
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Neurosci. , 26 July 2019
Sec. Cellular Neurophysiology
Volume 13 - 2019 | https://doi.org/10.3389/fncel.2019.00340
This article is part of the Research Topic Purinergic Signaling in Health and Disease View all 23 articles
P2X receptors are ATP-gated cations channels formed by the homo or hetero-trimeric association from the seven cloned subunits (P2X1-7). P2X receptors are widely distributed in different organs and cell types throughout the body including the nervous system and are involved in a large variety of physiological but also pathological processes in adult mammals. However, their expression and function during embryogenesis remain poorly understood. Here, we report the cloning and the comparative expression map establishment of the entire P2X subunit family in the clawed frog Xenopus. Orthologous sequences for 6 mammalian P2X subunits were identified in both X. laevis and X. tropicalis, but not for P2X3 subunit, suggesting a potential loss of this subunit in the Pipidae family. Three of these genes (p2rx1, p2rx2, and p2rx5) exist as homeologs in the pseudoallotetraploid X. laevis, making a total of 9 subunits in this species. Phylogenetic analyses demonstrate the high level of conservation of these receptors between amphibian and other vertebrate species. RT-PCR revealed that all subunits are expressed during the development although zygotic p2rx6 and p2rx7 transcripts are mainly detected at late organogenesis stages. Whole mount in situ hybridization shows that each subunit displays a specific spatio-temporal expression profile and that these subunits can therefore be grouped into two groups, based on their expression or not in the developing nervous system. Overlapping expression in the central and peripheral nervous system and in the sensory organs suggests potential heteromerization and/or redundant functions of P2X subunits in Xenopus embryos. The developmental expression of the p2rx subunit family during early phases of embryogenesis indicates that these subunits may have distinct roles during vertebrate development, especially embryonic neurogenesis.
The nucleotide ATP, as the universal source of energy, is an essential intracellular molecule to the survival of cells and whole organisms. However, the discovery of the first P2 purinergic receptor at the beginning of the 1990s confirmed Geoffrey Burnstock’s hypothesis that ATP acts also as an extracellular signaling molecule (Burnstock, 2014). In the last 25 years, numerous studies demonstrated the fundamental importance of extracellular ATP in the physiology of all organs through the activation of ionotropic P2X and metabotropic P2Y receptors (Ralevic and Burnstock, 1998). Purinergic signaling pathway is not restricted to extracellular ATP actions. The metabolic breakdown of extracellular ATP by ectonucleotidases is a source of ADP and adenosine. Both ATP derivatives act as extracellular signaling molecule through the activation of P2Y and adenosine G-protein coupled receptors (Adora) respectively (Yegutkin, 2014). It is nowadays recognized that the purinergic signaling in various organs play important physiological roles but also in diseases, placing purinergic receptors and ectonucleotidases as theurapeutic targets (Burnstock, 2018). In particular, in the nervous system, ATP and adenosine are involved in neuromodulation, glial-neuron interaction, and sensory transmission but also in neuropathic pain, neurodegenerative diseases, and multiple sclerosis (Burnstock et al., 2011; Khakh and North, 2012; Burnstock, 2016, 2017; Boué-Grabot and Pankratov, 2017; Domercq et al., 2018). Although the purinergic signaling has received less attention in a developmental context, several recent studies have shown a role of extracellular ATP and its derivatives in progenitor cell proliferation, migration, differentiation and synaptogenesis (Del Puerto et al., 2013; Oliveira et al., 2016; Fumagalli et al., 2017; Rodrigues et al., 2018). Furthermore, the description of the ectonucleotidases and purinoreceptors embryonic expression profile in vivo provides indication of their implication during nervous system development (Massé and Dale, 2012).
Due to the high number of actors, their large and overlapping expression profile and the very broad range of the purinergic receptor sensitivities, deciphering in vivo the specific embryonic functions of each extracellular purine is quite challenging. We decided to tackle this by studying each member of purinergic receptors and ectonucleotidases family using the clawed frog Xenopus embryo due to its numerous advantages, e.g., large size, large number production, relative transparency, external development and rapid generation times. Moreover, as its developmental cycle is temperature dependent, it is possible to modulate the length of the early stages such a neurulation allowing to access easily to the embryos at all developmental stages. Although having a pseudo-allotetraploid genome, the recent development of Xenopus genetics has broadened the use of this model especially as animal model for human pathologies (Sater and Moody, 2017; Tandon et al., 2017). We have previously established the embryonic expression pattern of enpp and entpd ectonucleotidases and adenosine signaling pathway members in Xenopus embryos. The specific spatio-temporal expression profile of each member of ectonucleotidases and adenosine receptors, suggested that ATP, ADP and adenosine may have distinct and specific functions during vertebrate embryogenesis (Massé et al., 2006, 2010; Tocco et al., 2015). Indeed, we demonstrated that ADP, via the P2Y1 receptor, triggers eye development by regulating the expression of the eye field transcription factors, such as pax6 (Massé et al., 2007). Xenopus is therefore an ideal model to address the functional roles of the purinergic pathway during the early phases of vertebrate embryogenesis, phases that are difficult to study in other vertebrate models (Massé and Dale, 2012).
Here we characterized the Xenopus P2X receptor subunit family. P2X receptors are ATP-gated cation channels formed by homomeric or heteromeric association of three subunits, which are involved in numerous physiological and pathological processes (North, 2002; Khakh and North, 2006; North, 2016). Seven genes for P2X subunits have been cloned and shared a common topology with two membrane-spanning domains, a large extracellular domain containing the ATP binding sites and intracellular N and C termini (Kawate et al., 2009; Habermacher et al., 2016). We report the cloning of the entire family in Xenopus laevis and tropicalis and reveal the absence of the p2rx3 gene in both of these species. Our phylogenetic data demonstrate a high degree of conservation of these p2rx sequences during vertebrate evolution. Analysis of the temporal and spatial expression of each amphibian subunit during X. laevis development showed that all identified subunits are expressed during development. Based on their expression or not in the nervous system, two groups of subunits can be proposed. Transcripts of all p2rx subunits, except p2rx2.S and p2rx5.L, are detected in the central and peripheral nervous system and their overlapping expression suggests potential hetero-oligomerization or redundant functions in the sensory system. A group of three subunits, p2rx2.S, p2rx4.L, and p2rx5, display specific expression in mesodermal derivatives suggesting these subunits may regulate renal and muscle development.
Sequences were identified on the NCBI and Xenbase databases. BLAST (Basic Local Alignment Search Tool) searches were performed on the NBCI Nucleotide and on the Xenbase Xenopus laevis 9.1 genome databases (Altschul et al., 1990). This identification was based on p2rx subunits orthologs nucleotide alignments, using either predicted (from automated computational analysis) mRNA sequences from Xenopus genomic sequences or validated mRNA sequences from different species such as Homo sapiens, Mus musculus, Rattus norvegicus, Danio rerio, Gallus gallus, Takifugu rubripes, Xenopus tropicalis, and Xenopus laevis. Conceptual translation of cDNA and protein sequence analysis was performed on the ExPASy internet site using the program Translate Tool1 (Artimo et al., 2012). Accession numbers of all sequences used in this study are given in Supplementary Table S1. Alignments were performed on the EMBL-EBI internet site using the program Pairwise sequence global alignment EMBOSS Needle and Multiple Sequence alignment Clustal Omega program (Needleman and Wunsch, 1970; Thompson et al., 1994). A phylogenetic tree was created on the Phylogeny.fr platform using muscle for multiple alignments, PhyML for tree building and TreeDyn for tree rendering (Dereeper et al., 2008).
Xenopus laevis males and females were purchased from the CNRS Xenopus breeding Center (Rennes, France). Embryos were obtained by in vitro fertilization of eggs collected in 1X MMR (Marc’s Modified Ringers) saline solution [100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 5 mM Hepes (pH 7.8), pH 7.4], from a hormonally [hCG (Agripharm), 750 units] stimulated X. laevis female by adding crushed testis isolated from a sacrificed male. Fertilized eggs were dejellied in 3% L-cysteine hydrochloride, pH 7.6 (Sigma-Aldrich), and washed several times with 0.1X MMR. Embryos were then cultured to the required stage in 0,1X MMR in presence of 50 mμ/mL of gentamycin sulfate (Sigma-Aldrich). The embryos were staged according to the Nieuwkoop and Faber table of X. laevis development (Nieuwkoop and Faber, 1994).
RNA extraction from whole embryos and cDNA synthesis were performed as previously described (Massé et al., 2010). For each subunit, specific primers were designed, when possible, on two different exons with at least one primer positioned at exon junctions (Supplementary Table S2), in order to discriminate genomic from cDNA amplification. Furthermore, those chosen primers were selected to differentiate homeologs expression, with a restrictive annealing possibility on the other homeologous mRNA sequence. After optimization of the PCR conditions using a gradient PCR machine (Bio-Rad), PCR products were verified by sequencing (Beckman Coulter Genomics Company). To assess semi-quantitative PCR experiments, the quantity of input cDNA was determined by equalization of the samples with the constant gene odc (ornithine decarboxylase) (Bassez et al., 1990). Linearity of the signal was controlled by carrying out PCR reactions on doubling dilutions of cDNA. Negative controls (-RNA, -RT, and -cDNA) were also performed.
Whole-mount in situ hybridizations were carried out as previously described (Massé et al., 2006, 2010; Blanchard and Massé, 2019). In order to detect the expression of each p2rx subunit, fragments of their cDNA, except for p2rx4, were subcloned into plasmid pBSKS, as described in Supplementary Table S3, in order to generate specific antisense and sense probes. Riboprobes were generated by in vitro transcription using a DIG RNA labeling kit following manufacturer’s recommendations (Roche) after linearization of the corresponding plasmid (see Supplementary Table S3 for details). Templates were designed in order to generate 250–600 bases riboprobes; when using larger riboprobes, a limited alkaline hydrolysis was performed. Riboprobe hybridization detection was carried out with an anti-DIG Alkaline Phosphatase antibody (Roche, Reference 11093274910) and the BM-Purple AP substrate (Roche Reference, 11442074001). After bleaching, embryos were photographed using the SMZ18 binocular (Nikon).
After whole-mount in situ hybridization, embryos were gradually dehydrated in 100% gelatin/sucrose-PBS solution before an overnight incubation in 100% gelatin/sucrose solution. Identified embryos were embedded in Tissue-Tek OCT, frozen on dry ice and conserved at −80°C. The frozen blocks were sectioned on a cryostat at 10 μm thickness. Transverse sections were also performed with a razor blade on fixed embryos. Sections were photographed using the SMZ18 binocular (Nikon).
The p2rx subunit sequences were identified by bioinformatics analysis, this work having benefited from the recent X. tropicalis and X. laevis genome sequencing (Hellsten et al., 2010; Session et al., 2016). The accession numbers of the protein sequences of the amphibian subunits can be found in Supplementary Table S1A. Unexpectedly, no genomic sequences were identified for p2rx3 subunit in both X. laevis and X. tropicalis genomes.
All Xenopus tropicalis sequences are available on the Xenbase website, although only as annotated and predicted by automated computational analysis from genomic sequences. Their identity as P2X subunits was confirmed by conducting two complementary analyses: (1) Blastx search of these predicted X. tropicalis sequences on the human protein database (NCBI) was first conducted. (2) X. tropicalis cDNA sequences were deduced from the genomic sequences and corrected by reference to the human sequences according to the Breathnach and Chambon rule (Breathnach and Chambon, 1981). The protein sequences were deduced from conceptual translation and aligned with their potential orthologs.
Regarding the Xenopus laevis p2rx subunits, the p2rx4.L and p2rx7.L sequences have been previously published (Juranka et al., 2001; Paukert et al., 2002). P2rx1.L, p2rx2.L, p2rx5.L, and p2rx5.S sequences were available on the Xenbase site, but only as annotated sequences. Sequences verification was performed as described above for the X. tropicalis sequences. P2rx1 and p2rx6 X. tropicalis protein sequences blast on the Xenopus laevis protein database (NCBI website) allowed us to identify the p2rx1.L sequence and two X. laevis hypothetical proteins, whose sequences were derived from genome annotation. The identity of these sequences as p2rx1.S and p2rx6.L subunits was confirmed as described above. A p2rx2.S genomic sequence was identified but no cDNA or protein sequences were available on the Xenbase or NCBI website. From this genomic sequence, we retrieved the cDNA sequence after checking the exon/intron boundary by comparison to the human p2rx2 ones according to the Breathnach and Chambon rule (Breathnach and Chambon, 1981) and carried out several blasts on Xenopus laevis databases (NCBI website). Alignment of these different sequences allowed us to generate the consensus and complete sequence of p2rx2.S subunit cDNA. The protein sequence was then obtained by conceptual translation and checked by blast on the protein databases (NCBI website).
In order to characterize the evolution of P2X receptor subunits in vertebrates, we performed an alignment of all Xenopus protein sequences (Supplementary Figure S1). In X. laevis, p2rx1, p2rx2, p2rx5 have two homeologs (L and S), whose protein sequences display high degree of identity (more than 90%, Table 1A). However, the percentage of identity between the different X.laevis p2rx subunit sequences is less than 50% with p2rx4.L sharing the highest identity percentage with p2rx5 homeologs. The identity percentage between p2rx7.L and its X. laevis paralogs sequences is less than 30%. This low percentage is certainly due to the presence of the long and most divergent C terminus domain of p2rx7.L in this analysis.
To address the evolutionary diversification of the P2X subunits in vertebrates, a phylogenetic analysis of P2X proteins was carried out (Figure 1). This resulting tree reveals a clear separation between the p2rx6 and p2rx5 proteins and the other p2rx family members, which could be subdivided into three groups, p2rx3 group, p2rx1 and p2rx2 group and p2rx4 and p2rx7 group. Each member is more related to its orthologs than to the other family members in the same species, suggesting that any function identified in X. laevis may well be conserved in other vertebrates (Table 1B). The percentage of identity between Xenopus and its orthologous proteins is between 44 and 76%, with p2rx7.L being the least conserved protein as it only shares 38–46% of identity with zebrafish and chick p2rx7 subunit respectively. P2rx4.L is the most conserved member during vertebrate evolution, with a percentage of identity higher to 49% with the other P2X4 vertebrate proteins.
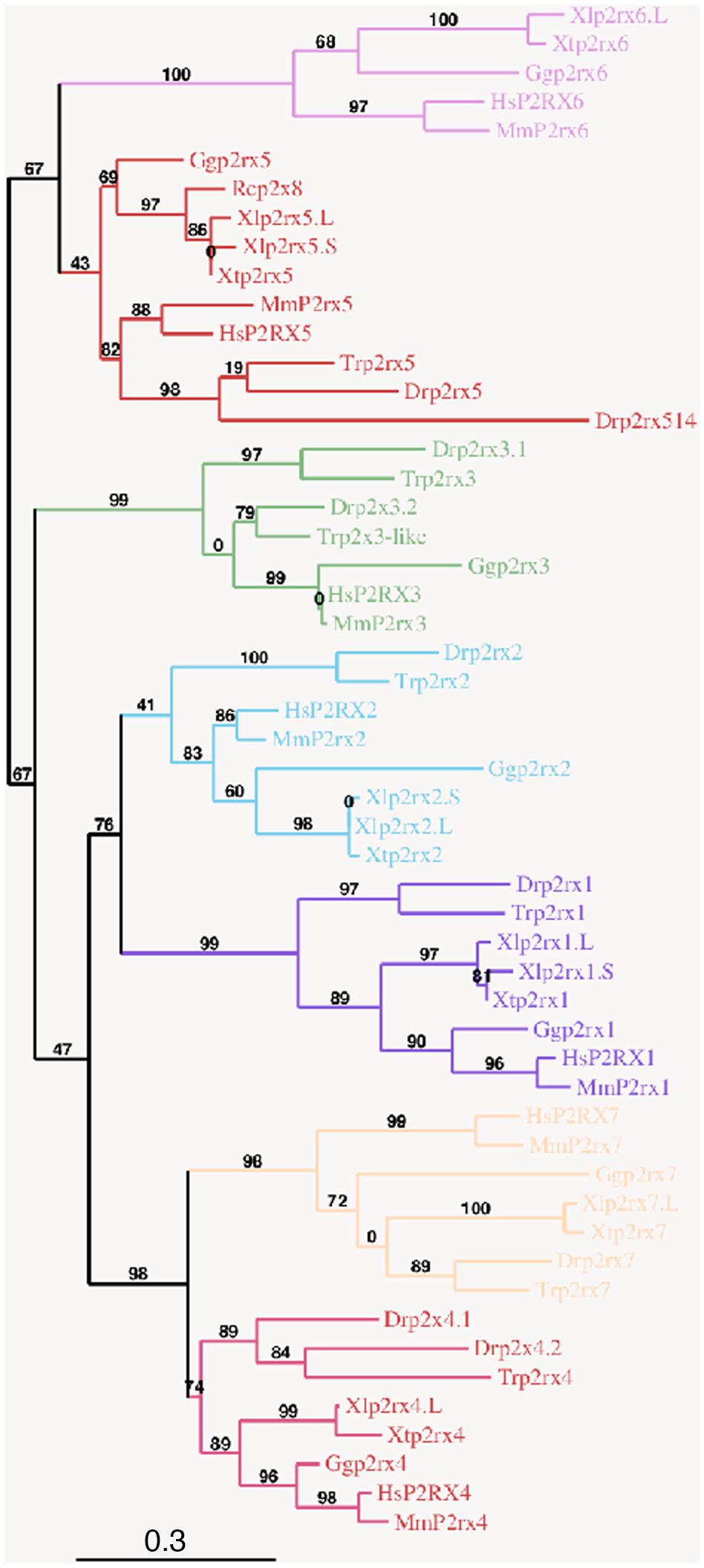
Figure 1. Phylogenetic conservation of the vertebrate p2x receptor subunits. A phylogenetic tree of the p2rx subunits family based on African clawed frogs (Xenopus laevis, Xl and Xenopus tropicalis, Xt), human (Homo sapiens, Hs), mouse (Mus musculus, Mm), chicken (Gallus gallus, Gg), zebrafish (Danio rerio, Dr), Japanese puffer (Takifugu rubripes, Tr) and American bullfrog (Rana catesbeiana, Rc) was constructed on the plastform phylogeny.fr, using the Maximum Likelihood method. Bootstrap percentages (ranging from 0 to 100) are indicated at each node of the tree while branch lengths are representative of sequence substitution rates. P2rx1, 2, 3, 4, 5, 6, 7 subunit subfamilies are respectively colored in purple, blue, green, dark pink, red, light pink, and orange. The Genbank accession numbers of the different receptors are given in the Supplementary Tables S1A,B except for Rcp2rx8 (Accession Number: AAL24075.1).
The temporal expression of the p2rx genes during embryonic development was assessed by RT-PCR (Figure 2). Adult brain cDNA was used as positive control. Each subunit displays a specific expression profile; however, the level of their expression increases during development to reach a maximum at stage 45, last embryonic stage tested during this experiment. The p2rx1.L subunit is expressed from late neurula stage whereas expression of its homeolog p2rx1.S is weakly detected from late segmentation to mid-neurulation and at a higher level during late organogenesis. The two p2rx2 subunits display similar expression profile: both of these subunits are not expressed maternally and their zygotic expression can be detected from late segmentation stage, stage 9, although p2rx2.L seems more expressed during gastrulation. The p2rx4.L gene displays a specific expression profile, as being the only one subunit whose expression is detected at all stages tested, with a high level of maternal expression. This is in agreement with its previously published expression in oocytes (Juranka et al., 2001). The homeologs p2rx5.L and p2rx5.S are not maternally expressed. Zygotic expression of p2rx5.L is visible during neurulation whereas p2rx5.S expression is weakly detected at stage 33. Expression of these both homeologs is detected at a higher level from stage 41. The p2rx6.L subunit is the second p2rx gene being expressed maternally; however, its zygotic expression is only detected at late organogenesis stages. The p2rx7.L gene displays another distinct expression profile with its expression only clearly visible at late organogenesis stages, from stage 41, suggesting no functional roles for this subunit at early embryogenesis stages.
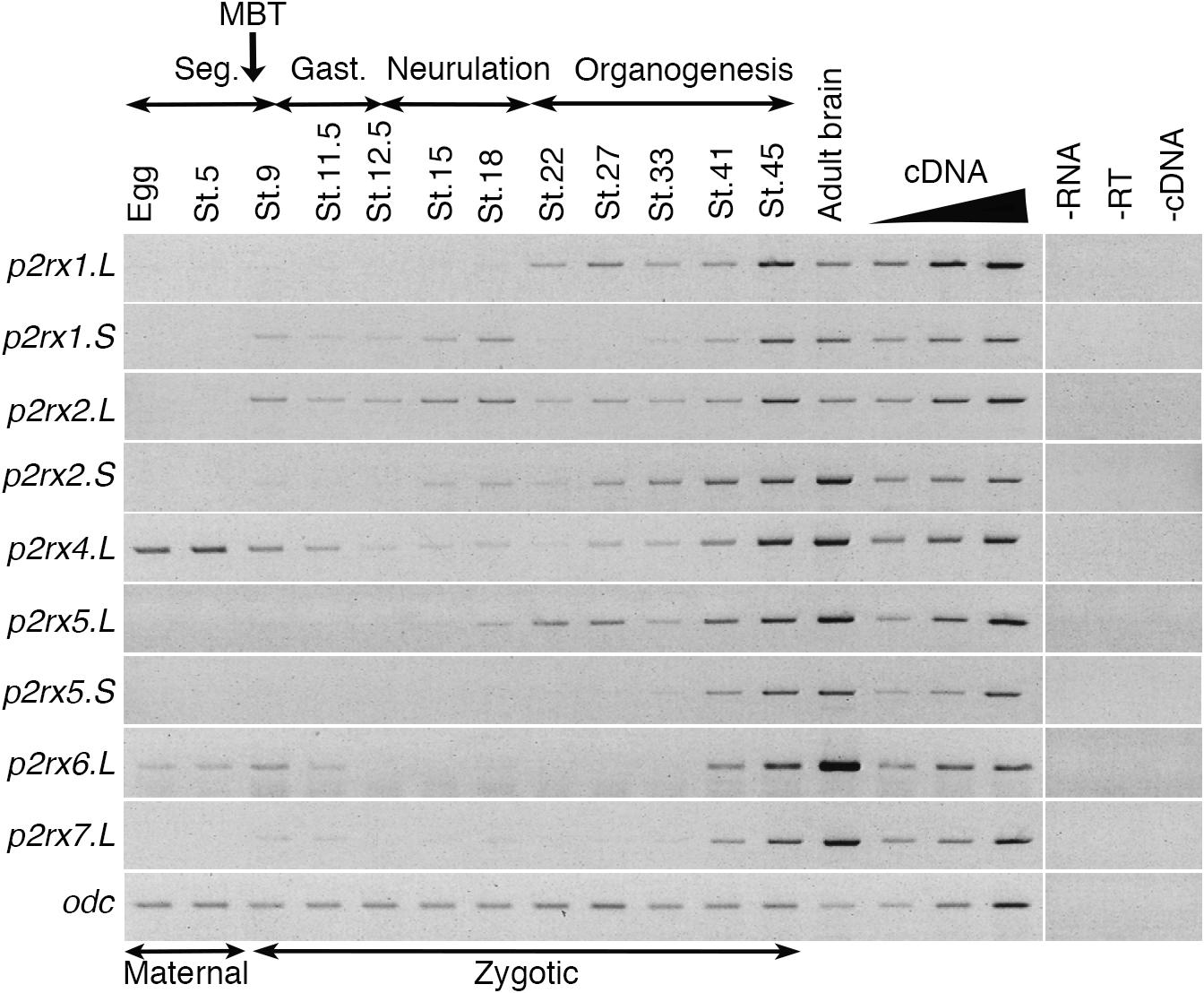
Figure 2. Temporal expression profile of p2rx genes during embryogenesis. RT-PCR analysis showing the expression pattern of p2rx subunits in X. laevis unfertilized egg and embryos at different stages (St.). Adult brain cDNA was used as positive control and three negative controls (-RT, -RNA, -cDNA) were also performed. The linearity of the signal was controlled by carrying out PCR reactions on doubling dilutions of cDNA from stage 45 embryo, illustrated by the triangle. Odc (ornithine decarboxylase) was used as a loading control. The different phases of embryogenesis and the Mid-Blastula transition (MBT), the switch between maternal and zygotic expression, are indicated above the different embryonic stages. Seg, segmentation; Gast, Gastrulation.
The spatial expression of these genes in the embryo was assessed by whole-mount in situ hybridization, performed from stage 5 to stage 41 with specific antisense riboprobes (Figure 3). Probes were designed in order to discriminate the expression of the homeologs except for p2rx5 genes. Probes were designed in the 3′UTR region except for p2rx5, p2rx6, p2rx7 for which genes probes are located in the coding region. For each subunit, controls were carried out with sense probes to discriminate specific and unspecific signals (see Figure 4). Sectioning of representative p2rx stained embryos was performed in order to further characterize their expression profile (Figure 5). As shown in Figures 3, 5, p2rx subunits display specific and different, but also overlapping expression domains during X. laevis development. Expression profile of the different p2rx subunits is summarized in Table 2.
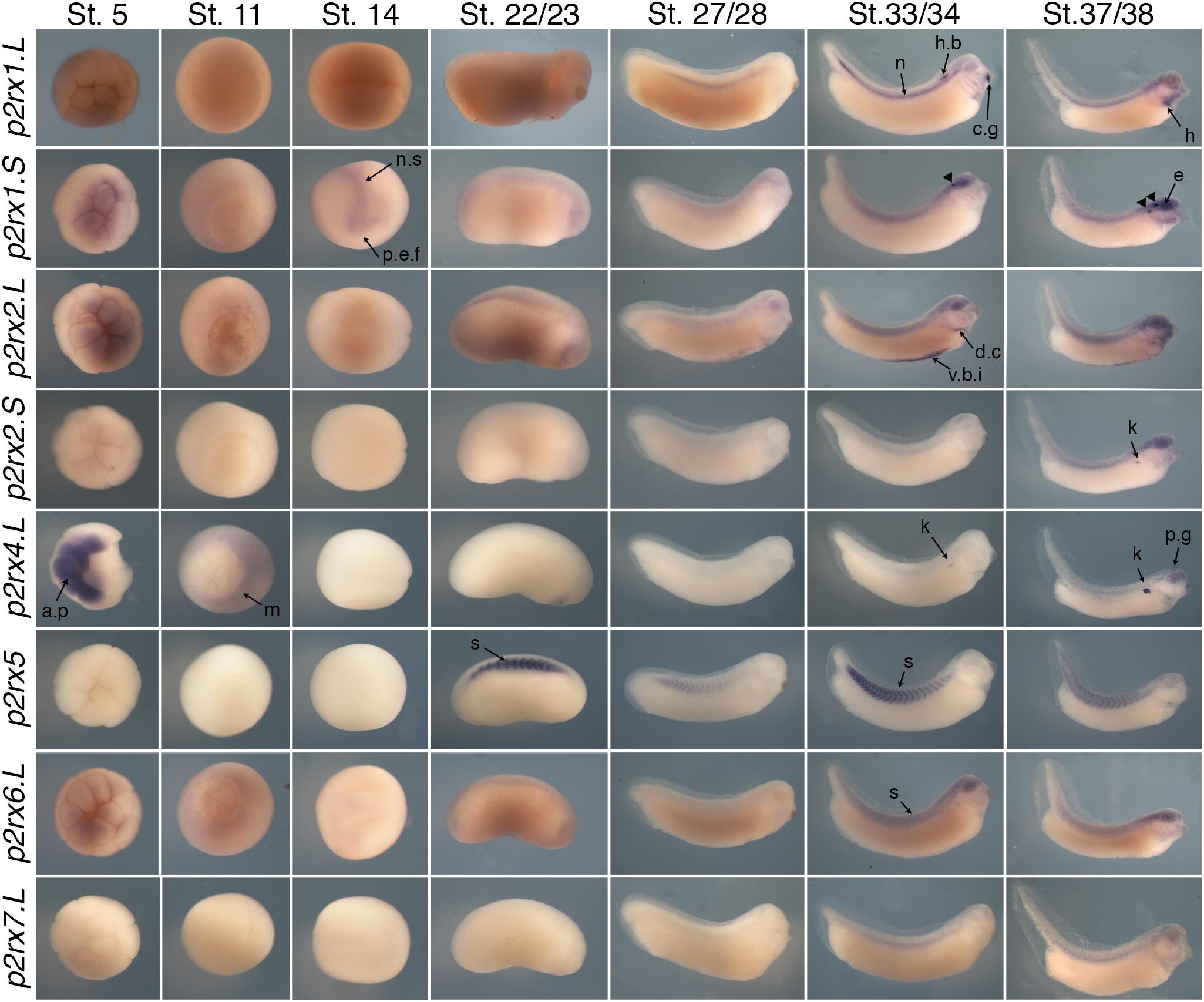
Figure 3. Comparative spatial expression profile of p2rx genes during embryogenesis. Whole-mount in situ hybridization with specific DIG-labeled antisense RNA probes was performed on embryos from stages (St.) 5 to 37/38. Representative embryos were photographed at the magnification X20 for stages 5, 11, and 14 and X16 for later stages. Stage 5: animal pole view, stage 11: vegetal pole view, stage 14: dorsal view except for p2rx1.S (anterior view), later stages: lateral views, with dorsal is up and anterior is right. The arrowheads indicate the staining in the neurogenic placodes and peripheral nervous system detailed in Figure 5. a.p, animal pole; c.g, cement gland; d.c, duct of Cuvier; e, eye; h, heart; h.b, hindbrain; k, kidney; m, mesoderm; n, notochord; n.s, nervous system; p.e.f, presumptive eye field; p.g, pineal gland; s, somite; t.n, trigeminal nerve; v.b.i, ventral blood island.
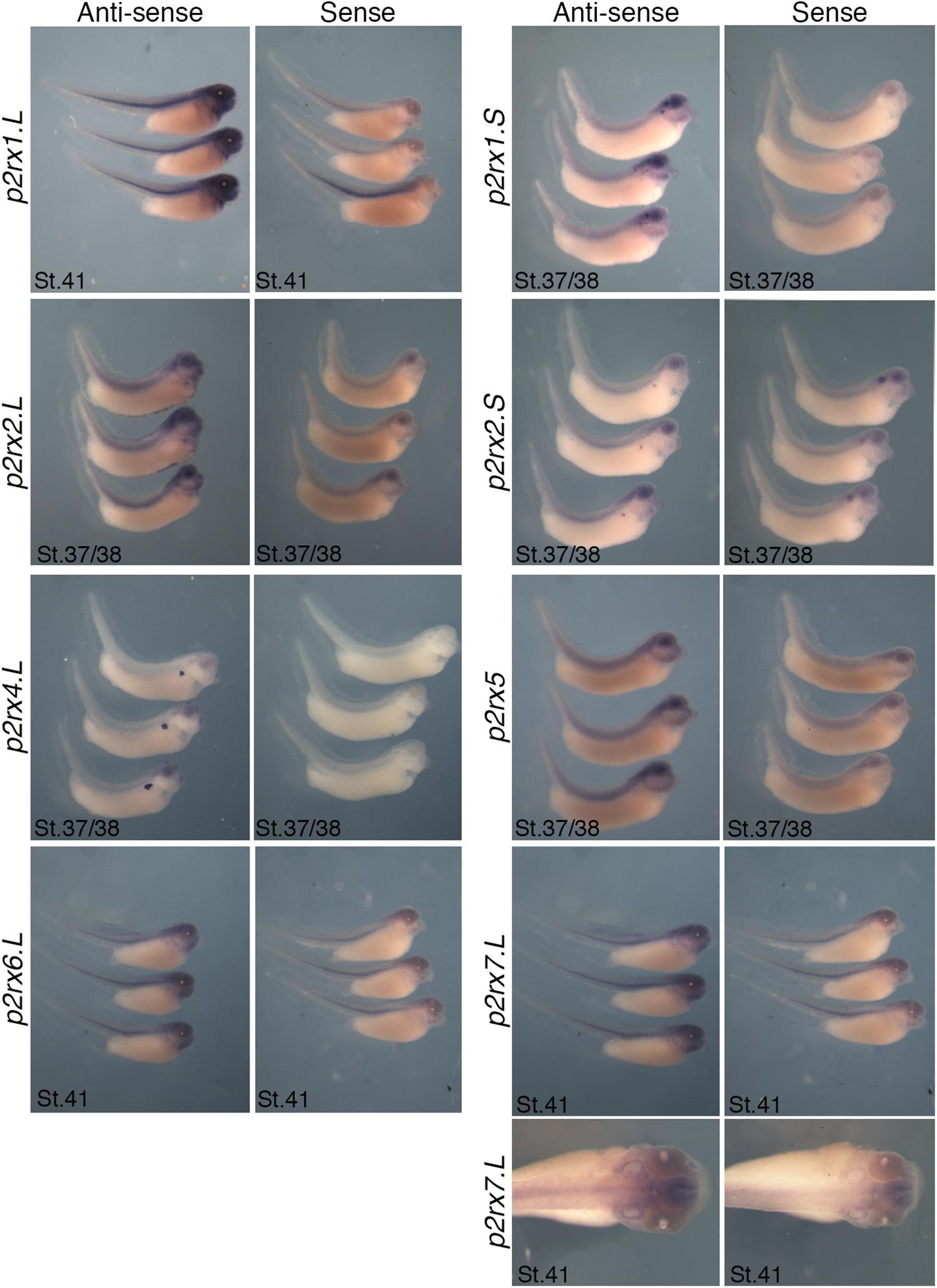
Figure 4. Comparative staining obtained with the antisense and sense probes of p2rx genes. Lateral views of three representative embryos at stage 37/38 or stage 41stained with the antisense and sense probes for p2rx1.L, p2rx1.S, p2rx2.L, p2rx2.S, p2rx4.L, p2rx6.L, and p2rx7.L. subunits and p2rx5 homeologs. Some unspecific staining, due to probe trapping, can be observed in the head region (brain ventricles, otic vesicle) and in the notochord. For p2r7.L subunit, a dorsal view of a representative embryo completes the lateral views. For lateral views, dorsal is up and anterior is right. For dorsal views, anterior is up.
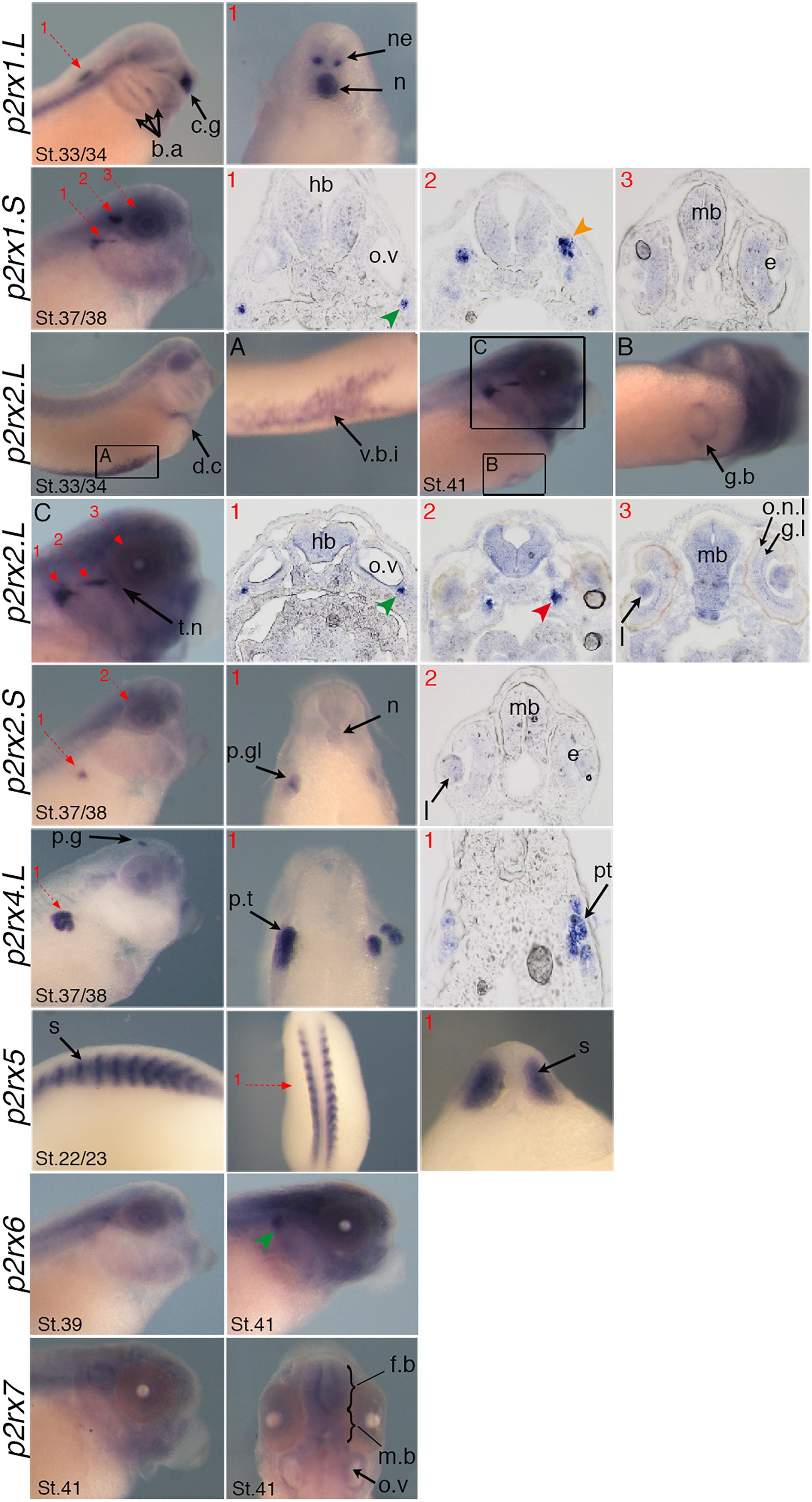
Figure 5. Specific expression domains of p2rx genes. Representative embryos from stages (St.) 22/23–41 displaying p2rx subunit expression detected by whole-mount in situ hybridization. To further characterize their expression profile, embryos stained for p2rx genes, except p2r6.L and p2rx7.L, were selected for scalpel sections and cryostat sections. Sections were performed on stage 22/23 for p2rx5 stained embryos, on stage 33/34 for p2rx1.L stained embryos, on stage 37/38 for p2rx1.S, p2rx2.S, and p2rx4.L stained embryos and stage 41 for p2rx2.L stained embryos. The red numbered dotted arrows indicate the plan of the sections. For whole mount pictures, dorsal is up and anterior is right except for the p2rx5 and p2rx7 subunits for which a dorsal view completes the lateral view. For section pictures, dorsal is up. The green arrowhead indicates the fused ganglion of middle lateral line and glossopharyngeal nerves, the orange one shows the trigeminal ganglion and ganglion of anterodorsal lateral line nerve and the red one the ganglia of anteroventral lateral line and facial nerves. c.g, cement gland; e, eye; fb, forebrain; g.b, gall bladder; g.l, ganglion layer; h, heart; hb, hindbrain; l, lens; mb, midbrain; n, notochord; o.n.l, outer nuclear layer; o.v, otic vesicle; p.g, pineal gland; p.gl, pronephric glomus; p.t, pronephric tubules; s, somite; t.n, trigeminal nerve; v.b.i, ventral blood island.
P2rx1 homeologs display distinct expression profile. The p2rx1.L subunit is not detected before stage 27 (Figure 3). At this stage and stage 33/34, its transcripts are localized in a very specific region of the hindbrain, in a two rows of laterally symmetrical neurons, in a ventral position along the dorso-ventral axis (Figure 5). At stage 33/34, p2rx1.L expression is also detected in the head region, in the branchial arches and in the cement gland, a mucus-secreting ectodermal structure. At later stages, staining is also found in the heart and in the vascular network between the heart and kidney (Figure 3). However, its expression in the notochord is unspecific as detected with the sense probe (Figure 4). Expression of the p2rx1.S homeolog is detected from cleavage stage (Figure 3). At stage 5, its expression is observed in some blastomeres at the animal pole. No specific signal was detected during gastrulation. During neurulation, its transcripts are detected in the developing nervous system and in the presumptive eye field. Its expression remains in the developing eye until stage 41. Transverse sections show expression in the retina layers (Figure 5). From stage 27/28, weak diffuse expression is observed in the head and dorsal region of the embryo. At later stages, strong p2rx1.S expression is observed into two specific domains of the neurogenic placodes and peripheral nervous system: in the fused ganglion of middle lateral line and glossopharyngeal nerves as well as in the trigeminal ganglion and ganglion of anterodorsal lateral line nerve (Figure 5).
The homeologs p2rx2 also display different expression pattern. P2rx2.L transcripts are only detected in specific manner from early organogenesis stages, the staining during segmentation phase being observed with the sense probe (Figure 3). During organogenesis, p2rx2.L expression is mostly detected in developing mesoderm derivatives and nervous system. At early tailbud phase (st. 22/23), weak staining can be observed in the somites. At tailbud stages, staining is found in the ventral blood island from stage 27, in the duct of Cuvier from stage 33/34 and in the heart from stage 37 (Figures 3, 5). At stage 41, expression can be observed in the gall bladder (Figure 5). Transcripts are also found in the head region, in the brain and the sensory organs such as eye and otic vesicule. At stage 41, staining in the eye is detected both in the retina, in the retinal ganglion and outer nuclear layers, and in the lens (Figure 5). From stage 37/38, p2rx2.L transcripts are detected in the trigeminal nerve (ophthalmic and maxillary branches) and strong expression is detected into specific regions of neurogenic placodes and peripheral nervous system : in the fused ganglion of middle lateral line and glossopharyngeal nerves and in ganglia of anteroventral lateral line and facial nerves (Figures 3, 5). The p2rx2.S subunit displays a very tissue specific expression domain, with undetectable neural expression (Figure 5). No expression can be detected before stage 37 (Figure 3). From stage 37–41, its expression is only detected in the kidney region, in the pronephric glomus, as signal in the head and notochord is unspecific (Figures 3–5).
The p2rx4.L subunit also displays a very specific expression domain. As expected from the RT-PCR results, strong staining is observed in the animal pole in blastula and in the involuting mesoderm in gastrula embryos (Figure 3). However, no zygotic expression is detected before stage 33/34. At this stage, its transcripts are mostly found in the kidney. At stage 37/38, p2rx4.L is highly expressed in the proximal tubules (Figure 5). Expression is also observed in the pineal gland (epiphysis) (Figure 3).
The staining domain observed with the common p2rx5 riboprobe, which does not discriminate the two homeologs, is also very specific, only in mesodermal and not neural tissues (Figure 3). From stage 22/23, p2rx5 genes are only and highly expressed in the developing somites (Figures 3–5). This restricted muscle expression is unique to these members of the family.
The p2rx6.L and p2rx7.L genes are more weakly expressed in the embryo (Figure 3). At stage 33/34, weak p2rx6.L expression is observed in the somites. At stage 41, strong expression is detected in the head region and dorsal region of the embryo but this staining is unspecific as observed with the sense probe (Figure 4). However, the expression observed in the fused ganglion of middle lateral line and glossopharyngeal nerves is specific (Figure 5). The p2rx7.L subunit is only weakly but specifically detected at stage 41, in the head region, in the forebrain and midbrain and sensory organs (eye and otic vesicule) (see Figures 4, 5).
We here report the cloning of the complete p2rx gene family in Xenopus in addition to p2rx4 and p2rx7 members previously identified (Juranka et al., 2001; Paukert et al., 2002). Moreover, we provide a complete expression map of all p2rx genes throughout X. laevis embryonic life. This is the first study to compare the expression of the entire p2rx family during the early phases of development, notably the earliest ones such as gastrulation and neurulation, corresponding to the specification and formation of the neural tube.
Our study reveals that all members of the p2rx family, except p2rx3, are present in Xenopus laevis and tropicalis. Phylogenetic analysis demonstrates the high degree of conservation during vertebrate evolution both at the protein and genomic level. The two transmembrane domains positions are conserved in all amphibian P2X subunits and the exons/introns boundaries are also conserved except for the boundary 11 (separating exons 11 and 12), which is only conserved between homeologs and orthologs. The full length of cDNAs, proteins and genomic sequences of all P2X subunits was used for our phylogenetic analysis. C-terminus tail sequences, encoded by exon 12 or exons 12 and 13 for p2rx7.L, were retained in our analysis although they correspond to the most variable domain in terms in length and sequences. The percentage of identity between the different X. laevis P2X subunits is similar to those shared by mammalian ones (North, 2002), the most conserved region being the transmembrane domains and the extracellular loop. Sixty four amino acids (AA) are conserved in all 15 Xenopus p2rx sequences, with 56/64 being located in the extracellular domain sequence. In the intracellular N-terminal domain, only a motif of 3 amino acids (Thr-X-Arg/Lys, with X being any amino acid) is conserved. This motif is a phosphorylation site by the Protein Kinase C modulating the desensitization rate of mammalian P2X receptors (Boué-Grabot et al., 2000b). In the intracellular C-terminal domain, the consensus motif Tyr-XXX-Lys (with X being any amino acid) involved in surface targeting of mammalian P2X receptors is also conserved in all Xenopus P2X subunits (Chaumont et al., 2004). In the extracellular domain, the 10 cysteines (located at position 113, 124, 130, 147, 158, 164, 214, 224, 258, 267 in rat P2X2 and indicated with an asterisk on the Supplementary Figure S1) involved in tertiary structure of P2X subunit by disulphide bonds formation are conserved in Xenopus (North, 2002). Furthermore, 7 of the 8 AA involved in ATP binding are also conserved [Lys70, Lys72, Phe188, Thr189, Asn296, Phe297, Arg298, Lys316 in zebrafish P2X4 (Habermacher et al., 2016); indicated with an black triangle on Supplementary Figure S1]. The Phe297 amino acid (indicated with an open triangle on Supplementary Figure S1) is conserved in all, except P2X2, Xenopus sequences. This is quite surprising as this phenylalanine is present in the mammalian P2X2 orthologous sequences.
Intriguingly, no genomic sequence for p2rx3 was identified suggesting the loss of this subunit in Pipidae. Search on Ensembl and NCBI databases shows that this subunit is present in the different classes of vertebrates [mammalia, reptilia, aves, fish (cartilaginous, ray-finned fishes and coelacanths)] but not in agnatha. In vertebrates, P2X3 receptor is predominantly expressed in nociceptor sensory neurons of dorsal root and trigeminal ganglia and plays a key role in pain transduction (Vulchanova et al., 1997; Xiang et al., 1998; Boué-Grabot et al., 2000a; Norton et al., 2000; Kucenas et al., 2003; Appelbaum et al., 2007). In zebrafish or pufferfish, P2X3 is specifically expressed in trigeminal neurons and spinal cord Rohon-Beard cells and functional studies using transgenic animals showed its implication in sensory circuit formation during development (Kucenas et al., 2006, 2009; Palanca et al., 2013). In rodent P2X3 is similarly expressed in trigeminal and dorsal root ganglia sensory neurons and P2X3KO mice demonstrated its implication in nociception as well as sensory transduction (Cockayne et al., 2000; Souslova et al., 2000). Zebrafish p2rx3.2 is co-expressed with the ectonucleotidase entpdase3 in spinal cord Rohon-Beard cells (Appelbaum et al., 2007). We previously published entpdase3 expression in Xenopus Rohon-Beard cells (Massé et al., 2006), making amphibian p2rx3 gene absence even more unexpected. Our in situ hybridization data show that p2rx1.S, p2rx2.L, and p2rx6.L are expressed in cranial neurogenic placodes, in the trigeminal ganglion and sensory lateral line system. Future work will have to address if one of these subunits could fulfill the functions of P2X3.
Xenopus laevis p2rx genes are all expressed during embryogenesis although their expression patterns differ during frog development. Only two subunits, p2rx4.L and p2rx6.L, are maternally expressed. P2rx4 displays the highest level of expression in egg and early embryo stages (before the MBT), which is in agreement with its expression in oocytes (Juranka et al., 2001). Based on their zygotic expression, p2rx subunits can be divided into two groups: those which are not expressed in the nervous system, p2rx2.S and p2rx5, and those which are expressed in the nervous system, although their expression can not be restricted to this tissue. For example, p2rx4.L, expressed in the pineal gland, is also highly expressed in the pronephros, p2rx2.L transcripts, found in sensory system, are also detected in the cardiovascular system or p2rx1.L expression detected at high level in an interneuron population in the hindbrain is also found in the cement gland. These distinct expression profiles suggest specific functions for these receptors during embryogenesis, during the formation of neural and non-neural tissues. However, as in mouse and zebrafish embryos, p2rx7.L does not seem to be involved during early embryogenesis as its expression is only detected weakly at late organogenesis stages (Kucenas et al., 2003; Appelbaum et al., 2007; MGI website). The absence of gross phenotype at birth of null P2X7 mice is in agreement with this result (Solle et al., 2001), although its activity has been reported, in vitro, as essential for mouse embryonic stem cells survival (Thompson et al., 2012).
Several p2rx subunits display specific and almost restricted expression profile in two mesodermic derivatives, the kidney and somites. These expression patterns have been conserved during evolution. Our data demonstrate that p2xr5 subunit is highly expressed in the somitic tissues. In rat and chick embryos, P2X5 is the earliest P2X receptors expressed in somites (Meyer et al., 1999; Ruppelt et al., 2001; Ryten et al., 2001). Somitic expression of this subunit has also been reported in zebrafish (Kucenas et al., 2003). In vitro studies demonstrated that P2X5 is involved in myogenesis and muscle progenitor cells (satellite cells) differentiation (Ryten et al., 2002; Araya et al., 2004; Martinello et al., 2011). Its possible implication during muscle regeneration makes this receptor a therapeutic target for musculoskeletal pathologies (Ryten et al., 2004; Burnstock et al., 2013). In rat, P2X2 and P2X6 are also expressed in developing skeletal muscles (Ryten et al., 2001). We observed p2rx2.L and p2xr6.L expression in Xenopus somites, although their transcripts are only detected at a low level compared to p2rx5 somitic expression level. This suggests that P2X5 may be the major P2 receptor regulating somitogenesis. The easiness of Xenopus as an in vivo model for myogenesis makes this vertebrate embryo an ideal model to apprehend the functional roles of P2X5 during myogenesis (Sabillo et al., 2016).
P2rx4.L and p2rx2.S are both expressed at high level in the embryonic functional kidney, the pronephros, but in different regions: p2rx2.S is expressed in the glomus, the filtration unit, whereas p2rx4 is expressed in the proximal tubules. No p2rx transcripts were detected in the distal pronephric region, e.g., the distal, and connecting tubules. In the rat metanephros, P2X2 is found in the glomerulus and collecting duct where it may be involved in ATP inhibitory effects on AVP-induced water permeability (Burnstock et al., 2014). In mammalian metanephros P2RX4 is one of the major P2X receptors and its expression is found along the entire nephron (glomerulus, proximal tubules, descending and ascending limb of Henle, distal tubules, and collecting duct) (reviewed in Bailey et al., 2000; Burnstock et al., 2014; Menzies et al., 2017). P2X4 and P2X4/6 receptors are involved in diverse physiological renal processes, depending on its nephron localization and but also in renal pathology, such as diabetic nephropathy (Chen et al., 2013; Menzies et al., 2017). Several evidence suggest that P2X4 may form complexes with P2X7 in the metanephros (Craigie et al., 2013) but we failed to detect any p2rx6 or p2xr7 expression in the developing amphibian pronephros, suggesting no renal hetero-oligomerization between P2X4 and these receptors in Xenopus. As in mammals, the Xenopus kidney is one of the major sites of expression of the purinergic receptors and ectonucleotidases. We previously published the renal expression of the ectonucleotidases entpdase 5 and enpp2, enpp4, enpp6 (Massé et al., 2010, 2006). Due to its simple structural organization, composed of a single enormous nephron, Xenopus pronephros may be an ideal model to address the functions of the P2X receptors and the purinergic signaling during kidney formation, renal physiology but also pathophysiology (Lienkamp, 2016; Krneta-Stankic et al., 2017).
The developing nervous system is the major site of expression of the p2rx genes, with 7 out of the 9 genes displaying an expression in the central or peripheral nervous system. Surprisingly, only the expression of p2rx1.S was detected by in situ hybridization at stage neurula in developing nervous system, although p2rx2.L, p2rx2.S transcripts were amplified during neurulation by RT-PCR, a more sensitive technique, suggesting they may also be involved in neural tube formation. Little information is available regarding p2rx expression at neurula stages in other vertebrate models, although P2X3 seems to be the earliest subunit expressed during central nervous system formation (Massé and Dale, 2012; Burnstock and Dale, 2015). Developing central nervous system is a major site of expression of the p2rx genes, with 5 out of the 9 expressed in this tissue displaying diffuse and/or weak expresssion, for example p2rx7.L, or specific and strong expression. Such discrete domains are the pineal gland that expresses p2rx4.L or the subset of hindbrain neurons labeled with the p2rx1.L antisense probe. The ventral localization of these neurons suggests they might be a subset of interneurons or motor neurons (Thélie et al., 2015).
Several p2xr subunits are expressed in the cranial sensory peripheral nervous system at later organogenesis stages after the neurogenic placode differentiation (Schlosser and Northcutt, 2000). P2rx2.L is expressed in trigeminal ganglia. p2rx1.S, p2rx2.S, and p2rx6.L are expressed in Xenopus ganglia of the anterodorsal, anteroventral and middle lateral line nerves, which are mechanosensory organs, composed of mechanoreceptive neuromasts involved in the detection of water displacements and current and electric fields (Shelton, 1971; Winklbauer, 1989). Although, p2xr3, the prototypic sensory P2X subtypes, is absent in Xenopus laevis, our data suggest that P2X receptors might have functional implication in sensory transmission rather than in neurogenic placode differentiation (Nakatsuka and Gu, 2006). p2rx1.S, p2rx2.S, and p2rx6.L mRNAs are colocalized into the ganglia of glossopharyngeal and middle lateral line nerves, suggesting co-expression of these subunits. Several biochemical evidences suggested the existence of P2X1/2 complexes, but still not confirmed in vivo (reviewed in Saul et al., 2013). However, existence of functional P2X2/6 channels and the co-expression of these two subunits in several rat neuronal populations have been demonstrated (Collo et al., 1996; King et al., 2000). Future work will need to address in vivo the potential oligo-heteromerization of these subunits during vertebrate neurogenesis.
Taken together, this work is a fundamental prerequisite to apprehend the developmental functions of P2X receptors during vertebrate embryogenesis. Specific gain or loss of function studies of the P2X subunits identified in this work should allow to decipher their functions during the early and late phases of the formation of the central and peripheral nervous system.
The datasets generated for this study can be found in Genbank, ID 2203640.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the European Community and approved by the ethical committee of Bordeaux.
CB performed the experiments, analyzed the data, and corrected the manuscript. EB-G made the figures and wrote the manuscript. KM supervised the work, analyzed the data, and wrote the manuscript.
This work was supported by funding from the University of Bordeaux and the CNRS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2019.00340/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Appelbaum, L., Skariah, G., Mourrain, P., and Mignot, E. (2007). Comparative expression of p2x receptors and ecto-nucleoside triphosphate diphosphohydrolase 3 in hypocretin and sensory neurons in zebrafish. Brain Res. 1174, 66–75. doi: 10.1016/j.brainres.2007.06.103
Araya, R., Riquelme, M. A., Brandan, E., and Sáez, J. C. (2004). The formation of skeletal muscle myotubes requires functional membrane receptors activated by extracellular ATP. Brain Res. Brain Res. Rev. 47, 174–188. doi: 10.1016/j.brainresrev.2004.06.003
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., de Castro, E., et al. (2012). ExPASy: sIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Bailey, M. A., Hillman, K. A., and Unwin, R. J. (2000). P2 receptors in the kidney. J. Auton. Nerv. Syst. 81, 264–270. doi: 10.1016/s0165-1838(00)00125-9
Bassez, T., Paris, J., Omilli, F., Dorel, C., and Osborne, H. B. (1990). Post-transcriptional regulation of ornithine decarboxylase in Xenopus laevis oocytes. Development 110, 955–962.
Blanchard, C., and Massé, K. (2019). “Developmental expression of ectonucleotidase and purinergic receptors detection by whole-mount in situ hybridization in embryos in the methods molecular biology,” in Purinergic Signaling. Springer: Nature. doi: 10.1007/978-1-4939-9717-6_6
Boué-Grabot, E., Akimenko, M. A., and Séguéla, P. (2000a). Unique functional properties of a sensory neuronal P2X ATP-gated channel from zebrafish. J. Neurochem. 75, 1600–1607. doi: 10.1074/jbc.275.14.10190
Boué-Grabot, E., Archambault, V., and Séguéla, P. (2000b). A protein kinase C site highly conserved in P2X subunits controls the desensitization kinetics of P2X2 ATP-gated channels. J. Biol. Chem. 275, 10190–10195. doi: 10.1074/jbc.275.14.10190
Boué-Grabot, E., and Pankratov, Y. (2017). Modulation of central synapses by astrocyte-released atp and postsynaptic P2X receptors. Neural Plast. 2017:9454275. doi: 10.1155/2017/9454275
Breathnach, R., and Chambon, P. (1981). Organization and expression of eucaryotic split genes coding for proteins. Annu. Rev. Biochem. 50, 349–383. doi: 10.1146/annurev.bi.50.070181.002025
Burnstock, G. (2014). Purinergic signalling: from discovery to current developments. Exp. Physiol. 99, 16–34. doi: 10.1113/expphysiol.2013.071951
Burnstock, G. (2016). Purinergic mechanisms and pain. Adv. Pharmacol. 75, 91–137. doi: 10.1016/bs.apha.2015.09.001
Burnstock, G. (2017). Purinergic signalling and neurological diseases: an update. CNS Neurol. Disord. Drug Targets 16, 257–265. doi: 10.2174/1871527315666160922104848
Burnstock, G. (2018). The therapeutic potential of purinergic signalling. Biochem. Pharmacol. 151, 157–165. doi: 10.1016/j.bcp.2017.07.016
Burnstock, G., Arnett, T. R., and Orriss, I. R. (2013). Purinergic signalling in the musculoskeletal system. Purinergic Signal. 9, 541–572. doi: 10.1007/s11302-013-9381-4
Burnstock, G., and Dale, N. (2015). Purinergic signalling during development and ageing. Purinergic Signal. 11, 277–305. doi: 10.1007/s11302-015-9452-9
Burnstock, G., Evans, L. C., and Bailey, M. A. (2014). Purinergic signalling in the kidney in health and disease. Purinergic Signal. 10, 71–101. doi: 10.1007/s11302-013-9400-5
Burnstock, G., Krügel, U., Abbracchio, M. P., and Illes, P. (2011). Purinergic signalling: from normal behaviour to pathological brain function. Prog. Neurobiol. 95, 229–274. doi: 10.1016/j.pneurobio.2011.08.006
Chaumont, S., Jiang, L. H., Penna, A., North, R. A., and Rassendren, F. (2004). Identification of a trafficking motif involved in the stabilization and polarization of P2X receptors.: trafficking motif of P2X ATP-gated channels. J. Biol. Chem. 279, 29628–29638. doi: 10.1074/jbc.M403940200
Chen, K., Zhang, J., Zhang, W., Zhang, J., Yang, J., Li, K., et al. (2013). ATP-P2X4 signaling mediates NLRP3 inflammasome activation: a novel pathway of diabetic nephropathy. Int. J. Biochem. Cell Biol. 45, 932–943. doi: 10.1016/j.biocel.2013.02.009
Cockayne, D. A., Hamilton, S. G., Zhu, Q. M., Dunn, P. M., Zhong, Y., Novakovic, S., et al. (2000). Urinary bladder hyporeflexia and reduced pain-related behaviour in P2X3-deficient mice. Nature 407, 1011–1015. doi: 10.1038/35039519
Collo, G., North, R. A., Kawashima, E., Merlo-Pich, E., Neidhart, S., Surprenant, A., et al. (1996). Cloning OF P2X5 and P2X6 receptors and the distribution and properties of an extended family of ATP-gated ion channels. J. Neurosci. 16, 2495–2507. doi: 10.1523/jneurosci.16-08-02495.1996
Craigie, E., Birch, R. E., Unwin, R. J., and Wildman, S. S. (2013). The relationship between P2X4 and P2X7: a physiologically important interaction? Front. Physiol. 4:216. doi: 10.3389/fphys.2013.00216
Del Puerto, A., Wandosell, F., and Garrido, J. J. (2013). Neuronal and glial purinergic receptors functions in neuron development and brain disease. Front. Cell Neurosci. 7:197. doi: 10.3389/fncel.2013.00197
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Domercq, M., Zabala, A., and Matute, C. (2018). Purinergic receptors in multiple sclerosis pathogenesis. Brain Res. Bull. doi: 10.1016/j.brainresbull.2018.11.018 [Epub ahead of print].
Fumagalli, M., Lecca, D., Abbracchio, M. P., and Ceruti, S. (2017). Pathophysiological role of purines and pyrimidines in neurodevelopment: unveiling new pharmacological approaches to congenital brain diseases. Front. Pharmacol. 8:941. doi: 10.3389/fphar.2017.00941
Habermacher, C., Dunning, K., Chataigneau, T., and Grutter, T. (2016). Molecular structure and function of P2X receptors. Neuropharmacology 104, 18–30. doi: 10.1016/j.neuropharm.2015.07.032
Hellsten, U., Harland, R. M., Gilchrist, M. J., Hendrix, D., Jurka, J., Kapitonov, V., et al. (2010). The genome of the Western clawed frog Xenopus tropicalis. Science 328, 633–636. doi: 10.1126/science.1183670
Juranka, P. F., Haghighi, A. P., Gaertner, T., Cooper, E., and Morris, C. E. (2001). Molecular cloning and functional expression of Xenopus laevis oocyte ATP-activated P2X4 channels. Biochim. Biophys. Acta 1512, 111–124. doi: 10.1016/s0005-2736(01)00313-3
Kawate, T., Michel, J. C., Birdsong, W. T., and Gouaux, E. (2009). Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature 460, 592–598. doi: 10.1038/nature08198
Khakh, B. S., and North, R. A. (2006). P2X receptors as cell-surface ATP sensors in health and disease. Nature 442, 527–532. doi: 10.1038/nature04886
Khakh, B. S., and North, R. A. (2012). Neuromodulation by extracellular ATP and P2X receptors in the CNS. Neuron 76, 51–69. doi: 10.1016/j.neuron.2012.09.024
King, B. F., Townsend-Nicholson, A., Wildman, S. S., Thomas, T., Spyer, K. M., and Burnstock, G. (2000). Coexpression of rat P2X2 and P2X6 subunits in Xenopus oocytes. J. Neurosci. 20, 4871–4877.
Krneta-Stankic, V., DeLay, B. D., and Miller, R. K. (2017). Xenopus: leaping forward in kidney organogenesis. Pediatr. Nephrol. 32, 547–555. doi: 10.1007/s00467-016-3372-y
Kucenas, S., Cox, J. A., Soto, F., Lamora, A., and Voigt, M. M. (2009). Ectodermal P2X receptor function plays a pivotal role in craniofacial development of the zebrafish. Purinergic Signal. 5, 395–407. doi: 10.1007/s11302-009-9165-z
Kucenas, S., Li, Z., Cox, J. A., Egan, T. M., and Voigt, M. M. (2003). Molecular characterization of the zebrafish P2X receptor subunit gene family. Neuroscience. 121, 935–945. doi: 10.1016/s0306-4522(03)00566-9
Kucenas, S., Soto, F., Cox, J. A., and Voigt, M. M. (2006). Selective labeling of central and peripheral sensory neurons in the developing zebrafish using P2X3 receptor subunit transgenes. Neuroscience 138, 641–652. doi: 10.1016/j.neuroscience.2005.11.058
Lienkamp, S. S. (2016). Using Xenopus to study genetic kidney diseases. Semin. Cell Dev. Biol. 51, 117–124. doi: 10.1016/j.semcdb.2016.02.002
Martinello, T., Baldoin, M. C., Morbiato, L., Paganin, M., Tarricone, E., Schiavo, G., et al. (2011). Extracellular ATP signaling during differentiation of C2C12 skeletal muscle cells: role in proliferation. Mol. Cell. Biochem. 351, 183–196. doi: 10.1007/s11010-011-0726-4
Massé, K., Bhamra, S., Allsop, G., Dale, N., and Jones, E. A. (2010). Ectophosphodiesterase/nucleotide phosphohydrolase (Enpp) nucleotidases: cloning, conservation and developmental restriction. Int. J. Dev. Biol. 54, 181–193. doi: 10.1387/ijdb.092879km
Massé, K., Bhamra, S., Eason, R., Dale, N., and Jones, E. A. (2007). Purine-mediated signalling triggers eye development. Nature 449, 1058–1062. doi: 10.1038/nature06189
Massé, K., and Dale, N. (2012). Purines as potential morphogens during embryonic development. Purinergic Signal. 8, 503–521. doi: 10.1007/s11302-012-9290-y
Massé, K., Eason, R., Bhamra, S., Dale, N., and Jones, E. A. (2006). Comparative genomic and expression analysis of the conserved NTPDase gene family in Xenopus. Genomics 87, 366–381. doi: 10.1016/j.ygeno.2005.11.003
Menzies, R. I., Tam, F. W., Unwin, R. J., and Bailey, M. A. (2017). Purinergic signaling in kidney disease. Kidney Int. 91, 315–323. doi: 10.1016/j.kint.2016.08.029
Meyer, M. P., Gröschel-Stewart, U., Robson, T., and Burnstock, G. (1999). Expression of two ATP-gated ion channels, P2X5 and P2X6, in developing chick skeletal muscle. Dev. Dyn. 216, 442–449. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<442::AID-DVDY12>3.0.CO;2-Z
Nakatsuka, T., and Gu, J. G. (2006). P2X purinoceptors and sensory transmission. Pflugers Arch. 452, 598–607. doi: 10.1007/s00424-006-0057-6
Needleman, S. B., and Wunsch, C. D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48, 443–453. doi: 10.1016/0022-2836(70)90057-4
Nieuwkoop, P. D., and Faber, J. (1994). Normal Table of Xenopus Laevis (Daudin). New York, NY: Garland Publishing, Inc.
North, R. A. (2002). Molecular physiology of P2X receptors. Physiol. Rev. 82, 1013–1067. doi: 10.1152/physrev.00015.2002
North, R. A. (2016). P2X receptors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150427. doi: 10.1098/rstb.2015.0427
Norton, W. H., Rohr, K. B., and Burnstock, G. (2000). Embryonic expression of a P2X(3) receptor encoding gene in zebrafish. Mech. Dev. 99, 149–152. doi: 10.1016/s0925-4773(00)00472-x
Oliveira, Á, Illes, P., and Ulrich, H. (2016). Purinergic receptors in embryonic and adult neurogenesis. Neuropharmacology 104, 272–281. doi: 10.1016/j.neuropharm.2015.10.008
Palanca, A. M., Lee, S. L., Yee, L. E., Joe-Wong, C., Trinh le, A., Hiroyasu, E., et al. (2013). New transgenic reporters identify somatosensory neuron subtypes in larval zebrafish. Dev. Neurobiol 73, 152–167. doi: 10.1002/dneu.22049
Paukert, M., Hidayat, S., and Gründer, S. (2002). The P2X(7) receptor from Xenopus laevis: formation of a large pore in Xenopus oocytes. FEBS Lett. 513, 253–258. doi: 10.1016/s0014-5793(02)02324-4
Ralevic, V., and Burnstock, G. (1998). Receptors for purines and pyrimidines. Pharmacol. Rev. 50, 413–492.
Rodrigues, R. J., Marques, J. M., and Cunha, R. A. (2018). Purinergic signalling and brain development. Semin. Cell Dev. Biol. doi: 10.1016/j.semcdb.2018.12.001 [Epub ahead of print].
Ruppelt, A., Ma, W., Borchardt, K., Silberberg, S. D., and Soto, F. (2001). Genomic structure, developmental distribution and functional properties of the chicken P2X5 receptor. J. Neurochem. 77, 1256–1265. doi: 10.1046/j.1471-4159.2001.00348.x
Ryten, M., Dunn, P. M., Neary, J. T., and Burnstock, G. (2002). ATP regulates the differentiation of mammalian skeletal muscle by activation of a P2X5 receptor on satellite cells. J. Cell Biol. 158, 345–355. doi: 10.1083/jcb.200202025
Ryten, M., Hoebertz, A., and Burnstock, G. (2001). Sequential expression of three receptor subtypes for extracellular ATP in developing rat skeletal muscle. Dev. Dyn. 221, 331–341. doi: 10.1002/dvdy.1147
Ryten, M., Yang, S. Y., Dunn, P. M., Goldspink, G., and Burnstock, G. (2004). Purinoceptor expression in regenerating skeletal muscle in the mdx mouse model of muscular dystrophy and in satellite cell cultures. FASEB J. 18, 1404–1406. doi: 10.1096/fj.03-1175fje
Sabillo, A., Ramirez, J., and Domingo, C. R. (2016). Making muscle: morphogenetic movements and molecular mechanisms of myogenesis in Xenopus laevis. Semin. Cell Dev. Biol . 51, 80–91. doi: 10.1016/j.semcdb.2016.02.006
Sater, A. K., and Moody, S. A. (2017). Using Xenopus to understand human disease and developmental disorders. Genesis 55:e22997. doi: 10.1002/dvg.22997
Saul, A., Hausmann, R., Kless, A., and Nicke, A. (2013). Heteromeric assembly of P2X subunits. Front. Cell. Neurosci. 7:250. doi: 10.3389/fncel.2013.00250
Schlosser, G., and Northcutt, R. G. (2000). Development of neurogenic placodes in Xenopus laevis. J. Comp. Neurol. 418, 121–146. doi: 10.1002/(SICI)1096-9861(20000306)418:2<121::AID-CNE1>3.0.CO;2-M
Session, A. M., Uno, Y., Kwon, T., Chapman, J. A., Toyoda, A., Takahashi, S., et al. (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343. doi: 10.1038/nature19840
Shelton, P. M. (1971). The structure and function of the lateral line system in larval Xenopus laevis. J. Exp. Zool. 178, 211–231. doi: 10.1002/jez.1401780207
Solle, M., Labasi, J., Perregaux, D. G., Stam, E., Petrushova, N., Koller, B. H., et al. (2001). Altered cytokine production in mice lacking P2X(7) receptors. J. Biol. Chem. 276, 125–132. doi: 10.1074/jbc.M006781200
Souslova, V., Cesare, P., Ding, Y., Akopian, A. N., Stanfa, L., Suzuki, R., et al. (2000). Warm-coding deficits and aberrant inflammatory pain in mice lacking P2X3 receptors. Nature 407, 1015–1017. doi: 10.1038/35039526
Tandon, P., Conlon, F., Furlow, J. D., and Horb, M. E. (2017). Expanding the genetic toolkit in Xenopus: approaches and opportunities for human disease modeling. Dev. Biol. 426, 325–335. doi: 10.1016/j.ydbio.2016.04.009
Thélie, A., Desiderio, S., Hanotel, J., Quigley, I., Van Driessche, B., Rodari, A., et al. (2015). Prdm12 specifies V1 interneurons through cross-repressive interactions with Dbx1 and Nkx6 genes in Xenopus. Development 142, 3416–3428. doi: 10.1242/dev.121871
Thompson, B. A., Storm, M. P., Hewinson, J., Hogg, S., Welham, M. J., and MacKenzie, A. B. (2012). A novel role for P2X7 receptor signalling in the survival of mouse embryonic stem cells. Cell. Signal. 24, 770–778. doi: 10.1016/j.cellsig.2011.11.012
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequences weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tocco, A., Pinson, B., Thiébaud, P., Thézé, N., and Massé, K. (2015). Comparative genomic and expression analysis of the adenosine signaling pathway members in Xenopus. Purinergic Signal. 11, 59–77. doi: 10.1007/s11302-014-9431-6
Vulchanova, L., Riedl, M. S., Shuster, S. J., Buell, G., Surprenant, A., North, R. A., et al. (1997). Immunohistochemical study of the P2X2 and P2X3 receptor subunits in rat and monkey sensory neurons and their central terminals. Neuropharmacology 36, 1229–1242. doi: 10.1016/s0028-3908(97)00126-3
Winklbauer, R. (1989). Development of the lateral line system in Xenopus. Prog. Neurobiol. 32, 181–206. doi: 10.1016/0301-0082(89)90016-6
Xiang, Z., Bo, X., and Burnstock, G. (1998). Localization of ATP-gated P2X receptor immunoreactivity in rat sensory and sympathetic ganglia. Neurosci. Lett. 256, 105–108. doi: 10.1016/s0304-3940(98)00774-5
Keywords: P2 receptors, purinergic signaling, embryogenesis, neurogenic placodes, sensory neurons, nervous system, Xenopus laevis
Citation: Blanchard C, Boué-Grabot E and Massé K (2019) Comparative Embryonic Spatio-Temporal Expression Profile Map of the Xenopus P2X Receptor Family. Front. Cell. Neurosci. 13:340. doi: 10.3389/fncel.2019.00340
Received: 20 March 2019; Accepted: 10 July 2019;
Published: 26 July 2019.
Edited by:
Christian Lohr, Universität Hamburg, GermanyReviewed by:
Ivan Manzini, Justus Liebig University Giessen, GermanyCopyright © 2019 Blanchard, Boué-Grabot and Massé. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karine Massé, S2FyaW5lLm1hc3NlQHUtYm9yZGVhdXguZnI=
†Present address: Camille Blanchard, INSERM, U 1215, Neurocentre Magendie, Université de Bordeaux, Bordeaux, France
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.