- 1Institut de Génomique Fonctionnelle, CNRS UMR 5203, Universités Montpellier 1&2, Montpellier, France
- 2INSERM, U 661, Montpellier, France
- 3LabEx 'Ion Channel Science and Therapeutics', Montpellier, France
Ion channels are crucial components of cellular excitability and are involved in many neurological diseases. This review focuses on the sodium leak, G protein-coupled receptors (GPCRs)-activated NALCN channel that is predominantly expressed in neurons where it regulates the resting membrane potential and neuronal excitability. NALCN is part of a complex that includes not only GPCRs, but also UNC-79, UNC-80, NLF-1 and src family of Tyrosine kinases (SFKs). There is growing evidence that the NALCN channelosome critically regulates its ion conduction. Both in mammals and invertebrates, animal models revealed an involvement in many processes such as locomotor behaviors, sensitivity to volatile anesthetics, and respiratory rhythms. There is also evidence that alteration in this NALCN channelosome can cause a wide variety of diseases. Indeed, mutations in the NALCN gene were identified in Infantile Neuroaxonal Dystrophy (INAD) patients, as well as in patients with an Autosomal Recessive Syndrome with severe hypotonia, speech impairment, and cognitive delay. Deletions in NALCN gene were also reported in diseases such as 13q syndrome. In addition, genes encoding NALCN, NLF- 1, UNC-79, and UNC-80 proteins may be susceptibility loci for several diseases including bipolar disorder, schizophrenia, Alzheimer's disease, autism, epilepsy, alcoholism, cardiac diseases and cancer. Although the physiological role of the NALCN channelosome is poorly understood, its involvement in human diseases should foster interest for drug development in the near future. Toward this goal, we review here the current knowledge on the NALCN channelosome in physiology and diseases.
Introduction
Ion channels are integral membrane proteins that allow specific ions to pass through lipid membranes following a concentration gradient (Hille, 2001). More than 400 genes are known that encode ion channel subunits. In addition, alternative splicing and heteromeric assembly of different subunits increase tremendously the variety of ion channels. They are involved in many signaling and control processes in the cell as well as in pathologies referred to as “channelopathies” (reviewed in Ashcroft, 2006; Camerino et al., 2008). In addition, pharmaceutical companies view ion channels as therapeutic targets of choice (Kaczorowski et al., 2008; Clare, 2010). In the present review, we focus on the Na+-leak channel (NALCN), a major player in determining the influence of extracellular Na+ on a neuron's basal excitability and its modulation by hormones and neurotransmitters.
Structure of NALCN
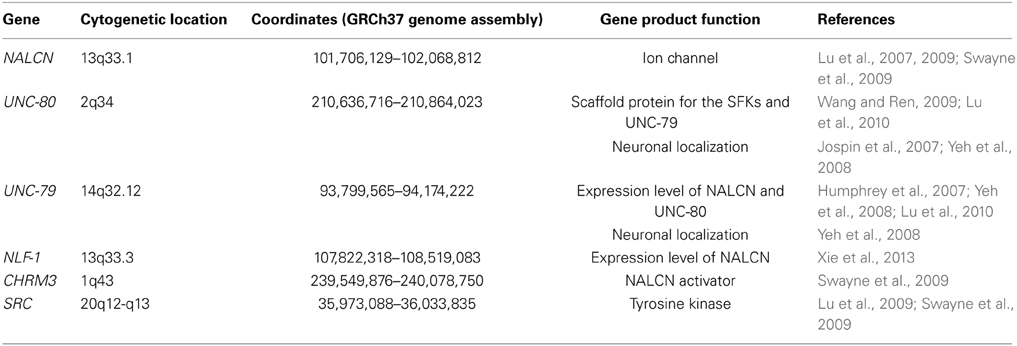
NALCN (also named Rb21, VGCNL-1, NA in Drosophila melanogaster and NCA-1/2 in Caenorhabditis elegans) was first cloned from rat brain and described by Perez-Reyes and colleagues who named it Rb21 (Lee et al., 1999). With the exception of the nematode Caenorhabditis, the cnidarian Nematostella and the sponge Amphimedon that have two related channels, there is only one gene encoding NALCN in other organisms (Liebeskind et al., 2012; Senatore et al., 2013). In mammals, NALCN is a 1738 amino-acids protein that forms the channel pore of the complex and has a predicted topology similar to voltage-gated sodium and calcium channels (Snutch and Monteil, 2007)(Figure 1A). Unlike other members of the four-domain ion channel family, the S4 transmembrane segments have fewer positive residues, especially in domains 2 and 4, possibly explaining NALCN's voltage insensitivity. The predicted pore region is also unique in that its ionic selectivity motif differs from that of calcium channels (EEEE or EEDD) and sodium channels (DEKA). NALCN's ion selectivity motif (EEKE) is implicated in its specific permeation properties. With the exception of C. elegans and D. melanogaster, alternative splicing events in the pore-forming region of invertebrate NALCN result in a calcium channel-like EEEE motif or a sodium channel-like EEKE (or EKEE) motif that remain to be explored at the functional level (Senatore et al., 2013). These findings suggest that NALCN could behave as a sodium or calcium channel depending on the expressed isoform (Senatore et al., 2013).
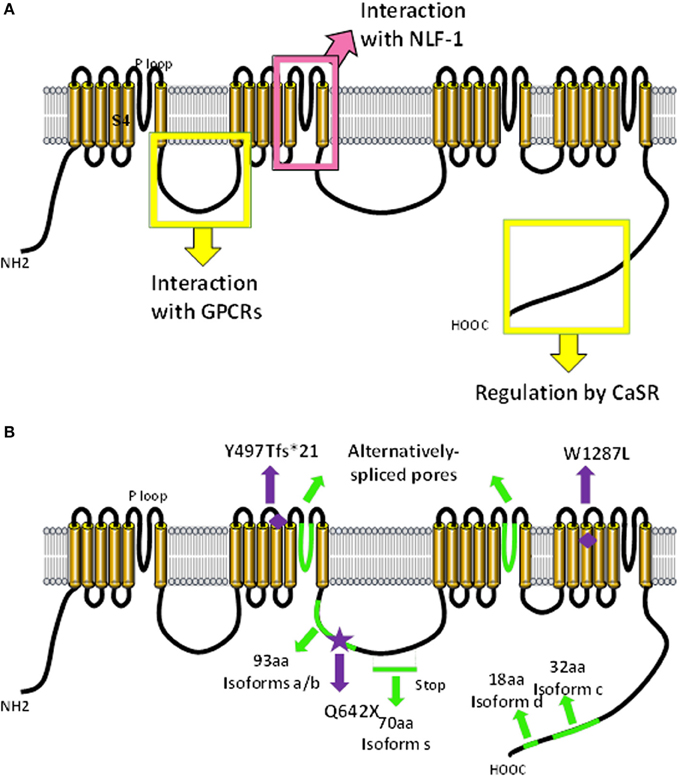
Figure 1. Predicted molecular make-up of NALCN. The predicted structure of NALCN is similar to α/α1 subunits of voltage-gated Na+ and Ca2+ channels (Lee et al., 1999). (A) It has four homologs repeats (domains I–IV) each composed of six transmembrane segments (S1–6). Four pore-forming loops (P-loops) spanning from S5 to S6 make up the ion selectivity filter. The intracellular loop linking domains I and II of NALCN interacts with M3R and molecular determinants involved in the regulation by CaSR are located in the carboxy-terminus (Swayne et al., 2009; Lu et al., 2010). The S5/P loop/S6 segment of transmembrane domain II is required for the interaction with NLF-1 (Xie et al., 2013). (B) Alternative events (shown in green) are detected in the intracellular loop linking domains II and III, in the P-loops of domains II and III, as well as in the carboxy-terminus (Senatore et al., 2013; data not shown). Mutations found in patients with INAD and an autosomal recessive syndrome with severe hypotonia, speech impairment, and cognitive delay are shown (Al-Sayed et al., 2013; Koroglu et al., 2013).
In humans, the gene encoding NALCN is located on chromosome 13q33.1 and comprises at least 44 exons (43 coding exons). Several splice variants were identified during our cloning step and by database scanning (unpublished results Figure 1B). Alternative splicing events were found in the intracellular loop linking domains 2 and 3 and in the carboxy-terminus region. Interestingly, there is a long non-coding RNA (lncRNA) gene named NALCN-AS1 partially overlapping with NALCN on the reverse strand (Gene ID: 100885778; http://www.ncbi.nlm.nih.gov/). lncRNAs are thought to function in various cellular contexts, including post-transcriptional regulation, post-translational regulation of protein activity, organization of protein complexes, cell–cell signaling, as well as recombination (reviewed in Geisler and Coller, 2013; Sabin et al., 2013). Importantly, lncRNAs would be associated with a wide range of neurodevelopmental, neurodegenerative and psychiatric diseases, as well as brain cancer (reviewed in Barry, 2014). The specific function of NALCN-AS1 remains to be elucidated.
NALCN Function
Gating Properties
The first functional characterization of NALCN was described by Ren and colleagues, who identified NALCN as the channel responsible for a tetrodotoxin (TTX)-resistant sodium leak current in mouse hippocampal neurons (Lu et al., 2007). In Nalcn knockout mice, they found that hippocampal neurons were hyperpolarized by ~10 mV compared to wild-type mice (Lu et al., 2007). They concluded that NALCN would contribute to the resting membrane potential in these neurons by eliciting a depolarizing current to counterbalance the hyperpolarizing current induced by two-pore potassium channels (reviewed in Ren, 2011; Lu and Feng, 2012). A similar NALCN-like channel activity was described in neurons from L. stagnalis and C. elegans (Lu and Feng, 2011; Xie et al., 2013). Contrasting with these data, NALCN does not conduct a background current but rather drives an acetylcholine-activated sodium current in the MIN-6 cell line, a pancreatic β-cell model (Swayne et al., 2009). This acetylcholine-activated NALCN current requires the M3 muscarinic receptor (M3R) and occurs though a G protein-independent, Src family of tyrosine kinases (SFK)-dependent pathway. Similarly, NALCN current was found to be activated by substance P (SP) and neurotensin through a SFK-dependent pathway in mouse hippocampal and ventral tegmental area neurons (Lu et al., 2009). It is not clear why NALCN behaves as a leak channel (e.g., looks like spontaneously active) in neuronal cells and not in MIN-6 cells and whether NALCN may be considered as a GPCR-activated channel with a cell type-dependent basal activity or as a constitutively open channel regulated by GPCRs. Although these studies provide the first demonstration for the functional properties and regulation of NALCN channels, much remains to be determined about the functionality of NALCN and the mechanism(s) responsible for its activation. Indeed, it has also been hypothesized that NALCN may not be an ion channel per se but rather an ion sensor (Senatore and Spafford, 2013). Discordance in the field indicates that more work is clearly required to reveal the real “channel” identity of NALCN. In the context of this review, we will consider NALCN as a pore-forming subunit.
The NALCN Channelosome
Like many ion channels, NALCN is associated with several proteins to form a larger channel complex (Figure 2, Table 1). These interacting proteins are involved in the folding, stabilization, cellular localization, and activation of NALCN.
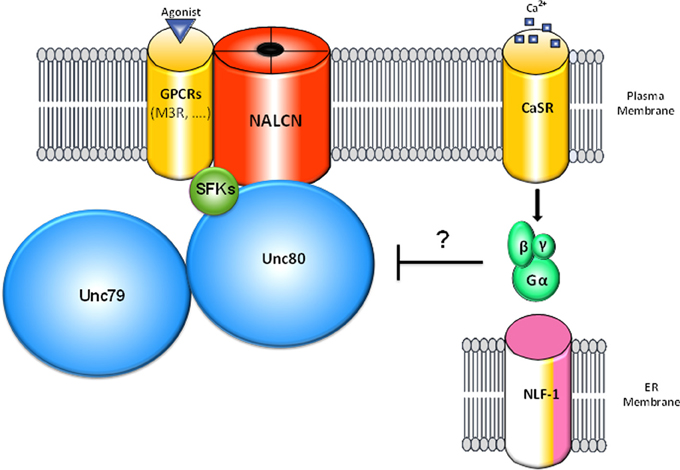
Figure 2. Schematic representation of the NALCN channelosome. The NALCN ion channel interacts with the M3 muscarinic receptor (M3R) that activates the channel through a G protein-independent and Src Family of Tyrosine Kinases (SFK)-dependent pathway upon activation by acetylcholine (Swayne et al., 2009). UNC-80 interacts with NALCN and SFKs and acts as a scaffolding protein (Wang and Ren, 2009; Lu et al., 2010). In addition, UNC-80 is involved in the expression levels of NALCN and UNC-79 as well as their neuronal localization (Jospin et al., 2007; Yeh et al., 2008). UNC-79 interacts with UNC-80 but not NALCN and is involved in the expression levels and the neuronal localization of NALCN and UNC-80 (Yeh et al., 2008). CaSR was found to regulate NALCN channel activity by an uncharacterized mechanism involving G-proteins activation and UNC-80 (Lu et al., 2010). For clarity, NLF-1, an endoplasmic reticulum-resident protein that interacts with NALCN and is possibly involved in its folding is shown apart. It is not known whether the interactions mentioned above are direct or not.
UNC-80
UNC-80 (also named KIAA1843, c2orf21) is located on human chromosome 2q34 and has at least 45 exons. UNC-80 is a large protein of about 3300 amino acids without any predicted transmembrane segments or particular functional domains. UNC-80 contributes to the neuronal localization and/or stabilization of NALCN channel in C. elegans and D. melanogaster (Jospin et al., 2007; Yeh et al., 2008; Lear et al., 2013). In addition, UNC-80 acts as a scaffold protein for the SFKs and UNC-79. In fact, NALCN can interact with UNC-80 in the absence of UNC-79 and UNC-79 can interact with UNC-80 in the absence of NALCN. The interaction of NALCN and UNC-80 is required for the NALCN activation/inhibition by GPCRs in neurons (Wang and Ren, 2009; Lu et al., 2010). Similar to NALCN, databases indicate the existence of several UNC-80 splice variants.
UNC-79
UNC-79 (also named KIAA1409) is located on human chromosome 14q32.12 and has at least 48 exons. UNC-79 is also a large protein (~2800 amino acids) without any predicted transmembrane segments or particular functional domains. UNC-79 was found to be involved in regulating the neuronal localization of the NALCN channel complex both in C. elegans and D. melanogaster (Humphrey et al., 2007; Yeh et al., 2008; Lear et al., 2013). UNC-79 has been found to regulate the expression level of UNC-80 and, as a pair, UNC-79 and UNC-80 regulate the expression level of NALCN. This likely occurs through a post-transcriptional mechanism as no difference in the corresponding mRNA levels was found (Humphrey et al., 2007; Yeh et al., 2008). This regulation may not be conserved in mammals. As a matter of fact, Unc-79 mutant mice lack detectable levels of the UNC-80 protein but NALCN is still present (Lu et al., 2010; Speca et al., 2010). Another difference between invertebrates and mammals concerns the redundant functions of these proteins. Indeed, in D. melanogaster, transgenic expression of NALCN, UNC-79, or UNC-80 does not relieve the requirement for the other subunits while, in mouse primary neurons, UNC-80 can bypass the requirement for UNC-79 (Lu et al., 2010; Lear et al., 2013).
NLF-1
NLF-1 (NCA Localization Factor 1, also named FAM155A) is an endoplasmic reticulum (ER) resident protein of around 438-468 amino-acids, depending on the species, that interacts with NALCN and promotes its neuronal localization in C. elegans (Xie et al., 2013). The gene encoding NLF-1 is located on human chromosome 13q33.3 and contains 3 exons. An intronic and non-coding transcript gene, FAM155A-IT1, is described on the same strand (Gene ID: 100874375; http://www.ncbi.nlm.nih.gov/) with no known function. In C. elegans, the loss of function of nlf-1 results in a reduced leak current and a hyperpolarized resting membrane potential in premotor interneurons. NLF-1 function is conserved across species as the mouse homolog functionally substitutes to the C. elegans one. In addition, knockdown of the D. melanogaster ortholog gene, CG33988 results in similar phenotypes as for na (see below; Ghezzi et al., 2014). Also, knockdown of Nlf-1 in primary mouse cortical cultures effectively reduced the background Na+ current. It has been postulated that NLF-1 could function as a chaperone to facilitate the folding and assembly of NALCN channels. This interaction involves the second transmembrane domain of NALCN and requires its S5/P loop/S6 segment. The topology of NLF-1 is predicted to have one single transmembrane domain within 40 residues of the carboxy-terminus and lack known amino-terminal signal sequences. There are putative ER retention motifs (RXR) at the amino-terminus region. nlf-1 is expressed in sensory neurons, interneurons, and excitatory motor neurons in C. elegans. Endogenous UNC-79 level is also significantly decreased in nlf-1 mutants but the NLF-1 level is unaffected by the absence of UNC-79 and UNC-80. In humans, there is another gene homologous to NLF-1, FAM155B that is localized to Xq13.1 and encodes for a putative protein of 473 amino-acids. It is not known if this putative protein belongs to the NALCN channel complex. Both NLF-1 and FAM155B have a cysteine-rich domain (CRD) involved in signal transduction in other proteins (Pei and Grishin, 2012). In addition, these two proteins are related to Mid1, a fungal stretch-activated calcium channel component that forms a complex with the transmembrane calcium channel Cch1 (Maruoka et al., 2002).
G protein-coupled receptors (GPCRs)
In addition to its baseline activity, NALCN activity is enhanced/modulated by several GPCRs. Acetylcholine-induced NALCN current requires the interaction of NALCN and M3R proteins through the 1–2 loop of NALCN and the i3 and carboxy-terminus of M3R (Swayne et al., 2009). Other GPCRs, the Neurokinin 1 receptor (NK1R) and a neurotensin receptor that remains to be identified, activate NALCN upon the binding of their ligands SP and neurotensin in primary mouse pyramidal hippocampal neurons and dopaminergic neurons from the ventral tegmental area (Lu et al., 2009). However, whether NK1R interacts with NALCN remains to be shown. This raises the possibility that several GPCRs may activate NALCN and the challenge to determine the repertoire of NALCN-activating GPCRs persists. A full analysis of the literature gives some clues on this point. For example, it has been reported that 5-HT modulates a non-selective leak current involved in the resting membrane potential in the pre-Bötzinger complex and motor neurons likely through activation of the 5-HT2A receptor (Ptak et al., 2009). Also, a cation conductance activated by glutamatergic metabotropic receptors through a G protein-independent pathway was described in CA3 hippocampal pyramidal neurons (Guerineau et al., 1995). In addition to its activation by several GPCRs, NALCN was found to be inhibited by another GPCR, the calcium-sensing receptor (CaSR) (Lu et al., 2010). This regulation is G-protein-dependent and SFK-independent, requires UNC-80 and involves molecular determinants in the carboxy-terminal portion of NALCN. It is not known if CaSR belongs to the NALCN channel complex.
Expression Pattern
NALCN is mainly expressed in the central nervous system (CNS) but also in heart, adrenal gland, thyroid gland, lymph node, and islets of Langerhans (both in α- and β-cells) (Lee et al., 1999; Kutlu et al., 2009; Swayne et al., 2009; Koroglu et al., 2013; Rorsman and Braun, 2013; see http://t1dbase.org). In the CNS, NALCN is expressed mainly in neurons, to a lesser extent in oligodendrocytes, and at a very low level in astrocytes (Cahoy et al., 2008). The temporal expression of NALCN is strongly correlated with the expression of genes involved in synapse development and with synaptic density in several areas of the human brain (Kang et al., 2011; see http://hbatlas.org/). The spatial expression of NALCN is also correlated with genes expressed in specific neural cell types such as CALB1 (Cortical GABA interneurons), CTGF and UNC-5C (Subplate cortical glutamatergic neurons). In invertebrates, NALCN-orthologs are concentrated at the synapse in D. melanogaster, and along the axon in C. elegans and L. stagnalis (Nash et al., 2002; Humphrey et al., 2007; Yeh et al., 2008; Senatore et al., 2013). The cellular sub-localization of NALCN in mammals is not known.
Physiological Roles of NALCN: Lessons from Animal Models
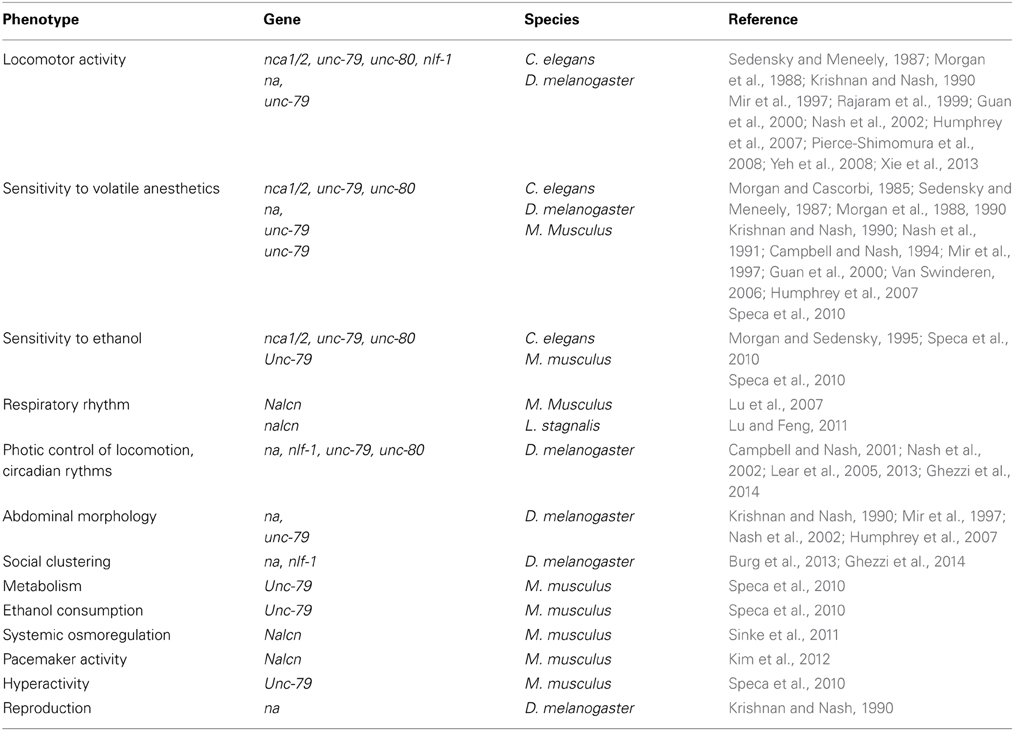
Mutations in genes coding for NALCN channel proteins, both in mammals and invertebrates, yield a wide range of phenotypes. Functional knock-out or hypomorphic mutations in these genes produce viable offspring (but fewer compared to wild-type) in C. elegans and D. melanogaster, whereas it is post-embryonic lethal in homozygous null mice (Krishnan and Nash, 1990; Nakayama et al., 2006; Lu et al., 2007; Speca et al., 2010). While no obvious developmental defects, including neuronal development, were found in Nalcn and Unc-79 mutant mice, these mice die as a result of disrupted respiratory rhythm (Lu et al., 2007, see below). The depletion of nca1/2, unc-79 or unc-80 in C. elegans does not result in any gross abnormality in neuronal cell body position, neuronal processes, or fasciculation suggesting that these mutations do not interfere with the nervous system development (Pierce-Shimomura et al., 2008). We present below the phenotypes observed for the animal mutants in NALCN channel complex (Table 2).
Locomotor Activity
Wild-type C. elegans travels on a culture plate through the continuous and rhythmic propagation of sinusoidal body bends. A simultaneous loss of both nca-1 and nca-2, or the individual loss of unc-79, unc-80 or nlf-1 results in fainting, a unique motor deficit characterized by periodic halting during movement (Sedensky and Meneely, 1987; Morgan et al., 1988; Rajaram et al., 1999; Humphrey et al., 2007; Jospin et al., 2007; Xie et al., 2013). A dysfunction of premotor interneurons was found to be the primary cause of the failure in the initiation and maintenance of rhythmic locomotion exhibited by fainters (Xie et al., 2013). A “hesitant” walking phenotype (e.g., the walk is interrupted by pauses) is also observed when na or unc-79 are invalidated in D. melanogaster (Krishnan and Nash, 1990; Mir et al., 1997; Guan et al., 2000; Nash et al., 2002; Humphrey et al., 2007). Conversely, gain-of-function mutations in nca-1 lead to exaggerated body bending, termed coiling, in C. elegans (Yeh et al., 2008).
When transitioned between solid and liquid environments, C. elegans switch between two patterns of rhythmic locomotion, crawling and swimming that are distinct in both kinematics and pattern of muscle activity (Pierce-Shimomura et al., 2008). A genetic screen was performed in order to find mutants capable of normal crawling but incapable of normal swimming. unc-79, unc-80 and nca-1;nca-2 mutants were found to be paralyzed upon immersion in liquid. The paralytic defect in swimming does not seem to be explained by general defects in neuronal excitability, synaptic function, or development because the fainting phenotype was not observed in mutants defective in voltage-gated calcium channels or major synaptic proteins.
Sensitivity to General Volatile Anesthetics (GAs) and Ethanol
Several studies reported an altered sensitivity to GAs both in invertebrates and mice mutants with some discrepancies. In C. elegans, nca-1;nca-2, unc-79, and unc-80 mutants are hypersensitive to the immobilizing effect of halothane (~2–3 fold increase compared to controls) and other anesthetic agents (Sedensky and Meneely, 1987; Morgan et al., 1988, 1990; Humphrey et al., 2007). In fact, unc-79 mutants are hypersensitive to thiomethoxuflurane, methoxyflurane, chloroform, and halothane. They are also more resistant to flurothyl and enflurane, insensitive to fluroxene and isoflurane, and mildly more sensitive to diethylether (Morgan and Cascorbi, 1985; Morgan et al., 1988, 1990; Humphrey et al., 2007). By contrast, hypomorphic nahar38 and nahar85 drosophila mutants were first described as resistant to halothane, methoxyflurane, chloroform and trichloroethylene but not to diethylether, isoflurane, and enflurane (Krishnan and Nash, 1990; Nash et al., 1991; Campbell and Nash, 1994; Mir et al., 1997). However, it was then reported that unc-79 and na mutants are hypersensitive to halothane but not enflurane (Guan et al., 2000; Humphrey et al., 2007). The fact that na mutants were first described as resistant instead of hypersensitive may be explained by the method used to score anesthetic sensitivity but in some cases also by the fact that different alleles were tested. In another study, nahar38 mutation was found to be hypersensitive to isoflurane and to increase the potency with which isoflurane alters local field potentials recorded directly from fly brains (Van Swinderen, 2006). It was also shown that halothane presynaptically depresses synaptic transmission in wild-type drosophila larvae but not in nahar38 and nahar85 mutants (Nishikawa and Kidokoro, 1999). In mammals, heterozygous Lightweight mice, that carry a hypomorphic mutation in Unc-79, exhibit no alteration in Minimum Alveolar Concentration (MAC) in response to halothane, cyclopropane, or sevoflurane. However, a significant resistance to isoflurane-induced anesthesia was described (Speca et al., 2010).
It remains to be determined if the NALCN channel complex is a direct target for GAs, is important for the function of cells that contain such targets, or influences anesthesia more indirectly. GAs produce a widespread neurodepression in the CNS by enhancing inhibitory neurotransmission and reducing excitatory neurotransmission. However, the action mechanisms of GAs are not completely understood. Several ion channels and GPCRs are affected by these compounds (reviewed in Chau, 2010; Minami and Uezono, 2013). Of interest, it is known for a long time that GAs such as halothane, hyperpolarize neurons by acting on the potassium currents likely by activating two-pore potassium channels (Patel et al., 1999; Gruss et al., 2004; Liu et al., 2004). Considering that neurons from Nalcn knockout animals exhibit a ~10 mV hyperpolarization of their resting membrane potential, it is tempting to hypothesize that, in the case of an observed hypersensitivity, the absence of NALCN could remove a kind of brake for some GAs to act on their targets such as two-pore potassium channels. In this scheme, it would explain why, contrasting with C. elegans and D. melanogaster unc-79 and unc-80 mutants, heterozygous lightweight mice do not display any hypersensitivity to halothane. In fact, NALCN current and protein were found to be still present in Unc-79 deficient mice whereas this is not the case in C. elegans and D. melanogaster (Yeh et al., 2008; Lu et al., 2010; Speca et al., 2010; Lear et al., 2013). An observed resistance to other GAs implies that the NALCN channel complex is a direct target while an absence of phenotype to other GAs would indicate that other targets beside NALCN are involved.
In addition to GAs, alteration in ethanol sensitivity was described in animal mutants for the NALCN channelosome. Morgan and Sedensky, in 1995, found mutations in several genes in C. elegans which seem to control the sensitivity to ethanol, including unc-79 (Morgan and Sedensky, 1995). As a matter of fact, unc-79 mutants showed a decrease by about 25% in ethanol sensitivity. By contrast, it was recently reported that unc-79, unc-80, and nca-1;nca-2 mutants show hypersensitivity to ethanol (Speca et al., 2010). This hypersensitivity seems to be conserved in mammals as heterozygous Lightweight mice exhibit a highly significant increase in the sensitivity to the acute sedative effects of ethanol (Speca et al., 2010). Heterozygous Lightweight mice also present an increased ethanol preference and consumption, compared to wild-type mice, particularly at higher alcohol concentrations (Speca et al., 2010). As with GAs, the nature of the interaction between ethanol and NALCN physiology remains to be studied.
Respiratory Rhythm
In 2007, it was shown that the Nalcn gene is crucial for survival in mammals (Lu et al., 2007). Homozygous Nalcn knockout mice pups appear normal up to 12 h after birth and then die within 12 h due to severely disrupted respiratory rhythms (Lu et al., 2007). While wild-type mice had no abnormalities, knockout animals' respiration was highly sporadic. Breathing was characterized by ~5 s of apnea, followed by a burst of deep breathing for ~5 s, and this occurred at a rate of ~5 apnea events/minute. Interestingly, this pattern is reminiscent of the periodic breathing of Cheyne-Stokes respiration found in humans with CNS damage (reviewed in Strohl, 2003). Electrophysiological recording from the fourth cervical nerve root that innervates the diaphragm revealed that rhythmic electrical activity present in wild-type mice was largely absent. Thus, the defects observed in the respiratory rhythm in knockout mice are likely to reflect defects in electrical signaling in the nervous system (Lu et al., 2007).
More recently, Lu and Feng, 2011, investigated the properties of a NALCN ortholog in the snail L. stagnalis and its role in the activity of a respiratory pacemaker neuron (Lu and Feng, 2011). An in vivo investigation of its role on regulating respiratory behavior was performed by using RNA interference approaches. Animals in which nalcn was knocked down showed a reduced total breathing time compared to the naïve control. The resting membrane potential of the right pedal dorsal 1 (RPeD1) neuron that initiates the respiratory rhythm was found to be hyperpolarized by ~15 mV and its rhythmic firing was abolished. Thus, NALCN also plays a role in maintaining the respiratory activity in adult animal. It remains to be demonstrated if NALCN plays the same role in mammals by studying its functional properties in the preBötzinger complex and the retrotrapezoidnucleus/parafacial respiratory group that are involved in the respiratory rhythmogenesis (reviewed in Feldman et al., 2013).
Photic Control of Locomotion, Circadian Rhythms
Several studies performed with D. melanogaster mutants revealed the NALCN channelosome as an important player of circadian rhythms. Indeed, null na mutants display disrupted circadian rhythm. Typically, wild-type D. melanogaster are diurnal, with a greater proportion of their activity occurring during the daytime but null na mutants exhibit most of their activity at night (Nash et al., 2002). Furthermore, light enhances the climbing deficit that is induced by GAs in these flies (Campbell and Nash, 2001). Interestingly, the amount of NA protein does not fluctuate over time or light cycles indicating that expression of NA is not under circadian regulation. The phenotype seems to be the result of a broad aberrant motor response to photic input. Central to this, na is expressed in circadian pacemaker neurons and regulates the output of these neurons (Lear et al., 2005). An accumulation of Pigment-Dispersing factor (PDF), a neuropeptide essential for robust circadian behavior, is observed in these neurons from na mutants. This suggests that na may be required for PDF release. It is not surprising to observe a disturbance of circadian rhythms in na null mutants as electrical activity is an important regulator of clock function both in invertebrates and mammals (Nitabach et al., 2002; Belle et al., 2009). Null mutations of unc-79 and unc-80 in D. melanogaster also display severe defects in circadian locomotor rhythmicity that are indistinguishable from na mutant phenotypes, and support the role of this channelosome in circadian rhythms (Lear et al., 2013). Knockdown of nlf-1 in D. melanogaster was also found to phenocopy na knockdown for the circadian locomotion phenotype (Ghezzi et al., 2014).
Social Clustering
Involvement of the NALCN channelosome in social clustering, the natural tendency of animals of the same species to congregate in close proximity within a group, was recently described. Indeed, in a study aiming to investigate resource-independent local enhancement (RILE) in D. melanogaster, Burg et al., 2013, reported that nahar38 mutants display a deficient social phenotype compared to wild-type flies (Burg et al., 2013). This deficiency is rescued by the expression of na under the control of its native promoter. In addition, a complete rescue of the phenotype is also observed when na is expressed in cholinergic neurons and a partial rescue is obtained when na is expressed in glutamatergic neurons. na is strongly expressed in the mushroom body, a structure known to play a role in olfactory and visual learning and memory and the specific blockade of na expression in this structure affects RILE behavior. Knockdown of nlf-1 in D. melanogaster was also found to significantly suppress the social clustering phenotype as for na (Ghezzi et al., 2014).
Abdominal Morphology (Narrow Abdomen)
The na mutant flies are noticeably smaller than controls and their abdomens are more slender and elongated but no obvious deformity has been identified (Krishnan and Nash, 1990; Mir et al., 1997; Nash et al., 2002). D. melanogaster bearing mutations in unc-79 also exhibit a cylindric shaped abdomen (Humphrey et al., 2007). It is not known if this alteration represents a developmental defect or reflects altered physiology. However, Nalcn knockout mice neonates do not display gross abnormalities in embryonic development, righting responses, spontaneous limb movement, and toe/tail pinch responses (Lu et al., 2007).
Metabolism (Body Composition and Food Consumption)
Heterozygous Lightweight mice are smaller (shorter in length and lower in body weight) and have a leaner body composition (increased lean tissue and decreased body fat) than wild-type mice (Speca et al., 2010). Interestingly, heterozygous Lightweight mice display an increased food intake in comparison with wild-type mice of the same weight.
Systemic Osmoregulation (Serum Sodium Concentration)
A genetic analysis performed in mice demonstrated that Nalcn is involved in systemic osmoregulation by controlling the serum sodium concentration (Sinke et al., 2011). Furthermore, this study reported that heterozygous Nalcn knockout mice exhibit a significant hypernatremia.
Pacemaker Activity (Interstitial Cells of Cajal)
With others channels, such as transient receptor potential canonical (TRPC) channels, NALCN is partly responsible for the SP-induced depolarization and regulation of the intestinal pacemaking activity in the interstitial cells of Cajal (Kim et al., 2012). This activity generates the phasic contractions of the gastrointestinal muscles. Of note, in this study, the authors found that NALCN is not required for the basal pacemaking activity in ICCs.
Possible Implications of NALCN in Human Diseases
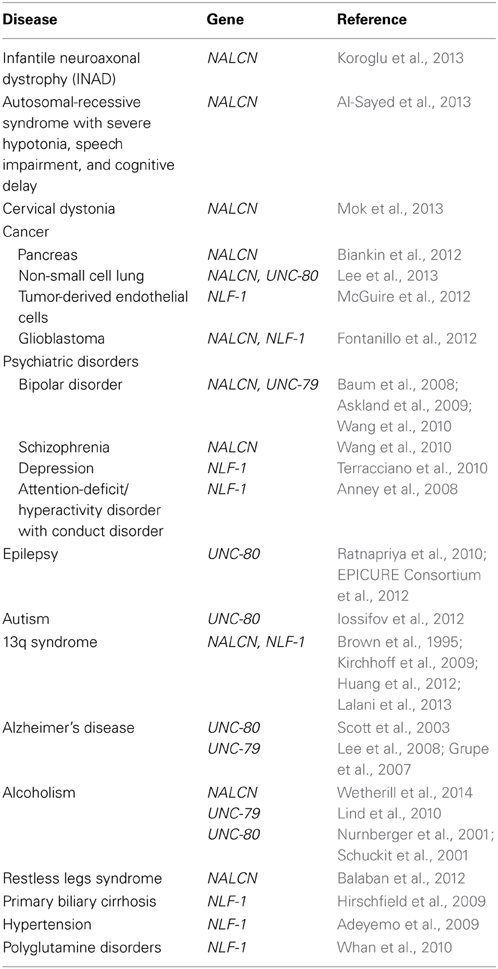
The NALCN channelosome was shown to be vital in mammals (Nakayama et al., 2006; Lu et al., 2007). Considering its role in regulating neuronal resting membrane potential, it is expected that polymorphisms, copy number variations (CNVs) and mutations in the corresponding genes may significantly impact neuronal physiology and lead to diseases. In this section, we review our current knowledge on data involving the NALCN channel complex in human diseases. We have also included a review of genetic data that loosely link the NALCN channel complex genes to diseases, which may provide insights into candidate genes involved in neuronal diseases (Table 3).
Infantile Neuroaxonal Dystrophy (INAD)
Infantile neuroaxonal dystrophy (INAD) is a rare neurodegenerative disease characterized by progressive motor, mental and visual deterioration that begins in infancy. Onset is usually between the age of 6 months and 3 years and death typically ensues before the age of 10 years (reviewed in Gregory et al., 1993). Early clinical manifestations include bilateral pyramidal tract signs, truncal hypotonia, cognitive decline and optic atrophy. Most cases present with distal axonal swelling and spheroid bodies in tissue biopsies (Cowen and Olmstead, 1963; Morgan et al., 2006). Originally, this recessive disorder was linked solely to mutations in the PLA2G6 gene, which encodes the calcium-independent phospholipase A2β (iPLA2β) enzyme, also designated iPLA2-VIA (Khateeb et al., 2006; Morgan et al., 2006). Recently, a study reported a mutation in the NALCN gene in two affected siblings with additional atypical features including facial dysmorphism, pectus carinatum, scoliosis, pes varus, zygodactyly and bilateral cryptorchidism. The patients also exhibit cerebellar atrophy and seizures and are still alive at 18 years-old (Koroglu et al., 2013; also see Seven et al., 2002). The identified mutation was a C to T conversion in NALCN coding exon 16 (c.1924C > T), creating a premature translational termination signal at codon 642 (Q642X) and truncated NALCN channel (Figure 1B). Interestingly, our unpublished data indicate that this exon is alternatively spliced resulting in the loss of one isoform but not NALCN in its entirety and may explain why the mutation is not lethal. It remains to be established whether this mutation results in a mRNA decay or expression of a truncated two-domain isoform. The fact that PL2G6 and NALCN genes mutations result in a similar disease raises the possibility that iPLA2β could be a regulator of NALCN.
Autosomal Recessive Syndrome with Severe Hypotonia, Speech Impairment, and Cognitive Delay
Mutations in the NALCN gene were recently reported in six patients with an autosomal-recessive syndrome characterized by severe hypotonia, speech impairment, and cognitive delay from two large consanguineous families (Al-Sayed et al., 2013). Three male patients (ranging from 4 to 7 years-old) from one family are homozygous for a single nucleotide deletion c.1489delT located in the coding region for the S4 helix from domain II (Figure 1B). This mutation results in a frame shift creating a stop codon 21 amino-acids downstream, which likely results in the production of a truncated protein or mRNA decay. Unless the involved exon is alternatively spliced, it is not clear why this deletion is not lethal as reported for Nalcn knockout mice (Lu et al., 2007). These patients have an absence of speech development, severe hypotonia, chronic constipation, global developmental delay, and facial dysmorphism which is reminiscent of infantile hypotonia with psychomotor retardation and characteristic facies (IHPRF, OMIM #615419). Three female patients (ranging from 9 to 17 years-old) from a second family are homozygous for a missense mutation c.3860G > T (W1287L) in exon 34 that encodes for the S4 helix from domain IV (Figure 1B). These patients have a milder but similar phenotype as the first family with additional features such as seizure disorder and hyperactivity.
Cervical Dystonia
Dystonia is a “syndrome of sustained muscle contractions, frequently causing twisting and repetitive movements or abnormal postures” (reviewed in Fahn et al., 1998). Estimates of the prevalence of dystonia range from ~1:10,000 to more than 1:200 (Nutt et al., 1988; Nakashima et al., 1995; Epidemiological Study of Dystonia in Europe Collaborative, G., 2000; Muller et al., 2002). Clinical presentation of dystonia is heterogeneous, from focal involvement such as cervical dystonia to generalized torsional dystonia. Cervical dystonia is the most common form of dystonia (Butler et al., 2004; Jankovic et al., 2007; Groen et al., 2013). The majority of cervical dystonia is transmitted in a non-Mendelian pattern, which suggests cervical dystonia is likely a complex disease rather than monogenic form. A recent genome-wide association study (GWAS) in 212 patients with this form of dystonia revealed a possible association with NALCN (Mok et al., 2013). However, this study did not find any single nucleotide polymorphisms (SNPs) with genome-wide significance (defined as P < 5 × 10−8) but a few clusters of possible association (defined as P < 5 × 10−6). Six SNPs within or in the vicinity of the NALCN gene with P-values ranging from 2.54 × 10−6 to 9.76 × 10−7 were found. Additional GWAS with larger cohorts of patients are required since the small sample size for this initial study was underpowered to detect loci with small disease effects.
Psychiatric Disorders
Schizophrenia and bipolar disorder
Schizophrenia (SCZ) is a chronic, severe disabling brain disorder characterized by abnormalities in the perception of reality. It most commonly manifests as auditory hallucinations, delusions, disorganized speech and thinking with significant social or occupational dysfunction (reviewed in Silveira et al., 2012). Bipolar disorder (BD), also known as manic-depressive illness, is a serious medical illness that causes shifts in a person's mood, energy, and ability to function (reviewed in Smith et al., 2012). Different from the normal ups and downs that everyone goes through, the symptoms of BD are severe. SCZ and BD affect around 1% of the population each. Both diseases have strong inherited components and growing evidence indicates that BD and SCZ may be closely related.
Interestingly, the NALCN gene lies within a region on chromosome 13q that has shown linkage to both BD and SCZ (reviewed in Detera-Wadleigh and McMahon, 2006). More specifically, the correlation between NALCN mRNA expression and the SCZ-associated gene GABRB2 in human brain (Kang et al., 2011), along with the association between the Drosophila homolog, na with circadian rhythms (disruptions of which are a hallmark of BD) suggest that NALCN may play a role in these two disorders (reviewed in Lenox et al., 2002). Importantly, while some genetic studies point out NALCN as a susceptibility locus in SCZ and BD, other studies have not concurred with these findings. A GWAS performed by genotyping more than 550,000 SNPs in two independent cohorts of European origin (for a total of 1233 patients and 1439 controls) revealed a significant association between NALCN and BD (SNP rs9513877; Baum et al., 2008). This finding was confirmed by Askland et al. (2009). A genome-wide meta-analysis also identified a significant association of NALCN (SNP rs2044117) with both SCZ and BD in cohorts of 1172 (SCZ) and 653 (BD) European-American patients (Wang et al., 2010). However, no significant association between BD and NALCN was found in a Finnish cohort of 723 individuals from 180 families with type I BD (Ollila et al., 2009). Furthermore, there was no association of NALCN with SCZ in a cohort of 583 patients (Souza et al., 2011), nor in a group of 240 individuals with treatment-resistant SCZ (Teo et al., 2012). The discrepancies in candidate gene studies may partly result from the clinical and genetic heterogeneity of BD and SCZ and their complex inheritance model, but also from methodological issues such as the definition of clinical phenotype. More work is needed to confirm that NALCN is a susceptibility locus in BD and SCZ.
Several findings suggest that UNC-79 and UNC-80 are also associated with these disorders, further implicating the NALCN channelosome with SCZ and BD. The UNC-79 encoding gene lies within a region on chromosome 14q that has shown linkage to BD. Askland et al., 2009, also suggested an association between SNP rs17125698 located at around 4Mb from the UNC-79 gene and BD (Askland et al., 2009). Another study also reported an association of SNP rs11622475 located in 14q32 in BD (Wellcome Trust Case Control, C, 2007). However, this SNP is located at around 100Mb from UNC-79. A proteomic study suggested that UNC-80 could belong to the same protein complex as dysbindin, a SCZ susceptibility gene product (Mead et al., 2010).
Major depressive and attention-deficit/hyperactivity disorders
NLF-1 has been loosely associated with both major depressive disorder and attention-deficit/hyperactivity disorder. Major depressive disorder (MDD) is a syndrome characterized by a number of behavioral, cognitive and emotional features. It is most commonly associated with a sad or depressed mood, a reduced capacity to feel pleasure, hopelessness, loss of energy, altered sleep patterns, weight fluctuations, difficulty in concentrating and suicidal ideation (reviewed in Uher et al., 2013). A GWAS of depression traits found a possible association between MDD and SNPs in the vicinity of NLF-1 (SNPs rs9634463, rs7329003, rs713548, rs9301191, and rs1924397), but statistical significance was not reached for any gene (Terracciano et al., 2010). Attention-deficit/hyperactivity disorder (ADHD) is characterized by inattention, excessive motor activity, impulsivity and distractibility and affects 8–12% of school-age children worldwide (reviewed in Sharma and Couture, 2014). Individuals with ADHD show high co-morbidity with a wide range of psychiatric disorders. In a study aiming to identify susceptibility loci for ADHD with conduct disorder, Anney et al., 2008, did not find any significant association with any SNP but two of the strongest associations were found with SNPs rs10492664 and rs8002852 that are located in the vicinity of NLF-1 (Anney et al., 2008). Interestingly, phenotypes observed for Nk1R knockout mice strongly parallel abnormalities expressed by patients with ADHD and heterozygous Lightweight mice are modestly but significantly hyperactive (Speca et al., 2010; Yan et al., 2010).
Epilepsy
Epilepsy, a very common neurological disorder, is defined by the occurrence of unprovoked seizures caused by the synchronous discharge of large number of neurons (reviewed in Sander, 2003; Khan and Al Baradie, 2012). Abnormal expression or function of ion channels may be involved in the pathophysiology of both acquired and inherited epilepsy (reviewed in Mantegazza et al., 2010; Lerche et al., 2013). Considering the importance of the NALCN channel complex in setting the resting membrane potential of neurons, the NALCN channelosome gene mutations are clear candidates for epilepsy. As mentioned in this review, patients with INAD or an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay have seizures (Al-Sayed et al., 2013; Koroglu et al., 2013). Seizures were also reported for patients with chromosome 13q deletions in regions that contain the NALCN gene (Kirchhoff et al., 2009; Lalani et al., 2013). Recently, a whole genome linkage analysis in three-generations of a south Indian family who had multiple members affected with juvenile myoclonic epilepsy (JME) found a critical genetic interval of 24 Mb between markers D2S116 and D2S2390 (Ratnapriya et al., 2010). The UNC-80 gene lies in this region. In addition, a region-wide linkage analysis carried out in 118 European families with an aggregation of JME indicated a significant linkage with marker rsD2S143 located on chromosome 2q34 at around 4Mb of the UNC-80 gene (EPICURE Consortium et al., 2012).
Autism
Autism Spectrum Disorders (ASDs) are neurodevelopmental disorders characterized by impairments in social interaction and communication, and the presence of restrictive and repetitive behaviors (reviewed in Jones et al., 2014). Exome sequencing of 343 families, each with a single child on the autism spectrum and at least one unaffected sibling, revealed de novo small indels and point substitutions in the affected sibling (Iossifov et al., 2012). One of the discovered mutations resulted in a truncating R518X mutation in one copy of UNC-80, suggesting that UNC-80 is a susceptibility locus in autistic spectrum.
13q Syndrome
The 13q syndrome is caused by structural and functional monosomy of the 13q chromosomal region and was first described in 1963 and consequently delineated as a specific syndrome in 1969 (Lele et al., 1963; Allderdice et al., 1969). Large numbers of patients with deletions of the long arm of chromosome 13, which includes NALCN and NLF-1, have been described in the literature. The phenotypes of these patients varied widely. Some showed severe malformations involving the brain, heart, kidneys, lungs, other organ systems, and digits while others were only mildly affected with minor dysmorphic features, developmental delay, and growth failure. The characteristic features of 13q deletions include growth retardation with microcephaly, facial dysmorphism, congenital heart, brain, and kidney defects (Brown et al., 1995). Several studies addressed karyotype-phenotype correlations depending on the extent of the deletions (Brown et al., 1995; Luo et al., 2000; McCormack et al., 2002; Alanay et al., 2005; Garcia et al., 2006; Ballarati et al., 2007; Walczak-Sztulpa et al., 2008; Kirchhoff et al., 2009; Quelin et al., 2009). It was reported that the severely malformed phenotype results from the deletion of a critical region lying between markers D13S136 and D13S147 (Brown et al., 1995). This region of around 2.3 Mb contains several genes including NALCN. Recently, a molecular-karyotyping study reported that a female with congenital heart defects, facial anomalies, developmental delay, and intellectual disability had a deletion of 12.75 Mb extending from 13q33.1 to 13q34 (Huang et al., 2012). The deleted region harbors 55 genes, including NALCN and NLF-1. Another study further refined a critical region for all CNS anomalies and neural tube defects taken together, to a region of 1.6 Mb (Kirchhoff et al., 2009). Only seven known genes are located within this region, including NALCN. A recent study aiming to identify rare DNA copy number variants in cardiovascular malformations with extracardiac abnormalities revealed the deletion of 641 kb in the NALCN gene region in 3 patients (Lalani et al., 2013). The extracardiac phenotypes of these patients include agenesis of corpus callosum, growth retardation, facial dysmorphism, and seizures.
Alzheimer's Disease
Alzheimer's disease (AD) is the most common form of dementia and the most frequent degenerative brain disorder encountered in old age. The risk of developing AD substantially increases after 65 years of age (reviewed in Nussbaum and Ellis, 2003). With the exception of rare cases of early onset familial Alzheimer's disease caused by mutations in the amyloid precursor protein or presenilins genes, the etiology of the vast majority of cases remains misunderstood. There is evidence for substantial genetic influence (reviewed in St. George-Hyslop and Petit, 2005; Tanzi and Bertram, 2005). A linkage analysis to detect novel AD loci from 437 families (1252 individuals) revealed a statistically significant linkage with marker D2S2944 located in 2q34 in 31 American families with a minimum age at onset between 50 and 60 years (Scott et al., 2003). This marker is located at less than 4Mb from the UNC-80 gene. Another study reported the existence of a susceptibility locus in 14q32.12 near marker D14S617 in a Caribbean Hispanic cohort of 1161 individuals from 209 families (Lee et al., 2008). The UNC-79 gene lies in a region at less than 3Mb from this marker. The existence of a susceptibility locus for AD in the vicinity of the UNC-79 gene (SNP rs11622883) was also reported by Grupe et al., 2007, from a cohort of 1808 American-European patients (Grupe et al., 2007).
Alcoholism
Alcohol dependence is one of the most common and costly public health problems. Several studies suggest a role of genetic factors and few genes have been shown to be associated with alcohol dependence (reviewed in Morozova et al., 2014). A recent GWAS performed in 118 European-American families (2322 individuals) demonstrated a significant linkage of SNP rs17484734, located in the NALCN gene, and high-risk of alcohol dependence (Wetherill et al., 2014). Interestingly, mice that heterologously carry a hypomorphic mutation in the Unc-79 gene voluntarily consume more ethanol than wild-type littermates (Speca et al., 2010). A GWAS on comorbid alcohol/nicotine dependence in 599 cases and 488 controls found SNP rs12882384 as one of the three top findings, which is located within the UNC-79 gene (Lind et al., 2010). Genome-wide significance was found for joint alcohol/nicotine comorbidity but not for nicotine or alcohol dependence alone suggesting the existence of genes that mediate the combined effect of these two addictive substances. In addition, two previous studies suggest the existence of a susceptibility locus located on chromosome 2, near the UNC-80 gene, linked to alcohol tolerance (markers D2S425, D2S434, D2S424, D2S1323, D2S1333) and the comorbidy of alcoholism and depression (marker DS1371) respectively (Nurnberger et al., 2001; Schuckit et al., 2001).
Restless Legs Syndrome
Restless legs syndrome (RLS) is a sensorimotor disorder characterized by abnormal sensations in the limbs that are both dependent on activity and time of day, such that symptoms are promoted by rest and relieved by activity and peak in the evening or at night (reviewed in Winkelman et al., 2013). RLS is a genetically heterogeneous complex trait with high prevalence but large phenotype variability. Current theories of RLS pathophysiology emphasize brain iron deficiency with abnormal dopaminergic consequences, together with a strong underlying genetic background (reviewed in Dauvilliers and Winkelmann, 2013). A recent study analyzing 11 individuals from the same family indicated the existence of a locus on chromosome 13q, between SNPs rs2182885 and rs7333498 (around 5.3 Mb in size; Balaban et al., 2012). The NALCN gene lies in this region.
Primary Biliary Cirrhosis
Primary biliary cirrhosis is a chronic granulomatous cholangitis, characteristically associated with antimitochondrial antibodies. Analysis of both aggregation data from families and concordance data from twins revealed a genetic predisposition for the disease (reviewed in Hirschfield and Gershwin, 2013). A GWAS with a cohort of 2072 North-American individuals (536 patients and 1536 controls) was carried out by genotyping more than 300,000 SNPs (Hirschfield et al., 2009). Although this study reported the strongest association of the disease with HLA-related genes, a significant association was found with SNP rs2211312 in the NLF-1 gene.
Hypertension and Blood Pressure
Hypertension is a common human disease affecting over one billion people world-wide and a major contributor to cerebrovascular accidents, myocardial infarction, congestive cardiac failure and chronic renal failure (reviewed in Zhao et al., 2013). A GWAS performed by genotyping more than 800,000 SNPs in a sample of 1017 African-American individuals led to the identification of several potential loci involved in the regulation of blood pressure (Adeyemo et al., 2009). A significant association was found between systolic blood pressure and SNP rs9301196 located within the NLF-1 gene.
Polyglutamine Disorders
Polyglutamine (polyQ)-expansions in different proteins cause at least eleven neurodegenerative diseases including Huntington disease and spinocerebellar ataxias (reviewed in Todd and Lim, 2013). These human diseases are caused by extreme expansion of the repeats, which adversely impact protein structure, often causing intracellular protein aggregation and altered protein function. Bovine NLF-1 exhibits a polymorphic polyQ tract in its sequence (Whan et al., 2010). This polyQ tract is also found in other species including human beings suggesting that aggregation of NLF-1 could be involved in neurodegenerative diseases.
Cancer
Biankin et al. examined pancreatic cancer genomes using exome sequencing and copy number analysis from a cohort of 142 patients of early (stages I and II) sporadic pancreatic ductal adenocarcinoma (Biankin et al., 2012). Analysis of genes with non-silent mutations occurring in 2 or more individual cancers identified 16 genes including NALCN (mutated in 4 patients). Pathway analysis allowed not only the identification of genes involved in mechanisms already known to be involved in cancer but also genes known to be involved in axon guidance. This latter finding was strengthened by the inclusion of data from another study (Jones et al., 2008). It is tempting to speculate that NALCN could also be involved in axon guidance.
An association of NALCN with non-small cell lung cancer was also suggested by a study where 217,817 SNPs were genotyped in 348 advanced patients who received chemotherapy (Lee et al., 2013). These genetic association studies revealed that SNP rs9557635, located in the genomic regions of the NALCN gene, was associated with this disease. Interestingly, this study also reported a strongly significant association with SNP rs2371030 located 25 kb downstream to the CPS1 gene on chromosome 2q34, which contains the UNC-80 gene. Further linking the channelosome to cancer, a recent study reported an NLF-1 copy number increase in populations of tumor-derived endothelial cells that are resistant to anti-angiogenic cancer therapies (McGuire et al., 2012). Finally, in a search for key altered genomic regions in human glioblastomas, Fontanillo et al., 2012, found a high correlation between the malignant state and copy number alterations in the NALCN and NLF-1 regions with a decrease in expression level (Fontanillo et al., 2012).
Concluding Remarks and Future Directions
Is NALCN Really a Channel per se?
From a fundamental point of view, it remains unclear whether NALCN is truly an ion channel. The early findings by Ren and colleagues supported that NALCN was an ion channel, based on mutations in the putative selectivity filter (EEKE) that alter the permeation properties and the gadolinium block (EEKA) or inhibit the observed currents in HEK-293 cells (EEEE)(Lu et al., 2007). Further supporting this, in D. melanogaster, the expression of a naEEEE mutant in na neurons using the UAS-GAL4 system did not restore the studied phenotype as nawt does (Lear et al., 2005). However, functional expression of NALCN channel in heterologous expression systems is difficult to obtain and these data do not demonstrate unambiguously that NALCN is a pore forming subunit. A clear proof of its channel activity would come from electrophysiological data exploring NALCN channel activity when rebuilt in lipid bilayers.
What are the Gating Properties of NALCN?
The mechanism(s) that gate NALCN have yet to be established. On one hand, NALCN channel is described as conducting a sodium leak current in neurons both in mammals and invertebrates (Lu et al., 2007; Lu and Feng, 2011; Xie et al., 2013). On the other hand, NALCN channel also conducts a sodium current activated by acetylcholine though the M3R in a pancreatic β-cell line without any evidence for a leak current (Swayne et al., 2009). One may hypothesize that the NALCN-mediated leak current observed in neurons may result from a constitutive activity of some GPCRs (Swayne et al., 2009). In this case, it would require UNC-80 and SFKs. However, this hypothesis may not be valid for two main reasons. First, considering that HEK-293 cells do not express UNC-79 and UNC-80 (Swayne et al., 2009), the expression of NALCN alone is sufficient to observe a sodium leak current in these cells (Lu et al., 2007). Second, it was shown that a sodium leak current mediated by NALCN is still present in hippocampal neurons from Unc-79 knockout mice where UNC-80 is not detected (Lu et al., 2010). Thus, the gating properties of NALCN remain mysterious and should be investigated further. In keeping with this idea, the search for additional proteins that belong to the NALCN channelosome could give clues regarding modulation of the NALCN gating mechanism.
What is the Repertoire of GPCRs Capable of Modulating NALCN Channel?
Recent studies have established that NALCN can be activated/potentiated or inhibited by different GPCRs (Lu et al., 2009; Swayne et al., 2009; Lu et al., 2010). Therefore, one may expect additional GPCRs to be involved in these processes. Identification of this repertoire is an important challenge and understanding why some GPCRs modulate NALCN, and some others do not, would help to understand further how NALCN is activated/regulated in neurons and how neurotransmitters and hormones affect neuronal electrophysiological properties.
What are the Functional Consequences of Alternative Splicing on NALCN Channelosome Genes?
Alternative splicing events were demonstrated in many ion channels and can significantly impact their biophysical properties, cellular localization, and functional regulations (reviewed in Noel et al., 2011; Jan and Jan, 2012; Lipscombe et al., 2013). Alternative splicing was identified in NALCN channelosome genes (Senatore et al., 2013; Monteil unpublished data). The physiological consequences of these alternative splicing events remain to be elucidated.
Are UNC-79, UNC-80, and NLF-1 Specific NALCN-Ancillary Subunits?
In C. elegans, nlf-1, unc-79, unc-80, and nca-1;nca-2 mutants exhibit exactly similar phenotypes and both Unc-79 and Nalcn knockout mice die soon after birth. However, the post-natal lethality observed in these mouse models could mask phenotypes more specific to one subunit compared to the others. As a matter of fact, we cannot exclude that UNC-79, UNC-80, and NLF-1 may have NALCN-independent physiological roles. Conditional knockout mice for each component of the NALCN channelosome would be very helpful to clarify this point.
Are there More Proteins in the NALCN Channelosome?
The NALCN channelosome is probably not restricted to NALCN, UNC-79, UNC-80, NLF-1, SFKs, and GPCRs. As an example, there is another gene highly related to NLF-1, FAM155B in mammals (accession number NG_021282.1). Whether the FAM155B gene product belongs to the NALCN channel complex should be investigated. Also, the mechanisms involved in the regulation of the NALCN channel by CaSR are not known and could imply a co-inclusion in the same protein complex. Some studies indicate genetic interactions between nca-1;nca-2 and other genes in C. elegans. As a matter of fact, null mutants of stomatins (unc-1 and unc-24) and innexins (unc-7 and unc-9) were found to restore the altered sensitivity to GAs and ethanol (Sedensky and Meneely, 1987; Morgan et al., 1990; Morgan and Sedensky, 1995; Sedensky et al., 2001; Humphrey et al., 2007). It was also found that nca-1;nca-2 mutants exhibit a reduced evoked post-synaptic current at the neuromuscular junction and this phenotype is rescued by unc-7 mutants (Bouhours et al., 2011). However, in this latter study, co-immunofluorescent staining experiments do not show obvious neuronal co-localization of UNC-7 and NCA-1/NCA-2. Thus, the reported genetic interaction between unc-7 and nca-1;nca-2 probably reflects a functional, rather than a physical interaction. A genetic interaction was also described between unc-26 null mutants and unc-80 and nca-1;nca-2 (Jospin et al., 2007). unc-26 encodes for Synaptojanin, a lipid phosphatase required to degrade phosphatidylinositol 4,5 bisphosphate (PIP2) at cell membranes during synaptic vesicle recycling (Cremona et al., 1999). It was found that unc-80 and nca-1;nca-2 mutants ameliorate the recycling defects of synaptic vesicles as well as the synaptic transmission in unc-26 null mutants and it was suggested that nca-1;nca-2 function could be regulated by PIP2 (Jospin et al., 2007). Whether Synaptojanin belongs to the NALCN channelosome remains to be investigated. Interestingly, NALCN and PLA2G6 mutations results in similar phenotypes in humans suggesting that iPLA2β could be a regulator of NALCN. Complementary techniques such as two-hybrid and immunoprecipitation studies followed by mass spectrometry approaches will certainly lead to the discovery of new components of the NALCN channelosome.
How is NALCN Functions Modulated?
In addition to their regulation by binding partners, ion channels are known to be post-translationally modulated. For example, ion channel complexes can be phosphorylated by a variety of kinases which regulate their physiological roles (reviewed in Smart, 1997; Dai et al., 2009; Cerda et al., 2011; Scheuer, 2011; Vacher and Trimmer, 2011). An in silico analysis of primary amino-acid sequences of NALCN, UNC-79, UNC-80, and NLF-1 reveals the existence of several predicted phosphorylation sites that are conserved in mammals and for some of these sites, are also conserved in C. elegans, D. Melanogaster, and L. Stagnalis (not shown). The physiological relevance of the NALCN channelosome phosphorylation, as well as the identification of the kinases involved, remains to be explored.
In Which Neuronal Subtype(s) is NALCN Expressed?
NALCN, and probably UNC-79, UNC-80, and NLF-1, is widely expressed throughout the CNS in mammals (Lee et al., 1999; Swayne et al., 2009; Kang et al., 2011). To better assess the physiological role of NALCN, it is crucial to identify whether expression is restricted to some specific neuronal populations. In addition, NALCN is localized along the axons in C. elegans and in synaptic regions in D. melanogaster (Nash et al., 2002; Humphrey et al., 2007; Yeh et al., 2008), but the neuronal localization of NALCN channel proteins in mammals is unknown. The development of specific antibodies for immunohistochemistry experiments will be crucial for this purpose.
Lack of Pharmacology of NALCN but Relevance of NALCN as a Drug Target
To date, no specific pharmacology for NALCN has been reported and this clearly represents a significant hurdle in the study of NALCN properties and activity. The discovery of specific NALCN agonists/antagonists is critically needed. Such molecules would be tools to investigate NALCN's physiological roles and may also hold interest to develop innovative therapeutic strategies. Whether the NALCN channelosome represents a drug target of interest remains unclear at the present time. Considering that NALCN channel is expressed in pancreatic islets, both in α- and β-cells, and is possibly involved in the regulation of insulin secretion (Kutlu et al., 2009 and http://t1dbase.org; Swayne et al., 2010), targeting NALCN might be relevant to treat type 2 diabetes. It is tempting to speculate that agonists of NALCN would have beneficial effects in pancreatic β-cells on insulin secretion (reviewed in Gilon and Rorsman, 2009). NALCN blockers might be relevant to treat pain considering that SP plays a pivotal role in the transmission of noxious stimuli in the spinal cord (reviewed in Seybold, 2009) and that NALCN is highly expressed in the dorsal spinal cord compared to the ventral spinal cord (Sun et al., 2002). Based on the potential implication of NALCN channel in human physiology and diseases, its pharmacological targeting might also be relevant to treat epilepsy, bowell motility disorders, osmoregulation disorders and psychiatric disorders.
Summary
The NALCN channelosome is vital in mammals: homozygous mutant mice exhibit neonatal lethal phenotype and NALCN plays a crucial role in neuronal excitability. NALCN is involved in the regulation of resting membrane potential and is modulated by hormones and neurotransmitters. In the present article, we provide an extensive review of the literature describing the molecular and functional properties of the NALCN channelosome, the various phenotypes observed in NALCN animal models, as well as the recent implications of NALCN in human diseases. We also discuss how NALCN could represent a therapeutic target to treat a wide variety of human diseases, especially neurological and psychiatric diseases. Unfortunately, the lack of specific pharmacology is clearly a brake to all the NALCN investigations. In the coming years, the expected development of selective agonists/antagonists, as well as new animal models, should foster our understanding of the functional properties and physiological roles of the NALCN channelosome.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Stephanie Knapp who participated to the early writing steps of this review article. We thank Sarah England, Erin Reinl, and Natalia Prevarskaya for critical readings and comments on the manuscript and Drs. Céline Lemmers, Isabelle Bidaud, Samer Khoury-Hanna, Hamid Gholamipour-Badie, and Adriano Senatore for their contribution to the work performed in the laboratory. Our laboratory is supported by a grant from “Agence Nationale de la Recherche” 11 BSV1 004 01 and is part of the Laboratory of Excellence “Ion Channel Science and Therapeutics” (http://www.labex-icst.fr/en; ANR-11-LABX-0015-01_ICST).
References
Adeyemo, A., Gerry, N., Chen, G., Herbert, A., Doumatey, A., Huang, H., et al. (2009). A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 5:e1000564. doi: 10.1371/journal.pgen.1000564
Alanay, Y., Aktas, D., Utine, E., Talim, B., Onderoglu, L., Caglar, M., et al. (2005). Is Dandy-Walker malformation associated with “distal 13q deletion syndrome?” Findings in a fetus supporting previous observations. Am. J. Med. Genet. A 136, 265–268. doi: 10.1002/ajmg.a.30808
Allderdice, P. W., Davis, J. G., Miller, O. J., Klinger, H. P., Warburton, D., Miller, D. A., et al. (1969). The 13q-deletion syndrome. Am. J. Hum. Genet. 21, 499–512.
Al-Sayed, M. D., Al-Zaidan, H., Albakheet, A., Hakami, H., Kenana, R., Al-Yafee, Y., et al. (2013). Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am. J. Hum. Genet. 93, 721–726. doi: 10.1016/j.ajhg.2013.08.001
Anney, R. J., Lasky-Su, J., O'Dushlaine, C., Kenny, E., Neale, B. M., Mulligan, A., et al. (2008). Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 1369–1378. doi: 10.1002/ajmg.b.30871
Askland, K., Read, C., and Moore, J. (2009). Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum. Genet. 125, 63–79. doi: 10.1007/s00439-008-0600-y
Balaban, H., Bayrakli, F., Kartal, U., Pinarbasi, E., Topaktas, S., and Kars, H. Z. (2012). A novel locus for restless legs syndrome on chromosome 13q. Eur. Neurol. 68, 111–116. doi: 10.1159/000338779
Ballarati, L., Rossi, E., Bonati, M. T., Gimelli, S., Maraschio, P., Finelli, P., et al. (2007). 13q Deletion and central nervous system anomalies: further insights from karyotype-phenotype analyses of 14 patients. J. Med. Genet. 44, e60. doi: 10.1136/jmg.2006.043059
Barry, G. (2014). Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatry. 19, 410–416. doi: 10.1038/mp.2013.196
Baum, A. E., Akula, N., Cabanero, M., Cardona, I., Corona, W., Klemens, B., et al. (2008). A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol. Psychiatry 13, 197–207. doi: 10.1038/sj.mp.4002012
Belle, M. D., Diekman, C. O., Forger, D. B., and Piggins, H. D. (2009). Daily electrical silencing in the mammalian circadian clock. Science 326, 281–284. doi: 10.1126/science.1169657
Biankin, A. V., Waddell, N., Kassahn, K. S., Gingras, M. C., Muthuswamy, L. B., Johns, A. L., et al. (2012). Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405. doi: 10.1038/nature11547
Bouhours, M., Po, M. D., Gao, S., Hung, W., Li, H., Georgiou, J., et al. (2011). A co-operative regulation of neuronal excitability by UNC-7 innexin and NCA/NALCN leak channel. Mol. Brain 4:16. doi: 10.1186/1756-6606-4-16
Brown, S., Russo, J., Chitayat, D., and Warburton, D. (1995). The 13q- syndrome: the molecular definition of a critical deletion region in band 13q32. Am. J. Hum. Genet. 57, 859–866.
Burg, E. D., Langan, S. T., and Nash, H. A. (2013). Drosophila social clustering is disrupted by anesthetics and in narrow abdomen ion channel mutants. Genes Brain Behav. 12, 338–347. doi: 10.1111/gbb.12025
Butler, A. G., Duffey, P. O., Hawthorne, M. R., and Barnes, M. P. (2004). An epidemiologic survey of dystonia within the entire population of northeast England over the past nine years. Adv. Neurol. 94, 95–99.
Cahoy, J. D., Emery, B., Kaushal, A., Foo, L. C., Zamanian, J. L., Christopherson, K. S., et al. (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278. doi: 10.1523/JNEUROSCI.4178-07.2008
Camerino, D. C., Desaphy, J. F., Tricarico, D., Pierno, S., and Liantonio, A. (2008). Therapeutic approaches to ion channel diseases. Adv. Genet. 64, 81–145. doi: 10.1016/S0065-2660(08)00804-3
Campbell, D. B., and Nash, H. A. (1994). Use of Drosophila mutants to distinguish among volatile general anesthetics. Proc. Natl. Acad. Sci. U.S.A. 91, 2135–2139. doi: 10.1073/pnas.91.6.2135
Campbell, J. L., and Nash, H. A. (2001). Volatile general anesthetics reveal a neurobiological role for the white and brown genes of Drosophila melanogaster. J. Neurobiol. 49, 339–349. doi: 10.1002/neu.10009
Cerda, O., Baek, J. H., and Trimmer, J. S. (2011). Mining recent brain proteomic databases for ion channel phosphosite nuggets. J. Gen. Physiol. 137, 3–16. doi: 10.1085/jgp.201010555
Chau, P. L. (2010). New insights into the molecular mechanisms of general anaesthetics. Br. J. Pharmacol. 161, 288–307. doi: 10.1111/j.1476-5381.2010.00891.x
Cowen, D., and Olmstead, E. V. (1963). Infantile neuroaxonal dystrophy. J. Neuropathol. Exp. Neurol. 22, 175–236. doi: 10.1097/00005072-196304000-00001
Cremona, O., Di Paolo, G., Wenk, M. R., Luthi, A., Kim, W. T., Takei, K., et al. (1999). Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 99, 179–188. doi: 10.1016/S0092-8674(00)81649-9
Dai, S., Hall, D. D., and Hell, J. W. (2009). Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol. Rev. 89, 411–452. doi: 10.1152/physrev.00029.2007
Dauvilliers, Y., and Winkelmann, J. (2013). Restless legs syndrome: update on pathogenesis. Curr. Opin. Pulm. Med. 19, 594–600. doi: 10.1097/MCP.0b013e328365ab07
Detera-Wadleigh, S. D., and McMahon, F. J. (2006). G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol. Psychiatry 60, 106–114. doi: 10.1016/j.biopsych.2006.01.019
EPICURE Consortium, Leu, C., De Kovel, C. G., Zara, F., Striano, P., Pezzella, M., et al. (2012). Genome-wide linkage meta-analysis identifies susceptibility loci at 2q34 and 13q31.3 for genetic generalized epilepsies. Epilepsia 53, 308–318. doi: 10.1111/j.1528-1167.2011.03379.x
Epidemiological Study of Dystonia in Europe Collaborative, G. (2000). A prevalence study of primary dystonia in eight European countries. J. Neurol. 247, 787–792. doi: 10.1007/s004150070094
Fahn, S., Bressman, S. B., and Marsden, C. D. (1998). Classification of dystonia. Adv. Neurol. 78, 1–10.
Feldman, J. L., Del Negro, C. A., and Gray, P. A. (2013). Understanding the rhythm of breathing: so near, yet so far. Annu. Rev. Physiol. 75, 423–452. doi: 10.1146/annurev-physiol-040510-130049
Fontanillo, C., Aibar, S., Sanchez-Santos, J. M., and De Las Rivas, J. (2012). Combined analysis of genome-wide expression and copy number profiles to identify key altered genomic regions in cancer. BMC Genomics 13(Suppl. 5):S5. doi: 10.1186/1471-2164-13-S5-S5
Garcia, N. M., Allgood, J., Santos, L. J., Lonergan, D., Batanian, J. R., Henkemeyer, M., et al. (2006). Deletion mapping of critical region for hypospadias, penoscrotal transposition and imperforate anus on human chromosome 13. J. Pediatr. Urol. 2, 233–242. doi: 10.1016/j.jpurol.2006.03.006
Geisler, S., and Coller, J. (2013). RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14, 699–712. doi: 10.1038/nrm3679
Ghezzi, A., Liebeskind, B. J., Thompson, A., Atkinson, N. S., and Zakonm, H. H. (2014). Ancient association between cation leak channels and Mid1 proteins is conserved in fungi and animals. Front. Mol. Neurosci. 7:15. doi: 10.3389/fnmol.2014.00015
Gilon, P., and Rorsman, P. (2009). NALCN: a regulated leak channel. EMBO Rep. 10, 963–964. doi: 10.1038/embor.2009.185
Gregory, A., Kurian, M. A., Maher, E. R., Hogarth, P., and Hayflick, S. J. (1993). “PLA2G6-associated neurodegeneration,” in GeneReviews, eds R. A. Pagon, M. P. Adam, T. D. Bird, C. R. Dolan, C. T. Fong, and K. Stephens (Seattle, WA). Available online at: http://www.ncbi.nlm.nih.gov/books/NBK1675/
Groen, J. L., Simon-Sanchez, J., Ritz, K., Bochdanovits, Z., Fang, Y., Van Hilten, J. J., et al. (2013). Cervical dystonia and genetic common variation in the dopamine pathway. Parkinsonism Relat. Disord. 19, 346–349. doi: 10.1016/j.parkreldis.2012.08.016
Grupe, A., Abraham, R., Li, Y., Rowland, C., Hollingworth, P., Morgan, A., et al. (2007). Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Hum. Mol. Genet. 16, 865–873. doi: 10.1093/hmg/ddm031
Gruss, M., Bushell, T. J., Bright, D. P., Lieb, W. R., Mathie, A., and Franks, N. P. (2004). Two-pore-domain K+ channels are a novel target for the anesthetic gases xenon, nitrous oxide, and cyclopropane. Mol. Pharmacol. 65, 443–452. doi: 10.1124/mol.65.2.443
Guan, Z., Scott, R. L., and Nash, H. A. (2000). A new assay for the genetic study of general anesthesia in Drosophila melanogaster: use in analysis of mutations in the X-chromosomal 12E region. J. Neurogenet. 14, 25–42. doi: 10.3109/01677060009083475
Guerineau, N. C., Bossu, J. L., Gahwiler, B. H., and Gerber, U. (1995). Activation of a nonselective cationic conductance by metabotropic glutamatergic and muscarinic agonists in CA3 pyramidal neurons of the rat hippocampus. J. Neurosci. 15, 4395–4407.
Hirschfield, G. M., and Gershwin, M. E. (2013). The immunobiology and pathophysiology of primary biliary cirrhosis. Annu. Rev. Pathol. 8, 303–330. doi: 10.1146/annurev-pathol-020712-164014
Hirschfield, G. M., Liu, X., Xu, C., Lu, Y., Xie, G., Lu, Y., et al. (2009). Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N. Engl. J. Med. 360, 2544–2555. doi: 10.1056/NEJMoa0810440
Huang, C., Yang, Y. F., Yin, N., Chen, J. L., Wang, J., Zhang, H., et al. (2012). Congenital heart defect and mental retardation in a patient with a 13q33.1-34 deletion. Gene 498, 308–310. doi: 10.1016/j.gene.2012.01.083
Humphrey, J. A., Hamming, K. S., Thacker, C. M., Scott, R. L., Sedensky, M. M., Snutch, T. P., et al. (2007). A putative cation channel and its novel regulator: cross-species conservation of effects on general anesthesia. Curr. Biol. 17, 624–629. doi: 10.1016/j.cub.2007.02.037
Iossifov, I., Ronemus, M., Levy, D., Wang, Z., Hakker, I., Rosenbaum, J., et al. (2012). De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285–299. doi: 10.1016/j.neuron.2012.04.009
Jan, L. Y., and Jan, Y. N. (2012). Voltage-gated potassium channels and the diversity of electrical signalling. J. Physiol. 590, 2591–2599. doi: 10.1113/jphysiol.2011.224212
Jankovic, J., Tsui, J., and Bergeron, C. (2007). Prevalence of cervical dystonia and spasmodic torticollis in the United States general population. Parkinsonism Relat. Disord. 13, 411–416. doi: 10.1016/j.parkreldis.2007.02.005
Jones, E. J., Gliga, T., Bedford, R., Charman, T., and Johnson, M. H. (2014). Developmental pathways to autism: a review of prospective studies of infants at risk. Neurosci. Biobehav. Rev. 39C, 1–33. doi: 10.1016/j.neubiorev.2013.12.001
Jones, S., Zhang, X., Parsons, D. W., Lin, J. C., Leary, R. J., Angenendt, P., et al. (2008). Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806. doi: 10.1126/science.1164368
Jospin, M., Watanabe, S., Joshi, D., Young, S., Hamming, K., Thacker, C., et al. (2007). UNC-80 and the NCA ion channels contribute to endocytosis defects in synaptojanin mutants. Curr. Biol. 17, 1595–1600. doi: 10.1016/j.cub.2007.08.036
Kaczorowski, G. J., McManus, O. B., Priest, B. T., and Garcia, M. L. (2008). Ion channels as drug targets: the next GPCRs. J. Gen. Physiol. 131, 399–405. doi: 10.1085/jgp.200709946
Kang, H. J., Kawasawa, Y. I., Cheng, F., Zhu, Y., Xu, X., Li, M., et al. (2011). Spatio-temporal transcriptome of the human brain. Nature 478, 483–489. doi: 10.1038/nature10523
Khan, S., and Al Baradie, R. (2012). Epileptic encephalopathies: an overview. Epilepsy Res. Treat. 2012:403592. doi: 10.1155/2012/403592
Khateeb, S., Flusser, H., Ofir, R., Shelef, I., Narkis, G., Vardi, G., et al. (2006). PLA2G6 mutation underlies infantile neuroaxonal dystrophy. Am. J. Hum. Genet. 79, 942–948. doi: 10.1086/508572
Kim, B. J., Chang, I. Y., Choi, S., Jun, J. Y., Jeon, J. H., Xu, W. X., et al. (2012). Involvement of Na(+)-leak channel in substance P-induced depolarization of pacemaking activity in interstitial cells of Cajal. Cell. Physiol. Biochem. 29, 501–510. doi: 10.1159/000338504
Kirchhoff, M., Bisgaard, A. M., Stoeva, R., Dimitrov, B., Gillessen-Kaesbach, G., Fryns, J. P., et al. (2009). Phenotype and 244k array-CGH characterization of chromosome 13q deletions: an update of the phenotypic map of 13q21.1-qter. Am. J. Med. Genet. A 149A, 894–905. doi: 10.1002/ajmg.a.32814
Koroglu, C., Seven, M., and Tolun, A. (2013). Recessive truncating NALCN mutation in infantile neuroaxonal dystrophy with facial dysmorphism. J. Med. Genet. 50, 515–520. doi: 10.1136/jmedgenet-2013-101634
Krishnan, K. S., and Nash, H. A. (1990). A genetic study of the anesthetic response: mutants of Drosophila melanogaster altered in sensitivity to halothane. Proc. Natl. Acad. Sci. U.S.A. 87, 8632–8636. doi: 10.1073/pnas.87.21.8632
Kutlu, B., Burdick, D., Baxter, D., Rasschaert, J., Flamez, D., Eizirik, D. L., et al. (2009). Detailed transcriptome atlas of the pancreatic beta cell. BMC Med. Genomics 2:3. doi: 10.1186/1755-8794-2-3
Lalani, S. R., Shaw, C., Wang, X., Patel, A., Patterson, L. W., Kolodziejska, K., et al. (2013). Rare DNA copy number variants in cardiovascular malformations with extracardiac abnormalities. Eur. J. Hum. Genet. 21, 173–181. doi: 10.1038/ejhg.2012.155
Lear, B. C., Darrah, E. J., Aldrich, B. T., Gebre, S., Scott, R. L., Nash, H. A., et al. (2013). UNC79 and UNC80, putative auxiliary subunits of the NARROW ABDOMEN ion channel, are indispensable for robust circadian locomotor rhythms in Drosophila. PLoS ONE 8:e78147. doi: 10.1371/journal.pone.0078147
Lear, B. C., Lin, J. M., Keath, J. R., McGill, J. J., Raman, I. M., and Allada, R. (2005). The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron 48, 965–976. doi: 10.1016/j.neuron.2005.10.030
Lee, J. H., Barral, S., Cheng, R., Chacon, I., Santana, V., Williamson, J., et al. (2008). Age-at-onset linkage analysis in Caribbean Hispanics with familial late-onset Alzheimer's disease. Neurogenetics 9, 51–60. doi: 10.1007/s10048-007-0103-3
Lee, J. H., Cribbs, L. L., and Perez-Reyes, E. (1999). Cloning of a novel four repeat protein related to voltage-gated sodium and calcium channels. FEBS Lett. 445, 231–236. doi: 10.1016/S0014-5793(99)00082-4
Lee, Y., Yoon, K. A., Joo, J., Lee, D., Bae, K., Han, J. Y., et al. (2013). Prognostic implications of genetic variants in advanced non-small cell lung cancer: a genome-wide association study. Carcinogenesis 34, 307–313. doi: 10.1093/carcin/bgs356
Lele, K. P., Penrose, L. S., and Stallard, H. B. (1963). Chromosome Deletion in a Case of Retinoblastoma. Ann. Hum. Genet. 27, 171–174. doi: 10.1111/j.1469-1809.1963.tb00209.x
Lenox, R. H., Gould, T. D., and Manji, H. K. (2002). Endophenotypes in bipolar disorder. Am. J. Med. Genet. 114, 391–406. doi: 10.1002/ajmg.10360
Lerche, H., Shah, M., Beck, H., Noebels, J., Johnston, D., and Vincent, A. (2013). Ion channels in genetic and acquired forms of epilepsy. J. Physiol. 591, 753–764. doi: 10.1113/jphysiol.2012.240606
Liebeskind, B. J., Hillis, D. M., and Zakon, H. H. (2012). Phylogeny unites animal sodium leak channels with fungal calcium channels in an ancient, voltage-insensitive clade. Mol. Biol. Evol. 29, 3613–3616. doi: 10.1093/molbev/mss182
Lind, P. A., Macgregor, S., Vink, J. M., Pergadia, M. L., Hansell, N. K., De Moor, M. H., et al. (2010). A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res. Hum. Genet. 13, 10–29. doi: 10.1375/twin.13.1.10
Lipscombe, D., Andrade, A., and Allen, S. E. (2013). Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim. Biophys. Acta 1828, 1522–1529. doi: 10.1016/j.bbamem.2012.09.018
Liu, C., Au, J. D., Zou, H. L., Cotten, J. F., and Yost, C. S. (2004). Potent activation of the human tandem pore domain K channel TRESK with clinical concentrations of volatile anesthetics. Anesth. Analg. 99, 1715–1722. doi: 10.1213/01.ANE.0000136849.07384.44
Lu, B., Su, Y., Das, S., Liu, J., Xia, J., and Ren, D. (2007). The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell 129, 371–383. doi: 10.1016/j.cell.2007.02.041
Lu, B., Su, Y., Das, S., Wang, H., Wang, Y., Liu, J., et al. (2009). Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature 457, 741–744. doi: 10.1038/nature07579
Lu, B., Zhang, Q., Wang, H., Wang, Y., Nakayama, M., and Ren, D. (2010). Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron 68, 488–499. doi: 10.1016/j.neuron.2010.09.014
Lu, T. Z., and Feng, Z. P. (2011). A sodium leak current regulates pacemaker activity of adult central pattern generator neurons in Lymnaea stagnalis. PLoS ONE 6:e18745. doi: 10.1371/journal.pone.0018745
Lu, T. Z., and Feng, Z. P. (2012). NALCN: a regulator of pacemaker activity. Mol. Neurobiol. 45, 415–423. doi: 10.1007/s12035-012-8260-2
Luo, J., Balkin, N., Stewart, J. F., Sarwark, J. F., Charrow, J., and Nye, J. S. (2000). Neural tube defects and the 13q deletion syndrome: evidence for a critical region in 13q33-34. Am. J. Med. Genet. 91, 227–230. doi: 10.1002/(SICI)1096-8628(20000320)91:3<227::AID-AJMG14>3.0.CO;2-I
Mantegazza, M., Rusconi, R., Scalmani, P., Avanzini, G., and Franceschetti, S. (2010). Epileptogenic ion channel mutations: from bedside to bench and, hopefully, back again. Epilepsy Res. 92, 1–29. doi: 10.1016/j.eplepsyres.2010.08.003
Maruoka, T., Nagasoe, Y., Inoue, S., Mori, Y., Goto, J., Ikeda, M., et al. (2002). Essential hydrophilic carboxyl-terminal regions including cysteine residues of the yeast stretch-activated calcium-permeable channel Mid1. J. Biol. Chem. 277, 11645–11652. doi: 10.1074/jbc.M111603200
McCormack, W. M. Jr., Shen, J. J., Curry, S. M., Berend, S. A., Kashork, C., Pinar, H., et al. (2002). Partial deletions of the long arm of chromosome 13 associated with holoprosencephaly and the Dandy-Walker malformation. Am. J. Med. Genet. 112, 384–389. doi: 10.1002/ajmg.10659
McGuire, T. F., Sajithlal, G. B., Lu, J., Nicholls, R. D., and Prochownik, E. V. (2012). In vivo evolution of tumor-derived endothelial cells. PLoS ONE 7:e37138. doi: 10.1371/journal.pone.0037138
Mead, C. L., Kuzyk, M. A., Moradian, A., Wilson, G. M., Holt, R. A., and Morin, G. B. (2010). Cytosolic protein interactions of the schizophrenia susceptibility gene dysbindin. J. Neurochem. 113, 1491–1503. doi: 10.1111/j.1471-4159.2010.06690.x
Minami, K., and Uezono, Y. (2013). The recent progress in research on effects of anesthetics and analgesics on G protein-coupled receptors. J. Anesth. 27, 284–292. doi: 10.1007/s00540-012-1507-2
Mir, B., Iyer, S., Ramaswami, M., and Krishnan, K. S. (1997). A genetic and mosaic analysis of a locus involved in the anesthesia response of Drosophila melanogaster. Genetics 147, 701–712.
Mok, K. Y., Schneider, S. A., Trabzuni, D., Stamelou, M., Edwards, M., Kasperaviciute, D., et al. (2013). Genomewide association study in cervical dystonia demonstrates possible association with sodium leak channel. Mov. Disord. 29, 245–251. doi: 10.1002/mds.25732
Morgan, N. V., Westaway, S. K., Morton, J. E., Gregory, A., Gissen, P., Sonek, S., et al. (2006). PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat. Genet. 38, 752–754. doi: 10.1038/ng1826
Morgan, P. G., and Cascorbi, H. F. (1985). Effect of anesthetics and a convulsant on normal and mutant Caenorhabditis elegans. Anesthesiology 62, 738–744. doi: 10.1097/00000542-198506000-00007
Morgan, P. G., Sedensky, M., and Meneely, P. M. (1990). Multiple sites of action of volatile anesthetics in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 87, 2965–2969. doi: 10.1073/pnas.87.8.2965
Morgan, P. G., and Sedensky, M. M. (1995). Mutations affecting sensitivity to ethanol in the nematode, Caenorhabditis elegans. Alcohol. Clin. Exp. Res. 19, 1423–1429. doi: 10.1111/j.1530-0277.1995.tb01002.x
Morgan, P. G., Sedensky, M. M., Meneely, P. M., and Cascorbi, H. F. (1988). The effect of two genes on anesthetic response in the nematode Caenorhabditis elegans. Anesthesiology 69, 246–251. doi: 10.1097/00000542-198808000-00015
Morozova, T. V., Mackay, T. F., and Anholt, R. R. (2014). Genetics and genomics of alcohol sensitivity. Mol. Genet. Genomics. doi: 10.1007/s00438-013-0808-y. [Epub ahead of print].
Muller, J., Kiechl, S., Wenning, G. K., Seppi, K., Willeit, J., Gasperi, A., et al. (2002). The prevalence of primary dystonia in the general community. Neurology 59, 941–943. doi: 10.1212/01.WNL.0000026474.12594.0D
Nakashima, K., Kusumi, M., Inoue, Y., and Takahashi, K. (1995). Prevalence of focal dystonias in the western area of Tottori Prefecture in Japan. Mov. Disord. 10, 440–443. doi: 10.1002/mds.870100406
Nakayama, M., Iida, M., Koseki, H., and Ohara, O. (2006). A gene-targeting approach for functional characterization of KIAA genes encoding extremely large proteins. FASEB J. 20, 1718–1720. doi: 10.1096/fj.06-5952fje
Nash, H. A., Campbell, D. B., and Krishnan, K. S. (1991). New mutants of Drosophila that are resistant to the anesthetic effects of halothane. Ann. N.Y. Acad. Sci. 625, 540–544. doi: 10.1111/j.1749-6632.1991.tb33885.x
Nash, H. A., Scott, R. L., Lear, B. C., and Allada, R. (2002). An unusual cation channel mediates photic control of locomotion in Drosophila. Curr. Biol. 12, 2152–2158. doi: 10.1016/S0960-9822(02)01358-1
Nishikawa, K., and Kidokoro, Y. (1999). Halothane presynaptically depresses synaptic transmission in wild-type Drosophila larvae but not in halothane-resistant (har) mutants. Anesthesiology 90, 1691–1697. doi: 10.1097/00000542-199906000-00026
Nitabach, M. N., Blau, J., and Holmes, T. C. (2002). Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109, 485–495. doi: 10.1016/S0092-8674(02)00737-7
Noel, J., Sandoz, G., and Lesage, F. (2011). Molecular regulations governing TREK and TRAAK channel functions. Channels (Austin). 5, 402–409. doi: 10.4161/chan.5.5.16469
Nurnberger, J. I. Jr., Foroud, T., Flury, L., Su, J., Meyer, E. T., Hu, K., et al. (2001). Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am. J. Psychiatry 158, 718–724. doi: 10.1176/appi.ajp.158.5.718
Nussbaum, R. L., and Ellis, C. E. (2003). Alzheimer's disease and Parkinson's disease. N. Engl. J. Med. 348, 1356–1364. doi: 10.1056/NEJM2003ra020003
Nutt, J. G., Muenter, M. D., Aronson, A., Kurland, L. T., and Melton, L. J. 3rd. (1988). Epidemiology of focal and generalized dystonia in Rochester, Minnesota. Mov. Disord. 3, 188–194. doi: 10.1002/mds.870030302
Ollila, H. M., Soronen, P., Silander, K., Palo, O. M., Kieseppa, T., Kaunisto, M. A., et al. (2009). Findings from bipolar disorder genome-wide association studies replicate in a Finnish bipolar family-cohort. Mol. Psychiatry 14, 351–353. doi: 10.1038/mp.2008.122
Patel, A. J., Honore, E., Lesage, F., Fink, M., Romey, G., and Lazdunski, M. (1999). Inhalational anesthetics activate two-pore-domain background K+ channels. Nat. Neurosci. 2, 422–426. doi: 10.1038/8084
Pei, J., and Grishin, N. V. (2012). Cysteine-rich domains related to Frizzled receptors and Hedgehog-interacting proteins. Protein Sci. 21, 1172–1184. doi: 10.1002/pro.2105
Pierce-Shimomura, J. T., Chen, B. L., Mun, J. J., Ho, R., Sarkis, R., and McIntire, S. L. (2008). Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc. Natl. Acad. Sci. U.S.A. 105, 20982–20987. doi: 10.1073/pnas.0810359105
Ptak, K., Yamanishi, T., Aungst, J., Milescu, L. S., Zhang, R., Richerson, G. B., et al. (2009). Raphe neurons stimulate respiratory circuit activity by multiple mechanisms via endogenously released serotonin and substance P. J. Neurosci. 29, 3720–3737. doi: 10.1523/JNEUROSCI.5271-08.2009
Quelin, C., Bendavid, C., Dubourg, C., De La Rochebrochard, C., Lucas, J., Henry, C., et al. (2009). Twelve new patients with 13q deletion syndrome: genotype-phenotype analyses in progress. Eur. J. Med. Genet. 52, 41–46. doi: 10.1016/j.ejmg.2008.10.002
Rajaram, S., Spangler, T. L., Sedensky, M. M., and Morgan, P. G. (1999). A stomatin and a degenerin interact to control anesthetic sensitivity in Caenorhabditis elegans. Genetics 153, 1673–1682.
Ratnapriya, R., Vijai, J., Kadandale, J. S., Iyer, R. S., Radhakrishnan, K., and Anand, A. (2010). A locus for juvenile myoclonic epilepsy maps to 2q33-q36. Hum. Genet. 128, 123–130. doi: 10.1007/s00439-010-0831-6
Ren, D. (2011). Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron 72, 899–911. doi: 10.1016/j.neuron.2011.12.007
Rorsman, P., and Braun, M. (2013). Regulation of insulin secretion in human pancreatic islets. Annu. Rev. Physiol. 75, 155–179. doi: 10.1146/annurev-physiol-030212-183754
Sabin, L. R., Delas, M. J., and Hannon, G. J. (2013). Dogma derailed: the many influences of RNA on the genome. Mol. Cell 49, 783–794. doi: 10.1016/j.molcel.2013.02.010
Sander, J. W. (2003). The epidemiology of epilepsy revisited. Curr. Opin. Neurol. 16, 165–170. doi: 10.1097/01.wco.0000063766.15877.8e
Scheuer, T. (2011). Regulation of sodium channel activity by phosphorylation. Semin. Cell Dev. Biol. 22, 160–165. doi: 10.1016/j.semcdb.2010.10.002
Schuckit, M. A., Edenberg, H. J., Kalmijn, J., Flury, L., Smith, T. L., Reich, T., et al. (2001). A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol. Clin. Exp. Res. 25, 323–329. doi: 10.1111/j.1530-0277.2001.tb02217
Scott, W. K., Hauser, E. R., Schmechel, D. E., Welsh-Bohmer, K. A., Small, G. W., Roses, A. D., et al. (2003). Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am. J. Hum. Genet. 73, 1041–1051. doi: 10.1086/379083
Sedensky, M. M., and Meneely, P. M. (1987). Genetic analysis of halothane sensitivity in Caenorhabditis elegans. Science 236, 952–954. doi: 10.1126/science.3576211
Sedensky, M. M., Siefker, J. M., and Morgan, P. G. (2001). Model organisms: new insights into ion channel and transporter function. Stomatin homologues interact in Caenorhabditis elegans. Am. J. Physiol. Cell Physiol. 280, C1340–C1348.
Senatore, A., Monteil, A., Van Minnen, J., Smit, A. B., and Spafford, J. D. (2013). NALCN ion channels have alternative selectivity filters resembling calcium channels or sodium channels. PLoS ONE 8:e55088. doi: 10.1371/journal.pone.0055088
Senatore, A., and Spafford, J. D. (2013). A uniquely adaptable pore is consistent with NALCN being an ion sensor. Channels (Austin). 7, 60–68. doi: 10.4161/chan.23981
Seven, M., Ozkilic, A., and Yuksel, A. (2002). Dysmorphic face in two siblings with infantile neuroaxonal dystrophy. Genet. Couns. 13, 465–473.
Seybold, V. S. (2009). The role of peptides in central sensitization. Handb. Exp. Pharmacol. 451–491. doi: 10.1007/978-3-540-79090-7_13
Sharma, A., and Couture, J. (2014). A review of the pathophysiology, etiology, and treatment of attention-deficit hyperactivity disorder (ADHD). Ann. Pharmacother. 48, 209–225. doi: 10.1177/1060028013510699
Silveira, C., Marques-Teixeira, J., and De Bastos-Leite, A. J. (2012). More than one century of schizophrenia: an evolving perspective. J. Nerv. Ment. Dis. 200, 1054–1057. doi: 10.1097/NMD.0b013e318275d249
Sinke, A. P., Caputo, C., Tsaih, S. W., Yuan, R., Ren, D., Deen, P. M., et al. (2011). Genetic analysis of mouse strains with variable serum sodium concentrations identifies the Nalcn sodium channel as a novel player in osmoregulation. Physiol. Genomics 43, 265–270. doi: 10.1152/physiolgenomics.00188.2010
Smart, T. G. (1997). Regulation of excitatory and inhibitory neurotransmitter-gated ion channels by protein phosphorylation. Curr. Opin. Neurobiol. 7, 358–367. doi: 10.1016/S0959-4388(97)80063-3
Smith, D. J., Whitham, E. A., and Ghaemi, S. N. (2012). Bipolar disorder. Handb. Clin. Neurol. 106, 251–263. doi: 10.1016/B978-0-444-52002-9.00015-2
Snutch, T. P., and Monteil, A. (2007). The sodium “leak” has finally been plugged. Neuron 54, 505–507. doi: 10.1016/j.neuron.2007.05.005
Souza, R. P., Rosa, D. V., Romano-Silva, M. A., Zhen, M., Meltzer, H. Y., Lieberman, J. A., et al. (2011). Lack of association of NALCN genetic variants with schizophrenia. Psychiatry Res. 185, 450–452. doi: 10.1016/j.psychres.2010.07.009
Speca, D. J., Chihara, D., Ashique, A. M., Bowers, M. S., Pierce-Shimomura, J. T., Lee, J., et al. (2010). Conserved role of unc-79 in ethanol responses in lightweight mutant mice. PLoS Genet. 6:1057. doi: 10.1371/journal.pgen.1001057
St. George-Hyslop, P. H., and Petit, A. (2005). Molecular biology and genetics of Alzheimer's disease. C. R. Biol. 328, 119–130. doi: 10.1016/j.crvi.2004.10.013
Strohl, K. P. (2003). Periodic breathing and genetics. Respir. Physiol. Neurobiol. 135, 179–185. doi: 10.1016/S1569-9048(03)00036-3
Sun, H., Xu, J., Della Penna, K. B., Benz, R. J., Kinose, F., Holder, D. J., et al. (2002). Dorsal horn-enriched genes identified by DNA microarray, in situ hybridization and immunohistochemistry. BMC Neurosci. 3:11. doi: 10.1186/1471-2202-3-11
Swayne, L. A., Mezghrani, A., Lory, P., Nargeot, J., and Monteil, A. (2010). The NALCN ion channel is a new actor in pancreatic beta-cell physiology. Islets 2, 54–56. doi: 10.4161/isl.2.1.10522
Swayne, L. A., Mezghrani, A., Varrault, A., Chemin, J., Bertrand, G., Dalle, S., et al. (2009). The NALCN ion channel is activated by M3 muscarinic receptors in a pancreatic beta-cell line. EMBO Rep. 10, 873–880. doi: 10.1038/embor.2009.125
Tanzi, R. E., and Bertram, L. (2005). Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell 120, 545–555. doi: 10.1016/j.cell.2005.02.008
Teo, C., Zai, C., Borlido, C., Tomasetti, C., Strauss, J., Shinkai, T., et al. (2012). Analysis of treatment-resistant schizophrenia and 384 markers from candidate genes. Pharmacogenet. Genomics 22, 807–811. doi: 10.1097/FPC.0b013e3283586c04
Terracciano, A., Tanaka, T., Sutin, A. R., Sanna, S., Deiana, B., Lai, S., et al. (2010). Genome-wide association scan of trait depression. Biol. Psychiatry 68, 811–817. doi: 10.1016/j.biopsych.2010.06.030
Todd, T. W., and Lim, J. (2013). Aggregation formation in the polyglutamine diseases: protection at a cost? Mol. Cells 36, 185–194. doi: 10.1007/s10059-013-0167-x
Uher, R., Payne, J. L., Pavlova, B., and Perlis, R. H. (2013). Major depressive disorder in Dsm-5: implications for clinical practice and research of changes from Dsm-Iv. Depress. Anxiety. doi: 10.1002/da.22217. [Epub ahead of print].
Vacher, H., and Trimmer, J. S. (2011). Diverse roles for auxiliary subunits in phosphorylation-dependent regulation of mammalian brain voltage-gated potassium channels. Pflugers Arch. 462, 631–643. doi: 10.1007/s00424-011-1004-8
Van Swinderen, B. (2006). A succession of anesthetic endpoints in the Drosophila brain. J. Neurobiol. 66, 1195–1211. doi: 10.1002/neu.20300
Walczak-Sztulpa, J., Wisniewska, M., Latos-Bielenska, A., Linne, M., Kelbova, C., Belitz, B., et al. (2008). Chromosome deletions in 13q33-34: report of four patients and review of the literature. Am. J. Med. Genet. A 146, 337–342. doi: 10.1002/ajmg.a.32127
Wang, H., and Ren, D. (2009). UNC80 functions as a scaffold for Src kinases in NALCN channel function. Channels (Austin). 3, 161–163. doi: 10.4161/chan.3.3.8853
Wang, K. S., Liu, X. F., and Aragam, N. (2010). A genome-wide meta-analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr. Res. 124, 192–199. doi: 10.1016/j.schres.2010.09.002
Wellcome Trust Case Control, C. (2007). Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678. doi: 10.1038/nature05911
Wetherill, L., Kapoor, M., Agrawal, A., Bucholz, K., Koller, D., Bertelsen, S. E., et al. (2014). Family-based association analysis of alcohol dependence criteria and severity. Alcohol. Clin. Exp. Res. 38, 354–366. doi: 10.1111/acer.12251
Whan, V., Hobbs, M., McWilliam, S., Lynn, D. J., Lutzow, Y. S., Khatkar, M., et al. (2010). Bovine proteins containing poly-glutamine repeats are often polymorphic and enriched for components of transcriptional regulatory complexes. BMC Genomics 11:654. doi: 10.1186/1471-2164-11-654
Winkelman, J. W., Gagnon, A., and Clair, A. G. (2013). Sensory symptoms in restless legs syndrome: the enigma of pain. Sleep Med. 14, 934–942. doi: 10.1016/j.sleep.2013.05.017
Xie, L., Gao, S., Alcaire, S. M., Aoyagi, K., Wang, Y., Griffin, J. K., et al. (2013). NLF-1 delivers a sodium leak channel to regulate neuronal excitability and modulate rhythmic locomotion. Neuron 77, 1069–1082. doi: 10.1016/j.neuron.2013.01.018
Yan, T. C., McQuillin, A., Thapar, A., Asherson, P., Hunt, S. P., Stanford, S. C., et al. (2010). NK1 (TACR1) receptor gene ‘knockout’ mouse phenotype predicts genetic association with ADHD. J. Psychopharmacol. 24, 27–38. doi: 10.1177/0269881108100255
Yeh, E., Ng, S., Zhang, M., Bouhours, M., Wang, Y., Wang, M., et al. (2008). A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 6:e55. doi: 10.1371/journal.pbio.0060055
Keywords: NALCN, UNC-79, UNC-80, ion channel, excitability
Citation: Cochet-Bissuel M, Lory P and Monteil A (2014) The sodium leak channel, NALCN, in health and disease. Front. Cell. Neurosci. 8:132. doi: 10.3389/fncel.2014.00132
Received: 05 March 2014; Accepted: 28 April 2014;
Published online: 20 May 2014.
Edited by:
Christophe Altier, University of Calgary, CanadaReviewed by:
Zhong-Ping Feng, University of Toronto, CanadaLaurens Bosman, Erasmus MC, Netherlands
David Speca, University of California, Davis, USA
Copyright © 2014 Cochet-Bissuel, Lory and Monteil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arnaud Monteil, Institut de Génomique Fonctionnelle, Université Montpellier 1 & 2, CNRS UMR 5203, 141 rue de la Cardonille, Montpellier F-34094, France e-mail:YXJuYXVkLm1vbnRlaWxAaWdmLmNucnMuZnI=
 Maud Cochet-Bissuel
Maud Cochet-Bissuel Philippe Lory
Philippe Lory Arnaud Monteil
Arnaud Monteil