- 1Ear Institute, University College London, London, United Kingdom
- 2The Francis Crick Institute, London, United Kingdom
- 3Centre for Mathematics and Physics in the Life Sciences and Experimental Biology (CoMPLEX), University College London, London, United Kingdom
- 4Department of Cell and Developmental Biology, University College London, London, United Kingdom
A vital task for every organism is not only to decide what to do but also when to do it. For this reason, “circadian clocks” have evolved in virtually all forms of life. Conceptually, circadian clocks can be divided into two functional domains; an autonomous oscillator creates a ~24 h self-sustained rhythm and sensory machinery interprets external information to alter the phase of the autonomous oscillation. It is through this simple design that variations in external stimuli (for example, daylight) can alter our sense of time. However, the clock’s simplicity ends with its basic concept. In metazoan animals, multiple external and internal stimuli, from light to temperature and even metabolism have been shown to affect clock time. This raises the fundamental question of cue integration: how are the many, and potentially conflicting, sources of information combined to sense a single time of day? Moreover, individual stimuli, are often detected through various sensory pathways. Some sensory cells, such as insect chordotonal neurons, provide the clock with both temperature and mechanical information. Adding confusion to complexity, there seems to be not only one central clock in the animal’s brain but numerous additional clocks in the body’s periphery. It is currently not clear how (or if) these “peripheral clocks” are synchronized to their central counterparts or if both clocks “tick” independently from one another. In this review article, we would like to leave the comfort zones of conceptual simplicity and assume a more holistic perspective of circadian clock function. Focusing on recent results from Drosophila melanogaster we will discuss some of the sensory, and computational, challenges organisms face when keeping track of time.
Introduction
A good decision is not absolute. It varies depending on context. Foraging for food can be a good or bad decision depending on the presence of a looming predator. The key is to optimize behavior for the current context. But what if the context is in perpetual flux? Owing to the spin of our planet on its longitudinal axis, the vast majority of life on earth exists in 24-h cycles of environmental change. The sun rises and sets. Dawn comes and goes. Temperatures change, and both predator and prey alike sleep and wake. Thus, good decisions depend as much on when to do, as they do on what to do.
This critical importance of time for decision-making necessitates the existence of internal clocks. These “circadian” oscillators give temporal structure to behavior. The circadian system of Drosophila melanogaster presents a remarkable tool to investigate how external environmental changes can impact internal timekeeping that best prepares an organism for time-appropriate tasks. The circadian clock has a set period (i.e., one full cycles takes ~24 h) and its time, or phase, can be adjusted by incoming sensory information. This flexibility is paramount to a system that is orchestrating numerous behavioral and physiological processes while external stimuli are constantly changing.
In this review article we set out to highlight the complexities of the Drosophila melanogaster circadian clock, in which “decisions” must be made with regard to external environmental cues. These fundamental timing decisions are made by individual neurons in the context of multimodal sensory information. By discussing the complexities that exist on the molecular, cellular, network and behavioral level, we propose computational approaches that may be fruitful to gain further insight into the nuances of circadian systems.
The Molecular Clock
To understand how the wider circadian system keeps time, we must first begin with the oscillatory building blocks that comprise it. Time is computed at the level of individual cells and integral to this cellular timekeeping is a molecular clock that is driven by the autonomous oscillations of so called “clock genes”. The autonomous oscillators are driven by a series of molecular transcriptional/translational feedback loops (TTFL) of which components autoregulate their own expression (Figure 1), reviewed in Hardin (2011).
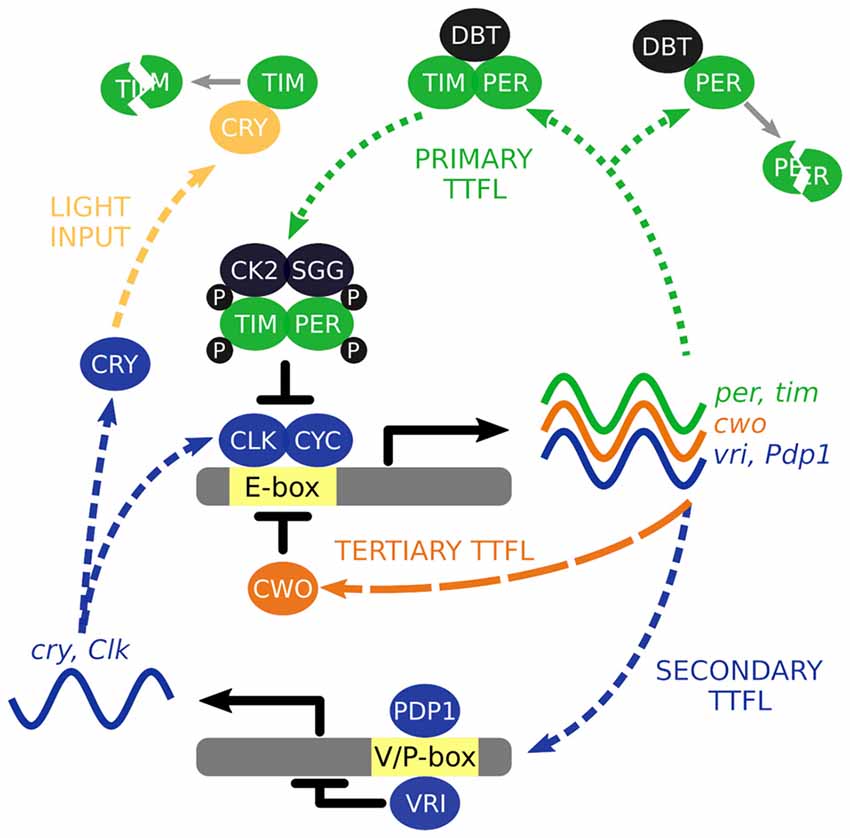
Figure 1. The interlocked core transcription/translation feedback loops (TTFL) of Drosophila melanogaster. The primary TTFL is initiated by the CLK-CYC mediated transcription of the per and tim genes. The PER protein is inherently unstable and DBT will direct it for degradation unless PER is stabilized by TIM. PER and TIM are phosphorylated (P) by CK2 and SGG to negatively regulate their own expression by repressing CLK-CYC activity. VRI and PDP1 are also produced by CLK-CYC activity, each of which have potentially opposing effects and offset timing to modulate Clk and cry transcription. Light activation of CRY leads to the degradation of TIM acting as a molecular reset of the TTFL cycling. CWO is a part of third TTFL that also acts as a repressor of CLK-CYC activity.
The molecular clock starts with the transcription factors, CLOCK (CLK) and CYCLE (CYC), activating transcription of the primary TTFL genes, per and tim, through binding to E-box regions in their gene promoters (Hao et al., 1997; Allada et al., 1998; Darlington et al., 1998; Rutila et al., 1998). Transcription occurs between ~ZT4 to ~ZT18 (where ZT0 is lights on and ZT12 is lights off) and translation of these mRNAs generate the protein products PERIOD (PER) and TIMELESS (TIM). The PER protein is inherently unstable, and is rapidly targeted for proteasomal degradation by the kinase DOUBLETIME (DBT; Kloss et al., 1998, 2001; Price et al., 1998). However, the PER-DBT dimer can be stabilized through dimerization with TIM allowing the PER-TIM-DBT complex to accumulate in the cytosol, around 6–8 h after per and tim transcription activation or ~ZT12 (Curtin et al., 1995; Gekakis et al., 1995; Price et al., 1995; Zeng et al., 1996).
Phosphorylation of PER and TIM by the kinases, CK2 and SGG (Martinek et al., 2001; Lin et al., 2002; Akten et al., 2003), initiates nuclear accumulation of PER and TIM (Nawathean and Rosbash, 2004; Top et al., 2016). Inside the nucleus, the active form of PER inhibits CLK-CYC activation of per and tim transcription between ~ZT18 to ~ZT4 (Lee et al., 1998, 1999; Bae et al., 2000; Menet et al., 2010). Consequently, PER and TIM concentrations decline in the cytosol, reducing nuclear abundance of PER and TIM, and ultimately removing the inhibition of CLK-CYC activity. The molecular cycle then starts again, circa 24 h later.
The primary TTFL is bolstered by additional interlocked loops. In a second TTFL CLK-CYC also activates transcription of vrille and Pdp1ɛ/δ between ~ZT4 and ZT16 (Blau and Young, 1999; Cyran et al., 2003). VRILLE (VRI) protein concentration peaks several hours before PDP1ɛ/δ (~ZT14 vs. ~ZT18) and both act at VRI/PDP1-boxes to modulate Clk and cry transcription (Cyran et al., 2003; Glossop et al., 2003). PER-TIM-DBT activity from the primary TTFL inhibits CLK-CYC activity interlinking the two loops. The cry gene encodes a blue-light photoreceptor that upon photic activation promotes TIM degradation essentially acting as a molecular reset switch of the primary TTFL (Emery et al., 1998; Stanewsky et al., 1998). In the third TTFL, CLK-CYC promotes expression of clockwork orange that competitively binds to the same E-box regions to negatively regulate CLK-CYC mediated transcription (Kadener et al., 2007; Lim et al., 2007; Matsumoto et al., 2007).
The Central Clock—A Network of Coupled Oscillators
If the oscillating proteins of the TTFLs are the gears that direct timekeeping, the central pacemaker neurons are analogous to a synchronizer that sets a coherent time in context with the external environment. The architecture of this central pacemaker in flies consist of ~150 neurons comprising distinct subgroups identified in the brain through cytological staining of clock genes (Figure 2; Helfrich-Förster and Homberg, 1993; Helfrich-Förster, 1995). The subgroups are named after their morphology and anatomical location; each brain hemisphere has four small ventral lateral neurons (s-LNv) that express the pigment dispersing factor (PDF), a fifth PDF-negative s-LNv, four large PDF-positive ventral lateral neurons (l-LNv), six dorsal lateral neurons (LNd), 17 group one dorsal neurons (DN1), two group two dorsal neurons (DN2), ~40 group three dorsal neurons (DN3) and three lateral posterior neurons (LPN; Helfrich-Förster et al., 2007; Nitabach and Taghert, 2008; Schubert et al., 2018).
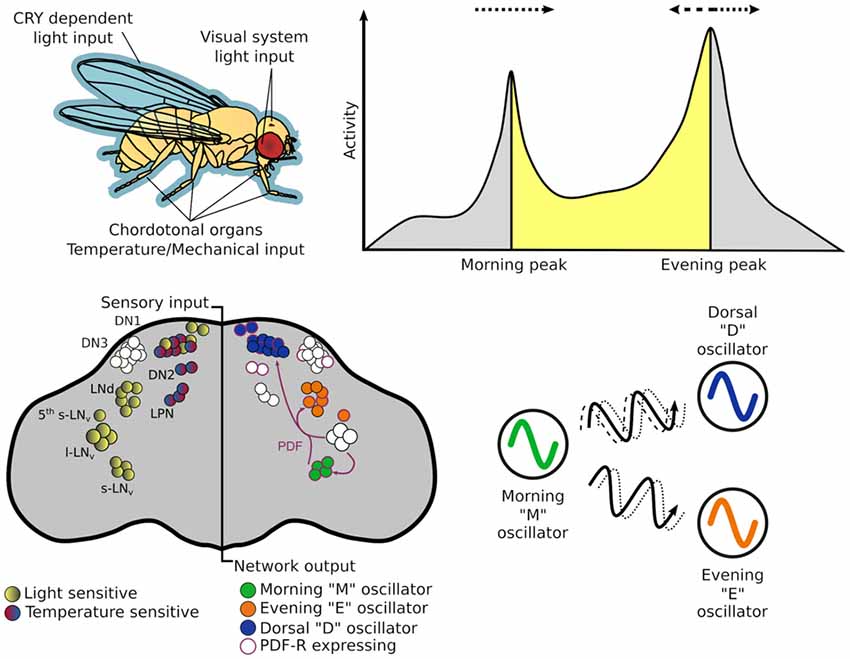
Figure 2. The circadian system of Drosophila melanogaster from input stimulus to output behavior. Light information may enter the circadian system through the cuticle of the fly to directly act on CRY, which is expressed in many central and peripheral clock cells. Alternatively, it may enter via sensory cells of the visual systems—compound eye, H-B eyelets or the ocelli that make direct—or indirect — synaptic contact with central clock neurons. While flies have many thermo - and mechano-sensing organs, chordotonal organs (ChOs) have been directly implicated in communicating both temperature and mechanical stimuli to the circadian system. The neuronal populations of central pacemakers differ in their sensitivity to external stimuli as well as their neurochemical and electrical connectivity. Pacemaker neurons expressing CRY preferentially entrain to light cues while others not expressing CRY have demonstrated preference for temperature cues. Neurochemical differences can denote distinct functional groups—for example the M oscillator neurons express and release pigment dispersing factor (PDF) that can lead to phase changes in molecular oscillations of cells expressing PDF-R. This can result in phase advances or delays of downstream processes that in turn can advance or delay behavioral outputs.
The dual oscillator model, originally hypothesized for nocturnal rodents (Pittendrigh and Daan, 1976), has historically been used to also describe the Drosophila crepuscular activity patterns, proposing separate autonomous oscillators that control activity peaks observed at dusk and dawn. The PDF+ LNv are required for the fly’s morning activity peak (and free-running rhythms; Renn et al., 1999; Grima et al., 2004; Stoleru et al., 2004), while the LNd and 5th PDF- LNv are important for the evening peak (Grima et al., 2004), thus, these two groups are labeled the morning (M) and evening (E) oscillators respectively. In practice these designations are too strict as the roles and hierarchy of some cellular oscillators change under different environmental conditions (Rieger et al., 2006, 2009; Murad et al., 2007; Picot et al., 2007).
Current pacemaker models depict a variable—both neurochemically and electrically—coupled neuronal network that differentially responds to incoming multimodal stimuli (Yao and Shafer, 2014; Schlichting et al., 2016; Yao et al., 2016) reviewed in detail in Hermann-Luibl and Helfrich-Förster (2015); Top and Young (2018).
Peripheral Clocks
In addition to the central pacemaker clocks, circadian oscillators also exist in several tissues outside the central nervous system and are likely to regulate organ or tissue specific functions. Peripheral clocks have largely been characterized through clock TTFL protein rhythms identified either by immunohistochemistry or by luciferase reporter expression being driven by clock gene promoters. Located in a number of tissues, peripheral clocks are heterogeneous in both their molecular machinery and their relationship to the central clock (Ito and Tomioka, 2016).
At the molecular level, CRY appears to act not only as a photoreceptor but also as a core component of the primary TTFL. In some peripheral clocks of cry mutants—in which CRY normally fulfils both roles—light entrainment in lost, however they also lose molecular oscillations of per and tim (Ivanchenko et al., 2001; Krishnan et al., 2001). In these peripheral clocks it appears CRY not only integrates photic information into the clock, but it also plays a role in driving oscillations of the clock, possibly acting as a repressor (Collins et al., 2006). Alternatively, CRY may act exclusively as a photoreceptor with persistent free running rhythms observed in certain tissues of cry mutants. This indicates that in these peripheral clocks CRY is not required to drive oscillations of the clock (Ito et al., 2008).
Phase relations between central clocks and peripheral clocks are also heterogeneous. For example, peripheral clocks in the Malpighian tubules and chemosensory sensilla can directly entrain to external stimuli and maintain entrained rhythms even without information from central clock neurons (Hege et al., 1997; Krishnan et al., 1999). A different phase relationship is observed between oenocytes and the central clock. PDF release from clock neurons into the hemeolymph can set the phase of the distally located oenocytes, which control rhythmic release of mating pheromones (Krupp et al., 2013). The different phase relationships identified between central and peripheral clocks add yet another layer of complexity to the timekeeping system.
Locomotor Activity Rhythms
Circadian rhythms exist at the cellular and network levels, yet it is the appropriate timing of behaviors that makes this system adaptive for the organism. Behavioral output of a circadian clock is typically measured as the organism’s activity pattern; this is no different for Drosophila. Simple infra-red beam-breaking monitors record the activity of individual flies isolated in small glass tubes with enough food to last the duration of the experiment. These monitors are placed inside incubators in which the environmental conditions can be accurately controlled according to the test paradigm. In the simplest experiments, transitions between environmental conditions, or Zeitgebers (“time giver” in German), occur almost instantaneously (e.g., 12 h of light followed by 12 h of dark—LD). Here, Drosophila exhibits the previously mentioned bimodal activity pattern, with M and E activity peaks occurring at the Zeitgeber transition points. If the flies are then released into constant, free-running conditions, devoid of temporal information, rhythmic activity persists. However, the rhythms become almost exclusively unimodal possibly due to the merging of the previous M and E activity peaks (Wheeler et al., 1993; Helfrich-Förster, 2000). The timing of this free-running activity peak is a common read-out of “clock time” or circadian time. By adjusting the timing of cue onset/offset, the entrainability of the autonomous oscillator, which adjusts subsequent free-running rhythms to new schedules, can be tested. The speed of entrainment and relative amplitudes of autonomous oscillations depend on the strength and mode (e.g., light, temperature, vibration) of the specific Zeitgeber signal. Indeed, the precise activity profile observed depends on both the environmental conditions and the genetic background of the fly (Schlichting and Helfrich-Förster, 2015).
Entrainment
Photic information is communicated to clock neurons both directly via CRY, and indirectly via visual photoreceptors residing in the compound eyes, ocelli and Hofbauer-Buchner eyelets. CRY mediates cell-autonomous perception of light in a number of tissues that harbor circadian rhythms via its interaction with TIM in the primary TTFL (Plautz et al., 1997; Emery et al., 2000; Lin et al., 2001). This direct input pathway makes the Drosophila circadian system particularly sensitive to light, capable of entraining to low light intensities (0.03 lux; Bachleitner et al., 2007), and can exhibit significant phase shifts following brief light pulses (Levine et al., 1994; Egan et al., 1999; Vinayak et al., 2013). Visual pathways to the clock are not as well defined and do not reset the clock with the same efficiency as CRY dependent pathways (Emery et al., 2000; Helfrich-Förster et al., 2001). However, it appears that a subset of rhodopsin photoreceptors communicate visual photic information to the clock by a novel phototransduction pathway (Stanewsky et al., 1998; Ogueta et al., 2018).
Temperature cycles (TC) can also entrain the circadian clock, and although this can occur in a tissue autonomous fashion in peripheral clocks, signaling from peripheral sensors play an important role in entraining the central clock (Glaser and Stanewsky, 2005; Sehadova et al., 2009). Thermoreceptors, expressed in chordotonal organs (ChOs) have been identified in the thermotransduction pathway, relaying information to the clock across different TC regimes (Wolfgang et al., 2013; Chen et al., 2015). Furthermore, genes important to the structural integrity of ChOs have also shown importance for entrainment to TC (Glaser and Stanewsky, 2005; Sehadova et al., 2009). Specifically, nocte (no circadian temperature entrainment) was identified in a screen for mutants that could entrain to LD but not to TC (Glaser and Stanewsky, 2005). Interestingly, the nocte mutant cannot entrain to combined LD and TC, hinting at a role in a sensory integration pathway that appears to eventuate in the DN1 central clock neurons (Chen et al., 2018).
ChOs also play a role in entrainment to mechanical inputs and this appears dependent on where the ChO is located. ChOs are stretch sensitive organs that populate almost every exoskeletal joint mediating proprioception in the legs as well as hearing in the antennae (Kavlie and Albert, 2013). Flies with no functional ChOs fail to entrain to 12-h vibration/12-h silence cycles, whereas flies lacking only antennal ChOs entrain better than their wild type controls (Simoni et al., 2014). This may be another feedback loop in the circadian system where output activity feeds information back to the clock.
Natural Conditions
In order to perform more ecologically relevant experiments, environmental conditions can also be shifted in relation to each other in order to closely mimic natural conditions. This can be taken even further by placing the activity monitors outside the predictable conditions of the laboratory in the real-world environment. Under semi-natural “Summer” conditions an extra activity peak is observed that coincides with the temperature maxima termed the afternoon (A) peak (Vanin et al., 2012). Evidence suggests that A peak activity is induced by activation of the TRPA1 thermosensor in the AC neurons, rather than the circadian clock pacemakers (Tang et al., 2013; Green et al., 2015). The ecological relevance of the A peak is hypothesized to be an escape response from afternoon heat which is made evident by the occurrence of this peak being largely environmentally controlled. Closer observations of the fly activity support this hypothesis. Inactive flies at a preferable temperature (28°C) quickly retreat to the relative coolness of their food at an uncomfortable temperature (31°C) and are subsequently induced to erratic hyperactivity at noxious temperatures (35°C; Menegazzi et al., 2012). The phase of the A peak, however, still appears to be modulated by both the clock and environmental conditions (Menegazzi et al., 2012; Vanin et al., 2012). Interestingly, a functional clock appears to suppress the A peak at non-noxious temperatures. Clock mutant flies, deficient in either PER or CLK, exhibit A peak activity under milder TC in which wild-type flies exhibit bimodal activity patterns (Currie et al., 2009; Menegazzi et al., 2012; Vanin et al., 2012). This could be the clock mutant’s lack of time perception being unaware of the daily afternoon peaks of temperature, whereas wild-type flies anticipate the regular afternoon increase in temperature knowing it is nothing to be concerned about until a critical threshold is crossed at noxious temperatures. This is a more explicit decision-making process governed by the circadian clock—knowing what stimuli to respond to and what stimuli can safely be ignored.
Conflict Conditions
The complexity of the number of potential interactions that could occur between the central clock pacemakers and the peripheral clocks is only now starting to be realized. How many clocks contribute to the animal’s overall sense of time? The historical view that one master pacemaker clock actively sets the phase of all the downstream clocks is being challenged. Perhaps the central pacemaker sets the time of central processes and regional peripheral clocks set time locally? To test this, offset environmental conditions have been used to understand the molecular, and subsequent behavioral repercussions, of receiving potentially conflicting environmental cues. During antiphasic conflict of light and temperature (a 12-h maximal misalignment between the two cues), the activity patterns of the flies demonstrate preferential entrainment to light (Yoshii et al., 2010; Harper et al., 2016). An assessment of the relative strength of photic and thermic input under these conditions suggests light to be the victor. However, observation of fly activity under other misalignment schedules reveals that flies appear to follow temperature cues when the misalignment is short (2–4 h) and light cues when the misalignment is long (>7 h). Misalignment between this (5–7 h) produces a novel behavior of sustained activity over the course of misalignment (termed “plateau” or P behavior; Harper et al., 2016). P behavior was not observed in the clock-less per mutants or the light-input pathway impaired cry mutants. The per mutant generally displayed arrhythmic behavior with brief startle responses to environmental changes while, due to their lack of the CRY photoreceptor, cry mutants entrained preferentially to temperature cues. The lack of the P behavior in these two clock mutants during sensory conflict demonstrates that the behavior depends on a functional clock and is not the result of environmental masking. Furthermore, a severe dampening of PER oscillations in the central pacemakers under conflict conditions support the hypothesis that sensory conflict is causing the abnormal P behavior. This contrasts with results from a subsequent study in peripheral clocks. During similar conflict conditions, bioluminescent PER reporters expressed in peripheral clocks, revealed peripheral molecular rhythms remained entrained to the light cue (Harper et al., 2017). Again, CRY expression appears to be key for this light cue entrainment in these tissues as peripheral molecular rhythms in cry mutants follow the temperature cue under conflict conditions. The fact that molecular rhythms in these peripheral clocks do not collapse as observed in the central clock could indicate that these peripheral clocks do not contribute to locomotor rhythms or alternatively that asynchrony between central and peripheral clocks results in abnormal behavior. Further studies are required to unpack this relationship.
Probabilistic Considerations and Future Computational Approaches
The “circadian clock” is a complex, highly interconnected system, which attempts to determine—or predict—the state of the world from noisy or incomplete data. Probabilistic modeling provides a powerful conceptual framework for the theoretical, and experimental, analysis of such a system.
In a circadian setting, we can think of Zeitgebers as observable variables, from which the clock must compute the otherwise unobservable time of day. Bayesian integration provides an optimal algorithmic method by which to combine different sources of information. Here, the goal of a Bayesian observer (e.g., the “circadian clock”) is to compute the conditional density function that specifies the probability of the time of day from the given external cues (e.g., light and temperature). Further complexity is added by the temporal structure of circadian data, requiring that probability distributions be calculated over sequences of observations. Hidden markov models (HMMs) are one way of representing such temporal distributions and are used widely in tasks such as speech recognition (Rabiner, 1990), computational genomics (Eddy, 2004) and decoding neural spike data (Escola et al., 2011). More recently, HMMs have begun making their way into the circadian field, effectively modeling rhythms of both molecular (Bieler et al., 2014) and behavioral data (Harper, 2017). We expect such probabilistic frameworks to be crucial for understanding the relationships between circadian clock components across all levels of the system.
Discussion
Complexity exists at all levels of the circadian system. Multiple TTFL maintain the ticking of the individual cellular clocks. This timing can then be communicated, neurochemically or electrically, to other cells that use the information to set the phase of their own internal clocks and orchestrate downstream processes. However, the flow of this information is not unidirectional. Multiple central oscillators exchange timing information set by incoming sensory information to compute time. This raises a further question of how the circadian system weights each oscillator’s contribution (a problem well-suited to probabilistic modeling).
Existing methods used to quantify circadian function are imperfect. They are typically measured in free running conditions in order to avoid masking effects that occur as a stimulus response to environmental condition transitions. This necessarily removes the context in which circadian clocks operate and results in rapidly dampened rhythms—both molecular and behavioral. Ideally the phase of the circadian system would be measured during the entrainment period to alleviate the rapid dampening effects of free run, and also reducing the overall length of the experiment. Again, probabilistic models pose an interesting tool for extracting key circadian metrics—such as phase, period and rhythm strength—from noisy time series data in the presence of changing Zeitgebers.
Finally, a better quantitative understanding of the computations—and possible conflicts—occurring in the circadian system is also relevant in the context of the multitude of clock-related pathologies in humans (Roenneberg and Merrow, 2016). These pathologies may be the result of the circadian system simply making the wrong decisions, e.g., due to conflicting data (light at night, social jetlag, trans-meridian travels, etc.). A deeper understanding of where the errors in the decision making processes occur may be a decisive first step toward identifying and treating these pathologies.
Author Contributions
JS, RH and JA together wrote and designed different aspects of the article.
Funding
RH received funding from the Engineering and Physical Sciences Research Council (EP/F500351/1) through UCL CoMPLEX. JS was supported by a European Research Council grant to JA (H2020-ERC-2014-CoG/648709/Clock Mechanics). JA was supported by grants from the Biotechnology and Biological Sciences Research Council (BB/L02084X/1) and the European Research Council (H2020-ERC-2014-CoG/648709/Clock Mechanics).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the valuable inputs that resulted from discussions with Ralf Stanewsky.
References
Akten, B., Jauch, E., Genova, G. K., Kim, E. Y., Edery, I., Raabe, T., et al. (2003). A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 6, 251–257. doi: 10.1038/nn1007
Allada, R., White, N. E., So, W. V., Hall, J. C., and Rosbash, M. (1998). A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804. doi: 10.1016/s0092-8674(00)81440-3
Bachleitner, W., Kempinger, L., Wülbeck, C., Rieger, D., and Helfrich-Förster, C. (2007). Moonlight shifts the endogenous clock of Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A 104, 3538–3543. doi: 10.1073/pnas.0606870104
Bae, K., Lee, C., Hardin, P. E., and Edery, I. (2000). dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J. Neurosci. 20, 1746–1753. doi: 10.1523/JNEUROSCI.20-05-01746.2000
Bieler, J., Cannavo, R., Gustafson, K., Gobet, C., Gatfield, D., and Naef, F. (2014). Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol. Syst. Biol. 10:739. doi: 10.15252/msb.20145218
Blau, J., and Young, M. W. (1999). Cycling vrille expression is required for a functional Drosophila clock. Cell 99, 661–671. doi: 10.1016/s0092-8674(00)81554-8
Chen, C., Buhl, E., Xu, M., Croset, V., Rees, J. S., Lilley, K. S., et al. (2015). Drosophila ionotropic receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520. doi: 10.1038/nature16148
Chen, C., Xu, M., Anantaprakorn, Y., Rosing, M., and Stanewsky, R. (2018). nocte is required for integrating light and temperature inputs in circadian clock neurons of Drosophila. Curr. Biol. 28, 1595.e3–1605.e3. doi: 10.1016/j.cub.2018.04.001
Collins, B., Mazzoni, E. O., Stanewsky, R., and Blau, J. (2006). Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 16, 441–449. doi: 10.1016/j.cub.2006.01.034
Currie, J., Goda, T., and Wijnen, H. (2009). Selective entrainment of the Drosophila circadian clock to daily gradients in environmental temperature. BMC Biol. 7:49. doi: 10.1186/1741-7007-7-49
Curtin, K. D., Huang, Z. J., and Rosbash, M. (1995). Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14, 365–372. doi: 10.1016/0896-6273(95)90292-9
Cyran, S. A., Buchsbaum, A. M., Reddy, K. L., Lin, M.-C., Glossop, N. R. J., Hardin, P. E., et al. (2003). vrille, Pdp1 and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112, 329–341. doi: 10.1016/s0092-8674(03)00074-6
Darlington, T. K., Wager-Smith, K., Ceriani, M. F., Staknis, D., Gekakis, N., Steeves, T. D. L., et al. (1998). Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603. doi: 10.1126/science.280.5369.1599
Eddy, S. R. (2004). What is a hidden Markov model? Nat. Biotechnol. 22, 1315–1316. doi: 10.1038/nbt1004-1315
Egan, E. S., Franklin, T. M., Hilderbrand-Chae, M. J., McNeil, G. P., Roberts, M. A., Schroeder, A. J., et al. (1999). An extraretinally expressed insect cryptochrome with similarity to the blue light photoreceptors of mammals and plants. J. Neurosci. 19, 3665–3673. doi: 10.1523/JNEUROSCI.19-10-03665.1999
Emery, P., So, W. V., Kaneko, M., Hall, J. C., and Rosbash, M. (1998). CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679. doi: 10.1016/s0092-8674(00)81637-2
Emery, P., Stanewsky, R., Helfrich-Förster, C., Emery-Le, M., Hall, J. C., and Rosbash, M. (2000). Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26, 493–504. doi: 10.1016/s0896-6273(00)81181-2
Escola, S., Fontanini, A., Katz, D., and Paninski, L. (2011). Hidden Markov models for the stimulus-response relationships of multistate neural systems. Neural Comput. 23, 1071–1132. doi: 10.1162/NECO_A_00118
Gekakis, N., Saez, L., Delahaye-Brown, A. M., Myers, M. P., Sehgal, A., Young, M. W., et al. (1995). Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 270, 811–815. doi: 10.1126/science.270.5237.811
Glaser, F. T., and Stanewsky, R. (2005). Temperature synchronization of the Drosophila circadian clock. Curr. Biol. 15, 1352–1363. doi: 10.1016/j.cub.2005.06.056
Glossop, N. R. J., Houl, J. H., Zheng, H., Ng, F. S., Dudek, S. M., and Hardin, P. E. (2003). VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 37, 249–261. doi: 10.1016/s0896-6273(03)00002-3
Green, E. W., O’Callaghan, E. K., Hansen, C. N., Bastianello, S., Bhutani, S., Vanin, S., et al. (2015). Drosophila circadian rhythms in seminatural environments: summer afternoon component is not an artifact and requires TrpA1 channels. Proc. Natl. Acad. Sci. U S A 112, 8702–8707. doi: 10.1073/pnas.1506093112
Grima, B., Chélot, E., Xia, R., and Rouyer, F. (2004). Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873. doi: 10.1038/nature02935
Hao, H., Allen, D. L., and Hardin, P. E. (1997). A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol. 17, 3687–3693. doi: 10.1128/mcb.17.7.3687
Hardin, P. E. (2011). Molecular genetic analysis of circadian timekeeping in Drosophila. Adv. Genet. 74, 141–173. doi: 10.1016/B978-0-12-387690-4.00005-2
Harper, R. E. F. (2017). “Time flies” multisensory processing by circadian clocks in Drosophila melanogaster. Available online at: http://discovery.ucl.ac.uk/10037959/. [Accessed on June 29, 2018].
Harper, R. E. F., Dayan, P., Albert, J. T., and Stanewsky, R. (2016). Sensory conflict disrupts activity of the Drosophila circadian network. Cell Rep. 17, 1711–1718. doi: 10.1016/j.celrep.2016.10.029
Harper, R. E. F., Ogueta, M., Dayan, P., Stanewsky, R., and Albert, J. T. (2017). Light dominates peripheral circadian oscillations in Drosophila melanogaster during sensory conflict. J. Biol. Rhythms 32, 423–432. doi: 10.1177/0748730417724250
Hege, D. M., Stanewsky, R., Hall, J. C., and Giebultowicz, J. M. (1997). Rhythmic expression of a PER-reporter in the Malpighian tubules of decapitated Drosophila: evidence for a brain-independent circadian clock. J. Biol. Rhythms 12, 300–308. doi: 10.1177/074873049701200402
Helfrich-Förster, C. (1995). The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A 92, 612–616. doi: 10.1073/pnas.92.2.612
Helfrich-Förster, C. (2000). Differential control of morning and evening components in the activity rhythm of Drosophila melanogaster—sex-specific differences suggest a different quality of activity. J. Biol. Rhythms 15, 135–154. doi: 10.1177/074873040001500208
Helfrich-Förster, C., and Homberg, U. (1993). Pigment-dispersing hormone-immunoreactive neurons in the nervous system of wild-type Drosophila melanogaster and of several mutants with altered circadian rhythmicity. J. Comp. Neurol. 337, 177–190. doi: 10.1002/cne.903370202
Helfrich-Förster, C., Winter, C., Hofbauer, A., Hall, J. C., and Stanewsky, R. (2001). The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30, 249–261. doi: 10.1016/s0896-6273(01)00277-x
Helfrich-Förster, C., Yoshii, T., Wülbeck, C., Grieshaber, E., Rieger, D., Bachleitner, W., et al. (2007). The lateral and dorsal neurons of Drosophila melanogaster: new insights about their morphology and function. Cold Spring Harb. Symp. Quant. Biol. 72, 517–525. doi: 10.1101/sqb.2007.72.063
Hermann-Luibl, C., and Helfrich-Förster, C. (2015). Clock network in Drosophila. Curr. Opin. Insect Sci. 7, 65–70. doi: 10.1016/j.cois.2014.11.003
Ito, C., Goto, S. G., Shiga, S., Tomioka, K., and Numata, H. (2008). Peripheral circadian clock for the cuticle deposition rhythm in Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A 105, 8446–8451. doi: 10.1073/pnas.0800145105
Ito, C., and Tomioka, K. (2016). Heterogeneity of the peripheral circadian systems in Drosophila melanogaster: a review. Front. Physiol. 7:8. doi: 10.3389/fphys.2016.00008
Ivanchenko, M., Stanewsky, R., and Giebultowicz, J. M. (2001). Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J. Biol. Rhythms 16, 205–215. doi: 10.1177/074873040101600303
Kadener, S., Stoleru, D., McDonald, M., Nawathean, P., and Rosbash, M. (2007). Clockwork orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 21, 1675–1686. doi: 10.1101/gad.1552607
Kavlie, R. G., and Albert, J. T. (2013). Chordotonal organs. Curr. Biol. 23, R334–R335. doi: 10.1016/j.cub.2013.03.048
Kloss, B., Price, J. L., Saez, L., Blau, J., Rothenfluh, A., Wesley, C. S., et al. (1998). The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94, 97–107. doi: 10.1016/s0092-8674(00)81225-8
Kloss, B., Rothenfluh, A., Young, M. W., and Saez, L. (2001). Phosphorylation of period is influenced by cycling physical associations of double-time, period and timeless in the Drosophila clock. Neuron 30, 699–706. doi: 10.1016/s0896-6273(01)00320-8
Krishnan, B., Dryer, S. E., and Hardin, P. E. (1999). Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature 400, 375–378. doi: 10.1038/22566
Krishnan, B., Levine, J. D., Lynch, M. K. S., Dowse, H. B., Funes, P., Hall, J. C., et al. (2001). A new role for cryptochrome in a Drosophila circadian oscillator. Nature 411, 313–317. doi: 10.1038/35077094
Krupp, J. J., Billeter, J.-C., Wong, A., Choi, C., Nitabach, M. N., and Levine, J. D. (2013). Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in Drosophila. Neuron 79, 54–68. doi: 10.1016/j.neuron.2013.05.019
Lee, C., Bae, K., and Edery, I. (1998). The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation and interactions with the PER-TIM complex. Neuron 21, 857–867. doi: 10.1016/s0896-6273(00)80601-7
Lee, C., Bae, K., and Edery, I. (1999). PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol. 19, 5316–5325. doi: 10.1128/mcb.19.8.5316
Levine, J. D., Casey, C. I., Kalderon, D. D., and Jackson, F. R. (1994). Altered circadian pacemaker functions and cyclic AMP rhythms in the Drosophila learning mutant dunce. Neuron 13, 967–974. doi: 10.1016/0896-6273(94)90262-3
Lim, C., Lee, J., Choi, C., Kim, J., Doh, E., and Choe, J. (2007). Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol. Cell. Biol. 27, 4876–4890. doi: 10.1128/mcb.02155-06
Lin, J.-M., Kilman, V. L., Keegan, K., Paddock, B., Emery-Le, M., Rosbash, M., et al. (2002). A role for casein kinase 2α in the Drosophila circadian clock. Nature 420, 816–820. doi: 10.1038/nature01235
Lin, F.-J., Song, W., Meyer-Bernstein, E., Naidoo, N., and Sehgal, A. (2001). Photic signaling by cryptochrome in the Drosophila circadian system. Mol. Cell. Biol. 21, 7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001
Martinek, S., Inonog, S., Manoukian, A. S., and Young, M. W. (2001). A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105, 769–779. doi: 10.1016/s0092-8674(01)00383-x
Matsumoto, A., Ukai-Tadenuma, M., Yamada, R. G., Houl, J., Uno, K. D., Kasukawa, T., et al. (2007). A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 21, 1687–1700. doi: 10.1101/gad.1552207
Menegazzi, P., Yoshii, T., and Helfrich-Förster, C. (2012). Laboratory versus nature: the two sides of the Drosophila circadian clock. J. Biol. Rhythms 27, 433–442. doi: 10.1177/0748730412463181
Menet, J. S., Abruzzi, K. C., Desrochers, J., Rodriguez, J., and Rosbash, M. (2010). Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 24, 358–367. doi: 10.1101/gad.1883910
Murad, A., Emery-Le, M., and Emery, P. (2007). A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron 53, 689–701. doi: 10.1016/j.neuron.2007.01.034
Nawathean, P., and Rosbash, M. (2004). The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol. Cell 13, 213–223. doi: 10.1016/s1097-2765(03)00503-3
Nitabach, M. N., and Taghert, P. H. (2008). Organization of the Drosophila circadian control circuit. Curr. Biol. 18, R84–R93. doi: 10.1016/j.cub.2007.11.061
Ogueta, M., Hardie, R. C., and Stanewsky, R. (2018). Non-canonical phototransduction mediates synchronization of the Drosophila melanogaster circadian clock and retinal light responses. Curr. Biol. 28, 1725.e3–1735.e3. doi: 10.1016/j.cub.2018.04.016
Picot, M., Cusumano, P., Klarsfeld, A., Ueda, R., and Rouyer, F. (2007). Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 5:e315. doi: 10.1371/journal.pbio.0050315
Pittendrigh, C. S., and Daan, S. (1976). A functional analysis of circadian pacemakers in nocturnal rodents. J. Comp. Physiol. 106, 333–355. doi: 10.1007/bf01417859
Plautz, J. D., Kaneko, M., Hall, J. C., and Kay, S. A. (1997). Independent photoreceptive circadian clocks throughout Drosophila. Science 278, 1632–1635. doi: 10.1126/science.278.5343.1632
Price, J. L., Blau, J., Rothenfluh, A., Abodeely, M., Kloss, B., and Young, M. W. (1998). double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94, 83–95. doi: 10.1016/s0092-8674(00)81224-6
Price, J. L., Dembinska, M. E., Young, M. W., and Rosbash, M. (1995). Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 14, 4044–4049.
Rabiner, L. R. (1990). “A tutorial on hidden markov models and selected applications in speech recognition,” in Readings in Speech Recognition, eds A. Waibel and K.-F. Lee (San Francisco: Morgan Kaufmann), 267–296.
Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C., and Taghert, P. H. (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. doi: 10.1016/s0092-8674(00)81676-1
Rieger, D., Shafer, O. T., Tomioka, K., and Helfrich-Förster, C. (2006). Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J. Neurosci. 26, 2531–2543. doi: 10.1523/JNEUROSCI.1234-05.2006
Rieger, D., Wülbeck, C., Rouyer, F., and Helfrich-Förster, C. (2009). Period gene expression in four neurons is sufficient for rhythmic activity of Drosophila melanogaster under dim light conditions. J. Biol. Rhythms 24, 271–282. doi: 10.1177/0748730409338508
Roenneberg, T., and Merrow, M. (2016). The circadian clock and human health. Curr. Biol. 26, R432–R443. doi: 10.1016/j.cub.2016.04.011
Rutila, J. E., Suri, V., Le, M., So, W. V., Rosbash, M., and Hall, J. C. (1998). CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814. doi: 10.1016/s0092-8674(00)81441-5
Schlichting, M., and Helfrich-Förster, C. (2015). “Chapter five—photic entrainment in Drosophila assessed by locomotor activity recordings,” in Methods in Enzymology Circadian Rhythms and Biological Clocks, Part B., ed. A. Sehgal (Academic Press), 105–123. doi: 10.1016/bs.mie.2014.10.017
Schlichting, M., Menegazzi, P., Lelito, K. R., Yao, Z., Buhl, E., Dalla Benetta, E., et al. (2016). A neural network underlying circadian entrainment and photoperiodic adjustment of sleep and activity in Drosophila. J. Neurosci. 36, 9084–9096. doi: 10.1523/JNEUROSCI.0992-16.2016
Schubert, F. K., Hagedorn, N., Yoshii, T., Helfrich-Förster, C., and Rieger, D. (2018). Neuroanatomical details of the lateral neurons of Drosophila melanogaster support their functional role in the circadian system. J. Comp. Neurol. 526, 1209–1231. doi: 10.1002/cne.24406
Sehadova, H., Glaser, F. T., Gentile, C., Simoni, A., Giesecke, A., Albert, J. T., et al. (2009). Temperature entrainment of Drosophila’s circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron 64, 251–266. doi: 10.1016/j.neuron.2009.08.026
Simoni, A., Wolfgang, W., Topping, M. P., Kavlie, R. G., Stanewsky, R., and Albert, J. T. (2014). A mechanosensory pathway to the Drosophila circadian clock. Science 343, 525–528. doi: 10.1126/science.1245710
Stanewsky, R., Kaneko, M., Emery, P., Beretta, B., Wager-Smith, K., Kay, S. A., et al. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692. doi: 10.1016/s0092-8674(00)81638-4
Stoleru, D., Peng, Y., Agosto, J., and Rosbash, M. (2004). Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868. doi: 10.1038/nature02926
Tang, X., Platt, M. D., Lagnese, C. M., Leslie, J. R., and Hamada, F. N. (2013). Temperature integration at the AC thermosensory neurons in Drosophila. J. Neurosci. 33, 894–901. doi: 10.1523/JNEUROSCI.1894-12.2013
Top, D., Harms, E., Syed, S., Adams, E. L., and Saez, L. (2016). GSK-3 and CK2 kinases converge on timeless to regulate the master clock. Cell Rep. 16, 357–367. doi: 10.1016/j.celrep.2016.06.005
Top, D., and Young, M. W. (2018). Coordination between differentially regulated circadian clocks generates rhythmic behavior. Cold Spring Harb. Perspect. Biol. 10:a033589. doi: 10.1101/cshperspect.a033589
Vanin, S., Bhutani, S., Montelli, S., Menegazzi, P., Green, E. W., Pegoraro, M., et al. (2012). Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature 484, 371–375. doi: 10.1038/nature10991
Vinayak, P., Coupar, J., Hughes, S. E., Fozdar, P., Kilby, J., Garren, E., et al. (2013). Exquisite light sensitivity of Drosophila melanogaster cryptochrome. PLoS Genet. 9:e1003615. doi: 10.1371/journal.pgen.1003615
Wheeler, D. A., Hamblen-Coyle, M. J., Dushay, M. S., and Hall, J. C. (1993). Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J. Biol. Rhythms 8, 67–94. doi: 10.1177/074873049300800106
Wolfgang, W., Simoni, A., Gentile, C., and Stanewsky, R. (2013). The Pyrexia transient receptor potential channel mediates circadian clock synchronization to low temperature cycles in Drosophila melanogaster. Proc. Biol. Sci. 280:20130959. doi: 10.1098/rspb.2013.0959
Yao, Z., Bennett, A. J., Clem, J. L., and Shafer, O. T. (2016). The Drosophila clock neuron network features diverse coupling modes and requires network-wide coherence for robust circadian rhythms. Cell Rep. 17, 2873–2881. doi: 10.1016/j.celrep.2016.11.053
Yao, Z., and Shafer, O. T. (2014). The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343, 1516–1520. doi: 10.1126/science.1251285
Yoshii, T., Hermann, C., and Helfrich-Förster, C. (2010). CRYPTOCHROME-positive and -negative clock neurons in Drosophila entrain differentially to light and temperature. J. Biol. Rhythms 25, 387–398. doi: 10.1177/0748730410381962
Keywords: circadian clock, biological oscillator, multisensory integration, bayesian modeling, Drosophila melanogaster, sensory conflict
Citation: Somers J, Harper REF and Albert JT (2018) How Many Clocks, How Many Times? On the Sensory Basis and Computational Challenges of Circadian Systems. Front. Behav. Neurosci. 12:211. doi: 10.3389/fnbeh.2018.00211
Received: 06 July 2018; Accepted: 21 August 2018;
Published: 11 September 2018.
Edited by:
Gérard Manière, Université de Bourgogne, FranceReviewed by:
Charlotte Helfrich-Förster, Universität Würzburg, GermanyFrancois Rouyer, UMR9197 Institut des Neurosciences Paris Saclay (Neuro-PSI), France
Copyright © 2018 Somers, Harper and Albert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joerg T. Albert, am9lcmcuYWxiZXJ0QHVjbC5hYy51aw==
Jason Somers, ai5zb21lcnNAdWNsLmFjLnVr
 Jason Somers
Jason Somers Ross E. F. Harper
Ross E. F. Harper Joerg T. Albert
Joerg T. Albert