
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 24 November 2017
Sec. Cellular Neurophysiology
Volume 11 - 2017 | https://doi.org/10.3389/fncel.2017.00374
This article is part of the Research Topic Neuron-Astrocyte Communication at Synapses and Circuits View all 9 articles
Synaptic connections between individual nerve cells are fundamental to the process of information transfer and storage in the brain. Over the past decades a third key partner of the synaptic machinery has been unveiled: ultrathin processes of electrically passive astroglia which often surround pre- and postsynaptic structures. The recent advent of super-resolution (SR) microscopy has begun to uncover the dynamic nanoworld of synapses and their astroglial environment. Here we overview and discuss the current progress in our understanding of the synaptic nanoenvironment, as gleaned from the imaging methods that go beyond the diffraction limit of conventional light microscopy. We argue that such methods are essential to achieve a new level of comprehension pertinent to the principles of signal integration in the brain.
Information processing in the brain relies on synaptic connections between nerve cells. It has been, however, somewhat a revelation that the process of signal formation and handling by neural circuits also involves an intimate relationship between neurons and nearby astroglia. Initially, astrocytes were thought of as exclusively structure-supporting cells. Later, two key roles of astroglia in signal transmission have emerged, rapid uptake of neurotransmitters such as glutamate (Bergles and Jahr, 1997; Diamond, 2001), and extracellular potassium buffering (Hertz, 1965; Orkand et al., 1966). Finally, the discovery of Ca2+ waves within and among astrocytes (Cornell-Bell et al., 1990; Nedergaard, 1994; Parpura et al., 1994) has prompted the concept of astroglial excitability, hence the ability to integrate and transfer physiological signals.
Most astrocytes feature sponge-like morphology in vivo, with thousands of fine protrusions filling up the extracellular space, whereas individual cells occupy adjacent, non-overlapping tissue territories (Bushong et al., 2004; Halassa et al., 2007). At many excitatory synapses in the brain, fine perisynaptic astrocytic processes (PAPs) are in close contact with pre- or postsynaptic specializations. It has been found that PAPs and synapses are in a continued exchange of molecular signals regulating synaptic transmission and its use-dependent plasticity (for recent reviews see Hamilton and Attwell, 2010; Araque et al., 2014; Rusakov et al., 2014; Bazargani and Attwell, 2016). This exchange has prompted the notion of the tripartite synapse (Haydon, 2001), acknowledging astroglia as an important third partner in signal transfer between presynaptic and postsynaptic cells. More recently, this terminology has been extended to the “tetrapartite synapse” which includes the extracellular matrix (Dityatev and Rusakov, 2011) or the “quad-partite synapse” which incorporates microglial processes (Schafer et al., 2013).
The intricate interactions between synapses and astrocytes have been difficult to visualize using conventional microscopy because most astroglial compartments are smaller than the diffraction limit of light. The advance of electron microscopy (EM), and in particular when equipped with three-dimensional (3D) reconstruction from serial sections, has revealed the tripartite synapse structure in some intricate detail (Špaček, 1985; Ventura and Harris, 1999; Lehre and Rusakov, 2002; Witcher et al., 2010; Patrushev et al., 2013). Subsequent advances in immuno-EM have uncovered a plethora of signaling molecules that are located in PAPs (reviewed in Heller and Rusakov, 2015; Bazargani and Attwell, 2016). Moreover, EM studies have indicated that PAPs preferably approach thin spines rather than large ones (Medvedev et al., 2014). More recently, high-throughput 3D EM methods, such as serial blockface-scanning or focused ion beam scanning EM, have further moved the goalposts of synaptic 3D reconstruction studies, by increasing considerably (z-stack) sectioning resolution, and thus improving quantitative description of synaptic elements, compared to traditional serial-section TEM (Blazquez-Llorca et al., 2015; Bosch et al., 2015).
One important limitation of EM studies, however, is that they can only be carried out in fixed tissue preparations, thus providing correlational comparisons between different samples. In turn, existing immuno-EM methods have not been able to provide a contiguous 3D representation of the (immunogold-labeled) signaling proteins expressed in PAPs. Nonetheless, EM studies have discovered that the astroglial coverage of synapses may depend on the physiological state, local neuronal activity, induction of synaptic plasticity, or certain behaviors (Wenzel et al., 1991; Jones and Greenough, 1996; Oliet et al., 2001; Lushnikova et al., 2009; Bernardinelli et al., 2014; Ostroff et al., 2014). Subsequent studies in live tissue that employed confocal and two-photon excitation microscopy have indicated that the spatial relationship between PAPs and the adjacent synapses is indeed dynamic (Hirrlinger et al., 2004; Haber et al., 2006; Bernardinelli et al., 2014; Perez-Alvarez et al., 2014), light diffraction being the limiting factor.
More recently, the successful implementation of fluorescence recovery after photobleaching (FRAP) and single-particle tracking (SPT) techniques paved the way for monitoring dynamic changes in nanoscopic cellular compartments at super-resolution (SR), i.e., beyond the diffraction limit of conventional optical microscopy. In brief, the time course of FRAP depends on the diffusion rate (and the proportion of immobile fraction) for the fluorescent label of interest (Axelrod et al., 1976). In contrast, SPT methods rely on the labeling of individual molecules in live cells using bright fluorescent nanoparticles, such as quantum dots. The nanoscale localization and tracking of labeled molecules then proceeds through the registration of intermittent quantum dot “blinking” followed by the deconvolution of the signal point-spread function (PSF; reviewed in Choquet and Triller, 2003). Stimulated-emission depletion (STED) microscopy uses a second excitation light channel in which light forms a doughnut-shaped PSF to damp (deplete) excitation at the periphery of the first channel PSF: this narrows the emission spots enhancing resolution beyond the diffraction limit (Klar et al., 2000). In structured illumination microscopy (SIM), bar code-like excitation patterns are shifted and rotated in the excitation path, creating high-frequency information, which can be extracted using specialized algorithms to reconstruct a super-resolved image (Gustafsson, 2000; Gustafsson et al., 2008; Schermelleh et al., 2008). Single molecule localization microscopy (SMLM), which encompasses two generic approaches (and their specific applications), photo-activated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), is based on localizing the point source of fluorescence (which can be one molecule) from its detected image (Betzig et al., 2006; Rust et al., 2006; Fölling et al., 2008). Similar to the SPT technique, the exact position of the fluorescent label is reconstructed through repeated stochastic excitation of a small, sparsely distributed fraction of fluorescent molecules per imaging cycle and the subsequent PSF deconvolution.
Each of the SR techniques has certain advantages and shortcomings. For instance, resolution in STED microscopy depends directly on the intensity of the excitation and depletion laser light, which if excessive might produce phototoxic effects. Nonetheless, high imaging speed and advanced optical sectioning makes STED imaging applications feasible in live tissue. Another advantage of the technique is the applicability of common fluorescent proteins and dyes.
SIM is another highly flexible SR imaging method. It does not require specific fluorophores, and it allows for multi-color imaging in living cells and organisms. Importantly, it does not require high intensity excitation light thus carrying relatively low risk of photo-damage. However, maximal resolution of SIM is about three times lower than that of STED or SMLM.
STORM imaging has mainly been used in fixed cells and tissue, and it relies on labeling of endogenous or overexpressed proteins equipped with antibody constructs. Therefore, it requires good tissue penetration for the antibodies and dyes. Even though STORM enables multi-color imaging, it is only specific dye combinations that could work in certain buffer formulations.
In general, SMLM methods provide the greatest enhancement of resolution, which is effectively limited only by the size of the fluorophore molecule and the opto-mechanical stability of the system. Importantly, not only can various SR techniques be used in living cells (Jones et al., 2011; Owen et al., 2012; Bethge et al., 2013), they could also document the dynamics of molecular movement. One type of SMLM (sptPALM) uses the expression of a photoactivatable fluorescent protein fused to the protein of interest, thus enabling reconstruction of hundreds of molecular trajectories (Manley et al., 2008; Rossier et al., 2012). Another method, universal point accumulation for imaging in nanoscale topography (uPAINT), relies on the addition of fluorophores during the imaging process, which provides the dynamic imaging of continuously labeled, arbitrary membrane biomolecules in living cells (Giannone et al., 2010). In contrast to the nanoparticle-based SPT techniques, in which a few molecules are followed for prolonged periods of time, sptPALM and uPAINT image single-molecule trajectories at very high densities, for short periods of time (Cognet et al., 2014; Sibarita, 2014). A detailed description of these and related techniques could be found in some excellent recent reviews (Schermelleh et al., 2010; Sydor et al., 2015; Turkowyd et al., 2016; Minoshima and Kikuchi, 2017).
Our aim here is to overview and discusses some notable recent findings that took advantage of SR microscopy in visualizing synaptic and astrocytic structures in mammalian cells. We refer the reader elsewhere for the description of some important advances made in the model organism Drosophila (Liu et al., 2011; Ehmann et al., 2014; Bosch et al., 2015; Böhme et al., 2016; Bademosi et al., 2017). Similarly, in-depth reviews focusing on the synaptic receptor machinery, as revealed with SR microscopy, could be found in the recent literature (Ribrault et al., 2011; Czöndör et al., 2012; Choquet and Triller, 2013; Compans et al., 2016). Our focus here will be on the progress in our understanding of the relationship between molecular machineries acting at the presynaptic terminal, the postsynaptic dendritic spine, and the perisynaptic astroglia—as being discovered with SR methods. The rationale behind this approach is to bring together and illustrate SR-generated data coming from inter-disciplinary research groups that do not always communicate among themselves and have not always been directly related to synaptic physiology research.
The presynaptic specialization, or the “presynapse”, is a crowded yet highly dynamic axonal compartment, in which the finely tuned regulation of molecular interactions is essential for the function and integrity of synapses. Some key aspects pertinent to the molecular organization of the presynapse have been revealed using EM and mass spectrometry (Südhof, 2006; Takamori et al., 2006) and, more recently, with STED (Figure 1A; Wilhelm et al., 2014). The essential functions of presynapses include vesicular release and recycling of neurotransmitters, of which some critical aspects remain under debate. Clearly, single molecule imaging techniques could provide important clues in this regard (Rizzoli, 2014; Maidorn et al., 2016).
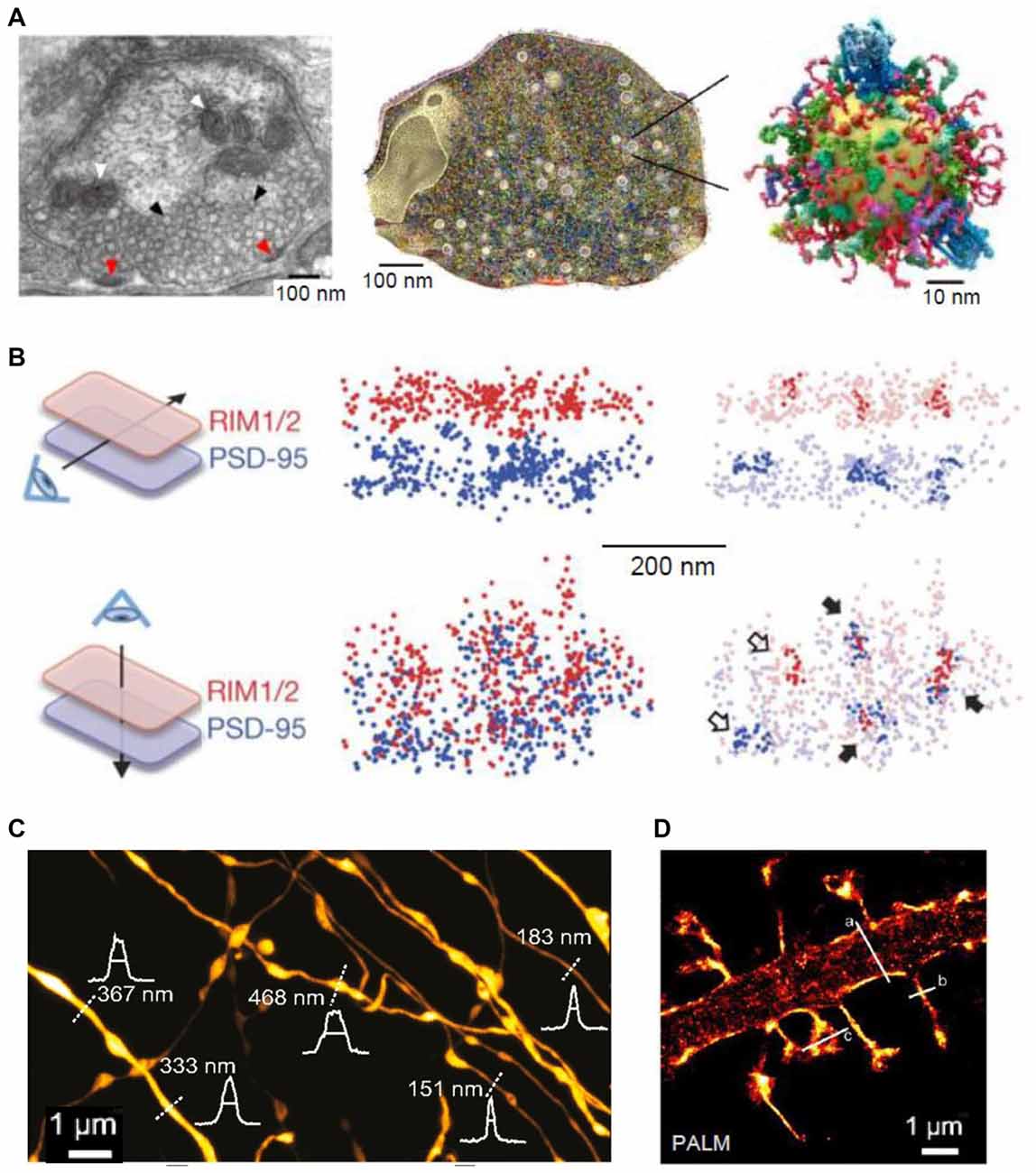
Figure 1. Super-resolution (SR) imaging of pre- and postsynaptic connections. (A) Left: electron micrograph depicting the ultrastructure of a presynaptic bouton; mitochondria (white arrowheads) appear positioned near the synaptic vesicles (black arrowheads). Fusion of synaptic vesicles takes place at the active zone (red arrowheads). Middle: three-dimensional (3D) reconstruction of the presynapse considering the copy numbers and localization of 60 synaptic proteins. Right: illustration of one synaptic vesicle; modified from Maidorn et al. (2016) with permission. (B) Trans-synaptic nanoscale alignment of active zone and postsynaptic density (PSD) proteins. Distributions of synaptic RIM1/2 and PSD-95 pair as the original localizations (left) and with nanoclusters highlighted (right); filled arrows, aligned nanoclusters; open arrows, non-aligned nanoclusters; modified from Tang et al. (2016) with permission. (C) Stimulated-emission depletion (STED) image of axons in the stratum radiatum in the CA1 area. Intensity profiles (white) and FWHM measurements (straight lines) of axon and bouton diameters were taken at the locations indicated by the white dotted lines; modified from Chéreau et al. (2017) with permission. (D) Photo-activated localization microscopy (PALM) of dendritic spines, acquired with a frame rate of 50 ms (8000 frames); modified from Izeddin et al. (2011) with permission.
Various SR techniques have been used to image synaptic vesicles, their molecular composition and recycling mechanisms (Willig et al., 2006; Westphal et al., 2008; Kamin et al., 2010; Lehmann et al., 2015). STED imaging has revealed that synaptotagmin molecules remain clustered in isolated presynaptic membrane patches after synaptic vesicle fusion, suggesting that at least some vesicle constituents remain together during recycling (Willig et al., 2006). Another application of STED imaging showed that the plasma membrane SNAREs form clusters of ~60 nm diameter containing about 75 densely packed molecules in PC12 cells (Sieber et al., 2007; Maglione and Sigrist, 2013). FRAP experiments detected freely diffusing molecules that dynamically move between such clusters (Sieber et al., 2007). Direct STORM (dSTORM) imaging showed that, indeed, unclustered syntaxin as well as SNAP-25 molecules reside next to these clusters in PC12 cells (Bar-On et al., 2012). Furthermore, it has been shown that subsequent stimulation leads to endocytosis of a “readily retrievable” pool of synaptic vesicles (Hua et al., 2011). Assemblies of synaptotagmin molecules in the plasma membrane that might control membrane integrity or serve as molecular platforms for the formation of SNARE complexes have been identified in recent studies with STED (Milovanovic and Jahn, 2015; Maidorn et al., 2016). STED was also used to visualize, for the first time, vesicular trafficking on the nanoscale in real time (Westphal et al., 2008). The buffer zone for endocytosis was also imaged in a study combining confocal and STED microscopy, mechanical force measurements, pharmacology and gene knockout to investigate the dynamic assembly of filamentous actin that mediates Ω-profile vesicle merging at synapses (Wen et al., 2016). Additionally, actin dynamics were found to modulate presynaptic structure and function by dynamically controlling the active zone organization following local neuronal activity (Glebov et al., 2017). A recent study detected individual vesicle fusion events with ~27 nm localization precision at single hippocampal synapses under physiological conditions (Maschi and Klyachko, 2017). The researchers found that multiple distinct release sites could exist within individual synapses. These sites were distributed throughout the active zone and underwent repeated reuse in a dynamic and activity-dependent manner (Maschi and Klyachko, 2017). A recent study has documented trans-synaptic nanocolumns that regulate the positioning of pre- and postsynaptic scaffolding proteins (Figure 1B; Tang et al., 2016). Therefore, one possible explanation for the distinct release sites reported recently (Maschi and Klyachko, 2017) are the nanoclusters of presynaptic proteins (especially RIM1/2) at the active zone (Tang et al., 2016). Hence, RIM1/2 clusters could be seen as markers for release sites (Tang et al., 2016).
The notion that individual presynaptic proteins form nanoclusters regulating the aforementioned nanocolumns is further supported by studies using EM and STED. Such studies reveal the clustering of voltage-gated Ca2+ channels at synapses of auditory hair cells (Frank et al., 2010) and at the neuromuscular junction in adult and aged mice (Nishimune et al., 2016), by bassoon. Moreover, a recent STED-based study found that the presynaptic clustering of active zone proteins RIM-BP2 was required for the clustering of calcium channels and bassoon at hippocampal CA3-CA1 synapses (Grauel et al., 2016).
In addition to revealing the nanoclustering of active zone organizers, SR microscopy has been employed to confirm the presence of brain-derived neurotrophic factor (BDNF) in small granules in the presynaptic face of excitatory synapses in cultured neurons, allowing for a regulated anterograde release from glutamatergic synapses (Andreska et al., 2014). Moreover, STORM imaging of hippocampal slices demonstrated that Δ9-tetrahydrocannabinol (THC) treatment induces internalization and disappearance of type 1 cannabinoid (CB1) receptors from γ-aminobutyric acid (GABA)-ergic axon terminals (Dudok et al., 2015). A combination of whole-cell recordings and STORM was employed to investigate the role of local protein synthesis in rodent hippocampal slices. They found that presynaptic protein synthesis is required for long-term, but not short-term, plasticity of GABA release from CB1-expressing axons and might play a general role in controlling long-term plasticity in the mature mammalian brain (Younts et al., 2016). Another recent study investigated the distribution of dopamine transporters in cultured dopaminergic neurons using PALM and STORM (Rahbek-Clemmensen et al., 2017). The authors have found that the transporters cluster in multiple irregular nanodomains and that the clustering is dependent on cholesterol. Furthermore, the transporter nanodomains are non-overlapping with the loci of tyrosine hydroxylase and vesicular monoamine transporter 2 in potential presynaptic transmitter release sites (Rahbek-Clemmensen et al., 2017). In addition to imaging individual molecules in presynaptic sites, STED has been employed to visualize the structure and plasticity of axon shafts and boutons in mice hippocampal sections (Figure 1C; Chéreau et al., 2017).
Both STED and SMLM have been implemented to analyze the structure of dendritic spines and the post-synapse. For example, in cultured neurons, STORM revealed the dynamics of thin filopodia and dendritic spines as well as of the endoplasmic reticulum and mitochondria (Shim et al., 2012). Additionally, STED has also been used to image the fine structure of dendritic spines (Nägerl et al., 2008; Ding et al., 2009; Tønnesen et al., 2011, 2014; Berning et al., 2012; Bethge et al., 2013). In addition to revealing macroscopic changes of dendrites and spines, STED (Urban et al., 2011; D’Este et al., 2015; Sidenstein et al., 2016) and SMLM techniques (such as sptPALM) have been used to investigate actin filaments and their dynamics in spines and dendrites and their role in spine maturation and function (Figure 1D; Tatavarty et al., 2009, 2012; Frost et al., 2010; Izeddin et al., 2011; Chazeau et al., 2014; MacGillavry et al., 2016; Wang et al., 2016). It has been found that dendritic actin polymerization is more complex and heterogeneous than previously thought, with different actin flows and random motions (Tatavarty et al., 2009; Frost et al., 2010). Moreover, enhanced actin polymerization was revealed in subdomains of the dendritic spine including the postsynaptic density (PSD) and the spine neck, which might play a role in the repositioning of glutamate receptors (Frost et al., 2010). Furthermore, the nucleation of actin extension seemed to start from a single point within the PSD, raising the possibility that changes in the environment are sensed by the actin filaments and in turn are translated into the PSD to mediate spine maturation (Izeddin et al., 2011; Chazeau et al., 2014). Among the organizing proteins of mature spine structure is synaptopodin, which exerts an indirect effect, via F-actin, on the diffusion of membrane proteins in the spine neck (Wang et al., 2016). The lattice-like structures of filamentous actin were also imaged in vivo, revealing a structure similar to that seen in vitro (Berning et al., 2012; Willig et al., 2014). A recent study used a far-red emitting fluorescent protein, mNeptune2, as a STED probe and achieved to resolve actin filaments in the living brain at a resolution of ~80 nm (Blazquez-Llorca et al., 2015). The researchers did not find any phototoxicity, confirming the advantage of far-red light for in vivo applications (Blazquez-Llorca et al., 2015). Altogether, SR techniques have substantially extended our view of the organization of the spine cytoskeleton revealing a much more crowded and organized cell compartment than previously thought (MacGillavry and Hoogenraad, 2015; Chazeau and Giannone, 2016).
In addition to the macroscopic structure of spines and the dynamics of their cytoskeleton, SR techniques have helped to decipher the nanoscopic organization of the PSD. The first study investigating the nanoscale structure of excitatory synapses was performed in fixed rat brain tissue (Dani et al., 2010). The group studied the nanoscopic distribution of pre- and postsynaptic scaffolding proteins and established a detailed 3D map of the synapses. Furthermore, they demonstrated variations in the organization of N-methyl-D-aspartate receptors (NMDARs) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) across the main olfactory bulb (MOB) and the accessory OB (AOB; Dani et al., 2010). The researchers found an increased ratio of the NMDAR subunit NR2B to the AMPAR subunit GluR1 in the AOB, possibly representing a larger number of “immature” synapses and hence suggesting the potential for significant activity-dependent plasticity in the adult AOB. The authors confirmed this hypothesis using optogenetics to specifically mimic vomeronasal organ (VMO) activation, also detecting significant synapse maturation (an increase in the synaptic content of GluR1 and Homer1 and a moderate decrease in NR2B) upon light stimulation (Dani et al., 2010). The activity of AMPARs is key to memory formation and synaptic plasticity, and their insertion into and removal from synapses are tightly regulated (Newpher and Ehlers, 2008; MacGillavry et al., 2011). In 2013, three independent groups used different SR methods to discover that at excitatory synapses AMPARs accumulate into nanoscopic clusters (Fukata et al., 2013; MacGillavry et al., 2013; Nair et al., 2013). One group used PALM of several postsynaptic scaffolding molecules such as PSD95 and Homer1, as well as dSTORM of AMPARs and NMDARs in live and fixed neuronal cultures (MacGillavry et al., 2013). Another group employed a combination of techniques in live cultures investigating the dynamics of palmitoylated PSD95 (Fukata et al., 2013). The researchers used FRAP and PALM and found that the PSD was organized through subdomains of PSD95, which in turn are regulated through DHHC2-dependent palmitoylation (Fukata et al., 2013). Finally, uPAINT, sptPALM, dSTORM and STED were used to discover that AMPARs are immobile at nanodomains but diffuse freely between them, which could be due to the nanoorganization of PSD95 scaffolds (Nair et al., 2013). All the groups have concluded that AMPARs are organized at synapses in nanoclusters that are much smaller than the PSD (0–4 AMPAR nanodomains per PSD, with an average size of ~80 nm; Compans et al., 2016). Furthermore, the number of the AMPAR nanodomains depended on the size of the underlying PSD. The average center-to-center distance between the nanodomains is 500 nm suggesting that a single glutamate release cannot act on more than one domain (Nair et al., 2013). In these studies, dSTORM data revealed that 20–25 AMPARs are located in each nanodomain.
The organization of PSD95 molecules within the PSD was investigated further. The results revealed that, even though PSD95 can be found throughout the PSD, this protein, too, clusters in nanodomains of ~150 nm in diameter (Fukata et al., 2013; MacGillavry et al., 2013; Nair et al., 2013). These PSD95 nanodomains were also found in brain slices (Broadhead et al., 2016; Tang et al., 2016). Using a single transmembrane domain with a PDZ binding motif, the impact of the tight protein packing within the PSD was investigated leading to a conclusion that both crowding and binding dynamics limit diffusion and escape of AMPARs from the synapse (Li et al., 2016). Blocking the interaction of AMPARs with stargazin, which regulates AMPAR trafficking and gating, increased the rate of AMPAR diffusion as measured with sptPALM but revealed an interaction of the receptors with other proteins outside of the PSD (Hoze et al., 2012). Even though the diffusion properties of neuronal receptors had been investigated already in live cells using nanoparticle-based SPT, these novel studies were the first to describe the nanoorganization of AMPARs and of scaffolding proteins at PSDs.
The nanoorganization and its dynamic rearrangement opposing similar nanostructures of presynaptic priming and docking factors such as RIM1/2 was later confirmed in the aforementioned study by Tang et al. (2016). A similar alignment was seen at the neuromuscular junction in mice (York and Zheng, 2017). Using SIM and STORM, the authors located acetylcholine receptors to the edges of crests surrounding the opening of folds, therefore displaying a trans-synaptic alignment with the presynaptic active zones and enabling effective synaptic transmission (York and Zheng, 2017).
STED and SMLM have also been applied to assess the synaptic localization and the dynamics of other receptors, channels and scaffolding proteins. For example, STORM revealed that the A-kinase-anchoring protein 79/150 prompts the clustering of different ion channels (M-type K+, L-type Ca2+ and TRPV1) into functional domains, together with G protein-coupled receptors (Zhang et al., 2016). STED was used to analyze the size distribution, the association into nanoclusters and the long-range interactions of nicotinic acetyl-choline receptor nanoclusters and their cholesterol dependence (Kellner et al., 2007). The number and the nanoorganization of the Na+K+-ATPase at excitatory synapses, as well as clustering and colocalization with dopamine D1 receptor and DARPP-32, were assessed using STED and PALM (Blom et al., 2011, 2012, 2013, 2016). It has been found that the P2X7 receptor is stabilized in nanoclusters of ~100 nm diameter and that its trafficking shows confinement in synaptic areas (Shrivastava et al., 2013). Moreover, STED and sptPALM were employed to study the surface dynamics of calcium channel subunits in cultured cells (Voigt et al., 2016). A novel monomeric streptavidin dSTORM approach was employed to investigate the relative distribution of presynaptic neurexins (Nrx) 1β and its postsynaptic binding partners neuroligin (Nlg) 1 and LRRTM2 in primary hippocampal neurons (Chamma et al., 2016a,b).
The sub-synaptic distribution of molecular components in inhibitory (GABA-ergic) synapses has also been visualized using SR imaging techniques. For instance, quantitative 3D-PALM analysis has revealed the gephyrin organization in inhibitory synapses in cultured spinal cord neurons: the ensuring calculations estimated the postsynaptic clusters of 40–500 gephyrin molecules, packed at a density of about 5000 molecules/μm2, whereas synapses in situ contained about three times as many molecules and were packed more densely (Specht et al., 2013). The same group recently used quantitative PALM to show that alpha-1 and alpha-3 containing glycine receptors display the same α3:β2 stoichiometry and gephyrin binding (Patrizio et al., 2017). However, the authors found that alpha-1 containing receptors are more mobile and less densely packed (1100 molecules/μm2 vs. 1500 molecules/μm2; alpha-3 containing receptors) and that their number at synapses was reduced in response to interleukin 1β. This highlights an alpha subunit-specific mechanism of receptor-gephyrin binding as well as a cell-type dependent regulation of glycinergic currents (Patrizio et al., 2017). The nanoorganization of the postsynaptic scaffolding proteins gephyrin and PSD95 in cultured hippocampal neurons was suggested using STED imaging (Dzyubenko et al., 2016). The researchers counted nanodomains of PSD95 and gephyrin and found that their number is significantly increased when the postsynaptic puncta are paired with presynaptic puncta compared to unpaired clusters (Dzyubenko et al., 2016). Another elegant study combined STORM with array tomography to reconstruct the entire inhibitory synaptic input field of individual retinal ganglion and amacrine cells in mice and determined the spatial distribution and neurotransmitter receptor identity of the synapses therein (Sigal et al., 2015). Whilst such SR approaches cannot achieve spatial resolution of EM, they could reveal the 3D distribution of individual protein molecules on a larger, contiguous scale, the task unattainable by EM techniques.
The visualization of live astrocytic processes and their signaling protein complexes has been technically challenging, mainly due to their nanoscopic size and the fact that their spatial separation could also be nanoscopic (Rusakov, 2015), as evidenced by the visualization of 3D-reconstructed images of perisynaptic astroglia (Figure 2A). Therefore, even though substantial advances have been made in the imaging of synaptic molecules, little is known about the molecular nanostructure of synaptic microenvironment associated with astroglia. Astrocyte cultures have been used to visualize cytoskeletal and receptor localization (reviewed in Heller and Rusakov, 2015) and to correlate subcellular cytoskeletal organization with cell morphology and membrane stiffness (Curry et al., 2017). The localization of the endoplasmic reticulum and mitochondria in live cultured cortical astrocytes has been assessed using SIM revealing their dynamic arrangements close to the basal plasma membrane, across the whole cultured cell (Brunstein et al., 2013). Implementing PALM and dSTORM has revealed the clustering behavior of AQP4 and Kir4.1 in cultured cells as well as in brain sections (Rossi et al., 2012; Smith A. J. et al., 2014; Smith and Verkman, 2015). Some ubiquitous astrocytic metabolic proteins, such as glutamine synthetase and S100β, have been used to reveal the nanoscopic structure of thin astroglial processes in fixed cultured cells and brain sections with STED and dSTORM (Volterra et al., 2014; Heller et al., 2017). Similarly, genetically-encoded fluorescent proteins that fill whole cells were used to dissect tripartite synapses with the help of STED (Figure 2B; Panatier et al., 2014). Another approach achieved near SR resolution using the Zeiss Airyscan method to reveal thin astrocytic processes in cleared tissue (Miller and Rothstein, 2016). A combination of EM and STED confirmed the presence of astrocytic and NG2 cell processes at nodes of Ranvier (Serwanski et al., 2017).
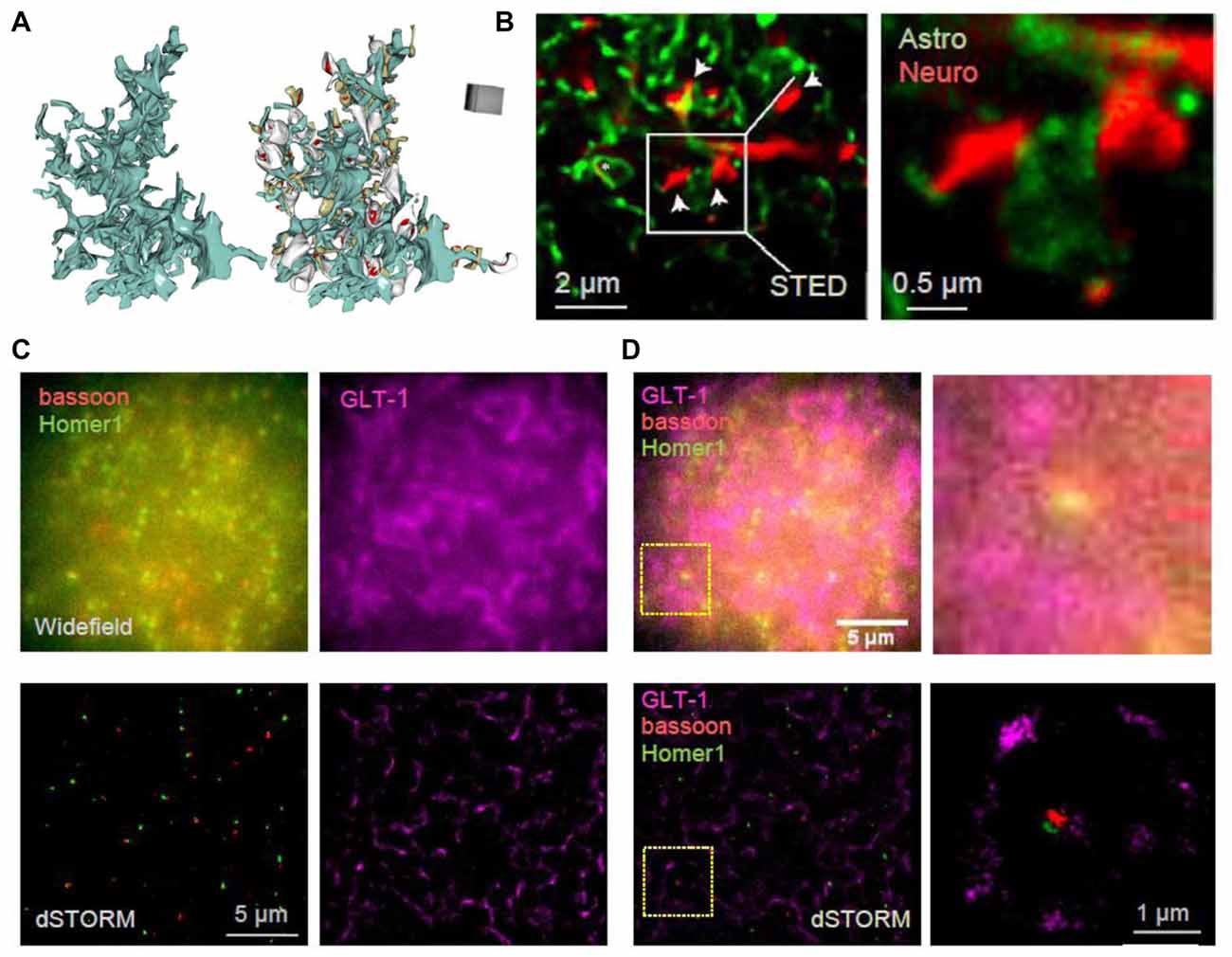
Figure 2. Towards deciphering the nanoscale architecture of tripartite synapses. (A) 3D reconstruction of an astrocyte fragment (blue) shown with and without adjacent thin (gray) and mushroom (dark yellow) dendritic spines equipped with PSDs (red); adapted from Medvedev et al. (2014) with permission. (B) STED images showing CA1 stratum radiatum astrocytic processes (green) adjacent to synaptic structures in organotypic slices (red; dendritic spines, arrows) at lower (left) and higher (right) magnification; square (left), magnified area; modified from Panatier et al. (2014) with permission. (C) In-house example: widefield (top) and direct STORM (dSTORM; bottom) images of bassoon (CF568-tagged, red), Homer1 (Atto488-tagged, green) and GLT-1 (Alexa647-tagged, magenta) of a 20 × 20 μm ROI in hippocampal area CA1; 40 μm thick hippocampal sections (500 g male rat); photoswitching buffer containing 100 mM cysteamine and oxygen scavengers (glucose oxidase and catalase; Metcalf et al., 2013); recorded with a Vutara 350 microscope (Bruker Corp., Billerica, MA, USA). (D) Same example as in (C) but shown as combined three-color widefield (top) and dSTORM (bottom) images, at lower (left) and higher (right) magnification; dotted squares (left), magnified area.
Intriguingly, two recent studies asked whether SR microscopy was technically feasible or advantageous when applied to cryo-cut human brain tissue. The first study revealed serotonin receptor clusters using a dSTORM approach on neuronal and astrocytic elements (Jacak et al., 2015). The second study compared STORM and SR optical fluctuation imaging (SOFI) as means to reveal axons and astrocytic processes (Hainsworth et al., 2017). It was found that STORM underestimated the width of axons and astrocytic processes and even though SOFI has lower spatial resolution it provided more accurate width measurements (Hainsworth et al., 2017). A recent elegant study investigated local translation in astrocytic processes (Sakers et al., 2017) using STORM to illuminate ribosomes in the close proximity of synapses where the locally produced proteins may alter synapse plasticity.
Imaging the tripartite synapses in fixed brain sections using dSTORM has also been progressing. Three-color 3D dSTORM could reveal the positioning of glial glutamate transporter GLT-1 around excitatory synapses delineated by the tight pre- and postsynaptic clusters of bassoon and Homer1, respectively (Figures 2C,D). In addition to imaging vesicle dynamics in presynaptic structures, SR methods have been used to assess the merging of vesicles with the plasma membrane in astrocytes, revealing similar release mechanisms (Singh et al., 2014; Li et al., 2015; Guček et al., 2016; Jorgačevski et al., 2017). Recently, SIM and high-resolution cell-attached capacitance measurements have been used to show that dynamin regulates fusion pore geometry and kinetics of endo- and exocytotic vesicles in cultured astrocytes (Lasic et al., 2017).
Clearly, electrophysiological or optical readout of synaptic function is essential for achieving reliable interpretation of SMLM data pertinent to the molecular dynamics of synaptic (and extra-synaptic) components (Hosy et al., 2014; Compans et al., 2016). One of the earliest several recent studies have assessed dynamic changes of receptor clustering upon synaptic stimulation. Following induction of long-term potentiation (LTP) of inhibitory transmission (iLTP), gephyrin was found to accumulate at inhibitory synapses leading to an increased number of synaptic GABAA receptors (Petrini et al., 2014; Pennacchietti et al., 2017). The authors found that the nanoscale organization of inhibitory synaptic proteins determined inhibitory synaptic plasticity, thus adding a further level of complexity to the regulation of the neuronal network activity by plastic inhibitory synaptic signals (Pennacchietti et al., 2017). PALM and STORM were used to investigate the trafficking of CamKII in spines and revealed that the protein exists in three kinetic states, slow (interaction with immobile substrates), intermediate (binding to actin), and fast (CamKII alone), whereas NMDAR activation slowed down diffusion rates (Lu et al., 2014). Furthermore, it was found that the phosphorylation of AMPARs by CamKII was crucial for P2X2-mediated AMPAR internalization and ATP-driven synaptic depression (Pougnet et al., 2014, 2016).
SR was used to discover that ankyrin-G accumulates in dendritic spines after induction of chemical LTP and that its knockdown prevents spine head enlargement (Smith K. R. et al., 2014). Chemical LTP also induces a matrix metalloproteinase-dependent enlargement of a subset of small spines and the immobilization, synaptic accumulation and clustering of AMPARs at dendritic spines (Szepesi et al., 2014). Interestingly, the induction of long-term depression (LTD) led to an increased nanodomain size of the cell adhesion molecule SynCAM 1 (Perez De Arce et al., 2015), and ephrin B3 was found to regulate the localization of PSD95 to stable synapses and in stable nanodomains (Hruska et al., 2015). Interestingly, presynaptic RIM1/2 molecules cluster in a similar way to postsynaptic PSD95 molecules, and both nanodomains co-align across the synaptic cleft (Figure 1B; Tang et al., 2016). Furthermore, after LTP induction these nanocolumns remain, with an increase in PSD95 clusters (Figures 3A,B), thus providing further evidence for the trans-synaptic organization of scaffolding molecules. The mechanisms that mediate this clustering are yet to be discovered. A recent elegant study combined PALM and optogenetics to control the insertion of PSD95 molecules and hence AMPARs into established PSDs (Sinnen et al., 2017). It appears that simple insertion of the receptors was not enough to drive synaptic potentiation. Therefore, more complex changes in the nanodomain composition and characteristics are required for spine maturation and the gating of synaptic strength. STED has been used mainly to visualize the morphology of neuronal structures and how they change upon plasticity. For example, STED revealed an increased spine head volume and a decreased spine neck length after LTP induction (Figures 3C,D; Tønnesen et al., 2014). Furthermore, STED imaging has been used to investigate morphological plasticity of synaptic boutons and axon shafts which dynamically fine-tune action potential conduction velocity (Figures 1C, 3E,F; Chéreau et al., 2017). Another study used a combination of AMPAR immobilization approaches together with FRAP and uPAINT to evaluate the impact of AMPAR surface diffusion on hippocampal LTP and contextual learning (Penn et al., 2017). The authors have shown, in brain slices and in vivo, that AMPAR lateral diffusion and synaptic trapping is an integral part of the early phases of hippocampal-dependent fear learning (Penn et al., 2017).
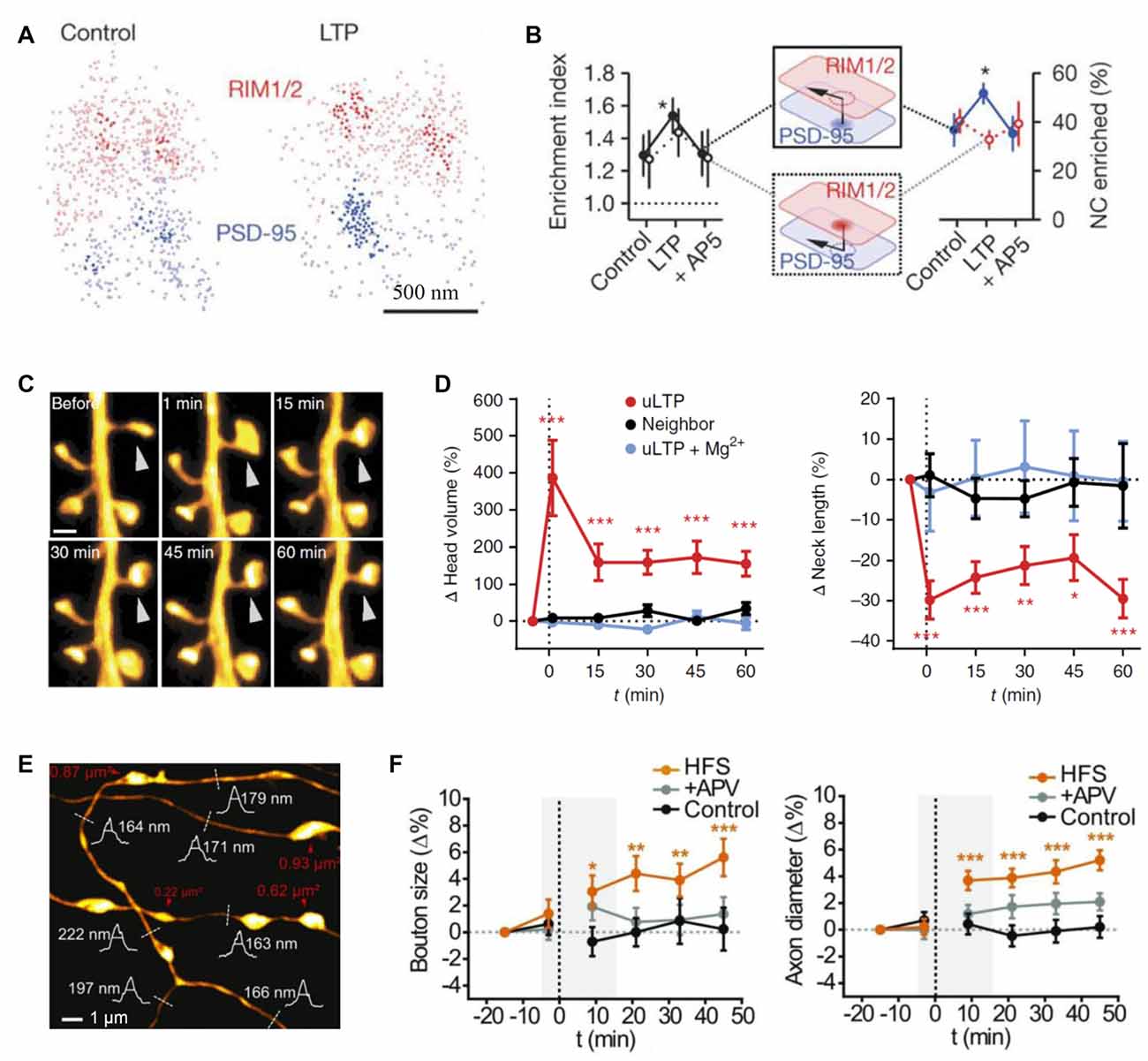
Figure 3. Probing synaptic plasticity on the nanoscale. (A) Space distributions of synaptic RIM1/2 and PSD-95 proteins, in control and post-long-term potentiation (LTP) conditions, with nanoclusters highlighted. (B) Across-condition comparison of enrichment index and the percentage of nanoclusters enriched (n = 45, 87 and 42 synapses for control, LTP and AP5, respectively); modified from Tang et al. (2016) with permission. (C) Monitoring dendritic spine morphology following glutamate uncaging-induced LTP (uLTP) protocol; arrowhead, uncaging spot. (D) The dynamics of spine head size (left) and the neck length (right) following uLTP protocol; modified from Tønnesen et al. (2014) with permission. (E) An example of stimulated-emission depletion (STED) imaging employed to monitor long-term dynamics of axonal morphology following high frequency stimulation (HFS, a protocol to induce LTP). (F) Time source of the (normalized) axonal bouton size (left) and axon diameter (right) in three conditions: control, HFS and APV (N-methyl-D-aspartate receptors, NMDA receptor blockade); dotted line, HFS onset; gray shadows, time window of short-term observations (as in Figure 2B); modified from Chéreau et al. (2017) with permission. *p < 0.5, **p < 0.1, ***p < 0.01.
It appears that only one study to date has used SR microscopy to investigate astrocyte dynamic during plasticity conditions. The researchers evaluated the role of BDNF as a gliotransmitter in vivo using EM and SIM in combination with LTP-induction (Vignoli et al., 2016). It was found that BDNF recycling by glial cells was required for memory consolidation, the latter being followed by highly localized TrkB phosphorylation on adjacent neurons (Vignoli et al., 2016).
There have been a growing number of elegant studies that employed SR technology to shed light on the nanoscopic events pertinent to synaptic function and plasticity. However, the majority of them have been conducted either in cultured cells or in fixed brain sections. Whilst these data have moved the field forward, the goal post is now to achieve SR imaging in organized, preferably live, brain tissue. This is particularly important for astrocytes whose morphology in culture is drastically different from that in situ.
In this respect, STED imaging has been ahead of the field providing live imaging in situ (Urban et al., 2011; Berning et al., 2012; Gao et al., 2012; Chen et al., 2014; Panatier et al., 2014; Chéreau et al., 2017) including intact animals (Berning et al., 2012; Willig et al., 2014; Blazquez-Llorca et al., 2015). Clearly, achieving reliable SR results in situ faces technical challenges, such as restricted optical access, sample movement, or optical aberrations induced by the specimen. In addition, organized brain tissue is packed with nanoscopic cellular compartments of neuronal or astroglial origin that have to be optically separated from the structures of interest. One possible way towards overcoming these difficulties is adaptive optics (Booth et al., 2015), which improves the signal-to-noise ratio, axial resolution, and depth penetration as shown using in vivo two-photon excitation imaging (Rueckel et al., 2006; Ji et al., 2012; Wang et al., 2014) or STED (Gould et al., 2012; Patton et al., 2016). In parallel, further improvements in performing multi-color imaging are needed, which depends on the progress in achieving the required spectral properties of fluorescent labels and the optics equipment and analyses involved. So far only large cellular structures such as dendritic spines and their actin dynamics at a resolution of just under 100 nm have been visualized in the living brain (Berning et al., 2012; Willig et al., 2014; Blazquez-Llorca et al., 2015). Nevertheless, advances in reducing phototoxicity have enabled longer-term imaging. One common drawback of these approaches is that it is difficult to label endogenous proteins in situ, due to the limited access, which prompted the use of overexpressed proteins.
At present, 3D SR microscopy can be employed efficiently in many laboratories using custom build or commercial microscopes helping to reveal structures that are beyond the diffraction limit of light (Huang et al., 2008; Juette et al., 2008; Heller et al., 2017). The development and adaption of novel SR techniques such as qPAINT, which permits quantitative SR imaging in cellular structures (Jungmann et al., 2016), integrating exchangeable single-molecule localization (IRIS; Kiuchi et al., 2015) and 4Pi single-molecule switching nanoscopy, which allows volumetric reconstruction with 10- to 20-nm isotropic resolution of ~10-μm-thick samples (Huang et al., 2016), should increase the available resolution even further. The development of novel labeling techniques and probes is an important factor in improving resolution. For example, the use of photoactivatable, genetically encoded calcium indicators (Berlin et al., 2015) opens a door into nanoscopic calcium events in the cells studied. Similarly, the use of smaller labeling probes such as nanobodies (Pleiner et al., 2015) and aptamers (de Castro et al., 2016), monomeric streptavidin (Chamma et al., 2017), or the pore-forming bacterial toxin streptolysin O to label intracellular proteins in living mammalian cells for SR microscopy (Teng et al., 2016, 2017) should accelerate this progress. In parallel, the recent invention of other methods such as expansion microscopy (Chen et al., 2015; Chozinski et al., 2016; Tillberg et al., 2016) should make SR probing more affordable for many laboratories. Finally, SR imaging has already been applied to investigate established models of neurological disease, such as Alzheimer’s (Šišková et al., 2014; Schedin-Weiss et al., 2016), intellectual disabilities (Wijetunge et al., 2014; Barnes et al., 2015), amyotrophic lateral sclerosis, and frontotemporal dementia (Schoen et al., 2016). The new discoveries in SR should certainly help to translate these findings into the quest for the efficient therapeutic intervention.
The continued emergence of SR techniques has been opening new horizons in our exploration of the perisynaptic nano-world. Whilst the traditional EM methods have been able to reveal the fine architectural features of synaptic coverage by PAPs, SR can provide us with unique tools to answer some fundamental questions pertinent to the synaptic machinery and its microenvironment. How are the key signaling molecules (neurotransmitter transporters, potassium channels, aquaporins, ion exchangers, etc.) distributed in neuronal and astroglial membranes outside synapses? Do PAPs and their protein complexes re-arrange upon neural activity and/or the induction of synaptic plasticity? From what perisynaptic astroglial loci are gliotransmitters released? Where do mitochondria and internal calcium stores, if any, occur in PAPs? Does the synaptic nano-architecture reflect functional identities of individual synapses? Answering these and related questions should give us a previously unattainable quantitative basis to decipher molecular mechanisms acting at and around synapses, at the level of biophysical interactions among individual molecules.
JPH and DAR wrote the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the Wellcome Trust (Principal Fellowship 101896), European Research Council (AdG 323113-NETSIGNAL; PoC 767372-NEUROCLOUD), European Commission FP7 ITN 606950 EXTRABRAIN, Russian Science Foundation (15-14-30000, EM reconstructions) (all DAR).
Andreska, T., Aufmkolk, S., Sauer, M., and Blum, R. (2014). High abundance of BDNF within glutamatergic presynapses of cultured hippocampal neurons. Front. Cell. Neurosci. 8:107. doi: 10.3389/fncel.2014.00107
Araque, A., Carmignoto, G., Haydon, P. G., Oliet, S. H., Robitaille, R., and Volterra, A. (2014). Gliotransmitters travel in time and space. Neuron 81, 728–739. doi: 10.1016/j.neuron.2014.02.007
Axelrod, D., Koppel, D. E., Schlessinger, J., Elson, E., and Webb, W. W. (1976). Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16, 1055–1069. doi: 10.1016/s0006-3495(76)85755-4
Bademosi, A. T., Lauwers, E., Padmanabhan, P., Odierna, L., Chai, Y. J., Papadopulos, A., et al. (2017). In vivo single-molecule imaging of syntaxin1A reveals polyphosphoinositide- and activity-dependent trapping in presynaptic nanoclusters. Nat. Commun. 8:13660. doi: 10.1038/ncomms14492
Barnes, S. A., Wijetunge, L. S., Jackson, A. D., Katsanevaki, D., Osterweil, E. K., Komiyama, N. H., et al. (2015). Convergence of hippocampal pathophysiology in syngap+/− and Fmr1−/y mice. J. Neurosci. 35, 15073–15081. doi: 10.1523/JNEUROSCI.1087-15.2015
Bar-On, D., Wolter, S., van de Linde, S., Heilemann, M., Nudelman, G., Nachliel, E., et al. (2012). Super-resolution imaging reveals the internal architecture of nano-sized syntaxin clusters. J. Biol. Chem. 287, 27158–27167. doi: 10.1074/jbc.M112.353250
Bazargani, N., and Attwell, D. (2016). Astrocyte calcium signaling: the third wave. Nat. Neurosci. 19, 182–189. doi: 10.1038/nn.4201
Bergles, D. E., and Jahr, C. E. (1997). Synaptic activation of glutamate transporters in hippocampal astrocytes. Neuron 19, 1297–1308. doi: 10.1016/s0896-6273(00)80420-1
Berlin, S., Carroll, E. C., Newman, Z. L., Okada, H. O., Quinn, C. M., Kallman, B., et al. (2015). Photoactivatable genetically encoded calcium indicators for targeted neuronal imaging. Nat. Methods 12, 852–858. doi: 10.1038/nmeth.3480
Bernardinelli, Y., Randall, J., Janett, E., Nikonenko, I., König, S., Jones, E. V., et al. (2014). Activity-dependent structural plasticity of perisynaptic astrocytic domains promotes excitatory synapse stability. Curr. Biol. 24, 1679–1688. doi: 10.1016/j.cub.2014.06.025
Berning, S., Willig, K. I., Steffens, H., Dibaj, P., and Hell, S. W. (2012). Nanoscopy in a living mouse brain. Science 335:551. doi: 10.1126/science.1215369
Bethge, P., Chéreau, R., Avignone, E., Marsicano, G., and Nägerl, U. V. (2013). Two-photon excitation STED microscopy in two colors in acute brain slices. Biophys. J. 104, 778–785. doi: 10.1016/j.bpj.2012.12.054
Betzig, E., Patterson, G. H., Sougrat, R., Lindwasser, O. W., Olenych, S., Bonifacino, J. S., et al. (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645. doi: 10.1126/science.1127344
Blazquez-Llorca, L., Hummel, E., Zimmerman, H., Zou, C., Burgold, S., Rietdorf, J., et al. (2015). Correlation of two-photon in vivo imaging and FIB/SEM microscopy. J. Microsc. 259, 129–136. doi: 10.1111/jmi.12231
Blom, H., Bernhem, K., and Brismar, H. (2016). Sodium pump organization in dendritic spines. Neurophotonics 3:041803. doi: 10.1117/1.NPh.3.4.041803
Blom, H., Rönnlund, D., Scott, L., Spicarova, Z., Rantanen, V., Widengren, J., et al. (2012). Nearest neighbor analysis of dopamine D1 receptors and Na+-K+-ATPases in dendritic spines dissected by STED microscopy. Microsc. Res. Tech. 75, 220–228. doi: 10.1002/jemt.21046
Blom, H., Rönnlund, D., Scott, L., Spicarova, Z., Widengren, J., Bondar, A., et al. (2011). Spatial distribution of Na+-K+-ATPase in dendritic spines dissected by nanoscale superresolution STED microscopy. BMC Neurosci. 12:16. doi: 10.1186/1471-2202-12-16
Blom, H., Ronnlund, D., Scott, L., Westin, L., Widengren, J., Aperia, A., et al. (2013). Spatial distribution of DARPP-32 in dendritic spines. PLoS One 8:e75155. doi: 10.1371/journal.pone.0075155
Böhme, M. A., Beis, C., Reddy-Alla, S., Reynolds, E., Mampell, M. M., Grasskamp, A. T., et al. (2016). Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca2+ channel-vesicle coupling. Nat. Neurosci. 19, 1311–1320. doi: 10.1038/nn.4364
Booth, M., Andrade, D., Burke, D., Patton, B., and Zurauskas, M. (2015). Aberrations and adaptive optics in super-resolution microscopy. Microscopy 64, 251–261. doi: 10.1093/jmicro/dfv033
Bosch, C., Martinez, A., Masachs, N., Teixeira, C. M., Fernaud, I., Ulloa, F., et al. (2015). FIB/SEM technology and high-throughput 3D reconstruction of dendritic spines and synapses in GFP-labeled adult-generated neurons. Front. Neuroanat. 9:60. doi: 10.3389/fnana.2015.00060
Broadhead, M. J., Horrocks, M. H., Zhu, F., Muresan, L., Benavides-Piccione, R., Defelipe, J., et al. (2016). PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci. Rep. 6:24626. doi: 10.1038/srep24626
Brunstein, M., Wicker, K., Herault, K., Heintzmann, R., and Oheim, M. (2013). Full-field dual-color 100-nm super-resolution imaging reveals organization and dynamics of mitochondrial and ER networks. Opt. Express 21, 26162–26173. doi: 10.1364/OE.21.026162
Bushong, E. A., Martone, M. E., and Ellisman, M. H. (2004). Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. Int. J. Dev. Neurosci. 22, 73–86. doi: 10.1016/j.ijdevneu.2003.12.008
Chamma, I., Letellier, M., Butler, C., Tessier, B., Lim, K. H., Gauthereau, I., et al. (2016a). Mapping the dynamics and nanoscale organization of synaptic adhesion proteins using monomeric streptavidin. Nat. Commun. 7:10773. doi: 10.1038/ncomms10773
Chamma, I., Levet, F., Sibarita, J. B., Sainlos, M., and Thoumine, O. (2016b). Nanoscale organization of synaptic adhesion proteins revealed by single-molecule localization microscopy. Neurophotonics 3:041810. doi: 10.1117/1.nph.3.4.041810
Chamma, I., Rossier, O., Giannone, G., Thoumine, O., and Sainlos, M. (2017). Optimized labeling of membrane proteins for applications to super-resolution imaging in confined cellular environments using monomeric streptavidin. Nat. Protoc. 12, 748–763. doi: 10.1038/nprot.2017.010
Chazeau, A., and Giannone, G. (2016). Organization and dynamics of the actin cytoskeleton during dendritic spine morphological remodeling. Cell. Mol. Life Sci. 73, 3053–3073. doi: 10.1007/s00018-016-2214-1
Chazeau, A., Mehidi, A., Nair, D., Gautier, J. J., Leduc, C., Chamma, I., et al. (2014). Nanoscale segregation of actin nucleation and elongation factors determines dendritic spine protrusion. EMBO J. 33, 2745–2764. doi: 10.15252/embj.201488837
Chen, B. C., Legant, W. R., Wang, K., Shao, L., Milkie, D. E., Davidson, M. W., et al. (2014). Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346:1257998. doi: 10.1126/science.1257998
Chen, F., Tillberg, P. W., and Boyden, E. S. (2015). Optical imaging. Expansion microscopy. Science 347, 543–548. doi: 10.1126/science.1260088
Chéreau, R., Saraceno, G. E., Angibaud, J., Cattaert, D., and Nägerl, U. V. (2017). Superresolution imaging reveals activity-dependent plasticity of axon morphology linked to changes in action potential conduction velocity. Proc. Natl. Acad. Sci. U S A 114, 1401–1406. doi: 10.1073/pnas.1607541114
Choquet, D., and Triller, A. (2003). The role of receptor diffusion in the organization of the postsynaptic membrane. Nat. Rev. Neurosci. 4, 251–265. doi: 10.1038/nrn1077
Choquet, D., and Triller, A. (2013). The dynamic synapse. Neuron 80, 691–703. doi: 10.1016/j.neuron.2013.10.013
Chozinski, T. J., Halpern, A. R., Okawa, H., Kim, H. J., Tremel, G. J., Wong, R. O., et al. (2016). Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488. doi: 10.1038/nmeth.3833
Cognet, L., Leduc, C., and Lounis, B. (2014). Advances in live-cell single-particle tracking and dynamic super-resolution imaging. Curr. Opin. Chem. Biol. 20, 78–85. doi: 10.1016/j.cbpa.2014.04.015
Compans, B., Choquet, D., and Hosy, E. (2016). Review on the role of AMPA receptor nano-organization and dynamic in the properties of synaptic transmission. Neurophotonics 3:041811. doi: 10.1117/1.nph.3.4.041811
Cornell-Bell, A. H., Finkbeiner, S. M., Cooper, M. S., and Smith, S. J. (1990). Glutamate induces calcium waves in cultured astrocytes: long-range glial signaling. Science 247, 470–473. doi: 10.1126/science.1967852
Curry, N., Ghézali, G., Kaminski Schierle, G. S., Rouach, N., and Kaminski, C. F. (2017). Correlative STED and atomic force microscopy on live astrocytes reveals plasticity of cytoskeletal structure and membrane physical properties during polarized migration. Front. Cell. Neurosci. 11:104. doi: 10.3389/fncel.2017.00104
Czöndör, K., Mondin, M., Garcia, M., Heine, M., Frischknecht, R., Choquet, D., et al. (2012). Unified quantitative model of AMPA receptor trafficking at synapses. Proc. Natl. Acad. Sci. U S A 109, 3522–3527. doi: 10.1073/pnas.1109818109
Dani, A., Huang, B., Bergan, J., Dulac, C., and Zhuang, X. (2010). Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856. doi: 10.1016/j.neuron.2010.11.021
de Castro, M. A., Rammner, B., and Opazo, F. (2016). Aptamer stainings for super-resolution microscopy. Methods Mol. Biol. 1380, 197–210. doi: 10.1007/978-1-4939-3197-2_17
D’Este, E., Kamin, D., Göttfert, F., El-Hady, A., and Hell, S. W. (2015). STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep. 10, 1246–1251. doi: 10.1016/j.celrep.2015.02.007
Diamond, J. S. (2001). Neuronal glutamate transporters limit activation of NMDA receptors by neurotransmitter spillover on CA1 pyramidal cells. J. Neurosci. 21, 8328–8338.
Ding, J. B., Takasaki, K. T., and Sabatini, B. L. (2009). Supraresolution imaging in brain slices using stimulated-emission depletion two-photon laser scanning microscopy. Neuron 63, 429–437. doi: 10.1016/j.neuron.2009.07.011
Dityatev, A., and Rusakov, D. A. (2011). Molecular signals of plasticity at the tetrapartite synapse. Curr. Opin. Neurobiol. 21, 353–359. doi: 10.1016/j.conb.2010.12.006
Dudok, B., Barna, L., Ledri, M., Szabó, S. I., Szabadits, E., Pinter, B., et al. (2015). Cell-specific STORM super-resolution imaging reveals nanoscale organization of cannabinoid signaling. Nat. Neurosci. 18, 75–86. doi: 10.1038/nn.3892
Dzyubenko, E., Rozenberg, A., Hermann, D. M., and Faissner, A. (2016). Colocalization of synapse marker proteins evaluated by STED-microscopy reveals patterns of neuronal synapse distribution in vitro. J. Neurosci. Methods 273, 149–159. doi: 10.1016/j.jneumeth.2016.09.001
Ehmann, N., van de Linde, S., Alon, A., Ljaschenko, D., Keung, X. Z., Holm, T., et al. (2014). Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 5:4650. doi: 10.1038/ncomms5650
Fölling, J., Bossi, M., Bock, H., Medda, R., Wurm, C. A., Hein, B., et al. (2008). Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat. Methods 5, 943–945. doi: 10.1038/nmeth.1257
Frank, T., Rutherford, M. A., Strenzke, N., Neef, A., Pangršič, T., Khimich, D., et al. (2010). Bassoon and the synaptic ribbon organize Ca2+ channels and vesicles to add release sites and promote refilling. Neuron 68, 724–738. doi: 10.1016/j.neuron.2010.10.027
Frost, N. A., Shroff, H., Kong, H., Betzig, E., and Blanpied, T. A. (2010). Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron 67, 86–99. doi: 10.1016/j.neuron.2010.05.026
Fukata, Y., Dimitrov, A., Boncompain, G., Vielemeyer, O., Perez, F., and Fukata, M. (2013). Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J. Cell Biol. 202, 145–161. doi: 10.1083/jcb.201302071
Gao, L., Shao, L., Higgins, C. D., Poulton, J. S., Peifer, M., Davidson, M. W., et al. (2012). Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell 151, 1370–1385. doi: 10.1016/j.cell.2012.10.008
Giannone, G., Hosy, E., Levet, F., Constals, A., Schulze, K., Sobolevsky, A. I., et al. (2010). Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys. J. 99, 1303–1310. doi: 10.1016/j.bpj.2010.06.005
Glebov, O. O., Jackson, R. E., Winterflood, C. M., Owen, D. M., Barker, E. A., Doherty, P., et al. (2017). Nanoscale structural plasticity of the active zone matrix modulates presynaptic function. Cell Rep. 18, 2715–2728. doi: 10.1016/j.celrep.2017.02.064
Gould, T. J., Burke, D., Bewersdorf, J., and Booth, M. J. (2012). Adaptive optics enables 3D STED microscopy in aberrating specimens. Opt. Express 20, 20998–21009. doi: 10.1364/OE.20.020998
Grauel, M. K., Maglione, M., Reddy-Alla, S., Willmes, C. G., Brockmann, M. M., Trimbuch, T., et al. (2016). RIM-binding protein 2 regulates release probability by fine-tuning calcium channel localization at murine hippocampal synapses. Proc. Natl. Acad. Sci. U S A 113, 11615–11620. doi: 10.1073/pnas.1605256113
Guček, A., Jorgačevski, J., Singh, P., Geisler, C., Lisjak, M., Vardjan, N., et al. (2016). Dominant negative SNARE peptides stabilize the fusion pore in a narrow, release-unproductive state. Cell. Mol. Life Sci. 73, 3719–3731. doi: 10.1007/s00018-016-2213-2
Gustafsson, M. G. (2000). Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87. doi: 10.1046/j.1365-2818.2000.00710.x
Gustafsson, M. G., Shao, L., Carlton, P. M., Wang, C. J., Golubovskaya, I. N., Cande, W. Z., et al. (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970. doi: 10.1529/biophysj.107.120345
Haber, M., Zhou, L., and Murai, K. K. (2006). Cooperative astrocyte and dendritic spine dynamics at hippocampal excitatory synapses. J. Neurosci. 26, 8881–8891. doi: 10.1523/JNEUROSCI.1302-06.2006
Hainsworth, A. H., Lee, S., Foot, P., Patel, A., Poon, W. W., and Knight, A. E. (2017). Super-resolution imaging of subcortical white matter using stochastic optical reconstruction microscopy (STORM) and super-resolution optical fluctuation imaging (SOFI). Neuropathol. Appl. Neurobiol. doi: 10.1111/nan.12426 [Epub ahead of print].
Halassa, M. M., Fellin, T., Takano, H., Dong, J. H., and Haydon, P. G. (2007). Synaptic islands defined by the territory of a single astrocyte. J. Neurosci. 27, 6473–6477. doi: 10.1523/JNEUROSCI.1419-07.2007
Hamilton, N. B., and Attwell, D. (2010). Do astrocytes really exocytose neurotransmitters? Nat. Rev. Neurosci. 11, 227–238. doi: 10.1038/nrn2803
Haydon, P. G. (2001). GLIA: listening and talking to the synapse. Nat. Rev. Neurosci. 2, 185–193. doi: 10.1038/35058528
Heller, J. P., Michaluk, P., Sugao, K., and Rusakov, D. A. (2017). Probing nano-organization of astroglia with multi-color super-resolution microscopy. J. Neurosci. Res. 95, 2159–2171. doi: 10.1002/jnr.24026
Heller, J. P., and Rusakov, D. A. (2015). Morphological plasticity of astroglia: understanding synaptic microenvironment. Glia 63, 2133–2151. doi: 10.1002/glia.22821
Hertz, L. (1965). Possible role of neuroglia: a potassium-mediated neuronal—neuroglial—neuronal impulse transmission system. Nature 206, 1091–1094. doi: 10.1038/2061091a0
Hirrlinger, J., Hulsmann, S., and Kirchhoff, F. (2004). Astroglial processes show spontaneous motility at active synaptic terminals in situ. Eur. J. Neurosci. 20, 2235–2239. doi: 10.1111/j.1460-9568.2004.03689.x
Hosy, E., Butler, C., and Sibarita, J. B. (2014). Organization and dynamics of AMPA receptors inside synapses-nano-organization of AMPA receptors and main synaptic scaffolding proteins revealed by super-resolution imaging. Curr. Opin. Chem. Biol. 20, 120–126. doi: 10.1016/j.cbpa.2014.05.017
Hoze, N., Nair, D., Hosy, E., Sieben, C., Manley, S., Herrmann, A., et al. (2012). Heterogeneity of AMPA receptor trafficking and molecular interactions revealed by superresolution analysis of live cell imaging. Proc. Natl. Acad. Sci. U S A 109, 17052–17057. doi: 10.1073/pnas.1204589109
Hruska, M., Henderson, N. T., Xia, N. L., Le Marchand, S. J., and Dalva, M. B. (2015). Anchoring and synaptic stability of PSD-95 is driven by ephrin-B3. Nat. Neurosci. 18, 1594–1605. doi: 10.1038/nn.4140
Hua, Y., Sinha, R., Thiel, C. S., Schmidt, R., Hüve, J., Martens, H., et al. (2011). A readily retrievable pool of synaptic vesicles. Nat. Neurosci. 14, 833–839. doi: 10.1038/nn.2838
Huang, F., Sirinakis, G., Allgeyer, E. S., Schroeder, L. K., Duim, W. C., Kromann, E. B., et al. (2016). Ultra-high resolution 3D imaging of whole cells. Cell 166, 1028–1040. doi: 10.1016/j.cell.2016.06.016
Huang, B., Wang, W., Bates, M., and Zhuang, X. (2008). Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813. doi: 10.1126/science.1153529
Izeddin, I., Specht, C. G., Lelek, M., Darzacq, X., Triller, A., Zimmer, C., et al. (2011). Super-resolution dynamic imaging of dendritic spines using a low-affinity photoconvertible actin probe. PLoS One 6:e15611. doi: 10.1371/journal.pone.0015611
Jacak, J., Schaller, S., Borgmann, D., and Winkler, S. M. (2015). Characterization of the distance relationship between localized serotonin receptors and glia cells on fluorescence microscopy images of brain tissue. Microsc. Microanal. 21, 826–836. doi: 10.1017/s1431927615013513
Ji, N., Sato, T. R., and Betzig, E. (2012). Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc. Natl. Acad. Sci. U S A 109, 22–27. doi: 10.1073/pnas.1109202108
Jones, T. A., and Greenough, W. T. (1996). Ultrastructural evidence for increased contact between astrocytes and synapses in rats reared in a complex environment. Neurobiol. Learn. Mem. 65, 48–56. doi: 10.1006/nlme.1996.0005
Jones, S. A., Shim, S. H., He, J., and Zhuang, X. (2011). Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 8, 499–508. doi: 10.1038/nmeth.1605
Jorgačevski, J., Potokar, M., Kreft, M., Guček, A., Mothet, J. P., and Zorec, R. (2017). Astrocytic vesicle-based exocytosis in cultures and acutely isolated hippocampal rodent slices. J. Neurosci. Res. 95, 2152–2158. doi: 10.1002/jnr.24051
Juette, M. F., Gould, T. J., Lessard, M. D., Mlodzianoski, M. J., Nagpure, B. S., Bennett, B. T., et al. (2008). Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5, 527–529. doi: 10.1038/nmeth.1211
Jungmann, R., Avendano, M. S., Dai, M., Woehrstein, J. B., Agasti, S. S., Feiger, Z., et al. (2016). Quantitative super-resolution imaging with qPAINT. Nat. Methods 13, 439–442. doi: 10.1038/nmeth.3804
Kamin, D., Lauterbach, M. A., Westphal, V., Keller, J., Schonle, A., Hell, S. W., et al. (2010). High- and low-mobility stages in the synaptic vesicle cycle. Biophys. J. 99, 675–684. doi: 10.1016/j.bpj.2010.04.054
Kellner, R. R., Baier, C. J., Willig, K. I., Hell, S. W., and Barrantes, F. J. (2007). Nanoscale organization of nicotinic acetylcholine receptors revealed by stimulated emission depletion microscopy. Neuroscience 144, 135–143. doi: 10.1016/j.neuroscience.2006.08.071
Kiuchi, T., Higuchi, M., Takamura, A., Maruoka, M., and Watanabe, N. (2015). Multitarget super-resolution microscopy with high-density labeling by exchangeable probes. Nat. Methods 12, 743–746. doi: 10.1038/nmeth.3466
Klar, T. A., Jakobs, S., Dyba, M., Egner, A., and Hell, S. W. (2000). Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. U S A 97, 8206–8210. doi: 10.1073/pnas.97.15.8206
Lasic, E., Stenovec, M., Kreft, M., Robinson, P. J., and Zorec, R. (2017). Dynamin regulates the fusion pore of endo- and exocytotic vesicles as revealed by membrane capacitance measurements. Biochim. Biophys. Acta 1861, 2293–2303. doi: 10.1016/j.bbagen.2017.06.022
Lehmann, M., Gottschalk, B., Puchkov, D., Schmieder, P., Schwagerus, S., Hackenberger, C. P., et al. (2015). Multicolor caged dSTORM resolves the ultrastructure of synaptic vesicles in the brain. Angew. Chem. Int. Ed. Engl. 54, 13230–13235. doi: 10.1002/anie.201505138
Lehre, K. P., and Rusakov, D. A. (2002). Asymmetry of glia near central synapses favors presynaptically directed glutamate escape. Biophys. J. 83, 125–134. doi: 10.1016/s0006-3495(02)75154-0
Li, D., Herault, K., Zylbersztejn, K., Lauterbach, M. A., Guillon, M., Oheim, M., et al. (2015). Astrocyte VAMP3 vesicles undergo Ca2+ -independent cycling and modulate glutamate transporter trafficking. J. Physiol. 593, 2807–2832. doi: 10.1113/JP270362
Li, T. P., Song, Y., MacGillavry, H. D., Blanpied, T. A., and Raghavachari, S. (2016). Protein crowding within the postsynaptic density can impede the escape of membrane proteins. J. Neurosci. 36, 4276–4295. doi: 10.1523/JNEUROSCI.3154-15.2016
Liu, K. S., Siebert, M., Mertel, S., Knoche, E., Wegener, S., Wichmann, C., et al. (2011). RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science 334, 1565–1569. doi: 10.1126/science.1212991
Lu, H. E., MacGillavry, H. D., Frost, N. A., and Blanpied, T. A. (2014). Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J. Neurosci. 34, 7600–7610. doi: 10.1523/JNEUROSCI.4364-13.2014
Lushnikova, I., Skibo, G., Muller, D., and Nikonenko, I. (2009). Synaptic potentiation induces increased glial coverage of excitatory synapses in CA1 hippocampus. Hippocampus 19, 753–762. doi: 10.1002/hipo.20551
MacGillavry, H. D., and Hoogenraad, C. C. (2015). The internal architecture of dendritic spines revealed by super-resolution imaging: what did we learn so far? Exp. Cell Res. 335, 180–186. doi: 10.1016/j.yexcr.2015.02.024
MacGillavry, H. D., Kerr, J. M., and Blanpied, T. A. (2011). Lateral organization of the postsynaptic density. Mol. Cell. Neurosci. 48, 321–331. doi: 10.1016/j.mcn.2011.09.001
MacGillavry, H. D., Kerr, J. M., Kassner, J., Frost, N. A., and Blanpied, T. A. (2016). Shank-cortactin interactions control actin dynamics to maintain flexibility of neuronal spines and synapses. Eur. J. Neurosci. 43, 179–193. doi: 10.1111/ejn.13129
MacGillavry, H. D., Song, Y., Raghavachari, S., and Blanpied, T. A. (2013). Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78, 615–622. doi: 10.1016/j.neuron.2013.03.009
Maglione, M., and Sigrist, S. J. (2013). Seeing the forest tree by tree: super-resolution light microscopy meets the neurosciences. Nat. Neurosci. 16, 790–797. doi: 10.1038/nn.3403
Maidorn, M., Rizzoli, S. O., and Opazo, F. (2016). Tools and limitations to study the molecular composition of synapses by fluorescence microscopy. Biochem. J. 473, 3385–3399. doi: 10.1042/bcj20160366
Manley, S., Gillette, J. M., Patterson, G. H., Shroff, H., Hess, H. F., Betzig, E., et al. (2008). High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157. doi: 10.1038/nmeth.1176
Maschi, D., and Klyachko, V. A. (2017). Spatiotemporal regulation of synaptic vesicle fusion sites in central synapses. Neuron 94, 65.e3–73.e3. doi: 10.1016/j.neuron.2017.03.006
Medvedev, N., Popov, V., Henneberger, C., Kraev, I., Rusakov, D. A., and Stewart, M. G. (2014). Glia selectively approach synapses on thin dendritic spines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20140047. doi: 10.1098/rstb.2014.0047
Metcalf, D. J., Edwards, R., Kumarswami, N., and Knight, A. E. (2013). Test samples for optimizing STORM super-resolution microscopy. J. Vis. Exp. 79:e50579. doi: 10.3791/50579
Miller, S. J., and Rothstein, J. D. (2016). Astroglia in thick tissue with super resolution and cellular reconstruction. PLoS One 11:e0160391. doi: 10.1371/journal.pone.0160391
Milovanovic, D., and Jahn, R. (2015). Organization and dynamics of SNARE proteins in the presynaptic membrane. Front. Physiol. 6:89. doi: 10.3389/fphys.2015.00089
Minoshima, M., and Kikuchi, K. (2017). Photostable and photoswitching fluorescent dyes for super-resolution imaging. J. Biol. Inorg. Chem. 22, 639–652. doi: 10.1007/s00775-016-1435-y
Nägerl, U. V., Willig, K. I., Hein, B., Hell, S. W., and Bonhoeffer, T. (2008). Live-cell imaging of dendritic spines by STED microscopy. Proc. Natl. Acad. Sci. U S A 105, 18982–18987. doi: 10.1073/pnas.0810028105
Nair, D., Hosy, E., Petersen, J. D., Constals, A., Giannone, G., Choquet, D., et al. (2013). Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 33, 13204–13224. doi: 10.1523/JNEUROSCI.2381-12.2013
Nedergaard, M. (1994). Direct signaling from astrocytes to neurons in cultures of mammalian brain cells. Science 263, 1768–1771. doi: 10.1126/science.8134839
Newpher, T. M., and Ehlers, M. D. (2008). Glutamate receptor dynamics in dendritic microdomains. Neuron 58, 472–497. doi: 10.1016/j.neuron.2008.04.030
Nishimune, H., Badawi, Y., Mori, S., and Shigemoto, K. (2016). Dual-color STED microscopy reveals a sandwich structure of Bassoon and Piccolo in active zones of adult and aged mice. Sci. Rep. 6:27935. doi: 10.1038/srep27935
Oliet, S. H. R., Piet, R., and Poulain, D. A. (2001). Control of glutamate clearance and synaptic efficacy by glial coverage of neurons. Science 292, 923–926. doi: 10.1126/science.1059162
Orkand, R. K., Nicholls, J. G., and Kuffler, S. W. (1966). Effect of nerve impulses on the membrane potential of glial cells in the central nervous system of amphibia. J. Neurophysiol. 29, 788–806.
Ostroff, L. E., Manzur, M. K., Cain, C. K., and Ledoux, J. E. (2014). Synapses lacking astrocyte appear in the amygdala during consolidation of Pavlovian threat conditioning. J. Comp. Neurol. 522, 2152–2163. doi: 10.1002/cne.23523
Owen, D. M., Williamson, D., Magenau, A., and Gaus, K. (2012). Optical techniques for imaging membrane domains in live cells (live-cell palm of protein clustering). Methods Enzymol. 504, 221–235. doi: 10.1016/B978-0-12-391857-4.00011-2
Panatier, A., Arizono, M., and Nägerl, U. V. (2014). Dissecting tripartite synapses with STED microscopy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130597. doi: 10.1098/rstb.2013.0597
Parpura, V., Basarsky, T. A., Liu, F., Jeftinija, K., Jeftinija, S., and Haydon, P. G. (1994). Glutamate-mediated astrocyte-neuron signalling. Nature 369, 744–747. doi: 10.1038/369744a0
Patrizio, A., Renner, M., Pizzarelli, R., Triller, A., and Specht, C. G. (2017). α subunit-dependent glycine receptor clustering and regulation of synaptic receptor numbers. Sci. Rep. 7:10899. doi: 10.1038/s41598-017-11264-3
Patrushev, I., Gavrilov, N., Turlapov, V., and Semyanov, A. (2013). Subcellular location of astrocytic calcium stores favors extrasynaptic neuron-astrocyte communication. Cell Calcium 54, 343–349. doi: 10.1016/j.ceca.2013.08.003
Patton, B. R., Burke, D., Owald, D., Gould, T. J., Bewersdorf, J., and Booth, M. J. (2016). Three-dimensional STED microscopy of aberrating tissue using dual adaptive optics. Opt. Express 24, 8862–8876. doi: 10.1364/OE.24.008862
Penn, A. C., Zhang, C. L., Georges, F., Royer, L., Breillat, C., Hosy, E., et al. (2017). Hippocampal LTP and contextual learning require surface diffusion of AMPA receptors. Nature 549, 384–388. doi: 10.1038/nature23658
Pennacchietti, F., Vascon, S., Nieus, T., Rosillo, C., Das, S., Tyagarajan, S. K., et al. (2017). Nanoscale molecular reorganization of the inhibitory postsynaptic density is a determinant of gabaergic synaptic potentiation. J. Neurosci. 37, 1747–1756. doi: 10.1523/JNEUROSCI.0514-16.2016
Perez-Alvarez, A., Navarrete, M., Covelo, A., Martin, E. D., and Araque, A. (2014). Structural and functional plasticity of astrocyte processes and dendritic spine interactions. J. Neurosci. 34, 12738–12744. doi: 10.1523/JNEUROSCI.2401-14.2014
Perez De Arce, K., Schrod, N., Metzbower, S. W., Allgeyer, E., Kong, G. K., Tang, A. H., et al. (2015). Topographic mapping of the synaptic cleft into adhesive nanodomains. Neuron 88, 1165–1172. doi: 10.1016/j.neuron.2015.11.011
Petrini, E. M., Ravasenga, T., Hausrat, T. J., Iurilli, G., Olcese, U., Racine, V., et al. (2014). Synaptic recruitment of gephyrin regulates surface GABAA receptor dynamics for the expression of inhibitory LTP. Nat. Commun. 5:3921. doi: 10.1038/ncomms4921
Pleiner, T., Bates, M., Trakhanov, S., Lee, C. T., Schliep, J. E., Chug, H., et al. (2015). Nanobodies: site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. Elife 4:e11349. doi: 10.7554/elife.15597
Pougnet, J. T., Compans, B., Martinez, A., Choquet, D., Hosy, E., and Boue-Grabot, E. (2016). P2X-mediated AMPA receptor internalization and synaptic depression is controlled by two CaMKII phosphorylation sites on GluA1 in hippocampal neurons. Sci. Rep. 6:31836. doi: 10.1038/srep31836
Pougnet, J. T., Toulme, E., Martinez, A., Choquet, D., Hosy, E., and Boué-Grabot, E. (2014). ATP P2X receptors downregulate AMPA receptor trafficking and postsynaptic efficacy in hippocampal neurons. Neuron 83, 417–430. doi: 10.1016/j.neuron.2014.06.005
Rahbek-Clemmensen, T., Lycas, M. D., Erlendsson, S., Eriksen, J., Apuschkin, M., Vilhardt, F., et al. (2017). Super-resolution microscopy reveals functional organization of dopamine transporters into cholesterol and neuronal activity-dependent nanodomains. Nat. Commun. 8:740. doi: 10.1038/s41467-017-00790-3
Ribrault, C., Sekimoto, K., and Triller, A. (2011). From the stochasticity of molecular processes to the variability of synaptic transmission. Nat. Rev. Neurosci. 12, 375–387. doi: 10.1038/nrn3025
Rizzoli, S. O. (2014). Synaptic vesicle recycling: steps and principles. EMBO J. 33, 788–822. doi: 10.1002/embj.201386357
Rossi, A., Moritz, T. J., Ratelade, J., and Verkman, A. S. (2012). Super-resolution imaging of aquaporin-4 orthogonal arrays of particles in cell membranes. J. Cell Sci. 125, 4405–4412. doi: 10.1242/jcs.109603
Rossier, O., Octeau, V., Sibarita, J. B., Leduc, C., Tessier, B., Nair, D., et al. (2012). Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat. Cell Biol. 14, 1057–1067. doi: 10.1038/ncb2588
Rueckel, M., Mack-Bucher, J. A., and Denk, W. (2006). Adaptive wavefront correction in two-photon microscopy using coherence-gated wavefront sensing. Proc. Natl. Acad. Sci. U S A 103, 17137–17142. doi: 10.1073/pnas.0604791103
Rusakov, D. A. (2015). Disentangling calcium-driven astrocyte physiology. Nat. Rev. Neurosci. 16, 226–233. doi: 10.1038/nrn3878
Rusakov, D. A., Bard, L., Stewart, M. G., and Henneberger, C. (2014). Diversity of astroglial functions alludes to subcellular specialisation. Trends Neurosci. 37, 228–242. doi: 10.1016/j.tins.2014.02.008
Rust, M. J., Bates, M., and Zhuang, X. (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795. doi: 10.1038/nmeth929
Sakers, K., Lake, A. M., Khazanchi, R., Ouwenga, R., Vasek, M. J., Dani, A., et al. (2017). Astrocytes locally translate transcripts in their peripheral processes. Proc. Natl. Acad. Sci. U S A 114, E3830–E3838. doi: 10.1073/pnas.1617782114
Schafer, D. P., Lehrman, E. K., and Stevens, B. (2013). The “quad-partite” synapse: microglia-synapse interactions in the developing and mature CNS. Glia 61, 24–36. doi: 10.1002/glia.22389
Schedin-Weiss, S., Caesar, I., Winblad, B., Blom, H., and Tjernberg, L. O. (2016). Super-resolution microscopy reveals γ-secretase at both sides of the neuronal synapse. Acta Neuropathol. Commun. 4:29. doi: 10.1186/s40478-016-0296-5
Schermelleh, L., Carlton, P. M., Haase, S., Shao, L., Winoto, L., Kner, P., et al. (2008). Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336. doi: 10.1126/science.1156947
Schermelleh, L., Heintzmann, R., and Leonhardt, H. (2010). A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190, 165–175. doi: 10.1083/jcb.201002018
Schoen, M., Reichel, J. M., Demestre, M., Putz, S., Deshpande, D., Proepper, C., et al. (2016). Super-resolution microscopy reveals presynaptic localization of the ALS/FTD related protein fus in hippocampal neurons. Front. Cell. Neurosci. 9:496. doi: 10.3389/fncel.2015.00496
Serwanski, D. R., Jukkola, P., and Nishiyama, A. (2017). Heterogeneity of astrocyte and NG2 cell insertion at the node of ranvier. J. Comp. Neurol. 525, 535–552. doi: 10.1002/cne.24083
Shim, S. H., Xia, C., Zhong, G., Babcock, H. P., Vaughan, J. C., Huang, B., et al. (2012). Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. U S A 109, 13978–13983. doi: 10.1073/pnas.1201882109
Shrivastava, A. N., Rodriguez, P. C., Triller, A., and Renner, M. (2013). Dynamic micro-organization of P2X7 receptors revealed by PALM based single particle tracking. Front. Cell. Neurosci. 7:232. doi: 10.3389/fncel.2013.00232
Sibarita, J. B. (2014). High-density single-particle tracking: quantifying molecule organization and dynamics at the nanoscale. Histochem. Cell Biol. 141, 587–595. doi: 10.1007/s00418-014-1214-1
Sidenstein, S. C., D’Este, E., Böhm, M. J., Danzl, J. G., Belov, V. N., and Hell, S. W. (2016). Multicolour multilevel STED nanoscopy of actin/spectrin organization at synapses. Sci. Rep. 6:26725. doi: 10.1038/srep26725
Sieber, J. J., Willig, K. I., Kutzner, C., Gerding-Reimers, C., Harke, B., Donnert, G., et al. (2007). Anatomy and dynamics of a supramolecular membrane protein cluster. Science 317, 1072–1076. doi: 10.1126/science.1141727
Sigal, Y. M., Speer, C. M., Babcock, H. P., and Zhuang, X. (2015). Mapping synaptic input fields of neurons with super-resolution imaging. Cell 163, 493–505. doi: 10.1016/j.cell.2015.08.033
Singh, P., Jorgačevski, J., Kreft, M., Grubišić, V., Stout, R. F. Jr., Potokar, M., et al. (2014). Single-vesicle architecture of synaptobrevin2 in astrocytes. Nat. Commun. 5:3780. doi: 10.1038/ncomms4780
Sinnen, B. L., Bowen, A. B., Forte, J. S., Hiester, B. G., Crosby, K. C., Gibson, E. S., et al. (2017). Optogenetic control of synaptic composition and function. Neuron 93, 646.e5–660.e5. doi: 10.1016/j.neuron.2016.12.037
Šišková, Z., Justus, D., Kaneko, H., Friedrichs, D., Henneberg, N., Beutel, T., et al. (2014). Dendritic structural degeneration is functionally linked to cellular hyperexcitability in a mouse model of Alzheimer’s disease. Neuron 84, 1023–1033. doi: 10.1016/j.neuron.2014.10.024
Smith, A. J., Jin, B. J., Ratelade, J., and Verkman, A. S. (2014). Aggregation state determines the localization and function of M1- and M23-aquaporin-4 in astrocytes. J. Cell Biol. 204, 559–573. doi: 10.1083/jcb.201308118
Smith, K. R., Kopeikina, K. J., Fawcett-Patel, J. M., Leaderbrand, K., Gao, R., Schurmann, B., et al. (2014). Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron 84, 399–415. doi: 10.1016/j.neuron.2014.10.010
Smith, A. J., and Verkman, A. S. (2015). Superresolution imaging of aquaporin-4 cluster size in antibody-stained paraffin brain sections. Biophys. J. 109, 2511–2522. doi: 10.1016/j.bpj.2015.10.047
Špaček, J. (1985). Three-Dimensional analysis of dendritic spines. III. Glial sheath. Anat. Embryol. 171, 245–252. doi: 10.1007/bf00341419
Specht, C. G., Izeddin, I., Rodriguez, P. C., El Beheiry, M., Rostaing, P., Darzacq, X., et al. (2013). Quantitative nanoscopy of inhibitory synapses: counting gephyrin molecules and receptor binding sites. Neuron 79, 308–321. doi: 10.1016/j.neuron.2013.05.013
Südhof, T. C. (2006). Synaptic vesicles: an organelle comes of age. Cell 127, 671–673. doi: 10.1016/j.cell.2006.10.033
Sydor, A. M., Czymmek, K. J., Puchner, E. M., and Mennella, V. (2015). Super-Resolution microscopy: from single molecules to supramolecular assemblies. Trends Cell Biol. 25, 730–748. doi: 10.1016/j.tcb.2015.10.004
Szepesi, Z., Hosy, E., Ruszczycki, B., Bijata, M., Pyskaty, M., Bikbaev, A., et al. (2014). Synaptically released matrix metalloproteinase activity in control of structural plasticity and the cell surface distribution of GluA1-AMPA receptors. PLoS One 9:e98274. doi: 10.1371/journal.pone.0098274
Takamori, S., Holt, M., Stenius, K., Lemke, E. A., Grønborg, M., Riedel, D., et al. (2006). Molecular anatomy of a trafficking organelle. Cell 127, 831–846. doi: 10.1016/j.cell.2006.10.030
Tang, A. H., Chen, H., Li, T. P., Metzbower, S. R., MacGillavry, H. D., and Blanpied, T. A. (2016). A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210–214. doi: 10.1038/nature19058
Tatavarty, V., Das, S., and Yu, J. (2012). Polarization of actin cytoskeleton is reduced in dendritic protrusions during early spine development in hippocampal neuron. Mol. Biol. Cell 23, 3167–3177. doi: 10.1091/mbc.E12-02-0165
Tatavarty, V., Kim, E. J., Rodionov, V., and Yu, J. (2009). Investigating sub-spine actin dynamics in rat hippocampal neurons with super-resolution optical imaging. PLoS One 4:e7724. doi: 10.1371/journal.pone.0007724
Teng, K. W., Ishitsuka, Y., Ren, P., Youn, Y., Deng, X., Ge, P., et al. (2016). Labeling proteins inside living cells using external fluorophores for microscopy. Elife 5:e20378. doi: 10.7554/eLife.20378
Teng, K. W., Ishitsuka, Y., Ren, P., Youn, Y., Deng, X., Ge, P., et al. (2017). Labeling proteins inside living cells using external fluorophores for fluorescence microscopy. Elife 6:e25460. doi: 10.7554/elife.25460
Tillberg, P. W., Chen, F., Piatkevich, K. D., Zhao, Y., Yu, C. C., English, B. P., et al. (2016). Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992. doi: 10.1038/nbt.3625
Tønnesen, J., Katona, G., Rózsa, B., and Nägerl, U. V. (2014). Spine neck plasticity regulates compartmentalization of synapses. Nat. Neurosci. 17, 678–685. doi: 10.1038/nn.3682
Tønnesen, J., Nadrigny, F., Willig, K. I., Wedlich-Söldner, R., and Nägerl, U. V. (2011). Two-color STED microscopy of living synapses using a single laser-beam pair. Biophys. J. 101, 2545–2552. doi: 10.1016/j.bpj.2011.10.011
Turkowyd, B., Virant, D., and Endesfelder, U. (2016). From single molecules to life: microscopy at the nanoscale. Anal. Bioanal. Chem. 408, 6885–6911. doi: 10.1007/s00216-016-9781-8
Urban, N. T., Willig, K. I., Hell, S. W., and Nägerl, U. V. (2011). STED nanoscopy of actin dynamics in synapses deep inside living brain slices. Biophys. J. 101, 1277–1284. doi: 10.1016/j.bpj.2011.07.027
Ventura, R., and Harris, K. M. (1999). Three-dimensional relationships between hippocampal synapses and astrocytes. J. Neurosci. 19, 6897–6906.
Vignoli, B., Battistini, G., Melani, R., Blum, R., Santi, S., Berardi, N., et al. (2016). Peri-synaptic glia recycles brain-derived neurotrophic factor for LTP stabilization and memory retention. Neuron 92, 873–887. doi: 10.1016/j.neuron.2016.09.031
Voigt, A., Freund, R., Heck, J., Missler, M., Obermair, G. J., Thomas, U., et al. (2016). Dynamic association of calcium channel subunits at the cellular membrane. Neurophotonics 3:041809. doi: 10.1117/1.nph.3.4.041809
Volterra, A., Liaudet, N., and Savtchouk, I. (2014). Astrocyte Ca2+ signalling: an unexpected complexity. Nat. Rev. Neurosci. 15, 327–335. doi: 10.1038/nrn3725
Wang, L., Dumoulin, A., Renner, M., Triller, A., and Specht, C. G. (2016). The role of synaptopodin in membrane protein diffusion in the dendritic spine neck. PLoS One 11:e0148310. doi: 10.1371/journal.pone.0148310
Wang, K., Milkie, D. E., Saxena, A., Engerer, P., Misgeld, T., Bronner, M. E., et al. (2014). Rapid adaptive optical recovery of optimal resolution over large volumes. Nat. Methods 11, 625–628. doi: 10.1038/nmeth.2925
Wen, P. J., Grenklo, S., Arpino, G., Tan, X., Liao, H. S., Heureaux, J., et al. (2016). Actin dynamics provides membrane tension to merge fusing vesicles into the plasma membrane. Nat. Commun. 7:12604. doi: 10.1038/ncomms12604
Wenzel, J., Lammert, G., Meyer, U., and Krug, M. (1991). The influence of long-term potentiation on the spatial relationship between astrocyte processes and potentiated synapses in the dentate gyrus neuropil of rat-brain. Brain Res. 560, 122–131. doi: 10.1016/0006-8993(91)91222-m
Westphal, V., Rizzoli, S. O., Lauterbach, M. A., Kamin, D., Jahn, R., and Hell, S. W. (2008). Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science 320, 246–249. doi: 10.1126/science.1154228
Wijetunge, L. S., Angibaud, J., Frick, A., Kind, P. C., and Nägerl, U. V. (2014). Stimulated emission depletion (STED) microscopy reveals nanoscale defects in the developmental trajectory of dendritic spine morphogenesis in a mouse model of fragile X syndrome. J. Neurosci. 34, 6405–6412. doi: 10.1523/JNEUROSCI.5302-13.2014
Wilhelm, B. G., Mandad, S., Truckenbrodt, S., Kröhnert, K., Schäfer, C., Rammner, B., et al. (2014). Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028. doi: 10.1126/science.1252884
Willig, K. I., Rizzoli, S. O., Westphal, V., Jahn, R., and Hell, S. W. (2006). STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature 440, 935–939. doi: 10.1038/nature04592
Willig, K. I., Steffens, H., Gregor, C., Herholt, A., Rossner, M. J., and Hell, S. W. (2014). Nanoscopy of filamentous actin in cortical dendrites of a living mouse. Biophys. J. 106, L01–L03. doi: 10.1016/j.bpj.2013.11.1119
Witcher, M. R., Park, Y. D., Lee, M. R., Sharma, S., Harris, K. M., and Kirov, S. A. (2010). Three-dimensional relationships between perisynaptic astroglia and human hippocampal synapses. Glia 58, 572–587. doi: 10.1002/glia.20946
York, A. L., and Zheng, J. Q. (2017). Super-Resolution microscopy reveals a nanoscale organization of acetylcholine receptors for trans-synaptic alignment at neuromuscular synapses. eNeuro 4:ENEURO.0232-17.2017. doi: 10.1523/ENEURO.0232-17.2017
Younts, T. J., Monday, H. R., Dudok, B., Klein, M. E., Jordan, B. A., Katona, I., et al. (2016). Presynaptic protein synthesis is required for long-term plasticity of GABA release. Neuron 92, 479–492. doi: 10.1016/j.neuron.2016.09.040
Keywords: synapses, astroglia, tripartite synapse, super-resolution microscopy, plasticity
Citation: Heller JP and Rusakov DA (2017) The Nanoworld of the Tripartite Synapse: Insights from Super-Resolution Microscopy. Front. Cell. Neurosci. 11:374. doi: 10.3389/fncel.2017.00374
Received: 08 August 2017; Accepted: 10 November 2017;
Published: 24 November 2017.
Edited by:
Yukiko Goda, RIKEN Brain Science Institute (BSI), JapanReviewed by:
Mathieu Letellier, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2017 Heller and Rusakov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janosch P. Heller, ai5oZWxsZXJAdWNsLmFjLnVr
Dmitri A. Rusakov, ZC5ydXNha292QHVjbC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.