- 1Shandong Collaborative Innovation Center for Diagnosis, Treatment and Behavioral Interventions of Mental Disorders, Institute of Mental Health, Jining Medical University, Jining, China
- 2Shandong Key Laboratory of Behavioral Medicine, School of Mental Health, Jining Medical University, Jining, China
- 3Research Center for Quality of Life and Applied Psychology, School of Humanities and Management, Guangdong Medical University, Dongguan, China
The ubiquitin-proteasome system (UPS) mediated protein degradation is crucial to maintain quantitive and functional homeostasis of diverse proteins. Balanced cellular protein homeostasis controlled by UPS is fundamental to normal neurological functions while impairment of UPS can also lead to some neurodevelopmental and neurodegenerative disorders. Functioning as the substrate recognition component of the SCF-type E3 ubiquitin ligase, FBXW7 is essential to multiple aspects of cellular processes via targeting a wide range of substrates for proteasome-mediated degradation. Accumulated evidence shows that FBXW7 is fundamental to neurological functions and especially implicated in neurodevelopment and the nosogenesis of neurodegeneration. In this review, we describe general features of FBXW7 gene and proteins, and mainly present recent findings that highlight the vital roles and molecular mechanisms of FBXW7 in neurodevelopment such as neurogenesis, myelination and cerebral vasculogenesis and in the pathogenesis of some typical neurodegenerative disorders such as Alzheimer’s disease, Parkinson’s disease and Huntington’s disease. Additionally, we also provide a prospect on focusing FBXW7 as a potential therapeutic target to rescue neurodevelopmental and neurodegenerative impairment.
Introduction
Proteolysis plays critical roles in diverse cellular processes including cell division, growth, differentiation and senescence. The ubiquitin–proteasome system (UPS) spatially and temporally controls a vast majority of protein degradation (Pohl and Dikic, 2019). Proteasomal degradation pathway is regulated by targeted ubiquitylation which undergoes a multi-step process participated by three key enzymes: an ubiquitin-activating enzyme (E1), an ubiquitin-conjugating enzyme (E2) and an ubiquitin ligase (E3). Ubiquitin, an evolutionally conserved protein of 76 amino acids is firstly activated by E1 with ATP causing a thioester bond between E1 and ubiquitin. Then, the ubiquitin is transferred to E2, and sequentially covalently binds to the ε-amino group of specific lysine residue on target protein by E3 (Li et al., 2018). E3 ligase determines the specificity of the ubiquitylation to target protein, and estimated over 600 E3 ligase genes are identified in human genome (Li et al., 2008).
UPS play vital roles in maintaining neurological functions while dysregulation and dysfunction of UPS components are involved in several neurodevelopmental and neurodegenerative disorders. For example, mutations, deletions, and duplications of UBE3A E3 ligase gene can lead to three human neurodevelopmental disorders: Prader-Willi syndrome (PWS), Angelman syndrome (AS) and Dup15q syndrome (LaSalle et al., 2015). E3 ligase HERC1 (regulator of chromosome condensation 1-like domain-containing protein 1) deficiency presents with delayed and abnormal brain development in mouse model (Bachiller et al., 2015), and patients with HERC1 mutations present with thicker corpus callosum, seizures, intellectual disability, and other autism-resembling clinical symptoms (Ortega-Recalde et al., 2015; Aggarwal et al., 2016; Utine et al., 2017). Meanwhile, neurodegenerative diseases which can be commonly featured with aberrant aggregation of neurotoxic proteins in the central nervous system (CNS) have also been widely recognized to be associated with impairment of ubiquitin-proteasome system (McKinnon and Tabrizi, 2014; Harrigan et al., 2018). These evidence highlight the possibility that UPS components especially specific E3 ligases may be valid therapeutic targets for the treatment of neurodevelopmental and neurodegenerative disorders.
Broadly, E3 ligases are typically grouped into three major classes: the homologous to the E6AP carboxyl terminus (HECT) domain containing E3s, the really interesting new gene (RING) domain containing E3s and RING-between-RING (RBR) family E3s (Berndsen and Wolberger, 2014). Cullin-RING ubiquitin ligases (CRLs) belong to RING type E3 ligases. CRL1, also termed as the Skp1-Cullin 1-F-box protein (SCF) complex, is the most studied member among CRLs. The SCF complex consists of the S-phase kinase-associated protein 1 (Skp1), ring-box 1 (Rbx1), and Cullin 1 (Cul1), as well as a variable F-box protein which is responsible for substrate recognition (Figure 1). F-box and WD repeat domain containing 7 (FBXW7), also known as FBW7, AGO, hCDC4, and SEL-10, is one of the F-box proteins composing SCF type of E3 ubiquitin ligases (Shimizu et al., 2018). FBXW7 has been well studied for its crucial suppressive roles in tumorigenesis (Yumimoto and Nakayama, 2020). Meanwhile, mounting studies have focused on its functional roles in nervous system, especially suggesting that FBXW7 is crucial to neurodevelopment and neurodegeneration. In this review, we present evidence on the functional implications of FBXW7 in crucial neurodevelopmental processes and in the pathogenesis of some neurodegenerative disorders and also discuss the critical issues for drug development by targeting FBXW7, aiming to propose the therapeutic potential of targeting FBXW7 to ameliorate neurodevelopmental and neurodegenerative impairments.
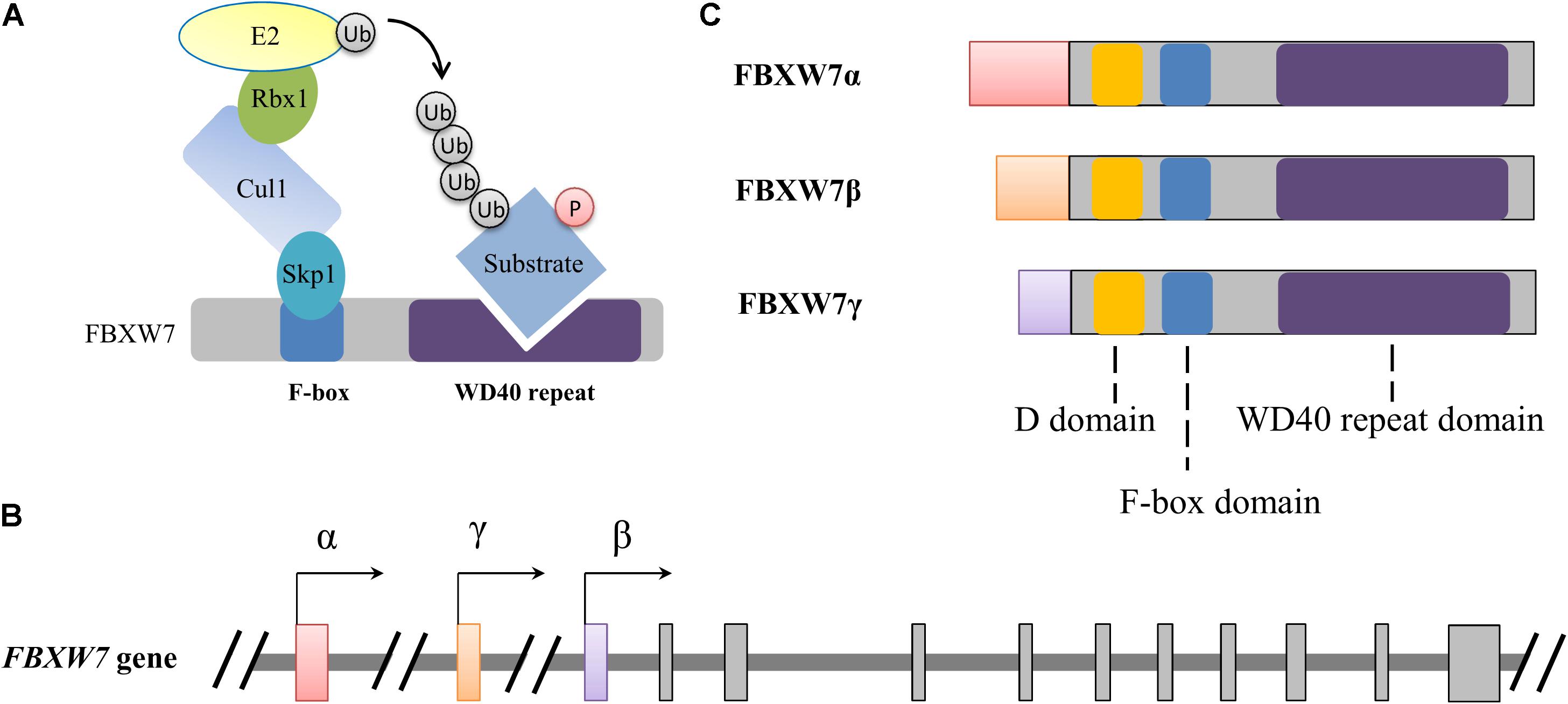
Figure 1. Schematic illustration of the SCFFBXW7 E3 ubiquitin ligase complex (A) and FBXW7 gene (B) and proteins (C). (A) Functional model of FBXW7 involved in SCF complex-mediated substrate ubiquitylation. Ubiquitin and phosphorylation is indicated with Ub and P with a circle, respectively. (B,C) Alternative transcriptional initiations from different promoters generate three distinct FBXW7 transcripts and corresponding proteins with conserved D, F-box and WD40 repeat domains. All three FBXW7 transcripts (or isoforms) share 10 common exons which were shown with gray rectangles.
The FBXW7 Gene and Proteins
The FBXW7 gene, spanning 216,330 bp of genomic DNA is located on chromosome 4q31.3. Alternative transcriptional initiations from different promoters and selective splicing generate three distinct FBXW7 transcripts (Spruck et al., 2002). These transcripts share 10 common exons which are responsible for the encoding of conserved C-terminal region of FBXW7 proteins (FBXW7α, FBXW7β, and FBXW7γ). Alternative transcription determines the distinguished distribution of the three subtypes in tissues: FBXW7α widely expresses in almost all tissues, FBXW7β only exists in the brain and testes, and FBXW7γ is mainly detected in the heart and skeletal muscle (Jin et al., 2004; Matsumoto et al., 2006). All these isoforms share three crucial functional domains: a D domain mediating FBXW7 dimerization, a F-box domain interacting with the SKP1-CUL1 complex, and a tryptophan-aspartic acid 40 (WD40)-repeat domain which is responsible for substrate recognition and binding (Figure 1). The distinct N-terminal sequence of each isoform determines subcellular localization, with FBXW7α primarily in the nucleoplasm, FBXW7β in cytoplasm (or more precisely on the endoplasmic reticulum membrane), and FBXW7γ in the nucleolus (Welcker et al., 2004; Matsumoto et al., 2011b). As the result of wide distribution and dominant level of FBXW7α, plenty of known functions of FBXW7 are attributable to the α isoform while the β and γ subtypes may also irreplaceably contribute distinct roles in some specific physiological processes (Matsumoto et al., 2011b; Welcker et al., 2011; Xu et al., 2020).
Regulation of FBXW7 Expression
The expression of FBXW7 is regulated at transcriptional, translational and post-translational levels. The CCAAT/enhancer binding protein-δ (C/EBPδ), an inflammatory response transcription factor, targets the FBXW7α promoter and directly inhibits FBXW7α transcription (Balamurugan et al., 2010). A functional p53-binding site was also identified in the 1st exon of β transcript of FBXW7 gene, and p53 was confirmed to directly promote FBXW7β transcription (Kimura et al., 2003). Similarly, bHLH transcription factor 5 (HES5), a member of the HES family, was found to bind to FBXW7β promoter and suppress the FBXW7β transcription, although the specific binding sites have not been identified (Sancho et al., 2013; Chen et al., 2021). Additionally, the CpG sequences in the promoter region of FBXW7β were also proven to be methylated, leading to FBXW7β transcriptional suppression (Gu et al., 2008).
Multiple non-coding microRNAs (miRNAs) can modulate FBXW7 translation via interacting with the 3′ untranslated region of the mRNA. miR-24 (Zhao et al., 2016), miR-25 (Xiang et al., 2015; Hua et al., 2017; Peng et al., 2019), miR-27 (Wu et al., 2015; Liu Z. et al., 2018), miR-32 (Hua et al., 2016; Xia et al., 2017), miR-92 (Yang et al., 2015; Zhou et al., 2015), miR-129-5p (Hasler et al., 2012), miR-155-3p (Cao et al., 2016; Tang et al., 2016), miR-182 (Jeon et al., 2013; Li et al., 2014; Chang et al., 2018), miR-195-5p (Wang et al., 2019), miR-223 (Xu et al., 2010; Mansour et al., 2013), miR-367 (Xiao et al., 2017; Xu et al., 2017), miR-424 (miR-322 in mice) (Chen et al., 2019), miR-503 (Li et al., 2014), miR-544a (Liu X. et al., 2018), miR-586 (Zhang et al., 2016), and miR-1290 (Zhang et al., 2021) were shown to reduce FBXW7 protein level in different cancer cells. Adversely, some long non-coding RNAs (lncRNAs), such as MIF, TINCR, CASC2, MALAT1, and MT1JP can block the inhibition of FBXW7 expression acting as miRNA “sponges” (Cao et al., 2016; Wang et al., 2017; Liu X. et al., 2018; Zhang et al., 2018). Moreover, FBXW7 translation is also regulated by mRNA modification. N6-methyladenosine (m6A) modification mediated by METTL3 (methyltransferase-like 3) was proven to enhance FBXW7 translation (Wu et al., 2021).
Post-translational regulation of FBXW7 includes ubiquitination, phosphorylation and dimerization of the proteins. First, ubiquitination and deubiquitination of FBXW7 regulate its proteasomal degradation. COP9 signalosome complex subunit 6 promotes FBXW7 autoubiquitination and proteasome-mediated degradation. Ubiquitin specific peptidase 28 (USP28), a deubiquitinating enzyme can also repress autocatalytic ubiquitination and degradation of FBXW7 (Diefenbacher et al., 2014; Schulein-Volk et al., 2014). Moreover, the stability and function of FBXW7 are regulated by multiple kinases. For example, extracellular signal–regulated kinase (ERK) and Polo-like kinase 1 and 2 (PLK1 and PLK2) directly interact and thereby mediate phosphorylation of FBXW7, resulting in its ubiquitination and proteasomal degradation (Cizmecioglu et al., 2012; Ji et al., 2015; Xiao et al., 2016). In contrast, phosphorylation of FBXW7α by phosphoinositide 3-kinase (PI3K) and serumand glucocorticoid-regulated kinase 1 (SGK1) were demonstrated to inhibit its autocatalytic ubiquitin transfer and stabilization (Mo et al., 2011; Schulein et al., 2011). Besides, the dimerization mediated by the D domain in all three FBXW7 isoforms also affects the stability of FBXW7. Peptidylprolyl cis/trans isomerase NIMA-interacting 1 (Pin1) has been demonstrated to destabilize FBXW7 by repressing dimerization and thereby promoting FBXW7 autoubiquitination (Min et al., 2012). Similarly, FBXW7 monomers were found to be stable as dimerization could destabilize the protein because of accelerated autoubiquitylation (Welcker et al., 2013).
FBXW7 in Neurodevelopment
Mounting studies indicate the homeostasis of FBXW7 is crucial for neurodevelopment. In this section, we will present evidence of functional involvement of FBXW7 in neurodevelopment and review the underlying mechanisms of FBXW7 in the processes of neurogenesis, myelination, and cerebral vasculogenesis. Additionally, potential therapeutic effect on neurodevelopmental disorders treatment via targeting FBXW7 or its substrates is also discussed.
Structural and Functional Abnormalities in FBXW7-Deficient Mouse Brain
Mice with specific deletion of exon 5 of Fbxw7 in brain die in a short while after birth with substantial changes and morphological abnormities in brain structure. Neurogenesis was found to be defective while astrogenesis was enhanced, leading to the tendentious differentiation toward astrocytes in these conditional Fbxw7-deficient brain. These newborn Fbxw7-deficient mice also show defective suckling behavior which may be associated with the hypoplasia of the brain stem although the underlying cause of the defective behavior remains to be explored (Matsumoto et al., 2011a). Similarly, it was also reported that conditional inactivation of Fbxw7 in the nervous system resulted in severely defective stem cell differentiation and anabatic progenitor cell death. Neurospheres from Fbxw7 deficient embryos were generally smaller in size, and significantly lower in number (Hoeck et al., 2010). Moreover, conditional Fbxw7-knockout in the cerebellar anlage of mouse leaded to reduced Purkinje cell number, decreased cerebellar size and defects in axonal arborization. Fbxw7-deficient cerebella presented with reduced vermis size and aberrant migration of progenitor cells (Jandke et al., 2011). Besides, primary cultures of neurons prepared from the mice only lacking β isoform of Fbxw7 were more vulnerable to oxidative stress although rare morphological abnormalities exhibit in brain development (Matsumoto et al., 2011b).
FBXW7 and Neurogenesis
Initial expansion of the progenitor cell by symmetrical division and subsequent generation of differentiated cells such as neurons, astrocytes, and oligodendrocytes through asymmetrical division are crucial to brain development (de la Pompa et al., 1997; Lutolf et al., 2002). The functional implication of FBXW7 in brain development is crucially mediated by Notch and c-Jun, both of which are substrates of FBXW7. The Notch signaling pathway acts via a process of lateral inhibition to play a fundamental role in neuronal and glial differentiation. Dll1, a ligand of Notch can trigger Notch signaling and suppress the expression of the proneural genes via inducing Hes1 expression and thereby block neuronal differentiation (Kageyama et al., 2009). Notch signaling inactivation promotes premature neurogenesis, leading to exhaustion of the progenitor pool and decreased number of mature neurons (de la Pompa et al., 1997; Lutolf et al., 2002). FBXW7 regulates Notch protein stability in this process, thereby controlling the maintenance and differentiation orientation of neural stem cells. Notch accumulation caused by FBXW7 deficiency results in aberrant activation of Notch target genes, resulting to excessive proliferation of neural stem cells and aberrant differentiation toward the astrocytes (Matsumoto et al., 2011a). c-Jun is another important regulator of neuronal viability. Restraining of c-Jun activation prominently rescued the cellularity defect caused by Fbxw7 deletion in the mantle layer of the midbrain tectum. Likewise, compared to Fbxw7 single mutant, Fbxw7/Jun mutant cells exhibited substantially elevated neurosphere formation in vitro, accompanied by a considerable reduction of apoptotic cells in neurospheres, indicating that c-Jun–mediated cell death is functionally implicated in defective neurosphere formation under Fbxw7 deficiency background (Hoeck et al., 2010). Besides, it was also reported that deletion of c-Jun or specific abrogation of c-Jun N-terminal phosphorylation could rescue Purkinje cell numbers and arborization in the Fbxw7 knockout background indicating phosphorylated c-Jun is an important substrate of Fbxw7 in neurogenesis during cerebellar development (Jandke et al., 2011). WD repeat domain 62 (WDR62) is crucial to promoting c-Jun N-terminal kinase signaling in the control of neurogenesis (Wasserman et al., 2010; Cohen-Katsenelson et al., 2011). It was also reported that FBXW7 controls self-renewal and differentiation of neural progenitor cells (NPCs) during brain development by regulating WDR62 degradation (Xu et al., 2018).
FBXW7 and Myelination
Myelin, a specialized proteolipid-rich membrane surrounding neuronal axons, is crucial for axons protection and insulation. Myelination is a sophisticated solution for efficient conduction velocity of potential actions along axons. Myelin is formed by glial cells which are oligodendrocytes and Schwann cells in the central nervous system and peripheral nervous system, respectively.
During myelin development in central nervous system, specified oligodendrocyte precursor cells (OPCs) migrate to target axons before they begin to differentiate into premyelinating oligodendrocytes which wrap axons and synthesize multiple myelin proteins and lipids comprising the myelin sheath. Consequently, oligodendrocytes can form multitudinous myelin sheaths with substantial variability in lengths and thicknesses (Kessaris et al., 2006; Hughes et al., 2013; Hughes and Appel, 2019). Notch signaling mediated by Notch protein and its receptors plays a vital role in balancing development of neurons and glia. Dysfunction of Notch pathway in vertebrate embryos generally leads to reduced neural precursors, excessive early born neurons and a deficit of glial cells, oligodendrocytes included (Chitnis et al., 1995; de la Pompa et al., 1997; Park and Appel, 2003). It was reported that fbxw7 mutation leaded to excessive differentiation of neural precursors toward oligodendrocyte progenitor cells in zebrafish embryos nearly identical to that of the mutant with constitutive activation of Notch (Park and Appel, 2003). Hyperactive Notch signaling was found in fbxw7 mutant embryos while pharmacological inhibition of Notch proteins under fbxw7 mutant background inhibited formation of excess oligodendrocyte progenitors indicating that Notch signaling are functional target of Fbxw7 in the process of oligodendrocyte specification (Snyder et al., 2012). Similarly, mTOR (mammalian target of rapamycin) is another target of FBXW7 in regulating oligodendrocyte differentiation. mTOR signaling is also promoted in oligodendrocyte lineage cells of fbxw7 mutant zebrafish larvae. Both genetic and pharmacological inhibition of mTOR signaling are beneficial to rescue aberrant profiles of myelin genes caused by dysfunction of Fbxw7, indicating that mTOR is a functional target of Fbxw7 in oligodendrocytes (Kearns et al., 2015).
In peripheral nervous system, myelin is fundamentally developed from Schwann cells (SCs). Neural crest precursor cells initially proliferate and differentiate into SC precursors which then differentiate into immature SCs. The maturation of SC comes up around birth via the process of radial sorting, during which the cytoplasmic components of individual SCs extend into bundles of axons, progressively separates them into smaller bundles, and finally surrounds a single larger diameter axon (Martin and Webster, 1973; Webster et al., 1973; Chernousov et al., 2008). Meanwhile non-myelinating SCs will form into Remak bundles by ensheathing multiple small diameter axons (Jessen and Mirsky, 2005). Conditional knockout of Fbxw7 specifically in SC precursors at approximately embryonic day (E) 12.5 results in thicker myelin sheaths and a higher proportion of myelinated axons compared to control nerves. More intriguingly, Fbxw7 mutant SCs sometimes appear to myelinate multiple axons in a fashion reminiscent of oligodendrocytes. It is identified that Fbxw7 regulates mTOR to control SC number, myelination, and Remak bundle organization during myelination peripheral nervous system. The activation of c-Jun is also found in Fbxw7 mutant SCs while the potential role of c-Jun in regulating SCs needs further demonstration (Harty et al., 2019). These evidences indicate that FBXW7 functionally regulate plasticity of SCs during myelination and may be a beneficial target for myelin repair.
FBXW7 and Cerebral Vasculogenesis
FBXW7 also control vasculogenesis in brain. Apart from metabolic functions of ensuring adequate supply of oxygen and nutrients to maintain homeostasis of neuronal networks, vessels have also been considered to serve as niches and scaffolds for neuronal migration and expansion during brain development and neurogenesis (Attwell et al., 2010; Segarra et al., 2015, 2018). Fbxw7-null mice (Fbxw7–/– with disruptions of all three isoforms) die in utero at embryonic day around 10.5 as a result of impaired vascular development in the brain and yolk sac. Fbxw7–/–embryos shows defects of vessels along the entire length of their neural tubes, indicating FBXW7 is potentially fundamental for brain function by regulating construction of neurovascular architecture during development (Tetzlaff et al., 2004; Tsunematsu et al., 2004). However, the mechanism remains elusive at present.
FBXW7 and Neurodevelopmental Disorders
It is undoubted that FBXW7 and its substrates function in different vital neurodevelopmental processes, but the potential roles of FBXW7 in the pathogenesis of neurodevelopmental disorders still remain to be investigated. There is no FBXW7 mutation identified to be associated with any neurodevelopmental disorders at present, however, functional implications or mutations of its substrate have been widely reported in relevant diseases. For example, c-Jun was aberrantly increased in an autism mouse model (Engrailed-2 knockout) (Tripathi et al., 2009), and c-Jun activation was possibly involved in the autism via inducing disordered inflammatory response in the brain (Shimoyama et al., 2019; Bjorklund et al., 2020). Also, some neurodevelopmental disorders such as focal cortical dysplasias, tuberous sclerosis complex and syndromic autism spectrum disorder (ASD) are thought to arise due to the effects of mTOR mutations during fetal development (Sato, 2016; Iffland and Crino, 2017; Salussolia et al., 2019). Similarly, mutations in Notch 3 can also lead to cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (Mizuno et al., 2020). Considering functional implications of FBXW7 in neurodevelopment, it may be beneficial to explore effective methods via targeting FBXW7 or its substrate for treatment of neurodevelopmental disorders. For instance, everolimus, an inhibitor of mTOR which is used for treatment of tumor manifestations in patients with tuberous sclerosis complex also provide impressive therapeutic effect on improving neuropsychiatric symptoms (Kilincaslan et al., 2017). Rapamycin, another mTOR inhibitor was also reported to prevent the pathological and behavioral deficits in ASD (Tsai et al., 2012). These emerging evidence suggests mTOR inhibitors could be a potential pharmacotherapy for ASD (Sato, 2016).
FBXW7 in Neurodegeneration
Despite of dysgenopathy in nervous system, anabatic neurodegeneration is another prominent characteristic caused by FBXW7 dysfunction. A large body of evidence indicates FBXW7 may be implicated in the pathogenesis of some typical neurodegenerative diseases. Potential underlying pathways of FBXW7 involved in Alzheimer’s disease, Parkinson’s disease and Huntington’s disease are discussed in this part.
Alzheimer’s Disease
Alzheimer’s disease (AD), the most common form of dementia especially in the old, affects more than 50 million people worldwide. AD patients are typically characterized with amyloid plaque and neurofibrillary tangles in the brain, both of which are regarded as two hallmarks of this disease (Drew, 2018). At present, dysregulation or dysfunction of FBXW7 has not been reported in AD patients or animal models, but some evidence may support the issue that FBXW7 is involved in the pathogenesis of AD.
Firstly, FBXW7 potentially regulates amyloid-β (Aβ) generation. Overexpression of FBXW7 in HEK293 cells could alter APP metabolism and lead to an increase in the production of Aβ (Li et al., 2002). However, the effect and mechanism of FBXW7 on Aβ production remains elusive so far. It has been widely recognized that the generation of Aβ, especially Aβ42, which aggregates into bioactive conformational species, likely initiates the toxicity in AD (O’Brien and Wong, 2011; Potter et al., 2013; Szaruga et al., 2015; Selkoe and Hardy, 2016; Makin, 2018). Aβ is generated from APP, successively processed by β-secretase and γ-secretase complex. BACE1 (β-site cleaving enzyme 1), a membrane-located aspartyl peptidase, acts as the dominated β-secretase which provides the first cleavage of APP at β-site (Zhang et al., 2021). Hypoxia-inducible factor 1 subunit α (HIF-1α), a factor induced by hypoxia, was reported to directly regulate BACE1 transcription and is contributing to BACE1 upregulation in response to hypoxia in the pathogenesis of AD (Sun et al., 2006; Zhang et al., 2007). It indicates that FBXW7 may be implicated in Aβ generation by regulating BACE1 level in a HIF-1α dependent pathway. Moreover, Li et al. (2002) also showed that FBXW7 interacted with Presenilin 1 (PS1), a crucial component of γ-secretase, revealing FBXW7 may alter γ-secretase activity by binding to PS1 protein, thus promoting processing of APP and Aβ generation.
Furthermore, FBXW7 may regulate neuronal apoptosis which seems to be inordinate in AD brain (Obulesu and Lakshmi, 2014; Fricker et al., 2018). FBXW7 is known to be implicated in neuronal apoptosis. For example, FBXW7 can mitigate neuronal apoptosis by mediating c-Jun proteolysis in response to glutamate-induced excitotoxicity (Ko et al., 2019). c-Jun is known as a substrate of SCFFBXW7 and plays a crucial role in accelerating cell apoptosis (Bossy-Wetzel et al., 1997). Consistently, FBXW7 has also been confirmed to bind parkin in neurons and to collaborate with parkin to ubiquitylate and destabilize the target cyclin E1 (Staropoli et al., 2003). Excessive cyclin E1 accumulation in neurons can lead to neuronal apoptosis (Padmanabhan et al., 1999), especially under conditions of excitotoxicity, suggesting a neuroprotective role for FBXW7. Besides, Ko et al. (2020) demonstrated that Fbxw7 was cleaved by activated calpain in the ipsilateral cortex in the rat model with middle cerebral artery occlusion. Negative regulation of Fbxw7 by calpain leaded to neuronal cell death while the preservation of Fbxw7 by the inhibition of calpain or other strategies may provide a novel protective mechanism against aberrant cell apoptosis in response to excitotoxicity (Ko et al., 2020). Similarly, Fbxw7 level was significantly reduced in mice spinal cord tissues in response to spinal cord injury while enhanced Fbxw7 expression can effectively moderate the progression of spinal cord injury by repressing microglial inflammation and neuronal death (Chen et al., 2020). Additionally, FBXW7 possibly mitigate neuronal apoptosis via mediating proteasome-dependent degradation of regulator of calcineurin 1 (RCAN1). RCAN1, a crucial endogenous regulator of calcineurin (Wang S. et al., 2020), is highly expressed in human brain and is particularly aberrantly elevated in the brains of AD patients (Ermak et al., 2001; Harris et al., 2007). Several lines of evidence suggest that RCAN1 functions in neuronal apoptosis (Lee et al., 2007; Sun et al., 2011; Ermak et al., 2012; Wu and Song, 2013) and the degradation of RCAN1 proteins is mediated by both UPS and the chaperone-mediated autophagy pathways (Liu et al., 2009). RCAN1 is a specific target of FBXW7 in the process of ubiquitin-proteasome-mediated degradation (Lee et al., 2012; Hong et al., 2015). It suggests that FBXW7 may be responsible for dysregulation of RCAN1 in AD but the mechanism needs to be further investigated. However, the functional role of FBXW7 in neuronal apoptosis remains controversial. For instance, FBXW7β was also reported to promote neuronal apoptosis via mediating ubiquitylation-dependent proteolysis of Mcl-1. Mcl-1, a specific SCFFbxw7 target in neurons, functions as a mitochondrial prosurvival factor in neuronal apoptosis (Ekholm-Reed et al., 2013). It suggests that functional implication of FBXW7 in neuronal apoptosis potentially depends on its subcellular localization and specific target affinity.
In addition, FBXW7 may also be involved in AD by modulating cell senescence. It is known that aging is the dominated risk factor for AD. Cell senescence is irreversible programed process which determines the aging process of the body. Telomere shortening plays a crucial role in cell senescence therefore telomere dysfunction is always associated with aging-related diseases (Blackburn et al., 2015; Tian et al., 2019). A recent work reported that FBXW7 mediated cell senescence through telomere uncapping. FBXW7 interacts with telomere protection protein 1 (TPP1), promotes TPP1 multisite ubiquitylation and degradation, and thereby triggers telomere uncapping and DNA damage response (Wang L. et al., 2020). However, the potential role of FBXW7 in neuronal senescence remains to be confirmed.
Parkinson Disease
Parkinson’s disease (PD) is another most common neurodegenerative disorders, affecting over 1% of the population older than 60 years of age. PD is characterized by progressive degeneration of nigrostriatal dopaminergic (DA) neurons in the midbrain and clinically diagnosed as motor abnormalities including bradykinesia, resting tremor, and cogwheel rigidity (Duvoisin, 1992; Abou-Sleiman et al., 2006). Mutations in the PARK2 gene are involved in a small portion of the cases which is known as autosomal recessive juvenile parkinsonism (ARJP) (Saito et al., 2000). It has been reported that FBXW7β levels are elevated in the cortexes of PD patients with a biallelic PARK2 mutation. Parkin, the product of PARK2 gene, is responsible for polyubiquitylating FBXW7β and targeting it for proteasomal degradation. Parkin deficiency-mediated FBXW7β elevation in some PD cases accelerates Mcl-1 degradation, subsequently leading to aberrant neuronal apoptosis (Ekholm-Reed et al., 2013). Intriguingly, it was also reported that FBXW7β protein level did not change in postmortem sporadic PD brains but FBXW7β was highly oxidized with excessive carbonyl formation. Similarly in the 6-hyroxydopamine (6-OHDA) induced PD mouse model, both of the total and oxidation level of FBXW7β decreased in the substantia nigra compacta. 6-OHDA enhanced the binding of FBXW7β with Hsc70, another fundamental regulator of chaperone-mediated autophagy (CMA), enabling the delivery of FBXW7β to LAMP2A and accelerating FBXW7β degradation mediated by CMA. However, the functional implication of oxidation-mediated FBXW7β reduction in the pathogenesis of PD deserves further investigation (Wang et al., 2018).
Huntington’s Disease
Huntington’s disease (HD) is an inherited autosomal dominant neurodegenerative disorder caused by accumulated mutant Huntingtin (Htt) protein with a poly-glutamine expansion (encoded by CAG trinucleotide repeat) (Ross and Tabrizi, 2011). Mutant Htt upregulates CK2α kinase and FBXW7, which phosphorylates and ubiquitylates heat shock transcription factor 1 (HSF1), respectively, thus promoting its proteasome-mediated degradation. Consistently, HSF1 was downregulated in striatum and cortex from patients with HD, causally leading to neuronal dysfunction. It indicates that blocking FBXW7-mediated HSF1 degradation may effectively ameliorate defects in neuronal function and promote survival in HD (Gomez-Pastor et al., 2017). Moreover, FBXW7 is also implicated in HD by targeting p53 for degradation. A potential causal role of impaired mitochondrial fission caused by dysfunction of dynamin-related protein 1 (Drp1) in neuronal damage of HD has widely been suggested (Song et al., 2011; Shirendeb et al., 2012). p53, a stress sensor involved in HD pathogenesis, interacts with DRP1 to promote DRP1-induced mitochondrial and neuronal damage (Guo et al., 2013). p53 can be phosphorylated by GSK3 and ATM at serine 33, then ubiquitylated by SCFFBXW7 and degraded in the proteasomal pathway (Galindo-Moreno et al., 2019; Cui et al., 2020). This suggests that targeting FBXW7 for inhibiting p53 may prevent the progression of HD by suppressing DRP1-dependent excessive mitochondrial fission and neuronal damage.
Challenges and Prospects on Drug Development by Targeting FBXW7
Considering the crucial roles of FBXW7 in neurodevelopment and neurodegeneration, FBXW7 may be a potential therapeutic target for neurodevelopmental and neurodegenerative disorders treatment. However, drug development by targeting FBXW7 is also faced with several challenges which may result from the following reasons. Firstly, FBXW7 functions in multiple physiological processes by targeting a variety of substrates therefore potential side effects of FBXW7 modulation should be considered. For example, FBXW7 is also regarded as a tumor suppressor and inactivation of FBXW7 can increase resistance to anti-tubulin drugs and promote tumorigenesis (Yumimoto and Nakayama, 2020). Moreover, a broad spectrum of the tissue-specific regulatory mechanisms and substrate selectivity of FBXW7 further increase the requirement for precise drug development. Besides, the functional heterogeneity of FBXW7 isoforms remains elusive although it would be more precise to target specific isoform of FBXW7 for amelioration of neurodevelopmental and neurodegenerative impairments.
However, instead of directly modulating FBXW7 level or activity, interventions on interactions between FBXW7 and its targets may provide a more feasible therapeutic strategy. For example, some specific oligopeptides which are designed based on the degron motif within the substrate are confirmed to effectively inhibit substrates degradation via competing binding with FBXW7 (Yalla et al., 2018; Wang L. et al., 2020). As a consequence, the development of chemical inhibitors or oligopeptides targeting FBXW7 by inhibiting SCFFBXW7 mediated substrate degradation should shed light on the therapeutic potential of targeting FBXW7-mediated degradation for the treatment of neurodevelopmental and neurodegenerative disorders. For example, blocking the FBXW7 mediated HSF1 and Mcl-1 degradation may effectively ameliorate defects in neuronal function in HD and PD, respectively, even though effective acceleration of HIF-1α, PS-1 and RCAN1 by targeting FBXW7-mediated degradation possibly provides curative effect on AD treatment (Figure 2). Meanwhile, considering the therapeutic effect of mTOR inhibitors on ameliorating autism-like symptoms, appropriate induction of FBXW7-mediated degradation of mTOR may also be beneficial for autism treatment but the effect still remains to be demonstrated.
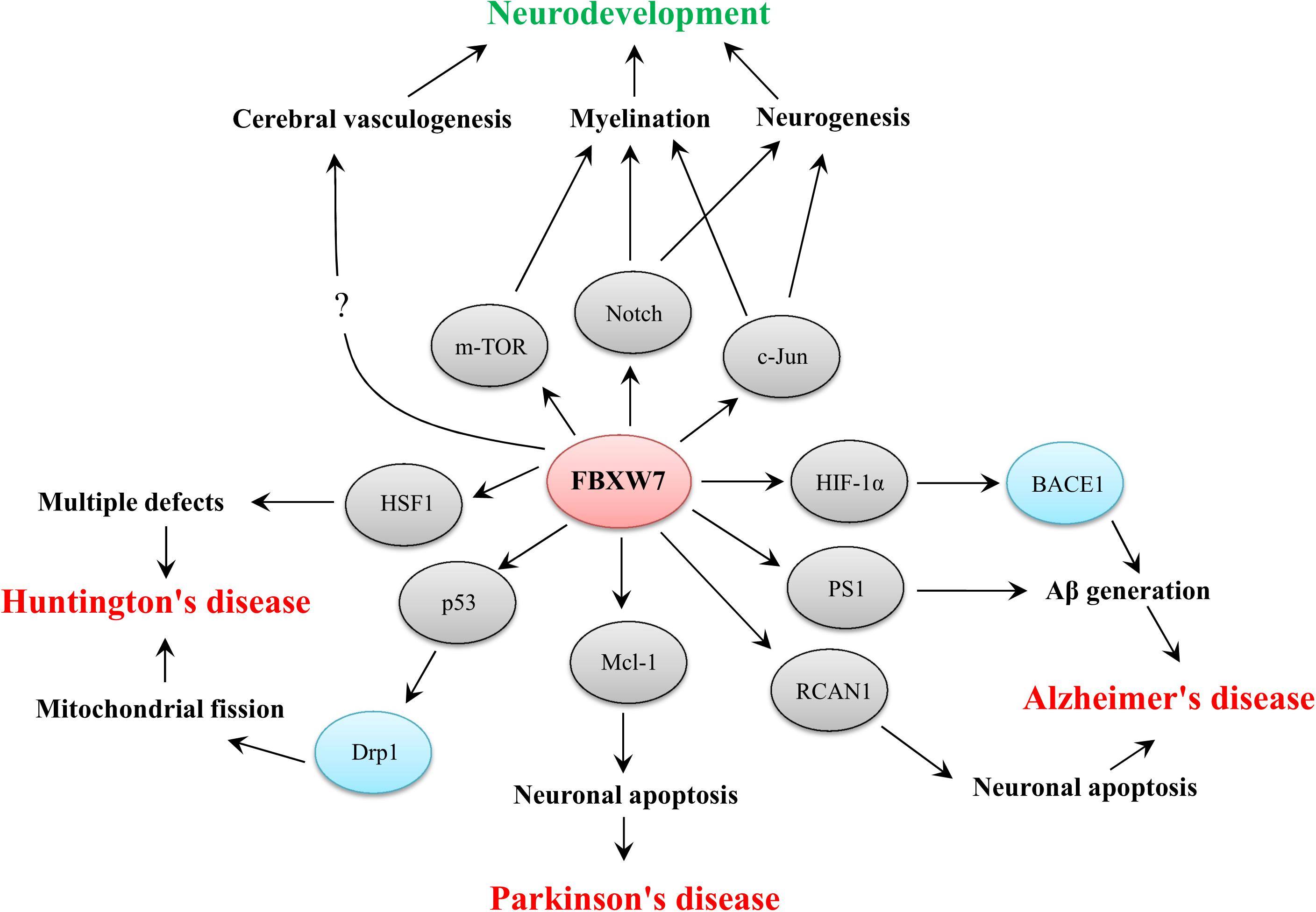
Figure 2. Functional implications of FBXW7 in neurodevelopment and three typical neurodegenerative diseases. The substrates of FBXW7 and other downstream factors are presented by gray or blue ellipses, respectively.
Conclusion
Accumulating evidence has shown that FBXW7 functions in neurodevelopment and neurodegeneration. In summary, FBXW7 is not only implicated in neurodevelopment by regulating neurogenesis, myelin development and cerebral vasculogenesis but also involved in the pathogenesis of some neurodegenerative disorders, such as AD, PD, and HD (Figure 2). Thus, targeting FBXW7 or FBXW7-substrate interaction may offer the opportunities for drug development against neurodevelopmental and neurodegenerative impairments even though some challenges also deserve further consideration.
Author Contributions
YY and SW wrote the manuscript and made the figures. XZ, XL, RS, and YG revised and approved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was granted from the Natural Science Foundation of Shandong Province of China (ZR2019PC038) to SW, the Research Fund for Academician Lin He New Medicine (JYHL2019MS01) to SW, Supporting Fund for Teachers’ Research of Jining Medical University (JYFC2018JS009) to YY, and Undergraduate Innovation Training Project (S202010443013) to XL.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abou-Sleiman, P. M., Muqit, M. M., and Wood, N. W. (2006). Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat. Rev. Neurosci. 7, 207–219. doi: 10.1038/nrn1868
Aggarwal, S., Bhowmik, A. D., Ramprasad, V. L., Murugan, S., and Dalal, A. (2016). A splice site mutation in HERC1 leads to syndromic intellectual disability with macrocephaly and facial dysmorphism: further delineation of the phenotypic spectrum. Am. J. Med. Genet. A 170, 1868–1873. doi: 10.1002/ajmg.a.37654
Attwell, D., Buchan, A. M., Charpak, S., Lauritzen, M., Macvicar, B. A., and Newman, E. A. (2010). Glial and neuronal control of brain blood flow. Nature 468, 232–243. doi: 10.1038/nature09613
Bachiller, S., Rybkina, T., Porras-Garcia, E., Perez-Villegas, E., Tabares, L., Armengol, J. A., et al. (2015). The HERC1 E3 Ubiquitin Ligase is essential for normal development and for neurotransmission at the mouse neuromuscular junction. Cell Mol. Life Sci. 72, 2961–2971. doi: 10.1007/s00018-015-1878-2
Balamurugan, K., Wang, J. M., Tsai, H. H., Sharan, S., Anver, M., Leighty, R., et al. (2010). The tumour suppressor C/EBPdelta inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J. 29, 4106–4117. doi: 10.1038/emboj.2010.280
Berndsen, C. E., and Wolberger, C. (2014). New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307. doi: 10.1038/nsmb.2780
Bjorklund, G., Tinkov, A. A., Hosnedlova, B., Kizek, R., Ajsuvakova, O. P., Chirumbolo, S., et al. (2020). The role of glutathione redox imbalance in autism spectrum disorder: a review. Free Radic. Biol. Med. 160, 149–162. doi: 10.1016/j.freeradbiomed.2020.07.017
Blackburn, E. H., Epel, E. S., and Lin, J. (2015). Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198. doi: 10.1126/science.aab3389
Bossy-Wetzel, E., Bakiri, L., and Yaniv, M. (1997). Induction of apoptosis by the transcription factor c-Jun. EMBO J. 16, 1695–1709. doi: 10.1093/emboj/16.7.1695
Cao, S., Wang, Y., Li, J., Lv, M., Niu, H., and Tian, Y. (2016). Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am. J. Cancer Res. 6, 2561–2574.
Chang, H., Liu, Y. H., Wang, L. L., Wang, J., Zhao, Z. H., Qu, J. F., et al. (2018). MiR-182 promotes cell proliferation by suppressing FBXW7 and FBXW11 in non-small cell lung cancer. Am. J. Transl. Res. 10, 1131–1142.
Chen, L. J., Hu, B., Han, Z. Q., Liu, W., Zhu, J. H., Chen, X. X., et al. (2021). Repression of FBXW7 by HES5 contributes to inactivation of the TGF-beta signaling pathway and alleviation of endometriosis. FASEB J. 35:e20938. doi: 10.1096/fj.202000438RRR
Chen, M., Gao, Y. T., Li, W. X., Wang, J. C., He, Y. P., Li, Z. W., et al. (2020). FBW7 protects against spinal cord injury by mitigating inflammation-associated neuronal apoptosis in mice. Biochem. Biophys. Res. Commun. 532, 576–583. doi: 10.1016/j.bbrc.2020.08.071
Chen, Z., Su, X., Shen, Y., Jin, Y., Luo, T., Kim, I. M., et al. (2019). MiR322 mediates cardioprotection against ischemia/reperfusion injury via FBXW7/notch pathway. J. Mol. Cell Cardiol. 133, 67–74. doi: 10.1016/j.yjmcc.2019.05.020
Chernousov, M. A., Yu, W. M., Chen, Z. L., Carey, D. J., and Strickland, S. (2008). Regulation of Schwann cell function by the extracellular matrix. Glia 56, 1498–1507. doi: 10.1002/glia.20740
Chitnis, A., Henrique, D., Lewis, J., Ish-Horowicz, D., and Kintner, C. (1995). Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature 375, 761–766. doi: 10.1038/375761a0
Cizmecioglu, O., Krause, A., Bahtz, R., Ehret, L., Malek, N., and Hoffmann, I. (2012). Plk2 regulates centriole duplication through phosphorylation-mediated degradation of Fbxw7 (human Cdc4). J. Cell Sci. 125, 981–992. doi: 10.1242/jcs.095075
Cohen-Katsenelson, K., Wasserman, T., Khateb, S., Whitmarsh, A. J., and Aronheim, A. (2011). Docking interactions of the JNK scaffold protein WDR62. Biochem. J. 439, 381–390. doi: 10.1042/BJ20110284
Cui, D., Xiong, X., Shu, J., Dai, X., Sun, Y., and Zhao, Y. (2020). FBXW7 Confers Radiation Survival by Targeting p53 for Degradation. Cell Rep. 30:e494. doi: 10.1016/j.celrep.2019.12.032
de la Pompa, J. L., Wakeham, A., Correia, K. M., Samper, E., Brown, S., Aguilera, R. J., et al. (1997). Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 124, 1139–1148.
Diefenbacher, M. E., Popov, N., Blake, S. M., Schulein-Volk, C., Nye, E., Spencer-Dene, B., et al. (2014). The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J. Clin. Invest. 124, 3407–3418. doi: 10.1172/JCI73733
Duvoisin, R. C. (1992). Overview of Parkinson’s disease. Ann. N. Y. Acad. Sci. 648, 187–193. doi: 10.1111/j.1749-6632.1992.tb24537.x
Ekholm-Reed, S., Goldberg, M. S., Schlossmacher, M. G., and Reed, S. I. (2013). Parkin-dependent degradation of the F-box protein Fbw7beta promotes neuronal survival in response to oxidative stress by stabilizing Mcl-1. Mol. Cell Biol. 33, 3627–3643. doi: 10.1128/MCB.00535-13
Ermak, G., Morgan, T. E., and Davies, K. J. (2001). Chronic overexpression of the calcineurin inhibitory gene DSCR1 (Adapt78) is associated with Alzheimer’s disease. J. Biol. Chem. 276, 38787–38794. doi: 10.1074/jbc.M102829200
Ermak, G., Sojitra, S., Yin, F., Cadenas, E., Cuervo, A. M., and Davies, K. J. (2012). Chronic expression of RCAN1-1L protein induces mitochondrial autophagy and metabolic shift from oxidative phosphorylation to glycolysis in neuronal cells. J. Biol. Chem. 287, 14088–14098. doi: 10.1074/jbc.M111.305342
Fricker, M., Tolkovsky, A. M., Borutaite, V., Coleman, M., and Brown, G. C. (2018). Neuronal Cell Death. Physiol. Rev. 98, 813–880. doi: 10.1152/physrev.00011.2017
Galindo-Moreno, M., Giraldez, S., Limon-Mortes, M. C., Belmonte-Fernandez, A., Reed, S. I., Saez, C., et al. (2019). SCF(FBXW7)-mediated degradation of p53 promotes cell recovery after UV-induced DNA damage. FASEB J. 33, 11420–11430. doi: 10.1096/fj.201900885R
Gomez-Pastor, R., Burchfiel, E. T., Neef, D. W., Jaeger, A. M., Cabiscol, E., McKinstry, S. U., et al. (2017). Abnormal degradation of the neuronal stress-protective transcription factor HSF1 in Huntington’s disease. Nat. Commun. 8:14405. doi: 10.1038/ncomms14405
Gu, Z., Inomata, K., Mitsui, H., and Horii, A. (2008). Promoter hypermethylation is not the major mechanism for inactivation of the FBXW7 beta-form in human gliomas. Genes Genet. Syst. 83, 347–352. doi: 10.1266/ggs.83.347
Guo, X., Disatnik, M. H., Monbureau, M., Shamloo, M., Mochly-Rosen, D., and Qi, X. (2013). Inhibition of mitochondrial fragmentation diminishes Huntington’s disease-associated neurodegeneration. J. Clin. Invest. 123, 5371–5388. doi: 10.1172/JCI70911
Harrigan, J. A., Jacq, X., Martin, N. M., and Jackson, S. P. (2018). Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78. doi: 10.1038/nrd.2017.152
Harris, C. D., Ermak, G., and Davies, K. J. (2007). RCAN1-1L is overexpressed in neurons of Alzheimer’s disease patients. FEBS J. 274, 1715–1724. doi: 10.1111/j.1742-4658.2007.05717.x
Harty, B. L., Coelho, F., Pease-Raissi, S. E., Mogha, A., Ackerman, S. D., Herbert, A. L., et al. (2019). Myelinating Schwann cells ensheath multiple axons in the absence of E3 ligase component Fbxw7. Nat. Commun. 10:2976. doi: 10.1038/s41467-019-10881-y
Hasler, R., Jacobs, G., Till, A., Grabe, N., Cordes, C., Nikolaus, S., et al. (2012). Microbial pattern recognition causes distinct functional micro-RNA signatures in primary human monocytes. PLoS One 7:e31151. doi: 10.1371/journal.pone.0031151
Hoeck, J. D., Jandke, A., Blake, S. M., Nye, E., Spencer-Dene, B., Brandner, S., et al. (2010). Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat. Neurosci. 13, 1365–1372. doi: 10.1038/nn.2644
Hong, A., Lee, J. E., and Chung, K. C. (2015). Ubiquitin-specific protease 22 (USP22) positively regulates RCAN1 protein levels through RCAN1 de-ubiquitination. J. Cell Physiol. 230, 1651–1660. doi: 10.1002/jcp.24917
Hua, J., Ding, T., and Yang, L. (2016). Dysfunction of microRNA-32 regulates ubiquitin ligase FBXW7 in multiple myeloma disease. Onco Targets Ther. 9, 6573–6579. doi: 10.2147/OTT.S105945
Hua, Y., Zhao, K., Tao, G., Dai, C., and Su, Y. (2017). miR-25 promotes metastasis via targeting FBXW7 in esophageal squamous cell carcinoma. Oncol. Rep. 38, 3030–3038. doi: 10.3892/or.2017.5995
Hughes, A. N., and Appel, B. (2019). Oligodendrocytes express synaptic proteins that modulate myelin sheath formation. Nat. Commun. 10:4125. doi: 10.1038/s41467-019-12059-y
Hughes, E. G., Kang, S. H., Fukaya, M., and Bergles, D. E. (2013). Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat. Neurosci. 16, 668–676. doi: 10.1038/nn.3390
Iffland, P. H. II, and Crino, P. B. (2017). Focal Cortical Dysplasia: gene Mutations, Cell Signaling, and Therapeutic Implications. Annu. Rev. Pathol. 12, 547–571. doi: 10.1146/annurev-pathol-052016-100138
Jandke, A., Da Costa, C., Sancho, R., Nye, E., Spencer-Dene, B., and Behrens, A. (2011). The F-box protein Fbw7 is required for cerebellar development. Dev. Biol. 358, 201–212. doi: 10.1016/j.ydbio.2011.07.030
Jeon, T. I., Esquejo, R. M., Roqueta-Rivera, M., Phelan, P. E., Moon, Y. A., Govindarajan, S. S., et al. (2013). An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 18, 51–61. doi: 10.1016/j.cmet.2013.06.010
Jessen, K. R., and Mirsky, R. (2005). The origin and development of glial cells in peripheral nerves. Nat. Rev. Neurosci. 6, 671–682. doi: 10.1038/nrn1746
Ji, S., Qin, Y., Shi, S., Liu, X., Hu, H., Zhou, H., et al. (2015). ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell Res. 25, 561–573. doi: 10.1038/cr.2015.30
Jin, J., Cardozo, T., Lovering, R. C., Elledge, S. J., Pagano, M., and Harper, J. W. (2004). Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580. doi: 10.1101/gad.1255304
Kageyama, R., Ohtsuka, T., Shimojo, H., and Imayoshi, I. (2009). Dynamic regulation of Notch signaling in neural progenitor cells. Curr. Opin. Cell Biol. 21, 733–740. doi: 10.1016/j.ceb.2009.08.009
Kearns, C. A., Ravanelli, A. M., Cooper, K., and Appel, B. (2015). Fbxw7 Limits Myelination by Inhibiting mTOR Signaling. J. Neurosci. 35, 14861–14871. doi: 10.1523/JNEUROSCI.4968-14.2015
Kessaris, N., Fogarty, M., Iannarelli, P., Grist, M., Wegner, M., and Richardson, W. D. (2006). Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 9, 173–179. doi: 10.1038/nn1620
Kilincaslan, A., Kok, B. E., Tekturk, P., Yalcinkaya, C., Ozkara, C., and Yapici, Z. (2017). Beneficial Effects of Everolimus on Autism and Attention-Deficit/Hyperactivity Disorder Symptoms in a Group of Patients with Tuberous Sclerosis Complex. J. Child Adolesc. Psychopharmacol. 27, 383–388. doi: 10.1089/cap.2016.0100
Kimura, T., Gotoh, M., Nakamura, Y., and Arakawa, H. (2003). hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 94, 431–436. doi: 10.1111/j.1349-7006.2003.tb01460.x
Ko, Y. U., Kim, C., Lee, J., Kim, D., Kim, Y., Yun, N., et al. (2019). Site-specific phosphorylation of Fbxw7 by Cdk5/p25 and its resulting decreased stability are linked to glutamate-induced excitotoxicity. Cell Death Dis. 10:579. doi: 10.1038/s41419-019-1818-4
Ko, Y. U., Song, H. Y., Kim, W. K., Yune, T. Y., Yun, N., and Oh, Y. J. (2020). Calpain-mediated cleavage of Fbxw7 during excitotoxicity. Neurosci. Lett. 736:135265. doi: 10.1016/j.neulet.2020.135265
LaSalle, J. M., Reiter, L. T., and Chamberlain, S. J. (2015). Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics 7, 1213–1228. doi: 10.2217/epi.15.70
Lee, E. J., Lee, J. Y., Seo, S. R., and Chung, K. C. (2007). Overexpression of DSCR1 blocks zinc-induced neuronal cell death through the formation of nuclear aggregates. Mol. Cell Neurosci. 35, 585–595. doi: 10.1016/j.mcn.2007.05.003
Lee, J. W., Kang, H. S., Lee, J. Y., Lee, E. J., Rhim, H., Yoon, J. H., et al. (2012). The transcription factor STAT2 enhances proteasomal degradation of RCAN1 through the ubiquitin E3 ligase FBW7. Biochem. Biophys. Res. Commun. 420, 404–410. doi: 10.1016/j.bbrc.2012.03.007
Li, J., Pauley, A. M., Myers, R. L., Shuang, R., Brashler, J. R., Yan, R., et al. (2002). SEL-10 interacts with presenilin 1, facilitates its ubiquitination, and alters A-beta peptide production. J. Neurochem. 82, 1540–1548. doi: 10.1046/j.1471-4159.2002.01105.x
Li, L., Sarver, A. L., Khatri, R., Hajeri, P. B., Kamenev, I., French, A. J., et al. (2014). Sequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. J. Pathol. 234, 488–501. doi: 10.1002/path.4407
Li, W., Bengtson, M. H., Ulbrich, A., Matsuda, A., Reddy, V. A., Orth, A., et al. (2008). Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One 3:e1487. doi: 10.1371/journal.pone.0001487
Li, X., Elmira, E., Rohondia, S., Wang, J., Liu, J., and Dou, Q. P. (2018). A patent review of the ubiquitin ligase system: 2015-2018. Expert Opin. Ther. Pat. 28, 919–937. doi: 10.1080/13543776.2018.1549229
Liu, H., Wang, P., Song, W., and Sun, X. (2009). Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J. 23, 3383–3392. doi: 10.1096/fj.09-134296
Liu, X., Ma, J., Xu, F., and Li, L. (2018). TINCR suppresses proliferation and invasion through regulating miR-544a/FBXW7 axis in lung cancer. Biomed. Pharmacother. 99, 9–17. doi: 10.1016/j.biopha.2018.01.049
Liu, Z., Liu, X., Liu, S., and Cao, Q. (2018). Cholesterol promotes the migration and invasion of renal carcinoma cells by regulating the KLF5/miR-27a/FBXW7 pathway. Biochem. Biophys. Res. Commun. 502, 69–75. doi: 10.1016/j.bbrc.2018.05.122
Lutolf, S., Radtke, F., Aguet, M., Suter, U., and Taylor, V. (2002). Notch1 is required for neuronal and glial differentiation in the cerebellum. Development 129, 373–385.
Makin, S. (2018). The amyloid hypothesis on trial. Nature 559, S4–S7. doi: 10.1038/d41586-018-05719-4
Mansour, M. R., Sanda, T., Lawton, L. N., Li, X., Kreslavsky, T., Novina, C. D., et al. (2013). The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J. Exp. Med. 210, 1545–1557. doi: 10.1084/jem.20122516
Martin, J. R., and Webster, H. D. (1973). Mitotic Schwann cells in developing nerve: their changes in shape, fine structure, and axon relationships. Dev. Biol. 32, 417–431. doi: 10.1016/0012-1606(73)90251-0
Matsumoto, A., Onoyama, I., and Nakayama, K. I. (2006). Expression of mouse Fbxw7 isoforms is regulated in a cell cycle- or p53-dependent manner. Biochem. Biophys. Res. Commun. 350, 114–119. doi: 10.1016/j.bbrc.2006.09.003
Matsumoto, A., Onoyama, I., Sunabori, T., Kageyama, R., Okano, H., and Nakayama, K. I. (2011a). Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. J. Biol. Chem. 286, 13754–13764. doi: 10.1074/jbc.M110.194936
Matsumoto, A., Tateishi, Y., Onoyama, I., Okita, Y., Nakayama, K., and Nakayama, K. I. (2011b). Fbxw7beta resides in the endoplasmic reticulum membrane and protects cells from oxidative stress. Cancer Sci. 102, 749–755. doi: 10.1111/j.1349-7006.2011.01851.x
McKinnon, C., and Tabrizi, S. J. (2014). The ubiquitin-proteasome system in neurodegeneration. Antioxid. Redox Signal. 21, 2302–2321. doi: 10.1089/ars.2013.5802
Min, S. H., Lau, A. W., Lee, T. H., Inuzuka, H., Wei, S., Huang, P., et al. (2012). Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol. Cell 46, 771–783. doi: 10.1016/j.molcel.2012.04.012
Mizuno, T., Mizuta, I., Watanabe-Hosomi, A., Mukai, M., and Koizumi, T. (2020). Clinical and Genetic Aspects of CADASIL. Front. Aging Neurosci. 12:91. doi: 10.3389/fnagi.2020.00091
Mo, J. S., Ann, E. J., Yoon, J. H., Jung, J., Choi, Y. H., Kim, H. Y., et al. (2011). Serum- and glucocorticoid-inducible kinase 1 (SGK1) controls Notch1 signaling by downregulation of protein stability through Fbw7 ubiquitin ligase. J. Cell Sci. 124, 100–112. doi: 10.1242/jcs.073924
O’Brien, R. J., and Wong, P. C. (2011). Amyloid precursor protein processing and Alzheimer’s disease. Annu. Rev. Neurosci. 34, 185–204. doi: 10.1146/annurev-neuro-061010-113613
Obulesu, M., and Lakshmi, M. J. (2014). Apoptosis in Alzheimer’s disease: an understanding of the physiology, pathology and therapeutic avenues. Neurochem. Res. 39, 2301–2312. doi: 10.1007/s11064-014-1454-4
Ortega-Recalde, O., Beltran, O. I., Galvez, J. M., Palma-Montero, A., Restrepo, C. M., Mateus, H. E., et al. (2015). Biallelic HERC1 mutations in a syndromic form of overgrowth and intellectual disability. Clin. Genet. 88, e1–e3. doi: 10.1111/cge.12634
Padmanabhan, J., Park, D. S., Greene, L. A., and Shelanski, M. L. (1999). Role of cell cycle regulatory proteins in cerebellar granule neuron apoptosis. J. Neurosci. 19, 8747–8756.
Park, H. C., and Appel, B. (2003). Delta-Notch signaling regulates oligodendrocyte specification. Development 130, 3747–3755. doi: 10.1242/dev.00576
Peng, G., Yang, C., Liu, Y., and Shen, C. (2019). miR-25-3p promotes glioma cell proliferation and migration by targeting FBXW7 and DKK3. Exp. Ther. Med. 18, 769–778. doi: 10.3892/etm.2019.7583
Pohl, C., and Dikic, I. (2019). Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 366, 818–822. doi: 10.1126/science.aax3769
Potter, R., Patterson, B. W., Elbert, D. L., Ovod, V., Kasten, T., Sigurdson, W., et al. (2013). Increased in vivo amyloid-beta42 production, exchange, and loss in presenilin mutation carriers. Sci. Transl. Med. 5:189ra177. doi: 10.1126/scitranslmed.3005615
Ross, C. A., and Tabrizi, S. J. (2011). Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98. doi: 10.1016/S1474-4422(10)70245-3
Saito, M., Maruyama, M., Ikeuchi, K., Kondo, H., Ishikawa, A., Yuasa, T., et al. (2000). Autosomal recessive juvenile parkinsonism. Brain Dev. 22, S115–S117. doi: 10.1016/s0387-7604(00)00137-6
Salussolia, C. L., Klonowska, K., Kwiatkowski, D. J., and Sahin, M. (2019). Genetic Etiologies, Diagnosis, and Treatment of Tuberous Sclerosis Complex. Annu. Rev. Genomics Hum. Genet. 20, 217–240. doi: 10.1146/annurev-genom-083118-015354
Sancho, R., Blake, S. M., Tendeng, C., Clurman, B. E., Lewis, J., and Behrens, A. (2013). Fbw7 repression by hes5 creates a feedback loop that modulates Notch-mediated intestinal and neural stem cell fate decisions. PLoS Biol. 11:e1001586. doi: 10.1371/journal.pbio.1001586
Sato, A. (2016). mTOR, a Potential Target to Treat Autism Spectrum Disorder. CNS Neurol. Disord. Drug Targets 15, 533–543. doi: 10.2174/1871527315666160413120638
Schulein, C., Eilers, M., and Popov, N. (2011). PI3K-dependent phosphorylation of Fbw7 modulates substrate degradation and activity. FEBS Lett. 585, 2151–2157. doi: 10.1016/j.febslet.2011.05.036
Schulein-Volk, C., Wolf, E., Zhu, J., Xu, W., Taranets, L., Hellmann, A., et al. (2014). Dual regulation of Fbw7 function and oncogenic transformation by Usp28. Cell Rep. 9, 1099–1109. doi: 10.1016/j.celrep.2014.09.057
Segarra, M., Aburto, M. R., Cop, F., Llao-Cid, C., Hartl, R., Damm, M., et al. (2018). Endothelial Dab1 signaling orchestrates neuro-glia-vessel communication in the central nervous system. Science 361:eaao2861. doi: 10.1126/science.aao2861
Segarra, M., Kirchmaier, B. C., and Acker-Palmer, A. (2015). A vascular perspective on neuronal migration. Mech. Dev. 138, 17–25. doi: 10.1016/j.mod.2015.07.004
Selkoe, D. J., and Hardy, J. (2016). The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608. doi: 10.15252/emmm.201606210
Shimizu, K., Nihira, N. T., Inuzuka, H., and Wei, W. (2018). Physiological functions of FBW7 in cancer and metabolism. Cell. Signal. 46, 15–22. doi: 10.1016/j.cellsig.2018.02.009
Shimoyama, S., Furukawa, T., Ogata, Y., Nikaido, Y., Koga, K., Sakamoto, Y., et al. (2019). Lipopolysaccharide induces mouse translocator protein (18 kDa) expression via the AP-1 complex in the microglial cell line, BV-2. PLoS One 14:e0222861. doi: 10.1371/journal.pone.0222861
Shirendeb, U. P., Calkins, M. J., Manczak, M., Anekonda, V., Dufour, B., McBride, J. L., et al. (2012). Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet. 21, 406–420. doi: 10.1093/hmg/ddr475
Snyder, J. L., Kearns, C. A., and Appel, B. (2012). Fbxw7 regulates Notch to control specification of neural precursors for oligodendrocyte fate. Neural. Dev. 7:15. doi: 10.1186/1749-8104-7-15
Song, W., Chen, J., Petrilli, A., Liot, G., Klinglmayr, E., Zhou, Y., et al. (2011). Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 17, 377–382. doi: 10.1038/nm.2313
Spruck, C. H., Strohmaier, H., Sangfelt, O., Muller, H. M., Hubalek, M., Muller-Holzner, E., et al. (2002). hCDC4 gene mutations in endometrial cancer. Cancer Res. 62, 4535–4539.
Staropoli, J. F., McDermott, C., Martinat, C., Schulman, B., Demireva, E., and Abeliovich, A. (2003). Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37, 735–749. doi: 10.1016/s0896-6273(03)00084-9
Sun, X., He, G., Qing, H., Zhou, W., Dobie, F., Cai, F., et al. (2006). Hypoxia facilitates Alzheimer’s disease pathogenesis by up-regulating BACE1 gene expression. Proc. Natl. Acad. Sci. U. S. A. 103, 18727–18732. doi: 10.1073/pnas.0606298103
Sun, X., Wu, Y., Chen, B., Zhang, Z., Zhou, W., Tong, Y., et al. (2011). Regulator of calcineurin 1 (RCAN1) facilitates neuronal apoptosis through caspase-3 activation. J. Biol. Chem. 286, 9049–9062. doi: 10.1074/jbc.M110.177519
Szaruga, M., Veugelen, S., Benurwar, M., Lismont, S., Sepulveda-Falla, D., Lleo, A., et al. (2015). Qualitative changes in human gamma-secretase underlie familial Alzheimer’s disease. J. Exp. Med. 212, 2003–2013. doi: 10.1084/jem.20150892
Tang, B., Lei, B., Qi, G., Liang, X., Tang, F., Yuan, S., et al. (2016). MicroRNA-155-3p promotes hepatocellular carcinoma formation by suppressing FBXW7 expression. J. Exp. Clin. Cancer Res. 35:93. doi: 10.1186/s13046-016-0371-6
Tetzlaff, M. T., Yu, W., Li, M., Zhang, P., Finegold, M., Mahon, K., et al. (2004). Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. U. S. A. 101, 3338–3345. doi: 10.1073/pnas.0307875101
Tian, Y., Wang, S., Jiao, F., Kong, Q., Liu, C., and Wu, Y. (2019). Telomere Length: a Potential Biomarker for the Risk and Prognosis of Stroke. Front. Neurol. 10:624. doi: 10.3389/fneur.2019.00624
Tripathi, P. P., Sgado, P., Scali, M., Viaggi, C., Casarosa, S., Simon, H. H., et al. (2009). Increased susceptibility to kainic acid-induced seizures in Engrailed-2 knockout mice. Neuroscience 159, 842–849. doi: 10.1016/j.neuroscience.2009.01.007
Tsai, P. T., Hull, C., Chu, Y., Greene-Colozzi, E., Sadowski, A. R., Leech, J. M., et al. (2012). Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature 488, 647–651. doi: 10.1038/nature11310
Tsunematsu, R., Nakayama, K., Oike, Y., Nishiyama, M., Ishida, N., Hatakeyama, S., et al. (2004). Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279, 9417–9423. doi: 10.1074/jbc.M312337200
Utine, G. E., Taskiran, E. Z., Kosukcu, C., Karaosmanoglu, B., Guleray, N., Dogan, O. A., et al. (2017). HERC1 mutations in idiopathic intellectual disability. Eur. J. Med. Genet. 60, 279–283. doi: 10.1016/j.ejmg.2017.03.007
Wang, L., Chen, R., Li, G., Wang, Z., Liu, J., Liang, Y., et al. (2020). FBW7 Mediates Senescence and Pulmonary Fibrosis through Telomere Uncapping. Cell Metab. 32, 860–877.e9. doi: 10.1016/j.cmet.2020.10.004
Wang, S., Wang, Y., Qiu, K., Zhu, J., and Wu, Y. (2020). RCAN1 in cardiovascular diseases: molecular mechanisms and a potential therapeutic target. Mol. Med. 26:118. doi: 10.1186/s10020-020-00249-0
Wang, L., Qin, D., Shi, H., Zhang, Y., Li, H., and Han, Q. (2019). MiR-195-5p Promotes Cardiomyocyte Hypertrophy by Targeting MFN2 and FBXW7. Biomed. Res. Int. 2019:1580982. doi: 10.1155/2019/1580982
Wang, X., Zhai, H., and Wang, F. (2018). 6-OHDA Induces Oxidation of F-box Protein Fbw7beta by Chaperone-Mediated Autophagy in Parkinson’s Model. Mol. Neurobiol. 55, 4825–4833. doi: 10.1007/s12035-017-0686-0
Wang, Y., Liu, Z., Yao, B., Li, Q., Wang, L., Wang, C., et al. (2017). Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol. Cancer 16:123. doi: 10.1186/s12943-017-0702-z
Wasserman, T., Katsenelson, K., Daniliuc, S., Hasin, T., Choder, M., and Aronheim, A. (2010). A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell 21, 117–130. doi: 10.1091/mbc.E09-06-0512
Webster, H. D., Martin, R., and O’Connell, M. F. (1973). The relationships between interphase Schwann cells and axons before myelination: a quantitative electron microscopic study. Dev. Biol. 32, 401–416. doi: 10.1016/0012-1606(73)90250-9
Welcker, M., Larimore, E. A., Frappier, L., and Clurman, B. E. (2011). Nucleolar targeting of the fbw7 ubiquitin ligase by a pseudosubstrate and glycogen synthase kinase 3. Mol. Cell Biol. 31, 1214–1224. doi: 10.1128/MCB.01347-10
Welcker, M., Larimore, E. A., Swanger, J., Bengoechea-Alonso, M. T., Grim, J. E., Ericsson, J., et al. (2013). Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 27, 2531–2536. doi: 10.1101/gad.229195.113
Welcker, M., Orian, A., Grim, J. E., Eisenman, R. N., and Clurman, B. E. (2004). A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 14, 1852–1857. doi: 10.1016/j.cub.2004.09.083
Wu, X. Z., Wang, K. P., Song, H. J., Xia, J. H., Jiang, Y., and Wang, Y. L. (2015). MiR-27a-3p promotes esophageal cancer cell proliferation via F-box and WD repeat domain-containing 7 (FBXW7) suppression. Int. J. Clin. Exp. Med. 8, 15556–15562.
Wu, Y., Chang, N., Zhang, Y., Zhang, X., Xu, L., Che, Y., et al. (2021). METTL3-mediated m(6)A mRNA modification of FBXW7 suppresses lung adenocarcinoma. J. Exp. Clin. Cancer Res. 40:90. doi: 10.1186/s13046-021-01880-3
Wu, Y., and Song, W. (2013). Regulation of RCAN1 translation and its role in oxidative stress-induced apoptosis. FASEB J. 27, 208–221. doi: 10.1096/fj.12-213124
Xia, W., Zhou, J., Luo, H., Liu, Y., Peng, C., Zheng, W., et al. (2017). MicroRNA-32 promotes cell proliferation, migration and suppresses apoptosis in breast cancer cells by targeting FBXW7. Cancer Cell Int. 17:14. doi: 10.1186/s12935-017-0383-0
Xiang, J., Hang, J. B., Che, J. M., and Li, H. C. (2015). MiR-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int. J. Clin. Exp. Pathol. 8, 9147–9153.
Xiao, D., Yue, M., Su, H., Ren, P., Jiang, J., Li, F., et al. (2016). Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol. Cell 64, 493–506. doi: 10.1016/j.molcel.2016.09.016
Xiao, G., Gao, X., Sun, X., Yang, C., Zhang, B., Sun, R., et al. (2017). miR-367 promotes tumor growth by inhibiting FBXW7 in NSCLC. Oncol. Rep. 38, 1190–1198. doi: 10.3892/or.2017.5755
Xu, D., Yao, M., Wang, Y., Yuan, L., Hoeck, J. D., Yu, J., et al. (2018). MEKK3 coordinates with FBW7 to regulate WDR62 stability and neurogenesis. PLoS Biol. 16:e2006613. doi: 10.1371/journal.pbio.2006613
Xu, J., Wu, W., Wang, J., Huang, C., Wen, W., Zhao, F., et al. (2017). miR-367 promotes the proliferation and invasion of non-small cell lung cancer via targeting FBXW7. Oncol. Rep. 37, 1052–1058. doi: 10.3892/or.2016.5314
Xu, Y., Sengupta, T., Kukreja, L., and Minella, A. C. (2010). MicroRNA-223 regulates cyclin E activity by modulating expression of F-box and WD-40 domain protein 7. J. Biol. Chem. 285, 34439–34446. doi: 10.1074/jbc.M110.152306
Xu, Z., Zhuang, L., Wang, X., Li, Q., Sang, Y., and Xu, J. (2020). FBXW7gamma is a tumor-suppressive and prognosis-related FBXW7 transcript isoform in ovarian serous cystadenocarcinoma. Future Oncol. 16, 1921–1930. doi: 10.2217/fon-2020-0371
Yalla, K., Elliott, C., Day, J. P., Findlay, J., Barratt, S., Hughes, Z. A., et al. (2018). FBXW7 regulates DISC1 stability via the ubiquitin-proteosome system. Mol. Psychiatry 23, 1278–1286. doi: 10.1038/mp.2017.138
Yang, W., Dou, C., Wang, Y., Jia, Y., Li, C., Zheng, X., et al. (2015). MicroRNA-92a contributes to tumor growth of human hepatocellular carcinoma by targeting FBXW7. Oncol. Rep. 34, 2576–2584. doi: 10.3892/or.2015.4210
Yumimoto, K., and Nakayama, K. I. (2020). Recent insight into the role of FBXW7 as a tumor suppressor. Semin. Cancer Biol. 67, 1–15. doi: 10.1016/j.semcancer.2020.02.017
Zhang, G., Li, S., Lu, J., Ge, Y., Wang, Q., Ma, G., et al. (2018). LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer 17:87. doi: 10.1186/s12943-018-0829-6
Zhang, J., Chen, Z., Liu, X., Yang, C., and Xie, D. (2021). Gain of circBRAF Represses Glioma Progression by Regulating miR-1290/FBXW7 Axis. Neurochem. Res. 46, 1203–1213. doi: 10.1007/s11064-021-03259-4
Zhang, P., Cao, L., Fan, P., Mei, Y., and Wu, M. (2016). LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 17, 1204–1220. doi: 10.15252/embr.201642067
Zhang, X., Zhou, K., Wang, R., Cui, J., Lipton, S. A., Liao, F. F., et al. (2007). Hypoxia-inducible factor 1alpha (HIF-1alpha)-mediated hypoxia increases BACE1 expression and beta-amyloid generation. J. Biol. Chem. 282, 10873–10880. doi: 10.1074/jbc.M608856200
Zhao, J., Hu, C., Chi, J., Li, J., Peng, C., Yun, X., et al. (2016). miR-24 promotes the proliferation, migration and invasion in human tongue squamous cell carcinoma by targeting FBXW7. Oncol. Rep. 36, 1143–1149. doi: 10.3892/or.2016.4891
Keywords: FBXW7, E3 ubiquitin ligase, neurodevelopment, neurodegenerative disorders, therapeutic approach
Citation: Yang Y, Zhou X, Liu X, Song R, Gao Y and Wang S (2021) Implications of FBXW7 in Neurodevelopment and Neurodegeneration: Molecular Mechanisms and Therapeutic Potential. Front. Cell. Neurosci. 15:736008. doi: 10.3389/fncel.2021.736008
Received: 04 July 2021; Accepted: 04 August 2021;
Published: 25 August 2021.
Edited by:
Sharon DeMorrow, University of Texas at Austin, United StatesReviewed by:
Timothy J. Jarome, Virginia Tech, United StatesA. Raquel Esteves, University of Coimbra, Portugal
Copyright © 2021 Yang, Zhou, Liu, Song, Gao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Wang, d2FuZ3NodWFpenFAMTI2LmNvbQ==; d2FuZ3NodWFpanNAbWFpbC5qbm1jLmVkdS5jbg==
 Yu Yang
Yu Yang Xuan Zhou
Xuan Zhou Xinpeng Liu
Xinpeng Liu Ruying Song1,2
Ruying Song1,2 Shuai Wang
Shuai Wang