
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 10 March 2020
Sec. Cellular Neuropathology
Volume 14 - 2020 | https://doi.org/10.3389/fncel.2020.00044
This article is part of the Research Topic Cellular Neuropathology Editor’s Pick 2021 View all 24 articles
Peroxisomes exist in most cells, where they participate in lipid metabolism, as well as scavenging the reactive oxygen species (ROS) that are produced as by-products of their metabolic functions. In certain tissues such as the liver and kidneys, peroxisomes have more specific roles, such as bile acid synthesis in the liver and steroidogenesis in the adrenal glands. In the brain, peroxisomes are critically involved in creating and maintaining the lipid content of cell membranes and the myelin sheath, highlighting their importance in the central nervous system (CNS). This review summarizes the peroxisomal lifecycle, then examines the literature that establishes a link between peroxisomal dysfunction, cellular aging, and age-related disorders that affect the CNS. This review also discusses the gap of knowledge in research on peroxisomes in the CNS.
Peroxisomes are small, nearly ubiquitous organelles found in almost all cell types, except mature red blood cells (Gronowicz et al., 1984). Their major functions include the beta-oxidation of very-long-chain fatty acids and lipid peroxidation; as a result of this metabolism, they secrete reactive oxygen species (ROS) as by-products (Reddy and Hashimoto, 2001; Poirier et al., 2006; Lodhi et al., 2015; Park et al., 2019). Peroxisomes also possess enzymes that break down ROS, such as catalase and glutathione peroxidase, which breaks down hydrogen peroxide, and superoxide dismutase, which breaks down superoxide (Nordgren and Fransen, 2014). They degrade prostaglandins, amino acids, polyamines, and purines, and are commonly enriched in the kidneys, liver, pancreas and adrenal glands, which are involved in fat metabolism and detoxification (Magalhães and Magalhães, 1997; Bradford, 2007; Ferdinandusse et al., 2009; Hasegawa et al., 2010; Smith and Aitchison, 2013; Vasko, 2016; Baboota et al., 2019). Furthermore, they are implicated in lipogenic and ROS signaling roles in the heart and intestines (Colasante et al., 2015; Morvay et al., 2017). In the central nervous system (CNS) in particular, peroxisomes synthesize lipids that make up the myelin sheath and cellular membranes, as well as ether phospholipids in neurons and glia; peroxisome dysfunction is also known to impair neuronal migration and membranes (Farooqui and Horrocks, 2001; Powers, 2001; Bottelbergs et al., 2010; Kassmann, 2014). They also play a critical role in breaking down D-serine via D-amino acid oxidase (DAO), important in glutamatergic signaling (Sasabe et al., 2014; Figure 1). Certain diseases, such as peroxisomal biogenesis disorders, underscore the importance of functional peroxisomes in the CNS. Peroxisomal biogenesis disorders are a subset of diseases where: (1) peroxisomes are either not present, leading to severe neurological phenotypes (as seen in neonatal adrenoleukodystrophy, where seizures, hypotonia, and loss of vision and hearing occur) and a short lifespan; or (2) genes coding for a single peroxisomal protein are defective, where the symptoms are not as severe (Fujiki et al., 2012; Aubourg et al., 2013). To conclude, peroxisomes are small, but important organelles that play supportive, yet critical roles in maintaining cellular health, especially in the CNS.
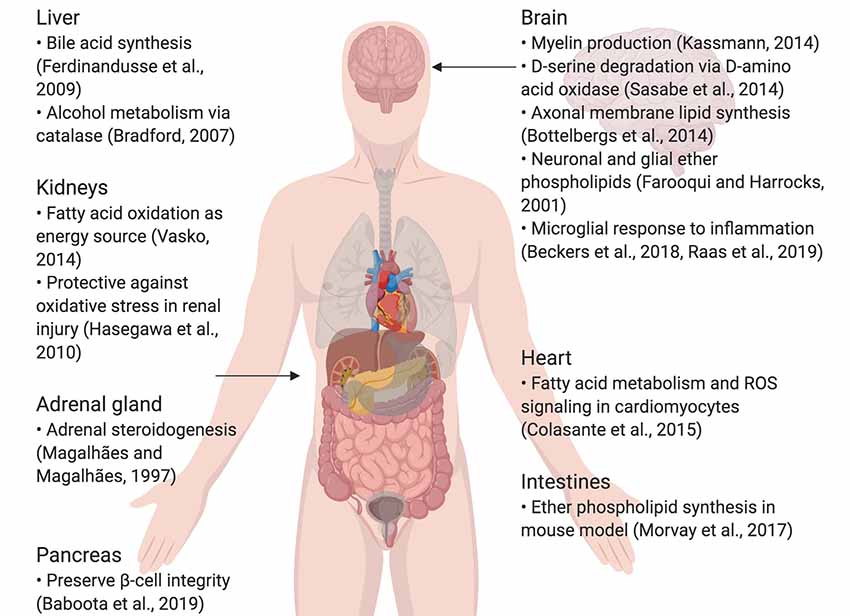
Figure 1. Summary of specialized roles of peroxisomes in some organs, including the brain. Created using BioRender.
This review summarizes peroxisomal biogenesis, and yeast and mammalian pexophagy, with an extended focus on peroxisomes in cellular senescence models, and the peroxisomal dysfunction shared by both age-related neurodegenerative diseases and peroxisomal biogenesis disorders. In all of these conditions, functional peroxisomes move from understudied, secondary organelles to critical sustainers of cellular homeostasis that are disrupted by disease. Future studies will elucidate the role of peroxisomes in aging and CNS function in other diseases and models.
Peroxisomes begin their lifecycle by budding off the endoplasmic reticulum in response to peroxisome proliferator-activated receptor (PPAR) activation due to signaling of the PPAR gamma coactivator-1α (PGC-1α) protein (Bagattin et al., 2010). Unlike mitochondrial proteins, peroxisomal proteins are synthesized on free ribosomes in the cytosol (Koehler, 2000; Jan et al., 2014). After this, peroxisomal proteins are inserted into peroxisomal membranes and matrices by the peroxisomal protein Pex5 (Smith and Aitchison, 2013). Pex5 recognizes the peroxisomal targeting sequence (PTS1) serine-lysine-leucine (SKL), which is found on the C-terminal of many peroxisomal proteins (Brocard and Hartig, 2006). After proteins are inserted, peroxisomes are considered mature and functional. For peroxisomal maintenance, division and maturation, peroxisomes are known to make contact with the endoplasmic reticulum (Hua et al., 2017). To conclude, peroxisomal division and maintenance are modulated by the endoplasmic reticulum, and peroxisomes mature due to peroxisomal protein import into their matrices and membranes.
The peroxisomal lifespan in mammalian cells lasts about 2 to 3 days (Poole et al., 1969; Huybrechts et al., 2009; Moruno-Manchon et al., 2018b). Peroxisomes are then degraded by a selective form of macroautophagy: macropexophagy, which specifically targets peroxisomes (Yang and Klionsky, 2010; Bartoszewska et al., 2012; Cho et al., 2018). A lesser-known form of pexophagy micropexophagy exists, but has only been, so far, observed in yeast models (Strømhaug et al., 2001; Mukaiyama et al., 2004). In macroautophagy, targets for degradation are recognized by a phagophore, which matures to form an autophagosome (Reggiori and Tooze, 2009; Mizushima et al., 2011; Feng et al., 2014; Biazik et al., 2015; Moruno Manchon et al., 2015, 2016). The autophagosome envelops the targets and then fuses with an acidic structure known as the lysosome. Together, they form the autophagolysosome, which degrades the target (Figure 2A; Nakamura and Yoshimori, 2017; Sasaki et al., 2017). Pexophagy itself uses the same process; however, peroxisomes are targeted via particular proteins on their membrane (Jin et al., 2013; Cho et al., 2018). Once recognized, peroxisomes are enveloped by the phagophore, and eventually degraded by the autophagolysosome. Recently, a study in HeLa SH-SY5Ycells and mutant Drosophila flies unearthed a novel pexophagy inducer: HSPA9, a heat shock protein which responds to cellular changes such as glucose deprivation (Jo et al., 2020). In summary, peroxisomes that have reached the end of their life cycle are degraded through a selective autophagic process known as pexophagy, due to the enzymatic action of the autophagolysosome.
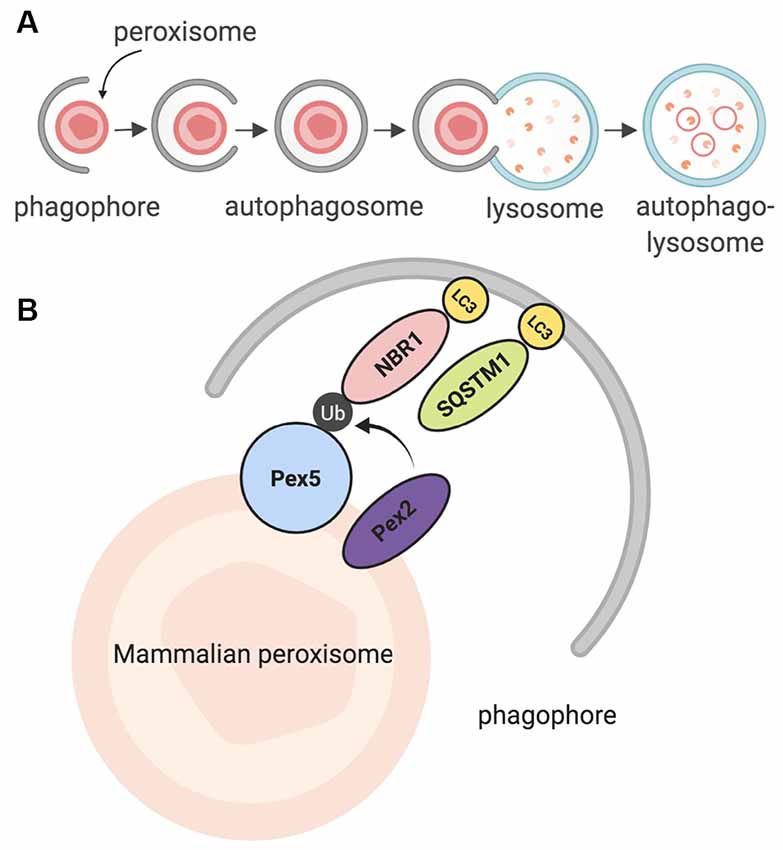
Figure 2. Summary of pexophagy. (A) In macropexophagy, a form of macroautophagy selective for peroxisomes, a single membrane known as a phagophore engulfs a peroxisome for degradation. The phagophore matures into an autophagosome, which then fuses with a lysosome. Their fusion creates an autophagolysosome that degrades the target. (B) In mammalian systems, pexophagy occurs when Pex2 ubiquitinates Pex5. As a result, autophagy adaptor proteins NBR1 or SQSTM1 (p62) bind to ubiquitinated Pex5, and then eventually bind to LC3 on the phagophore. Due to this process, pexophagy occurs. Created using BioRender.
The foundation of the pexophagy mechanism (and peroxisome biology) began in studies using yeast and plants as models (Lefevre et al., 2013; Williams and van der Klei, 2013; Kao and Bartel, 2015; Young and Bartel, 2016). The Saccharomyces cerevisiae yeast homolog of PEX5, Pex5, recognizes cytosolic peroxisomal matrix proteins and delivers them to the peroxisomal membrane (Carvalho et al., 2007). In this model, pexophagy involves Pex3, which is recognized and bound by phosphorylated Atg36, which is itself recognized by Atg8 or Atg11, which are bound to the phagophore (Motley et al., 2012a,b; Farré et al., 2013; Yamashita et al., 2014). In some cases, such as when mitochondria and peroxisomes interact, peroxisomal fission occurs before pexophagy, modulated by Dnm1 and Vps1 (Mao et al., 2014).
Further studies in mammalian models revealed that in order for mammalian pexophagy to begin, PEX5 has to be monoubiquitinated by PEX2, an E3 ubiquitin ligase (Nordgren et al., 2015; Sargent et al., 2016; Germain and Kim, 2020). Previously, it was not clear what directly induces pexophagy; however, recent evidence has shown that increased ROS in the cytosol can stimulate this monoubiquitination, allowing PEX5 to act as a ROS sensor, leading to an increase in peroxisome degradation (Kim et al., 2008; Zhang et al., 2015; Walton et al., 2017). After PEX5 is monoubiquitinated, it is recognized by one of two LC3 adaptor proteins: NBR1, or p62 (SQSTM1); these proteins are then bound to LC3, which is bound to the autophagosome (Figure 2B; Kabeya et al., 2000; Kirkin et al., 2009; Deosaran et al., 2013). Together, the interaction of these proteins induces pexophagy in the mammalian cell.
In the CNS, neurons rely on different forms of autophagy (general and selective) to clear organelles and proteins that are no longer of use; this use of autophagy is due to neurons being post-mitotic and unable to divide, making them more vulnerable than cells that can divide and dilute toxic protein build-up (Moore and Holzbaur, 2016; Evans and Holzbaur, 2019; Stavoe and Holzbaur, 2019a,b). Neuronal autophagy is compartment-specific: it begins at the distal axon, after which axonal autophagosomes then move into the cell soma; the soma also contains its own autophagosomes (Maday and Holzbaur, 2016; Kulkarni et al., 2018; Moruno-Manchon et al., 2018a). Neurons also respond to autophagy inducers differently than other neural cells, underscoring the uniqueness of neuronal autophagy among other forms of autophagy (Ferguson et al., 2009; Pamenter et al., 2012; Bordi et al., 2016; Moruno Manchon et al., 2016; Kulkarni et al., 2019; Sung and Jimenez-Sanchez, 2020).
Interestingly, a common trait of neurodegenerative diseases is the impairment of protein and organelle turnover. Alzheimer disease (AD) is the most common form of dementia in elderly people, with patients exhibiting symptoms such as memory loss and mood changes; the disease eventually destroys neurons in the hippocampus and the cortex (Liang et al., 2008; GBD 2013 Mortality and Causes of Death Collaborators, 2015). In AD, beta-amyloid and tau accumulate, and senescent mitochondria are also present (Zilka et al., 2006; Mitchell, 2009; Nilsson et al., 2013; Shi et al., 2016; Harada et al., 2018). While Parkinson’s disease (PD) has a lower prevalence than AD, the number of people with PD has increased over time, as the number of aged people has increased (Dorsey et al., 2018). In PD, neurodegeneration occurs in the substantia nigra, leading to tremors, bradykinesia, postural instability, and rigidity (Jagadeesan et al., 2017). Huntington disease (HD) occurs due to the mutated huntingtin gene and affects the medium spiny neurons in the striatum as well as neurons in the cortex, leading to symptoms such as chorea (jerky movements), rigidity and progressive motor failure (Ehrlich, 2012; Wyant et al., 2017). In Parkinson disease and HD, damaged mitochondria and causative proteins (alpha-synuclein and to a much smaller extent, tau in PD, and mutant huntingtin in HD) accumulate in affected neurons, indicating a problem with autophagy or the ubiquitin/proteasome system (Bloom, 2014; Atik et al., 2016; Zhao et al., 2016; Chiasseu et al., 2017; Zhang et al., 2018; Finkbeiner, 2019; Harrison et al., 2019). Amyotrophic lateral sclerosis (ALS) can be familial or sporadic, leading to neurodegeneration of motor neurons in the CNS; a wide range of genetic mutations can induce this neurodegeneration, including the SOD1 gene, which codes for superoxide dismutase (Peters et al., 2015). Inducing autophagy improves survival in neuronal ALS models (Barmada et al., 2014). In aging neurons, mitochondrial senescence is observed (Gilmer et al., 2010; Menzies et al., 2017). However, not much is known about how pexophagy, or how peroxisomal proteins are affected by these diseases. First, we will summarize the present data on peroxisomes and pexophagy in neurodegenerative disease studies, then review cases where global peroxisomal disturbances lead to neurodegenerative phenotypes.
In some neurodegenerative diseases, the amount and/or function of peroxisomes may be compromised. In Alzheimer’s disease, in which beta-amyloid and tau accumulate in neurons, peroxisomes may be affected. In one study, rat hippocampal cultures with beta-amyloid overexpression were treated with Wy-14.463, a peroxisomal proliferator. This treatment increased peroxisomal number and catalase activity reduced ROS production, and overall, reduced the degenerative effects of beta-amyloid such as the instability of beta-catenin and the increase of calcium (Santos et al., 2005). In a clinical study, plasmalogens (which peroxisomes synthesize) were negatively affected in post-mortem samples of Alzheimer patients’ brains, suggesting a reduction in peroxisomal activity, or a shorter half-life of plamalogens (Goodenowe and Senanayake, 2019). ALS, a disease in which motor neurons degenerate, is linked to peroxisome dysfunction through a genetic mutation that codes for DAO, a peroxisomal enzyme that specifically breaks down D-serine (Kondori et al., 2017, 2018).
In other cases, peroxisome dysfunction, as seen in peroxisome biogenesis disorders, may lead to degenerative neurological symptoms. Peroxisome biogenesis disorders occur due to peroxisome genetic defects, either resulting in single peroxisomal enzyme dysfunction, or in rare cases, the absence of peroxisomes themselves (Braverman et al., 2016). Two groupings of peroxisome biogenesis disorders exist under the Zellweger spectrum (neonatal adrenoleukodystrophy, Zellweger syndrome and infantile Refsum disease), and those outside of it. In Zellweger syndrome, which is inherited in an autosomal recessive manner, one of 13 peroxin (PEX) genes is mutated (PEX1, PEX2, PEX3, PEX5, PEX6, PEX10, PEX11β, PEX12, PEX13, PEX14, PEX16, PEX19, PEX26), leading to issues with neuronal migration, myelination and brain development (Waterham and Ebberink, 2012; Klouwer et al., 2015; Wang et al., 2015). A cellular model of Zellweger syndrome, particularly of a Pex5 mutation, has shown an increase in alpha-synuclein Lewy bodies; alpha-synuclein is thought to be a causative agent in Parkinson disease, particularly in familial cases (Yakunin et al., 2010; Riederer et al., 2019). In vivo, Pex5−/− mouse brain samples exhibited an increase in alpha-synuclein oligomers in comparison to control, suggesting a correlation between peroxisome dysfunction and PD (Yakunin et al., 2010). Neonatal adrenoleukodystrophy is also an autosomal recessive PBD, but with multiple peroxisomal enzymes affected; infant patients exhibit neurological symptoms such as hearing loss, neuropathy, and demyelination (Aubourg et al., 1986). The last PBD under the Zellweger spectrum is infantile Refsum disease, where a build-up of phytanic acid and other very-long-chain fatty acids in the body (a result of mutated PEX genes) leads to neurological symptoms such as mixed neuropathy and hearing loss (Warren et al., 2018). Outside the Zellweger spectrum, adult Refsum disease has similar symptoms to infantile Refsum disease, but the adult-onset disease is due to a mutation in the PHYH gene that codes for the peroxisomal enzyme phytanoyl-CoA dioxygenase, which peroxisomes use to break down phytanic acid into pristanic acid (Wanders et al., 2011; Wanders and Poll-The, 2017; Gettelfinger and Dahl, 2018).
Rhizomelic chondrodysplasia punctata (RCDP) is a set of peroxisome biogenesis disorders where peroxisomal genes coding for proteins involved in plasmalogen synthesis are mutated (Barøy et al., 2015). Of note is RCDP type 1, which is due to the mutation of PEX7, which codes for PEX7, a peroxisomal receptor that inserts proteins into the peroxisomal membrane that carries peroxisome targeting signal 2 (PTS2; Purdue et al., 1999). This mutation results in severe neurological symptoms such as epilepsy and age-related conditions such as cataracts (Purdue et al., 1999; Malheiro et al., 2015; Landino et al., 2017). In conclusion, peroxisomal dysfunction in the CNS is shared by both neurodegenerative and peroxisomal disorders, leading to disrupted cellular homeostasis that contributes to the pathogenesis of those diseases (Figure 3 and Table 1).
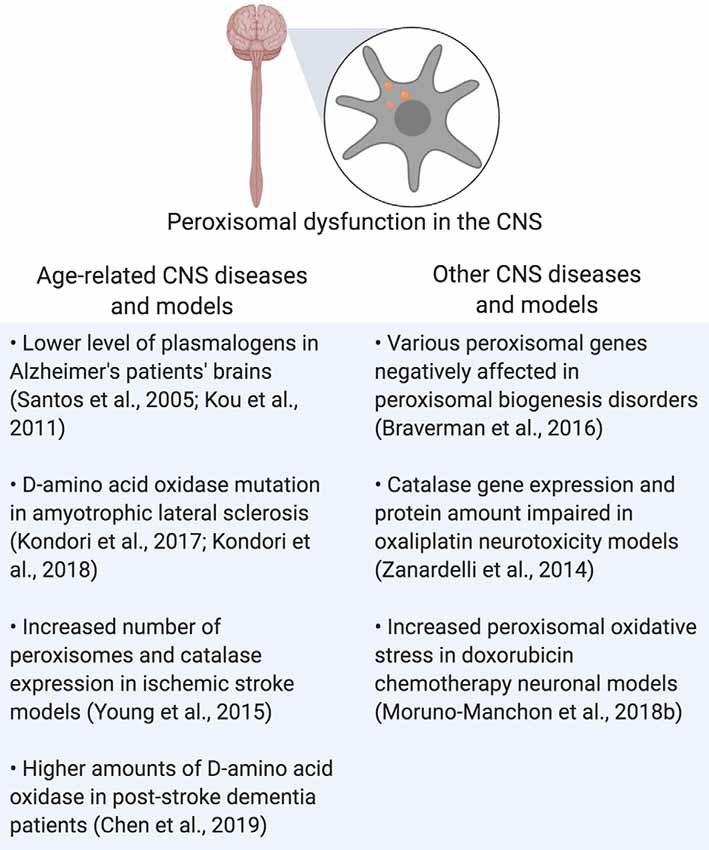
Figure 3. Summary of peroxisomal dysfunction in age-related diseases in the CNS, and other diseases that affect the CNS. Created using BioRender.
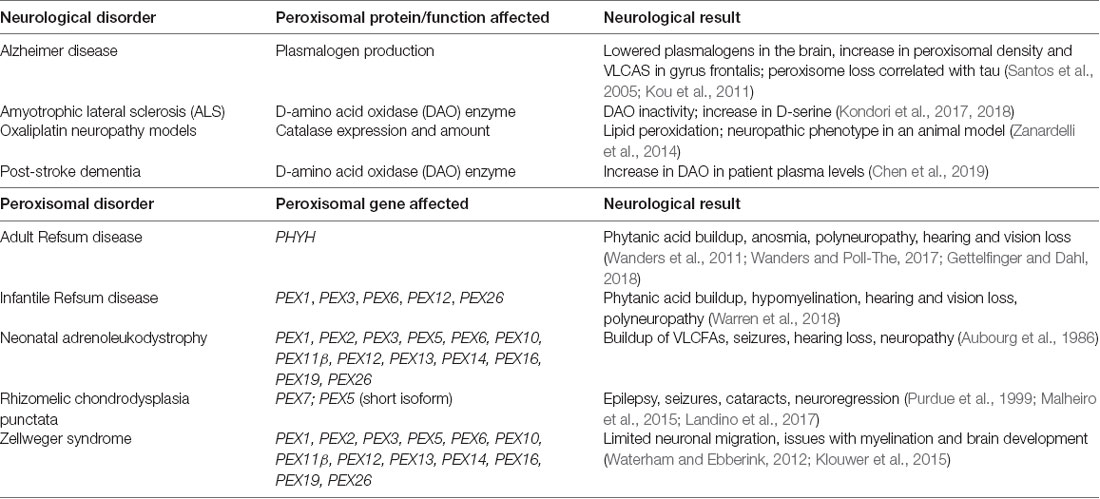
Table 1. Summary of neurological symptoms in neurological and peroxisomal disorders that arise as a result of peroxisomal dysfunction.
Peroxisomal dysfunction also contributes to neuropathies. For instance, oxaliplatin, a chemotherapy drug for colorectal cancer, is known to cause peripheral neuropathies in patients (Grothey, 2003; Banach et al., 2018). A study uncovered the role of peroxisomes in this mechanism using primary rat astrocyte cultures, a human colon cancer cell line and ex vivo analysis of an oxaliplatin neuropathy rat model: peroxisomal catalase expression and levels were impaired with oxaliplatin treatment of cell cultures, and in the dorsal root ganglia and spinal cords of treated animals; this change was also linked with lipid peroxidation in the spinal cord of treated animals (Zanardelli et al., 2014). More recent research has strengthened the role of peroxisome function in neuropathies: the peripheral nerves in peroxisomal mutation mouse models exhibited various abnormalities, such as impaired lysosomal function, accumulation of ganglioside, and a changed redistribution of Kv1 channels and their anchoring proteins that may lead to impaired signaling (Kleinecke et al., 2017). These studies, in conclusion, highlight the important, but previously hidden role that peroxisomal function plays, not only in the CNS but in peripheral nerves as well.
As previously mentioned, peroxisomes are negatively affected by disorders that affect the CNS, leading to undesirable consequences. Some characterization of basal peroxisomal pathways has been made in oligodendrocytes and astrocytes in the CNS (Chistyakov et al., 2014; Di Cesare Mannelli et al., 2014; Aguirre-Rueda et al., 2015; Nury et al., 2018). In the case of neurons, there has also been a focus on peroxisomes (Ballister et al., 2015; Olenick et al., 2016). In hippocampal neurons, it was discovered that preventing tuberous sclerosis complex 2 (TSC2; a regulator of mTORC1 activity) from localizing to peroxisomes led to several axons extending from the neuronal body, indicating a change in morphology (Zhang et al., 2013). In studies of noise-induced hair loss, neurons in mice deficient in pevjakin (a protein associated with neuronal peroxisomes in the auditory pathway), exhibited less peroxisomal proliferation in response to loud sounds in comparison to their wild-type counterparts; peroxisomal proliferation is protective against oxidative stress produced by loud sounds (Defourny et al., 2019). We recently discovered that in neuronal models of doxorubicin treatment (a chemotherapy drug that leads to chemobrain), peroxisomes exhibited increased oxidative stress, which eventually damaged neurons (Kesler, 2014; Wefel et al., 2015; Kesler and Blayney, 2016; Manchon et al., 2016; Moruno-Manchon et al., 2016, 2018b). A more positive link has been found between peroxisomes and ischemic stroke; peroxisomal volume in in vitro and in vivo models of ischemia increased after injury, leading to an increased number of peroxisomes, as well as increased expression of peroxisomal catalase (Young et al., 2015). Inhibiting catalase or dynamin-related protein 1 (Drp1), a protein needed for peroxisomal fission, led to increased neuronal susceptibility to death from oxygen-glucose deprivation (OGD), a cellular model of ischemic stroke (Young et al., 2015). These findings inspired a clinical study, which investigated the link between post-stroke dementia (PSD) and peroxisomal DAO, an enzyme that oxidizes D-serine; plasma levels of PSD patients had higher levels of DAO, indicating its role in stroke and stroke-related dementia (Chen et al., 2019). In conclusion, these neuronal studies show that peroxisomal dysfunction can contribute to changes in neuronal morphology, increased oxidative stress, and even death in the CNS. Therefore, it is crucial to keep the negative side effects of treatments on various metabolic pathways, including those that involve peroxisomes, in mind.
The link between peroxisomal function and inflammation has been established in non-CNS models; however, a few microglial studies have shed light on potential peroxisomal dysfunction mechanisms in the brain (Di Cara et al., 2019). For one, deleting the MFP2 peroxisomal enzyme (which is responsible for β-oxidation) in mouse microglia, switched their state to a pro-inflammatory one, but this change did not affect neuronal health or the microglial response to injury (Beckers et al., 2019). Another study looked at a neuron-specific form of MFP2 deletion and discovered that unlike constitutive Mfp2−/− knockouts, Nestin-Mfp2−/− knockout brains possessed microglia that were not primed for an inflammatory response (Beckers et al., 2018). Microglial peroxisomal dysfunction, as seen in a microglial model deficient in acyl-CoA oxidase 1 (ACOX1), has also been shown to affect catalase activity, the peroxisome, lipid droplet and mitochondrial number in microglia, as well as the induction of interleukin-1β (IL-1β), the repression of interleukin-6 (IL-6) and the increased expression of Trem2, which codes for a cell surface protein that plays a role in microglial phagocytosis (Raas et al., 2019). Taking these studies together, it can be assumed that microglial peroxisomal dysfunction affects the inflammatory response of microglia in the brain, directly and indirectly. The results of these microglial studies stress the importance of the peroxisomal role in inflammation of the CNS: peroxisomal dysfunction in microglia may lead to a pro-inflammatory response that negatively affects the whole system.
Nonetheless, one gap in the literature exists regarding peroxisomes in the aging CNS, that is unaffected by neurodegenerative disease. Non-neuronal senescence studies have shed some light on peroxisomes in aging cells, such as in senescent human fibroblasts, where there is a reported reduction in the import of PTS1-tagged proteins, an increase in hydrogen peroxide and peroxisomal number, and changes in peroxisomal appearance (Legakis et al., 2002). Proteomic analysis of C. elegans also shows a reduction of peroxisomal protein import, as well as a reduction in about 30 peroxisomal proteins, including PRX-5, the nematode homolog of PEX5; PRX-5 was also found to be mislocalized in the aged animals, suggesting that peroxisomal proteins were not properly localized (Narayan et al., 2016). Knocking it out reduced brood size, implicating a potential role of PRX-5 in both development and aging (Narayan et al., 2016). Cell type-specific ribosome profiling of Drosophila melanogaster oenocytes (cells involved in liver-like processes) revealed that peroxisomal pathways were downregulated with aging (Huang et al., 2019). Some related evidence exists in post-mortem Alzheimer’s studies, where there is an increase in peroxisomal density and very-long-chain fatty acids (but a reduction in plasmalogen levels) in neurons in the gyrus frontalis of AD patients, and a loss of peroxisomes in neuronal processes where phosphorylated tau is present (Kou et al., 2011). However, a search of the literature does not currently reveal evidence of peroxisomal perturbations in the normal aging brain. Another gap in the literature is present when investigating how sex, particularly in age-related neurological disease, affects peroxisomes. For instance, there is evidence that a sex difference exists in response to cerebral ischemia, or ischemic stroke, but it is unknown how these sex-associated differences may affect peroxisomes specifically (Siegel and McCullough, 2013; Mirza et al., 2015; Ritzel et al., 2017). Future studies on age-related neurological changes should investigate how peroxisomal pathways are affected, given the important roles that peroxisomes play in the brain, and how they are affected in other related diseases.
As small and understudied as they are, there is ample evidence that peroxisomes play a supportive, yet critical role in the maintenance of the CNS; future studies should investigate the treatment of neurological diseases while keeping the peroxisomal role in maintaining cellular homeostasis in mind.
N-EU wrote the manuscript. All authors contributed to manuscript revision and references, read and approved the submitted version.
This work was supported by the 2017–2018 Russell and Diana Hawkins Family Foundation Discovery Fellowship and the National Institutes of Health, Grant No. R01NS094543-02.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures were created in BioRender.
Aguirre-Rueda, D., Guerra-Ojeda, S., Aldasoro, M., Iradi, A., Obrador, E., Ortega, A., et al. (2015). Astrocytes protect neurons from Aβ1–42 peptide-induced neurotoxicity increasing TFAM and PGC-1 and decreasing PPAR-γ and SIRT-1. Int. J. Med. Sci. 12, 48–56. doi: 10.7150/ijms.10035
Atik, A., Stewart, T., and Zhang, J. (2016). α-synuclein as a biomarker for Parkinson’s disease. Brain Pathol. 26, 410–418. doi: 10.1111/bpa.12370
Aubourg, P., Scotto, J., Rocchiccioli, F., Feldmann-Pautrat, D., and Robain, O. (1986). Neonatal adrenoleukodystrophy. J. Neurol. Neurosurg. Psychiatry 49, 77–86. doi: 10.1136/jnnp.49.1.77
Aubourg, P., Wanders, R., Dulac, O., Lassonde, M., and Sarnat, H. B. (2013). “Chapter 163 - Peroxisomal disorders,” in Handbook of Clinical Neurology, eds O. Dulac, M. Lassonde and H. B. Sarnat (Amsterdam, Netherlands: Elsevier), 1593–1609.
Baboota, R. K., Shinde, A. B., Lemaire, K., Fransen, M., Vinckier, S., Van Veldhoven, P. P., et al. (2019). Functional peroxisomes are required for β-cell integrity in mice. Mol. Metab. 22, 71–83. doi: 10.1016/j.molmet.2019.02.001
Bagattin, A., Hugendubler, L., and Mueller, E. (2010). Transcriptional coactivator PGC-1α promotes peroxisomal remodeling and biogenesis. Proc. Natl. Acad. Sci. U S A 107, 20376–20381. doi: 10.1073/pnas.1009176107
Ballister, E. R., Ayloo, S., Chenoweth, D. M., Lampson, M. A., and Holzbaur, E. L. F. (2015). Optogenetic control of organelle transport using a photocaged chemical inducer of dimerization. Curr. Biol. 25, R407–R408. doi: 10.1016/j.cub.2015.03.056
Banach, M., Zygulska, A. L., and Krzemieniecki, K. (2018). Oxaliplatin treatment and peripheral nerve damage in cancer patients: a Polish cohort study. J. Cancer Res. Ther. 14, 1010–1013. doi: 10.4103/jcrt.jcrt_971_16
Barmada, S. J., Serio, A., Arjun, A., Bilican, B., Daub, A., Ando, D. M., et al. (2014). Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat. Chem. Biol. 10, 677–685. doi: 10.1038/nchembio.1563
Barøy, T., Koster, J., Strømme, P., Ebberink, M. S., Misceo, D., Ferdinandusse, S., et al. (2015). A novel type of rhizomelic chondrodysplasia punctata, RCDP5, is caused by loss of the PEX5 long isoform. Hum. Mol. Genet. 24, 5845–5854. doi: 10.1093/hmg/ddv305
Bartoszewska, M., Williams, C., Kikhney, A., Opaliński, Ł., van Roermund, C. W. T., de Boer, R., et al. (2012). Peroxisomal proteostasis involves a lon family protein that functions as protease and chaperone. J. Biol. Chem. 287, 27380–27395. doi: 10.1074/jbc.m112.381566
Beckers, L., Geric, I., Stroobants, S., Beel, S., Van Damme, P., D’Hooge, R., et al. (2019). Microglia lacking a peroxisomal β-oxidation enzyme chronically alter their inflammatory profile without evoking neuronal and behavioral deficits. J. Neuroinflammation 16:61. doi: 10.1186/s12974-019-1442-3
Beckers, L., Stroobants, S., D’Hooge, R., and Baes, M. (2018). Neuronal dysfunction and behavioral abnormalities are evoked by neural cells and aggravated by inflammatory microglia in peroxisomal β-oxidation deficiency. Front. Cell. Neurosci. 12:136. doi: 10.3389/fncel.2018.00136
Biazik, J., Ylä-Anttila, P., Vihinen, H., Jokitalo, E., and Eskelinen, E.-L. (2015). Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy 11, 439–451. doi: 10.1080/15548627.2015.1017178
Bloom, G. S. (2014). Amyloid-β and tau: the trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 71, 505–508. doi: 10.1001/jamaneurol.2013.5847
Bordi, M., Berg, M. J., Mohan, P. S., Peterhoff, C. M., Alldred, M. J., Che, S., et al. (2016). Autophagy flux in CA1 neurons of Alzheimer hippocampus: increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy 12, 2467–2483. doi: 10.1080/15548627.2016.1239003
Bottelbergs, A., Verheijden, S., Hulshagen, L., Gutmann, D. H., Goebbels, S., Nave, K.-A., et al. (2010). Axonal integrity in the absence of functional peroxisomes from projection neurons and astrocytes. Glia 58, 1532–1543. doi: 10.1002/glia.21027
Bradford, B. U. (2007). Role of peroxisomes in the swift increase in alcohol metabolism. J. Gastroenterol. Hepatol. 22, S28–S30. doi: 10.1111/j.1440-1746.2006.04641.x
Braverman, N. E., Raymond, G. V., Rizzo, W. B., Moser, A. B., Wilkinson, M. E., Stone, E. M., et al. (2016). Peroxisome biogenesis disorders in the Zellweger spectrum: an overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol. Genet. Metab. 117, 313–321. doi: 10.1016/j.ymgme.2015.12.009
Brocard, C., and Hartig, A. (2006). Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573. doi: 10.1016/j.bbamcr.2006.08.022
Carvalho, A. F., Pinto, M. P., Grou, C. P., Alencastre, I. S., Fransen, M., Sá-Miranda, C., et al. (2007). Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267–31272. doi: 10.1074/jbc.M706325200
Chen, Y.-C., Chou, W.-H., Tsou, H.-H., Fang, C.-P., Liu, T.-H., Tsao, H.-H., et al. (2019). A post hoc study of d-amino acid oxidase in blood as an indicator of post-stroke dementia. Front. Neurol. 10:402. doi: 10.3389/fneur.2019.00402
Chiasseu, M., Alarcon-Martinez, L., Belforte, N., Quintero, H., Dotigny, F., Destroismaisons, L., et al. (2017). Tau accumulation in the retina promotes early neuronal dysfunction and precedes brain pathology in a mouse model of Alzheimer’s disease. Mol. Neurodegener. 12:58. doi: 10.1186/s13024-017-0199-3
Chistyakov, D. V., Aleshin, S., Sergeeva, M. G., and Reiser, G. (2014). Regulation of peroxisome proliferator-activated receptor β/δ expression and activity levels by toll-like receptor agonists and MAP kinase inhibitors in rat astrocytes. J. Neurochem. 130, 563–574. doi: 10.1111/jnc.12757
Cho, D.-H., Kim, Y. S., Jo, D. S., Choe, S.-K., and Jo, E.-K. (2018). Pexophagy: molecular mechanisms and implications for health and diseases. Mol. Cells 41, 55–64. doi: 10.14348/molcells.2018.2245
Colasante, C., Chen, J., Ahlemeyer, B., and Baumgart-Vogt, E. (2015). Peroxisomes in cardiomyocytes and the peroxisome/peroxisome proliferator-activated receptor-loop. Thromb. Haemost. 113, 452–463. doi: 10.1160/th14-06-0497
Defourny, J., Aghaie, A., Perfettini, I., Avan, P., Delmaghani, S., and Petit, C. (2019). Pejvakin-mediated pexophagy protects auditory hair cells against noise-induced damage. Proc. Natl. Acad. Sci. U S A 116, 8010–8017. doi: 10.1073/pnas.1821844116
Deosaran, E., Larsen, K. B., Hua, R., Sargent, G., Wang, Y., Kim, S., et al. (2013). NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126, 939–952. doi: 10.1242/jcs.114819
Di Cara, F., Andreoletti, P., Trompier, D., Vejux, A., Bülow, M. H., Sellin, J., et al. (2019). Peroxisomes in immune response and inflammation. Int. J. Mol. Sci. 20:E3877. doi: 10.3390/ijms20163877
Di Cesare Mannelli, L., Zanardelli, M., Micheli, L., and Ghelardini, C. (2014). PPAR-γ impairment alters peroxisome functionality in primary astrocyte cell cultures. Biomed Res. Int. 2014, 546453–546453. doi: 10.1155/2014/546453
Dorsey, E. R., Elbaz, A., Nichols, E., Abd-Allah, F., Abdelalim, A., Adsuar, J. C., et al. (2018). Global, regional, and national burden of Parkinson’s disease, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet Neurol. 17, 939–953. doi: 10.1016/S1474-4422(18)30295-3
Ehrlich, M. E. (2012). Huntington’s disease and the striatal medium spiny neuron: cell-autonomous and non-cell-autonomous mechanisms of disease. Neurotherapeutics 9, 270–284. doi: 10.1007/s13311-012-0112-2
Evans, C. S., and Holzbaur, E. L. F. (2019). Quality control in neurons: mitophagy and other selective autophagy mechanisms. J. Mol. Biol. 432, 240–260. doi: 10.1016/j.jmb.2019.06.031
Farooqui, A. A., and Horrocks, L. A. (2001). Book review: plasmalogens: workhorse lipids of membranes in normal and injured neurons and glia. Neuroscientist 7, 232–245. doi: 10.1177/107385840100700308
Farré, J.-C., Burkenroad, A., Burnett, S. F., and Subramani, S. (2013). Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 14, 441–449. doi: 10.1038/embor.2013.40
Feng, Y., He, D., Yao, Z., and Klionsky, D. J. (2014). The machinery of macroautophagy. Cell Res. 24, 24–41. doi: 10.1038/cr.2013.168
Ferdinandusse, S., Denis, S., Faust, P. L., and Wanders, R. J. A. (2009). Bile acids: the role of peroxisomes. J. Lipid Res. 50, 2139–2147. doi: 10.1194/jlr.R900009-JLR200
Ferguson, C. J., Lenk, G. M., and Meisler, M. H. (2009). Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 18, 4868–4878. doi: 10.1093/hmg/ddp460
Finkbeiner, S. (2019). The autophagy lysosomal pathway and neurodegeneration. Cold. Spring Harbor. Perspect. Biol. doi: 10.1101/cshperspect.a033993 [Epub ahead of print].
Fujiki, Y., Yagita, Y., and Matsuzaki, T. (2012). Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease. Biochim. Biophys. Acta 1822, 1337–1342. doi: 10.1016/j.bbadis.2012.06.004
GBD 2013 Mortality and Causes of Death Collaborators. (2015). Global, regional and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385, 117–171. doi: 10.1016/S0140-6736(14)61682-2
Germain, K., and Kim, K. P. (2020). Pexophagy: a model for selective autophagy. Int. J. Mol. Sci. 21:E578. doi: 10.3390/ijms21020578
Gettelfinger, J. D., and Dahl, J. P. (2018). Syndromic hearing loss: a brief review of common presentations and genetics. J. Pediatr. Genet. 7, 1–8. doi: 10.1055/s-0037-1617454
Gilmer, L. K., Ansari, M. A., Roberts, K. N., and Scheff, S. W. (2010). Age-related changes in mitochondrial respiration and oxidative damage in the cerebral cortex of the Fischer 344 rat. Mech. Ageing Dev. 131, 133–143. doi: 10.1016/j.mad.2009.12.011
Goodenowe, D. B., and Senanayake, V. (2019). Relation of serum plasmalogens and APOE genotype to cognition and dementia in older persons in a cross-sectional study. Brain Sci. 9:E92. doi: 10.3390/brainsci9040092
Gronowicz, G., Swift, H., and Steck, T. L. (1984). Maturation of the reticulocyte in vitro. J. Cell Sci. 71, 177–197.
Grothey, A. (2003). Oxaliplatin-safety profile: neurotoxicity. Semin. Oncol. 30, 5–13. doi: 10.1016/s0093-7754(03)00399-3
Harada, R., Ishiki, A., Kai, H., Sato, N., Furukawa, K., Furumoto, S., et al. (2018). Correlations of 18F-THK5351 PET with postmortem burden of tau and astrogliosis in Alzheimer disease. J. Nucl. Med. 59, 671–674. doi: 10.2967/jnumed.117.197426
Harrison, T. M., La Joie, R., Maass, A., Baker, S. L., Swinnerton, K., Fenton, L., et al. (2019). Longitudinal tau accumulation and atrophy in aging and Alzheimer disease. Ann. Neurol. 85, 229–240. doi: 10.1002/ana.25406
Hasegawa, K., Wakino, S., Yoshioka, K., Tatematsu, S., Hara, Y., Minakuchi, H., et al. (2010). Kidney-specific overexpression of Sirt1 protects against acute kidney injury by retaining peroxisome function. J. Biol. Chem. 285, 13045–13056. doi: 10.1074/jbc.m109.067728
Hua, R., Cheng, D., Coyaud, É., Freeman, S., Di Pietro, E., Wang, Y., et al. (2017). VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 216, 367–377. doi: 10.1083/jcb.201608128
Huang, K., Chen, W., Zhu, F., Li, P. W.-L., Kapahi, P., and Bai, H. (2019). RiboTag translatomic profiling of Drosophila oenocytes under aging and induced oxidative stress. BMC Genomics 20:50. doi: 10.1186/s12864-018-5404-4
Huybrechts, S. J., Van Veldhoven, P. P., Brees, C., Mannaerts, G. P., Los, G. V., and Fransen, M. (2009). Peroxisome dynamics in cultured mammalian cells. Traffic 10, 1722–1733. doi: 10.1111/j.1600-0854.2009.00970.x
Jagadeesan, A. J., Murugesan, R., Vimala Devi, S., Meera, M., Madhumala, G., Vishwanathan Padmaja, M., et al. (2017). Current trends in etiology, prognosis and therapeutic aspects of Parkinson’s disease: a review. Acta Biomed. 88, 249–262. doi: 10.23750/abm.v88i3.6063
Jan, C. H., Williams, C. C., and Weissman, J. S. (2014). Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346:1257521. doi: 10.1126/science.1257521
Jin, M., Liu, X., and Klionsky, D. J. (2013). SnapShot: selective autophagy. Cell 152, 368.e2–368.e2. doi: 10.1016/j.cell.2013.01.004
Jo, D. S., Park, S. J., Kim, A.-K., Park, N. Y., Kim, J. B., Bae, J.-E., et al. (2020). Loss of HSPA9 induces peroxisomal degradation by increasing pexophagy. Autophagy doi: 10.1080/15548627.2020.1712812 [Epub ahead of print].
Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., et al. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728. doi: 10.1093/emboj/19.21.5720
Kao, Y.-T., and Bartel, B. (2015). Elevated growth temperature decreases levels of the PEX5 peroxisome-targeting signal receptor and ameliorates defects of Arabidopsis mutants with an impaired PEX4 ubiquitin-conjugating enzyme. BMC Plant Biol. 15:224. doi: 10.1186/s12870-015-0605-3
Kassmann, C. M. (2014). Myelin peroxisomes - essential organelles for the maintenance of white matter in the nervous system. Biochimie 98, 111–118. doi: 10.1016/j.biochi.2013.09.020
Kesler, S. R. (2014). Default mode network as a potential biomarker of chemotherapy-related brain injury. Neurobiol. Aging 35, S11–S19. doi: 10.1016/j.neurobiolaging.2014.03.036
Kesler, S. R., and Blayney, D. W. (2016). Neurotoxic effects of anthracycline- vs. nonanthracycline-based chemotherapy on cognition in breast cancer survivors. JAMA Oncol. 2, 185–192. doi: 10.1001/jamaoncol.2015.4333
Kim, P. K., Hailey, D. W., Mullen, R. T., and Lippincott-Schwartz, J. (2008). Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U S A 105, 20567–20574. doi: 10.1073/pnas.0810611105
Kirkin, V., Lamark, T., Sou, Y.-S., Bjørkøy, G., Nunn, J. L., Bruun, J.-A., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516. doi: 10.1016/j.molcel.2009.01.020
Kleinecke, S., Richert, S., de Hoz, L., Brügger, B., Kungl, T., Asadollahi, E., et al. (2017). Peroxisomal dysfunctions cause lysosomal storage and axonal Kv1 channel redistribution in peripheral neuropathy. Elife 6:e23332. doi: 10.7554/elife.23332
Klouwer, F. C. C., Berendse, K., Ferdinandusse, S., Wanders, R. J. A., Engelen, M., and Poll-The, B. T. (2015). Zellweger spectrum disorders: clinical overview and management approach. Orphanet J. Rare Dis. 10:151. doi: 10.1186/s13023-015-0368-9
Koehler, C. M. (2000). Protein translocation pathways of the mitochondrion. FEBS Lett. 476, 27–31. doi: 10.1016/s0014-5793(00)01664-1
Kondori, N. R., Paul, P., Robbins, J. P., Liu, K., Hildyard, J. C. W., Wells, D. J., et al. (2017). Characterisation of the pathogenic effects of the in vivo expression of an ALS-linked mutation in D-amino acid oxidase: phenotype and loss of spinal cord motor neurons. PLoS One 12:e0188912. doi: 10.1371/journal.pone.0188912
Kondori, N. R., Paul, P., Robbins, J. P., Liu, K., Hildyard, J. C. W., Wells, D. J., et al. (2018). Focus on the role of D-serine and D-amino acid oxidase in amyotrophic lateral sclerosis/motor neuron disease (ALS). Front. Mol. Biosci. 5:8. doi: 10.3389/fmolb.2018.00008
Kou, J., Kovacs, G. G., Höftberger, R., Kulik, W., Brodde, A., Forss-Petter, S., et al. (2011). Peroxisomal alterations in Alzheimer’s disease. Acta Neuropathol. 122, 271–283. doi: 10.1007/s00401-011-0836-9
Kulkarni, A., Chen, J., and Maday, S. (2018). Neuronal autophagy and intercellular regulation of homeostasis in the brain. Curr. Opin. Neurobiol. 51, 29–36. doi: 10.1016/j.conb.2018.02.008
Kulkarni, A., Dong, A., Kulkarni, V. V., Chen, J., Laxton, O., Anand, A., et al. (2019). Differential regulation of autophagy during metabolic stress in astrocytes and neurons. Autophagy doi: 10.1080/15548627.2019.1703354 [Epub ahead of print].
Landino, J., Jnah, A. J., Newberry, D. M., and Iben, S. C. (2017). Neonatal rhizomelic chondrodysplasia punctata type 1: weaving evidence into clinical practice. J. Perinat. Neonatal Nurs. 31, 350–357. doi: 10.1097/JPN.0000000000000282
Lefevre, S. D., van Roermund, C. W., Wanders, R. J. A., Veenhuis, M., and van der Klei, I. J. (2013). The significance of peroxisome function in chronological aging of Saccharomyces cerevisiae. Aging Cell 12, 784–793. doi: 10.1111/acel.12113
Legakis, J. E., Koepke, J. I., Jedeszko, C., Barlaskar, F., Terlecky, L. J., Edwards, H. J., et al. (2002). Peroxisome senescence in human fibroblasts. Mol. Biol. Cell 13, 4243–4255. doi: 10.1091/mbc.e02-06-0322
Liang, W. S., Dunckley, T., Beach, T. G., Grover, A., Mastroeni, D., Ramsey, K., et al. (2008). Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol. Genomics 33, 240–256. doi: 10.1152/physiolgenomics.00242.2007
Lodhi, I. J., Wei, X., Yin, L., Feng, C., Adak, S., Abou-Ezzi, G., et al. (2015). Peroxisomal lipid synthesis regulates inflammation by sustaining neutrophil membrane phospholipid composition and viability. Cell Metab. 21, 51–64. doi: 10.1016/j.cmet.2014.12.002
Maday, S., and Holzbaur, E. L. F. (2016). Compartment-specific regulation of autophagy in primary neurons. J. Neurosci. 36, 5933–5945. doi: 10.1523/jneurosci.4401-15.2016
Magalhães, M. M., and Magalhães, M. C. (1997). Peroxisomes in adrenal steroidogenesis. Microsc. Res. Tech. 36, 493–502. doi: 10.1002/(sici)1097-0029(19970315)36:6<493::aid-jemt6>3.0.co;2-j
Malheiro, A. R., da Silva, T. F., and Brites, P. (2015). Plasmalogens and fatty alcohols in rhizomelic chondrodysplasia punctata and Sjögren-Larsson syndrome. J. Inherit. Metab. Dis. 38, 111–121. doi: 10.1007/s10545-014-9795-3
Manchon, J. F. M., Dabaghian, Y., Uzor, N.-E., Kesler, S. R., Wefel, J. S., and Tsvetkov, A. S. (2016). Levetiracetam mitigates doxorubicin-induced DNA and synaptic damage in neurons. Sci. Rep. 6:25705. doi: 10.1038/srep25705
Mao, K., Liu, X., Feng, Y., and Klionsky, D. J. (2014). The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 10, 652–661. doi: 10.4161/auto.27852
Menzies, F. M., Fleming, A., Caricasole, A., Bento, C. F., Andrews, S. P., Ashkenazi, A., et al. (2017). Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034. doi: 10.1016/j.neuron.2017.01.022
Mirza, M. A., Ritzel, R., Xu, Y., McCullough, L. D., and Liu, F. (2015). Sexually dimorphic outcomes and inflammatory responses in hypoxic-ischemic encephalopathy. J. Neuroinflammation 12, 32–32. doi: 10.1186/s12974-015-0251-6
Mitchell, A. J. (2009). CSF phosphorylated tau in the diagnosis and prognosis of mild cognitive impairment and Alzheimer’s disease: a meta-analysis of 51 studies. J. Neurol. Neurosurg. Psychiatry 80, 966–975. doi: 10.1136/jnnp.2008.167791
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Moore, A. S., and Holzbaur, E. L. F. (2016). Spatiotemporal dynamics of autophagy receptors in selective mitophagy. Autophagy 12, 1956–1957. doi: 10.1080/15548627.2016.1212788
Moruno Manchon, J. F., Uzor, N.-E., Dabaghian, Y., Furr-stimming, E. E., Finkbeiner, S., and Tsvetkov, A. S. (2015). Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Sci. Rep. 5:15213. doi: 10.1038/srep15213
Moruno Manchon, J. F., Uzor, N.-E., Finkbeiner, S., and Tsvetkov, A. S. (2016). SPHK1/sphingosine kinase 1-mediated autophagy differs between neurons and SH-SY5Y neuroblastoma cells. Autophagy 12, 1418–1424. doi: 10.1080/15548627.2016.1183082
Moruno-Manchon, J. F., Uzor, N.-E., Ambati, C. R., Shetty, V., Putluri, N., Jagannath, C., et al. (2018a). Sphingosine kinase 1-associated autophagy differs between neurons and astrocytes. Cell Death Dis. 9, 521–521. doi: 10.1038/s41419-018-0599-5
Moruno-Manchon, J. F., Uzor, N.-E., Kesler, S. R., Wefel, J. S., Townley, D. M., Nagaraja, A. S., et al. (2018b). Peroxisomes contribute to oxidative stress in neurons during doxorubicin-based chemotherapy. Mol. Cell. Neurosci. 86, 65–71. doi: 10.1016/j.mcn.2017.11.014
Moruno-Manchon, J. F., Uzor, N.-E., Kesler, S. R., Wefel, J. S., Townley, D. M., Nagaraja, A. S., et al. (2016). TFEB ameliorates the impairment of the autophagy-lysosome pathway in neurons induced by doxorubicin. Aging 8, 3507–3519. doi: 10.18632/aging.101144
Morvay, P. L., Baes, M., and Van Veldhoven, P. P. (2017). Differential activities of peroxisomes along the mouse intestinal epithelium. Cell Biochem. Funct. 35, 144–155. doi: 10.1002/cbf.3255
Motley, A. M., Nuttall, J. M., and Hettema, E. H. (2012a). Atg36: the Saccharomyces cerevisiae receptor for pexophagy. Autophagy 8, 1680–1681. doi: 10.4161/auto.21485
Motley, A. M., Nuttall, J. M., and Hettema, E. H. (2012b). Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31, 2852–2868. doi: 10.1038/emboj.2012.151
Mukaiyama, H., Baba, M., Osumi, M., Aoyagi, S., Kato, N., Ohsumi, Y., et al. (2004). Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 15, 58–70. doi: 10.1091/mbc.e03-05-0340
Nakamura, S., and Yoshimori, T. (2017). New insights into autophagosome-lysosome fusion. J. Cell Sci. 130, 1209–1216. doi: 10.1242/jcs.196352
Narayan, V., Ly, T., Pourkarimi, E., Murillo, A. B., Gartner, A., Lamond, A. I., et al. (2016). Deep proteome analysis identifies age-related processes in C. elegans. Cell Syst. 3, 144–159. doi: 10.1016/j.cels.2016.06.011
Nilsson, P., Loganathan, K., Sekiguchi, M., Matsuba, Y., Hui, K., Tsubuki, S., et al. (2013). Aβ secretion and plaque formation depend on autophagy. Cell Rep. 5, 61–69. doi: 10.1016/j.celrep.2013.08.042
Nordgren, M., Francisco, T., Lismont, C., Hennebel, L., Brees, C., Wang, B., et al. (2015). Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy 11, 1326–1340. doi: 10.1080/15548627.2015.1061846
Nordgren, M., and Fransen, M. (2014). Peroxisomal metabolism and oxidative stress. Biochimie 98, 56–62. doi: 10.1016/j.biochi.2013.07.026
Nury, T., Sghaier, R., Zarrouk, A., Ménétrier, F., Uzun, T., Leoni, V., et al. (2018). Induction of peroxisomal changes in oligodendrocytes treated with 7-ketocholesterol: attenuation by α-tocopherol. Biochimie 153, 181–202. doi: 10.1016/j.biochi.2018.07.009
Olenick, M. A., Tokito, M., Boczkowska, M., Dominguez, R., and Holzbaur, E. L. F. (2016). Hook adaptors induce unidirectional processive motility by enhancing the dynein-dynactin interaction. J. Biol. Chem. 291, 18239–18251. doi: 10.1074/jbc.m116.738211
Pamenter, M. E., Perkins, G. A., McGinness, A. K., Gu, X. Q., Ellisman, M. H., and Haddad, G. G. (2012). Autophagy and apoptosis are differentially induced in neurons and astrocytes treated with an in vitro mimic of the ischemic penumbra. PloS One 7:e51469. doi: 10.1371/journal.pone.0051469
Park, H., He, A., Tan, M., Johnson, J. M., Dean, J. M., Pietka, T. A., et al. (2019). Peroxisome-derived lipids regulate adipose thermogenesis by mediating cold-induced mitochondrial fission. J. Clin. Invest. 129, 694–711. doi: 10.1172/jci120606
Peters, O. M., Ghasemi, M., and Brown, R. H. Jr. (2015). Emerging mechanisms of molecular pathology in ALS. J. Clin. Invest. 125, 1767–1779. doi: 10.1172/JCI71601
Poirier, Y., Antonenkov, V. D., Glumoff, T., and Hiltunen, J. K. (2006). Peroxisomal β-oxidation—A metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763, 1413–1426. doi: 10.1016/j.bbamcr.2006.08.034
Poole, B., Leighton, F., and De Duve, C. (1969). The synthesis and turnover of rat liver peroxisomes. II. Turnover of peroxisome proteins. J. Cell Biol. 41, 536–546. doi: 10.1083/jcb.41.2.536
Powers, J. M. (2001). Normal and defective neuronal membranes: structure and function. J. Mol. Neurosci. 16, 285–287. doi: 10.1385/jmn:16:2-3:285
Purdue, P. E., Skoneczny, M., Yang, X., Zhang, J.-W., and Lazarow, P. B. (1999). Rhizomelic chondrodysplasia punctata, a peroxisomal biogenesis disorder caused by defects in Pex7p, a peroxisomal protein import receptor: a minireview. Neurochem. Res. 24, 581–586. doi: 10.1023/a:1023957110171
Raas, Q., Saih, F. E., Gondcaille, C., Trompier, D., Hamon, Y., Leoni, V., et al. (2019). A microglial cell model for acyl-CoA oxidase 1 deficiency. Biochim. Biophys. Acta 1864, 567–576. doi: 10.1016/j.bbalip.2018.10.005
Reddy, J. K., and Hashimoto, T. (2001). Peroxisomal β-oxidation and peroxisome proliferator-activated receptor α: an adaptive metabolic system. Annu. Rev. Nutr. 21, 193–230. doi: 10.1146/annurev.nutr.21.1.193
Reggiori, F., and Tooze, S. A. (2009). The EmERgence of autophagosomes. Dev. Cell 17, 747–748. doi: 10.1016/j.devcel.2009.12.004
Riederer, P., Berg, D., Casadei, N., Cheng, F., Classen, J., Dresel, C., et al. (2019). α-synuclein in Parkinson’s disease: causal or bystander? J. Neural. Transm 126, 815–840. doi: 10.1007/s00702-019-02025-9
Ritzel, R. M., Patel, A. R., Spychala, M., Verma, R., Crapser, J., Koellhoffer, E. C., et al. (2017). Multiparity improves outcomes after cerebral ischemia in female mice despite features of increased metabovascular risk. Pro. Natl. Acad. Sci. U S A 114, E5673–E5682. doi: 10.1073/pnas.1607002114
Santos, M. J., Quintanilla, R. A., Toro, A., Grandy, R., Dinamarca, M. C., Godoy, J. A., et al. (2005). Peroxisomal proliferation protects from β-amyloid neurodegeneration. J. Biol. Chem. 280, 41057–41068. doi: 10.1074/jbc.m505160200
Sargent, G., van Zutphen, T., Shatseva, T., Zhang, L., Di Giovanni, V., Bandsma, R., et al. (2016). PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J. Cell Biol. 214, 677–690. doi: 10.1083/jcb.201511034
Sasabe, J., Suzuki, M., Imanishi, N., and Aiso, S. (2014). Activity of D-amino acid oxidase is widespread in the human central nervous system. Front. Synaptic Neurosci. 6:14. doi: 10.3389/fnsyn.2014.00014
Sasaki, T., Lian, S., Khan, A., Llop, J. R., Samuelson, A. V., Chen, W., et al. (2017). Autolysosome biogenesis and developmental senescence are regulated by both Spns1 and v-ATPase. Autophagy 13, 386–403. doi: 10.1080/15548627.2016.1256934
Shi, C., Zhu, J., Leng, S., Long, D., and Luo, X. (2016). Mitochondrial FOXO3a is involved in amyloid β peptide-induced mitochondrial dysfunction. J. Bioenerg. Biomembr. 48, 189–196. doi: 10.1007/s10863-016-9645-0
Siegel, C. S., and McCullough, L. D. (2013). NAD+ and nicotinamide: sex differences in cerebral ischemia. Neuroscience 237, 223–231. doi: 10.1016/j.neuroscience.2013.01.068
Smith, J. J., and Aitchison, J. D. (2013). Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14, 803–817. doi: 10.1038/nrm3700
Stavoe, A. K. H., and Holzbaur, E. L. F. (2019a). Autophagy in neurons. Ann. Rev. Cell Dev. Biol. 35, 477–500. doi: 10.1146/annurev-cellbio-100818-125242
Stavoe, A. K. H., and Holzbaur, E. L. F. (2019b). Axonal autophagy: mini-review for autophagy in the CNS. Neurosci. Lett. 697, 17–23. doi: 10.1016/j.neulet.2018.03.025
Strømhaug, P. E., Bevan, A., and Dunn, W. A. (2001). GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in pichia pastoris. J. Biol. Chem. 276, 42422–42435. doi: 10.1074/jbc.m104087200
Sung, K., and Jimenez-Sanchez, M. (2020). Autophagy in astrocytes and its implications in neurodegeneration. J. Mol. Biol. doi: 10.1016/j.jmb.2019.12.041 [Epub ahead of print].
Vasko, R. (2016). Peroxisomes and kidney injury. Antioxid. Redox Signal. 25, 217–231. doi: 10.1089/ars.2016.6666
Walton, P. A., Brees, C., Lismont, C., Apanasets, O., and Fransen, M. (2017). The peroxisomal import receptor PEX5 functions as a stress sensor, retaining catalase in the cytosol in times of oxidative stress. Biochim. Biophys. Acta 1864, 1833–1843. doi: 10.1016/j.bbamcr.2017.07.013
Wanders, R. J. A., Komen, J., and Ferdinandusse, S. (2011). Phytanic acid metabolism in health and disease. Biochim. Biophys. Acta 1811, 498–507. doi: 10.1016/j.bbalip.2011.06.006
Wanders, R. J. A., and Poll-The, B. T. (2017). “Role of peroxisomes in human lipid metabolism and its importance for neurological development”. Neurosci. Lett. 637, 11–17. doi: 10.1016/j.neulet.2015.06.018
Wang, X.-M., Yik, W. Y., Zhang, P., Lu, W., Huang, N., Kim, B. R., et al. (2015). Induced pluripotent stem cell models of Zellweger spectrum disorder show impaired peroxisome assembly and cell type-specific lipid abnormalities. Stem Cell Res. Ther. 6, 158–158. doi: 10.1186/s13287-015-0149-3
Warren, M., Mierau, G., Wartchow, E. P., Shimada, H., and Yano, S. (2018). Histologic and ultrastructural features in early and advanced phases of Zellweger spectrum disorder (infantile Refsum disease). Ultrastruct. Pathol. 42, 220–227. doi: 10.1080/01913123.2018.1440272
Waterham, H. R., and Ebberink, M. S. (2012). Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim. Biophys. Acta 1822, 1430–1441. doi: 10.1016/j.bbadis.2012.04.006
Wefel, J. S., Kesler, S. R., Noll, K. R., and Schagen, S. B. (2015). Clinical characteristics, pathophysiology and management of noncentral nervous system cancer-related cognitive impairment in adults. CA Cancer J. Clin. 65, 123–138. doi: 10.3322/caac.21258
Williams, C., and van der Klei, I. J. (2013). Pexophagy-linked degradation of the peroxisomal membrane protein Pex3p involves the ubiquitin-proteasome system. Biochem. Biophys. Res. Commun. 438, 395–401. doi: 10.1016/j.bbrc.2013.07.086
Wyant, K. J., Ridder, A. J., and Dayalu, P. (2017). Huntington’s disease—update on treatments. Curr. Neurol. Neurosci. Rep. 17:33. doi: 10.1007/s11910-017-0739-9
Yakunin, E., Moser, A., Loeb, V., Saada, A., Faust, P., Crane, D. I., et al. (2010). α-synuclein abnormalities in mouse models of peroxisome biogenesis disorders. J. Neurosci. Res. 88, 866–876. doi: 10.1002/jnr.22246
Yamashita, S.-I., Abe, K., Tatemichi, Y., and Fujiki, Y. (2014). The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 10, 1549–1564. doi: 10.4161/auto.29329
Yang, Z., and Klionsky, D. J. (2010). Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822. doi: 10.1038/ncb0910-814
Young, P. G., and Bartel, B. (2016). Pexophagy and peroxisomal protein turnover in plants. Biochim. Biophys. Acta 1863, 999–1005. doi: 10.1016/j.bbamcr.2015.09.005
Young, J. M., Nelson, J. W., Cheng, J., Zhang, W., Mader, S., Davis, C. M., et al. (2015). Peroxisomal biogenesis in ischemic brain. Antioxid. Redox Signal. 22, 109–120. doi: 10.1089/ars.2014.5833
Zanardelli, M., Micheli, L., Cinci, L., Failli, P., Ghelardini, C., and Di Cesare Mannelli, L. (2014). Oxaliplatin neurotoxicity involves peroxisome alterations. PPARγ agonism as preventive pharmacological approach. PLoS One 9, e102758–e102758. doi: 10.1371/journal.pone.0102758
Zhang, X., Gao, F., Wang, D., Li, C., Fu, Y., He, W., et al. (2018). Tau pathology in Parkinson’s disease. Front. Neurol. 9, 809–809. doi: 10.3389/fneur.2018.00809
Zhang, J., Kim, J., Alexander, A., Cai, S., Tripathi, D. N., Dere, R., et al. (2013). A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat. Cell Biol. 15, 1186–1196. doi: 10.1038/ncb2822
Zhang, J., Tripathi, D. N., Jing, J., Alexander, A., Kim, J., Powell, R. T., et al. (2015). ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 17, 1259–1269. doi: 10.1038/ncb3230
Zhao, T., Hong, Y., Li, S., and Li, X.-J. (2016). Compartment-dependent degradation of mutant huntingtin accounts for its preferential accumulation in neuronal processes. J. Neurosci. 36, 8317–8328. doi: 10.1523/jneurosci.0806-16.2016
Keywords: neuronal peroxisomes, peroxisome biogenesis disorders, aging peroxisomes, neurodegenerative disease, peroxisomal dysfunction
Citation: Uzor N-E, McCullough LD and Tsvetkov AS (2020) Peroxisomal Dysfunction in Neurological Diseases and Brain Aging. Front. Cell. Neurosci. 14:44. doi: 10.3389/fncel.2020.00044
Received: 05 December 2019; Accepted: 18 February 2020;
Published: 10 March 2020.
Edited by:
Chao Deng, University of Wollongong, AustraliaReviewed by:
Björn Spittau, University Hospital Rostock, GermanyCopyright © 2020 Uzor, McCullough and Tsvetkov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrey S. Tsvetkov, YW5kcmV5LnMudHN2ZXRrb3ZAdXRoLnRtYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.