
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Neurosci., 20 February 2018
Sec. Cellular Neurophysiology
Volume 12 - 2018 | https://doi.org/10.3389/fncel.2018.00041
This article is part of the Research TopicHair Cells: From Molecules to FunctionView all 17 articles
 Clive P. Morgan1†
Clive P. Morgan1† Hongyu Zhao1†
Hongyu Zhao1† Meredith LeMasurier1†
Meredith LeMasurier1† Wei Xiong2
Wei Xiong2 Bifeng Pan3
Bifeng Pan3 Piotr Kazmierczak2
Piotr Kazmierczak2 Matthew R. Avenarius1
Matthew R. Avenarius1 Michael Bateschell1
Michael Bateschell1 Ruby Larisch1
Ruby Larisch1 Anthony J. Ricci3
Anthony J. Ricci3 Ulrich Müller2‡
Ulrich Müller2‡ Peter G. Barr-Gillespie1*
Peter G. Barr-Gillespie1*Hair cells of the inner ear transduce mechanical stimuli like sound or head movements into electrical signals, which are propagated to the central nervous system. The hair-cell mechanotransduction channel remains unidentified. We tested whether three transient receptor channel (TRP) family members, TRPV6, TRPM6 and TRPM7, were necessary for transduction. TRPV6 interacted with USH1C (harmonin), a scaffolding protein that participates in transduction. Using a cysteine-substitution knock-in mouse line and methanethiosulfonate (MTS) reagents selective for this allele, we found that inhibition of TRPV6 had no effect on transduction in mouse cochlear hair cells. TRPM6 and TRPM7 each interacted with the tip-link component PCDH15 in cultured eukaryotic cells, which suggested they might be part of the transduction complex. Cochlear hair cell transduction was not affected by manipulations of Mg2+, however, which normally perturbs TRPM6 and TRPM7. To definitively examine the role of these two channels in transduction, we showed that deletion of either or both of their genes selectively in hair cells had no effect on auditory function. We suggest that TRPV6, TRPM6 and TRPM7 are unlikely to be the pore-forming subunit of the hair-cell transduction channel.
A central mystery in auditory neuroscience is the identity of the molecules making up the pore of the hair cell’s transduction channel, a cation-selective channel that responds to mechanical stimuli and produces a receptor potential in the cell. Transduction channels are gated by tension in tip links, thin extracellular filaments made from the cadherins CDH23 and PCDH15 (Siemens et al., 2004; Sollner et al., 2004; Alagramam et al., 2011). PCDH15 is located at the base of the tip link (Kazmierczak et al., 2007), where the transduction channel is located (Beurg et al., 2009). Theoretical modeling suggests that the tip link binds to the transduction channel complex (Powers et al., 2012), so PCDH15 might interact directly with the transduction channel. The ion channel HCN1 has been reported to bind to PCDH15 (Ramakrishnan et al., 2009, 2012), but Hcn1 knockouts have normal hearing (Horwitz et al., 2010). By contrast, strong evidence suggests that the channel complex contains the transmembrane proteins TMC1 and TMC2 (Kawashima et al., 2011; Pan et al., 2013), TMIE (Zhao et al., 2014) and LHFPL5 (Xiong et al., 2012); moreover, PCDH15 interacts with both TMC1 and TMC2 (Maeda et al., 2014; Beurg et al., 2015). Nevertheless, while the TMCs do influence transduction-channel conductance and permeability properties (Pan et al., 2013; Beurg et al., 2014), evidence that any of these proteins contributes directly to the ion-permeation pathway is lacking.
Because they carry out a wide variety of cellular functions, including sensory transduction, the 33-member transient receptor potential (TRP) family might well contain the transduction-channel pore molecule (Venkatachalam and Montell, 2007; Zanini and Göpfert, 2014). Many TRP channels are present in the inner ear; a comprehensive quantitative RT-PCR investigation of TRP channel expression during cochlear development detected transcripts for 30 different TRP genes (Asai et al., 2010). Several TRP channels have been advanced specifically as candidates for the transduction channel, but all evidence so far has ruled out these candidates (Fettiplace and Kim, 2014).
Our preliminary evidence raised the possibility that TRPV6, TRPM6, or TRPM7 might be part of the transduction channel. Interestingly, TRPV channels play key roles in transduction in fly mechanoreceptors (Gong et al., 2004; Kernan, 2007; Lehnert et al., 2013; Zhang et al., 2013). As it is highly selective for Ca2+ (Owsianik et al., 2006), TRPV6 is less likely as a candidate for the transduction channel than other channels; nevertheless, the transduction channel’s permeability and conductance can change substantially depending on other components expressed (Xiong et al., 2012; Beurg et al., 2015). TRPM7, a member of the melanostatin subfamily, has been implicated in mechanosensation (Bessac and Fleig, 2007; Numata et al., 2007a,b; Wei et al., 2009; Xiao et al., 2015). TRPM7 and its closely related paralog TRPM6 uniquely possess a C-terminal kinase domain; these channels are thought to regulate Mg2+ homeostasis as well as cell death, proliferation, differentiation, and migration (Chubanov and Gudermann, 2014; Fleig and Chubanov, 2014). Trpv6, Trpm6 and Trpm7 transcripts were all detected in cochlea (Asai et al., 2010); Trpm6 levels increased during development, while Trpv6 and Trpm7 decreased. The Trpv6 paralog Trpv5 was seen in only a fraction of the samples and may not be significantly expressed in the cochlea. Both Trpm6 and Trpm7 were detected in a cochlear cDNA library using conventional PCR, but Trpv5 and Trpv6 were not (Cuajungco et al., 2007).
Because a cysteine-substitution mutation in TRPV6 (M527C) renders this channel sensitive to methanethiosulfonate (MTS) reagents applied from the extracellular solution (Voets et al., 2004a), we generated M527C-Trpv6 knock-in mice. Mechanotransduction was insensitive to MTS reagents, however, in both heterozygote and homozygote hair cells from this line. Similarly, mechanotransduction in hair cells was not affected by conditions that should alter TRPM6 or TRPM7 function, and conditional deletion of the Trpm6 and Trpm7 genes in hair cells did not affect auditory function. We suggest that TRPV6, TRPM6, and TRPM7 should be added to the long list of ion channels that have been shown to not be part of the transduction channel (Horwitz et al., 2010).
This study was carried out in compliance with the Animal Welfare Act regulations and the Office of Laboratory Animal Welfare—Public Health Service Policy on Humane Care and Use of Laboratory Animals. Protocols were approved by the Institutional Animal Care and Use Committees (IACUC) of Oregon Health & Science University, Stanford University, and Scripps Research Institute.
For M527-Trpv6, we generated a targeted construct that contained (5′ to 3′) a short homology arm (1.8 kb) with an added loxP-flanked neomycin resistance cassette, exon 13 including the codon encoding M527 to a cysteine and an introduced ScaI site for screening, a long homology arm (5.3 kb), and a diphtheria toxin cassette. The targeting construct was linearized with XhoI for electroporation into ES cells. Screening was carried out on the 5′ and 3′ arms using PCR, with TOPO-cloning of PCR products to verify that the correct genomic DNA region was targeted. Mice were generated from the correctly-targeted ES cells by the University of Cincinnati Gene-Targeted Mouse Service. After blastocyst injections, founders were identified by coat color chimerism and were bred to C57BL/6 mice for >6 generations. Mice were genotyped using PCR, with AAATGGGAACCAGATTCATCTCA as the forward primer and ACTATACAAAAGGGTAACCTACCCACA as the reverse primer. The amplified samples were digested with BamHI (the knock-in removes a BamHI site) and analyzed by electrophoresis.
We obtained Trpm6tm1a(KOMP)Wtsi ES cells from the UC Davis KOMP repository (clones EPD0741_2_G10 and EPD0741_2_G11). After blastocyst injections, founders were identified by coat color chimerism and targeting was verified by PCR analysis. Founders were bred to C57BL/6, and the FRT-neo cassette was removed by crossing with the Flp deleter line B6;SJL-Tg(ACTFLPe)9205Dym/J (Jackson Laboratories). PCR was used to verify loss of the cassette. These mice were referred to as Trpm6fl. To produce Trpm6CKO mice, we used the Atoh1-Cre mouse line (Matei et al., 2005), which recombines floxed genes in hair cells with >99% efficiency (Avenarius et al., 2017). Floxed mice were genotyped using PCR, with GCTCCTCAGGGTTCCTCCAGTCTGT as the forward primer and GCAAGGACAAGAGGG-CGTCAGAGC as the reverse primer. The amplified samples were analyzed by electrophoresis; a wild-type allele produces a band of 670 bp, while the floxed-allele band is 833 bp.
We generated a mouse allele with the putative ion-conductance pore of Trpm7 flanked by loxP sites. The 5′ arm (4948 bp) was PCR-amplified using a forward primer that introduced an ApaI site (GCGGGGCCCTGGGTGATTGACATTTCATTCCAAGT) and a reverse primer that introduced a SalI site (GCGGTCGACTGTCAACTAGCAATGGAAATGCAGACTT). This fragment was introduced into the pBS-FRT-Neo-FRT plasmid. The 3′ arm (2987 bp) was PCR-amplified using the forward primer GCGCTCGAGGTGTATATAAGAATTGTCTCAGGATAGT and reverse primer GCGCCGCGGCC-TCTTATCCTGTTTCTCTACATGTGT. A separate middle piece was prepared as a SalI/NotI fragment flanked by loxP sites; it was PCR-amplified using the forward primer GCGGTCGACGGTTTT-GCCTTATATTTGCAAGGCATA and reverse primer GCGGCGGCCGCCCATTACCATCATTCCTT-GAAGTGGCTTT. The middle piece was cloned into the 5′ arm vector at SalI and NotI sites; the 3′ arm was then cloned into this latter piece (5′ and middle). Founders were bred to C57BL/6, and the FRT-neo cassette was removed as above (producing Trpm7fl mice). Trpm7CKO mice were generated with Atoh1-Cre as above. Floxed mice were genotyped using PCR, with CCATACTGGATGATTTTTGGTG-AAGTTTATGCA as the forward primer and CACAAACAAGGAAGGGAAGAGTTTTAATATCCA as the reverse primer. The amplified samples were analyzed by electrophoresis; a wild-type allele produces a band of 514 bp, while the floxed-allele band is 633 bp.
For co-immunoprecipitation assays, we used HEK293T cells grown in six-well tissue culture plates. Cells were transfected with Effectene transfection reagent (Qiagen, #301425) by preparing the DNA complex according to the manufacturer’s suggested protocol in PCR tubes, then adding the DNA complex to the cells. Cells were harvested at 48 h, centrifuged briefly (16,000 g for 5 s), and the supernatant removed. To the pellet, 300 μl of cold lysis buffer containing PBS, 1% Triton X-100, 0.5% NP-40, and protease inhibitor cocktail (Sigma, #P8340) was added, and the cells were then sonicated on ice using a tip sonicator (Ultracell Sonicator, Sonics) set at 25% power. The lysate was then centrifuged at 16,000 g for 20 min at 4°C, and the supernatant was separated from the pellet. A total of 10 μl of HA agarose (Sigma, #A2095, 20 μl slurry) was added and the mixture was rotated overnight at 4°C. The immunoprecipitate mixture was centrifuged at 16,000 g for 1 s, then washed 2× with 300 μl lysis buffer and 1X with PBS. After the washes, 32 μl of 2× SDS sample buffer (Invitrogen) was added to the agarose beads, and the mixture was incubated for 10 min at 70°C. The agarose beads were removed using a spin filter (Costar #8163), and 5 μl of 10× reducing agent (Invitrogen, #NP0007) was added to the protein solution. The mixture was again incubated at 70°C for 10 min. The protein samples were separated on a 3%–8% Tris-acetate gel (Invitrogen, #EA03755BOX) for 30–40 min. Proteins in the gel were then transferred to PVDF membrane (Millipore, #IPVH00010) using a semi-dry setup (Bio-Rad, Transblot SD). Towbin buffer (25 mM Tris, 192 mM glycine, pH 8.3) with 10% MeOH and without SDS were used as transfer buffer. After transferring for 40–50 min, the blot was washed 1× in PBS and blocked in blocking buffer (PBS containing 10% FBS, 0.1% Tween) for 30 min, during which primary antibody were diluted in blocking buffer (1:1000) and incubated at room temperature for 10–20 min. The blot was incubated with primary antibody for 2 h at room temperature. After washing the blot, HRP-coupled secondary antibody, diluted 1:100 in blocking buffer, was incubated with the blot for 30 min. After washing, SuperSignal West Pico Chemiluminescent Substrate (Pierce, #34077) was used for detection.
Anti-HA agarose or anti-V5 agarose (Sigma, #A7345) were used to pull down HA-tagged TRP channels. For the PCDH15 pull down, we used 10 μg of PB811, a rabbit antibody against PCDH15 (Kazmierczak et al., 2007) or 10 μg of anti-HRP antibody (Jackson Immunoresearch), precipitating with 10 μl of Ultralink immobilized protein A (Pierce, #53139). For protein immunoblotting, PCDH15 was detected with PB811 and TRP channels were detected with monoclonal anti-HA antibody (Applied Biological Materials, #G036).
For these experiments, electrophysiological recordings from outer hair cells (OHCs) were carried out as previously described (Peng et al., 2016). For tissue preparation, organ of Corti explants from the mid-apex region were dissected from mice of either sex at ages P6–P9. The tissue was placed into the recording chamber and held in place with single strands of dental floss. Apical perfusion with external solution was used to protect the hair bundles from internal solution. The perfusion was carried out at a rate of ~0.1 ml/min using pipettes with tip size of ~50 μm. The external solution contained (in mM) 140 NaCl, 2 KCl, 2 CaCl2, 2 MgCl2, 10 (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; HEPES), 6 glucose, 2 sodium pyruvate, and 2 creatine phosphate, and was balanced to a pH of 7.4 and osmolality of 300–310 osmol/l.
Borosilicate patch pipettes (2.5–3.5 MΩ) were used to establish whole-cell recordings. The internal solution contained (in mM) 125 CsCl, 10 HEPES, 1 1,2-bis(o-aminophenoxy)ethane-N,N,N′, N′-tetraacetic acid (BAPTA), 5 ATP, 5 creatine phosphate, and 3.5 MgCl2; the pH was adjusted to 7.0 and osmolality 290–295 osmol/l. An Axopatch 200b amplifier was used to control electrode voltage and monitor current; the amplifier was controlled via JClamp software coupled to an IOtech interface. Cells were clamped at −80 mV. Recordings were included for data analysis if MET current amplitudes were stable prior to drug application and if the leak currents were less than 100 pA. For 70 cells recorded, the series resistance was 14.6 ± 7 MΩ (and was uncompensated); capacitance was 5.8 ± 0.8 pF. Data were included for analysis only if the series resistance changed less than 10% during recording. Only one cell was recorded per mouse and recordings were done blindly to animal genotype.
Hair bundles were stimulated in either of two ways (Peng et al., 2016). A glass probe affixed to a piezoelectric stack actuator (Thorlabs AE0505D08F or APC International PSt 150/7x7/7) controlled by JClamp software was placed in the central region of the hair bundle. The tip was shaped to that of the angled outer hair cell bundle. Driving voltage was filtered at 3 kHz to limit piezo resonance. The displacement was calibrated using an eyepiece reticule. Hair bundles were also stimulated as described (Peng et al., 2016) with fluid motion driven by a Picospritzer III.
Inhibitors, including [2-(trimethylammonium)ethyl] MTS bromide (MTSET; Toronto Research Chemicals #T795900) or sodium (2-sulfonatoethyl) MTS (MTSES; Toronto Research Chemicals #S672000), were aliquoted into single-use samples and kept frozen until a recording was obtained. The inhibitor was put into solution and applied to cells within 3 min of dissolving to avoid any breakdown of the compound; the inhibitor was hand pipetted into the recording chamber with enough exchanges so that the chamber volume was replaced 3×. The inhibitor was typically applied 10–15 min after breakthrough and the post-inhibitor measurements reported here were within 10 min of drug application. Longer application led to slow declines in current for both WT and mutant hair cells.
For these experiments, electrophysiological recordings from OHCs were carried out as previously described (Xiong et al., 2012). For tissue preparation, organ of Corti explants from the mid-apex region were dissected from mice of either sex at ages P6–P7. The external solution contained (in mM): 144 NaCl, 0.7 NaH2PO4, 5.8 KCl, 1.3 CaCl2, 0.9 MgCl2, 5.6 glucose and 10 H-HEPES, pH 7.4.
Borosilicate patch pipettes (3–5 MΩ) were used to establish whole-cell recordings. The internal solution contained (in mM; in mM) 140 KCl, 0 or 3 MgCl2, 0.1 EGTA, 2 Na-ATP, 0.3 Na-GTP and 10 H-HEPES, pH 7.2. Transducer currents were sampled at 100 KHz with a patch-clamp amplifier operated by Patchmaster 2.35 software (EPC10-USB, HEKA). OHCs were clamped at −70 mV. An HEKA EPC10–2 USB amplifier with integrated interface and PatchMaster software was used to control electrode voltage and monitor current. Series resistance of 5–10 MΩ was automatically compensated by the PatchMaster software.
Hair bundles were deflected with a glass pipette mounted on a piezoelectric stack actuator (P-885, Physik Instruments). The tip of the pipette was fire-polished to 4–6 μm in diameter to fit the shape of OHC bundles. The actuator was driven with voltage steps that were low-pass filtered at 10 KHz with an eight-pole Bessel filter (900CTF, Frequency Devices). The displacement was calibrated with a high-magnification objective and high-resolution camera as described (Grillet et al., 2009).
Yeast two-hybrid assays were conducted as described (Zhao et al., 2014). RT-PCR was carried out on whole vestibular system using previously described methods (Schwander et al., 2009) Gene-gun transfection and visualization of fluorescence protein-tagged constructs was carried using an optimized procedure (Zhao et al., 2012). For biolistic transfection, mouse cochleae were dissected at P6, subjected to gene-gun transfection, and then cultured for 1 day. Surface biotinylation was carried out with NHS-LC-Sulfo-biotin (Elia, 2008). Shotgun mass spectrometry on immunoprecipitates was carried out using a Thermo Velos ion-trap mass spectrometer; peptides and proteins were identified using the PAW pipeline and SEQUEST searches (Wilmarth et al., 2009; Krey et al., 2014). Auditory brainstem response (ABR) measurements were carried out as described (Schwander et al., 2007; Ebrahim et al., 2016). The Student’s t-test was used for all pairwise comparisons (two-sided, two-sample equal variance). Data distribution was assumed to be normal but this was not formally tested.
While localization of PCDH15 to stereocilia tips does not depend on USH1C (Lefevre et al., 2008), PCDH15 and USH1C can bind directly under some conditions (Adato et al., 2005; Reiners et al., 2005), suggesting that USH1C might couple the transduction channel to PCDH15. Sequences of 33 TRP channel genes were scanned for PDZ binding interfaces (PBIs); seven that had candidate PBIs were tested in a yeast two-hybrid assay with USH1C as the bait (Figure 1A). Of these, only TRPV6 interacted with USH1C (Figure 1B). We carried out RT-PCR for Trpv6 transcripts and found significant expression in vestibular tissues (Figure 1C). We noted that Trpv6 was also detected in the cochlea (Asai et al., 2010). Although immunocytochemistry experiments did not reliably detect TRPV6, the low levels of ion channels like TRP channels make detection very difficult (Gilliam and Wensel, 2011). These data raised the possibility that TRPV6 interacts with USH1C in hair cells, thus making TRPV6 a plausible transduction-channel candidate.
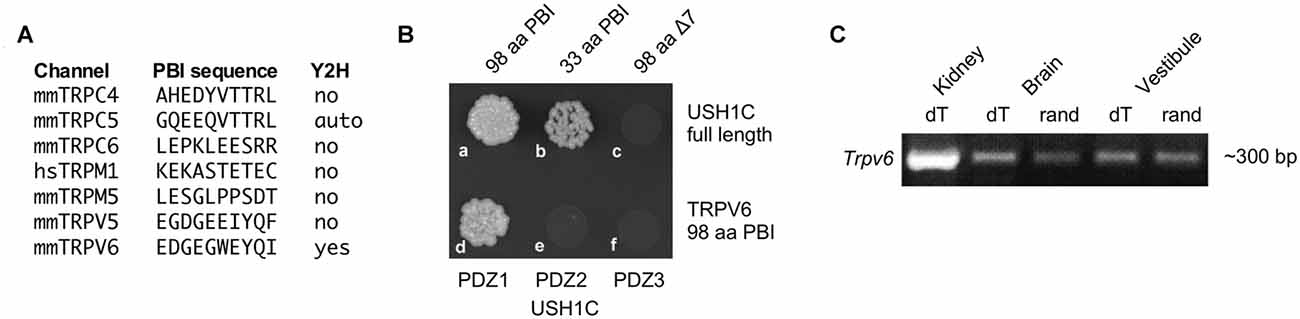
Figure 1. Characterization of TRPV6. (A) Seven transient receptor channel (TRP) channels (of 33) with candidate PDZ-binding interface motifs. Yeast two-hybrid (Y2H) results examining interactions with USH1C (harmonin) are indicated. (B) The 33 terminal amino acids of the TRPV6 PBI interact with USH1C, but deletion of the C-terminal seven amino acids abolishes binding (top row). The interaction of TRPV6 is through PDZ1 of USH1C (bottom row). (C) RT-PCR (with reverse transcription primed either with oligo dT or random hexamers) showing that mRNA for TRPV6 is present in vestibular RNA.
Individual knockouts of Trpv6 or its close paralog Trpv5 had no reported effects on auditory or vestibular function (Hoenderop et al., 2003; Bianco et al., 2007). These two genes are closely linked on human chromosome 7 or mouse chromosome 6 (Hoenderop and Bindels, 2008), so double knockouts cannot easily be created by breeding. To determine whether TRPV6 participates in hair-cell transduction, we developed a method whereby inhibition of a single channel subunit should lead to a dominant-negative effect on the tetrameric channel (Voets et al., 2004a). We used cysteine-substitution mutagenesis to create a Trpv6 allele that was sensitive to MTS reagents. Residue-scanning experiments with cysteine substitution and MTS inhibition demonstrated that the M527C mutation rendered TRPV6 sensitive to sulfhydryl reagents (Voets et al., 2004a). We used standard gene-targeting methods to introduce mutations in the mouse genome that produced expression of the M527C-TRPV6 protein (Figure 2).
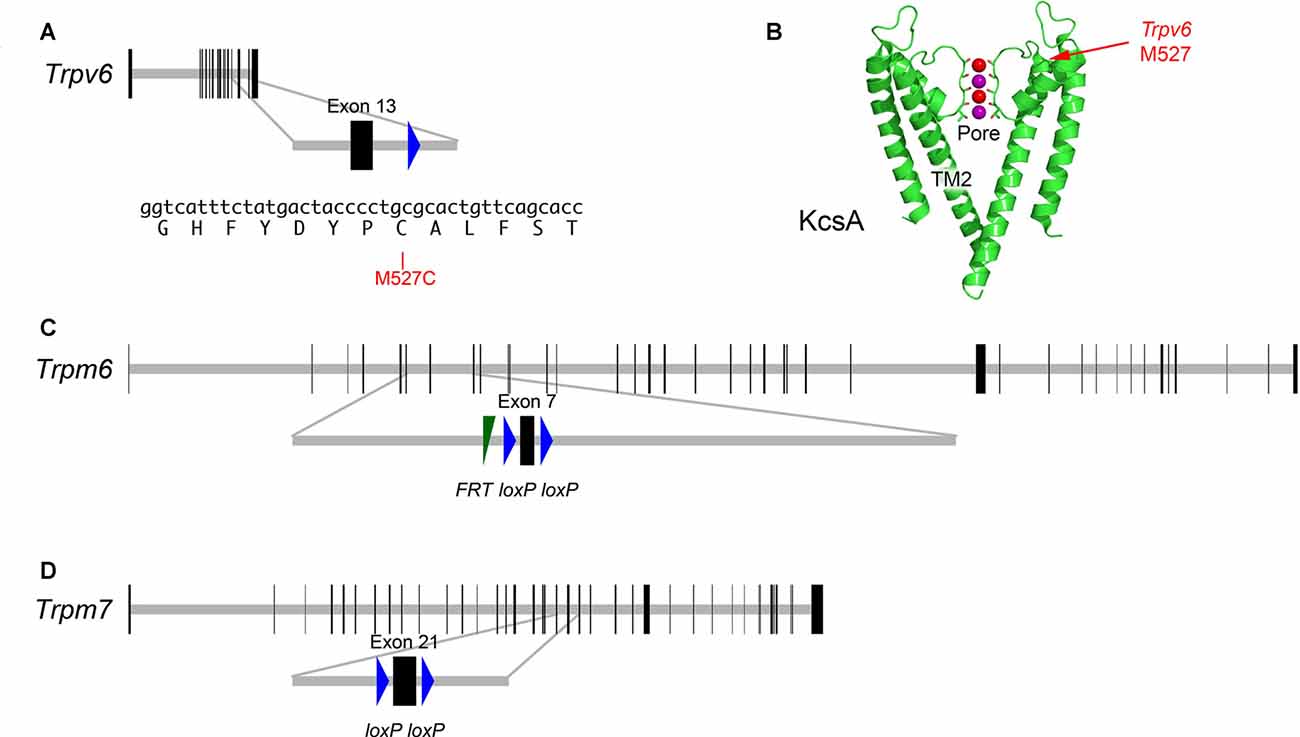
Figure 2. Targeted mutations in mouse Trpv6, Trpm6 and Trpm7 genes. (A) Top, Trpv6 gene structure. Horizontal gray line indicates scaled length of gene with coding exons, which are shown as vertical black rectangles. Middle, magnification of targeted exon. The loxP site remaining after excision of the neo cassette is shown in blue. Bottom, nucleotides and protein translation for region targeted. The M527C mutation is indicated. (B) Structure of the KcsA ion channel (image from https://commons.wikimedia.org/wiki/File:1K4C.png), along with the locations of the residue analogous to M527C-Trpv6. The pore and transmembrane domain 2 (TM2) are indicated as well. Two of the four subunits are shown. Color coding: protein, green; backbone carbonyl groups, oxygen in red and carbon in green; potassium ions (occupying S2 and S4), purple spheres; and oxygen atoms of water molecules (S1 and S3), red spheres. (C) Structure of Trpm6fl allele; exon 7 is flanked by loxP recombination sites; a residual FRT site remaining after excision of the Flp cassette is indicated. (D) Structure of Trpm7fl allele; exon 21 is flanked by loxP sites.
M527C-Trpv6 mice were viable and exhibited no apparent behavioral abnormalities as heterozygotes or homozygotes. Transduction currents of heterozygous M527C-Trpv6 OHCs under control conditions appeared normal in their amplitudes, kinetics, and adaptation (Figure 3). For example, maximum transduction currents were 527 ± 46 pA (mean ± SEM; n = 21) in WT hair cells and 514 ± 26 pA (n = 31) in M527C-Trpv6 hair cells (p = 0.8 by Student’s t-test). In wild-type mice, MTS reagents like MTSET and MTSES applied in the external solution had no effect on mechanotransduction in the absence of the sensitizing Trpv allele (Figure 3). This result shows that the cysteine-substitution approach should be suitable for probing the transduction channel; if MTS reagents had inhibited transduction currents of wild-type mice, we would not have be able to determine whether this or any other mutation had any impact.
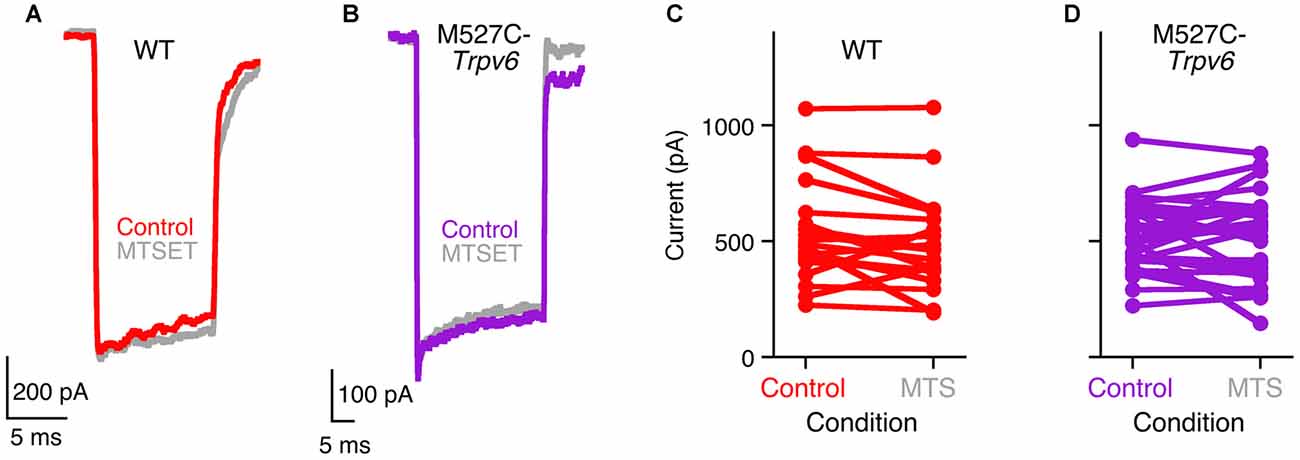
Figure 3. Methanethiosulfonate (MTS) reagents do not inhibit hair-cell transduction in M527C-Trpv6 hair cells. (A,B) Hair-cell transduction currents from wild-type (A) and heterozygous M527C-Trpv6 (B) mice before and after treatment with 1 mM MTSET, applied in the extracellular solution. Transduction currents were recorded with a holding potential of −80 mV. (C,D) Summarized data; each line connects a cell’s control maximum current (before application of inhibitor) and its maximum current after application of 1 mM MTS reagents (MTSET or MTSES).
Unfortunately, extracellular treatment with 1 mM MTSET or MTSES, which inhibit TRPV6 activity by >95% in tissue culture cells (Voets et al., 2004a), also had no significant effects on mechanotransduction in heterozygous or homozygous M527C-Trpv6 hair cells (Figure 3) when MTS reagents were applied in the external solution. There were no consistent changes in the maximal transduction current under these conditions for either wild-type or M527C-Trpv6 heterozygotes over the course of the recording (Figures 3C,D). For WT hair cells, the maximum transduction current went from 527 ± 46 pA before application of MTS reagents to 474 ± 46 pA after (not significant; p = 0.4 by Student’s t-test). For M527C-Trpv6, hair cells, the maximum transduction current went from 514 ± 26 pA before application of MTS reagents to 509 ± 33 pA after (p = 0.9). We conclude that TRPV6 does not participate in hair cell transduction.
Based on its participation in mechanotransduction in other cell types, TRPM7 (and its close paralog TRPM6) is another plausible transduction channel candidate. We used heterologous expression of mouse proteins in HEK cells to investigate whether the TRPM channels can interact with PCDH15 (Figure 4). We cloned full-length Trpm6 and Trpm7 from cDNA prepared from mouse inner ear, tagged them with the HA epitope tag, and co-expressed them with native, full-length PCDH15 in HEK293 cells. After solubilizing proteins and immunoprecipitating, when HA-tagged TRPMs were expressed, we could immunoprecipitate PCDH15 with anti-HA beads but not with negative-control anti-V5 beads (Figure 4A; Supplementary Figure S1). Likewise, immunoprecipitation with the anti-PCDH15 antibody PB811 selectively co-precipitated TRPM6 and TRPM7 (Figure 4B; Supplementary Figure S2). Co-expression of TRPM6 and TRPM7 consistently led to substantially more PCDH15 immunoprecipitated than with either alone (Figure 4A).
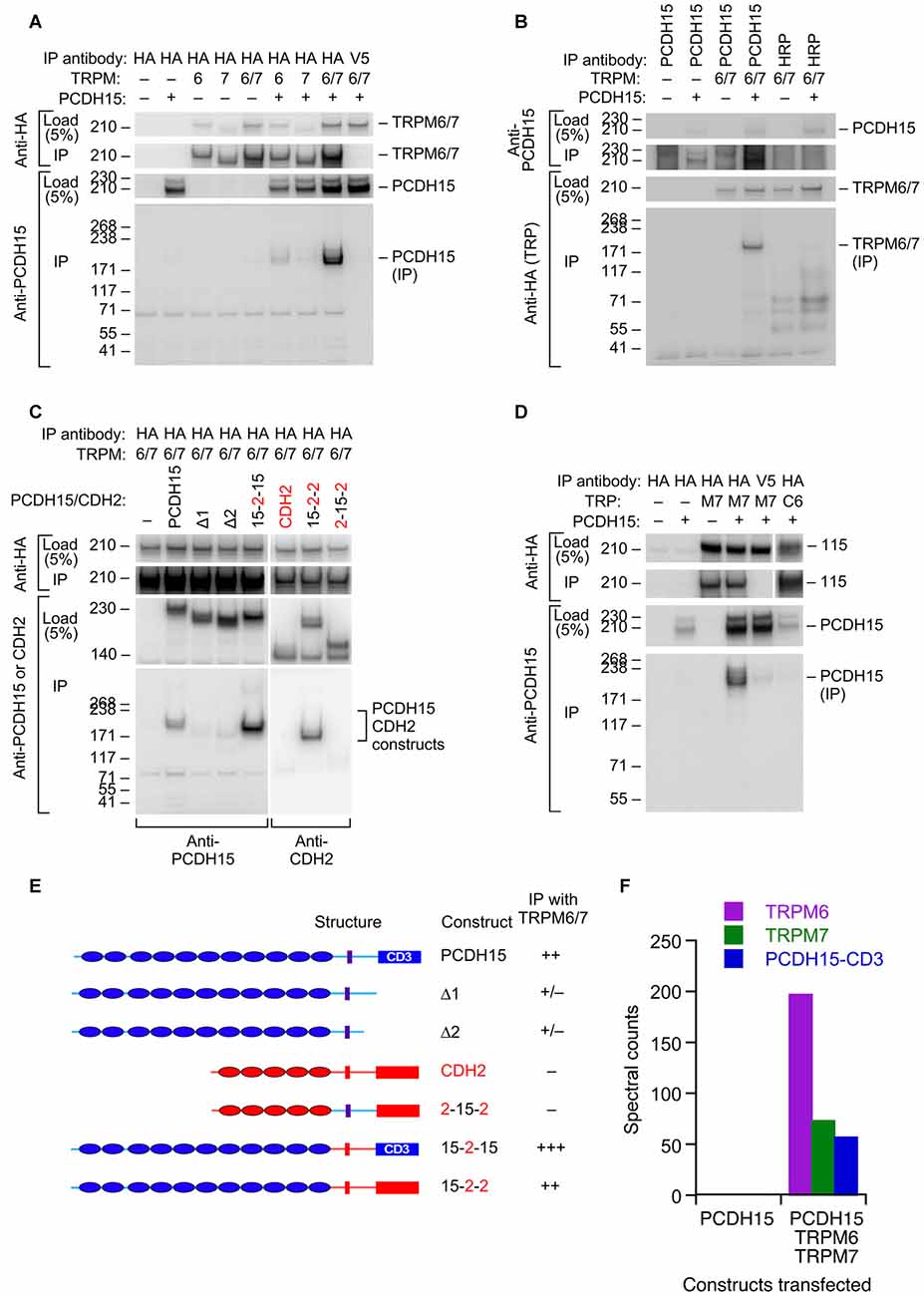
Figure 4. Interaction of PCDH15 with TRPM6 and TRPM7. (A) Lysates were immunoprecipitated with anti-HA agarose, and probed with anti-HA (top panels) or anti-PCDH15 antibody (bottom panels). PCDH15 was only immunoprecipitated by anti-HA if TRPM6 or RPM7 were present. No PCDH15 was precipitated by the control antibody (V5). (B) Lysates were immunoprecipitated with protein A/G agarose and either PB811 antibody (for PCDH15) or HRP antibody (control), then probed for TRP channels with anti-HA antibody. TRPM6 and TRPM7 were only precipitated with co-expressed with PCDH15 and precipitated with an anti-PCDH15 antibody. (C) Lysates were immunoprecipitated with anti-HA agarose, and probed with anti-PCDH15 antibody (left panels) or CDH2 antibody (right). Two C-terminal deletion proteins of PCDH15, Δ1 and Δ2, did not immunoprecipitate with TRPM6/7. Chimeras that have N-terminal extracellular domains of PCDH15 (15–2–15 and 15–2–2) immunoprecipitated with TRPM6/7 more strongly than PCDH15. The chimera that does not contain extracellular domains of PCDH15 (2–15–2) did not immunoprecipitate with TRPM6/7. (D) Lysates were immunoprecipitated with anti-HA agarose and probed with an anti-PCDH15 antibody (PB811). The last two lanes are two controls: anti-V5 agarose as a control for non-specific binding of the agarose with PCDH15, and TRPC6 as a TRP channel outside the TRPM family. Both controls have much lower levels of PCDH15 immunoprecipitated. (E) Summary of PCDH15 constructs (N- to C-terminal) used in panel (C). Chimeras with CDH2 used the CD3 splice form of PCDH15. (F) Mass spectrometry spectral counts for TRPM6, TRPM7, and PCDH15 in large-scale immunoprecipitation experiments using ant-HA agarose. No PCDH15 was immunoprecipitated when it was expressed alone, but large amounts were co-precipitated with TRPM6 and TRPM7.
To demonstrate the selectivity of the interaction, we showed that a different cadherin, CDH2 (N-cadherin), was not immunoprecipitated with HA-tagged TRPM6 or TRPM7 (Figure 4C; Supplementary Figure S3), nor was PCDH15 immunoprecipitated with HA-TRPC6, a different TRP channel (Figure 4D; Supplementary Figure S4). Co-expression was important; if HA-tagged TRPMs were expressed in separate samples from those of PCDH15 and the cell extracts were mixed, no PCDH15 was immunoprecipitated with HA-tagged TRPMs (data not shown). Deletion of the C-terminus of PCDH15 eliminated the interaction, but chimeras of PCDH15 and CDH2 that contained only the N-terminal extracellular domain of PCDH15 robustly co-precipitated TRPM6 and TRPM7 (Figure 4C).
To determine whether any endogenous HEK proteins mediated the interaction between the TRPMs and PCDH15, we carried out a larger scale immunoprecipitation and subjected the final eluates to tandem mass spectrometry. When expressed by itself and immunoprecipitated by HA-agarose, no PCDH15 was detected in eluates. By contrast, when TRPM6 and TRPM7 were co-expressed with PCDH15, all three proteins were detected at high levels in the immune pellet eluates (Figure 4F). Besides human heat shock protein cognate 71, no additional proteins co-immunoprecipitated at apparent stoichiometric levels, suggesting that the TRPM-PCDH15 interaction is direct.
Localization of TRP channels by immunocytochemistry is generally unreliable (Gilliam and Wensel, 2011). Instead, we generated a tdTomato-Trpm7 plasmid and transfected it into hair-cell progenitors at P6 using gene-gun transfection. Transfection efficiency was poor, presumably because of the large size of the expression construct. We were able to identify several transfected cells using co-expressed ZsGreen (Figure 5A), and found tdTomato signal that was largely localized to the lateral membranes of hair cells, close to the adherens junctions (Figure 5B). While not detected in stereocilia, the location of tdTomato-TRPM7 close to the hair cell’s apical domain raised the possibility that small amounts of TRPM7 could be transported to stereocilia membranes.
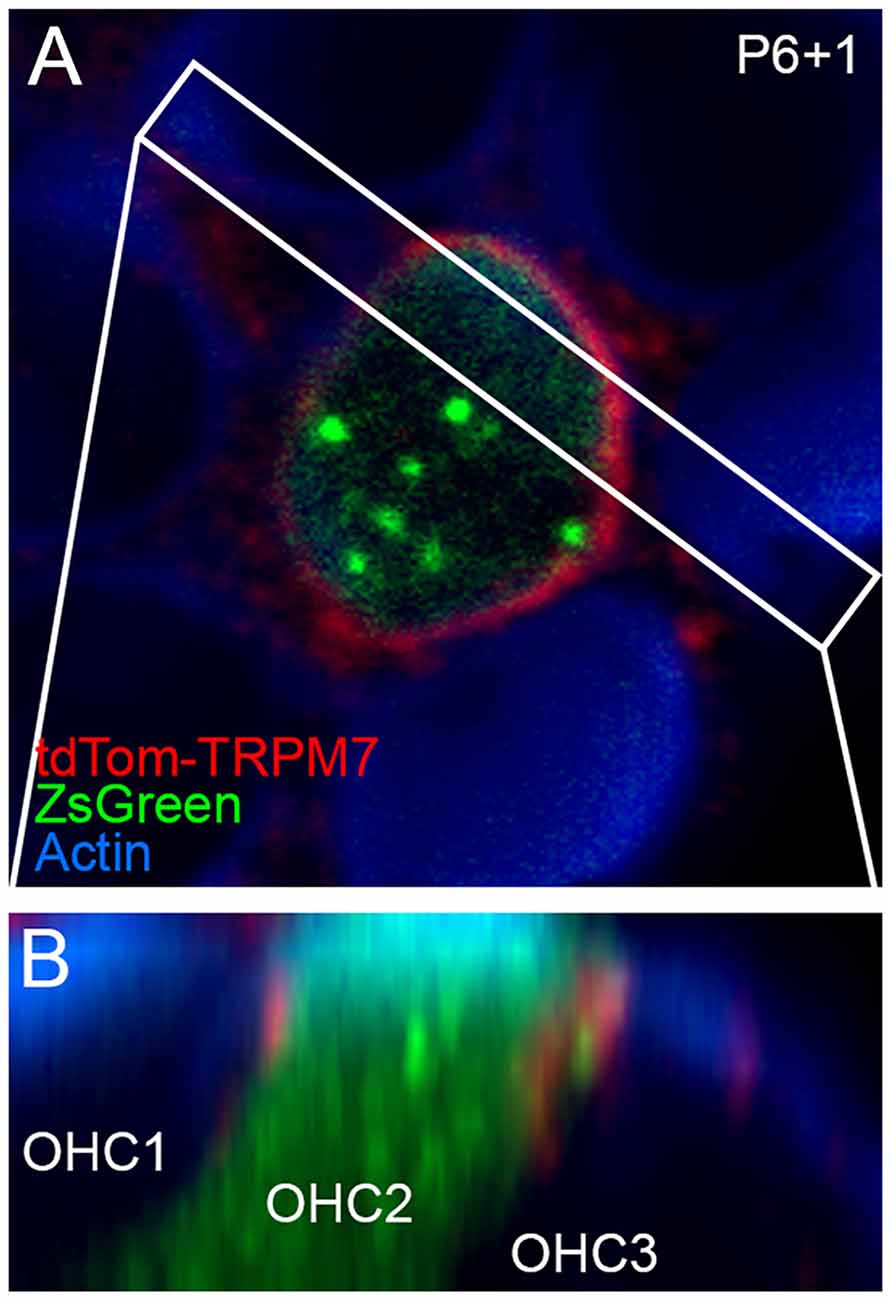
Figure 5. Localization of TRPM7 in the inner ear. (A) Single x-y slice through apical region of outer hair cells. One transfected cell, identified by ZsGreen signal, shows tdTomato signal at the lateral membrane. Box indicates region used for x-z reslice in (B). P6+1, cochlea dissected at P6 and maintained in culture for 1 day. (B) Reslice of stack shows lateral view of transfected hair cell (in row 2; OHC2) and two other untransfected cells. tdTomato-TRPM7 signal was located near apex of cell. Red signal outside of OHC2 is presumably background fluorescence. Panel full widths in (A,B) are 15 μm.
When cloning full-length Trpm6 and Trpm7 from cDNA prepared from mouse inner ear, we also identified novel splice forms that skipped exon 20 of each gene, which in both genes encodes transmembrane helix 2 (TM2). Using RT-PCR analysis, we found that while the canonical Trpm7 splice form was expressed in all mouse tissues, Dex27 (the Trpm7 splice form lacking exon 20) was restricted to inner ear, heart, and liver. Dex26 was expressed in a variety of tissues, including the inner ear (Figure 6A).
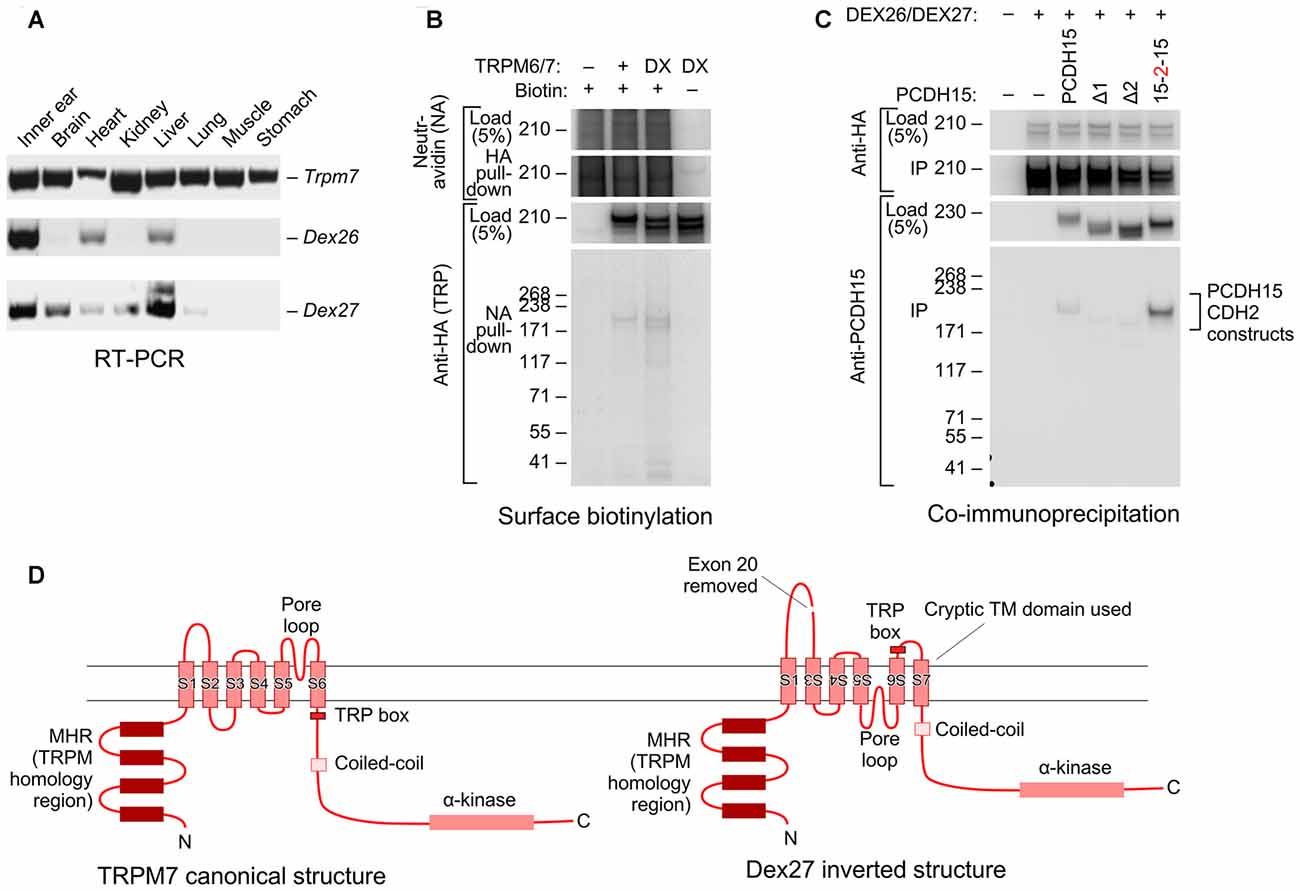
Figure 6. Novel splice forms of Trpm6 and Trpm7 interact with PCDH15. (A) Tissue expression profiles of mouse Trpm6, Dex27 and Dex26. Primer sets for Dex27 and Dex26 were designed such that they bind to the junctions of Trpm7 and Trpm6 exons 19 and 21; signal was only seen if exon 20 was absent. Both Dex27 and Dex26 were highly expressed in the ear (first lane on the left). (B) Both TRPM6/7 and DEX26/7 proteins traveled to the cell surface. HEK293T cells were transfected with TRPM6/7 or DEX26/7; before harvest, the cells were surface biotinylated. Cell lysates were then immunoprecipitated with neutravidin-agarose, then probed with anti-HA antibody. (C) DEX26 and DEX27 interacted with PCDH15 in a similar fashion with TRPM6 and TRPM7. (D) Predicted membrane topology for canonical TRPM7 sequence (left) and hypothetical inverted DEX27 sequence (right). The loss of TM2 when exon 20 is spliced out could force inversion of TM3–TM6; a cryptic transmembrane domain (S7 here) is predicted by some topology algorithms to be used, maintaining the N- and C-termini intracellular.
Interestingly, both DEX26 and DEX27 were readily expressed in HEK293 cells and were present on the cell surface, as assessed by a surface biotinylation assay (Figure 6B; Supplementary Figure S5). DEX26 and DEX27 both co-immunoprecipitated PCDH15 as efficiently as the canonical splice forms (Figure 6C; Supplementary Figure S6), and could be co-immunoprecipitated with the canonical forms.
Our protein interaction results raised the possibility that TRPM6 or TRPM7 mediates mechanotransduction by hair cells. A characteristic of TRPM6 and TRPM7 conductances is that they are blocked by intracellular Mg2+ (Nadler et al., 2001; Voets et al., 2004b). Hair-cell transduction was not affected by manipulations of Mg2+, however; currents appeared identical if recorded in the presence of 0 or 3 mM intracellular MgCl2 in the internal solution (Figure 7A). In recording from five cells for each condition, maximum transduction currents were reduced slightly but not significantly (p = 0.6; Student’s t-test) comparing cells recorded with 3 mM Mg2+ (599 ± 32 pA; mean ± SEM) with those with 0 mM Mg2+ (637 ± 60 pA; Figure 7B). Parameter values for three-state Boltzmann fits to displacement-Popen data were within experimental error for 0 and 3 mM Mg2+ (Figure 7C).
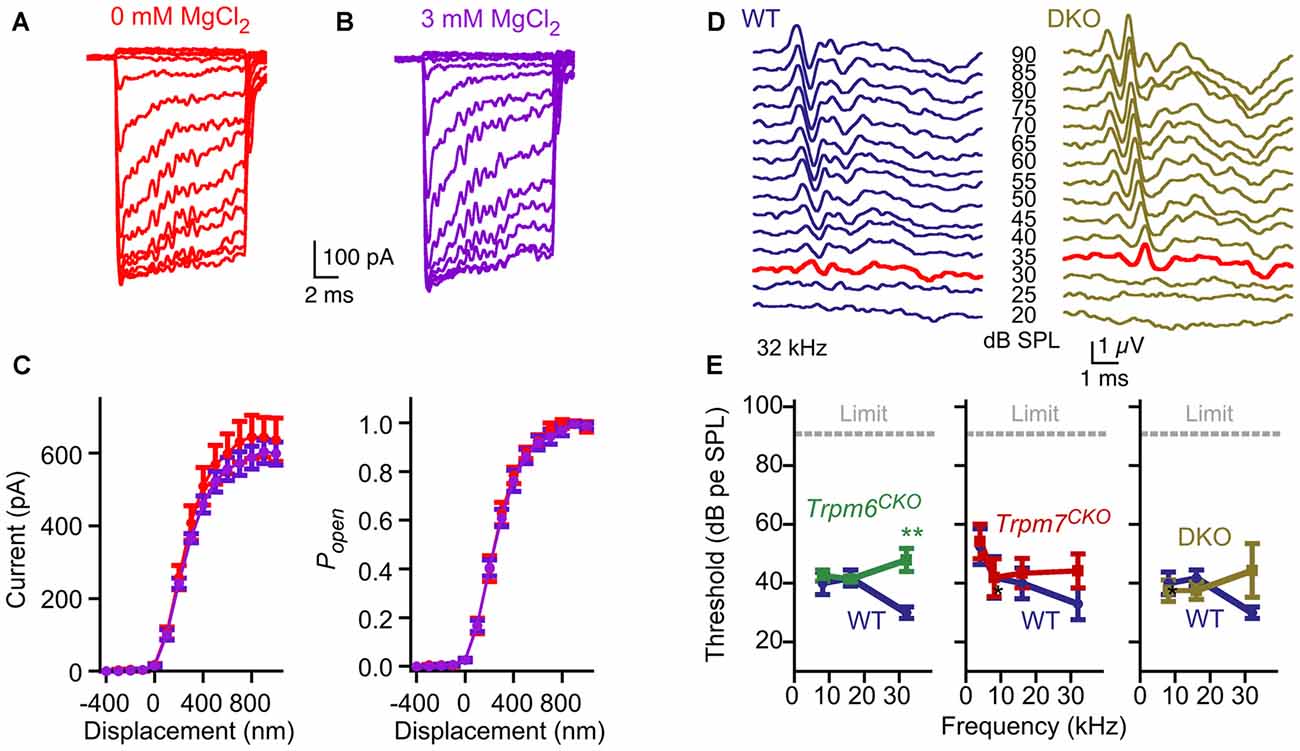
Figure 7. No evidence for contribution of TRPM6 or TRPM7 conductances to hair-cell transduction or auditory function. (A–C) No effect of internal Mg2+. (A) Representative transduction currents from hair cells that were dialyzed with internal solution containing 0 or 3 mM MgCl2. (B) No difference in transduction current amplitudes were observed. Mean ± SEM are plotted and fit with a three-state Boltzmann relation; n = 5 for each. (C) Replotted as displacement-Popen relationship. Boltzmann fits are nearly identical. (D) Examples of auditory brainstem response (ABR) waveforms at 32 kHz for WT (Trpm6fl/fl; Trpm7fl/fl; Atoh1-CRE-negative) and Trpm6CKO; Trpm7CKO DKO mice. The bold red trace for each indicates the threshold (30 and 35 dB SPL, respectively). (E) Summarized ABR measurements for Trpm6CKO, Trpm7CKO, and Trpm6CKO; Trpm7CKO DKO mice. Mean ± SEM are plotted. Trpm6CKO : WT, n = 11; Trpm6CKO, n = 17. **p < 0.01. Trpm7CKO: WT, n = 10; Trpm7CKO, n = 14. Trpm6CKO; Trpm7CKO DKO: WT, n = 11; Trpm6CKO; Trpm7CKO DKO, n = 8.
To determine definitively whether TRPM6 or TRPM7 participate in hair-cell mechanotransduction, we produced Trpm6fl and Trpm7fl mice (Figure 2) and expressed CRE recombinase in hair cells to delete essential exons for each gene. Global-null Trpm6Δ/Δ mice rarely survive past birth due to Mg2+ insufficiency (Walder et al., 2009; Woudenberg-Vrenken et al., 2011), and global-null Trpm7Δ/Δ mice have an embryonic lethality phenotype (Jin et al., 2008). We therefore used the Atoh1-Cre transgenic line to restrict Trpm6 and Trpm7 deletion to hair cells and a few other cell types (Matei et al., 2005; Pan et al., 2012). Mice with both alleles recombined by CRE are referred to here as Trpm6CKO or Trpm7CKO.
Trpm6CKO, Trpm7CKO and Trpm6CKO;Trpm7CKO (double knockout, or DKO) mice were behaviorally normal, with no indication of gross auditory or vestibular disruption. We assessed auditory function using ABR measurements (Figures 7D,E). While ABR thresholds at 32 kHz were slightly but significantly elevated for Trpm6CKO mice, thresholds were not statistically significant different for Trpm7CKO or Trpm6CKO;Trpm7CKO DKO mice as compared to controls at all frequencies (Figure 7E). These results suggest that neither TRPM6 nor TRPM7 is a major contributor the mechanotransduction.
Our experimental results suggest that TRPV6, TRPM6 and TRPM7 are not involved in hair-cell transduction. While each of these TRP channels had plausible initial evidence supporting their involvement, direct experiments—allele-specific inhibition for TRPV6, and single and double knockouts for TRPM6 and TRPM7—gave no support for a role of any of them in forming the hair cell’s transduction pore. While these results do not provide direct evidence for the participation of TMC1 and TMC2 in forming the pore, the paucity of viable alternative candidates lends indirect support for the two TMCs.
When applied in the external solution, MTS reagents did not inhibit transduction currents measured in hair cells of M527C-Trpv6 mice. In cell culture, each of these residues can be robustly inhibited from the extracellular side of the membrane (Dodier et al., 2004; Voets et al., 2004a), so the lack of MTS inhibition of transduction in mutant hair cells suggests that TRPV6 does not contribute to the transduction channel.
Like allele-selective inhibition of myosins (Gillespie et al., 1999; Holt et al., 2002), this strategy provides strong evidence for the role of a protein in a biological function; three control conditions (wild-type, wild-type plus inhibitor, mutant) are matched with one experimental condition (mutant plus inhibitor), strengthening the interpretation of an inhibitory effect. To definitively establish the viability of the cysteine substitution/MTS inhibition approach with this channel, future experiments could determine whether Ca2+ transport in intestine, mediated by TRPV6 (Nijenhuis et al., 2003), is inhibited when the M527C allele is present and MTS reagents are applied.
Single and double knockouts of Trpm6 and Trpm7 had no effect on mouse hearing. While demonstrating conclusively that Trpm6 and Trpm7 transcripts are eliminated in hair cells is difficult because of the low level of expression (making in situ mRNA localization challenging) and expression in other cochlear cell types (preventing analysis of whole cochlea to assess transcript loss), recombination by Atoh1-CRE occurs robustly during hair cell development (Matei et al., 2005; Pan et al., 2012). We assessed ABRs at 4–8 weeks of age, and it is very unlikely that Trpm7 mRNA or TRPM7 protein would be stable for so long after the genes were recombined. Nevertheless, even though there are limitations to our experiments, they nonetheless strongly suggest that TRPM6 and TRPM7 are not part of the transduction channel.
Our results suggest that under some circumstances, TRPM6 and TRPM7 can bind to PCDH15, a component of the tip link. Because TRPM6 and TRPM7 are not essential for hearing, these results suggest that TRPM6 and TRPM7 might interact with PCDH15 elsewhere besides the transduction complex. One possibility is that the TRPMs are present in kinocilia, and interact with the PCDH15 molecules that contribute to the kinocilial links (Goodyear et al., 2010). If so, the TRPMs are unlikely to play an essential role in kinocilia-link function, as loss of kinocilial links leads to deafness (Webb et al., 2011) but loss of the TRPMs does not affect hearing. Alternatively, RNA-seq experiments show that PCDH15 is expressed in inner-ear cell types besides hair cells (Burns et al., 2015), and so these channels could form complexes with PCDH15 there.
An alternative view is that PCDH15 or the TRPMs bind membrane proteins promiscuously, which may call into question interactions described based on similar immunoprecipitation experiments (Ramakrishnan et al., 2009, 2012; Maeda et al., 2014; Beurg et al., 2015; Cunningham et al., 2017; Erickson et al., 2017). While our experiments were well controlled, including the use of alternative cadherins and TRP channels, reliance simply on cell-culture immunoprecipitation experiments is fraught. Such evidence should always be backed up by alternative experiments that lack some of the ambiguity of in vitro experiments.
We identified splice forms of Trpm6 and Trpm7 that each lack exon 20, which encodes most of transmembrane domain 2 in each channel. DEX26 and DEX27 were targeted to the surface of HEK293 cells, and, like the canonical versions of TRPM6 and TRPM7, interacted with PCDH15 via its extracellular domain. Membrane topology programs suggested the possibility that DEX26 and DEX27 have partially inverted transmembrane domain regions, so that the pore of the channel is a re-entrant loop coming from the inside rather than the outside (Figure 6D). This topology is speculative at this moment, however, as we have no experimental evidence that the pore is flipped. If this topology is correct, however, it limits the regions of the TRPM proteins that PCDH15 can interact with from the N-terminus through TM2 and from the cryptic TM7 to the C-terminus. Regardless of whether the channel’s pores are inverted, the loss of TM2 will force an unusual topology on this channel, however, and it will be interesting to follow up the role of these splice forms in the organism.
Our results rule out several transduction channel candidates, adding to the list of channels disproven that includes P2RX2, SCNN1A, TRPN1, TRPA1, TRPV4, TRPML3, TRPC3, TRPC5, TRPC6, TRPM1, TRPM2, TRPM3, PKD1, PKD1L3, PKD2, PKD2L1 and PKD2L2 (Rüsch and Hummler, 1999; Vollrath et al., 2007; Steigelman et al., 2011; Fettiplace and Kim, 2014; Wu et al., 2016). Because of their essential role in mechanotransduction and their size, the TMCs remain the best candidate for the transduction channel’s pore. Nonetheless, the TMCs still have not been conclusively demonstrated to form ion channels, necessary for showing that a given molecule forms the transduction pore.
The cysteine-substitution/MTS inhibition approach could serve as a useful tool for testing additional transduction channel candidates, although the accurate knowledge of the location of the modified residues from cysteine-scanning mutagenesis (Dodier et al., 2004; Voets et al., 2004a) is essential, as is confirmation of the location of the modified residues in the protein structure (Saotome et al., 2016). Moreover, as modification of cysteines could allosterically affect channel function, perhaps even mediated through protein-protein interactions, the cysteine/MTS approach does not definitively prove that the modified residue is in the tested channel’s pore. Perhaps the most conclusive test for the transduction channel would be to change the ion selectivity of the pore based on knowledge of channel structure, and then show that hair-cell transduction ion selectivity changes in the predicted manner.
CPM carried out initial mass spectrometry experiments implicating TRPM6 and TRPM7, and conducted mass spectrometry analysis of immunoprecipitates. HZ demonstrated interaction of TRPM6 and TRPM7 with PCDH15 using immunoprecipitation. ML developed the Trpv6 knock-in mice. WX carried out electrophysiological assays of wild-type mouse hair cells. BP carried out electrophysiological recordings of Trpv6 mice. PK acquired evidence for TRPV6 expression in hair cells, and developed the Trpm7 floxed allele. MRA carried out experiments localizing TRPM7. MB assisted in development of Trpv6 knock-in mice, developed genotyping assays for all Trp alleles and conducted ABR measurements. RL was responsible for mouse husbandry and ABRs. AJR supervised all TRPV6 electrophysiology, carried out recordings of Trpv6 mice and analyzed data. UM supervised TRPV6 molecular characterization and development of the Trpm7 floxed allele and analyzed data. PGB-G supervised development of Trpv6 knock-in mice, development of Trpm6 and Trpm7 knockout mice, and protein interaction experiments; he analyzed data and also wrote the article with contributions from all authors. All authors agree to be accountable for the content of the work.
PGB-G was supported by NIH grants R01 DC002368 and P30 DC005983; AJR was supported by NIH R01 DC003896; UM was supported by NIH grant R01 DC005965.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Thomas F. Wagner and Nicolas Grillet for participating in the early phases of this project. We received support from the following core facilities: mouse generation from the Transgenic Mouse Models Core (OHSU), Trpm6fl embryonic stem cells from KOMP (the Knockout Mouse Project, University of California, Davis), and confocal microscopy from the OHSU Advanced Light Microscopy Core @ The Jungers Center.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2018.00041/full#supplementary-material
Adato, A., Michel, V., Kikkawa, Y., Reiners, J., Alagramam, K. N., Weil, D., et al. (2005). Interactions in the network of Usher syndrome type 1 proteins. Hum. Mol. Genet. 14, 347–356. doi: 10.1093/hmg/ddi031
Alagramam, K. N., Goodyear, R. J., Geng, R., Furness, D. N., van Aken, A. F., Marcotti, W., et al. (2011). Mutations in protocadherin 15 and cadherin 23 affect tip links and mechanotransduction in mammalian sensory hair cells. PLoS One 6:e19183. doi: 10.1371/journal.pone.0019183
Asai, Y., Holt, J. R., and Geleoc, G. S. (2010). A quantitative analysis of the spatiotemporal pattern of transient receptor potential gene expression in the developing mouse cochlea. J. Assoc. Res. Otolaryngol. 11, 27–37. doi: 10.1007/s10162-009-0193-8
Avenarius, M. R., Krey, J. F., Dumont, R. A., Morgan, C. P., Benson, C. B., Vijayakumar, S., et al. (2017). Heterodimeric capping protein is required for stereocilia length and width regulation. J. Cell Biol. 216, 3861–3881. doi: 10.1083/jcb.201704171
Bessac, B. F., and Fleig, A. (2007). TRPM7 channel is sensitive to osmotic gradients in human kidney cells. J. Physiol. 582, 1073–1086. doi: 10.1113/jphysiol.2007.130534
Beurg, M., Fettiplace, R., Nam, J. H., and Ricci, A. J. (2009). Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat. Neurosci. 12, 553–558. doi: 10.1038/nn.2295
Beurg, M., Kim, K. X., and Fettiplace, R. (2014). Conductance and block of hair-cell mechanotransducer channels in transmembrane channel-like protein mutants. J. Gen. Physiol. 144, 55–69. doi: 10.1085/jgp.201411173
Beurg, M., Xiong, W., Zhao, B., Müller, U., and Fettiplace, R. (2015). Subunit determination of the conductance of hair-cell mechanotransducer channels. Proc. Natl. Acad. Sci. U S A 112, 1589–1594. doi: 10.1073/pnas.1420906112
Bianco, S. D., Peng, J. B., Takanaga, H., Suzuki, Y., Crescenzi, A., Kos, C. H., et al. (2007). Marked disturbance of calcium homeostasis in mice with targeted disruption of the Trpv6 calcium channel gene. J. Bone. Miner. Res. 22, 274–285. doi: 10.1359/jbmr.061110
Burns, J. C., Kelly, M. C., Hoa, M., Morell, R. J., and Kelley, M. W. (2015). Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat. Commun. 6:8557. doi: 10.1038/ncomms9557
Chubanov, V., and Gudermann, T. (2014). TRPM6. Handb. Exp. Pharmacol. 222, 503–520. doi: 10.1007/978-3-642-54215-2_20
Cuajungco, M. P., Grimm, C., and Heller, S. (2007). TRP channels as candidates for hearing and balance abnormalities in vertebrates. Biochim. Biophys. Acta 1772, 1022–1027. doi: 10.1016/j.bbadis.2007.01.002
Cunningham, C. L., Wu, Z., Jafari, A., Zhao, B., Schrode, K., Harkins-Perry, S., et al. (2017). The murine catecholamine methyltransferase mTOMT is essential for mechanotransduction by cochlear hair cells. Elife 6:e33307. doi: 10.7554/eLife.33307
Dodier, Y., Banderali, U., Klein, H., Topalak, O., Dafi, O., Simoes, M., et al. (2004). Outer pore topology of the ECaC-TRPV5 channel by cysteine scan mutagenesis. J. Biol. Chem. 279, 6853–6862. doi: 10.1074/jbc.M310534200
Ebrahim, S., Avenarius, M. R., Grati, M., Krey, J. F., Windsor, A. M., Sousa, A. D., et al. (2016). Stereocilia-staircase spacing is influenced by myosin III motors and their cargos espin-1 and espin-like. Nat. Commun. 7:10833. doi: 10.1038/ncomms10833
Elia, G. (2008). Biotinylation reagents for the study of cell surface proteins. Proteomics 8, 4012–4024. doi: 10.1002/pmic.200800097
Erickson, T., Morgan, C. P., Olt, J., Hardy, K., Busch-Nentwich, E., Maeda, R., et al. (2017). Integration of Tmc1/2 into the mechanotransduction complex in zebrafish hair cells is regulated by Transmembrane O-methyltransferase (Tomt). Elife 6:e28474. doi: 10.7554/eLife.28474
Fettiplace, R., and Kim, K. X. (2014). The physiology of mechanoelectrical transduction channels in hearing. Physiol. Rev. 94, 951–986. doi: 10.1152/physrev.00038.2013
Fleig, A., and Chubanov, V. (2014). TRPM7. Handb. Exp. Pharmacol. 222, 521–546. doi: 10.1007/978-3-642-54215-2_21
Gillespie, P. G., Gillespie, S. K., Mercer, J. A., Shah, K., and Shokat, K. M. (1999). Engineering of the myosin-Iβ nucleotide-binding pocket to create selective sensitivity to N6-modified ADP analogs. J. Biol. Chem. 274, 31373–31381. doi: 10.1074/jbc.274.44.31373
Gilliam, J. C., and Wensel, T. G. (2011). TRP channel gene expression in the mouse retina. Vision Res. 51, 2440–2452. doi: 10.1016/j.visres.2011.10.009
Gong, Z., Son, W., Chung, Y. D., Kim, J., Shin, D. W., McClung, C. A., et al. (2004). Two interdependent TRPV channel subunits, inactive and Nanchung, mediate hearing in Drosophila. J. Neurosci. 24, 9059–9066. doi: 10.1523/jneurosci.1645-04.2004
Goodyear, R. J., Forge, A., Legan, P. K., and Richardson, G. P. (2010). Asymmetric distribution of cadherin 23 and protocadherin 15 in the kinocilial links of avian sensory hair cells. J. Comp. Neurol. 518, 4288–4297. doi: 10.1002/cne.22456
Grillet, N., Xiong, W., Reynolds, A., Kazmierczak, P., Sato, T., Lillo, C., et al. (2009). Harmonin mutations cause mechanotransduction defects in cochlear hair cells. Neuron 62, 375–387. doi: 10.1016/j.neuron.2009.04.006
Hoenderop, J. G., and Bindels, R. J. (2008). Calciotropic and magnesiotropic TRP channels. Physiology 23, 32–40. doi: 10.1152/physiol.00039.2007
Hoenderop, J. G., van Leeuwen, J. P., van der Eerden, B. C., Kersten, F. F., van der Kemp, A. W., Mérillat, A. M., et al. (2003). Renal Ca2+ wasting, hyperabsorption, and reduced bone thickness in mice lacking TRPV5. J. Clin. Invest. 112, 1906–1914. doi: 10.1172/jci19826
Holt, J. R., Gillespie, S. K., Provance, D. W., Shah, K., Shokat, K. M., Corey, D. P., et al. (2002). A chemical-genetic strategy implicates myosin-1c in adaptation by hair cells. Cell 108, 371–381. doi: 10.1016/s0092-8674(02)00629-3
Horwitz, G. C., Lelli, A., Géléoc, G. S., and Holt, J. R. (2010). HCN channels are not required for mechanotransduction in sensory hair cells of the mouse inner ear. PLoS One 5:e8627. doi: 10.1371/journal.pone.0008627
Jin, J., Desai, B. N., Navarro, B., Donovan, A., Andrews, N. C., and Clapham, D. E. (2008). Deletion of Trpm7 disrupts embryonic development and thymopoiesis without altering Mg2+ homeostasis. Science 322, 756–760. doi: 10.1126/science.1163493
Kawashima, Y., Géléoc, G. S., Kurima, K., Labay, V., Lelli, A., Asai, Y., et al. (2011). Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest. 121, 4796–4809. doi: 10.1172/JCI60405
Kazmierczak, P., Sakaguchi, H., Tokita, J., Wilson-Kubalek, E. M., Milligan, R. A., Müller, U., et al. (2007). Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–91. doi: 10.1038/nature06091
Kernan, M. J. (2007). Mechanotransduction and auditory transduction in Drosophila. Pflugers Arch. 454, 703–720. doi: 10.1007/s00424-007-0263-x
Krey, J. F., Wilmarth, P. A., Shin, J. B., Klimek, J., Sherman, N. E., Jeffery, E. D., et al. (2014). Accurate label-free protein quantitation with high- and low-resolution mass spectrometers. J. Proteome Res. 13, 1034–1044. doi: 10.1021/pr401017h
Lefevre, G., Michel, V., Weil, D., Lepelletier, L., Bizard, E., Wolfrum, U., et al. (2008). A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development 135, 1427–1437. doi: 10.1242/dev.012922
Lehnert, B. P., Baker, A. E., Gaudry, Q., Chiang, A. S., and Wilson, R. I. (2013). Distinct roles of TRP channels in auditory transduction and amplification in Drosophila. Neuron 77, 115–128. doi: 10.1016/j.neuron.2012.11.030
Maeda, R., Kindt, K. S., Mo, W., Morgan, C. P., Erickson, T., Zhao, H., et al. (2014). Tip-link protein protocadherin 15 interacts with transmembrane channel-like proteins TMC1 and TMC2. Proc. Natl. Acad. Sci. U S A 111, 12907–12912. doi: 10.1073/pnas.1402152111
Matei, V., Pauley, S., Kaing, S., Rowitch, D., Beisel, K. W., Morris, K., et al. (2005). Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev. Dyn. 234, 633–650. doi: 10.1002/dvdy.20551
Nadler, M. J., Hermosura, M. C., Inabe, K., Perraud, A. L., Zhu, Q., Stokes, A. J., et al. (2001). LTRPC7 is a Mg.ATP-regulated divalent cation channel required for cell viability. Nature 411, 590–595. doi: 10.1038/35079092
Nijenhuis, T., Hoenderop, J. G., Nilius, B., and Bindels, R. J. (2003). Patho)physiological implications of the novel epithelial Ca2+ channels TRPV5 and TRPV6. Pflugers Arch. 446, 401–409. doi: 10.1007/s00424-003-1038-7
Numata, T., Shimizu, T., and Okada, Y. (2007a). Direct mechano-stress sensitivity of TRPM7 channel. Cell. Physiol. Biochem. 19, 1–8. doi: 10.1159/000099187
Numata, T., Shimizu, T., and Okada, Y. (2007b). TRPM7 is a stretch- and swelling-activated cation channel involved in volume regulation in human epithelial cells. Am. J. Physiol. Cell Physiol. 292, C460–C467. doi: 10.1152/ajpcell.00367.2006
Owsianik, G., Talavera, K., Voets, T., and Nilius, B. (2006). Permeation and selectivity of TRP channels. Annu. Rev. Physiol. 68, 685–717. doi: 10.1146/annurev.physiol.68.040204.101406
Pan, B., Geleoc, G. S., Asai, Y., Horwitz, G. C., Kurima, K., Ishikawa, K., et al. (2013). TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515. doi: 10.1016/j.neuron.2013.06.019
Pan, N., Jahan, I., Kersigo, J., Duncan, J. S., Kopecky, B., and Fritzsch, B. (2012). A novel Atoh1 “self-terminating” mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS One 7:e30358. doi: 10.1371/journal.pone.0030358
Peng, A. W., Gnanasambandam, R., Sachs, F., and Ricci, A. J. (2016). Adaptation independent modulation of auditory hair cell mechanotransduction channel open probability implicates a role for the lipid bilayer. J. Neurosci. 36, 2945–2956. doi: 10.1523/JNEUROSCI.3011-15.2016
Powers, R. J., Roy, S., Atilgan, E., Brownell, W. E., Sun, S. X., Gillespie, P. G., et al. (2012). Stereocilia membrane deformation: implications for the gating spring and mechanotransduction channel. Biophys. J. 102, 201–210. doi: 10.1016/j.bpj.2011.12.022
Ramakrishnan, N. A., Drescher, M. J., Barretto, R. L., Beisel, K. W., Hatfield, J. S., and Drescher, D. G. (2009). Calcium-dependent binding of HCN1 channel protein to hair cell stereociliary tip link protein protocadherin 15 CD3. J. Biol. Chem. 284, 3227–3238. doi: 10.1074/jbc.M806177200
Ramakrishnan, N. A., Drescher, M. J., Khan, K. M., Hatfield, J. S., and Drescher, D. G. (2012). HCN1 and HCN2 proteins are expressed in cochlear hair cells: HCN1 can form a ternary complex with protocadherin 15 CD3 and F-actin-binding filamin A or can interact with HCN2. J. Biol. Chem. 287, 37628–37646. doi: 10.1074/jbc.M112.375832
Reiners, J., Märker, T., Jürgens, K., Reidel, B., and Wolfrum, U. (2005). Photoreceptor expression of the Usher syndrome type 1 protein protocadherin 15 (USH1F) and its interaction with the scaffold protein harmonin (USH1C). Mol. Vis. 11, 347–355. Available online at: http://www.molvis.org/molvis/v11/a41/
Rüsch, A., and Hummler, E. (1999). Mechano-electrical transduction in mice lacking the α-subunit of the epithelial sodium channel. Hear Res. 131, 170–176. doi: 10.1016/s0378-5955(99)00030-1
Saotome, K., Singh, A. K., Yelshanskaya, M. V., and Sobolevsky, A. I. (2016). Crystal structure of the epithelial calcium channel TRPV6. Nature 534, 506–511. doi: 10.1038/nature17975
Schwander, M., Lopes, V., Sczaniecka, A., Gibbs, D., Lillo, C., Delano, D., et al. (2009). A novel allele of myosin VIIa reveals a critical function for the C-terminal FERM domain for melanosome transport in retinal pigment epithelial cells. J. Neurosci. 29, 15810–15818. doi: 10.1523/JNEUROSCI.4876-09.2009
Schwander, M., Sczaniecka, A., Grillet, N., Bailey, J. S., Avenarius, M., Najmabadi, H., et al. (2007). A forward genetics screen in mice identifies recessive deafness traits and reveals that pejvakin is essential for outer hair cell function. J. Neurosci. 27, 2163–2175. doi: 10.1523/JNEUROSCI.4975-06.2007
Siemens, J., Lillo, C., Dumont, R. A., Reynolds, A., Williams, D. S., Gillespie, P. G., et al. (2004). Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature 428, 950–955. doi: 10.1038/nature02483
Sollner, C., Rauch, G. J., Siemens, J., Geisler, R., Schuster, S. C., Müller, U., et al. (2004). Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature 428, 955–959. doi: 10.1038/nature02484
Steigelman, K. A., Lelli, A., Wu, X., Gao, J., Lin, S., Piontek, K., et al. (2011). Polycystin-1 is required for stereocilia structure but not for mechanotransduction in inner ear hair cells. J. Neurosci. 31, 12241–12250. doi: 10.1523/JNEUROSCI.6531-10.2011
Venkatachalam, K., and Montell, C. (2007). TRP channels. Annu. Rev. Biochem. 76, 387–417. doi: 10.1146/annurev.biochem.75.103004.142819
Voets, T., Janssens, A., Droogmans, G., and Nilius, B. (2004a). Outer pore architecture of a Ca2+-selective TRP channel. J. Biol. Chem. 279, 15223–15230. doi: 10.1074/jbc.M312076200
Voets, T., Nilius, B., Hoefs, S., van der Kemp, A. W., Droogmans, G., Bindels, R. J., et al. (2004b). TRPM6 forms the Mg2+ influx channel involved in intestinal and renal Mg2+ absorption. J. Biol. Chem. 279, 19–25. doi: 10.1074/jbc.M311201200
Vollrath, M. A., Kwan, K. Y., and Corey, D. P. (2007). The micromachinery of mechanotransduction in hair cells. Annu. Rev. Neurosci. 30, 339–365. doi: 10.1146/annurev.neuro.29.051605.112917
Walder, R. Y., Yang, B., Stokes, J. B., Kirby, P. A., Cao, X., Shi, P., et al. (2009). Mice defective in Trpm6 show embryonic mortality and neural tube defects. Hum. Mol. Genet. 18, 4367–4375. doi: 10.1093/hmg/ddp392
Webb, S. W., Grillet, N., Andrade, L. R., Xiong, W., Swarthout, L., Della Santina, C. C., et al. (2011). Regulation of PCDH15 function in mechanosensory hair cells by alternative splicing of the cytoplasmic domain. Development 138, 1607–1617. doi: 10.1242/dev.060061
Wei, C., Wang, X., Chen, M., Ouyang, K., Song, L. S., and Cheng, H. (2009). Calcium flickers steer cell migration. Nature 457, 901–905. doi: 10.1038/nature07577
Wilmarth, P. A., Riviere, M. A., and David, L. L. (2009). Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Infor. 2, 223–234. doi: 10.1007/s12177-009-9042-6
Woudenberg-Vrenken, T. E., Sukinta, A., van der Kemp, A. W., Bindels, R. J., and Hoenderop, J. G. (2011). Transient receptor potential melastatin 6 knockout mice are lethal whereas heterozygous deletion results in mild hypomagnesemia. Nephron Physiol. 117, p11–p19. doi: 10.1159/000320580
Wu, X., Indzhykulian, A. A., Niksch, P. D., Webber, R. M., Garcia-Gonzalez, M., Watnick, T., et al. (2016). Hair-cell mechanotransduction persists in TRP channel knockout mice. PLoS One 11:e0155577. doi: 10.1371/journal.pone.0155577
Xiao, E., Yang, H. Q., Gan, Y. H., Duan, D. H., He, L. H., Guo, Y., et al. (2015). Brief reports: TRPM7 Senses mechanical stimulation inducing osteogenesis in human bone marrow mesenchymal stem cells. Stem Cells 33, 615–621. doi: 10.1002/stem.1858
Xiong, W., Grillet, N., Elledge, H. M., Wagner, T. F., Zhao, B., Johnson, K. R., et al. (2012). TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell 151, 1283–1295. doi: 10.1016/j.cell.2012.10.041
Zanini, D., and Göpfert, M. C. (2014). TRPs in hearing. Handb. Exp. Pharmacol. 223, 899–916. doi: 10.1007/978-3-319-05161-1_7
Zhang, W., Yan, Z., Jan, L. Y., and Jan, Y. N. (2013). Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae. Proc. Natl. Acad. Sci. U S A 110, 13612–13617. doi: 10.1073/pnas.1312477110
Zhao, H., Avenarius, M. R., and Gillespie, P. G. (2012). Improved biolistic transfection of hair cells. PLoS One 7:e46765. doi: 10.1371/journal.pone.0046765
Keywords: hair cells, stereocilia, mechanotransduction, TRP channels, auditory brainstem response (ABR)
Citation: Morgan CP, Zhao H, LeMasurier M, Xiong W, Pan B, Kazmierczak P, Avenarius MR, Bateschell M, Larisch R, Ricci AJ, Müller U and Barr-Gillespie PG (2018) TRPV6, TRPM6 and TRPM7 Do Not Contribute to Hair-Cell Mechanotransduction. Front. Cell. Neurosci. 12:41. doi: 10.3389/fncel.2018.00041
Received: 13 December 2017; Accepted: 01 February 2018;
Published: 20 February 2018.
Edited by:
David Z. He, Creighton University School of Medicine, United StatesReviewed by:
Corne Kros, University of Sussex, United KingdomCopyright © 2018 Morgan, Zhao, LeMasurier, Xiong, Pan, Kazmierczak, Avenarius, Bateschell, Larisch, Ricci, Müller and Barr-Gillespie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter G. Barr-Gillespie, Z2lsbGVzcHBAb2hzdS5lZHU=
†These authors have contributed equally to this work.
‡ Present address: Ulrich Müller, Department of Neuroscience, School of Medicine, Johns Hopkins University, Baltimore, MD, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.