
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci., 02 November 2020
Sec. Non-Neuronal Cells
Volume 14 - 2020 | https://doi.org/10.3389/fncel.2020.573278
This article is part of the Research TopicNext-Generation Genetically-Encoded Fluorescent SensorsView all 10 articles
Understanding how building blocks of life contribute to physiology is greatly aided by protein identification and cellular localization. The two main labeling approaches developed over the past decades are labeling with antibodies such as immunoglobulin G (IgGs) or use of genetically encoded tags such as fluorescent proteins. However, IgGs are large proteins (150 kDa), which limits penetration depth and uncertainty of target position caused by up to ∼25 nm distance of the label created by the chosen targeting approach. Additionally, IgGs cannot be easily recombinantly modulated and engineered as part of fusion proteins because they consist of multiple independent translated chains. In the last decade single domain antigen binding proteins are being explored in bioscience as a tool in revealing molecular identity and localization to overcome limitations by IgGs. These nanobodies have several potential benefits over routine applications. Because of their small size (15 kDa), nanobodies better penetrate during labeling procedures and improve resolution. Moreover, nanobodies cDNA can easily be fused with other cDNA. Multidomain proteins can thus be easily engineered consisting of domains for targeting (nanobodies) and visualization by fluorescence microscopy (fluorescent proteins) or electron microscopy (based on certain enzymes). Additional modules for e.g., purification are also easily added. These nanobody-based probes can be applied in cells for live-cell endogenous protein detection or may be purified prior to use on molecules, cells or tissues. Here, we present the current state of nanobody-based probes and their implementation in microscopy, including pitfalls and potential future opportunities.
Defining protein identity and visualizing protein localization is fundamental in biology. Uncovering dynamics of protein localization and function were boosted when green fluorescent protein (GFP) and other fluorescent proteins (FPs) were developed and used to tag proteins of interest (Tsien, 1998; Giepmans et al., 2006; Rodriguez et al., 2017). Advantages of these chimeric fusion proteins include the lack of distance between protein of interest and label, thereby improving the resolution, as well as the specificity of labeling derived from the genetic fusion. Disadvantages include modification of the target protein, with the consequence that unmodified endogenous proteins cannot be studied (Giepmans et al., 2006). To detect endogenous proteins, immunolabeling using antibodies (immunoglobulins, mostly of the IgG isotype; IgGs) conjugated with small fluorophores are typically applied. However, for intracellular targeting IgGs require plasma membrane permeabilization leading to a damaged ultrastructure (Schnell et al., 2012). Furthermore, IgGs are large (∼150 kDa; ∼14 nm long; Table 1). This may result in a distance greater than 25 nm between target and label in indirect conventional immunolabeling, the so-called linkage error (Muyldermans, 2013; Mikhaylova et al., 2015). In addition, IgGs are multidomain proteins which require post-translational modifications (Muyldermans, 2013) and therefore preclude routine controlled genetic modification and modular expression in conjunction with e.g., GFP.
Nanobodies are single variable domains of heavy-chain only antibodies(hcAB) derived from Camelidae species (Hamers-Casterman et al., 1993; Muyldermans, 2013; Helma et al., 2015; Van Audenhove and Gettemans, 2016), but do not compromise in the binding-affinity compared to IgGs, due to its complementarity-determining region (CDR) organization (Muyldermans et al., 2001; Muyldermans, 2013; Beghein and Gettemans, 2017). Nanobodies have been explored since 2006 as labeling tools in light microscopy (LM) (Rothbauer et al., 2006), because of the several potential advantages of nanobodies over other labeling techniques. Nanobody-mediated targeting for protein identification is more precise than IgG targeting, as nanobodies are only ∼15 kDa with a diameter of 2–3 nm (Table 1) and can be encoded by a relative short stretch single cDNA of 360 base pairs (Van Audenhove and Gettemans, 2016; Traenkle and Rothbauer, 2017; Carrington et al., 2019). This cDNA can genetically be fused to FPs cDNAs for intracellular (live-cell) imaging or tags can be added for purification and chemical modifications. Like IgGs, customized nanobodies can be created against a protein of interest and the cDNA can be shared free of charge, as opposed to IgGs (Zuo et al., 2017; McMahon et al., 2018). Here, an overview is given about the past and potential future of nanobody application in microscopy.
Nanobodies (see Box 1 for terminology) can be expressed in cells conjugated to a detection module (like GFP) to target endogenous intracellular proteins, or they can be expressed, purified and then applied in immunolabeling resembling traditional immunofluorescence approach. Conventional immunolabeling is performed using IgGs, but for an improved penetration nanobodies can be used as an alternative (Fang et al., 2018). The improved penetration of nanobodies is illustrated in nuclear labeling of anti-GFP labeling targeting Histone2B (H2B)-GFP. Note that, in an equal labeling time, the nanobodies are colocalizing in the nucleus with the GFP, whereas the IgGs are mainly localized in the cytoplasm (Figure 1A). Though the anti-GFP is the best characterized and most used nanobody to date (Table 2), nanobodies are also used for visualizing endogenous proteins (Figure 1B and Table 2). Finally, nanobodies can be applied for live-cell imaging, both as purified proteins typically targeting extracellular antigens or being expressed from its introduced cDNA and targeted to intracellular antigens (Rothbauer et al., 2006; Ries et al., 2012).
Box 1 Nanobody terminology Nanobody
Synonyms: single domain antibody (sdAB), variable domain of heavy-chain only antibody (VhH), nAbs.
Small antigen binding protein, derived from heavy-chain only antibodies (hcAB). These are produced in cell culture or in bacteria.
Intrabody
The cDNA of this probe is expressed in the cells for intracellular antigen targeting.
Chromobody
Synonyms: Fluobody, fluorescent nanobody.
Genetic fusion of intrabody and fluorescent protein. Direct visualization with microscopy is possible.
Labeled nanobodies
Synonyms: fluobody, fluorescent nanobody.
Nanobodies protein purified and tagged in vitro with e.g., a chemical dye.
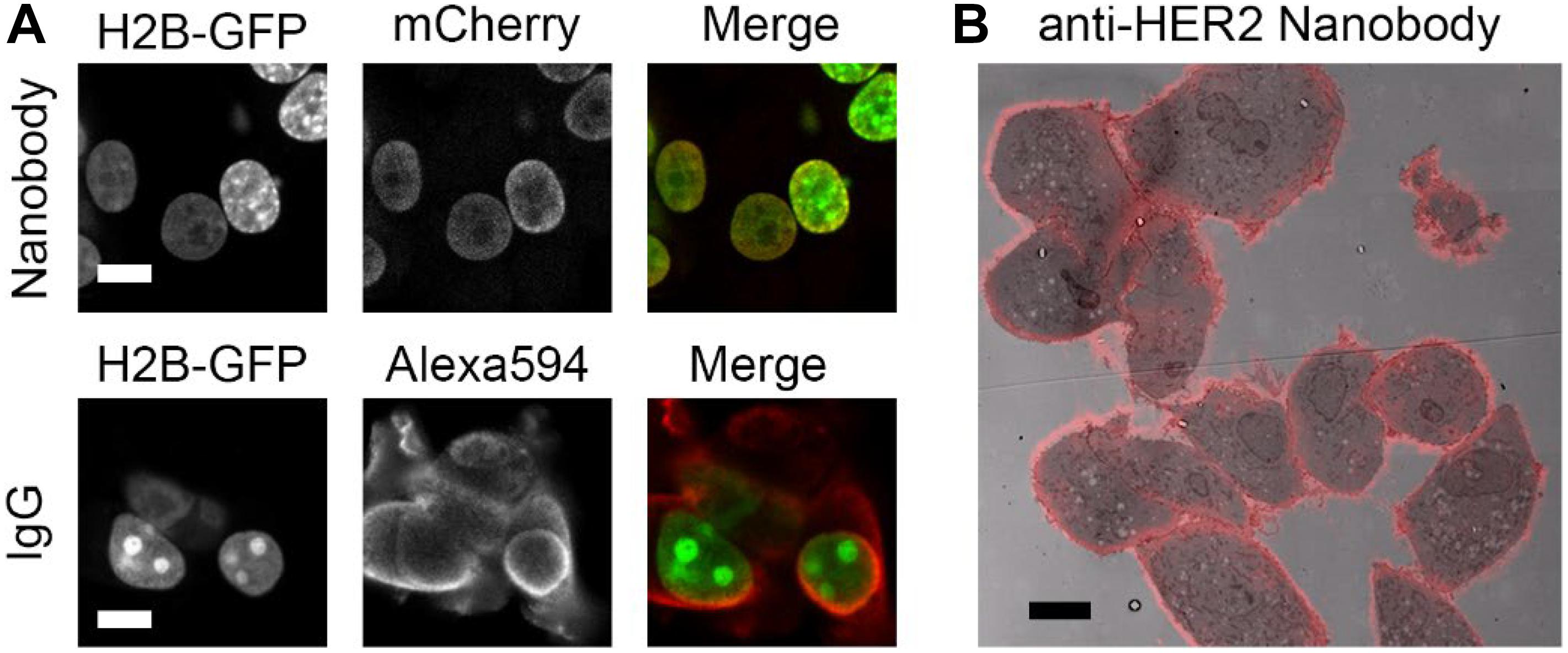
Figure 1. Nanobodies improve penetration and detect endogenous proteins. (A) Anti-GFP nanobody labeling (mCherry and peroxidase fused) and IgG labeling in H2B-GFP expressing cells. Cells permeabilized for 5 min with 0.1% Triton before labeling. Nanobodies and primary and secondary antibodies incubated for 1 h each. Note the colozalization between GFP and mCherry (nanobody), most prominently in the low-expressing cells, while Alexa Fluor 594 (IgG) mainly localizes in the cytoplasm. (B) High HER2 expressing cells, SkBr3, labeled with nanobodies targeting HER2. Overlay of nanobody fluoresence and EM image. Note the positive labeling at cell-cell contact sites. Bars: 10 μm. Reproduced from De Beer et al. (2018), http://creativecommons.org/licenses/by/4.0/.
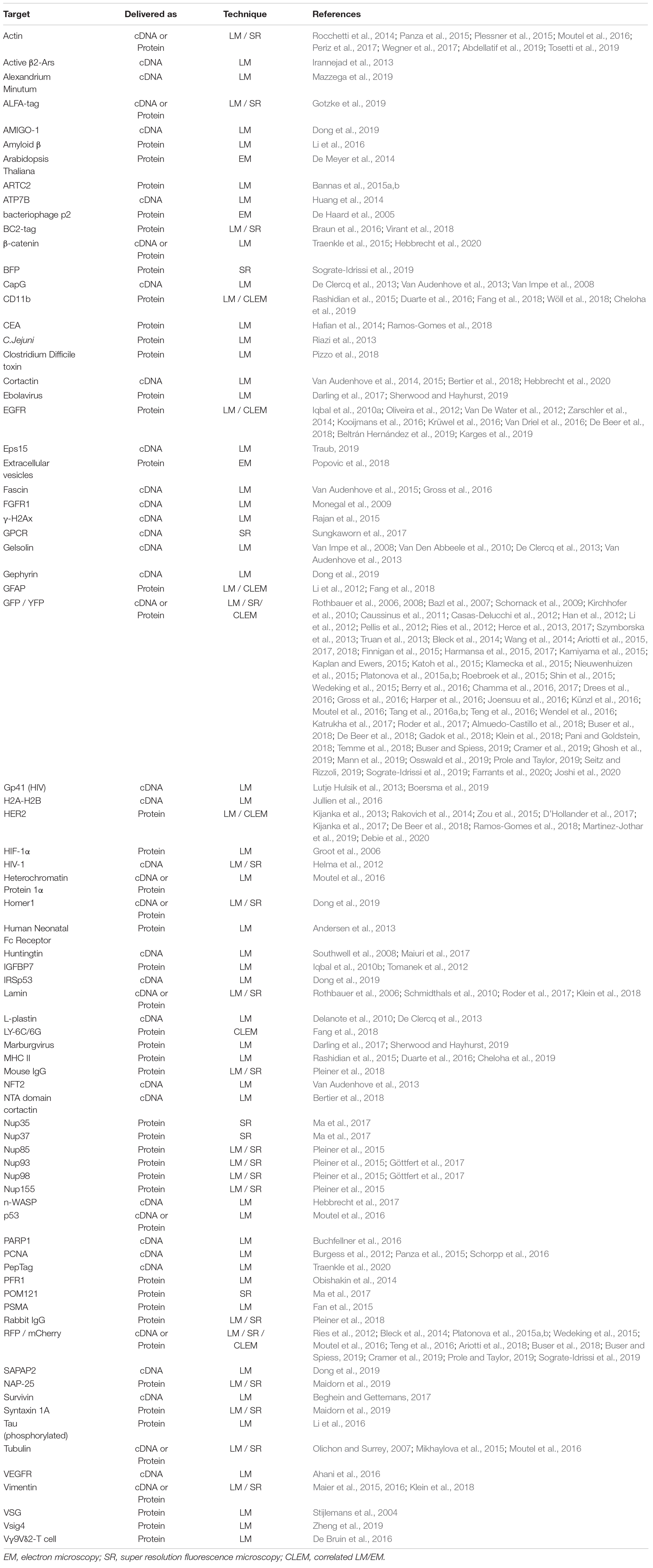
Table 2. Nanobody implemented in microscopy – An overview of targets that have been visualized using nanobodies and microscopy.
The first nanobody-based visualization of intracellular targets was achieved by the fusion to FPs (“chromobodies” Rothbauer et al., 2006; Table 1), being expressed in the target cells. Since then more nanobodies where developed targeting proteins inside cells (Table 2, delivered as cDNA). These intracellular chromobodies have been applied in small organisms like Danio rerio (Panza et al., 2015), Drosophila melanogaster (Harmansa et al., 2017), Caenorhabditis elegans (Pani and Goldstein, 2018) and Toxoplasma gondii (Tosetti et al., 2019) allowing live-cell and intravital microscopy. Intracellular expression of chromobodies in mice was successfully achieved by infecting the mice with adeno-associated viral particles containing the coding sequence of the chromobody (Figure 2A) (Wegner et al., 2017). Importantly, these chromobody approaches opens the opportunity for intravital imaging of endogenous proteins.
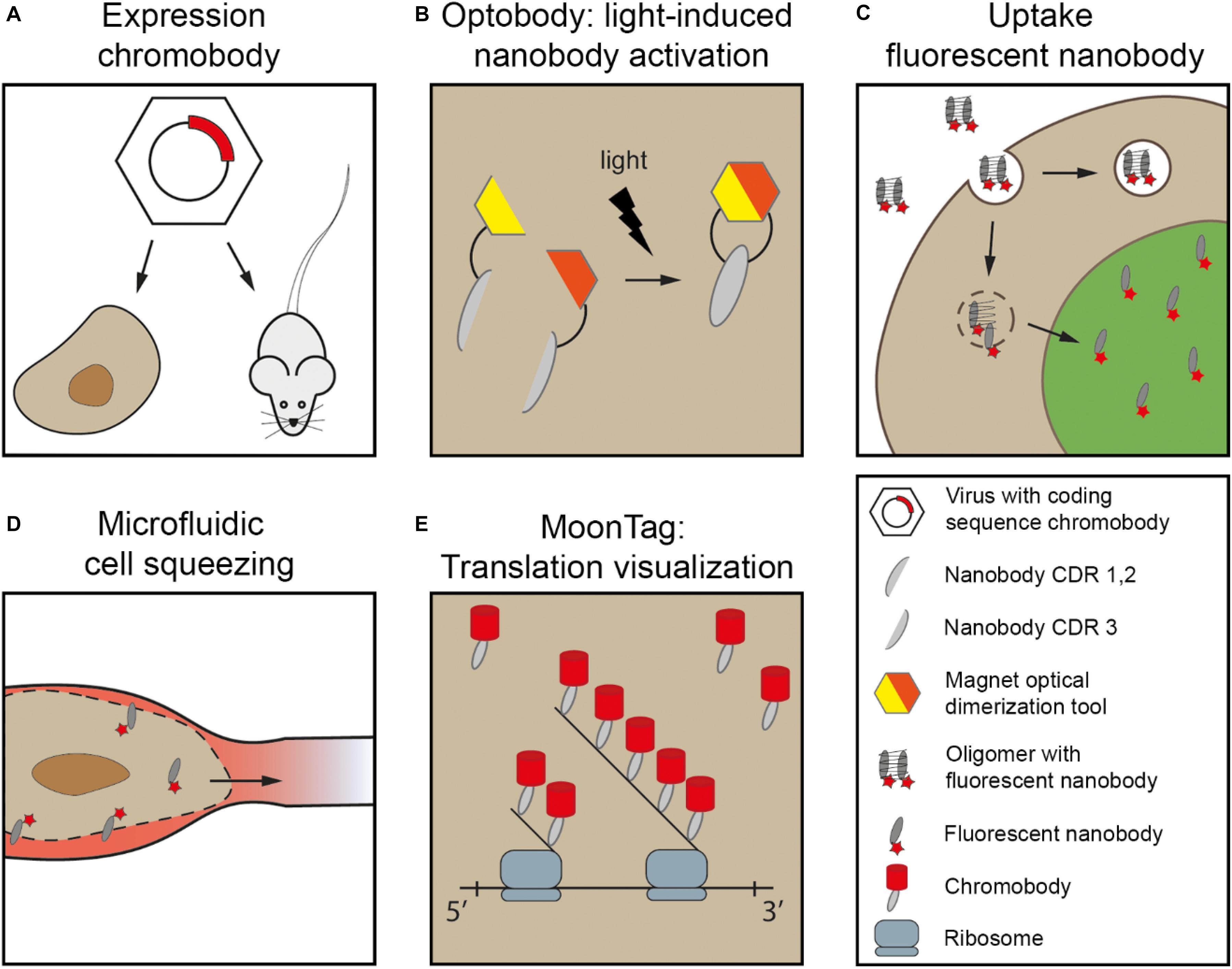
Figure 2. Nanobodies delivered for intracellular live-cell imaging. (A) Chromobodies cDNA loaded in e.g., adeno-associated viral particles (AAV). These viruses are used to infect cells in culture or in animals e.g., mouse. (B) Magnetic optical dimerization tool exists of two nMagHigh1 and pMagHigh1. Here, the nMagHigh1 is fused to nanobody fragment containing CDR1/2 and pMagHigh1 is fused to nanobody fragment containing CDR3. Upon stimulation of light, the magnetic tool paired together, resulting in restoring the nanobody. (C) Fluorescent nanobodies in e.g., oligomers can be taken up via endocytosis. Here, when endosomal rupture is induced, the free fluorescent nanobodies can bind to their target. (D) Microfluidic cell squeezing for temporary cell permeabilization. Purified fluorescent nanobodies are present in the medium, and diffuse into the accessible cytoplasm. (E) MoonTag: array of 24 peptide sequence repeat allows visualization of single molecules directly following translation by signal amplification. Chromobodies accumulate at the peptide chain. Major inspiration for this cartoon is from Roder et al. (2017), Klein et al. (2018), Boersma et al. (2019), Yu et al. (2019).
Next to defining protein localization, chromobodies can also relay functional changes in cells by fusing the nanobody to a fluorescent sensor for e.g., Ca2+ or pH (Prole and Taylor, 2019). For instance, anti-GFP nanobodies are fused to a Ca2+ sensor targeting GFP labeled mitochondria (Zhao et al., 2011). The nanobody facilitates the Ca2+ sensor to be in close proximity of the mitochondria to allow for Ca2+ dependent fluorescence readout. This results in the imaging of the local Ca2+ concentrations upon different stimuli. In the same study, anti-GFP nanobodies were conjugated to a SNAP-tag, a 20 kDa protein, modified from the human DNA repair protein O6-alkylguanine-DNA alkyltransferase (Keppler et al., 2003). The SNAP-tag is used to recruit a chemical dye, which facilitates live-cell imaging for Chromophore-Assisted Light Inactivation (CALI) (Jacobson et al., 2008): laser-induced subcellular destruction of a protein of interest. Thus nanobodies allow precise molecular targeting and enable analysis of the function of biomolecules, as well as precise protein manipulation with CALI allowing a direct cause/consequence study in living cells.
Despite the success of chromobodies as intracellular probes, a disadvantage is their continued presence. Chromobodies fluoresce whether or not they bind to their target, as opposed to immunolabeling techniques that include multiple wash-out steps for unbound reagents or genetic fusions between target and FPs. To reduce the signal from non-bound chromobodies, conditionally stable chromobodies have been developed (Tang et al., 2016b). These modified anti-GFP chromobodies are instable and rapidly degraded by the proteasome. When stabilized, however, these chromobodies will bind their target and consequently will no longer be degraded. Indeed, engineering and application of conditionally stable anti-GFP chromobodies resulted in a reduced background fluorescence (Tang et al., 2016b). The mutations in the genetically modified nanobodies are highly conserved within nanobodies and therefore the switching from stable to instable nanobodies is generically applicable.
Non-targeted fluorescence can also be reduced by enhancer nanobodies (Roebroek et al., 2015). When the enhancer nanobodies bind with GFP, it increases the fluorescence and stability of the GFP. Enhancer nanobodies were also applied in a method to track single molecules in live-cell imaging (Ghosh et al., 2019). Here, an array of nanobodies is fused to the protein of interest, and expressed in cells with cytosolic monomeric GFP. Upon GFP binding to the array of nanobodies, the GFP molecules increase in fluorescent intensity, resulting in an increased signal-to-noise ratio. This binding can in the microscope be seen as a single dot, that represents a single protein of interest. In conclusion, the signal-to-noise ratio of chromobodies can be improved by degrading unbound chromobodies or by enhancing the fluorescence of bound FPs, which both will be beneficial in the detection of proteins.
Modifying cellular systems, in any way, may of course result in altered biology (Schnell et al., 2012). Interactions between endogenous proteins and ectopically expressed chromobodies can potentially influence the localization and function of the protein of interest. Binding of nanobodies to their target during or post-translationally may disrupt proper protein folding, macromolecule organization and/or transport. Despite the often-highlighted benefits of nanobody-technology, like any other technique their use should be validated including the effect on protein localization. Artificial modifying localization of proteins can be used on purpose for targeted interference. Chromobodies targeting endogenous actin-binding proteins, like gelsolin and cortactin, led to a disturbed actin distribution (Van Audenhove et al., 2013; Hebbrecht et al., 2017; Wegner et al., 2017; Bertier et al., 2018) paralleled with a decrease in both the number of invadopodia as well as extracellular matrix degradation. These factors are important in cell migration (Wolf and Friedl, 2009). Thus, chromobodies-assisted protein modulation allows to study the contribution of specific proteins of biology, including cell migration or some of its consequences, like the delay of metastasis (Bertier et al., 2018).
Spatiotemporal control of chromobody function is desirable in several assays. To initiate functionalized chromobodies in a controlled manner, chemogenetics or light stimuli can be applied to influence the binding capacity of the nanobody during and after synthesis. Such chemogenetic control employs ligand-modulated antibody fragments (LAMAs): a circular permutated bacterial dihydrofolate reductase (cpDHFR) linked to the nanobody is in such a conformation that it recognizes and binds to the nanobody target. In the presence of cell permeable DHFR inhibitors, the conformation changes precluding the antigen binding site of the nanobody binding the target, and thereby loss of association of nanobody and target. This process can be reversed to activate the nanobody binding (Farrants et al., 2020).
The light-dependent nanobody, termed photobody, uses a genetic photocaged tyrosine variant that results in the inactivation of the antigen-binding site (Arbely et al., 2012; Mootz et al., 2019). The photocaged tyrosine is photo-labile, and upon light induction (365 nm) the antigen-binding properties of the chromobodies are restored. Optobody, a second light-dependent tool, uses a split nanobody with a N-terminal fragment containing CDR1 and CDR2, and a C-terminal fragment containing CDR3 (Figure 2B; Yu et al., 2019). When both fragments are genetically fused to an optical-induced dimerization tool [MagHigh (Kawano et al., 2015)], the complete nanobody folds upon light stimulus and thereby forms the antigen-binding site. However, a generic position to split the nanobody is lacking, and thus for every different nanobody optimization and validation is needed. Overall, the activation of nanobodies using light or chemogenetic stimuli gives spatiotemporal control over the nanobodies, allowing precise subcellular modulation followed by direct readout of the biological consequences on targets studied.
Nanobodies can also be generated in cellular systems, subsequently purified and/or modified and then used in bioassays. Typically, these are secreted by mammalian cells or produced in high yields by bacteria (Harmsen and De Haard, 2007). After purification, the nanobody can for instance be coupled to chemical dyes (Beghein and Gettemans, 2017) to create fluorescent nanobodies (Tables 1, 2). Dyes suitable for super-resolution microscopy (SRM), i.e., LM beyond the diffraction limit resulting in typically 20 – 100 nm lateral resolution [reviewed in (Schermelleh et al., 2019)], will increase the resolution when using nanobodies compared to IgGs because of the smaller size of the reagents used, reducing the linkage error discussed above (Ries et al., 2012). Alternative to conjugation of purified nanobody with small fluorescent molecules, chromobodies can be expressed and purified. These chromobodies can then be directly used in fluorescent microscopy studies because they contain both the targeting module as well as the fluorescent module (Figure 1). The benefit of using cDNA encoding protein modules is the ease to switch target or color with molecular cloning tools. To employ these fluorescent nanobodies in live-cells different delivery mechanisms for extracellular or intracellular targets have been created.
The extracellular domain of peripheral membrane proteins in cultured cells is well accessible to ectopic added reagents and therefore straightforward to target in live-cell imaging. Nanobodies that target extracellular receptors may trigger receptor specific endocytosis, which can be important for e.g., drug delivery. Endocytosis can be triggered via binding with the human epidermal growth factor receptor 2 (HER2). Anti-HER2 nanobodies (Kijanka et al., 2013) coupled to fluorescent, drug containing nanoparticles (Martinez-Jothar et al., 2019), were indeed able to trigger endocytosis. After the trigger, uptake and cell viability was visualized to examine the effect of the therapeutic nanoparticles. So, receptor mediated endocytosis can be activated using fluorescent nanobodies to study therapeutic agents coupled to the nanobodies.
Super resolution localization of GFP surface-exposed by cells has been achieved while studing dynamic changes at the plasma membrane: The glycosylphosphatidylinositol (GPI)-anchored GFP reporter was further probed with Alexa647-conjugated nanobodies to enable SRM based on the blinking of the Alexa-dye. This resulted in higher resolution imaging of dynamic changes and detection of protein enrichments in the plasma membrane (Ries et al., 2012; Virant et al., 2018). Indirect visualization enables newly displayed proteins at the plasma membrane. Here, all available antigens first are blocked by unconjugated nanobody. Upon exocytosis stimuli, at the plasma membrane, new extracellular exposed antigens can be detected with fluorescent nanobodies (Seitz and Rizzoli, 2019). This pulse-chase approach allows dynamic studies, e.g., protein turnover, of receptors and other cell surface proteins.
The plasma membrane is a physical barrier for the nanobodies to target intracellular proteins in live-cells. Therefore, custom delivery methods are needed to target endogenous proteins in (living) cells without permeabilizing the plasma membrane. If nanobody expression is not an option because it first needs chemical modification or the concentration should be well-controlled, a purified nanobody may be delivered to cells. Fluorescent nanobodies can enter cells via endocytosis, when they formed non-covalent complexes with oligomers (Figure 2C; Roder et al., 2017) or they undergo lipid-based protein transfection (Oba and Tanaka, 2012; Virant et al., 2018). After endocytosis, the nanobodies need to escape the endosomal system and the formed complexes need to be degraded. However, using this strategy one has to take into account that the efficiency of endosomal escape is low (Stewart et al., 2016) and the nanobodies in the endosomes are already fluorescent, resulting in localized labeling of the endosomal system.
To prevent cellular uptake via endocytosis, cell-permeable nanobodies were generated by the addition of a cyclic cell-penetrating peptide [cCPP; (Herce et al., 2017)]. cCPPs are arginine-rich peptides that facilitates direct penetration of the plasma membrane to enter directly into the cytoplasm, independent of endocytosis. The efficiency of the labeling is expected to be increased because endosomal escape after endocytic uptake is omitted. Another advantage of using the cell-penetrating peptide is the ability to co-transport recombinant proteins, e.g., GFP, inside the cells, when both are bound to the nanobody. Although the efficiency of this co-transport was low, this cell-permeable nanobody can be further explored to serve as a drug delivery vehicle. A major disadvantage of the cCPPs however, is the strong tendency to accumulate in the nucleolus.
Generating temporary permeable plasma membranes is another approach to artificially deliver cargo inside cells. Different methods to temporary permeabilize the membrane have been developed: (i) Electroporation to deliver nanobodies linked to fluorescent quantum dots (QDs) into cells (Shi et al., 2018). The QDs were used for single particle visualization of intercellular transport by targeting kinesin motor proteins (Katrukha et al., 2017); (ii) Artificial plasma membrane channels that allow chromobody delivery into cells can be formed by bacterial Streptolysin O (Teng et al., 2016). Unbound nanobodies are removed during rinsing and the channels are closed upon switching to a recovery medium; (iii) Photoporation in which a laser-induced transfection enables the delivery of the nanobodies intracellular, and when these are fluorescently labeled, these can be directly detected (Hebbrecht et al., 2020). (iv) Another method to induce temporary damage to the plasma membrane is cell squeezing through the small capillaries of a microfluidic system resulting in fragility of the plasma membrane (Figure 2D; Klein et al., 2018). While the cells are squeezed, extracellular proteins can diffuse into the cells. When cells leave the small capillary the plasma membrane recovers to its normal state. Obviously, the effect of any temporary permeabilization approach used to enable nanobodies entrance into cells, is that endogenous molecules might diffuse out or targets may delocalize.
Genetic fusion with FPs cDNA is the widely used technique for protein visualization in living systems, but sometimes smaller peptide tags are preferred. Currently, there are no nanobodies available against common generic peptide tags (Muyldermans et al., 2001; Stijlemans et al., 2004; De Genst et al., 2006; Braun et al., 2016). Hence, three new small tags have been developed along with their respective targeting nanobodies. (i) The BC2-tag is a 12 amino acid peptide sequence originating from β-catenin (Braun et al., 2016; Virant et al., 2018), but the nanobody does not recognize endogenous β-catenin making it fairly specific for the tag only. (ii) The ALFA-tag (13 amino acids) forms an α-helix and is naturally absent in eukaryotes (Gotzke et al., 2019), also making it a specific target for its nanobody, which also counts for the (iii) Pep-Tag (15 amino acids) (Traenkle et al., 2020).
An array of peptides fused to the protein of interest can amplify the fluorescent signal, and thus increase the signal-to-noise ratio. A peptide-repeat called MoonTag (Boersma et al., 2019) was created to visualize active translation (Figure 2E). Here, an array of a 15 amino acid peptide sequence was added N-terminal of the protein of interest. The newly formed peptide chain forms a docking site for chromobodies. The MoonTag can be combined with the SunTag; an intracellular expressed single-chain variable fragment (scFV; Tanenbaum et al., 2014). Combining the two tags will allow visualization of different reading frames within a single mRNA or can be used to amplify the signal from different targets. Given the mechanism of probing the target with a cDNA encoded peptide repeat, making use of the same antibodies, this is a highly versatile enhancer system.
While resolution of targets in LM is improved by using nanobodies because of a reduced linkage error compared to IgG targeting, the ultrastructural remains unexplored. In electron microscopy (EM), the ultrastructure is revealed, but localizing the protein of interest within this structure also requires probes. EM-visualization of targets benefits from the nanobody-technology because the probe is small and thus penetrates better. Therefore, milder permeabilization is needed, better preserving the ultrastructure. Moreover, the small size improves the resolution compared to traditional immuno-EM because the target and identifiable tag are in close proximity. Also in EM proteins can be specifically identified using genetically encoded or affinity-based probes. Genetically encoded probes may be based on peroxidases that creates black precipitates in the presence of diaminobenzidine (DAB) and H2O2. Affinity-based probes include electron dense nanoparticles like nanogold and QDs (De Boer et al., 2015). Of course the genetically encoded probes form good candidates to use in conjunction with nanobodies as a multi-modular probe for EM studies.
Intracellular nanobodies fused with soybean ascorbate peroxidase (APEX2) (Lam et al., 2015) can target GFP or mCherry to add an electron dense mark to the protein of interest at EM level (Ariotti et al., 2015, 2017). This APEX2-nanobody can be applied as general CLEM (correlated LM/EM) probe because many cells and small organisms have already been engineered to express GFP or mCherry. Using conditionally stable nanobodies (section Nanobodies In Light Microscopy), unbound APEX2-nanobody is degraded and thereby improve EM detection of the target proteins (Ariotti et al., 2018). The conditionally stable APEX2-nanobody enables studying protein-protein interactions by making use of splitFPs as target (Figure 3A; Ghosh et al., 2000; Hu and Kerppola, 2003). Here, a protein of interest and its potential interacting protein are genetically fused with different segment of e.g., GFP. Anti-GFP APEX2-nanobody can only bind to the folded GFP, representing the interaction between the two proteins of interest. When GFP is absent, the conditionally stable APEX2-nanobodies are degraded. Another alternative to study protein-protein interactions would be by using nanobodies fused to splitHRP or splitAPEX2 (Martell et al., 2016; Han et al., 2019). Two distinct nanobodies recognizing proteins within close proximity will facilitate the refolding of the peroxidase, which will result in DAB precipitates at that location in the sample. The split peroxidase thus reduces the signal of unbound nanobodies, leading to a more conclusive picture. In conclusion, where chromobodies allow dynamic studies of endogenous proteins, APEX2-nanobodies enable high resolution analysis in the ultrastructural context.
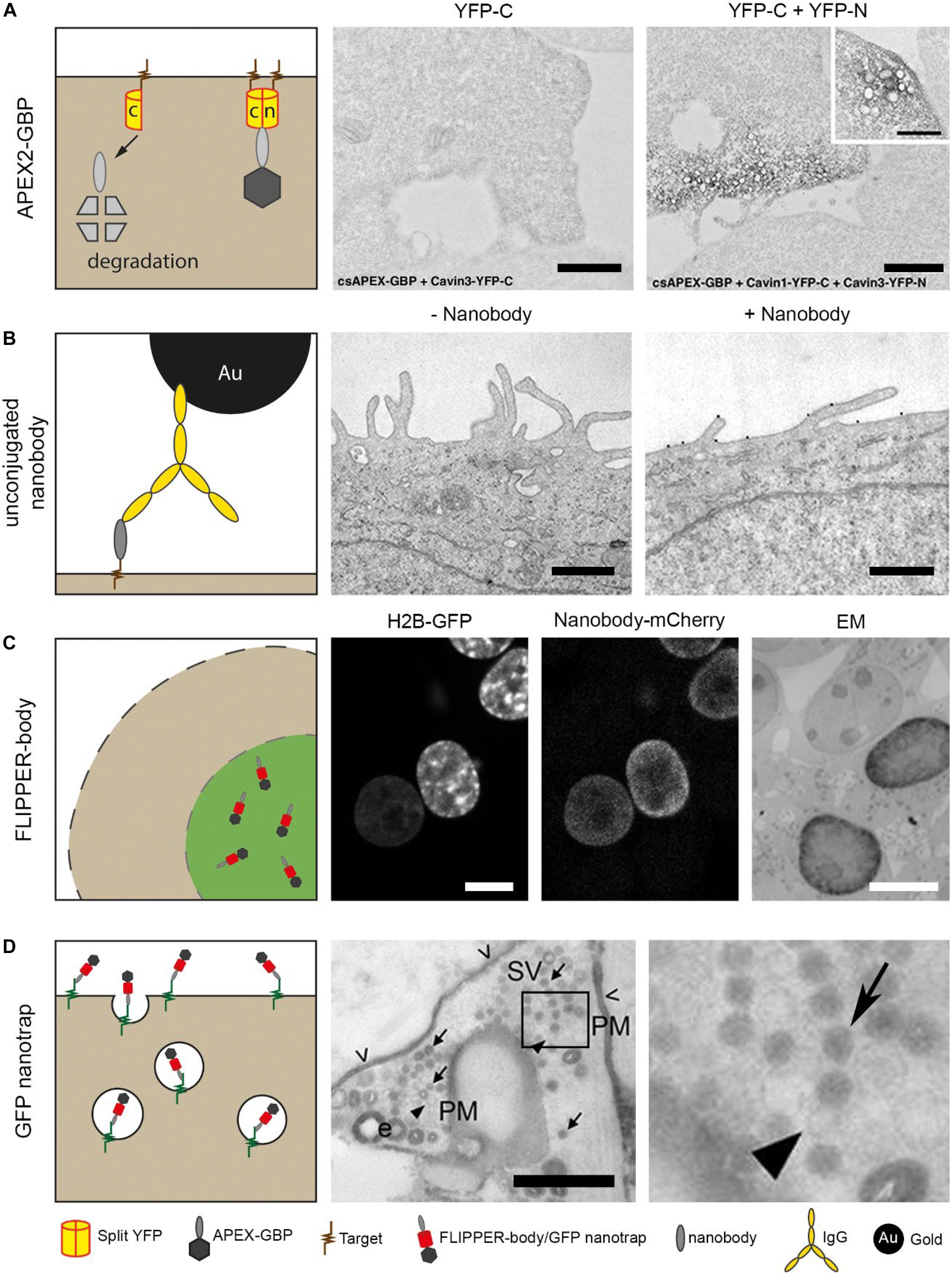
Figure 3. Nanobody technology for EM. (A) Cells were expressing only C-terminus YFP or both C- and N-terminus YFP. The cells also expressed a conditionally stable anti-YFP, for proteasomal degradation of unbound probe. The nanobodies were genetically fused with peroxidase, APEX2, for EM detection. Note that only black precipitation is visible in cells expressing C-terminus YFP and N-terminus YFP. This confirmed the degradation of the nanobody with the peroxidase. Bars: 1 μm. (B) Purified nanobodies were used as primary antigen binding protein to reduce the distance between label and antigen. Nanobodies can be detected using an anti-VhH IgG conjugated to gold for EM visualization. Bars: 0.5 μm. (C) Cells express H2B-GFP from Figure 1A, and are permeabilized after fixation followed by labeling with anti-GFP FLIPPER-bodies. Note the colocalization in LM and the dark, positive nucleus in EM versus an unlabeled nucleus. Bars: 10 μm. (D) Neuronal cells expressing pHluorin on the plasma membrane. Added anti-GFP nanobodies bind to the pHluorin, and after a pulse stimulation, synaptic vesicles are formed. In EM, a population of unlabeled and labeled synaptic vesicles is detected. Arrows indicate synaptic vesicles, arrow heads indicate unstained vesicles and open arrow heads indicate residual staining (PM, plasma membrane, SV, Synaptic vesicle). Bar: 0.5 μm. The images in (A–D) have been reproduced from the following studies (Joensuu et al., 2016; Kijanka et al., 2017; Ariotti et al., 2018; De Beer et al., 2018), all of which have been published under a Creative Commons Attribution License.
Purified nanobodies can be applied for affinity-based immunolabeling, and thereby replace conventional IgGs. Immunolabeling with unconjugated nanobodies was used to label HER2 in breast cancer cells, followed by a secondary anti-nanobody IgG conjugated with nanogold (Figure 3B; Kijanka et al., 2017). Although this already improved the resolution, it could benefit more by replacing the IgG a direct labeling method using conjugated nanobodies.
Reagents used in CLEM that are readily detectable both in the FLM and EM, like nanogold or QDs, are relatively large and/or bulky in comparison to 15–30 kDa range FPs or even smaller fluorescent dyes like FITC. These inorganic nanoparticles whether or not conjugated to nanobodies, will have limited penetration (Giepmans et al., 2005; Schnell et al., 2012). Therefore, we developed a completely protein-based probe, called “FLIPPER”-body (Kuipers et al., 2015; De Beer et al., 2018) with (i) a nanobody as targeting module; (ii) a FP for LM analysis; and (iii) a horseradish peroxidase (HRP) for EM analysis. The FLIPPER-bodies were produced by mammalian cells applied as immunoreagent to label e.g., intracellular GFP (Figure 3C). Other targets were generated by simple molecular cloning interchanging the modules of the probe. FLIPPER-bodies improve penetration due to its size and flexibility comparted to IgGs-targeted nanoparticles and thus lead to a better target detection while maintaining reasonable ultrastructure.
In conventional EM glutaraldehyde is used to fix the samples for ultrastructural preservation (Schnell et al., 2012). However, cells fixed with paraformaldehyde become permeable for small molecules like e.g., small fluorescent nanobodies (Fang et al., 2018). This labeling without adding permeabilization reagents method was further developed to stain intracellular targets in fixed mouse tissue slices. When targeting an intracellular target with both nanobody and IgG, the nanobody penetrated up to 100 μm into a brain tissue slice, while the IgGs only stained the surface of the slice. Subsequently, the same cells were imaged with EM and ultrastructural details were well preserved due to glutaraldehyde fixation after performing the nanobody labeling. So, detergent treatment is not required when using small nanobody probes and thus cellular structure is better preserved. The technique needs to mature further to establish the ratio of positives and false negatives to determine if the recognition of the targets is generically reliable. Moreover, the accessibility of targets within organelles, like mitochondria or nucleus, remains to be studied further.
Alternatively to expressing nanobody-based probes in the cellular system of interest, they also can first be purified and then applied to cells. An elegant example of this approach has been used to visualize the formation of synaptic vesicles in living cells (Joensuu et al., 2016). Ultrastructural localization of newly formed synaptic vesicles can be examined using peroxidase-fused nanobodies. Vesicle-associated membrane protein 2 (VAMP2) mediates the formation of synaptic vesicles and is expressed at the plasma membrane of neuronal cells. Anti-GFP nanobodies were applied to target pHluorin, a GFP derivative, fused with VAMP2 (Figure 3D). Upon K+ stimulation, pHluorin-VAMP2 with bound nanobody is endocytosed, and new synaptic vesicles are formed. After EM preparation, the newly formed synaptic vesicles were stained black, as a result of the peroxidase attached to the nanobody, whereas older synaptic vesicles remained unstained. Thus, the endocytic route of specific proteins of interest can be visualized.
Purified nanobodies, fused to a FP and a peroxidase, were used to analyze the retrograde transport system in live-cells (Buser and Spiess, 2019) or within ultrastructural context (Buser et al., 2018) to study the transport from the cell surface to the Golgi complex. Cells with GFP-modified cycling reporter proteins at the plasma membrane captured the nanobodies extracellular and transported them in the cells. CLEM revealed the dynamic behavior of different cycling reporter proteins, and showed the ultrastructural localization of the reporter proteins. Overall, these nanobody-based CLEM probes can reveal the localization of plasma membrane proteins and visualize their dynamics, together with the ultrastructural context.
Structural analysis of proteins at atomic resolution is achieved by cryo-EM [reviewed in (Kühlbrandt, 2014; Egelman, 2016; Cheng, 2018)]. However different conformations of proteins may be present, hindering the structural determination of specific states. Here, nanobodies can help to stabilize the proteins into a certain conformational state [reviewed in (Uchański et al., 2020)], especially as the targeting module in rigid chimeras termed megabodies. These stabilizing nanobody-chimeras recently have been used to solve the type A γ-aminobutyric (GABAA) structure in membranes-like structures in presence and absence of natural ligands and antagonists (Laverty et al., 2019; Masiulis et al., 2019). Using the megabodies, no longer the target proteins themselves need to be engineered and thus structure on the endogenous proteins is being revealed. Likely, nanobody technology will further contribute to structural biology on many other target proteins by facilitating single conformational protein states and better understand the dynamic regulation of biomolecules by their ligands without directly altering the proteins at study.
Nanobodies have clear benefits over conventional IgGs antibodies or genetically encoded probes. Nanobodies (i) have a small diameter resulting in better resolution and better penetration; (ii) can visualize endogenous proteins in live-cell imaging; (iii) are encoded by a one cDNA, which enables easy molecular cloning; (iv) allow researchers to create and produce custom multi-modal probes. Potential artifacts specific for nanobody technology in microscopy include signal from chromobodies independent of target binding and modified localization of the protein target, as detailed above.
Nanobodies are still limited used in research, mainly due to the limitation in the availability of nanobodies for general targets compared to IgGs and researchers are not aware of the possibilities of nanobody in microscopy. Here, we aim to increase the visibility of opportunities and benefits highlighted in the main text, but also exemplified by the successful new insights and new nanobody-based reagents by many (Table 2). Other ways to improve the use of nanobodies in research are open access sharing of nanobodies cDNA to allow researchers to create and manipulate their own labeling reagent and to increase the nanobody database of general targets. Currently, most nanobodies are generated via Camelidae immunization, a route that may be a (seemingly) hurdle for scientists. However, these days the generation of nanobodies is both economically and time-wise competitive with generation of newly synthesized rabbit IgG against specific targets. The novice user obviously will benefit of expertise and accessibility to the needed infrastructure by more experienced users mentioned throughout this review. In addition, a fast method based on a yeast display platform to select nanobodies in vitro has been established (McMahon et al., 2018).
Protein identification in microscopy has been greatly aided by immunolabeling using IgGs as well as the application of genetically encoded tags. Nanobody technology, being explored for approximately 25 years is a great additional tool. Like IgGs, endogenous proteins can be easily studied without direct modification. Moreover, the single domain properties and therefore the easy use as a module in other genetically encoded probes allow freedom of use of tags and targeting module. In line with SRM techniques, nanobodies have the added benefit over IgGs that they hardly provide distance between target and label (linkage error). The major limitation in the use of nanobodies in general is the sparse availability of high affinity nanobodies for specific targets. Increasing the availability for different targets using immunization and/or micro-organism-based libraries will increase the variation in the decade to come. When more targets can be studied with nanobodies these will become a common tool in the lab and share the top-3 podium with genetically encoded tags as well as IgGs since they share benefits of both approaches to identify and visualize targets of interest in dynamic systems and with high precision.
MB performed the experiment and made the figures. MB and BG wrote the manuscript. Both authors contributed to the article and approved the submitted version.
All sources of funding received for the research have been submitted. Open access publication fee was partially waived by FiCN.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank our lab members for discussions; financial support by Netherlands Organization for scientific research (STW Microscopy Valley 12718, NWO 175-010-2009-023, and ZonMW 91111.006) and de Cock-Hadders Stichting.
Abdellatif, M. E. A., Hipp, L., Plessner, M., Walther, P., and Knöll, B. (2019). Indirect visualization of endogenous nuclear actin by correlative light and electron microscopy (CLEM) using an actin-directed chromobody. Histochem. Cell Biol. 152, 133–143. doi: 10.1007/s00418-019-01795-3
Ahani, R., Roohvand, F., Cohan, R. A., Etemadzadeh, M. H., Mohajel, N., Behdani, M., et al. (2016). Sindbis virus-pseudotyped lentiviral vectors carrying VEGFR2-specific nanobody for potential transductional targeting of tumor vasculature. Mol. Biotechnol. 58, 738–747. doi: 10.1007/s12033-016-9973-7
Almuedo-Castillo, M., Bläβle, A., Mörsdorf, D., Marcon, L., Soh, G. H., Rogers, K. W., et al. (2018). Scale-invariant patterning by size-dependent inhibition of Nodal signalling. Nat. Cell Biol. 20, 1032–1042. doi: 10.1038/s41556-018-0155-7
Andersen, J. T., Gonzalez-Pajuelo, M., Foss, S., Landsverk, O. J. B., Pinto, D., Szyroki, A., et al. (2013). Selection of nanobodies that target human neonatal Fc receptor. Sci.Rep. 3:1118.
Arbely, E., Torres-Kolbus, J., Deiters, A., and Chin, J. W. (2012). Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J. Am. Chem. Soc. 134, 11912–11915. doi: 10.1021/ja3046958
Ariotti, N., Hall, T. E., and Parton, R. G. (2017). Correlative light and electron microscopic detection of GFP-labeled proteins using modular APEX. Methods Cell Biol. 140, 105–121. doi: 10.1016/bs.mcb.2017.03.002
Ariotti, N., Hall, T. E., Rae, J., Ferguson, C., McMahon, K. A., Martel, N., et al. (2015). Modular detection of GFP-labeled proteins for rapid screening by electron microscopy in cells and organisms. Dev. Cell 35, 513–525. doi: 10.1016/j.devcel.2015.10.016
Ariotti, N., Rae, J., Giles, N., Martel, N., Sierecki, E., Gambin, Y., et al. (2018). Ultrastructural localisation of protein interactions using conditionally stable nanobodies. PLoS Biol. 16:e2005473. doi: 10.1371/journal.pbio.2005473
Bannas, P., Lenz, A., Kunick, V., Fumey, W., Rissiek, B., Schmid, J., et al. (2015a). Validation of nanobody and antibody based in vivo tumor xenograft NIRF-imaging experiments in mice using ex vivo flow cytometry and microscopy. J. Vis. Exp 98:e52462.
Bannas, P., Lenz, A., Kunick, V., Well, L., Fumey, W., Rissiek, B., et al. (2015b). Molecular imaging of tumors with nanobodies and antibodies: timing and dosage are crucial factors for improved in vivo detection. Contrast Media Mol. Imaging 10, 367–378. doi: 10.1002/cmmi.1637
Bazl, M. R., Rasaee, M. J., Foruzandeh, M., Rahimpour, A., Kiani, J., Rahbarizadeh, F., et al. (2007). Production of chimeric recombinant single domain antibody-green fluorescent fusion protein in Chinese hamster ovary cells. Hybridoma 26, 1–9. doi: 10.1089/hyb.2006.037
Beghein, E., and Gettemans, J. (2017). Nanobody technology: a versatile toolkit for microscopic imaging, protein-protein interaction analysis, and protein function exploration. Front. Immunol. 8:771.
Beltrán Hernández, I., Rompen, R., Rossin, R., Xenaki, K. T., Katrukha, E. A., Nicolay, K., et al. (2019). Imaging of tumor spheroids, dual-isotope SPECT, and autoradiographic analysis to assess the tumor uptake and distribution of different nanobodies. Mol. Imaging Biol. 21, 1079–1088. doi: 10.1007/s11307-019-01320-x
Berry, L. K., Ölafsson, G., Ledesma-Fernández, E., and Thorpe, P. H. (2016). Synthetic protein interactions reveal a functional map of the cell. eLife 5:e13053.
Bertier, L., Hebbrecht, T., Mettepenningen, E., De Wit, N., Zwaenepoel, O., Verhelle, A., et al. (2018). Nanobodies targeting cortactin proline rich, helical and actin binding regions downregulate invadopodium formation and matrix degradation in SCC-61 cancer cells. Biomed. Pharmacother. 102, 230–241. doi: 10.1016/j.biopha.2018.03.064
Bleck, M., Itano, M. S., Johnson, D. S., Thomas, V. K., North, A. J., Bieniasz, P. D., et al. (2014). Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc. Natl. Acad. Sci. U.S.A. 111, 12211–12216. doi: 10.1073/pnas.1321655111
Boersma, S., Khuperkar, D., Verhagen, B. M. P., Sonneveld, S., Grimm, J. B., Lavis, L. D., et al. (2019). Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178, 458–472.e19.
Braun, M. B., Traenkle, B., Koch, P. A., Emele, F., Weiss, F., Poetz, O., et al. (2016). Peptides in headlock–a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy. Sci. Rep. 6:19211.
Buchfellner, A., Yurlova, L., Nuske, S., Scholz, A. M., Bogner, J., Ruf, B., et al. (2016). A New nanobody-based biosensor to study endogenous PARP1 in vitro and in live human cells. PLoS One. 11:e0151041. doi: 10.1371/journal.pone.0151041
Burgess, A., Lorca, T., and Castro, A. (2012). Quantitative live imaging of endogenous DNA replication in mammalian cells. PLoS One. 7:e45726. doi: 10.1371/journal.pone.0045726
Buser, D. P., Schleicher, K. D., Prescianotto-Baschong, C., and Spiess, M. A. (2018). versatile nanobody-based toolkit to analyze retrograde transport from the cell surface. Proc. Natl. Acad. Sci. U.S.A. 115, E6227–E6236.
Buser, D. P., and Spiess, M. (2019). Analysis of endocytic uptake and retrograde transport to the trans-golgi network using functionalized nanobodies in cultured cells. J. Vis. Exp. 144:e59111.
Carrington, G., Tomlinson, D., and Peckham, M. (2019). Exploiting nanobodies and Affimers for superresolution imaging in light microscopy. Mol. Biol. Cell 30, 2737–2740. doi: 10.1091/mbc.e18-11-0694
Casas-Delucchi, C., Becker, A., Bolius, J. J., and Cardoso, M. C. (2012). Targeted manipulation of heterochromatin rescues MeCP2 Rett mutants and re-establishes higher order chromatin organization. Nucleic Acids Res. 40:e176. doi: 10.1093/nar/gks784
Caussinus, E., Kanca, O., and Affolter, M. (2011). Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–121. doi: 10.1038/nsmb.2180
Chamma, I., Letellier, M., Butler, C., Tessier, B., Lim, K., Gauthereau, I., et al. (2016). Mapping the dynamics and nanoscale organization of synaptic adhesion proteins using monomeric streptavidin. Nat. Commun. 7:10773.
Chamma, I., Rossier, O., Giannone, G., Thoumine, O., and Sainlos, M. (2017). Optimized labeling of membrane proteins for applications to super-resolution imaging in confined cellular environments using monomeric streptavidin. Nat. Protoc. 12, 748–763. doi: 10.1038/nprot.2017.010
Cheloha, R. W., Li, Z., Bousbaine, D., Woodham, A. W., Perrin, P., Volarić, J., et al. (2019). Internalization of Influenza virus and cell surface proteins monitored by site-specific conjugation of protease-sensitive probes. ACS Chem. Biol. 14, 1836–1844. doi: 10.1021/acschembio.9b00493
Cheng, Y. (2018). Single-particle cryo-EM-How did it get here and where will it go. Science 361, 876–880. doi: 10.1126/science.aat4346
Cramer, K., Bolender, A., Stockmar, I., Jungmann, R., Kasper, R., and Shin, J. Y. (2019). Visualization of bacterial protein complexes labeled with fluorescent proteins and nanobody binders for STED microscopy. Int. J. Mol. Sci. 20:3376. doi: 10.3390/ijms20143376
Darling, T. L., Sherwood, L. J., and Hayhurst, A. (2017). Intracellular crosslinking of filoviral nucleoproteins with xintrabodies restricts viral packaging. Front. Immunol. 8:1197.
De Beer, M. A., Kuipers, J., Van Bergen En Henegouwen, P. M. P., and Giepmans, B. N. G. (2018). A small protein probe for correlated microscopy of endogenous proteins. Histochem. Cell Biol. 149, 261–268. doi: 10.1007/s00418-018-1632-6
De Boer, P., Hoogenboom, J. P., and Giepmans, B. N. (2015). Correlated light and electron microscopy: ultrastructure lights up! Nat. Methods 12, 503–513. doi: 10.1038/nmeth.3400
De Bruin, R. C. G., Lougheed, S. M., Van Der Kruk, L., Stam, A. G., Hooijberg, E., Roovers, R. C., et al. (2016). Highly specific and potently activating Vγ9Vδ2-T cell specific nanobodies for diagnostic and therapeutic applications. Clin. Immunol. 169, 128–138. doi: 10.1016/j.clim.2016.06.012
De Clercq, S., Zwaenepoel, O., Martens, E., Vandekerckhove, J., Guillabert, A., and Gettemans, J. (2013). Nanobody-induced perturbation of LFA-1/L-plastin phosphorylation impairs MTOC docking, immune synapse formation and T cell activation. Cell Mol. Life Sci. 70, 909–922. doi: 10.1007/s00018-012-1169-0
De Genst, E., Silence, K., Decanniere, K., Conrath, K., Loris, R., Kinne, J., et al. (2006). Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. U.S.A. 103, 4586–4591. doi: 10.1073/pnas.0505379103
De Haard, H. J. W., Bezemer, S., Ledeboer, A. M., Müller, W. H., Boender, P. J., Moineau, S., et al. (2005). Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection. J. Bacteriol. 187, 4531–4541. doi: 10.1128/jb.187.13.4531-4541.2005
De Meyer, T., Eeckhout, D., De Rycke, R., De Buck, S., Muyldermans, S., and Depicker, A. (2014). Generation of VHH antibodies against the Arabidopsis thaliana seed storage proteins. Plant Mol. Biol. 84, 83–93. doi: 10.1007/s11103-013-0118-0
Debie, P., Lafont, C., Defrise, M., Hansen, I., Van Willigen, D. M., Van Leeuwen, F. W. B., et al. (2020). Size and affinity kinetics of nanobodies influence targeting and penetration of solid tumours. J. Control Release 317, 34–42. doi: 10.1016/j.jconrel.2019.11.014
Delanote, V., Vanloo, B., Catillon, M., Friederich, E., Vandekerckhove, J., and Gettemans, J. (2010). An alpaca single-domain antibody blocks filopodia formation by obstructing L-plastin-mediated F-actin bundling. FASEB J. 24, 105–118. doi: 10.1096/fj.09-134304
D’Hollander, A., Jans, H., Velde, G. V., Verstraete, C., Massa, S., Devoogdt, N., et al. (2017). Limiting the protein corona: a successful strategy for in vivo active targeting of anti-HER2 nanobody-functionalized nanostars. Biomaterials 123, 15–23. doi: 10.1016/j.biomaterials.2017.01.007
Dong, J. X., Lee, Y., Kirmiz, M., Palacio, S., Dumitras, C., Moreno, C. M., et al. (2019). A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons. Elife 8:48750. doi: 10.7554/eLife.48750
Drees, C., Raj, A. N., Kurre, R., Busch, K. B., Haase, M., and Piehler, J. (2016). Engineered upconversion nanoparticles for resolving protein interactions inside living cells. Angew. Chem. Int. Ed. Engl. 55, 11668–11672. doi: 10.1002/anie.201603028
Duarte, J. N., Cragnolini, J. J., Swee, L. K., Bilate, A. M., Bader, J., Ingram, J. R., et al. (2016). Generation of immunity against pathogens via single-domain antibody-antigen constructs. J. Immunol. 197, 4838–4847. doi: 10.4049/jimmunol.1600692
Egelman, E. H. (2016). The current revolution in cryo-EM. Biophys. J. 110, 1008–1012. doi: 10.1016/j.bpj.2016.02.001
Fan, X., Wang, L., Guo, Y., Tu, Z., Li, L., Tong, H., et al. (2015). Ultrasonic nanobubbles carrying Anti-PSMA nanobody: construction and application in prostate cancer-targeted imaging. PLoS One 10:e0127419. doi: 10.1371/journal.pone.0127419
Fang, T., Lu, X., Berger, D., Gmeiner, C., Cho, J., Schalek, R., et al. (2018). Nanobody immunostaining for correlated light and electron microscopy with preservation of ultrastructure. Nat. Methods 15, 1029–1032. doi: 10.1038/s41592-018-0177-x
Farrants, H., Tarnawski, M., Müller, T. G., Otsuka, S., Hiblot, J., Koch, B., et al. (2020). Chemogenetic control of nanobodies. Nat. Methods 17, 279–282. doi: 10.1038/s41592-020-0746-7
Finnigan, G. C., Booth, E. A., Duvalyan, A., Liao, E. N., and Thorner, J. (2015). The carboxy-terminal tails of tins Cdc11 and Shs1 recruit myosin-II binding factor Bni5 to the bud neck in Saccharomyces cerevisiae. Genetics 200, 843–862. doi: 10.1534/genetics.115.176503
Gadok, A. K., Zhao, C., Meriwether, A. I., Ferrati, S., Rowley, T. G., Zoldan, J., et al. (2018). The display of single-domain antibodies on the surfaces of connectosomes enables gap junction-mediated drug delivery to specific cell populations. Biochemistry (N. Y.) 57, 81–90. doi: 10.1021/acs.biochem.7b00688
Ghosh, I., Hamilton, A. D., and Regan, L. (2000). Antiparallel leucine zipper-directed protein reassembly:?application to the green fluorescent protein. J. Am. Chem. Soc. 122, 5658–5659. doi: 10.1021/ja994421w
Ghosh, R. P., Franklin, J. M., Draper, W. E., Shi, Q., Beltran, B., Spakowitz, A. J., et al. (2019). A fluorogenic array for temporally unlimited single-molecule tracking. Nat. Chem. Biol. 15, 401–409. doi: 10.1038/s41589-019-0241-6
Giepmans, B. N., Adams, S. R., Ellisman, M. H., and Tsien, R. Y. (2006). The fluorescent toolbox for assessing protein location and function. Science 312, 217–224. doi: 10.1126/science.1124618
Giepmans, B. N., Deerinck, T. J., Smarr, B. L., Jones, Y. Z., and Ellisman, M. H. (2005). Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat. Methods 2, 743–749. doi: 10.1038/nmeth791
Göttfert, F., Pleiner, T., Heine, J., Westphal, V., Görlich, D., Sahl, S. J., et al. (2017). Strong signal increase in STED fluorescence microscopy by imaging regions of subdiffraction extent. Proc. Natl. Acad. Sci. U.S.A. 114, 2125–2130. doi: 10.1073/pnas.1621495114
Gotzke, H., Kilisch, M., Martinez-Carranza, M., Sograte-Idrissi, S., Rajavel, A., Schlichthaerle, T., et al. (2019). The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 10:4403.
Groot, A. J., Verheesen, P., Westerlaken, E. J., Gort, E. H., Van Der Groep, P., Bovenschen, N., et al. (2006). Identification by phage display of single-domain antibody fragments specific for the ODD domain in hypoxia-inducible factor 1alpha. Lab. Invest. 86, 345–356. doi: 10.1038/labinvest.3700395
Gross, C., Wiesmann, V., Millen, S., Kalmer, M., Wittenberg, T., Gettemans, J., et al. (2016). The tax-inducible actin-bundling protein fascin is crucial for release and cell-to-cell transmission of human T-cell leukemia virus type 1 (HTLV-1). PLoS Pathog. 12:e1005916. doi: 10.1371/journal.ppat.1005916
Hafian, H., Sukhanova, A., Turini, M., Chames, P., Baty, D., Pluot, M., et al. (2014). Multiphoton imaging of tumor biomarkers with conjugates of single-domain antibodies and quantum dots. Nanomedicine 10, 1701–1709. doi: 10.1016/j.nano.2014.05.014
Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hamers, C., Songa, E. B., et al. (1993). Naturally occurring antibodies devoid of light chains. Nature 363, 446–448. doi: 10.1038/363446a0
Han, Y., Branon, T. C., Martell, J. D., Boassa, D., Shechner, D., Ellisman, M. H., et al. (2019). Directed evolution of split APEX2 peroxidase. ACS Chem. Biol. 14, 619–635. doi: 10.1021/acschembio.8b00919
Han, Z., Zhang, B., Wang, Y. E., Zuo, Y. Y., and Su, W. W. (2012). Self-assembled amyloid-like oligomeric-cohesin Scaffoldin for augmented protein display on the saccharomyces cerevisiae cell surface. Appl. Environ. Microbiol. 78, 3249–3255. doi: 10.1128/aem.07745-11
Harmansa, S., Alborelli, I., Bieli, D., Caussinus, E., and Affolter, M. (2017). A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. eLife 6:e22549.
Harmansa, S., Hamaratoglu, F., Affolter, M., and Caussinus, E. (2015). Dpp spreading is required for medial but not for lateral wing disc growth. Nature 527, 317–322. doi: 10.1038/nature15712
Harmsen, M. M., and De Haard, H. J. (2007). Properties, production, and applications of camelid single-domain antibody fragments. Appl. Microbiol. Biotechnol. 77, 13–22. doi: 10.1007/s00253-007-1142-2
Harper, C. B., Papadopulos, A., Martin, S., Matthews, D. R., Morgan, G. P., Nguyen, T. H., et al. (2016). Botulinum neurotoxin type-A enters a non-recycling pool of synaptic vesicles. Sci.Rep. 6:19654.
Hebbrecht, T., Liu, J., Zwaenepoel, O., Boddin, G., Van Leene, C., Decoene, K., et al. (2020). Nanobody click chemistry for convenient site-specific fluorescent labelling, single step immunocytochemistry and delivery into living cells by photoporation and live cell imaging. N. Biotechnol. 59, 33–43. doi: 10.1016/j.nbt.2020.05.004
Hebbrecht, T., Van Audenhove, I., Zwaenepoel, O., Verhelle, A., and Gettemans, J. V. C. A. (2017). nanobodies target N-WASp to reduce invadopodium formation and functioning. PLoS One 12:e0185076. doi: 10.1371/journal.pone.0185076
Helma, J., Cardoso, M. C., Muyldermans, S., and Leonhardt, H. (2015). Nanobodies and recombinant binders in cell biology. J. Cell Biol. 209, 633–644. doi: 10.1083/jcb.201409074
Helma, J., Schmidthals, K., Lux, V., Nuske, S., Scholz, A. M., Krausslich, H. G., et al. (2012). Direct and dynamic detection of HIV-1 in living cells. PLoS One 7:e50026. doi: 10.1371/journal.pone.0050026
Herce, H. D., Deng, W., Helma, J., Leonhardt, H., and Cardoso, M. C. (2013). Visualization and targeted disruption of protein interactions in living cells. Nat.Commun. 4:2660.
Herce, H. D., Schumacher, D., Schneider, A. F. L., Ludwig, A. K., Mann, F. A., Fillies, M., et al. (2017). Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem. 9, 762–771. doi: 10.1038/nchem.2811
Hu, C. D., and Kerppola, T. K. (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21, 539–545. doi: 10.1038/nbt816
Huang, Y., Nokhrin, S., Hassanzadeh-Ghassabeh, G., Yu, C. H., Yang, H., Barry, A. N., et al. (2014). Interactions between metal-binding domains modulate intracellular targeting of Cu(I)-ATPase ATP7B, as revealed by nanobody binding. J. Biol. Chem. 289, 32682–32693. doi: 10.1074/jbc.m114.580845
Iqbal, U., Albaghdadi, H., Luo, Y., Arbabi, M., Desvaux, C., Veres, T., et al. (2010a). Molecular imaging of glioblastoma multiforme using anti-insulin-like growth factor-binding protein-7 single-domain antibodies. Br. J. Cancer 103, 1606–1616. doi: 10.1038/sj.bjc.6605937
Iqbal, U., Trojahn, U., Albaghdadi, H., Zhang, J., O’Connor-McCourt, M., Stanimirovic, D., et al. (2010b). Kinetic analysis of novel mono- and multivalent VHH-fragments and their application for molecular imaging of brain tumours. Br. J. Pharmacol. 160, 1016–1028. doi: 10.1111/j.1476-5381.2010.00742.x
Irannejad, R., Tomshine, J. C., Tomshine, J. R., Chevalier, M., Mahoney, J. P., Steyaert, J., et al. (2013). Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538. doi: 10.1038/nature12000
Jacobson, K., Rajfur, Z., Vitriol, E., and Hahn, K. (2008). Chromophore-assisted laser inactivation in cell biology. Trends Cell Biol. 18, 443–450. doi: 10.1016/j.tcb.2008.07.001
Joensuu, M., Padmanabhan, P., Durisic, N., Bademosi, A. T., Cooper-Williams, E., Morrow, I. C., et al. (2016). Subdiffractional tracking of internalized molecules reveals heterogeneous motion states of synaptic vesicles. J. Cell Biol. 215, 277–292. doi: 10.1083/jcb.201604001
Joshi, B. S., de Beer, M. A., Giepmans, B. N. G., and Zuhorn, I. S. (2020). Endocytosis of extracellular vesicles and release of their cargo from endosomes. ACS Nano. 14, 4444–4455. doi: 10.1021/acsnano.9b10033
Jullien, D., Vignard, J., Fedor, Y., Béry, N., Olichon, A., Crozatier, M., et al. (2016). Chromatibody, a novel non-invasive molecular tool to explore and manipulate chromatin in living cells. J. Cell Sci. 129, 2673–2683. doi: 10.1242/jcs.183103
Kamiyama, D., McGorty, R., Kamiyama, R., Kim, M. D., Chiba, A., and Huang, B. (2015). Specification of dendritogenesis site in Drosophila aCC motoneuron by membrane enrichment of Pak1 through Dscam1. Dev. Cell 35, 93–106. doi: 10.1016/j.devcel.2015.09.007
Kaplan, C., and Ewers, H. (2015). Optimized sample preparation for single-molecule localization-based superresolution microscopy in yeast. Nat. Protoc. 10, 1007–1021. doi: 10.1038/nprot.2015.060
Karges, J., Jakubaszek, M., Mari, C., Zarschler, K., Goud, B., Stephan, H., et al. (2019). Synthesis and characterization of an epidermal growth factor receptor-selective Ru(II) Polypyridyl-nanobody conjugate as a photosensitizer for photodynamic therapy. Chembiochem 4, 531–542. doi: 10.1002/cbic.201900419
Katoh, Y., Nozaki, S., Hartanto, D., Miyano, R., and Nakayama, K. (2015). Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128, 2351–2362. doi: 10.1242/jcs.168740
Katrukha, E. A., Mikhaylova, M., Van Brakel, H. X., Van Bergen En Henegouwen, P. M., Akhmanova, A., Hoogenraad, C. C., et al. (2017). Probing cytoskeletal modulation of passive and active intracellular dynamics using nanobody-functionalized quantum dots. Nat. Commun. 8:14772.
Kawano, F., Suzuki, H., Furuya, A., and Sato, M. (2015). Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 6:6256.
Keppler, A., Gendreizig, S., Gronemeyer, T., Pick, H., Vogel, H., and Johnsson, K. A. (2003). general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89. doi: 10.1038/nbt765
Kijanka, M., Van Donselaar, E. G., Muller, W. H., Dorresteijn, B., Popov-Celeketic, D., El Khattabi, M., et al. (2017). A novel immuno-gold labeling protocol for nanobody-based detection of HER2 in breast cancer cells using immuno-electron microscopy. J. Struct. Biol. 199, 1–11. doi: 10.1016/j.jsb.2017.05.008
Kijanka, M., Warnders, F. J., El Khattabi, M., Lub-de Hooge, M., Van Dam, G. M., Ntziachristos, V., et al. (2013). Rapid optical imaging of human breast tumour xenografts using anti-HER2 VHHs site-directly conjugated to IRDye 800CW for image-guided surgery. Eur. J. Nucl. Med. Mol. Imaging 40, 1718–1729. doi: 10.1007/s00259-013-2471-2
Kirchhofer, A., Helma, J., Schmidthals, K., Frauer, C., Cui, S., Karcher, A., et al. (2010). Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138. doi: 10.1038/nsmb.1727
Klamecka, K., Severin, P. M., Milles, L. F., Gaub, H. E., and Leonhardt, H. (2015). Energy profile of nanobody-GFP complex under force. Phys. Biol. 12:056009. doi: 10.1088/1478-3975/12/5/056009
Klein, A., Hank, S., Raulf, A., Joest, E. F., Tissen, F., Heilemann, M., et al. (2018). Live-cell labeling of endogenous proteins with nanometer precision by transduced nanobodies. Chem. Sci. 9, 7835–7842. doi: 10.1039/c8sc02910e
Kooijmans, S. A. A., Aleza, C. G., Roffler, S. R., Van Solinge, W. W., Vader, P., and Schiffelers, R. M. (2016). Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles 5:31053. doi: 10.3402/jev.v5.31053
Krüwel, T., Nevoltris, D., Bode, J., Dullin, C., Baty, D., Chames, P., et al. (2016). In vivo detection of small tumour lesions by multi-pinhole SPECT applying a (99m)Tc-labelled nanobody targeting the Epidermal Growth Factor Receptor. Sci. Rep. 6:21834.
Kuipers, J., Van Ham, T. J., Kalicharan, R. D., Veenstra-Algra, A., Sjollema, K. A., Dijk, F., et al. (2015). FLIPPER, a combinatorial probe for correlated live imaging and electron microscopy, allows identification and quantitative analysis of various cells and organelles. Cell Tissue Res. 360, 61–70. doi: 10.1007/s00441-015-2142-7
Künzl, F., Früholz, S., Fäβler, F., Li, B., and Pimpl, P. (2016). Receptor-mediated sorting of soluble vacuolar proteins ends at the trans-Golgi network/early endosome. Nat. Plants 2:16017.
Lam, S. S., Martell, J. D., Kamer, K. J., Deerinck, T. J., Ellisman, M. H., Mootha, V. K., et al. (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54. doi: 10.1038/nmeth.3179
Laverty, D., Desai, R., Uchański, T., Masiulis, S., Stec, W. J., Malinauskas, T., et al. (2019). Cryo-EM structure of the human α1β3γ2 GABA(A) receptor in a lipid bilayer. Nature 565, 516–520.
Li, T., Bourgeois, J., Celli, S., Glacial, F., Le Sourd, A., Mecheri, S., et al. (2012). Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB J. 26, 3969–3979. doi: 10.1096/fj.11-201384
Li, T., Vandesquille, M., Koukouli, F., Dudeffant, C., Youssef, I., Lenormand, P., et al. (2016). Camelid single-domain antibodies: a versatile tool for in vivo imaging of extracellular and intracellular brain targets. J. Control Release. 243, 1–10. doi: 10.1016/j.jconrel.2016.09.019
Lutje Hulsik, D., Liu, Y., Strokappe, N. M., Battella, S., El Khattabi, M., McCoy, L. E., et al. (2013). A gp41 MPER-specific llama VHH requires a hydrophobic CDR3 for neutralization but not for antigen recognition. PLoS Pathog. 9:e1003202. doi: 10.1371/journal.ppat.1003202
Ma, J., Kelich, J. M., Junod, S. L., and Yang, W. (2017). Super-resolution mapping of scaffold nucleoporins in the nuclear pore complex. J. Cell Sci. 130, 1299–1306. doi: 10.1242/jcs.193912
Maidorn, M., Olichon, A., Rizzoli, S. O., and Opazo, F. (2019). Nanobodies reveal an extra-synaptic population of SNAP-25 and Syntaxin 1A in hippocampal neurons. MAbs 11, 305–321. doi: 10.1080/19420862.2018.1551675
Maier, J., Traenkle, B., and Rothbauer, U. (2015). Real-time analysis of epithelial-mesenchymal transition using fluorescent single-domain antibodies. Sci. Rep. 5:13402.
Maier, J., Traenkle, B., and Rothbauer, U. (2016). Visualizing epithelial-mesenchymal transition using the chromobody technology. Cancer Res. 76, 5592–5596. doi: 10.1158/0008-5472.can-15-3419
Maiuri, T., Mocle, A. J., Hung, C. L., Xia, J., Van Roon-Mom, W. M., and Truant, R. (2017). Huntingtin is a scaffolding protein in the ATM oxidative DNA damage response complex. Hum. Mol. Genet. 26, 395–406.
Mann, F. A., Lv, Z., Groβhans, J., Opazo, F., and Kruss, S. (2019). Nanobody-conjugated nanotubes for targeted near-infrared in vivo imaging and sensing. Angew. Chem. Int. Ed. Engl. 58, 11469–11473. doi: 10.1002/anie.201904167
Martell, J. D., Yamagata, M., Deerinck, T. J., Phan, S., Kwa, C. G., Ellisman, M. H., et al. (2016). A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 34, 774–780. doi: 10.1038/nbt.3563
Martinez-Jothar, L., Beztsinna, N., Van Nostrum, C. F., Hennink, W. E., and Oliveira, S. (2019). Selective cytotoxicity to HER2 positive breast cancer cells by saporin-loaded nanobody-targeted polymeric nanoparticles in combination with photochemical internalization. Mol. Pharm. 16, 1633–1647. doi: 10.1021/acs.molpharmaceut.8b01318
Masiulis, S., Desai, R., Uchański, T., Serna Martin, I., Laverty, D., Karia, D., et al. (2019). GABA(A) receptor signalling mechanisms revealed by structural pharmacology. Nature 565, 454–459. doi: 10.1038/s41586-018-0832-5
Mazzega, E., Beran, A., Cabrini, M., and De Marco, A. (2019). In vitro isolation of nanobodies for selective Alexandrium minutum recognition: a model for convenient development of dedicated immuno-reagents to study and diagnostic toxic unicellular algae. Harmful Algae 82, 44–51. doi: 10.1016/j.hal.2019.01.002
McMahon, C., Baier, A. S., Pascolutti, R., Wegrecki, M., Zheng, S., Ong, J. X., et al. (2018). Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 25, 289–296. doi: 10.1038/s41594-018-0028-6
Mikhaylova, M., Cloin, B. M., Finan, K., Van Den Berg, R., Teeuw, J., Kijanka, M. M., et al. (2015). Resolving bundled microtubules using anti-tubulin nanobodies. Nat. Commun. 6:7933.
Monegal, A., Ami, D., Martinelli, C., Huang, H., Aliprandi, M., Capasso, P., et al. (2009). Immunological applications of single-domain llama recombinant antibodies isolated from a naïve library. Protein. Eng. Des. Sel. 22, 273–280. doi: 10.1093/protein/gzp002
Mootz, H. D., Jedlitzke, B., Yilmaz, Z., and Dorner, W. (2019). Photobodies: light-activatable single-domain antibody fragments. Angew. Chem. Int. Ed. Engl. 59, 1506–1510. doi: 10.1002/anie.201912286
Moutel, S., Bery, N., Bernard, V., Keller, L., Lemesre, E., De Marco, A., et al. (2016). NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife 5:e16228.
Muyldermans, S. (2013). Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797. doi: 10.1146/annurev-biochem-063011-092449
Muyldermans, S., Cambillau, C., and Wyns, L. (2001). Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem. Sci. 26, 230–235. doi: 10.1016/s0968-0004(01)01790-x
Nieuwenhuizen, R. P. J., Bates, M., Szymborska, A., Lidke, K. A., Rieger, B., and Stallinga, S. (2015). Quantitative localization microscopy: effects of photophysics and labeling stoichiometry. PLoS One 10:e0127989. doi: 10.1371/journal.pone.0127989
Oba, M., and Tanaka, M. (2012). Intracellular internalization mechanism of protein transfection reagents. Biol. Pharm. Bull. 35, 1064–1068. doi: 10.1248/bpb.b12-00001
Obishakin, E., Stijlemans, B., Santi-Rocca, J., Vandenberghe, I., Devreese, B., Muldermans, S., et al. (2014). Generation of a nanobody targeting the paraflagellar rod protein of trypanosomes. PLoS One 9:e115893. doi: 10.1371/journal.pone.0115893
Olichon, A., and Surrey, T. (2007). Selection of genetically encoded fluorescent single domain antibodies engineered for efficient expression in Escherichia coli. J. Biol. Chem. 282, 36314–36320. doi: 10.1074/jbc.m704908200
Oliveira, S., Van Dongen, G. A., Stigter-Van Walsum, M., Roovers, R. C., Stam, J. C., Mali, W., et al. (2012). Rapid visualization of human tumor xenografts through optical imaging with a near-infrared fluorescent anti-epidermal growth factor receptor nanobody. Mol. Imaging 11, 33–46.
Osswald, M., Santos, A. F., and Morais-de-Sá, E. (2019). Light-induced protein clustering for optogenetic interference and protein interaction analysis in Drosophila S2 cells. Biomolecules 9:61. doi: 10.3390/biom9020061
Pani, A. M., and Goldstein, B. (2018). Direct visualization of a native Wnt in vivo reveals that a long-range Wnt gradient forms by extracellular dispersal. eLife 7:e38325.
Panza, P., Maier, J., Schmees, C., Rothbauer, U., and Sollner, C. (2015). Live imaging of endogenous protein dynamics in zebrafish using chromobodies. Development 142, 1879–1884. doi: 10.1242/dev.118943
Pellis, M., Pardon, E., Zolghadr, K., Rothbauer, U., Vincke, C., Kinne, J., et al. (2012). A bacterial-two-hybrid selection system for one-step isolation of intracellularly functional Nanobodies. Arch Biochem. Biophys. 526, 114–123. doi: 10.1016/j.abb.2012.04.023
Periz, J., Whitelaw, J., Harding, C., Gras, S., Del Rosario Minina, M. I., Latorre-Barragan, F., et al. (2017). Toxoplasma gondii F-actin forms an extensive filamentous network required for material exchange and parasite maturation. Elife 6:e24119.
Pizzo, M. E., Wolak, D. J., Kumar, N. N., Brunette, E., Brunnquell, C. L., Hannocks, M., et al. (2018). Intrathecal antibody distribution in the rat brain: surface diffusion, perivascular transport and osmotic enhancement of delivery. J. Physiol. (Lond.) 596, 445–475. doi: 10.1113/jp275105
Platonova, E., Winterflood, C. M., and Ewers, H. (2015a). A simple method for GFP- and RFP-based dual color single-molecule localization microscopy. ACS Chem. Biol. 10, 1411–1416. doi: 10.1021/acschembio.5b00046
Platonova, E., Winterflood, C. M., Junemann, A., Albrecht, D., Faix, J., and Ewers, H. (2015b). Single-molecule microscopy of molecules tagged with GFP or RFP derivatives in mammalian cells using nanobody binders. Methods 88, 89–97. doi: 10.1016/j.ymeth.2015.06.018
Pleiner, T., Bates, M., and Gorlich, D. (2018). A toolbox of anti-mouse and anti-rabbit IgG secondary nanobodies. J. Cell Biol. 217, 1143–1154. doi: 10.1083/jcb.201709115
Pleiner, T., Bates, M., Trakhanov, S., Lee, C., Schliep, J. E., Chug, H., et al. (2015). Nanobodies: site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. eLife 4:e11349.
Plessner, M., Melak, M., Chinchilla, P., Baarlink, C., and Grosse, R. (2015). Nuclear F-actin formation and reorganization upon cell spreading. J. Biol. Chem. 290, 11209–11216. doi: 10.1074/jbc.m114.627166
Popovic, M., Mazzega, E., Toffoletto, B., and De Marco, A. (2018). Isolation of anti-extra-cellular vesicle single-domain antibodies by direct panning on vesicle-enriched fractions. Microb. Cell Fact. 17:6.
Prole, D. L., and Taylor, C. W. (2019). A genetically encoded toolkit of functionalized nanobodies against fluorescent proteins for visualizing and manipulating intracellular signalling. BMC Biol. 17:41.
Rajan, M., Mortusewicz, O., Rothbauer, U., Hastert, F. D., Schmidthals, K., Rapp, A., et al. (2015). Generation of an alpaca-derived nanobody recognizing γ-H2AX. FEBS Open Bio. 5, 779–788. doi: 10.1016/j.fob.2015.09.005
Rakovich, T. Y., Mahfoud, O. K., Mohamed, B. M., Prina-Mello, A., Crosbie-Staunton, K., Van Den Broeck, T., et al. (2014). Highly sensitive single domain antibody-quantum dot conjugates for detection of HER2 biomarker in lung and breast cancer cells. ACS Nano 8, 5682–5695. doi: 10.1021/nn500212h
Ramos-Gomes, F., Bode, J., Sukhanova, A., Bozrova, S. V., Saccomano, M., Mitkovski, M., et al. (2018). Single- and two-photon imaging of human micrometastases and disseminated tumour cells with conjugates of nanobodies and quantum dots. Sci. Rep. 8:4595.
Rashidian, M., Keliher, E. J., Bilate, A. M., Duarte, J. N., Wojtkiewicz, G. R., Jacobsen, J. T., et al. (2015). Noninvasive imaging of immune responses. Proc. Natl. Acad. Sci. U.S.A. 112, 6146–6151.
Riazi, A., Strong, P. C. R., Coleman, R., Chen, W., Hirama, T., Van Faassen, H., et al. (2013). Pentavalent single-domain antibodies reduce Campylobacter jejuni motility and colonization in chickens. PLoS One 8:e83928. doi: 10.1371/journal.pone.0083928
Ries, J., Kaplan, C., Platonova, E., Eghlidi, H., and Ewers, H. A. (2012). simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584. doi: 10.1038/nmeth.1991
Rocchetti, A., Hawes, C., and Kriechbaumer, V. (2014). Fluorescent labelling of the actin cytoskeleton in plants using a cameloid antibody. Plant Methods 10:12. doi: 10.1186/1746-4811-10-12
Roder, R., Helma, J., Preiss, T., Radler, J. O., Leonhardt, H., and Wagner, E. (2017). Intracellular delivery of nanobodies for imaging of target proteins in live cells. Pharm Res. 34, 161–174. doi: 10.1007/s11095-016-2052-8
Rodriguez, E. A., Campbell, R. E., Lin, J. Y., Lin, M. Z., Miyawaki, A., Palmer, A. E., et al. (2017). The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111–129. doi: 10.1016/j.tibs.2016.09.010
Roebroek, T., Duwé, S., Vandenberg, W., and Dedecker, P. (2015). Reduced fluorescent protein switching fatigue by binding-induced emissive state stabilization. Int. J. Mol. Sci. 2017:5869.
Rothbauer, U., Zolghadr, K., Muyldermans, S., Schepers, A., Cardoso, M. C., and Leonhardt, H. (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7, 282–289. doi: 10.1074/mcp.m700342-mcp200
Rothbauer, U., Zolghadr, K., Tillib, S., Nowak, D., Schermelleh, L., Gahl, A., et al. (2006). Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889. doi: 10.1038/nmeth953
Schermelleh, L., Ferrand, A., Huser, T., Eggeling, C., Sauer, M., Biehlmaier, O., et al. (2019). Super-resolution microscopy demystified. Nat. Cell Biol. 21, 72–84. doi: 10.1038/s41556-018-0251-8
Schmidthals, K., Helma, J., Zolghadr, K., Rothbauer, U., and Leonhardt, H. (2010). Novel antibody derivatives for proteome and high-content analysis. Anal. Bioanal. Chem. 397, 3203–3208. doi: 10.1007/s00216-010-3657-0
Schnell, U., Dijk, F., Sjollema, K. A., and Giepmans, B. N. (2012). Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 9, 152–158. doi: 10.1038/nmeth.1855
Schornack, S., Fuchs, R., Huitema, E., Rothbauer, U., Lipka, V., and Kamoun, S. (2009). Protein mislocalization in plant cells using a GFP-binding chromobody. Plant J. 60, 744–754. doi: 10.1111/j.1365-313x.2009.03982.x
Schorpp, K., Rothenaigner, I., Maier, J., Traenkle, B., Rothbauer, U., and Hadian, K. (2016). A multiplexed high-content screening approach using the chromobody technology to identify cell cycle modulators in living cells. J. Biomol. Screen 21, 965–977. doi: 10.1177/1087057116641935
Seitz, K. J., and Rizzoli, S. O. (2019). GFP nanobodies reveal recently-exocytosed pHluorin molecules. Sci. Rep. 9:7773.
Sherwood, L. J., and Hayhurst, A. (2019). Periplasmic nanobody-APEX2 fusions enable facile visualization of ebola, marburg, and mănglà virus nucleoproteins, alluding to similar antigenic landscapes among marburgvirus and dianlovirus. Viruses 11:364. doi: 10.3390/v11040364
Shi, J., Ma, Y., Zhu, J., Chen, Y., Sun, Y., Yao, Y., et al. (2018). A review on electroporation-based intracellular delivery. Molecules 23:3044. doi: 10.3390/molecules23113044
Shin, Y. J., Park, S. K., Jung, Y. J., Kim, Y. N., Kim, K. S., Park, O. K., et al. (2015). Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 5:14269.
Sograte-Idrissi, S., Oleksiievets, N., Isbaner, S., Eggert-Martinez, M., Enderlein, J., Tsukanov, R., et al. (2019). Nanobody detection of standard fluorescent proteins enables multi-target DNA-PAINT with high resolution and minimal displacement errors. Cells 8:48. doi: 10.3390/cells8010048
Southwell, A. L., Khoshnan, A., Dunn, D. E., Bugg, C. W., Lo, D. C., and Patterson, P. H. (2008). Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J. Neurosci. 28, 9013–9020. doi: 10.1523/jneurosci.2747-08.2008
Stewart, M. P., Lorenz, A., Dahlman, J., and Sahay, G. (2016). Challenges in carrier-mediated intracellular delivery: moving beyond endosomal barriers. Wiley Interdiscip Rev. Nanomed. Nanobiotechnol. 8, 465–478. doi: 10.1002/wnan.1377
Stijlemans, B., Conrath, K., Cortez-Retamozo, V., Van Xong, H., Wyns, L., Senter, P., et al. (2004). Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm. J. Biol. Chem. 279, 1256–1261. doi: 10.1074/jbc.m307341200
Sungkaworn, T., Jobin, M., Burnecki, K., Weron, A., Lohse, M. J., and Calebiro, D. (2017). Single-molecule imaging reveals receptor-G protein interactions at cell surface hot spots. Nature 550, 543–547. doi: 10.1038/nature24264
Szymborska, A., De Marco, A., Daigle, N., Cordes, V. C., Briggs, J. A. G., and Ellenberg, J. (2013). Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658. doi: 10.1126/science.1240672
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S., and Vale, R. D. A. (2014). protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646. doi: 10.1016/j.cell.2014.09.039
Tang, A., Chen, H., Li, T. P., Metzbower, S. R., MacGillavry, H. D., and Blanpied, T. A. (2016a). A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214. doi: 10.1038/nature19058
Tang, J. C., Drokhlyansky, E., Etemad, B., Rudolph, S., Guo, B., Wang, S., et al. (2016b). Detection and manipulation of live antigen-expressing cells using conditionally stable nanobodies. Elife 5:e15312.
Temme, S., Baran, P., Bouvain, P., Grapentin, C., Krämer, W., Knebel, B., et al. (2018). Synthetic cargo internalization receptor system for nanoparticle tracking of individual cell populations by fluorine magnetic resonance imaging. ACS Nano 12, 11178–11192. doi: 10.1021/acsnano.8b05698
Teng, K. W., Ishitsuka, Y., Ren, P., Youn, Y., Deng, X., Ge, P., et al. (2016). Labeling proteins inside living cells using external fluorophores for microscopy. Elife 5:e20378.
Tomanek, B., Iqbal, U., Blasiak, B., Abulrob, A., Albaghdadi, H., Matyas, J. R., et al. (2012). Evaluation of brain tumor vessels specific contrast agents for glioblastoma imaging. Neuro Oncol. 14, 53–63. doi: 10.1093/neuonc/nor183
Tosetti, N., Dos Santos Pacheco, N., Soldati-Favre, D., and Jacot, D. (2019). Three F-actin assembly centers regulate organelle inheritance, cell-cell communication and motility in Toxoplasma gondii. Elife 8:e42669.
Traenkle, B., Emele, F., Anton, R., Poetz, O., Haeussler, R. S., Maier, J., et al. (2015). Monitoring interactions and dynamics of endogenous beta-catenin with intracellular nanobodies in living cells. Mol. Cell Proteomics 14, 707–723. doi: 10.1074/mcp.m114.044016
Traenkle, B., and Rothbauer, U. (2017). Under the microscope: single-domain antibodies for live-cell imaging and super-resolution microscopy. Front. Immunol. 8:1030.
Traenkle, B., Segan, S., Fagbadebo, F. O., Kaiser, P. D., and Rothbauer, U. (2020). A novel epitope tagging system to visualize and monitor antigens in live cells with chromobodies. Sci. Rep. 10:14267.
Traub, L. M. (2019). A nanobody-based molecular toolkit provides new mechanistic insight into clathrin-coat initiation. Elife 8:e41768.
Truan, Z., Tarancón Díez, L., Bönsch, C., Malkusch, S., Endesfelder, U., Munteanu, M., et al. (2013). Quantitative morphological analysis of arrestin2 clustering upon G protein-coupled receptor stimulation by super-resolution microscopy. J. Struct. Biol. 184, 329–334. doi: 10.1016/j.jsb.2013.09.019
Uchański, T., Pardon, E., and Steyaert, J. (2020). Nanobodies to study protein conformational states. Curr. Opin. Struct. Biol. 60, 117–123. doi: 10.1016/j.sbi.2020.01.003
Van Audenhove, I., Boucherie, C., Pieters, L., Zwaenepoel, O., Vanloo, B., Martens, E., et al. (2014). Stratifying fascin and cortactin function in invadopodium formation using inhibitory nanobodies and targeted subcellular delocalization. FASEB J. 28, 1805–1818. doi: 10.1096/fj.13-242537
Van Audenhove, I., Debeuf, N., Boucherie, C., and Gettemans, J. (2015). Fascin actin bundling controls podosome turnover and disassembly while cortactin is involved in podosome assembly by its SH3 domain in THP-1 macrophages and dendritic cells. Biochim. Biophys. Acta 1853, 940–952. doi: 10.1016/j.bbamcr.2015.01.003
Van Audenhove, I., and Gettemans, J. (2016). Nanobodies as versatile tools to understand, diagnose, visualize and treat cancer. EBioMedicine 6, 40–48. doi: 10.1016/j.ebiom.2016.04.028
Van Audenhove, I., Van Impe, K., Ruano-Gallego, D., De Clercq, S., De Muynck, K., Vanloo, B., et al. (2013). Mapping cytoskeletal protein function in cells by means of nanobodies. Cytoskeleton (Hoboken). 70, 604–622. doi: 10.1002/cm.21122
Van De Water, J. A. J. M., Bagci-Onder, T., Agarwal, A. S., Wakimoto, H., Roovers, R. C., Zhu, Y., et al. (2012). Therapeutic stem cells expressing variants of EGFR-specific nanobodies have antitumor effects. Proc. Natl. Acad. Sci. U.S.A. 109, 16642–16647. doi: 10.1073/pnas.1202832109
Van Den Abbeele, A., De Clercq, S., De Ganck, A., De Corte, V., Van Loo, B., Soror, S. H., et al. (2010). A llama-derived gelsolin single-domain antibody blocks gelsolin-G-actin interaction. Cell Mol. Life Sci. 67, 1519–1535. doi: 10.1007/s00018-010-0266-1
Van Driel, P. B. A. A., Boonstra, M. C., Slooter, M. D., Heukers, R., Stammes, M. A., Snoeks, T. J. A., et al. (2016). EGFR targeted nanobody-photosensitizer conjugates for photodynamic therapy in a pre-clinical model of head and neck cancer. J. Control Release 229, 93–105. doi: 10.1016/j.jconrel.2016.03.014
Van Impe, K., Hubert, T., De Corte, V., Vanloo, B., Boucherie, C., Vandekerckhove, J., et al. (2008). A new role for nuclear transport factor 2 and Ran: nuclear import of CapG. Traffic 9, 695–707. doi: 10.1111/j.1600-0854.2008.00720.x
Virant, D., Traenkle, B., Maier, J., Kaiser, P. D., Bodenhofer, M., Schmees, C., et al. (2018). A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat. Commun. 9:930.
Wang, Y., Cai, E., Rosenkranz, T., Ge, P., Teng, K. W., Lim, S. J., et al. (2014). Small quantum dots conjugated to nanobodies as immunofluorescence probes for nanometric microscopy. Bioconjug Chem. 25, 2205–2211. doi: 10.1021/bc5004179
Wedeking, T., Löchte, S., Richter, C. P., Bhagawati, M., Piehler, J., and You, C. (2015). Single cell GFP-trap reveals stoichiometry and dynamics of cytosolic protein complexes. Nano Lett. 15, 3610–3615. doi: 10.1021/acs.nanolett.5b01153
Wegner, W., Ilgen, P., Gregor, C., Van Dort, J., Mott, A. C., Steffens, H., et al. (2017). In vivo mouse and live cell STED microscopy of neuronal actin plasticity using far-red emitting fluorescent proteins. Sci. Rep. 7:11781.
Wendel, S., Fischer, E. C., Martínez, V., Seppälä, S., and Nørholm, M. H. H. (2016). A nanobody:GFP bacterial platform that enables functional enzyme display and easy quantification of display capacity. Microb. Cell Fact. 15:71.
Wolf, K., and Friedl, P. (2009). Mapping proteolytic cancer cell-extracellular matrix interfaces. Clin. Exp. Metastasis 26, 289–298. doi: 10.1007/s10585-008-9190-2
Wöll, S., Bachran, C., Schiller, S., Schröder, M., Conrad, L., Swee, L. K., et al. (2018). Sortaggable liposomes: evaluation of reaction conditions for single-domain antibody conjugation by Sortase-A and targeting of CD11b(+) myeloid cells. Eur. J. Pharm. Biopharm. 133, 138–150. doi: 10.1016/j.ejpb.2018.09.017
Yu, D., Lee, H., Hong, J., Jung, H., Jo, Y., Oh, B. H., et al. (2019). Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins. Nat. Methods 16, 1095–1100. doi: 10.1038/s41592-019-0592-7
Zarschler, K., Prapainop, K., Mahon, E., Rocks, L., Bramini, M., Kelly, P. M., et al. (2014). Diagnostic nanoparticle targeting of the EGF-receptor in complex biological conditions using single-domain antibodies. Nanoscale 6, 6046–6056. doi: 10.1039/c4nr00595c
Zhao, Y., Araki, S., Wu, J., Teramoto, T., Chang, Y., Nakano, M., et al. (2011). An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891. doi: 10.1126/science.1208592
Zheng, F., Luo, S., Ouyang, Z., Zhou, J., Mo, H., Schoonooghe, S., et al. (2019). NIRF-molecular imaging with synovial macrophages-targeting vsig4 nanobody for disease monitoring in a mouse model of arthritis. Int. J. Mol. Sci. 20:3347. doi: 10.3390/ijms20133347
Zou, T., Dembele, F., Beugnet, A., Sengmanivong, L., Trepout, S., Marco, S., et al. (2015). Nanobody-functionalized PEG-b-PCL polymersomes and their targeting study. J. Biotechnol. 214, 147–155. doi: 10.1016/j.jbiotec.2015.09.034
Keywords: nanobody, chromobody, fluobody, probes, light microscopy, super-resolution microscopy, electron microscopy, tagging
Citation: de Beer MA and Giepmans BNG (2020) Nanobody-Based Probes for Subcellular Protein Identification and Visualization. Front. Cell. Neurosci. 14:573278. doi: 10.3389/fncel.2020.573278
Received: 16 June 2020; Accepted: 05 October 2020;
Published: 02 November 2020.
Edited by:
Shai Berlin, Technion Israel Institute of Technology, IsraelReviewed by:
Serge Muyldermans, Vrije University Brussel, BelgiumCopyright © 2020 de Beer and Giepmans. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ben N. G. Giepmans, Yi5uLmcuZ2llcG1hbnNAdW1jZy5ubA==
†Present address: Marit A. de Beer, Department of Biochemistry, Radboud University Medical Center, Nijmegen, Netherlands
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.