- Departamento de Química Biológica, Instituto de Química y Fisicoquímica Biológicas, Facultad de Farmacia y Bioquímica, Consejo Nacional de Investigaciones Científicas y Técnicas-Universidad de Buenos Aires, Buenos Aires, Argentina
The plasma membrane Ca2+ pumps (PMCA) are P-ATPases that control Ca2+ signaling and homeostasis by transporting Ca2+ out of the eukaryotic cell. Humans have four genes that code for PMCA isoforms (PMCA1-4). A large diversity of PMCA isoforms is generated by alternative mRNA splicing at sites A and C. The different PMCA isoforms are expressed in a cell-type and developmental-specific manner and exhibit differential sensitivity to a great number of regulatory mechanisms. PMCA4 has two A splice variants, the forms “x” and “z”. While PMCA4x is ubiquitously expressed and relatively well-studied, PMCA4z is less characterized and its expression is restricted to some tissues such as the brain and heart muscle. PMCA4z lacks a stretch of 12 amino acids in the so-called A-M3 linker, a conformation-sensitive region of the molecule connecting the actuator domain (A) with the third transmembrane segment (M3). We expressed in yeast PMCA4 variants “x” and “z”, maintaining constant the most frequent splice variant “b” at the C-terminal end, and obtained purified preparations of both proteins. In the basal autoinhibited state, PMCA4zb showed a higher ATPase activity and a higher apparent Ca2+ affinity than PMCA4xb. Both isoforms were stimulated by calmodulin but PMCA4zb was more strongly activated by acidic lipids than PMCA4xb. The results indicate that a PMCA4 intrinsically more active and more responsive to acidic lipids is produced by the variant “z” of the splicing site A.
Introduction
The Ca2+ transporting P-ATPases known as Ca2+ pumps are key elements for the control of intracellular Ca2+ homeostasis (Chen et al., 2020). Animal cells possess three types of Ca2+ pumps, the endoplasmic-sarcoplasmic reticulum (SERCA), the secretory pathway (SPCA), and the plasma membrane Ca2+ pump (PMCA). By taking Ca2+ out of the cell, the PMCA modulates the resting cytosolic Ca2+ and is fundamental for shaping and ending the Ca2+ signals (Lopreiato et al., 2014; Stafford et al., 2017). The relative contribution of the PMCA to these processes depends on the cell type and the activity of other Ca2+ transporters. In excitable cells, the SERCA pump and the plasma membrane Na+/Ca2+ exchanger (NCX) are mostly responsible for removing Ca2+ from the cytosol, while the function of PMCA seems more suited for the control of Ca2+ dependent microdomain processes (Strehler, 2015). Consistent with this idea, in excitable cells, PMCA is a part of protein networks that by including voltage-gated Ca2+ channels, couple influx, and clearance of Ca2+ (Müller et al., 2010). The loss of PMCA function has been associated with several human diseases, including neurologic pathologies, such as familial spastic paraplegia (Mohamed et al., 2011, 2016; Li et al., 2014).
In humans four genes code for PMCA proteins (PMCA1-4) with a calculated molecular mass of 135 kDa and an average sequence identity of about 80% (Lopreiato et al., 2014). The expression profile of the PMCA isoforms varies depending on the tissue and the stage of development. PMCA1 and PMCA4 have ubiquitously expressed forms while the expression of PMCA2 and PMCA3 is more restricted and particularly abundant in nervous tissues (Strehler and Zacharias, 2001). While neurons are rich in all PMCA isoforms and variants, PMCA4 constitutes a major one at the early stages of the differentiation process and is markedly enriched, together with PMCA1, and PMCA2 at the postsynaptic membrane through the interaction with the postsynaptic density protein 95 (Kim et al., 1998; Boczek et al., 2019).
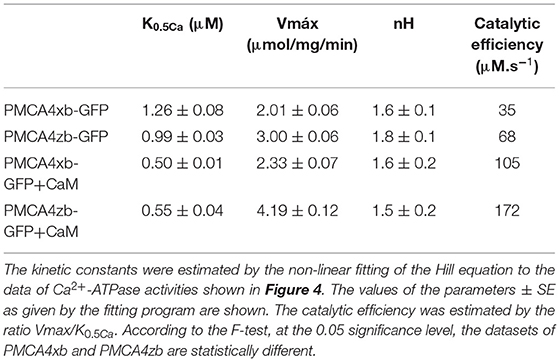
Recently, the Cryo-EM structure of the human PMCA1 in complex with neuroplastin was reported (Gong et al., 2018). As expected, the general architecture of the PMCA protein closely resembles that of other P-ATPases with the characteristic N, nucleotide binding, P phosphorylation, and A, action domain (Figure 1A). PMCA forms heteromers with the glycoproteins neuroplastin or basigin in different cell types (Korthalsa et al., 2017; Schmidt et al., 2017). Contacts between PMCA and neuroplastin were observed at the PMCA extracellular M8-M9 linker and with the M10 transmembrane segment. The interaction of the PMCA with neuroplastin takes place in the ER and affects PMCA stability, function, and plasma membrane targeting. These findings open the interesting possibility that the activity of the PMCA can influence neuroplastin functions, for example, synaptic plasticity and neuritogenesis (Beesley et al., 2014).
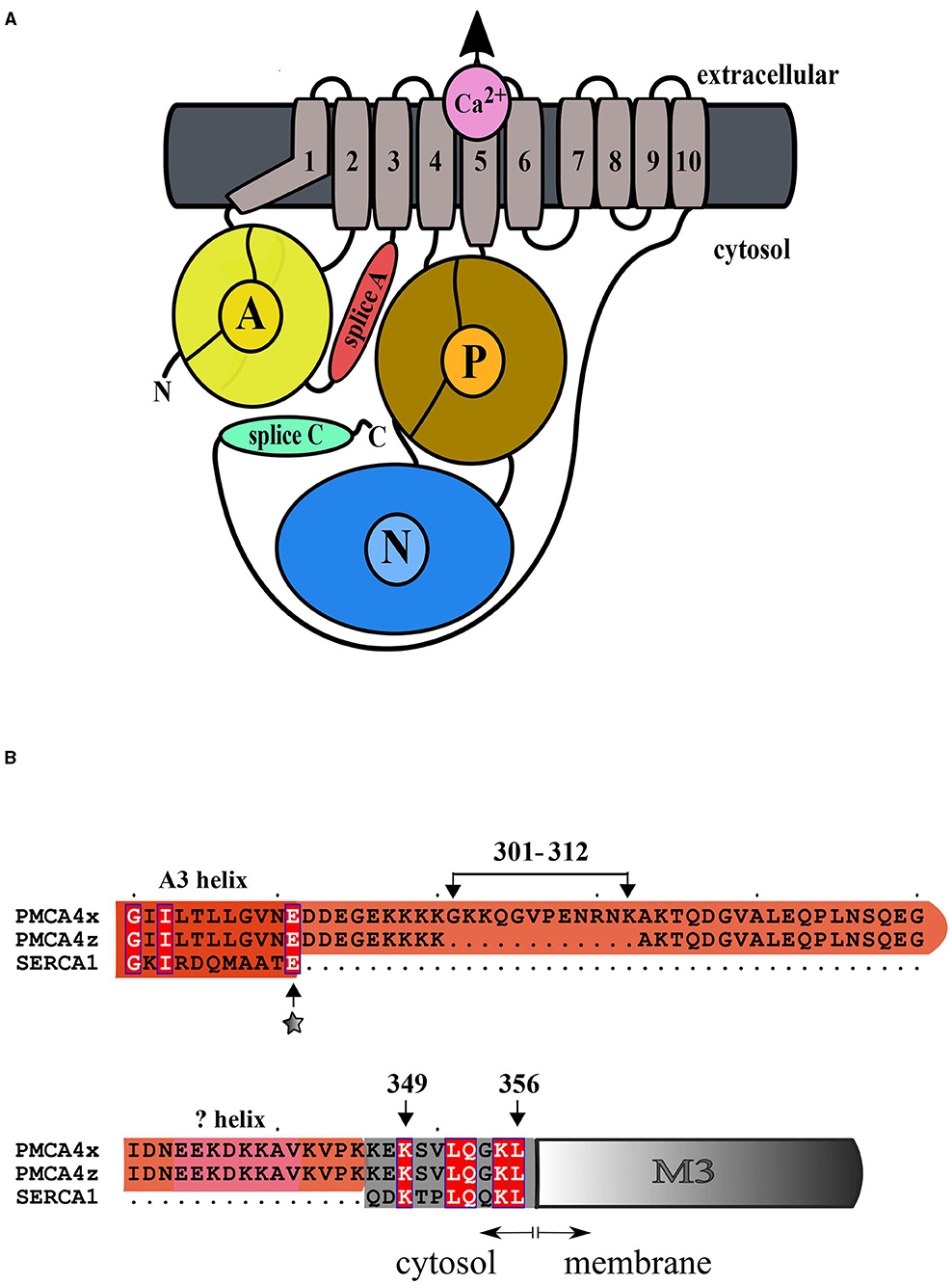
Figure 1. (A) General architecture of the plasma membrane Ca2+ (PMCA) P-ATPase. Only the catalytic subunit is represented. Helices M1-M6 constitute the transport domain. The action domain (A) is shown in yellow, the phosphorylation domain (P) is brown and the nucleotide-binding domain (N) is light blue. The A-M3 linker, which is modified by mRNA splicing at site A, is shown in red and the regulatory C-terminal domain, which is modified by mRNA splicing at site C, is shown in green. (B) Alignment of the amino acid sequence of the A-M3 linker of PMCA4x, PMCA4z, and SERCA1. Conserved residues are in white bold. The “x” insertion of amino acids 301–312 is shown. The position of the A3 (red) and AL helices which are predicted by modeling (pink) are indicated. The transmembrane M3 helix is colored gray and its membrane inserted portion is shown as a box. The site of SERCA mutational insertion (site 1) described by Nyholm Holdensen and Andersen (2009) is indicated by a star.
A distinctive characteristic of the PMCA family of Ca2+ pumps is the autoinhibitory mechanism and the direct stimulation by calmodulin (CaM) (Hegedus et al., 2020). Autoinhibition is mediated by the interaction of the regulatory domain, located in the extended C-terminal tail with the central catalytic core of the protein. The binding of Ca2+-CaM to the C-terminal regulatory domain releases the PMCA from the autoinhibition (Adamo and Grimaldi, 1998; Bredeston and Adamo, 2004; Corradi and Adamo, 2007; Mazzitelli and Adamo, 2014; Hegedus et al., 2020).
A second important mechanism of PMCA regulation is the Ca2+-independent activation by acidic lipids (Niggli et al., 1981; Enyedi et al., 1987; Brodin et al., 1992; Filoteo et al., 1992; de Tezanos Pinto and Adamo, 2002, 2006). Although this mechanism has not been explained, part of the stimulatory effect of acidic lipids is due to their interaction with the C-terminal regulatory domain (Brodin et al., 1992; Filoteo et al., 1992). However, because some acidic lipids, in particular phosphoinositides, were found more effective than calmodulin to activate the PMCA at low concentrations of Ca2+ (Enyedi et al., 1987) a second site was proposed, the so-called acidic lipid (AL) responsive region, in the segment that links domain A with the transmembrane helix M3 (Brodin et al., 1992; Filoteo et al., 1992; de Tezanos Pinto and Adamo, 2002, 2006).
Besides the four PMCA genes, many PMCA variants are generated by the combination of the products of alternative splicing of mRNAs at the two sites called sites A and C. While these splice variations do not affect the basic Ca2+ transport function, they are strategically located in the regulatory regions of the PMCA molecule and hence they may affect the maximal transport capacity and Ca2+ affinity as well as the interaction of the PMCA with the other regulatory proteins (Strehler and Zacharias, 2001; Lopreiato et al., 2014). Splicing at site C changes the C-terminal regulatory tail, generating the most common variants “a” and “b” and others identified as “c” through “g”, some of which have not yet been confirmed at the protein level. The degree of autoinhibition and the kinetics of CaM activation are unique features of each PMCA isoform and are modified by the alternative C-terminal sequences of each isoform variant (Enyedi et al., 1994; Strehler, 2015).
In humans, splicing at site A results in three PMCA variants, the forms “z”, “x”, and “w”, which have insertions of 0, 12, and 45 extra amino acids in the AL region, respectively. PMCA1 contains only the “x” variant, while in PMCA2-4 variants “x” and “z” have been detected. In humans, only PMCA2 has the longest “w” insertion (Strehler and Zacharias, 2001; Hegedus et al., 2020) while in the rat in addition to “w”, an “y” variant with the insertion of the first 31 residues has been detected (Adamo and Penniston, 1992). Because these insertions modify the AL region, previous studies have suggested that different splice A variants may differ in their sensitivity to acidic lipid activation (Brodin et al., 1992; Filoteo et al., 1992; de Tezanos Pinto and Adamo, 2002, 2006).
At present, only few PMCA forms have been individually characterized and the functional consequences of some splice variants are still controversial. Furthermore, the effect of the splicing variants may depend on the context of the particular PMCA isoform considered. In this study, we have characterized the enzymatic activity of purified preparations of the site A splice variants of PMCA4 which comprise two forms (Figure 1B), the ubiquitously expressed form “x” and the shorter variant “z” showing a more restricted expression profile, particularly, in the brain and heart. We found that PMCA4z is more active and more responsive to activation by acidic lipids than the PMCA4x.
Materials and Methods
Chemicals
Yeast synthetic drop-out media supplement without leucine, yeast nitrogen base without amino acids, dextrose, polyoxyethylene 10 lauryl ether (C12E10), L-α-phosphatidylcholine type XVI-E (Sigma) from fresh egg yolk, brain extract (BE) Type I Folch Fraction I from bovine brain containing approximately (w/w) 10% PI, 50% PS, and other lipids, phosphodiesterase 3′, 5′ cyclic nucleotide activator (calmodulin) from bovine brain, calmodulin-agarose, ATP (disodium salt, vanadium-free), sodium dodecyl sulfate (SDS), and all other chemicals were obtained from Sigma.
Construction of DNA Coding for PMCA4zb
The cDNA coding for PMCA4zb was obtained by a two-step PCR using Pfu DNA polymerase and the PMCA4xb cDNA as template. During the first step, two PCR reactions were run, one using primer His: 5′CTGGAGGTCGACATGACGCATCACCATCACCATCACAACCCATCAGACCGTGTCTTGCCTGCCAA3′, and N-term 5′TTTCTTCTTTTTCTCCCCTTCGTCATC3′, and the other using primer 2096: 5′TGACAACATCAACACAGCCCGGGCCATTGCCACCA3′ and C-term 5′GATGACGAAGGGGAGAAAAAGAAAGCAAAGACCCAAGACGGAGTG3′. The primer His contains a restriction site for nuclease SalI that matches a unique site for SalI at the 5′position of the PMCA4xb cDNA, while primer 2096 anneals to the PMCA4xb DNA downstream of a naturally occurring BspEI unique site. The 930-bp His-N-term and 1210-bp C-term-2096 products were combined in the next PCR step along with primers His and 2096. The amplified 2119-bp His-2096 fragment was digested with SalI and BspEI and used to replace the corresponding sequence of PMCA4xb GFP DNA cloned into pMP625 (Bredeston and Adamo, 2004) or pYX112 (Mazzitelli and Adamo, 2014) expression vectors.
Complementation Assay on K616 Saccharomyces cerevisiae Strain
K616 cells (MATα pmr1::HIS3 pmc::TRP1 cnb1::LEU2, ura3) (Cunningham and Fink, 1994) were transformed using a lithium acetate/polyethylene glycol method (Elble, 1992) with expression vector pYX112 containing different PMCA constructs, and the transformants were selected for their ability to grow in the absence of uracil on plates containing 6.7% YNB, 0.67% complete supplemented medium minus Ura (Ura-), 2.2% dextrose, 0.05% 2N-morpholino-ethanesulfonic acid (MES) pH = 6.0, 10 mM Cl2Ca, and 1.5% agar. For complementation studies, 15 μl of log-phase was streaked onto plates lacking uracil and containing either 10 mM Cl2Ca or 10 mM ethylene glycol-bis-N,N,N′,N′-tetra acetic acid (EGTA). The plates were allowed to grow for 72 h at 28°C.
Plasma Membrane Ca2+ Pump Expression and Purification
Saccharomyces cerevisiae strain DBY2062 (MATa his4-619 leu2-3,112) (Bauer and Kolling, 1996) was used for expression and purification. Yeast cells were transformed with vector pMP625 containing a Leu+ marker and the PMA1 promoter. Yeast cells were transformed and selected in the absence of leucine. PMCA4xb-GFP and PMCA4zb-GFP were purified as described previously (Bredeston and Adamo, 2004; Mazzitelli and Adamo, 2014). Briefly, 4 L of yeast (A600 ~ 4.0–5.0) were solubilized with C12E10 and the solubilizate was loaded onto a calmodulin-agarose chromatography column. After washing with a purification buffer containing Ca2+, the PMCA protein was eluted with EGTA. Lipids were not added during the purification.
Calcium-ATPase Activity
The Ca2+-ATPase activity of the purified preparations of PMCA4xb-GFP and PMCA4zb-GFP was estimated from the release of [32P]Pi from [γ-32P]ATP at 28°C as described previously (Mazzitelli and Adamo, 2014). The reaction medium contained 20 mM HEPES-K (pH = 7.2 at 28°C), 100 mM KCl, 4 mM MgCl2, 3 mM ATP, 500 μM EGTA, and enough CaCl2 to give the Ca2+ concentrations indicated in each experiment. The concentration of free Ca2+ in the reaction medium was calculated using the WebmaxC Standard program. Before starting the ATPase reaction, 1 μg of the enzyme (50 μl) was gently stirred for 5 min with 5 μl of a sonicated mixture of 57 mg/ml C12E10 with 29 mg/ml PC or the indicated amount of BE. The suspension was thoroughly mixed and preincubated for at least 10 min on ice before being added to the ATPase reaction. The PMCA was incubated for 3 min at 37°C with the reaction medium before the initiation of the reaction by adding 3 mM [γ-32P]ATP. The reaction was finished by acid denaturation after 30 min. During this period, the amount of Pi released from ATP increased linearly with time.
Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis, Protein Quantitation and Western Blotting
Proteins were electrophoresed on a 7.5% acrylamide gel according to Laemmli (Laemmli, 1970) and revealed by Coomassie Blue staining as previously described (Mazzitelli and Adamo, 2014). The protein concentration was initially estimated by the method of Bradford (Bradford, 1976) using bovine serum albumin as standard. The content of PMCA protein in each preparation was estimated by SDS–PAGE comparing the intensity of the bands of PMCA protein with different amounts of bovine serum albumin as standard, stained with Coomassie Blue.
Western blotting was performed as previously described (Cura et al., 2008). Briefly, after SDS–PAGE the proteins were electrotransferred onto Millipore Immobilon P membranes and the non-specific binding was blocked by incubating the membranes overnight at 4°C in a solution of 160 mM NaCl, 0.05% Tween 20, and 1% non-fat dry milk. The membranes were incubated for 1 h with anti-PMCA monoclonal antibodies (Adamo et al., 1992; Caride et al., 1996) from ascitic fluid (dilution 1:1,000) and developed using biotinylated antimouse immunoglobulin G and avidin-horseradish peroxidase conjugate.
Data Analysis
Data presented in this work are representative of at least three independent experiments performed in duplicate. The kinetic data were analyzed by non-linear curve fitting using the SigmaPlot 10 scientific data analysis and graphing software (Systat Software Inc.). The same model based on the Hill equation was fitted to PMCA4xb and PMCA4zb and the statistical significance of the difference between datasets was assessed by the F-test as determined by the OriginLab2019 software.
Results
The Isoform h4z Complements the Mutant Yeast K616
In the S. cerevisiae strain K616, the genes coding for Ca2+ ATPases PMR1, PMC1, and the regulatory subunit of calcineurin (CNB) have been inactivated (Cunningham and Fink, 1994). K616 cells cannot proliferate in media containing low concentrations of Ca2+ (10 mM EGTA), but this phenotype can be rescued by the expression of a high-affinity Ca2+ transport system. We have previously shown that the expression of PMCA4xb does not allow the growth of K616 cells in Ca2+ depleted media and successful complementation requires the expression of hyperactive forms of the protein, in which the autoinhibitory mechanism has been disrupted Activated versions of PMCA4xb can be produced either by introducing mutations in the A domain or by the removal of the C-terminal autoinhibitory segment, as in the case of the mutant CT120 lacking the C-terminal 120 amino acid residues (Enyedi et al., 1993; Bredeston and Adamo, 2004; Mazzitelli and Adamo, 2014). As shown in Figure 2, K616 cells transformed with the empty vector, and a vector containing DNA coding for PMCA4xb, PMCA4xCT120, and PMCA4zb, successfully grew in selective media containing 10 mM CaCl2. However, only cells transformed with PMCA4xCT120 and PMCA4zb were able to grow in the presence of 10 mM EGTA. This result suggested that, such as the PMCA4xCT120 mutant, the PMCA4zb is hyperactive and has a higher Ca2+ pumping activity than PMCA4xb.
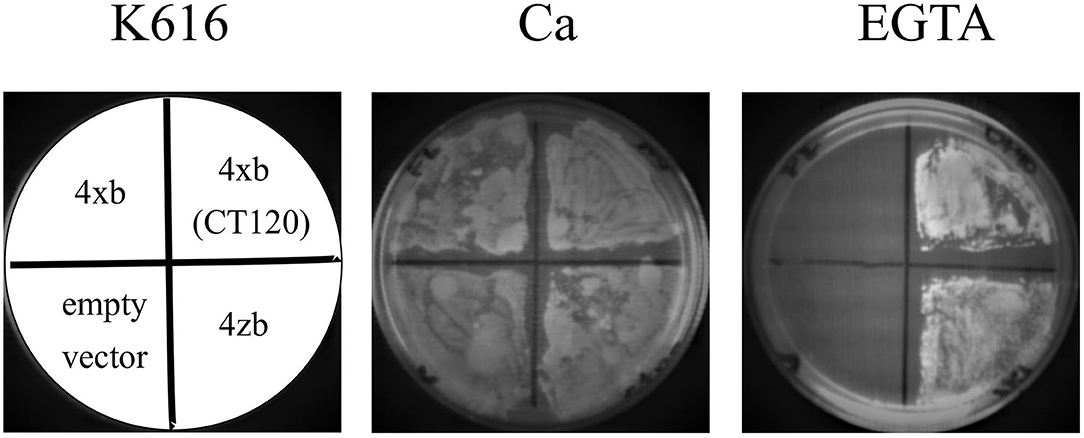
Figure 2. PMCA4xb-GFP complements the K616 yeast strain lacking endogenous Ca2+ ATPases. K616 cells were transformed with the empty pYX112 vector, or vector containing PMCA4xb-GFP, PMCA4zb-GFP, or the hyperactive mutant PMCA4x(CT120)-GFP lacking the C-terminal 120 amino acids segment containing the autoinhibitory domain. Cultures of K616 yeast expressing the recombinant PMCA proteins were grown for 72 h at 28°C on SC-Ura− plates containing 10 mM CaCl2 or 10 mM EGTA.
Subcellular Localization of the Human PMCA4xb and PMCAzb Expressed in Yeast Cells
Using the GFP fusions, we determined the localization of the expressed PMCA4 proteins by confocal microscopy. As shown in Figure 3A, both PMCA4xb and PMCA4zb exhibited a similar pattern of fluorescence intensity in the cell periphery and around the nucleus, suggesting that they localized in the membranes of the endoplasmic reticulum. Confirming this idea, PMCA fluorescence showed significant colocalization with the endoplasmic reticulum protein-membrane ELO3 and very little with PIL1 at the plasma membrane (Figure 3B).
Figure 3. (A) Expression of PMCA4xb and PMCA4zb-GFP in yeast cells. Saccharomyces cerevisiae cells (strain DBY2062) were transformed with the pMP625 vector containing the DNA coding for PMCA4xb-GFP and PMCA4zb-GFP and selected in a Leu− media. The confocal fluorescent image (EGFP filter, LUT 2300, amplification x60) is shown on the left. The prominent yeast vacuoles can be distinguished in the visible image shown on the right. (B) Intracellular localization of the expressed PMCA4zb. On top, PMCA4zb-GFP was coexpressed with the ER membrane protein Elo3-mCherry. The observed distribution suggests that both proteins localize in the perinuclear and peripheral ER membranes. Some background mCherry fluorescence is also detected in vacuoles. On the bottom, mCherry-PMCA4zb was coexpressed with the plasma membrane protein Pil1-GFP. The mCherry fluorescence suggests that most of the PMCA protein in the cell periphery does not reach the plasma membrane.
Purified Preparations of PMCA4xb and PMCA4zb
The PMCA proteins were purified from S. cerevisiae membranes by calmodulin affinity chromatography as described previously (Bredeston and Adamo, 2004; Corradi and Adamo, 2007; Cura et al., 2008; Mazzitelli and Adamo, 2014). Analysis by SDS–PAGE showed that both purified preparations contained a major peptide of an estimated Mr 180 kDa corresponding to the PMCA-GFP fusions and small amounts of two faster-migrating fragments of about 140 and 125 kDa. These shorter fragments were likely the result of the PMCA-GFP proteolysis at the C-terminal region because they lacked GFP fluorescence (Figure 4A). Both PMCA4xb-GFP and PMCAzb-GFP showed similar proteolytic peptides. We further analyzed the purified preparations by Western blot using anti-PMCA specific antibodies 5F10, JA3, and JA9 (Adamo et al., 1992; Caride et al., 1996). The 5F10 epitope is between PMCA4xb residues 719–738 in the central cytosolic loop of the molecule, while JA3 and JA9 recognize the C and N-terminal segments at residues 1156–1180 and 51–75, respectively. Figure 4B shows that antibodies 5F10 and JA3 detected three PMCA peptides that, based on their migration, corresponded to the full-length PMCA-GFP and two shorter proteolytic products. JA9 recognized the PMCA-GFP full-length peptide and only the larger proteolytic fragment. This proteolysis pattern suggests that the large proteolytic fragment of 140 kDa that was recognized by the three antibodies corresponded to the PMCA proteins and was produced by the removal of GFP from the PMCA-GFP. The proteolytic peptide of 125 kDa, would be produced by the cleavage of a segment from the N-terminal region of PMCA because it was no longer detected by JA9.
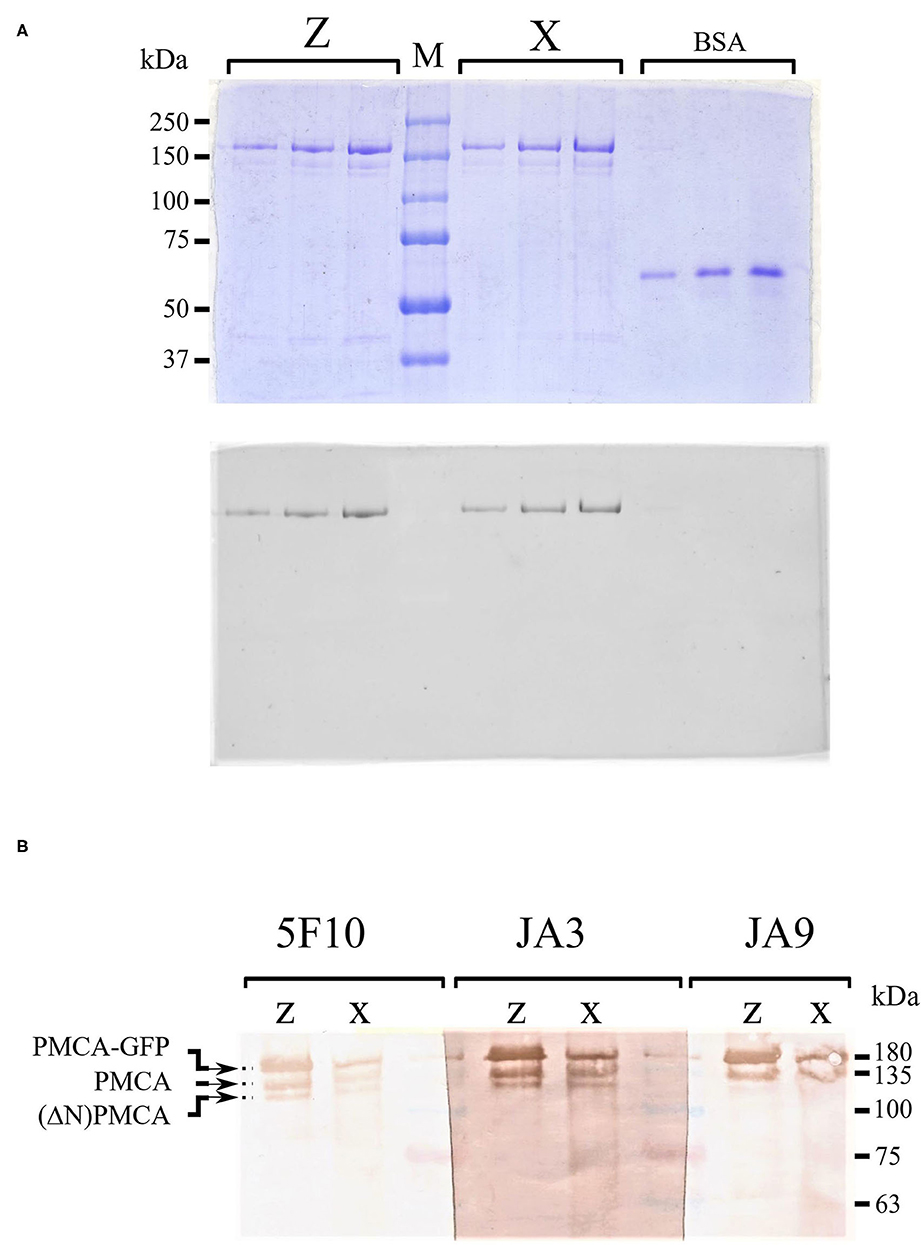
Figure 4. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) of purified PMCA4xb-GFP and PMCA4zb-GFP. The recombinant PMCA proteins were purified as described in “Materials and methods.” Aliquots of the eluate containing the purified proteins were submitted to electrophoresis in a 7.5% SDS–PAGE. Three lanes containing 0.25, 0.5, and 0.75 μg of PMCA4xb-GFP and PMCA4zb-GFP are shown. For comparison 0.3, 0.6, and 0.9 μg of bovine serum albumin were run on the right side of the gel. On top, the gel was stained with Coomassie-blue. On the bottom, the GFP-fluorescence of the same gel is shown. (B) Western blots of purified PMCA4xb-GFP and PMCA4zb-GFP. Western blots of purified PMCA4 preparations with antibodies 5F10, JA3, and JA9. Aliquots of 6 μl (about 1 μg of protein) of PMCA4xb-GFP and PMCA4zb-GFP eluates were loaded.
Adenosine Triphosphatase Activity and Ca2+ Dependency of PMCA4xb and PMCA4zb
The purified and partially delipidated PMCA4xb and PMCAzb were reactivated by the addition of phosphatidylcholine before the Ca2+-ATPase activity was measured at increasing concentrations of Ca2+. As shown in Figure 5, the activity of PMCA4zb was higher than that of PMCA4xb in all the range of Ca2+ concentrations tested. Kinetic analysis indicated that the higher activity of PMCA4zb was a consequence of the increase of the Vmax and the apparent affinity for Ca2+ (Table 1). The addition to the reaction media of a high saturating amount of CaM increased the Vmax and decreased the Ca2+ apparent affinity of PMCA4xb and PMCA4zb. Both, in basal conditions and the CaM-activated state, PMCA4zb exhibited 1.5–2-fold higher maximal activity than PMCA4xb. However, in the CaM-activated state, the apparent affinity for Ca2+ of both variants was similar.
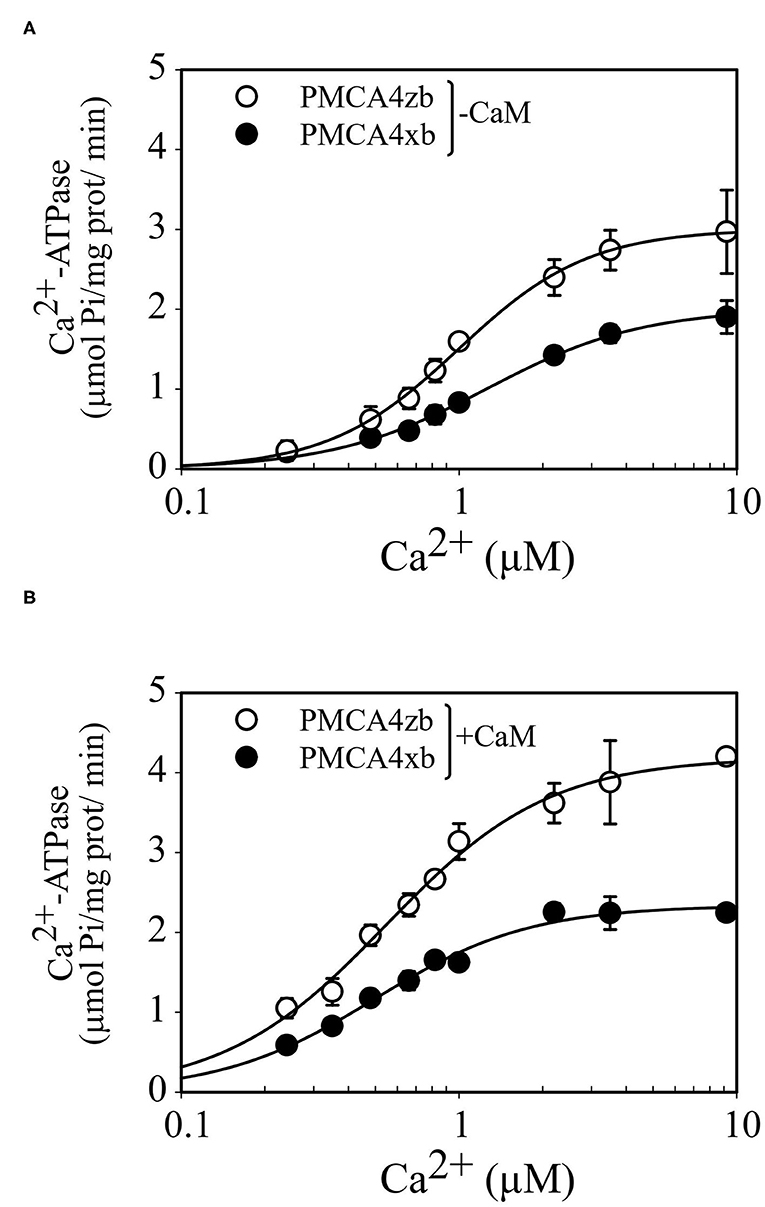
Figure 5. Ca2+ dependence of the Ca2+-ATPase activity of purified PMCA4zb-GFP and PMCA4xb-GFP in the absence (A) and the presence of calmodulin (B). The ATPase reaction was measured at 28°C as described previously (Mazzitelli and Adamo, 2014). The reaction media contained 1 μg of PMCA supplemented with PC, 50 mM HEPES-K (pH = 7.2 at 28°C), 100 mM KCl, 4 mM MgCl2, 3 mM ATP, 500 μM EGTA, and enough CaCl2 to give the indicated concentrations of Ca2+. When calmodulin was present its final concentration was 200 nM. The reaction time was 15 min. The data points are from a representative experiment conducted in duplicate. Error bars show the SD. The lines are the best fit to the data given by the Hill equation v = Vmax *[Ca]nH/([K0.5Ca]nH + [Ca]nH) using the values of the parameters shown in Table 1.
Dependence With Acidic Lipids
To compare the responses of PMCA4xb and PMCAzb to acidic lipids, we supplemented the purified proteins with different amounts of a mixture of acidic lipids extracted from the porcine brain (BE) before measuring the Ca2+-ATPase activity. As shown in Figure 6, the activity of both PMCA4xb and PMCAzb increased with increasing amounts of BE following a sigmoidal curve. At saturating, amounts of BE PMCA4zb had about two times the activity of PMCAxb, an increment somewhat similar to that obtained with calmodulin. However, at lower amounts of acidic lipids, PMCA4zb was nearly 8-fold more active than PMCA4xb, and its activity increased steeply, reaching maximal activity at lower amounts of BE.
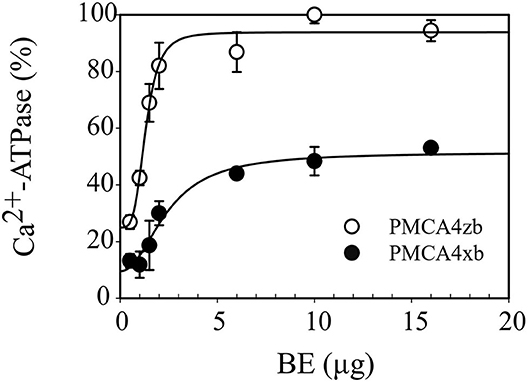
Figure 6. Activation of purified PMCA4xb-GFP and PMCA4zb-GFP by acidic lipids. The Ca2+-ATPase activity was measured as indicated in Figure 5. PMCA4xb-GFP and PMCA4zb-GFP proteins (1 μg) were supplemented with the indicated amount of acidic lipid brain extract (BE). The concentration of Ca2+ was 10 μM. The continuous lines represent the best fitting of the data to a Hill equation v = v0 + vBE *[BE]nH /([K0.5BE]nH + [BE]nH) with the following parameters. For PMCA4xb, v0 = 9 ± 5%, vBE = 42 ± 8%, K0.5BE = 2.4 ± 0.5 μg, nH = 2 ± 1; for PMCA4zb; v0 = 25 ± 7%, vBE = 69 ± 7%, K0.5BE = 1.3 ± 0.1 μg, nH = 4 ± 1.
Discussion
The existence of a great diversity of PMCA isoforms and splice variants is likely the result of cell-specific requirements for controlling the spatial and temporal magnitude of the cytosolic Ca2+ signaling. Most cells express simultaneously a variety of PMCA variants, and for understanding the role of PMCA, it is necessary to advance the knowledge of each PMCA protein, their basal activity and how they respond to activators (Brini et al., 2003; Streheler et al., 2007). PMCA4 is a ubiquitously expressed isoform. Previous studies of PMCA4 used the protein from natural sources, for example, red blood cells which in addition contain PMCA1, or with the recombinant PMCA4x protein (Enyedi et al., 1994; Adamo and Grimaldi, 1998; de Tezanos Pinto and Adamo, 2002, 2006; Bredeston and Adamo, 2004; Corradi and Adamo, 2007; Cura et al., 2008; Mazzitelli and Adamo, 2014). In this study, we have focused on the two PMCA4 variants “x” and “z” produced by the splice at site A, we compared the kinetics properties of purified micellar preparations of the PMCA4xb and PMCA4zb obtained by expression in yeast cells.
Recent studies showed that PMCA associates with IgG-domain proteins neuroplastin and basigin (Korthalsa et al., 2017; Schmidt et al., 2017). The interaction with these auxiliary subunits influences PMCA function, stability, and membrane targeting. The PMCA-neuroplastin interaction seems quite sensitive to the detergents used for PMCA purification, a fact that may explain why the neuroplastin peptide was not previously identified in purified preparations of PMCA despite the proposed equimolar association PMCA-neuroplastin (Schmidt et al., 2017). In this work, as in previous studies of recombinant PMCA, we expressed only the catalytic PMCA4 polypeptide.
PMCA Expression in Yeast
As we reported previously, yeasts are capable of functional PMCA expression (Bredeston and Adamo, 2004; Cura et al., 2008; Mazzitelli and Adamo, 2014). Active PMCA can complement the K616 yeast, and the purified recombinant protein is fully functional. This seems somewhat puzzling because yeasts lack an obvious neuroplastin homolog. However, we observed that expressed PMCA4xb and PMCA4zb localized in the yeast ER membrane, a fact that might be related to the lack of yeast neuroplastin. Further studies of the importance of neuroplastin for PMCA targeting, function, and regulation seem necessary.
The expression of the wild-type autoinhibited PMCA4xb does not rescue the yeast K616 phenotype (Bredeston and Adamo, 2004; Mazzitelli and Adamo, 2014). Successful K616 complementation requires the activation of PMCA4xb by mutations that disrupt the autoinhibitory mechanism. On the contrary, we found that K616 cells expressing the variant PMCA4zb did proliferate in Ca2+ depleted media. Thus, according to the K616 complementation assay, PMCA4zb has a higher Ca2+ transporting activity than PMCA4xb in the yeast cell. In addition, we determined that the purified PMCA4zb had higher Ca2+-ATPase activity and higher apparent affinity for Ca2+ than PMCA4xb in the absence of any activator, confirming the idea that PMCA4zb is a naturally hyperactive PMCA4 variant. Calmodulin activated both PMCA4 variants and increased their affinity for Ca2+ to a high and similar value. This result suggests that the different Ca2+ affinity of the two variants in the absence of calmodulin should be related to the mechanism of autoinhibition and does not involve the modification of the intrinsic affinity of the Ca2+ binding site.
Acidic–Lipid stimulation
Yeast expression provides preparations of purified PMCA, which are very sensitive to acidic lipids (Cura et al., 2008). We found that acidic lipids stimulated both yeast recombinant PMCA4 variants but the activity of PMCA4zb increased much more rapidly with low amounts of acidic lipids. The activation by acidic lipids would involve the interaction at two distinct sites of the PMCA molecule, one in the calmodulin-binding site at the C-terminal regulatory segment, and the second in the AL region of the A-M3 linker (Enyedi et al., 1994; de Tezanos Pinto and Adamo, 2002, 2006). In PMCA4xb, the linker A-M3 region comprises the segment of amino acid residues 300–356. A peptide from this region (residues 339–363) binds acidic lipids, although with a lower affinity than the calmodulin-binding site (Brodin et al., 1992; Filoteo et al., 1992). Thus, the primary site of acidic lipid activation would overlap with that of calmodulin.
We have previously characterized mutants of PMCA4xb containing deletions in the AL region using microsomes of CHO cells expressing the recombinant proteins (de Tezanos Pinto and Adamo, 2002, 2006). The deletion mutant d300-314, with a sequence similar to the natural splicing A variant PMCA4zb, exhibited a higher Ca2+ transport activity due to the higher apparent affinity for Ca2+. On the contrary, deletion of the downstream region (residues 339–360), which was indicated as the acidic lipid-binding site (Brodin et al., 1992), did not promote the activation of the PMCA. While the results presented here show that splice A alters the acidic–lipid stimulation of PMCA4, there is no conclusive evidence that this effect was mediated by the direct binding of lipids to residues 339–360.
The atomic model of PMCA1d was recently reported, but unfortunately, the structure of the A-M3 linker could not be determined (Gong et al., 2018). This segment is enriched in polar and charged residues, and as judged by its susceptibility to protease cleavage, is unusually accessible (Brodin et al., 1992). In this line, it is worth considering that PMCA is very sensitive to cellular proteases and most PMCA preparations exhibit some proteolysis (Adamo et al., 2000), a fact that has led to propose that proteolysis is a natural mechanism for regulating PMCA (Filoteo et al., 1997). We detected a small number of proteolytic fragments in both the PMCA4xb and the PMCAzb preparations. By using the GFP fusions and antibodies with defined epitopes, we observed that the endogenous proteolytic pattern of the PMCA4xb and the PMCA4zb was similar. In addition, the main proteolytic fragment corresponded to the full-length proteins lacking GFP suggesting that a cleavage near the C-terminus of the protein occurred without affecting the C-terminal regulatory region.
It seems appropriate to notice that the variability produced by the splicing site A may have different consequences depending on the PMCA isoform considered. Indeed, in polarized cells, the splice variant “w” targets PMCA2 to apical membranes, while variants “x” and “z” remain basolateral (Chicka and Strehler, 2003; Antalffy et al., 2012). On the other hand, PMCA4x is targeted to apical membranes in pancreatic acinar cells indicating that the targeting capacity of splice A variants may be isoform and cell-specific (Baggaley et al., 2007). Furthermore, membranes from CHO cells expressing PMCA2wb and PMCA2xb exhibit similar Ca2+-ATPase activity and acidic lipid stimulation suggesting that the “w” insertion in PMCA2 had minimal functional consequences (Brini et al., 2010). Interestingly, the PMCA2zb was recognized as a very active pump capable of decreasing the Ca2+ signal peak in about half the time needed by the PMCA2wb (Ficarella et al., 2007).
A Possible Molecular Mechanism of PMCA4zb Activation
The A-M3 linker where the splicing A variants of PMCA occur is an important functional region of the P-ATPases. Indeed, the rotation of the A domain that accompanies the formation of an intramembrane exit pathway of the bound transported substrate is triggered by the strain of the A-M3 (Nagarajan et al., 2012). The functional consequences of the A-M3 linker length were analyzed in detail in SERCA (Nyholm Holdensen and Andersen, 2009). SERCA possesses a shorter A-M3 linker compared with the PMCA (refer to Figure 1). A series of insertional SERCA mutants containing up to 41 glycine and proline residues at the corresponding site of splicing A of PMCA, decreased the SERCA turnover to 30% of the WT. These results show that, in analogy with SERCA, the length of the A-M3 linker influenced the activity of the PMCA and made PMCAzb with a shorter linker more active than PMCA4x.
In comparison with SERCA, the PMCA A-M3 linker shows, in addition to the conserved A3 helix, a possible helical structure highly enriched in charged amino acids formed by PMCA4x residues E335-V342. By using a combination of homology modeling and structure prediction (Källberg et al., 2012), it can be observed that in PMCA4x these helical structures are deeply inserted in the catalytic core, where they may impair the interaction of the A and P domains while in PMCA4z these helices would move toward the surface of the protein (Figure 7).
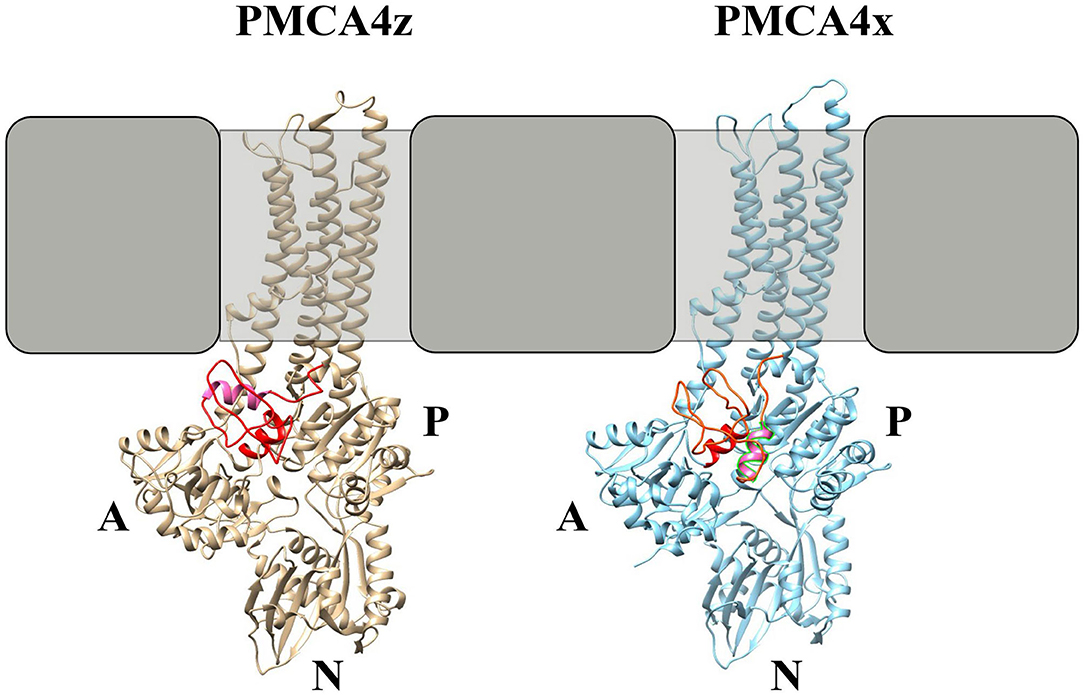
Figure 7. Molecular modeling of PMCA4x and PMCA4z. Homology models of PMCA4 were obtained using the partial structure of PMCA1d (Gong et al., 2018) as a template. Only the portion of the PMCA from the amino-terminal region to the end of M6 was modeled. The tertiary structure and contact prediction model of the A-M3 linker were obtained using the RaptorX web service (Källberg et al., 2012). The SERCA-conserved A3 helix is colored red while the putative AL helix in PMCA is colored pink.
A clear difference between SERCA and PMCA pumps is the C-terminal autoinhibitory domain of PMCA that binds to the A and N domains. A lower affinity of the autoinhibitory domain for its sites in the core of the protein may explain the basal activated state of PMCA4zb. It seems reasonable to consider that two activating mechanisms, one involving the modification of the A-M3 linker (splice site A) and the other resulting in the release of the C-terminal autoinhibition (splice site C) are not independent but cooperate for regulating the activity of the PMCA.
In this study, we found that PMCA4z a previously not characterized variant of PMCA originated by splicing at site A is more active and more sensitive to activation by acidic lipids than the more common form of PMCA4x. Future work will be needed to assess the relevance of this finding for the control of the Ca2+ signal in cells expressing the variant “z” in normal and pathological conditions.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
GRC, LRM, GDP, LR, and FTP performed experiments and analyzed data. GRC, FTP, LRM, and HPA designed research and analyzed data. HPA conceived the project and wrote the manuscript with comments from all the authors. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge funding from Agencia Nacional de Promoción Científica y Tecnológica PICT 2017-1690, Consejo Nacional de Investigaciones Científicas y Tecnológicas PIP 2015-773 and Universidad de Buenos Aires UBACyT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Pablo S. Aguilar at the Institut Pasteur, Uruguay for the gift of yeasts expressing Elo3-mCherry and Pil1-GFP.
Abbreviations
PMCA, plasma membrane Ca2+ pump; PMCA4xb, plasma membrane Ca2+ pump derived from the human gene 4, spliced form x at the splicing site “A” and spliced form b at the splicing site “C”; PMCA4zb, plasma membrane Ca2+ pump derived from the human gene 4, spliced form z at the splicing site “A” and spliced form b at the splicing site “C”; GFP, green fluorescent protein, CaM, calmodulin; PC, phosphatidylcholine; PI, phosphatidylinositol; PS, phosphatidylserine, PA, phosphatidic acid; C12E10, polyoxyethylene10-laurylether; E1 and E2, conformers of the Ca2+-ATPase; EP, phosphoenzyme; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; BE, extract from bovine brain containing acidic lipids.
References
Adamo, H. P., Caride, A. J., and Penniston, J. T. (1992). Use of expression mutants and monoclonal antibodies to map the erythrocyte Ca2+ pump. J. Biol. Chem. 267, 14244–14249. doi: 10.1016/S0021-9258(19)49704-4
Adamo, H. P., and Grimaldi, M. E. (1998). Functional consequences of relocating the C-terminal calmodulin-binding autoinhibitory domains of the plasma membrane Ca2+ pump near the N-terminus. Biochem. J. 331, 763–766. doi: 10.1042/bj3310763
Adamo, H. P., Grimaldi, M. E., and Bredeston, L. M. (2000). The N-terminal region of the plasma membrane Ca(2+) pump does not separate from the main catalytic fragments after proteolysis. Biochim. Biophys. Acta. 1464, 127–134. doi: 10.1016/S0005-2736(99)00253-9
Adamo, H. P., and Penniston, J. T. (1992). New Ca2+ pump isoforms generated by alternative splicing of rPMCA2 mRNA. Biochem. J. 283, 355–359. doi: 10.1042/bj2830355
Antalffy, G., Mauer, A. S., Pászty, K., Hegedus, L., Padányi, R., Enyedi, A., and Strehler, E. E. (2012). Plasma membrane calcium pump (PMCA) isoform 4 is targeted to the apical membrane by the w-splice insert from PMCA2. Cell Calcium 51, 171–178. doi: 10.1016/j.ceca.2011.12.010
Baggaley, E., McLarnon, S., Demeter, I., Varga, G., and Bruce, J. I. E. (2007). Differential regulation of the apical plasma membrane Ca2+-ATPase by protein kinase A in parotid acinar cells. J. Biol. Chem. 282, 37678–37693. doi: 10.1074/jbc.M703416200
Bauer, A., and Kolling, R. (1996). The SAC3 gene encodes a nuclear protein required for normal progression of mitosis. Yeast 12, 965–975. doi: 10.1002/(SICI)1097-0061(199608)12:10<965::AID-YEA999>3.0.CO;2-Q
Beesley, P. W., Herrera-Molina, R., Smalla, K. H., and Seidenbecher, C. (2014). The neuroplastin adhesion molecules: key regulators of neuronal plasticity and synaptic function. J. Neurochem. 131, 268–283. doi: 10.1111/jnc.12816
Boczek, T., Radzik, T., Ferenc, B., and Zylinska, L. (2019). The puzzling role of neuron-specific PMCA isoforms in the aging process. Int. J. Mol. Sci. 20:6338. doi: 10.3390/ijms20246338
Bradford, M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–256. doi: 10.1016/0003-2697(76)90527-3
Bredeston, L. M., and Adamo, H. P. (2004). Loss of autoinhibition of the plasma membrane Ca2+ pump by substitution of Aspartic 170 by Asparagine: Activation of plasma membrane calcium ATPase 4 without disruption of the interaction between the catalytic core and the C-terminal regulatory domain. J. Biol. Chem. 279, 41619–41625. doi: 10.1074/jbc.M403116200
Brini, M., Coletto, L., Pierobon, N., Kraev, N., Guerini, D., and Carafoli, E. (2003). A comparative functional analysis of plasma membrane Ca2+ pump isoforms in intact cells. J. Biol. Chem. 278, 24500–24508. doi: 10.1074/jbc.M300784200
Brini, M., Di Leva, F., Ortega, C. K., Domi, T., Ottolini, D., Leonardi, E., Tosatto, S. C., and Carafoli, E. (2010). Deletions and mutations in the acidic lipid-binding region of the plasma membrane Ca2+ pump: a study on different splicing variants of isoform 2. J. Biol. Chem. 285, 30779–30791. doi: 10.1074/jbc.M110.140475
Brodin, P., Falchetto, R., Vorherr, T., and Carafoli, E. (1992). Identification of two domains which mediate the binding of activating phospholipids to the plasma-membrane Ca2+ pump. Eur. J. Biochem. 204, 939–946. doi: 10.1111/j.1432-1033.1992.tb16715.x
Caride, A. J., Filoteo, A. G., Enyedi, A., Verma, A. K., and Penniston, J. T. (1996). Detection of isoform 4 of the plasma membrane calcium pump in human tissues by using isoform-specific monoclonal antibodies. Biochem. J. 316, 353–359. doi: 10.1042/bj3160353
Chen, J., Sitsel, A., Benoy, V., Sepúlveda, M. R., and Vangheluwe, P. (2020). Primary active Ca2+ transport systems in health and disease. Cold Spring Harb. Perspect. Biol. 12:a035113. doi: 10.1101/cshperspect.a035113
Chicka, M. C., and Strehler, E. E. (2003). Alternative splicing of the first intracellular loop of plasma membrane Ca2+-ATPase isoform 2 alters its membrane targeting. J. Biol. Chem. 278, 18464–18470. doi: 10.1074/jbc.M301482200
Corradi, G. R., and Adamo, H. P. (2007). Intramolecular fluorescence resonance energy transfer between fused autofluorescent proteins reveals rearrangements of the N- and C-terminal segments of the plasma membrane Ca2+ pump involved in the activation. J. Biol. Chem. 282, 35440–35448. doi: 10.1074/jbc.M703377200
Cunningham, K. W., and Fink, G. R. (1994). Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J.Cell. Biol. 124, 351–363 doi: 10.1083/jcb.124.3.351
Cura, C. I., Corradi, G. R., Rinaldi, D. E., and Adamo, H. P. (2008). High sensibility to reactivation by acidic lipids of the recombinant human plasma membrane Ca2+-ATPase isoform 4xb purified from Saccharomyces cerevisiae. Biochim. Biophys. Acta Biomembr. 1778, 2757–2764. doi: 10.1016/j.bbamem.2008.08.020
de Tezanos Pinto, F., and Adamo, H. P. (2002). Deletions in the acidic lipid-binding region of the plasma membrane Ca2+ pump. A mutant with high affinity for Ca2+ resembling the acidic lipid-activated enzyme. J. Biol. Chem. 277, 12784–12789. doi: 10.1074/jbc.M111055200
de Tezanos Pinto, F., and Adamo, H. P. (2006). Deletions in the AL region of the h4xb plasma membrane Ca2+ pump. High apparent affinity for Ca2+ of a deletion mutant resembling the alternative spliced form h4zb. FEBS Lett. 580, 1576–1580. doi: 10.1016/j.febslet.2006.01.088
Elble, R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechnology 13, 18–20.
Enyedi, A., Flura, M., Sarkadi, B., Gardos, G., and Carafoli, E. (1987). The maximal velocity and the calcium affinity of the red cell calcium pump may be regulated independently. J.Biol. Chem. 262, 6425–6430. doi: 10.1016/S0021-9258(18)45587-1
Enyedi, A., Verma, A. K., Adamo, H. P., Filoteo, A. G., Streheler, E. E., and Penniston, J. T. (1994). The Ca2+ affinity of the plasma membrane Ca2+ pump is controlled by alternative splicing. J. Biol. Chem. 269, 41–43. doi: 10.1016/S0021-9258(17)42307-6
Enyedi, A., Verma, A. K., Filoteo, A. G., and Penniston, J. T. (1993). A highly active 120-kDa truncated mutant of the plasma membrane Ca2+ pump. J. Biol. Chem. 268, 10621–10626. doi: 10.1016/S0021-9258(18)82243-8
Ficarella, R., Di Leva, F., Bortolozzi, M., Ortolano, S., Donaudy, F., Petrillo, M., et al. (2007). A functional study of plasma-membrane calcium-pump isoform 2 mutants causing digenic deafness. Proc. Natl. Acad. Sci. U. S. A. 104, 1516–1521. doi: 10.1073/pnas.0609775104
Filoteo, A. G., Elwess, N. L., Enyedi, A., Caride, A., Aung, H. H., and Penniston, J. T. (1997). Plasma membrane Ca2+ pump in rat brain. Patterns of alternative splices seen by isoform-specific antibodies. J. Biol. Chem. 272, 23741–23747. doi: 10.1074/jbc.272.38.23741
Filoteo, A. G., Enyedi, A., and Penniston, J. T. (1992). The lipid-binding peptide from the plasma membrane Ca2+ pump binds calmodulin, and the primary calmodulin-binding domain interacts with lipid. J. Biol. Chem. 267, 11800–11805. doi: 10.1016/S0021-9258(19)49769-X
Gong, D., Chi, X., Ren, K., Huang, G., Zhou, G., Yan, N., Lei, J., and Zhou, Q. (2018). Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin. Nat. Commun. 9:3623. doi: 10.1038/s41467-018-06075-7
Hegedus, L., Zámbó, B., Pászty, K., Padányi, R., Varga, K., Penniston, J. T., and Enyedi, Á. (2020). Molecular diversity of plasma membrane Ca(2+) transporting ATPases: their function under normal and pathological conditions. Adv. Exp. Med. Biol. 1131, 93–129. doi: 10.1007/978-3-030-12457-1_5
Källberg, M., Wang, H., Wang, S., Peng, J., Wang, Z., Lu, H., and Xu, J. (2012). Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522. doi: 10.1038/nprot.2012.085
Kim, E., DeMarco, S. J., Marfatia, S. M., Chishti, A. H., Sheng, M., and Strehler, E. E. (1998). Plasma membrane Ca2+-ATPase isoform 4b binds to membrane-associated guanylate kinase (MAGUK) proteins via their PDZ (PSD-95/Dlg/ZO-1) domains. J. Biol. Chem. 273, 1591–1595. doi: 10.1074/jbc.273.3.1591
Korthalsa, M., Langnaese, K., Smalla, K. H., Kähne, T., Herrera-Molina, R., Handschuh, J., et al. (2017). A complex of neuroplastin and plasma membrane calcium ATPase controls T cell activation. Sci. Rep. 7:8358. doi: 10.1038/s41598-017-08519-4
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Li, M., Ho, P. W., Pang, S. Y., Tse, Z. H., Kung, M. H., Sham, P. C., and Ho, S. L. (2014). PMCA4(ATP2B4) mutation in familial spastic paraplegia. PLoS ONE 9:e104790. doi: 10.1371/journal.pone.0104790
Lopreiato, R., Giacomello, M., and Carafoli, E. (2014). The plasma membrane calcium pump: new ways to look at an old enzyme. J. Biol. Chem. 289, 10261–10268. doi: 10.1074/jbc.O114.555565
Mazzitelli, L. R., and Adamo, H. P. (2014). Hyperactivation of the human plasma membrane Ca2+ pump PMCA h4xb by mutation of Glu99 to Lys”. J. Biol. Chem. 289, 10761–10768. doi: 10.1074/jbc.M113.535583
Mohamed, T. M., Abou-Leisa, R., Stafford, N., Maqsood, A., Zi, M., Prehar, S., et al. (2016). The plasma membrane calcium ATPase 4 signaling in cardiac fibroblasts mediates cardiomyocyte hypertrophy. Nat. Commun. 7:11074. doi: 10.1038/ncomms11074
Mohamed, T. M., Oceandy, D., Zi, M., Prehar, S., Alatwi, N., Wang, Y., et al. (2011). Plasma membrane calcium pump (PMCA4)-neuronal nitric-oxide synthase complex regulates cardiac contractility through modulation of a compartmentalized cyclic nucleotide microdomain. J. Biol. Chem. 286, 41520–41529. doi: 10.1074/jbc.M111.290411
Müller, C. S., Haupt, A., Bildl, W., Schindler, J., Knaus, H.-G., Meissner, M., et al. (2010). Quantitative proteomics of the Cav2 channel nano-environments in the mammalian Brain 107, 14959–14957. doi: 10.1073/pnas.1005940107
Nagarajan, A., Andersen, J. P., and Woolf, T. B. (2012). The role of domain:domain interactions vs domain:water interactions in the coarse-grained simulations of the E1P to E2P transitions in Ca-ATPase (SERCA). Proteins 80, 1929–1947. doi: 10.1002/prot.24070
Niggli, V., Adunyah, E. S., Penniston, J. T., and Carafoli, E. (1981). Purified (Ca2+-Mg2+)-ATPase of the erythrocyte membrane. Reconstitution and effect of calmodulin and phospholipids. J. Biol. Chem. 256, 395–401. doi: 10.1016/S0021-9258(19)70149-5
Nyholm Holdensen, A., and Andersen, J. P. (2009). The length of the A-M3 linker is a crucial determinant of the rate of the Ca2+-transport cycle of sarcoplasmic reticulum Ca2-ATPase. J. Biol. Chem. 284, 12258–12265. doi: 10.1074/jbc.M900977200
Schmidt, N., Kollewe, A., Constantin, C. E., Henrich, S., Ritzau-Jost, A., Bildl, W., Saalbach, A., Hallermann, S., Kulik, A., Fakler, B., and Schulte, U. (2017). Neuroplastin and basigin are essential auxiliary subunits of plasma membrane Ca2+-ATPases and key regulators of Ca2+- clearance. Neuron 96, 827–838. doi: 10.1016/j.neuron.2017.09.038
Stafford, N., Wilson, C., Oceandy, D., Neyses, L., and Cartwright, E. J. (2017). The plasma membrane calcium ATPases and their role as major new players in human disease. Physiol. Rev. 97, 1089–1125. doi: 10.1152/physrev.00028.2016
Streheler, E. E., Caride, A. J., Filoteo, A. G., Xiong, Y., Penniston, J. T., and Enyedi, A. (2007). Plasma membrane Ca2+ ATPases as dynamic regulators of cellular calcium handling. Ann. N. Y. Acad. Sci.1099, 226–236. doi: 10.1196/annals.1387.023
Strehler, E. E. (2015). Plasma membrane calcium ATPases: from generic Ca2+ sump pumps to versatile systems for fine-tuning cellular Ca2+. Biochem. Biophys. Res. Commun. 460, 26–33. doi: 10.1016/j.bbrc.2015.01.121
Keywords: calcium transport, calcium ATPase, PMCA isoforms, neuronal PMCA, heart PMCA
Citation: Corradi GR, Mazzitelli LR, Petrovich GD, de Tezanos Pinto F, Rochi L and Adamo HP (2021) Plasma Membrane Ca2+ Pump PMCA4z Is More Active Than Splicing Variant PMCA4x. Front. Cell. Neurosci. 15:668371. doi: 10.3389/fncel.2021.668371
Received: 16 February 2021; Accepted: 22 July 2021;
Published: 26 August 2021.
Edited by:
Tatiana Adasme, Centro de Investigación Clínica Avanzada (CICA), ChileReviewed by:
Luís F. Ribeiro, VIB and KU Leuven Center for Brain and Disease Research, BelgiumFrancesco Lodola, University of Milano-Bicocca, Italy
Pablo Munoz, Universidad de Valparaiso, Chile
Copyright © 2021 Corradi, Mazzitelli, Petrovich, de Tezanos Pinto, Rochi and Adamo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerardo R. Corradi, Z2NvcnJhZGlAcWIuZmZ5Yi51YmEuYXI=; Hugo P. Adamo, aHBhZGFtb0BxYi5mZnliLnViYS5hcg==
In Memoriam: This paper is dedicated to the memory of Dr. Alcides F. Rega.
 Gerardo R. Corradi
Gerardo R. Corradi Luciana R. Mazzitelli
Luciana R. Mazzitelli Guido D. Petrovich
Guido D. Petrovich Felicitas de Tezanos Pinto
Felicitas de Tezanos Pinto Hugo P. Adamo
Hugo P. Adamo