- 1Laboratory of Neurodegeneration, International Institute of Molecular and Cell Biology in Warsaw, Warsaw, Poland
- 2Department of Ichthyology and Biotechnology in Aquaculture, Institute of Animal Sciences, Warsaw University of Life Sciences, Warsaw, Poland
Niemann-Pick type C (NPC) disease is an autosomal recessive lysosomal storage disease that is caused by a mutation of the NPC1 or NPC2 gene, in which un-esterified cholesterol and sphingolipids accumulate mainly in the liver, spleen, and brain. Abnormal lysosomal storage leads to cell damage, neurological problems, and premature death. The time of onset and severity of symptoms of NPC disease are highly variable. The molecular mechanisms that are responsible for NPC disease pathology are far from being understood. The present study generated and characterized a zebrafish mutant that lacks Npc2 protein that may be useful for studies at the organismal, cellular, and molecular levels and both small-scale and high-throughput screens. Using CRISPR/Cas9 technology, we knocked out the zebrafish homolog of NPC2. Five-day-old npc2 mutants were morphologically indistinguishable from wildtype larvae. We found that live npc2–/– larvae exhibited stronger Nile blue staining. The npc2–/– larvae exhibited low mobility and a high anxiety-related response. These behavioral changes correlated with downregulation of the mcu (mitochondrial calcium uniporter) gene, ppp3ca (calcineurin) gene, and genes that are involved in myelination (mbp and mpz). Histological analysis of adult npc2–/– zebrafish revealed that pathological changes in the nervous system, kidney, liver, and pancreas correlated with inflammatory responses (i.e., the upregulation of il1, nfκβ, and mpeg; i.e., hallmarks of NPC disease). These findings suggest that the npc2 mutant zebrafish may be a model of NPC disease.
Introduction
Cholesterol is a lipid-type organic molecule that is critical for life. It builds, maintains, and modulates cell membranes, where it forms lipid rafts and is involved in endocytosis. It also serves as a precursor of the biosynthesis of essential biomolecules (e.g., vitamin D, sex hormones, and corticosteroids). Alterations of cholesterol metabolism were linked to arteriosclerosis in cardiovascular diseases and neurodegenerative diseases, such as Alzheimer’s disease, and Niemann-Pick type C (NPC) disease.
Niemann-Pick type C disease is a recessive lysosomal storage disease that is caused by a mutation of the intracellular cholesterol transporters NPC1 (in 95% of cases) or NPC2 (in 5% of cases), in which un-esterified cholesterol and glycolipids accumulate in lysosomes and late endosomes in the liver, spleen, and brain. The onset of the disease may vary from early childhood (most common) to adulthood, and patients present a wide range of symptoms (Vanier, 2010; Geberhiwot et al., 2018; Seker Yilmaz et al., 2020). Cholestatic jaundice and visceromegaly are often observed as the first signs of this disease. Progressive neurodegeneration is the most devastating and fatal outcome of NPC disease. Neurological symptoms of NPC disease resemble other neurodegenerative disorders, such as Alzheimer’s disease, frontotemporal dementia, and some mitochondrial disorders1. In NPC disease, advanced hypotonia, locomotor dysfunction, ataxia, spasticity, dystonia, dysphagia, and dementia, together with developmental delay and regression, often develop in the first decade of life. These changes coincide with a lower volume of the cerebellum, hippocampus, basal ganglia, and thalamus (Geberhiwot et al., 2018). Histological analyses of mouse models of NPC disease identified the loss of Purkinje cells in the cerebellum (Sarna et al., 2003; Ko et al., 2005; Elrick et al., 2010). Aberrant myelination of the central nervous system, coinciding with a decrease in the expression of genes that are associated with oligodendrocyte differentiation (e.g., Olig1 and Olig2), has been observed in Npc1–/– mice (Yan et al., 2011; Yang et al., 2018). NPC1 disease has other hallmarks, including immune dysfunction (Baudry et al., 2003; Platt et al., 2016), early neuroinflammation, and microglia activation (Cologna et al., 2014; Kavetsky et al., 2019).
The clinical spectrum of NPC disease symptoms ranges from a rapidly progressing prenatal or neonatal form to a chronic form during adolescence. Clinical management guidelines have been developed to help physicians diagnose NPC disease (Geberhiwot et al., 2018). Cholesterol-specific Filipin staining is used as the gold standard for diagnosing NPC disease. Although very useful, this test requires culturing skin fibroblasts, which is rather demanding and cannot be performed in every laboratory. This is why Filipin staining is not recommended as a primary tool for diagnosis. In recent years, several other possible candidate stains and biomarkers have been proposed (Geberhiwot et al., 2018), but there is a need for a more robust method for NPC diagnosis that can be accomplished by genetic analyses.
Although last decade brought progress in the field of NPC, many gaps still need to be filled to improve our understanding of this disease. A further advance in understanding and managing the mechanisms that lead to neurodegeneration that is associated with NPC disease requires the use of high-throughput models, especially based on vertebrates. Zebrafish are a small tropical fish that has been used in laboratories worldwide for more than 30 years. Its significant advantages are genetic similarity to humans (over 70%), its small size, transparent body, and an externally developing body that allows in vivo observations at the cellular and organism levels from fertilization until natural death. Their rapid development, high fecundity, and relatively easy husbandry and breeding are other notable advantages. One month of fish life corresponds to approximately 2 years of human life. Most zebrafish studies that are equivalent to the age of patients can be conducted using embryos and early larval stages. The use of zebrafish as a model of human diseases also has economic value. Experiments on embryos are generally inexpensive and fast and can be readily scaled up for high-throughput screening. Zebrafish have been proven to be an excellent model for studies of lipid metabolism (Anderson et al., 2011), neuroinflammation, and neurodegenerative diseases (de Araujo Boleti et al., 2020). A wide range of tools and techniques has been established that allow assessments of somatic and cognitive functions and developmental and pathological processes at both the cellular and organismal levels.
The human NPC1 and NPC2 genes have homologs in zebrafish. npc1 mutants were recently created (Lin et al., 2018; Tseng et al., 2018). We applied CRISPR/Cas9 technology and created a zebrafish line with a mutation of the npc2 gene (Figure 1). The homozygous npc2 mutant (npc2–/–) reflects NPC disease, which is a starting point for further studies of NPC disease. We found that Nile blue staining of the external olfactory organ can be used as a robust method for identifying npc2–/– larvae, the olfactory organ of which is affected during development.
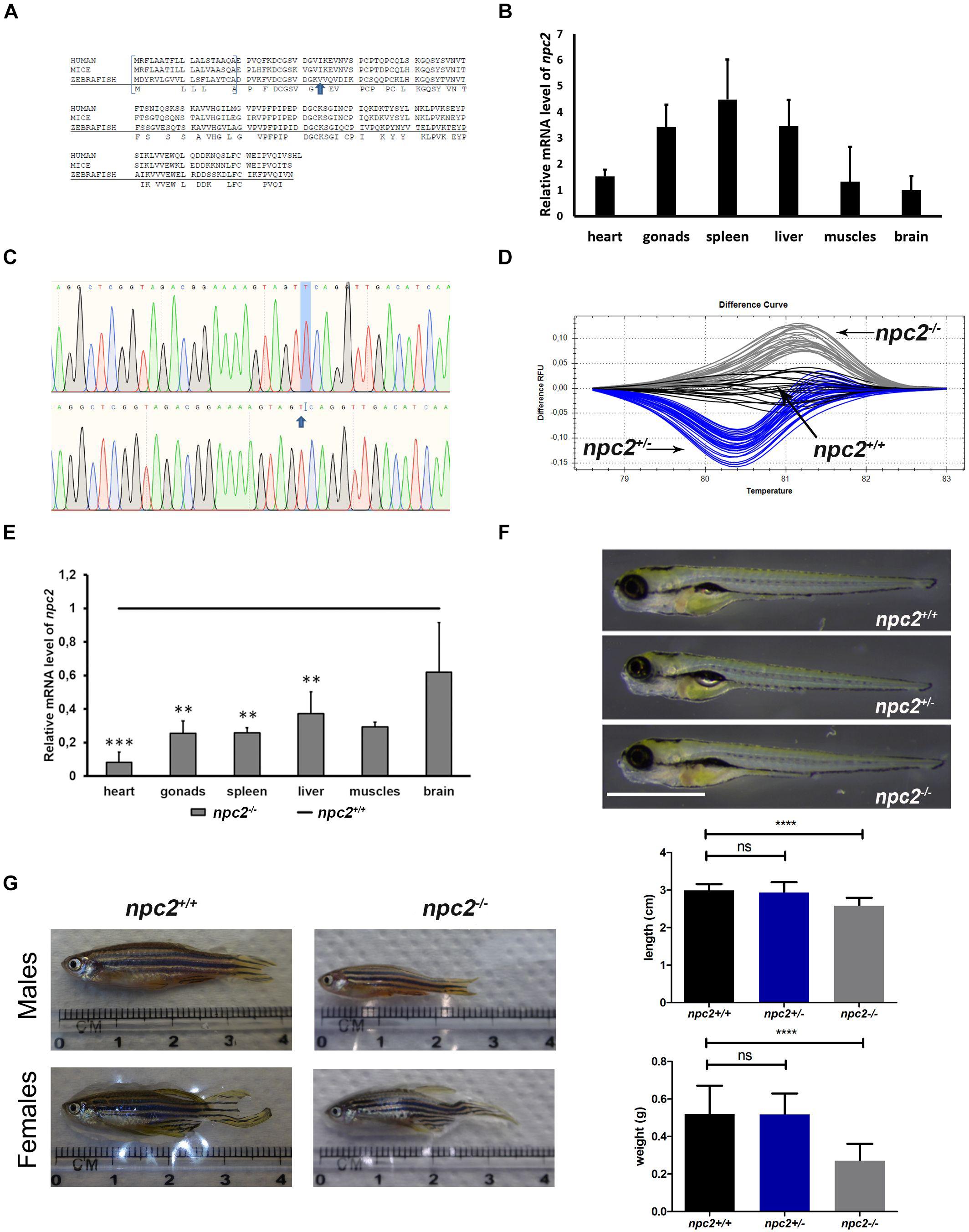
Figure 1. Zebrafish npc2 mutant. (A) Multiple species alignments demonstrated high conservation of the NPC2 protein among vertebrates. Arrows indicate positions of the mutation. (B) Expression levels of the npc2 gene in different tissues in adult fish. Expression was normalized to tissue with the lowest expression (brain). (C) Chromatograms confirmed a small deletion in the npc2 mutant. (D) Graphical representation of melt profiles in wildtypes (black), heterozygous mutants (blue), and homozygous mutants (gray) that originated from fish after incrossing npc2±. (E) mRNA levels of the npc2 gene in various organs in npc2–/– zebrafish relative to wildtype controls. The data are expressed as the mean ± SEM of three 9-month-old fish per group. Error bars represent the SEM. ***p < 0.001 and **p < 0.01. (F) Wildtype, heterozygous, and homozygous npc2 mutants had indistinguishable phenotypes at 5 dpf. Scale = 1 mm. (G) Morphology of adult fish. Smaller body size and weight in 8-month-old npc2–/– fish and indistinguishable phenotype of npc2±. Error bars represent the SD.
Materials and Methods
Animals
Zebrafish (Danio rerio), TL line, were bred and housed at the Zebrafish Core Facility (ZCF) at the International Institute of Molecular and Cell Biology in Warsaw, Poland (license no. PL14656251 from the District Veterinary Inspectorate in Warsaw; license no. 064 and 0051 from the Ministry of Science and Higher Education in Poland). All of the animal procedures were performed in accordance with fundamental ethical principles for the protection of animals that are used for scientific or educational purposes (Act of January 15, 2015; Directive 2010/63/EU). Embryos at 5–13 days postfertilization (dpf) were kept in static tanks with rotifer (Brachionus plicatilis) culture (maintained inhouse; rotifer and the rotifer diet were originally purchased from Varicon Aqua Solutions Ltd, United Kingdom). At 2 weeks of age, the fish were kept in circulating water. Fish at 13–30 dpf were fed Gemma Micro three times per day, artemia once a day, and also received rotifers at least once a day. Thereafter, rotifers were excluded while the diet remained unchanged (dry feed three times a day and artemia once a day. The facility maintains a 14 h light/10 h dark photoperiod. The fish were kept in groups of >5 individuals. Larvae and fish up to 6 weeks of age were kept at a maximal density of 50 fish in 3.5 L tanks. From 4 to 6 weeks of age, the fish were kept at a maximal density of 24 fish in 3.5 L tanks. Average values of water quality were the following: salinity of 800 ± 10 μS, pH 7.0 ± 0.2, temperature of 28 ± 0.4°C, undetectable NO2– and NH3/NH4+ levels, GH 4, KH 6, dissolved O2 of 8.6 ± 0.1 mg/L, and 20% of daily water change. The chosen donor fish were outcrossed to the TL line. Some embryos were sacrificed for genotyping, and the rest were left to grow and used to establish new stable lines with a mutation of the npc2 gene.
CRISPR/Cas9
For CRISPR prediction, the following free web tools were used to select target sites: CHOPCHOP2, E-CRISP3, CRISPRscan4, and CCTop5. Sequences that were common in at least three predictions and were scored as low risk for off-targets were chosen. The correctness of the sequence in the targeted area in the TL fish at the ZCF was confirmed by Sanger sequencing. Mutagenesis was performed according to the protocol of Gagnon et al. (2014), with modifications (Gagnon et al., 2014). The following gene-specific oligos with T7 overlaps were used: npc2b_(86/34aa) 5′-taatacgactcactataGGTAG ACGGAAAAGTAGTTCgttttagagctagaa-3′ (to create guide IT8). During gRNA preparation, annealing and filling in steps were combined, and the template was prepared for polymerase chain reaction (PCR) using 10 μl of PCR Mix Plus (A&A Biotechnology), 2 μl of gene-specific and constant oligo (5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAA CGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAA AC-3; each at 100 mM stock), 2 μl dNTPs (100 mM stock), and water up to 20 μl. The PCR conditions were the following: pre-incubation at 95°C for 3 min, followed by 35 cycles of 95°C for 15 s, 40°C for 15 s, and 68°C for 15 s. sgRNA was prepared using 1 μl of gene-cleaned template (145 ng), 0.5 μl T7 RNA polymerase (20 U/μl, A&A Biotechnology), 2 μl NTPs (75 mM each, A&A Biotechnology), 1 μl of buffer (5×, A&A Biotechnology), and water up to 5 μl. The reaction was incubated for 2 h at 37°C. Thereafter, a mixture of 1 μl of RNA-free DNAse (Qiagen) and 14 μl of water was added to each sample. After 15 min of incubation, 10 μl of ammonium acetate (5 M stock) and 60 μl of pure ethanol were added, and the samples were left for precipitation over night at -20°C. gRNA was suspended in water, and the concentration was adjusted to 500 ng/μl. Cas9 protein (14 mg/ml stock, made in-house) was diluted in KCl/HEPES (200 mM/10 mM, pH 7.5) buffer to a final concentration of 600 ng/μl. The injection mixture was assembled fresh by mixing 2 μl of gRNA, 2 μl of gRNA, and 0.4 μl phenol red (Sigma, catalog no. P0290) and left for complex formation at room temperature for 5–10 min. Thereafter, the samples were kept on ice.
Injections
Microneedles were pulled from borosilicate glass capillaries (Sutter; catalog no. BF 100-50-10) using a P-1000 Flaming/Brown micropipette puller (Sutter) set to the following parameters: heat 542, pull 80, velocity 80, time 170, pressure 500, and RAMP 552. The needle tip was chipped with Dumont no. 5 ceramic-coated fine forceps (Dumostar). One picoliter of the gRNA/Cas9/phenol red mixture was injected into the yolk (just below the zygote) at 1–2 cell-stage zebrafish embryos using a PV 820 Pneumatic PicoPump (World Precision Instruments). Injected embryos were sorted 50/plate and incubated up to 5 dpf in 9 cm diameter Petri dishes that were two-thirds filled with E3 medium in an HPP110 incubator (Memmert) set to 28.5°C, a 14 h/10 h light/dark cycle, 20% light intensity (cold/warm light, 1:1), and 60% humidity.
DNA Extraction
Fin clipping of adult zebrafish was performed under anesthesia with tricaine (Sigma, catalog no. A-5040). The fish were immersed in 0.7 mM tricaine solution in system water until they stop moving and exhibited no response to tough. Euthanasia of - larvae was performed by an overdose of tricaine. Fish tissue (fragment of the tail fin or pulls of 3–5 dpf embryos) was dehydrated in pure ethanol, incubated at 80°C until complete dryness, soaked in 50–150 μl of TE 10-1 buffer [aqueous solution of 10 mM Tris (pH 8.0) and 1 mM ethylenediaminetetraacetic acid (pH 8.0)] and cooked for 10 min at 95°C. Thereafter, an equal volume of TE 10-1 with 20–60 μg proteinase K (10 mg/ml aqueous stock) was added to each sample. The samples were incubated for 1 h at 55°C. Proteinase K was then inactivated by heat (incubation for 15 min at 95°C). Samples were stored at −20°C before use.
DNA Sequencing
For sequencing, the amplicon was amplified using PCR Mix Plus (catalog no. 2005–100P, A&A Biotechnology) and npc2_a_F1 5′-GCATATTCGCTGTCATGTGATT-3′ and npc2_b_R1 5′-GTAGGATTGTCCCTTGTGAAGC-3′ primers. The PCR conditions were preincubation at 95°C for 3 min and 35 amplification cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s, followed by 5 min of incubation at 72°C. The PCR products were purified using EPPiC Fast (catalog no. 1021–100F, A&A Biotechnology) according to the manufacture’s protocol.
RNA Isolation and cDNA Synthesis
The fish were anesthetized with an overdose of tricaine. RNA was isolated using TRI reagent (Sigma, catalog no. T9424) according to the manufacture’s instructions. The tissue was shredded in 1 ml of solution with a 23-gauge needle. After the addition of 0.2 ml of chloroform, the sample was vortexed and centrifuged at 13,000 rotations per minute (rpm) for 15 min in 4°C. The supernatant was transferred to a new tube, and RNA was precipitated with pure isopropanol overnight at −20°C. After centrifugation at 13,000 rpm for 30 min at 4°C, the pellet was washed with 70% ethanol, air-dried, and resuspended in 15–25 μl of sterile water.
Total RNA was extracted from 5 dpf larvae (n = 9/group from at least two independent cohorts), and various organs (brain, liver, heart, spleen, skeletal muscles, and gonads) were collected from 9-month-old male fish (n = 4 homozygotes and n = 4 wildtypes). Additionally, to test the influence of 2-hydroksypropylo-β-cyclodextrin (2HPβCD) treatment (a drug that is used for the treatment of Niemann-Pick disease) on the expression of selected genes, total RNA was extracted from two groups of treated and untreated zebrafish larvae (5 dpf) from two independent experiments. The RNA template (1,000 ng) was used for cDNA synthesis using the iScript cDNA Synthesis Kit (Bio-Rad).
High-Resolution Melting (HRM) Analysis
The reaction mixture was composed of (i) 5 μl of LightCycler 480 High Resolution Melting Master mix (Roche), 1 μl of MgCl2 (25 mM stock), 0.3 μl of each primer (10 μM stock), 0.5 μl of template, and water up to 10 μl or (ii) 5 μl of Precision Melt Supermix (Bio-Rad), 0.2 μl of each primer (10 μM stock), 1.0 μl of template, and water up to 10 μl. Two primer pairs were used. npc2a HRM F1 (5′-AACTCTAGTGTGTTGGTTCCTAAC-3′) and npc2a HRM R1 (5′-CAAGTGTACGCGAGAAAAGAAAGTA-3′) were used as controls, which amplified the region upstream of the target site. npc2b HRM F1 (5′-TAATTTCCACTTTCATCTTACAGGC-3′), and npc2b HRM R1 (5′-GGATTGTCCCTTGTGAAGCTTG-3′), which span the mutation site, were used to detect the mutation. HRM analysis was performed on either LightCycler 480 System PCR (Roche) or CFX96 (Bio-Rad), both according to the manufacturer’s protocol. For the Bio-Rad system, the conditions were preincubation at 95°C for 2 min, 35 amplification cycles of 95°C for 10 s and 60°C for 30 s, heteroduplex formation at 95°C for 30 s, followed by 60°C for 1 min, and HRM at 65–95°C with ramp at 0.2°C/10 s. Genotypes were automatically assigned by LightCycler 96 software’s HRM module for the Roche system or Precision Melt Analysis software for the Bio-Rad system and manually annotated as wildtype (black), heterozygous mutant (blue), and homozygous mutant (gray; Figure 1).
Quantitative PCR Gene Expression Analysis
Gene expression levels were analyzed using the CFX Connect Real-time PCR (RT-PCR) Detection System (Bio-Rad, Hercules, CA, United States). RT-PCR was performed in duplicate using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, catalog no. 1725274). The data were analyzed using Bio-Rad CFX Maestro 1.0 software. The specificity of the reactions was determined based on dissociation curve analysis. Fold changes were calculated using the ΔΔCq method as described before (Majewski et al., 2019). Expression levels were compared between groups using analysis of variance (ANOVA) followed by Tukey’s Honestly Significant Difference (HSD) post hoc test. The 18S ribosomal gene was used as a reference. The sets of primers that were used in the analysis are shown in Table 1.
LysoTracker, Filipin, Neutral Red and Nile Blue Staining
Nile blue (B&K, Bytom, Poland) was prepared as a 1% aqueous stock solution. Stock solution of Neutral red (Sigma N7005) was prepared as 2.5 mg/ml solution in water. LysoTracker Red DND-99 (Invitrogen, catalog no. L7528), Neutral red or Nile blue (each at 1:1000 dilution) was added directly into the plates with live 5-day-old zebrafish larvae. After 1 h of incubation at 28°C in the dark, the medium was exchanged to fresh E3 three times over a 15 min period.
The stock of filipin complex (Sigma, catalog no. F9765) was prepared by dissolving 25 mg filipin in 1 ml of dimethylsulfoxide. Aliquoted stocks were stored at −20°C. 5-day-old zebrafish larvae were fixed in 4% paraformaldehyde (Sigma, catalog no. 47608) in 1 × phosphate-buffered saline (PBS; Sigma, catalog no. D5652) for 1 h at room temperature and washed for 5 min three times with 1 × PBS with 1.5% glycine (Sigma, catalog no. G7126). Embryos were stained with Filipin solution (final Filipin concentration of 0.5 mg/ml) in 1 × PBS with 10% bovine calf serum (Sigma, catalog no. 12133C) in the dark for 2 h at room temperature.
Stained larvae were rinsed with 1 × PBS, immersed in 0.7 mM tricaine, transferred to a plate with 3% methylcellulose, and imaged in the ultraviolet channel (Filipin), RFP channel (Lysotracker), and bright field (Nile blue) under a Nikon SMZ25 fluorescent stereomicroscope. The genotype of the imaged fish was confirmed by HRM analysis.
Treatments
The stock solutions were prepared by dissolving the powdered compound in Milli-Q water to a final concentration of 100 mM for (2-hydroxypropyl)-β-cyclodextrin (2HPβCD; Sigma, catalog no. C0926) and 10 mM for 3-β-[2-(diethylamino)ethoxy]androst-5-en-17-one (U18666A; Sigma, catalog no. 662015). Good-quality eggs were placed in 9 cm diameter Petri dishes filled with 25 ml of E3 medium and kept under static condition at 28 ± 0.5°C. At 3 dpf, the medium was changed to fresh E3 medium supplemented with U18666A (8 μM), 2HPβCD (2 mM), and U18666A (8 μM) together with 2HPβCD (2 mM). As a control, an equal volume of the solvent was used. Embryos were kept in static condition for 2 days. At 5 dpf, the fish were stained with Nile blue and collected for RNA extraction.
Histology
Adult zebrafish were fixed in neutral buffered 4% formaldehyde solution (Sigma, catalog no. 47608–250ML-F), dehydrated in ethylene, and embedded in paraffin. Longitudinal and transverse 5–6 μm sections were cut using a Leica microtome (RM2265, Leica, Bensheim, Germany) and stained with standard hematoxylin and eosin (H&E). For glycogen and lipofuscin inclusions, periodic acid-Schiff (PAS) reactions were performed according to the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO, United States). Perl Prussian blue staining to detect hemosiderin was performed according to Orchard (2019). Luxol Fast Blue (LFB), combined with H&E, was used to stain myelin sheaths. The immunohistochemical frequency of CD3 and proliferating cells in the intestinal epithelium and liver was determined in samples that were stained according to the manufacture’s protocol for anti-CD3 (Bond Ready-to-Use Primary Antibody CD3, LN10, Leica Newcastle, United Kingdom) and anti- proliferating cell nuclear antigen (PCNA; 1:400 dilution; clone PC10, DAKO, Poland) antibodies. The colorimetric detection of these cells was performed using DAB (Novolink Polymer Detection Kit, Novocastra, Leica, Newcastle, United Kingdom). The microscopic analysis was performed using NIS Elements software and Nikon Ni-E with NIS Elements software.
Behavioral Analysis
The behavioral studies of zebrafish larvae were performed as described previously (Wasilewska et al., 2020). Before the experiment, the larvae were kept in a Petri dish (∼50 larvae/dish) that was two-thirds filled with E3 medium in an HPP110 incubator (Memmert) that was set to 28.5°C under a 14 h/10 h light/dark cycle with 20% light intensity (cold/warm light, 1:1) and 60% humidity. The larvae (5 dpf) were transferred to 12-well plates and placed in a ZebraBox high-throughput monitoring system (ViewPoint Life Sciences, Lyon, France). The experiments were performed in a volume of 2 ml of E3 medium, and the light intensity was set to 70%. Locomotor activity was recorded for 15 min using ZebraBox. A total of 40 wildtype and 32 npc2–/– larvae from two independent cohorts were used in the experiment.
The video files were further analyzed using EthoVision XT software (Noldus, Wageningen, Netherlands). The data were exported to Microsoft Excel files and further analyzed using Excel (Microsoft, Redmond, WA, United States) and R software (R Foundation for Statistical Computing, Vienna, Austria, R package version 3.6.2). The experiment was divided into three 5 min periods, and the mean total distance traveled (mm), mean velocity (mm/s), and mean movement duration were calculated and compared independently for these time bins. The Wilcoxon rank-sum test was used for comparisons between wildtype and npc2–/– larvae.
Additionally, the area of the well was divided into borders and a central area. This allowed analyses of the level of thigmotaxis that was calculated according to the following formula: [duration of movement (in borders or center) + duration of no movement (in borders or center)]/[duration of movement (total) + duration of no movement (total)] × 100% (Wasilewska et al., 2020). The data are expressed as medians with first and third quartiles using boxplots, and dots represent data outliers. Heatmaps that represent average traces of wildtype and npc2–/– larvae were generated using EthoVision XT software.
Statistical Analysis
In quantitative PCR gene expression analysis, expression levels were compared between groups using ANOVA followed by Tukey’s HSD post hoc test. In the behavioral studies the Wilcoxon rank-sum test was used for comparisons between wildtype and npc2–/– larvae.
Results
Generation and Phenotype of the npc2 Zebrafish Mutant
The zebrafish npc2 gene (ENSDARG00000090912) maps to chromosome 17 and encodes a 149 amino acid protein that shares over 64% sequence identity with its human ortholog (Figure 1A). Expression of the npc2 gene could be found in each tested tissue (i.e., brain, heart, liver, muscles, skin, and reproductive organs; Figure 1B). Using CRISPR/Cas9 technology and gRNAs that targeted the npc2 gene, we generated F0 carriers of different mutations. After an initial screening by HRM analysis, six donor fish with the most distinct HRM patterns were bred, and their F2 offspring were analyzed by HRM and sequencing. A mutant line with a 1 bp deletion in the third exon of the npc2 gene (ENSDARG00000090912) that is predicted to introduce a premature stop codon was selected for further studies (Figure 1C). The deletion caused a characteristic shift in the melting temperature of the amplicon that was identified by HRM (Figure 1D). In the mutant, qPCR analysis confirmed a significantly lower level of npc2 transcripts (Figure 1E). At 5 dpf, the morphology and length of the hetero- and homozygousnpc2 mutants were indistinguishable from wildtype siblings (Figure 1F), similar to npc1 zebrafish knockouts (Lin et al., 2018; Tseng et al., 2018). Over time, the differences became more apparent. The 2-month-old npc2–/– fish were smaller than their siblings, and the sex ratio of homozygotes was skewed toward males (data not shown). When separated from their siblings, the npc2–/– fish did not need to compete for food and grew faster. From 4 months of age, their motor functions were clearly impaired (Supplementary Video 1) as compared to wildtype siblings (Supplementary Video 2). The malnutrition and progressive loss of motor skills continued to worsen, but not all npc2–/– fish were equally affected. Some fish had to be euthanized at the age of 5 months because their eating and swimming skills were severely impaired, whereas others were fit and survived four more months. The length and weight of the 9-month-old npc2–/– fish were significantly reduced (Figure 1G). The npc2–/– fish bred normally, whereas inbreeding of the npc2–/– fish was unsuccessful (data not shown). Interestingly, we could obtain offspring by crossing npc2–/– males with either wildtype or npc2± females.
Nile Blue Staining Identifies npc2–/– Larvae
Although, nowadays not recommended as a primary tool, the cholesterol-specific filipin staining for many years was used as a gold standard in NPC diagnostics (Geberhiwot et al., 2018). That is why we applied this stain to the mutant fish and confirmed the presence of blighter signals in the npc2–/– mutant (Supplementary Figure 1). This finding indicates that as expected, un-esterified cholesterol accumulates in homozygous npc2 mutant.
Niemann-Pick type C disease is a lysosomal storage disease. We used the LysoTracker red probe to visualize acidic organelles in vivo to quickly identify the mutant. We observed increases in LysoTracker red staining in neuromasts in some npc2–/– larvae (Supplementary Figure 2). However, using this staining, we failed to reliably distinguish wildtype from npc2–/–.
We found that Nile blue staining allowed the robust and reliable detection of npc2–/– larvae. When added to the water in the dish, Nile blue reversibly stained live larvae in a few minutes. At 5 dpf, strong staining in the peripheral olfactory organ was specific (certainty > 95%) to npc2–/– fish, whereas staining in other parts of the body appeared random (Figure 2).

Figure 2. Nile blue staining distinguishes npc2–/– fish from wildtype and heterozygous siblings. The strong signal could be detected in the peripheral olfactory organ in 5-day-old npc2–/– larvae (B–B”), unlike their siblings (A–A”). (A,B) lateral view; (A’,A”,B’,B”) dorsal view; and (A”,B”) enlarged fragments of A’,B’, respectively. Arrows indicate the position of the olfactory organ. Scale bar = 0.5 mm.
Nile blue stains lipids. It has been primarily used to detect melanin and lipofuscin (Lillie, 1958). Nile blue-based dyes were later discovered to localize in lysosomes. Their lysosomotropic photosensitizer properties could be used to target and kill tumor cells (Gattuso et al., 2016; Martinez and Henary, 2016). To verify the specificity of Nile blue staining, we applied it to larvae that were treated with 2HPβCD (a drug that is used in clinical trials to treat NPC patients) and U18666A (an inhibitor of cholesterol synthesis that is used to chemically model NPC disease). 2HPβCD treatment reduced the intensity of blue staining in the nose in npc2–/– fish, whereas U18666A treatment increased this signal (Figure 3). The intensity of staining in wildtype fish and heterozygous npc2 mutant fish followed the same trend, in which staining was weaker in 2HPβCD-treated larvae and stronger in U18666A-treated larvae (data not shown).

Figure 3. Specificity of Nile blue staining in the olfactory organ in npc2–/– larvae at 5 dpf. (A–A”) untreated npc2–/– larvae; (B–B”) treatment with 2 mM 2-hydroxypropyl-β-cyclodextrin (2HPβCD; a drug that is used in clinical trials to treat NPC patients) decreased staining intensity to wildtype levels (Figures 2A–A”); and (C–C”) treatment with 8 mM (3β)-3-(2-[diethylamino]ethoxy)androst-5-en-17-one hydrochloride (U18666A, an inhibitor of cholesterol synthesis that is used to model NPC chemically) increased staining intensity in npc2–/– larvae. (A–C) lateral view; (A’,A”,B’,B”,C’,C”), dorsal view; and (A”,B”,C”) enlarged fragments of A’,B’,C’, respectively. Scale bar = 0.5 mm.
Decrease in Locomotor Activity in npc2–/– Zebrafish Larvae
To investigate the effect of npc2 mutant on behavior, locomotor activity in npc2+/+ and npc2–/– zebrafish larvae was analyzed. A significant reduction of mobility was observed in npc2–/– larvae compared with their wildtype siblings (Figure 4A). Mutants traveled a shorter distance (Figure 4B), and they remained mobile for a shorter period of time (Figure 4C). The mean velocity during the first 5 min of the experiment was lower in npc2–/– larvae compared with npc2+/+ larvae (Figure 4D). During the initial 5 min of the experiment, mutant larvae exhibited a stronger tendency to stay in close proximity to the borders of the well (thigmotaxis; Figure 4E). This phenomenon is a typical anxiety-like response in zebrafish larvae (Lundegaard et al., 2015), suggesting a stronger anxiety-related response to a stressor in this group.
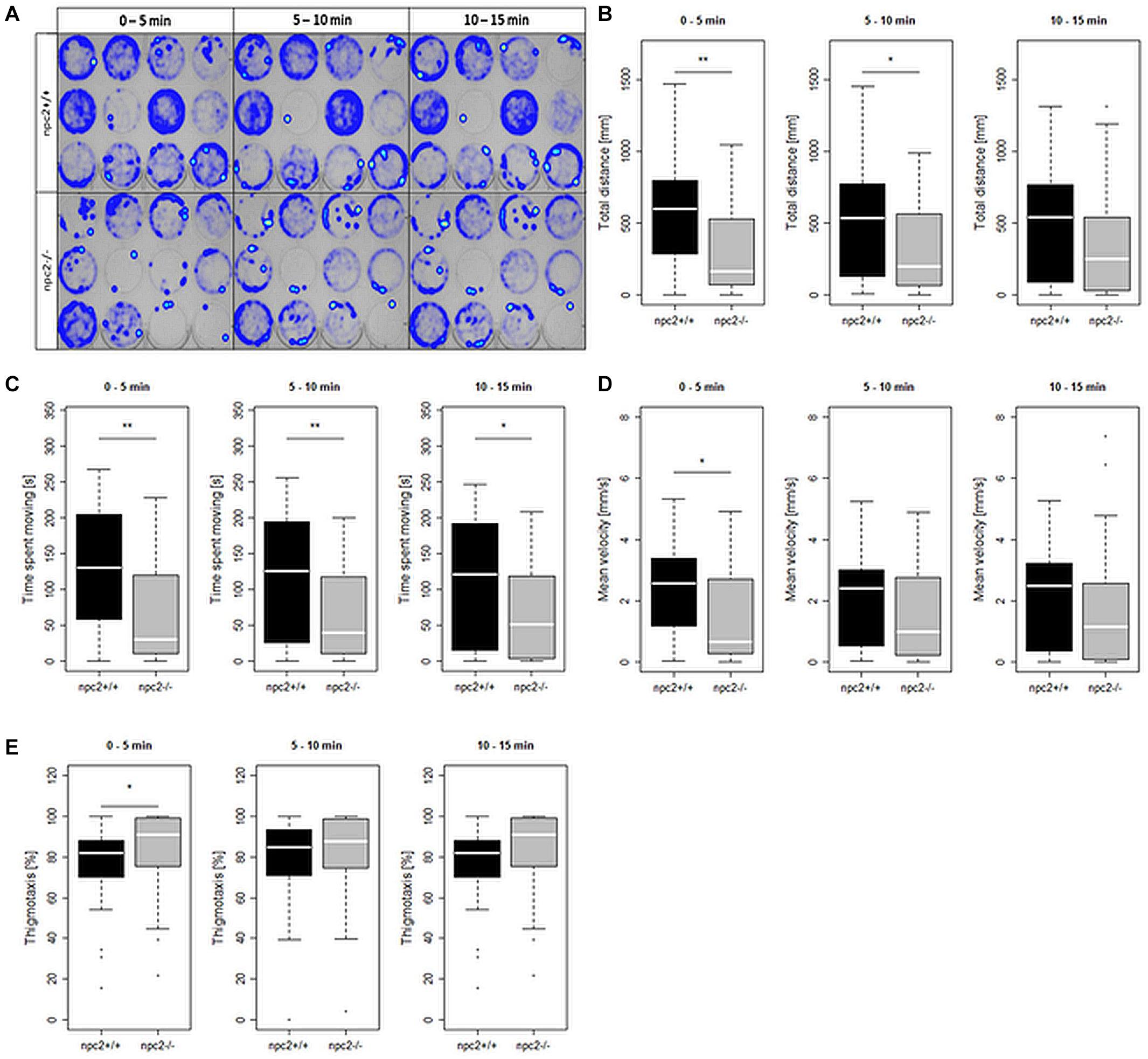
Figure 4. Decrease in mobility and increase in thigmotaxis in npc2–/– larvae. (A) Heatmaps of mean traces of npc2+/+ and npc2–/– larvae during each 5 min period. (B–E) Boxplots of total distance traveled, time spent moving, mean velocity, and thigmotaxis in npc2+/+ and npc2–/– larvae. (E) *p < 0.05 and **p < 0.01.
Pathological Changes in Internal Organs in Adult npc2–/– Fish
The histopathological analysis of adult npc2–/– larvae revealed lesions in the spleen, liver, and pancreas. In the liver in npc2–/– fish, numerous foam hepatocytes, hepatocytes that contained fat droplets, and cells with enlarged nuclei were observed (Figure 5A). An atypical interstitial group of foam cells was observed in the anterior part of the npc2–/– kidney (Figure 5B). The structure of the pancreas was disturbed. Acinar cells in the pancreas in npc2–/– larvae had no characteristic zymogen granules but had multiple circular cholesterol inclusions that were not observed in the control group (Figure 5C).
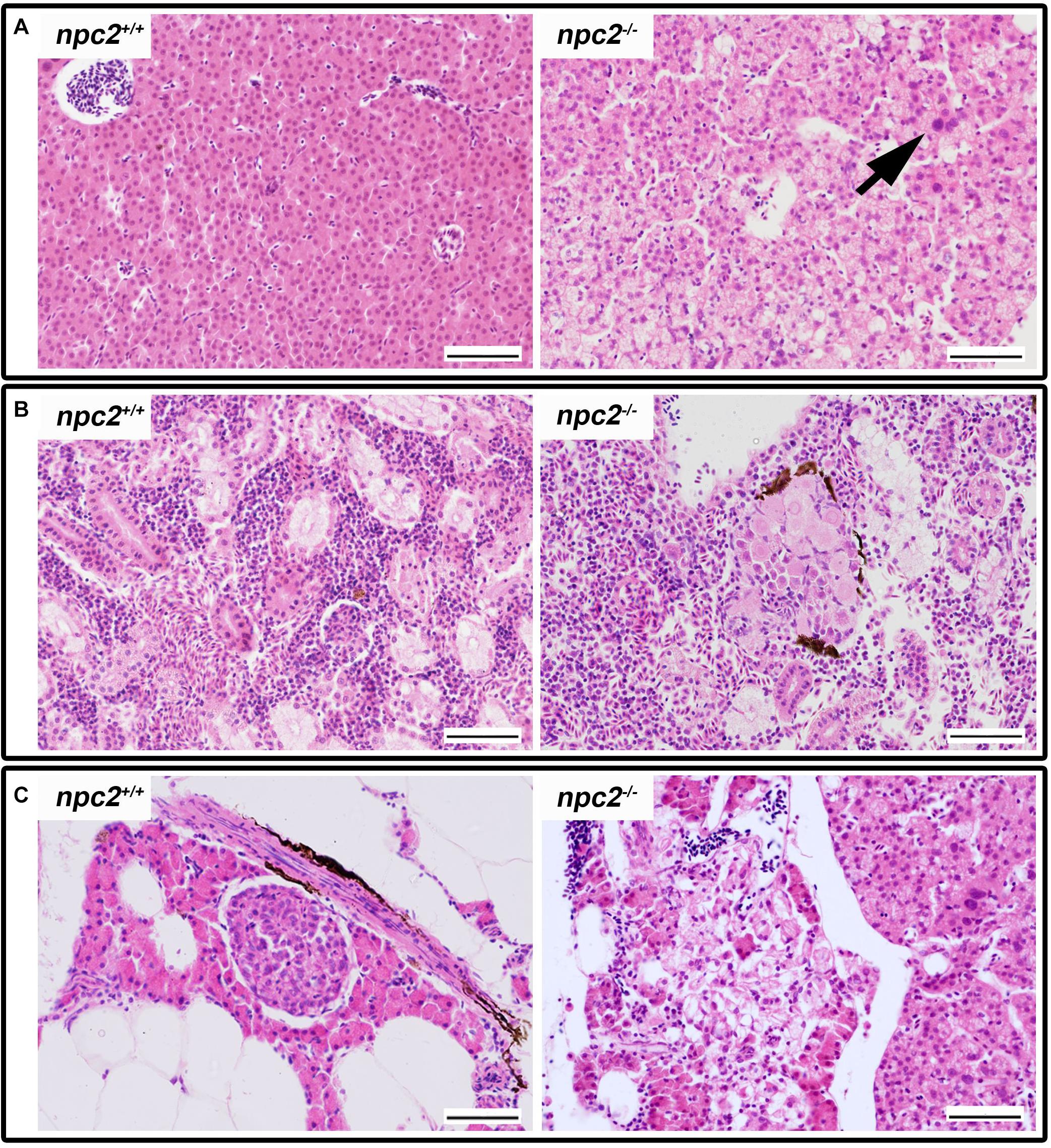
Figure 5. Pathological changes in soft tissues in npc2–/– revealed by H&E staining. (A) Cells with enlarged nuclei (black arrow), hepatosteatosis with different type of fatty droplet accumulation (open arrowhead) and foam cytoplasm (asterix) were found in npc2-deficient livers. (B) Kidneys with degenerating cells (DMc) between ductal (DT) and proximal (PT) tubules. (C) Pancreas with degeneration of zymogen granules in acinar cells and cholesterol deposits. The paraffin sections are from adult npc2–/– fish and wildtype npc2+/+ control. Scale bar = 50 μm.
Morphological and histopathological changes were also observed in the brain in npc2–/– fish (Supplementary Figure 3 and Figure 6). The myelination of axons in npc2–/– fish was affected (Figure 6A), especially in the habenular commissure (Figures 6B,C). The habenula’s characteristic layered structure was affected. The fascicular retroflexus, interpeduncular nucleus, and raphe were vacuolated, and the myelination of axonal tracts decreased (Figures 6B,C). The two mechanoreceptive areas with well-differentiated axonal tracts (lateral and medial longitudinal fascicle) were observed only in wildtype fish compared with npc2–/– fish (Figure 6C). In the cerebellum, Purkinje cells were more diffuse than in the control group, and the structure on these cells differed from control fish (Figures 6C, 7A). Purkinje cells had spheroid, highly non-condensed chromatin, enlarged nuclei, and a distended perikaryon (Figure 7A). In this region, no PCNA-positive cells were observed, although proliferation was detected in the parvocellular preoptic nucleus and caudal zone of the periventricular hypothalamus compared with the control group (Supplementary Figure 4A).
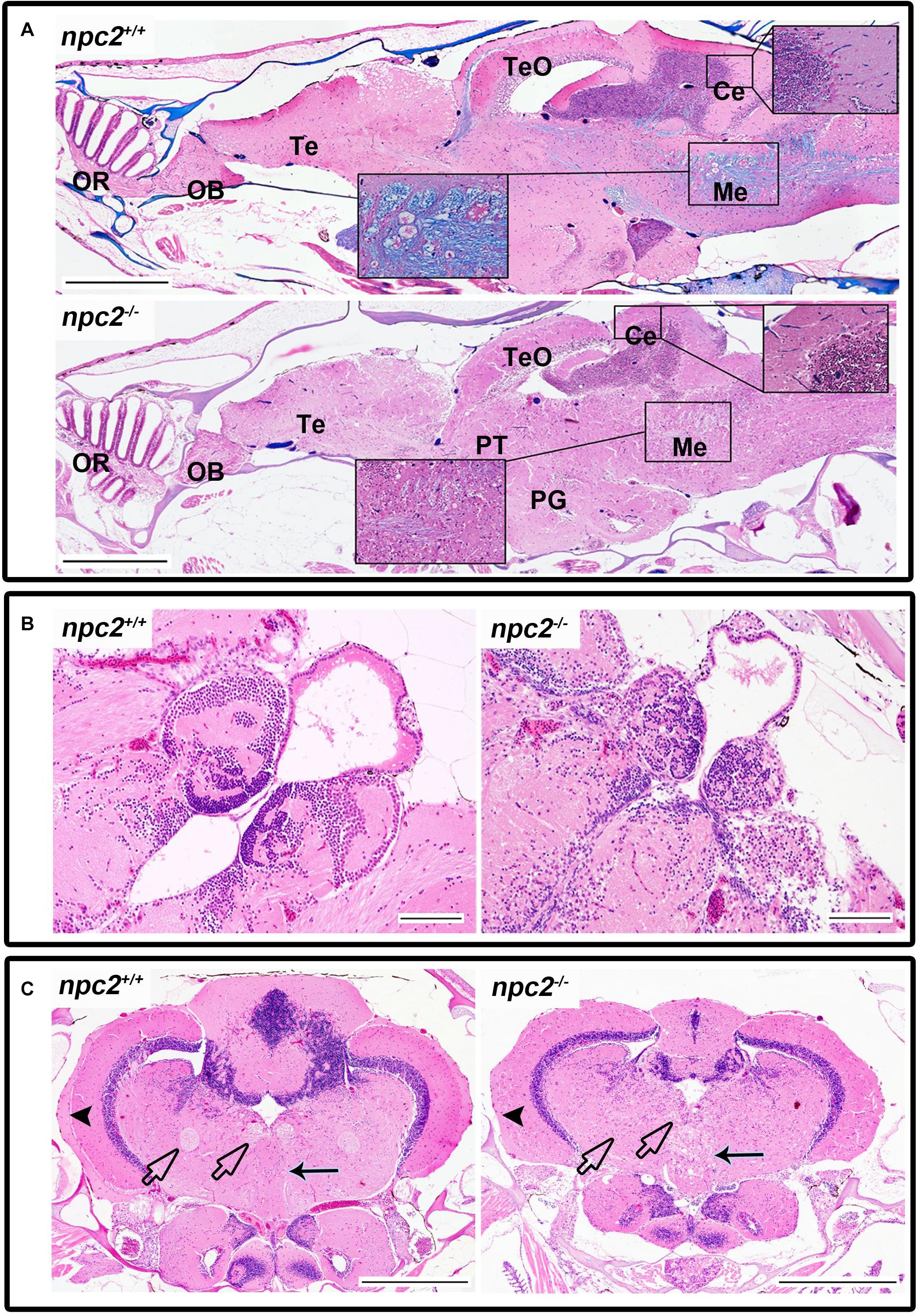
Figure 6. Pathological changes in the central nervous system in adult npc2–/– zebrafish. (A) Luxol fast blue staining showed differences in myelination between wildtype and npc2–/– fish. (B) Changes in habenula structure were present in some npc2–/– fish, which lacked the characteristic structure of this part of the habenular tract. (C) Midbrain with vacuolization of the habenula tract (blue arrow) with fascicular retroflexus (blue arrowhead) and degenerative changes in the medial and lateral longitudinal fascicle. OB, olfactory bulb; OR, olfactory rosette; Te, telencephalon; TeO, tectum opticum; Ce, cerebellum; Me, medulla; PG, preglomerular complex; and PT, posterior tuberculum. (A) LFB staining. Scale bar = 500 μm. (B,C) H&E staining. Scale bar = 100 μm. (C) scale bar = 500 μm.
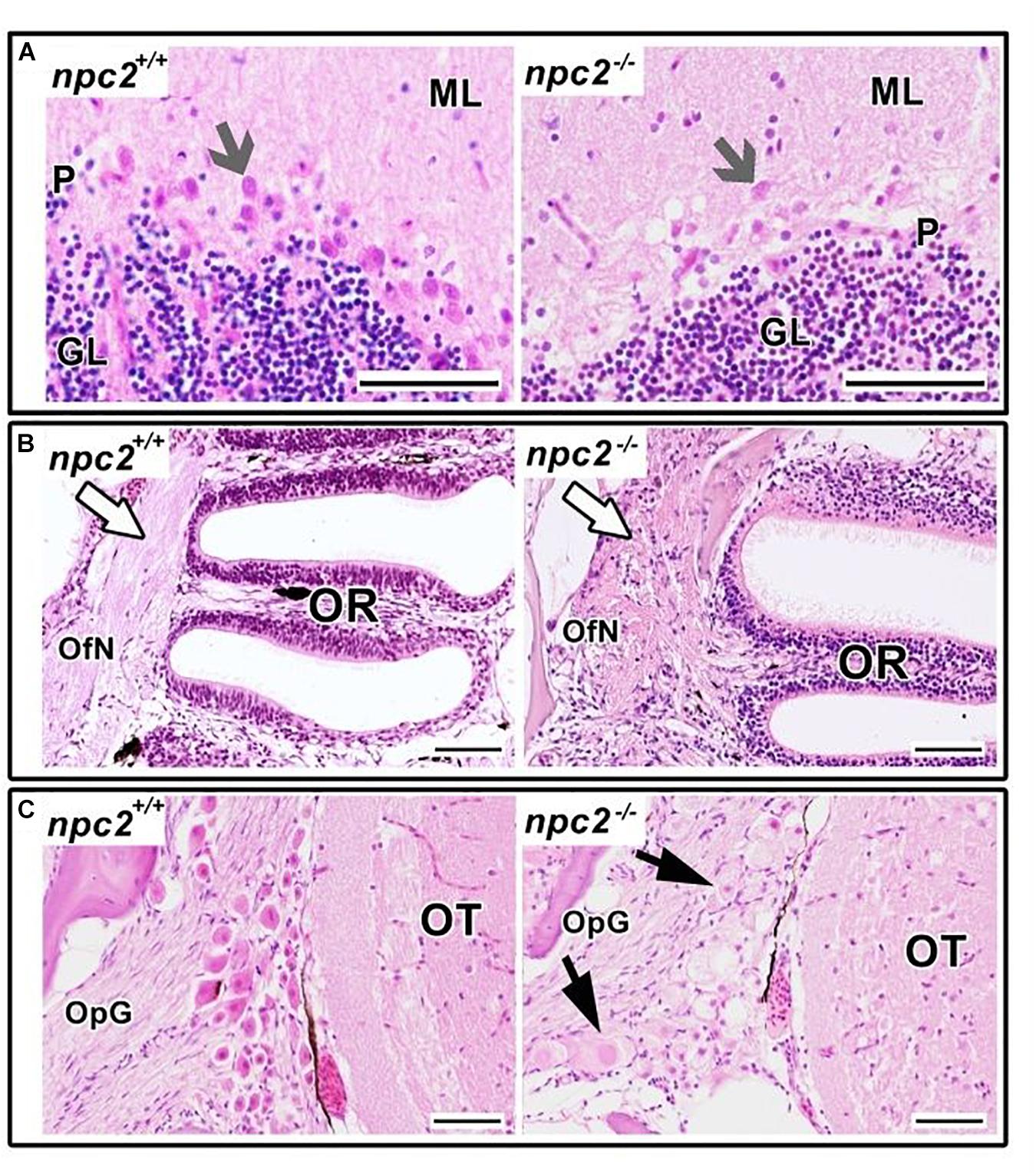
Figure 7. Pathological changes in npc2–/– sensory organs. (A) Cerebellum structure with Purkinje cells degeneration in npc2 mutant (gray arrow), (B) Olfactory rosette with olfactory nerves (white arrow). (C) Optic nerve ganglion. Cells with a foamy, pale cytoplasm, eosinophilic nuclei, and signs of degeneration (black arrows) within the optic tectum. (A) ML, molecular layer; P, Purkinje cell leyer; GL, granule cell layer; OfN, olfactory nerves; and OR, olfactory rosette; (B) OT, optic tectum; OpG, optic nerve ganglion. H&E staining. Scale bar = 50 μm.
In the olfactory sensory epithelium, no histopathological changes were observed, although the olfactory nerve had some degenerative attributes, characterized by a shrunken cytoplasm of axons that were visible in transversal sections. Similar histopathological changes were observed in the telencephalon and rhombencephalon (Figure 6A). In the external and internal cellular layers of the olfactory bulb, multiple vacuolations were observed in intracellular matter, but no neural cytopathological changes were found. Widespread foamy vacuolation of the cytoplasm of neurons and glia and neuronal spheroids were present in the npc2–/– telencephalon and optic tectum (mostly in the stratum fibroetgricialem). Similar changes were observed in the olfactory bulb and optic nerve (Figures 7B,C). CD3-positive cells were observed in npc2–/– fish, whereas no signal was detected in the wildtype group (Supplementary Figure 4B).
We determined the level of gene expression in wildtype and npc2–/– fish at two developmental stages. Homozygous larvae were preselected based on Nile blue staining. The genotype of both 5-day-old larvae and adult fish was confirmed by HRM analysis. In whole larvae, the mbp, cldn, and olig1 genes were down-regulated, whereas expression of the mpz, plp1, and sox10 genes was unchanged. In the npc2–/– adult brain, a significant reduction of expression was detected only for the mbp and mpz genes (Figure 8). The downregulation of myelination markers was consistent with the decrease in LFB staining (Figure 6A).
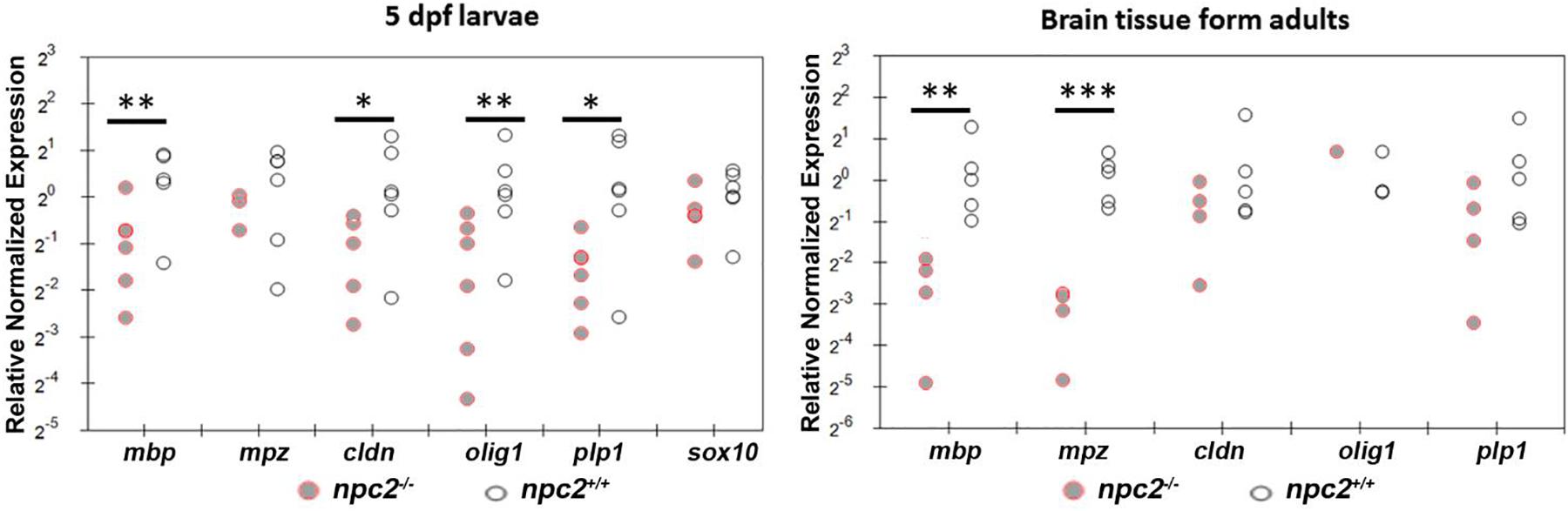
Figure 8. Decrease in myelination in npc2-deficient larvae and adult fish. Scatter plots show the normalized expression of selected genes in 5 dpf larvae and in the brain in npc–/– and wildtype zebrafish. Each circle corresponds to one zebrafish. The 18S ribosomal gene was used as a reference. At least three samples were analyzed. ***p < 0.001; **p < 0.01; and *p < 0.05.
Alterations of Expression of Genes That Shape Inflammation, Autophagy and Ca2+ Signaling in npc2–/– Fish
The downregulation of mbp expression is associated with the upregulation of interleukin-1 signaling in npc1–/– animal models (Mannie et al., 1987; Qiao et al., 2018). To determine whether npc2–/– zebrafish exhibit hallmarks of inflammatory responses that are associated with NPC disease, we profiled the expression of selected genes in the brain and liver in 9-month-old fish. In the brain, the upregulation of il1, nfκβ, mpeg, and capn2a was detected (Figure 9). In the liver, il1 expression but not nfκβ, mpeg, or capn2a expression was also up-regulated. However, in contrast to the brain, mpx, apoE, and ppp3ca expression was up-regulated (Figure 9).
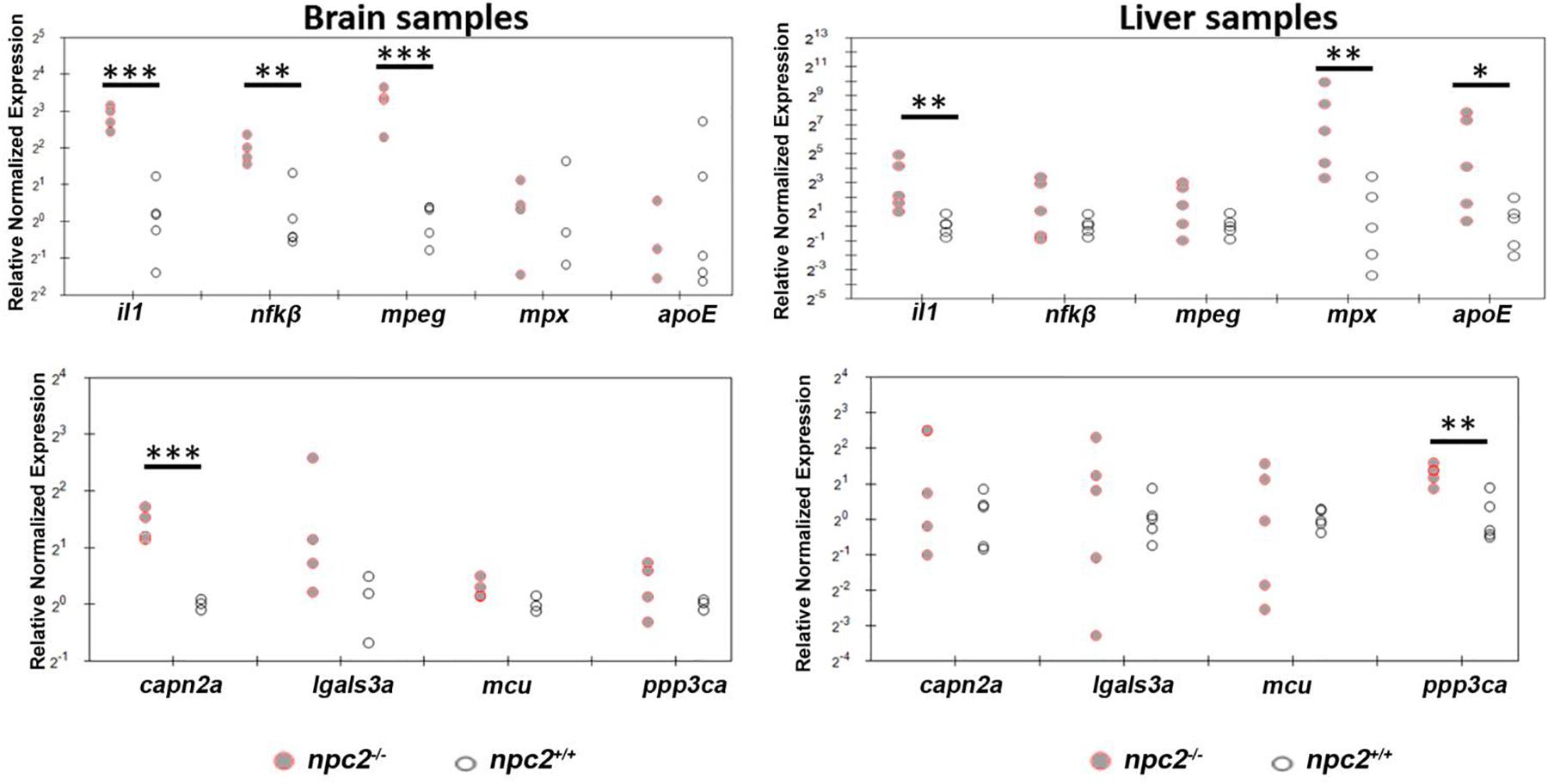
Figure 9. Signs of inflammation and alterations of Ca2+ homeostasis in npc2–/– zebrafish. Scatterplots show the normalized expression of selected genes in brain and liver tissue from 9-month-old npc2–/– homozygotes and wildtype zebrafish. Each circle corresponds to one zebrafish. The 18S ribosomal gene was used as a reference. Samples from at least three fish were analyzed. ***p < 0.001, **p < 0.01, and *p < 0.05.
In addition, we stained zebrafish larvae with Neutral red, a dye which specifically labels microglia in the larval zebrafish brain. In 5 dpf npc2–/– larvae, we found numerous red-labeled cells with dark red aggregates having typical appearance of over-activated micro-microglia, i.e., soma size was enlarged and cells had amoeboid shape (Supplementary Figure 5). Beclin-1 plays an important role autophagy during neurodegeneration. We found that npc2–/– larvae show slightly increased expression level of becn1, which is in line with previous finding of Pacheco and colleagues who demonstrated that enhanced basal autophagy in NPC1 deficiency is mediated by increased expression of Beclin-1 (Pacheco et al., 2007). We did not detect significant changes in the expression level of markers of ubiquitin-dependent stress-induced autophagy (atg5, atg7, and p62; Supplementary Figure 6).
We also found that apoE, capn2a, lgals3a, mcu, mpeg, and ppp3ca expression were down-regulated in 5 dpf npc2–/– larvae compared with wildtype fish (Figure 10A). The most striking difference was related to the level of mcu, with very high statistical significance. These changes appeared to be specific to NPC disease because 2HPβCD treatment rescued the levels of mcu and ppp3ca expression and the expression of genes that are associated with myelination (olig1, mbp, cldn, and plp1; Figures 8, 10B).
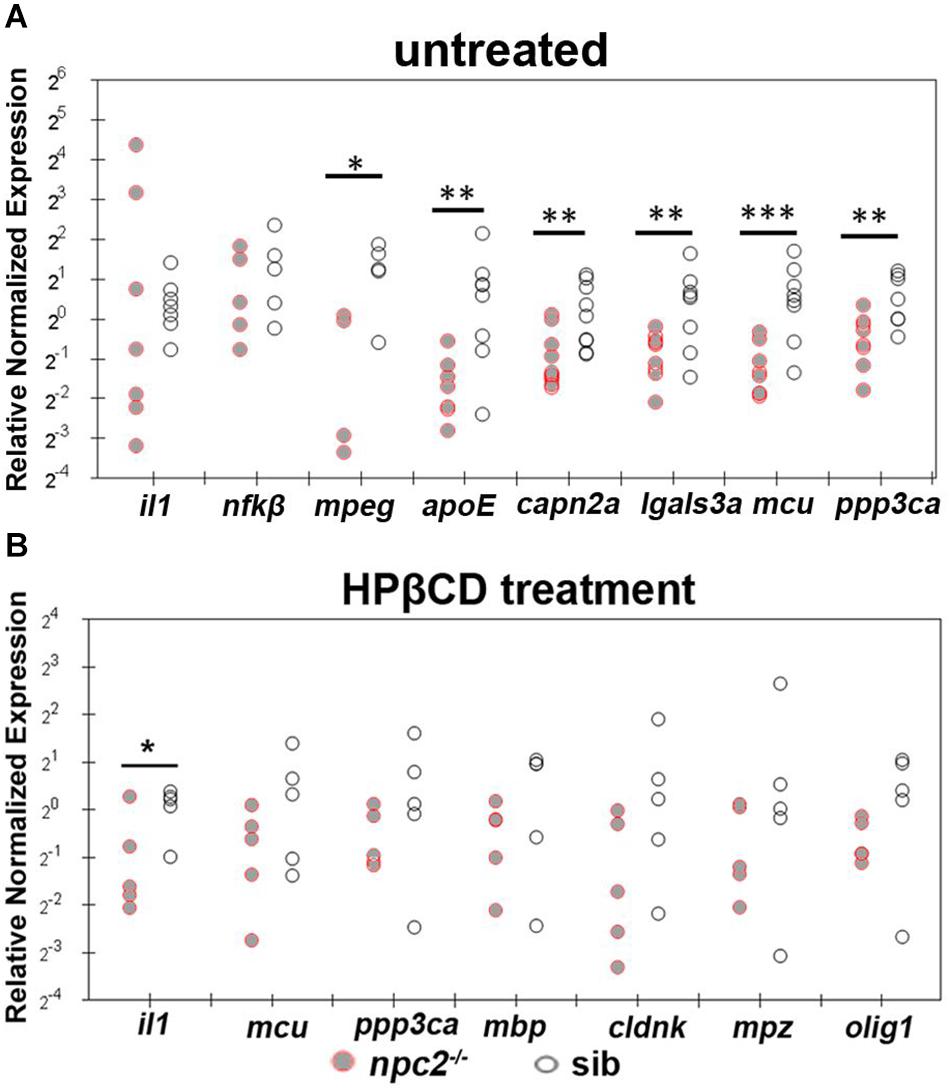
Figure 10. Treatment with 2-hydroxypropyl-β-cyclodextrin increased the expression of genes that are related to inflammation and Ca2+ homeostasis in 5 dpf npc2–/– larvae to wildtype levels. Scatterplots show the normalized expression of selected genes in untreated larvae (A) and larvae that were treated with 2 mM 2HPβCD (B) at 5 dpf. Each circle corresponds to one zebrafish. The 18S ribosomal gene was used as a reference. The experiments were performed in triplicate. ***p < 0.001, **p < 0.01, *p < 0.05.
Discussion
Niemann-Pick type C disease is a rare disease. The elucidation of its pathogenesis can help understand the molecular mechanisms that underlie more common disorders that are related to impairments in cholesterol homeostasis, autophagy, inflammation, and neurodegeneration. Accumulating evidence links cholesterol transport inhibition with lower susceptibility to viral infection, including SARS-CoV-2 (Gong et al., 2016; Wec et al., 2016; Stoeck et al., 2018; Ballout et al., 2020; Sturley et al., 2020; Vial et al., 2020), and studies of NPC genes are highly relevant. Of two human NPC genes, NPC1 and its homologs have been the most studied (Fog and Kirkegaard, 2019; Wheeler and Sillence, 2020). Various tools have been created and successfully applied to advance our knowledge of NPC1 function in normal and pathological processes (Lin et al., 2018; Tseng et al., 2018; Fog and Kirkegaard, 2019). Surprisingly, little is known about NPC2, a soluble lysosomal protein that delivers cholesterol to NPC1 (Deffieu and Pfeffer, 2011). Homologs of both the human NPC1 and NPC2 genes are conserved in zebrafish. Zebrafish npc1 mutants were recently described (Lin et al., 2018; Tseng et al., 2018). To our knowledge, no zebrafish mutant is available for npc2. To fill this gap in the literature, we created and characterized homozygous npc2–/– fish.
Clinical manifestations in humans who are deficient in NPC1 or NPC2 are similar. Hence, unsurprising is that npc2–/– zebrafish in many ways resemble the phenotype of npc1–/– fish. In the present study, we found that the manifestation of the npc2 mutation resembles changes that are seen in human patients. NPC disease can be highly heterogeneous in patients (Vanier, 2010; Reunert et al., 2016). In npc2 mutant fish, we observed variability in symptoms and their intensity. Similar to patients, npc2–/– fish exhibited clear signs of neuropathy and cytological changes in the liver and pancreas. Fish do not have bone marrow, and hematopoietic processes are conducted by the thymus, spleen, and anterior kidney. These three major hematopoietic organs were affected, and cells with a characteristic foam cytoplasm were observed, similar to several human diseases (Eom et al., 2015) and animal models (Kuemmel et al., 1997) of renal degeneration. Foam cells within the npc2–/– brain were also present in NPC patients and mouse models. Defects in Purkinje cells in the cerebellum were found in adult npc2–/– fish. In addition to histological changes, npc2–/– fish also recapitulated molecular changes, including alterations of the expression of genes that are involved in myelination and inflammation. However, gene expression was not changed in the same manner in larvae and adults. This is unsurprising because adult fish were severely affected, whereas the disease just started to progress in 5-day-old larvae.
Links between mitochondria, lysosomes, and neurodegeneration have been confirmed previously (Saffari et al., 2017; Torres et al., 2017). Mitochondrial dysfunctions have also been described in human cells with a mutation the human NPC1 gene and in corresponding mouse models (Woś et al., 2016; Erickson et al., 2020). We found a significant decrease in expression of the mcu gene in npc2–/– larvae. 2HPβCD treatment rescued expression levels. To our knowledge, this is the first study that found that impairments in calcium uptake into mitochondria may contribute to NPC disease. We previously reported that mcu knockout can rescue pink1 mutant zebrafish, a model of Parkinson’s disease, from the loss of dopaminergic neurons (Soman et al., 2019). Alterations of calcium homeostasis and signaling are not restricted to mitochondria. NPC disease was linked to a decrease in the expression of Ppp3ca in the mouse cerebellum (Reddy et al., 2006). Significant reductions of ppp3ca expression were also observed in npc2–/– larvae, whereas expression in the adult mutant brain appeared to be unaffected. We also found an increase in ppp3ca expression in the npc2–/– liver. The regulation of expression levels may be tissue-dependent and vary within more complex structures. In such a case, the analysis of the whole brain could have masked changes that occurred in the cerebellum.
In humans and zebrafish, the loss of one allele of the NPC1 or NPC2 gene is insufficient to cause NPC disease. The homozygous mutation of either npc gene leads to premature death. Some individuals can live longer, whereas some die at a young age. The prevalence of the early infantile form of NPC disease is much higher in children with a mutation of the NPC1 gene compared with patients with the mutation of NPC2 (Seker Yilmaz et al., 2020). The majority of npc2–/– fish live longer and can reach the age of 4 months, whereas the majority of npc1–/– fish die within the first month of life. Assuming that 1 month of zebrafish life corresponds to 2 years of human life, the lifespan of patients and npc2–/– zebrafish is similarly affected.
A link between Niemann-Pick disease and growth restriction has been established for Niemann-Pick type B disease, which is a lysosomal storage disease that is associated with the accumulation of sphingolipids (Wasserstein et al., 2003). This link has not yet been established for NPC disease. Homozygous mutations of npc1 (Lin et al., 2018; Tseng et al., 2018) and npc2 (present study) in zebrafish result in significant reductions of body length and weight compared with wildtype and heterozygous siblings. Tseng et al. suggested that “the growth defect observed in the npc1 mutant zebrafish may be due to impaired feeding” (Tseng et al., 2018). Lawrence et al. reported that the sex ratio in zebrafish is strongly influenced by the feeding regimen, and fish that eat more are more likely to become female (Lawrence et al., 2008). npc2–/– fish and their heterozygous and wildtype siblings occupied the same tank. The sex ratio is skewed in npc2 mutants, and npc2–/– fish tend to be males, whereas their wildtype siblings tend to be mostly females. Thus, growth restriction and male-rich population of npc2–/– fish may be at least partially attributable to an inability to take food. Moreover, anatomical and histological changes in the olfactory and optic neuron tracts and degenerative changes in mechanosensory fibers may hinder the ability of npc2–/– fish to find food.
Despite the smaller body size, adult npc2–/– fish produced gametes, but our attempts to inbreed these fish were unsuccessful. A similar problem was reported for Npc2–/– mice (Busso et al., 2010, 2014) and npc1–/– zebrafish (Tseng et al., 2018). However, we found that male npc2–/– fish could occasionally mate with npc2± female fish, resulting in viable offspring. This finding is consistent with the observation by Busso et al. (2014) that male Npc2–/– mice generated sperm that could fertilize eggs in vitro, although they did not mate. These authors suggested that locomotor dysfunction prevents male mice from breeding, but other factors might contribute to this problem. Abnormal cerebellar morphogenesis could be one of the factors (Caporali et al., 2016). Mating behavior involves social integrations, in which odor communication plays an important role. Impairments in the sense of smell is an early marker of neurodegenerative diseases. We found pathological changes in the olfactory system in npc2–/– fish. Sensory deficits that are associated with pronounced neurodegeneration in the olfactory system have also been described in Npc1–/– mice (Hovakimyan et al., 2013). Therefore, tremor, defective movements, and an inability to respond to chemical cues likely prevented animals with NPC disease from successfully breeding.
Niemann-Pick type C disease patients often exhibit neuropsychiatric symptoms (Evans and Hendriksz, 2017; Mengel et al., 2017; Patterson et al., 2017). In behavioral tests, npc2–/– larvae exhibited a range of behavioral changes, including an increase in anxiety-like behavior upon stress exposure. Studies showing decreased exploratory activity, hyperactivity and reduced anxiety were performed on mice, but only on adult animals which motor skills already were impaired and hence their performance may have been affected (Võikar et al., 2002; Gómez-Grau et al., 2017). Similar to fish, behavioral changes can be quantified in 5-day-old tremor-free animals, and disease progression can be followed over time. The test may be scalable. The zebrafish model of NPC disease presents unique features that may help efficiently and reliably detect early changes, monitor disease progression, and assay drug efficiency.
Although several NPC models exist, many gaps still need to be filled to improve our understanding of this disease. The npc2 mutant makes an important contribution to the range of existing models that allow further studies of NPC disease and other neurodegenerative cholesterol-dependent disorders at the organismal, cellular, and molecular levels. Experiments in zebrafish are scalable and suited for high-throughput drug screens. The zebrafish npc2 mutant may be utilized as a helpful model for the drug development pipeline in early stages of NPC disease. Another issue is the clinical diagnostic value of Nile blue, a dye that specifically provides a strong stain in the nose in npc2–/– larvae.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
All activities were performed in compliance with fundamental ethical principles (Protection of animals used for scientific or educational purposes, Act of January 15, 2015; Directive 2010/63/EU). The majority of the experiments were performed in 5-day-old zebrafish larvae (a stage at which zebrafish are not considered protected animals). The experiments did not qualify as a subject of review by the local ethical committee. In the case of adult fish, recording their natural behavior was performed without any interference. The fish remained in their home tank, and no handling or any other intervention that could cause any pain or distress was applied to them. Consequently, this action did not classify as a procedure and did not require any permit from the ethics committee.
Author Contributions
MW, LM, IW, and JK conceived and planned the experiments. MW, LM, IW, and DA-U performed the experiments. MW, LM, IW, and DA-U contributed to sample preparation. MW, LM, IW, DA-U, and JK contributed to interpretation of the results. MW took the lead in writing the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Funding
Statutory funding was received from the International Institute of Molecular and Cell Biology in Warsaw.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Maria Teresa Fiorenza and Giampiero Palladino from the Sapienza University of Rome for suggesting to generate the npc2 mutant fish and designing the IT8 gRNAs, Michal Pastor and Matthias Bochtler from IIMCB for providing Cas9 protein, Vladimir Korzh and Tomasz Wegierski from the Laboratory of Neurodegeneration at IIMCB for critically reading the manuscript, Michael Arends from Arends Editing for linguistic proofreading, and staff members of the Zebrafish Core Facility at IIMCB for daily care of the zebrafish. IW is a student at the Postgraduate School of Molecular Medicine, Warsaw Medical University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2021.647860/full#supplementary-material
Supplementary Image 1 | Un-esterified cholesterol accumulation in the npc2 mutant.
Supplementary Image 2 | LysoTracker red staining of the npc2 mutant.
Supplementary Image 3 | Morphology of the npc2–/– brain.
Supplementary Image 4 | Immunohistochemical localization of proliferating nuclear antigen and CD3 in central nervous system tissue in npc2–/– fish. A.
Supplementary Image 5 | Neutral red staining of the npc2 mutant at 5 dpf.
Supplementary Image 6 | Expression of the autophagy markers in npc2-deficient larvae and adult fish.
Supplementary Video 1 | Natural behavior in adult npc2–/– fish differs from wildtype controls. Similar to human patients, locomotor functions were impaired in npc2-deficient fish, reflected by uncoordinated swimming.
Supplementary Video 2 | Natural behavior in adult wildtype fish.
Footnotes
- ^ https://rarediseases.org/rare-diseases/niemann-pick-disease-type-c/
- ^ http://chopchop.cbu.uib.no/
- ^ http://www.e-crisp.org/E-CRISP/
- ^ http://www.crisprscan.org/?page=gene
- ^ https://crispr.cos.uni-heidelberg.de/index.html
References
Anderson, J. L., Carten, J. D., and Farber, S. A. (2011). Zebrafish lipid metabolism: from mediating early patterning to the metabolism of dietary fat and cholesterol. Methods Cell Biol. 101, 111–141. doi: 10.1016/B978-0-12-387036-0.00005-0
Ballout, R. A., Sviridov, D., Bukrinsky, M. I., and Remaley, A. T. (2020). The lysosome: a potential juncture between SARS-CoV-2 infectivity and Niemann-Pick disease type C, with therapeutic implications. FASEB J. 34, 7253–7264. doi: 10.1096/fj.202000654R
Baudry, M., Yao, Y., Simmons, D., Liu, J., and Bi, X. (2003). Postnatal development of inflammation in a murine model of Niemann-Pick type C disease: immunohistochemical observations of microglia and astroglia. Exp. Neurol. 184, 887–903. doi: 10.1016/S0014-4886(03)00345-5
Busso, D., Oñate-Alvarado, M. J., Balboa, E., Castro, J., Lizama, C., Morales, G., et al. (2014). Spermatozoa from mice deficient in Niemann-Pick disease type C2 (NPC2) protein have defective cholesterol content and reduced in vitro fertilising ability. Reprod. Fertil. Dev. 26, 609–621. doi: 10.1071/rd12059
Busso, D., Oñate-Alvarado, M. J., Balboa, E., Zanlungo, S., and Moreno, R. D. (2010). Female infertility due to anovulation and defective steroidogenesis in NPC2 deficient mice. Mol. Cell. Endocrinol. 315, 299–307. doi: 10.1016/j.mce.2009.10.011
Caporali, P., Bruno, F., Palladino, G., Dragotto, J., Petrosini, L., Mangia, F., et al. (2016). Developmental delay in motor skill acquisition in Niemann-Pick C1 mice reveals abnormal cerebellar morphogenesis. Acta Neuropathol. Commun. 4:94. doi: 10.1186/s40478-016-0370-z
Cologna, S. M., Cluzeau, C. V., Yanjanin, N. M., Blank, P. S., Dail, M. K., Siebel, S., et al. (2014). Human and mouse neuroinflammation markers in Niemann-Pick disease, type C1. J. Inherit. Metab. Dis. 37, 83–92. doi: 10.1007/s10545-013-9610-6
de Araujo Boleti, A. P., de Oliveira Flores, T. M., Moreno, S. E., Anjos, L. D., Mortari, M. R., and Migliolo, L. (2020). Neuroinflammation: an overview of neurodegenerative and metabolic diseases and of biotechnological studies. Neurochem. Int. 136:104714. doi: 10.1016/j.neuint.2020.104714
Deffieu, M. S., and Pfeffer, S. R. (2011). Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc. Natl. Acad. Sci. U.S.A. 108, 18932–18936. doi: 10.1073/pnas.1110439108
Elrick, M. J., Pacheco, C. D., Yu, T., Dadgar, N., Shakkottai, V. G., Ware, C., et al. (2010). Conditional Niemann-Pick C mice demonstrate cell autonomous Purkinje cell neurodegeneration. Hum. Mol. Genet. 19, 837–847. doi: 10.1093/hmg/ddp552
Eom, M., Hudkins, K. L., and Alpers, C. E. (2015). Foam cells and the pathogenesis of kidney disease. Curr. Opin. Nephrol. Hypertens. 24, 245–251. doi: 10.1097/mnh.0000000000000112
Erickson, R. P., Aras, S., Purandare, N., Hüttemann, M., Liu, J., Dragotto, J., et al. (2020). Decreased membrane cholesterol in liver mitochondria of the point mutation mouse model of juvenile Niemann-Pick C1, Npc1(nmf164). Mitochondrion 51, 15–21. doi: 10.1016/j.mito.2019.12.003
Evans, W. R., and Hendriksz, C. J. (2017). Niemann-Pick type C disease - the tip of the iceberg? A review of neuropsychiatric presentation, diagnosis and treatment. BJPsych. Bull. 41, 109–114. doi: 10.1192/pb.bp.116.054072
Fog, C. K., and Kirkegaard, T. (2019). Animal models for Niemann-Pick type C: implications for drug discovery & development. Expert Opin. Drug Discov. 14, 499–509. doi: 10.1080/17460441.2019.1588882
Gagnon, J. A., Valen, E., Thyme, S. B., Huang, P., Akhmetova, L., Pauli, A., et al. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9:e98186. doi: 10.1371/journal.pone.0098186
Gattuso, H., Besancenot, V., Grandemange, S., Marazzi, M., and Monari, A. (2016). From non-covalent binding to irreversible DNA lesions: nile blue and nile red as photosensitizing agents. Sci. Rep. 6:28480. doi: 10.1038/srep28480
Geberhiwot, T., Moro, A., Dardis, A., Ramaswami, U., Sirrs, S., Marfa, M. P., et al. (2018). Consensus clinical management guidelines for niemann-pick disease type C. Orphanet. J. Rare Dis. 13:50. doi: 10.1186/s13023-018-0785-7
Gómez-Grau, M., Albaigès, J., Casas, J., Auladell, C., Dierssen, M., Vilageliu, L., et al. (2017). New murine Niemann-Pick type C models bearing a pseudoexon-generating mutation recapitulate the main neurobehavioural and molecular features of the disease. Sci. Rep. 7:41931. doi: 10.1038/srep41931
Gong, X., Qian, H., Zhou, X., Wu, J., Wan, T., Cao, P., et al. (2016). Structural insights into the niemann-pick C1 (NPC1)-mediated cholesterol transfer and ebola infection. Cell 165, 1467–1478. doi: 10.1016/j.cell.2016.05.022
Hovakimyan, M., Meyer, A., Lukas, J., Luo, J., Gudziol, V., Hummel, T., et al. (2013). Olfactory deficits in Niemann-Pick type C1 (NPC1) disease. PLoS One 8:e82216. doi: 10.1371/journal.pone.0082216
Kavetsky, L., Green, K. K., Boyle, B. R., Yousufzai, F. A. K., Padron, Z. M., Melli, S. E., et al. (2019). Increased interactions and engulfment of dendrites by microglia precede Purkinje cell degeneration in a mouse model of Niemann Pick Type-C. Sci. Rep. 9:14722. doi: 10.1038/s41598-019-51246-1
Ko, D. C., Milenkovic, L., Beier, S. M., Manuel, H., Buchanan, J., and Scott, M. P. (2005). Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet. 1:e0010007. doi: 10.1371/journal.pgen.0010007
Kuemmel, T. A., Thiele, J., Schroeder, R., and Stoffel, W. (1997). Pathology of visceral organs and bone marrow in an acid sphingomyelinase deficient knock-out mouse line, mimicking human Niemann-Pick disease type A. A light and electron microscopic study. Pathol. Res. Pract. 193, 663–671. doi: 10.1016/s0344-0338(97)80025-8
Lawrence, C., Ebersole, J. P., and Kesseli, R. V. (2008). Rapid growth and out-crossing promote female development in zebrafish (Danio rerio). Environ. Biol. Fish. 81, 239–246. doi: 10.1007/s10641-007-9195-8
Lillie, R. D. (1958). The Nile blue reaction of peptic gland zymogen granules: the effect of methylation and alkali demethylation. J. Histochem. Cytochem. 6, 130–132. doi: 10.1177/6.2.130
Lin, Y., Cai, X., Wang, G., Ouyang, G., and Cao, H. (2018). Model construction of Niemann-Pick type C disease in zebrafish. Biol. Chem. 399, 903–910. doi: 10.1515/hsz-2018-0118
Lundegaard, P. R., Anastasaki, C., Grant, N. J., Sillito, R. R., Zich, J., Zeng, Z., et al. (2015). MEK inhibitors reverse cAMP-mediated anxiety in Zebrafish. Chem. Biol. 22, 1335–1346. doi: 10.1016/j.chembiol.2015.08.010
Majewski, L., Wojtas, B., Maciag, F., and Kuznicki, J. (2019). Changes in calcium homeostasis and gene expression implicated in epilepsy in hippocampi of mice overexpressing ORAI1. Int. J. Mol. Sci. 20:5539. doi: 10.3390/ijms20225539
Mannie, M. D., Dinarello, C. A., and Paterson, P. Y. (1987). Interleukin 1 and myelin basic protein synergistically augment adoptive transfer activity of lymphocytes mediating experimental autoimmune encephalomyelitis in Lewis rats. J. Immunol. 138, 4229–4235.
Martinez, V., and Henary, M. (2016). Nile red and nile blue: applications and syntheses of structural analogues. Chemistry 22, 13764–13782. doi: 10.1002/chem.201601570
Mengel, E., Pineda, M., Hendriksz, C. J., Walterfang, M., Torres, J. V., and Kolb, S. A. (2017). Differences in Niemann-Pick disease Type C symptomatology observed in patients of different ages. Mol. Genet. Metab. 120, 180–189. doi: 10.1016/j.ymgme.2016.12.003
Orchard, G. E. (2019). “Pigments and minerals,” in Bancroft’s Theory and Practice of Histological Techniques, 8th Edn, eds S. K. Suvarna, C. Layton, and J. D. Bancroft (New York, NY: Elsevier), 198–230.
Pacheco, C. D., Kunkel, R., and Lieberman, A. P. (2007). Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum. Mol. Genet. 16, 1495–1503. doi: 10.1093/hmg/ddm100
Patterson, M. C., Clayton, P., Gissen, P., Anheim, M., Bauer, P., Bonnot, O., et al. (2017). Recommendations for the detection and diagnosis of Niemann-Pick disease type C: an update. Neurol. Clin. Pract. 7, 499–511. doi: 10.1212/cpj.0000000000000399
Platt, N., Speak, A. O., Colaco, A., Gray, J., Smith, D. A., Williams, I. M., et al. (2016). Immune dysfunction in niemann-pick disease type C. J. Neurochem. 136(Suppl. 1), 74–80. doi: 10.1111/jnc.13138
Qiao, L., Yang, E., Luo, J., Lin, J., and Yan, X. (2018). Altered myelination in the niemann-pick type C1 mutant mouse. Histol. Histopathol. 33, 1311–1321. doi: 10.14670/hh-18-017
Reddy, J. V., Ganley, I. G., and Pfeffer, S. R. (2006). Clues to neuro-degeneration in Niemann-Pick type C disease from global gene expression profiling. PLoS One 1:e19. doi: 10.1371/journal.pone.0000019
Reunert, J., Fobker, M., Kannenberg, F., Du Chesne, I., Plate, M., Wellhausen, J., et al. (2016). Rapid diagnosis of 83 patients with niemann pick type C disease and related cholesterol transport disorders by cholestantriol screening. EBioMedicine 4, 170–175. doi: 10.1016/j.ebiom.2015.12.018
Saffari, A., Kölker, S., Hoffmann, G. F., and Ebrahimi-Fakhari, D. (2017). Linking mitochondrial dysfunction to neurodegeneration in lysosomal storage diseases. J. Inherit. Metab. Dis. 40, 631–640. doi: 10.1007/s10545-017-0048-0
Sarna, J. R., Larouche, M., Marzban, H., Sillitoe, R. V., Rancourt, D. E., and Hawkes, R. (2003). Patterned Purkinje cell degeneration in mouse models of Niemann-Pick type C disease. J. Comp. Neurol. 456, 279–291. doi: 10.1002/cne.10522
Seker Yilmaz, B., Baruteau, J., Rahim, A. A., and Gissen, P. (2020). Clinical and molecular features of early infantile niemann pick type C disease. Int. J. Mol. Sci. 21:5059. doi: 10.3390/ijms21145059
Soman, S. K., Bazala, M., Keatinge, M., Bandmann, O., and Kuznicki, J. (2019). Restriction of mitochondrial calcium overload by mcu inactivation renders a neuroprotective effect in zebrafish models of Parkinson’s disease. Biol. Open 8:bio044347. doi: 10.1242/bio.044347
Stoeck, I. K., Lee, J. Y., Tabata, K., Romero-Brey, I., Paul, D., Schult, P., et al. (2018). Hepatitis C virus replication depends on endosomal cholesterol homeostasis. J. Virol. 92:e01196-17. doi: 10.1128/jvi.01196-17
Sturley, S. L., Rajakumar, T., Hammond, N., Higaki, K., Márka, Z., Márka, S., et al. (2020). Potential COVID-19 therapeutics from a rare disease: weaponizing lipid dysregulation to combat viral infectivity. J. Lipid Res. 61, 972–982. doi: 10.1194/jlr.R120000851
Torres, S., Balboa, E., Zanlungo, S., Enrich, C., Garcia-Ruiz, C., and Fernandez-Checa, J. C. (2017). Lysosomal and mitochondrial liaisons in niemann-pick disease. Front. Physiol. 8:982. doi: 10.3389/fphys.2017.00982
Tseng, W. C., Loeb, H. E., Pei, W., Tsai-Morris, C. H., Xu, L., Cluzeau, C. V., et al. (2018). Modeling Niemann-Pick disease type C1 in zebrafish: a robust platform for in vivo screening of candidate therapeutic compounds. Dis. Model. Mech. 11:dmm034165. doi: 10.1242/dmm.034165
Vanier, M. T. (2010). Niemann-Pick disease type C. Orphanet. J. Rare Dis. 5:16. doi: 10.1186/1750-1172-5-16
Vial, C., Calderón, J. F., and Klein, A. D. (2020). NPC1 as a modulator of disease severity and viral entry of Sars-CoV-2. Curr. Mol. Med. 21, 2–4. doi: 10.2174/1566524020666200713175426
Võikar, V., Rauvala, H., and Ikonen, E. (2002). Cognitive deficit and development of motor impairment in a mouse model of Niemann-Pick type C disease. Behav. Brain Res. 132, 1–10. doi: 10.1016/s0166-4328(01)00380-1
Wasilewska, I., Gupta, R. K., Wojtas, B., Palchevska, O., and Kuznicki, J. (2020). stim2b knockout induces hyperactivity and susceptibility to seizures in Zebrafish larvae. Cells 9:1285. doi: 10.3390/cells9051285
Wasserstein, M. P., Larkin, A. E., Glass, R. B., Schuchman, E. H., Desnick, R. J., and McGovern, M. M. (2003). Growth restriction in children with type B niemann-pick disease. J. Pediatr. 142, 424–428. doi: 10.1067/mpd.2003.113
Wec, A. Z., Nyakatura, E. K., Herbert, A. S., Howell, K. A., Holtsberg, F. W., Bakken, R. R., et al. (2016). A “Trojan horse” bispecific-antibody strategy for broad protection against ebolaviruses. Science 354, 350–354. doi: 10.1126/science.aag3267
Wheeler, S., and Sillence, D. J. (2020). Niemann-Pick type C disease: cellular pathology and pharmacotherapy. J. Neurochem. 153, 674–692. doi: 10.1111/jnc.14895
Woś, M., Szczepanowska, J., Pikuła, S., Tylki-Szymańska, A., Zabłocki, K., and Bandorowicz-Pikuła, J. (2016). Mitochondrial dysfunction in fibroblasts derived from patients with niemann-pick type C disease. Arch. Biochem. Biophys. 593, 50–59. doi: 10.1016/j.abb.2016.02.012
Yan, X., Lukas, J., Witt, M., Wree, A., Hubner, R., Frech, M., et al. (2011). Decreased expression of myelin gene regulatory factor in niemann-pick type C 1 mouse. Metab. Brain Dis. 26, 299–306. doi: 10.1007/s11011-011-9263-9
Keywords: Niemann-Pick type C, npc2, zebrafish model, Nile blue, lysosomal storage disease, neurodegeneration, inflammation, myelin
Citation: Wiweger M, Majewski L, Adamek-Urbanska D, Wasilewska I and Kuznicki J (2021) npc2-Deficient Zebrafish Reproduce Neurological and Inflammatory Symptoms of Niemann-Pick Type C Disease. Front. Cell. Neurosci. 15:647860. doi: 10.3389/fncel.2021.647860
Received: 30 December 2020; Accepted: 26 March 2021;
Published: 27 April 2021.
Edited by:
Grzegorz Kreiner, Polish Academy of Sciences (IF PAS), PolandReviewed by:
Martin Witt, Centre of Transdisciplinary Neuroscience Rostock, GermanyEwa Trojan, Polish Academy of Sciences (IF PAS), Poland
Copyright © 2021 Wiweger, Majewski, Adamek-Urbanska, Wasilewska and Kuznicki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Malgorzata Wiweger, bXdpd2VnZXJAaWltY2IuZ292LnBs
 Malgorzata Wiweger
Malgorzata Wiweger Lukasz Majewski
Lukasz Majewski Dobrochna Adamek-Urbanska
Dobrochna Adamek-Urbanska Iga Wasilewska1
Iga Wasilewska1