
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci., 23 April 2014
Sec. Cellular Neurophysiology
Volume 8 - 2014 | https://doi.org/10.3389/fncel.2014.00096
This article is part of the Research TopicRegulatory RNAs in the Nervous SystemView all 35 articles
Hox genes specify the structures that form along the anteroposterior (AP) axis of bilateria. Within the genome, they often form clusters where, remarkably enough, their position within the clusters reflects the relative positions of the structures they specify along the AP axis. This correspondence between genomic organization and gene expression pattern has been conserved through evolution and provides a unique opportunity to study how chromosomal context affects gene regulation. In Drosophila, a general rule, often called “posterior dominance,” states that Hox genes specifying more posterior structures repress the expression of more anterior Hox genes. This rule explains the apparent spatial complementarity of Hox gene expression patterns in Drosophila. Here we review a noticeable exception to this rule where the more-posteriorly expressed Abd-B Hox gene fails to repress the more-anterior abd-A gene in cells of the central nervous system (CNS). While Abd-B is required to repress ectopic expression of abd-A in the posterior epidermis, abd-A repression in the posterior CNS is accomplished by a different mechanism that involves a large 92 kb long non-coding RNA (lncRNA) encoded by the intergenic region separating abd-A and Abd-B (the iab8ncRNA). Dissection of this lncRNA revealed that abd-A is repressed by the lncRNA using two redundant mechanisms. The first mechanism is mediated by a microRNA (mir-iab-8) encoded by intronic sequence within the large iab8-ncRNA. Meanwhile, the second mechanism seems to involve transcriptional interference by the long iab-8ncRNA on the abd-A promoter. Recent work demonstrating CNS-specific regulation of genes by ncRNAs in Drosophila, seem to highlight a potential role for the iab-8-ncRNA in the evolution of the Drosophila Hox complexes.
Hox genes specify the structures that form along the anteroposterior (AP) axis of bilateria. They are strikingly conserved between invertebrates and vertebrates. This conservation extends past the gene sequences and into their relative positioning along the chromosome, as Hox genes are generally found in clusters (or complexes) in which the individual Hox genes are aligned along the chromosome in the same order as the structures they specify along the AP axis (McGinnis and Krumlauf, 1992). While this correspondence between genomic organization and body axis is suggestive of a fundamental mechanism of activation that has been conserved through evolution, thus far, no common overlying principle can completely explain the evolutionary conservation of the collinear alignment of the genes. In fact, clustering does not seem to be absolutely necessary for proper Hox gene regulation in Drosophila, the place where Hox genes were first discovered. Indeed, the Hox gene cluster in fruit flies has been split at different location during the evolution of the Drosophila lineage (Lewis et al., 2003; Negre et al., 2003; Negre and Ruiz, 2007). In D. melanogaster, the Hox genes have been split into two clusters separated between the Antennapedia (Antp) and Ultrabithorax (Ubx) Hox genes (forming the Antp complex, and the bithorax complex, BX-C). Meanwhile, in Drosophila virilis, the complex is split between the Ubx and abd-A genes (Von Allmen et al., 1996). However, the fact that the Drosophila Hox complex has been split does not mean that the remaining collinear arrangement of the Drosophila Hox genes plays no role in their regulation. In fact, based on genetic rearrangement experiments, we know that the collinear arrangement of the Drosophila Hox genes is important for their proper expression (Maeda and Karch, 2010). Thus, the breaks found in the Drosophila Hox complexes may be exceptional cases of rearrangements that bypassed deleterious effects.
Based on our current understanding of Hox gene regulation in vertebrates and invertebrates, it now seems likely that at least some of the reason for preserving collinearity diverged during the evolutionary history of the two lineages. In mammals, collinearity seems to be preserved primarily due to the sharing of distal enhancer elements. Within the mouse Hoxd cluster, for example, it has been shown that Hox gene expression is controlled by shared remote enhancers located, 5′ and 3′ to the Hox complex. This sharing of enhancers presumably provides evolutionary pressure to keep the Hox genes clustered. Furthermore, it seems that distance from these enhancers controls the timing and ultimate location of Hox gene expression, providing pressure to preserve collinearity. However, this is not the case in invertebrates. In Drosophila, Hox gene expression is controlled by gene-specific enhancers located within the complex itself. It is perhaps for this reason that invertebrate Hox complexes are generally larger than their vertebrate counterparts and why the Drosophila Hox complex could be split in two.
Work on non-coding RNAs (ncRNAs) has provided an additional aspect regarding the conservation of the Hox gene clusters. Two microRNA genes (miRNA) have been found at similar positions within the Hox clusters of vertebrates and arthropods (Lagos-Quintana et al., 2003). The conserved miR-10 miRNA lies between the Drosophila Hox genes Deformed and Sex-comb-reduced. These fly Hox genes correspond to mammalian orthologs Hox4 and Hox5, respectively. Remarkably, the vertebrate miR-10b miRNA can be found between the Hox4 and Hox5 paralogs in the HoxB complex. A second miRNA gene in vertebrates (miR-196) is located between the Hox9 and Hox10 paralogs in the HoxA complex. These genes correspond to the fly genes abd-A and Abd-B. As in the case of miR10, a miRNA gene is found at a similar location in arthropods, though the primary sequence of the miRNA genes differ between the two lineages. In Drosophila, this miRNA gene is transcribed on both strands, giving rise to miR-iab-4 on one strand, and miR-iab-8 on the other strand. The miR-iab-8 template is embedded in a very large transcription unit of >92 kb (the iab-8ncRNA). Recent work from our lab on the iab-8-ncRNA has led to a number of interesting results, and provide additional reasons for the preservation of Hox clustering.
Hox genes were discovered through mutations that affect the identities of the segments that form along the AP axis of the fly. Many of these mutations were identified within the posterior Hox complex of the fly, called the BX-C (Lewis, 1978 for review, see Maeda and Karch, 2006). The BX-C encodes three Hox genes, Ubx, abd-A, and Abd-B (Figure 1), which are responsible for the identities of parasegments 8 to 13. These parasegments form the posterior thorax and all the abdominal segments of the fly (posterior T2, T3 and all eight abdominal segments A1–A8)1. Before the molecular genetic era, classical genetic analysis revealed the existence of mutations that affect the identities of each of the segments under the control of the BX-C. These mutations defined nine segment-specific functions. By genetic mapping, Ed Lewis discovered that these nine segments-specific functions are aligned along the chromosome in the same order as the segments they specify along the AP axis. This was the first identification of colinearity. Molecular analysis later revealed that the BX-C encoded only three, homeotic genes and that the genetically identified segment-specific functions were probably regulatory in nature. This was confirmed by antibody staining in mutant embryos. Antibody staining showed that Ubx, abd-A, and Abd-B are expressed in overlapping domains in the posterior half of the embryo (see also below). These expression patterns are intricate and finely tuned from one parasegment to the next (see for example Figure 2). By staining various mutant embryos it was shown that the segment-specific functions correspond to cis-regulatory regions that regulate the expression of Ubx. abd-A, or Abd-B in a parasegment-specific fashion. Thus the abx/bx and bxd/pbx cis- regulatory regions direct Ubx expression in PS5 and PS6, respectively. Similarly the iab-2 through iab-4 cis-regulatory regions direct the parasegment-specific expression patterns of abd-A in PS7, PS8, and PS9 (Figures 1 and 2; for review, see Maeda and Karch, 2006). And finally, the iab-5 trough iab-8 cis-regulatory regions regulate Abd-B in PS10 to PS13, respectively. Thus, the collinearity that exists in flies extends beyond the genes themselves to the cis-regulatory elements that drive the Hox gene expression.
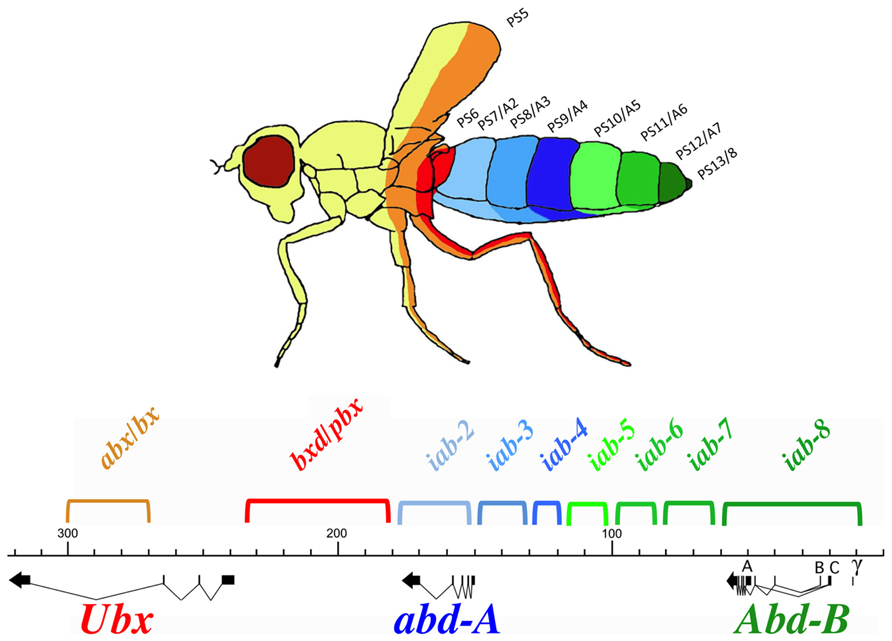
FIGURE 1. Synopsis of the BX-C. The genomic region of the BX-C is marked off in kilobases according to the numbering of (Martin et al., 1995). The three transcription units Ubx, abd-A, and Abd-B with their exons marked as thick lines and the arrows showing the transcription polarity are drawn below the DNA map. The horizontal and colored brackets above the DNA line indicate the extends of the segment-specific cis- regulatory regions with the following color code. Orange and red (abx/bx and bxd/pb) regulate expression of Ubx in PS5/T3 and PS6/A1, respectively. The blue iab-2, iab-3, and iab-4 regions regulate abd-A expression in PS7/A2, PS8/A3, and PS9/A4. Finally, the green iab-5, iab-6, iab-7, and iab-8 regulate Abd-B expression in PS10/A5, PS11/A6, PS12/A7, and PS13/A8, respectively. These segmental boundaries are depicted with the same colors on the fly above the BX-C map. Note that the parasegmental boundaries are visible in the thoracic segments where PS5 corresponds to the posterior part of T2 and the anterior part of T3. PS6 corresponds to posterior T3 and anterior A1.
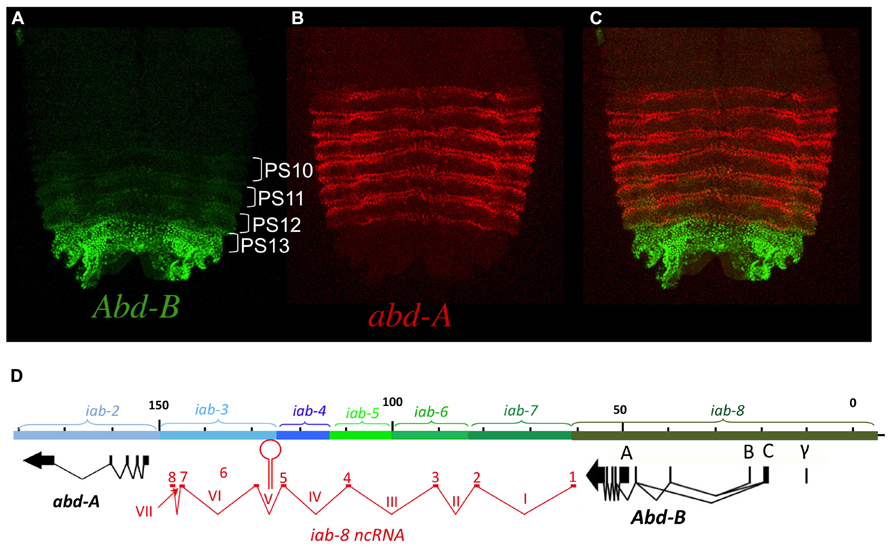
FIGURE 2. abd-A and Abd-B are expressed in broad domains. Panels A, B, and C show pelts of stage 13 embryos. In these preparations, embryos were cut along the dorsal midline and flattened on a slide. Anterior is at the top. In stage 13 embryos, Hox gene expression is mostly visible in the epidermis with abd-A displayed in red and Abd-B in green. In panel A, Abd-B appears in a graded fashion from PS10 to PS13 (parasegments are marked by brackets). In these parasegments, Abd-B is produced from promoter A under the regulation of, respectively the iab-5, iab-6, iab-7, and iab-8 regulatory regions (see also text). In PS14 an alternative form of Abd-B is produced from promoters B, C, and γ. The abd-A expression pattern in PS7 to PS12 is shown in panel B. Both abd-A and Abd-B are displayed in panel C. Note that their overall expression domains appear complementary to each other. Original observations published in (Celniker et al., 1990; Karch et al., 1990; Gummalla et al., 2012) The abdominal part of the BX-C is shown in panel D with the same map coordinates as in Figure 1 (Martin et al., 1995) and with the same color code for the abd-A, Abd-B genes and their respective regulatory domains. The structure of the iab-8ncRNA is shown in red under te DNA map. Introns are numbered with latin numbers and exons with regular numbering. Note that the polarity of transcription is the same as that for abd-A and Abd-B. Note also the presence of one exon for each of the iab cis-regulatory regions to the exception of 2 exons in iab-3. The location of miRiab-4/iab-8 in intron V is shown.
Like in vertebrates, most Drosophila Hox genes are expressed in broad domains along the AP axis. This is the case for the Antp gene that specifies the identity of PS4. While its segmental specification role is restricted to this single parasegments, Antp remains expressed in all the more posterior parasegments, until PS12 (Hafen et al., 1984) Similarly, the Ubx gene that specifies PS5 and PS6 identities remains expressed up to PS12 (White and Wilcox, 1984; Akam and Martinez-Arias, 1985; Beachy et al., 1985). Finally, abd-A that specifies PS7 to PS9 remains expressed up to PS12 (Karch et al., 1990; Macias et al., 1990; see Figure 2B). Thus these three Hox genes remain expressed posterior to the parasegments they specify respectively (though, in each parasegment, expression is limited to a subset of cells, see below).
Abd-B organization is a bit more complex than its counterparts of the BX-C, Ubx, and abd-A. While Abd-B is also expressed in a broad domain (Figure 2A), it is expressed as a parasegmental step-wise gradient and plays a visible specification role in all the parasegments where it is expressed (from PS10/A5 to PS13/A8; Kuziora and McGinnis, 1988; Celniker et al., 1990; Delorenzi and Bienz, 1990). Also, there is an alternatively spliced, truncated form of Abd-B originating from upstream promoters (B, C, and γ). This alternatively spliced isoform produces a truncated protein called Abd-Br, which is expressed in PS14 (see Figures 2D, 4E, 5B, 6A, 8A or 9A) and where it plays a role in specifying PS14 identity (see below for more details).
Looking at the overall parasegment-specific expression pattern of Ubx and abd-A, or that of abd-A and Abd-B (Figure 2), their respective expression domains appear complementary to each other. These complementary appearances result from a general rule referred as to as “posterior dominance” in which a posterior Hox gene represses the expression of the immediately adjacent anterior Hox gene. For instances, abd-A represses Ubx in PS7 to PS12 (Struhl and White, 1985), and Abd-B represses abd-A in PS10 to PS13 (Karch et al., 1990). It should be noted that abd-A repression is not easily visible in PS10, PS11, and PS12, as Abd-B is expressed in only a few cells in these parasegments. A similar negative, trans-regulatory interaction exists between Ubx and Antp, the Hox gene responsible for PS4 specification. In this case, Ubx is known to repress Antp (Hafen et al., 1984; Carroll et al., 1986).
As a result of these negative cross-regulatory interactions, each parasegement is a mosaic of cells expressing different combinations of Hox genes. In Peifer et al. (1987) proposed that parasegmental identity was the readout of the unique mosaicism in each parasegments. This model predicts that each cell within a parasegment expresses a single Hox gene. In order to test his hypothesis, we carefully reexamined Hox gene expression in the Drosophila embryo using confocal microscopy analysis with antibodies directed against Ubx, abd-A, and Abd-B. The general rule that a given Hox gene represses expression of the immediately anterior expressed Hox gene appears mostly true. However, there is a notable exception with abd-A and Abd-B in the central nervous system (CNS), where both proteins are found co-expressed in many cells (Figure 3). Interestingly, we often found that cells with the highest levels of Abd-A protein also express high levels of Abd-B protein (Figure 3).
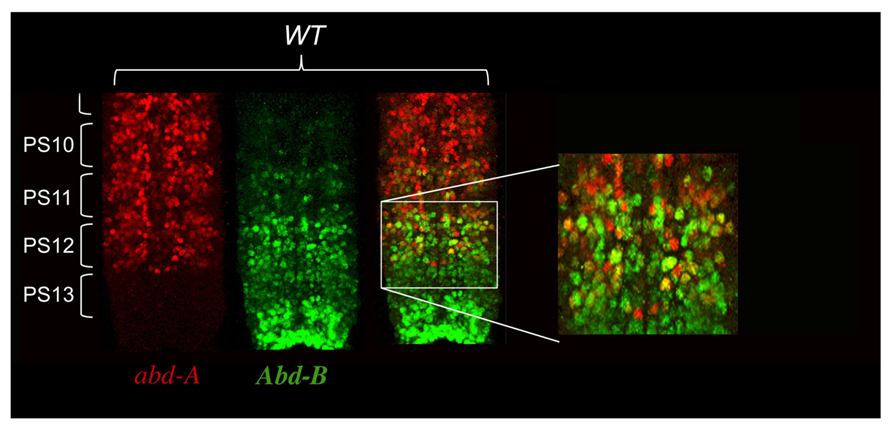
FIGURE 3. abd-A and Abd-B are both co-expressed in some cells of the central nervous system. CNS of stage 15 embryos stained for abd-A (red) and Abd-B (green) were dissected and mounted on a slide with anterior on top. Parasegments boundaries are shown. Note the presence of neurons in PS10 to PS12 expressing both proteins as seen by the yellow color. Often the neurons expressing high level of abd-A also express Abd-B. (original observation published in Gummalla et al., 2012).
The finding of cells expressing both abd-A and Abd-B contradicted the posterior transcriptional dominance rule of Hox genes as established by previous experiments. This prompted us to reexamine some of these experiments in more detail. Previously, it was shown that in the absence of Abd-B protein, abd-A protein becomes ectopically expressed in more posterior parasegments (Karch et al., 1990). This finding supported the idea that Abd-B and the posterior dominance rule restricted abd-A to more anterior abdominal parasegments. When we examined Abd-B null mutants in detail, we found that while we do indeed observe an extension of abd-A expression in PS13 in the epidermis, expression in the CNS remains unaffected (Figure 4B). This can, perhaps, be more easily seen in Abd-BD14 mutants (Abd-BD14; Figure 5). As mentioned above, the Abd-B transcription unit displays some complexity, harboring multiple promoters (marked A, B, C, and γ; Figures 1–8; Zavortink and Sakonju, 1989; Boulet et al., 1991). Transcription initiating from the A promoter encodes the long isoform of the Abd-B protein referred as to the “m” isoform (for morphogenetic function; Casanova et al., 1986). The ABD-B m isoform is expressed from PS10/A5 to PS13/A8, thereby assigning identities to these parasegments/segments. Promoters B, C, and γ are only active in PS14. Splicing of these transcripts lead to the generation of a shorter isoform of Abd-B lacking the N terminal sequences of the m isoform. This shorter isoform is referred as to the “r” isoform (for regulatory function; Casanova et al., 1986; Kuziora and McGinnis, 1988; Boulet et al., 1991). In Abd-BD14, a deletion removes the “A” promoter along with the N-terminal coding sequences of the ABD-B m isoform (Karch et al., 1985; Zavortink and Sakonju, 1989). As a consequence, there is no detectable expression of Abd-B in PS10 through PS13 (Figure 5). In agreement with this observation, the few emerging escaper flies have their fifth through eighth abdominal segments transformed into the fourth abdominal segment (Karch et al., 1985). In PS14, however, note the presence of the truncated “r” ABD-B isoform that is encoded by transcripts initiating from the B, C, and γ promoters. While Abd-B is absent in PS10-13, there is no extension of abd-A expression in the CNS into PS13 in the context of the Abd-BD14 mutant background (as illustrated by the gap between the red and green staining in Figure 4). This indicates that Abd-B is probably not responsible (or at least, not exclusively responsible) for abd-A repression in PS13.
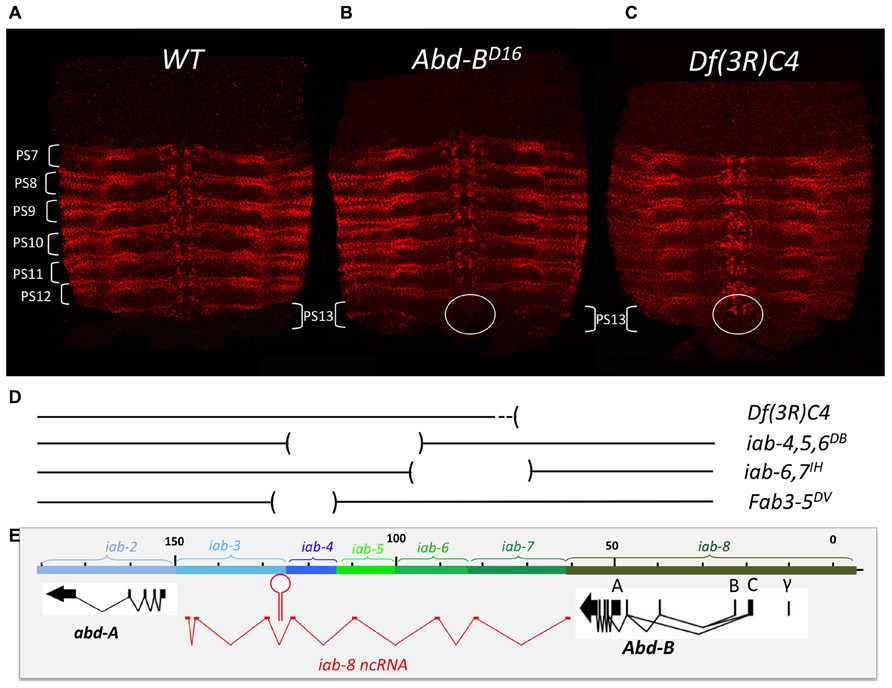
FIGURE 4. abd-A expansion in PS13 in Abd-B mutant context is restricted to the epidermis. Pelts of stage 15 embryos stained for abd-A were prepared as in Figure 1. The WT expression pattern from PS7 to PS12 is shown in panel A. Panel B shows the pattern of abd-A expression in a homozygous Abd-BD16 mutant embryo. Note the expansion of abd-A expression in PS13 in the epidermis. In the CNS, however, (circled) there is no expansion. Panel C shows a homozygous Df(3R)C4 mutant embryo in which abd-A expansion in PS13 occurs in both epidermis and CNS (circled; original observation published in Gummalla et al., 2012). Panel D depicts the extend of the various deficiencies we used in our unsuccessful attempts to locate a second discrete repressive mechanism (see page 13). Panel E, same as panel D in Figure 2.
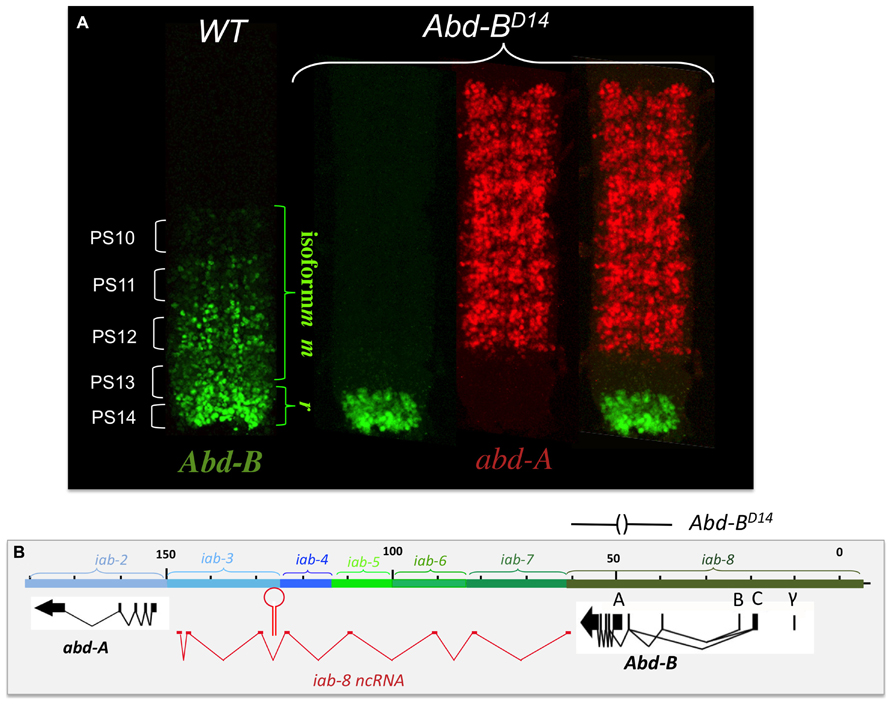
FIGURE 5. abd-A is still repressed in the CNS of Abd-BD14 mutant embryo. Abd-BD14 removes the promoter A of the Abd-B transcription unit (indicated above panel B). As the A promoter is regulated by the iab-5, iab-6, iab-7, and iab-8 regulatory domains, there is no Abd-B expression in PS10 to PS13 (see panel A). In PS14, however, a truncated version of Abd-B (cross-reacting with the antibody) is expressed in PS14 from the B, C, and γ promoters (panel A). This result indicates the existence of alternate mechanism(s) (than Abd-B repression) to keep abd-A off in PS13 (original observation published in Gummalla et al., 2012).
We further confirmed this finding by asking if ectopic Abd-B could repress abd-A in the CNS. If PS13 like levels of Abd-B could repress abd-A, then ectopically activating Abd-B to PS13 levels in another PS, should repress abd-A expression. To do this, we used the Fab-8205 mutation (Barges et al., 2000). Fab-8205 is a mutation that removes a cis-regulatory domain boundary between iab-7 and iab-8. Through a mechanism that is too complex to explain here, this deletion results in iab-8, normally driving PS13 levels of Abd-B expression, being activated in PS12. As expected of such a mutation, Fab-8205 results in a homeotic transformation of PS12/A7 into PS13/A8. Staining of Fab-8205 for abd-A showed normal levels of abd-A protein in PS12, indicating that PS13 levels of Abd-B cannot repress abd-A in the CNS(data not shown).
Based on these results, two possibilities can be imagined to account for the lack of abd-A expression in PS13 of the CNS. The simplest possibility is that abd-A may not be expressed in PS13 simply because it is never turned on. This would imply that the iab cis-regulatory domains act differently on abd-A in the epidermis versus the CNS. Alternatively, the lack of abd-A in PS13 of the CNS could results from a different, not-yet-identified repressive mechanism.
Mutation analysis points to the latter hypothesis as being correct. Df(3R)C4 is a large deficiency that removes the entire Abd-B transcription unit as well as iab-8 and about half of the of iab-7 (Figure 4D). Staining for abd-A protein in Df(3R)C4 embryos demonstrates that abd-A can be expressed in the CNS of PS13 (Figure 4C), suggesting that a repressive mechanism is involved in limiting abd-A expression. As we know Abd-B is not involved in this repression, we must assume that Df(3R)C4 must delete additional sequences essential for the this second repressive mechanism. Previously, a large, 92 kb ncRNA spanning the intergenic region between abd-A and Abd-B was discovered emanating from a region in iab-8 near the Fab-8 boundary (see below and Figures 2D, 4E, 5B, 6A, 8A, and 9A). We wondered if this long non-coding RNA (lncRNA), called the iab-8-ncRNA, could be involved in abd-A repression. As the promoter for the iab-8-ncRNA mapped to a region in iab-8 just next to the Fab-8 boundary, we examined abd-A expression in a larger Fab-8 deletion (Fab-864) that also removes the ncRNA promoter. Interestingly, we found that in Fab-864 mutants, we could see ectopic abd-A in PS13 even though Abd-B was expressed in both PS12 and PS13 at PS13 levels (Figure 8C). In fact, the levels of abd-A protein in PS13 resembled the levels of expression normally seen in PS12. Thus, these results pointed to the long iab-8ncRNA as the probable source of abd-A repression in PS13 of the CNS.
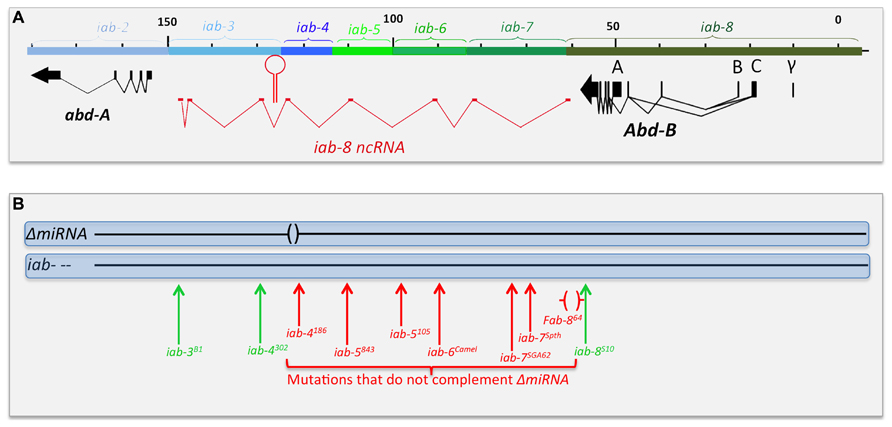
FIGURE 6. Chromosomal breaks to the right of the miR-iab-8 fail to complement ΔmiRNA. Panel A shows the genomic map of the abdominal region of the BX-C as described in Figure 2. Panel B symbolizes the two homologs chromosomes of heterozygotes between ΔmiRNA and various rearrangement breakpoints that disrupt the abdominal region of the BX-C. Breaks in red fail to complement the sterility phenotype of ΔmiRNA, while break in green are fully fertile over ΔmiRNA. The Fab-864 deletion removing the promoter of the iab-8ncRNA is indicated by red brackets.
The first evidence for the existence of a large transcription unit spanning the abd-A/Abd-B intergenic region arose with the emergence of in situ hybridization techniques. Already, Sanchez-Herrero and Akam (1989) noticed the presence of a signal at the posterior end of the embryos detected with many large genomic probes. Then, several studies reported similar embryonic expression patterns in the CNS and epidermis in PS13 and 14 with strand-specific probes detecting transcripts oriented from Abd-B toward abd-A (Bae et al., 2002; Drewell et al., 2002; Hogga and Karch, 2002; Rank et al., 2002; Schmitt et al., 2005). The similarity between the expression patterns reported in these various studies was evident, but it was only in 2008 that it became clear that they reflected the existence of a very large transcription unit active in PS13 and PS14 (Bender, 2008). In Bender (2008) used gene conversion to generate a surgical deletion of a miRNA located between abd-A and Abd-B. At the time, it was known that the miRNA was expressed from both DNA strands and were called miR iab3-4 and miR iab 4-3 respectively, based on the orientation of the transcription unit producing the miRNA. The deletion created was only 45 nucleotides long (henceforth called ΔmiRNA) to remove only the sequence encoding the two miRNAs. Although both miRNAs were predicted to target the Ubx and abd-A Hox genes, flies homozygous for the deletion did not harbor any segmental abnormalities, indicating that both miRNAs probably do not have a strict “homeotic function.” While the body structure and anatomy of these flies appeared completely normal, both females and males deleted for these miRNAs are sterile. However, this sterility does not seem to stem from a physical problem with their reproductive organs (gonads and/or the genitalia). Instead, the sterility phenotype present in ΔmiRNA flies seems to stem from a neuronal defect that makes them either unable to copulate (males) or unable to deposit eggs (females).
In as much as the miRNA gene is transcribed on both strands, Bender (2008) used a classical complementation test to determine if the sterility resulted from failure in the production of one or the other (or both) miRNA. Drawing from the vast collections of bithorax alleles that interrupt the chromosomal continuity of the abdominal region of the BX-C, he determined that a 65 kb-long region between the miRNAs and Abd-B was required for the production of the miRNA (Figure 6B), as any chromosomal break within this region (the red vertical arrows in Figure 6B) failed to complement ΔmiRNA. Breaks further to the left of the site of the miRNAs (Figure 6B) or to the right of Fab-864 (green vertical arrows) are fertile when in trans to ΔmiRNA. These observations indicate that the sterility phenotype is caused by loss of the miRNA produced from sense stand (relative to abd-A and Abd-B transcription) and define the region of DNA required for the production of this template RNA that spans from the region just downstream of the Abd-B transcription unit and extending to, at least, the site of the miRNA. As the position of the promoter lies within the iab-8 regulatory domain, the transcript was named the iab-8-ncRNA and the miRNA was renamed miR-iab-8 (Bender, 2008). RACE and RNAseq data later led to the precise definition of the iab-8- ncRNA as a 92 kb-long transcription unit spanning the entire abd-A-Abd-B intergenic region (Enderle et al., 2011; Graveley et al., 2011; Gummalla et al., 2012). Remarkably, the pri-miRNA transcript is spiced, with an exon derived from each of the iab cis-regulatory domains. A comparison with the genomic sequence data from 13 Drosophila species revealed that the transcript is conserved. Intriguingly, it is not the exonic sequences that are the most conserved, but the intron/exon junctions, as if it was the act of spicing that matters for the function of the iab-8-ncRNA. At present, there is no hint at the function of the spliced product or at the role of spicing in the function of the iab-8-ncRNA and/or miR-iab-8.
The expression pattern of the iab-8-ncRNA (and thus miR-iab-8) is consistent with the location of the promoter in iab-8, which controls the expression of Abd-B in PS13. The iab-8-ncRNA transcripts first appear at the posterior end of the embryo 3 h after fertilization, at the cellular blastoderm stage (Figure 7A). When the first signs of segmentation are visible (during germband elongation, Figure 7B), expression is restricted to PS13 and PS14 and mostly visible in the epidermis. After germband retraction, at the developmental stage where the nerve chord become visible, expression decays rapidly in the epidermis and become predominantly expressed in the CNS in PS13 and PS14, where it remains until for some time (Figure 7C). In fact, PS13/14 expression can even be seen in the CNS of third instar larvae (unpublished).
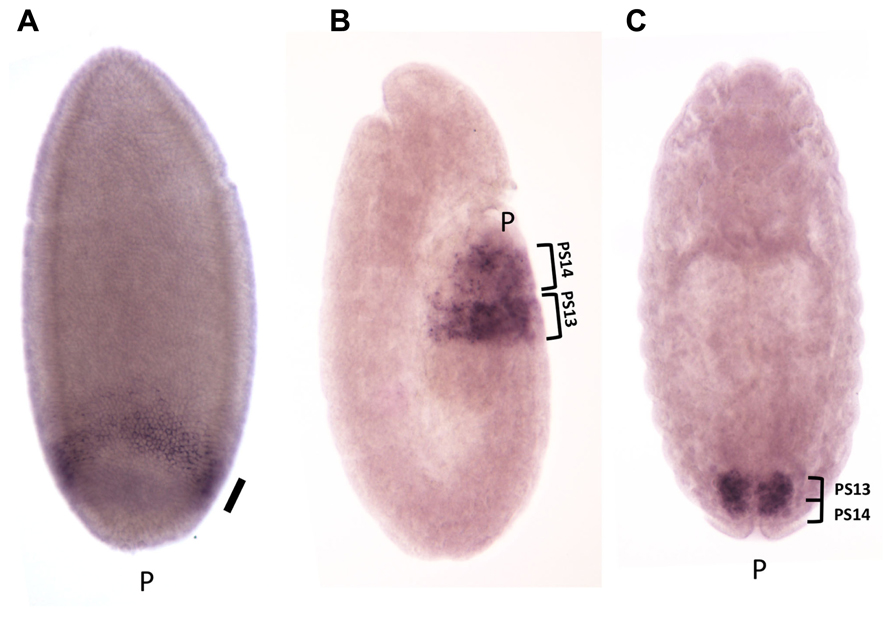
FIGURE 7. Expression pattern of the iab-8ncRNA. Embryos were hybridized with a strand-specific probe derived from the iab-6 region, to detect transcription in the same polarity than abd-A and Abd-B. Panel A shows an embryo 3 h after fertilization at the cellular blastoderm stage. A uniform band is visible at the posterior end of the embryo (shown by the thick oblique bar). At this stage, transient transcription from the iab-6 regulatory regions is detectable in PS11 (oblique arrow). At the elongated germ band stage (B), transcription is visible in the epidermis in PS13 and PS14. Panel C show a stage 15 embryo, after germ band contraction. Transcription is restricted to the CNS in PS13 and PS14 (original observation published in Bender, 2008).
Several features of miR-iab-8 made it the prime candidate to be the repressor of abd-A expression in PS13 of the CNS. First, bioinformatics analysis predicted abd-A as a probable target of miR-iab-8. Second, it was strongly expressed in the cells where abd-A is repressed (PS13 of the CNS). Third, deletion of its promoter leads to a strong derepression of abd-A. And finally, reporter and ectopic expression studies showed that the abd-A 3′ UTR could in fact be targeted by the miRNA for translational repression (Stark et al., 2008; Tyler et al., 2008). Based on these findings, it seemed obvious that deletion of the miRNA would lead to abd-A derepression.
Examining abd-A expression in the CNS of ΔmiRNA mutant embryos showed that there is indeed a misexpression of abd-A in animals lacking miR-iab8. Surprisingly, however, this misexpression is limited to only a few neurons (Figure 8B). Furthermore, the misexpression appears stochastic as the pattern of derepression varies between different nerve chords. This observation was unexpected as the deletion of the promoter caused much more drastic derepression (Figure 8C). Based on this result, we hypothesized the existence of a second, partially redundant mechanism involving the iab-8-ncRNA to keep abd-A repressed.
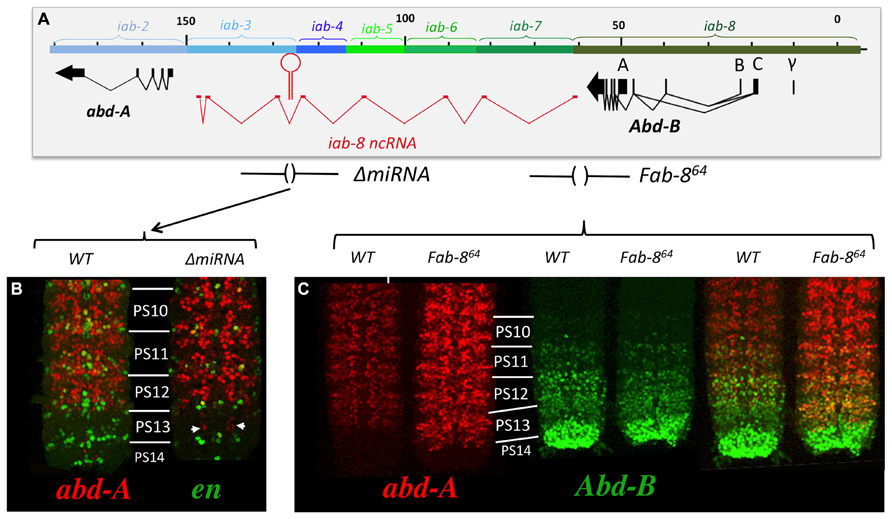
FIGURE 8. abd-A is only de-repressed in a few cells in PS13 in ΔmiRNA. Panel A shows the genomic map of the abdominal region of the BX-C as described in Figure 2 with the ΔmiRNA deletion drawn above. CNSs were dissected out from stage 15 embryos Note in panel B that abd-A is de-repressed in only few neurons in PS13. Panel C show the abd-A(red) and Abd-B (green) expression patterns in WT and Fab-864 homozygotes. Note the complete de-repression of abd-A in PS13 (original observation published in Gummalla et al., 2012).
As mentioned earlier, deletion of the iab-8-ncRNA promoter resulted in a complete derepression of abd-A in PS13. We used this phenotype to map additional elements in the iab-8-ncRNA that were important for abd-A repression. To do this, we first stained embryos, homozygous for various internal deficiencies in the iab-8-ncRNA sequence, thinking that if something like a second miRNA existed in the transcript, we might be able to identify it in this manner (Mihaly et al., 2006). Unfortunately, all deficiencies tested, with the exception of Fab-3-5DV, which removes the iab-8-miRNA, show no phenotype in this assay (see Figure 4D for the extend of the deficiencies). It must be noted, however, that while most of the ncRNA sequence has been tested by deletion analysis, we have no deficiencies spanning the 3′ end of the RNA (a region of about 15 kb) that do not also remove the abd-A promoter.
Therefore, to continue this analysis, we next decided to stain embryos from flies homozygous for chromosomal rearrangements that break the continuity of the iab-8-ncRNA. Using these lines, we found that all breaks lying in between the miRNA and its promoter showed complete derepression of abd-A in PS13 of the CNS (Figure 9). For example, break iab-4186, which breaks just upstream if the miRNA, shows a complete derepression of abd-A in PS13 of the CNS, much like an iab-8-ncRNA promoter deletion. Meanwhile, breaks lying between the miRNA and its 3′ end, which presumably still make the miRNA, showed a much milder, but visible derepression of abd-A in PS13 (Figure 9). This phenotype was reminiscent of ΔmiRNA embryos (see, for example iab-35022 in Figure 9). Based on the 3′-most rearrangement that causes a derepression of abd-A, we can limit the area where this second element must lie to a sequence of, at most, 5 kb (due to the resolution of the mutation mapping). This area contains two exons of the lncRNA and lies just 5′ to the abd-A transcriptional start site.
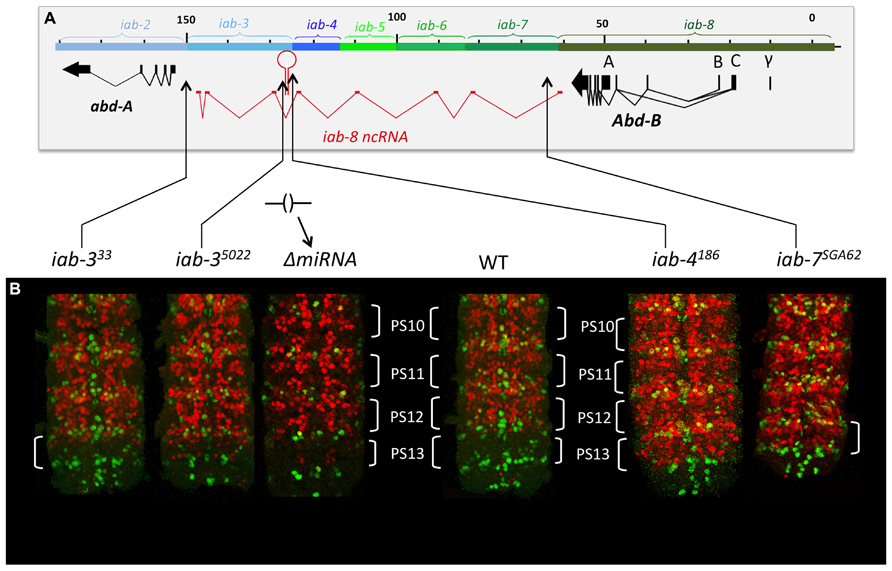
FIGURE 9. abd-A expression in the CNS in mutant that truncate the iab-8ncRNA. Panel A, show the molecular map of the abdominal region of the BX-C as in the figure above. The various rearrangement breaks truncating the iab-8ncRNA are shown below the map, along with the ΔmiRNA. Panel B show the posterior CNS of embryos that were stained for abd-A (red) and engrailed (en, green). The engrailed stripes mark each of the parasegments. Note that rearrangements disrupting the iab-8ncRNA upstream from miR-iab-8 lead to a complete de-repression of abd-A in the CNS in PS13 (iab-6186, iab-7SGA62). Rearrangements breaks disrupting the iab-8-ncRNA downstream from the site of miR-iab-8 result in only a partial de-repression of abd-A in PS13. A ΔmiRNA CNS is also shown for comparisons (original observation published in Gummalla et al., 2012).
As stated above, we have no deficiencies covering most of this area that do not also remove the abd-A promoter. Therefore, we have had difficulty identifying the exact mechanism of this repression. However, a number of observations make us believe that the second mechanism does not involve a diffusible molecule, but simply depends on the transcription of the region around the abd-A promoter. First, no miRNAs have been predicted bioinformatically, or found from any miRNA screens, derived from the area in question. Second, although the transcript is spliced and polyadenylated, no known polypeptides are encoded by this transcript. Here, it must be noted that our colleague, Bender (2008) has studied the resulting cDNA from the spliced iab-8-ncRNA transcript in the fly. While he has found a conserved sequence in the eighth exon that could encode a micropeptide (Gummalla et al., 2012), overexpression of the iab-8 cDNA has no affect on abd-A expression.
Based on our mapping experiments, we know that the second repressive function must be located in the last ~5 kb of the iab-8-ncRNA. Much of this sequence makes up the final two exons of the iab-8-ncRNA, whose spliced product seems to play no role in abd-A regulation. As this region also includes the upstream promoter area of abd-A, we wondered if the act of transcribing this area could provide the repressive function. This was a difficult thing to test because of the lack of genetic tools in the area. Still, we thought about what such a mechanism would imply. We reasoned that diffusible molecules should work both in cis and in trans, meaning that if one copy of the element is mutated, the product of the other copy of the element should be able to compensate for its loss, since it is a diffusible molecule. Indeed, loss of one copy of the iab-8-miRNA shows no effect on abd-A expression (it is recessive). However, if the mechanism was transcription across the abd-A promoter, then this mode of repression should only worked in cis, as the wild-type copy of the element on one chromosome should not be able to compensate for its loss on the other. We tested this by staining heterozygous rearrangement break mutants whose breaks were downstream of the miRNA. In all of the lines previously shown to derepress abd-A as homozygotes, we observed weaker but still noticeable derepression of abd-A as heterozygotes (Figure 10). The fact that a deficiency that removes the entire BX-C (including the abd-A and the iab-8-ncRNA) does not show a similar phenotype (Gummalla et al., 2012) means that this derepression is not due to simple haploinsufficiency for this second element and points to a cis-dominant effect, consistent with our model that transcription across the abd-A promoter causes abd-A repression.
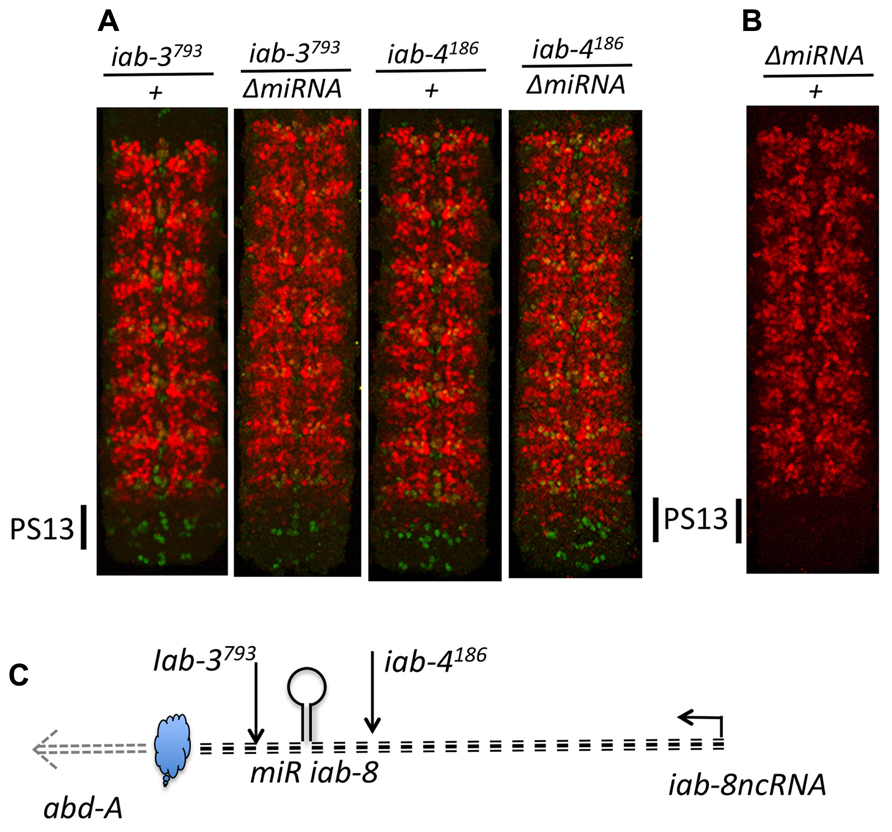
FIGURE 10. Haplo-insufficiency of breaks disrupting the iab-8ncRNA. Panel A show CNSs stained for abd-A (red) and engrailed (green) from embryos heterozygous for mutations disrupting the iab-8ncRNA. Panel B dispalys a CNS from a heterozygous ΔmiRNA/+ embryo. Note that while one dose of miR-iab-8 is sufficient to keep abd-A repressed in PS13 (B), de-repression of abd-A in PS13 is observed in each of the four genotypes displayed in panel A. Panel C summarizes the relative positions of the trans-acting repression mechanism (miRiab-8) and cis-acting repression mechanism symbolized as a cloud. The level of de-repressions depends on the position of the disrupting break (upstream or downstream of miRiab-8). De-repression increases when the disrupting break is over ΔmiRNA. In iab-4186/ΔmiRNA PS13 abd-A expression reaches a level as if only one of the two homologs produces abd-A.
This type of repressive mechanism is generally called transcriptional interference. Although some instances of this phenomenon have been reported in metazoans, it has mostly been observed in yeast where one gene is inhibited by the transcription of its promoter region by a polymerase transcribing from an upstream gene (Greger and Proudfoot, 1998; Martens et al., 2005; Kim et al., 2012). This is very similar to the situation we observe at the abd-A locus, where the iab-8-ncRNA, though its promoter lies 93 kb away, is transcribed across the intervening sequence until within about 1 kb of the abd-A transcriptional start site. Though we have not proved this, we imagine that this transcription would then prevent promoter proximal enhancer elements from initiating transcription at the abd-A promoter.
We started this review by explaining how abd-A regulation in the CNS seems to break the posterior dominance rule of the Drosophila Hox genes. Now, with this new data, we realize that this may not be the case. The transcriptional control of abd-A by the iab-8-ncRNA can simply be viewed as a modified example of posterior dominance. In this case, the repression occurs not through a transcription factor, but through two, completely different mechanisms: a miRNA-based repression mechanism and what is most likely a transcriptional interference-based repression mechanism. If we think of the iab-8-ncRNA as a Hox complex “gene,” then a more-posterior “gene” is still inhibiting a more anterior Hox gene, which fits with the posterior dominance rule.
The transcriptional interference model also provides another reason to explain the clustering of Hox genes in the fly. Transcriptional interference relies on having two genes in close proximity, so that the transcription of one interferes with the promoter of the other. Here, this seems to have been accomplished by the transcription of a lncRNA interfering with the promoter of the abd-A gene. The fact that loss of the ncRNA causes sterility and that it is initiated from a promoter in the iab-8cis-regulatory domain (which controls Abd-B expression) means that there will be selective pressure to keep the abd-A and Abd-B genes clustered.
But why create such a complex mechanism to control abd-A expression in PS13 and 14 of the CNS? Although we cannot truly answer this question, we can provide some thoughts on the issue. First, we must assume that, in the CNS, there is a reason to eliminate the standard cross-regulatory interactions between Abd-B and abd-A to allow co-expression of the two Hox genes in the same cell. While fate mapping work has been extensively done in the CNS, we have not identified all of the neurons that express abd-A, or Abd-B or both, to know if the combinatorial expression of Hox genes leads to modification of cell fate. However, having said that, we do have some indication that co-expressing at least some Hox genes might affect cell viability. Work from the lab of Alex Gould showed that expression of abd-A in larval abdominal neuroblasts was required for the cessation cell division and eventual apoptosis of these cells (Bello et al., 2003). This work stemmed from the idea that there must be something to control neuroblast division in the brain, and from the initial observation that the there were ~10× fewer neuroblasts in the abdominal segments than in the thoracic segments. Based on the abdominal localization of this phenomenon, Bello et al. (2003) asked if abd-A could mediate this loss of neuroblasts. Their experiments showed both that the loss of abd-A in abdominal neuroblast led to an increase in the pool of neuroblasts and that the ectopic expression of abd-A in thoracic neuroblasts led to a decrease in the pool of neuroblasts. These phenotypes eventually could all be attributed to a pulse of abd-A expression during the third instar stage that caused the neuroblasts to undergo apoptosis. As the loss of neuroblasts affects A1–A7, it seems likely that abd-A and Abd-B might have to be expressed in the same cell to have this phenomenon occur in A5–A7.
Next, we must ask why this new type of regulation happens in the CNS. As it turns out, regulation by miRNAs may be a common feature for neuronal genes in Drosophila. Work by the Levine lab has shown that the function of the common pan-neuronal gene ELAV is to bind to the 3′ UTR sequences of certain transcripts and to prevent normal polyadenylation. The result of this activity is the extension of 3′ UTR sequences for many neuronal genes (Hilgers et al., 2011, 2012). In agreement with this finding it has been long known that many Hox genes with extended 3′ UTR (Antp, Ubx, abd-A, and Abd-B) are specifically expressed in the nervous system (Garber et al., 1983; Scott et al., 1983; Akam and Martinez-Arias, 1985; Kuziora and McGinnis, 1988; O’Connor et al., 1988). Supporting this work, the lab of Claudio Alonso has recently shown that many Hox genes, including abd-A, possess CNS-specific 3′ UTR extensions. The result of these extensions is often an increase in the number of miRNA target sites. In the case of abd-A, the extension adds two additional targets for the iab-8-miRNA. Alonso and colleagues propose that these 3′ extensions could indicate a need to lose miRNA regulation in the epidermis or a need to augment miRNA regulation in the CNS (Thomsen et al., 2010). The fact that the iab-8-ncRNA is expressed primarily in the CNS would definitely support the latter hypothesis with regards to abd-A regulation.
Lastly, we must discuss why such a long transcript has been conserved to perform these functions when a much smaller transcript might be able to do the same. Indeed a transcript starting just upstream of the miRNA could, if expressed in the right place, perform the same function. We know, for example, that artificially starting a transcript downstream of the actual iab-8-ncRNA promoter can inhibit abd-A expression in anterior segments (Gummalla, 2011). However, if the iab-8-ncRNA is required only in the posterior parasegments, then how could such a smaller RNA be expressed only in PS13 and 14 within the context of a more-anterior cis-regulatory domain. Although gene regulation in the BX-C is a little too complex to explain in this review, we can say that, in general, promoters located in a specific cis-regulatory domain, gain regulation by that cis-regulatory domain. Thus, a promoter located in iab-4 would probably be expressed in a pattern driven by the iab-4cis-regulatory domain (meaning that expression would start in PS9/A4) and would be expressed too anterior to be viable. Of course, one could simply imagine the cells of the CNS making a specific transcription factor or miRNA from another locus to inhibit abd-A expression in certain places, but then the issue becomes a matter of cost from where the system originated. Given that the fly has a system to elongate neuronal transcripts to provide more miRNA targets, and has a perfect place to obtain PS13 and 14 expression, we imagine that it was simpler to evolve the current ncRNA system than a secondary repressor. Given the large amount of ncRNAs currently being found in the cells of most organisms, it now seems clear that the energetic cost of transcription is probably not prohibitively high.
But this all assumes that transcriptional interference was added after the other mechanisms of Hox gene repression. This is still far from clear. It is possible that the first Hox genes were regulated by transcriptional interference. This is not an absurd notion to entertain. We know that the Hox genes were probably derived from tandem duplication events. Based on the similarity in construction of different cis-regulatory domains it seems likely that they too were made by duplication events happening later in evolution of an ancestral cis-regulatory region. Thus, the ancestral Hox complex contained just two very similar Hox genes, each probably controlled by small cis-regulatory domains. Each gene would probably express in a very similar pattern, having been duplicated from the same gene. Assuming a perfect duplication event, then the only differing feature with regards to these genes would be a slight difference in location on the chromosome and their neighboring genes. One can therefore imagine that if the 5′ gene could interfere with the transcription of its downstream brother, then this could have been one of the first events differentiating the two genes and allowing divergent functions to evolve.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Eva Favre Benjamin Barendun and Jorge Faustino for excellent technical assistance. This work was supported by the State of Geneva, the Swiss National Fund for Research and the Fonds Claraz.
Akam, M. E., and Martinez-Arias, A. (1985). The distribution of Ultrabithorax transcripts in Drosophila embryos. EMBO J. 4, 1689–1700.
Bae, E., Calhoun, V. C., Levine, M., Lewis, E. B., and Drewell, R. A. (2002). Characterization of the intergenic RNA profile at abdominal-A and abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. U.S.A. 99, 16847–16852. doi: 10.1073/pnas.222671299
Barges, S., Mihaly, J., Galloni, M., Hagstrom, K., Müller, M., Shanower, G., et al. (2000). The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127, 779–790.
Beachy, P. A., Helfand, S. L., and Hogness, D. S. (1985). Segmental distribution of bithorax complex proteins during Drosophila development. Nature 313, 545–551. doi: 10.1038/313545a0
Bello, B. C., Hirth, F., and Gould, A. P. (2003). A pulse of the Drosophila Hox protein abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron 37, 209–219. doi: 10.1016/S0896-6273(02)01181-9
Bender, W. (2008). MicroRNAs in the Drosophila bithorax complex. Genes Dev. 22, 14–19. doi: 10.1101/gad.1614208
Boulet, A., Lloyd, A., and Sakonju, S. (1991). Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development 111, 393–405.
Carroll, S. B., Laymon, R. A., McCutcheon, M. A., Riley, P. D., and Scott, M. P. (1986). The localization and regulation of Antennapedia protein expression in Drosophila embryos. Cell 47, 113–122. doi: 10.1016/0092-8674(86)90372-7
Casanova, J., Sanchez-Herrero, E., and Morata, G. (1986). Identification and characterization of a parasegment specific regulatory element of the abdominal-B gene of Drosophila. Cell 47, 627–636. doi: 10.1016/0092-8674(86)90627-6
Celniker, S. E., Sharma, S., Keelan, D. J., and Lewis, E. B. (1990). The molecular genetics of the bithorax complex of Drosophila: cis- regulation in the abdominal-B domain. EMBO J. 9, 4277–4286.
Delorenzi, M., and Bienz, M. (1990). Expression of abdominal-B homeoproteins in Drosophila embryos. Development 108, 323–329.
Drewell, R. A., Bae, E., Burr, J., and Lewis, E. B. (2002). Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. U.S.A. 99, 16853–16858. doi: 10.1073/pnas.222671199
Enderle, D., Beisel, C., Stadler, M. B., Gerstung, M., Athri, P., and Paro, R. (2011). Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res. 21, 216–226. doi: 10.1101/gr.114348.110
Garber, R. L., Kuroiwa, A., and Gehring, W. J. (1983). Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. EMBO J. 2, 2027–2036.
Graveley, B. R., Brooks, A. N., Carlson, J. W., Duff, M. O., Landolin, J. M., Yang, L., et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479. doi: 10.1038/nature09715
Greger, I. H., and Proudfoot, N. J. (1998). Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17, 4771–4779. doi: 10.1093/emboj/17.16.4771
Gummalla, M. (2011). abd-A regulation by the iab-8 noncoding RNA. Ph.D. thesis, Thesis no. 4379, Faculty of Science, University of Geneva, Geneva.
Gummalla, M., Maeda, R. K., Castro Alvarez, J. J., Gyurkovics, H., Singari, S., Edwards, K. A., et al. (2012). abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 8:e1002720. doi: 10.1371/journal.pgen.1002720
Hafen, E., Levine, M., and Gehring, W. J. (1984). Regulation of Antennapedia transcript distribution by the bithorax complex in Drosophila. Nature 307, 287–289. doi: 10.1038/307287a0
Hilgers, V., Lemke, S. B., and Levine, M. (2012). ELAV mediates 3’ UTR extension in the Drosophila nervous system. Genes Dev. 26, 2259–2264. doi: 10.1101/gad.199653.112
Hilgers, V., Perry, M. W., Hendrix, D., Stark, A., Levine, M., and Haley, B. (2011). Neural-specific elongation of 3’ UTRs during Drosophila development. Proc. Natl. Acad. Sci. U.S.A. 108, 15864–15869. doi: 10.1073/pnas.1112672108
Hogga, I., and Karch, F. (2002). Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development 129, 4915–4922.
Karch, F., Bender, W., and Weiffenbach, B. (1990). abdA expression in Drosophila embryos. Genes Dev. 4, 1573–1587. doi: 10.1101/gad.4.9.1573
Karch, F., Weiffenbach, B., Peifer, M., Bender, W., Duncan, I., Celniker, S., et al. (1985). The abdominal region of the bithorax complex. Cell 43, 81–96. doi: 10.1016/0092-8674(85)90014-5
Kim, T., Xu, Z., Clauder-Munster, S., Steinmetz, L. M., and Buratowski, S. (2012). Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150, 1158–1169. doi: 10.1016/j.cell.2012.08.016
Kuziora, M. A., and McGinnis, W. (1988). Different transcripts of the Drosophila Abd-B gene correlate with distinct genetic sub-functions. EMBO J. 7, 3233–3244.
Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A., and Tuschl, T. (2003). New microRNAs from mouse and human. RNA 9, 175–179. doi: 10.1261/rna.2146903
Lewis, E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. doi: 10.1038/276565a0
Lewis, E. B., Pfeiffer, B. D., Mathog, D. R., and Celniker, S. E. (2003). Evolution of the homeobox complex in the Diptera. Curr. Biol. 13, R587–R588. doi: 10.1016/S0960-9822(03)00520-7
Macias, A., Casanova, J., and Morata, G. (1990). Expression and regulation of the abd-A gene of Drosophila. Development 110, 1197–1207.
Maeda, R. K., and Karch, F. (2006). The ABC of the BX-C: the bithorax complex explained. Development 133, 1413–1422. doi: 10.1242/dev.02323
Maeda, R. K., and Karch, F. (2010). Cis-regulation in the Drosophila bithorax complex. Adv. Exp. Med. Biol. 689, 17–40. doi: 10.1007/978-1-4419-6673-5_2
Martens, J. A., Wu, P. Y., and Winston, F. (2005). Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19, 2695–2704. doi: 10.1101/gad.1367605
Martin, C. H., Mayeda, C. A., Davis, C. A., Ericsson, C. L., Knafels, J. D., Mathog, D. R., et al. (1995). Complete sequence of the bithorax complex of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 92, 8398–8402. doi: 10.1073/pnas.92.18.8398
Martinez-Arias, A., and Lawrence, P. (1985). Parasegments and compartments in the Drosophila embryo. Nature 313, 639–642. doi: 10.1038/313639a0
McGinnis, W., and Krumlauf, R. (1992). Homeobox genes and axial patterning. Cell 68, 283–302. doi: 10.1016/0092-8674(92)90471-N
Mihaly, J., Barges, S., Sipos, L., Maeda, R., Cleard, F., Hogga, I., et al. (2006). Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 133, 2983–2993. doi: 10.1242/dev.02451
Negre, B., Ranz, J. M., Casals, F., Caceres, M., and Ruiz, A. (2003). A new split of the Hox gene complex in Drosophila: relocation and evolution of the gene labial. Mol. Biol. Evol. 20, 2042–2054. doi: 10.1093/molbev/msg238
Negre, B., and Ruiz, A. (2007). HOM-C evolution in Drosophila: is there a need for Hox gene clustering? Trends Genet. 23, 55–59. doi: 10.1016/j.tig.2006.12.001
O’Connor, M. B., Binari, R., Perkins, L. A., and Bender, W. (1988). Alternative RNA products from the Ultrabithorax domain of the bithorax complex. EMBO J. 7, 435–445.
Peifer, M., Karch, F., and Bender, W. (1987). The bithorax complex: control of segmental identity. Genes Dev. 1, 891–898. doi: 10.1101/gad.1.9.891
Rank, G., Prestel, M., and Paro, R. (2002). Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 22, 8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002
Sanchez-Herrero, E., and Akam, M. (1989). Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107, 321–329.
Schmitt, S., Prestel, M., and Paro, R. (2005). Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 19, 697–708. doi: 10.1101/gad.326205
Scott, M. P., Weiner, A. J., Hazelrigg, T. I., Polisky, B. A., Pirrotta, V., Scalenghe, F., et al. (1983). The molecular organization of the Antennapedia locus of Drosophila. Cell 35, 763–776. doi: 10.1016/0092-8674(83)90109-5
Stark, A., Bushati, N., Jan, C. H., Kheradpour, P., Hodges, E., Brennecke, J., et al. (2008). A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 22, 8–13. doi: 10.1101/gad.1613108
Struhl, G., and White, R. A. (1985). Regulation of the Ultrabithorax gene of Drosophila by other bithorax complex genes. Cell 43, 507–519. doi: 10.1016/0092-8674(85)90180-1
Thomsen, S., Azzam, G., Kaschula, R., Williams, L. S., and Alonso, C. R. (2010). Developmental RNA processing of 3’UTRs in Hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development 137, 2951–2960. doi: 10.1242/dev.047324
Tyler, D. M., Okamura, K., Chung, W. J., Hagen, J. W., Berezikov, E., Hannon, G. J., et al. (2008). Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 22, 26–36. doi: 10.1101/gad.1615208
Von Allmen, G., Hogga, I., Spierer, A., Karch, F., Bender, W., Gyurkovics, H., et al. (1996). Splits in fruitfly Hox gene complexes. Nature 380, 116. doi: 10.1038/380116a0
White, R. A., and Wilcox, M. (1984). Protein products of the bithorax complex in Drosophila. Cell 39, 163–171. doi: 10.1016/0092-8674(84)90202-2
Keywords: Hox genes, abd-A, ncRNA, miRNA, bithorax-complex
Citation: Gummalla M, Galetti S, Maeda RK and Karch F (2014) Hox gene regulation in the central nervous system of Drosophila. Front. Cell. Neurosci. 8:96. doi: 10.3389/fncel.2014.00096
Received: 26 February 2014; Accepted: 14 March 2014;
Published online: 23 April 2014.
Edited by:
Tommaso Pizzorusso, Università degli Studi di Firenze, ItalyReviewed by:
Yuri Bozzi, University of Trento, ItalyCopyright © 2014 Gummalla, Galetti, Maeda and Karch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert K. Maeda and François Karch, Department of Genetics and Evolution, University of Geneva, 30 quai Ernest Ansermet, 1211 Geneva 4, Switzerland e-mail:cm9iZXJ0Lm1hZWRhQHVuaWdlLmNo;ZnJhbmNvaXMua2FyY2hAdW5pZ2UuY2g=
†Maheshwar Gummalla, Institute of Biochemistry, University of Medicine – University of Göttingen, Göttingen, Germany
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.