- 1Department of Pediatric Rehabilitation Medicine, Kunming Children’s Hospital, Kunming, China
- 2State Key Laboratory of Primate Biomedical Research, Institute of Primate Translational Medicine, Kunming University of Science and Technology, Kunming, China
- 3School of Basic Medical Sciences, Yunnan University of Chinese Medicine, Kunming, China
Autism Spectrum Disorder (ASD) is a heterogeneous neurodevelopmental disorder with deficient social skills, communication deficits and repetitive behaviors. The prevalence of ASD has increased among children in recent years. Children with ASD experience more sleep problems, and sleep appears to be essential for the survival and integrity of most living organisms, especially for typical synaptic development and brain plasticity. Many methods have been used to assess sleep problems over past decades such as sleep diaries and parent-reported questionnaires, electroencephalography, actigraphy and videosomnography. A substantial number of rodent and non-human primate models of ASD have been generated. Many of these animal models exhibited sleep disorders at an early age. The aim of this review is to examine and discuss sleep disorders in children with ASD. Toward this aim, we evaluated the prevalence, clinical characteristics, phenotypic analyses, and pathophysiological brain mechanisms of ASD. We highlight the current state of animal models for ASD and explore their implications and prospects for investigating sleep disorders associated with ASD.
Introduction
Sleep appears to be essential for most living organisms’ survival and integrity, especially for typical synaptic development and brain plasticity (Fogel et al., 2012; Hartsock and Spencer, 2020). Over the past decades, the role of sleep in learning and memory has been probed by many studies at behavioral, systemic, cellular, and molecular levels (Walker and Stickgold, 2006; Rawashdeh et al., 2007; Sawangjit et al., 2018; Kim et al., 2019). The American Academy of Sleep Medicine (AASM) has recently released the third edition of the International Classification of Sleep Disorders (ICSD-3) in 2014. This guideline grouped sleep disorders into seven basic types: insomnia disorders, central disorders of hypersomnolence, circadian rhythm sleep-wake disorders, sleep-disordered breathing, movement disorders, parasomnias, and other sleep disorders (Sateia, 2014; Ito and Inoue, 2015). There are universal physiologic changes during sleep, and some biologic, environmental, psychological, social as well as genetic factors can affect change in the sleep pattern (Jenni and O’Connor, 2005; Bathory and Tomopoulos, 2017). The sleep-wake circadian rhythm is regulated through both circadian and homeostatic processes. Arousal and sleep are active and involved in neurophysiologic processes, including both activation and suppression of neural pathways. Sleep disorders can be an early symptom of the disease, and the presence of rapid eye movement (REM) sleep behavior disorder (RBD) can be used as an early diagnostic indicator for neurodegenerative diseases (Galland et al., 2012; Kotterba, 2015). Besides, sleep disorders have observable effects on physical and mental health of children with autism spectrum disorder (ASD) and their parents (Zhang et al., 2021).
Autism spectrum disorder is a neurodevelopmental disorder and the prevalence of ASD is increasing, with 1 in 59 children in the United States. diagnosed with ASD (Orefice, 2019). ASD is approximately four times more prevalent among males than females (Christensen et al., 2016; Satterstrom et al., 2020). According to the DSM-V, the previous categories of pervasive developmental disorders, pervasive developmental disorder-not otherwise specified (PDD-NOS) and Asperger disorder were combined into ASD. The new diagnostic criteria from DSM-V defined ASD as a heterogeneous spectrum disorder with deficits in social interaction and communication, restricted and repetitive interests, and stereotyped behaviors (American Psychiatric Association, 2013; Devnani and Hegde, 2015).
It is not surprising when considering the numerous health and behavioral issues that sleep disturbance are commonly observed in the clinical progression of ASD. Children with ASD experience more sleep problems compared with the general population, particularly insomnia. Sleep-wake cycle abnormalities are associated with communication deficits, stereotyped behaviors, and autism severity (Tudor et al., 2012). Disrupted sleep may exacerbate the daily dysfunction of ASD children, such as social and communication skills, behavioral performance, stereotypical behaviors, and motor output on non-verbal performance tasks (Schreck et al., 2004; Limoges et al., 2013).
As we gain deeper knowledge of the neural mechanisms of ASD and sleep, more contributions from sleep-related biomarkers to the study of neurophysiology in ASD. Prospectively, the emergence of digital technologies and devices is making studies of sleep physiology more flexible and convenient. The sleep study provided new insights for research on the children with ASD when compared with the other behavioral tests currently used in human subjects and animal experimental models. This review’s main objective is to explore animal models’ role, especially non-human primate (NHP) models, as a useful tool to investigate sleep disorders in ASD children. Firstly, we present data on sleep disorders in autistic children, emphasizing their prevalence, clinical characteristics, phenotypic analyses, and pathophysiological mechanisms. Next, we highlight the current state of animal models for ASD and explore their implications and future prospects in translational research. We suggest that using NHP animal models may provide insights into sleep disorders in ASD.
The Role of Sleep
In most mammalian species, sleep amounts are highest during the neonatal period (Weber and Dan, 2016). Sleep loss can significantly affect a child’s health-related quality and activities of daily living (Maski and Kothare, 2013). The brain is one of the organs most impacted by sleep or the lack thereof while adolescence is a critical period for brain reorganization. It is beyond doubt that sleep disorders during this period exert irreversible effects on children’s brain development (Roffwarg et al., 1966; Klein et al., 2000; Weber et al., 2018). REM sleep can prune newly formed postsynaptic dendritic spines during motor learning (Li et al., 2017), and the balance of newly formed and original dendritic spines is crucial for neuronal circuit development and behavioral improvement in children. Two studies found that sleep enhance cortical plasticity in the visual cortex during the developmental critical period (Frank et al., 2001; Aton et al., 2009). In conclusion, sleep seems to be important for brain development, learning, and memory consolidation by selectively eliminating and maintaining newly formed synapses (Li et al., 2017).
Sleep deprivation may cause physical diseases and developmental problems. During early life, sleep deprivation has been shown to have long-term implications for social behaviors in adulthood (Hudson et al., 2020). Neural substrates can be affected by sleep deprivation, including the prefrontal cortex, basal ganglia, and amygdala. Furthermore, sleep deprivation may cause difficulties in executive functioning, reward learning as well as emotional reactivity. Such issues may contribute to difficulties in judgment, resolution of problems, challenging behaviors, emotional control, and public health concerns, such as depression, suicide, and risk-taking behavior (Maski and Kothare, 2013). These findings indicate that insufficient sleep during early life has persistent effects on brain development and later behavioral performance.
It has been assumed that sleep can clear out brain’s toxins, such as beta-amyloid which was associated with Alzheimer’s disease (Xie et al., 2013). Sleep is essential for maintaining the body’s physical health and is associated with neurodegeneration, metabolic diseases, cancer, and aging. The processes of growth and development are related to sleep quality. The abnormal sleep and circadian also affect hormones and metabolism (Li et al., 2018a). Getting adequate sleep can help the immune system to better react against infection (Grigg-Damberger, 2009; Herculano-Houzel, 2013; Welberg, 2013).
Clinical Characteristics of Sleep Disorders in ASD Children
Many neurodevelopmental processes have been reported in the children with ASD, such as synaptic plasticity, neurogenesis and migration of neuron (Gilbert and Man, 2017). About 40–80% of children with ASD exhibit at least one sleep-related problems (Verhoeff et al., 2018), including irregular sleeping and waking patterns, decreased sleep efficiency, reductions in total sleep time and REM sleep time, sleep onset delays, decreased sleep efficiency, increased wakening after sleep onset, bedtime resistance, and daytime sleepiness (Humphreys et al., 2014). Studies utilizing Actigraphy (ACT) and Polysomnogram (PSG) have found that increased sleep latency, and decreased sleep duration and sleep efficiency in ASD children (Elrod and Hood, 2015). A comprehensive review in children with ASD reported that insomnia is one of the most common sleep problems (Souders et al., 2009). Another study also documented that the predominant sleep disorder included insomnia, difficulty falling, and staying asleep (Malow et al., 2006). Mutluer et al. (2016) found that the most common symptoms reported were troubles falling asleep, sleep after waking up and tired after sleeping.
Prevalence of Sleep Problems in Children With ASD
Childhood sleep disorders which are mostly reported by parents are associated with emotional, cognitive, and behavioral disturbances. Sleep disturbances occur in approximately 20–30% of preschool children, including bedtime resistance, sleep onset delays, night terrors or nightmares, and repetitive rhythmic behaviors (Lozoff et al., 1985; Krakowiak et al., 2008; Knappe et al., 2020). The abnormalities of ASD may predispose children to various threaten of sleep and make them especially susceptible to sleep problems (Morgenthaler et al., 2007; Maxwell-Horn and Malow, 2017). Sleep problems have become one of the most common symptoms among ASD children (Richdale, 1999; Wiggs, 2001; Liu et al., 2006; Uren et al., 2019). Two studies compared sleep behaviors of ASD with typically developing (TD) children, they found that 66% of ASD children exhibited moderate sleep disturbances (Souders et al., 2009) and 71% in another study (Malow et al., 2016). A parent-reported study found that 35% of ASD children had at least one sleep dysfunction (Krakowiak et al., 2008). The risk of sleep disturbance is 2.8-fold higher in children with ASD (Köse et al., 2017). A recent study repeated sleep measures at different age in 5,151 children, and found that ASD children have an increase in sleep problems with age, whereas TD children decrease (Verhoeff et al., 2018).
Phenotype Analyses for Sleep Disorders
Over the past years, many different sleep analysis methods have been reported (Lomeli et al., 2008; Kelly et al., 2012; Ibáñez et al., 2018; de Zambotti et al., 2019). Infection, pain as well as trauma can disrupt sleep and activity (Vandekerckhove and Cluydts, 2010; Doufas et al., 2012), even some issues that might seem minor to us are often very significant to a child. As children progress from infancy to adolescence, sleep structure, sleep behavior and sleep duration will also change (Williams et al., 2013), it is crucial to take into account the specificity of different ages of children when investigating sleep states. Some sleep studies require an intimate contact of the electrode with the skin and even require surgical implantation of electrodes, which are difficult to apply in freely moving animals and humans, particularly in children related to the lively side of their nature. Even though several new technological developments have been brought to reduce inconvenience, pain, and further damage of these methods, expensive and burdensome must also be considered, especially for long-term studies that include large samples. Some assessments were developed to monitor sleep through observation of body motion and posture. These methods could obviate the need for direct contact and even avoid surgery or electrode implantation. It is non-invasive and low cost. Nevertheless, the behavioral observation does not provide sufficient information compared to those provided by electroencephalogram (EEG) and electromyogram (EMG). In general, both humans’ and animals’ sleep analyses include sleep patterns, locomotor activity, temperature, and food intake. The current study summarized phenotype analyses for sleep disorders obtained from sleep diaries, parent-reported questionnaires, electroencephalography, actigraphy, and videosomnography. We summarized the main types of experimental approaches applicable to assessment methods of sleep studies and all of these methods have advantages and disadvantages (Figure 1). The selection of clinical sleep assessment should be tailored to children’s unique characteristics, and safety and feasibility must also be taken into consideration.
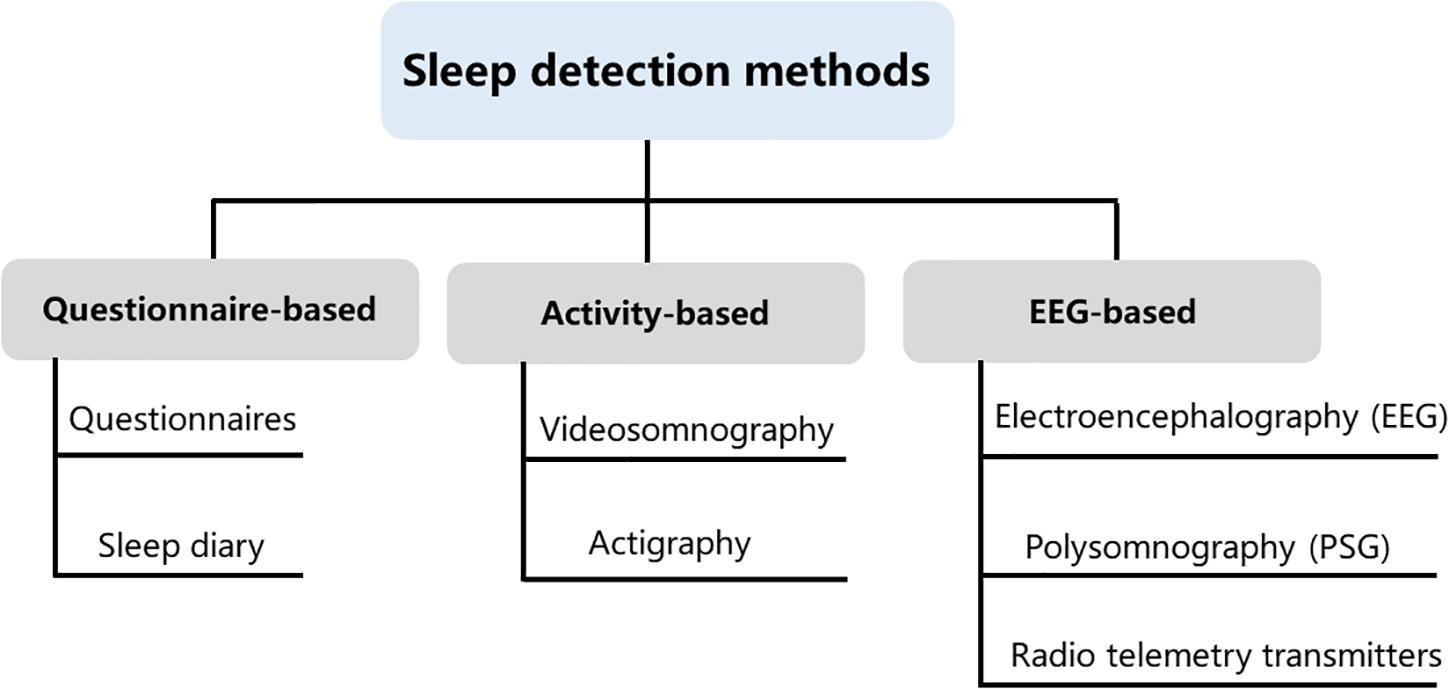
Figure 1. Sleep detection methods. Current methods available for measuring sleep in young children include questionnaire-based, activity-based, and electroencephalogram (EEG)-based methods. These three types of assessments are not interchangeable, as each method contains its own idiosyncrasies that can influence the quality and meaning of the data that are collected.
Sleep Diaries and Parent-Reported Questionnaires
Subjective measures including sleep diaries and parent-reported questionnaires are the most common analyses in human studies. They have several advantages such as the non-invasive ease of acquisition and low cost. Parents are usually quick to recognize any changes in their child’s behavior and mood, and these observations should be recorded (Bhargava, 2011). One of the most common parent-reported questionnaires is Children’s Sleep Habits Questionnaire (CSHQ), a parent-report sleep screening instrument designed for school-aged children. The CSHQ score has eight measures, evaluating the behavioral and psychological symptoms of sleep disorders in children (Owens et al., 2000). Many factors affect the reliability of sleep analysis. These factors include the aspect of sleep assessed, the period of sleep aggregated, and the sample population and so forth (Acebo et al., 1999; Camerota et al., 2018). Most retrospective studies on sleep are reported by proxies, such data are limited by problems related to recall bias and subject selection bias (Short et al., 2017). When parents have to estimate the sleep habits of their children, it has been shown that parents tended to estimate with more accurate sleep schedule variables than time awake in bed (Werner et al., 2008). Moreover, the consistency of their reports decreased when the monitoring lasted a long time (Short et al., 2013). Although biases are introduced when utilizing these methods, sleep diaries, and parent reports are commonly used in monitoring children with sleep problems because of their low cost and ease of administration (Honomichl et al., 2002).
Electroencephalography
The influence of technology advances becomes increasingly evident in the study of neuroscience. EEG can provide the temporal and spatial characteristics of subject. A sleep EEG is a recording of the electrical activity of the brain while you are awake and then asleep. It involves having small disks (electrodes) which record the activity attached to the subject’s scalp (Feinberg et al., 1967; Weber and Dan, 2016). Compared with a single-channel EEG, one technique named polysomnogram (PSG) is considered as the gold standard to objectively assess sleep (O’Donnell et al., 2018). PSG can be used in a diverse range of monitoring, such as brain electrical activity, muscle activity, eye movements, respiratory rate and other channels relying on experimental design (Boulos et al., 2019). It integrates both normal and abnormal physiological indicators of brain electrical activity, sleeps architecture, sleep stages, quality of sleep, eye movements, and physical activities during the sleep period. Wakefulness, NREM sleep, and REM sleep can be clearly distinguished making sleep a directly quantifiable behavior, which could be introduced more easily into clinical routine and less stressful for patients (Blume et al., 2015). The main drawback of PSG is the need of electrodes attached to the skin surface, and not convenient to use in clinical sleep monitoring for children (Lucey et al., 2016). The children cannot be sedated by given medicine such as tranquilizers or sleep aids during the PSG sleep study, and thus doctors may use a blanket or papoose board to keep the child from rolling around on the bed or pulling on the wires. However, this issue may restrict children’s normal sleep as we don’t expect. And also, PSG instruments are bulky and expensive and may be difficult to monitor changes in patients for long-term studies (Stepnowsky et al., 2013; Qin et al., 2020). Recently, telemetry transmitters have been used for long-term measuring of EEG and electromyography signals in rodent and NHP animals, it could collect data from conscious, freely moving laboratory animals without skin-electrode contact impedance and reduce animals’ stress (Ishikawa et al., 2017; Qiu et al., 2019). This strategy can be potentially applied for future clinical applications.
Actigraphy
Actigraphy is a non-invasive method that measures limb movement by a watch-size accelerometer to determine sleep and wake episodes. It allows for multiple-day data collection in natural environments. One study compared the validity of actigraphy and PSG, found that intraclass correlations between PSG and actigraphy variables were strong (>0.80) for sleep latency, sleep duration, and sleep efficiency (Bélanger et al., 2013). Nevertheless, lack of correspondence of circadian sleep-wake cycles between actigraphy and PSG was confirmed in school-age children (Meltzer et al., 2016). Actigraphy assessments may severely underestimate the true sleep statements in children with significantly elevated sleep disorders (Sadeh, 2011).
The next generation multisensory consumer sleep trackers are different from the first motion-based generation of consumer wearables (actigraphy). New generation sleep trackers apply algorithms to achieve functions approximately similar to PSG. Fitbit (Montgomery-Downs et al., 2012; Meltzer et al., 2015; de Zambotti et al., 2016; Mantua et al., 2016; Cook et al., 2017; de Zambotti et al., 2018) and Jawbone (de Zambotti et al., 2015; Toon et al., 2016; Cook et al., 2018) sleep trackers are most frequently tested wearables and their performance has always been compared with PSG. Boe et al. (2019) recently presented a wireless, wearable sensor measuring hand acceleration, electrocardiography (ECG), and skin temperature that outperforms the ActiWatch (one common equipment of actigraphy), detecting wake and sleep with a recall of 74.4 and 90.0%, respectively.
Videosomnography
For centuries, many videosomnography monitoring systems have been used to measure predefined daily activities continuously (von Ziegler et al., 2021). Like actigraphy, the advantages of videosomnography lie in its objective documentation for long-term interval (Goodlin-Jones et al., 2001; Burnham et al., 2002). It can also be used for capturing abnormal events such as parasomnias during night. However, there are several challenges using videosomnography in sleep research for children. First of all, the portable systems that capture time-lapse video recording are expensive and often need laborious and subjective human labeling. Additionally, camera is placed in fixed positions, the angle of review and the motion of children may affect the quality of video recording. Finally, ethical concerns and privacy issues of videosomnography surveillance system must be considered (Sadeh, 2015; Schwichtenberg et al., 2018). Videosomnography is now widely used in animal sleep research. Most non-invasive rodent sleep assessments depend on gross body movement (Pack et al., 2007; Fisher et al., 2012). Three-state sleep staging can be recorded by using electric field sensors to capture both gross body movement and respiration-related measures (McShane et al., 2012; Mingrone et al., 2020). For NHP study, sleep states are judged by focusing on two major behavioral features: whether the eyes were open or closed, and whether gross movements were present or absent (Prechtl, 1974; Mizuno et al., 2006; Chen et al., 2017). Over the past years, software packages based on deep learning/neural networks allow marker less tracking of multiple, hand-picked body points with astonishing performance.
Animal Models Used in the Study of ASD
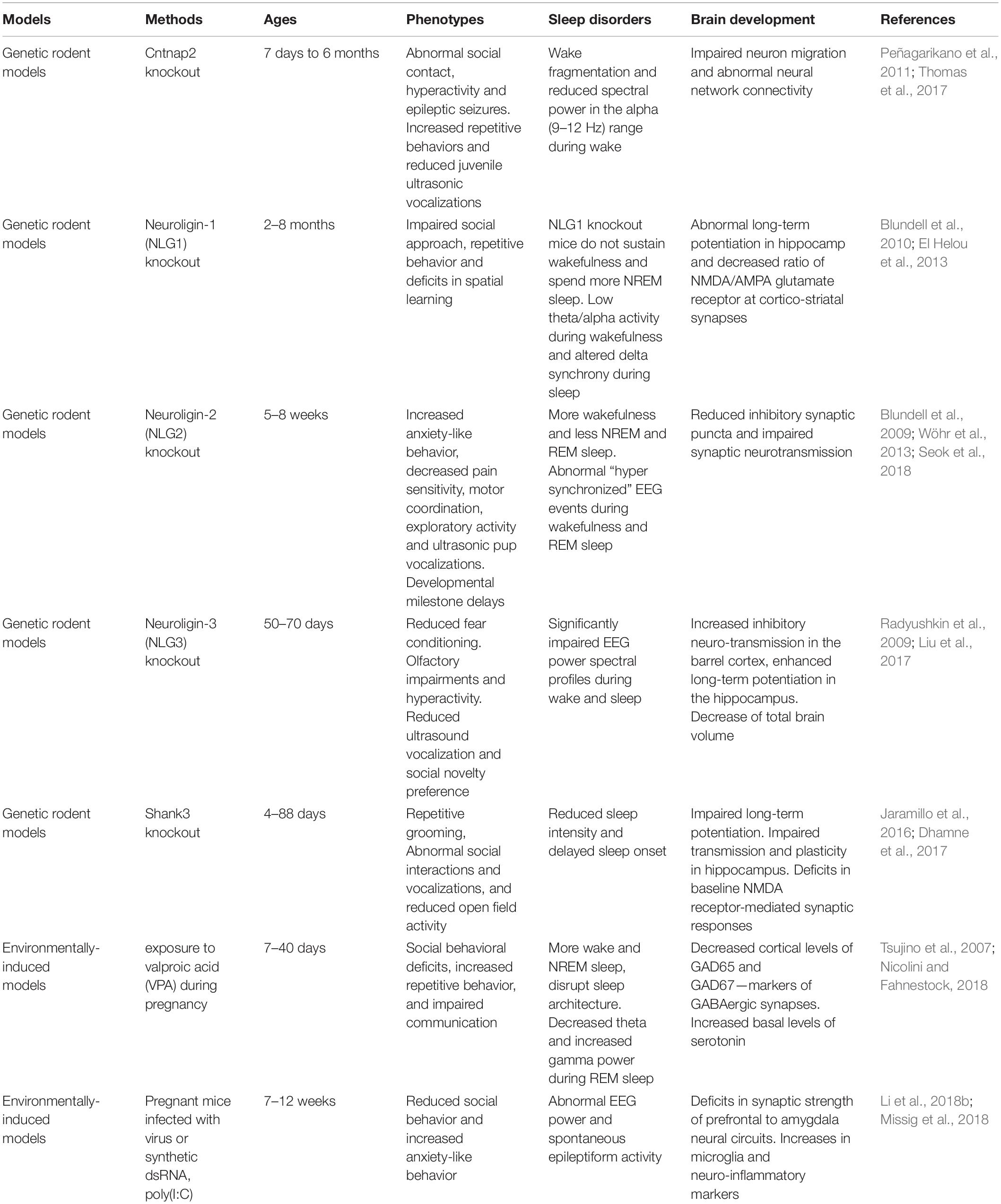
Numerous animal models of ASD have been generated in the last decade (Peñagarikano et al., 2011; Li et al., 2015; Kazdoba et al., 2016; Sacai et al., 2020). Many ASD-associated genes such as Neuroligins play a crucial role in regulation of synaptic adhesion and keeping imbalance between excitatory and inhibitory control in brain circuits (Wintler et al., 2020). The gene editing tools have rapidly been adopted by scientists to parse the role of genetic abnormalities in the etiology and symptomology of ASD. Because the more established gene editing technologies were used in the mice, mice have become the primary animal model of genetic diseases (Crawley, 2012). Growing studies of NHP models have been generated because their close phylogenetic relatedness to humans (Gadad et al., 2013). Moreover, mounting evidence suggests that environmental factors during early development is important. Animal models of maternal exposure to valproic acid and maternal immune activation appear to be the most commonly used. Frequent blood draws and PSG recordings, which are difficult procedures for children with ASD, also make the ASD models becoming ideal candidates. Here, we summarize some rodent (Table 1) and NHP (Table 2) models of ASD, which may have potential value to investigate the causes and effects of ASD, as well as their effects on brain development and sleep disorders.
Rodent Models for ASD
The CNTNAP2 gene encodes cortactin-associated protein-like 2 (CASPR2), which is a cell adhesion molecule and receptor (Jackman et al., 2009). Research of CNTNAP2 demonstrated a connection between genetic risk for autism and specific brain structures (Alarcón et al., 2008). A linkage study reported an increased familial risk for autism with mutations of the CNTNAP2 gene (Arking et al., 2008). Cntnap2 knockout (KO) mice have very similar presentations as with ASD including hyperactivity and epileptic seizures. Analyses of these mice indicated abnormal neuronal migration and synchrony (Peñagarikano et al., 2011).
Neuroligins (NLs) are a diverse class of proteins distributed molecules with functions of excitatory or inhibitory synapse specification (Ichtchenko et al., 1995; Ichtchenko et al., 1996; Graf et al., 2004; Blundell et al., 2010). Neuroligin-1 (NLG-1) is enriched preferentially at excitatory synapses (Song et al., 1999), neuroligin-2 (NLG-2) is enriched at inhibitory synapses (Varoqueaux et al., 2004; Levinson and El-Husseini, 2005), and neuroligin-3 (NLG-3) appears to be present at both (Fekete et al., 2015). The activity of NLG1 is impaired by prolonged wakefulness. Neuroligin-1 is related to neuronal activity and associated with regulation of sleep and wake (El Helou et al., 2013). Janine et al. found that NLG-1 knockout mice can hardly sustain wakefulness and spend more time in NREM sleep. Neuroligin-2 knock-out mice have less total sleep time and exhibit abnormal spike and wave discharges and behavioral arrests characteristic of absence seizures (Cao et al., 2020). Neuroligin-3 knock-out mice exhibit reduced fear conditioning, olfactory impairments and hyperactivity, as well as reduced ultrasound vocalization and social novelty preference (Radyushkin et al., 2009; Liu et al., 2017).
It has been proven that SHANK3 may induce sleep difficulties in patients with ASD. SHANK proteins are important organizers for signaling proteins in the post-synapse of excitatory neurons. In neurons, SHANK2 and SHANK3 have a positive effect on the induction and maturation of dendritic spines, whereas SHANK1 induces the enlargement of spine heads. Patients with an ASD-associated condition called Phelan-McDermid syndrome (PMS) are often missing the SHANK3 gene and they also often have sleep problems (Phelan and McDermid, 2012; Bro et al., 2017; De Rubeis et al., 2018). A recent meta-analysis of SHANK mutations suggested that SHANK3 mutations have a higher frequency and penetrance in individuals with ASD, compared to SHANK1 and SHANK2 (Leblond et al., 2014). Shank3 mutant mice show a variety of features of both ASD and PMS (Jaramillo et al., 2017; Ingiosi et al., 2019). In Shank3 heterozygous mice, there was a reduction in basal neurotransmission (Bozdagi et al., 2010). Shank3 knockout mice exhibit many autistic-like behaviors such as repetitive grooming, social deficits, reduced activity, anxiety-related behavior, as well as learning and memory impairments (Jaramillo et al., 2016; Dhamne et al., 2017). Shank3 KO mice have reduced sleep intensity and delayed sleep onset.
Overall, there were many other rodent models of ASD displaying reduced sleep time: 16p11.2, Fmr1, Mecp2, Ube3a, Rims1, Scn1a, Scn8a, Disc1, Gabrb3,Camk2a, Cacna1g, and Npas2 (Dudley et al., 2003; Lee et al., 2004; Anderson et al., 2005; Lonart et al., 2008; Kimura et al., 2010; Papale et al., 2010; Johnston et al., 2014; Zhou et al., 2014; Ehlen et al., 2015; Kalume et al., 2015; Kumar et al., 2015; Jaramillo et al., 2016; Tatsuki et al., 2016; Dittrich et al., 2017; Lu et al., 2019). Although the majority of these mutant rodent models exhibit reduced activity, which could be indicative of decrease sleep duration, the prevalence of serious sleep problems such as sleep fragmentation is far less than what has been observed in the clinical population.
While there is strong genetic effect, the etiology of ASD seems to be multifactorial. Environmental factors including toxins, pesticides, infection, and drugs also have a strong correlation. Environmental exposure during preconception, prenatal, and postnatal pregnancy can impact the immune system and the developing nervous system, and may cause neurodevelopmental disorders including ASD.
Valproic acid (VPA) is a drug used in humans primarily for epilepsy and seizure control. VPA is currently considered to be a risk factor for ASD and is also known teratogenicity (Balfour and Bryson, 1994). It has been demonstrated that exposure to VPA during pregnancy would increase the risk of autism in children based on several studies in humans (Laegreid et al., 1993; Christianson et al., 1994) and experimental evidence in animals (Lin et al., 2013). Furthermore, rodents prenatally exposed to this drug exhibit autism-like behavior including social behavioral deficits, repetitive and stereotypic behaviors, and impaired communication (Mychasiuk et al., 2012; Nicolini and Fahnestock, 2018). Intraperitoneal injection of VPA to rats with pregnancy would make their offspring exhibiting autism relevant behavioral and physiological indicators (Schneider et al., 2008).
Several studies have reported correlation between maternal antibody reactivity toward fetal brain proteins and ASD in the children (Braunschweig et al., 2008; Croen et al., 2008; Brimberg et al., 2013). In the rodent maternal immune activation model of ASD (Smith et al., 2007; Malkova et al., 2012; Choi et al., 2016; Kim et al., 2017), offspring from pregnant mice which were infected with virus or injected intra-peritoneally with synthetic dsRNA [poly(I: C)], exhibited behavioral symptoms such as social deficits, communication deficits, and repetitive behaviors. For brain neuropathology, the offspring of maternally infected mice displayed significantly fewer Purkinje cells. These data are quite similar to both ASD behavioral and neuropathological phenotypes.
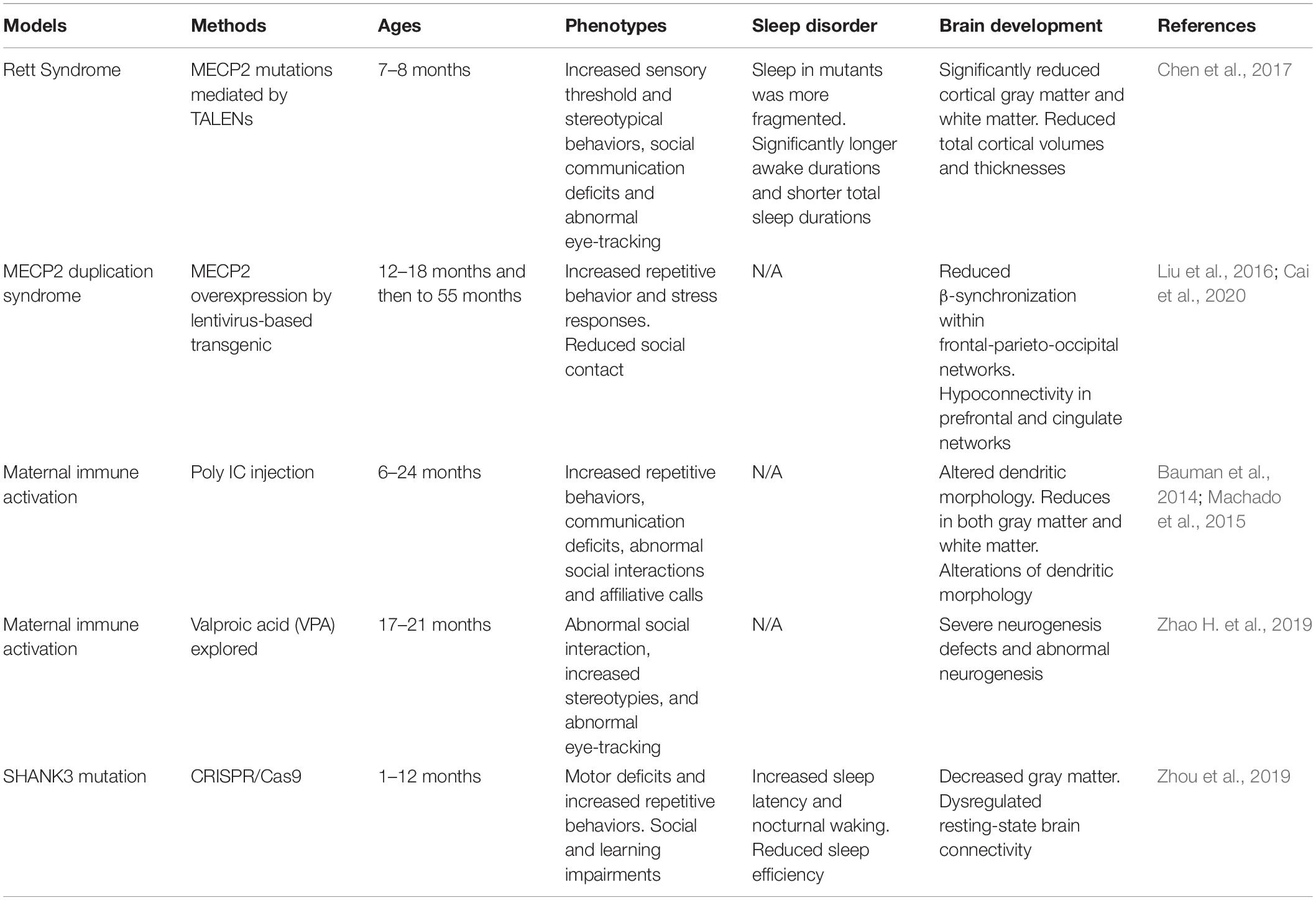
Non-human Primate Models
Non-human primates are among the optimal animal models, in large part because of their close phylogenetic relatedness with humans (Zhang et al., 2014; Nunn and Samson, 2018). With the rapid advances in gene-editing technologies, researchers have established several NHP models for ASD (Liu et al., 2016; Sato et al., 2016; Chen et al., 2017; Tu et al., 2019). It would be valuable for researchers to be attentive to study of many kinds of disease by using NHP animal models (Anderson, 2000).
MECP2 duplication syndrome is an X-linked recessive syndrome resulting from abnormal genomic rearrangement. The two major clinical symptoms are intellectual disability and anxiety. MECP2 overexpressed monkey models exhibited characteristic features of ASD such as social deficits, repetitive behaviors, and increased anxiety (Liu et al., 2016). Cai et al. reported a combination of gene-circuit-behavior analyses, including MECP2 co-expression network, locomotive and cognitive behaviors, and EEG and fMRI findings in MECP2 overexpressed monkeys. Whole-genome expression analysis revealed MECP2 co-expressed genes were significantly enriched in GABA-related signaling pathways, whereby reduced β-synchronization within frontal-parietal-occipital networks was associated with abnormal locomotive behaviors (Cai et al., 2020).
Rett syndrome caused by mutations in MECP2 is a prototypical neurodevelopmental disorder. Researchers demonstrated that MECP2 mutant monkeys could well mimic autism-associated abnormalities in physiology and social behavior (Chen et al., 2017). The mutant monkeys exhibited significantly increased total awake time and more fragmental sleep during night, which have also been found in Mecp2 mutant mice (Li et al., 2015).
Feng et al. used CRISPR/Cas9 to generate SHANK3, a top autism gene mutant monkey. SHANK3 mutant monkeys tend to be less active and have troubles sleeping that they take longer time to fall asleep and wake up more often. Monkeys in this study have severe repetitive movement, deficient social skills, and show brain-activity patterns similar to those seen in autistic people (Le Bras, 2019; Tu et al., 2019). SHANK3-deficient monkeys showed reduced spine density and impaired development of mature neurons in the prefrontal cortex (Zhao et al., 2017). It has also been found that some rhesus macaques carried spontaneous mutation of SHANK3 (Vegué and Roxin, 2015). Spontaneous mutations in NHPs may have the potential to be used as a suitable animal model to figure out the relationships between genetic variants and behaviors (Haus et al., 2014; Zhao et al., 2018).
Rodent animal models of maternal exposure to VPA provided evidence that environmental risk factors in ASD. Recently, Zhao H. et al., 2019 reported the neurodevelopmental and behavioral outcomes of maternal VPA exposure in NHP for the first time. Offspring from maternal exposure to VPA has significantly impaired neuronal development. VPA-exposed monkey offspring showed impaired social interaction, communication disabilities, and abnormal eye-tracking (Zhao H. et al., 2019).
When rhesus monkeys were given the viral mimicking synthetic double-stranded RNA (polyinosinic:polycytidylic acid stabilized with poly-L-lysine) during pregnancy, and their offspring could exhibit abnormal repetitive behaviors, altered communication, impaired social interactions and abnormal gaze patterns to salient social information (Bauman et al., 2014; Machado et al., 2015). These offspring with autism-like behaviors also have reduced gray matter in most of the cortex and decreased white matter in the parietal cortex (Short et al., 2010). Novel evidence implicating MIA exposure with alterations of NHP dendritic morphology have been found (Weir et al., 2015). The mother and the fetus exploit several mechanisms in order to avoid fetal rejection and to maintain an immunotolerant environment during pregnancy. The placenta is an important organ that facilitates nutrient exchange. It has been reported that the anatomy of the placenta is varied across species, and it is highest in humans, intermediate in rhesus macaques, and minimal in rodents (Carter, 2007). Thus, the role of the NHP animal model in this field of research is important.
Monkeys as an Ideal Animal Model for Studying Sleep in ASD
An ideal animal model of human disease should show tight junctions with clinical characteristics of the disease. The statistics from United States government in 2010 indicated that almost 90% of the laboratory animals used in science research are mice, rats, and other rodents. NHP only represents 0.28% among all animals (Phillips et al., 2014). However, rodents diverged from humans by more than 70 million years of evolution. There are significantly evolutionary differences in brain anatomy, cognitive capacity, and social behavior between humans and rodents (Kumar and Hedges, 1998; Gibbs et al., 2004). Compared with rodents, rhesus macaque (Macaca mulatta), most common NHP used in study, are separated from humans approximately 25 million years ago and are more similar to humans in genetics, neurobiology, and behavior. Thus, NHP have reasonable behavioral correlates to the characteristics of patients in ASD, such as repetitive behaviors, communication deficits, and stereotyped behavior (Watson and Platt, 2012; Parker et al., 2018). As mentioned previously, prenatal environment and gestational timing may impact neurodevelopment of offspring. The gestational period of rhesus monkeys (165 days) and humans (280 days) is much longer than mouse (18–23 days) (Clancy et al., 2001). Besides, the prenatal immune challenge and neuron development of primates occur mostly during the third trimester of prenatal and during early postnatal period (Careaga et al., 2017). The mouse is becoming increasingly popular for genetic studies. However, the mouse’s brain weighs a few grams, and ours weighs one and a half kilos. Can we use the mouse to learn something about our brain? The region of the neocortex is almost 80% in the human brain, which is just 28% in the rat (Roberts and Clarke, 2019). Human prefrontal cortex includes granular and agranular cortex, while rat prefrontal cortex only contains agranular cortex (Ongür and Price, 2000; Uylings et al., 2003). It has been proposed that the prefrontal cortex has a substantial role in social processing, and its potential dysfunction may cause ASD (Sugranyes et al., 2011). The temporal lobe is a morphological brain region which is unique to primates (Colombo et al., 2000; Bryant and Preuss, 2018). The major areas of the human brain classified by Brodmann have also been identified in NHP. Structure and function of the amygdala are nearly the same in the human and non-human primate (Gowen et al., 2007; Rutishauser et al., 2015; Schumann et al., 2016), but remarkably different from the rodent brain (Chareyron et al., 2011). The close relationship of development and evolution between NHP and human show that great prospects to mimic clinical realities by designing NHP animal models.
As mentioned previously, sleep problems in children with ASD are caused by multi-factorial risks such as abnormal neurodevelopment and environmental factors (Bourgeron, 2007; Owens and Mindell, 2011). Modeling clinical disorders in animals provide an opportunity to improve translational research although the human disorder’s clinical phenotype is complex and heterogeneous and lacks objective homologous endpoints across species (Missig et al., 2020). Many previous studies of ASD animal models exhibited several hallmark features which have been documented in humans.
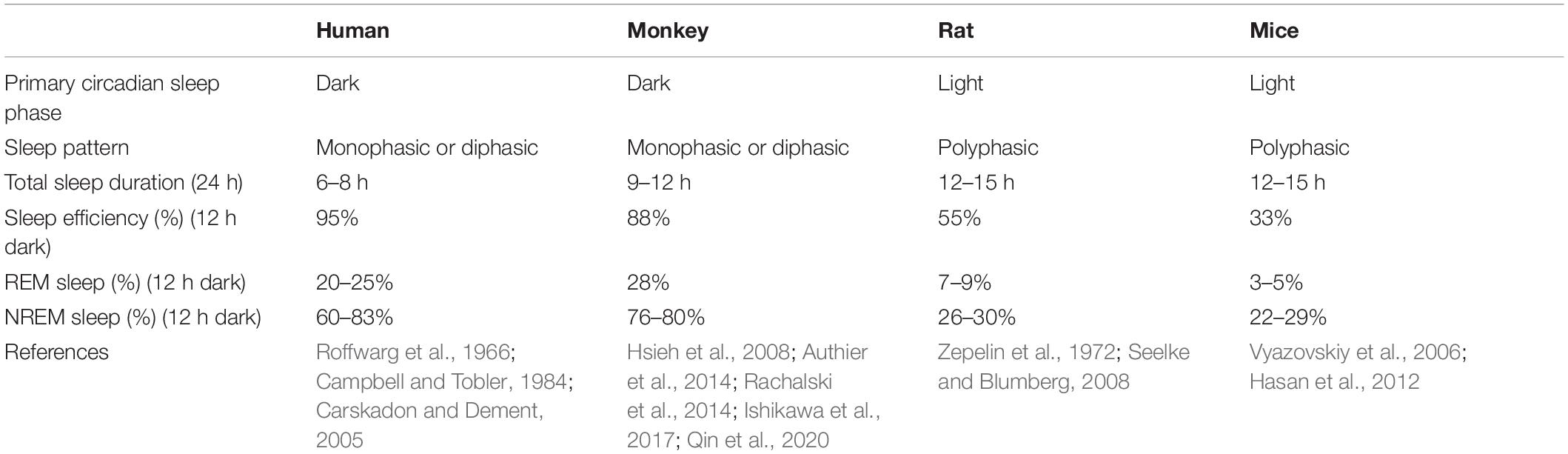
Sleeping studies in humans must be done in accessible samples, predominantly saliva or blood, and confounded by environmental factors. Species-specific differences including light and biological rhythm, as well as sleep features have been noted in studies of sleep (Campbell and Tobler, 1984). For example, rodents are commonly thought to awake during the dark phase and asleep during the light phase. However, researchers found that mice are not explicitly nocturnal, and they have diurnal feeding activity. Researchers also reported that seasonal influences were demonstrated to be more potent on activity than specific genes which was generally considered to control sleep (Daan et al., 2011). Effective use of animals to study normal sleep and sleep disorders must consider known similarities and differences between human and animals. Likewise, sleep is important to keep health and can significantly influence daily activity schedules in NHP (Fruth et al., 2018; Qiu et al., 2019). Sleep structure and EEG patterns of NHP are closely related to the consolidated and monophasic organization observed in humans (Reite et al., 1965; Hsieh et al., 2008), which contrasts with the more fragmented sleep patterns in rodents (Fifel and Cooper, 2014) (Table 3). NHP is also a diurnal animal to better recapitulate clinical conditions with behavioral and metabolic properties closer to humans. Humans pass through 4–6 cycles of NREM and REM within a night’s sleep, which are much shorter in rats and mice (Fuller et al., 2006; Toth and Bhargava, 2013). Nunn and Samson compared sleep patterns in 30 different species of primates, including humans. Most species generally sleep between 9 and 15 h, while humans averaged just 7 h (Nunn and Samson, 2018). In summary, measuring behavioral and sleep states in NHP may provide a better understanding of sleep disorders in children with ASD compared with rodents.
Current Challenges
Autism spectrum disorder is a neurodevelopmental disorder and the origins of ASD remain unresolved. The potential estimates including genetic, maternal, and environmental effects (Bai et al., 2019). At first, there are various types of genetic variation, such as single-nucleotide polymorphism (SNP) or rare genetic mutations (Weiner et al., 2017). Animal model studies have shown that the impact of genetic on behavior is complex and not completely correspond to specific behavior. Brain development can be influenced by not only the expression of genes, but also modified by environmental factors during the pregnancy and postnatal period. Therefore, the application of gene editing technology in animal models of disease may not completely mimic clinical phenotypes of humans. Secondly, although the NHP animal model has been used to study the impact of environmental modifications on the brain development. Genetic influences may also affect individual responses to different situations and different types of environmental challenges (Machado and Bachevalier, 2003). In this area, rodent models may be more appropriate which have more identical genetic backgrounds compared with NHPs. Besides, NHP exhibit significant and stable individual differences in social commination (Capitanio, 1999; Capitanio and Widaman, 2005).
Non-human primates have much longer reproductive cycle and lower reproduction efficiency compared with rodents, which may bring the difficulties to prepare an adequate quantity of experimental animals. Furthermore, the giant body size of NHP may cause a significant challenge for experimental design. Limitations associated with the gene-editing technique, including editing efficiency, chimeras, and off-target effects, should also be brought to attention (Niu et al., 2014; Chen et al., 2016). Lastly, NHP, rather than other animals, require more significant ethical consideration because its significant cognitive capacity and complex social behavior. Researchers have moral responsibility to ensure that experimental animals receive reduced negative effects and suffering (Bentham, 1996; Beauchamp and Frey, 2011). Animal experiments should follow the principles of the 3Rs, including replacement, reduction, and refinement (Russell and Burch, 1959; Jennings et al., 2009).
Conclusion and Perspectives
Autism spectrum disorder is a neurodevelopmental disorder and with the increasing incidence of ASD, it is essential to understand what has changed in our genes and environments that may contribute to these disorders. It has been showed that ASD is not a single disease, but rather several conditions including genetic, maternal, and environmental effects that ultimately cause similar behavioral impairments. The abnormalities of ASD may predispose children to various threaten of sleep and make them especially susceptible to sleep problems. Sleep disorders have been reported as one of the most common symptoms and in up to 80% of children with ASD have sleep problems which may even contribute to the altered brain structure and activity (Blakemore et al., 2006). Thus, understanding how sleep affected children with ASD by specific mechanisms such as brain development and synaptic plasticity will enable a broader understanding of the disorders’ causes and provide insights into specific treatments. Over the years, many different sleep analysis methods have been reported. The selection of sleep assessment method should be tailored to specific subjects and taken into consideration of their unique characteristics.
Animal models hold great potential values to investigate the causes and treatments for sleep problems in children with ASD. Numerous animal models of ASD have been generated in the last decade. An ideal animal model should show tight junctions with clinical characteristics of the disease. Rodents are the most common experimental animals and growing studies of NHP models have been generated because their close phylogenetic relatedness to humans.
Because of the complexity and heterogeneity of the ASD, it is still inadequate to understand how genes control and influence complex behavior. The animal models of ASD are currently oversimplified and have many issues. Recently, Liu et al. (2019) demonstrated that an approach to generate cloned monkeys by somatic cell nuclear transfer (SCNT), which can creatively solve the problem of generating NHP models with uniform genetic backgrounds. This study is profoundly improving the overall reproducibility of the model. Continued research used this technique to generate five BMAL1 knockout monkeys for sleeping study, and these monkeys exhibited more activities and reduced sleep during night (Qiu et al., 2019). The development of genome editing technologies (such as CRISPR/Cas9 and base editing, etc.) has opened up the revolutionary ways to directly target and modify genomic sequences in animals (Kang et al., 2019; Zhao J. et al., 2019; Li et al., 2020). We anticipate greater numbers of applications will materialize shortly, such as genome-edited NHP combined with SCNT. Although some issues still need to be solved, studying the sleep disorder across multiple biological scales can offer the hope in the field of translational medicine for ASD and other human diseases. The role of NHP animal model in this process is irreplaceable and must be recognized.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the National Natural Science Foundation of China (31960178), the Applied Basic Research Programs of Science and Technology Commission Foundation of Yunnan Province (2018FB053 and 2019FA007), the Key Realm R&D Program of Guangdong Province (2019B030335001), the China Postdoctoral Science Foundation (2018M631105), and the Yunnan Provincial Academician and Expert Workstation (202005AF150017 and 2019IC051).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Acebo, C., Sadeh, A., Seifer, R., Tzischinsky, O., Wolfson, A. R., Hafer, A., et al. (1999). Estimating sleep patterns with activity monitoring in children and adolescents: how many nights are necessary for reliable measures? Sleep 22, 95–103. doi: 10.1093/sleep/22.1.95
Alarcón, M., Abrahams, B. S., Stone, J. L., Duvall, J. A., Perederiy, J. V., Bomar, J. M., et al. (2008). Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 82, 150–159. doi: 10.1016/j.ajhg.2007.09.005
American Psychiatric Association (2013). Diagnostic and Statistical Manual of Mental Disorders (DSM-5§). Virginia: American Psychiatric Pub.
Anderson, J. R. (2000). Sleep-related behavioural adaptations in free-ranging anthropoid primates. Sleep Med. Rev. 4, 355–373. doi: 10.1053/smrv.2000.0105
Anderson, M. P., Mochizuki, T., Xie, J., Fischler, W., Manger, J. P., Talley, E. M., et al. (2005). Thalamic Cav3.1 T-type Ca2+ channel plays a crucial role in stabilizing sleep. Proc. Natl. Acad. Sci. U.S.A. 102, 1743–1748. doi: 10.1073/pnas.0409644102
Arking, D. E., Cutler, D. J., Brune, C. W., Teslovich, T. M., West, K., Ikeda, M., et al. (2008). A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 82, 160–164. doi: 10.1016/j.ajhg.2007.09.015
Aton, S. J., Seibt, J., Dumoulin, M., Jha, S. K., Steinmetz, N., Coleman, T., et al. (2009). Mechanisms of sleep-dependent consolidation of cortical plasticity. Neuron 61, 454–466. doi: 10.1016/j.neuron.2009.01.007
Authier, S., Bassett, L., Pouliot, M., Rachalski, A., Troncy, E., Paquette, D., et al. (2014). Effects of amphetamine, diazepam and caffeine on polysomnography (EEG, EMG, EOG)-derived variables measured using telemetry in Cynomolgus monkeys. J. Pharmacol. Toxicol. Methods 70, 86–93. doi: 10.1016/j.vascn.2014.05.003
Bai, D., Yip, B. H. K., Windham, G. C., Sourander, A., Francis, R., Yoffe, R., et al. (2019). Association of genetic and environmental factors with Autism in a 5-country cohort. JAMA Psychiatry 76, 1035–1043. doi: 10.1001/jamapsychiatry.2019.1411
Bathory, E., and Tomopoulos, S. (2017). Sleep regulation, physiology and development, sleep duration and patterns, and sleep hygiene in infants, toddlers, and preschool-age children. Curr. Probl. Pediat. Adolesc. Health Care 47, 29–42. doi: 10.1016/j.cppeds.2016.12.001
Bauman, M. D., Iosif, A. M., Smith, S. E., Bregere, C., Amaral, D. G., and Patterson, P. H. (2014). Activation of the maternal immune system during pregnancy alters behavioral development of rhesus monkey offspring. Biol. Psychiatry 75, 332–341. doi: 10.1016/j.biopsych.2013.06.025
Beauchamp, T. L., and Frey, R. G. (2011). The Oxford Handbook of Animal Ethics. Oxford: Oxford University Press.
Bélanger, M., Bernier, A., Paquet, J., Simard, V., and Carrier, J. (2013). Validating actigraphy as a measure of sleep for preschool children. J. Clin. Sleep Med. 9, 701–706. doi: 10.5664/jcsm.2844
Bentham, J. (1996). An Introduction to the Principles of Morals and Legislation: The Collected Works of Jeremy Bentham. Oxford: Oxford University Press.
Bhargava, S. (2011). Diagnosis and management of common sleep problems in children. Pediatr. Rev. 32, 91–98. doi: 10.1542/pir.32-3-91
Blakemore, S.-J., Tavassoli, T., Calò, S., Thomas, R. M., Catmur, C., Frith, U., et al. (2006). Tactile sensitivity in Asperger syndrome. Brain Cogn. 61, 5–13. doi: 10.1016/j.bandc.2005.12.013
Blume, C., Del Giudice, R., Wislowska, M., Lechinger, J., and Schabus, M. (2015). Across the consciousness continuum-from unresponsive wakefulness to sleep. Front. Hum. Neurosci. 9:105. doi: 10.3389/fnhum.2015.00105
Blundell, J., Blaiss, C. A., Etherton, M. R., Espinosa, F., Tabuchi, K., Walz, C., et al. (2010). Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J. Neurosci. 30, 2115–2129. doi: 10.1523/jneurosci.4517-09.2010
Blundell, J., Tabuchi, K., Bolliger, M. F., Blaiss, C. A., Brose, N., Liu, X., et al. (2009). Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 8, 114–126. doi: 10.1111/j.1601-183x.2008.00455.x
Boe, A. J., McGee Koch, L. L., O’Brien, M. K., Shawen, N., Rogers, J. A., Lieber, R. L., et al. (2019). Automating sleep stage classification using wireless, wearable sensors. NPJ Digital Med. 2:131.
Boulos, M. I., Jairam, T., Kendzerska, T., Im, J., Mekhael, A., and Murray, B. J. (2019). Normal polysomnography parameters in healthy adults: a systematic review and meta-analysis. Lancet Respir. Med. 7, 533–543. doi: 10.1016/s2213-2600(19)30057-8
Bourgeron, T. (2007). The possible interplay of synaptic and clock genes in autism spectrum disorders. Cold Spring Harb. Symp. Quant. Biol. 72, 645–654. doi: 10.1101/sqb.2007.72.020
Bozdagi, O., Sakurai, T., Papapetrou, D., Wang, X., Dickstein, D. L., Takahashi, N., et al. (2010). Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol. Autism 1:15. doi: 10.1186/2040-2392-1-15
Braunschweig, D., Ashwood, P., Krakowiak, P., Hertz-Picciotto, I., Hansen, R., and Croen, L. A. I, et al (2008). Autism: maternally derived antibodies specific for fetal brain proteins. Neurotoxicology 29, 226–231.
Brimberg, L., Sadiq, A., Gregersen, P. K., and Diamond, B. (2013). Brain-reactive IgG correlates with autoimmunity in mothers of a child with an autism spectrum disorder. Mol. Psychiatry 18, 1171–1177. doi: 10.1038/mp.2013.101
Bro, D., O’Hara, R., Primeau, M., Hanson-Kahn, A., Hallmayer, J., and Bernstein, J. A. (2017). Sleep disturbances in individuals with phelan-McDermid syndrome: correlation with caregivers’ sleep quality and daytime functioning. Sleep 40:zsw062.
Bryant, K., and Preuss, T. (2018). A Comparative Perspective on the Human Temporal Lobe BT-Digital Endocasts: From Skulls to Brains. Cham: Springer, 239–258.
Burnham, M. M., Goodlin-Jones, B. L., Gaylor, E. E., and Anders, T. F. (2002). Nighttime sleep-wake patterns and self-soothing from birth to one year of age: a longitudinal intervention study. J. Child Psychol. Psychiatry 43, 713–725. doi: 10.1111/1469-7610.00076
Cai, D. C., Wang, Z., Bo, T., Yan, S., Liu, Y., Liu, Z., et al. (2020). MECP2 duplication causes aberrant GABA pathways, circuits and behaviors in transgenic monkeys: neural mappings to patients with autism. J. Neurosci. 40, 3799–3814. doi: 10.1523/jneurosci.2727-19.2020
Camerota, M., Tully, K. P., Grimes, M., Gueron-Sela, N., and Propper, C. B. (2018). Assessment of infant sleep: how well do multiple methods compare? Sleep 41, zsy146.
Campbell, S. S., and Tobler, I. (1984). Animal sleep: a review of sleep duration across phylogeny. Neurosci. Biobehav. Rev. 8, 269–300. doi: 10.1016/0149-7634(84)90054-x
Cao, F., Liu, J. J., Zhou, S., Cortez, M. A., Snead, O. C., Han, J., et al. (2020). Neuroligin 2 regulates absence seizures and behavioral arrests through GABAergic transmission within the thalamocortical circuitry. Nat. Commun. 11:3744.
Capitanio, J. P. (1999). Personality dimensions in adult male rhesus macaques: prediction of behaviors across time and situation. Am. J. Primatol. 47, 299–320. doi: 10.1002/(sici)1098-2345(1999)47:4<299::aid-ajp3>3.0.co;2-p
Capitanio, J. P., and Widaman, K. F. (2005). Confirmatory factor analysis of personality structure in adult male rhesus monkeys (Macaca mulatta). Am. J. Primatol. 65, 289–294. doi: 10.1002/ajp.20116
Careaga, M., Murai, T., and Bauman, M. D. (2017). Maternal immune activation and autism spectrum disorder: from rodents to nonhuman and human primates. Biol. Psychiatry 81, 391–401. doi: 10.1016/j.biopsych.2016.10.020
Carskadon, M. A., and Dement, W. C. (2005). Normal human sleep: an overview. Principles Pract. Sleep Med. 4, 13–23. doi: 10.1016/b0-72-160797-7/50009-4
Carter, A. M. (2007). Animal models of human placentation–a review. Placenta 28, (Suppl. A), S41–S47.
Chareyron, L. J., Banta Lavenex, P., Amaral, D. G., and Lavenex, P. (2011). Stereological analysis of the rat and monkey amygdala. J. Comp. Neurol. 519, 3218–3239. doi: 10.1002/cne.22677
Chen, Y., Niu, Y., and Ji, W. (2016). Genome editing in nonhuman primates: approach to generating human disease models. J. Intern. Med. 280, 246–251. doi: 10.1111/joim.12469
Chen, Y., Yu, J., Niu, Y., Qin, D., Liu, H., Li, G., et al. (2017). Modeling rett syndrome using TALEN-edited MECP2 mutant cynomolgus monkeys. Cell 169, 945.e10–955.e10.
Choi, G. B., Yim, Y. S., Wong, H., Kim, S., Kim, H., Kim, S. V., et al. (2016). The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939. doi: 10.1126/science.aad0314
Christensen, D. L., Baio, J., Van Naarden Braun, K., Bilder, D., Charles, J., Constantino, J. N., et al. (2016). Prevalence and characteristics of autism spectrum disorder among children aged 8 years–autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill. Summ. 65, 1–23. doi: 10.15585/mmwr.ss6802a1
Christianson, A. L., Chester, N., and Kromberg, J. G. (1994). Fetal valproate syndrome: clinical and neuro-developmental features in two sibling pairs. Dev. Med. Child Neurol. 36, 361–369. doi: 10.1111/j.1469-8749.1994.tb11858.x
Clancy, B., Darlington, R. B., and Finlay, B. L. (2001). Translating developmental time across mammalian species. Neuroscience 105, 7–17. doi: 10.1016/s0306-4522(01)00171-3
Colombo, J. A., Fuchs, E., Härtig, W., Marotte, L. R., and Puissant, V. (2000). Rodent-like and primate-like types of astroglial architecture in the adult cerebral cortex of mammals: a comparative study. Anat. Embryol. 201, 111–120. doi: 10.1007/pl00008231
Cook, J. D., Prairie, M. L., and Plante, D. T. (2017). Utility of the Fitbit Flex to evaluate sleep in major depressive disorder: a comparison against polysomnography and wrist-worn actigraphy. J. Affect. Disord. 217, 299–305. doi: 10.1016/j.jad.2017.04.030
Cook, J. D., Prairie, M. L., and Plante, D. T. (2018). Ability of the multisensory jawbone UP3 to quantify and classify sleep in patients with suspected central disorders of hypersomnolence: a comparison against polysomnography and actigraphy. J. Clin. Sleep Med. 14, 841–848. doi: 10.5664/jcsm.7120
Crawley, J. N. (2012). Translational animal models of autism and neurodevelopmental disorders. Dialog. Clin. Neurosci. 14, 293–305. doi: 10.31887/dcns.2012.14.3/jcrawley
Croen, L. A., Braunschweig, D., Haapanen, L., Yoshida, C. K., Fireman, B., Grether, J. K., et al. (2008). Maternal mid-pregnancy autoantibodies to fetal brain protein: the early markers for autism study. Biol. Psychiatry 64, 583–588. doi: 10.1016/j.biopsych.2008.05.006
Daan, S., Spoelstra, K., Albrecht, U., Schmutz, I., Daan, M., Daan, B., et al. (2011). Lab mice in the field: unorthodox daily activity and effects of a dysfunctional circadian clock allele. J. Biol. Rhythms 26, 118–129. doi: 10.1177/0748730410397645
De Rubeis, S., Siper, P. M., Durkin, A., Weissman, J., Muratet, F., Halpern, D., et al. (2018). Delineation of the genetic and clinical spectrum of Phelan-McDermid syndrome caused by SHANK3 point mutations. Mol. Autism 9:31.
de Zambotti, M., Baker, F. C., and Colrain, I. M. (2015). Validation of sleep-tracking technology compared with polysomnography in adolescents. Sleep 38, 1461–1468. doi: 10.5665/sleep.4990
de Zambotti, M., Baker, F. C., Willoughby, A. R., Godino, J. G., Wing, D., Patrick, K., et al. (2016). Measures of sleep and cardiac functioning during sleep using a multi-sensory commercially-available wristband in adolescents. Physiol. Behav. 158, 143–149. doi: 10.1016/j.physbeh.2016.03.006
de Zambotti, M., Cellini, N., Goldstone, A. I, Colrain, M., and Baker, F. C. (2019). Wearable sleep technology in clinical and research settings. Med. Sci. Sports Exerc. 51, 1538–1557. doi: 10.1249/mss.0000000000001947
de Zambotti, M., Goldstone, A., Claudatos, S. I, Colrain, M., and Baker, F. C. (2018). A validation study of Fitbit Charge 2TM compared with polysomnography in adults. Chronobiol. Int. 35, 465–476. doi: 10.1080/07420528.2017.1413578
Devnani, P. A., and Hegde, A. U. (2015). Autism and sleep disorders. J. Pediatr. Neurosci. 10, 304–307.
Dhamne, S. C., Silverman, J. L., Super, C. E., Lammers, S. H. T., Hameed, M. Q., Modi, M. E., et al. (2017). Replicable in vivo physiological and behavioral phenotypes of the Shank3B null mutant mouse model of autism. Mol. Autism 8:26.
Dittrich, L., Petese, A., and Jackson, W. S. (2017). The natural Disc1-deletion present in several inbred mouse strains does not affect sleep. Sci. Rep. 7:5665.
Doufas, A. G., Panagiotou, O. A., and Ioannidis, J. P. (2012). Concordance of sleep and pain outcomes of diverse interventions: an umbrella review. PLoS One 7:e40891. doi: 10.1371/journal.pone.0040891
Dudley, C. A., Erbel-Sieler, C., Estill, S. J., Reick, M., Franken, P., Pitts, S., et al. (2003). Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science 301, 379–383. doi: 10.1126/science.1082795
Ehlen, J. C., Jones, K. A., Pinckney, L., Gray, C. L., Burette, S., Weinberg, R. J., et al. (2015). Maternal Ube3a loss disrupts sleep homeostasis but leaves circadian rhythmicity largely intact. J. Neurosci. 35, 13587–13598. doi: 10.1523/jneurosci.2194-15.2015
El Helou, J., Bélanger-Nelson, E., Freyburger, M., Dorsaz, S., Curie, T., La Spada, F., et al. (2013). Neuroligin-1 links neuronal activity to sleep-wake regulation. Proc. Natl. Acad. Sci. U.S.A. 110, 9974–9979. doi: 10.1073/pnas.1221381110
Elrod, M. G., and Hood, B. S. (2015). Sleep differences among children with autism spectrum disorders and typically developing peers: a meta-analysis. J. Dev. Behav. Pediatr. 36, 166–177. doi: 10.1097/dbp.0000000000000140
Feinberg, I., Koresko, R. L., and Heller, N. (1967). EEG sleep patterns as a function of normal and pathological aging in man. J. Psychiatr. Res. 5, 107–144. doi: 10.1016/0022-3956(67)90027-1
Fekete, C. D., Chiou, T. T., Miralles, C. P., Harris, R. S., Fiondella, C. G., Loturco, J. J., et al. (2015). In vivo clonal overexpression of neuroligin 3 and neuroligin 2 in neurons of the rat cerebral cortex: differential effects on GABAergic synapses and neuronal migration. J. Comp. Neurol. 523, 1359–1378. doi: 10.1002/cne.23740
Fifel, K., and Cooper, H. M. (2014). Loss of dopamine disrupts circadian rhythms in a mouse model of Parkinson’s disease. Neurobiol. Dis. 71, 359–369. doi: 10.1016/j.nbd.2014.08.024
Fisher, S. P., Godinho, S. I., Pothecary, C. A., Hankins, M. W., Foster, R. G., and Peirson, S. N. (2012). Rapid assessment of sleep-wake behavior in mice. J. Biol. Rhythms 27, 48–58. doi: 10.1177/0748730411431550
Fogel, S., Martin, N., Lafortune, M., Barakat, M., Debas, K., Laventure, S., et al. (2012). NREM sleep oscillations and brain plasticity in aging. Front. Neurol. 3:176. doi: 10.3389/fneur.2012.00176
Frank, M. G., Issa, N. P., and Stryker, M. P. (2001). Sleep enhances plasticity in the developing visual cortex. Neuron 30, 275–287. doi: 10.1016/s0896-6273(01)00279-3
Fruth, B., Tagg, N., and Stewart, F. (2018). Sleep and nesting behavior in primates: a review. Am. J. Phys. Anthropol. 166, 499–509. doi: 10.1002/ajpa.23373
Fuller, P. M., Gooley, J. J., and Saper, C. B. (2006). Neurobiology of the sleep-wake cycle: sleep architecture, circadian regulation, and regulatory feedback. J. Biol. Rhythms 21, 482–493. doi: 10.1177/0748730406294627
Gadad, B. S., Hewitson, L., Young, K. A., and German, D. C. (2013). Neuropathology and animal models of autism: genetic and environmental factors. Autism Res. Treat. 2013:731935.
Galland, B. C., Taylor, B. J., Elder, D. E., and Herbison, P. (2012). Normal sleep patterns in infants and children: a systematic review of observational studies. Sleep Med. Rev. 16, 213–222. doi: 10.1016/j.smrv.2011.06.001
Gibbs, R. A., Weinstock, G. M., Metzker, M. L., Muzny, D. M., Sodergren, E. J., Scherer, S., et al. (2004). Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521. doi: 10.1038/nature02426
Gilbert, J., and Man, H. Y. (2017). Fundamental elements in autism: from neurogenesis and neurite growth to synaptic plasticity. Front. Cell Neurosci. 11:359. doi: 10.3389/fncel.2017.00359
Goodlin-Jones, B. L., Burnham, M. M., Gaylor, E. E., and Anders, T. F. (2001). Night waking, sleep-wake organization, and self-soothing in the first year of life. J. Dev. Behav. Pediatr. 22, 226–233. doi: 10.1097/00004703-200108000-00003
Gowen, E., Abadi, R. V., Poliakoff, E., Hansen, P. C., and Miall, R. C. (2007). Modulation of saccadic intrusions by exogenous and endogenous attention. Brain Res. 1141, 154–167. doi: 10.1016/j.brainres.2007.01.047
Graf, E. R., Zhang, X., Jin, S. X., Linhoff, M. W., and Craig, A. M. (2004). Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119, 1013–1026. doi: 10.1016/j.cell.2004.11.035
Grigg-Damberger, M. M. (2009). The AASM scoring manual: a critical appraisal. Curr. Opin. Pulm. Med. 15, 540–549. doi: 10.1097/mcp.0b013e328331a2bf
Hartsock, M. J., and Spencer, R. L. (2020). Memory and the circadian system: identifying candidate mechanisms by which local clocks in the brain may regulate synaptic plasticity. Neurosci. Biobehav. Rev. 118, 134–162. doi: 10.1016/j.neubiorev.2020.07.023
Hasan, S., Dauvilliers, Y., Mongrain, V., Franken, P., and Tafti, M. (2012). Age-related changes in sleep in inbred mice are genotype dependent. Neurobiol. Aging 33, 195.e113–195.e126.
Haus, T., Ferguson, B., Rogers, J., Doxiadis, G., Certa, U., Rose, N. J., et al. (2014). Genome typing of nonhuman primate models: implications for biomedical research. Trends Genet. 30, 482–487. doi: 10.1016/j.tig.2014.05.004
Honomichl, R. D., Goodlin-Jones, B. L., Burnham, M., Gaylor, E., and Anders, T. F. (2002). Sleep patterns of children with pervasive developmental disorders. J. Autism Dev. Disord. 32, 553–561.
Hsieh, K. C., Robinson, E. L., and Fuller, C. A. (2008). Sleep architecture in unrestrained rhesus monkeys (Macaca mulatta) synchronized to 24-hour light-dark cycles. Sleep 31, 1239–1250.
Hudson, A. N., Van Dongen, H. P. A., and Honn, K. A. (2020). Sleep deprivation, vigilant attention, and brain function: a review. Neuropsychopharmacology 45, 21–30. doi: 10.1038/s41386-019-0432-6
Humphreys, J. S., Gringras, P., Blair, P. S., Scott, N., Henderson, J., Fleming, P. J., et al. (2014). Sleep patterns in children with autistic spectrum disorders: a prospective cohort study. Arch. Dis. Child 99, 114–118. doi: 10.1136/archdischild-2013-304083
Ibáñez, V., Silva, J., and Cauli, O. (2018). A survey on sleep assessment methods. PeerJ 6:e4849. doi: 10.7717/peerj.4849
Ichtchenko, K., Hata, Y., Nguyen, T., Ullrich, B., Missler, M., Moomaw, C., et al. (1995). Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell 81, 435–443. doi: 10.1016/0092-8674(95)90396-8
Ichtchenko, K., Nguyen, T., and Südhof, T. C. (1996). Structures, alternative splicing, and neurexin binding of multiple neuroligins. J. Biol. Chem. 271, 2676–2682. doi: 10.1074/jbc.271.5.2676
Ingiosi, A. M., Schoch, H., Wintler, T., Singletary, K. G., Righelli, D., Roser, L. G., et al. (2019). Shank3 modulates sleep and expression of circadian transcription factors. eLife 8:e42819.
Ishikawa, A., Sakai, K., Maki, T., Mizuno, Y., Niimi, K., Oda, Y., et al. (2017). Investigation of sleep-wake rhythm in non-human primates without restraint during data collection. Exp. Anim. 66, 51–60. doi: 10.1538/expanim.16-0073
Ito, E., and Inoue, Y. (2015). [The International Classification of Sleep Disorders, third edition. American Academy of Sleep Medicine. Includes bibliographies and index]. Nihon Rinsho 73, 916–923.
Jackman, C., Horn, N. D., Molleston, J. P., and Sokol, D. K. (2009). Gene associated with seizures, autism, and hepatomegaly in an Amish girl. Pediatr. Neurol. 40, 310–313. doi: 10.1016/j.pediatrneurol.2008.10.013
Jaramillo, T. C., Speed, H. E., Xuan, Z., Reimers, J. M., Escamilla, C. O., Weaver, T. P., et al. (2017). Novel Shank3 mutant exhibits behaviors with face validity for autism and altered striatal and hippocampal function. Autism Res. 10, 42–65. doi: 10.1002/aur.1664
Jaramillo, T. C., Speed, H. E., Xuan, Z., Reimers, J. M., Liu, S., and Powell, C. M. (2016). Altered striatal synaptic function and abnormal behaviour in Shank3 Exon4-9 deletion mouse model of autism. Autism Res. 9, 350–375. doi: 10.1002/aur.1529
Jenni, O. G., and O’Connor, B. B. (2005). Children’s sleep: an interplay between culture and biology. Pediatrics 115, (Suppl. 1), 204–216. doi: 10.1542/peds.2004-0815b
Jennings, M., Prescott, M. J., Buchanan-Smith, H. M., Gamble, M. R., Gore, M., Hawkins, P., et al. (2009). Refinements in husbandry, care and common procedures for non-human primates: ninth report of the BVAAWF/FRAME/RSPCA/UFAW joint working group on refinement. Lab. Anim. 43, (Suppl. 1), 1–47. doi: 10.1258/la.2008.007143
Johnston, M. V., Ammanuel, S., O’Driscoll, C., Wozniak, A., Naidu, S., and Kadam, S. D. (2014). Twenty-four hour quantitative-EEG and in-vivo glutamate biosensor detects activity and circadian rhythm dependent biomarkers of pathogenesis in Mecp2 null mice. Front. Syst. Neurosci. 8:118. doi: 10.3389/fnsys.2014.00118
Kalume, F., Oakley, J. C., Westenbroek, R. E., Gile, J., de la Iglesia, H. O., Scheuer, T., et al. (2015). Sleep impairment and reduced interneuron excitability in a mouse model of Dravet Syndrome. Neurobiol. Dis. 77, 141–154. doi: 10.1016/j.nbd.2015.02.016
Kang, Y., Chu, C., Wang, F., and Niu, Y. (2019). CRISPR/Cas9-mediated genome editing in nonhuman primates. Dis. Models Mech. 12:dmm039982. doi: 10.1242/dmm.039982
Kazdoba, T. M., Leach, P. T., and Crawley, J. N. (2016). Behavioral phenotypes of genetic mouse models of autism. Genes Brain Behav. 15, 7–26. doi: 10.1111/gbb.12256
Kelly, J. M., Strecker, R. E., and Bianchi, M. T. (2012). Recent developments in home sleep-monitoring devices. ISRN Neurol. 2012:768794.
Kim, J., Gulati, T., and Ganguly, K. (2019). Competing roles of slow oscillations and delta waves in memory consolidation versus forgetting. Cell 179, 514.e3–526.e3.
Kim, S., Kim, H., Yim, Y. S., Ha, S., Atarashi, K., Tan, T. G., et al. (2017). Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532. doi: 10.1038/nature23910
Kimura, M., Müller-Preuss, P., Lu, A., Wiesner, E., Flachskamm, C., Wurst, W., et al. (2010). Conditional corticotropin-releasing hormone overexpression in the mouse forebrain enhances rapid eye movement sleep. Mol. Psychiatry 15, 154–165. doi: 10.1038/mp.2009.46
Klein, C. H., Brügner, G., Foerster, F., Müller, W., and Schweickhardt, A. (2000). The gap effect in pro-saccades and anti-saccades in psychometric schizotypes. Biol. Psychol. 55, 25–39. doi: 10.1016/s0301-0511(00)00062-4
Knappe, S., Pfarr, A. L., Petzoldt, J., Härtling, S., and Martini, J. (2020). Parental cognitions about sleep problems in infants: a systematic review. Front. Psychiatry 11:554221. doi: 10.3389/fpsyt.2020.554221
Köse, S., Yılmaz, H., Ocakğlu, F. T., and Özbaran, N. B. (2017). Sleep problems in children with autism spectrum disorder and intellectual disability without autism spectrum disorder. Sleep Med. 40, 69–77. doi: 10.1016/j.sleep.2017.09.021
Krakowiak, P., Goodlin-Jones, B., Hertz-Picciotto, I., Croen, L. A., and Hansen, R. L. (2008). Sleep problems in children with autism spectrum disorders, developmental delays, and typical development: a population-based study. J. Sleep Res. 17, 197–206. doi: 10.1111/j.1365-2869.2008.00650.x
Kumar, D., Dedic, N., Flachskamm, C., Voulé, S., Deussing, J. M., and Kimura, M. (2015). Cacna1c (Cav1.2) modulates electroencephalographic rhythm and rapid eye movement sleep recovery. Sleep 38, 1371–1380. doi: 10.5665/sleep.4972
Kumar, S., and Hedges, S. B. (1998). A molecular timescale for vertebrate evolution. Nature 392, 917–920. doi: 10.1038/31927
Laegreid, L., Kyllerman, M., Hedner, T., Hagberg, B., and Viggedahl, G. (1993). Benzodiazepine amplification of valproate teratogenic effects in children of mothers with absence epilepsy. Neuropediatrics 24, 88–92. doi: 10.1055/s-2008-1071520
Le Bras, A. (2019). Gene-edited SHANK-3 mutant macaques display autism behavior. Lab. Anim. 48, 236–236. doi: 10.1038/s41684-019-0366-y
Leblond, C. S., Nava, C., Polge, A., Gauthier, J., Huguet, G., Lumbroso, S., et al. (2014). Meta-analysis of SHANK mutations in autism spectrum disorders: a gradient of severity in cognitive impairments. PLoS Genet. 10:e1004580. doi: 10.1371/journal.pgen.1004580
Lee, J., Kim, D., and Shin, H. S. (2004). Lack of delta waves and sleep disturbances during non-rapid eye movement sleep in mice lacking alpha1G-subunit of T-type calcium channels. Proc. Natl. Acad. Sci. U.S.A. 101, 18195–18199. doi: 10.1073/pnas.0408089101
Levinson, J. N., and El-Husseini, A. (2005). Building excitatory and inhibitory synapses: balancing neuroligin partnerships. Neuron 48, 171–174. doi: 10.1016/j.neuron.2005.09.017
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., and Zhao, X. (2020). Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal. Transduct. Target. Therapy 5:1.
Li, Q., Loh, D. H., Kudo, T., Truong, D., Derakhshesh, M., Kaswan, Z. M., et al. (2015). Circadian rhythm disruption in a mouse model of Rett syndrome circadian disruption in RTT. Neurobiol. Dis. 77, 155–164. doi: 10.1016/j.nbd.2015.03.009
Li, W., Ma, L., Yang, G., and Gan, W. B. (2017). REM sleep selectively prunes and maintains new synapses in development and learning. Nat. Neurosci. 20, 427–437. doi: 10.1038/nn.4479
Li, Y., Hao, Y., Fan, F., and Zhang, B. (2018a). The role of microbiome in insomnia, circadian disturbance and depression. Front. Psychiatry 9:669. doi: 10.3389/fpsyt.2018.00669
Li, Y., Missig, G., Finger, B. C., Landino, S. M., Alexander, A. J., Mokler, E. L., et al. (2018b). Maternal and early postnatal immune activation produce dissociable effects on neurotransmission in mPFC–amygdala circuits. J. Neurosci. 38, 3358–3372. doi: 10.1523/jneurosci.3642-17.2018
Limoges, É, Bolduc, C., Berthiaume, C., Mottron, L., and Godbout, R. (2013). Relationship between poor sleep and daytime cognitive performance in young adults with autism. Res. Dev. Disabil. 34, 1322–1335. doi: 10.1016/j.ridd.2013.01.013
Lin, H.-C., Gean, P.-W., Wang, C.-C., Chan, Y.-H., and Chen, P. S. (2013). The amygdala excitatory/inhibitory balance in a valproate-induced rat autism model. PLos One 8:e55248. doi: 10.1371/journal.pone.0055248
Liu, J. J., Grace, K. P., Horner, R. L., Cortez, M. A., Shao, Y., and Jia, Z. (2017). Neuroligin 3 R451C mutation alters electroencephalography spectral activity in an animal model of autism spectrum disorders. Mol. Brain 10, 10–10.
Liu, X., Hubbard, J. A., Fabes, R. A., and Adam, J. B. (2006). Sleep disturbances and correlates of children with autism spectrum disorders. Child Psychiatry Hum. Dev. 37, 179–191. doi: 10.1007/s10578-006-0028-3
Liu, Z., Cai, Y., Liao, Z., Xu, Y., Wang, Y., Wang, Z., et al. (2019). Cloning of a gene-edited macaque monkey by somatic cell nuclear transfer. Natl. Sci. Rev. 6, 101–108. doi: 10.1093/nsr/nwz003
Liu, Z., Li, X., Zhang, J. T., Cai, Y. J., Cheng, T. L., Cheng, C., et al. (2016). Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature 530, 98–102. doi: 10.1038/nature16533
Lomeli, H. A., Pérez-Olmos, I., Talero-Gutiérrez, C., Moreno, C. B., González-Reyes, R., Palacios, L., et al. (2008). Sleep evaluation scales and questionaries: a review. Actas Esp Psiquiatr. 36, 50–59.
Lonart, G., Tang, X., Simsek-Duran, F., Machida, M., and Sanford, L. D. (2008). The role of active zone protein Rab3 interacting molecule 1 alpha in the regulation of norepinephrine release, response to novelty, and sleep. Neuroscience 154, 821–831. doi: 10.1016/j.neuroscience.2008.03.047
Lozoff, B., Wolf, A. W., and Davis, N. S. (1985). Sleep problems seen in pediatric practice. Pediatrics 75, 477–483.
Lu, H. C., Pollack, H., Lefante, J. J., Mills, A. A., and Tian, D. (2019). Altered sleep architecture, rapid eye movement sleep, and neural oscillation in a mouse model of human chromosome 16p11.2 microdeletion. Sleep 42:zsy253.
Lucey, B. P., McLeland, J. S., Toedebusch, C. D., Boyd, J., Morris, J. C., Landsness, E. C., et al. (2016). Comparison of a single-channel EEG sleep study to polysomnography. J. Sleep Res. 25, 625–635. doi: 10.1111/jsr.12417
Machado, C. J., and Bachevalier, J. (2003). Non-human primate models of childhood psychopathology: the promise and the limitations. J. Child Psychol. Psychiatry 44, 64–87. doi: 10.1111/1469-7610.00103
Machado, C. J., Whitaker, A. M., Smith, S. E. P., Patterson, P. H., and Bauman, M. D. (2015). Maternal immune activation in nonhuman primates alters social attention in juvenile offspring. Biol. Psychiatry 77, 823–832. doi: 10.1016/j.biopsych.2014.07.035
Malkova, N. V., Yu, C. Z., Hsiao, E. Y., Moore, M. J., and Patterson, P. H. (2012). Maternal immune activation yields offspring displaying mouse versions of the three core symptoms of autism. Brain Behav. Immun. 26, 607–616. doi: 10.1016/j.bbi.2012.01.011
Malow, B. A., Katz, T., Reynolds, A. M., Shui, A., Carno, M., Connolly, H. V., et al. (2016). Sleep difficulties and medications in children with autism spectrum disorders: a registry study. Pediatrics 137, (Suppl. 2), S98–S104.
Malow, B. A., Marzec, M. L., McGrew, S. G., Wang, L., Henderson, L. M., and Stone, W. L. (2006). Characterizing sleep in children with autism spectrum disorders: a multidimensional approach. Sleep 29, 1563–1571. doi: 10.1093/sleep/29.12.1563
Mantua, J., Gravel, N., and Spencer, R. M. (2016). Reliability of sleep measures from four personal health monitoring devices compared to research-based actigraphy and polysomnography. Sensors 16:646. doi: 10.3390/s16050646
Maski, K. P., and Kothare, S. V. (2013). Sleep deprivation and neurobehavioral functioning in children. Int. J. Psychophysiol. 89, 259–264. doi: 10.1016/j.ijpsycho.2013.06.019
McShane, B. B., Galante, R. J., Biber, M., Jensen, S. T., Wyner, A. J., and Pack, A. I. (2012). Assessing REM sleep in mice using video data. Sleep 35, 433–442. doi: 10.5665/sleep.1712
Meltzer, L. J., Hiruma, L. S., Avis, K., Montgomery-Downs, H., and Valentin, J. (2015). Comparison of a commercial accelerometer with polysomnography and actigraphy in children and adolescents. Sleep 38, 1323–1330. doi: 10.5665/sleep.4918
Meltzer, L. J., Wong, P., Biggs, S. N., Traylor, J., Kim, J. Y., Bhattacharjee, R., et al. (2016). Validation of actigraphy in middle childhood. Sleep 39, 1219–1224. doi: 10.5665/sleep.5836
Mingrone, A., Kaffman, A., and Kaffman, A. (2020). The promise of automated home-cage monitoring in improving translational utility of psychiatric research in rodents. Front. Neurosci. 14:618593. doi: 10.3389/fnins.2020.618593
Missig, G., McDougle, C. J., and Carlezon, W. A. (2020). Sleep as a translationally-relevant endpoint in studies of autism spectrum disorder (ASD). Neuropsychopharmacology 45, 90–103. doi: 10.1038/s41386-019-0409-5
Missig, G., Mokler, E. L., Robbins, J. O., Alexander, A. J., McDougle, C. J., and Carlezon, W. A. (2018). Perinatal immune activation produces persistent sleep alterations and epileptiform activity in male mice. Neuropsychopharmacology 43, 482–491. doi: 10.1038/npp.2017.243
Mizuno, Y., Takeshita, H., and Matsuzawa, T. (2006). Behavior of infant chimpanzees during the night in the first 4 months of life: smiling and suckling in relation to behavioral state. Infancy 9, 221–240. doi: 10.1207/s15327078in0902_7
Montgomery-Downs, H. E., Insana, S. P., and Bond, J. A. (2012). Movement toward a novel activity monitoring device. Sleep Breath 16, 913–917. doi: 10.1007/s11325-011-0585-y
Morgenthaler, T., Alessi, C., Friedman, L., Owens, J., Kapur, V., Boehlecke, B., et al. (2007). Practice parameters for the use of actigraphy in the assessment of sleep and sleep disorders: an update for 2007. Sleep 30, 519–529. doi: 10.1093/sleep/30.4.519
Mutluer, T., Karakoc Demirkaya, S., and Abali, O. (2016). Assessment of sleep problems and related risk factors observed in Turkish children with Autism spectrum disorders. Autism Res. 9, 536–542. doi: 10.1002/aur.1542
Mychasiuk, R., Richards, S., Nakahashi, A., Kolb, B., and Gibb, R. (2012). Effects of rat prenatal exposure to valproic acid on behaviour and neuro-anatomy. Dev. Neurosci. 34, 268–276. doi: 10.1159/000341786
Nicolini, C., and Fahnestock, M. (2018). The valproic acid-induced rodent model of autism. Exp. Neurol. 299(Pt A), 217–227. doi: 10.1016/j.expneurol.2017.04.017
Niu, Y., Shen, B., Cui, Y., Chen, Y., Wang, J., Wang, L., et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843. doi: 10.1016/j.cell.2014.01.027
Nunn, C. L., and Samson, D. R. (2018). Sleep in a comparative context: investigating how human sleep differs from sleep in other primates. Am. J. Phys. Anthropol. 166, 601–612. doi: 10.1002/ajpa.23427
O’Donnell, S., Beaven, C. M., and Driller, M. W. (2018). From pillow to podium: a review on understanding sleep for elite athletes. Nat. Sci. Sleep 10, 243–253. doi: 10.2147/nss.s158598
Ongür, D., and Price, J. L. (2000). The organization of networks within the orbital and medial prefrontal cortex of rats, monkeys and humans. Cereb. Cortex 10, 206–219. doi: 10.1093/cercor/10.3.206
Orefice, L. L. (2019). Outside-in: rethinking the etiology of autism spectrum disorders. Science 366, 45–46. doi: 10.1126/science.aaz3880
Owens, J. A., and Mindell, J. A. (2011). Pediatric insomnia. Pediatr. Clin. N. Am. 58, 555–569. doi: 10.1016/j.pcl.2011.03.011
Owens, J. A., Spirito, A., and McGuinn, M. (2000). The Children’s Sleep Habits Questionnaire (CSHQ): psychometric properties of a survey instrument for school-aged children. Sleep 23, 1043–1051.
Pack, A. I., Galante, R. J., Maislin, G., Cater, J., Metaxas, D., Lu, S., et al. (2007). Novel method for high-throughput phenotyping of sleep in mice. Physiol. Genom. 28, 232–238. doi: 10.1152/physiolgenomics.00139.2006
Papale, L. A., Paul, K. N., Sawyer, N. T., Manns, J. R., Tufik, S., and Escayg, A. (2010). Dysfunction of the Scn8a voltage-gated sodium channel alters sleep architecture, reduces diurnal corticosterone levels, and enhances spatial memory. J. Biol. Chem. 285, 16553–16561. doi: 10.1074/jbc.m109.090084
Parker, K. J., Garner, J. P., Oztan, O., Tarara, E. R., Li, J., Sclafani, V., et al. (2018). Arginine vasopressin in cerebrospinal fluid is a marker of sociality in nonhuman primates. Sci. Transl. Med. 10:eaam9100. doi: 10.1126/scitranslmed.aam9100
Peñgarikano, O., Abrahams, B. S., Herman, E. I., Winden, K. D., Gdalyahu, A., Dong, H., et al. (2011). Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 147, 235–246. doi: 10.1016/j.cell.2011.08.040
Phelan, K., and McDermid, H. E. (2012). The 22q13.3 deletion syndrome (phelan-mcdermid syndrome). Mol. Syndromol. 2, 186–201.
Phillips, K. A., Bales, K. L., Capitanio, J. P., Conley, A., Czoty, P. W., t Hart, B. A., et al. (2014). Why primate models matter. Am. J. Primatol. 76, 801–827. doi: 10.1002/ajp.22281
Prechtl, H. F. R. (1974). The behavioural states of the newborn infant (a review). Brain Res. 76, 185–212. doi: 10.1016/0006-8993(74)90454-5
Qin, D. D., Feng, S. F., Zhang, F. Y., Wang, N., Sun, W. J., Zhou, Y., et al. (2020). Potential use of actigraphy to measure sleep in monkeys: comparison with behavioral analysis from videography. Zool. Res. 41, 437–443.
Qiu, P., Jiang, J., Liu, Z., Cai, Y., Huang, T., Wang, Y., et al. (2019). BMAL1 knockout macaque monkeys display reduced sleep and psychiatric disorders. Natl. Sci. Rev. 6, 87–100. doi: 10.1093/nsr/nwz002
Rachalski, A., Authier, S., Bassett, L., Pouliot, M., Tremblay, G., and Mongrain, V. (2014). Sleep electroencephalographic characteristics of the Cynomolgus monkey measured by telemetry. J. Sleep Res. 23, 619–627. doi: 10.1111/jsr.12189
Radyushkin, K., Hammerschmidt, K., Boretius, S., Varoqueaux, F., El-Kordi, A., Ronnenberg, A., et al. (2009). Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 8, 416–425. doi: 10.1111/j.1601-183x.2009.00487.x
Rawashdeh, O., de Borsetti, N. H., Roman, G., and Cahill, G. M. (2007). Melatonin suppresses nighttime memory formation in zebrafish. Science 318, 1144–1146. doi: 10.1126/science.1148564
Reite, M. L., Rhodes, J. M., Kavan, E., and Adey, W. R. (1965). Normal sleep patterns in macaque monkey. Arch. Neurol. 12, 133–144. doi: 10.1001/archneur.1965.00460260023003
Richdale, A. L. (1999). Sleep problems in autism: prevalence, cause, and intervention. Dev. Med. Child Neurol. 41, 60–66. doi: 10.1111/j.1469-8749.1999.tb00012.x
Roberts, A. C., and Clarke, H. F. (2019). Why we need nonhuman primates to study the role of ventromedial prefrontal cortex in the regulation of threat- and reward-elicited responses. Proc. Natl. Acad. Sci. 116, 26297–26304. doi: 10.1073/pnas.1902288116
Roffwarg, H. P., Muzio, J. N., and Dement, W. C. (1966). Ontogenetic development of the human sleep-dream cycle. Science 152, 604–619. doi: 10.1126/science.152.3722.604
Russell, W. M. S., and Burch, R. L. (1959). The Principles of Humane Experimental Technique. London: Methuen.
Rutishauser, U., Mamelak, A. N., and Adolphs, R. (2015). The primate amygdala in social perception - insights from electrophysiological recordings and stimulation. Trends Neurosci. 38, 295–306. doi: 10.1016/j.tins.2015.03.001
Sacai, H., Sakoori, K., Konno, K., Nagahama, K., Suzuki, H., Watanabe, T., et al. (2020). Autism spectrum disorder-like behavior caused by reduced excitatory synaptic transmission in pyramidal neurons of mouse prefrontal cortex. Nat. Commun. 11:5140.
Sadeh, A. (2011). The role and validity of actigraphy in sleep medicine: an update. Sleep Med. Rev. 15, 259–267. doi: 10.1016/j.smrv.2010.10.001
Sadeh, A. (2015). Iii. Sleep assessment methods. Monogr. Soc. Res. Child Dev. 80, 33–48. doi: 10.1111/mono.12143
Sateia, M. J. (2014). International classification of sleep disorders-third edition: highlights and modifications. Chest 146, 1387–1394. doi: 10.1378/chest.14-0970
Sato, K., Oiwa, R., Kumita, W., Henry, R., Sakuma, T., Ito, R., et al. (2016). Generation of a nonhuman primate model of severe combined immunodeficiency using highly efficient genome editing. Cell Stem Cell 19, 127–138. doi: 10.1016/j.stem.2016.06.003
Satterstrom, F. K., Kosmicki, J. A., Wang, J., Breen, M. S., De Rubeis, S., An, J. Y., et al. (2020). Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568.e23–584.e23.
Sawangjit, A., Oyanedel, C. N., Niethard, N., Salazar, C., Born, J., and Inostroza, M. (2018). The hippocampus is crucial for forming non-hippocampal long-term memory during sleep. Nature 564, 109–113. doi: 10.1038/s41586-018-0716-8
Schneider, T., Roman, A., Basta-Kaim, A., Kubera, M., Budziszewska, B., Schneider, K., et al. (2008). Gender-specific behavioral and immunological alterations in an animal model of autism induced by prenatal exposure to valproic acid. Psychoneuroendocrinology 33, 728–740. doi: 10.1016/j.psyneuen.2008.02.011
Schreck, K. A., Mulick, J. A., and Smith, A. F. (2004). Sleep problems as possible predictors of intensified symptoms of autism. Res. Dev. Disabil. 25, 57–66. doi: 10.1016/j.ridd.2003.04.007
Schumann, C., Vargas, M., Lee, A., Amaral, D., and Adolphs, R. (2016). Living Without an Amygdala. New York, NY: Guilford Publications.
Schwichtenberg, A. J., Choe, J., Kellerman, A., Abel, E. A., and Delp, E. J. (2018). Pediatric videosomnography: can signal/video processing distinguish sleep and wake states? Front. Pediatr. 6:158. doi: 10.3389/fped.2018.00158
Seelke, A. M., and Blumberg, M. S. (2008). The microstructure of active and quiet sleep as cortical delta activity emerges in infant rats. Sleep 31, 691–699. doi: 10.1093/sleep/31.5.691
Seok, B. S., Bélanger-Nelson, E., Provost, C., Gibbs, S., and Mongrain, V. (2018). The effect of Neuroligin-2 absence on sleep architecture and electroencephalographic activity in mice. Mol. Brain 11:52.
Short, M. A., Arora, T., Gradisar, M., Taheri, S., and Carskadon, M. A. (2017). How many sleep diary entries are needed to reliably estimate adolescent sleep? Sleep 40:zsx006.
Short, M. A., Gradisar, M., Lack, L. C., Wright, H. R., and Chatburn, A. (2013). Estimating adolescent sleep patterns: parent reports versus adolescent self-report surveys, sleep diaries, and actigraphy. Nat. Sci. Sleep 5:23. doi: 10.2147/nss.s38369
Short, S. J., Lubach, G. R., Karasin, A. I., Olsen, C. W., Styner, M., Knickmeyer, R. C., et al. (2010). Maternal influenza infection during pregnancy impacts postnatal brain development in the rhesus monkey. Biol. Psychiatry 67, 965–973. doi: 10.1016/j.biopsych.2009.11.026
Smith, S. E., Li, J., Garbett, K., Mirnics, K., and Patterson, P. H. (2007). Maternal immune activation alters fetal brain development through interleukin-6. J. Neurosci. 27, 10695–10702. doi: 10.1523/jneurosci.2178-07.2007
Song, J. Y., Ichtchenko, K., Südhof, T. C., and Brose, N. (1999). Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc. Natl. Acad. Sci. U.S.A. 96, 1100–1105. doi: 10.1073/pnas.96.3.1100
Souders, M. C., Mason, T. B. A., Valladares, O., Bucan, M., Levy, S. E., Mandell, D. S., et al. (2009). Sleep behaviors and sleep quality in children with autism spectrum disorders. Sleep 32, 1566–1578.
Stepnowsky, C., Levendowski, D., Popovic, D., Ayappa, I., and Rapoport, D. M. (2013). Scoring accuracy of automated sleep staging from a bipolar electroocular recording compared to manual scoring by multiple raters. Sleep Med. 14, 1199–1207. doi: 10.1016/j.sleep.2013.04.022
Sugranyes, G., Kyriakopoulos, M., Corrigall, R., Taylor, E., and Frangou, S. (2011). Autism spectrum disorders and schizophrenia: meta-analysis of the neural correlates of social cognition. PLoS One 6:e25322. doi: 10.1371/journal.pone.0025322
Tatsuki, F., Sunagawa, G. A., Shi, S., Susaki, E. A., Yukinaga, H., Perrin, D., et al. (2016). Involvement of Ca(2+)-dependent hyperpolarization in sleep duration in mammals. Neuron 90, 70–85. doi: 10.1016/j.neuron.2016.02.032
Thomas, A. M., Schwartz, M. D., Saxe, M. D., and Kilduff, T. S. (2017). Cntnap2 knockout rats and mice exhibit epileptiform activity and abnormal sleep-wake physiology. Sleep 40:zsw026.
Toon, E., Davey, M. J., Hollis, S. L., Nixon, G. M., Horne, R. S., and Biggs, S. N. (2016). Comparison of commercial wrist-based and smartphone accelerometers, actigraphy, and PSG in a clinical cohort of children and adolescents. J. Clin. Sleep Med. 12, 343–350. doi: 10.5664/jcsm.5580
Tsujino, N., Nakatani, Y., Seki, Y., Nakasato, A., Nakamura, M., Sugawara, M., et al. (2007). Abnormality of circadian rhythm accompanied by an increase in frontal cortex serotonin in animal model of autism. Neurosci. Res. 57, 289–295. doi: 10.1016/j.neures.2006.10.018
Tu, Z., Zhao, H., Li, B., Yan, S., Wang, L., Tang, Y., et al. (2019). CRISPR/Cas9-mediated disruption of SHANK3 in monkey leads to drug-treatable autism-like symptoms. Hum. Mol. Genet. 28, 561–571. doi: 10.1093/hmg/ddy367
Tudor, M. E., Hoffman, C. D., and Sweeney, D. P. (2012). Children with autism: sleep problems and symptom severity. Focus Autism Other Dev. Disabil. 27, 254–262. doi: 10.1177/1088357612457989
Uren, J., Richdale, A. L., Cotton, S. M., and Whitehouse, A. J. O. (2019). Sleep problems and anxiety from 2 to 8 years and the influence of autistic traits: a longitudinal study. Eur. Child Adolesc. Psychiatry 28, 1117–1127. doi: 10.1007/s00787-019-01275-y
Uylings, H. B., Groenewegen, H. J., and Kolb, B. (2003). Do rats have a prefrontal cortex? Behav. Brain Res. 146, 3–17. doi: 10.1016/j.bbr.2003.09.028
Vandekerckhove, M., and Cluydts, R. (2010). The emotional brain and sleep: an intimate relationship. Sleep Med. Rev. 14, 219–226. doi: 10.1016/j.smrv.2010.01.002
Varoqueaux, F., Jamain, S., and Brose, N. (2004). Neuroligin 2 is exclusively localized to inhibitory synapses. Eur. J. Cell. Biol. 83, 449–456. doi: 10.1078/0171-9335-00410
Vegué, M., and Roxin, A. (2015). Program No. 94.11, Neuroscience Meeting Planner. Chicago, IL: Society for Neuroscience.
Verhoeff, M. E., Blanken, L. M. E., Kocevska, D., Mileva-Seitz, V. R., Jaddoe, V. W. V., White, T., et al. (2018). The bidirectional association between sleep problems and autism spectrum disorder: a population-based cohort study. Mol. Autism 9:8.
von Ziegler, L., Sturman, O., and Bohacek, J. (2021). Big behavior: challenges and opportunities in a new era of deep behavior profiling. Neuropsychopharmacology 46, 33–44. doi: 10.1038/s41386-020-0751-7
Vyazovskiy, V. V., Kopp, C., Wigger, E., Jones, M. E., Simpson, E. R., and Tobler, I. (2006). Sleep and rest regulation in young and old oestrogen-deficient female mice. J. Neuroendocrinol. 18, 567–576. doi: 10.1111/j.1365-2826.2006.01452.x
Walker, M. P., and Stickgold, R. (2006). Sleep, memory, and plasticity. Annu. Rev. Psychol. 57, 139–166.
Watson, K. K., and Platt, M. L. (2012). Of mice and monkeys: using non-human primate models to bridge mouse- and human-based investigations of autism spectrum disorders. J. Neurodev. Disord. 4:21.
Weber, F., and Dan, Y. (2016). Circuit-based interrogation of sleep control. Nature 538, 51–59. doi: 10.1038/nature19773
Weber, F., Do, J. P. Hoang, Chung, S., Beier, K. T., Bikov, M., Saffari Doost, M., et al. (2018). Regulation of REM and Non-REM sleep by periaqueductal GABAergic neurons. Nat. Commun. 9:354.
Weiner, D. J., Wigdor, E. M., Ripke, S., Walters, R. K., Kosmicki, J. A., Grove, J., et al. (2017). Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat. Genet. 49, 978–985. doi: 10.1038/ng.3863
Weir, R. K., Forghany, R., Smith, S. E., Patterson, P. H., McAllister, A. K., Schumann, C. M., et al. (2015). Preliminary evidence of neuropathology in nonhuman primates prenatally exposed to maternal immune activation. Brain Behav. Immun. 48, 139–146. doi: 10.1016/j.bbi.2015.03.009
Werner, H., Molinari, L., Guyer, C., and Jenni, O. G. (2008). Agreement rates between actigraphy, diary, and questionnaire for children’s sleep patterns. Archiv. Pediatr. Adolesc. Med. 162, 350–358. doi: 10.1001/archpedi.162.4.350
Wiggs, L. (2001). Sleep problems in children with developmental disorders. J. R. Soc. Med. 94, 177–179.
Williams, J. A., Zimmerman, F. J., and Bell, J. F. (2013). Norms and trends of sleep time among US children and adolescents. JAMA Pediatr. 167, 55–60. doi: 10.1001/jamapediatrics.2013.423
Wintler, T., Schoch, H., Frank, M. G., and Peixoto, L. (2020). Sleep, brain development, and autism spectrum disorders: insights from animal models. J. Neurosci. Res. 98, 1137–1149. doi: 10.1002/jnr.24619
Wöhr, M., Silverman, J. L., Scattoni, M. L., Turner, S. M., Harris, M. J., Saxena, R., et al. (2013). Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav. Brain Res. 251, 50–64. doi: 10.1016/j.bbr.2012.07.024
Xie, L., Kang, H., Xu, Q., Chen, M. J., Liao, Y., Thiyagarajan, M., et al. (2013). Sleep drives metabolite clearance from the adult brain. Science 342, 373–377. doi: 10.1126/science.1241224
Zepelin, H., Whitehead, W. E., and Rechtschaffen, A. (1972). Aging and sleep in the albino rat. Behav. Biol. 7, 65–74. doi: 10.1016/s0091-6773(72)80189-5
Zhang, X. L., Pang, W., Hu, X. T., Li, J. L., Yao, Y. G., and Zheng, Y. T. (2014). Experimental primates and non-human primate (NHP) models of human diseases in China: current status and progress. Dongwuxue Yanjiu 35, 447–464.
Zhang, Z., Adamo, K. B., Ogden, N., Goldfield, G. S., Okely, A. D., Kuzik, N., et al. (2021). Longitudinal correlates of sleep duration in young children. Sleep Med. 78, 128–134. doi: 10.1016/j.sleep.2020.12.023
Zhao, H., Jiang, Y.-H., and Zhang, Y. Q. (2018). Modeling autism in non-human primates: opportunities and challenges. Autism Res. 11, 686–694. doi: 10.1002/aur.1945
Zhao, H., Tu, Z., Xu, H., Yan, S., Yan, H., Zheng, Y., et al. (2017). Altered neurogenesis and disrupted expression of synaptic proteins in prefrontal cortex of SHANK3-deficient non-human primate. Cell Res. 27, 1293–1297. doi: 10.1038/cr.2017.95
Zhao, H., Wang, Q., Yan, T., Zhang, Y., Xu, H.-J., Yu, H.-P., et al. (2019). Maternal valproic acid exposure leads to neurogenesis defects and autism-like behaviors in non-human primates. Transl. Psychiatry 9:267.
Zhao, J., Lai, L., Ji, W., and Zhou, Q. (2019). Genome editing in large animals: current status and future prospects. Natl. Sci. Rev. 6, 402–420. doi: 10.1093/nsr/nwz013
Zhou, L., Bryant, C. D., Loudon, A., Palmer, A. A., Vitaterna, M. H., and Turek, F. W. (2014). The circadian clock gene Csnk1e regulates rapid eye movement sleep amount, and nonrapid eye movement sleep architecture in mice. Sleep 37, 785–793. doi: 10.5665/sleep.3590
Keywords: autism, sleep, non-human primate, brain development, animal model
Citation: Feng S, Huang H, Wang N, Wei Y, Liu Y and Qin D (2021) Sleep Disorders in Children With Autism Spectrum Disorder: Insights From Animal Models, Especially Non-human Primate Model. Front. Behav. Neurosci. 15:673372. doi: 10.3389/fnbeh.2021.673372
Received: 27 February 2021; Accepted: 16 April 2021;
Published: 20 May 2021.
Edited by:
Rafael S. Maior, University of Brasilia, BrazilReviewed by:
Kyungmin Lee, Kyungpook National University, South KoreaHiroki Toyoda, Osaka University, Japan
Copyright © 2021 Feng, Huang, Wang, Wei, Liu and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Liu, bGl1eXVuQGV0eXkuY24=; Dongdong Qin, cWluZG9uZzEwOEAxNjMuY29t
†These authors have contributed equally to this work
 Shufei Feng
Shufei Feng Haoyu Huang1†
Haoyu Huang1† Yuanyuan Wei
Yuanyuan Wei Dongdong Qin
Dongdong Qin