
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 21 April 2021
Sec. Cellular Neuropathology
Volume 15 - 2021 | https://doi.org/10.3389/fncel.2021.664151
This article is part of the Research Topic Molecular Mechanisms Underlying C9orf72 Neurodegeneration View all 8 articles
Since the discovery of the C9orf72 repeat expansion mutation as causative for chromosome 9-linked amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) in 2011, a multitude of cellular pathways have been implicated. However, evidence has also been accumulating for a key mechanism of cellular compartmentalization—phase separation. Liquid-liquid phase separation (LLPS) is fundamental for the formation of membraneless organelles including stress granules, the nucleolus, Cajal bodies, nuclear speckles and the central channel of the nuclear pore. Evidence has now accumulated showing that the formation and function of these membraneless organelles is impaired by both the toxic arginine rich dipeptide repeat proteins (DPRs), translated from the C9orf72 repeat RNA transcript, and the repeat RNA itself. Both the arginine rich DPRs and repeat RNA themselves undergo phase separation and disrupt the physiological phase separation of proteins involved in the formation of these liquid-like organelles. Hence abnormal phase separation may explain a number of pathological cellular phenomena associated with C9orf72-ALS/FTD. In this review article, we will discuss the principles of phase separation, phase separation of the DPRs and repeat RNA themselves and how they perturb LLPS associated with membraneless organelles and the functional consequences of this. We will then discuss how phase separation may impact the major pathological feature of C9orf72-ALS/FTD, TDP-43 proteinopathy, and how LLPS may be targeted therapeutically in disease.
The C9orf72 mutation is an expansion of a GGGGCC (G4C2) repeat in intron 1 of the gene. In unaffected individuals the G4C2 is repeated 2 to 23 times, whereas in those with the mutation, the sequence is expanded to contain hundreds to thousands of repeats (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Due to its location upstream of the coding region, the mutation can lead to a reduction in the levels of the protein that it encodes (Xiao et al., 2015; Sivadasan et al., 2016), which is involved in the regulation of endo-lysosomal trafficking and autophagy (Farg et al., 2014; Sellier et al., 2016; Webster et al., 2016). However, a common finding from murine C9orf72 knockout models is the lack of neurodegeneration or TDP-43 pathology—a key pathological feature of C9orf72-ALS/FTD (O’Rourke et al., 2015; Atanasio et al., 2016; Burberry et al., 2016; Jiang et al., 2016; Sudria-Lopez et al., 2016; Sullivan et al., 2016). These findings indicate that loss of protein function is not sufficient to cause the disease. However, effects of reduced C9orf72 in autophagy, immune dysregulation (O’Rourke et al., 2015; Atanasio et al., 2016; Burberry et al., 2016; Jiang et al., 2016; Sellier et al., 2016; Sudria-Lopez et al., 2016; Sullivan et al., 2016; Ugolino et al., 2016; Webster et al., 2016; Shi et al., 2018; Zhu et al., 2020), axon growth (Sivadasan et al., 2016) and stress granule dynamics (Maharjan et al., 2017) propose a modulatory role in disease pathogenesis.
However, similar to other non-coding repeat expansion mutations, the G4C2 repeat produces repetitive RNA which is translated into repetitive polypeptides, which have both been proposed to cause pathogenesis (Mizielinska and Isaacs, 2014). The G4C2 repeat is transcribed in both directions generating G4C2 (sense) and C4G2 (antisense) repeat RNA. These RNA species both form small RNA aggregates called RNA foci in patient brain (DeJesus-Hernandez et al., 2011; Gendron et al., 2013; Lagier-Tourenne et al., 2013; Mizielinska et al., 2013; Mackenzie et al., 2014; DeJesus-Hernandez et al., 2017), which may cause dysfunction by sequestering RNA-binding proteins (Almeida et al., 2013; Donnelly et al., 2013; Lee et al., 2013; Sareen et al., 2013; Cooper-Knock et al., 2014, 2015; Haeusler et al., 2014; Rossi et al., 2015; Mori et al., 2016; Celona et al., 2017). Both sense and antisense repeat RNA also undergo non ATG-dependent translation into repetitive polypeptides. The polypeptides produced consist of two alternating amino acids (due to the repetitive RNA code) and are thus termed dipeptide repeat proteins (DPRs). Translation occurs in all six possible frames, three sense and three antisense, producing five different DPRs as one sense and antisense frame are the same. These are polypeptides of glycine-proline (poly-GP), glycine-alanine (poly-GA), glycine-arginine (poly-GR), proline-arginine (poly-PR) and proline-alanine (poly-PA). These DPRs are also all found to form inclusions in patient brain (Ash et al., 2013; Mori et al., 2013a), but seem largely distinct from the classic TDP-43 pathology. In vitro and in vivo models show that the arginine rich DPRs poly-GR and poly-PR and the most aggregation prone DPR poly-GA can induce significant toxicity (Kwon et al., 2014; May et al., 2014; Mizielinska et al., 2014; Wen et al., 2014; Zhang et al., 2014, 2019a; Tao et al., 2015; Lee et al., 2016; Schludi et al., 2017; Choi et al., 2019; Hao et al., 2019; Cook et al., 2020; LaClair et al., 2020; Zhou et al., 2020). In vivo studies have also suggested a direct role for the repeat RNA, albeit with associations with cytoplasmic RNA rather than the classic nuclear foci (Burguete et al., 2015; Swinnen et al., 2018).
A number of cellular processes have been shown to be impaired by the repeat RNA and/or the arginine rich DPRs including the regulation of transcription, ribosomal biogenesis and translation, nucleocytoplasmic transport and RNA granules (Balendra and Isaacs, 2018; Mandrioli et al., 2020). All of these cellular processes are associated with membraneless organelles—multicomponent, viscous liquid-like structures that lack a lipid bilayer. These membraneless assemblies are found in both the nucleus and cytoplasm and typically contain both RNA and protein molecules. Examples of such membraneless organelles include the nucleolus, nuclear pore complex, stress granules, nuclear speckles, paraspeckles, p-bodies and Cajal bodies (Brangwynne et al., 2015; Freibaum and Taylor, 2017). The altered assembly, dynamics, and function of membraneless organelles may account for many of the widespread cellular abnormalities observed in C9orf72-ALS/FTD and can explain several of the mechanisms associated with both G4C2 repeat RNA and arginine rich DPR toxicity.
In cell biology liquid-liquid phase separation (LLPS) is a process in which a homogenous liquid solution consisting of RNA and/or protein separates into two different phases; with one of these separated phases containing an increased concentration of RNA and protein—the dense phase, and the other phase—known as the dilute phase—now depleted of them (Alberti and Dormann, 2019; Figures 1A,B). The dense phase usually resembles liquid droplets and indeed shows liquid-like properties including fusion, coalescence, dripping and a rapid exchange of molecules (Alberti and Dormann, 2019; Alberti et al., 2019). This phase separation of organic molecules into droplets through LLPS is known as coacervation and the resulting droplets can also be referred to as coacervates (Deshpande et al., 2019). Membraneless organelle formation most frequently occurs through spontaneous LLPS in which protein laden RNAs separate themselves from the surrounding aqueous nucleoplasm or cytoplasm forming a reversible state which can equally quickly dissolve (Brangwynne et al., 2015; Taylor et al., 2016).
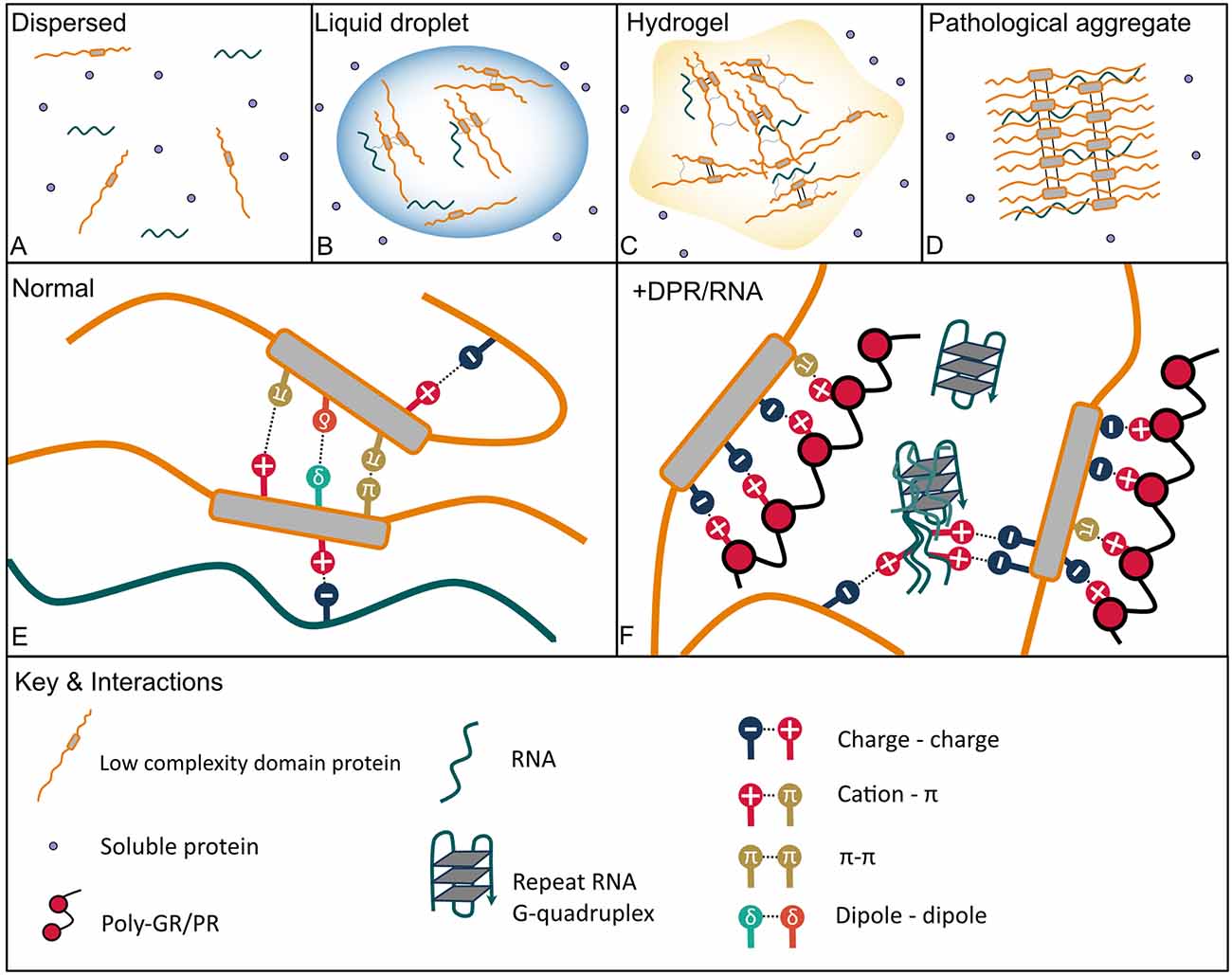
Figure 1. Protein phase transition states and interactions with C9orf72 arginine rich dipeptide repeat proteins (DPRs) and GGGGCC (G4C2) repeat RNA that lead to aberrant phase separation. (A) Dispersed—soluble proteins are freely dispersed in a dilute phase. (B) Proteins with intrinsically disordered regions such as low complexity domains (LCDs) can de-mix from a dilute phase often in the presence of RNA through liquid-liquid phase separation (LLPS) to form dynamic liquid droplets in which molecules can still diffuse in and out and disperse rapidly. (C) Liquid droplets can transition into more solid-like hydrogels which consist of amyloid-like fibrils, which are less dynamic but still reversable. (D) Aberrant phase separation facilitates the formation of pathological permanent amyloid fibrils which are the constituents of aggregates found in neurodegenerative disease brain. (E) Physiological interactions that mediate LLPS include pi (Π)–Π, cation–Π, polar (dipole-dipole), charge-charge interactions in addition to hydrophobic and Π-sp2 interactions, and hydrogen bonding (not shown). (F) The arginine-rich DPRs mimic physiological interactions driving aberrant phase separation with the arginine residues acting as the cation in electrostatic interactions, both charge-charge and with aromatic side chains within LCDs (cation-Π). The G4C2 repeat RNA can also undergo charge-charge interactions with LCDs; further G4C2 G-quadruplexes may enhance RNA-RNA and RNA-LCD interactions and nucleate droplet formation by cellular RNA.
The formation of membraneless organelles has been described as a dynamic liquid demixing process in which cells actively generate phase boundaries to confine functional entities for a temporary period (Aguzzi and Altmeyer, 2016). Once LLPS has occurred it is thought the proteins within the dense phase compartment find themselves in a different solvent environment compared to their surroundings, which may promote specific biochemical interactions and thus functionalities (Aguzzi and Altmeyer, 2016). Most of the proteins that drive intracellular phase separation and the formation of membraneless organelles show strong conformational heterogeneity and are referred to together as intrinsically disordered proteins (Brangwynne et al., 2015; Boeynaems et al., 2018). These proteins do not have well defined protein folding and are highly flexible due to significantly reduced numbers of aliphatic and aromatic residues (Alberti and Dormann, 2019). However, many of these proteins still contain structured regions and only segments that do not form a well-defined tertiary three-dimensional structure; these disordered regions of the protein are referred to as intrinsically disordered regions. The structural plasticity of these disordered proteins and regions allows them to dynamically adopt different confirmations such as energetically favorable higher-order protein assemblies and undergo a multitude of promiscuous multivalent interactions (Aguzzi and Altmeyer, 2016). Such factors dictate the behavior of disordered proteins within complex liquids like the intracellular milieu and facilitate phase separation (Hyman et al., 2014; Brangwynne et al., 2015; Aguzzi and Altmeyer, 2016; Boeynaems et al., 2018). Thus, these multivalent interactions are important for the formation of dynamic heterogeneous assembles, such as membraneless organelles (Mitrea and Kriwacki, 2016; Freibaum and Taylor, 2017).
Intrinsically disordered regions typically contain repetitive sequence strings biased towards specific amino acids (Boeynaems et al., 2018) and are categorized based upon the composition of their sequence and motifs (Alberti and Dormann, 2019). One important example is low complexity sequence domains (LCD)—amino acid sequences between 75 and 300 amino acids in length with a high evolutionary conservation, present in one-third of the human proteome (Freibaum and Taylor, 2017). These domains often consist of a high number of uncharged polar amino acids such as glycine, asparagine and serine interspersed with aromatic and charged residues (Wang et al., 2018b). The low sequence diversity generates multiple short motifs and repeats such as, glycine/serine-phenylalanine/tyrosine-glycine/serine, arginine-glycine, phenylalanine-glycine, poly-glutamine, poly-asparagine, and blocks of positive or negative charges important for the formation of ribonucleoprotein granules and other biomolecular condensates (Banani et al., 2017; Feng et al., 2019). Specific LCDs such as prion-like domains and glycine-arginine rich (RGG) domains found in RNA-binding proteins are also named for their composition patterns with prion-like domains enriched in glutamine-asparagine and aromatic residues, similar to yeast prion sequences. The residues within these domains mediate several important interactions that mediate phase separation including charge-charge, cation–pi, pi–pi, and polar (dipole-dipole) in addition to hydrophobic and pi/sp2 interactions, and hydrogen bonding (Murthy et al., 2019; Peran and Mittag, 2020; Figure 1E). Pi (Π)-stacking interactions occur between delocalized pi electrons in aromatic rings but also between planar non-aromatic residues, such as arginine, glutamine, asparagine, aspartic acid and glutamic acid (Vernon et al., 2018). Positively charged residues, most commonly arginine, can also form cation-pi interactions with electron-rich aromatic residues. Oppositely charged residues arranged in like-charged clusters also undergo charge neutralization by electrostatic charge-charge interactions. These side-chain interactions have been shown to mediate LCD phase separation in a number of condensate biomolecules (Feng et al., 2019; Spannl et al., 2019) and post-translational modifications, missense mutations or scrambling charge clustering inhibits droplet formation by disrupting relevant weak interactions (Feng et al., 2019). These multivalent, low affinity associations mediate interactions that can be rapidly rearranged, including both protein-protein and protein-nucleic acid interactions (Aguzzi and Altmeyer, 2016; Boeynaems et al., 2018; Alberti and Dormann, 2019). Indeed, RNA plays a key role in the formation of the majority of cellular membraneless organelles via phase separation acting as a scaffold for LCD interactions and thus RNA concentration can determine the phase separation behavior of LCD proteins depending on their subcellular location (Langdon and Gladfelter, 2018; Maharana et al., 2018; Garcia-Jove Navarro et al., 2019; Rhine et al., 2020). It is suggested that the ability of RNA to undergo numerous multivalent interactions along with its flexible structure means that RNA essentially imitates the LCD of proteins (Rhine et al., 2020). Further RNA itself has been shown to phase separate via RNA-RNA interactions mediated by electrostatic forces (Jain and Vale, 2017).
Some dense protein solutions have the potential to further phase transition into structures with properties resembling a solid—a process that has been referred to as gelation (Alberti and Dormann, 2019; Figure 1C). Indeed, whilst many ribonucleoprotein granules are liquid like, other membraneless organelles feature solid-like properties, such as the central channel of the nuclear pore (Frey et al., 2006; Schmidt and Görlich, 2015). Indeed, numerous RNA binding proteins, including FUS and hnRNPA1, transition into reversable hydrogels composed of amyloid-like cross-β fibrils (Kato et al., 2012; Molliex et al., 2015; Murray et al., 2017; Gui et al., 2019). These fibrils however differ from the highly stable amyloid fibrils seen in pathological aggregates formed by proteins associated with neurodegenerative disease (Figure 1D) as the amyloid formation associated with hydrogel formation of LCDs is reversable (Murray et al., 2017; Gui et al., 2019). This reversibility is associated with short motifs that allow amyloid-like β-strand interactions known as kinked β sheets, termed LARKs (low-complexity aromatic-rich kinked sequences). LARKs form weakly stabilizing fibrils (Gomes and Shorter, 2019) unlike cross-β sheets of amyloid fibrils which form stable pathogenic steric “zippers” due to interdigitated side chain amino acids. LCDs of RNA binding proteins that undergo LLPS to form membraneless organelles are enriched in LARKs, suggesting a possible role in LLPS (Hughes et al., 2018). Further the hydrogel forming phenylalanine-glycine domain of nuclear pore proteins is also thought to form LARKs (Hughes et al., 2018). Other motifs that form reversible amyloid fibrils have also been identified in the LCDs of FUS, hnRNPA1 and hnRNPA2 (Luo et al., 2018; Gui et al., 2019; Lu et al., 2020; Sun et al., 2020b). Hence amyloid formation may have a practical function in membraneless organelle formation, for more stable, less transient structures. However, this acquisition of solid-like properties has also been associated with the formation of protein aggregates in neurodegenerative disease (Elbaum-Garfinkle, 2019). Disease mutations in the LCD of proteins have been shown to alter phase separation behavior, generally promoting phase separation and reducing droplet dynamics: ALS associated mutations in FUS lead to the formation of more solid-like droplets and irreversible fibrillar hydrogels which can trap other RNA binding proteins (Murakami et al., 2015; Patel et al., 2015). Similarly, phase separation of hnRNPA1 with the disease mutation D262V causes enhanced amyloid fibril formation (Lin et al., 2015). ALS causing mutations in TIA1 lead to phase separated droplets with reduced mobility and faster fibrilization (Mackenzie et al., 2017). Finally, mutations in the C-terminal domain of TDP-43 enhance its phase separation and show reduced droplet fluidity (Conicella et al., 2020). Thus, phase separation underlies the localization and function of many LCD containing proteins, and disruption in the physiological equilibrium of this process can lead to pathological behavior of the membraneless organelles they are associated with.
In relation to phase separation, there is particular interest in the arginine rich C9orf72 DPRs due to the well-established role of intrinsically disordered arginine rich regions as promoters of phase separation. Arginine and glycine rich motifs (RGG/RG) are an important class of LCD sequence found within numerous RNA binding proteins that undergo LLPS, including those associated with neurodegenerative diseases such as FUS, hnRNPA1, and FMRP (Thandapani et al., 2013; Chong et al., 2018). Proteins with these motifs contain varying numbers of RGG/RG repeats interspaced typically with aromatic residues (Thandapani et al., 2013), with repeat length influencing multivalency and phase separation (Chong et al., 2018); phase separation of these domains also correlates with the number of arginine residues present (Chong et al., 2018; Wang et al., 2018b). Within FUS, interactions between aromatic residues in the prion-like domain and positively charged residues in the RNA binding domain determine the saturation concentration, which appeared specific to tyrosine-arginine interactions (Wang et al., 2018b). Increasing the negative charge of the prion-like domain increased phase separation mediated by interaction with the RNA binding domain, but reduced phase separation in isolation. Hence electrostatic attractions between arginine residues and negatively charged residues in the prion-like domain additionally regulate phase separation by bolstering the interaction between aromatic tyrosine residues and positively charged arginines in the RNA binding domain, whereas electrostatic repulsions within the prion-like domain help prevent unfruitful self-interactions. This is reflected in the toxicity of FUS in HeLa cells, where the severity of FUS toxicity correlates with the strength of the interaction between the prion-like domain and RNA binding domain, modulated by arginine residues in the RNA binding domain (Wang et al., 2018b). Functionally, the arginine residues in the C-terminal RNA binding domain of FUS are crucial for the maturation of the protein in stress granules, recruitment to sites of DNA damage and also regulate its toxicity in Drosophila (Bogaert et al., 2018).
The C9orf72 arginine rich DPRs poly-GR and poly-PR, translated from the G4C2 repeat expansion, undergo phase separation (in the presence of a crowding agent) in vitro similarly to RGG/RG domains (Boeynaems et al., 2017). Further analysis of the phase separation of poly-PR revealed that the droplets formed have liquid-like properties—with the droplets being circular in shape, showing recovery upon photobleaching (due to fast internal rearrangement) and deformation under stress, fusing together and being reversible upon dilution or changes in temperature or solute. Poly-PR phase separation was also impaired by elevating salt concentration, indicating that arginine-driven electrostatic forces modulate poly-PR LLPS. Normally, the coacervation of molecules into droplets is inhibited when proteins consist primarily of a single charge due to charge repulsion and require the presence of counter ions (Pak et al., 2016; Boeynaems et al., 2017). Indeed, phase separation of poly-GR and poly-PR does not result from electrostatic repulsion between the arginine residues of the dipeptide, but rather due to anions in the buffer, with anions with more valence driving droplet formation whereas monovalent anions which exist in a single charge state being inhibitory (Boeynaems et al., 2017; Jafarinia et al., 2020). Biological polyvalent anions such as poly-uracil RNA can even overcome the need for crowding agents in poly-PR LLPS (Boeynaems et al., 2017). Furthermore, like arginine residues in the RNA binding domain of FUS, poly-PR also engages in cation-pi interactions with tyrosine. Therefore, the phase separation of the arginine rich DPRs poly-GR and poly-PR and likely their interactions with other molecules that contain LCDs are similar to that of arginine rich RNA binding proteins like FUS: complex coacervation driven by electrostatic interactions, both charge-charge and charge-pi, with the arginine residues providing the cation for these weak, multivalent interactions (Nedelsky and Taylor, 2019; Figure 1F).
Further analysis of poly-PR phase separation via complex coacervation revealed that the process is also governed by the chemistry of associated polyanions (Boeynaems et al., 2019). Negatively charged protein assemblies such as microtubules provide a scaffold for poly-PR recruitment whereas flexible polyanions such as RNA caused spherical droplet formation. The latter is in line with LLPS via complex coacervation with the dynamics of droplets varying depending on the polyanion used. Poly-PR-RNA LLPS is dependent on the structure of the RNA; all homopolymeric RNAs, except for poly-rGuanine lead to poly-PR liquid droplet formation. The properties of these droplets, such as viscosity, are dependent on the specific RNA molecules involved in RNA-PR and RNA-RNA interactions. The lack of coacervation of poly-PR in presence of poly-rGuanine is connected to higher-order RNA structures formed by poly-rGuanine. Poly-rGuanine unlike the other RNA bases can form a highly stable secondary structure known as a G-quadruplex, which appears to kinetically stall poly-PR phase separation. Boeynaems et al. (2019) interpret this as poly-PR being unable to outcompete base stacking interactions within G-quadruplexes. The length of the DPR in addition to the length of its interacting polyanion also influences droplet size due to increased multivalency; with modeling showing that larger PR molecules and larger polyanions produce smaller droplets of a higher concentration (Jafarinia et al., 2020). Hence variations in the size of droplets owe to variations in the robustness of poly-PR interactions and the molecules it is phase separating with. Similar findings have been observed for poly-GR post-translational modifications; poly-GR aggregates have been found to be methylated in patient brain and synthetic dimethylated poly-GR peptides form larger but reduced numbers of droplets compared to unmethylated poly-GR, indicative of reduced LLPS (Gittings et al., 2020). Arginine methylation does not disrupt charge but may affect cation-pi interactions, as it does when disrupting the LLPS of several RNA-binding proteins (Qamar et al., 2018; Ryan et al., 2018; Hofweber and Dormann, 2019), and thereby would be expected to weaken pathological interactions of the arginine rich DPRs and LCD containing proteins.
One question that arises from these studies is, whether the ability to undergo phase separation is what causes the severe toxicity of poly-PR and poly-GR. When peptides of different composition were studied, expression of a poly-arginine only peptide located to the cytoplasm and showed no toxicity (Meloni et al., 2013, 2015, 2017) whereas PR12 with the same number of arginine residues was highly toxic and localized to the nucleus (Kanekura et al., 2018). Indeed poly-PR peptides and constructs in model systems typically show higher toxicity than poly-GR of similar lengths and number of arginine residues (Wen et al., 2014). Hence arginine content and the ability to undergo LLPS are not the only determining factors in the toxicity of these DPRs. One theory of FUS separation proposed by Wang et al. (2018b) uses the concept of stickers and spacers, in which the stickers (such as arginine) determine the phase separation properties of the protein; and the spacers (such as glycine or proline) which separate the stickers determine the flexibility of the peptide. The different spacers found in poly-GR (glycine) and poly-PR (proline) may influence the interactions undergone by the peptides thereby influencing their relative toxicities. Additionally, the methylation of poly-GR reduces its propensity for LLPS and decreases toxicity in neurons, and symmetric dimethylation of poly-GR inclusions in C9orf72-FTD/ALS patient brain positively correlates with disease duration, suggesting methylation of poly-GR and reduced LLPS could be protective (Gittings et al., 2020).
As already mentioned, RNA plays an important regulatory role in driving phase separation of intrinsically disordered proteins and the physical properties of resultant droplets, dependent on concentration, secondary structure and sequence of RNA (Langdon and Gladfelter, 2018; Zhang et al., 2019b). The guanine (G)-rich DNA and RNA of the C9orf72 repeat expansion forms highly complex secondary structures including unimolecular and multimolecular G-quadruplexes (Fratta et al., 2012; Reddy et al., 2013; Haeusler et al., 2014; Zhou et al., 2015; Conlon et al., 2016). G-quadruplexes are stable four strand structures formed of planar guanine tetramers stacked on top of one another (Zhou et al., 2015). G-quadruplex containing RNA has been reported to itself undergo phase separation (Zhang et al., 2019b), as has triplet repeat RNAs of CAG and CUG (Jain and Vale, 2017). Indeed, similarly to CAG and CUG repeat RNA, C9orf72 G4C2, but not antisense C4G2, repeat RNA directly formed gels in vitro in a repeat-length dependent manner and was disrupted by monovalent cations and antisense oligonucleotides (ASOs), indicating both electrostatic interactions and base-pairing interactions (Jain and Vale, 2017; Figure 1F). Interestingly, the authors speculate that the increased valency that comes with repeat length may explain the length-dependent threshold that exists in disease (Langbehn et al., 2010; Rohrer et al., 2015). In cells, CAG repeat RNA formed nuclear foci that had liquid properties, which was proposed to be due to the presence of RNA-binding proteins such as helicases which remodel RNA-base pairing. Whereas, G4C2 RNA foci showed only partial recovery after photobleaching, indicating that they are less dynamic than CAG foci. In another study, G4C2 repeat RNA LLPS only occurred in the presence of cell lysate and required cellular RNA but was equally dependent on repeat-length and electrostatic interactions (Fay et al., 2017). In this study, G4C2 RNA precipitated stress granule proteins, and thus will be discussed in the relevant section below.
As already mentioned, phase separation is key to the formation of membraneless organelles, including stress granules, the nucleolus, Cajal bodies, nuclear speckles, and the central channel of the nuclear pore. These liquid organelles regulate several important cellular functions, include splicing, protein translation, nucleocytoplasmic transport and the cellular stress response. A disruption in any one of these molecular functions is likely to be catastrophic for a cell. In the following sections, we review the evidence that the C9orf72 arginine rich DPRs and G4C2 RNA disrupt the phase separation of these membraneless organelles and the cellular pathways associated with them (Figure 2).
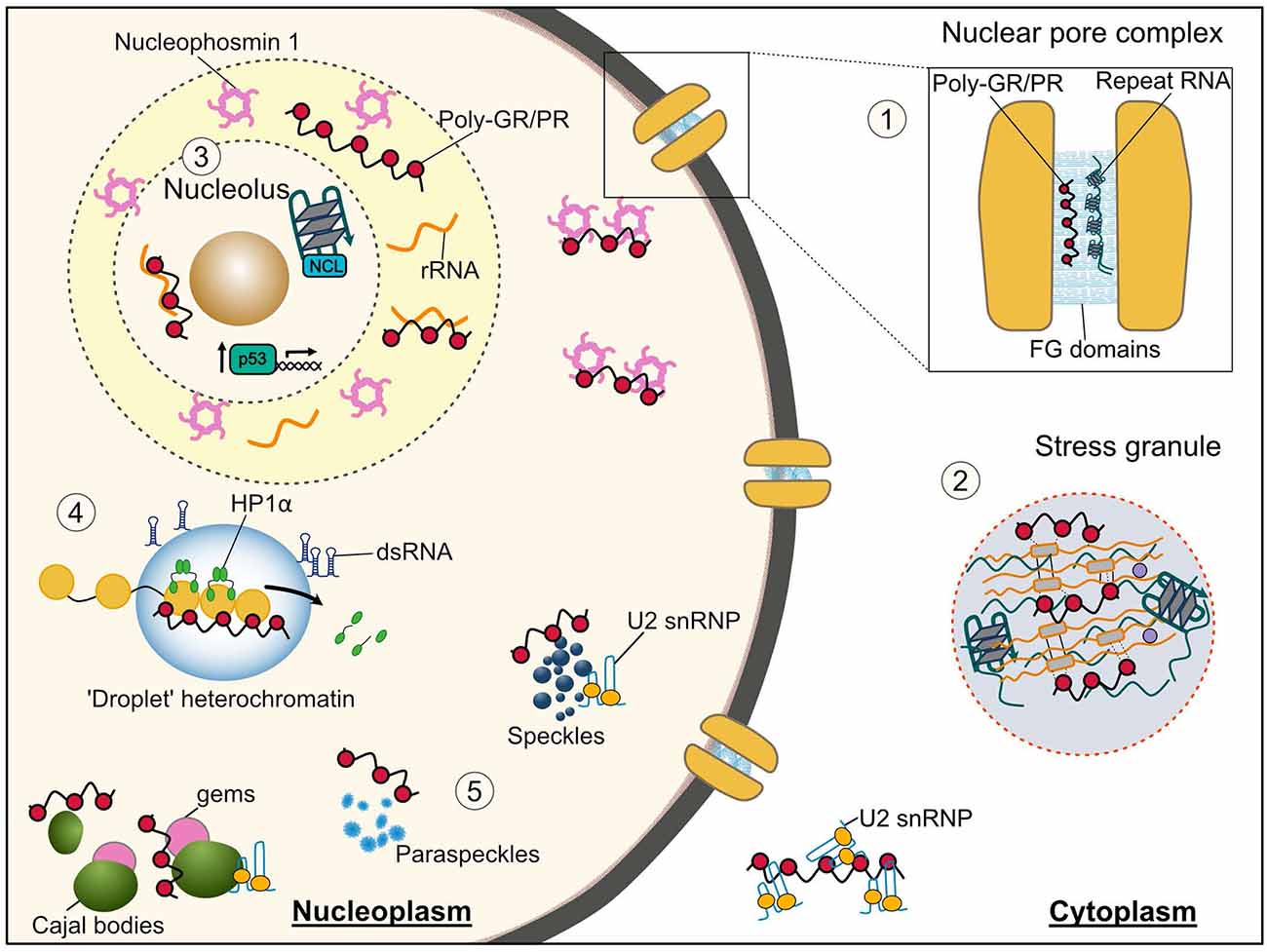
Figure 2. Membraneless organelles and associated functions that are impaired by the C9orf72 arginine rich DPRs and G4C2 repeat RNA. (1) Nuclear pore complex—Phenylalanine-glycine low complexity domains (LCDs) of nuclear pore proteins (nucleoporins) phase separate into a selective hydrogel in the central channel of the nuclear pore. Both the arginine rich DPRs and G4C2 RNA have been shown to interact with these nucleoporins, thus their altered phase separation may underlie the nucleocytoplasmic transport dysfunction seen in C9orf72-ALS/FTD. (2) Stress granules—both the arginine rich DPRs and repeat RNA bind and cause the aberrant phase separation of LCD containing stress granule proteins inducing formation of poorly dynamic stress granules, which likely contributes to the translational repression seen in C9orf72-ALS/FTD models. (3) Nucleolus—Poly-PR disrupts the phase separation of the nucleolar protein nucleophosmin with ribosomal RNA leading to its mislocalization from nucleoli to the nucleoplasm, and an impairment in ribosomal biogenesis; this nucleolar stress may also lead to the observed aberrant activation of the p53 pathway. The G-quadruplex forming G4C2 RNA has also been shown to associate with the nucleolar protein nucleolin (NCL) and cause nucleolar dysfunction. (4) Heterochromatin—poly-PR perturbs phase separation of heterochromatin protein 1α (HP1α) displacing it from heterochromatin and resulting in its degradation and an upregulation of repetitive RNA elements which form double-stranded RNA and initiate pathological interferon signaling. (5) Nuclear structures—proteins associated with nuclear speckles, paraspeckles, Cajal bodies and Gems interact with both the arginine rich DPRs and G4C2 RNA. The arginine rich DPRs have specifically been shown alter the liquid-like properties of these organelles and binding to LCD containing U2 snRNP proteins results in their mislocalization from nuclear speckles to the cytosol and reduced splicing activity.
In the nucleus, there are a variety of different membraneless structures formed by LLPS including nuclear speckles, paraspeckles, Cajal bodies and Gems. These are multiprotein-RNA organelles that play an essential role in transcriptional regulation and the formation and function of the spliceosome, a large ribonucleoprotein complex where pre-mRNA splicing is catalyzed (Lamond and Spector, 2003; Will and Lührmann, 2011). Cajal bodies are organelles whose major function is the modification and assembly of uridine-rich small nuclear ribonucleoproteins (U snRNPs; Morris, 2008), and Gems (Gemini of Cajal bodies) localize adjacent to Cajal bodies and are characterized by the presence of the SMN (survival of motor neuron) protein (Liu and Dreyfuss, 1996). Mature U snRNPs accumulate in nuclear speckles, also known as splicing speckles, which function as the site for the storage and modification of pre-mRNA splicing factors and pre-mRNA splicing itself (Spector and Lamond, 2011; Galganski et al., 2017; Gruss et al., 2017). Paraspeckles primarily regulate gene expression through sequestration of RNAs and proteins (Fox et al., 2018).
Nuclear speckles and Cajal body proteins have been shown to interact with the arginine rich DPRs poly-PR and poly-GR, and speckle proteins can modify their toxicity (Lee et al., 2016; Boeynaems et al., 2017; Yin et al., 2017; Hartmann et al., 2018; Moens et al., 2019). The nuclear speckle protein SRSF7 is specifically affected by overexpression of poly-PR but not GR, displaying reduced recovery after photobleaching (Lee et al., 2016), an indication of perturbed LLPS. Poly-PR can also increase the levels of nuclear paraspeckles by direct interaction with paraspeckle proteins and RNA (Suzuki et al., 2018, 2019). In cells, expression of poly-PR, GR or GA leads to a dramatic reduction in Cajal bodies and Gems (Lee et al., 2016; Rossi et al., 2020); a few poly-GR expressing cells displayed increased numbers but were noticeably smaller than controls, indicating that the phase separation properties of Cajal bodies may also be perturbed. Functionally, these changes may explain the changes in gene expression and splicing-associated with expression of both poly-PR and poly-GR (Kwon et al., 2014; Yin et al., 2017; Kramer et al., 2018; Sun et al., 2020a). Relating to Cajal body function, in a proteomic interaction study of poly-GR and poly-PR all known U2 snRNP proteins were found, with six of these as top interactors (Yin et al., 2017). Interestingly, this was specific to the U2 snRNP complex, as only three of eight U5 snRNP components were identified. Poly-GR was further found to inhibit assembly of the spliceosome, a key function of the U2 snRNP complex, and lead to downstream mis-splicing. Perturbation in the U2 snRNP has also been observed in patient cells. Using C9orf72 iPSC-derived motor neurons which have been shown to produce detectable levels of poly-GR (Lopez-Gonzalez et al., 2016) the U2 snRNP associated proteins SNRPB2 and S3Fa were mislocalized to the cytoplasm, with SNRPB2 mislocalization in around 40–60% of neurons, whereas it was completely nuclear in controls (Yin et al., 2017). This was also recapitulated in cell lines exposed to poly-PR peptide, confirming the link to the DPR. Furthermore, in patient cells with cytoplasmic accumulation of SNRPB2, levels of the protein in nuclear speckles and the nucleoplasm were decreased. Again, this was a specific effect as U1 snRNP proteins were unaffected. This was also associated with a preferential mis-splicing of U2 snRNP dependent exons in patient lines, which is also dominant in patient tissue. The disassembly of Cajal bodies by poly-GR and PR in cells could be rescued by co-expression of the nuclear transport receptor importin β1 (Rossi et al., 2020). As importin β1 mediates, nucleocytoplasmic transport of snRNPs and nucleocytoplasmic transport is impaired during stress (Boeynaems et al., 2016b), the arginine rich DPRs may also impair Cajal body assembly indirectly by disrupting the transport of U2 snRNPs into the nucleus.
A number of the LCD containing RNA binding proteins that bind to the repeat RNA have important functions in RNA splicing, including the nuclear speckle protein SRSF1 (Hautbergue et al., 2017). SRSF1 is a nuclear speckle protein that facilities pre-mRNA splicing factor assembly and plays a role in nuclear export (Huang et al., 2003; Tripathi et al., 2012). SRSF1 could also be found to colocalize with G4C2 RNA foci in motor neurons of patient spinal cord sections. Knockdown of SRSF1 can also rescue neurodegeneration in G4C2 repeat Drosophila and reduce toxicity from C9orf72 patient-derived astrocytes on control motor neurons. Importantly, this rescue was specific to the SRSF1-G4C2 RNA interaction as SRSF1 knockdown in arginine rich DPR flies resulted in no rescue. Mechanistically, SRSF1 depletion reduced nuclear export of G4C2 mRNA via its interaction with nuclear export receptor NXF1, and subsequent DPR production. Thus, it is proposed that the sequestration of SRSF1 into RNA foci leads to increased nuclear export of G4C2 RNA and thereby increasing RAN translation, enhancing DPR toxicity in C9orf72-ALS/FTD. The effect on the properties of nuclear speckles, however, has not yet been investigated.
The nucleolus is a nuclear organelle whose primary function is ribosome biogenesis, including both the synthesis and processing of ribosomal RNA via RNA polymerase I and the assembly of ribosomes (Boisvert et al., 2007; Iarovaia et al., 2019). The nucleolus consists of three defined liquid-like phases: the fibrillary centre(s) surrounded by a dense fibrillar component rich in fibrillarin, all encompassed by a granule component enriched in nucleophosmin; the latter is an LCD-containing protein known to undergo phase separation (Mitrea et al., 2018; Frottin et al., 2019).
An early observation in cell models of the C9orf72 DPRs poly-GR and poly-PR was a strong nucleolar localization with nucleolar swelling and the displacement of nucleolar proteins to the nucleoplasm (Haeusler et al., 2014; Kwon et al., 2014; May et al., 2014; Wen et al., 2014; Zhang et al., 2014; Tao et al., 2015; Callister et al., 2016; Lee et al., 2016). They have also been shown to directly interact in vitro (Lee et al., 2016; Boeynaems et al., 2017; Hartmann et al., 2018). Both poly-GR and poly-PR are recruited to the liquid-like granular component of the nucleolus, and poly-GR could additionally interact with the dense fibrillar components (Lee et al., 2016). Poly-GR and poly-PR peptides facilitate phase separation of nucleophosmin in vitro, which is accompanied by a reduction in the mobility of the nucleolar proteins nucleophosmin and nucleolin in the nucleolus of cells expressing either poly-GR or poly-PR (Lee et al., 2016). However, when in molar excess the arginine rich DPRs can inhibit LLPS (Lee et al., 2016; White et al., 2019). They can also outcompete SURF6, an arginine-tract containing protein and native binding partner of nucleophosmin (Lee et al., 2016) and have been found to specifically bind via the third acidic tract (A3) within its intrinsically disordered region (White et al., 2019). In this latter study, by disturbing the nucleophosmin-SURF6 interaction which is required for physiological phase separation and causing a dissolution of nucleophosmin particles, poly-PR leads to the sequestration of nucleophosmin into soluble nucleophosmin/poly-PR complexes (White et al., 2019). Poly-PR was also able to outcompete nucleophosmin for ribosomal RNA (rRNA) binding leading to the accumulation of rRNA in rRNA/poly-PR puncta which persisted even at high concentrations of the DPR. These findings were recapitulated in cells where increasing poly-PR concentration caused the delocalization of nucleophosmin from nucleoli into the nucleoplasm and a sequestration of rRNA with poly-PR in the nucleolus; increasing poly-PR peptide length displayed a similar increase in dissolution effect. Hence a disruption of the phase separation of nucleophosmin explains the nucleolar dysfunction observed in cellular and animal models of DPR toxicity.
Functionally, studies have shown that numerous nucleolar functions are perturbed by the arginine rich DPRs such as ribosomal protein transport, the processing of rRNA and the assembly of ribosomes (Kwon et al., 2014; Jovičić et al., 2015; Tao et al., 2015; Kanekura et al., 2018; Suzuki et al., 2018). Co-expression of nucleophosmin can rescue arginine rich DPR toxicity in cells (Farg et al., 2017), and knockout or overexpression of genes encoding proteins in ribosomal rRNA processing can modify poly-PR toxicity in yeast (Jovičić et al., 2015; Chai and Gitler, 2018) and Drosophila (Boeynaems et al., 2016a). Notably, homologs of the nucleolar protein nucleolin (NCL), identified in multiple studies, could suppress PR-toxicity upon deletion. In C9orf72-ALS/FTD brain tissue, the majority of neuronal nucleoli were smaller in comparison to age-matched controls, however neurons which contained a cytoplasmic poly-GR inclusion had a significantly increased nucleolar volume (Mizielinska et al., 2017). Overexpression of poly-GR (and poly-GA but to a much lesser extent) could also cause nucleolar enlargement in Drosophila neurons (Mizielinska et al., 2017), however this has not been recapitulated in mouse models of the arginine rich DPRs (Zhang et al., 2018b, 2019a); these differences have been proposed to differ depending on whether the DPRs expressed can enter the nucleus.
Although to a lesser extent, nucleolar dysfunction has also been associated with C9orf72 repeat RNA toxicity. G4C2 repeat RNA can bind several nucleolar proteins, predominantly in its G-quadruplex form; indeed, NCL could be found in association with RNA foci in the motor cortex of expansion carriers (Haeusler et al., 2014; Cooper-Knock et al., 2015). In cells, this caused a deficit in the production of mature ribosomes and a build-up of untranslated mRNAs in the cytoplasm (Haeusler et al., 2014). A subtle increase in nucleolar volume has also been observed in frontal cortex neurons containing sense RNA foci in C9orf72-FTD patient brain (Mizielinska et al., 2017). Interestingly, although in vitro the C4G2 antisense RNA does not bind nucleolar proteins (Haeusler et al., 2014), they are found to more frequently associate and surround the nucleolus in patient tissue (Mizielinska et al., 2013; Vatsavayai et al., 2016; Aladesuyi Arogundade et al., 2019); note, nucleolar proteins do not however associate with antisense RNA foci outside the nucleolus (Cooper-Knock et al., 2015).
Stress granules are transient, dynamic, cytoplasmic assemblies which form reversibly under conditions of acute cellular stress, such as heat shock, oxidative stress or nutrient depletion, to sequester non-translating mRNA, translation initiation complexes and related RNA binding proteins. By protecting and temporarily storing stalled translation complexes until stress dissipates, stress granules effectively regulate translation of housekeeping mRNA, while promoting translation of cytoprotective proteins such as chaperones (Baradaran-Heravi et al., 2020). Similarities in the dynamic behavior and liquid-like properties of in vivo cellular ribonucleoprotein granules and in vitro granule components support the assertion that these compartments form via LLPS and interactions between proteins with LCDs and RNA (Hyman et al., 2014; Kroschwald et al., 2015).
The arginine rich C9orf72 DPR interactomes are enriched in LCD-containing proteins including RNA-binding proteins and components of stress granules, such as TDP-43, FUS, hnRNPA1, TIA1 and G3BP1 (Lee et al., 2016; Lin et al., 2016; Moens et al., 2019), highlighting a role for disrupted stress granule function in DPR toxicity. In vitro poly-PR and GR selectively associate with and decrease the saturation concentration at which droplets form with hnRNPA1, TIA1 and FUS, unlike the non-arginine rich DPRs which have no effect (Lee et al., 2016; Boeynaems et al., 2017). Upon overexpression in cells, poly-GR and PR also increase the formation of stress granules (Lee et al., 2016; Boeynaems et al., 2017). However, the cellular systems vary in their DPR localization with one showing colocalization of stress granules with PR100 (Boeynaems et al., 2017) and the other with GR50 but not PR50 (Lee et al., 2016); this is likely due to the length difference in the PR polypeptides with the shorter version being limited to the nucleus.
Interestingly, numerous disease-linked mutations have been found to enhance LCD polymer stability (Kato and McKnight, 2017; Boeynaems et al., 2018) and a hnRNAP1 variant harboring a mutation which diminishes LCD polymerization (F291S) prevents immunoprecipitation by a PR20 peptide (Lin et al., 2016), suggesting that poly-PR interacts with LCDs in a polymeric conformation. PR30 treatment also increases the β-sheet content of FUS LCD droplets, as shown by the increase in thioflavin-T fluorescence with increasing poly-PR concentration (Boeynaems et al., 2017). Therefore, DPR interactions with stress granule proteins may increase β-sheet content and enhance stability of stress granule protein LCD polymers, rendering stress granules less dynamic. Indeed, liquid droplets of hnRNPA1, TIA1 or FUS display reduced liquid-like properties when treated with arginine rich DPR peptides, including fewer wetting and fusion events, and reduced recovery from photobleaching (Lee et al., 2016; Boeynaems et al., 2017). This is also recapitulated in cells where arginine rich DPRs induce formation of poorly dynamic stress granules. Live imaging of HeLa cells showed that DPR-induced stress granule G3BP1 puncta increase in number over time and do not appear to disassemble. Photobleaching analysis further confirms that the recovery rate of G3BP1 in DPR-induced stress granules is significantly reduced, compared to arsenite-induced stress granules (Lee et al., 2016; Boeynaems et al., 2017). Poly-PR induced stress granules are also enriched in disease-linked proteins, including ataxin-2 and TDP-43 (Boeynaems et al., 2017), which may reflect their entrapment due to reduced diffusivity or their recruitment may contribute to this process. In mice, the stress granule marker TIA1 remained nuclear and diffuse upon GFP-GR100 expression but formed cytoplasmic puncta which colocalized with poly-GR in (G4C2)149 mice using postnatal adenovirus expression (Zhang et al., 2018b). This correlates with the formation of cytoplasmic poly-GR inclusions but not diffuse poly-GR, suggesting that inclusions specifically induce stress granules. Similarly, diffuse GFP-GR100 expression in cells did not change the number of stress granules upon heat shock but more stress granules were retained upon recovery, corroborating previous findings in the impairment of disassembly (Zhang et al., 2018b). Thus, arginine rich DPRs nucleate phase separation of LCD-containing stress granule proteins and promote assembly of poorly dynamic stress granules with reduced disassembly compared to adaptive stress granules, highlighting a pathological outcome. The contribution of arginine residues in this process demonstrates that expression of DPRs does not only trigger the stress response, but actively mediates phase separating interactions with stress granule components through high multivalency.
When G4C2 RNA is incubated with cellular lysates in vitro phase separated particles are formed which are enriched in stress granule components (Fay et al., 2017). Constituents such as G3BP1 and FUS precipitated with all repeat lengths studied, but others including TIA1 only condensed with longer lengths. Particles exhibited classical features of LLPS, including dependence on concentration, temperature, molecular crowding and salt; the latter indicating a role for electrostatic interactions. Phase separation was repeat length dependent and associated with the presence of G-quadruplex forming sequences. Notably, when assays were performed with equal weight of different length G4C2 repeat RNAs rather than equimolar (where the number of repeat units should be equal) similar condensation was observed, suggesting that repeat number is important, but these can be in cis or trans. It also required the presence of cellular RNA, suggesting that G-quadruplex G4C2 RNA enhances intermolecular RNA interactions and thereby promotes the nucleation behavior of RNA that causes protein condensation. Indeed, transfection of G4C2 RNA in cells leads to the formation of stress granules (Fay et al., 2017). In granules containing both mCherry-tagged G3BP1 and FAM-labeled G4C2 RNA, photobleaching results in recovery of the G3BP1 signal whereas G4C2 RNA signal does not. This indicates that within these granules the stress granule protein can rapidly internally rearrange, but the RNA forms a stable component which cannot be replaced. In another study transfection of G4C2 DNA in cells also induced formation of stress granules, although in this system they did not contain G4C2 RNA and thus may be induced by the DPRs translated from repeat RNA as detailed above (Rossi et al., 2015).
An important consideration for assessing the relevance of DPRs and RNA to aberrant stress granule phase separation in the wider context of C9orf72-ALS/FTD will be evaluating the contribution of each proposed mechanism. Interestingly, the C9orf72 protein has additionally been implicated in stress granule formation (Maharjan et al., 2017). Thus it is likely that there may be interplay amongst mechanisms. Perhaps, loss of C9orf72 protein inhibits the appropriate physiological stress response, then arginine rich DPRs and repeat RNA promote stress granule assembly by nucleating phase separation, and the DPRs subsequently interact with stress granules to reduce dynamics and inhibit disassembly, promoting a state of chronic stress and eventual neurodegeneration.
The nuclear pore is a large multiprotein complex comprised of transmembrane nucleoporins that anchor the pore to the nuclear envelope, structural nucleoporins that act as a scaffold and nucleoporins which make up the central channel of the nuclear pore (Cautain et al., 2015). Notably for this review, several nucleoporins contain low complexity phenylalanine-glycine (FG) repeats. These domains primarily localize to the central channel where they form a permeability barrier via LLPS critical for nuclear pore selectivity (Frey et al., 2006). In vitro these domains spontaneously phase separate into hydrogels that exclude inert macromolecules but allow the entry of nuclear transport receptors, mimicking the selectivity of the nuclear pore in cells (Schmidt and Görlich, 2016).
DPRs have been associated with defective nucleocytoplasmic transport by the ability of transport factors to modify their toxicity or via resultant mislocalization of transport factors in cultured neurons, yeast and Drosophila (Jovičić et al., 2015; Boeynaems et al., 2016a; Chai and Gitler, 2018; Solomon et al., 2018). The arginine rich DPRs can also directly interact in vitro with different types of transport factors (Lee et al., 2016; Lin et al., 2016; Boeynaems et al., 2017). The arginine rich DPRs bind the low complexity FG domains of some nucleoporins (Lin et al., 2016), being specifically bound when the FG domains are in a polymeric state (Shi et al., 2017). The authors of the latter finding suggest that FG domains of nucleoporins in the nuclear pore exist in equilibrium between polymerized and unpolymerized states, and upon binding poly-PR shifts this equilibrium by stabilizing the polymeric state leading to a less permeable nuclear pore barrier and disruption of transport. The relation of this equilibrium to phase separation has not yet been investigated. Of note, the interaction of the arginine rich DPRs with nucleoporins may not be specific to FG domains, as another study found that binding was only partially reduced when phenylalanines in the FG domain were substituted for alanines (Hayes et al., 2020). Indeed, it has been noted that other low complexity sequences found within the FG domains of nucleoporins (repetitions of the tripeptide sequence glycine/serine-tyrosine-glycine/serine) are similar to the LCDs found in RNA binding proteins (Shi et al., 2017), and thus binding may occur here. Poly-PR (PR20) was shown to accumulate in the central channel of the nuclear pore in isolated nuclei from Xenopus oocytes (Shi et al., 2017). However, when GR200 was overexpressed using an adenovirus in mice, FG-nucleoporins were found to co-aggregate with poly-GR in the cytoplasm of cortical neurons (Cook et al., 2020). This occurred with concomitant loss of nuclear and cytoplasmic aggregation of TDP-43, indicating that the arginine rich DPR-induced defects of FG-nucleoporins can lead to TDP-43 mislocalization, a key marker of nucleocytoplasmic transport dysfunction and neurodegeneration.
A recent study has provided evidence that C9orf72 G4C2 RNA specifically disrupts the function of the nuclear pore via by inducing loss of a specific subset of eight nucleoporins driven by loss of one key nucleoporin POM121 (Coyne et al., 2020). Notably, changes were specifically attributed to G4C2 RNA and not DPRs as overexpression of either poly-GR or poly-PR were insufficient to induce similar changes, but expression of RNA-only G4C2 repeats with stop codons inserted to prevent RAN translation were. POM121 is an FG-domain containing nucleoporin that can phase separate into a hydrogel that mimics active and passive nucleocytoplasmic transport (Labokha et al., 2013). Given the ability of the G4C2 RNA itself to phase separate and promote the phase separation of other LCD containing proteins (Fay et al., 2017; Jain and Vale, 2017), it will be important to study whether G4C2 RNA can disrupt the phase separation of POM121.
Since the discovery of the C9orf72 repeat expansion mutation in 2011, major progress has been made in elucidating the underlying pathogenic mechanisms. Both the G4C2 repeat RNA and DPRs have been shown to be neurotoxic and disrupt a number of cellular processes including nucleocytoplasmic transport, the stress response, nucleolar dysfunction and RNA processing. All these processes require the correct assembly, dynamics, and function of membraneless organelles formed by physiological phase separation of LCD-containing proteins or domains. Both the repeat RNA and arginine rich DPRs themselves undergo phase separation and can disrupt the phase separation of other LCD proteins required for membraneless organelle formation and function. These disturbed phase transitions account for widespread cellular abnormalities observed in C9orf72-ALS/FTD and may be a target for therapeutic intervention.
In repeat expansion disorders a unique circumstance is produced whereby repetitive RNA and repetitive polypeptides are produced, which due to their repetitive nature are domains of low complexity. This is the most overt in disorders where the expansion is large, as is the case for C9orf72-ALS/FTD. Intriguingly, in the evidence highlighted above both the C9orf72 DPRs and G4C2 repeat RNA affect many of the same pathways although the RNA and polypeptides have dramatically different structures and thus you would anticipate different interaction partners. It is likely that the arginine rich DPRs result in the convergence of these molecules as the strong positive charge from their arginine content lends them to interact with nucleic acids and their binding partners. The combination of low complexity proteins and RNA is also a key driver of the multivalent interactions that lead to phase separation, present in the majority of the membraneless organelles discussed above. Both the DPRs and repeat RNA may act as molecular seeds initiating aberrant phase transitions by binding to the LCD of intrinsically disordered proteins and causing liquid demixing. Furthermore, for the DPRs, due to the strength of their interactions with LCDs, proteins will no longer interact with their normal binding partners thus disrupting the assembly of membraneless organelles and their physiological function. Hence the majority of observed pathological phenomena and perturbed pathways associated with C9orf72-ALS/FTD converge on a disruption of LLPS of membraneless organelles.
One of these convergences is on RNA splicing. As detailed above, the arginine rich DPRs can bind the LCD containing U2 snRNP proteins leading to their mislocalization from nuclear speckles to the cytosol, an alteration in the liquid-like properties of the nuclear speckles and reduced splicing activity (Lee et al., 2016; Freibaum and Taylor, 2017; Yin et al., 2017). Poly-PR can also increase the levels of nuclear paraspeckles by direct interaction with paraspeckle proteins and RNA (Suzuki et al., 2018, 2019). G4C2 repeat RNA also binds to the nuclear speckle protein SRSF1, sequestering it into RNA foci and this interaction leads to increased nuclear export of G4C2 RNA, thereby increasing RAN translation and enhancing DPR toxicity (Hautbergue et al., 2017). The transcriptional regulators Pur-α, a binding partner of SRSF1, and Matrin-3 can also bind G4C2 repeat RNA and modify toxicity; both are also mislocalized to the cytoplasm upon G4C2 RNA expression (Xu et al., 2013; Ramesh et al., 2020), with Pur-α being recruited to stress granules (Rossi et al., 2015). Similarly, to SRSF1, Matrin-3 also reduced levels of RAN-translation products (Ramesh et al., 2020). HnRNP H is also a strong interactor of G4C2 repeat RNA and is sequestered into RNA foci in patients where its depletion results in the reduction in alternative splicing of its targets (Lee et al., 2013; Conlon et al., 2016). The interaction of these DPR and RNA gain of function mechanisms has not yet been studied.
Translation is also targeted. As detailed above, nucleolar function can be impacted by both DPRs and G4C2 repeat RNA. The arginine rich DPRs bind nucleolar proteins, particularly nucleophosmin, and can both facilitate and inhibit their phase separation depending on DPR concentration; this results in consequent reduced mobility of proteins in the nucleolus, displacement of proteins and ribosomal RNA away from the nucleolus and functional impairment of ribosome biogenesis (Lee et al., 2016; White et al., 2019). Both poly-GR and PR have also been shown to directly bind both cytoplasmic and mitochondrial ribosomal proteins and translation initiation and elongations factors (Kanekura et al., 2016; Lee et al., 2016; Lin et al., 2016; Lopez-Gonzalez et al., 2016; Boeynaems et al., 2017; Yin et al., 2017; Hartmann et al., 2018; Moens et al., 2019; Radwan et al., 2020); the translation initiation factor eIF1A was able to rescue neuronal toxicity by enhancing translation in Drosophila (Moens et al., 2019). Ribosomal proteins are also found in a poly-GR mouse model and in cytoplasmic poly-GR and PR aggregates in patient tissue (Hartmann et al., 2018; Zhang et al., 2018b), suggesting that they may also sequester ribosomal proteins or assembled ribosomes as well. The arginine rich DPRs could also bind cellular RNAs rendering them insoluble and inaccessible to translation factors (Kanekura et al., 2016). An accumulation of nuclear mRNA has also been observed from expression of G4C2 repeats and proposed to be due to G4C2 repeat RNA sequestration of the nuclear export factor Aly/REF or the poly(A) binding protein PABPC (Cooper-Knock et al., 2014; Rossi et al., 2015). Both the arginine rich DPRs and G4C2 repeat RNA also facilitate the phase separation of stress granules, whose role is to temporarily stall translation of sequestered mRNAs when required by the cell. Poly-GR and PR increase β-sheet content and enhance stability of stress granule protein LCD polymers, increasing the number of stress granules in a cell by reducing their dynamics and preventing disassembly (Lee et al., 2016; Lin et al., 2016; Boeynaems et al., 2017). Within G4C2 RNA-induced stress granules, the protein component G3BP1 maintained mobility whereas the repeat RNA did not (Fay et al., 2017), showing that alterations in phase separation can have varying impact on different constituents. Additionally, the C9orf72 protein, which is reduced in C9orf72-FTD/ALS, has been implicated in physiological stress granule formation (Maharjan et al., 2017), showing co-operative pathology from the C9orf72 mutation. In addition, G4C2 repeat RNA has been identified in neuritic transport granules in murine and patient-derived neurons and in Drosophila where it colocalized with translational regulators and resulted in branching defects (Burguete et al., 2015). Thus, both DPR and G4C2 repeat RNA effects on the nucleolus, stress granules and other processes involved in translation can contribute to the translation repression seen in C9orf72-ALS/FTD.
We have detailed above, how both the arginine rich DPRs and G4C2 repeat RNA can disrupt the nuclear pore. Poly-GR and poly-PR bind FG-domain containing nucleoporins, stabilizing their polymeric form, and thus may change the biophysical properties of the central channel (Shi et al., 2017). G4C2 repeat RNA can also lead to a loss of a selective group of nucleoporins with dysfunction of the FG-nucleoporin POM121 central to its pathogenic effect (Coyne et al., 2020), however the mechanism is yet unknown. In addition to nucleoporins, nucleocytoplasmic transport involves nuclear transport receptors and proteins involved in the Ran cycle. Further studies have provided evidence that the impact of the arginine rich DPRs on nucleocytoplasmic transport can also occur via transport receptor (also known as karyopherins) interaction. Poorly dynamic stress granules induced by expression of the arginine rich DPRs can sequester both nucleoporins and the transport receptors and thereby disrupt nucleocytoplasmic transport (Zhang et al., 2018a); indeed, transport receptors are known components of physiological stress granules (Chang and Tarn, 2009; Fujimura et al., 2010; Mahboubi et al., 2013). Transport receptor-mediated nuclear import is also impaired by the arginine rich DPRs (Jovičić et al., 2015; Solomon et al., 2018; Hayes et al., 2020; Cook et al., 2020; Hutten et al., 2020). The arginine rich DPRs bind several transport receptors in vitro (Lee et al., 2016; Hutten et al., 2020). Interaction with importin-β occurs via competition with the binding of arginine rich nuclear localization signals on cargo and thus inhibits their transport (Hayes et al., 2020), whereas polyPR-transportin-1 binding has recently been pinpointed to the nuclear localization recognition domain of transportin-1 (Nanaura et al., 2019, preprint). Arginine rich DPR binding reduces transport receptor solubility and drives their oligomerization and LLPS with transportin-1, importin-α or an importin-α/β complex (as occurs for physiological transport) being more susceptible than importin-β alone to these effects; exportin-1 remained unaffected in solubility and biophysical assays (Hutten et al., 2020). Condensates formed by poly-GR and transportin-1 showed no recovery after photobleaching showing that proteins are immobile in these structures. DPR binding was generally stronger for poly-GR than PR, which was reflected in the selective impairment of a classic NLS reporter or TDP-43 import by poly-GR. Disruption of transport receptor-dependent nucleocytoplasmic transport is however not observed in all studies, likely due to differences in assay sensitivities, transport factors or cell lines studied (Khosravi et al., 2017; Vanneste et al., 2019). Nucleocytoplasmic transport factors are also found with poly-GR, GA and GP aggregates in patient tissue (Khosravi et al., 2017; Solomon et al., 2018). Transport factors seem to be non-specifically prone to be sequestered into inclusions, as even those formed by artificial β-sheet containing proteins can induce this, and this process is specific to cytoplasmic and not nuclear accumulations (Woerner et al., 2016).
C9orf72 G4C2 repeat RNA can also bind directly with nucleocytoplasmic transport factors, including Ran GTPase-activating protein 1 (RanGAP1; Donnelly et al., 2013). RanGAP is a key regulator of the Ran cycle which maintains the directionality of nucleocytoplasmic transport; it is anchored to the cytoplasmic face of the nuclear pore where it hydrolyzes RanGTP into RanGDP causing the dissociation of transport complexes releasing RanGDP and transport receptors to either release cargo into the cytoplasm for nuclear export or leave receptors available to bind cargo for import (Stewart, 2007). RanGAP overexpression was one of the strongest suppressors of toxicity from expression of 30 G4C2 repeats in a Drosophila screen and was found to mislocalize in patient brain tissue and in patient-derived neurons (Zhang et al., 2015), although the former finding in patient tissue has been disputed (Saberi et al., 2018). Cells expressing 30 G4C2 repeats and patient neurons also showed concomitant disturbances in the nuclear/cytoplasmic ratio of Ran, which could again be rescued by overexpression of RanGAP. Cytosolic accumulation of the Drosophila homolog of TDP-43, TBPH, was also found in Drosophila salivary gland cells expressing 30 G4C2 repeats, indicative of an imbalance in its nucleocytoplasmic shuttling. Although RanGAP does not contain any low complexity domains, interestingly its enzymatic activity can be dramatically enhanced when artificially targeted to liquid droplets (Peeples and Rosen, 2020, preprint). Noting that although the conclusions drawn implicate the C9orf72 G4C2 repeat RNA, it is possible that effects were also mediated by low undetectable levels of DPRs translated from the RNA, as abnormalities in RanGAP distribution have been observed as a result of poly-PR expression in the cortex of mice (Zhang et al., 2019a). In summary, both DPRs and G4C2 repeat RNA can disrupt nucleocytoplasmic transport by affecting the nuclear pore directly, but also via transport receptors and the Ran cycle, much of which involves interactions with LCD containing proteins and alterations in phase separation behavior.
Additional evidence also relates C9orf72 pathogenesis and phase separation to genomic homeostasis. DNA damage has been associated with arginine rich DPR toxicity, in particular aberrant activation of the p53 pathway (Lopez-Gonzalez et al., 2016). Poly-PR has recently been shown to lead to a stabilization of p53 and the transcription of its targets, and p53 reduction or knockdown rescues both poly-GR and poly-PR toxicity in neurons and mice, and in G4C2 repeat expressing Drosophila and C9orf72 patient-derived motor neurons (Maor-Nof et al., 2021). Interestingly, the nucleolus mediates the stabilization of p53 during DNA damage and regulates both its export and degradation (Rubbi and Milner, 2003; Boyd et al., 2011) and the nucleolar stress response pathway results in p53 accumulation (Rubbi and Milner, 2003; Yuan et al., 2005). As discussed above, both the arginine rich DPRs and G4C2 repeat RNA are associated with nucleolar dysfunction, with disruption in the phase separation of nucleolar proteins (White et al., 2019), and thus these pathologies may intersect in this pathway. In a mouse model expressing GFP-PR50 and in C9orf72-FTD/ALS patient tissue, poly-PR was found to colocalize with heterochromatin, highly condensed regions of chromatin which are transcriptionally inactive (Zhang et al., 2019a). These changes were not seen in GFP-(GR)100 expressing mice, likely due to the restricted cytoplasmic distribution of the DPR in this model, confirming that the actions of poly-PR are due to its nuclear localization. Poly-PR was subsequently found to disrupt the phase separation of heterochromatin protein 1α (HP1α) causing solid compartments to form within HP1α droplets and their bursting, and resulting in a significantly reduced number of droplets in vitro. This was recapitulated by reduced HP1α protein levels in the mouse model with functionally disrupted H3 histone post-translational modifications and upregulation of repetitive RNA elements, known to localize to heterochromatin, and double-stranded RNA which form from these which can initiate interferon signaling and cell death in neurons; these changes could also be induced by HP1α knockdown in cells. Interestingly, these changes occurred concurrently with irregularities in the nuclear lamina, which can also cause heterochromatin dysregulation (Scaffidi and Misteli, 2005; Shumaker et al., 2006). Indeed, changes in the nuclear lamina have been observed in C9orf72-FTD/ALS models (Zhang et al., 2019a), and loss of a Drosophila lamin enhanced C9orf72 repeat toxicity (Freibaum et al., 2015). Thus, DPRs may also contribute to DNA damage and reduced transcription via alterations in genomic homeostasis.
Perturbed phase separation and membraneless organelle formation appears to explain one of the key pathological features of C9orf72-ALS/FTD, the nuclear depletion and cytoplasmic aggregation of TDP-43 present in 97% and 45% of ALS and FTD cases respectively (Ling et al., 2013). Unlike the widespread DPR and RNA foci pathology, TDP-43 pathology is highly correlated with brain areas showing the highest levels of neurodegeneration and clinical symptoms (Mackenzie et al., 2013; DeJesus-Hernandez et al., 2017). Based on this it has been proposed that TDP-43 mislocalization and/or aggregation are the most likely effectors of toxicity from the C9orf72 mutation (Edbauer and Haass, 2016). Overt TDP-43 pathology is absent in many in vivo models of the C9orf72 mutation (possibly due to the short lifespan of model organisms) but increased cytoplasmic accumulation, increased biochemical insolubility or phosphorylation have been observed in Drosophila, murine and cellular models expressing poly-GR and poly-GA (Khosravi et al., 2017, 2020; Schludi et al., 2017; Solomon et al., 2018; Cook et al., 2020; Hutten et al., 2020; LaClair et al., 2020; Park et al., 2020; West et al., 2020). There have also been reports of association between TDP-43 and DPR pathologies: dendritic-like aggregates of poly-GR co-localized almost completely with phosphorylated TDP-43 in C9orf72-ALS motor cortex but formed only a small proportion of these TDP-43 aggregates in total (Saberi et al., 2018); similarly, a proportion of both inclusions of poly-GR and poly-GA have been found to colocalize with TDP-43 in C9orf72-FTD/ALS hippocampus (Cook et al., 2020). In agreement with associations between these pathologies, the burden of TDP-43 and poly-GA, poly-GP or poly-GR inclusions correlates with neurodegeneration (Mackenzie et al., 2013, 2015; Gendron et al., 2015; Saberi et al., 2018; Sakae et al., 2018). Soluble poly-GP and poly-GR levels also correlate with clinical severity (Quaegebeur et al., 2020). Interestingly, there were also associations with the methylation status of poly-GR, which can affect its phase separation properties (Sakae et al., 2018; Gittings et al., 2020). C9orf72 RNA foci composed of antisense C4G2 but not sense G4C2 transcripts have also been associated with the cytoplasmic mislocalization of TDP-43 in patient tissue (Cooper-Knock et al., 2015; Aladesuyi Arogundade et al., 2019).
Disrupted stress granule dynamics have been proposed to both directly and indirectly underlie the characteristic TDP-43 proteinopathy in ALS/FTD. Chronic stress can lead to an accumulation of TDP-43 in cytoplasmic stress granules, which become less dynamic by alterations in phase separation behavior from liquid to non-fluid gel states (Boeynaems and Gitler, 2018; McGurk et al., 2018b). Thus, the direct recruitment of TDP-43 to stress granules with impaired dynamics caused by the arginine rich DPRs and G4C2 repeat RNA may similarly explain pathological TDP-43 aggregation in DPR models and C9orf72-ALS/FTD. However, TDP-43 itself harbors an LCD and can also undergo LLPS in vitro (Molliex et al., 2015; Conicella et al., 2016, 2020; Schmidt et al., 2019) and form cytoplasmic droplets in cells independently of stress granules (Gasset-Rosa et al., 2019; Mann et al., 2019). RNA appears to be essential for these differences, with its presence being essential for recruitment of TDP-43 to stress granules and maintaining its solubility within them, and mutants lacking the ability to bind RNA forming immobile homomeric inclusions (Mann et al., 2019). This finding is consistent with TDP-43 inclusions in ALS/FTD patient tissue which do not contain stress granule proteins or RNA (Mann et al., 2019). Alterations in stress granule dynamics may thus indirectly prevent TDP-43 recruitment and promote homomeric TDP-43 inclusions. As the arginine rich DPRs and G4C2 repeat RNA perturb stress granule dynamics, their effect on TDP-43 aggregation may also similarly be indirect. A pathological retention of mRNA to poorly dynamic stress granules and DPR or repeat RNA-induced nuclear mRNA retention via impaired export (Freibaum et al., 2015; Rossi et al., 2015) would also further exacerbate this pathway by resulting in a cytoplasm deficient in mRNA preventing recruitment of TDP-43 to stress granules and promoting homomeric TDP-43 interactions in the cytoplasm resulting in further LLPS and the formation of more solid-like hydrogels of TDP-43 (Guenther et al., 2018) and its eventual aggregation. However, poly-GR interaction with TDP-43 does not require the RNA binding capability of TDP-43 (Cook et al., 2020) and can directly promote TDP-43 phase separation in vitro and reduce its solubility (Hutten et al., 2020). The direct effect of G4C2 RNA on TDP-43 phase separation has not yet been studied. In addition, DPR induced stress granule accumulation has been shown to specifically enhance RAN translation (Green et al., 2017; Cheng et al., 2018; Westergard et al., 2019), which would exacerbate any DPR induced mechanism discussed including cytoplasmic mislocalization of TDP-43; which itself has been shown to further enhance RAN translation (Solomon et al., 2018). Thus, both direct and indirect mechanisms relating to stress granules may account for pathological TDP-43 aggregation in DPR models and C9orf72-ALS/FTD (Figure 3).
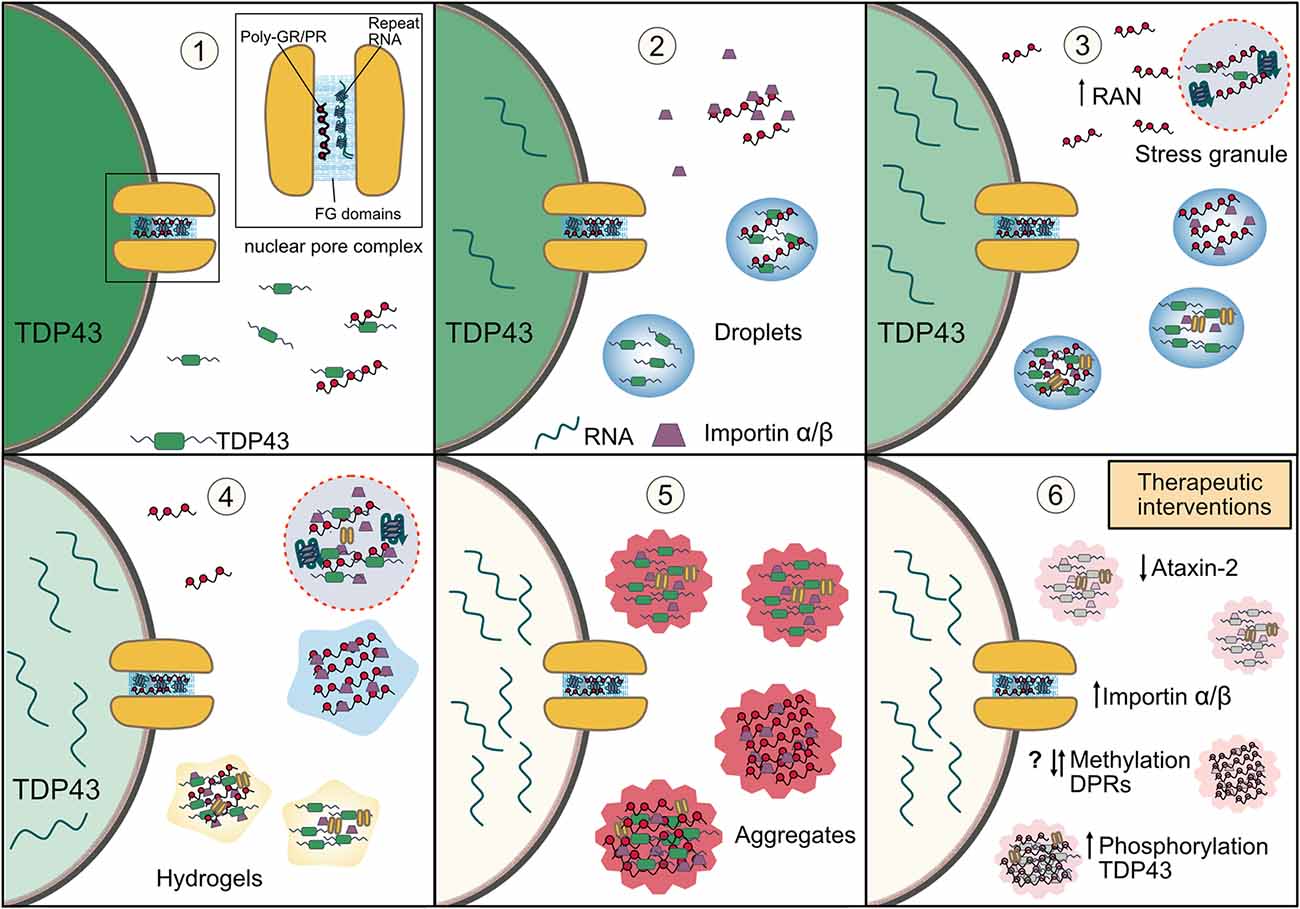
Figure 3. How disruptions in phase separation and membraneless organelles may lead to TDP-43 aggregation in C9orf72-ALS/FTD and possible therapeutic strategies. (1) C9orf72 arginine rich DPRs and G4C2 repeat RNA bind nuclear pore proteins with phenylalanine rich repeats (FG domains) and result in nucleocytoplasmic transport dysfunction and mislocalization of TDP-43 to the cytoplasm. (2) Interaction between cytoplasmic TDP-43 and the arginine rich DPRs results in the LLPS of TDP-43 in the cytoplasm. Impaired nucleocytoplasmic transport also results in an accumulation of the importin-α/β complex (the import receptor for TDP-43) in the cytoplasm where it is also bound by the arginine rich DPRs and results in their reduced solubility. This begins a vicious feedback loop as impaired nuclear import of TDP-43 further increases levels of cytoplasmic TDP-43, whose LLPS is potentiated by a nuclear retention of mRNA from impaired nuclear export. (3) Cellular stress and the direct interaction of arginine rich DPRs and G4C2 RNA with stress granule proteins (including TDP-43) promotes phase separation and the formation of stress granules. The arginine rich DPRs also induce condensation of importin-α/β. TDP-43 droplets recruit importin-α/β complexes and nuclear pore proteins further impairing nucleocytoplasmic transport, resulting in more TDP-43 accumulation in the cytoplasm and depletion of nuclear TDP-43. Both stress granule accumulation and cytoplasmic TDP-43 also enhance RAN translation of the arginine rich DPRs. (4) The stress granules induced by the arginine rich DPRs and repeat RNA have reduced dynamics which entraps TDP-43, import receptors and nuclear pore proteins. Persistent TDP-43 and DPR-importin-α/β droplets are likely to mature into more solid-like states such as hydrogels, further immobilizing these proteins. (5) TDP-43 in solid-like states and within stress granules, and also DPRs, mature into pathological insoluble aggregates which further sequester proteins involved in nucleocytoplasmic transport. Thus, the disruption of phase separation and membraneless organelles leads to a cascade of vicious feedback loops which result in depletion of nuclear TDP-43 and its accumulation and aggregation in the cytoplasm in disease. (6) Therapeutic targeting of stress granules by reducing ataxin-2 levels, manipulating post-translation modifications such as methylation of the DPRs and phosphorylation of TDP-43, or increasing importin-α/β to reduce excessive LLPS may enhance the solubility of TDP-43, help to reduce its aggregation and ameliorate the pathological cascade in C9orf72-ALS/FTD and other TDP-43 proteinopathies.
In cells, although RNA-binding capacity was essential for TDP-43 aggregation, the formation of cytoplasmic inclusions (as predominates in disease) only occurred with a disrupted nuclear localization signal which localizes TDP-43 to the cytoplasm but not wildtype TDP-43 which remains in the nucleus (Mann et al., 2019). Similarly, TDP-43 recruitment to stress granules induced by poly-GR was also dependent on its prior cytoplasmic mislocalization with the same mutants (Cook et al., 2020). Together these indicate that nucleocytoplasmic transport pathology lies upstream of TDP-43 pathology, as has been previously suggested (Dormann and Haass, 2011; Boeynaems et al., 2016b). Described in detail above, both arginine rich DPRs and G4C2 RNA induced deficits in nucleocytoplasmic transport may occur through the sequestration or demixing of nucleocytoplasmic transport factors by inclusions of either DPRs or G4C2 RNA or DPR or RNA induced stress granules, or by direct disruption of FG nucleoporins in the nuclear pore, through which all transport occurs. Indeed, poly-PR induced mislocalization of nucleocytoplasmic transport factors was seen in the absence of TDP-43 pathology in a mouse model (Zhang et al., 2019a). It is currently unclear how a disruption in the nuclear pore structure or biophysical properties may lead to a directional imbalance in nucleocytoplasmic shuttling and cytoplasmic mislocalization of TDP-43. Cytoplasmic mislocalization of TDP-43 may be caused by either or both a reduction in nuclear import and enhancement of nuclear export. TDP-43 nuclear import is governed by active transport via importin-β binding to its nuclear localization signal, and thus reduced availability of transport receptors or Ran cycle proteins may have a major impact, whereas its export may either be passive or via redundant export receptors (Archbold et al., 2018; Ederle et al., 2018; Pinarbasi et al., 2018), and thus changes in the nuclear pore may have greater effect. Further both cytoplasmic TDP-43 (Solomon et al., 2018; Gasset-Rosa et al., 2019) and TDP-43 aggregates (Chou et al., 2018) can also sequester or lead to cytoplasmic demixing of proteins including nucleocytoplasmic transport factors which would again exacerbate these mechanisms– further enhancing TDP-43 mislocalization and aggregation. Together, through disruptions in phase separation behavior, stress granules and nucleocytoplasmic transport in combination with loss of nuclear TDP-43 autoregulation (Ayala et al., 2011; White et al., 2018), TDP-43 pathology may become independent of the initial DPR insult and maintain its own pathological cascade in a vicious feedback cycle (Solomon et al., 2018; Figure 3), which may explain the segregation of DPR and TDP-43 pathology in patient tissue.
We have described the evidence for the ability of the C9orf72 arginine rich DPRs and G4C2 RNA to disrupt LLPS and perturb membraneless organelles leading to key pathomechanisms in disease, but what can be done to target these therapeutically to prevent TDP-43 aggregation and toxicity? One potential avenue would be to attempt to ameliorate the enhanced propensity for proteins to phase separate in the presence of pathological molecules and the resulting change in the biophysical properties and functionality of the droplets or organelles formed (Elbaum-Garfinkle, 2019). So far, the majority of treatments in this area relating to C9orf72-FTD/ALS and TDP-43 proteinopathy target stress granules. A high throughput screen identified a diverse range of small-molecule compounds that could alter stress granule properties; around half of these were compounds with planar moieties, such as Mitoxantrone which reduced both the size and number of stress granules formed, prevented recruitment of RNA-binding proteins (including TDP-43) to stress granules and decreased persistent TDP-43 puncta in cells treated with cellular stressors, and also ameliorated toxicity from overexpression of a mutant TDP-43 in primary neurons (Fang et al., 2019). Planar compounds intercalate into nucleic acids, and thus may act by preventing the RNA-dependent recruitment of TDP-43 to aberrant stress granules. Interestingly, another planar compound identified in this screen, doxorubicin, can alter the phase separation of CAG and CUG repeat RNAs and reduces G4C2 repeat RNA foci formation in cells (Jain and Vale, 2017). In the former screen, doxorubicin could also ameliorate toxicity from mutant TDP-43 but had differing cell line-dependent effects on stress granules and did not prevent the recruitment of TDP-43 (Fang et al., 2019). Thus, compounds with planar moieties act on multiple and varying mechanisms and maybe of therapeutic potential in C9orf72-FTD/ALS pathology. Another target of note is the stress granule protein ataxin-2, as its knockout or ASOs targeting ataxin-2 could significantly decrease TDP-43 aggregation and increase survival in a mouse model overexpressing wildtype TDP-43 (Becker et al., 2017). Ataxin-2 harbors two intrinsically disordered regions which both modulate phase separation behavior in vitro (Bakthavachalu et al., 2018). Ataxin-2 knockdown significantly decreases both the number of stress granules and the recruitment of endogenous TDP-43 (Becker et al., 2017). The arginine rich DPRs also directly interact with ataxin-2 (Lee et al., 2016; Hayes et al., 2020), colocalizing with poly-GR aggregates and TDP-43 in GR200 expressing mice (Cook et al., 2020). Knockdown of ataxin-2 can rescue mislocalization of Ran (part of the driving force for active nucleocytoplasmic transport) in C9orf72 patient-derived neurons (Zhang et al., 2018a) and was the strongest suppressor of poly-GR toxicity in a Drosophila RNA inhibitor screen (Lee et al., 2016). This suppression could also be recapitulated when either intrinsically disordered region of ataxin-2 was deleted (Bakthavachalu et al., 2018), suggesting that its LLPS ability facilities C9orf72 DPR toxicity. Hence it is possible that stress granule assembly promotes an environment for poly-GR to aggregate, undergo aberrant interactions (such as with TDP-43) and enhance toxicity, therefore modulating this process via reducing ataxin-2 levels may provide a novel therapeutic target in C9orf72-ALS/FTD (Figure 3, panel 6). Post translational modifications can also regulate the LLPS of proteins and the properties of the resulting condensates (Hofweber and Dormann, 2019) and thus could be targets for intervention. The post-translational modification poly-ADPribose (PAR) binds within the NLS of TDP-43 promoting its LLPS and is required for recruitment to stress granules (McGurk et al., 2018b). Knockdown of the PAR polymerases (PARPs) tankyrase 1/2 or small molecule inhibition of PARP-1/2 could both reduce toxicity from overexpression of TDP-43 in Drosophila or spinal cord cultures, respectively (McGurk et al., 2018a, b). Interestingly, the small molecule inhibitors of PARP-1/2 reduce the formation of TDP-43-positive stress granules in cells (McGurk et al., 2018a), whereas the tankyrase-1/2 inhibitors do not affect stress granules but prevent TDP-43 recruitment to them (McGurk et al., 2018b), suggesting that the latter may act more specifically on TDP-43. Indeed, recent data demonstrates a novel binding site for the tankyrases on TDP-43 and suggests that binding may prevent its ubiquitination and proteasomal turnover in the nucleus, stabilizing TDP-43 in the cytoplasm (McGurk et al., 2020). Thus, either direct or indirect pathways associated with LLPS and stress granules are showing promise as therapies for C9orf72-FTD/ALS.
Targeting DPRs or TDP-43 directly to prevent their aberrant phase separation and aggregation may also be effective. It has been recently shown that anti-cancer drugs partition into droplets formed by their molecular targets and change the properties of the condensate (Klein et al., 2020). Two molecules which have recently shown promise for ALS are lipoamide and lipoic acid which prevent FUS aggregation via influencing FUS phase separation and reduce FUS toxicity in vivo (Wheeler et al., 2019). The identification of small molecules and compounds that can modulate the charge-charge and cation-pi interactions essential for phase separation of LCD containing proteins is an interesting avenue for future research, although a greater understanding is needed into the physiochemical mechanisms as to how molecules such as lipoamide and lipoic acid disrupt these interactions inhibiting LLPS (Wheeler, 2020). Again, post-translational modifications can be targeted. As detailed above symmetric dimethylation of poly-GR reduces its propensity for LLPS and appears to be protective (Gittings et al., 2020; Pakravan et al., 2020). The enzymes responsible for arginine methylation belong to the protein arginine methyltransferase (PRMT) family (Yang and Bedford, 2013). Knockdown of several PRMTs enhance toxicity in Drosophila models of poly-PR toxicity, with PRMT1 also colocalizing with poly-PR upon co-transfection in cell lines (Boeynaems et al., 2016a), in agreement with the protective role suggested by human studies. However, small molecule inhibition of type I PRMTs, which produce asymmetrically dimethylated arginine, alleviated the toxicity of both poly-GR and PR in cells (Premasiri et al., 2020). This could align with the human studies as symmetric dimethylation would protect the arginine residues from asymmetric demethylation, although it does not explain why knockdown of the type I PRMT PRMT1 exacerbated toxicity in the Drosophila studies. These opposing roles do not however align with their effect on LLPS, as both symmetric and asymmetric demethylation of poly-GR reduce propensity to undergo LLPS. Interestingly, a rapid demethylation of the stress granule protein G3BP1 is critical for stress granule assembly (Tsai et al., 2016), showing that methylation can modulate LLPS behavior in different contexts and that the process can also be dynamic. Further work will be required to assimilate these findings and determine whether modulating the phase separation behavior of the arginine rich DPRs by altering their methylation state is a valid therapeutic target (Figure 3, panel 6). Phosphorylation can also both enhance and hinder LLPS of ribonuclear granules in vitro (Hofweber et al., 2018). In the context of TDP-43, phosphorylation of its N-terminal domain inhibits LLPS (Wang et al., 2018a) and C-terminal domain phosphorylation has been shown to reduce TDP-43 aggregation and toxicity in cells (Li et al., 2011), suggesting that the associated kinases or phosphatases could be targeted (Figure 3, panel 6). However, as is commonly found in neurodegeneration (Hofweber and Dormann, 2019), TDP-43 inclusions are hyperphosphorylated in patient brain which may suggest that a role in aggregate formation (Hasegawa et al., 2008). Also, post-translation modifications regulate a vast number of protein and cellular functions, hence targeting them on a specific molecule of interest is incredibly challenging and complicating this even further is the fact numerous post-translational modification enzymes have been associated with ALS (Guo et al., 2020).
It has also been proposed that chaperones exist whose normal function is to modulate LLPS within a cellular environment that is particularly concentrated, unstable and oversaturated in order to prevent abnormal fibrillization (Elbaum-Garfinkle, 2019). Such chaperones may provide more specific therapeutic targets, rather than modifying the LLPS of membraneless organelles or post-translational modifications which are both vitally important in normal cell function. Heat shock proteins are a classic example of cellular chaperones and the master transcriptional regulator of heat shock protein expression HSF1 can prevent cytoplasmic accumulation and toxicity from over-expression of wildtype or mutant TDP-43; the associated reduction in TDP-43 solubility was found to be enacted by the heat shock protein DNAJB2a (Chen et al., 2016). Variants of the heat shock protein Hsp104 engineered to potentiate disaggregation are also able to dissolve both FUS and TDP-43 aggregates in yeast (Jackrel and Shorter, 2014; Jackrel et al., 2014) and FUS aggregates in cells (Yasuda et al., 2017). Interestingly, nuclear import receptors, associated with both DPR and TDP-43 toxicity in models and post-mortem brain (Nishimura et al., 2010; Jovičić et al., 2015; Solomon et al., 2018; Gasset-Rosa et al., 2019; Hayes et al., 2020; Cook et al., 2020; Hutten et al., 2020; Park et al., 2020) have also been shown to prevent the pathological phase separation of RNA binding proteins (Springhower et al., 2020). Transportin-1 can both prevent phase separation, hydrogel formation and fibrillization of FUS and hnRNP A1 and A2, and importantly dissolve already formed hydrogels and fibrillar aggregates (Guo et al., 2018; Qamar et al., 2018; Yoshizawa et al., 2018; Hofweber et al., 2018). ALS-associated mutations in the NLS of FUS disrupt binding to transportin-1, preventing its solubilizing effects on FUS and leading to FUS accumulation in stress granules (Hofweber et al., 2018). Functionally, increasing transportin-1 expression could also restore nuclear localization of FUS or hnRNPA1/2, restore proteins synthesis in hypomethylated FUS neurons, and improve lifespan and muscle degeneration phenotypes in Drosophila models of mutant FUS and hnRNP A2, respectively (Guo et al., 2018; Qamar et al., 2018). Similarly, an importin-α/β complex reduces in vitro LLPS and fibril formation of TDP-43 and could also disassemble preformed fibrils (Guo et al., 2018). TDP-43 is transported into the nucleus via its classic NLS recognized by the importin-α/β complex, whilst FUS and the hnRNPs contain a PY-NLS which is bound and transported by transportin-1 (Mihevc et al., 2017). If the PY-NLS of FUS is substituted for a classic NLS, the importin-α/β complex can now inhibit its phase separation (Yoshizawa et al., 2018). Hence, nuclear import receptors modulate the phase separation and fibril formation of cargo in their respective nucleocytoplasmic transport pathway, indicating an additional physiological chaperone function of transport receptors in maintaining solubility of their cargo. Thus, potential disease therapies will likely need to reflect the major pathology involved. In the case of C9orf72-ALS/FTD where TDP-43 is the major pathology, the arginine rich DPRs bind to importin-α and β, reducing their solubility and lead to their precipitation and condensation; this results in defective nuclear import of TDP-43, and thus likely contributes to the dominant TDP-43 pathology (Hutten et al., 2020). High concentrations of importin-α/β can suppress poly-GR induced TDP-43 phase separation in vitro and its reduced solubility in cells, and inhibit RNA-driven LLPS of poly-GR. Therefore, therapies boosting levels of importin-α and β could ameliorate both DPR and TDP-43 pathologies and prevent other pathogenic interactions, which may be beneficial for the wide range of cellular abnormalities observed in C9orf72-ALS/FTD (Figure 3, panel 6). Notably, both heat shock proteins and import receptors are reduced in ALS/FTD patient tissue (Kinoshita et al., 2009; Nishimura et al., 2010; Chen et al., 2016; Solomon et al., 2018), and thus supplementation strategies will also restore physiological roles lost in disease.
A complementary strategy to all of the above strategies is to target the C9orf72 G4C2 repeat RNA and DPRs directly to reduce their levels and prevent pathological intra and intermolecular associations. An approach that has gained much traction and is being pursued in clinical trials is using ASOs to the C9orf72 repeat which can reduce the formation of repeat RNA and DPRs in C9orf72 repeat-expressing neurons and mice and patient-derived neurons with an associated reduction in glutamate-induced toxicity in cultured neurons and behavioral and cognitive phenotypes in mice (Donnelly et al., 2013; Lagier-Tourenne et al., 2013; Sareen et al., 2013; Jiang et al., 2016; Gendron et al., 2017). Small molecules which bind G4C2 G-quadruplexes can reduce the production of DPRs and rescue toxicity (Zhang et al., 2015; Simone et al., 2018), potentially by destabilizing and ablating G-quadruplex dependent interactions (Zamiri et al., 2014), which may prevent its nucleation behavior in relation to RNA-dependent protein condensation and stress granule formation discussed above. Endogenous targets that modulate the production of repeat RNA and/or RAN translation have also been identified. Reducing the transcription elongation factor Spt4 or PAF1C complex, a transcriptional regulator of RNA polymerase II, reduce production of expanded G4C2 repeat RNA and thus the translation of DPRs and their associated toxicity in model systems (Liu et al., 2012; Kramer et al., 2016; Goodman et al., 2019a). Conversely, the RNA-binding protein hnRNPA3 can bind G4C2 repeat RNA and its depletion increases production of repeat RNA and DPRs (Mori et al., 2013b, 2016). Genetic screens have also identified regulators of RAN translation, including the small ribosomal protein RPS25 and the eukaryotic translation initiation factors eIF4B and eIF4H, whose loss could reduce the production of DPRs and extend lifespan in Drosophila models and the former also in C9orf72 patient cells (Goodman et al., 2019b; Yamada et al., 2019). The integrated stress response also promotes RAN translation (Green et al., 2017; Cheng et al., 2018; Westergard et al., 2019), and inhibition of one part of this—the double-stranded RNA-dependent protein kinase (PKR) pathway—can reduce RAN translation and mitigate gliosis, motor neuron loss and behavioral phenotypes in a C9orf72 mutation mouse model (Zu et al., 2020). Of note, hnRNPA3 and eIF4H are reduced in C9orf72-FTD/ALS patient brain (Mori et al., 2016), and therefore supplementation may provide therapeutic benefit, whereas components of the PAF1C complex are upregulated and the PKR pathway is aberrantly activated in disease and thus inhibition of these pathways may normalize levels (Goodman et al., 2019a; Zu et al., 2020).
There is now substantial and clear evidence that a disruption in the phase separation behavior of proteins and RNA involved in the formation of liquid-like membraneless organelles explains much of the major pathological phenomena associated with C9orf72-ALS/FTD. Gain-of-function mechanisms associated with the G4C2 repeat expansion in C9orf72–G4C2 repeat RNA and the arginine rich DPRs poly-GR and poly-PR—undergo phase separation themselves and perturb the phase separation of LCD containing proteins, resulting in abnormal membraneless organelle formation and dissolution, impairing their physiological functions and leading to neurodegeneration. Further pathological phase separation induced by the arginine rich DPRs is strongly associated with TDP-43 dysfunction and aggregation, the major pathological hallmark of C9orf72-ALS/FTD correlating with neuronal cell death. The targeting of abnormally phase separated condensates using small molecules or gene therapy provides a novel strategy for future therapeutics, although a greater understanding is needed of phase separation in order to design targets which are both beneficial and precise.
DS and SM conceived the review. DS, RS and SM wrote sections of the manuscript. MR and DS designed and produced figures. All authors contributed to the article and approved the submitted version.
This work was supported by the UK Dementia Research Institute which receives its funding from DRI Limited, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aguzzi, A., and Altmeyer, M. (2016). Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 26, 547–558. doi: 10.1016/j.tcb.2016.03.004
Aladesuyi Arogundade, O., Stauffer, J. E., Saberi, S., Diaz-Garcia, S., Malik, S., Basilim, H., et al. (2019). Antisense RNA foci are associated with nucleoli and TDP-43 mislocalization in C9orf72-ALS/FTD: a quantitative study. Acta Neuropathol. 137, 527–530. doi: 10.1007/s00401-018-01955-0
Alberti, S., and Dormann, D. (2019). Liquid-liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194. doi: 10.1146/annurev-genet-112618-043527
Alberti, S., Gladfelter, A., and Mittag, T. (2019). Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434. doi: 10.1016/j.cell.2018.12.035
Almeida, S., Gascon, E., Tran, H., Chou, H. J., Gendron, T. F., Degroot, S., et al. (2013). Modeling key pathological features of frontotemporal dementia with C9orf72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 126, 385–399. doi: 10.1007/s00401-013-1149-y
Archbold, H. C., Jackson, K. L., Arora, A., Weskamp, K., Tank, E. M. H., Li, X., et al. (2018). TDP43 nuclear export and neurodegeneration in models of amyotrophic lateral sclerosis and frontotemporal dementia. Sci. Rep. 8:4606. doi: 10.1038/s41598-018-22858-w
Ash, P. E. A., Bieniek, K. F., Gendron, T. F., Caulfield, T., Lin, W. L., DeJesus-Hernandez, M., et al. (2013). Unconventional translation of C9orf72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646. doi: 10.1016/j.neuron.2013.02.004
Atanasio, A., Decman, V., White, D., Ramos, M., Ikiz, B., Lee, H. C., et al. (2016). C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci. Rep. 6:23204. doi: 10.1038/srep23204
Ayala, Y. M., De Conti, L., Avendaño-Vázquez, S. E., Dhir, A., Romano, M., D’Ambrogio, A., et al. (2011). TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 30, 277–288. doi: 10.1038/emboj.2010.310
Bakthavachalu, B., Huelsmeier, J., Sudhakaran, I. P., Hillebrand, J., Singh, A., Petrauskas, A., et al. (2018). RNP-granule assembly via ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98, 754.e4–766.e4. doi: 10.1016/j.neuron.2018.04.032
Balendra, R., and Isaacs, A. M. (2018). C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 14, 544–558. doi: 10.1038/s41582-018-0047-2
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. doi: 10.1038/nrm.2017.7
Baradaran-Heravi, Y., Van Broeckhoven, C., and van der Zee, J. (2020). Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol. Dis. 134:104639. doi: 10.1016/j.nbd.2019.104639
Becker, L. A., Huang, B., Bieri, G., Ma, R., Knowles, D. A., Jafar-Nejad, P., et al. (2017). Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371. doi: 10.1038/nature22038
Boeynaems, S., and Gitler, A. D. (2018). Pour some sugar on TDP(-43). Mol. Cell 71, 649–651. doi: 10.1016/j.molcel.2018.08.032
Boeynaems, S., Alberti, S., Fawzi, N. L., Mittag, T., Polymenidou, M., Rousseau, F., et al. (2018). Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435. doi: 10.1016/j.tcb.2018.02.004
Boeynaems, S., Bogaert, E., Kovacs, D., Konijnenberg, A., Timmerman, E., Volkov, A., et al. (2017). Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055. doi: 10.1016/j.molcel.2017.02.013
Boeynaems, S., Bogaert, E., Michiels, E., Gijselinck, I., Sieben, A., Jovičić, A., et al. (2016a). Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 6:20877. doi: 10.1038/srep20877
Boeynaems, S., Bogaert, E., Van Damme, P., and Van Den Bosch, L. (2016b). Inside out: the role of nucleocytoplasmic transport in ALS and FTLD. Acta Neuropathol. 132, 159–173. doi: 10.1007/s00401-016-1586-5
Boeynaems, S., Holehouse, A. S., Weinhardt, V., Kovacs, D., Van Lindt, J., Larabell, C., et al. (2019). Spontaneous driving forces give rise to protein−RNA condensates with coexisting phases and complex material properties. Proc. Natl. Acad. Sci. U S A 116, 7889–7898. doi: 10.1073/pnas.1821038116
Bogaert, E., Boeynaems, S., Kato, M., Guo, L., Caulfield, T. R., Steyaert, J., et al. (2018). Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 24, 529.e4–537.e4. doi: 10.1016/j.celrep.2018.06.070
Boisvert, F. M., van Koningsbruggen, S., Navascués, J., and Lamond, A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585. doi: 10.1038/nrm2184
Boyd, M. T., Vlatković, N., and Rubbi, C. P. (2011). The nucleolus directly regulates p53 export and degradation. J. Cell Biol. 194, 689–703. doi: 10.1083/jcb.201105143
Brangwynne, C. P., Tompa, P., and Pappu, R. V. (2015). Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899–904. doi: 10.1038/nphys3532
Burberry, A., Suzuki, N., Wang, J. Y., Moccia, R., Mordes, D. A., Stewart, M. H., et al. (2016). Loss-of-function mutations in the C9orf72 mouse ortholog cause fatal autoimmune disease. Sci. Transl. Med. 8:347ra93. doi: 10.1126/scitranslmed.aaf6038
Burguete, A. S., Almeida, S., Gao, F. B., Kalb, R., Akins, M. R., and Bonini, N. M. (2015). GGGGCC microsatellite RNA is neuritically localized, induces branching defects, and perturbs transport granule function. eLife 4:e08881. doi: 10.7554/eLife.08881
Callister, J. B., Ryan, S., Sim, J., Rollinson, S., and Pickering-Brown, S. M. (2016). Modelling C9orf72 dipeptide repeat proteins of a physiologically relevant size. Hum. Mol. Genet. 25, 5069–5082. doi: 10.1093/hmg/ddw327
Cautain, B., Hill, R., de Pedro, N., and Link, W. (2015). Components and regulation of nuclear transport processes. FEBS J. 282, 445–462. doi: 10.1111/febs.13163
Celona, B., von Dollen, J., Vatsavayai, S. C., Kashima, R., Johnson, J. R., Tang, A. A., et al. (2017). Suppression of C9orf72 RNA repeat-induced neurotoxicity by the ALS-associated RNA-binding protein Zfp106. eLife 6:e19032. doi: 10.7554/eLife.19032
Chai, N., and Gitler, A. D. (2018). Yeast screen for modifiers of C9orf72 poly(glycine-arginine) dipeptide repeat toxicity. FEMS Yeast Res. 18:foy024. doi: 10.1093/femsyr/foy024
Chang, W.-L., and Tarn, W.-Y. (2009). A role for transportin in deposition of TTP to cytoplasmic RNA granules and mRNA decay. Nucleic Acids Res. 37, 6600–6612. doi: 10.1093/nar/gkp717
Chen, H.-J., Mitchell, J. C., Novoselov, S., Miller, J., Nishimura, A. L., Scotter, E. L., et al. (2016). The heat shock response plays an important role in TDP-43 clearance: evidence for dysfunction in amyotrophic lateral sclerosis. Brain 139, 1417–1432. doi: 10.1093/brain/aww028
Cheng, W., Wang, S., Mestre, A. A., Fu, C., Makarem, A., Xian, F., et al. (2018). C9orf72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun. 9:51. doi: 10.1038/s41467-017-02495-z
Choi, S. Y., Lopez-Gonzalez, R., Krishnan, G., Phillips, H. L., Li, A. N., Seeley, W. W., et al. (2019). C9ORF72-ALS/FTD-associated poly(GR) binds Atp5a1 and compromises mitochondrial function in vivo. Nat. Neurosci. 22, 851–862. doi: 10.1038/s41593-019-0397-0
Chong, P. A., Vernon, R. M., and Forman-Kay, J. D. (2018). RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665. doi: 10.1016/j.jmb.2018.06.014
Chou, C.-C., Zhang, Y., Umoh, M. E., Vaughan, S. W., Lorenzini, I., Liu, F., et al. (2018). TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 21, 228–239. doi: 10.1038/s41593-017-0047-3
Conicella, A. E., Dignon, G. L., Zerze, G. H., Schmidt, H. B., D’Ordine, A. M., Kim, Y. C., et al. (2020). TDP-43 α-helical structure tunes liquid-liquid phase separation and function. Proc. Natl. Acad. Sci. U S A 117, 5883–5894. doi: 10.1073/pnas.1912055117
Conicella, A. E., Zerze, G. H., Mittal, J., and Fawzi, N. L. (2016). ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-Terminal domain. Structure 24, 1537–1549. doi: 10.1016/j.str.2016.07.007
Conlon, E. G., Lu, L., Sharma, A., Yamazaki, T., Tang, T., Shneider, N. A., et al. (2016). The C9orf72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife 5:e17820. doi: 10.7554/eLife.17820
Cook, C. N., Wu, Y., Odeh, H. M., Gendron, T. F., Jansen-West, K., del Rosso, G., et al. (2020). C9orf72 poly(GR) aggregation induces TDP-43 proteinopathy. Sci. Transl. Med. 12:eabb3774. doi: 10.1126/scitranslmed.abb3774
Cooper-Knock, J., Bury, J. J., Heath, P. R., Wyles, M., Higginbottom, A., Gelsthorpe, C., et al. (2015). C9orf72 GGGGCC expanded repeats produce splicing dysregulation which correlates with disease severity in amyotrophic lateral sclerosis. PLoS One 10:e0127376. doi: 10.1371/journal.pone.0127376
Cooper-Knock, J., Walsh, M. J., Higginbottom, A., Highley, J. R., Dickman, M. J., Edbauer, D., et al. (2014). Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 137, 2040–2051. doi: 10.1093/brain/awu120
Coyne, A. N., Zaepfel, B. L., Hayes, L., Fitchman, B., Salzberg, Y., Luo, E. C., et al. (2020). G4C2 repeat RNA initiates a POM121-mediated reduction in specific nucleoporins in C9orf72 ALS/FTD. Neuron 107, 1124.e11–1140.e11. doi: 10.1016/j.neuron.2020.06.027
DeJesus-Hernandez, M., Finch, N. C. A., Wang, X., Gendron, T. F., Bieniek, K. F., Heckman, M. G., et al. (2017). In-depth clinico-pathological examination of RNA foci in a large cohort of C9orf72 expansion carriers. Acta Neuropathol. 134, 255–269. doi: 10.1007/s00401-017-1725-7
DeJesus-Hernandez, M., Mackenzie, I. R., Boeve, B. F., Boxer, A. L., Baker, M., Rutherford, N. J., et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9orf72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. doi: 10.1016/j.neuron.2011.09.011
Deshpande, S., Brandenburg, F., Lau, A., Last, M. G. F., Spoelstra, W. K., Reese, L., et al. (2019). Spatiotemporal control of coacervate formation within liposomes. Nat. Commun. 10:1800. doi: 10.1038/s41467-019-09855-x
Donnelly, C. J., Zhang, P. W., Pham, J. T., Heusler, A. R., Mistry, N. A., Vidensky, S., et al. (2013). RNA toxicity from the ALS/FTD C9orf72 expansion is mitigated by antisense intervention. Neuron 80, 415–428. doi: 10.1016/j.neuron.2013.10.015
Dormann, D., and Haass, C. (2011). TDP-43 and FUS: a nuclear affair. Trends Neurosci. 34, 339–348. doi: 10.1016/j.tins.2011.05.002
Edbauer, D., and Haass, C. (2016). An amyloid-like cascade hypothesis for C9orf72 ALS/FTD. Curr. Opin. Neurobiol. 36, 99–106. doi: 10.1016/j.conb.2015.10.009
Ederle, H., Funk, C., Abou-Ajram, C., Hutten, S., Funk, E. B. E., Kehlenbach, R. H., et al. (2018). Nuclear egress of TDP-43 and FUS occurs independently of Exportin-1/CRM1. Sci. Rep. 8:7084. doi: 10.1038/s41598-018-25007-5
Elbaum-Garfinkle, S. (2019). Matter over mind: liquid phase separation and neurodegeneration. J. Biol. Chem. 294, 7160–7168. doi: 10.1074/jbc.REV118.001188
Fang, M. Y., Markmiller, S., Vu, A. Q., Javaherian, A., Dowdle, W. E., Jolivet, P., et al. (2019). Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent tdp-43 accumulation in ALS/FTD. Neuron 103, 802.e11–819.e11. doi: 10.1016/j.neuron.2019.05.048
Farg, M. A., Konopka, A., Soo, K. Y., Ito, D., and Atkin, J. D. (2017). The DNA damage response (DDR) is induced by the C9orf72 repeat expansion in amyotrophic lateral sclerosis. Hum. Mol. Genet. 26, 2882–2896. doi: 10.1093/hmg/ddx170
Farg, M. A., Sundaramoorthy, V., Sultana, J. M., Yang, S., Atkinson, R. A. K., Levina, V., et al. (2014). C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum. Mol. Genet. 23, 3579–3595. doi: 10.1093/hmg/ddu068
Fay, M. M., Anderson, P. J., and Ivanov, P. (2017). ALS/FTD-associated C9orf72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 21, 3573–3584. doi: 10.1016/j.celrep.2017.11.093
Feng, Z., Chen, X., Wu, X., and Zhang, M. (2019). Formation of biological condensates via phase separation: characteristics, analytical methods, and physiological implications. J. Biol. Chem. 294:294. doi: 10.1074/jbc.REV119.007895
Fox, A. H., Nakagawa, S., Hirose, T., and Bond, C. S. (2018). Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 43, 124–135. doi: 10.1016/j.tibs.2017.12.001
Fratta, P., Mizielinska, S., Nicoll, A. J., Zloh, M., Fisher, E. M. C., Parkinson, G., et al. (2012). C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep. 2:1016. doi: 10.1038/srep01016
Freibaum, B. D., and Taylor, J. P. (2017). The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci. 10:35. doi: 10.3389/fnmol.2017.00035
Freibaum, B. D., Lu, Y., Lopez-Gonzalez, R., Kim, N. C., Almeida, S., Lee, K.-H., et al. (2015). GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133. doi: 10.1038/nature14974
Frey, S., Richter, R. P., and Görlich, D. (2006). FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817. doi: 10.1126/science.1132516
Frottin, F., Schueder, F., Tiwary, S., Gupta, R., Körner, R., Schlichthaerle, T., et al. (2019). The nucleolus functions as a phase-separated protein quality control compartment. Science 365, 342–347. doi: 10.1126/science.aaw9157
Fujimura, K., Suzuki, T., Yasuda, Y., Murata, M., Katahira, J., and Yoneda, Y. (2010). Identification of importin α1 as a novel constituent of RNA stress granules. Biochim. Biophys. Acta 1803, 865–871. doi: 10.1016/j.bbamcr.2010.03.020
Galganski, L., Urbanek, M. O., and Krzyzosiak, W. J. (2017). Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Research 45, 10350–10368. doi: 10.1093/nar/gkx759
Garcia-Jove Navarro, M., Kashida, S., Chouaib, R., Souquere, S., Pierron, G., Weil, D., et al. (2019). RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat. Commun. 10:3230. doi: 10.1038/s41467-019-11241-6
Gasset-Rosa, F., Lu, S., Yu, H., Chen, C., Melamed, Z., Guo, L., et al. (2019). Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339.e7–357.e7. doi: 10.1016/j.neuron.2019.02.038
Gendron, T. F., Bieniek, K. F., Zhang, Y.-J., Jansen-West, K., Ash, P. E. A., Caulfield, T., et al. (2013). Antisense transcripts of the expanded C9orf72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844. doi: 10.1007/s00401-013-1192-8
Gendron, T. F., Chew, J., Stankowski, J. N., Hayes, L. R., Zhang, Y. J., Prudencio, M., et al. (2017). Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci. Transl. Med. 9:eaai7866. doi: 10.1126/scitranslmed.aai7866
Gendron, T. F., van Blitterswijk, M., Bieniek, K. F., Daughrity, L. M., Jiang, J., Rush, B. K., et al. (2015). Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9orf72 repeat expansion carriers. Acta Neuropathol. 130, 559–573. doi: 10.1007/s00401-015-1474-4
Gittings, L. M., Boeynaems, S., Lightwood, D., Clargo, A., Topia, S., Nakayama, L., et al. (2020). Symmetric dimethylation of poly-GR correlates with disease duration in C9orf72 FTLD and ALS and reduces poly-GR phase separation and toxicity. Acta Neuropathol. 139, 407–410. doi: 10.1007/s00401-019-02104-x
Gomes, E., and Shorter, J. (2019). The molecular language of membraneless organelles. J. Biol. Chem. 294, 7115–7127. doi: 10.1074/jbc.TM118.001192
Goodman, L. D., Prudencio, M., Kramer, N. J., Martinez-Ramirez, L. F., Srinivasan, A. R., Lan, M., et al. (2019a). Toxic expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nat. Neurosci. 22, 863–874. doi: 10.1038/s41593-019-0396-1
Goodman, L. D., Prudencio, M., Srinivasan, A. R., Rifai, O. M., Lee, V. M. Y., Petrucelli, L., et al. (2019b). eIF4B and eIF4H mediate GR production from expanded G4C2 in a Drosophila model for C9orf72-associated ALS. Acta Neuropathol. Commun. 7:62. doi: 10.1186/s40478-019-0711-9
Green, K. M., Glineburg, M. R., Kearse, M. G., Flores, B. N., Linsalata, A. E., Fedak, S. J., et al. (2017). RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun. 8:2005. doi: 10.1038/s41467-017-02200-0
Gruss, O. J., Meduri, R., Schilling, M., and Fischer, U. (2017). UsnRNP biogenesis: mechanisms and regulation. Chromosoma 126, 577–593. doi: 10.1007/s00412-017-0637-6
Guenther, E. L., Cao, Q., Trinh, H., Lu, J., Sawaya, M. R., Cascio, D., et al. (2018). Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat. Struct. Mol. Biol. 25, 463–471. doi: 10.1038/s41594-018-0064-2
Gui, X., Luo, F., Li, Y., Zhou, H., Qin, Z., Liu, Z., et al. (2019). Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat. Commun. 10:2006. doi: 10.1038/s41467-019-09902-7
Guo, L., Kim, H. J., Wang, H., Monaghan, J., Freyermuth, F., Sung, J. C., et al. (2018). Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677.e20–692.e20. doi: 10.1016/j.cell.2018.03.002
Guo, W., Vandoorne, T., Steyaert, J., Staats, K. A., and Van Den Bosch, L. (2020). The multifaceted role of kinases in amyotrophic lateral sclerosis: genetic, pathological and therapeutic implications. Brain 143, 1651–1673. doi: 10.1093/brain/awaa022
Haeusler, A. R., Donnelly, C. J., Periz, G., Simko, E. A. J., Shaw, P. G., Kim, M.-S., et al. (2014). C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200. doi: 10.1038/nature13124
Hao, Z., Liu, L., Tao, Z., Wang, R., Ren, H., Sun, H., et al. (2019). Motor dysfunction and neurodegeneration in a C9orf72 mouse line expressing poly-PR. Nat. Commun. 10:2906. doi: 10.1038/s41467-019-10956-w
Hartmann, H., Hornburg, D., Czuppa, M., Bader, J., Michaelsen, M., Farny, D., et al. (2018). Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity. Life Sci. Alliance 1:e201800070. doi: 10.26508/lsa.201800070
Hasegawa, M., Arai, T., Nonaka, T., Kametani, F., Yoshida, M., Hashizume, Y., et al. (2008). Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70. doi: 10.1002/ana.21425
Hautbergue, G. M., Castelli, L. M., Ferraiuolo, L., Sanchez-Martinez, A., Cooper-Knock, J., Higginbottom, A., et al. (2017). SRSF1-dependent nuclear export inhibition of C9orf72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nat. Commun. 8:16063. doi: 10.1038/ncomms16063
Hayes, L. R., Duan, L., Bowen, K., Kalab, P., and Rothstein, J. D. (2020). C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. eLife 9:e51685. doi: 10.7554/eLife.51685
Hofweber, M., and Dormann, D. (2019). Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150. doi: 10.1074/jbc.TM118.001189
Hofweber, M., Hutten, S., Bourgeois, B., Spreitzer, E., Niedner-Boblenz, A., Schifferer, M., et al. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706.e13–719.e13. doi: 10.1016/j.cell.2018.03.004
Huang, Y., Gattoni, R., Stévenin, J., and Steitz, J. A. (2003). SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11, 837–843. doi: 10.1016/s1097-2765(03)00089-3
Hughes, M. P., Sawaya, M. R., Boyer, D. R., Goldschmidt, L., Rodriguez, J. A., Cascio, D., et al. (2018). Atomic structures of low-complexity protein segments reveal kinked B sheets that assemble networks. Science 359, 698–701. doi: 10.1126/science.aan6398
Hutten, S., Usluer, S., Bourgeois, B., Simonetti, F., Odeh, H. M., Fare, C. M., et al. (2020). Nuclear import receptors directly bind to arginine-rich dipeptide repeat proteins and suppress their pathological interactions. Cell Rep. 33:108538. doi: 10.1016/j.celrep.2020.108538
Hyman, A. A., Weber, C. A., and Jülicher, F. (2014). Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58. doi: 10.1146/annurev-cellbio-100913-013325
Iarovaia, O. V., Minina, E. P., Sheval, E. V., Onichtchouk, D., Dokudovskaya, S., Razin, S. V., et al. (2019). Nucleolus: a central hub for nuclear functions. Trends Cell Biol. 29, 647–659. doi: 10.1016/j.tcb.2019.04.003
Jackrel, M. E., and Shorter, J. (2014). Reversing deleterious protein aggregation with re-engineered protein disaggregases. Cell Cycle 13, 1379–1383. doi: 10.4161/cc.28709
Jackrel, M. E., Desantis, M. E., Martinez, B. A., Castellano, L. M., Stewart, R. M., Caldwell, K. A., et al. (2014). Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156, 170–182. doi: 10.1016/j.cell.2013.11.047
Jafarinia, H., van der Giessen, E., and Onck, P. R. (2020). Phase separation of toxic dipeptide repeat proteins related to C9orf72 ALS/FTD. Biophys. J. 119, 843–851. doi: 10.1016/j.bpj.2020.07.005
Jain, A., and Vale, R. D. (2017). RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. doi: 10.1038/nature22386
Jiang, J., Zhu, Q., Gendron, T. F., Saberi, S., McAlonis-Downes, M., Seelman, A., et al. (2016). Gain of toxicity from ALS/FTD-linked repeat expansions in C9orf72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90, 535–550. doi: 10.1016/j.neuron.2016.04.006
Jovičić, A., Mertens, J., Boeynaems, S., Bogaert, E., Chai, N., Yamada, S. B., et al. (2015). Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 18, 1226–1229. doi: 10.1038/nn.4085
Kanekura, K., Harada, Y., Fujimoto, M., Yagi, T., Hayamizu, Y., Nagaoka, K., et al. (2018). Characterization of membrane penetration and cytotoxicity of C9orf72-encoding arginine-rich dipeptides. Sci. Rep. 8:12740. doi: 10.1038/s41598-018-31096-z
Kanekura, K., Yagi, T., Cammack, A. J., Mahadevan, J., Kuroda, M., Harms, M. B., et al. (2016). Poly-dipeptides encoded by the C9orf72 repeats block global protein translation. Hum. Mol. Genet. 25, 1803–1813. doi: 10.1093/hmg/ddw052
Kato, M., and McKnight, S. L. (2017). Cross-β polymerization of low complexity sequence domains. Cold Spring Harb. Perspect. Biol. 9:a023598. doi: 10.1101/cshperspect.a023598
Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. doi: 10.1016/j.cell.2012.04.017
Khosravi, B., Hartmann, H., May, S., Möhl, C., Ederle, H., Michaelsen, M., et al. (2017). Cytoplasmic poly-GA aggregates impair nuclear import of TDP-43 in C9orf72 ALS/FTLD. Hum. Mol. Genet. 26, 790–800. doi: 10.1093/hmg/ddw432
Khosravi, B., LaClair, K. D., Riemenschneider, H., Zhou, Q., Frottin, F., Mareljic, N., et al. (2020). Cell-to-cell transmission of C9orf72 poly-(Gly-Ala) triggers key features of ALS/FTD. EMBO J. 39:e102811. doi: 10.15252/embj.2019102811
Kinoshita, Y., Ito, H., Hirano, A., Fujita, K., Wate, R., Nakamura, M., et al. (2009). Nuclear contour irregularity and abnormal transporter protein distribution in anterior horn cells in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 68, 1184–1192. doi: 10.1097/NEN.0b013e3181bc3bec
Klein, I. A., Boija, A., Afeyan, L. K., Hawken, S. W., Fan, M., Dall’Agnese, A., et al. (2020). Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392. doi: 10.1126/science.aaz4427
Kramer, N. J., Carlomagno, Y., Zhang, Y.-J., Almeida, S., Cook, C. N., Gendron, T. F., et al. (2016). Spt4 selectively regulates the expression of C9orf72 sense and antisense mutant transcripts. Science 353, 708–712. doi: 10.1126/science.aaf7791
Kramer, N. J., Haney, M. S., Morgens, D. W., Jovičić, A., Couthouis, J., Li, A., et al. (2018). CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9orf72 dipeptide-repeat-protein toxicity. Nat. Genet. 50, 603–612. doi: 10.1038/s41588-018-0070-7
Kroschwald, S., Maharana, S., Mateju, D., Malinovska, L., Nüske, E., Poser, I., et al. (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4:e06807. doi: 10.7554/eLife.06807
Kwon, I., Xiang, S., Kato, M., Wu, L., Theodoropoulos, P., Wang, T., et al. (2014). Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145. doi: 10.1126/science.1254917
Labokha, A. A., Gradmann, S., Frey, S., Hülsmann, B. B., Urlaub, H., Baldus, M., et al. (2013). Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 32, 204–218. doi: 10.1038/emboj.2012.302
LaClair, K. D., Zhou, Q., Michaelsen, M., Wefers, B., Brill, M. S., Janjic, A., et al. (2020). Congenic expression of poly-GA but not poly-PR in mice triggers selective neuron loss and interferon responses found in C9orf72 ALS. Acta Neuropathol. 140, 121–142. doi: 10.1007/s00401-020-02176-0
Lagier-Tourenne, C., Baughn, M., Rigo, F., Sun, S., Liu, P., Li, H.-R., et al. (2013). Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U S A 110, E4530–E4539. doi: 10.1073/pnas.1318835110
Lamond, A. I., and Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612. doi: 10.1038/nrm1172
Langbehn, D. R., Hayden, M. R., Paulsen, J. S., Johnson, H., Aylward, E., Biglan, K., et al. (2010). CAG-repeat length and the age of onset in Huntington disease (HD): a review and validation study of statistical approaches. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153, 397–408. doi: 10.1002/ajmg.b.30992
Langdon, E. M., and Gladfelter, A. S. (2018). A new lens for RNA localization: liquid-liquid phase separation. Annu. Rev. Microbiol. 72, 255–271. doi: 10.1146/annurev-micro-090817-062814
Lee, K.-H., Zhang, P., Kim, H. J., Mitrea, D. M., Sarkar, M., Freibaum, B. D., et al. (2016). C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774.e17–788.e17. doi: 10.1016/j.cell.2016.10.002
Lee, Y.-B., Chen, H.-J., Peres, J. N., Gomez-Deza, J., Attig, J., Štalekar, M., et al. (2013). Hexanucleotide repeats in ALS/FTD form length-dependent RNA Foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 5, 1178–1186. doi: 10.1016/j.celrep.2013.10.049
Li, H.-Y., Yeh, P. -A., Chiu, H.-C., Tang, C.-Y., and Tu, B. P.-H. (2011). Hyperphosphorylation as a defense mechanism to reduce TDP-43 aggregation. PLoS One 6:e23075. doi: 10.1371/journal.pone.0023075
Lin, Y., Mori, E., Kato, M., Xiang, S., Wu, L., Kwon, I., et al. (2016). Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789.e12–802.e12. doi: 10.1016/j.cell.2016.10.003
Lin, Y., Protter, D. S. W. S. W., Rosen, M. K. K., and Parker, R. (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219. doi: 10.1016/j.molcel.2015.08.018
Ling, S.-C., Polymenidou, M., and Cleveland, D. W. (2013). Converging mechanisms in als and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438. doi: 10.1016/j.neuron.2013.07.033
Liu, C.-R., Chang, C.-R., Chern, Y., Wang, T.-H., Hsieh, W.-C., Shen, W. C., et al. (2012). Spt4 is selectively required for transcription of extended trinucleotide repeats. Cell 148, 690–701. doi: 10.1016/j.cell.2011.12.032
Liu, Q., and Dreyfuss, G. (1996). A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15, 3555–3565. doi: 10.1002/j.1460-2075.1996.tb00725.x
Lopez-Gonzalez, R., Lu, Y., Gendron, T. F., Karydas, A., Tran, H., Yang, D., et al. (2016). Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron 92, 383–391. doi: 10.1016/j.neuron.2016.09.015
Lu, J., Cao, Q., Hughes, M. P., Sawaya, M. R., Boyer, D. R., Cascio, D., et al. (2020). CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid. Nat. Commun. 11:4090. doi: 10.1038/s41467-020-17905-y
Luo, F., Gui, X., Zhou, H., Gu, J., Li, Y., Liu, X., et al. (2018). Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat. Struct. Mol. Biol. 25, 341–346. doi: 10.1038/s41594-018-0050-8
Mackenzie, I. R. A., Frick, P., and Neumann, M. (2014). The neuropathology associated with repeat expansions in the C9orf72 gene. Acta Neuropathol. 127, 347–357. doi: 10.1007/s00401-013-1232-4
Mackenzie, I. R. A., Frick, P., Grässer, F. A., Gendron, T. F., Petrucelli, L., Cashman, N. R., et al. (2015). Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9orf72 mutation carriers. Acta Neuropathol. 130, 845–861. doi: 10.1007/s00401-015-1476-2
Mackenzie, I. R., Arzberger, T., Kremmer, E., Troost, D., Lorenzl, S., Mori, K., et al. (2013). Dipeptide repeat protein pathology in C9orf72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 126, 859–879. doi: 10.1007/s00401-013-1181-y
Mackenzie, I. R., Nicholson, A. M., Sarkar, M., Messing, J., Purice, M. D., Pottier, C., et al. (2017). TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95, 808–816.e9. doi: 10.1016/j.neuron.2017.07.025
Maharana, S., Wang, J., Papadopoulos, D. K., Richter, D., Pozniakovsky, A., Poser, I., et al. (2018). Molecular biology RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921. doi: 10.1126/science.aar7366
Maharjan, N., Künzli, C., Buthey, K., and Saxena, S. (2017). C9orf72 regulates stress granule formation and its deficiency impairs stress granule assembly, hypersensitizing cells to stress. Mol. Neurobiol. 54, 3062–3077. doi: 10.1007/s12035-016-9850-1
Mahboubi, H., Seganathy, E., Kong, D., and Stochaj, U. (2013). Identification of novel stress granule components that are involved in nuclear transport. PLoS One 8:e68356. doi: 10.1371/journal.pone.0068356
Mandrioli, J., Mediani, L., Alberti, S., and Carra, S. (2020). ALS and FTD: where rna metabolism meets protein quality control. Semin. Cell Dev. Biol. 99, 183–192. doi: 10.1016/j.semcdb.2019.06.003
Mann, J. R., Gleixner, A. M., Mauna, J. C., Gomes, E., DeChellis-Marks, M. R., Needham, P. G., et al. (2019). RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e8. doi: 10.1016/j.neuron.2019.01.048
Maor-Nof, M., Shipony, Z., Lopez-Gonzalez, R., Nakayama, L., Zhang, Y.-J., Couthouis, J., et al. (2021). p53 is a central regulator driving neurodegeneration caused by C9orf72 poly(PR). Cell 184, 689–708. doi: 10.1016/j.cell.2020.12.025
May, S., Hornburg, D., Schludi, M. H., Arzberger, T., Rentzsch, K., Schwenk, B. M., et al. (2014). C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 128, 485–503. doi: 10.1007/s00401-014-1329-4
McGurk, L., Mojsilovic-Petrovic, J., Van Deerlin, V. M., Shorter, J., Kalb, R. G., Lee, V. M., et al. (2018a). Nuclear poly(ADP-ribose) activity is a therapeutic target in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 6, 1–15. doi: 10.1186/s40478-018-0586-1
McGurk, L. G. E., Guo, L., Mojsilovic-Petrovic, J., Tran, V., Kalb, R. G., Shorter, J., et al. (2018b). Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9. doi: 10.1016/j.molcel.2018.07.002
McGurk, L., Rifai, O., and Bonini, N. M. (2020). TDP-43 a protein central to amyotrophic lateral sclerosis is destabilized by Tankyrase-1/2. J. Cell Sci. 133:jcs245811. doi: 10.1242/jcs.245811
Meloni, B. P., Brookes, L. M., Clark, V. W., Cross, J. L., Edwards, A. B., Anderton, R. S., et al. (2015). Poly-arginine and arginine-rich peptides are neuroprotective in stroke models. J. Cereb. Blood Flow Metab. 35, 993–1004. doi: 10.1038/jcbfm.2015.11
Meloni, B. P., Craig, A. J., Milech, N., Hopkins, R. M., Watt, P. M., and Knuckey, N. W. (2013). The neuroprotective efficacy of cell-penetrating peptides TAT, penetratin, Arg-9 and Pep-1 in glutamic acid, kainic acid and in vitro ischemia injury models using primary cortical neuronal cultures. Cell. Mol. Neurobiol. 34, 173–181. doi: 10.1007/s10571-013-9999-3
Meloni, B. P., Milani, D., Cross, J. L., Clark, V. W., Edwards, A. B., Anderton, R. S., et al. (2017). assessment of the neuroprotective effects of arginine-rich protamine peptides, poly-arginine peptides (R12-Cyclic, R22) and arginine-tryptophan-containing peptides following in vitro excitotoxicity and/or permanent middle cerebral artery occlusion in rats. NeuroMolecular Med. 19, 271–285. doi: 10.1007/s12017-017-8441-2
Mihevc, S. P., Darovic, S., Kovanda, A., Česnik, A. B., Župunski, V., and Rogelj, B. (2017). Nuclear trafficking in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Brain 140, 13–26. doi: 10.1093/brain/aww197
Mitrea, D. M., and Kriwacki, R. W. (2016). Phase separation in biology; Functional organization of a higher order short linear motifs - The unexplored frontier of the eukaryotic proteome. Cell Commun. Signal. 14:1. doi: 10.1186/s12964-015-0125-7
Mitrea, D. M., Cika, J. A., Stanley, C. B., Nourse, A., Onuchic, P. L., Banerjee, P. R., et al. (2018). Self-interaction of nucleophosmin modulates multiple mechanisms of liquid-liquid phase separation. Nat. Commun. 9, 1–13. doi: 10.1038/s41467-018-03255-3
Mizielinska, S., Gronke, S., Niccoli, T., Ridler, C. E., Clayton, E. L., Devoy, A., et al. (2014). C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194. doi: 10.1126/science.1256800
Mizielinska, S., and Isaacs, A. M. (2014). C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia. Curr. Opin. Neurol. 27, 515–523. doi: 10.1097/WCO.0000000000000130
Mizielinska, S., Lashley, T., Norona, F. E., Clayton, E. L., Ridler, C. E., Fratta, P., et al. (2013). C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 126, 845–857. doi: 10.1007/s00401-013-1200-z
Mizielinska, Sarah., Ridler, C. E., Balendra, R., Thoeng, A., Woodling, N. S., Grässer, F. A., et al. (2017). Bidirectional nucleolar dysfunction in C9orf72 frontotemporal lobar degeneration. Acta Neuropathol. Commun. 5:5. doi: 10.1186/s40478-017-0432-x
Moens, T. G., Niccoli, T., Wilson, K. M., Atilano, M. L., Birsa, N., Gittings, L. M., et al. (2019). C9orf72 arginine-rich dipeptide proteins interact with ribosomal proteins in vivo to induce a toxic translational arrest that is rescued by eIF1A. Acta Neuropathol. 137, 487–500. doi: 10.1007/s00401-018-1946-4
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. doi: 10.1016/j.cell.2015.09.015
Mori, K., Arzberger, T., Grässer, F. A., Gijselinck, I., May, S., Rentzsch, K., et al. (2013a). Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893. doi: 10.1007/s00401-013-1189-3
Mori, K., Lammich, S., Mackenzie, I. R. A., Forné, I., Zilow, S., Kretzschmar, H., et al. (2013b). HnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 125, 413–423. doi: 10.1007/s00401-013-1088-7
Mori, K., Nihei, Y., Arzberger, T., Zhou, Q., Mackenzie, I. R., Hermann, A., et al. (2016). Reduced hn RNPA 3 increases C9orf72 repeat RNA levels and dipeptide-repeat protein deposition. EMBO Rep. 17, 1314–1325. doi: 10.15252/embr.201541724
Morris, G. E. (2008). The Cajal body. Biochim. Biophys. Acta Mol. Cell Res. 1783, 2108–2115. doi: 10.1016/j.bbamcr.2008.07.016
Murakami, T., Qamar, S., Lin, J. Q., Schierle, G. S. K., Rees, E., Miyashita, A., et al. (2015). ALS/FTD mutation-induced phase transition of fus liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690. doi: 10.1016/j.neuron.2015.10.030
Murray, D. T., Kato, M., Lin, Y., Thurber, K. R., Hung, I., McKnight, S. L., et al. (2017). Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16. doi: 10.1016/j.cell.2017.08.048
Murthy, A. C., Dignon, G. L., Kan, Y., Zerze, G. H., Parekh, S. H., Mittal, J., et al. (2019). Molecular interactions underlying liquid−liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 26, 637–648. doi: 10.1038/s41594-019-0250-x
Nanaura, H., Kawamukai, H., Fujiwara, A., Uehara, T., Nakanishi, M., Shiota, T., et al. (2019). Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target Kapβ2 and dysregulate phase separation of low-complexity domains. bioRxiv [Preprint]. doi: 10.1101/812099
Nedelsky, N. B., and Taylor, J. P. (2019). Bridging biophysics and neurology: aberrant phase transitions in neurodegenerative disease. Nat. Rev. Neurol. 15, 272–286. doi: 10.1038/s41582-019-0157-5
Nishimura, A. L., Upunski, V., Troakes, C., Kathe, C., Fratta, P., Howell, M., et al. (2010). Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133, 1763–1771. doi: 10.1093/brain/awq111
O’Rourke, J. G., Bogdanik, L., Muhammad, A. K. M. G., Gendron, T. F., Kim, K. J., Austin, A., et al. (2015). C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron 88, 892–901. doi: 10.1016/j.neuron.2015.10.027
Pak, C. W., Kosno, M., Holehouse, A. S., Padrick, S. B., Mittal, A., Ali, R., et al. (2016). Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 63, 72–85. doi: 10.1016/j.molcel.2016.05.042
Pakravan, D., Orlando, G., Bercier, V., and Van Den Bosch, L. (2020). Role and therapeutic potential of liquid—liquid phase separation in amyotrophic lateral sclerosis. J. Mol. Cell Biol. 25:mjaa049. doi: 10.1093/jmcb/mjaa049
Park, J. H., Chung, C. G., Park, S. S., Lee, D., Kim, K. M., Jeong, Y., et al. (2020). Cytosolic calcium regulates cytoplasmic accumulation of tdp-43 through calpain-a and importin α3. eLife 9, 1–31. doi: 10.7554/eLife.60132
Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. doi: 10.1016/j.cell.2015.07.047
Peeples, W., and Rosen, M. K. (2020). Phase separation can increase enzyme activity by concentration and molecular organization. bioRxiv [Preprint]. doi: 10.1101/2020.09.15.299115
Peran, I., and Mittag, T. (2020). Molecular structure in biomolecular condensates. Curr. Opin. Struct. Biol. 60, 17–26. doi: 10.1016/j.sbi.2019.09.007
Pinarbasi, E. S., Cağatay, T., Fung, H. Y. J., Li, Y. C., Chook, Y. M., and Thomas, P. J. (2018). Active nuclear import and passive nuclear export are the primary determinants of TDP-43 localization. Sci. Rep. 8. doi: 10.1038/s41598-018-25008-4
Premasiri, A. S., Gill, A. L., and Vieira, F. G. (2020). Type I PRMT inhibition protects against C9orf72 arginine-rich dipeptide repeat toxicity. Front. Pharmacol. 11:569661. doi: 10.3389/fphar.2020.569661
Qamar, S., Wang, G. Z., Randle, S. J., Ruggeri, F. S., Varela, J. A., Lin, J. Q., et al. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-Π interactions. Cell 173, 720–734.e15. doi: 10.1016/j.cell.2018.03.056
Quaegebeur, A., Glaria, I., Lashley, T., and Isaacs, A. M. (2020). Soluble and insoluble dipeptide repeat protein measurements in C9orf72-frontotemporal dementia brains show regional differential solubility and correlation of poly-GR with clinical severity. Acta Neuropathol. Commun. 8:8. doi: 10.1186/s40478-020-01036-y
Radwan, M., Ang, C. S., Ormsby, A. R., Cox, D., Daly, J. C., Reid, G. E., et al. (2020). Arginine in C9orf72 dipolypeptides mediates promiscuous proteome binding and multiple modes of toxicity. Mol. Cell. Proteomics. 19, 640–654. doi: 10.1074/mcp.RA119.001888
Ramesh, N., Daley, E. L., Gleixner, A. M., Mann, J. R., Kour, S., Mawrie, D., et al. (2020). RNA dependent suppression of C9orf72 ALS/FTD associated neurodegeneration by Matrin-3. Acta Neuropathol. Commun. 8:177. doi: 10.1186/s40478-020-01060-y
Reddy, K., Zamiri, B., Stanley, S. Y. R., Macgregor, R. B., and Pearson, C. E. (2013). The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 288, 9860–9866. doi: 10.1074/jbc.C113.452532
Renton, A. E., Majounie, E., Waite, A., Simón-Sánchez, J., Rollinson, S., Gibbs, J. R., et al. (2011). A hexanucleotide repeat expansion in C9orf72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268. doi: 10.1016/j.neuron.2011.09.010
Rhine, K., Vidaurre, V., and Myong, S. (2020). RNA droplets. Annu. Rev. Biophys. 49, 247–265. doi: 10.1146/annurev-biophys-052118-115508
Rohrer, J. D., Isaacs, A. M., Mizlienska, S., Mead, S., Lashley, T., Wray, S., et al. (2015). C9orf72 expansions in frontotemporal dementia and amyotrophic lateral sclerosis. Lancet Neurol. 14, 291–301. doi: 10.1016/S1474-4422(14)70233-9
Rossi, S., Rompietti, V., Antonucci, Y., Giovannini, D., Scopa, C., Scaricamazza, S., et al. (2020). UsnRNP trafficking is regulated by stress granules and compromised by mutant ALS proteins. Neurobiol. Dis. 138:104792. doi: 10.1016/j.nbd.2020.104792
Rossi, S., Serrano, A., Gerbino, V., Giorgi, A., Di Francesco, L., Nencini, M., et al. (2015). Nuclear accumulation of mRNAs underlies G4C2-repeat-induced translational repression in a cellular model of C9orf72 ALS. J. Cell Sci. 128, 1787–1799. doi: 10.1242/jcs.165332
Rubbi, C. P., and Milner, J. (2003). Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22, 6068–6077. doi: 10.1093/emboj/cdg579
Ryan, V. H., Dignon, G. L., Zerze, G. H., Chabata, C. V., Silva, R., Conicella, A. E., et al. (2018). Mechanistic view of hnRNPA2 low-complexity domain structure, interactions and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479.e7. doi: 10.1016/j.molcel.2017.12.022
Saberi, S., Stauffer, J. E., Jiang, J., Garcia, S. D., Taylor, A. E., Schulte, D., et al. (2018). Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 135, 459–474. doi: 10.1007/s00401-017-1793-8
Sakae, N., Bieniek, K. F., Zhang, Y. J., Ross, K., Gendron, T. F., Murray, M. E., et al. (2018). Poly-GR dipeptide repeat polymers correlate with neurodegeneration and clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathol. Commun. 6:6. doi: 10.1186/s40478-018-0564-7
Sareen, D., O’Rourke, J. G., Meera, P., Muhammad, A. K. M. G., Grant, S., Simpkinson, M., et al. (2013). Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9orf72 repeat expansion. Sci. Transl. Med. 5:208ra149. doi: 10.1126/scitranslmed.3007529
Scaffidi, P., and Misteli, T. (2005). Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 11, 440–445. doi: 10.1038/nm1204
Schludi, M. H., Becker, L., Garrett, L., Gendron, T. F., Zhou, Q., Schreiber, F., et al. (2017). Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol. 134, 241–254. doi: 10.1007/s00401-017-1711-0
Schmidt, H. B., and Görlich, D. (2015). Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife 4:e04251. doi: 10.7554/eLife.04251
Schmidt, H. B., and Görlich, D. (2016). Transport Selectivity of Nuclear Pores, Phase Separation and Membraneless Organelles. Trends Biochem. Sci. 41, 46–61. doi: 10.1016/j.tibs.2015.11.001
Schmidt, H. B., Barreau, A., and Rohatgi, R. (2019). Phase separation-deficient TDP43 remains functional in splicing. Nat. Commun. 10, 1–14. doi: 10.1038/s41467-019-12740-2
Sellier, C., Campanari, M., Julie Corbier, C., Gaucherot, A., Kolb-Cheynel, I., Oulad-Abdelghani, M., et al. (2016). Loss of C9orf72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 35, 1276–1297. doi: 10.15252/embj.201593350
Shi, K. Y., Mori, E., Nizami, Z. F., Lin, Y., Kato, M., Xiang, S., et al. (2017). Toxic PR n poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl. Acad. Sci. U S A 114, E1111–E1117. doi: 10.1073/pnas.1620293114
Shi, Y., Lin, S., Staats, K. A., Li, Y., Chang, W. H., Hung, S. T., et al. (2018). Haploinsufficiency leads to neurodegeneration in C9orf72 ALS/FTD human induced motor neurons. Nat. Med. 24, 313–325. doi: 10.1038/nm.4490
Shumaker, D. K., Dechat, T., Kohlmaier, A., Adam, S. A., Bozovsky, M. R., Erdos, M. R., et al. (2006). Mutant nuclear lamin a leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U S A 103, 8703–8708. doi: 10.1073/pnas.0602569103
Simone, R., Balendra, R., Moens, T. G., Preza, E., Wilson, K. M., Heslegrave, A., et al. (2018). G-quadruplex-binding small molecules ameliorate C9orf72 FTD/ALS pathology in vitro and in vivo. EMBO Mol. Med. 10, 22–31. doi: 10.15252/emmm.201707850
Sivadasan, R., Hornburg, D., Drepper, C., Frank, N., Jablonka, S., Hansel, A., et al (2016). C9orf72 interaction with cofilin modulates actin dynamics in motor neurons. Nat. Neurosci. 19, 1610–1618. doi: 10.1038/nn.4407
Solomon, D. A., Stepto, A., Au, W. H., Adachi, Y., Diaper, D. C., Hall, R., et al (2018). A feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-α mediates C9orf72-related neurodegeneration. Brain 141, 2908–2924. doi: 10.1093/brain/awy241
Spannl, S., Tereshchenko, M., Mastromarco, G. J., Ihn, S. J., and Lee, H. O. (2019). Biomolecular condensates in neurodegeneration and cancer. Traffic 20, 890–911. doi: 10.1111/tra.12704
Spector, D. L., and Lamond, A. I. (2011). Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3:a000646. doi: 10.1101/cshperspect.a000646
Springhower, C. E., Rosen, M. K., and Chook, Y. M. (2020). Karyopherins and condensates. Curr. Opin. Cell Biol. 64, 112–123. doi: 10.1016/j.ceb.2020.04.003
Stewart, M. (2007). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208. doi: 10.1038/nrm2114
Sudria-Lopez, E., Koppers, M., de Wit, M., van der Meer, C., Westeneng, H. J., Zundel, C. A. C., et al (2016). Full ablation of C9orf72 in mice causes immune system-related pathology and neoplastic events but no motor neuron defects. Acta Neuropathol. 132, 145–147. doi: 10.1007/s00401-016-1581-x
Sullivan, P. M., Zhou, X., Robins, A. M., Paushter, D. H., Kim, D., Smolka, M. B., et al (2016). The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol. Commun. 4:51. doi: 10.1186/s40478-016-0324-5
Sun, Y., Eshov, A., Zhou, J., Isiktas, A. U., and Guo, J. U. (2020a). C9orf72 arginine-rich dipeptide repeats inhibit UPF1-mediated RNA decay via translational repression. Nat. Commun. 11:3354. doi: 10.1038/s41467-020-17129-0
Sun, Y., Zhao, K., Xia, W., Feng, G., Gu, J., Ma, Y., et al (2020b). The nuclear localization sequence mediates hnRNPA1 amyloid fibril formation revealed by cryoEM structure. Nat. Commun. 11:6349. doi: 10.1038/s41467-020-20227-8
Suzuki, H., Shibagaki, Y., Hattori, S., and Matsuoka, M. (2018). The proline-arginine repeat protein linked to C9-ALS/FTD causes neuronal toxicity by inhibiting the DEAD-box RNA helicase-mediated ribosome biogenesis. Cell Death Dis. 9:975. doi: 10.1038/s41419-018-1028-5
Suzuki, H., Shibagaki, Y., Hattori, S., and Matsuoka, M. (2019). C9-ALS/FTD-linked proline-arginine dipeptide repeat protein associates with paraspeckle components and increases paraspeckle formation. Cell Death Dis. 10:746. doi: 10.1038/s41419-019-1983-5
Swinnen, B., Bento-Abreu, A., Gendron, T. F., Boeynaems, S., Bogaert, E., Nuyts, R., et al (2018). A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathol. 135, 427–443. doi: 10.1007/s00401-017-1796-5
Tao, Z., Wang, H., Xia, Q., Li, K., Li, K., Jiang, X., et al. (2015). Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet. 24, 2426–2441. doi: 10.1093/hmg/ddv005
Taylor, J. P., Brown, R. H., and Cleveland, D. W. (2016). Decoding ALS: From genes to mechanism. Nature 539, 197–206. doi: 10.1038/nature20413
Thandapani, P., O’Connor, T. R., Bailey, T. L., and Richard, S. (2013). Defining the RGG/RG motif. Mol. Cell 50, 613–623. doi: 10.1016/j.molcel.2013.05.021
Tripathi, V., Song, D. Y., Zong, X., Shevtsov, S. P., Hearn, S., Fu, X. D., et al. (2012). SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol. Biol. Cell 23, 3694–3706. doi: 10.1091/mbc.E12-03-0206
Tsai, W. C., Gayatri, S., Reineke, L. C., Sbardella, G., Bedford, M. T., and Lloyd, R. E. (2016). Arginine demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 291, 22671–22685. doi: 10.1074/jbc.M116.739573
Ugolino, J., Ji, Y. J., Conchina, K., Chu, J., Nirujogi, R. S., Pandey, A., et al (2016). Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet. 12:e1006443. doi: 10.1371/journal.pgen.1006443
Vanneste, J., Vercruysse, T., Boeynaems, S., Sicart, A., Van Damme, P., Daelemans, D., et al (2019). C9orf72-generated poly-GR and poly-PR do not directly interfere with nucleocytoplasmic transport. Sci. Rep. 9:15728. doi: 10.1038/s41598-019-52035-6
Vatsavayai, S. C., Yoon, S. J., Gardner, R. C., Gendron, T. F., Vargas, J. N. S., Trujillo, A., et al (2016). Timing and significance of pathological features in C9orf72 expansion-associated frontotemporal dementia. Brain 139, 3202–3216. doi: 10.1093/brain/aww250
Vernon, R. M. C., Chong, P. A., Tsang, B., Kim, T. H., Bah, A., Farber, P., et al (2018). Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7:e31486. doi: 10.7554/eLife.31486
Wang, A., Conicella, A. E., Schmidt, H. B., Martin, E. W., Rhoads, S. N., Reeb, A. N., et al (2018a). A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation and RNA splicing. EMBO J. 37. doi: 10.15252/embj.201797452
Wang, J., Choi, J. M., Holehouse, A. S., Lee, H. O., Zhang, X., Jahnel, M., et al (2018b). A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, e16.688–e16.699. doi: 10.1016/j.cell.2018.06.006
Webster, C. P., Smith, E. F., Bauer, C. S., Moller, A., Hautbergue, G. M., Ferraiuolo, L., et al (2016). The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 35, 1656–1676. doi: 10.15252/embj.201694401
Wen, X., Tan, W., Westergard, T., Krishnamurthy, K., Markandaiah, S. S., Shi, Y., et al (2014). Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225. doi: 10.1016/j.neuron.2014.12.010
West, R. J. H., Sharpe, J. L., Voelzmann, A., Munro, A. L., Hahn, I., Baines, R. A., et al (2020). Co-expression of C9orf72 related dipeptide-repeats over 1000 repeat units reveals age-A nd combination-specific phenotypic profiles in Drosophila. Acta Neuropathol. Commun. 8:158. doi: 10.1186/s40478-020-01028-y
Westergard, T., McAvoy, K., Russell, K., Wen, X., Pang, Y., Morris, B., et al (2019). Repeat-associated non- AUG translation in C9orf72- ALS/FTD is driven by neuronal excitation and stress. EMBO Mol. Med. 11:e9423. doi: 10.15252/emmm.201809423
Wheeler, R. J., Lee, H. O., Poser, I., Pal, A., Doeleman, T., Kishigami, S., et al (2019). Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv [Preprint]. doi: 10.1101/721001
Wheeler, R. J. (2020). Therapeutics-how to treat phase separation-associated diseases. Emerg. Top. Life Sci. 11, 331–342. doi: 10.1042/ETLS20190176
White, M. A., Kim, E., Duffy, A., Adalbert, R., Phillips, B. U., Peters, O. M., et al (2018). TDP-43 gains function due to perturbed autoregulation in a Tardbp knock-in mouse model of ALS-FTD. Nat. Neurosci. 21, 552–563. doi: 10.1038/s41593-018-0113-5
White, M. R., Mitrea, D. M., Zhang, P., Stanley, C. B., Cassidy, D. E., Nourse, A., et al (2019). C9orf72 poly(PR) dipeptide repeats disturb biomolecular phase separation and disrupt nucleolar function. Mol. Cell 74, e6.713–e6.728. doi: 10.1016/j.molcel.2019.03.019
Will, C. L., and Lührmann, R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3:a003707. doi: 10.1101/cshperspect.a003707
Woerner, A. C., Frottin, F., Hornburg, D., Feng, L. R., Meissner, F., Patra, M., et al (2016). Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351, 173–176. doi: 10.1126/science.aad2033
Xiao, S., MacNair, L., McGoldrick, P., McKeever, P. M., McLean, J. R., Zhang, M., et al (2015). Isoform-specific antibodies reveal distinct subcellular localizations of C9orf72 in amyotrophic lateral sclerosis. Ann. Neurol. 78, 568–583. doi: 10.1002/ana.24469
Xu, Z., Poidevin, M., Li, X., Li, Y., Shu, L., Nelson, D. L., et al. (2013). Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl. Acad. Sci. U S A 110, 7778–7783. doi: 10.1073/pnas.1219643110
Yamada, S. B., Gendron, T. F., Niccoli, T., Genuth, N. R., Grosely, R., Shi, Y., et al. (2019). RPS25 is required for efficient RAN translation of C9orf72 and other neurodegenerative disease-associated nucleotide repeats. Nat. Neurosci. 22, 1383–1388. doi: 10.1038/s41593-019-0455-7
Yang, Y., and Bedford, M. T. (2013). Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50. doi: 10.1038/nrc3409
Yasuda, K., Clatterbuck-Soper, S. F., Jackrel, M. E., Shorter, J., and Mili, S. (2017). FUS inclusions disrupt RNA localization by sequestering kinesin-1 and inhibiting microtubule detyrosination. J. Cell Biol. 216, 1015–1034. doi: 10.1083/jcb.201608022
Yin, S., Lopez-Gonzalez, R., Kunz, R. C., Gangopadhyay, J., Borufka, C., Gygi, S. P., et al (2017). Evidence that C9orf72 dipeptide repeat proteins associate with U2 snRNP to cause mis-splicing in ALS/FTD patients. Cell Rep. 19, 2244–2256. doi: 10.1016/j.celrep.2017.05.056
Yoshizawa, T., Ali, R., Jiou, J., Fung, H. Y. J., Burke, K. A., Kim, S. J., et al (2018). Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 173, e22.693–e22.705. doi: 10.1016/j.cell.2018.03.003
Yuan, X., Zhou, Y., Casanova, E., Chai, M., Kiss, E., Gröne, H. J., et al (2005). Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest and p53-mediated apoptosis. Mol. Cell 19, 77–87. doi: 10.1016/j.molcel.2005.05.023
Zamiri, B., Reddy, K., Macgregor, R. B., and Pearson, C. E. (2014). TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNAbinding proteins. J. Biol. Chem. 289, 4653–4659. doi: 10.1074/jbc.C113.502336
Zhang, K., Daigle, J. G., Cunningham, K. M., Coyne, A. N., Ruan, K., Grima, J. C., et al (2018a). Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, e17.958–e17.971. doi: 10.1016/j.cell.2018.03.025
Zhang, K., Donnelly, C. J., Haeusler, A. R., Grima, J. C., Machamer, J. B., Steinwald, P., et al (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61. doi: 10.1038/nature14973
Zhang, Y. J., Gendron, T. F., Ebbert, M. T. W., O’Raw, A. D., Yue, M., Jansen-West, K., et al (2018b). Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 24, 1136–1142. doi: 10.1038/s41591-018-0071-1
Zhang, Y. J., Guo, L., Gonzales, P. K., Gendron, T. F., Wu, Y., Jansen-West, K., et al (2019a). Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 363:eaav2606. doi: 10.1126/science.aav2606
Zhang, Y. J., Jansen-West, K., Xu, Y. F., Gendron, T. F., Bieniek, K. F., Lin, W. L., et al (2014). Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 128, 505–524. doi: 10.1007/s00401-014-1336-5
Zhang, Y., Yang, M., Duncan, S., Yang, X., Abdelhamid, M. A. S., Huang, L., et al (2019b). G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res. 47, 11746–11754. doi: 10.1093/nar/gkz978
Zhou, B., Liu, C., Geng, Y., and Zhu, G. (2015). Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci. Rep. 5:16673. doi: 10.1038/srep16673
Zhou, Q., Mareljic, N., Michaelsen, M., Parhizkar, S., Heindl, S., Nuscher, B., et al. (2020). Active poly-GA vaccination prevents microglia activation and motor deficits in a C9orf72 mouse model. EMBO Mol. Med. 12:e10919. doi: 10.15252/emmm.201910919
Zhu, Q., Jiang, J., Gendron, T. F., McAlonis-Downes, M., Jiang, L., Taylor, A., et al (2020). Reduced C9orf72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat. Neurosci. 23, 615–624. doi: 10.1038/s41593-020-0619-5
Keywords: phase separation, C9orf72, membraneless organelles, TDP-43, therapeutics
Citation: Solomon DA, Smikle R, Reid MJ and Mizielinska S (2021) Altered Phase Separation and Cellular Impact in C9orf72-Linked ALS/FTD. Front. Cell. Neurosci. 15:664151. doi: 10.3389/fncel.2021.664151
Received: 04 February 2021; Accepted: 19 March 2021;
Published: 21 April 2021.
Edited by:
Annakaisa Haapasalo, University of Eastern Finland, FinlandReviewed by:
Isabella Zanella, University of Brescia, ItalyCopyright © 2021 Solomon, Smikle, Reid and Mizielinska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah Mizielinska,c2FyYWgubWl6aWVsaW5za2FAa2NsLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.