- 1Institute of Life Sciences, Fuzhou University, Fuzhou, China
- 2Department of Biology, University of Nevada, Reno, Reno, NV, United States
- 3Institute of Applied Ecology, Fujian Agriculture and Forestry University, Fuzhou, China
The circadian system, which has a period of about 24 h, is import for organismal health and fitness. The molecular circadian clock consists of feedback loops involving both transcription and translation, and proper function of the circadian system also requires communication among intracellular organelles. As important hubs for signaling in the cell, mitochondria integrate a variety of signals. Mitochondrial dysfunction and disruption of circadian rhythms are observed in neurodegenerative diseases and during aging. However, how mitochondrial dysfunction influences circadian rhythm is largely unknown. Here, we report that Drosophila prune (pn), which localizes to the mitochondrial matrix, most likely affects the function of certain clock neurons.Deletion of pn in flies caused decreased expression of mitochondrial transcription factor TFAM and reductions in levels of mitochondrial DNA, which resulted in mitochondrial dysfunction. Loss of pn decreased the amplitude of circadian rhythms.In addition, we showed that depletion of mtDNA by overexpression of a mitochondrially targeted restriction enzyme mitoXhoI also decreased the robustness of circadian rhythms. Our work demonstrates that pn is important for mitochondrial function thus involved in the regulation of circadian rhythms.
Introduction
Circadian clocks drive rhythms in animal physiology and behavior, thus enabling anticipation of daily environmental changes. Cellular processes occur in compartments including the nucleus, endoplasmic reticulum, and mitochondria. These organelles display diurnal rhythms in structural properties, functions, and responses to stress (Chedid and Nair, 1972; Chaix et al., 2016). Diurnal rhythms in intracellular organelles serve critical cellular functions. For instance, the circadian clock can regulate transcription, the DNA damage response, and the general architecture of the nucleus (Gery et al., 2006; Koike et al., 2012; Chaix et al., 2016). Temporal coordination of protein folding, the unfolded protein response, and secretion likely reduces proteotoxicity (Zhu et al., 2014; Chaix et al., 2016). Mitochondria generate most of the cell’s supply of adenosine triphosphate (ATP), used as a source of chemical energy. In addition to supplying cellular energy, mitochondria are also involved in other processes such as cell signaling (Chandel, 2014). The anterograde (nuclear to mitochondrial) and retrograde (mitochondrial to nuclear) signaling pathways mediate communications between nucleus and mitochondria (Butow and Avadhani, 2004; Chandel, 2014; Ng et al., 2014).
Recent studies have suggested that circadian clocks control aspects of mitochondrial function including oxidative metabolism, nutrient utilization, and mitochondrial dynamics (Manella and Asher, 2016; de Goede et al., 2018). However, how mitochondrial function feeds back to the circadian clock mechanism has not been well characterized. Aging processes are associated with declines in mitochondrial function and accumulation of abnormal mitochondria (Sun et al., 2016). Decreases in circadian amplitude and other disruptions of circadian rhythms are also observed in aged animals (Brown et al., 2011; Duffy et al., 2015). Moreover, mitochondrial dysfunction is a characteristic of neurodegenerative diseases such as Parkinson’s disease, Huntington’s disease, and amyotrophic lateral sclerosis (Quintanilla et al., 2008; Shi et al., 2010; Bose and Beal, 2016), and disruption of circadian rhythms is also a common feature and an early symptom of neurodegeneration. This indicates that there are potential connections between mitochondrial dysfunction and circadian rhythms.
The fruit fly, Drosophila melanogaster, is a powerful model for studies of biological rhythms. In organisms, behavioral or physiological traits can be entrained by environmental cues (i.e., light and temperature) that cycle during 24 h, and cycling of these traits persists in the absence of environmental cues due to the self-sustaining clock (Allada and Chung, 2010; Dubowy and Sehgal, 2017). Under conditions of 12 h of light and 12 h of dark cycling (LD), flies have bimodal activity patterns that peak around dawn and dusk (Allada and Chung, 2010; Dubowy and Sehgal, 2017). Their locomotor activity gradually increases before the lights-on and lights-off transitions, a phenomenon termed anticipation. In constant dark conditions (DD), the evening activity peak persists and reoccurs with a period of near 24 h (Allada and Chung, 2010; Dubowy and Sehgal, 2017). The small ventral lateral neurons (sLNvs), which express the neuropeptide PDF, are the master pacemaker neurons responsible for generating the locomotor rhythms under DD (Helfrich-Förster, 1998; Renn et al., 1999; Stoleru et al., 2007; Nitabach and Taghert, 2008; Yao and Shafer, 2014). The PDF-negative circadian neurons, such as the dorsal lateral neurons (LNds) and the 5th sLNvs, control the evening peak of activity (Allada and Chung, 2010; Dubowy and Sehgal, 2017). The dorsal neuron 1 (DN1s) can integrate light and also sense temperature to regulate locomotor activity and sleep (Zhang et al., 2010; Guo et al., 2016; Yadlapalli et al., 2018).
Periodogram analysis is usually used for the analysis of periodic data (Klarsfeld et al., 2003; Rosato and Kyriacou, 2006). Rhythmic data can be considered as a wave, and mainly parameters of period and power are analyzed (Klarsfeld et al., 2003; Rosato and Kyriacou, 2006). The period refers to the time interval at which a rhythmic event repeat. The power, or amplitude, of a locomotor rhythm is the difference in activity between the peaks and troughs and reflects the robustness of the endogenous rhythm.
Here, we report that prune (pn), which encodes a cyclic nucleotide phosphodiesterase that localizes to the mitochondrial matrix, is involved in the regulation of circadian rhythm in Drosophila. Loss of pn in circadian cells reduced levels of the mitochondrial transcription factor TFAM and decreased the mitochondrial DNA (mtDNA) level resulting in mitochondrial dysfunction and a decrease in the amplitude of the endogenous clock rhythms. In addition, we found that overexpression of a restriction enzyme, mitoXhoI, which cuts mtDNA, in circadian neurons decreases the robustness of circadian rhythm. Our work demonstrates a potential link between mitochondrial dysfunction and circadian rhythm.
Materials and Methods
Fly Strains
Fly stocks were raised at 25°C on standard cornmeal-molasses-yeast medium. The mutants of pn1 (BL174) and pn2 (BL81) were obtained from the Bloomington Drosophila Stock Center. These two lines were outcrossed into the iso31 wild-type line for five generations to change the genetic background. iso31 is a wild-type strain widely used for circadian behavior studies (Ryder et al., 2004). The following lines were also obtained from the Bloomington Drosophila Stock Center: UAS-pn-RNA interference (RNAi; BL65920), UAS-tfam-RNAi (BL26744), act5C-Gal4 (BL4414), elav-Gal4; UAS-Dicer2 (BL25750), sqh-mitoEYFP (BL7194), GMR-Gal4 (BL1104), repo-Gal4 (BL7415) and tub-Gal80ts (BL7018). UAS-mitoXhoI and hs-pn were kindly provided by Hong Xu (National Heart, Lung and Blood Institute, Bethesda, MD, USA; Zhang et al., 2015). UAS-tfam was a gift from Joseph Bateman (King’s College London, London, UK; Cagin et al., 2015). NP1-Gal4; UAS-mitoAT1.03 was provided by Yufeng Yang (Institute of Life Sciences, Fuzhou University, China). tim-Gal4 (Kaneko and Hall, 2000), pdf-Gal4 (Renn et al., 1999; Park et al., 2000) and tim-Gal4;pdf-Gal80 (Murad et al., 2007) were used to drive in circadian cells.
Plasmids Construction and Transgenesis
To generate the pnΔ fly line using CRISPR/Cas9 system, a single-stranded oligonucleotide, Pn-ssODN, was used: 5′-GCCGGAAATTACATCTGGTAATGGGCAACGAATCGTGTGACTTG
GACTCCGCCGTTTCGGCCGTCAGATAAGCTTCATCGGGAGCACGACTATGTACCT
ATACTGAACATTCCTCGCCGGGACTACCCGTTGAAA-3′. A HindIII recognition site and a stop codon were introduced into the DDH domain (a domain of predicated phosphoesterases) of pn locus with this oligonucleotide.
For chiRNAs (a vector for guide RNA expression under the control of U6 promoter) construction, the two target site sequences were cloned into the pU6-BbsI-chiRNA plasmid (Addgene, #45946) as previously described (Gratz et al., 2014) using the following oligonucleotides:
• PAM1-sense: 5′-CTTCGTAGACAAAAGCCAAAGTGA-3′
• PAM1-antisense: 5′-AAACTCACTTTGGCTTTTGTCTAC-3′
• PAM2-sense: 5′-CTTCGTGTCTACGCGCAGCGTCAT-3′
• PAM2-antisense: 5′-AAACATGACGCTGCGCGTAGACAC-3′
The Pn-ssODN and pU6-BbsI-chiRNA plasmids were mixed and injected into embryos of the act5C-Cas9 (BL#54590, y M{w[+mC] = Act5C-Cas9.P}ZH-2A w[*]) fly line (Rainbow Transgenic Flies).
Screen for Integration
After injection, a single F0 fly was collected and crossed with a FM7 balancer fly. Ten to fifteen larval or pupal flies from the F1 generation were pooled for genome extraction. The following primers were used for amplification and sequencing:
• F1: 5′- CGAATCGATACGCGCAACTGTTG-3′
• R1: 5′- TCGTGCTCCCGATGAAGCTTATC-3′
The F1 primer flanks the left homologous arm of Pn-ssODN, and R1 hybridizes to the HindIII site and stop codon region. For the F1R1 polymerase chain reaction (PCR)-positive vials, 10–20 F1 flies were crossed with the FM7 balancer fly by single cross. When eggs were present in the food, the genome of a single F1 fly was extracted and amplified using F1 and R1 primers. The F1 primer plus another primer R2 that flanks the right homologous arm were used for amplification and sequencing:
• R2: 5′-TGCTTGAAGGATGGACTGCTGTC-3′
The pnΔ fly line and its control line act5C-Cas9 were outcrossed into the iso31 wild-type line for five generations to change the genetic background.
Immunostaining
Adult flies (3–5 days old) were collected and fixed at room temperature for 1 h in 4% formaldehyde diluted in PBT (PBS with 0.1% Triton X-100). The brains were dissected in PBT at indicated time points and fixed in 4% formaldehyde in PBT for 20 min at room temperature. The brains were rinsed and washed with PBT three times (15 min each) and then were blocked in 10% normal goat serum in PBT for 2 h at room temperature. The brains were incubated with primary antibody, mouse anti-PDF C7 (Developmental Studies Hybridoma Bank), diluted 1:200 or rabbit anti-PERIOD (1:1,000) in PBT overnight at 4°C. After three 15-min washes, brains were incubated with secondary goat anti-mouse Cy5 (Jackson Immuno Research) or goat anti-rabbit Alexa Fluor488 (ThermoFisher, Waltham, MA, USA) antibody at 1:200 dilution overnight at 4°C, followed by extensive washes with PBT. Finally, the brains were mounted in the VectaMount Permanent Mounting Medium. Imaging was performed on a Leica Confocal Microscope SP8 system. Confocal images were obtained at an optical section thickness of 1–2 μm.
Quantitative RT-PCR
Real-time quantitative reverse transcription PCR (qPCR) was conducted with total RNA extracted from 20 to 30 male fly heads using TRIzol as per the manufacturer’s protocol (Life Technologies, Carlsbad, CA, USA). cDNA from a reverse-transcription reaction or genomic DNA containing mtDNA were used as a starting material. qPCR was performed using SYBR Select Master Mix for CFX (Life Technologies, Carlsbad, CA, USA) on the CFX96 Real-Time System (BIO-RAD, Hercules, CA, USA). The following primers were used:
• Actin5C-f: 5′-CAGAGCAAGCGTGGTATCCT-3′
• Actin5C-r: 5′-CTCATTGTAGAAGGTGTGGTGC-3′
• pn-f: 5′-TTTCTACGATTTTTGGCCCAGG-3′
• pn-r: 5′-TCGTTGCCCATTACCAGATGTA-3′
• tfam-f: 5′-TGCAACAAGTTCCCCGTGAT-3′
• tfam-r: 5′-GCTAGGGGCCTGACTTTGTT-3′
• mito-f: 5′-GCTCATCATATATTTACCGTTGGA-3′
• mito-r: 5′-AAAATTTTAATTCCAGTAGGAACTGC-3′
• Gapdh1-f: 5′-GACGAAATCAAGGCTAAGGTCG-3′
• Gapdh1-r: 5′-AATGGGTGTCGCTGAAGAAGTC-3′
Locomotor Behavior Assay
Unless noted otherwise, all the flies were raised on cornmeal/agar medium as indicated at 25°C under LD. Male flies were monitored using Trikinetics Drosophila Activity Monitors at temperature indicated under LD for 3–4 days, followed by 6–7 days in DD. Activity records were collected in 1-min bins and analyzed using the Fly Activity Analysis suite software developed by M. Boudinot as part of the Brandeis Rhythm Package (Klarsfeld et al., 2003). Circadian periods were determined by periodogram analysis. The periodograms correspond to 5 days of activity in constant darkness, beginning 24 h after the last light-off transition. Amplitude of the rhythm is defined as the height of the main peaks above the line corresponding to the 95% confidence limit (Klarsfeld et al., 2003). Rhythmic flies were defined by periodogram analysis with the following criteria (filter on): amplitude ≥20, width ≥2 h. Amplitude and width are the height and width of the periodogram peak, respectively. A Matlab-based signal-processing toolbox, flyplot, was used to generate figures (Levine et al., 2002).
Survival Analysis for Paraquat Treatment
The paraquat toxicity assay was performed on 15-day-old male flies and kept on sucrose agar food (5% sucrose and 2% agar and 10 mM paraquat). For the control and pn mutant flies, we made three vials at a time for each genotype. Each vial contains 10–20 flies. The flies were transfer from normal food to paraquat containing food at ZT0 and then counted for living flies every 12 h until all death. Two independent repeats were performed and combined to analysis together. Gehan-Breslow-Wilcoxon test was performed to do curve comparison.
Statistical Analysis
All statistical analyses were conducted using Graph Pad Prism 7. Normally distributed data were analyzed with two-tailed, unpaired Student’s t-test.
Results
Loss of pn Decreases the Robustness of Circadian Rhythm
pn was previously shown as an oscillating gene in fly head (Claridge-Chang et al., 2001). It is required for mtDNA maintenance in mitochondria matrix (Zhang et al., 2015). mtDNA is the small circular DNA located in mitochondria. It encodes 37 proteins that cooperate with the proteins encoded in nuclear DNA to maintain normal mitochondria function (Anderson et al., 1981). To determine the effects of mitochondrial dysfunction on circadian rhythms, we obtained previously described pn mutant flies, pn1 and pn2 (Frolov et al., 1994; Simmons et al., 1994). To rule out potential effects of genetic background, we also employed CRISIPR/Cas9 to generate a new pn mutant line (pnΔ) by introducing a stop codon in the DDH domain (Figure 1A). As previously reported, both the pn1 and pn2 mutants, as well as our newly generated pnΔ flies have brownish-purple compound eye phenotype (Figure 1A) that was due to the reduction of drosopterins (Evans and Howells, 1978; Kim et al., 2013). Since there are no Pn antibodies available, we used quantitative RT-PCR to determine pn abundance in the fly head. We found that pn was significantly reduced in both pn1 and pnΔ mutants (Supplementary Figures S1A,B). To minimize the effect of genetic background on behavior rhythms, we crossed the pn mutants into iso31, a wild-type strain widely used for circadian behavior studies (Ryder et al., 2004). We then tested the circadian locomotor behavior rhythms and found that pn mutant flies have normal bimodal activity patterns and anticipation under LD (Figures 1B,C). Under DD, the circadian periods and percentages of rhythmic flies were indistinguishable between mutants and control flies. However, the amplitudes of rhythms were reduced in pn mutants compared to control, which indicates loss of the robustness of circadian rhythm (Figure 1C). Together, these results indicate that Pn is involved in circadian rhythm regulation and loss of Pn decreases the robustness of the endogenous clock.
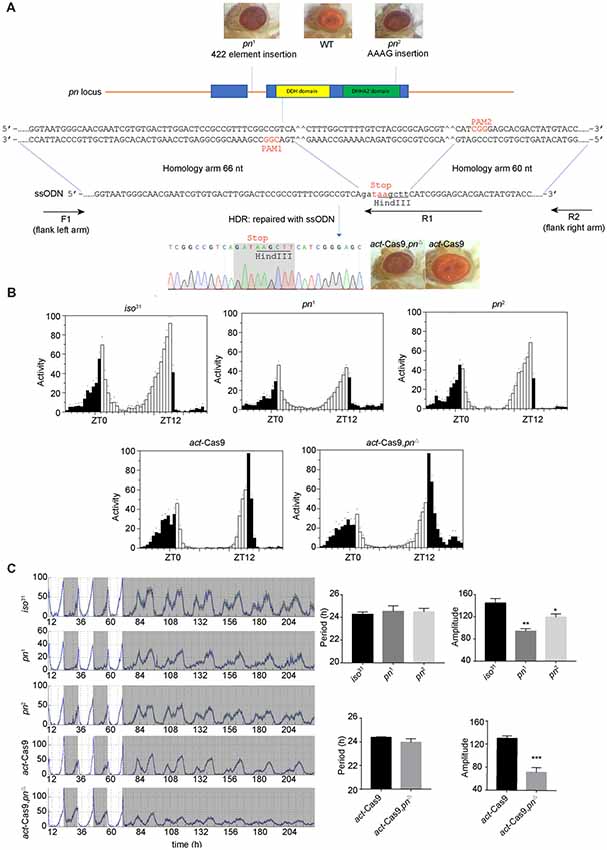
Figure 1. Loss of prune (pn) decreases the robustness of circadian rhythm. (A) Strategy for generating pnΔ mutant. The two target sites located in the DDH domain (a domain of predicated phosphoesterases) are shown. The pn-ssODN donor containing two homology arms was designed to introduce a stop codon and delete some bases of pn locus. pn1 is an allele that contains a 422 elements insertion in the first intron of pn. pn2 is an allele that contains an AAAG insertion at the end of DHHA2 domain (a domain often associated with the DHH domain and is diagnostic of DHH subfamily two members). F1, R1, and R2 are the primers used in molecular screening. (B) Averaged activity profiles of pn mutants during LD. White bars represent day, black bars represent night, Zeitgeber time (ZT) is indicated on the x axes. The dots above the bars indicate standard errors of the means (SEM). (C) Actograms, period, and amplitude of pn mutants and controls. The period and amplitude are shown as means ± SEM (n = 32–40). *P < 0.05, **P < 0.01, ***P < 0.001.
Adulthood-Specific Overexpression of pn Rescues the Decrease in Amplitude of Circadian Rhythms in pn Mutants
To characterize when pn is required for the control of circadian rhythm, we performed an adulthood-specific rescue experiment. We employed a transgenic line expressing pn under the control of a temperature-inducible hsp70 promoter (hs-pn) and did behavior assay as described in a previous study (Ewer et al., 1988). The flies were reared at 18°C, and then were either transferred to 29°C or kept at the low temperature for locomotor activity behavior assay at eclosion (Figure 2A). At 29°C, hs-pn increased the pn expression about two fold relative to levels in the control (Supplementary Figure 1C). When the pn1; hs-pn flies were raised at a low temperature, transferred to 29°C, and monitored at this higher temperature under DD, the amplitude of locomotor activity rhythms was partially rescued (Figure 2B). In contrast, when flies were raised and tested at the low temperature, the locomotor activity was similar in pn1; hs-pn and pn1 flies (Figure 2C).
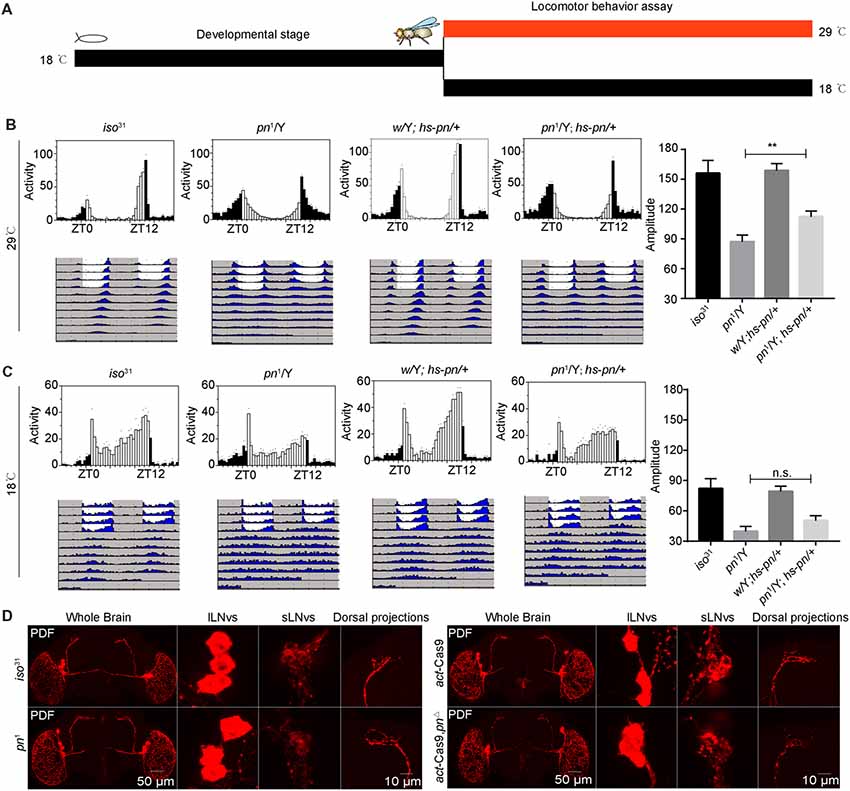
Figure 2. Pn functional in adulthood to maintain the amplitude of the circadian rhythm. (A) A schematic to show adulthood specific rescue experiments. The flies were reared at 18°C, and then were either transferred to 29°C or kept at the low temperature, and a locomotor activity behavior assay was performed at eclosion. (B) Averaged activity profiles and amplitude analysis of flies transferred to 29°C. (C) Averaged activity profiles and amplitude analysis of flies maintained at 18°C. White bars represent day, black bars represent night, ZT is indicated on the x axes. The dots above the bars indicate SEM. The period and amplitude are shown as means ± SEM (n = 30–48). **P < 0.01, n.s. no significance. At least three independent repeats were performed. (D) pn1, pnΔ mutant and control brains stained with PDF demonstrate that development of ventral lateral neurons (LNvs) and dorsal projections are normal (n = 8–10).
Next, we stained the pn1 and pnΔ mutant flies with anti-PDF to check the PDF neurons and their projections. We observed that the cell bodies and axonal projections of PDF-expressing circadian pacemaker neurons were indistinguishable to those of controls (Figure 2D). This indicates that abnormal development of PDF neurons is not responsible for the phenotype of the pn mutant. Together these results suggest that pn is necessary for sustaining robust rhythms in free-running circadian behaviors in the adult.
Depletion of pn in Circadian Tissues Decreases the Robustness of Circadian Rhythm
To further identify the cellular requirement of pn for circadian rhythms, we used the Gal4-UAS system to drive expression of a mediator of RNAi to downregulate pn in specific groups of neurons. When we used actin5C-Gal4, a ubiquitous Gal4 driver to express the shRNA targeting pn, flies phenocopied the pn mutants. These flies had brownish-purple eye color (Figure 3A), which indicates that pn expression was suppressed by the shRNA. Furthermore, RT-PCR results showed that pn levels were decreased by approximately 60% in the elav >pn-RNAi flies compared to the GAL4 control flies (Supplementary Figure S1D). As expected, we observed a significant decrease in amplitudes of circadian rhythms when pn expression was downregulated (Figures 3A,B).
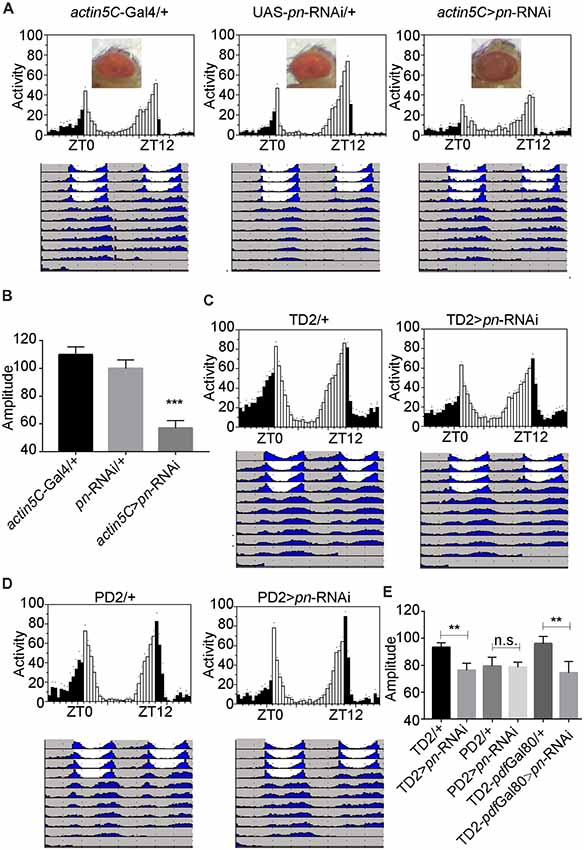
Figure 3. Pn expression in circadian tissues affects the robustness of circadian rhythm. (A) Averaged activity profiles and actograms of control flies and actin5C >pn-RNA interference (RNAi) flies. (B) Amplitudes of circadian rhythms in control flies and actin5C >pn-RNAi flies. (C–E) Averaged activity profiles, actograms and amplitudes of control flies and flies in which pn expression was inhibited by expression of shRNA with different circadian neuron-specific drivers. TD2 indicates tim-Gal4, UAS-Dicer2. PD2 indicates pdf-Gal4, UAS-Dicer2. White bars represent day, black bars represent night, ZT is indicated on the x axes. The dots above the bars indicate SEM. The period and amplitude are shown as means ± SEM (n = 33–50). At least three independent repeats were performed. **P < 0.01, ***P < 0.001, n.s. no significance.
Downregulation of pn in all circadian cells using tim-Gal4 resulted in decreased circadian amplitudes, similar to ubiquitous inhibition of expression, which indicates that pn is required in circadian cells for the amplitude of rhythms (Figures 3C–E). However, when we depleted pn using the pdf-Gal4 driver, which is expressed specifically in PDF-positive neurons, no obvious decrease in amplitude was observed (Figures 3C–E). These results suggest that pn is required in PDF-negative circadian cells. To confirm this, we combined tim-Gal4 with pdf-Gal80 to inhibit pn expression in PDF-negative circadian neurons. pdf-Gal80 is expressed in all PDF-positive LNvs and represses transcription of the UAS in these cells (Murad et al., 2007). We found that pn reduction in PDF-negative circadian cells resulted in a decrease in amplitude of circadian rhythms (Figure 3E). Considering that pn mutant may impact on visual system and tim-Gal4 also expresses in other cells including glia cells, we employed the eye-specific (GMR-Gal4) and glia-specific (repo-Gal4) Gal4 to knockdown pn. No significant reduction of amplitude was observed when pn was disrupted in the eye or glia cells (Supplementary Figure S2). These results indicate that Pn is not likely required in fly eyes or glial cells for circadian rhythms.
The decrease of amplitude might be effects of circadian locomotor output or dampening of molecular pacemaker. The molecular pacemaker consists of a negative transcriptional translational feedback loop, in which PERIOD (PER) is one of the main pacemaker proteins (Rosbash, 2009; Hardin and Panda, 2013; Tataroglu and Emery, 2015). Next, we entrained the flies under LD and detected PER abundance in the first day of DD. Our results showed that the PER abundance and oscillation in major circadian neurons of pn mutant and control flies are comparable. By contrast, there might be slight change of PER phase in PDF negative circadian neurons, especially in DN1s and LNds in pn mutants (Supplementary Figure S3).
Taken together, these results indicate that Pn is required in PDF negative circadian cells and may function in the output of pacemaker to maintain the robustness of endogenous clock.
Loss of pn Causes Mitochondrial Dysfunction
Pn depletion has been reported to significantly reduce the mtDNA levels in Drosophila S2 cells (Zhang et al., 2015). To determine whether the decrease of pn causes mtDNA reduction in vivo, we analyzed mtDNA levels in three different body segments. We found that there was a significant decline of mtDNA in the head, thorax, and abdomen of pn mutant flies relative to control flies (Figure 4A). We next used a ubiquitously expressed EYFP tagged with a mitochondrial-targeting sequence (sqh-mitoEYFP; Thomas et al., 2014) to detect the mitochondrial content in pn mutant flies. Since we detected significant decreases in mtDNA in the abdomen, we analyzed the foregut cells from the digestive tract, which are large making it easy to detect the mitochondria. In the proventriculus cells from the foregut of the 3rd larval stage, we found that the percentage of cellular area marked for mitochondria was significantly reduced in pn mutant flies compared to controls (Figures 4B,C). Moreover, we also employed the ATP sensor (AT1.03) to detect the changes of ATP levels in the midgut cells of pn mutant flies (Figures 4D,E). AT1.03 is a fluorescence resonance energy transfer (FRET)-based indicator for ATP that were composed of the ε subunit of the bacterial F0F1-ATP synthase sandwiched by the cyan- and yellow-fluorescent proteins (Imamura et al., 2009). The ratio of YFP/CFP indicates the relative abundance of ATP. As expected, we observed a slightly but significant reduction of ATP levels in the pn mutants (Figures 4D,E). Furthermore, pn mutant flies were more sensitive than control flies to treatment with paraquat (Figure 4F), a quaternary nitrogen herbicide, which causes oxidative stress and induce mitochondrial dysfunction (Hwang et al., 2012; Kohyama-Koganeya et al., 2017). These data indicate that Pn deficiency causes mitochondrial dysfunction in flies.
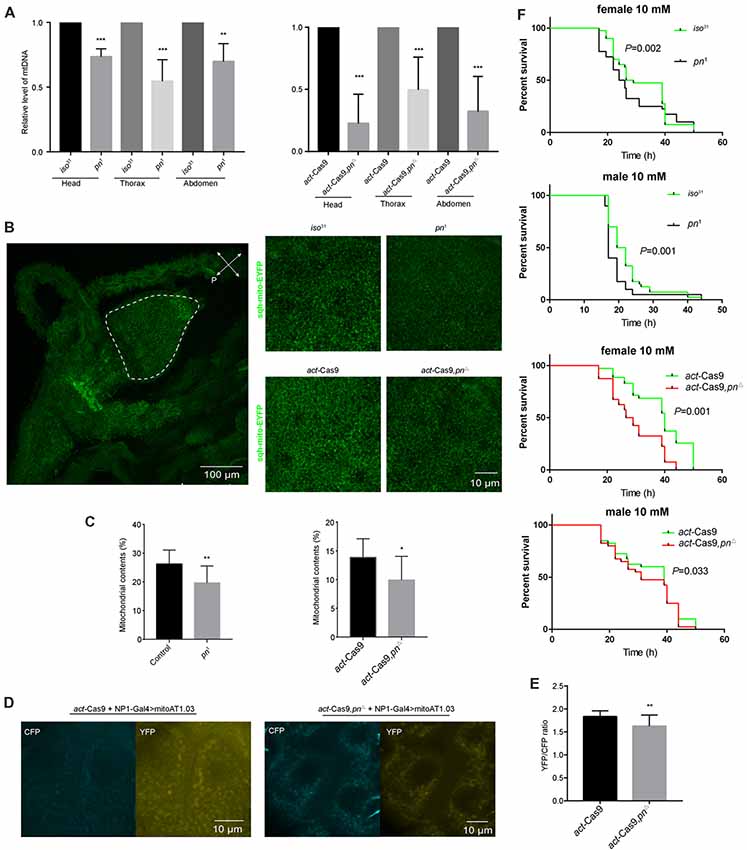
Figure 4. Loss of pn causes mitochondrial dysfunction. (A) Analysis of mitochondrial DNA (mtDNA) in head, thorax, and abdomen of pn1, pnΔ mutant and control flies. (B) Left: image of representative proventriculus cell from the foregut of the 3rd larval of a control fly that expresses mitochondrial label sqh-mitoEYFP. Right: representative images of higher magnification proventriculus cells from pn1, pnΔ mutant and control flies labeled with sqh-mitoEYFP. (C) Comparison of the mitochondrial contents in pn1, pnΔ mutant and control flies. Means ± SD are shown (n = 10). *P < 0.05, **P < 0.01, ***P < 0.001. (D) Image of representative midgut cells from the 3rd larval of control and pnΔ mutant flies labeled with mitoAT1.03 under the driver of NP1-Gal4. (E) Comparison of YFP/CFP ratio of mitoAT1.03 in control and pnΔ mutant flies. Means ± SD are shown (n = 8). **P < 0.01. (F) Pn mutant and control flies were treated with 10 mM paraquat and were tested for survival curve. Percent survival and times are indicated on the y and x axes, respectively. Two independent repeats were performed. P value are shown on the figures (n = 60–80).
Mitochondrial Dysfunction Decreases the Robustness of Circadian Rhythm
The mitochondrial transcription factor TFAM is critical for mtDNA replication (Zhang et al., 2015). Cells depleted of Pn and pn mutant flies have lower levels of TFAM protein than relevant control cells or flies, and tfam silencing in post-mitotic tissues driven by GMR-Gal4 leads to severely deformed eyes in pn mutant flies (Zhang et al., 2015). In pn mutant fly heads, there were lower levels of tfam mRNA than observed in control fly heads (Figure 5A), which is consistent with previous results (Zhang et al., 2015). In addition, when we inhibited expression of pn by expression of an shRNA targeting pn with a pan-neuronal driver, we also observed decreases in tfam (Figure 5B). This indicates that pn mutants may cause mitochondrial dysfunction in neurons by influencing TFAM levels.
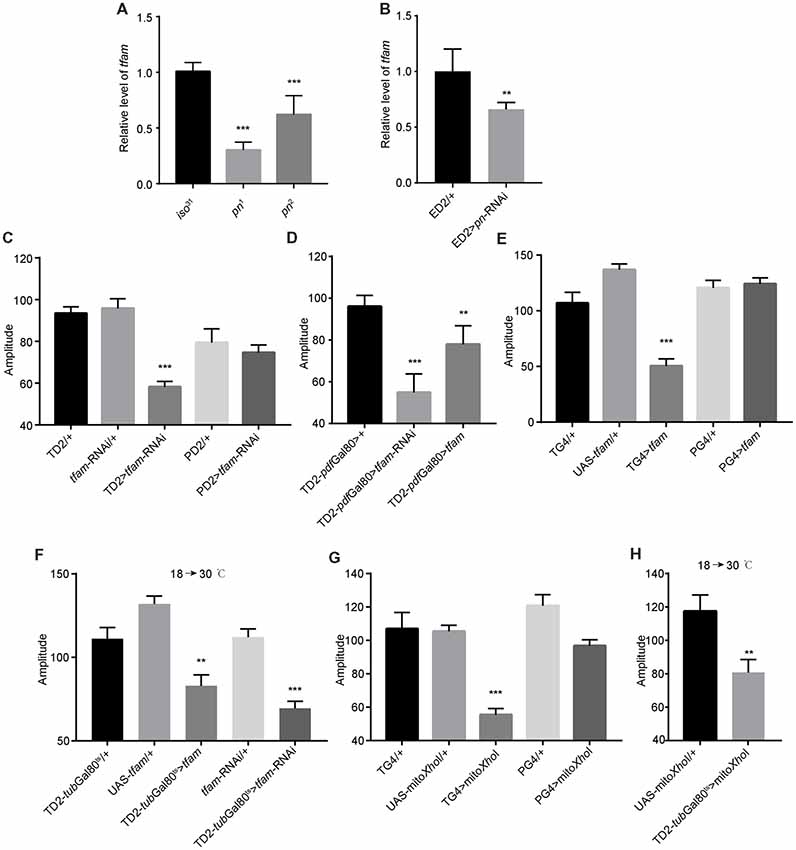
Figure 5. Mitochondrial dysfunction decreases the robustness of circadian rhythm. (A) tfam mRNA levels in the heads of pn mutants and controls. (B) tfam mRNA levels in the heads of flies in which pn is silenced with a pan-neuronal driver and control flies. ED2 indicates elav-Gal4; UAS-Dicer2. (C) Amplitudes of circadian rhythms in flies in which tfam has been silenced in the circadian neurons with TD2 or PD2 drivers. (D) Amplitudes of circadian rhythms in flies in which tfam has been silenced or overexpressed in PDF-negative circadian tissues. (E) Amplitudes of circadian rhythms in flies in which tfam has been overexpressed in circadian neurons with tim-Gal4 or pdf-Gal4 drivers. (F) Amplitudes of circadian rhythms in flies in which tfam was silenced or overexpressed in adulthood stage with tim-Gal4 driver. Flies were crossed and reared under 18°C and monitored behavior under 30°C. (G) Amplitudes of circadian rhythms in flies in which mitoXhoI has been overexpressed in circadian neurons with tim-Gal4 or pdf-Gal4 drivers. (H) Amplitudes of circadian rhythms in flies in which mitoXhoI was overexpressed in adulthood stage with tim-Gal4 driver. Flies were crossed and reared under 18°C and monitored behavior under 30°C. The amplitudes are shown as means ± SEM (n = 30–45). At least two independent repeats were performed. **P < 0.01, ***P < 0.001.
In order to test this, we downregulated tfam in all clock cells by driving tfam-targeted shRNA expression with tim-Gal4. As expected, we found that depletion of TFAM significantly decreased circadian rhythm amplitude (Figure 5C and Supplementary Figure S1E). Consistent with effects of pn downregulation, there was no effect when we restricted tfam downregulation to PDF-positive cells using the pdf-Gal4 driver (Figure 5C). Moreover, we found that tfam reduction in PDF-negative circadian cells also resulted in a decrease in amplitude of circadian rhythms (Figure 5D).
It has been shown that excessive TFAM also causes mitochondrial dysfunction by inhibiting mitochondrial gene expression (Falkenberg et al., 2002; Maniura-Weber et al., 2004; Pohjoismäki et al., 2006; Ylikallio et al., 2010). Interestingly, tfam overexpression in TIM-positive and PDF-negative neurons also reduced the robustness of clock (Figures 5D,E and Supplementary Figure S1E). In addition, we confirmed that tfam dysfunction in adults but not in earlier developmental stages altered locomotor activity (Figure 5F). Thus, TFAM depletion and overexpression both cause mitochondrial dysfunction and reduce the clock robustness.
To further confirm whether other inducers of mitochondrial dysfunction cause the same phenotype as altered TFAM abundance, we manipulated mtDNA levels in vivo by expression of a mitochondrially targeted restriction enzyme, mitoXhoI, that cuts the mtDNA once in the cox I locus (Cagin et al., 2015). mitoXhoI expression has been demonstrated to cause mitochondrial dysfunction in mice and Drosophila (Xu et al., 2008; Ylikallio et al., 2010). We found that mitoXhoI expression driven by tim-Gal4 significantly decreased the level of mtDNA in fly heads (Supplementary Figure S1F). Interestingly, mitoXhoI expression in TIM-positive and PDF-negative cells caused reductions in amplitudes of circadian rhythms in the mutants compared to appropriate controls (Figure 5G), a result consistent with the TFAM manipulations. Moreover, we found that mitochondrial dysfunction in the adult but not at earlier developmental stages reduced rhythm robustness (Figure 5H). Thus, mitochondrial dysfunction decreases the robustness of the endogenous circadian rhythm in Drosophila.
Discussion
Circadian systems have three main parts: the core clock, input pathways, and output pathways (Allada and Chung, 2010; Dubowy and Sehgal, 2017). The core clock receives signals from circadian input pathways and then controls the behavior and physiology outputs (Allada and Chung, 2010; Dubowy and Sehgal, 2017). Here, we found that mitochondrial dysfunction in PDF-negative circadian cells affects the amplitude of circadian rhythm. Since PDF-positive cells are the pacemaker neurons, this finding indicates that mitochondrial dysfunction may modulate the output of circadian clock rather than controlling the core circadian system to influence circadian amplitude. Previous work demonstrated that aging is associated with reduced expression of core clock genes in peripheral head clocks in mammals, although similar reductions are not observed in central clock neurons (Duffy et al., 2015). There are two pn orthologs PRUNE1 and PRUNE2 in mammals, whether they are involved in the regulation of circadian rhythms need to be studied. It has also been shown that the molecular circadian clock is dampened in peripheral tissues of old flies (Luo et al., 2012; Rakshit et al., 2012; Long and Giebultowicz, 2018). With age, circadian rhythms may lose proper synchronization and become less pronounced. Although we tried different tissue-specific drivers to knockdown pn, which cells are involved in the effect of pn on the circadian amplitude change need to be determined in the future.
Rhythmic mitochondrial function is known to feed back into the circadian clock mechanism. Changes in mitochondrial function affect the cell at multiple levels, but how the circadian system responds to homeostatic changes is poorly understood. The enzyme SIRT1, an NAD+ dependent deacetylase appears to be one link between the clock and aging (Orozco-Solis and Sassone-Corsi, 2014). SIRT1 modulates mitochondrial function by regulating NAD+ levels. Interestingly, SIRT1 modulates nuclear-mitochondrial communication and may maintain mitochondrial homeostasis during aging (Gomes et al., 2013; Orozco-Solis and Sassone-Corsi, 2014). In future experiments, it will be interesting to test whether mitochondrial dysfunction is communicated to the circadian clock through SIRT1.
We have confirmed that mitochondrial dysfunction in neurons adversely affects Drosophila circadian rhythms. We found that depletion of TFAM specifically in TIM-positive and PDF-negative cells resulted in decreases in circadian amplitude. Moreover, loss of pn, which resulted in tfam decreases, also caused the same circadian phenotype as tfam silencing. Interestingly, both expression of mitoXhoI, which causes degradation of mtDNA, and tfam overexpression caused mitochondrial dysfunction and decrease circadian amplitude. This indicates that TFAM mis-expression or direct mtDNA depletion can be used to study the relationship between mitochondria and circadian rhythms.
Author Contributions
YZ and WC conceived the project, designed the experiments and wrote the manuscript. WC, YX, LS and DW performed the experiments. WC performed the analysis.
Funding
WC’s work is supported by the National Natural Science Foundation of China (grant number 31601894), the Fujian Natural Science Foundation (grant number 2017J0106) and the Chinese Government Scholarship (201706655001). YZ’s lab is supported by the National Institutes of Health COBRE Grant P20 GM103650.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Bloomington Drosophila Stock Center for providing fly stocks. We also thank Dr. Xu, Dr. Bateman and Dr. Yang for providing flies. The anti-PER antibody is a generous gift from Dr. Michael Rosbash. The PDF antibodies, developed by Justin Blau was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA, United States.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2019.00076/full#supplementary-material
FIGURE S1 | The results of qPCR analysis. (A,B) pn mRNA levels in the heads of pn mutants and controls. (C) pn mRNA levels expressed from hs-pn heads at indicated temperatures. (D) pn mRNA levels in the heads of flies in which pn is silenced with a pan-neuronal driver and control flies. (E) tfam mRNA levels in the heads of flies in which tfam was silenced or overexpression with the tim-Gal4 driver. (F) mtDNA levels in the heads of flies in which mitoXhoI was overexpressed. Three independent repeats were performed. *P < 0.05, ***P < 0.001. n.s. no significance.
FIGURE S2 | Pn expression in eyes and glia cells does not affect the robustness of circadian rhythm. (A) Averaged activity profiles and actograms of control flies and GMR>pn-RNAi flies. (B) Amplitudes of circadian rhythms in control flies and GMR>pn-RNAi flies. (C) Averaged activity profiles and actograms of control flies and repo >pn-RNAi flies. (D) Amplitudes of circadian rhythms in control flies and repo >pn-RNAi flies. n.s. no significance. White bars represent day, black bars represent night, Zeitgeber time (ZT) is indicated on the x axes. The dots above the bars indicate standard errors of the means (SEM). The amplitudes are shown as means ± SEM (n = 40–45). Three independent repeats were performed. n.s. no significance.
FIGURE S3 | Oscillation of PERIOD in the circadian neurons of control and pnΔ mutant flies. Adult brains were dissected at CT 1, 5, 9, 13, 17, 21 at DD1 and circadian neurons were immunostained with anti-PERIOD. Quantification of average intensity in each clock cell group (n = 10) was shown as means ± SEM. Signal was normalized to the value at CT1 which was set as 100%. CT is indicated on the x axes. Two independent repeats were performed. *P < 0.05, **P < 0.01, ***P < 0.001.
References
Allada, R., and Chung, B. Y. (2010). Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 72, 605–624. doi: 10.1146/annurev-physiol-021909-135815
Anderson, S., Bankier, A. T., Barrell, B. G., de Bruijn, M. H., Coulson, A. R., Drouin, J., et al. (1981). Sequence and organization of the human mitochondrial genome. Nature 290, 457–465. doi: 10.1038/290457a0
Bose, A., and Beal, M. F. (2016). Mitochondrial dysfunction in Parkinson’s disease. J. Neurochem. 139, 216–231. doi: 10.1111/jnc.13731
Brown, S. A., Schmitt, K., and Eckert, A. (2011). Aging and circadian disruption: causes and effects. Aging 3, 813–817. doi: 10.18632/aging.100366
Butow, R. A., and Avadhani, N. G. (2004). Mitochondrial signaling: the retrograde response. Mol. Cell 14, 1–15. doi: 10.1016/S1097-2765(04)00179-0
Cagin, U., Duncan, O. F., Gatt, A. P., Dionne, M. S., Sweeney, S. T., and Bateman, J. M. (2015). Mitochondrial retrograde signaling regulates neuronal function. Proc. Natl. Acad. Sci. U S A 112, E6000–E6009. doi: 10.1073/pnas.1522518112
Chaix, A., Zarrinpar, A., and Panda, S. (2016). The circadian coordination of cell biology. J. Cell Biol. 215, 15–25. doi: 10.1083/jcb.201603076
Chandel, N. S. (2014). Mitochondria as signaling organelles. BMC Biol. 12:34. doi: 10.1186/1741-7007-12-34
Chedid, A., and Nair, V. (1972). Diurnal rhythm in endoplasmic reticulum of rat liver: electron microscopic study. Science 175, 176–179. doi: 10.1126/science.175.4018.176
Claridge-Chang, A., Wijnen, H., Naef, F., Boothroyd, C., Rajewsky, N., and Young, M. W. (2001). Circadian regulation of gene expression systems in the Drosophila head. Neuron 32, 657–671. doi: 10.1016/s0896-6273(01)00515-3
de Goede, P., Wefers, J., Brombacher, E. C., Schrauwen, P., and Kalsbeek, A. (2018). Circadian rhythms in mitochondrial respiration. J. Mol. Endocrinol. 60, R115–R130. doi: 10.1530/jme-17-0196
Dubowy, C., and Sehgal, A. (2017). Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205, 1373–1397. doi: 10.1534/genetics.115.185157
Duffy, J. F., Zitting, K. M., and Chinoy, E. D. (2015). Aging and circadian rhythms. Sleep Med. Clin. 10, 423–434. doi: 10.1016/j.jsmc.2015.08.002
Evans, B. A., and Howells, A. J. (1978). Control of drosopterin synthesis in Drosophila melanogaster: mutants showing an altered pattern of GTP cyclohydrolase activity during development. Biochem. Genet. 16, 13–26. doi: 10.1007/bf00484381
Ewer, J., Rosbash, M., and Hall, J. C. (1988). An inducible promoter fused to the period gene in Drosophila conditionally rescues adult per-mutant arrhythmicity. Nature 333, 82–84. doi: 10.1038/333082a0
Falkenberg, M., Gaspari, M., Rantanen, A., Trifunovic, A., Larsson, N. G., and Gustafsson, C. M. (2002). Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31, 289–294. doi: 10.1038/ng909
Frolov, M. V., Zverlov, V. V., and Alatortsev, V. E. (1994). The mRNA product of the Drosophila gene prune is spliced and encodes a protein containing a putative transmembrane domain. Mol. Gen. Genet. 242, 478–483. doi: 10.1007/bf00281800
Gery, S., Komatsu, N., Baldjyan, L., Yu, A., Koo, D., and Koeffler, H. P. (2006). The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell 22, 375–382. doi: 10.1016/j.molcel.2006.03.038
Gomes, A. P., Price, N. L., Ling, A. J., Moslehi, J. J., Montgomery, M. K., Rajman, L., et al. (2013). Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 155, 1624–1638. doi: 10.1016/j.cell.2013.11.037
Gratz, S. J., Ukken, F. P., Rubinstein, C. D., Thiede, G., Donohue, L. K., Cummings, A. M., et al. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. doi: 10.1534/genetics.113.160713
Guo, F., Yu, J., Jung, H. J., Abruzzi, K. C., Luo, W., Griffith, L. C., et al. (2016). Circadian neuron feedback controls the Drosophila sleep—activity profile. Nature 536, 292–297. doi: 10.1038/nature19097
Hardin, P. E., and Panda, S. (2013). Circadian timekeeping and output mechanisms in animals. Curr. Opin. Neurobiol. 23, 724–731. doi: 10.1016/j.conb.2013.02.018
Helfrich-Förster, C. (1998). Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J. Comp. Physiol. A 182, 435–543. doi: 10.1007/s003590050192
Hwang, A. B., Jeong, D. E., and Lee, S. J. (2012). Mitochondria and organismal longevity. Curr. Genomics 13, 519–532. doi: 10.2174/138920212803251427
Imamura, H., Nhat, K. P., Togawa, H., Saito, K., Iino, R., Kato-Yamada, Y., et al. (2009). Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. U S A 106, 15651–15656. doi: 10.1073/pnas.0904764106
Kaneko, M., and Hall, J. C. (2000). Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 422, 66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2
Kim, H., Kim, K., and Yim, J. (2013). Biosynthesis of drosopterins, the red eye pigments of Drosophila melanogaster. IUBMB Life 65, 334–340. doi: 10.1002/iub.1145
Klarsfeld, A., Leloup, J. C., and Rouyer, F. (2003). Circadian rhythms of locomotor activity in Drosophila. Behav. Processes 64, 161–175. doi: 10.1016/s0376-6357(03)00133-5
Kohyama-Koganeya, A., Kurosawa, M., and Hirabayashi, Y. (2017). Loss of BOSS causes shortened lifespan with mitochondrial dysfunction in Drosophila. PLoS One 12:e0169073. doi: 10.1371/journal.pone.0169073
Koike, N., Yoo, S. H., Huang, H. C., Kumar, V., Lee, C., Kim, T. K., et al. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354. doi: 10.1126/science.1226339
Levine, J. D., Funes, P., Dowse, H. B., and Hall, J. C. (2002). Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3:1. doi: 10.1186/1471-2202-3-1
Long, D. M., and Giebultowicz, J. M. (2018). Age-related changes in the expression of the circadian clock protein PERIOD in Drosophila glial cells. Front. Physiol. 8:1131. doi: 10.3389/fphys.2017.01131
Luo, W., Chen, W. F., Yue, Z., Chen, D., Sowcik, M., Sehgal, A., et al. (2012). Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell 11, 428–438. doi: 10.1111/j.1474-9726.2012.00800.x
Manella, G., and Asher, G. (2016). The circadian nature of mitochondrial biology. Front. Endocrinol. 7:162. doi: 10.3389/fendo.2016.00162
Maniura-Weber, K., Goffart, S., Garstka, H. L., Montoya, J., and Wiesner, R. J. (2004). Transient overexpression of mitochondrial transcription factor A (TFAM) is sufficient to stimulate mitochondrial DNA transcription, but not sufficient to increase mtDNA copy number in cultured cells. Nucleic Acids Res. 32, 6015–6027. doi: 10.1093/nar/gkh921
Murad, A., Emery-Le, M., and Emery, P. (2007). A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron 53, 689–701. doi: 10.1016/j.neuron.2007.01.034
Ng, S., De Clercq, I., Van Aken, O., Law, S. R., Ivanova, A., Willems, P., et al. (2014). Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development and stress. Mol. Plant 7, 1075–1093. doi: 10.1093/mp/ssu037
Nitabach, M. N., and Taghert, P. H. (2008). Organization of the Drosophila circadian control circuit. Curr. Biol. 18, R84–R93. doi: 10.1016/j.cub.2007.11.061
Orozco-Solis, R., and Sassone-Corsi, P. (2014). Circadian clock: linking epigenetics to aging. Curr. Opin. Genet. Dev. 26, 66–72. doi: 10.1016/j.gde.2014.06.003
Park, J. H., Helfrich-Forster, C., Lee, G., Liu, L., Rosbash, M., and Hall, J. C. (2000). Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci. U S A 97, 3608–3613. doi: 10.1073/pnas.070036197
Pohjoismäki, J. L., Wanrooij, S., Hyvärinen, A. K., Goffart, S., Holt, I. J., Spelbrink, J. N., et al. (2006). Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 34, 5815–5828. doi: 10.1093/nar/gkl703
Quintanilla, R. A., Jin, Y. N., Fuenzalida, K., Bronfman, M., and Johnson, G. V. (2008). Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-γ (PPARγ) in the pathogenesis of Huntington disease. J. Biol. Chem. 283, 25628–25637. doi: 10.1074/jbc.m804291200
Rakshit, K., Krishnan, N., Guzik, E. M., Pyza, E., and Giebultowicz, J. M. (2012). Effects of aging on the molecular circadian oscillations in Drosophila. Chronobiol. Int. 29, 5–14. doi: 10.3109/07420528.2011.635237
Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C., and Taghert, P. H. (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. doi: 10.1016/s0092-8674(00)81676-1
Rosato, E., and Kyriacou, C. P. (2006). Analysis of locomotor activity rhythms in Drosophila. Nat. Protoc. 1, 559–568. doi: 10.1038/nprot.2006.79
Rosbash, M. (2009). The implications of multiple circadian clock origins. PLoS Biol. 7:e62. doi: 10.1371/journal.pbio.1000062
Ryder, E., Blows, F., Ashburner, M., Bautista-Llacer, R., Coulson, D., Drummond, J., et al. (2004). The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167, 797–813. doi: 10.1534/genetics.104.026658
Shi, P., Gal, J., Kwinter, D. M., Liu, X., and Zhu, H. (2010). Mitochondrial dysfunction in amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1802, 45–51. doi: 10.1016/j.bbadis.2009.08.012
Simmons, G. M., Kwok, W., Matulonis, P., and Venkatesh, T. (1994). Polymorphism and divergence at the prune locus in Drosophila melanogaster and D. simulans. Mol. Biol. Evol. 11, 666–671. doi: 10.1093/oxfordjournals.molbev.a040145
Stoleru, D., Nawathean, P., Fernández, M. P., Menet, J. S., Ceriani, M. F., and Rosbash, M. (2007). The Drosophila circadian network is a seasonal timer. Cell 129, 207–219. doi: 10.1016/j.cell.2007.02.038
Sun, N., Youle, R. J., and Finkel, T. (2016). The mitochondrial basis of aging. Mol. Cell 61, 654–666. doi: 10.1016/j.molcel.2016.01.028
Tataroglu, O., and Emery, P. (2015). The molecular ticks of the Drosophila circadian clock. Curr. Opin. Insect Sci. 7, 51–57. doi: 10.1016/j.cois.2015.01.002
Thomas, R. E., Andrews, L. A., Burman, J. L., Lin, W. Y., and Pallanck, L. J. (2014). PINK1-Parkin pathway activity is regulated by degradation of PINK1 in the mitochondrial matrix. PLoS Genet. 10:e1004279. doi: 10.1371/journal.pgen.1004279
Xu, H., DeLuca, S. Z., and O’Farrell, P. H. (2008). Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science 321, 575–577. doi: 10.1126/science.1160226
Yadlapalli, S., Jiang, C., Bahle, A., Reddy, P., Meyhofer, E., and Shafer, O. T. (2018). Circadian clock neurons constantly monitor environmental temperature to set sleep timing. Nature 555, 98–102. doi: 10.1038/nature25740
Yao, Z., and Shafer, O. T. (2014). The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343, 1516–1520. doi: 10.1126/science.1251285
Ylikallio, E., Tyynismaa, H., Tsutsui, H., Ide, T., and Suomalainen, A. (2010). High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 19, 2695–2705. doi: 10.1093/hmg/ddq163
Zhang, L., Chung, B. Y., Lear, B. C., Kilman, V. L., Liu, Y., Mahesh, G., et al. (2010). DN1p circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr. Biol. 20, 591–599. doi: 10.1016/j.cub.2010.02.056
Zhang, F., Qi, Y., Zhou, K., Zhang, G., Linask, K., and Xu, H. (2015). The cAMP phosphodiesterase Prune localizes to the mitochondrial matrix and promotes mtDNA replication by stabilizing TFAM. EMBO Rep. 16, 520–527. doi: 10.15252/embr.201439636
Keywords: prune, TFAM, mitochondrial dysfunction, circadian rhythms, Drosophila
Citation: Chen W, Xue Y, Scarfe L, Wang D and Zhang Y (2019) Loss of Prune in Circadian Cells Decreases the Amplitude of the Circadian Locomotor Rhythm in Drosophila. Front. Cell. Neurosci. 13:76. doi: 10.3389/fncel.2019.00076
Received: 08 October 2018; Accepted: 14 February 2019;
Published: 01 March 2019.
Edited by:
Dieter Wicher, Max Planck Institute for Chemical Ecology, GermanyReviewed by:
Masayuki Ikeda, University of Toyama, JapanLezi E, Duke University, United States
Charlotte Helfrich-Förster, Universität Würzburg, Germany
Copyright © 2019 Chen, Xue, Scarfe, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Zhang, eW9uZ3poYW5nQHVuci5lZHU=
 Wenfeng Chen
Wenfeng Chen Yongbo Xue
Yongbo Xue Lisa Scarfe
Lisa Scarfe Danfeng Wang
Danfeng Wang Yong Zhang
Yong Zhang