
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Neurosci. , 24 February 2014
Sec. Cellular Neuropathology
Volume 8 - 2014 | https://doi.org/10.3389/fncel.2014.00047
This article is part of the Research Topic Regulatory RNAs in the Nervous System View all 35 articles
 Bharat R. Iyengar1,2
Bharat R. Iyengar1,2 Ashwani Choudhary3
Ashwani Choudhary3 Mayuresh A. Sarangdhar3
Mayuresh A. Sarangdhar3 K. V. Venkatesh2
K. V. Venkatesh2 Chetan J. Gadgil1
Chetan J. Gadgil1 Beena Pillai3*
Beena Pillai3*The human brain is one of the most complex biological systems, and the cognitive abilities have greatly expanded compared to invertebrates without much expansion in the number of protein coding genes. This suggests that gene regulation plays a very important role in the development and function of nervous system, by acting at multiple levels such as transcription and translation. In this article we discuss the regulatory roles of three classes of non-protein coding RNAs (ncRNAs)—microRNAs (miRNAs), piwi-interacting RNA (piRNAs) and long-non-coding RNA (lncRNA), in the process of neurogenesis and nervous function including control of synaptic plasticity and potential roles in neurodegenerative diseases. miRNAs are involved in diverse processes including neurogenesis where they channelize the cellular physiology toward neuronal differentiation. miRNAs can also indirectly influence neurogenesis by regulating the proliferation and self renewal of neural stem cells and are dysregulated in several neurodegenerative diseases. miRNAs are also known to regulate synaptic plasticity and are usually found to be co-expressed with their targets. The dynamics of gene regulation is thus dependent on the local architecture of the gene regulatory network (GRN) around the miRNA and its targets. piRNAs had been classically known to regulate transposons in the germ cells. However, piRNAs have been, recently, found to be expressed in the brain and possibly function by imparting epigenetic changes by DNA methylation. piRNAs are known to be maternally inherited and we assume that they may play a role in early development. We also explore the possible function of piRNAs in regulating the expansion of transposons in the brain. Brain is known to express several lncRNA but functional roles in brain development are attributed to a few lncRNA while functions of most of the them remain unknown. We review the roles of some known lncRNA and explore the other possible functions of lncRNAs including their interaction with miRNAs.
The complexity of the nervous system is evident at the organ-system level, cellular level and at the molecular level. The systems-level complexity of the neuronal system is due to the highly connected neuronal network wherein each neuron connects to many other neurons by establishing synapses. At the cellular level, the complexity of the neuronal system arises from the cellular heterogeneity of the vertebrate brain, carrying at least four distinctly different cell types-neurons including several biochemically diverse sub-classes, astrocytes, oligodendrocytes and microglia; that have specialized but interdependent functions. At the molecular level, the complexity arises from the ability to create functional diversity from the genome through mechanisms like alternative splicing and RNA editing that are more prevalent in the nervous system as compared to other tissues; which makes the brain have the most complex transcriptome (Ramsköld et al., 2009), compared to other organs. The fact that brain capacity and cognitive abilities have greatly expanded from invertebrates to humans with a much lesser increase in the number of protein coding genes, indicates that gene regulation plays a major role in the development and function of the nervous system.
It was a long held idea that proteins are the versatile catalysts of life processes and RNAs serve the role of relaying the genetic message for the protein output. However, some of the key cellular processes including the very process of protein synthesis, is controlled by RNAs and studies, over the years, have shown that the functional ability of RNAs is much more than what was previously assumed. The diverse class of non-protein coding RNAs (ncRNAs), including microRNAs (miRNA), piwi-interacting RNA (piRNA), long-non-coding RNA (lncRNA) etc, are mainly involved in regulation of gene expression and are thus integral parts of the gene regulatory network (GRN). The recent evidences demonstrating wide-spread transcription throughout the genome suggest that the non-coding RNA form a sizeable component of the transcriptome of a eukaryotic cell (Jacquier, 2009; Clark et al., 2011). Several recent reviews have comprehensively cataloged these regulatory ncRNAs and provide a wealth of emerging evidence regarding their biogenesis, interacting partners and ability to regulate key target genes (Liu and Paroo, 2010).
Here, after a brief introduction to these classes of non-coding RNAs, we focus on two critical aspects of non-coding RNA function in the development of the vertebrate nervous system: their expression pattern and the network architecture of their interactions with other genes. The regulatory nature of these interactions is derived as much from the inherent features of the network as from the identity and functions of the genes that form the network. This review therefore draws on the principles of systems biology to explore the network of non-coding RNA and target interactions in the context of the nervous system.
Currently, these non-coding RNAs are classified into functional groups, on the basis of the rather arbitrary criteria of size and a limited knowledge of their functional roles in the cell. The major classes of ncRNAs are lncRNA (Mercer et al., 2009), piRNA, endogeneous small interfering RNA (endo-siRNA) and miRNAs.
miRNAs in their mature form are, on an average, 22 nt long and repress mRNAs which harbor miRNA “target sites” or miRNA recognition elements (MRE) in their 3′UTRs. These target sites are partially complementary to the miRNA and interact with the latter by Watson-Crick type base pairing. Functional miRNAs are bound to Argonaute (Ago) proteins and constitute a ribonucleoprotein complex called RNA Induced Silencing Complex (RISC), that is tethered to mRNAs by the miRNA-mRNA interaction. miRNAs arise from longer transcripts, called pri-miRNAs (primary miRNAs). Stem-loop like structures on pri-miRNA are identified by Drosha-DGCR8 (microprocessor) complex and are liberated from the long transcript by ribonucleolytic cleavage, to give rise to precursor miRNA (pre-miRNA). Pre-miRNA is exported to cytoplasm by Exportin-5 where it is again cleaved by Dicer to produce an imperfect RNA duplex. One or both the strands of the duplex is incorporated into Ago proteins to form a functional RISC (Figure 1A). Alternatively, miRNAs can also arise from introns of other transcripts as a result of splicing and subsequent cleavage by Dicer and these intronic regions are referred to as mirtrons (Filipowicz et al., 2008). The expression of miRNA can, therefore, be regulated at transcriptional or post-transcriptional levels (Winter et al., 2009). Even though the MREs are partially complementary to the miRNA, a 7-8mer region, called the seed, corresponding to 2nd–9th position of the miRNA, is essential for miRNA mediated regulation (Filipowicz et al., 2008). However, the partial complementarity allows the miRNA to target several mRNAs simultaneously (Chi et al., 2009; Helwak et al., 2013), forming a network motif called single input module (SIM) (Figure 1D). This may help in canalization of the gene regulation program (Hornstein and Shomron, 2006).
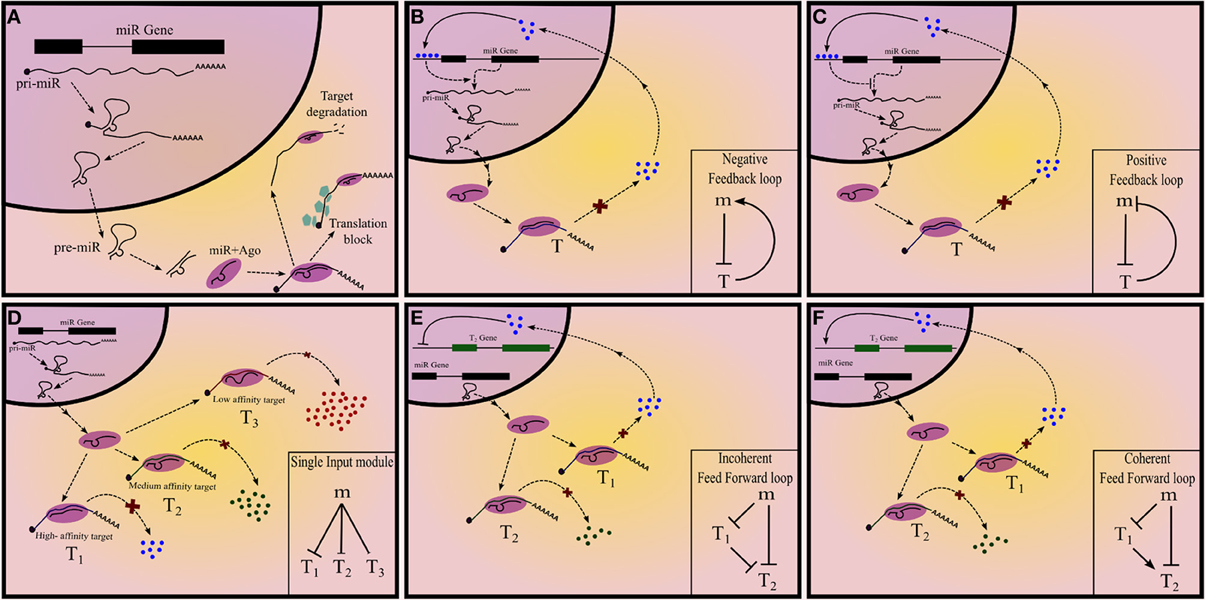
Figure 1. Summary of different types of network motifs in miRNA mediated gene regulatory networks. (A) Summary of miRNA biogenesis and mechanism of action. (B) Positive and (C) negative feedback loops, with a miRNA and a transcription factor. (D) Single input module with miRNA regulating three target nodes. The extent of regulation can differ between different targets. Feed-forward loops (E) Incoherent and (F) coherent feedforward loops with a miRNA regulating the target-T2.
piRNAs are slightly longer than miRNA (24–32 nt long) and associate with Piwi family of proteins which include Piwi, Aubergine (Aub) and Ago3 in Drosophila and MIWI, MILI, and MIWI2 in mouse. Even though the binding partners of piRNAs are structurally similar to the miRNA-binding Ago proteins, the piRNA biosynthetic pathway is very different from that of miRNA. piRNAs arise from specific genomic loci known as piRNA clusters and check transposon expansion. Initially piRNAs were reported in the Drosophila and Mouse germline (Ishizu et al., 2012); subsequently they were also reported in somatic tissues like ovarian follicular cells of Drosophila and nervous system of mouse (Lee et al., 2011) and Aplysia (Rajasethupathy et al., 2012). Another class of piRNAs is predominantly expressed in mouse spermatids during pachytene stage of meiosis I. This class of piRNA is expressed in high abundance in these cells but its functions and putative targets, remain unknown. piRNA clusters store transposon derived sequences; transcripts arising from these clusters associate with Piwi-proteins (Piwi and Aub in Drosophila and MIWI and MILI in mouse) and are trimmed from the 3′ end to generate primary piRNAs. The 3′ end is 2′-O-methylated which makes the piRNA stable. Primary piRNA can undergo a cycle of amplification, called ping-pong cycle, to generate secondary piRNAs (Ishizu et al., 2012). Unlike Drosophila where cleavage is sufficient to repress transposons, piRNA-mediated transposon repression predominantly happens via DNA methylation in mouse (Aravin et al., 2008; Watanabe et al., 2011). The exact mechanism of this process is yet to be explored.
lncRNAs are a diverse set that are loosely defined as ncRNAs longer than 200 nt that lack apparent protein coding ability due to the absence of a relatively long uninterrupted open reading frame. Therefore, unlike miRNAs or piRNAs, the lncRNAs differ greatly from each other with respect to size, interacting partners and mode of action. Most lncRNA regulate gene expression by affecting chromatin dynamics or providing scaffold/tether to regulatory proteins (Mercer et al., 2009). lncRNAs typically have the same structural features as mRNAs such as the 5′ cap, polyadenylated 3′ tail and undergo splicing to give rise to the final product. They are localized both to the nucleus and cytoplasm, but the signals that drive their localization are not known. The genomic loci of lncRNA can provide clues to their regulatory targets as it is observed that genes that are regulated by an lncRNA are usually located very proximal to it on the genome. Unlike miRNAs, they are not highly conserved and their primary sequence has not provided much information about their function. Recently, it has been shown that several lncRNAs maybe spliced at their 5′ and 3′ ends to form circular RNAs (Memczak et al., 2013). However the functional importance of circularization, presumably for increased stability, has not been established.
miRNAs can interact with lncRNA (Jalali et al., 2013) and circular RNAs (Hansen et al., 2013; Memczak et al., 2013) besides their mRNA targets suggesting that in the cell, these ncRNAs may exist as a network of mutually regulating entities. They can possibly serve as sinks that sequester each other temporarily, or even target each other for degradation by the formation of dsRNA.
There are several lines of evidence which suggest that some of these ncRNAs have functional roles in the brain. Regulation by miRNAs is well known in the brain where they regulate neurogenesis and synaptic plasticity. Moreover, mRNAs in brain have extended 3′UTRs, which suggests that the component of regulation by miRNA or other forms of post-transcriptional mechanisms is higher in the brain compared to other tissues (Ramsköld et al., 2009; Miura et al., 2013). Brain also expresses several lncRNAs (Mercer et al., 2008) and is the only non-germline associated somatic tissue presently known to express piRNAs (Lee et al., 2011; Rajasethupathy et al., 2012). In this article we discuss how these three classes of ncRNAs regulate development of nervous system and maintenance of its activities. We primarily emphasize on the role of these ncRNAs as a part of the GRN and how their connectivity determines the functional output. We shed light on the effects of miRNA mediated regulation when they are present in certain types of network motifs.
In network terminology, each interacting entity is called a node and the interactions are called edges. A GRN is a directed network of gene products (including proteins and RNAs), in which certain genes control the expression of other genes. Since the interaction can either activate (positive) or repress (negative) the “target” gene, the edges of a GRN carry a sign along with direction. It is evident that for a given set of nodes several theoretically possible networks can be defined by taking all the combinations of edges from one gene to the other. However, real networks form a small subset of this large number of theoretically possible networks. Network motifs are the patterns of connections that are highly prevalent in real networks compared to what would be expected in a random network with the same set of nodes. This indicates that network motifs were selected over other patterns of connections because of the unique functions that the former can perform as a result of their structure. Network motifs form functional modules in a real network such as GRN: some common network motifs in the GRN include feedback loops (FBL) (autoregulation), feedforward loops (FFL), SIM, multiple input modules and dense overlapping regulons (Alon, 2007).
A gene “X” can regulate a target gene “Y” via multiple intermediates; the sign of the path from X to Y is the product of the signs of all the intermediate edges. If there is a path from a gene back to itself then the resultant network is called a FBL and based on the sign of this path there are two types of FBL: positive and negative, which perform distinct functions (Figures 1B,C).
In a FFL a node targets another node via two parallel branches. If the sign of both the branches are similar then the FFL is called coherent; otherwise incoherent. There are four types each of coherent and incoherent FFLs. Some types of FFL are more commonly observed than the others and are therefore well studied (Figures 1E,F). Like in the case of FBL, each type of FFL is associated with a unique function.
SIM is a network motif in which one node targets at least two nodes (Figure 1D). This is exemplified by one transcription factor or miRNA regulating many genes simultaneously. (Readers are encouraged to refer to Alon, 2007 for details on network motifs in GRN).
Since a gene is a part of a network, its regulation (or the regulation it exerts) cannot be studied in isolation. The study of the entire GRN would be cumbersome and intensive; however, because of the modular nature of the network motifs, they can be studied in isolation and their dynamic effect on the GRN can be predicted. Most of the network motifs discussed in this article are local; these in turn can be a part of a larger motif.
miRNAs and transcription factors are the best studied components of the GRNs that underlie many developmental gene expression programs. Both of them can modulate the expression of multiple targets, alter cell fate and are often engaged in mutually reinforcing functions. However, miRNAs differ from transcription factors in many critical ways (Hobert, 2008). Firstly, almost all the known miRNAs are repressors while transcription factors are either repressors or activators and in some rare cases can act as both depending on the target and interacting partners. Secondly, miRNAs usually bring about downregulation of their targets by a post-transcriptional mechanism, by degrading the target RNA or blocking its translation. Transcription factor interaction with target DNA is largely mediated through structural elements while miRNA interaction with targets is largely governed by the rules of nucleic acid complementarity and are therefore more easily predicted. When a particular gene is targeted by a transcription factor there is usually a single or at most a few tandemly repeated sites present at that locus. However, a typical miRNA-target interaction is characterized by miRNA molecules that have to bind to several messenger RNA molecules. Lastly, transcription factors are usually not consumed in the transcription factor-DNA interactions and may indeed engage in multiple rounds of regulation. The fate of the miRNA engaged in miRNA-target complexes is not understood with similar clarity.
A general principle derived from empirical studies is that, usually, the lifetime of a response is inversely correlated with the response speed. Cell signaling responses, which usually rely on post-translational modifications or conformational changes of proteins, are fast but transient whereas transcriptional or epigenetic responses are long-lived but have a slow response. Post-transcriptional mechanisms, such as miRNA-mediated regulation, fall between these two extremes; faster than transcriptional regulation and relatively stable compared to cell signaling responses. The slow response time of transcriptional repression also results because of the continued presence of stable messenger RNAs since the already existing mRNAs continue to produce proteins. Indeed RNA degradation signals have evolved to ensure rapid turn-over of certain messenger RNAs but they do not allow regulation of the turn-over. miRNAs, can accelerate the response by rapidly clearing these mRNAs along with minimizing the effect of leaky transcription (Hornstein and Shomron, 2006). Therefore miRNA mediated regulation may be preferred over transcriptional regulation in certain situations while, in other situations it may by used along with the latter as a reinforcement.
The miRNAs that play an important role in regulation of the neuronal system act at three levels that in turn correspond to three different developmental time scales. During the earliest stages of development of the nervous system, the numbers of neural stem cells are determined through a wave of neural stem cell proliferation followed by a phase of massive apoptosis (Buss and Oppenheim, 2004). Several miRNAs regulate the development of nervous system by either promoting cell division by repressing pro-apoptotic genes or later promote apoptosis by shutting down pro-survival signals during this early phase of development. miR-29b targets multiple BH3 family of pro-apoptotic genes such as Bim, Bmf, Puma, Bak, and Hrk during early neural differentiation (Figure 2). miR-29b mediated repression of apoptosis in the surviving healthy neurons is essential for neural development (Kole et al., 2011).
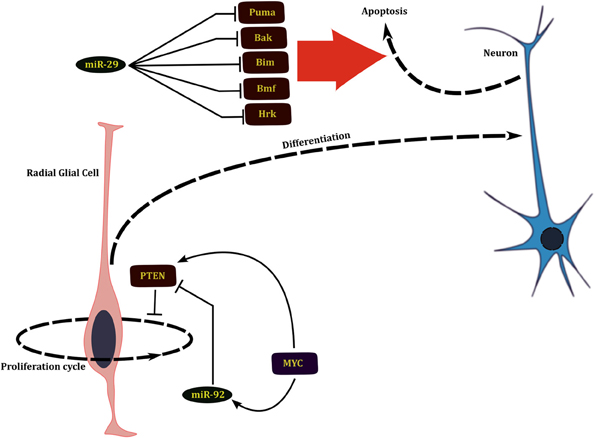
Figure 2. miRNAs affect the development of nervous system by regulating proliferation of neural progenitors and apoptosis of young neurons.
A second set of miRNAs regulate the development of nervous system by either promoting differentiation (developmental function) or allowing the initial expansion of neural progenitors (maintenance function). miR-124 and miR-9 are classical examples of miRNAs associated with developmental functions. miR-124 targets several genes including Poly-Pyrimidine Tract Binding Protein-1 (PTBP1) (Makeyev et al., 2007), Small C-terminal domain Phosphatase 1 (SCP1/CTDSP1) (Visvanathan et al., 2007), Laminin-γ (LAMC1) and Integrin-β1 (ITGB1) (Cao et al., 2007), to promote neuronal differentiation (Figure 3). This type of interaction (also the previously discussed case of miR-29b) forms a network motif called SIM in which a single factor acts on multiple downstream genes. This network architecture allows coordinated regulation of a set of targets that are perhaps simultaneously required to be cleared. Each of these targets in turn may target several genes thus amplifying the scope of regulation. An interesting case is that of PTBP1, a splicing factor, whose downregulation by miR-124 causes a global change in alternative splicing patterns, leading to expression of several neuronal transcript variants. In transcription factor based networks, it has been shown that as the concentration of the transcription factor increases over time following an activating signal, the targets maybe switched on, one after the other in the order of their affinity for the factor (Kalir et al., 2001). Although such an evidence of temporal regulation does not exist for miRNA encoded SIMs, the understanding of the properties of a SIM allows a general extrapolation. When the expression of an miRNA is knocked down partially, the targets with the lowest affinity are likely to be relieved of repression while high affinity targets may continue to be repressed. Attempts to target miRNAs for therapeutic applications have to consider this aspect of miRNA mediated targeting.
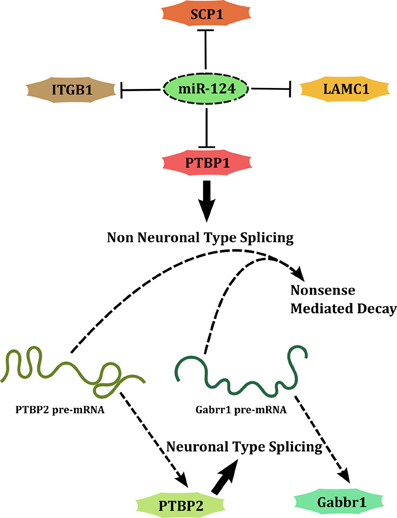
Figure 3. miR-124 promotes neuronal differentiation by acting through a SIM. PTBP1 is downstream of miR-124 and is a part of another SIM.
The products of the miR-9 precursor—miR-9-3p and miR-9-5p—target the transcription factors REST and CoREST, respectively (Packer et al., 2008). In the proliferating progenitors, REST-CoREST, transcriptionally repress all the miR-9 genes, miR-9-1/2/3, and other neuronal genes. This mutual repression between miR-9 and REST-CoREST gives rise to a positive FBL (PFBL). miR-9 is also involved in a PFBL with TLX (Figure 4); a factor that regulates proliferation of neural progenitors (Zhao et al., 2009). It can be observed that developmental functions of miRNAs are associated with network motifs like SIM and PFBL. Multiple targets of a miRNA in a SIM allows simultaneous action on several genes thereby canalizing the cell toward differentiation. PFBLs give rise to bistability; existence of two stable steady states, i.e., differentiated and stem cell state. Examples of PFBL mediated switching behavior is also evident in case of transcriptional regulation; the most famous example would be the switching between lytic and lysogenic phenotypes in λ-phage.
Neural development is also indirectly affected by regulation of self-renewal and expansion of neural progenitors. Phosphatidylinsositide 3-kinase (PI3K)/Akt pathway is involved in growth and self renewal of stem cells whose dysregulation is implicated in several cancers. This pathway is also involved in regulation of neurogenesis; brains of Akt3 knockout mice are greatly reduced in size (Easton et al., 2005). PTEN, which is a negative regulator of this pathway, is in turn regulated by the miR-17–92 cluster of miRNAs and loss of these miRNAs leads to suppression of neural stem cell expansion (Bian et al., 2013). Network analysis by El Baroudi et al. (2011) reveals that both the miR-17–2 cluster and PTEN are positively regulated by MYC, constituting a type 1 incoherent FFL (1I-FFL) (Figure 2). One of the functions that miRNAs perform as a part of 1I-FFL is to buffer transcriptional noise (Osella et al., 2011).
miRNA are also involved in maintenance of neuronal function by regulating synaptic plasticity. Since neurons are highly polarized cells with the nucleus quite distant from the dendritic spines, a local regulatory mechanism is required near the synapses to control protein synthesis at these regions. In other words, a transcriptional regulation in response to synaptic signals would be delayed and therefore miRNA mediated regulation is of great importance in neurons. miR-134 (Schratt et al., 2006) and miR-132 (Wayman et al., 2008; Mellios et al., 2011) are known to regulate synaptic plasticity and the morphology of dendritic spines. miR-134 is also shown to localize at the dendritic spines and repress LIMK1 (Schratt et al., 2006). It is to be noticed that under these conditions, the miRNA mediated regulation doesn't cause transcript degradation but rather, causes a translational repression. RNA binding proteins such as Dnd1 (Kedde et al., 2007) and HuR (Kundu et al., 2012) are reported to reverse miRNA mediated translational repression in germline and liver tissue. Banerjee et al. (2009) have reported that MOV10, a component of RISC, is rapidly degraded via NMDA-receptor mediated signaling, in dendritic spines. This relieves certain mRNAs, including LIMK1 and LYPA1, from miRNA mediated regulation. miRNAs have also been shown to specifically localize in the axons (Sasaki et al., 2013; Hancock et al., 2014); however, the list of axonally enriched miRNAs reported by these two studies are non-overlapping. Hancock et al. have found that miR-132 promotes axonal extension in mouse dorsal root ganglionic (DRG) neurons, by targeting Rasa1. In other studies it has been shown that certain miRNAs like miR-9 (Dajas-Bailador et al., 2012) and miR-138 (Liu et al., 2013) inhibit axonal extension by targeting Map1b and SIRT1, respectively. miRNAs are also implicated in regulation of axon regeneration, post-injury. Injury to sciatic nerve leads to upregulation of miR-21 and miR-431 in the DRG. Also, it was shown that miR-21 and miR-431 promote neurite outgrowth in cultured DRG neurons by targeting Spry2 and Kremen1, respectively (Strickland et al., 2011; Wu and Murashov, 2013a). Taken together, these facts indicate that miRNAs can perform contrasting roles in axonal regulation. Kaplan et al. (2013) and Wu and Murashov (2013b) have extensively reviewed this aspect of miRNA function in the nervous system. Many miRNA are known to co-express with their targets in the neurons suggesting that they might be controlled by a common regulator (Tsang et al., 2007). The fact that miRNAs and their targets are spatiotemporally co-expressed suggests that, in neurons, miRNAs are preferred over transcriptional mechanisms for dynamic gene regulation.
piRNAs are a relatively new class of small non-coding RNAs originally discovered in mouse germline tissues. piRNAs are known to suppress transposable elements in the germline tissues, thereby protecting the germline DNA from deleterious mutations; dysregulation of piRNA pathway results in defects in germ cell proliferation and hence resulting in the loss of fertility. Although most studies on piRNA have been on its role in the germline, a few studies have reported their presence and function in somatic cells (Malone et al., 2009; Lee et al., 2011; Rajasethupathy et al., 2012).
The first evidence of piRNA in nervous system was reported by Lee et al. in which piRNAs in the mouse hippocampus were detected by deep sequencing of small RNA libraries and applying stringent criteria to filter other small RNA sequences including miRNAs (Lee et al., 2011). Some candidate sequences were also found to be associated with MIWI by real-time PCR. Further they showed the presence of the abundant piRNA complexes in the dendritic spines and the knockdown of piRNAs resulted in reduced spine density in the axons.
In another report it was shown that approximately 300 genomic regions encode for piRNAs in the neurons of the Aplysia (Rajasethupathy et al., 2012). It was shown that certain piRNAs are induced by serotonin (5-HT) signaling. Subsequently, using Piwi knockdown studies, it was found that piRNA pathway leads to methylation of CREB2, thereby reducing its expression and promoting memory formation. Even though Rajasethupathy et al. argue that the previous report of piRNA in the brain by Lee et al. is a misclassification and may have arisen because of RNA impurities, a careful analysis from multiple model systems is required to fully comprehend the role of piRNAs in the nervous system.
Despite these reports, the presence of piRNA in the brain is not fully justified. A possible clue about the functions of piRNA in the brain comes from the discovery of the L1 retrotransposons in the brain. These elements have been shown to be regulated during neuronal differentiation and hypothesized to give rise to neuronal heterogeneity and somatic mosacism in brain (Muotri et al., 2005; Coufal et al., 2009). It has been already shown that L1 retrotransposons are regulated by piRNAs in the germline tissues. The presence of both piRNAs and L1 retrotransposons in the brain suggests that the former may regulate the latter in the brain as well.
Recently, it has been shown that the there is extensive transposon expression in the αβ neurons of mushroom bodies in the Drosophila brain, compared to the adjacent neurons (Perrat et al., 2013). In contrast Ago3 and Aub show a low expression in the αβ neurons compared to the rest of the brain. Moreover, mutants of different piRNA pathway components, i.e., Aub, Ago3, and Armi showed elevated levels of transposon expression in the brain. Considering all these observations it can be proposed, though not conclusively stated, that piRNAs are present in the brain for controlling transposons and repeat derived RNAs, and thereby regulating somatic heterogeneity.
lncRNAs have been reported in the brain and known to be associated with specific regions (Mercer et al., 2008). Many of these are found to be specifically expressed during the development of the brain and neural cell differentiation (Ng et al., 2012).
Although it is difficult to classify lncRNAs based on their structural or mechanistic features, lncRNAs are associated with specific roles that they play in the development and functioning of the nervous system therefore allowing a functional classification. There are two functional groups of lncRNAs associated with the development of nervous system—lncRNAs that promote either the self-renewal of neural stem cells or neural differentiation. Another group of lncRNAs are involved in the maintenance of the functioning of nervous system, such as regulation of synaptic activity.
In the previously mentioned study by Ng et al., it was found that three lncRNAs, designated as lncRNA_ES1, lncRNA_ES2, and lncRNA_ES3, are specifically associated with pluripotent stem cells (Ng et al., 2012) lncRNA_ES1 and lncRNA_ES3 had binding sites for the pluripotency associated transcription factors OCT4 and NANOG, and just NANOG respectively; it was further confirmed that knockdown of these proteins reduced the levels of these lncRNAs. The role of these lncRNAs in maintenance of pluripotency was confirmed when their knockdown resulted in reduced percentage of pluripotent cells. These lncRNA_ES1/2 were found to be associated with the PRC2 component SUZ12, suggesting that they indirectly regulate pro-differentiation genes by repressing their transcription.
In the same study four lncRNAs- RMST, lncRNA_N1, lncRNA_N2, and lncRNA_N3, were identified to be associated with neuronal differentiation and their knockdown resulted in reduction in number of neurons in culture along with a reduced expression of neuronal markers and increased expression of glial markers. Out of these, lncRNA_N2 was found to harbor neurogenesis associated miRNAs- miR-125b and let-7 in its intronic regions. In a subsequent study by the same group it was shown that RMST is transcriptionally repressed by REST (Figure 4). Also, it was shown that RMST associates with SOX2 and tethers it to promoters of neurogenetic genes like DLX1, NEUROG2 and ASCL1, thereby inducing their expression (Ng et al., 2013). DLX1 locus also encodes an antisense-lncRNA, DLX1AS which is expressed during neurogenesis in the subventricular zone of hippocampus, and positively regulates the expression of DLX1 and DLX2 (Ramos et al., 2013).
lncRNAs that are involved in maintenance activities regulate the general functioning of neurons and control processes such as synaptic signaling. BC1 which is a cytoplasmic lncRNA, localizes to dendrites and is involved in regulating the post-synaptic signaling, by repressing metabotropic glutamate receptor signaling (mGluR) induced local protein synthesis. BC1 represses translation initiation by interacting with eIF4A and poly-A binding protein (PABP), thereby disallowing the recruitment of the small ribosomal subunit to the mRNA (Wang et al., 2005). Loss of BC1 results in hyperexcitation of the neurons (Zhong et al., 2009) and is implicated in the mouse models of epilepsy (Gitaí et al., 2011). One of the genes induced by mGluR is Fragile-X mental retardation protein (FMRP), which is significantly upregulated in BC1 knockouts (Zhong et al., 2009). However, FMRP is also involved in controlling hyperexcitability of neurons by regulating protein synthesis and it has been shown that double knockout of FMRP and BC1 results in a more severe epileptic phenotype (Zhong et al., 2010) This suggests that FMRP supplements BC1 and may rescue any imbalance caused by fluctuations in BC1 activity.
BACE1 is a membrane protease which is implicated in Alzheimer's Disease by promoting cleavage of Amyloid Precursor Protein (APP) to form Amyloid-Beta 1-42 (Abeta 1-42) peptide. BACE1-Anti Sense (BACE1AS) RNA is partially complementary to BACE1 coding region (CDS) and renders stability to the mRNA, thereby leading to upregulation of Abeta 1-42. BACE1AS is in turn upregulated because of the cellular stress due to Abeta 1-42, thereby further increasing Abeta 1-42 via BACE1. This results in a PFBL (Faghihi et al., 2008), however, in this study the authors have called it a feedforward regulation. It had been subsequently shown that BACE1AS stabilizes BACE1 mRNA by masking a non-canonical target site for miR-485-5p in the CDS of BACE1 mRNA (Faghihi et al., 2010).
In another contrasting case of antisense lncRNA mediated regulation, BDNF-AS represses Brain Derived Neurotrophic Factor (BDNF) and results in restriction of neurite growth. Also, this regulation occurs at the level of transcription where BDNF-AS helps in recruiting EZH2, which is a part of Polycomb Repressive Complex 2 (PRC2) and marks histones with repressive lysine methylation- H3k4me3 (Modarresi et al., 2012).
There are many other lncRNAs implicated in the development and function of nervous system but the targets and mechanisms are unknown for a majority of them; Table 1 summarizes these different lncRNAs. In many cases lncRNAs emerge from the same genomic locus as their targets; this targeting is based on simple base pairing can happen at the level of either DNA or RNA. Some lncRNAs such as RMST act on several genes by tethering transcription factors to their promoters or serving as a scaffold for the assembly of RNA-binding proteins.
Recent studies have revealed the importance of RNA mediated gene regulation in diverse biological processes. There are some advantages of having ncRNAs as gene expression regulators compared to an exclusively protein-regulated GRN. Since there is no step of translation, the cost as well the time required for making an ncRNA would be less compared to that of a protein. ncRNAs can target genes based on simple base pairing interactions; therefore evolution of such a regulator has a higher likelihood.
miRNA based regulation has a special importance in the highly polarized neurons as they control local translation at the dendrites thereby preventing the delay that can arise in transcriptional responses because of RNA synthesis and transport to distant cellular regions. Even though the exact importance of piRNAs in the brain is not understood it is highly likely that they regulate retrotransposons. Unlike miRNAs and piRNAs that are mostly repressors, lncRNAs can either activate or repress a gene. Since the dsDNA has limited ability to adopt distinctive structures, the DNA binding specificity of transcription factors and other DNA binding proteins relies on a limited repertoire of DNA binding domains. Long RNAs on the other hand are structured and proteins can bind to specific structures more effectively. The sequence information in the RNA can provide additional specificity to the binding. Thus lncRNAs may efficiently bridge the interactions between protein and DNA. lncRNAs can also mark large genomic regions for epigenetic regulation as in the case of XIST. It has also been argued that it is the act of transcription and not the product per-se, that causes the regulation (Berretta and Morillon, 2009).
Although these regulatory RNAs act via different mechanisms, usually their roles are convergent with that of the protein based transcriptional regulation to ensure an efficient and foolproof control of gene expression. As evident in many studies, blocking the activity of these regulatory RNAs affects the gene regulation to different extents. Therefore it is undeniable that ncRNAs supplement the gene regulation by proteins and are not merely redundant pathways. A systems-level analysis of ncRNAs is essential to understand their precise roles and the ability to confer robustness to the GRN.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Bharat R. Iyengar and Ashwani Choudhary acknowledge the Council of Scientific and Industrial Research (CSIR), India and Mayuresh A. Sarangdhar acknowledges the University Grants Commission (UGC), India for research fellowships. The authors acknowledge CSIR-Institute of Genomics and Integrative Biology projects BSC0123 and BSC0124 for funding and for approving the manuscript for publication.
Alon, U. (2007). Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461. doi: 10.1038/nrg2102
Aravin, A. A., Sachidanandam, R., Bourc'his, D., Schaefer, C., Pezic, D., Toth, K. F., et al. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799. doi: 10.1016/j.molcel.2008.09.003
Banerjee, S., Neveu, P., and Kosik, K. S. (2009). A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 64, 871–884. doi: 10.1016/j.neuron.2009.11.023
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. doi: 10.1038/emboj.2010.199
Berretta, J., and Morillon, A. (2009). Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 10, 973–982. doi: 10.1038/embor.2009.181
Bian, S., Hong, J., Li, Q., Schebelle, L., Pollock, A., Knauss, J. L., et al. (2013). MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 3, 1398–1406. doi: 10.1016/j.celrep.2013.03.037
Bond, A. M., Vangompel, M. J., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA Is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027. doi: 10.1038/nn.2371
Buss, R. R., and Oppenheim, R. W. (2004). Role of programmed cell death in normal neuronal development and function. Anat. Sci. Int. 79, 191–197. doi: 10.1111/j.1447-073x.2004.00088.x
Cao, X., Pfaff, S. L., and Gage, F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536. doi: 10.1101/gad.1519207
Chi, S. W., Zang, J. B., Mele, A., and Darnell, R. B. (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486. doi: 10.1038/nature08170
Clark, M. B., Amaral, P. P., Schlesinger, F. J., Dinger, M. E., Taft, R. J., Rinn, J. L., et al. (2011). The reality of pervasive transcription. PLoS Biol. 9:e1000625. doi: 10.1371/journal.pbio.1000625
Coufal, N. G., Garcia-Perez, J. L., Peng, G. E., Yeo, G. W., Mu, Y., Lovci, M. T., et al. (2009). L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131. doi: 10.1038/nature08248
Dajas-Bailador, F., Bonev, B., Garcez, P., Stanley, P., Guillemot, F., and Papalopulu, N. (2012). microRNA-9 regulates axon extension and branching by targeting map1b in mouse cortical neurons. Nat. Neurosci. 15, 697–699. doi: 10.1038/nn.3082
Easton, R. M., Cho, H., Roovers, K., Shineman, D. W., Mizrahi, M., Forman, M. S., et al. (2005). Role for Akt3/protein kinase Bγ in attainment of normal brain size. Mol. Cell. Biol. 25, 1869–1878. doi: 10.1128/MCB.25.5.1869-1878.2005
El Baroudi, M., Corà, D., Bosia, C., Osella, M., and Caselle, M. (2011). A curated database of miRNA mediated feed-forward loops involving MYC as master regulator. PLoS ONE 6:e14742. doi: 10.1371/journal.pone.0014742
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of [beta]-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
Faghihi, M. A., Zhang, M., Huang, J., Modarresi, F., Van der Brug, M. P., Nalls, M. A., et al. (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 11, R56. doi: 10.1038/nm1784
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114. doi: 10.1038/nrg2290
Gitaí, D. L., Fachin, A. L., Mello, S. S., Elias, C. F., Bittencourt, J. C., Leite, J. P., et al. (2011). The Non-coding RNA BC1 Is down-regulated in the hippocampus of Wistar Audiogenic Rat (WAR) strain after audiogenic kindling. Brain Res. 1367, 114–121. doi: 10.1016/j.brainres.2010.10.069
Hancock, M. L., Preitner, N., Quan, J., and Flanagan, J. G. (2014). MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J. Neurosci. 34, 66–78. doi: 10.1523/JNEUROSCI.3371-13.2014
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Helwak, A., Kudla, G., Dudnakova, T., and Tollervey, D. (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665. doi: 10.1016/j.cell.2013.03.043
Hobert, O. (2008). Gene regulation by transcription factors and microRNAs. Science 319, 1785–1786. doi: 10.1126/science.1151651
Hornstein, E., and Shomron, N. (2006). Canalization of development by microRNAs. Nat. Genet. 38, S20–S24. doi: 10.1038/ng1803
Ishizu, H., Siomi, H., and Siomi, M. C. (2012). Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 26, 2361–2373. doi: 10.1101/gad.203786.112
Jacquier, A. (2009). The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 10, 833–844. doi: 10.1038/nrg2683
Jalali, S., Bhartiya, D., Lalwani, M. K., Sivasubbu, S., and Scaria, V. (2013). Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS ONE 8:e53823. doi: 10.1371/journal.pone.0053823
Kalir, S., McClure, J., Pabbaraju, K., Southward, C., Ronen, M., Leibler, S., et al. (2001). Ordering Genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292, 2080–2083. doi: 10.1126/science.1058758
Kaplan, B. B., Kar, A. N., Gioio, A., and Aschrafi, A. (2013). MicroRNAs in the axon and presynaptic nerve terminal. Front. Cell. Neurosci. 7:126. doi: 10.3389/fncel.2013.00126
Kedde, M., Strasser, M. J., Boldajipour, B., Oude Vrielink, J. A. F., Slanchev, K., le Sage, C., et al. (2007). RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273–1286. doi: 10.1016/j.cell.2007.11.034
Kole, A. J., Swahari, V., Hammond, S. M., and Deshmukh, M. (2011). miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 25, 125–130. doi: 10.1101/gad.1975411
Korneev, S. A., Korneeva, E. I., Lagarkova, M. A., Kiselev, S. L., Critchley, G., and O'Shea, M. (2008). Novel noncoding antisense RNA transcribed from human anti-NOS2A locus is differentially regulated during neuronal differentiation of embryonic stem cells. RNA 14, 2030–2037. doi: 10.1261/rna.1084308
Kundu, P., Fabian, M. R., Sonenberg, N., Bhattacharyya, S. N., and Filipowicz, W. (2012). HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 40, 5088–5100. doi: 10.1093/nar/gks148
Lee, E. J., Banerjee, S., Zhou, H., Jammalamadaka, A., Arcila, M., Manjunath, B. S., et al. (2011). Identification of piRNAs in the central nervous system. RNA 17, 1090–1099. doi: 10.1261/rna.2565011
Lin, M., Pedrosa, E., Shah, A., Hrabovsky, A., Maqbool, S., Zheng, D., et al. (2011). RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE 6:e23356. doi: 10.1371/journal.pone.0023356
Liu, C.-M., Wang, R.-Y., Saijilafu, Jiao, Z.-X., Zhang, B.-Y., and Zhou, F.-Q. (2013). MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev. 27, 1473–1483. doi: 10.1101/gad.209619.112
Liu, Q., and Paroo, Z. (2010). Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 79, 295–319. doi: 10.1146/annurev.biochem.052208.151733
Makeyev, E. V., Zhang, J., Carrasco, M. A., and Maniatis, T. (2007). The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448. doi: 10.1016/j.molcel.2007.07.015
Malone, C. D., Brennecke, J., Dus, M., Stark, A., McCombie, W. R., Sachidanandam, R., et al. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137, 522–535. doi: 10.1016/j.cell.2009.03.040
Mellios, N., Sugihara, H., Castro, J., Banerjee, A., Le, C., Kumar, A., et al. (2011). miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat. Neurosci. 14, 1240–1242. doi: 10.1038/nn.2909
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Mercer, T., Qureshi, I., Gokhan, S., Dinger, M., Li, G., Mattick, J., et al. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11:14. doi: 10.1186/1471-2202-11-14
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. doi: 10.1038/nrg2521
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105, 716–721. doi: 10.1073/pnas.0706729105
Miura, P., Shenker, S., Andreu-Agullo, C., Westholm, J. O., and Lai, E. C. (2013). Widespread and extensive lengthening of 3' UTRs in the mammalian brain. Genome Res. 23, 812–825. doi: 10.1101/gr.146886.112
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459. doi: 10.1038/nbt.2158
Muotri, A. R., Chu, V. T., Marchetto, M. C. N., Deng, W., Moran, J. V., and Gage, F. H. (2005). Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910. doi:10.1038/nature03663
Ng, S.-Y., Bogu, G. K., Soh, B. S., and Stanton, L. W. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. doi:10.1016/j.molcel.2013.07.017
Ng, S.-Y., Johnson, R., and Stanton, L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. doi: 10.1038/emboj.2011.459
Osella, M., Bosia, C., Corá, D., and Caselle, M. (2011). The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput. Biol. 7:e1001101. doi: 10.1371/journal.pcbi.1001101
Packer, A. N., Xing, Y., Harper, S. Q., Jones, L., and Davidson, B. L. (2008). The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci. 28, 14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008
Perrat, P. N., DasGupta, S., Wang, J., Theurkauf, W., Weng, Z., Rosbash, M., et al. (2013). Transposition-driven genomic heterogeneity in the Drosophila brain. Science 340, 91–95. doi: 10.1126/science.1231965
Pollard, K. S., Salama, S. R., Lambert, N., Lambot, M.-A., Coppens, S., Pedersen, J. S., et al. (2006). An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443, 167–172. doi: 10.1038/nature05113
Qureshi, I. A., Mattick, J. S., and Mehler, M. F. (2010). Long non-coding RNAs in nervous system function and disease. Brain Res. 1338, 20–35. doi:10.1016/j.brainres.2010.03.110
Rajasethupathy, P., Antonov, I., Sheridan, R., Frey, S., Sander, C., Tuschl, T., et al. (2012). A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 149, 693–707. doi: 10.1016/j.cell.2012.02.057
Ramos, A. D., Diaz, A., Nellore, A., Delgado, R. N., Park, K.-Y., Gonzales-Roybal, G., et al. (2013). Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12, 616–628. doi: 10.1016/j.stem.2013.03.003
Ramsköld, D., Wang, E. T., Burge, C. B., and Sandberg, R. (2009). An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 5:e1000598. doi: 10.1371/journal.pcbi.1000598
Rapicavoli, N., Poth, E., Zhu, H., and Blackshaw, S. (2011). The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 6:32. doi: 10.1186/1749-8104-6-32
Sasaki, Y., Gross, C., Xing, L., Goshima, Y., and Bassell, G. J. (2013). Identification of axon-enriched microRNAs localized to growth cones of cortical neurons. Dev. Neurobiol. doi: 10.1002/dneu.22113. [Epub ahead of print].
Schratt, G. M., Tuebing, F., Nigh, E. A., Kane, C. G., Sabatini, M. E., Kiebler, M., et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289. doi: 10.1038/nature04367
Strickland, I. T., Richards, L., Holmes, F. E., Wynick, D., Uney, J. B., and Wong, L.-F. (2011). Axotomy-induced miR-21 promotes axon growth in adult dorsal root ganglion neurons. PLoS ONE 6:e23423. doi: 10.1371/journal.pone.0023423
Tochitani, S., and Hayashizaki, Y. (2008). Nkx2.2 antisense RNA overexpression enhanced oligodendrocytic differentiation. Biochem. Biophys. Res. Commun. 372, 691–696. doi: 10.1016/j.bbrc.2008.05.127
Tsang, J., Zhu, J., and van Oudenaarden, A. (2007). MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 26, 753–767. doi:10.1016/j.molcel.2007.05.018
Tsuiji, H., Yoshimoto, R., Hasegawa, Y., Furuno, M., Yoshida, M., and Nakagawa, S. (2011). Competition between a noncoding exon and introns: gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 16, 479–490. doi: 10.1111/j.1365-2443.2011.01502.x
Visvanathan, J., Lee, S., Lee, B., Lee, J. W., and Lee, S.-K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749. doi: 10.1101/gad.1519107
Wang, H., Iacoangeli, A., Lin, D., Williams, K., Denman, R. B., Hellen, C. U. T., et al. (2005). Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 171, 811–821. doi: 10.1083/jcb.200506006
Watanabe, T., Tomizawa, S.-I., Mitsuya, K., Totoki, Y., Yamamoto, Y., Kuramochi-Miyagawa, S., et al. (2011). Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse rasgrf1 locus. Science 332, 848–852. doi: 10.1126/science.1203919
Wayman, G. A., Davare, M., Ando, H., Fortin, D., Varlamova, O., Cheng, H.-Y. M., et al. (2008). An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. U.S.A. 105, 9093–9098. doi: 10.1073/pnas.0803072105
Winter, J., Jung, S., Keller, S., Gregory, R. I., and Diederichs, S. (2009). Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11, 228–234. doi: 10.1038/ncb0309-228
Wu, D., and Murashov, A. K. (2013a). MicroRNA-431 regulates axon regeneration in mature sensory neurons by targeting the Wnt antagonist kremen1. Front. Mol. Neurosci. 6:35. doi:10.3389/fnmol.2013.00035
Wu, D., and Murashov, A. K. (2013b). Molecular mechanisms of peripheral nerve regeneration: emerging roles of microRNAs. Front. Physiol. 4:55. doi: 10.3389/fphys.2013.00055
Young, T. L., Matsuda, T., and Cepko, C. L. (2005). The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 15, 501–512. doi: 10.1016/j.cub.2005.02.027
Zhao, C., Sun, G., Li, S., and Shi, Y. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16, 365–371. doi: 10.1038/nsmb.1576
Zhong, J., Chuang, S.-C., Bianchi, R., Zhao, W., Lee, H., Fenton, A. A., et al. (2009). BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 29, 9977–9986. doi: 10.1523/JNEUROSCI.3893-08.2009
Keywords: miRNA, piRNA, lncRNA, network-motifs, gene expression regulation
Citation: Iyengar BR, Choudhary A, Sarangdhar MA, Venkatesh KV, Gadgil CJ and Pillai B (2014) Non-coding RNA interact to regulate neuronal development and function. Front. Cell. Neurosci. 8:47. doi: 10.3389/fncel.2014.00047
Received: 03 December 2013; Paper pending published: 09 January 2014;
Accepted: 03 February 2014; Published online: 24 February 2014.
Edited by:
Alessandro Cellerino, Scuola Normale Superiore, ItalyReviewed by:
Alexander K. Murashov, East Carolina University, USACopyright © 2014 Iyengar, Choudhary, Sarangdhar, Venkatesh, Gadgil and Pillai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beena Pillai, Functional Genomics, CSIR-Institute of Genomics and Integrative Biology, Mathura Road, Delhi 110020, India e-mail:YmVlbmFAaWdpYi5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.