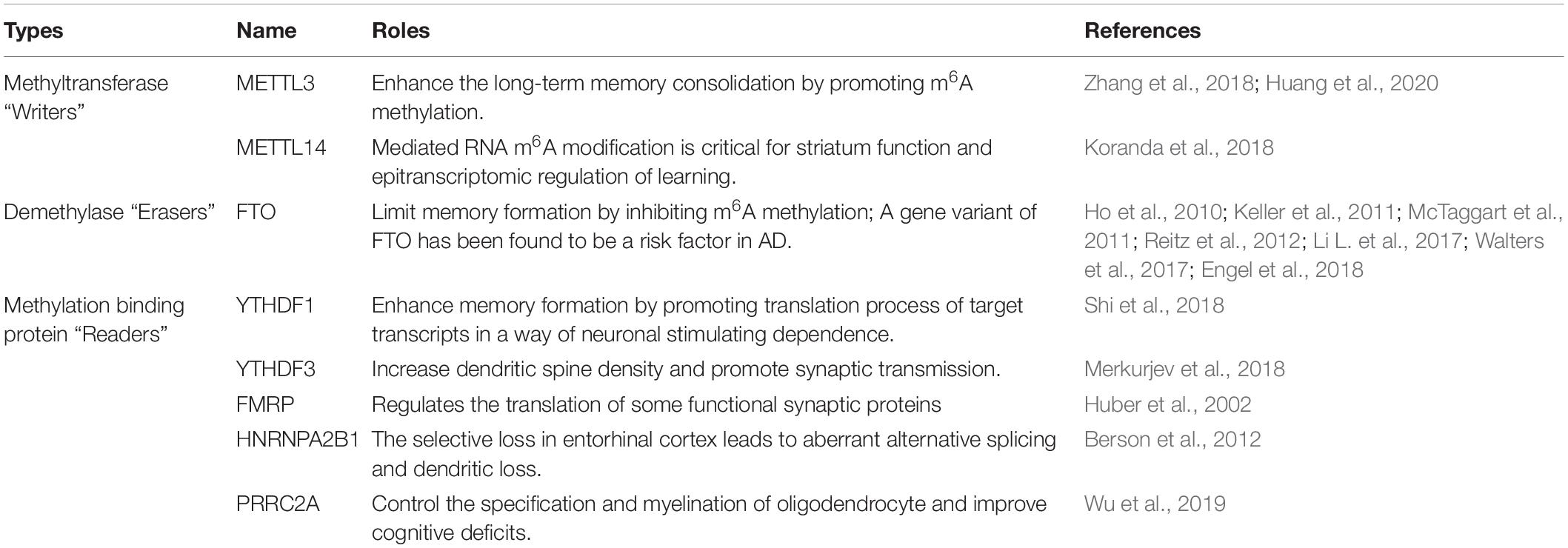
Table 1. The function of RNA N6-methyladenosine methylation modification enzyme in learning and memory.
N6-Methyladenosine Demethylase and Memory Disorders
Precise RNA m6A modification is necessary for memory formation, thereby implicating m6A demethylase concurs with memory regulation. Despite being ubiquitously expressed, FTO, the best characterized “Eraser,” is enriched in the nuclei and dendrites, and near dendritic spines of mouse dorsal hippocampal CA1 neurons (McTaggart et al., 2011). The expression of FTO protein decreased shortly after a situational fear reflex, which implies that FTO typically limits memory formation (Table 1; Walters et al., 2017). Indeed, knocking out FTO in the prefrontal cortex of mice was found to enhance fear memory consolidation, with m6A modification on several fear-related genes significantly increased (Li L. et al., 2017; Engel et al., 2018). However, a gene variant of FTO was found to be a possible risk factor for AD (Table 1; Ho et al., 2010; Keller et al., 2011; Reitz et al., 2012). A prospective cohort study by Keller et al. (2011) suggested that the FTO AA-genotype had a higher risk for AD compared to TT-carriers. Reitz et al. (2012) used 1,877 Caucasian cases and controls from the NIA-LOAD study and 1,093 Caribbean Hispanics to further explore the association of FTO with AD. They found that genetic variation in Introns 1 and 2 of the FTO gene may contribute to AD risk (Reitz et al., 2012). The aforementioned studies suggested that maintaining a low and basic expression level of FTO in AD might be necessary for precise RNA m6A modification.
RNA N6-Methyladenosine Methylation-Binding Protein and Memory Disorders
N6-methyladenosine methylation-binding proteins also make marked contributions to memory storage. Interrupting m6A-mediated function via knockdown of m6A “Readers” in hippocampal neurons resulted in synaptic dysfunction, including immature spine morphology, and destroyed excitatory synaptic transmission, accompanied by decreased clusters of PSD-95 and reduced surface expression of the AMPA receptor subunit GluA1 (Merkurjev et al., 2018).
YTHDF1 is mainly expressed in the hippocampus and promotes the translation of target transcripts through neuronal stimulation. YTHDF1-knockout mice showed impaired learning and memory, reduced synaptic transmission, and decreased long-term potentiation (Table 1; Shi et al., 2018). Interestingly, these phenotypes were similar to those obtained with METTL3 depletion (Shi et al., 2018), suggesting “Readers” and “Writers” can, to some extent, phenocopy each other in the brain. In YTHDF3-knockdown neurons, excessive dendritic filopodia in place of mature spines were observed, and a decreased percentage of spines containing a PSD-95 cluster and surface GluA1 expression was observed (Table 1; Merkurjev et al., 2018). In addition, FMRP was found to regulate the translation of some functional synaptic proteins (Table 1; Huber et al., 2002). The absence of FMRP in Fragile-X Syndrome causes excessive and persistent protein synthesis in dendrites, leading to an excess number of dendritic spines and synaptic dysfunction (Bassell and Warren, 2008; Richter et al., 2015). HNRNP was found to be relatively highly expressed in brains with a high metabolism. The selective loss of HNRNPA2B1 in the entorhinal cortex led to aberrant alternative splicing and dendritic loss (Table 1; Berson et al., 2012). PRRC2A has been shown to control the specifications and myelination of oligodendrocytes. PRRC2A-knockout mice showed cognitive deficits (Table 1; Wu et al., 2019).
Conclusion and Outlook
RNA m6A methylation, which is abundant in the mammalian brain, is a significant epitranscriptomic modification. m6A has a wide range of effects on AD and can be precisely regulated. The emergence of cross-talk between m6A “Writers,” “Erasers,” and “Readers” makes it more complicated. The most appropriate example is METTL3. METTL3 is a key component of the m6A methyltransferase complex, with its eminent methyltransferase activity, but has also been reported to function as an m6A “Reader.” Studies have reported that METTL3 directly promotes the translation of several m6A-modified mRNAs, such as the Hippo pathway effector TAZ and the epidermal growth factor receptor, by interacting with translation initiation machinery, independent of its methyltransferase and downstream m6A “Reader” activity (Lin et al., 2016). Hence, METTL3 might be both an m6A “Writer” that methylates mRNA along with other members of the methyltransferase complex, by identifying unmethylated mRNA, and an m6A “Reader” that enhances mRNA translation alone, by identifying methylated mRNA. The second pertinent example is METTL16, which interacts with MAT2A hairpins to regulate MAT2A through two mechanisms: reducing mRNA stability in SAM-sufficient conditions and promoting pre-mRNA splicing in SAM-limiting conditions. The former relies on METTL16 recognition of methylated MAT2A pre-mRNA. As the sites of m6A on MAT2A pre-mRNA are occupied, METTL16 is quickly separated from MAT2A pre-mRNA, increases retention of the last intron in MAT2A pre-mRNA, and reduces its stability. However, in SAM-limiting conditions, METTL16 binds to unmethylated MAT2A pre-mRNA to promote the splicing of MAT2A pre-mRNA by recruiting the cleavage factor Im complex (CFIm), and finally increases the expression of MAT2A mature mRNA (Scarborough et al., 2021). In brief, the effects of METTL3 and METTL16 likely reflect the typical “Writer—Reader” paradigm. More importantly, their effects are precisely regulated. The activity of METTL3 is controlled by post-translational modifications, such as SUMOylation (Liu et al., 2020), and METTL16 is regulated by intracellular SAM levels, which makes the regulatory network extremely intricate.
In addition, differences in experimental conditions and animal models increase discrepancies in research results. Some studies have shown a reduction in m6A modification in AD (Shafik et al., 2021) and Parkinson’s disease models (Chen et al., 2019). However, Han et al. (2020) found that m6A methylation was elevated in the cortex and hippocampus of an AD model. The variation tendency of METTL3 and FTO in AD brain were also contradictory between the studies by Han et al. (2020), Shafik et al. (2021). Shafik et al. (2021) found METTL3 was downregulated and FTO was upregulated, which was in contrast to the study by Han et al. (2020) These discrepancies might be due to differences in the animal models employed. 9-month-old APP/PS1 mice and 6-month-old mice were used by Han et al. (2020), Shafik et al. (2021), respectively. Given that Shafik et al. also observed significantly more m6A sites as age increases during the aging process in both mouse and human brain areas, we speculate the increase in m6A methylation in that the study of Han et al. (2020) may be more likely to be compensatory and early changes in AD. But, of course, more comprehensive experiments are required to elucidate the changes of RNA m6A methylation modification in the various stages of AD.
Author Contributions
RZ contributed to the design of the review and drafted the manuscript. YZ and FG contributed to revision of the manuscript. SL and HC contributed to the design of the review and critical revision of the manuscript and had primary responsibility for the final content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82171582 and 91849134) and the Science and Technology Research Project of Higher Education Institutions in Hebei Province (ZD2020105).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adhikari, S., Xiao, W., Zhao, Y. L., and Yang, Y. G. (2016). m(6)A: signaling for mRNA splicing. RNA Biol. 13, 756–759. doi: 10.1080/15476286.2016.1201628
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 162, 1299–1308. doi: 10.1016/j.cell.2015.08.011
Alzheimer, A. (1907). Uber eine eigenartige Erkrankung der Hirnride. Allg. Z. Psychiatr. 64, 146–148.
Amor, S., Peferoen, L. A., Vogel, D. Y., Breur, M., van der Valk, P., Baker, D., et al. (2014). Inflammation in neurodegenerative diseases–an update. Immunology 142, 151–166. doi: 10.1111/imm.12233
Aoyama, T., Yamashita, S., and Tomita, K. (2020). Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 48, 5157–5168. doi: 10.1093/nar/gkaa227
Arguello, A., DeLiberto, A., and Kleiner, R. (2017). RNA Chemical Proteomics Reveals the N-Methyladenosine (mA)-Regulated Protein-RNA Interactome. J. Am. Chem. Soc. 139, 17249–17252. doi: 10.1021/jacs.7b09213
Ash, P. E., Vanderweyde, T. E., Youmans, K. L., Apicco, D. J., and Wolozin, B. (2014). Pathological stress granules in Alzheimer’s disease. Brain Res. 1584, 52–58. doi: 10.1016/j.brainres.2014.05.052
Bartosovic, M., Molares, H., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res. 45, 11356–11370. doi: 10.1093/nar/gkx778
Bassell, G. J., and Warren, S. T. (2008). Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60, 201–214. doi: 10.1016/j.neuron.2008.10.004
Berson, A., Barbash, S., Shaltiel, G., Goll, Y., Hanin, G., Greenberg, D. S., et al. (2012). Cholinergic-associated loss of hnRNP-A/B in Alzheimer’s disease impairs cortical splicing and cognitive function in mice. EMBO Mol. Med. 4, 730–742. doi: 10.1002/emmm.201100995
Bonda, D. J., Wang, X., Perry, G., Nunomura, A., Tabaton, M., Zhu, X., et al. (2010). Oxidative stress in Alzheimer disease: a possibility for prevention. Neuropharmacology 59, 290–294. doi: 10.1016/j.neuropharm.2010.04.005
Cai, D. J., Aharoni, D., Shuman, T., Shobe, J., Biane, J., Song, W., et al. (2016). A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534, 115–118. doi: 10.1038/nature17955
Chang, M., Lv, H., Zhang, W., Ma, C., He, X., Zhao, S., et al. (2017). Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 7:170166. doi: 10.1098/rsob.170166
Chen, K., Zhao, B. S., and He, C. (2016). Nucleic Acid Modifications in Regulation of Gene Expression. Cell Chem. Biol. 23, 74–85. doi: 10.1016/j.chembiol.2015.11.007
Chen, T., Hao, Y. J., Zhang, Y., Li, M. M., Wang, M., Han, W., et al. (2015). m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16, 289–301. doi: 10.1016/j.stem.2015.01.016
Chen, X., Yu, C., Guo, M., Zheng, X., Ali, S., Huang, H., et al. (2019). Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem. Neurosci. 10, 2355–2363. doi: 10.1021/acschemneuro.8b00657
Chen, Y., Lin, Y., Shu, Y., He, J., and Gao, W. (2020). Interaction between N-methyladenosine (mA) modification and noncoding RNAs in cancer. Mol. Cancer 19:94. doi: 10.1186/s12943-020-01207-4
Chiang, V. S. C. (2021). MicroRNAs as Potential Regulators of Docosahexaenoic Acid Benefits in Alzheimer’s Disease. Arch. Neurol. Neurol. Disord. 4:125. doi: 10.1179/1476830515Y.0000000014
Chokkalla, A. K., Mehta, S. L., Kim, T., Chelluboina, B., Kim, J., and Vemuganti, R. (2019). Transient Focal Ischemia Significantly Alters the m(6)A Epitranscriptomic Tagging of RNAs in the Brain. Stroke 50, 2912–2921. doi: 10.1161/strokeaha.119.026433
Cockova, Z., Honc, O., Telensky, P., Olsen, M. J., and Novotny, J. (2021). Streptozotocin-Induced Astrocyte Mitochondrial Dysfunction Is Ameliorated by FTO Inhibitor MO-I-500. ACS Chem. Neurosci. 12, 3818–3828. doi: 10.1021/acschemneuro.1c00063
Darnell, J., Van Driesche, S., Zhang, C., Hung, K., Mele, A., Fraser, C., et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. doi: 10.1016/j.cell.2011.06.013
Dasdag, O., Adalier, N., and Dasdag, S. (2020). Electromagnetic radiation and alzheimer’s disease. Biotechnol. Biotechnol. Equip. 34, 1087–1094.
Deng, X., Su, R., Weng, H., Huang, H., Li, Z., and Chen, J. (2018). RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 28, 507–517. doi: 10.1038/s41422-018-0034-6
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71, 3971–3975. doi: 10.1073/pnas.71.10.3971
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Doxtader, K. A., Wang, P., Scarborough, A. M., Seo, D., Conrad, N. K., and Nam, Y. (2018). Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol. Cell 71, 1001–1011.e4. doi: 10.1016/j.molcel.2018.07.025
Du, Y., Hou, G., Zhang, H., Dou, J., He, J., Guo, Y., et al. (2018). SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 46, 5195–5208. doi: 10.1093/nar/gky156
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP Modulates Neural Differentiation through m(6)A-Dependent mRNA Nuclear Export. Cell Rep. 28, 845–854.e5. doi: 10.1016/j.celrep.2019.06.072
Engel, M., and Chen, A. (2018). The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav. 17:e12428. doi: 10.1111/gbb.12428
Engel, M., Eggert, C., Kaplick, P. M., Eder, M., Röh, S., Tietze, L., et al. (2018). The Role of m(6)A/m-RNA Methylation in Stress Response Regulation. Neuron 99, 389–403.e9. doi: 10.1016/j.neuron.2018.07.009
Feng, C., Liu, Y., Wang, G., Deng, Z., Zhang, Q., Wu, W., et al. (2014). Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J. Biol. Chem. 289, 11571–11583. doi: 10.1074/jbc.M113.546168
Feng, Z., Li, Q., Meng, R., Yi, B., and Xu, Q. (2018). METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell. Mol. Med. 22, 2558–2568. doi: 10.1111/jcmm.13491
Fu, Y., and Zhuang, X. (2020). mA-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 16, 955–963. doi: 10.1038/s41589-020-0524-y
Geissler, R., Simkin, A., Floss, D., Patel, R., Fogarty, E., Scheller, J., et al. (2016). A widespread sequence-specific mRNA decay pathway mediated by hnRNPs A1 and A2/B1. Genes Dev. 30, 1070–1085. doi: 10.1101/gad.277392.116
Gerken, T., Girard, C., Tung, Y., Webby, C., Saudek, V., Hewitson, K., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. doi: 10.1126/science.1151710
Gershoni-Emek, N., Mazza, A., Chein, M., Gradus-Pery, T., Xiang, X., Li, K. W., et al. (2016). Proteomic Analysis of Dynein-Interacting Proteins in Amyotrophic Lateral Sclerosis Synaptosomes Reveals Alterations in the RNA-Binding Protein Staufen1. Mol. Cell. Proteom. 15, 506–522. doi: 10.1074/mcp.M115.049965
Gong, G., and O’Bryant, S. E. (2010). The arsenic exposure hypothesis for Alzheimer disease. Alzheimer Dis. Assoc. Disord. 24, 311–316. doi: 10.1097/WAD.0b013e3181d71bc7
Grilli, M., Toninelli, G. F., Uberti, D., Spano, P., and Memo, M. (2003). Alzheimer’s disease linking neurodegeneration with neurodevelopment. Funct. Neurol. 18, 145–148.
Guo, T., Zhang, D., Zeng, Y., Huang, T. Y., Xu, H., and Zhao, Y. (2020). Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol. Neurodegener. 15:40. doi: 10.1186/s13024-020-00391-7
Han, M., Liu, Z., Xu, Y., Liu, X., Wang, D., Li, F., et al. (2020). Abnormality of m6A mRNA Methylation Is Involved in Alzheimer’s Disease. Front. Neurosci. 14:98. doi: 10.3389/fnins.2020.00098
Han, Z., Niu, T., Chang, J., Lei, X., Zhao, M., Wang, Q., et al. (2010). Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 464, 1205–1209. doi: 10.1038/nature08921
Hess, M., Hess, S., Meyer, K., Verhagen, L., Koch, L., Brönneke, H., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16, 1042–1048. doi: 10.1038/nn.3449
Ho, A. J., Stein, J. L., Hua, X., Lee, S., Hibar, D. P., Leow, A. D., et al. (2010). A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc. Natl. Acad. Sci. U. S. A. 107, 8404–8409. doi: 10.1073/pnas.0910878107
Holtmaat, A., and Caroni, P. (2016). Functional and structural underpinnings of neuronal assembly formation in learning. Nat. Neurosci. 19, 1553–1562. doi: 10.1038/nn.4418
Hsu, P., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi: 10.1038/cr.2017.99
Hu, Y., Chen, J., Sun, J., Wang, Y., Huang, P., Feng, J., et al. (2020). FTO alleviates Aβ1-40 induced retinal pigment epithelium degeneration via PKA/CREB signaling pathway. Res. Square [Preprint] doi: 10.21203/rs.3.rs-30549/v1
Huang, H., Camats-Perna, J., Medeiros, R., Anggono, V., and Widagdo, J. (2020). Altered Expression of the m6A Methyltransferase METTL3 in Alzheimer’s Disease. eNeuro 7:ENEURO.0125-20.2020. doi: 10.1523/eneuro.0125-20.2020
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Huber, K. M., Gallagher, S. M., Warren, S. T., and Bear, M. F. (2002). Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl. Acad. Sci. U. S. A. 99, 7746–7750. doi: 10.1073/pnas.122205699
Jaouen, F., and Gascon, E. (2016). Understanding the Role of miR-33 in Brain Lipid Metabolism: implications for Alzheimer’s Disease. J. Neurosci. 36, 2558–2560. doi: 10.1523/JNEUROSCI.4571-15.2016
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Karunakaran, S., Chowdhury, A., Donato, F., Quairiaux, C., Michel, C. M., and Caroni, P. (2016). PV plasticity sustained through D1/5 dopamine signaling required for long-term memory consolidation. Nat. Neurosci. 19, 454–464. doi: 10.1038/nn.4231
Ke, S., Alemu, E. A., Mertens, C., Gantman, E. C., Fak, J. J., Mele, A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 29, 2037–2053. doi: 10.1101/gad.269415.115
Ke, S., Pandya-Jones, A., Saito, Y., Fak, J. J., Vågbø, C. B., Geula, S., et al. (2017). m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006. doi: 10.1101/gad.301036.117
Keller, L., Xu, W., Wang, H. X., Winblad, B., Fratiglioni, L., and Graff, C. (2011). The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: a prospective cohort study. J. Alzheimers Dis. 23, 461–469. doi: 10.3233/jad-2010-101068
Knuckles, P., and Bühler, M. (2018). Adenosine methylation as a molecular imprint defining the fate of RNA. FEBS Lett. 592, 2845–2859. doi: 10.1002/1873-3468.13107
Knuckles, P., Lence, T., Haussmann, I., Jacob, D., Kreim, N., Carl, S., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the mA machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429. doi: 10.1101/gad.309146.117
Kolisnyk, B., Al-Onaizi, M., Soreq, L., Barbash, S., Bekenstein, U., Haberman, N., et al. (2017). Cholinergic Surveillance over Hippocampal RNA Metabolism and Alzheimer’s-Like Pathology. Cereb. Cortex 27, 3553–3567. doi: 10.1093/cercor/bhw177
Koranda, J., Dore, L., Shi, H., Patel, M., Vaasjo, L., Rao, M., et al. (2018). Mettl14 Is Essential for Epitranscriptomic Regulation of Striatal Function and Learning. Neuron 99, 283.e–292.e. doi: 10.1016/j.neuron.2018.06.007
Lakk, M., Yarishkin, O., Baumann, J. M., Iuso, A., and Križaj, D. (2017). Cholesterol regulates polymodal sensory transduction in Müller glia. Glia 65, 2038–2050. doi: 10.1002/glia.23213
Lane, C. A., Hardy, J., and Schott, J. M. (2018). Alzheimer’s disease. Eur. J. Neurol. 25, 59–70. doi: 10.1111/ene.13439
Leighton, L., Ke, K., Zajaczkowski, E., Edmunds, J., Spitale, R., and Bredy, T. (2018). Experience-dependent neural plasticity, learning, and memory in the era of epitranscriptomics. Genes Brain Behav. 17:e12426. doi: 10.1111/gbb.12426
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447. doi: 10.1038/cr.2017.10
Li, L., Zang, L., Zhang, F., Chen, J., Shen, H., Shu, L., et al. (2017). Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411. doi: 10.1093/hmg/ddx128
Li, F., Zhao, D., Wu, J., and Shi, Y. (2014). Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 24, 1490–1492. doi: 10.1038/cr.2014.153
Li, H., Ren, Y., Mao, K., Hua, F., Yang, Y., Wei, N., et al. (2018). FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem. Biophys. Res. Commun. 498, 234–239. doi: 10.1016/j.bbrc.2018.02.201
Li, J., Yang, X., Qi, Z., Sang, Y., Liu, Y., Xu, B., et al. (2019). The role of mRNA mA methylation in the nervous system. Cell Biosci. 9:66. doi: 10.1186/s13578-019-0330-y
Li, N., Zhang, D., Cao, S., Qiao, M., Zhang, P., Zhao, Q., et al. (2021). The effects of folic acid on RNA m6A methylation in hippocampus as well as learning and memory ability of rats with acute lead exposure. J. Funct. Foods 76:104276. doi: 10.1016/j.jff.2020.104276
Li, Y., Bedi, R. K., Wiedmer, L., Sun, X., Huang, D., and Caflisch, A. (2021). Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m(6)A) Recognition by the Reader Domain of YTHDC1. J. Chem. Theory Comput. 17, 1240–1249. doi: 10.1021/acs.jctc.0c01136
Li, Q., Wen, S., Ye, W., Zhao, S., and Liu, X. (2021). The potential roles of m(6)A modification in regulating the inflammatory response in microglia. J. Neuroinflammation 18:149. doi: 10.1186/s12974-021-02205-z
Li, W., Jiang, M., Zhao, S., Liu, H., Zhang, X., Wilson, J. X., et al. (2015). Folic Acid Inhibits Amyloid β-Peptide Production through Modulating DNA Methyltransferase Activity in N2a-APP Cells. Int. J. Mol. Sci. 16, 25002–25013. doi: 10.3390/ijms161025002
Li, Y., Bedi, R. K., Moroz-Omori, E. V., and Caflisch, A. (2020). Structural and Dynamic Insights into Redundant Function of YTHDF Proteins. J. Chem. Inf. Model. 60, 5932–5935. doi: 10.1021/acs.jcim.0c01029
Liao, S., Sun, H., and Xu, C. (2018). YTH Domain: a Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteomics Bioinformatics 16, 99–107. doi: 10.1016/j.gpb.2018.04.002
Lim, J. W., Lee, J., and Pae, A. N. (2020). Mitochondrial dysfunction and Alzheimer’s disease: prospects for therapeutic intervention. BMB Rep. 53, 47–55. doi: 10.5483/BMBRep.2020.53.1.279
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 62, 335–345. doi: 10.1016/j.molcel.2016.03.021
Lin, S., Liu, J., Jiang, W., Wang, P., Sun, C., Wang, X., et al. (2019). METTL3 Promotes the Proliferation and Mobility of Gastric Cancer Cells. Open Med. 14, 25–31. doi: 10.1515/med-2019-0005
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, S., Zhuo, L., Wang, J., Zhang, Q., Li, Q., Li, G., et al. (2020). METTL3 plays multiple functions in biological processes. Am. J. Cancer Res. 10, 1631–1646.
Lu, Z., Ma, Y., Li, Q., Liu, E., Jin, M., Zhang, L., et al. (2019). The role of N(6)-methyladenosine RNA methylation in the heat stress response of sheep (Ovis aries). Cell Stress Chaperon. 24, 333–342. doi: 10.1007/s12192-018-00965-x
Ma, C., Liao, S., and Zhu, Z. (2019). Crystal structure of human YTHDC2 YTH domain. Biochem. Biophys. Res. Commun. 518, 678–684. doi: 10.1016/j.bbrc.2019.08.107
McTaggart, J., Lee, S., Iberl, M., Church, C., Cox, R., and Ashcroft, F. (2011). FTO is expressed in neurones throughout the brain and its expression is unaltered by fasting. PLoS One 6:e27968. doi: 10.1371/journal.pone.0027968
Merkurjev, D., Hong, W. T., Iida, K., Oomoto, I., Goldie, B. J., Yamaguti, H., et al. (2018). Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21, 1004–1014. doi: 10.1038/s41593-018-0173-6
Meyer, K., Saletore, Y., Zumbo, P., Elemento, O., Mason, C., and Jaffrey, S. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Meyer, K. D., and Jaffrey, S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326. doi: 10.1038/nrm3785
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Miao, Z., Zhang, T., Qi, Y., Song, J., Han, Z., and Ma, C. (2020). Evolution of the RNA N (6)-Methyladenosine Methylome Mediated by Genomic Duplication. Plant Physiol. 182, 345–360. doi: 10.1104/pp.19.00323
Myrick, L. K., Hashimoto, H., Cheng, X., and Warren, S. T. (2015). Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum. Mol. Genet. 24, 1733–1740. doi: 10.1093/hmg/ddu586
Nance, D. J., Satterwhite, E. R., Bhaskar, B., Misra, S., Carraway, K. R., and Mansfield, K. D. (2020). Characterization of METTL16 as a cytoplasmic RNA binding protein. PLoS One 15:e0227647. doi: 10.1371/journal.pone.0227647
Pakavathkumar, P., Noël, A., Lecrux, C., Tubeleviciute-Aydin, A., Hamel, E., Ahlfors, J. E., et al. (2017). Caspase vinyl sulfone small molecule inhibitors prevent axonal degeneration in human neurons and reverse cognitive impairment in Caspase-6-overexpressing mice. Mol. Neurodegener. 12:22. doi: 10.1186/s13024-017-0166-z
Patil, D. P., Pickering, B. F., and Jaffrey, S. R. (2018). Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol. 28, 113–127. doi: 10.1016/j.tcb.2017.10.001
Peña-Bautista, C., Álvarez-Sánchez, L., Cañada-Martínez, A. J., Baquero, M., and Cháfer-Pericás, C. (2021). Epigenomics and Lipidomics Integration in Alzheimer Disease: pathways Involved in Early Stages. Biomedicines 9:1812. doi: 10.3390/biomedicines9121812
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 169, 824–835.e14. doi: 10.1016/j.cell.2017.05.003
Ping, X., Sun, B., Wang, L., Xiao, W., Yang, X., Wang, W., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Popugaeva, E., Pchitskaya, E., and Bezprozvanny, I. (2018). Dysregulation of Intracellular Calcium Signaling in Alzheimer’s Disease. Antioxid. Redox Signal. 29, 1176–1188. doi: 10.1089/ars.2018.7506
Reitz, C., Tosto, G., Mayeux, R., Luchsinger, J. A., NIA-LOAD/NCRAD Family Study Group, and Alzheimer’s Disease Neuroimaging Initiative (2012). Genetic variants in the Fat and Obesity Associated (FTO) gene and risk of Alzheimer’s disease. PLoS One 7:e50354. doi: 10.1371/journal.pone.0050354
Richter, J. D., Bassell, G. J., and Klann, E. (2015). Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci. 16, 595–605. doi: 10.1038/nrn4001
Rosen, L. B., Ginty, D. D., Weber, M. J., and Greenberg, M. E. (1994). Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron 12, 1207–1221. doi: 10.1016/0896-6273(94)90438-3
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6:e31311. doi: 10.7554/eLife.31311
Růžička, K., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., et al. (2017). Identification of factors required for m(6) A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172. doi: 10.1111/nph.14586
Scarborough, A., Flaherty, J., Hunter, O., Liu, K., Kumar, A., Xing, C., et al. (2021). SAM homeostasis is regulated by CFI-mediated splicing of MAT2A. Elife 10:e64930. doi: 10.7554/eLife.64930
Schwartz, S., Mumbach, M., Jovanovic, M., Wang, T., Maciag, K., Bushkin, G., et al. (2014). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8, 284–296. doi: 10.1016/j.celrep.2014.05.048
Shafik, A. M., Zhang, F., Guo, Z., Dai, Q., Pajdzik, K., Li, Y., et al. (2021). N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 22:17. doi: 10.1186/s13059-020-02249-z
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Shi, H., Wei, J., and He, C. (2019). Where, When, and How: context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 74, 640–650. doi: 10.1016/j.molcel.2019.04.025
Shi, H., Zhang, X., Weng, Y. L., Lu, Z., Liu, Y., Lu, Z., et al. (2018). m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563, 249–253. doi: 10.1038/s41586-018-0666-1
Shi, W., Yang, F., Dai, R., Sun, Y., Chu, Y., Liao, S., et al. (2021). METTL3-Mediated N6-Methyladenosine Modification Is Involved in the Dysregulation of NRIP1 Expression in Down Syndrome. Front. Cell. Dev. Biol. 9:621374. doi: 10.3389/fcell.2021.621374
Shima, H., Matsumoto, M., Ishigami, Y., Ebina, M., Muto, A., Sato, Y., et al. (2017). S-Adenosylmethionine Synthesis Is Regulated by Selective N-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cell Rep. 21, 3354–3363. doi: 10.1016/j.celrep.2017.11.092
Slobodin, B., Han, R., Calderone, V., Vrielink, J., Loayza-Puch, F., Elkon, R., et al. (2017). Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation. Cell 169, 326–337.e12. doi: 10.1016/j.cell.2017.03.031
Suhl, J., Chopra, P., Anderson, B., Bassell, G., and Warren, S. (2014). Analysis of FMRP mRNA target datasets reveals highly associated mRNAs mediated by G-quadruplex structures formed via clustered WGGA sequences. Hum. Mol. Genet. 23, 5479–5491. doi: 10.1093/hmg/ddu272
Walters, B., Mercaldo, V., Gillon, C., Yip, M., Neve, R., Boyce, F., et al. (2017). The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology 42, 1502–1510. doi: 10.1038/npp.2017.31
Wang, J., Ishfaq, M., Xu, L., Xia, C., Chen, C., and Li, J. (2019). METTL3/mA/miRNA-873-5p Attenuated Oxidative Stress and Apoptosis in Colistin-Induced Kidney Injury by Modulating Keap1/Nrf2 Pathway. Front. Pharmacol. 10:517. doi: 10.3389/fphar.2019.00517
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578. doi: 10.1038/nature18298
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2017). Corrigendum: structural basis of N-adenosine methylation by the METTL3-METTL14 complex. Nature 542:260. doi: 10.1038/nature21073
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Warda, A., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Höbartner, C., et al. (2017). NHuman METTL16 is a -methyladenosine (mA) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. doi: 10.15252/embr.201744940
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 Regulates Nuclear RNA mA Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 69, 1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015
Wen, L., Sun, W., Xia, D., Wang, Y., Li, J., and Yang, S. (2020). The m6A methyltransferase METTL3 promotes LPS-induced microglia inflammation through TRAF6/NF-κB pathway. Neuroreport doi: 10.1097/wnr.0000000000001550 [Epub Online ahead of print].
Weng, Y., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018). Epitranscriptomic mA Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 97, 313–325.e6. doi: 10.1016/j.neuron.2017.12.036
Widagdo, J., and Anggono, V. (2018). The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J. Neurochem. 147, 137–152. doi: 10.1111/jnc.14481
Widagdo, J., Zhao, Q. Y., Kempen, M. J., Tan, M. C., Ratnu, V. S., Wei, W., et al. (2016). Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J. Neurosci. 36, 6771–6777. doi: 10.1523/jneurosci.4053-15.2016
Wu, B., Li, L., Huang, Y., Ma, J., and Min, J. (2017). Readers, writers and erasers of N(6)-methylated adenosine modification. Curr. Opin. Struct. Biol. 47, 67–76. doi: 10.1016/j.sbi.2017.05.011
Wu, B., Su, S., Patil, D. P., Liu, H., Gan, J., Jaffrey, S. R., et al. (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9:420. doi: 10.1038/s41467-017-02770-z
Wu, R., Li, A., Sun, B., Sun, J. G., Zhang, J., Zhang, T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29, 23–41. doi: 10.1038/s41422-018-0113-8
Xiang, Y., Laurent, B., Hsu, C. H., Nachtergaele, S., Lu, Z., Sheng, W., et al. (2017). RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576. doi: 10.1038/nature21671
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 61, 507–519. doi: 10.1016/j.molcel.2016.01.012
Xu, C., Liu, K., Ahmed, H., Loppnau, P., Schapira, M., and Min, J. (2015). Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins. J. Biol. Chem. 290, 24902–24913. doi: 10.1074/jbc.M115.680389
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929. doi: 10.1038/nchembio.1654
Yoon, K. J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 171, 877–889.e17. doi: 10.1016/j.cell.2017.09.003
Yu, J., Chen, M., Huang, H., Zhu, J., Song, H., Zhu, J., et al. (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423. doi: 10.1093/nar/gkx1182
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential mA mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10. doi: 10.1038/s41421-018-0019-0
Yue, Y., Liu, J., and He, C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355. doi: 10.1101/gad.262766.115
Zaccara, S., and Jaffrey, S. R. (2020). A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell 181, 1582–1595.e18. doi: 10.1016/j.cell.2020.05.012
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016). Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U. S. A. 113, E2047–E2056. doi: 10.1073/pnas.1602883113
Zhang, X., Li, X., Jia, H., An, G., and Ni, J. (2021). The m6A methyltransferase METTL3 modifies PGC-1α mRNA promoting mitochondrial dysfunction and oxLDL-induced inflammation in monocytes. J. Biol. Chem. 297:101058. doi: 10.1016/j.jbc.2021.101058
Zhang, Z., Wang, M., Xie, D., Huang, Z., Zhang, L., Yang, Y., et al. (2018). METTL3-mediated N(6)-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 28, 1050–1061. doi: 10.1038/s41422-018-0092-9
Zhao, F., Xu, Y., Gao, S., Qin, L., Austria, Q., Siedlak, S. L., et al. (2021). METTL3-dependent RNA m6A dysregulation contributes to neurodegeneration in Alzheimer’s disease through aberrant cell cycle events. Mol. Neurodegener. 16:70. doi: 10.1186/s13024-021-00484-x
Zhao, T., Li, X., Sun, D., and Zhang, Z. (2019). Oxidative stress: one potential factor for arsenite-induced increase of N(6)-methyladenosine in human keratinocytes. Environ. Toxicol. Pharmacol. 69, 95–103. doi: 10.1016/j.etap.2019.04.005
Zhao, Y., Cong, L., and Lukiw, W. J. (2017). Lipopolysaccharide (LPS) Accumulates in Neocortical Neurons of Alzheimer’s Disease (AD) Brain and Impairs Transcription in Human Neuronal-Glial Primary Co-cultures. Front. Aging Neurosci. 9:407. doi: 10.3389/fnagi.2017.00407
Zheng, G., Dahl, J., Niu, Y., Fedorcsak, P., Huang, C., Li, C., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zheng, L., Tang, X., Lu, M., Sun, S., Xie, S., Cai, J., et al. (2020). microRNA-421-3p prevents inflammatory response in cerebral ischemia/reperfusion injury through targeting m6A Reader YTHDF1 to inhibit p65 mRNA translation. Int. Immunopharmacol. 88:106937. doi: 10.1016/j.intimp.2020.106937
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., and Qian, S. B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. doi: 10.1038/nature15377
Keywords: Alzheimer’s disease, demethylase, methyltransferase, methylation-binding protein, memory disorder, N6-methyladenosine
Citation: Zhang R, Zhang Y, Guo F, Li S and Cui H (2022) RNA N6-Methyladenosine Modifications and Its Roles in Alzheimer’s Disease. Front. Cell. Neurosci. 16:820378. doi: 10.3389/fncel.2022.820378
Received: 12 January 2022; Accepted: 28 February 2022;
Published: 24 March 2022.
Edited by:
Daniela Tropea, Trinity College Dublin, IrelandReviewed by:
Francesco Rusconi, University of Milan, ItalyBrandon Walters, University of Toronto Mississauga, Canada
Copyright © 2022 Zhang, Zhang, Guo, Li and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sha Li, bGlzaGFAaGVibXUuZWR1LmNu; Huixian Cui, Y3VpaHhAaGVibXUuZWR1LmNu
 Runjiao Zhang1,2
Runjiao Zhang1,2 Yizhou Zhang
Yizhou Zhang Sha Li
Sha Li