
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 30 June 2017
Sec. Cellular Neuropathology
Volume 11 - 2017 | https://doi.org/10.3389/fncel.2017.00175
This article is part of the Research Topic Gene Silencing and Editing Strategies for Neurodegenerative Diseases View all 10 articles
Accumulating studies have revealed that the human genome encodes tens of thousands of long non-coding RNAs (lncRNAs), which participate in multiple biological networks modulating gene expression via transcriptional, post-transcriptional and epigenetic regulation. Strikingly, a large fraction of tissue-specific lncRNAs are expressed in the Central Nervous System (CNS) with precisely regulated temporal and spatial expression patterns. These brain-specific lncRNAs are also featured with the cell-type specificity, the highest signals of evolutionary conservation, and their preferential location adjacent to brain-expressed protein-coding genes. Mounting evidence has indicated dysregulation or mutations in lncRNA gene loci are associated with a variety of CNS-associated neurodegenerative disorders, such as Alzheimer’s, Parkinson’s, Huntington’s diseases, Amyotrophic Lateral Sclerosis and others. However, how lncRNAs contribute to these disorders remains to be further explored and studied. In this review article, we systematically and comprehensively summarize the current studies of lncRNAs, demonstrate the specificity of lncRNAs expressed in the brain, their functions during neural development and expression profiles in major cell types of the CNS, highlight the regulatory mechanisms of several studied lncRNAs that may play essential roles in the pathophysiology of neurodegenerative diseases, and discuss the current challenges and future perspectives of lncRNA studies involved in neurodegenerative and other diseases.
For decades, people have considered that “genes and gene-encoded proteins” play crucial roles in regulating diverse cellular processes. However, with the completion of the human genome project, it was observed that less than 5% of the genome is comprised of coding sequences, whereas the majority of human genes are non-protein-coding genes which basically include abundant pseudogenes and comparably numerous non-coding RNAs (ncRNAs; Lander et al., 2001; ENCODE Project Consortium, 2012; FANTOM Consortium and the RIKEN PMI and CLST (DGT), 2014). NcRNAs are broadly defined as all types of RNA that are not translated into proteins due to lack of open reading frames (ORFs). They are also considered to be generated from sections of pseudogenes, which are DNA copies of protein-coding genes with high sequence similarity but have lost at least some of the functions relative to their parental genes over the course of evolution (Milligan and Lipovich, 2014; Ji et al., 2015).
Generally, ncRNAs can be classified into small ncRNAs and long ncRNAs (lncRNAs) based on whether their transcripts are less or larger than 200 nucleotides as a cutoff value (Elling et al., 2016). Small ncRNAs are usually defined as regulatory RNAs with a length ranging from 18 to 35 nucleotides. According to their diverse regulatory functions, small ncRNAs can be divided into several species, including transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), piwi-interacting RNAs (piRNAs) and endogenous small interfering RNAs (siRNAs), as well as microRNAs (miRNAs; Lander et al., 2001; Costa, 2005; Grivna et al., 2006; Sosinska et al., 2015; Elling et al., 2016). For many short RNAs, their functions have been well-characterized in gene expression control. Apart from these, many of them have been shown to be involved in specific pathologies, including neurodegenerative diseases and cancers. An increasing number of studies have reported that short ncRNAs are involved in Alzheimer’s, Parkinson’s and Huntington’s diseases (AD, PD and HD; Lee et al., 2011; Gstir et al., 2014).
LncRNAs are the largest class of longer (≥200 nt) non-protein coding RNA and their gene number was recently estimated at approximately 9000 within the human genome according to the GENECODE project (ENCODE Project Consortium, 2012). Other lncRNA studies even suggested there are more than 50,000 in the human genome (Managadze et al., 2013). The discovery of large numbers of lncRNAs genes that are redefined as a gene into a transcriptional unit was initially described by the FANTOM Consortium on the mouse transcriptome study (Carninci et al., 2005). Later on, studies of metazoan lncRNA repertoires further demonstrated the ubiquity of lncRNAs, which are however relatively lower-expressed, more tissue-specific and with greater variability from one tissue to another in comparison to protein-coding genes (Derrien et al., 2012; Milligan and Lipovich, 2014). Roles performed by lncRNAs have been evidenced by their participation in multiple networks controlling gene expression in transcriptional, post-transcriptional or epigenetic levels (Batista and Chang, 2013; Kung et al., 2013; Qureshi and Mehler, 2013). However, the biological significance of the majority of lncRNAs is yet to be further elucidated.
It is well known that RNA biology is of foremost significance in the central neural system (CNS) since neural cells are highly transcriptionally active and exhibit a robust expression of ncRNAs (Cherubini et al., 2006; Kapranov et al., 2010; Qureshi and Mehler, 2012). Remarkably, the brain is the organ where a large proportion of tissue-specific lncRNAs are preferentially expressed in particular regions or different cell types (Mercer et al., 2008; Derrien et al., 2012). These lncRNAs in the CNS participate in many aspects of brain functions and their roles in the pathologies of brain-related neurodegenerative diseases have been intensively and comprehensively investigated (Qureshi and Mehler, 2012, 2013). In this review article, we systematically and comprehensively summarize the diverse mechanisms reported for lncRNAs, describe the specificity of lncRNAs expressed in the brain and their functions during neural development as well as their expression profile in major cell types of the CNS. Meanwhile, we present those intensively studied lncRNAs that may play essential roles in the pathophysiology of neurodegenerative diseases, and discuss current challenges and future perspectives of lncRNA studies that are involved in neurodegenerative and other diseases. Hopefully, this review will broaden insights for future research in the field of lncRNAs in the CNS and associated neurodegenerative diseases.
Studies by Derrien T and team (Derrien et al., 2012) aiming at analyzing of lncRNAs from GENCODE V7 catalog revealed that, lncRNAs are produced in a similar way as that of protein-coding genes, whereas they display a striking bias toward two-exon transcripts and they are predominately localized in the chromatin and nucleus, expressed at relatively low levels (Guttman et al., 2009; Quinn and Chang, 2016). In comparison to the protein-coding genes, lncRNAs are under secondary structure conservation, and therefore they are believed to have arisen from different evolutionary pathways (Ponting et al., 2009; Kaessmann, 2010). Yet, origins of lncRNAs are not well understood so far. Due to the fact that lncRNAs harbor low sequence conservations and rapid evolution among mammals, several evolutionary hypotheses could be proposed such as: (1) lncRNAs might be generated by the metamorphosis of protein-coding genes through a gene duplication process; (2) lncRNAs might have evolved from segmental or whole gene duplication of other ncRNA genes; (3) lncRNAs might have originated via de novo generation, such as alternations in genomes including chromosomal rearrangement, generation of splice sites and promoters might transform nonfunctional genomic sequences to functional lncRNAs; and (4) transposable elements (TEs) insertions might be another origin of lncRNAs (Ponting et al., 2009; Kaessmann, 2010; Kapusta et al., 2013; Kazemzadeh et al., 2015). However, it was observed that rarely or only a minority (~15%) of lncRNAs showed significant sequence similarity to other lncRNAs or protein-coding genes on positions other than the shared repetitive elements, suggesting that novel lncRNAs genes are basically originated rather from de novo non-exonic sequences and/or from TEs than duplication (Derrien et al., 2012; Kapusta et al., 2013).
LncRNAs and associated lncRNA transcripts have quite heterogeneousgenomic context, regulation, life cycles, mechanism of action and functional profiles. Broadly, lncRNAs can be classified based on their genomic localization and orientation relative to protein coding genes into several categories: (1) long intergenic noncoding RNAs (LincRNAs), consisting of separate transcript units that are located between but do not overlap with protein-coding genes; (2) intronic transcripts, that are located within intron regions of protein-coding genes (sense or antisense); (3) overlapping lncRNAs that are overlapping with other genes either divergently or convergently transcribed; and (4) bidirectional ncRNAs (BincRNAs) with transcripts that are transcribed from divergent bidirectional promoters (see Figure 1; Guttman et al., 2009; Li and Ramchandran, 2010; Mattick and Rinn, 2015). Despite the diversities of lncRNAs, they share some common features, including: (1) most lncRNAs are transcribed by RNA polymerase II, spliced and modified with a 5′-cap and a poly-A tail, which makes them undistinguishable from protein-coding mRNAs; (2) they are poorly conserved at the sequence level, have a relatively low expression level and display a much more cell-tissue-specific pattern; and (3) they are generally regulated by transcription factors (Xiong et al., 2016). In addition, another subgroup of lncRNAs, the circular RNAs (circRNAs) have recently come into focus with the discovery of their pervasiveness and evolutionary conservation in mammalian and human cells (Jeck and Sharpless, 2014). Most circRNAs are generated during splicing either by spliceosomal machinery or by ribozymes I and II which thus splice out non-coding sequences from exons (exonic circRNAs), introns (intronic circRNAs), or a combination of introns and exons (exon-intron circRNAs; Abdelmohsen et al., 2015). CircRNAs can be differentiated from their linear counterparts by their adoption of a circular form and their lack of 5′ and 3′ ends (Vicens and Westhof, 2014).
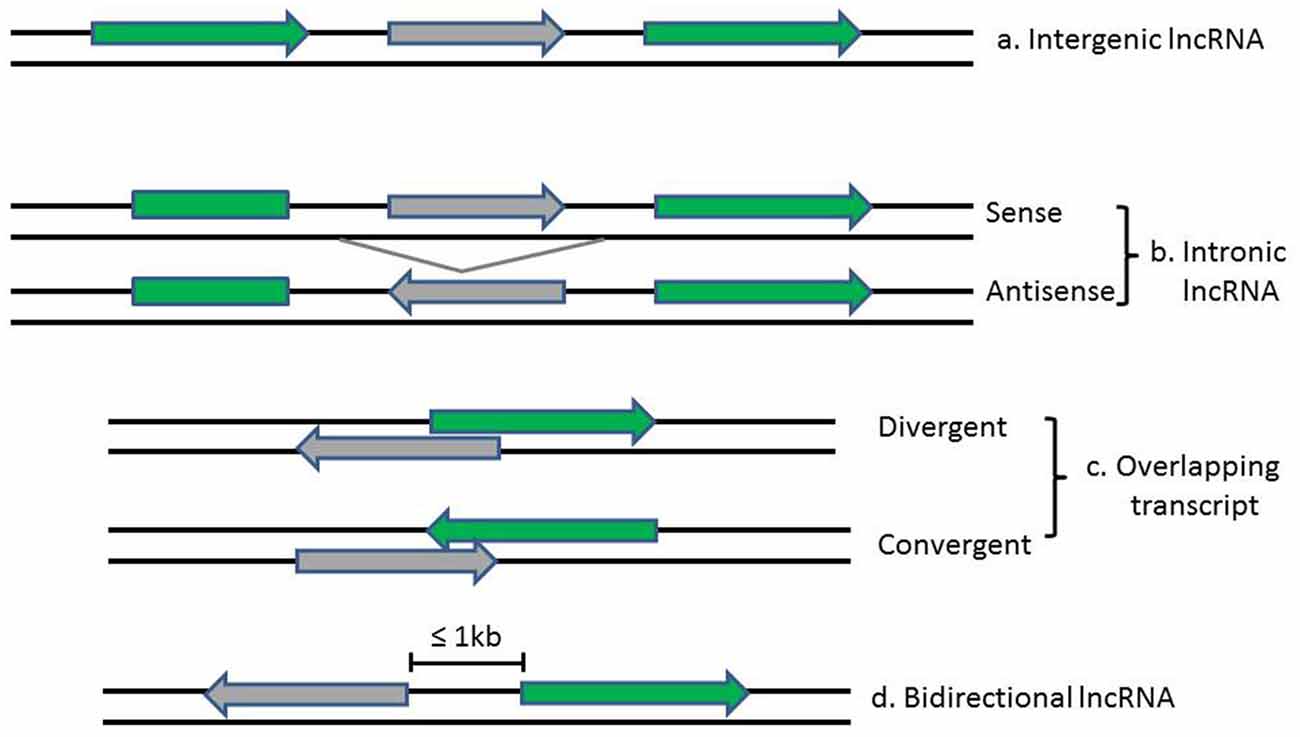
Figure 1. Diagrams show the classification of lncRNAs (in gray) according to their position relative to neighboring protein-coding genes (in green). Arrows indicate direction of transcription.
LncRNAs have been best described for their participation in regulating gene and genome activity at various levels (see Figure 2). LncRNAs can regulate the expression of nearby genes on the same allele in cis, or in trans to control genes at other genomic locations on different chromosomes, through which they can regulate gene expression at diverse levels, such as transcription, RNA processing and translation (Elling et al., 2016). The majority of lncRNAs are localized in the nucleus, in which they can fulfill their regulatory functions via acting as scaffolds for chromatin modifiers by interacting with chromatin-modifying complexes or as transcriptional co-regulators by binding to transcription factors (Rinn and Chang, 2012; Ulitsky and Bartel, 2013).
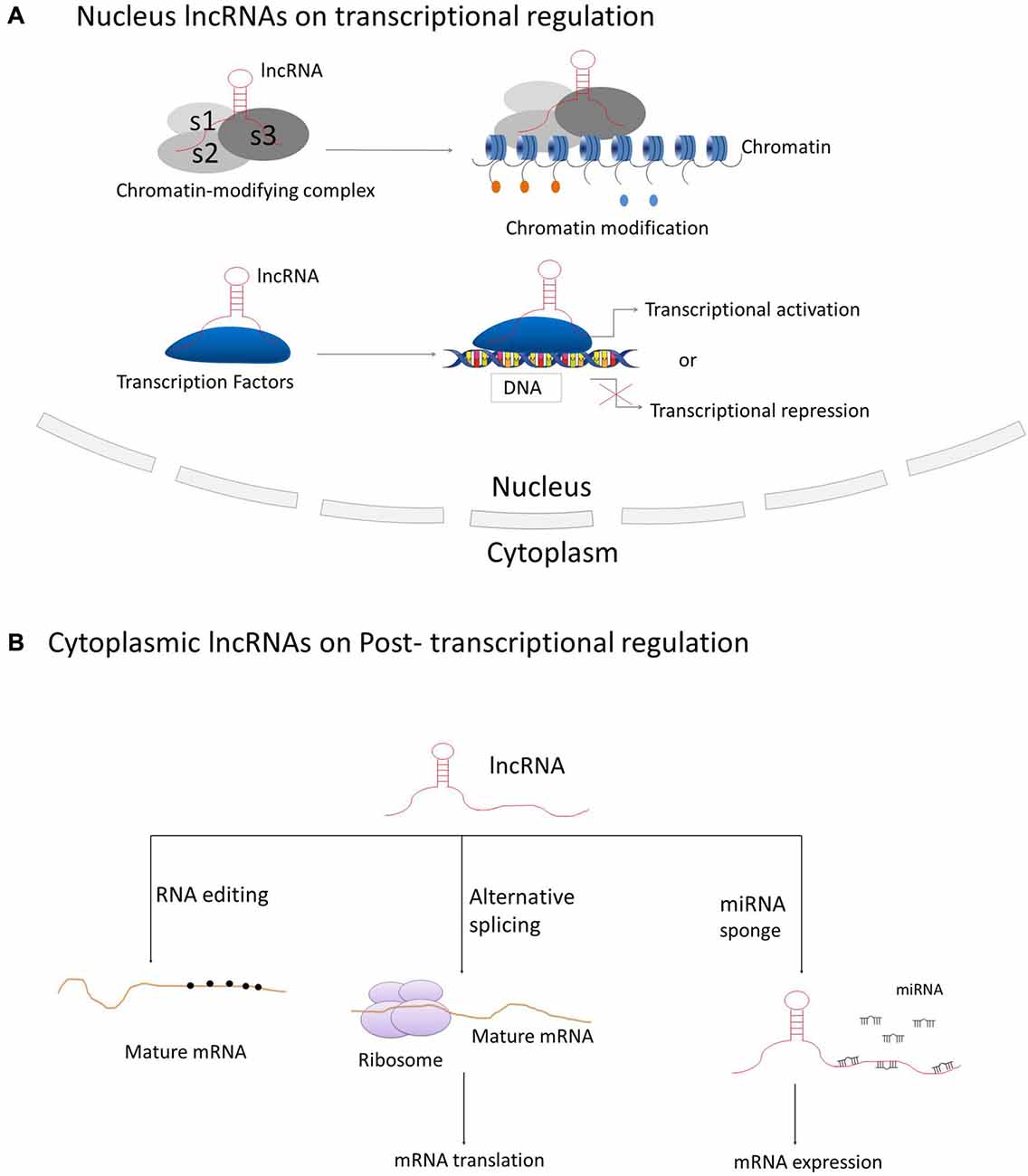
Figure 2. Principle mechanisms of lncRNAs on regulation of gene and genome activity. (A) LncRNAs located in the nucleus are basically functioning in transcriptional regulation through interacting with chromatin-modifying complexes or transcription factors; (B) Cytoplasmic lncRNAs are generally acting as regulators on RNA processing, such as RNA editing, alternative splicing and miRNA-mediated mRNA expression.
The best-known case of lncRNA that regulates transcription mediated through chromatin modification is Xist, a 17 kb lincRNA generated from the inactive X-chromosome (Clemson et al., 1996). It mediates the silencing of the inactive X-chromosome in human female cells through recruiting Polycomb Repressive Complex 2 (PRC2) by the Repeat A motif (RepA) on Xist and thus initiating chromosome-wide silencing via catalyzing Lysine 27 trimethylation on histone H3 (H3K27; Pinter et al., 2012; Jiang et al., 2013; Bergmann and Spector, 2014). Hotair, a 2.2 kb conserved lncRNA transcribed from the human HOXC locus on chromosome 12, is involved in repressing transcription in trans across the HOXD locus. Hotair was shown to physically interact with PRC2 to ensure the PRC2 occupancy and histone H3 lysine-27 trimethylation of HOXD locus (Rinn et al., 2007). The lncRNA Braveheart, prominently expressed in the mouse heart, can interact with Suz12, a subunit of PRC2 (Klattenhoff et al., 2013). The lncRNA Fendrr (Fetal-lethal developmental regulatory RNA), that is also related to cardiac development and heart function, can bind to PRC2 and WDR5, a member of the MLL histone methyl-transferase complex (Grote and Herrmann, 2013; Grote et al., 2013).
There are also cases that many lncRNAs interact with transcription factors. The definitive endoderm-associated lncRNA1 (DEANR1) is crucial for human endoderm differentiation via interaction and upregulation of the endoderm factor FOXA2 (Jiang et al., 2015). The lnc-DC (Lnc dendritic cells) is a lncRNA exclusively expressed in human conventional dendritic cells. It was revealed that lnc-DC can bind to the transcription factor signal transducer and activator of transcription 3 (STAT3) directly in the cytoplasm and induce its phosphorylation on Tyrosine-75 by inhibiting its binding to and dephosphorylation by SHP1, thereby leading to the activation of STAT3 on dendritic cell differentiation (Wang et al., 2014). In addition, the lncRNA breast cancer anti-estrogen resistance 4 (BCAR4), functioning in breast cancer metastasis, was discovered to directly interact with Smad nuclear-interacting protein 1 (SNIP1) and Serine/threonine-protein phosphatase one regulatory subunit 10 (PPP1R10 or PNUTS), thus activating phosphor-GLI2 dependent gene expression (Xing et al., 2014).
Cytoplasmic lncRNAs are also acting as modulators on post-transcriptional regulation of genes through various mechanisms during RNA processing, such as mRNA editing, alternative splicing and others. The antisense intronic lncRNA prostate cancer antigen 3 (PCA3), acting as a dominant-negative oncogene, was demonstrated to interact with and down-regulate an as yet to be determined tumor suppressor gene PRUNE2, by the formation of PRUNE2/PCA3 double-stranded RNAs that allow the adenosine deaminase to edit RNAs via adenosine to inosine editing (A-to-I editing; Salameh et al., 2015). Non-coding RNA activated by DNA damage (NORAD) is an abundant and highly conserved human lncRNA that acts as a multivalent binding platform for RNA binding proteins in the PUMILIO family in order to maintain genomic stability (Lee et al., 2016). Furthermore, the lncRNA-asFGFR2 is an evolutionarily conserved nuclear antisense lncRNA that was generated from within the human FGFR2 locus. It was found to modulate the epithelial- specific alternative splicing of FGFR2 by recruiting PRC2 and histone demethylase KDM2a in PNT2 cells (Gonzalez et al., 2015).
Recent studies also revealed certain lncRNAs acting as “miRNA sponges” that they have the potential to sponge and compete with miRNA target genes for the binding of miRNA response elements (MREs) to relieve miRNA-mediated target mRNA repression (Ebert and Sharp, 2010). The circRNA ITCH (cir-ITCH) was newly discovered to be functionally sponging miR-7, miR-17 and miR-214 and inducing the expression level of ITCH, which induced the ubiquitination and degradation of phosphorylated Dvl2 and thereby the inhibition of the Wnt/β-catenin pathway (Li et al., 2016). The lncRNA urothelial carcinoma-associated 1 (UCA1) was shown to work as an endogenous sponge that can down-regulate miR-216b expression by directly binding to miR-216b (Wang et al., 2015). The lncRNA human ovarian cancer-specific transcript 2 (HOST2) was verified to be a molecular sponge that can modulate the availability of miR let-7b (a potent tumor suppressor) and inhibit miR let-7b functions, thus post-transcriptionally suppressing target gene expressions (Gao et al., 2015).
Based on studies from the GENCODE project in 2012 that 10,000–50,000 lncRNAs in the human genome have been annotated to date, it was remarkably revealed that approximately 40% of lncRNAs (which is about 4000–20,000 lncRNAs) are tissue-specifically expressed in the brain (Derrien et al., 2012). This number is strikingly large in comparison to the number of protein-coding genes in human genome which is approximately 20,000–25,000 in general (Briggs et al., 2015). These brain-specific lncRNAs display the highest signals of evolutionary conservation in comparison with those expressed in other tissues. Studies also found that brain-expressed lncRNAs are enriched in predicted, conserved RNA structures and thus are more likely to possess conserved functions (Ponjavic et al., 2009). In addition, brain-expressed lncRNAs show greater brain region, temporal and spatial specificity than mRNAs. Numerous transcriptome analysis have revealed that various lncRNAs are expressed differentially over time and/or in those brain regions, such as cortex, cerebellum and hippocampus during development and adulthood (Lipovich et al., 2014; Kadakkuzha et al., 2015). LncRNAs are also found to be expressed in a more cell-type-specific manner than protein-coding genes. The transcriptome studies on cortical pyramidal neurons have uncovered 806 of 5195 lncRNAs have differential expression across neuronal types, of which, 55% of lncRNAs are annotated as cell-type signature cluster, 32% of lncRNAs are related to cell-type independent clusters, while around 10% of lncRNAs are found in the mixed cell-type clusters, indicating their role on the specification and maintenance of cell identity (Molyneaux et al., 2015). Another feature of brain-specific lncRNAs is their preferential location adjacent to brain-expressed protein-coding genes that are active in transcriptional modulation or in the nervous system development (Ponjavic et al., 2009). Transcriptome sequencing analysis in the corticogenesis displayed that most lncRNAs overlap with neurogenic genes and share with them a similar expression pattern, indicating that lncRNAs regulate corticogenesis through the tuning of the expression of nearby cell fate determinants (Aprea et al., 2013). Considering the complexity and heterogeneity of the mammalian CNS, the brain would be considered as the largest repertoire of lncRNAs in comparison to other somatic tissues and the tissue and cell-type specificity of these lncRNAs make them greatly contribute to cell fate, lineage specification and maintenance of cell identity during the development of the mammalian brain (Hart and Goff, 2016).
The development of the CNS is a complicated and highly stereotyped process that requires elaborate spatiotemporal regulation of stem/progenitor cell proliferation and differentiation. LncRNAs have been demonstrated to play indispensable roles in CNS development from early neural differentiation to late-stage synaptogenesis (see Figure 3; Briggs et al., 2015).
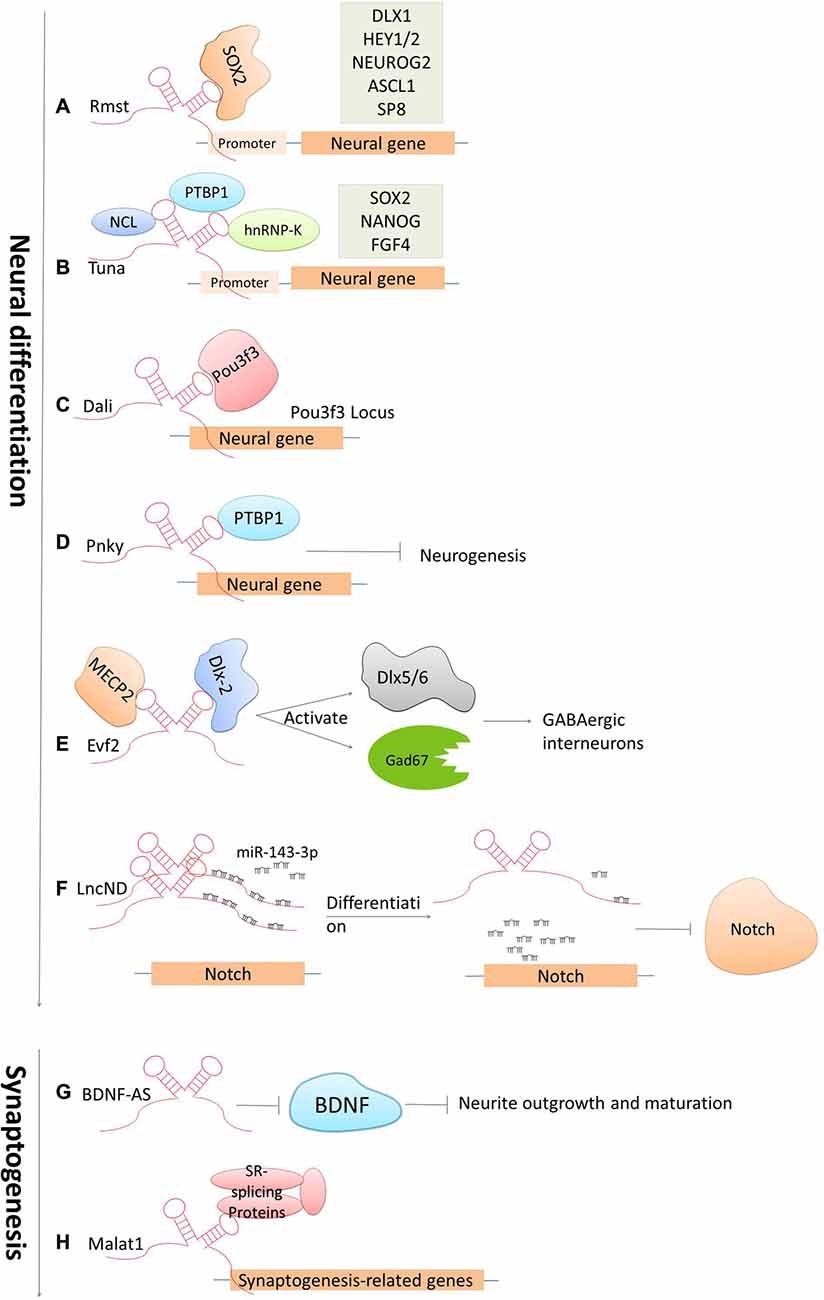
Figure 3. Cases of lncRNAs functioning in neuronal development. (A–F) LncRNAs function in early neural commitment through the recruitment of the transcriptional machinery to regulate neural-related gene expressions (Rmst, Tuna, Dali and Pnky). Evf2 is particularly acting in regulating GABAergic interneuron specification. LncND functions in regulating Notch signaling pathways via sequestering miR143-3p. (G–H) LncRNAs function in late developmental processes, such as neurite outgrowth and maturation mediated by BDNF-AS, synapse function by Malat1.
Accumulated studies have confirmed that dozens of lncRNAs are identified to be functional in establishing pluripotency or driving neural lineage entry in the widely used in vitro model systems—mouse embryonic stem cells (ESCs; Guttman et al., 2011; Ng et al., 2012). These lncRNAs are functional at various stages along the progression from pluripotent cells in the early embryo to the terminal cell types in the mature mammalian brain. The regulatory mechanisms are generally based on modulating these lncRNAs by canonical pluripotency transcription factors, which in turn perform their regulatory effects by directing transcription factors or chromatin remodeling complexes to specific lineage-specifying genes. For examples, the lncRNA Rhabdomyosarcoma 2-associated transcript (Rmst), specifically expressed in the brain and regulated by the transcription repressor REST, was found to modulate neural differentiation in vitro. It was shown that Rmst interacts with SOX2 to co-regulate a large pool of downstream genes (such as DLX1, HEY2 and SP8) implicated in neurogenesis. The absence of Rmst can block the exit from the ESC state and the initiation of neural differentiation (Ng et al., 2013). Tuna, is also a lncRNA that regulates neuronal gene expression in a similar mechanism as Rmst. Tuna interacts with three RNA-binding proteins (NCL, PTBP1 and hnRNP-K) and together target promoter regions of neural genes in differentiating mouse ESCs. The knockdown of Tuna or any of the three RNA-binding proteins can sufficiently suppress neural differentiation (Lin et al., 2014). This is also conserved from relatively related vertebrates, mice and zebrafish, indicating that the neural lineage commitment driven by lncRNAs displays the highly evolutionary conservation (Briggs et al., 2015). Furthermore, the lncRNA Dali, transcribed downstream of the transcription factor Pou3f3, was shown to drive the expression of an essential gene involved in neuronal differentiation in neuroblastoma cells. Dali can regulate transcription of the Pou3f3 locus locally and physically interact with Pou3f3 protein to regulate the expression of neural differentiation genes distally (Chalei et al., 2014). Paupar, a chromatin-associated intergenic lncRNA expressed in the CNS, is divergently transcribed from a locus upstream from the gene encoding the transcription factor Pax6. It was shown that the knockdown of Paupar destroys the normal cell cycle profile of neuroblastoma cells, thus enhancing neuronal differentiation. The function of Paupar is performed via locally interacting with and transcriptionally regulating Pax6, as well as via distally controlling neural gene expression on a large scale (such as SOX2, HES1 and EVF2), which in part requires physical association with Pax6 protein (Vance et al., 2014). Besides that, one of the latest studies has identified an lncRNA, termed LncND (Neurodevelopment) that harbors 16 miRNA response elements for miR-143-3p in primates. It performs its role in neuronal development by sequestering miR-143-3p, and in doing so, modulates Notch signaling pathways via regulating expression of Notch receptors, NOTCH-1 and NOTCH-2. Also, NOTCH-1 and NOTCH-2 show the same expression pattern as LncND in early neurogenesis process when Notch expression is indispensable. While, later in neural differentiation process, reduction of NOTCH expression is followed by LncND, leading to the release of miR-143-3p and the decrease of Notch signaling specifically in LncND-expressing cells (Rani et al., 2016). Interestingly, knocking down LncND in neuroblastoma cells can inhibit NOTCH-1 and NOTCH-2 and make cells differentiate to neurons, which show similar phenotypes as what can be observed by miR-143-3p overexpression. These findings suggested a role of LncND in miR-mediated regulation of Notch signaling to sustain the neural progenitor pool during cerebral cortex expansion in primates (Rani et al., 2016).
There are also several important lncRNAs being identified to control stem cell turnover and the specification of particular lineages in the embryonic mouse brain in vivo. For example, Evf2, the firstly identified nervous system-specific lncRNA, has been shown to have a significant regulatory role in neural development. Depletion in EVF2 can cause imbalance of the excitatory to inhibitory neurons in the postnatal hippocampus and dentate gyrus. This imbalance is caused by the failure of GABAergic interneuron specification (Bond et al., 2009). It was revealed that Evf2 recruits DLX and methyl CpG-binding protein (MECP2) to modulate the transcriptional activity of two transcription factors Dlx5/6 and the glutamate decarboxylase 67 (Gad67, which is required for the conversion of glutamate to GABA), and thereby regulates the gene expression of GABAergic interneurons in the developing mouse forebrain (Feng et al., 2006; Bond et al., 2009). Pnky is an evolutionally conserved and neural-specific lncRNA that modulates neurogenesis of neural stem cells (NSC) in the embryonic and postnatal brain. The Ramos AD group (Ramos et al., 2013) has unveiled that knockdown of Pnky can increase neuronal differentiation and deplete the NSC population in the embryotic mouse cortex. In addition, it was shown that Pnky interacts with PTBP1, a splicing regulator expressed in NSCs that represses the inclusion of neural exons in non-neural cells. Knockdown of PTBP1 also reinforce neurogenesis, indicating that Pnky and PTBP1 function similarly to modulate the alternative splicing of a group of transcripts that are involved with cellular phenotypes (Aprea and Calegari, 2015; Ramos et al., 2015). Together, these studies indicate that lncRNAs regulate cell-fate determination and progenitor cell turnover during neural development both in vitro and in vivo.
Another key process during CNS development is the late-stage synaptogenesis. Accordingly, several lncRNAs have been discovered to make great contributions to this process (Briggs et al., 2015). Malat1 is a well-known lncRNA that have been implicated in the regulation of neurite elaboration. Malat1 is expressed in multiple tissues but is highly abundant in neurons. It was revealed that Malat1 can actively recruit SR-family splicing proteins to transcription sites in order to regulate synaptogenesis-related gene expression. The knockdown of MALAT1 leads to a decreased level of synaptic density; while in contrast, overexpression can reciprocally increase synaptic density (Bernard et al., 2010). Primate-specific BC200 RNA (Brain cytoplasmic RNA, 200 nt) and its rodent counterpart BC1 are evolutionarily conserved lncRNAs that have been firstly identified to be expressed in both developing and adult nervous systems (Muslimov et al., 1997). Studies later reported that BC200/BC1 is functional in the neuronal excitation-repression equilibria via protein-synthesis-dependent implementation (Zhong et al., 2009). In addition, several antisense lncRNAs have been recently presented as core protein regulators, for example Brain-derived neurotrophic factor (BDNF), glial-derived neurotrophic factor (GDNF) and ephrin receptor B2 (EPHB2) that control neurite elaboration. The expression of BDNF is repressed by its antisense lncRNA BDNF-AS. The inhibition of BDNF-AS can release and induce the expression of BDNF by 2- to 7-folds, which is in line with the reduction of EZH2 recruitment and an alternation of the chromatin state at the BDNF locus. This correspondingly induces neuronal outgrowth, differentiation, survival and proliferation both in vitro and in vivo (Modarresi et al., 2012). These observations, along with the current understanding of lncRNA mechanisms of action, imply that lncRNAs have a critical role in regulating neural gene expression and brain development.
LncRNAs have been proposed to play diverse roles in the CNS, such as in neural differentiation, in synaptogenesis and others. As a result, the expression profiles of lncRNAs in the CNS were recently studied. The CNS has prominent cellular diversities owning hundreds of distinct cell types. In particular, neurons and neuroglia cells (mainly including astrocytes and oligodendrocytes) are the major cell types. A recent study has successfully isolated nuclear RNAs from different CNS cell types (neurons, astrocytes and oligodendrocytes) and compared both protein-coding and noncoding nuclear transcriptome profiles in these three cell types (Reddy et al., 2017). For non-coding transcriptome, it was revealed that approximately 300 transcripts at a level of >5 CPM (counts per million) can be observed in one of the three different cell types, and the majority is transcribed at the level of >1 CPM. Several other highly expressed transcripts are also found in all three cell types, such as Xist and its regulators Ftx and Jpx (Tian et al., 2010; Chureau et al., 2011; Reddy et al., 2017). Nevertheless, 169 lincRNAs were shown to have a >10-fold difference in one of the pairwise comparisons, such as Mirg and other adjacent maternally expressed lncRNAs (Meg3 and Rian) that are found to be highly enriched in neuronal nuclei. Mirg was found to be precisely localized within bright subnuclear puncta in neurons; the expression of Meg3 was shown as neuronal selective that it was enhanced in gray matter where neurons are found but depleted from white matter areas where only glia are found; and Rian was found to have similar expression pattern as Meg3 (Zhang et al., 2003; Balik et al., 2013; Reddy et al., 2017). In astrocytes, various transcripts were found but most of them do not possess known functions. Whereas, the lncRNA Rmst, a key co-regulator of neurogenesis with the SOX2 transcription factor, is an exception that it has been shown to be expressed robustly in the nuclei of astrocytes (Ng et al., 2013; Reddy et al., 2017). Meanwhile, it was shown that there are at least 16 lncRNAs with >5 CPM found in oligodendrocytes, such as Neat1 and DLeu2. Notably, although Neat1 was shown to have the highest expression level in oligodendrocytes, it was also present in astrocytes (Klein et al., 2010; Reddy et al., 2017). These data demonstrated different lncRNA expression patterns in three major cell types in the brain, providing a crucial clue for evaluating functions of various lncRNAs in the brain and CNS-associated disorders.
The importance of lncRNAs in the brain has been asserted by their association with various brain functions, including the maintenance of pluriotency, neuroectodermal differentiation, neuron-glial cell fate determination, synaptogenesis and so on (Roberts et al., 2014). Considering that, it is not surprising that dysregulation or mutation of lncRNAs is tightly related to various neurological disorders. Genome-wide association studies and comparative transcriptome analysis have implied that lncRNAs are involved in a variety of neurological disorders, such as psychiatric disorders, neurodegenerative diseases (like Alzheimer’s disease-AD, Parkinson disease-PD, Huntington disease-HD, Frontotemporal lobar degeneration-FTLD and Amyotrophic lateral sclerosis-ALS, etc.), and others. Similarly, as described above, lncRNAs contribute to these diseases in diverse ways, from the regulation of transcription to the modulation of RNA processing and translation (see Table 1). Here, we briefly exemplified several studied cases of lncRNAs that have been identified to be associated with neurodegenerative diseases.
Huntingtin (HTT) is an essential gene for HD, a CAG trinucleotide repeat expansion in exon 1, and the main cause of HD. The HTT antisense (HTT-AS), a natural antisense transcript at the HD repeat locus containing the repeat tract, was identified and characterized with 5′ capped, poly A tailed, three exons maintained and alternatively being spliced into HTTAS-v1 (exons 1 and 3) and HTTAS-v2 (exons 2 and 3). Cell studies revealed that the overexpression of HTTAS-v1 specifically decreases endogenous HTT transcript levels, whereas the siRNA knockdown of HTTAS-v1 induces HTT transcript levels. What’s more, HTTAS-v1 expression was found to be downregulated in frontal cortex of HD patients, strongly suggesting the existence of a gene antisense to HTT acting as a regulator for its own expression and its contribution to the development of HD (Chung et al., 2011).
Beta-secretase 1 (BACE1) is the key enzyme that produces β-amyloid peptide (Aβ) which aggregates and forms into amyloid plaques as a main pathological hallmark of AD. Recent studies have identified a conserved noncoding antisense transcript of BACE1, BACE1-AS, that regulates BACE1 mRNA and subsequently BACE1 protein expression both in vivo and in vitro. Studies have highlighted that the knockdown of BACE1-AS can reduce BACE1 levels, as well as Aβ1–40 and Aβ1–42 production correspondingly. Exposure to various cell stressors (including Aβ1–42) that have been implicated in the pathogenesis of AD, was found to induce BACE1-AS levels. This induction is led by the formation of a RNA duplex with BACE1 mRNA, which in turn increases BACE1 mRNA stability and BACE1 protein, and consequently generates additional Aβ1–42 through a post-transcriptional free-forward mechanism (Faghihi et al., 2008). Furthermore, it was found that increased BACE1-AS levels exist in various brain regions in subjects with AD in comparison to control individuals, indicating the possibility of BACE1-AS being a potential biomarker of AD (Faghihi et al., 2008; Modarresi et al., 2011).
Recently, 17A was described as an antisense transcript of the human G-protein-coupled receptor 51 gene (GPR51, GABA B2 receptor) that is RNA polymerase III-dependent and embedded in the GPR5. In neuroblastoma cells, the stable expression of 17A can promote the synthesis of an alternative splicing isoform for GABA R2, resulting in the elimination of GABA B2 intracellular signaling and the enhancement of Aβ secretion and the ratio of Aβ42/Aβ40. Furthermore, 17A is expressed in the human brain and upregulated in cerebral tissues derived from AD patients, indicating its potential contribution to the development of AD (Massone et al., 2011).
NAT-Rad18, a natural antisense transcript against Rad18 (a gene encoding DNA repair protein), was investigated and considered to play potential roles in the DNA damage repair system in AD. RNA quantitative and immunohistochemistry analysis revealed that NAT-Rad18 is widely distributed in the adult rat brain, but with high levels in the cerebellum, brainstem and cortex where neurons are specifically expressed. Upon Aβ-induced apoptosis in cortical neurons, the expression of NAT-Rad18 was shown to be up-regulated, whereas Rad18 was post-transcriptionally down-regulated. This observation suggested NAT-Rad18 might reduce the ability of neurons and increase their apoptosis susceptibility via the post-transcriptional modulation of Rad18 to reduce their response to DNA damage stress (Parenti et al., 2007; Wu et al., 2013).
GDNFOS, transcribed from the opposite strand of GDNF, was demonstrated to be associated with neurodegenerative diseases, like AD. GDNFOS contains four exons that therefore are spliced into different isoforms, including GDNFOS1/2 acting as lncRNAs and GDNFOS3 encoding a protein of 105 amino acids (Airavaara et al., 2011). It was revealed that the mature GDNF peptide was reduced while the transcript GDNFOS1 upregulated in the postmortem middle temporal gyrus of patients suffering from AD when compared with those of normal controls, indicating the dysregulation of GDNF and GDNFOS might further implicate in other human brain diseases (Airavaara et al., 2011).
One brain-specific circRNA, ciRS-7 (circular RNA sponge for miR-7), also known as CDR1as, is transcribed antisense to the cerebellar degeneration-related protein 1 transcript (CDR1) that is highly expressed (even more than the sense transcript) in the mouse and human CNS. The ciRS-7 functions as a miR-7 sponge that strongly quenches miR-7 activity, thus causing induced levels of miR-7 targets. Recent studies also indicated an endogenous interaction between ciRS-7 and miR-7 based on the observation of the co-expression of ciRS-7 and miR-7 in the mouse brain (Hansen et al., 2013; Memczak et al., 2013). The ciRS-7 has been identified to be related to the sporadic AD, that dysregulation of ciRS-7 was evidenced in the hippocampal CA1 region of the sporadic AD (Lukiw, 2013). The ciRS-7 deficiency was expected to induce ambient miR-7 levels in AD-affected brain cells, which is probably responsible for down-regulating AD-associated targets, such as, the ubiquitin protein ligase A (UBE2A; Cogswell et al., 2008; Lukiw, 2013).
BC200 RNA is tightly related to AD development and its expression has been demonstrated to be substantially up-regulated in tested AD brain tissues (Brodmann’s area 9) in comparison to that in age-matched normal brain samples (Mus et al., 2007). Further analysis also demonstrated that the increase in levels of BC200 RNA only occurs in specific regions of the AD-brain and is accompanied by changes in BC200 RNA neuronal distribution, including dendritic mislocalization and gradual accumulation of BC200 RNA to the perikaryon (Mus et al., 2007). The role of BC200 RNA in regulating gene expression at translational level during the development of AD has been reported in many studies as being through mechanisms of interaction with many different proteins. It was shown that BC200 RNA interacts with the human synaptotagmin-binding cytoplasmic RNA interacting protein (SYNCRIP), a component of large mRNA transport granules in neurons and functioning in local protein synthesis at post-synaptic sites, through mediation by the N-terminal RNA recognition motifs and the central A-rich region of BC200 RNA (Duning et al., 2008). In addition, the polyA-binding protein (PABP1), a regulator of translation initiation, was also identified to bind to BC200 RNA mediated by its central A-rich region, leading to the hypothesis that BC200 RNA is associated with protein translation in neuronal dendrites (Muddashetty et al., 2002). The interactions between BC200 RNA and proteins involved in local protein synthesis in neurons indicated the important role of BC200 RNA in AD pathology.
The Uchl1-AS is a nuclear-enriched lncRNA that is transcribed antisense to the mouse ubiquitin carboxy-terminal hydrolase L1 (Uchl1). Uchl1 is a neuron-restricted protein acting as a de-ubiquitinating enzyme or a monoubiquitin stabilizer. UCHL1 gene mutations have been discovered to be related to familial PD, and the oxidative inactivation of Uchl1 protein has been reported in PD and AD brains (Choi et al., 2004; Barrachina et al., 2006). Uchl1-AS can increase the protein synthesis of UCHL1 at post-transcriptional level, which depends on the combined activities of two domains, the 5′ antisense region that provides specificity for the sense target gene and the embedded repetitive SINEB2 element (short interspersed nuclear element of B3 subclass) that confers the protein synthesis activation domain (Nishihara et al., 2006; Carrieri et al., 2012). In addition, the activity of Uchl1-AS is under the control of signaling pathways. Uchl1 mRNA is predominantly localized in the cytoplasm while Uchl1-AS is abundant in the nucleus of dopaminergic neurons. Intriguingly, the mTOR inhibitor-Rapamycin treatment resulted in the induction of Uchl1 protein by association of shuttling Uchl1-AS from the nucleus to the cytoplasm, indicating the interplay among Uchl1-ncRNA-mTOR might be crucial for the development of PD (Carrieri et al., 2012; Vučičevič et al., 2014).
C9ORF72, chromosome 9 ORF72, contains a hexanucleotide (GGGGCC, G4C2) repeat expansion in its non-coding region, which was found as the causative mutation for both ALS and FTLD (DeJesus-Hernandez et al., 2011). The C9ORF72 expansion mutation can be transcribed bidirectionally that produces unexpected proteins via repeat-associated non-ATG (RAN) translational mechanism. Recent discovery found that antisense C9ORF72 transcripts containing the G4C2 expansion are increased in ALS patients’ brains (Zu et al., 2013), indicating the vital role of antisense C9ORF72 transcripts on fundamental pathologies of ALS/FTLD.
In addition, the lncRNA nuclear-enriched abundant transcript 1–2 (NEAT1-2), containing nuclear bodies named as “paraspeckles”, has also been shown to be associated with ALS/FTLD. TAR DNA binding protein 43 (TDP-43) inclusions or that are fused in sarcoma/translocated in liposarcoma (FUS/TLS) are displayed and characterized as the major pathology of ALS/FTLD (Nishimoto et al., 2010). Studies revealed that TDP-43 and FUS/TLS are enriched in paraspeckles and bound to NEAT1-2 directly. Furthermore, the expression of NEAT1-2 was shown to be enriched and the specific assembly of NEAT1-2 as paraspeckles be formed in spinal motor neurons at an early stage of the ALS pathogenesis (Nishimoto et al., 2013; Lourenco et al., 2015). These suggested NEAT1-2 might be functional in modulation of ALS-associated RNA-binding proteins at the early stage of ALS.
Along with intensive studies of lncRNAs, there are still challenges for our understanding of lncRNAs. On one side, most investigations of lncRNA are limited to single studies, and some of them are only an in vitro study. Kohtz (2014) has stated that the roles of lncRNAs in cell lines can be distinguished with the ones examined in animal models (in vivo). Taking Evf2 as an example, Evf2 cell line assays indicated the in trans mechanism of lncRNA functioning as activation enhancers, whereas the knockdown of Evf2 in Evf2TS/TS mice associated lncRNA with repression in cis (Feng et al., 2006; Bond et al., 2009). On the other side, some lncRNAs are only studied in mouse models with genomic deletions. With respect to the characters of lncRNAs that are located in the nucleus and chromatin-associated that might be cis-acting transcriptional regulators, or in cytoplasm that might be predicted to act in trans, the phenotypes produced by deletion of an entire genomic locus apparently cannot be equivalently the same as the loss of lncRNA per se or as the associated loss of other overlapping DNA regulatory elements (Bassett et al., 2014). In these cases, it will be difficult to distinguish from the effects caused by loss of a lncRNA transcript to the general effects caused by the loss of the genomic region itself (Kohtz, 2014; Aprea and Calegari, 2015). Considering these challenges, researchers have proposed the importance of optimizing currently employed techniques along with developing advanced ones as tools to help differentiating between the influences of lncRNAs when they act as molecular species compared to when they act as gene regulatory elements. For example, fluorescent in situ hybridization (FISH) has been widely applied for the analysis of lncRNA localization in tissues and subcellular levels (Chakraborty et al., 2012). Also, a series of interaction assays for the identification of protein or nucleic acids with lncRNAs are applied, such as protein-RNA (crosslinking immunoprecipitation, CLIP), RNA-RNA (crosslinking analysis of synthetic hybrids, CLASH) or RNA-DNA (capture hybridization analysis of RNA targets, CHART; Helwak et al., 2013; Huppertz et al., 2014; Vance and Ponting, 2014). Apart from these technical updates, the Clustered regularly interspersed palindromic repeats (CRISPR/Cas9) system, functioning as a mechanistic tool, is popularly used for targeted genome engineering (Cheng et al., 2013; Bassett et al., 2014). With regards to this, the landscape of lncRNAs with proven functions in various biological processes have been substantially increasing and this range is expected to be further expanding in the coming years.
The brain is the organ where more lncRNAs are abundantly expressed, comprising the highest proportion of brain-specific lncRNAs (Derrien et al., 2012). Although limited numbers of lncRNAs have been identified to be associated with the complexity of the brain functions, lncRNAs are involved in brain functions in both a normal and diseased state. It is implied that brain-specific lncRNAs may be innovatively evolved, involved in human brain development and related to neurodegenerative diseases (Vučičevič et al., 2014). Based on studies of lncRNAs functioning in brain development and pathophysiology of neurological disorders, the definition criteria include: (1) genetic variation in lncRNA genes leads to disease and influences susceptibility; (2) epigenetic deregulation of lncRNAs is associated with disease; (3) genomic context binds lncRNAs to disease-related genes and pathways; and (4) lncRNAs are interconnected with known pathogenic mechanisms (Qureshi and Mehler, 2013). However, it is worth noting that phenomena, like some lncRNAs either might show different expression levels in healthy brains compared to diseased brains, or they might interact with certain proteins that function in brain disorders, could be a result of multiple reasons that are not related to the disease or caused by unspecific side effects (Vučičevič et al., 2014). Apart from studies of lncRNA functions in cell lines that would reveal basic molecular mechanisms of lncRNAs, the generation of lncRNAs-knockdown or mutation in mouse models would be more effective to study the functional relevance of lncRNAs during brain development and in the physiological conditions of neurodegenerative disease.
In fact, as proposed by many studies, more and more attention has been given to lncRNAs as being disease biomarkers or potential targets for therapeutic strategies. Indeed, several commercial entities (e.g., OPKO-CURNA and RaNA therapeutics), which target lncRNAs have been developed to design and develop oligoneucleotide therapeutics for the treatment of CNS-related neurological disorders (Qureshi and Mehler, 2013). However, due to the large size of many lncRNAs, it became evident that the crossing of lncRNAs through the blood-brain barrier (BBB) will be a significant issue to be considered. Exosomes, 20–100 nm membrane nanovesicles of endocytic origin secreted by most cell types in vivo and in vitro, are shown to be natural carriers of functional RNAs (such as mRNA, miRNA, rRNA and lncRNAs) and proteins (Raposo et al., 1996; Valadi et al., 2007). Exosomes can also transfer these genetic materials between cells and subsequently modulate the functions of targeted cells. These characteristics indicate their important role in communications between cells, and therefore make them great potentials for therapeutic delivery (Kawikova and Askenase, 2015; Barile and Vassalli, 2017). Moreover, exosomes were found to have an impact on the pathophysiology of the brain due to the fact that they can also be released by CNS cells (Faure et al., 2006; Kawikova and Askenase, 2015). As suggested, utilization of exosomes as delivery cargo would be an efficient strategy for helping to bypass the BBB (Lakhal and Wood, 2011; Katakowski et al., 2013). Hereby, further investigation on the role of lncRNAs will provide a better understanding of how the brain functions and how diseases develop, and lead to greater insights into further therapeutic development for neurodegenerative diseases based on manipulations of lncRNA functions.
ZQ and HQ organized the article and ZQ drafted the manuscript; ZQ and DZ designed the pictures and prepared the draft; ZQ and HQ approved the final version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work is funded by the National Natural Science Foundation of China under grant No. 81671268 and the China Postdoctoral Science Foundation under grant No. 2016M600931. We are grateful to Dr. Zhang N, Dr. Dikicioglu D, Zhao J, Xu Y, Qazi T J and Duru L for their valuable comments and language editing of this manuscript.
Abdelmohsen, K., Panda, A. C., De, S., Grammatikakis, I., Kim, J., Ding, J., et al. (2015). Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY) 7, 903–910. doi: 10.18632/aging.100834
Airavaara, M., Pletnikova, O., Doyle, M. E., Zhang, Y. E., Troncoso, J. C., and Liu, Q. R. (2011). Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J. Biol. Chem. 286, 45093–45102. doi: 10.1074/jbc.M111.310250
Aprea, J., and Calegari, F. (2015). Long non-coding RNAs in corticogenesis: deciphering the non-coding code of the brain. EMBO J. 34, 2865–2884. doi: 10.15252/embj.201592655
Aprea, J., Prenninger, S., Dori, M., Ghosh, T., Monasor, L. S., Wessendorf, E., et al. (2013). Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 32, 3145–3160. doi: 10.1038/emboj.2013.245
Balik, V., Srovnal, J., Sulla, I., Kalita, O., Foltanova, T., Vaverka, M., et al. (2013). MEG3: a novel long noncoding potentially tumour-suppressing RNA in meningiomas. J. Neurooncol. 112, 1–8. doi: 10.1007/s11060-012-1038-6
Barile, L., and Vassalli, G. (2017). Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 174, 63–78. doi: 10.1016/j.pharmthera.2017.02.020
Barrachina, M., Castaño, E., Dalfó, E., Maes, T., Buesa, C., and Ferrer, I. (2006). Reduced ubiquitin C-terminal hydrolase-1 expression levels in dementia with Lewy bodies. Neurobiol. Dis. 22, 265–273. doi: 10.1016/j.nbd.2005.11.005
Bassett, A. R., Akhtar, A., Barlow, D. P., Bird, A. P., Brockdorff, N., Duboule, D., et al. (2014). Considerations when investigating lncRNA function in vivo. Elife 3:e03058. doi: 10.7554/eLife.03058
Batista, P. J., and Chang, H. Y. (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307. doi: 10.1016/j.cell.2013.02.012
Bergmann, J. H., and Spector, D. L. (2014). Long non-coding RNAs: modulators of nuclear structure and function. Curr. Opin. Cell Biol. 26, 10–18. doi: 10.1016/j.ceb.2013.08.005
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. doi: 10.1038/emboj.2010.199
Bond, A. M., Vangompel, M. J., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027. doi: 10.1038/nn.2371
Briggs, J. A., Wolvetang, E. J., Mattick, J. S., Rinn, J. L., and Barry, G. (2015). Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 88, 861–877. doi: 10.1016/j.neuron.2015.09.045
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. doi: 10.1126/science.1112014
Carrieri, C., Cimatti, L., Biagioli, M., Beugnet, A., Zucchelli, S., Fedele, S., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. doi: 10.1038/nature11508
Chakraborty, D., Kappei, D., Theis, M., Nitzsche, A., Ding, L., Paszkowski-Rogacz, M., et al. (2012). Combined RNAi and localization for functionally dissecting long noncoding RNAs. Nat. Methods 9, 360–362. doi: 10.1038/nmeth.1894
Chalei, V., Sansom, S. N., Kong, L., Lee, S., Montiel, J. F., Vance, K. W., et al. (2014). The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 3:e04530. doi: 10.7554/eLife.04530
Cheng, A. W., Wang, H., Yang, H., Shi, L., Katz, Y., Theunissen, T. W., et al. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23, 1163–1171. doi: 10.1038/cr.2013.122
Cherubini, E., Gustincich, S., and Robinson, H. (2006). The mammalian transcriptome and the cellular complexity of the brain. J. Physiol. 575, 319–320. doi: 10.1113/jphysiol.2006.118364
Choi, J., Levey, A. I., Weintraub, S. T., Rees, H. D., Gearing, M., Chin, L. S., et al. (2004). Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J. Biol. Chem. 279, 13256–13264. doi: 10.1074/jbc.M314124200
Chung, D. W., Rudnicki, D. D., Yu, L., and Margolis, R. L. (2011). A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum. Mol. Genet. 20, 3467–3477. doi: 10.1093/hmg/ddr263
Chureau, C., Chantalat, S., Romito, A., Galvani, A., Duret, L., Avner, P., et al. (2011). Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 20, 705–718. doi: 10.1093/hmg/ddq516
Clemson, C. M., McNeil, J. A., Willard, H. F., and Lawrence, J. B. (1996). XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275. doi: 10.1083/jcb.132.3.259
Cogswell, J. P., Ward, J., Taylor, I. A., Waters, M., Shi, Y., Cannon, B., et al. (2008). Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 14, 27–41. doi: 10.3233/jad-2008-14103
Costa, F. F. (2005). Non-coding RNAs: new players in eukaryotic biology. Gene 357, 83–94. doi: 10.1016/j.gene.2005.06.019
DeJesus-Hernandez, M., Mackenzie, I. R., Boeve, B. F., Boxer, A. L., Baker, M., Rutherford, N. J., et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. doi: 10.1016/j.neuron.2011.09.011
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Duning, K., Buck, F., Barnekow, A., and Kremerskothen, J. (2008). SYNCRIP, a component of dendritically localized mRNPs, binds to the translation regulator BC200 RNA. J. Neurochem. 105, 351–359. doi: 10.1111/j.1471-4159.2007.05138.x
Ebert, M. S., and Sharp, P. A. (2010). Emerging roles for natural microRNA sponges. Curr. Biol. 20, R858–R861. doi: 10.1016/j.cub.2010.08.052
Elling, R., Chan, J., and Fitzgerald, K. A. (2016). Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur. J. Immunol. 46, 504–512. doi: 10.1002/eji.201444558
ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi: 10.1038/nature11247
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
FANTOM Consortium and the RIKEN PMI and CLST (DGT), Forrest, A. R., Kawaji, H., Rehli, M., Baillie, J. K., de Hoon, M. J., et al. (2014). A promoter-level mammalian expression atlas. Nature 507, 462–470. doi: 10.1038/nature13182
Faure, J., Lachenal, G., Court, M., Hirrlinger, J., Chatellard-Causse, C., Blot, B., et al. (2006). Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci. 31, 642–648. doi: 10.1016/j.mcn.2005.12.003
Feng, J., Bi, C., Clark, B. S., Mady, R., Shah, P., and Kohtz, J. D. (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484. doi: 10.1101/gad.1416106
Gao, Y., Meng, H., Liu, S., Hu, J., Zhang, Y., Jiao, T., et al. (2015). LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum. Mol. Genet. 24, 841–852. doi: 10.1093/hmg/ddu502
Gonzalez, I., Munita, R., Agirre, E., Dittmer, T. A., Gysling, K., Misteli, T., et al. (2015). A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 22, 370–376. doi: 10.1038/nsmb.3005
Grivna, S. T., Beyret, E., Wang, Z., and Lin, H. (2006). A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714. doi: 10.1101/gad.1434406
Grote, P., and Herrmann, B. G. (2013). The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 10, 1579–1585. doi: 10.4161/rna.26165
Grote, P., Wittler, L., Hendrix, D., Koch, F., Währisch, S., Beisaw, A., et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214. doi: 10.1016/j.devcel.2012.12.012
Gstir, R., Schafferer, S., Scheideler, M., Misslinger, M., Griehl, M., Daschil, N., et al. (2014). Generation of a neuro-specific microarray reveals novel differentially expressed noncoding RNAs in mouse models for neurodegenerative diseases. RNA 20, 1929–1943. doi: 10.1261/rna.047225.114
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. doi: 10.1038/nature07672
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G., et al. (2011). lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. doi: 10.1038/nature10398
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hart, R. P., and Goff, L. A. (2016). Long noncoding RNAs: central to nervous system development. Int. J. Dev. Neurosci. 55, 109–116. doi: 10.1016/j.ijdevneu.2016.06.001
Helwak, A., Kudla, G., Dudnakova, T., and Tollervey, D. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665. doi: 10.1016/j.cell.2013.03.043
Huppertz, I., Attig, J., D’Ambrogio, A., Easton, L. E., Sibley, C. R., Sugimoto, Y., et al. (2014). iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65, 274–287. doi: 10.1016/j.ymeth.2013.10.011
Jeck, W. R., and Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–461. doi: 10.1038/nbt.2890
Ji, Z., Song, R., Regev, A., and Struhl, K. (2015). Many lncRNAs, 5’UTRs and pseudogenes are translated and some are likely to express functional proteins. Elife 4:e08890. doi: 10.7554/eLife.08890
Jiang, J., Jing, Y., Cost, G. J., Chiang, J. C., Kolpa, H. J., Cotton, A. M., et al. (2013). Translating dosage compensation to trisomy 21. Nature 500, 296–300. doi: 10.1038/nature12394
Jiang, W., Liu, Y., Liu, R., Zhang, K., and Zhang, Y. (2015). The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 11, 137–148. doi: 10.1016/j.celrep.2015.03.008
Kadakkuzha, B. M., Liu, X. A., McCrate, J., Shankar, G., Rizzo, V., Afinogenova, A., et al. (2015). Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front. Cell. Neurosci. 9:63. doi: 10.3389/fncel.2015.00063
Kaessmann, H. (2010). Origins, evolution, and phenotypic impact of new genes. Genome Res. 20, 1313–1326. doi: 10.1101/gr.101386.109
Kapranov, P., St Laurent, G., Raz, T., Ozsolak, F., Reynolds, C. P., Sorensen, P. H., et al. (2010). The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol 8:149. doi: 10.1186/1741-7007-8-149
Kapusta, A., Kronenberg, Z., Lynch, V. J., Zhuo, X., Ramsay, L., Bourque, G., et al. (2013). Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9:e1003470. doi: 10.1371/journal.pgen.1003470
Katakowski, M., Buller, B., Zheng, X., Lu, Y., Rogers, T., Osobamiro, O., et al. (2013). Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 335, 201–204. doi: 10.1016/j.canlet.2013.02.019
Kawikova, I., and Askenase, P. W. (2015). Diagnostic and therapeutic potentials of exosomes in CNS diseases. Brain Res. 1617, 63–71. doi: 10.1016/j.brainres.2014.09.070
Kazemzadeh, M., Safaralizadeh, R., and Orang, A. V. (2015). LncRNAs: emerging players in gene regulation and disease pathogenesis. J. Genet. 94, 771–784. doi: 10.1007/s12041-015-0561-6
Klattenhoff, C. A., Scheuermann, J. C., Surface, L. E., Bradley, R. K., Fields, P. A., Steinhauser, M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. doi: 10.1016/j.cell.2013.01.003
Klein, U., Lia, M., Crespo, M., Siegel, R., Shen, Q., Mo, T., et al. (2010). The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 17, 28–40. doi: 10.1016/j.ccr.2009.11.019
Kohtz, J. D. (2014). Long non-coding RNAs learn the importance of being in vivo. Front. Genet. 5:45. doi: 10.3389/fgene.2014.00045
Kung, J. T., Colognori, D., and Lee, J. T. (2013). Long noncoding RNAs: past, present, and future. Genetics 193, 651–669. doi: 10.1534/genetics.112.146704
Lakhal, S., and Wood, M. J. (2011). Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. Bioessays 33, 737–741. doi: 10.1002/bies.201100076
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Lee, S. T., Chu, K., Im, W. S., Yoon, H. J., Im, J. Y., Park, J. E., et al. (2011). Altered microRNA regulation in Huntington’s disease models. Exp. Neurol. 227, 172–179. doi: 10.1016/j.expneurol.2010.10.012
Lee, S., Kopp, F., Chang, T. C., Sataluri, A., Chen, B., Sivakumar, S., et al. (2016). Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164, 69–80. doi: 10.1016/j.cell.2015.12.017
Li, K., and Ramchandran, R. (2010). Natural antisense transcript: a concomitant engagement with protein-coding transcript. Oncotarget 1, 447–452. doi: 10.18632/oncotarget.178
Li, J., Tian, H., Yang, J., and Gong, Z. (2016). Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol. 35, 459–470. doi: 10.1089/dna.2015.3187
Lin, N., Chang, K. Y., Li, Z., Gates, K., Rana, Z. A., Dang, J., et al. (2014). An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1005–1019. doi: 10.1016/j.molcel.2014.01.021
Lipovich, L., Tarca, A. L., Cai, J., Jia, H., Chugani, H. T., Sterner, K. N., et al. (2014). Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cereb. Cortex 24, 1451–1459. doi: 10.1093/cercor/bhs414
Lourenco, G. F., Janitz, M., Huang, Y., and Halliday, G. M. (2015). Long noncoding RNAs in TDP-43 and FUS/TLS-related frontotemporal lobar degeneration (FTLD). Neurobiol. Dis. 82, 445–454. doi: 10.1016/j.nbd.2015.07.011
Lukiw, W. J. (2013). Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet. 4:307. doi: 10.3389/fgene.2013.00307
Managadze, D., Lobkovsky, A. E., Wolf, Y. I., Shabalina, S. A., Rogozin, I. B., and Koonin, E. V. (2013). The vast, conserved mammalian lincRNome. PLoS Comput. Biol. 9:e1002917. doi: 10.1371/journal.pcbi.1002917
Massone, S., Vassallo, I., Fiorino, G., Castelnuovo, M., Barbieri, F., Borghi, R., et al. (2011). 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 41, 308–317. doi: 10.1016/j.nbd.2010.09.019
Mattick, J. S., and Rinn, J. L. (2015). Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22, 5–7. doi: 10.1038/nsmb.2942
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U S A 105, 716–721. doi: 10.1073/pnas.0706729105
Milligan, M. J., and Lipovich, L. (2014). Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front. Genet. 5:476. doi: 10.3389/fgene.2014.00476
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459. doi: 10.1038/nbt.2158
Modarresi, F., Faghihi, M. A., Patel, N. S., Sahagan, B. G., Wahlestedt, C., and Lopez-Toledano, M. A. (2011). Knockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesis. Int. J. Alzheimers Dis. 2011:929042. doi: 10.4061/2011/929042
Molyneaux, B. J., Goff, L. A., Brettler, A. C., Chen, H. H., Brown, J. R., Hrvatin, S., et al. (2015). DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288. doi: 10.1016/j.neuron.2014.12.024
Muddashetty, R., Khanam, T., Kondrashov, A., Bundman, M., Iacoangeli, A., Kremerskothen, J., et al. (2002). Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 321, 433–445. doi: 10.1016/s0022-2836(02)00655-1
Mus, E., Hof, P. R., and Tiedge, H. (2007). Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. U S A 104, 10679–10684. doi: 10.1073/pnas.0701532104
Muslimov, I. A., Santi, E., Homel, P., Perini, S., Higgins, D., and Tiedge, H. (1997). RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J. Neurosci. 17, 4722–4733.
Ng, S. Y., Bogu, G. K., Soh, B. S., and Stanton, L. W. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. doi: 10.1016/j.molcel.2013.07.017
Ng, S. Y., Johnson, R., and Stanton, L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. doi: 10.1038/emboj.2011.459
Nishihara, H., Smit, A. F., and Okada, N. (2006). Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 16, 864–874. doi: 10.1101/gr.5255506
Nishimoto, Y., Ito, D., Yagi, T., Nihei, Y., Tsunoda, Y., and Suzuki, N. (2010). Characterization of alternative isoforms and inclusion body of the TAR DNA-binding protein-43. J. Biol. Chem. 285, 608–619. doi: 10.1074/jbc.M109.022012
Nishimoto, Y., Nakagawa, S., Hirose, T., Okano, H. J., Takao, M., Shibata, S., et al. (2013). The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 6:31. doi: 10.1186/1756-6606-6-31
Parenti, R., Paratore, S., Torrisi, A., and Cavallaro, S. (2007). A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during beta-amyloid-induced apoptosis. Eur. J. Neurosci. 26, 2444–2457. doi: 10.1111/j.1460-9568.2007.05864.x
Pinter, S. F., Sadreyev, R. I., Yildirim, E., Jeon, Y., Ohsumi, T. K., Borowsky, M., et al. (2012). Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 22, 1864–1876. doi: 10.1101/gr.133751.111
Ponjavic, J., Oliver, P. L., Lunter, G., and Ponting, C. P. (2009). Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5:e1000617. doi: 10.1371/journal.pgen.1000617
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Qureshi, I. A., and Mehler, M. F. (2012). Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 13, 528–541. doi: 10.1038/nrn3234
Qureshi, I. A., and Mehler, M. F. (2013). Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics 10, 632–646. doi: 10.1007/s13311-013-0199-0
Ramos, A. D., Andersen, R. E., Liu, S. J., Nowakowski, T. J., Hong, S. J., Gertz, C. C., et al. (2015). The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447. doi: 10.1016/j.stem.2015.02.007
Ramos, A. D., Diaz, A., Nellore, A., Delgado, R. N., Park, K. Y., Gonzales-Roybal, G., et al. (2013). Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12, 616–628. doi: 10.1016/j.stem.2013.03.003
Rani, N., Nowakowski, T. J., Zhou, H., Godshalk, S. E., Lisi, V., Kriegstein, A. R., et al. (2016). A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron 90, 1174–1188. doi: 10.1016/j.neuron.2016.05.005
Raposo, G., Nijman, H. W., Stoorvogel, W., Liejendekker, R., Harding, C. V., Melief, C. J., et al. (1996). B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172. doi: 10.1084/jem.183.3.1161
Reddy, A. S., O’Brien, D., Pisat, N., Weichselbaum, C. T., Sakers, K., Lisci, M., et al. (2017). A comprehensive analysis of cell type-specific nuclear RNA from neurons and glia of the brain. Biol. Psychiatry 81, 252–264. doi: 10.1016/j.biopsych.2016.02.021
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi: 10.1146/annurev-biochem-051410-092902
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Roberts, T. C., Morris, K. V., and Wood, M. J. (2014). The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130507. doi: 10.1098/rstb.2013.0507
Salameh, A., Lee, A. K., Cardó-Vila, M., Nunes, D. N., Efstathiou, E., Staquicini, F. I., et al. (2015). PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl. Acad. Sci. U S A 112, 8403–8408. doi: 10.1073/pnas.1507882112
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 73, 2491–2509. doi: 10.1007/s00018-016-2174-5
Sosinska, P., Mikula-Pietrasik, J., and Ksiazek, K. (2015). The double-edged sword of long non-coding RNA: the role of human brain-specific BC200 RNA in translational control, neurodegenerative diseases, and cancer. Mutat. Res. Rev. Mutat. Res. 766, 58–67. doi: 10.1016/j.mrrev.2015.08.002
Tian, D., Sun, S., and Lee, J. T. (2010). The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143, 390–403. doi: 10.1016/j.cell.2010.09.049
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. doi: 10.1016/j.cell.2013.06.020
Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J. J., and Lötvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659. doi: 10.1038/ncb1596
Vance, K. W., and Ponting, C. P. (2014). Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 30, 348–355. doi: 10.1016/j.tig.2014.06.001
Vance, K. W., Sansom, S. N., Lee, S., Chalei, V., Kong, L., Cooper, S. E., et al. (2014). The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 33, 296–311. doi: 10.1002/embj.201386225
Vicens, Q., and Westhof, E. (2014). Biogenesis of circular RNAs. Cell 159, 13–14. doi: 10.1016/j.cell.2014.09.005
Vučičevič, D., Schrewe, H., and Orom, U. A. (2014). Molecular mechanisms of long ncRNAs in neurological disorders. Front. Genet. 5:48. doi: 10.3389/fgene.2014.00048
Wang, F., Ying, H. Q., He, B. S., Pan, Y. Q., Deng, Q. W., Sun, H. L., et al. (2015). Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget 6, 7899–7917. doi: 10.18632/oncotarget.3219
Wang, P., Xue, Y., Han, Y., Lin, L., Wu, C., Xu, S., et al. (2014). The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313. doi: 10.1126/science.1251456
Wu, P., Zuo, X., Deng, H., Liu, X., Liu, L., and Ji, A. (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 97, 69–80. doi: 10.1016/j.brainresbull.2013.06.001
Xing, Z., Lin, A., Li, C., Liang, K., Wang, S., Liu, Y., et al. (2014). lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110–1125. doi: 10.1016/j.cell.2014.10.013
Xiong, X. D., Ren, X., Cai, M. Y., Yang, J. W., Liu, X., and Yang, J. M. (2016). Long non-coding RNAs: an emerging powerhouse in the battle between life and death of tumor cells. Drug Resist. Updat. 26, 28–42. doi: 10.1016/j.drup.2016.04.001
Zhang, X., Zhou, Y., Mehta, K. R., Danila, D. C., Scolavino, S., Johnson, S. R., et al. (2003). A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab. 88, 5119–5126. doi: 10.1210/jc.2003-030222
Zhong, J., Chuang, S. C., Bianchi, R., Zhao, W., Lee, H., Fenton, A. A., et al. (2009). BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 29, 9977–9986. doi: 10.1523/JNEUROSCI.3893-08.2009
Keywords: long non-coding RNAs (lncRNAs), central nervous system (CNS), neurodegenerative disease, gene expression, transcriptional regulation
Citation: Quan Z, Zheng D and Qing H (2017) Regulatory Roles of Long Non-Coding RNAs in the Central Nervous System and Associated Neurodegenerative Diseases. Front. Cell. Neurosci. 11:175. doi: 10.3389/fncel.2017.00175
Received: 13 April 2017; Accepted: 09 June 2017;
Published: 30 June 2017.
Edited by:
Sandro Alves, Brainvectis Therapeutics, FranceReviewed by:
Grzegorz Kreiner, Institute of Pharmacology (PAS), PolandCopyright © 2017 Quan, Zheng and Qing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Qing, aHFpbmdAYml0LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.