- 1Department of Neurosurgery, The Second Affiliated Hospital Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
- 2Department of Neurology, The Second Affiliated Hospital Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
- 3Key Laboratory of Precise Treatment and Clinical Translational Research of Neurological Diseases, Hangzhou, Zhejiang, China
- 4Clinical Research Center for Neurological Diseases of Zhejiang Province, Hangzhou, China
- 5Department of Neurology, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
- 6Department of Pharmacy, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, China
Recent studies have revealed the critical role of AQP4 in the occurrence and development of gliomas. However, the role of AQP4 in immune regulation has not yet been reported. Many recent reports have identified the lymphatic system’s occurrence within the central nervous system (CNS) and the vital role of immune regulation in treating brain tumors. Therefore, the present study aimed to explore the role of AQP4 in the immune regulation of glioma. We used bioinformatics analysis to investigate the immunoregulatory function of AQP4, including its correlation with immunity, anti-tumor immune processes, immunotherapy, immune infiltration, tumor mutational burden (TMB), stemness, mutation, and pan-cancer. The results revealed that AQP4 was significantly associated with the expression of multiple immune checkpoints, immune cells, as well as multiple immune cell effector genes, and antigen presentation and processing abilities. Although no significant correlation was found between the AQP4 gene and IDH mutation and MGMT, AQP4 demonstrated substantial expression differences in different immunophenotypes and molecular types. Using the TTD database, we discovered that EGFR, ABAT, and PDGFRA are strongly associated with AQP4 expression in the glioblastoma (GBM) classification, and these factors could be the potential AQP4-related immunotherapy targets. Afterward, we screened the differential genes in the high and low AQP4 gene expression group, the high and low immune score group, and the high and low matrix score group and took the intersection as the candidate factor. Finally, univariate Cox analysis was used to find eight prognostic variables with significant differences across the candidate genes. After lasso dimensionality reduction, three genes built the model (RARRES1, SOCS3, and TTYH1). The scoring model generated by the three genes was eventually obtained after the multi-factor screening of the three genes. Finally, combined with clinical information and cox regression analysis, it was further confirmed that the model score could be used as an independent prognostic factor.
Introduction
Glioma is the most common primary central nervous system (CNS) tumor, accounting for about 60–70% of primary brain tumors, and the overall survival of patients after diagnosis is about 15–18 months (Smith et al., 2022). Glioblastoma (GBM) is the most malignant type. GBM is a major problem in the field of neurosurgery at present. The main treatment methods are surgery, combined with radiotherapy and chemotherapy. The treatment effect is poor due to its rapid development, high invasiveness, and resistance to chemotherapeutic medications, and novel treatments are urgently needed to improve patient prognosis. Simultaneously, if novel treatment targets can be discovered, they will undoubtedly benefit most GBM patients.
Currently, 13 aquaporins (AQP0–AQP12) have been identified in mammals, of which AQP4 is a class of aquaporins expressed explicitly in the CNS. A significant number of neurological disorders are linked to changes in AQP4 expression or location. AQP4 plays a role in brain inflammation, glial lymphoid clearance, synaptic plasticity, memory development, extracellular space (ECS) volume modulation, and potassium homeostasis (Papadopoulos and Verkman, 2012; Nagelhus and Ottersen, 2013; Hubbard et al., 2018). The research on AQP4 and various diseases is mainly based on the pathological analysis of dead brain tissue, in vitro experimental research, and the use of AQP4-deficient rodent models (Mader and Brimberg, 2019). Only a few studies use extensive data analysis, and bioinformatics research on AQP4 will assist in a more systematic and thorough investigation of AQP4’s role in numerous disorders.
The role of AQP4 in the occurrence and development of glioma is still unclear. Our earlier study systematically summarized the critical role of AQP4 in the malignant progression of glioma and its significance in the study of anti-tumor drug resistance (Lan et al., 2017). In glioma, the expression of the AQP4 protein is elevated, and inhibiting AQP4 can significantly reduce glioma malignant proliferation (Kahlert et al., 2013). The most recent research indicates that temozolomide can suppress the growth of malignant glioma by inhibiting the expression of AQP4, implying that the AQP4 pathway’s activity substantially impacts the chemotherapeutic effectiveness of GBM (Chen et al., 2017). Our two newly published studies further confirmed the great potential of AQP4 in treating GBM (Lan et al., 2020; Zou et al., 2021). We found for the first time that inhibition of AQP4 can significantly improve the sensitivity of GBM drug therapy (Lan et al., 2020). More intriguingly, current research has indicated that AQP4 protein aggregation state could be a determinant for glioma cell fate (Amiry-Moghaddam, 2019; Simone et al., 2019), strengthening the potential role of AQP4 as a target in the treatment of GBM. It’s known that AQP4 forms heterotetramers in the plasma membrane made of the M23-AQP4 and M1-AQP4 isoforms (Smith and Verkman, 2015). The isoform ratio controls AQP4 protein aggregation into supramolecular structures called orthogonal arrays of particles (AQP4-OAP). The role of AQP4 protein aggregation into OAP in malignant gliomas is still unclear. The authors (Simone et al., 2019) found that AQP4 protein aggregation/disaggregation into OAP influences the biology of glioma cells, demonstrating that AQP4 protein disaggregation may potentiate invasiveness potential, whereas AQP4 protein aggregation may activate the apoptotic path. Besides, Valente et al. (2022) found that AQP4ex, the new read through isoform of AQP4, could be associated with the degree of vasogenic brain edema and GBM cell progression. They found that the reduction in AQP4ex, leading to reduction and delocalization of AQP4, could be likely to undermine the integrity of the BBB, indicating that AQP4ex could be considered as a potential new early biomarker of GBM progression and a target for AQP4 modulation (Frigeri et al., 2007). Various studies have confirmed that AQP4 plays a vital role in GBM’s malignant progression and drug resistance (Valente et al., 2022), but its molecular mechanism still needs more in-depth and comprehensive research (Peng et al., 2020; Behnam et al., 2022).
Tumor immunotherapy is a prominent topic in anti-tumor research, and the significance of AQP4 in immune modulation has yet to be discovered. Our previous research has indicated that AQP4 exerted carcinogenic effects via various pathways in glioma (Lan et al., 2017). Furthermore, regulatory T-cell development has been found to be dependent on AQP4 expression (Chi et al., 2011). Mice lacking AQP4 receptors had suppressed levels of CD4+/CD25+ regulatory T-cells. This leads to an abnormally overactive microglial inflammatory response (Chi et al., 2011). In recent years, many reports have identified the occurrence of the lymphatic system within the CNS and its essential role in immune regulation in brain tumor therapy (Lan et al., 2022; Li et al., 2022). As a result, the involvement of AQP4 in the immune regulation of glioma and the immunotherapy process was investigated further in this work. This study aimed to examine the role of AQP4 in the CNS immune system and find out how important it is in the glioma immunotherapy process. More importantly, we anticipate developing an AQP4-related prognostic model, which would serve as a critical theoretical research foundation for improving the effect of glioma immunotherapy.
Materials and methods
Data retrieval and preprocessing
Expression profile data, phenotype data, and mutation data of GBM were downloaded from the Xena1 database. The GBM data packet named “01A,” which including 114 samples (Table 1), was selected. The data itself has been log2(data + 1). The information uses FPKM (Fragments Per Kilobase of transcript per Million mapped reads) without the normalization between arrays of the limma package. Twenty immune checkpoints and 122 immunomodulator-related genes were obtained from the document “Siglec15 shapes a non-inflamed tumor microenvironment and predict the molecular subtype in bladder cancer.” GBM data were obtained from CGGA (Chinese Glioma Genome Atlas) database. Data samples from two different data packets contain 693 cancer data and 325 cancer data, respectively. The number of GBM and rGBM in data packet of 693 data samples is 209, and the number of GBM and rGBM in data packet of 325 samples is 109. The raw data is RSEM, which has been converted to log2(data + 1).
Correlation analysis between AQP4 gene and immune checkpoints
The Hmisc package of R was used to calculate the correlation analysis of 20 immunological checkpoints in the data of the gene AQP4 and TCGA. Spearman’s technique was utilized for correlation analysis.
Correlation between AQP4 gene and immune infiltration
The R’s CIBERSORT was applied to assess the proportion of infiltrating 22 immune cells in cancer samples. The correlation between the gene AQP4 and the 22 immune cells implanted ratio was calculated. The same analysis was performed in low grade glioma (LGG), GBM, LGG + GBM, lung squamous cell carcinoma, lung adenocarcinoma, and breast cancer.
Differential analysis of immunomodulator-related genes between high and low AQP4 gene expression groups
The limma package in R was used to do the difference analysis. The median expression of the AQP4 gene served as the grouping criterion; individuals with expression levels higher than the median were classified into high expression groups, while those with expression levels lower than the median were divided into low expression groups. The screening criteria for differential genes were P-value < 0.05, |log2FC| > 0.5853. The differential genes were intersected with 122 immunomodulator-related genes to obtain differential immunomodulatory genes, plotted by R’s heatmap package.
The anti-cancer immune process activity is different in the high and low expression of the AQP4 gene
Immune circulation data for GBM were obtained from the TIP (Trip Medical Database) database and plotted using the ggplot2 package for R.
AQP4 gene-related functional enrichment analysis
The differential genes of AQP4 high and low groups were analyzed for functional enrichment by the cluster profile package of R.
Heatmap of the relationship between AQP4 gene high and low groups and tumor-associated immune cell effector genes
Heatmaps of tumor-associated immune cell effector genes were created using the R package pheatmap and data from the literature “Siglec15 shapes a non-inflamed tumor microenvironment and predict the molecular subtype in bladder cancer.”
Differential expression of AQP4 gene in clinical features
The patients were grouped according to clinical characteristics, including age, sex, immunophenotype, molecular type, IDH mutation, and MGMT, and the expression of the AQP4 gene in different groups was compared.
Correlation of AQP4 gene expression with tumor mutational burden, immune score, stromal score, and tumor purity
Immune scores, stromal scores, and tumor purity were calculated through the estimate package for R. The R package maftools were used to calculate TMB. The R package Hmisc was then used to calculate the connection between the expression value of gene AQP4 and TMB, immunological score, stromal score, and tumor purity.
AQP4 gene high and low expression groups, enrichment scores of immunotherapy prediction-related pathways, and differences in AQP4 gene expression among different immune response groups in the immune dataset
The R’s GSVA package was used to perform the pathway enrichment scores of AQP4 gene high and low groups, and the AQP4 gene expression in different immune groups in IMvigor210 (bladder cancer) and GSE91061 (melanoma) were analyzed. GSE126045 (NSCLC) was excluded from data analysis due to a lack of data.
Correlation analysis of AQP4 gene mutation, copy number variation, methylation, and expression
The number of AQP4 mutations is low. Copy number variation and methylation data were obtained from the Xena database.2
Mutations in AQP4 gene expression high and low groupings
The GenVisR package for R displayed high and low-grouped mutations in AQP4 gene expression.
Database screening for differential expression display of cancer-type therapeutic targets
Screening of cancer-type therapeutic targets was performed using data from the TTD (Therapeutic Target Database) database.3
Screening the intersection of differential genes of AQP4 gene expression group, immune score group, and matrix score group as candidate factors
As possible factors, the intersection of differential genes in high and low AQP4 gene groups, differential genes in high and low immunological score groups, and differential genes in high and low matrix score groups was used. The R package used is limma, and the filtering conditions are P-value < 0.05, |log2FC| > 0.585.
Screening of prognostic factors by univariate cox regression based on differential genes
The intersection of the above differential genes was subjected to single factor cox analysis by R survival. For subsequent analysis, genes with P-values less than 0.05 were screened as prognostic factors.
Lasso dimensionality reduction removes redundant factors and builds a related prognostic risk scoring model
Lasso analysis was performed by R’s glmnet package, followed by multivariate cox regression to obtain model scores.
Evaluation of the predictive power of prognostic models
The survival package in R was used to perform KM (Kaplan-Meier) survival analysis on the models. The survival ROC package in R was used to examine the AUC value of the model score and the different analyses of the genes in the high and low model score groups.
Validation of prognostic models with external datasets
The above analysis was carried out using ICGC (The International Cancer Genome Consortium) data.
Univariate and multivariate cox regression analyses
Combined with clinical information, univariate and multivariate cox regression analysis verified that risk scores were independent prognostic factors (binary classification method, at least stage, age, and gender were included). Univariate and multivariate separate predictive analyses were performed on risk scores.
Combined with clinical characteristics, a nomogram was developed to assist in clinical practice
The R’s rms package, foreign, and survival packages were used to construct a nomogram for clinical features and risk scores.
Human tissue microarray and immunohistochemistry
The human tissue microarrays were purchased from Outdo Biotech Company (Shanghai, China). A total of 77 glioma tissue samples and three normal samples were included. Slides were deparaffinized and rehydrated and were then immersed in target retrieval solution (pH 6) and boiled at medium heat three times for 10 min each in a microwave. After the slides were blocked with 3% BSA, the sections were incubated with primary antibody against AQP4 (Santa Cruz Biotechnology, Dallas, TX, USA) followed by HRP-labeled anti-rabbit IgG secondary antibody. The specimens were counterstained with hematoxylin. The negative control was obtained by replacing the primary antibody with a regular rabbit IgG. Target-positive cells were counted in 3-4 different fields and imaged using an Olympus microscope, and the immunoreactions were evaluated independently by two pathologists blinded to the clinicopathologic information in order to ensure unbiased assessment of tissue morphology. The antibody staining intensity was classified as follows: no staining, 0; weak staining, 1; moderate staining, 2; and strong staining, 3. A five-point scale was used to classify the percentage of cells stained: 0 (no positive cells), 1 (<25% positive cells), 2 (25–50% positive cells), 3 (50–75% positive cells), and 4 (>75% positive cells). The score for each tissue was calculated by multiplying the intensity index by the percentage index, yielding a score between 0 and 12. The median AQP4 score was employed to determine the cutoff value. Tumors with AQP4 scores less than or equal to the median were designated as having “low expression,” whereas those with scores greater than the median were designated as having “high expression.”
Statistical analysis
The Spearman test was utilized in every correlation analysis. The ns denotes that there is no significant difference, * means p-value < 0.05, ** means p-value < 0.01, *** means p-value < 0.001, **** means p-value < 0.0001.
Results
AQP4 protein expression may predict prognosis in glioma patients
The expression of AQP4 protein in tumor samples from 77 glioma patients was evaluated by immunohistochemical (IHC) staining of a tissue microarray. The expression of AQP4 protein in each IHC sample was classified as either high (score > 6) or low (score ≤ 6), using the median of the IHC scores as the cutoff value. The correlations between AQP4 protein expression and the clinicopathologic variables of 77 glioma patients are shown in Figure 1A. The results indicated that low AQP4 expression was significantly correlated with lower clinical grade glioma (grade 1/2), while high AQP4 expression predicted higher clinical grade glioma (grade 3/4) in patients (Figure 1A) (X 2 = 12.434, p < 0.001). In addition, younger age was significantly correlated with low expression of AQP4 (X 2 = 4.812, p = 0.028), and female patients tended to have lower expression of AQP4 than male patients (X 2 = 4.434, p = 0.035). More importantly, we also found that AQP4 was highly expressed in glioma tissues and that the expression level of AQP4 was negatively correlated with the glioma grade (Figures 1B,C) (Spearman’s rank correlation rs = 0.297, p = 0.013). Kaplan-Meier OS analyses revealed that the low AQP4 expression group (IHC staining indicating strong and moderate expression) had a better prognosis than the high AQP4 group (IHC staining indicating weak and negative expression) (p < 0.01) (Figure 1D). Overall, these data demonstrated the prognostic significance of AQP4 in glioma.
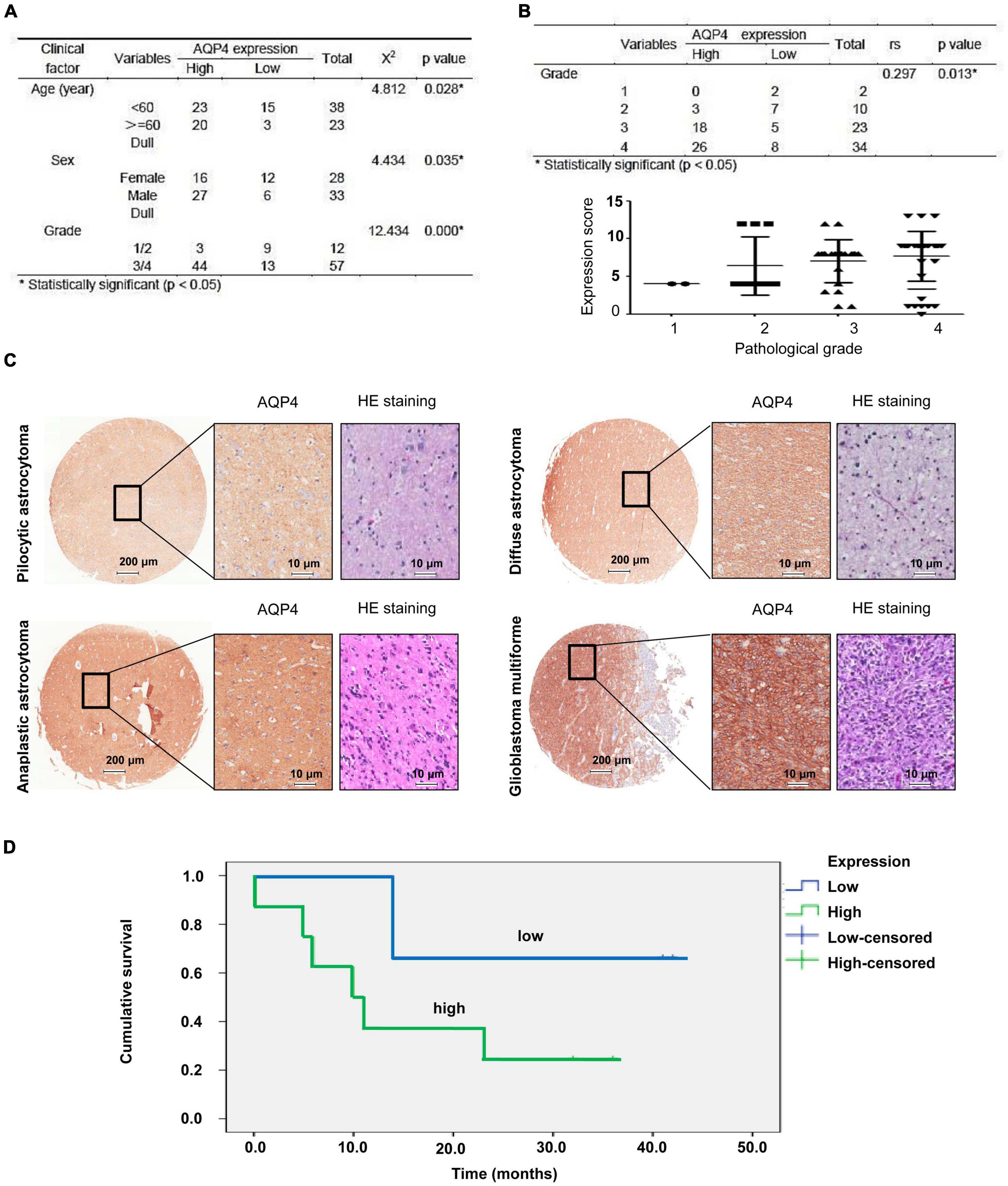
Figure 1. Clinical data analysis of AQP4 expression in human tissue microarrays from glioma patients. (A) Correlation analyses of AQP4 protein expression in relation to clinicopathologic variables of 77 glioma patients by a X 2 test. (B) Quantitative analysis of the relative AQP4 expression level in all groups was performed. Furthermore, the correlation between AQP4 expression and different glioma grades was also assessed by Spearman’s rank correlation analysis. (C) Representative images of immunohistochemical staining to assess AQP4 protein levels in the glioma tissue microarray. (D) Kaplan-Meier analysis of OS for glioma cancer patients with different expression levels of AQP4 by a log-rank test. “Low” indicates a low level of AQP4 expression; “high” indicates a high level of AQP4 expression.
Correlation of AQP4 gene with mainstream immune checkpoints and immune infiltrating cells
In the correlation analysis between the gene AQP4 and 20 immune checkpoints, only five immune checkpoints were significantly correlated. CD276, LAG3, VTCN1, and PVR were negatively correlated with the gene AQP4, and BTLA was negatively correlated with the gene AQP4 (Figures 2A,B and Supplementary Tables 1–3), and the correlations were all statistically significant. We also looked into the relationship between AQP4 expression and the number of immune invading cells (Figures 2C,D). The AQP4 gene was strongly connected with five immune cells, including B cells, out of 22 immune invading cells. Macrophages are immature. M0, plasma cells, T-cells, CD4 memory resting, and monocytes are some terms used to describe M0, plasma cells, T-cells, and monocytes (Supplementary Tables 4–6). The tumor types we included in the study mainly included LGG, GBM, LGG + GBM, lung squamous cell carcinoma, lung adenocarcinoma, and breast cancer. In Macrophages M0, all cancers except LGG were significantly associated with the gene AQP4. Only breast cancer had no significant correlation with the gene AQP4 in monocytes and T-cells, memory resting. In contrast, LGG, GBM, LGG + GBM, lung squamous cell carcinoma, and lung adenocarcinoma all had significant correlations (Supplementary Tables 7, 8).
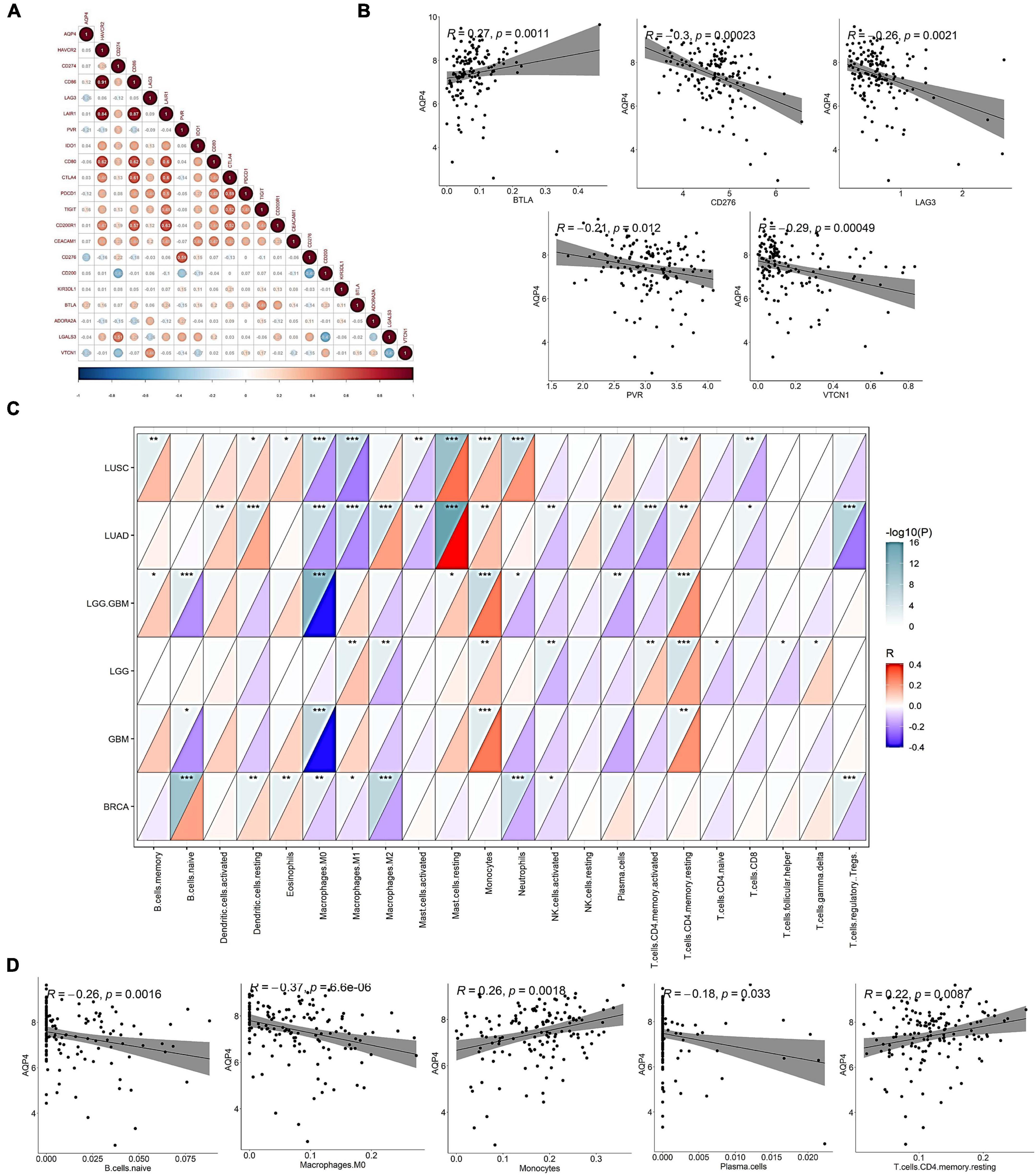
Figure 2. Correlation of AQP4 gene with mainstream immune checkpoints and immune infiltrating cells. (A) Association analysis of the gene AQP4 with 20 immune checkpoints. (B) Correlation analysis of AQP4 gene with mainstream immune checkpoints CD276, LAG3, PVR, BTLA, and VTCN1. (C) Correlation analysis of AQP4 gene with immune infiltrating cells in pan-cancer. (D) Correlation analysis of AQP4 gene with immune infiltrating cells, B cells. Naive macrophages M0, plasma cells, T-cells, CD4, memory, resting, and monocytes. *p < 0.05, **p < 0.01, ***p < 0.001.
The effect of AQP4 expression level on the anti-tumor immune process
Initially, we looked at how AQP4 gene expression affects immune cells’ ability to present and process antigens. A total of 122 genes were collected from the literature, 32 of which were substantially different in the AQP4 high and low expression groups, with the majority of the differential genes being low in the AQP4 high expression group. This indicated that antigen presentation and processing were attenuated in the high AQP4 group (Figure 3A and Supplementary Table 9). We further analyzed the anti-cancer immune process activity differences between high and low AQP4 gene expression groups (Figure 3B). In the high and low AQP4 expression groups, only the following five processes were significantly different between the high and low groups, including the release of cancer cell antigens, tumor antigen presentation, initiation and activation, eosinophil recruiting, and cancer cell clearance. The other 18 immunological mechanisms, on the other hand, did not differ significantly between the high and low groups (Figure 3B and Supplementary Table 10). Following that, we examined the relationships between the AQP4 gene, immunotherapy-related prediction pathway enrichment scores, and anti-cancer immune process activity (Figure 3C).
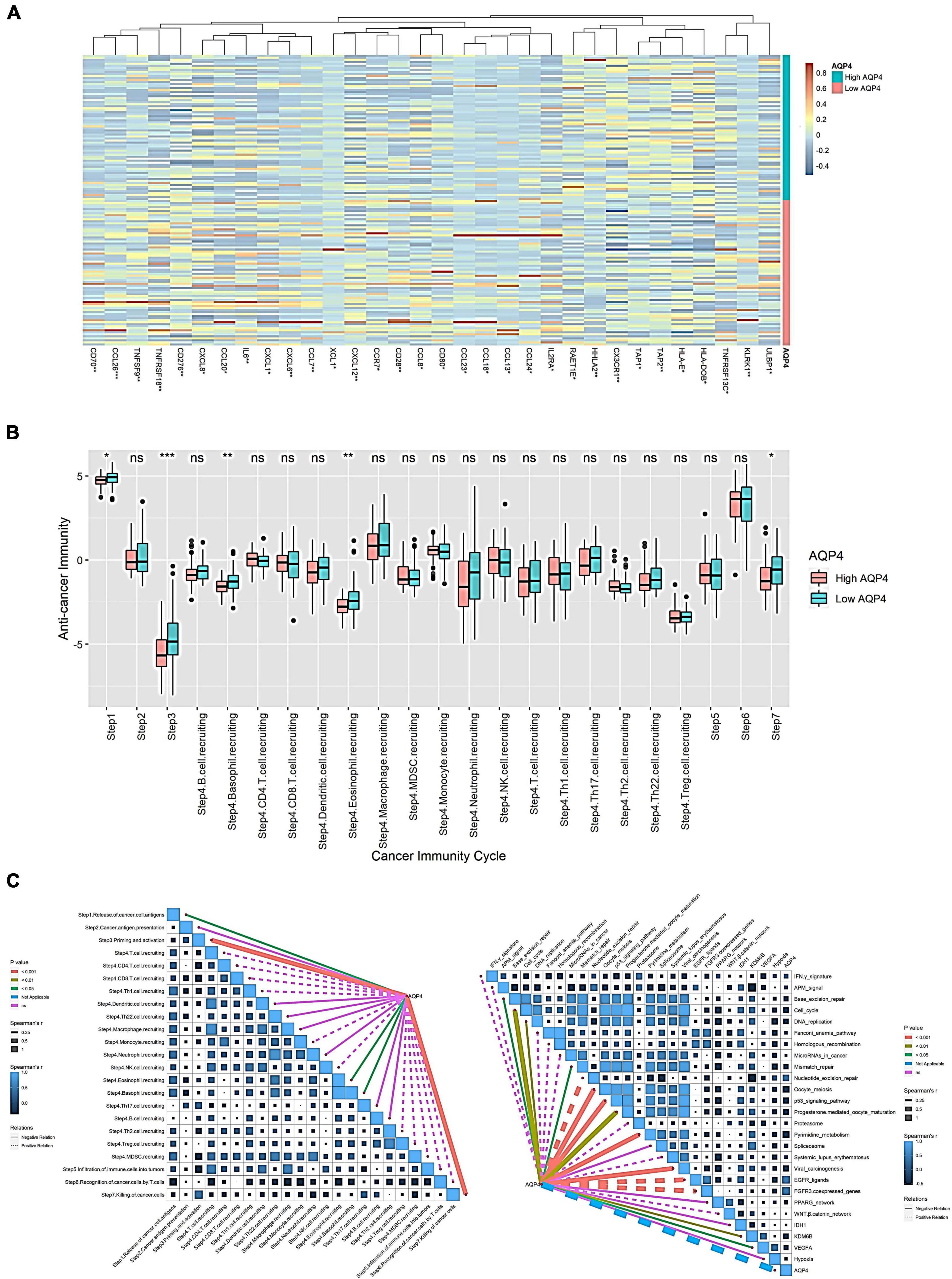
Figure 3. Effect of AQP4 expression level on immune cell anti-tumor immune process. (A) Differential gene heatmap of immunomodulator-related genes in high and low AQP4 expression groups. (B) The anti-cancer immune process activity was grouped differently in the high and low expression of the AQP4 gene. (C) Correlation analysis of AQP4 gene with immunotherapy-related predictive pathway enrichment score and anti-cancer immune process activity. *p < 0.05, **p < 0.01, ***p < 0.001.
We obtained 26 immunotherapy-related predictive pathways from the literature. The three groups with the strongest correlation in the anti-cancer immune process activity correlation analysis are Step 3 Priming and activation, Step 7 killing cancer cells, and Step 4 eosinophil recruiting, the correlations are −0.373, −0.288, and −0.186. The enrichment score was obtained by GSVA among the immunotherapy-related prediction pathways. Finally, we found that 14 pathways were significantly correlated with AQP4. The three groups with the strongest correlation were FGFR3-coexpressed genes, EGFR ligands, and pyrimidine metabolism, with correlations of 0.432, 0.422, and −0.366, respectively (Figure 3C and Supplementary Table 11).
Analysis of differential gene function, pathway enrichment, and tumor-related immune cell effector genes downstream of AQP4 gene in glioma
Among the biological process (BP) pathways, extracellular matrix organization, extracellular structure organization, external encapsulating structure organization, organic anion transport, and vascular process in the circulatory system have the most significant differences. The cellular component (CC) pathway showed substantial differences in the collagen-containing extracellular matrix, basolateral plasma membrane, basal plasma membrane, the basal section of the cell, and collagen trimer. In the MF pathway, the extracellular matrix structural constituents, monocarboxylic acid transmembrane transporter activity, carboxylic acid transmembrane transporter activity, organic acid transmembrane transporter activity, and growth factor binding had the most significant differences. Among the KEGG pathways, Proximal tubule bicarbonate reclamation, Bile secretion, Protein digestion and absorption, Gastric acid secretion, and AGE-RAGE signaling pathway in diabetic complications showed the most significant differences (Figures 4A–D and Supplementary Tables 12–15). The relationship between AQP4 gene levels and tumor-associated immune cell effector genes was also investigated (Figure 4E). Seven genes were significantly altered (p < 0.05) among the 35 immune cell effector genes. MARCO, SPON2, TBX21, MMP8, and SLAMF8 were among the genes with low expression in the AQP4 high expression group, while PRR5L and FGL2 were substantially expressed in the AQP4 high expression group (Figure 4E and Supplementary Table 16).

Figure 4. Analysis of differential gene functions, pathway enrichment, and tumor-related immune cell effector genes downstream of the AQP4 gene in glioma. (A–D) Differential gene function and pathway enrichment analysis of AQP4 gene between high and low-risk groups. (E) Heat map of the relationship between high and low AQP4 gene groups and tumor-associated immune cell effector genes. *p < 0.05, **p < 0.01.
Effects of AQP4 expression levels on tumor clinical features and immunotherapy targets
The AQP4 gene did not differ significantly in age, sex, IDH mutation, and MGMT. There were significant differences between C1 and C4 in immunophenotyping. There were significant differences in molecular typing between Classical and G-CIMP, Classical and Mesenchymal, Classical and Proneural, G-CIMP and Neural, Mesenchymal and Neural, and Neural and Proneural (Figure 5A). AQP4 gene expression was found to be substantially linked with microsatellite instability (MSI) but not with tumor mutational burden (TBI), immunological score, stromal score, or tumor purity (Figure 5B and Supplementary Table 17). In addition, we further analyzed the effect of AQP4 gene expression level on the enrichment score of immunotherapy prediction-related pathways, as well as differences in AQP4 gene expression among different immune response groups in the immune dataset (Figure 5C). According to the findings, only eight groups of immunotherapy predicted pathway enrichment scores were significantly different between the high and low expression groups of the AQP4 gene. Other immunological groups exhibited significant differences in progressive disease (PD) and partial response (PR)/complete response (CR) when GSE91061 was analyzed using the AQP4 gene (Figure 5C).
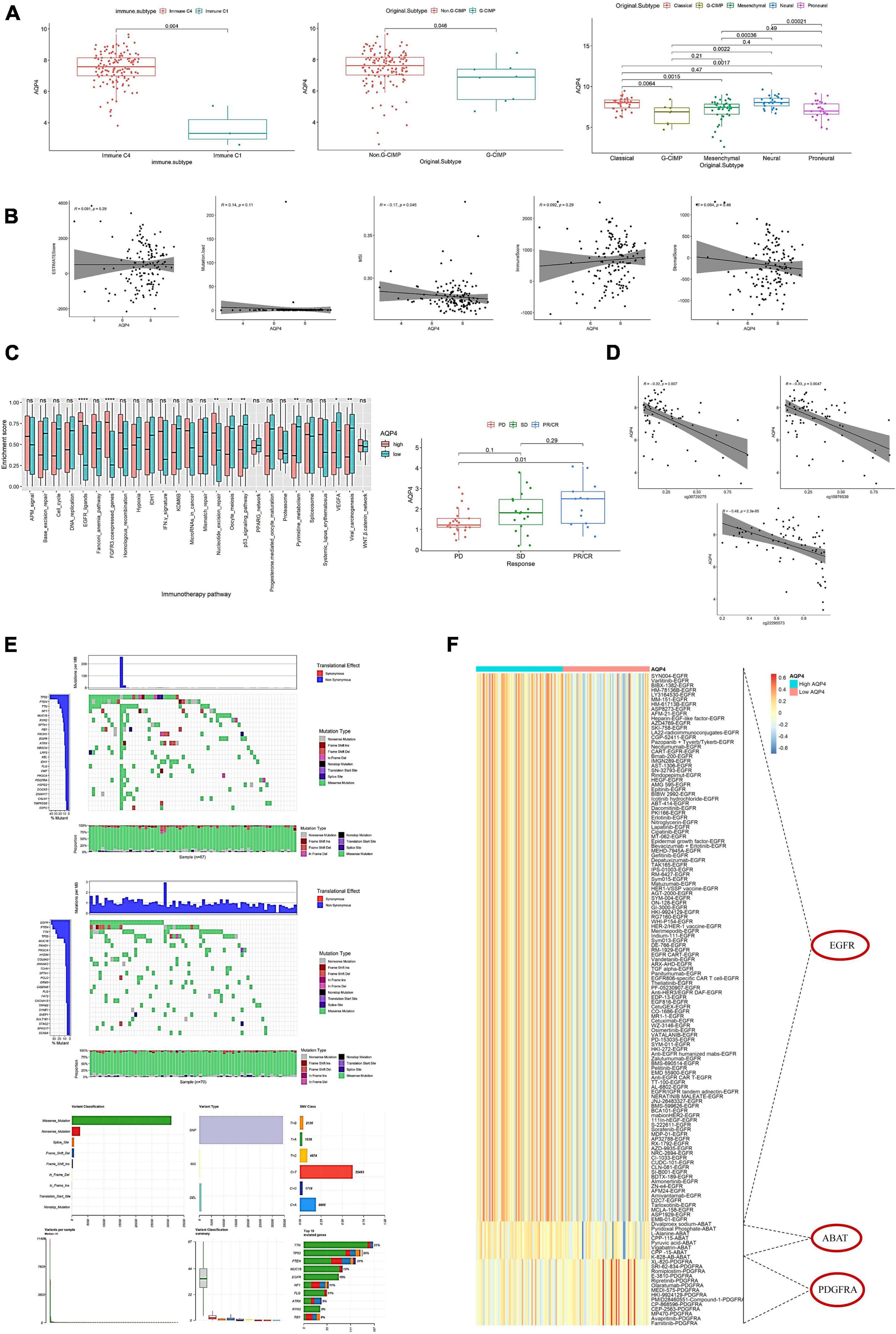
Figure 5. Effects of AQP4 expression levels on clinical tumor characteristics and immunotherapy targets. (A) Differential expression of AQP4 gene in clinical features. (B) Correlation of AQP4 gene expression with tumor mutational burden (TMB), immune score, MSI, stromal score, and tumor purity. (C) Differential analysis of pathway enrichment scores for immunotherapy prediction. The left figure shows the difference in the enrichment scores of immunotherapy prediction pathways between the high and low expression groups of the AQP4 gene, and the right figure shows the difference analysis of different immune groups in the GSE91061 dataset. (D) Methylation and expression correlation analysis. (E) AQP4 gene expression high and low grouped mutation display. The upper figure is the AQP4 low expression group, the middle figure is the AQP4 high expression group, and the lower figure is the summary of the maf file. (F) Screening for differential expression of cancer-type therapeutic targets. *p < 0.05, **p < 0.01, ****p < 0.0001.
The relationship between methylation and expression of the AQP4 gene was also investigated. AQP4 has three methylation sites, three of which have a strong relationship with the gene’s expression (Figure 5D). We also analyzed the mutation status in the high and low AQP4 gene expression groups and displayed the top 20 genes with mutation rates in the AQP4 low and high expression groups (Figure 5E). In addition, we evaluated AQP4-related therapeutic targets in GBM. To extract data for analysis, we used the TDD database. The findings revealed that three genes, EGFR, ABAT, and PDGFRA, significantly differed between high and low AQP4 expression groups in GBM (Figure 5F).
Construction of AQP4 gene-related clinical prognosis model
As a preliminary step, we searched for possible factors in the intersection of differential genes in the AQP4 gene high and low expression groups, the immunological score high and low group, and the matrix score group. The AQP4 gene high and low expression group had 291 differential genes, the immunological score high and low group had 883 divergent genes, the matrix score group had 792 differential genes, and the intersection of the three groups had 42 difference genes (Figures 6A–C and Supplementary Tables 18–20). Subsequently, we performed a univariate cox analysis on the 42 differential genes and obtained eight prognostic factors with significant differences (Figure 6D and Supplementary Table 21). In addition, duplicate elements were eliminated using LASSO dimensionality reduction to develop a useful predictive risk scoring model. After lasso dimension reduction, we obtained a model consisting of three genes from the eight prognostic factors. Then perform multi-factor screening on these three genes to get the final scoring model constructed by the three genes (Figure 6E and Supplementary Table 22), that is, Risk score = RARRES1*(0.06121847) + SOCS3*(0.16404808) + TTYH1*(−0.07449025).
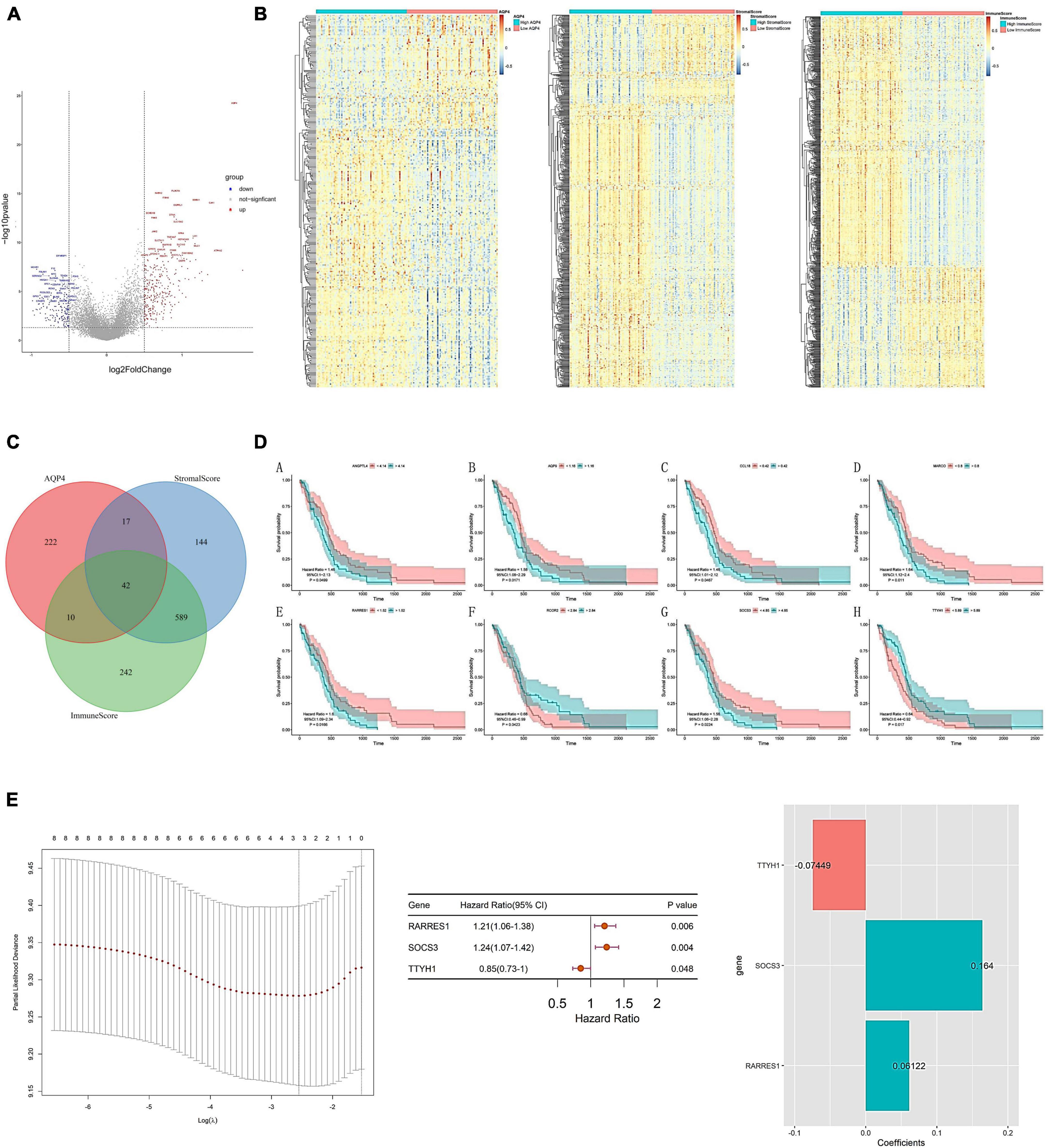
Figure 6. Construction of AQP4 gene-related clinical prognosis model. (A) Volcano plot of differential genes between AQP4 high and low expression groups. (B) The differential gene heat map of the high and low AQP4 gene expression group, the high and low immune score group, and the high and low matrix score group. (C) The intersection of differential genes in high and low AQP4 gene expression group, high and low immune score group, and high and low matrix score group. (D) Survival analysis of eight (A–H) prognostic factors. (E) Construction of a predictive risk scoring model.
Predictive efficacy evaluation of clinical prognostic models and validation on external datasets
We further evaluated the predictive power of the prognostic model. The high and low groups are divided according to the median value of the risk score of the prediction model. In evaluating the predictive model from the TCGA data, there was a significant difference in the survival analysis, with an AUC value of 0.774 for the ROC curve in the third year (Figures 7A–F). Following that, we validate the dataset with an external source. The survival analysis was significantly different across the two CGGA data sets, with AUC values of 0.602 and 0.695 for the 3-year ROC curves, respectively (Figures 7G–J).
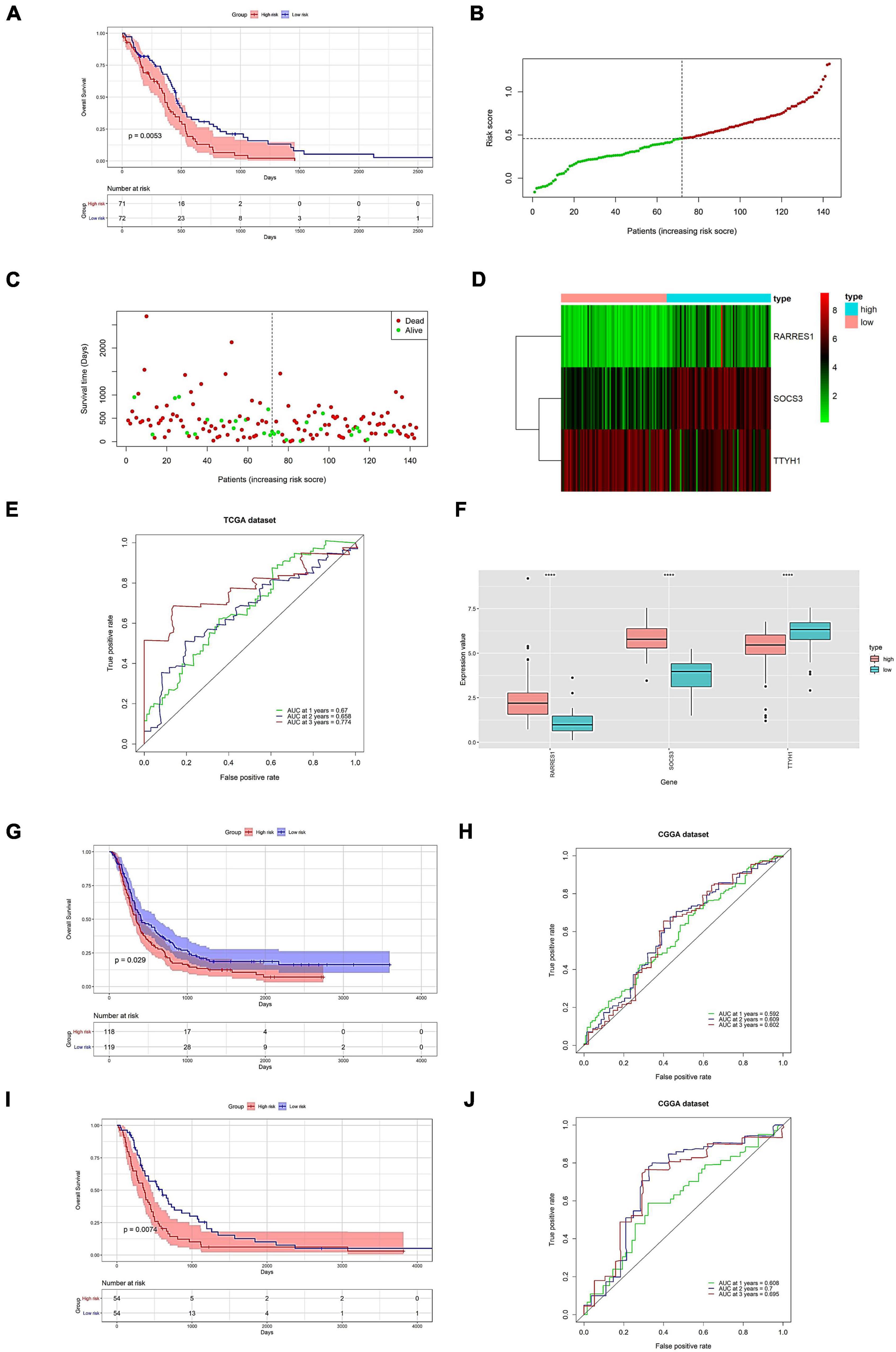
Figure 7. Prognostic model prediction efficacy assessment and validation on external datasets. Evaluation of predictive power (A–F). (A) Survival analysis of high and low expression groups. (B) Risk score distribution. (C) Distribution of patient survival. (D) The expression of the hub gene. (E) Predicted ROC curve for 1–3 years. (F) Model genes are shown differentially in high-risk and low-risk. Validation of prognostic models on external datasets (G–J). (G,I) Survival analysis of high and low expression groups. (H,J) Predicted 1-, 2-, and 3-year ROC curves. (G,H) Data for 693 samples in CGGA. (I,J) Data for 325 samples in CGGA. ****p < 0.0001.
Clinical information validates risk score as an independent prognostic factor
Initially, both univariate and multivariate independent prognostic assessments of risk scores revealed substantial differences (Figures 8A,B). Then, using a nomogram and clinical features, we created a clinical practice guide. In the constructed nomogram, risk score and age had a more significant impact on prognosis, and the combined AUC value of risk score and clinical features in the ROC curve was 0.777 (Figures 8C–G).
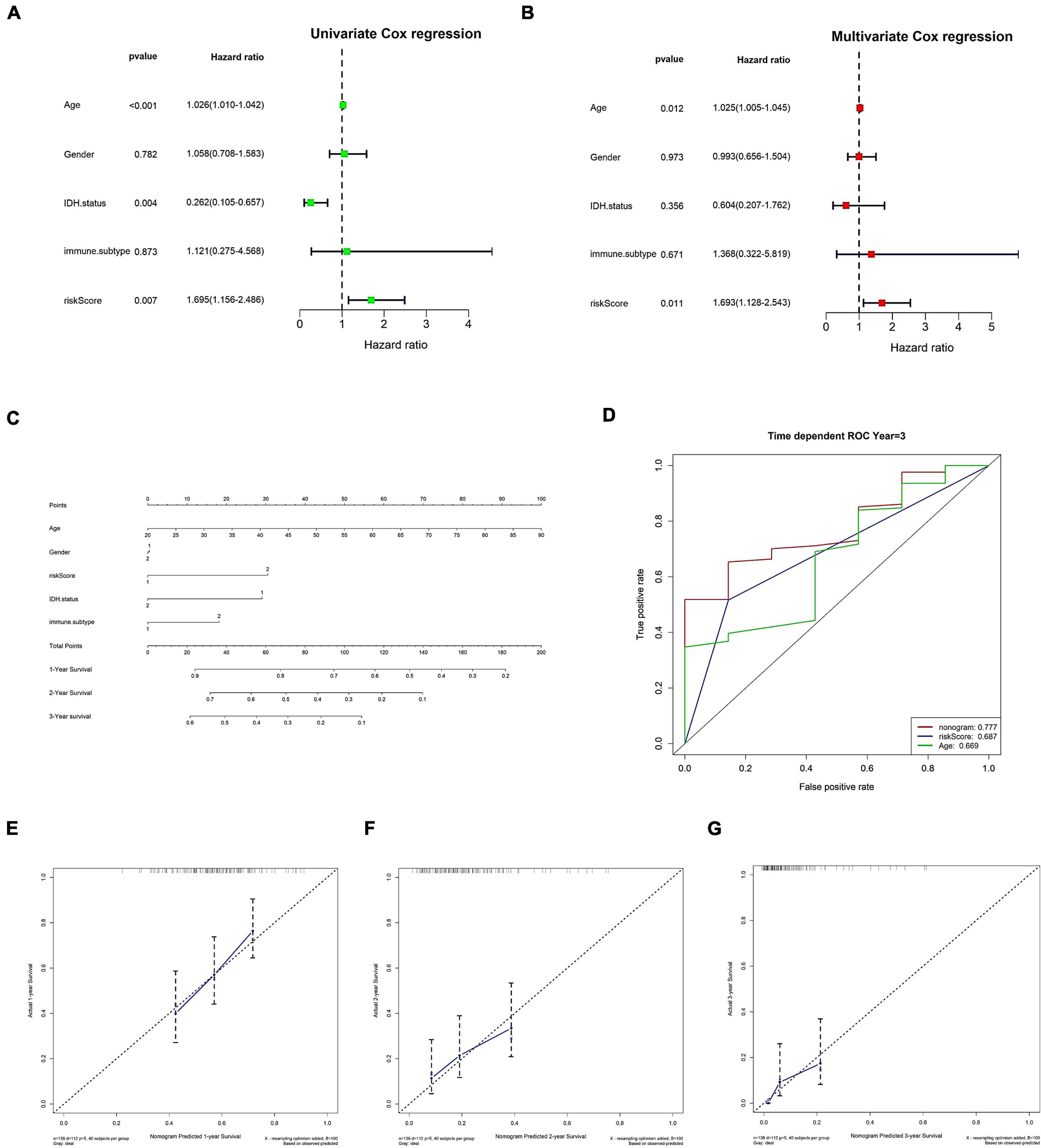
Figure 8. Clinical information was used to verify risk scores as an independent prognostic factor. (A,B) Independent prognostic factor analysis of risk scores. (A) Univariate Cox analysis. (B) Multivariate Cox analysis. (C–G) Nomograms. (C) Nomogram. (D) The nomogram’s 3-year risk score, stage, and overall ROC curve. (E–G) Calibration curves for 1, 2, and 3 years, respectively. In panel (C), immune C1 in the immune. The subtype is 1, and immune C4 is 2; in gender, the male is 1 and female is 2; in IDH mutation, WT is 1 and mutant is 2; in risk score, low is 1 and high is 2.
Discussion
One of the main characteristics of GBM is its great invasiveness of the surrounding nervous tissues, thus the investigation regarding certain proteins specifically expressed in the CNS, like the AQP4, should be more emphasized. Recent research indicate that GBM displays a pronounced genetic and biological heterogeneity between different tumors and even within the same tumor, which makes the tumor behavior highly variable and resistant to conventional therapy (Vitovcova et al., 2020). Thus, to find potential factors involved in tumor invasiveness and aggressiveness, it could be significantly important and informative to consider potential variation of molecular expression in human tumor samples. In the present study we evaluated the AQP4-related immunological pattern in various cancers, especially glioma. In particular, current bioinformatics analysis could be highly informative for further more in-depth research.
Previously, AQP4 was well-descripted as brain-specific regulator and in recent years, there has emerged an intriguing and surprising link between AQP4 and brain tumors (Ding et al., 2010; Yang et al., 2012; Zhao et al., 2012; Papadopoulos and Saadoun, 2015). Our previous research have also indicated the close involvement of AQP4 in glioma cell invasion and migration, as well as drug resistance (Lan et al., 2017). In our previous study, we have also confirmed that AQP4 was highly expressed in GBM tissues and that AQP4 could impact glioma patients’ overall survival (Lan et al., 2020). However, the roles of AQP4 in immunity have not been identified systematically. Current research provides a novel, vital AQP4-related immune status and prognostic model in glioma.
Tumor immunotherapy is a hot spot in the research field of various anti-tumor treatments (Chen and Mellman, 2017). The role of AQP4 in immune regulation has not yet been reported. In recent years, many reports have identified the occurrence of the lymphatic system within the CNS and its essential role in immune regulation in brain tumor therapy. Therefore, this study further explored the role of AQP4 in the immune regulation of glioma and the process of immunotherapy. Our study found that AQP4 was significantly associated with the expression of multiple immune checkpoints and immune cells, and the AQP4 expression level was negatively correlated with antigen presentation and processing capacity. Further, we combined literature reports and obtained enrichment scores through GSVA analysis and received the most relevant immunotherapy-related predictive pathways FGFR3 coexpressed genes, EGFR ligands, and pyrimidine metabolism. Furthermore, we found multiple immune cell effector genes significantly associated with AQP4 expression levels. Although no significant correlation was found between the AQP4 gene and IDH mutation and MGMT, AQP4 had considerable expression differences in different immunophenotypes and molecular types. In addition, we further analyzed AQP4-related immunotherapy targets through the TTD database and found that in the GBM classification, the therapeutic targets significantly associated with AQP4 expression were EGFR, ABAT, and PDGFRA. Finally, we screened the intersection of differential genes in the high and low AQP4 gene expression group, the high and low immune score group, and the high and low matrix score group as candidate factors. The candidate genes were subjected to univariate cox analysis to obtain eight prognostic factors with significant differences. A clinical prognosis model constructed by three genes (RARRES1, SOCS3, TTYH1) was obtained through lasso dimensionality reduction. Finally, combined with clinical information and cox regression analysis, it was further confirmed that the model score could be used as an independent prognostic factor. The results of this study have a significant reference value for the prognosis research of AQP4-related glioma patients, and have indicated that AQP4 could be considered as a potential new early biomarker of glioma progression (Frigeri et al., 2007; Du et al., 2020; Valente et al., 2022).
AQP4 protein is expressed as a particular morphological feature called orthogonal array of particles (OAPs). These structures are aggregates of the tetrameric unit (Valente et al., 2022). AQP4 protein is expressed as two major isoforms that differ in regard to methionine (M) starting codon. The shorter and more abundant form is called M23 and the longer and less abundant form is called M1 (Amiry-Moghaddam, 2019). The role of AQP4 isoforms in GBM biology has been addressed in earlier studies, which have shown that high-grade gliomas display higher expression of AQP4 than low-grade tumors (Warth et al., 2007). Furthermore, OAPs have been shown to be disintegrated or absent in GBM, and more recently an inverse correlation between OAP prevalence and malignancy was demonstrated (Fallier-Becker et al., 2016). The study by Simone et al. (2019) found that M1-AQP4 contributes to the invasiveness of glioma cells, while aggregation in OAPs by M23-AQP4 is deleterious and promotes apoptosis, interestingly indicating that the increased invasiveness was because of the increased activity of matrix metalloproteinase-9 (MMP9), which is associated with glioma cell proliferation and patient survival rate (Xue et al., 2017). Besides, AQP4 protein is expressed in different isoforms: two canonical M23 and M1 and two extended M23ex and M1ex, which influence expression, function and assembly in OAPs (Jin et al., 2011; Pisani et al., 2011; De Bellis et al., 2014). The extended isoforms are generated by the translational read through mechanism and are expressed in human CNS. Interestingly, Palazzo et al. (2019) found that research of AQP4ex-KO mice revealed that AQP4ex is indispensable to anchor AQP4 protein at the perivascular astrocytic end foot membrane domains. Indeed, large OAPs made of M1 and M23 canonical isoforms, still abundantly expressed in the AQP4ex mouse, are delocalized and confined at the astrocytic processes facing the brain neuropile. Thus AQP4ex may be supposed to be involved in the AQP4 alteration observed in the GBM (Valente et al., 2022). In the study of Valente et al. (2022), they evaluated the difference of expression and spatial distribution of AQP4 in grossly tumoral, peritumoral or non-tumoral brain regions. All these suggest that the AQP4ex isoform is critical in the triggering event of progressive downregulation and mislocalization of AQP4 in GBM, which may affect the integrity of the BBB, indicating that AQP4ex could be a potential early biomarker of GBM progression.
Currently, although kinds of immunotherapies have achieved remarkable success in cancer treatment, only limited number of patients could exhibit long-lasting anti-tumor response, where tumor immune infiltration status played a significant role (Chen et al., 2017). Identification of cancer patients with abundant infiltration of immune cells is of great importance to screen out potential candidates for immunotherapy. Our results of GBM and LGG cohorts highlighted immune-related GO and KEGG pathways in AQP4 low- and high-expression groups, which along with results of the estimated immune infiltration level based on five algorithms could contribute to the distinction of “cold” and “hot” tumors. Our study then examined the immunological pattern of AQP4 in gliomas, as well we the potential prognostic value of the AQP4-related signatures in the response of commonly used drugs and drug resistance of chemotherapy in different databases. Although AQP4-related prognostic model was consolidated with different public datasets, some more prospective and updated data are still necessary, and many genes for other traits that are of prognostic value may be excluded from the present study. We can conclude that AQP4 could play a key role in the malignant proliferation and immunological regulation of human brain tumor. Besides, the barrier losing its normal microenvironment conformation may also be involved in the accumulation of edema in the peritumoral tissue (Valente et al., 2022). Thus AQP4 could be considered as a potential new early biomarker of GBM progression (Frigeri et al., 2007; Du et al., 2020; Valente et al., 2022). The results presented in this study that could serve as a basis for increased research interest and warrant more detailed exploration. Recognition of AQP4-related therapy may open a new avenue for developing more specific targeted treatment for brain cancers. Our study provides robust evidence supporting AQP4 as a new candidate for cancer treatment. Despite the prognostic value of the signature, this study still encountered several limitations which must be considered. Our report was retrospective and based on public databases, devoid of certain crucial clinicopathological information. Further biochemical experiments need to be conducted to confirm the findings. Furthermore, more work should be done to find factors determining AQP4 protein relocalization and AQP4 aggregation state in the near future, and this would accelerate definitive evaluation of the role of AQP4 in the treatment of glioma and various other neurological diseases.
Conclusion
The promoting role of AQP4 in GBM cell invasion still need more research. Currently the significant roles of AQP4 in combating drug resistance during glioma chemotherapy, as well as the potential AQP4 pharmacological blockers require additional research. We should pay close attention to the various unresolved questions regarding AQP4 functions in brain tumors and various other CNS neurological diseases. It is noteworthy that most findings presented in this research are based on the bioinformatics techniques. Although the observations summarized in current study should be confirmed with more in-depth research, we believe that they could be critically informative for the design of more focused research in our future research, which will lead to definitive evaluation of the role of AQP4 in the treatment of glioma and various other neurological diseases. For the first time, current research has examined the role of AQP4 in the CNS immune system and find out how important it is in the glioma immunotherapy process. More importantly, we anticipated developing an AQP4-related prognostic model, which would serve as a critical theoretical research foundation for improving the effect of glioma immunotherapy. More effort should be directed toward clarifying the newly discovered functions and molecular mechanisms of AQP4 in malignant gliomas. Furthermore, more in-depth explorations should be done to elucidate the roles of AQP4 protein relocalization in glioma in our future research.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
Y-LL and SZ contributed to conception, design, data processing, analysis, and interpretation. TN collected the data. SZ contributed to revision of the manuscript. All authors contributed to the manuscript and approved the submitted version.
Funding
This work was supported by grants from National Natural Science Foundation of China (No. 82103480) and Zhejiang Provincial Natural Science Foundation (No. LQ22H090018).
Acknowledgments
We acknowledge TCGA and CGGA database for providing their platform and contributors for uploading their meaningful datasets.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2022.1061428/full#supplementary-material
Supplementary Table 1 | Twenty immune checkpoints used in current research.
Supplementary Table 2 | The correlation analysis between the gene AQP4 and 20 immune checkpoints.
Supplementary Table 3 | The correlation analysis between the gene AQP4 and 20 immune checkpoints (adjusted).
Supplementary Table 4 | The original data regarding the connection between AQP4 gene and 22 immune invading cells.
Supplementary Table 5 | The connection between AQP4 gene and 22 immune invading cells.
Supplementary Table 6 | The connection between AQP4 gene and 22 immune invading cells (adjusted).
Supplementary Table 7 | The original data regarding the association between the gene AQP4 and immune cells in various cancers.
Supplementary Table 8 | The association between the gene AQP4 and immune cells in various cancers.
Supplementary Table 9 | One hundred and twenty two genes collected from literature that potentially affects immune cells’ ability to present and process antigens.
Supplementary Table 10 | The original data regarding the association between various immunological mechanisms and AQP4 gene expression.
Supplementary Table 11 | Fourteen immunotherapy-related predictive pathways that could be significantly correlated with AQP4 expression.
Supplementary Table 12 | The original data regarding the analysis of biological process downstream of AQP4 gene in glioma.
Supplementary Table 13 | The original data regarding the analysis of cellular components downstream of AQP4 gene in glioma.
Supplementary Table 14 | The original data regarding the analysis of molecular functions downstream of AQP4 gene in glioma.
Supplementary Table 15 | The original data regarding the analysis of KEGG pathways downstream of AQP4 gene in glioma.
Supplementary Table 16 | The original data regarding the association between tumor-associated immune cell effector genes and AQP4 gene levels.
Supplementary Table 17 | The original data regarding the association between AQP4 expression levels and tumor clinical features.
Supplementary Table 18 | The original data regarding the differential genes between the gene AQP4 high and low expression groups.
Supplementary Table 19 | The original data regarding the divergent genes between the immunological score high and low groups.
Supplementary Table 20 | The original data regarding the differential genes between the matrix score high and low groups.
Supplementary Table 21 | AQP4 gene-related eight prognostic factors with great significance upon univariate cox analysis.
Supplementary Table 22 | AQP4 gene-related three genes constructing the final scoring model upon multi-factor screening.
Footnotes
References
Amiry-Moghaddam, M. (2019). AQP4 and the fate of gliomas. Cancer Res. 79, 2810–2811. doi: 10.1158/0008-5472.CAN-19-1185
Behnam, M., Motamedzadeh, A., Aalinezhad, M., Dadgostar, E., Rashidi Noshabad, F. Z., Pourfridoni, M., et al. (2022). The role of aquaporin 4 in brain tumors: Implications for pathophysiology, diagnosis and therapy. Mol. Biol. Rep. 49, 10609–10615. doi: 10.1007/s11033-022-07656-y
Chen, D. S., and Mellman, I. (2017). Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330. doi: 10.1038/nature21349
Chen, Y., Gao, F., Jiang, R., Liu, H., Hou, J., Yi, Y., et al. (2017). Down regulation of AQP4 expression via p38 MAPK signaling in temozolomide induced glioma cells growth inhibition and invasion impairment. J. Cell. Biochem. 118, 4905–4913. doi: 10.1002/jcb.26176
Chi, Y., Fan, Y., He, L., Liu, W., Wen, X., Zhou, S., et al. (2011). Novel role of aquaporin-4 in CD4+ CD25+ T regulatory cell development and severity of Parkinson’s disease. Aging Cell 10, 368–382. doi: 10.1111/j.1474-9726.2011.00677.x
De Bellis, M., Pisani, F., Mola, M. G., Basco, D., Catalano, F., Nicchia, G. P., et al. (2014). A novel human aquaporin-4 splice variant exhibits a dominant-negative activity: A new mechanism to regulate water permeability. Mol. Biol. Cell. 25, 470–480. doi: 10.1091/mbc.E13-06-0331
Ding, T., Gu, F., Fu, L., and Ma, Y. J. (2010). Aquaporin-4 in glioma invasion and an analysis of molecular mechanisms. J. Clin. Neurosci. 17, 1359–1361. doi: 10.1016/j.jocn.2010.02.014
Du, L., Xing, Z., Tao, B., Li, T., Yang, D., Li, W., et al. (2020). Both IDO1 and TDO contribute to the malignancy of gliomas via the Kyn-AhR-AQP4 signaling pathway. Signal. Transduct. Target. Ther. 5:10. doi: 10.1038/s41392-019-0103-4
Fallier-Becker, P., Nieser, M., Wenzel, U., Ritz, R., and Noell, S. (2016). Is upregulation of aquaporin 4-M1 isoform responsible for the loss of typical orthogonal arrays of particles in astrocytomas? Int. J. Mol. Sci. 17:1230. doi: 10.3390/ijms17081230
Frigeri, A., Nicchia, G. P., and Svelto, M. (2007). Aquaporins as targets for drug discovery. Curr. Pharm. Des. 13, 2421–2427. doi: 10.2174/138161207781368738
Hubbard, J. A., Szu, J. I., and Binder, D. K. (2018). The role of aquaporin-4 in synaptic plasticity, memory and disease. Brain Res. Bull. 136, 118–129.
Jin, B. J., Rossi, A., and Verkman, A. S. (2011). Model of aquaporin-4 supramolecular assembly in orthogonal arrays based on heterotetrameric association of M1-M23 isoforms. Biophys. J. 100, 2936–2945. doi: 10.1016/j.bpj.2011.05.012
Kahlert, U. D., Nikkhah, G., and Maciaczyk, J. (2013). Epithelial-to-mesenchymal(-like) transition as a relevant molecular event in malignant gliomas. Cancer Lett. 331, 131–138. doi: 10.1016/j.canlet.2012.12.010
Lan, Y. L., Chen, C., Wang, X., Lou, J. C., Xing, J. S., Zou, S., et al. (2020). Gamabufotalin induces a negative feedback loop connecting ATP1A3 expression and the AQP4 pathway to promote temozolomide sensitivity in glioblastoma cells by targeting the amino acid Thr794. Cell Prolif. 53:e12732. doi: 10.1111/cpr.12732
Lan, Y. L., Wang, H., Chen, A., and Zhang, J. (2022). Update on the current knowledge of lymphatic drainage system and its emerging roles in glioma management. Immunology Epub ahead of print. doi: 10.1111/imm.13517
Lan, Y. L., Wang, X., Lou, J. C., Ma, X. C., and Zhang, B. (2017). The potential roles of aquaporin 4 in malignant gliomas. Oncotarget 8, 32345–32355.
Li, X., Qi, L., Yang, D., Hao, S., Zhang, F., Zhu, X., et al. (2022). Meningeal lymphatic vessels mediate neurotropic viral drainage from the central nervous system. Nat. Neurosci. 25, 577–587. doi: 10.1038/s41593-022-01063-z
Mader, S., and Brimberg, L. (2019). Aquaporin-4 water channel in the brain and its implication for health and disease. Cells 8:90.
Nagelhus, E. A., and Ottersen, O. P. (2013). Physiological roles of aquaporin-4 in brain. Physiol. Rev. 93, 1543–1562. doi: 10.1152/physrev.00011.2013
Palazzo, C., Buccoliero, C., Mola, M. G., Abbrescia, P., Nicchia, G. P., Trojano, M., et al. (2019). AQP4ex is crucial for the anchoring of AQP4 at the astrocyte end-feet and for neuromyelitis optica antibody binding. Acta Neuropathol. Commun. 7:51. doi: 10.1186/s40478-019-0707-5
Papadopoulos, M. C., and Saadoun, S. (2015). Key roles of aquaporins in tumor biology. Biochim. Biophys. Acta 1848, 2576–2583. doi: 10.1016/j.bbamem.2014.09.001
Papadopoulos, M. C., and Verkman, A. S. (2012). Aquaporin 4 and neuromyelitis optica. Lancet Neurol. 11, 535–544. doi: 10.1016/S1474-4422(12)70133-3
Peng, Y., Wu, W., Shang, Z., Li, W., and Chen, S. (2020). Inhibition of lncRNA LINC00461/miR-216a/aquaporin 4 pathway suppresses cell proliferation, migration, invasion, and chemoresistance in glioma. Open Life Sci. 15, 532–543. doi: 10.1515/biol-2020-0048
Pisani, F., Rossi, A., Nicchia, G. P., Svelto, M., and Frigeri, A. (2011). Translational regulation mechanisms of aquaporin-4 supramolecular organization in astrocytes. Glia 59, 1923–1932.
Simone, L., Pisani, F., Mola, M. G., De Bellis, M., Merla, G., Micale, L., et al. (2019). AQP4 aggregation state is a determinant for glioma cell fate. Cancer Res. 79, 2182–2194.
Smith, A. J., and Verkman, A. S. (2015). Superresolution imaging of aquaporin-4 cluster size in antibody-stained paraffin brain sections. Biophys. J. 109, 2511–2522. doi: 10.1016/j.bpj.2015.10.047
Smith, H. L., Wadhwani, N., and Horbinski, C. (2022). Major features of the 2021 WHO classification of CNS tumors. Neurotherapeutics Epub ahead of print. doi: 10.1007/s13311-022-01249-0
Valente, O., Messina, R., Ingravallo, G., Bellitti, E., Zimatore, D. S., de Gennaro, L., et al. (2022). Alteration of the translational readthrough isoform AQP4ex induces redistribution and downregulation of AQP4 in human glioblastoma. Cell. Mol. Life Sci. 79:140. doi: 10.1007/s00018-021-04123-y
Vitovcova, B., Skarkova, V., Rudolf, K., and Rudolf, E. (2020). Biology of glioblastoma multiforme-exploration of mitotic catastrophe as a potential treatment modality. Int. J. Mol. Sci. 21:5324. doi: 10.3390/ijms21155324
Warth, A., Simon, P., Capper, D., Goeppert, B., Tabatabai, G., Herzog, H., et al. (2007). Expression pattern of the water channel aquaporin-4 in human gliomas is associated with blood-brain barrier disturbance but not with patient survival. J. Neurosci. Res. 85, 1336–1346. doi: 10.1002/jnr.21224
Xue, Q., Cao, L., Chen, X. Y., Zhao, J., Gao, L., Li, S. Z., et al. (2017). High expression of MMP9 in glioma affects cell proliferation and is associated with patient survival rates. Oncol. Lett. 13, 1325–1330. doi: 10.3892/ol.2017.5567
Yang, L., Wang, X., Zhen, S., Zhang, S., Kang, D., and Lin, Z. (2012). Aquaporin-4 upregulated expression in glioma tissue is a reaction to glioma-associated edema induced by vascular endothelial growth factor. Oncol. Rep. 28, 1633–1638. doi: 10.3892/or.2012.1973
Zhao, W. J., Zhang, W., Li, G. L., Cui, Y., Shi, Z. F., and Yuan, F. (2012). Differential expression of MMP-9 and AQP4 in human glioma samples. Folia Neuropathol. 50, 176–186.
Keywords: aquaporin 4, glioma, immune, target, treatment
Citation: Lan Y-L, Nie T and Zou S (2022) Identification of the prognostic and immunological roles of aquaporin 4: A potential target for survival and immunotherapy in glioma patients. Front. Cell. Neurosci. 16:1061428. doi: 10.3389/fncel.2022.1061428
Received: 04 October 2022; Accepted: 16 November 2022;
Published: 29 November 2022.
Edited by:
Jenny Szu, Saint John’s Health Center, United StatesReviewed by:
Jiacheng Lou, The Second Affiliated Hospital of Dalian Medical University, ChinaXun Wang, Northern Theater General Hospital, China
Copyright © 2022 Lan, Nie and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Long Lan, bGFueXVsb25nQHpqdS5lZHUuY24=; Shuang Zou, em91c2h1YW5nMjAxN0AxNjMuY29t
 Yu-Long Lan
Yu-Long Lan Tian Nie5
Tian Nie5