
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 31 March 2021
Sec. Non-Neuronal Cells
Volume 15 - 2021 | https://doi.org/10.3389/fncel.2021.660523
This article is part of the Research Topic Non-neuronal Cell Heterogeneity in the Nervous System During Health and Disease View all 10 articles
Microglia are the resident immune effector cells of the central nervous system (CNS) rapidly reacting to any perturbation in order to maintain CNS homeostasis. Although their outstanding reactive properties have been elucidated over the last decades, their heterogeneity in healthy tissue, such as across brain regions, as well as their diversity in the development and progression of brain diseases, are currently opening new avenues to understand the cellular and functional states of microglia subsets in a context-dependent manner. Here, we review the main breakthrough studies that helped in elucidating microglia heterogeneity in the healthy and diseased brain and might pave the way to critical functional screenings of the inferred cellular diversity. We suggest that unraveling the cellular and molecular mechanisms underlying specific functionalities of microglial subpopulations, which may ultimately support or harm the neuronal network in neurodegenerative diseases, or may acquire pro- or anti-tumorigenic phenotypes in brain tumors, will possibly uncover new therapeutic avenues for to date non-curable neurological disorders.
Microglial cells are the innate immune cells of the brain and key players in maintaining the homeostasis of the central nervous system (CNS) (Sierra et al., 2016). Microglia originate from erythro-myeloid precursors in the yolk sac and migrate to the brain around embryonic day 9.5 in mouse (Ginhoux et al., 2010), while they colonize the human cerebrum between the 4th and 24th week of gestation (Menassa and Gomez-Nicola, 2018). Their ontogeny, together with their slow turnover, which differentiate them from most other hematopoietic lineages in adult individuals (Réu et al., 2017), as well as the local environment in the CNS, make microglia a distinct immune cell population (Sousa et al., 2017). Until approximately 20 years ago, microglia have been considered as a resident resting cell type of the healthy CNS able to react to pathogens or toxic elements. However, this paradigm has shifted into the concept of “surveillant” and “supporting” microglia exerting additional multiple functional roles, such as neuromodulation and phagocytosis (Gomez-Nicola and Perry, 2015). For example, during development microglia contribute to building the neuronal circuit through synaptic pruning and stripping, phagocytosis of dying neurons and secretion of neurotrophic factors (Paolicelli et al., 2011; Ekdahl, 2012; Schafer et al., 2012; Paolicelli and Ferretti, 2017; Scott-Hewitt et al., 2020). Further, it has recently been shown that an ATP-dependent microglia-driven negative feedback mechanism operates similarly to inhibitory neurons and is essential for protecting the brain from an excessive activation (Badimon et al., 2020). Taken together, due to their multiple critical functional roles in the homeostatic brain, various neurological disorders, including neurodegenerative diseases and brain tumors, implicate microglia. Briefly, in Parkinson’s disease (PD) and Alzheimer’s disease (AD), a mix of beneficial and detrimental roles of microglia have been suggested (Wyss-Coray and Mucke, 2002; Glass et al., 2010; Bodea et al., 2014; Joers et al., 2017; Salter and Stevens, 2017; Wolf et al., 2017; Duffy et al., 2018). For example, in AD microglial cells have been associated with the phagocytosis and degradation of amyloid-ß plaques, but the subsequent excessive release of cytokines is supposed to contribute to neuronal loss (Salter and Stevens, 2017). Similarly, in PD, where microglia are able to recognize and engulf alpha-synuclein, but the concomitant release of reactive oxygen species (ROS) or pro-inflammatory mediators can actively contribute to neurodegeneration (Glass et al., 2010). In brain tumors, microglia, along with tumor-infiltrating macrophages, constitute the predominant immunological cell types (Graeber et al., 2002) and have been shown to affect tumor progression as well as patient survival (Morimura et al., 1990; Gieryng et al., 2017; Sorensen et al., 2018). Indeed, tumor-associated microglia/macrophages (TAMs) are key players along tumor development by contributing to the establishment of a tumor-supporting microenvironment (Hambardzumyan et al., 2016; Grabowski et al., 2020; Maas et al., 2020).
In this context, the hypothesis that several microglial cell subsets exist in the brain has gain momentum in the recent years and microglial heterogeneity has been addressed from different points of views, including morphology, cellular density, proliferation capacity as well as transcriptional and proteomic signatures (De Biase and Bonci, 2018; Silvin and Ginhoux, 2018). Additionally, significant advances have been made by taking advantage of the recently developed single-cell technologies, including RNA-sequencing and mass cytometry (CyTOF). Indeed, several studies using these approaches have now confirmed that microglial cells represent a complex population constituted by different subsets, both in the healthy and diseased brain, displaying specific neuroimmunological adaptations in a context-dependent manner (Stratoulias et al., 2019; Masuda et al., 2020; Provenzano et al., 2020).
Here, we will review the main breakthrough studies that helped to elucidate microglia heterogeneity in the healthy and diseased brain. We suggest that unraveling the cellular and molecular mechanisms underlying specific microglia subpopulations might contribute to uncover new therapeutic targets for brain disorders with an immunological component, including neurodegenerative diseases and tumors.
An extensive characterization of microglial heterogeneity encompassing fundamental aspects, including development, gender specificities and spatial distribution has been conducted in the healthy brain. Hence, along this chapter, we chronologically review the main studies that contributed to acquire the current knowledge of microglia diversity under homeostatic conditions (Figures 1, 2).
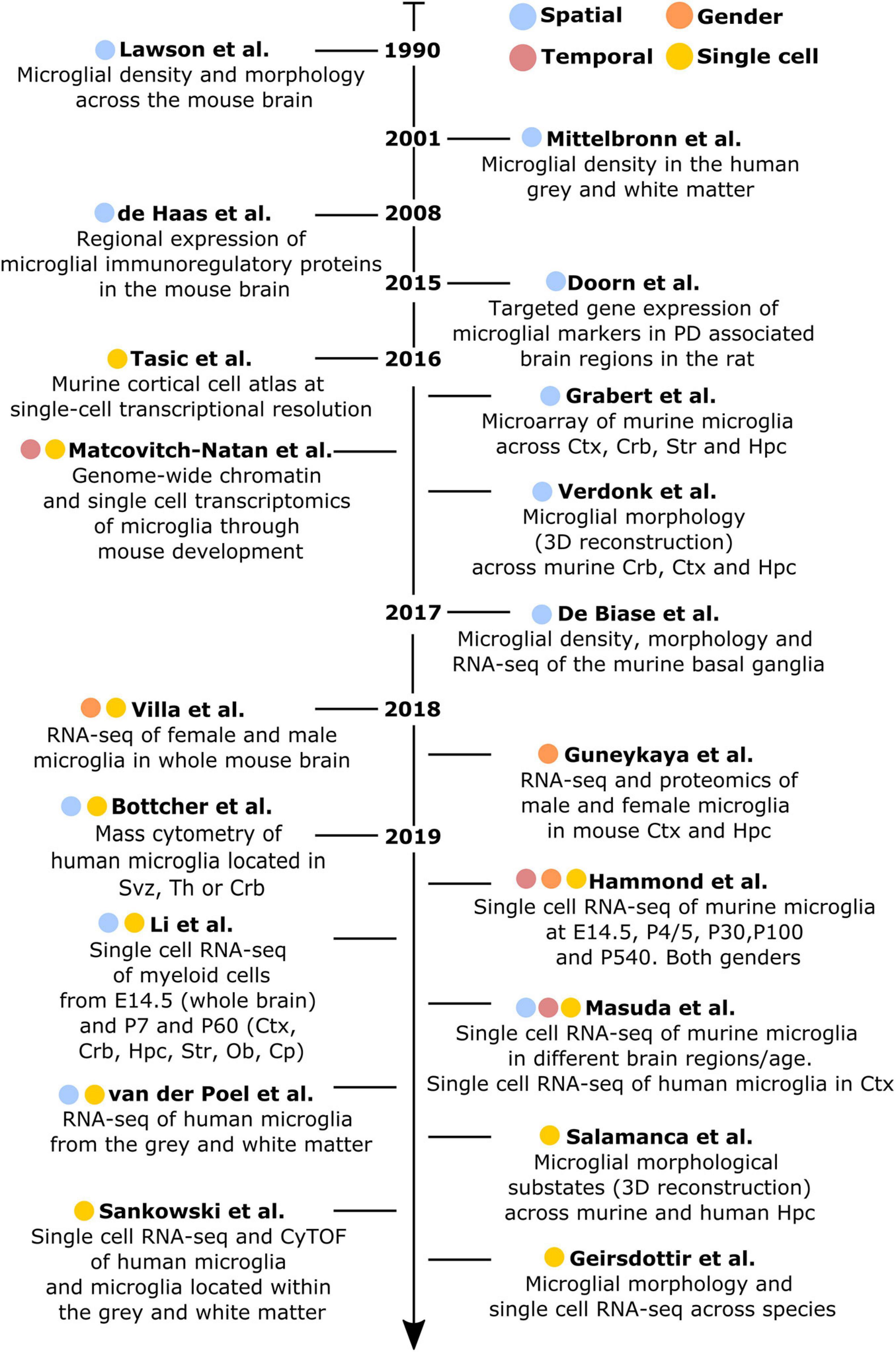
Figure 1. Chronologically ordered list of the main studies that have contributed to elucidate microglial diversity in the healthy brain. Studies are color-coded based on their corresponding addressed topics: spatial (blue), temporal (red), gender (orange), or single-cell resolution (yellow). Ctx, cortex; Crb, cerebellum; Str, striatum; Hpc, hippocampus; Svz, subventricular zone; Th, thalamus; Ob, olfactory bulb; Cp, corpus callosum.
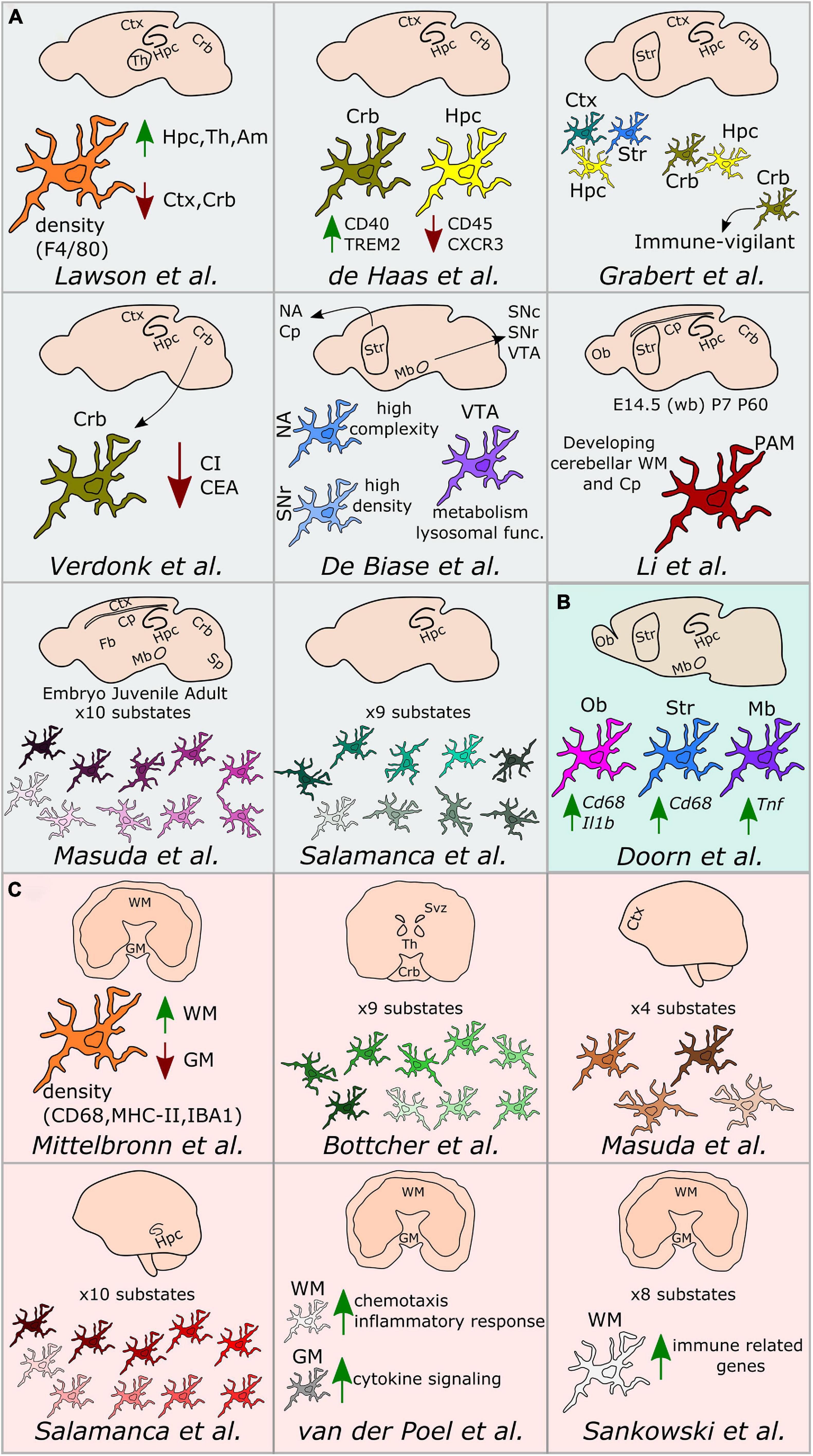
Figure 2. Regional microglial heterogeneity in the healthy brain. Schematic representation of the main findings over the main studies addressing microglial heterogeneity across mouse (A), rat (B), and human (C) brain regions. Ctx, cortex; Crb, cerebellum; Str, striatum; Hpc, hippocampus; Svz, subventricular zone; Th, thalamus; Ob, olfactory bulb; Cp, corpus callosum; Mb, midbrain; NA, nucleus accumbens; VTA, ventral tegmental area; SNc, substantia nigra pars compacta; SNr, substantia nigra pars reticulata; Sp, spinal cord; Fb, forebrain; WM, white matter; GM, gray matter; PAM, proliferative-region-associated microglia; WB, whole brain; CI, complexity index; CEA, covered environment area.
Lawson et al. (1990) have conducted the first study addressing microglial heterogeneity in the 90’s. The authors showed that, in the mouse brain, microglial ramifications and cell shapes were region-dependent. By using the macrophage marker F4/80, they also reported divergent microglial densities across specific brain regions, with higher density in the hippocampus, thalamus and amygdala compared to the cortex and cerebellum (Lawson et al., 1990). Concomitantly to this study in mice, Mittelbronn et al. (2001) studied microglial density across human brain regions using myeloid-specific-immunological markers, including CD68, MHC-II, and IBA1. In this study, the authors described a higher content of microglial cells within the white-matter when compared to gray-matter (Mittelbronn et al., 2001). Seven years later, de Haas et al. (2008) took advantage of ex vivo flow cytometry analyses to investigate regional differences in the expression of immunoregulatory proteins across the mouse spinal cord and various brain regions, such as the cerebral cortex, hippocampus, cerebellum or striatum. Interestingly, the authors described that CD40 was overexpressed by microglia located in the cerebellum when compared to cortical microglia. Similarly, CD45 or CXCR3 were described to be less expressed in the hippocampus in comparison to the other regions, while TREM2 was differentially expressed between microglia in the cerebellum and cortex (de Haas et al., 2008). Later on, Doorn et al. (2015) investigated baseline differences in microglial expression of genes in brain regions associated with PD, including substantia nigra, striatum, olfactory bulb, hippocampus, or amygdala of rats. The authors did not detect differences in the expression levels of Aif1, Cd11b, or Tlr2 genes in microglia isolated from those regions. However, the expression levels of the phagocytic and pro-inflammatory markers Cd68 and Il1b were higher in the olfactory bulb compared to the other brain regions. Besides, Cd68 expression was higher in the striatum compared to the amygdala and Tnf was overexpressed in the substantia nigra (Doorn et al., 2015).
Along with a deeper understanding of microglial heterogeneity across brain regions, the raise of single-cell transcriptomic technologies has been a breakthrough toward further revealing the cellular diversity of the brain at single-cell resolution. In this context, a pioneer work from Tasic et al. (2016) enabled to build a cell taxonomy atlas of the murine cortex identifying up to 49 transcriptomic cell types, 7 of them being non-neuronal cell types, including microglia. In the same year, two key studies helped to understand microglia diversity across brain regions, although not at single-cell resolution yet. More specifically, Grabert et al. (2016) conducted a large RNA-sequencing analysis of isolated microglial cells from the mouse cortex, hippocampus, striatum, and cerebellum. This study demonstrated that the most variable gene ontology terms discriminating the analyzed brain regions were related to metabolism and immune regulation. Besides, microglial transcriptomic heterogeneity clustered into three different signatures: the “cortex, hippocampus, and striatum,” the “cerebellum and hippocampus,” and the “cerebellum” transcriptomes. Importantly, the authors showed that microglia from the cerebellum exhibit a specific “immune-vigilant” transcriptional signature when compared to the others brain regions (Grabert et al., 2016). In addition, Verdonk et al. (2016) expanded the knowledge on regional heterogeneity by shedding light on microglia morphology using an automated method based on 3D reconstruction. With this technique, microglial morphologies have been analyzed according to the complexity of primary ramifications (CI, complexity index) and the total 2D area covered by ramifications (CEA, covered environment area). In line with the transcriptomic findings obtained by Grabert et al. (2016), microglial morphology from the cerebellum was the most diverse. Indeed, cerebellar microglia had smaller CI and CEA in comparison to microglia from the hippocampus, frontal cortex and striatum, which exhibit similar cell body and cellular area (Verdonk et al., 2016). Still in 2016, the first single cell study addressing microglia diversity along development was published by Matcovitch-Natan et al. (2016) who specifically defined early-stage cycling (e.g., Dab2, Mcm5, Lyz2), synaptic pruning (e.g., Crybb1, Csf1, Cxcr2), and adult immune surveillant (e.g., MafB, Cd14, Mef2a) microglia.
It was in 2017 when De Biase et al. (2017) investigated microglia diversity across different areas of the basal ganglia, including the nucleus accumbens, ventral tegmental area, substantia nigra pars compacta, and substantia nigra pars reticulata, using a combination of morphological and transcriptomic analyses. The authors observed that microglia in the striatum and the nucleus accumbens displayed a higher complexity when compared to the others sub-regions. Further, they reported variability in terms of microglial density within the basal ganglia, detecting a higher number of microglial cells within the substantia nigra pars reticulata when compared with the substantia nigra pars compacta and the ventral tegmental area. Interestingly, they also reported a uniform density of all the other cell types within the basal ganglia, interrogating the origin of the region-specific microglial proliferation capacities. Lastly, they described different microglial transcriptional signatures, especially within the ventral tegmental area, where genes related to mitochondrial function, metabolism, oxidative signaling, or lysosomal function were differentially expressed when compared to the other regions (De Biase et al., 2017).
In 2018, two major studies addressing sexual dichotomy at the transcriptional level have been conducted. In both cases, biological processes related to the immune phenotype (e.g., cytokine production, expression of antigen-presenting cell markers and purinergic receptors) were over-represented in male microglia (Guneykaya et al., 2018; Villa et al., 2018). Further, female microglia have been linked to neuronal processes (e.g., perpetuation of neuronal transmission or promotion of neuroprotective mechanisms), as well as have been described to be more susceptible to microbiota modifications (Villa et al., 2019).
Doubtless, 2019 has been the year when microglial heterogeneity has been mostly elucidated at single-cell resolution. For example, Bottcher et al. (2019), by using mass-cytometry, described nine different human microglial substates across the subventricular zone, thalamus, cerebellum, and the temporal and lateral lobes, with the subventricular zone displaying a more distinct signature compared with the other brain regions. In mice, Hammond et al. (2019), by using single cell technology, found high microglial heterogeneity in young mice (E14.5 and P5), with eight different substates of microglia, for which the expression of markers such as Arg1, Rrm2, Hmox1, or Spp1 differed. In addition, the authors aimed to identify gender-specific differences of microglia across three different ages (E14.5, P4/P5, and P100), but did not detect main differences between males and females, except for the expression of chromosome-specific genes, such as Eif2s3y and Xist (Hammond et al., 2019). In addition to the study of Hammond et al. (2019), Li et al. (2019) described the so-called proliferative-region-associated microglia (PAM), a specific microglial subtype located in the developing cerebellar white matter and corpus callosum, which is characterized by the expression of Spp1 and Gpnmb. Lastly, Masuda et al. (2019) conducted a more detailed single-cell study, mainly focused on microglia, in which their heterogeneity has been addressed across specific brain regions, such as the corpus callosum, cerebellum, cortex, hippocampus, and facial nucleus at different mouse ages. The authors identified 10 main substates during development with differences in the expression of microglial markers, such as Tmem119, Malat1, Lamp1, or Apoe (Masuda et al., 2019). Notably, in the same study, four different substates of microglia have also been described in the cortex of the human brain (Masuda et al., 2019). In parallel, microglial diversity between the gray and white matter has been studied at the transcriptomic level in the human brain (van der Poel et al., 2019). While microglia located in the gray matter were enriched in genes related to cytokine signaling, microglia in the white matter were characterized by genes involved in chemotaxis and inflammatory responses (van der Poel et al., 2019). In the same year, an automated method named MIC-MAC (Microglia and Immune Cells Morphologies Analyzer and Classifier) enabled to evaluate microglial density and morphologies at single-cell level in the hippocampi of human and mouse brains. This method allowed the clustering of microglial subpopulations based on their similarities in a 3D environment and assisted the identification of 10 different microglial subsets within the mouse and the human hippocampus, revealing a unique subset of human microglia (Salamanca et al., 2019). Still regarding microglia diversity in the human brain, Sankowski et al. (2019) recently applied high dimensional techniques and identified up to eight clusters with differential expression of microglia core genes (e.g., CX3CR1 and TMEM119), genes related to major histocompatibility complex II (e.g., HLA-DRA and CD74) as well as chemokines and cytokines (e.g., CCL2 and IL1B). In the same study, the authors explored regional-associated differences in the temporal lobe between the gray and white matter and detected higher expression levels of immune genes in microglia located in the latter (Sankowski et al., 2019). Later in 2019, Geirsdottir et al. (2019) studied microglia diversity across a various range of species across the evolutionary tree, including mouse and human. The authors used a combination of single cell transcriptomics and 3D reconstruction to study microglia morphology. In this particular study, the authors showed a conserved morphological pattern of parenchymal microglial cells across the species. However, they called attention on variations observed in terms of dendrite length, number of segments, branch points, and terminal points between the cortical and cerebellum microglia in mice and humans. From the transcriptomic point of view, microglia express a set of core genes that are conserved across species, with human microglia displaying a more pronounced heterogeneity than other mammals. Lastly, microglial genes related to the complement pathway, phagocytosis, or genes implicated in neurodegenerative diseases differ between rodents and primates (Geirsdottir et al., 2019).
Microglial cells scan the brain parenchyma and react to specific threats to avoid a disturbance of the critical, fine-tuned activities of the CNS. As mentioned previously, the phenotypic analysis of microglia in the healthy brain parenchyma revealed specific poised subsets, which might eventually support or harm the neuronal network under specific vulnerabilities. For example, this is all the more important for the understanding of CNS disorders exhibiting regional-specific and cellular pathological hallmarks, as seen in various neurodegenerative diseases, including AD (entorhinal cortex and hippocampus) and PD (nigrostriatal pathway). Therefore, in this chapter we extend the study of microglial heterogeneity in a disease-associated context. More specifically, we tackle microglia diversity associated with neuroinflammatory and neurodegenerative diseases as well as with brain tumors (Figure 3).
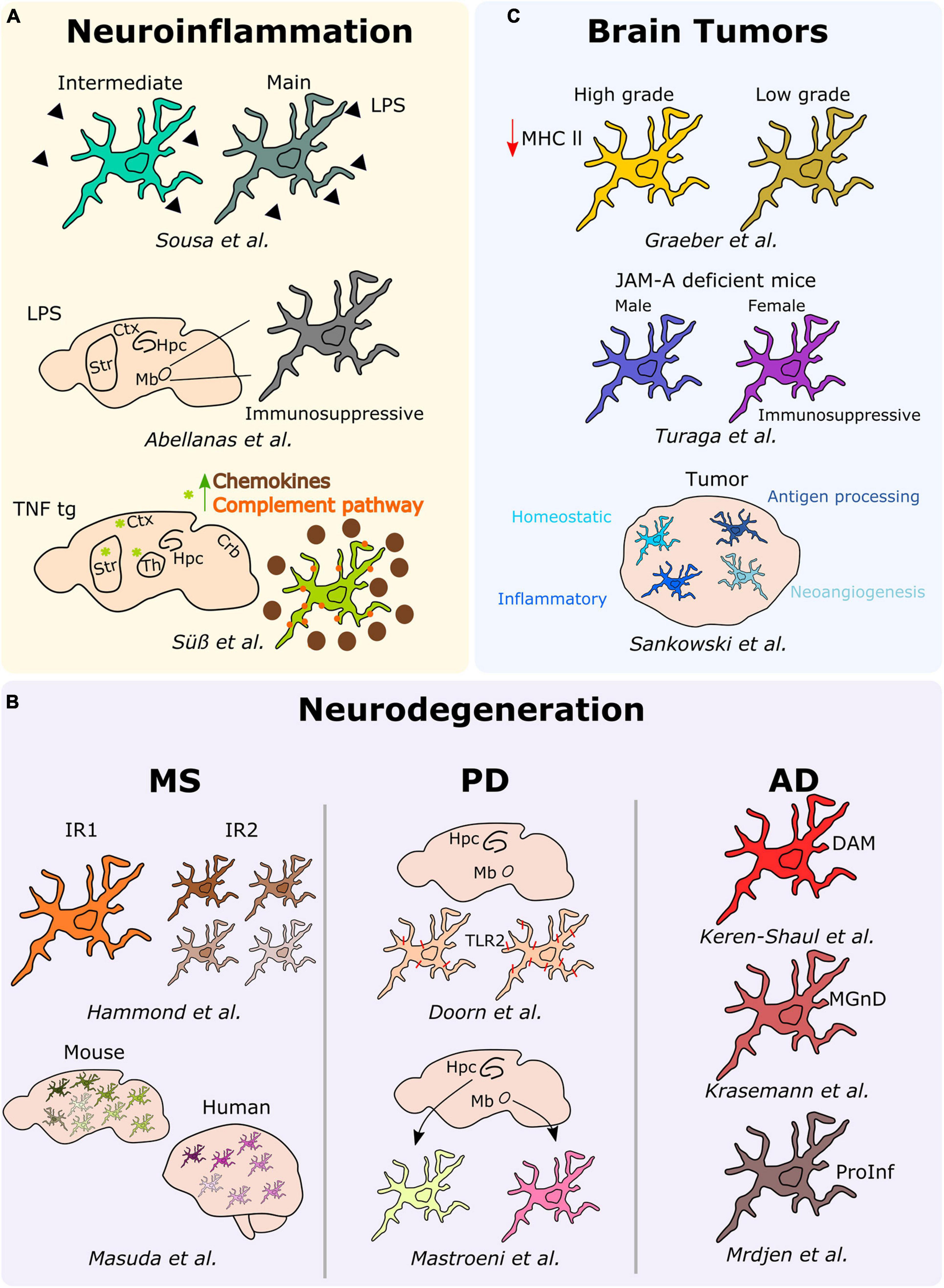
Figure 3. Microglial heterogeneity in different neurological diseases. Schematic description of the main results over the main studies addressing microglia heterogeneity in neuroinflammation, neurodegeneration (MS, PD, and AD), and brain tumors. Hpc, hippocampus; Mb, midbrain; Ctx, cortex; Str, striatum; Th, thalamus; DAM, disease-associated microglia; MGnD, microglial neurodegenerative phenotype; ProInf, pro-inflammatory; IR1, injury responsive 1; IR2, injury responsive 2.
In an attempt to elucidate potential heterogeneous responses of microglia under neuroinflammatory conditions, we analyzed ex vivo pre-sorted cells from the mouse brain following a peripheral acute endotoxin challenge, a common model used as a paradigm to study the effect of systemic bacterial infections, ultimately leading or not to neurodegeneration (Bodea et al., 2014). By applying single-cell RNA-sequencing, we demonstrated that the microglial response associated with a peripheral injection of lipopolysaccharide (LPS), most probably induced by a transient serum cytokine storm, rather than stimulated by the response to the TLR ligand that might not reach the brain parenchyma (Banks and Robinson, 2010; Shemer et al., 2020) is heterogeneous. Specifically, we identified two discrete reactive states characterized by various levels of activation and showed that inflammation-induced microglia signatures are distinct from neurodegenerative disease-associated profiles (Sousa et al., 2018). Notably, a similar study conducted a year later showed that microglia isolated from the midbrain of peripherally LPS injected mice adopt an immunosuppressive phenotype in comparison to microglia located within the striatum (Abellanas et al., 2019), thus suggesting that distinct microglia reactions toward neuroinflammatory threats might be region-dependent. In this context, the analysis of microglia phenotypes associated with chronic peripheral inflammation in TNF transgenic mice revealed distinct signatures across different brain regions, including the cortex, striatum, hippocampus, thalamus, and cerebellum. Indeed, microglia located in the cortex, striatum, and thalamus of the transgenic mice clustered together, and their transcriptome significantly differed from the other brain regions. More specifically, microglial cells located within the cortex, striatum and thalamus were characterized by the overexpression of inflammatory genes, such as Cxcl13, Ccl2, C3, and C4b, thus suggesting a more pronounced reactive state of microglia under persistent inflammation in these specific regions (Süß et al., 2020).
In the context of neuroinflammatory diseases, multiple sclerosis (MS) is the most common inflammatory, demyelinating and neurodegenerative disorder of the CNS. In this perspective, Hammond et al. (2019) studied microglia diversity in the white matter of mice injected with lysolecithin (LPC), a commonly used mouse model of MS. In this model, microglial cells initially segregated into two clusters, with the so-called injury responsive 1 (IR1) cluster mainly composed by microglial cells of the control group, whereas the IR2 cluster constituted by microglia from LCP injured mice. Further, the authors revealed that IR2 subset was composed by four sub-clusters representing different microglial subtypes or responses in LCP mice differing in the expression levels of proliferative (e.g., Birc5) and inflammatory (e.g., Cxcl10 or Ccl4) markers, suggesting that microglia acquire different phenotypes to respond to demyelination (Hammond et al., 2019). Similarly, Masuda et al. (2019) investigated microglial heterogeneity in a MS and neurodegeneration-associated mouse model, respectively the cuprizone and the unilateral facial nerve axotomy models. In this study, the authors describe up to nine different subtypes of microglia displaying differences in the expression levels of inflammatory (e.g., Spp1, Ccl4, Cybb) or MHC-II-related (e.g., Cd74, H2-Aa, H2-Ab1) markers among others (Masuda et al., 2019). Remarkably, a similar microglial heterogeneity has been confirmed in MS patients. More specifically, microglial cells segregated into seven different subtypes, with differential expression levels of chemokines and cytokines (CCL4 or ERG2), MHC-II-related proteins (CD74 or HLA-DRA), and activation markers (SPP1 or CTSD) (Masuda et al., 2019).
In the context of neurodegenerative diseases, Doorn et al. (2014) took advantage of immunohistochemical analyses to examine the expression of TLR2, a known receptor involved in the activation of microglia following its interaction with alpha-synuclein in the substantia nigra and hippocampus of patients with PD and incidental Lewy Body Disease (iLBD), a prodromal state of PD. In iLBD patients, TLR2 microglia expression regionally differed between the substantia nigra and hippocampus. Additionally, Mastroeni et al. (2018) studied the human regional microglial profile associated with PD and AD. More specifically, they conducted RNA-sequencing of isolated microglia from the two most vulnerable brain regions affected in PD and AD, the substantia nigra and the hippocampus CA1, respectively (Mastroeni et al., 2018). They uncovered regional differences highlighting 313 differentially expressed genes between microglia located within the substantia nigra of PD samples and the corresponding cells located in the hippocampus CA1. These differential expressed genes reflected changes in behavior, synaptic transmission or regulation of transport. In the AD samples, 104 differential expressed genes associated with synaptic transmission, cell-cell signaling, or metal ion transport have been detected between microglia located in the hippocampus CA1 and substantia nigra (Mastroeni et al., 2018). In a similar context, Keren-Shaul et al. (2017) used a classical mouse model of AD, the 5XFAD, to study microglial subsets associated with AD at the single-cell level. Notably, the authors depicted a specific population of microglial cells associated with AD, namely disease-associated microglia (DAM). They uncovered that the Trem2 associated pathway, which confers them a higher phagocytic capacity, drives the acquisition of the DAM phenotype. This specific population is supposed to be located around the amyloid-ß plaques and has been confirmed to be also present in the brains of AD patients (Keren-Shaul et al., 2017). Consistently, a transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases, termed “microglial neurodegenerative phenotype” (MGnD), driven by the TREM2-APOE pathway, has been concomitantly described (Krasemann et al., 2017). Further, Mrdjen et al. (2018) took advantage of the CyTOF to identify a microglial subtype in another mouse model of AD, the APP/PS1. Indeed, they identified a subset of microglial cells characterized by the overexpression of phagocytic (e.g., Cd11c and Cd14), activation (e.g., Cd86 and Cd44) as well as MHC-II-associated markers. In this subset, in line with a pro-inflammatory phenotype, the expression levels of the homeostatic microglia markers (e.g., Cx3cr1 or Siglec-H) were downregulated (Mrdjen et al., 2018).
In the past, inter-tumor microglial morphological heterogeneity has been described, for example, across different gliomas, where microglia display a more pronounced amoeboid morphology in high-grade tumors, while they are ramified in low-grade tumors. These differences were also associated with lower levels of MHC-II expression in high-grade gliomas when compared to their low-grade counterparts (Graeber et al., 2002). Interestingly, microglia diversity associated with brain tumors, and specifically in GBM, has been investigated also taking into account gender specificities. Turaga et al. (2020), while conducting a study regarding the implication of junctional adhesion molecule-A (JAM-A), noticed a poorer prognosis in GBM implanted female JAM-A deficient mice when compared to the corresponding implanted males. Notably, the authors reported an upregulation of the anti-inflammatory genes Fizz1 and Ifi202b in microglia from female JAM-A deficient mice compared to their male counterparts (Turaga et al., 2020). Sankowski et al. (2019), by combining scRNA-seq and CyTOF analyses on human brain samples, including samples obtained from GBM patients, discovered various subpopulations of microglial cells within the tumor. Indeed, they defined a continuum from control-enriched clusters to glioma-associated microglial clusters, the latest being characterized by a decreased expression of core microglial signature genes (CX3CR1, SELPLG, P2RY12, CSFR1) and an increased expression of metabolic, inflammatory and interferon-associated genes (CD163, APOE, LPL, IFI27, IFI44, SPP1). Interestingly, the in-between clusters were exhibiting differential expression levels of the previous cited genes, but also hypoxia-related (VEGFA, HIF1A) and antigen processing MHCI-related genes. Additionally, CyTOF analyses enabled the detection of differential proportions of HLA-DR, TREM2, APOE, and GPR56, confirming major differences between control and glioma-associated microglia also at the protein level (Sankowski et al., 2019). Although only poorly investigated to date, also secondary brain metastases can alter microglial properties, or—vice versa—microglia may even pave the way for an enhanced cerebral dissemination of peripheral tumor cells via increased secretion of IGF-1 and CCL20 together with a reduced expression of SIRP-alpha, the latter leading to impaired phagocytic properties (Simon et al., 2020; Wu et al., 2020). Very recently, the first experimental studies addressing the composition of primary and secondary myeloid cell populations at single-cell level have been performed in mouse models indicating that the genetic programming of brain metastasis-associated myeloid cells is a very early and stable event (Schulz et al., 2020).
Undoubtedly, the ability to acquire different resting and activated phenotypes confers microglia the advantage to be a plastic and adaptive cell type in the CNS. Along with this review, we have highlighted a large number of studies demonstrating that microglia are far from being a resting or homogenous cell population. For example, in the healthy mouse brain, microglia heterogeneity across various regions has been described in terms of density, morphology, molecular signatures, and metabolism (Tan et al., 2020). For translational purposes, it will be critical to consider that microglia heterogeneity has been suggested to be even higher in the human brain compared to mouse (Prinz et al., 2019). Notably, regarding neurodegenerative diseases little is known about microglial heterogeneity in PD. Hence, future efforts will need to be directed at understanding if specific microglia subsets might differently contribute to PD pathology. In the context of brain tumors, with the difficulty to define reliable markers deciphering macrophages and microglia, the majority of the transcriptomic studies have been focusing on defining a glioma-associated microglial signature in different models, but not assessing their heterogeneity within the tumor mass (Darmanis et al., 2017; Walentynowicz et al., 2018; Maas et al., 2020). The study by Sankowski et al. (2019), being the first one highlighting specific heterogeneous signatures of microglial cells in GBM, calls attention on the lack of knowledge on microglial heterogeneity in that context. Hence, further studies would need to be directed at understanding the implication of different microglial subsets in glioma development and progression (Sankowski et al., 2019). Similarly detrimental, however, much more frequently than primary brain tumors, also brain metastases, at least experimentally, show a relevant contribution of microglia in the establishment and progression of secondary brain tumors, therefore constituting a target for future treatment strategies.
Taken together, microglial variety embraces fundamental aspects, such as spatial-temporal organization, which is present in the healthy and diseased brain. Thanks to the development of high-throughput technologies, including single-cell approaches, different microglial subsets have been unraveled, indicating that microglia are able to adapt to specific environments across particular niches in the healthy brain. In addition, single-cell analyses have been conducted to study microglia associated with inflammation and neurological disorders untangling specific subsets of cells that might differently contribute to each specific pathology (Masuda et al., 2020). Notwithstanding, a crucial aspect that would need to be tackled in future studies would be to understand the functional implication of specific microglial subpopulations across particular neurological diseases, which might enable to explore novel avenues to target neuroinflammation and microglial cells in a specialized context-dependent manner.
OUH and AM conceived the manuscript. OUH, LR, and AM wrote the manuscript. OUH created the figures. OUH, LR, MM, and AM critically revised and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abellanas, M. A., Zamarbide, M., Basurco, L., Luquin, E., Garcia-Granero, M., Clavero, P., et al. (2019). Midbrain microglia mediate a specific immunosuppressive response under inflammatory conditions. J. Neuroinflamm. 16:233. doi: 10.1186/s12974-019-1628-8
Badimon, A., Strasburger, H. J., Ayata, P., Chen, X., Nair, A., Ikegami, A., et al. (2020). Negative feedback control of neuronal activity by microglia. Nature 586, 417–423. doi: 10.1038/s41586-020-2777-8
Banks, W. A., and Robinson, S. M. (2010). Minimal penetration of lipopolysaccharide across the murine blood-brain barrier. Brain Behav. Immun. 24, 102–109. doi: 10.1016/j.bbi.2009.09.001
Bodea, L.-G., Wang, Y., Linnartz-Gerlach, B., Kopatz, J., Sinkkonen, L., Musgrove, R., et al. (2014). Neurodegeneration by activation of the microglial complement-phagosome pathway. J. Neurosci. 34, 8546–8556. doi: 10.1523/JNEUROSCI.5002-13.2014
Bottcher, C., Schlickeiser, S., Sneeboer, M. A. M., Kunkel, D., Knop, A., Paza, E., et al. (2019). Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci. 22, 78–90. doi: 10.1038/s41593-018-0290-2
Darmanis, S., Sloan, S. A., Croote, D., Mignardi, M., Chernikova, S., Samghababi, P., et al. (2017). Single-Cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human Glioblastoma. Cell Rep. 21, 1399–1410. doi: 10.1016/j.celrep.2017.10.030
De Biase, L. M., and Bonci, A. (2018). Region-specific phenotypes of microglia: the role of local regulatory cues. Neuroscientist 2018:107385841880099. doi: 10.1177/1073858418800996
De Biase, L. M., Schuebel, K. E., Fusfeld, Z. H., Jair, K., Hawes, I. A., Cimbro, R., et al. (2017). Local cues establish and maintain region-specific phenotypes of basal ganglia Microglia. Neuron 95, 341–356.e6. doi: 10.1016/j.neuron.2017.06.020
de Haas, A. H., Boddeke, H. W. G. M., and Biber, K. (2008). Region-specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia 56, 888–894. doi: 10.1002/glia.20663
Doorn, K. J., Breve, J. J. P., Drukarch, B., Boddeke, H. W., Huitinga, I., Lucassen, P. J., et al. (2015). Brain region-specific gene expression profiles in freshly isolated rat microglia. Front. Cell. Neurosci. 9:84. doi: 10.3389/fncel.2015.00084
Doorn, K. J., Moors, T., Drukarch, B., van de Berg, W. D., Lucassen, P. J., and van Dam, A.-M. (2014). Microglial phenotypes and toll-like receptor 2 in the substantia nigra and hippocampus of incidental Lewy body disease cases and Parkinson’s disease patients. Acta Neuropathol. Commun. 2:90. doi: 10.1186/s40478-014-0090-1
Duffy, M. F., Collier, T. J., Patterson, J. R., Kemp, C. J., Luk, K. C., Tansey, M. G., et al. (2018). Lewy body-like alpha-synuclein inclusions trigger reactive microgliosis prior to nigral degeneration. J. Neuroinflamm. 15:129. doi: 10.1186/s12974-018-1171-z
Ekdahl, C. T. (2012). Microglial activation—Tuning and pruning adult neurogenesis. Front. Pharmacol. 3:41. doi: 10.3389/fphar.2012.00041
Geirsdottir, L., David, E., Keren-Shaul, H., Weiner, A., Bohlen, S. C., Neuber, J., et al. (2019). Cross-Species single-cell analysis reveals divergence of the primate microglia program. Cell 179, 1609–1622.e16. doi: 10.1016/j.cell.2019.11.010
Gieryng, A., Pszczolkowska, D., Walentynowicz, K. A., Rajan, W. D., and Kaminska, B. (2017). Immune microenvironment of gliomas. Lab. Invest. 97, 498–518. doi: 10.1038/labinvest.2017.19
Ginhoux, F., Greter, M., Leboeuf, M., Nandi, S., See, P., Gokhan, S., et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. doi: 10.1126/science.1194637
Glass, C. K., Saijo, K., Winner, B., Marchetto, M. C., and Gage, F. H. (2010). Mechanisms underlying inflammation in neurodegeneration. Cell 140, 918–934. doi: 10.1016/j.cell.2010.02.016
Gomez-Nicola, D., and Perry, V. H. (2015). Microglial dynamics and role in the healthy and diseased brain. Neuroscientist 21, 169–184. doi: 10.1177/1073858414530512
Grabert, K., Michoel, T., Karavolos, M. H., Clohisey, S., Baillie, J. K., Stevens, M. P., et al. (2016). Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 19, 504–516. doi: 10.1038/nn.4222
Grabowski, M. M., Sankey, E. W., Ryan, K. J., Chongsathidkiet, P., Lorrey, S. J., Wilkinson, D. S., et al. (2020). Immune suppression in gliomas. J. Neuro Oncol. 151, 3–12. doi: 10.1007/s11060-020-03483-y
Graeber, M. B., Scheithauer, B. W., and Kreutzberg, G. W. (2002). Microglia in brain tumors. Glia 40, 252–259. doi: 10.1002/glia.10147
Guneykaya, D., Ivanov, A., Hernandez, D. P., Haage, V., Wojtas, B., Meyer, N., et al. (2018). Transcriptional and translational differences of microglia from male and female brains. Cell Rep. 24, 2773–2783.e6. doi: 10.1016/j.celrep.2018.08.001
Hambardzumyan, D., Gutmann, D. H., and Kettenmann, H. (2016). The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 19, 20–27. doi: 10.1038/nn.4185
Hammond, T. R., Dufort, C., Dissing-Olesen, L., Giera, S., Young, A., Wysoker, A., et al. (2019). Single-Cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6. doi: 10.1016/j.immuni.2018.11.004
Joers, V., Tansey, M. G., Mulas, G., and Carta, A. R. (2017). Microglial phenotypes in Parkinson’s disease and animal models of the disease. Prog. Neurobiol. 155, 57–75. doi: 10.1016/j.pneurobio.2016.04.006
Keren-Shaul, H., Spinrad, A., Weiner, A., Matcovitch-Natan, O., Dvir-Szternfeld, R., Ulland, T. K., et al. (2017). A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17. doi: 10.1016/j.cell.2017.05.018
Krasemann, S., Madore, C., Cialic, R., Baufeld, C., Calcagno, N., El Fatimy, R., et al. (2017). The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9. doi: 10.1016/j.immuni.2017.08.008
Lawson, L. J., Perry, V. H., Dri, P., and Gordon, S. (1990). Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39, 151–170.
Li, Q., Cheng, Z., Zhou, L., Darmanis, S., Neff, N. F., Okamoto, J., et al. (2019). Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10. doi: 10.1016/j.neuron.2018.12.006
Maas, S. L. N., Abels, E. R., Van De Haar, L. L., Zhang, X., Morsett, L., Sil, S., et al. (2020). Glioblastoma hijacks microglial gene expression to support tumor growth. J. Neuroinflamm. 17:120. doi: 10.1186/s12974-020-01797-2
Mastroeni, D., Nolz, J., Sekar, S., Delvaux, E., Serrano, G., Cuyugan, L., et al. (2018). Laser-captured microglia in the Alzheimer’s and Parkinson’s brain reveal unique regional expression profiles and suggest a potential role for hepatitis B in the Alzheimer’s brain. Neurobiol. Aging 63, 12–21. doi: 10.1016/j.neurobiolaging.2017.10.019
Masuda, T., Sankowski, R., Staszewski, O., Bottcher, C., Amann, L., and Saga, et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. doi: 10.1038/s41586-019-0924-x
Masuda, T., Sankowski, R., Staszewski, O., and Prinz, M. (2020). Microglia heterogeneity in the single-cell era. Cell Rep. 30, 1271–1281. doi: 10.1016/j.celrep.2020.01.010
Matcovitch-Natan, O., Winter, D. R., Giladi, A., Vargas Aguilar, S., Spinrad, A., Sarrazin, S., et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353:aad8670. doi: 10.1126/science.aad8670
Menassa, D. A., and Gomez-Nicola, D. (2018). Microglial dynamics during human brain development. Front. Immunol. 9:1014. doi: 10.3389/fimmu.2018.01014
Mittelbronn, M., Dietz, K., Schluesener, H. J., and Meyermann, R. (2001). Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol. 101, 249–255. doi: 10.1007/s004010000284
Morimura, T., Neuchrist, C., Kitz, K., Budka, H., Scheiner, O., Kraft, D., et al. (1990). Monocyte subpopulations in human gliomas: expression of Fc and complement receptors and correlation with tumor proliferation. Acta Neuropathol. 80, 287–294. doi: 10.1007/BF00294647
Mrdjen, D., Pavlovic, A., Hartmann, F. J., Schreiner, B., Utz, S. G., Leung, B. P., et al. (2018). High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395.e6. doi: 10.1016/j.immuni.2018.01.011
Paolicelli, R. C., Bolasco, G., Pagani, F., Maggi, L., Scianni, M., Panzanelli, P., et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. doi: 10.1126/science.1202529
Paolicelli, R. C., and Ferretti, M. T. (2017). Function and dysfunction of Microglia during brain development: consequences for synapses and neural circuits. Front. Synapt. Neurosci. 9:9. doi: 10.3389/fnsyn.2017.00009
Prinz, M., Jung, S., and Priller, J. (2019). Microglia biology: one century of evolving concepts. Cell 179, 292–311. doi: 10.1016/j.cell.2019.08.053
Provenzano, F., Pérez, M. J., and Deleidi, M. (2020). Redefining microglial identity in health and disease at single-cell resolution. Trends Mol. Med. 27, 47–59. doi: 10.1016/j.molmed.2020.09.001
Réu, P., Khosravi, A., Bernard, S., Mold, J. E., Salehpour, M., Alkass, K., et al. (2017). The lifespan and turnover of Microglia in the human brain. Cell Rep. 20, 779–784. doi: 10.1016/j.celrep.2017.07.004
Salamanca, L., Mechawar, N., Murai, K. K., Balling, R., Bouvier, D. S., and Skupin, A. (2019). MIC-MAC: an automated pipeline for high-throughput characterization and classification of three-dimensional microglia morphologies in mouse and human postmortem brain samples. Glia 67, 1496–1509. doi: 10.1002/glia.23623
Salter, M. W., and Stevens, B. (2017). Microglia emerge as central players in brain disease. Nat. Med. 23, 1018–1027. doi: 10.1038/nm.4397
Sankowski, R., Böttcher, C., Masuda, T., Geirsdottir, L., Sagar, Sindram, E., et al. (2019). Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 22, 2098–2110. doi: 10.1038/s41593-019-0532-y
Schafer, D. P., Lehrman, E. K., Kautzman, A. G., Koyama, R., Mardinly, A. R., Yamasaki, R., et al. (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. doi: 10.1016/j.neuron.2012.03.026
Schulz, M., Michels, B., Niesel, K., Stein, S., Farin, H., Rödel, F., et al. (2020). Cellular and molecular changes of brain metastases-associated myeloid cells during disease progression and therapeutic response. IScience 23:101178. doi: 10.1016/j.isci.2020.101178
Scott-Hewitt, N., Perrucci, F., Morini, R., Erreni, M., Mahoney, M., Witkowska, A., et al. (2020). Local externalization of phosphatidylserine mediates developmental synaptic pruning by microglia. EMBO J. 39:e105380. doi: 10.15252/embj.2020105380
Shemer, A., Scheyltjens, I., Frumer, G. R., Kim, J.-S., Grozovski, J., Ayanaw, S., et al. (2020). Interleukin-10 prevents pathological Microglia hyperactivation following peripheral endotoxin challenge. Immunity 53, 1033–1049.e7. doi: 10.1016/j.immuni.2020.09.018
Sierra, A., de Castro, F., Del Rio-Hortega, J., Rafael Iglesias-Rozas, J., Garrosa, M., and Kettenmann, H. (2016). The ‘Big-Bang’ for modern glial biology: translation and comments on Pío del Río-Hortega 1919 series of papers on microglia. Glia 64, 1801–1840. doi: 10.1002/glia.23046
Silvin, A., and Ginhoux, F. (2018). Microglia heterogeneity along a spatio-temporal axis: more questions than answers. Glia 66, 2045–2057. doi: 10.1002/glia.23458
Simon, A., Yang, M., Marrison, J. L., James, A. D., Hunt, M. J., O’Toole, P. J., et al. (2020). Metastatic breast cancer cells induce altered microglial morphology and electrical excitability in vivo. J. Neuroinflamm. 17:87. doi: 10.1186/s12974-020-01753-0
Sorensen, M. D., Dahlrot, R. H., Boldt, H. B., Hansen, S., and Kristensen, B. W. (2018). Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol. Appl. Neurobiol. 44, 185–206. doi: 10.1111/nan.12428
Sousa, C., Biber, K., and Michelucci, A. (2017). Cellular and molecular characterization of Microglia: a unique immune cell population. Front. Immunol. 8:198. doi: 10.3389/fimmu.2017.00198
Sousa, C., Golebiewska, A., Poovathingal, S. K., Kaoma, T., Pires-Afonso, Y., Martina, S., et al. (2018). Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep. 19:e46171. doi: 10.15252/embr.201846171
Stratoulias, V., Venero, J. L., Tremblay, M.-E., and Joseph, B. (2019). Microglial subtypes: diversity within the microglial community. EMBO J. 38:e101997. doi: 10.15252/embj.2019101997
Süß, P., Hoffmann, A., Rothe, T., Ouyang, Z., Baum, W., Staszewski, O., et al. (2020). Chronic peripheral inflammation causes a region-specific myeloid response in the central nervous system. Cell Rep. 30, 4082–4095.e6. doi: 10.1016/j.celrep.2020.02.109
Tan, Y.-L., Yuan, Y., and Tian, L. (2020). Microglial regional heterogeneity and its role in the brain. Mol. Psychiatry 25, 351–367. doi: 10.1038/s41380-019-0609-8
Tasic, B., Menon, V., Nguyen, T. N., Kim, T. K., Jarsky, T., Yao, Z., et al. (2016). Adult mouse cortical cell taxonomy by single cell transcriptomics. Nat. Neurosci. 19, 335–346. doi: 10.1038/nn.4216
Turaga, S. M., Silver, D. J., Bayik, D., Paouri, E., Peng, S., Lauko, A., et al. (2020). JAM-A functions as a female microglial tumor suppressor in glioblastoma. Neuro Oncol. 22, 1591–1601. doi: 10.1093/neuonc/noaa148
van der Poel, M., Ulas, T., Mizee, M. R., Hsiao, C.-C., Miedema, S. S. M., and Adelia, et al. (2019). Transcriptional profiling of human microglia reveals grey-white matter heterogeneity and multiple sclerosis-associated changes. Nat. Commun. 10:1139. doi: 10.1038/s41467-019-08976-7
Verdonk, F., Roux, P., Flamant, P., Fiette, L., Bozza, F. A., Simard, S., et al. (2016). Phenotypic clustering: a novel method for microglial morphology analysis. J. Neuroinflamm. 13:153. doi: 10.1186/s12974-016-0614-7
Villa, A., Della Torre, S., and Maggi, A. (2019). Sexual differentiation of microglia. Front. Neuroendocrinol. 52, 156–164. doi: 10.1016/j.yfrne.2018.11.003
Villa, A., Gelosa, P., Castiglioni, L., Cimino, M., Rizzi, N., Pepe, G., et al. (2018). Sex-specific features of microglia from adult mice. Cell Rep. 23, 3501–3511. doi: 10.1016/j.celrep.2018.05.048
Walentynowicz, K. A., Ochocka, N., Pasierbinska, M., Wojnicki, K., Stepniak, K., Mieczkowski, J., et al. (2018). In search for reliable markers of glioma-induced polarization of Microglia. Front. Immunol. 9:1329. doi: 10.3389/fimmu.2018.01329
Wolf, S. A., Boddeke, H. W. G. M., and Kettenmann, H. (2017). Microglia in physiology and disease. Annu. Rev. Physiol. 79, 619–643. doi: 10.1146/annurev-physiol-022516-034406
Wu, S.-Y., Xing, F., Sharma, S., Wu, K., Tyagi, A., Liu, Y., et al. (2020). Nicotine promotes brain metastasis by polarizing microglia and suppressing innate immune function. J. Exper. Med. 217:e20191131. doi: 10.1084/jem.20191131
Keywords: microglia, heterogeneity, brain regions, neuroinflammation, neurodegenerative diseases, brain tumors
Citation: Uriarte Huarte O, Richart L, Mittelbronn M and Michelucci A (2021) Microglia in Health and Disease: The Strength to Be Diverse and Reactive. Front. Cell. Neurosci. 15:660523. doi: 10.3389/fncel.2021.660523
Received: 29 January 2021; Accepted: 15 March 2021;
Published: 31 March 2021.
Edited by:
Jason R. Plemel, University of Alberta, CanadaCopyright © 2021 Uriarte Huarte, Richart, Mittelbronn and Michelucci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessandro Michelucci, YWxlc3NhbmRyby5taWNoZWx1Y2NpQGxpaC5sdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.